Abstract
In the United States, horses are used for a variety of purposes including recreation, exhibition, and racing. As farm, performance, and companion animals, horses are a unique species from a zoonotic disease risk perspective, and the risks of subclinical infections spreading among horses can pose challenges. Using a nanoscale real-time PCR platform, we investigated the prevalence of 14 enteric pathogens, 11 Escherichia coli genes, and 9 respiratory pathogens in fecal samples from 97 apparently healthy horses at a multi-day horse event. In addition, sugar flotation test was performed for fecal parasites. E. coli f17 was commonly detected, prevalent in 59% of horses, followed closely by Streptococcus equi subsp. zooepidemicus (55%). Additional pathogens recognized included betacoronavirus, Campylobacter jejuni, Cryptosporidium sp., E. coli O157, equine adenovirus 1, equine rhinitis B virus, and others. The use of PCR data may overestimate the true prevalence of these pathogens but provides a sensitive overview of common pathogens present in healthy horses. Our results prompt the continued need for practical biosecurity measures at horse shows, both to protect individuals interacting with these horses and to minimize transmission among horses.
Keywords: equine infectious diseases, equine surveillance, horses
Introduction
In the United States, there are >7.2 million privately owned horses, which are used for a variety of purposes including recreation, exhibition, and racing.1 Participation in these activities means horses are often trailered to new locations and brought into close proximity to unfamiliar horses, raising the potential risk for disease transmission.35 The spread of infectious disease agents between horses poses threats to animal welfare, increases the risk of lost training or competition time, and highlights the need for enhanced biosecurity.26 Additionally, each year, it is estimated that ~150 million people attend state and county fairs, where they may come into close contact with horses and other animals.28 The mixing of people and animals in settings with minimal sanitation undeniably raises concerns for zoonotic diseases caused by pathogens such as Cryptosporidium sp., Clostridioides difficile, and Salmonella.39 For example, a 1999 outbreak of Escherichia coli O157:H7 was traced to a county fair and also led to the identification of several patients with Campylobacter jejuni,30 presumably from fair attendance. In another outbreak of E. coli, horse manure and other animal feces were considered the most likely source of the bacteria.25 Interestingly, in a petting zoo setting, 2 of 12 horses tested positive for E. coli O157:H7.15 In a sample of 25 horses used for 4-H, all of the animals were negative for Salmonella, Campylobacter, and enterohemorrhagic E. coli; 2 horses, however, were positive for C. difficile.27 Understanding the prevalence of various infectious disease agents in apparently healthy horses has the potential to protect both horses and people in contact with horses as well as aid in the identification and prediction of disease outbreaks.
Questions have been raised regarding whether subclinical viral infections pose risks for equine health and performance.3 The intent of our study was not to correlate pathogen burden and show performance, but rather to quantify the pathogen shedding via feces in apparently healthy horses. However, all of the horses in our study were subject to travel and other potential stressors, which have the potential to dampen the immune system and allow pathogen shedding.7 Using a nanoscale real-time PCR (rtPCR) platform, we investigated the prevalence of 14 enteric pathogens, 11 E. coli genes, and 9 respiratory pathogens in fecal samples of 97 apparently healthy horses at a multi-day horse show. The enteric pathogens included: betacoronavirus, Campylobacter coli, C. jejuni, Clostridium perfringens, C. difficile, Cryptosporidium parvum, Cryptosporidium sp., Giardia lamblia, Listeria monocytogenes, Listeria sp., equine rotavirus A (VP4), bovine rotavirus (VP6), Salmonella sp., and Salmonella enterica subsp. enterica serovar Typhimurium. Specific E. coli genes or virulence factors screened for included O157, sta, stx1, stx2, 987p, cnf1, eae, f17, f41, and k99. Additionally, the presence of Shigella and enteroinvasive E. coli (EIEC) was evaluated. Respiratory pathogens included: influenza A virus, equine adenovirus 1 (EAdV-1; Equine mastadenovirus A), equine adenovirus 2 (EAdV-2; Equine mastadenovirus B), equine rhinitis A and B viruses (ERBV; Equine rhinitis A virus), equine herpesvirus 1 (EHV-1; Equid alphaherpesvirus 1), equine herpesvirus 4 (EHV-4; Equid alphaherpesvirus 4), Streptococcus equi subsp. equi, and Streptococcus equi subsp. zooepidemicus. Many of these pathogens have been associated with disease in horses, for example E. coli f17 has been suggested as a cause of equine diarrhea11; are potential emerging pathogens, such as betacoronavirus33; or pose potential zoonotic disease risks.37 Additionally, using a sugar flotation test, we screened for endoparasites, including strongyles, Strongyloides westeri, Oxyuris equi, and Parascaris sp.
Materials and methods
Participant recruitment and sample collection
Convenience, noninvasive fecal samples were obtained from 100 horses at a show in August 2018 in the northeastern United States. Permission from owners and/or handlers was obtained orally, and information collected on each horse included age, sex, breed, and number of days at the show. Information regarding most recent deworming, distance traveled, diet, or where individual animals were normally housed was not collected. No information regarding individual farms and/or barns was collected. All horses were considered clinically healthy, as they had met the requirement for exhibition, which included a certificate of veterinary inspection (CVI) if coming from out of state; a rabies vaccine if >105 d old, administered prior to arrival; and a negative equine infectious anemia virus (Coggins) test within the previous year for all horses >6 mo old. Individual fecal samples were collected from the horse’s stall and aliquoted into 2 plastic bags. Samples were stored at −20°C for PCR quantification and at 4°C for parasite detection.
Laboratory methods
A routine Wisconsin double-centrifugation sugar flotation test was performed on each sample, following the standard protocol of the Parasitology Laboratory at the New York State Veterinary Diagnostic Laboratory/Cornell Animal Health Diagnostic Center (AHDC; Ithaca, NY).
For direct molecular analysis, a 400-mg subsample of feces from each horse was homogenized in 800 μL of phosphate-buffered saline (PBS). Homogenates were centrifuged for 5 min at 18,000 × g, and 175 µL of the supernatants were extracted (MagMAX total nucleic acid isolation kit, Applied Biosystems; Kingfisher Flex, Thermo Fisher Scientific). The manufacturer’s instructions were altered with an added mechanical lysis step of 2 × 2.5 min with zirconia beads at 2,100 oscillations per min in a Mini-Beadbeater-96 (BioSpec Products), with a 5-min rest between. Bacteriophage MS2 was added to the lysis buffer as an internal control to monitor for inhibition and extraction efficiency.9,41 Two negative extraction controls, consisting of PBS, were included on each extraction plate. Nanoliter-scale rtPCR was performed (QuantStudio 12K Flex OpenArray platform; Thermo Fisher Scientific) using a previously described customized respiratory panel13 and a modified version of another panel5 (Suppl. Table 1). Positive amplification control pools consisting of in vitro transcribed RNA for betacoronavirus, ERVB, equine rotavirus A, and influenza A virus; genomic DNA from purified C. parvum oocysts (Waterborne); genomic DNA from S. enterica serovar Cerro, Campylobacter sp., and Shigella flexneri; and ~450-bp long synthetic gBlock DNA fragments for all other targets (Integrated DNA Technologies) were run on each plate along with a negative amplification control (purified water). Each nanoscale PCR reaction was performed in duplicate (enteric panel) or triplicate (respiratory panel), and samples were considered positive if at least 1 (enteric) or 2 (respiratory) technical replicates produced a properly shaped amplification curve.
Salmonella enrichment PCR was performed on all samples as described previously.14 E. coli culture using gram-negative broth enrichment and eosin methylene blue agar was also performed on the 2 cryopreserved fecal samples that had the highest abundance of the gene encoding the E. coli O157 antigen. Whole-genome sequencing (WGS) was performed from a single colony and also attempted from enrichment broth from each sample. DNA was extracted (MagMAX CORE kit; Thermo Fisher Scientific). Barcoded libraries were prepared (Nextera XT library preparation kit; Illumina). Reads from WGS of colonies were assembled using SKESA.34 Serovar prediction was performed both on assembled and unassembled reads using ECtyper (https://github.com/phac-nml/ecoli_serotyping) and SRST219 with the EcOH18 database, respectively. Virulence factors were searched using the NCBI Pathogen Detection isolates browser (https://www.ncbi.nlm.nih.gov/pathogens/). The raw data from each of the sequenced colonies was uploaded to NCBI under Biosamples SAMN13429602 and SAMN13429521. SRST2 was also used to identify serotype markers in unassembled reads from the enrichment broth sequencing.
Data analysis
All analyses were performed in R Studio (v.3.6.0; https://rstudio.com/). Three horses, of 100 sampled horses, were eliminated from analysis because of incomplete data. Given the large number of breeds that were present in the study (15), the decision was made to classify horses as draft breed or light breed. The draft breeds included Percherons, Belgians, Shires, and Clydesdales. Light breeds included Miniature Horses, Saddlebreds, Morgans, Quarter Horses, a Thoroughbred, a Shetland Pony, a Paso Fino, a Paint, a Friesian, and 2 mixed breeds. The number of days at the show were grouped into a dichotomous variable, which included ≤3 d and >3 d. The horse ages were also classified as a dichotomous variable, consisting of horses ≤10 y old and those >10 y old. Additionally, all parasites were considered either as positive or below the limits of detection. Likewise, all other pathogens were considered either as positive or not detected. A point prevalence was established for each pathogen, and a 95% confidence interval was determined using the prop.test function in R Studio. Parasite or pathogen presence was additionally stratified by breed classification, sex, the dichotomous days at event variable, and the dichotomous age variable to identify nonrandom associations, with p values calculated using a 2-sided Fisher exact test. A p ≤ 0.05 was considered significant.
Results
Demographics
We included 97 horses in our study. The mean age was 10.2 y (SD = 7.2). Most horses were <10 y old (63%); 34% were ≥10 y old. Geldings were more common (65%) than mares (35%). The mean days at the event was 3.8 (SD = 1.4), with 43% of the horses present at the event ≤3 d and remainder present >3 d. The most common breeds included Percherons (20), Belgians (19), Miniature Horses (17), and American Saddlebreds (12); 49% of the horses were draft breeds and 51% were light breeds. The breed classification and dichotomous age variable were highly associated (p = 0.00007). In the draft breeds, 87% were classified in the younger age category, compared to the light breeds in which only 45% were in the younger age category. The average age among the draft breeds was 6.5 y; the average age of the light breeds was 13.8 y. No other demographic variables were considered to be significantly associated (p > 0.05).
Pathogen burden
Most of the horses in our study were positive for at least one pathogen (Fig. 1). On average, each horse had 3.26 pathogens (SD = 1.73), with the median number of pathogens being 3. A small cluster of horses was positive for 7 pathogens.
Figure 1.
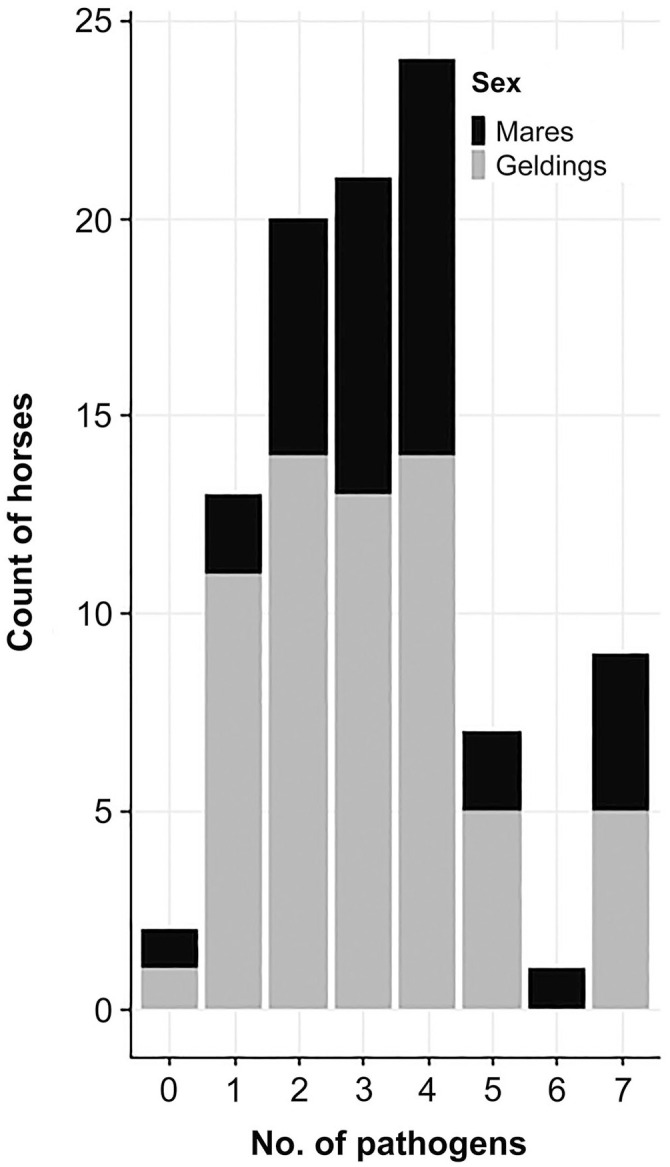
Total pathogen burden in horses participating in a multi-day event (n = 97). The mean number of pathogens detected was 3.26.
Four parasites were detected via sugar flotation: Eimeria leuckarti, Anoplocephala sp., strongyles, and Parascaris sp. Cryptosporidium sp., Strongyloides westeri, Oxyuris equi, and Dictyocaulus sp. were not detected in any of the samples. Strongyles were the most common parasite identified, detected in 46 of 97 horses (95% CI: 37.3–57.8%). Among the samples in which strongyles were detected, the average egg count was 912 (SD = 1,110) eggs/g (epg) and the median fecal egg count was 318 epg. Strongyles were more common among those in the younger age category (p = 0.011). Strongyles were also detected more frequently in mares (odds ratio [OR] = 2.46), but this association was not significant (p = 0.055). Adjusting for breed classification, age was weakly associated with strongyles (Mantel–Haenszel OR: 2.79; 95% CI: 1.06–7.37). E. leuckarti was identified only in 4 geldings (95% CI: 1.3–10.8%), and an association between sex and E. leuckarti was apparent (p = 0.013). Parascaris sp., identified in 10 horses (95% CI: 5.3–18.6%) was associated with draft breeds (p = 0.0077), with 9 of the positive animals classified as draft breeds. Adjusting for age, however, this association was not statistically significant (p = 0.066). Anoplocephala sp. was identified in 5 horses (95% CI: 1.9–12.2%) and did show any apparent associations with factors assessed.
No samples were positive for Salmonella either on direct feces or from enrichment. A total of 7 enteric pathogens were detected: betacoronavirus, C. coli, C. jejuni, C. perfringens, Cryptosporidium sp., G. lamblia, and equine rotavirus A. Pathogens that were tested for, but not amplified, included C. difficile, C. parvum, Listeria spp., bovine rotavirus, and Salmonella sp. Additionally, 5 E. coli virulence genes were identified via direct fecal PCR (Table 1). Specific pathogens or genera included sta, cnf1, eae, and f17, and the O-antigen gene encoding E. coli serotype O157. E. coli factors that were tested, but not amplified, included stx1, stx2, 987p, f41, and k99. Shigella/EIEC was not detected. E. coli f17 was commonly detected; 59% of horses tested positive. The O157 serotype was also detected frequently, with matches in 32% of the horses. Days at event did not appear to impact shedding of any of the enteric pathogens (all p > 0.18). E. coli cnf1 was associated with draft breeds (p = 0.0075; age-adjusted p = 0.023). The exogenous internal control was detected in all samples, with a SD of 1.5 detection cycles.
Table 1.
Prevalence of enteric pathogens detected in 97 equine fecal samples by nanoscale real-time PCR.
| Enteric PCR target | Count | 95% CI |
|---|---|---|
| Betacoronavirus | 1 | 0.05–6.4 |
| Campylobacter coli | 1 | 0.05–6.4 |
| Campylobacter jejuni | 16* | 10.1–25.7 |
| Clostridium perfringens | 2 | 0.4–8.1 |
| Cryptosporidium sp. | 1 | 0.05–6.4 |
| Giardia lamblia | 17 | 10.8–26.9 |
| Equine rotavirus (VP4) | 1 | 0.05–6.4 |
| E. coli O157 | 31† | 23.1–42.3 |
| E. coli Sta | 1 | 0.05–6.4 |
| E. coli cnf1 | 13 | 7.6–22.2 |
| E. coli eae | 18 | 11.7–28.0 |
| E. coli f17 | 57‡ | 48.3–68.5 |
All samples were negative for Clostridioides difficile, Cryptosporidium parvum, Listeria, Salmonella, Shigella, and E. coli stx1, stx2, 987p, f41, k99, and LT.
7 of 16 were positive in 1 of 2 replicates.
1 of 31 was positive in 1 of 2 replicates.
1 of 57 was positive in 1 of 2 replicates.
E. coli colonies from the 2 horses with the highest O157 detection from feces (horses 23 and 55 with relative cycle threshold values of 13.9 and 12.2) did not match the O157 serotype by WGS. Instead, they were predicted to be O115:H10 and O93:H19, respectively. Similarly, a sequence corresponding to the O157 serotype was not detected in the aerobic enrichment broth incubated with feces from each of these horses. Both isolates encoded lpfA and espX1, and one also had a partial match to fdeC.29 No other E. coli strains in the NCBI Pathogen Detection database clustered phylogenetically with these 2 isolated strains.
Three pathogens with respiratory tropism were detected in feces directly: EAdV-1 in 1 horse (95% CI: 0.05–6.4%), ERBV in 38 horses (30.5–50.7%), and S. equi subsp. zooepidemicus in 53 horses (44.2–64.7%). Additional pathogens that were tested for, but not detected, included influenza A, EAdV-2, equine rhinitis A, EHV-1, EHV-4, and S. equi subsp. equi. No relationship between age, days at event, or sex was identified. S. equi subsp. zooepidemicus was associated with light-breed horses (p = 0.042; age-adjusted p = 0.023).
Discussion
The novel application of nanoscale rtPCR for molecular enteric pathogen detection in feces allows for efficient assessment of a large group of pathogens and to assess the prevalence of these pathogens in apparently healthy horses. The top 3 breeds in our study were Percherons (20 of 97), Belgians (19 of 97), and Miniature Horses (17 of 97), whereas the most common horse breeds in the United States are Quarter Horses, Paints, and Thoroughbreds.21 Thus, this was a unique sample to understand potential pathogen differences in less common breeds of horses, including breeds that may be used for special purposes. Draft breeds appeared to be associated with higher levels of the gene encoding E. coli cytotoxic necrotizing factor 1 (age-adjusted OR: 6.92; 95% CI: 1.2–38.9) and potentially lower levels of S. equi subsp. zooepidemicus than the light breeds in our study (age-adjusted OR: 0.15; 95% CI: 0.03–0.81). The only horse that had betacoronavirus detected was an 8-y-old draft mare, present at the event for 3 d; this was also the only animal positive for Cryptosporidium sp. and was additionally positive for strongyles, ERBV, C. jejuni, and S. equi subsp. zooepidemicus. Whether this high pathogen burden was the result of increased exposure or an underlying immune deficiency is unknown.
Sex did not appear to be highly associated with fecal pathogen shedding in our study. Geldings may be more likely to have E. leuckarti (OR: 5.2). Interestingly, the E. leuckarti prevalence observed in our study is considerably lower than previously reported prevalences of 27.5–59.1% in studies conducted in Kentucky and Montana.10 The prevalence of E. leuckarti of 4.12% that we observed follows most closely the studies in Albania, the Czech Republic, Greece, and Turkey.10 These differences may be the result of a variety of factors, including age.
Although the Wisconsin double-centrifugation sugar fecal flotation test is an excellent method for identifying most parasites that void their life stages in host feces, limitations exist for detecting certain parasites including Giardia. Our use of nanoscale rtPCR is apparently more sensitive than classical parasitology techniques, and it amplified genetic material from Cryptosporidium sp. and G. lamblia, both of which are of zoonotic significance.
S. equi subsp. zooepidemicus has often been considered a commensal respiratory pathogen in horses, although it can be present in high levels in respiratory fluids.2 A specific strain was implicated in one outbreak of upper respiratory disease.24 Additionally, concerns have grown about the potential spread of this subspecies from horses to people.31 The high prevalence of S. equi subsp. zooepidemicus may prompt a commitment to minimizing group housing across species of animals. S. equi subsp. zooepidemicus has previously been reported to cause disease in an alpaca,16 cats,4 dogs,32 and dairy goats.22
Of 11 total E. coli genes that we analyzed in direct feces, 5 were detected. E. coli f17 was the most common, with a 59% prevalence. Enteropathogenic E. coli in horses was initially characterized as associated with F41-type pili.38 F17 and other fimbriae are associated with pathogenic infections in ruminants. In a small study, 3 of 10 horses with diarrhea and 0 of 14 healthy horses were f17 positive.11 Importantly, that study characterized isolated colonies as opposed to direct PCR on feces.11 A wide diversity of E. coli strains is expected to exist in the healthy gut, as opposed to clinical cases that may have a predominant strain causing disease. Even multiple colonies chosen from a plate may not be representative of the overall diversity of this highly variable species in one host. At the time of sampling, no horses were experiencing diarrhea. If involved in pathogenesis of E. coli diarrhea in horses, our findings suggest that f17 may not act alone to elicit diarrhea and may require comorbidities.
Other E. coli genes detected included O157, cnf1, eae, and Sta. Genes that were not detected included stx1, stx2, 987p, f41, k99, and Shigella/EIEC. In our study, 32% of the horses were positive for the gene encoding E. coli O157; given that H antigen detection was not performed, it is unclear what proportion of these samples (or proportion of strains within each animal) were positive for E. coli O157:H7. Colonies selected for further characterization serotyped differently, which was not unexpected. Sampling of horses in Ohio for E. coli O157:H7 demonstrated a low prevalence of the pathogen (~0.4%).23 The clinical significance of cnf1 remains unknown; this virulence factor has been noted in both a healthy horse and a horse experiencing diarrhea,11 but also in an equine patient with bronchopneumonia.8 In humans, E. coli cnf1 is often considered uropathogenic.36 The eae gene encoding the adhesin intimin has been reported in up to half of isolates from diarrheic foals, but also in some healthy horses.6,17,20 The presence of E. coli O157 harboring eae but lacking the Stx-encoding bacteriophage is a potential zoonotic concern, especially if clinical testing is focused solely on detection of the stx genes.12
Most of the draft horses in our study were ≤10 y old, compared to the lighter breeds, which were more evenly aged. This may in part be the result of the competition purposes for which these different breeds are used. Draft breeds in our study were most likely to be participating in driving and pulling events and thus handled by adults. Light breeds participated in a range of events, including events in which youth participants may be competing and for which it would be desired to have an older, calmer horse. Understanding the individuals who may be interacting with specific breeds or certain aged horses may be important when considering the risks of zoonotic disease potential. This may be even more important to consider for those who are immune compromised. The basics of biosecurity and personal hygiene may be the best method to protect other horses and individual people, regardless of the scenario.
We did not detect EHV-1, EHV-4, or influenza A virus. Although these are common equine pathogens, their presence in fecal samples is not expected. The respiratory picornavirus ERBV, however, is acid-stable and can be found in feces.40 Additionally, the use of voided fecal matter is a convenient opportunity for sampling; however, the use of a nasal swab or wash, utilizing nanoscale rtPCR, may be better suited for diagnostic evaluation. We did not detect Salmonella, indicating that subclinical shedding may be infrequent. Nonetheless, in the event of concerns about any of these pathogens, appropriate testing would be warranted. Although our study sample is meant to be representative of healthy horses, the prevalence of these pathogens may vary between geographic locations. The presence of other animals, water sources, individual immune status, and travel history could be other factors contributing to prevalence differences.
Supplemental Material
Supplemental material, Supplemental_material for Infectious disease surveillance of apparently healthy horses at a multi-day show using a novel nanoscale real-time PCR panel by Alison E. Stout, Hayley G. Hofmar-Glennon, Nicole M. André, Laura B. Goodman, Renee R. Anderson, Patrick K. Mitchell, Belinda S. Thompson, Manigandan Lejeune, Gary R. Whittaker and Erin L. Goodrich in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank the following personnel for technical laboratory support: John Beeby, Brittany Cronk, Rebecca Franklin-Guild, Ava Jarvis, Derek Rothenheber, James Ryan, and Rebecca Tallmadge. We are also grateful to Marcia Slater for assistance in design of the enteric nanoscale PCR panel and Colleen Mock for logistic support in production of the panel. We also thank Dr. Ana Cristina Barsallo Cochez and Hannah Pambianchi for help in sample collection.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Development of the nanoscale enteric PCR panel was funded (FOA PA-13-244) and performed in collaboration with the Food and Drug Administration’s Veterinary Laboratory Investigation and Response Network (FDA Vet-LIRN) under grant 1U18FD005144. Sequencing capacity support for this project was funded (FOA PAR-18-604) by FDA Vet-LIRN under grant 1U18FD006716. A. E. Stout is supported by the NIH Comparative Medicine Training Program T32OD011000.
ORCID iDs: Alison E. Stout  https://orcid.org/0000-0001-7902-2509
https://orcid.org/0000-0001-7902-2509
Nicole M. André  https://orcid.org/0000-0002-3703-5026
https://orcid.org/0000-0002-3703-5026
Laura B. Goodman  https://orcid.org/0000-0002-8327-3092
https://orcid.org/0000-0002-8327-3092
Patrick K. Mitchell  https://orcid.org/0000-0001-6848-0846
https://orcid.org/0000-0001-6848-0846
Supplementary material: Supplementary material for this article is available online.
Contributor Information
Alison E. Stout, Departments of Microbiology and Immunology, Cornell University, Ithaca, NY.
Hayley G. Hofmar-Glennon, College of Veterinary Medicine, and Master of Public Health Program, Cornell University, Ithaca, NY.
Nicole M. André, Departments of Microbiology and Immunology, Cornell University, Ithaca, NY
Laura B. Goodman, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY
Renee R. Anderson, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY
Patrick K. Mitchell, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY
Belinda S. Thompson, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY
Manigandan Lejeune, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY.
Gary R. Whittaker, Departments of Microbiology and Immunology, Cornell University, Ithaca, NY; College of Veterinary Medicine, and Master of Public Health Program, Cornell University, Ithaca, NY.
Erin L. Goodrich, Population Medicine and Diagnostic Sciences, Cornell University, Ithaca, NY
References
- 1. American Horse Council. Economic impact of the United States horse industry [Internet]. Am Horse Counc 2017. [cited 2019 Nov 5]. https://www.horsecouncil.org/resources/economics/
- 2. Anzai T, et al. Comparison of the phenotypes of Streptococcus zooepidemicus isolated from tonsils of healthy horses and specimens obtained from foals and donkeys with pneumonia. Am J Vet Res 2000;61:162–166. [DOI] [PubMed] [Google Scholar]
- 3. Back H, et al. A longitudinal study of poor performance and subclinical respiratory viral activity in Standardbred trotters. Vet Rec Open 2015;2:e000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blum S, et al. Outbreak of Streptococcus equi subsp. zooepidemicus infections in cats. Vet Microbiol 2010;144:236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brooks YM, et al. Fecal indicator bacteria, fecal source tracking markers, and pathogens detected in two Hudson River tributaries. Water Res 2020;171:115342. [DOI] [PubMed] [Google Scholar]
- 6. Chandran A, Mazumder A. Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl Environ Microbiol 2013;79:7371–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coumbe K. Equine travel: minimising the risk of illness and disease spread [Internet]. Vet Pract 2017. [cited 2019 Nov 5]. https://veterinary-practice.com/article/equine-travel-minimising-the-risk-of-illness-and-disease-spread
- 8. DebRoy C, et al. Bronchopneumonia associated with extraintestinal pathogenic Escherichia coli in a horse. J Vet Diagn Invest 2008;20:661–664. [DOI] [PubMed] [Google Scholar]
- 9. Dreier J, et al. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol 2005;43:4551–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubey JP, Bauer C. A review of Eimeria infections in horses and other equids. Vet Parasitol 2018;256:58–70. [DOI] [PubMed] [Google Scholar]
- 11. Duijkeren E van, et al. Characterization of Escherichia coli isolated from adult horses with and without enteritis. Vet Q 2000;22:162–166. [DOI] [PubMed] [Google Scholar]
- 12. Ferdous M, et al. Is Shiga toxin-negative Escherichia coli O157:H7 enteropathogenic or enterohemorrhagic Escherichia coli? Comprehensive molecular analysis using whole-genome sequencing. J Clin Microbiol 2015;53:3530–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goodman LB, et al. High-throughput detection of respiratory pathogens in animal specimens by nanoscale PCR. J Vis Exp 2016:e54781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodman LB, et al. Detection of Salmonella spp. in veterinary samples by combining selective enrichment and real-time PCR. J Vet Diagn Invest 2017;29:844–851. [DOI] [PubMed] [Google Scholar]
- 15. Hamzah A, et al. Isolation of Escherichia coli 0157:H7 strain from fecal samples of zoo animal. ScientificWorldJournal 2013:843968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hewson J, Cebra CK. Peritonitis in a llama caused by Streptococcus equi subsp. zooepidemicus. Can Vet J 2001;42:465–467. [PMC free article] [PubMed] [Google Scholar]
- 17. Holland RE, et al. Characterization of Escherichia coli isolated from foals. Vet Microbiol 1996;48:243–255. [DOI] [PubMed] [Google Scholar]
- 18. Ingle DJ, et al. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb Genomics 2016;2:e000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inouye M, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014;6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishii S, et al. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl Environ Microbiol 2007;73:5703–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kilby ER. The demographics of the U.S. equine population. In: Salem DJ, Rowan AN, eds., The State of the Animals IV. Humane Society Press, 2007:175–205. [Google Scholar]
- 22. Las Heras A, et al. Unusual outbreak of clinical mastitis in dairy sheep caused by Streptococcus equi subsp. zooepidemicus. J Clin Microbiol 2002;40:1106–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lengacher B, et al. Low prevalence of Escherichia coli O157:H7 in horses in Ohio, USA. J Food Protect 2010;73:2089–2092. [DOI] [PubMed] [Google Scholar]
- 24. Lindahl SB, et al. Outbreak of upper respiratory disease in horses caused by Streptococcus equi subsp. zooepidemicus ST-24. Vet Microbiol 2013;166:281–285. [DOI] [PubMed] [Google Scholar]
- 25. Luna S, et al. Outbreak of E. coli O157:H7 infections associated with exposure to animal manure in a rural community—Arizona and Utah, June–July 2017. MMWR Morb Mortal Wkly Rep 2018;67:659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lunn DP, Traub-Dargatz J. Managing infectious disease outbreaks at events and farms; challenges and the resources for success. Proc Am Assoc Eq Pract 2007;53:1–12. [Google Scholar]
- 27. McNamara SE, et al. Carriage of Clostridium difficile and other enteric pathogens among a 4-H avocational cohort. Zoonoses Public Health 2011;58:192–199. [DOI] [PubMed] [Google Scholar]
- 28. National Assembly of State Animal Health Officials, National Association of State Public Health Veterinarians. Measures to minimize influenza transmission at swine exhibitions, 2014. [cited 2019 Nov 5]. https://www.usaha.org/upload/Disease%20Info/SwineExhibitions2014.pdf
- 29. Nesta B, et al. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 2012;3:e00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Novello A. Outbreak of Escherichia coli O157:H7 and Campylobacter among attendees of the Washington County Fair-New York, 1999. MMWR Morb Mortal Wkly Rep 1999;48:803–805. [PubMed] [Google Scholar]
- 31. Pelkonen S, et al. Transmission of Streptococcus equi subspecies zooepidemicus infection from horses to humans. Emerg Infect Dis 2013;19:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Priestnall S, Erles K. Streptococcus zooepidemicus: an emerging canine pathogen. Vet J Lond Engl 1997. 2011;188:142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pusterla N, et al. Equine coronavirus: an emerging enteric virus of adult horses. Equine Vet Educ 2016;28:216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Souvorov A, et al. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol 2018;19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spence KL, et al. Descriptive analysis of horse movement networks during the 2015 equestrian season in Ontario, Canada. PLoS One 2019;14:e0219771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spurbeck RR, Mobley HLT. Chapter 9. Uropathogenic Escherichia coli. In: Donnenberg MS, ed. Escherichia coli. 2nd ed. London: Academic Press, 2013:275–304. [Google Scholar]
- 37. U.S. Food and Drug Administration. Bad Bug Book: Handbook of Foodborne Pathogenic Microorganisms and Natural Toxins. 2nd ed. 2012. https://www.fda.gov/media/83271/download
- 38. Ward AC, et al. Isolation of piliated Escherichia coli from diarrheic foals. Vet Microbiol 1986;12:221–228. [DOI] [PubMed] [Google Scholar]
- 39. Weese JS. A review of equine zoonotic diseases: risks in veterinary medicine. Proc Am Assoc Eq Pract 2002;48:362–369. [Google Scholar]
- 40. Woo PCY, et al. Equine rhinitis B viruses in horse fecal samples from the Middle East. Virol J 2016;13:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yan L, et al. Inhibition monitoring in veterinary molecular testing. J Vet Diagn Invest 2020;32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_material for Infectious disease surveillance of apparently healthy horses at a multi-day show using a novel nanoscale real-time PCR panel by Alison E. Stout, Hayley G. Hofmar-Glennon, Nicole M. André, Laura B. Goodman, Renee R. Anderson, Patrick K. Mitchell, Belinda S. Thompson, Manigandan Lejeune, Gary R. Whittaker and Erin L. Goodrich in Journal of Veterinary Diagnostic Investigation


