Abstract
Aquatic environments with high levels of dissolved ferrous iron and low levels of sulfate serve as an important systems for exploring biogeochemical processes relevant to the early Earth. Boreal Shield lakes, which number in the tens of millions globally, commonly develop seasonally anoxic waters that become iron rich and sulfate poor, yet the iron–sulfur microbiology of these systems has been poorly examined. Here we use genome-resolved metagenomics and enrichment cultivation to explore the metabolic diversity and ecology of anoxygenic photosynthesis and iron/sulfur cycling in the anoxic water columns of three Boreal Shield lakes. We recovered four high-completeness and low-contamination draft genome bins assigned to the class Chlorobia (formerly phylum Chlorobi) from environmental metagenome data and enriched two novel sulfide-oxidizing species, also from the Chlorobia. The sequenced genomes of both enriched species, including the novel “Candidatus Chlorobium canadense”, encoded the cyc2 gene that is associated with photoferrotrophy among cultured Chlorobia members, along with genes for phototrophic sulfide oxidation. One environmental genome bin also encoded cyc2. Despite the presence of cyc2 in the corresponding draft genome, we were unable to induce photoferrotrophy in “Ca. Chlorobium canadense”. Genomic potential for phototrophic sulfide oxidation was more commonly detected than cyc2 among environmental genome bins of Chlorobia, and metagenome and cultivation data suggested the potential for cryptic sulfur cycling to fuel sulfide-based growth. Overall, our results provide an important basis for further probing the functional role of cyc2 and indicate that anoxygenic photoautotrophs in Boreal Shield lakes could have underexplored photophysiology pertinent to understanding Earth’s early microbial communities.
Subject terms: Water microbiology, Microbial ecology, Biogeochemistry, Limnology, Bacterial genetics
Introduction
Precambrian Shield regions in northern areas globally contain millions of small humic lakes [1–3]. Many of these Boreal Shield lakes develop a seasonally anoxic water column that becomes ferruginous, dominated by dissolved ferrous iron compared to sulfate and sulfide [4, 5]. However, little is known about the iron and sulfur microbiology of seasonally anoxic and ferruginous Boreal Shield lake systems. Several studies have examined the role of phototrophic members of the class Chlorobia (formerly phylum Chlorobi [6]; also called “green sulfur bacteria”) in boreal lake waters (e.g., [7]), because these organisms are commonly found at high relative abundances in the illuminated and anoxic water columns of boreal lakes. Chlorobia members are typically assumed to use a reduced sulfur compound, such as sulfide or thiosulfate, as their photosynthetic electron donor [8, 9]. However, recent cultivation-based discoveries have expanded the known metabolic diversity of anoxygenic photosynthesis to include the photosynthetic oxidation of compounds such as ferrous iron, arsenate, and nitrite, and these alternative metabolic modes of photosynthesis challenge classical interpretations of how anoxic ecosystems function [10–13].
Among the Chlorobia class, diverse ferrous iron-oxidizing phototrophs, also called photoferrotrophs, have been cultivated recently. Previously, the only known photoferrotroph in this group was Chlorobium ferrooxidans strain KoFox, which was enriched from freshwater lake sediments [14]. Recently, Chlorobium phaeoferrooxidans strain KB01, a bacteriochlorophyll e-containing member of the class Chlorobia, was isolated from the anoxic water column of the meromictic and ferruginous Kabuno Bay [15]. In addition, Chlorobium sp. strain N1 was enriched from marine sediments [16]. Both Chl. ferrooxidans and Chl. phaeoferrooxidans oxidize ferrous iron as their sole photosynthetic electron donor, assimilating sulfur as sulfate [14, 15]. Chl. sp. N1 is capable of oxidizing either ferrous iron or sulfide as the photosynthetic electron donor and can also utilize organic compounds [16]. A fourth recently isolated photoferrotroph, Chlorobium sp. BLA1, was shown to also be capable of oxidizing sulfide, although genome data are not yet available for this strain [17]. Phylogenetically, Chl. ferrooxidans and Chl. phaeoferrooxidans are sister groups to one another, whereas Chl. sp. N1 is closely related to Chl. luteolum, a sulfide-oxidizing species of Chlorobia hypothesized to be able to switch to photoferrotrophic growth [15, 16, 18]. These cultivation-based discoveries have highlighted the potential metabolic diversity and flexibility of Chlorobia members. Discovery of additional photoferrotrophs has also stimulated discussion around the potential involvement of photoferrotrophs in early Earth microbial communities, such as the ferruginous water columns of Archaean Eon oceans (ca. 3.8–2.5 Ga) [15, 19, 20].
Despite growing evidence for the metabolic diversity of anoxygenic photosynthesis, the ecology of photoferrotrophy and other alternative photosynthetic modes remains poorly understood. One of the key challenges in assessing the modern importance of photoferrotrophy has been a limited understanding of the biochemistry and genetics of iron oxidation. Recent work has begun to unravel the molecular basis for iron oxidation in photoferrotrophs and other neutrophilic iron-oxidizing bacteria (reviewed in [21, 22]). Genes encoding for porin-cytochrome c protein complexes (PCC), such as pioAB and mtoAB, have been identified as potentially useful markers for iron oxidation in some species [22–24]. In addition, genes encoding for outer-membrane monoheme c-type cytochromes, such as cyc2, have received recent interest for their role in extracellular electron transfer, with proteomic and RNA-seq studies indicating a role for such genes in microaerophilic iron oxidation [25–27]. Among the Chlorobia, genome sequences available for Chl. ferrooxidans, Chl. phaeoferrooxidans, Chl. sp. N1, and Chl. luteolum show that their genomes encode cyc2 homologs, unlike all other sulfide-oxidizing Chlorobia members [18, 28–30], providing preliminary evidence that cyc2 might serve as a genomic marker for photoferrotrophy within the class Chlorobia. Confirming the validity of candidate iron oxidation gene markers such as cyc2 for photoferrotrophic metabolism would greatly expand the potential to survey for photoferrotrophy and other forms of extracellular electron transfer in nature.
Here, we combine genome-resolved metagenomics and enrichment cultivation to probe the metabolic diversity of anoxygenic phototrophs in seasonally anoxic and ferruginous Boreal Shield lakes, particularly examining the potential for photoferrotrophy. Following up initial 16S ribosomal RNA (16S rRNA) gene sequencing and iron isotope data that led us to hypothesize photoferrotrophy as an important photosynthetic process in the water columns of two Boreal Shield lakes, we recover high-completeness, low-contamination draft genomes of key populations of Chlorobia, the dominant anoxygenic phototrophs, from the same two lakes. We also report the enrichment of two novel species of Chlorobia from these lakes and from a third Boreal Shield lake, and we analyze the genetic and functional potential of these species for phototrophic sulfide and iron oxidation. Lastly, we explore the usage of cyc2 as a functional gene marker for photoferrotrophy compared to other phototrophic modes. Through this first usage of genome-resolved metagenomics to explore the metabolic diversity of Chlorobia in a ferruginous water column, we aim to extend sparse knowledge of iron–sulfur microbiology in ferruginous lake systems, with implications for understanding both Archaean microbial communities and modern ferruginous environments.
Materials and methods
Lake sampling, metagenome sequencing, assembly, and binning
An initial round of lake sampling was performed previously from 2011 to 2014 at the International Institute for Sustainable Development Experimental Lakes Area (IISD-ELA; near Kenora, Canada), which is located at 49°40′N, 93°45′W [4]. We studied Lake 227, an experimentally eutrophied lake, and Lake 442, a nearby and pristine reference lake [31, 32]. The IISD-ELA sampling site, lake geochemistry, and sample collection methods were described in detail previously [4]. Based on preliminary 16S rRNA gene data, we selected six water column genomic DNA samples collected from Lakes 227 and 442 for re-sequencing via shotgun metagenomics. All six samples were collected at or beneath the oxic–anoxic zone boundary, at depths where low light penetration occurs, and where we had previously observed high relative abundances of anoxygenic phototrophs, dominated by populations of Chlorobia. Samples from Lake 227 were selected at 6 and 8 m in 2013 and 2014, and samples from Lake 442 were selected at 16.5 m in 2011 and 13 m in 2014. The sequencing library was prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs; Ipswich, Massachusetts, USA) and was sequenced on a single lane of a HiSeq 2500 (Illumina; San Diego, California, USA) in Rapid Run Mode with 2 × 200 base paired-end reads, generating 26.3–39.5 million reads per sample. Library preparation and sequencing was performed by the McMaster Genome Facility (Hamilton, Ontario, Canada).
Raw metagenome reads were quality-trimmed, assembled, binned, and annotated using the ATLAS pipeline, version 1.0.22 [33, 34]. The configuration file with all settings for the pipeline is available in Supplementary File 1. To improve genome bin completeness and reduce bin redundancy, QC processed reads from related samples (i.e., L227 2013 6 and 8 m, L227 2014 6 and 8 m, and L442 samples) were co-assembled and re-binned using differential abundance information via a simple wrapper around ATLAS, as described in the Supplementary Methods.
Identification of cyc2 genes and Chlorobia genome bins
Assembled contigs were assessed for the presence of cyc2 gene homologs by building a custom profile hidden Markov model (HMM). The predicted primary sequences of cyc2 were recovered from the four available genomes of Chlorobia known to possess the gene, Chl. ferrooxidans, Chl. phaeoferrooxidans, Chl. sp. N1, and Chl. luteolum, as well as from the genomes of reference microaerophilic iron-oxidizing bacteria as described elsewhere [21]. A cytochrome 572 gene from Leptospirillum sp. (EDZ39515.1) was omitted from the reference dataset, due to its high divergence from other sequences, to build a more robust alignment for the cyc2 clade relevant to the Chlorobia [35]. Collected sequences were aligned using Clustal Omega, version 1.2.3 [36], and the alignment was used to build a profile HMM using the hmmbuild function of HMMER3, version 3.2.1 [37]. Recovered cyc2 genes over the course of the study were added to the alignment and HMM. The final HMM is provided in Supplementary File 2 and sequence alignments are available in the code repository associated with this work.
Recovered cyc2 genes were compared to cyc2 reference sequences by building a maximum-likelihood phylogeny. The alignment was masked using Gblocks, version 0.91b [38], as described in the Supplementary Methods. The maximum-likelihood phylogeny was then prepared from the masked sequence alignment via IQ-TREE, version 1.6.10 [39], using the LG + F + I + G4 sequence evolution model as determined by the ModelFinder module of IQ-TREE [40]. To build the consensus tree, 1000 bootstrap replicates were performed, each requiring ~100–110 tree search iterations for phylogeny optimization.
Genome bins were dereplicated using dRep version 2.0.5 [41] with default settings. All genome bins from individual assembles and co-assemblies were pooled for dereplication to help ensure that the highest quality bins, whether from individual assemblies or co-assemblies, would be retained for downstream analyses. Genome bins of Chlorobia with >90% completeness and <10% contamination based on CheckM statistics [42] were selected for further study. These genome bins were imported into Anvi’o version 4 [43] and were manually examined for contigs improperly binned based on read mapping, tetranucleotide frequencies, and contig taxonomic classification. Import of ATLAS data into Anvi’o was performed using the atlas-to-anvi.sh script, version 1.0.22-co-assembly-r4, available in the atlas-extensions GitHub repository at https://github.com/jmtsuji/atlas-extensions. Contigs in curated genome bins of Chlorobia were then ordered via the Mauve Contig Mover development snapshot 2015-02-13 for Linux [44] using the Chl. luteolum genome to guide ordering. To assess bin quality, tRNA genes were predicted using Prokka v1.13.3 [45].
Enrichment cultivation, sequencing, and assembly
Lake 227 and Lake 442 were sampled again in June 2016 and July 2017, along with the nearby Lake 304 in July 2017. Basic sampling information for each lake is summarized in Supplementary Table 1. Lake 304 has been described previously and also has seasonally anoxic bottom waters [46, 47]. The lake was experimentally fertilized with phosphate in 1971–72 and 1975–76, with ammonium in 1971–74, and with nitrate in 1973–74, but the lake has not been manipulated since then and rapidly returned to its non-eutrophied state once additions ceased, having a water residence time of ~2.7 years [48–50]. Lake 304 was added to our sampling efforts for enrichment culturing to explore the broader distribution of photoferrotrophy among IISD-ELA lakes, because preliminary 16S rRNA gene sequencing data (not shown) indicated high relative abundances of Chlorobia populations in this lake. Anoxic water was collected from Lakes 227 and 304 at a depth of 6 m and Lake 442 at 15 m, where trace levels of light are generally detectable (i.e., in the range of 0.01–1 µmol photons m−2 s−1 between 400 and 700 nm wavelengths, as commonly measured when field sampling). Lake water was pumped to the surface using a gear pump and directly injected into N2-filled 160 mL glass serum bottles that were sealed with blue butyl rubber stoppers (Bellco Glass; Vineland, New Jersey, USA), using a secondary needle to vent N2 gas until the bottles were full. Water was kept cold (~4 °C) and dark after collection and during shipping.
Enrichment cultures were grown using sulfide-containing Pfennig’s medium, prepared as described by Imhoff [8, 9], or ferrous iron-containing freshwater medium, prepared as described by Hegler et al. [51]. The ferrous iron-containing medium contained 8 mM ferrous chloride (FeCl2), without filtration of precipitates, and used trace element solution SLA [8]. Initial enrichments contained 10–20% lake water at a total volume of 50 mL and were inoculated into 120–160 mL glass serum bottles sealed with blue butyl rubber stoppers (Bellco Glass) or black rubber stoppers (Geo-Microbial Technology Company; Ochelata, Oklahoma, USA). The remaining headspace was flushed with a 90:10 N2:CO2 gas mix at left at a pressure of 150 kPa. Several bottles additionally had anoxic DCMU (i.e., Diuron or 3-(3,4-dichlorophenyl)-1,1-dimethylurea; Sigma-Aldrich; St. Louis, Missouri, USA) added to a concentration of 50 µM to block activity of oxygenic phototrophs [52, 53]. Bottles were incubated at 22 °C and ~50 cm away from far red PARSource PowerPAR LED Bulbs (LED Grow Lights Depot; Portland, Oregon, USA) as the light source.
Two enrichment cultures survived repeated subculture or dilution-to-extinction and contained green sulfur bacteria based on pigment and marker gene sequence analyses. Biomass from these cultures was collected by centrifugation, and genomic DNA was extracted using the DNeasy UltraClean Microbial Kit (Qiagen; Venlo, The Netherlands). Genomic DNA was prepared into metagenome sequencing libraries using the Nextera DNA Flex Library Prep Kit (Illumina), and the library was sequenced on a fraction of a HiSeq 2500 (Illumina) lane in High Output Run Mode with 2 × 125 base paired-end reads. Library preparation and sequencing was performed by The Center for Applied Genomics (TCAG; The Hospital for Sick Children, Toronto, Canada), generating ~6 million total reads per sample. Metagenomes were assembled using ATLAS version 1.0.22 without co-assembly. Genome bins of Chlorobia were manually refined as described above, using Anvi’o version 5 [43] via atlas-to-anvi.sh commit 99b85ac; contigs of curated bins were subsequently ordered, and tRNA genes counted, as described above.
Comparative genomics of Chlorobia genomes
Refined genome bins of Chlorobia, which belonged to the Chlorobium genus, were compared to genomes of reference strains from the Chlorobiaceae family. Genomes of all available type strains (as of 2017) and photoferrotrophs from the family were downloaded from the NCBI (Supplementary File 3). Average nucleotide identity (ANI) between genomes was calculated using FastANI version 1.1 [54]. The phylogenetic relationship between genomes was determined by constructing a concatenated ribosomal protein alignment based on the rp1 set of 16 ribosomal protein genes using GToTree version 1.1.10 [55, 56]. IQ-TREE version 1.6.9 [39] was used to construct the maximum-likelihood phylogeny from the alignment. The LG + F + R4 model of sequence evolution was identified as optimal by the IQ-TREE ModelFinder module [33], and phylogeny construction used 1000 bootstrap replicates, each requiring ~100–110 tree search iterations for optimization.
The presence or absence of genes implicated in iron or sulfur cycling metabolism in the collection of Chlorobiaceae genomes was assessed using bidirectional BLASTP [57]. Genes were selected from the genomes of Chl. ferrooxidans and Chl. clathratiforme based on the genome comparison of Frigaard and Bryant [18]. Predicted primary sequences of these genes were used to identify putative homologs across other genomes of Chlorobia using the BackBLAST pipeline, version 2.0.0-alpha2 (10.5281/zenodo.3465955) [58]. The e value cutoff for BLASTP hits was set at 10−40, based on empirical testing of gene clusters known to be present or absent in reference genomes of Chlorobia, and the identity cutoff was set to 20%. Selected genes (e.g., qmoA, dsrJ) were omitted due to poor homology between reference genomes. Genes associated with photosynthesis and carbon fixation were similarly assessed using BackBLAST, except that most reference genes were selected from the genome of Chl. tepidum according to Bryant et al. [59] and Tourova et al. [60].
Metagenome taxonomic and functional profiling
Environmental relative abundances of microbial populations were estimated by read mapping to dereplicated genome bins (further details in Supplementary Methods). Taxonomy and functional gene information were then overlaid onto relative abundance data. Genome bin taxonomy was determined based on the Genome Taxonomy Database (GTDB) using GTDB-Tk, version 0.2.2, relying on GTDB release 86, version 3 (April 2019) [6, 61]. As such, GTDB taxonomy names are used throughout this paper. Genomes were tested for the presence of the cyc2 and dsrA functional genes using the HMM developed in this study and an HMM available on FunGene [62], respectively. Gene amino acid translations were predicted for all genome bins using prodigal version 2.6.3 [63], via the GTDB-Tk, and amino acid translations were searched via HMMs using the MetAnnotate pipeline, version 0.9.2 [64]. An e value cutoff of 10−40 was used to filter low-quality gene hits. In case of bias due to unassembled or incorrectly binned genes, genome bin-based environmental abundance metrics were cross compared to abundance metrics generated by directly scanning unassembled metagenome reads, as described in the Supplementary Methods. Genome bins were further assessed for their broader iron- and sulfur-cycling genomic potential using additional gene markers and FeGenie [65] commit 30174bb, as described in the Supplementary Methods.
Assessment of ferrous iron oxidation potential of Chlorobia enrichments
After initial sequencing, cultures containing Chlorobia spp. continued to be purified in the laboratory. A single culture, which was enriched on sulfide-containing medium, survived continued cultivation and was provisionally named “Candidatus Chlorobium canadense strain L304-6D” (ca.na.den’se N.L. neut. adj. canadense from or belonging to Canada). “Ca. Chl. canadense” was purified through multiple rounds of incubation in deep agar dilution series and picking of isolated green colonies [66]. For deep agar shakes, Pfennig’s medium or modified Chloracidobacterium thermophilum Midnight medium [67] containing 1–3 mM buffered sulfide feeding solution were used [8]. Eventually, in preparation for growth in ferrous iron-containing medium, cultures of “Ca. Chl. canadense” were transferred to growth in liquid freshwater medium as described by Hegler et al. [51] (here termed, “Hegler freshwater medium”) but containing 0.5–1 mM buffered sulfide feeding solution, 10–20 µM FeCl2, and 0.5 mg/L resazurin to monitor the medium’s redox status.
To test their ability to oxidize ferrous iron phototrophically, “Ca. Chl. canadense” cells were inoculated into Hegler freshwater medium containing low levels of ferrous iron and lacking sulfide. The base Hegler freshwater medium (before addition of ferrous iron or sulfide) was aliquoted into 120 mL glass serum bottles under flow of sterile 90:10 N2:CO2 gas, with 50 mL of medium per bottle. Bottles were sealed with black rubber stoppers (Geo-Microbial Technology Company) and were bubbled for at least 9 min with additional 90:10 N2:CO2 gas to purge trace oxygen. The headspace of each bottle was left at 150 kPa pressure. A sterile solution of FeCl2 was added to each bottle to reach a ferrous iron content of 100 μM, and to some bottles, a sterile and anoxic solution of the chelator ethylenediaminetetraacetic acid (EDTA) was added to a concentration of 120 μM as done by Peng et al. [68] to explore whether this chelator could enhance photoferrotrophic iron oxidation. Bottles were incubated at room temperature in the dark overnight to allow for complexation of ferrous iron and EDTA. All media was confirmed to have a pH of 6.5–7.
“Ca. Chl. canadense” was grown in 100 mL Hegler freshwater medium using a total of ~2 mM sulfide, fed incrementally in 0.4 mM doses, until sulfide was completely oxidized and the resazurin in the bottle went slightly pink. At the same time, a culture of Chl. ferrooxidans KoFox was grown in the same freshwater medium containing ~8 mM FeCl2 until ferrous iron was completely oxidized. The entire 100 mL “Ca. Chl. canadense” culture was pelleted by centrifugation at 7000 × g for 13 min, and the pellet was washed twice with unamended Hegler freshwater medium. The ~200 mg of wet biomass recovered was suspended in 1 mL of unamended freshwater medium, and 0.1 mL was inoculated into each relevant incubation bottle. Similarly, 100 mL of the Chl. ferrooxidans reference culture was pelleted by centrifugation at 7000 × g for 5 min, washed twice, and suspended in 1 mL of unamended freshwater media, and 0.1 mL was added to relevant bottles. Only the surfaces of Chl. ferrooxidans cell pellets, which contained mostly green cells and not brown ferric iron precipitate, were suspended with each wash, allowing for ~3 mg of nearly iron-free wet biomass to be recovered. Once cultures were inoculated, they were transferred to a 22 °C incubator without shaking where they received 30 μmol photons m−2 s−1 white light from a mix of fluorescent (F48T12/CW/VHO; Osram Sylvania; Wilmington, Massachusetts, USA) and incandescent (60 W) bulbs.
Cultures were sampled regularly over a 21-day incubation period to monitor iron concentrations and iron oxidation states. At each sampling time point, an aliquot of culture was removed from each bottle using a sterile 90:10 N2:CO2-flushed syringe and 25 G needle, and 330 μL of this culture aliquot was acidified immediately with 30 μL of 6 N hydrochloric acid (HCl). Acidified samples were stored for no more than 2 days at 4 °C (with the exception of samples collected on day 14; see Supplementary Methods) before being assessed for iron species concentrations via the ferrozine assay [69], with 5% ascorbic acid being used as the reducing agent to determine total iron species [70].
Results
Recovery of Chlorobium genome bins
Binning of assembled contigs from the lake and enrichment culture metagenomes, followed by dereplication of highly similar bins and manual curation, allowed for the recovery of six highly complete, low-contamination genome bins that classified within the genus Chlorobium (Table 1). Three of the genome bins recovered from lake metagenomes had best representatives selected by dRep from the Lake 227 (2013 sample) co-assembly, and one had its best representative from the Lake 442 (2011/2014 sample) co-assembly. In addition, a single Chlorobium genome bin was recovered from each enrichment culture. Recovered bins had at least 90.1% completeness and a maximum of 2.8% contamination based on analysis by CheckM [42]. Recovered genome bins were between 1.9 and 2.6 Mb in size, within the approximate length range of reference genomes associated with the family Chlorobiaceae (2.0–3.3 Mb; average 2.7 Mb), and were represented by 13–208 contigs. Other lower-quality genome bins classified as belonging to the Chlorobia were also recovered (Supplementary File 4), but these were not considered for gene pathway analysis in order to minimize the risk of including false positives. In particular, L442 Bin 74 was excluded from the set of curated bins due to its high contig count (282) and elevated predicted contamination (4.1%) and strain heterogeneity (25%).
Table 1.
Quality statistics for genome bins of Chlorobia recovered in this study.
| Bin ID | Contigs | Length (Mb) | N50 (kb) | L50 | GC content (%) | Genes | tRNA genes | Completeness (%) | Contamination (%) |
|---|---|---|---|---|---|---|---|---|---|
| L227 2013 Bin 55 | 208 | 2.38 | 16.7 | 42 | 48.5 | 2397 | 39 | 97.4 | 0.1 |
| L227 2013 Bin 56 | 140 | 2.09 | 27.6 | 21 | 43.9 | 2009 | 38 | 96.7 | 0.6 |
| L227 2013 Bin 22 | 57 | 2.55 | 79.5 | 12 | 45.9 | 2471 | 37 | 98.5 | 2.8 |
| L442 Bin 64 | 128 | 1.90 | 28.6 | 22 | 48.3 | 1871 | 35 | 90.1 | 0.0 |
| L227 enr. S-6D Bin 1 | 13 | 2.50 | 306.7 | 3 | 46.8 | 2385 | 44 | 99.5 | 1.0 |
| Ca. Chl. canadense L304-6D | 15 | 2.61 | 271.5 | 3 | 48.7 | 2504 | 45 | 99.4 | 0.0 |
Completeness and contamination were calculated using CheckM (see “Methods”). Note that no ribosomal RNA genes were recovered for the bins. A summary of bin statistics for lower-quality genome bins is included in Supplementary File 4.
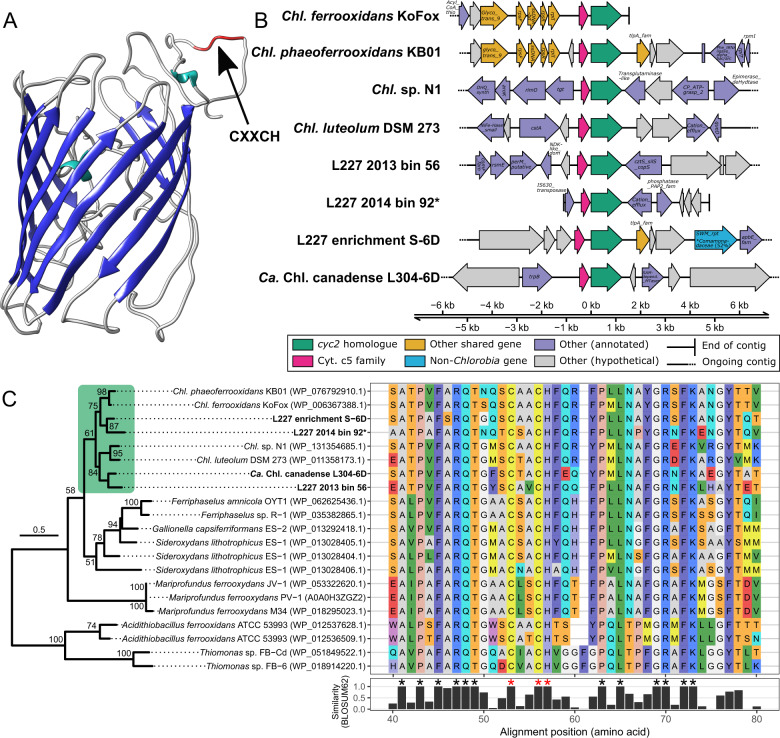
The constructed HMM for the cyc2 gene enabled the recovery of five potential cyc2 homologs associated with Chlorobia from assembled contigs (Fig. 1). Three of these cyc2 homologs were found in the set of six manually curated genome bins described above, and another homolog was detected in one of the lower-quality genome bins of Chlorobia (L227 2014 Bin 92). The final homolog was detected on a short contig in the same lower-quality genome bin and was removed from subsequent analyses due to its lack of gene neighbors. Detected cyc2 homologs were full-length genes, and their predicted primary sequences were detected in the HMM search with a maximum e value of ~10−140. All recovered cyc2 homologs contained the heme-binding CXXCH motif near the N-terminus of their predicted primary sequence, and all homologs were predicted by I-TASSER [71] to contain a porin-like beta barrel structure for membrane binding (Fig. 1a; one example shown) [13]. Aside from the closely related genomes of Chl. ferrooxidans and Chl. phaeoferrooxidans (Supplementary Fig. 1), all other genomes showed substantial genome rearrangement around the cyc2 homolog (Fig. 1b). However, detected cyc2 homologs were always adjacent to a c5 family cytochrome (Fig. 1b). These c5 family cytochromes had low sequence identity to one another (i.e., as low as 30%) yet also contained a conserved CXXCH motif (Supplementary Fig. 2). All recovered cyc2 homologs grouped monophyletically with cyc2 predicted primary sequences belonging to reference genomes of Chlorobia compared to the cyc2 of other reference strains (Fig. 1c). Overall, the combination of HMM search specificity, genomic context, predicted gene motifs, and phylogenetic placement is strong evidence that the identified cytochrome genes are cyc2 homologs.
Fig. 1. Recovered cyc2 homologs of Chlorobia from metagenomes.
a Homology model of the Cyc2 protein from Chl. phaeoferrooxidans KB01, generated by I-TASSER based on sequence homology (with leading 18 signal peptides trimmed according to SignalP [88]) and visualized using UCSF Chimera [89]. The porin-like beta barrel structure is highlighted in dark blue, and the location of the single N-terminal heme-binding motif on the outer-membrane portion of the structure is indicated by a black arrow. b Genomic context of the recovered cyc2 homologs. A ~10 kb region is shown of assembled contigs surrounding the homologs. The cyc2 and adjacent c5 family cytochrome genes are highlighted in green and pink, respectively. Other shared genes between contigs are highlighted in yellow. Remaining genes are colored gray if they lacked a functional annotation (e.g., no significant BLASTP hit, or hit to a domain of unknown function) or are colored purple if their closest BLASTP hit corresponded to a gene associated with the Chlorobiaceae family. One gene in the 10 kb region had its closest BLASTP hit to a gene associated with the Comamonadaceae family (Betaproteobacteria; ~51% amino acid identity); this gene is highlighted in blue. c Sequence comparison of recovered cyc2 homologs to cyc2 genes of known iron-oxidizing microorganisms. The phylogenetic tree was built from a 223 residue masked Cyc2 amino acid sequence alignment (see “Methods”). Bootstrap values over 50/100 are shown, and the scale bar represents the proportion of residue changes along the alignment. Due to the uncertain evolutionary history of cyc2, the phylogeny is midpoint rooted. A green box highlights the monophyletic Chlorobia clade. Adjacent to the phylogeny, a subset of the unmasked Cyc2 sequence alignment is shown (positions 40–80 of 609; N-terminus end). Alignment positions having 100% sequence identity in the predicted heme-binding site (CXXCH motif) are marked with red asterisks. Other positions with 100% sequence identity are marked with black asterisks. Note that L227 2014 Bin 92 (name marked with an asterisk) was included in this figure for comparison of its cyc2 gene despite the bin quality being lower than the main Chlorobia genome bin set. Chl. Chlorobium.
Enrichment cultivation of Chlorobia
Enrichment cultivation was attempted with anoxic lake water using both sulfide- and ferrous iron-containing anoxic media. For Lake 442, one enrichment in a ferrous iron-containing medium grew with evidence of light-driven iron oxidation, but 16S rRNA gene sequencing showed that the culture was dominated by a member of the genus Rhodopseudomonas, which did not represent a detectable genus in environmental sequencing data. Given the known metabolic versatility of Rhodopseudomonas spp. and the negligible contribution of members of this genus to the studied lake microbial communities, this culture was not pursued further. No members of the Chlorobia were detected in any enrichment culture from Lake 442.
For Lakes 227 and 304, growth was observed for both media tested. Enrichments grown in sulfide-containing medium developed green color after ~12–15 weeks of incubation and contained populations of Chlorobia based on Sanger sequencing of both the V3–V4 region of the 16S rRNA gene and the partial dsrAB gene (using Chlorobia-targeted PCR primers 341f/GSB822r [72] and PGdsrAF/PGdsrAR [73], respectively). However, the appearances of cultures grown in ferrous iron-containing medium for these lakes differed from those of reference photoferrotrophic Chlorobia members. Instead of forming a reddish-brown ferric iron precipitate, the enrichments blackened, potentially indicative of sulfate reduction, and metal sulfide formation. After blackening, enrichments developed a green color (Supplementary Fig. 3). Suspecting that sulfate reduction was occurring in the ferrous iron enrichment bottles to support sulfide-oxidizing phototrophy, one Lake 227 enrichment was transferred into sulfide-containing media and still developed green pigmentation, albeit slowly (Supplementary Fig. 3). Amplification and Sanger sequencing of the partial dsrAB gene cluster from this enrichment showed that the gene sequence only differed by one ambiguous base from, and thus was essentially identical to, a corresponding Lake 227 sulfide enrichment. Ferrous iron enrichments eventually stopped being followed due to long growth times, low biomass, and instability of the cultures. Enrichments on sulfide-containing media were continued instead.
Metagenome sequencing of the two successful enrichments of Chlorobia grown on sulfide (L227 enrichment S-6D and L304 enrichment S-6D) showed that genome bins of the Chlorobia from both enrichments encoded cyc2 homologs. Although L227 enrichment S-6D ceased to grow in laboratory culture, the L304 enrichment S-6D continued to be purified and was named “Candidatus Chlorobium canadense strain L304-6D”. At the time of running the ferrous iron oxidation test (below), the “Ca. Chl. canadense” culture consisted of >90% cells of Chlorobia based on epifluorescence microscopy (Supplementary Fig. 4), and other contaminating cells were known to be chemoorganoheterotrophs and sulfate-reducing bacteria based on previous 16S rRNA gene amplicon sequencing data (Supplementary File 5).
Metabolic diversity and phylogeny of Chlorobium genome bins
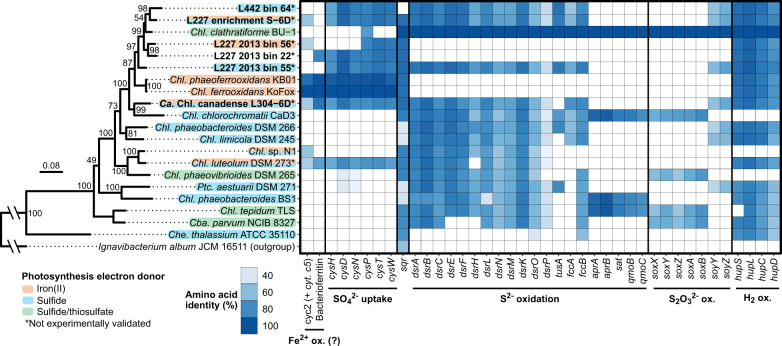
Recovered genome bins of Chlorobia were genetically diverse with respect to the surveyed photosynthesis-associated genes (Fig. 2). One of the lake-recovered bins (L227 2013 Bin 56) lacked all assessed genes involved in sulfide, elemental sulfur, and thiosulfate oxidation but contained the cyc2 gene homolog. Conversely, two other lake-recovered bins (L227 2013 Bin 55 and L442 Bin 64) contained all assessed genes in the sulfide oxidation pathway but lacked the cyc2 gene. Although the final lake-recovered bin (L227 2013 Bin 22) lacked both cyc2 and genes for sulfur oxidation, this lack of key metabolism genes might be due to incomplete genome binning. Lastly, the two enrichment culture bins encoded both cyc2 and the sulfide oxidation gene pathway, similarly to Chl. sp. N1 and Chl. luteolum. Like some cultured photoferrotrophs, L227 2013 Bin 22 and the “Ca. Chl. canadense” genome also encoded putative homologs to a bacterioferritin hypothesized by Frigaard and Bryant [18] to play a role in photoferrotrophy. Genes for hydrogen oxidation were detected in all of the genome bins (Fig. 2), along with marker genes for photosynthesis and bacteriochlorophyll biosynthesis (Supplementary Fig. 5). Marker genes for carbon fixation (via the reverse TCA cycle) were detected in all genome bins, with the exception of L227 2013 Bin 56, which was missing one of the two assessed gene clusters for the process (aclAB; Supplementary Fig. 5). Although L227 2013 Bin 56 was also missing half of the cys gene pathway for sulfate uptake (Fig. 2), the cys gene cluster occurred at the very end of a contig for this genome bin, implying that the bin likely ought to contain the complete cys pathway but that an assembly break occurred.
Fig. 2. Iron- and sulfur-oxidizing genetic potential in recovered genome bins of Chlorobia compared to reference strains.
The left side of the figure shows a maximum-likelihood phylogenetic tree of the Chlorobia based on concatenated ribosomal protein amino acid sequences (see “Methods”). Bootstrap values over 50/100 are shown, and the scale bar represents the percentage of residue changes across the 2243 amino acid alignment. Species of Chlorobia are shaded based on their known or hypothesized metabolic potential. Genome bins of Chlorobia recovered from this study are bolded. On the right side, a heatmap is shown displaying the presence/absence of genes associated with iron and sulfur metabolism among Chlorobia based on bidirectional BLASTP. Heatmap tiles are shaded based on the percent amino acid identity of an identified gene compared to the reference sequence (Chl. ferrooxidans for putative iron metabolism-associated genes, and Chl. phaeoclathratiforme for sulfur metabolism-associated genes). Although the cytochrome c5 family gene upstream of cyc2 was not searched for directly due to poor sequence homology, the gene was verified manually to be adjacent to all hits of cyc2. Chl. Chlorobium, Cba. Chlorobaculum, Che. Chloroherpeton, Ptc. Prosthecochloris.
Phylogenetic analysis of the recovered Chlorobium genome bins based on concatenated ribosomal protein sequences showed that all four lake-recovered bins and the bin from the Lake 227 enrichment formed a monophyletic subgroup within the Chlorobiaceae, including Chl. phaeoclathratiforme BU-1 (Fig. 2). Directly basal to this subgroup was a branch containing Chl. ferrooxidans and Chl. phaeoferrooxidans. By comparison, the bin of “Ca. Chl. canadense” grouped sister to Chl. chlorochromatii, separately from the above group and from the group containing Chl. luteolum and Chl. sp. strain N1. Comparison of the concatenated ribosomal protein phylogeny to the cyc2 predicted primary sequence phylogeny reveals some congruency between the two phylogenies for potentially photoferrotrophic members of the Chlorobia, with the exception of L227-S-6D and “Ca. Chl. canadense” (Supplementary Fig. 6). Overall, all six recovered Chlorobium genome bins appear to represent novel species, having an ANI of <79.6% compared to the genomes of all available type strains (Supplementary Fig. 1).
Functional and taxonomic profiling
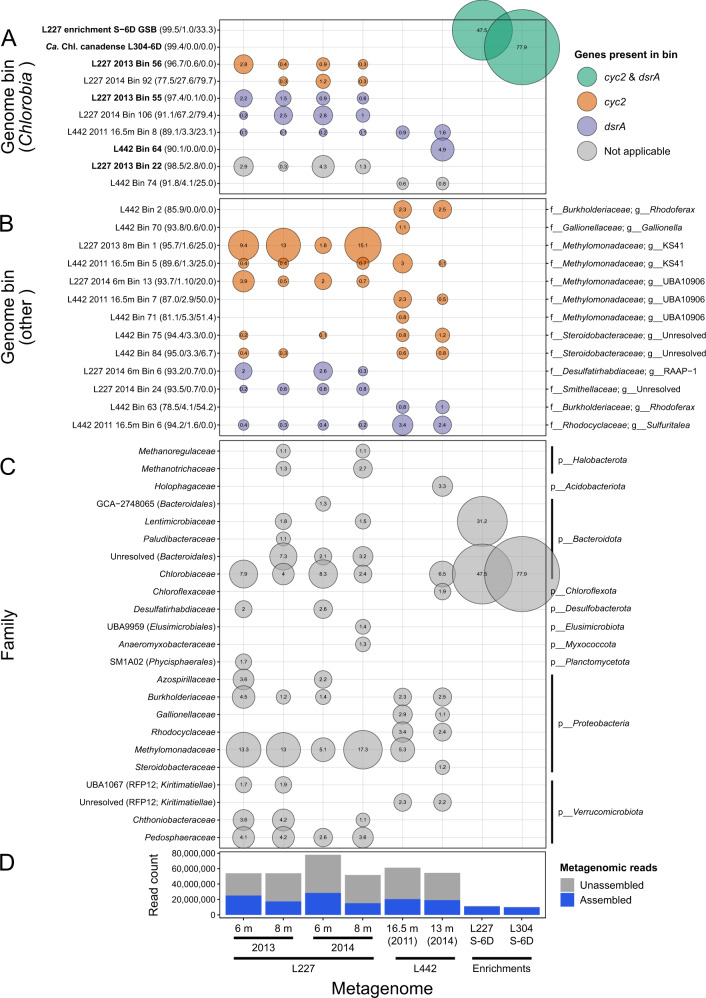
Read mapping to genome bins showed that several distinct populations of Chlorobia were relevant to the lake environment (Fig. 3). Bins containing cyc2 (no dsrA) and bins containing dsrA (no cyc2) were both found at similar relative abundances in Lake 227 samples in 2013 and 2014 (Fig. 3a). Generally, genome bins of Chlorobia containing cyc2 accounted for 0.6–2.8% of the Lake 227 microbial community data in the assessed samples, whereas bins of Chlorobia containing dsrA accounted for 1.7–4.1% of the data. Analyses of unassembled metagenome read data indicated similar relative abundances (Supplementary Fig. 7). In contrast, only genome bins encoding sulfide oxidation were detected in Lake 442 metagenome assemblies. Even when unassembled metagenome data were scanned, only a single HMM hit from the Lake 442 metagenomes matched cyc2 classified as belonging to a member of Chlorobia, compared to 27,000–37,000 total rpoB hits for these same metagenomes, showing that the lack of cyc2 genes detected in genome bins of Chlorobia from L442 was not a result of incomplete assembly or genome binning. One genome bin of Chlorobia overlapped between L227 and L442 (L442 2011 16.5 m Bin 8); the rest were distinct between the two lakes. In addition, the bins of Chlorobia recovered from the enrichment culture metagenomes were not detected at high relative abundances in any of the six lake metagenomes.
Fig. 3. Bubble plot showing predicted relative abundances of recovered genome bins in the lake and enrichment culture metagenomes.
The size of each bubble corresponds to the relative abundance of a genome bin or taxon within metagenome data based on mapping of assembled reads. Each bubble is also labeled with its percent relative abundance for clarity. a Bins of Chlorobia colored by their metabolic potential based on functional gene markers described in this study. Bolded names correspond to the higher-quality bins described in Table 1 and Fig. 2. In parentheses beside each name are bin quality statistics reported by CheckM—the predicted % completion, % contamination, and % strain heterogeneity, respectively. The displayed bins all classify to the Chlorobium genus based on GTDB taxonomy. b Non-Chlorobia bins (≥0.01% relative abundance) found to have the same functional gene markers (cyc2 or dsrA) in their genome sequences. Quality statistics are reported as in a. On the right side of the panel, the GTDB family and genus classifications of each bin are shown. c Family-level taxonomic composition of the metagenomes based on GTDB classifications of genome bins with ≥1% relative abundance. The phylum of each family is displayed on the right side of the panel. For unresolved or non-Latin family names, the order (and class, if needed) is displayed in parentheses beside the family name for clarity. d Assembly statistics for the metagenome data. The total number of quality-trimmed metagenome reads is represented by the total height of each bar. Reads that mapped to the filtered sequence assembly (i.e., excluding short contigs, see “Methods”) are highlighted in blue and are considered “assembled” reads. Assembled read totals were used to determine relative abundances of genome bins in a–c. Supplementary Figure 7 shows relative abundances based on predictions from unassembled metagenome reads for comparison.
Several non-Chlorobia genome bins were also found to contain the cyc2 or dsrA gene through sequence searches (Fig. 3b). Genome bins containing potential cyc2 homologs grouped into the Rhodoferax or Gallionella genera known to include iron-cycling bacteria [23, 74] or into the Steroidobacteraceae or Methylomonadaceae families. Given that both genomes associated with the Steroidobacteraceae (L442 Bin 75 and L442 Bin 84) also encoded the PCC-type mtrABC operon implicated in iron reduction (Supplementary Fig. 8) [21], it is possible that these genomes represent novel, neutrophilic iron-reducing bacterial populations related to the acidophile Acidibacter ferrireducens, for which genome sequencing data are not yet available [75]. The genome bins classified to the Methylomonadaceae occurred at high relative abundances in the lake metagenomes, having a cumulative relative abundance of up to ~17% in Lake 227 (Fig. 3c), and encoded particulate methane monooxygenase (pmoA) genes, with the exception of L442 2011 16.5 m Bin 5. These genome bins encoded a “repCluster2” cyc2 variant based on analysis by FeGenie, unlike the “repCluster1” variants detected in genomes of Chlorobia and of Gallionella, but they encoded no detectable cyc1 gene adjacent to the cyc2 sequences as seen in the “repCluster2” cyc2 sequences of Rhodoferax (Supplementary Fig. 8). Three other genome bins belonging to the phyla Myxococcota and Verrucomicrobiota encoded PCC-type gene clusters potentially involved in extracellular electron transfer (Supplementary Fig. 8). Several genomes, including all detected genomes of Chlorobium, were found based on FeGenie to encode a “repCluster3” cyc2 variant (Supplementary Fig. 8). This distant homolog of the photoferrotrophy-associated “repCluster1” cyc2 variant (having ~20% amino acid identity to “repCluster1” sequences) could potentially play a role in extracellular electron transfer but has not been associated with photoferrotrophy in reference cultures and was not detected by the custom HMM developed in this study.
Genome bins encoding dsrA, which is involved in either sulfide oxidation or sulfate reduction, included a bin related to known chemolithotrophic sulfide-oxidizing bacteria of the genus Sulfuritalea and a bin related to heterotrophic sulfate-reducing bacteria of the family Desulfatirhabdiaceae [76, 77]. A bin classified to the family Smithellaceae of the order Syntrophales, along with a bin classified to the genus Rhodoferax, also encoded dsrA. Three of these four genomes bins were also found to encode the aprAB operon involved in sulfite metabolism (Supplementary Fig. 8). The two bins related to the Sulfuritalea and Rhodoferax genera were found to encode the soxB gene involved in thiosulfate oxidation (Supplementary Fig. 8), which suggests that both genome bins can oxidize sulfur compounds [18]. No other genome bins in the dataset were found to encode these key sulfur-cycling gene markers except for two genome bins that could not be confidently classified as sulfur cyclers due to limited gene content (Supplementary Fig. 8).
Chlorobia members appeared to represent the dominant phototrophs in the lake anoxic zone samples. In total, genome bins classified to the Chlorobia represented as high as 8.3% of lake microbial communities based on read mapping (Fig. 3c). Predicted relative abundances of populations of Chlorobia were 1.2–1.8 times higher when unassembled reads were assessed directly (Supplementary Fig. 7), with a predicted maximum relative abundance of 12.3%, showing that high relative abundance is not a result of assembly bias (Fig. 3d). The only other potential chlorophototroph detected in the dataset at >1% relative abundance was a single genome bin classified to the family Chloroflexaceae (L442 Bin 82; 92.9/1.2% completeness/contamination), which made up 1.9% of the L442 microbial community in 2014 at 15 m depth (Fig. 3c) and contained the bchL and pufM photosynthesis genes (data not shown). This genome bin was undetected in all other samples.
Assessment of ferrous iron oxidation potential of “Ca. Chl. canadense”
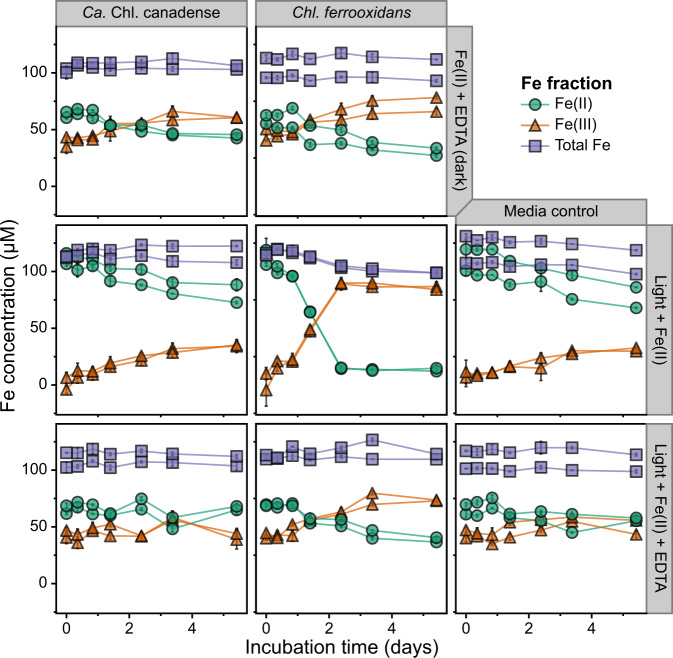
Cultures of Chl. ferrooxidans showed the expected behavior when incubated in ferrous iron containing medium. After a short lag phase, both Chl. ferrooxidans cultures containing 100 μM FeCl2 (no EDTA) and exposed to light began oxidizing ferrous iron, and the cultures had nearly completely oxidized all ferrous iron within 2 days of the initial setup (Fig. 4). Similar bottles incubated in the dark showed consistent iron oxidation rates of ~5 μM d−1 that corresponded to rates observed in uninoculated media controls; this iron oxidation was inferred to be abiotic, for example due to small amounts of oxygen contamination during sampling. No ferrous iron oxidation outside of this baseline effect was observed in the Chl. ferrooxidans cultures supplemented with 120 μM EDTA, in contrast to the stimulatory effect of EDTA reported for Rhodopseudomonas palustris TIE-1 [68], which uses a different gene system (pioAB) for extracellular electron transfer [24]. The EDTA-amended medium consistently appeared to have ~50% ferrous iron oxidation at the beginning of the experiment, based on the ferrozine assay, but this apparent oxidation has been observed previously with EDTA and could potentially be due to ineffective binding of ferrozine to EDTA-chelated iron [70].
Fig. 4. Iron oxidation activity of two Chlorobia cultures under different growth conditions over time.
The concentrations of ferrous iron, total iron, and ferric iron (determined by the subtraction of ferrous iron from total iron) are shown over a 10 day incubation period for “Ca. Chl. canadense” (enriched in this study) and Chl. ferrooxidans. The activity of the cultures in iron-amended media is shown when incubated in the dark with EDTA, in the light, and in the light with EDTA. Each panel depicts the results of biological duplicates, with error bars representing the standard deviation of technical duplicates in the ferrozine assay. The same results are shown for the full 21-day incubation period in Supplementary Fig. 9.
In contrast to Chl. ferrooxidans, “Ca. Chl. canadense” showed no photoferrotrophic iron oxidation activity over the course of the experiment (Fig. 4; Supplementary Fig. 9), and the iron oxidation profiles of the cultures closely matched those of the uninoculated media control bottles. A reference bottle of Hegler freshwater medium prepared in the same batch as those in the main experiment but amended with 600 μM buffered sulfide feeding solution, in place of ferrous iron, received the same inoculum of “Ca. Chl. canadense”. Complete oxidation of sulfide in the bottle was observed within 5 days of inoculation and incubation under light (not shown), indicating that the media and inoculum were not the cause of the lack of observed photoferrotrophic activity.
Discussion
In this study, we explored the diversity of anoxygenic photosynthesis in the ferruginous water columns of Boreal Shield lakes and evaluated the potential for photoferrotrophy using a combination of genome-resolved metagenomics and enrichment cultivation. Although we were unable to induce photoferrotrophy in laboratory cultures, we detected robust homologs of the cyc2 gene associated with photoferrotrophy in the genus Chlorobium and found evidence for both iron cycling and cryptic sulfur cycling in the lakes. Overall, our findings provide genomic context to previous 16S rRNA gene data supporting the existence of Chlorobia-driven photoferrotrophy in Boreal Shield lakes [4], adding nuance to the interpretation of these data while expanding sparse knowledge of the diversity of cyc2 within the Chlorobia. Our research reinforces the need for additional genetic work to understand the function and regulation of cyc2 in photoferrotrophy.
We detected several novel variants of cyc2 in the genome bins of Chlorobia recovered in this study from lake and enrichment culture metagenomes (Fig. 1). The cyc2 gene, which is putatively implicated in ferrous iron oxidation among microaerophilic iron-oxidizing bacteria [27], has been hypothesized to play a role in photoferrotrophy based on its presence in the genomes of cultured photoferrotrophs within the Chlorobia and its absence in the genomes of strict sulfide-oxidizing Chlorobia members. Our detection of four additional high-confidence homologs of cyc2 doubles the known diversity of this gene among members of the Chlorobia compared to cultured representatives, and we also developed a robust method combining sequence composition, genomic context, and phylogenetic placement for detection of additional cyc2 homologs compared to related cytochromes. Our work suggests that cyc2-encoding Chlorobia members, although only cultivated in the laboratory recently, play an important role in seasonally anoxic Boreal Shield lake microbial communities based on their high relative abundances in lake metagenomes (Fig. 3a).
Homologs of cyc2 also appear to be associated with other bacteria potentially involved in iron cycling in Lakes 227 and 442. One genome bin classified to the genus Gallionella, L442 Bin 70, encoded cyc2, and could represent a microaerophilic iron-oxidizing bacterium. In addition, genomes such as L442 Bin 2 classified to the genus Rhodoferax (related to Rhodoferax ferrireducens) and L442 Bins 75 and 84 belonging to the Steroidobacteraceae (related to Acidibacter ferrireducens) encoded an alternative cyc1/cyc2 gene cluster and could correspond to known iron-reducing bacteria. Other bacterial genomes encoding PCC-type gene systems involved in extracellular electron transfer were also detected, hinting at a broader iron-based biogeochemical cycle in the lakes. At the same time, genome bins classified to the Methylomonadaceae that are unrelated to known iron-cycling bacteria also encoded an alternative cyc2 variant. It is possible that the cyc2 variant might be involved in a form of extracellular electron transfer for these potential methanotrophs, which are found in the anoxic zones of Lakes 227 and 442 despite the oxygen requirement for particulate methane monooxygenase, as recently observed for other lakes [78, 79]. However, extracellular electron transfer by bacterial methanotrophs has not been reported in the literature previously, leaving the role of these cyc2 homologs unclear. Altogether, our data suggest that iron metabolism plays an important biogeochemical role in the seasonally anoxic and ferruginous water columns of these lakes.
Along with cyc2, we also detected genomic potential for sulfide oxidation among genome bins of Chlorobia recovered in this study. A higher number of genome bins of Chlorobia encoded the genetic potential for sulfide oxidation (based on the marker gene dsrA) than encoded cyc2, and in addition, cyc2 associated with Chlorobia members was entirely undetectable in Lake 442 metagenome data. Combined with our observations of blackening of ferrous iron-containing medium during initial enrichment culturing before phototrophic growth (Supplementary Fig. 3), our data suggest that sulfide is an important electron donor for anoxygenic phototrophs in ferruginous Boreal Shield lake waters. We detected at least one genome bin at high relative abundance in Lake 227 that likely represented a sulfate-reducing bacterium (Fig. 3c; Supplementary Fig. 8) and might thus provide the required sulfide to sustain phototrophic sulfide oxidation, even if sulfide is at low levels overall in the lakes. Such “cryptic sulfur cycling”, where sulfur turnover is rapid at low measured concentrations of sulfur redox species, has been observed previously in other sulfide-limited systems [80], including the permanently anoxic and ferruginous Lake La Cruz [81], and might have also played an important role in Archaean oceans once sufficient organic carbon was available. In addition, because all detected genomes of Chlorobia encoded the hupLS genes (Fig. 2), it is possible that oxidation of molecular hydrogen, as observed in laboratory cultures of Chlorobia [14], could also play a role in the metabolism of the populations of Chlorobia in the lakes.
We were unable to induce photoferrotrophy in “Ca. Chl. canadense” (Fig. 4), which we enriched in a sulfide-containing medium, despite the fact that this organism encoded a cyc2 homolog and a bacterioferritin in its genome (Fig. 2). The lack of observed photoferrotrophic growth in “Ca. Chl. canadense” could be due to several reasons. Firstly, it is possible that, although cyc2 is associated with reference photoferrotrophic Chlorobia members, the cyc2 gene plays a peripheral role in the metabolism of these organisms and is not directly involved in photoferrotrophy. Secondly, cyc2 could represent a single step in a larger iron oxidation gene pathway that is missing in the “Ca. Chl. canadense” genome. For example, the c5 family cytochrome detected directly upstream of all cyc2 homologs identified among genomes of Chlorobia (Fig. 1b) could potentially be involved in iron electron transfer. Unlike the primary sequences of all other c5 family cytochromes adjacent to cyc2 in reference genomes or environmental genome bins, the c5 family cytochrome of “Ca. Chl. canadense” contains an additional cysteine residue in the heme-binding motif in its primary sequence (i.e., CCXCH rather than the typical CXXCH; Supplementary Fig. 2) that could have deleterious consequences for iron oxidation [82].
As a third possibility, cyc2 could be associated with alternative cellular roles, including electron exchange with humic substances as hypothesized by He et al. [83] or even direct electron uptake from solid-phase conductive substances, as has been observed in the photoferrotroph Rps. palustris TIE-1 [84]. However, direct links between Cyc2 and oxidation of alternate substrates to dissolved ferrous iron have not been demonstrated experimentally. Lastly, the cyc2 gene product in “Ca. Chl. canadense” might allow for iron oxidation but might not be readily expressed under our laboratory conditions. This option would align with preliminary reports that the cyc2-encoding but sulfide-oxidizing Chl. luteolum is capable of changing to photoferrotrophic growth (Kate Thompson and Sean Crowe, personal communication), despite previous difficulties inducing this behavior [16]. The “Ca. Chl. canadense” culture can be used to probe the regulation and function of cyc2 within the class Chlorobia in future work, especially given its atypical cytochrome c5 sequence and given that all other cyc2-encoding and cultured Chlorobia members to date have photoferrotrophic activity. However, we are unable to draw conclusive links between cyc2 and photoferrotrophy from the present study.
Despite being physically nearby (~13.5 km apart) and having similar overall physico-chemistry, Lakes 227 and 442 differed substantially in the microbial community composition of their anoxic water columns based on metagenome data. Most genome bins examined in this study (Fig. 3a, b) were specific to one of the two lakes based on read mapping counts. Such single-lake specificity is lost when summarizing data at higher taxonomic levels (e.g., Fig. 3c) and was not observed for the same DNA samples when analyzed previously using 16S rRNA gene amplicon sequencing and clustering of sequences at a 97% identity threshold [4]. Our population-level analyses showed that cyc2 was not encoded by Lake 442 genome bins of Chlorobia despite being encoded by genome bins of Chlorobia in Lake 227 metagenome data. It is possible that physicochemical differences between the lakes, such as ferrous iron and sulfate concentration, hydrogen and bicarbonate concentrations (observed to impact photoferrotrophy rates in Rps. palustris TIE-1 [85]), lake bathymetry, anoxia timing, pH, light quality, or dissolved organic carbon levels, could drive this and other microbial community differences between the two lakes. Understanding key factors governing the favourability of cyc2 among Boreal Shield lakes could prove valuable for understanding the ecology and function of cyc2.
The strong iron isotope fractionation observed between the oxic and anoxic zone boundaries of Lakes 227 and 442 previously by Schiff et al. [4] appears to occur regardless of whether Chlorobia-affiliated cyc2 genes are detectable. It is possible that microaerophilic iron oxidation could contribute to the observed iron isotope fractionation in Lake 442 given the detection of a cyc2-encoding genome bin within the Gallionella in Lake 442 metagenomic data (Fig. 3b). Partial iron reduction by iron-reducing bacteria might also contribute to the observed fractionation. However, other yet-unknown iron cycling processes might also be at work. Sampling for iron isotopes at higher spatial and temporal resolution, combined with knowledge of additional iron-cycling processes in the lakes, could allow for the sources of the observed isotopic fractionation in both lakes to be determined, with implications for reconstructing ancient biogeochemical cycles from the rock record.
Our research provides insight into the metabolic diversity and ecology of anoxygenic photoautotrophs in seasonally anoxic Boreal Shield lakes. We show that high relative abundance genome bins of Chlorobia in lake metagenomes can encode the cyc2 gene associated with iron oxidation but that phototrophic sulfide oxidation is more widespread and reproducible in the laboratory. Although we were unable to induce photoferrotrophic growth, we enriched the novel cyc2-encoding species “Ca. Chl. canadense”, which could be used in future research exploring the function and regulation of cyc2 as genetic studies continue to progress in linking the cyc2 gene product to its cellular role [86]. Probing the metabolic diversity of anoxygenic phototrophs in Boreal Shield lakes could lead to novel inferences about photosynthesis in early Earth oceans, including the role of cryptic sulfur cycling compared to photoferrotrophic activity. Novel phototrophic metabolisms, such as recently reported phototrophic oxidation of manganese(II) [87], could also be explored with implications for both modern and prehistoric biogeochemical cycling. Altogether, our findings serve as an important basis for future work probing the biogeography and activity of anoxygenic phototrophs in natural waters. Understanding the metabolic diversity of phototrophy in Boreal Shield lakes will enhance our comprehension of modern boreal ecosystems with additional implications for the evolution of early life on Earth.
Supplementary information
Acknowledgements
We thank R. Henderson and staff at the IISD-ELA for assistance in field sampling and interpretation of the dynamics of ELA lakes. In addition, we thank S.A. Crowe for valuable feedback on the paper, K.J. Thompson for helpful discussion related to the work, L.H. Bergstrand for assistance with bioinformatics, and K.E. Engel, R.C. Beaver, and N.A. Shaw for assistance with laboratory tasks. JMT thanks M.S.M. Jetten, C.U. Welte, and staff at the Soehngen Institute of Anaerobic Microbiology, for providing helpful training for anaerobic microbiology methods. We also thank the editor and anonymous peer reviewers for helpful feedback that enhanced the quality and scope of this work. This research was supported by Discovery Grants and a Strategic Partnership Grant for Projects from the National Sciences and Engineering Research Council of Canada.
Author contributions
SLS, JJV, and LM were involved in the overall lake sampling project that led to this work and provided input and assistance with sample collection and data interpretation. JMT and JDN designed the experimental work in this study. JMT, NT, and MT performed enrichment cultivation with support from SH. JMT analyzed the sequence data and drafted the paper with the edits and feedback of all authors.
Data availability
Raw metagenome reads for the freshwater lake and enrichment culture metagenomes are available in the NCBI sequence read archive (SRA) under BioProjects PRJNA518727 and PRJNA534305, respectively. Six curated genome bins of Chlorobia are available under the same BioProjects, along with all assembled contigs from freshwater lake metagenomes. The complete set of uncurated genome bins were uploaded to a Zenodo repository at 10.5281/zenodo.2720705. Code for downloading these data and performing the analyses presented in this paper, along with intermediate data files, are available at https://github.com/jmtsuji/Chlorobia-cyc2-genomics (10.5281/zenodo.3228523).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0725-0) contains supplementary material, which is available to authorized users.
References
- 1.Keller W. Implications of climate warming for Boreal Shield lakes: a review and synthesis. Environ Rev. 2007;15:99–112. [Google Scholar]
- 2.Teodoru CR, del Giorgio PA, Prairie YT, Camire M. Patterns in p CO2 in boreal streams and rivers of northern Quebec, Canada. Glob Biogeochem Cycles. 2009;23:GB2012. [Google Scholar]
- 3.Anas MUM, Scott KA, Wissel B. Carbon budgets of boreal lakes: state of knowledge, challenges, and implications. Environ Rev. 2015;23:275–87. [Google Scholar]
- 4.Schiff SL, Tsuji JM, Wu L, Venkiteswaran JJ, Molot LA, Elgood RJ, et al. Millions of Boreal Shield lakes can be used to probe Archaean Ocean biogeochemistry. Sci Rep. 2017;7:46708. doi: 10.1038/srep46708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulton SW, Canfield DE. Ferruginous conditions: a dominant feature of the ocean through Earth’s history. Elements. 2011;7:107–12. [Google Scholar]
- 6.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 7.Karhunen J, Arvola L, Peura S, Tiirola M. Green sulphur bacteria as a component of the photosynthetic plankton community in small dimictic humic lakes with an anoxic hypolimnion. Aquat Microb Ecol. 2013;68:267–72. [Google Scholar]
- 8.Imhoff JF. The family Chromatiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin Heidelberg: Springer; 2014. pp. 151–78. [Google Scholar]
- 9.Imhoff JF. The family Chlorobiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin Heidelberg: Springer; 2014. pp. 501–14. [Google Scholar]
- 10.Ehrenreich A, Widdel F. Anaerobic oxidation of ferrous iron by purple bacteria, a new type of phototrophic metabolism. Appl Environ Microbiol. 1994;60:4517–26. doi: 10.1128/aem.60.12.4517-4526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin BM, Schott J, Schink B. Nitrite, an electron donor for anoxygenic photosynthesis. Science. 2007;316:1870. doi: 10.1126/science.1139478. [DOI] [PubMed] [Google Scholar]
- 12.Kulp TR, Hoeft SE, Asao M, Madigan MT, Hollibaugh JT, Fisher JC, et al. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science. 2008;321:967–70. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- 13.Thiel V, Tank M, Bryant DA. Diversity of chlorophototrophic bacteria revealed in the omics era. Annu Rev Plant Biol. 2018;69:21–49. doi: 10.1146/annurev-arplant-042817-040500. [DOI] [PubMed] [Google Scholar]
- 14.Heising S, Richter L, Ludwig W, Schink B. Chlorobium ferrooxidans sp. nov., a phototrophic green sulfur bacterium that oxidizes ferrous iron in coculture with a “Geospirillum” sp. strain. Arch Microbiol. 1999;172:116–24. doi: 10.1007/s002030050748. [DOI] [PubMed] [Google Scholar]
- 15.Llirós M, García–Armisen T, Darchambeau F, Morana C, Triadó–Margarit X, Inceoğlu Ö, et al. Pelagic photoferrotrophy and iron cycling in a modern ferruginous basin. Sci Rep. 2015;5:13803. doi: 10.1038/srep13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laufer K, Niemeyer A, Nikeleit V, Halama M, Byrne JM, Kappler A. Physiological characterization of a halotolerant anoxygenic phototrophic Fe(II)-oxidizing green-sulfur bacterium isolated from a marine sediment. FEMS Microbiol Ecol. 2017;93:fix054. doi: 10.1093/femsec/fix054. [DOI] [PubMed] [Google Scholar]
- 17.Lambrecht N. Insights into early Earth ocean biogeochemistry from intensive monitoring of two ferruginous meromictic lakes. PhD thesis. Iowa, USA:Iowa State University;2019.
- 18.Frigaard N-U, Bryant DA. Genomic insights into the sulfur metabolism of phototrophic green sulfur bacteria. In: Hell R, Dahl DC, Knaff D, Leustek T, editors. Sulfur metabolism in phototrophic organisms. Netherlands: Springer; 2008. pp. 337–55. [Google Scholar]
- 19.Camacho A, Walter XA, Picazo A, Zopfi J. Photoferrotrophy: remains of an ancient photosynthesis in modern environments. Front Microbiol. 2017;8:323. doi: 10.3389/fmicb.2017.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koeksoy E, Halama M, Konhauser KO, Kappler A. Using modern ferruginous habitats to interpret Precambrian banded iron formation deposition. Int J Astrobiol. 2016;15:205–17. [Google Scholar]
- 21.He S, Barco RA, Emerson D, Roden EE. Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front Microbiol. 2017;8:1584. doi: 10.3389/fmicb.2017.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melton ED, Swanner ED, Behrens S, Schmidt C, Kappler A. The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle. Nat Rev Microbiol. 2014;12:797–808. doi: 10.1038/nrmicro3347. [DOI] [PubMed] [Google Scholar]
- 23.Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M, et al. Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol. 2015;6:1265. doi: 10.3389/fmicb.2015.01265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta D, Sutherland MC, Rengasamy K, Meacham JM, Kranz RG, Bose A. Photoferrotrophs produce a PioAB electron conduit for extracellular electron uptake. mBio. 2019;10:e02668–19. doi: 10.1128/mBio.02668-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castelle C, Guiral M, Malarte G, Ledgham F, Leroy G, Brugna M, et al. A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J Biol Chem. 2008;283:25803–11. doi: 10.1074/jbc.M802496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barco RA, Emerson D, Sylvan JB, Orcutt BN, Meyers MEJ, Ramírez GA, et al. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol. 2015;81:5927–37. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAllister SM, Polson SW, Butterfield DA, Glazer BT, Sylvan JB, Chan CS. Validating the Cyc2 neutrophilic iron oxidation pathway using meta-omics of Zetaproteobacteria iron mats at marine hydrothermal vents. mSystems. 2020;5:e00553–19. doi: 10.1128/mSystems.00553-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crowe SA, Hahn AS, Morgan-Lang C, Thompson KJ, Simister RL, Llirós M, et al. Draft genome sequence of the pelagic photoferrotroph Chlorobium phaeoferrooxidans. Genome Announc. 2017;5:e01584–16. doi: 10.1128/genomeA.01584-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson KJ, Simister RL, Hahn AS, Hallam SJ, Crowe SA. Nutrient acquisition and the metabolic potential of photoferrotrophic Chlorobi. Front Microbiol. 2017;8:1212. doi: 10.3389/fmicb.2017.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryce C, Blackwell N, Straub D, Kleindienst S, Kappler A. Draft genome sequence of Chlorobium sp. strain N1, a marine Fe(II)-oxidizing green sulfur bacterium. Microbiol Resour Announc. 2019;8:e00080–19. doi: 10.1128/MRA.00080-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindler DW, Armstrong FAJ, Holmgren SK, Brunskill GJ. Eutrophication of Lake 227, Experimental Lakes Area, northwestern Ontario, by addition of phosphate and nitrate. J Fish Res Board Can. 1971;28:1763–82. [Google Scholar]
- 32.Campbell P. Phosphorus budgets and stoichiometry during the open-water season in two unmanipulated lakes in the Experimental Lakes Area, northwestern Ontario. Can J Fish Aquat Sci. 1994;51:2739–55. [Google Scholar]
- 33.White RA, III, Brown J, Colby S, Overall CC, Lee J-Y, Zucker J, et al. ATLAS (Automatic Tool for Local Assembly Structures) - a comprehensive infrastructure for assembly, annotation, and genomic binning of metagenomic and metatranscriptomic data. PeerJ Prepr. 2017;5:e2843v1. [Google Scholar]
- 34.Kieser S, Brown J, Zdobnov EM, Trajkovski M, McCue LA. ATLAS: a Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinform. 2020;21:257. doi: 10.1186/s12859-020-03585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeans C, Singer SW, Chan CS, VerBerkmoes NC, Shah M, Hettich RL, et al. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2008;2:542–50. doi: 10.1038/ismej.2008.17. [DOI] [PubMed] [Google Scholar]
- 36.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–8. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve Aligner. Bioinformatics. 2009;25:2071–3. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 46.Armstrong FAJ, Schindler DW. Preliminary chemical characterization of waters in the Experimental Lakes Area, northwestern Ontario. J Fish Res Board Can. 1971;28:171–87. [Google Scholar]
- 47.Schindler DW. Eutrophication and recovery in experimental lakes: implications for lake management. Science. 1974;184:897–9. doi: 10.1126/science.184.4139.897. [DOI] [PubMed] [Google Scholar]
- 48.Schindler D. The coupling of elemental cycles by organisms: evidence from whole-lake chemical perturbations. In: Stumm W, editor. Chemical processes in lakes. New York, NY: John Wiley and Sons; 1985. pp. 225–250. [Google Scholar]
- 49.Schindler DW. The dilemma of controlling cultural eutrophication of lakes. Proc R Soc B Biol Sci. 2012;279:4322–33. [DOI] [PMC free article] [PubMed]
- 50.Curtis PJ, Schindler DW. Hydrologic control of dissolved organic matter in low-order Precambrian Shield lakes. Biogeochemistry. 1997;36:125–38. [Google Scholar]
- 51.Hegler F, Posth NR, Jiang J, Kappler A. Physiology of phototrophic iron(II)-oxidizing bacteria: implications for modern and ancient environments. FEMS Microbiol Ecol. 2008;66:250–60. doi: 10.1111/j.1574-6941.2008.00592.x. [DOI] [PubMed] [Google Scholar]
- 52.Milucka J, Kirf M, Lu L, Krupke A, Lam P, Littmann S, et al. Methane oxidation coupled to oxygenic photosynthesis in anoxic waters. ISME J. 2015;9:1991–2002. doi: 10.1038/ismej.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clavier CGJ, Boucher G. The use of photosynthesis inhibitor (DCMU) for in situ metabolic and primary production studies on soft bottom benthos. Hydrobiologia. 1992;246:141–5. [Google Scholar]
- 54.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee MD. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. 2019;35:4162–4. doi: 10.1093/bioinformatics/btz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 57.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 58.Bergstrand LH, Cardenas E, Holert J, Hamme JDV, Mohn WW. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio. 2016;7:e00166–16. doi: 10.1128/mBio.00166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bryant DA, Liu Z, Li T, Zhao F, Costas AMG, Klatt CG, et al. Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria. In: Burnap R, Vermaas W, et al., editors. Functional genomics and evolution of photosynthetic systems. Netherlands, Dordrecht: Springer; 2012. pp. 47–102. [Google Scholar]
- 60.Tourova TP, Kovaleva OL, Gorlenko VM, Ivanovsky RN. Use of genes of carbon metabolism enzymes as molecular markers of Chlorobi phylum representatives. Microbiology. 2013;82:784–93. [PubMed] [Google Scholar]
- 61.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, et al. FunGene: the functional gene pipeline and repository. Front Microbiol. 2013;4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:1–11. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrenko P, Lobb B, Kurtz DA, Neufeld JD, Doxey AC. MetAnnotate: function-specific taxonomic profiling and comparison of metagenomes. BMC Biol. 2015;13:1–8. doi: 10.1186/s12915-015-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garber AI, Nealson KH, Okamoto A, McAllister SM, Chan CS, Barco RA, et al. FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37. doi: 10.3389/fmicb.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfennig N. Rhodocyclus purpureus gen. nov. and sp. nov., a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int J Syst Evol Microbiol. 1978;28:283–8. [Google Scholar]
- 67.Tank M, Bryant DA. Nutrient requirements and growth physiology of the photoheterotrophic Acidobacterium, Chloracidobacterium thermophilum. Front Microbiol. 2015;6:226. [DOI] [PMC free article] [PubMed]
- 68.Peng C, Bryce C, Sundman A, Borch T, Kappler A. Organic matter complexation promotes Fe(II) oxidation by the photoautotrophic Fe(II)-oxidizer Rhodopseudomonas palustris TIE-1. ACS Earth Space Chem. 2019;3:531–6.
- 69.Stookey LL. Ferrozine—a new spectrophotometric reagent for iron. Anal Chem. 1970;42:779–81. [Google Scholar]
- 70.Verschoor MJ, Molot LA. A comparison of three colorimetric methods of ferrous and total reactive iron measurement in freshwaters. Limnol Oceanogr Methods. 2013;11:113–25. [Google Scholar]
- 71.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Overmann J, Coolen MJL, Tuschak C. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch Microbiol. 1999;172:83–94. doi: 10.1007/s002030050744. [DOI] [PubMed] [Google Scholar]
- 73.Mori Y, Purdy KJ, Oakley BB, Kondo R. Comprehensive detection of phototrophic sulfur bacteria using PCR primers that target reverse dissimilatory sulfite reductase gene. Microbes Environ. 2010;25:190–6. doi: 10.1264/jsme2.me10109. [DOI] [PubMed] [Google Scholar]
- 74.Finneran KT, Johnsen CV, Lovley DR. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III) Int J Syst Evol Microbiol. 2003;53:669–73. doi: 10.1099/ijs.0.02298-0. [DOI] [PubMed] [Google Scholar]
- 75.Falagán C, Johnson DB. Acidibacter ferrireducens gen. nov., sp. nov.: an acidophilic ferric iron-reducing gammaproteobacterium. Extremophiles. 2014;18:1067–73. doi: 10.1007/s00792-014-0684-3. [DOI] [PubMed] [Google Scholar]
- 76.Watanabe T, Kojima H, Fukui M. Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst Appl Microbiol. 2014;37:387–95. doi: 10.1016/j.syapm.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Kuever J. The family Desulfobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria. Berlin Heidelberg: Springer; 2014. pp. 45–73. [Google Scholar]
- 78.van Grinsven S, Damsté JSS, Asbun AA, Engelmann JC, Harrison J, Villanueva L. Methane oxidation in anoxic lake water stimulated by nitrate and sulfate addition. Environ Microbiol. 2019;22:766–82. doi: 10.1111/1462-2920.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Grinsven S, Damsté JSS, Harrison J, Villanueva L. Impact of electron acceptor availability on methane-influenced microorganisms in an enrichment culture obtained from a stratified lake. Front Microbiol. 2020;11:715. doi: 10.3389/fmicb.2020.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kappler A, Bryce C. Cryptic biogeochemical cycles: unravelling hidden redox reactions. Environ Microbiol. 2017;19:842–6. doi: 10.1111/1462-2920.13687. [DOI] [PubMed] [Google Scholar]
- 81.Walter XA, Picazo A, Miracle MR, Vicente E, Camacho A, Aragno M, et al. Phototrophic Fe(II)-oxidation in the chemocline of a ferruginous meromictic lake. Front Microbiol. 2014;5:713. doi: 10.3389/fmicb.2014.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen JWA, Sawyer EB, Ginger ML, Barker PD, Ferguson SJ. Variant c-type cytochromes as probes of the substrate specificity of the E. coli cytochrome c maturation (Ccm) apparatus. Biochem J. 2009;419:177–86. doi: 10.1042/BJ20081999. [DOI] [PubMed] [Google Scholar]
- 83.He S, Lau MP, Linz AM, Roden EE, McMahon KD. Extracellular electron transfer may be an overlooked contribution to pelagic respiration in humic-rich freshwater lakes. mSphere. 2019;4:e00436–18. doi: 10.1128/mSphere.00436-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guzman MS, Rengasamy K, Binkley MM, Jones C, Ranaivoarisoa TO, Singh R, et al. Phototrophic extracellular electron uptake is linked to carbon dioxide fixation in the bacterium Rhodopseudomonas palustris. Nat Commun. 2019;10:1355. doi: 10.1038/s41467-019-09377-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Croal LR, Jiao Y, Kappler A, Newman DK. Phototrophic Fe(II) oxidation in an atmosphere of H2: implications for Archean banded iron formations. Geobiology. 2009;7:21–4. doi: 10.1111/j.1472-4669.2008.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chan C, McAllister SM, Garber A, Hallahan BJ, Rozovsky S. Fe oxidation by a fused cytochrome-porin common to diverse Fe-oxidizing bacteria. bioRxiv. 2018. 10.1101/228056. [DOI] [PMC free article] [PubMed]
- 87.Daye M, Klepac-Ceraj V, Pajusalu M, Rowland S, Farrell-Sherman A, Beukes N, et al. Light-driven anaerobic microbial oxidation of manganese. Nature. 2019;576:311–4. doi: 10.1038/s41586-019-1804-0. [DOI] [PubMed] [Google Scholar]
- 88.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 89.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw metagenome reads for the freshwater lake and enrichment culture metagenomes are available in the NCBI sequence read archive (SRA) under BioProjects PRJNA518727 and PRJNA534305, respectively. Six curated genome bins of Chlorobia are available under the same BioProjects, along with all assembled contigs from freshwater lake metagenomes. The complete set of uncurated genome bins were uploaded to a Zenodo repository at 10.5281/zenodo.2720705. Code for downloading these data and performing the analyses presented in this paper, along with intermediate data files, are available at https://github.com/jmtsuji/Chlorobia-cyc2-genomics (10.5281/zenodo.3228523).