Abstract
Plant nutrition is a crucial factor that is usually underestimated when designing plant in vitro culture protocols of unexploited plants. As a complex multifactorial process, the study of nutritional imbalances requires the use of time-consuming experimental designs and appropriate statistical and multiple regression analysis for the determination of critical parameters, whose results may be difficult to interpret when the number of variables is large. The use of machine learning (ML) supposes a cutting-edge approach to investigate multifactorial processes, with the aim of detecting non-linear relationships and critical factors affecting a determined response and their concealed interactions. Thus, in this work we applied artificial neural networks coupled to fuzzy logic, known as neurofuzzy logic, to determine the critical factors affecting the mineral nutrition of medicinal plants belonging to Bryophyllum subgenus cultured in vitro. The application of neurofuzzy logic algorithms facilitate the interpretation of the results, as the technology is able to generate useful and understandable “IF-THEN” rules, that provide information about the factor(s) involved in a certain response. In this sense, ammonium, sulfate, molybdenum, copper and sodium were the most important nutrients that explain the variation in the in vitro culture establishment of the medicinal plants in a species-dependent manner. Thus, our results indicate that Bryophyllum spp. display a fine-tuning regulation of mineral nutrition, that was reported for the first time under in vitro conditions. Overall, neurofuzzy model was able to predict and identify masked interactions among such factors, providing a source of knowledge (helpful information) from the experimental data (non-informative per se), in order to make the exploitation and valorization of medicinal plants with high phytochemical potential easier.
Keywords: Kalanchoe, ANNs, media mineral nutrition, plant in vitro culture, artificial intelligence algorithms, structural risk minimization (SRM), ASMOD
Introduction
Recent biotechnological reports highlighted that medicinal plants constitute the source for more than the 25% of drugs officially approved by the Food and Drug Administration (FDA; Marchev et al., 2020). Furthermore, medicinal plant-derived products are effectively used in the primary healthcare systems for around the 90% of developing countries (El Sheikha, 2017). Taking this into account, the exploitation of medicinal plants has emerged as one of the major challenges in the biotechnological and pharmacological industries for this decade. Additionally, in response to current global demands, increasing efforts are being made to satisfy the future expectations of plant-derived food and drug production worldwide, thus requiring surface maximization for agricultural and nutritional purposes (Pastor et al., 2019). Consequently, novel approaches must be undertaken in the field of medicinal plant research with the aim of meeting the requirements for the large-scale exploitation of medicinal plants. In this sense, plant tissue culture (PTC) constitutes an efficient pool of methodologies becoming a sustainable platform to achieve true-to-type products with added-value properties (Eibl et al., 2018; Chandran et al., 2020) and requires much less space for achieving the same yields than conventional open field agriculture, due to their ability for scaling-up in bioreactors. This methodology confers an absolute independence of climatic threats, plant pathogens and harsh agriculture management and large storage facilities of plant materials. Nevertheless, PTC should cope with their own difficulties, as it requires the investment for specialized equipment and consumables and the recruitment of trained staff to develop the associated methodologies (Bridgen et al., 2018; Patra et al., 2020).
One of the crucial factors that impact the success of PTC establishment is the optimization of growth conditions and the mineral composition of culture medium (Isah et al., 2018). Although a high number of publications have focused on the study of in vitro growth conditions for many species (Batista et al., 2016; Golkar et al., 2019; Hoang et al., 2019), culture medium composition is a paramount factor usually underestimated during the design of plant in vitro culture protocols (Nezami-Alanagh et al., 2014; García-Pérez et al., 2020a). Under PTC conditions, the ingredients included in the culture medium constitute the only source of nutrients available for plants and subsequent nutritional imbalances may occur discretely affecting culture development (Niedz and Evens, 2007), reflecting substantial physiological symptoms (Nezami-Alanagh et al., 2019). Thus, PTC media formulations contain a wide spectrum of mineral and organic nutrients that interact in a complex, multifactorial, nonlinear and non-deterministic way, without considering the individual susceptibilities and requirements that discrete species present, leading to the existence of nutritional imbalances, causing underlying deleterious effects that may be easily detectable (Nezami-Alanagh et al., 2019; Phillips and Garda, 2019).
As a rule, culture media components contain, as average, 18 different mineral nutrients, some required at high concentrations (macronutrients) such as nitrogen, potassium, calcium, phosphorus, sulfur and magnesium, while others are required at lower concentrations (micronutrients), such as manganese, zinc, boron, molybdenum, copper and iron, among others, being all essential for certain physiological processes (Twaij et al., 2020). Together with mineral nutrients, a source of carbon, normally sucrose, as well as other organic molecules, such as vitamins and amino acids, some plant growth regulators, are supplied to media to ensure a healthy plant growth and development (Saad and Elshahed, 2012). In addition, there are additional factors that show a significant impact on mineral nutrition, such as the genotype, because even closely related species have been shown to present differential behaviors toward certain media ingredients (Gago et al., 2011; Nezami-Alanagh et al., 2014).
Due to the high heterogeneity of ingredients that make part of culture media formulations and other additional factors, such as plant genotype or growth conditions, the study of nutritional requirements applied to unknown medicinal plants leads to the design of complex multivariate experimental designs (Nezami-Alanagh et al., 2017; Teixeira da Silva et al., 2020). In the last decade (Hesami and Jones, 2020; Niazian and Niedbala, 2020), several ML algorithms have been successfully employed as alternative to traditional statistical methods and/or response surface methodology (RSM) to identify factors and interactions on complex, non-linear and non-deterministic process such as PTC (Landin et al., 2009; Gago et al., 2010a,c; Nezami-Alanagh et al., 2017). Therefore, revealing all the information encrypted over the large amount of experimental results derived from this type of multifactorial processes becomes a highly challenging task. In such cases, machine learning (ML) offers a cutting-edge computer-based methodology with the ability of handling very complex multivariate datasets, in which there are unknown patterns between inputs and outputs or large amount of uncategorized or different kind of data relating with complex processes, being able to transform data into useful information and knowledge (Gago et al., 2010c; Ertel, 2017; Bini, 2018; Freiesleben et al., 2020). On this purpose, different ML algorithms such as artificial neural networks (ANNs); deep neural networks (DNNs); convolutional neural networks (CNNs); support vector machines (SVMs) or random forest (RF) has been used in plant biotechnology (Niazian and Niedbala, 2020) and, particularly, in PTC (Gago et al., 2010a). Among all of them, ANNs have been successfully applied with the aim of establishing robust predictive models that contribute to the optimization and characterization of multifactorial processes (Landin and Rowe, 2013; Gago et al., 2014; Arteta et al., 2018; Villarrubia et al., 2018; Nezami-Alanagh et al., 2019). In addition, the combination of ANNs with fuzzy logic, the so-called neurofuzzy logic, confers several advantages in the search of critical factors that impact plant nutrition, by providing “IF-THEN” rules that make result interpretation easier, in other words, understandable for the human brain (Landin et al., 2009; Gago et al., 2010b; Gallego et al., 2011). Successful applications of neurofuzzy logic in the field of PTC for seed germination (Ayuso et al., 2017), the identification of physiological disorders associated to nutritional imbalances (Nezami-Alanagh et al., 2018, 2019), improvement of bioactive compounds accumulation (García-Pérez et al., 2020b) and revealing the role of phytohormones on plant in vitro organogenesis (García-Pérez et al., 2020a) have been already performed successfully.
Bryophyllum (genus Kalanchoe, Crassulaceae family) constitutes a subgenus with more than 25 plant species that have been used in the traditional medicine across both the American and African continents (Stefanowicz-Hajduk et al., 2020). Pharmacognostical and phytochemical analyses have highlighted that phenolic compounds and bufadienolides were the bioactive compounds that develop their therapeutic effects, since Bryophyllum spp. have been largely applied to treat infections and chronic diseases, such as diabetes, cardiovascular diseases and cancer (García-Pérez et al., 2018a). The knowledge derived from the combination of ML and PTC will be highly valuable for considering the biotechnological exploitation of Bryophyllum spp. in order to take advantage of their added-value properties as a potential source of bioactive compounds.
In this work, we applied the ML (ANNs algorithms), to model and provide insight about the critical factors and their interactions that drive mineral nutrition of three medicinal plants from the subgenus Bryophyllum cultured in vitro, by focusing on the content in macronutrients and micronutrients from culture media formulations, with the aim of revealing masked nutritional imbalances and interactions that may occur between nutrients that impact plant growth-related parameters.
Materials and Methods
Plant Material and Culture Conditions
The establishment of in vitro culture was conducted for three different Bryophyllum species, namely: Bryophyllum daigremontianum Raym. - Hamet et Perr. (syn. Kalanchoe daigremontinana, BD), Bryophyllum × houghtonii D.B. Ward (Bryophyllum daigremontianum × tubiflorum, syn. Kalanchoe × houghtonii, BH) and Bryophyllum tubiflorum Harv. (syn. Kalanchoe tubiflora, BT).
Epiphyllous plantlets from these three species were used for the disinfection and transference to in vitro conditions as described in previous works (García-Pérez et al., 2019). After surface disinfection, plantlets were cultured by groups of three in glass culture vessels containing 25 mL of previously autoclaved MS medium (Murashige and Skoog, 1962) supplemented with 3% sucrose and solidified with 0.8% agar at pH = 5.8. Cultures were then transferred to growth chambers and placed randomly in the shelves at 25 ± 1°C under a photoperiod of 16 h light (55 μmol m−2 s−1) and 8 h dark and subcultured every 12 weeks by using newly formed epiphyllous plantlets as the explants for next subculture.
Experimental Design
Spontaneously rooted epiphyllous plantlets from the three Bryophyllum species, proceeding from 12-week-old plants grown on MS medium, were subjected to nutrition experiments. Plantlets were transferred by pairs into 10 glass culture vessels, grown and subcultured under the same conditions stated above, making a total of 20 replicates per treatment.
For nutrition experiment, nine different culture media formulations, based on MS medium were used. Due to the low mineral requirements associated to Crassulaceae plants, as it is the case of Bryophyllum spp. (Phillips and Garda, 2019; García-Pérez et al., 2020b), each media contained proportional reduced contents of both either MS macronutrients (M) or MS micronutrients (μ). Thus, half-concentrations (1/2MSM and 1/2MSμ), quarter-concentrations (1/4MSM and 1/4MSμ), eighth-concentrations (1/8MSM and 1/8MSμ) and macronutrient and micronutrient absence (0MSM, 0MSμ) and, as control, full MS medium was tested (Table 1). EDTA-chelated iron, vitamins and organic molecules were supplied in all media at same concentration than in the original MS formulation. All media were also supplemented with 3% sucrose and solidified with 0.8% agar at pH = 5.8.
Table 1.
Mineral salt composition included in cultured media formulations used in this study.
| Salt |
MS (mg L−1) |
1/2MSM (mg L−1) |
1/4MSM (mg L−1) |
1/8MSM (mg L−1) |
0MSM (mg L−1) |
1/2MSμ (mg L−1) |
1/4MSμ (mg L−1) |
1/8MSμ (mg L−1) |
0MSμ (mg L−1) |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Macronutrients | KNO3
NH4NO3 CaCl2 2H2O MgSO4 7H2O KH2PO4 |
1,900 1650 440 370 170 |
950 825 220 185 85 |
475 412.5 110 92.5 42.5 |
237.5 206.3 55 46.3 21.30 |
0 | 1,900 1650 440 370 170 |
|||
| Micronutrients | MnSO4 4H2O ZnSO4 7H2O H3BO3 KI Na2MoO4 2H2O CuSO4 5H2O CoCl2 6H2O |
22.3 8.6 6.2 0.83 0.25 0.025 0.025 |
11.2 4.3 3.1 0.42 0.13 0.013 0.013 |
5.6 2.2 1.6 0.21 0.063 0.0063 0.0063 |
2.8 1.1 0.78 0.10 0.031 0.0031 0.0031 |
0 | ||||
| Iron source | Na2EDTA FeSO4 7H2O |
37.25 27.85 |
||||||||
In order to observe the nutritional long-term impact on Bryophyllum growth parameters, four subcultures were performed. At the end of each subculture (every 12 weeks) two newly-formed epiphyllous plantlets per vessel were randomly selected, transferred to fresh medium of the same media (treatment) in which were cultured, and grown in the same conditions. In the treatments in which epiphyllous plantlet formation was not observed (such as 0MS), two rooted and newly-formed epiphyllous plantlets from the control treatment (MS) were selected and cultured in that media. In total, 20 replicates (10 vessels with two plantlets) were kept in each subculture per treatment.
Thus, the experimental design included 3 different genotypes (BD, BH and BT) × 9 different culture media formulations (MS control + 4 macronutrient-reduced formulations + 4 micronutrient-reduced formulations) × 4 subcultures, accounting for a total of 108 different treatments with 20 replicates each.
At the end of each subculture (12 weeks) six physiological parameters were determined in the new epiphyllous plantlets: shoot length (SL, expressed as cm), longest root length (RL, expressed as cm), plantlet number (PN), leaf number (LN), aerial parts fresh weight (AFW, expressed in g) and root fresh weight (RFW, expressed in g).
After experimental data collection from nutrition experiment, all data were merged into one large multifactorial database including 108 treatments following a factorial design for 18 factors (Supplementary Table 1). Salts included in culture media were split into their containing ions with the aim of avoiding ion confounding (Niedz and Evens, 2006). As a result, the eighteen factors were selected as the inputs (genotype, subculture number and 16 ions) plus the six physiological parameters as outputs (SL, RL, PN, LN, AFW, and RFW) for building the model. In all cases, results were expressed as the mean ± standard error (Supplementary Table 1).
Statistical Analysis
Initially, data derived from the nutrition experiments (SL, RL, PN, LN, AFW, and RFW) were analyzed statistically in order to evaluate the significance of each factor and their interactions (significance level: α = 0.01) on the parameters studied. To that end, factorial ANOVA was performed to elucidate the effect of genotype, subculture and culture media and their interactions, followed by Tukey's HSD post hoc test (α = 0.01). Data normality and homoscedasticity was assessed by Kolmogorov-Smirnov's and Levene's tests, respectively. Count data (PN and LN) parameters should be analyzed by Poisson Regression, but as number of replicates was large (n = 20) and, thus, ANOVA had the same inference than Poisson Regression (α = 0.01; Mize et al., 1999). ANOVA was also applied to those parameters as in previous works (Ayuso et al., 2019). In both cases, the software used was STATISTICA v.12 (StatSoft Inc., 2014, Street Tulsa, OK, USA).
Modeling Tools
Data modeling was performed by using the commercial FormRules® v.4.03 software (Intelligensys LTD, UK) as described elsewhere (Nezami-Alanagh et al., 2018; García-Pérez et al., 2020a). Briefly, FormRules® performed the adaptive-spline-modeling-of-data (ASMOD) algorithm to minimize the number of relevant inputs and to reduce the model complexity and facilitating accuracy with fewer parameters (Shao et al., 2006). Several statistical fitness criteria including cross validation (CV), leave one out cross validation (LOOCV), minimum description length (MDL), Bayesian information criterion (BIC) and structural risk minimization (SRM) were investigated to obtain the model that gave the best Train Set R2. Two of these, CV and LOOCV, split the data into subsets that are either used for training and testing (validation method), while the others (MDL, BIC and SRM) are statistical significance methods, which use all the data for training. These are designed to avoid overtraining, minimizing a criterion that contains two terms: (i) the number of parameters in the model (the variance) and (ii) the prediction errors computed on the data set (the bias). The best results were found for SRM, which ensured the highest predictability with the minimum generalization error and provided the generation of the simplest and more intelligible rules (Vapnik, 1992).
The training process was conducted as described in detail elsewhere (Shao et al., 2006) and training parameters are summarized in Table 2. The quality of submodels (predictability and accuracy), independently generated for every output, were assessed according to the ANOVA f-ratio, mean square error (MSE; Equation 1) and the coefficient of determination (Train Set R2; Equation 2):
Table 2.
Training parameters for the construction of neurofuzzy model used by FormRules®.
| Minimization Parameters (ASMOD) |
|---|
| Ridge regression factor: 1 × 10−6 |
| MODEL SELECTION CRITERIA |
| Structural risk minimization (SRM) |
| C1 = 0.95, C2 = 4.8 |
| Number of set densities: 2 |
| Set densities: 2 |
| Adapt nodes: TRUE |
| Maximum inputs per submodel: 3 |
| Maximum nodes per input: 15 |
| (1) |
| (2) |
Where yi represents the experimental value from the data set, represents the predicted value by the model, and represents the mean of the dependent variable. MSE are calculated to provide information about the random error component of the built model, thus indicating the usefulness of model fitting for prediction due to a smaller incidence of random error (Hesami and Jones, 2020). Models with high Train Set R2 (>70%) and an f-ratio (>4) assess model accuracy and no statistical differences among predicted and experimental values. Models with higher values than 99.9% should be rejected due to model over-fitting (Colbourn and Rowe, 2005; Landin et al., 2009).
The results provided by the application of neurofuzzy logic were expressed as “IF-THEN” rules, thus making their interpretation easier, and they were given a range level (from low to high), combined with a corresponding membership degree value, that ranges between 0 and 1 (Gallego et al., 2011). Supplementary Figure 1 is attached to facilitate the understanding of the linguistic expressions of the variables obtained by the neurofuzzy logic model (Low, Medium and High).
Results
Traditionally plant tissue researchers used statistical method such as factorial ANOVA to analyze data and test if two or more treatment differ significantly from each other due to the effect of some independent variables (factors), but also serves to infer cause-effect relationships. Here, data analysis using factorial ANOVA reflected that all factors studied (genotype, subculture number and culture media) and all interactions among them, caused a significance effect on all parameters studied, except for the interaction genotype × subculture on RL, PN, AFW and RFW parameters (α = 0.01; Supplementary Table 2). The combined effect of the interactions between the number of subcultures and the culture medium is graphically exemplified for SL and RL in Figure 1, depending on each genotype studied: BD, BH and BT, respectively.
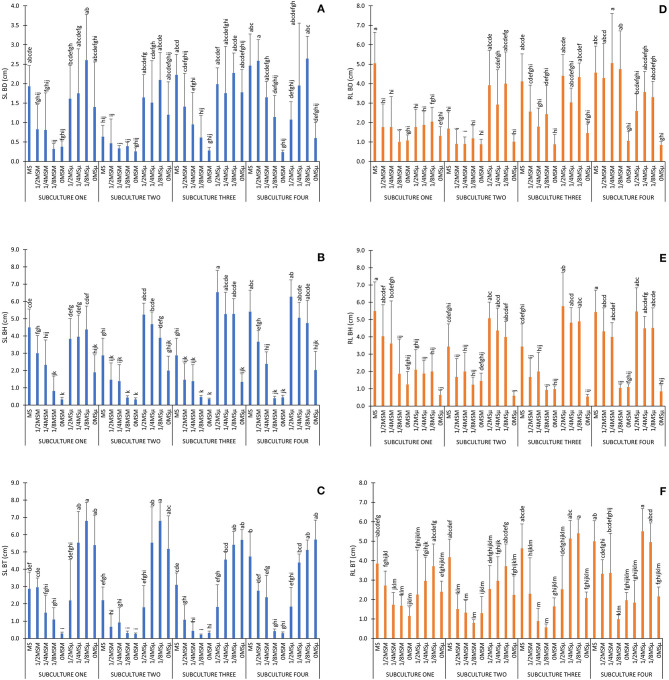
Figure 1.
Experimental data obtained for SL and RL. Values are expressed as the mean and vertical bars indicate standard deviation. Different letters indicate significant differences (α = 0.01). (A) SL BD (cm); (B) SL BH (cm); (C) SL BT (cm); (D) RL BD (cm); (E) RL BH (cm); (F) RL BT (cm).
In general, MS full strength media promoted high SL and RL in all genotypes, although the final shoot and root length varied significantly depending on the genotype and subculture (Figure 1). Those treatments with reduced micronutrients (1/2MSμ, 1/4MSμ, and 1/8MSμ) promoted more length than those with reduced macronutrients (1/2MSM, 1/4MSM, and 1/8MSM), and the treatments with absence of minerals in the media, particularly without macronutrients (0MSM), promoted the worst results overall (Figure 1). The same effect, can be observed for the rest of parameters (see Supplementary Table 1), particularly for PN, which measures the organogenesis capacity, since the absence of mineral nutrients causes the total inhibition of the generation of new epiphyllous plantlets in the leaf margins of all species (Supplementary Table 1: treatments 9, 18, 27, 36 for BD; 45, 54, 63, 72 for BH, and 81, 90, 99, 108 for BT). All together, these ANOVA results showed that MS media composition can be modified dramatically by reducing the amount of macro and micronutrients and obtain exactly the same results. But little information was obtained about the effect of each mineral nutrient or its role on the effect observed.
Data modeling by neurofuzzy logic emerges as a solution to provide useful knowledge on Bryophyllum mineral nutrition after training and learning from the experimental data. The model showed high predictability with Train Set R2 values higher than 70%, f-ratio >4 and low values of MSE in all submodels (Table 3). In addition, model accuracy was assessed by ANOVA f-critical, which proved that predicted values from the model did not show statistically significant differences with respect to the experimental values for any of the outputs (α = 0.05; Table 3).
Table 3.
Quality parameters and critical factors detected by neurofuzzy logic model.
| Output | Submodel | Significant inputs | MSE | Train Set R2 | f ratio | df1, df2 | f critical (α = 0.05) |
|---|---|---|---|---|---|---|---|
| SL | 1 | 0.92 | 74.97 | 42.79 | 7, 100 | 2.10 | |
| 2 | Genotype × Cu2+ | ||||||
| RL | 1 | × × Genotype | 0.74 | 76.97 | 10.91 | 25, 82 | 1.64 |
| 2 | Subculture × Na+ | ||||||
| PN | 1 | × × Genotype | 60.60 | 72.85 | 8.76 | 25, 82 | 1.64 |
| 2 | Subculture × Na+ | ||||||
| LN | 1 | Genotype × | 9.00 | 72.90 | 45.29 | 6, 101 | 2.19 |
| AFW | 1 | × × Genotype | 0.04 | 89.89 | 43.66 | 18, 89 | 1.72 |
| RFW | 1 | Genotype × Cu2+ × | 0.001 | 79.69 | 25.75 | 14, 93 | 1.80 |
| 2 |
Bold inputs indicate the strongest effect associated to each output.
For SL, the model generated two submodels being the interaction between genotype and copper the one with the highest contribution (Table 3). As previously stated, neurofuzzy model had the ability of unraveling masked interactions between different factors, once the salt media composition of each treatment was formulated as their ion composition, to avoid ion confounding. An additional submodel showed that concentration also caused a significant effect but with lower contributions to SL prediction, than the interaction of genotype × Cu2+ (Table 3). In the case of RL, two submodels were found, being the triple interaction between the genotype, and the one presenting the major contribution to RL. Additionally, a second submodel for RL included the interaction of number of subcultures with sodium (Table 3). Exactly the same submodels were also predicted for PN and, interestingly, this finding suggests that sulfur and molybdenum play a crucial role not only on Bryophyllum nutrition, but on controlling its asexual reproduction. The second submodel spotted the interaction of number of subcultures with sodium for PN, too. Concerning LN only one model was generated, being predicted by the interaction between the genotype and concentration (Table 3), what indicates that this output is closely related and highly influenced by this nitrogen-containing macronutrient ion. In the case of AFW, the interaction between genotype, and was the only factor spotted as the most significant affecting this output, in the same way than RL and PN. Finally, RFW was predicted by two submodels, showing the interaction between the genotype, Cu2+ and the major contribution (Table 3), being the only output that was dependent on copper besides SL. An additional submodel for RFW was predicted by molybdate concentration.
In general, the application of neurofuzzy logic identified all the significant factors on all the outputs related to Bryophyllum in vitro growth and their concealed interactions. Nevertheless, the information conferred by this machine learning-based tool was useful, thanks to the establishment of “IF-THEN” rules, which described how these inputs influenced their corresponding outputs. The full set of rules can be found in Supplementary Table 3, while the rules including the highest membership degrees for each output were summarized in Table 4. In order to make result interpretation easier, all factors were ranged as low, mid and high at the same time, according to their effect on every output and the experimental space tested. The graphical ranging of each ion can be visualized in Supplementary Figure 1.
Table 4.
Summary of “IF-THEN” rules generated by ANN modeling.
| Rules | Genot | Subcult | Na+ | Cu2+ | SL | RL | PN | LN | AFW | RFW | Membership | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I F |
Low | T H E N |
Low | 0.98 | |||||||||||
| 2 | High | High | 0.83 | |||||||||||||
| 5 | BD | Low | Low | 1.00 | ||||||||||||
| 6 | BD | High | Low | 1.00 | ||||||||||||
| 7 | BT | Low | High | 0.77 | ||||||||||||
| 8 | BT | High | Low | 1.00 | ||||||||||||
| 9 | I F |
BH | Low | Low | T H E N |
Low | 1.00 | |||||||||
| 10 | BD | Low | Low | Low | 1.00 | |||||||||||
| 11 | BT | Low | Low | Low | 1.00 | |||||||||||
| 12 | BH | Low | Mid | High | 1.00 | |||||||||||
| 13 | BD | Low | Mid | High | 1.00 | |||||||||||
| 14 | BT | Low | Mid | High | 1.00 | |||||||||||
| 15 | BH | Low | High | Low | 1.00 | |||||||||||
| 16 | BD | Low | High | Low | 1.00 | |||||||||||
| 17 | BT | Low | High | Low | 1.00 | |||||||||||
| 18 | BH | High | Low | High | 1.00 | |||||||||||
| 19 | BD | High | Low | High | 1.00 | |||||||||||
| 20 | BT | High | Low | High | 1.00 | |||||||||||
| 21 | BH | High | Mid | High | 1.00 | |||||||||||
| 22 | BD | High | Mid | High | 1.00 | |||||||||||
| 23 | BT | High | Mid | Low | 1.00 | |||||||||||
| 24 | BH | High | High | Low | 1.00 | |||||||||||
| 25 | BD | High | High | Low | 1.00 | |||||||||||
| 26 | BT | High | High | Low | 1.00 | |||||||||||
| 27 | ONE | Low | Low | 1.00 | ||||||||||||
| 28 | ONE | High | High | 1.00 | ||||||||||||
| 29 | TWO | Low | Low | 1.00 | ||||||||||||
| 30 | TWO | High | High | 1.00 | ||||||||||||
| 31 | THREE | Low | Low | 1.00 | ||||||||||||
| 32 | THREE | High | High | 1.00 | ||||||||||||
| 33 | FOUR | Low | Low | 1.00 | ||||||||||||
| 34 | FOUR | High | High | 1.00 | ||||||||||||
| 35 | I F |
BH | Low | Low | T H E N |
Low | 1.00 | |||||||||
| 36 | BD | Low | Low | Low | 1.00 | |||||||||||
| 37 | BT | Low | Low | Low | 1.00 | |||||||||||
| 38 | BH | Low | Mid | Low | 1.00 | |||||||||||
| 39 | BD | Low | Mid | High | 1.00 | |||||||||||
| 40 | BT | Low | Mid | High | 1.00 | |||||||||||
| 41 | BH | Low | High | Low | 1.00 | |||||||||||
| 42 | BD | Low | High | Low | 1.00 | |||||||||||
| 43 | BT | Low | High | Low | 1.00 | |||||||||||
| 44 | BH | High | Low | High | 1.00 | |||||||||||
| 45 | BD | High | Low | High | 1.00 | |||||||||||
| 46 | BT | High | Low | High | 1.00 | |||||||||||
| 47 | BH | High | Mid | High | 1.00 | |||||||||||
| 48 | BD | High | Mid | Low | 1.00 | |||||||||||
| 49 | BT | High | Mid | Low | 1.00 | |||||||||||
| 50 | BH | High | High | Low | 1.00 | |||||||||||
| 51 | BD | High | High | Low | 1.00 | |||||||||||
| 52 | BT | High | High | Low | 1.00 | |||||||||||
| 53 | ONE | Low | Low | 1.00 | ||||||||||||
| 54 | ONE | High | High | 1.00 | ||||||||||||
| 55 | TWO | Low | Low | 1.00 | ||||||||||||
| 56 | TWO | High | High | 1.00 | ||||||||||||
| 57 | THREE | Low | Low | 1.00 | ||||||||||||
| 58 | THREE | High | High | 1.00 | ||||||||||||
| 59 | FOUR | Low | Low | 1.00 | ||||||||||||
| 60 | FOUR | High | High | 1.00 | ||||||||||||
| 63 | I F |
BD | Low | T H E N |
Low | 0.96 | ||||||||||
| 66 | BT | High | High | 0.74 | ||||||||||||
| 67 | I F |
BH | Low | Low | T H E N |
Low | 1.00 | |||||||||
| 68 | BD | Low | Low | Low | 1.00 | |||||||||||
| 69 | BT | Low | Low | Low | 1.00 | |||||||||||
| 70 | BH | Low | Mid | Low | 1.00 | |||||||||||
| 71 | BD | Low | Mid | Low | 1.00 | |||||||||||
| 72 | BT | Low | Mid | High | 1.00 | |||||||||||
| 73 | BH | Low | High | Low | 1.00 | |||||||||||
| 75 | BT | Low | High | Low | 1.00 | |||||||||||
| 76 | BH | High | Low | High | 1.00 | |||||||||||
| 77 | BD | High | Low | High | 1.00 | |||||||||||
| 78 | BT | High | Low | High | 1.00 | |||||||||||
| 79 | BH | High | Mid | High | 1.00 | |||||||||||
| 80 | BD | High | Mid | High | 1.00 | |||||||||||
| 81 | BT | High | Mid | Low | 1.00 | |||||||||||
| 85 | I F |
BH | Low | Low | T H E N |
Low | 1.00 | |||||||||
| 86 | BH | Low | High | High | 1.00 | |||||||||||
| 87 | BH | High | Low | High | 1.00 | |||||||||||
| 88 | BH | High | High | High | 1.00 | |||||||||||
| 89 | BD | Low | Low | Low | 1.00 | |||||||||||
| 90 | BD | Low | High | High | 1.00 | |||||||||||
| 91 | BD | High | Low | High | 1.00 | |||||||||||
| 92 | BD | High | High | High | 1.00 | |||||||||||
| 93 | BT | Low | Low | Low | 1.00 | |||||||||||
| 94 | BT | Low | High | High | 1.00 | |||||||||||
| 95 | BT | High | Low | High | 1.00 | |||||||||||
| 96 | BT | High | High | High | 1.00 | |||||||||||
| 97 | Low | Low | 1.00 | |||||||||||||
| 98 | Mid | Low | 1.00 | |||||||||||||
| 99 | High | Low | 1.00 |
Inputs with the highest membership degree, showing the major contibutions for each response on every output, are indicated in bold. Only the rules with the highest membership degree were selected Genot: genotype; Subcult: number of subcultures. SL and RL were expressed in cm. AFW and RFW were expressed in g.
As previously stated, the observed changes in the output SL was mainly caused by the interaction between genotype and Cu2+ and, besides that, the model generated the corresponding rules. The rules for SL indicate that high values were obtained in the case of BT when cultured under Low Cu2+ concentrations (<0.05 μM) with a membership degree of 0.77 (rule 7; Table 4). In fact, it was the only case that presented a High SL response related to copper. In contrast, the lowest SL value (showing a membership degree of 1.00) was obtained for BD under Low Cu2+ concentrations (rule 5; Table 4). Concerning the rules associated to the other submodels, High SL values were obtained under High concentrations (>10.31 mM) with high membership (0.83, rule 2; Table 4). These results suggest a predominant effect of genotype and copper, over the , on SL.
RL, PN and AFW were predicted mainly by the interaction of three factors: genotype, and . Thus, the response with the highest contribution to High RL values (membership 1.00) is the interaction between Low concentrations (<1.01 mM) with Mid concentrations (0.25–0.75 μM) for BT (rule 14 for RL, rule 40 for PN, and rule 72 for AFW; Table 4). On the contrary, the interaction between Low concentrations (<1.01 mM) with Low concentrations (<0.25 μM) for BH showed the highest contribution to Low RL, PN and AFW values (rules 9, 35, and 67, respectively; Table 4). In addition, RL and PN presented another submodel, based on the interaction between the number of subcultures and sodium (Table 3). In all cases, the model rules showed that High Na+ concentrations (0.22 mM) caused High RL and PN in all subcultures (rules 27–34 for RL, and 53–60 for PN; Table 4).
LN was exclusively predicted by the interaction between the genotype and concentrations. High LN values were predicted only by High concentrations for BT (>10.31 mM, membership degree 0.74, rule 66; Table 4), showing Low values for the rest of cases, specially BD at Low concentrations (membership degree 0.96, rule 63; Table 4). These results suggest a predominant role of genotype, as LN was exclusively favored on BT, only if High concentrations were supplied into the media.
Finally, the major submodel predicting RFW included the interaction between genotype, and Cu2+ concentrations. The High RFW values with the highest membership degree (1.00) were obtained for BH cultured under High concentrations (>0.88 mM) and Low Cu2+ concentrations (<0.05 μM, rule 87; Table 4). Meanwhile, Low values were predicted by Low concentrations of both ions (membership degree 1.00, rule 85; Table 4), independently of the genotype used (rules 85, 89 and 93; Table 4). In addition, the second submodel generated for RFW pointed at molybdate concentrations as the significant output, causing Low RFW values in all cases (rules 97–99; Table 4). These results suggest a predominant role of genotype, favored when High and/or Cu2+ concentrations were included into the media.
Discussion
The combination of artificial neural networks (ANNs) with fuzzy logic, called neurofuzzy logic, constitutes ML algorithms used for predicting and identifying critical factors of multifactorial nonlinear systems (Shihabudheen and Pillai, 2017), as it is the case of plant in vitro nutrition (Gallego et al., 2011). Advantages of ANNs algorithms over traditional statistics have been pointed out previously (Landin et al., 2009; Gago et al., 2010a,c). In this work, the application of neurofuzzy confers a simple and efficient solution about which factors determined the effects found on each Bryophyllum growth parameter, by extracting the knowledge among the deep interactions learnt after data training.
Genotype was a widely distributed factor identified for the prediction of all outputs either alone or in combination with one or two additional factors. This indicates that, although the three species of the Bryophyllum subgenus are considered closely genetically related, each species shows different nutritional requirements, including macronutrients and micronutrients. These differences may probably be due to the transcriptional regulation of uptake systems, such as the primary response to nutrient limitation conditions, since they are highly inducible by environmental conditions (Bird, 2015). Thus, the establishment of in vitro culture results in an effective system to test nutritional imbalances, as it eliminates the influence of side biotic or abiotic factors that impact mineral acquisition, such as pathogen and soil-mediated interactions (Comerford, 2005; Ferrante et al., 2017). These differential patterns for Bryophyllum species have already been related to leaf morphology (Chernetskyy et al., 2018) and other discrepancies in physiological processes, such as the biosynthesis of phenolic compounds (Fürer et al., 2016; Bogucka-Kocka et al., 2018; García-Pérez et al., 2020a) and organogenesis (García-Pérez et al., 2020b). Furthermore, the specific growth responses predicted by the ANN model, denote that Bryophyllum spp. present a tight range of concentrations to achieve an efficient mineral nutrition (Shrivastav et al., 2020).
Another factor, associated to PTC technology and identified by the model as critical, was the number of subcultures. The number of subcultures was identified in combination with sodium to be significant in a secondary submodel for RL and PN. The differential number of subcultures required to achieve certain responses reveals that nutrient deficiencies may be sensed at different periods during the culture time. The delay in responses under nutritional deficiencies, may be explained as a consequence of the induced stress triggered by the increased synthesis of signaling molecules, such as nitric oxide and reactive oxygen species (ROS), mainly driven by macronutrient limitations and micronutrient limitation to a lesser degree (Hajiboland, 2012; Pérez-Pérez et al., 2012; Buet et al., 2019). Its importance relies on the decrease in the rate of epigenetic variation after successive subcultures (Smulders and de Klerk, 2011). This factor becomes crucial to assess the genetical stability of in vitro-cultured plants, making their valorization easier. Moreover, long-term subcultures constitute an efficient strategy to improve interesting phenomena under in vitro conditions, such as rooting (Mendonça et al., 2019; Wang and Yao, 2020), plant regeneration (Konar et al., 2019) and callus induction (Nakasha et al., 2016), very useful for biotechnological production of by-products from medicinal plants.
Among the nutrients, the model has identified as critical factors, the macronutrients and , and the micronutrients Cu2+, Na+, and . The effect of ammonia, the source of nitrogen along with nitrate in most culture media, is clear on the SL parameter. Furthermore, its effect varies depending on the genotype for LN (Table 3). Nitrogen plays a controversial role on crassulacean species, such as Bryophyllum spp. Differential rates of crassulacean acid metabolism (CAM) have been observed as a function of nitrogen source (Pereira et al., 2017). Thus, two groups are distinguished: nitrate-enhanced CAM species and ammonium-enhanced CAM species (Rodrigues et al., 2014). Nevertheless, there is no general rule for this classification, since different Bryophyllum species show particular preferences toward both nitrogen sources (Pereira and Cushman, 2019). Our results suggest that nitrogen source preferences is species-dependent. The effects caused by on CAM activity are mainly negative, due to the inhibition of nocturnal transport rates of organic acids into the vacuole and the cost, in terms of energy, required for ammonium mobilization (Lüttge et al., 2000; Britto et al., 2001). However, it could be noted that such paradigm has been established for soil-grown plants and nitrogen influence may not be the same under in vitro conditions. In this sense, only BT presented high LN values under high concentrations (rule 66), while BD and BH always showed low values, whatever the ammonium supply was within the limits of the study (rules 61–65; Supplementary Table 3). Secondarily, high SL values were obtained under high concentrations (rule 2; Table 4). In addition to being an essential nutrient for plant development, has recently been identified as a signal molecule that triggers both, physiological and morphological responses (Liu and von Wirén, 2017). A recent report has shown that ammonium concentration lower than 15 mM improves the biosynthesis of phenolic compounds in the aerial parts of Bryophyllum spp. cultured in vitro as a consequence of a physiological response induced by abiotic stress (García-Pérez et al., 2020a). Consequently, in order to characterize the impact of ammonium on Bryophyllum spp., further studies at a molecular level are required.
The model reveals a close relationship between sulfur and molybdenum, whose effects on RL, PN and AFW parameters depend on the genotype (Table 3). This close interaction between both nutrients has been already reported in other species, since molybdenum is a crucial component of molybdoenzymes, involved in sulfur metabolism, being both nutrients essential for the development of aerial tissues and roots (Mendel and Hänsch, 2002; Naqib and Jahan, 2017; Blasco et al., 2018; Bouranis et al., 2020). Furthermore, due to the identical chemical configuration of both ions, the uptake of molybdate and sulfate by roots, may take place via sulfur-specific receptors found in root tissues, thus enabling the co-absorption of molybdate with sulfate (Ali et al., 2020). This could be suggested according to the model results, since low RL, PN and AFW values were observed under low concentrations and high concentrations, and conversely, RL, PN and AFW values were high under high concentrations and low concentrations (Table 4). In the case of RL, sulfate plays a positive role, by promoting root biomass accumulation and nutrient uptake during root growth (Alarcón-Poblete et al., 2018), in agreement with the results describe here for RFW (Table 3). These observations indicate that sulfur is essential for in vitro root development on Bryophyllum spp., although molybdate at mid concentrations may assist to its function when sulfate is limited, as demonstrated for BT (rules 14, 40, and 72; Table 4), and reported by other authors (Alhendawi et al., 2005; Shinmachi et al., 2010). Moreover, similar results were obtained for PN.
PN was the only output associated to reproduction of Bryophyllum spp., since this process constitutes the mechanism developed by these species for their asexual reproduction. It combines several processes belonging to both organogenesis and embryogenesis phenomena, that have not been fully elucidated to date (Garcês et al., 2014). Such process takes place at the margin of adult leaves and is considered the major mechanism driving Bryophyllum clonal invasiveness, as it has been reported for BD, BH and BT (Guerra-García et al., 2015). Fully developed plantlets require the formation of their proper root systems before detaching from mother plants to form new functional clones. The development of such process may explain the close relationship between RL and PN according to their same critical factors spotted by the model (Table 3), since rooting should occur at both adult plants and newly-formed plantlets. Additionally, RL and PN presented a secondary submodel, indicating that high sodium concentrations were required for high values of both parameters from the first subculture (Table 4; >0.223 mM), which is in accordance to the sodium requirements previously reported for BT and other Crassulaceae species (George et al., 2008). In this sense, this asexual reproductive mechanism makes Bryophyllum spp. a suitable biological system for the establishment of in vitro culture, thanks to their constitutive plantlet formation. Furthermore, PN was strongly dependent on genotype, and it could be explained because of the mechanisms of plantlet formation: BH and BD develop this process along the whole leaf margins (Garcês and Sinha, 2009; Herrando-Moraira et al., 2020), whereas PN is restricted to the distal leaf end in BT (Guerra-García et al., 2018), thus potentially causing such genotype influence (Figure 2).
Figure 2.
Plantlet formation in Bryophyllum spp. cultured in vitro. (A) Plantlets forming along the leaf margins on BD. (B) Plantlets forming along the leaf margins on BH. (C) Plantlets forming at the distal leaf end on BT.
Copper was revealed as the most influential micronutrient on Bryophyllum spp. cultured in vitro, since it was selected as a critical factor for SL, interacting with genotype, and RFW, interacting with genotype and sulfate concentrations (Table 3). The results from the neurofuzzy logic model indicate that this nutrient affects Bryophyllum physiology in a species-dependent manner, as it was always found in combination with genotype. In this sense, the interaction between genotype and copper was the major factor that influenced SL. In fact, this nutrient played a differential role among the three genotypes. Only BT showed high SL values cultured under Cu2+ concentrations below 0.05 μM (rule 7). This indicates that BT was the genotype most affected negatively by copper, suggesting that toxicity events may occur for this species. These findings reveal that a fine-tuned control of copper homeostasis is required to prevent its toxicity due to a copper excess, since this nutrient is essential for the correct cell function, being part of highly important metalloproteins as a cofactor (Printz et al., 2016; García-Pérez et al., 2018b). Its importance relies on its contribution to basic physiological processes in plants, such as early plant growth, photosynthetic efficiency, mitochondrial respiration and the impairment of oxidative stress (Schulten and Kramer, 2017; Blasco et al., 2018). This wide influence on plant physiology could aid explaining why this nutrient was crucial for SL, associated to aerial part development, SL, and linked to root formation and growth, RFW. Such hypothesis is reinforced by the sophisticated mechanism of copper distribution within plant tissues that contains preventive molecular mechanisms enabling its accumulation by preventing eventual toxic effects at root level (Castro et al., 2018; Migocka and Malas, 2018). In the case of RFW, the coordinate action of sulfate with copper, as described by the ML model (Table 3), may indicate that sulfur contributes to such copper-induced toxic prevention, as it was earlier stated to other metals. In addition, RFW presented a second submodel that included molybdate as a critical factor, that was shown as a negative factor on this parameter, according to the model rules (rules 97–99). This observation can be justified by the effects reported for molybdenum excess, including a severe impairment of photosynthetic efficiency and the inhibition of rooting in other species (Arif et al., 2016). Thus, our results suggest that a minimum copper supplementation (<0.05 μM) may efficiently contribute to in vitro-cultured Bryophyllum plant growth.
In conclusion, our results show that the lack of specific culture media forces the use of universal formulations, as MS medium. Although such formulations contain complete combinations of mineral essential nutrients, may suppose supra-optimal concentrations for the cultivation of many species (Phillips and Garda, 2019), particularly for little-studied medicinal plants with high phytochemical potential. The nutritional imbalances spotted by ML offered a source of knowledge for the prediction of critical factors affecting Bryophyllum spp. plant in vitro culture. Through the ion split approach, neurofuzzy logic model was able to shed light on the masked interactions that take place during the in vitro culture of three different Bryophyllum species, by additionally highlighting the importance of related factors, such as the genotype and the number of subcultures. The essentiality of achieving an enhanced nutritional profiling for the correct development of medicinal plants, as it is the case of Bryophyllum spp., is a paramount feature that should be successfully accomplished in order to get a sustainable exploitation of such species with the aim of reaching large-scale applications in several fields, such as food, cosmetics and nutraceutical industries.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
PG-P and EL-M performed the experiments. PG contributed with reagents and materials. PG-P, ML, and PG contributed with modeling and analysis tools. PG-P and PG conceived the experimental design and wrote the manuscript. All authors contributed in the revision and acceptance version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors declare their sincere acknowledgments to the Spanish Ministry of Education for the FPU grant awarded to PG-P (Grant Number FPU15/04849) and Clinical Research Center ADICAM for kindly providing the plant material used in this work.
Footnotes
Funding. This study was funded by Xunta de Galicia through Red de Uso Sostenible de los Recursos Naturales y Agroalimentarios (REDUSO, Grant Number ED341D 2017/2018) and Cluster of Agricultural Research and Development (CITACA Strategic Partnership, Grant Number ED431E 2018/07).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.576177/full#supplementary-material
References
- Alarcón-Poblete E., Inostroza-Blancheteau C., Alberdi M., Rengel Z., Reyes-Díaz M. (2018). Molecular regulation of aluminum resistance and sulfur nutrition during root growth. Planta 247, 27–39. 10.1007/s00425-017-2805-6 [DOI] [PubMed] [Google Scholar]
- Alhendawi R. A., Kirkby E. A., Pilbeam D. J. (2005). Evidence that sulfur deficiency enhances molybdenum transport in xylem sap of tomato plants. J. Plant Nutr. 28, 1347–1353. 10.1081/PLN-200067449 [DOI] [Google Scholar]
- Ali A., Bhat B. A., Rather G. A., Malla B. A., Ganie S. A. (2020). Proteomic studies of micronutrient deficiency and toxicity, in Plant Micronutrients, eds Aftab T., Hakeem K. R. (Cham: Springer Nature Switzerland AG; ), 257–284. [Google Scholar]
- Arif N., Yadav V., Singh S., Singh S., Ahmad P., Mishra R. K., et al. (2016). Influence of high and low levels of plant-beneficial heavy metal ions on plant growth and development. Front. Environ. Sci. 4:69 10.3389/fenvs.2016.00069 [DOI] [Google Scholar]
- Arteta T. A., Hameg R., Landin M., Gallego P. P., Barreal M. E. (2018). Neural networks models as decision-making tool for in vitro proliferation of hardy kiwi. Eur. J. Horticult. Sci. 83, 259–265. 10.17660/eJHS.2018/83.4.6 [DOI] [Google Scholar]
- Ayuso M., García-Pérez P., Ramil-Rego E., Gallego P. P., Barreal M. E. (2019). In vitro culture of the endangered plant Eryngium viviparum as dual strategy for its ex situ conservation and source of bioactive compounds. Plant Cell Tissue Organ Cult. 138, 427–435. 10.1007/s11240-019-01638-y [DOI] [Google Scholar]
- Ayuso M., Ramil-Rego P., Landin M., Gallego P. P., Barreal M. E. (2017). Computer-assisted recovery of threatened plants: keys for breaking seed dormancy of Eryngium viviparum. Front. Plant Sci. 8:2092. 10.3389/fpls.2017.02092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista D. S., de Castro K. M., da Silva A. R., Teixeira M. L., Sales T. A., Soares L. I., et al. (2016). Light quality affects in vitro growth and essential oil profile in Lippia alba (Verbenaceae). In Vitro Cell. Dev. Biol. Plant 52, 276–282. 10.1007/s11627-016-9761-x [DOI] [Google Scholar]
- Bini S. A. (2018). Artificial intelligence, machine learning, deep learning, and cognitive computing: what do these terms mean and how will they impact health care? J. Anthroplasty 33, 2358–2361. 10.1016/j.arth.2018.02.067 [DOI] [PubMed] [Google Scholar]
- Bird A. J. (2015). Cellular sensing and transport of metal ions: implications in micronutrient homeostasis. J. Nutr. Biochem. 26, 1103–1115. 10.1016/j.jnutbio.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B., Navarro-León E., Ruiz J. M. (2018). Oxidative stress in relation with micronutrient deficiency or toxicity, in Plant Micronutrient Use Efficiency, eds Hossain M. A., Kamiya T., Burritt D.J., Tran L.-S. P., Fujiwara T. (London: Academic Press, Elsevier; ), 181–194. [Google Scholar]
- Bogucka-Kocka A., Zidorn C., Kasprzycka M., Szymczak G., Szewczyk K. (2018). Phenolic acid content, antioxidant and cytotoxic activities of four Kalanchoë species. Saudi J. Biol. Sci. 25, 622–630. 10.1016/j.sjbs.2016.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouranis D. L., Malagoli M., Avice J. C., Bloem E. (2020). Advances in plant sulfur research. Plants 9, 1–6. 10.3390/plants9020256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen M. P., Van Houtven W., Eeckhaut T. (2018). Plant tissue culture techniques for breeding, in Ornamental Crops, ed J. Van Huylenbroeck (Amsterdam: Springer International Publishing AG; ), 127–144. [Google Scholar]
- Britto D. T., Siddiqi M. Y., Glass A. D. M., Kronzucker H. J. (2001). Futile transmembrane NH4+ cycling: a cellular hypothesis to explain ammonium toxicity in plants. Proc. Natl. Acad. Sci. U. S. A. 98, 4255–4258. 10.1073/pnas.061034698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buet A., Galatro A., Ramos-Artuso F., Simontacchi M. (2019). Nitric oxide and plant mineral nutrition: current knowledge. J. Exp. Bot. 70, 4461–4476. 10.1093/jxb/erz129 [DOI] [PubMed] [Google Scholar]
- Castro P. H., Lilay G. H., Assunção A. G. L. (2018). Regulation of micronutrient homeostasis and deficiency response in plants, in Plant Micronutrient Use Efficiency, eds Hossain M.A., Kamiya T., Burritt D.J., Tran L.-S.P., Fujiwara T. (London: Academic Press, Elsevier; ), 1–15. [Google Scholar]
- Chandran H., Meena M., Barupal T., Sharma K. (2020). Plant tissue culture as a perpetual source for production of industrially important bioactive compounds. Biotechnol. Rep. 26:e00450. 10.1016/j.btre.2020.e00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernetskyy M., Wozniak A., Skalska-Kamińska A., Zuraw B., Blicharska E., Rejdak R., et al. (2018). Structure of leaves and phenolic acids in Kalanchoë daigremontinana Raym.-Hamet & H. Perrier. Acta Sci. Polonorum Hortorum Cultus 17, 137–155. 10.24326/asphc.2018.4.13 [DOI] [Google Scholar]
- Colbourn E., Rowe R. C. (2005). Neural computer and pharmaceutical formulation, in Encyclopedia of Pharmaceutical Technology, eds Swarbrick J., Boylan J. C. (New York, NY: Marcel Dekker; ). [Google Scholar]
- Comerford N. B. (2005). Soil factors affecting nutrient bioavailability, in Nutrient Acquisition by Plants: An Ecological Perspective, ed H. Bassirirad (Berlin: Springer Verlag Berlin Heidelberg; ), 1–14. [Google Scholar]
- Eibl R., Meier P., Stutz I., Schildberger D., Hühn T., Eibl D. (2018). Plant cell culture technology in the cosmetic and food industries: current state and future trends. Appl. Microbiol. Biotechnol. 102, 8661–8675. 10.1007/s00253-018-9279-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sheikha A. F. (2017). Medicinal plants: ethno-uses to biotechnology era, in Biotechnology and Production of Anti-Cancer Compounds, ed S. Malik (Cham: Springer International Publishing AG; ), 1–38. [Google Scholar]
- Ertel W. (2017). What is artificial intelligence?” in Introduction to Artificial Intelligence, ed I. Mackie (Cham: Springer International Publishing AG; ), 1–20. [Google Scholar]
- Ferrante A., Nocito F. F., Morgutti S., Sacchi G. A. (2017). Plant breeding for improving nutrient uptake and utilization efficiency, in Advances in Research on Fertilization Management of Vegetable Crops, eds Tei F., Nicola S., Benincasa P. (Cham: Springer International Publishing AG; ), 221–246. [Google Scholar]
- Freiesleben J., Keim J., Grutsch M. (2020). Machine learning and design of experiments: alternative approaches or complementary methodology for quality improvement? Qual. Reliabil. Eng. Int. 36, 1837–1848. 10.1002/qre.2579 [DOI] [Google Scholar]
- Fürer K., Simões-Wüst A. P., von Mandach U., Hamburger M., Potterat O. (2016). Bryophyllum pinnatum and related species used in anthroposophic medicine: constituents, pharmacological activities, and clinical efficacy. Planta Med. 82, 930–941. 10.1055/s-0042-106727 [DOI] [PubMed] [Google Scholar]
- Gago J., Landin M., Gallego P. P. (2010a). Strengths of artificial neural networks in modeling complex plant processes. Plant Signal. Behav. 5, 743–745. 10.4161/psb.5.6.11702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago J., Landin M., Gallego P. P. (2010b). A neurofuzzy logic approach for modeling plant processes: A practical case of in vitro direct rooting and acclimatization of Vitis vinifera L. Plant Sci. 179, 241–249. 10.1016/j.plantsci.2010.05.009 [DOI] [Google Scholar]
- Gago J., Martínez-Núñez L., Landin M., Flexas J., Gallego P. P. (2014). Modelling the effects of light and sucrose on in vitro propagated plants: a multiscale system analysis using artificial intelligence technology. PLoS ONE 9:e85989. 10.1371/journal.pone.0085989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago J., Martínez-Núñez L., Landin M., Gallego P. P. (2010c). Artificial neural networks as an alternative to the traditional statistical methodology in plant research. J. Plant Physiol. 167, 23–27. 10.1016/j.jplph.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Gago J., Pérez-Tornero O., Landin M., Burgos L., Gallego P. P. (2011). Improving knowledge of plant tissue culture and media formulation by neurofuzzy logic: a practical case of data mining using apricot databases. J. Plant Physiol. 168, 1858–1865. 10.1016/j.jplph.2011.04.008 [DOI] [PubMed] [Google Scholar]
- Gallego P. P., Gago J., Landin M. (2011). Artificial neural networks technology to model and predict plant biology process, in Artificial Neural Networks – Methodological Advances and Biomedical Applications, ed K. Suzuki (Rijeka: InTech Open; ), 197–216. [Google Scholar]
- Garcês H. M. P., Koenig D., Townsley B. T., Kim M., Sinha N. R. (2014). Truncation of LEAFY COTYLEDON1 protein is required for asexual reproduction in Kalanchoë daigremontinana. Plant Physiol. 165, 196–206. 10.1104/pp.114.237222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês H. M. P., Sinha N. (2009). The ‘Mother of Thousands’ (Kalanchoë daigremontiana): a plant model for asexual reproduction and CAM studies. Cold Spring Harbor Protoc. 4, 1–9. 10.1101/pdb.emo133 [DOI] [PubMed] [Google Scholar]
- García-Pérez P., Barreal M. E., Rojo-de Dios L., Cameselle-Teijeiro J. F., Gallego P. P. (2018a). Bioactive natural products from the genus Kalanchoe as cancer chemopreventive agents: a review, in Studies in Natural Products Chemistry, ed A. ur-Rahman (Amsterdam: Elsevier; ), 49–84. [Google Scholar]
- García-Pérez P., Losada-Barreiro S., Gallego P. P., Bravo-Díaz C. (2019). Adsorption of gallic acid, propyl gallate and polyphenols from Bryophyllum extracts on activated carbon. Sci. Rep. 9:14830. 10.1038/s41598-019-51322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pérez P., Lozano-Milo E., Gallego P. P., Tojo C., Losada-Barreiro S., Bravo-Díaz C. (2018b). Plant antioxidants in food emulsions, in Some New Aspects of Colloidal Systems in Foods, ed J. Milani (Rijeka: InTech Open; ), 11–29. [Google Scholar]
- García-Pérez P., Lozano-Milo E., Landin M., Gallego P. P. (2020a). Machine learning technology reveals the concealed interactions of phytohormones on medicinal plant in vitro organogenesis. Biomolecules 10:746. 10.3390/biom10050746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pérez P., Lozano-Milo E., Landin M., Gallego P. P. (2020b). Combining medicinal plant in vitro culture with machine learning technologies for maximizing the production of phenolic compounds. Antioxidants 9:210. 10.3390/antiox9030210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E. F., Hall M. A., De Klerk G.-J. (2008). The components of plant tissue culture media I: macro- and micro-nutrients, in Plant Propagation by Tissue Culture, eds George E. F., Hall M. A., De Klerk G.-J. (Dordrecht: Springer; ), 65–114. [Google Scholar]
- Golkar P., Hadian F., Dehkordi M. K. (2019). Production of a new mucilage compound in Lepidium sativum callus by optimizing in vitro growth conditions. Nat. Prod. Res. 33, 130–135. 10.1080/14786419.2018.1437426 [DOI] [PubMed] [Google Scholar]
- Guerra-García A., Barrales-Alcal,á D., Argueta-Guzmán M., Cruz A., Mandujano M. C., Arévalo-Ramírez J. A., et al. (2018). Biomass allocation, plantlet survival, and chemical control of the invasive chandelier plant (Kalanchoe delagoensis) (Crassulaceae). Invas. Plant Sci. Manag. 11, 33–39. 10.1017/inp.2018.6 [DOI] [Google Scholar]
- Guerra-García A., Golubov J., Mandujano M. C. (2015). Invasion of Kalanchoe by clonal spread. Biol. Invas. 17, 1615–1622. 10.1007/s10530-014-0820-0 [DOI] [Google Scholar]
- Hajiboland R. (2012). Effect of micronutrient deficiencies on plants stress responses, in Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability, eds Ahmad P., Prasad M. N. V. (New York, NY: Springer; ), 283–329. [Google Scholar]
- Herrando-Moraira S., Vitales D., Nualart N., Gómez-Bellver C., Ibáñez N., Mass,ó S., et al. (2020). Global distribution patterns and niche modelling of the invasive Kalanchoe × houghtonii (Crassulaceae). Sci. Rep. 10:3143. 10.1038/s41598-020-60079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesami M., Jones A. M. P. (2020). Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 104, 9449–9485. 10.1007/s00253-020-10888-2 [DOI] [PubMed] [Google Scholar]
- Hoang N. N., Kitaya Y., Shibuya T., Endo R. (2019). Effects of suppoting materials in in vitro acclimatization stage on ex vitro growth of wasabi plants. Sci. Horticult. 261:109042 10.1016/j.scienta.2019.109042 [DOI] [Google Scholar]
- Isah T., Umar S., Mujib A., Sharma M. P., Rajasekharan P. E., Zafar N., et al. (2018). Secondary metabolism of pharmaceuticals in the plant in vitro cultures: strategies, approaches, and limitations to achieving higher yield. Plant Cell Tissue Organ Cult. 132, 239–265. 10.1007/s11240-017-1332-2 [DOI] [Google Scholar]
- Konar S., Adhikari S., Karmakar J., Ray A., Bandyopadhyay T. K. (2019). Evaluation of subculture ages on organogenic response from root callus and SPAR based genetic fidelity assessment in the regenerants of Hibiscus sabdariffa L. Industr. Crops Prod. 135, 321–329. 10.1016/j.indcrop.2019.04.018 [DOI] [Google Scholar]
- Landin M., Rowe R. C. (2013). Artificial neural networks technology to model, understand, and optimize drug formulations, in Formulation Tools for Pharmaceutical Development (Cambridge: Woodhead Publishing; ), 7–37. [Google Scholar]
- Landin M., Rowe R. C., York P. (2009). Advantages of neurofuzzy logic against conventional experimental design and statistical analysis in studying and developing direct compression formulations. Eur. J. Pharm. Sci. 38, 325–331. 10.1016/j.ejps.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Liu Y., von Wirén N. (2017). Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 68, 2581–2592. 10.1093/jxb/erx086 [DOI] [PubMed] [Google Scholar]
- Lüttge U., Pfeifer T., Fischer-Schliebs E., Ratajczak R. (2000). The role of vacuolar malate-transport capacity in crassulacean acid metabolism and nitrate nutrition. Higher malate-transport capacity in ice plant after crassulacean acid metabolism-induction and in tobacco under nitrate nutrition. Plant Physiol. 124, 1335–1348. 10.1104/pp.124.3.1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchev A. S., Zhenya P. Y., Georgiev M. I. (2020). Green (cell) factories for advanced production of plant secondary metabolites. Crit. Rev. Biotechnol. 40, 443–458. 10.1080/07388551.2020.1731414 [DOI] [PubMed] [Google Scholar]
- Mendel R. R., Hänsch R. (2002). Molybdoenzymes and molybdenum. J. Exp. Bot. 53, 1689–1698. 10.1093/jxb/erf038 [DOI] [PubMed] [Google Scholar]
- Mendonça E. G., Batista T. R., Stein V. C., Balieiro F. P., de Abreu J. R., Pires M. F., et al. (2019). In vitro serial subculture to improve rooting of Eucalyptus urophylla. New Forests 51, 801–816. 10.1007/s11056-019-09761-6 [DOI] [Google Scholar]
- Migocka M., Malas K. (2018). Plant responses to copper: molecular and regulatory mechanisms of copper uptake, distribution and accumulation in plants, in Plant Micronutrient Use Efficiency, eds Hossain M. A., Kamiya T., Burritt D. J., Tran L.-S. P., Fujiwara T. (London: Academic Press, Elsevier; ), 71–86. [Google Scholar]
- Mize C. W., Koehler K. J., Compton M. E. (1999). Statistical considerations for in vitro research: II – Data to presentation. In Vitro Cell. Dev. Biol. Plant 35, 122–126. 10.1007/s11627-999-0021-1 [DOI] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Nakasha J. J., Sinniah U. R., Kemat N., Mallappa K. S. (2016). Induction, subculture cycle, and regeneration of callus in safed musli (Chlorophytum borivilianum) using different types of phytohormones. Pharmacogn. Mag. 12, S460–S464. 10.4103/0973-1296.191457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqib S. A., Jahan M. S. (2017). The function of molybdenum and boron on the plants. J. Agric. Res. 2, 000136. [Google Scholar]
- Nezami-Alanagh E., Garoosi G.-A., Landin M., Gallego P. P. (2019). Computer-based tools provide new insight into the key factors that cause physiological disorders of pistachio rootstocks cultured in vitro. Sci. Rep. 9:9740. 10.1038/s41598-019-46155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami-Alanagh E., Garoosi G. A., Haddad R., Maleki S., Landin M., Gallego P. P. (2014). Design of tissue culture media for efficient Prunus rootstock micropropagation using artificial intelligence models. Plant Cell Tissue Organ Cult. 117, 349–359. 10.1007/s11240-014-0444-1 [DOI] [Google Scholar]
- Nezami-Alanagh E., Garoosi G. A., Landin M., Gallego P. P. (2018). Combining DOE with neurofuzzy logic for healthy mineral nutrition of pistachio rootstocks in vitro culture. Front. Plant Sci. 9:1474. 10.3389/fpls.2018.01474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami-Alanagh E., Garoosi G. A., Maleki S., Landin M., Gallego P. P. (2017). Predicting optimal in vitro culture medium for Pistacia vera micropropagation using neural networks models. Plant Cell Tissue Organ Cult. 129, 19–33. 10.1007/s11240-016-1152-9 [DOI] [Google Scholar]
- Niazian M., Niedbala G. (2020). Machine learning for plant breeding and biotechnology. Agriculture 10:436 10.3390/agriculture10100436 [DOI] [Google Scholar]
- Niedz R. P., Evens T. J. (2006). A solution to the problem of ion confounding in experimental biology. Nat. Methods 3:417. 10.1038/nmeth0606-417 [DOI] [PubMed] [Google Scholar]
- Niedz R. P., Evens T. J. (2007). Regulating plant tissue growth by mineral nutrition. In Vitro Cell. Dev. Biol. Plant 43, 370–381. 10.1007/s11627-007-9062-5 [DOI] [Google Scholar]
- Pastor A. V., Palazzo A., Havlik P., Blemans H., Wada Y., Obersteiner M., et al. (2019). The global nexus of food-trade-water sustaining environmental flows by 2050. Nat. Sustain. 2, 499–507. 10.1038/s41893-019-0287-1 [DOI] [Google Scholar]
- Patra J. K., Das G., Das S. K., Thatoi H. (2020). Plant tissue culture techniques and nutrient analysis, in A Practical Guide to Environmental Biotechnology, eds Patra J. K., Das G., Das S. K., Thatoi H. (Singapore: Springer Nature Singapore Pte Ltd.), 135–164. [Google Scholar]
- Pereira P. N., Cushman J. C. (2019). Exploring the relationship between crassulacean acid metabolism (cam) and mineral nutrition with a special focus on nitrogen. Int. J. Mol. Sci. 20:4363. 10.3390/ijms20184363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P. N., Smith J. A. C., Mercier H. (2017). Nitrate enhancement of CAM activity in two Kalanchoë species is associated with increased vacuolar proton transport capacity. Physiol. Plant. 160, 361–372. 10.1111/ppl.12578 [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez M. E., Lemaire S. D., Crespo J. L. (2012). Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 160, 156–164. 10.1104/pp.112.199992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. C., Garda M. (2019). Plant tissue culture media and practices: an overview. In Vitro Cell. Dev. Biol. Plant 55, 242–257. 10.1007/s11627-019-09983-5 [DOI] [Google Scholar]
- Printz B., Lutts S., Hausman J.-F., Sergeant K. (2016). Copper trafficking in plants and its implication on cell wall dynamics. Front. Plant Sci. 7:601. 10.3389/fpls.2016.00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M. A., Freschi L., Pereira P. N., Mercier H. (2014). Interaction between nutrients and crassulacean acid metabolism, in Progress in Botany, eds Lüttge U., Beyschlag W., Cushman J. (London: Springer-Verlag Berlin Heidelberg; ), 167–186. [Google Scholar]
- Saad A. I. M., Elshahed A. M. (2012). Plant tissue culture media, in Recent Advances in Plant in vitro Culture, ed A. Leva (Rijeka: InTech Open; ), 29–40. [Google Scholar]
- Schulten A., Kramer U. (2017). Interaction between copper homeostasis and metabolism in plants, in Progress in Botany, eds Cánovas F., Lüttge U., Matyssek R. (Cham: Springer International Publishing AG; ), 111–146. [Google Scholar]
- Shao Q., Rowe R. C., York P. (2006). Comparison of neurofuzzy logic and neural networks in modelling experimental data of an immediate release tablet formulation. Eur. J. Pharm. Sci. 28, 394–404. 10.1016/j.ejps.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Shihabudheen K. V., Pillai G. N. (2017). Regularized extreme learning adaptive neurofuzzy algorithm for regression and classification. Knowled. Based Syst. 127, 100–113. 10.1016/j.knosys.2017.04.007 [DOI] [Google Scholar]
- Shinmachi F., Buchner P., Stroud J. L., Parmar S., Zhao F.-J., McGrath S. P., et al. (2010). Influence of sulfur deficiency on the expression of specific sulfate transporters and the distribution of sulfur, selenium and molybdenum in wheat. Plant Physiol. 153, 327–336. 10.1104/pp.110.153759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav P., Prasad M., Singh T. B., Yadav A., Goyal D., Ali A., et al. (2020). Role of nutrients in plant growth and development, in Contaminants in Agriculture, eds Naeem M., Ansari A., Gill S. (Cham: Springer International Publishing AG; ), 43–59. [Google Scholar]
- Smulders M. J. M., de Klerk G. J. (2011). Epigenetics in plant tissue culture. Plant Growth Regul. 63, 137–146. 10.1007/s10725-010-9531-4 [DOI] [Google Scholar]
- Stefanowicz-Hajduk J., Asztemborska M., Krauze-Baranowska M., Godlewska S., Gucwa M., Moniuszko-Szajwaj B., et al. (2020). Identification of flavonoids and bufadienolides and cytotoxic effects of Kalanchoe daigremontiana extracts on human cancer cell lines. Planta Med. 86, 239–246. 10.1055/a-1099-9786 [DOI] [PubMed] [Google Scholar]
- Teixeira da Silva J. A., Nezami-Alanagh E., Barreal M. E., Kher M. M., Wicaksono A., Gulyás A., et al. (2020). Shoot tip necrosis of in vitro plant cultures: a reappraisal of possible causes and solutions. Planta 252, 1–35. 10.1007/s00425-020-03449-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twaij B. M., Jazar Z. H., Hasan M. N. (2020). Trends in the use of tissue culture, applications and future aspects. Int. J. Plant Biol. 11 10.4081/pb.2020.8385 [DOI] [Google Scholar]
- Vapnik V. (1992). Principles of risk minimization for learning theory, in Proceedings of the Advances in Neural Information Processing Systems (Denver, CO: ), 831–838. [Google Scholar]
- Villarrubia G., de Paz J. F., Chamoso P., de la Pietra F. (2018). Artificial neural networks used in optimization problems. Neurocomputing 272, 10–16. 10.1016/j.neucom.2017.04.075 [DOI] [Google Scholar]
- Wang Y., Yao R. (2020). Increased endogenous gibberellin levels inhibits root growth of Pinus massoniana Lamb. plantlets during long-term subculture. In Vitro Cell. Dev. Biol. Plant 56, 470–479. 10.1007/s11627-020-10067-y [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.