Abstract
N6-methyladenosine (m6A) is an abundant epitranscriptomic modification that plays important roles in many aspects of RNA metabolism. While m6A is thought to mainly function by recruiting reader proteins to specific RNA sites, the modification can also reshape RNA-protein and RNA-RNA interactions by altering RNA structure mainly by destabilizing base pairing. Little is known about how m6A and other epitranscriptomic modifications might affect the kinetic rates of RNA folding and other conformational transitions that are also important for cellular activity. Here, we used NMR R1ρ relaxation dispersion and chemical exchange saturation transfer to non-invasively and site-specifically measure nucleic acid hybridization kinetics. The methodology was validated on two DNA duplexes and then applied to examine how a single m6A alters the hybridization kinetics in two RNA duplexes. The results show that m6A minimally impacts the rate constant for duplex dissociation, changing koff by ~1-fold but significantly slows the rate of duplex annealing, decreasing kon by ~7-fold. A reduction in the annealing rate was observed robustly for two different sequence contexts at different temperatures, both in the presence and absence of Mg2+. We propose that rotation of the N6-methyl group from the preferred syn conformation in the unpaired nucleotide to the energetically disfavored anti conformation required for Watson-Crick pairing is responsible for the reduced annealing rate. The results help explain why in mRNA, m6A slows down tRNA selection, and more generally suggest that m6A may exert cellular functions by reshaping the kinetics of RNA conformational transitions.
Graphical abstract
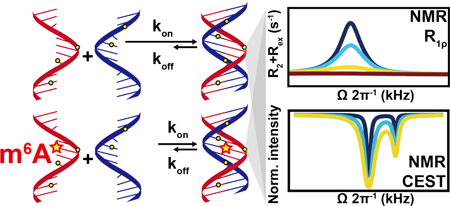
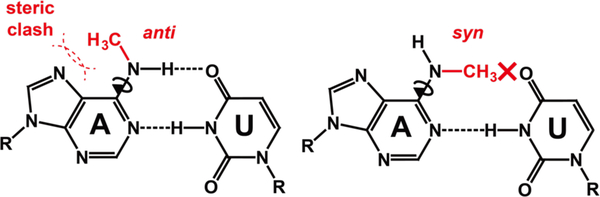
N6-methyladenosine (m6A) is an abundant reversible epitranscriptomic modification found in coding and noncoding RNAs1–4. It plays important roles in RNA metabolism5–8 and is implicated in a growing number of cellular processes9–15. While the modification is thought to primarily exert its function by recruiting reader proteins to specific RNA sites, it can also reshape RNA-RNA and RNA-protein interactions by modulating RNA structure16–20. A single m6A destabilizes RNA duplexes by 0.5–1.7 kcal/mol21–22, enhancing binding to single-stranded RNA (ssRNA) binding proteins16. m6A destabilizes A-U base pairs (bps) because hydrogen bonding requires that the N6-methyl group adopts the energetically unfavorable anti conformation21–22 (Figure 1).
Figure 1. N6-methyl adenosine (m6A) destabilizes m6-A-U pairing and RNA duplexes.
The methyl group has to adopt an anti conformation to form the Watson-Crick H6--O4 hydrogen bond but this leads to unfavorable steric contacts with N7.
The activities of many RNAs also depends on the kinetic rates of folding, protein-RNA, RNA-RNA, and RNA-ligand association/dissociation and conformational transitions23–29. Surprisingly little is known about how m6A and other epitranscriptomic modifications impact these kinetic properties of RNA. Compelling evidence for such a kinetic effect comes from a study showing that in mRNA, m6A slows down tRNA selection during translation20. Here, we developed an approach based on NMR spin relaxation dispersion (RD) in the rotating frame (R1ρ)30–32 and Chemical Exchange Saturation Transfer (CEST)33–34 to site-specifically and non-invasively measure hybridization kinetics of nucleic acid duplexes and then used the approach to examine how a single m6A impacts RNA duplex hybridization kinetics.
The melting and annealing of RNAs occurs in a wide variety of biochemical reactions27, 35. Relative to other methods for studying hybridization kinetics36–45, the NMR approach does not require a potentially perturbing label, which could obscure the impact of a small chemical modification, and kinetics can be measured at atomic resolution32, 46 to enable characterization of any intermediates that may form at the modified site.
We first evaluated the R1ρ RD methodology on DNA duplexes whose hybridization kinetics has been extensively characterized previously38, 41, 44–45, 47–51. R1ρ RD relies on measuring the exchange contribution (Rex) to transverse spin relaxation (R2) due to chemical exchange between a major ground-state (GS) and a low-abundance and short-lived ‘excited-state’ (ES)52–53.
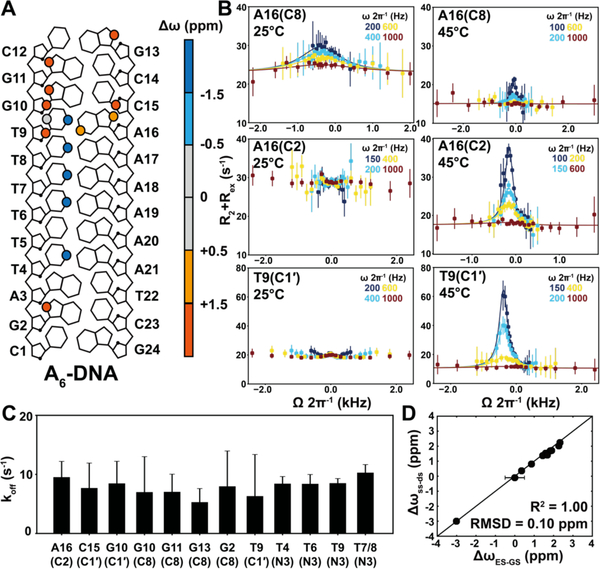
Prior R1ρ studies on RNA and DNA duplexes were carried out at temperatures below the melting temperature (Tm)46, 54–56. Under these conditions, the population (pss) of the single-stranded (ss) species falls below detection (< 0.1%)31, enabling studies of bp dynamics. For example, at T=25°C, the R1ρ profiles measured for various sites in the A6-DNA duplex55, 57 (Tm~51°C and [A6-DNA] ~ 0.9 mM) reflect exchange between a major Watson-Crick GS and minor Hoogsteen ES55 (Figure 2A, 2B, S1). There is no evidence for a transient ss species, which is estimated to have a pss ~ 0.1% based on UV melting experiments (Table S1).
Figure 2. Site-specific characterization of A6-DNA hybridization kinetics using NMR R1ρ RD.
(A) The A6-DNA duplex. Δω = ωES – ωGS obtained from global fitting of the R1ρ RD profiles is color-coded on each atom. Sites which are not colored indicates that no measurements were done. (B) Off-resonance R1ρ (13C) RD profiles measured in A6-DNA at 25°C (left) and 45°C (right). T9(C1′) RD at 25°C were reprinted by permission from58. Buffer conditions were 25 mM NaCl, 15 mM sodium phosphate, 0.1 mM EDTA and 10% D2O at pH 6.8. (C) The site-specific koff values obtained from 2-state fitting of the R1ρ RD profiles measured for A6-DNA at 45°C. (D) Comparison of ΔωES-GS = ωES – ωGS measured by RD with Δωss-ds = ωss – ωds values obtained from the major and minor resonance observed in 2D [13C,1H], [15N,1H] and [15N, 13C] HSQC spectra of A6-DNA at 45°C.
Based on simulations50, increasing the temperature so that pss>1.0% should bring hybridization kinetics within R1ρ detection (Figure S2). Indeed, the R1ρ profiles for A6-DNA changed when increasing the temperature to T=45 °C (pss~10%). RD is now apparent at A16(C2) and T9(C1′), which are otherwise flat at T=25°C (Figure 2B). A single peak was observed in all cases consistent with two-state exchange (GS⇌ES). Fitting the R1ρ data to a 2-state exchange model yielded very similar k1 = koff (differences < 2-fold; koff is the rate constant for dissociation) for different sites as expected for concerted melting and annealing of the duplex (Figure 2C). This is in stark contrast to Hoogsteen exchange at T=25°C, in which k1 varies 50-fold across sites reflecting sequence-specific differences in bp dynamics59. The ES chemical shifts measured for various sites were also in excellent agreement with those measured for the isolated ss, confirming that the ES is the ss species (Figure 2D, S3).
In the ‘zip-up’ model48, 60, DNA annealing proceeds through a slow nucleation step followed by a fast zipping step occurring on the ns-μs timescale which is too fast for RD detection. Since the Hoogsteen exchange at higher temperatures is likely too fast for RD detection, ‘all-or-nothing’ behavior is observed with strands either being fully annealed or fully unzipped. These results establish the utility of R1ρ RD to measure hybridization kinetics in DNA duplexes with site-specific resolution.
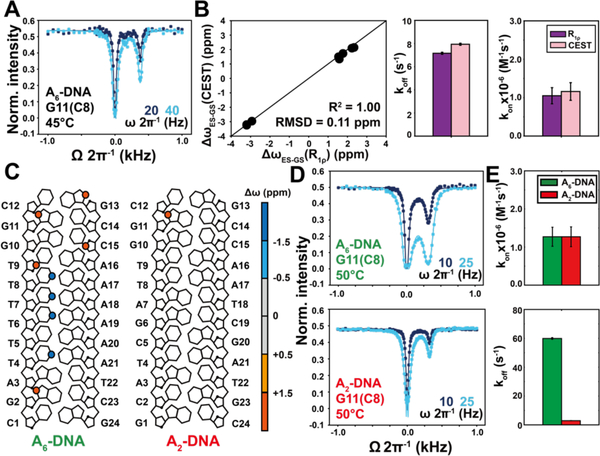
The backward rate constant k-1 = kon × [ss] (kon is the rate constant for duplex annealing) was ill-defined when fitting the R1ρ RD data (Figure S4). Such a degeneracy is expected when the exchange is slow on the NMR timescale and when using spin lock powers (ω1) in the R1ρ experiment that exceed the exchange rate (kex = k1 + k-1)61–62. Indeed, in the slow exchange limit, the line broadening of the GS resonance only depends on the forward rate. To address this degeneracy, we used CEST experiments which can employ much lower spin locking fields more suitable for characterizing systems in slow exchange33–34. CEST relies on measuring the resonance intensity of the GS as a function of the power and offset of an applied weak radio frequency (rf) field. At T=45°C, the CEST profiles for A6-DNA revealed a dip at the chemical shift of the ss ES (Figure 3A, 3C, S5). Fitting the CEST profiles allowed the reliable determination of all exchange parameters including kon (Figure S4), resulting in values (Figure 3B) that are in good agreement with those previously reported values for similar DNA duplexes45, 50.
Figure 3. Site-specific characterization of hybridization kinetics using CEST.
(A) 13C CEST profile for G11(C8) measured in A6-DNA at 45°C. (B) Comparison of ΔωES-GS, koff and kon values obtained from R1ρ and CEST (fits of the R1ρ profiles were preformed fixing pss to the value measured using CEST). Buffer conditions were 25 mM NaCl, 15 mM sodium phosphate, 0.1 mM EDTA and 10% D2O at pH 6.8. (C) The sequence of A2-DNA and A6-DNA. Δω = ωES – ωGS obtained from CEST fitting is color-coded on each atom. (D) 13C CEST profiles for G11(C8) measured in A2-DNA and A6-DNA at 50°C. (E) Comparison of kon and koff values measured for A2-DNA (red) and A6-DNA (green).
Fixing pss to the CEST determined value, the R1ρ RD profiles could be satisfactorily globally fitted (Figure S5), yielding exchange parameters (k1 = koff, k-1 = kon × [ss] and ΔωES-GS that are in excellent agreement with the CEST derived values (Figure 3B, Table S3, S4). This mutual consistency further supports the validity of the approach. Finally, we further evaluated the CEST methodology by comparing the hybridization kinetics of A6-DNA with another A2-DNA duplex, which has higher stability (Tm~60°C and [A2-DNA] ~ 0.8 mM) (Figure 3C, 3D). Consistent with prior studies47–49, the two duplexes have similar kon values but koff is 20-fold faster for the less stable A6-DNA duplex (Figure 3E).
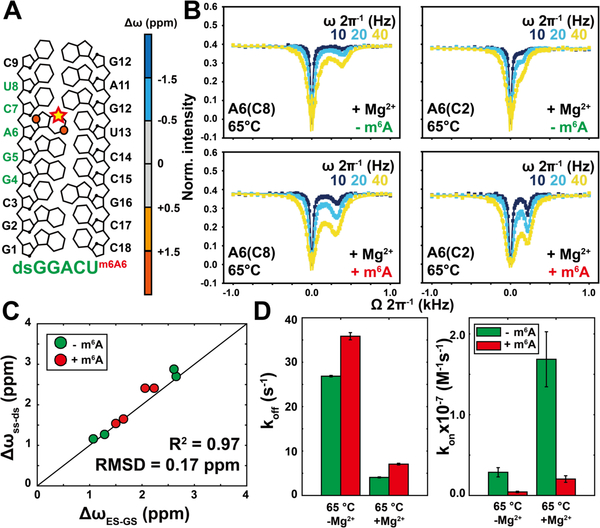
Next, we applied the methodology to examine how m6A impacts hybridization kinetics in an RNA duplex containing the most abundant m6A consensus sequence (GGACU) in eukaryotic mRNA1–2 (Tm~80°C and [dsGGACU] ~ 0.7 mM with Mg2+). In canonical RNA duplexes, there are no contributions from Hoogsteen exchange or any other process as verified for Watson-Crick bps in a variety of sequence and structural contexts54. However, since m6A could induce local melting of the duplex, it was important to carry out measurements on the m6A residue itself. To this end, two dsGGACU duplexes were chemically synthesized containing 13C2/C8 labeled m6A or A near the center of the duplex (Figure 4A, S1, S6) (see methods). m6A destabilized the dsGGACU duplex by ~1 kcal/mol (Table S1), consistent with prior studies21–22.
Figure 4. Measuring the impact of m6A on dsGGACU hybridization kinetics using CEST.
(A) The dsGGACU sequence. Δω = ωES – ωGS obtained from global fitting of CEST is color-coded on each atom. (B) 13C CEST profiles measured for A6 in unmodified (left, green) and m6A modified (right, red) dsGGACU at 65°C in the presence of 3 mM Mg2+ (profiles in the absence of Mg2+ are shown in Figure S7). Buffer conditions were 25 mM NaCl, 15 mM sodium phosphate, 3 mM Mg2+, 0.1 mM EDTA and 10% D2O at pH 6.8. (C) Comparison of ΔωES-GS = ωES – ωGS measured by CEST with Δωss-ds = ωss – ωds values obtained from the major and minor resonance observed in 2D [13C,1H] HSQC spectra of dsGGACU with (red) and without (green) m6A at 65°C. (D) Comparison of kon and koff measured for unmodified (green) and m6A modified (red) dsGGACU.
The CEST and R1ρ profiles for both unmodified and modified dsGGACU duplex at T=65°C revealed a single peak/dip consistent with 2-state exchange (Figure 4B, S7). However, the profiles for the modified duplex differed markedly from its unmodified counterpart (Figure 4B, S7). In both cases, global fitting of the CEST and R1ρ data yielded ES chemical shifts that are in excellent agreement with those measured for the isolated ss (Figure 4C, S7, S8). Fitting the CEST data revealed that m6A changes koff by 0.7–1.7 fold but decreases kon by 4–9 fold (Figure 4D, S7). This m6A induced slowdown of annealing was observed robustly with or without Mg2+ (Figure 4D, S7), for a different sequence derived from Hepatitis C virus (HCV)15 (Tm~76°C and [dsHCV] ~ 0.7 mM with Mg2+) (Figure S1, S7), at a higher concentration of monovalent ions (Figure S7), and when using the R1ρ RD data (Figure S7).
When unpaired, the N6-methyl group favors the syn conformation, while the anti conformation required for Watson-Crick pairing and duplex annealing is unfavorable with an estimated population of ~5%63. Rotation of the N6-methyl group is likely responsible for the reduced annealing rate. Mismatches have also been shown to reduce kon by up to 50-fold27, 64 through mechanisms that are not fully understood. Further studies are needed to dissect the kinetic mechanism by which m6A slows the annealing rate and how this varies with position and sequence context64.
In conclusion, we have described an NMR strategy for site-specifically resolving duplex hybridization kinetics. The ease and throughput of these experiments can be improved in the future by using longitudinal optimized 1H-CEST experiments65 as well as other approaches for optimal data collection34, 66. The approach can also be applied to mismatch containing duplexes ideally by targeting remote sites that are not involved in any local mismatch dynamics and to use multi-site exchange models as needed to fit data56. Our results show that in the middle of a duplex, m6A minimally affects the melting rate but substantially decreases the rate of annealing. This may help explain why tRNA selection during translation is slower for mRNAs containing m6A20. m6A is also found in the seed sequence of microRNAs and in their mRNA target sites67 and mismatches that slowdown microRNA:mRNA annealing have substantial effects on gene expression64. Thus, m6A could similarly affect gene expression by altering the kinetics of annealing. m6A may also affect the kinetics of RNA-protein and RNA-ligand association and also reshape co-transcriptional RNA folding pathways68–71 by prolonging the lifetime of the unpaired conformation25, 72–73 perhaps in a manner analogous to cis-trans proline isomerization in proteins74–75.
Supplementary Material
ACKNOWLEDGMENT
We thank Nicole Orlovsky for critical comments on the manuscript and Prof Lewis Kay (University of Toronto) for helpful discussions. We acknowledge the technical support and resources from the Duke Magnetic Resonance Spectroscopy Center. We thank Dr. Richard Brennan for use of the UV-Vis spectrophotometer.
Funding Sources
This work was supported by US National Institute for General Medical Sciences (1R01GM132899 to H.M.A.), the Austrian Science Fund (FWF, project P28725 and P30370 to C.K.), and the Austrian Research Promotion Agency FFG (West Austrian BioNMR, 858017 to C.K.)
The research reported in this article was performed by the Duke University faculty and students and was funded by a U.S. National Institutes of Health contract to H.M.A.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
Details of sample preparation, NMR experiments, NMR R1ρ RD and CEST profiles (PDF)
The authors declare the following competing financial interest(s): H.M.A. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company.
REFERENCES
- 1.Meyer KD; Saletore Y; Zumbo P; Elemento O; Mason CE; Jaffrey SR, Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149 (7), 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D; Moshitch-Moshkovitz S; Schwartz S; Salmon-Divon M; Ungar L; Osenberg S; Cesarkas K; Jacob-Hirsch J; Amariglio N; Kupiec M; Sorek R; Rechavi G, Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature 2012, 485 (7397), 201–U84. [DOI] [PubMed] [Google Scholar]
- 3.Desrosiers R; Friderici K; Rottman F, Identification of Methylated Nucleosides in Messenger-Rna from Novikoff Hepatoma-Cells. P Natl Acad Sci USA 1974, 71 (10), 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X; Xiong X; Yi C, Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 2016, 14 (1), 23–31. [DOI] [PubMed] [Google Scholar]
- 5.Wang X; Zhao BS; Roundtree IA; Lu ZK; Han DL; Ma HH; Weng XC; Chen K; Shi HL; He C, N-6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161 (6), 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X; Lu ZK; Gomez A; Hon GC; Yue YN; Han DL; Fu Y; Parisien M; Dai Q; Jia GF; Ren B; Pan T; He C, N-6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505 (7481), 117-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X; Yang Y; Sun BF; Shi Y; Yang X; Xiao W; Hao YJ; Ping XL; Chen YS; Wang WJ; Jin KX; Wang X; Huang CM; Fu Y; Ge XM; Song SH; Jeong HS; Yanagisawa H; Niu YM; Jia GF; Wu W; Tong WM; Okamoto A; He C; Danielsen JMR; Wang XJ; Yang YG, FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24 (12), 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao W; Adhikari S; Dahal U; Chen YS; Hao YJ; Sun BF; Sun HY; Li A; Ping XL; Lai WY; Wang X; Ma HL; Huang CM; Yang Y; Huang N; Jiang GB; Wang HL; Zhou Q; Wang XJ; Zhao YL; Yang YG, Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61 (4), 507–519. [DOI] [PubMed] [Google Scholar]
- 9.Zhao BXS; Roundtree IA; He C, Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol 2017, 18 (1), 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree IA; Evans ME; Pan T; He C, Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169 (7), 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer KD; Jaffrey SR, Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol 2017, 33, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng XL; Su R; Feng XS; Wei MJ; Chen JJ, Role of N-6-methyladenosine modification in cancer. Curr. Opin. Genet. Dev 2018, 48, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T; Hao Y-J; Zhang Y; Li M-M; Wang M; Han W; Wu Y; Lv Y; Hao J; Wang L; Li A; Yang Y; Jin K-X; Zhao X; Li Y; Ping X-L; Lai W-Y; Wu L-G; Jiang G; Wang H-L; Sang L; Wang X-J; Yang Y-G; Zhou Q, m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell stem cell 2015, 16 (3), 289–301. [DOI] [PubMed] [Google Scholar]
- 14.Weng Y-L; Wang X; An R; Cassin J; Vissers C; Liu Y; Liu Y; Xu T; Wang X; Wong SZH; Joseph J; Dore LC; Dong Q; Zheng W; Jin P; Wu H; Shen B; Zhuang X; He C; Liu K; Song H; Ming G. l., Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97 (2), 313–325.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gokhale NS; McIntyre ABR; McFadden MJ; Roder AE; Kennedy EM; Gandara JA; Hopcraft SE; Quicke KM; Vazquez C; Willer J; Ilkayeva OR; Law BA; Holley CL; Garcia-Blanco MA; Evans MJ; Suthar MS; Bradrick SS; Mason CE; Horner SM, N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection . Cell Host Microbe 2016, 20 (5), 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N; Dai Q; Zheng G; He C; Parisien M; Pan T, N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L; Ashraf S; Wang J; Lilley DMJ, Control of box C/D snoRNP assembly by N(6)-methylation of adenine. EMBO Rep. 2017, 18 (9), 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitale RC; Flynn RA; Zhang QC; Crisalli P; Lee B; Jung J-W; Kuchelmeister HY; Batista PJ; Torre EA; Kool ET; Chang HY, Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B; Merriman DK; Choi SH; Schumacher MA; Plangger R; Kreutz C; Horner SM; Meyer KD; Al-Hashimi HM, A potentially abundant junctional RNA motif stabilized by m(6)A and Mg(2). Nat. Commun 2018, 9 (1), 2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J; Ieong K-W; Demirci H; Chen J; Petrov A; Prabhakar A; O’Leary SE; Dominissini D; Rechavi G; Soltis SM; Ehrenberg M; Puglisi JD, N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol 2016, 23, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roost C; Lynch SR; Batista PJ; Qu K; Chang HY; Kool ET, Structure and Thermodynamics of N6-Methyladenosine in RNA: A Spring-Loaded Base Modification. J. Am. Chem. Soc 2015, 137 (5), 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kierzek E; Kierzek R, The thermodynamic stability of RNA duplexes and hairpins containing N-6-alkyladenosines and 2-methylthioN-6-alkyladenosines. Nucleic Acids Res. 2003, 31 (15), 4472–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dethoff EA; Petzold K; Chugh J; Casiano-Negroni A; Al-Hashimi HM, Visualizing transient low-populated structures of RNA. Nature 2012, 491 (7426), 724–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganser LR; Kelly ML; Herschlag D; Al-Hashimi HM, The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol 2019, 20 (8), 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinert H; Sochor F; Wacker A; Buck J; Helmling C; Hiller F; Keyhani S; Noeske J; Grimm S; Rudolph MM; Keller H; Mooney RA; Landick R; Suess B; Furtig B; Wohnert J; Schwalbe H, Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker WR; Ober-Reynolds B; Jouravleva K; Jolly SM; Zamore PD; Greenleaf WJ, High-Throughput Analysis Reveals Rules for Target RNA Binding and Cleavage by AGO2. Mol. Cell 2019, 75 (4), 741–755 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cisse II; Kim H; Ha T, A rule of seven in Watson-Crick base-pairing of mismatched sequences. Nat. Struct. Mol. Biol 2012, 19 (6), 623-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gleitsman KR; Sengupta RN; Herschlag D, Slow molecular recognition by RNA. Rna 2017, 23 (12), 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen KP; Choi J; Prabhakar A; Puglisi EV; Puglisi JD, Relating Structure and Dynamics in RNA Biology. Cold Spring Harbor Perspect. Biol 2019, 11 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massi F; Johnson E; Wang CY; Rance M; Palmer AG, NMR R-1 rho rotating-frame relaxation with weak radio frequency fields. J. Am. Chem. Soc 2004, 126 (7), 2247–2256. [DOI] [PubMed] [Google Scholar]
- 31.Rangadurai A; Szymaski ES; Kimsey IJ; Shi H; Al-Hashimi HM, Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1ρ relaxation dispersion. Prog. Nucl. Magn. Reson. Spectrosc 2019, 112–113, 55–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer AG; Massi F, Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev 2006, 106 (5), 1700–1719. [DOI] [PubMed] [Google Scholar]
- 33.Zhao B; Hansen AL; Zhang Q, Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R(1rho) NMR spectroscopy. J. Am. Chem. Soc 2014, 136 (1), 20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallurupalli P; Bouvignies G; Kay LE, Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc 2012, 134 (19), 8148–61. [DOI] [PubMed] [Google Scholar]
- 35.Steitz J, RNA-RNA base-pairing: theme and variations. Rna 2015, 21 (4), 476–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung J; Van Orden A, Folding and Unfolding Kinetics of DNA Hairpins in Flowing Solution by Multiparameter Fluorescence Correlation Spectroscopy. J. Phys. Chem. B 2005, 109 (8), 3648–3657. [DOI] [PubMed] [Google Scholar]
- 37.Jung J; Van Orden A, A Three-State Mechanism for DNA Hairpin Folding Characterized by Multiparameter Fluorescence Fluctuation Spectroscopy. J. Am. Chem. Soc 2006, 128 (4), 1240–1249. [DOI] [PubMed] [Google Scholar]
- 38.Chen XD; Zhou Y; Qu P; Zhao XS, Base-by-Base Dynamics in DNA Hybridization Probed by Fluorescence Correlation Spectroscopy. J. Am. Chem. Soc 2008, 130 (50), 16947–16952. [DOI] [PubMed] [Google Scholar]
- 39.Nayak RK; Peersen OB; Hall KB; Van Orden A, Millisecond Time-Scale Folding and Unfolding of DNA Hairpins Using Rapid-Mixing Stopped-Flow Kinetics. J. Am. Chem. Soc 2012, 134 (5), 2453–2456. [DOI] [PubMed] [Google Scholar]
- 40.He G; Li J; Ci HN; Qi CM; Guo XF, Direct Measurement of Single-Molecule DNA Hybridization Dynamics with Single-Base Resolution. Angew. Chem., Int. Ed 2016, 55 (31), 9036–9040. [DOI] [PubMed] [Google Scholar]
- 41.Wetmur JG; Davidson N, Kinetics of Renaturation of DNA. J. Mol. Biol 1968, 31 (3), 349–&. [DOI] [PubMed] [Google Scholar]
- 42.Wetmur JG, Hybridization and Renaturation Kinetics of Nucleic-Acids. Annu. Rev. Biophys. Bioeng 1976, 5, 337–361. [DOI] [PubMed] [Google Scholar]
- 43.Bonnet G; Krichevsky O; Libchaber A, Kinetics of conformational fluctuations in DNA hairpin-loops. P Natl Acad Sci USA 1998, 95 (15), 8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams AP; Longfellow CE; Freier SM; Kierzek R; Turner DH, Laser Temperature-Jump, Spectroscopic, and Thermodynamic Study of Salt Effects on Duplex Formation by Dgcatgc. Biochemistry 1989, 28 (10), 4283–4291. [DOI] [PubMed] [Google Scholar]
- 45.Howorka S; Movileanu L; Braha O; Bayley H, Kinetics of duplex formation for individual DNA strands within a single protein nanopore. P Natl Acad Sci USA 2001, 98 (23), 12996–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimsey IJ; Szymanski ES; Zahurancik WJ; Shakya A; Xue Y; Chu CC; Sathyamoorthy B; Suo ZC; Al-Hashimi HM, Dynamic basis for dG.dT misincorporation via tautomerization and ionization. Nature 2018, 554 (7691), 195-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauzan B; McMichael E; Cave R; Sevcik LR; Ostrosky K; Whitman E; Stegemann R; Sinclair AL; Serra MJ; Deckert AA, Kinetics and Thermodynamics of DNA, RNA, and Hybrid Duplex Formation. Biochemistry 2013, 52 (5), 765–772. [DOI] [PubMed] [Google Scholar]
- 48.Craig ME; Crothers DM; Doty P, Relaxation Kinetics of Dimer Formation by Self Complementary Oligonucleotides. J. Mol. Biol 1971, 62 (2), 383-&. [DOI] [PubMed] [Google Scholar]
- 49.Pörschke D; Uhlenbeck OC; Martin FH, Thermodynamics and kinetics of the helix-coil transition of oligomers containing GC base pairs. Biopolymers 1973, 12 (6), 1313–1335. [Google Scholar]
- 50.Wyer JA; Kristensen MB; Jones NC; Hoffmann SV; Nielsen SB, Kinetics of DNA duplex formation: A-tracts versus AT-tracts. Phys. Chem. Chem. Phys 2014, 16 (35), 18827–39. [DOI] [PubMed] [Google Scholar]
- 51.Braunlin WH; Bloomfield VA, H-1-Nmr Study of the Base-Pairing Reactions of D(Ggaattcc) - Salt Effects on the Equilibria and Kinetics of Strand Association. Biochemistry 1991, 30 (3), 754–758. [DOI] [PubMed] [Google Scholar]
- 52.Mulder FA; Mittermaier A; Hon B; Dahlquist FW; Kay LE, Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Mol. Biol 2001, 8 (11), 932–5. [DOI] [PubMed] [Google Scholar]
- 53.Xue Y; Kellogg D; Kimsey IJ; Sathyamoorthy B; Stein ZW; McBrairty M; Al-Hashimi HM, Characterizing RNA Excited States Using NMR Relaxation Dispersion. Methods Enzymol. 2015, 558, 39–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H; Kimsey IJ; Nikolova EN; Sathyamoorthy B; Grazioli G; McSally J; Bai T; Wunderlich CH; Kreutz C; Andricioaei I; Al-Hashimi HM, m1A and m1G disrupt A-RNA structure through the intrinsic instability of Hoogsteen base pairs. Nat. Struct. Mol. Biol 2016, 23 (9), 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikolova EN; Kim E; Wise AA; O’Brien PJ; Andricioaei I; Al-Hashimi HM, Transient Hoogsteen base pairs in canonical duplex DNA. Nature 2011, 470 (7335), 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimsey IJ; Petzold K; Sathyamoorthy B; Stein ZW; AlHashimi HM, Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 2015, 519 (7543), 315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathyamoorthy B; Shi H; Zhou H; Xue Y; Rangadurai A; Merriman DK; Al-Hashimi HM, Insights into Watson– Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res. 2017, 45 (9), 5586–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi H; Clay MC; Rangadurai A; Sathyamoorthy B; Case DA; Al-Hashimi HM, Atomic structures of excited state A–T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations. J. Biomol. NMR 2018, 70 (4), 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alvey HS; Gottardo FL; Nikolova EN; Al-Hashimi HM, Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat. Commun 2014, 5, 4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pörschke D; Eigen M, Co-operative non-enzymatic base recognition III. Kinetics of the helix—coil transition of the oligoribouridylic · oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol 1971, 62 (2), 361–381. [DOI] [PubMed] [Google Scholar]
- 61.Vallurupalli P; Sekhar A; Yuwen TR; Kay LE, Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: a primer. J. Biomol. NMR 2017, 67 (4), 243–271. [DOI] [PubMed] [Google Scholar]
- 62.Bouvignies G; Hansen DF; Vallurupalli P; Kay LE, Divided-Evolution-Based Pulse Scheme for Quantifying Exchange Processes in Proteins: Powerful Complement to Relaxation Dispersion Experiments. J. Am. Chem. Soc 2011, 133 (6), 1935–1945. [DOI] [PubMed] [Google Scholar]
- 63.Engel JD; Von Hippel PH, Effects of methylation on the stability of nucleic acid conformations. Monomer level. Biochemistry 1974, 13 (20), 4143–4158. [DOI] [PubMed] [Google Scholar]
- 64.Miller CL; Haas U; Diaz R; Leeper NJ; Kundu RK; Patlolla B; Assimes TL; Kaiser FJ; Perisic L; Hedin U; Maegdefessel L; Schunkert H; Erdmann J; Quertermous T; Sczakiel G, Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014, 10 (3), e1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuwen T; Kay LE, Longitudinal relaxation optimized amide (1)H-CEST experiments for studying slow chemical exchange processes in fully protonated proteins. J. Biomol. NMR 2017, 67 (4), 295–307. [DOI] [PubMed] [Google Scholar]
- 66.Bouvignies G; Kay LE, A 2D C-13-CEST experiment for studying slowly exchanging protein systems using methyl probes: an application to protein folding. J. Biomol. NMR 2012, 53 (4), 303–310. [DOI] [PubMed] [Google Scholar]
- 67.Linder B; Grozhik AV; Olarerin-George AO; Meydan C; Mason CE; Jaffrey SR, Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12 (8), 767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heilman-Miller SL; Woodson SA, Effect of transcription on folding of the Tetrahymena ribozyme. Rna 2003, 9 (6), 722–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer FR; Mills DR, Secondary structure formation during RNA synthesis. Nucleic Acids Res. 1981, 9 (19), 5109–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan T; Artsimovitch I; Fang XW; Landick R; Sosnick TR, Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. P Natl Acad Sci USA 1999, 96 (17), 9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong TN; Sosnick TR; Pan T, Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. P Natl Acad Sci USA 2007, 104 (46), 17995–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao B; Guffy SL; Williams B; Zhang Q, An excited state underlies gene regulation of a transcriptional riboswitch. Nat. Chem. Biol 2017, 13 (9), 968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breaker RR, Riboswitches and Translation Control. Cold Spring Harbor Perspect. Biol 2018, 10 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brandts JF; Halvorson HR; Brennan M, Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 1975, 14 (22), 4953–4963. [DOI] [PubMed] [Google Scholar]
- 75.Wedemeyer WJ; Welker E; Scheraga HA, Proline cistrans isomerization and protein folding. Biochemistry 2002, 41 (50), 14637–14644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.