Abstract
NAD+ and its derivatives NADH, NADP+, and NADPH are essential cofactors in redox reactions and electron transport pathways. NAD serves also as substrate for an extensive series of regulatory enzymes including cyclic ADP-ribose hydrolases, mono(ADP-ribosyl)transferases, poly(ADP-ribose) polymerases, and sirtuin deacetylases which are O-acetyl-ADP-ribosyltransferases. As a result of the numerous and diverse enzymes that utilize NAD as well as depend on its synthesis and concentration, significant interest has developed in its role in a variety of physiologic and pathologic processes, and therapeutic initiatives have focused both on augmenting its levels as well as inhibiting some of its pathways. In this article, we examine the biosynthesis of NAD, metabolic processes in which it is involved, and its role in aging, cancer, and other age-associated comorbidities including neurodegenerative, cardiovascular, and metabolic disorders. Therapeutic interventions to augment and/or inhibit these processes are also discussed.
Impact statement
NAD is a central metabolite connecting energy balance and organismal growth with genomic integrity and function. It is involved in the development of malignancy and has a regulatory role in the aging process. These processes are mediated by a diverse series of enzymes whose common focus is either NAD’s biosynthesis or its utilization as a redox cofactor or enzyme substrate. These enzymes include dehydrogenases, cyclic ADP-ribose hydrolases, mono(ADP-ribosyl)transferases, poly(ADP-ribose) polymerases, and sirtuin deacetylases. This article describes the manifold pathways that comprise NAD metabolism and promotes an increased awareness of how perturbations in these systems may be important in disease prevention and/or progression.
Keywords: NAD, poly(ADP-Ribose), mono(ADP-ribose), sirtuins, aging, cancer
Introduction
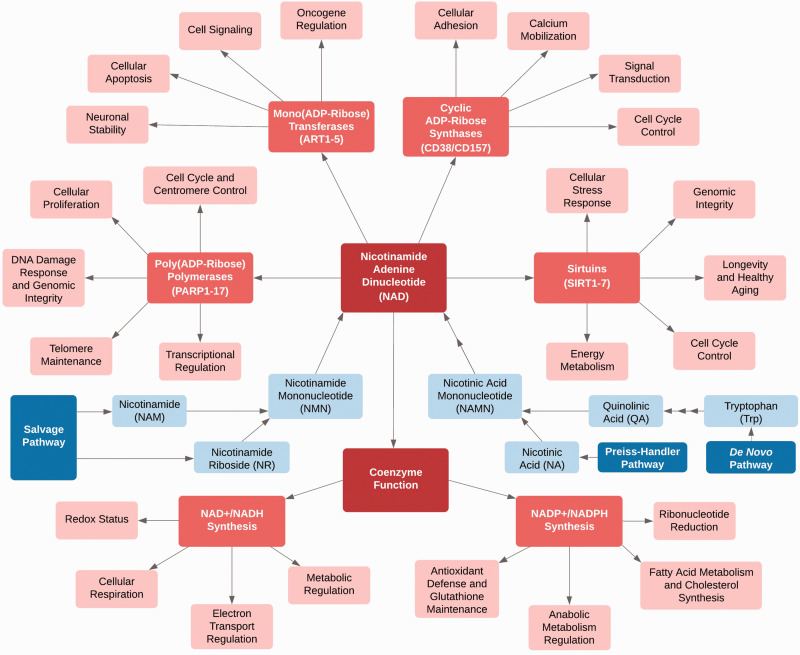
Nicotinamide adenine dinucleotide (NAD) is a soluble, multifunctional metabolite consisting of the covalent linkage of two mononucleotides: adenosine monophosphate (AMP) and nicotinamide mononucleotide (NMN).1 The dinucleotide NAD has a fascinating biomolecular, epidemiologic, and translational history. NAD, along with its reduced counterpart, NADH, plays a critical role in the regulation of cellular respiration, electron transport, and redox status.2 Phosphorylation of NAD yields the triphosphorylated metabolite, NADP, which, along with its reduced counterpart, NADPH, plays important regulatory roles in anabolic metabolism and is essential for lipid and cholesterol synthesis and fatty acid elongation.2–4 In addition to its function as a coenzyme in oxidation reduction reactions, NAD was found to be a substrate for catalytic cleavage yielding nicotinamide and ADP-ribose moieties, the latter being used for covalent modification of protein structure and function, and for synthesis of the poly nucleotide, poly(ADP-ribose), which may contain multiple ADP-ribose units, covalently linked in either linear or branch chain fashion.4–6 Polymer synthesis and degradation are involved in modulating the DNA damage response, transcription, translation, and a multitude of other biological and physiologic processes.5,6 More recently, NAD has been identified as an essential cofactor for longevity-promoting sirtuins, where it facilitates deacetylation and is cleaved to generate nicotinamide and o-acetyl ADP-ribose.7,8 Given NAD’s manifold and diverse functions and ubiquitous presence, it is not surprising that organisms have developed multiple systems for regulation of its synthesis, utilization, and distribution, and that glitches in these systems may have profound biologic and disease-related effects (Figure 1).
Figure 1.
Central role of NAD, biosynthesis, metabolism, and function.
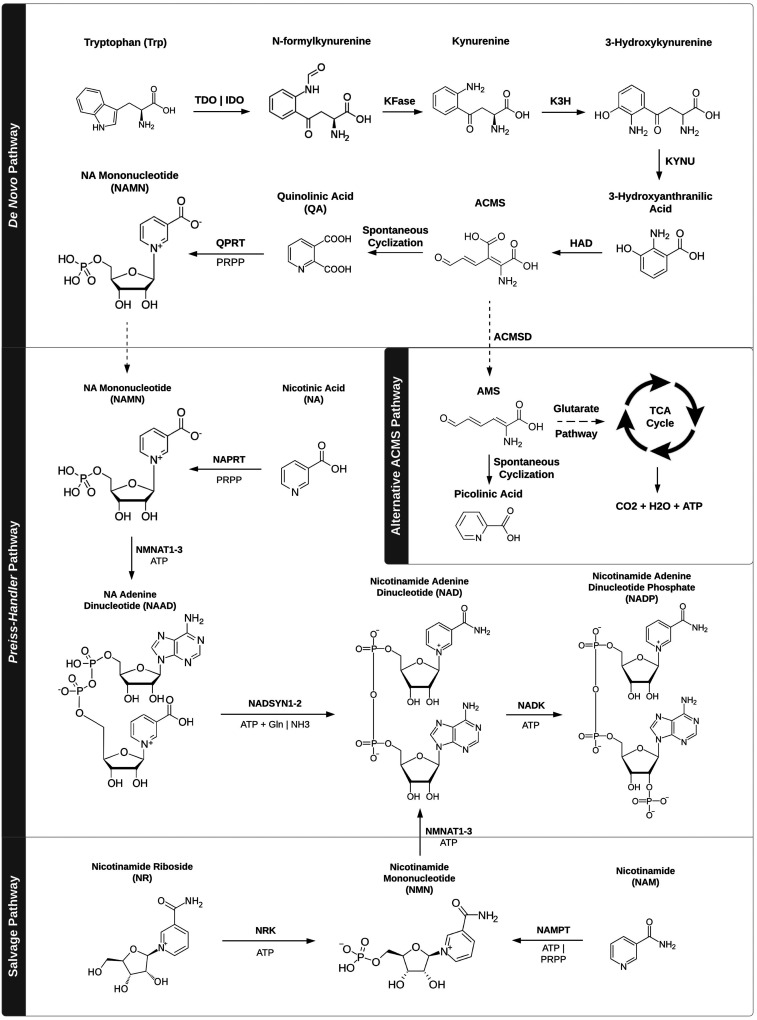
An early indication of the biomedical importance of NAD was realized when the disease pellagra was shown to result from nicotinamide deficiency and to be cured by dietary nicotinamide supplementation.9,10 Recent observations have suggested that aberrations in NAD levels and/or its utilization may contribute to multiple disorders including the aging process, carcinogenesis, neurodegenerative diseases, ischemia reperfusion injury, and inflammatory disorders.11–16 As a consequence, supplementation of NAD and/or its precursors has become a popular approach and strategic target to retard the aging process and ameliorate some of its comorbidities; however, this approach may enhance the risk of carcinogenesis.11–16 Conversely, NAD synthesis and its utilization have become an important target for cancer chemotherapy, as well as a potential target for some neurodegenerative and age-related disorders.17 In this review, we outline many of the molecular processes mediated by NAD synthesis, metabolism, and utilization, particularly as they relate to aging and cancer, as well as highlight areas where these processes may be therapeutically targeted. Since augmentation of NAD levels has become a popular therapeutic objective, we start with a review of its synthesis and precursors (Figure 2).
Figure 2.
NAD biosynthetic pathways
Biosynthesis of NAD
The de novo pathway
In de novo NAD synthesis, tryptophan (Trp), via the first rate-limiting step of the pathway, is aerobically converted to N-formylkynurenine by either tryptophan-2,3-dioxygenase (TDO) or indoleamine-2,3-dioxygenase (IDO). IDO has shown expression in a diverse range of cell types within the small intestine, lung, heart, and less so in neuronal tissue, whereas TDO activity is mainly localized to the liver.18,19 In addition, TDO and IDO have been implicated in the pathogenesis of a variety of diseases, including cancer, diabetes, and a number of inflammatory disorders,20–23 highlighting the potential that NAD biosynthetic pathways may serve as targets for therapeutic development. N-formylkynurenine is subsequently converted to kynurenine by kynurenine formamidase (KFase), then hydroxylated to form 3-hydroxykynurenine via the NADPH-dependent action of kynurenine-3-hydroxylase (K3H). The subsequent two enzymatic reactions convert 3-hydroxykynurenine to a high-energy intermediate, 2-amino-3-carboxy-muconate-semialdehyde (ACMS). ACMS can then react with ACMS decarboxylase (ACMSD), forming 2-amino-3-muconate-semialdehyde (AMS) and exit the de novo pathway. Following its anabolism, AMS will either react spontaneously to form picolinic acid or follow the glutarate pathway, eventually leading to entrance into the tricarboxylic acid (TCA) cycle to form CO2, H2O, and ATP. However, ACMS can also undergo spontaneous cyclization to quinolinic acid (QA), which can be converted to nicotinic acid mononucleotide (NAMN) by quinolinate phosphoribosyltransferase (QPRT). The action of QPRT is comparatively inefficient relative to that of ACMSD, and only when ACMSD becomes saturated is QA converted to NAMN, making this reaction the second rate-limiting step of the de novo synthetic pathway.24 This infrequent reaction remains a key contributor to the relative inefficiency of de novo NAD synthesis,25 with prior study indicating an intake of 60 mg of dietary Trp to be equivalent to 1 mg of niacin.26 NAMN, a key precursor of NAD, will enter as an intermediate of the Preiss-Handler pathway after conversion from QA.
The Preiss-Handler pathway
In 1958, Jack Preiss and Philip Handler utilized an in vivo rat model and cultured human erythrocytes to discover nicotinic acid’s (NA) conversion to NAD via a three-step synthetic pathway.27,28 Similar to QA, NA enters the Preiss-Handler pathway as NAMN through its metabolism by NA phosphoribosyltransferase (NAPRT), which uses phosphoribosyl pyrophosphate (PRPP) as a substrate. NAPRT is allosterically regulated by ATP, being stimulated at concentrations <100 µM and inhibited at concentrations of 100–640 µM. In addition, the activity of NAPRT is regulated by a number of molecules involved in aerobic respiration, including stimulation by pyruvate and dihydroxyacetone phosphate (DHAP), and inhibition by fructose-1,6-bisphosphate (F1,6BP), glyceraldehyde-3-phosphate (G3P), CoA, succinyl-CoA, and acetyl-CoA.29 NAMN is then converted to NA adenine dinucleotide (NAAD) by the ATP-dependent action of a variety of NMN adenylyltransferases (NMNAT1-3). NMNAT expression is characterized by varied subcellular and tissue-specific distributions, indicating the compartmentalization of NAD metabolism.30–35 NMNAT1 is located solely in the nucleus, and is highly expressed in the skeletal muscle and heart, moderately in the liver and kidney, and slightly in neuronal tissue.30,32 NMNAT2, located in the Golgi apparatus and cytosol, has shown activity largely in the brain with lower level expression in skeletal muscle and heart.31,34 Lastly, NMNAT3 is expressed in the mitochondria and cytosol, and is mainly located in the lung and spleen.35 To conclude the Preiss-Handler pathway, NAAD is converted to NAD by NAD synthetases (NADSYN1-2), which, similar to NMNATs, are also differentially compartmentalized. NADSYNs synthesize NAD through catalysis of ATP and transfer of an amide group from glutamine or ammonia. NADSYN1, which can utilize either amide group as a donor to NAAD, is located largely in the small intestine, liver, kidney, and testicles, and weakly expressed in the skeletal muscle and heart.36 Conversely, NADSYN2 activity is exclusively ammonia-dependent, and its expression is low in the small intestine, liver, and kidney, and high in skeletal muscle and heart.37
The salvage pathway
Apart from NA, additional metabolite recycling and the intake of other nicotinic precursors can be used to synthesize NAD through the salvage pathway.38 Nicotinamide (NAM) is converted to NAM mononucleotide (NMN) by nicotinamide phosphoribosyltransferases (NAMPTs) in the first rate-limiting step of the pathway which uses PRPP as a cofactor.39 NAMPTs exist in both intracellular (iNAMPT) and extracellular (eNAMPT) forms, of which the extracellular variant demonstrates an increased propensity towards NMN biosynthesis. To this end, a relatively high concentration of both NAMPT and NMN have been found circulating in human plasma.40 Alternatively represented as pre-B cell colony-enhancing factor (PBEF), eNAMPT was originally discovered as a cytokine implicated in cell colony formation.41 Additionally, eNAMPT production has been connected to the action of both leukocytes and adipocytes, and deacetylation of iNAMPT via SIRT1 results in stimulated adipocyte eNAMPT secretion in mice.42 iNAMPT is located both in the nucleus and cytosol, and demonstrates diverse tissue expression with activity mainly occurring in the bone marrow, liver, and skeletal muscle.43–45 NAMPTs have also been linked to the pathogenesis of a variety of human diseases including cancer, diabetes mellitus, stroke, atherosclerosis, and rheumatoid arthritis.46,47 NAM riboside (NR), another NAD precursor, may also enter the salvage pathway and be converted to NMN by NAM riboside kinases (NRK1-2), a highly conserved enzymatic family in eukaryotes.48 NRK1 is universally expressed across human tissue, whereas NRK2 exhibits variable expression largely in the skeletal muscle, heart, and brain.49 Following formation from either NAM or NR, NMN is converted to NAD by the same NMNAT enzymes of the Preiss-Handler pathway which adenylate NAMN to NAAD.2,50
Biosynthesis of NADP
A phosphorylated analogue of NAD, NADP is synthesized by NAD kinases (NADK), which transfer a phosphate from ATP to the 2ʹ-hydroxyl group on the ribose portion of NAD.51 NADK has also shown high selectivity towards its substrates ATP and NAD, as well as the use of Ca2+, Mg2+, and Mn2+ as cofactors, mechanisms originating both from evolutionary divergence and conservation.4 In addition, the process may be modulated by calmodulin kinase-dependent phosphorylation, as well as the possibility of regulation via feedback inhibition.52 NADK is ubiquitously expressed in human tissue although the small intestine and skeletal muscle demonstrate minimal activity.53 Recent findings have also indicated the varied subcellular compartmentation of NADK isoforms: a cytoplasmic form and a mitochondrial form (MNADK).54 Although both variants elicit the same tissue-specific activity, MNADK has demonstrated significantly higher expression than its cytosolic counterpart.55
Compartmentalization of NAD
In alignment with its diverse functionality, NAD demonstrates varied tissue-specific and subcellular compartmentalization. While the intracellular concentration of NAD is usually conserved between 0.2 and 0.5 mM, NAD levels can shift approximately 2-fold depending on its location in tissue or the cell, as well as in the presence of various physiological stimuli. Because of NAD’s role as a redox carrier in cellular respiration, many metabolic conditions associated with energy stress (i.e. exercise and fasting) will increase NAD formation. In addition, NAD has been shown to be a key metabolite in the mediation of circadian rhythm.56 Since NAD lacks the ability to diffuse across cell membranes,57 the mechanisms by which intracellular levels of NAD are compartmentally regulated play a major role in the molecule’s metabolism.
The localization of NMNATs, as well as the presence of NAD-dependent enzymes (i.e. SIRTs and PARPs), in the cytosol, mitochondria, and nucleus, indicate the requirement for NAD synthesis in these subcellular compartments. In addition, each member of the NMNAT enzymatic family demonstrate unique cell-specificity: NMNAT1 to the nucleus, NMNAT2 to the Golgi apparatus and cytosol, and NMNAT3 to the mitochondria and cytosol.58 This distribution suggests that NAD metabolism is differentially regulated to meet the individual demands of each compartment. Thus, Yang et al.59 found that mitochondrial NAD levels can remain homeostatically balanced despite depletion of nuclear and cytoplasmic NAD pools, suggesting that compartment-specific NAD metabolism may be independent of NAD-centered activity in other compartments. Studies have also demonstrated unequal compartmental distribution, with mitochondrial NAD concentrations being ≥250 µM and nuclear concentrations being significantly lower at approximately 70 µM.60 While estimating cytosolic levels of NAD has proven difficult, analyses have suggested levels similar to those found in mitochondrion.61,62
Although it is widely viewed that NAD does not cross the mammalian cell membrane, recent evidence has suggested the presence of an NAD/NADH transporter.63 However, because of the purported cellular impermeability to NAD, the molecule’s homeostatic maintenance is solely dependent on derivation from NAMN or NMN synthesized via the salvage and Preiss-Handler pathways, or formation from the NAM by-product of NAD-dependent enzymes. Treatment with exogenous NAD has been shown to increase mitochondrial, cytosolic, and nuclear NAD levels, resulting in a comparatively high elevation of mitochondrial NAD, thus indicating the capacity for nicotinic precursors to permeate the mitochondrial membrane. The administration of NR has also resulted in increased levels of mitochondrial NAD.64 In addition, supplementation of NR has been shown to enhance oxidative metabolism and protect against high-fat diet-induced obesity via the activation of SIRT1 and SIRT3.50 Because NRK is not expressed in the mitochondria to convert NR to NMN, it is likely that NMN has the ability to cross the mitochondrial membrane (following cytosolic conversion from NR) for the synthesis of NAD.65 The work of Grozio et al.66 identified the first NMN transporter, which is encoded by the Slc12a8 gene and requires the presence of sodium ions for activity. These observations indicate an underlying complexity behind not only the compartments which segregate NAD pools, but also the interactions of biosynthetic and transport pathways that regulate pyridine nucleotide metabolism.
NAD functions as redox cofactor
NAD plays a crucial role as a redox carrier in eukaryotic energy metabolism, acting as a hydrogen acceptor to generate its reduced form, NADH, and contributing to processes such as glycolysis, the TCA cycle, and oxidative phosphorylation. It is these compartmentally segregated pathways that make the independence of subcellular NAD pools necessary. Located in the cytosol, the glycolytic pathway metabolizes glucose through a series of enzymatic reactions, resulting in the synthesis of two molecules of NADH, ATP, and pyruvate per glucose molecule catalyzed; a process consuming two molecules of NAD and ADP. Following conversion of glucose to fructose-1,6-bisphosphate (F1,6BP) via the first three steps of the pathway, F1,6BP is cleaved to form two molecules of glyceraldehyde-3-phosphate (G3P). Next, G3P is oxidized and subsequently phosphorylated to make 1,3-bisphosphoglycerate by glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which couples this favorable reaction to the reduction of NAD to NADH.67 The energy harnessed in NADH is used in the production of ATP from oxidative phosphorylation, but due to the impermeability of the mitochondrial membrane to NADH,57 the action of the malate-aspartate or glycerol-3 phosphate shuttles is used to transport a reducing equivalent of NADH into the mitochondrial matrix. The TCA cycle, which functions in the mitochondrion, is another contributor of reducing equivalents for oxidative phosphorylation, producing six molecules of NADH per turn of the cycle.
Mitochondrial NADH, following synthesis from either the glycolytic pathway or TCA cycle, are oxidized by Complex I (NADH:ubiquinone reductase), the first of four multiprotein structures which compose the electron transport chain (ETC).68 The two electrons received from oxidizing NADH are transported through a series of redox reactions relayed through Complex I, ubiquinone (Coenzyme Q), Complex III (Coenzyme Q-cytochrome c oxidoreductase), cytochrome c, and finally Complex IV (cytochrome c oxidase). The oxidation of NADH (as well as succinate and FAD) is coupled to the transport of protons across the mitochondrial inner membrane to the intermembrane space, creating an osmotic gradient. Protons then diffuse back to the matrix via osmotic flux, a process whose energy release is coupled to the synthesis of ATP by ATP synthase.69 NAD’s role in oxidative phosphorylation contributes to the molecule’s high mitochondrial concentration.70 At times of nutritional abundance, the energy producing metabolic balance shifts the ratio of NAD+/NADH to increase NADH pools. In contrast, at times of nutritional restriction and/or heavy energy utilization, the oxidized to reduced ratio is shifted to favor increased NAD+.71 In addition, the levels of NAD can be limiting to the glycolytic pathway, TCA cycle, and ETC,72 meaning their function may contribute to homeostatic imbalances in both nuclear and cytosolic NAD pools and vice versa.61,73 For example, DNA damage, which activates poly(ADP-ribose) polymerase (PARP1) to synthesize ADP-ribose polymers, causes systemic NAD depletion via the molecule’s function as a PARP substrate, which induces downstream inhibition of glycolysis, oxidative phosphorylation, and eventually cell death.74,75 Inhibition of PARP1 increases SIRT1 activity, suggesting a reciprocal relationship between these NAD-dependent enzymes,76 as well as the need to understand the mechanisms regulating the interdependence of compartmentalized NAD pools. NADP and its reduced counterpart, NADPH, likewise play a vital role in electron transport and biosynthetic pathways, particularly for fatty acid and cholesterol synthesis and for reduction of ribonucleotides to deoxynucleotides. In addition, NADPH is important for maintaining reduced glutathione pools for protection against oxidative stress.2,3
NAD functions as substrate
Mono-ADP-ribosylation
The covalent addition of an adenosine diphosphate ribose (ADP-ribose) moiety to proteins is a post-translational modification (PTM) catalyzed both by mono(ADP-ribosyl)transferases and/or poly(ADP-ribosyl)transferases.77–79 Both sets of enzymes cleave NAD at the N-glycosidic bond between nicotinamide and ADP-ribose, liberating free nicotinamide and covalently attaching the ADP-ribose moiety to protein amino acid side chains, including arginine, asparagine, glutamate, lysine, serine, and/or cysteine residues.80,81 As discussed below, the poly(ADP-ribose) polymerases continue to cleave NAD and attach subsequent ADP-ribose moieties to growing chains of poly(ADP-ribose).5
The mono(ADP-ribosyl)transferases (mARTs) are widely distributed among prokaryotes and eukaryotes where their function and regulatory selectivity appear to be related to their organ and subcellular distribution.79,81 mARTs serve as a common mechanism for several prokaryotic toxins.82 ADP-ribosylation of an arginine residue on a membrane-bound GTP-binding protein by cholera toxin or E. coli heat-labile enterotoxin increases the activity of adenylyl cyclase resulting in intestinal fluid and electrolyte flux leading to severe fluid loss and dehydration.83,84 Both diphtheria toxin and pseudomonas toxin ADP-ribosylate a modified histidine residue in eukaryotic elongation factor 2, which leads to cell death through the inhibition of protein synthesis.85 Some of the effects of these toxins can be modulated by (ADP-ribosyl)hydrolases (ARHs) which remove the protein bound ADP-ribose residues.84
At least five mARTs (ART1–5) have been identified in mammalian tissues,5 including cardiac and skeletal muscle, brain, testes, and hematologic, including erythrocytes, platelets, and lymphocytes, where their function may be further defined by subcellular location.5,81,85 A variety of acceptor proteins have likewise been identified suggesting a regulatory role for mono-ADP-ribosylation in a multitude of metabolic processes and structural activities. These ADP-ribose acceptor proteins include such diverse proteins as glutamate synthetase, dinitrogen reductase, and glyceraldehyde-3-phosphate dehydrogenase. Mono-ADP-ribosylation has also been noted to modulate histone protein and oncogene functions.86 For example, mono-ADP-ribosylation of histone H3 at arginine 117 (H3R117) promotes proliferation, migration, and colonization by cancer cell lines through a mechanism involving activation of P300 and the up-regulation of β-catenin, c-Myc, and cyclin D1.86 These results suggest the possibility that mono-ADP-ribosylated proteins may be a target for control of the malignant phenotype.79,86
Several of the eukaryotic mARTs are located at cell surface where they function as ecto-ARTs.87 Proteins shown to be ADP-ribosylated by these ecto-enzymes include integrins and P2X7, the latter involved in calcium flux leading to apoptotic cell death.87–89 Overall, the ecto-ARTs are considered to regulate cell–cell communication and signal transduction. The ecto-(ADP-ribosyl)transferases have also been implicated in regulation of multiple soluble growth factors including fibroblast growth factor 2 (FGF2), platelet-derived growth factor (PDGF), insulin-like growth factor (IGF), and α-defensin 1 (DEFA1).87 Interestingly, the Km for NAD of the ecto-ARTs is approximately 0.6 mM, whereas free NAD in plasma is 0.1 to 0.3 µM. However, extracellular NAD may be significantly increased at times of cell damage, lysis due to trauma, and/or stress. Accordingly, the function of these enzymes may be restricted to times of stress and/or trauma.87
Regulatory roles of mono-ADP-ribosyl PTMs are further controlled by a series of (ADP-ribosyl)hydrolases (ARHs), which can remove ADP-ribose moieties restoring protein structure and function90–92 as well as from ADP-ribosylated sites on DNA and RNA.81 As suggested, the ARHs are composed of a group of enzymes including MacroD1 and D2, terminal ADP-ribose protein glycohydrolase (TARG1), and the ADP-ribosyl-acceptor hydrolases, ARH 1 and 3.81,90
Similar to the ARTs, the selectivity of the ARHs is dependent on tissue and subcellular distribution. Thus, different ARHs are distributed among different subcellular organelles.81,90 Of particular interest to carcinogenesis is the demonstration that ARH1-deficient MEF cells injected into nude mice developed tumors in multiple organs and increased metastasis.90 Moreover, mice genetically engineered to be defective in ARH1 developed increased spontaneous malignancies including lymphoma, hepatocellular carcinoma, sarcoma and tumors in the lungs, GI tract, uterus, and mammary tissue.90
A role for ARHs in the neurologic system is shown by a family where homozygous offspring of individuals heterozygous for a mutation that inactivates the ARH3 gene showed variable degrees of neurodegeneration with histologic signs of hippocampus, cerebellum, and cortical degeneration.93 In mice, this defect showed accumulation of poly(ADP-ribose) in the cytoplasm associated with neurodegeneration. The PARP inhibitor, veloparib prevented the histologic and neurodegenerative changes in ARH3-knockout mice.93 Thus, the (ADP-ribosyl)transferases and hydrolases constitute important regulatory elements for multiple diverse systems and defects in their function can result in pathologic conditions.
Cyclic ADP-ribosylation
Cyclic ADP-ribose (cADP-ribose) is a secondary messenger involved in signal transduction, cell adhesion, calcium mobilization, and cell cycle control.94–96 cADP-ribosylation, catalyzed through the cyclization of NAD, occurs via the action of the cADP-ribose hydrolase family of enzymes, specifically CD38 and its homologue, CD157.97 Although first observed within T lymphocytes and thymocytes, CD38 and CD157’s tissue- and cell-specific expression is now considered virtually ubiquitous with comparatively high expression in cell types of the immune system.98 In addition to its function in secondary messenger synthesis, CD38 plays a major role in the hydrolysis of NAD, suggesting its role as a regulator of cellular NAD levels.99 Specifically, the NADase activity of CD38 has been shown to increase with age, and is implicated as a key mediator of age-related NAD decline and mitochondrial function. The mechanism by which CD38 regulates metabolism has been connected to the modulation of the NAD-dependent sirtuin family. SIRT1, which is implicated in aging, cell protection, and energy metabolism, and SIRT3, which functions in oxidative metabolism, have both demonstrated significantly increased expression in Cd38-KO mice.100,101 Through activation of the SIRT3/FOXO3-mediated antioxidative stress pathway, CD38-deficient mice become protected from diet-induced obesity, highlighting the enzyme’s role in energy expenditure.102,103 CD38 deficiency also activates the SIRT1/FOXOs pathway, which protects cardiac muscle from ischemia-reperfusion injury (IRI).104 Conversely, Hu et al.105 utilized proteomic analysis to demonstrate that overexpression of CD38 decreases cellular NAD levels, thus inhibiting a variety of proteins involved in antioxidative function and metabolic regulation. It has been shown that CD38 expression is induced by both lipopolysaccharides and inflammatory cytokines like TNF-α.106–108 From a cancer perspective, nasopharyngeal carcinoma (NPC) cell lines overexpressing CD38 promoted cellular proliferation and metastasis, as well as an inhibited senescence response.109 In addition, CD38 has implicated functions in mediating psychosocial behavior via its role in hormonal regulation,110 as well as distinct activity in the immune response.111 These varied effects of CD38 activity present the potential impact on numerous conditions by therapeutic interventions of the NAD metabolome.
Poly-ADP-ribosylation
Poly-ADP-ribosylation, another NAD-dependent reversible PTM catalyzed by the poly(ADP-ribose) polymerase (PARP1-17) family of enzymes is heavily involved in signal transduction, particularly in the DNA damage response (DDR), transcriptional regulation, cellular proliferation, centrosome and mitotic spindle organization, telomere maintenance, and cell cycle control.5,6 Like the mono(ADP-ribosyl)transferases, poly(ADP-ribose) polymerase cleaves NAD at the N-glycosidic bond releasing nicotinamide, a proton, and ADP-ribose, with the latter becoming covalently linked to amino acid side chains, especially glutamate or aspartate residues. Once activated, the enzyme continues to cleave NAD with subsequent ADP-ribose moieties being joined by ribosyl-ribosyl linkages to form a growing linear or branched chain, unique polymer, poly(ADP-ribose), which may reach hundreds of units in length.112,113 The sole nucleoside in poly(ADP-ribose) is adenosine, while the backbone is composed of ribose–ribose bonds alternating with diphosphate ester bonds.6,78,112 Extensive studies have shown that the polymer can be attached directly to PARP as well as to a host of other signaling molecules including p53, p21, MSH6, DNA ligase III, X-ray repair cross-complementing 1 (XRCC1), DNA polymerase ε, DNA PK’s, Ku70, NF-κB, inducible nitric oxide synthase (iNOS), caspase-activated DNAse, and telomerase.114
While 17 proteins have been identified with DNA sequence homology to PARPs, only four have been clearly shown to be capable of synthesizing ADP-ribose polymers.6 PARP1 and PARP2 have been extensively investigated in the DDR, where PARP facilitates base excision repair and provides a mechanism for repairing DNA double strand breaks, especially when cellular capacity for homologous recombination is defective or exceeded as in BRCA1 and 2 deficiency. PARP1 is activated by single and double DNA strand breaks, causing the enzyme to auto-poly-ADP-ribosylate and recruit protein complexes involved in DNA repair such as XRCC1.6,78,112 In addition, PARP1 has displayed a role in regulating the activation of the p53 tumor suppressor pathway, as well as alternative activation from both heat shock proteins (HSPs) and phosphorylated extra signal-regulated kinases (ERKs) during DNA stress responses.6,78,112 PARP1 and PARP2, which is also involved in DNA repair, both exhibit association with mammalian centromeres during mitosis and meiosis, indicating these enzymes’ function in cell cycle regulation.6,78,112 In addition, PARP2 functions in telomere maintenance through inhibition of telomeric repeat-binding factor 2 (TRF2), thus modulating DNA binding functions to promote telomeric integrity. In addition to modifying the activity of multiple signaling proteins, the diphosphate polymer backbone of PARP is negatively charged and by virtue of its interaction with negatively charged DNA and positively charged chromatin proteins, it serves to decondense chromatin.78
PARP activity is dependent on and proportional to the amount of DNA strand breaks. Upon PARP activation by DNA strand breaks, poly(ADP-ribose) is rapidly synthesized and the polymer is rapidly degraded by the enzyme poly(ADP-ribose) glycohydrolase (PARG), yielding free ADP-ribose. The latter can be re-used in an ATP dependent process to synthesize NAD. Depending on number and/or duration of DNA strand breaks, PARP may be sufficiently activated to deplete cellular NAD.74 Attempts to resynthesize NAD can subsequently consume ATP, and the rapid turnover of polymer and attempts to resynthesize NAD can deplete both NAD and ATP leading to cell necrosis. The importance and relevance of poly-ADP-ribosylation in the DDR are shown by the demonstration that genetically modified mice with deletions of either PARP1 or the 110 kDa isoform of PARG, each show normal growth and development, but each is hypersensitive to alkylating agents and/or ionizing radiation and each is hypersensitive to endotoxic shock.115–117
Studies of PARP activity in peripheral blood mononuclear leukocytes from a series of mammalian species have shown a positive correlation with longevity; however, no correlation was observed with the amount of PARP protein.118,119 PARP activity has been noted to decrease with chronologic age in both rats and humans. In contrast, Epstein-Barr virus-transformed lymphocytes from centenarians were found to have greater PARP activity than control cells prepared from 20- to 70-year-old individuals.120 PARP has been described as a guardian of the genome protecting against accumulation of DNA damage.121 Taken together, these reports of association of its activity with longevity, its decline with aging, yet elevated levels in centenarians, suggest that an age-associated decrease in enzyme activity renders the genome more susceptible to DNA damage, whereas higher levels may be associated with greater longevity, promoted via protection of genetic integrity.121–124
In addition to its involvement in cellular proliferation and the DDR, PARP mediates immune function of T-cells, B-cells, and dendritic cells by activation of IKK and NF-κB, as well by altering nuclear retention of NF-κB.125 PARP has also been shown to play an important role in organizing chromatin during spermatogenesis, and the enzyme’s normal function may be required for male fertility.123,126 Poly-ADP-ribosylation has been shown to modify and decrease activity of alcohol dehydrogenase in model organisms and human cells in tissue culture leading to the suggestion that this process may stimulate the accumulation of toxic aldehydes and subsequent neurodegenerative diseases.127
PARP inhibitors in malignant, inflammatory, and neurodegenerative diseases
Because of its role in the DDR, and since enzyme inhibitors were shown to potentiate the cytotoxicity of alkylating agents and ionizing radiation, PARP was identified as a potential therapeutic target to amplify the effect of cancer chemotherapy. However, early clinical trials showed these combinations to be too toxic. Subsequent studies showed that PARP inhibitors (PARPi) alone could be useful in treating tumors in patients with defective homologous recombination (HR), where the predisposing genetic defect in HR, in combination with the PARPi, created a synthetically lethal combination which was initially shown to be effective in treatment of breast and ovarian cancer in patients with BRCA1 or 2 deficiency.128–130 Agents now approved for clinical use in treating oncologic disorders include niraparib, olaparib, rucaparib, and talase. These agents have been found to be useful in first line and maintenance therapy as well as treatment of advanced disease.131 More recently, PARPis have been extended to patients with BRCA mutations in pancreatic and prostate cancers132,133 as well as in patients with metastatic disease.134
PARP inhibitors have shown some efficacy in treating Parkinson’s disease in animal models where inhibition of PARP interferes with synthesis of poly(ADP-ribose), slows α-synuclein fibrillation, and sequesters the development of neurotoxicity.135,136 Age-associated diseases, such as ischemia reperfusion, Alzheimer’s and others have been suggested to result in PARP activation, providing an opportunity to examine repurposing of PARPi for non-oncologic disorders.137 In model systems, PARPis are able to downregulate immunologic-mediated diseases including rheumatoid arthritis, colitis, experimental autoimmune encephalomyelitis, and allergic airway inflammation.125
Acetylation/deacetylation reactions and sirtuins
Acetylation reactions, in which an acetyl group (CH3CO) becomes covalently linked to a target protein, are another form of post-translational modification. Like other forms of PTM including protein methylation, phosphorylation, ADP ribosylation, ubiquitination, and others, acetylation has the capacity to regulate both protein structure and function. Each of these processes has their own unique specificity for alteration of molecular structure and function. Accordingly, each of these PTM systems has a unique set of regulatory enzyme locations and molecular donors, such that, just as kinases and phosphatases control protein phosphorylation, so too acetylation is reciprocally controlled by acetylation and deacetylation. While protein acetylation takes place in the nucleus, cytoplasm, and mitochondria, the major focus of this activity is in the nucleus and mitochondria. For example, 20% of liver mitochondrial proteins may be reversibly acetylated, with high levels noted on proteins of the TCA cycle and beta-oxidation pathways.138
Acetylating enzymes use acetyl CoA as the donor of acetyl groups, which are then transferred to covalent linkage at the α-amino group or ammo-terminal residues or to ε amino groups of lysine, resulting in the neutralization of their positive charge and consequent changes in protein structure and/or function.139 Acetylation of proteins may also result from non-enzymatic processes. Accordingly, both enzymatic and non-enzymatic acetylation of proteins is partially dependent on acetyl CoA concentration, which is a common product of carbohydrate, amino acid, and fatty acid catabolism. Generated in mitochondria and increased during periods of nutritional abundance,140,141 acetyl CoA can be converted to citrate, which can then be transported from the mitochondria to the cytoplasm to be re-converted to acetyl CoA via the ATP-dependent enzymes, acetyl CoA synthetases 1 and 2.140,141
Increased production of acetyl CoA and protein acetylation activity in the presence of nutrient excess may contribute to increased transcriptional activity by multiple mechanisms, including the neutralization of histone charges resulting in transcriptionally open and active euchromatin as well as the activation of specific transcription activators. Interaction of positively charged lysine residues in histone proteins with negatively charged phosphates in the DNA backbone contributes to the tight packing of chromatin in heterochromatin, which is transcriptionally inactive. Charge neutralization via the acetylation of lysine residues in chromatin proteins is associated with the transition from a tightly packed heterochromatin to transcriptionally active euchromatin. While acetylation usually reduces the activity of modified enzymes, in some cases, acetylation activates function. For example, acetylation of histone H3 serves as a transcriptional promoter.142
Removal of acetyl residues is carried out by several different classes of enzymes known as deacetylases, which include histone deacetylases (HDAC)143 and sirtuins.7 The reason for discussing acetylations and deacetylations in this review is because the major group of deacetylases, sirtuins, are (ADP-ribosyl)transferases that transfer the acetyl groups from proteins to ADP-ribose-acceptors in a reaction requiring NAD and yielding 2′-O-acetyl-ADP-ribose, nicotinamide, and the deacetylated protein.7 Mammalian tissue contains a series of sirtuins, which, as described below, are differentially distributed on a subcellular and tissue basis, and have multiple regulatory functions. Accordingly, NAD is an essential substrate for this class of deacetylases, and increased NAD levels resulting from nutrient restriction result in decreased acetylation, leading to inactive heterochromatin with decreased transcriptional activity. Thus, increased generation of acetyl CoA during nutritional abundance, in contrast to increased NAD during times of nutritional restriction, provides a reciprocal regulatory mechanism for mitochondrial metabolism to regulate chromatin structure and activity, as well as nutrient metabolism based on metabolic production of intermediary metabolites. Increased NAD and its utilization by sirtuins in deacetylation reactions during periods of scarce nutrition, such as exercise, fasting, or calorie restriction are considered to partially mediate the longevity-promoting effects of these activities as well as protect against some of the age-associated comorbidities such as cancer and heart failure. This likely contributes to the decreased acetylation of histone protein during aging in human fibroblasts, and in part provides the rationale for increasing NAD levels to promote healthier aging.7
As noted above, sirtuins (SIRT1-7), also known as silent information regulator proteins, are a family of highly conserved, NAD-dependent deacetylases. They have been extensively studied for their roles in cancer development and aging, which are connected to their widespread activity in genomic integrity, energy metabolism, cell cycle control, inflammation, circadian rhythm, cardiac physiology, and neuronal function. Originally discovered in Saccharomyces cerevisiae, Sir2 (the equivalent of mammalian SIRT1) was identified through its role in silencing all heterochromatin-like regions, such as telomeres, rDNA, and the cryptic mating type loci, HML/HMR.144,145 Subsequently, four Sir2 homologues (Hst1, Hst2, Hst3, and Hst4) were located, although deletions of these genes did not result in the inhibition of HML/HMR silencing.146,147 Caloric restriction (CR) also plays a role in modulating sirtuins and aging in yeast, with a decrease in media glucose from 2% to 0.5% resulting in significant longevity promotion, a phenotype initiated either via a Sir2-dependent mechanism through decreasing levels of NADH, a competitive inhibitor of Sir2, or via the action of the nicotinic phosphoribosyltransferase 1 (Npt1), a component of NAD salvage which converts NAM, a non-competitive inhibitor of sirtuins, to NAD.148–150 To this end, pyrazinamidase/nicotinamidase 1 (Pnc1), an enzyme upstream of Npt1, is required for extending lifespan via CR.151 These pathways can also be stimulated via exogenous treatment of NAD precursors, with treatment of NR in yeast cells resulting in stimulated Sir2 activity and extended lifespan via an NRK-dependent pathway of NAD replenishment.152 These in vitro findings not only implicate the cooperation between the NAD metabolome and sirtuins in the aging process, but also highlight the importance of low-intensity stress responses in the modulation of these cellular processes.
The function of Pnc1 in yeast is largely analogous to the role of NAMPT in mammalian systems, which is not only nutrient- and stress-sensitive, but also a mediator of nuclear and mitochondrial sirtuin activity through its function in NAD biosynthesis.153,154 However, regulation of mammalian Nampt expression is uniquely circadian through the transcription factors CLOCK/BMAL. NAD biosynthesis and sirtuin activity modulation via CLOCK/BMAL are also affected by a sirtuin-based feedback loop in both peripheral tissue and the suprachiasmatic nucleus (SCN) of the hypothalamus, whereby SIRT1 and SIRT6 positively regulate circadian phase in a reciprocal pathway with Period 2 (Per2), whose activity suppresses Sirt1 and Sirt6 transcription.155 SIRT1, which forms a complex with peroxisome proliferator-activated receptor-gamma coactivator (PGC-1α) and binds directly to the BMAL promoter, activates transcription and amplifies a robust circadian response. SIRT1 and BMAL were found to decrease in the SCN of aged mice, owing to the characteristic, age-associated decline in circadian control. The Per2 promoter, a primary target of SIRT1, was found to be epigenetically modified during the aging process, leading to an increased level of Per2 protein in aged mice.156 SIRT1 has also been found to mediate the secretion of eNAMPT from adipose tissue following the enzyme’s deacetylation of iNAMPT at lysine 53, which affects hypothalamic NAD production and neural function.42 This implicates both adipose tissue and sirtuins as central communication units for the modulation of systemic NAD biosynthesis, as well as potential hypothalamic regulators. Unlike SIRT1, SIRT6’s interaction with circadian control is through stimulating chromatin recruitment of both CLOCK/BMAL1 and SREBP-1, additionally implicating SIRT6 in fatty acid metabolism and explaining the disruption of fatty acid and cholesterol metabolism in SIRT6 knockout mice.157 SIRT6 deficiency also results in a compromised and dysregulated circadian rhythm, making the enzyme an integral component of circadian function.158 SIRT3, which deacetylates a variety of mitochondrial enzymes involved in oxidative metabolism, is also mediated by the effect of the circadian-linked fluctuations of NAD bioavailability, with exogenous treatment of NAD resulting in restored SIRT3 function and improved oxygen consumption in circadian mutant, Bmal1−/− mice.159
SIRT1 displays an especially diverse functional profile. Brain-specific SIRT1-overexpressing (BRASTO) transgenic mice demonstrated significantly increased longevity, a phenotype likely connected to stimulated neural activity in the hypothalamus.160 Conversely, systemic upregulation of SIRT1 in mice had no effect on aging, indicating brain-localized SIRT1 is likely the primary sirtuin mediator of mammalian longevity. However, whole-body overexpression of SIRT1 actually promoted healthy aging through downregulation of p16 gene expression, as well as reduced liver cancer susceptibility and metabolic damage in mice.161 Hydrogen peroxide-treated human diploid fibroblast cells demonstrated stimulated senescence due to an accumulation of acetylated p53, which resulted from a significant downregulation of SIRT1 activity.162 To this end, SIRT1 expression was diminished in atherosclerotic vascular smooth muscle cells, reducing DNA repair and allowing the cells to undergo replicative senescence.163 SIRT1 has demonstrated differential expression in a wide variety of cancer types,164 indicating not only dual roles in tumor promotion and suppression, but also the need for further study and improved understanding of the enzyme’s varied functions. SIRT1’s pleiotropic capabilities implicate the enzyme in the aging process, cancer development, and a number of diseases and comorbidities.
SIRT2, which is primarily localized to the cytoplasm, also has the ability to deviate to the nucleus where it is known to deacetylate histone-based lysine residues H4K16 and H3K56 to initiate DNA repair.165,166 In addition, the enzyme shares a number of non-histone substrates with SIRT1, such as FOXO1, FOXO3, and p53,167–169 which modulate cell cycle progression and apoptosis, as well as, uniquely, α-tubulin, which contributes to oligodendrocyte differentiation.170 In terms of carcinogenic functionality, SIRT2 has demonstrated roles both in tumor development as well as suppression. Murine SIRT2 deficiency leads to a robust tumorigenic response,171 and human melanomas and gliomas have been found to be associated with various abnormalities and expression changes in the SIRT2 locus,172,173 indicating the enzyme’s tumor-suppressive capability. Conversely, SIRT2 has also demonstrated diverse oncogenic effects, with in vitro expression positively correlating to the proliferation of acute myeloid leukemia, prostate, glioblastoma, pancreatic, and colorectal cancer (CRC) cells.174–178 In addition, silencing of SIRT2 has been found to inhibit tumor angiogenesis by inactivating STAT3/VEGFA signalling, resulting in heavily impeded tumor progression.179 Similar to SIRT1, due to SIRT2’s varied involvement in a number of both carcinogenic and tumor suppressive pathways, enhanced understanding of the enzyme’s functionality is required to elucidate SIRT2’s exact impact in cancer biology.
SIRT3, the primary mitochondrial sirtuin,180 acts as both a deacetylase and ADP-ribosyltransferase to regulate mitochondrial-based biogenesis, energy metabolism, and thermogenesis, as well as inhibit rates of nuclear gene transcription through deacetylating H4K16, H3K9, and most recently H3K56.181 Unlike SIRT1 and SIRT2, SIRT3 has largely been considered a tumor suppressive enzyme due to its defined role in the scavenging of reactive oxygen species (ROS) and antioxidant defense.182,183 SIRT3 also inhibits the metabolic reprogramming characteristic of cancer cells through the destabilization of HIF1α, a key transcription factor involved in the cellular response to systemic oxygen and, transitively, energy metabolism.184 However, it is noteworthy that the implication of SIRT3 in carcinogenesis is both cell- and tumor-specific. While expression of SIRT3 is downregulated in a variety of human malignancies such as breast, lung, colon, prostate, pancreatic, and ovarian cancers, as well as osteosarcoma and glioblastoma,185–188 increased expression has been observed in oral, thyroid, pancreatic, and node-positive breast cancer, as well as melanoma,189–193 suggesting the protein’s capability to promote cancer development. While the mechanisms by which SIRT3 promotes tumorigenesis remain undefined, the enzyme’s expression can be linked to defending cancer cells from genotoxicity and stress-mediated apoptosis.194
Unlike its other enzymatic family members, SIRT4 lacks observed deacetylase functionality, instead primarily catalyzing mono-ADP-ribosylation and regulating metabolic activity in the mitochondria. SIRT4 largely ADP-ribosylates glutamate dehydrogenase (GDH), inhibiting the catalysis of glutamate to α-ketoglutarate and downregulating the flux of amino acids into energy production. Through regulating the stress-induced inhibition of glutamine catabolism, SIRT4 has demonstrated a clear role in attenuating carcinogenesis, with decreased expression levels having been observed in T-cell leukemia as well as lung, gastric, colorectal, and breast cancers.195–199
Expressed in the mitochondria and cytoplasm,200 SIRT5 functions as both an NAD-dependent deacetylase as well as a demalonylase and desuccinylase,201 with the enzyme’s main substrate being carbamoyl phosphate synthetase 1 (CPS1), which catalyzes the rate-limiting step of the urea cycle.60 SIRT5’s implications in carcinogenesis are poorly understood, but the enzyme’s involvement in energy metabolism may play a role in the metabolic reprogramming characteristic of cancer cells. Having been shown to deacylate glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as well as other key enzymes of the glycolytic pathway, SIRT5 may facilitate aerobic glycolysis and regulate carbon flux into the mitochondria, potentially aiding cell growth,202 which may account for its overexpression in hepatocellular and ovarian carcinomas, as well as in non-small cell lung cancer.203–205 SIRT5 has also demonstrated potential functionality as a tumor suppressor, with the enzyme being underexpressed in both endometrial and squamous cell carcinomas.206,207
Similar to SIRT3 and SIRT4, SIRT6 is largely regarded as a tumor suppressor due to its roles in DNA repair and cancer metabolism. Bound to chromatin and localized to the nucleoplasm,208 SIRT6 is an NAD-dependent deacetylase and ADP-ribosyltransferase whose main substrates are histones H3K9 and H3K56.209,210 Overexpression of SIRT6 results in the activation of both the p53 and p73 pathways leading to widespread apoptosis of cancer cells,211 and SIRT6−/− mice displayed a number of aging-associated complications such as lymphopenia, lordokyphosis, and acute metabolic defects resulting from genomic instability.212 To this end, SIRT6 expression has been shown to be negatively correlated with colon, pancreatic, liver, breast, and nasopharyngeal cancer cell growth.213–217 SIRT6 has also demonstrated the potential to promote cancer development, with expression being increased in ovarian, prostate, and skin cancer.218–220 In addition to its roles in carcinogenesis, SIRT6, similar to SIRT1, plays a profound role in lifespan determination. The recent studies by Onn et al.221 found SIRT6 to share a role with PARP1 as a DNA double-strand break sensor, a finding which, in conjunction with the enzyme’s function in the recruitment of proteins of the homologous recombination and non-homologous end joining pathways, may contribute to increased SIRT6 activity and efficiency of DSB repair in long-lived species.222 To this end, SIRT6-deficient cynomolgus monkeys (Macaca fascicularis) displayed severe developmental retardation and dramatically reduced survival, emphasizing the protein’s role in primate development.223
Considerably less studied than other sirtuins, SIRT7 is an NAD-dependent deacetylase which targets H3K18 as well as non-histone substrates such as p53, PGK1, and FOXO3.224 SIRT7, which is primarily localized to the nucleolus, has additionally been shown to act through its role in desuccinylating H3K122.225 Currently, SIRT7 has been purported to have oncogenic potential through its implications in defending cancer cells from genotoxicity. Colorectal, ovarian, and liver cancer cells have all shown increased expression levels of SIRT7,226–229 lending to the likelihood of the enzyme’s function as a tumor promoter. While further study is necessary to understand its direct implications in cancer development, the diverse functional profile of SIRT7 highlights numerous possibilities for the enzyme’s involvement in carcinogenesis.
Altered NAD levels and associated diseases
As NAD synthesis, compartmentalization, utilization, and response to stress is subject to multiple levels of control and regulation, it is not surprising that variations in these processes have been identified to accompany a variety of physiologic changes and disease states. Accordingly, therapeutic attempts have been targeted at correcting NAD levels with the expectation of improving comorbidities, especially those that are aging-associated. Several possibilities need to be considered regarding a role for NAD in the aging process. First, it is possible that NAD levels are primarily altered through decreased synthesis and/or increased utilization, that changes in NAD levels affect processes requiring NAD, and that NAD levels may be improved by targeting the synthetic or utilization pathways. Alternatively, it is possible that some age-associated molecular pathologies may alter NAD levels, which in-turn may affect other processes requiring NAD. In the first proposal, where the primary defect is synthesis or degradation of NAD, the correction of NAD levels might completely correct any associated disorders. For example, a primary, age-associated defect in the de novo NAD synthetic pathway could affect a number of processes, all of which might be corrected through promoting other pathways of NAD biosynthesis. In the second proposal, NAD levels could be secondarily affected by an indirect pathway. For example, a primary defect in DNA repair could sufficiently activate poly(ADP-ribose) polymerase to continuously consume NAD, resulting in lowered cellular NAD levels and decreased availability for co-enzymatic functionality in oxidation-reduction reactions. In the latter case, it might be possible to augment NAD biosynthesis to keep up with its rapid utilization. This process might correct the side effects of NAD depletion, but might not correct its primary mutagenic consequences. The following sections review some of the systems and comorbidities where altered NAD levels are considered to be etiologically associated with disease and/or their comorbidities.
Pellagra
Pellagra is a disease caused by severe niacin or tryptophan deficiencies, resulting in the development of clinical neurological, gastrointestinal, and dermatologic manifestations, specifically dementia, diarrhea, and dermatitis.230 Although pellagra is not an age-related disorder, some of its consequences have important implications for age-related problems. While 1914 marked Joseph Goldberger’s discovery that development of pellagra is due to dietary inadequacy,10 it was not until 1937 that Conrad Elvehjem demonstrated the therapeutic effect of NAM and NA supplementation in malnourished dogs that symptomatically recapitulated pellagra.231 Following exogenous NAM or NA treatment, subjects of Elvejehm’s study demonstrated attenuated dermatitis as well as stimulated growth. In conjunction with the findings of Katsyuba et al.,232 who demonstrated the role of NAD supplementation in health, aging, and mitochondrial homeostasis, the enhanced growth of Elvejehm’s specimens can likely be connected to inhibited mitochondrial function via niacin deficiency. While these phenotypes were reversible through therapeutic intervention, prolonged dietary deficiency and pellagra result in irreversible neurological complications. Although pellagra has become practically eradicated due to the fortification of flour products with niacin, the biochemical implications of NAD deficiency can be correlated to a variety of other pathological situations. Research has linked flaky paint dermatosis, a condition involving the flaking and pigmentation of photo-exposed skin regions (not to be confused with pellagra-associated dermatitis), to both increased IDO expression as well as decreased urinary excretion of N1-methylnicotinamide, a biomarker of pellagra.233 These observations represent the potential clinical prevalence of niacin and NAD metabolism in disease pathophysiology.
Aging
NAD has been implicated in the aging process since Braidy et al.234 in 2011 observed significant age-associated intracellular NAD decreases in rats. Subsequently, systemic NAD levels were clinically demonstrated to experience age-linked decline, with significant decreases observed in adults from ages 36 to 77.235 Chini et al. have connected the age-linked decline of NAD to the function of CD38, the primary regulator of NAD homeostasis through its role in both NMN and NAD degradation.99,101
Aging is a process characterized by a number of physiological changes such as metabolic dysfunction, genomic instability, chronic inflammation, and an increased susceptibility to disease. The suppression of mitochondrial energy production, widely regarded as a main aspect of aging, is linked to a mitochondrial-specific loss of protein complexes involved in oxidative phosphorylation. This process is impacted by a decline in nuclear NAD levels, which is facilitated by an alternate peroxisome proliferator activated receptor gamma coactivator 1 alpha/beta (PGC-1α/β)-independent pathway of nuclear-mitochondrial communication.236 Decreases in NAD also lead to the suppression of SIRT1 and SIRT3, whose inhibited roles in oxidative stress defense and mitochondrial function lead to downregulated energy production.100,101 Due to NAD’s diverse subcellular and tissue-specific expression, CD38 mediation of age-related NAD decline may be further impacted by other pathways, such as with PARP activity, remains open, for example, PARP activities have been noted to increase with aging.122 In addition, the chronic inflammatory characteristic of aging results in the production of bacterial endotoxins and pro-inflammatory cytokines, which have both been shown to stimulate CD38 expression.106–108
Decreasing NAD levels may impact regenerative capacity and the aging process through effects at the stem cell level. For example, Zhang et al. showed that muscle stem cells from 24-month-old mice compared to 3-month-old mice demonstrated lower NAD levels, lower levels of TCA cycle and oxidative phosphorylation enzymes, increased senescence pathway enzymes, reduced rates of oxidative phosphorylation, loss of mitochondrial membrane potential, and a reduced ability to replenish muscle stem cells in a mouse model of Duchene muscular dystrophy, a disease characterized by the loss of muscle stem cells.16 The older mice also showed more phosphorylated γH2AX and DNA strand breaks on comet assays, indicating increased DNA damage. These observations suggest that ongoing or sustained DNA damage-induced activation of PARP may contribute to the decrease in NAD levels demonstrated by these cells. Notably, all of the above abnormalities improved with dietary supplementation of NR, which also improved endurance and led to a slight increase (4.7%) in average life span, although the authors noted that NR supplementation was only administered late in life. Importantly, the improvement in muscle stem cell regeneration upon NR supplementation was abolished in SIRT1 knockdown mice. In addition, it is notable that the authors reported no toxicity nor change in spontaneous malignancies due to the administration of exogenous NR.16
Cancer
As noted above, many of the processes are affected by the enzymes of ADP-ribosylation and its removal function as tumor suppressors and in some cases, tumor promoters. To the extent that these enzymes are affected by NAD levels, these metabolites may play an important role in maintaining genome integrity, fidelity of the signaling process, and energy balance in support of normal and/or tumor growth. A specific aging and cancer-related issue not yet covered is the senescence-associated secretory phenotype (SASP), in which damaged cells lose their proliferative capacity, undergo stable growth arrest, and upregulate secretion of proinflammatory cytokines as an age-related process. SASP is thought to contribute to the low-grade inflammation that accompanies aging, and may be associated with age-dependent comorbidities237 such as insulin resistance, neurodegenerative disease, heart failure, progressive accumulation of DNA damage, cognitive decline, and unfolded protein response. It is noteworthy that many of these processes can be partially slowed or reversed through upregulating NAD levels via the supplementation of NAD precursors.237
This replicative senescence may be oncogene-induced senescence (OIS), in which NAD and NAD/NADH ratios are increased, or mitochondrial dysfunction-associated senescence (MiDAS), which is associated with decreased NAD/NADH ratios.238 With OIS, the elevated NAD levels and NAD/NADH ratios are mediated by high mobility group (HMG) proteins that epigenetically enhance upregulation of NAMPT synthesis resulting in increased NAD and NAD/NADH ratios, as well as upregulation of NF-κB activity, leading to increased expression of IL1β, IL6, and IL8. These processes promote tumor growth in tissue culture and pancreatic neoplasia in a mouse model in response to elevated NAD, and can be prevented by genetic knockdown of NAMPT or pharmacologic inhibition with FK866. The latter agent prevents the increase in IL1β, IL6, and IL8. These results have raised the concern that, while strategies to boost NAD levels may retard the aging process, they may concurrently promote the carcinogenic impact of SASP cells. A theoretical strategy has been proposed to eliminate SASP cells using senolytic agents such as dasatinib, quercetin, and/or navitoclax before attempting to increase NAD levels.237,239,240
Brain and neurologic diseases
In pursuit of a role for NAD levels in regulation of age-associated disease and comorbidities, a great deal of research has been focused on the brain and neurodegenerative disorders, the heart and cardiovascular disorders and metabolic diseases such as type 2 diabetes. In a recent review of NAD+ in Brain, Aging, and Neurodegenerative Disorders, Lautrup et al.241 indicated the importance of NAD on synthesis of the neurotransmitters glutamate and acetylcholine, as well as the role of SIRT1 in promoting neuronal and axonal outgrowth. They also reported on NAD decreases in normal brain from c. elegans, mice, and humans, and showed their decreases were associated with cognitive impairment. Importantly, they reported that genetic knockdown of NAMPT-produced cognitive characteristics of old mice,242 and that NMN treatment could correct the phenotype. They suggested also that NAD decreases in aging brain could be associated with aging mitochondria and with accumulation of oxidatively damaged DNA. NAD levels have been implicated in facilitating neuronal stress response, maintaining neuroplasticity, suppressing neural inflammation, and maintaining neurogenesis. Mouse model studies using NR supplementation suggest these processes may be amenable to increasing NAD levels.
In mouse model studies of Alzheimer’s disease, NAD depletion has been suggested to contribute to neural inflammation, and deposition of neural fibrillary tangles, while NAD augmentation may slow some cognitive defects, and slow accumulation of DNA damage. In Parkinson’s disease rodent models, accumulation of α-synuclein fibrils has been suggested to occur with NAD deficits241 possibly associated with hyper activation of SARM1, and NADase, leading to axonal degeneration.243 Moreover, PARP1 is activated by α-synuclein leading to further consumption of NAD and consequent cell death.244 Other neurodegenerative diseases associated with decreased NAD levels include Huntington disease (HD) and amyotrophic lateral sclerosis.245,246 In HD, the primary molecular pathology is known to be associated with tri-nucleotide repeats leading to self-aggregation of the Huntington protein and impairment of the kynurenine pathway, potentially leading to NAD reduction. Although the mechanism of ALS is not clearly determined, the disease is simulated by SOD-Tg mice where a defect in NAM metabolism has been noted and NAM supplementation has corrective effects.247 Each of the above diseases appears to affect NAD levels by different mechanisms, suggesting that the NAD defect is a secondary consequence. In the case of HD, the primary etiologic defect has clearly been identified as amplification of the trinucleotide repeat. Nonetheless, mouse model studies and some early clinical trials suggest a beneficial effect of augmenting NAD synthetic pathways.
Cardiovascular disease
NAD levels and their regulation by sirtuins have been implicated in cardiovascular disorders including heart failure and accompanying cardiac hypertrophy,248 ischemia reperfusion,249 and cardiac arrhythmia.249 Much of this research has been conducted at the cellular and tissue culture level, with further investigation in rodent models, and now with increasing investigation in humans leading to clinical trials of supplementation with NAD+ and its precursors.248,249
Using genetically altered mouse models with deletion of serum response factor (SRFHKO), Diguet et al.248 demonstrated that the development of dilated cardiomyopathy (DCM), decreased left ventricular ejection (LVE) fraction, and heart failure were all associated with increased protein acetylation and a 30% decrease in NAD level. They further demonstrated a decrease in nicotinamide phosphoribosyltransferase with a concurrent increase in nicotinamide riboside kinase 2 (NMRK2). These morphologic and physiologic changes of cardiac remodeling were significantly prevented by intraperitoneal (IP) and/or oral administration of NR to stabilize myocardial NAD.248
Similar metabolic findings, but to a lesser degree, were observed with mechanically induced cardiac hypertrophy using transverse aortic constriction (TAC), where NR supplementation partially protected the myocardial NAD pool and the decrease in LVE fraction, but not the morphologic changes of DCM.250 An important indication that that these findings represent the condition in humans comes from the demonstration of reduced NAD levels in biopsies obtained from patients with failing hearts. In these studies, the NR-driven restoration of NAD appears to be based on increased synthesis, with the NR supplying sufficient precursors to the NMRK2 pathway to increase NAD, resulting in increased glycolysis, increased mitochondrial citrate synthase, decreased acetyl-ATP citrate lyase, and increased selected nuclear and cytoplasmic acetylated proteins. It is notable that there were no increases in ADP-ribosylated protein, poly-ADP-ribosylated protein, ADP-ribose, or NAM. On the other hand, NR supplements were shown to increase NAAD, methyl-NAM, and N1-methyl-4-pyridine-5-carboxamide, which provides the basis for biochemical monitoring of the NR effect, although there is potential for increased H2O2 production associated with Me4Py.250 Based on this and previous evidence that expanding NAD pools can improve a failing heart,250 there was a call for translational clinical trials to normalize NAD levels in patients with cardiac dysfunction while conducting careful pharmacokinetic, metabolomic, safety, and tolerability studies.
Multiple studies in mice and isolated cardiac myocytes show the impact of NAD and its precursors, as well as sirtuins on cardiac rhythm control based on regulatory improvement of metabolic defects, oxidative stress, sirtuin activation, and voltage channel signaling such as NAD channel Ca2+ signaling. Since both cardiac NAD and sirtuin activity studies in rodent models and isolated cardiac myocytes demonstrate an impact of NAD, its precursors, and sirtuins on cardiac rhythm control, these effects appear to be mediated by multiple pathways including metabolic and oxidative stress, voltage channel signaling, and SIRT activation, and modulating these channels by altering NAD and/or sirtuin levels has shown some efficacy in attenuating arrhythmias in murine models and has been suggested as a strategic approach for preventing the associated development of cardiac arrhythmias in patients.249,251
Ischemia reperfusion (IR) injury occurs in the myocardium, most commonly with aging-associated coronary artery obstruction resulting in acute cardiac ischemia and hypoxia, causing metabolic shifts with rapid depletion of energy metabolites such as NAD and ATP. Reperfusion associated with reoxygenation may then result in ROS, oxidative stress, and an increase in intracellular Ca2+, leading to apoptosis and extended injury.249,251 In vitro studies in rat cardiac myoblasts showed that elevated NAD levels increase survival in a NAD/SIRT1-dependent fashion.252 In vivo, these findings were averted by pretreatment with IP injections of NMN,104 resulting in increased SIRT1 activity and decreased infarct size.253 Moreover, intravenous NAD infusion has been shown to reduce infarct size as well as decrease troponin 1 levels and apoptosis in a murine model of IR.254 It is further notable that mice with deficiency of SIRT3 are more susceptible to IR injury.255 These studies are supportive of clinical trials to evaluate the contribution of NAD and its precursors on limiting IR toxicity in humans.
Metabolic disorders
Declining NAD levels and shifts in NAD/NADH ratios occur with age-associated mitochondrial dysfunction.236 In addition to the clinical disorders noted above, changes in NAD have been implicated in many of the comorbidities that accompany aging, including obesity and its consequences, diabetes mellitus, metabolic syndrome, and atherosclerotic cardiovascular disease, the latter of which involves both lipid abnormalities and alterations in vascular endothelial function.251 Many of these comorbidities appear to be mediated in association with quantitative and/or functional alterations in sirtuins, and most have been demonstrated in experimental systems, particularly genetically altered animal models. In general, these models support an anti-aging comorbidity role for NAD and sirtuins. For example, aging transgenic mice that overexpress SIRT in pancreatic β islets show increased insulin secretion and better glucose tolerance,256 while SIRT1 knockout mice show decreased insulin levels.257,258 Nonetheless, other studies suggest SIRT may contribute to insulin resistance.259 Thus, clinical trials in which SIRT activities and NAD levels are modulated alone and in combination are crucial to targeting these pathways to prevent and/or reverse aging phenomena. In conducting these studies, it will be important to determine the specific metabolic and genetic conditions in which they may be effective, and to further evaluate whether they may be effective in a preventive or therapeutic fashion. Studies on organ specific targeting will also be important in this regard. The critical question is whether utilization of precursors to increase NAD levels can compensate for mitochondrial underproduction, or whether underlying mitochondrial disorders will still result in age-associated disease.
Conclusion
NAD, its precursors, and its derivatives including NADH, NADP, NADPH, cyclic ADP-ribose, mono-ADP-ribosylated proteins, poly(ADP-ribose) and poly(ADP-ribosylated) proteins, and acetyl-ADP-ribose, as well as the enzymes and proteins involved in their synthesis and degradation, constitute a vast array of interconnected metabolic, regulatory, and structural processes, all impacted by their central dependence on NAD, yet each characterized by its own unique vulnerability. Based on demonstration of their roles in cancer, aging, and other disease states, as well as their unique vulnerabilities, they have become the focus for multiple targeted interventions. PARP inhibitors have undergone successful clinical trials, are now major components of the cancer control armamentarium, and are being further evaluated for therapy of inflammatory, neurodegenerative, and ischemia/reperfusion disorders.113,137
Elevating NAD levels by administration of its precursors to improve health, extend longevity, and ameliorate aging-associated comorbidities is supported by considerable experimental data11,12,14,16,260 and is already being commercially promoted. Although this approach may have considerable benefits, its real benefits and risks of adverse effects associated with the vast array of its dependent processes need to be evaluated with carefully controlled clinical trials. These trials should serve to demonstrate the following principles: (1) it should be possible to demonstrate altered NAD levels or interactions in tissue culture, animal models, and human samples of disease, (2) it should be possible to simulate the condition by molecular and/or pharmacologic manipulation of the defect, (3) it should be possible to translate the observation to correct the consequences by restoring NAD levels at both the individual and population levels, (4) for a primary disorder of NAD levels, it should be possible to protect against development of consequences by promoting normal NAD levels, and finally (5) it should be shown that there are no adverse effects associated with long-term administration of the agent.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data, and review of the manuscript; all authors contributed to writing the manuscript and designing both figures.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded in part by R25 CA225461, P30 CA043703, and the Hanna-Payne Professorship in Experimental Medicine
ORCID iDs
John WR Kincaid https://orcid.org/0000-0003-0660-9813
Nathan A Berger https://orcid.org/0000-0001-7086-9885
References
- 1.Pollak N, Dölle C, Ziegler M. The power to reduce: pyridine nucleotides – small molecules with a multitude of functions. Biochem J 2007; 402:205–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao W, Wang RS, Handy DE, Loscalzo J. NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid Redox Signal 2018; 28:251–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agledal L, Niere M, Ziegler M. The phosphate makes a difference: cellular functions of NADP. Redox Rep 2010; 15:2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love NR, Pollak N, Döllea C, Nierea M, Chen Y, Oliverid P, Amaya E, Patel S, Ziegler M. NAD kinase controls animal NADP biosynthesis and is modulated via evolutionarily divergent calmodulin-dependent mechanisms. Proc Natl Acad Sci U S A 2015; 112:1386–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 2012; 13:411–24 [DOI] [PubMed] [Google Scholar]
- 6.Schreiber V, Dantzer F, Amé JC, De Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7:517–28 [DOI] [PubMed] [Google Scholar]
- 7.Imai SI, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000; 403:795–800 [DOI] [PubMed] [Google Scholar]
- 8.Imai SI, Guarente L. It takes two to tango: NAD+ and sirtuins in aging/longevity control. Aging Mech Dis 2016; 2:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elvehjem CA, Madden RJ, Strong FM, Woolley DW. The isolation and identification of the anti-black tongue factor. Nutr Rev 1974; 32:48–50 [DOI] [PubMed] [Google Scholar]
- 10.Morabia A. Joseph goldberger's research on the prevention of pellagra. J R Soc Med 2008; 101:566–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol 2016; 17:679–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demarest TG, Babbar M, Okur MN, Dan X, Croteau DL, Fakouri NB, Mattson MP, Bohr VA. NAD+ metabolism in aging and cancer. Annu Rev Cancer Biol 2019; 3:105–30 [Google Scholar]
- 13.Guarente L. The resurgence of NAD+: restoring a mitochondrial metabolite slows stem cell loss and aging. Science 2016; 352:1396–7 [DOI] [PubMed] [Google Scholar]
- 14.Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, Liu F, Turner K, Yang H, Brunk E, Bi J, Furnari F, Bafna V, Ren B, Mischel PS. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 2019; 569:570–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaku K, Okabe K, Hikosaka K, Nakagawa T. NAD metabolism in cancer therapeutics. Front Oncol 2018; 8:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D'Amico D, Ropelle ER, Lutolf MP, Aebersold R, Schoonjans K, Menzies KJ, Auwerx J. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016; 352:1436–43 [DOI] [PubMed] [Google Scholar]
- 17.Curtin NJ. PARP inhibitors for cancer therapy. Expert Rev Mol Med 2005; 7:1–20 [DOI] [PubMed] [Google Scholar]
- 18.Salter M, Pogson CI. The role of tryptophan 2,3-dioxygenase in the hormonal control of tryptophan metabolism in isolated rat liver cells: effects of glucocorticoids and experimental diabetes. Biochem J 1985; 229:499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki F, Kuroiwa T, Takikawa O, Kido R. Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem J 1985; 230:635–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Van Valkenburgh J, Hong X, Conti PS, Zhang X, Chen K. Small molecules as theranostic agents in cancer immunology. Theranostics 2019; 9:7849–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen DJM, Theodoropoulos G, Li Y-Y, Wu C, Sha W, Feun LG, Lampidis TJ, Savaraj N, Wangpaichitr M. Targeting the kynurenine pathway for the treatment of cisplatin-resistant lung cancer. Mol Cancer Res 2020; 18:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z, Yue L, Shi J, Shao M, Wu T. Role of IDO and TDO in cancers and related diseases and the therapeutic implications. J Cancer 2019; 10:2771–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zahm CD, Johnson LE, McNeel DG. Increased indoleamine 2,3-dioxygenase activity and expression in prostate cancer following targeted immunotherapy. Cancer Immunol Immunother 2019; 68:1661–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci 2004; 61:19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youn HS, Gyun Kim T, Kim MK, Bu Kang G, Youn Kang J, Lee JG, Yop An J, Ryoung Park K, Lee Y, Jun Im Y, Hyuck Lee J, Hyun Eom S. Structural insights into the quaternary catalytic mechanism of hexameric human quinolinate phosphoribosyltransferase, a key enzyme in de novo NAD biosynthesis. Sci Rep 2016; 6. doi: 10.1038/srep19681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitt MK, Harvey CC, Rothwell WS, Cutler JL, Haffron D. Tryptophan-Niacin relationships in man. J Nutrition 1956; 60:1–4313385712 [Google Scholar]
- 27.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem 1958; 233:488–92 [PubMed] [Google Scholar]
- 28.Preiss J, Handler P. Biosynthesis of diphosphopyridine nucleotide. II. Enzymatic aspects. J Biol Chem 1958; 233:493–500 [PubMed] [Google Scholar]
- 29.Galassi L, Di Stefano M, Brunetti L, Orsomando G, Amici A, Ruggieri S, Magni G. Characterization of human nicotinate phosphoribosyltransferase: kinetic studies, structure prediction and functional analysis by site-directed mutagenesis. Biochimie 2012; 94:300–9 [DOI] [PubMed] [Google Scholar]
- 30.Balducci E, Emanuelli M, Magni G, Raffaelli N, Ruggieri S, Vita A, Natalini P. Nuclear matrix-associated NMN adenylyltransferase activity in human placenta. Biochem Biophys Res Commun 1992; 189:1275–9 [DOI] [PubMed] [Google Scholar]
- 31.Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 2005; 280:36334–41 [DOI] [PubMed] [Google Scholar]
- 32.Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem 2001; 276:406–12 [DOI] [PubMed] [Google Scholar]
- 33.Emanuelli M, Natalini P, Raffaelli N, Ruggieri S, Vita A, Magni G. NAD biosynthesis in human placenta: purification and characterization of homogeneous NMN adenylyltransferase. Arch Biochem Biophys 1992; 298:29–34 [DOI] [PubMed] [Google Scholar]
- 34.Raffaelli N, Sorci L, Amici A, Emanuelli M, Mazzola F, Magni G. Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem Biophys Res Commun 2002; 297:835–40 [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Kurnasov OV, Karthikeyan S, Grishin NV, Osterman AL, Zhang H. Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis. J Biol Chem 2003; 278:13503–11 [DOI] [PubMed] [Google Scholar]
- 36.Hara N, Yamada K, Terashima M, Osago H, Shimoyama M, Tsuchiya M. Molecular identification of human glutamine- and ammonia-dependent NAD synthetases: carbon-nitrogen hydrolase domain confers glutamine dependency. J Biol Chem 2003; 278:10914–21 [DOI] [PubMed] [Google Scholar]
- 37.Bieganowski P, Brenner C. The reported human NADsyn2 is ammonia-dependent NAD synthetase from a pseudomonad. J Biol Chem 2003; 278:33056–9 [DOI] [PubMed] [Google Scholar]
- 38.Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 2010; 31:194–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svoboda P, Krizova E, Sestakova S, Vapenkova K, Knejzlik Z, Rimpelova S, Rayova D, Volfova N, Krizova I, Rumlova M, Sykora D, Kizek R, Haluzik M, Zidek V, Zidkova J, Skop V. Nuclear transport of nicotinamide phosphoribosyltransferase is cell cycle–dependent in mammalian cells, and its inhibition slows cell growth. J Biol Chem 2019; 294:8676–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardell SJ, Hopf M, Khan A, Dispagna M, Hampton Sessions E, Falter R, Kapoor N, Brooks J, Culver J, Petucci C, Ma C-T, Cohen SE, Tanaka J, Burgos ES, Hirschi JS, Smith SR, Sergienko E, Pinkerton AB. Boosting NAD+ with a small molecule that activates NAMPT. Nat Commun 2019; 10. doi: 10.1038/s41467-019-11078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Lei H, Zhang Z. Pre-B cell colony enhancing factor (PBEF), a cytokine with multiple physiological functions. Cytokine Growth Factor Rev 2013; 24:433–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon MJ, Yoshida M, Johnson S, Takikawa A, Usui I, Tobe K, Nakagawa T, Yoshino J. Imai Si SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab 2015; 21:706–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friebe D, Neef M, Kratzsch J, Erbs S, Dittrich K, Garten A, Petzold-Quinque S, Blüher S, Reinehr T, Stumvoll M, Blüher M, Kiess W, Körner A. Leucocytes are a major source of circulating nicotinamide phosphoribosyltransferase (NAMPT)/pre-B cell colony (PBEF)/visfatin linking obesity and inflammation in humans. Diabetologia 2011; 54:1200–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitani T, Okuno S, Fujisawa H. Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett 2003; 544:74–8 [DOI] [PubMed] [Google Scholar]
- 45.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 1994; 14:1431–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang SN, Miao CY. Targeting NAMPT as a therapeutic strategy against stroke. Stroke Vasc Neurol 2019; 4:83–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang LQ, Heruth DP, Ye SQ. Nicotinamide phosphoribosyltransferase in human diseases. J Bioanal Biomed 2011; 3:13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004; 117:495–502 [DOI] [PubMed] [Google Scholar]
- 49.Bogan KL, Brenner C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD + precursor vitamins in human nutrition. Annu Rev Nutr 2008; 28:115–30 [DOI] [PubMed] [Google Scholar]
- 50.Cantó C, Menzies KJ, Auwerx J. NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 2015; 22:31–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lerner F, Niere M, Ludwig A, Ziegler M. Structural and functional characterization of human NAD kinase. Biochem Biophys Res Commun 2001; 288:69–74 [DOI] [PubMed] [Google Scholar]
- 52.Grose JH, Joss L, Velick SF, Roth JR. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc Natl Acad Sci U S A 2006; 103:7601–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li BB, Wang X, Tai L, Ma TT, Shalmani A, Liu WT, Li WQ, Chen KM. NAD kinases: metabolic targets controlling redox co-enzymes and reducing power partitioning in plant stress and development. Front Plant Sci 2018; 9. doi: 10.3389/fpls.2018.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohashi K, Kawai S, Murata K. Identification and characterization of a human mitochondrial NAD kinase. Nat Commun 2012; 3. doi: 10.1038/ncomms2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang R. MNADK, a long-awaited human mitochondrion-localized NAD kinase. J Cell Physiol 2015; 230:1697–701 [DOI] [PubMed] [Google Scholar]
- 56.Nakahata Y, Bessho Y. The circadian NAD+ metabolism: impact on chromatin remodeling and aging. BioMed Res Int 2016; 2016:3208429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J 1995; 14:3480–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryu KW, Nandu T, Kim J, Challa S, DeBerardinis RJ, Kraus WL. Metabolic regulation of transcription through compartmentalized NAD+ biosynthesis. Science 2018; 360:eaan5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, Cabo D, R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 2007; 130:1095–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 2009; 137:560–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Linden MR, Dölle C, Pettersen IKN, Kulikova VA, Niere M, Agrimi G, Dyrstad SE, Palmieri F, Nikiforov AA, Tronstad KJ, Ziegler M. Subcellular distribution of NAD+ between cytosol and mitochondria determines the metabolic profile of human cells. J Biol Chem 2015; 290:27644–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem 2006; 352:282–5 [DOI] [PubMed] [Google Scholar]
- 63.Davila A, Liu L, Chellappa K, Redpath P, Nakamaru-Ogiso E, Paolella LM, Zhang Z, Migaud ME, Rabinowitz JD, Baur JA. Nicotinamide adenine dinucleotide is transported into mammalian mitochondria. eLife 2018; 7. doi: 10.7554/eLife.33246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trammell SAJ, Schmidt MS, Weidemann BJ, Redpath P, Jaksch F, Dellinger RW, Li Z, Abel ED, Migaud ME, Brenner C. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun 2016; 7. doi: 10.1038/ncomms12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nikiforov A, Dölle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem 2011; 286:21767–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grozio A, Mills KF, Yoshino J, Bruzzone S, Sociali G, Tokizane K, Lei HC, Cunningham R, Sasaki Y, Migaud ME. Imai Si Slc12a8 is a nicotinamide mononucleotide transporter. Nat Metab 2019; 1:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dukes ID, McIntyre MS, Mertz RJ, Philipson LH, Roe MW, Spencer B, Worley JF., 3rd Dependence on NADH produced during glycolysis for beta-cell glucose signaling. J Biol Chem 1994; 269:10979–82 [PubMed] [Google Scholar]
- 68.Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Electron transport and oxidative phosphorylation. Molecular Cell Biology. New York, NY: W. H. Freeman, 2000. p. 666-73 [Google Scholar]
- 69.Guo R, Gu J, Zong S, Wu M, Yang M. Structure and mechanism of mitochondrial electron transport chain. Biomed J 2018; 41:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pirinen E, Cantó C, Jo YS, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L, Moullan N, Hagberg C, Li W, Timmers S, Imhof R, Verbeek J, Pujol A, Van Loon B, Viscomi C, Zeviani M, Schrauwen P, Sauve AA, Schoonjans K, Auwerx J. Pharmacological inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 2014; 19:1034–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y. Molecular links between caloric restriction and Sir2/SIRT1 activation. Diab Metab J 2014; 38:321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly DJ, Hughes NJ, Poole RK. Microaerobic physiology: aerobic respiration, anaerobic respiration, and carbon dioxide metabolism In: HLT Mobley, GL Mendz, SL Hazell. (eds) Helicobacter pylori: physiology and genetics. Washington, DC: ASM Press, 2001, p. 113. [PubMed] [Google Scholar]
- 73.Croteau DL, Fang EF, Nilsen H, Bohr VA. NAD+ in DNA repair and mitochondrial maintenance. Cell Cycle 2017; 16:491–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res 1985; 101:4. [PubMed] [Google Scholar]
- 75.Zhang D, Hu X, Li J, Liu J, Baks-Te Bulte L, Wiersma M, Malik NuA van Marion DMS, Tolouee M, Hoogstra-Berends F, Lanters EAH, van Roon AM, de Vries AAF, Pijnappels DA, de Groot NMS, Henning RH, Brundel B. DNA damage-induced PARP1 activation confers cardiomyocyte dysfunction through NAD + depletion in experimental atrial fibrillation. Nat Commun 2019; 10. doi: 10.1038/s41467-019-09014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai P, Cantó C, Oudart H, Brunyánszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-De Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 2011; 13:461–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banasik M, Komura H, Shimoyama M, Ueda K. Specific inhibitors of poly(ADP-ribose) synthetase and Mono(ADP-ribosyl)transferase. J Biol Chem 1992; 267:1569–75 [PubMed] [Google Scholar]
- 78.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev 2017; 31:101–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okazaki IJ, Moss J. Mono-ADP-ribosylation: a reversible posttranslational modification of proteins. Adv Pharmacol 1996; 35:247–80 [DOI] [PubMed] [Google Scholar]
- 80.Liu C, Yu X. ADP-ribosyltransferases and poly-ADP-ribosylation. Curr Protein Pept Sci 2015; 16:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stevens LA, Kato J, Kasamatsu A, Oda H, Lee D-Y, Moss J. The ARH and macrodomain families of α-ADP-ribose-acceptor hydrolases catalyze α-NAD+ hydrolysis. ACS Chem Biol 2019; 14:2576–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yahiro K, Ogura K, Terasaki Y, Satoh M, Miyagi S, Terasaki M, Yamasaki E, Moss J. Cholix toxin, an eukaryotic elongation factor 2 ADP-ribosyltransferase, interacts with prohibitins and induces apoptosis with mitochondrial dysfunction in human hepatocytes. Cell Microbiol 2019; 21:e13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vaughan M, Moss J. G proteins and toxin-catalyzed ADP-ribosylation. Biology of Cellular Transducing Signals. Boston, MA: Springer US, 1990. pp. 193-200 [Google Scholar]
- 84.Watanabe K, Kato J, Zhu J, Oda H, Ishiwata-Endo H, Moss J. Enhanced sensitivity to cholera toxin in female ADP-ribosylarginine hydrolase (ARH1)-deficient mice. PLoS One 2018; 13:e0207693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Foley BT, Moehring JM, Moehring TJ. Mutations In the elongation factor 2 gene which confer resistance to diphtheria toxin and pseudomonas exotoxin A: genetic and biochemical analyses. J Biol Chem 1995; 270:23218–25 [DOI] [PubMed] [Google Scholar]
- 86.Ling F, Tang Y, Li M, Li Q-S, Li X, Yang L, Zhao W, Jin C-C, Zeng Z, Liu C, Wu C-F, Chen W-W, Lin X, Wang Y-L, Threadgill MD. Mono-ADP-ribosylation of histone 3 at arginine-117 promotes proliferation through its interaction with P300. Oncotarget 2017; 8:72773–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zolkiewska A. Ecto-ADP-ribose transferases: cell-surface response to local tissue injury. Physiology 2005; 20:374–81 [DOI] [PubMed] [Google Scholar]
- 88.Adriouch S, Ohlrogge W, Haag F, Koch-Nolte F, Seman M. Rapid induction of naive T cell apoptosis by ecto-nicotinamide adenine dinucleotide: requirement for Mono(ADP-ribosyl)transferase 2 and a downstream effector. J Immunol 2001; 167:196–203 [DOI] [PubMed] [Google Scholar]
- 89.Kawamura H, Aswad F, Minagawa M, Malone K, Kaslow H, Koch-Nolte F, Schott WH, Leiter EH, Dennert G. P2X7 receptor-dependent and -independent T cell death is induced by nicotinamide adenine dinucleotide. J Immunol 2005; 174:1971–9 [DOI] [PubMed] [Google Scholar]
- 90.Bu X, Kato J, Moss J. Emerging roles of ADP-ribosyl-acceptor hydrolases (ARHs) in tumorigenesis and cell death pathways. Biochem Pharmacol 2019; 167:44–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mashimo M, Moss J. ADP-ribosyl-acceptor hydrolase activities catalyzed by the arh family of proteins. Methods Mol Biol 2018; 1813:187–204 [DOI] [PubMed] [Google Scholar]
- 92.Moss J, Jacobson MK, Stanley SJ. Reversibility of arginine-specific Mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-L-arginine cleavage enzyme. Proc Natl Acad Sci U S A 1985; 82:5603–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mashimo M, Bu X, Aoyama K, Kato J, Ishiwata-Endo H, Stevens LA, Kasamatsu A, Wolfe LA, Toro C, Adams D, Markello T, Gahl WA, Moss J. PARP1 inhibition alleviates injury in ARH3-deficient mice and human cells. JCI Insight 2019; 4:124519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee HC. Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu Rev Pharmacol Toxicol 2001; 41:317–45 [DOI] [PubMed] [Google Scholar]
- 95.Lee HC. Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J Biol Chem 2012; 287:31633–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev 2008; 88:841–86 [DOI] [PubMed] [Google Scholar]
- 97.Wei W, Graeff R, Yue J. Roles and mechanisms of the CD38/cyclic adenosine diphosphate ribose/Ca(2+) signaling pathway. World J Biol Chem 2014; 5:58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quarona V, Zaccarello G, Chillemi A, Brunetti E, Singh VK, Ferrero E, Funaro A, Horenstein AL, Malavasi F. CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytometry B Clin Cytom 2013; 84:207–17 [DOI] [PubMed] [Google Scholar]
- 99.Chini CCS, Tarragó MG, Chini EN. NAD and the aging process: role in life, death and everything in between. Mol Cell Endocrinol 2017; 455:62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN. Regulation of SIRT1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun 2006; 349:353–9 [DOI] [PubMed] [Google Scholar]
- 101.Camacho-Pereira J, Tarragó MG, Chini CCS, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A, Chini EN. CD38 dictates age-related NAD decline and mitochondrial dysfunction through a SIRT3-dependent mechanism. Cell Metab 2016; 23:1127–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbosa MTP, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, Chini EN. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J 2007; 21:3629–39 [DOI] [PubMed] [Google Scholar]
- 103.Wang L-F, Huang C-C, Xiao Y-F, Guan X-H, Wang X-N, Cao Q, Liu Y, Huang X, Deng L-B, Deng K-Y, Xin H-B. CD38 deficiency protects heart from high fat diet-induced oxidative stress via activating Sirt3/FOXO3 pathway. Cell Physiol Biochem 2018; 48:2350–63 [DOI] [PubMed] [Google Scholar]
- 104.Guan X-H, Liu X-H, Hong X, Zhao N, Xiao Y-F, Wang L-F, Tang L, Jiang K, Qian Y-S, Deng K-Y, Ji G, Fu M, Xin H-B. CD38 deficiency protects the heart from ischemia/reperfusion injury through activating SIRT1/FOXOs-mediated antioxidative stress pathway. Oxid Med Cell Longev 2016; 2016:7410257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu Y, Wang H, Wang Q, Deng H. Overexpression of CD38 decreases cellular NAD levels and alters the expression of proteins involved in energy metabolism and antioxidant defense. J Proteome Res 2014; 13:786–95 [DOI] [PubMed] [Google Scholar]
- 106.Barata H, Thompson M, Zielinska W, Han YS, Mantilla CB, Prakash YS, Feitoza S, Sieck G, Chini EN. The role of cyclic-ADP-ribose-signaling pathway in oxytocin-induced Ca 2+ transients in human myometrium cells. Endocrinology 2004; 145:881–9 [DOI] [PubMed] [Google Scholar]
- 107.Lee CU, Song EK, Yoo CH, Kwak YK, Han MK. Lipopolysaccharide induces CD38 expression and solubilization in J774 macrophage cells. Mol Cells 2012; 34:573–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Musso T, Deaglio S, Franco L, Calosso L, Badolato R, Garbarino G, Dianzani U, Malavasi F. CD38 expression and functional activities are up-regulated by IFN-gamma on human monocytes and monocytic cell lines. J Leukoc Biol 2001; 69:605–12 [PubMed] [Google Scholar]
- 109.Ge Y, Long Y, Xiao S, Liang L, He Z, Yue C, Wei X, Zhou Y. CD38 affects the biological behavior and energy metabolism of nasopharyngeal carcinoma cells. Int J Oncol 2019; 54:585–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin D, Liu HX, Hirai H, Torashima T, Nagai T, Lopatina O, Shnayder NA, Yamada K, Noda M, Seike T, Fujita K, Takasawa S, Yokoyama S, Koizumi K, Shiraishi Y, Tanaka S, Hashii M, Yoshihara T, Higashida K, Islam MS, Yamada N, Hayashi K, Noguchi N, Kato I, Okamoto H, Matsushima A, Salmina A, Munesue T, Shimizu N, Mochida S, Asano M, Higashida H. CD38 is critical for social behaviour by regulating oxytocin secretion. Nature 2007; 446:41–5 [DOI] [PubMed] [Google Scholar]
- 111.Lischke T, Heesch K, Schumacher V, Schneider M, Haag F, Koch-Nolte F, Mittrücker HW. CD38 controls the innate immune response against listeria monocytogenes. Infect Immun 2013; 81:4091–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPs, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008; 13:3046–82 [DOI] [PubMed] [Google Scholar]
- 113.Sodhi RK, Singh N, Jaggi AS. Poly(ADP-ribose) polymerase-1 (PARP-1) and its therapeutic implications. Vascul Pharmacol 2010; 53:77–87 [DOI] [PubMed] [Google Scholar]
- 114.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 2000; 275:40974–80 [DOI] [PubMed] [Google Scholar]
- 115.Cortes U, Tong W-M, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang Z-Q. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol 2004; 24:7163–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ménissier De Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, Lemeur M, Walztinger C, Chambon P, De Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A 1997; 94:7303–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat Res 2000; 460:1–15 [DOI] [PubMed] [Google Scholar]
- 118.Beneke S, Alvarez-Gonzalez R, Bürkle A. Comparative characterisation of poly(ADP-ribose) polymerase-1 from two mammalian species with different life span. Exp Gerontol 2000; 35:989–1002 [DOI] [PubMed] [Google Scholar]
- 119.Grube K, Burkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci U S A 1992; 89:11759–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muiras ML, Müller M, Schächter F, Bürkle A. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J Mol Med 1998; 76:346–54 [DOI] [PubMed] [Google Scholar]
- 121.Chatterjee S, Berger SJ, Berger NA. Poly(ADP-ribose) polymerase: a guardian of the genome that facilitates DNA repair by protecting against DNA recombination. Mol Cell Biochem 1999; 193:23–30 [PubMed] [Google Scholar]
- 122.Beneke S, Bürkle A. Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Res 2007; 35:7456–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol 2005; 37:920–6 [DOI] [PubMed] [Google Scholar]
- 124.Tong WM, Cortes U, Wang ZQ. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim Biophys Acta 2001; 1552:27–37 [DOI] [PubMed] [Google Scholar]
- 125.Rosado MM, Bennici E, Novelli F, Pioli C. Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology 2013; 139:428–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Agarwal A, Mahfouz RZ, Sharma RK, Sarkar O, Mangrola D, Mathur PP. Potential Biological role of poly (ADP-ribose) polymerase (PARP) in male gametes. Reprod Biol Endocrinol 2009; 7:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yamashita S, Tanaka M, Nodono H, Hamada A, Hamada T, Hasegawa M, Nishi Y, Moss J, Miwa M. Human alcohol dehydrogenase 1 is an acceptor protein for polyADP-ribosylation. Biochem Pharmacol 2019; 167:27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, Okamoto A, Moore KN, Efrat Ben-Baruch N, Werner TL, Cloven NG, Oaknin A, DiSilvestro PA, Morgan MA, Nam JH, Leath CA, Nicum S, Hagemann AR, Littell RD, Cella D, Baron-Hay S, Garcia-Donas J, Mizuno M, Bell-McGuinn K, Sullivan DM, Bach BA, Bhattacharya S, Ratajczak CK, Ansell PJ, Dinh MH, Aghajanian C, Bookman MA. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med 2019; 381:2403–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.González-Martín A, Pothuri B, Vergote I, DePont Christensen R, Graybill W, Mirza MR, McCormick C, Lorusso D, Hoskins P, Freyer G, Baumann K, Jardon K, Redondo A, Moore RG, Vulsteke C, O'Cearbhaill RE, Lund B, Backes F, Barretina-Ginesta P, Haggerty AF, Rubio-Pérez MJ, Shahin MS, Mangili G, Bradley WH, Bruchim I, Sun K, Malinowska IA, Li Y, Gupta D, Monk BJ. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019; 381:2391–402 [DOI] [PubMed] [Google Scholar]
- 130.Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017; 377:523–33 [DOI] [PubMed] [Google Scholar]
- 131.O'Cearbhaill RE, Gnjatic S, Aghajanian C, Iasonos A, Konner JA, Losada N, Sarasohn D, Rasalan T, Tew WP, Zamarin D, Wong P, Sabbatini P. A phase I study of concomitant galinpepimut-s (GPS) in combination with nivolumab (nivo) in patients (pts) with WT1+ ovarian cancer (OC) in second or third remission. J Clin Oncol 2018; 36:5553 [Google Scholar]
- 132.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, Park J-O, Hochhauser D, Arnold D, Oh D-Y, Reinacher-Schick A, Tortora G, Algül H, O’Reilly EM, McGuinness D, Cui KY, Schlienger K, Locker GY, Kindler HL. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019; 381:317–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Virtanen Paunu Ahlskog Varnai Sipeky, Sundvall PARP inhibitors in prostate cancer – the preclinical rationale and current clinical development. Genes 2019; 10:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sachdev E, Tabatabai R, Roy V, Rimel BJ, Mita MM. PARP inhibition in cancer: an update on clinical development. Target Oncol 2019; 14:657–79 [DOI] [PubMed] [Google Scholar]
- 135.Kam T-I, Mao X, Park H, Chou S-C, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL. Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease. Science 2018; 362:eaat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Olsen AL, Feany MB. PARP inhibitors and Parkinson's disease. N Engl J Med 2019; 380:492–94 [DOI] [PubMed] [Google Scholar]
- 137.Berger NA, Besson VC, Boulares AH, Bürkle A, Chiarugi A, Clark RS, Curtin NJ, Cuzzocrea S, Dawson TM, Dawson VL, Haskó G, Liaudet L, Moroni F, Pacher P, Radermacher P, Salzman AL, Snyder SH, Soriano FG, Strosznajder RP, Sümegi B, Swanson RA, Szabo C. Opportunities for the repurposing of PARP inhibitors for the therapy of non-oncological diseases. Br J Pharmacol 2018; 175:192–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 2006; 23:607–18 [DOI] [PubMed] [Google Scholar]
- 139.Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol 2002; 3:reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hillgartner FB, Charron T, Chesnut KA. Alterations in nutritional status regulate acetyl-CoA carboxylase expression in avian liver by a transcriptional mechanism. Biochem J 1996; 319:263–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 2015; 21:805–21 [DOI] [PubMed] [Google Scholar]
- 142.Roy D, Paul A, Roy A, Ghosh R, Ganguly P, Chaudhuri S. Differential acetylation of histone H3 at the regulatory region of OsDREB1b promoter facilitates chromatin remodelling and transcription activation during cold stress. PLoS One 2014; 9:e100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yoshida M, Shimazu T, Matsuyama A. Protein deacetylases: enzymes with functional diversity as novel therapeutic targets. Prog Cell Cycle Res 2003; 5:269–78 [PubMed] [Google Scholar]
- 144.Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol 1986; 6:688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 1987; 116:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 1995; 9:2888–902 [DOI] [PubMed] [Google Scholar]
- 147.Derbyshire MK, Weinstock KG, Strathern JN. HST1, a new member of the SIR2 family of genes. Yeast 1996; 12:631–40 [DOI] [PubMed] [Google Scholar]
- 148.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science 1999; 285:1390–93 [DOI] [PubMed] [Google Scholar]
- 149.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000; 289:2126–28 [DOI] [PubMed] [Google Scholar]
- 150.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 2004; 18:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 2003; 423:181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via NRK and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 2007; 129:473–84 [DOI] [PubMed] [Google Scholar]
- 153.Costford SR, Bajpeyi S, Pasarica M, Albarado DC, Thomas SC, Xie H, Church TS, Jubrias SA, Conley KE, Smith SR. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol 2010; 298:E117–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 2008; 14:661–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang RH, Zhao T, Cui K, Hu G, Chen Q, Chen W, Wang XW, Soto-Gutierrez A, Zhao K, Deng CX. Negative reciprocal regulation between Sirt1 and Per2 modulates the circadian clock and aging. Sci Rep 2016; 6. doi: 10.1038/srep28633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chang H-C, Guarente L. SIRT1 mediates Central circadian control in the SCN by a mechanism that decays with aging. Cell 2013; 153:1448–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, Baldi P, Sassone-Corsi P. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014; 158:659–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sun S, Liu Z, Feng Y, Shi L, Cao X, Cai Y, Liu B. Sirt6 deacetylase activity regulates circadian rhythms via Per2. Biochem Biophys Res Commun 2019; 511:234–38 [DOI] [PubMed] [Google Scholar]
- 159.Peek CB, Affinati AH, Ramsey KM, Kuo H-Y, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, Bass J. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 2013; 342:1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai SI. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab 2013; 18:416–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun 2010; 1. doi: 10.1038/ncomms1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Furukawa A, Tada-Oikawa S, Kawanishi S, Oikawa S. H2O2 accelerates cellular senescence by accumulation of acetylated p53 via decrease in the function of SIRT1 by NAD depletion. Cell Physiol Biochem 2007; 20:45–54 [DOI] [PubMed] [Google Scholar]
- 163.Gorenne I, Kumar S, Gray K, Figg N, Yu H, Mercer J, Bennett M. Vascular smooth muscle cell sirtuin 1 protects against DNA damage and inhibits atherosclerosis. Circulation 2013; 127:386–96 [DOI] [PubMed] [Google Scholar]
- 164.Wilking MJ, Ahmad N. The role of SIRT1 in cancer: the Saga continues. Am J Pathol 2015; 185:26–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009; 459:113–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2007; 2:e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Anwar T, Khosla S, Ramakrishna G. Increased expression of SIRT2 is a novel marker of cellular senescence and is dependent on wild type p53 status. Cell Cycle 2016; 15:1883–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 2007; 6:105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang F, Nguyen M, Qin FXF, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007; 6:505–14 [DOI] [PubMed] [Google Scholar]
- 170.Szabó A, Oláh J, Szunyogh S, Lehotzky A, Szénási T, Csaplár M, Schiedel M, Low P, Jung M, Ovádi J. Modulation of microtubule acetylation by the interplay of TPPP/p25, SIRT2 and new anticancer agents with anti-SIRT2 potency. Sci Rep 2017; 7. doi: 10.1038/s41598-017-17381-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, Ji J, Wang XW, Park SH, Cha YI, Gius D, Deng CX. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011; 20:487–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CGT, Kurimasa A, Oshimura M. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun 2003; 309:558–66 [DOI] [PubMed] [Google Scholar]
- 173.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wölfel C, Huber C, Wölfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A 2005; 102:16013–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Damodaran S, Damaschke N, Gawdzik J, Yang B, Shi C, Allen GO, Huang W, Denu J, Jarrard D. Dysregulation of sirtuin 2 (SIRT2) and histone H3K18 acetylation pathways associates with adverse prostate cancer outcomes. BMC Cancer 2017; 17. doi: 10.1186/s12885-017-3853-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dan L, Klimenkova O, Klimiankou M, Klusman JH, van den Heuvel-Eibrink MM, Reinhardt D, Welte K, Skokowa J. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica 2012; 97:551–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Funato K, Hayashi T, Echizen K, Negishi L, Shimizu N, Koyama-Nasu R, Nasu-Nishimura Y, Morishita Y, Tabar V, Todo T, Ino Y, Mukasa A, Saito N, Akiyama T. SIRT2-mediated inactivation of p73 is required for glioblastoma tumorigenicity. EMBO Rep 2018; 19:e45587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Quan S, Principe DR, Dean AE, Park SH, Grippo PJ, Gius D, Horikoshi N. Loss of Sirt2 increases and prolongs a caerulein-induced pancreatitis permissive phenotype and induces spontaneous oncogenic Kras mutations in mice. Sci Rep 2018; 8. doi: 10.1038/s41598-018-34792-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou Y, Wang Q, Rychahou P, Weiss HL, Mark Evers B. Differential role of SIRT2 protein in normal intestinal epithelium and colorectal cancer cells. Gastroenterology 2017; 152:S252 [Google Scholar]
- 179.Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X, Luo X, Hu J, Wang G. Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis 2018; 10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem J 2012; 444:1–10 [DOI] [PubMed] [Google Scholar]
- 181.Sengupta A, Haldar D. Human sirtuin 3 (SIRT3) deacetylates histone H3 lysine 56 to promote nonhomologous end joining repair. DNA Repair 2018; 61:1–16 [DOI] [PubMed] [Google Scholar]
- 182.Bause AS, Haigis MC. SIRT3 regulation of mitochondrial oxidative stress. Exp Gerontol 2013; 48:634–39 [DOI] [PubMed] [Google Scholar]
- 183.Liu J, Li D, Zhang T, Tong Q, Ye RD, Lin L. SIRT3 protects hepatocytes from oxidative injury by enhancing ROS scavenging and mitochondrial integrity. Cell Death Dis 2017; 8:e3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Finley LWS, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 2011; 19:416–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bell EL, Emerling BM, Ricoult SJH, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011; 30:2986–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep 2013; 30:1323–28 [DOI] [PubMed] [Google Scholar]
- 188.Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, Zhao S, Yang Y, Ling ZQ, Guan KL, Xiong Y, Ye D. SIRT3‐dependent GOT2 acetylation status affects the malate–aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J 2015; 34:1110–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 2011; 117:1670–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ashraf N, Zino S, MacIntyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 2006; 95:1056–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.George J, Nihal M, Singh CK, Zhong W, Liu X, Ahmad N. Pro-proliferative function of mitochondrial sirtuin deacetylase SIRT3 in human melanoma. J Invest Dermatol 2016; 136:809–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mei Y, Chunsong Y, Yuhua P. Effects of downregulation of SIRT3 expression on proliferation and apoptosis in esophageal squamous cell carcinoma EC9706 cells and its molecular mechanisms. Bio med Mater Eng 2014; 24:3883–90 [DOI] [PubMed] [Google Scholar]
- 193.Shackelford R, Hirsh S, Henry K, Abdel-Mageed A, Kandil E, Coppola D. Nicotinamide phosphoribosyltransferase and SIRT3 expression are increased in well-differentiated thyroid carcinomas. Anticancer Res 2013; 33:3047–52 [PubMed] [Google Scholar]
- 194.Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y, Huang J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis 2014; 5:e1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Choi YL, Tsukasaki K, O'Neill MC, Yamada Y, Onimaru Y, Matsumoto K, Ohashi J, Yamashita Y, Tsutsumi S, Kaneda R, Takada S, Aburatani H, Kamihira S, Nakamura T, Tomonaga M, Mano H. A genomic analysis of adult T-cell leukemia. Oncogene 2007; 26:1245–55 [DOI] [PubMed] [Google Scholar]
- 196.Fu L, Dong Q, He J, Wang X, Xing J, Wang E, Qiu X, Li Q. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene 2017; 36:2724–36 [DOI] [PubMed] [Google Scholar]
- 197.Jeong SM, Xiao C, Finley LWS, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 2013; 23:450–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Ogawa H, Hamabe A, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Doki Y, Mori M, Ishii H. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer 2015; 113:492–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sun H, Huang D, Liu G, Jian F, Zhu J, Zhang L. SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion. Onco Targets Ther 2018; 11:3959–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Du Y, Hu H, Hua C, Du K, Wei T. Tissue distribution, subcellular localization, and enzymatic activity analysis of human SIRT5 isoforms. Biochem Biophys Res Commun 2018; 503:763–69 [DOI] [PubMed] [Google Scholar]
- 201.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011; 334:806–09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, Verdin E. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell 2015; 59:321–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lu W, Zuo Y, Feng Y, Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 2014; 35:10699–705 [DOI] [PubMed] [Google Scholar]
- 204.Sun X, Wang S, Gai J, Guan J, Li J, Li Y, Zhao J, Zhao C, Fu L, Li Q. SIRT5 promotes cisplatin resistance in ovarian cancer by suppressing dna damage in a ROS-dependent manner via regulation of the Nrf2/HO-1 pathway. Front Oncol 2019; 9. doi: 10.3389/fonc.2019.00754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang R, Wang C, Tian Y, Yao Y, Mao J, Wang H, Li Z, Xu Y, Ye M, Wang L. SIRT5 promotes hepatocellular carcinoma progression by regulating mitochondrial apoptosis. J Cancer 2019; 10:3871–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bartosch C, Monteiro-Reis S, Almeida-Rios D, Vieira R, Castro A, Moutinho M, Rodrigues M, Graça I, Lopes JM, Jerónimo C. Assessing sirtuin expression in endometrial carcinoma and nonneoplastic endometrium. Oncotarget 2016; 7:1144–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lai C-C, Lin P-M, Lin S-F, Hsu C-H, Lin H-C, Hu M-L, Hsu C-M, Yang M-Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumor Biol 2013; 34:1847–54 [DOI] [PubMed] [Google Scholar]
- 208.Ardestani PM, Liang F. Sub-cellular localization, expression and functions of Sirt6 during the cell cycle in HeLa cells. Nucleus 2012; 3:442–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TLA, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008; 452:492–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009; 8:2664–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011; 10:3153–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006; 124:315–29 [DOI] [PubMed] [Google Scholar]
- 213.Bae JS, Park SH, Jamiyandorj U, Kim KM, Noh SJ, Kim JR, Park HJ, Kwon KS, Jung SH, Park HS, Park BH, Lee H, Moon WS, Sylvester KG, Jang KY. CK2α/CSNK2A1 phosphorylates SIRT6 and is involved in the progression of breast carcinoma and predicts shorter survival of diagnosed patients. Am J Pathol 2016; 186:3297–315 [DOI] [PubMed] [Google Scholar]
- 214.Kugel S, Sebastián C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, Ramaswamy S, Sadreyev RI, Goren A, Deshpande V, Bardeesy N, Mostoslavsky R. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell 2016; 165:1401–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ouyang L, Yi L, Li J, Yi S, Li S, Liu P, Yang X. SIRT6 overexpression induces apoptosis of nasopharyngeal carcinoma by inhibiting Nf-κB signaling. Onco Targets Ther 2018; 11:7613–24 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 216.Tian J, Yuan L. Sirtuin 6 inhibits Colon cancer progression by modulating PTEN/AKT signaling. Biomed Pharmacother 2018; 106:109–16 [DOI] [PubMed] [Google Scholar]
- 217.Zhang C, Yu Y, Huang Q, Tang K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol Med Rep 2019; 20:1575–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bae JS, Noh SJ, Kim KM, Park S-H, Hussein UK, Park HS, Park B-H, Ha SH, Lee H, Chung MJ, Moon WS, Cho DH, Jang KY. SIRT6 is involved in the progression of ovarian carcinomas via β-catenin-mediated epithelial to mesenchymal transition. Front Oncol 2018; 8. doi: 10.3389/fonc.2018.00538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Garcia-Peterson LM, Ndiaye MA, Singh CK, Chhabra G, Huang W, Ahmad N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer 2017; 8:701–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liu Y, Xie QR, Wang B, Shao J, Zhang T, Liu T, Huang G, Xia W. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell 2013; 4:702–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Onn L, Portillo M, Ilic S, Cleitman G, Stein D, Kaluski S, Shirat I, Slobodnik Z, Einav M, Erdel F, Akabayov B, Toiber D. SIRT6 is a DNA double-strand break sensor. eLife 2020; 9:e51636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, Tombline G, Tan R, Simon M, Henderson S, Steffan J, Goldfarb A, Tam J, Zheng K, Cornwell A, Johnson A, Yang JN, Mao Z, Manta B, Dang W, Zhang Z, Vijg J, Wolfe A, Moody K, Kennedy BK, Bohmann D, Gladyshev VN, Seluanov A, Gorbunova V. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 2019; 177:622–38.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, Ren R, Liu Z, Zhang L, Chen Z, Wang S, Zhao Y, Wang Z, Yuan Y, Zhou Q, Li W, Liu GH, Hu B. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018; 560:661–65 [DOI] [PubMed] [Google Scholar]
- 224.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani1 O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012; 487:114–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wu D, Li Y, Zhu KS, Wang H, Zhu W-G. Advances in cellular characterization of the sirtuin isoform, SIRT7. Front Endocrinol 2018; 9. doi: 10.3389/fendo.2018.00652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Deng Z, Wang X, Long X, Liu W, Xiang C, Bao F, Wang D. Sirtuin 7 promotes colorectal carcinoma proliferation and invasion through the inhibition of E-cadherin. Exp Ther Med 2018; 15:2333–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, Liu X, Tao Y, Yin H, Zhou H, Zhou L, Ye D, Ye Q, Gao D. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 2017; 65:515–28 [DOI] [PubMed] [Google Scholar]
- 228.Wang H-L, Lu R-Q, Xie S-H, Zheng H, Wen X-M, Gao X, Guo L. SIRT7 exhibits oncogenic potential in human ovarian cancer cells. Asian Pac J Cancer Prev 2015; 16:3573–7 [DOI] [PubMed] [Google Scholar]
- 229.Zhao J, Wozniak A, Adams A, Cox J, Vittal A, Voss J, Bridges B, Weinman SA, Li Z. SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J Exp Clin Cancer Res 2019; 38:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hegyi J, Schwartz RA, Hegyi V. Pellagra: dermatitis, dementia, and diarrhea. Int J Dermatol 2004; 43:1–5 [DOI] [PubMed] [Google Scholar]
- 231.Elvehjem CA, Madden RJ, Strong FM, Wolley DW. The isolation and identification of the anti-black tongue factor. 1937. J Biol Chem 2002; 277:e22. [PubMed] [Google Scholar]
- 232.Katsyuba E, Mottis A, Zietak M, De Franco F, van der Velpen V, Gariani K, Ryu D, Cialabrini L, Matilainen O, Liscio P, Giacchè N, Stokar-Regenscheit N, Legouis D, de Seigneux S, Ivanisevic J, Raffaelli N, Schoonjans K, Pellicciari R, Auwerx J. De novo NAD + synthesis enhances mitochondrial function and improves health. Nature 2018; 563:354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Maltos AL, Portari GV, Moraes GV, Rodrigues Monteiro MC, Vannucchi H, da Cunha DF. Niacin metabolism and indoleamine 2,3-dioxygenase activation in malnourished patients with flaky paint dermatosis. Nutrition 2015; 31:890–92 [DOI] [PubMed] [Google Scholar]
- 234.Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age-related changes in NAD+ metabolism oxidative stress and SIRT1 activity in wistar rats. PLoS One 2011; 6:e19194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One 2012; 7:e42357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gomes AP, Price NL, Ling AJY, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, De Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013; 155:1624–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nacarelli T, Lau L, Fukumoto T, Zundell J, Fatkhutdinov N, Wu S, Aird KM, Iwasaki O, Kossenkov AV, Schultz D, Noma Ki Baur JA, Schug Z, Tang HY, Speicher DW, David G, Zhang R. NAD+ metabolism governs the proinflammatory senescence-associated secretome. Nat Cell Biol 2019; 21:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wiley CD, Campisi J. From ancient pathways to aging cells – connecting metabolism and cellular senescence. Cell Metab 2016; 23:1013–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 2010; 5:99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rizza S, Filomeni G. Regulation of S-nitrosylation in aging and senescence. Rejuvenation Res 2019; 22:359–61 [DOI] [PubMed] [Google Scholar]
- 241.Lautrup S, Sinclair DA, Mattson MP, Fang EF. NAD+ in brain aging and neurodegenerative disorders. Cell Metab 2019; 30:630–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Johnson S, Wozniak DF, Imai S. CA1 Nampt knockdown recapitulates hippocampal cognitive phenotypes in old mice which nicotinamide mononucleotide improves. Aging Mech Dis 2018; 4. doi: 10.1038/s41514-018-0029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J. The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 2017; 93:1334–43.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL. Poly(ADP-ribose) drives pathologic a-synuclein neurodegeneration in Parkinson’s disease. Science 2018; 362:pii: eaat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Harlan BA, Pehar M, Sharma DR, Beeson G, Beeson CC, Vargas MR. Enhancing NAD+ salvage pathway reverts the toxicity of primary astrocytes expressing amyotrophic lateral sclerosislinked mutant superoxide dismutase 1 (SOD1). J Biol Chem 2016; 291:10836–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lloret A, Beal MF. PGC-1α, sirtuins and PARPs in Huntington’s disease and other neurodegenerative conditions: NAD+ to rule them all. Neurochem Res 2019; 44:2423–34 [DOI] [PubMed] [Google Scholar]
- 247.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, Zabari M, Brik RBZ, Kviatcovsky D, Zmora N, Cohen Y, Bar N, Levi I, Amar N, Mehlman T, Brandis A, Biton I, Kuperman Y, Tsoory M, Alfahel L, Harmelin A, Schwartz M, Israelson A, Arike L, Johansson MEV, Hansson GC, Gotkine M, Segal E, Elinav E. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 2019; 572:474–80 [DOI] [PubMed] [Google Scholar]
- 248.Diguet N, Trammell SAJ, Tannous C, Deloux R, Piquereau J, Mougenot N, Gouge A, Gressette M, Manoury B, Blanc J, Breton M, Decaux JF, Lavery GG, Baczkó I, Zoll J, Garnier A, Li Z, Brenner C, Mericskay M. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation 2018; 137:2256–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Matasic DS, Brenner C, London B. Emerging potential benefits of modulating NAD + metabolism in cardiovascular disease. Am J Physiol Heart Circ Physiol 2018; 314:H839–H52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Walker MA, Tian R. Raising NAD in heart failure: time to translate? Circulation 2018; 137:2274–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kane AE, Sinclair DA. Sirtuins and NAD+ in the development and treatment of metabolic and cardiovascular diseases. Circ Res 2018; 123:868–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Liu L, Wang P, Liu X, He D, Liang C, Yu Y. Exogenous NAD+ supplementation protects H9c2 cardiac myoblasts against hypoxia/reoxygenation injury via Sirt1-p53 pathway. Fundam Clin Pharmacol 2014; 28:180–89 [DOI] [PubMed] [Google Scholar]
- 253.Yamamoto T, Byun J, Zhai P, Ikeda Y, Oka S, Sadoshima J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS One 2014; 9:e98972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhang Y, Wang B, Fu X, Guan S, Han W, Zhang J, Gan Q, Fang W, Ying W, Qu X. Exogenous NAD+ administration significantly protects against myocardial ischemia/reperfusion injury in rat model. Am J Transl Res 2016; 8:3342–50 [PMC free article] [PubMed] [Google Scholar]
- 255.Porter GA, Urciuoli WR, Brookes PS, Nadtochiy SM. SIRT3 deficiency exacerbates ischemia-reperfusion injury: implication for aged hearts. Am J Physiol Heart Circ Physiol 2014; 306:H1602–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Méneur C, Permutt MA, Imai SI. Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab 2005; 2:105–17 [DOI] [PubMed] [Google Scholar]
- 257.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin S-J, Guarente L. Correction: Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol 2015; 13:e1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol 2006; 4:210–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 2008; 22:1753–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 2008; 10:179–206 [DOI] [PubMed] [Google Scholar]