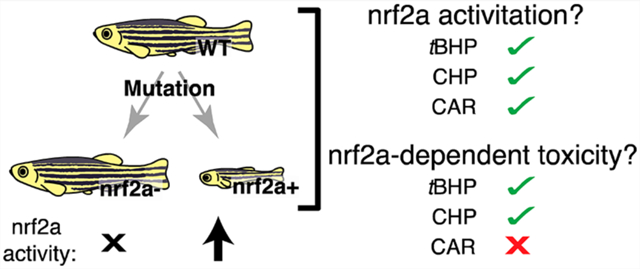
Abstract
The transcription factor Nrf2a induces a cellular antioxidant response and provides protection against chemical-induced oxidative stress, as well as playing a critical role in development and disease. Zebrafish are a powerful model to study the role of Nrf2a in these processes but have been limited by reliance on transient gene knockdown techniques or mutants with only partial functional alteration. We developed several lines of zebrafish carrying different null (loss of function, LOF) or hyperactive (gain of function, GOF) mutations to facilitate our understanding of the Nrf2a pathway in protecting against oxidative stress. The mutants confirmed Nrf2a dependence for induction of the antioxidant genes gclc, gstp, prdx1, and gpx1a and identified a role for Nrf2a in the baseline expression of these genes, as well as for sod1. Specifically, the 4-fold induction of gstp by tert-butyl hydroperoxide (tBHP) in wild type fish was abolished in LOF mutants. In addition, baseline gstp expression in GOF mutants increased by 12.6-fold and in LOF mutants was 0.8-fold relative to wild type. Nrf2a LOF mutants showed increased sensitivity to the acute toxicity of cumene hydroperoxide (CHP) and tBHP throughout the first 4 days of development. Conversely, GOF mutants were less sensitive to CHP toxicity during the first 4 days of development and were protected against the toxicity of both hydroperoxides after 4 dpf. Neither gain nor loss of Nrf2a modulated the toxicity of R-(−)-carvone (CAR), despite the ability of this compound to potently induce Nrf2a-dependent antioxidant genes. Similar to other species, GOF zebrafish mutants exhibited significant growth and survival defects. In summary, these new genetic tools can be used to facilitate the identification of downstream gene targets of Nrf2a, better define the role of Nrf2a in the toxicity of environmental chemicals, and further the study of diseases involving altered Nrf2a function.
Graphical Abstract
INTRODUCTION
Perturbations in reductive/oxidative balance are essential for redox signaling and thus for life, but excessive oxidant challenge is a major mechanism of cellular damage resulting from an imbalance of reactive oxygen species and antioxidant defenses.1 This resulting cellular oxidative stress (OS) is implicated in the etiology of a wide range of human diseases from cancer2,3 to heart disease4–6 to neurodegenerative disorders.7 It is also a significant mechanism of toxicity due to chemical exposure.8–11 Antioxidants directly play a major role in reducing OS toxicity, and a primary cellular antioxidant pathway is regulated by the transcription factor NRF2 (NF-E2 p45-related Factor 2, encoded by the gene NFE2l2 in humans; reviewed in refs 12 and 13). Under homeostatic conditions, the NRF2 protein is constantly translated but bound by the regulatory protein KEAP1 in the cytoplasm14,15 where it is polyubiquitinated by associated proteins and targeted for degradation.16–18 Under conditions of oxidative stress, this process is inhibited.19,20 As a result, newly synthesized NRF2 may escape degradation and translocate to the nucleus and heterodimerize with sMAF proteins to bind antioxidant response elements (AREs) within promoter sequences to drive transcription of target genes.21–23
The NRF2/ARE response pathway is well-conserved in living organisms from humans to Drosophila fruit flies24 and C. elegans worms25 (and reviewed in ref 26). The model fish Danio rerio (zebrafish) serves as a relevant model to study the structure and function of the NRF2 pathway, particularly in response to chemical exposure, due to high fecundity and rapid development. Furthermore, ease of genetic manipulation makes zebrafish a powerful system to test the genetics of responses to chemicals. Zebrafish have an intact NRF2 pathway, including the NRF2 paralog Nrf2a (encoded by the gene nfe2l2a),27 Keap1a and b,28,29 as well as a conserved set of antioxidant response genes. Two manipulations have been used to study the Nrf2a pathway in zebrafish, including the nfe2l2afh318 hypomorphic allele caused by a single amino acid change in the DNA binding domain of Nrf2a,30 and knockdown of endogenous nfe2l2a transcript by morpholino.9,31 However, both of these tools provide incomplete elimination of Nrf2a activity. The nfe2l2afh318-derived protein retains some DNA-binding activity,30,32 and morpholino effectiveness is limited to the first 4–5 days of development.33 Furthermore, both of these tools allow study on the reduction of Nrf2a activity, but not the increase of that activity, a question of great significance to a number of human cancers (reviewed in ref 34). This increase in activity results from elimination of KEAP1 regulation, either by mutation in KEAP1 itself or by deletion or mutation of the regulatory domains in NRF2 by which KEAP1 binds.
In the present study, we took advantage of the targeting power of CRISPR gene editing tools to create both fully null (loss of function, LOF) and hyperactive (gain of function, GOF) Nrf2a lines in zebrafish. Once developed, we utilized these tools to better understand downstream targets of Nrf2a in zebrafish, as well as to characterize the induction response by model oxidative stressors. We further tested the hypothesis that the potency of Nrf2a induction response by three model oxidative stressors would predict acute toxicity of exposure to these compounds.
EXPERIMENTAL PROCEDURES
Fish Care.
All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Washington. Adult zebrafish of the outbred Ekkwill (EKW) strain were housed in a recirculating system at 27 ± 1 °C, on a 14h:10h light:dark cycle. Adult fish were fed 2% of their body weight in flake food (84% AquaTox flake (Zeigler Brothers), 7% Calafin (Argent Aquaculture), 9% 300–500 μm Golden Pearls (Brine Shrimp Direct) per day, and received live newly hatched Artemia nauplii as a supplement at least once daily. Fish were allowed to spawn naturally at the beginning of the daily light cycle. Embryos were collected within several hours of fertilization, sorted for proper development and for developmental stage, and housed in E3 embryo medium (0.1 mM NaCl, 3.4 μM KCl, 6.6 μM CaCl2, 6.6 μM MgSO4, pH 7.2).
Genomic Mutagenesis.
The cas9 nuclease derived from Streptococcus pyogenes was used for RNA-guided genome editing of nfe2l2a. Two sites within Exon 2 were targeted simultaneously (site 1: GGATCTGGGCGCGGGCCGTGagg; site 2: ccgCGCAGCACACCGCTCACAC; PAM lower case). Targets were selected using the CRISPRscan algorithm.35 The corresponding single-guide RNA (sgRNA) templates were created by cloning annealed oligonucleotides into pT7-gRNA (gift from Wenbiao Chen, Addgene plasmid # 4 6 7 5 9) as described3,6 (site 1 pair: tag-gATCTGGGCGCGGGCCGTG and aaacCACGGCCCGCGCCCAGAT; site 2 pair: taggGGTGTGAGCGGTGTGCTGCG and aaacCGCAGCACACCGCTCACACC). After sequence confirmation, sgRNAs were transcribed in vitro using MEGAshortscript kit (ThermoFisher) and purified by phenol extraction and ethanol precipitation, then refolded by heating to 98 °C and slow cooling to room temperature.37
Fertilized EKW embryos were collected from spawning tanks within 1 h postfertilization and injected at the one-cell stage using an MPPI-2 pressure injector (ASI). Embryos were injected with sgRNAs in one of two combinations: as a ribonucleoprotein complex with cas9 protein (PNA Bio) (300 mM KCl, 500 ng/μL cas9 protein, 125 ng/μL sgRNA#1, 125 ng/μL sgRNA#2);38 or with cas9 mRNA in vitro transcribed from pCS2-nCas9n (Addgene plasmid #47929)36 with mMESSAGE mMACHINE kit (ThermoFisher; 100 ng/μL sgRNA#1, 100 ng/μL sgRNA#2, 600 ng/μL cas9 mRNA, 500 mM KCl).
Genotyping and Stock Maintenance.
Genomic DNA was obtained by sodium hydroxide extraction from tail biopsies of juvenile fish.39 This was screened by PCR using primers flanking the target exon (FP1: GTGCAGCCCTAGTGTGTGATG; RP1: GGCATTTGTGTCAAATCTCACAGG). Amplicons were then sequenced directly to determine genomic edits that would lead to loss of function alleles or large in-frame deletion alleles. Germline transmission approached 100% in F0 fish screened.
Once the mutant lines were identified, they were maintained as heterozygous stocks by outcrossing to wild type EKW. Carriers of mutant alleles were identified by PCR amplification and melt curve analysis on a CFX Connect (Bio-Rad) real-time PCR machine; melt curves were analyzed using the Precision Melt Analysis Program (Bio-Rad). Carriers of nfe2l2aw210 were identified using FP1 and RP2 (AGCTGAAGTCGAACACCTCA). Carriers of nfe2l2aw211 were identified using FP2 (AGGAGCAGGAGAAGACACTG) and RP1). Carriers of nfe2l2aw212, nfe2l2adw213, and nfe2l2adw214 were identified using FP1 and RP1. For out-of-frame deletion alleles (nfe2l2aw210, nfe2l2aw211, and nfe2l2aw212), paired homozygous mutant and wild type stocks were generated by incrossing heterozygous stocks and were genotyped as described above. The paired homozygous stocks were then incrossed to produce embryos for subsequent experiments. For nfe2l2adw213, paired homozygous stocks were generated in the same way but the few homozygous mutant fish that reached sexual maturity were all crossed to homozygous wild type siblings for experiments.
RNA Isolation and cDNA Synthesis.
RNA was obtained from whole 1 dpf homozygous embryos using the RNeasy kit (Qiagen). Gene-specific cDNA was generated by reverse transcribing with SuperScript III (Invitrogen) using a primer for the 3′ end of the nfe2l2a transcript (GCTAGATATTCTTCACAAGAGTC). cDNA was used as a template for PCR amplification (FP: GAGACATGATGGAGATTGAAATGTC; RP: GCTAGATATTCTTCACAAGAGTC) and then these amplicons were direct sequenced to confirm effects of genomic mutations on RNA transcripts.
Chemical Exposures.
These experiments build on our previous study that examined the comparative effects of a number of compounds from the EPA ToxCast database on oxidative damage in zebrafish and fathead minnow.40 As part of that work, we characterized a subset of seven industrial compounds for their effects on zebrafish antioxidant gene expression (Figure S1). Of the seven compounds analyzed in that study, three compounds elicited particularly strong Nrf2a activation responses, including R-(−)-carvone (CAR, Sigma-Aldrich), cumene hydroperoxide (CHP, Thermo-Fisher), and tert-butyl hydroperoxide (tBHP, VWR International). Accordingly, these compounds were selected to conduct a more detailed analysis on the role of the Nrf2a mutations in chemical-induced changes in antioxidant gene expression and survivorship. Chemical exposures followed the Fish Embryo Test from the Organization for Economic Cooperation and Development (FET OECD no. 236)41 with minor modifications to accommodate the large number of embryos for the study. Exposures were carried out in 20 mL glass scintillation vials (Kimball-Chase). Groups of embryos were added to 10 mL of chemical solution in each vial starting at 30–50% epiboly (roughly 5 hpf)42 and were maintained in these vials with a daily 90% water change at 28 °C until 96 hpf. LC50 concentrations for wild type fish exposed to CAR, CHP, and tBHP were reported in our previous paper.40
LC50 concentration comparisons for nfe2l2aw211, heterozygous nfe2l2adw213, and wild type controls were determined using the same 5 hpf-96 hpf exposure regime, exposing three replicates of 10 embryos each to five concentrations, each diluted 2-fold from the one before, plus a negative or solvent control. Measurements of CAR toxicity were repeated for confirmation, and these two separate experiments were combined for analysis. The U.S. Environmental Protection Agency’s Toxicity Relationship Analysis Program (TRAP), version 1.30a43 was used to calculate LC50 values.
To assess toxicity after 4 dpf, three replicates of 10 larvae each were exposed to intended concentrations of CHP and tBHP in 100 mm Petri dishes, 25 mL of chemical solution per dish. Survival was measured after 24 h (for tBHP) or 48 h (for CHP) since no acute toxicity was observed at 24 h for CHP. CAR exposure was not lethal to larvae of any genotype after 4 dpf up to the limit of solubility in embryo medium, so the effect of Nrf2a function on CAR toxicity after 4 dpf could not be assessed.
Gene Expression Analysis.
All gene expression was analyzed using QuantiGene Plex (ThermoFisher), a multiplexed branched DNA quantification assay that can be carried out on homogenized fish tissue without the need for RNA isolation or cDNA synthesis, as previously described.44 Gene expression analysis was carried out using either the full panel of antioxidant response gene probes described in ref 44 (Design ID M17013009) or a screening panel containing probes for gstp and actb1. Expression of the targeted genes of interest was normalized to the reference gene(s) present in the panel used: the geometric mean of three reference genes for the full panel or actb1 for the screening panel.
For analysis of nfe2l2a out-of-frame mutant and wild type gene expression (n = 5 biological replicates, 5 larvae per replicate), zebrafish were exposed from 5 hpf to 96 hpf to an intended concentration of 20% LC50 of each chemical and negative or solvent control. Larvae from each replicate were euthanized and then homogenized in 100 μL of homogenization buffer plus 1 μL of Proteinase K and then diluted 1:9 with homogenization buffer before use in the assay. For in-frame deletion alleles (nfe2l2adw213 and nfe2l2adw214), incrossed heterozygous clutches were used in gene expression experiments (n = 5 biological replicates, 12 larvae per replicate). Control fish were photographed for measurement of standard length,45 and then all euthanized larvae were divided into anterior and posterior portions with a vertical cut at the anus using a razor blade. Anterior (head/body) portions were stored in RNAlater (ThermoFisher), while posterior (tail) portions were individually genotyped as described in Genotyping and Stock Maintenance. Anterior portions of 1–3 larvae per replicate were pooled by genotype and homogenized in 100 μL of homogenization buffer plus 1 μL of Proteinase K per larva in the sample. Wild type samples were diluted to a concentration of 1 fish per 200 μL (the same final concentration used above). Heterozygous and homozygous in-frame deletion mutant samples were diluted to a concentration of 1 fish per 1000 μL with homogenization buffer before analysis; reference gene measurements in each sample allowed direct comparison with less-diluted wild type samples.
Statistical Analysis.
All statistical analysis was completed using GraphPad Prism version 6. Data are expressed as mean ± SEM with sample sizes noted for each experiment. Relative–fold change in gene expression was calculated by dividing the average normalized values of the treated samples by the average normalized value of the control samples. The effects of chemical exposure on gene expression were assessed using two-way ANOVA followed by Sidak’s correction for multiple comparisons. The effects of genotype on gene expression were assessed using two-way ANOVA followed by Dunnett’s correction for multiple comparisons. Treatment-related effects were considered significant at p < 0.05.
RESULTS
Generation and Characterization of Novel Nrf2a Mutant Zebrafish Lines.
We used CRISPR to generate Nrf2a-null and Nrf2a-hyperactive zebrafish by simultaneously targeting two different locations in exon 2 of the nfe2l2a gene (Figure S2a). This approach resulted in small mutations at either guide site as well as large deletions that spanned the two sites. Screens of outcrossed F1 offspring from CRISPR-injected fish identified five mutant alleles from separate carriers, which we name nfe2l2aw210, nfe2l2aw211, nfe2l2aw212, nfe2l2adw213, and nfe2l2adw214. The cDNA sequences from the first three lines revealed mutations that result in frame shifts and premature stop codons. Sequences from the last two lines revealed large in-frame deletions that remove negative regulatory domains while leaving the remainder intact (Figures 1a and S2b).
Figure 1.
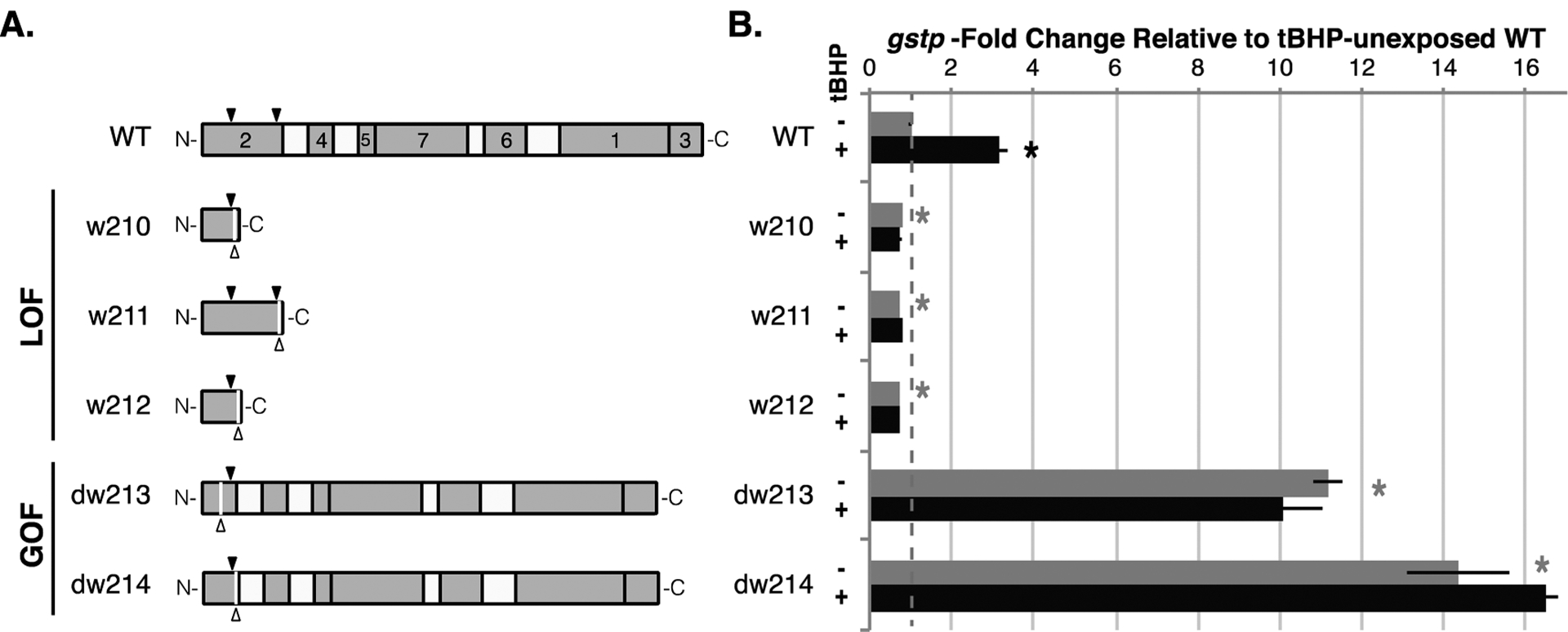
Independently isolated nfe2l2a mutant alleles encode null (LOF) or hyperactive (GOF) proteins. (a) Schematic of the wild type (WT) Nrf2a protein showing all canonical Neh domains (light gray, numbers 1–7) and keap1 contact sites in Neh 2 (black arrowheads above); schematics of the proteins encoded by each mutant allele showing deletion sites (white arrowheads below) and remaining portions of the WT protein (see Figure S2 for complete sequences). (b) Expression of the known Nrf2a-dependent gene gstp confirms effect of nfe2l2a mutation state on Nrf2a protein function: gstp is induced in WT larvae exposed to tBHP from 5 hpf to 96 hpf with daily renewal; baseline expression of gstp is altered in nfe2l2a mutants (lower in putative LOF mutants and higher in putative GOF mutants) and exposure to tBHP has no effect on these altered levels of expression. Unexposed WT expression is set to 1.0 (red dashed line). All expression data represent the mean ± SEM (n = 5 biological replicates, 5 larvae per replicate for WT and LOF alleles, 1–3 larvae per replicate for GOF alleles). Asterisks indicate significantly different expression (p < 0.001) relative to WT control.
We tested the functional effect of these five mutations, measuring expression of the known Nrf2a-dependent gene gstp46 at both baseline (unexposed) and activated Nrf2a (tBHP-exposed) levels in 4 dpf fish of each genotype (Figure 1b). Wild type fish showed induction of gstp expression following tBHP exposure as expected. In contrast, fish homozygous for all three out-of-frame mutations were refractory to gstp induction by tBHP, consistent with a lack of the transcription factor Nrf2a, and significantly lower baseline gstp expression than wild type fish. Conversely, fish homozygous for both in-frame mutations had significantly higher baseline gstp expression consistent with high Nrf2a activity in the absence of exogenous oxidative stress, and gstp was not further induced by tBHP exposure consistent with Nrf2a freed from negative regulation by Keap1 (Figure 1b). Based on these results we concluded that the three out-of-frame mutations were all null alleles, and that the two in-frame mutations were both hyperactive alleles. Moreover, these consistent observations in independently isolated lines established that the phenotypes observed were due to the identified mutations in nfe2l2a. We refer to the mutants as loss-of-function (LOF) and gain-of-function (GOF) for simplicity.
Phenotypic effects of nfe2l2a mutation differed dramatically between LOF and GOF alleles. None of the null nfe2l2a alleles had a negative effect on adult growth, survival, or fecundity under normal laboratory conditions, as reported for both NRF2-downregulated or -null mice,23,47 fruit flies,48 and zebrafish.30,49 All LOF alleles showed Mendelian ratios in adults, and homozygous LOF fish reached maturity at the same rate and attained the same size as their wild type siblings (Table 1). In contrast, the GOF alleles resulted in growth and survival deficits in the fish that carried them. Both GOF alleles displayed genotype frequencies consistent with Mendelian inheritance at embryonic and early larval stages, as well as embryonic growth and development indistinguishable from that of wild type fish (Table 1), but these similarities did not persist beyond larval stages. Fish homozygous for nfe2l2adw213 rarely reached 2 months of age, and only a few homozygous females ever grew large enough to spawn. Fish homozygous for nfe2l2adw214 died by 14 dpf, while even heterozygous carriers frequently reached no more than 10 mm standard length and only occasionally grew large enough to spawn. Those homozygous nfe2l2adw213 and heterozygous nfe2l2adw214 fish that did reach sexual maturity tended to die much sooner than wild type fish (data not shown). Furthermore, expression of gstp was significantly higher than wild type in both heterozygous nfe2l2adw213 (6.3-fold ±0.4, p < 0.0001) and nfe2l2adw214 (10-fold ±1, p < 0.0001) larvae, levels that were at least half of homozygous mutant expression from both lines (11.2-fold ±0.3, p < 0.0001; and 14-fold ±1, p < 0.0001, respectively; Figure 1b). These phenotypes are consistent with the dysregulation of Nrf2a even in heterozygous fish.
Table 1.
Effects of Zebrafish nfe2l2a Mutations on Genotype Frequency, Survival, and Growtha
| adults | larvae | ||||
|---|---|---|---|---|---|
| mutant line | genotype | genotype freq. | length | genotype freq. | length |
| w211 | WT/WT | 26.8% (n = 63) | 25 mm ±1 | ND | ND |
| mut/WT | 50.2% (n = 118) | 26 mm ±2 | ND | ND | |
| mut/mut | 23.0% (n = 54) | 25 mm ±1 | ND | ND | |
| dw213 | WT/WT | 36.2% (n = 125) | 30 mm ±1 | 21.7% (n = 15) | 3.6 mm ±0.1 |
| mut/WT | 59.7% (n = 206) | 30 mm ±2 | 47.8% (n = 33) | 3.6 mm ±0.1 | |
| mut/mut | 4.1% (n = 14) | 22 mm ±5 | 30.4% (n = 21) | 3.6 mm ±0.1 | |
| dw214 | WT/WT | 40.8% (n = 51) | 26 mm ±2 | 18.8% (n = 13) | 3.4 mm ±0.3 |
| mut/WT | 59.2% (n = 74) | 16 mm ±6 | 59.4% (n = 41) | 3.3 mm ±0.4 | |
| mut/mut | 0% (n = 0) | ND | 21.7% (n = 15) | 3.4 mm ±0.4 | |
ND: not determined. mut: mutant. Measurements from heterozygous incross clutches of each mutant line. The genotype frequency (raw numbers) and average standard length (±SD) of fish from each genotype as adults, and from both GOF lines as larvae. Metrics for adults were measured at 70 dpf (nfe2l2aw211 and nfe2l2adw214) or 160 dpf (nfe2l2adw213); metrics for larvae were measured at 4 dpf.
nfe2l2a Mutant Zebrafish Confirm Role of Nrf2a in Transcription of Several Oxidative Response Genes.
We selected the nfe2l2aw211 LOF line and the nfe2l2adw213 GOF line for subsequent experiments based on the size of the breeding populations available. For both mutant lines, we measured expression of a panel of antioxidant response genes44 to assess the necessity of Nrf2a activity in baseline transcription of each gene. Elimination of Nrf2a correlated significantly with lower baseline expression of gstp (0.77-fold, p < 0.05), as well as gclc (0.80-fold, p < 0.001), prdx1 (0.64-fold, p < 0.05), nqo1 (0.9-fold, p < 0.05), and sod1 (0.88-fold, p < 0.01) relative to wild type larvae (Figure 2a, Table S1). Conversely, GOF alleles correlated with higher baseline expression of gstp (12.6-fold, p < 0.001), as well as prdx1 (20-fold, p < 0.001), gclc (2.6-fold, p < 0.01), gpx1a (6.8-fold, p < 0.001), and nqo1 (1.9-fold, p <0.05) relative to wild type larvae (Figure 2a, Table S1).
Figure 2.
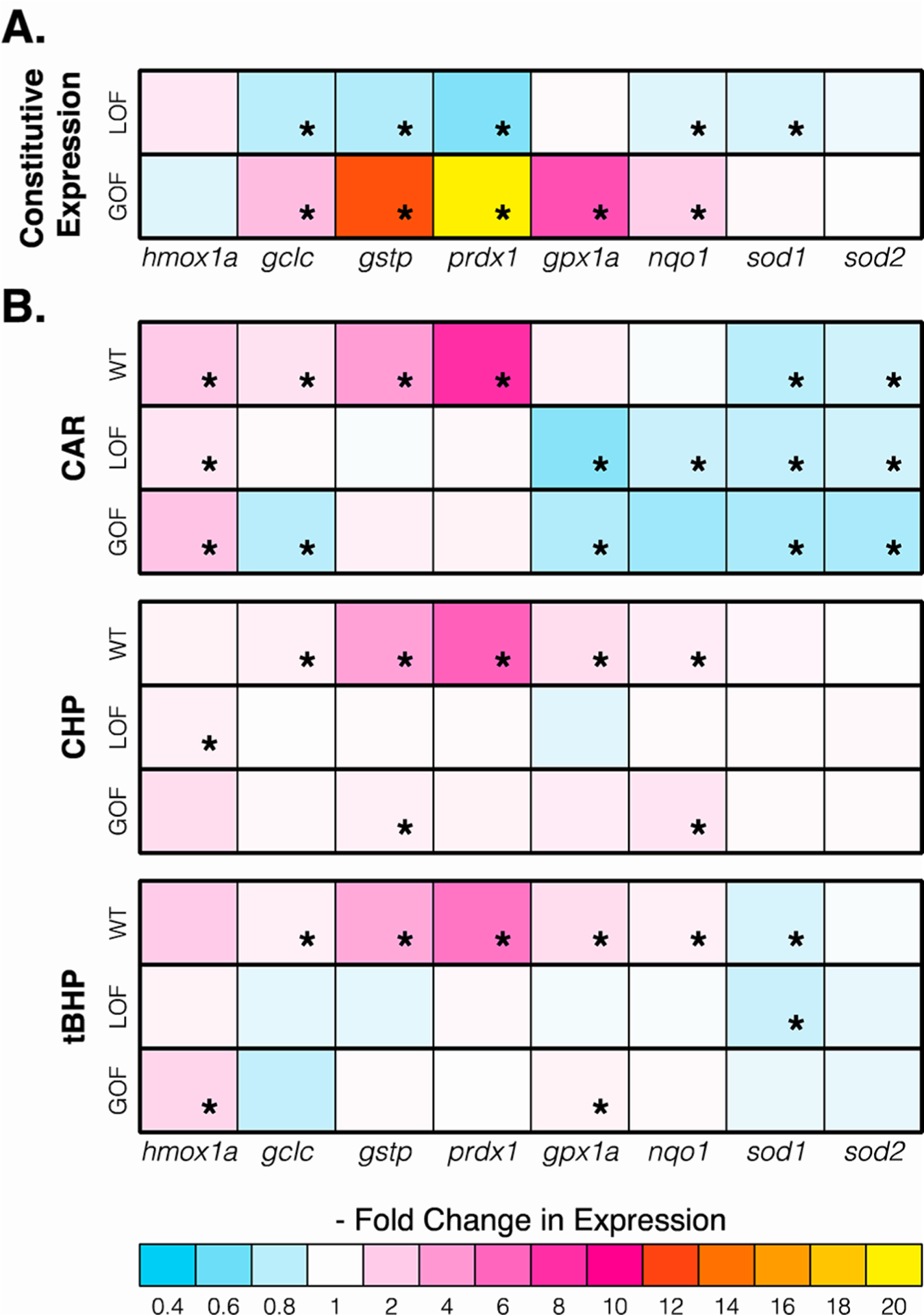
Altered Nrf2a function correlates with significant differences in expression of some, but not all, oxidative stress response genes. (a) Baseline expression of homozygous LOF (nfe2l2aw211) and homozygous GOF (nfe2l2adw213) relative to wild type (WT) larvae. (b) Expression of LOF, GOF, and WT following exposure to the same relative concentration (20% LC50) of three gstp-inducing compounds from 5 hpf to 96 hpf with daily renewal, relative to untreated larvae of the same genotype. Control expression in all cases is set to 1.0. All data represent the mean ± SEM (n = 5 biological replicates). Asterisks indicate conditions that result in significantly different expression (p < 0.05) from WT (a) or untreated (b) controls. See Table S1 for detailed–fold change and statistical significance values.
As observed in Figure 2b and Table S1, altered Nrf2a function largely eliminated the modulation in gene expression that all three compounds induced in wild type fish. Expression of gclc, gstp, and prdx1 in particular showed clear dependence on Nrf2a activity. Specifically, lack of Nrf2a eliminated the upregulation induced by all three chemicals, and constitutive Nrf2a activity eliminated nearly all further induction by chemical exposure. Induction of hmox1a, gpx1a, and nqo1 by the test compounds showed extensive variation in response to altered Nrf2a, suggesting joint transcriptional control of these genes by Nrf2a and other pathways. For example, induction of hmox1a by CAR exposure was reduced but not eliminated in either LOF fish (1.5-fold, p < 0.01) or GOF fish (2.3-fold, p <0.01), indicating that Nrf2a activation was responsible for some but not all of the CAR-induced hmox1a response. In contrast, the influence of chemical exposure on sod1 and sod2 genes was independent of nfe2l2a genotype. If an exposure resulted in decreased expression in wild type larvae, a similar result was observed in LOF and GOF larvae. Similarly, if expression in wild type larvae was unaffected by chemical exposure, the same was true of LOF and GOF larvae.
Effect of Nrf2a Mutation on Acute Toxicity of CHP, tBHP, and CAR.
Having established that Nrf2a plays a role in the antioxidant response of larval zebrafish to exposure by three model compounds, we tested the effect of Nrf2a activity and antioxidant gene expression on acute toxicity by both the hydroperoxides and the structurally dissimilar emerging contaminant CAR. We found that before 4 dpf, LOF larvae were more sensitive than wild type larvae to CHP (LC50 = 7.6 and 19 mg/L respectively; both survival curves were too steep to calculate SEM) and to tBHP (LC50 = 74 ± 3 and 155 ± 5 mg/L respectively). By contrast, there was no difference in CAR toxicity before 4 dpf in LOF larvae (LC50 = 35 ± 2 mg/L) relative to wild type larvae (37 mg/L; survival curve was too steep to calculate SEM; Figure 3a).
Figure 3.
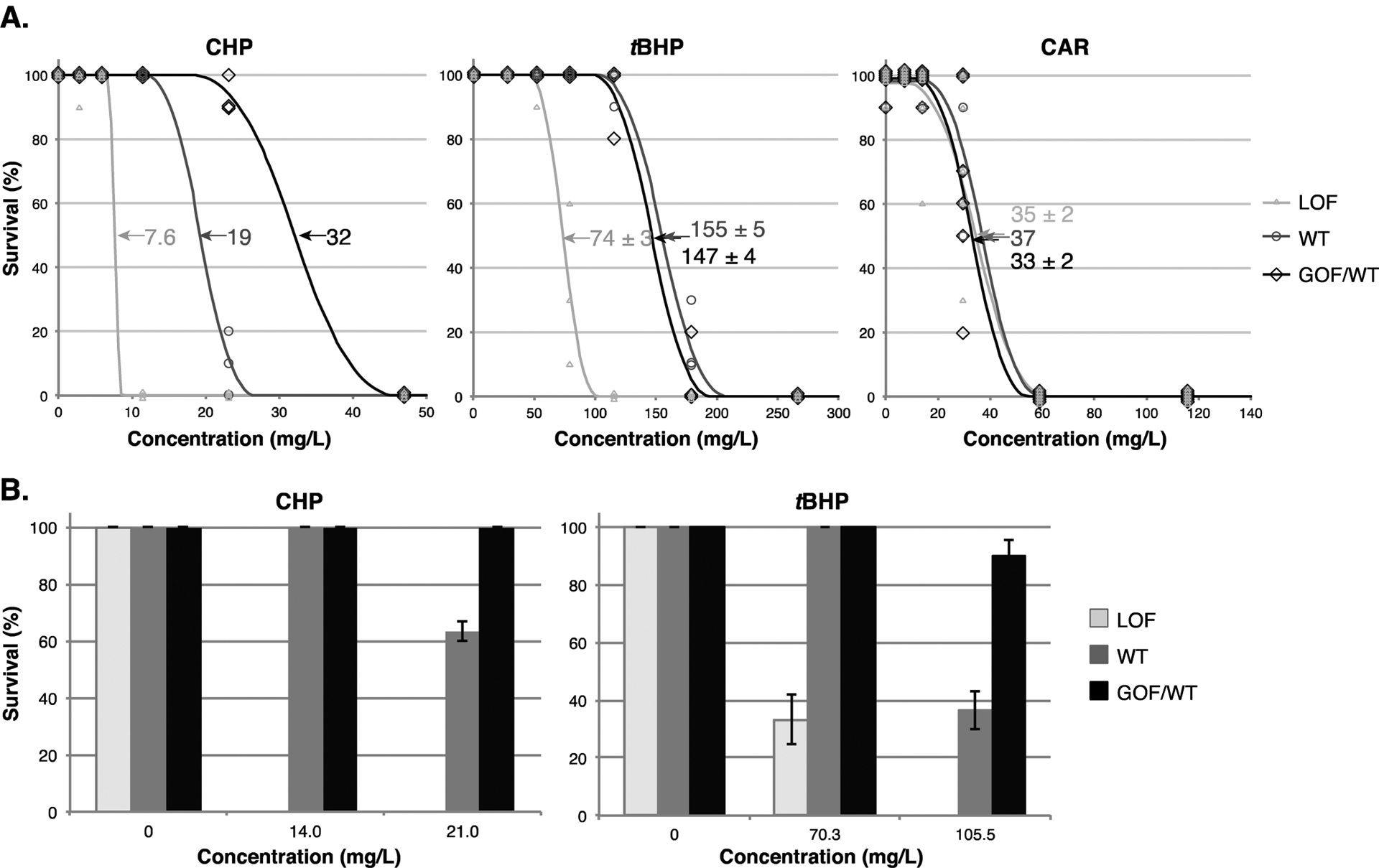
Mutation state of nfe2l2a correlates with chemical toxicity in larvae exposed to CHP and tBHP, but not those exposed to CAR. (a) LC50 exposures between 5 hpf and 96 hpf with daily renewal. Symbols show survival of larvae after exposure to varying concentrations of each chemical; lines show calculated toxicity curves; arrows and numbers indicate calculated LC50 values ± SEM (mg/L). (b) Survival of larvae in exposures starting at 4 dpf to CHP (for 48 h) and tBHP (for 24 h). In all experiments, genotypes are WT (gray), LOF (nfe2l2aw211, light gray), and GOF/WT (heterozygous nfe2l2adw213, black). For CAR, n = 6 biological replicates, 10 fish per replicate; for CHP and tBHP, n = 3 biological replicates, 10 fish per replicate.
To determine if the constitutive activity in GOF mutants provided protection from oxidative stress-mediated toxicity, we used heterozygous GOF (GOF/WT) larvae (offspring of homozygous mutant females crossed to wild type males), since only a few homozygous GOF fish reached sexual maturity. These GOF/WT larvae have higher gstp expression at 4 dpf than tBHP-exposed wild type fish at the same age (GOF/WT:6.3-fold, ± 0.4; tBHP-exposed wild type: 3.7-fold ±0.1). We found that GOF/WT larvae were less sensitive than wild type to CHP (32 mg/L; again, survival curve was too steep to calculate SEM), whereas there was no significant difference in toxicity between GOF/WT and wild type larvae for either tBHP or CAR in 5 hpf-96 hpf exposures (Figure 3b). However, because nfe2l2a transcription is quite low at 5 hpf,27 the constitutive activity associated with relatively low cellular Nrf2a protein levels may have been insufficient to provide protection against toxicity. We therefore compared toxicity to both hydroperoxides in wild type and Nrf2a mutant fish starting at 4 dpf, a time when nfe2l2a transcription is relatively high and Nrf2a activation provides protection from peroxide toxicity in wild type fish.30,49 At this developmental stage, our GOF/WT fish were less sensitive to both CHP and tBHP, and LOF larvae were more sensitive relative to wild type fish (Figure 3b).
DISCUSSION
Several studies targeting Nrf2a function utilizing morpholino knockdown, including from our laboratory, have advanced our understanding of this pathway in the context of chemical toxicity in zebrafish9,31,50 (and reviewed in ref 51). However, morpholino effectiveness is limited to roughly the first 5 days of development and may not totally abolish Nrf2a function, and accordingly, our LOF lines provide valuable tools to study the role of Nrf2a throughout zebrafish development. The new mutant lines also represent an improvement over the previously available hypomorphic nfe2l2afh318 allele30 which reduces but does not eliminate DNA binding by the transcription factor. Accordingly, fish carrying the aforementioned mutation show some but not all of the effects on Nrf2a-dependent gene expression that we measured in our LOF mutants, suggesting that even residual Nrf2a activity is sufficient to drive expression and induction of some Nrf2a-responsive genes.
For example, prdx1 baseline expression was unaffected in nfe2l2afh318 hypomorphic mutants and induction of prdx1 in H2O2-treated nfe2l2afh318 mutants was reduced but not eliminated compared to wild type fish.30 In contrast, we found that the prdx1 induction resulting from exposure to CAR, CHP, and tBHP was entirely eliminated in our LOF fish and that baseline expression was significantly reduced relative to controls. These data are consistent with results from a H2O2-treated null mutant reported by Yamashita and colleagues while this manuscript was in preparation.49 We were able to use our genetic lines in combination with Nrf2a-activating compounds identified in our previous study40 to provide a more robust approach to evaluate our assay panel containing purported Nrf2a-dependent, partially Nrf2a-dependent, and entirely Nrf2a-independent genes.44 Further, the lines allowed us to investigate the role of Nrf2a in the toxicity of two prototypical chemical oxidative stressors (tBHP and CHP) and a relatively understudied electrophilic emerging contaminant (CAR). Previous studies have shown that reduced Nrf2a activity increased the sensitivity of both zebrafish and mouse fibroblasts to peroxides27,30,31,52 but not to the superoxide-generating redox cycling pesticides paraquat and menadione.30,52 In contrast, loss of activity from the NRF2 orthologs CncC and SKN-1 increased the sensitivity of fruit flies and C. elegans respectively to paraquat,53,54 suggesting that care should be exercised in extrapolating species differences in Nrf2 status with respect to susceptibility to pro-oxidant chemicals that generate reactive oxygen species by different mechanisms. Relatedly, we found that complete loss of Nrf2a function increased sensitivity to the peroxides tBHP and CHP but not to CAR, despite similar potency of all three compounds to induce Nrf2a-dependent gene expression in wild type fish.
As opposed to the hydroperoxides tested in the present study, CAR is an electrophile due to of its enone functionality as a Michael acceptor.40 It is a terpenoid found in essential oils such as in seeds of caraway (Carum carvi), spearmint (Mentha spicata), and dill (Anethum graveolens). Carvone forms two enantiomers: R-(−)-carvone (or L-carvone, CAR) exhibits a minty odor similar to spearmint, whereas S-(+)-carvone (or D-carvone) exhibits an aroma similar to caraway seeds.55 Relatively large amounts of carvone enantiomers are consumed as food additives, although little information exists with regards to tissue or blood concentrations in humans. These compounds exhibit CNS-depressant effects in laboratory animals.56 The S-(+)-carvone enantiomer and similar essential oils also exhibit anticonvulsant activity in laboratory animals that has been associated with modulation of GABAergic neurotransmission and ion channels,57,58 but the R-(−)-carvone enantiomer does not have this anticonvulsant activity.56 CAR has also been used as a pesticide, feed additive, flavoring substance, component in certain cosmetics and personal care products, as well as to facilitate bioremediation of PCB-contaminated soils.59 Accordingly, this compound is of potential interest as an emerging environmental contaminant because little is known regarding its human health or environmental risk. The potent effects of CAR observed in the present study on both Nrf2a-related gene induction, as well as acute toxicity in zebrafish, indicate that CAR deserves further attention with respect to evaluation of environmental and human health risk.
In human cancers, dysregulation of NRF2 is correlated with increased drug resistance and poorer patient prognosis60–62 (and reviewed in ref 34). This dysregulation can result from removal or mutation of the domains by which KEAP1 binds NRF2 to keep it sequestered in the cytoplasm and targeted for degradation:15,63 the DLG and ETGE binding sites in the NRF2-ECH homology domain 2 (Neh2) of the NRF2 protein64,65 and the lysine residues between those sites that are ubiquitinated when NRF2 is targeted for degradation.16 Human and mouse NRF2 proteins have been shown to be regulated through other Neh domains as well. For example, mouse NRF2 function is inhibited by β-TrCP-mediated ubiquitination in Neh666,67 and/or by RXRa binding to Neh768 and transactivated by CBP binding to Neh4 and Neh5.69 As a result, our approach was to use CRISPR to simultaneously target two points in the Neh2 domain of zebrafish Nrf2a flanking the keap1a binding sites and screen targeted fish for large in-frame deletions of this region. We predicted that in-frame removal of much of the Neh2 domain would result in an Nrf2a protein maintaining other accessory protein interactions but with Keap1a-mediated regulation eliminated. These GOF fish exhibited baseline transcription of Nrf2a-responsive genes that far exceeded the level of those observed in wild type fish under conditions of significant toxicant-generated oxidative stress.
Interestingly, our heterozygous GOF/WT fish were more resistant to tBHP than their wild type siblings in exposures beginning at 4 dpf, but not at the earlier stage of development. This is consistent with previous reports that increased Nrf2a activity by sulforaphane exposure prior to peroxide challenge protects fish from toxicity after 4 dpf,30,49 as well as with protection provided by overexpression of the fruit fly ortholog CncC.53 The GOF/WT larvae further revealed a difference in the effects of the two hydroperoxides tested in that the additional Nrf2a activity provided protection from CHP exposure but not tBHP exposure prior to 4 dpf. These observations demonstrate the utility of our heterozygous GOF/WT mutant line as a valuable addition to study variation in the timing or strength of Nrf2a-dependent toxicity. While the mechanism underlying this developmental difference in Nrf2a overexpression between the two hydroperoxides tested needs further investigation, it is possible that a Keap1a-independent regulatory mechanism of nrf2a may be involved. Alternatively, differences in uptake of the two hydroperoxides and the efficiency of their reduction by proteins present at different points in embryonic and early larval development may be responsible.70
As discussed, increased Nrf2a activity in our zebrafish was correlated with reduced growth and survival of adult zebrafish. This observation contrasts with the increased lifespan of fruit flies with loss of Keap148,53 and also in C. elegans with increased expression of the NRF2 ortholog SKN-1,71 but is similar to the phenotype observed in homozygous KEAP1-null mice. The NRF2-hyperactive mice develop normally in utero, but die by 3 weeks of age from starvation, likely as a result of the hyperactive NRF2 effect on metabolic genes leading to hyperkeratosis in the esophagus.72,73 While the specific mechanism of the growth effects in our zebrafish are not known, the transcription factor Nrf2a plays an important role in cell signaling and development74,75 (and reviewed in ref 51), in addition to oxidative stress response, and substantial long-term changes in Nrf2a expression are likely to have effects on the expression and function of other important signaling pathways. For example, Nrf2a has been shown to influence the expression of other Nrf proteins like Nrf2a and Nfe276 and to upregulate Keap1,24,49,77 and it may therefore also increase the turnover of Nrf2b, which regulates a set of genes that only partially overlaps with the set controlled by Nrf2a and often functions as a transcriptional repressor.27,78 Nrf2a also competes with NF-κB for transcriptional cofactors79 so Nrf2a hyperactivity may result in reduced inflammatory response and immune function. Thus, caution should be used when relying on Nrf2a overexpressing mutants to assess inductive and protective responses to environmental compounds later in zebrafish development, and thus our comparative approach used both GOF and LOF models before GOF fish exhibit any negative effects of their genotype. Ultimately, the substantial variability in juvenile and adult growth rates within each GOF line observed herein suggest that the genetic diversity in the EKW background of these fish may allow identification of target genes and tissues that make hyperactive Nrf2a deleterious. Potential applications include using these mutants to examine the role of NRF2 in development and possibly cancer progression.
Of interest was the difference in phenotypic growth and survival effects, as well as differences in basal transcriptional activation activities exhibited by zebrafish carrying the two GOF alleles. For example, the nfe2l2adw214 mutation caused a much more potent induction of Nrf2a-responsive genes than did the nfe2l2adw213 mutation, and paradoxically this stronger transcriptional effect correlates with more dramatic lethality and reduced size in adult carriers. While it is possible that this difference is due to some residual Keap1a regulation of the Nrf2adw213 protein through its retained ETGE binding site, the chemical exposures failed to result in increased transcription of downstream genes. Thus, it is also possible that the difference in effect of the two GOF mutations is due to decreased stability of the Nrf2adw213 protein or to negative regulation by some Keap1a-independent mechanism.
In conclusion, the new zebrafish lines developed in the present study have enabled us to clarify downstream targets of nrf2a in zebrafish, and to clarify the role of Nrf2a in protecting against the toxicity of environmental chemicals. Our studies confirm dependence on Nrf2a for the inducibility of zebrafish genes gstp, prdx1, and gpx1a and suggest that other oxidative protective genes such as sod1 and sod2 may be less, or only partially, regulated by Nrf2a. Furthermore, studies with our GOF mutants confirmed how Nrf2a hyperactivity greatly modifies the expression of downstream antioxidant gene targets. Undoubtedly, this disruption of Nrf2a plays an important role in disease susceptibility as well as toxicological injury, as evidenced by the presence of drug-resistant cancers that show dysregulated NRF2. Finally, these tools may facilitate de novo sustainable molecular design, and the identification of industrial chemicals with reduced hazard to public and environmental health.40,80
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge the technical assistance of Patricia Janssen and Zahra Afshari with expression analysis. Madison Opp, Jamee Adams, Adele Lim, May Xie, Tammy Senior, Meghan Palmer, Yin Zhu, and Yoomin Jo provided excellent fish care.
Funding
Support for this study was provided by the U.S. National Science Foundation. [https://www.nsf.gov/] (CHE-1339637, the Molecular Design Research Network) and by the U.S. National Institutes of Health [https://www.nih.gov/] Super-fund research Program (P42-ES004696).
ABBREVIATIONS
- actb1
beta actin 1
- CAR
R-(−)-carvone
- CHP
cumene hydroperoxide
- dpf
days postfertilization
- EKW
Ekkwill wild type zebrafish strain
- gapdh
glyceraldehyde-3-phosphate dehydrogenase
- gclc
glutamate-cysteine ligase, catalytic subunit
- gpx1a
glutathione peroxidase 1a
- gstp
glutathione S-transferase pi
- hmox1a
heme oxygenase 1a
- hpf
hours postfertilization
- hprt1
hypoxanthine phosphoribosyltransferase 1
- nqo1
NAD(P)H quinone dehydrogenase 1
- prdx1
peroxiredoxin 1
- sod1
superoxide dismutase [Cu–Zn] or superoxide dismutase 1
- sod2
superoxide dismutase [Mn] or superoxide dismutase 2
- tBHP
tert-butyl hydroperoxide
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.9b00346.
Chemical structures, concentrations, and resulting gene expression changes in wild type fish for all chemicals used in the paper (Figure S1); detailed genomic and protein sequences for all mutant lines (Figure S2) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Sies H, Berndt C, and Jones DP (2017) Oxidative Stress. Annu. Rev. Biochem 86, 715–748. [DOI] [PubMed] [Google Scholar]
- (2).Jung B-J, Yoo H-S, Shin S, Park Y-J, and Jeon S-M (2018) Dysregulation of NRF2 in cancer: from molecular mechanisms to therapeutic opportunities. Biomol. Ther 26, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Toyokuni S (2016) The origin and future of oxidative stress pathology: From the recognition of carcinogenesis as an iron addiction with ferroptosis-resistance to non-thermal plasma therapy. Pathol. Int 66, 245–259. [DOI] [PubMed] [Google Scholar]
- (4).Chen QM, and Maltagliati AJ (2018) Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genomics 50, 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Montezano AC, Dulak-Lis M, Tsiropoulou S, Harvey A, Briones AM, and MD RMT (2015) Oxidative stress and human hypertension: vascular mechanisms, biomarkers, and novel therapies. Can. J. Cardiol 31, 631–641. [DOI] [PubMed] [Google Scholar]
- (6).Mimura J, and Itoh K (2015) Role of Nrf2 in the pathogenesis of atherosclerosis. Free Radical Biol. Med 88, 221–232. [DOI] [PubMed] [Google Scholar]
- (7).Calabrese V, Cornelius C, Dinkova-Kostova AT, Calabrese EJ, and Mattson MP (2010) Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid. Redox Signaling 13, 1763–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sun H, Brocato J, and Costa M (2015) Oral chromium exposure and toxicity. Curr. Envir Health Rpt 2, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang L, and Gallagher EP (2013) Role of Nrf2 antioxidant defense in mitigating cadmium-induced oxidative stress in the olfactory system of zebrafish. Toxicol. Appl. Pharmacol 266, 177–186. [DOI] [PubMed] [Google Scholar]
- (10).Williams CR, MacDonald JW, Bammler TK, Paulsen MH, Simpson CD, and Gallagher EP (2016) From the cover: cadmium exposure differentially alters odorant-driven behaviors and expression of olfactory receptors in juvenile coho salmon (Oncorhynchus kisutch). Toxicol. Sci 154, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Valavanidis A, Vlahogianni T, Dassenakis M, and Scoullos M (2006) Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf 64, 178–189. [DOI] [PubMed] [Google Scholar]
- (12).Sykiotis GP, and Bohmann D (2010) Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signaling 3, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tonelli C, Chio IIC, and Tuveson DA (2018) Transcriptional regulation by Nrf2. Antioxid. Redox Signaling 29, 1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, and Yamamoto M (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Canning P, Sorrell FJ, and Bullock AN (2015) Structural basis of Keap1 interactions with Nrf2. Free Radical Biol. Med 88, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhang DD, Lo SC, Cross JV, Templeton DJ, and Hannink M (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol 24, 10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, and Yamamoto M (2003) Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8, 379–391. [DOI] [PubMed] [Google Scholar]
- (18).Stewart D, Killeen E, Naquin R, Alam S, and Alam J (2003) Degradation of transcription factor Nrf2 via the ubiquitinproteasome pathway and stabilization by cadmium. J. Biol. Chem 278, 2396–2402. [DOI] [PubMed] [Google Scholar]
- (19).Sekhar KR, Rachakonda G, and Freeman ML (2010) Toxicology and Applied Pharmacology. Toxicol. Appl. Pharmacol 244, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kobayashi M, and Yamamoto M (2006) Nrf2–Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul 46, 113–140. [DOI] [PubMed] [Google Scholar]
- (21).Rushmore TH, Morton MR, and Pickett CB (1991) The antioxidant responsive element. J. Biol. Chem 266, 11632–11639. [PubMed] [Google Scholar]
- (22).Friling RS, Bensimon A, Tichauer Y, and Daniel V (1990) Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. U. S. A 87, 6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y-I (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun 236, 313–322. [DOI] [PubMed] [Google Scholar]
- (24).Lacher SE, Lee JS, Wang X, Campbell MR, Bell DA, and Slattery M (2015) Beyond antioxidant genes in the ancient Nrf2 regulatory network. Free Radical Biol. Med 88, 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Gacesa R, Dunlap WC, and Long PF (2015) Bioinformatics analyses provide insight into distant homology of the Keap1-Nrf2 pathway. Free Radical Biol. Med 88, 373–380. [DOI] [PubMed] [Google Scholar]
- (26).Fuse Y, and Kobayashi M (2017) Conservation of the Keap1-Nrf2 system: An evolutionary journey through stressful space and time. Molecules 22, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Timme-Laragy AR, Karchner SI, Franks DG, Jenny MJ, Harbeitner RC, Goldstone JV, McArthur AG, and Hahn ME (2012) Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2). J. Biol. Chem 287, 4609–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li L, Kobayashi M, Kaneko H, Nakajima-Takagi Y, Nakayama Y, and Yamamoto M (2008) Molecular evolution of Keap1: Two Keap1 molecules with distinctive intervening region structures are conserved among fish. J. Biol. Chem 283, 3248–3255. [DOI] [PubMed] [Google Scholar]
- (29).Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, and Yamamoto M (2009) The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol. Cell. Biol 29, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Mukaigasa K, Nguyen LTP, Li L, Nakajima H, Yamamoto M, and Kobayashi M (2012) Genetic evidence of an evolutionarily conserved role for Nrf2 in the protection against oxidative stress. Mol. Cell. Biol 32, 4455–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Timme-Laragy AR, Van Tiem LA, Linney EA, and Di Giulio RT (2009) Antioxidant responses and NRF2 in synergistic developmental toxicity of PAHs in zebrafish. Toxicol. Sci 109, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Rousseau ME, Sant KE, Borden LR, Franks DG, Hahn ME, and Timme-Laragy AR (2015) Regulation of Ahr signaling by Nrf2 during development: effects of Nrf2a deficiency on PCB126 embryotoxicity in zebrafish (Danio rerio). Aquat. Toxicol 167, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Schulte-Merker S, and Stainier DYR (2014) Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development (Cambridge, U. K.) 141, 3103–3104. [DOI] [PubMed] [Google Scholar]
- (34).Cloer EW, Goldfarb D, Schrank TP, Weissman BE, and Major MB (2019) NRF2 activation in cancer: from DNA to protein. Cancer Res. 79, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Moreno-Mateos MA, Vejnar CE, Beaudoin J-D, Fernandez JP, Mis EK, Khokha MK, and Giraldez AJ (2015) CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jao L-E, Wente SR, and Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U. S. A 110, 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Thyme SB, Akhmetova L, Montague TG, Valen E, and Schier AF (2016) Internal guide RNA interactions interfere with Cas9-mediated cleavage. Nat. Commun 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, Robinson MD, and Mosimann C (2016) Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development (Cambridge, U. K.) 143, 2025–2037. [DOI] [PubMed] [Google Scholar]
- (39).Meeker N, Hutchinson S, Ho L, and Trede N (2007) Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 43, 610–614. [DOI] [PubMed] [Google Scholar]
- (40).Corrales J, Kristofco LA, Steele WB, Saari GN, Kostal J, Williams ES, Mills M, Gallagher EP, Kavanagh TJ, Simcox N, Shen LQ, Melnikov F, Zimmerman JB, Voutchkova-Kostal AM, Anastas PT, and Brooks BW (2017) Toward the design of less hazardous chemicals: exploring comparative oxidative stress in two common animal models. Chem. Res. Toxicol 30, 893–904. [DOI] [PubMed] [Google Scholar]
- (41).OECD OECD guidelines for the testing of chemicals; OECD Publishing: Paris, 2013; pp 1–22. [Google Scholar]
- (42).Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, and Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev. Dyn 203, 253–310. [DOI] [PubMed] [Google Scholar]
- (43).The Toxicity Relationship Analysis Program (TRAP). https://archive.epa.gov/med/med_archive_03/web/html/trap.html.
- (44).Mills MG, and Gallagher EP (2017) A targeted gene expression platform allows for rapid analysis of chemical-induced antioxidant mRNA expression in zebrafish larvae. PLoS One 12, e0171025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Parichy DM, Elizondo MR, Mills MG, Gordon TN, and Engeszer RE (2009) Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn 238, 2975–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Suzuki T, Takagi Y, Osanai H, Takeuchi M, Katoh Y, Kobayashi M, and Yamamoto M (2005) Pi class glutathione S-transferase genes are regulated by Nrf2 through an evolutionarily conserved regulatory element in zebrafish. Biochem. J 388, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chan K, Lu R, Chang JC, and Kan YW (1996) NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. U. S. A 93, 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Spiers JG, Breda C, Robinson S, Giorgini F, and Steinert JR (2019) Drosophila Nrf2/Keap1 mediated redox signaling supports synaptic function and longevity and impacts on circadian activity. Front. Mol. Neurosci 12, 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yamashita A, Deguchi J, Honda Y, Yamada T, Miyawaki I, Nishimura Y, and Tanaka T (2019) Increased susceptibility to oxidative stress-induced toxicological evaluation by genetically modified nrf 2a-deficient zebrafish. J. Pharmacol. Toxicol. Methods 96, 34–45. [DOI] [PubMed] [Google Scholar]
- (50).Nakajima H, Nakajima-Takagi Y, Tsujita T, Akiyama S-I, Wakasa T, Mukaigasa K, Kaneko H, Tamaru Y, Yamamoto M, Kobayashi M, and Taylor CT (2011) Tissue-restricted expression of Nrf2 and its target genes in zebrafish with gene-specific variations in the induction profiles. PLoS One 6, e26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hahn ME, Timme-Laragy AR, Karchner SI, and Stegeman JJ (2015) Nrf and Nrf2-related proteins in development and developmental toxicity: insights from studies in zebrafish (Danio rerio). Free Radical Biol. Med 88, 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Higgins LG, Kelleher MO, Eggleston IM, Itoh K, Yamamoto M, and Hayes JD (2009) Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol 237, 267–280. [DOI] [PubMed] [Google Scholar]
- (53).Sykiotis GP, and Bohmann D (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).An JH, and Blackwell TK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Leitereg TJ, Guadagni DG, Harris J, Mon TR, and Teranishi R (1971) Chemical and sensory data supporting the difference between the odors of the enantiomeric carvones. J. Agric. Food Chem 19, 785–787. [Google Scholar]
- (56).De Sousa DP, Nóbrega FFF, and Almeida RN (2007) Influence of the chirality of (R)-(−)- and (S)-(+)-carvone in the central nervous system: A comparative study. Chirality 19, 264–268. [DOI] [PubMed] [Google Scholar]
- (57).Bahr TA, Rodriguez D, Beaumont C, and Allred K (2019) The effects of various essential oils on epilepsy and acute seizure: a systematic review. Evidence-Based Complementary and Alternative Medicine 2019, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Almeida RN, De Sousa DP, Nóbrega FFF, Claudino FS, Araújo DAM, Leite JR, and Mattei R (2008) Anticonvulsant effect of a natural compound α,β-epoxy-carvone and its action on the nerve excitability. Neurosci. Lett 443, 51–55. [DOI] [PubMed] [Google Scholar]
- (59).Singer AC, Gilbert ES, Luepromchai E, and Crowley DE (2000) Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl. Microbiol. Biotechnol 54, 838–843. [DOI] [PubMed] [Google Scholar]
- (60).Sparaneo A, Fabrizio FP, la Torre A, Graziano P, Di Maio M, Fontana A, Bisceglia M, Rossi A, Pizzolitto S, De Maglio G, Tancredi A, Grimaldi F, Balsamo T, Centra F, Manzorra MC, Trombetta D, Pantalone A, Bonfitto A, Maiello E, Fazio VM, and Muscarella LA (2019) Effects of KEAP1 silencing on the regulation of NRF2 activity in neuroendocrine lung tumors. Int. J. Mol. Sci 20, 2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Martinez VD, Vucic EA, Thu KL, Pikor LA, Lam S, and Lam WL (2015) Disruption of KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex components by multiple genetic mechanisms: Association with poor prognosis in head and neck cancer. Head Neck 37, 727–734. [DOI] [PubMed] [Google Scholar]
- (62).Jiang T, Chen N, Zhao F, Wang X-J, Kong B, Zheng W, and Zhang DD (2010) High levels of Nrf2 determine chemo-resistance in Type II endometrial cancer. Cancer Res. 70, 5486–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, Hartmaier RJ, Chmielecki J, Seshagiri S, Gentleman R, and Stokoe D (2016) Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 16, 2605–2617. [DOI] [PubMed] [Google Scholar]
- (64).Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takeagi Y, and Yamamoto M (2002) Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7, 807–820. [DOI] [PubMed] [Google Scholar]
- (65).McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD (2006) Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism. J. Biol. Chem 281, 24756–24768. [DOI] [PubMed] [Google Scholar]
- (66).McMahon M, Thomas N, Itoh K, Yamamoto M, and Hayes JD (2004) Redox-regulated turnover of Nrf2 Is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem 279, 31556–31567. [DOI] [PubMed] [Google Scholar]
- (67).Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD (2013) Nrf2 is controlled by two distinct b-TrCP recognition motifs in its Neh6 domain. Oncogene 32, 3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, Wang XJ, and Tang X (2013) RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 73, 3097–3108. [DOI] [PubMed] [Google Scholar]
- (69).Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, and Yamamoto M (2001) Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6, 857–868. [DOI] [PubMed] [Google Scholar]
- (70).Kanzok SM, Rahlfs S, Becker K, and Schirmer RH Thioredoxin, thioredoxin reductase, and thioredoxin peroxidase of malaria parasite Plasmodium falciparum Methods in Enzymology; Elsevier Masson SAS, 2002; Vol. 347, pp 370–381. [DOI] [PubMed] [Google Scholar]
- (71).Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, and Blackwell TK (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, and Yamamoto M (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet 35, 238–245. [DOI] [PubMed] [Google Scholar]
- (73).Fu J, Xiong Z, Huang C, Li J, Yang W, Han Y, Paiboonrungruan C, Major MB, Chen K-N, Kang X, and Chen X (2019) Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J. Biol. Chem 294, 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Ramkissoon A, and Wells PG (2013) Developmental role of nuclear factor E2-related factor 2 in mitigating methamphetamine fetal toxicity and postnatal neurodevelopmental deficits. Free Radical Biol. Med 65, 620–631. [DOI] [PubMed] [Google Scholar]
- (75).Bell KFS, Al-Mubarak B, Martel M-A, McKay S, Wheelan N, Hasel P, Márkus NM, Baxter P, Deighton RF, Serio A, Bilican B, Chowdhry S, Meakin PJ, Ashford MLJ, Wyllie DJA, Scannevin RH, Chandran S, Hayes JD, and Hardingham GE (2015) Neuronal development is promoted by weakened intrinsic antioxidant defences due to epigenetic repression of Nrf2. Nat. Commun 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Williams LM, Timme-Laragy AR, Goldstone JV, McArthur AG, Stegeman JJ, Smolowitz RM, Hahn ME, and Gibert Y (2013) Developmental expression of the Nfe2-related factor (Nrf) transcription factor family in the zebrafish, Danio rerio. PLoS ONE 8, e79574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Lee O-H, Jain AK, Papusha V, and Jaiswal AK (2007) An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem 282, 36412–36420. [DOI] [PubMed] [Google Scholar]
- (78).Ramsden R, and Gallagher EP (2016) Dual NRF2 paralogs in Coho salmon and their antioxidant response element targets. Redox Biol. 9, 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Bellezza I, Mierla AL, and Minelli A (2010) Nrf2 and NF-κB and their concerted modulation in cancer pathogenesis and progression. Cancers 2, 483–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Coish P, Brooks BW, Gallagher EP, Kavanagh TJ, Voutchkova-Kostal A, Zimmerman JB, and Anastas PT (2016) Current status and future challenges in molecular design for reduced hazard. ACS Sustainable Chem. Eng 4, 5900–5906. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


