Abstract
Protein lysine methylation is a prevalent post-translational modification (PTM) and plays critical roles in all domains of life. However, its extent and function in photosynthetic organisms are still largely unknown. Cyanobacteria are a large group of prokaryotes that carry out oxygenic photosynthesis and are applied extensively in studies of photosynthetic mechanisms and environmental adaptation. Here we integrated propionylation of monomethylated proteins, enrichment of the modified peptides, and mass spectrometry (MS) analysis to identify monomethylated proteins in Synechocystis sp. PCC 6803 (Synechocystis). Overall, we identified 376 monomethylation sites in 270 proteins, with numerous monomethylated proteins participating in photosynthesis and carbon metabolism. We subsequently demonstrated that CpcM, a previously identified asparagine methyltransferase in Synechocystis, could catalyze lysine monomethylation of the potential aspartate aminotransferase Sll0480 both in vivo and in vitro and regulate the enzyme activity of Sll0480. The loss of CpcM led to decreases in the maximum quantum yield in primary photosystem II (PSII) and the efficiency of energy transfer during the photosynthetic reaction in Synechocystis. We report the first lysine monomethylome in a photosynthetic organism and present a critical database for functional analyses of monomethylation in cyanobacteria. The large number of monomethylated proteins and the identification of CpcM as the lysine methyltransferase in cyanobacteria suggest that reversible methylation may influence the metabolic process and photosynthesis in both cyanobacteria and plants.
Keywords: Cyanobacteria, Lysine monomethylation, Lysine methyltransferase, Post-translational modification, Aspartate aminotransferase
Introduction
Lysine methylation is a reversible post-translational modification (PTM) in which mono-, di-, or tri-methyl groups are added to the ɛ-amino group of a lysine [1]. Since lysine methylation was first reported over half a century ago in Salmonella typhimurium [2], a wide range of lysine methylated proteins were discovered, suggesting that lysine methylation is a common and reversible PTM in both prokaryotes and eukaryotes. Despite the critical discoveries and intense research interest in histone lysine methylation, non-histone proteins also contain a large number of lysine methylations. The recent revelation that lysine methylation occurs in hundreds and potentially thousands of non-histone proteins suggested diverse functions of lysine methylation and ushered in a new area of research [3], [4]. For example, in eukaryotes, lysine methylation is involved in growth signaling and DNA damage response [3], [5]. In bacteria, lysine methylation could play a role in cell motility in Synechocystis sp. PCC 6803 (hereafter Synechocystis) [6]. In archaea, the helicase activity in Sulfolobus islandicus is regulated by methylation [7]. Such reports demonstrate that lysine methylation occurs in numerous proteins and influences diverse biological functions in all domains of life.
Lysine methylation is a reversible PTM triggered by protein lysine methyltransferases (PKMTs) and reversed by lysine demethylases (KDMs) [8]. The first PKMT responsible for lysine methylation of histone proteins was revealed in 2000 [9]. Since then, more PKMTs have been reported in diverse organisms. In humans, 52 genes have been predicted to encode PKMTs [10], which can methylate p53 [11], transcription factor TAF10 [12], and many other non-histone proteins. In prokaryotes, the first identified PKMT was the bacterial PrmA enzyme in Escherichia coli [13]. In Pseudomonas, EftM catalyzes the tri-methylation of elongation factor EF-Tu [14], and in Rickettsia, two PKMTs can catalyze the methylation of different lysine residues in outer membrane protein B (OmpB) [15], [16]. However, PKMT counterparts in cyanobacteria remain unknown. In the present study, we identified and characterized a PKMT, CpcM, in Synechocystis that can catalyze lysine methylation reactions. The function of the enzyme in cyanobacteria was discussed.
Cyanobacteria are a group of prehistoric prokaryotes that carry out oxygenic photosynthesis. They are of global economic importance owing to their roles in nitrogen and carbon cycles on Earth [17]. Synechocystis is the first sequenced cyanobacterium and a model organism for photosynthesis and carbon metabolism studies [18]. Lysine methylation has been reported to be one of the functional PTMs, which plays critical roles in signaling pathways and metabolism in diverse organisms [1], [3], [4]. Therefore, we speculate that lysine methylation may be involved in metabolic processes in cyanobacteria. However, the extent and function of lysine methylation in cyanobacteria remain largely unknown.
To elucidate the functions and regulatory mechanisms of lysine methylation in cyanobacteria, it is critical to establish a lysine methylome and identify the enzymes that control the methylation status of proteins. Recent developments in mass spectrometry (MS) technology have enabled high-throughput identification of lysine methylated proteins in Arabidopsis chloroplasts [19], S. islandicus [20], human cells [21], and Saccharomyces cerevisiae mitochondria [22]. However, detection of lysine monomethylation across the proteome is still a daunting technical challenge largely because of the minimal structural changes caused by a methyl group [23]. To address the challenge, we previously developed a approach for the identification of protein lysine monomethylation that combines propionylation of monomethylated proteins, enrichment of the propionyl-monomethylated peptides, and MS analysis [24]. Using this method, 446 lysine monomethylation sites in 398 proteins were identified from human liver tissues with high confidence [24]. Therefore, this approach offers a highly sensitive and accurate method for globally profiling lysine monomethylation. In the present study, we used this method to perform a proteomic analysis of lysine monomethylated proteins in Synechocystis. 376 lysine monomethylation sites on 270 proteins were identified in Synechocystis with high confidence, and many of the monomethylated proteins participate in photosynthetic and metabolic processes. Our results present an extensive dataset with regard to lysine monomethylation in a photosynthetic organism.
Results
Proteomic analysis of monomethylated proteins in Synechocystis
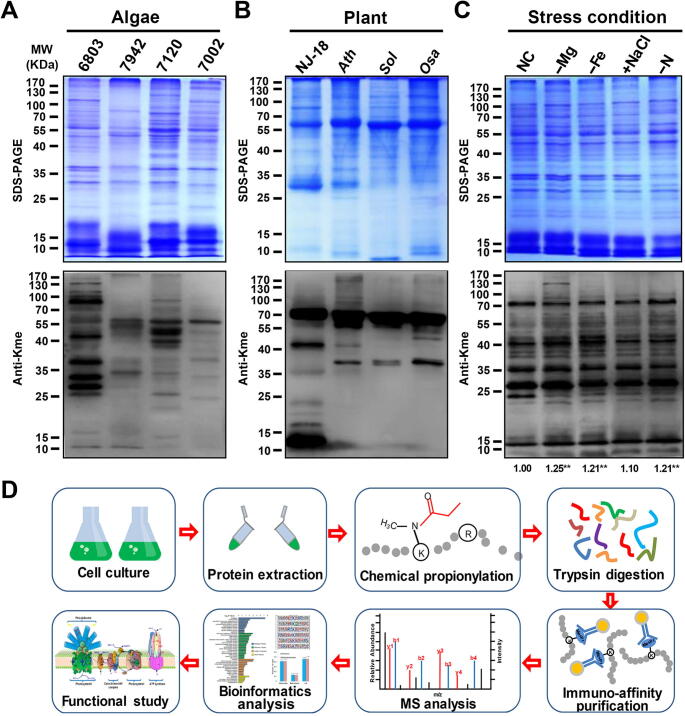
To test the extent of lysine methylation in photosynthetic organisms, we analyzed the methylation levels of whole-cell lysates from prokaryotic photosynthetic organisms (Synechocystis sp. PCC 6803, Synechococcus elongatus PCC 7942, Anabaena sp. PCC 7120, and Synechococcus sp. PCC 7002) and eukaryotic photosynthetic organisms (Chlorella sp. NJ-18, Arabidopsis thaliana, Spinacia oleracea, and Oryza sativa) using anti-monomethyllysine antibody. Multiple immunoblot signals were detected in the photosynthetic organisms, suggesting the prevalence of lysine methylation (Figure 1A and B). We further observed changes in lysine methylation levels in Synechocystis under different culture conditions (normal condition, magnesium deficiency, ferric deficiency, high salt concentration, and nitrogen deficiency), suggesting that lysine methylation may play regulatory roles in response to the environmental stresses (Figure 1C). To explore the functions of lysine monomethylation in Synechocystis, we employed a previously developed technique [24] to identify monomethylated proteins (Figure 1D). In total, 376 distinct lysine monomethylation sites in 270 proteins were confirmed with a false discovery rate (FDR) < 1%. Table S1 provides information on all identified modified peptides and proteins. The high quality of the identified peptide data was demonstrated by the following: the average site localization score was 100.4684, and the overall mass accuracy was 0.04365 (Figure S1A). Furthermore, we calculated the number of methylation sites identified per protein, and the results showed that most of the modified proteins contained a single methylation site, and the remaining proteins (22%) contained two or more methylation sites (Figure S1B). Our dataset provided a lysine monomethylome of a photosynthetic organism and confirmed that lysine monomethylation is a common and abundant PTM in cyanobacteria.
Figure 1.
Profiling lysine methylation in photosynthetic organisms
A. Detection of lysine methylation in Synechocystis sp. PCC 6803 (6803), Synechococcus sp. PCC 7942 (7942), Anabaena sp. PCC 7120 (7120), and Synechococcus sp. PCC 7002 (7002) using 20 μg of proteins and anti-monomethyllysine (anti-Kme) antibody. B. Detection of lysine methylation in Chlorella sp. NJ-18 (NJ-18), Arabidopsis thaliana (Ath), Spinacia oleracea (Sol), and Oryza sativa (Osa) using 10 μg of proteins and anti-Kme antibody. C. Profiling of lysine methylation in Synechocystis sp. PCC 6803 under various stress conditions. Proteins (20 μg) were extracted from the cells cultured under normal condition (NC), magnesium deficiency (−Mg), ferric deficiency (−Fe), sodium chloride enrichment (+NaCl), or nitrogen deficiency (−N). A densitometry analysis was performed using ImageJ software. Samples were standardized through comparisons with Coomassie-blue-stained gel. Data represent results from three independent experiments. The level of statistical significance was determined using two-sample Student’s t-test (**, P < 0.01). D. Flowchart illustrating the experimental procedure for lysine monomethylation proteomics.
Functional characterization of identified monomethylated proteins
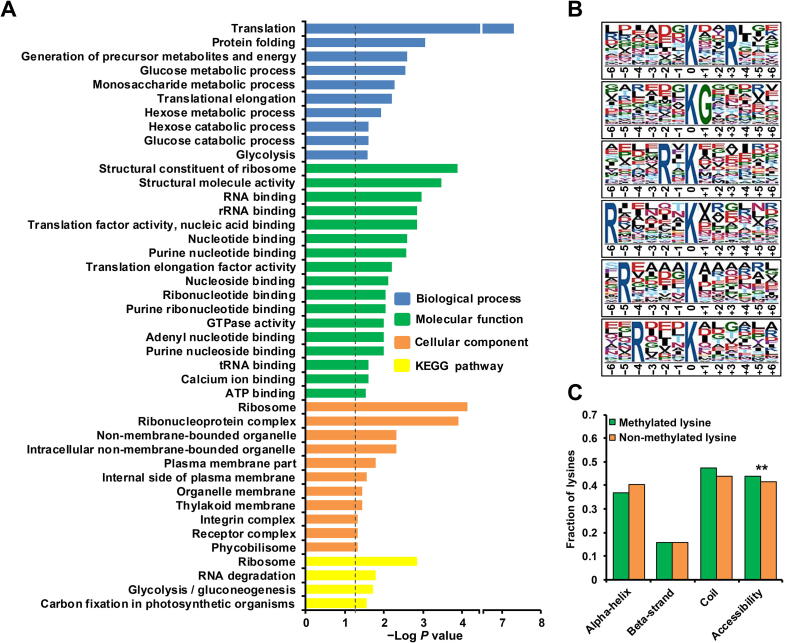
To explore the functions of all identified monomethylated proteins, we applied DAVID bioinformatics resources to investigate the Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in the lysine monomethylome compared to those in the entire proteome (Table S2). Figure 2A depicts the biological processes, molecular functions, cell organelles involved, and pathways of all identified monomethylated proteins. GO enrichment revealed that the modified proteins were markedly enriched in translation, protein folding, translational elongation, glycolysis, and other carbohydrate metabolic processes. Consistently, the modified proteins were overrepresented in ribosomal structures (P = 1.29E−04) and were enriched in binding and catalytic activities. On the basis of the enrichment analysis among cellular components, we obtained similar results, where a large proportion of modified proteins were enriched in ribosome (P = 7.39E−05), ribonucleoprotein complex (P = 1.26E−04), non-membrane-bound organelle (P = 4.68E−03), phycobilisome (PBS) (P = 4.72E−02), and numerous membrane structures (plasma membrane part, internal side of plasma membrane, organelle membrane, and thylakoid membrane). In addition, KEGG analysis presented similar results, where the methylated proteins were overrepresented in ribosome (P = 1.39E−03), RNA degradation (P = 1.65E−02), glycolysis/gluconeogenesis (P = 1.89E−02), and carbon fixation in photosynthetic organisms (P = 2.8E−02). Notably, similar distribution patterns of methylated proteins, including elongation factors and several ribosomal proteins, were also observed in human cells according to previous reports [24], [25]. Therefore, lysine methylation could play a regulatory role in protein translation and energy metabolism, which is similar to the case in lysine acetylome [26] and malonylome [27] in Synechocystis.
Figure 2.
Enrichment analysis of monomethylated proteins and characterization of lysine monomethylation sites
A. Bar graph showing the enrichment of monomethylated proteins for Gene Ontology (GO) categories (including biological processes, molecular functions, and cellular components) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The enrichment of GO categories and KEGG pathways was carried out using DAVID bioinformatics tools (P < 0.05). B. Sequence motifs for all lysine monomethylation sites. The significant motifs were identified by Motif-X software with a corresponding P < 0.000001. C. Distributions of methylated and non-methylated lysines in structured and unstructured regions of the proteins. Statistical significance was determined by two-sample Student’s t-test (**, P < 0.01).
We subsequently analyzed the position-specific sequence biases for the surrounding sequences of the methylated lysines using Motif-X software. Figure 2B illustrates a significant preference for a polar basic amino acid (arginine) in the modified peptide sequences, showing a site-specific methylation motif that is quite different from those reported in eukaryotes. In addition, we observed significant enrichment of a non-polar hydrophobic amino acid (glycine) next to the lysine. To the best of our knowledge, phenylalanine/leucine-rich motifs are highly overrepresented in the methylated peptide subset [28], and a methionine-rich region appears to be conserved through human and yeast cells [25], [29]. On the basis of the findings, we speculated that there could be a sequence-specific lysine methyltransferase or demethylase in prokaryotes.
We further predicted the secondary structure localization and solvent accessibility of the modified and non-modified lysines of the methylated proteins (Table S3). Our results showed that the methylated lysines have a higher side-chain solvent accessibility (relative surface accessibility = 0.44) than non-methylated lysines (relative surface accessibility = 0.42) (P = 0.00331), suggesting that methylation sites are enriched on the surface of the protein (Figure 2C). Overall, the methylated lysines could have a weak preference for coil structures, in which 47% of methylated lysines were predicted to be located, in comparison to 44% of non-methylated lysines. The methylation sites appeared to have non-structural preferences that are consistent with those of protein phosphorylation sites, indicating a strong correlation with disordered regions [30].
Lysine monomethylation in photosynthesis and metabolism
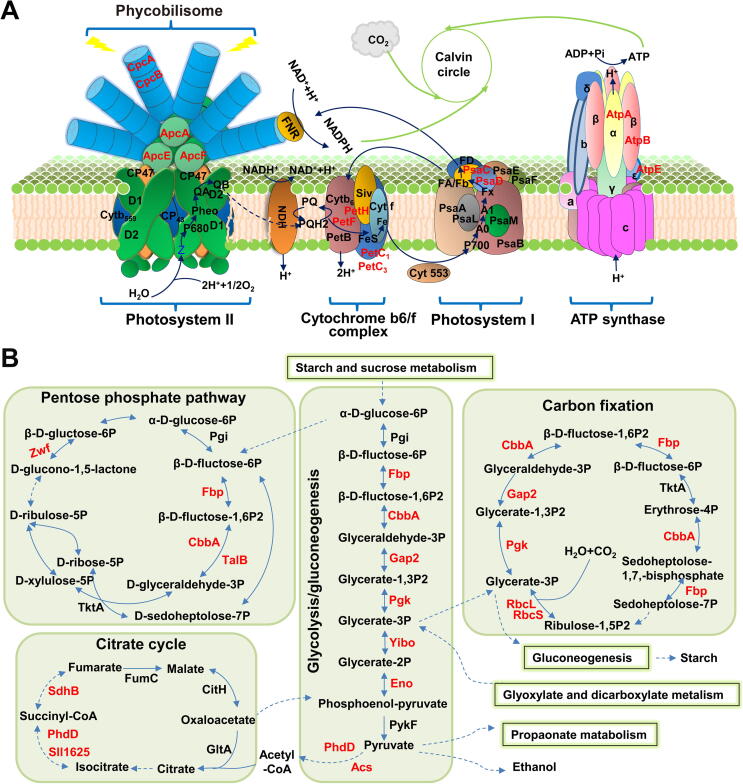
Increasing evidence indicates that diverse PTMs in both cyanobacteria [31] and plants [32] could be involved in the regulation and coordination of photosynthetic and metabolic processes, which are tightly linked [33]. To explore the potential functions of lysine methylation events in photosynthetic and metabolic processes, the methylated proteins were mapped to the KEGG pathways. Numerous methylated proteins were located in the thylakoid membrane, such as PBS (CpcA, CpcB, ApcA, ApcE, and ApcF), cytochrome b6/f complex (PetH, PetF, PetC1, and PetC3), photosystem I (PSI) components (PsaD and PsaC), and ATPase complex (AtpA, AtpB, and AtpE) (Figure 3A). Since the asparagine methylation in the β-subunits of the phycobiliproteins (CpcB and ApcB) is a critical factor in the maintenance of efficient energy transfer [34], we inferred that the lysine methylation events occurring in phycobiliproteins could also play regulatory roles in energy absorption and transfer in photosynthesis. It is critical to note that PSI subunit II (PsaD) was methylated at eight lysine sites, further implying the potential functions of lysine methylation in the photosynthetic complex.
Figure 3.
Synopsis of lysine monomethylation events involved in photosynthesis and carbon metabolism in Synechocystis
A. Schematic illustration of monomethylated enzymes involved in photosynthesis. B. Schematic illustration of monomethylated enzymes involved in the pentose phosphate pathway, carbon fixation, glycolysis/gluconeogenesis, and citrate cycle. The identified monomethylated proteins were marked light red.
We further revealed the occurrence of lysine methylation events in metabolic processes, such as glycolysis/gluconeogenesis, citrate cycle, carbon fixation, and the pentose phosphate pathway (Figure 3B). A high number of enzymes associated with glycolysis/gluconeogenesis (8), citrate cycle (3), and carbon fixation (8) were found to be methylated. Similarly, four of the identified methylated enzymes were involved in the pentose phosphate pathway. Our results suggested that the identified methylation sites on the key enzymes could play regulatory roles in the respective metabolic pathways.
Identification of Synechocystis lysine methyltransferase
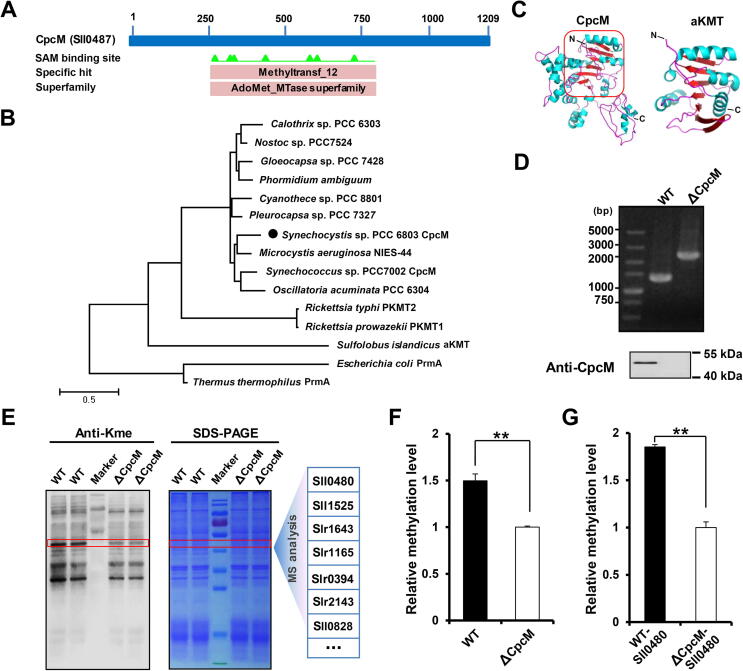
In the Synechocystis genome, 12 genes were predicated to encode putative methyltransferases (Table 1); however, no PKMT has been identified in cyanobacteria. To identify PKMTs in cyanobacteria, we performed conserved domain research of Synechocystis proteins and found out that a previously identified asparagine methyltransferase, CpcM [34], [35], contains S-adenosyl methionine (SAM) binding sites and belongs to the AdoMet_MTase superfamily (Figure 4A). CpcM can post-translationally methylate the amide nitrogen of asparagine in phycobiliproteins in Synechocystis [34], [35]. The maximum likelihood phylogram showed that CpcM is distantly related to the 7BS PKMTs, including PKMT1 in R. prowazekii, PKMT2 in R. typhi, aKMT in S. islandicus, and PrmA in E. coli and Thermus thermophiles, which belong to the superfamily of 7BS PKMTs (Figure 4B). In addition, structural modeling of CpcM using PKMT2 as a template revealed that the core structure of CpcM consists of seven β-strands, where the first six β-strands are parallel and the rest has an opposite orientation, suggesting a nearly identical core fold with the 7BS PKMT family (Figure 4C). Apart from the highly conserved core 7BS fold, the PKMTs usually contain different N-terminal structural elements that are part of the catalytically active enzyme (Figure S2). Therefore, our results suggested that CpcM may be a PKMT in Synechocystis.
Table 1.
Potential protein methyltransferases in Synechocystis sp. PCC6803
| ID | Description (prediction) | Length (aa) | MW (kDa) |
Domain |
E value | Accession | |
|---|---|---|---|---|---|---|---|
| From | To | ||||||
| Sll0487 | SAM-dependent methyltransferase | 402 | 45.85 | 58 | 155 | 8.56E−25 | pfam13649 |
| Sll1237 | N(5)-glutamine methyltransferase | 299 | 33.82 | 6 | 299 | 1.68E−131 | TIGR00536 |
| Sll0171 | Probable aminomethyltransferase | 372 | 41.02 | 7 | 367 | 0 | TIGR00528 |
| Sll0829 | Probable methyltransferase | 212 | 37.07 | 49 | 139 | 9.24E−29 | pfam13649 |
| Sll1300 | Guanine-N(7)-methyltransferase | 211 | 24.42 | 21 | 211 | 3.97E−94 | TIGR00091 |
| Sll1407 | Probable methyltransferase | 265 | 30.75 | 45 | 139 | 1.61E−31 | pfam08241 |
| Sll1909 | Ribosomal protein L11 methyltransferase | 300 | 33.70 | 7 | 290 | 2.02E−151 | TIGR00406 |
| Slr0095 | O-methyltransferase | 220 | 24.31 | 18 | 219 | 3.76E−121 | pfam01596 |
| Slr0309 | Uncharacterized methyltransferase | 473 | 54.48 | 3 | 406 | 6.00E−57 | COG1032 |
| Slr1115 | Probable methyltransferase | 257 | 28.76 | 74 | 173 | 8.49E−27 | pfam13649 |
| Slr1610 | Putative C-3 methyl transferase | 433 | 48.01 | 271 | 428 | 4.38E−92 | pfam08484 |
| 30 | 91 | 1.57E−26 | pfam08421 | ||||
| 108 | 265 | 9.37E−13 | pfam13489 | ||||
| Slr1763 | Probable methyltransferase | 400 | 45.36 | 240 | 395 | 4.55E−67 | pfam08484 |
| 3 | 63 | 8.16E−25 | pfam08421 | ||||
| 80 | 231 | 1.21E−14 | pfam13489 | ||||
Figure 4.
CpcM has a lysine methyltransferase activity in Synechocystis
A. CpcM was predicted to be a potential SAM-dependent methyltransferase. B. Evolutionary conservation of lysine methyltransferases. The maximum likelihood phylogenetic tree was built using MEGA 5.0.5 software. C. Homology model of CpcM and its ortholog aKMT from Sulfolobus islandicus. D. Verification of the mutant lacking CpcM by PCR and Western blot. E. Identification of lysine methylated substrates. Three pairs of bands from the total cell lysates of WT and ΔCpcM in Coomassie-blue-stained gel, which exhibited varying levels of lysine methylation by Western blot analysis, were cut from three replicate gels and trypsin-digested in-gel, followed by LC-MS/MS analysis. F. Quantitative analysis of the corresponding methylated proteins from the bands in (E). The intensities of modified peptides were normalized using the identified protein abundances determined by MaxQuant software. G. Quantitative analysis of methylated Sll0480 from the bands in (E). Data are presented as means ± SD from three independent experiments. A two-sample Student’s t-test was performed to determine the level of statistical significance (**, P < 0.01).
To test our hypothesis and investigate the functions of CpcM in Synechocystis, we deleted the cpcM gene by interposon mutagenesis, and the deletion was verified by PCR and immunoblot analysis with anti-CpcM antibody (Figure 4D). We then compared the global lysine monomethylation levels in the wild-type (WT) strain with those in the ΔCpcM strain using pan anti-monomethylated lysine antibodies. In contrast to the WT strain, the methylation levels of the whole proteins in the ΔCpcM strain significantly decreased (Figure 4E). To validate the decrease in protein lysine methylation and identify the potential methylated substrates, the gel bands with the apparent decreases in methylation levels were cut from three replicate gels and trypsin-digested for further MS analysis. As expected, methylated peptides were identified in both the WT and ΔCpcM strains. Table S4 provides information on all identified peptides (Table S4A), proteins (Table S4B), and methylated peptides (Table S4C). To estimate the stoichiometry of modified proteins, the methylation levels were then characterized by estimating the MS intensities of methylated peptides in the WT and ΔCpcM strains. Subsequently, we corrected the modified peptide intensity with protein abundances determined by the Label-Free Quantification (LFQ) method as previously described [36]. The abundance of methylated proteins and sites was higher in WT than in ΔCpcM (Figure 4F, Table S4D and E). Only the methylated peptides that were quantified in three replicates were used for relative quantification (Table S4E). All MS/MS spectra of methylated peptides that were used for relative quantification were uploaded onto the PeptideAtlas public database with the identifier PASS01127 (MS/MS spectra of Table S4E). One of the identified methylated proteins (Sll0480), a probable aspartate aminotransferase (AAT), showed approximately 50% reduction in methylation levels in ΔCpcM strain (Figure 4G, Table S4D–F). On the basis of the observations, we speculated that CpcM may be a PKMT and Sll0480 is one of its substrates in Synechocystis.
Effect of lysine methylation on Sll0480 activity
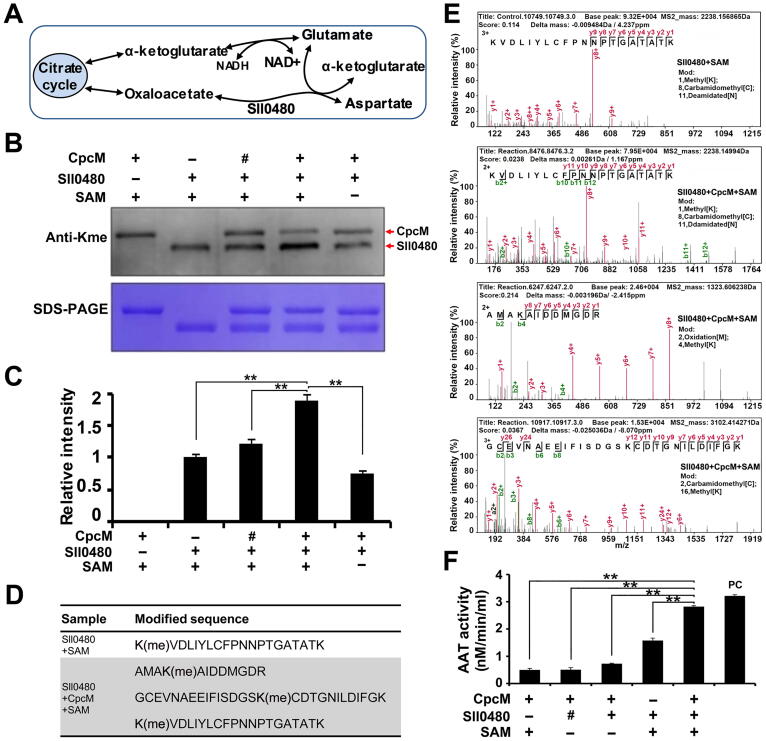
AAT can catalyze the conversion of oxaloacetate and glutamate to aspartate and α-ketoglutarate, respectively (Figure 5A). On the basis of the conservation analysis, we found out that Sll0480 exhibited limited evolutionary conservation and sequence identity with its probable orthologs (Figure S3A and B). However, structural modeling analysis demonstrated that Sll0480 and its orthologs, including AspC (E. coli) and Got1 (Homo sapiens), had similar 3D structures (Figure S3C). Therefore, it is likely that Sll0480 is an AAT in Synechocystis.
Figure 5.
Effect of lysine methylation on Sll0480 enzymatic activity
A. Schematic illustration of the metabolic pathway catalyzed by Sll0480. B. Western blot analysis of the methylation level of Sll0480 after incubation with CpcM and SAM using anti-Kme antibody. Samples were standardized through comparisons with Coomassie-blue-stained gel. The untreated Sll0480 (Sll0480+SAM) group and the heat-inactivated CpcM (represented by #) group were used as negative controls. SAM, S-adenosyl methionine. C. The densitometry analysis of methylated Sll0480 in the reactions from (B) using ImageJ software. D. List of identified methylated peptides from Sll0480 in the reactions from (B). The untreated Sll0480 (Sll0480+SAM) was used as a control, and the Sll0480 incubated with CpcM and SAM was tested. E. Representative MS/MS spectra of the methylated peptides in (D). A series of b-ions (green) and y-ions (red) were labeled in the spectra. F. Enzyme activity of Sll0480 after incubation with CpcM and SAM. The provided aspartate aminotransferase (AAT) in the kit was used as a positive control (PC), while the reactions without Sll0480 or heat-inactivated Sll0480 (represented by #) were used as negative controls. Data are presented as means ± SD from three independent experiments. The level of statistical significance was determined by two-sample Student’s t-test (**, P < 0.01).
To confirm that Sll0480 is the potential lysine methylated substrate catalyzed by CpcM, we first expressed and purified CpcM and Sll0480. Subsequently, Sll0480 was methylated by CpcM in the presence of SAM, and the methylation level of Sll0480 was detected by immunoblot analysis. As shown in Figure 5B, compared with the untreated Sll0480 (Sll0480 + SAM) group and the heat-inactivated CpcM group, the methylation level of Sll0480 significantly increased when it was incubated with CpcM and SAM, while there was no increase in the methylation level of Sll0480 when it was incubated with CpcM alone. These results were further confirmed by densitometry analysis (Figure 5C). In the present study, E. coli was used to overexpress CpcM and Sll0480 proteins. Since one or more PKMTs could exist in E. coli, the overexpressed CpcM and Sll0480 proteins from E. coli may be methylated by the endogenous PKMTs in E. coli and could be detected by the anti-monomethyllysine antibody (Figure 5B). We further performed an MS quantitative analysis and identified all methylated peptides of Sll0480 catalyzed by CpcM using pFind software [37]. Table S5A presents the results of all identified peptides and proteins. Consistent with the immunoblot analysis, we observed three unique methylated peptides from Sll0480 after incubation with CpcM in the presence of SAM, and only one methylated peptide was identified in the control group containing Sll0480 and SAM, indicating that CpcM could methylate Sll0480 (Figure 5D, Table S5B and C). Figure 5E illustrates the representative spectra of the identified methylated peptides that contained successive b- or y-type ion series, confirming the high accuracy of the identification of modified peptides. We further selected two methylated proteins from Table S4E, i.e., fructose-1, 6-bisphosphatase (Slr2094) and phosphoglycerate kinase (Slr0394), to test if they are substrates of CpcM. As shown in Figure S4, the lysine methylation level in Slr2094 was significantly enhanced when incubated with CpcM and SAM, while the methylation level of Slr0394 exhibited no significant difference.
To assess whether the lysine methylation catalyzed by CpcM could influence Sll0480 enzyme activity, the AAT activities of CpcM-treated and untreated Sll0480 were measured using an AAT activity assay kit. The AAT activity of Sll0480 was significantly enhanced in the presence of CpcM and SAM, when compared with the groups in which Sll0480, SAM or CpcM was absent or Sll0480 was heat-inactivated (Figure 5F). Consequently, it is plausible that Sll0480 is one of the substrates of CpcM and lysine methylation could regulate enzyme activity of Sll0480.
Effects of CpcM on photosynthesis in Synechocystis
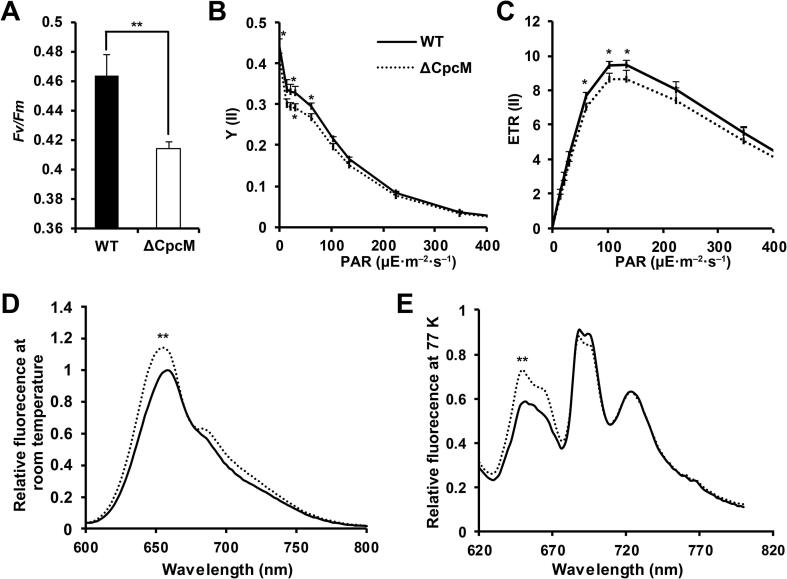
The ΔCpcM mutant has been reported to be sensitive to high light treatment, and its state transitions were affected [34]. To further explore the potential functions of CpcM in Synechocystis, we measured the photosynthetic electron flow in the thylakoid membrane in both WT and ΔCpcM strains. On the basis of the results of the chlorophyll fluorescence analysis, we detected significant decreases in the maximal photochemical efficiency of photosystem II (PSII) (Fv/Fm), actual photochemical efficiency of PSII [Y(II)], and PSII-mediated electron transport rate [ETR(II)] in the ΔCpcM strain relative to those in the WT strain (Figure 6A–C), suggesting that a deficiency in cpcM could influence energy transfer in photosystems. We further examined the energy transfer from the PBS to chlorophyll a (Chl a) in the thylakoid membrane by measuring the fluorescence emission spectra of WT and ΔCpcM strains at room temperature or low temperature (77 K). In contrast to the WT strain, an increase of fluorescence emission spectra was detected in the ΔCpcM mutant at room temperature when the PBS was excited using 590-nm light (Figure 6D). Since the fluorescence emission spectra at 77 K can reveal resolved emissions related to each of the phycobiliproteins, we observed large differences between the WT and ΔCpcM strains. As shown in Figure 6E, the ΔCpcM strain was characterized by increased emissions from phycocyanin (PC) at 650 nm as well as allophycocyanin (APC) at 660 nm relative to the WT strain. Conversely, the 695-nm emission peak related to PSII was slightly decreased in the ΔCpcM strain. Therefore, our 77 K emission data revealed that CpcM deletion could affect the efficiency of energy transfer from PBS to PSII Chl a.
Figure 6.
Functional effects of CpcM deletion in Synechocystis
A. Maximum photochemistry efficiency of PSII (FV/FM) in the WT and ΔCpcM strains. B. Light-intensity dependence of the actual photochemical efficiency of PSII [Y(II)] in the WT and ΔCpcM strains. PAR, photosynthetically active radiation. C. Light-intensity dependence of the PSII-mediated electron transport rate [ETR(II)] in the WT and ΔCpcM strains. Y(II) and ETR(II) were calculated using Dual-PAM software under different PARs. D. and E. Fluorescence emission spectra at room temperature (D) and 77 K (E) for the dark-adapted WT and ΔCpcM strains excited at 590 nm (phycobilisome excitation). 77 K fluorescence emission spectra were normalized at 730 nm for comparison of the fluorescence emission peak derived from phycobilisome. All depicted spectra represent averages of three biological replicates with five individual readings of each replicate. All data are presented as means ± SD from three independent experiments. Statistical significance was determined by two-sample Student’s t-test (*, P < 0.05; **, P < 0.01).
Discussion
Recently, we have analyzed the lysine acetylome [26] and malonylome [27] of Synechocystis and revealed previously unreported roles of the two acylations in photosynthesis and metabolism. Here, we report the first systematic study of lysine monomethylation in Synechocystis. The identified monomethylated proteins are involved in diverse pathways and processes (Figure 2A), including some major metabolic and photosynthetic pathways (Figure 3). Since lysine methylation is well recognized as a critical PTM and influences diverse cellular functions, our dataset revealed a large number of putative functional lysine methylation sites in Synechocystis. Our results also showed that lysine methylation is a common PTM in plants (Figure 1). Therefore, the identification of site-specific lysine monomethylation events in specific proteins is fundamental for further exploring the molecular mechanisms and functional roles in such a critical PTM. It should be noted that multiple factors may prevent the detection of certain methylated proteins in proteomics experiments, for example, the methylated proteins may be expressed under certain culture conditions, in very low quantities, or have very short half-lives. It could also be challenging to detect some hydrophobic peptides in membrane-bound proteins because of the technical limitations of MS.
Among the numerous identified proteins in the present study, the probable AAT, encoded by sll0480, was found to be methylated by a previously identified asparagine methyltransferase CpcM. Therefore, it was selected as a model for testing the effect of monomethylation on its enzyme activity. The enzyme AAT plays critical roles in carbon, nitrogen, and amino acid metabolism in diverse organisms [38]. In prokaryotes, AAT plays a key role in amino acid biosynthesis [39]. In eukaryotes, AAT plays essential roles in growth, reproduction, development, and stress responses [40], [41]. In the present study, several attempts were made to obtain AAT deletion mutants in Synechocystis using interposon mutagenesis, but all of them were unsuccessful. We concluded that deletion of AAT is lethal in Synechocystis, demonstrating the critical role of AAT in cyanobacteria. This lethal phenotype in Synechocystis is consistent with the results reported in Arabidopsis thaliana [42], indicating that AAT enzymes are indispensable for the viability of both cyanobacteria and plants. Notably, it has been reported that certain lysine residues in the active center can affect substrate recognition and AAT enzyme activity [43], [44]. Although monomethylation in lysine residues may not induce profound conformational changes in protein structures, it alters the charges in ε-amino groups and affects electrostatic interactions and hydrogen bonding in protein–protein interactions. Therefore, it may affect many features of the methylated protein, such as enzyme activity and enzyme substrate interactions. Consistent with this view, our functional results showed that lysine methylation could regulate AAT activity significantly (Figure 5F). Therefore, lysine methylation could be a novel regulatory mechanism for AAT activity in Synechocystis.
When investigating the functions of a PTM, it is critical to identify the enzymes catalyzing the modification. The dynamic balance of lysine methylation in vivo is regulated by PKMTs and KDMs. In recent years, more PKMTs and KDMs have been identified, and the enzymes are directly associated with human diseases and are pursued as therapeutic targets [45], [28]. However, to date, no PKMT has been identified in cyanobacteria. For the first time, we demonstrated a robust lysine methyltransferase activity of CpcM both in vitro and in vivo with AAT as one of its substrates. CpcM was initially identified as an asparagine methyltransferase in Synechocystis [34], [35]. Deletion of CpcM in Synechocystis reduced light resistance and impaired state transitions [34]. In the present study, we further demonstrated that deletion of CpcM could influence the maximal photochemical efficiency of PSII and energy transfer from PBS to PSII (Figure 6), suggesting the critical role of CpcM in the regulation of photosynthesis in Synechocystis.
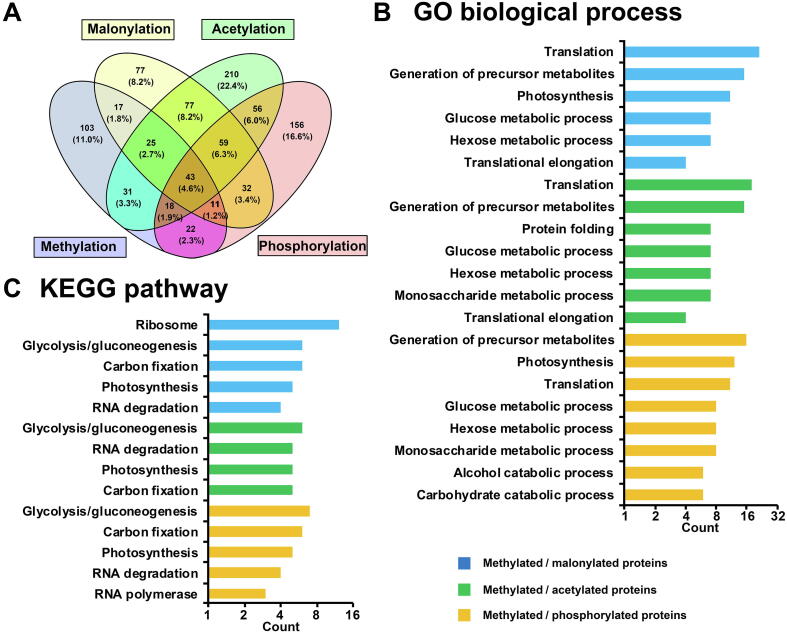
Lysine methylation is one of the numerous and different types of PTMs. Methyl lysine has been reported to provide docking sites for effector proteins or hinder other PTMs on the same lysine site [3]. Therefore, complicated crosstalk and interactions may exist between methylation and other PTMs. For example, lysine methylation could increase protein stability by competing with ubiquitination [29]. In addition, lysine methylation could interact with acetylation and phosphorylation to regulate E2F1-mediated cell death [46]. In the present study, to explore combinatorial activities between lysine monomethylation and other PTMs in Synechocystis, the lysine monomethylome was compared to the published Synechocystis malonylome [27], acetylome [26], and phosphoproteome (Table S6A) [47], [48], [49]. We observed that 61.9% of monomethylated proteins carry other types of PTMs (Figure 7A), and the overlapping proteins are largely categorized into translation and photosynthesis processes (Figure 7B, Table S6B). KEGG pathway analysis of the overlapping proteins revealed that they could be involved in glycolysis/gluconeogenesis, carbon fixation, photosynthesis, and RNA degradation (Figure 7C, Table S6C), prompting a complicated regulatory mechanism of diverse PTMs in the pathways. Previously, we showed that CpcB phosphorylation may change the energy transfer of PBS [47]. Interestingly, the inefficient PBS was also observed in cpcM deficient cells (Figure 6), and CpcB contains three lysine monomethylation sites in Synechocystis (Table S1), suggesting a crosstalk between protein methylation and phosphorylation, which is similar to that in their eukaryotic counterparts [46]. Notably, we observed 14 lysine sites that could be modified by three types of PTMs (including methylation, acetylation and malonylation) (Table S6D). Among these lysine sites, 35.7% (5/14) were located in photosynthetic proteins (Table S6D), suggesting the existence of complicated crosstalk and interactions among the three PTMs in regulating photosynthesis. Unraveling the crosstalk and interactions between lysine methylation and other PTMs would be critical for understanding the molecular mechanisms of metabolism and photosynthesis in cyanobacteria.
Figure 7.
Comparison of lysine monomethylome with Synechocystis malonylome, acetylome, and phosphoproteome
A. Venn diagram representation of the identified Synechocystis proteins containing methylation, acetylation, malonylation, or phosphorylation. B. and C. Biological process and KEGG pathway analysis of the overlapping proteins.
Although we identified a PKMT in Synechocystis, it is possible that more PKMTs exist in the organism. In addition to PKMTs, more KDMs such as lysine-specific demethylase 1 (LSD1) have been reported in diverse organisms [50]. It would be interesting to determine whether KDMs have counterparts in cyanobacteria.
In conclusion, we report the first lysine monomethylome and a PKMT in cyanobacteria, suggesting that lysine methylation is potentially one of the functional PTMs that regulates carbon metabolism and photosynthesis in photosynthetic organisms.
Materials and methods
Cell culture and preparation of cell lysates
The Synechocystis cells were photoautotrophically cultured in BG-11 medium at 30 °C, bubbled with filtrated air and under 40 μmol·photons·m−2·s−1 continuous illumination. Cells in logarithmic phase were collected and washed twice with PBS buffer, resuspended in ice-cold lysis buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1% Triton X-100, and 1 mM phenylmethanesulfonyl fluoride), and sonicated at an output of 135 W (3 s on, 3 s off) for 30 min on ice by a JY92-IIN sonicator (Scientz Biotechnology, Ningbo, China). The supernatant was collected after centrifugating at 5000 g for 5 min at 4 °C. The detection of protein concentration was carried out using butyleyanoacrylate (BCA) protein assay (Beyotime, Haimen, China). Nutrient deficiency experiments were performed as previously described [27]. For high salt treatment, exponentially growing cells were harvested and resuspended with BG-11 medium containing 1 M NaCl for 2 h. For protein extraction of plants, the whole plants including leaves, stems, and roots were washed and collected as previously described [51].
Protein digestion, liquid chromatography (LC) fractionation, and affinity purification
For the identification of expressed proteins, the whole Synechocystis proteins were directly digested with trypsin as described previously [52]. Before MS analysis, the C18 STAGE tips were used for peptide desalting. For the affinity purification of modified proteins, the Synechocystis proteins were chemically propionylated with propionic anhydride as described previously [24]. The chemical derivatization of the whole proteins was then precipitated with trichloroacetic acid and redissolved in 50 mM ammonium bicarbonate. The protein extracts were digested with sequencing grade modified trypsin according to a method described previously [24]. Finally, the digested peptides were fractionated by a Varian LC system (Agilent Technologies Inc., Palo Alto, CA) using an Xbridge C18 column (19 cm × 150 cm). The peptides were separated into 60 fractions using a salt gradient (2%–40%) of buffer B (10 mM ammonium formats in 85% acetonitrile, pH 8.5) at a flow rate of 10 ml/min in 70 min. The neighboring fractions were merged and combined into seven fractions, and each fraction was dried with a vacuum centrifuge and stored at −20 °C for further use. Modified peptides were isolated using anti-propionyl-methyllysine antibody (PTM Biolabs Inc., Chicago, IL) as previously reported [24].
LC–MS/MS analysis
LC–MS/MS analysis was carried out with a nLC-1000 system coupled to Q-Exactive plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA) [26]. MS data collection was performed by using Xcalibur 3.0 in data-dependent acquisition mode. Full MS survey with an m/z range of 350–1800 was acquired in the Orbitrap with resolution r = 70,000 at m/z 200, and the 20 most intense precursor ions with charge state 2–5 were sequentially fragmented in each scan cycle by higher energy C-trap dissociation (HCD) with normalized collision energy of 30%. The exclusion duration for the data-dependent scan was 15 s, the repeat count was 1, and the exclusion window was ±10 ppm.
Data analysis
All MS/MS spectra were analyzed by MaxQuant software (version 1.3.0.5) against the Synechocystis protein database (http://genome.annotation.jp/cyanobase/Synechocystis; 3672 sequences, released 2012) combined with the reverse decoy database and common contaminants [53]. The maximum missed cleavage for trypsin was set to 2. The variable modifications are acetylation (protein N-terminal), deamidation (Asn/Gln), oxidation (Met), propionylation (Lys), and propionyl-methylation (Lys). The fixed modification is carbamidomethylation (Cys). The MS tolerances were set to 10 ppm, and 0.02 Da were allowed for MS/MS tolerances. The estimated FDR threshold for modified peptide was <0.01, and the minimum peptide length was 6. All spectra were manually filtrated using strict criterions as previously described [54], and the peptides with modified lysine at C-terminus were removed.
Bioinformatics analysis
Functional enrichment analysis of all identified methylated proteins for GO terms and KEGG metabolic pathways were carried out using DAVID bioinformatics resources [55]. Motif-X was used for the analysis of amino acid sequence motifs [56]. The secondary structure of methylated lysine residues and all lysines in the methylated proteins were predicted by NetSurfP tool [57]. The P < 0.05 in a Student’s t-test was considered statistically significant in our data. To investigate the evolutionary conservation, the homologous proteins to Synechocystis proteins were determined via multi-sequence alignments. The phylogenetic tree was calculated and visualized using MEGA (version 5.05). Three-dimensional (3D) structure of CpcM module was generated by comparing with existing crystalline structure of PKMT2 in R. typhi using the online SWISS-MODEL Server and presented using PyMOL software (version 1.7.2).
Construction of the ΔCpcM mutant
The cpcM gene fragment was amplified from the genomic DNA of Synechocystis by PCR using the primers: 5′-ATGTTGTCCAACTCCGACCA-3′ and 5′-CTATGAGTTGCCGAGGGTCA-3′. Then the PCR product was inserted into PMD19-T vector (Takara, Dalian, China), and disrupted by inserting a kanamycin resistance cartridge to the internal EcoRI fragment of cpcM. Finally, the resultant plasmids were transformed into Synechocystis as described previously [58]. The transformants were screened on plates with solid BG-11 medium supplemented with 50 μg/ml kanamycin. The mutants were further confirmed by PCR and Western blot.
Production of anti-CpcM antibody
The generation of polyclonal antibody against CpcM was carried out by Hubei Proteingene Biotechnology (Wuhan, China). The CpcM protein was overexpressed and purified as described below. The following rabbit immunization and antisera sampling were performed by Proteingene (Wuhan, China). ELISA and Western blot were performed to confirm the specificity of the resulting antibodies.
Western blot analysis
20 μg of cell lysates from each cyanobacteria strain or 10 μg of plant protein extracts were separated by 12% SDS-PAGE, followed by transferring to polyvinylidene difluoride (PVDF) membrane, blocking with 5% bovine serum albumin (BSA) in TBS buffer (25 mM Tris-HCl, 150 mM NaCl, pH 8.0) at 20 °C for 2 h, and then incubating with primary rabbit polyclonal (anti-monomethyllysine antibody, Cell Signaling Technology, Danvers, MA) or anti-CpcM antibody at a 1:2000 dilution for 4 h at 4 °C. The membrane was washed three times with TBST buffer (150 mM NaCl, 25 mM Tris-HCl, 0.1% Tween20, pH 8.0) for 15 min each, and then incubated with horseradish peroxidase-conjugated anti-mouse IgG at a 1:5000 dilution for 1 h at room temperature. The electrochemiluminescence (ECL) system (Advansta Inc., San Jose, CA) was used for signal measurement with a luminescent image analyzer.
In-gel proteolytic digestion, protein identification and quantification analysis
The specific bands from the total cell lysates of WT and ΔCpcM strains in Coomassie-blue-stained gel, which represented different levels of lysine methylation by Western blot analysis, were cut from three replicate gels and trypsin-digested in-gel, followed by LC-MS/MS analysis. The in-gel proteolytic digestion and MS identification were performed as previously described [52]. The raw MS data were analyzed with MaxQuant. For protein identification, the variable modifications are acetylation (protein N-terminal), deamidation (Asn/Gln), oxidation (Met), methylation (Lys), di-methylation (Lys), and tri-methylation (Lys). The fixed modification is carbamidomethylation (Cys). Peptide tolerance was set to 20 ppm for the first search peptide and 6 ppm for the main search peptide. The maximum number of missed cleavages was set to 2. The estimated FDR threshold for peptide and site was <0.01. LFQ algorithm in MaxQuant was used to provide the value of LFQ intensity of each sample [59]. Then, protein ratio was computed based on the value of LFQ intensity and can be further used for the normalization of the modification stoichiometry. Perseus software (version 1.4.1.3) and Microsoft Excel were employed to perform the statistical analysis of data. Only the proteins that were identified and quantified in three biological replicates were allowed for relative quantification. The LFQ intensity was presented as the mean of three biological replicates. The two-sample Student’s t-test was used for the statistical evaluation and the coefficient of variation (CV) was computed from three biological replicates. To quantify the relative abundances of methylated proteins, the intensities of modified peptides were summed up and normalized with the abundance of proteins determined by the LFQ quantification as previously described [36]. All spectra were manually filtrated using strict criterions as previously described [54], and the peptides with modified lysine at C-terminus were removed.
Purification of His6-tagged proteins in vitro
The full-length cpcM, sll0480, slr2094, and slr0394 genes were amplified using the following primers: cpcM-F: 5′-GGAATTCCATATGATGTTGTCCAACTCCGACCA-3′ and cpcM-R: 5′-CCGCTCGAGTGAGTTGCCGAGGGTCAGTA-3′ for cpcM; sll0480-F: 5′-GGAATTCCATATGATGGCCAGTATCAACGACAA-3′ and sll0480-R: 5′-CCGCTCGAGACCCAATTTGAGGGTGGAAG-3′ for sll0480; slr2094-F: 5′-GGAATTCCATATGGTGGACAGCACCCTCGGTTT-3′ and slr2094-R: 5′-CCGCTCGAGATGCAGTTGGATTACAAAGG-3′ for slr2094; slr0394-F: 5′-GGAATTCCATATGTTGTCTAAGCAATCGATCGC-3′ and slr0394-R: 5′-CCGCTCGAGTCG GTCATCTAAAGCGGCAA-3′ for slr0394. The PCR products were then cloned into the expression vector pET-21b (Novagen, Darmstadt, Germany) to overexpress proteins in E. coli according to the procedures described in manufacturers. All recombinant strains were cultured to the exponential phase (OD600 = 0.4–0.6) in 500 ml Luria-Bertani (LB) medium at 37 °C, and then 0.5 mM isopropyl β-D-thiogalactoside (IPTG) was used for cell induction at 16 °C for 12 h, followed by cell collection and washing. The cells were disrupted by sonication, and the debris was discarded after centrifugating at 12,000 g for 10 min at 4 °C for two times. Then the supernatants were loaded onto Ni-NTA agarose column (Qiagen Inc., Germantown, MD) pre-equilibrated with the binding buffer (500 mM NaCl, 20 mM Tris-HCl, pH 8.0). After washing with washing buffer (20 mM Tris-HCl, 100 mM imidazole, 500 mM NaCl, pH 8.0), the binding proteins were eluted with 10 ml elution buffer (20 mM Tris-HCl, 250 mM imidazole, 500 mM NaCl, pH 8.0). The resulting proteins were then desalted by concentrating with a 30,000 MWCO concentrator (Millipore, Bedford, MA) in the storage buffer (50 mM NaCl, 20 mM Tris-HCl, pH 8.0). The BCA protein assay was performed to measure the concentration of purified proteins.
In vitro methylation assay and identification of lysine methylation sites
In vitro methylation assays were performed as previously described [60]. Briefly, the purified Sll0480 and CpcM were mixed with 50 μl reaction buffer (50 mM Tris-HCl, pH 8.5, 5 mM MgCl2, 4 mM DL-dithiothreitol) and 10 μg SAM (B903S, New England Biolabs, Ipswich, MA). The heat-inactivated CpcM protein was used as a negative control. After incubating at 37 °C for 60 min, the reaction products were subjected to SDS-PAGE and Western blot to detect methylation levels with the anti-monomethyllysine antibody. The methylation levels of substrates in the mixtures were also subjected to in-solution tryptic digestion and LC-MS/MS analysis as described previously [52]. The MS data were searched against the E. coli BL21 (DE3) database (NC_012892.2, https://www.ncbi.nlm.nih.gov/genome/167?Genome_assembly_id=161528) including the sequences of CpcM and Sll0480 by using pFind software [37]. The maximum missed cleavage for trypsin was set to 2. The precursor ion mass tolerance was set to 10 ppm, and the fragment ion mass tolerance was allowed to 20 ppm. The acetylation (protein N-terminal), deamidation (Asn/Gln), oxidation (Met), methylation (Lys), di-methylation (Lys), and tri-methylation (Lys) were set as variable modifications, and carbamidomethylation (Cys) as fixed modification. The estimated FDR of peptide identification was less than 1%. Finally, all spectra were manually filtrated using strict criterions as previously described [54], and the peptides with modified lysine at C-terminus were removed. The intensities of identified peptides were summed up and the relative abundance of proteins was calculated. A two-sample Student’s t-test analysis was used to evaluate the statistical significance.
AAT activity assay
For the AAT activity assay, the constant amounts of Sll0480 and Sll0480 treated by CpcM were incubated in the reaction buffer with or without 10 μg SAM at 37 °C for 60 min. The mixtures were then subjected to AAT activity assay according to the standard procedure described in the manufacturers of AAT activity assay kit (MAK055, Sigma-Aldrich Corp., St. Louis, MO). The standard AAT and purified CpcM with SAM were used as the positive and negative controls, respectively.
Chlorophyll fluorescence measurement
Chlorophyll fluorescence was measured at room temperature using a Dual-PAM-100 (Heinz Walz Gmbh, Effeltrich, Germany) [61]. The minimum fluorescence in dark-adapted state (F0), maximum fluorescence in dark-adapted state (Fm), minimum fluorescence in light-adapted state (F′0) and maximum fluorescence in light-adapted state (F′m) values were detected by measuring the chlorophyll fluorescence traces in the Dual-PAM software, and all fluorescence parameters were computed by standard equations as previously described [61]. Fv/Fm represents the maximal photochemical efficiency of PSII in the dark-adapted state, Y(II) represents the actual photochemical efficiency of PSII under different light intensities. The relative PSII ETR (II) was also calculated with maximal chlorophyll fluorescence by Dual-PAM software.
Spectroscopic measurement
The fluorescence emission spectra measurement was performed at room temperature or 77 K using the single channel fluorimeter (Photon Technology International, New Brunswick, NJ) equipped with a matched liquid nitrogen Dewar (4-mm inner diameter and 6-mm outer diameter). Cells under different conditions were adjusted to OD730 ≈ 0.8 with fresh BG-11 medium. For 77 K fluorescence emission spectra measurement, samples were dark-adapted at room temperature for 15 min and immediately frozen in liquid nitrogen before measurement. The excitation wavelength was set to 590 nm for phycocyanin excitation, both the excitation and emission were measured every 1 nm in the wavelength range from 600 nm to 800 nm. Each measurement consisting of three biological replicates was performed and averaged. The fluorescence emission spectra at 77 K were normalized at 730 nm for direct comparison of the fluorescence emission peak deriving from PBS (650 nm).
Data availability
The raw data and modified peptide spectra can be accessed at the public database PeptideAtlas (PASS: PASS01127).
CRediT author statement
Xiaohuang Lin: Methodology, Investigation, Writing - original draft. Mingkun Yang: Methodology, Investigation, Writing - original draft. Xin Liu: Methodology, Investigation. Zhongyi Cheng: Methodology. Feng Ge: Conceptualization, Methodology, Supervision, Writing - review & editing. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31570829), the Chinese Academy of Sciences Grant QYZDY-SSW-SMC004, the CAS Key Technology Talent Program (to MKY), and the Open Fund of Key Laboratory of Experimental Marine Biology, Chinese Academy of Sciences (Grant No. KF2017NO3). The authors would like to thank the Analysis and Testing Center of Institute of Hydrobiology for their helps in proteomic experiments and Hubei Proteingene Co., Ltd for their helps in data processing.
Handled by Yu Xue
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.04.005.
Supplementary material
The following are the Supplementary data to this article:
Overview of the proteomics data. A. Distribution of peptide score based on precursor mass deviations of methylated peptides. B. Distribution of the methylated sites identified per protein.
Overview of the tertiary structure of PKMTs. A. CpcM (Synechocystis). B. PKMT2 (Rickettsia typhi). C. PKMT1 (Rickettsia prowazekii). D. aKMT (Sulfolobus islandicus). E. PrmA (E. coli). F. PrmA (Thermus thermophiles).
Conservation analysis of aspartate aminotransferases in Synechocystis. A. Phylogenetic analysis of the aspartate aminotransferases from the prokaryotic and eukaryotic cells. The phylogenetic tree was generated by Maximum Likelihood method using MEGA 5.0.5 software. B. Multiple sequence alignment of Sll0480 and its orthologs. The identified methylation sites were marked by arrows. C. Structural modeling of the aspartate aminotransferases from Synechocystis (Sll0480), E. coli (AspC), and Homo sapiens (Got1).
Western blot analysis of the methylation levels of Slr2094 (A) and Slr0394 (B) after incubation with CpcM using anti-monomethyllysine antibody. Each sample was standardized by comparing to the Coomassie-blue-stained gel. The untreated Sll0480 (Sll0480+SAM) group and the heat-inactivated CpcM (represented by #) group were used as negative controls. The densitometry analysis of methylated substrates in each reaction was performed using ImageJ software (bottom). Data are presented as means ± SD from three independent experiments. Asterisk indicates significant differences from the negative controls. SAM, S-adenosyl methionine.
Detailed information on identified monomethylated peptides in Synechocystis sp. PCC 6803.
GO and pathway enrichment analysis of identified methylated proteins in Synechocystis sp. PCC 6803.
Secondary structure localization and solvent accessibility of the modified and non-modified sites of the methylated proteins in Synechocystis sp. PCC 6803.
Detailed list of peptides and proteins detected by MS and quantification analysis of methylated proteins from the protein bands of Figure 4E.
Detailed list of all identified peptides and proteins in the reaction mix.
The overlapped proteins and sites of methylation and other PTMs in Synechocystis sp. PCC 6803.
References
- 1.Moore K.E., Gozani O. An unexpected journey: lysine methylation across theproteome. Biochim Biophys Acta. 2014;1839:1395–1403. doi: 10.1016/j.bbagrm.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R.P., Rees M.W. Epsilon-N-methyl-lysine in bacterial flagellar protein. Nature. 1959;184:56–57. doi: 10.1038/184056b0. [DOI] [PubMed] [Google Scholar]
- 3.Lanouette S., Mongeon V., Figeys D., Couture J.F. The functional diversity of protein lysine methylation. Mol Syst Biol. 2014;10:724. doi: 10.1002/msb.134974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J., Berger S.L. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Carlson S.M., Gozani O. Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y.H., Park K.H., Kim S.Y., Ji E.S., Kim J.Y., Lee S.K. Identification of trimethylation at C-terminal lysine of pilin in the cyanobacterium Synechocystis PCC 6803. Biochem Biophys Res Commun. 2011;404:587–592. doi: 10.1016/j.bbrc.2010.11.133. [DOI] [PubMed] [Google Scholar]
- 7.Xia Y., Niu Y., Cui J., Fu Y., Chen X.S., Lou H. The helicase activity of hyperthermophilic archaeal MCM is enhanced at high temperatures by lysine methylation. Front Microbiol. 2015;6:1247. doi: 10.3389/fmicb.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falnes P.O., Jakobsson M.E., Davydova E., Ho A., Malecki J. Protein lysine methylation by seven-beta-strand methyltransferases. Biochem J. 2016;473:1995–2009. doi: 10.1042/BCJ20160117. [DOI] [PubMed] [Google Scholar]
- 9.Rea S., Eisenhaber F., O'Carroll N., Strahl B.D., Sun Z.W., Schmid M. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 10.Richon V.M., Johnston D., Sneeringer C.J., Jin L., Majer C.R., Elliston K. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 11.Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 12.Kouskouti A., Scheer E., Staub A., Tora L., Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 13.Colson C., Lhoest J., Urlings C. Genetics of ribosomal-protein methylation in Escherichia coli. III. Map position of two genes, Prma and Prmb, governing methylation of proteins L11 and L3. Mol Gen Genet. 1979;169:245–250. doi: 10.1007/BF00382270. [DOI] [PubMed] [Google Scholar]
- 14.Barbier M., Owings J.P., Martinez-Ramos I., Damron F.H., Gomila R., Blazquez J. Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. mBio. 2013;4:e00207–13. doi: 10.1128/mBio.00207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abeykoon A., Wang G., Chao C.C., Chock P.B., Gucek M., Ching W.M. Multimethylation of Rickettsia OmpB catalyzed by lysine methyltransferases. J Biol Chem. 2014;289:7691–7701. doi: 10.1074/jbc.M113.535567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abeykoon A.H., Chao C.C., Wang G., Gucek M., Yang D.C., Ching W.M. Two protein lysine methyltransferases methylate outer membrane protein B from Rickettsia. J Bacteriol. 2012;194:6410–6418. doi: 10.1128/JB.01379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nozzi N.E., Oliver J.W., Atsumi S. Cyanobacteria as a platform for biofuel production. Front Bioeng Biotechnol. 2013;1:7. doi: 10.3389/fbioe.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 19.Alban C., Tardif M., Mininno M., Brugiere S., Gilgen A., Ma S. Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vorontsov E.A., Rensen E., Prangishvili D., Krupovic M., Chamot-Rooke J. Abundant lysine methylation and N-terminal acetylation in Sulfolobus islandicus revealed by bottom-up and top-down proteomics. Mol Cell Proteomics. 2016;15:3388–3404. doi: 10.1074/mcp.M116.058073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore K.E., Carlson S.M., Camp N.D., Cheung P., James R.G., Chua K.F. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol Cell. 2013;50:444–456. doi: 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caslavka Zempel K.E., Vashisht A.A., Barshop W.D., Wohlschlegel J.A., Clarke S.G. Determining the mitochondrial methyl proteome in Saccharomyces cerevisiae using heavy methyl SILAC. J Proteome Res. 2016;15:4436–4451. doi: 10.1021/acs.jproteome.6b00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Z., Connolly J., Biggar K.K. Beyond histones - the expanding roles of protein lysine methylation. FEBS J. 2017;284:2732–2744. doi: 10.1111/febs.14056. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z.X., Cheng Z.Y., Sun M.W., Wan X.L., Liu P., He T.M. A chemical proteomics approach for global analysis of lysine monomethylome profiling. Mol Cell Proteomics. 2015;14:329–339. doi: 10.1074/mcp.M114.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao X.J., Arnaudo A.M., Garcia B.A. Large-scale global identification of protein lysine methylation in vivo. Epigenetics. 2013;8:477–485. doi: 10.4161/epi.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mo R., Yang M., Chen Z., Cheng Z., Yi X., Li C. Acetylome analysis reveals the involvement of lysine acetylation in photosynthesis and carbon metabolism in the model cyanobacterium Synechocystis sp. PCC 6803. J Proteome Res. 2015;14:1275–1286. doi: 10.1021/pr501275a. [DOI] [PubMed] [Google Scholar]
- 27.Ma Y., Yang M., Lin X., Liu X., Huang H., Ge F. Malonylome analysis reveals the involvement of lysine malonylation in metabolism and photosynthesis in cyanobacteria. J Proteome Res. 2017;16:2030–2043. doi: 10.1021/acs.jproteome.7b00017. [DOI] [PubMed] [Google Scholar]
- 28.Olsen J.B., Cao X.J., Han B.M., Chen L.H., Horvath A., Richardson T.I. Quantitative profiling of the activity of protein lysine methyltransferase smyd2 using silac-based proteomics. Mol Cell Proteomics. 2016;15:892–905. doi: 10.1074/mcp.M115.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang C., Gasteiger E., Wilkins M.R. Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications. BMC Genomics. 2010;11:92. doi: 10.1186/1471-2164-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iakoucheva L.M., Radivojac P., Brown C.J., O'Connor T.R., Sikes J.G., Obradovic Z. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong Q., Chen Z., Ge F. Proteomic analysis of post translational modifications in cyanobacteria. J Proteomics. 2016;134:57–64. doi: 10.1016/j.jprot.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 32.Canut H., Albenne C., Jamet E. Post-translational modifications of plant cell wall proteins and peptides: a survey from a proteomics point of view. Biochim Biophys Acta. 2016;1864:983–990. doi: 10.1016/j.bbapap.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 33.Padmasree K., Padmavathi L., Raghavendra A.S. Essentiality of mitochondrial oxidative metabolism for photosynthesis: optimization of carbon assimilation and protection against photoinhibition. Crit Rev Biochem Mol Biol. 2002;37:71–119. doi: 10.1080/10409230290771465. [DOI] [PubMed] [Google Scholar]
- 34.Shen G., Leonard H.S., Schluchter W.M., Bryant D.A. CpcM posttranslationally methylates asparagine-71/72 of phycobiliprotein beta subunits in Synechococcus sp. strain PCC 7002 and Synechocystis sp. strain PCC 6803. J Bacteriol. 2008;190:4808–4817. doi: 10.1128/JB.00436-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller C.A., Leonard H.S., Pinsky I.G., Turner B.M., Williams S.R., Harrison L., Jr Biogenesis of phycobiliproteins. III. CpcM is the asparagine methyltransferase for phycobiliprotein beta-subunits in cyanobacteria. J Biol Chem. 2008;283:19293–19300. doi: 10.1074/jbc.M802734200. [DOI] [PubMed] [Google Scholar]
- 36.Weinert B.T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S.A., Magnes C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chi H., Liu C., Yang H., Zeng W.F., Wu L., Zhou W.J. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat Biotechnol. 2018;36:1059–1061. doi: 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- 38.de la Torre F., Canas R.A., Pascual M.B., Avila C., Canovas F.M. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J Exp Bot. 2014;65:5527–5534. doi: 10.1093/jxb/eru240. [DOI] [PubMed] [Google Scholar]
- 39.Rastogi V.K., Watson R.J. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in Rhizobium meliloti. J Bacteriol. 1991;173:2879–2887. doi: 10.1128/jb.173.9.2879-2887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toney M.D. Aspartate aminotransferase: an old dog teaches new tricks. Arch Biochem Biophys. 2014;544:119–127. doi: 10.1016/j.abb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimmich G.A., Roussie J.A., Randles J. Aspartate aminotransferase isotope exchange reactions: implications for glutamate/glutamine shuttle hypothesis. Am J Physiol Cell Physiol. 2002;282:C1404–C1413. doi: 10.1152/ajpcell.00487.2001. [DOI] [PubMed] [Google Scholar]
- 42.Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- 43.Nobe Y., Kawaguchi S., Ura H., Nakai T., Hirotsu K., Kato R. The novel substrate recognition mechanism utilized by aspartate aminotransferase of the extreme thermophile Thermus thermophilus HB8. J Biol Chem. 1998;273:29554–29564. doi: 10.1074/jbc.273.45.29554. [DOI] [PubMed] [Google Scholar]
- 44.de la Torre F., Moya-Garcia A.A., Suarez M.F., Rodriguez-Caso C., Canas R.A., Sanchez-Jimenez F. Molecular modeling and site-directed mutagenesis reveal essential residues for catalysis in a prokaryote-type aspartate aminotransferase. Plant Physiol. 2009;149:1648–1660. doi: 10.1104/pp.108.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamey J.J., Wienert B., Quinlan K.G.R., Wilkins M.R. METTL21B is a novel human lysine methyltransferase of translation elongation factor 1A: discovery by CRISPR/Cas9 knock out. Mol Cell Proteomics. 2017;16:2229–2242. doi: 10.1074/mcp.M116.066308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kontaki H., Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z., Zhan J., Chen Y., Yang M., He C., Ge F. Effects of phosphorylation of beta subunits of phycocyanins on state transition in the model cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2015;56:1997–2013. doi: 10.1093/pcp/pcv118. [DOI] [PubMed] [Google Scholar]
- 48.Angeleri M., Muth-Pawlak D., Aro E.M., Battchikova N. Study of O-phosphorylation sites in proteins involved in photosynthesis-related processes in Synechocystis sp. strain PCC 6803: application of the SRM approach. J Proteome Res. 2016;15:4638–4652. doi: 10.1021/acs.jproteome.6b00732. [DOI] [PubMed] [Google Scholar]
- 49.Spat P., Macek B., Forchhammer K. Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front Microbiol. 2015;6:248. doi: 10.3389/fmicb.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anand R., Marmorstein R. Structure and mechanism of lysine-specific demethylase enzymes. J Biol Chem. 2007;282:35425–35429. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 51.Wang K., Zhao Y., Li M., Gao F., Yang M.K., Wang X. Analysis of phosphoproteome in rice pistil. Proteomics. 2014;14:2319–2334. doi: 10.1002/pmic.201400004. [DOI] [PubMed] [Google Scholar]
- 52.Yang M.K., Yang Y.H., Chen Z., Zhang J., Lin Y., Wang Y. Proteogenomic analysis and global discovery of posttranslational modifications in prokaryotes. Proc Natl Acad Sci U S A. 2014;111:E5633–E5642. doi: 10.1073/pnas.1412722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y., Kwon S.W., Kim S.C., Zhao Y.M. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 55.Jiao X., Sherman B.T., Huang da W., Stephens R., Baseler M.W., Lane H.C. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwartz D., Gygi S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 57.Petersen B., Petersen T.N., Andersen P., Nielsen M., Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frigaard N.U., Sakuragi Y., Bryant D.A. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol Biols. 2004;274:325–340. doi: 10.1385/1-59259-799-8:325. [DOI] [PubMed] [Google Scholar]
- 59.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed maxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X., Chen Z., Xu C.X., Leng X.Q., Cao H., Ouyang G. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res. 2015;43:5081–5098. doi: 10.1093/nar/gkv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell D., Hurry V., Clarke A.K., Gustafsson P., Oquist G. Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol Mol Biol Rev. 1998;62:667–683. doi: 10.1128/mmbr.62.3.667-683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the proteomics data. A. Distribution of peptide score based on precursor mass deviations of methylated peptides. B. Distribution of the methylated sites identified per protein.
Overview of the tertiary structure of PKMTs. A. CpcM (Synechocystis). B. PKMT2 (Rickettsia typhi). C. PKMT1 (Rickettsia prowazekii). D. aKMT (Sulfolobus islandicus). E. PrmA (E. coli). F. PrmA (Thermus thermophiles).
Conservation analysis of aspartate aminotransferases in Synechocystis. A. Phylogenetic analysis of the aspartate aminotransferases from the prokaryotic and eukaryotic cells. The phylogenetic tree was generated by Maximum Likelihood method using MEGA 5.0.5 software. B. Multiple sequence alignment of Sll0480 and its orthologs. The identified methylation sites were marked by arrows. C. Structural modeling of the aspartate aminotransferases from Synechocystis (Sll0480), E. coli (AspC), and Homo sapiens (Got1).
Western blot analysis of the methylation levels of Slr2094 (A) and Slr0394 (B) after incubation with CpcM using anti-monomethyllysine antibody. Each sample was standardized by comparing to the Coomassie-blue-stained gel. The untreated Sll0480 (Sll0480+SAM) group and the heat-inactivated CpcM (represented by #) group were used as negative controls. The densitometry analysis of methylated substrates in each reaction was performed using ImageJ software (bottom). Data are presented as means ± SD from three independent experiments. Asterisk indicates significant differences from the negative controls. SAM, S-adenosyl methionine.
Detailed information on identified monomethylated peptides in Synechocystis sp. PCC 6803.
GO and pathway enrichment analysis of identified methylated proteins in Synechocystis sp. PCC 6803.
Secondary structure localization and solvent accessibility of the modified and non-modified sites of the methylated proteins in Synechocystis sp. PCC 6803.
Detailed list of peptides and proteins detected by MS and quantification analysis of methylated proteins from the protein bands of Figure 4E.
Detailed list of all identified peptides and proteins in the reaction mix.
The overlapped proteins and sites of methylation and other PTMs in Synechocystis sp. PCC 6803.
Data Availability Statement
The raw data and modified peptide spectra can be accessed at the public database PeptideAtlas (PASS: PASS01127).