Abstract
Proposing a theory about the pathophysiology of cytokine storm in COVID19, we were to find the potential drugs to treat this disease and to find any effect of these drugs on the virus infectivity through an in silico study. COVID-19-induced ARDS is linked to a cytokine storm phenomenon not explainable solely by the virus infectivity. Knowing that ACE2, the hydrolyzing enzyme of AngII and SARS-CoV2 receptor, downregulates when the virus enters the host cells, we hypothesize that hyperacute AngII upregulation is the eliciting factor of this ARDS. We were to validate this theory through reviewing previous studies to figure out the role of overzealous activation of AT1R in ARDS. According to this theory losartan may attenuate ARDS in this disease. Imatinib, has previously been elucidated to be promising in modulating lung inflammatory reactions and virus infectivity in SARS and MERS. We did an in silico study to uncover any probable other unconsidered inhibitory effects of losartan and imatinib against SARS-CoV2 pathogenesis. Reviewing the literature, we could find that over-activation of AT1R could explain precisely the mechanism of cytokine storm in COVID19. Our in silico study revealed that losartan and imatinib could probably: (1) decline SARS-CoV2 affinity to ACE2. (2) inhibit the main protease and furin, (3) disturb papain-like protease and p38MAPK functions. Our reviewing on renin-angiotensin system showed that overzealous activation of AT1R by hyper-acute excess of AngII due to acute downregulation of ACE2 by SARS-CoV2 explains precisely the mechanism of cytokine storm in COVID-19. Besides, based on our in silico study we concluded that losartan and imatinib are promising in COVID19.
Electronic supplementary material
The online version of this article (10.1007/s40203-020-00058-7) contains supplementary material, which is available to authorized users.
Keywords: SARS-CoV2, Losartan, Imatinib, Cytokine storm, Papain-like protease, Main protease
Introduction
Since December 2019, a viral disease called COVID-19 has hunted thousands of people savagely all around the world and the death toll increases in a skyrocketing manner every day. It was elucidated that a coronavirus named SARS-CoV2 caused the disease eventually named as COVID-19 (Chan et al. 2020; Viruses 2020). Coronaviruses are classified into the order Nidovirales, the suborder Coronavirinae, Coronaviridae family, the subfamily Orthocoronavirinae with four genera: Alphacoronavirus (αCoV), Betacoronavirus (βCoV), Deltacoronavirus (δCoV), and Gammacoronavirus (γCoV). SARS-CoV2, along with SARS-CoV and MERS-CoV, belongs to β-genus (Chan et al. 2020; Gorbalenya et al. 2020).
These pathogens are large, enveloped and positive-sense single stranded RNA viruses (Cui et al. 2019; Chan et al. 2020). The genome of SARS-CoV2 consists of 29,891 nucleotides encoding 9860 amino acids with genomic sequence of Chan et al. (2020):
Two main open-reading frames (ORFs), ORF1a and ORF1b of the nucleotide sequence of the virus genome are translated into two co-terminal polyproteins ppl1a (ORF1a) and ppl1ab (ORF1a & ORF1b together). These polyproteins are cleaved into non-strucrtural proteins (NSPs) by two proteases: a papain-like protease (PLpro) and the main protease (Mpro) (Chan et al. 2020; Mielech et al. 2014; Fehr and Perlman 2015; Snijder et al. 2016).
Intriguingly, PLpro and some nsps (nsp3) apart from their crucial role in replication (Snijde et al. 2016; Lei et al. 2018; Chan et al. 2020), provide the virus with deubiquitinating and deISGylating properties (Nejat and Sadr 2019, Shin et al. 2020). These viral proteins inhibit Toll-like receptor3 (TLR3) and Toll-like receptor7 (TLR7), as well (Mielech et al. 2014; Snijder et al. 2016). In vivo, it has been demonstrated that type I and type III interferon responses against SARS-CoV2 is lower than that of seen in respiratory syncytial virus (RSV) and influenza A virus. It seems that SARS-CoV2 like SARS-CoV evade innate immunity successfully (Yoshikawa et al. 2010; Blanco-Melo et al. 2020).
Viral genomic and subgenomic RNA synthesis leads to production of membrane (M), nucleocapsid (N), and envelope (E) structural proteins (Fehr and Perlman 2015) which determine the different compartments and shape of the virus. Furthermore, a glycoprotein called spike (S) protein protruding from the surface of the virus determines the host range, tissue tropism and the host immune responses (Li 2016). As a class 1 fusion protein, S protein of coronaviruses is composed of two trimeric subunits, S1 and S2. S1, containing the receptor binding domain (RBD), mediates the attachment of the virus to its receptor on the host cells. S2, the stalk of S glycoprotein, is responsible for virus-cell fusion (White et al. 2008; Ou et al. 2020; Zhou et al. 2020). As a mandatory step in pathogenesis of SARS-CoV2, cleavage of S protein into S1 and S2 subunits as well as in S’2, immediately upstream to fusion peptide in S2, plays an essential part in entry of the virus to the host cell and in cell–cell fusion and transmission. Furin (found abundantly in the lungs) and transmembrane serine protease-2 (TMPRSS2) contribute to this cleaving process crucially (Coutard et al. 2020, Jaimes et al. 2020; Wrapp et al. 2020).
The receptor of SARS-CoV2 on the host cells, like SARS-CoV, is angiotensin converting enzyme 2 (ACE2), a member of renin-angiotensin system (RAS) (Li et al. 2003, 2005; Wan et al. 2020). RAS is a very complex network of systemic as well as local ligands and receptors (Marshall 2003). It contributes to the regulation of immune system and cytokine production, cardiovascular system, metabolism, cell growth, salt and electrolyte homeostasis and vascular resistance (Goossens et al. 2007; Parodi-Rullan et al. 2012; Satou et al. 2018).
ACE2 as a mono-carboxypeptidase which removes single amino acids from peptides of RAS is not inhibited by ACE inhibitors like captopril or lisinopril (Donoghue et al. 2000; Riviere et al. 2005; Reddy Gaddam et al. 2014). This mono-peptidase converts angiotensin I [1–10] and angiotensin II [1–8] to angiotensin [1–9] and angiotensin [1–7], respectively (Riviere et al. 2005; Arendse et al. 2019). ACE2 is a functional competitor of ACE as the former reduces available angiotensin I [1–10], the substrate of ACE, by converting it to a less active metabolite, angiotensin [1–9]. Opposing to ACE2, ACE degrades angiotensin [1–7] to inactive products like angiotensin [1–5] (Dilauro and Burns 2009; Bader 2013). Angiotensin [1–7] is considered as an active peptide in RAS with antioxidative, anti-inflammatory, antiproliferative/antifibrotic, potent vasodilatory, and anti-thrombotic properties which exerts most of its effect via Mas receptor (Watanabe et al. 2005; Uhal et al. 2012; Bader 2013; Reddy et al. 2019). On the other hand, ACE increases Ang II with its oxidative, proinflammatory, proliferative/fibrotic, vasoconstrictive and thrombotic effects which are mostly exerted through activation of angiotensin II type 1 receptor (AT1R) (Manabe et al. 2005; Benign et al. 2010; Lee et al. 2019). Another receptor for Ang II called angiotensin II type 2 receptor (AT2R) with cell protective and some opposing post-receptor effects to AT1R is distributed in a limited number of organs (Dandona et al. 2007). It is implicated that ACE2/Ang[1–7]/Mas axis plays a counter-regulatory role against ACE/Ang II[1–8]/AT1R signaling pathway (Imai et al. 2008; Santos et al. 2013; Dominici et al. 2014; Reddy Gaddam et al. 2014).
Attachment of protein S with ACE2 results in the downregulation of the latter (Glowacka et al. 2010; Jia 2016). Internalization of this receptor with the virus into the host cell or upregulation of constitutive physiologic shedding of ACE2 in the airways are potential causes of this phenomenon (Jia 2016). Downregulation of ACE2 deregulates the balance in local RAS pathways in favor of ACE/Ang II [1–8]/AT1R in the lungs. In this context, hyperacute upregulation of local intracrine Ang II [1–8]/AT1R in the setting of invasion of huge number of SARS-CoV2 is not encountered with appropriate negative physiological feedback with ACE2. Furthermore, AngII has been demonstrated to decline ACE2 expression and function via lysosomal degradation mediated by AT1R (Deshotels et al. 2014). Henceforth, overzealous stimulation of AT1R sets on fire locally to provoke lung inflammation through pro-inflammatory, cytokine inducing, proliferative, thrombotic and tissue destructive effects as well as activating platelet derived growth factor receptor (PDGFR) (Kelly et al. 2004; Gao et al. 2006; Imai et al. 2006).
It is implied that SARS-CoV2:
evades innate immunity via avoiding IRF3, TLR3 and TLR7 pathways by its nsp3 and papain-like proteases and elicits a moderate immune response,
downregulates its receptor (ACE2) after attachment, thereby, dysregulates the balance between two opposing axes of local RAS in the lungs in favor of AngII with all of its inflammatory, destructive and fibrotic properties in the tissues.
According to the fact that the virus does not elicit an initial innate immunity response to explain the eruption of cytokine storm and simultaneously deregulates RAS in favor of ACE/AngII[1–8]/AT1R axis, we hypothesized that overzealous increase in Ang II/angiotensin[1–7] ratio may erupt the cytokine storm. Therefore, blocking of AT1R with an angiotensin receptor blocker like losartan would suppress the post-receptor deleterious effects of AngII in favor of angiotensin[1–7] or activation of AT2R with tissue protective effects. Moreover, subsiding immunopathological changes by an immunomodulator such as imatinib which had been used previously in SARS and MERS might alleviate the severity of the disease and may reduce the morbidity and mortality rates in COVID 19. We also did an in silico study on different stages of SARS-CoV2 replication cycle to see if there are any other pharmacodynamic properties of these drugs that might be promising in reducing the infectivity of this virus.
Method
Proposing a new insight to SARS-CoV2 biology and the pathophysiology of cytokine storm and ARDS in COVID-19 to reach into a way to reduce the mortality and morbidity of this disease, we searched for review and original articles focused on COVID-19, SARS, MERS, RAS, the effect of AT1R and Ang II in ARDS and inflammatory reactions in search engines science direct, scopus database and google scholar from 1990 till now. After reaching into the conclusion that Ang II/AT1R pathway may explain cytokine storm and ARDS in COVID-19 and accordingly losartan and imatinib could attenuate ARDS in this disease, we did an in silico study to investigate whether losartan and imatinib have any probable unconsidered inhibiting effect against the replication of the virus.
In silico study
Preparation of the protein structures
The required protein structures were obtained from Protein Data Bank (Berman et al. 2000) (PDB) according to Table 1:
Table 1.
The protein crystal structures used in this study
| Macromolecule | Sequence Length | Organism | ID |
|---|---|---|---|
| SARS-CoV-2 spike receptor-RBD bound to ACE2 | 603/229 | Homosapiens/SARS-Cov2 | 6m0j (Lan et al. 2020) |
| Angiotensin Converting Enzyme 2 (ACE2) | 615 | Homo sapiens | 1r4l (Towler et al. 2004) |
| COVID-19 main protease | 306 | SARS-Cov2 | 6lu7 (Jin et al. 2020) |
| MAP Kinase p38 | 379 | Homo sapiens | 1a9u (Wang et al. 1998) |
| Furin | 482 | Homo sapiens | 6hzb (Van Lam van et al. 2019) |
| Papain-like protease | 316 | SARS-CoV | 3mj5 (Ghosh et al. 2010) |
| Angiotensin II type 1 receptor & Angiotensin II | 425/8 | Homo sapiens | 6os0 (Wingler et al. 2020) |
Proteins were studied for the date of publishing, crystallography techniques, the resolution, accompaniment of predefined inhibitor in crystal (“Pre-inh”) and any required reconstruction due to probable missing of amino acids in their sequence vs the sequence of reference protein. The structures were observed by visualizing softwares UCSF chimera (Pettersen et al. 2004), Pymol (DeLano 2002), Swiss-PdbViewer (Guex and Peitsch 1997) to determine their unique protein chains and whether the structure is accompanied by other undesired molecules like (water, ions….) and to purify selectively to achieve the most desirable structure.
The mutated new amino acids in the sequence of SARS-CoV2 RBD were replaced on SARS-CoV RBD; MD-simulation was performed for new structure
In the beginning of our study (mid of February 2020) due to the lack of crystasl structure of SARS-CoV2 RBD with complete amino acid sequence, we replaced 22 defined mutated amino acids in SARS-CoV2 RBD on the corresponding place on SARS-CoV RBD to achieve a RBD structure with the most similarity to the real crystal structure of SARS-CoV2 RBD, assuming that S proteins in these two viruses are 76% homologous. MD simulation of 100 ns was performed after accomplishing this replacement to achieve a persistently stable structure with the most homology to RBD-ACE2 complex of SARS-CoV2. RMSD, RMSF, H-bonding and radius of gyration diagrams are available. Due to the publishing of crystal structure of RBD-ACE2 complex of SARS-CoV2 by X-ray diffraction (resolution of 2.45 Å) we quitted using the achieved mutated RBD-ACE2 complex and continued our bioinformatic study on the new published SARS-CoV2 RBD (Fig S3).
Preparation of losartan and imatinib
The drug structures were obtained from Structure Data Bank such as Pubchem database (Kim et al. 2019) and Drug bank database (Table S1) (Wishart et al. 2018). The structures of the drugs were imported through gauss view, and then fully optimized geometries and properties of the electronic and structural properties of two molecules were derived by means of the density functional theory (DFT) method (Nagy 1998) with B3LYP (Becke 1988) and STO-3G basis sets (McKean et al. 1996). The calculations were carried out using the Gaussian 03 package (Frisch et al. 2004). The program Open Babel (O'Boyle et al. 2011) was used to generate SMILES strings from the optimized structure to be used for a similarity study by drug bank Chemical Structure Search with 0.5–0.7 Similarity threshold.
Molecular docking simulation
Docking study was performed by AutoDock4 (Morris et al. 2009) to find the suitable orientation of the molecules in the active site of the protein structures. AutoDockTools 1.5.4 (ADT) was used to prepare input PDBQT files and to calculate a grid box. A special grid map appropriate for each structural size (Table 2) around the active site of proteins was defined. The center of the grid boxes was aligned to the coordinates of the “Pre-inh” for each of the protein structures. For the case of ACE2 three grid boxes were defined in three dimensions as appeared in Table 2. The first ACE2 grid box was set around the “Pre-inh” position. The position of losartan and imatinib with the most affinity for ACE2 in grid box 1 vs the position of predefined inhibitor was determined.The second and the third ACE2 grid boxes encompassed RBD binding site around α-helix and the whole ACE2 molecule, respectively.
Table 2.
Grid box dimensions for each structure
| Macrmolcules | Steps | Grid points | Spacing | Grid Center |
|---|---|---|---|---|
| ACE2 | 1st Docking | |||
| ACE2 | 2nd Docking | |||
| ACE2 | 3rd Docking | |||
| Mpro | ||||
| p38MAPK | ||||
| PLpro | Before MD | |||
| PLpro | After MD | |||
| Furin |
As PLpro inhibitory active site changed in 100 ns MD simulation, two grid boxes were defined based on the position of active site; one before MD simulation and the second after 100 ns MD simulation. Binding energies for losartan and imatinib attachment with PLpro were calculated before and after MD simulation.
A Lamarckian genetic algorithm (LGA) was used for the searching of the status of binding sites. Every LGA job comprised of 250 runs. The final structures were grouped and classified according to the most favorable binding energy. This procedure was applied to the two drugs in a similar manner. A less negative score determines which of these drugs are more likely to dock with a protein structure (target protein) with subsequent more favorable interactions. The docking model of each protein complex with its “Pre-inh” was generated by AutoDock4 (Table 2). Reliability of the applied docking protocol was assessed by separation of “Pre-inh” from its protein structure and then redocking of each “Pre-inh” into the active site of the latter.
After exposing of the two drugs with the protein structures, we obtained docking energy for each [drug structure] complex. The clusters of docking energies were determined for 250 posing status and the relevant numerical tables for each complex was studied.
MD simulations
Molecular dynamic (MD) simulations, to study the dynamicity of the protein structures over a defined time period to characterize the behavior and stability of the structures were executed by Gromacs package for 100 nano-second (ns) in this study.
MD simulations of the protein-drug complexes following docking were performed with the GROMACS 2018 package using the GROMOS96 43a1 force field (Berendsen et al. 1995). The conformation status for ACE2 and PLpro complexes with their ligands with the highest affinity were selected as the initial conformation for MD simulations. First the topology parameters of protein were created and the complex was immersed in a cubic box of simple point charge (SPC) water molecules (van Gunsteren et al. 1998). The “solvated system” (protein, ions, small molecule and water) was neutralized by adding required Na+ or Cl− counter-ions. To equilibrate the system, the solutes (Proteins, counter-ions, and two drugs) were subjected to the position-restrained dynamic simulation (NVT and NPT) at 299.177 K for 1 ns. Finally, the full system was subjected to an MD run for 100 ns at 300 K temperature and 1 bar pressure.
MD simulation was performed for ACE2 crystal, ACE2-SARS-CoV2 RBD (refer to results), imatinib-ACE2, losartan-ACE2 and imatinib-PLpro, losartan-PLpro complexes.
New ACE2 after MD simulation of ACE2-losartan and ACE2-imatinib complexes
In order to study the persistency of losartan-ACE2 and imatinib-ACE2 complexes, we performed 100 ns MD simulation for each complex. Two new ACE2 (nACE2) under the influence of each of these ligands were exported: losartan (Lo,nACE2) and imatinib (Im,nACE2). RMSD, RMSF, radius of gyration and h-bonding diagrams of each complex were obtained (Fig. S2).
Super-imposition of ACE2 structures before and after 100 ns MD with losartan and imatinib
After MD simulations of ACE2-losartan and ACE2-imatinib complexes, ACE2 was separated from the drugs. The structure of ACE2 under the influence of losartan and imatinib were superimposed on the structure of ACE2 crystal prior to any docking to evaluate the degree of changes in conformational shape of ACE2 in each complex.
Losartan against other ARBs in docking with ACE2
ARBs were searched for on Kyoto Encyclopedia of Genes and Genomes (Kanehisa and Goto 2000). Docking was done for each member of ARBs with ACE2 crystal structure and the relevant energy binding for each item was obtained.
Redocking study after MD simulations
After MD simulations of complexes for 100 ns, the ligands were separated. Through redocking, the binding energies and the binding status of the two drugs with the protein structures were evaluated (Dolatkhah et al. 2017).
Analysis
RMSD, RMSF, hydrogen bonding and radius of gyration diagrams exported after performing MD simulations were analyzed by qtgrace (Turner 2005). Ligplot (Wallace et al. 1995) and poseview (Stierand and Rarey 2010) were used for determining the hydrogen bonding, hydrophobic and pi-pi interactions after docking and MD simulations. Visual analyzing was done with ucsf chimera and pymol.
Exposure of imatinib and losartan with Mpro, furin, p38MAPK and PLpro
We exposed imatinib and losartan to crystal structure of Mpro, furin, p38MAPK and PLpro to figure out if the drugs would pose the site of their relevant “Pre-inh”. It is worth mentioning that redocking of predefined inhibitor of furin in its crystal structure obtained from PDB was disturbed and showed error in the process. We optimized the structure and performed docking for the second round to earn a reliable reference for binding energy.
MD simulation of imatinib and losartan with PLpro
We performed MD simulation for 100 ns after exposure of imatinib and losartan with PLpro. RMSD, RMSF and radius of gyration were obtained.
Configurations of computational systems
In this study we used multiple computational systems by different configurations (Table S2).
Results
Losartan and imatinib bind to ACE2 with low energy (high affinity) compared to “Pre-inh”
Docking of losartan and imatinib molecules with ACE2 before MD simulations, showed that the binding energy in three grid boxes as appeared in (Table 3) for the two drugs is lower than that of “Pre-inh”. It is implied that losartan and imatinib attach to ACE2 with higher affinity (Fig. S1) in comparison with “Pre-inh”. It merits mentioning that affinity of losartan and imatinib around α-helix of ACE2 is lower than the other two grid boxes.
Table 3.
Docking Energies of Imatinib and Losartan with ACE2 in 3 distinct grid boxes
| Small Molecules | Grid Box1 | Grid Box2 | Grid Box3 |
|---|---|---|---|
| Imatinib | – 15.21 | – 11.79 | – 12.64 |
| Losartan | – 9.67 | – 8.44 | – 9.6 |
| Pre-inh | – 7.28 |
Losartan and imatinib could change the conformational structure of ACE2 persistently
The data showed that the conformational structure of ACE2 after binding with both losartan and imatinib changed significantly at the binding site of ACE2 and RBD (Fig. 1). It is expected that these two drugs lengthen the binding distance between α-helix of ACE2 and SARS-CoV2 RBD due to relocation of this part of ACE2 (Table S3).
Fig. 1.
a SARS-CoV2-ACE2 complex. b 1-Im,nACE2, 2-super-imposition of ACE2 and Im,nACE2, 3-super-imposition of α-helix of ACE2 and Im,nACE2. c 1-Lo,nACE2, 2-super-imposition of ACE2 and Lo.nACE2, 3-super-imposition of α-helix of ACE2 and Lo,nACE2. d Hydrophobic and hydrogen bonds of SARS-CoV2RBD-ACE2
The binding energy of losartan and imatinib with Lo,nACE2 and Im,nACE2 after performing 100 ns MD simulations changed
The binding energies among losartan and imatinib with Lo,nACE2 and Im,nACE2 after redocking of losartan with Lo,nACE2, imatinib with Im,nACE2, losartan with Im,nACE2 and imatinib with Lo,nACE2 showed that the affinity of both drugs have increased with Lo,nACE2 and Im,nACE2 (Table 4).
Table 4.
Docking energies for Lo,nACE2 and Im,nACE2 bound to imatinib and losartan after 100 ns MD simulation
| nACE2 | Binding energy (Kcal/mole) | |
|---|---|---|
| Losartan | Imatinib | |
| Lo,nACE2 | − 11.99 | − 14.25 |
| Im,nACE2 | − 8.47 | − 17.78 |
Available ARBs vs losartan could only bind to ACE2 with reasonable but lower affinity
Losartan in association with two unavailable ARBs, pratosartan and tasosartan, was in the upper three ranking of binding energies. Other ARBs bind with ACE2 with lower, yet reasonable energy (Wishart et al. 2018) (Table S4).
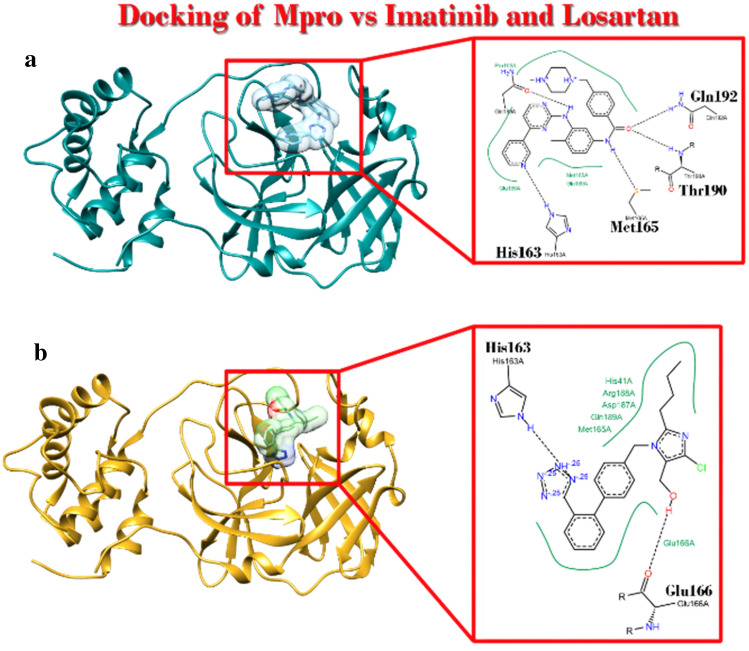
Imatinib and losartan could occupy the space where the “Pre-inh” in crystal structure of the main protease (Mpro) of SARS-CoV2 poses
It was elucidated that imatinib as well as losartan had higher affinity to Mpro. Considering CADD theories, it shows that these ligands based on their affinity probably behave as inhibitors of Mpro (Fig. 2).
Fig. 2.
Main Protease (Mpro) of SARS-CoV2 complex with imatinib and losartan. a Ribbon view of complex and Poseview of interaction with imatinib. b Ribbon view of complex and Poseview of interaction with losartan
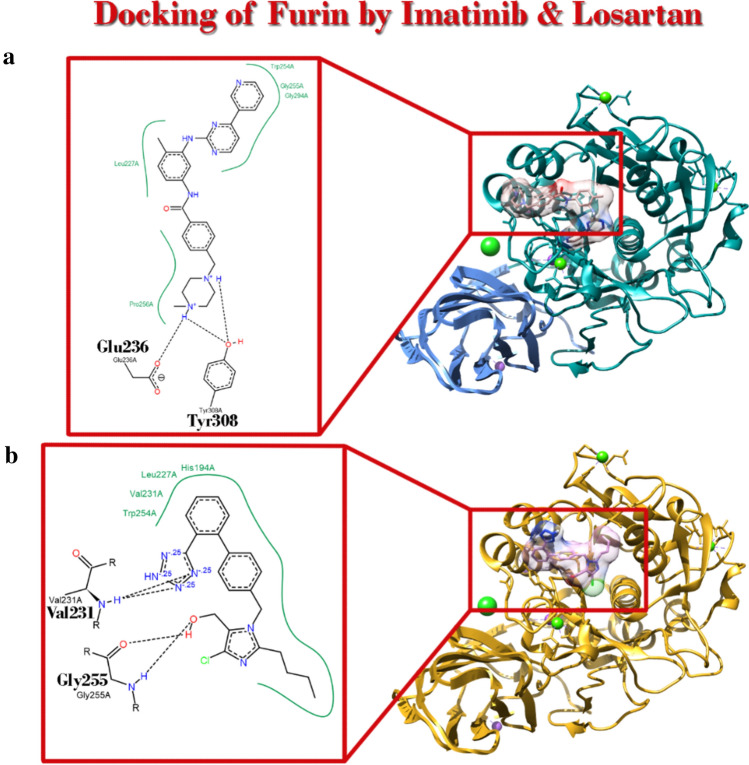
Imatinib and losartan could occupy the space where the “Pre-inh” in crystal structure of the furin of SARS-CoV2 poses
Our data demonstrated that imatinib had higher affinity for furin. But losartan showed up with lower but reasonable affinity. Considering CADD theories, it shows that imatinib based on its affinity will probably inhibit furin function (Fig. 3) (Table 5).
Fig. 3.
Position of Imatinib and Losartan in complex with furin with PDBID: 6hzb after Docking simulation. a Ribbon view of complex and Poseview of interaction with imatinib. b Ribbon view of complex and Poseview of interaction with Losartan
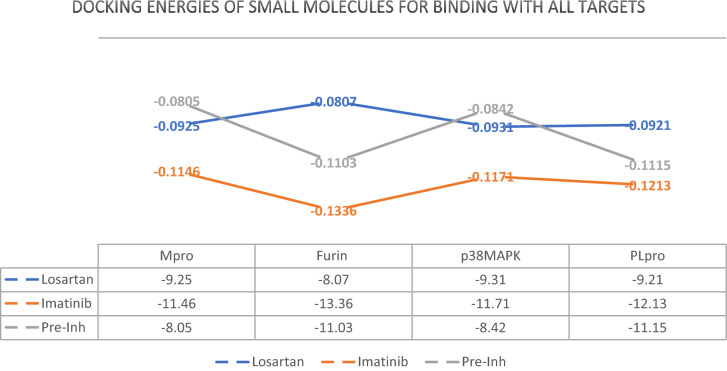
Table 5.
Docking Energies of losartan and imatinib for binding with the protein structures
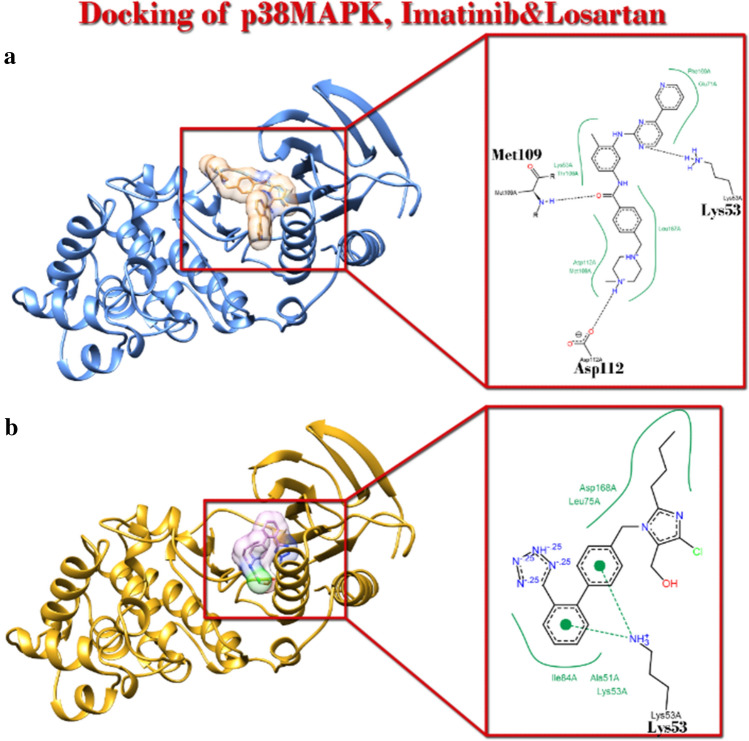
Imatinib and losartan could occupy with higher affinity the space where the “Pre-inh” in crystal structure of p38MAPK poses
It was implicated that imatinib and losartan had higher affinity to p38 MAPK. Considering CADD theories, it shows that imatinib and losartan based on their affinity will probably inhibit p38MAPK function (Fig. 4) (Table 5).
Fig. 4.
Position of Imatinib and losartan in complex with p38MAPK with PDBID: 1a9u after Docking simulation. a Ribbon view of complex and Poseview of interaction with imatinib. b Ribbon view of complex and Poseview of interaction with Losartan
Imatinib could occupy with higher affinity the space where “Pre-inh” in crystal structure of papin-like protease (PLpro) poses
Imatinib showed higher affinity and losartan has lower affinity for PLpro relative to its “Pre-inh”. Considering CADD theories imatinib might act as an inhibitor of PLpro function (Table 5) (Figs. 5, 6).
Fig. 5.
Position of Imatinib and Losartan in complex with PLpro with PDBID: 3mj5, before and after 100 ns MD simulation and redocking. a Ribbon and poseview of complex of PLpro with imatinib before 100 ns MD simulation, b ribbon and poseview of complex PLpro with imatinib after 100 ns MD simulation. c Ribbon and poseview of complex of PLpro with losartan before 100 ns MD simulation, d ribbon and poseview of complex PLpro with losartan after 100 ns MD simulation
Fig. 6.
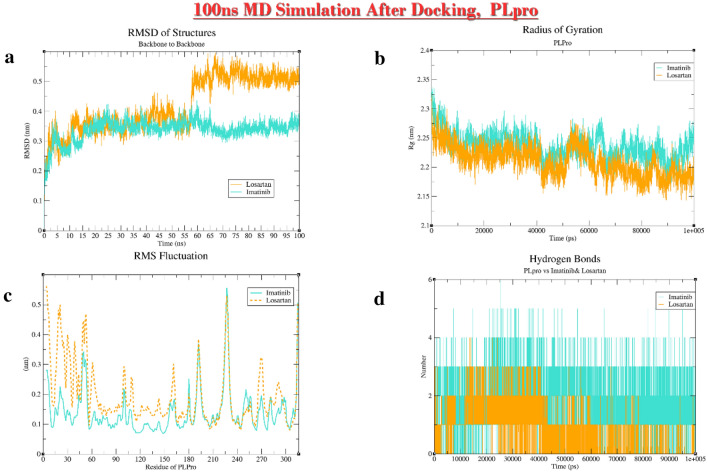
Diagrams of Losartan-PLpro and Imatinib-PLpro complexes after 100 ns MD simulation. a RMSD; b Rg; c RMSF; d H-Bonding
Losartan could change the conformational shape of PLpro
After 100 ns MD simulation exposure of losartan and PLpro, it was demonstrated that although affinity of losartan for PLpro is lower than that of imatinib and its “Pre-inh”, this ARB in 60 ns changes the radius of gyration and RMSD plots for at least 2.3 A°. It seems that losartan can disrupt PLpro.
Angiotensin II (Ang II) – Imatinib Interaction
We exposed imatinib and Ang II. The result shows that imatinib binds with Ang II with binding energy of Kcal/mol Fig. S4).
Discussion
A new insight to pathophysiology of ARDS in COVID19
The fatality and the change of the life style imposed by COVID19 have resulted in uprising death and economic burden in all countries. Despite the efforts done, the development of acute respiratory distress syndrome as the culprit of high morbidity and mortality in infection with SARS-CoV2 has remained elusive, yet attributed to a kind of cytokine storm. Apart from ARDS and cytokine storm, COVID19 seems to be a moderate illness, since SARS-CoV2 itself elicits a limited antiviral response in the hosts (Blanco-Melo et al. 2020).
Incubation of the virus in the upper respiratory tract results in the release of replicas into the lower airways and alveoli in huge number (about 10–1000 virion/μl in its peak on day 5–6 of infection) (Pan et al. 2020). ACE2, the virus receptor on host cells, besides to its expression on cardiovascular system, kidney, small intestine and adrenal, is distributed in the apical portion of the ciliated nasal and tracheobronchial cells as well as pneumocytes type I and type II (Hamming et al. 2004; Sims et al. 2005; Jia 2016). Consequently, these immune competent cells of the respiratory tract may harbor the virions easily. SARS-CoV2 entry to the cells is associated with downregulation of ACE2. This results in imbalance of ACE2/angiotensin[1–7]/Mas and ACE/Ang II/AT1R pathways in favor of the latter with pro-inflammatory, proliferative, prothrombotic, tissue destructive and pro-apoptotic properties (Glowacka et al. 2010; Kuba et al. 2005). In this context, hyperacute excess of Ang II seems to be the agent which can activate innate arm of the inflammatory cascades chaotically which results in sequential cytokine secretion. This process, as is not triggered directly by the antigens in a pathogen and originates from a collateral imbalance of hormonal milieu with immunological consequences, eventuates to the cytokine storm if not halted by negative feedback from activation of an adaptive immunity cascade(Tufet 2007; Tisoncik et al. 2012). Preliminary clues to this theory are:
Dependent to the viral load, circulating Ang II level has been reported to be higher in infected patients with SARS-CoV2 than in non-infected healthy people (Liu et al. 2020).
ACE2 is tissue protective for the lungs in acid- or sepsis-induced ARDS in mice (Imai et al. 2005).
Mechanical-stress induced ARDS as in ventilator-induced lung injury (VILI) is correlated to activation of ACE-dependent Nox1(NADPH-oxidase1)-MK(midkine)-Notch2 pathway and promoted by ACE/Ang-II/AT1R axis (Zhang et al. 2015; Wang et al. 2019).
Although contribution of RAS to evolving ARDS was previously suggested in SARS (Imai et al. 2006), it seems that many latent aspects of the involving pathways and the role they may play have recently been elucidated. A recent animal study in Karolinska University showed that Ang II infusion in healthy swines resulted in pathological changes identical to what happens in the lungs in COVID-19 (Rysz et al. 2020).
There are three orders of RAS in our body: systemic-hormonal, tissue-local (with paracrine and autocrine effects) and cellular-subcellular (with intracrine effects) (Marshall 2003; Abadir et al. 2011; Re 2018). Invasion of SARS-CoV2 in huge number downregulates ACE2 in the host cells outrageously so that the balance in tissue-local and cellular-subcellular RAS disrupts hyper-acutely. Accordingly, the host cells lose their ability to adapt to or defeat against the consequences of sudden rise in the content of Ang II with adequate negative feedback responses by ACE2. It is worth mentioning that in healthy people, intra-cellular Ang II content in some tissues may reach up to 1000 times higher than that of plasma (Fazeli et al. 2012). In ACE2 deficiency, Ang II is not hydrolized to angiotensin[1–7] (with its cytoprotective effect) or to other less active metabolites. Consequently, hyperacute excessive content of Ang II exerts rather untoward chaotic pathological effects in an intracrine (intracellular) and autocrine (cell to the same cell) manner and even through spilling over extracellularly, in a paracrine (cell to different neighboring endothelial, macrophages, monocytes, vascular smooth muscle cells or fibroblasts) and endocrine (cell to circulation) fashion. On the other hand, some studies demonstrated that Ang II itself downregulates ACE2 expression through internalization, lysosomal degradation and AT1R-mediated ROS activated ERK/p38 MAPK pathway which promotes TACE/ADAM17 activity, as well (Palau et al. 2020; Koka et al. 2008; Patel et al. 2014).
Ang II increases reactive oxygen species (ROS) through AT1R-dependent induction of NADPH oxidase (Nox), mostly Nox2 and Nox4 (Garrido and Griendling 2009; Bernard, et al. 2014; Rosenfeld, et al. 2008). Even though ROS as a signaling molecule contributes to cell homeostasis, its overproduction may lead to cell damage (Ray et al. 2012). Intriguingly, ROS upregulates the production of Ang II in a positive feedback response. In deficiency of ACE2, this amplifies the production of ROS dramatically (Zhang and Baker 2017; Chen et al. 2018). ROS causes DNA damage and mitochondrial dysfunction (Cooke et al. 2003; Cadet and Davies 2017). It has been reported in animal studies that Ang II through AT1R reduces mitochondrial number as well as pro-survival genes (Nampt and sirtuin3) (Benigni et al. 2009). ROS, in turn, through opening mitochondrial K-ATP channels and disturbing mitochondrial membrane potential, upregulates mitochondrial ROS (mtROS) production in a positive feedback response (Vajapey et al. 2014). mtROS functions as a triggering signaling molecule for production of pro-inflammatory cytokines (Naik and Dixit 2011). It was reported in influenza that regulated amount of mtROS induces interferon γ (IFNγ) to restrain infection (Kim et al. 2013a, b). But when mtROS rises up excessively, striking upregulation of pro-inflammatory cytokines must be expected. mtROS was shown to activate NLRP3 which induces IL-1 and IL-18 production (Shi et al. 2018). Furthermore, ROS activates inflammatory responses by inducing redox-sensitive transcriptional factors like NF-kB and activator protein 1 (AP1) (Re 2018; Bernstein et al. 2018).
Intriguingly, S protein in binding with ACE2 induces ADAM-17/TACE as a sheddase to separate the ectodomain subunit of this peptidase. Shedding of ACE2 is associated with the production of TNF-α which was argued to be the initiating cause of inflammation of the lung in SARS (Haga et al. 2008; Glowacka et al. 2010). Over-expression of TNF-α in the process of hyper-acute shedding of ACE2 by ADAM17 through synergism with Ang II may aggravate inflammatory milieu by inducing oxidative stress via NF-kB and p38 MAPK dependent pathways (Takahashi et al. 2008; Patel et al. 2014; May et al. 2016). Furthermore, activation of AT1R by AngII induces expression of TNF-α (presented already in the scene), IL-1β, IL-6, IL-8, MCP-1 and even IL-10 through NF-kB and activating protein 1 (AP-1) transcriptional factors (Guo et al. 2011; Saute et al. 2015).
Ang II through AT1R was also reported to amplify oxidative stress by distorting iron homeostasis and increasing labile ferrous iron as well as expression of ferritin in endothelial cells (Ishizaka et al. 2005; Tajima et al. 2010). Even though ferritin may show an antioxidative effect, it has been described in mice that ferritin may act as a local cytokine and activate NF-kB through MAPK-mediated pathway. This response results in a rise of inducible NO synthase for about 100-fold and IL-1β and RANTES for 50-fold with a small increase in intercellular adhesion molecule (ICAM). Ferritin may suppress adaptive immune response, as well (Kernan and Carcillo 2017).
Aggravating its pro-inflammatory effects, Ang II signaling through AT1R increases vascular permeability in the lung by releasing prostaglandins and vascular endothelial growth factor (VEGF) (Velazquez-Salinas et al. 2019). Disruption of endothelial-epithelial (blood-air) barrier in alveoli and increase in permeability of endothelium rises the fluid in the alveolar sacs that should be cleared out by epithelial Na channels (ENaCs). In rats, endogenous activation of AT1R by Ang II downregulates ENaC expression and disturbs pouring out the extra fluid (Deng et al. 2012).
Of the cytokines induced by Ang II, IL-6 plays a more special role in immunopathological effect of Ang II. IL-6 induces JAK2/STAT1/3 signaling pathway which promotes many genes contributing to the production of signaling molecules like cytokines, adaptors, receptors and protein kinases (Velazquez-Salinas et al. 2019; Weidanz et al. 2005). In this context, IL-6 is involved in regulation of differentiation of monocytes into macrophages, upregulation of B-cell IgG production, downregulation of dendritic cell maturation by activation of the STAT3 signaling pathway and the promotion of the Th2 response by inhibiting Th1 polarization (Velazquez-Salinas et al. 2019).
Ang II promotes production and release of IL-6 and IL-8 from human cultured adipocytes by NF-kB-mediated pathway to which AT1R rather than AT2R contributes. It is demonstrated that in the obese IL-6 plasma level is closely correlated to body mass index (BMI) (Skurk et al. 2004). Ang II-induced over expression of IL-6 in adipose tissue might be the reason why obesity is a risk factor in the severity of COVID19 (Lighter et al. 2020).
IL-6 has been found to increase platelet and immune cell aggregation through a T-cell dependent mechanism by Ang II (Senchenkova et al. 2019). In addition, an in vitro study showed Ang II to upregulate plasminogen activator inhibitors (PAIs) as well as tissue plasminogen activator (TPA) mRNA in vascular smooth muscle (VSM) cell culture both directly and indirectly through PDGF. In this manner the increase in PAIs not only neutralizes the effect of TPA and increases thrombophilia but the remains of TPA by resorbing extra-cellular matrix provides the possibility for the immune cell migration to the inflammation site (Van Leeuwen et al. 1994). Moreover, apart from the ability of Ang II to stimulate platelet aggregation in normotensive and hypertensive subjects, this vasoactive substance could change the shape of the platelets derived from healthy volunteers in in vitro studies; this effect of AngII on increasing of mean platelet volume (MPV) which is considered the first step of platelet activation does not respond to aspirin (Touyz and Schiffrin 1993; Jagroop and Mikhailidis 2000). Molecular and cellular endocrinology findings in previous studies show that the theory of Ang II-mediated immunopathology in COVID 19 could explain the thrombophilia observed in this disease (Klok et al. 2020).
Plasma IL-6 level has been correlated to the severity of COVID19 (Herold et al. 2020). This cytokine, in association with IL-1 and TNF-α, is the major inducer of CRP production in the liver (Bermudez et al. 2002; Eklund 2009). In addition, Ang II induces CRP expression in hepatocytes in a time- and dose-dependent manner through activation of AT1R and resulting from ROS-MAPK-(NF-kb) pathway independent of IL-1β and IL-6 (Zhao et al. 2013). Reciprocally, CRP increases expression of AT1R in vascular smooth muscle cells with subsequent upregulation of ROS (Wang et al. 2003).
Ang II through AT1R-PKA-proteosome pathway as well as activation of STAT1 and NF-kB promotes differentiation of Th0 to Th1. It has been demonstrated that in shifting from Th0 to Th1 or Th2, Ang II upregulates the production of IFNγ (tenfold), IL-2 (18-fold), IL-4 (3.5-fold) and IL-10 (1.5-fold). In addition, Ang II increases Tbox transcription factor mRNA (Tbet, marker for Th1) and GATA3 mRNA (marker for Th2) by 38 and 1.6-fold, respectively. Amazingly, losartan, an AT1R blocker, has been shown to inhibit Th1 differentiation without having any effect on that of Th2 (Qin et al. 2018). It is noteworthy that Th1 tends to differentiate to Th17 in the presence of IL-6 and TGF-β (Velazquez-Salinas et al. 2019; Qin et al. 2018). Th17 induces synthesis of IL-17, IL-21, IL-22 and TNF-α. TNF-α, itself in the presence of IL-6 and IL-1β may promote differentiation of T cells to Th17 (Zheng et al. 2014; Qin et al. 2018). High level of IL-17 in patients with ARDS suggests its contribution to this syndrome. This cytokine in a model of influenza and LPS induced acute lung injury has been associated with neutrophil recruitment and increased alveolar layer permeability (Rizzo et al. 2015a, b).
Local RAS has been found in DCs, T and NK cells with a complete enzymatic repertoire enabling them to synthesize and metabolize Ang II and even AT1R and AT2R (Lapteva et al. 2001; Hoch et al. 2009; Sun et al. 2009; Meng et al. 2017). It has also been described that these cells not only respond to Ang II but they have the tendency to migrate to this peptide. Thenceforth, Ang II may orchestrate recruitment of leukocytes to the site of inflammation. Besides, Ang II induces synthesis of CCL5/RANTES chemokine in T cells and NK cells. It shows that Ang II may direct chemotaxis of cells possessing CCR1, CCR3 and CCR5 and even regulate proliferation of T cells via CCR5 (Jurewicz et al. 2007)
Activation of AT1R by Ang II has been correlated with apoptosis in pneumocytes (Lukkarinen et al. 2005). Consequent alveolar and bronchial cell death contributes to the pathogenesis of SARS-CoV2. But cell loss would ultimately restrain the distribution of the virus. To make scene ready for the virus to continue its replication a reasonable percentage of the cells should survive. It merits mentioning that IL-6 in synergism with IL-17 (secreted by Th17) induces expression of pro-survival proteins Bcl-2 and Bcl-xl which inhibit cell destruction by CD8 + cytotoxic T cells and prevent apoptosis of pneumocytes (Hou et al. 2014). Moreover, IL-22, a member of IL-10 anti-inflammatory cytokine family, secreted by Th17 also prevents apoptosis of pulmonary endothelial cells and ameliorates ARDS through inducing JAK2/STAT3 pathway (Ren et al. 2017). But it should be taken into account that over-expression of Ang II upregulates pro-inflammatory cytokines [IFNγ (tenfold) and IL-2 (18 fold)] much more than anti-inflammatory cytokines [IL-4 (3.5-fold) and IL-10 (1.5-fold)] (Qin et al. 2018). It seems that in this milieu of highly complicated set of pro-inflammatory cytokines some anti-inflammatory molecules prevent apoptosis of the respiratory epithelial and may even help giant cells produced in COVID 19 to survive.
Ang II induced upregulation of matrix metalloproteinases (MMPs) in vascular smooth muscle cells results in destruction of the pulmonary tissue including the interstitial and basal collagen-elastin structures. AT1R-mediated extra-cellular signal-regulated kinase 1/2 (ERK1/2) and AT1R-ROS mediated NF-kB and AP-1 pathways lead to an increase in expression and tissue content of MMPs. These proteinases regulate remodeling and turn-over of extra-cellular matrix (ECM) and promote smooth muscle and endothelial cell proliferation and migration resulting in vascular wall fibrosis which eventually may end up in pulmonary hypertension if their presence lasts (Browatzki et al. 2005; Guo et al. 2015; Wang et al. 2015a, b, c). It has been reported in an animal study that ACE2 deficiency results in activation MMP and STAT3 pathway which may promote lung injuries (Hung et al. 2016).
In oxidative stress, macrophages exhibit more AT1R. Activating AT1R impairs efferocytosis (clearance of apoptotic cells) and interferes with the resolution of inflammatory cascades (Keidar et al. 2002; Yamamoto et al. 2011). MerTK is a tyrosine kinase which shifts DCs from pro-inflammatory to anti-inflammatory status (Anwar et al. 2009). Ang II reduces MerTK content of the cell membrane through shedding of MerTk in an AT1R/ROS/p38MAPK/ADAM17 mediated pathway (Zhang et al. 2019). Consequently, Ang II impairs switching M1 (with more pro-inflammatory abilities) to M2 (with more anti-inflammatory and tissue repairing properties). Continuation of pro-inflammatory status result in activation of MMPs and inducing of ECM remodeling processes. Failure of Tregs to show up due to the predominance of IL-6 promoting Th17 may prevent effective efferocytosis. In this milieu, pro-fibrotic IL-13 and TGF-β may lead to lung fibrosis. Ang II through AT1R induces fibrotic changes in the lungs with direct and indirect effects. It has been shown in transgenic mice that Ang II stimulates lung fibroblast/myofibroblast proliferation and synthesis of ECM. It induces production of TGF-β and connective tissue growth factor (CTGF) (Wang et al. 2015a, b, c; Proto et al. 2018). Ang II in synergism with TGF-β may promote fibrosis in many organs including the lungs. Moreover, AngII-induced oxidative stress may promote pulmonary fibrosis through downregulation of sirtuin3 (Murphy et al. 2015; Sosulski et al. 2017). On the other hand, angiotensin [1–7], the product of ACE2, has been described as an antifibrotic molecule in cardiomyocytes through stimulation of Sirtuin3-dependent deacetylation of Foxo3a (Uhal et al. 2012; Guo et al. 2017) In a recent study deletion of sirtuin3 accentuated AngII-mediated tissue stiffness and induced arterial pericyte to fibroblast transition through activation of AT1R (Chen and Zeng 2019).
Losartan in cytokine storm and ARDS in COVID-19
According to all above-mentioned studies it is rationally expected that losartan as an AT1R blocker might attenuate ARDS and cytokine storm in COVID-19. Ang II was previously considered as a factor that plays a role in ARDS and losartan is effective in ameliorating this pathology (Ruthman and Festic 2015; Khan et al. 2017; Kim et al. 2017). However, there are also some studies considering inconsistency of ARB effects in attenuating inflammatory reactions in vascular bed (Chang et al. 2007; Del Fiorentino Alessandra et al. 2009). But these studies were assessing ARB effects on the probable metabolic-based medium- to long-term atherosclerotic vascular inflammatory reactions but in the milieu where hydrolyzing of AngII by ACE2 was still possible. It merits mentioning that in some other studies anti-oxidant effect of losartan and its ability to reduce vascular inflammatory markers are discernible (Rajagopalan, et al. 2002; Graninger et al. 2004). Anyhow, it should be notified that what is encountered in COVID19 might hypothetically be a sudden dramatic downregulation of ACE2 due to invasion of huge number of virions and subsequent striking upregulation of Ang II followed by huge increase in AT1R content by Ang II in a positive feedback manner. Losartan was effective in many studies in suppressing pro-inflammatory effects of cytokines due to its immunomodulatory properties or even in preventing lung fibrosis.(Nahmod et al. 2003; Anwar et al. 2009; Mitra et al. 2010; Wang et al. 2014; Guo et al. 2015).
Moreover, losartan inhibits Ang II dependent change of shape (not responding to aspirin) and aggregation of platelets and also, independent to Ang II, shows antagonistic effect against thromboxane A2 more than irbesartan, telmisartan, valsartan, candesartan and olmesartan (Guerra-Cuesta et al. 1999; Jagroop and Mikhailidis 2000; Yamada et al. 2007).
Early in pandemic of COVID-19 it was suggested that ARBs might increase viral load through upregulation of ACE2 according to previous studies (Ferrario et al. 2005; Soler et al. 2009; Guo et al. 2020). It should be notified that in these studies overexpression of ACE2 after administration of ARBs needed 25–28 days to appear. However, according to an animal study this might be the upregulation of AT1R and PKCσ which increases the severity of COVID-19 in hypertensive patients with chronic ARBs intake (Song et al. 2015). On the other hand, entry of the virus to the cells needs that TMPRSS2 increases simultaneous to upregulation of ACE2. Otherwise, overexpression of ACE2 alone is capable of entrapping and decreasing the infectivity of the virus (Perico et al. 2020; Sanchis-Gomar et al. 2020; Vaduganathan et al. 2020). Accordingly, scientific societies in Europe and the USA recommended that ARBs be continued in patients with hypertension or heart diseases (South et al. 2020). Conclusively, BRACE CORONA trial recently showed safety of ARBs in COVID-19 (Lopes et al. 2020a,b).
As a preliminary clinical evidence of curing effect of losartan in COVID-19, two slices of lung CT-scan of a patient with COVID-19 whom was admitted to ICU, Bazarganan Hospital, Tehran, Iran, are shown. It is obviously seen that ground glass opacities cleared out significantly within four days after administration of losartan (6.25 mg twice a day). The patient’s dyspnea subsided drastically after three doses of losartan (Fig. 7).
Fig. 7.
Spiral lung CT-scan of a 50 years old patient with COVID19; a Apr 6, 2020; b Apr 10, 2020
Imatinib in cytokine storm and ARDS in COVID-19
As the patients with cytokine storm may experience hypotension due to superimposed infection in the extreme severity of the disease and losartan administration may reduce the blood pressure, it seems to be rational to recommend an immunomodulator in association with losartan to stop the cytokine storm more effectively. There is conflicting evidence that systemic corticosteroids may be hazardous to patients with COVID 19 (Veronese et al. 2020). As Abelson tyrosine-protein kinase 2 (Abl 2) is needed in replication of SARS-CoV and MERS-CoV., tyrosine kinase inhibitors have been introduced in the treatment of SARS and MERS. In this family of viruses, imatinib hinders the initial phases of the virion replication by inhibiting fusion of the virion at the endosomal membrane (Coleman et al. 2016; Ruella et al. 2017). Imatinib, with immunomodulatory effects, has also been suggested to have subsiding effects specially against ARDS and the vascular leak seen in this syndrome (Kim et al. 2013a, b; Rizzo et al. 2015a, b; Stephens et al. 2015). There is also a report that low dose imatinib was effective in reducing pulmonary blood pressure in dogs (Arita et al. 2013). In another report, inhaled imatinib was also used as a drug to subside pulmonary hypertension (Pitsiou et al. 2014).
Amazingly, Imatinib was found to have an inhibitory effect against Ang II impact on vascular smooth muscle cells in dissection of the aorta in mice (Sun et al. 2017). Furthermore, expression of MHC class I and II, production of co-stimulatory molecules and secretion of cytokines and chemokines in monocyte-derived dendritic cells decrease in the presence of imatinib. This tyrosine kinase inhibitor subsides phosphatidylinositol 3-kinase/Akt pathways and downregulates exhibition of NF-kB in the nucleus (Appel et al. 2005). Cultured human monocytes are morphologically and functionally suppressed in the presence on imatinib which reduces the ability of these cells to synthesize IL-6 and TNF-α and to respond efficiently to M-CSF and GM-CSF stimulation (Dewar et al. 2005). In an in vitro study, imatinib could inhibit expression of TNF-α, IL-6, IFNγ and IL-17 in cultured splenocyte of mice with arthritis in a dose dependent manner (Berlin and Lukacs 2005). In monocytes and macrophages, TNF-α production was reduced by imatinib while IL-10 expression did not change (Wolf et al. 2005). Imatinib in mice with hyper-reactive airway disease could subside peri-bronchial eosinophil accumulation and could also decrease secretion of IL-4 and IL-13 by Th2 as well as CCL2, CCL5 and CCL6 chemokines.(Akashi et al. 2011). In vitro, imatinib could impair immunosuppressive ability and expression of FoxP + in Tregs along with the subsiding of STAT3 and STAT5 pathways without affecting IL-10 and TGF-β in these cells (Larmonier et al. 2008). Besides, in low dosage, imatinib may elicit a physiologic innate immune response to infection called “emergency response” in which myelopoiesis, but not lymphopoiesis, is potentiated (Napier et al. 2015). Imatinib inhibits not only S protein-dependent virus-cell fusion but also prevents syncytia and giant cell formation in the lungs (Sisk et al. 2018.)
In silico study
We conducted an in silico study to investigate the probable inhibitory or modulatory effect of losartan and imatinib in some critical points of the replication cycle of SARS-CoV2. It was elucidated that both losartan and imatinib could bind to ACE2 with higher affinity relative to the “Pre-inh” (reference ligand, Table 2). It does not mean that these drugs could inhibit the catalytic property of this carboxypeptidase as it has not been reported, yet.
As a novel finding, we have demonstrated that losartan and imatinib could distort the binding site of SARS-CoV2 RBD to ACE2. According to our study there are seven points on the α-helix arm of ACE2 molecule located between glycine 24 and lysine 353 (Fig. 1a, d, Table S3) where SARS-CoV2 RBD (acceptor) and ACE2 (donor) may establish hydrogen (H) bonds. The distance between the acceptor–donor residues at these points are between 2.69 and 2.90 Å (Table S3). It means that hydrogen bond energies between SARS-CoV2 RBD and ACE2 based on the distance of acceptor and donor residues, without considering the hydrophobic bonds, are of moderate magnitude and mostly electrostatic (Jeffrey and Jeffrey 1997). The high affinity of losartan and imatinib to bind with ACE2 which increases even more after 100 ns of exposure of these two drugs to ACE2 in MD simulation, show that the bonds are stable enough. According to Table S3, α-helix of ACE2 was shown to be relocated for at least 1.80 Å in the presence of losartan and imatinib in 100 ns MD simulation. But in some binding residues the change after relocation was more significant (more than 3.00 Å) which may weaken H-bonds at these points:
for losartan in four out of seven binding points,
for imatinib in six out of seven binding points.
According to this modeling it is expected that the affinity of the virus to its receptor might decrease in the presence of these two drugs. In addition, our study showed that losartan among other available ARBs in the market has the highest affinity to ACE2 (Table S4). It is implicated that other ARBs might not be efficacious in docking to ACE2 to make stable similar conformational structural change in the receptor of the virus.
Furthermore, we could find that both losartan and imatinib could pose in Mpro, PLpro and MAPK molecules in the position of their “Pre-inh” with higher affinity. These docking energies in our model are important because they determine how these ligands may probably affect the behavior of the proteases.
Of all the proposed ligands, imatinib could pose favorably in furin structure with higher affinity relative to its “Pre-inh”. Considering high expression of furin in the lungs and importance of S’2 cleavage (Wrapp et al. 2020) in the entry of SARS-CoV2 to host cell, if the inhibitory effect of imatinib against furin is approved in in vivo studies it can be regarded as an inhibitor that hinders entry of SARS-CoV2 to the target cells, as well. Imatinib and losartan due to their effective docking to PLpro and Mpro with higher affinity relative to “Pre-inh” might be successful in preventing those proteases from letting the virus evade innate immunity or start replication. Although in the case of PLpro the docking energy for losartan were higher (lower affinity) than that of “Pre-inh”, the magnitude of energies (< − 9.21) was low enough to consider losartan as a probable inhibitor of PLpro. RMSD diagram for 100 ns MD simulation of losartan-PLpro complex showed that a sudden change of about 0.25 nm (2.5 Å) in the mean position of PLpro molecules occurred in about 60 ns that continued till the end of MD simulation for 100 ns with a fall in affinity or in other words rise in the docking energy [from -9.21 (Kcal/mole) before MD simulation to − 7.83 (Kcal/mole) post-hoc] (Fig. 6). This may indicate that losartan affects the conformational shape and probably the function of PLpro at the expense of losing its affinity to the protein to some degree (Oferkin et al. 2015; Pantsar and Poso 2018). Perhaps longer dynamic study is needed to explain the real behavior of PLpro in the presence of losartan.
Bradykinin induces allergic inflammatory responses with activation of airway fibroblast/myofibroblasts through MAPK pathway (Golias et al. 2007; Sabatini et al. 2013). In many of the pathophysiological destructive pathways in COVID 19 the signature of MAPK is evident. Surprisingly, in this modeling in silico study, losartan and imatinib showed to have significant tendency to bind with p38 MAPK. This might indicate that these drug ligands may have inhibitory effect against p38MAPK and its downstream pathways and may inhibit untoward bradykinin-dependent responses, as well. In our in silico study, imatinib also showed its tendency to bind to Ang II (Fig. S4). It should be investigated if imatinib could change the function of Ang II.
Conclusion
Through binding the scattered rings of previously found evidence, our reviewing the literature showed that hyper-acute activation of AT1R by a sudden rise of intracellular Ang II due to downregulation of ACE2 after showering of SARS-CoV2 to the lungs explains precisely the pathophysiology of cytokine storm and ARDS in COVID-19. It is implied that administration of losartan, an AT1R blocker, and imatinib, an immunomodulator and as an adjunctive to losartan might be promising in the attenuating ARDS in COVID-19.
Our in silico study showed that losartan and imatinib may decrease the affinity of the virus to ACE2 and might have inhibitory effects against furin, PLpro and MAPK to be validated in in-vitro, subclinial and clinical studies in the future.
According to the findings in this study and the preliminary clinical evidences, we suggest low dose systemic losartan and inhaled aerosolized low dose imatinib be studied in a subclinical setting in treating ARDS in COVID 19. In this manner, while losartan by blocking AT1R antagonizes Ang II effects, aerosolized imatinib may modulate the local immunological responses. Furthermore, this combination may reduce the probability of binding of the virus to ACE2 more effectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to extend their deep sense of gratitude to their family for their support and patience. Besides, they would like to express their greatest appreciation to all physicians including intensivists, anesthesiologists, radiologists, emergency medicine and infectious disease specialists and other healthcare workers including committed nurses and radiology technicians and all the personnel of hospitals, especially in Bazarganan Hospital, who sacrifice the lives with dignity in CORONA pandemic. Besides, the authors would like to thank High Performance Computing (HPC) Center, Institute for Research in Fundamental Sciences (IPM) and Computer Engineering Department at Sharif University of Technology.
Funding
This study was not funded by anybody.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reza Nejat and Ahmad Shahir Sadr contributed equally to this work.
Contributor Information
Reza Nejat, Email: rezanejat@yahoo.com.
Ahmad Shahir Sadr, Email: shahirsadr@cheragh.edu.af.
References
- Abadir PM, Foster DB, Crow M, Cooke CA, Rucker JJ, Jain A, Smith BJ, Burks TN, Cohn RD, Fedarko NS. Identification and characterization of a functional mitochondrial angiotensin system. Proc Natl Acad Sci. 2011;108(36):14849–14854. doi: 10.1073/pnas.1101507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi N, Matsumoto I, Tanaka Y, Inoue A, Yamamoto K, Umeda N, Tanaka Y, Hayashi T, Goto D, Ito S. Comparative suppressive effects of tyrosine kinase inhibitors imatinib and nilotinib in models of autoimmune arthritis. Mod Rheumatol. 2011;21(3):267–275. doi: 10.3109/s10165-010-0392-5. [DOI] [PubMed] [Google Scholar]
- Anwar A, Keating AK, Joung D, Sather S, Kim GK, Sawczyn KK, Brandao L, Henson PM, Graham DK. Mer tyrosine kinase (MerTK) promotes macrophage survival following exposure to oxidative stress. J Leukoc Biol. 2009;86(1):73–79. doi: 10.1189/jlb.0608334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel S, Rupf A, Weck MM, Schoor O, Brümmendorf TH, Weinschenk T, Grünebach F, Brossart P. Effects of imatinib on monocyte-derived dendritic cells are mediated by inhibition of nuclear factor-κB and Akt signaling pathways. Clin Cancer Res. 2005;11(5):1928–1940. doi: 10.1158/1078-0432.CCR-04-1713. [DOI] [PubMed] [Google Scholar]
- Arendse LB, Danser AJ, Poglitsch M, Touyz RM, Burnett JC, Llorens-Cortes C, Ehlers MR, Sturrock ED. Novel Therapeutic Approaches Targeting the Renin-Angiotensin System and Associated Peptides in Hypertension and Heart Failure. Pharmacol Rev. 2019;71(4):539–570. doi: 10.1124/pr.118.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita S, Arita N, Hikasa Y. Therapeutic effect of low-dose imatinib on pulmonary arterial hypertension in dogs. Can Vet J. 2013;54(3):255. [PMC free article] [PubMed] [Google Scholar]
- Bader M. ACE2, aniotensin-(1–7), and Mas: the other side of the coin. Pflüg Arch-Eur J Physioly. 2013;465(1):79–85. doi: 10.1007/s00424-012-1120-0. [DOI] [PubMed] [Google Scholar]
- Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A. 1988;38(6):3098. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Investig. 2009;119(3):524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benigni A, Cassis P, Remuzzi G. Angiotensin II revisited: new roles in inflammation, immunology and aging. EMBO Mol Med. 2010;2(7):247–257. doi: 10.1002/emmm.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJ, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1–3):43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Berlin AA, Lukacs NW. Treatment of cockroach allergen asthma model with imatinib attenuates airway responses. Am J Respir Crit Care Med. 2005;171(1):35–39. doi: 10.1164/rccm.200403-385OC. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez EA, Rifai N, Buring J, Manson JE, Ridker PM. Interrelationships among circulating interleukin-6, C-reactive protein, and traditional cardiovascular risk factors in women. Arterioscler Thromb Vasc Biol. 2002;22(10):1668–1673. doi: 10.1161/01.ATV.0000029781.31325.66. [DOI] [PubMed] [Google Scholar]
- Bernard K, Hecker L, Luckhardt TR, Cheng G, Thannickal VJ. NADPH oxidases in lung health and disease. Antioxid Redox Signal. 2014;20(17):2838–2853. doi: 10.1089/ars.2013.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KE, Khan Z, Giani JF, Cao D-Y, Bernstein EA, Shen XZ. Angiotensin-converting enzyme in innate and adaptive immunity. Nat Rev Nephrol. 2018;14(5):325. doi: 10.1038/nrneph.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant B, Liu W-C, Møller R, Panis M, Sachs D, Albrecht R (2020) SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. BioRxiv
- Browatzki M, Larsen D, Pfeiffer CA, Gehrke SG, Schmidt J, Kranzhöfer A, Katus HA, Kranzhöfer R. Angiotensin II stimulates matrix metalloproteinase secretion in human vascular smooth muscle cells via nuclear factor-κB and activator protein 1 in a redox-sensitive manner. J Vasc Res. 2005;42(5):415–423. doi: 10.1159/000087451. [DOI] [PubMed] [Google Scholar]
- Cadet J, Davies KJ. Oxidative DNA damage & repair: an introduction. Free Radical Biol Med. 2017;107:2–12. doi: 10.1016/j.freeradbiomed.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JF-W, Kok K-H, Zhu Z, Chu H, To KK-W, Yuan S, Yuen K-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L-T, Sun C-K, Chiang C-H, Wu C-J, Chua S, Yip H-K. Impact of simvastatin and losartan on antiinflammatory effect: in vitro study. J Cardiovasc Pharmacol. 2007;49(1):20–26. doi: 10.1097/FJC.0b013e31802ba4ec. [DOI] [PubMed] [Google Scholar]
- Chen JX, Zeng H. Deficiency of Sirtuin 3 Accentuates Angiotensin II-Induced Arterial/Myocardial Stiffness and Hypertension. FASEB J. 2019;33(S1):819814. [Google Scholar]
- Chen Q, Wang Q, Zhu J, Xiao Q, Zhang L. Reactive oxygen species: key regulators in vascular health and diseases. Br J Pharmacol. 2018;175(8):1279–1292. doi: 10.1111/bph.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CM, Sisk JM, Mingo RM, Nelson EA, White JM, Frieman MB. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and middle east respiratory syndrome coronavirus fusion. J Virol. 2016;90(19):8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah N, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandona P, Dhindsa S, Ghanim H, Chaudhuri A. Angiotensin II and inflammation: the effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockade. J Hum Hypertens. 2007;21(1):20–27. doi: 10.1038/sj.jhh.1002101. [DOI] [PubMed] [Google Scholar]
- Del Fiorentino Alessandra SC, Celi A, Dell’Omo G, Pedrinelli R. The effect of angiotensin receptor blockers on C-reactive protein and other circulating inflammatory indices in man. Vasc Health and Risk Manag. 2009;5:233. doi: 10.2147/vhrm.s4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL molecular graphics system." http://www.pymol.org
- Deng J, Wang D-X, Deng W, Li C-Y, Tong J. The effect of endogenous angiotensin II on alveolar fluid clearance in rats with acute lung injury. Can Respir J. 2012;19(5):311–318. doi: 10.1155/2012/951025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshotels MR, Xia H, Sriramula S, Lazartigues E, Filipeanu CM. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin ii type i receptor–dependent mechanism. Hypertension. 2014;64(6):1368–1375. doi: 10.1161/HYPERTENSIONAHA.114.03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar AL, Doherty KV, Hughes TP, Lyons AB. Imatinib inhibits the functional capacity of cultured human monocytes. Immunol Cell Biol. 2005;83(1):48–56. doi: 10.1111/j.1440-1711.2004.01296.x. [DOI] [PubMed] [Google Scholar]
- Dilauro M, Burns KD. Angiotensin-(1–7) and its effects in the kidney. Sci World J. 2009;9:522–535. doi: 10.1100/tsw.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolatkhah Z, Javanshir S, Sadr AS, Hosseini J, Sardari S. Synthesis, molecular docking, molecular dynamics studies, and biological evaluation of 4 h-chromone-1, 2, 3, 4-tetrahydropyrimidine-5-carboxylate derivatives as potential antileukemic agents. J Chem Inf Model. 2017;57(6):1246–1257. doi: 10.1021/acs.jcim.6b00138. [DOI] [PubMed] [Google Scholar]
- Dominici FP, Burghi V, Munoz MC, Giani JF. Modulation of the action of insulin by angiotensin-(1–7) Clin Sci. 2014;126(9):613–630. doi: 10.1042/CS20130333. [DOI] [PubMed] [Google Scholar]
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87(5):e1–e9. doi: 10.1161/01.RES.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Eklund CM. Proinflammatory cytokines in CRP baseline regulation. Adv Clin Chem. 2009;48:111–136. doi: 10.1016/S0065-2423(09)48005-3. [DOI] [PubMed] [Google Scholar]
- Fazeli G, Stopper H, Schinzel R, Ni C-W, Jo H, Schupp N. Angiotensin II induces DNA damage via AT1 receptor and NADPH oxidase isoform Nox4. Mutagenesis. 2012;27(6):673–681. doi: 10.1093/mutage/ges033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses, pp 1-23 [DOI] [PMC free article] [PubMed]
- Ferrario CM, Jessup J, Chappell MC, Averill DB, Brosnihan KB, Tallant EA, Diz DI, Gallagher PE. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111(20):2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Montgomery J, Jr, Vreven T, Kudin K, Burant J. "Gaussian 03, Revision C. 02. Wallingford: Gaussian. Inc.; 2004. [Google Scholar]
- Gao B-B, Hansen H, Chen H-C, Feener EP. Angiotensin II stimulates phosphorylation of an ectodomain-truncated platelet-derived growth factor receptor-β and its binding to class IA PI3K in vascular smooth muscle cells. Biochem J. 2006;397(2):337–344. doi: 10.1042/BJ20060095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido AM, Griendling KK. NADPH oxidases and angiotensin II receptor signaling. Mol Cell Endocrinol. 2009;302(2):148–158. doi: 10.1016/j.mce.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Takayama J, Rao KV, Ratia K, Chaudhuri R, Mulhearn DC, Lee H, Nichols DB, Baliji S, Baker SC. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein− ligand X-ray structure and biological evaluation. J Med Chem. 2010;53(13):4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golias C, Charalabopoulos A, Stagikas D, Charalabopoulos K, Batistatou A. The kinin system-bradykinin: biological effects and clinical implications. Multiple role of the kinin system-bradykinin. Hippokratia. 2007;11(3):124. [PMC free article] [PubMed] [Google Scholar]
- Goossens G, Blaak E, Arner P, Saris W, Van Baak M. Angiotensin II: a hormone that affects lipid metabolism in adipose tissue. Int J Obes. 2007;31(2):382–384. doi: 10.1038/sj.ijo.0803388. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graninger M, Reiter R, Drucker C, Minar E, Jilma B. Angiotensin receptor blockade decreases markers of vascular inflammation. J Cardiovasc Pharmacol. 2004;44(3):335–339. doi: 10.1097/01.fjc.0000137160.76616.cc. [DOI] [PubMed] [Google Scholar]
- Guerra-Cuesta JI, Montón M, Rodríguez-Feo JA, Jiménez AM, González-Fernández F, Rico LA, Gomez RGJ, Farré J, Casado S, López-Farré A. Effect of losartan on human platelet activation. J Hypertens. 1999;17(3):447–452. doi: 10.1097/00004872-199917030-00019. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guo F, Chen X-L, Wang F, Liang X, Sun Y-X, Wang Y-J. Role of angiotensin II type 1 receptor in angiotensin II-induced cytokine production in macrophages. J Interferon Cytokine Res. 2011;31(4):351–361. doi: 10.1089/jir.2010.0073. [DOI] [PubMed] [Google Scholar]
- Guo Y-S, Wu Z-G, Yang J-K, Chen X-J. Impact of losartan and angiotensin II on the expression of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 in rat vascular smooth muscle cells. Mol Med Rep. 2015;11(3):1587–1594. doi: 10.3892/mmr.2014.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Yin A, Zhang Q, Zhong T, O’Rourke ST, Sun C. Angiotensin-(1–7) attenuates angiotensin II-induced cardiac hypertrophy via a Sirt3-dependent mechanism. Am J Physiol-Heart Circ Physiol. 2017;312(5):H980–H991. doi: 10.1152/ajpheart.00768.2016. [DOI] [PubMed] [Google Scholar]
- Guo J, Huang Z, Lin L, Lv J. Coronavirus disease 2019 (covid-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J Am Heart Assoc. 2020;9(7):e016219. doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci. 2008;105(22):7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis M, Lely A, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold T, Arnreich C, Hellmuth J, von Bergwelt-Baildon M, Klein M, Weinberger T (2020) Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. medRxiv 2020." Google Scholar
- Hoch NE, Guzik TJ, Chen W, Deans T, Maalouf SA, Gratze P, Weyand C, Harrison DG. Regulation of T-cell function by endogenously produced angiotensin II. Am J Physiol-Regul Integr Comp Physiol. 2009;296:R208–R216. doi: 10.1152/ajpregu.90521.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Jin Y-H, Kang HS, Kim BS. Interleukin-6 (IL-6) and IL-17 synergistically promote viral persistence by inhibiting cellular apoptosis and cytotoxic T cell function. J Virol. 2014;88(15):8479–8489. doi: 10.1128/JVI.00724-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Y-H, Hsieh W-Y, Hsieh J-S, Liu C, Tsai C-H, Lu L-C, Huang C-Y, Wu C-L, Lin C-S. Alternative roles of STAT3 and MAPK signaling pathways in the MMPs activation and progression of lung injury induced by cigarette smoke exposure in ACE2 knockout mice. Int J Biol Sci. 2016;12(4):454. doi: 10.7150/ijbs.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Rao S, Huan Y, Guo F, Guan B, Yang P, Sarao R, Wada T, Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Penninger JM. The renin–angiotensin system in acute respiratory distress syndrome. Drug Discov Today Dise Mech. 2006;3(2):225–229. doi: 10.1016/j.ddmec.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kuba K, Penninger JM. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93(5):543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaka N, Saito K, Mori I, Matsuzaki G, Ohno M, Nagai R. Iron chelation suppresses ferritin upregulation and attenuates vascular dysfunction in the aorta of angiotensin II–infused rats. Arterioscler Thromb Vasc Biol. 2005;25(11):2282–2288. doi: 10.1161/01.ATV.0000181763.57495.2b. [DOI] [PubMed] [Google Scholar]
- Jagroop I, Mikhailidis D. Angiotensin II can induce and potentiate shape change in human platelets: effect of losartan. J Hum Hypertens. 2000;14(9):581–585. doi: 10.1038/sj.jhh.1001102. [DOI] [PubMed] [Google Scholar]
- Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. Iscience. 2020;23(6):101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey GA, Jeffrey GA. An introduction to hydrogen bonding. New York: Oxford University Press; 1997. [Google Scholar]
- Jia H. Pulmonary angiotensin-converting enzyme 2 (ACE2) and inflammatory lung disease. Shock. 2016;46(3):239–248. doi: 10.1097/SHK.0000000000000633. [DOI] [PubMed] [Google Scholar]
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C (2020) Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv. [DOI] [PubMed]
- Jurewicz M, McDermott DH, Sechler JM, Tinckam K, Takakura A, Carpenter CB, Milford E, Abdi R. Human T and natural killer cells possess a functional renin-angiotensin system: further mechanisms of angiotensin II–induced inflammation. J Am Soc Nephrol. 2007;18(4):1093–1102. doi: 10.1681/ASN.2006070707. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keidar S, Heinrich R, Kaplan M, Aviram M. Oxidative stress increases the expression of the angiotensin-II receptor type 1 in mouse peritoneal macrophages. J Renin Angiotensin Aldos Syst. 2002;3(1):24–30. doi: 10.3317/jraas.2002.004. [DOI] [PubMed] [Google Scholar]
- Kelly DJ, Cox AJ, Gow RM, Zhang Y, Kemp BE, Gilbert RE. Platelet-derived growth factor receptor transactivation mediates the trophic effects of angiotensin II in vivo. Hypertension. 2004;44(2):195–202. doi: 10.1161/01.HYP.0000132883.20764.12. [DOI] [PubMed] [Google Scholar]
- Kernan KF, Carcillo JA. Hyperferritinemia and inflammation. Int Immunol. 2017;29(9):401–409. doi: 10.1093/intimm/dxx031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21(1):234. doi: 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim C-H, Ryu J-H, Kim M-J, Park CY, Lee JM, Holtzman MJ, Yoon J-H. Reactive oxygen species induce antiviral innate immune response through IFN-λ regulation in human nasal epithelial cells. Am J Respir Cell Mol Biol. 2013;49(5):855–865. doi: 10.1165/rcmb.2013-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IK, Rhee CK, Yeo CD, Kang HH, Lee DG, Lee SH, Kim JW. Effect of tyrosine kinase inhibitors, imatinib and nilotinib, in murine lipopolysaccharide-induced acute lung injury during neutropenia recovery. Crit Care. 2013;17(3):R114. doi: 10.1186/cc12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Choi SM, Lee J, Park YS, Lee CH, Yim J-J, Yoo C-G, Kim YW, Han SK, Lee S-M. Effect of renin-angiotensin system blockage in patients with acute respiratory distress syndrome: a retrospective case control study. Korean J Crit Care Med. 2017;32(2):154. doi: 10.4266/kjccm.2016.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok FA, Kruip MJ, Van der Meer NJ, Arbous MS, Gommers DA, Kant KM, Kaptein FH, van Paassen J, Stals MA, Huisman MV, Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koka V, Huang XR, Chung AC, Wang W, Truong LD, Lan HY. Angiotensin II up-regulates angiotensin I-converting enzyme (ACE), but down-regulates ACE2 via the AT1-ERK/p38 MAP kinase pathway. Am J Pathol. 2008;172(5):1174–1183. doi: 10.2353/ajpath.2008.070762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lapteva N, Nieda M, Ando Y, Ide K, Hatta-Ohashi Y, Dymshits G, Ishikawa Y, Juji T, Tokunaga K. Expression of renin-angiotensin system genes in immature and mature dendritic cells identified using human cDNA microarray. Biochem Biophys Res Commun. 2001;285(4):1059–1065. doi: 10.1006/bbrc.2001.5215. [DOI] [PubMed] [Google Scholar]
- Larmonier N, Janikashvili N, LaCasse CJ, Larmonier CB, Cantrell J, Situ E, Lundeen T, Bonnotte B, Katsanis E. Imatinib mesylate inhibits CD4+ CD25+ regulatory T cell activity and enhances active immunotherapy against BCR-ABL− tumors. J Immunol. 2008;181(10):6955–6963. doi: 10.4049/jimmunol.181.10.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Fujioka S, Takahashi R, Oe T. Angiotensin II-induced oxidative stress in human endothelial cells: modification of cellular molecules through lipid peroxidation. Chem Res Toxicol. 2019;32(7):1412–1422. doi: 10.1021/acs.chemrestox.9b00110. [DOI] [PubMed] [Google Scholar]
- Lei J, Kusov Y, Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Ann Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong SK, Huang IC, Xu K, Vasilieva N. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO journal. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighter J, Phillips M, Hochman S, Sterling S, Johnson D, Francois F, Stachel A. Obesity in patients younger than 60 years is a risk factor for Covid-19 hospital admission. Clin Infect Dis. 2020;9(10):1093. doi: 10.1093/cid/ciaa415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Li J, Feng C. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RD, Macedo AVS, Moll-Bernardes RJ, Feldman A, Arruda GDAS, de Souza AS, de Albuquerque DC, Mazza L, Santos MF, Salvador NZ. Continuing versus suspending angiotensin-converting enzyme inhibitors and angiotensin receptor blockers: Impact on adverse outcomes in hospitalized patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–The BRACE CORONA Trial. Am Heart J. 2020;226:49–59. doi: 10.1016/j.ahj.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RD, Macedo AVS, Moll-Bernardes RJ, Feldman A (2020) Continuing Versus Suspending Angiotensin-Converting Enzyme Inhibitors and Angiotensin Receptor Blockers - BRACE CORONA. J Am College Cardiol [DOI] [PMC free article] [PubMed]
- Lukkarinen HP, Laine J, Aho H, Zagariya A, Vidyasagar D, Kääpä PO. Angiotensin II receptor inhibition prevents pneumocyte apoptosis in surfactant-depleted rat lungs. Pediatr Pulmonol. 2005;39(4):349–358. doi: 10.1002/ppul.20187. [DOI] [PubMed] [Google Scholar]
- Manabe S, Okura T, Watanabe S, Fukuoka T, Higaki J. Effects of angiotensin II receptor blockade with valsartan on pro-inflammatory cytokines in patients with essential hypertension. J Cardiovasc Pharmacol. 2005;46(6):735–739. doi: 10.1097/01.fjc.0000185783.00391.60. [DOI] [PubMed] [Google Scholar]
- Marshall RP. The pulmonary renin-angiotensin system. Curr Pharm Des. 2003;9(9):715–722. doi: 10.2174/1381612033455431. [DOI] [PubMed] [Google Scholar]
- Mayr M, Duerrschmid C, Medrano G, Taffet GE, Wang Y, Entman ML, Haudek SB. TNF/Ang-II synergy is obligate for fibroinflammatory pathology, but not for changes in cardiorenal function. Physiol Rep. 2016;4(8):e12765. doi: 10.14814/phy2.12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean D, Edwards H, Lewis I, Mastryukov V, Boggs JE, Leong M. Infrared and Raman spectra of 1, 1-dibromodisilanes and STO-3G∗ calculations. Spectrochim Acta Part A Mol Biomol Spectrosc. 1996;52(2):199–205. doi: 10.1016/0584-8539(95)01554-X. [DOI] [Google Scholar]
- Meng Y, Chen C, Liu Y, Tian C, Li H-H. Angiotensin II regulates dendritic cells through activation of NF-κB/p65, ERK1/2 and STAT1 pathways. Cell Physiol Biochem. 2017;42(4):1550–1558. doi: 10.1159/000479272. [DOI] [PubMed] [Google Scholar]
- Mielech AM, Chen Y, Mesecar AD, Baker SC. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra AK, Gao L, Zucker IH. Angiotensin II-induced upregulation of AT1 receptor expression: sequential activation of NF-κB and Elk-1 in neurons. Am J Physiol-Cell Physiol. 2010;299(3):C561–C569. doi: 10.1152/ajpcell.00127.2010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Wong AL, Bezuhly M. Modulation of angiotensin II signaling in the prevention of fibrosis. Fibrogen Tissue Repair. 2015;8(1):7. doi: 10.1186/s13069-015-0023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Á. Density functional. Theory and application to atoms and molecules. Phys Rep. 1998;298(1):1–79. doi: 10.1016/S0370-1573(97)00083-5. [DOI] [Google Scholar]
- Nahmod KA, Vermeulen ME, S. RAIDEN, G. Salamone, R. Gamberale, P. Fernández-Calotti, A. Alvarez, V. Nahmod, M. Giordano and J. R. Geffner, Control of dendritic cell differentiation by angiotensin II. FASEB J. 2003;17(3):491–493. doi: 10.1096/fj.02-0755fje. [DOI] [PubMed] [Google Scholar]
- Naik E, Dixit VM. Mitochondrial reactive oxygen species drive proinflammatory cytokine production. J Exp Med. 2011;208(3):417–420. doi: 10.1084/jem.20110367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier RJ, Norris BA, Swimm A, Giver CR, Harris WA, Laval J, Napier BA, Patel G, Crump R, Peng Z, Bornmann W. Low doses of imatinib induce myelopoiesis and enhance host anti-microbial immunity. PLoS Pathog. 2015;11(3):e1004770. doi: 10.1371/journal.ppat.1004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejat R, Sadr AS. SARS virus papain-like protease: a mysterious weapon. J Biostat Epidemiol. 2019;5(4):288–295. [Google Scholar]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oferkin IV, Katkova EV, Sulimov AV, Kutov DC, Sobolev SI, Voevodin VV, Sulimov VB (2015) Evaluation of docking target functions by the comprehensive investigation of protein-ligand energy minima. Adv Bioinform [DOI] [PMC free article] [PubMed]
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1–12. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palau V, Riera M, Soler MJ. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol Dial Transpl. 2020;35(6):1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhang D, Yang P, Poon LL, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantsar T, Poso A. Binding affinity via docking: fact and fiction. Molecules. 2018;23(8):1899. doi: 10.3390/molecules23081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi-Rullan R, Barreto-Torres G, Ruiz L, Casasnovas J, Javadov S. Direct renin inhibition exerts an anti-hypertrophic effect associated with improved mitochondrial function in post-infarction heart failure in diabetic rats. Cell Physiol Biochem. 2012;29(5–6):841–850. doi: 10.1159/000178526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VB, Clarke N, Wang Z, Fan D, Parajuli N, Basu R, Putko B, Kassiri Z, Turner AJ, Oudit GY. Angiotensin II induced proteolytic cleavage of myocardial ACE2 is mediated by TACE/ADAM-17: a positive feedback mechanism in the RAS. J Mol Cell Cardiol. 2014;66:167–176. doi: 10.1016/j.yjmcc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- Perico L, Benigni A, Remuzzi G. Should COVID-19 concern nephrologists? Why and to what extent? The emerging impasse of angiotensin blockade. Nephron. 2020;144(5):213–221. doi: 10.1159/000507305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pitsiou G, Zarogoulidis P, Petridis D, Kioumis I, Lampaki S, Organtzis J, Porpodis K, Papaiwannou A, Tsiouda T, Hohenforst-Schmidt W. Inhaled tyrosine kinase inhibitors for pulmonary hypertension: a possible future treatment. Drug Des Dev Ther. 2014;8:1753. doi: 10.2147/DDDT.S70277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proto JD, Doran AC, Gusarova G, Yurdagul A, Jr, Sozen E, Subramanian M, Islam MN, Rymond CC, Du J, Hook J. Regulatory T cells promote macrophage efferocytosis during inflammation resolution. Immunity. 2018;49(4):666–677.e666. doi: 10.1016/j.immuni.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X-Y, Zhang Y-L, Chi Y-F, Yan B, Zeng X-J, Li H-H, Liu Y. Angiotensin II regulates Th1 T cell differentiation through angiotensin II type 1 receptor-PKA-mediated activation of proteasome. Cell Physiol Biochem. 2018;45(4):1366–1376. doi: 10.1159/000487562. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Brook R, Mehta RH, Supiano M, Pitt B. Effect of losartan in aging-related endothelial impairment. Am J Cardiol. 2002;89(5):562–566. doi: 10.1016/S0002-9149(01)02297-4. [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang B-W, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24(5):981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re RN. Role of intracellular angiotensin II. Am J Physiol-Heart and Circ Physiol. 2018;314(4):H766–H771. doi: 10.1152/ajpheart.00632.2017. [DOI] [PubMed] [Google Scholar]
- Reddy R, Asante I, Liu S, Parikh P, Liebler J, Borok Z, Rodgers K, Baydur A, Louie SG (2019) Circulating angiotensin peptides levels in Acute Respiratory Distress Syndrome correlate with clinical outcomes: a pilot study. PLoS One 14(3) [DOI] [PMC free article] [PubMed]
- Reddy Gaddam R, Chambers S, Bhatia M. ACE and ACE2 in inflammation: a tale of two enzymes. Inflamm Allergy-Drug Targets. 2014;13(4):224–234. doi: 10.2174/1871528113666140713164506. [DOI] [PubMed] [Google Scholar]
- Ren W, Wang Z, Wu Z, Hu Z, Dai F, Chang J, Li B, Liu H, Ruan Y. JAK2/STAT3 pathway was associated with the protective effects of IL-22 on aortic dissection with acute lung injury. Dis Markers. 2017;2017:1917804. doi: 10.1155/2017/1917804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere G, Michaud A, Breton C, VanCamp G, Laborie C, Enache M, Lesage J, Deloof S, Corvol P, Vieau D. Angiotensin-converting enzyme 2 (ACE2) and ACE activities display tissue-specific sensitivity to undernutrition-programmed hypertension in the adult rat. Hypertension. 2005;46(5):1169–1174. doi: 10.1161/01.HYP.0000185148.27901.fe. [DOI] [PubMed] [Google Scholar]
- Rizzo AN, Aman J, van Nieuw Amerongen GP, Dudek SM. Targeting Abl kinases to regulate vascular leak during sepsis and acute respiratory distress syndrome. Arterioscler Thromb Vasc Biol. 2015;35(5):1071–1079. doi: 10.1161/ATVBAHA.115.305085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo AN, Sammani S, Esquinca AE, Jacobson JR, Garcia JG, Letsiou E, Dudek SM. Imatinib attenuates inflammation and vascular leak in a clinically relevant two-hit model of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2015;309(11):L1294–L1304. doi: 10.1152/ajplung.00031.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CR, Zagariya AM, Liu X-T, Willis BC, Fluharty S, Vidyasagar D. Meconium increases type 1 angiotensin II receptor expression and alveolar cell death. Pediatr Res. 2008;63(3):251–256. doi: 10.1203/PDR.0b013e318163a2b8. [DOI] [PubMed] [Google Scholar]
- Ruella M, Kenderian S, Shestova O, Klichinsky M, Melenhorst J, Wasik M, Lacey S, June C, Gill S. Kinase inhibitor ibrutinib to prevent cytokine-release syndrome after anti-CD19 chimeric antigen receptor T cells for B-cell neoplasms. Leukemia. 2017;31(1):246–248. doi: 10.1038/leu.2016.262. [DOI] [PubMed] [Google Scholar]
- Ruthman CA, Festic E. Emerging therapies for the prevention of acute respiratory distress syndrome. Ther Adv Respir Dis. 2015;9(4):173–187. doi: 10.1177/1753465815585716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rysz S, Al-Saadi J, Farm M. COVID-19 pathophysiology may be driven by a loss of inhibition of the renin-angiotensin-aldosterone system. Preprint (Version 2) available at Research Square, 10. 10.21203/rs.3.rs-32494/v2
- Sabatini F, Luppi F, Petecchia L, Di Stefano A, Longo AM, Eva A, Vanni C, Hiemstra PS, Sterk PJ, Sorbello V. Bradykinin-induced asthmatic fibroblast/myofibroblast activities via bradykinin B2 receptor and different MAPK pathways. Eur J Pharmacol. 2013;710(1–3):100–109. doi: 10.1016/j.ejphar.2013.03.048. [DOI] [PubMed] [Google Scholar]
- Sanchis-Gomar F, Lavie CJ, Perez-Quilis C, Henry BM, Lippi G (2020) Angiotensin-converting Enzyme 2 and antihypertensives (angiotensin receptor blockers and angiotensin-converting enzyme inhibitors) in Coronavirus Disease 2019. In: Mayo Clinic Proceedings, Elsevier. [DOI] [PMC free article] [PubMed]
- Santos R, Ferreira AJ, Verano-Braga T, Bader M. Angiotensin-converting enzyme 2, angiotensin-(1–7) and Mas: new players of the renin-angiotensin system. J Endocrinol. 2013;216(2):R1–R17. doi: 10.1530/JOE-12-0341. [DOI] [PubMed] [Google Scholar]
- Satou R, Penrose H, Navar LG. Inflammation as a regulator of the renin-angiotensin system and blood pressure. Curr Hypertens Rep. 2018;20(12):100. doi: 10.1007/s11906-018-0900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter NS, Thienel C, Plutino Y, Kampe K, Dror E, Traub S, Timper K, Bédat B, Pattou F, Kerr-Conte J. Angiotensin II induces interleukin-1β–mediated islet inflammation and β-cell dysfunction independently of vasoconstrictive effects. Diabetes. 2015;64(4):1273–1283. doi: 10.2337/db14-1282. [DOI] [PubMed] [Google Scholar]
- Senchenkova EY, Russell J, Yildirim A, Granger DN, Gavins FN. Novel Role of T Cells and IL-6 (Interleukin-6) in angiotensin II–induced microvascular dysfunction. Hypertension. 2019;73(4):829–838. doi: 10.1161/HYPERTENSIONAHA.118.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Lei Z, Cheng G, Li D, Wang Q, Luo S, Yang H, Jia H. Mitochondrial ROS activate interleukin-1β expression in allergic rhinitis. Oncol Lett. 2018;16(3):3193–3200. doi: 10.3892/ol.2018.8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Grewe D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol. 2005;79(24):15511–15524. doi: 10.1128/JVI.79.24.15511-15524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk JM, Frieman MB, Machamer CE. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J Gener Virol. 2018;99(5):619. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurk T, van Harmelen V, Hauner H. Angiotensin II stimulates the release of interleukin-6 and interleukin-8 from cultured human adipocytes by activation of NF-κB. Arterioscler Thromb Vasc Biol. 2004;24(7):1199–1203. doi: 10.1161/01.ATV.0000131266.38312.2e. [DOI] [PubMed] [Google Scholar]
- Snijder E, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler MJ, Ye M, Wysocki J, William J, Lloveras J, Batlle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am J Physiol-Renal Physiol. 2009;296(2):F398–F405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- Song MA, Dasgupta C, Zhang L. Chronic losartan treatment up-regulates AT 1 R and increases the heart vulnerability to acute onset of ischemia and reperfusion injury in male rats. PloS One. 2015;10(7):e0132712. doi: 10.1371/journal.pone.0132712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosulski ML, Gongora R, Feghali-Bostwick C, Lasky JA, Sanchez CG. Sirtuin 3 deregulation promotes pulmonary fibrosis. J Gerontol Ser A. 2017;72(5):595–602. doi: 10.1093/gerona/glw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South AM, Tomlinson L, Edmonston D, Hiremath S, Sparks MA (2020) Controversies of renin–angiotensin system inhibition during the COVID-19 pandemic. Nature Rev Nephrol 16(6):305–307. 10.1038/s41581-020-0279-4 [DOI] [PMC free article] [PubMed]
- Stephens RS, Johnston L, Servinsky L, Kim BS, Damarla M. The tyrosine kinase inhibitor imatinib prevents lung injury and death after intravenous LPS in mice. Physiol Rep. 2015;3(11):e12589. doi: 10.14814/phy2.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierand K, Rarey M. PoseView–molecular interaction patterns at a glance. J Cheminform. 2010;2(1):1–1. doi: 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Zhang W, Zhu W, Yan H, Zhu J. Expression of renin-angiotensin system on dendritic cells of patients with coronary artery disease. Inflammation. 2009;32(6):347. doi: 10.1007/s10753-009-9141-3. [DOI] [PubMed] [Google Scholar]
- Sun T, Chen S, Dong N, Zhou X, Li H. Imatinib inhibits angiotensin II-induced aortic dissection through the c-Abl signaling pathway. Int J Clin Exp Pathol. 2017;10(5):5316–5324. [Google Scholar]
- Tajima S, Tsuchiya K, Horinouchi Y, Ishizawa K, Ikeda Y, Kihira Y, Shono M, Kawazoe K, Tomita S, Tamaki T. Effect of angiotensin II on iron-transporting protein expression and subsequent intracellular labile iron concentration in human glomerular endothelial cells. Hypertens Res. 2010;33(7):713–721. doi: 10.1038/hr.2010.63. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Suzuki E, Takeda R, Oba S, Nishimatsu H, Kimura K, Nagano T, Nagai R, Hirata Y. Angiotensin II and tumor necrosis factor-α synergistically promote monocyte chemoattractant protein-1 expression: roles of NF-κB, p38, and reactive oxygen species. Am J Physiol-Heart Circ Physiol. 2008;294(6):H2879–H2888. doi: 10.1152/ajpheart.91406.2007. [DOI] [PubMed] [Google Scholar]
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touyz RM, Schiffrin EL. Effects of angiotensin II and endothelin-1 on platelet aggregation and cytosolic pH and free Ca2+ concentrations in essential hypertension. Hypertension. 1993;22(6):853–862. doi: 10.1161/01.HYP.22.6.853. [DOI] [PubMed] [Google Scholar]
- Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279(17):17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufet M. T cells calm the storm. Nat Rev Immunol. 2007;7(11):834–835. doi: 10.1038/nri2197. [DOI] [Google Scholar]
- Turner P. XMGRACE, Version 5.1. 19. Cent. Coast. land-margin Res. oregon Grad. Beaverton: Inst. Sci. Technol; 2005. [Google Scholar]
- Uhal BD, Li X, Piasecki CC, Molina-Molina M. Angiotensin signalling in pulmonary fibrosis. Int J Biochem Cell Biol. 2012;44(3):465–468. doi: 10.1016/j.biocel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduganathan M, Vardeny O, Michel T, McMurray JJ, Pfeffer MA, Solomon SD. Renin–angiotensin–aldosterone system inhibitors in patients with Covid-19. N Engl J Med. 2020;382(17):1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajapey R, Rini D, Walston J, Abadir P. The impact of age-related dysregulation of the angiotensin system on mitochondrial redox balance. Front Physiol. 2014;5:439. doi: 10.3389/fphys.2014.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren WF, Daura X, Mark AE. Encyclopedia of computational chemistry, chapter GROMOS force field. New York: John Wiley and Sons; 1998. [Google Scholar]
- Van Lam van T, Ivanova T, Hardes K, Heindl MR, Morty RE, Böttcher-Friebertshäuser E, Lindberg I, Than ME, Dahms SO, Steinmetzer T. Design, synthesis, and characterization of macrocyclic inhibitors of the proprotein convertase furin. ChemMedChem. 2019;14(6):673–685. doi: 10.1002/cmdc.201800807. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen R, Kol A, Andreotti F, Kluft C, Maseri A, Sperti G. Angiotensin II increases plasminogen activator inhibitor type 1 and tissue-type plasminogen activator messenger RNA in cultured rat aortic smooth muscle cells. Circulation. 1994;90(1):362–368. doi: 10.1161/01.CIR.90.1.362. [DOI] [PubMed] [Google Scholar]
- Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The role of interleukin 6 during viral infections. Front Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese N, Demurtas J, Yang L, Tonelli R, Barbagallo M, Lopalco P, Lagolio E, Celotto S, Pizzol D, Zou L. Use of corticosteroids in Coronavirus Disease 2019 pneumonia: a systematic review of the literature. Front Med. 2020;7:170. doi: 10.3389/fmed.2020.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viruses., C. S. G. o. t. I. C. o. T. o. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Version 2. Nature Microbiology. 2020;5(4):8. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng Des Sel. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Canagarajah BJ, Boehm JC, Kassisà S, Cobb MH, Young PR, Abdel-Meguid S, Adams JL, Goldsmith EJ. Structural basis of inhibitor selectivity in MAP kinases. Structure. 1998;6(9):1117–1128. doi: 10.1016/S0969-2126(98)00113-0. [DOI] [PubMed] [Google Scholar]
- Wang C-H, Li S-H, Weisel RD, Fedak PW, Dumont AS, Szmitko P, Li R-K, Mickle DA, Verma S. C-reactive protein upregulates angiotensin type 1 receptors in vascular smooth muscle. Circulation. 2003;107(13):1783–1790. doi: 10.1161/01.CIR.0000061916.95736.E5. [DOI] [PubMed] [Google Scholar]
- Wang F, Huang L, Peng Z-Z, Tang Y-T, Lu M-M, Peng Y, Mei W-J, Wu L, Mo Z-H, Meng J. Losartan inhibits LPS+ ATP-induced IL-1beta secretion from mouse primary macrophages by suppressing NALP3 inflammasome. Die Pharm Int J Pharm Sci. 2014;69(9):680–684. [PubMed] [Google Scholar]
- Wang C, Qian X, Sun X, Chang Q. Angiotensin II increases matrix metalloproteinase 2 expression in human aortic smooth muscle cells via AT1R and ERK1/2. Exp Biol Med. 2015;240(12):1564–1571. doi: 10.1177/1535370215576312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen L, Chen B, Meliton A, Liu SQ, Shi Y, Liu T, Deb DK, Solway J, Li YC. Chronic activation of the renin-angiotensin system induces lung fibrosis. Sci Rep. 2015;5:15561. doi: 10.1038/srep15561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shi K, Li J, Chen T, Guo Y, Liu Y, Yang Y, Yang S. Effects of angiotensin II intervention on MMP-2, MMP-9, TIMP-1, and collagen expression in rats with pulmonary hypertension. Genet Mol Res. 2015;14(1):1707–1717. doi: 10.4238/2015.March.6.17. [DOI] [PubMed] [Google Scholar]
- Wang D, Chai X-Q, Magnussen CG, Zosky GR, Shu S-H, Wei X, Hu S-S. Renin-angiotensin-system, a potential pharmacological candidate, in acute respiratory distress syndrome during mechanical ventilation. Pulm Pharmacol Ther. 2019;58:101833. doi: 10.1016/j.pupt.2019.101833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Barker TA, Berk BC. Angiotensin II and the endothelium: diverse signals and effects. Hypertension. 2005;45(2):163–169. doi: 10.1161/01.HYP.0000153321.13792.b9. [DOI] [PubMed] [Google Scholar]
- Weidanz J, Jacobson LM, Muehrer RJ, Djamali A, Hullett DA, Sprague J, Chiriva-Internati M, Wittman V, Thekkumkara TJ, Becker BN. AT1R blockade reduces IFN-γ production in lymphocytes in vivo and in vitro. Kidney Int. 2005;67(6):2134–2142. doi: 10.1111/j.1523-1755.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler LM, Skiba MA, McMahon C, Staus DP, Kleinhenz AL, Suomivuori C-M, Latorraca NR, Dror RO, Lefkowitz RJ, Kruse AC. Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR. Science. 2020;367(6480):888–892. doi: 10.1126/science.aay9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AM, Wolf D, Rumpold H, Ludwiczek S, Enrich B, Gastl G, Weiss G, Tilg H. The kinase inhibitor imatinib mesylate inhibits TNF-α production in vitro and prevents TNF-dependent acute hepatic inflammation. Proc Natl Acad Sci. 2005;102(38):13622–13627. doi: 10.1073/pnas.0501758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hirayama T, Hasegawa Y. Antiplatelet effect of losartan and telmisartan in patients with ischemic stroke. J Stroke Cerebrovasc Dis. 2007;16(5):225–231. doi: 10.1016/j.jstrokecerebrovasdis.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Yancey PG, Zuo Y, Ma L-J, Kaseda R, Fogo AB, Ichikawa I, Linton MF, Fazio S, Kon V. Macrophage polarization by angiotensin II-type 1 receptor aggravates renal injury-acceleration of atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31(12):2856–2864. doi: 10.1161/ATVBAHA.111.237198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome-associated coronavirus infection. PLoS One. 2010;5(1):e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Baker A. Recombinant human ACE2: acing out angiotensin II in ARDS therapy. Crit Care. 2017;21:305. doi: 10.1186/s13054-017-1882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Pan Y, Fanelli V, Wu S, Luo AA, Islam D, Han B, Mao P, Ghazarian M, Zeng W. Mechanical stress and the induction of lung fibrosis via the midkine signaling pathway. Am J Respir Crit Care Med. 2015;192(3):315–323. doi: 10.1164/rccm.201412-2326OC. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Zhou D, Zhang L-S, Deng F-X, Shu S, Wang L-J, Wu Y, Guo N, Zhou J. Angiotensin II deteriorates advanced atherosclerosis by promoting MerTK cleavage and impairing efferocytosis through the AT1R/ROS/p38 MAPK/ADAM17 pathway. Am J Physiol-Cell Physiol. 2019;317(4):C776–C787. doi: 10.1152/ajpcell.00145.2019. [DOI] [PubMed] [Google Scholar]
- Zhao J, Liu J, Pang X, Wang S, Wu D, Zhang X, Feng L. Angiotensin II induces C-reactive protein expression via AT1-ROS-MAPK-NF-κB signal pathway in hepatocytes. Cell Physiol Biochem. 2013;32(3):569–580. doi: 10.1159/000354461. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Sun L, Jiang T, Zhang D, He D, Nie H. TNFα promotes Th17 cell differentiation through IL-6 and IL-1β produced by monocytes in rheumatoid arthritis. J Immunol Res. 2014;2014:385352. doi: 10.1155/2014/385352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.