Abstract
Photoreceptors of the phytochrome family control a multitude of responses in plants. Phytochrome A (phyA) is essential for far-red light perception, which is important for germination and seedling establishment in strong canopy shade. Translocation of phyA from the cytosol into nucleus is a key step in far-red light signaling and requires FAR-RED ELONGATED HYPOCOTYL 1 (FHY1) and FHY1-LIKE (FHL). FHY1/FHL bind to phyA downstream signaling components. Therefore, it has been suggested that FHY1/FHL also have a function in assembling phyA transcription factor complexes in the nucleus. Yet, in this study, we show that constitutively nuclear-localized phyA is active in the absence of FHY1 and FHL. Furthermore, an artificial FHY1, consisting of an SV40 NLS, a phyA binding site, and a YFP tag as spacer between them, complements the fhy1-3 fhl-1 double mutant. These findings show that FHY1 and FHL are not required for phyA downstream signaling in the nucleus. However, we found that lines expressing phyA-NLS-YFP are hypersensitive to red and far-red light and that slightly increased levels of constitutively nuclear-localized phyA result in photomorphogenic development in the dark. Thus, restricting phyA to the cytosol and inducing nuclear transport in light by interaction with FHY1/FHL might be important to suppress photomorphogenesis in the dark.
Key words: phytochrome, phyA, FHY1, FHL, nuclear transport, far-red light
FHY1 and FHL transport light-activated phytochrome A from the cytosol into the nucleus, where it controls expression of light-regulated genes. This study shows that constitutively nuclear-localized phytochrome A is active in mutant seedlings lacking functional FHY1 and FHL. Therefore, we conclude that FHY1 and FHL are not essential for phytochrome A downstream signaling in the nucleus.
Introduction
Light plays an important role throughout the life of plants, which use light for photosynthesis and as a source of information. By measuring the direction, the intensity, the spectral composition, and temporal patterns of incident light, plants gain important information about their environment. As sessile organisms, plants rely on such information to adapt growth and development to the environmental conditions. For light perception, plants have different classes of photoreceptors. These include the blue light/UV-A-sensing cryptochromes, phototropins, and ZEITLUPE family proteins, the UV-B receptor UVB-RESISTANCE 8, and the phytochromes, which primarily work in the red (R) and far-red (FR) range of the light spectrum (Galvão and Fankhauser, 2015).
Phytochromes contain a linear tetrapyrrole as chromophore, which is covalently bound to a conserved cysteine residue. They can exist in two different states, the inactive Pr form and the biologically active Pfr form, which have absorption peaks in R and FR light, respectively. Phytochromes are synthesized in Pr and can reversibly interconvert between Pr and Pfr by absorption of light (Rockwell et al., 2006, Burgie and Vierstra, 2014). In Arabidopsis, the phytochrome family includes five members classified as phytochrome A to E (phyA-E). PhyA and phyB are most prominent and mediate a broad range of responses, whereas phyC-E possibly have more specific functions. PhyB is of particular importance in light-grown and adult plants. To initiate downstream signaling, phyB requires a high Pfr:Ptot (Ptot = Pr + Pfr) ratio, such as in R or white light. In contrast, phyA triggers signal transduction in response to very low Pfr:Ptot ratios typically established by irradiation with FR or weak light of any wavelength. Thus, phyA has a dual function, working as a receptor for weak light in the very low fluence response mode and as a sensor for FR light in the high irradiance response mode (Casal et al., 1998). Both response modes are of ecological relevance and it has been shown that phyA is essential for germination and seedling establishment in FR-rich environments, such as the understorey of forests (Nagy and Schäfer, 2002, Casal et al., 2014).
Phytochromes localize to the cytosol in the dark and translocate into the nucleus upon activation by light (Klose et al., 2015). PhyA and phyB use different molecular mechanisms for translocation into the nucleus. PhyA does not contain a nuclear localization signal (NLS) and its nuclear import depends on the functional homologs FHY1 (FAR-RED ELONGATED HYPOCOTOYL 1) and FHL (FHY1-LIKE) (Hiltbrunner et al., 2005, Hiltbrunner et al., 2006, Rösler et al., 2007, Genoud et al., 2008). In contrast, phyB employs an FHY1/FHL-independent mechanism and has been suggested to contain an NLS or to bind to transcription factors for transport into the nucleus (Chen et al., 2005, Hiltbrunner et al., 2005, Hiltbrunner et al., 2006, Pfeiffer et al., 2012). FHY1 and FHL are small plant-specific proteins containing an NLS and NES (nuclear export signal) motif at the N terminus, and a phyA binding site at the very C terminus. Both the NLS and the phyA binding motif are essential for proper FHY1/FHL function (Zeidler et al., 2004, Zhou et al., 2005, Hiltbrunner et al., 2006, Genoud et al., 2008). In contrast, the NES motif appears not to be essential, although this has only been investigated with transgenic Arabidopsis lines overexpressing FHY1 and it is possible that the NES is required when only wild-type levels are present (Zeidler et al., 2004). Consistent with their functional relevance, the NLS and the phyA binding site are highly conserved in FHY1/FHL homologs from monocots and dicots. It is interesting that FHY1/FHL-like proteins are also present in species that diverged from seed plants before the emergence of phyA and, for example, play a role in nuclear accumulation of PHY1 in Physcomitrella patens (Possart and Hiltbrunner, 2013, Inoue et al., 2016, Inoue et al., 2019, Han et al., 2019). Therefore, it is possible that FHY1/FHL-like proteins are components of an evolutionarily ancient phytochrome nuclear transport system.
Transcription factors such as LAF1 (LONG AFTER FAR-RED LIGHT 1) (Ballesteros et al., 2001), HFR1 (LONG HYPOCOTYL IN FAR-RED 1) (Fairchild et al., 2000, Fankhauser and Chory, 2000, Soh et al., 2000), HY5 (ELONGATED HYPOCOTYL 5) (Oyama et al., 1997), and PIF3 (PHYTOCHROME INTERACTING FACTOR 3) (Ni et al., 1998, Kim et al., 2003) are signaling components downstream of phyA involved in phyA-regulated gene expression. Interestingly, it has been shown that all these transcription factors can bind to FHY1 and/or FHL (Yang et al., 2009, Chen et al., 2012, Jang et al., 2013) and that phyA can associate with target promoters through FHY1 (Chen et al., 2012, Chen et al., 2014a, Chen et al., 2014b). Thus, it has been concluded that FHY1/FHL are not only essential for nuclear transport of phyA but also play a direct role in phyA-dependent gene expression by promoting the assembly of transcription factor complexes and guiding phyA to target promoters (Yang et al., 2009, Chen et al., 2012).
The NLS and in particular the C-terminal phyA binding site (FHY1 CT) are highly conserved in FHY1/FHL-like proteins, whereas the sequence between these two motifs is variable and only conserved in closely related species. We previously generated an artificial FHY1 that consisted of an SV40 NLS and the C-terminal phyA binding site of FHY1, and YFP as linker between these two motifs (Figure 1A) (Genoud et al., 2008). Even though artificial FHY1 does not contain any sequence from endogenous FHY1—except for the phyA binding site—it restored phyA signaling when expressed in fhy1-1 mutant background, suggesting that the NLS and the phyA binding site are sufficient for proper FHY1 function. Furthermore, expression of phyA-NLS-GFP, which is transported into the nucleus independently of FHY1/FHL, rescued the phyA fhy1 mutant phenotype, supporting the notion that FHY1 is not essential for phyA signaling if phyA is targeted into the nucleus by an NLS. However, these experiments have been done in FHL wild-type background and it is possible that FHL can compensate for the function of FHY1 in assembly of transcription factor complexes in the nucleus and guiding phyA to target promoters (Chen et al., 2012, Yang et al., 2009). Consistent with this idea, expression of YFP-NLS-FHY1 CT does not complement the fhy1-3 fhl-1 double mutant (Yang et al., 2009). However, YFP-NLS-FHY1 CT differs from artificial FHY1 regarding the position of the YFP tag (Figure 1A). In artificial FHY1, YFP replaces the non-conserved part between the NLS and the phyA binding site, not changing the general arrangement of the essential motifs; furthermore, YFP has roughly the same size as the non-conserved part between the NLS and the phyA binding site in endogenous FHY1/FHL-like proteins (Genoud et al., 2008). In contrast, in YFP-NLS-FHY1 CT, the NLS and the phyA binding site are directly fused and YFP is added as an N-terminal tag (Yang et al., 2009). Another difference between the approaches in Genoud et al. (2008) and Yang et al. (2009) is that fhy1-1 is in Ler background while fhy1-3 fhl-1 is in Col-0.
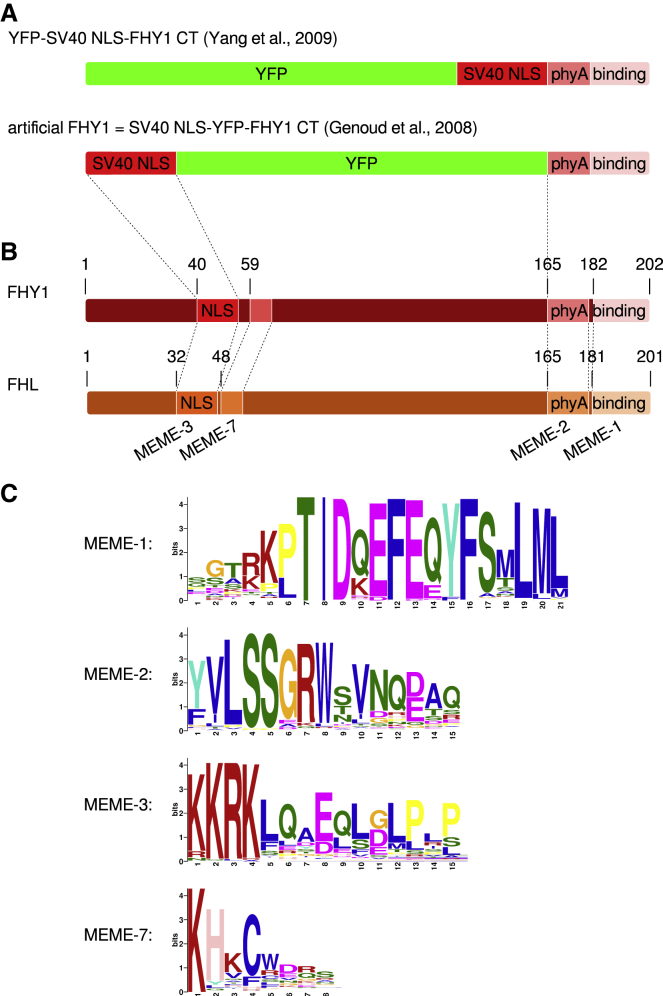
Figure 1.
Artificial FHY1 and Conserved Motifs in FHY1/FHL.
(A) Constructs used by Yang et al. (2009) and Genoud et al. (2008); we refer to the construct used by Genoud et al. (2008) as artificial FHY1.
(B) Schematic alignment of FHY1 and FHL. Numbers indicate amino acid positions. The NLS and the phyA binding site is indicated. MEME motifs are shown in different colors.
(C) Conserved motifs in FHY1/FHL-like proteins. MEME was used to identify conserved motifs in 116 FHY1/FHL-related sequences. MEME motifs 1, 2, 3, and 7 are present in Arabidopsis FHY1 and FHL.
Here, we investigated if artificial FHY1 (i.e., NLS-YFP-FHY1 CT) is active in fhy1-3 fhl-1 double-mutant background and if expression of phyA-NLS-YFP can restore FR responses in the phyA-211 fhy1-3 fhl-1 triple mutant.
Results
Artificial FHY1 Is Functional in the fhy1-3 fhl-1 Mutant
We have shown previously that an artificial FHY1 consisting of an SV40 NLS and the phyA binding site of Arabidopsis FHY1, and YFP as spacer between these motifs (NLS-YFP-FHY1 CT), is functional when expressed in fhy1-1 single-mutant background (Figure 1A) (Genoud et al., 2008). In contrast, a version in which YFP is placed at the N terminus (YFP-NLS-FHY1 CT) does not complement the fhy1-3 fhl-1 double mutant and, therefore, it has been suggested that the specific amino acid sequence of the spacer region of endogenous FHY1 is essential for FHY1 function (Figure 1A) (Yang et al., 2009). To identify conserved motifs in FHY1/FHL-like proteins, we used the consensus motif of the phyA binding site of FHY1/FHL-like proteins defined in Possart and Hiltbrunner (2013) to search the database at NCBI (see Materials and Methods for details). After removing redundant sequences and very similar sequences from closely related species, we obtained 116 entries containing a motif similar to the consensus sequence of the phyA binding site of FHY1/FHL-like proteins (Supplemental Data 1). In this dataset, MEME (http://meme-suite.org/; Bailey et al., 2009) identified eight motifs that are present in at least half of the sequences (Supplemental Data 2), but only four are present in Arabidopsis FHY1 and FHL (Figure 1B and 1C, Supplemental Figure 1, Supplemental Data 3). Two of these motifs (MEME-1 and -2) are included in the phyA binding site, and one motif (MEME-3) contains the NLS (Figure 1B). However, the MEME-3 motif also contains additional amino acid residues not part of the NLS that are conserved and there is another motif (MEME-7) that is present in most FHY1/FHL-like proteins, including Arabidopsis FHY1 and FHL (Figure 1B and 1C, Supplemental Figure 1, Supplemental Data 3). Thus, it is possible that these motifs are essential for full FHY1/FHL function and that endogenous FHL in fhy1-1 background expressing artificial FHY1 can compensate for functions depending on these motifs. Potential functions of these motifs are binding of HFR1 and/or LAF1, which have been shown to interact with the N-terminal half of FHY1/FHL not containing MEME-1 and -2 (Jang et al., 2013, Yang et al., 2009); furthermore, these motifs might also bind PIF3 and HY5, for which binding sites in FHY1 are still unknown (Chen et al., 2012). To investigate this hypothesis we generated several independent lines in the fhy1-3 fhl-1 double-mutant background that express artificial FHY1 under the control of the 35S promoter and measured hypocotyl growth in FR light. Normal phyA responsiveness is restored in all lines expressing 35S:artificial FHY1 in fhy1-3 fhl-1, very similar to the line expressing YFP-tagged wild-type FHY1 in fhy1-3 background (Figure 2A, Supplemental Figure 2) (Genoud et al., 2008). In contrast, when grown in the dark, there was no difference to Col-0 and fhy1-3 fhl-1 control seedlings. Also in terms of anthocyanin accumulation artificial FHY1 was active and restored wild-type anthocyanin levels when expressed in fhy1-3 fhl-1 mutant background (Figure 2B). FHY1 and FHL have been shown to form phyA-dependent photobodies in seedlings exposed to light (Hiltbrunner et al., 2005, Hiltbrunner et al., 2006). Similar to FHY1 and FHL, also artificial FHY1 expressed in fhy1-3 fhl-1 seedlings formed photobodies under conditions in which there are high levels of active phyA in the nucleus (6 h FR followed by 5 min; Figure 2C). We therefore conclude that artificial FHY1, which consists of an SV40 NLS, YFP, and the phyA binding motif of FHY1, is functional and behaves like native Arabidopsis FHY1.
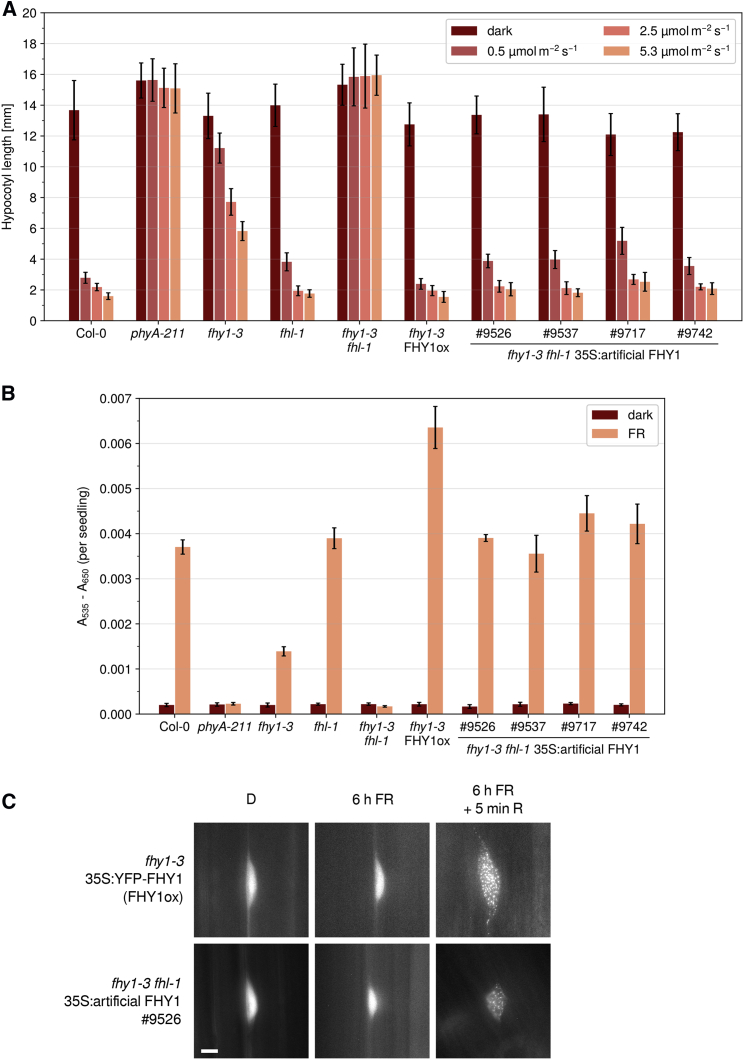
Figure 2.
Artificial FHY1 Restores Far-Red Light Responses in fhy1-3 fhl-1.
(A) Artificial FHY1 inhibits hypocotyl growth in FR light. Seedlings were grown in the dark or in FR light of different intensities. After 5 days, hypocotyl length was measured. Bars show mean hypocotyl length of ≥20 seedlings ± SD. Replicates are shown in Supplemental Figure 2.
(B) Artificial FHY1 promotes anthocyanin accumulation in FR light. Seedlings were grown in the dark or in continuous FR light (13 μmol m−2 s−1) for 5 days. Then, anthocyanin was extracted and relative amounts were quantified. Bars show mean values of A535–A650 per seedling of three replicates ± SD.
(C) Subcellular localization of artificial FHY1. Four-day-old dark-grown seedlings were used for fluorescence microscopy. The seedlings were analyzed directly (D), after 6 h irradiation with FR light (13 μmol m−2 s−1), or after 6 h FR light exposure followed by 5 min of R light (8 μmol m−2 s−1). Scale bar represents 5 μm.
(A–C) #9526, #9537, #9717, and #9742 are independent transgenic lines; #9526 was used in (C).
PhyA Signaling without FHY1 and FHL
Artificial FHY1 still contains the C-terminal 36 amino acid residues of wild-type FHY1 that are required for phyA binding. There is no indication or evidence suggesting that any protein except phyA binds to this motif, but we cannot rule out this possibility. Therefore, we tested if constitutively nuclear localized phyA expressed from the endogenous phyA promoter (ProPHYA:PHYA-NLS-YFP) would be active in the phyA-211 fhy1-3 fhl-1 triple mutant, i.e., in a background where there is no FHY1 and FHL. For this purpose we generated two independent lines (#10772 and #9494) by crossing phyA-211 phyA-NLS-YFP (#7129) and fhy1-3 fhl-1 phyA-NLS-YFP (#6900) into phyA-211 fhy1-3 fhl-1 and thereafter selecting siblings in the F2 generation that were homozygous for phyA-211 fhy1-3 fhl-1 and the transgene.
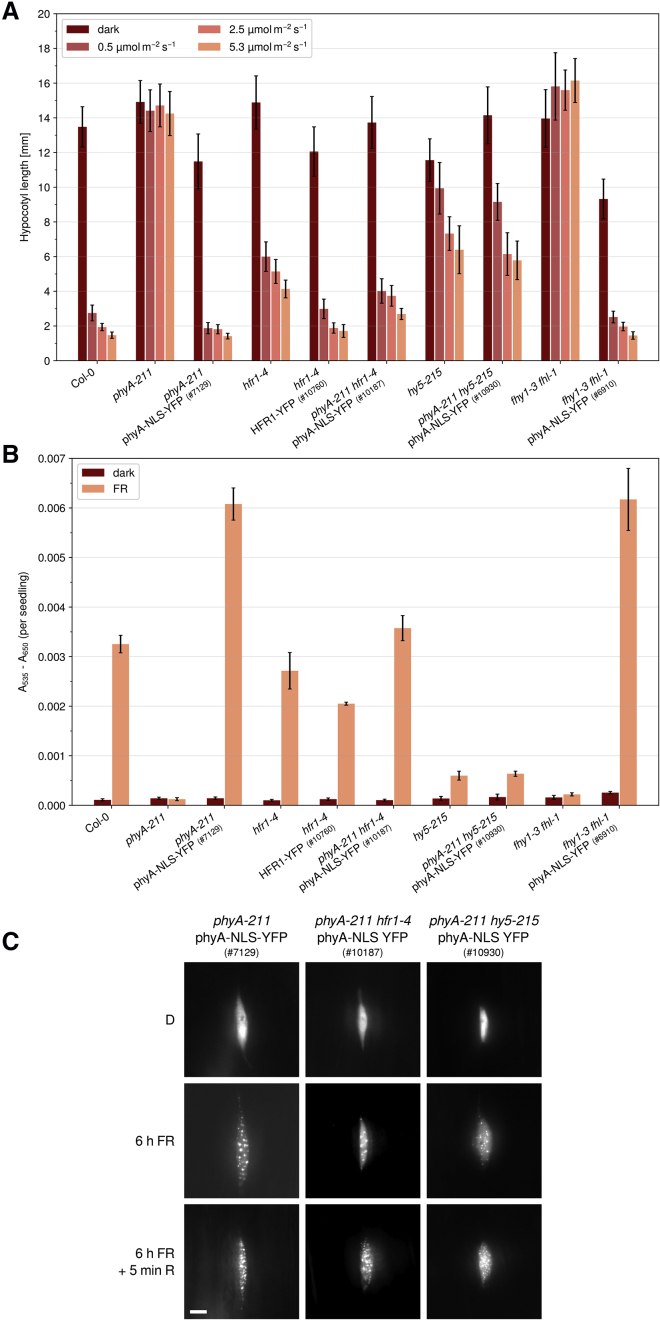
Expression of phyA-NLS-YFP restored inhibition of hypocotyl growth and accumulation of anthocyanin in FR light in phyA-211 single-, fhy1-3 fhl-1 double-, and phyA-211 fhy1-3 fhl-1 triple-mutant background (Figure 3A and 3B, Supplemental Figure 3). As expected, phyA-YFP is unable to translocate into the nucleus in the absence of FHY1 and FHL, while fusing an NLS to phyA renders phyA nuclear accumulation light and FHY1/FHL independent. Moreover, phyA-NLS-YFP formed photobodies in light similar to the control line expressing phyA-YFP in phyA-211 mutant background, demonstrating that recruitment of phyA into photobodies is independent of FHY1 and FHL (Figure 3C).
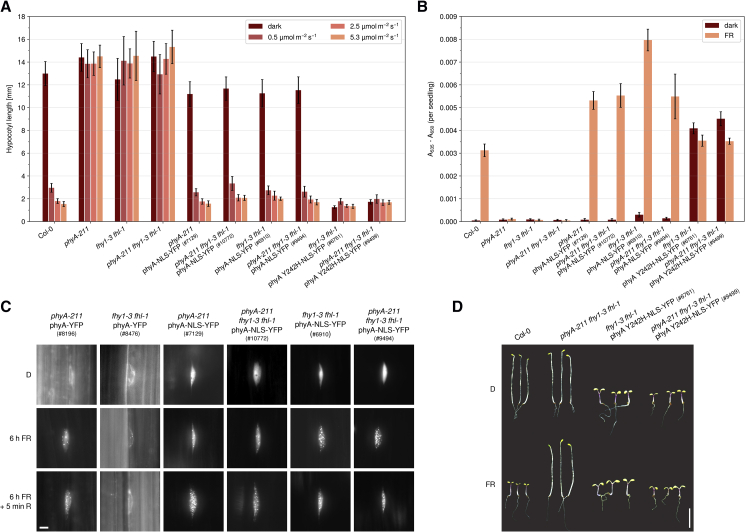
Figure 3.
Constitutively Nuclear Localized phyA Mediates Far-Red Light Responses without FHY1 and FHL.
(A) PhyA-NLS-YFP inhibits hypocotyl growth in FR light. Seedlings were grown in the dark or in FR light of different intensities. After 5 days, hypocotyl length was measured. Bars show mean hypocotyl length of ≥20 seedlings ± SD. Replicates are shown in Supplemental Figure 3.
(B) Constitutively nuclear localized phyA promotes anthocyanin accumulation in FR light. Seedlings were grown in the dark or in continuous FR light (13 μmol m−2 s−1) for 5 days. Anthocyanin was then extracted and relative amounts were quantified. Bars show mean values of A535–A650 per seedling of three replicates ± SD.
(C) Subcellular localization of phyA-YFP and phyA-NLS-YFP. Four-day-old dark-grown seedlings were used for fluorescence microscopy. The seedlings were analyzed directly (D), after 6 h irradiation with FR light (13 μmol m−2 s−1), or after 6 h FR light exposure followed by 5 min of R light (8 μmol m−2 s−1). Scale bar represents 5 μm.
(D) Seedlings expressing phyA Y242H-NLS-YFP in fhy1-3 fhl-1 are constitutively photomorphogenic. Seedlings were grown for 5 days in the dark (D) or continuous FR light (13 μmol m−2 s−1). Scale bar represents 5 mm.
The phyA Y242H mutant contains a Y-to-H amino acid substitution at position 242 and has been shown to be constitutively in a Pfr-like state (Su and Lagarias, 2007). Thus, it binds to FHY1/FHL and the phyA downstream signaling factors PIF1 and PIF3 in a light-independent fashion (Rausenberger et al., 2011). Constitutive binding of phyA Y242H possibly traps FHY1/FHL and inhibits recycling of FHY1/FHL from the nucleus into the cytosol, which interferes with efficient phyA nuclear transport (Rausenberger et al., 2011). Fusing an NLS to phyA Y242H-YFP overcomes this defect, and expression in wild-type background results in a strong constitutively photomorphogenic (cop) phenotype (Rausenberger et al., 2011). Here, we transformed a phyA Y242H-NLS-YFP construct into fhy1-3 fhl-1 mutant background and isolated a homozygous T2 line; we also crossed this line into the phyA-211 fhy1-3 fhl-1 triple mutant and selected in the F2 generation for siblings being homozygous for the transgene as well as for phyA-211, fhy1-3, and fhl-1. phyA Y242H-NLS-YFP was highly active in fhy1-3 fhl-1 double- and phyA-211 fhy1-3 fhl-1 triple-mutant background (Figure 3A and 3B, Supplemental Figure 3). Dark-grown fhy1-3 fhl-1 and phyA-211 fhy1-3 fhl-1 seedlings expressing phyA Y242H-NLS-YFP were fully de-etiolated and accumulated anthocyanin; hypocotyl length and anthocyanin in FR light were similar to the wild type (Figure 3A, 3B, and 3D, Supplemental Figure 3). These data further confirm that FHY1 and FHL are not required for phyA signaling in the nucleus.
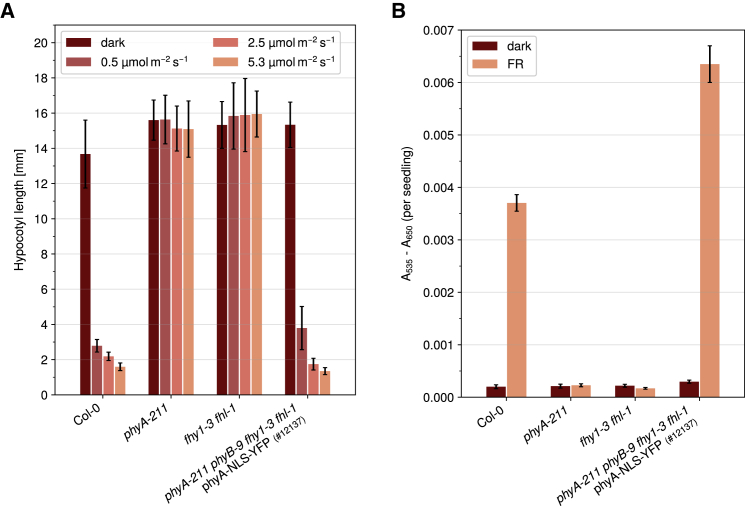
We also crossed fhy1-3 fhl-1 phyA-NLS-YFP into fhy1-3 fhl-1 phyB-9 and selected in the F2 generation for plants that are homozygous for the transgene and the mutant background, which we then crossed into phyA-211 fhy1-3 fhl-1. F2 seedlings being homozygous for the phyA-NLS-YFP transgene and phyA-211, phyB-9, fhy1-3, and fhl-1 were phenotypically very similar to the wild type and had strongly reduced hypocotyl growth in FR light compared with the dark and accumulated high levels of anthocyanin in FR light (Figure 4). Thus, as expected, FR light responses of seedlings expressing phyA-NLS-YFP do not depend on phyB.
Figure 4.
PhyB Is Not Required for Activity of phyA-NLS-YFP in FR Light.
(A) Inhibition of hypocotyl growth by phyA-NLS-YFP does not require phyB. Seedlings were grown in the dark or in FR light of different intensities. After 5 days, hypocotyl length was measured. Bars show mean hypocotyl length of ≥20 seedlings ± SD. Replicates are shown in Supplemental Figure 4.
(B) PhyB is not required for upregulation of anthocyanin levels by phyA-NLS-YFP. Seedlings were grown in the dark or in continuous FR light (13 μmol m−2 s−1) for 5 days. Anthocyanin was then extracted and relative amounts were quantified. Bars show mean values of A535–A650 per seedling of three replicates ± SD.
(A and B) Data for Col-0, phyA-211, and fhy1-3 fhl-1 are from Figure 2.
PhyA-NLS-YFP Relies on phyA Downstream Signaling Components for Induction of FR Light Responses
To analyze if fusing an NLS to phyA specifically overcomes defects in nuclear transport of phyA, but not in downstream signaling, we investigated the effect of constitutively nuclear localized phyA in mutant seedlings deficient in HFR1 or HY5. To this end we crossed the phyA-211 phyA-NLS-YFP line (#7129) into hfr1-4 and hy5-215 background and selected in the F2 generation for plants homozygous for the transgene, phyA-211 and hfr1-4 or hy5-215. Interestingly, expression of phyA-NLS-YFP only slightly reduced hypocotyl growth in FR light in hy5-215 and hfr1-4, while it fully complements the much stronger fhy1-3 fhl-1 double-mutant phenotype (Figure 5A, Supplemental Figure 5). The slight effect of phyA-NLS-YFP on hypocotyl growth in hy5-215 and hfr1-4 is possibly due to increased activation of HFR1- and HY5-independent signaling pathways. Thus, nuclear localized phyA still requires HFR1 and HY5 for downstream signaling, while FHY1 and FHL are not essential in presence of constitutively nuclear localized phyA (Figure 5A, Supplemental Figure 5). In terms of anthocyanin accumulation it is even more evident that expression of phyA-NLS-YFP in the phyA-211 hy5-215 double-mutant background is unable to restore the wild-type phenotype (Figure 5B). As reported previously, the hfr1-4 mutant has only a very weak phenotype regarding anthocyanin accumulation in FR light (Fankhauser and Chory, 2000, Soh et al., 2000); however, expression of phyA-NLS-YFP caused strongly increased anthocyanin levels in phyA-211, but not in phyA-211 hfr1-4 background. Recruitment of phyA-NLS-YFP into photobodies appears not to be affected in the absence of either HFR1 or HY5 (Figure 5C). Overall, these data show that adding an NLS to phyA specifically circumvents the need of FHY1/FHL for nuclear transport but not the requirement of downstream signaling factors, such HY5 and HFR1.
Figure 5.
FR Light Responses Mediated by phyA-NLS-YFP Depend on HFR1 and HY5.
(A) HY5 and HFR1 are required for phyA-NLS-YFP–mediated inhibition of hypocotyl growth. Seedlings were grown in the dark or in FR light of different intensities. After 5 days, hypocotyl length was measured. Bars show mean hypocotyl length of ≥20 seedlings ± SD. Replicates are shown in Supplemental Figure 5.
(B) Enhanced anthocyanin accumulation in phyA-NLS-YFP expressing seedlings depends on HY5 and HFR1. Seedlings were grown in the dark or in continuous FR light (13 μmol m−2 s−1) for 5 days. Then, anthocyanin was extracted and relative amounts were quantified. Bars show mean values of A535–A650 per seedling of three replicates ± SD.
(C) Subcellular localization of phyA-NLS-YFP in hfr1-4 and hy5-215. Four-day-old dark-grown seedlings were used for fluorescence microscopy. The seedlings were analyzed directly (D), after 6 h irradiation with FR light (13 μmol m−2 s−1), or after 6 h FR light exposure followed by 5 min of R light (8 μmol m−2 s−1). Scale bar represents 5 μm.
Plants Expressing phyA-NLS-YFP Are Hypersensitive to R and FR Light
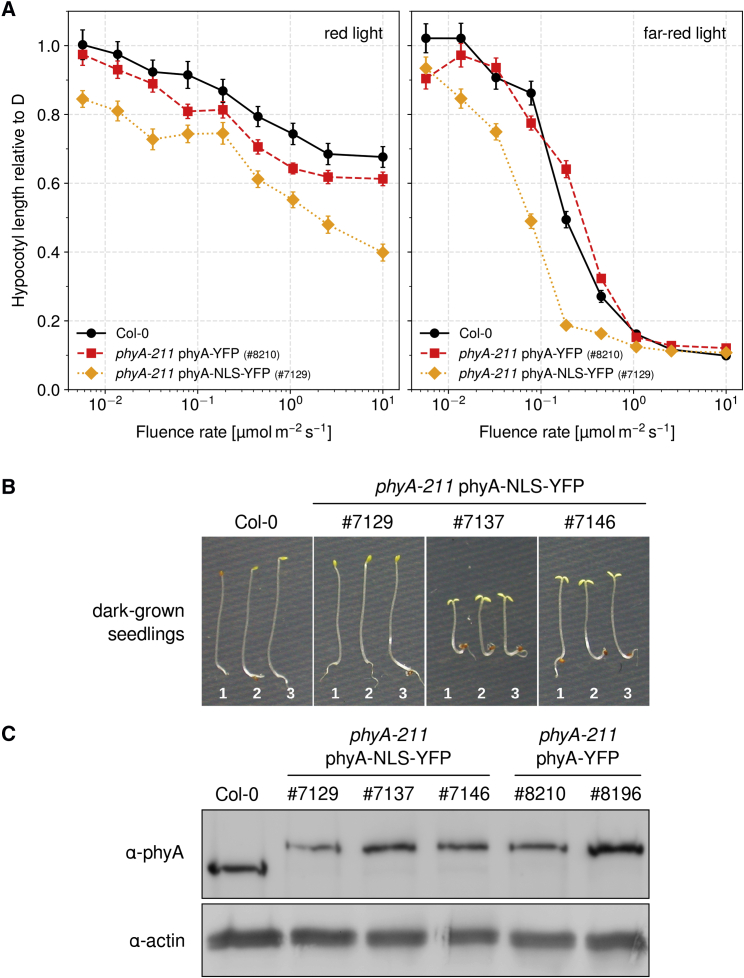
PhyB is the primary receptor for R light, but it is well-known that phyA also contributes to R light responses and even plays a dominant role in early R light signaling (Tepperman et al., 2006). Thus, we measured fluence rate response curves in R and FR light for the wild type and phyA-211 expressing either phyA-YFP or phyA-NLS-YFP. Hypocotyl length of phyA-YFP expressing seedlings was similar to the wild type in R and FR light conditions, while expression of phyA-NLS-YFP resulted in hypersensitivity to R and FR light over the full range of fluence rates (Figure 6A, Supplemental Figure 6).
Figure 6.
Differences between Wild Type and phyA-NLS-YFP Expressing Seedlings.
(A) PhyA-NLS-YFP expressing seedlings are hypersensitive to R and FR light. Seedlings were grown for 4 days in R light (left), FR light (right), or in the dark. Data show the mean hypocotyl length of ≥18 seedlings relative to dark-grown (D) seedlings ±SE. Absolute values are shown in Supplemental Figure 6.
(B) Seedlings expressing high levels of phyA-NLS-YFP are constitutively photomorphogenic. Col-0 and three independent transgenic lines expressing phyA-NLS-YFP in phyA-211 background (#7129, #7137, and #7146) were grown for 4 days in the dark. Germination was induced by different light treatments: (1) 5 min R light, 2 h 55 min dark, 5 min RG9 (long-wavelength FR light); (2) 3 h R light, 5 min RG9; (3) 3 h R light.
(C) PhyA protein levels in phyA-NLS-YFP and phyA-YFP expressing lines. Seedlings were grown for 4 days in the dark. Total protein was extracted, separated on an SDS–PAGE, and blotted onto PVDF membrane. A phyA-specific antibody was used to detect phyA; actin was detected as loading control.
(B and C) Line #7129 was used for experiments in Figures 3, 5, and 6.
High Expression Levels of phyA-NLS-YFP Result in a cop Phenotype
For all experiments with phyA-211 phyA-NLS-YFP we used transgenic line #7129. When grown in the dark, seedlings of this line occasionally had slightly unfolded cotyledons and reduced apical hook formation compared with wild-type seedlings. However, in most phyA-NLS-YFP-expressing lines that we isolated, this phenotype was much more pronounced than in line #7129. Many of these lines, including lines #7137 and #7146, had fully opened cotyledons in the dark and strongly reduced hypocotyl growth compared with the wild type (Figure 6B). We used different light treatments to induce germination (e.g., 3 h R, 3 h R followed by a long-wavelength FR [RG9] pulse, or 5 min R followed by 2 h 55 min D and an RG9 pulse [true-dark]; Leivar et al., 2008), but even under true-dark conditions lines #7137 and #7146 had an obvious cop phenotype (Figure 6B). Using a phyA-specific antibody for immunoblot analysis we compared the phyA-NLS-YFP levels in lines #7129, #7137, and #7146 (Figure 6C). Consistent with the strong cop phenotype of line #7137, this line has the highest expression levels, while phyA-NLS-YFP levels are only slightly higher in line #7146 than #7129. Yet, the slightly higher levels of phyA-NLS-YFP in line #7146 compared with #7129 appear to be sufficient to induce de-etiolation in the dark (Figure 6B and 6C). We conclude that high levels of constitutively nuclear localized phyA can induce downstream signaling in the absence of light, i.e., even if phyA is in the inactive Pr form.
Discussion
It has been widely accepted that FHY1 and its homolog FHL regulate a critical step in FR light signaling by controlling the nuclear import of phyA (Hiltbrunner et al., 2006, Rösler et al., 2007, Genoud et al., 2008). However, whether or not FHY1 and FHL also play an essential role in phyA downstream signaling in the nucleus, remained unclear. Here, we addressed this question in detail and provide conclusive evidence that FHY1 and FHL are essential for nuclear transport of phyA but not for downstream signaling of nuclear localized phyA.
Several transcription factors involved in phyA downstream signaling interact with FHY1 and/or FHL. HFR1 and LAF1 bind to FHY1 and FHL independently of the phyA binding site (Yang et al., 2009, Jang et al., 2013) and also HY5 and PIF3 have been shown to interact with FHY1 (Chen et al., 2012). Thus, it has been proposed that FHY1 and FHL are essential for the assembly of phyA signaling complexes in the nucleus and guiding phyA to target promoters (Yang et al., 2009, Chen et al., 2012). In contradiction to this model, we have previously shown that expression of either artificial FHY1 (SV40 NLS-YFP-FHY1 CT), which lacks the binding sites for HFR1 and LAF1, or phyA-NLS-GFP, fully complements the fhy1-1 mutant (Genoud et al., 2008). However, FHL transcript levels are three-fold upregulated in the absence of FHY1 (Yang et al., 2009) and therefore increased levels of FHL could compensate for the lack of FHY1 in lines expressing phyA-NLS-GFP or artificial FHY1 in fhy1-1 mutant background. Thus, it has not been possible to decide if FHY1 and FHL are required for phyA downstream signaling in the nucleus or if they only mediate phyA nuclear transport. Here, we expressed artificial FHY1 and phyA-NLS-YFP in fhy1-3 fhl-1 and phyA-211 fhy1-3 fhl-1 mutant background, respectively. Artificial FHY1 and constitutively nuclear localized phyA both restore accumulation of anthocyanin and inhibition of hypocotyl growth in FR light in the absence of functional FHY1 and FHL. Thus, we conclude that FHY1 and FHL are not essential for phyA downstream signaling in the nucleus. Our data do not exclude the possibility that FHY1 and FHL might promote the assembly of transcription factor complexes in the nucleus or guide phyA to target promoters but we demonstrate that these events either do not depend on FHY1 and FHL or are not essential for normal anthocyanin accumulation and inhibition of hypocotyl growth in response to FR light (Yang et al., 2009, Chen et al., 2012). In this study, we did not investigate other FR light-regulated responses and it is possible that other responses depend on FHY1/FHL-mediated assembly of transcription factor complexes or guiding phyA to target promoters.
Artificial FHY1 consists of an SV40 NLS fused to the N terminus of YFP and the phyA binding site of FHY1 (FHY1 167-202) attached to the C terminus. Thus, YFP in artificial FHY1 mimics the “spacer” in natural FHY1. This spacer region is roughly 150 to 250 amino acid residues in length without any annotated functional motifs and with only low sequence similarity between FHY1/FHL proteins from different species. However, it is interesting that this spacer region is present in all FHY1/FHL proteins and usually not shorter than around 150 amino acid residues. Despite the lack of similarity at the level of the primary sequence, the 3D structure of the spacer might be similar. Yet, we have shown that YFP can substitute for the spacer, suggesting that the 3D structure of the spacer is unlikely to be important for FHY1/FHL function. Alternatively, the spacer might be essential to provide sufficient flexibility for simultaneous binding of FHY1/FHL to phyA and importin alpha, which is required for phyA nuclear transport (Helizon et al., 2018). In this regard it is interesting that Yang et al. (2009) have shown that YFP-NLS-FHY1 167-202 is only very weakly active and does not complement the fhy1-3 fhl-1 double mutant. Our interpretation is that fusing the NLS directly to the phyA binding site and adding YFP at the very N terminus of the construct (instead of inserting YFP as a spacer between the NLS and the phyA binding site) does not allow phyA and importin alpha to bind simultaneously due to sterical hindrance and therefore results in a non-functional protein.
Using a mathematical modeling approach we previously explored potential molecular mechanisms involved in shifting the phyA action peak from R to FR light (Rausenberger et al., 2011). FHY1/FHL-dependent nuclear transport was identified as one potential mechanism but also the existence of FHY1/FHL-independent shifting modules working in parallel was predicted. Interestingly, lack of only one shifting module was predicted not to shift the phyA action peak but to result in a peak that extends into the R light range of the light spectrum. Adding an NLS directly to phyA renders phyA nuclear transport independent of FHY1 and FHL and therefore abolishes one shifting module. Here, we have shown that seedlings expressing phyA-NLS-YFP are hypersensitive to R and FR light to a similar extent, suggesting that they still have an action peak in FR light. This finding is in agreement with the prediction by Rausenberger et al. (2011); however, measurement of detailed action spectra is required to decide if expression of constitutively nuclear localized phyA results in extension of the action peak toward the R light range. Moreover, it is important to clarify if the increased response of phyA-NLS-YFP expressing seedlings in R light also requires phyB.
Dark-grown seedlings rapidly elongate, have folded cotyledons and an apical hook that protects the apical meristem from mechanical damage when the seedlings push through the soil. It is evident that, under natural conditions, this developmental program is important for the first phase in the life cycle following germination and that seedlings that are constitutively photomorphogenic have a much lower chance to reach the soil surface than wild-type seedlings. We have shown that moderate expression of phyA-NLS-YFP reinstates FR light-induced anthocyanin accumulation and inhibition of hypocotyl growth in phyA-211 fhy1-3 fhl-1, but that only slightly increased levels result in a constitutively photomorphogenic phenotype. In contrast, we have never observed such a cop phenotype for plants expressing wild-type phyA (i.e., phyA that relies on FHY1/FHL for nuclear transport), suggesting that restricting phyA to the cytosol in dark-grown seedlings contributes to ensure that seedlings do not de-etiolate in the dark. Weak but clearly detectable phyA-dependent induction of gene expression in fhy1-3 fhl-1 background has been observed, indicating that, even in the absence of FHY1 and FHL, residual amounts of phyA are present in nuclei (Kami et al., 2012, Pfeiffer et al., 2012); however, the amount of phyA in nuclei of dark-grown plants expressing phyA-NLS-YFP is possibly several orders of magnitude higher than in the wild type. We suggest that the activity of phyA-NLS-YFP in the dark is likely due to weak interaction of the inactive Pr form with downstream signaling factors, such as PIFs and SPAs, which could result in constitutive photomorphogenesis. Thus, in the hypothetical scenario that endogenous phyA would contain an NLS it might be very difficult for plants to regulate the levels of phyA precisely enough to ensure proper photomorphogenesis in light without losing skotomorphogenesis in the dark. In contrast, FHY1 and FHL provide a highly robust mechanism that reliably prevents phyA from accumulating in the nucleus in the dark, while efficiently transporting it into the nucleus in light. It is interesting that nuclear transport of phyB is much less tightly regulated than nuclear transport of phyA (Klose et al., 2015). The reason for this might be that there is simply no need for a more tight control of phyB nuclear transport. Induction of light signaling by phyB is much less sensitive than by phyA and therefore even comparably high levels of phyB Pr in the nucleus would not be sufficient to induce photomorphogenesis in dark-grown seedlings (Huq et al., 2003, Matsushita et al., 2003). Thus, there is no selection pressure that would drive the evolution of a phyB nuclear transport mechanism that is as selective as FHY1/FHL-dependent phyA nuclear transport. However, there is one caveat: even though there are no indications that YFP or other GFP-derived fluorescent proteins could disturb the normal function of phytochromes when fused to their C terminus, we cannot formally exclude the possibility that YFP fused to the C terminus of phyA-NLS is responsible for the signaling activity of phyA-NLS-YFP in the dark.
In summary, we have shown that FHY1 and FHL are required for phyA nuclear transport but that phyA, once it is in the nucleus, does not require FHY1 and FHL for downstream signaling. Furthermore, seedlings expressing phyA-NLS-YFP are hypersensitive to R and FR light to a similar extent, suggesting that they still have an action peak in FR light. Finally, FHY1/FHL provide a highly robust nuclear transport system for phyA, which prevents phyA from inducing light signaling in the dark.
Materials and Methods
Plasmid Constructs
Constructs coding for ProPHYA:PHYA-NLS-YFP and ProPHYA:PHYA Y242H-NLS-YFP have been described (Rausenberger et al., 2011).
pPPO72-FHY1 167-202 is a T-DNA vector containing a Pro35S:SV40 NLS-YFP-FHY1 167- 202:terRbcS cassette and PPO as selectable marker. It was obtained as follows. The PPO selection marker cassette was cut from pWCO35 (Hanin et al., 2001) using PvuII/PstI and ligated into the SbfI/PmlI site of pCHF72-FHY1 167-202 (Genoud et al., 2008) to replace the BASTA selectable marker.
The plant expression vector pPPO30v1HA (encoding Pro35S:BamHI-XbaI-YFP-HA:terRbcS and containing PPO as selection marker) was generated by first cutting pCHF5 (Hiltbrunner et al., 2005) with PmeI/NcoI, and ligating in a StuI/NcoI fragment from pYES2 (Invitrogen) to generate pCHF5v1. pWCO35 (Rausenberger et al., 2011) was then cut with PvuII/PstI, and this fragment ligated into PmlI/SbfI cut pCHF5v1 to generate pPPO5v1. Finally, EYFP was amplified by PCR from pPPO30 (Rausenberger et al., 2011) using the primers 5′-CGC GGA TCC CGC TCT AGA ATG GTG AGC AAG GGC GAG G-3′ and 5′-GTA CGT CGT ATG GGT AGC TAG CCT TGT ACA GCT CGT CCA TG-3′; the EYFP PCR fragment was used as template for another PCR using the primers 5′-CGC GGA TCC CGC TCT AGA ATG GTG AGC AAG GGC GAG G-3′ and 5′-GGA CTA GTT TAA GCG TAA TCT GGT ACG TCG TAT GGG TAG C-3′. The resulting PCR fragment was then cut with BamHI/SpeI and cloned into BamHI/XbaI cut pPPO5v1 to generate pPPO30v1HA. To obtain pPPO30v1HA-HFR1 (coding for Pro35S:HFR1-YFP-HA:terRbcS) we cut HFR1 from pBS II KS-HFR1 (Sheerin et al., 2015) using BamHI/SpeI and ligated it into the BamHI/XbaI site of pPPO30v1HA.
pPPO70-FHY1 is a T-DNA vector containing a Pro35S:YFP-FHY1:terRbcS cassette and PPO as selectable marker. It was obtained as follows. The PPO selection marker cassette was cut from pWCO35 (Hanin et al., 2001) using PvuII/PstI and ligated into the SbfI/PmlI site of pCHF70-FHY1 (Rausenberger et al., 2011) to replace the BASTA selectable marker.
Plant Material
Columbia (Col-0) ecotype of A. thaliana was used as wild type. The phyA-211, fhy1-3, fhl-1, fhy1-3 fhl-1, hfr1-4, and hy5-215 mutants have been described previously (Reed et al., 1994, Oyama et al., 1997, Zeidler et al., 2001, Sessa et al., 2005, Zhou et al., 2005, Rösler et al., 2007). The transgenic lines fhy1-3 Pro35S:YFP-FHY1 (FHY1ox; line #4810), fhy1-3 fhl-1 Pro35S:NLS-YFP-FHY1 167-202 (four independent lines: #9526, #9537, #9717, and #9742), phyA-211 ProPHYA:PHYA-NLS-YFP (lines #7129, #7137, and #7146), fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (two independent transgenic lines: #6900 and #6910), phyA-211 ProPHYA-PHYA-YFP (two independent transgenic lines: #8196 and #8210), fhy1-3 fhl-1 ProPHYA:PHYA Y242H-NLS-YFP (line #6761), fhy1-3 fhl-1 ProPHYA:PHYA-YFP (line #8476), and hfr1-4 Pro35S:HFR1-YFP-HA (line #10760) were obtained by Agrobacterium tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998).
The phyA-211 fhy1-3 fhl-1 line was obtained by crossing phyA-211 into fhl-1 and subsequently phyA-211 fhl-1 into fhy1-3 fhl-1 background. phyA-211 fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP lines #10772 and #9494 were generated by crossing phyA-211 ProPHYA:PHYA-NLS-YFP (line #7129) and fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #6900), respectively, into phyA-211 fhy1-3 fhl-1. phyA-211 fhy1-3 fhl-1 ProPHYA:PHYA Y242H-NLS-YFP (#line 9499) was obtained from fhy1-3 fhl-1 ProPHYA:PHYAY242H-NLS-YFP (line #6761) crossed into phyA-211 fhy1-3 fhl-1. By crossing phyA-211 ProPHYA:PHYA-NLS-YFP (line #7129) into hfr1-4 and hy5-215 we obtained phyA-211 hfr1-4 ProPHYA:PHYA-NLS-YFP (line #10187) and phyA-211 hy5-215 ProPHYA:PHYA-NLS-YFP (line #10930), respectively. phyA-211 phyB-9 fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #12137) was obtained as follows. We crossed phyB-9 into fhl-1 and subsequently phyB-9 fhl-1 into fhy1-3 fhl-1, resulting in phyB-9 fhy1-3 fhl-1. phyB-9 fhy1-3 fhl-1 was then crossed into fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #6900) to obtain phyB-9 fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #10593). Finally, we crossed phyB-9 fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #10593) into phyA-211 fhy1-3 fhl-1 resulting in phyA-211 phyB-9 fhy1-3 fhl-1 ProPHYA:PHYA-NLS-YFP (line #12137).
Seed Sterilization and Plating
Seeds were surface sterilized by shaking in 1 ml 70% ethanol with 0.05% Triton X-100 for 10 min followed by 5 min incubation in 100% ethanol. Seeds were then dried on sterile filter paper and spread onto Petri dishes containing 0.5× Murashige and Skoog salts (Duchefa) and 1% phytoagar (Duchefa). For measurement of anthocyanin accumulation sterilized seeds were plated on 0.5× MS/1% phytoagar supplemented with 1.5% sucrose.
Measurement of Hypocotyl Length and Anthocyanin Accumulation
Germination of Arabidopsis seeds was induced by incubating the plates for 4 days at 4°C followed by 4–8 h in white light. Then, plates were transferred to either complete darkness or exposed to continuous FR light (720 nm) of different intensities. For the analysis of hypocotyl length the 5-day-old seedlings were arranged on square plates containing 0.5× MS/1% phytoagar and scanned. The hypocotyl length was then measured using ImageJ.
Anthocyanin was extracted by collecting 60 seedlings from each light treatment/genotype into 700 μl extraction buffer (18% [v/v] 1-propanol, 0.37% [v/v] HCl). The samples were then heated to 95°C for 2 min, chilled on ice for 5 min, and incubated overnight in the dark at 4°C under continuous shaking. On the next day, the plant material was pelleted by centrifugation for 10 min and the supernatant was analyzed. Anthocyanin was quantified by measuring A535 and A650 using a spectrophotometer. The relative amount of anthocyanin per seedling was calculated by dividing (A535–A650) by the number of seedlings.
Microscopy
Image acquisition was performed with a Nikon ECLIPSE 90i microscope equipped with YFP filters and a 64× water objective. The 4-day-old dark-grown seedlings (dark condition) were directly observed under the microscope using safety green light. For the light conditions, the 4-day-old dark-grown seedlings were treated for 6 h with FR light (720 nm, 13 μmol m−2 s−1) either followed by a 5-min exposure to R light (670 nm, 8 μmol m−2 s−1) or not. All images were acquired using Metamorph (version 6.2r4). ImageJ (version 1.52p) and GIMP (version 2.8.16) software was used for image processing.
Protein Extraction and Immunoblot Analysis
Seedlings were grown in the dark on 0.5× MS/1% phytoagar. After 4 days, seedlings were harvested and used for extraction of total proteins as described previously (Kircher et al., 2002). Immunoblotting was done according to standard protocols. Commercially available antibodies were used for detection of phyA (Agrisera, no. AS07220) and actin (Sigma-Aldrich, no. A0480).
MEME Motifs
The consensus motif of the phyA binding site of FHY1/FHL-like proteins (Possart and Hiltbrunner, 2013) was used to search the protein database at NCBI (max target sequences, 1000; expected threshold, 1e-3; word size, 2; gap costs, existence 9, extension 1; otherwise default settings were used). Redundant sequences (i.e., sequences with ≥99% sequence identity) were removed. Remaining sequences were submitted to MEME (http://meme-suite.org/) to search for conserved motifs using default settings (Bailey et al., 2009).
Funding
This study was supported by the German Research Foundation (DFG) under Germany's Excellence Strategy (BIOSS—EXC-294; CIBSS—EXC-2189—Project ID 390939984) and by grants from the DFG (DFG HI 1369/4-1 and HI 1369/5-1) and the Human Frontier Science Program Organization (HFSP research grant RGP0025/2013) to A.H. C.K. was supported by the Ministry of Science, Research and the Arts Baden-Wuerttemberg. The article processing charge was funded by the German Research Foundation (DFG) and the Albert Ludwigs University Freiburg in the funding programme Open Access Publishing.
Author Contributions
Conceptualization, A.H. and C.M.; Methodology, A.H., C.M., and C.K.; Investigation, C.M., C.K., and A.H.; Resources, A.H. and C.M.; Writing – Orignal Draft, A.H. and C.M.; Writing – Review & Editing, A.H., C.K., and C.M.; Visualization, C.M., A.H., and C.K.; Supervision, A.H.; Project Administration, A.H.; Funding Acquisition, A.H. and C.K.
Acknowledgments
No conflict of interest declared.
Published: November 9, 2019
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Accession Numbers
FHL, At5g02200; FHY1, At2g37678; HFR1, At1g02340; HY5, At5g11260; LAF1, At4g25560; phyA, At1g09570; phyB, At2g18790; PIF3, At1g09530. Identifiers for FHY1- and FHL-like proteins from different species can be found in Supplemental Data 1.
Supplemental Information
References
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Bolle C., Lois L.M., Moore J.M., Vielle-Calzada J.P., Grossniklaus U., Chua N.H. LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie E.S., Vierstra R.D. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell. 2014;26:4568–4583. doi: 10.1105/tpc.114.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Sanchez R.A., Botto J.F. Modes of action of phytochromes. J. Exp. Bot. 1998;49:127–138. [Google Scholar]
- Casal J.J., Candia A.N., Sellaro R. Light perception and signalling by phytochrome A. J. Exp. Bot. 2014;65:2835–2845. doi: 10.1093/jxb/ert379. [DOI] [PubMed] [Google Scholar]
- Chen M., Tao Y., Lim J., Shaw A., Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- Chen F., Shi X., Chen L., Dai M., Zhou Z., Shen Y., Li J., Li G., Wei N., Deng X.W. Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell. 2012;24:1907–1920. doi: 10.1105/tpc.112.097733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Li B., Demone J., Charron J.-B., Shi X., Deng X.W. Photoreceptor partner FHY1 has an independent role in gene modulation and plant development under far-red light. Proc. Natl. Acad. Sci. U S A. 2014;111:11888–11893. doi: 10.1073/pnas.1412528111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Li B., Li G., Charron J.-B., Dai M., Shi X., Deng X.W. Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell. 2014;26:1949–1966. doi: 10.1105/tpc.114.123950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 2000;124:39–45. doi: 10.1104/pp.124.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão V.C., Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 2008;4:e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Chang X., Zhang Z., Chen H., He H., Zhong B., Deng X.W. Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant. 2019;12:847–862. doi: 10.1016/j.molp.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Hanin M., Volrath S., Bogucki A., Briker M., Ward E., Paszkowski J. Gene targeting in Arabidopsis. Plant J. 2001;28:671–677. doi: 10.1046/j.1365-313x.2001.01183.x. [DOI] [PubMed] [Google Scholar]
- Helizon H., Rösler-Dalton J., Gasch P., von Horsten S., Essen L.-O., Zeidler M. Arabidopsis phytochrome A nuclear translocation is mediated by a far-red elongated hypocotyl 1-importin complex. Plant J. 2018;96:1255–1268. doi: 10.1111/tpj.14107. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Quail P.H. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 2003;35:660–664. doi: 10.1046/j.1365-313x.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- Inoue K., Nishihama R., Kataoka H., Hosaka M., Manabe R., Nomoto M., Tada Y., Ishizaki K., Kohchi T. Phytochrome signaling is mediated by PHYTOCHROME INTERACTING FACTOR in the liverwort Marchantia polymorpha. Plant Cell. 2016;28:1406–1421. doi: 10.1105/tpc.15.01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Nishihama R., Araki T., Kohchi T. Reproductive induction is a far-red high irradiance response that is mediated by phytochrome and PHYTOCHROME INTERACTING FACTOR in Marchantia polymorpha. Plant Cell Physiol. 2019;60:1136–1145. doi: 10.1093/pcp/pcz029. [DOI] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Chua N.H. Three transcription factors, HFR1, LAF1 and HY5, regulate largely independent signaling pathways downstream of phytochrome A. Plant Cell Physiol. 2013;54:907–916. doi: 10.1093/pcp/pct042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Hersch M., Trevisan M., Genoud T., Hiltbrunner A., Bergmann S., Fankhauser C. Nuclear phytochrome A signaling promotes phototropism in Arabidopsis. Plant Cell. 2012;24:566–576. doi: 10.1105/tpc.111.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.-S., Choi G. Functional characterization of PHYTOCHROME INTERACTING FACTOR 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Müller T., Bauer D., Ádám É., Schäfer E., Nagy F. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell. 2002;14:1541–1555. doi: 10.1105/tpc.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C., Viczián A., Kircher S., Schäfer E., Nagy F. Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 2015;206:965–971. doi: 10.1111/nph.13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N., Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- Nagy F., Schäfer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Nagel M.-K., Popp C., Wüst F., Bindics J., Viczián A., Hiltbrunner A., Nagy F., Kunkel T., Schäfer E. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc. Natl. Acad. Sci. U S A. 2012;109:5892–5897. doi: 10.1073/pnas.1120764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A., Hiltbrunner A. An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell. 2013;25:102–114. doi: 10.1105/tpc.112.104331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146:813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.-S., Lagarias J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler J., Klein I., Zeidler M. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. U S A. 2007;104:10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Possenti M., Mittempergher F., Becker J., Morelli G., Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.-D., Huq E., Hiltbrunner A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015;27:189–201. doi: 10.1105/tpc.114.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh M.S., Kim Y.M., Han S.J., Song P.S. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell. 2000;12:2061–2074. doi: 10.1105/tpc.12.11.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Lagarias J.C. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J.M., Hwang Y.-S., Quail P.H. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Yang S.W., Jang I.-C., Henriques R., Chua N.-H. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21:1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M., Bolle C., Chua N.H. The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol. 2001;42:1193–1200. doi: 10.1093/pcp/pce177. [DOI] [PubMed] [Google Scholar]
- Zeidler M., Zhou Q., Sarda X., Yau C.-P., Chua N.-H. The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 2004;40:355–365. doi: 10.1111/j.1365-313X.2004.02212.x. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Hare P.D., Yang S.W., Zeidler M., Huang L.-F., Chua N.-H. FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 2005;43:356–370. doi: 10.1111/j.1365-313X.2005.02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.