Abstract
Cellular homeostasis in eukaryotic cells requires synchronized coordination of multiple organelles. A key role in this stage is played by mitochondria, which have recently emerged as highly interconnected and multifunctional hubs that process and coordinate diverse cellular functions. Beyond producing ATP, mitochondria generate key metabolites and are central to apoptotic and metabolic signaling pathways. Because most mitochondrial proteins are encoded in the nuclear genome, the biogenesis of new mitochondria and the maintenance of mitochondrial functions and flexibility critically depend upon effective mitonuclear communication. This review addresses the complex network of signaling molecules and pathways allowing mitochondria-nuclear communication and coordinated regulation of their independent but interconnected genomes, and discusses the extent to which dynamic communication between the two organelles has evolved for mutual benefit and for the overall maintenance of cellular and organismal fitness.
Keywords: Mitonuclear Communication, Mitochondrial Retrograde Signaling, Integrated Stress Response, Epigenetics
Graphical Abstract
Compounds:
Nicotinamide riboside; Superoxide anion; Acetyl-Coenzyme A; S-adenosylmethionine (SAM); Carbonyl cyanide m-chlorophenylhydrazone (CCCP); Oligoymycin; Antimycin; Humanin; alpha-Ketoglutarate
1 -. Introduction
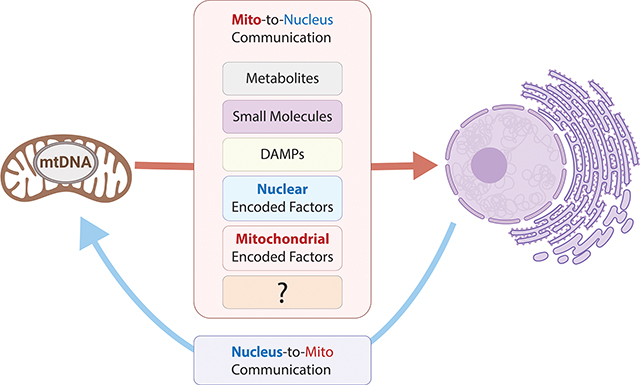
Eukaryotic cells presumably evolved from the union of a primordial ancestral cell and bacteria that each brought their independent genomes {Roger, 2017 #2333;Martijn, 2018 #2504}. The endosymbiotic bacteria gave rise to mitochondria, which, due to their bacterial origin, possess a unique circular genome. The mitochondrial and nuclear genomes together constitute a bi-genomic genetic system-Over time, much of the bacterial genome relocated to the host nucleus-through lateral gene transfer (Dyall et al., 2004)(Timmis et al., 2004) (Adams and Palmer, 2003; Burger et al., 2003; Gray, 2012). The transfer of genetic information represents an ongoing evolutionary process. Degenerate mitochondrial DNA segments or sequence insertions into the nuclear genome, referred to as nuclear mitochondrial DNAs (NUMTs) (Lopez et al., 1994), constitute genetic markers of the ongoing lateral gene transfer process (Caro-Quintero et al., 2011; Dayama et al., 2014; Lang et al., 2012; Thomas and Nielsen, 2005) and provide genetic material for gene regulation (Goldin et al., 2004; Schon et al., 2012; Turner et al., 2003; Willett-Brozick et al., 2001). NUMTs are present across species and exist in fragments of varying lengths and homology that are dispersed throughout the genome and can collectively cover the entire mitochondrial DNA sequence (Calabrese et al., 2017; Pereira and Baker, 2004; Richly and Leister, 2004; Simone et al., 2011). Although the evolutionary benefits of maintaining a bi-genomic system in the cell are not fully understood, a strong selection force appears to have favored it as the cost and complexity of maintaining two genomes within a cell is far higher than operating based on a single genome. As we discuss further below, one possible reason behind the dual-genome setup may be to optimize mitochondrial communication. Because mitochondria can exist in the thousands in a given cell, it is plausible that the inherent ability to respond without the need for the nucleus to keep track of all individual mitochondrion would be necessary. Nonetheless, mitochondria and the nucleus have co-evolved at multiple levels. As the two previously independent organisms integrated to generate a unified organism with two interdependent genomes, novel pathways evolved to promote communication between organelles and allow for coordinated and cross-regulated gene expression (Kim et al., 2018; Mottis et al., 2019), thus resulting in a unified genetic network that is built on and regulated by information contributed by both genomes.
Mitochondrial-nuclear coordination is mediated by a robust and sophisticated communication system, which includes a range of signaling factors that are increasingly being identified. Proper mitonuclear coordination is achieved through bi-directional transmission channels, traditionally referred to as anterograde (nucleus to mitochondria) and retrograde (mitochondria to nucleus) signaling. While much of the early work on mitonuclear communication focused on the nucleus-to-mitochondria direction, it is now well appreciated that mitochondria-to-nucleus signaling is also critical to regulating cellular homeostasis and specific processes, such as metabolism, proliferation, differentiation, and stress adaptation. For detailed discussion of the nuclear network of transcription factors and associated cofactors that regulate nucleus-to-mitochondria signaling through the expression of nuclear-encoded mitochondrial genes, we refer to other reviews (Fang et al., 2016; Hock and Kralli, 2009; Scarpulla et al., 2012; Whelan and Zuckerbraun, 2013; Wu et al., 1999). Here, we will provide a review of the mechanisms supporting mitochondria-to-nucleus signaling, including the exchange of mitochondrial- and nuclear-encoded factors, damaged mitochondrial components that elicit immune responses, and metabolic intermediates with important ties to epigenetic modifications, and discuss how these functional signaling networks are required for maintaining essential cellular communication, homeostatic balance, and prevention of disease through coordinated reprogramming of nuclear gene expression programs.
2-. Mitochondrial-derived signaling molecules
Advances in mitochondrial research in recent years have significantly expanded our view of their roles, from semi-independent organelles responsible for metabolic and apoptotic processes to key signaling hubs that are well integrated in multiple signal transduction pathways (Abate et al., 2020; Chandel, 2015; Rath et al., 2018). For excellent coverage of the many ways mitochondrial-derived signals contribute to regulate a variety of cellular functions, we refer to other recent reviews (Bohovych and Khalimonchuk, 2016; Mottis et al., 2019). Here, we will focus on discussing the many intricate mechanisms by which mitochondrial-derived signaling molecules contribute to the communication between mitochondria and nucleus, either directly through intraorganellar translocation, or indirectly by providing cofactors/substrates that provide the metabolic context for transcriptional and epigenetic regulation (Figure 1).
Figure 1.
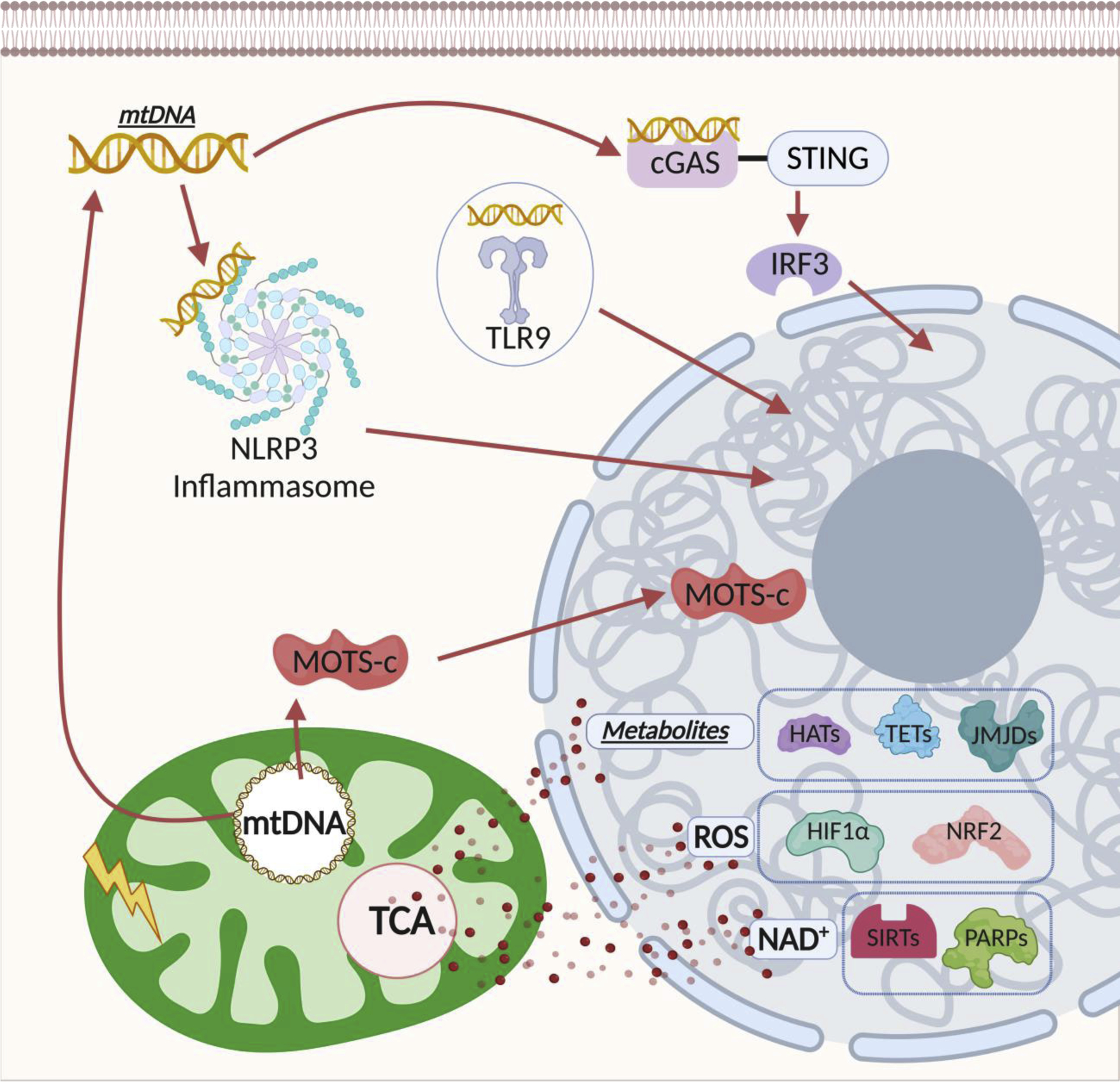
Mitochondrial-derived signaling molecules promoting mitonuclear communication. Mitochondria-derived signaling molecules promote signaling to the nucleus through both direct and indirect actions. They can be classified in three broad categories as discussed in the text: 1)Peptides, 2)mtDNA, and 3)Small molecules & Metabolites such as ROS, NAD+, Ca+, Acetyl-CoA, α-KG, 2-HG, succinate, fumarate.
2.1. Peptides
Human mitochondrial DNA (mtDNA) has been traditionally described to encode for 13 proteins, all of which are components of the electron transport chain (ETC). More recently, short open reading frames (sORFs) encoding for bioactive peptides, collectively referred to as mitochondrial-derived peptides (MDPs), have been identified in the mtDNA. MDPs appear to play a significant protective role in mitochondrial health and cell viability in a number of disease contexts including oxidative stress, atherosclerosis, and age-related macular degeneration (Nashine and Kenney, 2020). To date eight MDPs have been reported, including humanin {Hashimoto, 2001 #117;Guo, 2003 #133;Ikonen, 2003 #132}, six small humanin-like peptides (SHLP1–6)(Cobb et al., 2016), and mitochondrial ORF of the 12S ribosomal RNA type-c (MOTS-c)(Lee et al., 2015).
MOTS-c is a peptide encoded within the mitochondrial 12S ribosomal RNA. MOTS-c regulates metabolism, in part, via AMP-activated protein kinase (AMPK) and sirtuin 1 (SIRT1) and promotes cellular homeostasis. In mice, MOTS-c treatment prevented diet-induced obesity and insulin-resistance and reversed age-dependent insulin resistance (Lee et al., 2015). MOTS-c regulates adaptive nuclear gene expression by translocating to the nucleus in response to metabolic stress and interacting with transcriptional factors, including nuclear factor erythroid 2-related factor 2 (Nrf2) and activating transcription factor-1 (ATF1) (Kim et al., 2018). While the details of the nuclear actions of MOTS-c is a topic of active investigation, its identification as the first mitochondrial-encoded factor that directly regulates the nuclear genome reveals cross-regulation between the two genomes. This adds an unprecedented layer to the complexity of mitonuclear communication and raises the question whether other MDPs may facilitate communication between the two genomes. Notably, MOTS-c levels are induced in response to exercise stress in humans and its treatment in mice significantly improves physical performance at all age groups (Reynolds et al., 2019). RNA-seq analyses on aged mouse muscle and myoblasts indicate that MOTS-c regulates proteostasis under stress conditions (Reynolds et al., 2019), which is of special interest considering that MOTS-c is itself encoded within a ribosomal gene. Meanwhile, Tar1p, a small peptide encoded in the nuclear 25S ribosomal RNA gene in S. Cerevisiae, functions in mitochondria in response to respiratory demand and dysfunction (Bonawitz et al., 2008), which may reflect a coordinated mitonuclear ribosomal response.
Although it is unclear whether other MDPs directly regulate nuclear function, their diverse cellular functions entail potential interaction with and effect on nuclear gene expression. Humanin, the first reported MDP encoded within the mitochondrial 16S ribosomal RNA, was reported to have protective effects in neurons, brain, eyes, bone, the vascular and the cardiovascular system, (Lee et al., 2013; Tajima et al., 2002; Zarate et al., 2019). Its protective effects have been associated with inhibition of apoptosis and inflammation, oxidation, and regulation of mitochondrial function (Kuliawat et al., 2013; Yen et al., 2013; Zapala et al., 2010). Following humanin, SHLPs were discovered also within mitochondrial 16S ribosomal RNA. Whereas SHLP2 and SHLP3 were reported to have a cytoprotective effect similar to that of humanin, SHLP6 promotes apoptosis (Cobb et al., 2016). SHLPs are also involved in regulation of mitochondrial bioenergetics and chaperone-like function (Nashine et al., 2018; Okada et al., 2017).
2.2. Mitochondrial DNA
Damage-associated molecular patterns (DAMPs) derived from endogenous intracellular components that are released during cellular stress or damage can lead to activation of innate immune responses. In particular, stimulation of pro-inflammatory responses can be triggered by the release of mitochondrial-derived DAMPs (mtDAMPs), including N-formylated peptides and mtDNA as discussed in depth in other reviews (Grazioli and Pugin, 2018; West, 2017; West and Shadel, 2017). Relevant to this review is the fact that release of mtDNA in the cytosol indirectly induces signaling to the nucleus and reprogramming of nuclear gene expression to support the innate immune response activation. This is achieved through at least three different receptor-mediated pathways: toll-like receptors (TLRs), NOD-like receptors (NLRs), and interferon stimulatory DNA receptors, with each receptor supporting the regulation of different immune response arms. mtDNA recognition by TLRs, mainly TLR9, promotes gene expression through nuclear factor kappa B (NF-κB) activation (Kawai and Akira, 2010). mtDNA activation of the NLRP3 inflammasome is responsible for caspase-1 mediated processing and secretion of IL-1β and IL-8 (Kawai and Akira, 2010; Shimada et al., 2012). Furthermore, cytoplasmic mtDNA activates the cGAS-STING-TBK1-IRF3 pathway, resulting in interferon-stimulated gene expression that promotes antiviral immunity (Kwon and Bakhoum, 2020) and acts as a genotoxic stress sentinel that signals the nucleus to enhance DNA repair (Wu et al., 2019). Notably, signaling through mtDNA pattern recognition provides a means not only for intracellular mito-to-nuclear communication, but also for paracrine and organismal signaling, via release of mtDNA in the extracellular space and the circulation. Mitochondrial DNA found in the serum is referred to as circulating cell-free mtDNA (ccf-mtDNA). Several studies reported that ccf-mtDNA contributes to inflammation in type 2 diabetes (Bae et al., 2019) and neuro-immunological disorders (Gambardella et al., 2019). Moreover, not only physical stress but also psychological stress triggers increases of mtDNA in the circulating serum (Trumpff et al., 2019). Further studies are warranted for the understanding of this fine regulation in immune response, which may provide the opportunity for translational therapeutic development in the area of mtDNA signaling. For example, a key aspect of signaling through mtDNA release from damaged mitochondria involves the regulation of mtDNA export, including (i) formation of voltage-dependent anion channel (VDAC) oligomer pores (Kim et al., 2019), (ii) transient (non-lethal) and selective mitochondrial outer membrane permeabilization (MOMP), also referred to as minority MOMP (Brokatzky et al., 2019; Ichim et al., 2015; Xu et al., 2020), and (iii) mitochondrial permeability transition pore (mPTP) (Martinez-Abundis et al., 2007; Nakahira et al., 2011; Patrushev et al., 2004; Szczesny et al., 2018). Excessive interferon (IFN) response increases VDAC1/3 expression (Fernandez et al., 2009; Kim et al., 2019). MOMP has also been suggested to mediate mtDNA release and IFN response, commensurate to mitochondrial stress (Kim et al., 2019). Additionally, mPTP can mediate LPS-induced mtDNA release (Kim et al., 2019; Nakahira et al., 2011). As is the case with mtDNA-stimulated immune responses, mtDNA export mechanisms represent an area of potential therapeutic targets, particularly in the case of pathological conditions associated with uncontrolled inflammation.
2.3. Small Molecules
2.3.1. NAD+
Nicotinamide adenine dinucleotide (NAD) is present in the cell in two forms: an oxidized (NAD+) and a reduced (NADH) form. Because the ratio between this redox couple provides an indication of the metabolic status of the mitochondria, NAD serves as an important metabolic sensor or gauge. NAD+ functions as a key cofactor for multiple metabolic reactions and as a substrate for different classes of enzymes, including some with critical functions in the regulation of nuclear functions and gene expression, such as deacetylases and ADP-ribosyl transferases (ARTs). Thus, NAD+ levels reflect the cellular energetic status while providing metabolic input to adaptive gene regulation. This is favored by NAD+ being both tightly regulated and compartmentalized within the cell (Canto et al., 2009; Zhu et al., 2019). Distinct pools of NAD+ exist in the mitochondria, cytosol, and nucleus respectively, with the cytosolic pool serving as the hub that connects the others (Zhu et al., 2019). Even though the mechanism/s regulating the synthesis, compartmentalization and movement of NAD+ between the mitochondria and the cytosol/nucleus are not fully characterized, it is clear that each subcellular pool is regulated and utilized differently in terms of metabolic flux or NAD+-dependent signaling (Anderson et al., 2017; Davila et al., 2018; Zhu et al., 2019)
Deacetylation is a critical enzymatic process for the regulation of nuclear gene expression. Removal of an acetyl group from lysine residues on histone tails, as governed by histone deacetylases (HDACs), can directly affect chromatin compaction and result in gene repression (Seto and Yoshida, 2014; Yang and Seto, 2007). Moreover, as HDACs activity is not limited to histones, deacetylation of transcription factors and transcriptional regulators can be used to modulate their function, thus providing further specificity to transcriptional regulation (Choudhary et al., 2009; Park et al., 2015; Sterner and Berger, 2000; Thiagarajan et al., 2016). Among HDACs, sirtuins (Class II HDACs) are unique in that they require NAD+ as a cofactor. Overall, Sirtuins are well described as key regulators of cellular homeostasis and have been shown to be preventive of multiple diseases (Houtkooper et al., 2012; Kupis et al., 2016; Morigi et al., 2018; Pirinen et al., 2012; Vachharajani and McCall, 2020). The intricacies of the specific mechanism, utilizations, and biochemistry of NAD+ and its production are covered elsewhere (Anderson et al., 2017; Canto et al., 2015; Houtkooper et al., 2010; Kulkarni and Brookes, 2019; Sauve et al., 2006). Briefly, the NAD+/sirtuin enzymatic reaction functions such that NAD+ is consumed and nicotinamide (NAM), O-acetyl ADP ribose, and the deacetylated substrate (both histone and non-histone) are released. Given that NAD+ has a central role in energy metabolism, tricarboxylic acid (TCA) cycle flux, and nutrient sensing, changes in its availability can directly link the metabolic status of mitochondria with chromatin remodeling and nuclear reprogramming, in part, via sirtuin activity (Bosch-Presegue and Vaquero, 2015; North and Verdin, 2004). Phylogenetically, sirtuins have been around since prokaryotes and have emerged as key enzymatic regulators of nutrient and cell cycle stress during evolution (Bosch-Presegue and Vaquero, 2015; Frye, 2000). To date, seven mammalian sirtuins have been identified (SIRT1-SIRT7), each with unique subcellular localizations that allow for the utilization of specific NAD+ pools and the modulation of specific targets. SIRT3,4, & 5, for example, function as key regulators of metabolism and oxidative stress from within the mitochondria. This is not only based on the deacetylation of target proteins, as profiling of the acetylome regulated by the mitochondrial sirtuins has revealed a number of novel modifications targeted by sirtuins, including succinylation, malonylation and glutarylation (Carrico et al., 2018; Downey et al., 2015; Hirschey and Zhao, 2015; Park et al., 2013; Rardin et al., 2013; Rauh et al., 2013). The balance between spontaneous acylation and NAD-dependent de-acylation of mitochondrial proteins significantly contributes to the regulation of mitochondrial functions and metabolic pathways As a result, mitochondrial sirtuins have been associated with protective effects against aging, insulin resistance and other age-related pathologies. At the same time, the nuclear role of SIRT1 and its relationship with NAD+ have been widely studied in the context of mammalian physiology, stress response, aging and numerous diseases, including cancer and metabolic disorders (Alves-Fernandes and Jasiulionis, 2019; Haigis and Sinclair, 2010; Rahman and Islam, 2011; Tang, 2016; Xu et al., 2018). In addition to histones, the transcriptional cofactor PGC1α is a key target of NAD+-dependent deacetylation. Deacetylation of PGC1α by SIRT1 leads to its activation and the induction of downstream pathways that control mitochondrial gene expression, particularly those related to mitochondrial biogenesis, fatty acid oxidation, and oxidative stress response (Feige et al., 2008).
Other enzymes that utilize NAD+ as a substrate are ADP-ribosyl transferases. Among them, Poly(ADP-ribose) polymerase 1 (PARP1) plays key roles in transcriptional regulation, chromatin reorganization, nuclear organization and DNA damage repair (Ju and Rosenfeld, 2006; Kraus and Lis, 2003; Krishnakumar and Kraus, 2010). Changes in NAD+ levels can signal to the nucleus by affecting each of these functions. For example, under basal conditions, PARP-1 binds to nucleosomes and promotes chromatin compaction, which is impaired upon saturating NAD+ levels, leading to PARP1 auto-PARylation (Kim et al., 2004; Wacker et al., 2007). Notably, the need for NAD+ by multiple enzymes, including those discussed here and others (Anderson et al., 2017; Audrito et al., 2019; Koch-Nolte et al., 2011; Ma et al., 2012; Ma et al., 2015), sets the stage for crosstalk between them based on competition for NAD+ availability, as shown by studies with PARP-1 and SIRT1 (Canto et al., 2013; Kim et al., 2005; Luna et al., 2013; Zhang and Kraus, 2010).
2.3.2. ROS
Reactive oxygen species (ROS), including the superoxide anion (O2•−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH•), represent major byproducts of aerobic metabolism (Andreyev et al., 2005; Brieger et al., 2012; Liochev, 2013; Liu et al., 2018), with mitochondria being one of the major production sites in most cells (Brand et al., 2004; Turrens, 2003). Upon their initial discovery, it was immediately noted how excess levels of these superoxide species are damaging and associate with a variety of deleterious conditions including cell death, cancer, and degenerative aging (Balaban et al., 2005; Davalli et al., 2016; Droge, 2002; Evans et al., 2003; Harman, 1981). However, with time, our understanding of the role played by these small molecules in the cell has become more nuanced. Although it is clear that high levels of ROS play a major role in oxidative damage, it is now well appreciated that ROS-dependent redox signaling is physiologically necessary (Droge, 2002; Finkel, 2011; Sarsour et al., 2014; Sena and Chandel, 2012) Some of these include anti-microbial or tumoricidal defense coupled with phagocytic cells (Keisari et al., 1983), modulation of protein kinase cascades in vascular smooth muscle cell growth and migration (Griendling et al., 2000), maintenance of whole body homeostasis and metabolism through neuroendocrine connections (Shadel and Horvath, 2015), regulation of glucose-stimulated insulin secretion in pancreatic beta cells (Pi et al., 2007; Pi et al., 2010), and hormetic regulation of lifespan (Merry and Ristow, 2016; Santos et al., 2018; Schroeder et al., 2013; Scialo et al., 2013). At the molecular level, mitochondrial-derived ROS (mtROS) promote these responses through a variety of signaling pathways, often signaling to the nucleus through stabilization or regulated translocation of DNA-binding transcription factors. Increased ROS level under hypoxic conditions, for example, promote hypoxia-inducible factor 1 (HIF1)-mediated transcriptional regulation through inhibition of prolyl hydroxylases (PHDs) and stabilization of HIF1α (Chandel et al., 2000; Duranteau et al., 1998). Activation of the antioxidant protective response is similarly achieved through stabilization and nuclear translocation of Nrf2 (Kovac et al., 2015; Tonelli et al., 2018). mtROS also impact cell survival/cell death pathways and cytokine production through activation of NF-κB (Hamanaka and Chandel, 2010; Herb et al., 2019; Kuwabara et al., 2008; Morgan and Liu, 2011; Naik and Dixit, 2011). In parallel to specific regulation of DNA-binding transcription factors, oxidative stress can impact on nuclear gene expression via epigenetic changes induced through modulation of chromatin regulators, such as histone demethylases and deacetylases (Chervona and Costa, 2012; Niu et al., 2015). These examples, selected to highlight how ROS directly and indirectly impact upon nuclear functions and reprogramming of gene expression as signaling mediators, only scratch the surface of the diverse roles played by mtROS in cell signaling and physiology. Comprehensive reviews of mitochondrial-derived ROS outline how these small signaling molecules are crucial components of numerous signaling cascades, the likes of which work towards the maintenance of homeostasis and physiologic balance within the cell (Dan Dunn et al., 2015; Reczek and Chandel, 2015; Ristow and Schmeisser, 2011; Shadel and Horvath, 2015). One particularly intriguing aspect of ROS signaling, as relevant to the focus of this review, is the relationships between mitochondrial dynamics and nuclear functions of mtROS. In pulmonary artery endothelial cells, hypoxia was shown to trigger the increase of nuclear ROS levels and HIF1α-dependent regulation of vascular endothelial growth factor (VEGF) expression through accumulation of mitochondria in close proximity of the nucleus via microtubule-associated movement (Al-Mehdi et al., 2012). This observation suggests that reorganization of the subcellular distribution of mitochondria could play an important role in promoting local signaling to the nucleus, something that warrant additional research for ROS as well as other signaling mediators discussed below.
2.3.3. Calcium
As better discussed elsewhere, calcium (Ca2+) ions serve as signaling molecules in a vast number of cellular processes that are crucial for maintaining cellular homeostasis (Berridge et al., 2003; Bootman, 2012; Clapham, 2007; Islam, 2020). To allow for sensitivity and specificity of signaling, calcium levels and fluxes across cellular and intracellular membranes are tightly controlled by a complex and dynamic system of buffers and pumps, while changes are detected by calcium sensors that activate downstream signaling cascades (Berridge et al., 2003; Clapham, 2007; Park et al., 2019). Mitochondria are integral to this system as they both contribute to Ca2+ storage and flux regulation, and respond to changes in cellular Ca2+ levels via apoptotic responses and adaptation of mitochondrial metabolism (Contreras et al., 2010; Giorgi et al., 2018). Additionally, it has long been noted that Ca2+ is a regulator of TCA cycle dehydrogenase activity (specifically pyruvate dehydrogenase, isocitrate dehydrogenase, and α-ketoglutarate dehydrogenase), thus providing an important link between this small signaling molecule and metabolism (McCormack and Denton, 1980; Traaseth et al., 2004; Wan et al., 1989). In the context of mitochondrial mitochondria-to-nucleus signaling, an important facet of Ca2+ signaling is based on the effector protein calcineurin, a Ca2+-dependent serine-threonine phosphatase. Calcium-calcineurin mediated mitochondria-to-nucleus signaling is activated in response to cellular stress-inducing factors, including both genetic and metabolic mitochondrial stressors, that promote alteration in mtDNA copy number, loss of membrane potential and dysfunctional ETC. Signaling results into remodeling of nuclear gene expression either directly through activation of nuclear factor of activated T-cells (NFAT) transcriptional signaling and aspects of the NF-κB and cAMP responsive element binding protein (CREB) pathways, or indirectly via activation of other protein kinases and growth factor-dependent cell survival pathways (Biswas et al., 1999; Guha and Avadhani, 2013; Guha et al., 2007; Hogan, 2017; Park et al., 2019; Srinivasan et al., 2017; Tang et al., 2012). Taken together, this multifaceted signaling cascade is one that is conserved from yeast to humans, and serves a crucial role in adaptive homeostatic responses as required for organismal health.
2.4. Metabolites
Mitonuclear communication is facilitated not only by the translocation of functional proteins and small molecules, but also by the production and movement of metabolites. Classically, the TCA cycle is known to produce crucial metabolites for cell survival and proliferation, as well as production of bioenergetic intermediates responsible for feeding into the ETC (Chandel, 2015; Martinez-Reyes et al., 2016). Many of these metabolites also participate in mitochondria-to-nucleus signaling, whereby they are co-opted as secondary messengers with their levels providing a direct indication of mitochondrial health and metabolic status (Frezza, 2017). Because they often serve as substrates or regulators of enzymes involved in chromatin remodeling, changes in levels and availability of these metabolites are integrated into epigenetic regulatory strategies that drive transcriptional changes under differing stress conditions and physiologic states (Martinez-Reyes and Chandel, 2020). Two of the more common modifications directly linked to the functionality of mitochondrial metabolites are acetylation and methylation, which are mediated by enzymes responsive to changes in acetyl coenzyme A (acetyl-CoA) and α-ketoglutarate (α-KG)/succinate levels and affect nuclear gene expression through DNA methylation and post-translational modifications of histones that are central to the “histone code” (Allis and Jenuwein, 2016). Metabolite-induced changes to the epigenome or metabolic epigenetics contribute to reprogramming of gene expression under physiological and stress conditions, as well as disease progression (Shaughnessy et al., 2014). Changes in nuclear gene expression in response to accumulation of mtDNA mutations, a condition known as heteroplasmy, for example, are facilitated by abnormal metabolite production promoting specific epigenetic modifications, which in turn contribute to transcriptional regulation via modulation of chromatin dynamics (Fetterman and Ballinger, 2019; Kopinski et al., 2019). Changes in the methylation status of nuclear encoded genes upon defects in mtDNA copy number have been observed in the context of both breast cancer as well as osteosarcoma (Feeley et al., 2015; Smiraglia et al., 2008). These observations underscore the link between metabolite-induced epigenetic changes and transcriptional alterations associated with pathogenic phenotypes and disease progression, thus highlighting the important connection between mitochondrial retrograde signaling, metabolites, and genetics.
2.4.1. Acetyl-CoA
Acetyl-CoA generated by the breakdown of carbohydrates, through glycolysis, and the oxidation of fatty acids provides fuel for TCA cycle flux as well as ATP production in cells under oxygenated conditions. Acetyl-CoA levels are tightly associated with histone acetylation, a reversible process by which acetyl groups are either added or removed to/from histones and other target proteins, via the action of histone acetyltransferases (HATs) and histone deacetylases (HDACs) enzymes (Menzies et al., 2016). Acetyl-CoA is an essential substrate for acetyltransferases and histone acetylation is generally associated with increased chromatin relaxation and gene activation (Kaelin and McKnight, 2013; Mews et al., 2017; Pietrocola et al., 2015; Sivanand et al., 2018). Acetyl-CoA is exported out of the mitochondria in form of citrate, which is then converted back into acetyl-CoA and oxaloacetate, with the former becoming available for HATs utilization in the nucleus (Wellen et al., 2009). Several examples underscore the importance of acetyl-CoA and its mitochondria-to-nucleus signaling capabilities during instances of stress and nutrient sensing. During the “fed” and/or “growth” state, acetyl-CoA levels are high and found predominantly in the nucleus, where it aids in histone acetylation. In contrast, during the “fasted” and/or “survival” state, acetyl-CoA is directed into the mitochondria and is utilized by the organelle as part of the normal TCA cycle flux to generate ATP (Shi and Tu, 2015). Notably, metabolite-induced changes in the epigenome associate with highly specific transcriptional responses, as driven by the need to reprogram gene expression for adaptation to nutrients availability or environmental conditions (Schvartzman et al., 2018; Shaughnessy et al., 2014). Glucose-induced increase in histone acetylation, for example, is associated with the activation of glucose metabolism genes, whereas an increase in lipid-induced acetyl-CoA is reported to drive the upregulation of lipid metabolic genes (McDonnell et al., 2016; Wellen et al., 2009). In hypoxic cancer cells, acetyl-CoA generated from acetate promote histone acetylation for the upregulation of lipogenic genes (Gao et al., 2016). These tailored responses are likely achieved through integrated actions of nutrient-sensing transcription factors and coregulators driving histone acetylation to specific promoters and other regulatory regions. Local regulation of acetyl-CoA production at the site of utilization could also contribute to the specificity of gene regulation. Translocation of the pyruvate dehydrogenase complex (PDC) from mitochondria to nucleus in situations of mitochondrial stress and growth factor stimulation supports histone acetylation of cell cycle genes through the local production of acetyl-CoA (Sutendra et al., 2014). Local production of acetyl-CoA in the nucleus can also be driven by nuclear ATP-citrate lyase (ACLY), upon DNA damage, and acetyl-CoA synthase 2 (ACCS2), under hypoxic conditions. While a full mechanistic understanding of the relationship between these localized enzymatic activities and the regulation of specific gene programs is still lacking, translocation of the PDC was shown to be crucial for zygotic genome activation in both human and mouse embryos, thus emphasizing the importance of tight coordination in mitochondria-to-nucleus signaling and epigenetic modifications (Nagaraj et al., 2017).
2.4.2. Methyl groups
Methylation represents another well-characterized epigenetic modification that is tightly connected to the utilization of mitochondrial metabolites as enzymatic cofactors. Regulation of histone methylation is achieved through either methyltransferases (adding methyl groups) or demethylases (removing methyl groups), the likes of which more specifically repress or activate transcription by regulating the presence and number of methyl groups attached to specific lysine and/or arginine residues on the tails of histone 3 (H3) and histone 4 (H4) (Teperino et al., 2010). Methylation also occurs on DNA, and regulates chromatin structure and subsequent gene expression through single-nucleotide changes. Methylation mediated by DNA methyltransferases (Dnmt) typically occurs on clustered cytosine-guanine repeats, known as “CpG islands”, and the modification at these sites results in a 5-methylcytosine (5mC) (Moore et al., 2013)(Tajima et al., 2016). Overall, methyltransferases can be divided into three separate classes, all of which use S-adenosylmethionine (SAM) for the methyl group transfer process. This establishes a first important link to mitochondrial metabolism, in that although SAM is generated in the cytosol by the methionine-homocysteine cycle, this cycle is fueled by ATP and folate products stemming from mitochondrial metabolism (Ducker and Rabinowitz, 2017).
A more direct link is observed in the context of the reverse process, as removal of methyl groups by demethylation requires mitochondrial-produced cofactors. In the case of DNA methylation, the end result of this epigenetic mechanism is the triggering of base excision repair (BER), which serves to replace the modified cytosine with one devoid of any methyl marks (Moore et al., 2013). Prior to this step, ten-eleven translocation (TET) enzymes oxidize 5mC to 5-hydroxymethylcytosine (5hmC) in a process that requires mitochondrial-produced α-KG, Fe(II), and oxygen (O2), and can be inhibited by fumarate, succinate, and 2-hydroxyglutarate (2-HG) (Etchegaray and Mostoslavsky, 2016; Pastor et al., 2013; Rasmussen and Helin, 2016; Xu et al., 2011). This dependency on the TCA cycle/mitochondrial substrates establishes a firm connection between DNA methylation and mitonuclear signaling, while also showing similarities in the regulation of TET enzymes and Jumonji domain containing protein (JMJD) histone demethylases, which are discussed below (Matilainen et al., 2017).
Histone demethylases are grouped into two families, each with its own particular reliance upon a specific mitochondrial metabolite. The Lys-specific demethylases (LSDs) are flavin adenine dinucleotide (FAD)-dependent, with FAD being a B12 derived redox cofactor required for the TCA cycle as well as beta-oxidation processes (Matilainen et al., 2017). The second branch of the lysine demethylases are the Jumonji C (JmjC) domain demethylases (JMJDs/KDMs). KDMs are unique in that they use α-KG, an important TCA cycle intermediate, as a cofactor. Conversely, KDMs are inhibited by 2-hydroxyglutarate produced as a result of oncogenic mutations in isocitrate dehydrogenases (IDH1/2) or via non-canonical activity of lactate dehydrogenase (LDH) and malate dehydrogenase (MDH) under hypoxia conditions (Intlekofer et al., 2017; Lu et al., 2012; Turcan et al., 2012). Given this co-factor dependency, the interplay between the TCA cycle/mitochondrial function and the actions of demethylases within the nucleus is abundant across species. In S. Cerevisiae, Jumonji demethylase Jhd2-mediated regulation of histone methylation is tightly linked to α-KG/succinate ratio (Soloveychik et al., 2016). In C. elegans, the actions of H3K27 demethylases jmjd-1.2 and jmjd-3.1 also link mitochondrial stress with the activation of the mitochondrial unfolded protein response (mtUPR), a function conserved by their mammalian orthologs plant homeodomain finger (PHF) 8 and JMJD3 (Merkwirth et al., 2016). In mammals, this is associated with removal of repressive H3K9 methylation by JMJD2A/KDM4 from the promoters of nuclear-encoded mitochondrial genes and stress response genes as needed to support their upregulation upon mitochondria-to-nucleus signaling (Cardamone et al., 2018). The complexity of the roles covered by mammalian demethylases is further increased by the fact that modification of distinct histone residues can lead to opposing effects on gene activation or repression. For example, LSD1 can mediate the removal of both H3K4 and H3K9 methylation with the latter being important for LSD1-mediated regulation of energy expenditure, adaptive thermogenesis and oxidative metabolism in adipose tissue (Duteil et al., 2014; Inagaki, 2018; Nagaoka et al., 2015; Sambeat et al., 2016; Shi and Tu, 2015; Shi et al., 2004; Wang et al., 2020). Conversely, modulation of H3K4 methylation status by LSD1 (KDM1A) was associated with the shift from mitochondrial to glycolytic metabolism in human hepatocellular carcinoma cells (Sakamoto et al., 2015). Overall, the tight and intricate relationship between mitochondria-derived metabolites and histone demethylases provide dynamic and sensitive ways to integrate signals related to mitochondrial metabolism with the regulation of nuclear gene expression through methylation/demethylation of histones and DNA. Accordingly, α-KG, 2-HG and IDHs play central roles in mitochondrial signaling and their misregulation is linked to a variety of mitochondria-related diseases, including cancer, aging, neuronal dysfunction, autoimmune disease, and metabolic stress disorders (Bayliak et al., 2017; Fujii et al., 2016; Hunt et al., 2019; Raineri and Mellor, 2018; Salminen et al., 2014; Schulze and Harris, 2012; Ward et al., 2012; Weinberg et al., 2019).
3-. Nucleus-derived signaling molecules
In the previous section, we discussed how mitochondria contribute to epigenetic control of nuclear gene expression through the production of metabolites used as cofactors by chromatin remodeling enzymes. These processes affect gene expression at the genome-wide level rather than providing a means for the regulation of specific genes. Specificity for the activation/repression of dedicated gene expression programs is provided by the integration of these global changes in metabolites availability with the activity of specific DNA-binding transcription factors. Here, we will introduce the nuclear-encoded transcriptional regulators that contribute to the specificity of adaptive responses through sensing of triggering conditions, mitochondria-to-nucleus signaling and targeting of specific nuclear gene programs, and discuss how their conserved actions are integrated in the overall mitochondria-to-nucleus signaling process.
3.1. Retrograde Signaling in Yeast, Worms, and Flies
Initially described in yeast, the mitochondria-to-nucleus retrograde signaling or RTG pathway regulate the rerouting of carbon and nitrogen metabolism - through remodeling on nuclear gene expression - to counteract mitochondrial dysfunction. Key players in this pathway are the DNA-binding transcription factors RTG1-RTG3, and the regulatory factor RTG2, which drives RTG1-RTG3 translocation to the nucleus in response to changes in ATP availability and mitochondrial membrane potential (MMP) as reviewed in detail elsewhere (Borghouts et al., 2004; Jazwinski, 2005; Jazwinski and Kriete, 2012; Jia et al., 1997; Liu et al., 2003; Sekito et al., 2002; Torelli et al., 2015). Metabolic rerouting of yeast cells in conditions of impaired mitochondrial functions (as modeled by loss of mtDNA in ρ° petite cells) include the activation of nuclear genes to support the metabolism of two-carbon compounds through the glyoxylate cycle, promote the regeneration of NAD+ to restore oxidative balance in the cell, and ensure that the expression of the first four enzymes of the TCA cycle is preserved as needed to provide metabolic intermediates for anabolic biosynthesis (Chelstowska et al., 1999; Eisenberg-Bord and Schuldiner, 2017; Liao et al., 1991; Liu and Butow, 2006; Liu et al., 2003). Tight regulation of this adaptive pathway is guaranteed by positive and negative regulatory factors, such as Grr1p and Mks1p, and feedback regulatory loops, such as the negative regulation of Rtg1/3 by glutamate and glutamine, which production is upregulated by the retrograde response (Liu and Butow, 2006). Despite these common features, it should be noted that the response to different sources of mitochondrial dysfunction promotes the activation of distinct transcriptional programs as shown by genome-wide transcriptional profiling of yeast cells treated with either the mitochondrial uncoupler CCCP, the ATP synthase inhibitor oligomycin or the complex III inhibitor antimycin (Epstein et al., 2001). Taken together, these selected examples demonstrate the early work done in yeast elucidating a pathway of retrograde signaling, and the significant role it plays in metabolic reprogramming and stress defense under conditions of mitochondrial dysfunction.
Aside from yeast, the mitochondria-to-nucleus response to mitochondrial stress has been widely studied in worms. Most C. elegans studies have focused on the mtUPR, an arm of the mitochondrial stress response (MSR) which aims at resolving the stress induced by accumulation of unfolded proteins in the mitochondrial matrix (Mottis et al., 2019). The MSR can be triggered by any number of stimuli, including misfolded protein accumulation, mtDNA depletion and OXPHOS deficiencies, or toxin-induced mitochondrial dysfunctions (Pellegrino et al., 2013). Transcriptional outcomes of the MSR include increased expression of genes involved in restoring proteostasis (proteases and protein folding chaperones), preventing mtDNA damage (anti-oxidant enzymes) and recovering respiration functions (OXPHOS enzymes and assembly factors) (Jovaisaite and Auwerx, 2015; Jovaisaite et al., 2014; Nargund et al., 2015; Wu et al., 2018). The molecular players that mediate mtUPR signaling between the mitochondria and the nucleus have been addressed by a large body of work as discussed in excellent reviews (Anderson and Haynes, 2020; Haynes et al., 2007; Haynes et al., 2010; Liu et al., 2014; Melber and Haynes, 2018; Qureshi et al., 2017). Best characterized is the role of C. elegans Activating Transcription Factor associated with Stress-1 (ATFS-1), a DNA-binding transcription factor which kinetics are regulated through the alternative use of either a nuclear localization signal (NLS) or a mitochondrial targeting sequence, both located at the N-terminus of the protein. Under basal conditions, ATFS-1 is imported into the mitochondria and degraded. Conversely, import into the mitochondria is attenuated in conditions of mitochondrial stress, allowing for translocation into the nucleus and regulation of target genes expression along with other factors, such as transcription factors DVE-1 and LIN-65, ubiquitin-like protein UBL-5 and chromatin remodeling enzymes MET2, JMJD-3.1 and JMJD-1.2 (Merkwirth et al., 2016; Nargund et al., 2012; Shpilka and Haynes, 2018; Tian et al., 2016a).
Also important to mention is the work done in flies, which has provided key insights into the mechanism(s) of retrograde signaling, and its impact on organismal health. Early genetic studies in Drosophila melanogaster demonstrated that disruption of complex I of the ETC prevented cell cycle cycle progression through a ROS-induced cascade, mediated by ASK-1, JNK, FOXO, and the fly homolog to p27, Dacapo (Owusu-Ansah et al., 2008), thus revealing a pathway of mitochondria-to-nuclear signaling mediated by mitochondrial ROS production. Signaling mediated by JNK and FoxO in response to sublethal ROS levels is also important for promoting hematopoietic differentiation (Owusu-Ansah and Banerjee, 2009), whereas mild complex 1 stress in a model of muscle mitochondrial injury was shown to induce a two-pronged response, including the upregulation of genes controlling the mtUPR and the induction of the Drosophila ortholog of insulin-like growth factor binding protein 7 (IGFBP7), to preserve mitochondrial function, increase lifespan, and prevent muscle degeneration as a function of aging (Owusu-Ansah et al., 2013). Moreover, the mitochondrial retrograde response has been shown to be a crucial facet of Drosophila nervous system functionality, with implications for aging and age-related neurodegenerative disorders (Duncan and Bateman, 2016). Mitochondrial stress promotes reprogramming of gene expression in Drosophila neurons via activation of the UPR stress response ATF4, the Drosophila ortholog of HIFα, Sima, and Ras-ERK-ETS signaling (Cagin et al., 2015; Duncan et al., 2018; Hunt et al., 2019). Notably, in this context, retrograde signaling seems to contribute to neuronal dysfunction rather than playing a protective effect, as shown by the fact that downregulation of Sima was sufficient to restore neuronal function in different genetic models of mitochondrial dysfunction in absence of any impact on the mitochondrial defect per se (Cagin et al., 2015).
3.2. Mitochondria-to-nucleus Signaling in Mammals
Extensive searches for homologs of yeast and C. elegans retrograde genes in rodents and humans indicates that while overall goals and regulatory strategies of the MSR are conserved across species, higher organisms rely on more complex communication strategies involving an array of transcriptional regulators. Mammalian transcription factors associated with functions similar to those of Rtg1/Rtg3 or ATFS-1 include ATFs, FOXO, RXR, ETS, NF-κB and Myc (Arnould et al., 2015; Chae et al., 2013; Duncan et al., 2018; Fiorese et al., 2016; Jazwinski, 2013; Quiros et al., 2017; Zhao et al., 2016). This expansion has likely emerged to ensure that specific regulation of distinct subsets of mitochondrial and cellular genes can be achieved through the selective use of specific transcription factors (TFs), which functions are further integrated with a variety of chromatin remodelers that respond to mitochondrial-derived metabolites as discussed in the previous section. Accordingly, the mammalian mtUPR is comprised of different axes including a canonical mtUPR arm mediated by transcription factors ATF4, ATF5, and CHOP, a SIRT3/FOXO3A axis responsible for the activation of the anti-oxidant response and an ERα/NRF1 arm aimed at increasing protein quality control (Fiorese et al., 2016; Germain, 2016; He et al., 2016; Kenny and Germain, 2017; Munch, 2018; Naresh and Haynes, 2019; Papa and Germain, 2011; 2014). In addition to the increased complexity in the number of TFs involved, the mammalian mtUPR is fully integrated in a broader integrated stress response (ISR), which coordinates the response to mitochondrial stress with the regulation of cytosolic protein translation through phosphorylation of eIF2α and selective translation of mRNAs of stress genes, including mtUPR mediators (Costa-Mattioli and Walter, 2020; Garcia-Roves et al., 2008; Wu et al., 2002). As a result, the mammalian mtUPR is coordinately regulated with other growth and damage response pathways (Khan et al., 2017; Nikkanen et al., 2016). Mechanistically, a recent study indicates that activation of ATF4 downstream of mitochondrial stress is achieved through OMA1-dependent cleavage of DELE1 which in turn activates the eIF2α kinase activity of HRI (Fessler et al., 2020; Guo et al., 2020). Moreover, extensive characterization of the mammalian mitochondrial ISR (mtISR) in vivo shows that the ISR is not only cell- and stressor-specific, but it also progresses through distinct temporal stages with different ATF factors being involved in subsequent and interdependent waves of gene expression (Forsstrom et al., 2019; Mick et al., 2020; Suomalainen and Battersby, 2018).
As discussed above, integration of these transcription factor-driven responses with metabolite-induced epigenetic changes provides a multi-layered strategy that allows for pairing the metabolic changes occurring at mitochondria level with the activation of specific adaptive gene programs. While this can be achieved through translational regulation, as in the case of ATF4/5, or protein stabilization, as described for FOXO (Fasano et al., 2019; Lettieri-Barbato et al., 2019), there are also examples of direct mitonuclear communication mediated by actual mitochondria-to-nucleus translocation of nuclear-encoded factors residing on mitochondria (Monaghan et al., 2015a; Monaghan et al., 2015b). The oxidative stress response factor Nrf2, for example, translocates from mitochondria to the nucleus, where it regulates the expression of anti-oxidant genes, upon disruption of the KEAP1-PGAM5 complex that otherwise keeps it sequestered on the outer mitochondrial membrane (OMM) (O’Mealey et al., 2017). FOXO1 translocation from mitochondria to nucleus in response to starvation, instead, is induced by ROS-mediated activation of the mitochondrial phosphatase PTPMT1 (Lettieri-Barbato et al., 2019). Another example of mitochondria-to-nucleus signaling in response to increased ROS levels is provided by CLK-1 (also called COQ7), a mitochondrial monooxygenase playing a critical role in the biosynthesis of the ETC cofactor ubiquinone (Monaghan et al., 2015a). Because its presence in the nucleus, despite being controversial, has been associated with both upregulation of genes involved in ROS metabolism and dampening of the mtUPR, mitochondria-to-nucleus signaling in this case would provide a negative feedback loop to promote ROS homeostasis (Liu et al., 2017; Monaghan et al., 2015b).
To add another layer of increased complexity, direct mitonuclear signaling in mammalian systems is not limited to DNA-binding transcription factors, but rather extends to transcriptional regulators, such as cofactors and chromatin remodeling enzymes, as shown in the case of G-Protein Pathway Suppressor 2 (GPS2) (Cardamone et al., 2018). GPS2 functions as a mediator of mitochondria-to-nucleus signaling, metabolic reprogramming and chromatin remodeling, reminiscent of the yeast regulatory factor Rtg2. Its translocation from the OMM to the nucleus, as triggered by developmental cues or mitochondrial depolarization, is required for the activation of a large transcriptional program that includes nuclear-encoded mitochondrial genes and stress response genes that partially overlap with the programs downstream of the ATF4 and ATF5 mtUPR pathways (Cardamone et al., 2018; Quiros et al., 2017). Notably, recruitment of GPS2 to target promoters serves the purpose of remodeling the chromatin environment through stabilization of the H3K9me2/3 histone demethylase JMJD2A/KDM4A, thus effectively reinforcing the possibility to remove a repressive epigenetic mark only from specific genomic locations (Cardamone et al., 2018) (Figure 2). As discussed above, chromatin remodeling by specific methyltransferases/demethylases is also a key component of the mtUPR response in worms (Merkwirth et al., 2016; Tian et al., 2016b), thus suggesting that spatial regulation of histone methylation/demethylation, via metabolically-regulated enzymes working in cooperation with sequence-specific transcriptional regulators, plays a central role in metabolic adaptive responses across species.
Figure 2.
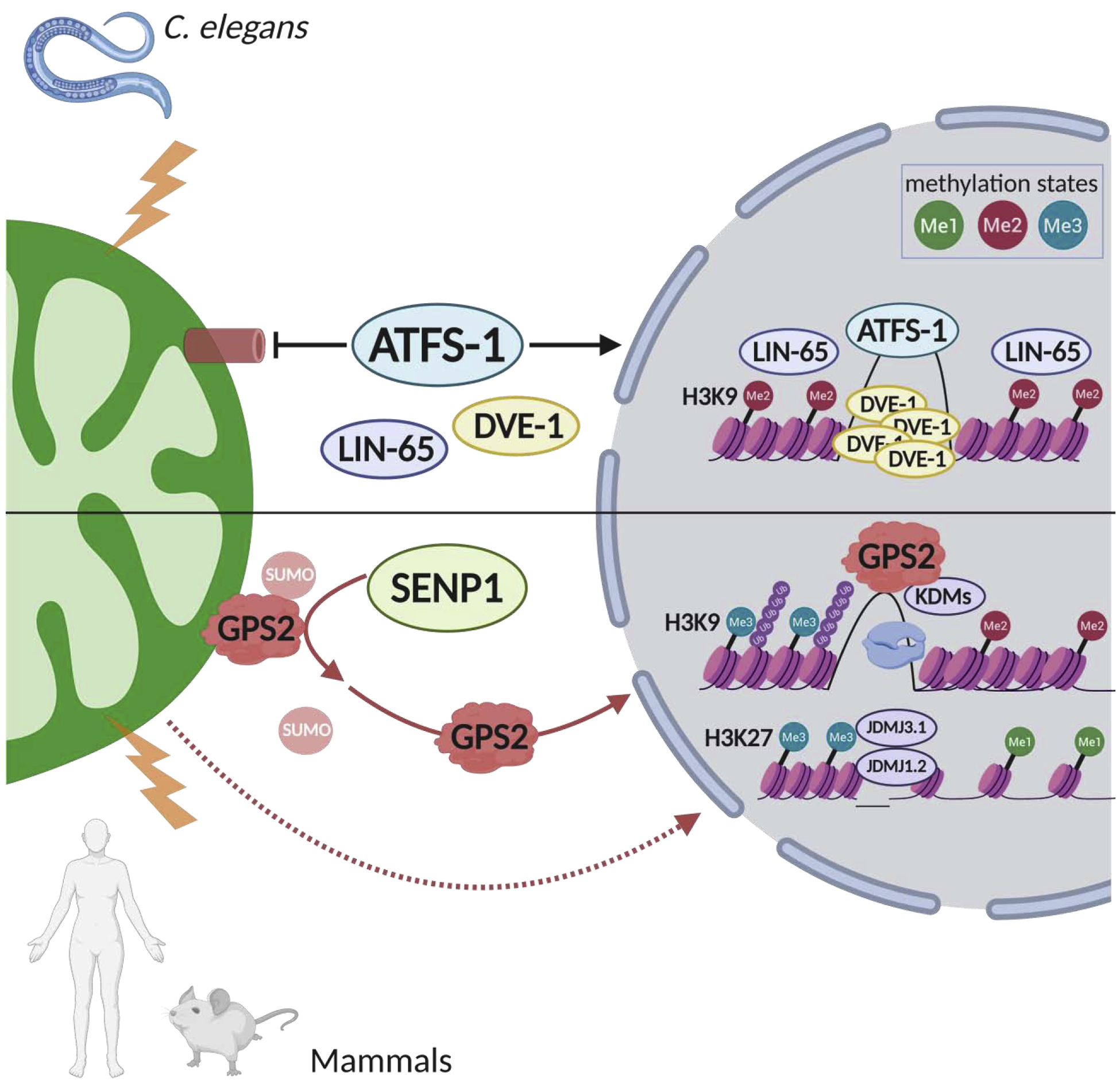
Mitochondria stress response (MSR) and remodeling on nuclear chromatin via histone methylation and demethylation. Chromatin remodeling through methylation/demethylation of histone tails supports the reprogramming of nuclear gene expression in response to mitochondria dysfunctions across species. In the top panel, it is outlined the mtUPR arm of the MSR in C. elegans. Nuclear translocation of transcription factors ATFS-1 and DVE-1 promotes the expression of stress response genes that are excluded from chromatin condensation mediated by H3K9 methyltransferase MET-2 and nuclear cofactor LIN-65. Accessibility of stress responsive promoters is guaranteed by the action of histone H3K9 and H3K27 demethylases, respectively KDM4A or jMJD1.2/PHF8 and JMJD3.1/JMJD3 as shown in mammals in the bottom panel. Mitochondria-to-nucleus translocation of GPS2, upon mitochondrial stress-induced de-sumoylation by SENP1, promotes demethylation of target promoters by stabilizing KDM4A through inhibition of Ubc13-dependent ubiquitination.
Similar to the regulation of C. elegans ATFS-1 described in the previous section, the balance between mitochondria and nuclear pools of nuclear-encoded retrograde factors can be effectively modulated by attenuation of mitochondrial matrix protein import, a step exquisitely sensitive to mitochondrial stress (Monaghan et al., 2015b). Other forms of regulation through post-translational modifications also contribute as in the case of SENP1-mediated de-sumoylation of GPS2 (Cardamone et al., 2018)(Figure 2). The molecular details of SENP1 activation in conditions of stress are not fully elucidated. However, recent findings indicate that SENP1 levels on the mitochondria fluctuate depending on the nutrition state, with SENP1 accumulation upon fasting driving fatty acid oxidation and energy expenditure via desumoylation of SIRT3 and deacetylation of mitochondrial proteins (Wang et al., 2019). De-sumoylation by SENP1 is also important for promoting the activity of PGC1α, a transcriptional cofactor central to the regulation of mitochondria biogenesis (Cai et al., 2012). Taken together, these observations suggest that SENP1 may be playing a central role as an effector of mitochondria adaptive responses.
4-. Mito-nuclear communication and cellular homeostasis
4.1. Genetic compatibility of bi-genomic system
Compatibility between the mitochondria and nuclear genomes is important for cellular fitness in the bi-genomic system. The Bateson-Dobzhansky-Muller model provides a framework for understanding how genes coevolved to stay compatible (Bateson, 1909; Dobzhansky, 1936; Muller, 1942), and recent studies show that delicate nuclear-mitochondrial compatibilities have an important role in optimizing mitochondrial function. Work by Rand and Dowling has shown that interaction of mitochondrial genome and the nucleus affects traits, reproductive success, rate of development, growth, behavior, and aging in fruit flies (Rand et al., 2006; Rand et al., 2004; Wolff et al., 2016). Mice engineered to host mitochondria from distant genetic backgrounds, providing a model of mitonuclear genomic mismatch, exhibit shifted cellular metabolism, mtROS levels, and resistance to cardiovascular burdens (Betancourt et al., 2014; Dunham-Snary and Ballinger, 2015; Fetterman et al., 2013). Moreover, Latorre-Pellicer et al compared mouse embryos of C57 strain nuclear background with different mitochondrial DNA haplotypes and found optimized compatibilities of nuclear and mitochondrial genomes affected mitochondrial function such as OXPHOS and mouse embryonic fibroblasts reprogramming efficiency (Latorre-Pellicer et al., 2019).
4.2. Mitonuclear Communication in Physiology and Disease
Given the complex strategies that have evolved to maintain efficient communication between mitochondria and nucleus across species, it is not surprising that their misregulation is involved in a number of pathologies. The disease state can be promoted by the inability to adapt to cellular and environmental stressors or by excessive/uncontrolled oxidative stress. At the same time, activation of mitochondria-to-nucleus signaling and transcriptional reprogramming of cellular functions in response to modest mitochondrial dysfunction can have beneficial effects on the metabolic health and longevity of an organism, through a process defined as “mitohormesis” (Barcena et al., 2018; Yi et al., 2018; Yun and Finkel, 2014). While we refer to other excellent reviews for a detailed summary of this expanding field (Mottis et al., 2014; Tan and Finkel, 2020), here we briefly touch upon the role of mitochondria-to-nucleus signaling within different disease contexts to highlight the importance of finely tuned mitonuclear communication to maintain cellular and organismal homeostasis.
4.2.1. Aging
Across species the aging process is associated with a decline in mitochondrial function and increase in oxidative stress, and mitochondrial dysfunction is widely regarded as a hallmark of aging (Bratic and Larsson, 2013; Jang et al., 2018; Lopez-Otin et al., 2013; Sun et al., 2016). It was thus surprising to observe that the downregulation of key mitochondrial genes in C elegans would promote extended lifespan, albeit associated with decreased respiration and developmental defects (Dillin et al., 2002; Felkai et al., 1999; Feng et al., 2001; Lee et al., 2003). Extensive studies in both worms and flies indicate that the increase in longevity is associated not only with decreased ROS production lowering the oxidative stress burden placed on the organism (Feng et al., 2001), but also with activation of the mtUPR, as triggered by mitochondrial dysfunction or mito-nuclear imbalance (Durieux et al., 2011; Houtkooper et al., 2012; Owusu-Ansah et al., 2013). Knockdown of components of complex IV across C elegans developmental stages, in particular, revealed that this is a temporally regulated phenotype whereby tissue-specific activation of the mtUPR during the larval stage triggers beneficial effects on lifespan in adulthood (Dillin et al., 2002; Durieux et al., 2011). The early triggering of a response to mild stress essentially primes the system, through retrograde-dependent reprogramming of gene expression, for improved handling of metabolic insults and oxidative stress throughout the duration of the extended lifespan (Schulz and Haynes, 2015). This is a finely tuned response not only in term of time, but also space. Adaptation is not limited to organ initially insulted, as activation of the mtUPR in one tissue is communicated to other parts of the organism through “mitokine” signaling (Zhang et al., 2018). In mammals, this role is at least partly played by the cytokine FGF21 which was shown to contribute to inter-organ communication in human patients with mitochondrial myopathies as well as mice models of mitochondrial dysfunction (Kim et al., 2013; Suomalainen et al., 2011). However, it is worth noting that mtUPR activation per se has not been observed in tissues other than those primarily affected by mtDNA deletions, despite the essential role played by FGF21 in mediating whole-body metabolic adaptation to muscle mitochondrial defects (Forsstrom et al., 2019). Mitochondrial-derived peptides (MDPs) may also have a role in aging. The levels of MDPs in certain tissues and blood have been shown to be age-dependent (Cobb et al., 2016; Kim and Koh, 2017; Lee et al., 2015; Muzumdar et al., 2009). Humanin is connected to the growth hormone/insulin-like growth factor-1 (GH/IGF-1) axis, a prominent endocrine longevity regulator. Blood humanin levels are higher in GH-deficient Ames mice, whereas the short-lived GH-transgenic mice had lower humanin levels compared to their wild-type counterparts (Lee et al., 2014). MOTS-c treatment significantly reversed age-dependent physical decline in old mice (22 mo.), allowing them to run twice as long on a treadmill and outcompete their middle-aged counterparts (Reynolds et al., 2019). Late-life initiated (23.5 mo.) intermittent MOTS-c treatment (3x/week) improved health- and life-span in mice (Reynolds et al., 2019). These studies suggest that aging is regulated by factors from both mitochondrial and nuclear genomes.
4.2.2. Obesity & Type 2 Diabetes
Given the high level of importance placed upon mitochondria in the maintenance of energy homeostasis and the balance between nutrient storage and utilization, both mitochondria-to-nucleus and nucleus-to-mitochondria signaling are widely implicated in metabolic diseases, such as obesity and type 2 diabetes. Moreover, the above discussion about the mitohormetic response to mild stress promoting cellular and organismal reprogramming towards extended lifespan directly ties into this discussion as improved metabolic performance represents a key aspect of increased longevity. Because of limited space we cannot discuss in details the numerous and complex interactions that link mitochondrial dysfunctions across different tissues to the development and progression of metabolic disorders, and refer to other reviews for a detailed discussion on these topics (Lin et al., 2005; Patti and Corvera, 2010; Sivitz and Yorek, 2010; Theurey and Rieusset, 2017; Wanet et al., 2015; Xu et al., 2019; Yi et al., 2018; Zorzano et al., 2009).
Here, we only mention a few examples relevant to our understanding of the role played by mitonuclear communication, or lack of thereof, in the development of obesity and associated metabolic diseases. Retrograde signaling by MOTS-c, one of the mitochondria-encoded peptides discussed above, regulates insulin sensitivity and metabolic homeostasis in skeletal muscle. An additional finding from this study shows that systemic treatment of high-fat diet (HFD)-fed mice with MOTS-c prevents age-dependent and diet-induced insulin resistance and obesity (Lee et al., 2015). Also in mice, adipose-specific deletion of the mammalian retrograde factor GPS2 leads to whitening of the brown adipose tissue (BAT), as associated with a significant reduction in mitochondrial content, and development of obesity (Cardamone et al., 2018; Cederquist et al., 2017). Notably, in chow-fed mice GPS2-AKO mice, obesity is uncoupled from inflammation and metabolic dysfunction, highly resembling a human condition known as “metabolically healthy” obesity (Bluher, 2010; Loos and Kilpelainen, 2018). However, work from us and others indicates that the protective effect is lost in condition of diet-induced obesity and that cold-induced browning of the tissue is impaired in absence of GPS2 (Cardamone et al., 2018; Drareni et al., 2018; Fan et al., 2016), suggesting that GPS2-mediated mitochondria-to-nucleus signaling may be critical for adapting to the stress imposed by cold or nutrients overload. Independent studies also show that the mammalian mtISR is induced in the BAT upon cold stimulation to promote ATF4-dependent expression of FGF21 and GD15 (Flicker et al., 2019).
Another area of significant interest in the context of metabolic disorders is mitochondrial lipid signaling. Excessive accumulation of long and medium chain acylcarnitines, as resulting from incomplete beta oxidation in presence of excess nutrients intake, for example is recognized as a driving factor in the development of insulin resistance (Koves et al., 2008; Muoio et al., 2012; Sarparanta et al., 2017; Yazici and Sezer, 2017). Carnitine lipid species have been proposed to impact upon insulin responsiveness, through retrograde signaling, in a similar manner to that increased cytosolic Ca2+ or increased ROS production alter nuclear-encoded mitochondrial gene expression under conditions of stress (Devarshi et al., 2017). Moreover, as discussed above, changes to the epigenome driven by metabolite-induced regulation of epigenetic modifiers (HDACs and methyltransferases) are emerging as important underlying events to the development of a variety of diseases, including metabolic disorders (Emamgholipour et al., 2020). According to this model, incomplete fatty acid oxidation could promote insulin resistance by modulating nuclear gene expression through regulation of acetyl-CoA availability and associated changes to the epigenome (Shi and Tu, 2015). Lastly, mitochondrial dysfunctions have been shown to affect not only nutrient handling but also obesity-associated inflammation via cytosolic release of mtDNA triggering mitochondria-to-nucleus signaling through TLR9 and/or cGAS/STING pathways (Bai et al., 2017; Kanneganti et al., 2015; Liu et al., 2016 Mao et al., 2017; Yuan et al., 2017; Zhong et al., 2019).
4.2.3. Cancer
Nutrient sensing is an essential cellular and overall life process heavily governed by mitochondrial signaling networks and epigenetic modifications, the likes of which need to be tightly regulated. In the context of cancer, metabolic flexibility provides transformed cells with a critical advantage for surviving in a nutrient-limited tumor microenvironment. Therefore, there are many instances in which cancer cells can co-opt metabolic and mitochondrial signaling pathways for their own benefit. Many of the metabolites highlighted in this review have been both conceptually and mechanistically shown to be involved in cancer development and progression, through participation in epigenetic modifications. Many of these modifications result in the upregulation of cell proliferation and survival, differential utilization of nutrient substrates based upon availability, and overall oncogenic reprogramming which altogether work for the benefit of the cancer cells. Specific examples of mitochondrial metabolites, their connection to chromatin modifications, and the resulting impact on malignancies through mitonuclear communication were recently reviewed by others (Campbell and Wellen, 2018; Deng and Haynes, 2017; Hirschey et al., 2015).
4.2.4. Neurological Disorders
Mitochondrial dysfunction is well regarded as a hallmark of many neurodegenerative and brain developmental disorders, but the actual characterization of retrograde signaling pathways in these disease contexts has just started to be explored within the last few years. Both inhibition of OXPHOS components, as well as mitochondrial dynamics in neurodegeneration are interconnected to retrograde mechanisms, with several different disease models in both Drosophila and rodent highlighting these signaling networks (Cagin et al., 2015; Celardo et al., 2016; Duncan et al., 2018; Kim et al., 2016; Krug et al., 2014; Meurers et al., 2009; Requejo-Aguilar et al., 2014; Ryu et al., 2002; Wu et al., 2014; Yap et al., 2013). Studies like these place a particular emphasis on the importance of understanding the connectivity of these networks at a basal level, and translating this into further research in neuro-mitochondrial disease treatment. Recent reviews have detailed mitochondrial retrograde signaling in neurological disease, as well as considerations for targeted therapies, and should be referred to for more insight and specifics (Granat et al., 2020; Hunt and Bateman, 2018).
5. Discussion and Future Directions
These are exciting times for mitochondrial research. Mitochondria are recognized as key signaling organelles playing a prominent role in the regulation of cellular homeostasis. This requires that their interaction with a variety of other cellular structures and organelles, including - but not limited to - the nucleus, is addressed as a whole and investigated through innovative approaches. In this review, we have discussed the emerging themes in the area of mitonuclear communication by specifically focusing on mitochondria-to-nucleus signaling. However, the pathways and signaling events described here are to be considered within a more holistic view of mitochondria contribution to whole cell signaling as recently done by others (Boos et al., 2020; Mottis et al., 2019).
One topic specific to mitochondria-nuclear communication is the interplay between metabolism and epigenetics, currently a very active area of investigation. As discussed above, a growing literature addresses how changes in cell metabolism are paired to epigenetic regulation of nuclear gene expression through the availability of metabolites serving as cofactors for chromatin remodeling enzymes. Intriguingly, conserved strategies for metabolic regulation of chromatin remodeling through the use of matching sets of metabolites/chromatin modifiers are revealed, with α-KG and histone demethylases emerging as a key regulatory dyad in mitochondria-to-nucleus signaling. However, it is still incompletely understood how metabolic metabolite-driven changes to the epigenome impinge upon the regulation of specific gene programs, rather than transcription at large. While we have highlighted a few examples where integration of mitochondria- and nuclear-encoded signals allows for the regulation of specific target genes, we expect that more will follow in coming years. It is also likely, that current studies have only scratched the surface of how changes in cellular metabolic pathways impact upon nuclear regulation through known and possibly uncharacterized metabolites. More examples may emerge as untargeted metabolomic approaches and use of isotope tracing become more widely employed.
Other open questions, more specifically related to the mechanistic aspect of mitochondria-to-nucleus signaling, include: first, how is the stress sensed and transmitted to the nucleus? Mitochondria-to-nucleus signaling is triggered by mitochondrial signals that are relayed to retrograde factors translocating to the nucleus to modulate gene expression. While the mechanistic details of the RTG pathway have been well elucidated in yeast, triggers and sensors of the mammalian mtISR are not fully defined. Also, is mitochondria-to-nucleus signaling mediated by translocation of isolated factors across the cytosol - and if so is the movement regulated? Or is mitochondria mobility and perinuclear clustering required for facilitating the translocation, possibly via the establishment of nucleus-mitochondria contact sites similar to those described for mitochondria tethering to the ER? As we answer these questions and others, new opportunities may become available for harnessing the beneficial effects of mitohormetic responses and developing novel therapeutics towards diseases with impaired mitochondrial functions.
ACKNOWLEDGMENTS
We are grateful to members of the Perissi and Lee labs for discussions and feedbacks. This work is funded by NIH R01GM127625 to VP, NIH R01AG052258 to CL, KGCRF to MDC, NIGMS 5T32GM008541-22 to JE, and an AFAR fellowship to JMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: C.L. is a consultant and shareholder of CohBar, Inc.
REFERENCES
- Abate M, Festa A, Falco M, Lombardi A, Luce A, Grimaldi A, Zappavigna S, Sperlongano P, Irace C, Caraglia M and Misso G (2020) Mitochondria as playmakers of apoptosis, autophagy and senescence. Semin Cell Dev Biol 98:139–153. [DOI] [PubMed] [Google Scholar]
- Adams KL and Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29:380–395. [DOI] [PubMed] [Google Scholar]
- Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF and Gillespie MN (2012) Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Science signaling 5:ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD and Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nature reviews Genetics 17:487–500. [DOI] [PubMed] [Google Scholar]
- Alves-Fernandes DK and Jasiulionis MG (2019) The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. International journal of molecular sciences 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Madsen AS, Olsen CA and Hirschey MD (2017) Metabolic control by sirtuins and other enzymes that sense NAD(+), NADH, or their ratio. Biochim Biophys Acta Bioenerg 1858:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NS and Haynes CM (2020) Folding the Mitochondrial UPR into the Integrated Stress Response. Trends in cell biology 30:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev AY, Kushnareva YE and Starkov AA (2005) Mitochondrial metabolism of reactive oxygen species. Biochemistry (Mosc) 70:200–214. [DOI] [PubMed] [Google Scholar]
- Arnould T, Michel S and Renard P (2015) Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals? International journal of molecular sciences 16:18224–18251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audrito V, Manago A, Gaudino F, Sorci L, Messana VG, Raffaelli N and Deaglio S (2019) NAD-Biosynthetic and Consuming Enzymes as Central Players of Metabolic Regulation of Innate and Adaptive Immune Responses in Cancer. Front Immunol 10:1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JH, Jo SI, Kim SJ, Lee JM, Jeong JH, Kang JS, Cho NJ, Kim SS, Lee EY and Moon JS (2019) Circulating Cell-Free mtDNA Contributes to AIM2 Inflammasome-Mediated Chronic Inflammation in Patients with Type 2 Diabetes. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Cervantes C, Liu J, He S, Zhou H, Zhang B, Cai H, Yin D, Hu D, Li Z, Chen H, Gao X, Wang F, O’Connor JC, Xu Y, Liu M, Dong LQ and Liu F (2017) DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proceedings of the National Academy of Sciences of the United States of America 114:12196–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S and Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120:483–495. [DOI] [PubMed] [Google Scholar]
- Barcena C, Mayoral P and Quiros PM (2018) Mitohormesis, an Antiaging Paradigm. Int Rev Cell Mol Biol 340:35–77. [DOI] [PubMed] [Google Scholar]
- Bateson (1909) Discussion on the Influence of Heredity on Disease, with special Reference to Tuberculosis, Cancer, and Diseases of the Nervous System: Introductory Address. Proc R Soc Med 2:22–30. [PMC free article] [PubMed] [Google Scholar]
- Bayliak MM, Lylyk MP, Shmihel HV, Sorochynska OM, Semchyshyn OI, Storey JM, Storey KB and Lushchak VI (2017) Dietary alpha-ketoglutarate promotes higher protein and lower triacylglyceride levels and induces oxidative stress in larvae and young adults but not in middle-aged Drosophila melanogaster. Comp Biochem Physiol A Mol Integr Physiol 204:28–39. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD and Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4:517–529. [DOI] [PubMed] [Google Scholar]
- Betancourt AM, King AL, Fetterman JL, Millender-Swain T, Finley RD, Oliva CR, Crowe DR, Ballinger SW and Bailey SM (2014) Mitochondrial-nuclear genome interactions in non-alcoholic fatty liver disease in mice. The Biochemical journal 461:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M and Avadhani NG (1999) Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J 18:522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluher M (2010) The distinction of metabolically ‘healthy’ from ‘unhealthy’ obese individuals. Curr Opin Lipidol 21:38–43. [DOI] [PubMed] [Google Scholar]
- Bohovych I and Khalimonchuk O (2016) Sending Out an SOS: Mitochondria as a Signaling Hub. Front Cell Dev Biol 4:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chatenay-Lapointe M, Wearn CM and Shadel GS (2008) Expression of the rDNA-encoded mitochondrial protein Tar1p is stringently controlled and responds differentially to mitochondrial respiratory demand and dysfunction. Curr Genet 54:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos F, Labbadia J and Herrmann JM (2020) How the Mitoprotein-Induced Stress Response Safeguards the Cytosol: A Unified View. Trends in cell biology 30:241–254. [DOI] [PubMed] [Google Scholar]
- Bootman MD (2012) Calcium signaling. Cold Spring Harb Perspect Biol 4:a011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J and Jazwinski SM (2004) Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics 166:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L and Vaquero A (2015) Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J 282:1745–1767. [DOI] [PubMed] [Google Scholar]
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL and Parker N (2004) Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med 37: 755–767. [DOI] [PubMed] [Google Scholar]
- Bratic A and Larsson NG (2013) The role of mitochondria in aging. The Journal of clinical investigation 123:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieger K, Schiavone S, Miller FJ Jr. and Krause KH (2012) Reactive oxygen species: from health to disease. Swiss Med Wkly 142:w13659. [DOI] [PubMed] [Google Scholar]
- Brokatzky D, Dorflinger B, Haimovici A, Weber A, Kirschnek S, Vier J, Metz A, Henschel J, Steinfeldt T, Gentle IE and Hacker G (2019) A non-death function of the mitochondrial apoptosis apparatus in immunity. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW and Lang BF (2003) Mitochondrial genomes: anything goes. Trends Genet 19:709–716. [DOI] [PubMed] [Google Scholar]
- Cagin U, Duncan OF, Gatt AP, Dionne MS, Sweeney ST and Bateman JM (2015) Mitochondrial retrograde signaling regulates neuronal function. Proceedings of the National Academy of Sciences of the United States of America 112:E6000–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Yu T, Huang C, Xia X, Liu X, Gu J, Xue S, Yeh ET and Cheng J (2012) SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. The Journal of biological chemistry 287:44464–44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese FM, Balacco DL, Preste R, Diroma MA, Forino R, Ventura M and Attimonelli M (2017) NumtS colonization in mammalian genomes. Sci Rep 7:16357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SL and Wellen KE (2018) Metabolic Signaling to the Nucleus in Cancer. Mol Cell 71:398–408. [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P and Auwerx J (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ and Auwerx J (2015) NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab 22:31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Sauve AA and Bai P (2013) Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med 34:1168–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, Rosenfeld MG, Liesa M and Perissi V (2018) Mitochondrial Retrograde Signaling in Mammals Is Mediated by the Transcriptional Cofactor GPS2 via Direct Mitochondria-to-Nucleus Translocation. Mol Cell 69:757–772 e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Quintero A, Deng J, Auchtung J, Brettar I, Hofle MG, Klappenbach J and Konstantinidis KT (2011) Unprecedented levels of horizontal gene transfer among spatially co-occurring Shewanella bacteria from the Baltic Sea. ISME J 5:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico C, Meyer JG, He W, Gibson BW and Verdin E (2018) The Mitochondrial Acylome Emerges: Proteomics, Regulation by Sirtuins, and Metabolic and Disease Implications. Cell metabolism 27:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist CT, Lentucci C, Martinez-Calejman C, Hayashi V, Orofino J, Guertin D, Fried SK, Lee MJ, Cardamone MD and Perissi V (2017) Systemic insulin sensitivity is regulated by GPS2 inhibition of AKT ubiquitination and activation in adipose tissue. Mol Metab 6:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celardo I, Costa AC, Lehmann S, Jones C, Wood N, Mencacci NE, Mallucci GR, Loh SH and Martins LM (2016) Mitofusin-mediated ER stress triggers neurodegeneration in pink1/parkin models of Parkinson’s disease. Cell death & disease 7:e2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D and Park KS (2013) A systems approach for decoding mitochondrial retrograde signaling pathways. Science signaling 6:rs4. [DOI] [PubMed] [Google Scholar]
- Chandel NS (2015) Evolution of Mitochondria as Signaling Organelles. Cell metabolism 22:204–206. [DOI] [PubMed] [Google Scholar]
- Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM and Schumacker PT (2000) Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 275:25130–25138. [DOI] [PubMed] [Google Scholar]
- Chelstowska A, Liu Z, Jia Y, Amberg D and Butow RA (1999) Signalling between mitochondria and the nucleus regulates the expression of a new D-lactate dehydrogenase activity in yeast. Yeast 15:1377–1391. [DOI] [PubMed] [Google Scholar]
- Chervona Y and Costa M (2012) The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic Biol Med 53:1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV and Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. [DOI] [PubMed] [Google Scholar]
- Clapham DE (2007) Calcium signaling. Cell 131:1047–1058. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N and Cohen P (2016) Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging 8:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras L, Drago I, Zampese E and Pozzan T (2010) Mitochondria: the calcium connection. Biochim Biophys Acta 1797:607–618. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M and Walter P (2020) The integrated stress response: From mechanism to disease. Science 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Dunn J, Alvarez LA, Zhang X and Soldati T (2015) Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol 6:472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalli P, Mitic T, Caporali A, Lauriola A and D’Arca D (2016) ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases. Oxid Med Cell Longev 2016:3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila A, Liu L, Chellappa K, Redpath P, Nakamaru-Ogiso E, Paolella LM, Zhang Z, Migaud ME, Rabinowitz JD and Baur JA (2018) Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayama G, Emery SB, Kidd JM and Mills RE (2014) The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res 42:12640–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P and Haynes CM (2017) Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol 47:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarshi PP, McNabney SM and Henagan TM (2017) Skeletal Muscle Nucleo-Mitochondrial Crosstalk in Obesity and Type 2 Diabetes. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J and Kenyon C (2002) Rates of behavior and aging specified by mitochondrial function during development. Science 298:2398–2401. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T (1936) Studies on Hybrid Sterility. II. Localization of Sterility Factors in Drosophila Pseudoobscura Hybrids. Genetics 21:113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Johnson JR, Davey NE, Newton BW, Johnson TL, Galaang S, Seller CA, Krogan N and Toczyski DP (2015) Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, gcn5, and esa1. Molecular & cellular proteomics : MCP 14:162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drareni K, Ballaire R, Barilla S, Mathew MJ, Toubal A, Fan R, Liang N, Chollet C, Huang Z, Kondili M, Foufelle F, Soprani A, Roussel R, Gautier JF, Alzaid F, Treuter E and Venteclef N (2018) GPS2 Deficiency Triggers Maladaptive White Adipose Tissue Expansion in Obesity via HIF1a Activation. Cell reports 24:2957–2971 e2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge W (2002) Free radicals in the physiological control of cell function. Physiol Rev 82:47–95. [DOI] [PubMed] [Google Scholar]
- Ducker GS and Rabinowitz JD (2017) One-Carbon Metabolism in Health and Disease. Cell Metab 25:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan OF and Bateman JM (2016) Mitochondrial retrograde signaling in the Drosophila nervous system and beyond. Fly (Austin) 10:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan OF, Granat L, Ranganathan R, Singh VK, Mazaud D, Fanto M, Chambers D, Ballard CG and Bateman JM (2018) Ras-ERK-ETS inhibition alleviates neuronal mitochondrial dysfunction by reprogramming mitochondrial retrograde signaling. PLoS genetics 14:e1007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Snary KJ and Ballinger SW (2015) GENETICS. Mitochondrial-nuclear DNA mismatch matters. Science 349:1449–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranteau J, Chandel NS, Kulisz A, Shao Z and Schumacker PT (1998) Intracellular signaling by reactive oxygen species during hypoxia in cardiomyocytes. J Biol Chem 273:11619–11624. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S and Dillin A (2011) The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteil D, Metzger E, Willmann D, Karagianni P, Friedrichs N, Greschik H, Gunther T, Buettner R, Talianidis I, Metzger D and Schule R (2014) LSD1 promotes oxidative metabolism of white adipose tissue. Nat Commun 5:4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Brown MT and Johnson PJ (2004) Ancient invasions: from endosymbionts to organelles. Science 304:253–257. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Bord M and Schuldiner M (2017) Ground control to major TOM: mitochondria-nucleus communication. FEBS J 284:196–210. [DOI] [PubMed] [Google Scholar]
- Emamgholipour S, Ebrahimi R, Bahiraee A, Niazpour F and Meshkani R (2020) Acetylation and insulin resistance: a focus on metabolic and mitogenic cascades of insulin signaling. Crit Rev Clin Lab Sci:1–19. [DOI] [PubMed] [Google Scholar]
- Epstein CB, Waddle JA, Hale Wt, Dave V, Thornton J, Macatee TL, Garner HR and Butow RA (2001) Genome-wide responses to mitochondrial dysfunction. Molecular biology of the cell 12:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP and Mostoslavsky R (2016) Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell 62:695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JL, Goldfine ID, Maddux BA and Grodsky GM (2003) Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes 52:1–8. [DOI] [PubMed] [Google Scholar]
- Fan R, Toubal A, Goni S, Drareni K, Huang Z, Alzaid F, Ballaire R, Ancel P, Liang N, Damdimopoulos A, Hainault I, Soprani A, Aron-Wisnewsky J, Foufelle F, Lawrence T, Gautier JF, Venteclef N and Treuter E (2016) Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nature medicine 22:780–791. [DOI] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL and Bohr VA (2016) Nuclear DNA damage signalling to mitochondria in ageing. Nature reviews Molecular cell biology 17:308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano C, Disciglio V, Bertora S, Lepore Signorile M and Simone C (2019) FOXO3a from the Nucleus to the Mitochondria: A Round Trip in Cellular Stress Response. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW and Welch DR (2015) Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential. Cancer Res 75:4429–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ and Auwerx J (2008) Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8:347–358. [DOI] [PubMed] [Google Scholar]
- Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG and Hekimi S (1999) CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. The EMBO journal 18:1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F and Hekimi S (2001) Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell 1:633–644. [DOI] [PubMed] [Google Scholar]
- Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, Phillips PE, Crow MK, Oess S, Muller-Esterl W and Perl A (2009) Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol 182:2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H and Jae LT (2020) A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetterman JL and Ballinger SW (2019) Mitochondrial genetics regulate nuclear gene expression through metabolites. Proc Natl Acad Sci U S A 116:15763–15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell’italia LJ, Darley-Usmar VM, Welch DR and Ballinger SW (2013) Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. The Biochemical journal 455:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T (2011) Signal transduction by reactive oxygen species. J Cell Biol 194:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW and Haynes CM (2016) The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Current biology : CB 26:2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicker D, Sancak Y, Mick E, Goldberger O and Mootha VK (2019) Exploring the In Vivo Role of the Mitochondrial Calcium Uniporter in Brown Fat Bioenergetics. Cell reports 27:1364–1375 e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsstrom S, Jackson CB, Carroll CJ, Kuronen M, Pirinen E, Pradhan S, Marmyleva A, Auranen M, Kleine IM, Khan NA, Roivainen A, Marjamaki P, Liljenback H, Wang L, Battersby BJ, Richter U, Velagapudi V, Nikkanen J, Euro L and suomalainen A (2019) Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions. Cell metabolism 30:1040–1054 e1047. [DOI] [PubMed] [Google Scholar]
- Frezza C (2017) Mitochondrial metabolites: undercover signalling molecules. Interface Focus 7:20160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273:793–798. [DOI] [PubMed] [Google Scholar]
- Fujii T, Khawaja MR, DiNardo CD, Atkins JT and Janku F (2016) Targeting isocitrate dehydrogenase (IDH) in cancer. Discov Med 21:373–380. [PubMed] [Google Scholar]
- Gambardella S, Limanaqi F, Ferese R, Biagioni F, Campopiano R, Centonze D and Fornai F (2019) ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Frontiers in immunology 10:1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lin SH, Ren F, Li JT, Chen JJ, Yao CB, Yang HB, Jiang SX, Yan GQ, Wang D, Wang Y, Liu Y, Cai Z, Xu YY, Chen J, Yu W, Yang PY and Lei QY (2016) Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat Commun 7:11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Roves PM, Osler ME, Holmstrom MH and Zierath JR (2008) Gain-of-function R225Q mutation in AMP-activated protein kinase gamma3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. The Journal of biological chemistry 283:35724–35734. [DOI] [PubMed] [Google Scholar]
- Germain D (2016) Sirtuins and the Estrogen Receptor as Regulators of the Mammalian Mitochondrial UPR in Cancer and Aging. Adv Cancer Res 130:211–256. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Marchi S and Pinton P (2018) The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol 19:713–730. [DOI] [PubMed] [Google Scholar]
- Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, Brady RO, Ellis JR and Schiffmann R (2004) Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited case of mucolipidosis IV. Hum Mutat 24:460–465. [DOI] [PubMed] [Google Scholar]
- Granat L, Hunt RJ and Bateman JM (2020) Mitochondrial retrograde signalling in neurological disease. Philos Trans R Soc Lond B Biol Sci 375:20190415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4:a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazioli S and Pugin J (2018) Mitochondrial Damage-Associated Molecular Patterns: From Inflammatory Signaling to Human Diseases. Frontiers in immunology 9:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griendling KK, Sorescu D, Lassegue B and Ushio-Fukai M (2000) Modulation of protein kinase activity and gene expression by reactive oxygen species and their role in vascular physiology and pathophysiology. Arterioscler Thromb Vasc Biol 20:2175–2183. [DOI] [PubMed] [Google Scholar]
- Guha M and Avadhani NG (2013) Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 13:577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha M, Srinivasan S, Biswas G and Avadhani NG (2007) Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J Biol Chem 282:14536–14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Aviles G, Liu Y, Tian R, Unger BA, Lin YT, Wiita AP, Xu K, Correia MA and Kampmann M (2020) Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC and Sinclair DA (2010) Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5:253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB and Chandel NS (2010) Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci 35:505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D (1981) The aging process. Proc Natl Acad Sci U S A 78:7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y and Ron D (2007) ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental cell 13:467–480. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA and Ron D (2010) The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell 37:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Hart PC, Germain D and Bonini MG (2016) SOD2 and the Mitochondrial UPR: Partners Regulating Cellular Phenotypic Transitions. Trends in biochemical sciences 41:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb M, Gluschko A, Wiegmann K, Farid A, Wolf A, Utermohlen O, Krut O, Kronke M and Schramm M (2019) Mitochondrial reactive oxygen species enable proinflammatory signaling through disulfide linkage of NEMO. Sci Signal 12. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, DeBerardinis RJ, Diehl AME, Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA, Locasale JW, Longo VD, Lyssiotis CA, McDonnell E, Mehrmohamadi M, Michelotti G, Muralidhar V, Murphy MP, Pedersen PL, Poore B, Raffaghello L, Rathmell JC, Sivanand S, Vander Heiden MG, Wellen KE and Target Validation T (2015) Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol 35 Suppl:S129–S150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD and Zhao Y (2015) Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Molecular & cellular proteomics : MCP 14:2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB and Kralli A (2009) Transcriptional control of mitochondrial biogenesis and function. Annual review of physiology 71:177–203. [DOI] [PubMed] [Google Scholar]
- Hogan PG (2017) Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 63:66–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Canto C, Wanders RJ and Auwerx J (2010) The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31:194–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E and Auwerx J (2012) Sirtuins as regulators of metabolism and healthspan. Nature reviews Molecular cell biology 13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RJ and Bateman JM (2018) Mitochondrial retrograde signaling in the nervous system. FEBS letters 592:663–678. [DOI] [PubMed] [Google Scholar]
- Hunt RJ, Granat L, McElroy GS, Ranganathan R, Chandel NS and Bateman JM (2019) Mitochondrial stress causes neuronal dysfunction via an ATF4-dependent increase in L-2-hydroxyglutarate. The Journal of cell biology 218:4007–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichim G, Lopez J, Ahmed SU, Muthalagu N, Giampazolias E, Delgado ME, Haller M, Riley JS, Mason SM, Athineos D, Parsons MJ, van de Kooij B, Bouchier-Haye L, Chalmers AJ, Rooswinkel RW, Oberst A, Blyth K, Rehm M, Murphy DJ and Tait SWG (2015) Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Molecular cell 57:860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T (2018) Histone demethylases regulate adipocyte thermogenesis. Diabetol Int 9:215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Wang B, Liu H, Shah H, Carmona-Fontaine C, Rustenburg AS, Salah S, Gunner MR, Chodera JD, Cross JR and Thompson CB (2017) L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol 13:494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS (2020) Calcium Signaling: From Basic to Bedside. Advances in experimental medicine and biology 1131:1–6. [DOI] [PubMed] [Google Scholar]
- Jang JY, Blum A, Liu J and Finkel T (2018) The role of mitochondria in aging. The Journal of clinical investigation 128:3662–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM (2005) Rtg2 protein: at the nexus of yeast longevity and aging. FEMS Yeast Res 5:1253–1259. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM (2013) The retrograde response: when mitochondrial quality control is not enough. Biochimica et biophysica acta 1833:400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM and Kriete A (2012) The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Frontiers in physiology 3:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J and Butow RA (1997) A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 17:1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V and Auwerx J (2015) The mitochondrial unfolded protein response-synchronizing genomes. Current opinion in cell biology 33:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V, Mouchiroud L and Auwerx J (2014) The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol 217:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG and Rosenfeld MG (2006) A breaking strategy for topoisomerase IIbeta/PARP-1-dependent regulated transcription. Cell Cycle 5:2557–2560. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr and McKnight SL (2013) Influence of metabolism on epigenetics and disease. Cell 153:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Kundu M and Green DR (2015) Innate immune recognition of mtDNA--an undercover signal? Cell metabolism 21:793–794. [DOI] [PubMed] [Google Scholar]
- Kawai T and Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology 11:373–384. [DOI] [PubMed] [Google Scholar]
- Keisari Y, Braun L and Flescher E (1983) The oxidative burst and related phenomena in mouse macrophages elicited by different sterile inflammatory stimuli. Immunobiology 165:78–89. [DOI] [PubMed] [Google Scholar]
- Kenny TC and Germain D (2017) mtDNA, Metastasis, and the Mitochondrial Unfolded Protein Response (UPR(mt)). Front Cell Dev Biol 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivela R, Pessia A, Velagapudi V and Suomalainen A (2017) mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell metabolism 26:419–428 e415. [DOI] [PubMed] [Google Scholar]
- Kim H, Yang J, Kim MJ, Choi S, Chung JR, Kim JM, Yoo YH, Chung J and Koh H (2016) Tumor Necrosis Factor Receptor-associated Protein 1 (TRAP1) Mutation and TRAP1 Inhibitor Gamitrinib-triphenylphosphonium (G-TPP) Induce a Forkhead Box O (FOXO)-dependent Cell Protective Signal from Mitochondria. The Journal of biological chemistry 291:1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Gupta R, Blanco LP, Yang S, Shteinfer-Kuzmine A, Wang K, Zhu J, Yoon HE, Wang X, Kerkhofs M, Kang H, Brown AL, Park SJ, Xu X, Zandee van Rilland E, Kim MK, Cohen JI, Kaplan MJ, Shoshan-Barmatz V and Chung JH (2019) VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science 366:1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, Ko T, Han J, Kim HL, Kim J, Back SH, Komatsu M, Chen H, Chan DC, Konishi M, Itoh N, Choi CS and Lee MS (2013) Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nature medicine 19:83–92. [DOI] [PubMed] [Google Scholar]
- Kim KH, Son JM, Benayoun BA and Lee C (2018) The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress. Cell metabolism 28:516–524 e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT and Kraus WL (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119:803–814. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T and Kraus WL (2005) Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes & development 19:1951–1967. [DOI] [PubMed] [Google Scholar]
- Kim S and Koh H (2017) Role of FOXO transcription factors in crosstalk between mitochondria and the nucleus. Journal of bioenergetics and biomembranes. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F, Fischer S, Haag F and Ziegler M (2011) Compartmentation of NAD+-dependent signalling. FEBS letters 585:1651–1656. [DOI] [PubMed] [Google Scholar]
- Kopinski PK, Janssen KA, Schaefer PM, Trefely S, Perry CE, Potluri P, Tintos-Hernandez JA, Singh LN, Karch KR, Campbell SL, Doan MT, Jiang H, Nissim I, Nakamaru-Ogiso E, Wellen KE, Snyder NW, Garcia BA and Wallace DC (2019) Regulation of nuclear epigenome by mitochondrial DNA heteroplasmy. Proceedings of the National Academy of Sciences of the United States of America 116:16028–16035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac S, Angelova PR, Holmstrom KM, Zhang Y, Dinkova-Kostova AT and Abramov AY (2015) Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochimica et biophysica acta 1850:794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, Newgard CB, Lopaschuk GD and Muoio DM (2008) Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell metabolism 7:45–56. [DOI] [PubMed] [Google Scholar]
- Kraus WL and Lis JT (2003) PARP goes transcription. Cell 113:677–683. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R and Kraus WL (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Molecular cell 39:8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug AK, Gutbier S, Zhao L, Poltl D, Kullmann C, Ivanova V, Forster S, Jagtap S, Meiser J, Leparc G, Schildknecht S, Adam M, Hiller K, Farhan H, Brunner T, Hartung T, Sachinidis A and Leist M (2014) Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP(+). Cell death & disease 5:e1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliawat R, Klein L, Gong Z, Nicoletta-Gentile M, Nemkal A, Cui L, Bastie C, Su K, Huffman D, Surana M, Barzilai N, Fleischer N and Muzumdar R (2013) Potent humanin analog increases glucose-stimulated insulin secretion through enhanced metabolism in the beta cell. FASEB journal : official publication of the federation of American Societies for Experimental Biology 27:4890–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni CA and Brookes PS (2019) Cellular Compartmentation and the Redox/Nonredox Functions of NAD(). Antioxidants & redox signaling 31:623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupis W, Palyga J, Tomal E and Niewiadomska E (2016) The role of sirtuins in cellular homeostasis. J Physiol Biochem 72:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M, Asanuma T, Niwa K and Inanami O (2008) Regulation of cell survival and death signals induced by oxidative stress. J Clin Biochem Nutr 43:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J and Bakhoum SF (2020) The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer discovery 10:26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O and Beatty JT (2012) Gene transfer agents: phage-like elements of genetic exchange Nat Rev Microbiol 10:472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Pellicer A, Lechuga-Vieco AV, Johnston IG, Hamalainen RH, Pellico J, Justo-Mendez R, Fernandez-Toro JM, Claveria C, Guaras A, Sierra R, Llop J, Torres M, Criado LM, Suomalainen A, Jones NS, Ruiz-Cabello J and Enriquez JA (2019) Regulation of Mother-to-Offspring Transmission of mtDNA Heteroplasmy. Cell metabolism 30:1120–1130 e1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A and Cohen P (2014) IGF-I regulates the age-dependent signaling peptide humanin. Aging cell 13:958–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Yen k P (2013) Humanin: a harbinger of mitochondrial-derived peptides? Trends in endocrinology and metabolism: TEM 24:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R and Cohen P (2015) The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell metabolism 21:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J and Ruvkun G (2003) A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics 33:40–48. [DOI] [PubMed] [Google Scholar]
- Lettieri-Barbato D, Ioannilli L, Aquilano K, Ciccarone F, Rosina M and Ciriolo MR (2019) FoxO1 localizes to mitochondria of adipose tissue and is affected by nutrient stress. Metabolism: clinical and experimental 95:84–92. [DOI] [PubMed] [Google Scholar]
- Liao XS, Small WC, Srere PA and Butow RA (1991) Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Molecular and cellular biology 11:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C and Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell metabolism 1:361–370. [DOI] [PubMed] [Google Scholar]
- Liochev SI (2013) Reactive oxygen species and the free radical theory of aging. Free Radic Biol Med 60:1–4. [DOI] [PubMed] [Google Scholar]
- Liu JL, Yee C, Wang Y and Hekimi S (2017) A single biochemical activity underlies the pleiotropy of the aging-related protein CLK-1. Scientific reports 7:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Feng M and Guan W (2016) Mitochondrial DNA sensing by STING signaling participates in inflammation, cancer and beyond. International journal of cancer Journal international du cancer 139:736–741. [DOI] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC and Ruvkun G (2014) Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508:406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z and Butow RA (2006) Mitochondrial retrograde signaling. Annual review of genetics 40:159–185. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ren Z, Zhang J, Chuang CC, Kandaswamy E, Zhou T and Zuo L (2018) Role of ROS and Nutritional Antioxidants in Human Diseases. Frontiers in physiology 9:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Spirek M, Thornton J and Butow RA (2003) Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Molecular cell 12:401–411. [DOI] [PubMed] [Google Scholar]
- Loos RJF and Kilpelainen TO (2018) Genes that make you fat, but keep you healthy. Journal of internal medicine 284:450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M and Kroemer G (2013) The hallmarks of aging. Cell 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W and O’Brien SJ (1994) Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39:174–190. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK and Thompson CB (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483:474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Aladjem MI KW (2013) SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome integrity 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Chen H, He X, Nie H, Hong Y, Sheng C, Wang Q, Xia W and Ying W (2012) NAD+ metabolism and NAD(+)-dependent enzymes: promising therapeutic targets for neurological diseases. Curr Drug Targets 13:222–229. [DOI] [PubMed] [Google Scholar]
- Ma Y, Nie H, Chen H, Li J, Hong Y, Wang B, Wang C, Zhang J, Cao W, Zhang M, Xu Y, Ding X, Yin SK, Qu X and Ying W (2015) NAD(+)/NADH metabolism and NAD(+)-dependent enzymes in cell death and ischemic brain injury: current advances and therapeutic implications. Curr Med Chem 22:1239–1247. [DOI] [PubMed] [Google Scholar]
- Mao Y, Luo W, Zhang L, Wu W, Yuan L, Xu H, Song J, Fujiwara K, Abe JI, LeMaire SA, Wang XL and Shen YH (2017) STING-IRF3 Triggers Endothelial Inflammation in Response to Free Fatty Acid-Induced Mitochondrial Damage in Diet-Induced Obesity. Arteriosclerosis, thrombosis, and vascular biology 37:920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Abundis E, Garcia N, Correa F, Franco M and Zazueta C (2007) Changes in specific lipids regulate BAX-induced mitochondrial permeability transition. The FEBS journal 274:6500–6510. [DOI] [PubMed] [Google Scholar]
- Martinez-Reyes I and Chandel NS (2020) Mitochondrial TCA cycle metabolites control physiology and disease. Nature communications 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, Dufour E, Spelbrink JN, Weinberg SE, Zhao Y, DeBerardinis RJ and Chandel NS (2016) TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular cell 61:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen O, Quiros PM and Auwerx J (2017) Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends in cell biology 27:453–463. [DOI] [PubMed] [Google Scholar]
- McCormack JG and Denton RM (1980) Role of calcium ions in the regulation of intramitochondrial metabolism. Properties of the Ca2+-sensitive dehydrogenases within intact uncoupled mitochondria from the white and brown adipose tissue of the rat. The Biochemical journal 190:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, Grimsrud PA and Hirschey MD (2016) Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell reports 17:1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A and Haynes CM (2018) UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell research 28:281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies KJ, Zhang H, Katsyuba E and Auwerx J (2016) Protein acetylation in metabolism - metabolites and cofactors. Nat Rev Endocrinol 12:43–60. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, Murillo V, Wolff SC, Shaw RJ, Auwerx J and Dillin A (2016) Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell 165:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry TL and Ristow M (2016) Mitohormesis in exercise training. Free Radic Biol Med 98:123–130. [DOI] [PubMed] [Google Scholar]
- Meurers BH, Zhu C, Fernagut PO, Richter F, Hsia YC, Fleming SM, Oh M, Elashoff D, Dicarlo CD, Seaman RL and Chesselet MF (2009) Low dose rotenone treatment causes selective transcriptional activation of cell death related pathways in dopaminergic neurons in vivo. Neurobiology of disease 33:182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T and Berger SL (2017) Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick E, Titov DV, Skinner OS, Sharma R, Jourdain AA and Mootha VK (2020) Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan RM, Barnes RG, Fisher K, Andreou T, Rooney N, Poulin GB and Whitmarsh AJ (2015a) A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nature cell biology 17:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan RM, Poulin GB and Whitmarsh AJ (2015b) A nuclear sensor of mitochondrial function. Oncotarget 6:15746–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LD, Le T and Fan G (2013) DNA methylation and its basic function. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MJ and Liu ZG (2011) Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell research 21:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morigi M, Perico L and Benigni A (2018) Sirtuins in Renal Health and Disease. J Am Soc Nephrol 29:1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottis A, Herzig S and Auwerx J (2019) Mitocellular communication: Shaping health and disease. Science 366:827–832. [DOI] [PubMed] [Google Scholar]
- Mottis A, Jovaisaite V and Auwerx J (2014) The mitochondrial unfolded protein response in mammalian physiology. Mamm Genome 25:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H (1942) Isolating mechanisms, evolution, and temperature, in Biol Symp pp 71–125. [Google Scholar]
- Munch C (2018) The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol 16:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, Covington JD, Bajpeyi S, Ravussin E, Kraus W, Koves TR and Mynatt RL (2012) Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell metabolism 15:764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar RH, Huffman DM, Atzmon G, Buettner C, Cobb LJ, Fishman S, Budagov T, Cui L, Einstein FH, Poduval A, Hwang D, Barzilai N and Cohen P (2009) Humanin: a novel central regulator of peripheral insulin action. PloS one 4:e6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka K, Hino S, Sakamoto A, Anan K, Takase R, Umehara T, Yokoyama S, Sasaki Y and Nakao M (2015) Lysine-specific demethylase 2 suppresses lipid influx and metabolism in hepatic cells. Molecular and cellular biology 35:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Sharpley MS, Chi F, Braas D, Zhou Y, Kim R, Clark AT and Banerjee U (2017) Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell 168:210–223 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik E and Dixit VM (2011) Mitochondrial reactive oxygen species drive proinflammatory cytokine production. The Journal of experimental medicine 208:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW and Choi AM (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh NU and Haynes CM (2019) Signaling and Regulation of the Mitochondrial Unfolded Protein Response. Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P and Haynes CM (2015) Mitochondrial and Nuclear Accumulation of the Transcription Factor ATFS-1 Promotes OXPHOS Recovery during the UPR(mt). Molecular cell 58:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM and Haynes CM (2012) Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashine S, Cohen P, Nesburn AB, Kuppermann BD and Kenney MC (2018) Characterizing the protective effects of SHLP2, a mitochondrial-derived peptide, in macular degeneration. Scientific reports 8:15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashine S and Kenney MC (2020) Effects of Mitochondrial-Derived Peptides (MDPs) on Mitochondrial and Cellular Health in AMD. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkanen J, Forsstrom S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamaki J, Roivainen A, Marjamaki P, Liljenback H, Ahola S, Buzkova J, Terzioglu M, Khan NA, Pirnes-Karhu S, Paetau A, Lonnqvist T, Sajantila A, Isohanni P, Tyynismaa H, Nomura DK, Battersby BJ, Velagapudi V, Carroll CJ and Suomalainen A (2016) Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. Cell metabolism 23:635–648. [DOI] [PubMed] [Google Scholar]
- Niu Y, DesMarais TL, Tong Z, Yao Y and Costa M (2015) Oxidative stress alters global histone modification and DNA methylation. Free Radic Biol Med 82:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ and Verdin E (2004) Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome biology 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mealey GB, Plafker KS, Berry WL, Janknecht R, Chan JY and Plafker SM (2017) A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking. Journal of cell science 130:3467–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada AK, Teranishi K, Lobo F, Isas JM, Xiao J, Yen K, Cohen P and Langen R (2017) The Mitochondrial-Derived Peptides, HumaninS14G and Small Humanin-like Peptide 2, Exhibit Chaperone-like Activity. Scientific reports 7:7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E and Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461:537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W and Perrimon N (2013) Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S and Banerjee U (2008) Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nature genetics 40:356–361. [DOI] [PubMed] [Google Scholar]
- Papa L and Germain D (2011) Estrogen receptor mediates a distinct mitochondrial unfolded protein response. Journal of cell science 124:1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L and Germain D (2014) SirT3 regulates the mitochondrial unfolded protein response. Molecular and cellular biology 34:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Lee SC, Cardenas ME and Heitman J (2019) Calcium-Calmodulin-Calcineurin Signaling: A Globally Conserved Virulence Cascade in Eukaryotic Microbial Pathogens. Cell Host Microbe 26:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB and Zhao Y (2013) SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Molecular cell 50:919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Jo SH, Kim MY, Kim TH and Ahn YH (2015) Role of transcription factor acetylation in the regulation of metabolic homeostasis. Protein Cell 6:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L and Rao A (2013) TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology 14:341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrushev M, Kasymov V, Patrusheva V, Ushakova T, Gogvadze V and Gaziev A (2004) Mitochondrial permeability transition triggers the release of mtDNA fragments. Cell Mol Life Sci 61:3100–3103. [DOI] [PubMed] [Google Scholar]
- Patti ME and Corvera S (2010) The role of mitochondria in the pathogenesis of type 2 diabetes. Endocrine reviews 31:364–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM and Haynes CM (2013) Signaling the mitochondrial unfolded protein response. Biochimica et biophysica acta 1833:410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SL and Baker AJ (2004) Low number of mitochondrial pseudogenes in the chicken (Gallus gallus) nuclear genome: implications for molecular inference of population history and phylogenetics. BMC evolutionary biology 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi J, Bai Y, Zhang Q, Wong V, Floering LM, Daniel K, Reece JM, Deeney JT, Andersen ME, Corkey BE and Collins S (2007) Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes 56:1783–1791. [DOI] [PubMed] [Google Scholar]
- Pi J, Zhang Q, Fu J, Woods CG, Hou Y, Corkey BE, Collins S and Andersen ME (2010) ROS signaling, oxidative stress and Nrf2 in pancreatic beta-cell function. Toxicol Appl Pharmacol 244:77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F and Kroemer G (2015) Acetyl coenzyme A: a central metabolite and second messenger. Cell metabolism 21:805–821. [DOI] [PubMed] [Google Scholar]
- Pirinen E, Lo Sasso G and Auwerx J (2012) Mitochondrial sirtuins and metabolic homeostasis. Best Pract Res Clin Endocrinol Metab 26:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP and Auwerx J (2017) Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MA, Haynes CM and Pellegrino MW (2017) The mitochondrial unfolded protein response: Signaling from the powerhouse. The Journal of biological chemistry 292:13500–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S and Islam R (2011) Mammalian Sirt1: insights on its biological functions. Cell Commun Signal 9:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri S and Mellor J (2018) IDH1: Linking Metabolism and Epigenetics. Frontiers in genetics 9:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A and sheldahl L (2006) Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA and Fry AJ (2004) Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol 19:645–653. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E and Gibson BW (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Sciences of the United States of America 110:6601–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD and Helin K (2016) Role of TET enzymes in DNA methylation, development, and cancer. Genes & development 30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath E, Moschetta A and Haller D (2018) Mitochondrial function - gatekeeper of intestinal epithelial cell homeostasis. Nat Rev Gastroenterol Hepatol 15:497–516. [DOI] [PubMed] [Google Scholar]
- Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M and Steegborn C (2013) An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nature communications 4:2327. [DOI] [PubMed] [Google Scholar]
- Reczek CR and Chandel NS (2015) ROS-dependent signal transduction. Current opinion in cell biology 33:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requejo-Aguilar R, Lopez-Fabuel I, Fernandez E, Martins LM, Almeida A and Bolanos JP (2014) PINK1 deficiency sustains cell proliferation by reprogramming glucose metabolism through HIF1. Nature communications 5:4514. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Lai RW, Woodhead JST, Joly JH, Mitchell CJ, Cameron-Smith D, Lu R, Cohen P, Graham NA, Benayoun BA, Merry TL and Lee C (2019) MOTS-c is an Exercise-Induced Mitochondrial-Encoded Regulator of Age-Dependent Physical Decline and Muscle Homeostasis. bioRxiv:2019.2012.2022.886432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly E and Leister D (2004) NUMTs in sequenced eukaryotic genomes. Molecular biology and evolution 21:1081–1084. [DOI] [PubMed] [Google Scholar]
- Ristow M and Schmeisser S (2011) Extending life span by increasing oxidative stress. Free Radic Biol Med 51:327–336. [DOI] [PubMed] [Google Scholar]
- Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D and Greene LA (2002) Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. J Neurosci 22:10690–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Hino S, Nagaoka K, Anan K, Takase R, Matsumori H, Ojima H, Kanai Y, Arita K and Nakao M (2015) Lysine Demethylase LSD1 Coordinates Glycolytic and Mitochondrial Metabolism in Hepatocellular Carcinoma Cells. Cancer research 75:1445–1456. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kauppinen A, Hiltunen M and Kaarniranta K (2014) Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process. Ageing research reviews 16:45–65. [DOI] [PubMed] [Google Scholar]
- Sambeat A, Gulyaeva O, Dempersmier J, Tharp KM, Stahl A, Paul SM and Sul HS (2016) LSD1 Interacts with Zfp516 to Promote UCP1 Transcription and Brown Fat Program. Cell reports 15:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AL, Sinha S and Lindner AB (2018) The Good, the Bad, and the Ugly of ROS: New Insights on Aging and Aging-Related Diseases from Eukaryotic and Prokaryotic Model Organisms. Oxidative medicine and cellular longevity 2018:1941285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarparanta J, Garcia-Macia M and Singh R (2017) Autophagy and Mitochondria in Obesity and Type 2 Diabetes. Current diabetes reviews 13:352–369. [DOI] [PubMed] [Google Scholar]
- Sarsour EH, Kalen AL and Goswami PC (2014) Manganese superoxide dismutase regulates a redox cycle within the cell cycle. Antioxidants & redox signaling 20:1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL and Boeke JD (2006) The biochemistry of sirtuins. Annu Rev Biochem 75:435–465. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB and Kelly DP (2012) Transcriptional integration of mitochondrial biogenesis. Trends in endocrinology and metabolism: TEM 23:459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, DiMauro S and Hirano M (2012) Human mitochondrial DNA: roles of inherited and somatic mutations. Nature reviews Genetics 13:878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N and Shadel GS (2013) Epigenetic silencing mediates mitochondria stress-induced longevity. Cell metabolism 17:954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz AM and Haynes CM (2015) UPR(mt)-mediated cytoprotection and organismal aging. Biochimica et biophysica acta 1847:1448–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A and Harris AL (2012) How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491:364–373. [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Thompson CB and Finley LWS (2018) Metabolic regulation of chromatin modifications and gene expression. J Cell Biol 217:2247–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialo F, Mallikarjun V, Stefanatos R and Sanz A (2013) Regulation of lifespan by the mitochondrial electron transport chain: reactive oxygen species-dependent and reactive oxygen species-independent mechanisms. Antioxidants & redox signaling 19:1953–1969. [DOI] [PubMed] [Google Scholar]
- Sekito T, Liu Z, Thornton J and Butow RA (2002) RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3]. Molecular biology of the cell 13:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA and Chandel NS (2012) Physiological roles of mitochondrial reactive oxygen species. Molecular cell 48:158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E and Yoshida M (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS and Horvath TL (2015) Mitochondrial ROS signaling in organismal homeostasis. Cell 163:560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy DT, McAllister K, Worth L, Haugen AC, Meyer JN, Domann FE, Van Houten B, Mostoslavsky R, Bultman SJ, Baccarelli AA, Begley TJ, Sobol RW, Hirschey MD, Ideker T, Santos JH, Copeland WC, Tice RR, Balshaw DM and Tyson FL (2014) Mitochondria, energetics, epigenetics, and cellular responses to stress. Environmental health perspectives 122:1271–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L and Tu BP (2015) Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Current opinion in cell biology 33:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA and Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953. [DOI] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T and Arditi M (2012) Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36:401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka T and Haynes CM (2018) The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nature reviews Molecular cell biology 19:109–120. [DOI] [PubMed] [Google Scholar]
- Simone D, Calabrese FM, Lang M, Gasparre G and Attimonelli M (2011) The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics 12:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S, Viney I and Wellen KE (2018) Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci 43:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WI Sivitz and Yorek MA (2010) Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxidants & redox signaling 12:537–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiraglia DJ, Kulawiec M, Bistulfi GL, Gupta SG and Singh KK (2008) A novel role for mitochondria in regulating epigenetic modification in the nucleus. Cancer biology & therapy 7:1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloveychik M, Xu M, Zaslaver O, Lee K, Narula A, Jiang R, Rosebrock AP, Caudy AA and Meneghini MD (2016) Mitochondrial control through nutritionally regulated global histone H3 lysine-4 demethylation. Scientific reports 6:37942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Guha M, Kashina A and Avadhani NG (2017) Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg 1858:602–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE and Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64:435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ and Finkel T (2016) The Mitochondrial Basis of Aging. Molecular cell 61:654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A and Battersby BJ (2018) Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nature reviews Molecular cell biology 19:77–92. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Elo JM, Pietilainen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, Pihko H, Darin N, Ounap K, Kluijtmans LA, Paetau A, Buzkova J, Bindoff LA, Annunen-Rasila J, Uusimaa J, Rissanen A, Yki-Jarvinen H, Hirano M, Tulinius M, Smeitink J and Tyynismaa H (2011) FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol 10:806–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E and Michelakis ED (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158:84–97. [DOI] [PubMed] [Google Scholar]
- Szczesny B, Marcatti M, Ahmad A, Montalbano M, Brunyanszki A, Bibli SI, Papapetropoulos A and Szabo C (2018) Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Scientific reports 8:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H, Niikura T, Hashimoto Y, Ito Y, Kita Y, Terashita K, Yamazaki K, Koto A, Aiso S and Nishimoto I (2002) Evidence for in vivo production of Humanin peptide, a neuroprotective factor against Alzheimer’s disease-related insults. Neuroscience letters 324:227–231. [DOI] [PubMed] [Google Scholar]
- Tajima S, Suetake I, Takeshita K, Nakagawa A and Kimura H (2016) Domain Structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA Methyltransferases. Advances in experimental medicine and biology 945:63–86. [DOI] [PubMed] [Google Scholar]
- Tan JX and Finkel T (2020) Mitochondria as intracellular signaling platforms in health and disease. The Journal of cell biology 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL (2016) Sirt1 and the Mitochondria. Mol Cells 39:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Chowdhury AR, Guha M, Huang L, Van Winkle T, Rustgi AK and Avadhani NG (2012) Silencing of IkBbeta mRNA causes disruption of mitochondrial retrograde signaling and suppression of tumor growth in vivo. Carcinogenesis 33:1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K and Auwerx J (2010) Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell metabolism 12:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurey P and Rieusset J (2017) Mitochondria-Associated Membranes Response to Nutrient Availability and Role in Metabolic Diseases. Trends in endocrinology and metabolism: TEM 28:32–45. [DOI] [PubMed] [Google Scholar]
- Thiagarajan D, Vedantham S, Ananthakrishnan R, Schmidt AM and Ramasamy R (2016) Mechanisms of transcription factor acetylation and consequences in hearts. Biochimica et biophysica acta 1862:2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM and Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. [DOI] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ and Dillin A (2016a) Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell 165:1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Merkwirth C and Dillin A (2016b) Mitochondrial UPR: A Double-Edged Sword. Trends in cell biology 26:563–565. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY and Martin W (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature reviews Genetics 5:123–135. [DOI] [PubMed] [Google Scholar]
- Tonelli C, Chio IIC and Tuveson DA (2018) Transcriptional Regulation by Nrf2. Antioxidants & redox signaling 29:1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torelli NQ, Ferreira-Junior JR, Kowaltowski AJ and da Cunha FM (2015) RTG1- and RTG2-dependent retrograde signaling controls mitochondrial activity and stress resistance in Saccharomyces cerevisiae. Free radical biology & medicine 81:30–37. [DOI] [PubMed] [Google Scholar]
- Traaseth N, Elfering S, Solien J, Haynes V and Giulivi C (2004) Role of calcium signaling in the activation of mitochondrial nitric oxide synthase and citric acid cycle. Biochimica et biophysica acta 1658:64–71. [DOI] [PubMed] [Google Scholar]
- Trumpff C, Marsland AL, Basualto-Alarcon C, Martin JL, Carroll JE, Sturm G, Vincent AE, Mosharov EV, Gu Z, Kaufman BA and Picard M (2019) Acute psychological stress increases serum circulating cell-free mitochondrial DNA. Psychoneuroendocrinology 106:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK and Chan TA (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN and Biesecker LG (2003) Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet 112:303–309. [DOI] [PubMed] [Google Scholar]
- Turrens JF (2003) Mitochondrial formation of reactive oxygen species. The Journal of physiology 552:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachharajani V and McCall CE (2020) Sirtuins: potential therapeutic targets for regulating acute inflammatory response? Expert Opin Ther Targets 24:489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T and Kraus WL (2007) The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Molecular and cellular biology 27:7475–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B, LaNoue KF, Cheung JY and Scaduto RC Jr. (1989) Regulation of citric acid cycle by calcium. The Journal of biological chemistry 264:13430–13439. [PubMed] [Google Scholar]
- Wanet A, Arnould T, Najimi M and Renard P (2015) Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev 24:1957–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu CD, Li HF, Tian ML, Pan JQ, Shu G, Jiang QY, Yin YL and Zhang L (2020) LSD1 mediates microbial metabolite butyrate-induced thermogenesis in brown and white adipose tissue. Metabolism: clinical and experimental 102:154011. [DOI] [PubMed] [Google Scholar]
- Wang T, Cao Y, Zheng Q, Tu J, Zhou W, He J, Zhong J, Chen Y, Wang J, Cai R, Zuo Y, Wei B, Fan Q, Yang J, Wu Y, Yi J, Li D, Liu M, Wang C, Zhou A, Li Y, Wu X, Yang W, Chin YE, Chen G and Cheng J (2019) SENP1-Sirt3 Signaling Controls Mitochondrial Protein Acetylation and Metabolism. Molecular cell 75:823–834 e825. [DOI] [PubMed] [Google Scholar]
- Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA and Thompson CB (2012) Identification of additional IDH mutations associated with oncometabolite R(−)-2-hydroxyglutarate production. Oncogene 31:2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martinez-Reyes I, Gao P, Helmin KA, Abdala-Valencia H, Sena LA, Schumacker PT, Turka LA and Chandel NS (2019) Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565:495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR and Thompson CB (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP (2017) Mitochondrial dysfunction as a trigger of innate immune responses and inflammation. Toxicology 391:54–63. [DOI] [PubMed] [Google Scholar]
- West AP and Shadel GS (2017) Mitochondrial DNA in innate immune responses and inflammatory pathology. Nature reviews Immunology 17:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP and Zuckerbraun BS (2013) Mitochondrial signaling: forwards, backwards, and in between. Oxidative medicine and cellular longevity 2013:351613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett-Brozick JE, Savul SA, Richey LE and Baysal BE (2001) Germ line insertion of mtDNA at the breakpoint junction of a reciprocal constitutional translocation. Hum Genet 109:216–223. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Tompkins DM, Gemmell NJ and Dowling DK (2016) Mitonuclear interactions, mtDNA-mediated thermal plasticity, and implications for the Trojan Female Technique for pest control. Scientific reports 6:30016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R and Williams RS (2002) Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296:349–352. [DOI] [PubMed] [Google Scholar]
- Wu L, Luo N, Zhao HR, Gao Q, Lu J, Pan Y, Shi JP, Tian YY and Zhang YD (2014) Salubrinal protects against rotenone-induced SH-SY5Y cell death via ATF4-parkin pathway. Brain research 1549:52–62. [DOI] [PubMed] [Google Scholar]
- Wu Z, Oeck S, West AP, Mangalhara KC, Sainz AG, Newman LE, Zhang XO, Wu L, Yan Q, Bosenberg M, Liu Y, Sulkowski PL, Tripple V, Kaech SM, Glazer PM and Shadel GS (2019) Mitochondrial DNA Stress Signalling Protects the Nuclear Genome. Nat Metab 1:1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC and Spiegelman BM (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124. [DOI] [PubMed] [Google Scholar]
- Wu Z, Senchuk MM, Dues DJ, Johnson BK, Cooper JF, Lew L, Machiela E, Schaar CE, DeJonge H, Blackwell TK and Van Raamsdonk JM (2018) Mitochondrial unfolded protein response transcription factor ATFS-1 promotes longevity in a long-lived mitochondrial mutant through activation of stress response pathways. BMC Biol 16:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Jackson CW, Khoury N, Escobar I and Perez-Pinzon MA (2018) Brain SIRT1 Mediates Metabolic Homeostasis and Neuroprotection. Front Endocrinol (Lausanne) 9:702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM and Xiong Y (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell 19:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Surman DR, Diggs L, Xi S, Gao S, Gurusamy D, McLoughlin K, Drake J, Feingold P, Brown K, Wangsa D, Wangsa D, Zhang X, Ried T, Davis JL, Hernandez J, Hoang CD, Souza RF, Schrump DS and Taylor Ripley R (2020) Bile acid-induced “Minority MOMP” promotes esophageal carcinogenesis while maintaining apoptotic resistance via Mcl-1. Oncogene 39:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Fu T, Guo Q, Sun W and Gan Z (2019) Mitochondrial quality orchestrates muscle-adipose dialog to alleviate dietary obesity. Pharmacol Res 141:176–180. [DOI] [PubMed] [Google Scholar]
- Yang XJ and Seto E (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310–5318. [DOI] [PubMed] [Google Scholar]
- Yap YW, Chen MJ, Peng ZF, Manikandan J, Ng JM, Llanos RM, La Fontaine S, Beart PM and Cheung NS (2013) Gene expression profiling of rotenone-mediated cortical neuronal death: evidence for inhibition of ubiquitin-proteasome system and autophagy-lysosomal pathway, and dysfunction of mitochondrial and calcium signaling. Neurochem Int 62:653–663. [DOI] [PubMed] [Google Scholar]
- Yazici D and Sezer H (2017) Insulin Resistance, Obesity and Lipotoxicity. Advances in experimental medicine and biology 960:277–304. [DOI] [PubMed] [Google Scholar]
- Yen K, Lee C, Mehta H and Cohen P (2013) The emerging role of the mitochondrial-derived peptide humanin in stress resistance. Journal of molecular endocrinology 50:R11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi HS, Chang JY and Shong M (2018) The mitochondrial unfolded protein response and mitohormesis: a perspective on metabolic diseases. Journal of molecular endocrinology 61:R91–R105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Mao Y, Luo W, Wu W, Xu H, Wang XL and Shen YH (2017) Palmitic acid dysregulates the Hippo-YAP pathway and inhibits angiogenesis by inducing mitochondrial damage and activating the cytosolic DNA sensor cGAS-STING-IRF3 signaling mechanism. The Journal of biological chemistry 292:15002–15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J and Finkel T (2014) Mitohormesis. Cell metabolism 19:757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapala B, Kaczynski L, Kiec-Wilk B, Staszel T, Knapp A, Thoresen GH, Wybranska I and Dembinska-Kiec A (2010) Humanins, the neuroprotective and cytoprotective peptides with antiapoptotic and anti-inflammatory properties. Pharmacol Rep 62:767–777. [DOI] [PubMed] [Google Scholar]
- Zarate SC, Traetta ME, Codagnone MG, Seilicovich A and Reines AG (2019) Humanin, a Mitochondrial-Derived Peptide Released by Astrocytes, Prevents Synapse Loss in Hippocampal Neurons. Front Aging Neurosci 11:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wu X, Chen P, Liu L, Xin N, Tian Y and Dillin A (2018) The Mitochondrial Unfolded Protein Response Is Mediated Cell-Non-autonomously by Retromer-Dependent Wnt Signaling. Cell 174:870–883 e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T and Kraus WL (2010) SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochimica et biophysica acta 1804:1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH, Yan C, Dong Z, Huang S, Zha Y, Yang L, Cui H and Ding HF (2016) KDM4C and ATF4 Cooperate in Transcriptional Control of Amino Acid Metabolism. Cell reports 14:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Liang S and Zhong Z (2019) Emerging Role of Mitochondrial DNA as a Major Driver of Inflammation and Disease Progression. Trends in immunology 40:1120–1133. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Liu J, Park J, Rai P and Zhai RG (2019) Subcellular compartmentalization of NAD(+) and its role in cancer: A sereNADe of metabolic melodies. Pharmacol Ther 200:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzano A, Liesa M and Palacin M (2009) Role of mitochondrial dynamics proteins in the pathophysiology of obesity and type 2 diabetes. The international journal of biochemistry & cell biology 41:1846–1854. [DOI] [PubMed] [Google Scholar]


