Abstract
The mechanisms underlying rootzone-localized responses to salinity during early stages of barley development remain elusive. In this study, we performed the analyses of multi-root-omes (transcriptomes, metabolomes, and lipidomes) of a domesticated barley cultivar (Clipper) and a landrace (Sahara) that maintain and restrict seedling root growth under salt stress, respectively. Novel generalized linear models were designed to determine differentially expressed genes (DEGs) and abundant metabolites (DAMs) specific to salt treatments, genotypes, or rootzones (meristematic Z1, elongation Z2, and maturation Z3). Based on pathway over-representation of the DEGs and DAMs, phenylpropanoid biosynthesis is the most statistically enriched biological pathway among all salinity responses observed. Together with histological evidence, an intense salt-induced lignin impregnation was found only at stelic cell wall of Clipper Z2, compared with a unique elevation of suberin deposition across Sahara Z2. This suggests two differential salt-induced modulations of apoplastic flow between the genotypes. Based on the global correlation network of the DEGs and DAMs, callose deposition that potentially adjusted symplastic flow in roots was almost independent of salinity in rootzones of Clipper, and was markedly decreased in Sahara. Taken together, we propose two distinctive salt tolerance mechanisms in Clipper (growth-sustaining) and Sahara (salt-shielding), providing important clues for improving crop plasticity to cope with deteriorating global soil salinization.
Key words: barley root, transcriptomics, metabolomics, lipidomics, omics integration, salinity stress
Integrative analyses of the multi-root-omes (transcriptomes, metabolomes, and lipidomes) of barley genotypes demonstrate two differential salt-induced modulations of apoplastic and symplastic flows between their seedling roots. As demonstrated by the localization of cell wall components (lignin, suberin, and callose), this study illustrates two distinctive rootzone-localized growth and tolerance mechanisms in response to salinity during the early stage of barley development.
Introduction
Salinity is one of the major abiotic stresses severely affecting cereal crop yields worldwide. Improving salinity tolerance of one of the most widely cultivated cereals, barley (Hordeum vulgare L.), is essential to increase grain yields on saline agricultural lands. Barley is an essential feed, food, and brewing crop, and a model system for temperate cereals. As a glycophyte, barley suffers substantial yield loss when grown under saline conditions, with roots acting as the first sensors and responders (Glenn et al., 1999). Increased soil salinity exposes the roots to sodium (Na+) and chloride (Cl−) ions, which triggers a cascade of responses leading to differential gene expression, metabolism, and protein activity, as well as altered ion transport pathways, cell wall composition, and root morphology. Differential responses at the level of either cell types or developmental zones are part of a strategy for the root to respond and acclimate to salinity (Dinneny et al., 2008, Sarabia et al., 2018, Sarabia et al., 2020). Although a large number of studies have investigated salinity responses of plants at the physiological and molecular level (Hill et al., 2013, Shelden et al., 2013), relatively little is known about the early rootzone-specific response to salt stress in barley roots. Integrative ‘omics approaches within large-scale experiments, including genomics, transcriptomics, ionomics, proteomics, and metabolomics, can help decipher the interplay of cellular functions at different levels.
In barley, several initial analyses indicate that different developmental zones within the root respond distinctly to salt stress in tolerant and sensitive genotypes. Two barley genotypes, Clipper (a domesticated cultivar) and Sahara (an African landrace) are of particular interest based on previously reported diversity in salt tolerance: with long-term salt exposure, Widodo et al. (2009) found that both Clipper and Sahara showed similar initial reductions in biomass after 3 weeks of 100 mM NaCl exposure when grown hydroponically under controlled conditions. Whereas, after 5 weeks of salinity treatment, Sahara was showing a recovery phenomenon and resumed growth and Clipper continued to show reduced growth relative to control despite containing a sodium exclusion locus (Shi et al., 2010). With shorter-term salt exposure (72 h post-germination of seedlings), Sahara showed the most significant inhibition of root elongation but not of root mass grown on 100 mM NaCl agar medium, whereas Clipper maintained its relative root growth rate (Shelden et al., 2013). Furthermore, barley salinity stress responses were shown to depend on the growth system (hydroponic or soil) (Tavakkoli et al., 2010). These observations suggest that developmental stage and growth environment as well as the timing and length of exposure can change the overall responses of barley to salinity stress. Consistent experimental setups are crucial to achieve reproducible and comparable outcomes in studies of salt stress in barley. In this work, we focus on the contrasting early growth responses to salt stress during the barley seedling development using agar medium, in which Clipper showed sustained seminal root growth, whereas Sahara showed decreased seminal root growth consistent to the previous work (Shelden et al., 2013).
In a subsequent study, two de novo transcriptome assemblies of Clipper and Sahara were constructed and generalized linear models (GLM) were applied to access spatial, treatment-related, and genotype-specific gene responses along the developmental gradient of barley roots (Hill et al., 2016). A gradual transition from transcripts related to sugar-mediated signaling at Z1 to those involved in cell wall metabolism in Z2 was observed. These findings are consistent with transcriptional analyses of salt treatments in other cereal crops and model plants, such as rice (Walia et al., 2005), maize (Zhao et al., 2014), and Arabidopsis thaliana (Hunter et al., 2019), which also show that the transcript levels of many cell wall- or callose deposition-related genes consistently change in response to salt stress. Changes in the chemical composition of cell walls as a result of salt stress are less well documented than the changes in gene expression. Previous studies have shown that salt stress induces changes in the root cell wall composition, including the increased deposition of lignin and suberin in endodermal and exodermal cells, which influence water and ion permeability and transport pathways (Byrt et al., 2018). For example, in two recent studies, (Kreszies et al., 2018b, Kreszies et al., 2020) found that cultivated barley varieties increased suberin levels in roots in response to osmotic stress as an adaptation to prevent water loss, whereas some wild barley varieties used suberization of specific root tissues or specific rootzones, while others showed no changes in suberization and more consistent water uptake rates compared with the barley cultivars.
De novo assembly approach is a powerful tool for studying transcriptomes, particularly when dealing with species with no reference genomes or with limited sequencing coverage. The assembled contigs resulting from the de novo approach, however, are prone to error and inevitably differ from the original transcriptomes (Engström et al., 2013). This had been the case for barley (International Barley Genome Sequencing Consortium, 2012), for which only a draft genome with limited sequencing depth and coverage was available before 2016. In the previous study (Hill et al., 2016), only around 23% of the de novo assemblies of Clipper and Sahara could retrieve positive functional annotations. Also, most of the sequencing reads could not be mapped to the assembled contigs and hence these reads were not taken into account during the subsequent differential gene expression analyses.
To take advantage of the latest version of the barley reference genome (Morex) (Mascher et al., 2017), in this study we built on our previous work (Hill et al., 2016) and re-visited the 12 transcriptomes using an improved bioinformatics pipeline with four major modifications: (i) rather than constructing the de novo assemblies of transcriptomes based only on the raw reads, the newly available Morex genome with increased sequencing depth and genome coverage was served as the mapping base of this study; (ii) instead of using Bowtie v2.1.0, we adopted the HISAT2, a mapping algorithm proven to have improved performance for gapped-read mapping of raw reads (Kim et al., 2015); (iii) for differential gene expression determination, we replaced EdgeR with limma (Ritchie et al., 2015) for better capacity to prevent type I and II errors; and (iv) we performed BLAST search against both databases of protein sequence (such as TAIR, Swissprot, TREMBL) and domain (InterPro) homology to maximize the functional annotations of the gene-of-interest. For this study, we also applied a novel combined targeted metabolomics and lipidomics approach to quantitatively determine the alteration of the corresponding primary metabolites and lipids in different rootzones of Clipper and Sahara with and without salinity treatment, to obtain further molecular insights into the impact of salinity at the metabolite level. We then designed a new GLM-based analysis approach to identify the treatment-, genotype-, and rootzone-specific differentially expressed genes (DEGs) concurrently with the differentially abundant metabolites and lipids (DAMs) in barley rootzones upon salt stress. Integrated pathway over-representation of the DEGs and DAMs showed that the salt treatment led to two differential modulations of phenylpropanoid biosynthesis, which likely contributed to the salinity-induced localization changes of cell wall components, such as lignin and suberin, in Clipper and Sahara. As a proof of concept, we further explored the interconnections between affected metabolites and gene expression pathways by construction of global coexpression-correlation networks specific to each barley genotype. Histochemical and immunochemical microscopy of both Clipper and Sahara roots to detect different depositions of lignin, suberin, and callose after salinity treatment proved the detected cell wall-related gene expression and metabolic changes in the phenylpropanoid pathway. Based on our system-wide exploration, we demonstrate that seedlings of Clipper and Sahara respond to salinity stress differentially, suggesting the distinctive dynamics underpinning the plasticity of different barley genotypes in response to salt stress.
Results
Barley Transcriptome-Processing and -Annotation Pipeline
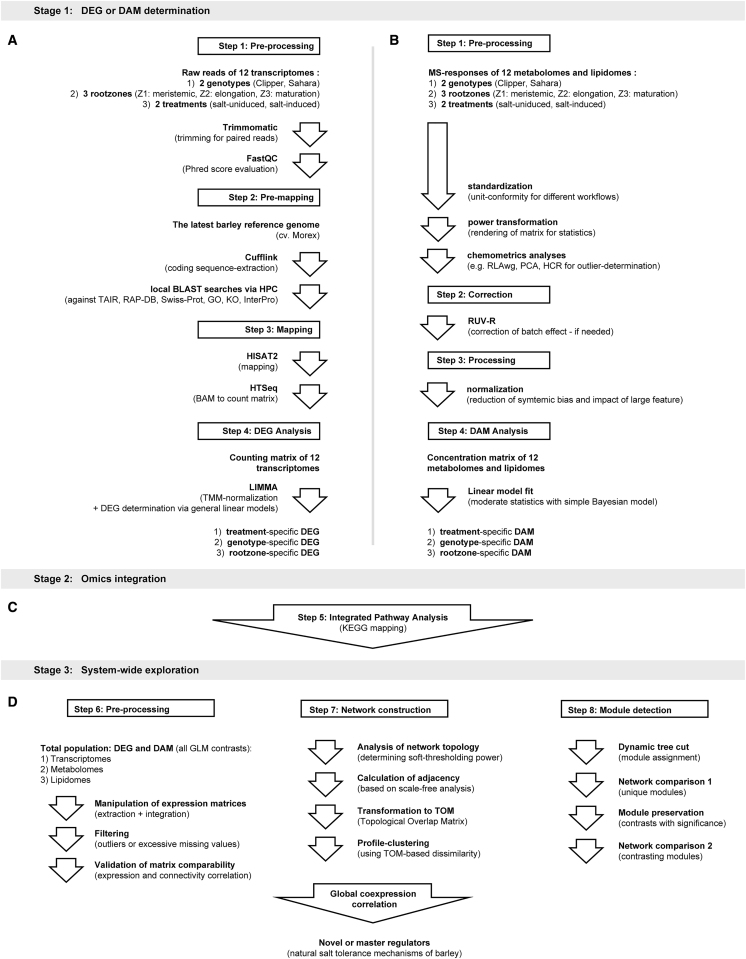
The improved workflow for transcriptome sequence pre-processing, pre-mapping, mapping, and transcript analyses is presented in Figure 1A. We achieved an average mapping efficiency of 95.7% ± 1.6% for the 192 sequenced libraries used in this study (Supplemental Figure 1A), demonstrating a high degree of sequence conservation among Morex, Clipper, and Sahara at the transcript level. In total, 247 281 out of the 333 926 predicted transcripts (74.1%) of the Morex genome were functionally annotated compared with around 37.4% and 40.1% annotation obtained for the de novo assemblies of Clipper and Sahara, respectively (Supplemental Figure 1B). From this, we constructed a new counting matrix composed of the trimmed mean of M values (TMM)-normalized counts per million (CPM) reads for the 12 transcriptomes (Supplemental Data 1).
Figure 1.
Overview of the Bioinformatics Pipelines Implemented in This Study.
(A) Pre-processing, pre-mapping, mapping, and DEG analysis of 12 transcriptomes.
(B) Pre-processing, data-correction, normalization, and DAM analysis of 12 metabolomes and lipidomes.
(C) KEGG-based integrated pathway analysis of transcriptomes, metabolomes, and lipidomes.
(D) Pre-processing, network construction, and module detection for global co-expression correlation analysis of multi-omes in barley.
DAM, differentially abundant metabolite; DEG, differentially expressed genes; GLM, general linear model; GO, Gene Ontology; HCR, hierarchical clustering; HPC, high performance computation; KO, Kyoto Encyclopedia of Genes and Genomes Ontology; MS, Mass Spectrometry; PCA, principal component analysis; RAP-DB, Rice Annotation Project - Database; RLAwg, within-group relative log adjustment; TAIR, The Arabidopsis Information Resource; TMM, trimmed mean normalization; Z1, zone 1 (meristematic zone); Z2, zone 2 (elongation zone); Z3, zone 3 (maturation zone).
Effects of Salinity on Barley Transcriptomes
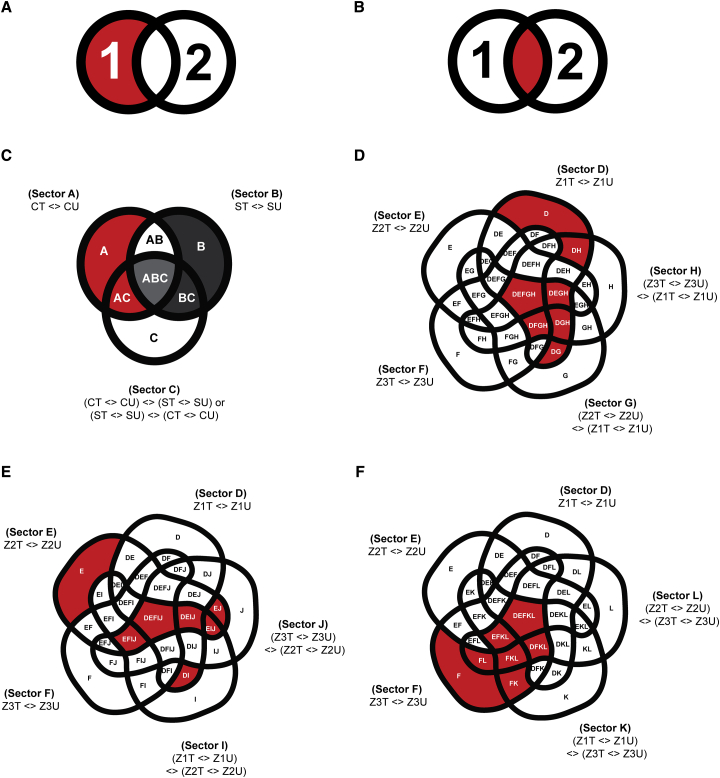
We determined the DEG specific to treatment (0 or 100 mM NaCl), genotype (Clipper or Sahara), and rootzone (meristematic [Z1], elongation [Z2], or maturation [Z3]). Here, specific GLMs taking the interactions among three factors, namely treatments, genotypes, and rootzones into account, were applied to determine genotype- and rootzone-specific DEGs. Notably, for explaining a phenotype specific to either a particular genotype or rootzone, two possibilities exist: differences could either be due to the effect of DEGs unique to a genotype or rootzone (Figure 2A), or of DEGs common to both genotypes and rootzones, but with significant differences in expression (Figure 2B). To this end, both uniqueness and significance of expression differences were addressed through specific GLM designs as described in Supplemental Note 1 with the logic illustrated in Figure 2C–2F. Outcomes of the GLM-based differential analyses was integrated and summarized in Supplemental Figure 2A (see Supplemental Data 2 for annotated DEG lists).
Figure 2.
Design of GLM and Subsetting for the DEG or DAM Determination.
(A and B) Two possible sets of DEGs or DAMs that could account for the genotype- or rootzone-specific phenotypes (colored in red): (A) DEGs or DAMs unique to one genotype or rootzone; (B) DEGs or DAMs common to both genotypes or rootzones, but showed significant contrast in expression or abundance between the two. Numbers 1 and 2 in the figures denote two sets of DEGs or DAMs from two different genotypes/rootzones in comparison.
(C) Genotype-specific DEGs or DAMs for each rootzone. Subsectors correspond to the Clipper-specific DEGs/DAMs (including subsectors A, AC) and Sahara-specific DEGs or DAMs (including subsectors B, BC) in each rootzone are highlighted in red and dark gray, respectively. Subsector ABC are common to both Clipper and Sahara (colored in light gray), but defined by GLM contrast in opposite directions: (CT <> CU) <> (ST <> SU), and (ST <> SU) <> (CT <> CU), respectively.
(D–F) Rootzone-specific DEGs or DAMs of Clipper/Sahara at (D) meristematic zone (Z1), (E) elongation zone (Z2), and (F) maturation zone (Z3), respectively, and with the corresponding subsectors highlighted in red. Symbol “< >” denotes a “contrast/comparison” being tested during differential analysis through fitting of GLM.
CT, salt-treated Clipper; CU, untreated Clipper; DEGs, differentially expressed genes; DAMs, differentially abundant metabolites; ST, salt-treated Sahara; SU, untreated Sahara; Z1T, salt-treated Z1; Z1U, untreated Z1; Z2T, salt-treated Z2; Z2U, untreated Z2; Z3T, salt-treated Z3; Z3U, untreated Z3; <>, contrast of GLM.
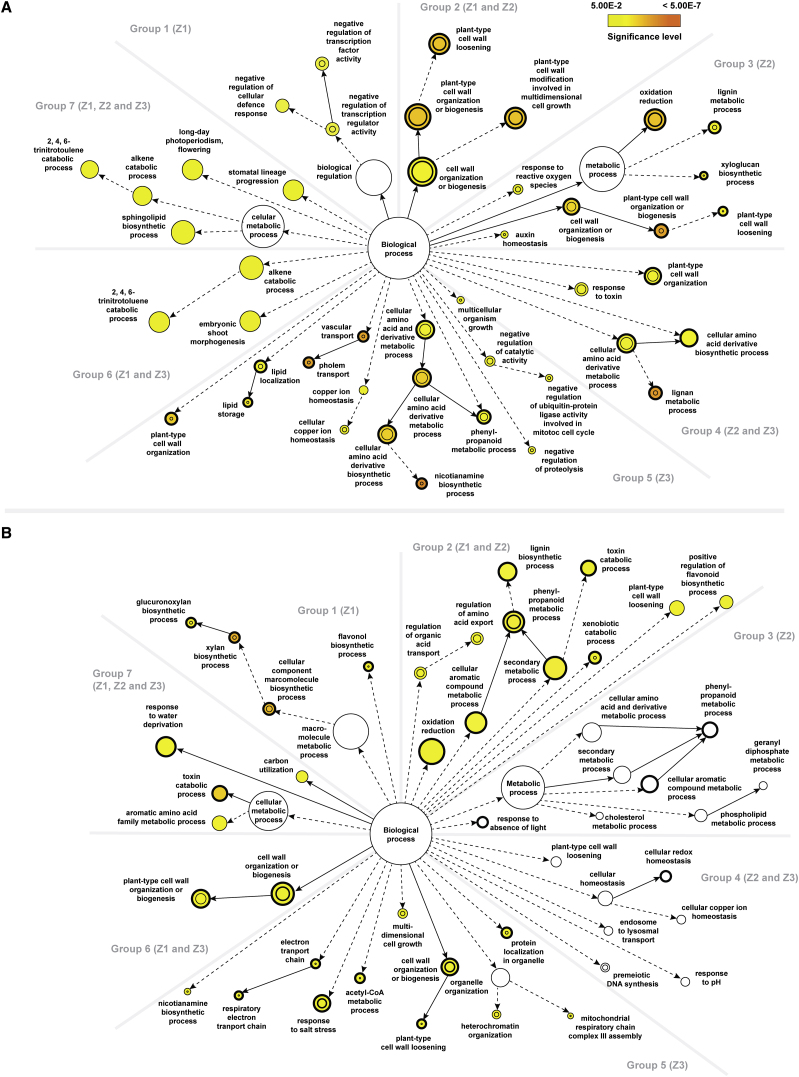
To determine which biological processes are most prominent in the two genotypes upon salt stress, treatment-specific DEGs in each genotype were classified into seven groups according to their spatial distribution in barley roots. Each group was then subject to enrichment analysis of gene ontology (GO) with a focus on the category of biological processes (Figure 3; Supplemental Data 3).
Figure 3.
Comparisons of the Statistical Over-Representation of GO Categories between Different Root Zones of the Two Barley Genotypes upon Salt Stress.
(A) Statistically over-represented GO categories unique to or shared between different rootzones of Clipper.
(B) Statistically over-represented GO categories unique to or shared between different rootzones of Sahara.
Group 1 composed of DEGs found only in Z1. GO analysis via BiNGO (Maere et al., 2005) and REVIGO (Supek et al., 2011) revealed that the most significant over-representation of this group were regulation of transcription and cellular defense response genes in Clipper, and biosynthesis of hemicelluloses, including xylan and its derivatives in Sahara. Group 2 included DEGs found in both Z1 and Z2. GO analysis indicated the genes to be mostly enriched in cell wall modification (in particular cell wall loosening) for Clipper, and phenylpropanoid metabolism for Sahara. Group 3 consisted of DEGs found only in Z2. While plant-type cell wall organization as well as lignin metabolism genes were strongly over-represented in Clipper, no significant enrichment of any GO category could be detected in Sahara. Group 4 represented DEGs found in Z2 and Z3. Lignan metabolism genes and related processes were drastically enriched in Clipper, but similar to Group 3, no significant over-representation was detected in Sahara. Group 5 consists of DEGs found only in Z3. In Clipper, nicotianamine metabolic process as well as vascular transport genes were ranked top in the overrepresentation list, compared with the enrichment of genes encoding proteins targeted to the mitochondrion, response to salt stress, and cell wall organization (xyloglucan metabolism) in Sahara. Group 6 represented DEGs found in both Z1 and Z3. This cluster was enriched in trinitrotoluene catabolism and related processes for Clipper, cell wall organization, or biosynthesis for Sahara. Group 7 contains DEGs found in all three rootzones. Sphingolipid biosynthesis genes were enriched in Clipper, whereas toxin metabolism was the most significantly overrepresented GO category in Sahara. Nodes represent GO categories and node-size is proportional to the number of detected genes for each node. Categories under the same GO hierarchy are linked by interconnected arrows (known as edges) and intensity of node-color indicates the significance level of statistical overrepresentation determined by Fisher's exact test with adjusted p < 0.05 as cutoff as per legend. For reference only, a threshold of 0.2 is set for those sectors showing no significant over-representation and white-colored nodes are used to visualize those ontologies closed to the threshold. Dotted edges indicate one or more hierarchies of GO, which have no statistical significance in the over-representation test and are determined as redundant via REVIGO, were not shown for clarity. DEGs statistically over-represented in both treatment-specific and genotype-specific analyses are denoted by nodes with thickened outlines. DEGs statistically over-represented in both treatment-specific and rootzone-specific batches are denoted by inner circle of nodes. Z1, zone 1 (meristemic zone); Z2, zone 2 (elongation zone); Z3, zone 3 (maturation zone).
Effects of Salinity on the Barley Metabolomes and Lipidomes
Next, we performed quantitative metabolomics and lipidomics analyses using the same root tissue samples to provide a complementary perspective to the early salt responses of barley seedling roots. A total of 154 compounds (22 sugars or sugar alcohols, 15 small organic acids, 32 amines or amino acids, 18 fatty acids, and 67 lipids) were quantified using four mass spectrometry-based metabolomics and lipidomics methods (Supplemental Data 4). The bioinformatics pipeline for elucidating the treatment, genotype, and rootzone-specific DAMs is illustrated in Figure 1B (see Supplemental Data 5 for annotated DAM lists). Notably, the same structure of GLM was used for the DAM and DEG determinations to facilitate the subsequent omics comparisons and integration.
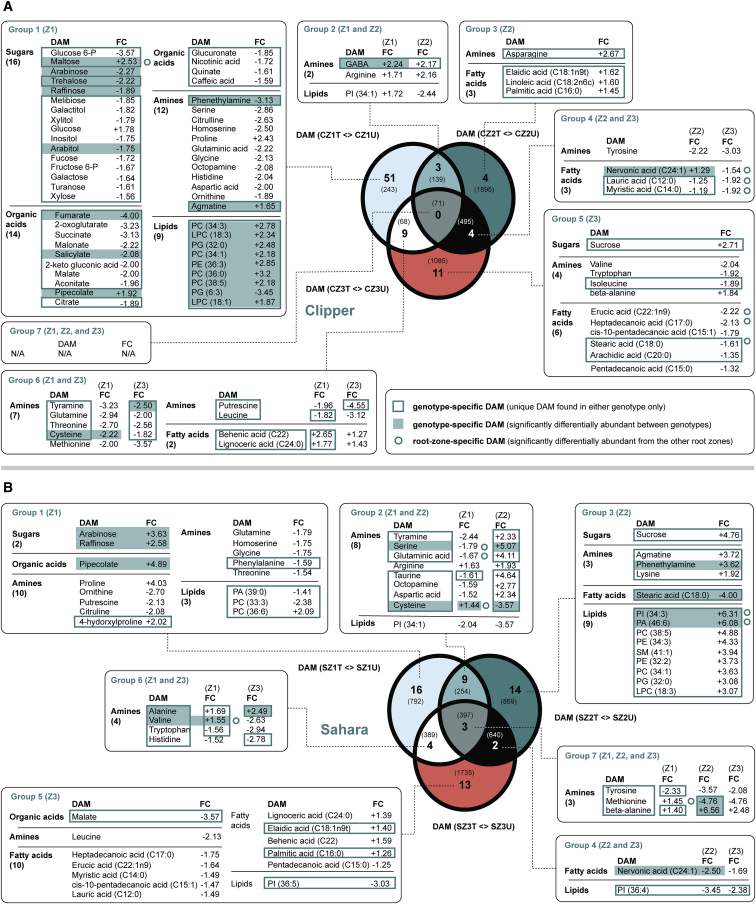
To provide insight as to which metabolic groups are most markedly different between the two genotypes upon salt stress, treatment-specific DAMs in each genotype were classified into seven groups according to their spatial distribution in barley roots (Figure 4). Each group was then subject to metabolite set enrichment analysis with results detailed in Supplemental Data 6.
Figure 4.
An Integrated View of the DAMs of the Two Barley Genotypes in Response to Salt Stress.
(A) DAMs unique to or shared among different rootzones of Clipper.
(B) DAMs unique to or shared among different rootzones of Sahara.
Parentheses in each text box refer to the number of metabolites within each metabolic subgroup. Parentheses in Venn diagrams: corresponding number of DEGs in each sector.
CZ1T, salt-treated Z1 in Clipper; CZ1U, untreated Z1 in Clipper; CZ2T, salt-treated Z2 in Clipper; CZ2U, untreated Z2in Clipper; CZ3T, salt-treated Z3 in Clipper; CZ3U, untreated Z3 in Clipper; DAMs, differentially abundant metabolites; DEGs, differentially expressed genes; FC, fold change; LPC, lysophosphatidylcholines; PA, phosphatidic acids; PC, phosphatidylcholines; PE, phosphatidylethanolamines; PG, phosphatidylglycerols; PI, phosphatidylinositols; PS, phosphatidylserines; SM, sphingomyelins; SZ1T, salt-treated Z1 in Sahara; SZ1U, untreated Z1 in Sahara; SZ2T, salt-treated Z2 in Sahara; SZ2U, untreated Z2 in Sahara; SZ3T, salt-treated Z3 in Sahara; SZ3U, untreated Z3 in Sahara; Z1, zone 1 (meristematic zone); Z2, zone 2 (elongation zone); Z3, zone 3 (maturation zone); -P, phosphate.
Effect of the Over-represented Salinity on the Barley Root-omes
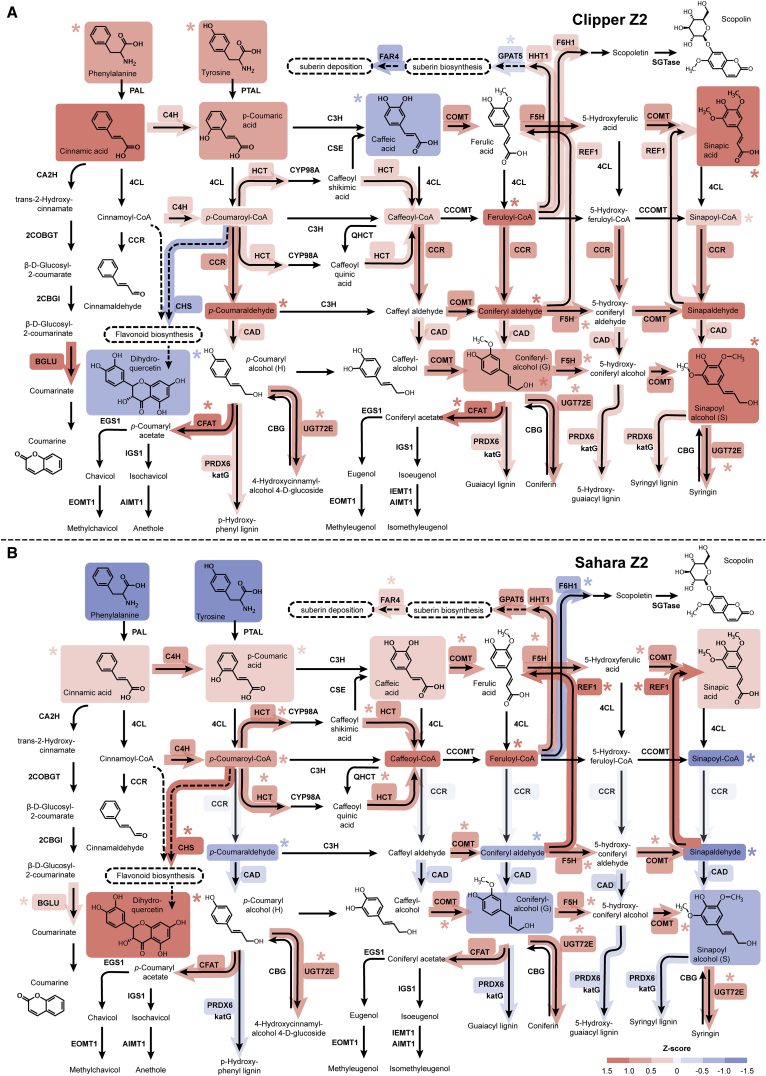
Next, we utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG) mapper to perform an integrated pathway analysis for the three omics datasets (Figure 1C) (Kanehisa et al., 2012). According to the number of matched DEG and DAM hits, biological pathways statistically over-represented at transcript and/or primary metabolite level in response to salinity were ranked in descending order, where the biosynthesis of phenylpropanoids (such as monolignols, flavonoids, lignins, and suberins) were enriched at both levels and identified at the top of the list (Supplemental Data 7). To visualize the specific post-salinity effect on the biosynthesis of phenylpropanoids, we calculated the Z scores of the TMM-normalized CPM for the transcripts involved and of the normalized concentration for the primary metabolites detected. In addition, we adopted an established method to perform a detailed quantification of the phenylpropanoid contents across our root samples (Supplemental Data 2) (Vanholme et al., 2012) and computed their Z scores. The relative abundance of transcripts, primary metabolites, and phenylpropanoids at different rootzones of the two barley genotypes were integrated and illustrated in the pathway frameworks modified based on the corresponding KEGG repository (Figure 5; Supplemental Figures 3 and 4).
Figure 5.
Standardized Abundance of Transcripts and Metabolites Involved in Phenylpropanoid Biosynthesis at the Elongation Zone (Z2) of the Two Barley Genotypes under Salt Stress.
(A) The abundance of transcripts and metabolites involved in the biosynthesis at Clipper Z2.
(B) The abundance of transcripts and metabolites involved in the biosynthesis at Sahara Z2.
Standardized abundances of transcripts and metabolites shown are the Z scores for TMM-normalized CPM and median-normalized concentration respectively. Level of the standardized abundance (i.e., positive, negative, and zero Z score) is indicated by intensity of shading in red, blue, and pale gray, respectively. Asterisks denote statistically significant differentiation of transcript- and metabolite-abundance (with Benjamini–Hochberg adjusted p < 0.05) after the salt stress compared with the untreated control. Standardized abundance of only the transcripts with significant degree of sequence similarities to the characterized homologs (E < 1.00E-3) and the metabolites within the limit of detection of methodologies and instrumentations adopted in this study are shown. Abundance details of these pathway components at different rootzones of the two barley genotypes before and after the salt treatment can be found in Supplemental Figures 5 and 6.
AIMT1, trans-anol O-methyltransferase; BGLU, beta-glucosidase; CA2H, cinnamic acid 2-hydroxylase; CBG, coniferin beta-glucosidase; 2CBGI, 2-coumarate β-D-glucoside isomerase; CAD, cinnamyl-alcohol dehydrogenase; CCOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CFAT, coniferyl alcohol acyltransferase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CHS, chalcone synthase; 4CL, 4-coumarate-CoA ligase; COBGT, 2-coumarate O-beta-glucosyltransferase; COMT, caffeic acid 3-O-methyltransferase; CSE, caffeoylshikimate esterase; CYP98A, coumaroylquinate(coumaroylshikimate) 3′-monooxygenase; EGS1, eugenol synthase; EOMT1, eugenol/chavicol O-methyltransferase; F5H, ferulate-5-hydroxylase; F6H1, feruloyl-CoA ortho-hydroxylase; FAR4, fatty acid reductase 4; GPAT5, glycerol-3-phosphate acyltransferase 5; HCT, shikimate O-hydroxycinnamoyltransferase; HHT1, hydroxyacid O-hydroxycinnamoyltransferase 1; IEMT1, (iso)eugenol O-methyltransferase; IGS1, isoeugenol synthase; katG, catalase-peroxidase; PAL, phenylalanine ammonia-lyase; PTAL, phenylalanine/tyrosine ammonia-lyase; PRDX6, peroxiredoxin 6; REF1, coniferyl-aldehyde dehydrogenase; SGTase, scopoletin glucosyltransferase; QHCT, quinate O-hydroxycinnamoyltransferase; UGT72E, coniferyl alcohol glucosyltransferase.
Salinity-Induced Abundance and Localization Shifts of Phenylpropanoids in the Barley Rootzones
The biosynthetic pathway of phenylpropanoids can be generally divided into three main stages: the general phenylpropanoid pathway from phenylalanine to CoA-esters, the monoligonol-specific pathway from CoA-esters to monolignols, and the lignin-specific pathway from monolignols to oligolignols or lignin polymers. For Z1 of both barley genotypes (Supplemental Figure 3), genes involved in all three stages of the biosynthesis remained weakly expressed as in the untreated controls. In line with the detection at the RNA level, negative standardized log2 concentration (Z scores) were recorded for almost all of the metabolic intermediates (Supplemental Figures 5 and 6). Histochemical staining also showed no observable difference in abundance and localization of phenylpropanoids, such as lignin and suberin after salt treatment (Supplemental Figures 7 and 8), implying that phenylpropanoid production in Z1 was not induced by salt.
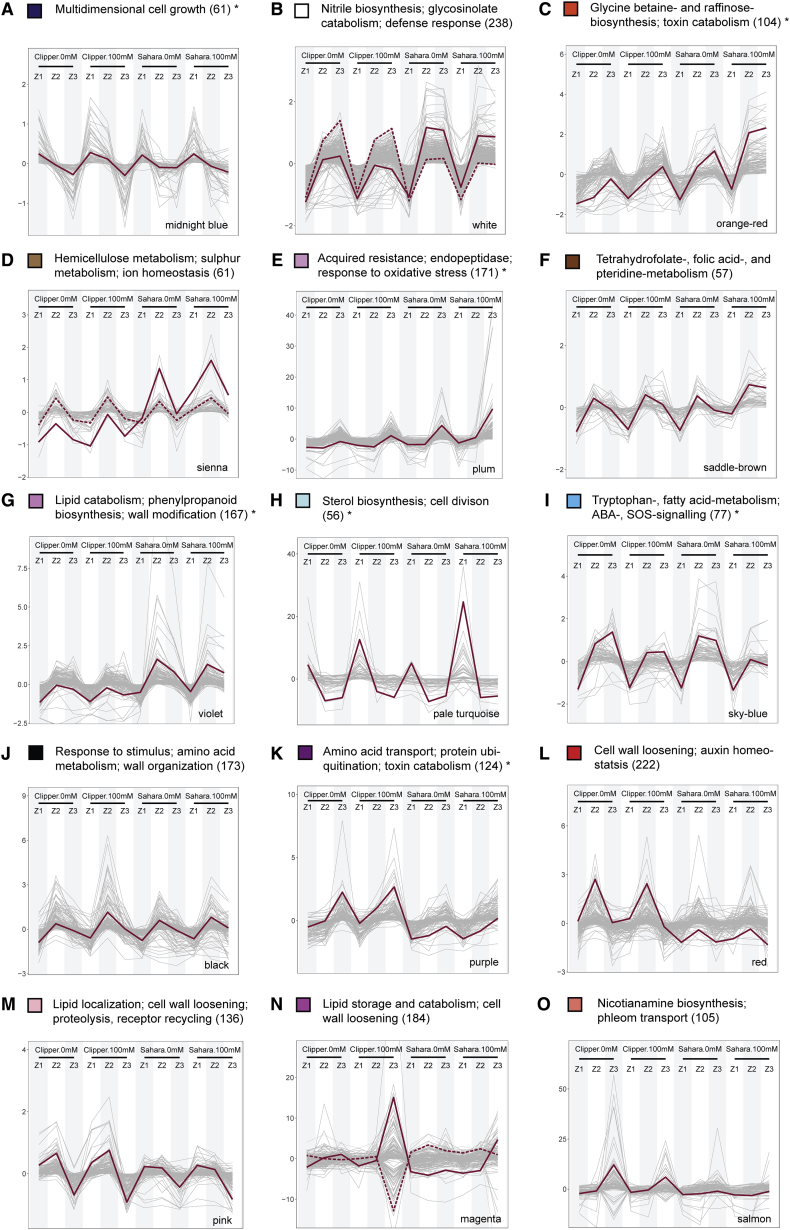
Figure 6.
Selected Modules of Weighted Coexpression Correlation Networks Showing Abundance Profiles of Transcripts and Metabolites.
(A) The abundance profile unique to Clipper.
(B–I) The abundance profiles unique to Sahara.
(J–O) The abundance profiles significantly contrast between the two barley genotypes.
Profiles showing either positive or negative correlations by clustering abundance into differently colored modules through weighted correlation networks. Additional profiles with less obvious differentiation between the two genotypes can be found in Supplemental Figure 11. The color of each module is consistent with Supplemental Figure 10. The most representative trend or centroid of each module represented by solid lines are determined by k-mean clustering (distance method: Pearson) with optimal number of clusters calculated from within-group sum of square method (Madsen and Browning, 2009). Second, the most representative centroid (if any) is indicated by a dotted line. Only expression profiles within 99th percentile are shown for clarity. Annotation of each co-expression clusters are determined by means of the statistical enrichment of GO categories below the cutoff (adjusted p ≤ 0.05) and specific biological role of each module specified here is designated by manual curation of the enrichment outcomes. Asterisks denote the clusters with no significant over-representation and annotations assigned to these clusters are the GO categories with the highest possible level of significance (Supplemental Data 8). Annotated lists of members for each module with significant match (E < 1.00E-4) against TAIR10 genome release (version: June 2016) ranked in descending order according to kME of members can be found in Supplemental Data 9.
In Clipper Z2 (Figure 5A), transcripts encoding for enzymes involved in the phenylpropanoid, monolignol, and lignin biosynthetic pathways were either increased in expression or maintained positive Z scores after salt treatment. The amount of the detected monolignols, including coniferyl alcohols (guaiacyl [G]-units of lignin) and sinapoyl alcohols (syringyl [S]-units of lignin), were significantly induced by salt (Supplemental Figure 6N and 6O). The active production of lignins at this rootzone was further supported by Basic Fuchsin staining, which showed a significant increase in lignin impregnation to cellulosic cell walls localized at the outer stelic regions (including endodermis, pericycle, and xylem) of Clipper roots after salt treatment (Supplemental Figure 7E, 7F, 7M, and 7N). Furthermore, gene products of CHALCONE SYNTHASE (CHS) are known to divert intermediates of the general phenylpropanoid pathway for flavonoid production (Heller and Hahlbrock, 1980). Weak expression of CHS and low levels of flavonoids, such as dihydroquercetin, were consistently detected in Clipper Z2. Notably, transcription of GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 5 (GPAT5) and FATTY ACID REDUCTASE 4 (FAR4), involved in biosynthesis and deposition of root suberin (Beisson et al., 2007, Domergue et al., 2010), were suppressed by the stress and maintained a negative Z score, respectively (Supplemental Figure 5I and 5J). These data are consistent with the observations of the salinity-induced decline in suberin levels visualized by Fluorol Yellow stain throughout Z2 (Supplemental Figure 8B and 8H).
For Sahara Z2 (Figure 5B), a significant increase of CHS expression diverted most phenylpropanoids toward the accumulation of dihydroquercetin (Supplemental Figure 6P). Together with the low expressions of CINNAMOYL-COA REDUCTASE (CCR) and CINNAMYL-ALCOHOL DEHYDROGENASE (CAD), the accumulation of monolignols and their precursors was restricted and no observable increase of lignin levels in the endodermal region could be detected histochemically after salt stress (Supplemental Figure 7G, 7H, 7O, and 7P). Furthermore, in contrast to Clipper Z2, there was higher abundance of HYDROXYACID O-HYDROXYCINNAMOYLTRANSFERASE 1 (HHT1), GPAT5, and FAR4 transcripts in Sahara Z2 (Supplemental Figure 5H–5J). The active biosynthesis of suberin inferred from the levels of biosynthetic enzyme transcripts was histologically confirmed, as increased levels of suberin were observed in the epidermis and across the subepidermal region of the rootzone (Supplemental Figure 8N and 8T).
In Z3 of Clipper (Supplemental Figure 4A), as in Z2, flavonoid production was inhibited. Salinity-induced accumulation of lignins was limited to G-units and localized at the endodermal and vascular regions (Supplemental Figure 7A and 7B). But, in contrast to Clipper Z2, an increased biosynthesis and deposition of suberin at the endodermal and stele regions was supported by the consistently higher abundance of GPAT5, and FAR4 transcripts (Supplemental Figure 5I and 5J) and by histochemical staining (Supplemental Figure 8A and 8G), respectively.
For Z3 of Sahara (Supplemental Figure 4B), relatively higher transcript abundance of CCR and CAD compared with Z2 upon salt treatment was detected. Intriguingly, the resulting metabolic changes led to higher accumulation of G-units of lignin and intense deposition of lignin mostly in the xylem vessels of this rootzone (Supplemental Figure 7C and 7D). Also, in line with the increased Z score for CHS and the suberin-related transcripts (such as HHT1, GPAT5, and FAR4) compared with Clipper Z3 (Supplemental Figure 5B and 5H–5J), a significant increase of dihydroquercetin and of endodermal and stele suberin deposition was recorded at Sahara Z3, respectively (Supplemental Figures 6P, 8M, and 8S). Taken together, the omics datasets at the transcriptional and metabolic levels combined with the histological observations suggest a strong differentiation in biosynthesis and localization of the phenylpropanoids between the two barley genotypes upon salt stress.
Global Intercorrelations of Salt Stress on the Barley Root-omes
Next, we extracted the abundance matrices of transcripts and metabolites that were significantly different in at least one of the GLM-based DEG or DAM determinations in Clipper (3802 transcripts, 83 metabolites) and Sahara (6477 transcripts, 61 metabolites). Global co-expression-correlation networks specific to the two barley genotypes were constructed via WGCNA (Langfelder and Horvath, 2008) to illustrate the system-wide consequences induced by salinity stress (Figure 1D). In these networks, each "leaf" or short vertical line represents an abundance profile of one transcript, metabolite, or lipid. Any interconnected lines within the same "branch" indicate profiles with highly correlated pattern of abundance. Based on the “guilt-by-association” principle, as defined in Saito et al. (2008), co-regregulated genes and metabolites among each co-expression cluster or branch are likely to have common functional roles. To systematically define the branch, we applied dynamic tree cut (Langfelder et al., 2008) to each network and the module assignment was performed to color code each highly correlated cluster (aka module) (Supplemental Figure 10).
For modules that were unique to either Clipper or Sahara, or common to both genotypes but with significantly contrast abundance patterns in response to salt stress (Supplemental Table 1), we generated parallel profiles to visualize their variations of abundance in response to salt stress (Figure 6 and Supplemental Figure 11). Co-expression clusters were annotated by statistically significant enrichment (adjusted p ≤ 0.05) of GO categories, and their specific biological roles were assigned through manual curation of the enrichment outcomes (Supplemental Data 8). Notably, the module eigengene (ME) corresponds to the first principal component of each module, and module membership (kME), a measure of the ME-based intramodular connectivity, is calculated by correlating the abundance profiles of modular members to their ME (Langfelder and Horvath, 2008). Providing that the importance of each regulator for a particular functional role is determined by its degree of contribution to the module variance and by its connection strength with the other intramodular members, ranking of members according to their kME in each module (Supplemental Data 9) can shed light on the key or master regulator(s) for a given biological role. Each cluster is categorized, explored, and discussed in detail in Supplemental Note 2. Biological processes in each rootzone and genotype, with module members being either induced or maintained at a high abundance level after the salt treatment, are summarized in Supplemental Table 2.
To validate the credibility of the global networks constructed for plants, we put one salt-induced biological process identified in Sahara, suppression of callose deposition, to the test and verified the callose abundance at four different tissue layers (focusing on epidermis, cortex, endodermis, and stele) using an immunochemical approach (Supplemental Figure 9). In contrast to the comparable amount of callose deposited in all layers of the three rootzones of Clipper after salt treatment (Supplemental Figure 9A–9F), as deduced from the global analysis, we detected declines of callose deposition throughout the layers underneath the epidermis of Sahara in all rootzones. Such declines (as indicated by the fading of orange fluorescence) were especially apparent at the plasmodesmata of cortical cell in Z3, plasmodesmata in stele and endodermis of Z2, and throughout the walls of stele and cortical cells in Z1) (Supplemental Figure 9G–9L). Furthermore, ABERRANT GROWTH AND DEATH 2 (AGD2) is a known suppressor of callose deposition (Rate and Greenberg, 2001). In line with the outcome of the immunochemical detection, we consistently showed that the expression of AGD2 was categorized in Module C of the correlation network, which is characterized by modular members with stronger salinity-induced abundance for all rootzones in Sahara than in Clipper (Figure 6C). Altogether, these findings support the precision and feasibility to apply this intercorrelation approach to understand the salinity responses in barley roots.
Discussion
The barley malting cultivar Clipper and landrace Sahara are two barley genotypes with known contrasting phenotypic traits in response to salt stress at an early stage of development: Clipper maintains root elongation, while in Sahara root elongation is significantly reduced in response to short-term salt stress (Shelden et al., 2013). In this study, we investigated system-wide responses of seedling roots of both genotypes to moderate salinity, and quantified spatial salt-induced perturbations in transcriptomes, metabolomes, and lipidomes of individual rootzones in each of the barley genotypes. By means of statistical over-representation of DEGs and DAMs (Figures 3 and 4), we investigated the datasets from the perspective of their "extremes" and illustrated the most differential salinity responses in three different rootzones of the two genotypes through integrated pathway analysis (Figure 5; Supplemental Figures 3 and 4). Using global co-expression correlation network analysis (Figures 6 and 7), we approached the datasets from the perspective of "intercorrelations" among the induced pathways to demonstrate the system-wide impacts on the genotypes triggered by salinity stress (Supplemental Table 2).
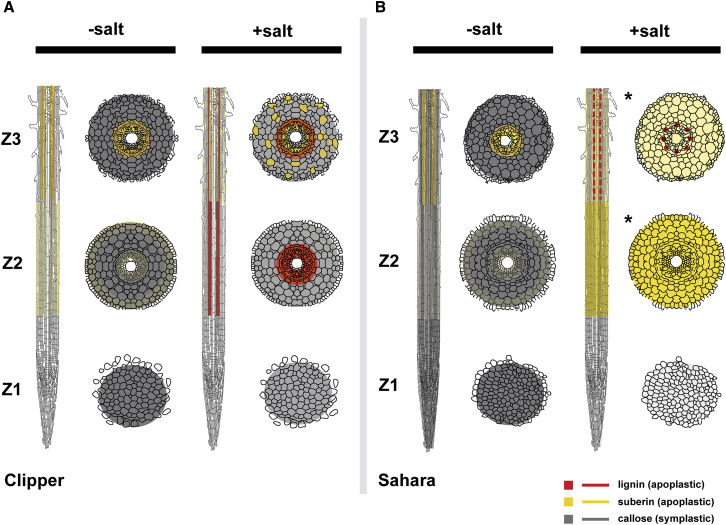
Figure 7.
A Model Deciphering Cell Wall Modifications in Roots of Clipper and Sahara in Presence or Absence of Salinity Stress.
(A and B) Longitudinal and the corresponding transverse sections of different rootzones for (A) Clipper and (B) Sahara with or without salinity stress.
Color intensities represent the relative levels of lignin, suberin, or callose, which were quantified based on their precursors detection through LC/GC-MS and/or RNA-seq with support of the direct detection of the compounds through histological methods. Localization of each compound was determined by the histo-/immuno-chemical stainings with proven specificity and optical filters applied for minimizing any autofluorescence or background signals. Red broken line (Sahara Z3, +salt) represents lignin deposition at only vasculature. Asterisks indicate rootzones at cortical, endodermal, and stelic regions with concurrent localizations of suberin and callose (where callose deposition was suppressed after the salt stress and only the intense suberin deposition is shown in this rootzone for clarity).
Through integration of the spatial omics information obtained from these approaches, we provide a novel and system-wide insight to the salt-induced modulations of apoplastic (lignin, suberin) and symplastic flows (callose) in barley roots (Figure 7). Besides providing a comprehensive multi-omics data resource allowing deep mining of salinity-induced changes in seedlings of barley at the rootzone level, we demonstrated seedling roots of different genotypes of barley could be in distinctive salinity response phases to cope with the stress, illustrating differential salt tolerance strategies could exist among the same plant species.
Salinity-Induced Lignin Precursor Production to Sustain Clipper Root Growth
Through modification and amplification of a very limited set of core structures derived from shikimate, phenylpropanoid metabolism generates an enormous array of plant secondary metabolites ranging from monomers (such as flavonoids, coumarins) to polymers (such as lignins, suberins) (Vogt, 2010). Upon short-term salt stress, our study shows that the building blocks of phenylpropanoids were diverted from the synthesis of flavonoids and suberins to the production of G- and S-units of lignins in Clipper Z2 (Figure 5A). Flavonoids, such as quercetin, are known as inhibitors of auxin efflux carriers in a variety of plant tissues (Jacobs and Rubery, 1988). Low levels of dihydroquercetin (Supplemental Figure 6P), an immediate upstream precursor of quercetin, may therefore, supress the inhibition of auxin efflux carriers in Clipper Z2 and facilitate the propagation of auxin signals, and auxin-mediated cell division and expansion. This finding is consistent with the previous phenotypic study of the two barley genotypes, in which Clipper maintains a greater root elongation rate than Sahara, even under moderate salt stress (Shelden et al., 2013). This also further validates our integrated pathway analysis approach to identify their molecular differences.
The Casparian strip is a specialized wall modification in the endodermis, which serves as a diffusion barrier to limit apoplastic flow and re-direct solute movement back to the symplastic stream through the plasma membrane (Steudle and Peterson, 1998). The strip is mainly composed of rings of lignin deposited around endodermal cells and interference in lignin biosynthesis has been shown to abrogate the early strip formation in Arabidopsis (Naseer et al., 2012). In monocotyledonous species, the lignin-like polymers of the Casparian strip are composed of a mixture of G- and S-units (Zeier et al., 1999). In Clipper Z2, both transcript and phenylpropanoid profiling results consistently show that the production and abundance of these units are significantly increased by the stress (Figure 5A; Supplemental Figure 6N and 6O). Also, an intense localization of lignins was detected at the outer stele region of the rootzone after the salt treatment (Supplemental Figure 7E and 7F). Similar to previous results in maize (Zea mays L.) (Shen et al., 2015), the salt-induced lignin production and its intense localization at the endodermis of Clipper Z2 likely contributed to the development of the Casparian strip closer to the root tip in response to the salt stress. There, passage of water and solutes have to undergo selective uptake via ion channels of the membranes (Apse and Blumwald, 2007). Filtering of excessive sodium ions might therefore be achieved in Clipper Z2 by this mechanism.
For most cereal crops and Arabidopsis thaliana, deposition of suberin can be induced at cell layers, such as the epidermis, outer cortex, and stele in response to salt and osmotic stresses (Schreiber et al., 2005, Kreszies et al., 2018a, Kreszies et al., 2018b, Kreszies et al., 2020). Intriguingly, our transcriptomic data shows that genes involved in suberin production were downregulated in Clipper Z2 relative to Sahara Z2 (Supplemental Figure 5H–5J). Reduced Fluorol Yellow staining in Clipper Z2 verifies this result and shows a decline in suberin levels throughout the whole rootzone after salinity treatment (Supplemental Figure 8B and 8H). The exodermis is a specialized outermost layer of the cortex in which Casparian strip development is inducible by salt and is found only in wild relatives of barley, such as Hordeum marinum (Byrt et al., 2018). In the absence of the exodermis in Hordeum vulgare L. genotypes, such as Clipper and Sahara, low suberization of cell layers surrounding the endodermis of Clipper Z2 would therefore imply that there is no additional barrier to sodium ion entry into its root epidermis and cortex under salt stress. Consistent with this hypothesis, whole seminal roots of Clipper were shown to have higher accumulation of sodium ions than Sahara when grown under the same salinity strength (Shelden et al., 2013).
Plasmodesmatal conductivity is known to be regulated by the controlled buildup of callose at the plasmodesmatal neck (De Storme and Geelen, 2014). In our study, immunochemical detection showed substantial callose deposition throughout Clipper Z2 independent of the salt stress (Supplemental Figure 9B and 9E). Assuming our observed callose deposition contributed to modulating the aperture size of the symplastic channels, this may suggest a persistent restriction of symplastic flow and hence accumulation of salt in cell walls of the epidermal and cortical regions. Indeed, the interwoven network of the cellulose microfibrils and pectin (such as homogalacturonan, rhamnogalacturonan I and II) is one of the major factors contributing to the cell wall strength with homogalacturonan chain interaction modulated by calcium ions (O'Neill et al., 2004). While barley root cell walls have also been suggested to be a "sodium ion trap" for restricting ionic movement from the root to the shoot (Flowers and Hajibagheri, 2001), the salt-tolerant varieties have shown to possess up to two-fold greater capacity of ion adsorption than sensitive ones, suggesting that the excessive amounts of sodium ions in the apoplast might displace calcium ions and thus weaken pectin chain calcium ion cross bridges (Ravanat and Rinaudo, 1980). To the best of our knowledge, there is no compelling evidence to support the presence of an active exclusion mechanism for the removal of an excess of sodium ions from the apoplast of Clipper. Assuming the root cell wall was under an optimal pH required for the interaction of sodium ions and uronic acids of pectin, presence of such a high level of apoplastic sodium ions would in turn weaken the cell wall strength of roots of Clipper, implying a shortcoming of this tolerance strategy for supporting the long-term development of this genotype.
Notably, production of lignin G-units was detected in both Z2 and Z3 of Clipper, but S-unit precursors of lignin were found only in Z2, not in Z3 under salt stress (Supplemental Figure 6N and 6O). Lignin G-units are known to be a major component of tracheary elements (Higuchi, 1990), which are the key components of xylem vessels that provide mechanical resistance in plants against the negative pressure associated with the transport of minerals and water to the aerial tissues in the rising sap (Turner, 1997). The continuance of synthesis of these units in Clipper Z3 is likely to reinforce and waterproof these cells. This suggests that the vital function for preventing the root structures from collapse and maintaining the hydro-mineral sap distribution to the whole plant served by the tracheary elements likely be independent of the salt stress. Taken together, roots of Clipper seedlings could adopt a "growth-sustaining" strategy, which maximizes root growth to increase the likelihood of overcoming the unfavorable saline conditions, but with the trade-off of developing a less effective epidermal or cortical barrier with suberin for preventing the subsequent salt accumulation in the root cortex (Figure 7A).
Salinity-Induced Flavonoids and Suberin Production to Shield Roots of Sahara
Unlike in Clipper Z2, our integrated pathway analysis also suggests that lignin production in Z2 of Sahara was not triggered by salinity. Instead, the building blocks of phenylpropanoids in this rootzone were in part diverted to suberin, and in part to the production of flavonoids, implying suppression of root cell elongation (Figure 5B).
Under normal growth conditions, suberization of root cells occurs in the endodermis subsequent to Casparian strip formation (Geldner, 2013). These wall modifications restrict the apoplastic uptake of water and solutes into endodermal cells. Under osmotic stress, increased numbers of suberized endodermal cells were observed at the late elongation zone of barley roots (Kreszies et al., 2018b). Under salinity stress, cereal crops, such as maize would, however, further expand their apoplastic diffusion barrier by inducing the suberization of cell walls in the entire root cortex to limit water loss from the cell layers and salt entry into xylem vessels (Andersen et al., 2015). In this study, we detected high levels of suberin synthesis-related gene expression and localization of suberin throughout Z2 of Sahara, but not of Clipper (Supplemental Figures 5H–5J, 8N, and 8T). This suggests that Sahara responds similarly as other cereal crops under salt stress by restricting apoplastic transport in Z2 via suberin deposition. Consistently, our global co-expression correlation study indicates high level of salt-induced AGD2 transcripts, a factor known for supressing callose deposition (Rate and Greenberg, 2001) (Supplemental Table 2: Sahara, AZ). Such inhibition across the subepidermal regions of Sahara Z2 (especially at the stele and endodermal regions) was confirmed by the immunochemical detection (Supplemental Figure 9H and 9K). Callose deposition is known to be crucial for regulating the closure of plasmodesmata (De Storme and Geelen, 2014). In the heavily suberized and cortical cells of Sahara Z2 with restricted apoplastic movement of nutrients taken up from the rhizosphere, inhibition of the callose deposition at plasmodesmata thus reduces the symplastic transport barrier allowing sharing and distribution of resources via the symplastic passages.
Furthermore, irrespective of the salt treatment, production of suberin (Supplemental Figure 5H–5J) and G-units of lignins (Supplemental Figure 6N) persisted in Sahara Z3, inferring the vital importance of these precursors in the maturation of the Casparian strip and tracheary elements, respectively. Notably, unlike the untreated control, Basic Fuchsin staining of Sahara Z3 showed an intense deposit of lignin around the meta- and proto-xylemic cell walls, accompanied by a small amount of lignins laid at the endodermis and pericycle after salt treatment (Supplemental Figure 7C and 7D). In the absence of the widespread salt-induced suberization of cells in epidermal and cortical layers observed in Sahara Z2 after salt stress (Supplemental Figure 8T and 8S), this lignin deposition at the stele of Sahara Z3 could serve as the last barrier of salt ions carried by apoplastic flows. Furthermore, similar to the response of the Sahara Z2, a boost in production of flavonoids was also observed in Sahara Z3 after the salt treatment (Supplemental Figure 6P). This implies that a comprehensive salt- or osmotic-induced growth restriction was triggered in both the zones of elongation (Z2) and maturation (Z3), which is consistent with the previous physiological data (Shelden et al., 2013). Taken together, seedling roots of Sahara appear to implement a "salt-shielding" strategy. Such a strategy restricts salt from being imported into the roots and minimizes water loss from root cells under the unfavorable salinity conditions, but at the expense of the rate of growth (Figure 7B).
Understanding the distinctive salt tolerance mechanisms adopted by seedling roots of different barley genotypes may help in designing plants to cope with the predicted increase in salinity stress, which will impact our ability to maintain yield in important food and feed crops in the near future.
Methods
Plant Materials
Genotypes of barley (Hordeum vulgare L.) were sourced from the University of Adelaide. Two genotypes of barley, the domesticated malting cultivar Clipper (Australia) and the landrace Sahara 3771 (North Africa), were used for the transcript, primary metabolite, lipid, and phenylpropanoid analyses in this study, and were selected based on previously reported physiological diversity in salt tolerance (Widodo et al., 2009, Shelden et al., 2013).
Growth Conditions and Sample Preparation
For study of the transcriptomes (RNA sequencing [RNA-seq]), we re-analyzed the raw sequencing reads of four biological replicates for each sample totaling 48 samples obtained from the previous study (Hill et al., 2016). No additional sample collection or extraction was performed. For study of the primary metabolomes and lipidomes gas chromatography-triple quadruple-mass spectrometry [GC-QqQ-MS] for sugar and organic acid quantification, liquid chromatography-triple-quadrupole-mass spectrometry [LC-QqQ-MS] for amine quantification, GC-quadruple-MS [GC-Q-MS] for fatty acid methyl ester quantification, and LC-QqQ-MS for lipid analysis), we made use of the exact same set of root material obtained from our large-scale root collection for the transcriptomics studies to side-by-side extract the corresponding metabolomes and lipidomes from three out of the four replicates totaling 36 samples. For detection of the phenylpropanoids, three biological replicates were prepared for each sample in three independent experiments with a total of 36 samples. All dissected seminal roots were collected into pre-chilled 1.5-ml tubes, immediately snap-frozen in liquid nitrogen, weighed, and then stored at –80°C until extraction of RNA, primary metabolites, lipids, and phenylpropanoids.
RNA Isolation and Sequencing
RNA isolation and sequencing were described previously (Hill et al., 2016). In short, the total RNA was extracted from 50 mg root tissue separately per genotype, treatment, and rootzone using the QIAGEN RNeasy kit following the manufacturer's protocol. All RNA-seq libraries were constructed and paired-end sequenced (100-bp) on an Illumina HiSeq 2000 system at the Australian Genome Research Facility (Melbourne, Australia). Four lanes were used for each genotype, and all 48 samples were run on a single flow cell. The RNA was sequenced to a depth of approximately 31 million read-pairs per sample per lane, giving a total of 1.48 billion reads (749 million read-pairs).
Metabolite and Lipid Quantification
Metabolites (sugars, organic acids) were quantified as described in (Dias et al., 2015). Amines and amino acids were quantified as described in (Boughton et al., 2011). Fatty acids were quantified as described in (Eder, 1995). Lipids were quantified as described in (Natera et al., 2016). Phenylpropanoids were extracted from three biological replicates of root tissues (10 mg) per genotype, treatment, and rootzone from exactly the same growth settings, using 500 μl of cold methanol each. After homogenization by CryoMill (Bertin Technologies), samples were agitated for 15 min at 70°C and 10 000 rpm in a thermoshaker (Eppendorf), then allowed to cool down the extract before being centrifuged for 5 min at room temperature at 14 000 rpm. Supernatant was transferred into a clean Eppendorf tube for further clean-up process using solid-phase extraction (SPE) cartridges. For the SPE clean-up process, 60 mg Agilent Bond Elut Plexa cartridges were conditioned using 1 ml of methanol, followed by 1 ml of water. The supernatant from root extracts was loaded and washed by passing 1 ml of methanol, then metabolites were eluted using 400 μl of methanol, followed by 400 μl of 5% formic acid in methanol. Combined elute was dried down in a speed vacuum and reconstituted in 100 μl of 50% methanol:water before LC/MS analysis.
Phenylpropanoids were analyzed using an Agilent 6490 triple quadrupole mass spectrometer coupled to an ultra-high-performance liquid chromatograph (LC-QqQ-MS) (Santa Clara, CA). An Agilent Luna C18 column (2.1 × 150 mm, 3 μm) was used for compound separation. The mobile phase composition included (A) 10 mM ammonium acetate in methanol/water/acetonitrile (10/85/5, v/v/v) and (B) 10 mM ammonium acetate in methanol/water/acetonitrile (85/10/5, v/v/v) with a gradient elution: 0–10 min, 45% A; 10–20 min, 55%–100% B; 20–22 min, 100% B; 22–25 min, 55% B to equilibrate the column to initial conditions. The flow rate of the mobile phase was maintained at 0.2 ml min−1 and the column temperature was maintained at 50°C. The needle wash was 20% (v/v) acetonitrile in water with sample injection volume of 5 μl. Analysis was performed using Agilent MassHunter acquisition software, version 7. Compounds were quantified based on calibration curves prepared using authentic standards.
MS detection was performed using an electrospray ionization source operated in positive ion mode. The source parameters were set as: capillary voltage 4.0 kV; iFunnel high pressure RF in positive and negative mode at 130V; low pressure RF in positive and negative mode at 60V; source temperature 200°C; sheath gas temperature 400°C; gas flow 12 l min−1; sheath gas flow 12 l min−1; fragmentor voltage 380 V; and cell accelerator 5V. Data were collected using in-house multiple reaction monitoring developed based on individual standards. Dwell time for each compound was set as 10 ms and data were quantified using MassHunter Quant software version 7.
Histochemical and Immunochemical Microscopy
Roots of Clipper and Sahara, grown on agar medium supplemented with either 0 or 100 mM NaCl for 3 days, were fixed in 4% paraformaldehyde overnight at 4°C and then washed in phosphate-buffered saline. For lignin and suberin staining, the roots were embedded in 6% agar followed by sectioning of 80-μm-thick sections using a VT1000 S vibratome (Leica Microsystems). Sections for lignin staining were cleared using Clearsee (Kurihara et al., 2015) and stained using 0.2% (w/v) Basic Fuchsin and 0.1% (w/v) Calcofluor White (general cell wall stain) (Ursache et al., 2018). Vibratome sections for suberin staining were placed in 0.01% (w/v) Fluorol Yellow 088 (Santa Cruz Biotechnology) in polyethylene glycol 200 for 1 h at 90°C (Brundrett et al., 1991) followed by counterstaining with 0.5% aniline blue for 30 min. Roots for callose labeling were dehydrated in an ethanol series followed by infiltration and embedding in London White Resin (ProSciTech). Sections (1 μm) were cut using an Ultracut S Ultramicrotome (Leica Microsystems) and labeled with the primary (1:3)-β-glucan antibody (Biosupplies Australia) (Meikle et al., 1991) at a concentration of 1:300 (Wilson et al., 2015) followed by the secondary anti-mouse 568 Alexa Fluor antibody (Thermo Fisher) at a 1:200 dilution.
A Nikon C2 confocal microscope (Coherent Scientific, Australia) equipped with a spectral detector was used to image the cell wall fluorescence using the following settings: Basic Fuchsin, ex 561 nm, em 600–650 nm; Calcofluor White, ex 405 nm, em 425–475 nm; anti-mouse Alexa Fluor 568 antibody, ex 561 nm, em 570–650 nm. Fluorol Yellow staining was imaged using a Leica DM6000 microscope equipped with a Leica DFC450 camera using the I3 (GFP/FITC) filter. Images were analyzed using FIJI (NIH).
Funding
This work was supported by funding from the Australian Research Council (Future Fellowship: FT130100326) and the University of Melbourne.
Author Contributions
C.B.H., M.S.D., M.C.S., A.B., and U.R. designed the experimental part of the research. C.B.H. performed the salinity experiment, collected and extracted samples, processed data from mass spectrometry, and performed initial metabolite data analyses. T.R. carried out the phenylpropanoid detection and processing of mass spectrometry data. A.V.D.M. performed histological work on barley root sections. W.W.H.H. designed and implemented the bioinformatics part of the research, and performed all subsequent statistical and computational analyses, including functional annotation of barley genome with high-performance computing, GLM-based differential analyses, and omics data integration via integrated pathway analysis and global expression correlation networks. W.W.H.H. and U.R. interpreted the data and wrote the article. All authors revised, edited and approved the manuscript.
Acknowledgments
The authors thank Ms Nirupama Jayasinghe (Metabolomics Australia, School of BioSciences, University of Melbourne) for providing excellent technical assistance for GC-QqQ-MS and LC-QqQ-MS analyses, Dr Daniel Sarabia and Mrs Cheong Bo Eng (School of BioSciences, University of Melbourne) for root sample collection for phenylpropanoid detection, as well as technical staff at the Australian Genome Research Facility (AGRF, Melbourne) for next-generation sequencing. The authors are grateful to Melbourne Bioinformatics (www.melbournebioinformatics.org.au) for access to the HPC and data storage facilities, and to the Victorian Node of Metabolomics Australia, which is funded through Bioplatforms Australia Pty. Ltd., a National Collaborative Research Infrastructure Strategy (NCRIS), 5.1 Biomolecular platforms and informatics investment, and co-investment from the Victorian State Government and The University of Melbourne. We would also like to acknowledge the provision of facilities from the BioSciences Microscopy Unit (BMU-05) and Ms Ewa Anna Reda for her pilot experiments on root sectioning and lignin staining. This research was supported by use of the Nectar Research Cloud, a collaborative Australian research platform supported by the Australian Government through the National Collaborative Research Infrastructure Strategy (∗NCRIS). U.R. is grateful to the Australian Research Council for funding this work through a Future Fellowship award.
Published: February 13, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Andersen T.G., Barberon M., Geldner N. Suberization––the second life of an endodermal cell. Curr. Opin. Plant Biol. 2015;28:9–15. doi: 10.1016/j.pbi.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Apse M.P., Blumwald E. Na+ transport in plants. FEBS Lett. 2007;581:2247–2254. doi: 10.1016/j.febslet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Beisson F., Li Y., Bonaventure G., Pollard M., Ohlrogge J.B. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton B.A., Callahan D.L., Silva C., Bowne J., Nahid A., Rupasinghe T., Tull D.L., McConville M.J., Bacic A., Roessner U. Comprehensive profiling and quantitation of amine group containing metabolites. Anal. Chem. 2011;83:7523–7530. doi: 10.1021/ac201610x. [DOI] [PubMed] [Google Scholar]
- Brundrett M.C., Kendrick B., Peterson C.A. Efficient lipid staining in plant material with Sudan red 7B or fluoral yellow 088 in polyethylene glycol-glycerol. Biotech. Histochem. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
- Byrt C.S., Munns R., Burton R.A., Gilliham M., Wege S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018;269:47–55. doi: 10.1016/j.plantsci.2017.12.012. [DOI] [PubMed] [Google Scholar]
- De Storme N., Geelen D. Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci. 2014;5:138. doi: 10.3389/fpls.2014.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias D.A., Hill C.B., Jayasinghe N.S., Atieno J., Sutton T., Roessner U. Quantitative profiling of polar primary metabolites of two chickpea cultivars with contrasting responses to salinity. J. Chromatogr. B. 2015;1000:1–13. doi: 10.1016/j.jchromb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Domergue F., Vishwanath S.J., Joubès J., Ono J., Lee J.A., Bourdon M., Alhattab R., Lowe C., Pascal S., Lessire R. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010;153:1539–1554. doi: 10.1104/pp.110.158238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder K. Gas chromatographic analysis of fatty acid methyl esters. J. Chromatogr. B Biomed. Sci. Appl. 1995;671:113–131. doi: 10.1016/0378-4347(95)00142-6. [DOI] [PubMed] [Google Scholar]
- Engström P.G., Steijger T., Sipos B., Grant G.R., Kahles A., Rätsch G., Goldman N., Hubbard T.J., Harrow J., Guigó R. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods. 2013;10:1185–1191. doi: 10.1038/nmeth.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T.J., Hajibagheri M.A. Salinity tolerance in Hordeum vulgare: ion concentrations in root cells of cultivars differing in salt tolerance. Plant Soil. 2001;231:1–9. [Google Scholar]
- Geldner N. The endodermis. Annu. Rev. Plant Biol. 2013;64:531–558. doi: 10.1146/annurev-arplant-050312-120050. [DOI] [PubMed] [Google Scholar]
- Glenn E.P., Brown J.J., Blumwald E. Salt tolerance and crop potential of halophytes. Crit. Rev. Plant Sci. 1999;18:227–255. [Google Scholar]
- Heller W., Hahlbrock K. Highly purified “flavanone synthase” from parsley catalyzes the formation of naringenin chalcone. Arch. Biochem. Biophys. 1980;200:617–619. doi: 10.1016/0003-9861(80)90395-1. [DOI] [PubMed] [Google Scholar]
- Higuchi T. Lignin biochemistry: biosynthesis and biodegradation. Wood Sci. Technol. 1990;24:23–63. [Google Scholar]
- Hill C.B., Cassin A., Keeble-Gagnère G., Doblin M.S., Bacic A., Roessner U. De novo transcriptome assembly and analysis of differentially expressed genes of two barley genotypes reveal root-zone-specific responses to salt exposure. Sci. Rep. 2016;6:31558. doi: 10.1038/srep31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C.B., Jha D., Bacic A., Tester M., Roessner U. Characterization of ion contents and metabolic responses to salt stress of different Arabidopsis AtHKT1;1 genotypes and their parental strains. Mol. Plant. 2013;6:350–368. doi: 10.1093/mp/sss125. [DOI] [PubMed] [Google Scholar]
- Hunter K., Kimura S., Rokka A., Tran H.C., Toyota M., Kukkonen J.P., Wrzaczek M. CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 2019;180:2004–2021. doi: 10.1104/pp.19.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- Jacobs M., Rubery P.H. Naturally occurring auxin transport regulators. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreszies T., Eggels S., Kreszies V., Osthoff A., Shellakkutti N., Baldauf J.A., Zeisler-Diehl V.V., Hochholdinger F., Ranathunge K., Schreiber L. Seminal roots of wild and cultivated barley differentially respond to osmotic stress in gene expression, suberization, and hydraulic conductivity. Plant Cell Environ. 2020;43:344–357. doi: 10.1111/pce.13675. [DOI] [PubMed] [Google Scholar]
- Kreszies T., Schreiber L., Ranathunge K. Suberized transport barriers in Arabidopsis, barley and rice roots: from the model plant to crop species. J. Plant Physiol. 2018;227:75–83. doi: 10.1016/j.jplph.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Kreszies T., Shellakkutti N., Osthoff A., Yu P., Baldauf J.A., Zeisler-Diehl V.V., Ranathunge K., Hochholdinger F., Schreiber L. Osmotic stress enhances suberization of apoplastic barriers in barley seminal roots: analysis of chemical, transcriptomic and physiological responses. New Phytol. 2018;221:180–194. doi: 10.1111/nph.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Zhang B., Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Madsen B.E., Brwoning S.R. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S.O., Wicker T., Radchuk V., Dockter C., Hedley P.E., Russell J. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- Meikle P.J., Bonig I., Hoogenraad N.J., Clarke A.E., Stone B.A. The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta. 1991;185:1–8. doi: 10.1007/BF00194507. [DOI] [PubMed] [Google Scholar]
- Naseer S., Lee Y., Lapierre C., Franke R., Nawrath C., Geldner N. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl. Acad. Sci. U S A. 2012;109:10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera S.H.A., Hill C.B., Rupasinghe T.W.T., Roessner U. Salt-stress induced alterations in the root lipidome of two barley genotypes with contrasting responses to salinity. Funct. Plant Biol. 2016;43:207. doi: 10.1071/FP15253. [DOI] [PubMed] [Google Scholar]
- O’Neill M.A., Ishii T., Albersheim P., Darvill A.G. Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 2004;55:109–139. doi: 10.1146/annurev.arplant.55.031903.141750. [DOI] [PubMed] [Google Scholar]
- Rate D.N., Greenberg J.T. The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 2001;27:203–211. doi: 10.1046/j.0960-7412.2001.1075umedoc.x. [DOI] [PubMed] [Google Scholar]
- Ravanat G., Rinaudo M. Investigation on oligo- and polygalacturonic acids by potentiometry and circular dichroism. Biopolym. Origin. Res. Biomol. 1980;19:2209–2222. [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. Decoding genes with coexpression networks and metabolomics––‘majority report by precogs’. Trends Plant Sci. 2008;13:36–43. doi: 10.1016/j.tplants.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Sarabia L.D., Boughton B.A., Rupasinghe T., van de Meene A.M.L., Callahan D.L., Hill C.B., Roessner U. High-mass-resolution MALDI mass spectrometry imaging reveals detailed spatial distribution of metabolites and lipids in roots of barley seedlings in response to salinity stress. Metabolomics. 2018;14:63. doi: 10.1007/s11306-018-1359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabia L.D., Boughton B.A., Rupasinghe T., Callahan D.L., Hill C.B., Roessner U. Comparative spatial lipidomics analysis reveals cellular lipid remodelling in different developmental zones of barley roots in response to salinity. Plant Cell Environ. 2020;43:327–343. doi: 10.1111/pce.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L., Franke R., Hartmann K.D., Ranathunge K., Steudle E. The chemical composition of suberin in apoplastic barriers affects radial hydraulic conductivity differently in the roots of rice (Oryza sativa L. cv. IR64) and corn (Zea mays L. cv. Helix) J. Exp. Bot. 2005;56:1427–1436. doi: 10.1093/jxb/eri144. [DOI] [PubMed] [Google Scholar]
- Shelden M.C., Roessner U., Sharp R.E., Tester M., Bacic A. Genetic variation in the root growth response of barley genotypes to salinity stress. Funct. Plant Biol. 2013;40:516. doi: 10.1071/FP12290. [DOI] [PubMed] [Google Scholar]
- Shen J., Xu G., Zheng H.Q. Apoplastic barrier development and water transport in Zea mays seedling roots under salt and osmotic stresses. Protoplasma. 2015;252:173–180. doi: 10.1007/s00709-014-0669-1. [DOI] [PubMed] [Google Scholar]
- Shi B.J., Sutton T., Collins N.C., Pallotta M., Langridge P. Construction of a barley bacterial artificial chromosome library suitable for cloning genes for boron tolerance, sodium exclusion and high grain zinc content. Plant Breed. 2010;129:291–296. [Google Scholar]
- Steudle E., Peterson C.A. How does water get through roots. J. Exp. Bot. 1998;49:775–788. [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E., Rengasamy P., McDonald G.K. High concentrations of Na+ and Cl– ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010;61:4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S.R. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R., Andersen T.G., Marhavý P., Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J.H., Goeminne G., Halpin C., Rohde A., Morreel K., Boerjan W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell. 2012;24:3506–3529. doi: 10.1105/tpc.112.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Walia H., Wilson C., Condamine P., Liu X., Ismail A.M., Zeng L., Wanamaker S.I., Mandal J., Xu J., Cui X. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widodo, Patterson J.H., Newbigin E., Tester M., Bacic A., Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009;60:4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Ho Y.Y., Lampugnani E.R., Van de Meene A.M., Bain M.P., Bacic A., Doblin M.S. Determining the subcellular location of synthesis and assembly of the cellwall polysaccharide (1,3; 1,4)-β-d-glucan in grasses. Plant Cell. 2015;27:754–771. doi: 10.1105/tpc.114.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J., Ruel K., Ryser U., Schreiber L. Chemical analysis and immunolocalisation of lignin and suberin in endodermal and hypodermal/rhizodermal cell walls of developing maize (Zea mays L.) primary roots. Planta. 1999;209:1–12. doi: 10.1007/s004250050601. [DOI] [PubMed] [Google Scholar]
- Zhao L., Wang P., Yan S., Gao F., Li H., Hou H., Zhang Q., Tan J., Li L. Promoter-associated histone acetylation is involved in the osmotic stress-induced transcriptional regulation of the maize ZmDREB2A gene. Physiol. Plant. 2014;151:459–467. doi: 10.1111/ppl.12136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.