Abstract
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous group of cancers. Collectively, HNSCC ranks sixth in incidence rate worldwide. Apart from classical risk factors like tobacco and alcohol, infection of human papillomavirus (HPV) is emerging as a discrete risk factor for HNSCC. HPV-positive HNSCC represent a distinct group of diseases that differ in their clinical presentation. These lesions are well-differentiated, occur at an early age, and have better prognosis. Epidemiological studies have demonstrated a specific increase in the proportions of the HPV-positive HNSCC. HPV-positive and HPV-negative HNSCC lesions display different disease progression and clinical response. For tumorigenic-transformation, HPV essentially requires a permissive cellular environment and host cell factors for induction of viral transcription. As the spectrum of host factors is independent of HPV infection at the time of viral entry, presumably entry of HPV only selects host cells that are permissive to establishment of HPV infection. Growing evidence suggest that HPV plays a more active role in a subset of HNSCC, where they are transcriptionally-active. A variety of factors provide a favorable environment for HPV to become transcriptionally-active. The most notable are the set of transcription factors that have direct binding sites on the viral genome. As HPV does not have its own transcription machinery, it is fully dependent on host transcription factors to complete the life cycle. Here, we review and evaluate the current evidence on level of a subset of host transcription factors that influence viral genome, directly or indirectly, in HNSCC. Since many of these transcription factors can independently promote carcinogenesis, the composition of HPV permissive transcription factors in a tumor can serve as a surrogate marker of a separate molecularly-distinct class of HNSCC lesions including those cases, where HPV could not get a chance to infect but may manifest better prognosis.
Keywords: oral cavity, transcription factor, oropharynx, prognosis, molecular markers, squamous cell carcinoma, human papillomavirus
Introduction
Squamous cell carcinoma (SCC) of head and neck (H&N) region is a set of highly heterogeneous group of malignancies that collectively pose a major health challenge worldwide. Last 50 years have witnessed a significant shift in the incidence and prevalence of SCC of different constituent subsites. These changes have been linked to changes in exposure to various known carcinogens and risk factors. Traditionally, head and neck squamous cell carcinoma (HNSCC) have been linked to tobacco and alcohol abuse; however, cancer registry data and several cohort studies have documented existence of infection of human papillomavirus (HPV) that have been abnormally detected in oral cavity and tumor tissues of H&N region (Syrjanen et al., 1983; Syrjanen et al., 1987; Syrjanen et al., 1992; Franceschi et al., 1996; Sankaranarayanan et al., 1998; Herrero, 2003; Herrero et al., 2003; Syrjanen et al., 2017). Large clinico-epidemiological studies have revealed that a subset of H&N tumors are specifically linked to HPV infection, which otherwise is traditionally associated with causation of SCC of the uterine cervix and other ano-genital organs (Berman and Schiller, 2017).
A deeper understanding of HPV-positive HNSCC tumors and their response to traditional chemo-radiotherapy revealed existence of a distinct disease type that do not associate with traditional risk factors like tobacco and alcohol abuse (Smith et al., 2004b; Chaturvedi et al., 2015). These are newly emerging tumors and showed increasing incidence. Due to strong association of sexual transmission of HPV in uterine cervix, HPV-positive tumors are associated with sexual behaviors in vastly changing societal practices; however, other modes by which HPV enters in oral cavity also contribute substantially.
Studies performed in initial two decades demonstrated that the affected individuals with HPV-positive HNSCC were of younger age group and showed typically well-differentiated tumors (Pintos et al., 1999; Gillison et al., 2000; Hammarstedt et al., 2006). These prominent clinico-epidemiological differences paved the way for detailed molecular studies to dissect differences in HPV-positive and HPV-negative HNSCC. Later studies subsequently pointed to salient differences at gene expression profiles among these two types of tumors (Cancer Genome Atlas, 2015). Mechanistic studies revealed that these differences can be traced back to the expression and activity of the host cell transcription factors, which work as molecular switches and control cellular behavior. Some of these transcription factors also control establishment of early phase of HPV infection in host cells (Mishra et al., 2006; Gaykalova et al., 2015; Verma et al., 2017). Present review is aimed to compile the available evidence that highlights the involvement and role of key transcription factors in HPV-positive and HPV-negative HNSCC that influence the pathological manifestation of the disease and thus may be useful in the prognostication and effective clinical management of H&N cancers.
HPV in H&N Region and Association With the Cancer Causation
With the exception of a small fraction of tumors of salivary gland, SCC collectively constitutes more than 90% of all cancers of the H&N region. HNSCC originate from squamous cells of different organ sites below the skull base and above the thoracic inlet. Because of the heterogeneity, HNSCC have been designated with a specific identifier under International Classification of Diseases-10 (ICD10) for individual anatomical site of origin of the tumor. Broadly, H&N region can be divided into oral cavity, nasal cavity, pharynx and larynx (Figure 1). Cancers of oral cavity comprise tongue, floor of the mouth, gingiva, gum, palate, lip mucosa and other sites of the mouth. Cancers of pharyngeal region include nasopharynx, oropharynx (base of tongue, palatine tonsils, soft palate, lingual tonsils and posterior and lateral oropharyngeal walls), and hypopharynx. In addition, cancers of larynx, paranasal sinuses, and nasal cavity are also included in HNSCC (Institute, 2017). Collectively, the total burden of cancers in H&N region (ICD-10: C00-13, and 32) sum up to 2,287,731 cases with an annual incidence and mortality rate of 887,659 and 453,307, respectively (Bray et al., 2018). These cancers are overrepresented in men (3:1) and are classically tobacco-associated. There is a significant variation in the incidence of cancers by subsites (Shield et al., 2017). The variation have been attributed to contribution of additional factors such as alcohol consumption, chewing of betel quid with or without tobacco, smoking of local cigars (Sankaranarayanan et al., 1998), environmental exposure to organic solvents, wood and steel dust (Shangina et al., 2006), poor oral hygiene (Rosenquist, 2005), dietary habits, genetic susceptibility (Louvanto et al., 2018), and exposure to infections of Epstein-Barr virus (Young et al., 1988), and of HPV (Herrero, 2003; Herrero et al., 2003). Among others, infection with HPV has emerged as an independent risk factor for a subset of H&N cancers. With changing lifestyle, HPV positive cancers are increasing at an alarming rate (Soto et al., 2000; Shah et al., 2017). Several excellent meta-analysis and reviews on HPV prevalence in H&N show significant variations in HPV positivity that range from 0–100% in different studies (Kreimer et al., 2005; Syrjanen et al., 2017). In a comprehensive analysis of 60 studies from different geographical regions, an overall HPV prevalence of 25.9% in HNSCC was reported (Kreimer et al., 2005). Among these, oropharyngeal cancers displayed the highest and increasing HPV positivity over time (Castellsague et al., 2016). Within oropharyngeal tumors, a comparatively higher HPV positivity is reported in tonsillar crypts and base of tongue (Haeggblom et al., 2017).
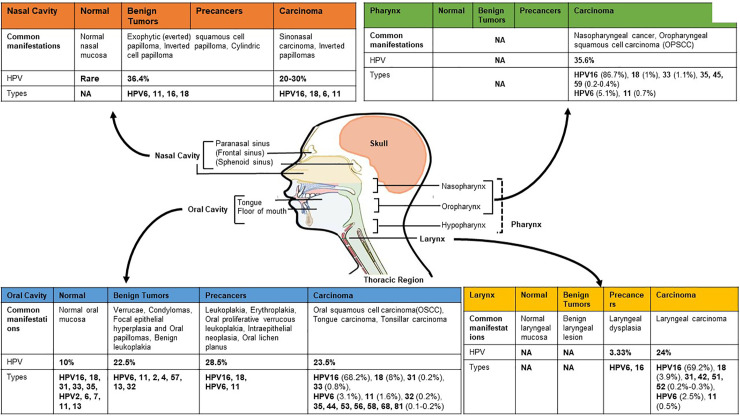
Figure 1.
HPV footprint in different anatomical sub-site of head and neck region. Site-wise HPV prevalence and type distribution in different regions of head and neck broadly classified as oral cavity (anterior and middle tongue, floor of the mouth, gingiva, gum, palate, lip mucosa and other sites of the mouth), pharynx (oropharynx with tonsils and base of tongue, nasopharynx, and hypopharynx), larynx and nasal cavity with paranasal sinus (frontal sinus, sphenoid sinus). Hypopharynx, which show low, or no HPV positivity, is sometimes included with laryngeal region for HPV prevalence studies. Data presented in the figure is derived from different systematic reviews and meta-analyses (Miller and Johnstone, 2001; Syrjanen, 2003; Kreimer et al., 2005; Sarkola et al., 2008; Rautava and Syrjanen, 2011; Syrjanen et al., 2017; Onerci Celebi et al., 2018). [Image originally produced by Macmillan Cancer Support and reused with permission].
HPV is a member of family Papillomaviridae that represents more than 205 different types of papillomaviruses (Bernard et al., 2010; Syrjanen, 2018). Apart from the classification on the basis of sequence divergence in HPV L1 gene into different genera, these double-stranded DNA viruses are also classified based on their epithelial tissue-tropism as mucosal and cutaneous types. Similarly, based on epidemiological correlates i.e. the frequency of occurrence in malignant or benign lesions of uterine cervix, HPV is grouped as high-risk (HPV16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, 66, 68, 69, 73, and 82) or low-risk types (HPV6, 11, 40, 42, 43, 44, 54, 61, 70 72, 81, and 89), respectively (Shukla et al., 2009). Among the high-risk HPV, HPV type 16 represents the most prominent HPV infection in both cervical and HNSCC, whereas, the low-risk HPV type 6 and type 11 are associated with benign lesions like papillomas and warts of H&N as well as genital regions. Different anatomical subsites reportedly have a specific spectrum of HPV infection and genotypes that differ with clinical manifestation of the lesion (Figure 1). Even though pharynx encompasses hypopharynx, oropharynx, nasopharynx, all the three sites show distinctive variations in susceptibility to HPV infection. Oropharynx has the highest occurrence of high-risk HPV infections. Nasopharynx is attributed more to Epstein-Barr virus than the HPV. The overall prevalence of HPV is higher in oropharyngeal cancer [Odds Ratio (OR): 14.66] than the oral cavity cancer (OR: 4.06) and laryngeal cancer (OR: 3.23) (Shaikh et al., 2015). Most cancers of hypopharynx and larynx are not related to HPV infection. The larynx is usually infected by low-risk HPVs with less than 2% infections leading to malignant transformation (Torrente et al., 2011; Gama et al., 2016). The type-specific contributions of HPV16/18 in H&N region is around 84.9%, and for all major HPV types (HPV6/11/16/18/31/33/45/52/58) collectively it is 89.7% (De Martel et al., 2017). In a site-wise distribution of HPV types in HPV-attributable HNSCC, HPV16 alone accounted for 86.7% oropharyngeal SCCs (OPSCC), 68.2% for oral SCCs and 69.2% for laryngeal SCCs (Kreimer et al., 2005).
Even though a number of studies have addressed issues related with HPV positivity in H&N region, unlike cervical neoplasia, an accurate estimate of HPV prevalence in H&N remain uncertain due to cumulative effects of a multitude of factors, such as: (i) the techniques that varied greatly in their sensitivity, (ii) all sites in H&N region are not equally susceptible to HPV infection and may not show productive HPV infection, and (iii) the differential risk of opportunistic exposure to HPV infection due to (a) variations in high-risk sexual behavior, or (b) vertical transmission for HPV-positive mothers leading to oro-genital transmission, that significantly differed among different populations.
HPV-Positive HNSCC as a Molecularly Distinct Subtype
Following detection of HPV antigens in H&N region (Syrjanen et al., 1983; Wookey et al., 2019), efforts in first 20 years were primarily directed towards detecting and enumerating HPV positivity and type-specific distribution in different H&N subsites (Syrjanen et al., 2017). These studies paved the way for large clinico-epidemiological surveys and meta-analyses directed to investigate various risk factors, natural history and thus clinically characterized the HNSCC with reference to their HPV status. These HPV-positive tumors were reported in early stage (Pintos et al., 1999; Smith et al., 2004b; Hammarstedt et al., 2006), well-differentiated histology (Pintos et al., 1999; Gupta et al., 2015), basaloid morphology (Gillison et al., 2000), larger tumors (Gletsou et al., 2018), and either no lymph node involvement (Pintos et al., 1999) or with cystic cervical lymph node positivity (Goldenberg et al., 2008). These tumors showed low risk of second primary malignant neoplasm (Adjei Boakye et al., 2018) with a better overall and disease-free survival (Ragin and Taioli, 2007; Fakhry et al., 2008; Ang et al., 2010; Rischin et al., 2010; Posner et al., 2011; Fakhry et al., 2014). Irrespective of the tissue subtype involved, HPV positivity in HNSCC emerged as strong biomarker associated with better prognosis (Gillison et al., 2000; Wookey et al., 2019).
Comprehensive molecular-genetic studies carried out on primary and metastatic tumors, and cell lines from HNSCC revealed a series of molecular differences among HPV-positive and HPV-negative tumors and the amount of molecular data is currently overwhelming (Leemans et al., 2018). As outlined in Figure 2, the differences are manifested through change in gene expression profile or appearance of somatic mutations in gene involved in cell survival and apoptosis, cell cycle, DNA replication, recombination and repair, cellular assembly and localization, nucleic acid metabolism, immune response, signal transduction, and transcriptional and post-transcriptional regulation through action of viral oncogenes or epigenetic silencing, which dominated in HPV-positive tumors (Slebos et al., 2006; Martinez et al., 2007; Lohavanichbutr et al., 2009). On the other hand, mutations either inactivating in tumor suppressor genes or gain of function in oncogenes predominated in HPV-negative HNSCC (Stransky et al., 2011; Lechner et al., 2013; Zhang P. et al., 2014). Since OPSCC sample prevailed in majority of the studies, the HPV-specific signatures can be directly associated with HPV-positive OPSCC.
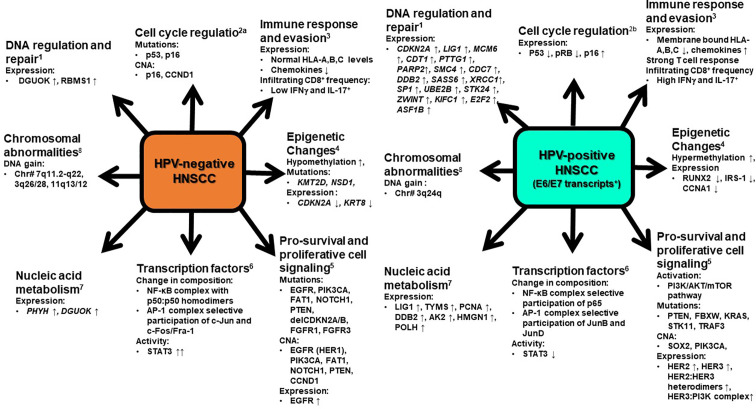
Figure 2.
A snapshot of molecular signatures that distinguish HPV-positive and HPV-negative HNSCC. HPV-positive HNSCC that display specified signatures typically possess transcriptionally-active HPV infections. The changes depicted are characteristically displayed by clinically advanced HNSCC. At an early stage or at the time of viral entry all these changes are not evident in potentially malignant cells. CNA, copy number alterations, ↑ - increase, ↓ -decrease. 1. Upregulation of genes involves in DNA regulation and repair (Lohavanichbutr et al., 2009). 2. Cell cycle regulation (Van Houten et al., 2001; Hafkamp et al., 2003). 3. Varied immune response by HPV positive and HPV negative HNSCC (Nasman et al., 2013; Partlova et al., 2015). 4. Epigenetic changes (Sartor et al., 2011; Lindsay et al., 2017; Leemans et al., 2018) 5. Pro-survival and proliferative cell signaling (Chung et al., 2015; Pollock et al., 2015; Seiwert et al., 2015) 6. Modification in expression and activation of different transcription factors (Mishra et al., 2006; Gaykalova et al., 2015; Verma et al., 2017; Gupta et al., 2018). 7. Upregulation of genes involved in nucleic acid metabolism (Lohavanichbutr et al., 2009). 8. Gain in DNA of chromosome (Dahlgren et al., 2003). At an early stage or at the time of viral entry all these changes are not evident in potentially malignant cells.
Changes in Cell Cycle Regulators
Maiden molecular differences between HPV-positive and HPV-negative HNSCC emerged from studies on p53, pRB and p16 that served as direct cellular targets of viral oncoproteins, E6 and E7 of high-risk HPVs (Van Houten et al., 2001; Hafkamp et al., 2003). HPV-positive tumors characteristically showed a wild type, functionally active p53 gene, whereas HPV-negative tumors invariably harbored inactivating p53 mutations. In addition, p53 mutations were also detected in HPV-positive tumors where the HPV was not transcriptionally-active. Preventing HPVE6 and E7 expression by experimental means restored functional p53 and pRb suppressor pathways in HPV-positive HNSCC (Rampias et al., 2009). E7-mediated pRb is associated with concomitant overexpression of cell cycle inhibitor p16 in HPV-positive tumors (Hafkamp et al., 2003). p16 showed a prognostic value in clinical follow up studies (Licitra et al., 2006; Chung et al., 2015), and it was subsequently referred as surrogate marker of active HPV infection. However, later comprehensive biomarker studies using p16 concordance with active HPV infection showed a false positivity rate of about 20% (Castellsague et al., 2016). Microarrary-based study consistently revealed a higher p16 expression in HPV-positive as compared to normal tissues, but p16 level remained indistinguishable from the normal counterpart in most of the HPV-negative tumors and the difference was not significant. The profile of overexpressed and downregulated genes in these categories significantly differed with a very few overlaps, which is indicative of entirely different tracks of cancer progression events in two HNSCC types (Slebos et al., 2006; Martinez et al., 2007; Lohavanichbutr et al., 2009). Reduced p53 transcript in HPV-positive tumors was associated with activation of oncogenic pathway genes such as CDKN2A/CCND1 and other candidate transcript genes such as SFRP1, CRCT1, DLG2, SYCP2, and CRNN with SYCP2, which contribute to genetic instability during development of HNSCC (Masterson et al., 2015).
Cytogenetic Alterations
Comparative genomic hybridization showed gross chromosomal abnormalities in 3q and 7q region (Dahlgren et al., 2003). HPV-positive tonsillar tumors displayed DNA gain of 3q (72%) with absence of any change in 7q loci, whereas HPV-negative tumors showed a specific gain of 3q (40%) and 7q (40%) loci in sizable number of cases. DNA gain in 3q24 region was associated with increased expression of corresponding genes in the locus (Slebos et al., 2006). Worth noting, the long arm of chromosome 3 (3q26) harbors the SOX2 and PIK3CA genes, which were implicated specifically in HPV-positive tumors (Yarbrough et al., 2007; Lechner et al., 2013).
Pro-Survival and Proliferative Cell Signaling Pathways
Mutations in genes such as PIK3CA, PTEN, DDX3X, FGFR2/3 and BRCA1/2, TRAF3/CYLD genes, and enriched copy number variations in PIK3CA, KRAS, MLL2/3, NOTCH1, and DNA repair genes were also reported in HPV-positive HNSCC (Chung et al., 2015; Seiwert et al., 2015; Hajek et al., 2017). Likewise HPV-negative HNSCC showed mutations in TP53, CDKN2A, PIK3CA, CUL3, NSD1, and NOTCH genes. Through a novel proximity based study using VeraTag assay, it was determined that HPV-positive HNSCC exhibited a significant elevated expression of total HER2, total HER3, HER2:HER3 heterodimers, and the HER3:PI3K complex. In contrast, HPV-negative HNSCC exhibited elevated expression of total EGFR(HER1), which contributes to the resistance to EGFR inhibitors in these tumors (Pollock et al., 2015). A cross talk occurs between PI3K signaling, HER3 receptor, and E6 and E7 expression. By RTK profiling, PI3K inhibition led to elevated expression of HER3 receptor, which in turn increased the abundance of E6 and E7 oncogenes to promote PI3K inhibitor resistance (Brand et al., 2018). HNSCC patients with wild type PIK3CA had higher 3-year disease free survival than PIK3CA mutant patients (Beaty et al., 2019).
DNA Replication and Repair
Differences in components of DNA replication and repair immensely contribute to higher sensitivity to radiation and chemotherapeutic agents displayed by the HPV-positive HNSCC (Liu C. et al., 2018). Owing to the existence of functional p53, strong double strand breaks in DNA override E6-mediated inhibition in p53, leading to cell cycle arrest and induction of apoptosis (Kimple et al., 2013); however, HPV-negative tumors sustain greater damage to DNA before entering the mitotic catastrophe. HPV-positive cells also showed impaired DNA repair (Rieckmann et al., 2013). Overexpressed p16 in HPV-positive HNSCC was found to block homologous recombination-mediated DNA repair by preventing RAD51 (Dok et al., 2014).
Immune Response
Reduced MHC class I antigens are the hallmark of HPV-positive HNSCC lesions (Nasman et al., 2013). E7 plays a key role in this process and increases susceptibility of HPV-positive HNSCC to natural killer cells (Georgopoulos et al., 2000). HPV-positive HNSCC exhibited enhanced and distinct pattern of immune activation to HPV-positive HNSCC, which included upregulation of genes associated with immune-associated processes, high infiltration rate of CD8+IFNγ+ T lymphocytes, Tc17 lymphocytes, naïve CD4+T lymphocytes, and myeloid DCs along with high level of production of chemokines CXCL9, CXCL10, CXCL12, CXCL17, and CXCL21 (Partlova et al., 2015; Chen et al., 2018). These lesions reported higher ratio of M1/M2 macrophages, lower expression of COX-2 mRNA, and higher expression of PD1 mRNA (Chen et al., 2018).
Epigenetic Modifications
HPV-negative HNSCC genomes, in general, are hypomethylated and showed higher loss-of-heterozygosity (LOH) and genomic instability as compared to HPV-positive tumors (Richards et al., 2009). HPV-positive tumors showed higher promoter methylation of polycomb repressive complex 2 target genes and increased expression of DNMT3A, whereas higher methylation and lower expression of RUNX2, IRS-1, and CCNA1 was noted in HPV-negative tumors (Sartor et al., 2011). These differences in methylation patterns with larger panels later were found to have prognostic value (Ren et al., 2018).
Transcriptional Regulation
Pioneering studies conducted by our group showed differential expression and activity of transcription factors in HNSCC with respect to the HPV status (Mishra et al., 2006; Gupta et al., 2015; Verma et al., 2017; Gupta et al., 2018). HNSCC in general showed constitutively active nuclear factor-κB (NF-κB); however, the active complex differed in the composition. HPV-negative tumors had p50:p50 homodimers, whereas p65 participation was detected in HPV-positive tumors (Mishra et al., 2006; Gupta et al., 2018). Similarly, constitutively active activator protein-1 (AP-1) differed in its composition. AP-1 with JunB and JunD participated with c-Fos & Fra-2 in HPV-positive tumors, whereas, c-Jun was the major binding partners forming the functional AP-1 in HPV-negative tumor (Gupta et al., 2015). Subtle changes in composition of these transcription factors have significant impact on the regulation of set of downstream genes (Gaykalova et al., 2015). In contrast to NF-κB and AP-1, signal transducer and activator of transcription-3 (STAT3) was negatively regulated with HPV positivity in HNSCC. These differences were reported in both retrospective and prospectively collected samples by simple IHC procedures (Verma et al., 2017). Analysis of Cancer Atlas data demonstrated defects in TRAF3 and CYLD, which correlated with activation of NF-κB (Hajek et al., 2017).
These evidences collectively suggest presence of molecularly distinct gene expression patterns in HPV-positive and HPV-negative HNSCC, which could potentially contribute to their favorable outcome and better prognosis. However, studies also emphasize essential requirement of HPV to be transcriptionally-active to have a prognostic relevance (Braakhuis et al., 2004; Jung et al., 2010; Janecka-Widla et al., 2020). Further, most of the studies that highlighted HPV-negative signatures also identified yet another subgroup of HNSCC that displayed favorable clinical outcome even in the absence of HPV. These findings emphasize need for greater understanding of the transcriptional state of the infected cells and the cellular players that perform driver role in coupling host transcription with the virus.
Entry and Establishment of HPV Infection in the Host Cell
In an in vitro culture system, the transforming potential of HPV in primary oral keratinocyte is well established (Chu, 2012). However, not all individuals positive for oral HPV DNA develop carcinoma, which is suggestive of cellular heterogeneity in H&N region and individualistic variations in cellular environment that may permit or restrict productive HPV infection (Smith et al., 2004a). Succeeding immunological escape, incident HPV can complete its life cycle in H&N tissues and exhibit genome maintenance, vegetative and replicative phases just like in cervix (Raff et al., 2013; Syrjanen, 2018). However, the exact mechanism and target cells in these tissues are poorly defined. Studies show gingival pockets as reservoirs of HPV infection (Hormia et al., 2005). Even though transcriptionally-active HPV infection is detected in different H&N sites, a well-defined HPV susceptible region in these tissues like squamocolumnar junction in cervix, do not exist. Being an epitheliotropic virus, it targets the basal layer cells of the epithelium that becomes accessible due to micro-abrasions. Cells of the basal layer that undergo keratinizing changes provide a suitable cellular environment for progression of viral life cycle (Harden and Munger, 2017). Transcriptionally active HPV, either detected by expression of viral oncoproteins or their transcripts, has been reported in almost all major H&N sites namely oral cavity (Shroyer and Greer, 1991), tonsillar (Snijders et al., 1992), and oropharyngeal region (Gillison et al., 2000) including larynx (Scheurlen et al., 1986).
HPV infection, if it has to be tumorigenic, has to overcome several barriers and challenges (Figure 3). Further, the cell division and differentiation program of infected host squamous epithelium controls the HPV life cycle (Harden and Munger, 2017). During establishment of HPV infection, the most critical contribution comes from the availability of a set of transcription factors that control expression of early HPV genes. In the absence of these host transcription factors, the virus is defunct and eliminated or just maintains its genome with host cell cycle till the required factors are expressed/activated during the cell differentiation stage or local inflammation. Chronic inflammation serves as a driving force in speeding up carcinogenesis in H&N region (Liu et al., 2015). HPV Infections incapable of undergoing transcriptional activation are non-productive and are either eliminated/cleared from the tissue or do not influence tumorigenic properties of the host cells (Boscolo-Rizzo et al., 2016). Therefore, for tumorigenic activation, HPV genome is entirely dependent on host factors (Kyo et al., 1997). Studies carried out by our group and others in last 5 years have clearly demonstrated the differential expression and activity of host transcription factors particularly AP1, NF-κB and STAT3 in HPV-positive HNSCC or the transcript profile of their downstream genes (Mishra et al., 2006; Gaykalova et al., 2015; Verma et al., 2017). Incidentally, the level of expression and activity of these transcription factors have independent oncogenic and predictive valve due to their more pragmatic role in regulation of variety of cellular functions, particularly in the regulation of inflammation (Iliopoulos et al., 2009). Therefore, HPV-related transcription factors provide a coupling action between inflammation, activation of HPV oncogenes, and development of cancer.
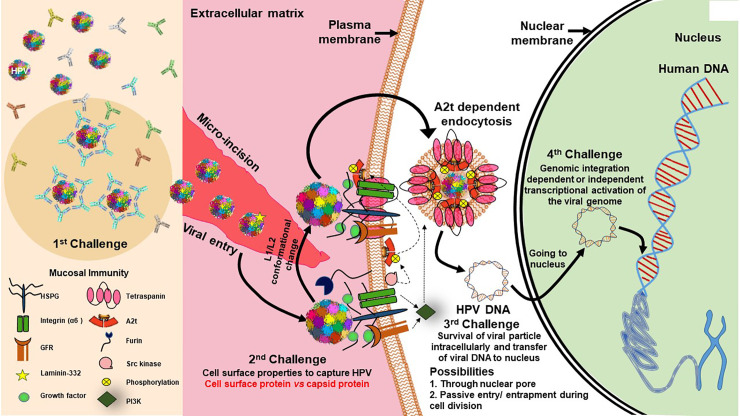
Figure 3.
Challenges for establishment of HPV infection in basal epithelial cells of H&N region. Establishment of transcriptionally-active HPV infection in epithelial tissues faces various challenges to establish in the host cell. First challenge is posed by the mucosal immunity against HPV coat proteins that results in clearance by HPV specific antibodies (Jenson et al., 1991). Second challenge is viral entry which is specifically facilitated through micro-incisions and interaction of HPV-HSPG (heparan sulfate proteoglycan)-growth factor complexes with growth factor receptors (Surviladze et al., 2012) that leads to rapid activation of signaling pathways, such as PI3K/Akt/mTOR, which further inhibit autophagy of cell for viral benefit (Surviladze et al., 2013). Further, src kinase phosphorylates annexin A2(AnxA2) at Tyr23 (A2t), HPV recognizes A2t and binds to it in Ca2+ dependent manner and A2t dependent endocytosis of HPV particle and support trafficking of virus in cell (Dziduszko and Ozbun, 2013). Third challenge is presented for the viral DNA for survival in intracellular milieu and its transport to the nucleus, which could likely be through a passive entry/entrapment during cell division. Last and most pivotal challenge is presented for its genomic integration and transcriptional activation of viral genome to produce functional E6/E7 transcripts. In the absence of transcriptionally activation of E6 and E7, the lesion is similar to HPV negative lesion. On the other forced expression of E6/E7 can independently derive keratinocyte transformation.
Transcriptionally-Active HPV Infections in HNSCC: Role of Long Control Region (LCR)
A ~850bp non-coding region present on viral DNA, designated as Long Control Region (LCR) or synonymously known as Upstream Regulatory Region, constitute over 10% of HPV genome. LCR contains most of the tissue/cell-specific enhancer region and is populated by (i) cis-responsive elements that regulate HPV life cycle, (ii) the replication origin on 3’ end where HPVE1 binds, and (iii) the early promoter (p97 in HPV16) that control the expression of oncogenes E6 and E7 (Figure 4) (Cripe et al., 1987; Gloss et al., 1987). LCR is the most variable region of HPV genome and regulates transcription and replication of the viral DNA (Pande et al., 2008; Xi et al., 2017; Ribeiro et al., 2018). The LCR of all clinically-relevant genital HPVs are organized similarly (O’connor et al., 1995). Viral oncogenes E6 and E7 produced as a fusion transcript are produced by a promoter which is directly regulated by the LCR. HPV16 LCR, which can be considered as a model for the HPVs, can be subdivided by two E2 binding sites, which serve as landmarks, into three functionally-distinct segments, named as 5’ or distal, the central, and 3’ proximal segment due to their positioning with respect to the early promoter (O’connor et al., 1995).
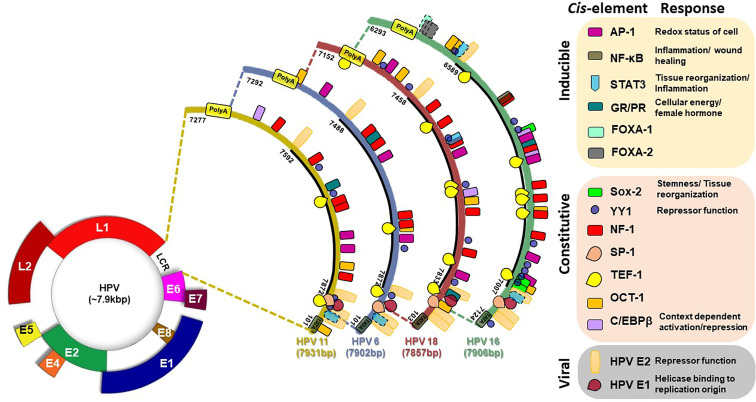
Figure 4.
Transcription factor binding site on LCR of HPV16 (831 bp), HPV18 (835), HPV6 (811 bp), and HPV11 (755 bp). Various consensus-binding sites for different transcription factors present on HPV LCR are represented as boxes. Solid line around the box indicate experimentally-validated site, broken line indicate putative site. Consensus-binding sites of AP-1 (Gloss et al., 1989; Chan et al., 1990; Chong et al., 1990; Nakshatri et al., 1990; Thierry et al., 1992; O’connor et al., 1995; O’connor et al., 1996; Wang et al., 2011), STAT3 (Arany et al., 2002), NF-κB (Fontaine et al., 2000), SOX-2 (Martinez-Ramirez et al., 2017), YY1 (O’connor et al., 1995; O’connor et al., 1996), NF-1 (Gloss et al., 1989; Chong et al., 1990; Nakshatri et al., 1990; O’connor et al., 1995), SP-1 (Gloss and Bernard, 1990), TEF1 (Ishiji et al., 1992; O’connor et al., 1995), Oct-1 (O’connor and Bernard, 1995; O’connor et al., 1995), FOXA1 (Sichero et al., 2012; Xi et al., 2017), on HPV16 (NC_001526.4), HPV18 (X05015.1), HPV6 (NC_001355.1), and HPV11 (M14119.1) have been reported in different studies and have been shown as per revised reference sequences in NCBI Nucleotide database.
As depicted in Figure 4, typical HPV16 LCR contains binding sites for various host transcription factors like AP-1, NF-κB, STAT3, Sox-2, Glucocorticoid Response Element (GRE)/Progesterone Response Element (PRE), YY-1, NF1 (CAAT binding Transcription Factor 1), SP1, TEF-1, Oct-1 (NFA), cEBP/β, FOXA1, FOXA2, KRF-1, GATA3, CDP, SOX9, TEF-2, and PHOX2A, which were identified by different bioinformatics and experimental approaches (Gloss et al., 1987; Gloss et al., 1989; Chan et al., 1990; Gloss and Bernard, 1990; Chong et al., 1991; Bauknecht et al., 1992; Ishiji et al., 1992; Thierry et al., 1992; O’connor and Bernard, 1995; Fontaine et al., 2000; Arany et al., 2002; Sichero et al., 2012; Martinez-Ramirez et al., 2017; Xi et al., 2017). Based on the functional role of the corresponding transcription factors, it is predicted that these elements assist the virus to fine-tune its response to the immediate environment such as redox status, local inflammation, cellular reorganization & reprogramming, cellular energy status, and most importantly the hormonal status among others. DNase footprinting, DNA binding and reporter assays helped to establish the functional role of different element detected in the LCR by bioinformatics approaches. Not all sites predicted from bioinformatics (underlined) could be validated in experimental system, whereas some of them either lacked functional activity (lacked DNA binding) (Martinez-Ramirez et al., 2017), or were inconsequential to the enhancer function of the LCR (Sichero et al., 2012). In addition, a few more transcription factors were found to interact with the LCR (Carson and Khan, 2006) even though their binding sites were not detectable by bioinformatics tools, and expected to be secondary interactions.
Minor changes in sequences within LCR region can lead to loss or addition of the binding sites for transcription factors or the efficiency of their interaction (Dong et al., 1994; Pande et al., 2008; Sichero et al., 2012; Xi et al., 2017). For example, NF-κB binding site in the HPV16 LCR promoter is localized in a small region exhibiting enhancer activity (Chong et al., 1990; Fontaine et al., 2000). However, the binding site of NF-κB contains two mismatches in comparison to the consensus NF-κB Igκ in the HPV16 LCR (Sen and Baltimore, 1986). The NF-κB site also overlaps with a NF-1 binding site. Therefore, it is possible that NF-κB competes/cooperate with NF-1 for DNA binding (Fontaine et al., 2000). Congregation of several sites in a small region also underlines a strong interaction, both positive and negative, among different transcription factors. Host factor YY-1 and viral E2 strikingly have repressor functions (Gloss and Bernard, 1990) and loss of their corresponding sites by mutations or by epigenetic mechanism promote the enhancer function of LCR (Bauknecht et al., 1992; Dong et al., 1994; Reuschenbach et al., 2015). Some of these transcription factors work on palindromic elements. In case of these elements, LCR conserves only half site whereas other half site is highly variable. Therefore, slight change in the non-conserved half site strongly influence transcription factor docking and resultant enhancer function of LCR.
AP-1, a redox sensitive transcription factor (Angel and Karin, 1991), specifically binds to the cis-regulatory elements identical or related to the consensus sequence TGAGTCA of HPV LCR. Four AP-1 binding sites on HPV16 LCR, three sites each on HPV18 and HPV6 LCR, and two sites on HPV11 LCR have been described (Gloss et al., 1989; Chan et al., 1990; Chong et al., 1990; Nakshatri et al., 1990; Thierry et al., 1992; O’connor et al., 1995; O’connor et al., 1996; Wang et al., 2011) (Table 1). Efficient activation of E6/E7 promoter requires the integrity of all the AP-1 elements of HPV16 LCR and mutation in any one cannot be complemented by other motifs (Nakshatri et al., 1990; Thierry et al., 1992). Similarly, binding of FOXA1 and MYC with the promoter and regulatory sequences of HPV16 and HPV18 has been shown in vivo. However, only overexpression of FOXA1 enhanced the LCR activity (Sichero et al., 2012). Two putative GATA3 binding sites within the LCR of HPV18 (7205–7210, TGATTG; 7532–7537 TAATCT) were also reported (Sichero et al., 2012). Three functional binding sites of SOX2 were found in the HPV16-LCR (Martinez-Ramirez et al., 2017). C/EBPβ element showed a context dependent action on HPV LCR. It acted as repressor for HPV11 in keratinocytes as levels of both HPV11 transcripts and HPV DNA increased after treatment with oligomers containing the c/EBPβ DNA binding motif (Wang et al., 1996). c/EBPβ-YY1 complex contributed to cell-type-specific HPV18 LCR activity (Bauknecht et al., 1996). c/EBP binding site overlaps with AP-1 in HPV16 LCR and mutation in this composite site that also eliminates AP-1 binding show increases transcription from early promoter (Liu et al., 2002). Together, these observations reflect a strong influence of host transcription factor composition in the target tissue on expression of viral oncogenes that in turn modify the cellular environment leading to manifestation of pathological characteristics of the HPV-positive tumors.
Table 1.
Transcription factor binding site on LCR of HPV16 (831 bp), HPV18 (835), HPV6 (811 bp), and HPV11 (755 bp).
| Transcription factor(Consensus*) | HPV16 | HPV18 | HPV6 | HPV11 | References | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence | Position | Sequence | Position | Sequence | Position | Sequence | Position | ||
|
AP1 (TGANTCA) |
TGAATCA | 6769–6775 | GTGGTATG | 7349–7356 | TGACTCA | 7435–7440 | TGCATGACTAAT | 7732–7743 | (Gloss et al., 1989; Chan et al., 1990; Chong et al., 1990; Nakshatri et al., 1990; Thierry et al., 1992; O’connor et al., 1995; O’connor et al., 1996; Wang et al., 2011) |
| TGTGTCA | 6786–6792 | TGGTATTA | 7603–7610 | TGTTTAA | 7518–7524 | TGGATTGCAGCCAA | 7776–7790 | ||
| TTAGTCA | 6949–6955 | TGACTAA | 7792–7798 | TTGTAGCA | 7781–7788 | ||||
| TTAGTAT | 7103–7109 | ||||||||
|
NF-κB (GGGRNNYYCC) |
TGCCAAATCCC | 6691–6702 | (Fontaine et al., 2000) | ||||||
|
STAT3 (TT(N)4-6AA) |
TTCAACCGAA | 6584–6594 | TTGAACAA | 7561–7568 | TTCAACCGAA | 46–55 | TTCAACCGAA | 46–55 | (Present study) (Seidel et al., 1995) |
| TTACAAGCAA | 7015–7024 | (Xi et al., 2017) | |||||||
|
SOX-2 (CAATGG/CATTGTT) |
CATTGTT | 6736–6742 | (Martinez-Ramirez et al., 2017) | ||||||
| CATTGTT | 6958–6964 | ||||||||
| CACATGG | 6979–6985 | ||||||||
| GRE/PRE (GGTACANNNTGTTCT) | TGTACATTGTGTCAT | 6779–6793 | AGCACATACTAT ACT | 7839–7853 | GGTACACATTG CCCT | 7630–7644 | GGTACATATTGCCCT | 7674–7688 | (Chan et al., 1989) |
|
YY1 (CCGCCATNTT) |
CCATTTTGTA | 6573–6582 | CCATTTT | 7343–7349 | ACATTTT | 7791–7797 | ACATTTT | 7620–7626 | (O’connor et al., 1995; O’connor et al., 1996) |
| CCATTCCATT | 6730–6739 | CCATTTT | 7443–7449 | ACATATT | 7885–7892 | ACATATT | 7677–7683 | ||
| CCATTGTTTT | 6736–6745 | ACATATT | 7554–7560 | ACATATT | 7914–7920 | ||||
| ACATGAACTG | 6931–6940 | ACATATT | 7692–7698 | ACATCTT | 8–15 | ||||
| TCATACATTG | 6953–6962 | ACATTCT | 7759–7765 | ||||||
| ACATTGTTCA | 6957–6966 | ACATAGT | 7808–7814 | ||||||
| TCATTTGTAA | 6964–6973 | ||||||||
| ACATGGGTGT | 6980–6989 | ||||||||
| GACATTTTATG | 7118–7127 | ||||||||
|
NF1 (TTGGC) |
TGCCAA | 6691–6696 | TTGGC | 7475–7479 | TTGCC | 7559–7563 | TTGCC | 7526–7530 | (Gloss et al., 1989; Chong et al., 1990; Nakshatri et al., 1990; O’connor et al., 1995) |
| TGCCAA | 6724–6729 | CTGGCA | 7513–7518 | TTGCC | 7655–7659 | TTGCC | 7603–7607 | ||
| CGCCAA | 6811–6818 | TTGGC | 7582–7586 | TTGCC | 7738–7742 | TTGCC | 7682–7687 | ||
| TTGGCT | 6849–6854 | TTGGC | 7732–7743 | TTGGC | 7776–7780 | TTGCC | 7699–7703 | ||
| TTGGCA | 6879–6885 | TTGGC | 7776–7790 | TTGGC | 7805–7809 | ||||
| GGCCAA | 6904–6909 | ||||||||
| SP1 (GGGNGG) | GGGCGT | 7070–7075 | GGGAGT | 35–40 | AGGAGG | 28–33 | AGGAGG | 28–33 | (Tan et al., 1992) |
|
TEF-1 (TACATACTTC) |
TGCATGCTTT | 6601–6610 | TGCATTGTAT | 7209–7218 | CACATTTTT | 7780–7788 | TACATATTGC | 7676–7685 | (Ishiji et al., 1992; O’connor et al., 1995) |
| TACATTGTGT | 6781–6790 | TACATATTTT | 7553–7562 | TACATATTTC | 7884–7893 | TACATATTTC | 7903–7922 | ||
| TACATACCGC | 6824–6832 | CACATATTTT | 7691–7700 | ||||||
| CACATATTTT | 6840–6849 | TGCATACTTG | 7704–7713 | ||||||
| TGCATATTTG | 6873–6882 | TACATAGTTT | 7807–7816 | ||||||
| TACATTGTTC | 6956–6965 | CACATACTAT | 7841–7850 | ||||||
|
OCT-1 (AATTGCAT) |
ATTTTGTAG | 6555–6563 | AACTGTAT | 7333–7340 | ATGTGTAT | 7385–7392 | AAAAGCAT | 7795–7802 | (O’connor and Bernard, 1995; O’connor et al., 1995) |
| ATTTTGTAG | 6575–6583 | AACTGCAC | 7403–7410 | AAAAGCAT | 7766–7773 | ATTAGCAG | 84–91 | ||
| AATTGCAT | 6870–6877 | AATTGCAT | 7721–7728 | ||||||
| AACTGCAC | 6974–6981 | AATTGTAG | 16–23 | ||||||
|
c/EBPβ (TKNNGNAAK) |
ACTACTGAAT | 6763–6773 | CTTAAGTAA | 7713–7722 | TTGTGCAAT | 7451–7459 | (Bauknecht et al., 1996; Wang et al., 1996; Liu et al., 2002) | ||
| ATTGTGTCA | 6784–6792 | ||||||||
|
FOXA-1 (MAWTRTTKRYTY) |
CACGTG | 6404–6409 | (Xi et al., 2017) | ||||||
|
FOXA-2 (MAWTRTTKRYTY) |
CACGTG | 6404–6409 | (Martinez-Ramirez et al., 2017) | ||||||
| CGTGTGTA | 6406–6413 | ||||||||
| GTGTATGTGTTT | 6409–6420 | ||||||||
*Consensus sequence represented with IUPAC codes to denote degeneracy. NA; Italic—Bioinformatics analysis; Bold—Experimental result; Underline—Putative binding sites.
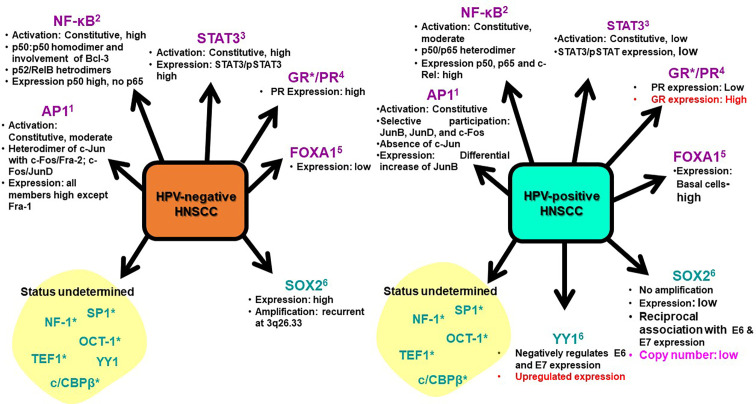
Expression and activation of regulatory host cell factors determine the magnitude, and duration of viral transcription. The transcription factors can be broadly classified as inducible and constitutive based on the fluctuations in their expression and activity levels in the cell. transcription factors AP-1, NF-κB, STAT3, GR, PR, and FOXA1/2 are inducible in nature and their aberrant expression and constitutive activation play an independent role in carcinogenic inflammation, transformation and maintenance of cancer stemness (Angel and Karin, 1991; Rinkenbaugh and Baldwin, 2016). Almost all of these transcription factors have pivotal cellular functions and their temporal expression or changed activation state is sufficient to alter the physiological state of the cell undergoing tumorigenic transformation. Research carried out in recent years have established differential expression and activation of these transcription factors among HPV-positive and negative HNSCC (Gaykalova et al., 2015; Gupta et al., 2015; Verma et al., 2017; Gupta et al., 2018) (Figure 5). Therefore, understanding the expression and activity of host transcription factors that could interact or alter the function of LCR in relation to active viral infection, have been an area of active research in HPV biology and development of anticancer therapeutics. Following section is aimed to review the evidence related to the level of expression and role of these transcription factors in HNSCC in general and HPV-driven HNSCC in particular (Table 2).
Figure 5.
Expression and activity of HPV-related transcription factors in HNSCC. HPV-positive and HPV-negative HNSCC displayed distinguishable transcriptional profiles. Transcription factors broadly classified as inducible (Purple) and constitutively-active (Blue) include: 1. AP1 (Gupta et al., 2015), 2. NF-κB (Fontaine et al., 2000; Bano et al., 2018; Zhang et al., 2018) 3. STAT3 (Gaykalova et al., 2015; Verma et al., 2017), 4. GR/PR (Mohamed et al., 2018; Kost et al., 2019) 5. FOXA1 (Karpathiou et al., 2017; Li et al., 2019) 6. SOX2 (Gut et al., 2018; Dogan et al., 2019) 7. YY1 (Ferris et al., 2005; He et al., 2011). *No specific study in HNSCC. Red text: Data from cervical cancer; Pink text: Data of vulvar carcinoma.
Table 2.
Expression and activity of key HPV-related transcription factors in normal and tumor tissues of H&N region.
| Transcription factor (family members) | Specimen | Transcription factor Activation status | Expression level of transcription factor family members | HPV-specific information | References |
|---|---|---|---|---|---|
|
AP-1 [c-Jun (39 kDa), JunB (36 kDa), JunD (35 kDa) c-Fos (41 kDa), FosB (36 kDa), Fra-1 (29 kDa), Fra-2 (35 kDa)] |
HNSCC tumors (n = 7) and adjacent control tissues | Increased binding of AP-1 in HIV LTR | Nil | Nil | (Zoumpourlis et al., 1994) |
| HNSCC cell lines UM-SCC-1, -9, -11A, -11B, and-38 | Active AP-1 with c-Jun, JunB, JunD and Fra-1 as major participants; Loss of AP-1 site reduced IL-8 promoter activity |
Nil | Nil | (Ondrey et al., 1999) | |
| HNSCC cell lines UM-SCC-9 and -11B | IL-1α-induced activation of AP-1; Loss of AP-1 site moderately reduced IL-8 promoter activity |
Nil | Nil | (Wolf et al., 2001) | |
| HPV16 E7 transgenic mouse model with dominant negative c-Jun under human K14 promoter | c-Jun associated AP-1 activation lost; Prevention of chemical-induced skin papillomas |
NIL | Increased HPV16 E7 expression in DN-c-Jun mice; more papillomas | (Young et al., 2002) | |
| HNSCC cell lines UM-SCC-9 and -11B | EGFR promoted AP-1 activity and downstream expression of VEGF | Nil | Nil | (Bancroft et al., 2002) | |
| HPV immortalized human oral keratinocytes- HOK-16B; | Acetaldehyde activates AP-1 containing c-Jun | Increased c-jun mRNA and protein accumulation | HPV-transformed cells | (Timmons et al., 2002) | |
| HNSCC cell lines: UM-SCC 1, 5, 6, 9,11A, 11B, 22A, 22B, 38, and 46 | Heterogeneity in cell lines with respect to AP-1 regulated gene expression | Nil | Nil, (Cell lines differed in their p53 expression | (Yan et al., 2007) | |
| FFPE-tissues of oral leukoplakias, with different degrees of epithelial dysplasia, and OSCC (n = 50); (HPV positivity-24%) | According to degree of oral dysplasia withing lesion c-Jun nuclear expression increased and greatest expression and nuclear localization in OSCC | Increased expression of c-Jun with increasing severity of the lesion | Malignant progression mediated by c-Jun is independent of the presence of HPV in oral carcinogenesis | (Acay et al., 2008) | |
| Fresh oral tissue specimens (n = 100) | Increased activation of AP-1 with increasing disease severity | Presence of JunD with c-Fos in AP-1 complex in majority of oral cancer; JunB showed sporadic increase in a subgroup | Nil | (Mishra et al., 2010) | |
| HPV immortalized human oral keratinocytes- HOK-16B HNSCC cell line- UM-SCC-9, 11A, 11B, 38 |
Tobacco-carcinogen induced AP-1 reporter gene activity; AP-1 activity dependent on c-Fos expression |
c-Fos expression induced by tobacco carcinogens | IL-8 and VEGF expressed both in HPV-transformed oral keratinocytes and HNSCC cell lines | (Swenson et al., 2011) | |
| Fresh tongue tissue biopsies (n-100), (HPV positivity-28%) Tongue cancer cell lines-HPV16 positive- UPCI:SCC090; HPV negative- AW13516 |
Increased activation of AP-1 with increasing disease severity; Upregulation of downstream target genes: cyclin D1, c-myc, Bcl-xl, MMP-9, EGFR Composition: c-Jun, c-Fos, Fra-2 in HPV-negative and JunD, JunB, c-Fos and Fra-2 in HPV-positive tumors |
Higher expression of all Jun and Fos family members except Fra-1 which showed reciprocal kinetics | Selective participation of JunD and JunB only in HPV16 positive tumors and cell lines; absence of c-Jun in HPV-positive | (Gupta et al., 2015) | |
| HNSCC cell line: HPV-positive- 93VU-147T | Inhibition of AP-1 activation and change in active AP-1 by curcumin | Curcumin induced loss of AP-1 c-Jun, JunD and JunB in HPV-positive oral cancer cells | Nil | (Mishra et al., 2015) | |
| HNSCC cell lines – 26; Mouse model of metastasis | Nil | JunB, Fos, Fra-1, JunD overexpressed in cells with metastatic potential | Nil | (Hyakusoku et al., 2016) | |
| Prospectively collected fresh biopsies - 116 and FFPE -30 from OSCC/OPSCC; Total tissue analyzed (n = 116), FFPE (n = 66) |
Nil | Overexpression of JunD, and c-Fos in SCC as compared to normal; Overexpression of JunB only in HPV-positive tissues | Differential increase in JunB in HPV-positive; Level of JunD upregulated but not differentially active only HPV-positive cancers |
(Verma et al., 2017) | |
| Human OSCC samples (n = 123) | Elevated expression of active AP-1 component phospho-c-Jun in resistant tumors; Active AP-1 strongly correlated with bcl-2 overexpression | c-Jun overexpression in chemo-radioresistant tumors | Nil | (Alam et al., 2017) | |
| HNSCC cell lines: HPV-negative- CAL33Res; HPV-positive- UM-SCC-47 | Overexpression of c-Jun target gene AXL | Silencing of c-JUN and c-FOS expression downregulated AXL expression and enhanced the sensitivity of HPV negative cells | Targeting AP-1 enhanced the antitumor efficacy of BYL719 against HPV positive HNSCC | (Badarni et al., 2019) | |
|
NF-κB [NF-κB1 (50 kDa), NF-κB2 (52 kDa) RelA (65 kDa), RelB (70 kDa) c-Rel (78 kDa)] |
HNSCC cell line: UM-SCC-9, -11B, and -38 | Constitutive and inducible NF-κB, and promoter activity | Overexpression of p65 in cell lines | Nil | (Duffey et al., 1999) |
| Human HNSCC cell lines: UM-SCC-1, 9, 11A, 11B, 38 | Constitutive DNA binding & promoter activity; Constituents p65/Rel A and p50; IL-8 induction: stronger role than AP-1 | Nil | Nil | (Ondrey et al., 1999) | |
| Laryngeal SCC: Tumor and non-tumor laryngeal tissues | Nuclear positivity of p65 | High levels of p65 in cytoplasm, moderate in nucleus | Nuclear p65 correlated with HPV16 E7 level | (Du et al., 2003) | |
| Oral tissue biopsies (n = 110). OSCC (n = 66) OPMD (n = 34), normal (n = 10); HPV positivity = 22%, HR-HPV16 = 18% |
Constitutively active NF-κB; increased with disease severity; Major participants:p50, p65, p52, c-Rel, RelB, and Bcl-3 | Upregulation of p50, p65 and c-Rel with increasing severity of lesion; immunoreactivity for p52, c-Rel and RelB in cancer tissues. | p50/p50 homodimerization common; Involvement of p65 only in NF-κB complex of HPV16-positive | (Mishra et al., 2006) | |
| HNSCC (n = 195); Control (n = 63; non cancer affected patients); Human HNSCC cell lines: Ho1N1, HSC2,and SKN3 | SiRNA mediated downregulation of NF-κB activity; NF-κB nuclear staining: 55.6% HPV-negatives (above median-12.85), HPV-positive below median | Overexpression of RELA, NF-κB1 | Differential gene signatures. Strong NF-κB activation in HPV-negative HNSCC |
(Gaykalova et al., 2015) | |
| OC and OPSCC Tissue biopsies (Fresh, n = 116; and FFPE, n = 66) |
Nil | Variable presence of p50 and p65 in HPV+ and HPV- tissues | p50: equal distribution among HPV-positive and HPV-negative tumors, p65: strong correlation with HPV-postive tumors |
(Verma et al., 2017) | |
| Tongue tissue biopsies (n = 100) TSCC cell line: UPCI:SCC090 and AW13516 |
Increase d activity with severity of disease; p50 and c-Rel forming NF-κB | Differential expression of NF-κB proteins | Selective participation of p65 in NF-κB complex of HPV16-positive HNSCC | (Gupta et al., 2018) | |
| STAT3 (88 kDa) | HNSCC Cell line:YCU-N861, YCU-H891; HNSCC tissues (n = 6) | Constitutive activation of EGFR and STAT3; STAT3DN66 and STAT3DN99 abrogate STAT3 activity |
Constitutive expression of STAT3 | Nil | (Masuda et al., 2002) |
| HNSCC tissues (n = 90) | High level of pSTAT3 in early stages (T1, T2), moderate in late stages (T3, T4); No STAT3 in normal samples | HNSCC - 82% with high or intermediate STAT3; No STAT3 in normal tssues | Nil | (Nagpal et al., 2002) | |
| HNSCC cell lines: HN6, HN12, HN13, HN30, HaCaT, HEK293T, HEK293FT cells; HNSCC FFPE tissues (n = 460) | pSTAT3 induction by IL-6 produced through active NF-κB signalling | STAT3 expression unaffected | Nil | (Squarize et al., 2006) | |
| HNSCC (n = 195); Control (n = 63; non cancer affected patients); Human HNSCC cell lines: Ho1N1, HSC2,and SKN3 | Transcript profiling of STAT3 with NF-κB pathway signature target genes (IRF1, CEBPD, CCND1, ICAM1, JAG1, JAK3, and NOS3); reporter gene expression | Indirect evidence | Specific signature for HPV positive and negative; 49 HPV negative, while 1 HPV positive patients stained for nuclear STAT3 above median | (Gaykalova et al., 2015) | |
| OC and OPSCC Tissue biopsies (Fresh, n = 116; and FFPE, n = 66) |
pSTAT3Y expression | Immunoblotting and Immunohistochemistry: inverse correlation between HPV positivity and STAT3/pSTAT3 expression. | HPV positive: low STAT3/pSTAT3; HPV negative: high STAT3/pSTAT3 | (Verma et al., 2017) | |
|
SOX2 (34 kDa) |
OSCC frozen tumor tissue samples (n = 40) | SOX2 overexpression associated with increased activity | High expression of SOX2 and CyclinE1 in OSCC specimens | Nil | (Freier et al., 2010) |
| HNSCC tumor tissues (n = 496) | – | SOX2 induced BCL-2 and enhanced chemo-resistance | SOX2 amplification in HPV-negative, but no amplification in HPV-positive | (Schrock et al., 2014) | |
| H&N tissue retrospectively collected (n = 94) | Nuclear positivity of SOX2 | SOX2 expression detected in 95% of laryngeal dysplasia | Nil | (Granda-Diaz et al., 2019) | |
| OPSCC (n = 157) | Nil | SOX2 overexpression with poor prognosis | Poorer overall survival in SOX2-amplified HPV-negative cases (p = 0.036) | (Dogan et al., 2019) | |
| Tongue SCC cell line- CAL‐27 | – | EGFR mediated stabilization and upregulation of SOX2 expression | Nil | (Lv et al., 2020) | |
| YY1 (44 kDa) | Laryngeal SCC Cell line - Hep-2; Control- HEK293T | Proliferation and migration with suppression of apoptosis as indicators of increased YY1 activity | YY1 upregulated in LSCC | Nil | (Qu et al., 2017) |
| NPC cases (n = 40); Stage I replication (297 cases and 611 controls); Stage II replication (768 cases and 1526 controls) | YY1-mediated repression of TRIM26 | Nil | Nil | (Lyu et al., 2018) | |
| OSCC tumor tissues (n-30); Cell lines- HEK293T, AW8507 | YY1 mediated reporter gene activation by CARM1 mediated arginine methylation | YY1 overexpressed in oral cancer | Nil | (Behera et al., 2019) | |
| SP1 (80.6 kDa) | Normal oral mucosa (n-8), OSCC (n-10); OSCC cell lines: SCC-15, YD-15 | SP-1 inhibitor downregulated expression of SP-1 and decrease tumor growth | SP-1 was overexpressed in oral tumors compared to normal | Nil | (Shin et al., 2013) |
| NPC tumors (n-82), Metastatic malignant tumors (n-60), Nonmetastatic malignant tumors (n-22) | miR-24 overexpression lead to reduced SP1 activity and inhibited proliferation | Reduced SP1 expression contributed to the reduction in radio-resistance | Nil | (Kang et al., 2016) | |
| OSCC patients (n-55); Cell line- CAL-27 and SCC9 |
Upregulated SP-1 in OSCC | SP-1 is overexpressed in OSCC and could promote cell invasion and migration in OSCC | Nil | (Liu et al., 2019) | |
| OCT-1 (76.4 kDa) | HNSCC cell line: PCI-04A | – | Ionizing radiation induce OCT-1 | Nil | (Meighan-Mantha et al., 1999) |
|
c/EBPβ (76.4kDa) |
HNSCC cell lines: UM-SCC-1, -9, -11A, -11B, and -38 | Constitutive activation | – | Nil | (Ondrey et al., 1999) |
| NPC and non-cancerous NPE (n-33); Cell line- HNE1 and 5–8F | Nil | (Wang et al., 2016) | |||
| FOXA1 (49.1 kDa)/FOXA2 (48.3 kDa) | HNSCC (n = 152) | Nil | FOXA1 expressed in basal cells of squamous epithelium, pre-invasive HNSCC lesions | Due to specificity of FOXA-1 positive site, it may have implication in HPV-mediated pathogenesis | (Karpathiou et al., 2017) |
| NPC tissues (n-114), Non-cancer inflammatory NPE tissues (n-64) | FOXA1 regulated TGF-β-stimulated transcriptome | FOXA1 protein was decreased in NPC cells; loss of FOXA1 associated with lymph node metastasis and poor prognosis. | Nil | (Li et al., 2019) |
AP-1, Activator Protein; DMBA, 7,12-dimethylbenz[a]anthracene; DN, Dominant Negative; EGFR, Epidermal Growth Factor Receptor; FFPE, Formalin-Fixed and Paraffin-Embedded; FOXA1, Forkhead Box A1; FOXA2, Forkhead Box A2; HPV, Human Papillomavirus; HNSCC, Head and Neck Squamous Cell Carcinoma; LSCC, Laryngeal Squamous Cell Carcinoma; NF-κB, Nuclear Factor-κB; NPC, Nasopharyngeal Carcinoma; NPE, Nasopharyngeal Epithelial; STAT3, Signal Transducer and Activator of Transcription – 3; SOX2, sex determining region Y (SRY)-box; SP1, Specificity Protein 1; TSCC, Tongue Squamous Cell Carcinoma; TPA, 12-O-tetradecanoylphorbol-13-acetate; OCT-1, Octamer Transcription Factor-1; OC- Oral Cancer; OSCC, Oral squamous Cell Carcinoma; OPSCC, Oropharyngeal Squamous Cell Carcinoma; OPMD, oral premalignant disease; VEGF, Vascular Endothelial Growth Factor; YY1, Yin Yang 1.
Role of HPV-Related Transcription Factors in H&N Carcinogenesis
AP-1
AP-1 was first discovered as TPA-inducible transcription factor that interacted selectively with sequences in the basal level enhancer of human metallothionein and showed binding to simian virus 40 enhancer region (Lee et al., 1987). Functional AP-1 is Lucine zipper family of transcription factors composed on homodimer of Jun family proteins (c-Jun, JunB and JunD) or its heterodimer with members of Fos family (c-Fos, FosB, Fra-1, and Fra-2). AP-1 is a redox sensor of the cell, which controls cell-proliferation, transformation, its members work as an oncogene [reviewed by (Angel and Karin, 1991; Shaulian and Karin, 2002)]. AP-1 proteins are primarily considered to be oncogenic but JunB, Fra-1 and Fra-2, have been shown to have tumor-suppressor activity (Eferl and Wagner, 2003). Therefore, oncogenic or tumor suppressive activity exhibited by distinct composition of AP-1 vary with cell context and the tumor genetic background. Due to the differential combination of the components, AP-1 is involved in a tissue-specific regulation of target genes. Considering the contrasting action of AP-1 complex, it has been referred as “double-edged sword” in tumorigenesis (Verde et al., 2007).
The first study that showed constitutively active AP-1 in human HNSCC tumors was incidentally meant to measure the activity of HIV LTR bearing AP-1 site (Zoumpourlis et al., 1994). An elevated AP-1 compared to adjacent normal tissues was reported in 7/7 tumor tissues studied. Later, AP-1 potentiated the action of NF-κB in basal and IL-1α-inducible expression of the pro-inflammatory cytokine, IL-8 in HNSCC cell lines (Ondrey et al., 1999; Wolf et al., 2001). AP-1 complex resulted in enhanced survival and proliferation of tumor cells (Wolf et al., 2001; Bancroft et al., 2002). EGFR-mediated signaling was found to contribute to expression and transactivation of AP-1, which participated more profoundly in the expression of angiogenic cytokine VEGF compared to IL-8 (Bancroft et al., 2001). Adverse effects of alcohol on oral epithelial cells are manifest by AP-1. Acetaldehyde, an ethanol metabolite, was found to activate c-Jun expression and dose-dependent activation of c-Jun containing AP-1 in human oral keratinocytes that were transformed by HPV oncoproteins (Timmons et al., 2002). Similarly, tobacco carcinogens independently promoted AP-1 activity, albeit through induction of c-Fos (Swenson et al., 2011). Metabolites of tobacco and alcohol together, promoted carcinogenesis with highly invasive AP-1 containing c-Jun and c-Fos in H&N region that block p53 (Schreiber et al., 1999). Our investigations on oral tissues revealed a weak/moderate level of most AP-1 members in oral mucosa except c-Fos, which was absent; or Fra-1 which showed high levels. AP-1 is seldom active in normal tissues (Mishra et al., 2010). While in precancer lesions, except Fra-1 expression of AP-1 proteins was upregulated and AP-1 activity was composed of JunD/JunD homodimers. Primary oral SCC (OSCC) showed heterodimers of c-Fos/JunD, which constituted the most prevalent AP-1 complex in cancer lesions (Mishra et al., 2010). In a murine metastasis model, HNSCC cell lines that showed lung metastatic potential were found to carry high differential expression of JunB, Fos, Fra-1 and JunD (Hyakusoku et al., 2016). Elevated expression of active phospho-c-Jun was typically reported in resistant tumors and it strongly correlated with bcl-2 overexpression (Alam et al., 2017). Although these studies emphasized the role of AP-1 in initiation, progression and chemoresistance of HNSCC, they did not address the heterogeneity within tumor types with respect to the HPV status and AP-1 components.
In a first landmark study carried out in mouse papilloma model that investigated the role c-Jun by expressing dominant negative c-Jun with HPV16 E7 showed that hyper-proliferative action of E7 was independent of c-Jun tumorigenic potential (Young et al., 2002). Similarly, c-Jun was found functional and got activated in HPV-transformed oral epithelial cells (Timmons et al., 2002). The observations showed that c-Jun, a common initiator in smoking-related AP-1 activation and HPV E7 action did not interfere with each other. Similarly, HPV and c-Jun expression did not show any correlation in premalignant and malignant oral tissues (Acay et al., 2008). Increased activation of AP-1 with increasing disease severity was seen in tongue SCC, which was accompanied with upregulation of downstream target genes: cyclin D1, c-myc, Bcl-xl, MMP-9, and EGFR (Gupta et al., 2015). With increasing disease severity, an increase in expression of all Jun and Fos family members was noted except Fra-1 which showed reciprocal kinetics. Selective participation of JunD and JunB was observed only in HPV16-positive tumors and cell lines, whereas c-Jun was absent in AP-1 complex of HPV-positive tumors. In a confirmatory immunohistochemical staining-based biomarker study on fresh and archival tissue, overexpression of JunD, and c-Fos was confirmed in SCC as compared to normal and an overexpression of JunB was seen only in HPV-positive tissues (Verma et al., 2017). These histological changes correlated well with p16 expression but showed no correlation to EGFR level of the tumor tissues. Overall these studies highlighted contrasting involvement of c-Jun in HPV-negative and JunB and JunD in HPV-positive tumors. Incidentally, c-Jun is known to downregulate p53 and p16 (Schreiber et al., 1999; Passegue and Wagner, 2000), whereas JunB was shown to upregulate the p16 expression (Passegue and Wagner, 2000). c-Jun also bypass PI3K signaling pathway and promote drug resistance by PI3K-independent AXL expression in HPV-negative tumors (Badarni et al., 2019). Further, HPV16 E6 was shown to contribute to AP-1 complex formation by inducing c-Fos after both ligand-dependent and independent EGFR activation (Liang et al., 2011). Independent of the HPV status, c-Jun has shown a poor prognosis in OSCC (Xu et al., 2017).
NF-κB
Nuclear factor-κB was initially described as a nuclear factor required for the transcription of immunoglobulin κ light chain in B lymphocytes (Sen and Baltimore, 1986). NF-κB is counted among a few top pro-carcinogenic transcription factors. Presence of a Rel homology domain is the characteristic feature of all NF-κB members. Rel domain contains a nuclear localization sequence, sequence-specific DNA binding signature, assist dimerization, and interact with the inhibitory protein IκB.There are five proteins in the mammalian NF-κB family that are evolutionarily and structurally conserved and grouped in two classes, namely, Class-I [NF-κB1(p50) and NF-κB2(p52)] and Class-II [RelA(p65), RelB, and c-Rel) [reviewed by (Nabel and Verma, 1993)]. NF-κB/Rel proteins act both in homo- and in heterodimeric form. Most of the NF-κB dimers are activators of transcription; however, the p50/p50 and p52/p52 homodimers can repress the transcription of their downstream target genes (Zhong et al., 2002). NF-κB is an inducible transcription factor that controls the expression of several genes which regulate cell cycle (cyclin D1), cell survival (Bcl-2, Bcl-xL, cIAP), differentiation (p21Cip/Waf1), cell adhesion (VCAM, ECAM-1), growth factors (VEGF), angiogenesis (MMPs), and stemness (Bharti and Aggarwal, 2002; Rinkenbaugh and Baldwin, 2016).
Aberrant activation and dysregulation of NF-κB signalling is reported in many cancer types including colon, breast, pancreatic, leukemia, myeloma, cervical, and HNSCC (Bours et al., 1994; Dokter et al., 1995; Sovak et al., 1997; Wang et al., 1999; Bharti et al., 2003; Prusty et al., 2005; Gupta et al., 2018). NF-κB has a multifaceted role in HNSCC and is reviewed elsewhere (Monisha et al., 2017). Constitutively active NF-κB has been documented in HNSCC cell lines (Ondrey et al., 1999), and primary (Mishra et al., 2006; Bano et al., 2018; Gupta et al., 2018) and recurrent metastatic HNSCC (Li Z. et al., 2015). The expression and activity of NF-κB proteins changed as a function of severity of the oral lesions during oral cancer development (Mishra et al., 2006; Bano et al., 2018; Gupta et al., 2018). NF-κB DNA binding was constituted primarily by the homodimerization of p50 in HPV-negative HNSCC. Gene polymorphisms at NFKB1 coding for p50 at locus rs28362491 have been found associated with susceptibility to HNSCC (Li and Zhang, 2019). Tobacco carcinogens were also found to induce NF-κB activity in HPV-transformed oral cavity cells (HOK 16B cells) (Rohrer et al., 2010).
Because of the overlapping role of NF-κB and AP-1 in inflammation and cancer, many studies investigated the role of these transcription factors together (Ondrey et al., 1999; Wolf et al., 2001; Chen et al., 2008) and their pharmacological targeting effectively attenuated expression of oncogenes HPV16 E6/E7 in HNSCC (Bancroft et al., 2002; Mishra et al., 2015). However, NF-κB showed a slightly skewed and stronger role in regulation of inflammatory mediators, whereas, AP-1 was found to control the angiogenic response. These observations fit well in the context of oral cavity. Local inflammation in periodontitis is associated with presence of soluble CD14 receptors in gingival crevicular fluid by oral epithelial cells (Nicu et al., 2009), which led to activation of NF-κB but not AP-1 (Feghali et al., 2011). This local inflammation and subsequent activation of NF-κB during an early HPV infection may promote LCR activity independent of AP-1 action. Gingival pockets have been referred as reservoirs of HPV and showed suitable environment including basal epithelium (Hormia et al., 2005). On the other hand, AP-1 gene network was reported to enhance epithelial repair in nasal polyposis by oral steroids (Li et al., 2009).
Dissection of NF-κB complex in HNSCC revealed a strong NF-κB activity composed of p50:p50 homodimer in HPV-negative lesions, whereas HPV16-positive lesions showed comparatively lower activity with additional participation of p65 and c-Rel (Mishra et al., 2006). Earlier a moderate intra-nuclear staining compared to high cytoplasmic p65 was noted in HPV16 E7 expressing laryngeal cancer cells earlier (Du et al., 2003). Incidentally, involvement of p65 also resulted in cisplatin-resistance observed in HNSCC (Li Z. et al., 2015). NF-κB activation was negatively influenced by HPV16 E7 by blocking upstream signaling (Spitkovsky et al., 2002) or induced by HPV16 E6 through its PDZ-binding motif (James et al., 2006). A detailed differential gene expression profile analysis revealed a STAT3 and NF-κB target gene signatures could effectively distinguish HPV-positive from HPV-negative HNSCC (Gaykalova et al., 2015). Later, we discovered that direct evaluation of NF-κB active members in association with active STAT3 in immunohistochemical staining can also discriminate HPV-positive from HPV-negative HNSCC. NF-κBp50 was associated with active STAT3 in HPV-negative HNSCC, whereas NF-κBp65 expression and lack of STAT3 was associated with HPV-positive HNSCC (Verma et al., 2017). Co-activated NF-κB and STAT3, often observed in HPV-negative tumors, strongly promoted cell survival through upregulation of BAX/BCL-XL expression in HNSCC (Lee et al., 2008).
Active NF-κB in HPV-positive lesions was found associated with better physical performance of HNSCC patients (Xiao et al., 2018). Coactivation of both classic and alternate NF-κB pathways collectively translates into inflammatory transcriptome in HNSCC (Yang et al., 2019). Recent reports suggest role of TRAF3, often found compromised in HPV-positive HNSCC (Hajek et al., 2017) in preventing nuclear localization of alternate NF-κB complex with p52/RelB constituents in HPV-negative HNSCC (Zhang et al., 2018). Thus, the presence of NF-κB is essential in all types of HNSCC; however, the subtle changes in their composition, activity, expression profile, and target genes correlate with HPV positivity and may also contribute to the prognosis.
STAT3
STAT3 belongs to the family of latent cytoplasmic proteins that are directly activated by tyrosine kinases (Schindler et al., 1992; Shuai et al., 1992) and regulate expression of key genes involved in development, inflammation and wound healing [reviewed by (Yu and Jove, 2004; Leeman et al., 2006; Lai and Johnson, 2010; Johnson et al., 2018)]. STAT3 particularly, is an established oncogene and its aberrant expression and activation is known to promote epithelial carcinogenesis (Zhong et al., 1994; Kim et al., 2007; D’amico et al., 2018). STAT3 has been associated with several types of epithelial and haematological malignancies. Constitutive activation of STAT3 is also a central feature of HNSCC (Song and Grandis, 2000). STAT3 modulates the expression of multitude of genes that are involved in cell proliferation, differentiation, apoptosis, cell cycle regulation, angiogenesis, EMT and immune invasion during development of HNSCC [reviewed by (Lai and Johnson, 2010; Masuda et al., 2010; Johnson et al., 2018)]. STAT3 activation occurs early during HNSCC carcinogenesis due to the autocrine activation of TGF/EGFR signalling (Grandis et al., 1998; Grandis et al., 2000).
Immunohistochemical analysis and protein micro-arrays studies in primary HNSCC tissue specimens have demonstrated constitutive activation of STAT3 (Nagpal et al., 2002; Shah et al., 2006; Weber et al., 2007). An overexpression with constitutively active STAT3 construct in HNSCC cells showed Cyclin D1 overexpression, increased proliferation in vitro and higher rates of in vivo tumor growth in mice (Kijima et al., 2002; Masuda et al., 2002). Inhibition of STAT3 using a dominant negative STAT3 construct, antisense oligonucleotides, a transcription factor decoy or siRNA resulted in decreased saturation density, increased serum dependence, growth inhibition and induction of apoptosis (Grandis et al., 1998; Grandis et al., 2000; Masuda et al., 2002; Leong et al., 2003; Gao et al., 2005; Gao et al., 2006). Further, siRNA administration against STAT3 in the xenograft model demonstrated a decrease in tumor volume and induction of apoptosis (Leong et al., 2003). In contrast, suppression of STAT3 in HNSCC cells promoted secretion of both pro-inflammatory chemokines and cytokines (Albesiano et al., 2010) thus suggestive of immunosuppressive function of STAT3 in HNSCC. Activation of STAT3 upregulated the immune checkpoints PD-1/PD-L1 expression, whereas downregulation of pSTAT3 in the absence of EGFR and PTEN decreased the PD-1/PD-L1 expression (Bu et al., 2017). A differential expression of STAT3 in subtypes of oral lichen planus have recently been noted to contribute to disease progression (Du et al., 2018).
Except in some studies (Pectasides et al., 2010; Mazibrada et al., 2014), presence of active nuclear STAT3 in HNSCC was invariably associated with poor prognosis (Masuda et al., 2010; Macha et al., 2011; Jinno et al., 2015; Gao et al., 2016; Lesinski et al., 2019). Emerging research on the functional regulatory role of STAT3 revealed its strong correlation with particularly HPV-negative HNSCC making it a compelling target particularly in HPV-negative HNSCC (Gaykalova et al., 2015). HPV positive HNSCC, however, showed a strong negative correlation with active STAT3 in HNSCC (Verma et al., 2017), which correlated well with the differential transcript profile of STAT3 target genes (Gaykalova et al., 2015). Elevated STAT3 and upstream signalling in HPV-positive HNSCC have been reported in some studies (Mazibrada et al., 2014; Chuerduangphui et al., 2016), though the reason behind such contrasting observations is unclear. In sharp contrast to HNSCC, STAT3 in cervical cancer was found aberrantly expressed and constitutively activated in HPV-induced cervical carcinogenesis (Shukla et al., 2010; Shukla et al., 2019) and the level of active STAT3 was found strongly correlated with the physical state of HPV genome in cervical precancer and cancer tissues (Shukla et al., 2019). Therefore, the involvement of STAT3 in HPV-positive epithelial malignancies is a tissue context dependent; however, its influence on pathological manifestation of HPV genome cannot be overlooked in HNSCC. It can be hypothesized that high STAT3 might upregulate HPV replication involved in an early phase of life cycle through HPV E6-driven NF-κB (Morgan et al., 2018; Morgan and Macdonald, 2019). However, such assumption needs to be tested rigorously in experimental systems.
SOX2
SOX2 gene was discovered in 1994 (Stevanovic et al., 1994). SOX2 transcription factor is a member of the SoxB1 group (Kijima et al., 2002; Leong et al., 2003). The protein family has a highly conserved DNA binding HMG (High mobility group) box consisting of about 80 amino acids. Sox proteins function majorly in embryonic development and cell fate determination. SOX2 is involved in stem cell maintenance. Amplification of SOX2 is common in different cancer types, which adversely effects cancer cell physiology via altering stem cell signaling pathways (Bass et al., 2009; Wuebben and Rizzino, 2017). SOX2 is activated by recurrent 3q26.33 amplifications in squamous cell carcinoma (Hussenet et al., 2010).
In HNSCC expression of SOX2 is correlated with poor prognosis (Leeman et al., 2006; Weber et al., 2007). SOX2 upregulates Cyclin B1 and promotes cell proliferation and leads to HNSCC cell dedifferentiation (Lee et al., 2014). In oral carcinogenesis, SOX2 is activated via gain of gene copy number (Freier et al., 2010). In laryngeal cancer, SOX2 promotes migration and invasion by inducing MMP-2 via the PI3K/Akt/mTOR pathway (Yang et al., 2014). SOX2 was found during early tumorigenesis of laryngeal cancer and its expression could be used as independent predictor of laryngeal cancer risk in patients with precancerous lesions (Granda-Diaz et al., 2019). On the contrary, EGFR in HNSCC enhances the stemness and progression of oral cancer through inhibition of SOX2 degradation (Lv et al., 2020). SOX2 is a marker for cancer stem cell in head and neck cancer (Dong et al., 2014), and is a prognostic marker with unfavorable outcome (Germain and Frank, 2007). SOX2 induced expression of the antiapoptotic protein BCL-2 and enhanced resistance to apoptosis-inducing chemotherapy agents including cisplatin, indicating SOX2 as a mediator of chemotherapy resistance in human HNSCC (Schrock et al., 2014). Further, HPV-positive HNSCC lacked SOX2 amplification and lower SOX2 protein expression as compared to HPV-negative tumors. SOX2 overexpression was associated with poor survival and negative HPV status of OPSCC (Dogan et al., 2019). Three putative sites of SOX2 were present exclusively in the enhancer region of HPV16-LCR and overexpression of this transcription factor led to repression of transcriptional activity of HPV16-LCR by decreasing the expression of E6 and E7 endogenous HPV oncogenes (Martinez-Ramirez et al., 2017). Notably, SOX2 is expressed in normal stem cells of gingiva (Soanca et al., 2018), was detectable in oral leukoplakia (Luiz et al., 2018) and have been implicated in oral carcinogenesis (Qiao et al., 2014).
Glucocorticoid Receptor (GR)/Progesterone Receptor (PR)
Glucocorticoid receptor is a hormone-inducible transcription factor (Ostlund Farrants et al., 1997). Glucocorticoids are a class of corticosteroids and act as ligand for glucocorticoid receptor (GR) (86 kDa) is an evolutionally conserved nuclear receptor superfamily protein and mediates diverse actions of glucocorticoid hormones by acting as a ligand-dependent transcription factor. In absence of ligand, GR reside primarily in cytoplasm of cell as a part of large multiprotein complex (receptor polypeptide and two molecules of hsp90 and other proteins), after interaction with ligand it dissociates from hsp and translocates to nucleus. In nucleus, GR binds to glucocorticoid receptor element (GRE) in promoter region of target genes (Tomoshige, 2017). In regulating stress response, a negative cross talk between AP-1 and GR has been reported which states that AP-1 increased level of circulating glucocorticoids, which reciprocally downregulated AP-1 transcriptional activity (Karin and Chang, 2001). A complex of AP-1 and GR function together as a composite site in HPV16 expression (Mittal et al., 1994). In nasal polyposis, steroids synergize with AP-1 to enhance epithelial repair through upregulation of AP-1 gene network (Li et al., 2009).
Glucocorticoids inhibit NF-κB activation as active GR binds to p65 subunit and prevents activation of inflammatory genes (Didonato et al., 1996; Barnes and Karin, 1997). GR interacts with NF-1, this interaction modulates activity of promoter in mouse mammary tumor virus (MMTV) (Hartig and Cato, 1994). GR inhibits p53 functions by forming a complex that mutually represses each other’s transcriptional activity (Sengupta et al., 2000). In HPV16 E6 and E7 oncogenes expression increased by steroid hormones and degraded p53 gene product and led to carcinogenesis (Moodley et al., 2003). GR has been reported to be a transcription factor associated with the regulation of HPV gene expression. HPV16 LCR contains enhancer element for GR (Moodley et al., 2003). Glucocorticoid hormone exerts regulatory action on HPV16 gene expression during growth and differentiation of cervical cancer (Khare et al., 1997). In prostate cancer GR increases radioresistance and triggers androgen hormone independence (Chen et al., 2019). However, there is no study so far on GR in relation to HNSCC.
Progesterone receptor (PR) (99 kDa) is a master regulator in female reproductive tissues that controls developmental processes, proliferation and differentiation during the reproductive cycle and pregnancy, however, its expression in low in males. GR and PR transcription factors bind to same consensus sequence (Jacobsen and Horwitz, 2012; Grimm et al., 2016). PR after engaging with the Progesterone dimmerizes and translocates to the nucleus. Hormone in oral contraceptive pills have been reported to enhance tumorigenesis in cervical region (De Villiers, 2003). Increased PR expression has been associated with the proliferation of normal cervical squamous epithelium, which is probably induced by HPV infection, however this is typical in neoplastic cervical squamous epithelium (Konishi et al., 1991; Monsonego et al., 1991). In contrast, high PR expression was associated with poor disease-specific and locoregional recurrence-free survival in OSCC in HPV-negative tumors (Mohamed et al., 2018).
Yin Yang 1 (YY1)
Yin Yang 1 is ubiquitously expressed, evolutionarily conserved zinc finger protein that belongs to the GLI-Krüppel gene family (Shi et al., 1991). It plays a critical regulatory role in expression of various gene sets and its function in cancerous environment is subtle (Thiaville and Kim, 2011). YY1 acts as a transcriptional repressor (Galvin and Shi, 1997), and plays oncogenic and proliferative role in carcinogenesis (Zhang et al., 2011). Oncogenic role of YY1 in different cancers have been shown (Seligson et al., 2005; Chinnappan et al., 2009; Lee et al., 2012; Wan et al., 2012; Zhang et al., 2013; Zhang J. J. et al., 2014; Liu D. et al., 2018). YY1 binds with transcriptional coactivator CREB-binding protein and represses AP-1 activity. Highly conserved AP-1 site was identified as a target of YY1 mediated repression in HPV LCR (O’connor et al., 1996). The binding of YY1 to its cognate site interferes with the formation of HPV16 transcription initiation complex at the P97 promoter for the E6/E7 genes (O’connor et al., 1996). Sequence analysis of the HPV16 LCR from in vitro HNSCC model expressing high level of E6 gene expression revealed a deletion of 163 bp that included two binding sites of YY1 known to negatively regulate HPV16 E6 and E7 expression (Ferris et al., 2005). YY1 promotes migration, proliferation and suppression of apoptosis in laryngeal cancer via directly inhibiting MYCT1, which can be used as target to prevent the progression of laryngeal cancer (Qu et al., 2017). YY1 attenuates viral early gene expression via CTCF-YY1-dependent looping in the HPV-infected undifferentiated cells. However, during cellular differentiation, YY1 protein expression is dramatically reduced and viral oncogenes E6 and E7 are upregulated, suggesting that CTCF (chromatin-organizing CCCTC-binding factor)-YY1 loop could be disrupted in HPV-mediated carcinogenesis (Pentland et al., 2018). Downregulation of YY1 has been correlated with low immunogenic role in nasopharyngeal carcinoma (Lyu et al., 2018). YY1 in association with CARM1 promotes oral cancer carcinogenesis (Behera et al., 2019). YY1 modulates inflammatory cytokines indirectly in gingival epithelial cells (Nakayama et al., 2019). Therefore, presence of high level of YY1 in pre-neoplastic tissues prevents transcriptional activation of HPV and takes the tumor to a fate resembling HPV-negative tumors.
Nuclear Factor–1 (NF-1)
NF-1 is a family of ubiquitous nuclear transcription factors with subtypes NF-1A (55 kDa), NF-1B(47 kDa), NF-1C(55.6 kDa), and NF-1X(48.8 kDa), which specifically bind as dimers to the palindromic consensus DNA sequence. NF-1 is also known as CAAT box-binding transcription factor (CTF). Oxidative inactivation plays an important role in the DNA-protein interaction, considerably reducing the affinity of the transcription factor NF-1 for DNA (Bandyopadhyay and Gronostajski, 1994). NF-1 family CTF/NF-1 transcription factors act as potent genetic insulators for integrating gene transfer vectors (Gaussin et al., 2012). The strict correlation between the activation or lack of activation of the HPV16 enhancer and cell-specific subsets of NF-1 proteins argues for the pivotal role of NF-1 binding sites in the epithelial cell-specific function of the viral enhancer (Apt et al., 1993). NF-1 and p53 play an important role in glioma initiation and progression (Gonzalez et al., 2018). In HPV16 genome NF-1 binding sites were found methylated in high grade disease, in cervical cancer methylation percentages of E6 and E7 CpG were elevated (Kottaridi et al., 2019). In HPV11, mutation in NF-1 binding site reduces promoter activity in differentiating cells (Zhao et al., 1999). Despite a strong redox regulated transcription factor related to HPV, status and role of NF-1 in HNSCC has not been evaluated till date.
Specificity Protein 1 (SP1)
Specificity Protein 1 (SP1) belongs to SP/KLF transcription factor family, which represents the major group of zinc-finger DNA binding proteins. It was first identified as a promoter‐specific binding factor that is essential for transcription of the SV40 major Immediate Early (IE) gene and shown to interact with GC and GT oligonucleotide sequences that are typically found in diverse viral and cellular gene promoter (Dynan and Tjian, 1983; Safe and Kim, 2004). It is estimated that at least 12,000 SP1 binding sites are present within human genome, and most studies support the idea that SP1 not only maintains basal transcription, but also contributes to the regulation, i.e., induction and inhibition, of transcription of a large number of cellular genes (Cawley et al., 2004; Oleaga et al., 2012).
SP1 is overexpressed in cancer cells and upregulates genes that enhance proliferation, invasion, metastasis stemness and chemoresistance. It is reported to be over‐expressed in a number of cancers, including breast (Oleaga et al., 2012), gastric (Yao et al., 2004), pancreatic (Jiang et al., 2008), lung (Hsu et al., 2012), glioma (Guan et al., 2012) and thyroid cancer (Chiefari et al., 2002). SP1 regulates the metastasis in LSCC, promotes cell migration and invasion in OSCC by upregulating Annexin A2 transcription, and downregulation of SP1 inhibits the growth of oral cancer cell SCC-15, YD-15 (Cai et al., 2012; Shin et al., 2013; Liu et al., 2019). Promoters of HPV are known to have the binding site of SP1 transcription factor which activates the transcription of HPV16 genes and regulate the replication of HPV18 (Tan et al., 1992; Demeret et al., 1995; Apt et al., 1996).
Overexpression of SP1 has been observed in HNSCC as compared to normal oral mucosa, and its inhibition downregulates growth in oral cancer cell lines (Shin et al., 2013). SP1 serves as a therapeutic target against oral cancer (Shin et al., 2012; Yu et al., 2013; Chae et al., 2014; Sun et al., 2016; Zhang et al., 2019; Zhao et al., 2019; Gao et al., 2020; Liu et al., 2020). SP1 is a functional target of miR-24. Nasopharyngeal carcinoma showed low level of miR-24. When miR24 was ectopically expressed in these cells, radiosensitivity was increased with a decline in level of SP1 (Kang et al., 2016). Involvement of SP1 with PI3K pathway was reported in EFGR expression by activin A in HNSCC (Tsai et al., 2019). Cell migration and invasion of OSCC was promoted by SP1 through Annexin A2. Annexin A2 is an oncogene, which has three putative binding sites of SP1 at its promoter region (Liu et al., 2019). Despite a critical transcription factor, there are only limited studies on this transcription factor with no HPV-specific information in HNSCC.
Transcriptional Enhancer Factor-1 (TEF-1)
TEF-1 (47.9 kDa) is also known as TEAD1- TEA domain family member 1. It was the first known transcription factor of TEAD family (Xiao et al., 1991). TEF-1 is a part of Hippo signaling pathway which is involved in organ size control and act as a tumor suppressor by restricting proliferation and promoting apoptosis. TEF-1 functions majorly in development of skeletal and cardiac muscle (Chen et al., 1994; Anbanandam et al., 2006). On HPV16 p97 promoter region, binding of TEF-1 is necessary for its activity, both TEF-1 and its co-activator are essential for transcription of HPV16 and are found active in human keratinocytes (Ishiji et al., 1992). A TATA-less promoter containing binding sites for ubiquitous transcription factors mediates cell type-specific regulation of the gene for TEF-1 (Boam et al., 1995). Even though TEF-1 is essential for HPV transcription, relation of TEF-1 in HNSCC has not been explored as yet.
OCT-1
Oct1 (POU2F1), stands for Octamer transcription factor-1 and is a OCT/POU family member, which was identified as a protein associated with regulatory DNA binding sites in animal viruses [adenovirus (Pruijn et al., 1986), SV-40 (Pruijn et al., 1986), and herpes simplex virus-1 (Gerster and Roeder, 1988)]. It binds to octamer motif (5’-ATTTGCAT-3’) and activates the promoters of variety of genes and of small nuclear RNAs (snRNA). OCT-1 regulates the genes associated with proliferation and immune modulation. Its recently identified targets are associated with oxidative and cytotoxic stress resistance, metabolic regulation, and stem cell function (Vazquez-Arreguin and Tantin, 2016). OCT-1 regulates gene expression via nuclear organization, controls the transcriptional regulation and affects tumor development (Malhas et al., 2009). OCT-1 mediates epithelial-mesenchymal transition in colorectal cancer and is associated with tumor progression and poor patient survival (Li Y. et al., 2015). Similar to some of basal transcription factors stated above, role of this transcription factor in HNSCC and HPV infection remains undefined.
CCAAT Enhancer Binding Protein Beta (c/EBPβ)
c/EBPβ/NF-IL6 belongs to basic leucine zipper family domain. It can also form heterodimers with the related proteins c/EBPα, c/EBPδ, and c/EBPγ. c/EBPβ transcription factor is involved in cell proliferation and tumorigenesis (Ondrey et al., 1999). c/EBPβ expression is stimulated with IL-6, glucogon, IL-1, lipopolysaccharide (LPS), and glucocorticoid, which suggests its role in the mediation of the inflammatory response (Akira et al., 1990; Alam et al., 1992; An et al., 1996; Matsuno et al., 1996). c/EBPβ is constitutively active in HNSCC cell lines (Ondrey et al., 1999). c/EBPβ act as repressor for HPV11 in keratinocytes as levels of both HPV11 transcripts and HPV DNA increased after treatment with oligomers containing the c/EBPβ DNA binding motif that neutralized endogenous c/EBPβ (Wang et al., 1996). A novel c/EBPβ-YY1 complex that binds the switch region and contributes to cell-type-specific HPV-18 LCR activity (Bauknecht et al., 1996). c/EBPβ is a repressor of E6/E7 promoter and its binding site overlaps with AP-1 in HPV16 LCR (Liu et al., 2002). HPVE2 interacts with c/EBP factors, and enhances keratinocyte differentiation in HPV-induced lesions (Hadaschik et al., 2003). The role of c/EBPβ in HNSCC has not been explored yet.
Forkhead Box A1 (FOXA1)/Forkhead Box A2 (FOXA2)
FOXA1 (Forkhead Box A1) and FOXA2 (48.3 kDa) are members of FOX (forkhead box) family. FOXA1 is also known as Heaptocyte Nuclear Factor 3 (HNF3α) and FOXA2 as (HNF3β) (Xanthopoulos et al., 1991; Bingle and Gowan, 1996). Forkhead proteins play an essential role in Estrogen receptor (ER) binding and ER-mediated gene transcription (Carroll et al., 2005). An oncogenic role of HNF3α (FOXA1) was observed in the progression and development of lung and esophageal adenocarcinoma (Yu and Jove, 2004). FOXA1 served as an important mediator for transcriptional activation of p16 gene induced by replicative and oncogene-induced senescence (Schindler et al., 1992). Role of FOXA1 has been elaborated in many cancers, prostate cancer cell proliferation and cancer progression was mediated through FOXA1 by targeting the tumor suppressor gene IGFBP-3 (Song and Grandis, 2000). Expression of FOXA1 can be used as a biomarker to differentiate the breast carcinoma from other carcinomas (Heinrich et al., 1998). Growth in lung adenocarcinoma was regulated by FOXA1 and FOXA2 activity (Grandis et al., 2000). FOXA1 is expressed in basal cells of squamous epithelium in dysplastic and invasive lesions of cervical and head and neck carcinomas (Yue and Turkson, 2009). FOXA1 is able to bind with HPV genome and, therefore, acts as a potential positive regulator (Shuai et al., 1992). FOXA1 is found in pre-cancerous lesions of head and neck origin supporting its role in HPV pathogenesis (Cripe et al., 1987).
Similarly, FOXA2 also has oncogenic potential and its role has been elucidated in many cancers. In uterine cancer, FOXA2 is frequently mutated and act as a pathogenic driver gene in the etiology of this cancer (Le Gallo et al., 2018). In pancreatic cancer, FOXA2 controls the cis-regulatory networks in a differentiation grade-specific manner (Milan et al., 2019). FOXA2 is overexpressed in ovarian cancer stem cells and regulates its autophagy activity (Peng et al., 2017). FOXA2 inhibited EMT in breast cancer cells by regulating the transcription of EMT-related genes such as E-cadherin and ZEB2 (Zhang et al., 2015). FOXA2 has a tumor suppressor function through inhibition of pancreatic cancer cell growth, migration, invasion, and colony formation (Vorvis et al., 2016). The role of FOXA2 in HNSCC has not been elucidated so far.
Discussion
Overall, the evidence presented above demonstrates that HPV positive and negative HNSCC are clearly two distinct entities that notably differ in their clinico-pathological and molecular characteristics. The striking difference in disease progression of two lesion types may be associated with a differential expression and activity profile of key transcription factors responsible for transcriptional activity of HPV oncogenes. Apart from regulation of oncogenic HPVs, the pro-carcinogenic role of many of these transcription factors is well-established. Any change/absence of this HPV-permissive profile in host cell milieu could be the rationale behind clearance of HPV infection or absence of transcriptionally-active HPV leading to a different pathologic manifestation and treatment response of the disease that resemble HPV-negative HNSCC. Therefore, such tumors even though are positive for HPV DNA, fail to show better prognosis.
Despite HPV positive HNSCC represent as a separate class, all tissues of H&N region or all individuals affected by HNSCC may not have the opportunity of oral HPV exposure during initiation or progression of the disease. However, there may be an underlying predisposition of the tumor tissues in terms of availability HPV-permissive transcription factor cocktail and downstream transcriptome (Figure 6). In the absence of HPV infection, such cases go unnoticed and clubbed with HPV-negative HNSCC and could be effectively treated with less aggressive treatment regimen. This special class of HPV-positive HNSCC is expected to have a better treatment outcome. However, this hypothesis remains to be tested in a clinical set-up.
Figure 6.
Transcription factor-based classification of HPV positive and HPV negative HNSCC. (A) Schematic representation of molecular sub-types of HNSCC. HPV negative HNSCC collectively represent tumors with availability of HPV-permissive transcription factors that are never exposed to HPV infection in the lifetime and HPV negative tumors, which are driven by genetic mutations in p53 or other such genes. The later represent tumors with poorer prognosis. Further, not all individuals that encounter HPV in H&N region develop cancer. HPV positivity is reported in oral rinse of normal individual. HPV-permissive transcription factor cocktail if present is expected to direct treatment outcome similar to the tumor-tissue with HPV-driven transcription profile. In the absence of HPV infection, such cases go unnoticed and clubbed with HPV negative HNSCC and could be effectively treated with less aggressive treatment regimen and likely to have a better treatment outcome. (B) Expression of viral oncogenes under the influence of HPV-permissive transcription factor milieu lead to functional inactivation of p53 and feeds to a positive feedback look that maintains the HPV-permissive transcription factor milieu. On the other hand, HPV negative tumors, particularly with mutations in p53, in consort with other procarcinogenic transcription factors like STAT3 drive tumors that show poorer response to anti-cancer therapies.
Present review and the evidence presented herein strongly support towards a need to collectively monitor activity and expression of these HPV-permissive transcription factors. However, required molecular diagnostic test to address these parameters is currently non-existent. In the absence of a direct test(s), the activity of these transcription factors can be assessed by evaluating each transcription factor separately (Mishra et al., 2006; Hussain et al., 2009; Shukla et al., 2010; Gupta et al., 2015; Mishra et al., 2015; Verma et al., 2017; Gupta et al., 2018), or by evaluating the transcripts of their respective downstream target genes by next generation sequencing, or similar platforms as described (Gaykalova et al., 2015). Apart from serving as prognostic tools, these potent procarcinogenic transcription factors may serve as molecular targets for pharmacological interventions in both HPV-positive and HPV-negative HNSCC.
Author Contributions
NA participated in study writing and manuscript preparation. JY, KT, RB, AC, and TT contributed in manuscript preparation. MK assisted in manuscript preparation. ACB conceived the presented idea and designed the manuscript, critically reviewed, and drafted and communicated the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by research funds to ACB from the Department of Biotechnology, Government of India (DBT:6242-P34/RGCB/PMD/DBT/ALCB/2015) and Senior Research Fellowship to KT, AB and MJ from Indian Council of Medical Research (5/13/38/2014 NCDIII-Eoffice73143), (2017-2834/CMB/BMS) and (3/2/2/278/2014-NCD III); Junior Research Fellowship to NA (09/045(1622)/2019-EMR-I), JY (09/045(1629)/2019-EMR-I), RB (09/045(1672)/2019-EMR-I) and US (09/045(1584)/2018-EMR-I) by Council of Scientific and Industrial Research (CSIR); Senior Research Fellowship to TS (2061430699 22/06/2014(i)EU-V), and Junior Research Fellowship to AC [573(CSIR-UGC NET JUNE 2017)] by University Grants Commission (UGC); Junior Research Fellowship to TT by Department of Science and Technology, India (EMR/2017/004018/BBM); and The Teachers Associateship for Research Excellence (TARE) from the Science and Engineering Research Board (SERB), India to MKK (TAR/2018/001054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AP-1, activator protein-1; APC, annual percentage change; CT, chemotherapy; DFS, disease free survival; ECOG, Eastern Cooperative Oncology Group; EGFR, epidermal growth factor receptor; FFPE, formalin-fixed, paraffin-embedded; HPV human papillomavirus; HR, hazard ratio; H&N, head and Neck; HNSCC, head and neck squamous cell carcinoma; NF-κB, nuclear factor-kappa B; OPSCC or OPC, oropharyngeal squamous cell carcinoma; OR-odds ratio; OS, overall survival; OSCC, oral squamous cell carcinoma; PCR, polymerase chain reaction; PFS, progression-free survival; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; R/M, recurrent and/or metastatic; SCC, squamous cell carcinoma; SEER, Surveillance, Epidemiology, and End Results program registries; STAT3-signal transducer and activator of transcription 3; TRAF3, TNF receptor associated factor 3; UADT, upper aero digestive tract; WDSCC, well differentiated squamous cell carcinoma.
References
- Acay R. R., Dos Santos E., De Sousa S. O. (2008). Correlation between c-Jun and human papillomavirus in oral premalignant and malignant lesions. Oral. Oncol. 44, 698–702. 10.1016/j.oraloncology.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Adjei Boakye E., Buchanan P., Hinyard L., Osazuwa-Peters N., Schootman M., Piccirillo J. F. (2018). Incidence and Risk of Second Primary Malignant Neoplasm After a First Head and Neck Squamous Cell Carcinoma. JAMA Otolaryngol. Head Neck Surg. 144, 727–737. 10.1001/jamaoto.2018.0993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., et al. (1990). A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9, 1897–1906. 10.1002/j.1460-2075.1990.tb08316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam T., An M. R., Papaconstantinou J. (1992). Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267, 5021–5024. 10.1007/s00784-017-2074-6 [DOI] [PubMed] [Google Scholar]
- Alam M., Kashyap T., Pramanik K. K., Singh A. K., Nagini S., Mishra R. (2017). The elevated activation of NFkappaB and AP-1 is correlated with differential regulation of Bcl-2 and associated with oral squamous cell carcinoma progression and resistance. Clin. Oral. Invest. 21, 2721–2731. 10.1007/s00784-017-2074-6 [DOI] [PubMed] [Google Scholar]
- Albesiano E., Davis M., See A. P., Han J. E., Lim M., Pardoll D. M., et al. (2010). Immunologic consequences of signal transducers and activators of transcription 3 activation in human squamous cell carcinoma. Cancer Res. 70, 6467–6476. 10.1158/0008-5472.CAN-09-4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M. R., Hsieh C. C., Reisner P. D., Rabek J. P., Scott S. G., Kuninger D. T., et al. (1996). Evidence for posttranscriptional regulation of C/EBPalpha and C/EBPbeta isoform expression during the lipopolysaccharide-mediated acute-phase response. Mol. Cell Biol. 16, 2295–2306. 10.1128/MCB.16.5.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbanandam A., Albarado D. C., Nguyen C. T., Halder G., Gao X., Veeraraghavan S. (2006). Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc. Natl. Acad. Sci. U. S. A. 103, 17225–17230. 10.1073/pnas.0607171103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang K. K., Harris J., Wheeler R., Weber R., Rosenthal D. I., Nguyen-Tan P. F., et al. (2010). Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 363, 24–35. 10.1056/NEJMoa0912217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Karin M. (1991). The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072, 129–157. 10.1016/0304-419X(91)90011-9 [DOI] [PubMed] [Google Scholar]
- Apt D., Chong T., Liu Y., Bernard H. U. (1993). Nuclear factor I and epithelial cell-specific transcription of human papillomavirus type 16. J. Virol. 67, 4455–4463. 10.1128/JVI.67.8.4455-4463.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt D., Watts R. M., Suske G., Bernard H. U. (1996). High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology 224, 281–291. 10.1006/viro.1996.0530 [DOI] [PubMed] [Google Scholar]
- Arany I., Grattendick K. G., Tyring S. K. (2002). Interleukin-10 induces transcription of the early promoter of human papillomavirus type 16 (HPV16) through the 5’-segment of the upstream regulatory region (URR). Antiviral Res. 55, 331–339. 10.1016/S0166-3542(02)00070-0 [DOI] [PubMed] [Google Scholar]
- Badarni M., Prasad M., Balaban N., Zorea J., Yegodayev K. M., Joshua B. Z., et al. (2019). Repression of AXL expression by AP-1/JNK blockage overcomes resistance to PI3Ka therapy. JCI Insight 5, 1–8. 10.1172/jci.insight.125341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft C. C., Chen Z., Dong G., Sunwoo J. B., Yeh N., Park C., et al. (2001). Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin. Cancer Res. 7, 435–442. [PubMed] [Google Scholar]
- Bancroft C. C., Chen Z., Yeh J., Sunwoo J. B., Yeh N. T., Jackson S., et al. (2002). Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-kappaB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. Int. J. Cancer 99, 538–548. 10.1002/ijc.10398 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Gronostajski R. M. (1994). Identification of a conserved oxidation-sensitive cysteine residue in the NFI family of DNA-binding proteins. J. Biol. Chem. 269, 29949–29955. [PubMed] [Google Scholar]
- Bano N., Yadav M., Mohania D., Das B. C. (2018). The role of NF-kappaB and miRNA in oral cancer and cancer stem cells with or without HPV16 infection. PloS One 13, e0205518. 10.1371/journal.pone.0205518 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Barnes P. J., Karin M. (1997). Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336, 1066–1071. 10.1056/NEJM199704103361506 [DOI] [PubMed] [Google Scholar]
- Bass A. J., Watanabe H., Mermel C. H., Yu S., Perner S., Verhaak R. G., et al. (2009). SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242. 10.1038/ng.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauknecht T., Angel P., Royer H. D., Zur Hausen H. (1992). Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: interaction with the transcriptional repressor YY1. EMBO J. 11, 4607–4617. 10.1002/j.1460-2075.1992.tb05563.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauknecht T., See R. H., Shi Y. (1996). A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J. Virol. 70, 7695–7705. 10.1128/JVI.70.11.7695-7705.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty B. T., Moon D. H., Shen C. J., Amdur R. J., Weiss J., Grilley-Olson J., et al. (2019). PIK3CA mutation in HPV-associated OPSCC patients receiving deintensified chemoradiation. J. Natl. Cancer Inst. 112, 855–858. 10.1093/jnci/djz224 [DOI] [PubMed] [Google Scholar]
- Behera A. K., Kumar M., Shanmugam M. K., Bhattacharya A., Rao V. J., Bhat A., et al. (2019). Functional interplay between YY1 and CARM1 promotes oral carcinogenesis. Oncotarget 10, 3709–3724. 10.18632/oncotarget.26984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman T. A., Schiller J. T. (2017). Human papillomavirus in cervical cancer and oropharyngeal cancer: One cause, two diseases. Cancer 123, 2219–2229. 10.1002/cncr.30588 [DOI] [PubMed] [Google Scholar]
- Bernard H. U., Burk R. D., Chen Z., Van Doorslaer K., Zur Hausen H., De Villiers E. M. (2010). Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401, 70–79. 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti A. C., Donato N., Singh S., Aggarwal B. B. (2003). Curcumin (diferuloylmethane) down-regulates the constitutive activation of nuclear factor-kappa B and IkappaBalpha kinase in human multiple myeloma cells, leading to suppression of proliferation and induction of apoptosis. Blood 101, 1053–1062. 10.1182/blood-2002-05-1320 [DOI] [PubMed] [Google Scholar]
- Bharti A. C., Aggarwal B. B. (2002). Nuclear factor-kappa B and cancer: its role in prevention and therapy. Biochem. Pharmacol. 64, 883–888. 10.1016/S0006-2952(02)01154-1 [DOI] [PubMed] [Google Scholar]
- Bingle C. D., Gowan S. (1996). Molecular cloning of the forkhead transcription factor HNF-3 alpha from a human pulmonary adenocarcinoma cell line. Biochim. Biophys. Acta 1307, 17–20. 10.1016/0167-4781(96)00058-9 [DOI] [PubMed] [Google Scholar]
- Boam D. S., Davidson I., Chambon P. (1995). A TATA-less promoter containing binding sites for ubiquitous transcription factors mediates cell type-specific regulation of the gene for transcription enhancer factor-1 (TEF-1). J. Biol. Chem. 270, 19487–19494. 10.1074/jbc.270.33.19487 [DOI] [PubMed] [Google Scholar]
- Boscolo-Rizzo P., Pawlita M., Holzinger D. (2016). From HPV-positive towards HPV-driven oropharyngeal squamous cell carcinomas. Cancer Treat Rev. 42, 24–29. 10.1016/j.ctrv.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Bours V., Dejardin E., Goujon-Letawe F., Merville M. P., Castronovo V. (1994). The NF-kappa B transcription factor and cancer: high expression of NF-kappa B- and I kappa B-related proteins in tumor cell lines. Biochem. Pharmacol. 47, 145–149. 10.1016/0006-2952(94)90448-0 [DOI] [PubMed] [Google Scholar]
- Braakhuis B. J., Snijders P. J., Keune W. J., Meijer C. J., Ruijter-Schippers H. J., Leemans C. R., et al. (2004). Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J. Natl. Cancer Inst. 96, 998–1006. 10.1093/jnci/djh183 [DOI] [PubMed] [Google Scholar]
- Brand T. M., Hartmann S., Bhola N. E., Li H., Zeng Y., O’keefe R. A., et al. (2018). Cross-talk Signaling between HER3 and HPV16 E6 and E7 Mediates Resistance to PI3K Inhibitors in Head and Neck Cancer. Cancer Res. 78, 2383–2395. 10.1158/0008-5472.CAN-17-1672 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Bu L. L., Yu G. T., Wu L., Mao L., Deng W. W., Liu J. F., et al. (2017). STAT3 Induces Immunosuppression by Upregulating PD-1/PD-L1 in HNSCC. J. Dent. Res. 96, 1027–1034. 10.1177/0022034517712435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G. M., Huang D. H., Dai Y. Z., Liu Y., Pi L. M., Tan H. L., et al. (2012). Analysis of transcriptional factors and regulation networks in laryngeal squamous cell carcinoma patients with lymph node metastasis. J. Proteome Res. 11, 1100–1107. 10.1021/pr200831g [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas, N. (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43. 10.1016/j.cell.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Carson A., Khan S. A. (2006). Characterization of transcription factor binding to human papillomavirus type 16 DNA during cellular differentiation. J. Virol. 80, 4356–4362. 10.1128/JVI.80.9.4356-4362.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellsague X., Alemany L., Quer M., Halec G., Quiros B., Tous S., et al. (2016). HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl. Cancer Inst. 108, djv403. 10.1093/jnci/djv403 [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H. H., Kapranov P., Sekinger E. A., Kampa D., et al. (2004). Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116, 499–509. 10.1016/S0092-8674(04)00127-8 [DOI] [PubMed] [Google Scholar]
- Chae J. I., Lee R., Cho J., Hong J., Shim J. H. (2014). Specificity protein 1 is a novel target of 2,4-bis (p-hydroxyphenyl)-2-butenal for the suppression of human oral squamous cell carcinoma cell growth. J. BioMed. Sci. 21, 4. 10.1186/1423-0127-21-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. K., Klock G., Bernard H. U. (1989). Progesterone and glucocorticoid response elements occur in the long control regions of several human papillomaviruses involved in anogenital neoplasia. J. Virol. 63, 3261–3269. 10.1128/JVI.63.8.3261-3269.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. K., Chong T., Bernard H. U., Klock G. (1990). Transcription of the transforming genes of the oncogenic human papillomavirus-16 is stimulated by tumor promotors through AP1 binding sites. Nucleic Acids Res. 18, 763–769. 10.1093/nar/18.4.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A. K., Graubard B. I., Broutian T., Pickard R. K., Tong Z. Y., Xiao W., et al. (2015). NHANES 2009-2012 Findings: Association of Sexual Behaviors with Higher Prevalence of Oral Oncogenic Human Papillomavirus Infections in U.S. Men. Cancer Res. 75, 2468–2477. 10.1158/0008-5472.CAN-14-2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Friedrich G. A., Soriano P. (1994). Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes Dev. 8, 2293–2301. 10.1101/gad.8.19.2293 [DOI] [PubMed] [Google Scholar]
- Chen Z., Ricker J. L., Malhotra P. S., Nottingham L., Bagain L., Lee T. L., et al. (2008). Differential bortezomib sensitivity in head and neck cancer lines corresponds to proteasome, nuclear factor-kappaB and activator protein-1 related mechanisms. Mol. Cancer Ther. 7, 1949–1960. 10.1158/1535-7163.MCT-07-2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yan B., Lou H., Shen Z., Tong F., Zhai A., et al. (2018). Immunological network analysis in HPV associated head and neck squamous cancer and implications for disease prognosis. Mol. Immunol. 96, 28–36. 10.1016/j.molimm.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Chen X., Chen F., Ren Y., Weng G., Keng P. C., Chen Y., et al. (2019). Glucocorticoid receptor upregulation increases radioresistance and triggers androgen independence of prostate cancer. Prostate 79, 1386–1398. 10.1002/pros.23861 [DOI] [PubMed] [Google Scholar]
- Chiefari E., Brunetti A., Arturi F., Bidart J. M., Russo D., Schlumberger M., et al. (2002). Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: a role in NIS expression regulation? BMC Cancer 2, 35. 10.1186/1471-2407-2-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnappan D., Xiao D., Ratnasari A., Andry C., King T. C., Weber H. C. (2009). Transcription factor YY1 expression in human gastrointestinal cancer cells. Int. J. Oncol. 34, 1417–1423. 10.1186/1471-2407-2-35 [DOI] [PubMed] [Google Scholar]
- Chong T., Chan W. K., Bernard H. U. (1990). Transcriptional activation of human papillomavirus 16 by nuclear factor I, AP1, steroid receptors and a possibly novel transcription factor, PVF: a model for the composition of genital papillomavirus enhancers. Nucleic Acids Res. 18, 465–470. 10.1093/nar/18.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong T., Apt D., Gloss B., Isa M., Bernard H. U. (1991). The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NF1, and AP-1 participate in epithelial cell-specific transcription. J. Virol. 65, 5933–5943. 10.1128/JVI.65.11.5933-5943.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S. (2012). Transcriptional regulation by post-transcriptional modification–role of phosphorylation in Sp1 transcriptional activity. Gene 508, 1–8. 10.1016/j.gene.2012.07.022 [DOI] [PubMed] [Google Scholar]
- Chuerduangphui J., Pientong C., Chaiyarit P., Patarapadungkit N., Chotiyano A., Kongyingyoes B., et al. (2016). Effect of human papillomavirus 16 oncoproteins on oncostatin M upregulation in oral squamous cell carcinoma. Med. Oncol. 33, 83. 10.1007/s12032-016-0800-6 [DOI] [PubMed] [Google Scholar]
- Chung C. H., Guthrie V. B., Masica D. L., Tokheim C., Kang H., Richmon J., et al. (2015). Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 26, 1216–1223. 10.1093/annonc/mdv109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripe T. P., Haugen T. H., Turk J. P., Tabatabai F., Schmid P. G.,. 3., Durst M., et al. (1987). Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 6, 3745–3753. 10.1002/j.1460-2075.1987.tb02709.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren L., Mellin H., Wangsa D., Heselmeyer-Haddad K., Bjornestal L., Lindholm J., et al. (2003). Comparative genomic hybridization analysis of tonsillar cancer reveals a different pattern of genomic imbalances in human papillomavirus-positive and -negative tumors. Int. J. Cancer 107, 244–249. 10.1002/ijc.11371 [DOI] [PubMed] [Google Scholar]
- De Martel C., Plummer M., Vignat J., Franceschi S. (2017). Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 141, 664–670. 10.1002/ijc.30716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Villiers E. M. (2003). Relationship between steroid hormone contraceptives and HPV, cervical intraepithelial neoplasia and cervical carcinoma. Int. J. Cancer 103, 705–708. 10.1002/ijc.10868 [DOI] [PubMed] [Google Scholar]
- Demeret C., Le Moal M., Yaniv M., Thierry F. (1995). Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 23, 4777–4784. 10.1093/nar/23.23.4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didonato J. A., Saatcioglu F., Karin M. (1996). Molecular mechanisms of immunosuppression and anti-inflammatory activities by glucocorticoids. Am. J. Respir. Crit. Care Med. 154, S11–S15. 10.1164/ajrccm/154.2_Pt_2.S11 [DOI] [PubMed] [Google Scholar]
- Dogan S., Xu B., Middha S., Vanderbilt C. M., Bowman A. S., Migliacci J., et al. (2019). Identification of prognostic molecular biomarkers in 157 HPV-positive and HPV-negative squamous cell carcinomas of the oropharynx. Int. J. Cancer 145, 3152–3162. 10.1002/ijc.32412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dok R., Kalev P., Van Limbergen E. J., Asbagh L. A., Vazquez I., Hauben E., et al. (2014). p16INK4a impairs homologous recombination-mediated DNA repair in human papillomavirus-positive head and neck tumors. Cancer Res. 74, 1739–1751. 10.1158/0008-5472.CAN-13-2479 [DOI] [PubMed] [Google Scholar]
- Dokter W. H., Tuyt L., Sierdsema S. J., Esselink M. T., Vellenga E. (1995). The spontaneous expression of interleukin-1 beta and interleukin-6 is associated with spontaneous expression of AP-1 and NF-kappa B transcription factor in acute myeloblastic leukemia cells. Leukemia 9, 425–432. [PubMed] [Google Scholar]
- Dong X. P., Stubenrauch F., Beyer-Finkler E., Pfister H. (1994). Prevalence of deletions of YY1-binding sites in episomal HPV 16 DNA from cervical cancers. Int. J. Cancer 58, 803–808. 10.1002/ijc.2910580609 [DOI] [PubMed] [Google Scholar]
- Dong Z., Liu G., Huang B., Sun J., Wu D. (2014). Prognostic significance of SOX2 in head and neck cancer: a meta-analysis. Int. J. Clin. Exp. Med. 7, 5010–5020. [PMC free article] [PubMed] [Google Scholar]
- D’amico S., Shi J., Martin B. L., Crawford H. C., Petrenko O., Reich N. C. (2018). STAT3 is a master regulator of epithelial identity and KRAS-driven tumorigenesis. Genes Dev. 32, 1175–1187. 10.1101/gad.311852.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Chen G. G., Vlantis A. C., Xu H., Tsang R. K., Van Hasselt A. C. (2003). The nuclear localization of NFkappaB and p53 is positively correlated with HPV16 E7 level in laryngeal squamous cell carcinoma. J. Histochem. Cytochem. 51, 533–539. 10.1177/002215540305100415 [DOI] [PubMed] [Google Scholar]
- Du G., Chen J., Wang Y., Cao T., Zhou L., Han X., et al. (2018). Differential expression of STAT-3 in subtypes of oral lichen planus: a preliminary study. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 125, 236–243.e231. 10.1016/j.oooo.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Duffey D. C., Chen Z., Dong G., Ondrey F. G., Wolf J. S., Brown K., et al. (1999). Expression of a dominant-negative mutant inhibitor-kappaBalpha of nuclear factor-kappaB in human head and neck squamous cell carcinoma inhibits survival, proinflammatory cytokine expression, and tumor growth in vivo. Cancer Res. 59, 3468–3474. [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. (1983). The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35, 79–87. 10.1016/0092-8674(83)90210-6 [DOI] [PubMed] [Google Scholar]
- Dziduszko A., Ozbun M. A. (2013). Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J. Virol. 87, 7502–7515. 10.1128/JVI.00519-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., Wagner E. F. (2003). AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868. 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Fakhry C., Westra W. H., Li S., Cmelak A., Ridge J. A., Pinto H., et al. (2008). Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl. Cancer Inst. 100, 261–269. 10.1093/jnci/djn011 [DOI] [PubMed] [Google Scholar]
- Fakhry C., Zhang Q., Nguyen-Tan P. F., Rosenthal D., El-Naggar A., Garden A. S., et al. (2014). Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J. Clin. Oncol. 32, 3365–3373. 10.1200/JCO.2014.55.1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghali K., Tanabe S., Grenier D. (2011). Soluble CD14 induces cytokine release by human oral epithelial cells. J. Periodontal. Res. 46, 147–152. 10.1111/j.1600-0765.2010.01311.x [DOI] [PubMed] [Google Scholar]
- Ferris R. L., Martinez I., Sirianni N., Wang J., Lopez-Albaitero A., Gollin S. M., et al. (2005). Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur. J. Cancer 41, 807–815. 10.1016/j.ejca.2004.11.023 [DOI] [PubMed] [Google Scholar]
- Fontaine V., Van Der Meijden E., De Graaf J., Ter Schegget J., Struyk L. (2000). A functional NF-kappaB binding site in the human papillomavirus type 16 long control region. Virology 272, 40–49. 10.1006/viro.2000.0363 [DOI] [PubMed] [Google Scholar]
- Franceschi S., Munoz N., Bosch X. F., Snijders P. J., Walboomers J. M. (1996). Human papillomavirus and cancers of the upper aerodigestive tract: a review of epidemiological and experimental evidence. Cancer Epidemiol. Biomarkers Prev. 5, 567–575. [PubMed] [Google Scholar]
- Freier K., Knoepfle K., Flechtenmacher C., Pungs S., Devens F., Toedt G., et al. (2010). Recurrent copy number gain of transcription factor SOX2 and corresponding high protein expression in oral squamous cell carcinoma. Genes Chromosomes Cancer 49, 9–16. 10.1002/gcc.20714 [DOI] [PubMed] [Google Scholar]
- Galvin K. M., Shi Y. (1997). Multiple mechanisms of transcriptional repression by YY1. Mol. Cell Biol. 17, 3723–3732. 10.1128/MCB.17.7.3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama R. R., Carvalho A. L., Longatto Filho A., Scorsato A. P., Lopez R. V., Rautava J., et al. (2016). Detection of human papillomavirus in laryngeal squamous cell carcinoma: Systematic review and meta-analysis. Laryngoscope 126, 885–893. 10.1002/lary.25738 [DOI] [PubMed] [Google Scholar]
- Gao L. F., Xu D. Q., Wen L. J., Zhang X. Y., Shao Y. T., Zhao X. J. (2005). Inhibition of STAT3 expression by siRNA suppresses growth and induces apoptosis in laryngeal cancer cells. Acta Pharmacol. Sin. 26, 377–383. 10.1111/j.1745-7254.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Gao L. F., Wen L. J., Yu H., Zhang L., Meng Y., Shao Y. T., et al. (2006). Knockdown of Stat3 expression using RNAi inhibits growth of laryngeal tumors in vivo. Acta Pharmacol. Sin. 27, 347–352. 10.1111/j.1745-7254.2006.00277.x [DOI] [PubMed] [Google Scholar]
- Gao J., Zhao S., Halstensen T. S. (2016). Increased interleukin-6 expression is associated with poor prognosis and acquired cisplatin resistance in head and neck squamous cell carcinoma. Oncol. Rep. 35, 3265–3274. 10.3892/or.2016.4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Zhang Y., Zhou H., Lv J. (2020). Baicalein inhibits the growth of oral squamous cell carcinoma cells by downregulating the expression of transcription factor Sp1. Int. J. Oncol. 56, 273–282. 10.3892/ijo.2019.4894 [DOI] [PubMed] [Google Scholar]
- Gaussin A., Modlich U., Bauche C., Niederlander N. J., Schambach A., Duros C., et al. (2012). CTF/NF1 transcription factors act as potent genetic insulators for integrating gene transfer vectors. Gene Ther. 19, 15–24. 10.1038/gt.2011.70 [DOI] [PubMed] [Google Scholar]
- Gaykalova D. A., Manola J. B., Ozawa H., Zizkova V., Morton K., Bishop J. A., et al. (2015). NF-kappaB and stat3 transcription factor signatures differentiate HPV-positive and HPV-negative head and neck squamous cell carcinoma. Int. J. Cancer 137, 1879–1889. 10.1002/ijc.29558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos N. T., Proffitt J. L., Blair G. E. (2000). Transcriptional regulation of the major histocompatibility complex (MHC) class I heavy chain, TAP1 and LMP2 genes by the human papillomavirus (HPV) type 6b, 16 and 18 E7 oncoproteins. Oncogene 19, 4930–4935. 10.1038/sj.onc.1203860 [DOI] [PubMed] [Google Scholar]
- Germain D., Frank D. A. (2007). Targeting the cytoplasmic and nuclear functions of signal transducers and activators of transcription 3 for cancer therapy. Clin. Cancer Res. 13, 5665–5669. 10.1158/1078-0432.CCR-06-2491 [DOI] [PubMed] [Google Scholar]
- Gerster T., Roeder R. G. (1988). A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc. Natl. Acad. Sci. U. S. A. 85, 6347–6351. 10.1073/pnas.85.17.6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison M. L., Koch W. M., Capone R. B., Spafford M., Westra W. H., Wu L., et al. (2000). Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92, 709–720. 10.1093/jnci/92.9.709 [DOI] [PubMed] [Google Scholar]
- Gletsou E., Papadas T. A., Baliou E., Tsiambas E., Ragos V., Armata I. E., et al. (2018). HPV infection in oropharyngeal squamous cell carcinomas: correlation with tumor size. J. BUON 23, 433–438. [PubMed] [Google Scholar]
- Gloss B., Bernard H. U., Seedorf K., Klock G. (1987). The upstream regulatory region of the human papilloma virus-16 contains an E2 protein-independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 6, 3735–3743. 10.1002/j.1460-2075.1987.tb02708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss B., Yeo-Gloss M., Meisterenst M., Rogge L., Winnacker E. L., Bernard H. U. (1989). Clusters of nuclear factor I binding sites identify enhancers of several papillomaviruses but alone are not sufficient for enhancer function. Nucleic Acids Res. 17, 3519–3533. 10.1093/nar/17.9.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloss B., Bernard H. U. (1990). The E6/E7 promoter of human papillomavirus type 16 is activated in the absence of E2 proteins by a sequence-aberrant Sp1 distal element. J. Virol. 64, 5577–5584. 10.1128/JVI.64.11.5577-5584.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg D., Begum S., Westra W. H., Khan Z., Sciubba J., Pai S. I., et al. (2008). Cystic lymph node metastasis in patients with head and neck cancer: An HPV-associated phenomenon. Head Neck 30, 898–903. 10.1002/hed.20796 [DOI] [PubMed] [Google Scholar]
- Gonzalez P. P., Kim J., Galvao R. P., Cruickshanks N., Abounader R., Zong H. (2018). p53 and NF 1 loss plays distinct but complementary roles in glioma initiation and progression. Glia 66, 999–1015. 10.1002/glia.23297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granda-Diaz R., Menendez S. T., Pedregal Mallo D., Hermida-Prado F., Rodriguez R., Suarez-Fernandez L., et al. (2019). The Novel Role of SOX2 as an Early Predictor of Cancer Risk in Patients with Laryngeal Precancerous Lesions. Cancers (Basel) 11, 1–11. 10.3390/cancers11030286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis J. R., Drenning S. D., Chakraborty A., Zhou M. Y., Zeng Q., Pitt A. S., et al. (1998). Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor- mediated cell growth In vitro. J. Clin. Invest. 102, 1385–1392. 10.1172/JCI3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis J. R., Drenning S. D., Zeng Q., Watkins S. C., Melhem M. F., Endo S., et al. (2000). Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 97, 4227–4232. 10.1073/pnas.97.8.4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. L., Hartig S. M., Edwards D. P. (2016). Progesterone Receptor Signaling Mechanisms. J. Mol. Biol. 428, 3831–3849. 10.1016/j.jmb.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Guan H., Cai J., Zhang N., Wu J., Yuan J., Li J., et al. (2012). Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int. J. Cancer 130, 593–601. 10.1002/ijc.26049 [DOI] [PubMed] [Google Scholar]
- Gupta S., Kumar P., Kaur H., Sharma N., Saluja D., Bharti A. C., et al. (2015). Selective participation of c-Jun with Fra-2/c-Fos promotes aggressive tumor phenotypes and poor prognosis in tongue cancer. Sci. Rep. 5, 16811. 10.1038/srep16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Kumar P., Kaur H., Sharma N., Gupta S., Saluja D., et al. (2018). Constitutive activation and overexpression of NF-kappaB/c-Rel in conjunction with p50 contribute to aggressive tongue tumorigenesis. Oncotarget 9, 33011–33029. 10.18632/oncotarget.26041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut A., Moch H., Choschzick M. (2018). SOX2 Gene Amplification and Overexpression is Linked to HPV-positive Vulvar Carcinomas. Int. J. Gynecol Pathol. 37, 68–73. 10.1097/PGP.0000000000000388 [DOI] [PubMed] [Google Scholar]
- Hadaschik D., Hinterkeuser K., Oldak M., Pfister H. J., Smola-Hess S. (2003). The Papillomavirus E2 protein binds to and synergizes with C/EBP factors involved in keratinocyte differentiation. J. Virol. 77, 5253–5265. 10.1128/JVI.77.9.5253-5265.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeggblom L., Ramqvist T., Tommasino M., Dalianis T., Nasman A. (2017). Time to change perspectives on HPV in oropharyngeal cancer. A systematic review of HPV prevalence per oropharyngeal sub-site the last 3 years. Papillomavirus Res. 4, 1–11. 10.1016/j.pvr.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafkamp H. C., Speel E. J., Haesevoets A., Bot F. J., Dinjens W. N., Ramaekers F. C., et al. (2003). A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5-8. Int. J. Cancer 107, 394–400. 10.1002/ijc.11389 [DOI] [PubMed] [Google Scholar]
- Hajek M., Sewell A., Kaech S., Burtness B., Yarbrough W. G., Issaeva N. (2017). TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 123, 1778–1790. 10.1002/cncr.30570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstedt L., Lindquist D., Dahlstrand H., Romanitan M., Dahlgren L. O., Joneberg J., et al. (2006). Human papillomavirus as a risk factor for the increase in incidence of tonsillar cancer. Int. J. Cancer 119, 2620–2623. 10.1002/ijc.22177 [DOI] [PubMed] [Google Scholar]
- Harden M. E., Munger K. (2017). Human papillomavirus molecular biology. Mutat. Res. Rev. Mutat. Res. 772, 3–12. 10.1016/j.mrrev.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig E., Cato A. C. (1994). In vivo binding of proteins to stably integrated MMTV DNA in murine cell lines: occupancy of NFI and OTF1 binding sites in the absence and presence of glucocorticoids. Cell Mol. Biol. Res. 40, 643–652. [PubMed] [Google Scholar]
- He G., Wang Q., Zhou Y., Wu X., Wang L., Duru N., et al. (2011). YY1 is a novel potential therapeutic target for the treatment of HPV infection-induced cervical cancer by arsenic trioxide. Int. J. Gynecol Cancer 21, 1097–1104. 10.1097/IGC.0b013e31821d2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich P. C., Behrmann I., Muller-Newen G., Schaper F., Graeve L. (1998). Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 334 ( Pt 2), 297–314. 10.1042/bj3340297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero R., Castellsague X., Pawlita M., Lissowska J., Kee F., Balaram P., et al. (2003). Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J. Natl. Cancer Inst. 95, 1772–1783. 10.1093/jnci/djg107 [DOI] [PubMed] [Google Scholar]
- Herrero R. (2003). Chapter 7: Human papillomavirus and cancer of the upper aerodigestive tract. J. Natl. Cancer Inst. Monogr. 31, 47–51. 10.1093/oxfordjournals.jncimonographs.a003482 [DOI] [PubMed] [Google Scholar]
- Hormia M., Willberg J., Ruokonen H., Syrjanen S. (2005). Marginal periodontium as a potential reservoir of human papillomavirus in oral mucosa. J. Periodontol. 76, 358–363. 10.1902/jop.2005.76.3.358 [DOI] [PubMed] [Google Scholar]
- Hsu T. I., Wang M. C., Chen S. Y., Yeh Y. M., Su W. C., Chang W. C., et al. (2012). Sp1 expression regulates lung tumor progression. Oncogene 31, 3973–3988. 10.1038/onc.2011.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Bharti A. C., Salam I., Bhat M. A., Mir M. M., Hedau S., et al. (2009). Transcription factor AP-1 in esophageal squamous cell carcinoma: alterations in activity and expression during human Papillomavirus infection. BMC Cancer 9, 329. 10.1186/1471-2407-9-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussenet T., Dali S., Exinger J., Monga B., Jost B., Dembele D., et al. (2010). SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PloS One 5, e8960. 10.1371/journal.pone.0008960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyakusoku H., Sano D., Takahashi H., Hatano T., Isono Y., Shimada S., et al. (2016). JunB promotes cell invasion, migration and distant metastasis of head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 35, 6. 10.1186/s13046-016-0284-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Hirsch H. A., Struhl K. (2009). An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706. 10.1016/j.cell.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute, N.C. (2017). Head and Neck Cancers Fact sheet. National Cancer Institute. Available at: https://www.cancer.gov/types/head-and-neck/head-neck-fact-sheet.
- Ishiji T., Lace M. J., Parkkinen S., Anderson R. D., Haugen T. H., Cripe T. P., et al. (1992). Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 11, 2271–2281. 10.1002/j.1460-2075.1992.tb05286.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen B. M., Horwitz K. B. (2012). Progesterone receptors, their isoforms and progesterone regulated transcription. Mol. Cell Endocrinol. 357, 18–29. 10.1016/j.mce.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M. A., Lee J. H., Klingelhutz A. J. (2006). Human papillomavirus type 16 E6 activates NF-kappaB, induces cIAP-2 expression, and protects against apoptosis in a PDZ binding motif-dependent manner. J. Virol. 80, 5301–5307. 10.1128/JVI.01942-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janecka-Widla A., Mucha-Malecka A., Majchrzyk K., Halaszka K., Przewoznik M., Slonina D., et al. (2020). Active HPV infection and its influence on survival in head and neck squamous-cell cancer. J. Cancer Res. Clin. Oncol. 146, 1677–1692. 10.1007/s00432-020-03218-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenson A. B., Kurman R. J., Lancaster W. D. (1991). Tissue effects of and host response to human papillomavirus infection. Dermatol. Clin. 9, 203–209. 10.1016/S0733-8635(18)30410-8 [DOI] [PubMed] [Google Scholar]
- Jiang N. Y., Woda B. A., Banner B. F., Whalen G. F., Dresser K. A., Lu D. (2008). Sp1, a new biomarker that identifies a subset of aggressive pancreatic ductal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 17, 1648–1652. 10.1158/1055-9965.EPI-07-2791 [DOI] [PubMed] [Google Scholar]
- Jinno T., Kawano S., Maruse Y., Matsubara R., Goto Y., Sakamoto T., et al. (2015). Increased expression of interleukin-6 predicts poor response to chemoradiotherapy and unfavorable prognosis in oral squamous cell carcinoma. Oncol. Rep. 33, 2161–2168. 10.3892/or.2015.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. E., O’keefe R. A., Grandis J. R. (2018). Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248. 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A. C., Briolat J., Millon R., De Reynies A., Rickman D., Thomas E., et al. (2010). Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int. J. Cancer 126, 1882–1894. 10.1002/ijc.24911 [DOI] [PubMed] [Google Scholar]
- Kang M., Xiao J., Wang J., Zhou P., Wei T., Zhao T., et al. (2016). MiR-24 enhances radiosensitivity in nasopharyngeal carcinoma by targeting SP1. Cancer Med. 5, 1163–1173. 10.1002/cam4.660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Chang L. (2001). AP-1–glucocorticoid receptor crosstalk taken to a higher level. J. Endocrinol. 169, 447–451. 10.1677/joe.0.1690447 [DOI] [PubMed] [Google Scholar]
- Karpathiou G., Da Cruz V., Casteillo F., Mobarki M., Dumollard J. M., Chauleur C., et al. (2017). FOXA1 in HPV associated carcinomas: Its expression in carcinomas of the head and neck and of the uterine cervix. Exp. Mol. Pathol. 102, 230–236. 10.1016/j.yexmp.2017.02.010 [DOI] [PubMed] [Google Scholar]
- Khare S., Pater M. M., Tang S. C., Pater A. (1997). Effect of glucocorticoid hormones on viral gene expression, growth, and dysplastic differentiation in HPV16-immortalized ectocervical cells. Exp. Cell Res. 232, 353–360. 10.1006/excr.1997.3529 [DOI] [PubMed] [Google Scholar]
- Kijima T., Niwa H., Steinman R. A., Drenning S. D., Gooding W. E., Wentzel A. L., et al. (2002). STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 13, 355–362. [PubMed] [Google Scholar]
- Kim D. J., Chan K. S., Sano S., Digiovanni J. (2007). Signal transducer and activator of transcription 3 (Stat3) in epithelial carcinogenesis. Mol. Carcinog. 46, 725–731. 10.1002/mc.20342 [DOI] [PubMed] [Google Scholar]
- Kimple R. J., Smith M. A., Blitzer G. C., Torres A. D., Martin J. A., Yang R. Z., et al. (2013). Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 73, 4791–4800. 10.1158/0008-5472.CAN-13-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi I., Fujii S., Nonogaki H., Nanbu Y., Iwai T., Mori T. (1991). Immunohistochemical analysis of estrogen receptors, progesterone receptors, Ki-67 antigen, and human papillomavirus DNA in normal and neoplastic epithelium of the uterine cervix. Cancer 68, 1340–1350. [DOI] [PubMed] [Google Scholar]
- Kost B. P., Beyer S., Schroder L., Zhou J., Mayr D., Kuhn C., et al. (2019). Glucocorticoid receptor in cervical cancer: an immunhistochemical analysis. Arch. Gynecol Obstet. 299, 203–209. 10.1007/s00404-018-4928-9 [DOI] [PubMed] [Google Scholar]
- Kottaridi C., Leventakou D., Pouliakis A., Pergialiotis V., Chrelias G., Patsouri E., et al. (2019). Searching HPV genome for methylation sites involved in molecular progression to cervical precancer. J. Cancer 10, 4588–4595. 10.7150/jca.30081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer A. R., Clifford G. M., Boyle P., Franceschi S. (2005). Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol. Biomarkers Prev. 14, 467–475. 10.1158/1055-9965.EPI-04-0551 [DOI] [PubMed] [Google Scholar]
- Kyo S., Klumpp D. J., Inoue M., Kanaya T., Laimins L. A. (1997). Expression of AP1 during cellular differentiation determines human papillomavirus E6/E7 expression in stratified epithelial cells. J. Gen. Virol. 78 ( Pt 2), 401–411. 10.1099/0022-1317-78-2-401 [DOI] [PubMed] [Google Scholar]
- Lai S. Y., Johnson F. M. (2010). Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: implications for future therapeutic approaches. Drug Resist. Update 13, 67–78. 10.1016/j.drup.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Le Gallo M., Rudd M. L., Urick M. E., Hansen N. F., National Institutes of Health Intramural Sequencing Center Comparative Sequencing, P. Merino M. J., et al. (2018). The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcomas and carcinomas. Cancer 124, 65–73. 10.1002/cncr.30971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M., Frampton G. M., Fenton T., Feber A., Palmer G., Jay A., et al. (2013). Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 5, 49. 10.1186/gm453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Haslinger A., Karin M., Tjian R. (1987). Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325, 368–372. 10.1038/325368a0 [DOI] [PubMed] [Google Scholar]
- Lee T. L., Yeh J., Friedman J., Yan B., Yang X., Yeh N. T., et al. (2008). A signal network involving coactivated NF-kappaB and STAT3 and altered p53 modulates BAX/BCL-XL expression and promotes cell survival of head and neck squamous cell carcinomas. Int. J. Cancer 122, 1987–1998. 10.1002/ijc.23324 [DOI] [PubMed] [Google Scholar]
- Lee M. H., Lahusen T., Wang R. H., Xiao C., Xu X., Hwang Y. S., et al. (2012). Yin Yang 1 positively regulates BRCA1 and inhibits mammary cancer formation. Oncogene 31, 116–127. 10.1038/onc.2011.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Oh S. Y., Do S. I., Lee H. J., Kang H. J., Rho Y. S., et al. (2014). SOX2 regulates self-renewal and tumorigenicity of stem-like cells of head and neck squamous cell carcinoma. Br. J. Cancer 111, 2122–2130. 10.1038/bjc.2014.528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman R. J., Lui V. W., Grandis J. R. (2006). STAT3 as a therapeutic target in head and neck cancer. Expert Opin. Biol. Ther. 6, 231–241. 10.1517/14712598.6.3.231 [DOI] [PubMed] [Google Scholar]
- Leemans C. R., Snijders P. J. F., Brakenhoff R. H. (2018). The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269–282. 10.1038/nrc.2018.11 [DOI] [PubMed] [Google Scholar]
- Leong P. L., Andrews G. A., Johnson D. E., Dyer K. F., Xi S., Mai J. C., et al. (2003). Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc. Natl. Acad. Sci. U. S. A. 100, 4138–4143. 10.1073/pnas.0534764100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesinski G. B., Nannapaneni S., Griffith C. C., Patel M., Chen W., Chen Z., et al. (2019). Interleukin-6/STAT3 Signaling is Prominent and Associated with Reduced Overall Survival in p16 Negative Oropharyngeal Squamous Cell Carcinoma. Head Neck Pathol. 13, 304–312. 10.1007/s12105-018-0962-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. W., Cheung W., Lin Z. B., Li T. Y., Lim J. T., Wang D. Y. (2009). Oral steroids enhance epithelial repair in nasal polyposis via upregulation of the AP-1 gene network. Thorax 64, 306–312. 10.1136/thx.2008.106096 [DOI] [PubMed] [Google Scholar]
- Li J., Wang W., Chen S., Cai J., Ban Y., Peng Q., et al. (2019). FOXA1 reprograms the TGF-beta-stimulated transcriptional program from a metastasis promoter to a tumor suppressor in nasopharyngeal carcinoma. Cancer Lett. 442, 1–14. 10.1016/j.canlet.2018.10.036 [DOI] [PubMed] [Google Scholar]
- Li Y., Dong M., Kong F., Zhou J. (2015). Octamer transcription factor 1 mediates epithelial-mesenchymal transition in colorectal cancer. Tumour Biol. 36, 9941–9946. 10.1007/s13277-015-3766-7 [DOI] [PubMed] [Google Scholar]
- Li Z., Yang Z., Lapidus R. G., Liu X., Cullen K. J., Dan H. C. (2015). IKK phosphorylation of NF-kappaB at serine 536 contributes to acquired cisplatin resistance in head and neck squamous cell cancer. Am. J. Cancer Res. 5, 3098–3110. 10.1007/s13277-015-3766-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang Z. T. (2019). Genetic Association between NFKBIA and NFKB1 Gene Polymorphisms and the Susceptibility to Head and Neck Cancer: A Meta-Analysis. Dis. Markers 2019, 6523837. 10.1155/2019/6523837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Kina S., Takemoto H., Matayoshi A., Phonaphonh T., Sunagawa N., et al. (2011). HPV16E6-dependent c-fos expression contributes to AP-1 complex formation in SiHa cells. Mediators Inflammation 2011, 263216. 10.1155/2011/263216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licitra L., Perrone F., Bossi P., Suardi S., Mariani L., Artusi R., et al. (2006). High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J. Clin. Oncol. 24, 5630–5636. 10.1200/JCO.2005.04.6136 [DOI] [PubMed] [Google Scholar]
- Lindsay C., Seikaly H., Biron V. L. (2017). Epigenetics of oropharyngeal squamous cell carcinoma: opportunities for novel chemotherapeutic targets. J. Otolaryngol. Head Neck Surg. 46, 9. 10.1186/s40463-017-0185-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mann D., Sinha U. K., Kokot N. C. (2018). The molecular mechanisms of increased radiosensitivity of HPV-positive oropharyngeal squamous cell carcinoma (OPSCC): an extensive review. J. Otolaryngol. Head Neck Surg. 47, 59. 10.1186/s40463-018-0302-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zhang J., Wu Y., Shi G., Yuan H., Lu Z., et al. (2018). YY1 suppresses proliferation and migration of pancreatic ductal adenocarcinoma by regulating the CDKN3/MdM2/P53/P21 signaling pathway. Int. J. Cancer 142, 1392–1404. 10.1002/ijc.31173 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li J. Z., Yuan X. H., Adler-Storthz K., Che Z. (2002). An AP-1 binding site mutation in HPV-16 LCR enhances E6/E7 promoter activity in human oral epithelial cells. Virus Genes 24, 29–37. 10.1023/A:1014081803232 [DOI] [PubMed] [Google Scholar]
- Liu X., Ma X., Lei Z., Feng H., Wang S., Cen X., et al. (2015). Chronic Inflammation-Related HPV: A Driving Force Speeds Oropharyngeal Carcinogenesis. PloS One 10, e0133681. 10.1145/2818302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. B., Wang J., Li K., Fan X. N. (2019). Sp1 promotes cell migration and invasion in oral squamous cell carcinoma by upregulating Annexin A2 transcription. Mol. Cell Probes 46, 101417. 10.1016/j.mcp.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang B., Jia Y., Fu M. (2020). SNHG17 enhances the malignant characteristics of tongue squamous cell carcinoma by acting as a competing endogenous RNA on microRNA-876 and thereby increasing specificity protein 1 expression. Cell Cycle 19, 1–15. 10.1080/15384101.2020.1838792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohavanichbutr P., Houck J., Fan W., Yueh B., Mendez E., Futran N., et al. (2009). Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch. Otolaryngol. Head Neck Surg. 135, 180–188. 10.1001/archoto.2008.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvanto K., Roger M., Faucher M. C., Syrjanen K., Grenman S., Syrjanen S. (2018). HLA-G and vertical mother-to-child transmission of human papillomavirus infection. Hum. Immunol. 79, 471–476. 10.1016/j.humimm.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Luiz S. T., Modolo F., Mozzer I., Dos Santos E. C., Nagashima S., Camargo Martins A. P., et al. (2018). Immunoexpression of SOX-2 in oral leukoplakia. Oral. Dis. 24, 1449–1457. 10.1111/odi.12922 [DOI] [PubMed] [Google Scholar]
- Lv X. X., Zheng X. Y., Yu J. J., Ma H. R., Hua C., Gao R. T. (2020). EGFR enhances the stemness and progression of oral cancer through inhibiting autophagic degradation of SOX2. Cancer Med. 9, 1131–1140. 10.1002/cam4.2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X. M., Zhu X. W., Zhao M., Zuo X. B., Huang Z. X., Liu X., et al. (2018). A regulatory mutant on TRIM26 conferring the risk of nasopharyngeal carcinoma by inducing low immune response. Cancer Med. 7, 3848–3861. 10.1002/cam4.1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macha M. A., Matta A., Kaur J., Chauhan S. S., Thakar A., Shukla N. K., et al. (2011). Prognostic significance of nuclear pSTAT3 in oral cancer. Head Neck 33, 482–489. 10.1002/hed.21468 [DOI] [PubMed] [Google Scholar]
- Malhas A. N., Lee C. F., Vaux D. J. (2009). Lamin B1 controls oxidative stress responses via Oct-1. J. Cell Biol. 184, 45–55. 10.1083/jcb.200804155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I., Wang J., Hobson K. F., Ferris R. L., Khan S. A. (2007). Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur. J. Cancer 43, 415–432. 10.1016/j.ejca.2006.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ramirez I., Del-Castillo-Falconi V., Mitre-Aguilar I. B., Amador-Molina A., Carrillo-Garcia A., Langley E., et al. (2017). SOX2 as a New Regulator of HPV16 Transcription. Viruses 9, 1–17. 10.3390/v9070175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson L., Sorgeloos F., Winder D., Lechner M., Marker A., Malhotra S., et al. (2015). Deregulation of SYCP2 predicts early stage human papillomavirus-positive oropharyngeal carcinoma: A prospective whole transcriptome analysis. Cancer Sci. 106, 1568–1575. 10.1111/cas.12809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M., Suzui M., Yasumatu R., Nakashima T., Kuratomi Y., Azuma K., et al. (2002). Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 62, 3351–3355. [PubMed] [Google Scholar]
- Masuda M., Wakasaki T., Suzui M., Toh S., Joe A. K., Weinstein I. B. (2010). Stat3 orchestrates tumor development and progression: the Achilles’ heel of head and neck cancers? Curr. Cancer Drug Targets 10, 117–126. 10.2174/156800910790980197 [DOI] [PubMed] [Google Scholar]
- Matsuno F., Chowdhury S., Gotoh T., Iwase K., Matsuzaki H., Takatsuki K., et al. (1996). Induction of the C/EBP beta gene by dexamethasone and glucagon in primary-cultured rat hepatocytes. J. Biochem. 119, 524–532. 10.1093/oxfordjournals.jbchem.a021273 [DOI] [PubMed] [Google Scholar]
- Mazibrada J., Longo L., Vatrano S., Cappia S., Giorcelli J., Pentenero M., et al. (2014). Differential expression of HER2, STAT3, SOX2, IFI16 and cell cycle markers during HPV-related head and neck carcinogenesis. New Microbiol. 37, 129–143. [PubMed] [Google Scholar]
- Meighan-Mantha R. L., Riegel A. T., Suy S., Harris V., Wang F. H., Lozano C., et al. (1999). Ionizing radiation stimulates octamer factor DNA binding activity in human carcinoma cells. Mol. Cell Biochem. 199, 209–215. 10.1023/A:1006958217143 [DOI] [PubMed] [Google Scholar]
- Milan M., Balestrieri C., Alfarano G., Polletti S., Prosperini E., Spaggiari P., et al. (2019). FOXA2 controls the cis-regulatory networks of pancreatic cancer cells in a differentiation grade-specific manner. EMBO J. 38, e102161. 10.15252/embj.2019102161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. S., Johnstone B. M. (2001). Human papillomavirus as a risk factor for oral squamous cell carcinoma: a meta-analysi-1997. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. Endod. 91, 622–635. 10.1067/moe.2001.115392 [DOI] [PubMed] [Google Scholar]
- Mishra A., Bharti A. C., Varghese P., Saluja D., Das B. C. (2006). Differential expression and activation of NF-kappaB family proteins during oral carcinogenesis: Role of high risk human papillomavirus infection. Int. J. Cancer 119, 2840–2850. 10.1002/ijc.22262 [DOI] [PubMed] [Google Scholar]
- Mishra A., Bharti A. C., Saluja D., Das B. C. (2010). Trans-activation and expression patterns of jun and fos / AP-1 super-family proteins in human oral cancer. Int. J. Cancer 126, 819–829. 10.1002/ijc.24807 [DOI] [PubMed] [Google Scholar]
- Mishra A., Kumar R., Tyagi A., Kohaar I., Hedau S., Bharti A. C., et al. (2015). Curcumin modulates cellular AP-1, NF-kB, and HPV16 E6 proteins in oral cancer. Ecancermedicalscience 9, 525. 10.3332/ecancer.2015.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R., Kumar K. U., Pater A., Pater M. M. (1994). Differential regulation by c-jun and c-fos protooncogenes of hormone response from composite glucocorticoid response element in human papilloma virus type 16 regulatory region. Mol. Endocrinol. 8, 1701–1708. 10.1210/me.8.12.1701 [DOI] [PubMed] [Google Scholar]
- Mohamed H., Aro K., Jouhi L., Makitie A., Remes S., Haglund C., et al. (2018). Expression of hormone receptors in oropharyngeal squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 275, 1289–1300. 10.1007/s00405-018-4949-9 [DOI] [PubMed] [Google Scholar]
- Monisha J., Roy N. K., Bordoloi D., Kumar A., Golla R., Kotoky J., et al. (2017). Nuclear Factor Kappa B: A Potential Target to Persecute Head and Neck Cancer. Curr. Drug Targets 18, 232–253. 10.2174/1389450117666160201112330 [DOI] [PubMed] [Google Scholar]
- Monsonego J., Magdelenat H., Catalan F., Coscas Y., Zerat L., Sastre X. (1991). Estrogen and progesterone receptors in cervical human papillomavirus related lesions. Int. J. Cancer 48, 533–539. 10.1002/ijc.2910480410 [DOI] [PubMed] [Google Scholar]
- Moodley M., Moodley J., Chetty R., Herrington C. S. (2003). The role of steroid contraceptive hormones in the pathogenesis of invasive cervical cancer: a review. Int. J. Gynecol Cancer 13, 103–110. 10.1136/ijgc-00009577-200303000-00001 [DOI] [PubMed] [Google Scholar]
- Morgan E. L., Wasson C. W., Hanson L., Kealy D., Pentland I., Mcguire V., et al. (2018). STAT3 activation by E6 is essential for the differentiation-dependent HPV18 life cycle. PloS Pathog. 14, e1006975. 10.1371/journal.ppat.1006975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. L., Macdonald A. (2019). Autocrine STAT3 activation in HPV positive cervical cancer through a virus-driven Rac1-NFkappaB-IL-6 signalling axis. PloS Pathog. 15, e1007835. 10.1371/journal.ppat.1007835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G. J., Verma I. M. (1993). Proposed NF-kappa B/I kappa B family nomenclature. Genes Dev. 7, 2063. 10.1101/gad.7.11.2063 [DOI] [PubMed] [Google Scholar]
- Nagpal J. K., Mishra R., Das B. R. (2002). Activation of Stat-3 as one of the early events in tobacco chewing-mediated oral carcinogenesis. Cancer 94, 2393–2400. 10.1002/cncr.10499 [DOI] [PubMed] [Google Scholar]
- Nakayama Y., Kobayashi R., Iwai Y., Noda K., Yamazaki M., Kurita-Ochiai T., et al. (2019). C/EBPbeta and YY1 bind and interact with Smad3 to modulate lipopolysaccharide-induced amelotin gene transcription in mouse gingival epithelial cells. FEBS Open Bio 9, 276–290. 10.1002/2211-5463.12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H., Pater M. M., Pater A. (1990). Ubiquitous and cell-type-specific protein interactions with human papillomavirus type 16 and type 18 enhancers. Virology 178, 92–103. 10.1016/0042-6822(90)90382-2 [DOI] [PubMed] [Google Scholar]
- Nasman A., Andersson E., Nordfors C., Grun N., Johansson H., Munck-Wikland E., et al. (2013). MHC class I expression in HPV positive and negative tonsillar squamous cell carcinoma in correlation to clinical outcome. Int. J. Cancer 132, 72–81. 10.1002/ijc.27635 [DOI] [PubMed] [Google Scholar]
- Nicu E. A., Laine M. L., Morre S. A., Van Der Velden U., Loos B. G. (2009). Soluble CD14 in periodontitis. Innate Immun. 15, 121–128. 10.1177/1753425908101577 [DOI] [PubMed] [Google Scholar]
- Oleaga C., Welten S., Belloc A., Sole A., Rodriguez L., Mencia N., et al. (2012). Identification of novel Sp1 targets involved in proliferation and cancer by functional genomics. Biochem. Pharmacol. 84, 1581–1591. 10.1016/j.bcp.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Ondrey F. G., Dong G., Sunwoo J., Chen Z., Wolf J. S., Crowl-Bancroft C. V., et al. (1999). Constitutive activation of transcription factors NF-(kappa)B, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Mol. Carcinog. 26, 119–129. [DOI] [PubMed] [Google Scholar]
- Onerci Celebi O., Sener E., Hosal S., Cengiz M., Gullu I., Guler Tezel G. (2018). Human papillomavirus infection in patients with laryngeal carcinoma. BMC Cancer 18, 1005. 10.1186/s12885-018-4890-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’connor M., Chan S. Y., Bernard H. U. (1995). “Transcription Factor Binding Sites in the Long Control Region of Genital HPVs,” in Human papillomaviruses, 1995 compendium. Eds. Meyers G., Bernard H. U., Delius H., Baker C., Icenogle J., Halpern A., Wheeler C. (Los Alamos, N.Mex: Los Alamos National Laboratory; ), 21–40. [Google Scholar]
- O’connor M. J., Tan S. H., Tan C. H., Bernard H. U. (1996). YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J. Virol. 70, 6529–6539. 10.1128/JVI.70.10.6529-6539.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’connor M., Bernard H. U. (1995). Oct-1 activates the epithelial-specific enhancer of human papillomavirus type 16 via a synergistic interaction with NFI at a conserved composite regulatory element. Virology 207, 77–88. 10.1006/viro.1995.1053 [DOI] [PubMed] [Google Scholar]
- Ostlund Farrants A. K., Blomquist P., Kwon H., Wrange O. (1997). Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell Biol. 17, 895–905. 10.1128/MCB.17.2.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S., Jain N., Prusty B. K., Bhambhani S., Gupta S., Sharma R., et al. (2008). Human papillomavirus type 16 variant analysis of E6, E7, and L1 genes and long control region in biopsy samples from cervical cancer patients in north India. J. Clin. Microbiol. 46, 1060–1066. 10.1128/JCM.02202-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partlova S., Boucek J., Kloudova K., Lukesova E., Zabrodsky M., Grega M., et al. (2015). Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology 4, e965570. 10.4161/21624011.2014.965570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegue E., Wagner E. F. (2000). JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 19, 2969–2979. 10.1093/emboj/19.12.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pectasides E., Egloff A. M., Sasaki C., Kountourakis P., Burtness B., Fountzilas G., et al. (2010). Nuclear localization of signal transducer and activator of transcription 3 in head and neck squamous cell carcinoma is associated with a better prognosis. Clin. Cancer Res. 16, 2427–2434. 10.1158/1078-0432.CCR-09-2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Qin J., Zhang Y., Cheng X., Wang X., Lu W., et al. (2017). Autophagy maintains the stemness of ovarian cancer stem cells by FOXA2. J. Exp. Clin. Cancer Res. 36, 171. 10.1186/s13046-017-0644-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentland I., Campos-Leon K., Cotic M., Davies K. J., Wood C. D., Groves I. J., et al. (2018). Disruption of CTCF-YY1-dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PloS Biol. 16, e2005752. 10.1371/journal.pbio.2005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos J., Franco E. L., Black M. J., Bergeron J., Arella M. (1999). Human papillomavirus and prognoses of patients with cancers of the upper aerodigestive tract. Cancer 85, 1903–1909. [DOI] [PubMed] [Google Scholar]
- Pollock N. I., Wang L., Wallweber G., Gooding W. E., Huang W., Chenna A., et al. (2015). Increased Expression of HER2, HER3, and HER2:HER3 Heterodimers in HPV-Positive HNSCC Using a Novel Proximity-Based Assay: Implications for Targeted Therapies. Clin. Cancer Res. 21, 4597–4606. 10.1158/1078-0432.CCR-14-3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner M. R., Lorch J. H., Goloubeva O., Tan M., Schumaker L. M., Sarlis N. J., et al. (2011). Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann. Oncol. 22, 1071–1077. 10.1093/annonc/mdr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn G. J., Van Driel W., Van Der Vliet P. C. (1986). Nuclear factor III, a novel sequence-specific DNA-binding protein from HeLa cells stimulating adenovirus DNA replication. Nature 322, 656–659. 10.1038/322656a0 [DOI] [PubMed] [Google Scholar]
- Prusty B. K., Husain S. A., Das B. C. (2005). Constitutive activation of nuclear factor -kB: preferntial homodimerization of p50 subunits in cervical carcinoma. Front. Biosci. 10, 1510–1519. 10.2741/1635 [DOI] [PubMed] [Google Scholar]
- Qiao B., He B., Cai J., Yang W. (2014). The expression profile of Oct4 and Sox2 in the carcinogenesis of oral mucosa. Int. J. Clin. Exp. Pathol. 7, 28–37. [PMC free article] [PubMed] [Google Scholar]
- Qu S. Y., Sun Y. Y., Li Y. H., Xu Z. M., Fu W. N. (2017). YY1 directly suppresses MYCT1 leading to laryngeal tumorigenesis and progress. Cancer Med. 6, 1389–1398. 10.1002/cam4.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff A. B., Woodham A. W., Raff L. M., Skeate J. G., Yan L., Da Silva D. M., et al. (2013). The evolving field of human papillomavirus receptor research: a review of binding and entry. J. Virol. 87, 6062–6072. 10.1128/JVI.00330-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragin C. C., Taioli E. (2007). Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int. J. Cancer 121, 1813–1820. 10.1002/ijc.22851 [DOI] [PubMed] [Google Scholar]
- Rampias T., Sasaki C., Weinberger P., Psyrri A. (2009). E6 and e7 gene silencing and transformed phenotype of human papillomavirus 16-positive oropharyngeal cancer cells. J. Natl. Cancer Inst. 101, 412–423. 10.1093/jnci/djp017 [DOI] [PubMed] [Google Scholar]
- Rautava J., Syrjanen S. (2011). Human papillomavirus infections in the oral mucosa. J. Am. Dent. Assoc. 142, 905–914. 10.14219/jada.archive.2011.0297 [DOI] [PubMed] [Google Scholar]
- Ren S., Gaykalova D., Wang J., Guo T., Danilova L., Favorov A., et al. (2018). Discovery and development of differentially methylated regions in human papillomavirus-related oropharyngeal squamous cell carcinoma. Int. J. Cancer 143, 2425–2436. 10.1002/ijc.31778 [DOI] [PubMed] [Google Scholar]
- Reuschenbach M., Huebbers C. U., Prigge E. S., Bermejo J. L., Kalteis M. S., Preuss S. F., et al. (2015). Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer 121, 1966–1976. 10.1002/cncr.29315 [DOI] [PubMed] [Google Scholar]
- Ribeiro A. L., Caodaglio A. S., Sichero L. (2018). Regulation of HPV transcription. Clinics (Sao Paulo) 73, e486s. 10.6061/clinics/2018/e486s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K. L., Zhang B., Baggerly K. A., Colella S., Lang J. C., Schuller D. E., et al. (2009). Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PloS One 4, e4941. 10.1371/journal.pone.0004941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann T., Tribius S., Grob T. J., Meyer F., Busch C. J., Petersen C., et al. (2013). HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother. Oncol. 107, 242–246. 10.1016/j.radonc.2013.03.013 [DOI] [PubMed] [Google Scholar]
- Rinkenbaugh A. L., Baldwin A. S. (2016). The NF-kappaB Pathway and Cancer Stem Cells. Cells 5, 1–19. 10.3390/cells5020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischin D., Young R. J., Fisher R., Fox S. B., Le Q. T., Peters L. J., et al. (2010). Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J. Clin. Oncol. 28, 4142–4148. 10.1200/JCO.2010.29.2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer J., Wuertz B. R., Ondrey F. (2010). Cigarette smoke condensate induces nuclear factor kappa-b activity and proangiogenic growth factors in aerodigestive cells. Laryngoscope 120, 1609–1613. 10.1002/lary.20972 [DOI] [PubMed] [Google Scholar]
- Rosenquist K. (2005). Risk factors in oral and oropharyngeal squamous cell carcinoma: a population-based case-control study in southern Sweden. Swed. Dent. J. Suppl. 179, 1–66. [PubMed] [Google Scholar]
- Safe S., Kim K. (2004). Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog. Nucleic Acid Res. Mol. Biol. 77, 1–36. 10.1016/S0079-6603(04)77001-4 [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan R., Masuyer E., Swaminathan R., Ferlay J., Whelan S. (1998). Head and neck cancer: a global perspective on epidemiology and prognosis. Anticancer Res. 18, 4779–4786. [PubMed] [Google Scholar]
- Sarkola M. E., Grenman S. E., Rintala M. A., Syrjanen K. J., Syrjanen S. M. (2008). Human papillomavirus in the placenta and umbilical cord blood. Acta Obstet. Gynecol Scand. 87, 1181–1188. 10.1080/00016340802468308 [DOI] [PubMed] [Google Scholar]
- Sartor M. A., Dolinoy D. C., Jones T. R., Colacino J. A., Prince M. E., Carey T. E., et al. (2011). Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 6, 777–787. 10.4161/epi.6.6.16216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurlen W., Stremlau A., Gissmann L., Hohn D., Zenner H. P., Zur Hausen H. (1986). Rearranged HPV 16 molecules in an anal and in a laryngeal carcinoma. Int. J. Cancer 38, 671–676. 10.1002/ijc.2910380509 [DOI] [PubMed] [Google Scholar]
- Schindler C., Shuai K., Prezioso V. R., Darnell J. E., Jr. (1992). Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257, 809–813. 10.1126/science.1496401 [DOI] [PubMed] [Google Scholar]
- Schreiber M., Kolbus A., Piu F., Szabowski A., Mohle-Steinlein U., Tian J., et al. (1999). Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 13, 607–619. 10.1101/gad.13.5.607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrock A., Bode M., Goke F. J., Bareiss P. M., Schairer R., Wang H., et al. (2014). Expression and role of the embryonic protein SOX2 in head and neck squamous cell carcinoma. Carcinogenesis 35, 1636–1642. 10.1093/carcin/bgu094 [DOI] [PubMed] [Google Scholar]
- Seidel H. M., Milocco L. H., Lamb P., Darnell J. E., Jr., Stein R. B., Rosen J. (1995). Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 92, 3041–3045. 10.1073/pnas.92.7.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiwert T. Y., Zuo Z., Keck M. K., Khattri A., Pedamallu C. S., Stricker T., et al. (2015). Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin. Cancer Res. 21, 632–641. 10.1158/1078-0432.CCR-13-3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson D., Horvath S., Huerta-Yepez S., Hanna S., Garban H., Roberts A., et al. (2005). Expression of transcription factor Yin Yang 1 in prostate cancer. Int. J. Oncol. 27, 131–141. 10.3892/ijo.27.1.131 [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. (1986). Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716. 10.1016/0092-8674(86)90346-6 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Vonesch J. L., Waltzinger C., Zheng H., Wasylyk B. (2000). Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells. EMBO J. 19, 6051–6064. 10.1093/emboj/19.22.6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N. G., Trivedi T. I., Tankshali R. A., Goswami J. A., Jetly D. H., Kobawala T. P., et al. (2006). Stat3 expression in oral squamous cell carcinoma: association with clinicopathological parameters and survival. Int. J. Biol. Markers 21, 175–183. 10.1177/172460080602100307 [DOI] [PubMed] [Google Scholar]
- Shah A., Malik A., Garg A., Mair M., Nair S., Chaturvedi P. (2017). Oral sex and human papilloma virus-related head and neck squamous cell cancer: a review of the literature. Postgrad. Med. J. 93, 704–709. 10.1136/postgradmedj-2016-134603 [DOI] [PubMed] [Google Scholar]
- Shaikh M. H., Mcmillan N. A., Johnson N. W. (2015). HPV-associated head and neck cancers in the Asia Pacific: A critical literature review & meta-analysis. Cancer Epidemiol. 39, 923–938. 10.1016/j.canep.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Shangina O., Brennan P., Szeszenia-Dabrowska N., Mates D., Fabianova E., Fletcher T., et al. (2006). Occupational exposure and laryngeal and hypopharyngeal cancer risk in central and eastern Europe. Am. J. Epidemiol. 164, 367–375. 10.1093/aje/kwj208 [DOI] [PubMed] [Google Scholar]
- Shaulian E., Karin M. (2002). AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136. 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L. S., Shenk T. (1991). Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388. 10.1016/0092-8674(91)90189-6 [DOI] [PubMed] [Google Scholar]
- Shield K. D., Ferlay J., Jemal A., Sankaranarayanan R., Chaturvedi A. K., Bray F., et al. (2017). The global incidence of lip, oral cavity, and pharyngeal cancers by subsite in 2012. CA Cancer J. Clin. 67, 51–64. 10.3322/caac.21384 [DOI] [PubMed] [Google Scholar]
- Shin J. A., Kim J. S., Kwon K. H., Nam J. S., Jung J. Y., Cho N. P., et al. (2012). Apoptotic effect of hot water extract of Sanguisorba officinalis L. @ in human oral cancer cells. Oncol. Lett. 4, 489–494. 10.3892/ol.2012.748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. A., Kim J. J., Choi E. S., Shim J. H., Ryu M. H., Kwon K. H., et al. (2013). In vitro apoptotic effects of methanol extracts of Dianthus chinensis and Acalypha australis L. targeting specificity protein 1 in human oral cancer cells. Head Neck 35, 992–998. 10.1002/hed.23072 [DOI] [PubMed] [Google Scholar]
- Shroyer K. R., Greer R. O., Jr. (1991). Detection of human papillomavirus DNA by in situ DNA hybridization and polymerase chain reaction in premalignant and malignant oral lesions. Oral. Surg. Oral. Med. Oral. Pathol. 71, 708–713. 10.1016/0030-4220(91)90279-L [DOI] [PubMed] [Google Scholar]
- Shuai K., Schindler C., Prezioso V. R., Darnell J. E., Jr. (1992). Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258, 1808–1812. 10.1126/science.1281555 [DOI] [PubMed] [Google Scholar]
- Shukla S., Bharti A. C., Mahata S., Hussain S., Kumar R., Hedau S., et al. (2009). Infection of human papillomaviruses in cancers of different human organ sites. Indian J. Med. Res. 130, 222–233. [PubMed] [Google Scholar]
- Shukla S., Shishodia G., Mahata S., Hedau S., Pandey A., Bhambhani S., et al. (2010). Aberrant expression and constitutive activation of STAT3 in cervical carcinogenesis: implications in high-risk human papillomavirus infection. Mol. Cancer 9, 282. 10.1186/1476-4598-9-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Jadli M., Thakur K., Shishodia G., Mahata S., Basir S. F., et al. (2019). Level of phospho-STAT3 (Tyr705) correlates with copy number and physical state of human papillomavirus 16 genome in cervical precancer and cancer lesions. PloS One 14, e0222089. 10.1371/journal.pone.0222089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichero L., Sobrinho J. S., Villa L. L. (2012). Identification of novel cellular transcription factors that regulate early promoters of human papillomavirus types 18 and 16. J. Infect. Dis. 206, 867–874. 10.1093/infdis/jis430 [DOI] [PubMed] [Google Scholar]
- Slebos R. J., Yi Y., Ely K., Carter J., Evjen A., Zhang X., et al. (2006). Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin. Cancer Res. 12, 701–709. 10.1158/1078-0432.CCR-05-2017 [DOI] [PubMed] [Google Scholar]
- Smith E. M., Ritchie J. M., Summersgill K. F., Hoffman H. T., Wang D. H., Haugen T. H., et al. (2004. a). Human papillomavirus in oral exfoliated cells and risk of head and neck cancer. J. Natl. Cancer Inst. 96, 449–455. 10.1093/jnci/djh074 [DOI] [PubMed] [Google Scholar]
- Smith E. M., Ritchie J. M., Summersgill K. F., Klussmann J. P., Lee J. H., Wang D., et al. (2004. b). Age, sexual behavior and human papillomavirus infection in oral cavity and oropharyngeal cancers. Int. J. Cancer 108, 766–772. 10.1002/ijc.11633 [DOI] [PubMed] [Google Scholar]
- Snijders P. J., Cromme F. V., Van Den Brule A. J., Schrijnemakers H. F., Snow G. B., Meijer C. J., et al. (1992). Prevalence and expression of human papillomavirus in tonsillar carcinomas, indicating a possible viral etiology. Int. J. Cancer 51, 845–850. 10.1002/ijc.2910510602 [DOI] [PubMed] [Google Scholar]
- Soanca A., Lupse M., Moldovan M., Pall E., Cenariu M., Roman A., et al. (2018). Applications of inflammation-derived gingival stem cells for testing the biocompatibility of dental restorative biomaterials. Ann. Anat. 218, 28–39. 10.1016/j.aanat.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Song J. I., Grandis J. R. (2000). STAT signaling in head and neck cancer. Oncogene 19, 2489–2495. 10.1038/sj.onc.1203483 [DOI] [PubMed] [Google Scholar]
- Soto U., Denk C., Finzer P., Hutter K. J., Zur Hausen H., Rosl F. (2000). Genetic complementation to non-tumorigenicity in cervical-carcinoma cells correlates with alterations in AP-1 composition. Int. J. Cancer 86, 811–817. [DOI] [PubMed] [Google Scholar]
- Sovak M. A., Bellas R. E., Kim D. W., Zanieski G. J., Rogers A. E., Traish A. M., et al. (1997). Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 100, 2952–2960. 10.1172/JCI119848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitkovsky D., Hehner S. P., Hofmann T. G., Moller A., Schmitz M. L. (2002). The human papillomavirus oncoprotein E7 attenuates NF-kappa B activation by targeting the Ikappa B kinase complex. J. Biol. Chem. 277, 25576–25582. 10.1074/jbc.M201884200 [DOI] [PubMed] [Google Scholar]
- Squarize C. H., Castilho R. M., Sriuranpong V., Pinto D. S., Jr., Gutkind J. S. (2006). Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia 8, 733–746. 10.1593/neo.06274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanovic M., Zuffardi O., Collignon J., Lovell-Badge R., Goodfellow P. (1994). The cDNA sequence and chromosomal location of the human SOX2 gene. Mamm. Genome 5, 640–642. 10.1007/BF00411460 [DOI] [PubMed] [Google Scholar]
- Stransky N., Egloff A. M., Tward A. D., Kostic A. D., Cibulskis K., Sivachenko A., et al. (2011). The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Liang J., Wang Q., Li Z., Du Y., Xu X. (2016). MicroRNA-137 suppresses tongue squamous carcinoma cell proliferation, migration and invasion. Cell Prolif. 49, 628–635. 10.1111/cpr.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surviladze Z., Dziduszko A., Ozbun M. A. (2012). Essential roles for soluble virion-associated heparan sulfonated proteoglycans and growth factors in human papillomavirus infections. PloS Pathog. 8, e1002519. 10.1371/journal.ppat.1002519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surviladze Z., Sterk R. T., Deharo S. A., Ozbun M. A. (2013). Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J. Virol. 87, 2508–2517. 10.1128/JVI.02319-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson W. G., Wuertz B. R., Ondrey F. G. (2011). Tobacco carcinogen mediated up-regulation of AP-1 dependent pro-angiogenic cytokines in head and neck carcinogenesis. Mol. Carcinog. 50, 668–679. 10.1002/mc.20775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrjanen K., Syrjanen S., Lamberg M., Pyrhonen S., Nuutinen J. (1983). Morphological and immunohistochemical evidence suggesting human papillomavirus (HPV) involvement in oral squamous cell carcinogenesis. Int. J. Oral. Surg. 12, 418–424. 10.1016/S0300-9785(83)80033-7 [DOI] [PubMed] [Google Scholar]
- Syrjanen S., Happonen R. P., Virolainen E., Siivonen L., Syrjanen K. (1987). Detection of human papillomavirus (HPV) structural antigens and DNA types in inverted papillomas and squamous cell carcinomas of the nasal cavities and paranasal sinuses. Acta Otolaryngol. 104, 334–341. 10.3109/00016488709107337 [DOI] [PubMed] [Google Scholar]
- Syrjanen S., Andersson B., Juntunen L., Syrjanen K. (1992). Polymerase chain reaction for producing biotinylated human papillomavirus DNA probes for in situ hybridization. Sex Transm. Dis. 19, 140–145. 10.1097/00007435-199205000-00006 [DOI] [PubMed] [Google Scholar]
- Syrjanen S., Rautava J., Syrjanen K. (2017). HPV in Head and Neck Cancer-30 Years of History. Recent Results Cancer Res. 206, 3–25. 10.1007/978-3-319-43580-0_1 [DOI] [PubMed] [Google Scholar]
- Syrjanen K. J. (2003). HPV infections in benign and malignant sinonasal lesions. J. Clin. Pathol. 56, 174–181. 10.1136/jcp.56.3.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrjanen S. (2018). Oral manifestations of human papillomavirus infections. Eur. J. Oral. Sci. 126 Suppl 1, 49–66. 10.1111/eos.12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. H., Gloss B., Bernard H. U. (1992). During negative regulation of the human papillomavirus-16 E6 promoter, the viral E2 protein can displace Sp1 from a proximal promoter element. Nucleic Acids Res. 20, 251–256. 10.1093/nar/20.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville M. M., Kim J. (2011). Oncogenic potential of yin yang 1 mediated through control of imprinted genes. Crit. Rev. Oncog. 16, 199–209. 10.1615/CritRevOncog.v16.i3-4.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry F., Spyrou G., Yaniv M., Howley P. (1992). Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J. Virol. 66, 3740–3748. 10.1128/JVI.66.6.3740-3748.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons S. R., Nwankwo J. O., Domann F. E. (2002). Acetaldehyde activates Jun/AP-1 expression and DNA binding activity in human oral keratinocytes. Oral. Oncol. 38, 281–290. 10.1016/S1368-8375(01)00056-2 [DOI] [PubMed] [Google Scholar]
- Tomoshige K. (2017). Glucocorticoid Receptor. In: Feingold KR, Anawalt B, Boyce AEA, editors. Endotext. South Dartmouth, MA: MDText.com, Inc. Available at: www.endotext.org. [Google Scholar]
- Torrente M. C., Rodrigo J. P., Haigentz M., Jr., Dikkers F. G., Rinaldo A., Takes R. P., et al. (2011). Human papillomavirus infections in laryngeal cancer. Head Neck 33, 581–586. 10.1002/hed.21421 [DOI] [PubMed] [Google Scholar]
- Tsai C. N., Tsai C. L., Yi J. S., Kao H. K., Huang Y., Wang C. I., et al. (2019). Activin A regulates the epidermal growth factor receptor promoter by activating the PI3K/SP1 pathway in oral squamous cell carcinoma cells. Sci. Rep. 9, 5197. 10.1038/s41598-019-41396-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten V. M., Snijders P. J., Van Den Brekel M. W., Kummer J. A., Meijer C. J., Van Leeuwen B., et al. (2001). Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int. J. Cancer 93, 232–235. 10.1002/ijc.1313 [DOI] [PubMed] [Google Scholar]
- Vazquez-Arreguin K., Tantin D. (2016). The Oct1 transcription factor and epithelial malignancies: Old protein learns new tricks. Biochim. Biophys. Acta 1859, 792–804. 10.1016/j.bbagrm.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde P., Casalino L., Talotta F., Yaniv M., Weitzman J. B. (2007). Deciphering AP-1 function in tumorigenesis: fra-ternizing on target promoters. Cell Cycle 6, 2633–2639. 10.4161/cc.6.21.4850 [DOI] [PubMed] [Google Scholar]
- Verma G., Vishnoi K., Tyagi A., Jadli M., Singh T., Goel A., et al. (2017). Characterization of key transcription factors as molecular signatures of HPV-positive and HPV-negative oral cancers. Cancer Med. 6, 591–604. 10.1002/cam4.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorvis C., Hatziapostolou M., Mahurkar-Joshi S., Koutsioumpa M., Williams J., Donahue T. R., et al. (2016). Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G1124–G1137. 10.1152/ajpgi.00035.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M., Huang W., Kute T. E., Miller L. D., Zhang Q., Hatcher H., et al. (2012). Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am. J. Pathol. 180, 2120–2133. 10.1016/j.ajpath.2012.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu K., Yuan F., Berdichevsky L., Taichman L. B., Auborn K. (1996). C/EBPbeta is a negative regulator of human papillomavirus type 11 in keratinocytes. J. Virol. 70, 4839–4844. 10.1128/JVI.70.7.4839-4844.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Abbruzzese J. L., Evans D. B., Larry L., Cleary K. R., Chiao P. J. (1999). The nuclear factor-kappa B RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin. Cancer Res. 5, 119–127. [PubMed] [Google Scholar]
- Wang W. M., Wu S. Y., Lee A. Y., Chiang C. M. (2011). Binding site specificity and factor redundancy in activator protein-1-driven human papillomavirus chromatin-dependent transcription. J. Biol. Chem. 286, 40974–40986. 10.1074/jbc.M111.290874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yi M., Chen S., Li J., Li G., Yang J., et al. (2016). Significance of the NOR1-FOXA1/HDAC2-Slug regulatory network in epithelial-mesenchymal transition of tumor cells. Oncotarget 7, 16745–16759. 10.18632/oncotarget.7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A., Hengge U. R., Stricker I., Tischoff I., Markwart A., Anhalt K., et al. (2007). Protein microarrays for the detection of biomarkers in head and neck squamous cell carcinomas. Hum. Pathol. 38, 228–238. 10.1016/j.humpath.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Wolf J. S., Chen Z., Dong G., Sunwoo J. B., Bancroft C. C., Capo D. E., et al. (2001). IL (interleukin)-1alpha promotes nuclear factor-kappaB and AP-1-induced IL-8 expression, cell survival, and proliferation in head and neck squamous cell carcinomas. Clin. Cancer Res. 7, 1812–1820. [PubMed] [Google Scholar]
- Wookey V. B., Appiah A. K., Kallam A., Ernani V., Smith L. M., Ganti A. K. (2019). HPV Status and Survival in Non-Oropharyngeal Squamous Cell Carcinoma of the Head and Neck. Anticancer Res. 39, 1907–1914. 10.21873/anticanres.13299 [DOI] [PubMed] [Google Scholar]
- Wuebben E. L., Rizzino A. (2017). The dark side of SOX2: cancer - a comprehensive overview. Oncotarget 8, 44917–44943. 10.18632/oncotarget.16570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthopoulos K. G., Prezioso V. R., Chen W. S., Sladek F. M., Cortese R., Darnell J. E., Jr. (1991). The different tissue transcription patterns of genes for HNF-1, C/EBP, HNF-3, and HNF-4, protein factors that govern liver-specific transcription. Proc. Natl. Acad. Sci. U. S. A. 88, 3807–3811. 10.1073/pnas.88.9.3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Chen J., Xu M., Yang H., Luo J., Pan Y., et al. (2017). Genetic variability and functional implication of the long control region in HPV-16 variants in Southwest China. PloS One 12, e0182388. 10.1371/journal.pone.0182388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. (1991). Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65, 551–568. 10.1016/0092-8674(91)90088-G [DOI] [PubMed] [Google Scholar]
- Xiao C., Beitler J. J., Higgins K. A., Wommack E. C., Saba N. F., Shin D. M., et al. (2018). Differential regulation of NF-kB and IRF target genes as they relate to fatigue in patients with head and neck cancer. Brain Behav. Immun. 74, 291–295. 10.1016/j.bbi.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Jin X., Yuan Y., Deng P., Jiang L., Zeng X., et al. (2017). Prognostic value from integrative analysis of transcription factors c-Jun and Fra-1 in oral squamous cell carcinoma: a multicenter cohort study. Sci. Rep. 7, 7522. 10.1038/s41598-017-05106-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Yang X., Lee T. L., Friedman J., Tang J., Van Waes C., et al. (2007). Genome-wide identification of novel expression signatures reveal distinct patterns and prevalence of binding motifs for p53, nuclear factor-kappaB and other signal transcription factors in head and neck squamous cell carcinoma. Genome Biol. 8, R78. 10.1186/gb-2007-8-5-r78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Hui L., Wang Y., Yang H., Jiang X. (2014). SOX2 promotes the migration and invasion of laryngeal cancer cells by induction of MMP-2 via the PI3K/Akt/mTOR pathway. Oncol. Rep. 31, 2651–2659. 10.3892/or.2014.3120 [DOI] [PubMed] [Google Scholar]
- Yang X., Cheng H., Chen J., Wang R., Saleh A., Si H., et al. (2019). Head and Neck Cancers Promote an Inflammatory Transcriptome through Coactivation of Classic and Alternative NF-kappaB Pathways. Cancer Immunol. Res. 7, 1760–1774. 10.1158/2326-6066.CIR-18-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J. C., Wang L., Wei D., Gong W., Hassan M., Wu T. T., et al. (2004). Association between expression of transcription factor Sp1 and increased vascular endothelial growth factor expression, advanced stage, and poor survival in patients with resected gastric cancer. Clin. Cancer Res. 10, 4109–4117. 10.1158/1078-0432.CCR-03-0628 [DOI] [PubMed] [Google Scholar]
- Yarbrough W. G., Whigham A., Brown B., Roach M., Slebos R. (2007). Phosphoinositide kinase-3 status associated with presence or absence of human papillomavirus in head and neck squamous cell carcinomas. Int. J. Radiat. Oncol. Biol. Phys. 69, S98–101. 10.1016/j.ijrobp.2007.04.082 [DOI] [PubMed] [Google Scholar]
- Young L. S., Dawson C. W., Clark D., Rupani H., Busson P., Tursz T., et al. (1988). Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J. Gen. Virol. 69 ( Pt 5), 1051–1065. 10.1099/0022-1317-69-5-1051 [DOI] [PubMed] [Google Scholar]
- Young M. R., Farrell L., Lambert P., Awasthi P., Colburn N. H. (2002). Protection against human papillomavirus type 16-E7 oncogene-induced tumorigenesis by in vivo expression of dominant-negative c-jun. Mol. Carcinog. 34, 72–77. 10.1002/mc.10050 [DOI] [PubMed] [Google Scholar]
- Yu H. J., Shin J. A., Nam J. S., Kang B. S., Cho S. D. (2013). Apoptotic effect of dibenzylideneacetone on oral cancer cells via modulation of specificity protein 1 and Bax. Oral. Dis. 19, 767–774. 10.1111/odi.12062 [DOI] [PubMed] [Google Scholar]
- Yu H., Jove R. (2004). The STATs of cancer–new molecular targets come of age. Nat. Rev. Cancer 4, 97–105. 10.1038/nrc1275 [DOI] [PubMed] [Google Scholar]
- Yue P., Turkson J. (2009). Targeting STAT3 in cancer: how successful are we? Expert Opin. Invest. Drugs 18, 45–56. 10.1517/13543780802565791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Stovall D. B., Inoue K., Sui G. (2011). The oncogenic role of Yin Yang 1. Crit. Rev. Oncog. 16, 163–197. 10.1615/CritRevOncog.v16.i3-4.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Li X., Wu C. W., Dong Y., Cai M., Mok M. T., et al. (2013). microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 32, 5078–5088. 10.1038/onc.2012.526 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Yang C., Gao W., Chen T., Qian T., Hu J., et al. (2015). FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 361, 240–250. 10.1016/j.canlet.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Zhang J., Chen T., Yang X., Cheng H., Spath S. S., Clavijo P. E., et al. (2018). Attenuated TRAF3 Fosters Activation of Alternative NF-kappaB and Reduced Expression of Antiviral Interferon, TP53, and RB to Promote HPV-Positive Head and Neck Cancers. Cancer Res. 78, 4613–4626. 10.1158/0008-5472.CAN-17-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jiang S., Guo L., Li X. (2019). MicroRNA-1258, regulated by c-Myb, inhibits growth and epithelial-to-mesenchymal transition phenotype via targeting SP1 in oral squamous cell carcinoma. J. Cell Mol. Med. 23, 2813–2821. 10.1111/jcmm.14189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. J., Zhu Y., Xie K. L., Peng Y. P., Tao J. Q., Tang J., et al. (2014). Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol. Cancer 13, 130. 10.1186/1476-4598-13-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Mirani N., Baisre A., Fernandes H. (2014). Molecular heterogeneity of head and neck squamous cell carcinoma defined by next-generation sequencing. Am. J. Pathol. 184, 1323–1330. 10.1016/j.ajpath.2014.01.028 [DOI] [PubMed] [Google Scholar]
- Zhao W., Chow L. T., Broker T. R. (1999). A distal element in the HPV-11 upstream regulatory region contributes to promoter repression in basal keratinocytes in squamous epithelium. Virology 253, 219–229. 10.1006/viro.1998.9478 [DOI] [PubMed] [Google Scholar]
- Zhao J., Ding D., Zhao G. (2019). Reduced miR-202 levels enhanced oral cancer development via targeting Sp1. Exp. Ther. Med. 18, 489–496. 10.3892/etm.2019.7603 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhong Z., Wen Z., Darnell J. E., Jr. (1994). Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98. 10.1126/science.8140422 [DOI] [PubMed] [Google Scholar]
- Zhong H., May M. J., Jimi E., Ghosh S. (2002). The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell 9, 625–636. 10.1016/S1097-2765(02)00477-X [DOI] [PubMed] [Google Scholar]
- Zoumpourlis V., Papadakis E., Delakas D., Cranidis A., Segas J., Papadakis H., et al. (1994). Human lung, bladder and head and neck tumors as compared to their adjacent normal-tissues have elevated ap-1 activity and recognize sequence elements of hiv-1 LTR. Oncol. Rep. 1, 403–409. 10.3892/or.1.2.403 [DOI] [PubMed] [Google Scholar]