Abstract
Sleep apnea is caused by several endophenotypic traits, namely pharyngeal collapsibility, poor muscle compensation, ventilatory instability (high loop gain), and arousability from sleep (low arousal threshold). Measures of these traits have shown promise for predicting outcomes of therapies (e.g. oral appliances, surgery, hypoglossal nerve stimulation, CPAP, and pharmaceuticals), which may become an integral part of precision sleep medicine. Currently, the methods Sands et al. developed for endotyping sleep apnea from polysomnography (PSG) are embedded in the original authors’ code, which is computationally expensive and requires technological expertise to run. We present a reimplementation and validation of the integrity of the original authors’ code by reproducing the endo-Phenotyping Using Polysomnography (PUP) method of Sands et al. The original MATLAB methods were reprogrammed in Python; efficient algorithms were developed to detect breaths, calculate normalized ventilation (moving time-average), and model ventilatory drive (intended ventilation). The new implementation (PUPpy) was validated by comparing the endotypes from PUPpy with the original PUP results. Both endotyping methods were applied to 38 manually scored polysomnographic studies. Results of the new implementation were strongly correlated with the original (p < 10–6 for all): ventilation at eupnea V̇ passive (ICC = 0.97), ventilation at arousal onset V̇ active (ICC = 0.97), loop gain (ICC = 0.96), and arousal threshold (ICC = 0.90). We successfully implemented the original PUP method by Sands et al. providing further evidence of its integrity. Additionally, we created a cloud-based version for scaling up sleep apnea endotyping that can be used more easily by a wider audience of researchers and clinicians.
Keywords: sleep apnea, endotype, phenotype, loop gain, collapsibility, arousal threshold, upper airway anatomy, personalized medicine
Statement of Significance.
It has been assumed that accurate endo-phenotyping of sleep apnea is integral to developing precision medicine in the field of sleep medicine. Despite this, sleep apnea endotyping has not become a part of the sleep clinicians’ toolkit due to the technical implementation challenges it entails. In this article, we present and validate a cloud-based reimplementation of the previously published method for endo-Phenotyping Using Polysomnography (PUP) Sands et al. The new cloud-based implementation confirms the reproducibility the PUP method and could be made available to researchers who are interested in endotyping but do not have the resources or expertise required to use the previously published method. This validation and improved access could allow scientists to further investigate the clinical relevance of sleep apnea endotypes.
Introduction
Sleep apnea is a chronic disorder where breathing is periodically interrupted during sleep. Diagnosis of sleep apnea is made via polysomnography (PSG), where a variety of physiological signals are measured during a night of sleep, including electroencephalogram, peripheral capillary oxygen saturation (SpO2), and ventilation. The sleep study is subsequently analyzed by a technician that labels sleep stages and events according to a standard rubric [1]. By counting the number of both partially and totally obstructed breathing events and dividing it by total sleep time, the apnea–hypopnea index (AHI) is calculated. The AHI can be interpreted as the average number of apneas and hypopneas per hour of sleep and has been traditionally used as the main indicator for sleep apnea severity [2]. A major limitation of current diagnostics techniques is that they do not provide information regarding the underlying cause or impact of sleep apnea in different individuals [3].
Respiratory endotyping is a methodology for identifying the pathophysiological traits of sleep apnea by better utilizing the wealth of data collected in a PSG. The endotypes of sleep apnea are loop gain, upper airway collapsibility, arousal threshold, and upper airway dilator muscle response (compensation) [4]. These parameters can be estimated by examining the characteristics of a patient’s ventilation during obstructed breathing periods [4, 5, 6, 7, 8, 9].
The respiratory endotypes correspond directly to the pathophysiological mechanisms underlying sleep apnea, making it an attractive method for guiding treatment options [6, 9]. For instance, patients with poor pharyngeal muscle responsiveness may respond well to hypoglossal nerve stimulation [10] and/or drugs that increase the upper airway dilator muscle activity [11]. Similarly, patients with an abnormally low arousal threshold may respond better to sedative drugs [12]. Other treatment options that have been shown to target specific endotypes are supplemental oxygen [13], upper airway surgery [14, 15], and oral appliances [16]. Endotyping could further help to guide combination therapy by identifying which traits to target with drugs or devices, for instance, individuals with a collapsible airway and a high loop gain might respond to the combination of oral appliance and acetazolamide [17]. A more complete review of endotype-driven OSA treatment options can be found in a recent article by Edwards et al. [18].
There are two established methods of determining the endotypes of sleep apnea during sleep. The first involves dropping CPAP pressure, creating a controlled apnea or hypopnea, and measuring the ventilation response when CPAP pressure is reestablished [4, 5, 17, 19]. The second method requires measuring the respiratory drive response to obstruction using esophageal manometry [9] or diaphragm electromyogram [8]. These methods have several shortcomings: they are not scalable, cause discomfort to the patient, and use equipment that is not a part of a standard PSG. To mitigate these problems, Sands et al. [6, 7] proposed a method for polysomnographic endotyping (implemented in a tool called “Phenotyping Using Polysomnography” or PUP) where the endotypes can be estimated from a standard PSG. The PUP method relies on an uncalibrated estimate of minute ventilation during sleep, derived from the measured flow signal, and it uses inverse modeling of the respiratory control system to estimate respiratory drive.
In this article, we present a python implementation (PUPpy) of the PUP method. With the reimplementation, we aim to reproduce and validate the PUP method with the goal of building a foundation for cloud-based software making the method accessible for a broader audience. The PUP method is reimplemented in a different programming language, building on its theoretical basis, with improvements with regards to efficiency. The cloud-based platform was chosen for its scalability, making it possible to run these computationally intensive methods at scale. This implementation represents an important step in making endotyping more accessible for both research and the clinic and lays a solid foundation for further developments of polysomnographic endotyping. Moreover, reimplementing the method from first principles and comparing the endotype results between the two versions serves as an independent validation of the integrity of the method itself since implementation errors are not likely to be replicated.
Methods
The control and regulation of ventilation can be described by a feedback system whereby a reduction in minute ventilation (V̇E) raises the pCO2 in the blood and causes a corresponding change in “chemical drive” to breathe (V̇chem). This feedback system can be formulated as a first-order linear model with a transport delay, equation (1) [4, 17, 20] which captures the magnitude (LG0, steady-state loop gain), response time (τ, time constant), and latency (δ, delay) of the chemical drive response to a drop in ventilation. The characteristics of the response depend on the model parameters: steady-state loop gain (LG0), the respiratory time constant (τ), and the feedback delay (δ). Engineers (or researchers using appropriate software) can analyze this model of the ventilatory control system by examining the governing equation for the feedback system, given by:
| (1) |
For example, with known parameters, equation (1) is used to calculate the “loop gain” (magnitude of chemical drive response for any ventilatory disturbance) that has been used to predict responses to therapies [14, 15, 17]. Polysomnographic endotyping employs equation (1) to generate a continuous chemical drive V̇chem signal (output of the ventilatory control system) based on the measured ventilatory fluctuations (V̇E). Parameters are adjusted through least squares regression (below). Once ventilatory drive is estimated, the remaining endotypes can be quantified: upper airway collapsibility, upper airway compensation, and the arousal threshold.
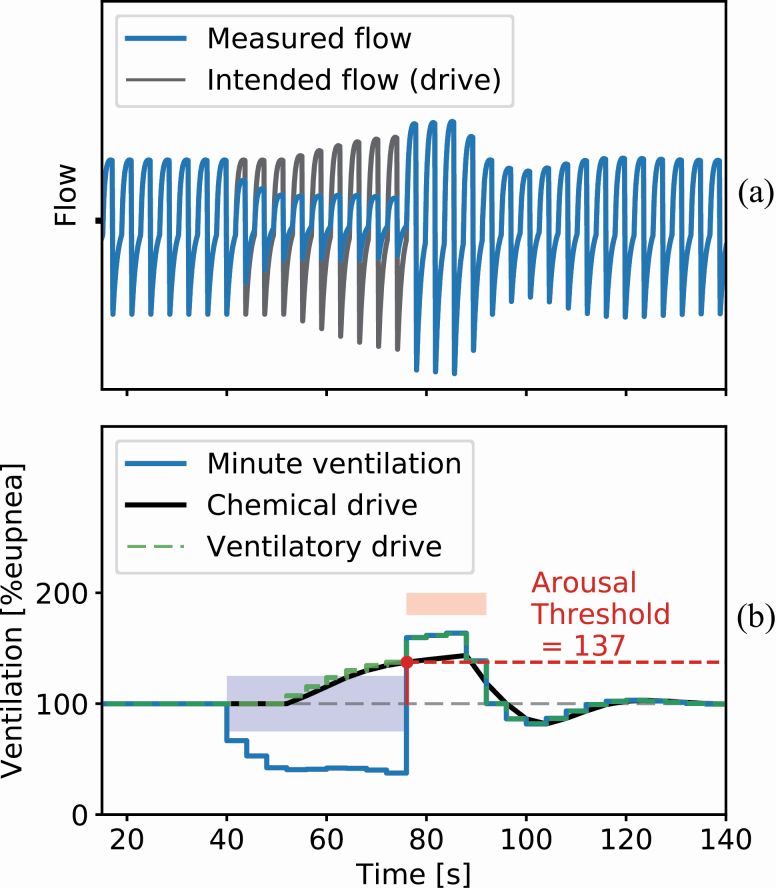
Figure 1 shows a simulation of the chemical drive response (V̇chem) to a loss of ventilation (V̇E) when a spontaneous hypopnea (blue area) interrupts normal (eupneic) breathing. Before the obstruction, the measured ventilation and chemical drive are identical. During the hypopnea, the loss of ventilation yields a gradual compensatory rise in chemical drive. When this drive reaches the arousal threshold, an arousal occurs (red area) and the obstruction is terminated. The open airway reveals the underlying elevation in chemical drive and yields a period of hyperventilation, which eventually converges back to the baseline quiet breathing. During arousal (red area), a nonchemical drive to breath (wakefulness drive, parameter ϒ) also raises the overall ventilatory drive independently of the chemical drive. Chemical drive plus wakefulness drive is referred to as the ventilatory drive. The arousal threshold is defined as the chemical drive at arousal onset, expressed as a percentage of eupnea.
Figure 1.
A simulated hypopnea for illustrating key concepts of the model underlying the endo- PUP method. (A) The blue trace shows the simulated flow during a hypopnea event and the black trace shows a simulation of the flow that the chemical drive would result in if there were no obstruction (intended flow). The hypopnea starts at around 40 s where the airflow is reduced. As the flow decreases the chemical drive increases, attempting to compensate for the reduced ventilation. The hypopnea terminates at the 75-s mark in an arousal, opening the airway. The buildup of intended flow results in large recovery breaths before stabilizing at eupnea. (B) The ventilation during the simulated hypopnea in (A). The hypopnea event, labeled by the blue square, terminates in an arousal, labeled by the red square. The blue trace shows the change in ventilation, calculated from the simulated flow signal in (A). The black and green traces show the chemical and ventilatory drives, respectively. Due to the blood circulatory delay, the drive only starts increasing 12 s after the ventilation is reduced and similarly continues to build up 12 s into the recovery period. The chemical drive is the V̇chem from equation (1) and the ventilatory drive is V̇chem plus an added drive contribution during the arousal (wakefulness drive, ϒ), simulated here as 20% eupnea. The arousal threshold can be read directly from the chemical drive estimate as the drive at arousal onset.
Minute ventilation
The model in equation (1) can be interpreted as an input–output system where the input (V̇E) is the minute ventilation and the output (V̇chem) is the chemical drive which is thought of as the intended minute ventilation and has the same units as V̇E. The minute ventilation is calculated from the measured flow signal on a breath-by-breath basis. The breath detection is carried out by integrating the flow trace, correcting for drift, and detecting the troughs and peaks of the resulting signal. When breaths have been identified, minute ventilation (V̇E) is calculated by integrating the inspired flow for each individual breath, yielding a volume estimate, and dividing by the breath duration.
The PSG data used for the validation of PUPpy include a flow signal recorded with a pneumotachograph equipped with a sealed oronasal mask. The pneumotachograph was calibrated at the start of the sleep study and the signal amplification set to provide a signal with a large amplitude without clipping. Although the pneumotachograph is calibrated at the beginning of the night, the eupneic ventilation can drift over the span of a sleep study (i.e. the data is nonstationary). To account for drift in signal amplitude, the continuous minute ventilation trace is normalized using a moving average window of 7 min. It is assumed that the amount of hyper- and hypoventilation even out over long periods and therefore the average is close to eupnea. This means that ventilation at 100% eupnea can be sustained indefinitely; values below 100% eupnea are interpreted as hypoventilation and above 100% eupnea as hyperventilation. All ventilation and drive estimates are expressed in the same way, as a percentage of eupnea. An additional upside of normalizing the minute ventilation signal is that semiquantitative, uncalibrated flow measurements can also be used.
In a clinical PSG study, changes in ventilation are typically measured using either a nasal cannula or two RIP belts measuring thoracoabdominal breathing movements. Both sensors have associated complications; the nasal cannula is prone to dislocations and confounded by oral ventilation while the RIP needs to be calibrated and is sensitive to movement artifacts. Previous research has proposed scaling the nasal cannula by an exponent; this transformation improves the correlation between endotypes derived from a sealed oronasal mask and nasal cannula [6, 7]. To the best of our knowledge, no research has yet been done on endotyping with RIP using different calibration techniques.
Inverse modeling
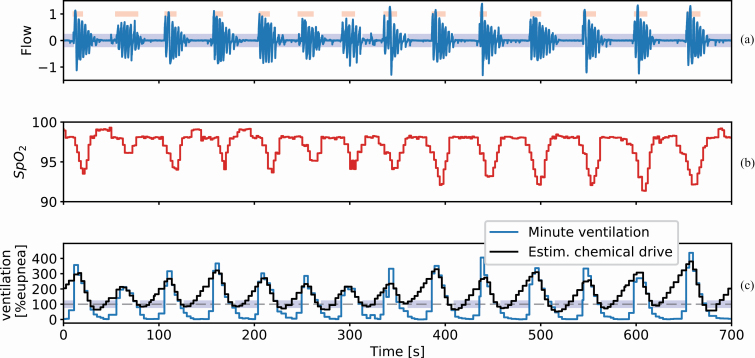
The model in equation (1) is algorithmically tuned such that it reflects the ventilatory patterns of the minute ventilation data for each subject. The implementation details of the model fitting process are detailed in the supplementary materials. The model is fit for an interval of 7 min at a time yielding both the model endotypes as well as the chemical drive estimate. The final product of the model fitting procedure for a single window of PSG data can be seen in Figure 2. Figure 2, A shows the measured flow for a period of repeated apneas (blue areas) and recovery breaths during cortical arousal (red areas). Figure 2, B shows the corresponding blood oxygen saturation. Figure 2, C shows the normalized minute ventilation and the estimated chemical drive, expressed as a percentage of eupnea.
Figure 2.
Example of the model fitting on PSG data during an episode of obstructed breathing. (A) The measured flow signal (blue trace) with scored obstructive apneas (blue areas) and arousals (red areas). Flow has been normalized to the [−1, 1] range where inspiration takes a positive value. (B) The oxygen saturation corresponding to the flow. (C) The normalized minute ventilation (V̇E, blue trace) is derived from the measured flow. Chemical drive (V̇chem, black trace) was estimated using the identified feedback model in equation (1). The chemical drive increases during the obstructed periods resulting in large recovery breaths when the obstruction is terminated.
Due to the physiological changes that occur during REM, it is reasonable to assume that endotypes are different between REM and NREM sleep. Loop gain has been shown to decrease during REM [21]. Similarly, upper airway muscle compensation is diminished during REM due to reduced muscle tone. Furthermore, unlike NREM sleep, during REM sleep the ventilation is not under the sole regulation of the chemical control system so it is not clear how well the model in equation (1) applies. To minimize the confounding effects of REM sleep, epochs scored as REM are traditionally omitted from the endotype analysis. More research is needed to explore the difference between REM and NREM sleep apnea endotypes.
Although the goal was to replicate PUP, some architectural decisions were made to make the code fast, testable, and maintainable. In addition, changes were made to the data processing and parameter fitting procedure. Firstly, a sliding window was used for eupnea normalization to account for slow changes in signal amplitude (gradual changes in average ventilation). Secondly, in the original article, a modified least-squares regression algorithm fit a third-order polynomial to the error and subtracted this prior to the mean squared error being calculated. This effectively high-pass filters the error, accounting for drift and nonzero-mean noise. To improve performance and avoid edge effects (outlier extremes at the start and end of the window), we opted to subtract a double-exponential moving average from the error instead of the polynomial, see implementation details in supplementary materials.
Deriving endotypes from model simulated drive
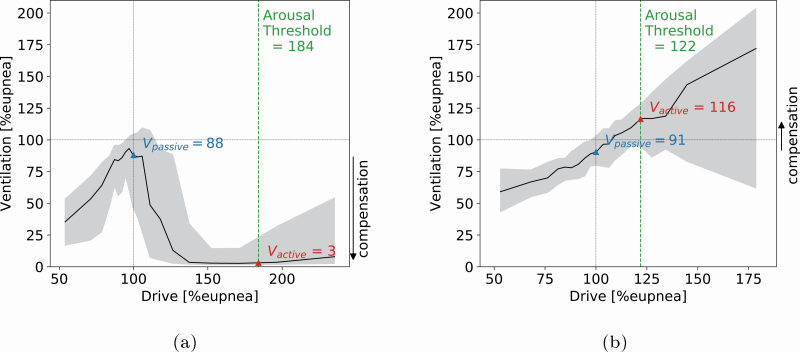
From the model fitting step, we directly get the loop gain endotype from the best-fit parameters, equation (1). The remaining endotypes are all based on chemical drive which can be estimated using the identified model. To derive the ventilatory endotypes of compensation and collapsibility, a ventilatory versus ventilatory drive (“endogram”) plot is generated (see Figure 3, A and B). Chemical drive is binned into quantile bins and the median ventilation is plotted for each bin, black line, and the interquartile range, gray area indicating the variance of the prediction. Figure 3, A and B shows the results from two subjects with different compensation endotype. From the figures, we can read the collapsibility (V̇passive) as the ventilation at 100% eupnic drive and V̇active as the ventilation at the arousal threshold which is defined as the chemical drive at arousal onset as shown in Figure 1 [8]. The upper airway compensation V̇comp is calculated using the following equation [6]:
Figure 3.
Drive-ventilation graph with derived endotypes. The black solid line shows the median ventilation as a function of a chemical drive for a single sleep study. The grey areas indicate the interquartile range of ventilation as a function of drive. V̇passive is the ventilation at eupnea drive and V̇active is the ventilation at the arousal threshold. This representation of drive and ventilation helps illustrate negative effort dependence (NED) as well as the endotype of upper airway compensation V̇comp,= V̇active − V̇passive, labeled by an arrow to the right of the figure. (A) The figure shows a patient with effort dependent reduction of airflow with low collapsibility (V̇passive is close to eupnea) and low compensation (V̇active< V̇passive). (B) The figure shows a patient with a similar collapsibility as the patient in (A) (V̇passive) but better compensation (since V̇active> V̇passive). This is a sign of a good functional response of airway muscles since the airway remains open as drive increases.
| (2) |
Loop gain
Loop gain is the magnitude component of the system in equation (1) and indicates the strength of the response to ventilatory disturbance (i.e. how high and fast the ventilatory drive increases following obstruction). The loop gain derived in the CPAP-drop method is the steady-state loop gain (LG0), For reasons detailed in the supplementary materials, LG0 cannot be accurately derived from naturally occurring apneas. Therefore, LG1 or the loop gain at 1 cycle per min is used as a surrogate.
While elevated LG1 (elevated chemoreflex sensitivity) will yield higher levels of drive for any reduction in ventilation, the chemoreflex delay also determines whether spontaneous periodic breathing (central sleep apnea) might occur. From the perspective of control theory, LGn or the loop gain at the natural frequency of the system (incorporates delay) is the parameter that determines ventilatory instability. Thus, we also examined LGn in the current analysis.
Deployment
The original PUP implementation was written in MATLAB as prototype software and has been made available online by the original authors [7]. The implementation documented in this article was written entirely in Python 3.7. Python was selected as a programming language due to its mature data science stack and strong cross-platform support. The system was deployed to a fully managed cloud architecture for high scalability and fault-tolerance with minimal operation burden and will be made available to researchers and clinicians as a closed source cloud service. The goal of the cloud implementation is to make the PUP method accessible to a larger audience of scientists to facilitate the clinical validation and acceptance of the method.
Cohort
The comparison between methods is done using retrospective data collected by Brigham and Women’s Hospital. The dataset contained 38 measurements, 23 males and 15 females (Table 1). A total of 16 of these had simultaneous measurements of nasal pressure and oronasal mask with a pneumotachograph. The data collection including polysomnographic setup and scoring criteria has been described elsewhere [6].
Table 1.
Summary statistics of the patient cohort
| Age (years) | BMI (kg/m2) | AHI (events/h) | |
|---|---|---|---|
| Mean | 55.18 | 33.74 | 31.08 |
| Std | 11.26 | 7.66 | 28.34 |
| Min | 22.25 | 21.83 | 5.28 |
| Max | 70.05 | 53.51 | 117.82 |
Statistics
The agreement between the PUP and PUPpy methods was analyzed using intraclass correlation (ICC), Pearson correlation coefficient (PCC), and Bland–Altman (BA) mean error and agreement. All statistical analysis was carried out using Python 3.7, using the SciPy 1.2.3 and NumPy 1.17.5 libraries. ICC was calculated based on two-way mixed effects, single rater, and absolute agreement (ICC(2.1)) [22]. Confidence intervals for ICC and PCC were calculated via bootstrapping where studies were randomly sampled with replacement over 10,000 iterations and the ICC and PCC coefficients calculated.
Results
We identify the endotypes in a dataset of 38 patients using both the MATLAB (PUP) and the new Python (PUPpy) implementation. This validates the entire pipeline, from data processing, breath detection, and minute ventilation calculations to model fitting and finally parameter derivation.
The validation was threefold: (1) Comparing the two implementations using oronasal pneumotachograph NREM sleep only. Using a gold standard measurement of ventilation and omitting REM, sleep minimizes the confounding effects of sensor noise and model uncertainty. (2) The two implementations compared include all sleep stages; the clinical relevance of lumping together REM and NREM endotypes is not clear but is done here in order to verify that the reimplementation is valid for all sleep stages. (3) To test the feasibility for usage in a clinical setting, endotypes derived from nasal cannula using PUPpy were compared to endotypes derived from oronasal pneumotachograph using PUPpy.
NREM sleep using oronasal mask
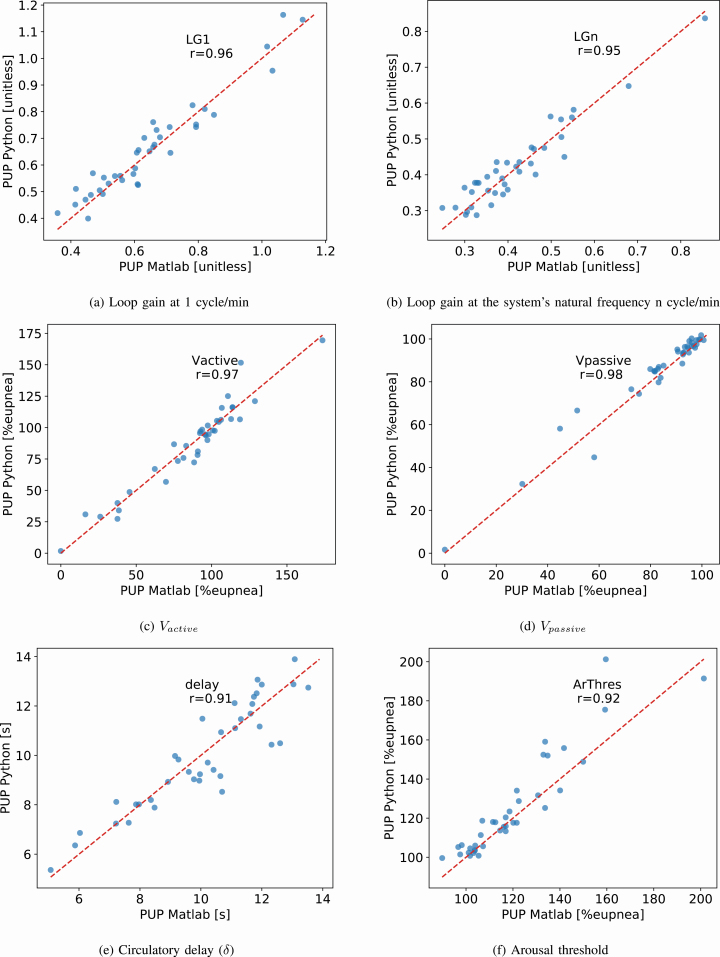
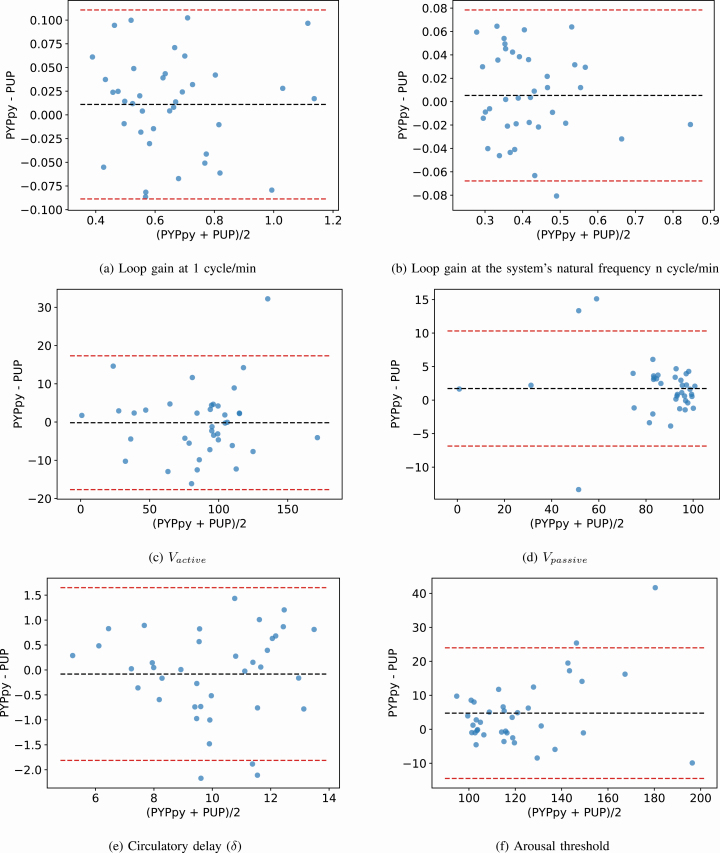
A list of summary metrics for the comparison of each endotype can be found in Table 2. All parameters derived using the PUPpy method correlate strongly with the PUP method (p < 10–6 for all) with ICC ≥ 0.90 (Pearson’s r > 0.90) and are unbiased compared to the original PUP method for each endotype. Scatterplots are shown in Figure 4 and the corresponding BA plots in Figure 5. In each subfigure of Figure 4, the PCC of the respective parameter is shown. Figure 4 shows the range in the endotype values as calculated by the PUP and PUPpy methods. The figures show that the endotype values cover broad ranges and the high correlations are not caused by single or few outliers or leverage points. Furthermore, the figures show that the endotype values sit around the 1:1 line plotted in a red dash line. Figure 5 shows the BA plots for the endotypes. The figures illustrate the bias and agreement intervals. A summary of the endotype results for the cohort for each implementation is presented in Tables 3 and 4.
Table 2.
Summary of results using NREM sleep only and an oronasal mask for flow
| ICC | PCC | Mean error | 95% agr. interv. | |
|---|---|---|---|---|
| LG 1 | 0.96 (0.92, 0.98) | 0.96 (0.93, 0.98) | 0.01 (−0.01, 0.03) | ±0.10 (0.08, 0.12) |
| LG n | 0.95 (0.86, 0.97) | 0.95 (0.86, 0.98) | 0.01 (−0.01, 0.02) | ±0.07 (0.06, 0.08) |
| Delay | 0.91 (0.83, 0.95) | 0.91 (0.84, 0.96) | −0.08 (−0.37, 0.19) | ±1.73 (1.31, 2.06) |
| Ar. Thresh. | 0.90 (0.82, 0.95) | 0.92 (0.90, 0.96) | 4.76 (1.91, 8.05) | ±19.23 (12.25, 25.74) |
| V passive | 0.97 (0.92, 0.99) | 0.98 (0.94 0.99) | 1.73 (0.35, 3.13) | ±8.59 (4.49, 11.77) |
| V active | 0.97 (0.92, 0.99) | 0.97 (0.93, 0.99) | −0.17 (−2.92, 2.77) | ±17.49 (11.65, 23.20) |
PCC, ICC, BA mean error, and agreement intervals (±1.96 SD). 95% confidence intervals in parenthesis. Confidence intervals are calculated using a bootstrap method.
Figure 4.
Scatter plots of the relevant endotype parameters identified using the two endo-PUP implementations. (A) loop gain at 1 cycle/min, (B) loop gain at the system’s natural frequency n cycle/min, (C) V̇active, (D) V̇passive, (E) circulatory delay (δ), (F) arousal threshold.
Figure 5.
BA plots of the relevant endotype parameters identified using the two endo-PUP implementations. The dotted centerline is the mean of the difference (PUPpy − PUP) and the red dotted lines signify the ±1.96 SD agreement interval. (A) loop gain at 1 cycle/min, (B) loop gain at the system’s natural frequency n cycle/min, (C) V̇active, (D) V̇passive, (E) circulatory delay (δ), (F) arousal threshold.
Table 3.
Summary of the endotype results for the cohort using the original PUP implementation, NREM sleep only, and an oronasal mask for flow
| LG1 | LGn | Delay (s) | Ar. Thresh. (% eupnea) | Vpassive (% eupnea) | Vactive (% eupnea) | |
|---|---|---|---|---|---|---|
| Mean | 0.65 | 0.42 | 10.05 | 119.86 | 83.58 | 86.86 |
| Std | 0.19 | 0.12 | 2.12 | 21.96 | 21.14 | 34.01 |
| Median | 0.61 | 0.39 | 10.32 | 116.50 | 92.49 | 94.76 |
| Min | 0.36 | 0.25 | 5.07 | 89.85 | 0.00 | 0.00 |
| Max | 1.13 | 0.86 | 13.52 | 201.33 | 100.68 | 173.55 |
Table 4.
Summary of the endotype results for the cohort using the reimplemented PUPpy, NREM sleep only, and an oronasal mask for flow
| LG1 | LGn | Delay (s) | Ar. Thresh. (% eupnea) | Vpassive (% eupnea) | Vactive (% eupnea) | |
|---|---|---|---|---|---|---|
| Mean | 0.66 | 0.42 | 9.97 | 124.62 | 85.31 | 86.69 |
| Std | 0.18 | 0.11 | 2.12 | 20.65 | 20.65 | 34.71 |
| Median | 0.65 | 0.40 | 9.77 | 117.64 | 93.54 | 94.60 |
| Min | 0.40 | 0.29 | 5.36 | 99.60 | 1.65 | 1.73 |
| Max | 1.16 | 0.84 | 13.89 | 201.24 | 101.76 | 169.47 |
All sleep stages, oronasal mask
To validate the integrity of the reimplementation for all sleep stages (including REM sleep), we calculated the endotype values using the PUP and PUPpy methods for all sleep periods. Scatter plots and BA plots comparing the endotype values from the PUP and PUPpy methods can be found in the Supplementary Figures S2 and S3. Table 5 shows the summary statistics comparing the two methods. The table shows that the correlation is still very high for all the derived parameters, ICC > 0.90 (Pearson’s r > 0.90). The table shows that including REM sleep does not change the agreement between the two implementations meaningfully compared with NREM only.
Table 5.
Summary of results using all sleep stages and an oronasal mask for flow
| ICC | PCC | Mean error | 95% agr. interv. | |
|---|---|---|---|---|
| LG1 | 0.96 (0.93, 0.98) | 0.96 (0.93, 0.98) | 0.00 (−0.01, 0.02) | ±0.10 (0.08, 0.11) |
| LGn | 0.96 (0.93, 0.98) | 0.94 (0.85, 0.98) | 0.00 (−0.01, 0.02) | ±0.07 (0.06, 0.08) |
| Delay | 0.93 (0.85, 0.97) | 0.93 (0.86 0.97) | −0.08 (−0.35, 0.19) | ±1.65 (1.20, 2.04) |
| Ar. Thresh. | 0.92 (0.86, 0.96) | 0.94 (0.91, 0.97) | 3.87 (1.26, 6.87) | ±17.35 (11.09, 23.14) |
| V passive | 0.98 (0.91, 0.99) | 0.98 (0.93, 0.99) | 1.60 (0.19, 2.98) | ±8.52 (4.88 11.53) |
| V active | 0.96 (0.91, 0.98) | 0.95 (0.91, 0.98) | 0.83 (−2.47, 4.17) | ±20.39 (13.70, 26.04) |
PCC, ICC, BA mean error, and agreement intervals (±1.96 SD). 95% confidence intervals in parenthesis. Confidence intervals are calculated using a bootstrap method.
NREM sleep nasal cannula
In clinical PSGs, nasal cannulas are used rather than oronasal mask and pneumotach. Here we compare the endotypes derived from the flow signals measured by the oronasal pneumotach and nasal cannula using the PUPpy method. In the dataset, 16 sleep studies included a simultaneous measurement of breathing using the oronasal pneumotach and nasal cannula. The nasal cannula signal was transformed using a scaling exponent of 0.67 as suggested by Sands et al. [6] and Mann et al. [23] The comparison was made in the same fashion as reported previously and the results are reported in Table 6 in a manner that is directly comparable to the same comparison done by Sands et al. [6]. The results show that the Pearson correlation between the pneumotach and cannula endotypes using the PUPpy method is high, r > 0.9. Furthermore, the mean error (i.e. bias) and mean absolute error are small or comparable to what was reported by Sands et al. [6].
Table 6.
Summary of results when comparing the oronasal mask endotypes to the nasal cannula using the PUPpy implementation
| Mean error (±SD) (% eupnea) | Mean absolute error (% eupnea) | PCC (95% CI) | |
|---|---|---|---|
| V passive | −0.81 ± 4.98 | 3.72 | 0.93 (0.7, 0.98) |
| V active | −0.24 ± 11.07 | 7.51 | 0.92 (0.73, 0.98) |
| V comp | 0.58 ± 10.65 | 8.59 | 0.94 (0.42, 0.99) |
Error is calculated for each of the three ventilatory endotypes as the endotype value using nasal cannula minus the endotype value when using oronasal pneumotachograph. PCC is reported with 95% confidence interval, calculated using a bootstrap method.
Discussion
Sleep apnea endotyping is a promising method of guiding OSA treatment but its exploration is still in the early stages. Making sleep apnea endotyping widely available to a broader range of researchers is the next major step for determining the clinical importance of the endotypes. Currently, due to the lack of large general population studies, there are no definitions of what constitutes clinically high or low endotype values, how the endotypes interact, and how to tailor treatment for a patient presenting with a combination of endotypic traits to alleviate the cause of their sleep apnea. Intra-subject endotype variability should also be explored, both for a single night and multiple nights.
It should be noted that even with excellent aggregate results between the PUP and PUPpy implementations (r > 0.9), there are individuals who deviate from the 1:1 line. This may be due to an individual demonstrating variability in endotype over a single night study, which would result in the endotype estimate being sensitive to slight algorithmic changes. Being able to provide a certainty estimate (i.e. a confidence interval) for each endotype may help in such cases. Confidence interval calculations could help detect misclassified endotypes due to sensor faults, such as oronasal mask leak, poor signal quality (including filter distortions, clipping, and sensor dislocation), incorrect RIP belt placement, or oral ventilation when flow is measured by a nasal cannula. It is, however, not clear how best to calculate these confidence intervals (simple measures of dispersion/variance versus state/position dependence) making it an interesting avenue for future research.
Since the model fitting relies on manual respiratory event scoring to mask out periods of ventilatory disturbance, it can be assumed that accurate scoring of apneas and hypopneas is important. The same is true for arousals and sleep stages as these annotations are also used as inputs to the fitting routine. This observation merits further investigation and scoring guidelines for endotyping should be subsequently created. Some new evidence indicates that scoring based on airflow that relies less on desaturation may be preferable [24].
When implementing complicated algorithms, there is always a risk of systematic biases and errors. This can be especially difficult to handle in medical signal analysis software, since biological signals have inherent variations that may either be interpreted as a physiological characteristic or a calculation error. In the case of complex algorithms, such as the one described in this article, these errors can be very subtle and hard to detect. An effective method to reveal such biases and errors is to reimplement the algorithm from a conceptual level and replicate previous experimental results. This is especially true when the reimplementation is done using a different programming language and software libraries, since the odds of repeating the errors or biases in the same way are low.
The reimplementation of the PUP method was done as a cloud-enabled service rather than as locally run software. Cloud-enabled services have many benefits, especially for computationally demanding algorithms where the processing time of a local computer quickly becomes a limiting factor. The scalability of a cloud computing makes it possible to quickly batch-process larger datasets than would be practical running in the local computer environment. In addition, this approach simplifies issues of algorithm version control and deployment to other researchers. We hope that the PUPpy implementation can serve as a platform for scientists who want to further investigate the clinical importance of the endotypes without having the expertise required to use the PUP implementation.
Conclusions
Respiratory endotyping is a promising method for differentiating physiological etiologies of sleep apnea and potential treatment guidance.
In this article, we have successfully reimplemented the endo-PUP method by Sands et al. [1] in a different software environment, validating the integrity of the original method. The outcome of the new implementation, PUPpy, was compared with the outcome of the original PUP and showed no systematic biases or errors. The primary validation was performed using flow measured by oronasal pneumotachography. Although this is not the standard method of measuring flow during a sleep study, the oronasal pneumotachography allowed us to validate the performance of the reimplementation without the influence of external factors caused by less reliable sensors. A thorough validation of the PUP and PUPpy implementations using breathing sensors such as nasal cannulas and RIP belts deserves its own publication is beyond the scope of the current work. Further investigation is needed to investigate how best to apply these methods on data from a standard sleep study with the presence of oral and nasal breathing, sensor movement, and other sources of signal disturbances found in routine sleep studies.
PUPpy was implemented in Python which allows for the deployment of respiratory endotyping in a scalable cloud environment. The reimplementation and its validation serve as a first step in developing a cloud service to provide access to the PUP method. As a result, sleep apnea endotyping can be offered to researchers who may not have the resources or expertise required to run the previously published MATLAB PUP method. This improved access will allow scientists to further investigate the clinical relevance of the endotypes.
Supplementary Material
Funding
This work was supported by the Icelandic Technology Development Fund (153523–613), the European Union’s Horizon 2020 SME Instrument (733461), the National Institutes of Health (R01-102321), and the American Heart Association (15SDG25890059).
Disclosure statement
Financial disclosure: The study was approved by the Brigham and Women’s Hospital Institutional Review Board, and all participants provided informed, written consent before participation in the study. Scott A. Sands and Andrew Wellman are consultants to Nox Medical. Dr. Sands also serves as a consultant for Merck and Apnimed, and has project grant support from Apnimed, Prosomnus, and Dynaflex. Andrew Wellman works as a consultant for Apnimed, Nox, Inspire, and Somnifix. He has received grants from Sanofi and Somnifix. He also has a financial interest in Apnimed Corp., a company developing pharmacologic therapies for sleep apnea. Dr. Wellman’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Non-financial disclosure: None.
References
- 1. Berry RB, et al. . The AASM Manual for the Scoring of Sleep and Associated Events; Rules, Terminology and Technical Specifications, version 2.5. United States, IL: American Academy of Sleep Medicine; 2018. [Google Scholar]
- 2. Rapoport DM. POINT: is the apnea-hypopnea index the best way to quantify the severity of sleep-disordered breathing? Yes. Chest. 2016;149(1):14–16. [DOI] [PubMed] [Google Scholar]
- 3. Punjabi NM. COUNTERPOINT: is the apnea-hypopnea index the best way to quantify the severity of sleep-disordered breathing? No. Chest. 2016;149(1):16–19. [DOI] [PubMed] [Google Scholar]
- 4. Wellman A, et al. . A method for measuring and modeling the physiological traits causing obstructive sleep apnea. J Appl Physiol (1985). 2011;110(6):1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wellman A, et al. . A simplified method for determining phenotypic traits in patients with obstructive sleep apnea. J Appl Physiol (1985). 2013;114(7):911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sands SA, et al. . Phenotyping pharyngeal pathophysiology using polysomnography in patients with obstructive sleep apnea. Am J Respir Crit Care Med. 2018;197(9):1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terrill PI, et al. . Quantifying the ventilatory control contribution to sleep apnoea using polysomnography. Eur Respir J. 2015;45(2):408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sands SA, et al. . Quantifying the arousal threshold using polysomnography in obstructive sleep apnea. Sleep. 2017;41(1). doi: 10.1093/sleep/zsx183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eckert DJ, et al. . Defining phenotypic causes of obstructive sleep apnea. Identification of novel therapeutic targets. Am J Respir Crit Care Med. 2013;188(8):996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eastwood PR, et al. . Treating obstructive sleep apnea with hypoglossal nerve stimulation. Sleep. 2011;34(11):1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taranto-Montemurro L, et al. . The combination of atomoxetine and oxybutynin greatly reduces obstructive sleep apnea severity. a randomized, placebo-controlled, double-blind crossover trial. Am J Respir Crit Care Med. 2019;199(10):1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eckert DJ, et al. . Eszopiclone increases the respiratory arousal threshold and lowers the apnoea/hypopnoea index in obstructive sleep apnoea patients with a low arousal threshold. Clin Sci (Lond). 2011;120(12):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sands SA, et al. . Identifying obstructive sleep apnoea patients responsive to supplemental oxygen therapy. Eur Respir J. 2018;52:1800674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Joosten SA, et al. . Loop gain predicts the response to upper airway surgery in patients with obstructive sleep apnea. Sleep. 2017;40(7). doi: 10.1093/sleep/zsx094 [DOI] [PubMed] [Google Scholar]
- 15. Li Y, et al. . The effect of upper airway surgery on loop gain in obstructive sleep apnea. J Clin Sleep Med. 2019;15(6):907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bamagoos AA, et al. . Polysomnographic endotyping to select patients with obstructive sleep apnea for oral appliances. Ann Am Thorac Soc. 2019;16(11):1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edwards BA, et al. . Acetazolamide improves loop gain but not the other physiological traits causing obstructive sleep apnoea. J Physiol. 2012;590(5):1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edwards BA, et al. . More than the sum of the respiratory events: personalized medicine approaches for obstructive sleep apnea. Am J Respir Crit Care Med. 2019;200(6):691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Messineo L, et al. . Phenotyping-based treatment improves obstructive sleep apnea symptoms and severity: a pilot study. Sleep Breath. 2017;21(4):861–868. [DOI] [PubMed] [Google Scholar]
- 20. Khoo MCK. Physiological Control Systems: Analysis, Simulation and Estimation. United States, NJ: Wiley-IEEE Press; 2000. ISBN: 0-7803-3408-6. [Google Scholar]
- 21. Messineo L, et al. . Loop gain in REM versus non-REM sleep using CPAP manipulation: a pilot study. Respirology. 2019;24(8):805–808. [DOI] [PubMed] [Google Scholar]
- 22. Koo TK, et al. . A Guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15(2):155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mann DL, et al. . Quantifying the magnitude of pharyngeal obstruction during sleep using airflow shape. Eur Respir J. 2019;54(1):1802262. doi: 10.1183/13993003.02262-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landry SA, et al. . Effect of hypopnea scoring criteria on noninvasive assessment of loop gain and surgical outcome prediction. Ann Am Thorac Soc. 2020;17(4):484–491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.