Abstract
Background
Biological invasions rank among the most significant threats to biodiversity and ecosystems. Correlative ecological niche modeling is among the most frequently used tools with which to estimate potential distributions of invasive species. However, when areas accessible to the species across its native distribution do not represent the full spectrum of environmental conditions that the species can tolerate, correlative studies often underestimate fundamental niches.
Methods
Here, we explore the utility of supraspecific modeling units to improve the predictive ability of models focused on biological invasions. Taking into account phylogenetic relationships in correlative ecological niche models, we studied the invasion patterns of three species (Aedes aegypti, Pterois volitans and Oreochromis mossambicus).
Results
Use of supraspecific modeling units improved the predictive ability of correlative niche models in anticipating potential distributions of three invasive species. We demonstrated that integrating data on closely related species allowed a more complete characterization of fundamental niches. This approach could be used to model species with invasive potential but that have not yet invaded new regions.
Keywords: Biological invasions, Invasive species, Supraspecific modeling units, Phylogenetic conservatism of ecological niches, Ecological niche models, Potential distribution
Introduction
Biological invasions are considered the second most serious cause of species extinctions (Richardson, Pyšek & Carlton, 2011). In recent decades, invasions have become more frequent because of globalization, since humans have re-located (accidentally or intentionally) many species far outside their native geographic ranges (Tatem & Hay, 2007; Wilson et al., 2009). In general, invasive species have direct negative impacts on native species and ecosystems via predation, competition, propagation of diseases, and changes in composition of trophic webs (Manchester & Bullock, 2000). Eradication of invasive species would be highly desirable before they become established, and the scientific community agrees broadly that prevention of invasions is the most effective and least expensive way to avoid negative impacts (Miller et al., 2005; Thuiller et al., 2005).
Mapping geographic regions presenting suitable environmental conditions for particular invasive species represents an important but challenging step in preventing establishment (Bomford, 2008). This goal implies, typically, estimating the set of abiotic conditions that allow the species to achieve positive population growth (i.e., its fundamental niche—NF) (Peterson & Soberón, 2012). Mechanistic niche modeling represents means by which to approximate and map a species’ NF, since experiments under controlled conditions are carried out to estimate its physiological tolerance (Kearney & Porter, 2004, 2009). However, these models require a lot of information obtained through long experimentation processes that can be quite costly (Gallien et al., 2010). Also, mechanistic models are usually generated in terms of one or two environmental variables, and may not capture all biologically relevant factors for the species (Aragón, Baselga & Lobo, 2010); interactions among variables are generally neglected; and application is limited to well-studied organisms (Larson et al., 2014).
Correlative ecological niche modeling (ENM) requires less information than mechanistic modeling, because it derives inferences from statistical associations between presence data and environmental dimensions. From these associations, ENMs can identify environmental conditions suitable for the species. By projecting these conditions into geographic space, one can generate hypotheses of species’ potential distributions (Peterson et al., 2011). Correlative ENM has been used widely to study biological invasions (Peterson, 2003; Thuiller et al., 2005), but its predictive capacity has been hindered by two factors. First, it is complicated to differentiate correlation from true causality (Dormann, 2007), and second, the environmental diversity across the area that has been accessible historically for the species (M; Soberón & Peterson, 2005) may not be sufficient to characterize fundamental niches (i.e., the set of abiotic conditions that allow a given species to survive and reproduce in the absence of biotic interactions) fully, which reduces the NF to what is termed the existing niche: the niche that one is able to observe and characterize. Hence, if M does not cover the whole environmental variation that the species tolerates, ENMs end up sub-characterizing NF.
This truncation of the fundamental niche imposes both conceptual and methodological challenges to anticipate the species’ success of establishment when facing novel environmental conditions, such as under climatic changes and biological invasions. Therefore, when a species is environmentally restricted due to geographical constraints, for example an insular species, it is difficult to determine the portion of its niche that is represented in the island and how much is missing. Some methodological proposals to improve the predictive capacity of correlative niche models include: (1) reducing numbers of predictors to avoid environmental combinations associated with presence data being unique and statistical regularities not observable (Peterson & Nakazawa, 2008), (2) controlling spatial autocorrelation to decrease overfitting to calibration data (Boria et al., 2014), (3) balancing complexity and generalization in niche models by varying parameter settings in algorithms (Warren & Seifert, 2011; Merow et al., 2014; Radosavljevic & Anderson, 2014), (4) analyzing transfer procedures in relation to strict extrapolation (Owens et al., 2013) and (5) using the most complete set of occurrences (presence records from both native and invaded areas) to achieve better characterization of the species’ NF (Jiménez-Valverde et al., 2011). This latter idea improves transfers (Escobar et al., 2016), but its application is limited to species with populations already established outside their native ranges. A possible alternative to modeling species with M areas that are not sufficiently representative environmentally could be based on the idea of phylogenetic conservatism of ecological niches (PNC, Peterson, Soberón & Sánchez-Cordero, 1999). Although niche conservatism obviously breaks down over evolutionary time, considerable evidence indicates the frequent absence of ecological niche differentiation at time scales comparable to those of invasions and speciation events (Peterson, 2011). That is, niche traits are often conserved among closely related species (Pavoine & Bonsall, 2011). If so, one may assume that sister species have identical or very similar NF’s, despite being distributed in geographic regions with different environmental characteristics. These differences in accessible environments mean that together they might be more representative of the NF of the species in a lineage. This strategy of modeling niches by grouping presence records above the species level, known as “lumping” (Smith et al., 2018), under certain circumstances, may be a more accurate way to characterize the niche of a lineage.
The objective of this study was to evaluate whether using supraspecific modeling units (including both the occurrences of a focal invasive species and those of sister species in their respective native ranges) can improve model capacity to predict the geographic potential of invasions. We chose three invasive species with multiple populations established outside of their native geographic ranges, which offer the advantage of abundant evaluation data. The similarity of the NF’s between closely related species may vary with respect to their genetic similarity, so we evaluated model performance as we created modeling units that included records of successively less closely-related species. We observed that supraspecific units improved the prediction of geographic potential of biological invasions in the three species, which suggests that niches are highly conserved and complementary among the species that composed the unit with better predictive ability.
Materials and Methods
We used QGIS 3.0.2 (QGIS Development Team, 2018) and R 3.4.4 (R Core Team, 2018) to carry out all the geographic information system and statistical procedures described in the following sections.
Presence data
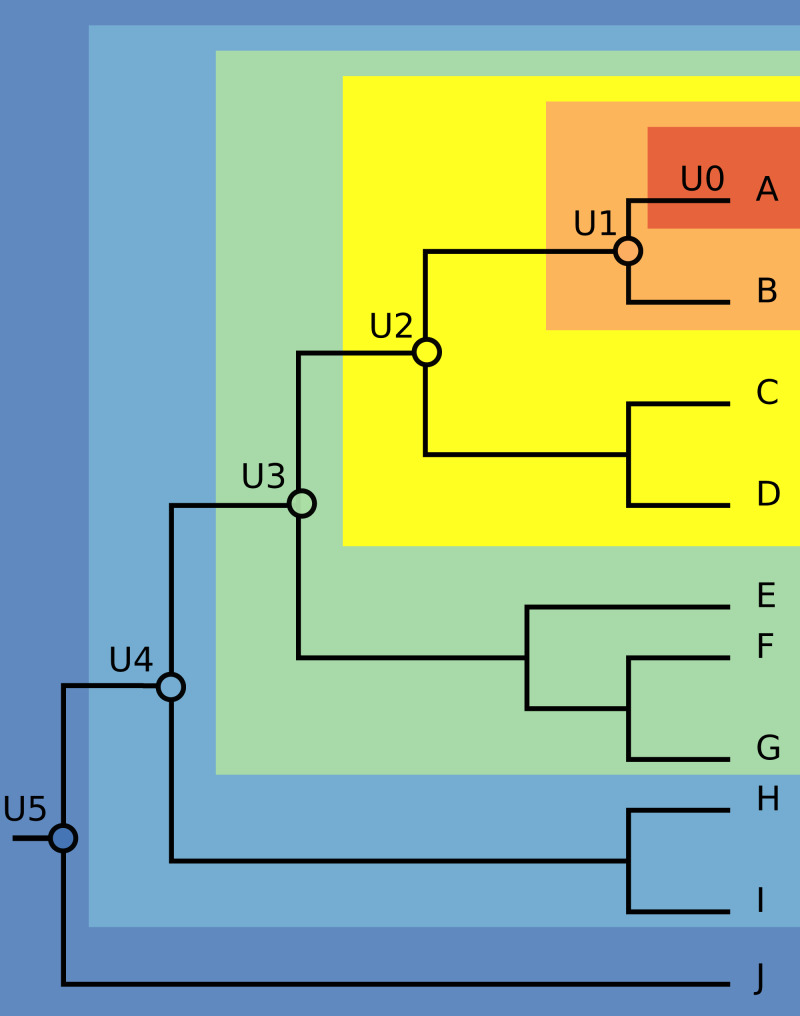
We chose three species with significant invasive potential that inhabit different environments: the mosquito Aedes aegypti (a terrestrial species), the Mozambique tilapia Oreochromis mossambicus (a freshwater fish), and the red lionfish Pterois volitans (a marine fish). For each species, we obtained a recently published phylogeny that allowed identification of supraspecific units corresponding to successively deeper nodes on the phylogenetic trees. The phylogenies that we used were those reported by Chu et al. (2018), He et al. (2011) and Kochzius et al. (2003), for the three species, respectively. Chu et al. (2018) used a portion of mitochondrial cytochrome oxidase to infer relationships of 34 taxa of mosquitoes, He et al. (2011) used mitogenomic data to explore relationships among 21 tilapia species, and Kochzius et al. (2003) estimated relationships of seven species of the Pterois genus based on mitochondrial DNA sequences of 16s rDNA and cytochrome b. We built the first unit (called U0) only with data from the native distribution of each focal species. The second supraspecific unit (U1) was composed of U0 plus the data from native distribution of the sister species. We constructed and named subsequent supraspecific units similarly, marching from the focal species down the phylogenetic tree toward its root. We also built a unit (UT) including all occurrences of the focal species, native and invasive, as a more full representation of its fundamental niche (Fig. 1).
Figure 1. Illustration of the modeling units’ construction.
The first unit (U0), depicted in the dark orange area, contains occurrences from native distribution of each focal species. The second unit (U1), depicted in the light orange area, is U0 plus the native occurrences of the sister species. The next units were built by adding the native occurrences of the species that compose the following nodes, marching from the focal species down the phylogenetic tree towards its root.
We obtained species occurrence data from the Global Biodiversity Information Facility (GBIF, 2019). We eliminated records without coordinates, duplicated records, and those with evident errors (e.g., records located at sea for terrestrial species or vice versa). In addition, we spatially filtered presence data by eliminating clusters of records in order to reduce sampling bias and model overfitting (Boria et al., 2014) with the R package spThin (Aiello-Lammens et al., 2015). Number of occurrence records used for each modeling unit are shown in Table S1.
We delimited the native historical accessible areas (M area, sensu Soberón & Peterson, 2005) for each species by selecting the ecoregions that included at least one of its presence records across their native geographic range. For continental species, M was delimited using terrestrial ecoregions (Olson et al., 2001), and for marine species, it was based on the bio-regionalization of coastal areas (Spalding et al., 2007).
Environmental data
We downloaded 23 global terrestrial environmental surfaces from the CliMond database (Kriticos et al., 2012), at a spatial resolution of 10′ (~17 km). These surfaces summarize annual trends, seasonality, and limiting environmental factors derived from monthly values of temperature, precipitation, radiation, and moisture for 1950–2000 (Table S2). To represent marine conditions, we downloaded 12 environmental surfaces from the Bio-Oracle database 2.0 (Assis et al., 2018). These surfaces were used at the same resolution as terrestrial ones, and represent annual patterns, seasonality and limiting factors of sea surface temperature and salinity for 2000–2014 (Table S3). We carried out a principal components analysis (PCA) to reduce multicollinearity and dimensionality using the PCARaster function of the R package ENMGadgets (Barve & Barve, 2013). We retained the first three components for both terrestrial and marine surfaces, which explained 88 and 93% of the global overall variance, respectively. The number of retained components was determined based on scree plots (terrestrial = Fig. S1; marine = Fig. S2).
Correlative ecological niche models
We estimated ecological niches as minimum-volume ellipsoids (MVE, Van Aelst & Rousseeuw, 2009). We chose this approach for two main reasons. First, in this method it is assumed that the NF presents a convex shape, as suggested by theoretical and physiological evidence (Maguire, 1973; Hooper et al., 2008; Angilletta, 2009; Soberón & Nakamura, 2009). Second, by using a simple-shaped envelope method we can measure directly the contribution of including presence records of the related species than in mathematically more complex algorithms. We also modeled with Maxent to assess whether the observed patterns were method-dependent (Maxent modeling protocol and results are presented in the Supplemental Information).
We fitted MVE’s in environmental space (E) taking into account 97.5% and 95% of the presence records. These percentages were based on the degree of confidence in the data, leaving out outlier points associated with atypical environmental conditions that could have been unidentified errors in the data-cleaning phase, or probably associated with sink population (Osorio-Olvera et al., 2020). We calculated both covariance matrix and centroid of MVE’s using the cov.rob function of the MASS package through the R package ntbox (Osorio-Olvera et al., 2016). From U1 up to the last unit, the final ellipsoids are the result of weighting (averaging centroids and covariances) the ellipsoids built with each group of occurrences units (e.g., we built U1 by weighting ellipsoid U0 and the ellipsoid built only with the presences of U1). To graph the MVEs, we used the R package rgl (Adler et al., 2018). In addition, we projected the models in geographic space as environmental suitability maps based on Mahalanobis distances from each environmental combination to the centroid of the ellipsoid (Osorio-Olvera et al., 2016). We estimated these distances using the R function mahalanobis with the ellipsoid covariance matrix, the vector of environmental variables, and the vector of means (the centroid). We converted environmental suitability models into binary maps (presence or absence of suitable conditions) using two thresholds of allowed omission: 2.5% and 5%. For visualization and manipulation of the resulting maps, we used the R package raster (Hijmans et al., 2018).
Niche models evaluation
To assess performance of the ENMs, we used the AUC ratio of the partial ROC technique (Peterson, Papeş & Soberón, 2008). This metric evaluates the relationship between omission error for independent presence records (here, occurrences of the focal species that fell outside its native range) and the proportion of area estimated as suitable for the species, but only under conditions of low omission error (Peterson, Papeş & Soberón, 2008). The AUC ratio varies from 0 to 2: values greater than 1 indicate that model predictions are better than null expectations. This analysis was carried out using the R script published by Barve (2008). Partial ROC test were computed based on the continuous suitability models and the presence records of the focal species from the invasion area (evaluation data); we allowed an omission error of 2.5% and 5%, and defined a bootstrapping 50% of the evaluation data 100 times to assess statistical significance of AUC ratio values.
Results
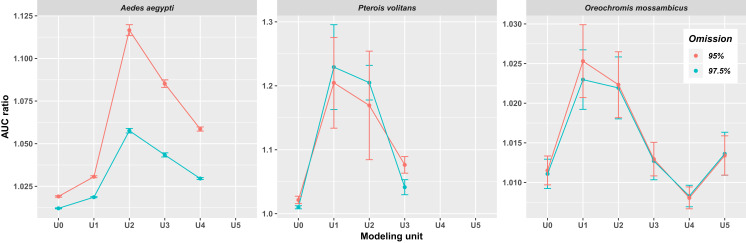
In all three species, the AUC ratio of the partial ROC test increased when occurrences of closely related species were included (Fig. 2). The increase was particularly pronounced in Ae. aegypti and P. volitans, whereas in O. mossambicus this pattern was not as conspicuous; indeed, in that species, the AUC ratio values of some supraspecific units were lower than those of U0. The highest values of AUC ratio were obtained for U1 in P. volitans and O. mossambicus and for U2 in Ae. aegypti. In addition, when we incorporated occurrences of successive related species, AUC ratios generally dropped, except for O. mossambicus, in which they remained high and even increased again in U5. Models including 97.5% and 95% of presence records were mostly very similar; however, the 95% models showed higher AUC ratios (Fig. 2), so hereafter we only show ellipsoids and thresholded maps derived from this modeling procedure.
Figure 2. Average AUC ratios of the ecological niche models obtained with the minimum-volume ellipsoid approach for each modeling unit.
Bars indicate the standard deviation.
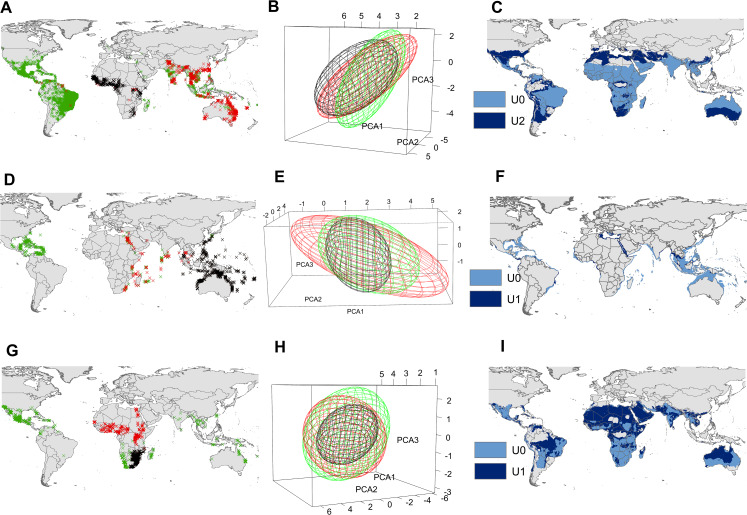
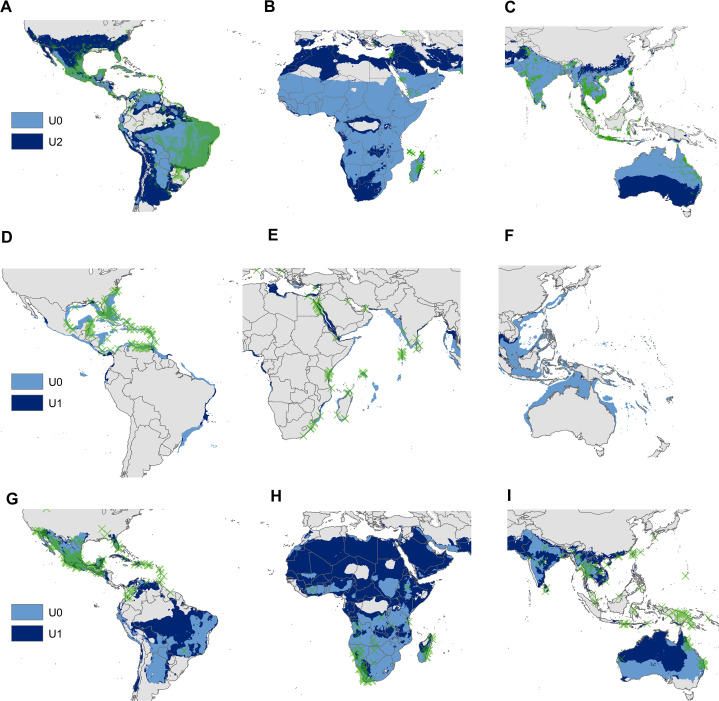
The UT ellipsoid was larger than the U0 ellipsoid in Ae. aegypti, but smaller than that of U2 (Fig. 3B). Thresholded models of U0 failed to predict a high number of presence records outside of the species’ native range. In contrast, U2 models correctly predicted many of these records (Fig. 3C), particularly in North America, some regions of South America (Fig. 4A) and Asia (Fig. 4C), the east coast of the Mediterranean Sea (Fig. 4B), and in the southeastern and southwestern coasts of Australia; although potential distribution obtained from the ellipsoid model that included 95% of presence records covered almost the entire surface of this country (Fig. 4C).
Figure 3. Input data and ecological niche models.
Presence records of the native range (black X’s), invasion range (red X’s), and those included in the supraspecific unit with the highest AUC ratio (blue X’s) of Aedes aegypti (A), Pterois volitans (D) and Oreochromis mossambicus (G). Ecological niche model constructed with the black X’s (black ellipsoid) (i.e., U0), ecological niche model constructed with de black X’s + blue X’s (blue ellipsoid) (i.e., U2 for Ae. aegypti and U1 for P. volitans and O. mossambicus), and ecological niche model constructed with the black X’s + red X’s (red ellipsoid) of Ae. aegypti (B), P. volitans (E) and O. mossambicus (H). Potential distribution obtained from the black ellipsoid (light blue), and from the blue ellipsoid (dark blue) of Ae. aegypti (C), P. volitans (F) and O. mossambicus (I). Minimum-volume ellipsoids presented in this figure included 95% of each input data.
Figure 4. Regional views of the potential distribution (obtained with the minimum-volume ellipsoids that include 95% of occurrences) across the invasion area of Aedes aegypti (A, B and C), Pterois volitans (D, E and F) and Oreochromis mossambicus (G, H and I).
Light blue model = distribution estimated from U0; Dark blue model = distribution estimated from the supraspecific unit with the highest AUC ratio; Green X’s = presence records of the invaded areas.
The UT ellipsoid of P. volitans was greater than that of U0, but smaller than U1 (Fig. 3E). Near the Bahamas and Florida, U0 and U1 predicted almost all presence records (Fig. 4D). Furthermore, U0 failed to predict some invasion records located at the east coast of the Mediterranean Sea and along the entire coast of the Red Sea, while in these regions U1 showed high predictability (Fig. 4E).
In O. mossambicus, UT ellipsoid contained U0 and U1 (Fig. 3H). Distribution maps derived from the U1 modeling unit predicted invaded regions that were not predicted by U0 (Fig. 3I). For instance, in eastern Florida, central and southern Mexico, Haiti, Dominican Republic, and the Colombian Andes in the Americas (Fig. 4G); Namibia, South Africa, and central Madagascar in Africa (Fig. 4H); central India, eastern Thailand, Southwestern Indonesia and Taiwan in Asia; and northeast Australia (Fig. 4I).
Discussion
As we hypothesized, creation of supraspecific modeling units improved the predictive ability of the ENMs for the three analyzed species. Specifically, by adding the occurrences of sister taxa of each focal species (that is, when we created the U1 or U2 modeling unit), omission rates decreased considerably in the areas of invasion. This result suggests that existing niches of the species were not fully representative of their fundamental niches before invading other regions; when we incorporated presences of the most closely related species, we added relevant and complementary information for the characterization of the fundamental niches (Godoy, Camargos & Lodi, 2018). However, in two of the three species evaluated, predictive power decreased when additional, less closely related lineages were added, probably because these lineages have somewhat diverged in niche characteristics from the focal species. Our results are consistent with Peterson (2011), who observed that niche conservatism tends to break down on temporal scales greater than those of speciation events.
In the case of O. mossambicus, predictive power decreased up to U4 but increased again when we added U5, which include less closely related lineages. This fluctuating result could indicate that some environments within the M’s of each lineage are less diverse than those contained in UT, or little ecological differentiation has occurred in this group. Previous studies have shown that ecological niches are frequently conserved in many freshwater fish clades (McNyset, 2009). The pattern observed in this species makes clear that, in some groups, identifying ideal modeling units may be challenging. For instance, predictive capacity in the O. mossambicus models reached a maximum at U1 and again went up at U5, the last unit that we evaluated, so the stability of niche characteristics has been particularly marked in this group.
Another important issue that should be taken with caution is that most of the supraspecific units overestimated the thermal tolerance limits reported in experimental studies (Ae. aegypti = 16.0–36.0 °C, De Almeida et al., 2010; Marinho et al., 2016; P. volitans = 22.0–31.0 °C, Barker, 2015; O. mossambicus = 6.0–43.3 °C, Fast, 1986; Del Rio Zaragoza, Rodriguez & Buckle-Ramirez, 2008). This may suggest that predictions outside environmental ranges associated with occurrences of the focal species could represent complementary conditions of their NF’s as well as environments not suitable for the survival of their populations. Differences between the thermal ranges could be also a consequence of certain methodological limitations. For example, experimental studies do not evaluate behavior and strategies of organisms to deal with suboptimal environments (Kearney & Porter, 2009; Soberón & Arroyo-Peña, 2017). Moreover, physiological thermal limits are usually determined based on a set of individuals that do not represent all the intraspecific variability (Marinho et al., 2016). On the other hand, our MVEs (constructed with an allowed omission of 2.5% and 5%) could still be including records of individuals observed in unsuitable environments but dispersed there through anthropic mechanisms (Bruno, Stachowicz & Bertness, 2003; Campbell et al., 2015) or that represent sink populations (Pulliam, 2000).
Despite the limitations and complications, taking into account phylogenetic relationships to carry out a “lumping” strategy (Smith et al., 2018) allowed us to build more informative niche models. Even if the presences of the sister species represent conditions outside the NF of the focal species, the models could be acceptable if commission errors are low (Qiao et al., 2017); in fact, these ‘breaks’ in complementarity of niches can be detected when the predictive capacity of the models decreases. However, the main reason for modeling above the species level in the case of biological invasions is to model across more environmentally diverse M areas, which leads to niche models that are more broadly representative and robust as regards the fundamental niche. Complementary predictions based on sister species could offer a preliminary hypothesis of areas with potential for invasion, but which should be considered after those represented in the observable, native-range niche. In a real application of “lumping”, the complementary niche areas should be explicitly indicated in G, together with an analysis of novel environments (e.g., MOP, Owens et al., 2013).
Modeling above the species level opens up many possibilities for research, particularly in species with BAM configurations where full characterization of fundamental niches is likely to prove difficult; for example, “Classic” BAM or “Wallace’s Dream” (Soberón & Peterson, 2005; Saupe et al., 2012). Beyond biological invasions, rigorous and systematized creation of supraspecific units in modeling ecological niches has potential to improve answers to interesting biological questions that depend on characterizing species’ fundamental niches. To mention some examples, transferring models to past or future scenarios of climate change based on closely related species may yield results distinct from those in which traditional modeling approaches are used. Research on niche structure (Martínez-Meyer et al., 2013; Pironon et al., 2018) could benefit from “lumping” since the centroids of the NF’s are informative about population abundance (Martínez-Meyer et al., 2013), population density (Yañez-Arenas et al., 2012), or genetic diversity (Lira-Noriega & Manthey, 2014).
Supplemental Information
Bars indicate the standard deviation.
Presence records of the native range (black X’s), invasion range (green X’s), and those included in the supraspecific unit with the highest AUC ratio (red X’s) of Aedes aegypti (A), Pterois volitans (C) and Oreochromis mossambicus (E). Potential distribution models obtained with Maxent using the black X’s as input presence data (light blue) (i.e., U0), and potential distribution models obtained with Maxent using the black X’s + red X’s as input presence data (dark blue) (i.e., U1) for Ae. aegypti (B), P. volitans (D) and O. mossambicus (F).
Light blue model = distribution estimated from U0; Dark blue model = distribution estimated from the supraspecific unit with the highest AUC ratio; Green X’s = presence records of the invaded areas.
Acknowledgments
We thank all the students of the ‘Laboratorio de Ecología Geográfica’ of the Universidad Nacional Autónoma de México (particularly Lina Jiménez for their support at the initial phase of the project). To Norberto Colín for the feedback on the choice of phylogenies and to Octavio Rojas for inspiring this study with his entertaining talks about ’concepts of species and niche modeling’.
Funding Statement
Luis Osorio-Olvera received funding from Consejo Nacional de Ciencia y Tecnología (CONACyT; postdoctoral fellowship number 740751; CVU: 368747) and PAPIIT IN116018. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Sandra Castaño-Quintero performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Jazmín Escobar-Luján performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Luis Osorio-Olvera performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
A. Townsend Peterson conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Xavier Chiappa-Carrara conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Enrique Martínez-Meyer conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Carlos Yañez-Arenas conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Data and code are available at OSF: Escobar-Luján, J. (2020, October 13). Dataset of Supraspecific units in correlative niche modeling improves the prediction of geographic potential of biological invasions. Retrieved from osf.io/7bpcg.
References
- Adler et al. (2018).Adler D, Murdoch D, Nenadic O, Urbanek S, Chen M, Gebhardt A, Bolker B, Csardi G, Strzelecki A, Senger A, Eddelbuettel D. 2018. https://r-forge.r-project.org/projects/rgl https://r-forge.r-project.org/projects/rgl
- Aiello-Lammens et al. (2015).Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography. 2015;38(5):541–545. doi: 10.1111/ecog.01132. [DOI] [Google Scholar]
- Angilletta (2009).Angilletta MJ. Thermal adaptation: a theoretical and empirical synthesis. New York: Oxford University Press; 2009. [Google Scholar]
- Aragón, Baselga & Lobo (2010).Aragón P, Baselga A, Lobo JM. Global estimation of invasion risk zones for the western corn rootworm Diabrotica virgifera virgifera: integrating distribution models and physiological thresholds to assess climatic favourability. Journal of Applied Ecology. 2010;47(5):1026–1035. doi: 10.1111/j.1365-2664.2010.01847.x. [DOI] [Google Scholar]
- Assis et al. (2018).Assis J, Tyberghein L, Bosch S, Verbruggen H, Serrão EA, De Clerck O. Bio-ORACLE v2.0: extending marine data layers for bioclimatic modeling. Global Ecology and Biogeography. 2018;27(3):277–284. doi: 10.1111/geb.12693. [DOI] [Google Scholar]
- Barker (2015).Barker B. Thermal preferences and critical temperature regimes of the Western North Atlantic Invasive Lionfish Complex (Pterois spp.) 2015. Master’s thesis. Nova Southeastern University.
- Barve (2008).Barve N. Tool for Partial-ROC (Biodiversity Institute, Lawrence, KS), v 1.0. 2008. https://kuscholarworks.ku.edu/handle/1808/10059 https://kuscholarworks.ku.edu/handle/1808/10059
- Barve & Barve (2013).Barve N, Barve V. ENMGadgets: tools for pre and post processing in ENM workflows. GitHub. 2013. https://github.com/vijaybarve/ENMGadgets https://github.com/vijaybarve/ENMGadgets
- Bomford (2008).Bomford M. Risk assessment models for establishment of exotic vertebrates in Australia and New Zealand. Canberra: Invasive Animals Cooperative Research Centre; 2008. [Google Scholar]
- Boria et al. (2014).Boria RA, Olson LE, Goodman SM, Anderson RP. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecological Modelling. 2014;275:73–77. doi: 10.1016/j.ecolmodel.2013.12.012. [DOI] [Google Scholar]
- Bruno, Stachowicz & Bertness (2003).Bruno JF, Stachowicz JJ, Bertness MD. Inclusion of facilitation into ecological theory. Trends in Ecology & Evolution. 2003;18(3):119–125. doi: 10.1016/S0169-5347(02)00045-9. [DOI] [Google Scholar]
- Campbell et al. (2015).Campbell LP, Luther C, Moo-Llanes D, Ramsey JM, Danis-Lozano R, Peterson AT. Climate change influences on global distributions of dengue and chikungunya virus vectors. Philosophical Transactions of the Royal Society B: Biological Sciences. 2015;370(1665):1–9. doi: 10.1098/rstb.2014.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu et al. (2018).Chu H, Li C, Guo X, Zhang H, Luo P, Wu Z, Wang G, Zhao T. The phylogenetic relationships of known mosquito (Diptera: Culicidae) mitogenomes. Mitochondrial DNA Part A. 2018;29(1):31–35. doi: 10.1080/24701394.2016.1233533. [DOI] [PubMed] [Google Scholar]
- De Almeida et al. (2010).De Almeida EAPC, De Mendonça EMS, Cavalcanti JC, De Ribeiro CMA. Impact of small variations in temperature and humidity on the reproductive activity and survival of Aedes aegypti (Diptera, Culicidae) Revista Brasileira de Entomologia. 2010;54(2):488–493. doi: 10.1590/S0085-56262010000200014. [DOI] [Google Scholar]
- Del Rio Zaragoza, Rodriguez & Buckle-Ramirez (2008).Del Rio Zaragoza O, Rodriguez MH, Buckle-Ramirez LF. Thermal stress effects on tilapia Oreochromis mossambicus (Pisces: Cichlidae) blood parameters. Marine and Freshwater Behaviour and Physiology. 2008;41(2):79–89. doi: 10.1080/10236240801896223. [DOI] [Google Scholar]
- Dormann (2007).Dormann CF. Promising the future? Global change projections of species distributions. Basic and Applied Ecology. 2007;8(5):387–397. doi: 10.1016/j.baae.2006.11.001. [DOI] [Google Scholar]
- Escobar et al. (2016).Escobar LE, Qiao H, Phelps NBD, Wagner CK, Larkin DJ. Realized niche shift associated with the Eurasian charophyte Nitellopsis obtusa becoming invasive in North America. Scientific Reports. 2016;6(1):1–15. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast (1986).Fast A. Pond production systems: water quality management practices. In: Lannan JE, Smitherman RO, Tchobanoglous G, editors. Principles and Practices of Pond Aquaculture. Corvallis: Oregon State University Press; 1986. pp. 141–167. [Google Scholar]
- Gallien et al. (2010).Gallien L, Münkemüller T, Albert CH, Boulangeat I, Thuiller W. Predicting potential distributions of invasive species: where to go from here? Diversity and Distributions. 2010;16(3):331–342. doi: 10.1111/j.1472-4642.2010.00652.x. [DOI] [Google Scholar]
- GBIF (2019).GBIF Global biodiversity information facility. 2019. https://www.gbif.org https://www.gbif.org
- Godoy, Camargos & Lodi (2018).Godoy BS, Camargos LM, Lodi S. When phylogeny and ecology meet: modeling the occurrence of Trichoptera with environmental and phylogenetic data. Ecology and Evolution. 2018;8(11):5312–5322. doi: 10.1002/ece3.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2011).He A, Luo Y, Yang H, Liu L, Li S, Wang C. Complete mitochondrial DNA sequences of the Nile tilapia (Oreochromis niloticus) and Blue tilapia (Oreochromis aureus): genome characterization and phylogeny applications. Molecular Biology Reports. 2011;38(3):2015–2021. doi: 10.1007/s11033-010-0324-7. [DOI] [PubMed] [Google Scholar]
- Hijmans et al. (2018).Hijmans RJ, van Etten J, Cheng J, Summer M, Mattiuzzi M. 2018. https://CRAN.R-project.org/package=raster https://CRAN.R-project.org/package=raster
- Hooper et al. (2008).Hooper HL, Connon R, Callaghan A, Fryer G, Yarwood-Buchanan S, Biggs J, Maund SJ, Hutchinson TH, Sibly RM. The ecological niche of Daphnia magna characterized using population growth rate. Ecology. 2008;89(4):1015–1022. doi: 10.1890/07-0559.1. [DOI] [PubMed] [Google Scholar]
- Jiménez-Valverde et al. (2011).Jiménez-Valverde A, Peterson AT, Soberón J, Overton JM, Aragón P, Lobo JM. Use of niche models in invasive species risk assessments. Biological Invasions. 2011;13(12):2785–2797. doi: 10.1007/s10530-011-9963-4. [DOI] [Google Scholar]
- Kearney & Porter (2004).Kearney M, Porter WP. Mapping the fundamental niche: physiology, climate, and the distribution of a nocturnal lizard. Ecology. 2004;85(11):3119–3131. doi: 10.1890/03-0820. [DOI] [Google Scholar]
- Kearney & Porter (2009).Kearney M, Porter W. Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecology Letters. 2009;12(4):334–350. doi: 10.1111/j.1461-0248.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- Kochzius et al. (2003).Kochzius M, Söller R, Khalaf MA, Blohm D. Molecular phylogeny of the lionfish genera Dendrochirus and Pterois (Scorpaenidae, Pteroinae) based on mitochondrial DNA sequences. Molecular Phylogenetics and Evolution. 2003;28(3):396–403. doi: 10.1016/S1055-7903(02)00444-X. [DOI] [PubMed] [Google Scholar]
- Kriticos et al. (2012).Kriticos DJ, Webber BL, Leriche A, Ota N, Macadam I, Bathols J, Scott JK. CliMond: global high-resolution historical and future scenario climate surfaces for bioclimatic modelling. Methods in Ecology and Evolution. 2012;3(1):53–64. doi: 10.1111/j.2041-210X.2011.00134.x. [DOI] [Google Scholar]
- Larson et al. (2014).Larson ER, Gallagher RV, Beaumont LJ, Olden JD. Generalized “avatar” niche shifts improve distribution models for invasive species. Diversity and Distributions. 2014;20(11):1296–1306. doi: 10.1111/ddi.12233. [DOI] [Google Scholar]
- Lira-Noriega & Manthey (2014).Lira-Noriega A, Manthey JD. Relationship of genetic diversity and niche centrality: a survey and analysis. Evolution. 2014;68(4):1082–1093. doi: 10.1111/evo.12343. [DOI] [PubMed] [Google Scholar]
- Maguire (1973).Maguire B. Niche response structure and the analytical potentials of its relationship to the habitat. American Naturalist. 1973;107(954):213–246. doi: 10.1086/282827. [DOI] [Google Scholar]
- Manchester & Bullock (2000).Manchester SJ, Bullock JM. The impacts of non-native species on UK biodiversity and the effectiveness of control. Journal of Applied Ecology. 2000;37(5):845–864. doi: 10.1046/j.1365-2664.2000.00538.x. [DOI] [Google Scholar]
- Marinho et al. (2016).Marinho RA, Beserra EB, Bezerra-Gusmão MA, de Porto VS, Olinda RA, dos Santos CAC. Effects of temperature on the life cycle, expansion, and dispersion of Aedes aegypti (Diptera: Culicidae) in three cities in Paraiba, Brazil. Journal of Vector Ecology. 2016;41(1):1–10. doi: 10.1111/jvec.12187. [DOI] [PubMed] [Google Scholar]
- Martínez-Meyer et al. (2013).Martínez-Meyer E, Díaz-Porras D, Peterson AT, Yáñez-Arenas C. Ecological niche structure and rangewide abundance patterns of species. Biology Letters. 2013;9(1):20120637. doi: 10.1098/rsbl.2012.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNyset (2009).McNyset KM. Ecological niche conservatism in North American freshwater fishes. Biological Journal of the Linnean Society. 2009;96(2):282–295. doi: 10.1111/j.1095-8312.2008.01121.x. [DOI] [Google Scholar]
- Merow et al. (2014).Merow C, Smith MJ, Edwards TC, Guisan A, Mcmahon SM, Normand S, Thuiller W, Wüest RO, Zimmermann NE, Elith J. What do we gain from simplicity versus complexity in species distribution models? Ecography. 2014;37(12):1267–1281. doi: 10.1111/ecog.00845. [DOI] [Google Scholar]
- Miller et al. (2005).Miller N, Estoup A, Toepfer S, Bourguet D, Lapchin L, Derridj S, Kim KS, Reynaud P, Furlan L, Guillemaud T. Multiple transatlantic introductions of the western corn rootworm. Science. 2005;310(5750):992. doi: 10.1126/science.1115871. [DOI] [PubMed] [Google Scholar]
- Olson et al. (2001).Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’amico JA, Itoua I, Strand HL, Morrison JC, Loucks CJ, Allnutt TF, Ricketts TH, Kura Y, Lamoreux JF, Wettengel WW, Hedao P, Kassem KR. Terrestrial ecoregions of the world: a new map of life on earth. Bioscience. 2001;51(11):933–938. doi: 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2. [DOI] [Google Scholar]
- Osorio-Olvera et al. (2016).Osorio-Olvera L, Barve V, Barve N, Soberón J. ntbox: from getting biodiversity data to evaluating species distribution models in a friendly GUI environment. 2016. https://rdrr.io/github/luismurao/ntbox https://rdrr.io/github/luismurao/ntbox
- Osorio-Olvera et al. (2020).Osorio-Olvera L, Yáñez-Arenas C, Martínez-Meyer E, Peterson AT. Relationships between population densities and niche-centroid distances in North American birds. Ecology Letters. 2020;23(3):555–564. doi: 10.1111/ele.13453. [DOI] [PubMed] [Google Scholar]
- Owens et al. (2013).Owens HL, Campbell LP, Dornak LL, Saupe EE, Barve N, Soberón J, Ingenloff K, Lira-Noriega A, Hensz SH, Myers CE, Peterson AT. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecological Modelling. 2013;263:10–18. doi: 10.1016/j.ecolmodel.2013.04.011. [DOI] [Google Scholar]
- Pavoine & Bonsall (2011).Pavoine S, Bonsall MB. Measuring biodiversity to explain community assembly: a unified approach. Biological Reviews. 2011;86(4):792–812. doi: 10.1111/j.1469-185X.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- Peterson (2003).Peterson AT. Predicting the geography of species’ invasions via ecological niche modeling. Quarterly Review of Biology. 2003;78(4):419–433. doi: 10.1086/378926. [DOI] [PubMed] [Google Scholar]
- Peterson (2011).Peterson AT. Ecological niche conservatism: a time-structured review of evidence. Journal of Biogeography. 2011;38(5):817–827. doi: 10.1111/j.1365-2699.2010.02456.x. [DOI] [Google Scholar]
- Peterson & Nakazawa (2008).Peterson AT, Nakazawa Y. Environmental data sets matter in ecological niche modelling: an example with Solenopsis invicta and Solenopsis richteri. Global Ecology and Biogeography. 2008;17:135–144. doi: 10.1111/j.1466-8238.2007.00347.x. [DOI] [Google Scholar]
- Peterson, Papeş & Soberón (2008).Peterson AT, Papeş M, Soberón J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecological Modelling. 2008;213(1):63–72. doi: 10.1016/j.ecolmodel.2007.11.008. [DOI] [Google Scholar]
- Peterson & Soberón (2012).Peterson AT, Soberón J. Species distribution modeling and ecological niche modeling: getting the concepts right. Natureza & Conservação. 2012;10(2):102–107. doi: 10.4322/natcon.2012.019. [DOI] [Google Scholar]
- Peterson et al. (2011).Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB. Ecological niches and geographic distributions. New Jersey: Princeton University Press; 2011. [Google Scholar]
- Peterson, Soberón & Sánchez-Cordero (1999).Peterson AT, Soberón J, Sánchez-Cordero V. Conservatism of ecological niches in evolutionary time. Science. 1999;285(5431):1265–1267. doi: 10.1126/science.285.5431.1265. [DOI] [PubMed] [Google Scholar]
- Pironon et al. (2018).Pironon S, Villellas J, Thuiller W, Eckhart VM, Geber MA, Moeller DA, García MB. The ‘Hutchinsonian niche’ as an assemblage of demographic niches: implications for species geographic ranges. Ecography. 2018;41(7):1103–1113. doi: 10.1111/ecog.03414. [DOI] [Google Scholar]
- Pulliam (2000).Pulliam HR. On the relationship between niche and distribution. Ecology Letters. 2000;3(4):349–361. doi: 10.1046/j.1461-0248.2000.00143.x. [DOI] [Google Scholar]
- QGIS Development Team (2018).QGIS Development Team QGIS geographic information system—open source geospatial found. 2018. https://qgis.org/en/site/ https://qgis.org/en/site/
- Qiao et al. (2017).Qiao H, Peterson AT, Ji L, Hu J. Using data from related species to overcome spatial sampling bias and associated limitations in ecological niche modeling. Methods and Ecology and Evolution. 2017;8(12):1804–1812. doi: 10.1111/2041-210X.12832. [DOI] [Google Scholar]
- R Core Team (2018).R Core Team . R: a language and environment for statistical computing. Vienna: The R Foundation for Statistical Computing; 2018. [Google Scholar]
- Radosavljevic & Anderson (2014).Radosavljevic A, Anderson RP. Making better M axent models of species distributions: complexity, overfitting and evaluation. Journal of Biogeography. 2014;41(4):629–643. doi: 10.1111/jbi.12227. [DOI] [Google Scholar]
- Richardson, Pyšek & Carlton (2011).Richardson DM, Pyšek P, Carlton JT. A compendium of essential concepts and terminology in biological invasions. In: Richardson D, editor. Fifty Years of Invasion Ecology: The Legacy of Charles Elton. Chichester: Wiley-Blackwell; 2011. pp. 409–442. [Google Scholar]
- Saupe et al. (2012).Saupe EE, Barve V, Myers CE, Soberón J, Barve N, Hensz CM, Peterson AT, Owens HL, Lira-Noriega A. Variation in niche and distribution model performance: the need for a priori assessment of key causal factors. Ecological Modelling. 2012;237:11–22. doi: 10.1016/j.ecolmodel.2012.04.001. [DOI] [Google Scholar]
- Smith et al. (2018).Smith AB, Godsoe W, Rodríguez-Sánchez F, Wang HH, Warren D. Niche estimation above and below the species level. Trends in Ecology & Evolution. 2018;34(3):260–273. doi: 10.1016/j.tree.2018.10.012. [DOI] [PubMed] [Google Scholar]
- Soberón & Arroyo-Peña (2017).Soberón J, Arroyo-Peña B. Are fundamental niches larger than the realized? Testing a 50-year-old prediction by Hutchinson. PLOS ONE. 2017;12(4):1–14. doi: 10.1371/journal.pone.0175138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón & Nakamura (2009).Soberón J, Nakamura M. Niches and distributional areas: concepts, methods, and assumptions. Proceedings of the National Academy of Sciences USA. 2009;106(Suppl. 2):19644–19650. doi: 10.1073/pnas.0901637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón & Peterson (2005).Soberón J, Peterson AT. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodiversity Informatics. 2005;2:1–10. doi: 10.17161/bi.v2i0.4. [DOI] [Google Scholar]
- Spalding et al. (2007).Spalding MD, Fox HE, Allen GR, Davidson N, Ferdaña ZA, Finlayson M, Halpern BJ, Jorge M, Lombana A, Lourie SA, Martin KD, McManus E, Molnar J, Recchia CA, Robertson J. Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience. 2007;57(7):573–583. doi: 10.1641/B570707. [DOI] [Google Scholar]
- Tatem & Hay (2007).Tatem AJ, Hay SI. Climatic similarity and biological exchange in the worldwide airline transportation network. Proceedings of the Royal Society B: Biological Sciences. 2007;274(1617):1489–1496. doi: 10.1098/rspb.2007.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller et al. (2005).Thuiller W, Richardson DM, Pyssek P, Midgley GF, Hughes GO, Rouget M. Niche-based modelling as a tool for predicting the risk of alien plant invasions at a global scale. Global Change Biology. 2005;11(12):2234–2250. doi: 10.1111/j.1365-2486.2005.001018.x. [DOI] [PubMed] [Google Scholar]
- Van Aelst & Rousseeuw (2009).Van Aelst S, Rousseeuw P. Minimum volume ellipsoid. Wiley Interdisciplinary Reviews: Computational Statistics. 2009;1(1):71–82. doi: 10.1002/wics.19. [DOI] [Google Scholar]
- Warren & Seifert (2011).Warren DL, Seifert SN. Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications. 2011;21(2):335–342. doi: 10.1890/10-1171.1. [DOI] [PubMed] [Google Scholar]
- Wilson et al. (2009).Wilson JRU, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends in Ecology & Evolution. 2009;24(3):136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Yañez-Arenas et al. (2012).Yañez-Arenas C, Martínez-Meyer E, Mandujano S, Rojas-Soto O. Modelling geographic patterns of population density of the white-tailed deer in central Mexico by implementing ecological niche theory. Oikos. 2012;121(12):2081–2089. doi: 10.1111/j.1600-0706.2012.20350.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bars indicate the standard deviation.
Presence records of the native range (black X’s), invasion range (green X’s), and those included in the supraspecific unit with the highest AUC ratio (red X’s) of Aedes aegypti (A), Pterois volitans (C) and Oreochromis mossambicus (E). Potential distribution models obtained with Maxent using the black X’s as input presence data (light blue) (i.e., U0), and potential distribution models obtained with Maxent using the black X’s + red X’s as input presence data (dark blue) (i.e., U1) for Ae. aegypti (B), P. volitans (D) and O. mossambicus (F).
Light blue model = distribution estimated from U0; Dark blue model = distribution estimated from the supraspecific unit with the highest AUC ratio; Green X’s = presence records of the invaded areas.
Data Availability Statement
The following information was supplied regarding data availability:
Data and code are available at OSF: Escobar-Luján, J. (2020, October 13). Dataset of Supraspecific units in correlative niche modeling improves the prediction of geographic potential of biological invasions. Retrieved from osf.io/7bpcg.