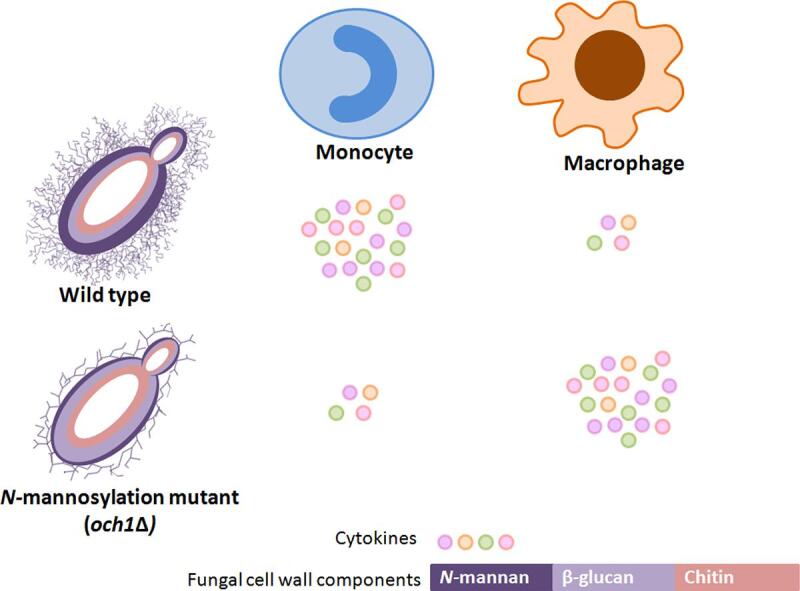
Graphical abstract
Keywords: Fungal cell wall, N-linked mannosylation, β-Glucan, Monocytes, Macrophages
Highlights
-
•
Cytokine response to N-mannan mutants was dependent on the immune cell type used.
-
•
N-mannan mutants stimulated less cytokines from monocytes but more from macrophages.
-
•
N-mannan can therefore act as both an immune agonist or an immune shield.
Abstract
We designed experiments to assess whether fungal cell wall mannans function as an immune shield or an immune agonist. Fungal cell wall β-(1,3)-glucan normally plays a major and dominant role in immune activation. The outer mannan layer has been variously described as an immune shield, because it has the potential to mask the underlying β-(1,3)-glucan, or an immune activator, as it also has the potential to engage with a wide range of mannose detecting PRRs. To resolve this conundrum we examined species-specific differences in host immune recognition in the och1Δ N-mannosylation-deficient mutant background in four species of yeast-like fungi. Irrespective of the fungal species, the cytokine response (TNFα and IL-6) induced by the och1Δ mutants in human monocytes was reduced compared to that of the wild type. In contrast, TNFα production induced by och1Δ was increased, relative to wild type, due to increased β-glucan exposure, when mouse or human macrophages were used. These observations suggest that N-mannan is not a major PAMP for macrophages and that in these cells mannan does shield the fungus from recognition of the inner cell wall β-glucan. However, N-mannan is a significant inducer of cytokine for monocytes. Therefore the metaphor of the fungal “mannan shield” can only be applied to some, but not all, myeloid cells used in immune profiling experiments of fungal species.
1. Introduction
We set out to understand some unresolved questions about the role of mannose-based outer cell wall components in fungi in either initiating or shielding immune recognition. The innate immune system forms the first line of defence against fungal infections and is essential for protective cell-mediated immunity (da Silva Dantas et al., 2016, Drummond et al., 2014, Netea et al., 2015, Qin et al., 2016, Salazar and Brown, 2018). Innate immune responses involve multiple immune cell types of which monocytes and macrophages are its vital components. These cells express germline-encoded pattern recognition receptors (PRRs) such as C-Type Lectin Receptors (CLRs), Toll-like-receptors (TLRs) and NOD-like receptors (NLRs) that sense conserved pathogen-specific molecular patterns (PAMPs) that, for fungi, are mainly located in the cell wall. These immune recognition events trigger intracellular signalling pathways leading to a release of pro-inflammatory cytokines and chemokines that orchestrate the recruitment of neutrophils and activate downstream innate and adaptive immune responses (Borriello et al., 2020, Dambuza and Brown, 2015, Erwig and Gow, 2016, Nikolakopoulou et al., 2020, Plato et al., 2015).
The fungal cell wall is the primary point of contact with the components of the host innate immune system (Erwig and Gow, 2016) and is a highly dynamic organelle essential for cell viability, morphogenesis, and pathogenesis (Garcia-Rubio et al., 2019, Gow et al., 2017, Gow and Yadav, 2017, Vendele et al., 2020). The composition and architecture of the fungal cell wall changes in response to different environmental and stress conditions and varies in different cell morphotypes, helping the fungi to adapt in different growth conditions (Brown et al., 2014, Cottier and Hall, 2019, Hopke et al., 2018). The best studied cell wall of a fungal pathogen is that of Candida albicans (Gow et al., 2012, Gow and Hube, 2012). It is a bilayer structure composed of an inner layer of β-(1,3)-glucan and chitin, which is interconnected through β-(1,6)-glucan to the outer layer rich in glycosylphosphatidylinositol-anchored mannoproteins, modified with N- and O-linked mannans (Hall and Gow, 2013, Klis et al., 2001). The outer cell wall mannan forms a fringe of microfibrils that represent a permeability barrier to high molecular weight molecules, but apparently not to liposomes (Walker et al., 2018). These polysaccharides comprising the cell wall are highly fungal specific and present important pathogen-associated molecular patterns (PAMPs) recognized by PRRs (Gow et al., 2012, Levitz, 2010, Netea et al., 2008, Rappleye and Goldman, 2008, Snarr et al., 2017, van de Veerdonk et al., 2008).
However, during evolution and species divergence, while some of the cell wall components have been conserved, others are species-specific. The inner cell wall polysaccharides have been shown to be largely conserved across the fungal kingdom, and studies have suggested the inner layer to be highly immunogenic and capable of eliciting a strong immune response (Snarr et al., 2017). The β-(1,3)-glucan-dectin-1 interaction is amongst the most extensively studied fungal PAMP–host PRR interactions (Brown and Gordon, 2001, Dambuza and Brown, 2015, Gow et al., 2007, Netea et al., 2006, Rogers et al., 2005, Taylor et al., 2004). β-(1,3)-Glucan is the major component of most fungal cell walls and comprises between 30 and 60% of the dry cell wall weight (Bowman and Free, 2006, Gow et al., 2017, Klis et al., 2001, Latgé, 2007). Although β-(1,3)-glucan is a common element of the cell wall of fungi there are differences in the extent of β-(1,6)-glucan cross linking of this component and some structural differences – for example in the yeast and hyphal β-(1,3)-glucans of C. albicans (Lowman et al., 2014).
Outer cell wall mannan comprises about 40% of dry cell wall weight in C. albicans (Gow et al., 2017, Klis et al., 2001) and has been shown across several studies to be important for host fungal interactions (Bates et al., 2006, Vendele et al., 2020). While O-mannan is mostly a linear structure of α-mannoses, with occasional β-(1–2) side chains, N-mannan is a highly branched structure of mannose residues in different linkages (Hall, 2015, Hall and Gow, 2013). It has a highly conserved oligosaccharide core synthesised in the endoplasmic reticulum, and a highly branched outer chain elaborated in the Golgi complex, which consists of a linear α-(1,6)-linked mannose backbone with numerous side chains composed of α-(1,2)-, α-(1,3)-, β-(1,2)-linked mannose residues and phosphomannan moieties (Mora-Montes et al., 2009, Shibata et al., 1992). The N-mannan branching patterns and patterns of linkages between mannose residues are fungal species-specific and contribute to the diversity of these structures.
Even though mannan has been considered as a mask for the underlying immunogenic cell wall components to escape immune recognition, numerous PRRs specific for mannans are involved in activating host defence responses, indeed there are more mannan-recognising immune receptors for fungi than for any other class of ligand (Erwig and Gow, 2016, Patin et al., 2019, Salazar and Brown, 2018). For example, it has been demonstrated that the CLRs mannose receptor (MR), DC-SIGN, dectin-2 and dectin-3 are involved in the sensing of N-linked mannans (Cambi et al., 2008, Netea et al., 2006, Saijo et al., 2010, Vendele et al., 2020, Yamamoto et al., 1997). It has also been established that TLR2 recognises the glycolipid phospholipomannan (Jouault et al., 2003) while TLR4 recognizes the O-linked mannan (Netea et al., 2006, Netea et al., 2002). Soluble receptors such as mannose-binding lectin (MBL) and galectin-3 are also involved in the recognition of the fungal mannose structures by the host (Snarr et al., 2017).
Significant progress has been made on understanding fungus-host interactions through studies of C. albicans, partly because of the availability of a comprehensive number of genetic mutants lacking, or altered in key immunologically active components of its cell wall. The study of C. albicans mutants with defects in the N-linked mannosylation pathway has helped to establish the importance of these cell wall structures in the recognition of C. albicans by innate immune cells (Netea et al., 2006, Hall, 2015, Hall et al., 2013, Hall and Gow, 2013, Zhang et al., 2016). Mutants of N-mannan biogenesis, (for example, mutants with defects in N-linked mannan processing enzyme α-glycosidase or mutants with loss of outer chain N-mannans), are affected in their ability to induce cytokine production by human peripheral blood mononuclear cells (hPBMCs), dendritic cells, and murine macrophages (Cambi et al., 2008, Hall et al., 2013, McKenzie et al., 2010, Mora-Montes et al., 2010, Mora-Montes et al., 2007, Netea et al., 2006, Zhang et al., 2016). They are also affected in their phagocytosis by human dendritic cells and murine macrophages (Bain et al., 2014, Bain et al., 2012, Cambi et al., 2008, Lewis et al., 2012, McKenzie et al., 2010). In contrast to the C. albicans glycosylation mutants, C. glabrata mannosylation mutants display enhanced virulence in a murine infection model (West et al., 2013).
However, the role of C. albicans N-linked mannans in the innate immune recognition cannot always be extrapolated to other fungi since there are considerable differences in the chemical structure of the N-mannan outer chains (Fabre et al., 2014, Jouault et al., 2006). These differences often define the species- and strain-specific serotype (Fukazawa, 1989, Kozel et al., 2004). Furthermore, the compactness of these structures can also affect the extent of masking or exposure of the inner immunogenic cell wall components and the overall organisation of the cell wall can be significantly affected by yeast-hypha morphogenesis, carbon source and local environmental stresses (Bain et al., 2014, Ballou et al., 2016, Graus et al., 2018, Mukaremera et al., 2017, Pradhan et al. 2018, Vendele et al., 2020).
The outer layer of fungal cell walls displays a large chemical diversity among fungal pathogens (Erwig and Gow, 2016, Garcia-Rubio et al., 2019) and has been said to conceal underlying immunogenic layers. By hiding β-(1,3)-glucan in particular the outer layers have the potential to contribute to immune evasion (Hernández-Chávez et al., 2017, Netea et al., 2008). In this regard α-(1,3)-glucan has been shown to mask immune recognition of underlying β-(1,3)-glucan in Histoplasma capsulatum (Garfoot et al., 2016, Rappleye et al., 2007) and outer cell wall mannans have been claimed to do the same for Candida species (Graus et al., 2018, Wheeler et al., 2008). However, a striking anomaly in the literature has been the variable assertions as to the immunological role of mannans. Both mannans and α-(1,3)-glucan have been suggested as acting primarily in shielding the underlying pro-inflammatory β-(1,3)-glucan layer from immune surveillance (Rappleye et al., 2007, Rappleye and Goldman, 2008, Wheeler and Fink, 2006). For example, S. cerevisiae wild type cells were shown to be poor in stimulating cytokine production, but mutant strains with defects in the N-linked mannan elaboration, such as the och1Δ null mutant, stimulated the mouse macrophage cell line RAW 264.7 to produce high levels of TNFα (Wheeler and Fink, 2006). These data contrast with the observations made using C. albicans och1Δ null mutant, where loss of the N-linked mannan outer chain led to reduced cytokine production by hPBMCs and dendritic cells (Bates et al., 2006, Cambi et al., 2008, Gow et al., 2007, Hall et al., 2013, Netea et al., 2006).
To further explore the apparent differences between N-mannan mediated immune recognition effects of different fungal species we used och1 mutants from four different fungal species, C. albicans, C. dubliniensis, C. glabrata and S. cerevisiae and compared the immune response induced by hPBMCs and mouse macrophages (RAW 264.7). Och1 is an α-(1,6)-mannosyl transferase catalyzing the addition of first α-(1,6)-mannose to the N-glycan core. The och1Δ mutants show a loss of the N-mannan fibrillar layer leading to the exposure of the inner cell wall components and also a compensatory increase in β-(1,3)-glucan and chitin. We also compared fungal cells prepared for the immune assays by killing by heat-treatment – which is known to expose inner cell wall ligands (Gow et al., 2007), and chemical fixation cells that better preserves the native cell wall structure. In some experiments we used heat killed fungal cells as the target for immune cell recognition. Heat killed cells have been used to prevent cell wall changes associated with yeast cell growth and yeast-hypha morphogenesis from interfering with cytokine induction assays. In our experiments heat-killing also permeabilises the cell wall, thus allowing PRR engagement with cell wall PAMPs that are not exposed in native cell walls (Gow et al., 2007). We also examined the relative porosity and cell wall composition of the cell walls of these species and their mutants.
We showed that loss of N-mannan led to a lower cytokine response (TNFα and IL-6) from hPBMCs, but a higher TNFα induction from RAW 264.7 and human macrophages compared to the controls, and that this was explained by recognition of fungal cells by macrophages being primarily mediated by dectin-1. But, irrespective of the fungal species, fungal cell wall N-mannan was important for fungal recognition by human monocytes. This provides an explanation for apparently contradictory results in the literature. Mannan acts only as an immune shield for cells such as macrophages that deploy dectin-1 as the primary immune recognition receptor, whilst other immune cell types depend strongly on recognition of N-mannan via a set of mannan detecting PRRs.
2. Methods
2.1. Strains and culture conditions
The strains constructed and used in this work are listed in Table 1. To prepare yeast cells for cytokine stimulation assays, cells were grown overnight at 30 °C in YPD broth with shaking at 200 rpm, transferred to fresh broth with a starting O.D.600 of 0.2, and further incubated for 4 h till they reached the mid-log growth. Candida strains were maintained at 30 °C in YPD, but S. cerevisiae cells were grown in SC medium (0.67% (w/v) yeast nitrogen base with ammonium sulfate without amino acids, 0.2% (w/v) complete supplement mixture minus uracil with 2% (w/v) glucose (SC-glc) or with 2% galactose and 3% (w/v) raffinose (SC-gal-raf). A supplement of 50 µg/ml uridine was added to media used for the growth of S. cerevisiae wild type and all Ura-och1Δ strains. C. glabrata strains were grown in YPD with and without 20 µg/ml doxycycline.
Table 1.
Strains used in this study.
| Species | Strain | Genotype | Source |
|---|---|---|---|
| C. albicans | CAI4 | ura3Δ::imm434/ura3Δ::imm434 | (Fonzi and Irwin, 1993) |
| C. albicans | CaWT (NGY152) | As CAI4 but RPS1/rps1Δ::CIp10 | (Brand et al., 2004) |
| C. albicans | Caoch1 (NGY357) | As CAI4 but och1Δ::hisG/och1Δ::hisG, RPS1/rps1Δ::CIp10 | (Bates et al., 2006) |
| C. albicans | Caoch1Δ/ CaOCH1 (NGY358) | As CAI4 but och1Δ::hisG/och1Δ::hisG, RPS1/rps1Δ::CIp10-CaOCH1 | (Bates et al., 2006) |
| C. albicans | Camnn2Δ6 (NGY 600) | As CAI4 but mnn2Δ::dpl200/mnn2Δ ::dpl200mnn22Δ::dpl200/mnn2Δ::dpl200 mnn23Δ::dpl200/mnn23Δ::dpl200mnn24Δ ::dpl200/mnn24Δ::dpl200mnn26Δ::dpl200/ mnn26Δ::dpl200mnn21Δ::dpl200/mnn21Δ ::dpl200 RPS1/rps1Δ::CIp10 | (Hall et al., 2013) |
| C. albicans | Camnn2Δ6 + CaMNN2 + CaMNN26(NGY 601) | As CAI4 but mnn2Δ::dpl200/mnn2Δ ::dpl200mnn22Δ::dpl200/mnn2Δ::dpl200 mnn23Δ::dpl200/mnn23Δ::dpl200mnn24Δ ::dpl200/mnn24Δ::dpl200mnn26Δ::dpl200/ mnn26Δ::dpl200mnn21Δ::dpl200/ mnn21Δ ::dpl200 RPS1/rps1Δ::CIp10-NN2/MNN26 | (Hall et al., 2013) |
| C. albicans | pmr1Δ(NGY355) | As CAI4 but pmr1Δ::hisG/pmr1Δ::hisG, RPS1/rps1Δ::CIp10 | (Bates et al., 2005) |
| C. albicans | pmr1Δ + PMR1(NGY356) | As CAI4 but pmr1Δ::hisG/pmr1Δ::hisG, RPS1/rps1Δ::CIp10-PMR1 | (Bates et al., 2005) |
| C. albicans | mnt1-mnt2Δ(NGY337) | As CAI4 but mnt1-mnt2Δ::hisG/mnt1-mnt2Δ::hisG, RPS1/rps1Δ::CIp10 | (Munro et al., 2005, p. 1) |
| C. albicans | mnt1-mnt2Δ + MNT1(NGY335) | As CAI4 but mnt1-mnt2Δ::hisG/mnt1-mnt2Δ::hisG, RPS1/rps1Δ::CIp10-MNT1 | (Munro et al., 2005) |
| C. albicans | mnn4Δ(CDH15) | As CAI4 but mnn4Δ::hisG/mnn4Δ::hisG, RPS1/rps1Δ::CIp10 | (Hobson et al., 2004) |
| C. albicans | mnn4Δ + MNN4(CDH13) | As CAI4 but mnn4Δ::hisG/mnn4Δ::hisG, RPS1/rps1Δ::(CIp10-MNN4)n | (Hobson et al., 2004) |
| C. dubliniensis | CdUM4A | As Wü284 but ura3Δ1::FRT/ ura3Δ2::FRT | (Staib et al., 2001) |
| C. dubliniensis | CdWT(NGY568) | As CdUM4A but RPS1/rps1Δ::CIp10 | This study |
| C. dubliniensis | Cdoch1Δ(NGY563) | As CDUM4A but och1Δ::dp1200/och1Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| C. dubliniensis | Cdoch1Δ/CdOCH1(NGY564) | As CDUM4A but och1Δ::dp1200/och1Δ::dp1200, RPS1/rps1Δ::CIp10-CdOCH1 | This study |
| C. glabrata | CgWT(2001) | HETS202 strain | (Ueno et al., 2007) |
| C. glabrata | PTET-CgOCH1 | As CgWT but PTET-OCH1 | This study |
| S.cerevisiae | ScWT(BY4741) | S. cerevisiae MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ATCC |
| S. cerevisiae | Scoch1Δ(Y04406) | As BY4741 but och1Δ::KanMx4 | Euroscarf |
| S. cerevisiae | Scoch1Δ + PGAL1-ScOCH1(NGS7) | Y04406 transformed with pYES2.1/V5-His-TOPO-ScOCH1 | This study |
Cells were harvested by centrifugation, and washed twice with PBS. The cells were heat-killed at 65 °C for 2 h. To compare the temperature-induced cell-wall changes in wild type strains, 2*107 cells/ml cells were killed with 100 mM thimerosal for 1 h, washed thrice with PBS, counted and cell suspensions of 2*106 cells/ml were exposed to 30, 37, 45, 50, 55, 65 and 75 °C for 2 h. The loss of cell viability was assessed by colony counting of viable cells on YPD plates.
2.2. Construction of null and conditional mutants
The mini-ura-blaster technique was used for C. dubliniensis OCH1 disruption as follows: the primer pair 5′-ATGCTACAACTAAGGGAACCCAAAATGGTTCATAGACATCTAAAACTAGCCATTTTAGGAATAATAGTCAGTTTTCCCAGTCACGACGTT-3′ and 5′-TTACTGCATTTCTGGCATACCATCATCTTTCCAACTACCCAGAAACATATGTTTTGCGTATGCCATTGGATGTGGAATTGTGAGCGGATA-3′ (underline bases correspond to complementary sequences to the 5′- and 3′- regions of ORF Cd36_86020 from the C. dubliniensis GeneDB, http://old.genedb.org/genedb/cdubliniensis/index.jsp) were used to amplify the disruption cassette from pDDB57 plasmid by PCR (Wilson et al., 2000). The strain CdUM4A, a Ura-mutant derived from the clinical isolate Wü284 (Staib et al., 2001), was sequentially transformed with the disruption cassette and the URA3 marker recycled by growing on SC medium supplemented with 1 mg/ml 5-fluoroorotic acid and uridine. The plasmid CIp10 was used to restore URA3 at the RPS1 locus as previously described (Murad et al., 2000).
Despite repeated attempts, a null mutant for OCH1 in C. glabrata could not be generated, indicating that this gene may be essential for the viability of this species, at least under conditions of transformation. Therefore a conditional mutant was generated by placing the OCH1 gene under the control of a tetracycline regulatable promoter 97t (Nakayama et al., 1998). The Tet-P cassette was amplified using plasmid pTK916 as a template, with the primer pairs 5′-TCCTGTTCTGAGACCAAATAGCAAACCGAAGCTGGCTTGATACAGTAAATTCAGTGggccgctgatcacg-3′ and 5′-AAGAATACCACCAGCACCAGCACCGCAAGCACTATGTGCCTCTTCTGCCATTTACTATcgtgaggctgg-3′. This amplicon was used to transform HETS202 strain (Ueno et al., 2007), thus replacing the wild type promoter of OCH1 in this strain with the conditional Tet-promoter. The mutants were confirmed by PCR and northern analysis (data not shown). OCH1 expression was repressed in the conditional mutant using 20 µg/ml doxycycline in the growth medium.
2.3. Complementation of och1Δ null mutants.
To generate a re-integrant control strain for C. dubliniensis och1Δ, the primer pair 5′-GCGGCCGCAAAATGAAAAATATTTACC TC and 5′-GCGGCCGCTTGTTAGATTTAATTTGGATT (with bases added to create a NotI site underlined) were used to amplify by PCR a 2907 bpDNA fragment containing the C. dubliniensis OCH1 ORF plus 995 bp of its promoter and 731 bp of its terminator regions, and the DNA fragment cloned into pCR®2.1-TOPO® vector (Invitrogen, Paisley, UK). The insert was released by digesting with NotI, and subcloned into the NotI site of CIp10, generating plasmid CIp10-CdOCH1. The StuI-digested plasmid was integrated at the RPS1 locus generating strain NGY565.
The S. cerevisiae optimized, galactose-inducible protein expression vector pYES2.1/V5-His-TOPO (Invitrogen, Paisley, UK) was used to express S. cerevisiae OCH1 in the S. cerevisiae och1Δ null mutant strain. The S. cerevisiae OCH1 ORF was amplified by PCR (primer pair 5′-ATGTCTAGGAAGTTGTCCCACCTGA and 5′- GATGCTGATAAAAATGCAGGTCATAAATAA) and ligated into the pYES2.1/V5-His-TOPO vector according to manufacturers’ instructions and the construction used to transform Escherichia coli TOP10 cells (Invitrogen, Paisley, UK). The construction of pYES2.1/V5-His-TOPO was confirmed by sequencing and used to transform S. cerevisiae och1Δ null mutant.
2.4. Analysis of N-linked mannosylation status and cell wall phosphomannan content
The electrophoretic mobility shift assay of secreted β-N-acetylglucosaminidase and the Alcian Blue binding assay were used to assess the N-linked mannosylation status and the phosphomannan content, respectively, as previously described (Bates et al., 2006).
2.5. Fluorescence microscopy
Mannan, chitin and β-glucan staining in the live fungal cell walls were done as previously described with minor adaptations (Hall et al., 2013).
For mannan staining, 2.5*106 exponentially growing yeast cells were washed three times with sterile PBS. The cells were then stained with 100 µg/ml ConA-Rh for 45 min in dark and washed three times in PBS. The cells were next stained for chitin using 100 µg/ml freshly prepared WGA-FITC for 45 min in dark. After staining, the cells were washed thrice with PBS, fixed in 4% formaldehyde for 1 h and observed under the Zeiss Axioplan 2 microscope (63X objective). The images were analysed using AxioVision ES64 Rel.4.9.1 software.
For β–glucan staining, 2.5*106 exponentially growing yeast cells were washed thrice with sterile PBS and blocked in Buffer A (0.5% BSA, 5.0 mM EDTA, 2.0 mM NaN3, 5% heat-inactivated goat serum) for 30 min at room temperature. The cells were next washed thrice with Buffer B (Buffer A without goat serum). An aliquot of 0.5 µg of purified Fc-dectin-1, (kind gift from Prof. Gordon Brown), was added to each of the samples and incubated in ice for 1 h. The cells were again washed three times with Buffer B. A 100.0 µl sample of TRITC-conjugated anti-human IgG Fc goat IgG (from Thermofisher Scientific, catalogue number A18822, diluted 1:200 in Buffer B) was added to each of the samples and incubated in ice for 45 min. The cells were next washed five times with Buffer A, fixed in 4% paraformaldehyde, and observed under the Zeiss Axioplan 2 microscope at 63X magnification. The images were analysed using AxioVision ES64 Rel.4.9.1 software.
2.6. Cell wall composition and porosity
Determination of cell wall mannan, chitin, and β-glucan content was achieved by acid-hydrolysing the polymers, and quantifying mannose, glucosamine, and glucose content, respectively, by high-performance anion-exchange chromatography with pulsed amperometric detection as previously described (Mora-Montes et al., 2007, Plaine et al., 2008).
The cell wall porosity was determine by relative porosity to polycations as described (De Nobel et al., 1990). Overnight-grown cells in Sabouraud broth were inoculated into fresh broth, further incubated at 30 °C for 4 h with 200 rpm, and washed twice with PBS. Cell pellets containing 1*108 cells were resuspended in 10 mM Tris-HCl, pH 7.4 (buffer A), buffer A plus 15 µg/ml poly-L-lysine (Mw 30–70 kDa, Sigma Cat. No. P2636) or buffer A plus 30 µg/ml DEAE-dextran (Mw 500 kDa, Sigma Cat. No. D9885), and incubated for 30 min at 30 °C, 200 rpm. Preparations were centrifuged to pellet cells, supernatants were collected, further centrifuged and absorbance at 260 nm measured. The relative cell wall porosity to DEAE-dextran was calculated as described (De Nobel et al., 1990).
2.7. Cytokine stimulation using human peripheral blood mononuclear cells (hPBMCs)
For hPBMC isolation, human blood was collected from healthy volunteers according to the local guidelines and regulations, as approved by the College Ethics Review Board of the University of Aberdeen (CERB/2012/11/676 and CERB/2016/8/1300). The isolation of hPBMCs was essentially performed as previously described (Endres et al., 1988). Samples of 5*105 freshly isolated hPBMCs were placed into round-bottom 96-well Nunclon plates (Nunc, Roskilde, Denmark) and stimulated with heat-killed 2*105 yeast cells for 24 h at 37 °C and 5% (v/v) CO2. For the preparation of glucan-phosphate-treated human PBMCs, cells were incubated with 10.0 µg of glucan-phosphate (kind gift from Prof. David L Williams) for 2 h at 37 °C and 5% (v/v) CO2 before challenge with fungal cells. Additionally, for understanding the temperature-induced cell wall changes in wild type strains, the hPBMCs were similarly challenged with thimerosal killed fungal cells exposed to different temperatures, as described in the material and methods section. The supernatants were collected after 24 h of challenge and stored at −20 °C till cytokine analysis was performed. Cytokine measurements were carried out by ELISA using commercially available kits from R&D systems (Abingdon, UK).
2.8. TNFα stimulation using mouse macrophages from the cell line RAW264.7
The RAW264.7 cell line was obtained from the European Collection of Cell Culture. Cells were grown at 37 °C and 5% (v/v) CO2in tissue culture flasks (Nagle Nunc. International, Hereford, UK) containing DMEM medium (Lonza Group Ltd, Braine-l’Alleud, Belgium), added with 1% (w/v) L-glutamine (Invitrogen, Paisley, UK), 10% (v/v) heat inactivated foetal calf serum (Biosera, Ringmer, UK), and 2% (w/v) penicillin/streptomycin antibiotics (Invitrogen Ltd, Paisley, UK). For cytokine stimulation, cells were gently and mechanically detached from bottles, and 2*105 cells/well were seeded in a 96-well flat-bottom plate. The cells were left to adhere overnight at 37 °C and 5% (v/v) CO2. 100 µl fresh medium was added to the wells and then stimulated with 2*105 heat-killed yeast cells for 24 h at 37 °C and 5% (v/v) CO2. For blocking with glucan-phosphate-treated, cells were incubated with 10.0 µg of the glucan-phosphate for 2 h at 37 °C and 5% (v/v) CO2 before challenge with fungal cells. To analyze the effect of temperature-induced cell wall changes in wild type strains, the RAW macrophages were similarly challenged with thimerosal killed fungal cells exposed to different temperatures, as described in the methods section. Upon stimulation, supernatants were collected as described above and used to quantify murine TNFα by ELISA using kits from R&D.
2.9. Statistical analysis
The cell wall analysis by HPLC was done using two independent cultures, performed in duplicate. Cytokine stimulations using human PMBCs were performed in duplicate with a total of six healthy donors; whereas assays with RAW264.7 macrophages were conducted four times in duplicate. Student t-test or one-way ANOVA (with posthoc correction using Tukey test) were used to establish statistical significance of data, as applicable, with a significance level set at p < 0.05.
3. Results
3.1. Cell wall structural differences between the fungal species results in their differential immune recognition
We first examined how cell wall differences between the different fungal species affected immune recognition. We chemically fixed yeast cells of C. albicans, C. dubliniensis, C. glabrata and S. cerevisiae with thimerosal before exposing them to different temperatures (from 30 °C to 75 °C) for 2 h. Heat treatment is known to increase exposure of the inner cell wall layers and is commonly used in immune assays to prevent unwanted fungal growth (for example yeast-hypha morphogenesis) during immune assays. These treated cells were exposed to hPBMCs as well as RAW264.7 macrophages and cytokine profiles were measured after 24 h.
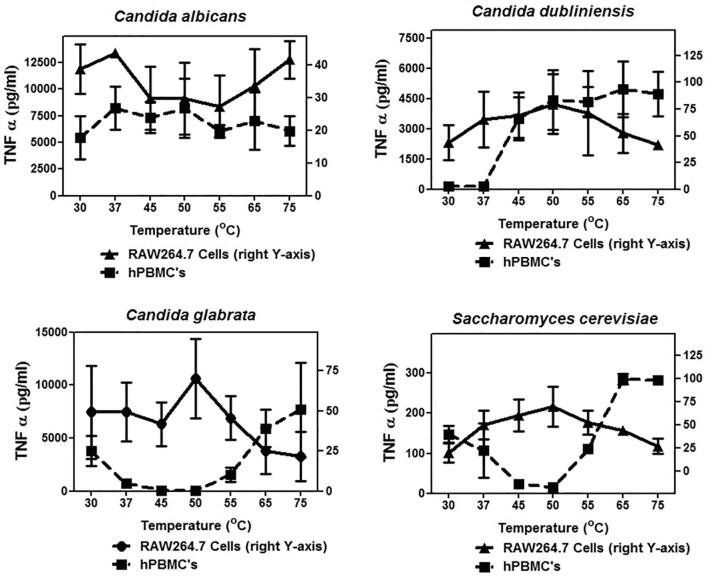
Interestingly despite the phylogenetic similarity between C. albicans and C. dubliniensis there was a marked difference in the temperature profile map of the TNFα response induced by yeast cells (Fig. 1). Induced TNFα levels did not vary significantly when the temperature exposure of the cells was increased. TNFα induced by cells of C. glabrata and S. cerevisiae displayed a similar pattern but the patterns were different to that for C. albicans and C. dubliniensis, showing a marked increase in TNFα induction in hPBMCs at higher temperatures. A different trend was observed in RAW264.7 macrophages, suggesting underlying differences in the thermostability of the cell wall of these species (Fig. 1). In the case of hPBMCs, patterns similar to TNFα secretion were observed for another cytokine, IL-6, as well (data not shown). IL-6 levels secreted by RAW macrophages were too low to be detected by our assay method. The microscopic assessment of the mannan, chitin or β-glucan levels with the increase in temperature did not reveal any striking differences in the relative exposure of β-glucan at different temperatures (data not shown). However, the thermal profiling experiments did suggest important underlying differences in the structure of the cell wall that were differentially affected by heat treatment.
Fig. 1.
TNFα production from hPBMCs and RAW264.7 macrophages upon stimulation with wild type cells: The wild type cells were thimerosal killed and exposed to different temperatures to expose the inner cell wall components. These fungal cells were then used to challenge hPBMCs and RAW macrophages for 24 h and the TNFα levels were monitored using ELISA. For hPBMCs the experiment was done with six donors, three independent fungal cultures, each time done in duplicates. For RAW macrophages, the experiment was repeated four times in duplicate. Data represent means ± SD.
3.2. Generation of C. dubliniensis and C. glabrata och1Δ null mutants
In order to investigate the impact of outer cell wall N-mannosylation defect in different fungal species on the immune recognition, we generated och1 mutant and re-integrant strains from the four fungal species (Table 1). The S. cerevisiae och1Δ null mutant strain was complemented with a galactose-inducible expression vector to express S. cerevisiae OCH1 in the S. cerevisiae och1Δ (ScOCH1). The C. dubliniensis CdUM4A, a ura- strain derived from the clinical isolate Wü284 (Staib et al., 2001), was used as genetic background for the sequential disruption of both OCH1 alleles, using the mini-ura-blaster strategy (Wilson et al., 2000). The Cdoch1Δ null mutant (NGY562) was transformed with the CIp10 plasmid (Murad et al., 2000), introducing URA3 at the neutral RPS1 locus (Brand et al., 2004). The re-integrant control strain (NGY564) had the CdOCH1 allele at the RPS1 locus as described in Methods section. Attempts to disrupt CgOCH1 in C. glabrata ATCC2001 were unsuccessful hence a conditional mutant using the tetracycline-regulatable system was generated.
Similar to Caoch1Δ and Scoch1Δ null mutants, yeast cells of the Cdoch1Δ null mutant and the Cgoch1 conditional mutant growing in presence of doxycycline were swollen, formed aggregates, and had small and crenulated colonies (data not shown). The defect in the N-linked glycosylation of Cdoch1Δ null mutant was assessed by electrophoretic mobility shift of secreted β-N-acetylhexosaminidase (HexNAcase) in native gels using an in situ activity assay (Bates et al., 2006). HexNAcase is a highly N-linked glycosylated enzyme (Molloy et al., 1994), and changes in its electrophoretic mobility have been used as indicators of N-linked glycosylation defects which lead to marked decreases in total molecular mass (Bates et al., 2006, Mora-Montes et al., 2010, 2007). The HexNAcase extracted from the null mutant ran faster than the enzyme obtained from wild type cells, while reintegration of CdOCH1 restored the enzyme mobility to wild type levels (Fig. S1A). Endoglycosidase H-treated HexNAcase from wild type, Cdoch1Δ null mutant and re-integrant control migrated faster and with the similar mobility (Fig. S1B). No HexNAcase activity was detectable in protein preparations from C. glabrata and native zymograms for invertase also failed to generate usable electrophoretic mobility shifts (data not shown). Hence, in this case cell wall phosphomannan levels was assessed by Alcian Blue binding to confirm that the N-linked glycosylation status of C. glabrata was altered in this mutant (Bates et al., 2006, Herrero et al., 2002, Mora-Montes et al., 2007, Nishikawa et al., 2002). C. glabrata wild type cells bound 82.7 ± 5.6 mg Alcian Blue, whereas the Cgoch1 conditional mutant bound 74.7 ± 5.4 (P value 0.15). When the mutant cells were grown in presence of 20 µg/ml doxycycline there was a significant reduction in the ability to bind Alcian Blue (22.2 ± 2.9 mg dye bound, P < 0.05). Therefore, these data confirmed that both the Cdoch1Δ null mutant and the Cgoch1 conditional mutant also had defects in N-linked glycosylation.
3.3. och1 mutants from different fungal species have altered cell wall composition and characteristics
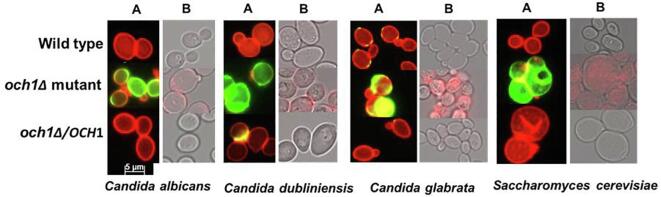
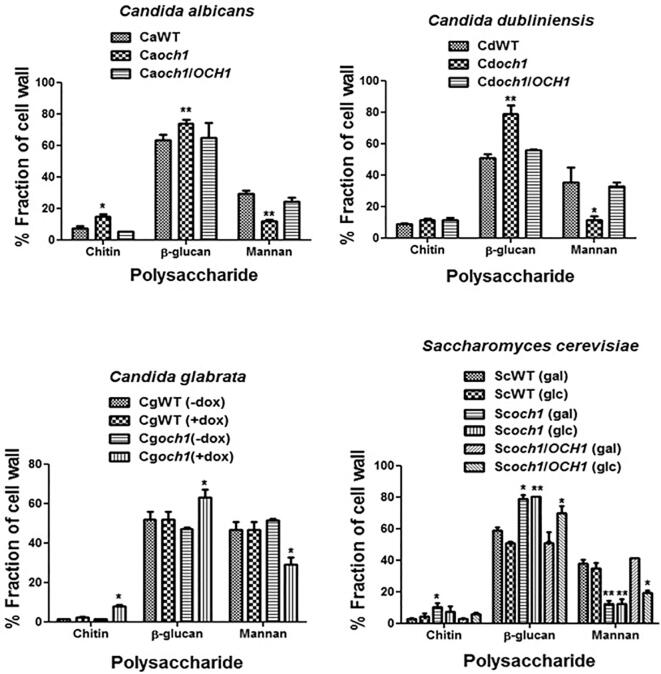
The cell wall organisation and sugar composition of the och1 mutants for all the fungal species, C. albicans, C. dubliniensis, C. glabrata, and S. cerevisiae, were assessed by fluorescence microscopy and HPLC respectively. The och1Δ mutants from all the fungal species displayed higher chitin and β-glucan levels compared to their wild type and re-integrant controls as assessed by microscopy and HPLC (Fig. 2, Fig. 3). Disruption of OCH1 resulted in gross reduction in the outermost N- mannan layer in the fungal cells, and exposure of the inner layers of chitin and β-glucan. As expected, all the mutants displayed reduced levels of mannan compared to the controls.
Fig. 2.
Mannan, chitin and β-glucan levels in live fungal cells: Cell wall structure was assessed in live cells by staining using fluorescence microscopy, as described in the methods section. (A) Mannan was stained using ConA-Rh (red), while cell surface chitin was stained using WGA-FITC (green). (B) β-glucan levels were detected using TRITC-conjugated anti-human IgG Fc goat IgG (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Cell wall composition analysis by HPLC: The acid hydrolysed cell walls of the strains were analyzed by HPLC to quantify glucosamine, glucose and mannose indicative of chitin, β-glucan and mannan contents respectively. Independent biological replicates were carried out in duplicate. Data represent means ± SD. *P < 0.05, **P < 0.005.
The walls of S. cerevisiae and C. glabrata were significantly more porous to DEAE-dextran than walls of C. albicans and C. dubliniensis (Fig. S2). Wall porosity was increased most in N-mannan mutants (och1Δ and pmr1Δ), intermediate in the O-mannan double mutant (mnt1Δ-mnt2Δ) and unaffected by loss of cell wall phosphomannan (mnn4Δ) (Fig. S2). The och1 conditional mutant of C. glabrata grown under repressing conditions did not show any further reduction in wall porosity – most likely because even wild cells were relatively porous (Fig. S2).
Thus, disruption of OCH1 in the four fungal species led to similar qualitative changes in the cell wall: reduced N-mannan, increased chitin content, increased internal wall exposure and increased cell wall porosity.
3.4. hPBMC’s recognize fungal N-mannosylation
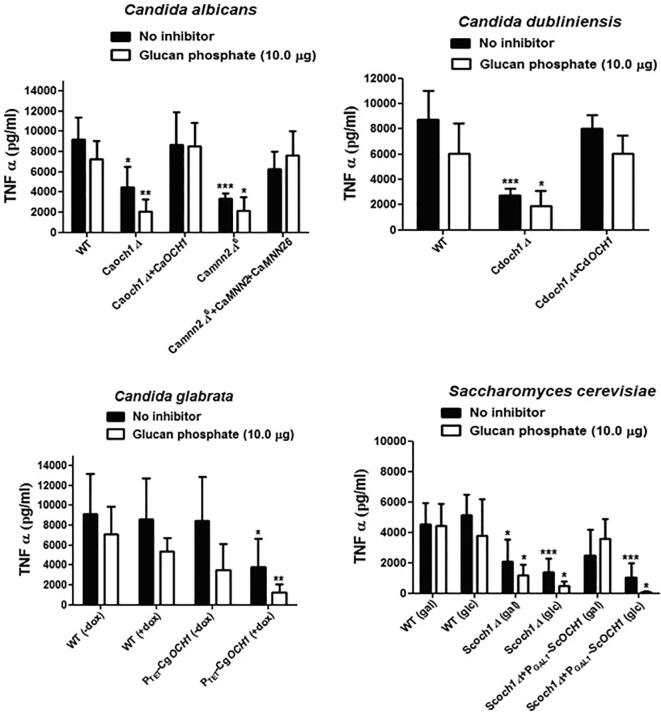
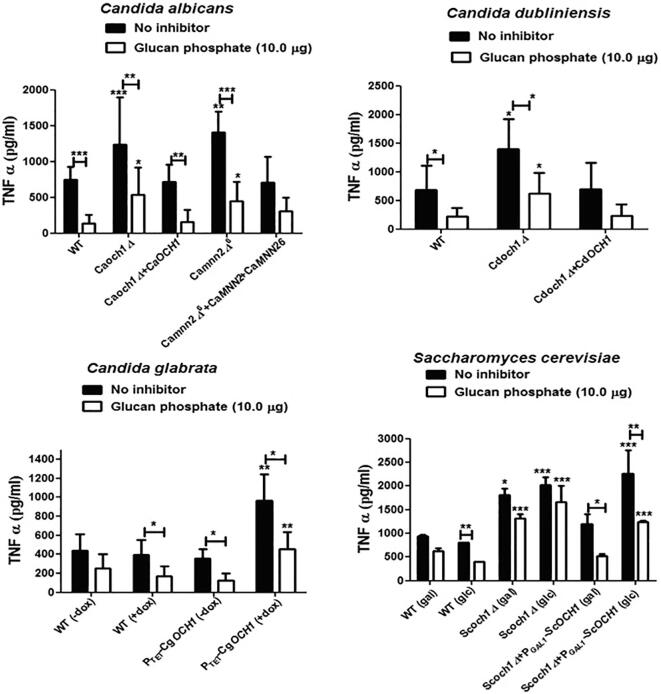
We demonstrated previously that hPBMCs produce low cytokine levels when stimulated with Caoch1Δ null mutant (e.g. Netea et al., 2006). We therefore assessed the impact of OCH1 disruption in C. dubliniensis, C. glabrata, and S. cerevisiae on the ability to stimulate cytokine production by human monocytes. TNFα stimulation was significantly lower upon co-incubation of hPBMCs with heat-killed och1Δ mutants from all the analyzed yeast species (Fig. 4). The levels of IL-6, were also found to be significantly lower in all species (Fig. S3). We showed above that the och1Δ mutants from all the four fungal species resulted in higher β-glucan and chitin exposure (Fig. 2). We therefore tested the effect of the four och1 mutants on immune recognition via dectin-1. TNFα or IL-6 levels were not affected significantly in the presence or absence of the dectin-1 blocker, glucan phosphate (Fig. 4), and thus dectin-1 did not have a major role in the recognition of fungal cells by hPBMCs. We also tested another N-glycosylation mutant from C. albicans, mnn2Δ6, that also had higher β-glucan exposure (Hall et al., 2013). This mutant also displayed reduced cytokine induction in hPBMCs, confirming the importance of N-mannan in immune recognition by human monocytes (Fig. 4 and Fig. S3). Chitin represents another important inner cell wall PAMP which has immunomodulatory effects (Elieh Ali Komi et al., 2018) and has been shown to induce the anti-inflammatory cytokine IL-10 (Wagener et al., 2014). To analyze the impact of exposed chitin in och1Δ mutants on IL-10 secretion by hPBMC’s, the same samples were also analyzed for IL-10 levels. Even though chitin levels were enhanced in och1Δ mutants of all the four fungal species, IL-10 levels were found to be reduced upon challenge with och1Δ mutants (data not shown), as for TNFα and IL-6, indicating a general reduced cytokine induction response by och1Δ mutants in hPBMCs. Therefore, even when the outer ‘masking’ cell wall layer is absent, and the underlying cell wall layers are exposed, human PBMCs recognition of the cell wall was strongly dependent on N-linked mannan.
Fig. 4.
TNFα stimulation by och1 mutants using the hPBMCs: 2*105 heat-killed fungal cells were co-incubated with 5*105 hPBMCs and TNFα levels were quantified by ELISA after 24 h of stimulation. For blocking dectin-1, hPBMCs were pre-treated with 10.0 µg glucan phosphate for 2 h, before challenge with fungal cells. The experiment used monocytes from six donors, with three independent biological replicates, each done in duplicate. Data represent means ± SD. *P < 0.05, **P < 0.005, **P < 0.001.
3.5. Macrophages primarily recognize fungal β-glucan
Next, we assessed the ability of the och1Δ mutants from the four fungal species to stimulate TNFα production in mouse macrophage cell line RAW264.7. We compared the cytokine induction by heat-killed wild type, och1Δ mutants and OCH1 re-integrant controls for these four fungal species. Starkly contrasting with the results obtained with hPBMCs, the heat-killed och1Δ null mutants from all four fungal species stimulated significantly more TNFα production in RAW macrophages than did the wild type and OCH1 re-integrant cells (Fig. 5). Also in contrast with the data using monocytes, TNFα induction in glucan phosphate- treated RAW macrophages was significantly reduced upon challenge with all fungal strains (as compared to the controls without inhibitor) indicating that recognition was dectin-1 dependent (Fig. 5). Interestingly, TNFα induction triggered by the och1 mutants in the presence of the dectin-1 blocking agent, although reduced, remained high compared to wild type and re-integrant control strains under the same conditions, suggesting other PRRs were also involved in cytokine stimulation.
Fig. 5.
TNFα stimulation by och1Δ mutants using the murine macrophage cell line RAW264.7: 2*105 RAW macrophages and 2*105 heat-killed yeast cells were co-incubated for 24 h at 37 °C, and TNFα concentration was determined by ELISA. For blocking dectin-1, RAW macrophages were pre-treated with 10.0 µg glucan phosphate for 2 h, before challenge with fungal cells. The experiment had four biological replicates. Data represent means ± SD. *P < 0.05, **P < 0.005, **P < 0.001.
These data suggest that the absence of N-mannosylation leads to a better recognition of fungal cells by RAW264.7 macrophages, due to unmasking of β-glucan in the inner cell wall layer. A similar increase in TNFα production by och1 mutants from C. albicans and S. cerevisiae was observed in human monocyte derived macrophages (Fig. S4A) and in the J774.1 murine macrophage cell line (Fig. S4B). The above data underline significant differences at interaction level of PAMPs with PRRs in macrophages and monocytes.
4. Discussion
The cell wall components play a central role in fungal sensing by the innate immune system (Erwig and Gow, 2016, Netea et al., 2015). The fungal kingdom displays large heterogeneity and diversity in cell wall structures but most walls consist of identifiable outer and inner cell wall layers (Erwig and Gow, 2016, Garcia-Rubio et al., 2019). β-(1,3)-glucan is a conserved component of most fungal walls and is a structural polymer residing predominately in the inner cell wall layer. β-(1,3)-glucan is a strong immune agonist but it is normally covered by outer wall components and therefore is not immediately accessible to its cognate PRR dectin-1. Therefore components of the outer cell wall have been considered as an impediment to host immune recognition by shielding or masking for the inner cell wall β-(1,3)-glucan. An example is the shielding effect of outer cell wall α-(1,3)-glucan of inner cell wall β-(1,3)-glucan in H. capsulatum (Rappleye et al., 2007, Rappleye and Goldman, 2008). In Candida species the fibrillar outer cell wall mannan layer that represents 30–40% of the wall mass has also been proposed to have this β-(1,3)-glucan shielding function (Bain et al., 2014, Ballou et al., 2016, Pradhan et al., 2019, Pradhan et al. 2018, Wheeler et al., 2008, Wheeler and Fink, 2006). Although cell wall mannan undoubtedly covers and obscures the inner cell wall PAMPs, β-(1,3)-glucan and chitin, multiple PRRs are potentially involved in mannan recognition including mannose receptor (MR), dectin-2, dectin-3, mincle, DC-SIGN, galectin-3, FcγR, CD14, CD23, TLR2, TLR4 and TLR6 (Erwig and Gow, 2016, Vendele et al., 2020) and simultaneous recognition of β-(1,3)-glucan and mannans can result in co-receptor amplification of immune responses (Dennehy et al., 2008). Therefore the superficial mannan layer has the potential to trigger immunity and amplify the inflammatory response as β-(1,3)-glucan recognition also becomes engaged.
N-mannan is the major cell wall protein post-translational modification of outer cell wall proteins and is a significant fraction of the total mass of the Candida cell wall. In the elaboration of N-mannan, Och1 is a conserved α-(1,6)-mannosyltransferase, that catalyses the addition of first α-(1,6)-mannose residue to the conserved core triantennary N-glycan structure (Hall and Gow, 2013). Mutants of Och1 display a severely truncated N-glycan that results in exposure of the inner cell wall layers. C. albicans och1Δ null mutant only stimulated about 20% of the cytokine levels in hPBMCs compared to the wild type control cells, stressing the importance of N-linked mannans for C. albicans immune recognition (Netea et al., 2006). In contrast, the S. cerevisiae och1Δ null mutant was reported to elicit higher levels of TNFα production in RAW264.7 macrophages than the wild type control (Wheeler and Fink, 2006). This marked difference in the phenotype of the och1 mutant in these yeasts could suggest that the mannan structures in S. cerevisiae are different from C. albicans. However, although there are differences in the cell walls of these two yeasts, we show here that the more likely explanation in this disparity lies in the immune cell deployed in these reports, and not the fungus. We demonstrate that although there are cell wall architectural differences between different yeast species such as wall porosity that could impact the immune response elicited, the apparent discrepancy can be explained by the fact that RAW264.7 macrophages predominantly recognize β–glucan and not N-mannan, whereas cytokine induction by hPBMCs is dependent on N-linked mannans. This observation was further supported by extending the analysis to two other Candida species – C. glabrata and C. dubliniensis (Butler et al., 2009, Jackson et al., 2009). For all four fungi, TNFα induction was reduced in the och1 mutant when using human monocytes but was increased when using RAW and other macrophage cell types.
Human PBMCs express most of the well characterised PRRs involved in fungal sensing, including dectin-1, TLR2, TLR4, and mannose receptor (Ferwerda et al., 2008, Netea et al., 2008, van de Veerdonk et al., 2009). Human PBMCs stimulated with C. albicans, C. dubliniensis, C. glabrata or S. cerevisiae och1Δ mutants induced less cytokine production than the wild type controls, indicating that even if the inner cell wall PAMPs β-glucan and chitin were exposed and available for recognition by PRRs, N-linked mannan recognition was required for maximum cytokine stimulation. This observation is in line with other reports indicating that N-linked mannans are required for fungal recognition by innate immune cells (Cambi et al., 2008, Keppler-Ross et al., 2010, Mora-Montes et al., 2010, Mora-Montes et al., 2007, Netea et al., 2006, van de Veerdonk et al., 2009).
Macrophages have a heterogeneous receptor expression and express most of the PRRs (Brown, 2011). Our results support the view that fungal β-glucan is the primary functional PAMP perceived by this cell line, hence och1Δ mutants that have more exposed β-(1,3)-glucan induce higher TNFα levels than the parent wild type. This recognition and TNFα induction was significantly reduced by pre-treatment of the RAW264.7 macrophages with glucan phosphate, a specific blocker of the major β-glucan receptor, dectin-1. In a previous study, macrophage migration had been shown to be enhanced towards C. albicans glycosylation mutants with exposed β-glucan, again suggesting better recognition of these mutants by macrophages (Lewis et al., 2012).
Inflammatory monocytes have been shown to have an essential and protective role in the first 48 h post fungal infection (Ngo et al., 2014). While in circulation, monocytes recognize fungi using a combination of TLRs and CTLs. Upon differentiation into macrophages, while expression of TLRs is maintained, there is an upregulation in the expression of CTLs (Netea et al., 2008). Thus, these results highlight the role of monocytes during the early fungal infection stage, when β-glucan is masked by the outer mannan layer (Wheeler et al., 2008). As the infection progresses, β-glucan is increasingly exposed (Hopke et al., 2018) and thus the dectin-1 mediated recognition of fungal cells by macrophages becomes increasingly relevant and important.
Several studies have demonstrated the cross-talk between the CLRs and TLRs and have shown that PRRs work co-operatively and synergistically in PAMP recognition. Dectin-1 collaboration with TLR2 and TLR4 upregulates cytokine production upon stimulation (Dennehy et al., 2008, Ferwerda et al., 2008, Gantner et al., 2003). Dectin-1 has also been shown to operate in conjunction with galectin-3 and this interaction has been shown to increase TNFα production by macrophages in response to β-(1,3)-glucan (Esteban et al., 2011).
Cooperative interactions of PRRs have been shown in multiple contexts. For example, cytokine production by human monocytes and macrophages was shown to be dependent on both N- and O-linked mannans (mediated by MR and TLR4, respectively), and was reduced when the fungal cells had either N- or O- mannosylation defect (Netea et al., 2006). Dectin-2 forms heterodimers with dectin-3 and these heterodimers were more potent stimulators of cytokine production than the homodimers (Zhu et al., 2013). Galectin-3 association with TLR2 is required for signalling upon recognition of C. albicans (Jouault et al., 2006).
A comparison of the expression of different PRRs in various macrophage and monocyte populations using Immunological Genome Project (Heng et al., 2008) showed that macrophage populations display a higher expression of dectin-1 and TLR-4 as compared to the monocytes. Hence a synergistic response from dectin-1 recognition of exposed β-(1,3)-glucan and TLR-4 recognition of O-mannan in och1Δ mutants could be responsible for higher cytokine production in macrophages. The macrophages also present a higher dectin-2 and dectin-3 expression compared to monocytes (Heng et al., 2008). Dectin-2 recognizes the core mannan structure (Vendele et al., 2020), which is exposed in och1Δ mutants, and hence a cross-talk between these two receptors could also contribute to an augmented cytokine response to och1Δ mutants.
To conclude, we observe that monocytes recognize fungal cells predominantly through N-mannan of the outer Candida cell wall, in contrast to RAW264.7 and other primary macrophages that primarily recognize β-(1,3)-glucan in the inner cell wall layer. These observations reconcile apparent contradictions in the literature about the importance of N-mannan which serves both as a shield of β-(1,3)-glucan recognition and an activator of a range of mannan detecting PRRs depending on the type of immune cell being tested. This highlights the importance of the type of immune cells chosen while studying fungal immune responses in general.
CRediT authorship contribution statement
Bhawna Yadav: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft, Writing - review & editing. Héctor M. Mora-Montes: Investigation, Methodology, Writing - review & editing. Jeanette Wagener: Data curation, Formal analysis, Investigation, Methodology, Writing - review & editing. Iain Cunningham: Investigation, Methodology, Writing - review & editing. Lara West: Data curation, Formal analysis, Writing - review & editing. Ken Haynes: Funding acquisition, Project administration, Conceptualization. Alistair J.P. Brown: Conceptualization, Writing - review & editing. Neil A.R. Gow: Conceptualization, Funding acquisition, Project administration, Resources, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Professor Gordon Brown for Fc-dectin-1 and Professor David Williams for glucan phosphate. We also thank Kevin MacKenzie, Debbie Wilkinson, Gillian Milne, and Lucy Wright at the University of Aberdeen Core Microscopy & Histology Facility.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2020.100042.
Contributor Information
Bhawna Yadav, Email: bhawnayadav1983@yahoo.com.
Héctor M. Mora-Montes, Email: hmora@ugto.mx.
Jeanette Wagener, Email: jeanette_wagener@oslerdiagnostics.com.
Iain Cunningham, Email: iaincunningham@abdn.ac.uk.
Lara West, Email: lara.edmonstone.west@gmail.com.
Alistair J.P. Brown, Email: a.j.p.brown@exeter.ac.uk.
Neil A.R. Gow, Email: n.gow@exeter.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bain J.M., Lewis L.E., Okai B., Quinn J., Gow N.A.R., Erwig L.-P. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet. Biol. FG B. 2012;49:677–678. doi: 10.1016/j.fgb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, J.M., Louw, J., Lewis, L.E., Okai, B., Walls, C.A., Ballou, E.R., Walker, L.A., Reid, D., Munro, C.A., Brown, A.J.P., Brown, G.D., Gow, N.A.R., Erwig, L.P., 2014. Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio 5, e01874. doi: 10.1128/mBio.01874-14. [DOI] [PMC free article] [PubMed]
- Ballou E.R., Avelar G.M., Childers D.S., Mackie J., Bain J.M., Wagener J., Kastora S.L., Panea M.D., Hardison S.E., Walker L.A., Erwig L.P., Munro C.A., Gow N.A.R., Brown G.D., MacCallum D.M., Brown A.J.P. Lactate signalling regulates fungal β-glucan masking and immune evasion. Nat. Microbiol. 2016;2:16238. doi: 10.1038/nmicrobiol.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S., Hughes H.B., Munro C.A., Thomas W.P.H., MacCallum D.M., Bertram G., Atrih A., Ferguson M.A.J., Brown A.J.P., Odds F.C., Gow N.A.R. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006;281:90–98. doi: 10.1074/jbc.M510360200. [DOI] [PubMed] [Google Scholar]
- Bates S., MacCallum D.M., Bertram G., Munro C.A., Hughes H.B., Buurman E.T., Brown A.J.P., Odds F.C., Gow N.A.R. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 2005;280:23408–23415. doi: 10.1074/jbc.M502162200. [DOI] [PubMed] [Google Scholar]
- Borriello F., Zanoni I., Granucci F. Cellular and molecular mechanisms of antifungal innate immunity at epithelial barriers: The role of C-type lectin receptors. Eur. J. Immunol. 2020;50:317–325. doi: 10.1002/eji.201848054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.M., Free S.J. The structure and synthesis of the fungal cell wall. BioEssays News Rev. Mol. Cell. Dev. Biol. 2006;28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- Brand A., MacCallum D.M., Brown A.J.P., Gow N.A.R., Odds F.C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J.P., Brown G.D., Netea M.G., Gow N.A.R. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014;22:614–622. doi: 10.1016/j.tim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.D. Innate antifungal immunity: the key role of phagocytes. Annu. Rev. Immunol. 2011;29:1–21. doi: 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.D., Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- Butler G., Rasmussen M.D., Lin M.F., Santos M.A.S., Sakthikumar S., Munro C.A., Rheinbay E., Grabherr M., Forche A., Reedy J.L., Agrafioti I., Arnaud M.B., Bates S., Brown A.J.P., Brunke S., Costanzo M.C., Fitzpatrick D.A., de Groot P.W.J., Harris D., Hoyer L.L., Hube B., Klis F.M., Kodira C., Lennard N., Logue M.E., Martin R., Neiman A.M., Nikolaou E., Quail M.A., Quinn J., Santos M.C., Schmitzberger F.F., Sherlock G., Shah P., Silverstein K.A.T., Skrzypek M.S., Soll D., Staggs R., Stansfield I., Stumpf M.P.H., Sudbery P.E., Srikantha T., Zeng Q., Berman J., Berriman M., Heitman J., Gow N.A.R., Lorenz M.C., Birren B.W., Kellis M., Cuomo C.A. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambi A., Netea M.G., Mora-Montes H.M., Gow N.A.R., Hato S.V., Lowman D.W., Kullberg B.-J., Torensma R., Williams D.L., Figdor C.G. Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J. Biol. Chem. 2008;283:20590–20599. doi: 10.1074/jbc.M709334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier F., Hall R.A. Face/off: the interchangeable side of Candida Albicans. Front. Cell. Infect. Microbiol. 2019;9:471. doi: 10.3389/fcimb.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Dantas A., Lee K.K., Raziunaite I., Schaefer K., Wagener J., Yadav B., Gow N.A. Cell biology of Candida albicans-host interactions. Curr. Opin. Microbiol. 2016;34:111–118. doi: 10.1016/j.mib.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza I.M., Brown G.D. C-type lectins in immunity: recent developments. Curr. Opin. Immunol. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobel J.G., Klis F.M., Munnik T., Priem J., van den Ende H. An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast Chichester Engl. 1990;6:483–490. doi: 10.1002/yea.320060605. [DOI] [PubMed] [Google Scholar]
- Dennehy K.M., Ferwerda G., Faro-Trindade I., Pyz E., Willment J.A., Taylor P.R., Kerrigan A., Tsoni S.V., Gordon S., Meyer-Wentrup F., Adema G.J., Kullberg B.-J., Schweighoffer E., Tybulewicz V., Mora-Montes H.M., Gow N.A.R., Williams D.L., Netea M.G., Brown G.D. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur. J. Immunol. 2008;38:500–506. doi: 10.1002/eji.200737741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond R.A., Gaffen S.L., Hise A.G., Brown G.D. Innate defense against fungal pathogens. Cold Spring Harb. Perspect. Med. 2014;5 doi: 10.1101/cshperspect.a019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elieh Ali Komi, D., Sharma, L., Dela Cruz, C.S., 2018. Chitin and Its Effects on Inflammatory and Immune Responses. Clin. Rev. Allergy Immunol. 54, 213–223. doi: 10.1007/s12016-017-8600-0. [DOI] [PMC free article] [PubMed]
- Endres S., Ghorbani R., Lonnemann G., van der Meer J.W., Dinarello C.A. Measurement of immunoreactive interleukin-1 beta from human mononuclear cells: optimization of recovery, intrasubject consistency, and comparison with interleukin-1 alpha and tumor necrosis factor. Clin. Immunol. Immunopathol. 1988;49:424–438. doi: 10.1016/0090-1229(88)90130-4. [DOI] [PubMed] [Google Scholar]
- Erwig L.P., Gow N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- Esteban A., Popp M.W., Vyas V.K., Strijbis K., Ploegh H.L., Fink G.R. Fungal recognition is mediated by the association of dectin-1 and galectin-3 in macrophages. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14270–14275. doi: 10.1073/pnas.1111415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Hurtaux T., Fradin C. Mannosylation of fungal glycoconjugates in the Golgi apparatus. Curr. Opin. Microbiol. 2014;20:103–110. doi: 10.1016/j.mib.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Ferwerda G., Meyer-Wentrup F., Kullberg B.-J., Netea M.G., Adema G.J. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell. Microbiol. 2008;10:2058–2066. doi: 10.1111/j.1462-5822.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Fonzi W.A., Irwin M.Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y. Antigenic structure of Candida albicans. Immunochemical basis of the serologic specificity of the mannans in yeasts. Immunol. Ser. 1989;47:37–62. [PubMed] [Google Scholar]
- Gantner B.N., Simmons R.M., Canavera S.J., Akira S., Underhill D.M. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio R., de Oliveira H.C., Rivera J., Trevijano-Contador N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2019;10:2993. doi: 10.3389/fmicb.2019.02993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfoot A.L., Shen Q., Wüthrich M., Klein B.S., Rappleye C.A. The Eng1 β-glucanase enhances histoplasma virulence by reducing β-glucan exposure. mBio. 2016;7:e01388–1315. doi: 10.1128/mBio.01388-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N.A.R., Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Gow N.A.R., Latge J.-P., Munro C.A. The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PubMed] [Google Scholar]
- Gow N.A.R., Netea M.G., Munro C.A., Ferwerda G., Bates S., Mora-Montes H.M., Walker L., Jansen T., Jacobs L., Tsoni V., Brown G.D., Odds F.C., Van der Meer J.W.M., Brown A.J.P., Kullberg B.J. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007;196:1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N.A.R., van de Veerdonk F.L., Brown A.J.P., Netea M.G. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 2012;10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N.A.R., Yadav B. Microbe Profile: Candida albicans: a shape-changing, opportunistic pathogenic fungus of humans. Microbiol. Read. Engl. 2017;163:1145–1147. doi: 10.1099/mic.0.000499. [DOI] [PubMed] [Google Scholar]
- Graus M.S., Wester M.J., Lowman D.W., Williams D.L., Kruppa M.D., Martinez C.M., Young J.M., Pappas H.C., Lidke K.A., Neumann A.K. Mannan molecular substructures control nanoscale glucan exposure in Candida. Cell Rep. 2018;24:2432–2442.e5. doi: 10.1016/j.celrep.2018.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A. Dressed to impress: impact of environmental adaptation on the Candida albicans cell wall. Mol. Microbiol. 2015;97:7–17. doi: 10.1111/mmi.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Bates S., Lenardon M.D., Maccallum D.M., Wagener J., Lowman D.W., Kruppa M.D., Williams D.L., Odds F.C., Brown A.J.P., Gow N.A.R. The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R.A., Gow N.A.R. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 2013;90:1147–1161. doi: 10.1111/mmi.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, T.S.P., Painter, M.W., Immunological Genome Project Consortium, 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed]
- Hernández-Chávez M.J., Pérez-García L.A., Niño-Vega G.A., Mora-Montes H.M. Fungal Strategies to Evade the Host Immune Recognition. J. Fungi Basel Switz. 2017;3 doi: 10.3390/jof3040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A.B., Uccelletti D., Hirschberg C.B., Dominguez A., Abeijon C. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot. Cell. 2002;1:420–431. doi: 10.1128/EC.1.3.420-431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson R.P., Munro C.A., Bates S., MacCallum D.M., Cutler J.E., Heinsbroek S.E.M., Brown G.D., Odds F.C., Gow N.A.R. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 2004;279:39628–39635. doi: 10.1074/jbc.M405003200. [DOI] [PubMed] [Google Scholar]
- Hopke A., Brown A.J.P., Hall R.A., Wheeler R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2018;26:284–295. doi: 10.1016/j.tim.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.P., Gamble J.A., Yeomans T., Moran G.P., Saunders D., Harris D., Aslett M., Barrell J.F., Butler G., Citiulo F., Coleman D.C., de Groot P.W.J., Goodwin T.J., Quail M.A., McQuillan J., Munro C.A., Pain A., Poulter R.T., Rajandream M.-A., Renauld H., Spiering M.J., Tivey A., Gow N.A.R., Barrell B., Sullivan D.J., Berriman M. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res. 2009;19:2231–2244. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouault T., El Abed-El Behi M., Martínez-Esparza M., Breuilh L., Trinel P.-A., Chamaillard M., Trottein F., Poulain D. Specific recognition of Candida albicans by macrophages requires galectin-3 to discriminate Saccharomyces cerevisiae and needs association with TLR2 for signaling. J. Immunol. Baltim. Md. 2006;1950(177):4679–4687. doi: 10.4049/jimmunol.177.7.4679. [DOI] [PubMed] [Google Scholar]
- Jouault T., Ibata-Ombetta S., Takeuchi O., Trinel P.-A., Sacchetti P., Lefebvre P., Akira S., Poulain D. Candida albicans phospholipomannan is sensed through toll-like receptors. J. Infect. Dis. 2003;188:165–172. doi: 10.1086/375784. [DOI] [PubMed] [Google Scholar]
- Keppler-Ross S., Douglas L., Konopka J.B., Dean N. Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot. Cell. 2010;9:1776–1787. doi: 10.1128/EC.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F.M., de Groot P., Hellingwerf K. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 2001;39(Suppl. 1):1–8. [PubMed] [Google Scholar]
- Kozel T.R., MacGill R.S., Percival A., Zhou Q. Biological activities of naturally occurring antibodies reactive with Candida albicans mannan. Infect. Immun. 2004;72:209–218. doi: 10.1128/iai.72.1.209-218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J.-P. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- Levitz S.M. Innate recognition of fungal cell walls. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L.E., Bain J.M., Lowes C., Gillespie C., Rudkin F.M., Gow N.A.R., Erwig L.-P. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman D.W., Greene R.R., Bearden D.W., Kruppa M.D., Pottier M., Monteiro M.A., Soldatov D.V., Ensley H.E., Cheng S.-C., Netea M.G., Williams D.L. Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J. Biol. Chem. 2014;289:3432–3443. doi: 10.1074/jbc.M113.529131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, C.G.J., Koser, U., Lewis, L.E., Bain, J.M., Mora-Montes, H.M., Barker, R.N., Gow, N. a. R., Erwig, L.P., 2010. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78, 1650–1658. doi: 10.1128/IAI.00001-10. [DOI] [PMC free article] [PubMed]
- Molloy C., Cannon R.D., Sullivan P.A., Shepherd M.G. Purification and characterization of two forms of N-acetylglucosaminidase from Candida albicans showing widely different outer chain glycosylation. Microbiol. Read. Engl. 1994;140(Pt 7):1543–1553. doi: 10.1099/13500872-140-7-1543. [DOI] [PubMed] [Google Scholar]
- Mora-Montes H.M., Bates S., Netea M.G., Castillo L., Brand A., Buurman E.T., Díaz-Jiménez D.F., Jan Kullberg B., Brown A.J.P., Odds F.C., Gow N.A.R. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010;285:12087–12095. doi: 10.1074/jbc.M109.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H.M., Bates S., Netea M.G., Díaz-Jiménez D.F., López-Romero E., Zinker S., Ponce-Noyola P., Kullberg B.J., Brown A.J.P., Odds F.C., Flores-Carreón A., Gow N.A.R. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell. 2007;6:2184–2193. doi: 10.1128/EC.00350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Montes H.M., Ponce-Noyola P., Villagómez-Castro J.C., Gow N.A., Flores-Carreón A., López-Romero E. Protein glycosylation in Candida. Future Microbiol. 2009;4:1167–1183. doi: 10.2217/fmb.09.88. [DOI] [PubMed] [Google Scholar]
- Mukaremera L., Lee K.K., Mora-Montes H.M., Gow N.A.R. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front. Immunol. 2017;8:629. doi: 10.3389/fimmu.2017.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro C.A., Bates S., Buurman E.T., Hughes H.B., Maccallum D.M., Bertram G., Atrih A., Ferguson M.A.J., Bain J.M., Brand A., Hamilton S., Westwater C., Thomson L.M., Brown A.J.P., Odds F.C., Gow N.A.R. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005;280:1051–1060. doi: 10.1074/jbc.M411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A.M., Lee P.R., Broadbent I.D., Barelle C.J., Brown A.J. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast Chichester Engl. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Nakayama H., Izuta M., Nagahashi S., Sihta E.Y., Sato Y., Yamazaki T., Arisawa M., Kitada K. A controllable gene-expression system for the pathogenic fungus Candida glabrata. Microbiol. Read. Engl. 1998;144(Pt 9):2407–2415. doi: 10.1099/00221287-144-9-2407. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Brown G.D., Kullberg B.J., Gow N.A.R. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Gow N.A.R., Munro C.A., Bates S., Collins C., Ferwerda G., Hobson R.P., Bertram G., Hughes H.B., Jansen T., Jacobs L., Buurman E.T., Gijzen K., Williams D.L., Torensma R., McKinnon A., MacCallum D.M., Odds F.C., Van der Meer J.W.M., Brown A.J.P., Kullberg B.J. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., van der Meer J.W.M., Kullberg B.-J., van de Veerdonk F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015;15:630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Van Der Graaf C.A.A., Vonk A.G., Verschueren I., Van Der Meer J.W.M., Kullberg B.J. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J. Infect. Dis. 2002;185:1483–1489. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- Ngo L.Y., Kasahara S., Kumasaka D.K., Knoblaugh S.E., Jhingran A., Hohl T.M. Inflammatory monocytes mediate early and organ-specific innate defense during systemic candidiasis. J. Infect. Dis. 2014;209:109–119. doi: 10.1093/infdis/jit413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakopoulou C., Willment J.A., Brown G.D. C-Type Lectin Receptors in Antifungal Immunity. Adv. Exp. Med. Biol. 2020;1204:1–30. doi: 10.1007/978-981-15-1580-4_1. [DOI] [PubMed] [Google Scholar]
- Nishikawa A., Poster J.B., Jigami Y., Dean N. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 2002;184:29–42. doi: 10.1128/JB.184.1.29-42.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patin E.C., Thompson A., Orr S.J. Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 2019;89:24–33. doi: 10.1016/j.semcdb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaine A., Walker L., Da Costa G., Mora-Montes H.M., McKinnon A., Gow N.A.R., Gaillardin C., Munro C.A., Richard M.L. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. FG B. 2008;45:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plato A., Hardison S.E., Brown G.D. Pattern recognition receptors in antifungal immunity. Semin. Immunopathol. 2015;37:97–106. doi: 10.1007/s00281-014-0462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A., Avelar G.M., Bain J.M., Childers D., Pelletier C., Larcombe D.E., Shekhova E., Netea M.G., Brown G.D., Erwig L., Gow N.A.R., Brown A.J.P. Non-canonical signalling mediates changes in fungal cell wall PAMPs that drive immune evasion. Nat. Commun. 2019;10:5315. doi: 10.1038/s41467-019-13298-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan, A., Avelar, G.M., Bain, J.M., Childers, D.S., Larcombe, D.E., Netea, M.G., Shekhova, E., Munro, C.A., Brown, G.D., Erwig, L.P., Gow, N.A.R., Brown, A.J.P., 2018. Hypoxia Promotes Immune Evasion by Triggering β-Glucan Masking on the Candida albicans Cell Surface via Mitochondrial and cAMP-Protein Kinase A Signaling. mBio 9. doi: 10.1128/mBio.01318-18. [DOI] [PMC free article] [PubMed]
- Qin Y., Zhang L., Xu Z., Zhang J., Jiang Y.-Y., Cao Y., Yan T. Innate immune cell response upon Candida albicans infection. Virulence. 2016;7:512–526. doi: 10.1080/21505594.2016.1138201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C.A., Eissenberg L.G., Goldman W.E. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye C.A., Goldman W.E. Fungal stealth technology. Trends Immunol. 2008;29:18–24. doi: 10.1016/j.it.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Rogers N.C., Slack E.C., Edwards A.D., Nolte M.A., Schulz O., Schweighoffer E., Williams D.L., Gordon S., Tybulewicz V.L., Brown G.D., Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Saijo S., Ikeda S., Yamabe K., Kakuta S., Ishigame H., Akitsu A., Fujikado N., Kusaka T., Kubo S., Chung S., Komatsu R., Miura N., Adachi Y., Ohno N., Shibuya K., Yamamoto N., Kawakami K., Yamasaki S., Saito T., Akira S., Iwakura Y. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Salazar F., Brown G.D. Antifungal innate immunity: a perspective from the last 10 years. J. Innate Immun. 2018;10:373–397. doi: 10.1159/000488539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Arai M., Haga E., Kikuchi T., Najima M., Satoh T., Kobayashi H., Suzuki S. Structural identification of an epitope of antigenic factor 5 in mannans of Candida albicans NIH B-792 (serotype B) and J-1012 (serotype A) as beta-1,2-linked oligomannosyl residues. Infect. Immun. 1992;60:4100–4110. doi: 10.1128/iai.60.10.4100-4110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snarr B.D., Qureshi S.T., Sheppard D.C. Immune recognition of fungal polysaccharides. J. Fungi Basel Switz. 2017;3 doi: 10.3390/jof3030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib P., Moran G.P., Sullivan D.J., Coleman D.C., Morschhäuser J. Isogenic strain construction and gene targeting in Candida dubliniensis. J. Bacteriol. 2001;183:2859–2865. doi: 10.1128/JB.183.9.2859-2865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.R., Brown G.D., Herre J., Williams D.L., Willment J.A., Gordon S. The role of SIGNR1 and the beta-glucan receptor (dectin-1) in the nonopsonic recognition of yeast by specific macrophages. J. Immunol. Baltim. Md. 2004;1950(172):1157–1162. doi: 10.4049/jimmunol.172.2.1157. [DOI] [PubMed] [Google Scholar]
- Ueno K., Uno J., Nakayama H., Sasamoto K., Mikami Y., Chibana H. Development of a highly efficient gene targeting system induced by transient repression of YKU80 expression in Candida glabrata. Eukaryot. Cell. 2007;6:1239–1247. doi: 10.1128/EC.00414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk F.L., Kullberg B.J., van der Meer J.W.M., Gow N.A.R., Netea M.G. Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr. Opin. Microbiol. 2008;11:305–312. doi: 10.1016/j.mib.2008.06.002. [DOI] [PubMed] [Google Scholar]
- van de Veerdonk F.L., Marijnissen R.J., Kullberg B.J., Koenen H.J.P.M., Cheng S.-C., Joosten I., van den Berg W.B., Williams D.L., van der Meer J.W.M., Joosten L.A.B., Netea M.G. The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell Host Microbe. 2009;5:329–340. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Vendele I., Willment J.A., Silva L.M., Palma A.S., Chai W., Liu Y., Feizi T., Spyrou M., Stappers M.H.T., Brown G.D., Gow N.A.R. Mannan detecting C-type lectin receptor probes recognise immune epitopes with diverse chemical, spatial and phylogenetic heterogeneity in fungal cell walls. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener J., Malireddi R.K.S., Lenardon M.D., Köberle M., Vautier S., MacCallum D.M., Biedermann T., Schaller M., Netea M.G., Kanneganti T.-D., Brown G.D., Brown A.J.P., Gow N.A.R. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, L., Sood, P., Lenardon, M.D., Milne, G., Olson, J., Jensen, G., Wolf, J., Casadevall, A., Adler-Moore, J., Gow, N.A.R., 2018. The Viscoelastic Properties of the Fungal Cell Wall Allow Traffic of AmBisome as Intact Liposome Vesicles. mBio 9. doi: 10.1128/mBio.02383-17. [DOI] [PMC free article] [PubMed]
- West L., Lowman D.W., Mora-Montes H.M., Grubb S., Murdoch C., Thornhill M.H., Gow N.A.R., Williams D., Haynes K. Differential virulence of Candida glabrata glycosylation mutants. J. Biol. Chem. 2013;288:22006–22018. doi: 10.1074/jbc.M113.478743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.T., Fink G.R. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006;2 doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.T., Kombe D., Agarwala S.D., Fink G.R. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.B., Davis D., Enloe B.M., Mitchell A.P. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast Chichester Engl. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Klein T.W., Friedman H. Involvement of mannose receptor in cytokine interleukin-1beta (IL-1beta), IL-6, and granulocyte-macrophage colony-stimulating factor responses, but not in chemokine macrophage inflammatory protein 1beta (MIP-1beta), MIP-2, and KC responses, caused by attachment of Candida albicans to macrophages. Infect. Immun. 1997;65:1077–1082. doi: 10.1128/iai.65.3.1077-1082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.Q., Zou Z., Shen H., Shen S.S., Miao Q., Huang X., Liu W., Li L.P., Chen S.M., Yan L., Zhang J.D., Zhao J.J., Xu G.T., An M.M., Jiang Y.Y. Mnn10 maintains pathogenicity in Candida albicans by extending α-1,6-mannose backbone to evade host dectin-1 mediated antifungal immunity. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.-L., Zhao X.-Q., Jiang C., You Y., Chen X.-P., Jiang Y.-Y., Jia X.-M., Lin X. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39:324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.