ABSTRACT
Microbiota, and the plethora of signalling molecules that they generate, are a major driving force that underlies a striking range of inter-individual physioanatomic and behavioural consequences for the host organism. Among the bacterial effectors, one finds peptidoglycan, the major constituent of the bacterial cell surface. In the steady-state, fragments of peptidoglycan are constitutively liberated from bacterial members of the gut microbiota, cross the gut epithelial barrier and enter the host system. The fate of these peptidoglycan fragments, and the outcome for the host, depends on the molecular nature of the peptidoglycan, as well the cellular profile of the recipient tissue, mechanism of cell entry, the expression of specific processing and recognition mechanisms by the cell, and the local immune context. At the target level, physiological processes modulated by peptidoglycan are extremely diverse, ranging from immune activation to small molecule metabolism, autophagy and apoptosis. In this review, we bring together a fragmented body of literature on the kinetics and dynamics of peptidoglycan interactions with the mammalian host, explaining how peptidoglycan functions as a signalling molecule in the host under physiological conditions, how it disseminates within the host, and the cellular responses to peptidoglycan.
Keywords: bacterial cell wall, peptidoglycan, muropeptides, gut microbiota, mammalian host, biodistribution
A comprehensive review of the diverse mechanisms acting in the mammalian host to facilitate the uptake, biodistribution, processing and recognition of the major bacterial cell wall constituent, peptidoglycan, a key microbiome effector molecule influencing host physiology during health and disease.
INTRODUCTION
The human body is home to 100 trillion bacteria, of which more than 95% reside in the large intestine. At least 40 000 bacterial species are harboured throughout this distal portion of the gastrointestinal tract, together comprising ∼150 times more genes than the human host (which contains, in turn, a relatively modest number of ∼26 000 functional genes) (Frank and Pace 2008; Qin et al. 2010). There is huge inter-individual variability regarding the composition of the host gut microbiome. Much in the same way as genetic and environmental variability accounts for striking inter-individual physioanatomic and behavioural differences, the gut microbiota composition is thought to significantly influence host physiology and behaviour. Accordingly, whilst the precise composition of a microbiome is specific to each individual, microbiome profiling has revealed ‘clusters’ specific to groups of individuals in both health and disease (Karlsson et al. 2013; Scher et al. 2015). Due to this inextricable and complex role as a mediator of host physiology, the human intestinal microbiota has come to be recognized as a host ‘organ’ (Guarner and Malagelada 2003; Bäckhed et al. 2005).
Variations in gut microbiota composition and gut microbiota-derived products have been shown to influence a multitude of mammalian host processes, including gastrointestinal, immune and neuronal development and function. However, the vast majority of this data is merely observational, and fails to pinpoint mechanistically how the microbiota and its molecular products actually influence host physiology. Specifically, whilst evidence clearly demonstrates that bacterial effectors disseminate from the gastrointestinal tract throughout the host organism, their view as constitutive signalling molecules is not yet well established. Among the plethora of such bacterial effectors, one finds a major, specific constituent of bacterial cell surfaces, the peptidoglycan. Peptidoglycan is the main component of the bacterial cell wall, comprised of long linear glycan strands cross-linked by short peptide stems. The glycan strands consist of repeating units of the disaccharide N-acetylglucosamine -β(1,4)-N-acetylmuramic acid [GlcNAc-β(1,4)-MurNAC], while the peptide stems are attached to the lactyl moiety of the N-acetylmuramic acid residues, and are typically composed of alternating L- and D-amino acids in the following order: L-Ala, D-Glx, a di-amino acid [most often meso-diaminopimelic acid (mDAP) or L-lysine], D-Ala, D-Ala (Fig. 1). Despite this common main structure, the mature peptidoglycan is highly heterogeneous and species-specific. Contributing factors to this diversity are the different degrees and patterns of crosslinking, the presence or absence of bridging peptides, variations in the type of di-amino acid present, hydrolases acting on the different bonds of the peptidoglycan, the presence of atypical amino acids and the diversity of possible chemical modifications (Vollmer, Blanot and De Pedro 2008).
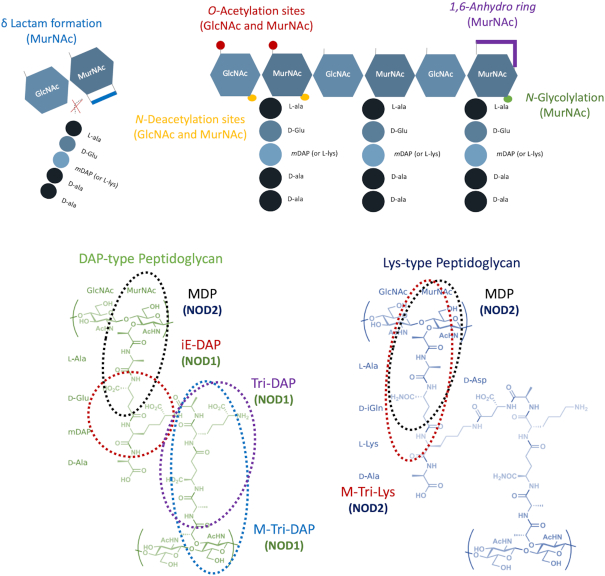
Figure 1.
Schematic representation of peptidoglycan structure with its modifiable sites and the minimal NOD1/2 Activating Motifs. Top: The archetypical structure of peptidoglycan consists of long linear chains of the alternating sugar residues GlcNAc and MurNAc. These chains are in turn crosslinked via peptide chains, which are typically synthesized with a pentapeptide core structure consisting of the amino acids L-Ala, D-Glu, mDAP or L-Lys, D-Ala, D-Ala. Variations in the amino acid composition are common, particularly at position 3 of the peptide stem. Typical modification sites together with the respective chemical modifications are represented as coloured areas on the top panel. Bottom: The chemical structures of the minimal NOD1 (iE-DAP, Tri-DAP and M-Tri-DAP) and NOD2 (MDP and M-Tri-Lys) activating motifs are represented, originating from either DAP-type (left) or Lys-type (right) peptidoglycan. GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; D-Glu, D-glutamic acid; D-Ala, D-alanine; L-Ala, L-alanine; L-Lys, L-lysine; mDAP, DL-2,6-diaminopimelic acid.
Peptidoglycan is not only essential for the vast majority of bacteria, it also comprises numerous different bioactive moieties that can exert critical developmental, physiological and pathological effects on the mammalian host (Fig. 1). Despite its essential and multifaceted role, and the way it has influenced the evolutionary development of eukaryotic cell signalling systems, we are not yet able to exploit the many biological properties of peptidoglycan in order to modulate host (patho)physiology. Reasons for this gap include the relative difficulty of isolating and working with pure peptidoglycan, its different composition depending on the source strains employed, limited means for the unambiguous detection of peptidoglycan in vivo, and mixed (sometimes conflicting) results regarding the host response to peptidoglycan.
In order to exploit peptidoglycan species as bioactive molecules with the aim of modulating host physiology, it is imperative to lay down the foundations of how peptidoglycan derived from our gut bacteria behaves inside the host organism. In other words, a mapping akin to that performed during ADME (absorption, distribution, metabolism and excretion) pharmacological studies on the possible fates of peptidoglycan originating from our gut bacteria, is required as a foundation for future peptidoglycan-mediated modulation of host physiology. This review attempts to bring together a somewhat large but fragmented body of literature on the kinetics and dynamics of peptidoglycan interactions within the mammalian host. It is our goal to highlight host- and peptidoglycan-specific factors governing such interactions, and to identify promising targets and gaps in our understanding of how one may fully exploit peptidoglycan in health and disease. A summary of host proteins reported to bind peptidoglycan is presented in Table 1.
Table 1.
Summary of proteins reported to bind peptidoglycan or facilitate peptidoglycan entry into host cells.
| Peptidoglycan binding protein | Minimal binding moieties identified | Biological roles | References |
|---|---|---|---|
| Toll-like receptor (TLR) 2 | MtriLys, and MtriDAP, with a strong impact of amide modifications on binding efficiency. Conflicting data regarding role in peptidoglycan recognition. | Classical innate immune pattern recognition receptor. Recognises specific PAMPs upon heterodimerisation with TLR1 or TLR6 | (Yoshimura et al. 1999b; Inohara et al. 2003; Dziarski and Gupta 2005; Asong et al. 2009) |
| Serotonin (5-HT) receptors | MDP proposed as a ligand | Neuropharmacological effects via the serotoninergic system. Induction of IL-1β | (Ševčík and Mašek 1999; Ševčík et al. 2000, 2002) |
| Solute carrier (SLC) 15 transporters (SLC15A1/PepT1, SLC15A2/PepT2, SLC15A4/PhT1, SLC15A3/PhT2) | triDAP (SLC15A2/PepT2); MDP and triDAP (SLC15A1/PepT1); SLC1514 triDAP Likely to happen almost exclusively for muropeptides containing peptide stems up to 3 peptides | Peptidoglycan translocation across the plasma and endosomal membrane | (Vavricka et al. 2004; Ismair et al. 2006; Swaan et al. 2008; Dalmasso et al. 2010; Smith, Clémençon and Hediger 2013; Nakamura et al. 2014) |
| SLC46 transporters (SLC46A2, SLC46A3) | Monomeric muropeptides including MDP and DAP-containing anhydro-muropeptide monomers (Tracheal cytotoxin; TCT) | Peptidoglycan translocation across late-endosome or endolysosome membranes | (Paik et al. 2017) |
| Platelet-activating factor receptor (PAFr) | Phosphorylcholine-decorated cell wall fragments | Peptidoglycan translocation across the plasma membrane, shuttling to lysosomal trafficking | (Cundell et al. 1995; Ring, Weiser and Tuomanen 1998; Yoshimura et al. 1999a; Rijneveld et al. 2004; Fillon et al. 2006; McLaughlin et al. 2006; Eckels et al. 2009; Loh, Gao and Tuomanen 2017) |
| CD14 | Insoluble and soluble DAP-type peptidoglycan | Immune activation characterized by NF-κB induction | (Gupta et al. 1996; Dziarski, Tapping and Tobias 1998; Dziarski et al. 2000) |
| Lysozyme | Polymeric peptidoglycan Glycan chain tetrasaccharide cleaved to GlcNAc-β-(1,4)-MurNAc disaccharides | Hydrolysis of the glycosidic bonds between N-Acetylmuramic acid (MurNAc) and N-Acetylglucosamine (GlcNAc), hydrolysis-independent antimicrobial activity | (Sharon 1967; Hammer et al. 1987; Cross et al. 1988; Ibrahim, Matsuzaki and Aoki 2001) |
| NOD1 | Dipeptide γ-iE-DAP and larger meso-DAP containing monomers (Gram-negative bacteria, some Gram-positive bacteria such as Listeria and Bacillus) | Immune activation characterized by NF-κB induction; p38, ERK1/ERK2 and c-Jun N-terminal kinase activation, apoptosis, autophagy, xenophagy | (Viala et al. 2004; Strober et al. 2006; Hasegawa et al. 2007; Allison et al. 2009; Watanabe et al. 2010) |
| NOD2 | MDP (Gram-positive and Gram-negative bacteria), MtriLys | Immune activation characterized by NF-κB induction; p38, ERK1/ERK2 and c-Jun N-terminal kinase activation, apoptosis, autophagy, xenophagy | (Girardin et al. 2003b, 2003c; Fernandez et al. 2011) |
| Calreticulin | MDP and larger polymers | Apoptosis induction | (Chen et al. 2004, 2005; Groenendyk, Lynch and Michalak 2004) |
| C-type lectins (mannose-binding lectins, M- L- and H ficolins) | Binding via GlcNAc moieties | Complement system activation, cytokines production, mast cells and basophils degranulation, increased vascular permeability | (Matsushita and Fujita 2001; Ma et al. 2004; Garlatti et al. 2007; Kilpatrick and Chalmers 2012; Bidula, Sexton and Schelenz 2019) |
| C-type lectins [Murine RegIIIγ and Human Hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein (HIP/PAP)] | Polymeric peptidoglycan | Direct antimicrobial activity via cell wall damage and cytoplasmic leakage | (Christa et al. 1996; Cash et al. 2006a,b) |
| LysM-domain proteins (LysMD1, LysMD2, LysMD3, LysMD4) | Binding not studied for mammalian proteins. Potentially bind to glycan chain, likely via GlcNAc residues | Assumed to be involved in immune responses against bacterial infection, based on homology | (Willmann et al. 2011; Yokoyama et al. 2018) |
| Peptidoglycan recognition proteins (PGRPs/PGLYRPs) [PGLYRP-1, PGLYRP-2, PGLYRP-3, PGLYRP-4] | PGLYRP-2: muramyl tripeptide and likely larger polymers; Polymeric peptidoglycan for the remaining PGRPs/PGLYRPs | Direct antimicrobial activity via membrane depolarization, hydroxyl radical production and synthetic pathways disruption; PGLYRP-2 additionally displays N-acetylmuramoyl-L-alanine amidase activity | (Wang et al. 2003; Girardin et al. 2003c; Royet and Dziarski 2007; Osanai et al. 2011; Read et al. 2015) |
| Hexokinase | GlcNAc | NLRP3 inflammasome activation, IL-1β production and secretion | (Jonas et al. 1989; Jonas and Jobe 1990; Pastorino, Shulga and Hoek 2002; Majewski et al. 2004; Chiara et al. 2008; Pastorino and Hoek 2008) |
A NOTE ON TERMINOLOGY
‘Bacterial cell envelope’ is a term used to encompass all bacterial components beginning at (and including) the cytoplasmic membrane and ending at the bacterial cell surface. Within the bacterial cell envelope, we find the peptidoglycan layer. The peptidoglycan is often referred to as ‘the cell wall’. However, whilst peptidoglycan is certainly the primary structure of the cell wall, it is nevertheless one of many components. For example, in Gram-positive bacteria, wall teichoic acids (WTA) are covalently attached to the peptidoglycan layer, and are estimated to comprise 27%–49% of the total cell wall mass in Lactobacillus plantarum strains (Tomita et al. 2010). Furthermore, Gram-positive bacteria possess lipoteichoic acids, which are anchored in the cytoplasmic membrane, and extrude into the peptidoglycan layer. Together, the teichoic acids contribute directly to the physical properties of the cell wall and to the regulation of its homeostasis. Other secondary cell wall structures are directly linked to, or in intimate interaction with, the peptidoglycan layer, including covalently anchored proteins, lipoproteins, S-layer and capsule.
In this review, we will use the term ‘cell wall’ when referring to phenomena that cannot be separated from, or are dependent on, the presence of secondary components described above. The term ‘peptidoglycan’ will refer specifically and exclusively to the ‘naked peptidoglycan macropolymer’, whilst the term ‘muropeptides’ refers to its derivative fragments, the soluble moieties typically generated by glycan hydrolysing enzymes.
It should be noted that due to the difficulty in purifying peptidoglycan from many bacteria (and in validating its purity), in particular from Gram-positive and Mycobacteria, preparations of peptidoglycan are frequently contaminated with secondary cell wall components, many of which also function as Microbe Associated Molecular Patterns (MAMPs) and are recognized by Pattern Recognition Receptors (PRRs), in particular the Toll-like receptors (TLRs). This includes peptidoglycan preparations that are commercially available (Travassos et al. 2004). It is our hope that other authors take care not to ascribe functions directly to the peptidoglycan when the presence of ‘contaminants’ has not been ruled out. For some studies, particularly in the field of immunology, the qualifying term ‘cell wall’ may be preferable, since it invites mechanistic validation studies that can reinforce or clarify published data and hypotheses. Indeed, it is likely that, in nature, host interactions with peptidoglycan begin with an encounter between a host cell and bacterial ‘cell wall’. Hence, the presence of these secondary cell wall fragments does not bring into question the importance and validity of studies on ‘cell wall’ activities within the mammalian host, as these most certainly confer important biological functions in association with peptidoglycan fragments, under physiological conditions.
DISSEMINATION THROUGHOUT THE HOST
Every organism possesses a large reservoir of peptidoglycan, our ‘peptidoglycome’, originating from the commensal bacteria, particularly those residing in the host gut. We now know that peptidoglycan from intestinal bacteria crosses the intestinal barrier under physiological conditions. For instance, in a seminal paper published in 2010, Clarke and colleagues demonstrated that upon oral gavage with living bacteria containing 3H-meso-DAP labelled peptidoglycan, the peptidoglycan released by these luminal bacteria disseminated via the bloodstream and homed to host organs where it triggered the priming of neutrophils within the bone marrow (Clarke et al. 2010). However, most of what we know regarding the behaviour of peptidoglycan inside the host comes from much older studies.
The study of how peptidoglycan molecules behave and disseminate inside the mammalian host was initiated forty years ago. At the time, the main driving motivation was to understand how and why peptidoglycan acts as a strong and targeted adjuvant agent for vaccines (Azuma et al. 1971; Adam et al. 1974). Of particular note, it was found that whilst peptidoglycan was very rapidly cleared from the host organism, its effects were widespread and long-lasting. In addition, it was found that peptidoglycan molecules and modified derivatives had antitumor effects, and successful cases of tumor shrinkage and remission were reported with the use of individual muropeptides (Sparks et al. 1973; Azuma et al. 1978; Mclaughlin et al. 1980). Unfortunately, for reasons that are hard to reconcile, most research and therapeutic use of muropeptides as vaccine adjuvants and antitumor agents was abandoned. In recent years, however, there has been some revival in this field (Sun et al. 2015). At the whole organism level, where does peptidoglycan travel to once it reaches the systemic circulation? Decades ago, in former Yugoslavia, research groups set out to establish the metabolic fate of peptidoglycan inside the mammalian host (Fig. 2) (Tomašić et al. 1980). In one description, the authors employed the 14C-labellled peptidoglycan monomer GlcNAc-MurNAC-L-Ala-D-isoGln-mDAP-D-Ala-D-Ala (MPP, a muropeptide containing the two sugars and the five stem amino acids) and administered it intravenously to mice at varying doses (25–500 µg, equivalent activity of 100,000–400,000 cpm/mg). It was observed that most of this muropeptide would be excreted via the urine, with 60–80% of the initially administered radioactivity recovered within the first hour, 4–8% within the next 3h and 2–7% in the following 18h. By labeling both one sugar ring and one amino acid with 14C, it was possible to determine that approximately 50% of the intravenously administered muropeptide was excreted unmodified. Intriguingly, approximately 20% was excreted as the peptide stem only (i.e. the sugar residues had been cleaved from the peptide, likely due to the activity of an amidase acting between the N-acetylmuramoyl residue and the first L-amino acid residue in the peptide stem). The remaining radioactivity could be accounted for by the sugar residues alone. However, these were not detected in the urine (Tomašić et al. 1980). Additionally, it was reported that these kinetics were not altered when the mice were previously ‘immunized’ with the same muropeptide by intravenous injection. The same group also showed that MPP was not metabolized by the liver, kidneys or spleen. In contrast, upon incubation with mouse or human blood, 10–50% of the MPP was hydrolyzed to its corresponding disaccharide and pentapeptide constituents. When incubated with plasma or serum, more than 90% of the MPP was hydrolyzed (∼30% in the first 10 min and almost complete hydrolysis within 1h). Only 0.25 mL of human or mouse serum could completely hydrolyze 5 mg of the muropeptide. Blood cells (intact or hemolyzed) had no metabolizing activity against the peptidoglycan monomer. In fact, the rate of hydrolysis of peptidoglycan in whole blood was slower than that of serum or plasma alone, and it was suggested that blood cells acted as an endogenous inhibitor of the apparent blood amidase. (Ladešić et al. 1981). This was the first evidence suggesting that circulating blood cells could serve as protective reservoirs (and potential shuttles) for circulating peptidoglycan.
Figure 2.
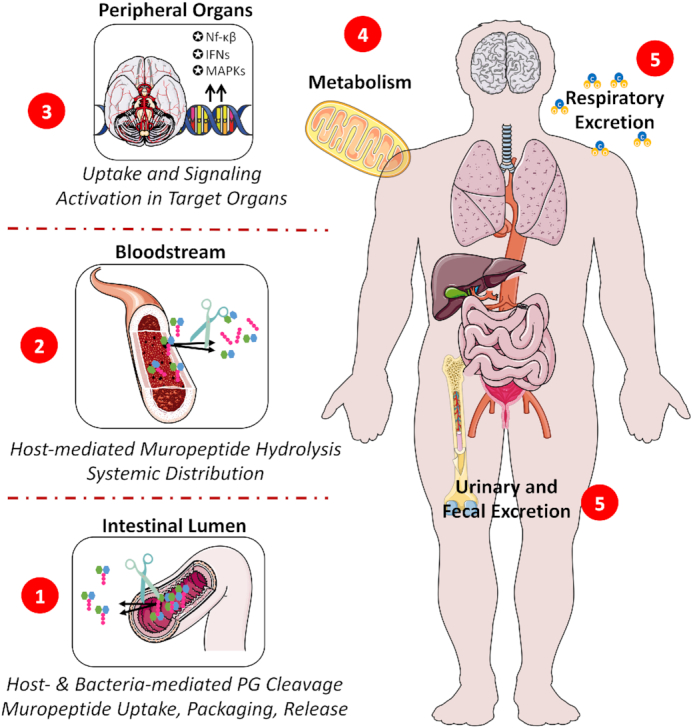
Systemic distribution of peptidoglycan from the intestinal lumen. (1) Peptidoglycan from the intestinal microbiota is cleaved during normal cell wall turnover by bacterial together with host-encoded hydrolytic enzymes, releasing a range of muropeptides with different sizes, structures and biological activities. (2) Upon translocation across the intestinal barrier via multiple possible uptake mechanisms, systemically circulating muropeptides rapidly undergo further hydrolysis, by the blood amidase activity, to their corresponding sugar moieties and peptides stems. (3) Free, intact muropeptides and their hydrolysis products may translocate to the parenchyma of several organs according to a tissue-specific tropism where they trigger different signalling responses depending on their nature and respective targets. (4) Peptidoglycan and its derived molecules may also undergo extensive metabolism by the host, being fully degraded or potentially recycled into endogenous metabolites. (5) Lastly, peptidoglycan metabolism products may be cleared from the host via the faeces, urine or respiration. IFNs: Interferons; MAPKs: Mitogen-activated protein kinases; NF-κB: nuclear factor-kappa B; PG: peptidoglycan.
In another set of experiments, 14C-labelled MPP (1 mg, 2–3 × 106 cpm) was administered intravenously to mice, and again shown to be rapidly excreted via the urine. It was observed that each organ behaves differently regarding its capacity to take up and dispose of peptidoglycan. Initially (30 min post-injection), most of the peptidoglycan circulating in the blood was found outside the cells. One hour post injection, the relative abundance of peptidoglycan present within the cellular compartment was increased, suggesting that free peptidoglycan is excreted faster than peptidoglycan sequestered inside blood cells. Accordingly, 6h post injection, more than 90% of the minute amounts of peptidoglycan still circulating were found inside blood cells (Ladešić, Perović and Hršak 1993). A comparable distribution profile has been reported when using 14C-labelled muramyl-tripeptide (MTP) or MPP alone (Yapo et al. 1982).
Later, this line of studies went on to show that the distribution and excretion patterns of radioactive peptidoglycan differ depending on the route of administration (i.e. subcutaneous, intravenous, oral gavage). While most muropentapeptide was excreted via the urine upon intravenous or subcutaneous injections, orally administered peptidoglycan remained in the GI tract for several hours and the main route of 14C excretion seemed to be via CO2 exhalation, revealing extensive metabolism and degradation of the peptidoglycan upon oral gavage. Accordingly, up to 30% of orally administered 14C-labelled peptidoglycan seemed to be degraded and cumulatively exhaled as 14CO2 48h later, while only 15% was excreted via the urine (Valinger et al. 1987). These data may also imply differential processing of the hydrolysis products of blood amidase activity, favouring clearance (peptide stems) or metabolism (MurNAc and GlcNAc).
The next question to be addressed was how the distinct host organs differentially take up circulating peptidoglycan. The kidneys took up relatively more peptidoglycan (0.5–1.5%) upon intravenous injection than other organs, followed by the intestines (0.2–0.8%). The spleen and the brain took up a much smaller amount immediately after injection (0.02–0.06%), but these figures changed over time. In the liver, maximal amounts of peptidoglycan were found 1h post-injection, but then seemed to remain stable for at least 6h. In the kidneys and the lungs, the peak in uptake of peptidoglycan was observed early post-injection, but was followed by a rapid and steady decline. The concentration in the spleen also peaked early, but like the liver it seemed to stabilize over several hours rather than rapidly decline. The most intriguing observations pertained to the intestines and the brain, where peptidoglycan seemed to accumulate over time (Ladešić, Perović and Hršak 1993).
Another group compared the fate of 3H-labelled muramyl dipeptide (MDP) upon intravenous, intraperitoneal and subcutaneous injection and were careful to measure the tissue intrinsic parenchyma activity only, by subtracting the value derived from the blood contained in the organs, using 125I-polyvinylpyrrolidone as a standard to determine the total blood volume (91.4 ml kg−1) and tissue blood volumes. While the presence of MDP in most of the organs peaked very soon after intravenous injection (e.g. 2 min), it then quickly decreased over time to almost undetectable levels by 60 min post-injection. In the liver, however, the concentration of intravenously administered MDP increased over time. Also, the liver was the only organ where significant metabolism of MDP was observed (as early as 2 min post-injection) (Ambler and Hudson 1984). However, the metabolites could not be found in the circulation, suggesting that they may be excreted directly by the liver via the bile.
Around the same time, one group set out to determine the fate of MDP and MTP (Fogler et al. 1985). This group measured the amount of muropeptide-derived radioactivity (muropeptides contained within the tissue parenchyma and inside the supplying microvasculature) as well as the amount of blood supplying each organ (via 51Cr-labeling of red blood cells). By doing so, it became evident that the organs receiving the largest blood supply (kidneys and lungs) were those also containing the largest amount of radioactive muropeptides upon intravenous injection. Such an observation revealed that the homing of intravenously administered muropeptides to different organs closely follows hemodynamic parameters and that most muropeptides remained within the vascular compartment. Possibly as a consequence of this retention within the vasculature (i.e. non-extensive crossing to the tissue parenchyma), it was found that most of the injected free muropeptides were quickly excreted via the urine. Remarkably, however, while the concentration of free muropeptides quickly declined for almost all organs, it remained stable in the brain for at least 24h (Fogler et al. 1985). Notably In the brain and intestines, the accumulation of peptidoglycan over time, following a single injection, could also be due to the presence of cells containing receptors for serotonin, which may bind to certain peptidoglycan derivates (Gershon et al. 1985; Silverman, Wu and Karnovsky 1985).
The authors next compared the intrinsic dissemination profiles to these molecules to derivatives of MDP and MTP made more lipophilic by the attachment of phosphatidylethanolamine (PE). In addition, the authors described what happens to MDP, MTP and their PE-containing counterparts, when they are encapsulated within liposomes. Making these muropeptides more lipophilic by addition of PE, increased their time in circulation and improved their homing to the host organs. When encapsulated, however, the distribution profiles more closely resembled those of the liposomes themselves, and extensive homing to the reticuloendothelial system was observed. Encapsulation of the native, hydrophilic muropeptides was not feasible (4% encapsulation) as they rapidly dissociated from the vesicles. Rendering them more lipophilic allowed for extensive (93%) and long-lasting encapsulation (31-fold higher concentrations after 4h). This enhanced encapsulation was reflected by much higher doses of PE-muropeptide homing to the host organs. Finally, the authors reported that intranasal administration allowed for a more than 12-fold increase in muropeptide homing to the brain, compared to intravenous injection (Fogler et al. 1985).
Another pharmacokinetic profiling study employed a more lipophilic, muramic acid-lacking, MDP-derivative (14C-labelled adamantylamide dipeptide, AdDP) (Walder et al. 1991). Very fast absorption rates were observed upon intravenous and subcutaneous injection of AdDP. In contrast with intravenous injection, however, subcutaneous administration was followed by prolonged distribution and elimination phases. Subcutaneous administration also reduced the bioavailability of AdDP due to deposit retention at the injection site. The authors also reported that the total radioactivity (and hence its concentration) was higher (1.7-fold) in the blood corpuscular elements than in plasma (Walder et al. 1991).
Recently, new light has been shed on this matter. A partial answer on how the host metabolizes peptidoglycan seems to lie in the kidneys. In the non-mammalian animal model Drosophila, renal filtration of microbiota-derived peptidoglycan at steady-state prevents aberrant immune activation (Troha et al. 2019). Nephrocytes, cells homologous to those of the kidney glomerulus, may have a critical role in metabolism and excretion of circulating microbiota-derived peptidoglycan, by degrading it inside lysosomes. Intriguingly, knockout of nephrocytes in flies led to a 3-fold increase in the concentration of circulating peptidoglycan, excessive immune activation and shortened lifespan. The decrease in lifespan was microbiota-dependent, and was the result of chronic inflammation caused by failure to clear peptidoglycan from the systemic circulation. Increased resistance to infection by several bacterial species (but not against all species tested, nor against fungi) was also observed in the absence of nephrocytes. Resistance to infection could be recapitulated by halting the endocytic capacity of mature nephrocytes, showing that this is a mechanism amenable to fine-tuning. The observed effect was the result of improved resistance at the early stages of infection, rather than increased tolerance to the burden associated with the overall infection process (Troha et al. 2019).
Multiple mechanisms are normally in place to prevent excessive immune activation in response to peptidoglycan. However, when peptidoglycan clearance pathways are compromised, it is thought that these immune checkpoints become saturated, leading to excessive immune activation (Hasegawa et al. 2008a; Marinis et al. 2011; Damgaard et al. 2012, 2013; Zhang et al. 2014; Zeissig et al. 2015). Considering that the gut is the main reservoir and source of circulating peptidoglycan, it is not surprising to find that germ-free status protects against the excessive immune activation observed in the absence of nephrocytes. However, mono-colonization with Gram-positive bacteria bearing Lys-type peptidoglycan, but not with DAP-containing bacteria, again allows for aberrant immune activation in the absence of nephrocytes (Troha et al. 2019). It must be noted that since insects do not express NLR family proteins, the observed effect is independent of NOD2 (Harton et al. 2002).
Even though renal clearance might be a major and conserved mechanism for peptidoglycan removal from the systemic circulation, it is not known whether specific subsets of circulating peptidoglycan molecules are preferentially selected for clearance. For example, it is possible that nephrocytes are specifically involved in the uptake and removal of Lys-type peptidoglycan, whilst other cells (e.g. hepatocytes) process other peptidoglycan structures (e.g. DAP-type containing muropeptides) (Troha et al. 2019).
CELLULAR HOMING AND ENTRY
In addition to the multitude of factors influencing the fate and function of peptidoglycan molecules inside the host at a systemic level (including the structural diversity of peptidoglycan and muropeptides, the different potential entry sites, hemodynamic factors, and others), multiple scenarios can be envisioned once peptidoglycan molecules actually reach the surface of host cells. When one considers the question of how, at the cellular and molecular level, peptidoglycan molecules of differing nature home to and enter distinct host cell types, we unveil another level of complexity regarding the interaction between peptidoglycan and the mammalian host. In this section, we dissect the different mechanisms through which peptidoglycan enters the host cells, discriminating these as direct (carrier-independent) or indirect (carrier-dependent) and as primarily bacteria- (e.g. secretion systems and membrane vesicles) or host cell-mediated (e.g. receptor-mediated, peptide transporters).
Bacteria-mediated mechanisms
Cellular invasion
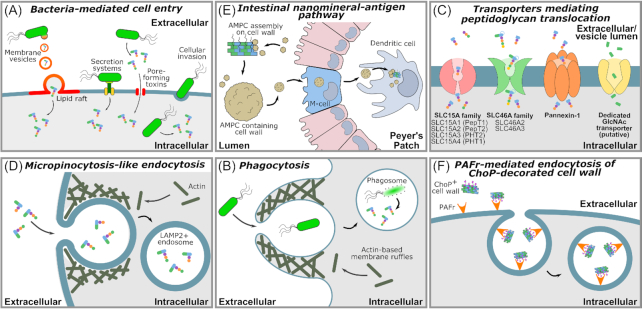
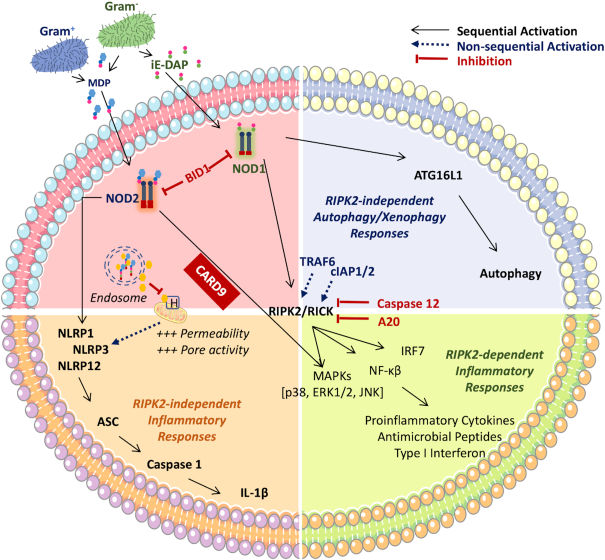
Intracellular bacterial pathogens, such as those belonging to the genera Shigella, Listeria, Francisella and Rickettsia, have evolved to evade the host adaptive immune response, by invading and surviving inside eukaryotic cells where they can find an abundance of nutrients, as well as reduced competition and immune surveillance. Nonetheless, the entry routes typically exploited by these pathogens (e.g. receptor-mediated endocytosis, phagocytosis) may lead to the formation of phagolysosomes and/or autophagy activation with consequent killing of the bacteria (Fredlund and Enninga 2014; Gomes and Dikic 2014; Case and Samuel 2016). Bacterial invasion may thus result in the release of peptidoglycan fragments (Shimada et al. 2009), whose detection via host intracellular receptors (described below) initiates appropriate defense responses against intracellular bacteria (Fig. 3A) (Branković et al. 2015). Among these responses, one finds autophagy directed against the invading pathogens (i.e. xenophagy), a process mediated by the recruitment of the key autophagy protein player Autophagy related 16 like 1 (ATG16L1) to the bacterial entry sites. Depending on the host cell type and the nature of the invading pathogen, this process may or may not require Receptor-interacting-serine/threonine-protein kinase 2 (RIP2), the main signalling adaptor protein of the Nucleotide-binding oligomerization domain (NOD) receptors, and activation of Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) (Travassos et al. 2010; Irving et al. 2014). However, many intracellular invading bacteria have developed ways of avoiding these immune responses, either by modifying the endocytic pathway and phagosome maturation to avoid bacterial killing, or by hijacking the host cell machinery in place to sense bacteria-derived ligands, thereby hampering the host response (Case and Samuel 2016).
Figure 3.
Peptidoglycan uptake mechanisms. Cell wall, Peptidoglycan, muropeptides and their metabolites may be taken up by cells via multiple different, potentially concomitant processes. The following mechanisms have been described: (A), Bacteria-mediated mechanisms of peptidoglycan delivery into host cells. These include direct cellular invasion, bacteria-encoded pore-forming toxins that facility muropeptide entry into the host cell cytosol, direct delivery of muropeptides via bacterial secretion systems, and delivery by bacteria-derived peptidoglycan-containing membrane vesicles that fuse with the host cell membranes. (B), Phagocytosis of bacteria via formation of membrane ruffles followed by lysis of the bacterium, generating peptidoglycan fragments. (C), Uptake of muropeptides or peptidoglycan derivatives via membrane spanning transporter proteins. (D), Micropinocytosis-like actin-dependent endocytic uptake. (E), A ‘Trojan horse’ mechanism where peptidoglycan- or cell wall-sequestering nanomineral particles are taken up by cells. (F), Receptor-mediated endocytosis of bacterial cell wall, mediated through recognition secondary call wall moieties. LAMP2: Lysosome-associated membrane protein 2; PG: peptidoglycan. MVs: membrane vesicles; SLC: solute carrier group of membrane transport proteins; PAFr: Platelet-activating factor receptor; AMCP: amorphous magnesium-substituted calcium phosphate.
Secretion systems
Bacterial secretion systems are highly sophisticated protein complexes used for the delivery of virulence factors across the bacterial cell envelope to the exterior environment. Six classes of secretion system (type I—VI) have been identified in Gram-negative bacteria. In Gram-positive and Gram-negative bacteria, type IV pili and flagella are related to the type II and type III secretion systems respectively, and may also serve a secretory function, whilst the type VII secretion system has been identified uniquely in Mycobacteria and other Gram-positive pathogens. Of these, the type III, IV and VI secretion systems can deliver effector molecules from the bacterium directly to the cytoplasm of eukaryotic host cells. Effectors classically take the form of proteins and, in the case of the type IV secretion system (T4SS), DNA. However, one pathogen has been described as transporting peptidoglycan via this system (Fig. 3A). The T4SS of the gastric pathogen Helicobacter pylori is encoded by the cag pathogenicity island (cagPAI), a ∼40 kb DNA insertion element containing approximately 32 genes (Akopyants et al. 1998; Backert, Tegtmeyer and Fischer 2015). While cagA- strains are found primarily in the mucus layer and apically to the gastric epithelial cells, strains carrying the cagPAI (cagA+) are able to penetrate the gastric mucus layer, and colonize in more intimate contact with the epithelial cells (Camorlinga-Ponce et al. 2004). Overall, one could hypothesize that H. pylori uses the cagPAI T4SS to target beneficial peptidoglycan bioactivities towards gastric epithelial cells, potentially promoting the epithelial environment to favour colonization. For the host however, cagA + strains are associated with increased risk of severe gastritis, atrophy, dysplasia and malignancy (Blaser et al. 1995; Kuipers et al. 1995; Peek et al. 1995). H. pylori was demonstrated to deliver radiolabeled-peptidoglycan directly into host gastric epithelial (AGS) cells via this T4SS, as AGS cell intracellular radioactivity was 50%–90% lower when exposed to H. pylori lacking a functional T4SS.(Viala et al. 2004) As such, this system probably contributes to most of the H. pylori peptidoglycan uptake by gastric epithelial cells. Peptidoglycan delivery through the cag T4SS is dependent on the activity of the lytic transglycosylase Slt, a glycoside hydrolase 23 family 1 protein that generates 1,6-anhydromuropeptides (Viala et al. 2004; Nagy et al. 2009). Slt is the major enzyme responsible for peptidoglycan turnover in H. pylori, and thus generates the muropeptides translocated via the cagPAI T4SS (Viala et al. 2004; Nagy et al. 2009).
Inside host cells, peptidoglycan binds the NOD1 receptor and leads to NF-κB, p38 and extracellular-signal-regulated kinase (Erk) activation (Strober et al. 2006). This activation is accompanied by increased production of pro-inflammatory cytokines such as macrophage inflammatory protein 2 (MIP-2), β-defensin and IL-8 (Viala et al. 2004; Allison et al. 2009). In addition, NOD1 activation by H. pylori peptidoglycan induces production of type I interferon (IFN), which is implicated in Th1 cell differentiation and further promotes gastric inflammation (Watanabe et al. 2010). The injected peptidoglycan also activates phosphoinositide 3-kinase (PI3K) and directly induces the migration of gastric epithelial cells. PI3K activation participates in gastric carcinogenesis and thus connects H. pylori peptidoglycan delivery with gastric malignancy.
The case of peptidoglycan trafficking via the T4SS of H. pylori raises a number of questions. Is trafficking of peptidoglycan via the T4SS a unique mechanism that has evolved in H. pylori, or is it common to pathogens employing T4SS? Do other types of secretion system transport muropeptides? Given that the major host receptors for Gram-negative peptidoglycan (NOD1 and NOD2) are cytoplasmic, muropeptide delivery by bacterial secretion systems across the host cell membrane is an enticing target to further understand how bacteria regulate the host immune response during infection.
Membrane vesicle secretion
Many bacteria release lipid membrane vesicles (MVs) that perform a variety of functions for the producing bacteria, including the delivery of cargo to other cells (Fig. 3A). MVs generated by didermal Gram-negative bacteria typically bud from the outer membrane, and are thus specifically termed outer membrane vesicles (OMVs). OMVs can act as shuttles for antigens and virulence factors, including peptidoglycan species, and are thus important mediators of host-pathogen interactions and peptidoglycan uptake (Cecil et al. 2019). OMVs from H. pylori, Pseudomonas aeruginosa and Neisseria gonorrhea carrying peptidoglycan have been shown to activate NOD1, culminating in downstream induction of NF-κB-dependent inflammatory phenomena and the development of adaptive immune responses against bacteria derived-membrane vesicles (Kaparakis et al. 2010). Accordingly, while epithelial cells are normally refractory to external stimulation with peptidoglycan, its incorporation within OMVs facilitates passage of peptidoglycan into the epithelial cell and presentation in a way that triggers the activation of cytosolic NOD1 (Kaparakis et al. 2010).
OMVs preferentially fuse with the host cell membrane via lipid rafts (Kesty et al. 2004; Mondal et al. 2016). Depletion of lipid rafts on the surface of epithelial cells by treatment with Fumonisin B1 (an inhibitor of sphingomyelin incorporation), or with the cholesterol-depleting agent methyl-ß-cyclodextrin, both reduced the fusion of OMVs containing peptidoglycan, and NOD1-dependent immune responses. In turn, cholesterol replenishment restores the fusion of OMVs and NOD1-mediated NF-κB responses. The same authors hypothesized that NOD1 may associate with the cytoplasmic face of the human epithelial cell membrane, and that the uptake of peptidoglycan-containing OMVs via lipid rafts may render peptidoglycan accessible to membrane-associated NOD1 (Kaparakis et al. 2010). In addition, lipophilic acryl residues produced by bacteria, associated with γ-D-glutamyl-mDAP acid (γ-iE-DAP, the recognition core of Nod1 stimulatory molecules) have been shown to potentiate the NOD1 stimulatory activity of iE-DAP, thus suggesting the presence of certain bacteria-derived lipid species to drive peptidoglycan-mediated NOD1 signalling (Hasegawa et al. 2007). Supporting the aforementioned hypothesis is the observation that NOD1 does localize to the cytoplasmic membrane in (at least) human epithelial cells, and that the addition of invasive Shigella to these cells leads to further recruitment of membrane-associated NOD1 to the specific lipid raft domain-containing focal points of bacterial entry (Kufer et al. 2008).
As a final note, innate and adaptive immune responses triggered by peptidoglycan-containing OMVs are, in fact, triggered by peptidoglycan-mediated NOD1 activation and not by TLR-activating species present in outer membrane vesicles. Myeloid differentiation primary response 88 (MyD88) and Myelin and lymphocyte protein (Mal) KO mice are defective in all known TLR pathways, but display no alterations in immune responses against such peptidoglycan-containing vesicles. In turn, NOD1 KO mice do not produce Cxcl2 mRNA responses, and do not display OMV-specific IgG antibodies (Kaparakis et al. 2010). How these adaptive immune responses against peptidoglycan-containing OMVs are mounted is not yet established. It is possible that antigen presenting cells also take up such vesicles, leading to surface antigen presentation and induction of antibody production by effector B cells. Alternatively, peptidoglycan molecules taken up at the apical side of epithelial cells, may be released by the basal side of these same cells via exosomes (as recently suggested in Bu et al. 2010), so that vesicle-mediated transportation of peptidoglycan may occur in vivo between host mammalian cells.
More recently, other types of MV have been identified. Gram-negative outer-inner membrane vesicles (OIMVs) have been isolated, and enclose both cytoplasmic material and a periplasm replete with peptidoglycan layer (Pérez-Cruz et al. 2015). OMVs produced by cell lysis may also contain cytoplasmic content. Monodermic Gram-positive bacteria have been found to release MVs, also called cytoplasmic membrane vesicles (CMVs) (Lee et al. 2009a; Andreoni et al. 2019). These CMVs have been associated with phage endolysin activity that destabilizes the peptidoglycan layer (Qing et al. 2019; Toyofuku, Nomura and Eberl 2019). Gram-positive CMVs and Gram-negative OIMVs are not well studied. However, given the presence of cytoplasm and cytoplasmic membrane, it is interesting to speculate that they may encapsulate peptidoglycan precursor muropeptides and mature muropeptides (undergoing turnover) captured from the bacterial cytoplasm. In turn, the CMV surface could be decorated with patches of polymeric peptidoglycan, whilst OIMVs have been shown to possess a periplasmic peptidoglycan layer. The presence of peptidoglycan precursors in MVs is of particular interest, since lipidated muropeptides generally have enhanced bioavailability and illicit stronger immune responses (Griffin et al. 2019). The potential consequences for uptake and immune activation of host cells by peptidoglycan, transported via these novel classes of MV, remains to be studied.
Host-mediated mechanisms
Phagocytosis
Upon encountering commensal and potentially pathogenic bacteria, the host professional phagocytes (including monocytes, macrophages, dendritic cells, osteoclasts, eosinophils and neutrophils) may engulf these bacteria via phagocytosis (Fig. 3B) (Rabinovitch 1995). Phagocytosis starts with the recognition and ingestion of potential pathogens followed by their engulfment into a plasma membrane-derived vesicle, the phagosome. The recognition step may occur directly via the recognition of pathogen-associated molecular patterns (PAMPs) by the host cell, or indirectly via recognition of opsonized bacteria (covered in antibodies, complement, fibronectin, mannose-binding lectin or other globulins) (Flannagan, Jaumouillé and Grinstein 2012). The bacteria-containing phagosome may then fuse with endosomes and lysosomes, leading to the formation of an enclosed environment capable of degrading the bacteria and generating polymeric peptidoglycan fragments and muropeptides (e.g. low pH, the presence of oxygen and nitrogen reactive oxygen species, nutrient starvation, antimicrobial peptides and enzymes). Phagocytosed bacteria can thus represent a long-lasting source of muropeptides which can be then be sensed by the host cell. Phagocytic cells may then directly activate other non-phagocytic cells in order to mount a response against potential invading pathogens.
While the host is endowed with mechanisms to degrade phagocytosed bacteria leading to the release of muropeptides from endosomal compartments, certain bacterial species may also actively put their own machinery for muropeptide release in place once phagocytosed. The pathobiont Streptococcus pneumoniae colonises the oro- and nasopharynx of up to 60% of target populations. Whilst colonisation is in most cases temporary and asymptomatic, it may also lead to life-threatening invasive infections such as community-acquired pneumonia and meningitis, accounting for more than 1 million infant deaths (even more among the elderly) worldwide every year (Henriques-Normark and Tuomanen 2013). Among the important S. pneumoniae virulence factors one finds the exotoxin pneumolysin, a member of the cholesterol-dependent cytolysins expressed across several Gram-positive bacterial species, which causes membrane rupture upon oligomerization on cholesterol-containing membranes (Kadioglu et al. 2008; Mitchell and Mitchell 2010; Henriques-Normark and Tuomanen 2013). Several different protein variants of pneumolysin exist, displaying a spectrum of hemolytic activity and accounting for differential invasiveness across the more than 90 S. pneumoniae serotypes (Weinberger et al. 2010). Streptococcus pneumoniae peptidoglycan is sensed by NOD2 in a pneumolysin-dependent way, as the enzyme facilitates the leakage of muropeptides into the cytosol upon bacterial phagocytosis and phagosomal degradation (Opitz et al. 2004; Davis, Nakamura and Weiser 2011; Koppe, Suttorp and Opitz 2012). Subsequently, a response characterized by the activation of NF-κB-induced genes, IL-1β and the NLRP3 inflammasome is put in place (see below for a detailed description of these signalling pathways). A similar mechanism of lysosomal rupture and leakage of bacterial contents into the cytosol, leading to the activation of the NLRP3 inflammasome, has also been reported for β-hemolysin in Streptococcus pyogenes infection (although peptidoglycan-mediated effects were not investigated) (Gupta et al. 2014).
A note on cell surface receptors
It seems logical that efficient uptake of free peptidoglycan and muropeptides by host cells would be initiated by a capture step, facilitated by cell surface receptor proteins. Indeed, fractionation studies performed in the 1980s suggested that peptidoglycan binds at the cell surface of lymphocytes in a tissue-dependent manner (Dziarski 1987). Nevertheless, few studies have definitively identified such receptors. Whilst the SLC15 transporter family is becoming well characterized for their muropeptide transporting activity, other receptors, such as CD14, have received little attention beyond their initial identification, and therefore remain to be solidly established as peptidoglycan receptors. Thus, this field is very much open for further investigation, and quite likely, new receptor discoveries. The putative peptidoglycan receptors identified so far are discussed in more detail under the relevant headings below.
One potential, major cell surface receptor of peptidoglycan is the PRR Toll-Like Receptor 2 (TLR2). A wealth of data is available that posit major immunological consequences of TLR2 activation by peptidoglycan. However, the recognition of peptidoglycan by TLR2 remains controversial. Carefully conducted biochemical studies, devoid of contaminating material, have explored the chemical nature of muropeptide recognition by TLR2. Nevertheless, these have generated conflicting data supporting or opposing a role for TLR2 in peptidoglycan recognition. A further complication is that immunological studies frequently involve the use of commercial preparations of peptidoglycan, some of which have been shown to contain other cell wall components that are known TLR2 ligands (Travassos et al. 2004; Hashimoto et al. 2006). Essentially, peptidoglycan could act as a carrier for presentation of other TLR2 ligands (i.e. by our previous definition, TLR2 is presented with and recognizes cell wall).
While some studies report TLR2 as a peptidoglycan receptor (Yoshimura et al. 1999; Dziarski and Gupta 2005), others have shown that TLR2 fails to recognize both muropeptides and highly purified polymeric peptidoglycan from several different bacteria species (Inohara et al. 2003; Travassos et al. 2004). However, more recent studies have provided strong evidence supporting that even contaminant-free, naked peptidoglycan can be bound by TLR2. For instance, it has been shown that isolated human TLR2 binds both synthetic DAP-type and Lys-type muropeptides (as measured by surface plasmon resonance), that modifications on the carboxylic acids of isoglutamine and mDAP modulate muropeptide binding to TLR2 (which may complicate matters when employing synthetic ligands), and that highly purified peptidoglycan from multiple species leads to TLR2-mediated cellular activation, although the ability of the synthetic muropeptides to activate cellular responses was not tested (Asong et al. 2009). Similarly, another study employed polymeric peptidoglycan isolated from Staphylococcus aureus Δlgt (defective in lipidation of pro-lipoproteins, the precursors of TLR2 ligands) in order to exclude contamination with lipoproteins and tested it on murine keratinocytes. Upon uptake via a NOD2- and TLR2-independent endocytosis-like process, naked polymeric peptidoglycan colocalized with, and independently activated, NOD2 and TLR2 to a similar extent. Knocking out either one of these receptors resulted in a 50% reduction in IL-6 and IL-1β production (Müller-Anstett et al. 2010). This synergistic activation is in agreement with that observed by others (Natsuka et al. 2008). However, in a subsequent study, the same S. aureus Δlgt mutant failed to induce TLR2-dependent costimulatory effects in murine bone marrow derived dendritic cells, murine macrophages (J774 cells) or human monocytes (MM6 cells). In contrast, polymeric peptidoglycan from the lipoprotein-replete SA113 parent strain stimulated TLR2-dependent and NOD2-dependent cytokine production, and enhanced maturation parameters (Schäffler et al. 2014).
One critical point seems pertinent in addressing the conflicting biochemical data: what is the physiologically relevant concentration at which TLR2 encounters activating-muropeptide structures in vivo, and at what concentration are they sufficient to initiate a host response? Note that the peptidoglycan potency is lower compared to that observed for typical contaminants present in commercial peptidoglycan preparations (including TLR2 agonists) (Müller-Anstett et al. 2010). Hence, it is possible that the peptidoglycan TLR2-activating capacity is negligible when compared side-by-side to that of ‘contaminated’ peptidoglycan. However, even if the peptidoglycan contribution to TLR2 activation in vivo is orders of magnitude smaller than that of the remaining TLR2 ligands, it should not be discarded. Studies have suggested that muropeptides are present in tissues and biofluids at sub-microgram to low microgram (per g or mL) quantities (Sen and Karnovsky 1984; Hoijer et al. 1995; Fox et al. 1996; Xu et al. 2008; Huang et al. 2019). Studies reporting TLR2 activation by peptidoglycan employ concentrations as low as 100 ng/mL (e.g. Dziarski and Gupta 2005).
One must also consider a complicating factor, that the overall concentration of muropeptides in biofluids may not reflect the local concentration. For example, proximity of receptors to transporters could increase local concentration of muropeptides, whilst vesicle encapsulation would mask the molecular cargo from nearby cognate receptors. It is also important to note that, even though TLR2 is typically considered as a surface receptor, its colocalization with peptidoglycan is found intracellularly (Müller-Anstett et al. 2010), which goes in line with its recruitment to endosomal compartments (Underhill et al. 1999; Eddie Ip et al. 2008; Dietrich et al. 2010). Thus, one should be careful when employing reporter assays using cells that would not otherwise express significant amounts of TLR2, as these could theoretically be defective in relevant recruitment pathways (as observed for other TLRs e.g. Koehn et al. 2007; Isnardi et al. 2008). By addressing these issues, and by understanding the context in which TLR2 encounters peptidoglycan in vivo, we should be able to clarify the uncertainty surrounding the TLR2 conundrum.
Peptide transporters
Integral transmembrane proton-coupled oligopeptide transporters shuttle dipeptides, tripeptides and peptidomimetics into cells. In general, eukaryotic cells employ solute carrier (SLC) family membrane proteins rather than ATP-binding cassette (ABC) transporters for the uptake of soluble molecules (Hediger et al. 2013). Of these, the SLC15 transporter family has a leading role in the translocation of NOD1 and NOD2 agonists into epithelial and dendritic cells (Fig. 3C) (Vavricka et al. 2004; Ismair et al. 2006; Dalmasso et al. 2010; Hediger et al. 2013). SLC15A1 (PepT1), SLC15A2 (PepT2), SLC15A4 (PhT1), and SLC15A3 (PhT2) (Lee et al. 2009b; Sasawatari et al. 2011; Nakamura et al. 2014; Wang et al. 2018) have all been implicated in the uptake of different muropeptide species (reviewed in Smith, Clémençon and Hediger 2013). For instance, SLC15A1 (PepT1) has been shown to transport MDP and triDAP molecules into human epithelial intestinal cells (Vavricka et al. 2004; Ismair et al. 2006; Dalmasso et al. 2010), whilst SLC15A2 (PepT2) transports triDAP into lung epithelial cells (Swaan et al. 2008). PepT2 was also shown to facilitate MDP uptake by bone marrow derived macrophages (BMDCs), although the authors noted that mechanisms other than transmembrane transport of MDP by PepT2 were possible (Hu et al. 2018). PhT1 (SLC15A4) and PhT2 (SLC15A3) have been implicated in MDP transport to the cytosol across endosomal membranes (Nakamura et al. 2014). PhT1 is additionally reported to transport triDAP (Lee et al. 2009b).
In terms of tissue expression, the expression pattern (at the mRNA and protein level) of PepT1 in the brain is particularly interesting as it is highest in the early postnatal period and declines in adulthood, which may correlate with the increased degree of colonization and expected increased load of circulating peptidoglycan fragments. In addition, its expression is dependent on colonization status (Arentsen et al. 2017). Both PhT1 and PhT2 are abundantly expressed in the brain (Yamashita et al. 1997) and lymphatic system (Sakata et al. 2001), being described as endosomal and lysosomal transporters with limited information regarding their substrate specificity for oligopeptides and histidine. The SLC15 family transporters may also be functionally differentiated depending on their cellular localisation. For example, PepT2 and PhT1 were proposed to act cooperatively in the transport of MDP into BMDCs, by differentially localising to the cytoplasmic membrane and endosomal compartments, respectively (Hu et al. 2018).
It should be noted that there is some conflict in the literature regarding the specific ligands that can or cannot be transported by the SLC15 family receptors. Such differences between studies may be attributed to the types of assay used and the specific study conditions (e.g. glycyl-sarcosine competition assay, fluorescent muropeptide probes, immune response readouts, differences in cell culture system, concentration of muropeptides). Thus, the true extent to which these transporters differ in their muropeptide substrate preference remains an open question. Members of the SLC15 transporter family cannot, however, mediate the uptake of all muropeptides. For instance, DAP-containing anhydro-muropeptide monomers, classically called ‘tracheal toxins’, are not taken up by SLC15 transporters, but by the SLC46 family of transporters (Paik et al. 2017). Humans and mice alike express three SLC46 transporters. SLC46A1 (proton-coupled folate transporter) is responsible for the intestinal uptake of folate/antifolates (Qiu et al. 2006). Human SLC46A2 (TSCOT) is abundantly expressed in the thymic cortical epithelium (Kim et al. 2000), and probably in the skin in both humans and mice (Harder and Nú̃ez 2009), and SLC46A3 is yet to be characterized. Both human and mouse SLC46A2, and mouse SLC46A3, have been shown to facilitate the uptake of DAP-containing monomers, as well as MDP. These receptors localize to acidic subcellular organelles (late-endosomes and/or endolysosomes) and, upon exposure to their muropeptide ligands, have been shown to aggregate and to trigger the NF-κB signalling pathway via NOD1/RIP2 activation (Paik et al. 2017).
It is intriguing that both SLC15 and SLC46 transporters facilitate the uptake of such similar muropeptides considering that, among the more than 390 SLCs present in humans, the SLC15s and SLC46s are considered to be relatively distinct when it comes to their cargos (Hediger et al. 2013), SLC15s being involved in amino acid/oligopeptide transport whilst SLC46s are implicated in shuttling of folic acid, and more closely related to the organic ion transporters (Schlessinger et al. 2010). It is tempting to speculate that other, unanticipated human proteins might be implicated in peptidoglycan recognition, shuttling and processing.
A third member of the SLC family is of interest as muropeptide transporter. A variety of Gram-negative bacteria encode a transmembrane permease, ampG, which participates in the recycling of muropeptides generated by lytic translgycosylase activity, by functioning as a transporter of GlcNAc-anhMurNAc disaccharide and GlcNAc-anhMurNAc-peptides (Cheng and Park 2002). An orthologue of E. coli ampG was identified in humans and other mammals—the major facilitator superfamily domain containing 3 (Mfsds3) (Park and Uehara 2008). MFSD3 has been reported as a new, putative member of the SLC family, most closely related to the acetyl-CoA transporter family SLC33 (Perland et al. 2017). It has a broad tissue expression in C57BL/6 mice, most notably in the liver, kidney and throughout the central nervous system, where immunofluorescence staining of mouse brain sections revealed a specific localization to the plasma membrane of neurons. Whether MFSD3 has in role in the transport of muropeptides, and the potential consequences for the host, is undoubtedly an enticing target for further study.
Pannexins are a family of vertebrate proteins that form hemichannels (hexamers of homomeric or heteromeric pannexins) that generate a pore in the membrane of cellular organelles, or the cytoplasmic membrane leading into the extracellular space. Their pore has a large internal diameter that allows the diffusion of relatively small molecules (∼1 kDa) (Bao, Locovei and Dahl 2004; Ma et al. 2012). The Pannexin-1 hemichannel has been shown to act as a transporter for muropeptides (Marina-García et al. 2008). When MDP-rhodamine was endocytosed in bone marrow-derived macrophages, it localized to acidified vesicles in a punctate granular pattern consistent with vesicular localization. Subsequently, ATP stimulation triggered Pannexin-1 association with the P2X7R protein, leading to channel formation and MDP release from the vesicular compartment to the cytosol. This rapid re-localization (<2 min) induced by ATP required a functional pannexin-1 protein and led to the activation of caspase-1 in a NLRP3-dependent but NOD2-independent manner (Marina-García et al. 2008).
Connexins are a family of proteins that form intercellular gap junctions, consisting of two juxtaposed hemichannels on the surface of two adjacent cells, forming a connected channel (Bennett et al. 2003; Sáez et al. 2003). Gap junctions are important not only for direct cellular activation (i.e. upon direct antigen exposure) but also for the propagation of antigen presentation, via gap junction-mediated intercellular antigen diffusion, in a process called cross-presentation (Neijssen et al. 2005; Matsue et al. 2006; Pang et al. 2009). Although they do not share sequence homology, connexins have an organization highly similar to that of pannexins (pannexins have a glycosylated extracellular domain that does not allow for the formation of cell-to-cell direct communication), and thus connexins are interesting targets for further exploration as muropeptide transporters. To our knowledge, transport of muropeptides by connexins has not been investigated, however the large pore size of hemichannels and gap junctions could easily accommodate the majority of monomeric muropeptides, and therefore connexins could potentially facilitate the propagation of muropeptide signals between cells (Boassa et al. 2007). It is interesting to note that whilst connexins may facilitate antigen diffusion and cross-presentation, drive immune response propagation and amplification, they may also allow for the development of tolerance phenomena (Mazzini et al. 2014). The potential for muropeptides to be propagated between cells, potentially contributing to inflammatory or tolerogenic outcomes, is undoubtedly worth further investigation. It is worth noting that peptidoglycan can regulate connexin expression. Peptidoglycan from S. aureus reduces connexin expression and functional gap junction formation in astrocytes via a p38/MAPK-dependent mechanism (Esen et al. 2007). In contrast, exposure of microglia cells to S. aureus peptidoglycan results in cellular activation and increased connexin 43 expression, allowing for the formation of functional intercellular communications via gap junctions. Peptidoglycan-induced gap junction formation may, therefore, be important for the mounting of effective immune responses, and for clearance and regenerative processes (Garg, Syed and Kielian 2005; Eugenin et al. 2012).
Actin-dependent endocytosis
Endocytosis is a process by which cells internalize material from the exterior environment, by invagination of the cytoplasmic membrane to form an intracellular vesicle (reviewed in McMahon and Boucrot 2011). An actin-dependent pathway for the endocytosis of bacterial cell wall fragments has been reported for epithelial and endothelial cells. This pathway is independent of caveolin-mediated endocytosis, as caveolin 1 (Cav1) depletion using siRNA does not inhibit cell wall uptake (Loh, Gao and Tuomanen 2017). However, it displays several features of the receptor-independent fluid uptake mechanism known as micropinocytosis, a generalized endocytic process through which nutrients and other molecules are acquired by eukaryotic cells. Micropinocytosis is dependent on actin dynamics, Cdc42, Rac1 and PI3K, but is independent of RhoA. Accordingly, inhibition of actin polymerization with cytochalasin D, inhibition of PI3K with wortmannin, inhibition of the GTPase Cdc42 with Pirl1, or inhibition of the GTPase Rac1 with EHT1864, all independently reduce cell wall internalization by 40–50% in Human Brain Microvascular Endothelial Cells (HBMEC), and in A549 (alveolar basal epithelium) cells (Loh, Gao and Tuomanen 2017). In turn, RhoA inhibition with rhosin does not alter cell wall uptake (Loh, Gao and Tuomanen 2017). Further similarities to micropinocytosis include the accumulation of an actin ring at the cell wall-binding sites (Loh, Gao and Tuomanen 2017). However, cell wall uptake is not accompanied by hallmarks of micropinocytosis such as enhanced fluid uptake or membrane ‘ruffles’. This pathway thus differs from classical micropinocytosis, whilst retaining several of its intracellular hallmarks, and bears similarity to the mechanism involved in human papillomavirus type 16 (HPV-16) endocytosis (Schelhaas et al. 2012). Upon cell wall internalization by this ‘micropinocytosis-like’ pathway, cell wall fragments localize to late endosome or lysosome compartments (lysosome-associate membrane glycoprotein 2 (LAMP2)-positive compartments), indicating that the actin-dependent pathway leads to cell wall trafficking within lysosomes (Fig. 3D).
It has been reported that cytoplasmic NOD1 and NOD2 are recruited to endosomes containing peptidoglycan. Such positioning allows for the detection of its ligands at localised sites of highest concentration, presumably contributing to a faster, more potent and more efficient receptor activation, and interaction with downstream mediators (e.g. RIP2) and respective biological responses (Irving et al. 2014; Nakamura et al. 2014). The NOD receptors localise to the endosome membrane, and may thereby directly detect muropeptides as they are released from the endosomal compartment via the endosomal peptide transporters or other membrane channels. However, if such release occurs for small muropeptides (e.g. MDP or MTP), it may not be possible for larger muropeptides or cell wall fragments containing NOD activating motifs. For instance, recognition of Streptococcus pneumoniae peptidoglycan by NOD2 in macrophages has been shown to require both hydrolysis of the peptidoglycan by lysozyme, and endosomal membrane puncture by pneumolysin (Davis, Nakamura and Weiser 2011).
Indirect mechanisms
Intestinal nanomineral-antigen pathway
Nano-sized amorphous magnesium-substituted calcium phosphate (AMCP) species are nanomineral particles constitutively formed through self-assembly in the intestinal lumen. These endogenous intestinal nanominerals also constitute an alternative route through which luminal antigens can be sampled in the gut, by acting as shuttling vehicles for minerals, proteins and microbial components including peptidoglycan (Pele et al. 2017). AMCP particles traverse specialized epithelial M cells and are released into the underlying Peyer patches, where they are scavenged by antigen-presenting cells (APCs; Fig. 3E). AMCPs and their cargo have been shown to reach the endosomal or lysosomal compartments of APCs in vivo, providing targeted co-delivery of antigens and additional immune modulators (e.g. cell wall or peptidoglycan) to APCs (Powell et al. 2015). Recognition and cellular handling of bacterial components carried by AMCPs display an immuno-regulatory role. Accordingly, a recent study demonstrated that whilst synthetic amorphous magnesium-substituted calcium phosphate (sAMCP) nanomineral particles are themselves largely devoid of immune activity, sAMCPs containing Staphylococcus aureus peptidoglycan shuttled the peptidoglycan to APCs and thereby act as immune-regulatory agents (Hewitt et al. 2017). The ability of AMCPs to adsorb biological materials has also been exploited for its use as adjuvants to achieve enhanced immune responses (Relyveld 1986; Heit et al. 2007; Slütter et al. 2009). In turn, some AMCP-shuttled MAMPs have been shown to induce tolerance under certain conditions (Hedl and Abraham 2011; Wolfle et al. 2011; Nahid et al. 2013), thereby reinforcing that the nature of such adsorbed materials (and not the AMCPs themselves) is responsible for dictating the form and extent of the modulated immune response.
AMCP-mediated co-delivery of antigen with bacterial peptidoglycan is believed to allow for tolerogenic signalling, whereby peptidoglycan attenuates the immune response against an otherwise strong immunogenic molecule. Accordingly, intestinal nanomineral carriers containing both antigen and peptidoglycan are capable of attenuating antigen-specific CD4+ T cell (adaptive) responses, resulting in reduced T cell proliferation to a cognate recall antigen (protein-purified derivative of tuberculin) (Hewitt et al. 2017). APCs that have taken up peptidoglycan via AMCPs in intestinal lymphoid tissues display upregulation of the immuno-inhibitory receptor Programmed death receptor ligand 1 (PD-L1) in a NOD1/NOD2-dependent manner (Powell et al. 2015). In vitro, the presence of peptidoglycan in AMCPs induces a shift in blood-derived monocytes towards a phenotype less suited to the promotion of Th1 type CD4+ T cell responses. This phenomenon is to a large extent regulated by the secretion of the anti-inflammatory cytokine IL-10 (Hewitt et al. 2017). IL-10 is secreted by APCs in the lamina propria and Peyer's patches, and it is known to be involved in the maintenance of intestinal tolerance (Hauer et al. 1998; Tsuji, Mizumachi and Kurisaki 2001; Denning et al. 2007; Takada et al. 2010). Early exposure to TLR agonists has been shown to block the differentiation of monocytes into mature APCs, resulting in a PD-L1+ tolerogenic APC phenotype that fails to induce Th1 cell proliferation, but which induces regulatory T cells (Wolfle et al. 2011). As peptidoglycan-containing AMCPs seem to lead to a similar attenuation of Th1 type CD4+ T cell responses, one can hypothesize that peptidoglycan is involved in the triggering/maintenance of a similar tolerogenic phenotype.
The down-regulation of cell surface MHC II molecules is one mechanism through which IL-10 suppresses adaptive immune responses (Koppelman et al. 1997; Thibodeau et al. 2008). In addition, cell surface expression of PD-L1 in monocytes might also dictate the type of T cell that proliferates, favouring the maturation of regulatory T cells (Francisco et al. 2009; Fukaya et al. 2010). Consistent with the suggestion that peptidoglycan has anti-inflammatory potential when taken up via the nanomineral pathway, is the observation that the immuno-inhibitory receptor PD-L1 fails to be upregulated when recognition of peptidoglycan (MDP) by NOD2 is defective, such as in the case of Crohn's Disease intestinal lymphoid tissue (Hewitt et al. 2012; Robertson et al. 2016). Thus, peptidoglycan delivered to APCs through the AMCP nanomineral pathway may act as an immune-modulator with predominantly anti-inflammatory potential. It is not yet known whether nucleation of AMCPs is affected by particular features of the peptidoglycan chemistry or other cell wall components, which could favour or oppose AMCP formation and uptake via this pathway.
Molecular mimicry
In mammals, the platelet-activating factor receptor (PAFr) is a G-protein-coupled receptor that induces endocytosis upon activation by the chemokine platelet-activating factor (PAF). Recognition of PAF by PAFr occurs via the phosphorylcholine (ChoP) moiety, and respiratory pathogens are thought to mimic PAF in order to induce PAFr-mediated endocytosis by decorating their surface with ChoP. However, this translocation is not restricted to live bacteria, as cell wall fragments decorated with ChoP are internalized by eukaryotic cells via PAFr recognition (Fig. 3F) (Cundell et al. 1995; Ring, Weiser and Tuomanen 1998; Rijneveld et al. 2004; Fillon et al. 2006). In vitro studies have shown that, when mediated by PAFr, endocytosis of cell wall fragments is β-arrestin-, clathrin- and dynamin-dependent (McLaughlin et al. 2006; Eckels et al. 2009; Loh, Gao and Tuomanen 2017). Treatment of human brain microvascular endothelial cells (HBMEC) and human adenocarcinoma lung epithelial cells (A549) with dynasore, a dynamin inhibitor, reduces the uptake of CypHer5E-labelled cell wall fragments from Streptococcus pneumonia by 50%. Similarly, small interfering RNA (siRNA)-mediated depletion of endogenous clathrin heavy chain and endophilin A2 (which reshapes the membranes before scission in the case of clathrin-independent endocytic events) in HBMEC and A549 cells reduces cell wall uptake by ∼20% (Loh, Gao and Tuomanen 2017). Hence, multiple membrane scission, endocytic and antigen uptake events occur in parallel, whereby endophilin A2, dynamin and actin all act together to drive independent but synergistic pulling forces.
Even though cell wall fragments from pneumococci interact with both TLR2 and PAFr in HBMEC and A549 cells (Cundell et al. 1995; Yoshimura et al. 1999), the internalization of pneumococcal cell wall fragments is still observed in the absence of TLR2 (Loh, Gao and Tuomanen 2017). Thus, there seems to be a division between the cell surface receptors involved in cell wall recognition leading to internalization, and cell surface receptors involved in recognition that triggers immune responses. Accordingly, cells treated in vitro with the PAFr antagonist CV-3988, PAFr knockouts and PAFr/TLR2 double knockouts, but not TLR2 single knockouts, all display a 40–50% reduction of cell wall internalization (Loh, Gao and Tuomanen 2017). When allowed to proceed, the PAFr-dependent pathway has been shown to account for only ∼40% of cell wall fragment endocytic events. Moreover, cell wall fragments of pathogens devoid of ChoP are internalized in PAFr knockout cells, highlighting the presence of host mechanisms other than PAFr activation for the internalization of cell wall fragments (Loh, Gao and Tuomanen 2017). As seen for actin-mediated uptake of cell wall fragments, PAFr-mediated uptake also seems to involve lysosomal trafficking because PAFr knockout cells display less cell wall within LAMP2 compartments (Loh, Gao and Tuomanen 2017). Thus, it is likely that these two uptake pathways converge at the level of the lysosomal compartment and share the same downstream intracellular processing pathways. The degree of overlap, and the receptors involved in these two different, downstream aspects of cell wall recognition are most likely cell type- and context-dependent.
PEPTIDOGLYCAN DETECTION IN MAMMALS
Upon reaching the intracellular compartment, peptidoglycan-mediated signalling is considered to be well established, as a result of comprehensive in vitro studies. A word of caution should be put forward, as these may not fully reflect the way host cells respond to peptidoglycan molecules in vivo. For example, in vitro cultures of immortalized cells may not necessarily express the same transporters as their primary cell counterparts, co-stimulation and co-activation phenomena are likely to take place in vivo, immune memory is hard to account for in vitro, and sequential presentation across different cell types may lead to chemical modifications of peptidoglycan-derived molecules. Nevertheless, in vitro studies have been valuable in elucidating the mechanisms through which peptidoglycan influences host (patho)physiological processes, and some of the players acting downstream of peptidoglycan recognition (discussed below) have well established roles in multiple disease phenotypes across a range of human and animal pathologies, thereby validating their role in sensing and responding to peptidoglycan.
Carbohydrate-binding domains
C-type lectins are a diverse superfamily of mainly Ca2+-dependent proteins that bind a variety of carbohydrates (including the glycan moieties of peptidoglycan) via carbohydrate recognition domains (CRDs), and function as innate immune PRRs (Hoving, Wilson and Brown 2014). The presence of at least one CRD is the defining feature of a C-type lectin. In mammals, C-type lectins secreted in the blood initiate the lectin-pathway of the complement cascade, via a general mechanism of binding to GlcNAc residues on bacterial peptidoglycan. The archetype protein player of this pathway is the collectin mannose-binding lectin (MBL), which in humans includes a serum-secreted form, and two lung surfactant proteins (SP-A and SP-D). The collectin CL-11 can also initiate the lectin pathway (Keshi et al. 2006; Hansen et al. 2010). Although these collectins bind GlcNAc, they have strong affinity for other sugar residues and therefore target a broad range of microorganisms.
Another group of C-type lectins that initiate the lectin-complement pathway through peptidoglycan recognition are the ficolins, a group of proteins containing one fibrinogen-like and one collagen-like domain. Almost all ficolins are capable of binding to GlcNAc and are arguably not true C-type lectins due to their dependency on acetyl groups for binding, rather than on the carbohydrate moiety itself (Bidula, Sexton and Schelenz 2019). Three ficolins, M- L- and H-ficolin, have been identified in humans, whereas mice have two functional ficolins, Ficolin A and B, and a third ficolin which is present as a pseudogene (Matsushita and Fujita 2001; Bidula, Sexton and Schelenz 2019). In humans, M-ficolin is found primarily in neutrophils and monocytes, although small amounts are also detected in the serum (Matsushita and Fujita 2001; Bidula, Sexton and Schelenz 2019). Both L- and H-ficolin are synthesized in the liver and secreted into the blood circulation. H-ficolin is additionally found in the gallbladder and in the lungs (Matsushita and Fujita 2001; Bidula, Sexton and Schelenz 2019). Whilst all three ficolins bind GlcNAc, M- and L-ficolin display less stringency than H-ficolin, as they are capable of binding to other acetylated sugars (Matsushita and Fujita 2001; Kilpatrick and Chalmers 2012; Bidula, Sexton and Schelenz 2019). Of particular note, the L-ficolin recognition groove is reminiscent of the peptidoglycan-binding proteins found in invertebrates, and it is expected to bind various acetylated and neutral carbohydrate moieties in extended polysaccharides such as peptidoglycan (Ma et al. 2004; Garlatti et al. 2007).
The murine C-type lectin RegIIIγ is secreted from Paneth cells in the intestine only in the presence of bacteria. Murine RegIIIγ binds the peptidoglycan of intestinal Gram-positive bacteria, and lacks the complement recruitment domains present in the above-mentioned collectins and ficolins (Cash et al. 2006). Murine RegIIIγ and its human counterpart Human hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein (HIP/PAP) act directly as antimicrobial peptides that directly bind to polymeric peptidoglycan carbohydrate moieties. The mechanism of bacterial killing involves cell wall damage and cytoplasmic leakage (Christa et al. 1996; Cash et al. 2006).
An alternative carbohydrate binding module conserved across viruses, bacteria, fungi, plants, invertebrates and vertebrates is the lysin motif, LysM. The LysM-domain is a highly conserved βααβ-fold that binds to β-1,4-linked polymers of GlcNAc (e.g. chitin) in eukaryotes or the glycan chain of peptidoglycan (Bateman and Bycroft 2000; Buist et al. 2008). Intriguingly, in Arabidopsis thaliana, three LysM domain proteins (membrane proteins Lym1 and Lym3, and the receptor kinase CERK1) interact with peptidoglycan to mount an immune response against bacterial infection (Willmann et al. 2011). However, the mammalian proteins containing LysM domains have received very little attention. Both mice and humans possess four LysM-domain proteins (LysMD1, LysMD2, LysMD3 and LysMD4). LysMD3 is a predicted membrane protein that localizes to the Golgi of HeLa cells, but no phenotype could be observed in a LysMD3-deficient mouse model, despite extensive testing (Yokoyama et al. 2018). It is interesting to speculate that one or more of these proteins could have a role as a peptidoglycan recognition protein in mammals.
The endotoxin/lipopolysaccharide surface receptor CD14 is a glycosylphosphatidylinositol (GPI)-linked 55-kDa protein expressed in macrophages and polymorphonuclear cells that has also been demonstrated to act as a sensor (via similar but not identical regions of its N-terminal domain) for both insoluble and soluble peptidoglycan from Gram-positive bacteria and mycobacterial lipoarabinomannan, thereby inducing the activation of NF-κB. Sensing of lipopolysaccharide and peptidoglycan occurs via distinct sequences of its N-terminal domain (Gupta et al. 1996). CD14 was further shown to bind the muropeptides MDP and GlcNAc-MDP using three different in vitro binding assays, and could be inhibited of N-terminal CD14 fragments and CD14 ligands (Dziarski, Tapping and Tobias 1998). Peptidoglycan binding by the soluble form of CD14 (sCD14) present in human serum, was found to enhance responses in cells expressing the membrane bound form of CD14 (mCD14), but could not replace the function of mCD14 (Dziarski et al. 2000). Whilst the authors data indicated a degree of overlap in the regions involved in binding of lipopolysaccharide and peptidoglycan by CD14, they also catalogued a series of differences between the parameters modulating the CD14-mediated response to these two MAMPS. The functional advantage (if any) of having both extracellular surface receptors (such as CD14) and intracellular sensors (such as NOD1 and NOD2 receptors) for peptidoglycan is open to debate, but it may allow for faster responses to peptidoglycan in the milieu, as evidenced by the detection of NF-κB activation within minutes of exposure to peptidoglycan (Gupta et al. 1996). It is also intriguing to note that CD14 is a well characterized interaction partner of TLR2, an interaction that has not received a great deal of attention in the context of peptidoglycan detection. Nevertheless, it was demonstrated that sCD14 enhanced the binding of a soluble TLR2 construct to insoluble peptidoglycan of Staphylococcus aureus (Iwaki et al. 2002). This interaction, and the specificity for peptidoglycan binding versus other cell wall components by these receptors, is certainly worthy of further validation and exploration.
Lysozyme
Lysozyme is a muramidase found in phagocytic granules, serum and body secretions, and it is the major class of peptidoglycan hydrolase found in mammals. Human lysozyme is encoded by a single gene lyz (also called lzm or lyzf1), whilst mice have two lysozyme isoforms encoded by the genes lyz1 and lyz2. Expression of lyz1 is restricted to intestinal Paneth cells, and the protein product is referred to as Lysozyme P (LysP). Lyz2 is expressed in myeloblasts, macrophages and neutrophils and its product is Lysozyme M (LysM) (Hammer et al. 1987; Cross et al. 1988; Cross and Renkawitz 1990). Lysozyme catalyzes the hydrolysis of peptidoglycan glycosidic bonds between MurNAc and GlcNAc. However, it also has an antimicrobial peptide activity independent of its peptidoglycan-hydrolytic activity (Ibrahim, Matsuzaki and Aoki 2001). For additional potential mechanisms, refer to (Bastos et al. 2017). Nevertheless, the hydrolytic activity of lysozyme generates soluble, bioactive peptidoglycan fragments. Therefore, its peptidoglycan-binding and hydrolytic activity become significant in the context of peptidoglycan-mediated bacterial recognition and modulation of host immune function (Boneca et al. 2007). As discussed previously, the presence of lysozyme within lysosomes is critical, as its glycan hydrolytic activity generates soluble muropeptides that might be transported out of the endosomal compartment by SLC15 family transporters or pannexin-1, and that may function as substrates of NOD1 and NOD2 receptors (Davis, Nakamura and Weiser 2011).
Peptidoglycan recognition proteins
Peptidoglycan recognition proteins (PGRPs/PGLYRPs) are PRRs first discovered as circulating antimicrobial agents in insects (Yoshida, Kinoshita and Ashida 1996). PGRPs are conserved from insects to mammals. Humans, mice and rats (and probably other mammals) all express four PGRPs (Kang et al. 1998). Mammalian PGRPs function as recognition proteins for bacteria and bacteria-derived products and act directly as antimicrobial agents. These were initially named PGRP-S, PGRP-L, PGRP-Iα and PGRP-Iβ, for short, long, and intermediate α and β transcripts, respectively. A new nomenclature has renamed them PGLYRP-1, PGLYRP-2, PGLYRP-3 and PGLYRP-4. All mammalian PGRPs are secreted and, excluding the case of PGLYRP-2, form disulphide-linked homodimers and heterodimers essential for their activity (Lu et al. 2006).
PGLYRP-1 is found in polymorphonuclear leukocytes (PMNs) and displays direct bactericidal activity (Lu et al. 2006; Wang et al. 2007). Similarly, both PGLYRP-3 and PGLYRP-4 display direct bactericidal activity, being produced in several tissues such as skin, sweat glands, sebaceous glands, salivary glands (PGLYRP-4 only), eyes and the intestinal tract (Lu et al. 2006; Wang et al. 2007). These PGRPs have been shown to bind to the bacterial cell wall or outer membrane, and exploit bacterial stress defense responses in order to kill the bacteria. In Gram-positive bacteria (e.g. Bacillus subtilis), PGRPs enter the cell wall at the site of daughter cell separation during cell division, activate the CssR-CssS two-component system (responsible for detection and disposal of misfolded proteins) and thereby cause membrane depolarization, production of hydroxyl radicals and cessation of peptidoglycan precursor, protein, RNA and DNA synthesis (Dziarski, Kashyap and Gupta 2012). In Gram-negative bacteria (e.g. Escherichia coli), PGRPs bind at the outer membrane and activate the functionally homologous CpxA-CpxR two-component system, causing bacterial death through a similar mechanism (Dziarski, Kashyap and Gupta 2012). Other potential bactericidal mechanisms for PGRPs such as membrane permeabilization, inhibition of extracellular peptidoglycan cross-linking, or peptidoglycan hydrolysis, were excluded by the same authors (Dziarski, Kashyap and Gupta 2012).
PGLYRP-2 (PGRP-L) is unique in that it displays N-acetylmuramoyl-L-alanine amidase activity, hydrolyzing the lactyl-amide bond between the MurNAc and the first amino acid (L-Ala) of the stem peptide of peptidoglycan. Its minimal peptidoglycan substrate is MTP (Wang et al. 2003). One cysteine residue (Cys530 in human PGLYRP-2) is substituted in non-amidase PGRPs by serine, and as such, the presence of a cysteine in this position predicts amidase activity of any given PGRP. Also in contrast with the other members of the human PGRP family, PGLYRP-2 has no direct bactericidal activity (Wang et al. 2003). It is constitutively produced by the liver and secreted into the blood circulation. In addition, its expression can be induced in skin keratinocytes, oral and intestinal epithelial cells (Zhang et al. 2005; Li et al. 2006). Differences in transcription regulation (alternative transcription factors and promoter binding regions) result in constitutive expression of PGLYRP-2 in the liver (e.g. c-Jun and ATF2 transcription factors) and inducible expression in epithelial cells (e.g. NF-κB and Sp1 transcription factors) (Li et al. 2006). PGLYRP-2 is proposed to prevent over-activation of the immune system and inflammation-induced tissue damage in response to NOD2 ligands, as these muropeptides can no longer be recognized by NOD2 upon separation of the peptide component from MurNAc. However, MurNAc is dispensable for NOD1 activation, and therefore PGLYRP-2 may act to shift the immune tone towards NOD1-mediated responses at the expense of NOD2 (Girardin et al. 2003c).
It is interesting to note that both NOD2 and PGLYRP-2 are required for peptidoglycan-induced acute arthritis in the ankle joints of BALB/c mice (Saha et al. 2009). In this model NOD2 acts upstream of PGLYRP-2 and stimulates PGLYRP-2 expression in the foot, together with cytokine and chemokine production that is required for recruitment of PMNs to the tissue, leading to arthritis. This NOD2-PGLYRP2-mediated arthritis pathway could be suppressed by PGLYRP-1. Furthermore, PGLYRP-1 was suggested to induce TNF during Listeria monocytogenes infection in mice (Osanai et al. 2011). PGLYRP-1 was recently shown to be the ligand for the transmembrane receptor Triggering Receptor Expressed on Myeloid cells (TREM)-1 (Read et al. 2015). The authors findings suggest that polymeric peptidoglycan, but not soluble muropeptides, may act as a scaffold for the presentation and multimerization of PGLYRP-1, required for activation of TREM-1. Whether innate immune receptor activation driven by peptidoglycan is unique to PGLYRP-1, or could be a function of the other mammalian PGLYRPs, remains to be investigated.
Serotonin
In the process of investigating the pyrogenic and somnogenic properties of muropeptides, Silverman et al. hypothesized that serotonin and muropeptides share a receptor on macrophages (Silverman, Wu and Karnovsky 1985). Binding of radiolabeled muropeptides to murine macrophages or PU5 cells was inhibited by similar concentrations of serotonin and unlabeled muropeptide, and also by treatment with the serotonin antagonists methysergide, cyproheptadine and spiperone. Furthermore, serotonin and muropeptides both enhanced superoxide induction by phorbol myristate acetate. Thus, it was proposed that muropeptides are recognized by one or more of the 5-HT (serotonin) receptors.
Outside the context of the central nervous system, high concentrations of MDP (100 μM) lead to ileal musculature relaxation via activation of 5-HT7 receptor on smooth muscle cells. However, low concentrations of MDP (5–500 nM) exert no effect (Ševčík et al. 2000). Also in the nanomolar range, MDP fails to activate either 5-HT4 or 5-HT1A receptors (Ševčík et al. 2002), and its low potency together with the fact that it doesn't seem to compete against other 5-HT receptor agonists have led some authors to suggest that MDP acts on other ‘functional determinants (Ševčík et al. 2000). In contrast, cells within the central nervous system may be more sensitive to MDP. Accordingly, lower MDP concentrations seem to effectively compete for 5-HT1 receptors in bovine cerebral cortical membranes (Takeuchi, Root-Bernstein and Shih 1990).
It should be noted that other proteins contain structurally similar ‘serotonin-binding sites’, including myelin basic protein, luteinizing hormone-releasing hormone, adrenocorticotropic hormone and α-melanocyte-stimulating hormone. Notably, it was reported that MDP specifically binds to these proteins, whilst substitutions of D-Ala or L-isoGln abolish binding, and muramic acid alone has no affinity (Root-Bernstein and Westall 1990). Thus, many conflicting observations regarding the serotonin-like effects of MDP may result from the fact that MDP can act on serotonin targets other than its formal receptors (Root-Bernstein and Westall 1990). In addition, note that each serotonin receptor may have multiple splice variants and the putative differential activation of these has not been taken into account. Thus, while we do believe enough evidences exist to consider MDP as a serotonergic system modulator, one should be cautious in considering MDP as a direct agonist of 5-HT receptors at physiologically relevant concentrations.
Intracellular receptors and responses
Whether due to host- or bacteria-mediated uptake mechanisms, delivery of peptidoglycan into the cytosol of eukaryotic cells allows peptidoglycan fragments to reach and interact with host intracellular receptors, such as the NOD-like receptors (NLRs). NLRs are characterized by (i) an N-terminal caspase recruitment domain (CARD), responsible for interaction with downstream effectors, (ii) a nucleotide-binding oligomerization domain (NBD) and (iii) a C-terminal leucine-rich repeat (LRR) domain which functions as a regulatory domain. The N-terminal segment can include pyrin domains (PYD), CARD, baculovirus inhibitor of apoptosis repeat (BIR), acidic transactivation (AD), or other unclassified domains (Proell et al. 2008). In terms of peptidoglycan sensing, NLRs are represented by the nucleotide-binding oligomerization domain (NOD) receptors NOD1 and NOD2, which are encoded by the card4 and card15 genes, and contain one and two N-terminal CARD domains, respectively. These domains allow NOD1 and NOD2 to interact with other CARD domain-containing molecules (e.g. homotypic interaction with RIP2/RICK) (Ogura et al. 2001). Even though NOD1 and NOD2 belong to a 22-member family of NLRs, they are the only NLRs with a well-established involvement in peptidoglycan recognition. Other NLRs have been reported to interact with MDP (NLRP3) or to associate with NOD2 when bound to peptidoglycan (NLRP1), but information regarding the role of such interactions other than promotion of IL-1β production and secretion is limited, and evidence for direct binding is not available (Martinon et al. 2004; Hsu et al. 2008). In contrast, direct binding of NOD2 to MDP with high affinity has been demonstrated (Grimes et al. 2012).
Human NOD1 senses cell wall fragments from Gram-negative bacteria in general and some Gram-positive bacteria (e.g. Listeria and Bacillus) by recognizing muropeptide monomers terminating in meso-diaminopimelic acid at the third position of the peptide stem. Both MurNAc and L-alanine are dispensable for NOD1 activation, so that the minimal motif required for NOD1 recognition is the dipeptide γ-iE-DAP (Chamaillard et al. 2003; Girardin et al. 2003a; Hasegawa et al. 2006), although recognition is less efficient than that of triDAP or MtriDAP (muramyl-triDAP). In mice, NOD1 is more prominently activated by the corresponding muramyltetrapeptide (MtetraDAP) terminating in D-alanine (Magalhaes et al. 2005). In contrast, the minimum required fragment for human and mouse NOD2 activation is N-acetylmuramyl-L-alanyl-D-isoglutamine (i.e. MDP), which is widely distributed across Gram-positive and Gram-negative bacteria (Girardin et al. 2003b, 2003c). NOD2 also recognizes lysine-containing muramyltripeptide (MtriLYS) (Fernandez et al. 2011). NOD1 and NOD2 differ at the level of their distribution in the host, as NOD1 is expressed across many tissues and cell types, while NOD2 is more restricted, being expressed in monocytes and macrophages (Ogura et al. 2001), epithelial intestinal cell lines (Hisamatsu et al. 2003), Paneth cells (Ogura et al. 2003), dendritic cells (Tada et al. 2005), osteoblasts (Marriott et al. 2005), keratinocytes (Voss et al. 2006), and other epithelial cell types (Uehara et al. 2007).
The canonical pathway of muropeptide-mediated NOD1/NOD2 activation ultimately culminates with the activation of the transcription factor NF-κB and the expression of its target genes (Fig. 4). Upon activation, CARD domains from NOD1 or NOD2 interact first through homotypic CARD-CARD interactions with the CARD-containing kinase RIPK2/RICK. The NBDs recruit additional players, including a variety of ubiquitination factors that regulate the signal transduction cascade that leads to NF-κB activation, modulating the strength of the host response. Among these essential players is the E3 ubiquitin ligase TNF Receptor Associated Factor 6 (TRAF6), which undergoes autoubiquitination to induce polyubiquitination of RIPK2. Additional E3 ubiquitin ligases mediate RIPK2 polyubiquitination. In particular, the cellular inhibitor of apoptosis proteins (cIAPs) 1 and 2 (cIAP1 and cIAP2) act as E3 ubiquitin ligases and are essential for both RIPK2 polyubiquitination and NOD signalling (Bertrand et al. 2009). Negatively regulating this process, the ubiquitin-editing enzyme A20 attenuates RIPK2 ubiquitination and thus prevents dramatic amplification of NOD2-dependent NF-κB activation (Hitotsumatsu et al. 2008). Similarly, caspase 12 binds RIPK2, displaces TRAF6 and thus prevents RIPK2 ubiquitination (LeBlanc et al. 2008).
Figure 4.
Signalling pathways activated by peptidoglycan upon stimulation of NOD-like receptors. The canonical pathway of muropeptide-mediated NOD1/NOD2 activation ultimately culminates with the activation of the transcription factor NF-κB and the expression of its target genes. The NOD1 or NOD2 CARD domains interact first through homotypic CARD-CARD interactions with the CARD-containing kinase RIPK2/RICK. The NBDs recruit additional players, including proteins that regulate the ubiquitination-mediated signal transduction cascade that leads NF-κB activation. These players may either positively (e.g. TRAF6, cIAP1 and 2) or negatively (e.g. Caspase 12, A20) modulate NOD receptor activation and subsequent signalling. In response to their muropeptide ligands, NOD1 and NOD2 activation can also signal through RIPK2/RICK-independent mechanisms. RIPK2/RICK-independent signalling occurs downstream of NOD2 activation upon binding to CARD9. Whilst this leads to the activation of MAPKs, other RIPK2/RICK-independent pathways exist which do not result in the activation of either MAPKs, NF-κB or IRF7. In this case, NOD1 and NOD2 interact with other NLRs. For instance, NOD2 has been shown to specifically and directly interact with and drive downstream signalling via NLRP1, NLRP3 and NLRP12. Interaction of NOD2 with NLRP3 activates ASC, which in turn activates caspase 1 and leads to the production of IL-1β. Similarly, NOD1 recruitment to the membrane is accompanied by ATG16L1 recruitment and activation, which in turn leads to activation of the autophagy process (which may be RIPK2/RICK-dependent or independent). Hexokinase is an enzyme that participates in glycolysis by catalyzing the conversion of glucose to glucose-6-phosphate. N-acetylglucosamine generated by peptidoglycan degradation (likely within endosomes/lysosomes) binds mitochondrial outer membrane-associated hexokinase (H), inducing its release and the liberation of its interacting partners (e.g. voltage-dependent anion channel VDAC, Akt). Following a multistep process that involves (among other events) the release of mitochondrial DNA, the NLRP3 inflammasome is activated, pro caspase-1 converted into caspase-1, and pro Il-1β converted into active IL-1β. This hexokinase-mediated NLRP3 inflammasome activation is independent of NOD1 and NOD2 receptors. A20: alpha-induced protein 3; ASC: apoptosis-associated speck-like protein containing a CARD; ATG16L1: autophagy related 16 like 1; cIAP: cellular inhibitor of apoptosis; eRK: extracellular signal-regulated kinase; IRF7: interferon regulatory transcription factor 7; JNK: c-Jun N-terminal kinase; MAPK: mitogen-activated protein kinase; NF-κB: nuclear factor-kappa B; NLRP: nucleotide-binding oligomerization domain, leucine rich repeat and pyrin domain containing; NOD: nucleotide oligomerization domain; RIPK2/RICK: receptor-interacting serine/threonine-protein kinase 2.
Transforming growth factor-beta-activated kinase 1 (TAK1) kinase is recruited following ubiquitination of RIPK2. Activated TAK1 complex then promotes K63 polyubiquitination, and activation of the IKK-β/IL2 kinase (a subunit of the NF-κB repressor IκB), causing phosphorylation and degradation of the NF-κB repressor IκB. The repressor IκB constitutively masks the nuclear localization signal of NF-κB and sequesters it in the cytoplasm. Upon IκB degradation, NF-κB becomes free to translocate into the nucleus, where it can activate the expression of more than 150 genes (Mercurio et al. 1997; Hasegawa et al. 2008b).
In parallel to this RIPK2/RICK-dependent pathway that culminates in NF-κB activation, alternative interactions can occur downstream of RIPK2 and TAK1 upon NOD1/NOD2 activation, resulting in the activation of the mitogen-activated protein kinases (MAPKs) p38 (Hsu et al. 2007), ERK1/ERK2 and c-Jun N-terminal kinase (JNK; Fig. 4) (Windheim et al. 2007). CARD-containing adaptor protein 9 (CARD9) has been shown to bind directly to NOD2 and to trigger the RIPK2/RICK-independent activation of p38 and JNK, thus driving NOD-mediated responses to MDP/iE-DAP. Accordingly, macrophages from Card9(-/-) mice show defects in the activation of p38 and JNK but not NF-κB when infected with the intracellular pathogen Listeria monocytogenes (Hsu et al. 2007). Similarly, the abundant pro-apoptotic protein BID (of the Bcl-2 family of apoptosis regulators) has been reported as required for muropeptide-induced NOD signalling, as deletion of BID was responsible for defective NOD activation and blunted NF-κB and ERK signalling (Yeretssian et al. 2011).
In response to their muropeptide ligands, NOD1 and NOD2 activation can signal through RIPK2/RICK-independent mechanisms that do not result in the activation of MAPKs or NF-κB (Fig. 4). In this case, NOD1 and NOD2 interact with other NLRs. For instance, NOD2 has been shown to specifically and directly interact with NLRP1, NLRP3 and NLRP12 (Hsu et al. 2008; Wagner et al. 2009), which implicates other NLRs in the indirect involvement of cellular responses triggered by MDP and γ-iE-DAP. For example, interaction of NOD2 with NLRP3 activates the apoptosis-associated speck-like protein containing a CARD (ASC), which in turn activates caspase 1, leading to the production of IL-1β.
The cell surface and endoplasmic reticulum multifunctional lectin-like chaperone protein calreticulin modulates cell sensitivity to apoptosis (Groenendyk, Lynch and Michalak 2004). In rabbit kidney RK13 cells (but not human HCC or HeLa cells), calreticulin was shown to directly bind MDP and peptidoglycan from Staphylococcus aureus and Bacillus subtilis, thereby inducing apoptosis (Chen et al. 2004). Binding was specific to the stereochemistry of (L,D)-MDP, native to peptidoglycan, as L,L- and D,D-MDP isoforms did not interact with calreticulin. At least in the case of kidney RK13 cells, MDP- and peptidoglycan-activated calreticulin induces apoptosis via the upregulation of tumour necrosis factor receptor 1 (TNFR1), TNFR-associated death domain protein (TRADD), the complex(es) of surface calreticulin with TNFR1 and/or TRADD and caspase 3, in a TLR2/4-, CD14- and NOD2-independent manner (Chen et al. 2005). Peptidoglycan of Lactobacillus paracasei subsp. paracasei X12 was also proposed to induce apoptosis and translocation of calreticulin from the endoplasmic reticulum to the plasma membrane, although in this study directly binding of peptidoglycan by calreticulin was not investigated (Tian et al. 2015).
Recently, it was demonstrated that NLRP3 is activated by peptidoglycan, through a mechanism that is independent of NOD1 and NOD2 (Wolf et al. 2016). Hexokinase is an enzyme that participates in glycolysis by catalyzing the conversion of glucose to glucose-6-phosphate. It associates to the mitochondrial outer membrane by interacting with the voltage-dependent anion channel (VDAC) (Pastorino and Hoek 2008). In macrophages, N-acetylglucosamine generated by peptidoglycan degradation was found to inhibit hexokinase activity and induce its release from the mitochondrial membrane. Dissociation of hexokinase from mitochondrial membrane protein partners such as VDAC and Akt, promotes NLRP3 inflammasome activation through a mechanism triggered by increased mitochondrial permeability pore activity (Pastorino, Shulga and Hoek 2002; Majewski et al. 2004; Chiara et al. 2008; Pastorino and Hoek 2008). The authors suggested that peptidoglycan-derived N-acetylglucosamine could be released from lysosomes into the cytosol via a dedicated N-acetylglucosamine/N-acetylgalactosamine transporter (Fig. 3C) (Jonas et al. 1989; Jonas and Jobe 1990). The precise mechanism by which N-acetylglucosamine is liberated from the peptidoglycan fragments remains unclear. It is possible that endogenous bacterial N-acetyl-β-D-glucosaminidase activity generates a glycan substrate with terminal N-acetylglucosamine residues that are released upon cleavage by lysozyme. Alternatively, host N-acetyl-β-D-glucosaminidase and di-N-acetyl chitobiase activities have been reported in lysosomal and thyroid microsome fractions isolated from the rat liver (Sellinger et al. 1960; Anumula and Spiro 1983; Kuranda and Aronson 1986). To our knowledge, these enzymes have not been tested for activity against peptidoglycan. Bacillus anthracis peptidoglycan, which is highly de-acetylated, was ineffective at stimulating NLRP3 inflammation, since glucosamine is a poor binder/inhibitor of hexokinase (Wolf et al. 2016). Several bacterial pathogens promote their survival in the host via deacetylation of N-acetylglucosamine residues, as this renders the peptidoglycan resistant to hydrolysis by lysozyme (Vollmer and Tomasz 2000; Boneca et al. 2007; Wolf et al. 2016). It remains to be demonstrated whether deacetylation of N-acetylglucosamine is also a general strategy to avoid NLRP3 activation.
CONCLUSION
In this review, we have described how peptidoglycan functions as a signalling molecule in the host under (patho)physiological conditions, how it disseminates within the host, and how cells respond to peptidoglycan. It is now known that the fate of peptidoglycan fragments depends on their structural/molecular nature, their origin, their uptake mechanism and their target. Different molecules will trigger profoundly different responses across a variety of tissues and cells under distinct conditions. However, aside from very few but remarkable cases (e.g. neutrophil priming in the bone marrow, brain development and behaviour, intestinal inflammation and susceptibility to infection), the developmental/physiological/pathological roles of peptidoglycan across most host organs and tissues is yet to be defined. Doing so is expected to provide access to untapped tools for disease modulation in the foreseeable future.
In order to exploit peptidoglycan/muropeptides as signalling molecules in mammals, it is required to characterize (potentially on a case-by-case basis) the ‘peptidoglycome’ of the host, in particular within the intestinal lumen, and at physiological levels circulating in the blood. Analysis of individual peptidoglycomes, together with genetic profiling, immune tone and the known health/disease status of the host, could likewise open up a whole new system for health and disease modulation. But before this system can be safely modulated, reference values must be defined, spanning host developmental stages, and across heterogenous human populations. Once these issues are addressed, such modulation may be achieved via a variety of strategies, such as exogenous administration of individual muropeptides, alterations in microbiota colonization status, or modulation of signalling pathways via inhibitors or drugs that mimicking specific muropeptide activities.
Looking more broadly, MAMPs, such as peptidoglycan, are largely overlooked by existing OMICs approaches. This is despite the fact that MAMPs and their signalling pathways are relatively well-defined. Understanding the intestinal MAMPome, for example, could provide a valuable insight into physiological, immunological and developmental phenomena that are known to be mediated by microbiota, but whose mechanisms remains obscure. The development of peptidoglycomics, alongside other ‘MAMPomic’ approaches, could be a valuable tool in understanding and predicting the risk, onset and severity of different types of disease, in particular chronic diseases. Empowered with this information, one may predict the targets and host responses to peptidoglycan and other MAMPs, and their effects in the steady-state and in disease contexts. Bringing together host genomics, transcriptomics, proteomics, metabolomics and ‘MAMPomics’ data is likely to provide a clearer and more complete picture of the mechanisms and molecular players driving disease expression and phenotypic variability.
ACKNOWLEDGEMENTS
PB is part of the Pasteur—Paris University (PPU) International PhD Program. Figures were prepared using Servier Medical Art, licensed under a Creative Common Attribution 3.0 Generic License (http://smart.servier.com/), and Inkscape 1.0 (https://inkscape.org).
Contributor Information
Paulo A D Bastos, Institut Pasteur, Biology and genetics of the bacterial cell wall Unit, 25-28 rue du Docteur Roux, Paris 75724, France; CNRS, UMR 2001 “Microbiologie intégrative et moléculaire”, Paris 75015, France; Université de Paris, Sorbonne Paris Cité, 12 rue de l'Ecole de Médecine, 75006, Paris, France.
Richard Wheeler, Institut Pasteur, Biology and genetics of the bacterial cell wall Unit, 25-28 rue du Docteur Roux, Paris 75724, France; CNRS, UMR 2001 “Microbiologie intégrative et moléculaire”, Paris 75015, France; Tumour Immunology and Immunotherapy, Institut Gustave Roussy, 114 rue Edouard-Vaillant, Villejuif 94800, France; INSERM UMR 1015, Villejuif 94800, France.
Ivo G Boneca, Institut Pasteur, Biology and genetics of the bacterial cell wall Unit, 25-28 rue du Docteur Roux, Paris 75724, France; CNRS, UMR 2001 “Microbiologie intégrative et moléculaire”, Paris 75015, France.
AUTHOR CONTRIBUTIONS
The authors contributed equally to all aspects of the article.
FUNDING
This work was supported by funding from the Institut Carnot Pasteur Microbes & Santé, the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie Grant agreement No 665807, an Agence Nationale de la Recherche (ANR) Grant [ANR-CE15-0021] and the RHU Torino Lumière [ANR-16-RHUS-0008].
Conflicts of interest
None declared.
REFERENCES
- Adam A, Ciorbaru R, Ellouz Fet al. Adjuvant activity of monomeric bacterial cell wall peptidoglycans. Biochem Biophys Res Commun. 1974;56:561–7. [DOI] [PubMed] [Google Scholar]
- Akopyants NS, Clifton SW, Kersulyte Det al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–53. [DOI] [PubMed] [Google Scholar]
- Allison CC, Kufer Ta, Kremmer Eet al. Helicobacter pylori induces MAPK phosphorylation and AP-1 activation via a NOD1-dependent mechanism. J Immunol. 2009;183:8099–109. [DOI] [PubMed] [Google Scholar]
- Ambler L, Hudson AM. Pharmacokinetics and metabolism of muramyl dipeptide and nor-muramyl dipeptide [3H-labelled] in the mouse. Int J Immunopharmacol. 1984;6:133–9. [DOI] [PubMed] [Google Scholar]
- Andreoni F, Toyofuku M, Menzi Cet al. Antibiotics stimulate formation of vesicles in staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob Agents Chemother. 2019, DOI: 10.1128/AAC.01439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anumula KR, Spiro RG. Release of glucose-containing polymannose oligosaccharides during glycoprotein biosynthesis. Studies with thyroid microsomal enzymes and slices. J Biol Chem. 1983;258:15274–82. [PubMed] [Google Scholar]
- Arentsen T, Qian Y, Gkotzis Set al. The bacterial peptidoglycan-sensing molecule Pglyrp2 modulates brain development and behavior. Mol Psychiatry. 2017;22:257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asong J, Wolfert MA, Maiti KKet al. Binding and cellular activation studies reveal that toll-like receptor 2 can differentially recognize peptidoglycan from gram-positive and gram-negative bacteria. J Biol Chem. 2009;284:8643–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma I, Kishimoto S, Yamamura Yet al. Adjuvanticity of mycobacterial cell walls. Jpn J Microbiol. 1971;15:193–7. [DOI] [PubMed] [Google Scholar]
- Azuma I, Sugimura K, Yamawaki Met al. Adjuvant activity of synthetic 6-O-’mycoloyl’-N-acetylmuramyl-L-Alanyl-D-isoglutamine and related compounds. Infect Immun. 1978;20:600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10:955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 2004;572:65–8. [DOI] [PubMed] [Google Scholar]
- Bastos P, Trindade F, da Costa Jet al. Human antimicrobial peptides in bodily fluids: current knowledge and therapeutic perspectives in the postantibiotic Era. Med Res Rev. 2017;0:1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol. 2000;299:1113–9. [DOI] [PubMed] [Google Scholar]
- Bennett MVL, Contreras JE, Bukauskas FFet al. New roles for astrocytes: Gap junction hemichannels have something to communicate. Trends Neurosci. 2003;26:610–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJM, Doiron K, Labbé Ket al. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. [DOI] [PubMed] [Google Scholar]
- Bidula S, Sexton DW, Schelenz S. Ficolins and the recognition of pathogenic microorganisms: An overview of the innate immune response and contribution of single nucleotide polymorphisms. J Immunol Res. 2019;2019:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Perez-Perez GI, Kleanthous Het al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–5. [PubMed] [Google Scholar]
- Boassa D, Ambrosi C, Qiu Fet al. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem. 2007;282:31733–43. [DOI] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes Det al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branković I, van Ess EF, Noz MPet al. NOD1 in contrast to NOD2 functional polymorphism influence Chlamydia trachomatis infection and the risk of tubal factor infertility. Pathog Dis. 2015;73:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu HF, Wang X, Tang Yet al. Toll-like receptor 2-mediated peptidoglycan uptake by immature intestinal epithelial cells from apical side and exosome-associated transcellular transcytosis. J Cell Physiol. 2010;222:658–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok Jet al. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–47. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JLet al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. [DOI] [PubMed] [Google Scholar]
- Camorlinga-Ponce M, Romo C, González-Valencia Get al. Topographical localisation of cagA positive and cagA negative Helicobacter pylori strains in the gastric mucosa; an in situ hybridisation study. J Clin Pathol. 2004;57:822–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case EDR, Samuel JE. Contrasting lifestyles within the host cell. Microbiol Spectr. 2016;4:DOI: 10.1128/microbiolspec.vmbf-0014-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham C V, Behrendt CLet al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (80-). 2006;313:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CLet al. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science. 2006b;313:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil JD, Sirisaengtaksin N, O'Brien-Simpson NMet al. Outer membrane vesicle-host cell interactions. Microbiol Spectr. 2019;7, DOI: 10.1128/microbiolspec.psib-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Hashimoto M, Horie Yet al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–7. [DOI] [PubMed] [Google Scholar]
- Chen D, Duggan C, Reden TBet al. Calreticulin is a binding protein for muramyl dipeptide and peptidoglycan in RK13 cells. Biochemistry. 2004;43:11796–801. [DOI] [PubMed] [Google Scholar]
- Chen D, Texada DE, Duggan Cet al. Surface calreticulin mediates muramyl dipeptide-induced apoptosis in RK13 cells. J Biol Chem. 2005;280:22425–36. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J Bacteriol. 2002;184:6434–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara F, Castellaro D, Marin Oet al. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS One. 2008;3:e1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christa L, Carnot F, Simon MTet al. HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am J Physiol. 1996;271:G993–1002. [DOI] [PubMed] [Google Scholar]
- Clarke TB, Davis KM, Lysenko ESet al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M, Mangelsdorf I, Wedel Aet al. Mouse lysozyme M gene: Isolation, characterization, and expression studies. Proc Natl Acad Sci USA. 1988;85:6232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M, Renkawitz R. Repetitive sequence involvement in the duplication and divergence of mouse lysozyme genes. EMBO J. 1990;9:1283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundell DR, Gerard NP, Gerard Cet al. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 1995;377:435–8. [DOI] [PubMed] [Google Scholar]
- Dalmasso G, Nguyen HT, Charrier-Hisamuddin Let al. PepT1 mediates transport of the proinflammatory bacterial tripeptide L-Ala-{gamma}-D-Glu-meso-DAP in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;299:G687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Fiil BK, Speckmann Cet al. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med. 2013;5:1278–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Nachbur U, Yabal Met al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell. 2012;46:746–58. [DOI] [PubMed] [Google Scholar]
- Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. J Clin Invest. 2011;121:3666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Wang Y, Patel SRet al. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17–producing T cell responses. Nat Immunol. 2007;8:1086–94. [DOI] [PubMed] [Google Scholar]
- Dietrich N, Lienenklaus S, Weiss Set al. Murine toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS One. 2010;5:e10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. Staphylococcus aureus peptidoglycan is a toll-like receptor 2 activator: A reevaluation. Infect Immun. 2005;73:5212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Kashyap DR, Gupta D. Mammalian peptidoglycan recognition proteins kill bacteria by activating two-component systems and modulate microbiome and inflammation. Microb Drug Resist. 2012;18:280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Tapping RI, Tobias PS. Binding of bacterial peptidoglycan to CD14. J Biol Chem. 1998;273:8680–90. [DOI] [PubMed] [Google Scholar]
- Dziarski R, Viriyakosol S, Kirkland TNet al. Soluble CD14 enhances membrane CD14-mediated responses to peptidoglycan: Structural requirements differ from those for responses to lipopolysaccharide. Infect Immun. 2000;68:5254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R. Binding sites for peptidoglycan on mouse lymphocytes. Cell Immunol. 1987;109:231–45. [DOI] [PubMed] [Google Scholar]
- Eckels PC, Banerjee A, Moore EEet al. Amantadine inhibits platelet-activating factor induced clathrin-mediated endocytosis in human neutrophils. Am J Physiol Cell Physiol. 2009;297:C886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddie Ip WK, Takahashi K, Moore KJet al. Mannose-binding lectin enhances Toll-like receptors 2 and 6 signaling from the phagosome. J Exp Med. 2008;205:169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen N, Shuffield D, Syed MMet al. Modulation of connexin expression and gap junction communication in astrocytes by the gram-positive bacterium S. aureus. Glia. 2007;55:104–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, Basilio D, Sáez JCet al. The role of gap junction channels during physiologic and pathologic conditions of the human central nervous system. J Neuroimmune Pharmacol. 2012;7:499–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez EM, Valenti V, Rockel Cet al. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut. 2011;60:1050–9. [DOI] [PubMed] [Google Scholar]
- Fillon S, Soulis K, Rajasekaran Set al. Platelet-activating factor receptor and innate immunity: uptake of gram-positive bacterial cell wall into host cells and cell-specific pathophysiology. J Immunol. 2006;177:6182–91. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Jaumouillé V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol Mech Dis. 2012;7:61–98. [DOI] [PubMed] [Google Scholar]
- Fogler WE, Wade R, Brundish DEet al. Distribution and fate of free and liposome-encapsulated [3H]nor-muramyl dipeptide and [3H]muramyl tripeptide phosphatidylethanolamine in mice. J Immunol. 1985;135:1372–7. [PubMed] [Google Scholar]
- Fox A, Fox K, Christensson Bet al. Absolute identification of muramic acid, at trace levels, in human septic synovial fluids in vivo and absence in aseptic fluids. Infect Immun. 1996;64:3911–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco LM, Salinas VH, Brown KEet al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. [DOI] [PubMed] [Google Scholar]
- Fredlund J, Enninga J. Cytoplasmic access by intracellular bacterial pathogens. Trends Microbiol. 2014;22:128–37. [DOI] [PubMed] [Google Scholar]
- Fukaya T, Takagi H, Sato Yet al. Crucial roles of B7-H1 and B7-DC expressed on mesenteric lymph node dendritic cells in the generation of antigen-specific CD4+Foxp3 + regulatory T cells in the establishment of oral tolerance. Blood. 2010;116:2266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S, Syed MM, Kielian T. Staphylococcus aureus-derived peptidoglycan induces Cx43 expression and functional gap junction intercellular communication in microglia. J Neurochem. 2005;95:475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlatti V, Belloy N, Martin Let al. Structural insights into the innate immune recognition specificities of L- and H-ficolins. EMBO J. 2007;26:623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon MD, Takaki M, Tamir Het al. The enteric neural receptor for 5-hydroxytryptamine. Experientia. 1985;41:863–8. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Carneiro LAMet al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science (80-). 2003a;300:1584–7. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Boneca IG, Viala Jet al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003b;278:8869–72. [DOI] [PubMed] [Google Scholar]
- Girardin SE, Travassos LH, Hervé Met al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003c;278:41702–8. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54:224–33. [DOI] [PubMed] [Google Scholar]
- Griffin ME, Hespen CW, Wang YCet al. Translation of peptidoglycan metabolites into immunotherapeutics. Clin Transl Immunol. 2019;8:e1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes CL, Ariyananda LDZ, Melnyk JEet al. The innate immune protein Nod2 binds directly to MDP, a bacterial cell wall fragment. J Am Chem Soc. 2012;134:13535–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendyk J, Lynch J, Michalak M. Calreticulin, Ca2+, and calcineurin - Signaling from the endoplasmic reticulum. Mol Cells. 2004;17:383–9. [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;360:512–9. [DOI] [PubMed] [Google Scholar]
- Gupta D, Kirkland TN, Viriyakosol Set al. CD14 is a cell-activating receptor for bacterial peptidoglycan. J Biol Chem. 1996;271:23310–6. [DOI] [PubMed] [Google Scholar]
- Gupta R, Ghosh S, Monks Bet al. RNA and β-hemolysin of group B streptococcus induce interleukin-1β (IL-1β) by activating NLRP3 inflammasomes in mouse macrophages. J Biol Chem. 2014;289:13701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Schilling JW, Prager EMet al. Recruitment of lysozyme as a major enzyme in the mouse gut: Duplication, divergence, and regulatory evolution. J Mol Evol. 1987;24:272–9. [DOI] [PubMed] [Google Scholar]
- Hansen S, Selman L, Palaniyar Net al. Collectin 11 (CL-11, CL-K1) Is a MASP-1/3-Associated Plasma Collectin with Microbial-Binding Activity. J Immunol. 2010;185:6096–104. [DOI] [PubMed] [Google Scholar]
- Harder J, Nú̃ez G. Functional expression of the intracellular pattern recognition receptor NOD1 in human keratinocytes. J Invest Dermatol. 2009;129:1299–302. [DOI] [PubMed] [Google Scholar]
- Harton JA, Linhoff MW, Zhang Jet al. Cutting Edge: CATERPILLER: A large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–93. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PCet al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 2008a;27:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto Y, Lucas PCet al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 2008b;27:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kawasaki A, Yang Ket al. A role of lipophilic peptidoglycan-related molecules in induction of Nod1-mediated immune responses. J Biol Chem. 2007;282:11757–64. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Yang K, Hashimoto Met al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281:29054–63. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Tawaratsumida K, Kariya Het al. Lipoprotein is a predominant toll-like receptor 2 ligand in staphylococcus aureus cell wall components. Int Immunol. 2006;18:355–62. [DOI] [PubMed] [Google Scholar]
- Hauer aC, Bajaj-Elliott M, Williams CBet al. An analysis of interferon gamma, IL-4, IL-5 and IL-10 production by ELISPOT and quantitative reverse transcriptase-PCR in human Peyer's patches. Cytokine. 1998;10:627–34. [DOI] [PubMed] [Google Scholar]
- Hediger MA, Clémençon B, Burrier REet al. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol Aspects Med. 2013;34:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl M, Abraham C. Secretory mediators regulate Nod2-induced tolerance in human macrophages. Gastroenterology. 2011;140:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heit A, Schmitz F, Haas Tet al. Antigen co-encapsulated with adjuvants efficiently drive protective T cell immunity. Eur J Immunol. 2007;37:2063–74. [DOI] [PubMed] [Google Scholar]
- Henriques-Normark B, Tuomanen EI. The pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb Perspect Med. 2013;3:a010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt RE, Pele LC, Tremelling Met al. Immuno-inhibitory PD-L1 can be induced by a Peptidoglycan/NOD2 mediated pathway in primary monocytic cells and is deficient in Crohn's patients with homozygous NOD2 mutations. Clin Immunol. 2012;143:162–9. [DOI] [PubMed] [Google Scholar]
- Hewitt RE, Robertson J, Haas CTet al. Reduction of T-helper cell responses to recall antigen mediated by codelivery with peptidoglycan via the intestinal nanomineral-antigen pathway. Front Immunol. 2017;8:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T, Suzuki M, Reinecker H-Cet al. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares Ret al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoijer MA, Melief MJ, Van Helden-Meeuwsen CGet al. Detection of muramic acid in a carbohydrate fraction of human spleen. Infect Immun. 1995;63:1652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoving JC, Wilson GJ, Brown GD. Signalling C-Type lectin receptors, microbial recognition and immunity. Cell Microbiol. 2014;16:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LC, Ali SR, McGillivray Set al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-MS, Zhang Y, You Yet al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wang J, Xu Xet al. Antibody neutralization of microbiota-derived circulating peptidoglycan dampens inflammation and ameliorates autoimmunity. Nat Microbiol. 2019;4:766–73. [DOI] [PubMed] [Google Scholar]
- Hu Y, Song F, Jiang Het al. SLC15A2 and SLC15A4 mediate the transport of bacterially derived Di/Tripeptides to enhance the nucleotide-binding oligomerization domain–dependent immune response in mouse bone marrow–derived macrophages. J Immunol. 2018;201:652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim HR, Matsuzaki T, Aoki T. Genetic evidence that antibacterial activity of lysozyme is independent of its catalytic function. FEBS Lett. 2001;506:27–32. [DOI] [PubMed] [Google Scholar]
- Inohara N, Ogura Y, Fontalba Aet al. Host recognition of bacterial muramyl dipeptide mediated through NOD2: Implications for Crohn's disease. J Biol Chem. 2003;278:5509–12. [DOI] [PubMed] [Google Scholar]
- Irving AT, Mimuro H, Kufer TAet al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014;15:623–35. [DOI] [PubMed] [Google Scholar]
- Ismair MG, Vavricka SR, Kullak-Ublick GAet al. hPepT1 selectively transports muramyl dipeptide but not Nod1-activating muramyl peptides. Can J Physiol Pharmacol. 2006;84:1313–9. [DOI] [PubMed] [Google Scholar]
- Isnardi I, Ng YS, Srdanovic Iet al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki D, Mitsuzawa H, Murakami Set al. The extracellular toll-like receptor 2 domain directly binds peptidoglycan derived from Staphylococcus aureus. J Biol Chem. 2002;277:24315–20. [DOI] [PubMed] [Google Scholar]
- Jonas AJ, Jobe H. N-acetyl-D-glucosamine countertransport in lysosomal membrane vesicles. Biochem J. 1990;268:41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas AJ, Speller RJ, Conrad PBet al. Transport of N-acetyl-D-glucosamine and N-acetyl-D-galactosamine by rat liver lysosomes. J Biol Chem. 1989;264:4953–6. [PubMed] [Google Scholar]
- Kadioglu A, Weiser JN, Paton JCet al. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. [DOI] [PubMed] [Google Scholar]
- Kang D, Liu G, Lundstrom Aet al. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci. 1998;95:10078–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis M, Turnbull L, Carneiro Let al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12:372–85. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew Iet al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- Keshi H, Sakamoto T, Kawai Tet al. Identification and characterization of a novel human collectin CL-K1. Microbiol Immunol. 2006;50:1001–13. [DOI] [PubMed] [Google Scholar]
- Kesty NC, Mason KM, Reedy Met al. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick DC, Chalmers JD. Human L-ficolin (ficolin-2) and its clinical significance. J Biomed Biotechnol. 2012;2012:138797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, Flomerfelt FA, Lee K-Net al. A putative 12 transmembrane domain cotransporter expressed in thymic cortical epithelial cells. J Immunol. 2000;164:3185–92. [DOI] [PubMed] [Google Scholar]
- Koehn J, Huesken D, Jaritz Met al. Assessing the function of human UNC-93B in Toll-like receptor signaling and major histocompatibility complex II response. Hum Immunol. 2007;68:871–8. [DOI] [PubMed] [Google Scholar]
- Koppelman B, Neefjes JJ, De Vries JEet al. Interleukin-10 down-regulates MHC class II αβ peptide complexes at the plasma membrane of monocytes by affecting arrival and recycling. Immunity. 1997;7:861–71. [DOI] [PubMed] [Google Scholar]
- Koppe U, Suttorp N, Opitz B. Recognition of Streptococcus pneumoniae by the innate immune system. Cell Microbiol. 2012;14:460–6. [DOI] [PubMed] [Google Scholar]
- Kufer TA, Kremmer E, Adam ACet al. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10:477–86. [DOI] [PubMed] [Google Scholar]
- Kuipers EJ, Pérez-pérez GI, Meuwissen SGMet al. Helicobacter pylori and atrophic gastritis: Importance of the cagA status. J Natl Cancer Inst. 1995;87:1777–80. [DOI] [PubMed] [Google Scholar]
- Kuranda MJ, Aronson NN. A di-N-acetylchitobiase activity is involved in the lysosomal catabolism of asparagine-linked glycoproteins in rat liver. J Biol Chem. 1986;261:5803–9. [PubMed] [Google Scholar]
- Ladešić B, Perović S, Hršak I. Pharmacokinetics of an immunomodulator peptidoglycan monomer in mice after intravenous administration. Int J Immunopharmacol. 1993;15:145–50. [DOI] [PubMed] [Google Scholar]
- Ladešić B, Tomašić J, Kveder Set al. The metabolic fate of 14C-labeled immunoadjuvant peptidoglycan monomer. II. In vitro studies. BBA - Gen Subj. 1981;678:12–17. [DOI] [PubMed] [Google Scholar]
- LeBlanc PM, Yeretssian G, Rutherford Net al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe. 2008;3:146–57. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DY, Kim DKet al. Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009a;9:5425–36. [DOI] [PubMed] [Google Scholar]
- Lee J, Tattoli I, Wojtal KAet al. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J Biol Chem. 2009b;284:23818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang S, Wang Het al. Differential expression of peptidoglycan recognition protein 2 in the skin and liver requires different transcription factors. J Biol Chem. 2006;281:20738–48. [DOI] [PubMed] [Google Scholar]
- Loh LN, Gao G, Tuomanen EI. Dissecting bacterial cell wall entry and signaling in eukaryotic cells: An actin-dependent pathway parallels platelet-activating factor receptor-mediated endocytosis. MBio. 2017;8:e02030–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wang M, Qi Jet al. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–907. [DOI] [PubMed] [Google Scholar]
- Magalhaes JG, Philpott DJ, Nahori MAet al. Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin. EMBO Rep. 2005;6:1201–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski N, Nogueira V, Bhaskar Pet al. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell. 2004;16:819–30. [DOI] [PubMed] [Google Scholar]
- Marina-García N, Franchi L, Kim Y-Get al. Pannexin-1-mediated intracellular delivery of muramyl dipeptide induces caspase-1 activation via Cryopyrin/NLRP3 independently of Nod2. J Immunol. 2008;180:4050–7. [DOI] [PubMed] [Google Scholar]
- Marinis JM, Homer CR, McDonald Cet al. A novel motif in the Crohn's disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. J Biol Chem. 2011;286:1938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott I, Rati DM, McCall SHet al. Induction of Nod1 and Nod2 intracellular pattern recognition receptors in murine osteoblasts following bacterial challenge. Infect Immun. 2005;73:2967–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Agostini L, Meylan Eet al. Identification of bacterial muramyl dipeptide as activator of the NALP3/Cryopyrin inflammasome. Curr Biol. 2004;14:1929–34. [DOI] [PubMed] [Google Scholar]
- Matsue H, Yao J, Matsue Ket al. Gap junction-mediated intercellular communication between dendritic cells (DCs) is required for effective activation of DCs. J Immunol. 2006;176:181–90. [DOI] [PubMed] [Google Scholar]
- Matsushita M, Fujita T. Ficolins and the lectin complement pathway. Immunol Rev. 2001;180:78–85. [DOI] [PubMed] [Google Scholar]
- Ma W, Compan V, Zheng Wet al. Pannexin 1 forms an anion-selective channel. Pflugers Arch Eur J Physiol. 2012;463:585–92. [DOI] [PubMed] [Google Scholar]
- Ma YG, Cho MY, Zhao Met al. Human mannose-binding lectin and L-ficolin function as specific pattern recognition proteins in the lectin activation pathway of complement. J Biol Chem. 2004;279:25307–12. [DOI] [PubMed] [Google Scholar]
- Mazzini E, Massimiliano L, Penna Get al. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity. 2014;40:248–61. [DOI] [PubMed] [Google Scholar]
- Mclaughlin CA, Schwartzman SM, Horner BLet al. Regression of tumors in guinea pigs after treatment with synthetic muramyl dipeptides and trehalose dimycolate. Science (80-). 1980;208:415–6. [DOI] [PubMed] [Google Scholar]
- McLaughlin NJD, Banerjee A, Kelher MRet al. Platelet-Activating Factor-Induced Clathrin-Mediated Endocytosis Requires Beta-Arrestin-1 Recruitment and Activation of the p38 MAPK Signalosome at the Plasma Membrane for Actin Bundle Formation. J Immunol. 2006;176:7039–50. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–33. [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BWet al. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–6. [DOI] [PubMed] [Google Scholar]
- Mitchell AM, Mitchell TJ. Streptococcus pneumoniae: Virulence factors and variation. Clin Microbiol Infect. 2010;16:411–8. [DOI] [PubMed] [Google Scholar]
- Mondal A, Tapader R, Chatterjee NSet al. Cytotoxic and inflammatory responses induced by outer membrane vesicle-associated biologically active proteases from Vibrio cholerae. Infect Immun. 2016;84:1478–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Anstett MA, Müller P, Albrecht Tet al. Staphylococcal peptidoglycan co-localizes with Nod2 and TLR2 and activates innate immune response via both receptors in primary murine keratinocytes. PLoS One. 2010;5:e13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy TA, Frey MR, Yan Fet al. Helicobacter pylori regulates cellular migration and apoptosis by activation of phosphatidylinositol 3-kinase signaling. J Infect Dis. 2009;199:641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahid MA, Yao B, Dominguez-Gutierrez PRet al. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J Immunol. 2013;190:1250–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Lill JR, Phung Qet al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–4. [DOI] [PubMed] [Google Scholar]
- Natsuka M, Uehara A, Yang Set al. A polymer-type water-soluble peptidoglycan exhibited both Toll-like receptor 2- and NOD2-agonistic activities, resulting in synergistic activation of human monocytic cells. Innate Immun. 2008;14:298–308. [DOI] [PubMed] [Google Scholar]
- Neijssen J, Herberts C, Drijfhout JWet al. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 2005;434:83–8. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Inohara N, Benito Aet al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–8. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Lala S, Xin Wet al. Expression of NOD2 in Paneth cells: A possible link to Crohn's ileitis. Gut. 2003;52:1591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B, Püschel A, Schmeck Bet al. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279:36426–32. [DOI] [PubMed] [Google Scholar]
- Osanai A, Sashinami H, Asano Ket al. Mouse peptidoglycan recognition protein PGLYRP-1 plays a role in the host innate immune response against Listeria monocytogenes infection. Infect Immun. 2011;79:858–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik D, Monahan A, Caffrey DRet al. SLC46 family transporters facilitate cytosolic innate immune recognition of monomeric peptidoglycans. J Immunol. 2017;199:263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B, Neijssen J, Qiao Xet al. Direct antigen presentation and gap junction mediated cross-presentation during apoptosis. J Immunol. 2009;183:1083–90. [DOI] [PubMed] [Google Scholar]
- Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol Mol Biol Rev. 2008;72:211–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008;40:171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277:7610–8. [DOI] [PubMed] [Google Scholar]
- Peek RM, Miller GG, Tham KTet al. Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab Invest. 1995;73:760–70. [PubMed] [Google Scholar]
- Pele LC, Haas CT, Hewitt REet al. Synthetic mimetics of the endogenous gastrointestinal nanomineral: Silent constructs that trap macromolecules for intracellular delivery. Nanomedicine Nanotechnology, Biol Med. 2017;13:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perland E, Hellsten SV, Lekholm Eet al. The novel membrane-bound proteins MFSD1 and MFSD3 are putative SLC transporters affected by altered nutrient intake. J Mol Neurosci. 2017;61:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JJ, Thomas-McKay E, Thoree Vet al. An endogenous nanomineral chaperones luminal antigen and peptidoglycan to intestinal immune cells. Nat Nanotechnol. 2015;10:361–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JHet al. The Nod-Like Receptor (NLR) family: A tale of similarities and differences. PLoS One. 2008;3:e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cruz C, Delgado L, López-Iglesias Cet al. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One. 2015;10:e0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Gong N, Chen Xet al. Natural and engineered bacterial outer membrane vesicles. Biophys Reports. 2019;5:184–98. [Google Scholar]
- Qin J, Li R, Raes Jet al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Jansen M, Sakaris Aet al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–28. [DOI] [PubMed] [Google Scholar]
- Rabinovitch M. Professional and non-professional phagocytes: an introduction. Trends Cell Biol. 1995;5:85–7. [DOI] [PubMed] [Google Scholar]
- Read CB, Kuijper JL, Hjorth SAet al. Cutting Edge: Identification of Neutrophil PGLYRP1 as a Ligand for TREM-1. J Immunol. 2015;194:1417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relyveld EH. Preparation and use of calcium phosphate adsorbed vaccines. Dev Biol Stand. 1986;65:131–6. [PubMed] [Google Scholar]
- Rijneveld AW, Weijer S, Florquin Set al. Improved host defense against pneumococcal pneumonia in platelet-activating factor receptor-deficient mice. J Infect Dis. 2004;189:711–6. [DOI] [PubMed] [Google Scholar]
- Ring A, Weiser JN, Tuomanen EI. Pneumococcal trafficking across the blood-brain barrier molecular analysis of a novel bidirectional pathway. J Clin Invest. 1998;102:347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J, Haas CT, Pele LCet al. Intestinal APCs of the endogenous nanomineral pathway fail to express PD-L1 in Crohn's disease. Sci Rep. 2016;6:26747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root-Bernstein RS, Westall FC. Serotonin binding sites. II. Muramyl dipeptide binds to serotonin binding sites on myelin basic protein, LHRH, and MSH-ACTH 4–10. Brain Res Bull. 1990;25:827–41. [DOI] [PubMed] [Google Scholar]
- Royet J, Dziarski R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defences. Nat Rev Microbiol. 2007;5:264–77. [DOI] [PubMed] [Google Scholar]
- Saha S, Qi J, Wang Set al. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe. 2009;5:137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Yamashita T, Maeda Met al. Cloning of a lymphatic peptide/histidine transporter. Biochem J. 2001;356:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasawatari S, Okamura T, Kasumi Eet al. The solute carrier family 15A4 regulates TLR9 and NOD1 functions in the innate immune system and promotes colitis in mice. Gastroenterology. 2011;140:1513–25. [DOI] [PubMed] [Google Scholar]
- Schelhaas M, Shah B, Holzer Met al. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog. 2012;8:e1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher JU, Ubeda C, Artacho Aet al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, Matsson P, Shima JEet al. Comparison of human solute carriers. Protein Sci. 2010;19:412–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffler H, Demircioglu DD, Kühner Det al. NOD2 stimulation by Staphylococcus aureus-derived peptidoglycan is boosted by toll-like receptor 2 costimulation with lipoproteins in dendritic cells. Infect Immun. 2014;82:4681–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellinger OZ, Beaufay H, Jacques Pet al. Tissue fractionation studies. 15. Intracellular distribution and properties of β-N-acetylglucos-aminidase and β-galactosidase in rat liver. Biochem J. 1960;74:450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen Z, Karnovsky ML. Qualitative detection of muramic acid in normal mammalian tissues. Infect Immun. 1984;43:937–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N. The chemical structure of lysozyme substrates and their cleavage by the enzyme. Proc R Soc London Ser B Biol Sci. 1967;167:402–15. [DOI] [PubMed] [Google Scholar]
- Shimada K, Chen S, Dempsey PWet al. The NOD/RIP2 pathway is essential for host defenses against chlamydophila pneumoniae lung infection. PLoS Pathog. 2009;5:e1000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman DHS, Wu H, Karnovsky ML. Muramyl peptides and serotonin interact at specific binding sites on macrophages and enhance superoxide release. Biochem Biophys Res Commun. 1985;131:1160–7. [DOI] [PubMed] [Google Scholar]
- Slütter B, Plapied L, Fievez Vet al. Mechanistic study of the adjuvant effect of biodegradable nanoparticles in mucosal vaccination. J Control Release. 2009;138:113–21. [DOI] [PubMed] [Google Scholar]
- Smith DE, Clémençon B, Hediger MA. Proton-coupled oligopeptide transporter family SLC15: Physiological, pharmacological and pathological implications. Mol Aspects Med. 2013;34:323–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks FC, Silverstein MJ, Hunt JSet al. Complications of BCG immunotherapy in patients with cancer. N Engl J Med. 1973;289:827–30. [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani Aet al. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9–20. [DOI] [PubMed] [Google Scholar]
- Sun Q, Fan J, Han Det al. Evaluation of toxicity and adjuvant effects of peptidoglycan microspheres orally administered to mice. J Microencapsul. 2015;32:46–53. [DOI] [PubMed] [Google Scholar]
- Swaan PW, Bensman T, Bahadduri PMet al. Bacterial peptide recognition and immune activation facilitated by human peptide transporter PEPT2. Am J Respir Cell Mol Biol. 2008;39:536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez JC, Berthoud VM, Brañes MCet al. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol Rev. 2003;83:1359–400. [DOI] [PubMed] [Google Scholar]
- Ševčík J, Mašek K. The interaction of immunomodulator muramyl dipeptide with peripheral 5-HT receptors: Overview of the current state. Int J Immunopharmacol. 1999;21:227–32. [DOI] [PubMed] [Google Scholar]
- Ševčík J, Růžička V, Slánský Jet al. MDP and 5-HT receptors. Does MDP interact with 5-HT7 receptors? Int J Immunopharmacol. 2000;22:587–95. [DOI] [PubMed] [Google Scholar]
- Ševčík J, Růžička V, Slánský Jet al. Muramyl dipeptide (MDP) and 5-HT receptors. Neuroimmunomodulatory effects of MDP are probably not mediated through 5-HT4 or 5-HT1a receptors. Immunopharmacol Immunotoxicol. 2002;24:43–53. [DOI] [PubMed] [Google Scholar]
- Tada H, Aiba S, Shibata K-Iet al. Synergistic effect of Nod1 and Nod2 agonists with toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun. 2005;73:7967–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y, Hisamatsu T, Kamada Net al. Monocyte chemoattractant protein-1 contributes to gut homeostasis and intestinal inflammation by composition of IL-10–producing regulatory macrophage subset. J Immunol. 2010;184:2671–6. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Root-Bernstein RS, Shih JC. Peptide displacement of [3H]5-hydroxytryptamine binding to bovine cortical membranes. Brain Res Bull. 1990;25:817–20. [DOI] [PubMed] [Google Scholar]
- Thibodeau J, Bourgeois-Daigneault MC, Hupp?? Get al. Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur J Immunol. 2008;38:1225–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian PJ, Li BL, Shan YJet al. Extraction of peptidoglycan from L. Paracasei subp. Paracasei X12 and its preliminary mechanisms of inducing immunogenic cell death in HT-29 cells. Int J Mol Sci. 2015;16:20033–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomašić J, Ladešić B, Valinger Zet al. The metabolic fate of 14C-labeled peptidoglycan monomer in mice I. Identification of the monomer and the corresponding pentapeptide in urine. BBA - Gen Subj. 1980;629:77–82. [DOI] [PubMed] [Google Scholar]
- Tomita S, Irisawa T, Tanaka Net al. Comparison of components and synthesis genes of cell wall teichoic acid among Lactobacillus plantarum strains. Biosci Biotechnol Biochem. 2010;74:928–33. [DOI] [PubMed] [Google Scholar]
- Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17:13–24. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LAM, Ramjeet Met al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Girardin SE, Philpott DJet al. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 2004;5:1000–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troha K, Nagy P, Pivovar Aet al. Nephrocytes remove microbiota-derived peptidoglycan from systemic circulation to maintain immune homeostasis. Immunity. 2019;51:625–637.e3. [DOI] [PubMed] [Google Scholar]
- Tsuji NM, Mizumachi K, Kurisaki JI. Interleukin-10-secreting Peyer's patch cells are responsible for active suppression in low-dose oral tolerance. Immunology. 2001;103:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara A, Fujimoto Y, Fukase Ket al. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–11. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AMet al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–5. [DOI] [PubMed] [Google Scholar]
- Valinger Z, Ladešić B, Hršak Iet al. Relationship of metabolism and immunostimulating activity of peptidoglycan monomer in mice after three different routes of administration. Int J Immunopharmacol. 1987;9:325–32. [DOI] [PubMed] [Google Scholar]
- Vavricka SR, Musch MW, Chang JEet al. hPepT1 transports muramyl dipeptide, activating NF-κB and stimulating IL-8 secretion in human colonic Caco2/bbe cells. Gastroenterology. 2004;127:1401–9. [DOI] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IGet al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–74. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, De Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–67. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Tomasz A. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J Biol Chem. 2000;275:20496–501. [DOI] [PubMed] [Google Scholar]
- Voss E, Wehkamp J, Wehkamp Ket al. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol Chem. 2006;281:2005–11. [DOI] [PubMed] [Google Scholar]
- Wagner RN, Proell M, Kufer TAet al. Evaluation of Nod-like receptor (NLR) effector domain interactions. PLoS One. 2009;4:e4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder P, Buchar E, Machková Zet al. Pharmacokinetic profile of the immunomodulating compound adamantylamide dipeptide (adDP), a muramyl dipeptide derivative in mice. Immunopharmacol Immunotoxicol. 1991;13:101–19. [DOI] [PubMed] [Google Scholar]
- Wang M, Liu L-H, Wang Set al. Human peptidoglycan recognition proteins require zinc to kill both Gram-positive and Gram-negative bacteria and are synergistic with antibacterial peptides. J Immunol. 2007;178:3116–25. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu Y, Li Pet al. Expression and regulation of proton-coupled oligopeptide transporters in colonic tissue and immune cells of mice. Biochem Pharmacol. 2018;148:163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZM, Li X, Cocklin RRet al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 2003;278:49044–52. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Fichtner-Feigl Set al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010;120:1645–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DM, Harboe ZB, Sanders EAMet al. Association of serotype with risk of death due to pneumococcal pneumonia: a meta‐analysis. Clin Infect Dis. 2010;51:692–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs Get al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA. 2011;108:19824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windheim M, Lang C, Peggie Met al. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem J. 2007;404:179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AJ, Reyes CN, Liang Wet al. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell. 2016;166:624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfle SJ, Strebovsky J, Bartz Het al. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol. 2011;41:413–24. [DOI] [PubMed] [Google Scholar]
- Xu XL, Lee RTH, Fang HMet al. Bacterial peptidoglycan triggers candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Shimada S, Guo Wet al. Cloning and functional expression of a brain peptide/histidine transporter. J Biol Chem. 1997;272:10205–11. [DOI] [PubMed] [Google Scholar]
- Yapo A, Petit JF, Lederer Eet al. Fate of two 14C labelled muramyl peptides: Ac-Mur-L-Ala-γ-D-Glu-meso-A2pm and Ac-Mur-L-Ala-γ-D-Glu-Meso-A2pm-D-Ala-D-Ala in mice. Evaluation of their ability to increase non specific resistance to Klebsiella infection. Int J Immunopharmacol. 1982;4:143–9. [DOI] [PubMed] [Google Scholar]
- Yeretssian G, Correa RG, Doiron Ket al. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474:96–9. [DOI] [PubMed] [Google Scholar]
- Yokoyama CC, Baldridge MT, Leung DWet al. LysMD3 is a type II membrane protein without an in vivo role in the response to a range of pathogens. J Biol Chem. 2018;293:6022–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kinoshita K, Ashida M. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J Biol Chem. 1996;271:13854–60. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Lien E, Ingalls RRet al. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- Zeissig Y, Petersen BS, Milutinovic Set al. XIAP variants in male Crohn's disease. Gut. 2015;64:66–76. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zhao J, Song Yet al. The E3 ligase RNF34 is a novel negative regulator of the NOD1 pathway. Cell Physiol Biochem. 2014;33:1954–62. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Van Der Fits L, Voerman JSet al. Identification of serum N-acetylmuramoyl-L-alanine amidase as liver peptidoglycan recognition protein 2. Biochim Biophys Acta - Proteins Proteomics. 2005;1752:34–46. [DOI] [PubMed] [Google Scholar]