Some microbes utilize different dissimilatory nitrate reduction (DNR) pathways, including DNR to ammonia (DNRA) and denitrification pathways, for anaerobic respiration in response to ambient carbon/nitrogen ratio changes. Large-scale industrial nitrogen fixation and fertilizer application raise the concern of emission of N2O, a stable gas with potent global warming potential, as consequence of microbial respiration, thereby aggravating global warming and climate change. However, little is known about the molecular mechanism underlying the choice of two competing DNR pathways. We demonstrate that the global regulator CRP1, which is widely encoded in bacteria, is required for DNRA in S. loihica PV-4 strain, while the CRP2 paralogue is required for transcription of the nitrite reductase gene nirK for denitrification. Sufficient carbon source lead to the predominance of DNRA, while carbon source/electron donor deficiency may result in an incomplete denitrification process, raising the concern of high levels of N2O emission from nitrate-rich and carbon source-poor waters and soils.
KEYWORDS: dissimilatory nitrate reduction to ammonia, denitrification, cyclic AMP receptor protein, carbon-to-nitrogen ratios, Shewanella loihica
ABSTRACT
Under anoxic conditions, many bacteria, including Shewanella loihica strain PV-4, could use nitrate as an electron acceptor for dissimilatory nitrate reduction to ammonium (DNRA) and/or denitrification. Previous and current studies have shown that DNRA is favored under higher ambient carbon-to-nitrogen (C/N) ratios, whereas denitrification is upregulated under lower C/N ratios, which is consistent with our bioenergetics calculations. Interestingly, computational analyses indicate that the common cyclic AMP receptor protein (designated CRP1) and its paralogue CRP2 might both be involved in the regulation of two competing dissimilatory nitrate reduction pathways, DNRA and denitrification, in S. loihica PV-4 and several other denitrifying Shewanella species. To explore the regulatory mechanism underlying the dissimilatory nitrate reduction (DNR) pathways, nitrate reduction of a series of in-frame deletion mutants was analyzed under different C/N ratios. Deletion of crp1 could accelerate the reduction of nitrite to NO under both low and high C/N ratios. CRP1 is not required for denitrification and actually suppresses production of NO and N2O gases. Deletion of either of the NO-forming nitrite reductase genes nirK or crp2 blocked production of NO gas. Furthermore, real-time PCR and electrophoretic mobility shift assays (EMSAs) demonstrated that the transcription levels of DNRA-relevant genes such as nap-β (napDABGH), nrfA, and cymA were upregulated by CRP1, while nirK transcription was dependent on CRP2. There are tradeoffs between the different physiological roles of nitrate/lactate, as nitrogen nutrient/carbon source and electron acceptor/donor and CRPs may leverage dissimilatory nitrate reduction pathways for maximizing energy yield and bacterial survival under ambient environmental conditions.
IMPORTANCE Some microbes utilize different dissimilatory nitrate reduction (DNR) pathways, including DNR to ammonia (DNRA) and denitrification pathways, for anaerobic respiration in response to ambient carbon/nitrogen ratio changes. Large-scale industrial nitrogen fixation and fertilizer application raise the concern of emission of N2O, a stable gas with potent global warming potential, as consequence of microbial respiration, thereby aggravating global warming and climate change. However, little is known about the molecular mechanism underlying the choice of two competing DNR pathways. We demonstrate that the global regulator CRP1, which is widely encoded in bacteria, is required for DNRA in S. loihica PV-4 strain, while the CRP2 paralogue is required for transcription of the nitrite reductase gene nirK for denitrification. Sufficient carbon source lead to the predominance of DNRA, while carbon source/electron donor deficiency may result in an incomplete denitrification process, raising the concern of high levels of N2O emission from nitrate-rich and carbon source-poor waters and soils.
INTRODUCTION
Industrial nitrogen fixation and chemical nitrogen fertilizers have not only dramatically increased crop yields, thereby relieving food shortage and supporting rapid global population growth in the past century, but have also enhanced the reactive nitrogen input into soils and surface and ground waters, accelerating the eutrophication of freshwaters and coastal marine environments in which cyanobacterial blooms and red tides occur frequently worldwide. More strikingly, the increasing emission of potent greenhouse gases, including nitrous oxide (N2O, with a global warming potential value as high as 268 over a period of 20 years), may have aggravated global warming and climate change (1). Microbes play a central role in the global biogeochemical cycles of carbon, nitrogen, phosphorus, sulfur, and other elements on Earth because of their ubiquitous distribution, huge biomass, high genetic diversity, and complex metabolic pathways (2, 3). Denitrification and dissimilatory nitrate reduction to ammonium (DNRA; also termed nitrate ammonification) are widespread in many bacteria (4). On the other hand, aerobic denitrification could occur in the presence of the oxygen-insensitive periplasmic nitrate reductase NapA in some heterotrophic facultative bacteria. Theoretically, dissimilatory nitrate ammonification and denitrification processes are considered to be the highest-energy-yielding respiration systems in anoxic environments after oxygen has been gradually depleted, and the corresponding free energy changes are only 35% and 7% lower than that of aerobic respiration (5). The DNRA pathway could increase the rate of ATP generation and thus the rate of growth, while the pathway of denitrification could maximize the energy yield and growth yield via multiple energy conservation steps. However, the growth yield of planktonic denitrifying bacteria was found to be actually lower than expected and was even lower than that of DNRA pathway-expressing strains (5).
Shewanella loihica strain PV-4, which harbors both competing dissimilatory nitrate reduction (DNR) pathways, DNRA and denitrification, had been utilized to address the issue of how and why these two DNR pathways are employed for anaerobic respiration (6). Denitrification dominated at low C/N ratios (the ratio of C atoms in the electron donor to N atoms in electron acceptor) (i.e., electron donor-limiting growth conditions), whereas the ammonium was the predominant product at high C/N ratios (i.e., electron acceptor-limiting growth conditions) (7). Although the ammonium is released into the ambient environment, it still could be utilized and assimilated by bacteria. However, nitric oxide (NO), nitrous oxide (N2O), and dinitrogen (N2) gases are completely lost and are unavailable as a nutrient. Nitrite (NO2−) is a relevant determinant for N retention (i.e., ammonification) versus N loss and greenhouse gas emission (i.e., denitrification) (7). When PV-4 was incubated with nitrate (NO3−) and N2O at a pH of 6.0, transient accumulation of N2O and no significant ammonium (NH4+) production were observed. At pH values of 7.0 and 8.0, the PV-4 strain served as a N2O sink, as N2O concentration decreased consistently without accumulation. Respiratory ammonification was upregulated at these higher pH values. When NO2− was used in place of NO3−, neither growth nor NO2− reduction was observed at a pH of 6.0. NH4+ was the exclusive product from NO2− reduction at both pH 7.0 and 8.0, and neither production nor consumption of N2O was observed, suggesting that NO2− regulation superseded pH effects on the nitrogen-oxide dissimilation reactions (8). In addition, temperature may also influence the dissimilatory nitrate reduction pathways (7). Furthermore, S. loihica PV-4 contains the copper nitrite reductase gene nirK and other genes for denitrification. These seemingly redundant respiratory chain components may be differentially expressed and functioning along environmental condition gradients such as different oxygen tensions and availability of electron acceptors and carbon source (electron donors) (9). These results could provide insights into the regulation of nitrate reduction pathways, but more detailed and mechanistic investigation are needed to address how the regulations are initiated by external and internal signals and are achieved in bacterial cells.
It is known that periplasmic nitrate reductase (Nap) is in charge for the first step of nitrate reduction for both DNRA and denitrification. Like most Shewanella species, PV-4 harbors two periplasmic nitrate reductase gene clusters, NapC-associated nap-α (napEDABC) and CymA-dependent nap-β (napDAGHB), for dissimilatory nitrate respiration. The denitrifying strain Shewanella denitrificans OS217 only harbors Nap-α for denitrification, and Shewanella oneidensis MR-1 encodes only Nap-β for DNRA (see Table S1 in the supplemental material). Therefore, Nap-α is considered to be mainly involved in denitrification, which is independent of CRP1 regulation, while Nap-β is mainly involved in DNRA and is regulated by CRP1 (10–12). Since the common cyclic AMP receptor protein CRP1 is an important regulator for carbon metabolism (13) and our previous study indicates that Nap-α, whose transcription starts earlier than that of Nap-β, is independent of CRP1 in the culture of Shewanella putrefaciens W3-18-1 under microaerobic conditions (cultivation without shaking) (9), we examined the influence of CRP1 on the nitrate reduction, including DNRA and denitrification, in S. loihica PV-4.
Like S. putrefaciens strain W3-18-1, S. loihica strain PV-4 encodes the PstI-like restriction endonuclease and is recalcitrant to genetic manipulations, which is due to DNA restriction mediated by prophage-borne restriction-modification systems and CRISPR (clustered regularly interspaced short palindromic repeat) elements (9, 14). To facilitate genetic manipulation, putative DNA endonuclease and DNA methylase genes were deleted (14). We generated a series of homogenic mutants to test the role played by a specific gene in the dissimilatory nitrate reduction pathways. The molecular mechanism underlying the choice of denitrification or DNRA based on different C/N ratios was further investigated by combining molecular genetics, physiology, and biochemistry approaches. Our results demonstrate that the common cyclic AMP receptor protein CRP1 is not required for denitrification, while its paralogue CRP2 is required for the transcription of nirK coding for the NO-generating nitrite reductase. It is clear that both CRP paralogues are involved in the bacterial choice of two competing dissimilatory nitrate reduction pathways, DNRA and denitrification. Our results could provide important insights into the molecular evolution and regulation of bacterial respirations in Shewanella species and in other bacteria.
RESULTS
Characteristics of nitrate reduction in S. loihica PV-4.
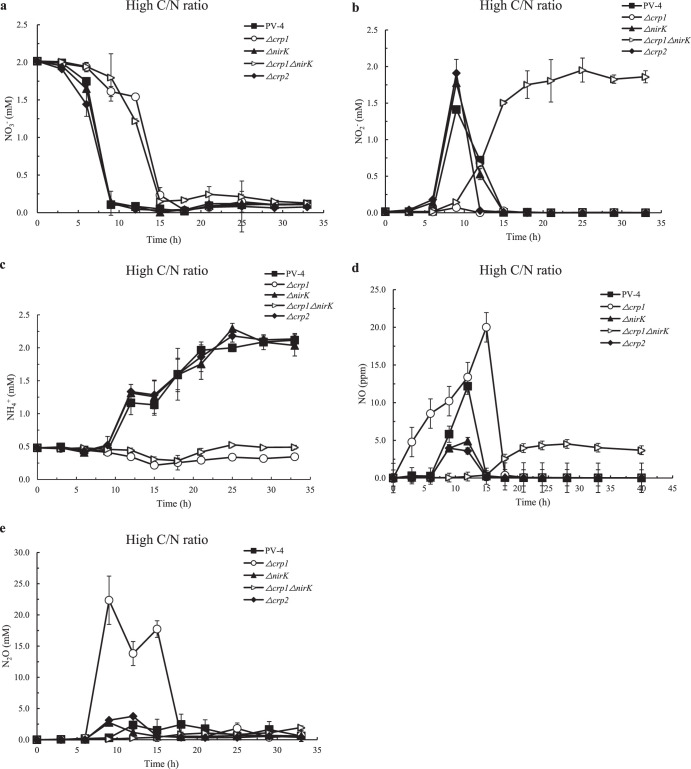
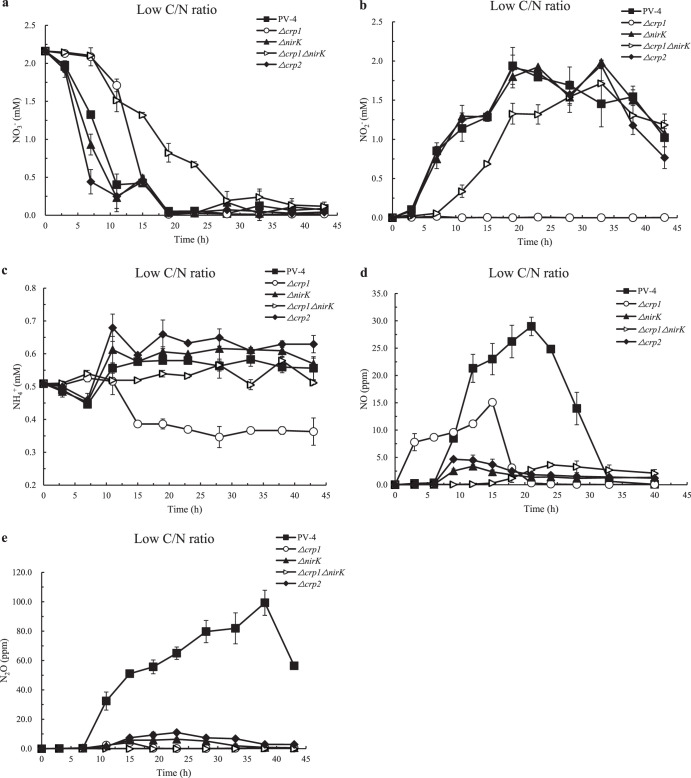
Shewanella oneidensis MR-1 and Shewanella putrefaciens W3-18-1 and CN-32 strains conduct dissimilatory nitrate ammonification, which exhibits sequential reduction of nitrate to nitrite and then to ammonium under anaerobic conditions (15, 16). In the S. loihica PV-4 wild-type strain, reduction of nitrate (Fig. 1a and 2a) and accumulation of nitrite (Fig. 1b and 2b) is observed before further reduction to ammonium under higher C/N ratios (12:1) (Fig. 1c) or to NO under lower C/N ratios (2:1) (Fig. 2d), which is consistent with previous studies (7). PV-4 Δcrp1 and PV-4 Δcrp1 ΔnirK mutants showed much lower reduction of nitrate and accumulation of nitrite. Under the higher C/N ratios, DNRA was the predominant DNR pathway in the culture of the wild-type PV-4 strain, and there was little N2O gas produced (Fig. 1e), which is also consistent with previous observations (7).
FIG 1.
The nitrate reduction of wild-type strain of S. loihica PV-4, the in-frame deletion mutants Δcrp1, Δcrp2, and ΔnirK, and the Δcrp1 ΔnirK double mutant under high carbon/nitrogen ratios. The concentrations of NO3− (a), NO2− (b), NH4+ (c), NO (d), and N2O (e) in the medium were measured under high carbon/nitrogen ratios (C/N = 12). Lactate (10 mM) and nitrate (2 mM) were added to phosphate-buffered basal salt medium supplemented with 0.5 mM ammonium chloride as a nitrogen source.
FIG 2.
The nitrate reduction of the wild-type strain of S. loihica PV-4, the in-frame deletion mutants Δcrp1, Δcrp2, ΔnirK, and the Δcrp1 ΔnirK double mutant under low carbon/nitrogen ratios. The concentrations of NO3− (a), NO2− (b), NH4+ (c), NO (d), and N2O (e) in the medium were measured under low carbon/nitrogen ratios (C/N = 2). Lactate (1.66 mM) and nitrate (2 mM) were added to phosphate-buffered basal salt medium supplemented with 0.5 mM ammonium chloride as a nitrogen source.
CRP1 is required for sequential reduction of nitrate and nitrite to ammonium.
By taking advantage of the easy genetic manipulation in the S. loihica PV-4 ΔpstI ΔpstM strain (9), we generated a series of in-frame deletion mutants in this strain (here called the wild-type strain) and tested their nitrate reduction. There are four CRP/FNR homologs encoded in the S. loihica PV-4 genome (see Table S2 in the supplemental material). The phylogenetic analysis indicated that these four CRP paralogues fall into four clusters with genetic divergence (see Fig. S1 in the supplemental material). Our results showed that the reduction of nitrate to nitrite remarkably was slowed down when the crp1 gene was deleted and the completion of nitrate reduction was postponed under the tested conditions (Fig. 1a and 2a). The low reduction rate of nitrate to nitrite was probably due to the loss of CRP1-dependent expression of nap-β and cymA in Shewanella strains, including in S. loihica PV-4, S. oneidensis MR-1, and S. putrefaciens W3-18-1 (16, 17). That is, CRP1 binding sites are present in the promoters of nap-β and cymA (see Fig. S2 in the supplemental material), which are responsible for the nitrate reduction to nitrite. In addition, the results clearly showed that accumulation of ammonia in PV-4 under high C/N ratios was absent in PV-4 under low C/N ratios (see Fig. S3 and S4 in the supplemental material), because the denitrification dominated at low C/N ratios, whereas the DNRA dominated at high C/N ratios. The accumulation of nitrite previously shown in S. oneidensis MR-1 (16), S. putrefaciens W3-18-1 (9, 17), and S. loihica PV-4 (14) was no longer detected in the culture of the PV-4 Δcrp1 mutant (Fig. 1b and 2b), and the ammonium level did not increase (Fig. 1c and 2c), although the nitrate levels gradually decreased at lower rates (Fig. 1a and 2a). There was almost no ammonia produced while the increase of NO production was observed because the nitrite ammonification was blocked by the deletion of the crp1 gene, and instead the denitrification was increased as more nitrite molecules could be diverted into this pathway (Fig. 1c and 2c), indicating that CRP1 is also required for DNRA in PV-4, as previously demonstrated in S. oneidensis MR-1 and S. putrefaciens W3-18-1 strains.
Although to various degrees, the accumulation of NO and N2O was observed in both wild-type PV-4 strain and the PV-4 Δcrp1 mutant under high C/N ratios. When comparing the two strains, relatively high levels of NO (Fig. 1d) and N2O (Fig. 1e) were detected in the culture of the PV-4 Δcrp1 mutant, and the release of NO and N2O also started several hours earlier than that of the wild-type strain (Fig. 1d), which is consistent with our observation that nitrite no longer accumulated because it was quickly reduced to NO by NirK (Fig. S3). Correspondingly, the nitrite levels remained at almost 2 mM (equivalent to the initial nitrate concentration) for a prolonged period and could not be further reduced by the PV-4 Δcrp1 ΔnirK double mutant (Fig. 1b and Fig. S3), indicating that reduction of nitrite to either ammonia under the high C/N ratio or NO under the low C/N ratios was blocked. These results suggest that CRP1 stimulates the DNRA and that dissimilatory nitrate reduction switched to denitrification from DNRA in the PV-4 Δcrp1 mutant.
Transcription of nirK, norBC, and nosZ is independent of CRP1 regulation.
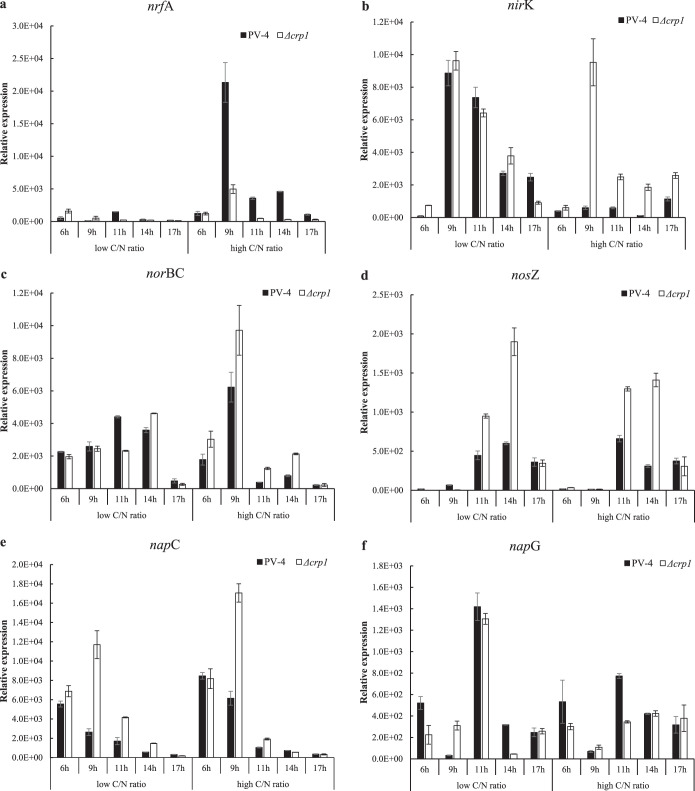
It is well-established that CRP1 plays a crucial role in the regulation of anaerobic respiration in the Shewanella species without denitrification (13). Our previous work also confirmed that nap-β was dependent of CRP-regulation, while nap-α was not (9). The regulatory role played by CRP1 in denitrification remains unclear since no genetic manipulation could have been conducted in the denitrifying Shewanella species, including in the PV-4 strain. In this study, we compared the transcription levels of the genes involved in nitrate reduction in the denitrifying wild-type PV-4 strain and the PV-4 Δcrp1 mutant by using real-time PCR. Deletion of crp1 not only severely affected the transcription of nrfA, encoding the nitrite reductase that catalyzes reduction of nitrite to ammonia under high C/N ratios, but also dramatically increased transcription of nirK, norBC, and nosZ involved in denitrification under these conditions (Fig. 3). In other words, deletion of crp1 not only switched off DNRA, as expected, but also turned on the denitrification pathway, which is not favored under the high C/N ratios, while it is upregulated under low C/N ratios in the wild-type strain. We also monitored the transcription of napC (CRP1-independent nap-α gene cluster) and napG (CRP1-dependent nap-β gene cluster), and these results were consistent with our chemical analyses of nitrogen compounds (Fig. 3; see also Fig. S5 in the supplemental material) and our previous results that showed that the transcription of napG, but not napC, was upregulated by CRP1 (9). The complete time course results showed that napG was upregulated by CRP1 under high C/N ratios at the 6th and 11th hours, although expression of napC and napG was not significantly different in the wild-type strain or the PV-4 Δcrp1 mutant at one time point (at the 14th hour) (see Fig. S6 in the supplemental material). These results are also consistent with previous findings that deletion of crp1 affected the transcription of nap-β gene cluster, cymA, and the nitrite reductase gene nrfA in S. oneidensis MR-1 and S. putrefaciens W3-18-1 (17). Furthermore, these results also indicate that the transcription of nirK, norBC, and nosZ is not dependent on CRP1.
FIG 3.
Quantitative PCR analyses on transcriptional profiles of nitrate reduction genes nrfA (a), nirK (b), norBC (c), nosZ (d), napC (e), and napG (f) in the wild-type strain S. loihica PV-4 and in-frame deletion mutant Δcrp1 under different carbon/nitrogen ratios (high carbon/nitrogen ratio means a C/N ratio of 12 and low carbon/nitrogen ratio means a C/N ratio of 2) and in different growth phases (at 6, 9, 11, 14, and 17 h after anaerobic cultivation). Gene expression was normalized against the 16S rRNA gene using the threshold cycle (2−ΔCT) calculation as follows: ΔCT = CTgene of interest − CT16S rRNA. Results are the means of three replicates, and error bars indicate standard deviation.
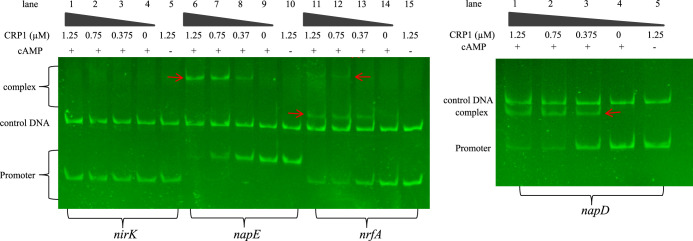
The binding of CRP1 to specific promoters in the presence and absence of cAMP.
Multiple alignment analyses had been conducted to reveal the conserved motifs upstream of the nap-α, nap-β, nrfA, cymA, and nirK genes as previously described for prediction of the potential regulators (Fig. S2) (9, 18), indicating that nap-α (napEDABC) and nirK are independent of CRP1 in the absence of CRP1-bound motifs, but nap-β (napDABGH), nrfA, and cymA genes are dependent on CRP1. Electrophoretic mobility shift assay (EMSA) analysis was further employed to assess the possible transcriptional regulation of nrfA, napD, and napE by CRP1. It is worth noting that purified CRP1-His (2.5 μM) in the range of 0 to 10 μl was initially employed to bind with the promoter regions of napE (PnapE), nrfA (PnrfA), and napD (PnapD) (Fig. 4), where the DNA was found to be completely bound with 10 μl of CRP1-His (2.5 μM), and this binding capacity was not observed without addition of cyclic AMP molecules (cAMP). Although there are no typical CRP1-bound motifs (aaaTGTGAtctagaTCACAttt) identified upstream of the nap-α (napEDABC) cluster (Fig. S2), in vitro CRP1 binding could still be observed in the EMSA analyses (Fig. 4). More importantly, there are no typical CRP1-binding motifs identified upstream of nirK (Fig. S2), and no CRP1 binding to the promoter sequence of nirK was observed in the EMSA analysis conducted as described above (Fig. 4), which is consistent with the chemical measurements of NO and N2O gases produced by the crp1 deletion mutant (Fig. 1 and 2) and with transcriptional profiling analysis of relevant genes (Fig. 3). These results indicate that the transcription of nirK is not directly regulated by CRP1.
FIG 4.
Electrophoretic mobility shift assay (EMSA) analysis on CRP1 binding to the promoter sequence of nap-α, nap-β, nrfA, and nirK. No binding of CRP1 with the nirK promoter sequence was observed in the presence of increasing levels of cyclic AMP. There was still binding observed in the nap-α promoter region, although there is no typical CRP1-recognized motif.
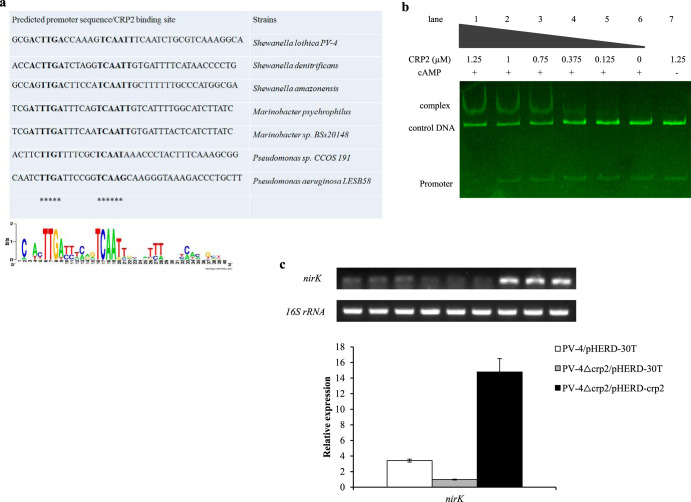
The binding of CRP2 to the promoter region of nirK.
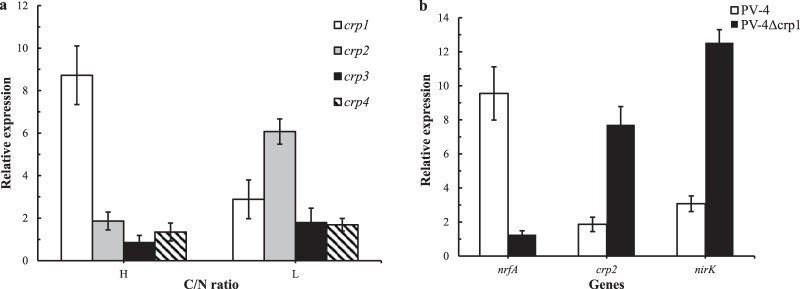
There are usually 2 to 4 CRP/FNR homologues encoded in all the sequenced genomes of different microbial species, and our comparative genomic analysis revealed that many Shewanella species contain CRP1 and CRP3. However, CRP2 and NirK/NirS are not encoded in most of the sequenced non-denitrification bacteria, such as Escherichia coli K-12, S. oneidensis MR-1, and S. putrefaciens W3-18-1. On the other hand, both CRP2 and NirK/NirS are encoded by the denitrifying bacteria such as S. loihica PV-4, S. denitrificans OS217, Shewanella amazonensis SB2B, Marinobacter psychrophilus, Marinobacter sp. BSs20148, Pseudomonas sp. CCOS 191, and Pseudomonas aeruginosa LESB58 (Table S2). Furthermore, we mapped the transcription start site (TSS) of nirK by using primer extension (see Fig. S7 in the supplemental material), and our sequence logo analyses of promoter sequences upstream of nirK have revealed the conserved motifs TTGA(N)6TCAATT upstream of nirK (Fig. 5a). In addition, we compared the transcription levels of CRP/FNR homologues in the denitrifying wild-type PV-4 strain under different C/N ratios by using real-time RCR and reverse transcription-PCR (RT-PCR). It was revealed that crp1 was upregulated under high C/N ratios, while crp2 was upregulated under low C/N ratios in the wild-type strain. When crp1 was deleted, the transcriptions of crp2 and nirK were increased dramatically, and the transcription of nrfA was not detected (Fig. 6), indicating that the transcription of nirK was dependent on CRP2. EMSA analysis was further employed to detect CRP2-promoter sequences interactions (Fig. 5b). Our results showed that when purified CRP2-His (2.5 μM) in the range of 0 to 10 μl was initially employed to bind with the promoter regions of nirK (PnirK) (Fig. 5b), the DNA was found to be completely bound with 10 μl of CRP2-His (2.5 μM), while this binding capacity was not observed without the addition of cyclic AMP molecules (cAMP). We conducted semiquantitative RT-PCR and real-time quantitative PCR analyses on the CRP2-induced transcription of nirK (Fig. 5c). Interestingly, the added inducer l-arabinose not only remarkably induced the expression of pHERD30T-borne crp2, but also in turn increased the transcription of nirK in the crp2 null mutant. These results indicate that nirK is transcriptionally dependent on CRP2/cAMP regulation.
FIG 5.
EMSA analysis on CRP2 binding to the promoter sequence of nirK. (a) Multiple alignment and sequence logo analyses of promoter sequences upstream of nirK in S. loihica PV-4, S. denitrificans, S. amazonensis, Marinobacter psychrophilus, Marinobacter sp. BSs20148, Pseudomonas sp. CCOS 191, and Pseudomonas aeruginosa LESB58. The conserved motifs are highlighted. The consensus sequence is TTGA(N)6TCAATT. (b) The binding of CRP2 with the nirK promoter sequence was observed in the presence of increasing levels of cyclic AMP. (c) Induced transcription of the plasmid-borne crp2 upregulated the expression of nirK in the in-frame deletion mutant Δcrp2, as revealed by reverse transcription-PCR (RT-PCR) (upper) and real-time PCR analyses (lower).
FIG 6.
(a) Quantitative PCR analyses on transcriptional profile of CRP/FNR homologues in the wild-type strain S. loihica PV-4 under high carbon/nitrogen ratios and low carbon/nitrogen ratios. H, C/N = 6; L, C/N = 2. (b) Quantitative PCR analyses of transcriptional profile of nrfA, crp2, and nirK in the wild-type strain S. loihica PV-4 and in-frame deletion mutant Δcrp under high carbon/nitrogen ratios. Gene expression was normalized against the 16S rRNA gene using the 2−ΔCT calculation as follows: ΔCT = CTgene of interest − CT16S rRNA. Results are the means of three replicates, and error bars indicate standard deviation.
Denitrification is upregulated under a lower C/N ratio.
Under the low C/N ratio (2:1, i.e., carbon source/electron donor limitation) condition, the reduction of nitrate to nitrite occurred quickly in the wild-type PV-4 strain (Fig. 2a). However, the reduction of nitrite by the wild-type strain, the PV-4ΔnirK mutant, and the double mutant was prolonged, probably due to the electron donor limitation (Fig. S3 and S4). Consistently, the ammonia levels remained around 0.6 mM in the culture of the PV-4 ΔnirK mutant or were even lower in other cultures under carbon source (also electron donor) limitation (Fig. 2c). The N2O emitted from the culture of wild-type PV-4 strain under the low C/N ratio condition (Fig. 2e) was almost 3-fold higher than those of a PV-4 Δcrp1 mutant culture under high C/N ratios typical of enough electron donor supply (Fig. 1e). On the other hand, the transcription of nrfA decreased under low C/N ratios in the wild-type PV-4 strain, while the expression of nirK, norBC, and nosZ increased (Fig. 3; see also Fig. S6 in the supplemental material), which was consistent with the increased release of NO and N2O in the wild-type PV-4 strain (Fig. 2d and e). By taking our real-time quantitative PCR results, chemical analyses, and previous study into account, we deduced that denitrification is upregulated under lower C/N ratios.
Bacterial growth of the wild-type strain and mutants under different C/N ratios.
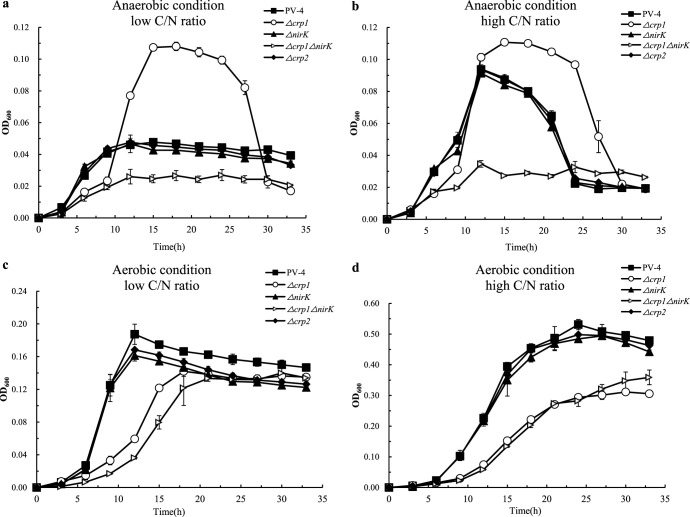
The growth rate and growth yield (maximum cell density) of the wild-type strain and the different mutants could be compared based on their population growth curves. The PV-4 ΔnirK Δcrp1 double mutant could hardly grow on nitrate because the reduction of nitrite to both ammonia and NO was blocked. Under different C/N ratios with anaerobic conditions, the wild-type strain (with DNRA and denitrification pathways) and the PV-4 ΔnirK mutant (with only the DNRA pathway) initially achieved higher growth rates and a growth yield comparable to that of the PV-4 Δcrp1 mutant (with only denitrification), although its initial growth yield was lower than that of the latter (Fig. 7a and b).
FIG 7.
Bacterial growth (optical density at 600 nm [OD600]) of the S. loihica PV-4, the in-frame deletion mutants Δcrp1, Δcrp2, and ΔnirK, and the Δcrp1 ΔnirK double mutant under different C/N ratios (high carbon/nitrogen ratio means a C/N ratio of 12 and low carbon/nitrogen ratio means a C/N ratio of 2) between anaerobic and aerobic conditions.
In other words, the DNRA pathway in the wild-type strain and the PV-4 ΔnirK mutant gave rise to higher growth rates but a slightly lower growth yield, while the denitrification pathway in the PV-4 Δcrp1 mutant, whose reduction of nitrite to ammonia was blocked in the absence of CRP1, led to slower initial growth and a higher growth yield. This trend was even more remarkable under both the low and high C/N ratio aerobic conditions (Fig. 7c and d) and was consistent with our bioenergetic calculations and analyses described below. Moreover, there was no growth yield difference under the low C/N ratio aerobic conditions, while under the high C/N ratio aerobic conditions, the growth yield of the wild-type PV-4 and the ΔnirK mutant were higher than those of the Δcrp1 mutant and the Δcrp1 ΔnirK double mutant. The Δcrp1 and Δcrp1 ΔnirK mutants only consume the ammonium in culture, and nitrate ammonification was blocked (see Fig. S8 in the supplemental material). Accordingly, there were no growth defects observed in the Δcrp1 or ΔnirK mutants or in the double mutant grown in the rich medium (LB broth) under aerobic condition (see Fig. S9 in the supplemental material). The deletion of crp1 did not affect the growth yields of the Δcrp1 mutant or the double mutant under the higher C (lactate)/N (ammonium) ratio conditions without supplement of nitrate (see Fig. S10 in the supplemental material). These results indicate that the CRP1/cAMP system may play an important role in sensing variation of C/N ratios.
Bioenergetics calculations and analyses.
Using a similar method as previously described (5), we calculated the free energy produced by DNRA and denitrification per molar nitrate (electron acceptor) and per molar lactate (electron donor), respectively (Equations 1 and 2). Theoretically, the free energy generated by denitrification is slightly lower than that of DNRA for ATP synthesis, but this process consumes fewer electrons (5 electrons per nitrogen and 10 electrons per N2) for reduction of nitrate (chemical valence, +5) to N2 (valence, 0). On the other hand, DNRA consumes more electrons (8 electrons per nitrogen and 16 electrons per two molecules of NH4+) for reduction of nitrate to ammonia (valence, −3) and generates slightly higher free energy at the expense of more carbon source (electron donor) (see Fig. S11 in the supplemental material). The free energy per molar lactate produced by denitrification (−1,236.5 kJ) is higher than that of DNRA (−836.6 kJ) by almost 30%, which is in agreement with their difference in the electron numbers consumed for reduction of equivalent amount of nitrate. When carbon source (and also electron donor) levels are relatively high, the DNRA pathway dominated, as shown by previous and current studies, because DNRA pathway generates more energy. On the other hand, the total energy generated by denitrification could be higher when carbon source (electron donor) is insufficient and becomes the limiting factor and nitrate (electron acceptor) is relatively high.
| (1) |
| (2) |
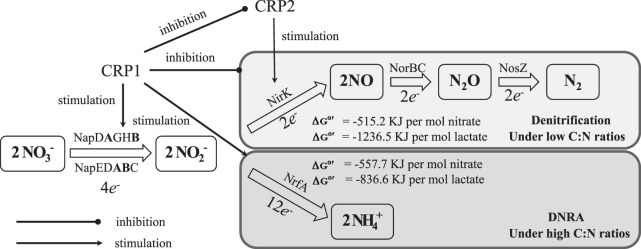
Taken together, our analyses demonstrate that an exquisite regulatory machinery, dependent on CRP paralogues and cellular cAMP, has been well evolved in bacteria to cope with changes of C/N ratios for a better choice between two different DNR pathways, DNRA and denitrification, and for the optimization and balance of both energy generation and cell growth (biosynthesis of biomolecules) under specific circumstances (Fig. 8). In other words, S. loihica and/or other bacteria could make the most of the limiting factor (either electron donor or electron acceptor) for maximal energy generation and growth yield by leveraging the different DNR pathways based on the immediate ambient C/N ratios.
FIG 8.
Schematic diagram of the regulation of CRP1 on the dissimilatory nitrate reduction to ammonia and denitrification. The positive regulation of CRP1 on transcription of nap-α and nrfA is mediated by its direct binding to specific motifs at the promoter region, while the negative regulation on nirK, norBC, and nosZ may be exerted via another unknown regulator, probably another CRP paralogue, CRP2. The ultimate goal is to make the most of the limiting factor.
DISCUSSION
A series of in-frame deletion mutants had been generated successfully in S. loihica PV-4 to study the regulatory role played by CRP paralogues in both the DNRA and denitrification pathways. By means of monitoring the nitrate, nitrite, ammonium, NO, and N2O levels and profiling relevant gene transcription in different test groups, including wild-type S. loihica PV-4 strain and a series of homogenic in-frame deletion mutants under both high and low C/N ratios, we have demonstrated that CRP paralogues play a central role in modulating the two competing dissimilatory nitrate reduction pathways, DNRA and denitrification. Our findings are consistent with previous analyses of the wild-type PV-4 strain under similar circumstances (7). It was demonstrated that DNRA was predominant and the final reduction product was mainly ammonia under the high C/N ratio condition, while denitrification took over under the low C/N ratio condition (carbon source and electron donor limitation), and high levels of N2O emission were detected. More importantly, we have further shown that the common cyclic AMP receptor protein CRP1, shared by Shewanella, Escherichia, Pseudomonas, and many other bacteria, is required for the relatively rapid reduction of nitrate to nitrite and further reduction of nitrite to ammonia (DNRA), but not for denitrification. Deletion of crp1 almost turned off DNRA and instead switched on denitrification, which was not active under high C/N ratios. Under the low C/N ratio condition, the wild-type PV-4 strain produced much more N2O, severalfold more than that of the PV-4 Δcrp1 mutant under the high C/N ratio condition (see Fig. S12 in the supplemental material). It seems that CRP1 not only activates DNRA, but also inhibits denitrification under high carbon source levels. CRP1 may titrate the two dissimilatory nitrate reduction pathways, DNRA and denitrification, based on the ambient C/N ratios, perhaps due to the differential biosynthesis of the second messenger cAMP. That is to say, high carbon source levels may lead to higher synthesis of cellular cAMP by the adenylate cyclases (18–20). cAMP molecules bind to CRP1 to activate the CRP1-dependent transcription of nap-β, cymA, and nrfA for DNRA, which requires more electrons per nitrogen atom (8 electrons), and to repress the expression of nirK stimulated by CRP2 for denitrification via a yet-unknown mechanism. The produced ammonium could still be assimilated later for synthesis of amino acids and other molecules for cell growth. On the other hand, under low C/N ratios, the carbon source (also electron donor) is limited, the synthesis of ATP and cAMP signaling molecules could be affected, and therefore CRP1 could not upregulate the transcription of the nap-β gene cluster, cymA, and nrfA for DNRA. Most likely, without enough cAMP molecules, CRP1 could not be activated and could no longer suppress the transcription of the denitrification-related genes, such as nirK, via another yet-unknown transcriptional regulator.
CRP1 represses transcription at the nirB promoter by blocking either the interaction between RNA polymerase and the promoter DNA or transcription from an alternative upstream promoter in E. coli (21). However, we have not found the typical CRP1-recognized motifs, which are present upstream of nap-β, nrfA, and cymA in Shewanella species (13), in the promoter region of nirK in S. loihica PV-4. Transcriptional suppression may be achieved indirectly via another regulator. The carbon source limitation may result in lower synthesis of cAMP and slower activation of DNRA. Nevertheless, the CRP1-mediated suppression of denitrification is relieved. Obviously, an efficient and precise regulatory circuit has evolved in S. loihica PV-4 for modulating the dissimilatory nitrate reduction pathways based on the availability of carbon source (energy source and electron donors) sensed by CRP1, as well as on oxygen availability sensed by FNR (20, 22) and availability of ambient nitrate and nitrite compounds (electron acceptors) sensed by the NarPQ two-component system (11, 23–25). The final bacterial choice-making is based on the integration of various signal inputs from CRP1, NarPQ, and FNR sensing their cognate signals, carbon source, nitrate/nitrite, and oxygen. Interestingly, our comparative genomic analysis and experimental evidence have revealed that another CRP/FNR paralogue, CRP2, is required for the transcription of nirK, and there are the typical CRP2-bound motifs (gacTTGAccaaagTCAATTt) identified upstream of nirK. Meanwhile, CRP2 and NirK/NirS coincidently coexist in most of the denitrifying bacteria. Our result suggested that CRP1 might repress the transcription of CRP2 transcription via a yet-unknown mechanism; therefore, we suspected that CRP2 might compete with CRP1 on cAMP when the synthesis of ATP and cAMP signaling molecules is limited under low C/N ratios. On the other hand, under high C/N ratios, cAMP molecules might bind to CRP1 to repress transcription of crp2, and nirK transcription is not activated. It was shown that the regulatory circuits involved in the nitrogen metabolism are more complex than previously expected.
To summarize, our results demonstrate that paralogues of CRP (CRP1 and CRP2), the widespread receptor protein for the second messenger cyclic AMP, play a crucial role in sensing changes of C/N ratios and regulating the bacterial choice of two competing dissimilatory nitrate reduction pathways, DNRA and denitrification, in Shewanella spp. for maximal energy generation and optimal adaption to specific ambient conditions (Fig. 6). There exists a tradeoff between the physiological functions of both lactate (or other carbon sources) and nitrate (or nitrite). In our experiments, lactate acted as both the sole carbon source for cell growth (biomass) and the electron donor for energy generation. Although nitrate was used as the electron acceptor and both DNRA and denitrification are dissimilatory nitrate reduction processes whose products are not immediately assimilated, the final product, ammonium, is still available subsequently for bacterial uptake as a nitrogen nutrient, whereas NO, N2O, and particularly N2 are unavailable for most of the bacteria. Moreover, there also exists a tradeoff between bacterial growth rate (favorable for competition) and growth yield (for survival), which may be favored under specific conditions. These results could shed light on the understanding of microbe-driven biogeochemistry cycles of nitrogen and provide important implications for efficient control of emission of N2O to the atmosphere from both the hydrosphere and the soil sphere (4). More importantly, our analyses provide an insight into the molecular evolution relevant to the bacterial respiration flexibility of Shewanella, a genus with potential applications in bioremediation and bioenergy.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. loihica PV-4 and mutants were cultured in Luria-Bertani broth/plates or modified M1 minimal medium at 28°C (supplemented with 15 μg/ml of gentamicin when necessary) (9). Since genetic manipulation of the wild-type PV-4 strain is difficult, possibly due to the presence of a PstI restriction-modification system, we created a ΔpstI ΔpstM double in-frame deletion mutant, which was used as a parental strain (PV-4 refers to the parental strain here) for the subsequent tests (14, 26). Escherichia coli strains were cultured at 37°C in LB broth. For the nitrate, nitrite, and ammonium detection and bacterial growth experiments with S. loihica strain PV-4, the completely synthetic, anoxic, phosphate-buffered basal salt medium adjusted to a pH of 7.0 was prepared as previously described (7, 27, 28). Sodium nitrate (2 mM), ammonium (0.5 mM), and various concentrations of sodium lactate were supplemented as electron acceptors and nitrogen and carbon sources, respectively. For N2O detection, the wild-type PV-4 strain and mutants were grown in 12-ml vials (Labco, UK) containing 12 ml of culture broth. The Hungate technique was used to dispense medium into N2-flushed vials, which were sealed with black butyl stoppers and autoclaved. For NO detection, the wild-type strain and mutants were grown in 300-ml glass bottles containing 100 ml of culture broth, and O2 was removed by 30 min of N2 flushing. The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli WM3064 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-1360 (araBAD)567 dapA1341::[erm pir(wt)] | W. Metcalf |
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| E. coli BL21(DE3) | F− ompT gal dcm lon hsdSB (rB−mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Novogene |
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF) U169, hsdR17(rK− mK+) phoA λ− | TaKaRa |
| Modified Shewanella loihica PV-4 | pstI (Shew_0993) and pstM (Shew_0992) double deletion mutant derived from PV-4 | 14 |
| Shewanella loihica PV-4 | Iron-rich microbial mat at a hydrothermal vent of Loihi Seamount, Pacific Ocean | 15 |
| PV-4 Δcrp2 | In-frame deletion mutant of crp2 gene (Shew_3331) derived from the modified PV-4 | This study |
| PV-4 ΔnirK | In-frame deletion mutant of nirK gene (Shew_3335) derived from modified PV-4 | This study |
| PV-4 Δcrp ΔnirK | In-frame deletion double mutant of crp (Shew_0585) and nirK (Shew_3335) derived from modified PV-4 | This study |
| Plasmids | ||
| pDS3.0 | Suicide vector derived from pCDV224; Ampr, Gmr, sacB | 18 |
| pDS3.0-PV-crpko | Suicide plasmid for deletion of crp derived from PV-4 | This study |
| pDS3.0-PV-nirKko | Suicide plasmid for deletion of nirK derived from PV-4 | This study |
| pET28a | Expression vector with T7 lac promoter | Novogene |
| pHERD30T | Shuttle vector with pBAD promoter, Gmr | 44 |
| pET28a-crp1 | Overexpression construct of crp | This study |
Gmr, gentamycin resistance; Ampr, ampicillin resistance; Smr, streptomycin resistance.
In-frame deletion and genetic complementation.
The two-step protocol of selection (single crossover and antibiotic resistance) and counterselection (double crossover and sucrose sensitivity) was applied for in-frame deletion of specific genes by using suicide vector pDS3.0 (R6K replicon, sacB, gentamycin resistance [Gmr])-based constructs carrying a fusion of upstream and downstream sequences of target genes as previously described (29). The suicide vector was introduced into S. loihica PV-4 by mating using E. coli WM3064 as the donor strain. The primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer or purpose | Oligonucleotide sequence (5′–3′) |
|---|---|
| Mutagenesis | |
| PV4-crp1_5O | 5′-GTGAGCTCCTGTCTACATGGAGGTGGCC-3′ |
| PV4-crp1_5I | 5′-CAAAAGCACTCTGATCCATGAGATTGGTCAGATCGTGG-3′ |
| PV4-crp1_3I | 5′-CCACGATCTGACCAATCTCATGGATCAGAGTGCTTTTG-3′ |
| PV4-crp1_3O | 5′-GTGAGCTCCTGCACGGCAAACAGGGCCAG-3′ |
| PV4-crp1_lf | 5′-GCGTTCGACCCAGTTGGCAC-3′ |
| PV4-crp1_lr | 5′-GCGTTCGACCCAGTTGGCAC-3′ |
| PV-4-crp2-3O | 5′-GTGAGCTCGCTCAAGGCTATCACTAGTC-3′ |
| PV-4-crp2-3I | 5′-GACTCGCCGATGATCCCTTAGGTGCGCCCCTCGCGCAC-3′ |
| PV-4-crp2-5I | 5′-GTGCGCGAGGGGCGCACCTAAGGGATCATCGGCGAGTC-3′ |
| PV-4-crp2-5O | 5′-GTGAGCTCGGACTATCTCACTGGGCTATG-3′ |
| PV4-nirK_5O | 5′-AGAGCTCTTGTCAGGAAGACAGGCGTAG-3′ |
| PV4-nirK_5I | 5′-TCAGGTTGTCATCCCACTCGACGACGTTGCTGATCGACATT-3′ |
| PV4-nirK_3I | 5′-AATGTCGATCAGCAACGTCGTCGAGTGGGATGACAACCTGA-3′ |
| PV4-nirK_3O | 5′-AGAGCTCGCGAGCGATAGGGGTGATAG-3′ |
| PV4-nirK_lf | 5′-GAGTTCGATGTGTTGGCGAC-3′ |
| PV4-nirK_lr | 5′-ACTCGTTGACCAGAGTGACG-3′ |
| qRT-PCRa | |
| QRT-napC-F | 5′-TTACCTTAGGCGGCTTCG-3′ |
| QRT-napC-R | 5′-GCATCTTACGGGCTATCTTAT-3′ |
| QRT-napG-F | 5′-GGTCAATCGCAATAGCAAAG-3′ |
| QRT-napG-R | 5′-CCAGTTTCAGGGTGTCGTAG-3′ |
| QRT-nrfA-F | 5′-AGCACCCTGGTTATGAAA-3′ |
| QRT-nrfA-R | 5′-GAGTATGACAAGTCGCACA-3′ |
| QRT-nirK-F | 5′-AAGCCGACTTCTATGTGCC-3′ |
| QRT-nirK-R | 5′-TGATGTGCGGACGGGTGT-3′ |
| QRT-norBC-F | 5′-AAGCCAGCCGAGCAAGAT-3′ |
| QRT-norBC-R | 5′-CCAGGTACGAGTAACCGAGT-3′ |
| QRT-nosZ-F | 5′-CAGATAAGATGATCACCATC-3′ |
| QRT-nosZ-R | 5′-CGCTCGGCGGTGATCATCTCG-3′ |
| QRT-crp1-F | 5′-AGGGTTCTGTTGCCGTCTT-3′ |
| QRT-crp1-R | 5′-CTTTCTGGCTGGTGCTTTG-3′ |
| QRT-crp2-F | 5′-GCGCTGATGTTTAGTAGGC-3′ |
| QRT-crp2-R | 5′-GGGAAAAAGTTTCTGGCTG-3′ |
| QRT-crp3-F | 5′-GTGATAGGTTTCGATGGCA-3′ |
| QRT-crp3-R | 5′-AGACGGAACTCTTTGGGCG-3′ |
| QRT-16S-F | 5′-GGTTGATTAAGCGAGATGTG-3′ |
| QRT-16S-R | 5′-TGAGCGTCAGTCTTTGTCC-3′ |
| Primer extension | |
| oligo(dT) | 5′-GCCAGTCGTTTTTTTTTTTTTTTTT-3′ |
| nirK-1R | 5′-CTCCACCTGCTTCTCTTCTAC-3′ |
| nirK-2R | 5′-CTTCACCTGCACCACCTTAG-3′ |
| Other purposes | |
| gfp-F | 5′-GCACTACTGGAAAACTACCT-3′ |
| gfp-R | 5′-CAAGAAGGACCATGTGGTCTC-3′ |
| PV-4-crp2-F | 5′-GGAATTCCTGTTCATTATGATCTAGA-3′ |
| PV-4-crp2-R | 5′-GCTGCAGAGGCTACACCATCTCAATAG-3′ |
| nrfA-promoter-F | 5′-GCCTTTCTTCATGTATTAG-3′ |
| nrfA-promoter-R | 5′-GTCTTAAAAACCACAACATC-3′ |
| napD-promoter-F | 5′-GTGTCATAGCAAGCTCCCTC-3′ |
| napD-promoter-R | 5′-CTTGAGAGGTAACTGGAGCT-3′ |
| napE-promoter-F | 5′-CGCCATTTTGTGACTATGT-3′ |
| napE-promoter-R | 5′-GCAGATAAAGCAGGCTAGC-3′ |
| cymA-promoter-F | 5′-CTCGAAAATCCAACAAAATC-3′ |
| cymA-promoter-R | 5′-CACTCTATCTCCAAAATAATATG-3′ |
qRT-PCR, reverse transcription-quantitative PCR.
Measurements of nitrate, nitrite, ammonium, nitric oxide, and nitrous oxide.
In S. loihica PV-4, nitrite could be produced from nitrate by NAP nitrate reductases. The concentrations of nitrate, nitrite, and ammonium in the medium were measured simultaneously using a standard colorimetric method (30). For measurement of produced N2O, 1 ml of culture broth was pipetted from the 12-ml vial into another empty 12-ml vial (Labco, UK). After liquid-and-air equilibrium was reached, the concentration of released NO gas in the headspace (31–39) and then that of the gas was measured by using the Agilent 7890B gas chromatography (GC) System (Santa Clara, CA) configured with a 63Ni electron capture detector (μECD); helium gas was used as the carrier gas at a flow rate of 6.5 ml · min−1 (31). The temperatures of the injector, column, and detector were 100, 70, and 320°C, respectively. NO gas concentration was determined by using a model 42i NO-NO2-NOx Analyzer (Thermo Scientific, Franklin, MA) according to the manufacturer’s protocol (31).
RNA extraction and real-time quantitative PCR and RT-PCR transcriptional analysis.
Total RNA was extracted using RNAiso Plus (TaKaRa) and an RNAprep Pure cell/bacteria kit (Tiangen Biotech Co., Ltd., Beijing, China). RNA was further purified using a DNase I treatment. The integrity of RNA was evaluated by agarose (1%) gel electrophoresis. The RNA concentration and purity were measured on a spectrophotometer (Nanodrop Technologies, Wilmington, DE), and reverse transcribed into cDNA using the PrimeScript reverse transcription (RT) reagent kit with genomic DNA (gDNA) Eraser (TaKaRa) according to the manufacturer’s protocol. The relative gene expression levels were quantified using SYBR Premix DimerEraser (TaKaRa) on a Roche LightCycler 480 II real-time PCR system (Roche Diagnostics, Penzberg, Germany). The expression levels of all of the genes were normalized to 16S rRNA gene expression as an internal standard and quantified according to a previously reported method (40, 41). Semiquantitative PCR analyses were carried out as described previously (14, 42). All experiments were performed in triplicate. The PCR products were also sequenced to confirm amplification of target genes. The primers used are listed in Table 2.
Expression and purification of His-tagged CRP recombinant protein.
The crp1 or crp2 fragment was amplified from genomic DNA of the PV-4 strain by using PCR and was cloned into pET-28a. The resulting plasmid pET28a-crp1 or pET28a-crp2 was transformed into E. coli BL21(DE3), and the His-tagged CRP1 or CRP2 recombinant protein (CRP1-His or CRP2-His) was overexpressed, extracted, and purified as previously described (42). The concentration of purified CRP1-His or CRP2-His was calculated using a protein assay kit (Jiancheng Biotech., Nanjing, China). The CRP1-His or CRP2-His proteins were stored in glycerol at −20°C.
Electrophoretic mobility shift assay.
An electrophoretic mobility shift assay (EMSA) was performed as previously described (43) with minor modifications. DNA templates containing respective promoters of nirK, napE, nrfA, and napD were obtained from genomic DNA of the PV-4 strain by PCR amplification. Different amounts (∼0 to 10 μl) of purified CRP1-His or CRP2-His (2.5 μM) were used in each reaction mixture. One DNA fragment (0.1 μM each), with approximately 500 bp of the gfp gene amplified from pMD18T-gfp used as the control DNA. After 30 to 45 min of incubation at room temperature, the samples were then loaded onto a 5% (wt/vol) native polyacrylamide gel at 4°C and electrophoresed in Tris-borate buffer for 1.5 h at 120 V. Subsequently, the gels were stained with ethidium bromide (Sangon Biotech Co., Shanghai, China) at 4°C. DNA and DNA-protein complexes were visualized using the Gel Doc XR+ system (Bio-Rad Laboratories, Inc., UK).
Determination of transcription start site.
Terminal deoxynucleotidyl transferase (TdT; TaKaRa) was used to catalyze the incorporation of single deoxynucleotides (dATPs) into the 3′-OH terminus of cDNA to make the dA-tailed cDNA according to the producer’s protocol. Touchdown and nested PCR were used to amplify the dA-tailed cDNA by using an oligo(dT) (5′-gccagtcTTTTTTTTTTTTTTTTT-3′) primer and a specific primer. The PCR product was cloned into the pMD18-T vector (TaKaRa, Dalian, China) for sequencing.
Bioinformatics tools.
The Clustal W2 package (https://www.ebi.ac.uk/Tools/msa/clustalw2/) was used for nucleotide sequence alignments and phylogenetic footprinting analyses of promoters, and WebLogo (http://weblogo.berkeley.edu/logo.cgi) was applied to nucleotide sequence motif identification.
Data availability.
All data are included in this article and the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Foundation for Natural Sciences of China (grant 91751106), the Chinese Academy of Science (grants XDA23040401 and Y15103-1-401), and a One-Hundred Scholar Award to D.Q.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jurado A, Borges AV, Brouyere S. 2017. Dynamics and emissions of N2O in groundwater: a review. Sci Total Environ 584-585:207–218. doi: 10.1016/j.scitotenv.2017.01.127. [DOI] [PubMed] [Google Scholar]
- 2.Sparacino-Watkins C, Stolz JF, Basu P. 2014. Nitrate and periplasmic nitrate reductases. Chem Soc Rev 43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson DJ, Watmough NJ. 1999. Inorganic nitrogen metabolism in bacteria. Curr Opin Chem Biol 3:207–219. doi: 10.1016/S1367-5931(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 4.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61:533–616. doi: 10.1128/.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strohm TO, Griffin B, Zumft WG, Schink B. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol 73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon S, Sanford RA, Löffler FE. 2015. Nitrite control over dissimilatory nitrate/nitrite reduction pathways in Shewanella loihica strain PV-4. Appl Environ Microbiol 81:3510–3517. doi: 10.1128/AEM.00688-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon S, Cruz-García C, Sanford R, Ritalahti KM, Löffler FE. 2015. Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3−/NO2− reduction pathways in Shewanella loihica strain PV-4. ISME J 9:1093–1104. doi: 10.1038/ismej.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H, Park D, Yoon S. 2017. pH Control enables simultaneous enhancement of nitrogen retention and N2O reduction in Shewanella loihica strain PV-4. Front Microbiol 8:1820. doi: 10.3389/fmicb.2017.01820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu D, Wei H, Tu Q, Yang Y, Xie M, Chen J, Pinkerton MH, Liang Y, He Z, Zhou J. 2013. Combined genomics and experimental analyses of respiratory characteristics of Shewanella putrefaciens W3-18-1. Appl Environ Microbiol 79:5250–5257. doi: 10.1128/AEM.00619-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Wang F, Xu J, Mehmood MA, Xiao X. 2011. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME J 5:843–855. doi: 10.1038/ismej.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y, Wang J, Fu H, Zhou G, Shi M, Gao H. 2012. A Crp-dependent two-component system regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS One 7:e51643. doi: 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson PJ, Richardson DJ, Codd R. 2010. The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology (Reading) 156:302–312. doi: 10.1099/mic.0.034421-0. [DOI] [PubMed] [Google Scholar]
- 13.Saffarini DA, Schultz R, Beliaev A. 2003. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol 185:3668–3671. doi: 10.1128/jb.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu D, Xie M, Dai J, An W, Wei H, Tian C, Kempher ML, Zhou A, He Z, Gu B, Zhou J. 2016. Differential regulation of the two ferrochelatase paralogues in Shewanella loihica PV-4 in response to environmental stresses. Appl Environ Microbiol 82:5077–5088. doi: 10.1128/AEM.00203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-García C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. 2007. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189:656–662. doi: 10.1128/JB.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH, Fredrickson JK, Tiedje JM, Zhou J. 2009. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- 17.Wei H, Dai J, Xia M, Romine MF, Shi L, Beliav A, Tiedje JM, Nealson KH, Fredrickson JK, Zhou J, Qiu D. 2016. Functional roles of CymA and NapC in reduction of nitrate and nitrite by Shewanella putrefaciens W3-18–1. Microbiology (Reading) 162:930–941. doi: 10.1099/mic.0.000285. [DOI] [PubMed] [Google Scholar]
- 18.Stewart V, Bledsoe PJ, Chen L-L, Cai A. 2009. Catabolite repression control of napF (periplasmic nitrate reductase) operon expression in Escherichia coli K-12. J Bacteriol 191:996–1005. doi: 10.1128/JB.00873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charania M, Brockman K, Zhang Y, Banerjee A, Pinchuk GE, Fredrickson JK, Beliaev AS, Saffarini D. 2009. Involvement of a membrane-bound class III adenylate cyclase in regulation of anaerobic respiration in Shewanella oneidensis MR-1. J Bacteriol 191:4298–4306. doi: 10.1128/JB.01829-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy JN, Durbin KJ, Saltikov CW. 2009. Functional roles of arcA, etrA, cyclic AMP (cAMP)-cAMP receptor protein, and cya in the arsenate respiration pathway in Shewanella sp. strain ANA-3. J Bacteriol 191:1035–1043. doi: 10.1128/JB.01293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng D, Constantinidou C, Hobman JL, Minchin SD. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res 32:5874–5893. doi: 10.1093/nar/gkh908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao H, Wang X, Yang ZK, Chen J, Liang Y, Chen H, Palzkill T, Zhou J. 2010. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS One 5:e15295. doi: 10.1371/journal.pone.0015295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darwin AJ, Tyson KL, Busby SJ, Stewart V. 1997. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K‐12 depends on DNA binding site arrangement. Mol Microbiol 25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- 24.Rabin RS, Stewart V. 1992. Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc Natl Acad Sci U S A 89:8419–8423. doi: 10.1073/pnas.89.18.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang RC, Cavicchioli R, Gunsalus RP. 1992. Identification and characterization of narQ, a second nitrate sensor for nitrate‐dependent gene regulation in Escherichia coli. Mol Microbiol 6:1913–1923. doi: 10.1111/j.1365-2958.1992.tb01364.x. [DOI] [PubMed] [Google Scholar]
- 26.Gao H, Obraztova A, Stewart N, Popa R, Fredrickson JK, Tiedje JM, Nealson KH, Zhou J. 2006. Shewanella loihica sp. nov., isolated from iron-rich microbial mats in the Pacific Ocean. Int J Syst Evol Microbiol 56:1911–1916. doi: 10.1099/ijs.0.64354-0. [DOI] [PubMed] [Google Scholar]
- 27.Myers CR, Nealson KH. 1990. Respiration-linked proton translocation coupled to anaerobic reduction of manganese (IV) and iron (III) in Shewanella putrefaciens MR-1. J Bacteriol 172:6232–6238. doi: 10.1128/jb.172.11.6232-6238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon S, Sanford RA, Löffler FE. 2013. Shewanella spp. use acetate as an electron donor for denitrification but not ferric iron or fumarate reduction. Appl Environ Microbiol 79:2818–2822. doi: 10.1128/AEM.03872-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan X-F, Verberkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu L, Liu X, Yan T, Leaphart A, Hettich RL, Zhou J, Thompson DK. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J Bacteriol 186:8385–8400. doi: 10.1128/JB.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ministry of Environmental Protection of the People’s Republic of China. 2002. Wastewater monitoring methods. Chinese Environmental Science Publishing House, Beijing, China: (In Chinese.) [Google Scholar]
- 31.Ma L, Tong W, Chen H, Sun J, Wu Z, He F. 2018. Quantification of N2O and NO emissions from a small-scale pond-ditch circulation system for rural polluted water treatment. Sci Total Environ 619-620:946–956. doi: 10.1016/j.scitotenv.2017.11.192. [DOI] [PubMed] [Google Scholar]
- 32.Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MS, Yin C. 2011. Anaerobic ammonia oxidation in a fertilized paddy soil. ISME J 5:1905–1912. doi: 10.1038/ismej.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Zhu G, Peng Y, Jetten MS, Yin C. 2012. Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environ Sci Technol 46:8834–8842. doi: 10.1021/es3017446. [DOI] [PubMed] [Google Scholar]
- 34.Zhu G, Wang S, Feng X, Fan G, Jetten MS, Yin C. 2011. Anammox bacterial abundance, biodiversity and activity in a constructed wetland. Environ Sci Technol 45:9951–9958. doi: 10.1021/es202183w. [DOI] [PubMed] [Google Scholar]
- 35.Zhu G, Wang S, Wang W, Wang Y, Zhou L, Jiang B, Op den Camp HJM, Risgaard-Petersen N, Schwark L, Peng Y, Hefting MM, Jetten MSM, Yin C. 2013. Hotspots of anaerobic ammonium oxidation at land-freshwater interfaces. Nature Geosci 6:103–107. doi: 10.1038/ngeo1683. [DOI] [Google Scholar]
- 36.Nie S, Li H, Yang X, Zhang Z, Weng B, Huang F, Zhu GB, Zhu YG. 2015. Nitrogen loss by anaerobic oxidation of ammonium in rice rhizosphere. ISME J 9:2059–2067. doi: 10.1038/ismej.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu G, Jetten MS, Kuschk P, Ettwig KF, Yin C. 2010. Potential roles of anaerobic ammonium and methane oxidation in the nitrogen cycle of wetland ecosystems. Appl Microbiol Biotechnol 86:1043–1055. doi: 10.1007/s00253-010-2451-4. [DOI] [PubMed] [Google Scholar]
- 38.Thamdrup B, Dalsgaard T. 2002. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol 68:1312–1318. doi: 10.1128/aem.68.3.1312-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu G, Wang S, Wang C, Zhou L, Zhao S, Li Y, Li F, Jetten MSM, Lu Y, Schwark L. 2019. Resuscitation of anammox bacteria after >10,000 years of dormancy. ISME J 13:1098–1109. doi: 10.1038/s41396-018-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 42.Dai J, Wei H, Tian C, Damron FH, Zhou J, Qiu D. 2015. An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol 15:34. doi: 10.1186/s12866-015-0357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J-P, Zhang W-M, Chao H-J, Zhou N-Y. 2017. PnpM, a LysR-type transcriptional regulator activates the hydroquinone pathway in para-nitrophenol degradation in Pseudomonas sp. strain WBC-3. Front Microbiol 8:1714. doi: 10.3389/fmicb.2017.01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in this article and the supplemental material.