Abstract
Objectives:
Osteoarthritis (OA) is the most common age-related joint disease. With aging and in OA, the expression of FOXO transcription factors is reduced, diminishing their chondroprotective actions. To elucidate the molecular mechanisms by which FOXO1 protects chondrocytes we identified the genome-wide occupancy profile of FOXO1.
Methods:
We performed FOXO1 chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) on human primary chondrocytes. ChIP-seq data were integrated with RNA-seq datasets. Bioinformatics results were confirmed in primary chondrocytes that were treated with a FOXO1 inhibitor.
Results:
Analysis of FOXO1 ChIP-seq on human primary chondrocytes showed that pathways implicated in OA pathogenesis are mainly regulated by FOXO1 binding to tissue-specific enhancers with suboptimal binding sites (20% of the peaks), while more ubiquitous FOXO1 pathways are regulated at the promoter level through interaction with its canonical binding motif (7% of the peaks) Integrating FOXO1 occupancy data with RNA-seq comparing OA and healthy human cartilage revealed 428 putative FOXO1 target genes that are dysregulated in OA. Pathway analysis showed enrichment for genes belonging to senescence (logP −6.73), extracellular matrix (ECM) (logP −12.97) and circadian clock (logP −6.30) pathways suggesting that FOXO1 dysregulation plays an important role in their abnormal expression in OA. Using an inhibitor of FOXO1, we confirmed that FOXO1 regulates these pathways in cultured human chondrocytes.
Conclusions:
FOXO1 regulates ubiquitous and cartilage-specific genes in chondrocytes by using different mechanisms. The FOXO1 transcriptional network has a direct role in regulating homeostasis, ECM and circadian clock genes and plays an important role in the abnormal expression of these pathways observed in OA pathogenesis.
INTRODUCTION
Dysregulated gene expression in chondrocytes compromises cellular and tissue homeostasis and promotes osteoarthritis (OA) pathogenesis. Recent studies have begun to investigate mechanisms that lead to this abnormal cell phenotype in aging and OA cartilage and examined cellular homeostasis and defense mechanisms against various forms of stress. This led to the discovery that anti-oxidant defensess1,2 and autophagy are deficient in aging and OA cartilage3.
FOXO proteins are evolutionarily conserved transcription factors with important functions in development, aging and longevity4. In mammals, the FOXO family is comprised of four members (FOXO1, FOXO3, FOXO4 and FOXO6) with distinct functions5. FOXO1, FOXO3 and FOXO4 are ubiquitously expressed whereas FOXO6 expression is largely restricted to the brain6. The role of FOXO in regulating lifespan is thought to be through control of cellular homeostasis and maintenance of stem/progenitor cells populations during aging5–8. FOXO proteins transcriptionally induce expression of antioxidant enzymes such as catalase and manganese superoxide dismutase9,10. They also regulate autophagy and the ubiquitin-proteasome system, to eliminate damaged and aggregated proteins8.
Dysregulation of FOXO expression or activity contributes to the pathogenesis of age-related diseases, including bone11,12 and muscle13,14. We have shown that FOXO mRNA and protein expression are reduced in human and mouse articular cartilage during aging and OA1. We also found that FOXO are important regulators of cartilage homeostasis, in part by promoting oxidative stress resistance and the expression of autophagy genes in human articular chondrocytes2. Studies on mice with cartilage specific deletion of FOXO revealed a role of FOXO in cartilage formation, maturation and maintenance15.
While these results establish a critical role of FOXO in cartilage homeostasis, FOXO target genes and mechanisms of FOXO-mediated gene regulation in chondrocytes are unknown. The study used FOXO1 ChIP-seq and an integrative analysis with data from RNA-seq and histone marks to reveal the role and mechanisms of FOXO1 in chondrocyte-specific gene expression.
RESULTS
FOXO1 regulates OA associated pathways by utilizing suboptimal binding motifs.
To better understand the role of FOXO1 in OA pathogenesis we performed FOXO1 ChIP-seq on primary human chondrocytes from OA cartilage samples. All three biological replicates showed high correlation scores >0.9 (Suppl Fig. S1). Peak calling was performed using PePr16, identifying 5459 consistent peaks among biological replicates. Suspected false positive peaks were identified as those not exhibiting the expected shift size, as well as overlapping with empirically determined artifact-prone regions (ENCODE blacklisted genomic regions), rendering a list of 4952 high-confidence peaks (GSE144026).
We performed de-novo motif discovery to investigate putative chondrocyte-specific chromatin binding mechanisms and interactors of FOXO1. As expected, the most over-represented motif was the canonical binding site for FOXO TFs (known as the Daf-16 family member-binding element (DBE): 5′-GTAAA(T/C)AA-3′) which was present in 64% of the peaks, followed by bZIP and ETS consensus binding motifs (Fig. 1A). Approximately half of the peaks without a canonical binding site for FOXO1 have a truncated version of the FOXO1 motif (5’-(A/C)AACA-3’), similar to the core sequence recognized by all forkhead proteins (5′-(A/C)AA(C/T)A-3′)17 (Suppl Fig. S2). FOXO proteins bind the DBE sequence with higher affinity mainly due to the more favorable interaction of Thy2 (5′-GTAAA(T/C)AA-3′) with His215 of the FOXO1 DNA binding domain18. This is in agreement with our data since FOXO1 peaks with a DBE motif have a higher average tag density than peaks without it (Fig. 1B). To assess if the chromatin binding mechanism plays a role in chondrocyte biology, we performed pathway analysis on the 2396 genes annotated exclusively with peaks harboring a FOXO1 canonical binding site, and on the 732 genes with peaks, having either a suboptimal or no FOXO1 motif. Interestingly, the top pathways over-represented in Reactome DB for genes bound exclusively by FOXO1 at loci without a canonical binding motif are chondrocyte-specific and directly associated with OA pathogenesis, with the “Collagen degradation” pathway showing the highest enrichment (Fig. 2A, Table S1). In contrast, the top pathways for genes bound by FOXO1 through its canonical binding motif involve more ubiquitous FOXO1 functions related to signaling with “Signaling by PTK6” as the most enriched pathway (Fig. 2A, Table S2).
Figure 1. de novo motif discovery at FOXO1 peaks.
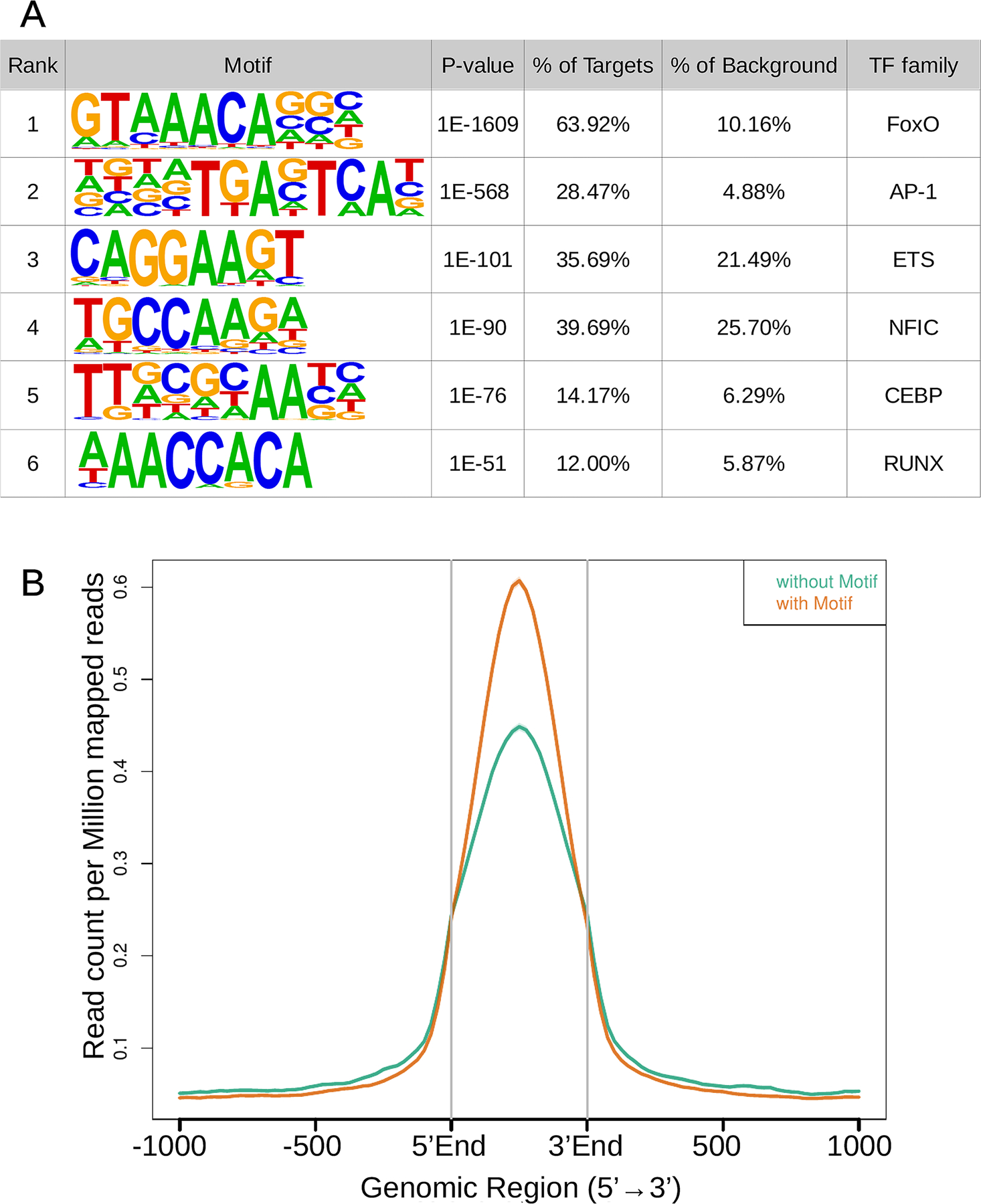
A) Over-representation of TF binding motifs at FOXO1 peaks. Motif logos display nucleotide frequencies at each position. Statistical significance of the over-representation is quantified as p-value relative to random sequences matched for GC% content.
B) Average tag density plotted over the center of peaks with and without a FOXO1 canonical binding motif.
Figure 2. Distinct over-represented pathways in genes bound by FOXO1 according to the transcriptional regulatory mechanism.
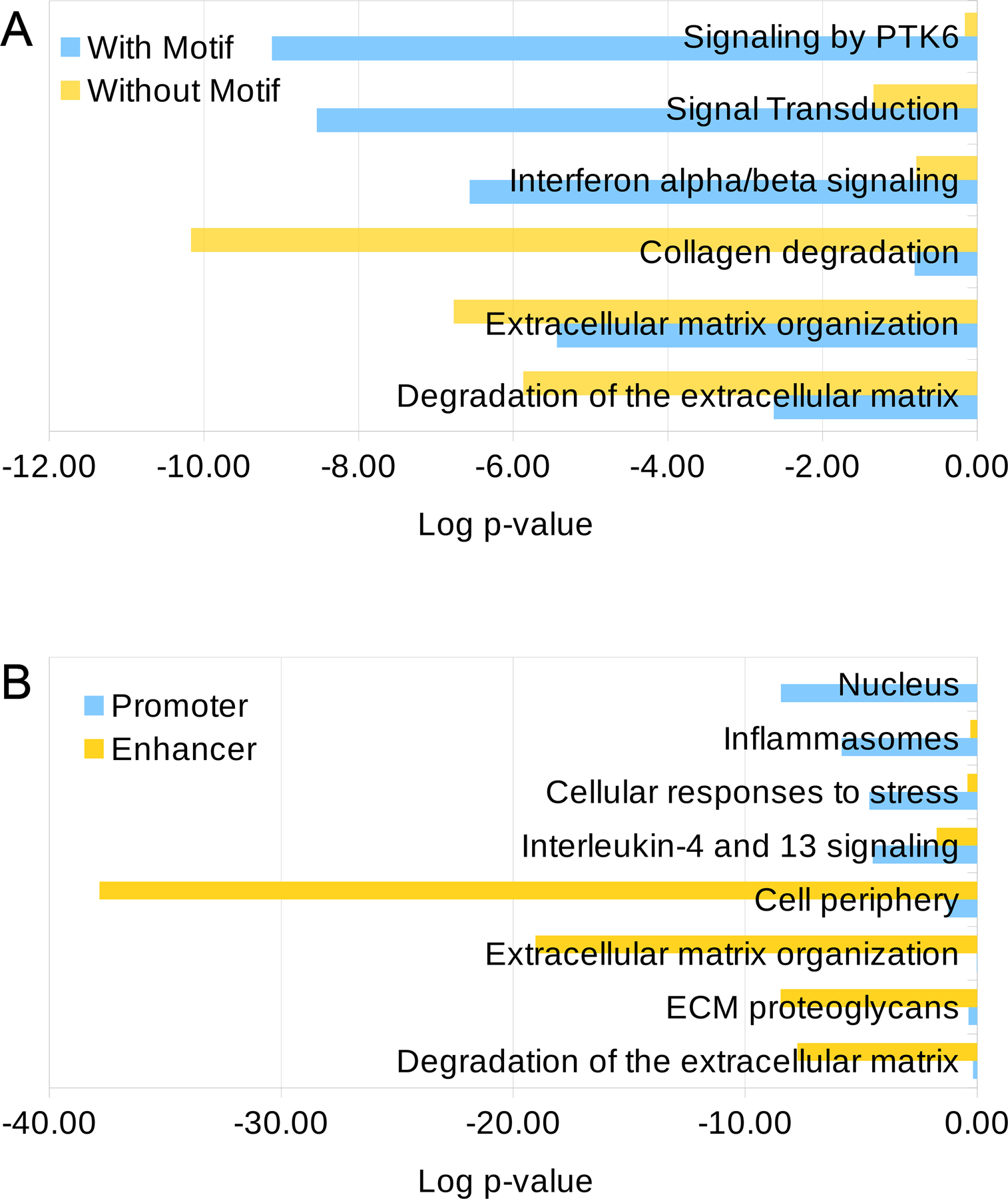
Results obtained using GO Cellular Component DB and Reactome DB.
A) Comparison of the 3 top terms from Reactome DB over-represented in lists of genes regulated exclusively by FOXO1 peaks with and without a canonical binding site.
B) Comparison of the top term from GO Cellular Component DB and the 3 top terms from Reactome DB over-represented in lists of genes regulated exclusively by FOXO1 peaks overlapping with enhancers or promoters.
FOXO1 occupies active regulatory regions.
To assess the functional role of the regions bound by FOXO1 in chondrocytes, we integrated our analysis with ChIP-seq data from 7 histone marks in chondrocytes (GSE17312). A striking 80% of the total FOXO1 peaks overlap with active regulatory regions marked by H3K27ac, H3K4me1, H3K4me3 and/or H3K9ac. This is in agreement with previous reports of FOXO1 binding in germinal center B-cells19. The most prevalent combination of histone marks overlapping FOXO1 peaks is H3K27ac and H3K4me1, which is a hallmark of active enhancers (Fig. 3A). FOXO1 binding at chondrocyte enhancer regions accounts for 61% of the total peaks and holds the highest enrichment score relative to the human genome (log2 enrichment = 3.53) (Fig. 3B). In contrast, promoter regions also show a high enrichment score (log2 enrichment = 3.04), but a marked reduction in the number of peaks (9.3%) (Fig. 3B).
Figure 3. FOXO1 occupies active regulatory regions.
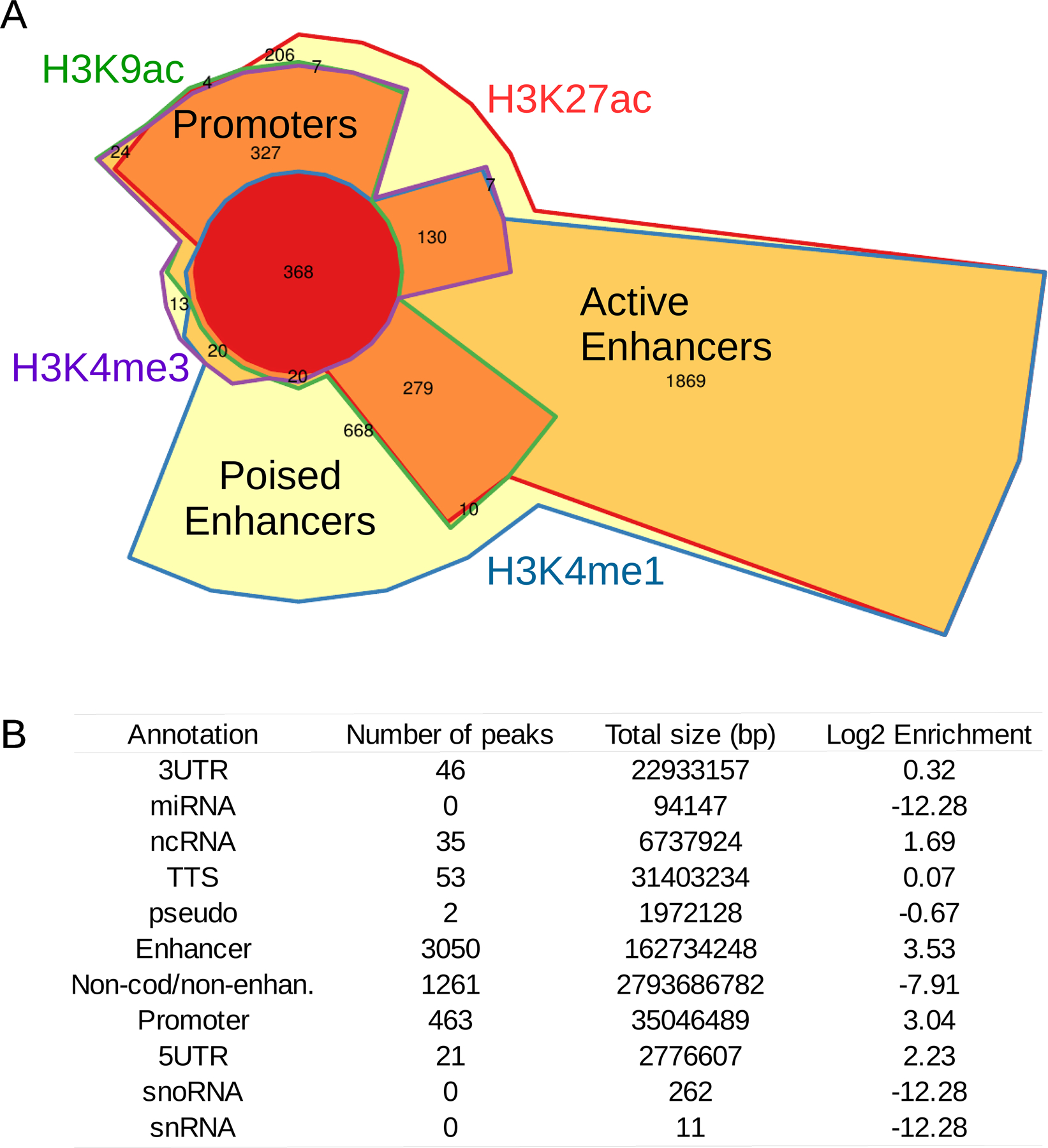
A) Chow-Ruskey plot showing FoxO1 occupancy overlapping with histone marks of active regulatory regions in human chondrocytes. The area of each region is proportional to the number of FOXO1 peaks overlapping with each histone mark combination. Each region is color-coded according to the number of histone marks overlapping with FOXO1 peaks: FOXO1 peaks overlapping with all four active histone marks (red), three histone marks (dark orange), two histone marks (light orange) and with only one histone mark (yellow). Histone marks are also color-coded: H3K9ac (green), H3K27ac (red), H3K4me1 (blue) and H3K4me3 (purple).
B) Enrichment of FOXO1 binding to predicted RefSeq genomic elements and chondrocyte enhancers identified using chondrocyte-derived histone marks for poised (H3K4me1) and active (H3K4me1+H3K27ac) enhancers. 3UTR: 3’ untranslated region; miRNA: micro RNA; ncRNA: non-coding RNA; TTS: transcription termination site; pseudo: pseudogene; Non-cod/non-enhan: non-coding regions which are not enhancers; 5UTR: 5’ untranslated region; snoRNA: small nucleolar RNA; snRNA: small nuclear RNA.
FOXO1 regulates distinct transcriptional programs at the promoter and enhancer levels.
We then investigated if FOXO1 peaks located at promoter regions have distinct motif composition as compared to peaks located at enhancers. Interestingly, the SP2 binding motifs are enriched at promoter regions, while motifs for AP-1 and RUNX TF families are more abundant at enhancer elements (Suppl Fig. S3). Other motifs like ETS and SMAD showed little difference between promoters and enhancers. Given the differential TF binding motif composition, we asked if genes bound by FOXO1 at the promoter level have distinct biological functions compared to those bound at the enhancer level. In line with our previous results, genes regulated exclusively by FOXO1 at the promoter level act mainly in the nucleus (1st GO cellular_component: logP = −8.46) and regulate conserved pathways, the top 3 in Reactome DB being Inflammasomes (1st, logP = −7.89), Cellular responses to stress (2nd, logP = −4.64) and Interleukin-4 and 13 signaling (3rd, logP = −4.50) (Fig. 2B, Table S3). On the other hand, genes regulated at the enhancer level act mainly in the cell periphery (1st GO cellular_component: logP = −37.83) and the top 3 pathways in Reactome DB are chondrocyte specific and dysregulated in OA, ie: ECM organization (1st, logP =−19.04), ECM proteoglycans (2nd, logP = −8.47), and degradation of the ECM (3rd, logP = −7.74) (Fig. 2B, Table S4). Representative examples of genome tracks of FOXO1 binding regulating chondrocyte-specific COL13A1 at the enhancer level and ubiquitous TXNIP at the promoter are shown in Suppl. Fig. S4.
These results suggest that FOXO1 regulates pathways implicated in OA pathogenesis, mainly by binding to tissue-specific enhancers, with suboptimal binding sites, while more ubiquitous FOXO1 pathways are regulated at the promoter level through interaction with its canonical binding motif.
FOXO1 occupancy is highly tissue-specific.
To better understand the characteristics of FOXO1 tissue-specific versus ubiquitous transcriptional programs we analyzed all currently available FOXO1 ChIP-seq datasets in human along with our chondrocyte dataset. We found 3 publicly available datasets that passed QC testing (Qtag >=0) i.e.: Pre-leukemia cells (GSM2136846), Endometrial stromal cells (GSE69542) and Germinal center B-cells (GSE68349). To minimize biases due to technical aspects all replicates were pooled, and peak calling was performed using HOMER with the same parameters for all samples. The top 5,000 peaks from each dataset were selected for further analyses.
We next determined the proportion of tissue-specific and ubiquitous FOXO1 peaks by performing a 4-way overlap between the 4 cell types. All cell types had a high proportion of non-overlapping peaks ranging from 81.22% for chondrocytes to 94.44% for pre-leukemia cells (Fig. 4A.) suggesting that FOXO1 occupancy is highly tissue-specific. Surprisingly, only 5 peaks (0.1%) are shared by all 4 cell types. Then, we assessed the distribution of FOXO1 occupancy respective to the nearest transcription start site (TSS) to investigate if one of the reasons for the low overlap in binding sites is the differential preference in occupancy of genomic elements. Indeed, in pre-leukemia cells FOXO1 binding is enriched at promoter regions (0–5 kb from TSS) in comparison to all 3 other cell types in which FOXO1 has a preference for distal regions (50–500 kb from TSS) (Fig. 4B). This suggests a distinct mechanism of action in pre-leukemia cells where the most enriched motif is RUNX1, whereas in all 3 other cell types the canonical FOXO1 binding motif is the most enriched.
Figure 4. FOXO1 binding is highly tissue-specific.
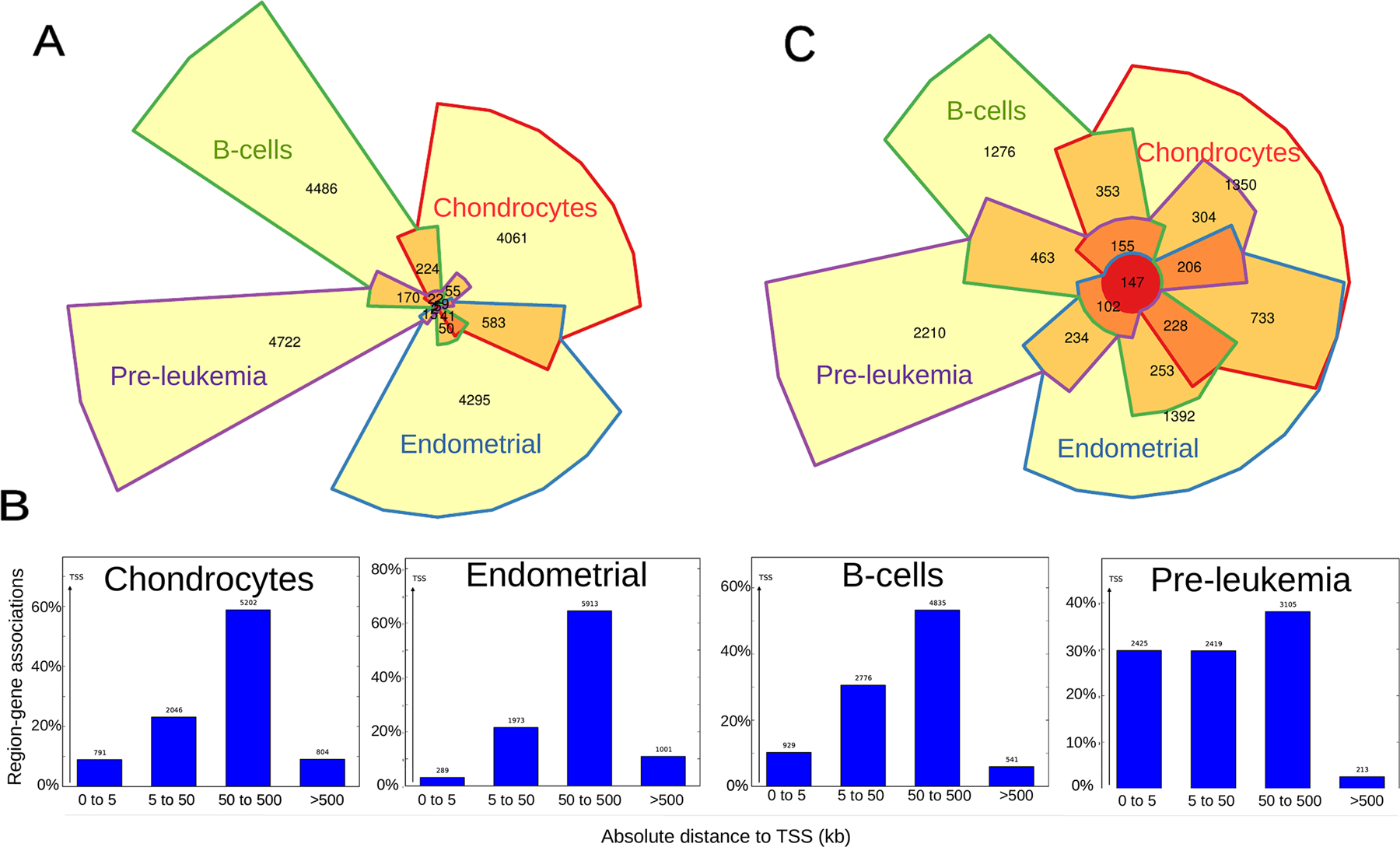
A) Chow-Ruskey diagram showing a 4-way overlap between FOXO1 peaks from 4 human cell types. The area of each region is proportional to the number of overlapping FOXO1 peaks. Each region is color-coded according to the number of overlapping FOXO1 peaks from each cell type: FOXO1 peaks overlapping in all four cell types (red), three cell types (dark orange), two cell types (light orange) and present in only one cell type (yellow). Cell types are also color-coded: B-cells (green), chondrocytes (red), endometrial (blue) and pre-leukemia (purple).
B) Histograms depicting the binned distribution of FOXO1 binding distance to the nearest TSS in 4 human cell types.
C) Chow-Ruskey diagram showing a 4-way overlap between FOXO1 putative target genes from 4 human cell types. The area of each region is proportional to the number of overlapping FOXO1 putative target genes. Each region is color-coded according to the number of overlapping FOXO1 putative target genes from each cell type: FOXO1 putative target genes shared by all four cell types (red), three cell types (dark orange), two cell types (light orange) and present at only one cell type (yellow). Cell types are also color-coded: B-cells (green), chondrocytes (red), endometrial (blue) and pre-leukemia (purple).
Since FOXO1 has conserved functions in homeostasis across tissues and species8, we associated each peak to a gene (nearest TSS) and assessed the overlap between putative FOXO1 target genes in the 4 cell types. As expected, the overlap between FOXO1 putative target genes is much greater than that of peaks, showing a total of 147 shared genes (~6.3%) (Fig. 4C). We then performed pathway analyses on the 147 conserved FOXO1 target genes using Reactome DB and found that all top 9 pathways are signaling cascades (Table S5). These results confirm that the overrepresented pathways regulated by FOXO1 in chondrocytes at the promoter level and/or using its canonical binding motif are similar to the ubiquitous gene network conserved between cell types (Fig. 2), while chondrocyte-specific pathways are regulated at the enhancer level, preferentially using non-canonical binding motifs.
FOXO1 pathway dysregulation in OA could lead to altered circadian rhythms and ECM homeostasis.
Previous results from our group have characterized the dysregulation in gene transcription occurring in OA pathogenesis by performing RNA-seq on healthy and OA human cartilage samples and identified 1307 differentially expressed genes (DEG)20. Importantly, the FOXO signaling pathway is significantly altered in OA compared to normal chondrocytes (logP = −6.57). Therefore, to gain further insight into the direct role of FOXO1 in OA pathogenesis we focused on the 428 genes that are bound by FOXO1 and significantly dysregulated in OA. Pathway analyses shows enrichment for genes from ubiquitous FOXO1 signaling pathways to maintain homeostasis like “Senescence-Associated Secretory Phenotype (SASP)” and “p53 signaling pathway”, chondrocyte-specific pathways like “ECM organization” and “ECM-receptor interaction” and surprisingly, “Circadian clock” and “Circadian rhythm” in Reactome (Table 1) and Kegg (Table S6) databases,.
Table 1.
Over-represented pathways in Reactome DB of FOXO1 bound genes differentially expressed in OA. All 2985 FOXO1 bound genes were used as background.
| Terms in Reactome DB | Enrichment | logP | Genes in Term | Target Genes in Term | Total Target Genes | Total Genes | Gene Symbols |
|---|---|---|---|---|---|---|---|
| Interleukin-4 and 13 signaling | 4.91E-07 | −14.53 | 37 | 18 | 236 | 1647 | TNFRSF1B, JUNB, F13A1, BCL6, MMP2, COL1A2, S1PR1, RORC, PIM1, IRF4, CDKN1A, VCAM1, CCND1, FOS, IL6R, MYC, SOCS3, RHOU |
| Extracellular matrix organization | 2.33E-06 | −12.97 | 77 | 27 | 236 | 1647 | COL6A3, SPP1, COL15A1, MMP2, COL1A2, COA1, ITGB3, VCAM1, SPARC, LTBP1, COL5A2, COL13A1, NCAM1, COL25A1, FBLN1, ITGB4, LUM, ADAMTS5, COL2A1, EFEMP1, ITGB8, HAPLN1, TNC, ASPN, HTRA1, P4HA3, ITGB5 |
| Collagen chain trimerization | 1.07E-04 | −9.14 | 13 | 8 | 236 | 1647 | COL5A2, COL13A1, COL25A1, COL15A1, COL6A3, COL1A2, COL2A1, COL3A1 |
| Collagen biosynthesis and modifying enzymes | 3.29E-04 | −8.02 | 18 | 9 | 236 | 1647 | COL13A1, COL5A2, COL25A1, P4HA3, COL1A2, COL6A3, COL15A1, COL3A1, COL2A1 |
| Collagen formation | 4.03E-04 | −7.82 | 22 | 10 | 236 | 1647 | COL5A2, COL13A1, P4HA3, COL25A1, COL15A1, ITGB4, COL6A3, COL1A2, COL2A1, COL3A1 |
| Class B/2 (Secretin family receptors) | 4.75E-04 | −7.65 | 12 | 7 | 236 | 1647 | WNT11, CD55, ADM, FZD8, WNT5A, FZD10, FZD5 |
| Assembly of collagen fibrils and other multimeric structures | 9.05E-04 | −7.01 | 13 | 7 | 236 | 1647 | COL3A1, COL2A1, COL1A2, COL6A3, COL5A2, ITGB4, COL15A1 |
| GPCR ligand binding | 9.38E-04 | −6.97 | 28 | 11 | 236 | 1647 | CCL28, CD55, FZD8, WNT5A, ADM, WNT11, AVPR1A, FZD5, FZD10, PENK, S1PR1 |
| Senescence-Associated Secretory Phenotype (SASP) | 1.20E-03 | −6.73 | 17 | 8 | 236 | 1647 | CDK6, CDKN2B, CDKN1B, FOS, UBC, CDKN1A, IGFBP7, CEBPB |
| Circadian Clock | 1.83E-03 | −6.30 | 30 | 11 | 236 | 1647 | NPAS2, BHLHE40, ARNTL, BHLHE41, NFIL3, UBC, KLF15, NR1D1, SREBF1, PER1, SIK1 |
To confirm our computational predictions, we validated FOXO1 ECM and circadian rhythm target genes using AS1842856, a selective FOXO1 inhibitor21 in primary human chondrocytes. Among ECM pathway genes that are differentially expressed in OA we observed a significant inhibition of COL2A1 and VCAM1, while MMP2 was significantly up-regulated (Fig. 5). AS1842856 also significantly inhibited the expression of ACAN and PRG4, which are bound by FOXO1 in human chondrocytes and are of central importance in cartilage biology and OA pathogenesis22,23. Other predicted ECM genes bound by FOXO1 (COL3A1, COL5A2) and non-bound genes (COL1A1) remained unchanged upon AS1842856 treatment. We also observed a significant inhibition of RORC and NR1D1, core clock circadian genes that are dysregulated in OA24. PER1 is also a core clock circadian gene dysregulated in OA, but no significant effect was observed upon treatment with AS1842856. Other predicted circadian FOXO1 target genes (BHLHE41, BMAL1, CRY2 and RORA) and non-target genes (PER2) remained unchanged upon AS1842856 treatment. We also measured SESN3 expression, a well know and validated FOXO1 target genes in chondrocytes15,25,26, and it was significantly repressed by AS1842856.
Figure 5. FOXO1 regulates ECM and circadian rhythm genes in human cartilage.
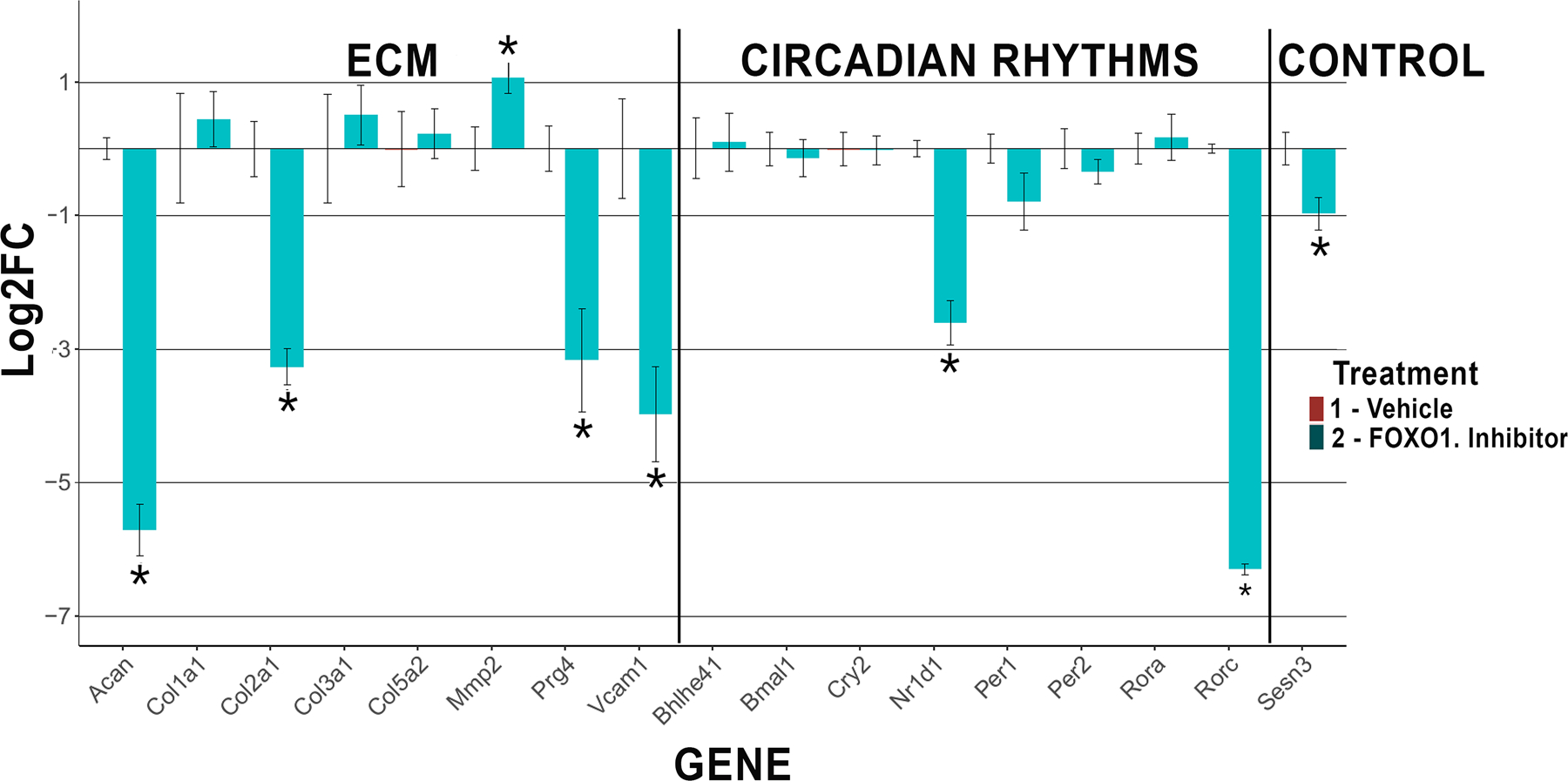
Human primary chondrocytes were treated with AS1842856, a selective FOXO1 inhibitor for 24 h, and FOXO1 target genes in the ECM and CR pathways were analyzed by RT-qPCR. SESN3, a known FOXO1 target gene, was used as a positive control. Results are averages from 3 human donors.
These results linking FOXO1 to circadian rhythm dysregulation in OA are described here for the first time and further confirm the central role of FOXO1 in maintaining cartilage homeostasis by sensing and regulating a wide array of biological processes.
DISCUSSION
Our previous findings that FOXO TFs regulate cartilage homeostasis and that FOXO pathways are dysregulated in OA1,2,15 led us to investigate the genome-wide occupancy profile of FOXO1 in human primary chondrocytes. Integration with RNA-seq data from healthy and OA human cartilage samples allowed us to describe distinct mechanisms of FOXO1 transcriptional regulation, as well as novel roles of FOXO1 in OA pathogenesis.
The first finding of our study is that FOXO1 uses distinct molecular mechanisms to drive ubiquitous or tissue-specific gene expression. The basis for the first distinct molecular mechanism of regulation is encoded in the DNA. We observe that 64% of FOXO1 peaks in chondrocytes have the FOXO1 canonical binding motif. Interestingly, this group is enriched for genes involved in signal transduction. These results are in line with our current understanding that FOXO1 functions as a sensor and effector in response to environmental changes to maintain homeostasis by modulating the activity of signal transduction cascades at the expression level27. The role of FOXO1 in maintaining homeostasis has been described in many tissues and is believed to be conserved throughout the body and evolution27,28. On the other hand, genes with FOXO1 occupancy harboring either a truncated FOXO motif, or completely lacking it, are highly enriched for pathways related to ECM organization, which are highly chondrocyte-specific. Chondrocytic lineage cells in cartilage produce and maintain the complex ECM necessary for biomechanical function29. We show that FOXO1 binds with lower affinity to sites lacking the canonical motif. Functional studies in model animals have shown that motif sub-optimization is essential for precise temporal and spatial gene expression since TF binding to sub-optimal motifs can only occur in the context of co-occupancy with other tissue-specific TFs30. Substituting the wild-type low-affinity sites with high-affinity sequences disrupts the level, location and timing of gene expression31,32. Altogether, our results suggest that FOXO1 utilizes low-affinity motifs to regulate cartilage-specific ECM pathways (Suppl Fig. S5) as an effective mechanism to produce precise tissue specificity and avoid ectopic expression, while ubiquitous functions of FOXO1 are regulated through its canonical binding motif (Suppl Fig. S5).
The second distinct molecular mechanism of gene regulation is associated with differential usage of regulatory elements between tissue-specific and conserved FOXO1 pathways. Epigenetic modifications laid out by a tissue-specific combination of developmental TFs give rise to regions, with precise boundaries within the genome, that can serve as regulatory elements33. Integration of our FOXO1 occupancy dataset with legacy ChIP-seq data from 7 histone marks in chondrocytes34 shows that FOXO1 occupancy is highly enriched at enhancer regions, followed by promoters. As observed for canonical versus sub-optimal FOXO1 binding motifs, we can also distinguish tissue-specific versus ubiquitous transcriptional programs regulated at the enhancer and promoter level, respectively. Genes regulated exclusively at the enhancer level are enriched for chondrocyte-specific pathways, whereas genes regulated exclusively at the promoter level are enriched in FOXO1 conserved pathways. Promoters occupy the region directly upstream of the TSS and therefore are more conserved between tissues than enhancer elements that can be present many hundred kbs away from the TSS and have less positional constraints. Therefore, preferential usage of highly conserved promoters regulates ubiquitous FOXO1 functions, while chondrocyte-specific functions are preferentially regulated at highly tissue-specific enhancers. This represents another regulatory layer to generate a robust but localized expression of FOXO1 target genes.
The third distinct mechanism regulating FOXO1 transcriptional programs involves differential functional interaction with other TFs. By performing motif discovery at chondrocyte FOXO1 peaks located enhancers or promoters, we observe co-occupying TF motifs with a marked preference of location, while others are similarly distributed. In particular, enhancer regions are enriched for motifs of the AP-1 and RUNX families, while promoters harbor more SP2 motifs. This differential motif composition likely translates into distinct functional partners of FOXO1 at promoters and enhancers that help to fine-tune ubiquitous and chondrocyte-specific transcriptional programs with a certain degree of independence. Distinct putative molecular interactors for FOXO1 are also observed between cell types by analyzing legacy data from other human FOXO1 ChIP-seq experiments. Particularly striking is the difference between pre-leukemia cells and chondrocytes. In pre-leukemia cells the most over-represented motif in the top 5000 FOXO1 peaks is the consensus binding motif for RUNX1 (present in 66% of the peaks), while only 18% of the peaks have a FOXO1 consensus binding site. In chondrocytes, we observe that 64% of the peaks have a FOXO1 consensus binding site. These results suggest that FOXO1 binds chromatin in chondrocytes mainly by direct DNA interaction, while in pre-leukemia cells it is tethered to DNA by protein-protein interactions involving RUNX1.
Currently, it is not fully understood how ubiquitously expressed TFs achieve precise tissue-specific expression of target genes. Here we describe how FOXO1 utilizes three levels of regulation to distinctively drive ubiquitous and chondrocyte-specific genes. More specifically, FOXO1 regulates chondrocyte-specific gene expression mainly by binding to tissue-specific enhancers (Suppl Fig. S5), enriched for suboptimal binding motifs and interacting with a defined set of functional partners. On the other hand, more ubiquitous FOXO1 pathways, like signal transduction and homeostasis, are regulated mainly at the promoter level, through interaction with its canonical binding motif and cooperating with a different set of TFs (Suppl Fig. S5).
The second main finding of this study is the identification and validation of novel FOXO1 target genes and pathways in chondrocytes and OA. Our group has recently described for the first time that several ECM genes are regulated by FOXO TFs in cartilage15. In the present study we reveal that the extent of FOXO1 regulation of ECM is not limited to a hand-full of genes but instead, it is the main tissue-specific function of FOXO1 in cartilage. We show that FOXO1 preferentially binds tissue-specific enhancers and that the most over-represented pathway regulated exclusively at the enhancer level in cartilage is ECM organization (a total of 77 ECM genes are bound by FOXO1 in human cartilage, out of which 53 are bound exclusively at the enhancer level). We also demonstrate in vitro, using a selective FOXO1 inhibitor in human primary chondrocyte cultures, that FOXO1 positively regulates the expression of ACAN, COL2A1 and PRG4, while repressing MMP2. Interestingly, our previous results from FOXO1/3/4 triple KO mouse cartilage also suggest a direct role of FOXO1 in up-regulating the expression of PRG4, although no differences are observed in the expression of ACAN, and FOXO-dependent repression is observed for COL2A115. This partial discrepancy suggests that the context in which the role of FOXO1 in ECM regulation is observed is key. Particularly, since FOXO1 is a repressor or activator depending on context35. The factors determining this is in chondrocytes is not known, nor how it differentially affects FOXO1 targets in mouse and human cartilage. What is increasingly clear from this study and our previous results is that FOXO1 is a key player regulating a wide array of ECM genes in cartilage.
Previous results from our group and others have shown that the circadian clock is disrupted in human OA20,36, and that perturbation of the circadian clock promotes OA pathogenesis24,37. Furthermore, FOXO pathways are disrupted in OA and have been linked to circadian rhythms24,38,39. Our study describes for the first time FOXO1-mediated transcriptional regulation of circadian clock genes. In chondrocytes, our genome-wide occupancy data suggests that FOXO1 has an important role in modulating the circadian clock. The most over-represented pathway among all 2985 chondrocyte genes bound by FOXO1 in Reactome DB is “Circadian Clock” (logP = −16.21), with a total of 30 genes including many core-clock genes (ie: BMAL1, CRY1, RORC, RORA, NR1D1, PER1). Importantly, we also found that the “Circadian Clock” (logP = −6.30) pathway is significantly over-represented among putative FOXO1 target genes that are differentially expressed in OA. Furthermore, FOXO1 itself is regulated by the clock denoting altered expression in cartilage in BMAL1-KO mice24. FOXO1 is a sensor and effector of environmental changes like ROS and metabolic stress, while ROS and metabolic stress have been shown to alter the expression of clock genes40,41. One possible mechanism could involve FOXO1 acting as a sensor of environmental change and feeding this information back to the clock. In this scenario, dysregulation of FOXO1 pathways in OA therefore, could contribute to the observed dysregulation of the circadian clock. In support of this hypothesis, we demonstrate that perturbation of FOXO1 in cultured human primary chondrocytes affects the expression of core-clock genes RORC and NR1D1.
Although FOXO1 occupancy data combined with differential gene expression of putative target genes in FOXO1 perturbation experiments do not unequivocally identify FOXO1 direct target genes, they do convincingly show that ECM and circadian rhythm pathways are modulated by FOXO1. Furthermore, in the present study we performed ChIP on isolated chondrocytes. A potential limitation is that the DNA binding pattern of FOXO1 and the present results may thus not represent a direct reflection of the native tissue state. To minimize this effect, cells in primary culture and not passaged cells were used. Nevertheless, integration with RNA-seq data from healthy and OA human fresh tissue samples mitigate these effects.
Altogether, using a combination of genome-wide approaches with in-vitro perturbation experiments, we identified the main FOXO1-dependent pathways that are dysregulated in OA and have described for the first time the distinct molecular mechanisms used by FOXO1 to regulate gene expression in cartilage. We believe that a better understanding of the diverse roles of FOXO1 in cartilage and the mechanisms by which it protects chondrocytes can accelerate the development of novel therapeutic approaches for OA.
MATERIALS AND METHODS
Tissue Processing and Primary Cell Culture.
Human cartilage was harvested from the tissue removed during knee replacement surgery from 2 female (age 65 and 77) donors and 1 male (age 66) donor. Cartilage was digested with 0.2% type 2 collagenase over-night. Isolated chondrocytes were maintained in T75 flasks in DMEM with 10% calf serum (CS) supplemented with 1X penicillin-streptomycin-glutamine (PSG).
Chondrocyte Treatment with AS1842856.
Seventy percent confluent primary chondrocytes were trypsinized and plated on 12 well plates at 70% confluency for 24 hrs in DMEM, 10% CS, 1X PSG. FOXO1-specific inhibitor21 AS1842856 (Sigma-Aldrich 344355) was added to a final concentration of 1 uM and incubated for 24 hrs. Cells were then synchronized with 100 nM dexamethasone (Sigma-Aldrich D2915) for 1 hr in DMEM 0.5% CS and 1X PSG. Then media was changed to CS-free DMEM, 1X PSG. RNA was collected 16 hrs after synchronization.
FOXO1 ChIP-seq.
Fifteen cm dishes at 90% confluency (~1.5×107 human primary chondrocytes) were cross-linked with 1% formaldehyde for 10 min. The crosslinking reaction was quenched by adding glycine to a final concentration of 125 μM. After two washes with cold PBS, cells were incubated 5 min on ice in 5 mL of Farnham lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40) containing 1x protease inhibitor cocktail (Thermo Scientific PI78425), and nuclei were collected by centrifugation and resuspended in 300 uL of nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 1% SDS, 10 mM EDTA) containing 1x protease inhibitor cocktail. Chromatin was fragmented with 3 rounds of 10 cycles, 30s/30s ON/OFF, using a Bioruptor (Diagenode). After centrifugation at 17,000 rpm for 15 minutes at 4°C, the fragmented chromatin was diluted 1:5 with IP dilution buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA) supplemented with 1x protease inhibitor cocktail, and incubated with 10 μg anti-FOXO1 (ab39670) overnight at 4°C. 100 μl of Dynabeads Protein A (Invitrogen 10002D) was added and incubated for another 3 h. Afterwards, the beads were washed sequentially with the following buffers: low salt and twice with 1X TE buffer. Two hundred μl of direct elution buffer (10mM Tris-HCl pH8, 0.3M NaCl, 5mM EDTA pH8, 0.5%SDS) was applied to the bead pellet along with RNAse A and reverse-crosslinked at 65°C overnight. Then proteinase K was applied and incubated at 55°C for 2 hrs. Finally, DNA was purified with QIAquick PCR Purification columns.
ChIP-seq Library Construction and Processing.
To construct libraries, 10 ng of each sample was prepared using the NEB Ultra DNA Library Prep Kit from Illumina following manufacturer’s instructions. Each library was dual size selected with 0.55X and 0.80X Ampure beads followed by PCR amplification with Kappa HiFi 2x PCR mix (15 cycles). Libraries were then gel purified on a 2% agarose gel to remove secondary PCR amplification artifacts, quantitated on the Qubit and loaded onto a NextSeq 500 (Illumina) for 1 × 75 base single-end sequencing. Approximately 20 million reads were generated for each sample.
Bioinformatic Analyses.
ChIP-seq tags of FOXO1 were aligned against the reference human genome hg19 using Bowtie2 and the peaks were called using PePr16. Mapped reads for histone marks from chondrocytes derived from bone marrow derived mesenchymal stem cells were obtained from GSE17312 and the peaks were called using HOMER42. bedGraph tracks, motif analyses and pathway analyses were obtained or performed using HOMER42. Overlapping peaks were identified using BEDOPS43 and the Chow-Ruskey diagrams were generated using the R package Vennerable.
Supplementary Material
Suppl Table 1. Top 10 over-represented pathways among the 147 putative FOXO1 target genes shared between 4 cell types.
Suppl Table 2. Over-represented pathways in Kegg DB of FOXO1 bound genes differentially expressed in OA.
Suppl Table 3. Pathway analysis of genes bound by FOXO1 exclusively at enhancer regions.
Suppl Table 4. Pathway analysis of genes bound by FOXO1 exclusively at promoter regions.
Suppl Table 5. Pathway analysis of the 147 FOXO1 putative genes shared by all 4 cell types.
Suppl Table 6. Pathway analysis using Kegg DB of the 428 genes bound by FOXO1 and DE in OA.
Suppl Figure S1. Heatmap of Pearson’s correlation between FOXO1 ChIP-seq replicates from human primary chondrocytes.
Suppl Figure S2. de novo motif discovery at FOXO1 peaks without a canonical FOXO1 binding motif. Fifty percent of the peaks without a canonical FOXO1 binding motif have a truncated FOXO1 binding motif (motif 2).
Suppl Figure S3. Distinct motif enrichment at FOXO1 occupied promoters and enhancers.
Suppl Figure S4. FOXO1 regulates distinct transcriptional programs at the promoter and enhancer levels.
A) Chondrocyte-specific COL13A1 is regulated at the enhancer level in the absence of a FOXO1 canonical binding motif.
B) Ubiquitous TXNIP is regulated at the promoter level with a FOXO1 binding motif.
Suppl Figure S5. Schematic for distinct mechanisms by which FOXO1 regulates tissue-specific and ubiquitous target genes.
ACKNOWLEDGEMENTS
This study was supported by NIH grants AG049617 and AG059418 to ML.
Footnotes
Competing Interests
No financial support or other benefits have been obtained from any commercial sources for this study and the authors declare that they have no competing financial interests.
REFERENCES
- 1.Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage 2014;22:162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol 2014;66:3349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum 2010;62:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahn AJ. FOXO3 and related transcription factors in development, aging, and exceptional longevity. J Gerontol A Biol Sci Med Sci 2015;70:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008;20:126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eijkelenboom A, Mokry M, de Wit E, Smits LM, Polderman PE, van Triest MH, et al. Genome-wide analysis of FOXO3 mediated transcription regulation through RNA polymerase II profiling. Mol Syst Biol 2013;9:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillejo Becerra CM, Mattson AM, Molstad DHH, Lorang IM, Westendorf JJ, Bradley EW. DNA methylation and FoxO3a regulate PHLPP1 expression in chondrocytes. J Cell Biochem 2018;119:7470–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 2014;39:159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 2002;295:2450–2. [DOI] [PubMed] [Google Scholar]
- 10.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 2002;419:316–21. [DOI] [PubMed] [Google Scholar]
- 11.Ambrogini E, Almeida M, Martin-Millan M, Paik JH, Depinho RA, Han L, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab 2010;11:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rached MT, Kode A, Xu L, Yoshikawa Y, Paik JH, Depinho RA, et al. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab 2010;11:147–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472–83. [DOI] [PubMed] [Google Scholar]
- 14.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004;117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuzaki T, Alvarez-Garcia O, Mokuda S, Nagira K, Olmer M, Gamini R, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Lin YH, Johnson TD, Rozek LS, Sartor MA. PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics 2014;30:2568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci U S A 2013;110:12349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta 2011;1813:1946–53. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez-Sola D, Kung J, Holmes AB, Wells VA, Mo T, Basso K, et al. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity 2015;43:1064–74. [DOI] [PubMed] [Google Scholar]
- 20.Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, et al. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage 2018;26:1531–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Zheng LD, Zou P, Brooke J, Smith C, Long YC, et al. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle 2016;15:2033–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mort JS, Geng Y, Fisher WD, Roughley PJ. Aggrecan heterogeneity in articular cartilage from patients with osteoarthritis. BMC Musculoskelet Disord 2016;17:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jay GD, Waller KA. The biology of lubricin: near frictionless joint motion. Matrix Biol 2014;39:17–24. [DOI] [PubMed] [Google Scholar]
- 24.Dudek M, Gossan N, Yang N, Im HJ, Ruckshanthi JP, Yoshitane H, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Invest 2016;126:365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, et al. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell 2010;18:592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Garcia O, Matsuzaki T, Olmer M, Masuda K, Lotz MK. Age-related reduction in the expression of FOXO transcription factors and correlations with intervertebral disc degeneration. J Orthop Res 2017;35:2682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 2013;14:83–97. [DOI] [PubMed] [Google Scholar]
- 28.Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell 2016;15:673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sophia Fox AJ, Bedi A, Rodeo SA. The basic science of articular cartilage: structure, composition, and function. Sports Health 2009;1:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016;167:1170–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. Suboptimization of developmental enhancers. Science 2015;350:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 2015;160:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell type-specific gene expression. Nature 2009;459:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 2010;28:1045–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langlet F, Haeusler RA, Linden D, Ericson E, Norris T, Johansson A, et al. Selective Inhibition of FOXO1 Activator/Repressor Balance Modulates Hepatic Glucose Handling. Cell 2017;171:824–35 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akagi R, Akatsu Y, Fisch KM, Alvarez-Garcia O, Teramura T, Muramatsu Y, et al. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-beta signaling in chondrocytes. Osteoarthritis Cartilage 2017;25:943–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krawetz RJ. Resetting the clock on arthritis. Arthritis Res Ther 2019;21:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luciano AK, Santana JM, Velazquez H, Sessa WC. Akt1 Controls the Timing and Amplitude of Vascular Circadian Gene Expression. J Biol Rhythms 2017;32:212–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaves I, van der Horst GT, Schellevis R, Nijman RM, Koerkamp MG, Holstege FC, et al. Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol 2014;24:1248–55. [DOI] [PubMed] [Google Scholar]
- 40.Wilking M, Ndiaye M, Mukhtar H, Ahmad N. Circadian rhythm connections to oxidative stress: implications for human health. Antioxid Redox Signal 2013;19:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinke H, Asher G. Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol 2019;20:227–41. [DOI] [PubMed] [Google Scholar]
- 42.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics 2012;28:1919–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl Table 1. Top 10 over-represented pathways among the 147 putative FOXO1 target genes shared between 4 cell types.
Suppl Table 2. Over-represented pathways in Kegg DB of FOXO1 bound genes differentially expressed in OA.
Suppl Table 3. Pathway analysis of genes bound by FOXO1 exclusively at enhancer regions.
Suppl Table 4. Pathway analysis of genes bound by FOXO1 exclusively at promoter regions.
Suppl Table 5. Pathway analysis of the 147 FOXO1 putative genes shared by all 4 cell types.
Suppl Table 6. Pathway analysis using Kegg DB of the 428 genes bound by FOXO1 and DE in OA.
Suppl Figure S1. Heatmap of Pearson’s correlation between FOXO1 ChIP-seq replicates from human primary chondrocytes.
Suppl Figure S2. de novo motif discovery at FOXO1 peaks without a canonical FOXO1 binding motif. Fifty percent of the peaks without a canonical FOXO1 binding motif have a truncated FOXO1 binding motif (motif 2).
Suppl Figure S3. Distinct motif enrichment at FOXO1 occupied promoters and enhancers.
Suppl Figure S4. FOXO1 regulates distinct transcriptional programs at the promoter and enhancer levels.
A) Chondrocyte-specific COL13A1 is regulated at the enhancer level in the absence of a FOXO1 canonical binding motif.
B) Ubiquitous TXNIP is regulated at the promoter level with a FOXO1 binding motif.
Suppl Figure S5. Schematic for distinct mechanisms by which FOXO1 regulates tissue-specific and ubiquitous target genes.


