Abstract
Viruses commonly use specifically folded RNA elements that interact with both host and viral proteins to perform functions important for diverse viral processes. Examples are found at the 3′ termini of certain positive-sense ssRNA virus genomes where they partially mimic tRNAs, including being aminoacylated by host cell enzymes. Valine-accepting tRNA-like structures (TLSVal) are an example that share some clear homology with canonical tRNAs but have several important structural differences. Although many examples of TLSVal have been identified, we lacked a full understanding of their structural diversity and phylogenetic distribution. To address this, we undertook an in-depth bioinformatic and biochemical investigation of these RNAs, guided by recent high-resolution structures of a TLSVal. We cataloged many new examples in plant-infecting viruses but also in unrelated insect-specific viruses. Using biochemical and structural approaches, we verified the secondary structure of representative TLSVal substrates and tested their ability to be valylated, confirming previous observations of structural heterogeneity within this class. In a few cases, large stem–loop structures are inserted within variable regions located in an area of the TLS distal to known host cell factor binding sites. In addition, we identified one virus whose TLS has switched its anticodon away from valine, causing a loss of valylation activity; the implications of this remain unclear. These results refine our understanding of the structural and functional mechanistic details of tRNA mimicry and how this may be used in viral infection.
Keywords: tRNA mimicry, viral RNA, RNA structure, aminoacylation
INTRODUCTION
RNAs adopt complex structures to accomplish a wide range of functions and often the same class of RNA appears in a variety of species but with substantial variation. For example, within a given class of RNA the primary sequence may vary, but conservation of base pair patterns and specific nucleotides preserves key tertiary interactions or forms critical functional elements. For some RNAs the overall global shape is not critical as long as a conserved functional core exists within the larger fold (e.g., to bind ligands or perform catalysis) (Doherty and Doudna 2001; Weinberg et al. 2015; McCown et al. 2017), while other RNAs rely almost entirely on their overall three-dimensional shape (e.g., to block a processive nuclease or mimic the structure of another RNA) (Pfingsten et al. 2006; Dreher 2010; Chapman et al. 2014; Akiyama et al. 2016; Pisareva et al. 2018). Finally, it is possible for different classes of RNAs with distinct secondary and tertiary structures to perform identical functions (Corbino et al. 2005; Perreault et al. 2011; Steckelberg et al. 2018a). Understanding the fundamental rules of the RNA structure-function relationship requires detailed explorations of diverse RNAs across and within functional classes.
An interesting class of functional RNA elements found in viruses are the transfer RNA (tRNA)-like structures (TLSs) found at the 3′ terminus of certain positive-sense single-stranded plant-infecting RNA viruses (Pinck et al. 1970; Yot et al. 1970; Rietveld et al. 1983; Mans et al. 1991; Dreher 2010). These RNAs were identified by their ability to induce in cis aminoacylation of the 3′ end of the viral RNAs by host cell aminoacyl tRNA synthetases (AARSes) (Pinck et al. 1970; Yot et al. 1970; Hall et al. 1972; Öberg and Philipson 1972). The implied structural mimicry makes them intriguing examples of RNA-based molecular mimicry (Rietveld et al. 1983; Hammond et al. 2009). The three known classes of TLSs have distinct secondary structures compared to one another and to canonical tRNAs, and each class is charged with a different amino acid: valine, histidine, or tyrosine (Mans et al. 1991; Hammond et al. 2009; Dreher 2010). TLSs play multiple roles that confer an advantage during infection (Hall 1979; Haenni et al. 1982; Mans et al. 1991; Dreher 2009, 2010), including in cis enhancement of translation of the viral RNA (Matsuda and Dreher 2004). While some portions of these viral TLSs resemble a tRNA, they are typically larger, and their positions at the 3′ end of the viral RNA mandate an alternate connectivity involving a 3′ pseudoknot structure, whereas a tRNA's 5′ and 3′ ends are paired in its acceptor stem (Rietveld et al. 1982, 1983; Pleij et al. 1985; Felden et al. 1994).
Valine-accepting TLSs (TLSVal) are on average the smallest of the three classes and share the most homology with tRNAs (Dreher and Goodwin 1998). This homology allows TLSVal to interact with many of the same host factors as tRNAs including the CCA-adding enzyme, valyl-tRNA synthetase (ValRS) and eukaryotic elongation factor 1A (eEF1A) (Pinck et al. 1970; Yot et al. 1970; Giege et al. 1978; Joshi et al. 1982; Dreher and Goodwin 1998; Dreher et al. 1999; Matsuda and Dreher 2004). In addition, the TLS contains the promoter for viral negative-strand synthesis and therefore also interacts with the viral RNA-dependent RNA polymerase (RdRp) (Deiman et al. 1998; Singh and Dreher 1998). While many of the interactions between TLSVal RNAs and host proteins have been characterized, numerous aspects of TLS function and mechanism of action remain unknown. For example, translation enhancement has been demonstrated for the prototype TLSVal from turnip yellow mosaic virus (TYMV), which acts synergistically with a 5′ cap (Matsuda and Dreher 2004). However, direct interactions between TLSs and the ribosome have long been debated, with specific modes of action being postulated (Haenni et al. 1973; Barends et al. 2003) and subsequently refuted (Haenni et al. 1982; Matsuda and Dreher 2007), leaving the mechanism of translation enhancement yet to be determined. Additional complexity in examining the functional roles of TLSs stems from reported differences in host factor interactions between the classes of TLS (e.g., TLSVal vs. TLSHis) or between TLS representatives of the same class in different viruses. For example, valylation of the TYMV TLSVal is required for TYMV infectivity (Tsai and Dreher 1991), while valylation of the peanut clump virus (PCV) TLSVal is not necessary and only provides a slight competitive advantage for infectivity (Matsuda et al. 2000). Additionally, despite both the TYMV and PCV TLSVal RNAs acting as highly efficient substrates for valylation, only the TYMV TLSVal can bind eEF1A with similar affinity to plant tRNAVal while the PCV TLSVal forms a weak ternary complex with eEF1A:GTP (Dreher and Goodwin 1998; Goodwin and Dreher 1998). Such functional variations within the TLSVal class motivate efforts to catalog and understand underlying sequence and structure variation.
The structure of the TYMV TLSVal RNA has been studied for decades using a variety of biochemical and biophysical techniques (Rietveld et al. 1982; Matsuda and Dreher 2004; Hammond et al. 2009, 2010), but only recently has high-resolution structural information become available. Specifically, two structures of the TYMV TLSVal, both on its own (Colussi et al. 2014) or attached to its 5′ upstream pseudoknot domain (UPD) (Hartwick et al. 2018), were solved by X-ray crystallography. These structures revealed similarities and differences between the TYMV TLSVal and tRNAs, identified key tertiary contacts needed to stabilize the fold, and showed how the TLS and UPD domains are oriented and how they communicate.
The structures of the TYMV TLS raise new questions about the TLSVal class as a whole and also enable new explorations. Specifically, while several dozen TLSVal are known in plant-infecting viruses (Dreher and Goodwin 1998; Goodwin and Dreher 1998; Dreher 2010), we do not have a full catalog of this type of tRNA mimicry. However, secondary and tertiary structural information from high-resolution structures (Colussi et al. 2014; Hartwick et al. 2018) can now be combined with new search tools to find additional examples. Because some features may be idiosyncratic, analyses of new examples help to understand the diversity within a class and therefore how different elements form tertiary interactions or contacts with viral or host factors. Comparative sequence analysis using homology searches is a powerful tool to expand knowledge of RNA classes such as riboswitches, ribozymes, exoribonuclease-resistant RNAs, and others of unknown function (Barrick et al. 2004; Weinberg et al. 2007, 2010, 2015, 2017; Roth et al. 2014; Steckelberg et al. 2018b). Therefore, we used structure-guided bioinformatic strategies to identify new examples of RNAs conforming to the TLSVal pattern. These used primary sequence and secondary structure conservation guided by information from three-dimensional crystal structures to iteratively search large databases for new sequences. Using chemical probing and functional assays, we verified putative novel examples and explored the extent of their conservation and diversity. Our findings reveal additional examples of structural and potentially functional variations in the TLSVal RNA class, including the verification of previously proposed TLSVal within insect-infecting viruses (Gordon et al. 1995, 1999).
RESULTS AND DISCUSSION
Homology searches reveal additional putative examples of valyl tRNA-like structures
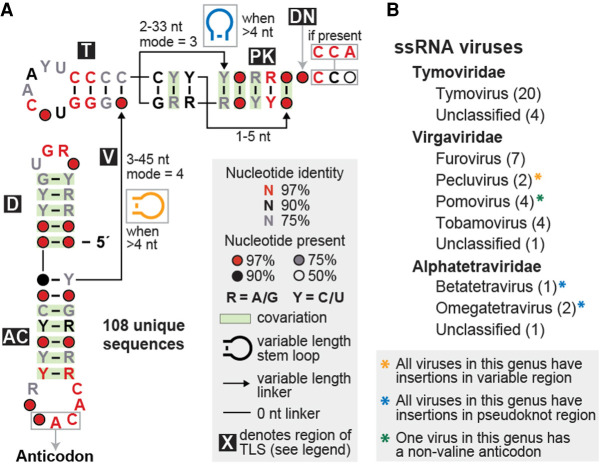
We conducted homology-based searches starting with a seed alignment containing 28 previously identified examples of TLSVal from the Rfam database (http://rfam.xfam.org/family/RF00233) (Kalvari et al. 2018). We made adjustments to this initial alignment based on the crystal structure of the TYMV TLS (Colussi et al. 2014) and previously proposed models for other members of this class (Goodwin and Dreher 1998). Homology searches using the program Infernal (Nawrocki and Eddy 2013) identified 108 unique sequences of putative TLSVal RNAs in 46 distinct viruses (Fig. 1, see also Supplemental Files 1, 2). Some of these viruses contain multiple RNA segments, which often contain similar but not identical putative TLSVal sequences at their 3′ ends.
FIGURE 1.
Structural conservation and phylogenetic distribution of all valyl tRNA-like structures in viruses. (A) Consensus sequence and secondary structure model of the 108 unique examples of valine-accepting tRNA-like structures. Each portion of the structure is labeled, mostly according to its homolog in a canonical tRNA (D: D-arm; AC: anticodon arm; V: variable region; T: T-arm; PK: pseudoknot region; DN: discriminator nucleotide). Stem–loop structure insertions are sometimes present in the linkers within the variable and pseudoknot regions. (B) Genera that contain valine-accepting tRNA-like structures, further organized by family. The number of individual viruses that contain these structures within each genus is listed in parentheses and all examples are derived from positive-sense single-stranded RNA viruses.
Most of the putative TLSVal RNAs are found in viruses of the Tymoviridae family with several other examples in the Virgaviridae family (Fig. 1B). It has previously been noted that TLSVal is more abundant in Tymoviridae and TLSHis is more abundant in Virgaviridae but each is occasionally found in the other family (Dreher 2010). We also identified four putative TLSVals in insect-infecting tetraviruses, which are positive-sense ssRNA viruses that are often segmented. It was previously proposed that the 3′ terminus of both Nudaurelia capensis beta virus (NCBV) and Helicoverpa armigera Stunt Tetravirus (HaSV), two of the four tetravirus examples, form a tRNA-like structure (Gordon et al. 1995, 1999), but these RNAs were not tested for aminoacylation and the proposed secondary structure model differed from known TLSVal. Specifically, these models proposed 5′ to 3′-end base pairing as in tRNAs instead of the 3′ pseudoknot arrangement that has since been demonstrated to form by other members of TLSVal (Gordon et al. 1995, 1999).
The compiled unique sequences were used in R-scape (Rivas et al. 2017, 2020) to calculate a consensus sequence and secondary structure model, which has several key features (Fig. 1A). All of the base-paired elements are supported by covarying mutations except the part homologous to the tRNA's T-arm. Except for three examples discussed below, the anticodon always encodes valine (generally “NAC”), the wobble position can be any of the 4 nt, and the 2 nt following the anticodon are similarly well conserved as “AC.” Within a set of tRNAs specific for a given amino acid, the “discriminator nucleotide” located just upstream of the 3′ terminal CCA is conserved (Crothers et al. 1972; Hou 1997). In contrast, in TLSVal the discriminator nucleotide is not significantly conserved although often it is an A, matching tRNAVal (Fig. 1A). Most examples from Tymoviridae and Tetraviridae have an A discriminator nucleotide while those from Virgaviridae have a C. Finally, because these viral genomes do not encode a terminal A, the viral genome is 3′-adenylated by the host cell's CCA-adding enzyme (Giege et al. 1978). This, and the technical limitation that many sequencing reads are truncated, causes the nucleotides at the 3′ terminus to be under-represented in the TLSVal alignments. Therefore, while the terminal “CCA” does not appear to be significantly conserved, when present these nucleotides are 100% conserved (see Supplemental File 1 for all aligned sequences).
The consensus model reflects the fact that the TLSVal T-loop is well conserved, to a similar extent as a canonical tRNA, and the stem is always capped by two noncovarying G–C pairs. It was proposed that the combined length of the T-arm stem and pseudoknotted acceptor stem is always equal to 12 bp, which can be composed of either a 4/3/5 or a 3/3/6 combination (Dreher and Goodwin 1998; Dreher 2010), and our consensus model agrees with this observation. The majority of examples have a T-arm stem four base pairs in length, corresponding to a 4/3/5 composition, thus this arrangement is the only one that is obviously apparent in the consensus model (Fig. 1A). However, approximately 30% of sequence examples appear to adhere to the 3/3/6 base pairing scheme. The examples with stem insertions, which are described in detail below, are slightly difficult to classify, but may use noncanonical pairing at the base of the T-arm stem and thus still adhere to the 4/3/5 pattern. Most representatives from Virgaviridae have a 4/3/5 arrangement while those from Tymoviridae representatives can have either the 4/3/5 or 3/3/6 arrangement. A few sequence representatives derived from strains of Clitoria yellow vein virus or Kennedya yellow mosaic virus have nucleotide mutations and insertions that are incompatible with the 4/3/5 arrangement observed for other strains of these viruses, and also with the 3/3/6 arrangement. The base pairing for these TLSs is ambiguous in the absence of high-resolution structural information or detailed mutational analyses, but a 3/4/5 arrangement base pairing scheme might be used by these examples (see Supplemental Files 1, 2 for additional details).
Overall, most new putative examples of TLSVal are predicted to closely resemble the prototypical TYMV, but a number contain variations including insertions in several linker regions as well as differences in the anticodon identity. These observations, and the presence of putative TLSVal in insect-infecting viruses, are explored and discussed here.
Most TLSVals conform to a shared secondary structure
Secondary structure predictions based on conservation across species provide compelling evidence for conserved stems and pseudoknots within a class of RNA, but it is critical to experimentally evaluate the structure of individual sequence representatives to ensure proper alignment. This is especially true in areas that are too highly conserved for covarying base pairs to be observed or in sites where substantial insertions are predicted. Thus, we performed in vitro chemical probing experiments for several different TLSVal RNAs. The relative reactivity of each nucleotide as determined by selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE) probing experiments resolved by capillary electrophoresis (Yoon et al. 2011; Kim et al. 2013; Cordero et al. 2014; Kladwang et al. 2014; Lee et al. 2015) was determined for each TLSVal RNA and mapped onto the corresponding secondary structure prediction (Fig. 2A,B; Supplemental Fig. S1). Using the prototype TYMV TLS as a positive control, we observed higher reactivity in known loops and variable regions and relatively low reactivity in base-paired elements, consistent with previous results and the crystal structure (Supplemental Fig. S1A; Hartwick et al. 2018). We then probed another representative TLSVal from Japanese soil-borne wheat mosaic virus (JSWMV), which conforms well to the consensus model. Probing data from this putative TLSVal matched TYMV, indicating the same overall fold (Supplemental Fig. S1B). These results strongly suggest that putative TLSVals that conform well to the consensus model share a common secondary structure and likely similar tertiary structures.
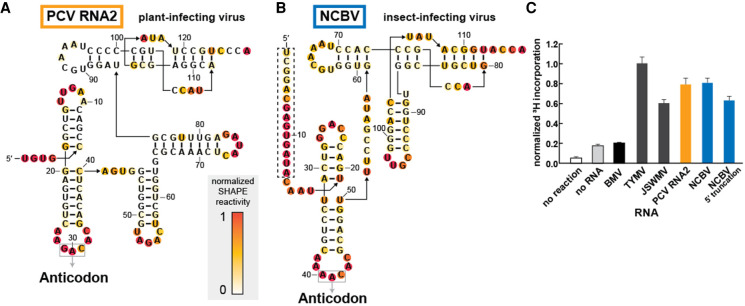
FIGURE 2.
Divergent TLSVal RNAs containing stem–loop insertions are competent substrates for valylation. (A,B) Chemical probing of TLS representatives from Peanut clump virus (A) and Nudaurelia capensis beta virus (B) using the SHAPE reagent NMIA. Reactivity was background subtracted and normalized according to the reactivity of loop regions in hairpin structures (not shown) flanking the TLS structure on both the 5′ and 3′ ends. (C) Activity of valyl tRNA synthetase (ValRS) on TLS RNAs as measured by the covalent addition of radiolabeled (3H) valine at their 3′ termini. The 3H incorporation, as measured by a scintillation counter, was normalized to the TYMV TLSVal construct, which had been previously tested and optimized under these reaction conditions. The truncated NCBV construct begins at nucleotide C15 (see panel B for sequence; the dashed box indicates the deleted region). The BMV TLS belongs to a separate class of tyrosine-accepting TLSs. Orange and blue bars correspond to the color schemes for PCV and NCBV, respectively, in panels A and B as well as in Figure 4.
Substantial insertions are tolerated in discrete locations within TLSVal
The main source of structural heterogeneity in the TLSVal is predicted insertions in the parts of the TLS that correspond to the “variable” region between the anticodon arm and T-arm, or within a loop of the 3′ acceptor stem pseudoknot (Fig. 1A). The former is found in examples from PCV RNAs, and the secondary structures predicted by our bioinformatic searches agree with a previously proposed model that places two large stem–loop structures in the variable region (Goodwin and Dreher 1998). The latter occurs in three of the four examples derived from insect-infecting Tetraviruses, but in those our predicted secondary structures were different than in previously published models (Gordon et al. 1995, 1999).
To determine the correct secondary structure of these divergent putative TLSVals, we applied chemical probing to RNAs from PCV and NCBV as representative of plant-infecting and insect-infecting viruses. The reactivity patterns were consistent with the bioinformatic predictions of an overall secondary structure pattern that matches the “typical” TLSVal, but with stem–loops inserted into the aforementioned regions (Fig. 2A,B). Because the NCBV RNA was previously proposed to have a different secondary structure, we mapped the reactivity data on both the predicted structure from this study (3′ pseudoknot model) and the previously proposed 5′ to 3′ pairing model (Gordon et al. 1999). The pattern is consistent with our new model proposed by the bioinformatic searches but does not match the previous model (Supplemental Fig. S1C).
Divergent TLSVal RNAs are aminoacylated in vitro
We then asked if the divergent TLSVals could serve as substrates for aminoacylation. While the TLSVal RNAs from plant-infecting TYMV, JSWMV, and PCV RNA1 had all been shown to undergo valylation in vitro (Dreher and Goodwin 1998; Goodwin and Dreher 1998), the TLS from PCV RNA2 was reported to have almost no valylation activity (Goodwin and Dreher 1998), and tests of putative examples from insect-infecting viruses had not been reported. We tested all of these RNAs for in vitro valylation using recombinantly purified ValRS from Thermus thermophilus and 3H-labeled valine under conditions previously used for the TYMV TLS (Hartwick et al. 2018). Negative control reactions containing no RNA or the TLSTyr from Brome mosaic virus (BMV) show low levels of valylation. However, all TLSVal RNAs, including PCV RNA2 and NCBV, show valine incorporation levels significantly above background (Fig. 2C). Our contradictory results for the PCV RNA2 TLS are almost certainly due to a difference in the tested sequences. Specifically, the first published sequence of the PCV RNA2 (Manohar et al. 1993) contains a deletion in the anticodon “G–C” and this TLS sequence was previously shown to have very low valylation activity (Goodwin and Dreher 1998). While this example appears in our alignment (Supplemental File 1), all other unique sequence representatives of PCV RNA2 have a canonical “GAC” valine anticodon. The PCV RNA2 TLSVal that we selected to test contained a “GAC” anticodon and showed no defect in valylation (Fig. 2A). It is unclear whether the deletion is present only in certain strains of PCV, or if this is the result of a sequencing error, but it seems likely that the TLSs in both RNAs of PCV are typically valylated.
In the above analysis we used the ValRS from T. thermophilus, which is not a natural host for any of the viruses that contain a TLSVal but that readily aminoacylated RNAs we tested. To explore the basis of this observation, we aligned several ValRS sequences using NCBI Cobalt (Papadopoulos and Agarwala 2007), including sequences from both bacterial and eukaryotic species (Supplemental Fig. S2). The eukaryotic ValRS has an expanded N-terminus, but the rest of the sequence was mostly conserved in both identity and physiochemical properties for each amino acid position. However, several insertions and areas of lower similarity were seen (Supplemental Fig. S2A–D). To relate the variable regions to tRNA binding, we used the Phyre2 (Kelley et al. 2015) server to build a homology model of the S. tuberosum ValRS based on the structure of the T. thermophilus ValRS (Supplemental Fig. S2D). Comparison of the model and structure shows that differing areas are not expected to affect tRNA recognition. Furthermore, mapping amino acids that are 95% conserved by identity, identified using Clustal Omega (Sievers et al. 2011; Sievers and Higgins 2018) onto the structure of the T. thermophilus ValRS–tRNAVal, shows the most conserved positions are placed near the anticodon and the 3′ CCA motif interaction sites (Supplemental Fig. S2E,F). Thus, while we cannot eliminate the possibility that some species of ValRS have idiosyncratic behavior matched to specific species of TLSVal, most ValRS—tRNAVal and ValRS—TLSVal combinations are predicted to be compatible and this may facilitate the spread of these viruses to new hosts.
The ability of the NCBV TLSVal to be valylated confirms that aminoacylatable TLSVal RNAs exist in insect-infecting viruses in addition to plant-infecting viruses, but we propose a different secondary structure compared to previous reports (Gordon et al. 1995, 1999). We used the valylation activity to further interrogate our bioinformatically predicted structural model (3′ pseudoknot) versus the previously reported model (5′ to 3′ paired). Because the 5′ to 3′ paired model includes ∼15 nt not involved in predicted structures in the 3′ pseudoknot model (Supplemental Fig. S1C; Gordon et al. 1999), we tested a truncated version that cannot form the 5′ to 3′ pairing (Fig. 2B). This 5′ truncated RNA is still valylated, which likely would not occur if the 5′ to 3′ paired model were valid. Note that 3H-valine incorporation is lower for the truncated construct as compared to the extended construct, which could be due to the absence of some optional substructures analogous to the TYMV TLS when the upstream pseudoknot domain is included (Dreher and Goodwin 1998; Hammond et al. 2010). Overall, the chemical probing and aminoacylation results show that the NCBV RNA forms a secondary structure similar to other TLSVal.
Taken together, our results verify that large insertions in some TLSVals are structurally and functionally tolerated. Whether these confer additional function is unknown. Our results also verified the proposal that TLSs are not exclusively a plant virus phenomenon but are also found in animal-infecting viruses. While our bioinformatic searches only revealed four examples of this, additional divergent TLSVal examples likely remain to be found in other viruses, and/or completely unrelated TLS classes may be discovered in other species.
TLSs with nonvaline anticodons fail to be aminoacylated
The bioinformatic analysis identified three putative TLSVals with a nonvaline anticodon in the three RNA segments of Colombian potato soil-borne virus (CPSbV), all of which have a UAA (leucine) in the anticodon loop (Gil et al. 2016). The TLS sequences from these three segments of CPSbV are similar (Supplemental Fig. S3A), but a few covarying mutations in base-paired regions and one other mutation in a variable region make the strict conservation of the UAA anticodon peculiar. A sequencing error seems unlikely since the UAA leucine anticodon was present in all of the RNAs in multiple isolates of this recently identified virus (Gil et al. 2016). This anticodon triplet is only 1 nt away from a valine anticodon (UAC is valine-compatible) yet none of the three examples contain a valine codon, raising the question of why all three RNA segments have converted to the same nonvaline anticodon.
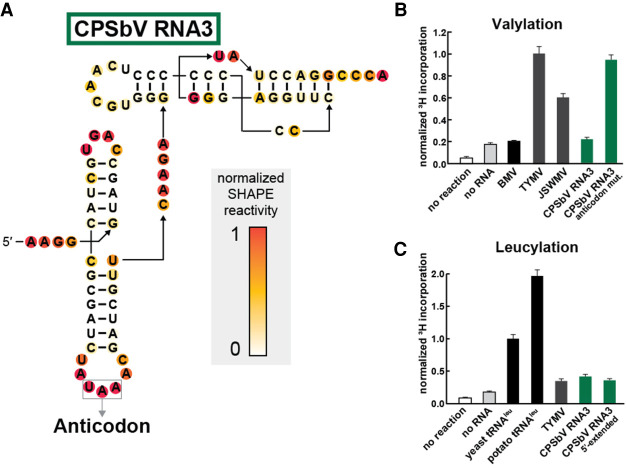
To verify the alignment that assigned the UAA triplet as the anticodon, we performed chemical probing of the putative TLS RNA from CPSbV. The pattern was consistent with the secondary structure model for TLSVal and supported that the UAA was properly placed, creating a leucine anticodon (Fig. 3A). We then tested the sequence from CPSbV RNA3 for in vitro valylation. Consistent with the nonvaline anticodon, the CPSbV RNA3 was not aminoacylated (Fig. 3B), nor was RNA2 (Supplemental Fig. S3B). However, mutation of the anticodon from UAA (leucine) to CAC (valine) in CPSbV RNA3 conferred the ability of this RNA to be valylated (Fig. 3B), hence other than their anticodons these RNAs have retained all other TLSVal features.
FIGURE 3.
The leucine anticodon of the CPSbV TLS prevents in vitro valylation activity. (A) Chemical probing of TLS representatives from Colombian potato soil-borne virus using the SHAPE reagent NMIA. Additional annotations and details are described in the legend to Figure 2. (B) Activity of valyl tRNA synthetase (ValRS) on CPSbV TLS RNAs as measured by the covalent addition of radiolabeled (3H) leucine at their 3′ termini. The WT sequence for CPSbV TLSs corresponds to a leucine anticodon, as depicted in panel A, while the anticodon mutation construct contains two mutations (UAA → CAC) that alter the anticodon identity to valine. For more experimental details, see the legend for Figure 2B. (C) Activity of leucyl tRNA synthetase (LeuRS) on TLS RNAs or tRNAs as measured by the covalent addition of radiolabeled (3H) leucine at their 3′ termini. Leucylation was measured by scintillation counter to determine 3H-Leucine incorporation and normalized to the yeast tRNALeu construct. The CPSbV 5′-extended construct includes an additional 65 nt upstream of the sequence depicted in panel A (see Supplemental Table S1 for sequence details). Green bars correspond to the color scheme in panels A and B as well as in Supplemental Figure S3.
It seemed possible that CPSbV RNAs might indicate a new class of TLSs with an overall secondary structure that conforms to the TLSVal class, but which are charged with leucine by the cognate synthetase. We tested this using recombinant leucyl-tRNA synthetase (LeuRS) from Saccharomyces cerevisiae and 3H-leucine. The S. cerevisiae LeuRS enzyme was active, shown by its ability to aminoacylate not only positive control in vitro-transcribed tRNALeu from S. cerevisiae, but also tRNALeu from Solanum tuberosum, the virus’ potato plant host (Fig. 3C). In vitro-transcribed TYMV TLSVal served as the negative control RNA and was not efficiently aminoacylated. Testing a CPSbV TLS, we did not find any conditions under which it was aminoacylated with leucine above the negative control (Fig. 3C). We then tested another CPSbV RNA3 construct with an additional 65 nt upstream of the beginning of the TLS structure. This includes the portion of the 3′-UTR conserved across all three genomic RNAs, as we speculated that additional structures in this region could be necessary for or increase the efficiency of leucylation. However, the extended CPSbV RNA also was not leucylated above the negative control. Variation of buffer conditions, RNA concentration, or Mg2+ and ATP concentration (see Materials and Methods for details) did not yield leucine incorporation signal above the negative control (Supplemental Fig. S3B). While the leucylation signal is slightly higher for some of these constructs compared to no RNA controls, the signal is still well below the positive controls (i.e., tRNALeu), and based on the precedent that the TYMV TLS is able to be valylated to the same levels as tRNAVal under certain conditions (Dreher and Goodwin 1998), we consider these low leucylation incorporation levels to be a negative result.
The inability of the S. cerevisiae LeuRS to aminoacylate the CPSbV TLS could be due to a high degree of specificity for the AARS of the host species S. tuberosum, but two lines of evidence argue against this. First, the S. cerevisiae LeuRS robustly leucylated a S. tuberosum tRNALeu, indicating compatibility between this LeuRS and tRNA from the host species. In fact, alignment of LeuRS sequences from several sources indicate that LeuRS nucleotide binding, editing, and activity are likely conserved between archaea, bacteria, and eukaryotes with subtle differences (Cusack et al. 2000; Fukunaga and Yokoyama 2005b, 2006). As with the ValRS, we aligned several representative LeuRS sequences and modeled the S. tuberosum version, observing that despite several differences the yeast and plant synthetases share homology in important domains (Supplemental Fig. S5). This further supports our observation of S. tuberosum tRNALeu activity with yeast LeuRS and not TLSVal representatives, such as those from CPSbV which lack the correct discriminator nucleotide identity (Supplemental Fig. S3A). Second, previous structural and functional characterization of LeuRS show that this synthetase does not appear to query the anticodon (Asahara et al. 1993; Fukunaga and Yokoyama 2005b). Modeling the TYMV TLS into a complex with the LeuRS enzyme supports the idea that the anticodon would not be recognized (Supplemental Fig. S4). Rather, LeuRS recognition of tRNALeu relies on other elements (Fukunaga and Yokoyama 2005a,b) that are absent from the putative CPSbV TLS. Specifically, a stem–loop in the variable region of tRNALeu is recognized by the LeuRS, though only acting as a weak discrimination element (Asahara et al. 1993), but the CPSbV TLS lacks this feature. The discriminator nucleotide for tRNALeu is an A that is important for recognition and discrimination by LeuRS (Fukunaga and Yokoyama 2005a) but all three CPSbV TLS RNAs contain a C at this position. Given these requirements, it is perhaps not surprising that the putative CPSbV TLSs would not be recognized by the LeuRS. Thus, while we cannot exclude the possibility that the CPSbV TLSs are leucylated in the context of the cell of their specific host, and therefore represent a separate class from the rest of the TLSVal RNAs, our assessment is that this is unlikely.
Why would the anticodon in the TLS of all three CPSbV RNAs specifically encode a leucine if the rest of the TLS structure matches a valine tRNA, especially if this prevents any aminoacylation? One possibility is that the CPSbV TLSs are a poorer substrate than the other TLSVal RNAs and it is beneficial for CPSbV to be aminoacylated inefficiently or not at all. There are a number of viruses related to those with TLSVal that lack substantial portions of the canonical TLSVal structure needed for aminoacylation and therefore do not have an amino acid added to the 3′ end of the genomic RNAs (Dreher and Goodwin 1998). However, in the case of CPSbV a small substitution mutation can convert them to true TLSVal, and it is odd that evolution would alter all three copies in such a targeted way if selection is just to eliminate aminoacylation. Indeed, this pattern suggests that these TLSs could readily switch to authentic TLSVal. It seems probable that this conversion occurred by a relatively recent mutation (i.e., UAC → UAA) in one of the three viral RNAs, followed by a recombination event between the genomic RNAs of CPSbV, which is a well-described and common phenomenon for multipartite plant viruses (Bujarski and Kaesberg 1986). While this likely explains the mechanism of this mutation appearing in all three RNA segments, it does not address why this conversion was selected for. Perhaps aminoacylation of some sort is still accomplished in one of the ways described above, or it is advantageous for the virus to have a specific species of uncharged tRNA-like element on its 3′ end, potentially activating stress-response pathways that in some way favor viral proliferation. Another possibility is that the CPSbV TLSs are aminoacylated by an entirely different host AARS, likely one that does not query the anticodon loop. Many members of the TLSVal class have been shown also to act as adequate substrates for HisRS, demonstrating modest levels of 3′ histidine incorporation (Rudinger et al. 1992; Goodwin and Dreher 1998). In the case of the PCV RNA2 TLS, an anticodon mutation that severely impacted its valylation efficiency did not affect its histidylation activity (Goodwin and Dreher 1998), indicating that the anticodon mutations of the CPSbV RNAs do not exclude the possibility that these TLSs act as substrates for HisRS or another AARS. Our data are not able to address these possibilities, but they hint at an intricate interplay between viral RNA and host machinery in CPSbV infection that will require infection-based experiments to explore.
The consensus model suggests protein binding strategies are conserved
Previous superpositions of the TYMV TLSVal structure onto the structure of an authentic tRNAVal-ValRS complex and a tRNA bound to elongation factor thermo unstable (EF-Tu) showed how it could bind those factors, as the overall geometry and key surfaces of a tRNA are maintained in the TYMV TLS (Colussi et al. 2014). Superimposing the TYMV TLS + UPD structure with the ValRS and eEF1A likewise shows the UPD does not interfere with TLS interactions with these proteins (Fig. 4) or with binding of the CCA-adding enzyme (Supplemental Fig. S6). Examination of the consensus model suggests that all TLSVals can make the same binding contacts. Specifically, the 3′ end acceptor stem pseudoknot is present and 12 bp are maintained from the T-loop to just before the 3′ CCA motif, as observed in tRNA and the crystallized TYMV crystal structures (Colussi et al. 2014; Hartwick et al. 2018). Thus, the length of the T-loop-acceptor stem helical stack is maintained across the class, whether in a 4/3/5 or 3/3/6 arrangement. In the tRNAVal-ValRS structure, the anticodon loop is highly distorted (Fukai et al. 2000) in order to be recognized by the synthetase. While the anticodon loop of the TYMV TLS does not adopt this conformation in the RNA-only crystal structure, it was previously hypothesized that the loop is flexible enough to adopt the same conformation as that of tRNAVal when bound by ValRS (Colussi et al. 2014). The anticodon loop is highly conserved among TLSVal RNAs and it is likely that all members of the class are recognized in this manner by ValRS. Put together, the consensus model is consistent with evolution constraining the structures to maintain features essential for interactions with ValRS, eEF1A, and the CCA-adding enzyme.
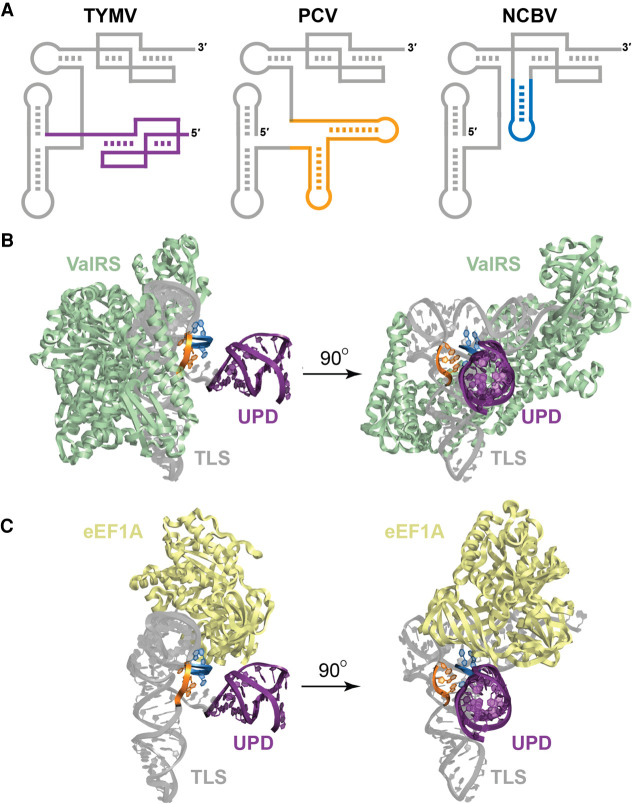
FIGURE 4.
Stem–loop insertions found in divergent TLSVals do not interfere with host factor interactions. (A) Cartoon models of the TLS representatives from TYMV, PCV, and NCBV. The upstream pseudoknot domain (UPD) of the TYMV TLS is drawn in purple (note that the PCV and NCBV TLSs are not preceded by a UPD in the context of the viral genomic RNA). The stems inserted within the variable region of the PCV TLS are orange and the insertion within the 3′ pseudoknot structure of NCBV is blue. (B) Modeling of the TYMV TLS (gray) with the valyl-tRNA synthetase (ValRS; green) from Thermus thermophilus. The nucleotides corresponding to the site of insertion for PCV (variable region) and NCBV (3′ pseudoknot) are colored orange and blue, respectively, and the UPD is colored purple to match panel A. (C) Modeling of the TYMV TLS (gray) with eEF1A (yellow) from Oryctoagus cuniculus. Colors are as for panel B.
Insertions are positioned to not interfere with protein binding
The presence of substantial stem–loop insertions in several TLSVals (Fig. 4A) are tolerated in terms of aminoacylation (Fig. 2) and thus do not seem to interfere with ValRS binding. To correlate this with structure, we mapped the location of these insertions onto the model of the TYMV TLS + UPD on ValRS and on eEF1A. Both insertion points lie on the face of the TYMV TLS where the UPD and genome connect and thus not where they will interfere with synthetase, eEF1A or CCA-adding enzyme binding (Fig. 4; Supplemental Fig. S6). Interestingly, the insertion points are very close to where the UPD is attached to the TYMV TLSVal, suggesting that the stem–loop insertions would clash with a UPD if one were present. However, we find no strong evidence that the PCV or NCBV TLSVals have UPDs 5′ of their predicted TLS regions and thus the inserted stem–loops can apparently occupy the same three-dimensional space as the UPD in TYMV. It is tempting to speculate that convergent evolution has resulted in several ways to place an RNA structural domain in this location to perform a similar function, but this remains untested.
The identity of the nucleotide at the discriminator position reflects the viral family
The discriminator nucleotide for tRNAVal is nearly always an A, while in the TLSVal it is not well conserved but is usually an A or sometimes a C. The ValRS–tRNAVal complex structure (Fukai et al. 2000) suggests no obvious interactions with the discriminator nucleobase and thus it could be a purine or pyrimidine. Previous mutational analysis of the discriminator base of the TYMV TLSVal showed that both G and C in this position caused a measurable yet modest decrease in the kinetic efficiency of valylation, indicating an A in this position serves as a weak identity element for ValRS (Dreher et al. 1992). In the TLSVal this nucleotide may play a role in interactions with a viral protein factor in addition to a host factor. Nearly all TLSVal RNAs with an A at the discriminator nucleotide position are of the Tymoviridae family, while a C is almost exclusively found in the Virgaviridae family. The preference for an A nucleotide in the discriminator position for the Tymoviridae TLSVals might be beneficial for interactions with both the host synthetase and the viral RdRp. This notion is supported by the observation that a C nucleotide at this position drastically lowered the transcription efficiency of the TYMV RdRp (Deiman et al. 1998). However, a parallel study of the TYMV RdRp did not reveal any defect in transcription upon mutation of the discriminator base identity (Singh and Dreher 1998), although in both cases activity was measured for truncated or minimal constructs (Deiman et al. 1998; Singh and Dreher 1998) and further studies will be necessary to determine the contribution of this nucleotide position in the context of the full TLS. In contrast, the RdRp from tobacco mosaic virus (TMV), a prototypical Virgaviridae member, is most efficient with a C at this position (Osman et al. 2000). Therefore, this change in identity from A to C for the Virgaviridae TLSVals might have a slightly negative impact on valylation while greatly improving reverse transcription efficiency. Hence, variations in the TLSVal evolve to match the specific needs of the virus, while maintaining the ability to interact with important host factors.
Concluding remarks
Consensus models based on the full set of RNAs of a given class help give a full understanding of the key characteristics necessary for function, contextualize the characteristics of each member, reveal the presence of interesting structural or functional variations, and suggest how evolution can fine-tune structure. Here, we built on the list of known TLSVals and also verified the presence and structure of interesting variants, including those with insertions, altered anticodons and in insect-infecting viruses. These discoveries open new questions regarding changes in function or advantage to the virus conferred by these differences as well as guide studies aimed at understanding the mechanisms of events such as translation enhancement and RdRp recruitment. Finally, observing the variations seen here underscores the possibility that other classes of tRNA-like structures outside of the known TLSVal, TLSHis, or TLSTyr remain to be identified, with architectures sufficiently different as to not appear in homology searches.
MATERIALS AND METHODS
Bioinformatic searches
A preliminary alignment of all known TYMV-like TLSs (Rfam ID: RF00233) was obtained from the Rfam database (Kalvari et al. 2018). Additional examples of this class of RNAs were identified using Infernal version 1.1 (Nawrocki and Eddy 2013) to query a compilation of all viral sequence reads in the National Center for Biotechnology Information (NCBI) nucleotide database (downloaded 01/22/2019). Iterative searches were performed and adjustments were made to the alignment based on information from the high-resolution crystal structure of the TYMV TLS (Colussi et al. 2014; Hartwick et al. 2018) as well as previously reported secondary structure models for the PCV TLS RNAs (Goodwin and Dreher 1998). Duplicate sequences were removed, resulting in 108 unique sequences derived from 46 unique viruses. The consensus sequence and secondary structure model as well as the statistical analysis of covariation were calculated using the RNA Significant Covariation Above Phylogenetic Expectation (R-scape) program (Rivas et al. 2017, 2020) visualized using R2R (Weinberg and Breaker 2011) and labeled in Adobe Illustrator.
RNA preparation
DNA templates were produced either by PCR amplification from a double-stranded DNA gene block fragment (Integrated DNA Technologies) or by combining two overlapping single-stranded DNA oligonucleotides (IDT) and using a SuperScript II Reverse Transcriptase (Invitrogen) reaction, according to manufacturer protocol, to fill in the remaining template. All DNA oligonucleotide sequences used in this study can be found in Supplemental Table S1. Reverse primers used to amplify the constructs for in vitro aminoacylation assays contained two 5′-terminal 2′OMe-modified bases to achieve a higher yield with the correct 3′ end of the construct, since the 3′ terminal adenosine nucleotide is the site of modification. All RNA oligonucleotides used in this study were produced by in vitro transcription using templates produced by PCR or RT. Each 200 µL transcription reaction contained: 30 mM Tris pH 8.0, 60 mM MgCl2, 8 mM each NTP, 10 mM DTT, 0.1% spermidine, 0.1% Triton X-100, and T7 RNA polymerase. Transcription reactions were incubated at 37°C overnight then purified by denaturing 10% PAGE. Gels were visualized by UV illumination and the gel piece containing the band of correct length was excised. Gel pieces were sliced into small pieces and soaked in ∼300 µL of diethylpyrocarbonate (DEPC)-treated milli-Q filtered water (Millipore) at 4°C overnight to elute the RNA. The supernatant containing RNA was subjected to ethanol precipitation then resuspended in DEPC-treated water and diluted to the appropriate concentration.
Protein expression and purification
The ValRS enzyme from T. thermophilus used in the current study was recombinantly expressed and purified previously and details can be found in the Materials and Methods section of Hartwick et al. (2018). The LeuRS enzyme from S. cerevisiae was purified for this study. LeuRS gene blocks (IDT) were cloned into a pET15b vector using Gibson ligation. The protein was expressed in BL21 (DE3) cells in LB containing ampicillin at 37°C until OD600 = 0.2–0.3, the temperature was reduced to 18°C and continued to grow until the OD600 = 0.6, then was induced with 0.25 mM IPTG overnight. The cells were harvested and spun at 5488g for 12 min at 6°C. The pellets were resuspended in lysis buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM β-mercaptoethanol, 5 mM MgCl2, 10% glycerol, and 1 mM EDTA-free protease inhibitor tablet (Roche), then sonicated for 2 min total processing time (20 sec on, 40 sec off). The lysate was centrifuged at 30,000g for 30 min at 4°C before being loaded into a gravity flow column (Bio-Rad) and purified by Ni-NTA resin (UBP Bio) followed by size exclusion using FPLC and a Sepax 300 SEC column. The protein was resuspended to 2 µM and stored in buffer containing 50 mM Tris-HCl pH 8.0, 5 mM MgCl2, 2 mM β-mercaptoethanol, and 5% glycerol.
Chemical probing of RNAs in vitro
Structure probing experiments using the chemical modifier N-methyl isatoic anhydride (NMIA) were performed according to the previously published protocol from reference: (Cordero et al. 2014). Briefly, RNAs were refolded by heating to 90°C for 3 min, cooled to room temperature, then modified by incubating for 20 min at room temperature with either NMIA or DMSO (final concentrations: 60 nM RNA, 3 mg/mL NMIA or DMSO, 50 mM HEPES-KOH pH 8.0, 10 mM MgCl2, 3 nM FAM-labeled DNA primer for reverse transcription, see Supplemental Table S1 for sequence). The probing reaction was quenched by adding NaCl (final concentration 500 mM) and Na-MES buffer (final concentration 50 mM, pH 6.0) and oligo(dT) magnetic beads (Invitrogen Poly(A)Purist MAG Kit), which hybridize to the poly(A) stretch contained in the RT DNA primer. Chemically modified RNAs were purified using the magnetic stand and washed with 70% ethanol. Reactions were resuspended in DEPC-treated water, then reverse transcription was performed using SuperScript III enzyme (Invitrogen). RNA ladders were produced by four separate reverse transcription reactions using ddNTPs. Reverse transcription reactions were incubated at 50°C for 45 min, then the RNA was degraded by adding NaOH (final concentration 200 mM), heating to 90°C for 5 min, then quenching with an acidic solution (final concentration: 250 mM sodium acetate pH 5.2, 250 mM HCl, 500 mM NaCl). The remaining DNA products were purified using the magnetic stand then washed with 70% ethanol. A solution containing HiDi formamide solution (ThermoFisher) and spiked with GeneScan 350 ROX Dye Size Standard (ThermoFisher) was added to elute DNA products from the magnetic beads. Labeled RT DNA products were analyzed by capillary electrophoresis using an Applied Biosystems 3500 XL system. Fragment size analysis, alignment, background subtraction, and normalization (based on reactivity in flanking stem–loop regions) were performed using the HiTrace RiboKit (https://ribokit.github.io/HiTRACE/) (Yoon et al. 2011; Kim et al. 2013; Kladwang et al. 2014; Lee et al. 2015) with MatLab (MathWorks), and figures were produced using RiboPaint (https://ribokit.github.io/RiboPaint/tutorial/) with Matlab subsequently labeled in Adobe Illustrator.
In vitro aminoacylation assays using 3H-labeled amino acids
In vitro-transcribed RNAs were resuspended in DEPC-treated water to 1 µM and refolded by incubating at 90°C for 3 min then cooling to room temperature. Aminoacylation reactions were set up by mixing 1 µL of RNA or water, 1 µL of standard buffer (10×: 300 mM HEPES-KOH pH 7.5, 20 mM ATP, 300 mM KCl, 50 mM MgCl2, 50 mM dithiothreitol), 1 µL 3H-labeled L-valine (60 Ci/mmol) or 3H-labeled L-leucine (100 Ci/mmol), 1 µL of ValRS or LeuRS enzyme (10×: 2 µM) and 6 µL of water (final volume = 10 µL). In additional experiments that attempted to optimize leucylation signal for the CPSbV TLS, the following conditions were altered: increased RNA concentration (1 µM RNA instead of 0.1 µM final concentration), increased Mg2+ and ATP concentration (10×: 40 mM ATP, 100 mM MgCl2), or completely altered buffer conditions according to previous studies (Goodwin and Dreher 1998)—IV buffer (10×: 25 mM Tris-HCl pH 8.0, 20 mM MgCl2, 10 mM ATP, 1 mM spermine) or TM buffer (10×: 300 mM HEPES-KOH pH 7.5, 1M potassium acetate, 25 mM magnesium acetate, 15 mM ATP). Aminoacylation reactions, each performed in triplicate, were incubated at 30°C for 2–3 h then immediately loaded onto a vacuum filter blotting apparatus. Filtering was achieved by using one layer each, ordered from bottom to top, of thick filter paper (BioRad gel dryer filter paper), HyBond positively charged membrane (GE Healthcare) and 0.45 µm Tuffryn membrane filter paper (PALL Life Sciences) and each layer was prewashed and equilibrated with a wash buffer (20 mM Bis-Tris pH 6.5, 10 mM NaCl, 1 mM MgCl2). After blotting, each reaction blot was washed five times with 200 µL of wash buffer that included trace xylene cyanol for visualization. After filtering and washing, the filters were dried, then the Hybond membrane was cut out, placed in a scintillation vial, and 3H incorporation was measured, taking two readings per sample, by a scintillation counter (Perkin-Elmer Tri-Carb 2910 TR). Data were analyzed and plotted using Microsoft Excel.
Modeling
A composite model of the TYMV 3′ UTR using both crystal structures for the complete TLS and UPD domains (PDB IDs: 4p5j [Colussi et al. 2014] and 6mj0 [Hartwick et al. 2018], respectively) was built using Coot (Emsley et al. 2010) as previously performed (Hartwick et al. 2018). Modeling and coloring of the TYMV 3′ UTR was performed using PyMOL (The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC) and Protein Data Bank (Berman et al. 2000; Burley et al. 2019) (PDB: rscb.org) deposited structures for ValRS (Fukai et al. 2000) (PDB ID: 1gax), eEF1A (Shao et al. 2016) (PDB ID: 5lzs), LeuRS (Fukunaga and Yokoyama 2005b) (PDB ID: 1wz2), and archaeal CCA-adding enzyme (Kuhn et al. 2015) (PDB ID: 4x4r). In each case, the TLS model was aligned to best match the tRNA present in each structure using PyMOL.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Kieft laboratory for many discussions and David Costantino and Dr. Quentin Vicens for critical reading of this manuscript. We thank Dr. Steve Bonilla Rosales for assistance with aminoacylation assays. This work was supported by National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) grant R35GM118070 to J.S.K. M.E.S. is a Jane Coffin Childs Postdoctoral Fellow. E.W.H. was a University of Colorado School of Medicine RNA BioScience Initiative Scholar.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.076968.120.
REFERENCES
- Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi P-Y, Nix JC, Beckham JD, Kieft JS. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 354: 1148–1152. 10.1126/science.aah3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M. 1993. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J Mol Biol 231: 219–229. 10.1006/jmbi.1993.1277 [DOI] [PubMed] [Google Scholar]
- Barends S, Bink HH, van den Worm SH, Pleij CW, Kraal B. 2003. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell 112: 123–129. 10.1016/S0092-8674(02)01256-4 [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. 10.1073/pnas.0308014101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res 28: 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski JJ, Kaesberg P. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321: 528–531. 10.1038/321528a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S. 2019. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 47: D464–D474. 10.1093/nar/gky1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. Elife 3: e01892 10.7554/eLife.01892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. 2014. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511: 366–369. 10.1038/nature13378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. 2005. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol 6: R70 10.1186/gb-2005-6-8-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero P, Kladwang W, VanLang CC, Das R. 2014. The mutate-and-map protocol for inferring base pairs in structured RNA. Methods Mol Biol 1086: 53–77. 10.1007/978-1-62703-667-2_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D, Seno T, Söll D. 1972. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci 69: 3063–3067. 10.1073/pnas.69.10.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. 2000. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J 19: 2351–2361. 10.1093/emboj/19.10.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiman B, Koenen A, Verlaan P, Pleij C. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J Virol 72: 3965–3972. 10.1128/JVI.72.5.3965-3972.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Doudna JA. 2001. Ribozyme structures and mechanisms. Annu Rev Biophys Biomol Struct 30: 457–475. 10.1146/annurev.biophys.30.1.457 [DOI] [PubMed] [Google Scholar]
- Dreher TW. 2009. Role of tRNA-like structures in controlling plant virus replication. Virus Res 139: 217–229. 10.1016/j.virusres.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW. 2010. Viral tRNAs and tRNA-like structures. Wiley Interdiscip Rev RNA 1: 402–414. 10.1002/wrna.42 [DOI] [PubMed] [Google Scholar]
- Dreher TW, Goodwin JB. 1998. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res 26: 4356–4364. 10.1093/nar/26.19.4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Tsai CH, Florentz C, Giege R. 1992. Specific valylation of turnip yellow mosaic virus RNA by wheat germ valyl-tRNA synthetase determined by three anticodon loop nucleotides. Biochemistry 31: 9183–9189. 10.1021/bi00153a010 [DOI] [PubMed] [Google Scholar]
- Dreher TW, Uhlenbeck OC, Browning KS. 1999. Quantitative assessment of EF-1α·GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem 274: 666–672. 10.1074/jbc.274.2.666 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B, Florentz C, Giege R, Westhof E. 1994. Solution structure of the 3′-end of brome mosaic virus genomic RNAs: conformational mimicry with canonical tRNAs. J Mol Biol 235: 508–531. 10.1006/jmbi.1994.1010 [DOI] [PubMed] [Google Scholar]
- Fukai S, Nureki O, Sekine SI, Shimada A, Tao J, Vassylyev DG, Yokoyama S. 2000. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103: 793–803. 10.1016/S0092-8674(00)00182-3 [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. 2005a. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol 12: 915–922. 10.1038/nsmb985 [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. 2005b. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J Mol Biol 346: 57–71. 10.1016/j.jmb.2004.11.060 [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S. 2006. Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J Mol Biol 359: 901–912. 10.1016/j.jmb.2006.04.025 [DOI] [PubMed] [Google Scholar]
- Giege R, Briand JP, Mengual R, Ebel JP, Hirth L. 1978. Valylation of the two RNA components of turnip-yellow mosaic virus and specificity of the tRNA aminoacylation reaction. Eur J Biochem 84: 251–256. 10.1111/j.1432-1033.1978.tb12163.x [DOI] [PubMed] [Google Scholar]
- Gil JF, Adams I, Boonham N, Nielsen SL, Nicolaisen M. 2016. Molecular and biological characterisation of two novel pomo-like viruses associated with potato (Solanum tuberosum) fields in Colombia. Arch Virol 161: 1601–1610. 10.1007/s00705-016-2839-2 [DOI] [PubMed] [Google Scholar]
- Goodwin JB, Dreher TW. 1998. Transfer RNA mimicry in a new group of positive-strand RNA plant viruses, the furoviruses: differential aminoacylation between the RNA components of one genome. Virology 246: 170–178. 10.1006/viro.1998.9193 [DOI] [PubMed] [Google Scholar]
- Gordon KH, Johnson KN, Hanzlik TN. 1995. The larger genomic RNA of Helicoverpa armigera stunt tetravirus encodes the viral RNA polymerase and has a novel 3'-terminal tRNA-like structure. Virology 208: 84–84. 10.1006/viro.1995.1132 [DOI] [PubMed] [Google Scholar]
- Gordon KH, Williams MR, Hendry DA, Hanzlik TN. 1999. Sequence of the genomic RNA of Nudaurelia β virus (Tetraviridae) defines a novel virus genome organization. Virology 258: 42–53. 10.1006/viro.1999.9677 [DOI] [PubMed] [Google Scholar]
- Haenni A, Prochiantz A, Bernard O, Chapeville F. 1973. TYMV valyl-RNA as an amino-acid donor in protein biosynthesis. Nat New Biol 241: 166–168. 10.1038/newbio241166a0 [DOI] [PubMed] [Google Scholar]
- Haenni A-L, Joshi S, Chapeville F. 1982. RNA-like structures in the genomes of RNA viruses. Prog Nucleic Acid Res Mol Biol 27: 85–104. 10.1016/S0079-6603(08)60598-X [DOI] [PubMed] [Google Scholar]
- Hall TC. 1979. Transfer RNA-like structures in viral genomes. Int Rev Cytol 60: 1 10.1016/S0074-7696(08)61257-7 [DOI] [PubMed] [Google Scholar]
- Hall TC, Shih D, Kaesberg P. 1972. Enzyme-mediated binding of tyrosine to brome-mosaic-virus ribonucleic acid. Biochem J 129: 969–976. 10.1042/bj1290969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Rambo RP, Filbin ME, Kieft JS. 2009. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA 15: 294–307. 10.1261/rna.1360709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Rambo RP, Kieft JS. 2010. Multi-domain packing in the aminoacylatable 3′ end of a plant viral RNA. J Mol Biol 399: 450–463. 10.1016/j.jmb.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwick EW, Costantino DA, MacFadden A, Nix JC, Tian S, Das R, Kieft JS. 2018. Ribosome-induced RNA conformational changes in a viral 3′-UTR sense and regulate translation levels. Nat Commun 9: 1–12. 10.1038/s41467-018-07542-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y-M. 1997. Discriminating among the discriminator bases of tRNAs. Chem Biol 4: 93–96. 10.1016/S1074-5521(97)90252-0 [DOI] [PubMed] [Google Scholar]
- Joshi S, Chapeville F, Haenni A-L. 1982. Length requirements for tRNA-specific enzymes and cleavage specificity at the 3′ end of turnip yellow mosaic virus RNA. Nucleic Acids Res 10: 1947–1962. 10.1093/nar/10.6.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46: D335–D342. 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Cordero P, Das R, Yoon S. 2013. HiTRACE-Web: an online tool for robust analysis of high-throughput capillary electrophoresis. Nucleic Acids Res 41: W492–W498. 10.1093/nar/gkt501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Mann TH, Becka A, Tian S, Kim H, Yoon S, Das R. 2014. Standardization of RNA chemical mapping experiments. Biochemistry 53: 3063–3065. 10.1021/bi5003426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C-D, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. 2015. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160: 644–658. 10.1016/j.cell.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim H, Tian S, Lee T, Yoon S, Das R. 2015. Automated band annotation for RNA structure probing experiments with numerous capillary electrophoresis profiles. Bioinformatics 31: 2808–2815. 10.1093/bioinformatics/btv282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar S, Guilley H, Dollet M, Richards K, Jonard G. 1993. Nucleotide sequence and genetic organization of peanut clump virus RNA 2 and partial characterization of deleted forms. Virology 195: 33–41. 10.1006/viro.1993.1343 [DOI] [PubMed] [Google Scholar]
- Mans RM, Pleij CW, Bosch L. 1991. tRNA-like structures: structure, function and evolutionary significance. Eur J Biochem 201: 303–324. 10.1111/j.1432-1033.1991.tb16288.x [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. 2004. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 321: 36–46. 10.1016/j.virol.2003.10.023 [DOI] [PubMed] [Google Scholar]
- Matsuda D, Dreher TW. 2007. Cap-and initiator tRNA-dependent initiation of TYMV polyprotein synthesis by ribosomes: evaluation of the Trojan horse model for TYMV RNA translation. RNA 13: 129–137. 10.1261/rna.244407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Dunoyer P, Hemmer O, Fritsch C, Dreher TW. 2000. The valine anticodon and valylatability of Peanut clump virus RNAs are not essential but provide a modest competitive advantage in plants. J Virol 74: 8720–8725. 10.1128/JVI.74.18.8720-8725.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. 2017. Riboswitch diversity and distribution. RNA 23: 995–1011. 10.1261/rna.061234.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öberg B, Philipson L. 1972. Binding of histidine to tobacco mosaic virus RNA. Biochem Biophys Res Commun 48: 927–932. 10.1016/0006-291X(72)90697-3 [DOI] [PubMed] [Google Scholar]
- Osman T, Hemenway C, Buck K. 2000. Role of the 3′ tRNA-like structure in tobacco mosaic virus minus-strand RNA synthesis by the viral RNA-dependent RNA polymerase in vitro. J Virol 74: 11671–11680. 10.1128/JVI.74.24.11671-11680.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. 2011. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol 7: e1002031 10.1371/journal.pcbi.1002031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. 2006. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314: 1450–1454. 10.1126/science.1133281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinck M, Yot P, Chapeville F, Duranton H. 1970. Enzymatic binding of valine to the 3′ end of TYMV-RNA. Nature 226: 954–956. 10.1038/226954a0 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Fernández IS. 2018. Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES. Elife 7: e34062 10.7554/eLife.34062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij CW, Rietveld K, Bosch L. 1985. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res 13: 1717–1731. 10.1093/nar/13.5.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Van Poelgeest R, Pleij CW, Van Boom J, Bosch L. 1982. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res 10: 1929–1946. 10.1093/nar/10.6.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Pleij C, Bosch L. 1983. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J 2: 1079–1085. 10.1002/j.1460-2075.1983.tb01549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nat Methods 14: 45–48. 10.1038/nmeth.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2020. Estimating the power of sequence covariation for detecting conserved RNA structure. Bioinformatics 36: 3072–3076. 10.1093/bioinformatics/btaa080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. 2014. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol 10: 56 10.1038/nchembio.1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Florentz C, Dreher T, Giege R. 1992. Efficient mischarging of a viral tRNA-like structure and aminoacylation of a minihelix containing a pseudoknot: histidinylation of turnip yellow mosaic virus RNA. Nucleic Acids Res 20: 1865–1870. 10.1093/nar/20.8.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Murray J, Brown A, Taunton J, Ramakrishnan V, Hegde RS. 2016. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167: 1229–1240.e1215. 10.1016/j.cell.2016.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27: 135–145. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, Dreher TW. 1998. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and-CCR-boxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA 4: 1083–1095. 10.1017/S1355838298980694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg A-L, Akiyama BM, Costantino DA, Sit TL, Nix JC, Kieft JS. 2018a. A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc Natl Acad Sci 115: 6404–6409. 10.1073/pnas.1802429115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckelberg A-L, Vicens Q, Kieft JS. 2018b. Exoribonuclease-resistant RNAs exist within both coding and noncoding subgenomic RNAs. MBio 9: e02461-18 10.1128/mBio.02461-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C-H, Dreher TW. 1991. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J Virol 65: 3060–3067. 10.1128/JVI.65.6.3060-3067.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3 10.1186/1471-2105-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 35: 4809–4819. 10.1093/nar/gkm487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31 10.1186/gb-2010-11-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. 2015. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11: 606 10.1038/nchembio.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Lünse CE, Corbino KA, Ames TD, Nelson JW, Roth A, Perkins KR, Sherlock ME, Breaker RR. 2017. Detection of 224 candidate structured RNAs by comparative analysis of specific subsets of intergenic regions. Nucleic Acids Res 45: 10811–10823. 10.1093/nar/gkx699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Kim J, Hum J, Kim H, Park S, Kladwang W, Das R. 2011. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics 27: 1798–1805. 10.1093/bioinformatics/btr277 [DOI] [PubMed] [Google Scholar]
- Yot P, Pinck M, Haenni A-L, Duranton HM, Chapeville F. 1970. Valine-specific tRNA-like structure in turnip yellow mosaic virus RNA. Proc Natl Acad Sci 67: 1345–1352. 10.1073/pnas.67.3.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.