SUMMARY
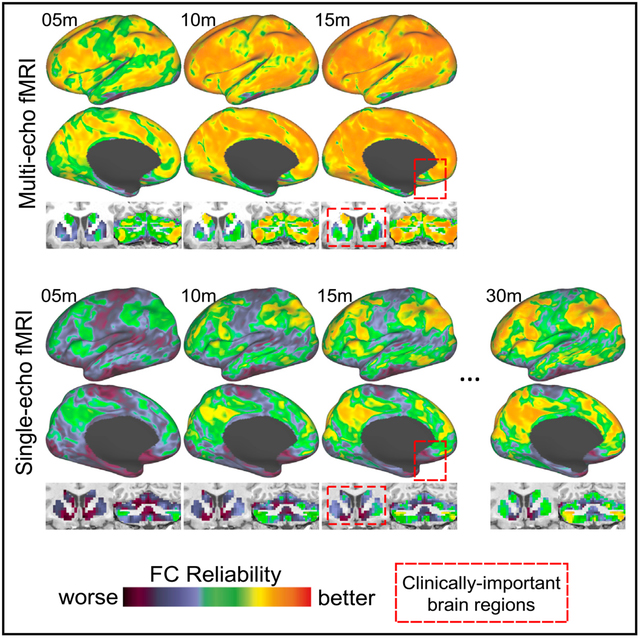
Resting-state functional magnetic resonance imaging (fMRI) is widely used in cognitive and clinical neuroscience, but long-duration scans are currently needed to reliably characterize individual differences in functional connectivity (FC) and brain network topology. In this report, we demonstrate that multi-echo fMRI can improve the reliability of FC-based measurements. In four densely sampled individual humans, just 10 min of multi-echo data yielded better test-retest reliability than 30 min of single-echo data in independent datasets. This effect is pronounced in clinically important brain regions, including the subgenual cingulate, basal ganglia, and cerebellum, and is linked to three biophysical signal mechanisms (thermal noise, regional variability in the rate of T2* decay, and S0-dependent artifacts) with spatially distinct influences. Together, these findings establish the potential utility of multi-echo fMRI for rapid precision mapping using experimentally and clinically tractable scan times and will facilitate longitudinal neuroimaging of clinical populations.
Graphical Abstract
In Brief
Lynch et al. demonstrate that the test-retest reliability of resting-state connectivity measurements can be improved using multi-echo fMRI. This effect is pronounced in clinically important brain regions and could help facilitate precision mapping of functional brain networks in healthy people and patient populations.
INTRODUCTION
The human brain is organized into functional networks that can be mapped non-invasively using resting-state functional magnetic resonance imaging (rsfMRI) (Smith et al., 2013; Snyder and Raichle, 2012), a technique that has evolved rapidly to become one of the most commonly used tools in cognitive and translational neuroscience. Pioneering studies have used rsfMRI to reveal the topology of functional brain networks (Biswal et al., 1995; Power et al., 2011; Yeo et al., 2011), and their associations with discrete cognitive processes and behaviors (Smith et al., 2009). Studies have also begun to define alterations within brain networks that are associated with psychosis (Anticevic et al., 2013; Baker et al., 2014; Karcher et al., 2019), depression (Downar et al., 2014; Drysdale et al., 2017; Fox et al., 2012; Oathes et al., 2015), autism (Di Martino et al., 2011, 2014; Hull et al., 2017; Padmanabhan et al., 2017), and other neuropsychiatric disease states (Castellanos et al., 2013; Menon, 2011; Xia et al., 2018). Promising clinical applications for rsfMRI (Fox and Greicius, 2010) include pre-operative mapping (Mitchell et al., 2013; Sair et al., 2016; Yahyavi-Firouz-Abadi et al., 2017), providing diagnostic and prognostic information (Drysdale et al., 2017; Dunlop et al., 2017; Fox et al., 2012), and mapping targets for neuromodulation therapies (Lynch et al., 2019; McMullen, 2018; Medaglia et al., 2020; Weigand et al., 2018). However, there are several obstacles to realizing its full clinical potential.
First, in the majority of rsfMRI studies, data acquired from many individuals are co-registered in a common atlas space, and functional brain networks and their relationships with clinical or behavioral variables are analyzed at the group level. But recent evidence indicates that this approach can obscure individual differences in the topology (size, shape, and spatial arrangement) of functional areas and networks in cortex (Gordon et al., 2017c; Laumann et al., 2015), in the cerebellum (Marek et al., 2018), and in subcortex (Greene et al., 2020), variability that could be both functionally meaningful and clinically useful (Kong et al., 2019; Seitzman et al., 2019; Wang et al., 2020). Efforts to develop neuroimaging tools for diagnosing neuropsychiatric disorders or predicting treatment response could benefit by accounting for these individual differences.
Second, resting-state functional connectivity (FC) measurements would ideally exhibit high reliability at the level of individual subjects (i.e., they would be similar across repeated assessments), especially in certain clinical contexts. However, obtaining reliable FC-based measurements throughout the brain at the individual subject level typically requires large amounts of per-subject rsfMRI data (on average, 45 min in cortex [Gordon et al., 2017c] and more than 90 min in the cerebellum and subcortex [Greene et al., 2020; Marek et al., 2018]), a practice referred to as “dense sampling” (Poldrack, 2017) or “precision functional mapping” (Gordon et al., 2017c; Gratton et al., 2020). It is thought that artifactual within-subject variation in FC is due primarily to two factors: (1) random sampling error (Laumann et al., 2015, 2017; Noble et al., 2017, 2019), in part because FC reflects low-frequency fluctuations in blood-oxygen-level-dependent (BOLD) signals, which may require long duration scans to assess accurately; and (2) the confounding influence of non-neurobiological artifacts, including those related to head motion (Power et al., 2012; Satterthwaite et al., 2012), and participant drowsiness (Laumann et al., 2017; Tagliazucchi and Laufs, 2014; Wang et al., 2017). Thus, long-duration scans may improve reliability by reducing sampling variability, but they also require that the subject remain still, awake, and alert throughout, which is a significant obstacle in both research and clinical populations that may not tolerate long (or multiple) scans. Low test-retest reliability may be less of a problem for some study designs, including those aimed at identifying effects at the group level in large samples. In contrast, low test-retest reliability is a fundamental obstacle to progress in other contexts, including intervention studies involving scans obtained before and after a treatment, biomarker discovery efforts, analyses of individual differences, and studying network plasticity at the individual level. Thus, there is a pressing need for techniques that can yield more reliable FC-based measurements from experimentally and clinically tractable scan times, as explained by recent commentaries on this topic (D’Esposito, 2019; Lynch and Liston, 2020; Satterthwaite et al., 2018).
Here, we tested whether multi-echo (ME) fMRI could enable more rapid and reliable mapping of functional brain networks in individuals, ameliorating the need for long scans. ME fMRI sequences (Posse, 2012; Posse et al., 1999) acquire multiple images at different echo times (i.e., at repeated intervals over tens of milliseconds) per volume, in contrast to standard single-echo (SE) sequences, which acquire a single image in the same period of time. Studies have previously demonstrated that ME fMRI increases BOLD signal sensitivity (Bhavsar et al., 2014; Posse et al., 1999) and can be used to identify and discard fMRI signals that cannot have originated from neurobiological activity (Kundu et al., 2012, 2014, 2015; Power et al., 2018). On first principles, either of these capabilities could enhance the reliability of FC at the individual subject level, but this possibility has not been tested directly.
To test this hypothesis, we acquired 6 h (24 × 14.5-min scans) of ME rsfMRI data from two adult participants over 9 months. We found that the reliability of FC measurements was enhanced nearly brain-wide in ME fMRI data, even when using limited amounts of per-subject data, compared to parallel analysis of SE data from the same participants and 14 densely sampled individuals from three SE datasets (n = 10 from the Midnight Scan Club [MSC] [Gordon et al., 2017c]; n = 3 from the Cast-induced Plasticity [CAST] study [Newbold et al., 2020]; n = 1 from the MyConnectome [MC] project [Laumann et al., 2015; Poldrack et al., 2015]). This effect was linked to at least three biophysical signal mechanisms with spatially distinct influences and replicated in two other less densely sampled participants.
RESULTS
Benchmarking the Reliability of FC Estimates in Three Independent SE Datasets
To begin, we quantified the reliability of FC at each point in the brains of 14 individuals from three independent datasets that were repeatedly scanned using SE sequences (MSC [Gordon et al., 2017c]: n = 10; 10 × 30-min scans acquired over 2 months; CAST [Newbold et al., 2020]: n = 3; 10 to 14 × 30-min scans acquired over consecutive days prior to casting of their upper right extremity; MC [Laumann et al., 2015; Poldrack et al., 2015]: n = 1; 84 × 10-min scans acquired over 18 months). A summary of the sequence parameters used to collect these datasets is provided in Table S1. The purpose of this first analysis was to obtain an independent benchmark against which our ME fMRI datasets could be evaluated.
We generated FC reliability maps for each subject using different amounts of data (ranging from 1 min to the full duration of scans in each dataset). In short, these maps index (using spatial correlation) how similar the FC of each point in the brain is when calculated using the specified amount of data from a single scan versus a large amount of independent data from the same participant. Values approaching 1 indicate better reliability. Reliability maps for example subjects from the MSC (MSC01 and MSC06 in Figure 1A), CAST (CAST01 in Figure 1B), and MC (MC01 in Figure 1C) datasets are shown in Figure 1. The two participants from the MSC dataset shown in Figure 1A were selected randomly and are ranked second (MSC06) and fourth (MSC01) of the n = 10 with respect to their average brain-wide reliability value. The FC reliability maps for all 14 subjects can be viewed in Video S1.
Figure 1. Obtaining Reliable Resting-State Functional Connectivity (FC) Estimates Can Require Large Quantities of Single-Echo (SE) fMRI Data Per Subject.
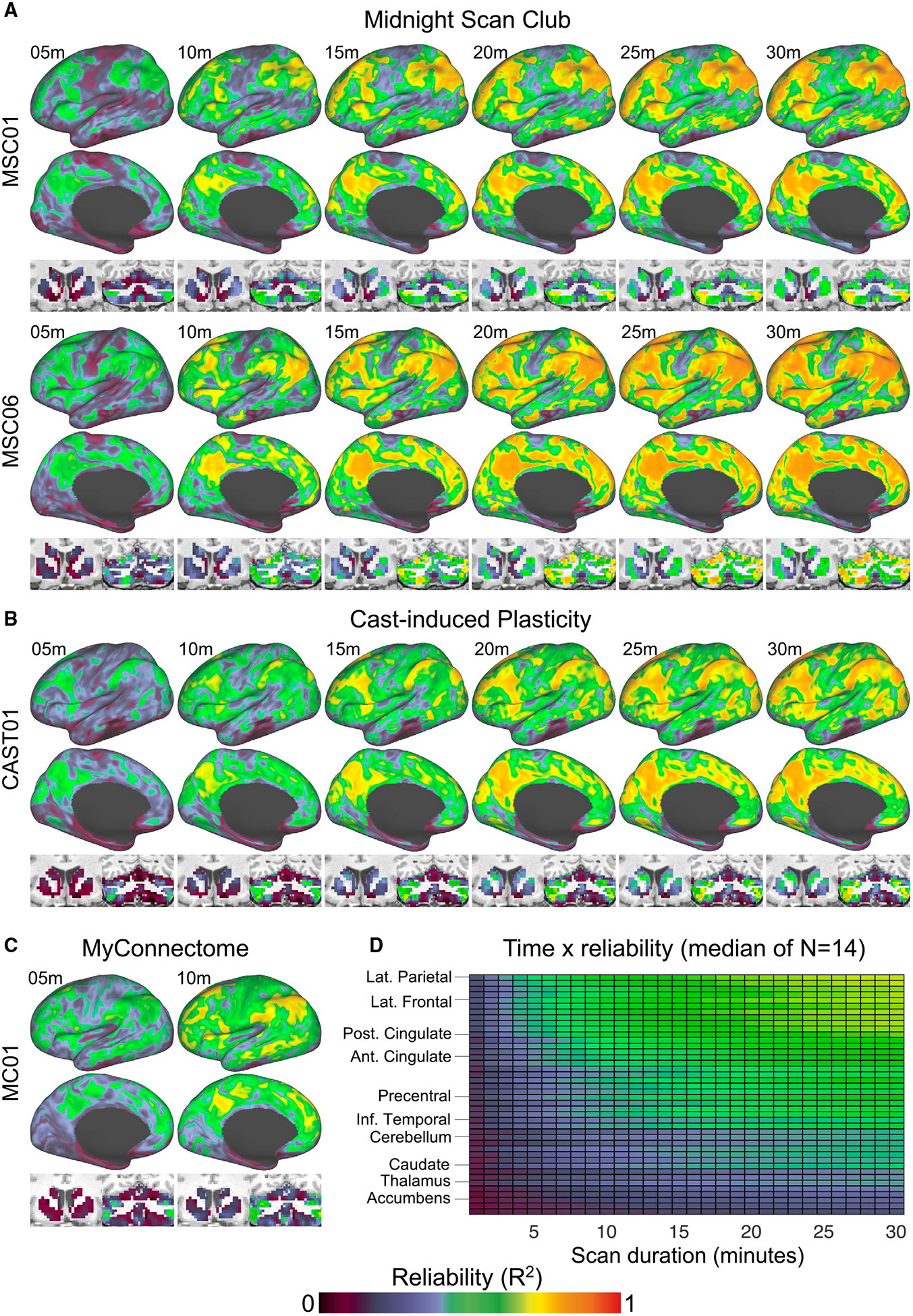
The reliability of resting-state FC was evaluated brain-wide in three independent SE rsfMRI datasets:
(A) The Midnight Scan Club (MSC) dataset (Gordon et al., 2017c), which consists of 10 individuals that underwent 10 × 30-min scans (three representative subjects are shown: MSC01, MSC04, and MSC06).
(B) The CAST dataset (Newbold et al., 2020), which consists of three individuals that underwent 10–14 × 30-min scans (one representative subject shown: CAST01),
(C) The MC dataset (Poldrack et al., 2015), which consists of a single individual that underwent 84 × 10-min scans.
(D) FC reliability maps index (using spatial correlation) how similar the FC of each point in the brain is when calculated using the specified amount of data from a single scan versus a large amount of independent data (all other scans available for that participant concatenated). Values approaching 1 indicate better reliability. The average reliability value within FreeSurfer defined cortical and subcortical regions of interest at each scan duration (the median value across the 14 subjects) is shown. Brain regions are ordered (in descending fashion) from most reliable to least reliable. m, minute.
This analysis yielded two results. First, reliability increased with scan duration, which is consistent with findings from other studies (Birn et al., 2013; Gordon et al., 2017c; Laumann et al., 2015), but relatively few points in the brain exhibited highly reliable FC. On average, only 28% (range: 0%–59%) of cortex exhibited high reliability (>0.7) in MSC and CAST subjects, even when using the full 30-min scan. Second, the rate that FC became reliable varied by brain region (Figure 1C). For example, FC was fairly reliable (>0.5) in lateral prefrontal, parietal cortex, posterior cingulate cortex, and other association areas with only 5–10 min of data. Other regions, including subcortical structures (basal ganglia, thalamus); the cerebellum; and somatomotor, inferior temporal, and subgenual cingulate cortex, exhibited relatively low reliability (<0.5) on average, even with 30 min of data. This finding is consistent with recent reports that more than 1 h of per-subject data may be necessary to obtain reliable FC estimates in subcortex (basal ganglia and thalamus; Greene et al., 2020) and in the cerebellum (Marek et al., 2018). We note that the brain regions exhibiting relatively low reliability—including the cerebellum (Schmahmann et al., 2009; Stoodley et al., 2016), subgenual cingulate (Fox et al., 2012; Mayberg et al., 2005; Pizzagalli, 2014), and basal ganglia (Dandekar et al., 2018; Morishita et al., 2014; Perlmutter and Mink, 2006)—are implicated in psychiatry and neurology, which underscores the potential impact of tools for enhancing the reliability of FC in these areas. In summary, this first analysis replicates previous reports of scan length influencing the reliability of FC, but it also highlights how FC reliability varies significantly by brain region, which has been reported elsewhere as well (Noble et al., 2017, 2019).
ME fMRI Improves BOLD Signal Sensitivity and Enables the Removal of Non-BOLD Artifacts
To test if ME fMRI could be used to obtain more reliable individual-specific FC-based measurements, we first acquired 6 h of ME fMRI data (24 × 14.5-min scans) from two individuals (referred to here as ME01 and ME02) over a period of 9 months.
In a SE fMRI sequence, like those used to collect the three independent datasets analyzed in Figure 1, images are acquired once per tissue excitation after a single fixed delay (“echo time”; usually 30–35 ms). A ME fMRI sequence acquires multiple images at different echo times spanning dozens of milliseconds, which affords two advantages. Below, we characterize these two advantages (readers are encouraged to see a review by Kundu et al. [2017] for a more complete discussion) before demonstrating how they additively enhance the reliability of FC and brain network topologies in the following sections.
The first advantage of ME fMRI is that images acquired at different echo times can be combined according to the rate of T2* decay at each voxel to create an “optimally combined” ME (OC-ME) time series (Posse et al., 1999). This is useful because BOLD contrast is optimal near the T2* of each voxel, and combining echoes in this manner can recover signal in brain regions with a short or long T2* compared to a single fixed echo. This effect is localized to specific brain regions. Consider, as an example, the subgenual cingulate (Figure 2A; purple), which has a short T2* (<20 ms in both ME01 and ME02; see T2* maps in Figure 2C) and exhibited relatively low reliability in Figure 1: in the OC-ME image, earlier echoes are weighted more heavily than later echoes (Figure 2B), which helps recover signal that would have otherwise been lost. Other brain regions, including portions of the basal ganglia and cerebellum, have a short T2* as well (see Figure 2C). Combining echoes also dampens thermal (random) noise (Caballero-Gaudes and Reynolds, 2017; Liu, 2016; Poser and Norris, 2009), which can be a large fraction of the recorded signal (Power, 2017). Because thermal noise is embedded in all fMRI signals, this effect will occur throughout the brain.
Figure 2. A Key Benefit of Multi-echo (ME) fMRI Is Improved BOLD Contrast and Reduced Signal Dropout after Echoes Are Combined.
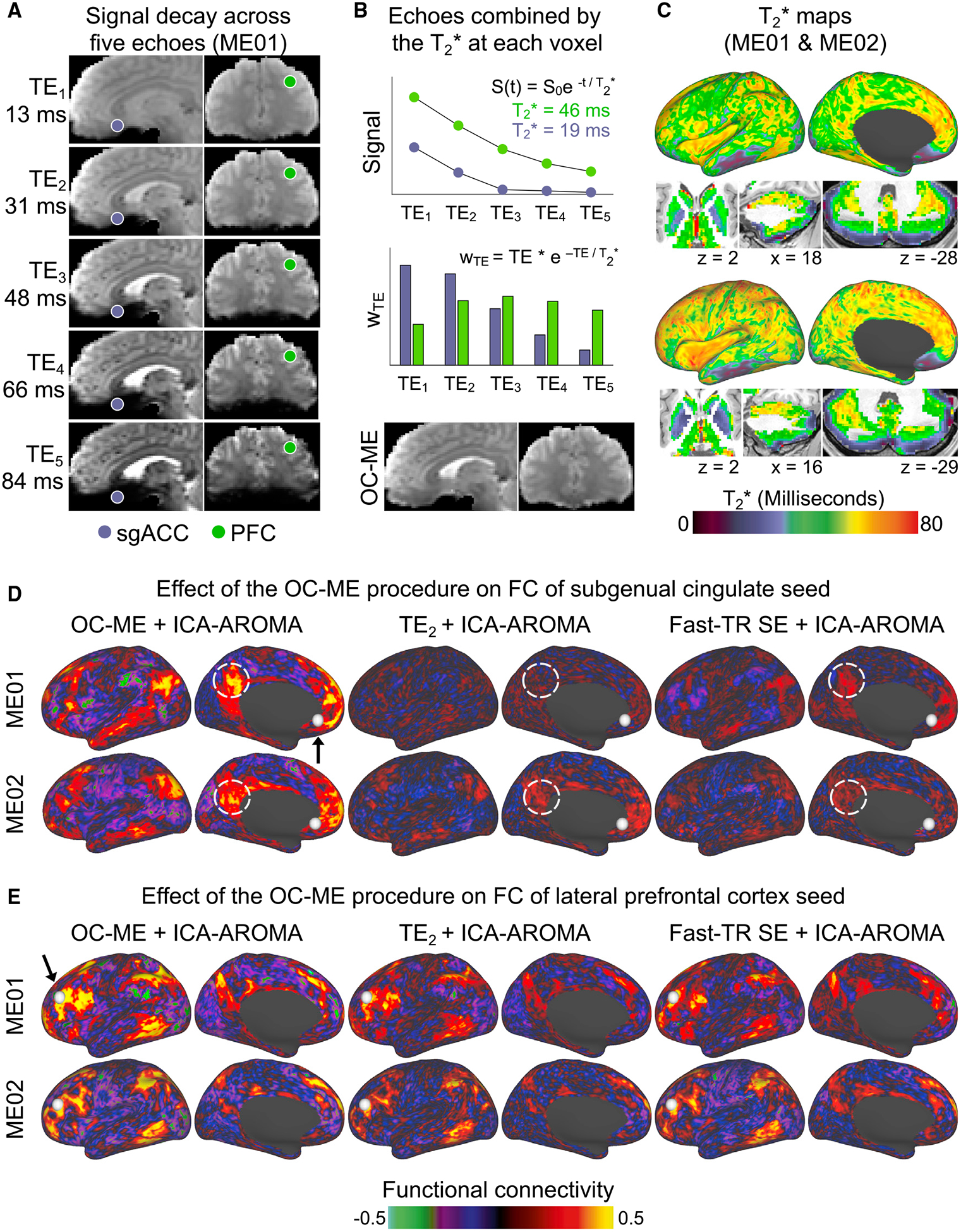
(A) A ME fMRI sequence acquires multiple images at different echo times (TE) spanning dozens of milliseconds.
(B) Signals decay more rapidly in brain regions with a short T2* value, such as the subgenual anterior cingulate cortex (sgACC; purple). Echoes are combined such that those near the estimated T2* at each voxel are weighted most heavily, yielding an “optimally combined” ME (OC-ME) time series with improved BOLD contrast, less signal dropout, and dampened thermal noise.
(C) WTE represents the optimal weight for each echo. T2* values are calculated at each point in the brains of ME01 and ME02. Differences in the FC of seed regions with different T2* values help to convey the region-specific effect of the OC-ME procedure. FC maps were created using OC-ME and two different kinds of SE data (the second echo of the ME scan and a separate fast-TR SE sequence with a faster sampling rate) that were collected from both ME01 and ME02. PFC, prefrontal cortex; TE2, second echo of the ME scan.
To demonstrate the effect that combining echoes has on FC, we generated FC maps using brain regions with relatively short (subgenual cingulate; Figure 2D) and long (lateral prefrontal cortex [PFC]; Figure 2E) T2* values as seeds. For both subjects ME01 and ME02, maps were first created using OC-ME data from a single representative ME scan denoised using an extensively validated and commonly used algorithm for cleaning SE fMRI data (ICA-AROMA; Ciric et al., 2017; Pruim et al., 2015). For comparison, the same maps were created using the second echo (TE2) of the ME scan and a separate connectome-style (faster sampling rate; TR = 800 ms versus TR = 1355 ms) SE acquisition, both of which were preprocessed and denoised in the same fashion as the OC-ME data. Two observations are notable. First, the seed placed in the short T2* subgenual cingulate exhibited stronger FC, with default mode network brain regions in OC-ME data, whereas this pattern of FC was absent in the TE2 and fast-TR SE comparison datasets (see dashed white circles in Figure 2D indicating weaker FC with the posterior cingulate). Second, as expected, differences among the OC-ME, TE2, and SE datasets were less pronounced for the seed in lateral PFC (Figure 2E), which has a longer T2* value and thus is less susceptible to signal dropout, but FC maps still appeared as if they were superimposed on static (thermal) noise. Collectively, this analysis demonstrates how ME fMRI improves BOLD signal sensitivity—particularly for short T2* brain regions—by accounting for regional differences in the rate T2* decay (Bandettini et al., 1994) and dampening thermal noise.
The second advantage of ME fMRI is that how signals decay across echoes can be used during denoising to identify and remove signals that cannot have originated from neurobiological activity, including those related to head motion, heating or instability of MRI hardware, and cerebrovascular pulsatility (Kundu et al., 2017). Discarding these kinds of artifacts is desirable, in part because they can produce spurious FC estimates (Power et al., 2012, 2015), but difficult to do in SE datasets because there is no ground truth for determining if a signal is indeed artifactual. ME fMRI provides this ground truth. For example, spatially structured signals in the OC-ME time series identified from a spatial ICA can be classified as neurobiological (and retained) or non-neurobiological (and discarded) on the basis of their signal-decay properties, an approach called ME-ICA (Kundu et al., 2012). Here, we confirmed that ME-ICA can be used to separate neurobiological and non-neurobiological signals using an instructed deep breathing task (see Figure S1 for the rationale behind using the event-related approach described in Power et al. [2020] to establish appropriate signal separation).
In summary, we reasoned that ME fMRI has the potential to improve the reliability of FC by at least two mechanisms: (1) by enhancing BOLD signal sensitivity through the weighted combination of echoes and (2) by removing non-neurobiological artifacts through a signal-decay-based denoising technique called ME-ICA.
Brain-wide Improvements in Connectivity Reliability in ME fMRI Data
Next, we evaluated the reliability of FC derived from OC-ME (Posse et al., 1999) data denoised using ME-ICA (Kundu et al., 2012) in ME01 and ME02, leveraging both advantages of ME fMRI detailed in the previous section. Reliability maps were created in the same way as was done for Figure 1. Consistent with our hypothesis, OC-ME data denoised using ME-ICA (OC-ME + ME-ICA) yielded reliable FC at nearly every point in the brain (Figure 3A; top row). Over 75% of cortex exhibited high (>0.7) reliability (82% in ME01 and 75% in ME02), compared to, on average, 28% (range: 0%–59%) in independent SE participants with twice as much data. Time × reliability curves (Figure 3B) show the average reliability value (calculated separately in cortex, subcortical structures, and cerebellum) given different scan durations. Scan duration is calculated before removal of high motion volumes. Curves from the three independent SE datasets are shown as comparators. Plotting reliability values relative to the amount of data retained after motion-censoring yielded a set of similar curves (with the exception of MSC participants exhibiting especially high levels of head motion; e.g., sub-MSC08) that can be viewed in Figure S2.
Figure 3. The Optimal Combination and ME Denoising Procedures Improve the Reliability of Resting-State FC Measurements in Two Densely Sampled Individuals.
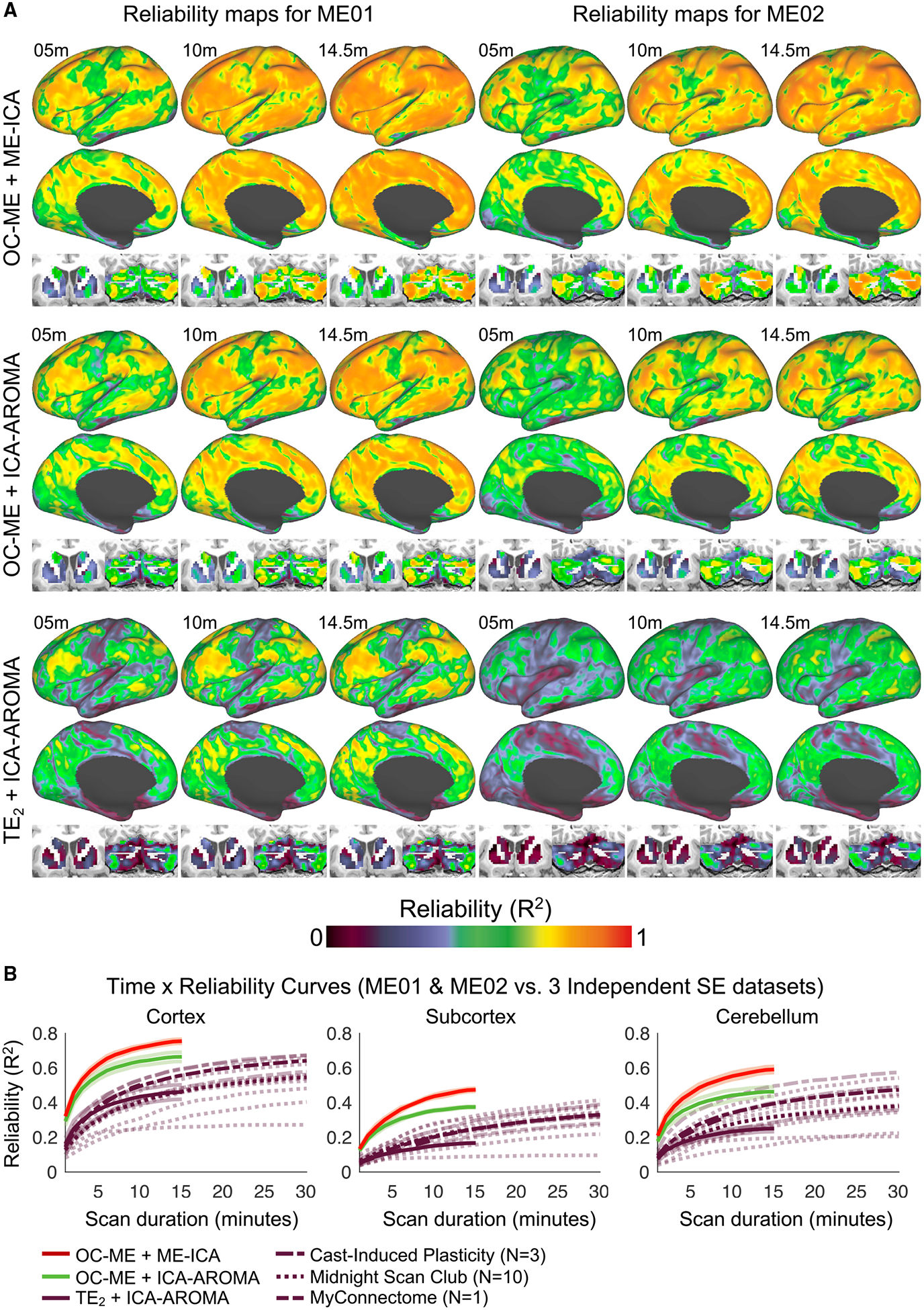
The reliability of FC estimates in ME01 and ME02 after repeated imaging using a ME fMRI sequence (6 h total; 24 × 14.5-min scans acquired over a 9-month period).
(A) Reliability maps were calculated using three different denoising strategies, leveraging both (OC-ME + ME-ICA), one (OC-ME + ICA-AROMA), or no (TE2 + ICA-AROMA) benefits of ME fMRI.
(B) Time × reliability curves show the average reliability obtained in gray matter, subcortical structures, and the cerebellum given different scan durations. Curves from the independent SE MC and MSC datasets are provided as comparators. Transparent curves represent individual subjects. Solid lines represent the median curve within datasets. Note that the purple lines representing the different independent SE datasets can be distinguished by their unique dash spacing patterns. m, minute.
One-sample t tests revealed that 10 min of OC-ME + ME-ICA data yielded FC estimates that were more reliable than those derived from 3 times as much independent SE data in cortex (t(12) = 5.52, p < 0.001, Cohen’s d = 1.46), in the cerebellum (t(12) = 4.89, p = 0.001, Cohen’s d = 1.38), and in subcortical structures (t(13) = 5.30, p = 0.001, Cohen’s d = 1.10). These findings, which were replicated in two other less densely sampled individuals (“ME03” and “ME04”; see Figure S3), indicate that FC reliability can be enhanced in OC-ME data when signal-decay-based denoising is used.
To better understand the unique contributions of the OC-ME procedure and signal-decay-based denoising (ME-ICA) to the enhanced reliability of FC observed in Figure 3A, we created reliability maps for OC-ME and TE2 data denoised using ICA-AROMA (Ciric et al., 2017; Pruim et al., 2015), which, unlike ME-ICA, does not leverage any signal-decay information. Two findings are notable. First, reliability was enhanced in OC-ME data denoised with ME-ICA, compared to ICA-AROMA, particularly in the cerebellum and basal ganglia (Figure 3A; top row versus middle row). One interpretation is that either the high density of vasculature (Vigneau-Roy et al., 2014) or close proximity to ventricles (Caballero-Gaudes and Reynolds, 2017) renders these areas more susceptible to certain kinds of physiological artifacts (e.g., those related to cardiac pulsation) that are discarded by ME-ICA, but not ICA-AROMA. Second, the FC reliability maps (bottom row in Figure 3A) and time × reliability curves associated with TE2 + ICA-AROMA data were similar to those in the three independent SE datasets (Figure 1). Reliability maps for all study participants can be viewed in Video S2.
A limitation of using TE2 as a stand-in for SE data is that contemporary SE sequences can acquire images at faster rates than ME sequences. To help address this concern, ME01 underwent 6 h of additional scanning (24 × 14.5-min scans) with a fast-TR (800 ms) SE sequence. Direct comparison of the FC reliability map derived from fast-TR SE + ICA-AROMA data to those shown in Figure 3A indicated better FC reliability brain-wide in OC-ME data when ME-ICA was used (Figures 4A and 4B). The fast-TR SE + ICA-AROMA time × reliability curve (dashed purple line in Figure 4C) resembled those in the MC and CAST datasets (see Figure 3B), indicating that faster sampling rates may be beneficial.
Figure 4. The Level of Reliability Obtained Using a ME Sequence Is Greater Than a SE Sequence with a Fast Sampling Rate.
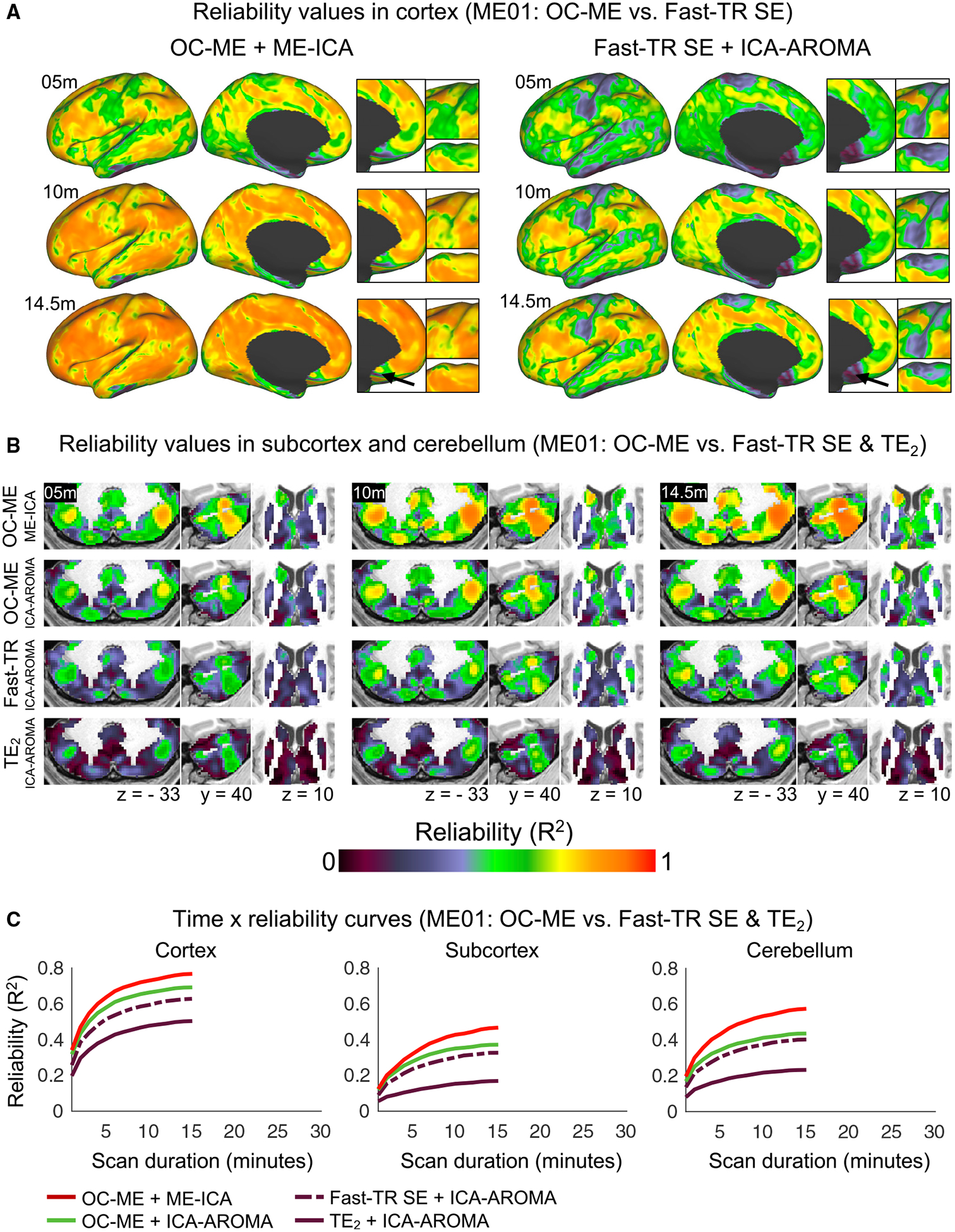
FC reliability maps derived from ME and fast-TR (800 ms) SE data acquired from the same individual (sub-ME01).
(A) Insets highlight regions of cortex where differences between the two sets of reliability maps were most pronounced.
(B) Reliability values in the cerebellum and in subcortex.
(C) Time × reliability curves show the average reliability value (calculated separately in cortex, subcortical structures, and cerebellum) given different amounts and kinds of data acquired from sub-ME01.
We conducted a follow-up analysis to further understand the biophysical signal mechanisms underlying better FC reliability in OC-ME data. Specifically, we attempted to separate the effect of recovering short and long T2* signals and the incidental dampening of thermal noise by shuffling the “optimal” weights assigned to each voxel. The rationale behind this analysis is that combining echoes in any manner can suppress thermal noise because it has no temporal or spatial structure (Liu, 2016; Power, 2017), but recovering short or long T2* signals requires appropriate weighting of early and late echoes in particular. Surprisingly, the reliability maps derived from OC-ME and “non-OC” data were nearly identical, with the exception of vertices with a very short T2* (<20 ms), which exhibited better reliability in OC-ME data than would be expected by chance after 1,000 random rotations of the T2* map on a spherical expansion of each subject’s cortical surface (all p values = 0.001, all Z scores > 6.53). This finding indicates that combining echoes according to the T2* at each voxel improves the reliability of FC primarily via the incidental dampening of thermal noise (a brain-wide effect), but also by enhancing BOLD sensitivity in brain regions with a very short T2* (another region-specific effect).
More Reliable Mapping of Individual-Specific Functional Topology in ME Data
The analyses performed to this point in our report have focused on the reliability of correlations in BOLD fMRI signals, which are the basis for most rsfMRI studies. However, there is increasing interest in higher-level descriptions of an individual’s functional brain organization, including the topology (size, shape, and spatial arrangement) of functional areas and brain networks, the latter of which is typically calculated using only the strongest of these correlations (Gordon et al., 2016, 2017c; Laumann et al., 2015; Power et al., 2011). Thus, we next tested whether ME fMRI could be used to obtain more reliable individual-specific network parcellations. We identified networks brain-wide in each individual using a widely used community detection algorithm, termed InfoMap (Rosvall and Bergstrom, 2008), and the precision mapping procedures developed collectively by Gordon et al., (2017c), Greene et al. (2020), and Marek et al. (2018). To aid in the interpretation of the maps produced by this routine, Figure 5A shows the networks identified in ME01 when using all 6 h of OC-ME + ME-ICA scans (concatenated). A seed (the gray sphere highlighted by a black arrow) placed in a fronto-parietal network patch highlights how seed FC is strongest within network. Brain networks were mapped in this manner in each of the 18 densely sampled individuals (the 4 individuals scanned using ME sequences + 14 individuals from independent SE datasets) using data from single scans and all other available scans concatenated (the latter of which serves as a kind of ground truth). Reliability was quantified using the adjusted Rand coefficient, which represents the fraction of points in the brain identified as belonging to the same network in single-scan and ground truth data (with values approaching 1 indicating better agreement after adjusting for chance).
Figure 5. Functional Brain Network Topology Is More Reliable in Individuals Scanned Using a ME Sequence.
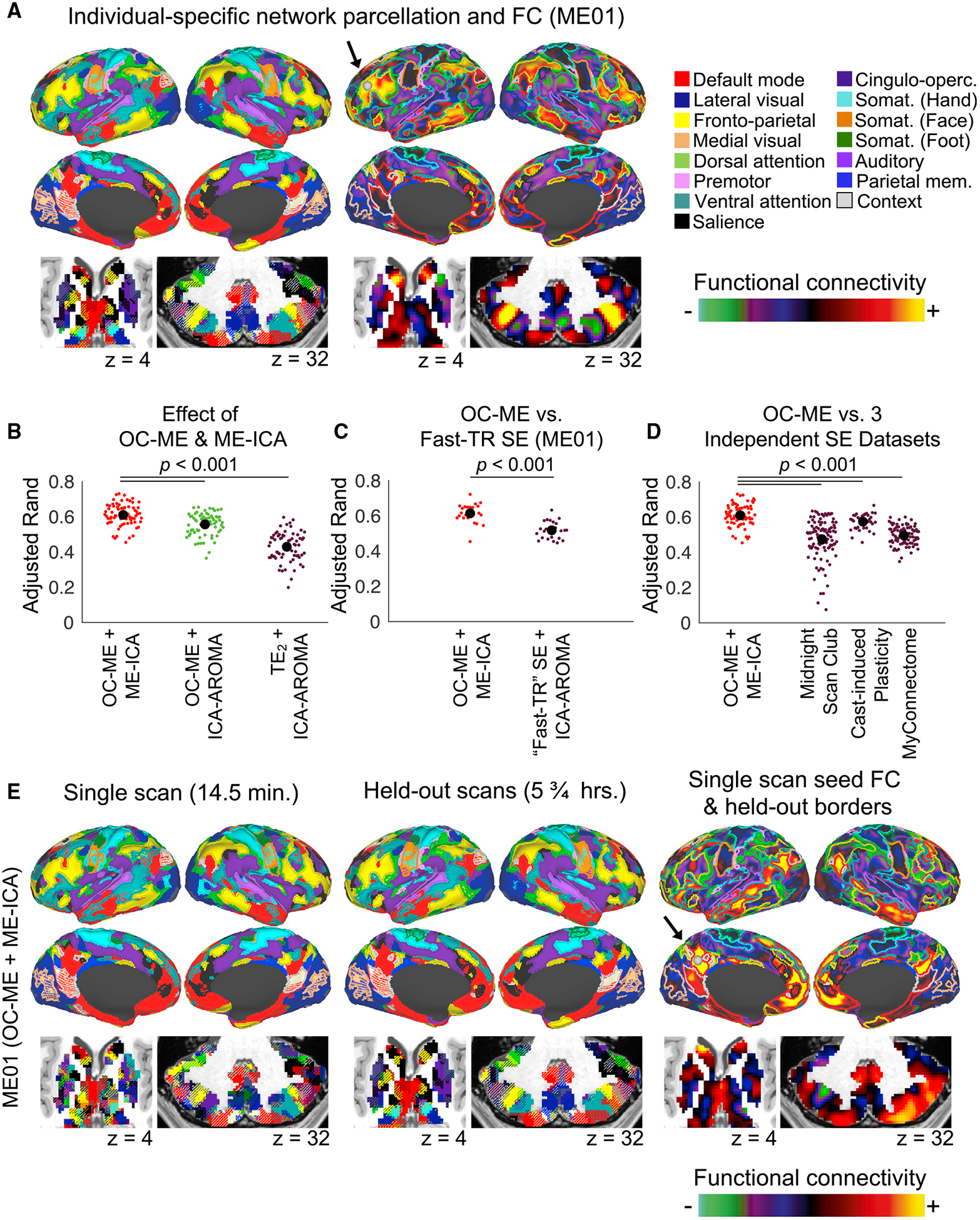
(A) Functional brain networks identified brain-wide in ME01 using a precision mapping routine and all 6 h of OC-ME + ME-ICA data. A seed (gray sphere) placed in a patch of fronto-parietal control network (yellow) in the left lateral PFC of ME01 highlights how FC is largely constrained within-network. The effect of the OC-ME and ME-ICA procedures on the reliability of individual-specific functional brain network topology was evaluated using a mixed-effects ANOVA model. The OC-ME procedure and ME-ICA denoising algorithm additively enhanced the reliability (indexed using the adjusted Rand coefficient comparing the similarity of network partitions defined using single-scan data and all other scans concatenated) of functional topology in the four densely sampled individuals.
(B and C) Comparison of adjusted Rand coefficients from OC-ME + ME-ICA data to those derived from fast-TR SE data (B) collected from the same study participant (ME01) and the three independent SE datasets (C).
(D) Functional brain networks mapped using data from a single OC-ME + ME-ICA scan and all other OC-ME + ME-ICA scans (concatenated; 5.75 h total) for sub-ME01. The resting-state FC (calculated using the single-scan data) of a seed (gray sphere; highlighted using an arrow) placed in a default mode network patch is constrained within the borders of this network defined using held-out data, indicating high reliability.
p denotes p value.
There was a significant main effect of the OC-ME and ME-ICA procedures (Figure 5B; F(2,187) = 161.65, p < 0.001), such that networks defined using OC-ME + ME-ICA data (red dots in Figure 5B) were more reliable than those defined in OC-ME (green dots in Figure 5B; t(142) = 5.42, p < 0.001, Cohen’s d = 0.82) and TE2 (purple dots in Figure 5B; t(142) = 15.4, p < 0.001, Cohen’s d = 1.57) data denoised using ICA-AROMA. Next, we compared the OC-ME and fast-TR SE data acquired from ME01 (the only participant scanned using both ME and fast-TR SE sequences). OC-ME + ME-ICA data yielded network maps that were more reliable than those defined using the SE acquisition with a faster sampling rate (Figure 5C; t(46) = 6.44, p < 0.001, Cohen’s d = 1.36). Independent SE datasets are shown for comparison in Figure 5D. Brain network topologies defined using OC-ME + ME-ICA data were significantly more reliable than those in the MSC (Figure 5C; t(170) = 9.21, p < 0.001, Cohen’s d = 1.16) and CAST (Figure 5C; t(170) = 3.35, p = 0.001, Cohen’s d = 0.65) participants, despite the 2-times difference in scan length. An example of functional brain networks defined using data from a single 14.5 OC-ME + ME-ICA scan and data from all other scans (5.75 h worth of scanning) is shown in Figure 5E. A seed (the gray sphere highlighted by a black arrow) placed in the posterior cingulate reveals that the FC (calculated using data from the single scan) of the seed is strongest within borders of the default mode network (borders defined using the large amount of independent data). Similar visualizations were created using a variety of cortical and subcortical seeds from subsets (5 to 14.5 min; in 5-min steps) of data from three randomly selected OC-ME, fast-TR SE, and TE2 scans (see Figures S5–S9). Collectively, these findings indicate that more reliable descriptions of individual-specific functional brain networks can be obtained from relatively small quantities of OC-ME data when signal-decay denoising is used.
DISCUSSION
In this report, we found enhanced reliability of FC and functional brain network topology in four densely sampled individuals after echoes were combined (OC-ME) and non-neurobiological artifacts were removed using a signal-decay-based denoising approach (ME-ICA). FC reliability increased rapidly with scan duration, such that less OC-ME data was necessary to achieve the same level of reliability in three independent SE datasets (n = 14) or in fast-TR SE data collected from the same individual (see time × reliability curves in Figures 3B and 4C). Several clinically important brain regions that exhibited relatively low FC reliability in SE data were more reliable in OC-ME data, especially when ME-ICA was used. The increased reliability of FC in OC-ME data was associated with more reliable descriptions of individual-specific functional brain networks (see Figure 5). Collectively, these findings indicate that ME fMRI is well suited for rapid precision mapping of functional networks at the individual level and for tracking changes within individuals over time. These enhancements, in turn, could help facilitate clinical neuroimaging research, particularly longitudinal studies of episodic forms of psychiatric illness, and for elucidating the neurobiological basis of individual differences in cognition and behavior.
The benefits of ME sequences have been studied for over two decades (Posse, 2012), beginning with seminal work by Posse et al. (1999) showing that combining echoes according to the T2* at each voxel increases BOLD sensitivity. For the most part, however, ME fMRI is not widely used—fewer than 1% of the more than 12,000 rsfMRI studies published in the last 10 years used a ME sequence. There are likely multiple reasons for this. For example, there may be the perception that the benefits of ME fMRI are relatively modest or that it is more important to optimize spatiotemporal resolution (i.e., smaller voxels and faster sampling rates). ME fMRI has historically entailed a compromise in either full-brain coverage or spatiotemporal resolution, but recent technological advances (high-density head coils, parallel imaging [Schmiedeskamp et al., 2010], and multi-band acceleration [Xu et al., 2013]) have made this trade-off less significant. Recent studies have leveraged these advances and found that how fMRI signals decay across echoes can be used to infer if they originate from neurobiological activity or not (Kundu et al., 2012, 2017), an approach that has been used to remove head-motion-related artifacts in rsfMRI scans (Power et al., 2018) and enhance statistical power by as much as 149% in task-based fMRI experiments (Lombardo et al., 2016). By comparing FC reliability maps derived from three versions of our ME fMRI datasets with key preprocessing and denoising procedures omitted or included (see Figure 3), our investigation found that ME fMRI can also be used to enhance FC reliability. It links this effect to three biophysical signal mechanisms with spatially distinct influences: BOLD signal sensitivity, which can be broken down further into thermal noise (a brain-wide effect) and regional differences in the rate of T2* decay (a region-specific effect); and S0-dependent artifacts (another region-specific effect). The extent to which ME-ICA improves FC reliability more than other denoising strategies that do not leverage signal-decay information may depend in part on how contaminated the data are by S0-dependent artifacts.
The significance of our investigation is 2-fold. First, our data indicate that ME fMRI could be used to obtain more stable individual-specific FC-based measurements with less data, which has implications for studies of individual differences in functional brain organization in both health and disease (Braga and Buckner, 2017; Braga et al., 2019; Finn et al., 2015; Gordon et al., 2017a, 2017b, 2017c; Kong et al., 2019; Laumann et al., 2015; Marek et al., 2018; Seitzman et al., 2019). Second, ME fMRI could be especially useful in clinical contexts, where the amount of per-subject data needed to obtain accurate assessments of FC and functional brain network topology is a significant bottleneck (O’Connor and Zeffiro, 2019). Importantly, some brain regions (e.g., lateral prefrontal, posterior parietal, and a subset of midline cortical areas) yielded generally reliable FC measurements (reliability values ranging from 0.5 to 0.8). However, other regions, including those implicated in neurologic and psychiatric illness (subgenual cingulate [Fox et al., 2012; Mayberg et al., 2005; Pizzagalli, 2014)] and cerebellum [Shakiba, 2014]) exhibited relatively low reliability in both our SE datasets and in three independent datasets (Figure 1). This was due in part to the fact that these brain regions are susceptible to rapid signal dropout and certain kinds of S0-dependent artifacts.
The improved test-retest reliability afforded by ME fMRI is not only statistically significant, but also scientifically useful. Low test-retest reliability of FC at the individual level is a fundamental obstacle to numerous within-subject study designs, especially in clinical neuroimaging. Consider, for example, that if an investigator wanted to understand the effect of an intervention on FC in individual patients, it is desirable to minimize artifactual variation in FC within subject over time (which can be driven by various kinds of S0-dependent artifacts, such as head movement levels or instabilities of MRI hardware, and removed by ME denoising), so that neurobiologically meaningful changes in FC associated with the intervention can be modeled. In other words, the enhanced reliability afforded by ME fMRI is a reduction in measurement error, which in turn should improve the power to detect brain-behavior effects in both within-subject and cross-sectional studies. ME fMRI could, therefore, help facilitate therapeutic intervention studies, biomarker discovery efforts, and longitudinal studies of functional network plasticity at the individual level.
Several aspects of this investigation warrant careful consideration. First, our conclusions are based on data acquired from four densely sampled individuals. Performing analyses separately in each individual enabled a 4-fold replication of all our major findings. However, smaller samples can have limited generality. Mitigating this concern to some degree, we found that the reliability maps and time × reliability curves derived from the second echo in all four ME subjects were comparable to those from 14 individuals from three independent SE datasets. It is also worth noting that head motion was relatively low for some of our study participants (see Figure S4), suggesting that they represent a best-case scenario with respect to their ability to remain still and awake during scanning. Second, there is no empirically tested best set of parameters for a ME fMRI scan, and systematically testing different combinations of parameters was outside of the scope of the present study. ME denoising itself is an active area of research and development (Caballero-Gaudes et al., 2019; Kundu et al., 2012), and the algorithms used here will likely be improved upon in the near future by other investigators. Third, we are not advocating for ME fMRI scans as a panacea for the challenges inherent to obtaining accurate descriptions of an individual’s functional brain organization. If time and funds permit, collecting more data will improve the reliability of FC measurements, regardless of the sequence or denoising strategy employed. However, an important caveat is that subjects are prone to drowsiness during long rsfMRI scans (Tagliazucchi and Laufs, 2014), and fluctuations in arousal or sleep state can reduce the stability of BOLD fMRI correlations (Laumann et al., 2017; Wang et al., 2017), which further underscores the attractiveness of obtaining reliable measurements from shorter scans, if possible.
Finally, it is plain that the rate FC estimates become reliable differs across the 14 individuals in the three independent SE datasets (see transparent purple lines representing individual participants in Figure 3B) and in our 4-participant ME dataset. What factors, other than baseline levels of head motion and other S0-dependent artifacts, could account for this variability? Our central hypothesis is that other factors including but not limited to certain kinds of breathing patterns (Lynch et al., 2020) (the effects of which cannot be removed using ME-ICA alone; see Figure S1), participant drowsiness (Laumann et al., 2017), and brain volume (which should affect signal-to-noise by virtue of the physical distance between the brain and the receive coil) all contribute to individual differences in FC reliability. A larger sample of densely sampled individuals is necessary to fully address this issue.
Obtaining reliable FC-based measurements of individuals has, to date, required collecting large quantities of per-subject data (D’Esposito, 2019; Poldrack, 2017; Satterthwaite et al., 2018), which may not always be feasible for the clinical applications that have been proposed for rsfMRI, including pre-operative mapping (Mitchell et al., 2013), gathering diagnostic and prognostic information (Drysdale et al., 2017; Dunlop et al., 2017; Fox et al., 2012), and mapping personalized targets for neuromodulation (McMullen, 2018; Medaglia et al., 2020; Weigand et al., 2018). Here, we demonstrate that ME fMRI enables more rapid and reliable FC-based measurements in individual subjects by increasing BOLD sensitivity and discarding fMRI signals that do not originate from neurobiological activity. By enabling more reliable measurements from shorter scan times, ME fMRI data may be especially useful for precision functional mapping of individual brains in clinical populations.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Charles J. Lynch (cjl2007@med.cornell.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
There are restrictions to the availability of neuroimaging data because the study participants did not consent to data sharing. Code for preprocessing multi-echo fMRI data is maintained in an online repository (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Participants and study design
The study sample consisted of four healthy adult participants. The first two participants (“ME01”; 29 year old male, “ME02”; 38 year old male) in this investigation underwent 24 × 14.5 min multi-echo fMRI scans. Both underwent additional scanning using a separate fast-TR single-echo sequence (SE01: 24 × 14.5 min scans, SE02: 2 × 14.5 min scans). Two other participants (“ME03”; 24 year old male and “ME04”; 31 year old male) underwent 12 × 14.5 min multi-echo fMRI scans. Participants ME01, ME02, and ME04 were study authors C.J.L, J.D.P, and M.A.S., respectively.
Three independent single-echo datasets were included in this investigation. Each of these datasets were analyzed “as is” (no additional preprocessing or denoising was performed). A brief summary of the sequence parameters used to collect each dataset is provided in the Supplementary Information (Table S1).
The Midnight Scan Club dataset (Gordon et al., 2017c) was obtained from OpenNeuro.org (https://openneuro.org/datasets/ds000224/versions/1.0.1). This dataset includes 5 h of (preprocessed, denoised, and surface registered) single-echo resting-state fMRI data (10 × 30 min scans acquired over two months) collected from ten participants aged 24–34 years (mean age = 29.1 ± 3.3 years, 5F/5M).
The Cast-induced Plasticity dataset (Newbold et al., 2020) was obtained from OpenfMRI.org (https://openneuro.org/datasets/ds002766/versions/3.0.0). The portion of this dataset used here included 5 to 7 h of (preprocessed, denoised, and surface registered) single-echo resting-state fMRI data (10 to 14 × 30 min scans acquired over consecutive days prior to the casting of their dominant upper extremity) collected from three healthy participants (mean age = 29 ± 5.29 years, 1F/2M). Two of these participants also participated in the Midnight Scan Club experiment (CAST01 is MSC02 and CAST02 is MSC06).
The MyConnectome dataset was obtained from the project’s website (http://myconnectome.org/wp/data-sharing/). This dataset included 14 h of (preprocessed, denoised, and surface registered) single-echo resting-state fMRI data (84 × 10 min scans acquired over 18 months) collected from a 45 year-old male participant.
METHOD DETAILS
MRI image acquisition
Data were acquired on a Siemens Magnetom Prisma 3T scanner at the Citigroup Biomedical Imaging Center of Weill Cornell’s medical campus using a Siemens 32-channel head coil. Multi-echo, multi-band resting-state fMRI scans were collected using a T2*-weighted echo-planar sequence covering the full brain (TR: 1355 ms; TE1: 13.40 ms, TE2: 31.11 ms, TE3: 48.82 ms, TE4: 66.53 ms, and TE5: 84.24 ms; FOV: 216 mm; flip angle: 68°; 2.4mm isotropic; 72 slices; AP phase encoding direction; in-plane acceleration factor: 2; and multi-band acceleration factor: 6) with 640 volumes acquired per scan for a total acquisition time of 14 min and 27 s. This sequence was generously provided by the Center for Magnetic Resonance Research (CMRR) at the University of Minnesota. Single-echo, multi-band resting-state fMRI scans (referred to in the text as the “fast-TR” single-echo sequence) were collected from participants ME01 and ME02 using T2*-weighted echo-planar sequences covering the full brain (TR: 800 ms; TE: 30 ms; FOV: 216 mm; flip angle: 49°; 2.4mm isotropic; 72 slices; AP phase encoding direction; and multi-band acceleration factor: 6) with 1084 volumes acquired per scan for a total acquisition time of 14 min and 27 s. A pair of spin echo EPI images with opposite phase encoding directions (AP and PA) but identical geometrical parameters and echo spacing were acquired to correct spatial distortions. High-resolution (MPRAGE) T1-weighted image (TR: 2400 ms; TE: 2.28 ms; FOV: 256; flip angle: 90°, and 208 sagittal slices with a 0.8 mm thickness) and T2-weighted anatomical images (TR: 3200 ms; TE: 563 ms; FOV: 256; flip angle: 8°, and 208 sagittal slices with a 0.8 mm thickness) were acquired. Custom headcases were obtained from Caseforge (https://caseforge.co) for each subject to improve comfort and minimize head motion during scanning (Power et al., 2019b).
Cortical surface generation
The average T1- and T2-weighted images were cropped to a smaller field of view (170mm in z plane), co-registered using FSL’s epi_reg tool (via a boundary-based cost function with 6 DOF), and corrected for intensity inhomogeneities (Glasser and Van Essen, 2011). The T1- and T2-weighted images were co-registered to an MNI atlas (hereafter referred to as “ACPC” alignment) using a rigid 6 DOF FLIRT transformation. Cortical surfaces were generated using Freesurfer’s “recon-all.v6.hires” pipeline. Pial surface placement was refined using the co-registered T2-weighted image by specifying the “-T2pial” option. Midthickness surfaces were obtained by averaging the pial and white surfaces. Fsaverage-registered left and right hemisphere surfaces (pial, white, and midthickness) were brought into register with each other in fs_LR space (Van Essen et al., 2012) and resampled to the computationally tractable resolution of 32k vertices using Connectome Workbench command line utilities.
Multi-echo fMRI preprocessing
Preprocessing of multi-echo data minimized spatial interpolation and volumetric smoothing while preserving the alignment of echoes. The single-band reference (SBR) images (five total; one per echo) for each scan were averaged. The resultant average SBR images were aligned, averaged, co-registered to the ACPC aligned T1-weighted anatomical image, and simultaneously corrected for spatial distortions using FSL’s topup and epi_reg programs. Freesurfer’s bbregister algorithm (Greve and Fischl, 2009) was used to refine this co-registration. For each scan, echoes were combined at each time point and a unique 6 DOF registration (one per volume) to the average SBR image was estimated using FSL’s MCFLIRT tool (Jenkinson et al., 2002), using a 4-stage (sinc) optimization. All of these steps (co-registration to the average SBR image, ACPC alignment, and correcting for spatial distortions) were concatenated using FSL’s convertwarp tool and applied as a single spline warp to individual volumes of each echo after correcting for slice time differences using FSL’s slicetimer program. All denoising was performed on these preprocessed, ACPC-aligned images.
Multi-echo fMRI denoising
Multi-echo ICA (ME-ICA; Dupre et al., 2020; Kundu et al., 2012, 2013) denoising designed to isolate spatially structured T2*- (neurobiological; “BOLD-like”) and S0-dependent (non-neurobiological; “not BOLD-like”) signals was performed using a modified version of the “tedana.py” workflow (https://tedana.readthedocs.io/en/latest/). In short, the preprocessed, ACPC-aligned echoes were first combined according to the average rate of T2* decay at each voxel across all time points by fitting the monoexponential decay, S(t) = S0e−t / T2*, using the “nlinfit.m” function in MATLAB with least-squares optimization and the initial coefficient values obtained from a linear model fit to the log of the data. From these T2* values, an optimally combined multi-echo (OC-ME) time-series was obtained by combining echoes using a weighted average (WTE = TE * e −TE/ T2*), as in Posse et al. (1999). The covariance structure of all voxel time-courses was used to identify major signals in the resultant OC-ME time-series using principal component and independent component analysis. Components were classified as either T2*-dependent (and retained) or S0-dependent (and discarded), primarily according to their decay properties across echoes following the decision tree described in Kundu et al. (2012). We found that a global influence of respiration (a T2*-dependent signal that is not of interest per se; see Figure S1) was retained after removing S0-dependent components. Mean gray matter time-series regression was subsequently performed to remove this spatially diffuse noise. Two other denoising strategies were performed. Specifically, the OC-ME and TE2 time-series were also submitted to ICA-AROMA (Pruim et al., 2015), a top performing algorithm for denoising single-echo fMRI data (Ciric et al., 2017), followed by mean gray matter time-series regression. Finally, temporal masks were generated for censoring high motion time-points using a frame-wise displacement (FD; Power et al., 2012) threshold of 0.3 mm and a backward difference of two TRs (2 * 1.355 = 2.77 s), for an effective sampling rate comparable to historical FD measurements (approximately 2 to 4 s; Power et al., 2019a). Prior to the FD calculation, head realignment parameters were filtered using a stopband Butterworth filter (0.2 – 0.35 Hz) to attenuate the influence of respiration (Power et al., 2019a). Concatenated filtered FD traces for each subject can be viewed in the Supplemental Information (Figure S4).
Surface processing and CIFTI generation of fMRI data
The denoised fMRI time-series was mapped to the midthickness surfaces (using the “-ribbon-constrained” method), combined into the Connectivity Informatics Technology Initiative (CIFTI) format, and spatially smoothed with geodesic (for surface data) and Euclidean (for volumetric data) Gaussian kernels (σ = 2.55 mm) using Connectome Workbench command line utilities (Glasser et al., 2013). Signals were normalized (z-scored). This yielded time courses representative of the entire cortical surface, subcortex (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, and thalamus), and cerebellum, but excluding non-gray matter tissue.
Functional connectivity reliability maps
Functional connectivity (FC) reliability maps were created using the following procedure. For each scan, FC maps were created using each point in the brain as a seed. This involved calculating the correlation between seed time-courses and the time-courses of all cortical vertices. FC maps were calculated using quantities of data corresponding to a range of scan durations (duration calculated before motion censoring), from 1 min to the full duration of individual scans in each dataset, in 1 min steps. Next, the same FC maps were created using all other scans (concatenated) available for that subject. This second set of FC maps served as a putative ground truth. Reliability was calculated at each seed point as the average spatial correlation (R2) between individual scan and ground truth FC maps. Thus, high and low reliability values indicate that the FC of a given vertex or voxel calculated using the specified amount of data from a single scan was similar or dissimilar to FC of that same vertex or voxel when calculated using a large amount of independent data. Correlations between pairs of vertices less than 10 mm apart in geodesic space were omitted from this calculation to avoid considering correlations due to spatial smoothing. Time × reliability curves were obtained by averaging reliability values within different anatomical compartments (cortex, subcortical structures, cerebellum) at each scan duration. Points in the time × reliability curves associated with single-echo and multi-echo datasets were compared statistically using the “ttest.m” function in MATLAB (null hypothesis being that the distribution of reliability values observed in independent single-echo data has a mean equivalent to the average value observed in multi-echo data). We note that this analysis differs from previous investigations of FC reliability in densely sampled individuals (Gordon et al., 2017c; Laumann et al., 2015) in two ways. First, the reliability values here are reported as R2 and not Pearson correlation r. Second, we evaluated the reliability of vertex-to-vertex (and voxel-to-vertex) FC, not parcel-to-parcel FC.
Vertex-wise mapping of functional brain networks
Functional brain networks were mapped brain-wide in individual subjects by following procedures developed collectively in Gordon et al. (2017c), Greene et al. (2020), and Marek et al. (2018). In short, a 59412 × 59412 functional connectivity matrix summarizing the temporal correlation between the time-courses of all cortical vertices was generated. Correlations between vertices with centroids less than 30 mm apart in geodesic space were set to zero. Community assignments were obtained over a range of graph densities (5% to 0.001%) using the InfoMap algorithm (Rosvall and Bergstrom, 2008). A template-matching procedure described in Gordon et al. (2017c) was used to assign 1 of 15 known brain network identities to the InfoMap communities identified at each graph density. Subcortical and cerebellar voxels were then assigned to a consensus brain network in cortex using the winner-take-all procedure described in Greene et al. (2020) and Marek et al. (2018). We identified “integrative” vertices and voxels (those exhibiting strong FC more than one network) brain-wide by testing if functional connectivity any other network was greater than 66.7% of the correlation with the consensus network assignment. Integrative regions were visualized on the surface and in the volume using stripes (with stripe colors representing the networks present at that point).
Assessing the reliability of functional brain networks
Functional brain networks were mapped in the manner described above using data from individual scans and concatenated data from all other scans available for each subject (the latter serving as a ground truth). Reliability was defined using the adjusted Rand index (calculated using the “zrand.m” function from the Network Community Toolbox; http://commdetect.weebly.com), which represents the fraction of points in the brain identified as belonging to the same network in single scan data and ground truth data after adjusting for chance. The effect of the OC-ME and ME-ICA procedures on the reliability of consensus brain network assignments within individuals was assessed using a mixed effects ANOVA model (via the “anovan.m” function in MATLAB), with data type treated as a fixed effect and subject identity treated as a random effect. Comparisons to the three independent single-echo datasets were performed using an independent sample t test (using the “ttest2.m” function in MATLAB). The reliability of functional brain networks derived from fast-TR single-echo and multi-echo data collected from participant ME01 was performed using a paired t test (using the “ttest.m” function in MATLAB).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and Algorithms | ||
| MATLAB | MathWorks | https://www.mathworks.com/ |
| Connectome Workbench | Marcus et al., 2011 | https://www.humanconnectome.org/software/connectome-workbench |
| Tedana | Dupre et al., 2020 | https://tedana.readthedocs.io/en/latest/ |
| FreeSurfer | Fischl, 2012 | https://surfer.nmr.mgh.harvard.edu/ |
| Infomap | Rosvall and Bergstrom, 2008 | https://www.mapequation.org/ |
Highlights.
Multi-echo fMRI improves the reliability of functional connectivity in individuals
Benefits of multi-echo fMRI are pronounced in clinically important brain regions
Enhanced reliability is due to better BOLD signal sensitivity and artifact removal
ACKNOWLEDGMENTS
We thank the staff at the Citigroup Biomedical Imaging Center for assistance with data collection. The developers of Tedana (“TE Dependent ANAlysis”) provided technical assistance. Dr. Evan Gordon shared templates used to assign known functional brain network identities to InfoMap communities and code for creating the appearance of stripes on the surface and in the volume. Dr. Andrew Breeden provided valuable feedback on an earlier version of this manuscript. C.L. and F.M.G. were supported by grants from NIMH, NIDA, the Rita Allen Foundation, and the Hope for Depression Research Foundation. C.J.L. was supported by an NIMH F32 National Research Service Award (F32MH120989).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108540.
DECLARATION OF INTERESTS
C.L. is listed as an inventor for Cornell University patent applications on neuroimaging biomarkers for depression that are pending or in preparation. The authors report no biomedical financial interests or other potential conflicts of interest.
REFERENCES
- Anticevic A, Brumbaugh MS, Winkler AM, Lombardo LE, Barrett J, Corlett PR, Kober H, Gruber J, Repovs G, Cole MW, et al. (2013). Global prefrontal and fronto-amygdala dysconnectivity in bipolar I disorder with psychosis history. Biol. Psychiatry 73, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JT, Holmes AJ, Masters GA, Yeo BT, Krienen F, Buckner RL, and Öngür D (2014). Disruption of cortical association networks in schizophrenia and psychotic bipolar disorder. JAMA Psychiatry 71, 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandettini PA, Wong EC, Jesmanowicz A, Hinks RS, and Hyde JS (1994). Spin-echo and gradient-echo EPI of human brain activation using BOLD contrast: a comparative study at 1.5 T. NMR Biomed. 7, 12–20. [DOI] [PubMed] [Google Scholar]
- Bhavsar S, Zvyagintsev M, and Mathiak K (2014). BOLD sensitivity and SNR characteristics of parallel imaging-accelerated single-shot multi-echo EPI for fMRI. Neuroimage 84, 65–75. [DOI] [PubMed] [Google Scholar]
- Birn RM, Molloy EK, Patriat R, Parker T, Meier TB, Kirk GR, Nair VA, Meyerand ME, and Prabhakaran V (2013). The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage 83, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, and Hyde JS (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med 34, 537–541. [DOI] [PubMed] [Google Scholar]
- Braga RM, and Buckner RL (2017). Parallel Interdigitated Distributed Networks within the Individual Estimated by Intrinsic Functional Connectivity. Neuron 95, 457–471.e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga RM, Van Dijk KRA, Polimeni JR, Eldaief MC, and Buckner RL (2019). Parallel distributed networks resolved at high resolution reveal close juxtaposition of distinct regions. J. Neurophysiol 121, 1513–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Gaudes C, and Reynolds RC (2017). Methods for cleaning the BOLD fMRI signal. Neuroimage 154, 128–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Gaudes C, Moia S, Panwar P, Bandettini PA, and Gonzalez-Castillo J (2019). A deconvolution algorithm for multi-echo functional MRI: Multi-echo Sparse Paradigm Free Mapping. Neuroimage 202, 116081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellanos FX, Di Martino A, Craddock RC, Mehta AD, and Milham MP (2013). Clinical applications of the functional connectome. Neuroimage 80, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric R, Wolf DH, Power JD, Roalf DR, Baum GL, Ruparel K, Shinohara RT, Elliott MA, Eickhoff SB, Davatzikos C, et al. (2017). Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. Neuroimage 154, 174–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Esposito M (2019). Are individual differences in human brain organization measured with functional MRI meaningful? Proc. Natl. Acad. Sci. USA 116, 22432–22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar MP, Fenoy AJ, Carvalho AF, Soares JC, and Quevedo J (2018). Deep brain stimulation for treatment-resistant depression: an integrative review of preclinical and clinical findings and translational implications. Mol. Psychiatry 23, 1094–1112. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Kelly C, Grzadzinski R, Zuo XN, Mennes M, Mairena MA, Lord C, Castellanos FX, and Milham MP (2011). Aberrant striatal functional connectivity in children with autism. Biol. Psychiatry 69, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, et al. (2014). The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 19, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downar J, Geraci J, Salomons TV, Dunlop K, Wheeler S, McAndrews MP, Bakker N, Blumberger DM, Daskalakis ZJ, Kennedy SH, et al. (2014). Anhedonia and reward-circuit connectivity distinguish nonresponders from responders to dorsomedial prefrontal repetitive transcranial magnetic stimulation in major depression. Biol. Psychiatry 76, 176–185. [DOI] [PubMed] [Google Scholar]
- Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, et al. (2017). Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med 23, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop BW, Rajendra JK, Craighead WE, Kelley ME, McGrath CL, Choi KS, Kinkead B, Nemeroff CB, and Mayberg HS (2017). Functional Connectivity of the Subcallosal Cingulate Cortex And Differential Outcomes to Treatment With Cognitive-Behavioral Therapy or Antidepressant Medication for Major Depressive Disorder. Am. J. Psychiatry 174, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre E, Taylor S, Markello RD, Kundu P, Whitaker KJ, and Hand-werker DA (2020). ME-ICA/tedana: 0.0.9a (Zenodo)
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, and Constable RT (2015). Functional connectome finger-printing: identifying individuals using patterns of brain connectivity. Nat. Neurosci 18, 1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B (2012). FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, and Greicius M (2010). Clinical applications of resting state functional connectivity. Front. Syst. Neurosci 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Buckner RL, White MP, Greicius MD, and Pascual-Leone A (2012). Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol. Psychiatry 72, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, and Van Essen DC (2011). Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci 31, 11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, et al. ; WU-Minn HCP Consortium (2013). The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, and Petersen SE (2016). Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb. Cortex 26, 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Gilmore AW, Nelson SM, Dosenbach NUF, and Petersen SE (2017a). Individual-specific features of brain systems identified with resting state functional correlations. Neuroimage 146, 918–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, and Petersen SE (2017b). Individual Variability of the System-Level Organization of the Human Brain. Cereb. Cortex 27, 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt-Drazen C, Gratton C, Sun H, et al. (2017c). Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807.e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Kraus BT, Greene DJ, Gordon EM, Laumann TO, Nelson SM, Dosenbach NUF, and Petersen SE (2020). Defining Individual-Specific Functional Neuroanatomy for Precision Psychiatry. Biol. Psychiatry 88, 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DJ, Marek S, Gordon EM, Siegel JS, Gratton C, Laumann TO, Gilmore AW, Berg JJ, Nguyen AL, Dierker D, et al. (2020). Integrative and Network-Specific Connectivity of the Basal Ganglia and Thalamus Defined in Individuals. Neuron 105, 742–758.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, and Fischl B (2009). Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48, 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JV, Dokovna LB, Jacokes ZJ, Torgerson CM, Irimia A, and Van Horn JD (2017). Resting-State Functional Connectivity in Autism Spectrum Disorders: A Review. Front. Psychiatry 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, and Smith S (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841. [DOI] [PubMed] [Google Scholar]
- Karcher NR, O’Brien KJ, Kandala S, and Barch DM (2019). Resting-State Functional Connectivity and Psychotic-like Experiences in Childhood: Results From the Adolescent Brain Cognitive Development Study. Biol. Psychiatry 86, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Li J, Orban C, Sabuncu MR, Liu H, Schaefer A, Sun N, Zuo XN, Holmes AJ, Eickhoff SB, and Yeo BTT (2019). Spatial Topography of Individual-Specific Cortical Networks Predicts Human Cognition, Personality, and Emotion. Cereb. Cortex 29, 2533–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Inati SJ, Evans JW, Luh WM, and Bandettini PA (2012). Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage 60, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Brenowitz ND, Voon V, Worbe Y, Vértes PE, Inati SJ, Saad ZS, Bandettini PA, and Bullmore ET (2013). Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proc. Natl. Acad. Sci. USA 110, 16187–16192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Santin MD, Bandettini PA, Bullmore ET, and Petiet A (2014). Differentiating BOLD and non-BOLD signals in fMRI time series from anesthetized rats using multi-echo EPI at 11.7 T. Neuroimage 102, 861–874. [DOI] [PubMed] [Google Scholar]
- Kundu P, Benson BE, Baldwin KL, Rosen D, Luh WM, Bandettini PA, Pine DS, and Ernst M (2015). Robust resting state fMRI processing for studies on typical brain development based on multi-echo EPI acquisition. Brain Imaging Behav. 9, 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Voon V, Balchandani P, Lombardo MV, Poser BA, and Bandettini PA (2017). Multi-echo fMRI: A review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage 154, 59–80. [DOI] [PubMed] [Google Scholar]
- Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen MY, Gilmore AW, McDermott KB, Nelson SM, Dosenbach NU, et al. (2015). Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, Gilmore AW, Nelson SM, Berg JJ, Greene DJ, et al. (2017). On the Stability of BOLD fMRI Correlations. Cereb. Cortex 27, 4719–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT (2016). Noise contributions to the fMRI signal: An overview. Neuroimage 143, 141–151. [DOI] [PubMed] [Google Scholar]
- Lombardo MV, Auyeung B, Holt RJ, Waldman J, Ruigrok ANV, Mooney N, Bullmore ET, Baron-Cohen S, and Kundu P (2016). Improving effect size estimation and statistical power with multi-echo fMRI and its impact on understanding the neural systems supporting mentalizing. Neuroimage 142, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, and Liston C (2020). Precision Functional Mapping of Cortico-striatal and Corticothalamic Circuits: Parallel Processing Reconsidered. Neuron 105, 595–597. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Breeden AL, Gordon EM, Cherry JBC, Turkeltaub PE, and Vaidya CJ (2019). Precision Inhibitory Stimulation of Individual-Specific Cortical Hubs Disrupts Information Processing in Humans. Cereb. Cortex 29, 3912–3921. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Silver BM, Dubin MJ, Martin A, Voss HU, Jones RM, and Power JD (2020). Prevalent and sex-biased breathing patterns modify functional connectivity MRI in young adults. Nat. Commun 11, 5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Harwell J, Olsen T, Hodge M, Glasser MF, Prior F, Jenkinson M, Laumann T, Curtiss SW, and Van Essen DC (2011). Informatics and data mining tools and strategies for the human connectome project. Front. Neuroinform 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S, Siegel JS, Gordon EM, Raut RV, Gratton C, Newbold DJ, Ortega M, Laumann TO, Adeyemo B, Miller DB, et al. (2018). Spatial and Temporal Organization of the Individual Human Cerebellum. Neuron 100, 977–993.e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, Schwalb JM, and Kennedy SH (2005). Deep brain stimulation for treatment-resistant depression. Neuron 45, 651–660. [DOI] [PubMed] [Google Scholar]
- McMullen DP (2018). Where to Target? The Precision Medicine Approach to Brain Stimulation. Biol. Psychiatry 84, e1–e2. [DOI] [PubMed] [Google Scholar]
- Medaglia JD, Erickson B, Zimmerman J, and Kelkar A (2020). Personalizing neuromodulation. Int. J. Psychophysiol 154, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V (2011). Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn. Sci 15, 483–506. [DOI] [PubMed] [Google Scholar]
- Mitchell TJ, Hacker CD, Breshears JD, Szrama NP, Sharma M, Bundy DT, Pahwa M, Corbetta M, Snyder AZ, Shimony JS, and Leuthardt EC (2013). A novel data-driven approach to preoperative mapping of functional cortex using resting-state functional magnetic resonance imaging. Neurosurgery 73, 969–982, discussion 982–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Fayad SM, Higuchi MA, Nestor KA, and Foote KD (2014). Deep brain stimulation for treatment-resistant depression: systematic review of clinical outcomes. Neurotherapeutics 11, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold DJ, Laumann TO, Hoyt CR, Hampton JM, Montez DF, Raut RV, Ortega M, Mitra A, Nielsen AN, Miller DB, et al. (2020). Plasticity and Spontaneous Activity Pulses in Disused Human Brain Circuits. Neuron 107, 580–589.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, and Scheinost D (2017). Influences on the Test-Retest Reliability of Functional Connectivity MRI and its Relationship with Behavioral Utility. Cereb. Cortex 27, 5415–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Scheinost D, and Constable RT (2019). A decade of test-retest reliability of functional connectivity: A systematic review and meta-analysis. Neuroimage 203, 116157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor EE, and Zeffiro TA (2019). Why is Clinical fMRI in a Resting State? Front. Neurol 10, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oathes DJ, Patenaude B, Schatzberg AF, and Etkin A (2015). Neurobiological signatures of anxiety and depression in resting-state functional magnetic resonance imaging. Biol. Psychiatry 77, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Lynch CJ, Schaer M, and Menon V (2017). The Default Mode Network in Autism. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter JS, and Mink JW (2006). Deep brain stimulation. Annu. Rev. Neurosci 29, 229–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli DA (2014). Depression, stress, and anhedonia: toward a synthesis and integrated model. Annu. Rev. Clin. Psychol 10, 393–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA (2017). Precision Neuroscience: Dense Sampling of Individual Brains. Neuron 95, 727–729. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen MY, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, et al. (2015). Long-term neural and physiological phenotyping of a single human. Nat. Commun 6, 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser BA, and Norris DG (2009). Investigating the benefits of multi-echo EPI for fMRI at 7 T. Neuroimage 45, 1162–1172. [DOI] [PubMed] [Google Scholar]
- Posse S (2012). Multi-echo acquisition. Neuroimage 62, 665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posse S, Wiese S, Gembris D, Mathiak K, Kessler C, Grosse-Ruyken ML, Elghahwagi B, Richards T, Dager SR, and Kiselev VG (1999). Enhancement of BOLD-contrast sensitivity by single-shot multi-echo functional MR imaging. Magn. Reson. Med 42, 87–97. [DOI] [PubMed] [Google Scholar]
- Power JD (2017). A simple but useful way to assess fMRI scan qualities. Neuroimage 154, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, and Petersen SE (2011). Functional network organization of the human brain. Neuron 72, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, and Petersen SE (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, and Petersen SE (2015). Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage 105, 536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Plitt M, Gotts SJ, Kundu P, Voon V, Bandettini PA, and Martin A (2018). Ridding fMRI data of motion-related influences: Removal of signals with distinct spatial and physical bases in multiecho data. Proc. Natl. Acad. Sci. USA 115, E2105–E2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Lynch CJ, Silver BM, Dubin MJ, Martin A, and Jones RM (2019a). Distinctions among real and apparent respiratory motions in human fMRI data. Neuroimage. 201, 116041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Silver BM, Silverman MR, Ajodan EL, Bos DJ, and Jones RM (2019b). Customized head molds reduce motion during resting state fMRI scans. Neuroimage 189, 141–149. [DOI] [PubMed] [Google Scholar]
- Power JD, Lynch CJ, Adeyemo B, and Petersen SE (2020). A Critical, Event-Related Appraisal of Denoising in Resting-State fMRI Studies. Cereb. Cortex 30, 5544–5559. [DOI] [PubMed] [Google Scholar]
- Pruim RHR, Mennes M, van Rooij D, Llera A, Buitelaar JK, and Beckmann CF (2015). ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage 112, 267–277. [DOI] [PubMed] [Google Scholar]
- Rosvall M, and Bergstrom CT (2008). Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 105, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sair HI, Yahyavi-Firouz-Abadi N, Calhoun VD, Airan RD, Agarwal S, Intrapiromkul J, Choe AS, Gujar SK, Caffo B, Lindquist MA, and Pillai JJ (2016). Presurgical brain mapping of the language network in patients with brain tumors using resting-state fMRI: Comparison with task fMRI. Hum. Brain Mapp 37, 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, and Gur RE (2012). Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neuro-development in youth. Neuroimage 60, 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Xia CH, and Bassett DS (2018). Personalized Neuroscience: Common and Individual-Specific Features in Functional Brain Networks. Neuron 98, 243–245. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Macmore J, and Vangel M (2009). Cerebellar stroke without motor deficit: clinical evidence for motor and non-motor domains within the human cerebellum. Neuroscience 162, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedeskamp H, Newbould RD, Pisani LJ, Skare S, Glover GH, Pruessmann KP, and Bammer R (2010). Improvements in parallel imaging accelerated functional MRI using multiecho echo-planar imaging. Magn. Reson. Med 63, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitzman BA, Gratton C, Laumann TO, Gordon EM, Adeyemo B, Dworetsky A, Kraus BT, Gilmore AW, Berg JJ, Ortega M, et al. (2019). Trait-like variants in human functional brain networks. Proc. Natl. Acad. Sci. USA 116, 22851–22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba A (2014). The role of the cerebellum in neurobiology of psychiatric disorders. Neurol. Clin 32, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, and Beckmann CF (2009). Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA 106, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Vidaurre D, Beckmann CF, Glasser MF, Jenkinson M, Miller KL, Nichols TE, Robinson EC, Salimi-Khorshidi G, Woolrich MW, et al. (2013). Functional connectomics from resting-state fMRI. Trends Cogn. Sci 17, 666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AZ, and Raichle ME (2012). A brief history of the resting state: the Washington University perspective. Neuroimage 62, 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley CJ, MacMore JP, Makris N, Sherman JC, and Schmahmann JD (2016). Location of lesion determines motor vs. cognitive consequences in patients with cerebellar stroke. Neuroimage Clin. 12, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, and Laufs H (2014). Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron 82, 695–708. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker DL, Harwell J, and Coalson T (2012). Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb. Cortex 22, 2241–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneau-Roy N, Bernier M, Descoteaux M, and Whittingstall K (2014). Regional variations in vascular density correlate with resting-state and task-evoked blood oxygen level-dependent signal amplitude. Hum. Brain Mapp 35, 1906–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Han J, Nguyen VT, Guo L, and Guo CC (2017). Improving the Test-Retest Reliability of Resting State fMRI by Removing the Impact of Sleep. Front. Neurosci 11, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Li M, Wang M, Schoeppe F, Ren J, Chen H,Öngür D, Brady RO Jr., Baker JT, and Liu H (2020). Individual-specific functional connectivity markers track dimensional and categorical features of psychotic illness. Mol. Psychiatry 25, 2119–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand A, Horn A, Caballero R, Cooke D, Stern AP, Taylor SF, Press D, Pascual-Leone A, and Fox MD (2018). Prospective Validation That Subgenual Connectivity Predicts Antidepressant Efficacy of Transcranial Magnetic Stimulation Sites. Biol. Psychiatry 84, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Ma Z, Ciric R, Gu S, Betzel RF, Kaczkurkin AN, Calkins ME, Cook PA, García de la Garza A, Vandekar SN, et al. (2018). Linked dimensions of psychopathology and connectivity in functional brain networks. Nat. Commun 9, 3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Moeller S, Auerbach EJ, Strupp J, Smith SM, Feinberg DA, Yacoub E, and Uğurbil K (2013). Evaluation of slice accelerations using multi-band echo planar imaging at 3 T. Neuroimage 83, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyavi-Firouz-Abadi N, Pillai JJ, Lindquist MA, Calhoun VD, Agarwal S, Airan RD, Caffo B, Gujar SK, and Sair HI (2017). Presurgical Brain Mapping of the Ventral Somatomotor Network in Patients with Brain Tumors Using Resting-State fMRI. AJNR Am. J. Neuroradiol 38, 1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollins-head M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, et al. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol 106, 1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are restrictions to the availability of neuroimaging data because the study participants did not consent to data sharing. Code for preprocessing multi-echo fMRI data is maintained in an online repository (https://github.com/cjl2007/Liston-Laboratory-MultiEchofMRI-Pipeline).


