Abstract
Generalist species able to exploit anthropogenic food sources are becoming increasingly common in urban environments. Coyotes (Canis latrans) are one such urban generalist that now resides in cities across North America, where diseased or unhealthy coyotes are frequently reported in cases of human-wildlife conflict. Coyote health and fitness may be related to habitat use and diet via the gut microbiome, which has far-reaching effects on animal nutrition and physiology. In this study, we used stomach contents, stable isotope analysis, 16S rRNA gene amplicon sequencing, and measures of body condition to identify relationships among habitat use, diet, fecal microbiome composition, and health in urban and rural coyotes. Three distinct relationships emerged: (1) Urban coyotes consumed more anthropogenic food, which was associated with increased microbiome diversity, higher abundances of Streptococcus and Enterococcus, and poorer average body condition. (2) Conversely, rural coyotes harbored microbiomes rich in Fusobacteria, Sutterella, and Anaerobiospirillum, which were associated with protein-rich diets and improved body condition. (3) Diets rich in anthropogenic food were associated with increased abundances of Erysipelotrichiaceae, Lachnospiraceae, and Coriobacteriaceae, which correlated with larger spleens in urban coyotes. Urban coyotes also had an increased prevalence of the zoonotic parasite Echinococcus multilocularis, but there were no detectable connections between parasite infection and microbiome composition. Our results demonstrate how the consumption of carbohydrate-rich anthropogenic food by urban coyotes alters the microbiome to negatively affect body condition, with potential relationships to parasite susceptibility and conflict-prone behavior.
Subject terms: Microbial ecology, Urban ecology
Introduction
Urbanization is causing dramatic changes to terrestrial ecosystems. For many species, the selective pressures created by the expanding urban landscape1,2 lead to local extirpations and consequent decreases in local biodiversity3, but several generalist species can thrive in urban environments4–6. The success of these urban generalists is largely enabled by behavioral adaptations, foremost of which is broadening their diet to exploit abundant but often variable sources of anthropogenic food4. Urban habitat use has been loosely associated with physical costs7, including unhealthy weight gain8 and increased cholesterol and corticosterone levels9,10. However, the direct physiological consequences of eating anthropogenic food remain less well understood, despite likely downstream effects on human-wildlife interactions including dependency, conflict-prone behavior, and the spread of zoonotic diseases11.
The gut microbiome may play a pivotal role linking the consumption of anthropogenic food by urban wildlife to changes in their physiology and ecology because it is necessarily altered by changes in diet and has far-reaching effects on nutrient assimilation, immune system function, and overall fitness12,13. Healthy, diverse gut microbiomes facilitate host digestion and nutrition14 and can act as a barrier to infection15. These effects have direct implications for host health and behavior. For example, variations in the animal gut microbiota have been linked to parasite susceptibility16 and body condition17, and an altered gut microbiota has been associated with aggression in neglected (and presumably malnourished) canines18. In some cases, specific bacterial taxa, such as Bifidobacterium and Lactobacillus in humans, have been identified for their beneficial effects on host health19.
Microbiome research continues to demonstrate the importance of the microbiome across diverse animal hosts in the context of environmental perturbation. Urban-induced alterations in the microbiome have been observed in wild birds20–23 and linked to physiological stress in squirrels24. Disturbances related to urbanization have been shown to affect microbiome diversity and composition in several animal species25–27. Some authors have suggested that these anthropogenic threats to host-associated microbial communities have important implications for wildlife management practices28. However, for information about the effects of microbiome alterations in urban wildlife to be meaningful to managers, it must also be accompanied by an understanding of the “healthy” microbiome in each species. Moreover, most current descriptions of the animal microbiome draw primarily from captive or model organisms, but captivity itself affects the microbiome29,30 and, due to natural physiological differences among animal taxa, results from one species cannot reliably be extrapolated to another31. It is therefore important to study directly how the complex process of urban adaptation affects the microbiome of different urban-adapted species.
In North America, coyotes (Canis latrans) are becoming a common resident of most major cities, which has coincided with increased reports of human-coyote conflict32. Their success in urban environments stems from their generalist and flexible diet32 and behavioral plasticity33. Although coyotes mainly consume insects, rodents, and young or diseased ungulates34, they may also consume fruit and anthropogenic food32,35. They are also a definitive host for the helminth parasite Echinococcus multilocularis, which is expanding its range in North America and can cause a rare but severe zoonosis in humans36. Previous work in our lab has shown that urban and conflict-prone coyotes eat less protein35, that consumption of anthropogenic food in urban coyotes is correlated with higher prevalence of parasites37,38, and that urban coyotes have an unusually high prevalence of E. multilocularis39. Knowledge of which microbial signatures are associated with diet, body condition, or parasite infection in coyotes would not only provide novel insights into host-microbiome relationships in wild canids but also have direct management implications for evaluating or monitoring host fitness, especially in the contexts of potential conflict or the spread of canid-borne zoonoses.
In this study, we tested the hypothesis that the consumption of anthropogenic food by urban coyotes causes a quantifiable shift in microbiome composition, with consequent declines in physiological condition. Using a sample of coyotes from urban and rural areas near Edmonton, Alberta, we identified specific relationships among diet, fecal bacterial communities, and host health, with an emphasis on generating a foundational understanding of which microbiome features most strongly connect diet to health in a wild canid. Our results support past evidence that urban coyotes eat a broader diet of lower quality, and additionally show that this is associated with poorer average body condition, increased immune stress, and a higher prevalence of a zoonotic parasite. Moreover, our results implicate specific bacterial taxa that characterize this shift in urban coyotes and provide an important basis for understanding how diet, microbiome composition, and health may interact to shape human-wildlife relationships in urban areas.
Results
We obtained 30 road-killed or lethally managed coyote carcasses from Edmonton, Alberta (“urban”) and 65 coyotes that were collected or lethally managed by local fur trappers working in the surrounding area (“rural”) (Table S1). For each coyote, we measured body condition and age, and we evaluated short-term diet (i.e., past 6–8 h) using stomach contents and long-term diet (i.e., past 6–8 months) using stable isotopes. We additionally used 16S rRNA gene amplicon sequencing to characterize fecal bacterial communities and PCR to test for E. multilocularis infection. Because many physiological measures were correlated with each other, we performed dimension reduction to generate a single index of physical condition. Spleen mass, which serves as an indicator of immune function in mammals40, was corrected for body mass and retained separately from the composite physical condition index. Our coyotes exhibited expected sex differences in size, with males being larger than females, but there were no sex- or age-based differences in diet, measured as either stomach contents or stable isotope values (Table 1; Table S2).
Table 1.
ANOVA results evaluating the effects of sex, age, and capture location on measures of diet, body condition, and microbiome alpha diversity.
| Mean Value | Sex | Age | Location | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Urban | Rural | F | df | p | F | df | p | F | df | P | |
| Mass (kg) | 10.30 | 10.85 | 23.48 | 1 | < 0.001 | 48.90 | 1 | < 0.001 | 0.34 | 1 | 0.725 |
| Body size (cm) | 84.87 | 87.31 | 8.08 | 1 | 0.006 | 31.94 | 1 | < 0.001 | 1.44 | 1 | 0.233 |
| Girth (cm) | 45.73 | 46.87 | 11.29 | 1 | < 0.001 | 25.70 | 1 | < 0.001 | 0.17 | 1 | 0.684 |
| Age (years) | 1.82 | 2.64 | 5.70 | 1 | 0.019 | – | – | – | 3.18 | 1 | 0.078 |
| Spleen size (g/kg) | 2.34 | 1.70 | 1.20 | 1 | 0.276 | 2.51 | 1 | 0.117 | 28.01 | 1 | < 0.001 |
| KFI | 0.32 | 0.57 | 2.06 | 1 | 0.150 | 1.68 | 1 | 0.198 | 15.14 | 1 | < 0.001 |
| Health index | − 0.02 | 0.01 | 13.27 | 1 | < 0.001 | 40.93 | 1 | < 0.001 | 0.01 | 1 | 0.316 |
| δ13C (‰) | − 21.48 | − 22.90 | 0.01 | 1 | 0.926 | 1.38 | 1 | 0.243 | 54.11 | 1 | < 0.001 |
| δ15N (‰) | 8.69 | 8.96 | 1.53 | 1 | 0.220 | 1.98 | 1 | 0.163 | 2.24 | 1 | 0.138 |
| Vol. anthro food (ml) | 34.42 | 24.61 | 0.07 | 1 | 0.793 | 3.21 | 1 | 0.076 | 0.24 | 1 | 0.627 |
| Vol. prey (ml) | 59.97 | 211.58 | 1.06 | 1 | 0.306 | 1.21 | 1 | 0.274 | 5.75 | 1 | 0.018 |
| ASV richness | 333.40 | 294.28 | 0.12 | 1 | 0.738 | 0.90 | 1 | 0.344 | 1.93 | 1 | 0.060 |
| Shannon index | 3.11 | 2.68 | 0.48 | 1 | 0.492 | 0.17 | 1 | 0.678 | 5.71 | 1 | 0.019 |
| Faith's PD | 44.27 | 31.34 | 0.37 | 1 | 0.546 | 1.25 | 1 | 0.270 | 38.68 | 1 | < 0.001 |
| NTI | 6.47 | 4.40 | 1.89 | 1 | 0.173 | 1.37 | 1 | 0.245 | 33.11 | 1 | < 0.001 |
All variables were tested using a linear regression, with the exception of ASV richness, which was tested using a negative binomial regression. Stomach contents were log-transformed prior to testing to meet the assumptions of a normal distribution.
KFI kidney fat index, ASV amplicon sequence variant, PD phylogenetic diversity, NTI nearest taxon index.
Coyote diet and health
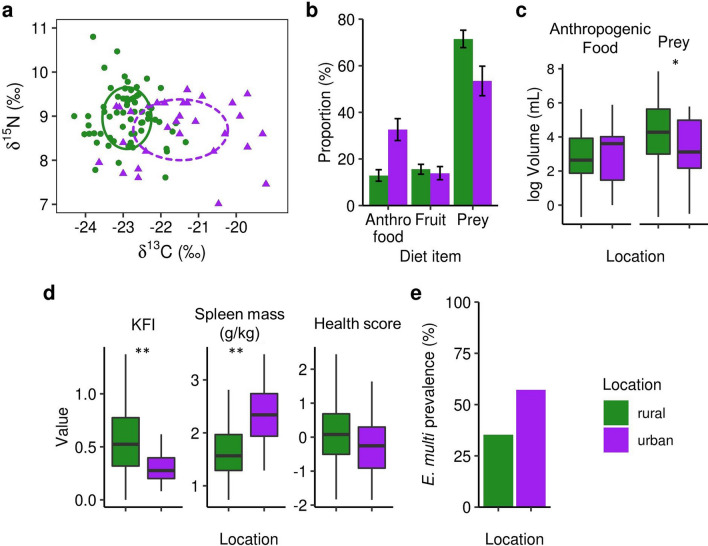
Urban coyotes consumed more anthropogenic food (e.g., compost and fast food waste) and less prey (e.g., small herbivores and ungulates) than rural coyotes. To assess habitual diet, we used stable isotope values (δ13C and δ15N) measured from claw samples. Stable isotopes can capture trends in anthropogenic food consumption because corn, which is ubiquitous in processed foods and livestock feed, has a distinctively high δ13C signature41. Higher δ15N signatures indicate greater protein consumption41. In our study, urban coyotes had a significantly higher mean δ13C signature than rural coyotes, as well as a lower mean δ15N signature, when controlling for sex and age (Fig. 1a; Table 1). Stable isotope mixing models estimated that urban coyotes consumed 2.5 times more anthropogenic food and 25% less prey than rural coyotes (Fig. 1b). We used stomach contents as a measure of short-term diet and found that urban coyotes were less likely to have natural prey in their stomach (70% vs. 84.6%), though this difference was only marginally significant (χ2 = 3.25, df = 1, p = 0.071). If natural prey was present, it was present at significantly lower volumes (Fig. 1c; Table 1). There were no significant differences in the presence or volume of anthropogenic food between urban and rural coyote stomachs (Table 1; Table S3).
Figure 1.
Diet and body condition in urban and rural coyotes. (a) Stable isotope isoscape based on δ13C and δ15N values, showing that urban coyotes have higher δ13C and lower δ15N signatures. (b) Results from a three-
source stable isotope mixing model predicting the proportions of prey, fruit, and anthropogenic food in coyote diets. (c) Mean volumes of stomach contents in urban and rural coyotes. (d) Urban coyotes have less kidney fat and larger spleens than rural coyotes and perform slightly worse on a composite index of physical condition. (e) Prevalence of E. multilocularis in urban and rural coyotes. In all figures, significant differences are indicated by asterisks (* for p < 0.05; ** for p < 0.01).
In addition to assimilating a more anthropogenic diet, urban coyotes exhibited poorer body condition than rural coyotes. Urban coyotes had half as much kidney fat, which we measured as the kidney fat index (KFI), an indicator of body fat reserves42 (Fig. 1d; Table 1). After controlling for body mass, urban coyotes also had 37% larger spleens (Fig. 1d; Table 1), suggesting they may be experiencing more challenges to their immune system40. We lastly found that urban coyotes were 50% more likely to carry the intestinal helminth E. multilocularis (Fig. 1e; 53% vs. 35% prevalence), and this increase was marginally significant (χ2 = 3.80, df = 1, p = 0.051). E. multilocularis was also more common in younger coyotes (Fig. S1). These changes in kidney fat, spleen mass, and infection prevalence were not confounded by any differences in body mass, size, or age (Table 1).
Fecal microbiome composition
We used 16S rRNA gene amplicon sequencing to investigate how the fecal microbiome may mediate the relationships between an anthropogenic diet and poor health in urban coyotes, and, equivalently, between protein-rich diets and good health in rural coyotes. Shannon diversity and phylogenetic diversity (PD) were significantly higher in urban coyotes (Fig. 2a, Table 1). ASV richness was also moderately higher in urban coyotes and, after controlling for location, in coyotes infected with E. multilocularis (Fig. S2), though these increases were only marginally significant (Table 1, Fig. S2). Generalized linear models (GLMs) confirmed that the best predictors of richness and Shannon diversity were coyote location and infection status (Fig. S3). No alpha diversity metrics were significantly associated with any measure of diet, but both richness and PD were moderately lower in healthier, older coyotes and in coyotes with empty stomachs (Fig. 2b; Figs. S3, S4). In addition, the nearest taxon index (NTI) was significantly lower in urban coyotes (Fig. 2a; Table 1). NTI can be used to assess phylogenetic clustering, or whether closely related species co-occur in an ecosystem; lower NTI values indicate that closely related species are less likely to co-occur, which suggests there may be more competitive interactions among taxa43. Aside from the effects of location, NTI was best predicted by the composite physical condition index and showed no relationship with diet (Fig. S3).
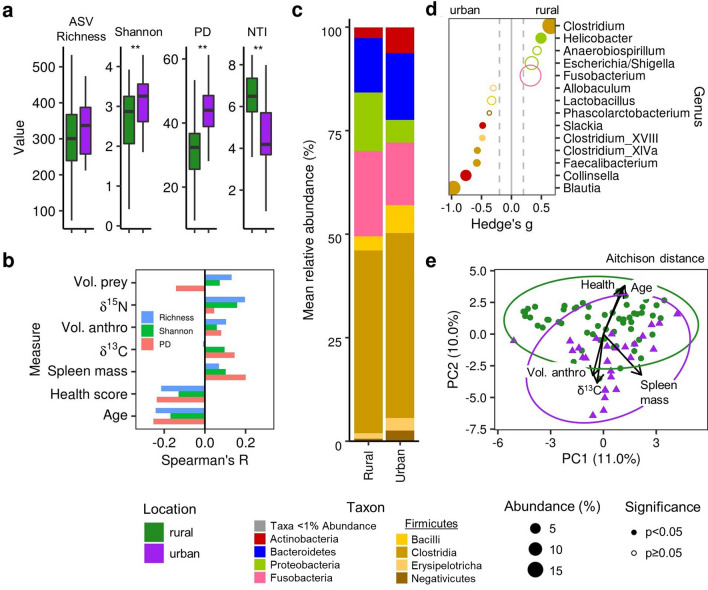
Figure 2.
Coyote fecal microbiome alpha-diversity, composition, and structure. (a) Differences in measures of ASV richness, Shannon diversity, Faith’s phylogenetic diversity (PD), and the nearest taxon index (NTI) between the fecal microbiome of urban and rural coyotes. Significant differences (p < 0.01) are indicated by asterisks. (b) Spearman correlation coefficients between richness, diversity, and PD and measures of coyote diet and body condition. (c) Average relative abundance of each taxon in urban and rural coyotes. Taxa with a mean relative abundance of < 1% are categorized as ‘other.’ Firmicutes (yellow shades) are separated to the class level; all other taxa appear at the phylum level. (d) Differentially abundant bacterial families between urban and rural coyotes, ranked by the effect-size difference between groups measured using Hedge’s g. Circle size indicates taxon relative abundance, and solid circles indicate significantly differentially abundant taxa (Benjamini–Hochberg adjusted p < 0.05). (e) Principal components analysis using the Aitchison distance, which is based on centered log-ratio transformed ASV abundances. Vectors indicate significant (p < 0.05) relationships with diet and health measures.
The differences in microbiome diversity between urban and rural samples were reflected by altered abundances of several bacterial taxa (Fig. 2c). Blautia, Collinsella, and Erysipelotrichaceae spp. were significantly more abundant in urban samples, whereas Clostridium was significantly more abundant in rural samples (Fig. 2d). No taxa at any taxonomic level were significantly differentially abundant based on sex, age, or E. multilocularis infection (Fig. S5). Random forest models were 86% effective at discriminating samples by location, with almost perfect classification of rural samples (96.7%) compared to urban (64.3%). ASVs from Lactobacillus and Fusobacterium had the most discriminatory power and were more abundant in urban and rural samples, respectively (Fig. S6). Urban samples that were misclassified as rural based on their microbiome composition had lower alpha diversity measures and smaller spleens than those that were classified correctly (Fig. S7).
Urban and rural coyote microbiomes also significantly differed in overall community composition, assessed based on an Aitchison distance-based PERMANOVA controlling for age and sex (F = 2.83, R2 = 0.032, df = 1, p = 0.001; Fig. 2e; Table S4). These location-specific differences were consistent when using alternative distance metrics that account for either taxon presence or abundance and were magnified when evaluated using the UniFrac distance metrics, which account for phylogenetic relatedness among taxa (Fig. S8; Table S4). Health, age, spleen size, and the consumption of anthropogenic food also influenced microbiome community structure, mirroring the differences in these measures between urban and rural coyotes (Fig. 2e), but sex and E. multilocularis infection did not (Fig. S9).
Taxa associated with health
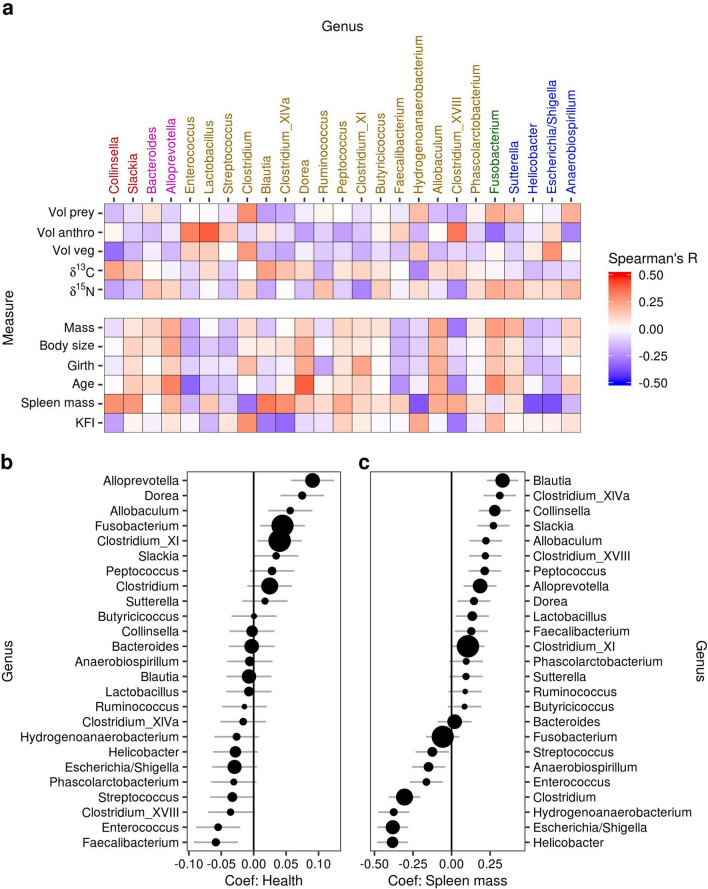
To examine how individual taxa responded to variation in coyote diet and body condition, we tested for correlations between taxon abundances and coyote metadata using both univariate and multivariate approaches. Based on Spearman’s correlation and rank-based regression models, Alloprevotella, Dorea, and Allobaculum were the best predictors of higher body condition, followed by the dominant genera Fusobacterium and Clostridium XI (Fig. 3a,b). These taxa were also positively correlated with age and both short- and long-term consumption of prey (Fig. 3a). Faecalibaterium, Enterococcus, Helicobacter, Clostridium cluster XVIII, and Streptococcus were the best predictors of poor body condition (Fig. 3a,b). Univariate approaches also showed that Blautia, Clostridium XIVa, Collinsella, and Slackia were associated with larger spleens, younger coyotes, and the consumption of anthropogenic food (Fig. 3a,c).
Figure 3.
Univariate correlations between taxon abundances, diet, and body condition. (a) Heat map indicating Spearman’s correlation coefficient between centered log-ratio transformed taxon abundances and coyote metadata. (b,c) Results from rank-based regression models that used taxon abundances to predict (b) physical condition and (c) spleen mass, while controlling for the effects of sex. Error bars indicate standard error.
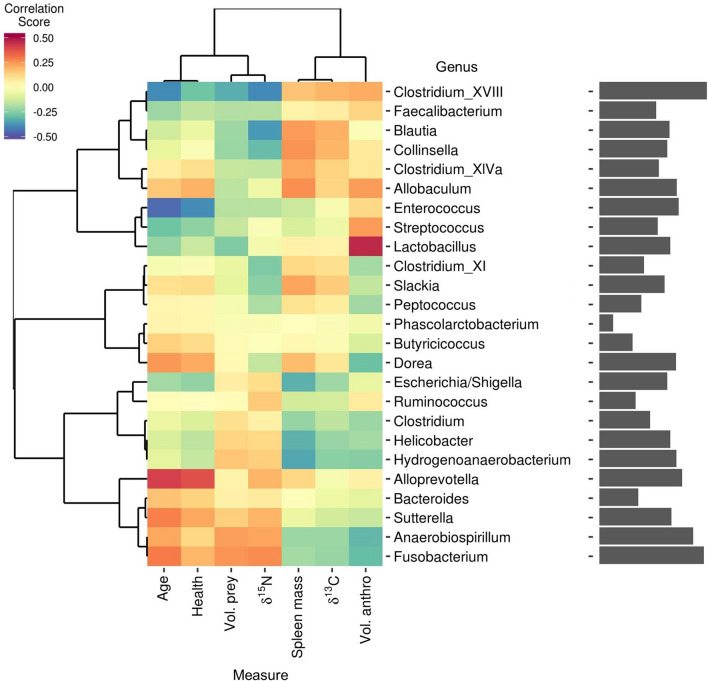
In a multivariate approach, we used clustering associations from a regularized canonical correlation analysis (rCCA) to identify taxa that responded to measures of diet and health (Fig. 4). Explanatory variables separated into two clusters: (1) age and physical condition, which grouped with measures of protein consumption separately from measures of (2) anthropogenic food consumption and spleen mass (Fig. 4). There was a strong network of positive associations connecting Fusobacterium, Anaerobiospirillum, Sutterella, and Bacteroides to older age, the consumption of prey, and higher body condition. A similar network linked Streptococcus, Enterococcus, and Lactobacillus to younger coyotes, the volume of anthropogenic food in the stomach, and poor body condition (Fig. 4). A third grouping connected several genera of Erysipelotrichaceae, Lachnospiraceae, and Coriobacteriaceae to spleen mass, volume of anthropogenic food in the stomach, and δ13C, with variable effects on health (Fig. 4). These networks were validated using GLMs predicting the abundance of these taxa based on available explanatory variables (Figs. S10–S12).
Figure 4.
Multivariate associations among taxon abundances, diet, and body condition. Clustered image map relating the 25 most prevalent and abundant bacterial genera to coyote health and diet information. Colors indicate similarity measures obtained from regularized canonical correlation analysis (rCCA). Dendrograms were obtained by correlation distance-based hierarchical clustering of the similarity measures. Bars on the right indicate the relative importance of each genus in the network, calculated as the sum of the absolute values of the similarity measures.
We lastly used structural equation models (SEM) to model causal relationships among dietary measures and taxon abundances in the context of our questions about coyote location and health. In general, the best-fitting structural equation models supported relationships where urban coyotes consumed more anthropogenic food, which had indirect negative effects on health via increased phylogenetic diversity and increased abundances of Enterococcus and Streptococcus (Fig. 5a; Table S5). The SEM framework also supported paths where health in rural coyotes was connected to protein-rich diets; in these models, Fusobacterium, Anaerobiospirillum, and Sutterella served as indicators of a protein-rich diet but did not directly predict health (Fig. 5b; Table S5). A third model supported connections between anthropogenic food consumption, Erysipelotrichaceae spp., and spleen mass, with lesser effects of Coriobacteriaceae spp. and Lachnospiraceae spp. (Fig. 5c; Table S5). Notably, the model based on dietary protein supported a path in which the volume of prey in the stomach responded negatively to spleen mass, suggesting that immune function may affect coyote foraging success. In addition, the long-term dietary measure δ15N was the stronger predictor of downstream effects in protein-based models, whereas the short-term measure, volume of anthropogenic food in the stomach, was the stronger predictor in anthropogenic food-based models. All models predicted E. multilocularis infection as a function of location only, with no effect of diet or microbiome.
Figure 5.
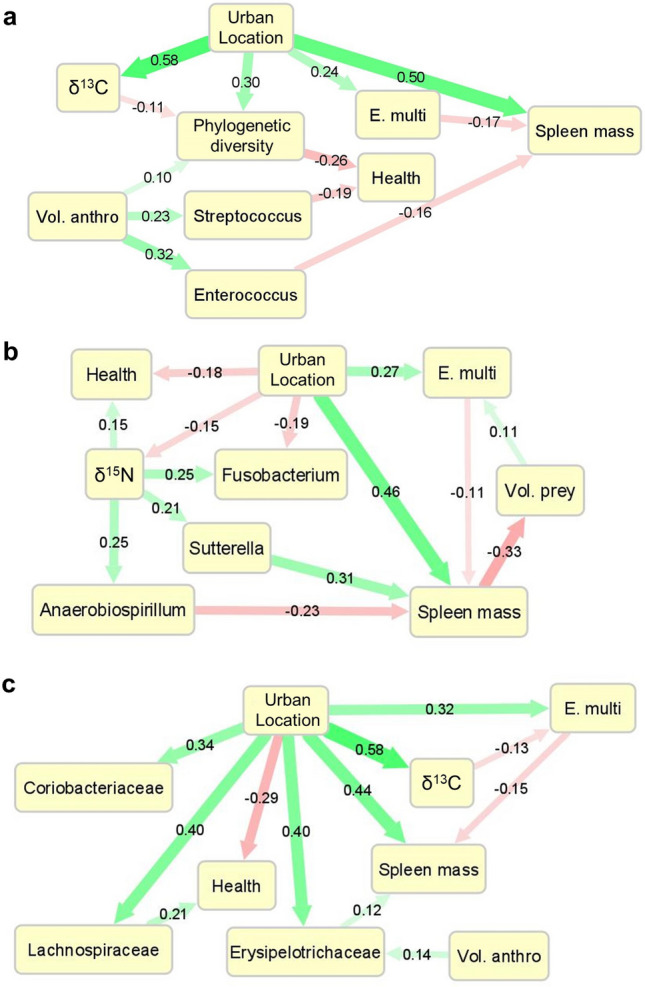
Structural equation models of factors associated with coyote health. Structural equation models were used to evaluate relationships among location, diet, microbiome composition, and health for microbiome features implicated in earlier analyses. The first model (a) focuses on the effects of anthropogenic food consumption; the second (b) focuses on the effects of protein consumption; and the third model (c) tests the taxa that were most strongly correlated with spleen mass. Model coefficients are standardized within each model.
Discussion
Generalist species able to survive on anthropogenic food are becoming increasingly common in urban environments, with important implications for human-wildlife interaction and conflict. Understanding the process of urban adaptation in these species requires understanding both how the gut microbiome responds to urban-associated changes in diet and how the microbiome relates to host fitness. In this study, urban coyotes consumed more anthropogenic food, had less assimilated kidney fat, displayed more signs of immune system stress, and were more likely to be infected with the zoonotic parasite E. multilocularis. These changes in diet and physiology were associated with distinct changes in fecal bacterial community composition. First, despite considerable evidence from humans and laboratory animals that gut microbiome diversity increases with age and health44, we found that the microbiomes of younger, less healthy urban coyotes were more diverse. In addition, we identified several bacterial taxa involved in the connection between diet and health, including some taxa known to be affected by diet in domestic dogs45. Specifically, Streptococcus and Enterococcus linked anthropogenic food consumption and poor health, whereas Fusobacteria, Anaerobiospirillum, and Sutterella were related to a protein-rich diet and improved body condition. Many of these microbial connections between diet and condition were preserved outside of the context of location and could therefore serve as non-invasive indicators of coyote health.
The relationships we observed linking the consumption of anthropogenic food by urban coyotes to increased microbiome diversity and increased abundances of Streptococcus and Enterococcus presumably stem from the altered macronutrient composition of an anthropogenic diet relative to conventional prey. Within a host species, much of the gut microbial alpha diversity is attributable to the range of microbial enzymatic capacities needed to degrade dietary nutrients46. This is why diets rich in plants and fibers lead to increased microbial diversity47,48. Anthropogenic foods contain a wider variety of macronutrients than prey46, particularly in the form of complex polysaccharides, and would therefore promote the higher phylogenetic diversity in the urban coyote gut that we observed here. These results disagree with previous studies in passerines that showed decreased microbiome diversity in urban birds20,23, but agree with another22, suggesting that how microbial diversity responds to urban habitat use may differ depending on host species, dietary components, or other aspects of an adaptation strategy.
The presumed increase in dietary carbohydrates associated with an anthropogenic diet would also contribute to the abundances of the lactic acid bacteria (LAB) Streptococcus and Enterococcus, with potential implications for overall condition. LAB are largely considered an important component of a healthy microbiome for omnivores and herbivores due to their role in digesting dietary carbohydrates19 and have been used as probiotics for many livestock species49. However, carnivore diets contain few carbohydrates12, and the increased abundance of LAB, as with the increase in diversity, may be a direct consequence of increased carbohydrate consumption. Despite these microbial responses to an altered diet, the digestive physiology of carnivores is not adapted to handle large amounts of carbohydrates, which presumably limits the amount of nutrition that can be effectively acquired from anthropogenic food and thus contributes to poor health50. Although this effect would be reflected, but not mediated, by the microbiome, select groups of Streptococcaceae and Enteroccaceae have been directly connected to negative health effects in humans51, and higher abundances of Streptococcus contribute to a dysbiosis index in dogs52. Taxon abundances were also better than diet at predicting condition in structural equation models, and it is therefore possible that some of these taxa may directly influence, rather than indicate, poor condition.
We suspect that these directional associations linking an anthropogenic diet to poor health via microbiome diversity and LAB abundance introduce a microbial element into what has previously been described as a “vicious cycle” of diet, body condition, and disease susceptibility53 (Fig. 6a). Animals in poor condition are more likely to become diseased and are less successful at capturing prey, thus increasing their reliance on anthropogenic food. This cycle explains why diseased coyotes are reported more frequently at compost sites37,38. However, not all urban coyotes depend on anthropogenic food subsidies: while some urban-dwelling coyotes range freely through developed land and consume anthropogenic food, others remain in urban natural areas and have diets nearly indistinguishable from rural coyotes37,54. In our study, urban coyotes that were classified as rural in random forest models likely represent this latter group, as misclassified coyotes had lower alpha diversity measures, lower LAB abundances, and appeared to be slightly healthier. Although microbiome diversity and LAB are frequently cited as a benefit to the host44, their negative effects in coyotes speak to the importance of understanding the functional value of commonly used microbiome measures in different host species living in different environments.
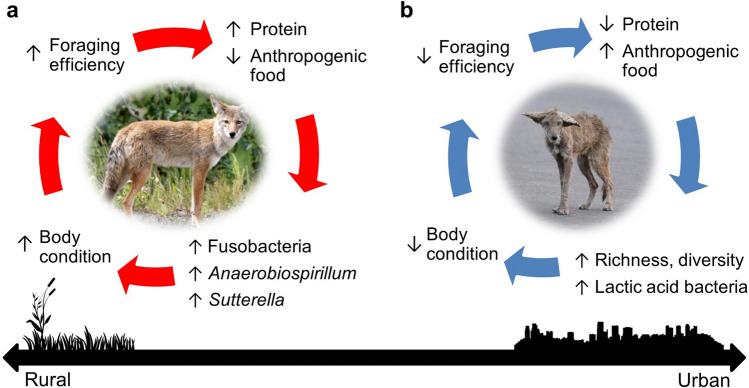
Figure 6.
Conceptual model of positive feedback loops among diet, microbiome composition, and health. (a) Coyotes consuming more protein, and less anthropogenic food, have more Fusobacteria, Anaerobiospirillum, and Sutterella in their microbiome and improved body condition. This, in theory, promotes the ability to continue capturing prey, and is the more common cycle in rural environments. (b) Increased diet subsidization with anthropogenic food leads to increased microbiome richness and diversity, higher abundances of select lactic acid bacteria such as Streptococcus and Enterococcus, and lower body condition. Poor health likely decreases foraging efficiency, thus increasing the reliance on anthropogenic food. This cycle predominates in urban-exploiting coyotes (images from Jitze Couperus, left, and Michael Renzi, right).
A complementary positive feedback loop characterizing rural coyotes revolved around Fusobacteria, Sutterella, and Anaerobiospirillum, in which coyotes consuming more prey harbored more of these taxa in their microbiome and were generally healthier. Healthy coyotes could, in turn, be expected to have better success obtaining protein-rich prey (Fig. 6b). Fusobacteria are considered a core member of the canine microbiome, despite their low abundance in other mammals55 and association with disease and cancer in humans56. Our observation that Fusobacteria are positively correlated with protein consumption agrees with previous observations that dogs fed protein-rich diets harbored more Fusobacteria than dogs fed carbohydrate-rich kibble57–60. Anaerobiospirillum and Sutterella are among the most abundant Proteobacterial genera in canines61, including wolves62, and may also play key roles in protein digestion61. Other general indicators of health in our study, including Alloprevotella, Allobaculum and Dorea, have also been connected to health in canines18,57,62. In one study, Alloprevotella and Sutterella were detected in healthy dogs but were absent in dogs infected with canine parvovirus63.
The families Erysipelotrichaceae, Coriobacteriaceae, and Lachnospiraceae were the most consistent indicators of spleen mass, which we used as a proxy for immune system function. Erysipelotrichiaceae are associated with carbohydrate consumption in dogs57,59 and correlates with colonic inflammatory responses in both humans and mice64. In humans, Coriobacteriaceae has also been connected to the abundance of pro-inflammatory cytokines65, and Lachnospiraceae, specifically Blautia, have been shown to decrease after splenectomy66. Interactions between the microbiome and the immune system are complex, and the interpretive power provided by the coarse measure of spleen mass alone is low; large spleens have alternately been considered indicators of immune stress67 or immune strength40. While we hypothesized that larger spleens reflect immune stress due to their association with urban areas and carbohydrate-degrading taxa, our results warrant more detailed studies of how the microbiome may regulate an immune response in urban coyotes. However, our data provide further evidence of the “vicious cycle” feedback loop in two ways: (1) spleen mass grouped with anthropogenic food consumption in rCCA models; and (2) larger spleens predicted low short-term prey consumption in structural equation models. This cycle predicts that unhealthy coyotes are less able to obtain prey and become more dependent on anthropogenic food subsidization, which may apply particularly to coyotes drawn to compost38.
An important additional observation in our study is that many of the relationships between microbiome features and the recent consumption of anthropogenic food became weaker when evaluated using long-term δ13C signatures. Conversely, relationships with prey consumption were stronger when evaluated with the long-term measure δ15N. We hypothesize that this observation reflects a combination of natural constraints on the microbiome and the rapidity with which the microbiome responds to perturbation. Gut microbiome composition is limited by host gut physiology31, and coyotes naturally harbor a gut microbiome shared by other carnivores with short, simple guts68. Diet-induced changes in the microbiome can occur within hours of a meal and revert equally quickly to a stable state69. Anthropogenic meals represent a novel and transient perturbation to a stable carnivorous microbiome, whereas protein consumption reflects the natural state of a carnivore over extended time. Indeed, the lower NTI values in urban coyotes provide evidence for more competitive microbiome structuring as the microbiome frequently adjusts to changes in diet. This continuous competition or adjustment could exacerbate the vicious cycle described above, insofar as instability in the microbiome induced by frequent alterations among food sources may reduce natural barriers to infection and prevent the microbiome from adapting to or metabolizing the immediate diet15.
Other aspects of health and behavior may additionally be affected by the microbiome in unpredictable ways. Although we found no strong connection between microbiome composition and the presence of E. multilocularis, it remains possible that disrupted co-evolutionary dynamics between urban coyotes and their gut microbiota provide a fertile environment for parasite establishment70. Further investigations using more sensitive measures, such as absolute worm counts, are needed to explore parasite/microbiome relationships in more detail. Behavior may likewise be affected by the microbiome: one urban coyote in our study was lethally managed because it attacked and killed a large domestic dog, an uncharacteristically aggressive behavior for an urban coyote. Low-protein diets and poor health have been cited as predictors of conflict35, but this coyote was above average in all measures of condition and prey consumption. Instead, this was the only coyote that did not contain any detectable Fusobacteria in its fecal microbiome. Documented relationships between the microbiome and behavior are becoming increasingly common in humans and other animals71, and microbiome composition, including fewer Fusobacteria, has been directly associated with aggression in dogs18. Aggressive behavior and the spread of E. multilocularis both have great implications for human-coyote interactions in urban areas, and we suspect urban-induced disruptions to the microbiome may play important but poorly understood roles in mediating those outcomes.
Conclusion
The urban landscape is highly heterogenous, and behavioral adaptations in urban animals generate a complex mosaic of spatially and temporally variable diets, lifestyle contexts, and environmental exposures among both individuals and species. Our results provide a foundational understanding of the gastrointestinal microbiome in urban-adapted coyotes and demonstrate how the consumption of carbohydrate-rich anthropogenic food alters a naturally carnivorous microbiome to negatively affect overall health. Moreover, we suggest mechanisms for how the microbiome participates in previously described positive feedback loops connecting diet, body condition, and disease susceptibility in urban animals. Further work may make it possible to use the bacterial taxa implicated in these feedback loops as indicators in non-invasive evaluations of coyote health via scat samples, providing similar information to urban wildlife managers as a dysbiosis index currently used for dogs in a veterinary setting52. We lastly speculate that urban-induced alterations in the microbiome may influence behavior and parasite susceptibility, though more work will be needed to untangle the exact mechanisms and magnitude of these relationships.
Methods
Sample collection and necropsy
We collected coyote carcasses from Edmonton (Alberta, Canada) and the surrounding area in the winters of 2017–2018 and 2018–2019. Samples were collected as roadkill, obtained from local fur trappers, or lethally managed after negative interactions with humans or domestic animals (Table S1). Coyotes were classified based on their location of death as either “urban” or “rural.” All samples, including roadkill, were collected and frozen within 24 h of death, and both collection seasons had similar temperature and precipitation patterns (see Supplementary Methods S1). Carcasses were stored at − 80 °C for 5 days to neutralize any zoonotic pathogens and then transferred to − 20 °C until necropsy.
At necropsy, we measured the mass, body size (snout to base of tail), and girth around the ribcage for each coyote, and we measured the kidney fat index (KFI) following published protocols42 and recorded the mass of the spleen. For analysis, spleen mass (in grams) was divided by body mass (in kilograms) to account for size differences among animals. We removed the lower mandible for age determination, clipped the left hind outer toenail for stable isotope analysis, and removed the stomach for dietary analysis. For microbiome analysis, we extruded fecal samples from the large intestine and stored them at − 80 °C.
Age determination
We determined the age of each coyote by counting cementum annuli. Lower canine teeth were removed from the mandible by soaking the mandible at 80 °C for 6–8 h. Teeth were fixed in a neutral solution of 10% formalin before being decalcified, sectioned, and stained following published methods72. We followed the aging criteria reported by Linhart et al.73. To increase precision, we assigned all coyotes a birth date of 1 May and used the difference between birth date and death date to determine age to the nearest month.
Stomach content analysis
To determine each coyote’s recent diet, we quantified stomach contents by volume. We rinsed stomach contents in an 850 μm sieve to remove mucus and then quantified the total food volume by measuring the amount of water displaced (in ml) when the contents were placed in a graduated cylinder. Discernable food items were then classified as prey (including rodents, small herbivores, ungulates, and insects), anthropogenic food (including fast food waste, compost, or other garbage), or vegetation (including grass, leaves, and sticks). The volume in each of these categories was measured using the same method as before.
Stable isotope analysis
We used stable isotope values (δ13C and δ15N) measured from claw samples to infer each coyote’s habitual diet. Whole claw samples were rinsed three times with a 2:1 chloroform:methanol solution to remove residual lipids and surface oils and then dried at 37 °C for 5 days. After drying, we manually homogenized the distal 5 mm of each claw and weighed 1.5 mg subsamples into tin capsules. Samples were combusted using a Vario Pyrocube and analyzed using an Isoprime Vision Mass Spectrometer at the Biogeochemical Analytical Service Laboratory (Dept. of Biological Sciences, Univ. of Alberta).
Isotopic values were used in stable isotope mixing models to estimate the proportion of different food items in coyote diets. Stable isotope values for various coyote diet items were obtained from a previous study in our lab37, and diet items were categorized as prey, fruit, or anthropogenic food. Isotopic data for anthropogenic food was supplemented with published values for beef and chicken74. We accounted for tissue-specific discrimination factors following previously described methods54 and ran mixing models using the default settings of the R package simmr75.
Microbiome analysis
We extracted total DNA from 100 mg of each fecal sample using the MP Bio FastDNA Spin Kit for Soil following the manufacturer’s instructions (MP Biomedicals, Santa Ana, CA)68. Fecal samples were thawed and manually homogenized using a pestle and mortar prior to extraction, and before the final elution we included a five-minute incubation at 50 °C to maximize DNA yield. Extracted DNA was submitted to Microbiome Insights (Vancouver, BC) for sequencing, where PCR amplification of the V4 region of the 16S rRNA gene was performed in 50 μl reactions with 2 μl of template DNA using previously described cycling conditions for the barcoded universal bacterial primers 515F and 806R76. Successful amplification was confirmed using agarose gel electrophoresis. Paired-end sequencing of equimolar concentrations of the PCR products was conducted on an Illumina MiSeq platform using V3 chemistry and 300 bp reads. All DNA extraction, PCR, and sequencing steps were performed alongside three negative control samples and a mock community (see Fig. S14).
Sequence data was processed to generate amplicon sequence variants, with taxonomic assignment and phylogenetic tree construction performed following a previously described protocol68 (see also Supplemental Methods). Five samples with fewer than 4,500 reads were excluded from downstream analysis, based on a visual examination of read count distributions across all samples and a sample completeness threshold of > 98% calculated using iNext77 (Fig. S13). In addition, we removed ASVs that were identified as chloroplasts, mitochondria, or protists in a BLAST search, as well as 21 putative contaminant ASVs detected using the prevalence-based detection method implemented in the package decontam78. No putative contaminant was either abundant (> 1% relative abundance) or prevalent (> 20% prevalence) across experimental samples (Fig. S13). These filtering procedures resulted in an average of 21,319 ± 9,477 reads per sample (range 4556–46,542).
Parasite survey
We used PCR to test each coyote for possible infection with E. multilocularis. DNA extracted from fecal samples was amplified in triplicate using the E. multilocularis-specific primers Cest1 and Cest279. PCR was performed in 25 μl reactions with 0.2 μM of each primer and 1 μl of template DNA using cycling conditions described previously79. We resolved PCR products using gel electrophoresis; a coyote was considered positive for E. multilocularis if the sample exhibited a 395 bp band in at least two replicates. Samples that tested negative were diluted and tested again to control for possible PCR inhibition.
Statistical analyses
All statistical analyses were performed in R 3.6.280 and a full description of our statistical workflow is available in the electronic supplementary material. In brief, a small number of missing physiological measurements were imputed using linear regressions. We then generated a single composite metric of physical condition by performing principal components analysis (PCA) on mass, body size, girth, and KFI and extracting the axis scores on the first principal component (Table S6). Spleen mass was retained as a separate measure of immune function because it did not load well on either of the first two components.
For each microbiome sample, we calculated total ASV richness, Shannon diversity, and Faith’s PD using iNext and iNextPD, which produce asymptotic estimates for these values based on rarefaction curves77,81. NTI was calculated using picante82. We used linear and logistic regressions to determine if any of our diet, microbiome, or condition measures varied with either coyote location or E. multilocularis infection status, while controlling for the effects of sex and age. Because there was a significant association between location and E. multilocularis infection, the effects of E. multilocularis infection were tested while also controlling for location.
To determine which measures were the best indicators of microbiome diversity, we used GLMs predicting each microbiome metric from the remaining diet and health measures. Continuous predictors were centered and standardized and all models subsets were examined. The relative importance of each predictor was determined by (1) summing the AIC model weights for each model in which the predictor appeared83 and (2) comparing model-averaged coefficients across models with a ΔAICc less than two. Model coefficients were standardized by their partial standard deviation prior to coefficient-averaging84.
We tested for differences in overall microbiome composition using three complementary approaches. First, differential abundance analyses were performed at all taxonomic levels using the ‘aldex.glm’ function in ALDEx285 while controlling for confounding variables. We used the Benjamini–Hochberg correction to control for multiple comparisons and the Hedge’s g statistic as a measure of effect size. Second, random forest models were trained to predict coyote location from centered log ratio (CLR)-transformed taxon abundances, and we tested for any differences in diet and health between correctly and incorrectly classified urban coyotes using the regression methods described above. Third, we used an Aitchison distance-based PERMANOVA to test for differences in overall microbiome composition, and we mapped continuous diet and health measures as vectors onto a PCA of microbiome composition.
We then tested for relationships linking diet or health to the abundance of individual taxa, limiting our analyses to the 25 most prevalent bacterial genera with a mean relative abundance greater than 0.1%. Univariate associations were evaluated using Spearman’s correlation. For each taxon, we additionally constructed rank regression models, with either the condition index or spleen mass as a response variable and taxon abundance as the predictor. Sex was included as a covariate. We did not control for age in these models because, in wild systems, only animals in good condition live to be old, and age and health were therefore considered redundant measures (Fig. S15). Multivariate associations were further investigated using rCCA. We used correlation distance-based hierarchical clustering to identify taxa that responded similarly to the various explanatory variables. Taxon abundances were CLR-transformed for all analyses.
Because of the inter-correlated nature of our variables, we lastly used structural equation modeling (SEM) to test for causal linkages among variables. Where necessary, variables included in each model were log-transformed or scaled to meet the model assumptions. For each taxon cluster identified in the rCCA, we constructed initial models that represented general hypotheses of causal linkages among variables and we specified residual correlations among all microbiome features. Additional paths were added as recommended by modification indices and non-significant paths (p > 0.1) were removed. The final models were selected when adding or removing an additional path either caused model AIC to increase or caused other fit parameters to exceed conventional thresholds, even if the path was non-significant.
Supplementary Information
Acknowledgements
Coyote carcasses were provided by Bill, Duncan, and Malcolm Abercrombie of Animal Damage, Inc., as well as the City of Edmonton Animal Care and Control Centre and the Edmonton Police Service. S.S. thanks Deanna Steckler and several undergraduate volunteers for assistance with coyote necropsies, Alvin Kwan for assistance with stable isotope analysis, and Arlene Oatway for assistance with cementum aging.
Author contributions
S.S., L.Y.S., and C.C.S.C. designed the study. S.S. did laboratory work, analyzed the data, and wrote the manuscript. K.F. and D.S. measured the stomach contents. L.Y.S. and C.C.S.C. assisted with data analysis and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This study was funded by Discovery Grants awarded to L.Y.S. (RGPIN-2019-04399) and C.C.S.C. (RGPIN-2017-05915) by the Natural Sciences and Engineering Research Council of Canada (NSERC). The funding body had no role in designing the study, interpreting the data, or writing the manuscript.
Data availability
The raw sequencing data supporting the conclusions of this article have been deposited in the NCBI Short Read Archive under project number PRJNA528764. The R scripts and workspace required to reproduce all analyses and figures are available in the GitHub repository https://github.com/sasugden/Coyote_microbiome.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78891-1.
References
- 1.Foley JA, et al. Global consequences of land use. Science. 2005;309:570–574. doi: 10.1126/science.1111772. [DOI] [PubMed] [Google Scholar]
- 2.Ellis EC, Goldewijk KK, Siebert S, Lightman D, Ramankutty N. Anthropogenic transformation of the biomes, 1700 to 2000. Glob. Ecol. Biogeogr. 2010;19:589–606. [Google Scholar]
- 3.Concepción ED, Moretti M, Altermatt F, Nobis MP, Obrist MK. Impacts of urbanisation on biodiversity: the role of species mobility, degree of specialisation and spatial scale. Oikos. 2015;124:1571–1582. doi: 10.1111/oik.02166. [DOI] [Google Scholar]
- 4.Lowry H, Lill A, Wong BBM. Behavioural responses of wildlife to urban environments. Biol. Rev. 2013;88:537–549. doi: 10.1111/brv.12012. [DOI] [PubMed] [Google Scholar]
- 5.Callaghan CT, et al. Generalists are the most urban-tolerant of birds: a phylogenetically controlled analysis of ecological and life history traits using a novel continuous measure of bird responses to urbanization. Oikos. 2019;128:845–858. doi: 10.1111/oik.06158. [DOI] [Google Scholar]
- 6.Ducatez S, Sayol F, Sol D, Lefebvre L. Are urban vertebrates city specialists, artificial habitat exploiters, or environmental generalists? Integr. Comp. Biol. 2018;58:929–938. doi: 10.1093/icb/icy101. [DOI] [PubMed] [Google Scholar]
- 7.Murray MH, et al. City sicker? A meta-analysis of wildlife health and urbanization. Front. Ecol. Environ. 2019;17:575–583. doi: 10.1002/fee.2126. [DOI] [Google Scholar]
- 8.Lyons J, Mastromonaco G, Edwards DB, Schulte-Hostedde AI. Fat and happy in the city: eastern chipmunks in urban environments. Behav. Ecol. 2017;28:1464–1471. doi: 10.1093/beheco/arx109. [DOI] [Google Scholar]
- 9.Meillère A, et al. Corticosterone levels in relation to trace element contamination along an urbanization gradient in the common blackbird (Turdus merula) Sci. Total Environ. 2016;566–567:93–101. doi: 10.1016/j.scitotenv.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Soto-Calderón, I., Acevedo-Garcés, Y., Álvarez-Cardona, J., Hernandez, C. & García, G. Physiological and parasitological implications of living in a city: the case of the white-footed tamarin (Saguinus leucopus). Am. J. Primatol.78, (2016). [DOI] [PubMed]
- 11.Sillero-Zubiri, C., Sukumar, R. & Treves, A. Living with wildlife: the roots of conflict and the solutions. In Key Topics in Conservation Biology (eds. MacDonald, D. & Service, K.) 255–272 (2006).
- 12.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanning I, Diaz-Sanchez S. The functionality of the gastrointestinal microbiome in non-human animals. Microbiome. 2015;3:51. doi: 10.1186/s40168-015-0113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 15.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mockler BK, Kwong WK, Moran NA, Koch H. Microbiome structure influences infection by the parasite Crithidia bombi in bumble bees. Appl. Environ. Microbiol. 2018;84:e02335–e2417. doi: 10.1128/AEM.02335-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki TA. Links between natural variation in the microbiome and host fitness in wild mammals. Integr. Comp. Biol. 2017;57:756–769. doi: 10.1093/icb/icx104. [DOI] [PubMed] [Google Scholar]
- 18.Kirchoff NS, Udell MA, Sharpton TJ. The gut microbiome correlates with conspecific aggression in a small population of rescued dogs (Canis familiaris) PeerJ. 2019;7:e6103. doi: 10.7717/peerj.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 2008;74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teyssier A, et al. Inside the guts of the city: urban-induced alterations of the gut microbiota in a wild passerine. Sci. Total Environ. 2018;612:1276–1286. doi: 10.1016/j.scitotenv.2017.09.035. [DOI] [PubMed] [Google Scholar]
- 21.Murray MH, et al. Gut microbiome shifts with urbanization and potentially facilitates a zoonotic pathogen in a wading bird. PLoS ONE. 2020;15:1–16. doi: 10.1371/journal.pone.0220926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips JN, Berlow M, Derryberry EP. The effects of landscape urbanization on the gut microbiome: an exploration into the gut of urban and rural white-crowned sparrows. Front. Ecol. Evol. 2018;6:148. doi: 10.3389/fevo.2018.00148. [DOI] [Google Scholar]
- 23.Teyssier, A. et al. Diet contributes to urban-induced alterations in gut microbiota: experimental evidence from a wild passerine. Proc. R. Soc. B Biol. Sci.287, (2020). [DOI] [PMC free article] [PubMed]
- 24.Stothart, M. R., Palme, R. & Newman, A. E. M. It’s what’s on the inside that counts: stress physiology and the bacterial microbiome of a wild urban mammal. Proc. R. Soc. B Biol. Sci.286, (2019). [DOI] [PMC free article] [PubMed]
- 25.Becker CG, Longo AV, Haddad CFB, Zamudio KR. Land cover and forest connectivity alter the interactions among host, pathogen and skin microbiome. Proc. R. Soc. B Biol. Sci. 2017;284:20170582. doi: 10.1098/rspb.2017.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bestion E, et al. Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat. Ecol. Evol. 2017;1:0161. doi: 10.1038/s41559-017-0161. [DOI] [PubMed] [Google Scholar]
- 27.Barelli C, et al. Habitat fragmentation is associated to gut microbiota diversity of an endangered primate: implications for conservation. Sci. Rep. 2015;5:14862. doi: 10.1038/srep14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trevelline, B. K., Fontaine, S. S., Hartup, B. K. & Kohl, K. D. Conservation biology needs a microbial renaissance: a call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B Biol. Sci.286, (2019). [DOI] [PMC free article] [PubMed]
- 29.Nelson TM, Rogers TL, Carlini AR, Brown MV. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 2013;15:1132–1145. doi: 10.1111/1462-2920.12022. [DOI] [PubMed] [Google Scholar]
- 30.Wasimuddin, et al. Gut microbiomes of free-ranging and captive Namibian cheetahs: diversity, putative functions and occurrence of potential pathogens. Mol. Ecol. 2017;26:5515–5527. doi: 10.1111/mec.14278. [DOI] [PubMed] [Google Scholar]
- 31.Amato KR, et al. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13:576–587. doi: 10.1038/s41396-018-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gehrt, S. D. & Riley, S. P. D. Coyotes (Canis latrans). in Urban Carnivores: Ecology, Conflict, and Conservation (eds. Gehrt, S. D., Riley, S. P. D. & Cypher, B. L.) 79–95 (2010).
- 33.Breck SW, Poessel SA, Mahoney P, Young JK. The intrepid urban coyote: a comparison of bold and exploratory behavior in coyotes from urban and rural environments. Sci. Rep. 2019;9:2104. doi: 10.1038/s41598-019-38543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gier, H. T. Coyotes in Kansas. (1968).
- 35.Murray MH, et al. Greater consumption of protein-poor anthropogenic food by urban relative to rural coyotes increases diet breadth and potential for human-wildlife conflict. Ecography. 2015;38:001–008. doi: 10.1111/ecog.01128. [DOI] [Google Scholar]
- 36.Massolo A, Liccioli S, Budke C, Klein C. Echinococcus multilocularis in North America: the great unknown. Parasite. 2014;21:73. doi: 10.1051/parasite/2014069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, M. H., Edwards, M. A., Abercrombie, B. & St. Clair, C. C. Poor health is associated with use of anthropogenic resources in an urban carnivore. Proc. R. Soc. B Biol. Sci.282, 20150009 (2015). [DOI] [PMC free article] [PubMed]
- 38.Murray, M. H., Hill, J., Whyte, P. & St. Clair, C. C. Urban compost attracts coyotes, contains toxins, and may promote disease in urban-adapted wildlife. Ecohealth13, 285–292 (2016). [DOI] [PubMed]
- 39.Luong, L. T., Chambers, J. L., Moizis, A., Stock, T. & St. Clair, C. Helminth parasites and zoonotic risk associated with urban coyotes (Canis latrans) in Alberta, Canada. J. Helminthol.94, e25 (2020). [DOI] [PubMed]
- 40.Corbin E, et al. Spleen mass as a measure of immune strength in mammals. Mamm. Rev. 2008;38:108–115. doi: 10.1111/j.1365-2907.2007.00112.x. [DOI] [Google Scholar]
- 41.Newsome, S. D., Ralls, K., Van Horn Job, C., Fogel, M. L. & Cypher, B. L. Stable isotopes evaluate exploitation of anthropogenic foods by the endangered San Joaquin kit fox (Vulpes macrotis mutica). J. Mammol.91, 1313–1321 (2010).
- 42.Huot J, Poulle M, Crate M. Evaluation of several indices for assessment of coyote (Canis latrans) body composition. Can. J. Zool. 1995;73:1620–1624. doi: 10.1139/z95-192. [DOI] [Google Scholar]
- 43.Tucker CM, et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 2016;92:698–715. doi: 10.1111/brv.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reese AT, Dunn RR. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. MBio. 2018;9:e01294–e1318. doi: 10.1128/mBio.01294-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilla R, Suchodolski JS. The role of the canine gut microbiome and metabolome in health and gastrointestinal disease. Front. Vet. Sci. 2020;6:498. doi: 10.3389/fvets.2019.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrition. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 48.Schnorr SL, et al. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieco-Saiz N, et al. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019;10:1–17. doi: 10.3389/fmicb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karasov WH, Douglas AE. Comparative digestive physiology. Comp. Physiol. 2013;3:741–783. doi: 10.1002/cphy.c110054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.AlShawaqfeh MK, et al. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiol. Ecol. 2017;93:1–8. doi: 10.1093/femsec/fix136. [DOI] [PubMed] [Google Scholar]
- 53.Beldomenico PM, Begon M. Disease spread, susceptibility and infection intensity: vicious circles? Trends Ecol. Evol. 2010;25:21–27. doi: 10.1016/j.tree.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 54.Newsome SD, Garbe HM, Wilson EC, Gehrt SD. Individual variation in anthropogenic resource use in an urban carnivore. Oecologia. 2015;178:115–128. doi: 10.1007/s00442-014-3205-2. [DOI] [PubMed] [Google Scholar]
- 55.Henderson G, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bermingham EN, Maclean P, Thomas DG, Cave NJ, Young W. Key bacterial families (Clostridiaceae, Erysipelotrichaceae and Bacteroidaceae) are related to the digestion of protein and energy in dogs. PeerJ. 2017;5:e3019. doi: 10.7717/peerj.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alessandri G, et al. Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ. Microbiol. 2019;21:1331–1343. doi: 10.1111/1462-2920.14540. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt M, et al. The fecal microbiome and metabolome differs between dogs fed Bones and Raw Food (BARF) diets and dogs fed commercial diets. PLoS ONE. 2018;13:e0201279. doi: 10.1371/journal.pone.0201279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandri M, Dal Monego S, Conte G, Sgorlon S, Stefanon B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet. Res. 2017;13:1–11. doi: 10.1186/s12917-017-0981-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moon CD, Cookson AL, Young W, Maclean PH, Bermingham EN. Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. Microbiologyopen. 2018;7:e677. doi: 10.1002/mbo3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, et al. Analysis and comparison of the wolf microbiome under different environmental factors using three different data of next generation sequencing. Sci. Rep. 2017;7:11332. doi: 10.1038/s41598-017-11770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang B, Wang X-L. Species diversity of fecal microbial flora in Canis lupus familiaris infected with canine parvovirus. Vet. Microbiol. 2019;237:108390. doi: 10.1016/j.vetmic.2019.108390. [DOI] [PubMed] [Google Scholar]
- 64.Chen L, et al. NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 2017;18:541–551. doi: 10.1038/ni.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martínez I, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, et al. Splenectomy leads to amelioration of altered gut microbiota and metabolome in liver cirrhosis patients. Front. Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Demas GE, Zysling DA, Beechler BR, Muehlenbein MP, French SS. Beyond phytohaemagglutinin: assessing vertebrate immune function across ecological contexts. J. Anim. Ecol. 2011;80:710–730. doi: 10.1111/j.1365-2656.2011.01813.x. [DOI] [PubMed] [Google Scholar]
- 68.Sugden, S. A., St. Clair, C. C. & Stein, L. Y. Individual and site-specific variation in a biogeographical profile of the coyote intestinal microbiota. Microb. Ecol. (2020). [DOI] [PubMed]
- 69.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung JM, Graham AL, Knowles SCL. Parasite-microbiota interactions with the vertebrate gut: synthesis through an ecological lens. Front. Microbiol. 2018;9:843. doi: 10.3389/fmicb.2018.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal behavior and the microbiome. Science. 2012;338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 72.Stewart REA, Stewart BE, Stirling I, Street E. Counts of growth layer groups in cementum and dentine in ringed seals. Mar. Mammal Sci. 1996;12:383–401. doi: 10.1111/j.1748-7692.1996.tb00591.x. [DOI] [Google Scholar]
- 73.Linhart SB, Knowlton FF. Determining age of coyotes by tooth cementum layers. J. Wildl. Manage. 1967;31:362–365. doi: 10.2307/3798334. [DOI] [Google Scholar]
- 74.Jahren AH, Kraft RA. Carbon and nitrogen stable isotopes in fast food: signatures of corn and confinement. Proc. Natl. Acad. Sci. 2008;105:17855–17860. doi: 10.1073/pnas.0809870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parnell, A. C. simmr: a stable isotope mixing model. (2019).
- 76.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsieh TC, Ma KH, Chao A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers) Methods Ecol. Evol. 2016;7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 78.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trachsel D, Deplazes P, Mathis A. Identification of taeniid eggs in the faeces from carnivores based on multiplex PCR using targets in mitochondrial DNA. Parasitology. 2007;134:911–920. doi: 10.1017/S0031182007002235. [DOI] [PubMed] [Google Scholar]
- 80.R Core Team. R: A language and environment for statistical computing. (2019).
- 81.Chao A, et al. Rarefaction and extrapolation of phylogenetic diversity. Methods Ecol. Evol. 2015;6:380–388. doi: 10.1111/2041-210X.12247. [DOI] [Google Scholar]
- 82.Kembel S, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 83.Giam X, Olden JD. Quantifying variable importance in a multimodel inference framework. Methods Ecol. Evol. 2016;7:388–397. doi: 10.1111/2041-210X.12492. [DOI] [Google Scholar]
- 84.Cade BS. Model averaging and muddled multimodel inferences. Ecology. 2015;96:2370–2382. doi: 10.1890/14-1639.1. [DOI] [PubMed] [Google Scholar]
- 85.Fernandes A, Macklaim JM, Linn T, Reid G, Gloor GB. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE. 2013;8:e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data supporting the conclusions of this article have been deposited in the NCBI Short Read Archive under project number PRJNA528764. The R scripts and workspace required to reproduce all analyses and figures are available in the GitHub repository https://github.com/sasugden/Coyote_microbiome.