Summary
Recent studies have begun to highlight the diverse and tumor-specific microbiomes across multiple cancer types. We believe this work raises the important question of whether the classical “Hallmarks of Cancer” should be expanded to include tumor microbiomes. To answer this question, the causal relationships and co-evolution of these microbiotic tumor ecosystems must be better understood. Because host-microbe interactions should be studied in a physiologically relevant context, animal models have been preferred. Yet these models are often poor mimics of human tumors and are difficult to interrogate at high spatiotemporal resolution. We believe that in vitro tissue engineered platforms could provide a powerful alternative approach that combines the high-resolution of in vitro studies with a high degree of physiological relevance. This review will focus on tissue engineered approaches to study host-microbe interactions and to establish their role as an emerging hallmark of cancer with potential as a therapeutic target.
Subject Areas: Microbiome, Bioengineering, Tissue Engineering, Cancer
Graphical Abstract
Microbiome; Bioengineering; Tissue Engineering; Cancer
Introduction
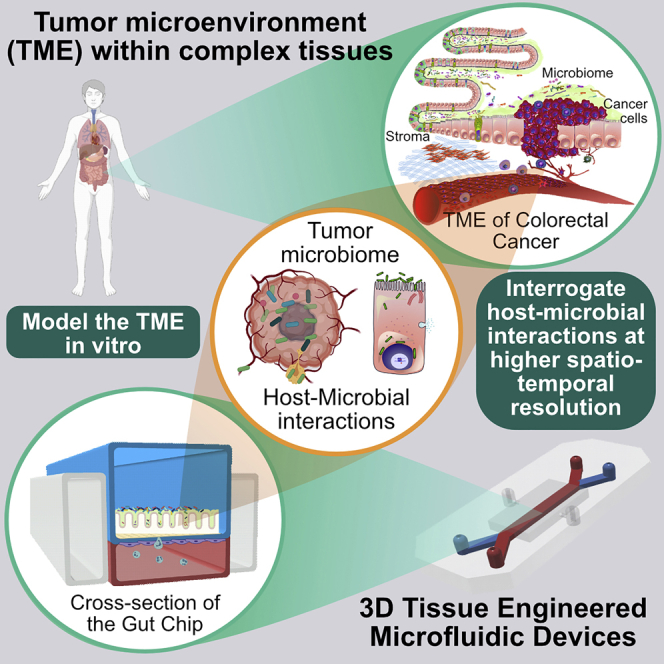
The involvement of tumor-specific bacteria, collectively termed the tumor microbiome, has garnered significant attention as a key potential regulator of the well-established “Hallmarks of Cancer.” These hallmarks include deregulated proliferation, replicative immortality, genomic instability, evasion of growth suppression, avoidance of immune surveillance, chronic inflammation, angiogenic induction, and the activation of metastatic pathways (Hanahan and Weinberg, 2011). Bacteria and their secreted metabolites have been implicated in influencing most, if not all, these host factors (Fulbright et al., 2017). Although many in vitro models have helped elucidate mechanisms related to tumorigenesis, there are only limited models that are amenable to directly investigate host-microbe interactions, and far fewer in the context of cancer. Similarly, although animal models have been an indispensable tool in microbiome studies associated with cancer, they exhibit significant variability in their resident bacterial species and immune profiles when compared with humans. To address these issues, we believe that tissue engineering provides a unique opportunity to bridge the gap between in vitro and animal models in analyzing these host-microbe interactions in the tumor microenvironment (TME) that are so critical for tumor progression and therapy response.
Tissue engineered models developed from human cells not only maintain the genetic constitution of the host but also do so in a physiologically relevant three-dimensional structure that consists of multiple, differentiated cell types functioning in synergism as in native tissue. Furthermore, these platforms are amenable to interrogation at high spatiotemporal resolutions, which is just not possible in larger animal models. For example, one can use these platforms to study the role of individual bacterial interactions with the host to distinguish correlation from causation in microbial impacts on cancer, which are normally obscured by multiple confounding factors within animal models.
Current tissue engineered platforms that have been developed to study host-microbiome interactions are predominantly based on recreating the gut epithelium, which harbors the majority of microorganisms in the human body (Sender et al., 2016). Historically, there have been several challenges limiting the development of these platforms. Each tissue type has its own specific engineering challenges including cellular spatial constraints, physical forces, biochemical cues, and cell growth and differentiation capacities that need to be addressed, and hence, a personalized and experimentally tailored approach is preferred to develop each tissue type. The convergence of technologies from multiple disciplines has enabled the possibility to incorporate advanced sensors and imaging modalities for real-time monitoring of oxygen, pH, barrier permeability, and other biological parameters to recapitulate and analyze host-microbial interactions. In this review, we highlight the latest insights derived from 2D, 3D, and organ-on-chip platforms that have been used to investigate the interactions within the host-microbial consortia and explore the potential for adapting these platforms to advance our understanding of the tumor microbiome.
The Tumor Microbiome
The human microbiome, consisting of trillions of microorganisms co-existing within the human body, has an enormous impact on maintaining health and normal physiology (Brestoff and Artis, 2013; Fan and Pedersen, 2020). A dysbiotic microbiome adversely impacts homeostasis, which leads to a number of unfavorable outcomes including inflammatory diseases, cardiovascular disease, obesity, and diabetes and can even potentiate cancer initiation and progression (Udayasuryan et al., 2019; Xavier et al., 2020). Studies of the microbiome often characterize microbial compositional alterations in disease conditions (Durack and Lynch, 2019). However, the inherent complexity and variability in these experiments makes it challenging to derive meaningful conclusions on the microbes' direct impact on cancer progression. This is further compounded by the fact that the gut microbiome can play a dual role by being both tumor promoting and tumor restricting. A striking recent example within a mouse model revealed that p53-disabling mutations exhibited divergent effects based on the location of the cells within the gut and its spatially segregated microbial composition, behaving oncogenic distally, but tumor-suppressive proximally (Kadosh et al., 2020). That microbiotic residents may regulate the action of such an archetypal tumor suppressor gene as p53 suggests that we are only scratching the surface of the role of tumor-localized microbiomes in cancer.
Our understanding of the role of microbes in tumor progression has been advanced by next-generation sequencing, specifically 16S rRNA sequencing, which has uncovered reproducible microbial signatures within a multitude of tumors. Most recently, Nejman et al. identified distinct intracellular bacteria within cancer and immune cells in 1,526 tumor samples (consisting of a mix of flash frozen and formalin-fixed paraffin-embedded samples) and their adjacent tissues of seven cancer types: breast, lung, ovary, pancreas, melanoma, bone, and brain tumors (Nejman et al., 2020). The number of bacterial species detected in most of the tumors averaged ~9. Interestingly, breast tissue was identified as having the most diverse tumor microbiome with an average of 16.4 species per sample (Figures 1A–1C). In our own work, we previously examined the impact of molecules secreted by a bacterium that had been identified as present in the breast TME (Balhouse et al., 2017). This more recent finding by Nejman et al. provides further evidence that local tumor microbiomes may play a critical role in multiple cancer types. However, critical interactions and modes of tumor regulation have largely only been studied in the context of gastrointestinal cancers. Building tissue engineered models recapitulating the TME of different cancer types that have the ability to support the growth and maintenance of tumor microbiomes will be a key next step to investigate the specific roles of microbes in cancer.
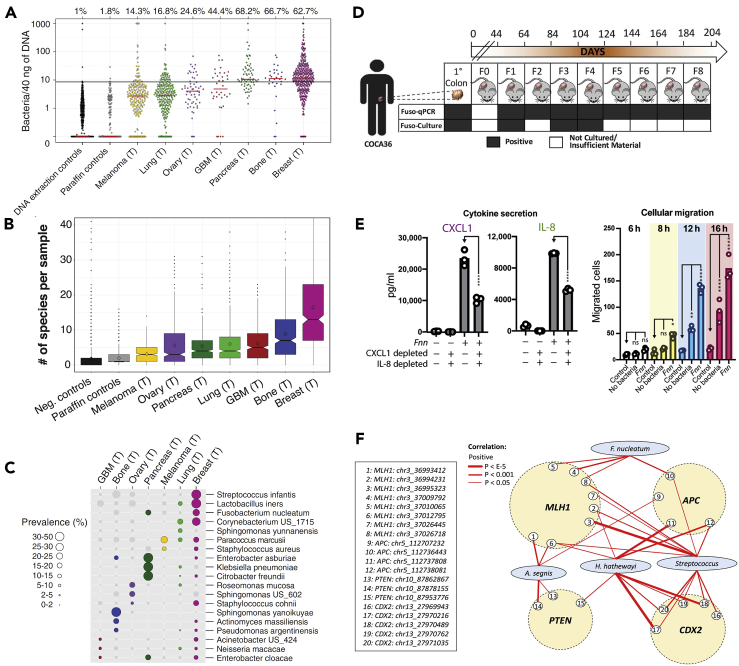
Figure 1.
Characterization of the Tumor Microbiome and Its Functional Relationship With Carcinogenesis
(A) Nejman et al. characterized the cancer microbiome profile of 1,526 human tumors across seven different tumor types. The presence of bacteria was assessed by bacterial 16S rDNA qPCR.
(B) There is high diversity of microbial species across the tumor types.
(C) Nejman et al. also characterized the prevalence of 19 bacterial species across the different tumor types. Reproduced from Nejman et al. (2020).
(D) Bullman et al. demonstrated persistence of F. nucleatum in patient-derived xenografts over a period of 204 days. Reproduced with permission from Bullman et al. (2017).
(E) Casasanta et al. showed Fusobacterium nucleatum-induced IL-8 and CXCL1 secretion from HCT116 colorectal cancer cells, driving cell migration in vitro using F. nucleatum-conditioned media and that could be blocked by inhibiting bacterial internalization. Reproduced with permission from (Casasanta et al., 2020).
(F) Xia et al. showed increased positive correlation of interactions involving Fusobacterium nucleatum and Hungatella hathewayi and the genes MLH1, APC, PTEN, and CDX2 in colorectal cancer (CC-BY-4.0 (https://creativecommons.org/licenses/by/4.0/)).
The Role of Specific Microbes in Tumorigenesis
Many seminal studies have shown that individual microbial species play a role in the onset and progression of multiple cancers. Well-known examples include Helicobacter pylori in gastric cancer (Correa and Piazuelo, 2011; Uemura et al., 2001) and MALT lymphoma (Farinha and Gascoyne, 2005), Salmonella typhi in gallbladder cancer (Ferreccio, 2012), Streptococcus bovis in colon cancer (Boleij et al., 2009), and Chlamydia pneumoniae in lung cancer (Zhan et al., 2011). Several associations including the discovery of Fusobacterium nucleatum within colorectal cancer (CRC) (Kostic et al., 2012), high abundance of Acidovorax temporans in lung cancers with TP53 mutations (Greathouse et al., 2018), and variations in the oral and gut microbiome of patients with melanoma undergoing PD-1 immunotherapy (Gopalakrishnan et al., 2018) indicate a correlative role of the human microbiome with cancer. In addition, intracellular organisms can also directly impact chemotherapy regimens. For example, Gammaproteobacteria within pancreatic ductal adenocarcinoma can metabolize the chemotherapeutic drug gemcitabine (Geller et al., 2017). The most dramatic consequence is the lowering of patient survival with the presence of these bacteria in the tumor.
Microbes within the TME can induce a mix of direct and indirect effects to impact tumorigenesis. From prior work largely on CRC, it is known that bacteria within tumors can cause chronic inflammation or produce and release toxins that impact the cell cycle and induce DNA damage that leads to tumor-initiating or -promoting mutations (van Elsland and Neefjes, 2018). Microbes in the TME can also influence tissue remodeling and deregulate mucosal immunity, creating a favorable niche for tumor cells to expand and migrate (Fares et al., 2020). Moreover, bacteria can induce epigenetic alterations upon gaining intracellular access that can activate dormant tumor-promoting genes (Niller and Minarovits, 2016).
As a prototypical oncomicrobe that has received significant attention, F. nucleatum's involvement in CRC has been extensively characterized in recent years and serves as a prime example to highlight the multiple mechanisms pathogens can use to impact cancer progression. High Fusobacterium levels in tumors correlate with decreased patient survival in CRC (Kunzmann et al., 2019; Mima et al., 2016), pancreatic cancer (Mitsuhashi et al., 2015), and esophageal cancers (Yamamura et al., 2016). An oral commensal microbe, it has been associated with periodontitis, gingivitis, and multiple extra-oral diseases. It's surface adhesin, Fap2, targets the host carbohydrate Gal/Gal-NAc (Abed et al., 2016; Parhi et al., 2020) that is overexpressed on many cancers (Shamsuddin et al., 1995) and may explain how these bacteria are found in higher abundance in CRC compared with the adjacent healthy tissue. Strikingly, it was found that Fusobacteria can travel intracellularly within a migrating host CRC cell leading to bacterial seeding at distant sites such as the liver (Bullman et al., 2017) (Figure 1D), yet it was unclear if these bacteria were active or passive participants in this process. We more recently provided an early clue as to the answer to this latter question, demonstrating that the cytokines IL-8 and CXCL1 are specifically secreted upon F. nucleatum invasion of HCT116 CRC cells and contribute to enhancing cancer cell migration directly (Casasanta et al., 2020) (Figure 1E). This bacterium is able to induce alterations even at the epigenomic level, where it was discovered that F. nucleatum infection, in conjunction with Hungatella hathewayi, induces the hypermethylation of tumor suppressor gene promoters in colonic epithelial tissue (Xia et al., 2020) (Figure 1F).
Unanswered Questions
These observations have generated a number of fundamental questions that we believe should be a broad focus of researchers across multiple cancer types beyond the gut, including:
-
•
Are bacteria seeded early on in tumorigenesis, thereby actively contributing to tumor initiation, or do they arrive at later stages?
-
•
Are specific features of the TME favorable for bacterial localization or survival/proliferation?
-
•
How do the bacteria modify the TME?
-
•
Are there cooperative relationships among multiple bacteria types within the TME, or between tumor and bacteria?
-
•
What factors govern the bacterial interactions with tumor-associated immune cells?
-
•
Does elimination of the internalized microbes reverse their effect on the tumor?
-
•
Can microbial signatures be used to identify the type or stage of the tumor and predict therapy response and toxicity?
-
•
Why do some tumors host more diverse microbiota than others?
-
•
Can we harness the tissue or niche-specific bacterial colonization of cancers to enable targeted delivery of therapeutics?
The consequences of these microbe-microbe and host-microbe interactions may materialize over long timescales. Nejman et al. suggest that there may be a low level of bacteria in every tissue and bacterial translocation increases after disruption of the epithelial barrier and increased vascular permeability (Nejman et al., 2020). Furthermore, evidence of alterations in metabolic profiles of host cells and internalized bacteria (Kasper et al., 2020), as well as the secretion of inflammatory cytokines, that may dramatically impact the hallmarks of cancer, raise questions on the role of secreted factors in influencing tumor progression. Answers to these questions will help identify novel therapeutic targets and reshape current cancer treatment procedures. However, many of these questions have yet to be addressed directly due to a lack of representative models to study tumor-resident bacteria.
In the next section, we discuss the challenges and limitations of existing methods to study host-microbial interactions and how tissue engineering can help model the TME.
Methods to Study Host-Microbial Interactions
To systematically interrogate host-microbe interactions, targeted questions must be defined to select appropriate experimental protocols (Fischbach, 2018). Experiments in animals and humans have generally been limited to overall population/compositional studies via 16S ribosomal RNA gene sequencing and shotgun metagenomics, due to difficulties in isolation and sampling and downstream culture of bacteria (Jovel et al., 2016). Although focusing on mechanistic studies of individual microbes may appear as a low-hanging fruit, microbes exhibit contrasting behaviors when studied within a multi-species community. In fact, many metabolites are only produced in the presence of other microbes (Bertrand et al., 2014). The secreted metabolites themselves may directly affect the tumor viability and proliferation. For example, we have previously shown that Pseudomonas aeruginosa found in breast cancer tissue secretes N-(3-oxododecanoyl)-L-homoserine lactone, which variably modulates viability of MDA-MB-231 and MCF-DCIS.com cells depending on the specific culture microenvironment (Balhouse et al., 2017).
Paradigmatic changes in experimental and conceptual approaches are needed to develop a comprehensive understanding of all the factors that influence host-microbiome interactions in cancer. Challenges that have hindered this goal include difficulties in:
-
•
Isolating causal microorganism(s)
-
•
Preventing over-proliferation of single species in a multi-species model
-
•
Accurately replicating in vivo physiological geometry and biochemical cues
-
•
Limiting variability in organoid structures and batch-to-batch extracellular matrix (ECM) composition
-
•
Developing cell culture medium supportive of all non-microbial cells within the model
-
•
Culturing anaerobic bacteria in oxygenated models
-
•
Real-time monitoring of host and microbial cells and their associated biochemical parameters
-
•
Recapitulating in vivo microbial community composition and immune cell interactions
Overcoming these bottlenecks is crucial to develop technologies to effectively dissect microbial interactions with the host and to target these therapeutically.
Limitations of Animal Models
Animal models are frequently employed due to the availability of powerful genetic tools and physiological relevance to humans. Moreover, with recent advances in whole-animal editing, animal models are becoming increasingly “humanized,” and are ideal for long-term compound studies (Hay et al., 2014). However, current animal models (i.e., xenograft tumor mice models) are poor representatives of human biology because they exhibit distinct bacterial compositions and immune profiles compared with humans (Mestas and Hughes, 2004). Furthermore, animal studies are expensive and not as scalable and accessible as other in vitro models. More specifically, when it comes to manipulating signaling molecules or growth factors, there is much less experimental control, making it challenging to add or tune elements that are necessary to mimic a physiological environment. Therefore, it is beneficial to utilize in vitro technologies that can provide valuable insights at a fraction of the cost of transgenic animals and can reduce dependence on animal experimentation at earlier screening or discovery stages of research, or to help dissect specific mechanism in later stages of study.
Embracing Tissue Engineered Models
Tissue engineering evolved as a strategy to build a tissue from the ground up and can prove to be a viable tool to reconstruct physiologically relevant in vitro models. These techniques begin from seeding cells in decellularized scaffolds, or ECM-based hydrogels. The use of stem cells and induced pluripotent stem cells (iPSCs) catapulted this field with organoid technology. Precise and tunable control with microfluidic devices and microelectromechanical systems has added further control and interrogation options to these technologies. Moreover, engineering principles guide the use of mechanical articulation to simulate biophysical cues that influence the differentiation of cells. However, a number of challenges still remain. While developing tissue engineered models, it is essential to recreate the natural homeostatic environment as well as the tumor-specific microenvironment, specifically for tumor-microbiome studies. Some challenges to this include developing the normal or physiological environment first, before incorporating the pathological tumor element.
Design Considerations for a Complex Tumor Microenvironment
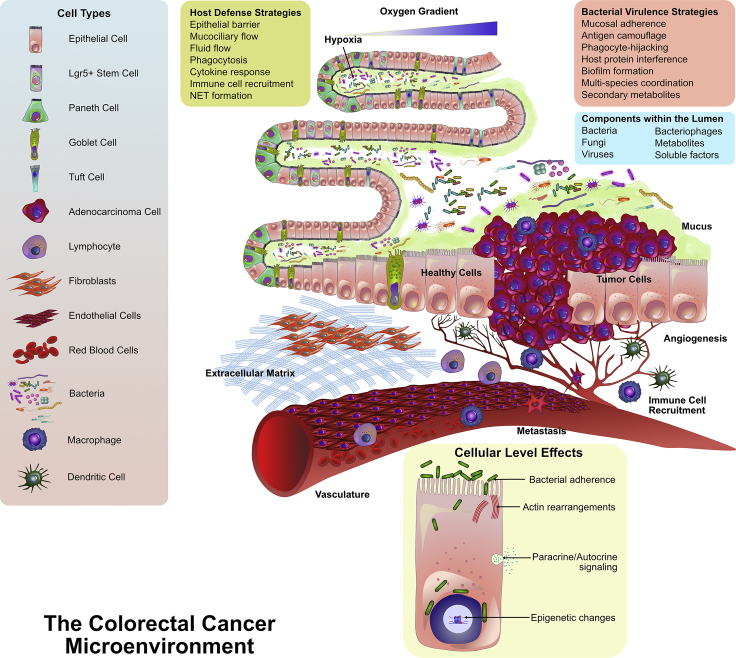
There are a host of factors within the TME that need to be considered for disease modeling. These factors markedly influence the type of microbes that colonize and infect the tumor. Figure 2 exemplifies the microenvironmental parameters to simulate the gut. There exists complex chemical, pH, nutrient, and oxygen gradients throughout the length of the gut that, for certain bacterial species, determine if the colonies are aerobic or anaerobic. The intestinal walls are composed of several different cell types including enterocytes, enteroendocrine cells, Paneth cells, goblet cells, M cells, and Tuft cells, each with unique functions. These cells help establish the epithelial barrier. The barrier itself has a high turnover rate, and any compromise to barrier may lead to microbial invasion and dissemination. Vascular and lymphatic networks skirt the walls of the intestine. In addition, cancer-associated fibroblasts, tumor-associated macrophages, stromal cells, and myriad immune cells create a highly intricate and dynamic microenvironment. Immune-host interactions are compartmentalized along the length of the intestinal tract, which additionally influences microbial diversity. A major factor to consider is mucus secreted by goblet cells, which significantly contributes to bacterial spatial aggregation (Schroeder, 2019). Biomechanical cues arising from peristalsis of the gut and mucociliary flow invariably influence host cell differentiation and the distribution of bacterial colonies. To further add to the tumor milieu are the microbes themselves, their virulence proteins, and synthesized metabolites, and a medley of host-secreted factors, cytokines, and gradients of soluble factors.
Figure 2.
A Multitude of Interacting Factors Within the Complex Tumor Microenvironment of CRC
The gut microenvironment is composed of several different cell types including epithelial, endothelial, and immune cells. Thousands of microbial species co-exist within the microenvironment. The host cells maintain the barrier integrity through multiple host defense strategies, whereas the microbes utilize multiple virulence strategies to target the host. There are variations in oxygen levels within the folds of the microvilli, and the tumor-specific microenvironment can exhibit changes in local ECM composition. Vascular networks can be hijacked by tumor cells via angiogenic signaling, which can enable cancer cell metastasis. Furthermore, many alterations can occur upon bacterial intracellular invasion that could contribute to tumor formation (figure panels adopted from Barkal et al., 2017).
Similar complex microarchitectures can be described for cancers of the breast (Bahcecioglu et al., 2020), pancreas (Ho et al., 2020), lung (Mittal et al., 2016), and other organs. Tissue engineering strategies endeavor to re-create these complex architectures and organ-type specificities to more accurately recapitulate host-microbial interactions in cancer. However, it is essential to note that not all components may be required to study a specific interaction and may additionally impact the reproducibility of studies.
In the following sections, we describe the utility of in vitro models that recapitulate specific features of the TME and demonstrate their feasibility to study host-microbial interactions.
In Vitro Models
There has been a logical evolution of in vitro models of increasing complexity that have enabled the interrogation of host-microbial crosstalk. Despite limitations present in two-dimensional (2D) and 3D cell culture systems, both strategies offer fundamental advantages to explore specific aspects influencing the host-microbiome dynamics. Far from comprehensive, the selected models highlight key features and findings in this field.
2D and 2.5D Models
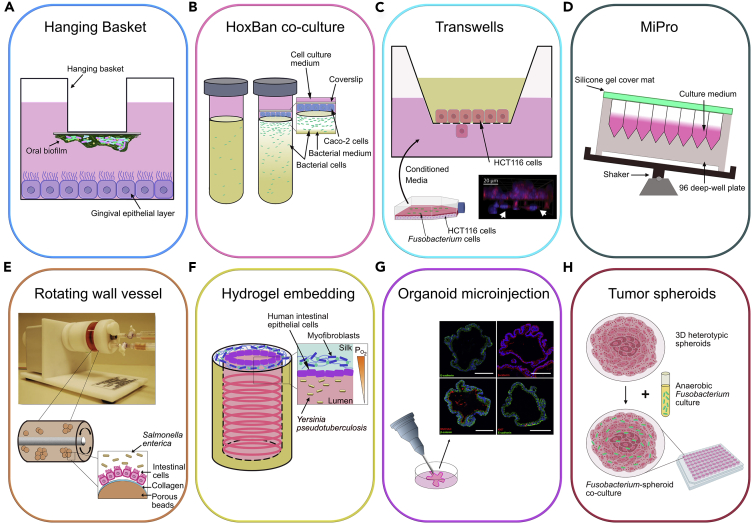
The simplest model used to study host-microbial interactions consists of human cell lines grown as 2-dimensional (2D) monolayers and inoculating microbes within the culture medium. An advancement to this system is the hanging basket model, which suspends a coverslip consisting of a pre-grown multi-species biofilm of pathogenic bacteria over a monolayer of epithelial cells to study gingival inflammation (Millhouse et al., 2014) (Figure 3A). Using a similar approach to simulate an aerobic-anaerobic interface naturally occurring in the gut, the “Human oxygen-Bacteria anaerobic” (HoxBan) system (Sadaghian Sadabad et al., 2015) utilized 50-mL culture tubes with host Caco-2 cells attached to a coverslip and positioned over a bacterial culture medium containing Faecalibacterium prausnitzii (Figure 3B).
Figure 3.
Techniques Used to Study Host-Microbiome Crosstalk
(A–H) 2D vitro static models include (A) the Hanging Basket model used to study gingival inflammation (Millhouse et al., 2014), (B) the HoxBan model that co-cultured Faecalibacterium prausnitzii in medium overlaid with Caco-2 cells attached to a coverslip (Sadaghian Sadabad et al., 2015), (C) the Transwell model that studied the migration of host HCT116 cells in response to conditioned media from Fusobacterium nucleatum-infected HCT116 cells (Image reproduced with permission from Casasanta et al., 2020), and (D) the MiPro model that utilized a shaking 96-deep well microplate format to culture microbiome samples. 3D host-microbe culture systems include (E) the rotating wall vessel (RWV) that induces continuous fluid rotation, ultimately enabling formation of 3D aggregates to model Salmonella enterica infection (Radtke et al., 2010); (F) hydrogel embedding, where 3D porous silk scaffolds was used to reconstitute human intestinal model (Chen et al., 2015); (G) 3D organoid models to mimic host-pathogen interactions (Schlaermann et al., 2016), CC-BY-4.0 (https://creativecommons.org/licenses/by/4.0/), and (H) tumor spheroids, used to co-culture intra-tumor Fusobacteria with colorectal tumor spheroids in a microplate platform (Kasper et al., 2020).
Transwell-based approaches (sometimes referred to as 2.5D) are widely used to assess migratory and invasive responses of the host cells (Casasanta et al., 2020; Park et al., 2017). This culture format consists of host cells seeded on top of a porous membrane or ECM-deposited membrane, with microbes most commonly introduced to the apical side or conditioned media obtained from infected cells to the lower chamber (Figure 3C). An advantage of Transwells is that they can be used to produce polarized, differentiated, multi-layer epithelial cultures. Moreover, because an epithelial monolayer harbors apical and basolateral compartments, this feature enables independent analysis of secretomes per direction, explaining this model's popularity.
Recently, Li et al. introduced a 96-deep well plate-based culturing model (MiPro) that conserved the functional and compositional profiles of individual gut microbiomes (Li et al., 2019). The MiPro setup consists of microbiome samples cultured in a 96-deep well plate in which the plate is covered with a silicone-gel cover and shaken at 500 rpm on a digital shaker (Figure 3D). The authors demonstrated the applicability of this model for high-throughput drug-microbiome interaction studies. More importantly, the MiPro system can be optimized for investigation of host-microbe crosstalk.
Although 2D models offer simple low-cost maintenance and high reproducibility, they fundamentally fail to mimic most of the natural 3D structures of tissues, resulting in the inability to induce complete cell differentiation and recapitulate key physicochemical parameters. More specifically, control of cellular bioactivities is insufficient due to cellular over-proliferation and lack of proper nutrient and oxygen gradients, which greatly affects the analysis of host-microbe dynamics. These limitations are similar to those that have contributed to the disappointing track record of 2D culture for screening of cancer drugs, due in part to the non-physiological hyperactive metabolism of cells cultured on hard plastic surfaces (Cox et al., 2015).
3D Models
3D cell culture systems provide a relatively young but rapidly maturing approach to addressing the inability of 2D models to reconstitute in vivo host-microbiome interactions. Representative examples include the rotating wall vessel, hydrogel scaffolds, organoids, and tumor spheroids. Although these have been in relatively wide use in tissue engineering broadly, and tumor engineering specifically, we believe there is still great potential to leverage as well as to advance previously developed approaches for tumor-microbiome interaction studies. In this section, we will outline some of the more traditional approaches that have already been used to study host-microbe interactions.
The rotating wall vessel (RWV) is a horizontally rotating cell culture chamber with suspended cells. These cells are usually adhered to ECM-coated microcarriers, and rotation results in cell aggregation. This construction induces continuous surface shear stress on the host cells to mimic physiological fluid forces (Barrila et al., 2010) (Figure 3E). Radtke et al. implemented this culture method to provide relevant pathological insights into human enteric salmonellosis (Radtke et al., 2010). Using the RWV, Ilhan et al. established a human endometrial epithelial cell (EEC) model. By incubating 13 Prevotella clinical strains isolated from the endometrium, vagina, amniotic fluid, and oral cavity with the EEC model, the authors explored species-specific effects of Prevotella on physiological and host defense responses in the human endometrial epithelium (Ilhan et al., 2020).
By leveraging biocompatible hydrogel materials and 3D bioprinting technologies, host tissue matrix and functionality can be rebuilt with increased accuracy and robustness. Indeed, recent work has shown successful replication of human intestinal epithelium microarchitecture with months-long function in vitro (Chen et al., 2015). Silk protein was employed to construct a 3D hollow lumen and to house human intestinal epithelial cells, which were supported and nourished by surrounding myofibroblasts (Figure 3F). The authors also showed the applicability of this model to microbiome studies by using it to model Yersinia pseudotuberculosis and Lactobacillus rhamnosus infections.
Organoid technology has greatly advanced the field of tissue engineering. As organoids more accurately reproduce the complexity of multi-cellular tissue, these systems provide a more precise picture of the host-microbiome interface. Organoids are generated from multiple sources including adult/fetal tissues, embryonic stem cells, iPSCs, and recently, patient-derived cells (Fujii et al., 2016; Shamir and Ewald, 2014; Yao et al., 2020). As organoids are typically embedded in extracellular matrices, they can receive matrix cues to facilitate self-organization and specific lineage commitment, resulting in production of near-native epithelial cell clusters (Bar-Ephraim et al., 2020). This technology has been used to develop infectious models of pathogenic H. pylori by microinjection into the organoid's lumen (Bartfeld et al., 2015; Schlaermann et al., 2016) (Figure 3G). Recently, Pleguezuelos-Manzano et al. exposed human intestinal organoids to genotoxic pks+ Escherichia coli by repeated luminal injection over 5 months (Pleguezuelos-Manzano et al., 2020). The authors were able to identify that colibactin secreted by pks+ E. coli directly caused distinct mutations in host epithelial cells, potentially putting individuals that harbor this E. coli strain at an increased risk of CRC. Combining organoid technology with the RWV, Barrila et al. demonstrated that incorporation of phagocytic macrophages into this 3D co-culture model revealed the contribution of distinct cell types during host-pathogen interactions of infection (Barrila et al., 2017).
Human intestinal enteroids, developed from Lgr5+ stem cells in the intestinal crypts, are increasingly used to study microbiome and host interactions. Enteroids contain most of the cell types normally found within the intestinal lining and can be grown as 3D spheroids or 2D monolayers based on experimental need. The Enteroid-Anaerobe Co-Culture system (EACC) developed by Fofanova et al. could re-create the steep oxygen gradients within the platform that are normally observed in vivo (Fofanova et al., 2019). This model was constructed by seeding enteroids on Transwells placed within modified gaskets sealed to a gas permeable 24-well plate and placed within an anaerobic chamber. Using the EACC, the authors demonstrated the co-culture of the anaerobes, Bacteroides thetaiotaomicron and Blautia sp. with patient-derived enteroids for 24 h. However, one specific limitation of enteroids is their limited cytokine secretion in response to pro-inflammatory stimuli, which may significantly impact microbial induced inflammatory responses. Recent refinements to the cell culture media that support enteroid growth endeavor to address this limitation (Ruan et al., 2020).
Finally, tumor spheroid models rely exclusively on cellular aggregation of either homotypic or heterotypic cells (Figure 3H), making this a non-scaffold-based culture method (Costa et al., 2014). Several techniques are available for spheroid production including liquid overlay, hanging drop, U-bottom microplates, microfluidic-based assembly, and spinner flasks (Nunes et al., 2019). Kasper et al. have introduced a tumor spheroid model that promotes the growth of anaerobic bacteria (Kasper et al., 2020). By directly co-culturing 28 Fusobacterium clinical isolates, the authors presented a unique model to study intra-tumor anaerobic bacteria and analyze subsequent effects including cancer-related gene expression and metabolomics.
Although 3D models offer a middle ground between 2D cell cultures and in vivo models and exhibit more accurate physiological response by recapitulating native cell-cell interactions, their applicability can be limiting due to limited differentiation capacities, the inability to provide biomechanical cues, constraints in real-time monitoring of host-microbial interactions, lack of oxygen control, and the long-term maintenance of a stably sustained host-microbiome ecosystem.
Microfluidic and Organ-On-Chip Models
Microfluidic organ-on-a-chip models are proving invaluable to dissect microenvironmental factors governing interactions between microbial and human cells, particularly via secreted soluble factors. Fluids are easily manipulated at the microscale allowing for precision tunability and the development of reproducible chemical gradients (Barkal et al., 2017). At this scale, diffusive forces dominate over convective mixing, which enables laminar flow profiles to regulate the subtle balances of chemicals and metabolites during infection. The basic template inscribing the fluid flow profile for these devices is constructed through soft lithography techniques predominantly using PDMS (polydimethylsiloxane) polymers. More sophisticated layered structures can be constructed using microporous membranes. Culture medium is typically perfused through the device using a syringe or peristaltic pump to maintain controlled flow rates that are used to manipulate fluid shear stress on cells, which has been shown to directly impact their differentiation and morphogenesis. Air and pressurized gas chambers have also been incorporated within these devices to recreate the physical cyclic compression of the tissue. The following sections describe the current state-of-the-art microfluidic models used to maintain co-cultures of host epithelial cells with microbes.
The HuMiX Model
The HuMiX device (Human-microbial crosstalk) mimics the human gut, and allows for the analysis of molecular crosstalk between the microbiome and human colorectal adenocarcinoma enterocytes (Shah et al., 2016). This system consists of three co-laminar microchannels for medium perfusion, human epithelial cell culture, and bacterial consortia. By providing a proximal 0.5- to 1-mm partition for human and microbial microchambers across a nanoporous membrane, this perfusion bioreactor reproduces a healthy intact epithelial barrier. Furthermore, the HuMiX setup integrates one dedicated inlet and outlet per microchannel, oxygen sensors, and a commercial chopstick style electrode for precise control of physicochemical parameters, accurate monitoring of oxygen concentrations, and valid measurement of transepithelial electrical resistance (TEER). Moreover, Shah et al. demonstrated how the HuMiX enabled recapitulation of transcriptional, metabolic, and immune responses in human Caco-2 cells after co-culturing with probiotic L. rhamnosus GG.
Gut-Chip Models
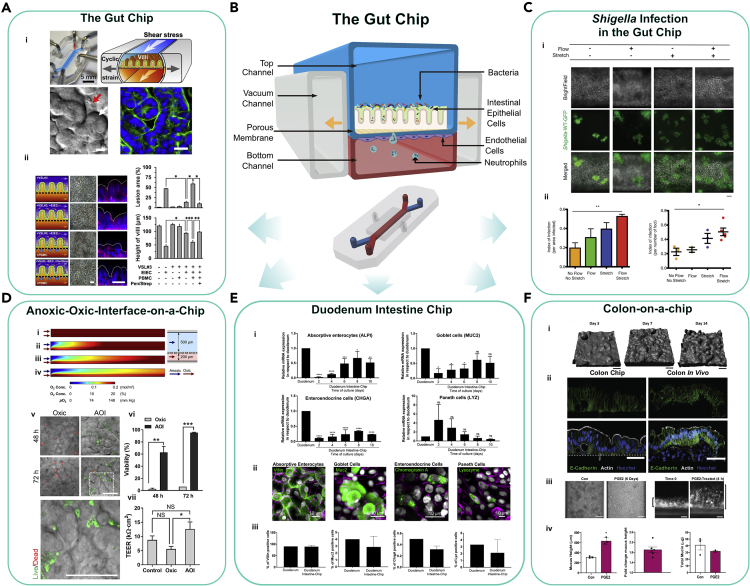
Organ-on-a-chip models, such as the Gut-Chip, have advanced the field by incorporating mechanical stimuli to boost cell differentiation and permit real-time monitoring and assessment of microbial contribution to intestinal disease exacerbation. One such representative biomimetic system (Kim et al., 2016a) contains a microfabricated porous elastic membrane sandwiched between an upper and lower chamber to house intestinal Caco-2 cells and endothelial cells (Figures 4A and 4B). The device is equipped with two hollow lateral vacuum chambers lining the chambers. By applying a cyclic suction, the design effectively induces peristaltic-like motions and trickling-like flow on the intestinal cells. Most importantly, this cyclic strain induced spontaneous villus morphogenesis of the Caco-2 cells and differentiated into four linages of small intestinal cells (absorptive, goblet, enteroendocrine, and Paneth). Using this platform, the authors demonstrated that the addition of lipopolysaccharide induced the secretion of the cytokines IL-1β, IL-6, IL-8, and TNFα into the microvascular chamber replicating chronic inflammatory diseases (Kim et al., 2016a). In a follow-up study, this system was co-cultured with a living microbiome and maintained viability for a week with a mixed population of eight different facultative or obligate anaerobic, probiotic bacteria (Bifidobacterium breve, Bifidobacterium longum, Bifidobacterium infantis, Lactobacillus acidophilus, Lactobacillus plantarum, Lactobacillus paracasei, Lactobacillus bulgaricus, and Streptococcus thermophiles) (Kim et al., 2016b). Employing the Gut-Chip, Grassart et al. revealed that Shigella in effect hijacked the host intestinal microarchitecture and mechanical forces to maximize its infectivity (Figure 4C) (Grassart et al., 2019).
Figure 4.
The Gut-Chip and Models Derived From It
(A) The first human-gut-on-a-chip (the Gut-Chip) was used to demonstrate that probiotic gut microbiota can protect against enteroinvasive E. coli (EIEC)-induced, immune cell-associated injury on chip with the presence of PBMCs (immune cells) Reproduced with permission from(Kim et al., 2016a).
(B) Schematic of the Gut-Chip.
(C) Shigella-WT-GFP (green) infections in the Intestine chip is dependent upon flow and stretch (Reproduced with permission from Grassart et al., 2019).
(D) The Anoxic-Oxic interface (AOI) on a chip generates an oxygen gradient by balancing the flow rates of anoxic and oxic culture medium. The authors co-cultured B. adolescentis with the 3D epithelial cells on the AOI chip, which show significantly increased viability and demonstrate enhanced barrier integrity quantified using TEER (Shin et al., 2019) CC-BY-4.0 (https://creativecommons.org/licenses/by/4.0/).
(E) The Duodenum Intestine chip shows multi-lineage differentiation of the human intestine and shows the expression of markers specific for each differentiated intestinal type (Kasendra et al., 2020) CC-BY-4.0 (https://creativecommons.org/licenses/by/4.0/).
(F) The Colon-on-chip was used to study prostaglandin E2 (PGE2)-induced mucous layer swelling on the chip (Sontheimer-Phelps et al., 2020) CC-BY-4.0 (https://creativecommons.org/licenses/by/4.0/).
The Gut-Chip platform demonstrates high adaptability and feasibility to study specific microenvironmental parameters that may influence host-microbe interactions. For example, to support a more diverse microbial community, the Gut-Chip was modified to enable precise oxygen control. In the “Anaerobic-intestine-on-a-chip,” Jalili-Firoozinezhad et al. established an oxygen gradient across the endothelial and epithelial interface of the Gut-Chip (Jalili-Firoozinezhad et al., 2019). Six oxygen-quenched fluorescent particles were embedded within the system to monitor oxygen levels. This enhancement increased the duration for a complex microbiota co-culture with patient-derived intestinal epithelium to at least 5 days. Furthermore, the introduction of Bacteroides fragilis to the chip was found to enhance the barrier function of the intestinal lining. Using a complementary approach, the Anoxic-Oxic Interface-on-a-chip (AOI) incorporated the simultaneous flow of anoxic and oxic culture medium through dedicated microchannels to re-create an oxygen gradient within the Gut-Chip (Figure 4D) (Shin et al., 2019). The authors employed TEER and platinum dendrimer-encapsulated nanoparticles to quantify barrier permeability and monitor oxygen gradient, respectively. The AOI chip was used to demonstrate increased viability of two obligate anaerobic bacteria (Bifidobacterium adolescentis and Eubacterium hallii) in co-culture with the gut epithelium. The Gut-Chip has also been used to model the colonic mucous layer structure and function with the Colon-on-a-chip (Sontheimer-Phelps et al., 2020) (Figure 4F) and has been used to study drug transport and metabolism in the Duodenum Intestine chip (Kasendra et al., 2020) (Figure 4E).
Other Microphysiological Models
Notably, microphysiological developments of Gut-Chip models also give rise to unique physiome-on-chip platforms such as “10-MPS” and “OrganoPlate” systems (Edington et al., 2018; Trietsch et al., 2017). Edington et al. developed microphysiological systems (MPS) supporting “4-way” (liver/immune, lung, gut/immune, and endometrium), “7-way” (4-MPS supplemented with brain, heart, and pancreas), and even “10-way” (7-MPS with kidney, skin, and skeletal muscles) interactions for weeks-long functional relevance. Together, their multi-MPS systems have enabled high-content preclinical drug screening pipeline and exhibit increasing appeal for studying the dynamics at host-microbe interface. The OrganoPlate, developed in 2017, showed, for the first time, a comprehensive approach to interrogate culture-perfused epithelia tubules that are exposed to an ECM. The OrganoPlate setup consists of 40 microfluidic channel networks integrated in the bottom of a 384-well plate format, wherein epithelial cells are introduced to a collagen ECM-housing lane adjacent to culture medium lanes. Lanz et al. demonstrated the translational utility of this model through therapy response testing of breast cancer (Lanz et al., 2017). Kramer et al. took it a step further by placing the plate on a tilted rocking platform to create a height difference, subsequently reproducing microfluidic interstitial flow to model intratumoral pressure in pancreatic ductal adenocarcinoma (Kramer et al., 2019).
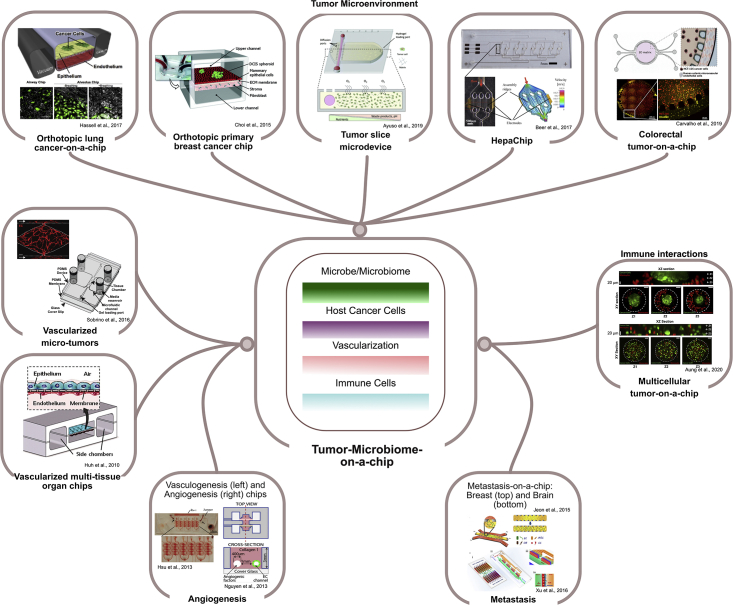
Tumor-on-a-Chip Models
Tissue- or organ-microbiome interaction models have shed light on normal physiological processes, whereas tumor-specific microbiomes have not been widely studied in such platforms. Tumor-on-chip models based on organ-on-chip biomimetic principles hold great potential for re-creation of human TMEs and adoption to study host-microbiome crosstalk, which may ultimately reveal both similarities and differences among different tumor types and stages. In this section, we will outline some models that have aimed to recapitulate the classical hallmarks of cancer, including angiogenic induction, immune interactions, biophysical alterations within the TME, and cell migration and metastasis. While we refer the reader to several excellent review topics that go into more depth (Cox et al., 2015; Ma et al., 2018; Shang et al., 2019; Trujillo-de Santiago et al., 2019; Tsai et al., 2017), here we will focus on some representative examples.
Vascularized multi-tissue organ models were the first to incorporate an endothelial layer juxtaposed with an epithelial layer, combined with mechanical stretch. The earliest application of this model was in the design of a lung-on-a-chip that accurately reconstituted the alveolar-capillary interface and its surrounding microenvironment (Huh et al., 2010). This biomimetic lung model expanded the capabilities to model other organs including the gut, breast, and pancreas, as well as in human cancers. Chips that re-created vasculogenesis and angiogenesis have helped elucidate the molecular mechanisms of angiogenic sprouting and serve as a foundation for future vascularized mechanical, biochemical, and cellular studies (Hsu et al., 2013; Nguyen et al., 2013). For instance, the in vitro vascularized microtumors (VMT) system encapsulates some of the complexity of the TME by the addition of an ECM and stromal cells with nutrients perfused through microvessels (Sobrino et al., 2016). Microvasculature embedded within 3D hydrogels is also commonly used in the construction of these devices (Morgan et al., 2013).
Immune interactions are ubiquitous within the TME and may influence microbial residents. In a multicellular tumor-on-a-chip platform, Aung et al. demonstrated that cancer cell-monocyte interactions increased T cell recruitment (Aung et al., 2020). Other platforms have studied the effects of macrophages and neutrophils migration and extravasation (Boussommier-Calleja et al., 2016).
Several microphysiological devices have also emphasized the importance of the TME in influencing cancer progression and treatment. 3D microengineered models of breast cancer have revealed insights into how the TME could contribute to an invasive phenotype (Choi et al., 2015). Using an orthotopic lung-on-a-chip, Hassell et al. identified that physical cues from breathing motions could influence lung cancer cell growth, invasion, and response to therapy (Hassell et al., 2017). In addition, the HepaChip integrated microfluidics and dielectrophoresis to better recapitulate the 3D microenvironment of pancreatic cancer and revealed that higher doses of cisplatin are needed to reduce the viability of Panc-1 pancreatic cancer cells when cultured in a more physiological context (Beer et al., 2017). Other models have also investigated starvation-induced tumor cell adaptations and resulting metabolic profiles that influence the development of necrotic cores in large tumors (Ayuso et al., 2019). Platforms such as the Colorectal-tumor-on-a-chip have further enabled studies in nanoparticle distribution in precision nanomedicine (Carvalho et al., 2019).
Finally, recent studies that implicate microbes impacting the metastatic potential of tumor make chips that study this phenomenon highly relevant (Coughlin and Kamm, 2020). Specific examples include devices that monitor in vitro metastatic breast and brain tumors and their extravasation to secondary tumor sites (Jeon et al., 2013; Xu et al., 2013).
Taken together, these microfluidic platforms are proving valuable to model intricate interactions to help better understand the development and progression of cancer. With increased relevance of the tumor microbiome in impacting these models, we believe that the development of an integrated tumor microbiome-on-chip will be especially crucial to advance future studies in this field (Figure 5).
Figure 5.
Incorporating Salient Features from Tumor-on-a-Chip Models to Develop a Tumor-Microbiome-on-a-Chip Model
Several tumor-on-a-chip platforms investigate the different aspects of the hallmarks of cancer. The technologies used to build these devices can be adopted to create a tumor-microbiome-on-a-chip that incorporates the necessary elements to characterize tumor-associated microbes.
Future Perspectives
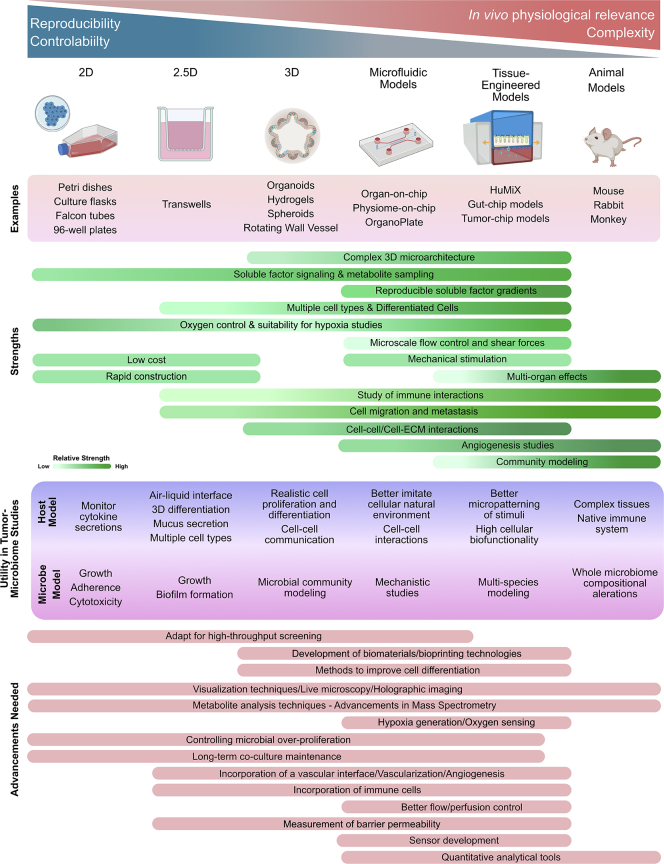
We believe that the future of host-microbiota studies in the context of cancer should focus on the development of next-generation platforms to overcome current challenges including stable culturing of user-defined bacterial communities, advancements in precise differentiation and patterning of cells, improved perfusion capabilities, and incorporating immune cells (Figure 6).
Figure 6.
Summary of Current Models
The strengths of current models, their utility in tumor-microbiome studies, and advancements needed to augment their current capabilities.
Multiplex devices merging engineering and biology will be critical in this field. Advances on the engineering front will go hand-in-hand with new biological insights. The development of effective biomaterials and scaffolds is critical for re-creating physiologically relevant tumor microbiome niches in vitro. For example, by making key advancements in collagen hydrogel biomimetic platforms and 3D printed platforms, multiple biomaterials and cell types can be patterned (Datta et al., 2020; Murphy et al., 2020), which could include tumor microbiotic participants. Gradients of soluble factors, and even bacteria, can be spatially patterned. However, some challenges to overcome are the development of practical bioinks with desired properties, as well as improved mechanical extrusion methods, as these can harm cells. Alternative polymers may be used to address the limitations of using PDMS for hypoxic studies.
Modular approaches need to be compatible with different analytical techniques, and modularity can enable the combination of multiple devices. Automated biosensors can be integrated to continually monitor and measure microenvironmental parameters. Furthermore, the commercialization of these technologies will accelerate scale-up, improve robustness, refine usability, and greatly reduce costs (Ramadan and Zourob, 2020). Advancements in analytical techniques including mass spectrometry for proteomic and metabolite analyses and epigenetic profiling of <~100 cells is now possible using techniques such as MOWChIP (Cox et al., 2019; Zhu et al., 2019) and will complement research in understanding the direct effects of bacteria on tumorigenesis. With respect to visualization, innovative genetic tools are needed to create bacterial mutant strains that express fluorescent proteins for visualization for live microscopy. Advanced imaging technologies such as holographic imaging and light sheet microscopy can improve resolution while live imaging in 3D.
Although complexity can be limitlessly extended and features added, the strengths of simpler 2D and 3D models should not be overlooked. To recapitulate the large number of variables and features to develop a complex, multi-dimensional TME is a daunting task. However, preliminary and pilot studies based on 2D and 3D culture can inform experimentation in more sophisticated platforms. Population models inevitably will have to be performed with animal models. Observations from individual bacterial species need to be connected to multi-species infection models, which may vastly alter the metabolome and infection dynamics. Organoids are increasing in complexity, and relevance and will play a huge role in studying immune effects on infected organoids as they are a highly tractable system to study immune regulation (Bar-Ephraim et al., 2020).
Hypoxia, angiogenesis, metastasis, and immune dysfunction are some of the leading hallmarks of cancer that are impacted by bacterial presence. The relatively immunosuppressed environment within the TME can define multiple cancer stages and therapy response. Without a vascular interface and immune modulation, the engineered models function in isolation greatly diminishing their physiological relevance. Shear stress, pressure, flow rate, oxygen gradients, vascular permeability, and tissue topography greatly affect bacterial localization and pathogenesis. Lymphatic vessels and interstitial fluid pressure are emerging themes in cancer (Munson and Shieh, 2014) and provide a route for host cell dissemination. Many microbes are thought to follow a hematogenous route to the tumor and thus, the incorporation of a microvascular and/or lymphatic network is critical to study host bacterial localization and homing. Endothelialized blood vessel models have already been developed for cancer studies. which need to be integrated into host-microbial platforms (Sontheimer-Phelps et al., 2019). The ability to develop and maintain hypoxic gradients is especially crucial to study the pathogenic mechanisms used by anaerobic bacteria in tumor cores and deep within the villus structure of the intestine. Real-time monitoring of oxygen concentrations will assist in validation of the mechanistic insights derived from these anaerobic models. The subsequent challenge will be co-culturing both aerobic and anaerobic species within the same platform.
As a final note, more advanced platforms and high-throughput technology yield enormous amounts of data that must be efficiently excavated. Here, computational techniques, bioinformatics, and mathematical models can help coalesce disparate data and define experimental parameters. Artificial intelligence and machine learning technologies could be more broadly applied to tissue engineering studies where the degree of complexity may be substantial yet defined in a fully deterministic manner (Fetah et al., 2019). New methods may need to be imported from disparate fields such as ecology and evolutionary biology, which may have useful quantitative analytical tools to make sense of such complex interacting systems (Cunningham, 2019).
Ultimately, just as the Hallmarks of Cancer paradigm has led to promising approaches for targeting these hallmarks as therapy, e.g., targeting the altered tumor metabolism, better understanding of tumor-microbe interactions may also reveal new targetable tumor hallmarks. Bacteria can be harnessed to target cancers directly using a Trojan horse approach to deliver drugs to the cells in bacteriotherapy (Suh et al., 2019). Although antibiotic therapy has shown promise for inhibiting tumor growth in animal models (Bullman et al., 2017), improved models need to be designed to advance such innovations to use for human patients. Important questions must still be answered, such as will targeting and killing intracellular bacteria help control a tumor once it has advanced beyond a certain point, or are there irreversible phenotypic changes driven by these bacteria that occur early in the history of a given tumor? It is likely that the concept of the tumor microbiome could be too intimately connected to the other hallmarks to be truly considered as distinct. In this case, it may be helpful instead to focus on how tumor microbiota influence each of the other hallmarks. For example, in our own work we have made key observations into mechanisms by which F. nucleatum may directly drive metastatic phenotypes (Casasanta et al., 2020).
Concluding Remarks
The systematic characterization of the tumor microbiome and mounting evidence implicating the role of “oncomicrobes” in cancer indicate a need to revise our current understanding of the hallmarks of cancer. Shifting from broad integrated microbiome studies to more focused studies that characterize the multiple mechanisms that individual or cohorts of pathogens employ to infect cells requires a conceptual shift to develop versatile experimental techniques to dissect host-microbe crosstalk. Despite significant progress in cancer-focused tissue engineering, current technologies do not completely re-create physiologically relevant systems, and hence there is still a preference for expensive and sometimes poorly representative animal models. However, with progress in microfluidic and tissue engineered devices there remains much promise in this field. In summary, recent tissue engineering advances in cancer have resulted in exciting new technologies and biomimetic platforms to characterize host-microbial interactions, thereby opening avenues of thought that could give rise to new paradigms of research and precision medicine.
Limitations of the Study
Not applicable.
Lead Contact
Further information and requests should be directed to and will be fulfilled by the Lead Contact, Scott S. Verbridge (sverb@vt.edu).
Materials Availability
Not applicable.
Data and Code Availability
Not applicable.
Acknowledgments
This work has been supported through the NSF Career Award (CBET-1652112 Verbridge), National Institutes of Health R21 Exploratory/Developmental Research Grant (1R21CA238630-01A1), and the Institute for Critical Technologies and Applied Sciences at Virginia Tech. Some figures were created using Biorender (Toronto).
Author Contributions
Writing – Original Draft: B.U. and T.T.D.N.; Writing – Review and Editing: B.U., T.T.D.N., D.J.S., and S.S.V.; Visualization: B.U. and T.T.D.N.; Supervision: D.J.S. and S.S.V.; Project Administration: D.J.S. and S.S.V.; Funding Acquisition: D.J.S. and S.S.V.
Declaration of Interests
The authors declare no competing interests.
References
- Abed J., Emgård J.E.M., Zamir G., Faroja M., Almogy G., Grenov A., Sol A., Naor R., Pikarsky E., Atlan K.A. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung A., Kumar V., Theprungsirikul J., Davey S.K., Varghese S. An engineered tumor-on-a-chip device with breast cancer–immune cell interactions for assessing T-cell recruitment. Cancer Res. 2020;80:263–275. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso J.M., Virumbrales-Munoz M., McMinn P.H., Rehman S., Gomez I., Karim M.R., Trusttchel R., Wisinski K.B., Beebe D.J., Skala M.C. Tumor-on-a-chip: a microfluidic model to study cell response to environmental gradients. Lab. Chip. 2019;19:3461–3471. doi: 10.1039/c9lc00270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahcecioglu G., Basara G., Ellis B.W., Ren X., Zorlutuna P. Breast cancer models: engineering the tumor microenvironment. Acta Biomater. 2020;106:1–21. doi: 10.1016/j.actbio.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhouse B.N., Patterson L., Schmelz E.M., Slade D.J., Verbridge S.S. N-(3-oxododecanoyl)-L-homoserine lactone interactions in the breast tumor microenvironment: implications for breast cancer viability and proliferation in vitro. PLoS One. 2017;12:e0180372. doi: 10.1371/journal.pone.0180372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ephraim Y.E., Kretzschmar K., Clevers H. Organoids in immunological research. Nat. Rev. Immunol. 2020;20:279–293. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- Barkal L.J., Berthier E., Theberge A.B., Keller N.P., Beebe D.J. Multikingdom microscale models. PLoS Pathog. 2017;13:e1006424. doi: 10.1371/journal.ppat.1006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrila J., Radtke A.L., Crabbé A., Sarker S.F., Herbst-Kralovetz M.M., Ott C.M., Nickerson C.A. Organotypic 3D cell culture models: using the rotating wall vessel to study host–pathogen interactions. Nat. Rev. Microbiol. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- Barrila J., Yang J., Crabbé A., Sarker S.F., Liu Y., Ott C.M., Nelman-Gonzalez M.A., Clemett S.J., Nydam S.D., Forsyth R.J. Three-dimensional organotypic co-culture model of intestinal epithelial cells and macrophages to study Salmonella enterica colonization patterns. NPJ Microgravity. 2017;3:10. doi: 10.1038/s41526-017-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S., Bayram T., van de Wetering M., Huch M., Begthel H., Kujala P., Vries R., Peters P.J., Clevers H. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer M., Kuppalu N., Stefanini M., Becker H., Schulz I., Manoli S., Schuette J., Schmees C., Casazza A., Stelzle M. A novel microfluidic 3D platform for culturing pancreatic ductal adenocarcinoma cells: comparison with in vitro cultures and in vivo xenografts. Sci. Rep. 2017;7:1325. doi: 10.1038/s41598-017-01256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand S., Bohni N., Schnee S., Schumpp O., Gindro K., Wolfender J.-L. Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014;32:1180–1204. doi: 10.1016/j.biotechadv.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Boleij A., Schaeps R.M.J., Tjalsma H. Association between Streptococcus bovis and colon cancer. J. Clin. Microbiol. 2009;47:516. doi: 10.1128/JCM.01755-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussommier-Calleja A., Li R., Chen M.B., Wong S.C., Kamm R.D. Microfluidics: a new tool for modeling cancer–immune interactions. Trends Cancer. 2016;2:6–19. doi: 10.1016/j.trecan.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho M.R., Barata D., Teixeira L.M., Giselbrecht S., Reis R.L., Oliveira J.M., Truckenmüller R., Habibovic P. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci. Adv. 2019;5:eaaw1317. doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasanta M.A., Yoo C.C., Udayasuryan B., Sanders B.E., Umaña A., Zhang Y., Peng H., Duncan A.J., Wang Y., Li L. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 2020;13:eaba9157. doi: 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lin Y., Davis K.M., Wang Q., Rnjak-Kovacina J., Li C., Isberg R.R., Kumamoto C.A., Mecsas J., Kaplan D.L. Robust bioengineered 3D functional human intestinal epithelium. Sci. Rep. 2015;5:13708. doi: 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Hyun E., Seo J., Blundell C., Kim H.C., Lee E., Lee S.H., Moon A., Moon W.K., Huh D. A microengineered pathophysiological model of early-stage breast cancer. Lab. Chip. 2015;15:3350–3357. doi: 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P., Piazuelo M.B. Helicobacter pylori infection and gastric adenocarcinoma. US Gastroenterol. Hepatol. Rev. 2011;7:59–64. [PMC free article] [PubMed] [Google Scholar]
- Costa E.C., Gaspar V.M., Coutinho P., Correia I.J. Optimization of liquid overlay technique to formulate heterogenic 3D co-cultures models. Biotechnol. Bioeng. 2014;111:1672–1685. doi: 10.1002/bit.25210. [DOI] [PubMed] [Google Scholar]
- Coughlin M.F., Kamm R.D. The use of microfluidic platforms to probe the mechanism of cancer cell extravasation. Adv. Healthc. Mater. 2020;9:1901410. doi: 10.1002/adhm.201901410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.C., Reese L.M., Bickford L.R., Verbridge S.S. Toward the broad adoption of 3D tumor models in the cancer drug pipeline. ACS Biomater. Sci. Eng. 2015;1:877–894. doi: 10.1021/acsbiomaterials.5b00172. [DOI] [PubMed] [Google Scholar]
- Cox M.C., Deng C., Naler L.B., Lu C., Verbridge S.S. Effects of culture condition on epigenomic profiles of brain tumor cells. ACS Biomater. Sci. Eng. 2019;5:1544–1552. doi: 10.1021/acsbiomaterials.9b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J.J. A call for integrated metastatic management. Nat. Ecol. Evol. 2019;3:996–998. doi: 10.1038/s41559-019-0927-x. [DOI] [PubMed] [Google Scholar]
- Datta P., Dey M., Ataie Z., Unutmaz D., Ozbolat I.T. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 2020;4:1–13. doi: 10.1038/s41698-020-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durack J., Lynch S.V. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edington C.D., Chen W.L.K., Geishecker E., Kassis T., Soenksen L.R., Bhushan B.M., Freake D., Kirschner J., Maass C., Tsamandouras N. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 2018;8:4530. doi: 10.1038/s41598-018-22749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsland D., Neefjes J. Bacterial infections and cancer. EMBO Rep. 2018;19:e46632. doi: 10.15252/embr.201846632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2020:1–17. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- Fares J., Fares M.Y., Khachfe H.H., Salhab H.A., Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal. Transduct. Target. Ther. 2020;5:1–17. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha P., Gascoyne R.D. Helicobacter pylori and MALT lymphoma. Gastroenterology. 2005;128:1579–1605. doi: 10.1053/j.gastro.2005.03.083. [DOI] [PubMed] [Google Scholar]
- Ferreccio C. Salmonella typhi and gallbladder cancer. In: Khan A.A., editor. Bacteria and Cancer. Springer Netherlands; Dordrecht: 2012. pp. 117–137. [Google Scholar]
- Fetah K.L., DiPardo B.J., Kongadzem E.-M., Tomlinson J.S., Elzagheid A., Elmusrati M., Khademhosseini A., Ashammakhi N. Cancer modeling-on-a-chip with future artificial intelligence integration. Small. 2019;15:e1901985. doi: 10.1002/smll.201901985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach M.A. Microbiome: focus on causation and mechanism. Cell. 2018;174:785–790. doi: 10.1016/j.cell.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofanova T.Y., Stewart C.J., Auchtung J.M., Wilson R.L., Britton R.A., Grande-Allen K.J., Estes M.K., Petrosino J.F. A novel human enteroid-anaerobe co-culture system to study microbial-host interaction under physiological hypoxia. BioRxiv. 2019:555755. [Google Scholar]
- Fujii M., Shimokawa M., Date S., Takano A., Matano M., Nanki K., Ohta Y., Toshimitsu K., Nakazato Y., Kawasaki K. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Fulbright L.E., Ellermann M., Arthur J.C. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017;13:e1006480. doi: 10.1371/journal.ppat.1006480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassart A., Malardé V., Gobaa S., Sartori-Rupp A., Kerns J., Karalis K., Marteyn B., Sansonetti P., Sauvonnet N. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting Shigella infection. Cell Host Microbe. 2019;26:435–444.e4. doi: 10.1016/j.chom.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Greathouse K.L., White J.R., Vargas A.J., Bliskovsky V.V., Beck J.A., von Muhlinen N., Polley E.C., Bowman E.D., Khan M.A., Robles A.I. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- Ho W.J., Jaffee E.M., Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 2020;17:527–540. doi: 10.1038/s41571-020-0363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-H., Moya M.L., Hughes C.C.W., George S.C., Lee A.P. A microfluidic platform for generating large-scale nearly identical human microphysiological vascularized tissue arrays. Lab. Chip. 2013;13:2990–2998. doi: 10.1039/c3lc50424g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhan Z.E., Łaniewski P., Tonachio A., Herbst-Kralovetz M.M. Members of Prevotella genus distinctively modulate innate immune and barrier functions in a human three-dimensional endometrial epithelial cell model. J. Infect. Dis. 2020;222:2082–2092. doi: 10.1093/infdis/jiaa324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili-Firoozinezhad S., Gazzaniga F.S., Calamari E.L., Camacho D.M., Fadel C.W., Bein A., Swenor B., Nestor B., Cronce M.J., Tovaglieri A. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., Zervantonakis I.K., Chung S., Kamm R.D., Charest J.L. In vitro model of tumor cell extravasation. PLoS One. 2013;8:e56910. doi: 10.1371/journal.pone.0056910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel J., Patterson J., Wang W., Hotte N., O’Keefe S., Mitchel T., Perry T., Kao D., Mason A.L., Madsen K.L. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh E., Snir-Alkalay I., Venkatachalam A., May S., Lasry A., Elyada E., Zinger A., Shaham M., Vaalani G., Mernberger M. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586:133–138. doi: 10.1038/s41586-020-2541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasendra M., Luc R., Yin J., Manatakis D.V., Kulkarni G., Lucchesi C., Sliz J., Apostolou A., Sunuwar L., Obrigewitch J. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. ELife. 2020;9:e50135. doi: 10.7554/eLife.50135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper S.H., Morell-Perez C., Wyche T.P., Sana T.R., Lieberman L.A., Hett E.C. Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 2020;10:5321. doi: 10.1038/s41598-020-62139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Li H., Collins J.J., Ingber D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U S A. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Lee J., Choi J.-H., Bahinski A., Ingber D.E. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. J. Vis. Exp. 2016;30:54344. doi: 10.3791/54344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., de Haan L., Vermeer M., Olivier T., Hankemeier T., Vulto P., Joore J., Lanz H.L. Interstitial flow recapitulates gemcitabine chemoresistance in A 3D microfluidic pancreatic ductal adenocarcinoma model by induction of multidrug resistance proteins. Int. J. Mol. Sci. 2019;20:4647. doi: 10.3390/ijms20184647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzmann A.T., Proença M.A., Jordao H.W., Jiraskova K., Schneiderova M., Levy M., Liska V., Buchler T., Vodickova L., Vymetalkova V. Fusobacterium nucleatum tumor DNA levels are associated with survival in colorectal cancer patients. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38:1891–1899. doi: 10.1007/s10096-019-03649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz H.L., Saleh A., Kramer B., Cairns J., Ng C.P., Yu J., Trietsch S.J., Hankemeier T., Joore J., Vulto P. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer. 2017;17:709. doi: 10.1186/s12885-017-3709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Abou-Samra E., Ning Z., Zhang X., Mayne J., Wang J., Cheng K., Walker K., Stintzi A., Figeys D. An in vitro model maintaining taxon-specific functional activities of the gut microbiome. Nat. Commun. 2019;10:4146. doi: 10.1038/s41467-019-12087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.-H.V., Middleton K., You L., Sun Y. A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsyst. Nanoeng. 2018;4:1–13. [Google Scholar]
- Mestas J., Hughes C.C.W. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Millhouse E., Jose A., Sherry L., Lappin D.F., Patel N., Middleton A.M., Pratten J., Culshaw S., Ramage G. Development of an in vitroperiodontal biofilm model for assessing antimicrobial and host modulatory effects of bioactive molecules. BMC Oral Health. 2014;14:80. doi: 10.1186/1472-6831-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K., Nishihara R., Qian Z.R., Cao Y., Sukawa Y., Nowak J.A., Yang J., Dou R., Masugi Y., Song M. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi K., Nosho K., Sukawa Y., Matsunaga Y., Ito M., Kurihara H., Kanno S., Igarashi H., Naito T., Adachi Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V., El Rayes T., Narula N., McGraw T.E., Altorki N.K., Barcellos-Hoff M.H. The microenvironment of lung cancer and therapeutic implications. In lung cancer and personalized medicine. In: Ahmad A., Gadgeel S.M., editors. Novel Therapies and Clinical Management. Springer International Publishing; Cham: 2016. pp. 75–110. [DOI] [PubMed] [Google Scholar]
- Morgan J.P., Delnero P.F., Zheng Y., Verbridge S.S., Chen J., Craven M., Choi N.W., Diaz-Santana A., Kermani P., Hempstead B. Formation of microvascular networks in vitro. Nat. Protoc. 2013;8:1820–1836. doi: 10.1038/nprot.2013.110. [DOI] [PubMed] [Google Scholar]
- Munson J.M., Shieh A.C. Interstitial fluid flow in cancer: implications for disease progression and treatment. Cancer Manag. Res. 2014;6:317–328. doi: 10.2147/CMAR.S65444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S.V., De Coppi P., Atala A. Opportunities and challenges of translational 3D bioprinting. Nat. Biomed. Eng. 2020;4:370–380. doi: 10.1038/s41551-019-0471-7. [DOI] [PubMed] [Google Scholar]
- Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.-H.T., Stapleton S.C., Yang M.T., Cha S.S., Choi C.K., Galie P.A., Chen C.S. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl. Acad. Sci. U S A. 2013;110:6712–6717. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niller H.H., Minarovits J. Patho-epigenetics of infectious diseases caused by intracellular bacteria. In: Minarovits J., Niller H.H., editors. Patho-Epigenetics of Infectious Disease. Springer International Publishing; 2016. pp. 107–130. [DOI] [PubMed] [Google Scholar]
- Nunes A.S., Barros A.S., Costa E.C., Moreira A.F., Correia I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019;116:206–226. doi: 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- Parhi L., Alon-Maimon T., Sol A., Nejman D., Shhadeh A., Fainsod-Levi T., Yajuk O., Isaacson B., Abed J., Maalouf N. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020;11:3259. doi: 10.1038/s41467-020-16967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.-S., Park M.H., Shin W., Zhao C., Sheikh S., Oh S.J., Kim H.J. Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro. Stem Cell Rev. Rep. 2017;13:321–334. doi: 10.1007/s12015-017-9739-z. [DOI] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C., Puschhof J., Rosendahl Huber A., van Hoeck A., Wood H.M., Nomburg J., Gurjao C., Manders F., Dalmasso G., Stege P.B. Mutational signature in colorectal cancer caused by genotoxic pks + E. coli. Nature. 2020;580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke A.L., Wilson J.W., Sarker S., Nickerson C.A. Analysis of interactions of Salmonella type three secretion mutants with 3-D intestinal epithelial cells. PLoS One. 2010;5:e15750. doi: 10.1371/journal.pone.0015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan Q., Zourob M. Organ-on-a-chip engineering: toward bridging the gap between lab and industry. Biomicrofluidics. 2020;14:041501. doi: 10.1063/5.0011583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W., Engevik M.A., Chang-Graham A.L., Danhof H.A., Goodwin A., Engevik K.A., Shi Z., Hall A., Rienzi S.C.D., Venable S. Enhancing responsiveness of human jejunal enteroids to host and microbial stimuli. J. Physiol. 2020;598:3085–3105. doi: 10.1113/JP279423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghian Sadabad M., von Martels J.Z.H., Khan M.T., Blokzijl T., Paglia G., Dijkstra G., Harmsen H.J.M., Faber K.N. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 2015;5:17906. doi: 10.1038/srep17906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaermann P., Toelle B., Berger H., Schmidt S.C., Glanemann M., Ordemann J., Bartfeld S., Mollenkopf H.J., Meyer T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.O. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019;7:3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Fritz J.V., Glaab E., Desai M.S., Greenhalgh K., Frachet A., Niegowska M., Estes M., Jäger C., Seguin-Devaux C. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat. Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir E.R., Ewald A.J. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 2014;15:647–664. doi: 10.1038/nrm3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsuddin A.M., Tyner G.T., Yang G.Y. Common expression of the tumor marker D-galactose-beta-[1-->3]-N-acetyl-D-galactosamine by different adenocarcinomas: evidence of field effect phenomenon. Cancer Res. 1995;55:149–152. [PubMed] [Google Scholar]
- Shang M., Soon R.H., Lim C.T., Khoo B.L., Han J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab. Chip. 2019;19:369–386. doi: 10.1039/c8lc00970h. [DOI] [PubMed] [Google Scholar]
- Shin W., Wu A., Massidda M.W., Foster C., Thomas N., Lee D.-W., Koh H., Ju Y., Kim J., Kim H.J. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front. Bioeng. Biotechnol. 2019;7:13. doi: 10.3389/fbioe.2019.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrino A., Phan D.T.T., Datta R., Wang X., Hachey S.J., Romero-López M., Gratton E., Lee A.P., George S.C., Hughes C.C.W. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- Sontheimer-Phelps A., Chou D.B., Tovaglieri A., Ferrante T.C., Duckworth T., Fadel C., Frismantas V., Sutherland A.D., Jalili-Firoozinezhad S., Kasendra M. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell. Mol. Gastroenterol. Hepatol. 2020;9:507–526. doi: 10.1016/j.jcmgh.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S., Jo A., Traore M.A., Zhan Y., Coutermarsh-Ott S.L., Ringel-Scaia V.M., Allen I.C., Davis R.M., Behkam B. Nanoscale bacteria-enabled autonomous drug delivery system (NanoBEADS) enhances intratumoral transport of nanomedicine. Adv. Sci. 2019;6:1801309. doi: 10.1002/advs.201801309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trietsch S.J., Naumovska E., Kurek D., Setyawati M.C., Vormann M.K., Wilschut K.J., Lanz H.L., Nicolas A., Ng C.P., Joore J. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017;8:262. doi: 10.1038/s41467-017-00259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-de Santiago G., Flores-Garza B.G., Tavares-Negrete J.A., Lara-Mayorga I.M., González-Gamboa I., Zhang Y.S., Rojas-Martínez A., Ortiz-López R., Álvarez M.M. The tumor-on-chip: recent advances in the development of microfluidic systems to recapitulate the physiology of solid tumors. Materials. 2019;12:2945. doi: 10.3390/ma12182945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.-F., Trubelja A., Shen A.Q., Bao G. Tumour-on-a-chip: microfluidic models of tumour morphology, growth and microenvironment. J. R. Soc. Interfaces. 2017;14:20170137. doi: 10.1098/rsif.2017.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayasuryan B., Slade D.J., Verbridge S.S. John Wiley & Sons, Ltd; 2019. Microfluidics in Microbiome and Cancer Research. [Google Scholar]
- Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- Xavier J.B., Young V.B., Skufca J., Ginty F., Testerman T., Pearson A.T., Macklin P., Mitchell A., Shmulevich I., Xie L. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer. 2020;6:192–204. doi: 10.1016/j.trecan.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Wu W.K.K., Wong S.H., Liu D., Kwong T.N.Y., Nakatsu G., Yan P.S., Chuang Y.-M., Chan M.W.-Y., Coker O.O. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome. 2020;8:108. doi: 10.1186/s40168-020-00847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gao Y., Hao Y., Li E., Wang Y., Zhang J., Wang W., Gao Z., Wang Q. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials. 2013;34:4109–4117. doi: 10.1016/j.biomaterials.2013.02.045. [DOI] [PubMed] [Google Scholar]
- Yamamura K., Baba Y., Nakagawa S., Mima K., Miyake K., Nakamura K., Sawayama H., Kinoshita K., Ishimoto T., Iwatsuki M. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016;22:5574–5581. doi: 10.1158/1078-0432.CCR-16-1786. [DOI] [PubMed] [Google Scholar]
- Yao Y., Xu X., Yang L., Zhu J., Wan J., Shen L., Xia F., Fu G., Deng Y., Pan M. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26.e6. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- Zhan P., Suo L., Qian Q., Shen X., Qiu L.-X., Yu L., Song Y. Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. Eur. J. Cancer Oxf. Engl. 2011;1990:742–747. doi: 10.1016/j.ejca.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Zhu B., Hsieh Y.-P., Murphy T.W., Zhang Q., Naler L.B., Lu C. MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications. Nat. Protoc. 2019;14:3366–3394. doi: 10.1038/s41596-019-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.