Abstract
The heterotrimeric eukaryotic Replication protein A (RPA) is a master regulator of numerous DNA metabolic processes. For a long time it has been viewed as an inert protector of ssDNA and a platform for assembly of various genome maintenance and signaling machines. Later, modular organization of the RPA DNA binding domains suggested a possibility for a dynamic interaction with ssDNA. This modular organization has inspired several models for the RPA-ssDNA interaction that aimed to explain how RPA, the high affinity ssDNA binding protein, is replaced by the downstream players in DNA replication, recombination and repair that bind ssDNA with much lower affinity. Recent studies, and in particular single-molecule observations of RPA-ssDNA interactions, led to the development of a new model for the ssDNA handoff from RPA to a specific downstream factor where not only stability and structural rearrangements, but also RPA conformational dynamics guide the ssDNA handoff. Here we will review the current knowledge of the RPA structure, its dynamic interaction with ssDNA, and how RPA conformational dynamics may be influenced by posttranslational modification and proteins that interact with RPA, as well as how RPA dynamics may be harnessed in cellular decision making.
Introduction
Replication protein A, RPA (sometimes also referred to as Replication Factor A, RF-A) is the major single-strand DNA (ssDNA) binding protein in eukaryotes (1–4). It was initially identified among a set of human proteins required for the initiation of simian virus 40 DNA replication in HeLa cell extracts (1). Subsequent identification of the S. cerevisiae RPA established a universal dependence of eukaryotic DNA replication on RPA protein (4). While RPA was first found to be required for replication, in both the steps of initiation and elongation (1,4,5), it has since been identified as an important factor in homologous recombination, nucleotide excision repair, and mismatch repair, among other repair processes (6–14). Thus, for over two decades it has been contemplated that R in RPA may stand not only for “Replication”, but rather for all 3Rs of genome maintenance, Replication, Recombination and Repair (2). RPA binds ssDNA with subnanomolar affinity and is highly abundant (~2 μM) in the cell (15–17). Due to RPA abundance and its high affinity, any exposed cellular ssDNA is rapidly bound by RPA. Not surprisingly, RPA depletion, haploinsufficiency or exhaustion can lead to DNA replication catastrophe, DNA repair defects, and genome instability (18–21). In addition to 3Rs of genome maintenance, the importance of RPA to RNA transcription can be illustrated by the interaction between RPA and histone chaperone HIRA, through which HIRA is recruited to promoters and enhances and regulates deposition of newly synthesized histone H3.3 (22). For some time, the capacity of RPA to quickly and efficiently coat ssDNA lead to the expectation that the function of RPA was simply to protect ssDNA from nucleolytic degradation and from unscheduled binding by downstream players in various DNA metabolic processes. Another property of RPA, an ability to destabilize duplex DNA and non-canonical ssDNA structures, such as G-quadruplexes (23–27), has been acknowledged as useful in preventing the formation of secondary structure in ssDNA. Indeed, the combination of tight ssDNA binding and duplex destabilization by RPA are at the core of its function in homologous recombination, where RPA plays both pre- and post-synaptic roles (28). Because RPA rapidly and tightly binds to exposed ssDNA and ssDNA is found in most DNA metabolic processes, the RPA-ssDNA complex is relevant in these processes. The high affinity of the RPA-ssDNA interaction, however, posits a question of how the ssDNA is handed off to the downstream players in various DNA metabolic and signaling pathways that start with the RPA-ssDNA complex. Another important question is how the “correct” downstream player is chosen to hand off the ssDNA.
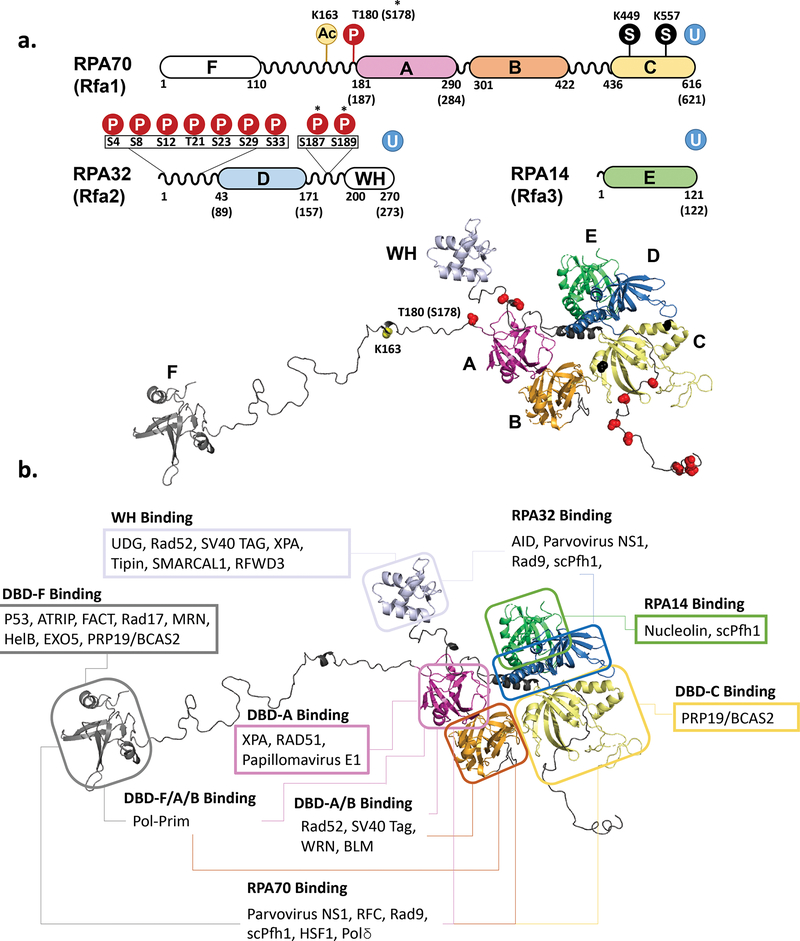
In most eukaryotic organisms, RPA is a heterotrimer made up of the subunits RPA70, RPA32, and RPA14 named for their respective sizes of ~70, 32, and 14 kDa or referred to as RPA1, RPA2, and RPA3 (and sometimes RFA1, RFA2, and, RFA3 in yeast), respectively (2) (Figure 1a&b). RPA is highly conserved across eukaryotic species. Yeast RPAs show ~45% sequence similarity in each of the subunits to their human counterparts, with greater conservation seen in DNA binding regions; the most conserved subunit is RPA70, which shows 31% identity and 44% similarity between S. cerevisiae and human proteins and 37% identity and 64% similarity between S. pombe and human proteins (2). While all bind ssDNA with high affinity, S. cerevisiae RPA exhibits higher cooperativity in comparison with human counterpart (16,29–31). In addition to the canonical RPA made up of these three subunits, an alternative form of RPA can be formed in human and other placental mammals with RPA2 replaced by a homologous subunit, RPA4. Human RPA2 and RPA4 show 47% identity on amino acid level (2). This alternative RPA appears to play a role in DNA repair, exhibiting higher affinity for damaged DNA (32). Many plants have copies of alternative RPA subunits resulting in multiple RPA heterotrimers with distinct specializations (33). Canonical and alternative eukaryotic RPAs contain 6 oligonucleotide binding (OB) folds, named DNA-binding domains (DBD) A-F. RPA70 contains DBDs A, B, C and F, RPA32 contains DBD-D, and RPA14 contains DBD-E (Figure 1a). The three subunits come together at the trimerization core, composed of DBDs C, D, and E (Figure 1b). Linking the connected DBDs together are flexible linker regions. Due to these flexible linkers between the DBDs, the RPA structure is dynamic. Structural rearrangements in the RPA architecture were first inferred from the modular organization of the DBDs and from accessibility of the RPA32 Thr-98 buried within the trimerization core to phosphorylation. This flexibility was later confirmed by NMR studies (34,35). The flexible structure of RPA and the recently characterized dynamic interaction with ssDNA have been suggested to play critical roles in ssDNA handoff to downstream proteins in replication, recombination, and repair (36–41).
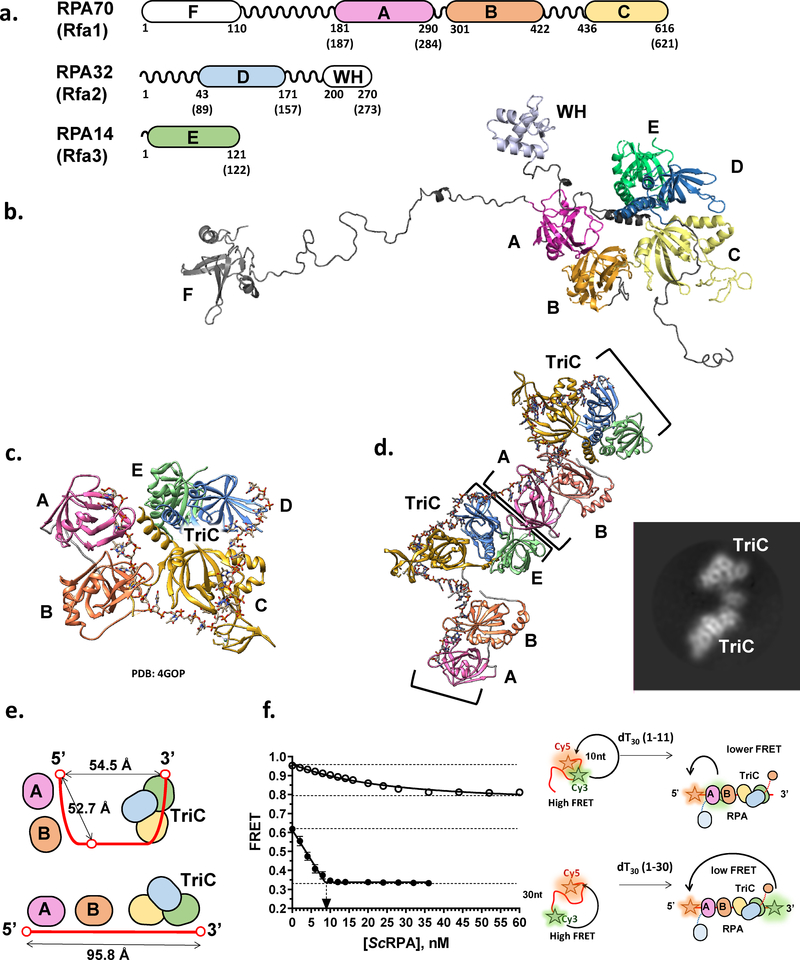
Figure 1. Structure of eukaryotic RPA-ssDNA complex.
(a) Primary structures of the three RPA subunits. The regions that make up DBDs/OB-folds A, B, C, D, E, and F, and the winged helix domain (WH) are shown as colored rectangles. The numbers below the map indicate residue numbers in the human RPA amino acid sequence where the domains start and end; in parentheses are the numbers for S. cerevisiae RPA. Areas between the domains are the flexible linkers. (b) Model of the full-length human RPA from Chen et al (36). Individual domains are shown in the same color scheme as in the primary structure. (c) Structure of the RPA-ssDNA complex from Fan and Pavletich ((98); PBD 4GOP). Ustilago maydis RPA construct consists of the DBDs A-E (depicted in the same color scheme as above); ssDNA is shown in elemental colors. TriC marks a trimerization core comprised DBDs C-D. (d) Cryo-EM reconstruction of the two S. cerevisiae RPA heterotrimers bound adjacently on ssDNA. The structure reveals an interaction between the DBDs A and E of the adjacent heterotrimers. This structure from Yates et al (50) also suggested that RPA extends ssDNA instead of folding it into a u-shaped structure as in the Ustilago maydis RPA crystal structure. Inset shows a representative 2D class average for the 2xRPA-ssDNA complex. (e) Schematic representation of the RPA-ssDNA complex in the u-shape and extended conformations. (f) FRET-based ssDNA binding experiment that suggests that in solution RPA induces ssDNA into a linear conformation (see text for details). Data are from (50).
Because of its central role in virtually all aspects of cellular DNA metabolism, a number of authoritative reviews (3,37,42–46) paralleled the development of the models for the regulation of the RPA-ssDNA interaction and the mechanisms underlying the handoff of ssDNA from RPA to correct ssDNA binding/processing proteins with functions in DNA replication, repair and homologous recombination. Recent single-molecule and structural studies (36,38–41,47,48) allow us to update the existing models. We believe, therefore, that it is time for a new comprehensive review that will incorporate a dynamics component into our understanding of the RPA function and regulation. Specifically, we would like to highlight the emerging importance of the dynamic interaction of the individual RPA DBDs with ssDNA (36).
The flexibility of RPA binding was first described as the binding of RPA in modes (17). The modes were described as conformations in which RPA was bound to 8–10, 18–20, or 28–30 nucleotides, each with different affinities (see below). These were expected to occur sequentially, as RPA became more engaged with the ssDNA. While RPA is sampling a variety of conformations, it is now clear that they are not simply the steps of sequential binding, but states that can be entered and exited readily, with RPA proceeding from less to more engaged states, and vice versa. In this review, we will summarize the current knowledge of the RPA conformational dynamics and will argue that the ability of RPA to sample more and less engaged conformations defines its ability to handoff ssDNA to proteins that function downstream of RPA in DNA metabolic processes.
The dynamics of RPA is important for allowing the handoff of ssDNA to other proteins, as well as a point of regulation. The dynamics of RPA ssDNA binding can be altered by protein-protein interactions, post-translational modifications, and the structure of ssDNA it is binding. Understanding the conformational flexibility of RPA, its dynamic ssDNA binding, and how the RPA conformational dynamics is modulated is revealing a new component in the regulation of DNA replication, recombination, and repair.
Flexible Structure
Canonical eukaryotic RPA is a heterotrimer comprised of RPA70, RPA32, and RPA14 (2) (Figure 1a&b). The majority of the RPA structure is made up of the oligonucleotide binding (OB) folds, referred to in RPA as DNA-binding domains (DBD). RPA70 contains DBD-A, B, C and F, with flexible linker regions connecting each of these DBDs (36,37,49). RPA32 contains DBD-D and a C-terminal winged helix domain. RPA14 contains DBD E. An OB fold from each of the subunits, DBD C, D and E, form the trimerization core, their interaction allowing formation of the stable heterotrimer. The high-resolution structure of the Ustilago maydis RPA-ssDNA complex (Figure 1c) containing most of the structural features with exception of DBD-F of RPA70 and WH domain of RPA32 shows a compact, horseshoe-like structure with ssDNA in a U-shape conformation and DBD-A, B, C and D making sequential contacts from a 5’ to 3’ direction (49). In contrast, the Cryo-EM structure of the three yeast RPA molecules bound sequentially to the 100 nucleotide long ssDNA molecule (50), as well as Förster resonance energy transfer (FRET)-based analyses of yeast and human proteins suggest an extended linear arrangement of the DBDs in RPA (Figure 1d–f) (50,51). In the latter, one can follow the change in the conformation of ssDNA decorated with two fluorescence dyes, the donor (e.g. Cy3) and the acceptor (e.g. Cy5) of FRET. In the presence of 10 mM Mg2+ and at a physiological ionic strength (e.g. 150 mM K+), the ssDNA assumes compact conformation resulting in an intermediate FRET between the Cy3 and Cy5 dyes placed 30 nucleotides apart. Straightening of the ssDNA upon RPA binding into a more linear conformation as observed by CryoEM predicts a gradual decrease in the measured FRET upon addition of RPA until all ssDNA is bound (Figure 1e). In the horseshoe conformation, the RPA binding should have a much smaller effect on FRET, if any. Moreover, the distance between the 5’ end of the RPA occluded ssDNA and its 3’ end are virtually the same as the distance between 5’ end the nucleotide 11th from the 5’ end when measured in the high-resolution Ustilago maydis RPA-ssDNA structure (Figure 1e). While slight discrepancies are possible due to the difference in the Cy3 dye position and associated photophysical effects (52), the horseshoe conformation would predict that FRET values obtained for the two constructs, one labeled at the ends and the other at the 5’ end and the middle should converge to the same value upon RPA binding. Note, that due to the high affinity of the RPA-ssDNA complex, these experiments are performed under stoichiometric binding conditions and therefore reflect on the binding stoichiometry (inflection point in the binding curve) and on the FRET value at saturation. Figure 1f shows a typical binding curve (50) consistent with a linear conformation of the RPA-ssDNA complex. Similar binding patterns were observed for human and yeast RPAs and for the substrates that can accommodate single or multiple RPAs (50,51). A non-stoichiometric binding curve observed for the DNA with internally positioned Cy3 dye is indicative either of a more transient interaction between the DBD-A and ssDNA or the RPA diffusion on the 30 nucleotide long ssDNA. In a recent single-molecule FRET study, Wang and colleagues (48) observed straightening of short (10 and 20 nucleotide) ssDNA overhangs of partial duplex substrates labeled with Cy3 at the end of the overhang and with Cy5 at the ssDNA/dsDNA junction, while intermittent exertions into high FRET values on longer ssDNA overhangs and more complex substrates were interpreted as RPA-induced ssDNA bending. It is important to note here, that these experiments were carried out under conditions different from those described above, specifically, in the absence of Mg2+, which effects RPA-ssDNA interactions, and at much higher RPA concentrations, suggesting a complex dependence of the RPA-ssDNA complex conformation and dynamics on experimental conditions.
A significant degree of flexibility in the RPA heterotrimer has been revealed by NMR and Cryo-EM studies (35,38,50,53). Brosey and colleagues demonstrated using NMR that the DBDs tethered by flexible linkers are not conformationally restrained by each other and are able to interact with ssDNA independently with a high degree of conformational freedom (35). Observation of DNA coated with RPA using AFM and EM also shows that RPA does not take on a single consistent conformation while bound to ssDNA, but that RPA-coated ssDNA maintains a similar level of flexibility (54,55).
OB-fold, a structural unit of ssDNA binding proteins
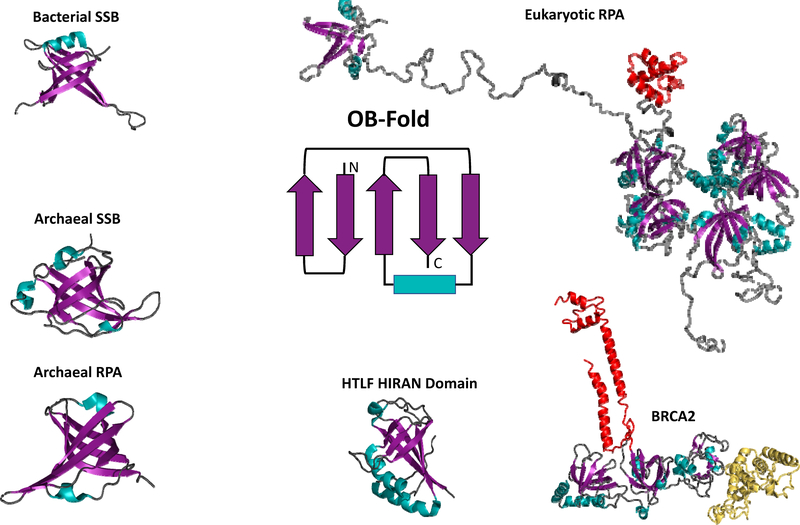
OB-folds (Figure 2) were originally identified as protein motifs that binds to oligonucleotides or oligosaccharides. The highly conserved OB-fold structure is found in many proteins in Archaea, Bacteria, and Eukarya. It is an approximately 120 aa structural motif composed of five beta sheets that form a mixed beta barrel, usually capped by an alpha helix, though there are some variations on this general structure (56,57). OB-fold containing proteins that bind ssDNA have been found across all domains of life, as RPA in eukaryotes and as single strand binding (SSB) proteins in bacteria. Both eukaryotic-like and bacterial-like OB-folds have been found in archaea (58). OB-folds comprising extant RPAs and SSBs, as well as OB-folds in other ssDNA binding proteins have been proposed to evolve from a common ancestral ssDNA binding protein through domain duplication and shuffling (59,60). Across eukaryotes, RPA is conserved as a heterotrimer made up of the 70, 32, and 14 kDa subunits (3). In bacteria, SSBs have been found that exist as homodimers or homotetramers containing four OB folds (61,62). Mitochondrial ssDNA binding proteins in eukaryotes are more akin to bacterial tetrameric SSBs (63). Unlike bacteria and eukaryotes, archaea possess a more diverse set of RPA and SSB proteins. While some archaeal organisms have a single, simple RPA made of a single OB-fold, others contain multiple variant forms that function independently, with some organisms containing both RPAs and SSBs, but with OB-folds more closely resembling eukaryotic rather than bacterial structures. Euryarchaeotes, and in particular several closely related methanogens have highly diverse, rapidly evolving RPA gene structures, which provide a glimpse into how multi OB-fold protein can evolve by domain duplication, fusion, fission and deletion (64–66).
Figure 2: Oligonucleotide/oligosaccharides binding folds (OB-folds) are conserved across all domains of life.
The cartoon at the center of the figure is the basic OB-fold motif. While there is some variation, the five beta sheets (purple) that form the mixed beta barrel and the alpha helix (teal) that caps the barrel are generally conserved. OB-folds are represented in each of the structures (bacterial SSB, archaeal SSB, archaeal RPA, and eukaryotic RPA and are also present in numerous ssDNA binding proteins represented here by the Hiran domain of HLTF and the ssDNA binding domain of human BRCA2) using the same purple beta sheet/teal alpha helix/grey loops scheme. Additional structural components are colored differently to highlight them. In the eukaryotic RPA structure the winged helix (WH) domain is depicted in red. In the BRCA2 structure the tower domain is depicted in red and the helix domain is depicted in yellow.
While all of the OB-fold containing ssDNA-binding proteins have structures based around OB-folds, they still exhibit mechanistic variability depending upon the organization of the modular domains. Eukaryotic RPA mostly extends ssDNA upon binding, and bacterial SSB wraps and condenses ssDNA (50,67). Some archaeal organisms contain multiple variants of RPA/SSB with some that induce RPA-like ssDNA extension while others induce SSB-like ssDNA wrapping and these wrapping/extending activities can be carried out by the same RPA under different conditions (66). The ubiquity of ssDNA binding proteins composed of OB-folds across all domains of life indicates the critical role these proteins play and suggests an ancestral RPA composed of a single OB-fold that has diverged over the course of evolution (66).
While ssDNA binding proteins are ubiquitous in all domains of life, this review will focus on heterotrimeric eukaryotic RPA, which contains six OB-folds (68) (Figure 1a&b). Four of the OB-folds of RPA (DBD-A, B, C, and D) exhibit significant binding to ssDNA (69). Nevertheless, all six OB-folds are often referred to as DBDs and we will apply these terms interchangeably because of the modular organization of the RPA’s DBDs/OB-folds. While all six DBDs/OB-folds are structurally similar, two aromatic residues in the DNA binding clefts of each DBD-A, B, C, and D have been identified that are highly conserved and contribute to the high affinity interaction with ssDNA through base stacking (36,70,71). These aromatic residues are a characteristic feature of all DNA-interacting OB-folds in eukaryotic and archaeal ssDNA binding proteins (see for example, DBD-A F238, F269, DBD-B W361 and F386 in human RPA numbering in Figure 3a). DBD-E and F lack these conserved aromatic residues. In addition to the conserved aromatic residues involved in base stacking, the DBDs also contain a less conserved loop comprised of basic residues. This basic loop is thought to act as a flexible clamp that interacts with the phosphate backbone upon RPA binding to ssDNA (43). The N-terminal OB-fold of RPA70, DBD-F, does not exhibit significant DNA binding activity, but instead is used as a hub for RPA protein-protein interactions (see below). DBD-E, the OB-fold found in RPA14, also does not exhibit ssDNA binding. DBD-E does play an important role in the formation of the trimerization core and is found in close proximity to DNA when the other DBDs are engaged (72). DBD-E also allows for interaction between RPA molecules that are bound adjacently on ssDNA (50) (Figure 1d). In addition to ssDNA, RPA can also interact with RNA, but at a much lower affinity (48,73).
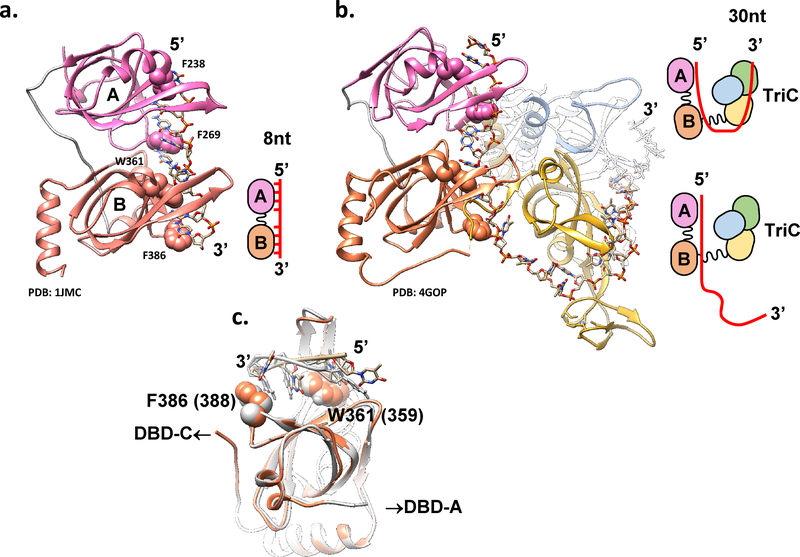
Figure 3: Proposed RPA-ssDNA binding modes.
Proposed structural basis for the distinct RPA-ssDNA binding modes. (a) PDB: 1JMC (76) structure of the human RPA DBD-A (pink) and DBD-B (orange) in complex with 8nt poly(dC) ssDNA represents an 8 nt binding mode schematically depicted on the right. Atoms of the four conserved aromatic residues F238 and F269 (in DBD-A) and W361 and F386 (in DBD-B) are shown as spheres. (b) PBD: 4GOP (50), which represents the high affinity (30 nt) ssDNA binding mode of Ustilago maydis RPA is shown with the same orientation of the DBDs A and B. Cartoon on the right represents the proposed transition between the 8nt and 30nt binding modes. (c) Overlap of the DBD-B from 4GOP (orange) and 1JMC (light grey) suggests different paths the ssDNA can take through this DBD likely due to the presence of the flexible linker connecting DBD-B and DBD-C. See text and (50) for extended discussion.
RPA and other OB-fold containing proteins are generally considered to bind ssDNA in a sequence independent manner. Both yeast and human RPAs, however, display preference for pyrimidines over purines with approximately 50-fold higher affinity (73). There are also sequences to which RPA binds preferentially, such as for example pyrimidine-rich strands of a replication origin (73). Systematic Evolution of Ligands by EXponential enrichment (SELEX) experiments revealed that the RPA trimerization core composed of DBD-C, DBD-D and DBD-E displays specificity for a G-rich motif capable of forming a G-quadruplex and is able to destabilize the quadruplex (74).
Each eukaryotic/archaeal OB-fold binds approximately 4–6 nucleotides of ssDNA. Due to the flexible regions linking the OB-folds, the DBDs of RPA are able to function as modular units. The individual DBDs are able to contact ssDNA, while others remain removed from ssDNA. There are multiple conformations available to RPA when it is bound to DNA. Because these conformations of RPA may result in different degrees of ssDNA engagement, these conformations may correlate to different binding modes. Linkers between the individual DBDs also play important roles in establishing the distinct DNA binding modes. A flexible linker between DBD-B and DBD-C, which can bind into the DNA-binding site of DBD-B can modulate the RPA binding mode between 8 nt and 30 nt (49) (Figure 3). While flexible in solution, the linker between DBD-A and DBD-B becomes ordered in the presence of ssDNA orienting the two DBDs (49,75,76).
Binding Modes and Dynamics
Modular architecture of RPA and other OB-fold containing ssDNA binding proteins implies that the affinity for ssDNA should scale with the number of OB folds engaging ssDNA. The lesson from numerous archaeal RPAs, however, suggests that the binding energies of the individual OB-folds are not simply additive. While there is a definite gain in affinity between F. acidarmanus RPA2 (a monomeric RPA containing a single OB fold) and RPA1 (a homodimer with each monomer containing two OB-folds and a Zn-finger) (65), RPAs from methanogenic archaea with the number of OB-folds ranging from three to five display similar affinities for ssDNA and similar binding site sizes. This suggests that in these larger RPAs, not all DBDs may be engaging the ssDNA (77). Similarly, the trimerization core of the eukaryotic RPA makes more extensive contacts with ssDNA than DBD-A and DBD-B (49), but when analyzed separately, the construct containing DBD-A, B and F (RPA-FAB) has much higher affinity for ssDNA than that of the trimerization core (DBD-C, D, and E) (see Table 1 and discussion below). The ability of RPA to bind to ssDNA in varied conformations, or binding modes, each with a different number of nucleotides engaged yields different affinities for ssDNA substrates of different lengths. These modes (Figure 3) were observed using biochemical and structural techniques. In the low-affinity mode, RPA binds to approximately 8 nucleotides of ssDNA and has an equilibrium dissociation constant of 50 nM. In this binding mode, DBD-A and DBD-B of RPA70 are believed to be the only DBDs engaged with the ssDNA (Figure 3a). In the high-affinity mode (Figure 3b), RPA binds to 28–30 nucleotides of ssDNA with a subnanomolar equilibrium dissociation constant. In the high-affinity binding mode, DBD-A, B, and C of RPA70 and RPA-D of RPA32, are all engaged in the binding of ssDNA (49). While DBD-A and DBD-B engage the ssDNA in both the 8nt and 30nt binding modes, by comparing structures representing the 8nt binding mode of human RPA (PDB: 1JMC (76)) and the high affinity binding mode of U. maydis RPA (PDB: 4GOP (49)) Fan and Pavletich proposed that the key contacts between the four aromatic or hydrophobic residues and ssDNA may be different between the two modes due to the difference in the DNA conformation (49) (Figure 3c). An additional medium-affinity binding mode has also been observed where RPA binds to 18–20 nucleotides. In this binding mode, three DBDs are expected to be engaged with ssDNA, proposed to be DBD-A, B, and C of RPA70 (17).
Table 1:
Dissociation constants of RPA constructs
| Construct | Kd | Method | Reference |
|---|---|---|---|
| RPA (full-length) | 0.4 nM | EMSA | (79) |
| RPA (full-length) binding to Gq23 (SELEX-derived preferred G-quadruplex forming sequence) | 40 nM | FPA | (73) |
| RPA (full-length) binding to PolyA | 80 nM | FPA | (73) |
| RPA (full-length) binding to PolyG | 200 nM | FPA | (73) |
| DBD-A | 2 μM | Heteronuclear NMR | (69) |
| DBD-B | 16.8 μM | Heteronuclear NMR | (69) |
| DBD-A/B | 52 nM | Fluorescence Quenching | (69) |
| DBD-A (185–292) | > 100 μM | EMSA | (79) |
| DBD-B (293–406) | > 100 μM | EMSA | (79) |
| DBD-A/B (185–406) | 13.7 nM | EMSA | (79) |
| DBD B/B (298–424:301–422) | > 10 μM | EMSA | (79) |
| DBD A/A (179–298:181–303) | 0.17 nM | EMSA | (79) |
| RPA70 (169–441) | 12.5 nM | EMSA | (62) |
| RPA70(R234A) | 15 nM | EMSA | (79) |
| RPA70(K263A) | 2.2 nM | EMSA | (79) |
| RPA70(E277A) | 4 nM | EMSA | (79) |
| RPA70(R382A) | 10 nM | EMSA | (79) |
| RPA70(F238A/F269A) | 1.8 nM | EMSA | (79) |
| RPA70(W361A/F386A) | 2.8 nM | EMSA | (79) |
| RPA14·32 | None/Low | EMSA | (80) |
| RPA14·32-(43–171) | 10–50 μM | EMSA | (80) |
| RPA70-(181–422) | 50–100 nM | EMSA | (80) |
| Trimerization Core | 5 μM | EMSA | (80) |
| Trimerization Core binding to Gq23 DNA | 0.64 μM | FPA | (73) |
| Trimerization Core binding to polyA | 6.6 μM | FPA | (73) |
| Trimerization Core binding to polyG | 10.04 μM | FPA | (73) |
| scRPA | 29.2 nM | Stopped flow fluorescence | (40) |
| scRPA DBD-FAB | 82.8 nM | smTIRFM | (40) |
| RPA WT | 680 pM (fast) 60.2 pM (slow) |
smTIRFM | (36) |
| RPA-AroA Mutant | 1.28 nM (fast) 81.9 pM (slow) |
smTIRFM | (36) |
| RPA-AroB Mutant | 926 pM (fast) 111 pM(slow) |
smTIRFM | (36) |
| RPA-AroA/B Mutant | 556 pM (fast) 49.0 pM (slow) |
smTIRFM | (36) |
| RPA/ssRNA | 15 nM | smTIRFM (FRET) | (48) |
The dissociation constants of various RPA constructs are listed. Constructs are human RPA, with the exception of two from saccharomyces cerevisiae, denoted with a ‘sc’. The monomer, DBDs, or residues are noted for each RPA construct that was not full-length, wild-type RPA. Mutations are noted where relevant. The methods used to determine the dissociations constants are included in the third column. NMR – nuclear magnetic resonance; EMSA – electrophoretic mobility shift assay; FPA – fluorescence polarization anisotropy; smTIRFM – single-molecule total internal reflection fluorescence microscopy.
In addition to testing RPA binding affinities using truncations composed of isolated DBDs, mutational studies have also informed the roles of the various DBDs in ssDNA binding and DNA replication and repair processes. When the aromatic residues of DBD-A or B were mutated, other domains were able to compensate, resulting in similar binding to longer ssDNA (25nt), but eliminated binding to short ssDNA (15nt) (36). Functionally, these aromatic residue mutants are less likely to form long-lived complexes with ssDNA, defective in duplex melting, and do not promote RAD51-mediated DNA strand exchange (36). The mutations of the aromatic residues are thought to cause defects in DNA repair not due to a loss of binding affinity, as that is largely maintained, but due to loss of ability to dynamically sample conformations within the ssDNA-RPA complex, a property of RPA seems to be necessary for DNA duplex destabilization (36,37). A recent computational study also suggests that aromatic residues may play a role in RPA diffusion on DNA (78).
The modular binding of RPA to ssDNA has been suggested to proceed sequentially, with the lowest affinity modes being the first step of RPA binding and progressing to more stable, engaged modes (69,81) (Figure 3a&b). This model was supported by observation of the low affinity mode only one short (<10 nucleotides) ssDNA substrates, while more engaged conformations were observed on longer (~30 nucleotides) ssDNA substrates (49,70,75,81). The model also offered an explanation for why RPA has higher affinity for longer ssDNA substrates. Additionally, constructs containing DBD-A and B show a relatively high affinity for ssDNA compared to the isolated trimerization core (DBD-C, D, and E), which suggested that the DBD-A and B may bind first, bringing the other domains into proximity, at which point all of the domains become engaged with the ssDNA and remain engaged (69). Perhaps the strongest evidence that pointed towards sequential transition between the 8nt and 30nt high affinity binding mode came from the experiments that demonstrated RPA loading by the SV40 T antigen (Tag) helicase (68). Here, Jiang and colleagues showed that the origin binding domain of SV40 Tag interacts with the DBD-A and DBD-B of human RPA. The site of the interaction is distant from the DNA binding site and allows formation of a ternary Tag-RPA-ssDNA complex on short oligonucleotides (i.e. the 8nt mode). Transition to the 30-nt binding mode, however, released the Tag-RPA interaction consistent with the sequence of events expected during RPA loading on the emerging ssDNA during activation of the SV40 pre-replication complex (68).
Recent single molecules studies have provided further support for the modular nature of the RPA-ssDNA interaction and for a potential sequential binding mechanism. Chen and colleagues employed total internal reflection fluorescence microscopy (TIRFM) to interrogate, at the single-molecule level, the interaction between surface tethered human RPA and ssDNA of various lengths (36). At least two distinct binding modes, characterized by different affinities, were observed (36). On longer (35nt) ssDNA molecules the calculated equilibrium dissociation constants for these complexes were 680 pM and 60 pM, respectively (Ka=1.47×109 M−1 (‘fast’ dissociating) and 1.66×1010 M−1 (‘slow’ dissociating complex)). The latter was consistent with the value previously obtained from the high affinity RPA-ssDNA complex (16). Two distinct affinities came from fitting the distributions of dwell times of the RPA-ssDNA complexes to two exponentials, which was interpreted as existence of complexes with distinct stabilities. Distributions of the times between binding events were best fit to a single exponential suggesting a single association rate constant for both complexes. In contrast, the dwell time distributions of the RPA molecules in complex with shorter (15nt) ssDNA fitted well to a single exponential, were shorter lived, and yielded an equilibrium dissociation constant of 606 pM (Ka=1.65×109 M−1), which was similar to the less stable complexes formed on 35 nt ssDNA. Thus, the data obtained on the longer ssDNA can be interpreted as a transition between the low affinity and high affinity modes. Another single-molecule study using FRET labeled DNA also found human RPA transitioning between two conformations: a more stable conformation in which the DNA was extended and a less stable conformation where the DNA may be bent (48). Notably, partial inactivation of the DBD-A or DBD-B by mutating one of the conserved aromatic residues in one or both high affinity DBDs also resulted in two distinct complexes, but the less stable complex was more prevalent than the high affinity one. None of the aromatic mutants, however, were able to bind to the 15nt ssDNA and displayed a single binding mode on the 20 nucleotide ssDNA (36). This is consistent with the idea that the DBD-A and DBD-B comprise the high affinity binding module of RPA, but the DNA can take a slightly different path within this module in 8nt vs. 30nt binding modes adapting to the lack of one of the aromatic residues. Notably, Chen and colleagues also demonstrated that the conserved aromatic residues are necessary for the ability of RPA to melt partial DNA duplexes, suggesting that engaging DBD-A and DBD-B in the 30nt mode is critical for duplex destabilization (36). Since the aromatic mutants of RPA are also separation of function mutants (82), access of RPA to the slow dissociation, high stability state is critical to RPA function in DNA repair, but not for its function in replication. Another single-molecule study by Nguyen and colleagues revealed that the ability of RPA to melt DNA hairpins depends on the location of the hairpin relative to the orientation of bound RPA (41). RPA binds ssDNA with a distinct polarity whereby DBD-A is at the 5’ end of the occluded ssDNA and DBD-D is at the 3’ end; the hairpin located at the 3’ of the bound RPA is melted much more efficiently than the hairpin located at the 5’ end (83–85), suggesting that RPA entered the duplex from the 5’ end of the hairpin, leading with DBD-D (41). In addition to hairpins, telomeric G-quadruplexes and duplexes are also preferentially dismantled in the 5’ to 3’ direction by RPA (86). The importance of the aromatic residues in DBD-A and DBD-B for duplex separation, therefore, is likely to stem from the role of the 30nt binding mode that engages the trimerization core.
While the modular binding model has explained observations of RPA binding with different affinities to different lengths of ssDNA, it does not fully explain how RPA is able to be rapidly displaced by other ssDNA interacting proteins that function downstream of RPA. Curiously, in some cases there is no apparent correlation between the affinity of the protein for ssDNA and its ability to displace RPA. For example, under conditions permitting ATP hydrolysis, human DNA strand exchange protein RAD51 has a modest ability to displace RPA and form a nucleoprotein filament, which is the active species in homology search and DNA strand exchange reactions that lay at the heart of homologous recombination and recombinational DNA repair (87). In the DNA strand exchange reactions biochemically reconstituted in vitro and in cells, RPA presents a kinetic block to the RAD51 nucleoprotein filament assembly thus preventing uncontrolled recombination (88). A specialized recombination mediator, such as Rad52 (in yeast) or BRCA2 (in human) is required to facilitate replacement of RPA with RAD51 (89–95). Surprisingly, the phosphomimetic version of RAD51, RAD51Y45pCMF, which displays reduced affinity for ssDNA, was capable of efficient RPA displacement (87). This observation is not confined only to RAD51. The “hand-off” model, developed by Fanning and colleagues, addressed the interaction of many downstream proteins with RPA and that these interactions may induce conformational changes that alter the ssDNA binding of RPA (42). While prior versions of the ‘hand-off’ model relied on pathways involving proteins that bound to RPA with sequentially increasing affinity, Fanning and colleagues proposed an updated model focused on the protein-protein interactions promoting RPA to adopt a less engaged binding mode, thus providing a landing site for the interacting protein to access the ssDNA (42).
More recent studies have continued to support that the interaction of RPA with ssDNA is highly dynamic (Figure 4a&b). Several single molecule studies of fluorescently labeled RPA on ssDNA curtains revealed that RPA is able to bind very tightly to ssDNA under conditions where there is no free RPA, remaining bound after 2 hours, and rapidly dissociate and exchange with free RPA at high concentrations (39). This difference in RPA dynamics and interaction with DNA in contexts where all RPA is DNA bound or free RPA is present are notable and may be important for the role of RPA as a sensor of accumulating DNA damage. These studies also revealed that interactions with other proteins involved in homologous recombination (Rad51, Rad52) modified the interaction of RPA with ssDNA, a function that was important in the regulation of pathway choice (11,39,96). NMR studies showed that due to the flexible linkers connecting the DBDs, RPA is able to sample a variety of functional conformations and that conformational changes are induced by the binding of ssDNA (35). A single-molecule magnetic tweezer study by Kimmerich and colleagues (47) demonstrated that a microscopic association of the individual DBDs of both human and yeast RPA creates a “toehold” that traps spontaneously melted DNA duplex at the ssDNA-dsDNA junction and promotes duplex destabilization by RPA (Figure 4c&d). Collectively, these studies provided the basis for a mechanism of the RPA replacement that evokes RPA conformational dynamics (Figure 5). Here, RPA as a whole remains macroscopically bound to ssDNA, but its individual DBDs undergo microscopic dissociation from and rebinding to ssDNA. RPA macroscopically dissociates from the ssDNA only when all of its DBDs dissociate simultaneously. Macroscopically bound RPA will thus continuously transition between different binding modes (Figure 5a). When no excess of ssDNA binding protein is present in the reaction, the probability of the DBDs’ rebinding is high resulting in a very stable RPA-ssDNA complex observed by Gibb and colleagues (39). The presence of free RPA in solution allows for protein exchange on ssDNA as the incoming RPA molecule may first bind in an 8nt mode resulting in two adjacent RPAs macroscopically bound to ssDNA, but with some of the DBDs microscopically dissociated (Figure 4b and Figure 5a). Similarly, if a downstream player is present in the reaction and its association rate and its binding site size match the probability of the free ssDNA of a sufficient length to be available due to the microscopic DBD dissociation, this downstream player can compete with rebinding of the dissociated DBD or binding of the RPA from solution (Figure 5b). In the case of human RPA and RAD51 proteins, the phosphomimetic RAD51Y54pCMF displays lower overall affinity for ssDNA than the wild type protein (87). This lower affinity, however, is offset by the higher cooperativity during the nucleation step of the RAD51 nucleoprotein filament formation, which Subramanyam and colleagues proposed to be responsible for an increased ability of the RAD51Y54pCMF to displace RPA pre-bound to ssDNA (87). Probability of the RPA-coated ssDNA to contain landing sites for the downstream players should not only depend on the RPA transition between different binding modes, but also on its one-dimensional diffusion along ssDNA (41,47), which in itself should depend on the nature of the ssDNA substrate and its occupancy by RPA and other proteins. The transient binding of individual RPA DBDs allows RPA to trap partially unwound forked DNA substrates, which provides an opening for more RPA DBDs to become engaged (47). One would expect RPA to behave differently if provided with a long ssDNA, such as present during the lagging strand DNA replication, end resection in homologous recombination, and homology-directed double strand break (DSB) repair, compared to relatively short stretches of ssDNA available to RPA on various DNA repair intermediates. Additionally, all atom molecular dynamic simulations also suggested that the mode of the RPA binding and its ability to melt the adjacent DNA duplex is different for a gapped ssDNA versus a bubble DNA (36).
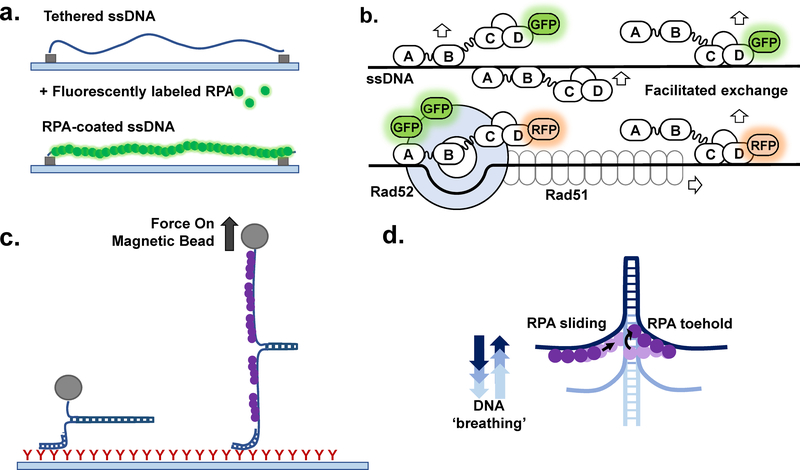
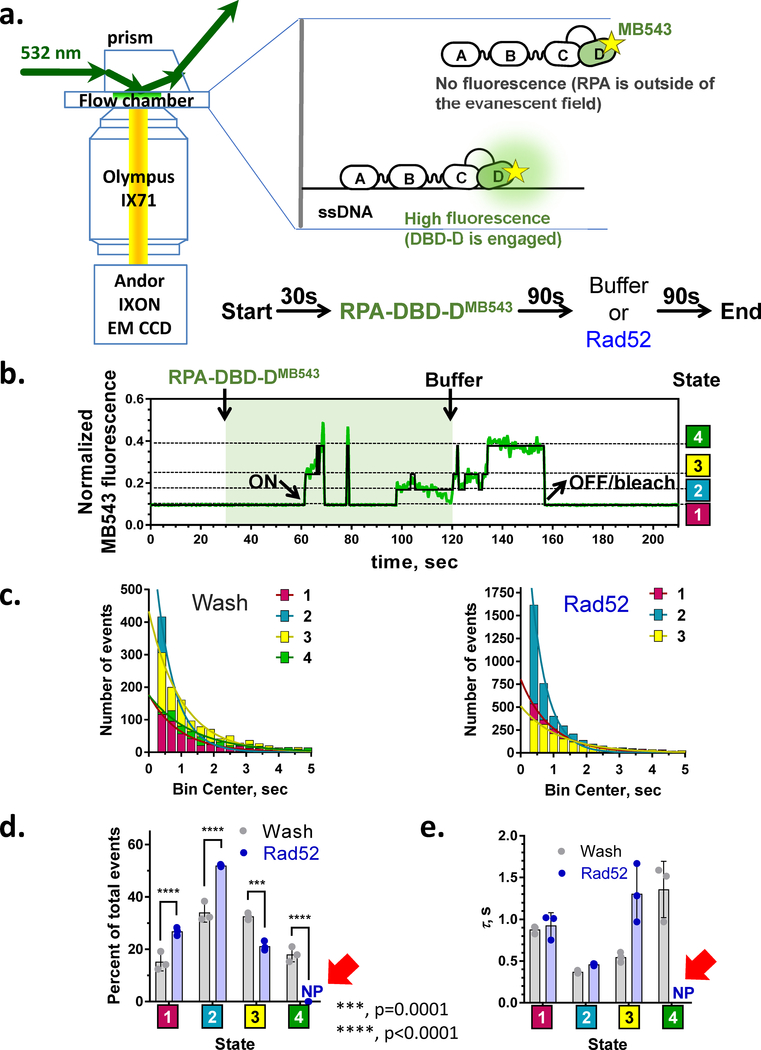
Figure 4: Single-molecule interrogation of the RPA-ssDNA complex.
(a) Schematic representation of an ssDNA curtain experiment (see (11,39,96) for details), side view. The ssDNA molecules are stretched parallel to each other over the surface of the TIRFM slide. Binding of the RPA-GFP (green) is visualized as appearance of the fluorescence along the DNA molecules. (b) The DNA curtains experiments allowed to propose the mechanism underlying facilitated exchange of RPA on ssDNA. When no additional RPA is present in the solution, RPA molecules remain stably bound to ssDNA even though their individual binding modules may microscopically dissociate from and rebind to ssDNA (e.g. transition between 8 nt and 30 nt binding modes). In the presence of unlabeled RPA in solution, microscopic dissociation of the trimerization core (30 nt → 8 nt transition) opens a landing spot for the RPA from solution and subsequent exchange of the GFP labeled protein on the ssDNA with the unlabeled counterpart. Two color experiments with RFP-labeled RPA and GFP-labeled Rad52 showed that the recombination mediator Rad52 stabilizes some RPA molecules on ssDNA resulting in the RPA-Rad52 clusters from which the Rad51 nucleoprotein filament grows. (c) Magnetic tweezer experiment to characterize the RPA-mediated DNA duplex melting (47). (d) These experiments suggested that a microscopic association of the individual RPA DBDs creates a “toehold” that traps spontaneously melted DNA duplex at the ssDNA-dsDNA junction and promotes duplex destabilization by RPA.
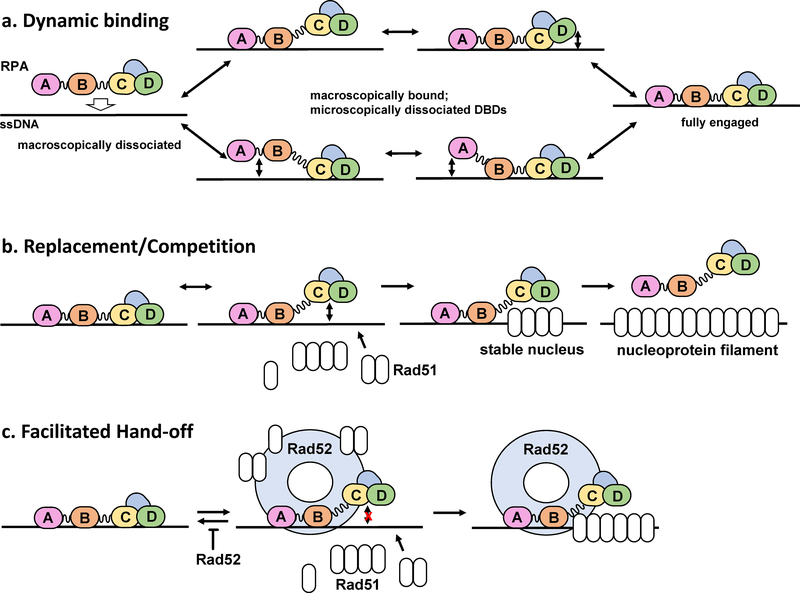
Figure 5. Model of RPA dynamic binding and the role in recruitment of weaker binding downstream proteins.
(a) The dynamic binding model differentiates between two types of binding referred to as macroscopic, where the molecule as a whole binds/dissociates, and microscopic, where the molecule as a whole remains bound while individual domains bind/dissociate within the bound RPA-ssDNA complex. In contrast to the original binding modes model, which implies that DBDs A and B constitute a high affinity binding module which associates with ssDNA first and remains associated in both 8 nt and 30 nt modes, the dynamic binding model suggests that the domain binding events do not occur in a sequential manner (40). (b) With the understanding that the individual domains of RPA can dissociate, the replacement/competition model treats these microscopic dissociation events are potential windows for smaller, weaker binding proteins to access ssDNA that is otherwise saturated with RPA. In this case, Rad51, the eukaryotic recombinase, would have an opportunity to nucleate and form a filament to outcompete RPA provides that some domains dissociate from the ssDNA (87). (c) Facilitated hand-off further explains how downstream proteins may outcompete RPA. Rad52, an RPA binding protein and recombination mediator, limits the ability of the 3’ RPA DBDs to access ssDNA. Rad52 also carries Rad51, potentially loading it in the opening created by preventing these RPA domains form accessing the DNA and promoting Rad51 activity in DNA repair (40). While these models address RPA replacement in homologous recombination, other DNA replication and repair processes may exhibit similar mechanisms as many proteins interact and compete with RPA for access to ssDNA.
While this model shares many similarities with the binding modes model described previously, the dissociation/rebinding of the individual DBDs does not necessarily have to occur in any specific sequence. More recent direct observations of RPA dynamics support dynamic binding over sequential binding. A recent single-molecule study by Pokhrel, Caldwell, and colleagues utilized electrostatically sensitive fluorophore MB543 site-specifically positioned on DBD-A or DBD-D of yeast RPA to reveal four distinct conformations within the RPA-ssDNA complex for each terminal DBD (40) (Figure 6). The lifetime of each conformation was on the order of a few seconds, with the RPA remaining macroscopically associated with ssDNA for many minutes. Thus, numerous transitions between these conformations were observed for each individual ssDNA bound RPA molecule. The RPA-FAB construct containing the DBD-F, MB543-labeled DBD-A and DBD-B displayed only two fluorescence states. RPA-FAB also formed much shorter lived complexes on ssDNA compared to the full-length heterotrimer and has an estimated equilibrium dissociation constant of 82.8nM, comparable to previously reported results for this construct (40,97). Notably, RPA DBDs in the RPA-ssDNA complex were able to proceed from less to more engaged states, as well as vice versa. Surprisingly and contradictory to the expectation of the binding modes model, no specific sequence was observed with respect to the binding of the individual RPA molecules to long (60nt) ssDNA. RPA was equally likely to initiate binding with either DBD-A or DBD-D fully engaged or in any of the other conformational states. The stopped flow experiments that monitored RPA binding to short ssDNA substrates in the same study, however, were more consistent with the DBD-A binding first followed by a transition into a binding mode where the DBD-A is detached, while the DBD-D is more fully engaged. This can be rationalized as the RPA-ssDNA interaction initiation at the DBA-A/DBA-B module in 8nt mode followed by a transition into a mode that has only the trimerization core bound to ssDNA. In the crystal structure of the U. maydis RPA (98), the trimerization core makes more extended contacts with ssDNA than the combined contacts made by the DBD-A and DBD-B. The linker between DBD-B and DBD-C may assume a position that competes with the ssDNA binding in the DBD-B. While Fan and Pavletich (98) proposed this as the mechanism for the transition between 8nt and 30nt binding mode, we would like to argue that the transition from engaged DBD-A and DBD-B to engaged trimerization core reflects the 8nt binding mode when only a short stretch of ssDNA is available. In contrast to the long ssDNA where RPA is free to fluctuate between all available conformations, the inability of the RPA aromatic mutants to form a complex with 15nt ssDNA despite having all the key residues in the trimerization core intact (36) suggests that the sequence of the DBDs’ engagement is important when short stretches of ssDNA are involved. The 8nt binding mode can also dominate under conditions when RPA is present in a large excess over available ssDNA (72,99).
Figure 6. Real time single-molecule observation of the RPA conformational dynamics.
(a) smTIRFM experiment that monitors microscopic association/dissociation of the individual DBD within the macroscopically bound RPA. The ssDNA is immobilized on the surface of TIRFM flow chamber. After 30 sec of observation, RPA labeled within the DBD-D with an environmentally sensitive MB543 dye is injected in the reaction chamber. RPA binding to surface-tethered ssDNA molecules is observed as appearance of the fluorescence signal in specific spots on the flow chamber surface. After 90 sec, the unbound RPA is removed by flashing in the buffer with or without Rad52. (b) A representative trajectory (time-based change in the fluorescence in a specific spot on the slide surface) depicting conformational dynamics of an individual RPA-DBD-DMB543 molecule. The data are from (40). “ON” marks the macroscopic association of the RPA molecule with ssDNA; “OFF/bleach” indicates the moment when the signal is lost due to either RPA dissociation or MB543 dye bleaching. Raw normalized fluorescence is shown in green; the black line corresponds to the idealized trajectory after global analysis of all trajectories using ebFRET (215), which has identified four distinct states interpreted as different degrees of ssDNA engagement by DBD-D (40). (c-e) Analysis of the dwell times of the RPA conformational states. (c) Exponential fits to the dwell time distributions after buffer wash (4 states) and in the presence of Rad52 (3 states). (d) Visitation frequencies for all states in the presence and absence of Rad52 show that state 4, which is the most engaged state of the DBD-D is lost in the presence of Rad52. Visitation of the state 3 also decreases in the presence of Rad52, while states 1 and 2 are visited more often. (e) Each of the four states exists on a second time scale (τ = 1/k, where k is the decay rate constant for each exponential fit); The presence of Rad52 results in disappearance of the state 4 and longer average dwell in state 3.
While the sequential, modular binding model explains why RPA binds to short ssDNA substrates in a lower affinity mode and longer ssDNA in a higher affinity mode, it does not explain how RPA is able to be efficiently displaced by other lower affinity DNA binding proteins that function in DNA replication, recombination, and repair. The dynamic model better explains how RPA is replaced by downstream proteins, with less engaged conformations of the ssDNA-RPA complex and RPA protein interactions playing important roles. Addition of Rad52, a recombination mediator protein that binds to RPA and functions downstream of RPA in homologous recombination, prevents RPA DBD-D from accessing the conformation in which it is fully engaged with ssDNA (40) (Figure 5c and Figure 6). The hand-off and dynamic models fit well together and with recent observations of RPA conformational flexibility and the modulation of RPA conformation by the binding of protein partners such as Rad52 (40,100).
Protein Interactions
RPA binds to a wide range of proteins that function in DNA replication, recombination, and repair downstream of RPA-coated ssDNA. While there are many protein-protein interactions that can occur with RPA, the interaction sites on RPA are largely confined to two domains, on RPA70 and RPA32. The interaction sites on RPA70 are the DBD-F, and DBD-A and B. The site on RPA32 is located at the C-terminus in the wing helix domain. Table 2 lists proteins known to interact with RPA and the location of the binding site on the RPA protein if it is known. The sheer number of interactions and the limited range of binding sites on RPA suggests that these interactions may be competitive. This is illustrated in Figure 7.
TABLE 2:
RPA Interacting Proteins
| RPA Interacting Protein | Region of interaction on RPA | Species | Reference |
|---|---|---|---|
| Activation induced Cytidine Deaminase (AID) | RPA32 | h | (101) |
| ATRIP | RPA70(DBD F) | h | (102–104) |
| Bloom Syndrome Helicase (BLM) | RPA70(168–308) | h | (105) |
| BRCA2 | ? | h | (106) |
| DDX11 | ? | h | (107) |
| DNA2 | RPA70(DBD F and C-term) | sc | (108,109) |
| EXO5 | RPA70(DBD-F) | h | (110) |
| FACT | RPA70(DBD F), RPA32 | sc | (111,112) |
| FancJ | ? | h | (113–116) |
| FBH1, F Box Helicase 1 | ? | h | (117) |
| H3-H3 | RPA70(DBD-F) | sc | (112) |
| Heat Shock Factor 1 | RPA70 | h | (118) |
| HELB | RPA70(DBD F) | h | (119,120) |
| HERC2 | RPA70 | h | (121) |
| HIRA | RPA70(DBD-C) | h | (22) |
| HLTF, Helicase Like Transcription Factor | RPA70? | h | (121,122) |
| Mre11-Rad50-Nbs1 | RPA70(DBD F) | h | (123–126) |
| Nucleolin | RPA14 | h | (127,128) |
| p53 | RPA70(1–120) | h | (129–132) |
| Papilomavirus E1 | RPA70(181–291) | h | (133,134) |
| Parvovirus NS1 | RPA70, RPA32 | h | (135) |
| PCNA | RPA70 | bo, h | (136,137) |
| Pol-prim | RPA70(1–327), RPA32 | bo,h | (5,138,139) |
| Polymerase delta | RPA70 | h | (140) |
| PP2A | ? | h | (141) |
| PRP19/BCAS2 | RPA70(DBD-F, DBD-C) | h | (121,142,143) |
| Rad17 | RPA70(DBD F) | h, sc | (144–146) |
| Rad18 | RPA70(167–452), RPA32 | sc | (147) |
| RAD51, Rad51 | RPA70(181–291) | h, sc | (148,149) |
| RAD52, Rad52 | RPA70(169–326), RPA32(224–271) | h, sc | (150–154) (155) |
| RAD9 | RPA70, RPA32 | h | (156) |
| RECQL1 | RPA70 | h | (113,157,158) |
| RECQL5B | ? | h | (159,160) |
| RFC | RPA70 | h, sc | (140,161) |
| RFWD3 | RPA70, RPA32 WH | h | (121,162,163) |
| RNF4 | ? | h | (164) |
| scPfh1 | RPA70, RPA32, RPA14 | sc | (165,166) |
| scPif1 | ? | sc | (167) |
| SMARCAL1 | WH | h | (168–170) |
| SV40 T antigen | RPA70(181–327), WH | h, sc | (133,134,171–173) |
| Tipin | WH | h | (174) |
| Uracil-DNA glycosylase (UDG) | RPA32(163–217) | h | (152,175,176) |
| Werner Syndrome Helicase (WRN) | RPA70(168–308) | h | (105,177) |
| XPA | RPA70(183–296), WH | h | (152,175,178–181) |
| XPF-ERCC1 | ? | h | (83,182,183) |
| XPG | ? | h | (83,183,184) |
A list of proteins that interact with RPA complied from a variety of reviews and sources (100,185,186). The site on RPA that the interaction occurs at, if it has been identified, is listed. The species in which the interaction was identified is noted (h:human, sc: saccharomyces cerevisiae, bo:bovine.
Figure 7. Protein-protein interaction and posttranslational modifications involving RPA.
(a) Schematic representation of the RPA primary structure with sites of posttranslational modifications and the same modifications mapped on the model of human RPA. Phosphorylation sites are marked with a P in a red circle and with the amino acid noted. SUMOylation sites are marked with an S in a black circle. Ubiquitinylation (U in a blue circle) has been identified on each of the RPA subunits, though specific site are not noted. *Asterisks note phosphorylation sites that were found in yeast RPA. S187/189 have only been noted in yeast. T180 is the corresponding site in human RPA that was identified as S187 in yeast. (b) Sites of Protein-Protein interactions mapped on the structural model of human RPA. Each box is labeled with the location on the RPA molecule that the listed proteins have been found to interact with. In the case of several proteins, multiple binding sites have been identified. While some proteins have only been tested for interaction with entire subunits of RPA, others have been found to bind to specific portions of a subunit, in which case the protein in only noted in the most specific region instead of the subunit. Table 2 indicates the specific amino acid regions where it is known.
While many of these interactions have been expected to primarily function to recruit downstream proteins to the site of DNA maintenance, these interactions may also be modulating the dynamics of RPA, as discussed in the previous section. For example, in yeast homologous recombination, Rad52 protein promotes loading of Rad51 recombinase on the RPA-coated ssDNA (90,92,93). Initial yeast two-hybrid analysis suggested that the Rad52-RPA interaction involves all three subunits of RPA, and also showed that two S. cerevisiae RAD52 mutants, rad52–34 and rad52–38, both within the conserved N-terminal domain of Rad52, were defective in binding RFA1 (RPA70), but not RFA2 (RPA32) (154). Later, an acidic region within the C-terminal domain of Rad52 (including residues Q308, D309, D310, D311) was identified as a site important for interaction with RPA (187). A multivalence of Rad52-RPA interaction is reflected in the observation that Rad52-Q308A/D309A/D310A/D311A was only partially defective in its recombination mediator activity in vitro (187). Further dissection of the Rad52-RPA interaction in homologous recombination suggested that the middle portion of conserved N-terminal domain of Rad52 associates with DNA-bound RPA and contributed to the recombination mediator activity, i.e. in Rad52 ability to facilitate replacement of RPA on ssDNA with the Rad51 recombinase (188). In an unexpected finding, Rad52 was observed to stabilize ssDNA-bound RPA (189). In a more recent single-molecule study of RPA Pokhrel, Caldwell and colleagues revealed that Rad52 binding prevented DBD-D from accessing ssDNA, providing an opening for downstream proteins like Rad51 to access ssDNA (40). In this way, Rad52 is able to both stabilize the ternary Rad52-RPA-ssDNA complex while allowing other proteins to access the ssDNA. Rad52 interacting sites have been identified in both RPA70 and RPA32, which may explain the ability of Rad52 to reduce the flexibility of RPA within the Rad52-RPA-ssDNA ternary complex (Figure 5 and Figure 6c). In human homologous recombination, BRCA2 plays the role of recombination mediator, but it is not known if the mechanism is similar to that of Rad52 in yeast.
Another example of a protein-protein interaction with RPA that results in a change in the dynamics of the ternary complex is xeroderma pigmentosum group A (XPA) (Figure 7b). XPA functions in the nucleotide excision repair pathway, playing an important role in the recognition of the bulky DNA adducts. Sites of interaction with XPA have been identified within RPA70 and the WH of RPA32. The formation of the RPA-XPA-damaged duplex DNA (ddDNA) complex results in behavior that is different from both the RPA-ddDNA and XPA-ddDNA complexes. The ternary complex shows greater specificity for ddDNA than with either RPA or XPA alone and an increase in the association rate and decrease in dissociation rate (190). This suggests that the formation and stability of the RPA-XPA-ddDNA complex is increased by the protein-protein interaction. Again, this effect is more complex than simply recruiting XPA to the site of DNA damage and may involve modulation of the RPA conformational dynamics.
In addition to interacting with proteins involved in DNA repair processes, RPA also interacts with proteins involved in DNA replication. One of these proteins, DNA polymerase α, was one of the three components required to reconstitute replication of SV40 DNA in vitro, along with RPA and T antigen (5). RPA inhibits priming activity in this reaction, while addition of T antigen partially diminished this inhibition (191). Notably, in a reconstituted of the yeast replisome encountering DNA damage, RPA inhibits priming by DNA polymerase α on the leading strand, but not the DNA synthesis from the annealed primer; in contrast lagging strand priming is stimulated by RPA (192). Thus, RPA depletion, which is expected under conditions of replication stress, can allow replication restart on the leading strand (192). A direct interaction was identified that occurs through the RPA32 and RPA70 subunits and is important for human RPA loading at the replication origin (Table 2 and Figure 7b). The interaction of DNA polymerase α with RPA was required for stimulation of polymerase activity and polymerase processivity, with RPA ssDNA binding activity necessary for the effect on processivity (138). The interactions resulting in loading of RPA are functionally different from the interactions that result in the ssDNA hand-off from RPA to downstream players, as the interactions with SV40 Tag and DNA polymerase α mediate the RPA deposition of ssDNA rather than its removal. One would expect a different type of RPA conformational dynamics to take place during these reactions.
It is important to distinguish protein-protein interactions that occur in solution and those that occur on or modulated by the DNA. Above, we mostly focused on the effects of interactions on the ssDNA-bound RPA. Some RPA interactions, such as the RPA interaction with yeast Rad52, are enhanced by or depend upon RPA being bound by ssDNA (187,193), though in contrast to yeast counterpart, human RAD52 binds RPA also in solution using an RQK motif (155) present in a number of RPA-interaction proteins including SMARCAL1, XPA and UNG2 (152,168). Other interactions occur primarily with free RPA. For example, p53 interaction with RPA was found to occur only with RPA not associated with DNA (131). The presence of ssDNA eliminates the p53:RPA interaction, while RPA inhibits p53 binding to ssDNA (194) and p53 inhibits RPA binding to ssDNA, with this p53:RPA interaction being necessary for p53 suppression of HR (195). In addition to p53, ssDNA was found to inhibit interactions with Papillomavirus E1 helicase, E2, human DNA polymerase α and SV40 T antigen; and this competition between RPA-protein and RPA-ssDNA interactions were proposed to play an important function in targeting RPA to viral origins of replication or an active replication forks both in human Papillomavirus and SV40 system (134). Other protein interactions occur with both free and ssDNA-bound RPA. Rad18 binds to free and ssDNA-bound RPA, though this interaction is enhanced by the presence of ssDNA (147). The relatively low-affinity Rad18 relies of ssDNA-bound RPA for recruitment, which occurs upstream of PCNA ubiquitylation that is necessary to recruit the polymerases responsible for DNA damage bypass by translesion synthesis (TLS) (147,196). Binding of Rad18 to free RPA inhibited this same process, suggesting that the RPA acts as a sensor, only promoting TLS in the context of stretches of RPA-coated ssDNA (197).
With the availability of single-molecule techniques that allow for the observation of RPA ssDNA binding dynamics, further investigation of known protein-protein interactions may reveal that the role of these interactions is much greater than simply recruiting downstream proteins to the site of damage. As has been observed with Rad52, XPA, and DNA polymerase α, these interactions can alter the RPA-ssDNA interaction, the interaction of the RPA-interacting protein with ssDNA, or the interactions of individual RPA DBDs with ssDNA. Inhibitors of RPA protein-protein interactions have already been developed and have potential as a starting point for cancer drug discovery (198). While this is promising, understanding the role of the protein-protein interactions with RPA in DNA repair would allow for a much more targeted approach to drug discovery. As RPA is ubiquitous in all processes involving ssDNA, from replication to repair, understanding how protein-protein interactions may drive pathway choice is key to understanding how cellular pathway choice proceeds from starting points that are seemingly similar.
Post-Translational Modifications
While the previous section discussed the effect that protein interactions have on RPA dynamics, this section will explore the effect of post-translational modifications (PTMs) not only on RPA dynamics, but also on protein interactions with RPA which have been more thoroughly studied. The most characterized site of PTM of RPA is the N-terminal portion of RPA32. Phosphorylation of RPA was first identified by Din and colleagues in 1990 in both human and yeast, showing a direct link between RPA, which is required for replication, and cell cycle dependent phosphorylation (199). Figure 7a shows a map of phosphorylation sites at the N-terminus of RPA32. This region of RPA32 in human and yeast is phosphorylated in a cell cycle dependent manner by cyclin-dependent kinases (CDK) with phosphorylation occurring during the transition from G1 to S-phase and dephosphorylation occurring after mitosis (46). Functionally, RPA phosphorylated during mitosis binds to ssDNA identically, but exhibits reduced binding to dsDNA as well as certain DNA replication and repair proteins (200). These cell cycle dependent phosphorylation events are also necessary for further modification to occur as part of the DNA damage response (201).
The RPA32 N-terminal region is also phosphorylated at other sites in response to DNA damage, shown in Figure 7a. The phosphorylation response in RPA to various genotoxic stressors that result in various types of DNA damage gives some hints to which pathways RPA hyperphosphorylation is relevant to. In response to agents, such as hydroxyurea (HU), or UV irradiation, that stall DNA replication and lead to one-ended DSBs (i.e. breakage of the replication fork and detachment of one of the fork arms), RPA is hyperphosphorylated at the N-terminus of RPA32 (202). On the other hand, γ-irradiation that causes conventional DSBs promotes a small increase in phosphorylation, but not hyperphosphorylation (202). RPA hyperphosphorylation is dependent upon CDKs, ATM, ATR and DNA-PK. One effect of hyperphosphorylation is a decrease in interaction with DNA replication proteins and inhibition of replication (203). While hyperphosphorylated RPA is not found associated with replication machinery, it is found at foci of DNA damage, suggesting that the role of hyperphosphorylation may be to direct RPA away from replication and to repair of DNA damage (204). Most studies of RPA phosphorylation have focused on the cell cycle dependent and DNA damage response phosphorylation of the N-terminus of RPA32. A recent single-molecule study by Soniat and colleagues has identified the hyperphosphorylated human RPA as a negative end resection factor in homologous recombination (205).
In yeast cells lacking Rad9, which accumulate resected ssDNA, RPA was found to be phosphorylated by Mec1 kinase at two new sites (RPA32 S187/189) that are separate from the sites typically found to be hyperphosphorylated by Mec1 under other conditions (206). These findings suggest that different types of RPA phosphorylation, even by the same kinase, may be critical for both promoting and suppressing resection and homologous recombination. While there have been many RPA phosphorylation sites found to affect the interaction of RPA with partner proteins, direct observation of the effect of phosphorylation on RPA dynamics has been more challenging. A study by Yates et al characterized yeast RPA with a phosphomimetic mutation at S178D of RPA70 (50). Phosphorylation of this site is Mec1-dependent in yeast (207,208), while the equivalent site (T180) in humans is phosphorylated in an ATM/ATR-dependent manner (209). Interestingly, on the three-dimensional structure of RPA, S178 of RPA70 is located in a close proximity to S187/189 of RPA32, other targets of Mec1 discussed above (Figure 7a). In yeast, S178 phosphorylation by Mec1 occurs in response to replication stress induced by hydroxyurea (HU), or exposure of the cells to UV radiation, ionizing radiation or the alkylating agent methyl methane sulfonate (207,208). S178 is located in the linker between DBD-F and A of the RPA70 very close to the beginning of the DBD-A. Mec1 (ATR homolog), the primary replication checkpoint kinase in budding yeast, is activated when DNA damage sensor proteins recognize accumulation of ssDNA indicative of DNA damage. This RPA-coated ssDNA serves as a platform for Mec1 checkpoint activation (210). The N-terminus of RPA70 interacts with Ddc2 (homolog of human ATRIP); Ddc2 then recruits Mec1 to the ssDNA bound RPA. RPA phosphorylation on S178 is one of the consequences of this recruitment. Unlike phosphorylation sites at the N-terminus of RPA32, the effect of this site has not been well characterized. The study by Yates et al focused on the structure of the multi RPA-ssDNA complexes found that the phosphomimetic RPA containing S178D substitution showed increased cooperativity of binding to long strands of ssDNA and an increased interaction between DBD-A and E on adjacent RPA molecules (50). Due to the presence of the charged residue near the DBD-A, the increased binding cooperativity was offset by the reduced binding affinity. One can speculate that despite lower affinity of individual RPA heterotrimers for ssDNA, cooperatively bound phosphorylated/phosphomimetic RPA may form more stable platform for the DNA damage signaling and have different preferences for the downstream partners for the ssDNA handoff.
In addition to phosphorylation, RPA can also be modified by SUMOylation, acetylation, and ubiquitinylation. In response to DNA damage, particularly that which results in DSBs, RPA is SUMOylated at RPA70 lysines, K449 and K577 (211,212). SUMOylation of RPA appears to be important in homologous recombination and stabilizes the interaction of RPA with Rad51, while lack of SUMOylation of recombination proteins, including RPA, resulted in less efficient formation of the Rad51 nucleoprotein filament (212). RPA acetylation is mediated by PCAF/GCN5 in response to UV-irradiation, promoting interaction with XPA and DNA repair through NER (213,214). Like phosphorylation and SUMOylation, RPA ubiquitylation is also stimulated by the accumulation of DNA damage (142). RPA interactions have been identified with several ubiquitin ligases (Table 2). In addition to RPA interaction with Rad18, which controls PCNA ubiquitylation (147,196,197), different studies have identified either PRP19 or RFWD3 as the ligase that ubiquitylates RPA (121,185).The study by Maréchal and Zou determined that ubiquitylation of RPA mediated by the PRP19 E3 ubiquitin ligase complex is associated with stimulation of RPA phosphorylation and promoted recombination (185), while the study by Elia and colleagues identified RFWD3 as the mediator of RPA ubiquitination and discovered an siRNA artifact caused their observation of what appeared to be PRP19 mediated ubiquitylation (121). It is possible that both RFWD3 and PRP19 act as mediators in different capacities. Interplay with other posttranslational modifications and DNA repair pathways is seen with many of the RPA posttranslational modifications, with some promoting recruiting the factors that allow for further modifications.
While there have been some studies that investigate the effects of posttranslational modifications on RPA interaction with DNA, they have largely focused on binding to alternative structures or damaged DNA. Current research has largely focused on the effect of these modifications on RPA interactions with partner proteins. While this is necessary to better understand the interplay of RPA and downstream proteins, it does not paint a full picture. With the recent finding that phosphorylation can change RPA binding to ssDNA and induce cooperativity (50), further studies of how modifications affect the basic DNA binding functions of RPA and conformational dynamics of the RPA-ssDNA complex are necessary.
Concluding Remarks
Recent advances in single-molecule biophysics allow real time observation of the protein-nucleic acid interactions at the level of individual complexes, and more importantly individual domains within the protein. These studies have clarified and updated the existing models for the handoff of ssDNA from the main eukaryotic ssDNA binding protein RPA to numerous downstream partners that bind ssDNA with much lower affinity than RPA. The model we currently favor builds upon previously proposed binding modes model, but suggest that the handoff involves matching the rates of the microscopic dynamics of binding/dissociation of the individual DBDs of RPA macroscopically bound to ssDNA with the association rate and the binding site size for the correct downstream partner. Various posttranslational modifications and proteins that interact with RPA can modify its conformational dynamics thus guiding the RPA-ssDNA complex to specific events in cellular DNA metabolism. Biochemical and single-molecule evidence (36,39,40,47) has validated this model for the ssDNA handoff during homologous recombination. It will be very interesting to see if the same holds for other transactions involving RPA and whether the DNA substrate itself can actively influence the RPA conformational dynamics and its modulation by protein partners and posttranslational modifications. The same experimental strategies can be applied to other dynamic, multi-subunit nucleoprotein complexes where physical concepts of stochasticity and conformational plasticity may drive subunit exchange and substrate DNA recognition. The ultimate challenge, however, would be to observe the behaviors consistent with this model in living cells.
Acknowledgements
We gratefully acknowledge the support by the NIH/NIGMS R35GM131704, NIH/NCI R01 CA232425, DOD/CDMRP BC180227P1 and 1836351 EAGER to M.S., and the support by an NIH T32 Pharmacological Sciences Training Grant (NIH T32 GM067795) to C.C.C. We thank Members of the Spies’ lab for critical reading of the manuscript and for valuable discussions. We thank Dr. Luke Yates and Prof. Xiaodong Zhang (Imperial College London) for sharing coordinates of the S. cerevisiae 2xRPA-ssDNA complex model and the CryoEM 2D class average depiction, and Prof. Adrian Elcock (University of Iowa) for the model of the complete human RPA heterotrimer. We also acknowledge Prof. Marc Wold (University of Iowa) for many valuable discussions regarding RPA dynamics, regulation, and functions.
References
- 1.Wold MS and Kelly T (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A, 85, 2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wold MS (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem, 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 3.Iftode C, Daniely Y and Borowiec JA (1999) Replication protein A (RPA): the eukaryotic SSB. Crit Rev Biochem Mol Biol, 34, 141–180. [DOI] [PubMed] [Google Scholar]
- 4.Brill SJ and Stillman B (1989) Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature, 342, 92–95. [DOI] [PubMed] [Google Scholar]
- 5.Nasheuer HP, von Winkler D, Schneider C, Dornreiter I, Gilbert I and Fanning E (1992) Purification and functional characterization of bovine RP-A in an in vitro SV40 DNA replication system. Chromosoma, 102, S52–59. [DOI] [PubMed] [Google Scholar]
- 6.Iyama T and Wilson DM (2013) DNA repair mechanisms in dividing and non-dividing cells. DNA Repair, 12, 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiricny J (2013) Postreplicative Mismatch Repair. Cold Spring Harbor Perspectives in Biology, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spies M and Fishel R (2015) Mismatch Repair during Homologous and Homeologous Recombination. Cold Spring Harbor Perspectives in Biology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schärer OD (2013) Nucleotide Excision Repair in Eukaryotes. Cold Spring Harbor Perspectives in Biology, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Symington LS (2014) End Resection at Double-Strand Breaks: Mechanism and Regulation. Cold Spring Harbor Perspectives in Biology, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng SK, Gibb B, de Almeida MJ, Greene EC and Symington LS (2014) RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nature Structural & Molecular Biology, 21, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasner DS, Daley JM, Sung P and Niu H (2015) Interplay between Ku and Replication Protein A in the Restriction of Exo1-mediated DNA Break End Resection. Journal of Biological Chemistry, 290, 18806–18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G-M (2008) Mechanisms and functions of DNA mismatch repair. Cell Research, 18, 85–98. [DOI] [PubMed] [Google Scholar]
- 14.Umezu K, Sugawara N, Chen C, Haber JE and Kolodner RD (1998) Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics, 148, 989–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK and Weissman JS (2003) Global analysis of protein expression in yeast. Nature, 425, 737–741. [DOI] [PubMed] [Google Scholar]
- 16.Kim C, Paulus BF and Wold MS (1994) Interactions of human replication protein A with oligonucleotides. Biochemistry, 33, 14197–14206. [DOI] [PubMed] [Google Scholar]
- 17.Kumaran S, Kozlov AG and Lohman TM (2006) Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry, 45, 11958–11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo LI, Altmeyer M, Rask MB, Lukas C, Larsen DH, Povlsen LK, Bekker-Jensen S, Mailand N, Bartek J and Lukas J (2013) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell, 155, 1088–1103. [DOI] [PubMed] [Google Scholar]
- 19.Hass CS, Gakhar L and Wold MS (2010) Functional characterization of a cancer causing mutation in human replication protein A. Molecular cancer research : MCR, 8, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng SK, Chen H and Symington LS (2015) Replication protein A prevents promiscuous annealing between short sequence homologies: Implications for genome integrity. BioEssays, 37, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E and Géli V (2004) RPA regulates telomerase action by providing Est1p access to chromosome ends. Nature genetics, 36, 46–54. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Gan H, Wang Z, Lee J-H, Zhou H, Ordog T, Wold MS, Ljungman M and Zhang Z (2017) RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Molecular cell, 65, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treuner K, Ramsperger U and Knippers R (1996) Replication Protein A Induces the Unwinding of Long Double-stranded DNA Regions. Journal of Molecular Biology, 259, 104–112. [DOI] [PubMed] [Google Scholar]
- 24.Lao Y, Lee CG and Wold MS (1999) Replication Protein A Interactions with DNA. 2. Characterization of Double-Stranded DNA-Binding/Helix-Destabilization Activities and the Role of the Zinc-Finger Domain in DNA Interactions. Biochemistry, 38, 3974–3984. [DOI] [PubMed] [Google Scholar]
- 25.Bartos JD, Willmott LJ, Binz SK, Wold MS and Bambara RA (2008) Catalysis of Strand Annealing by Replication Protein A Derives from Its Strand Melting Properties. Journal of Biological Chemistry, 283, 21758–21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny J-L, Favre A and Saintomé C (2006) Human replication protein A unfolds telomeric G-quadruplexes. Nucleic acids research, 34, 4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qureshi MH, Ray S, Sewell AL, Basu S and Balci H (2012) Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. The journal of physical chemistry. B, 116, 5588–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiyama T, Zaitseva EM and Kowalczykowski SC (1997) A Single-stranded DNA-binding Protein Is Needed for Efficient Presynaptic Complex Formation by the Saccharomyces cerevisiae Rad51 Protein. Journal of Biological Chemistry, 272, 7940–7945. [DOI] [PubMed] [Google Scholar]
- 29.Alani E, Thresher R, Griffith JD and Kolodner RD (1992) Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. Journal of Molecular Biology, 227, 54–71. [DOI] [PubMed] [Google Scholar]
- 30.Mitsis PG, Kowalczykowski SC and Lehman IR (1993) A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry, 32, 5257–5266. [DOI] [PubMed] [Google Scholar]
- 31.Kim C and Wold MS (1995) Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry, 34, 2058–2064. [DOI] [PubMed] [Google Scholar]
- 32.Kemp MG, Mason AC, Carreira A, Reardon JT, Haring SJ, Borgstahl GE, Kowalczykowski SC, Sancar A and Wold MS (2010) An alternative form of replication protein a expressed in normal human tissues supports DNA repair. J Biol Chem, 285, 4788–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aklilu BB and Culligan KM (2016) Molecular Evolution and Functional Diversification of Replication Protein A1 in Plants. Front Plant Sci, 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuss JE, Patrick SM, Oakley GG, Alter GM, Robison JG, Dixon K and Turchi JJ (2005) DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry, 44, 8428–8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brosey CA, Chagot ME, Ehrhardt M, Pretto DI, Weiner BE and Chazin WJ (2009) NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J Am Chem Soc, 131, 6346–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Subramanyam S, Elcock AH, Spies M and Wold MS (2016) Dynamic binding of replication protein a is required for DNA repair. Nucleic Acids Research, 44, 5758–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen R and Wold MS (2014) Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays, 36, 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosey CA, Soss SE, Brooks S, Yan C, Ivanov I, Dorai K and Chazin WJ (2015) Functional dynamics in replication protein A DNA binding and protein recruitment domains. Structure, 23, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, Sung P and Greene EC (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One, 9, e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pokhrel N, Caldwell CC, Corless EI, Tillison EA, Tibbs J, Jocic N, Tabei SMA, Wold MS, Spies M and Antony E (2019) Dynamics and selective remodeling of the DNA-binding domains of RPA. Nat Struct Mol Biol, 26, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen B, Sokoloski J, Galletto R, Elson EL, Wold MS and Lohman TM (2014) Diffusion of human replication protein A along single-stranded DNA. J Mol Biol, 426, 3246–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fanning E, Klimovich V and Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res, 34, 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochkarev A and Bochkareva E (2004) From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol, 14, 36–42. [DOI] [PubMed] [Google Scholar]
- 44.Oakley GG and Patrick SM (2010) Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci (Landmark Ed), 15, 883–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krasikova YS, Rechkunova NI and Lavrik OI (2016) Replication protein A as a major eukaryotic single-stranded DNA-binding protein and its role in DNA repair. Molecular Biology, 50, 649–662. [DOI] [PubMed] [Google Scholar]
- 46.Byrne BM and Oakley GG (2019) Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Seminars in Cell & Developmental Biology, 86, 112–120. [DOI] [PubMed] [Google Scholar]
- 47.Kemmerich FE, Daldrop P, Pinto C, Levikova M, Cejka P and Seidel R (2016) Force regulated dynamics of RPA on a DNA fork. Nucleic acids research, 44, 5837–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q-M, Yang Y-T, Wang Y-R, Gao B, Xi X and Hou X-M (2019) Human Replication protein A induces dynamic changes in single-stranded DNA and RNA structures. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan J and Pavletich NP (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev, 26, 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates LA, Aramayo RJ, Pokhrel N, Caldwell CC, Kaplan JA, Perera RL, Spies M, Antony E and Zhang X (2018) A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA. Nat Commun, 9, 5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grimme JM and Spies M (2011) FRET-based assays to monitor DNA binding and annealing by Rad52 recombination mediator protein. Methods in molecular biology (Clifton, N.J.), 745, 463–483. [DOI] [PubMed] [Google Scholar]
- 52.Rashid F, Raducanu VS, Zaher MS, Tehseen M, Habuchi S and Hamdan SM (2019) Initial state of DNA-Dye complex sets the stage for protein induced fluorescence modulation. Nat Commun, 10, 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park C-J, Lee J-H and Choi B-S (2005) Solution structure of the DNA-binding domain of RPA from Saccharomyces cerevisiae and its interaction with single-stranded DNA and SV40 T antigen. Nucleic Acids Research, 33, 4172–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Le S, Basu A, Chazin WJ and Yan J (2015) Mechanochemical regulations of RPA’s binding to ssDNA. Sci Rep, 5, 9296–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eckerich C, Fackelmayer FO and Knippers R (2001) Zinc affects the conformation of nucleoprotein filaments formed by replication protein A (RPA) and long natural DNA molecules. Biochim Biophys Acta, 1538, 67–75. [DOI] [PubMed] [Google Scholar]
- 56.Murzin AG (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J, 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arcus V (2002) OB-fold domains: a snapshot of the evolution of sequence, structure and function. Current Opinion in Structural Biology, 12, 794–801. [DOI] [PubMed] [Google Scholar]
- 58.Ishino Y and Ishino S (2012) Rapid progress of DNA replication studies in Archaea, the third domain of life. Science China Life Sciences, 55, 386–403. [DOI] [PubMed] [Google Scholar]
- 59.Chedin F, Seitz EM and Kowalczykowski SC (1998) Novel homologs of replication protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends in biochemical sciences, 23, 273–277. [DOI] [PubMed] [Google Scholar]
- 60.Lue NF (2018) Evolving Linear Chromosomes and Telomeres: A C-Strand-Centric View. Trends in biochemical sciences, 43, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams KR, Murphy JB and Chase JW (1984) Characterization of the structural and functional defect in the Escherichia coli single-stranded DNA binding protein encoded by the ssb-1 mutant gene. Expression of the ssb-1 gene under lambda pL regulation. Journal of Biological Chemistry, 259, 11804–11811. [PubMed] [Google Scholar]
- 62.Dąbrowski S, Olszewski M, Piątek R and Kur J (2002) Novel thermostable ssDNA-binding proteins from Thermus thermophilus and T. aquaticus—expression and purification. Protein Expression and Purification, 26, 131–138. [DOI] [PubMed] [Google Scholar]
- 63.Yang C, Curth U, Urbanke C and Kang C (1997) Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nature structural biology, 4, 153–157. [DOI] [PubMed] [Google Scholar]
- 64.Robbins JB, Murphy MC, White BA, Mackie RI, Ha T and Cann IK (2004) Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J Biol Chem, 279, 6315–6326. [DOI] [PubMed] [Google Scholar]
- 65.Pugh RA, Lin Y, Eller C, Leesley H, Cann IK and Spies M (2008) Ferroplasma acidarmanus RPA2 facilitates efficient unwinding of forked DNA substrates by monomers of FacXPD helicase. J Mol Biol, 383, 982–998. [DOI] [PubMed] [Google Scholar]
- 66.Robbins JB, McKinney MC, Guzman CE, Sriratana B, Fitz-Gibbon S, Ha T and Cann IKO (2005) The Euryarchaeota, Nature’s Medium for Engineering of Single-stranded DNA-binding Proteins. Journal of Biological Chemistry, 280, 15325–15339. [DOI] [PubMed] [Google Scholar]
- 67.Suksombat S, Khafizov R, Kozlov AG, Lohman TM and Chemla YR (2015) Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife, 4, e08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang X, Klimovich V, Arunkumar AI, Hysinger EB, Wang Y, Ott RD, Guler GD, Weiner B, Chazin WJ and Fanning E (2006) Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J, 25, 5516–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arunkumar AI, Stauffer ME, Bochkareva E, Bochkarev A and Chazin WJ (2003) Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J Biol Chem, 278, 41077–41082. [DOI] [PubMed] [Google Scholar]
- 70.Bastin-Shanower SA and Brill SJ (2001) Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. The Journal of biological chemistry, 276, 36446–36453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brill SJ and Bastin-Shanower S (1998) Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol Cell Biol, 18, 7225–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salas TR, Petruseva I, Lavrik O and Saintomé C (2009) Evidence for direct contact between the RPA3 subunit of the human replication protein A and single-stranded DNA. Nucleic acids research, 37, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim C, Snyder RO and Wold MS (1992) Binding properties of replication protein A from human and yeast cells. Mol Cell Biol, 12, 3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prakash A, Natarajan A, Marky LA, Ouellette MM and Borgstahl GE (2011) Identification of the DNA-Binding Domains of Human Replication Protein A That Recognize G-Quadruplex DNA. Journal of nucleic acids, 2011, 896947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bochkareva E, Belegu V, Korolev S and Bochkarev A (2001) Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J, 20, 612–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bochkarev A, Pfuetzner RA, Edwards AM and Frappier L (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- 77.Lin Y, Lin L-J, Sriratana P, Coleman K, Ha T, Spies M and Cann IKO (2008) Engineering of Functional Replication Protein A Homologs Based on Insights into the Evolution of Oligonucleotide/ Oligosaccharide-Binding Folds. Journal of bacteriology, 190, 5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mishra G, Bigman LS and Levy Y (2020) ssDNA diffuses along replication protein A via a reptation mechanism. Nucleic Acids Research, 48, 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wyka IM, Dhar K, Binz SK and Wold MS (2003) Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry, 42, 12909–12918. [DOI] [PubMed] [Google Scholar]
- 80.Bochkareva E, Frappier L, Edwards AM and Bochkarev A (1998) The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J Biol Chem, 273, 3932–3936. [DOI] [PubMed] [Google Scholar]
- 81.Bochkareva E, Korolev S, Lees-Miller SP and Bochkarev A (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J, 21, 1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hass CS, Lam K and Wold MS (2012) Repair-specific functions of replication protein A. J Biol Chem, 287, 3908–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG and Hoeijmakers JH (1998) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev, 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iftode C and Borowiec JA (2000) 5’ --> 3’ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry, 39, 11970–11981. [DOI] [PubMed] [Google Scholar]
- 85.Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A and Lavrik OI (2001) Polarity of human replication protein A binding to DNA. Nucleic Acids Res, 29, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Safa L, Gueddouda NM, Thiébaut F, Delagoutte E, Petruseva I, Lavrik O, Mendoza O, Bourdoncle A, Alberti P, Riou J-F et al. (2016) 5’ to 3’ Unfolding Directionality of DNA Secondary Structures by Replication Protein A: G-QUADRUPLEXES AND DUPLEXES. The Journal of biological chemistry, 291, 21246–21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanyam S, Ismail M, Bhattacharya I and Spies M (2016) Tyrosine phosphorylation stimulates activity of human RAD51 recombinase through altered nucleoprotein filament dynamics. Proc Natl Acad Sci U S A, 113, E6045–e6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baumann P, Benson FE and West SC (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- 89.Kojic M, Kostrub CF, Buchman AR and Holloman WK (2002) BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell, 10, 683–691. [DOI] [PubMed] [Google Scholar]
- 90.New JH, Sugiyama T, Zaitseva E and Kowalczykowski SC (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- 91.Benson FE, Baumann P and West SC (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- 92.Shinohara A and Ogawa T (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- 93.Sung P (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem, 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- 94.Jensen RB, Carreira A and Kowalczykowski SC (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature, 467, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Doty T, Gibson B and Heyer WD (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol, 17, 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gibb B, Ye LF, Kwon Y, Niu H, Sung P and Greene EC (2014) Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat Struct Mol Biol, 21, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walther AP, Gomes XV, Lao Y, Lee CG and Wold MS (1999) Replication Protein A Interactions with DNA. 1. Functions of the DNA-Binding and Zinc-Finger Domains of the 70-kDa Subunit. Biochemistry, 38, 3963–3973. [DOI] [PubMed] [Google Scholar]
- 98.Fan J and Pavletich NP (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev, 26, 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blackwell LJ and Borowiec JA (1994) Human replication protein A binds single-stranded DNA in two distinct complexes. Mol Cell Biol, 14, 3993–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fanning E, Klimovich V and Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res, 34, 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chaudhuri J, Khuong C and Alt FW (2004) Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature, 430, 992–998. [DOI] [PubMed] [Google Scholar]
- 102.Zou L and Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science, 300, 1542–1548. [DOI] [PubMed] [Google Scholar]
- 103.Ball HL, Myers JS and Cortez D (2005) ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell, 16, 2372–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Namiki Y and Zou L (2006) ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci U S A, 103, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK and Brosh RM Jr. (2005) Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J Biol Chem, 280, 29494–29505. [DOI] [PubMed] [Google Scholar]
- 106.Wong JM, Ionescu D and Ingles CJ (2003) Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene, 22, 28–33. [DOI] [PubMed] [Google Scholar]
- 107.Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS and Hurwitz J (2008) Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J Biol Chem, 283, 20925–20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bae S-H, Bae K-H, Kim J-A and Seo Y-S (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412, 456–461. [DOI] [PubMed] [Google Scholar]
- 109.Bae KH, Kim HS, Bae SH, Kang HY, Brill S and Seo YS (2003) Bimodal interaction between replication‐protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Research, 31, 3006–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sparks JL, Kumar R, Singh M, Wold MS, Pandita TK and Burgers PM (2012) Human exonuclease 5 is a novel sliding exonuclease required for genome stability. The Journal of biological chemistry, 287, 42773–42783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP and Formosa T (2006) The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell, 22, 363–374. [DOI] [PubMed] [Google Scholar]
- 112.Liu S, Xu Z, Leng H, Zheng P, Yang J, Chen K, Feng J and Li Q (2017) RPA binds histone H3-H4 and functions in DNA replication–coupled nucleosome assembly. Science, 355, 415. [DOI] [PubMed] [Google Scholar]
- 113.Sommers JA, Banerjee T, Hinds T, Wan B, Wold MS, Lei M and Brosh RM Jr. (2014) Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J Biol Chem, 289, 19928–19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB and Brosh RM Jr. (2007) FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood, 110, 2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu Y, Shin-ya K and Brosh RM Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol, 28, 4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS and Brosh RM Jr. (2009) FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem, 284, 18458–18470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jeong YT, Rossi M, Cermak L, Saraf A, Florens L, Washburn MP, Sung P, Schildkraut CL and Pagano M (2013) FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. J Cell Biol, 200, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura S, Natsume T and Nakai A (2012) RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell, 48, 182–194. [DOI] [PubMed] [Google Scholar]
- 119.Guler GD, Liu H, Vaithiyalingam S, Arnett DR, Kremmer E, Chazin WJ and Fanning E (2012) Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J Biol Chem, 287, 6469–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tkac J, Xu G, Adhikary H, Young JTF, Gallo D, Escribano-Diaz C, Krietsch J, Orthwein A, Munro M, Sol W et al. (2016) HELB Is a Feedback Inhibitor of DNA End Resection. Mol Cell, 61, 405–418. [DOI] [PubMed] [Google Scholar]
- 121.Elia AEH, Wang DC, Willis NA, Boardman AP, Hajdu I, Adeyemi RO, Lowry E, Gygi SP, Scully R and Elledge SJ (2015) RFWD3-Dependent Ubiquitination of RPA Regulates Repair at Stalled Replication Forks. Molecular cell, 60, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.MacKay C, Toth R and Rouse J (2009) Biochemical characterisation of the SWI/SNF family member HLTF. Biochem Biophys Res Commun, 390, 187–191. [DOI] [PubMed] [Google Scholar]
- 123.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ and Cortez D (2008) The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol, 28, 7345–7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Robison JG, Elliott J, Dixon K and Oakley GG (2004) Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem, 279, 34802–34810. [DOI] [PubMed] [Google Scholar]
- 125.Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H and Zou L (2013) Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep, 3, 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oakley GG, Tillison K, Opiyo SA, Glanzer JG, Horn JM and Patrick SM (2009) Physical interaction between replication protein A (RPA) and MRN: involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry, 48, 7473–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Daniely Y and Borowiec JA (2000) Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J Cell Biol, 149, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M and Borowiec JA (2005) Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol Cell Biol, 25, 2463–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS and Lowry DF (2001) The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res, 29, 3270–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH et al. (2005) Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A, 102, 15412–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dutta A, Ruppert JM, Aster JC and Winchester E (1993) Inhibition of DNA replication factor RPA by p53. Nature, 365, 79–82. [DOI] [PubMed] [Google Scholar]
- 132.Li R and Botchan MR (1993) The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell, 73, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 133.Han Y, Loo YM, Militello KT and Melendy T (1999) Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J Virol, 73, 4899–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Loo YM and Melendy T (2004) Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J Virol, 78, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Christensen J and Tattersall P (2002) Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J Virol, 76, 6518–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Loor G, Zhang S-J, Zhang P, Toomey NL and Lee MYWT (1997) Identification of DNA Replication and Cell Cycle Proteins That Interact with PCNA. Nucleic Acids Research, 25, 5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dianov GL, Jensen BR, Kenny MK and Bohr VA (1999) Replication Protein A Stimulates Proliferating Cell Nuclear Antigen-Dependent Repair of Abasic Sites in DNA by Human Cell Extracts. Biochemistry, 38, 11021–11025. [DOI] [PubMed] [Google Scholar]
- 138.Braun KA, Lao Y, He Z, Ingles CJ and Wold MS (1997) Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry, 36, 8443–8454. [DOI] [PubMed] [Google Scholar]
- 139.Dornreiter I, Erdile LF, Gilbert IU, von Winkler D, Kelly TJ and Fanning E (1992) Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J, 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yuzhakov A, Kelman Z, Hurwitz J and O’Donnell M (1999) Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J, 18, 6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Feng J, Wakeman T, Yong S, Wu X, Kornbluth S and Wang XF (2009) Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress. Mol Cell Biol, 29, 5696–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J and Zou L (2014) PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell, 53, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wan L and Huang J (2014) The PSO4 protein complex associates with replication protein A (RPA) and modulates the activation of ataxia telangiectasia-mutated and Rad3-related (ATR). J Biol Chem, 289, 6619–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ellison V and Stillman B (2003) Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5’ recessed DNA. PLoS Biol, 1, E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zou L, Liu D and Elledge SJ (2003) Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A, 100, 13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Majka J, Binz SK, Wold MS and Burgers PM (2006) Replication protein A directs loading of the DNA damage checkpoint clamp to 5’-DNA junctions. J Biol Chem, 281, 27855–27861. [DOI] [PubMed] [Google Scholar]
- 147.Davies AA, Huttner D, Daigaku Y, Chen S and Ulrich HD (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Molecular cell, 29, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stauffer ME and Chazin WJ (2004) Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J Biol Chem, 279, 25638–25645. [DOI] [PubMed] [Google Scholar]
- 149.Golub EI, Gupta RC, Haaf T, Wold MS and Radding CM (1998) Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res, 26, 5388–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Davis AP and Symington LS (2003) The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair (Amst), 2, 1127–1134. [DOI] [PubMed] [Google Scholar]
- 151.Jackson D, Dhar K, Wahl JK, Wold MS and Borgstahl GE (2002) Analysis of the human replication protein A:Rad52 complex: evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J Mol Biol, 321, 133–148. [DOI] [PubMed] [Google Scholar]
- 152.Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM and Chazin WJ (2000) Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell, 103, 449–456. [DOI] [PubMed] [Google Scholar]
- 153.Park MS, Ludwig DL, Stigger E and Lee SH (1996) Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J Biol Chem, 271, 18996–19000. [DOI] [PubMed] [Google Scholar]
- 154.Hays SL, Firmenich AA, Massey P, Banerjee R and Berg P (1998) Studies of the interaction between Rad52 protein and the yeast single-stranded DNA binding protein RPA. Mol Cell Biol, 18, 4400–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T and Spies M (2010) Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res, 38, 2917–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu X, Shell SM and Zou Y (2005) Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene, 24, 4728–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cui S, Arosio D, Doherty KM, Brosh RM Jr., Falaschi A and Vindigni A (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res, 32, 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cui S, Klima R, Ochem A, Arosio D, Falaschi A and Vindigni A (2003) Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem, 278, 1424–1432. [DOI] [PubMed] [Google Scholar]
- 159.Garcia PL, Liu Y, Jiricny J, West SC and Janscak P (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J, 23, 2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P et al. (2007) RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev, 21, 3073–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim HS and Brill SJ (2001) Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol Cell Biol, 21, 3725–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gong Z and Chen J (2011) E3 ligase RFWD3 participates in replication checkpoint control. J Biol Chem, 286, 22308–22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu S, Chu J, Yucer N, Leng M, Wang SY, Chen BP, Hittelman WN and Wang Y (2011) RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J Biol Chem, 286, 22314–22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Galanty Y, Belotserkovskaya R, Coates J and Jackson SP (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev, 26, 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McDonald KR, Sabouri N, Webb CJ and Zakian VA (2014) The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA Repair (Amst), 24, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McDonald KR, Guise AJ, Pourbozorgi-Langroudi P, Cristea IM, Zakian VA, Capra JA and Sabouri N (2016) Pfh1 Is an Accessory Replicative Helicase that Interacts with the Replisome to Facilitate Fork Progression and Preserve Genome Integrity. PLoS Genet, 12, e1006238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Boule JB and Zakian VA (2007) The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res, 35, 5809–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW and Elledge SJ (2009) The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev, 23, 2415–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bansbach CE, Betous R, Lovejoy CA, Glick GG and Cortez D (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev, 23, 2405–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yusufzai T, Kong X, Yokomori K and Kadonaga JT (2009) The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev, 23, 2400–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weisshart K, Taneja P and Fanning E (1998) The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J Virol, 72, 9771–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arunkumar AI, Klimovich V, Jiang X, Ott RD, Mizoue L, Fanning E and Chazin WJ (2005) Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome. Nat Struct Mol Biol, 12, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Park CJ, Lee JH and Choi BS (2005) Solution structure of the DNA-binding domain of RPA from Saccharomyces cerevisiae and its interaction with single-stranded DNA and SV40 T antigen. Nucleic Acids Res, 33, 4172–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A and Kaufmann WK (2007) The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol, 27, 3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nagelhus TA, Haug T, Singh KK, Keshav KF, Skorpen F, Otterlei M, Bharati S, Lindmo T, Benichou S, Benarous R et al. (1997) A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem, 272, 6561–6566. [DOI] [PubMed] [Google Scholar]
- 176.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O and Krokan HE (1999) Post-replicative base excision repair in replication foci. EMBO J, 18, 3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shen JC, Lao Y, Kamath-Loeb A, Wold MS and Loeb LA (2003) The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev, 124, 921–930. [DOI] [PubMed] [Google Scholar]
- 178.Yang ZG, Liu Y, Mao LY, Zhang JT and Zou Y (2002) Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry, 41, 13012–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stigger E, Drissi R and Lee SH (1998) Functional analysis of human replication protein A in nucleotide excision repair. J Biol Chem, 273, 9337–9343. [DOI] [PubMed] [Google Scholar]
- 180.Li L, Lu X, Peterson CA and Legerski RJ (1995) An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol, 15, 5396–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Daughdrill GW, Buchko GW, Botuyan MV, Arrowsmith C, Wold MS, Kennedy MA and Lowry DF (2003) Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res, 31, 4176–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bessho T, Sancar A, Thompson LH and Thelen MP (1997) Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem, 272, 3833–3837. [DOI] [PubMed] [Google Scholar]
- 183.Matsunaga T, Park CH, Bessho T, Mu D and Sancar A (1996) Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem, 271, 11047–11050. [DOI] [PubMed] [Google Scholar]
- 184.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM and Wood RD (1995) Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- 185.Maréchal A and Zou L (2015) RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Research, 25, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Awate S and Brosh RM Jr. (2017) Interactive Roles of DNA Helicases and Translocases with the Single-Stranded DNA Binding Protein RPA in Nucleic Acid Metabolism. Int J Mol Sci, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Plate I, Hallwyl SCL, Shi I, Krejci L, Müller C, Albertsen L, Sung P and Mortensen UH (2008) Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. The Journal of biological chemistry, 283, 29077–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P and Krejci L (2008) Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol Chem, 283, 12166–12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ma CJ, Kwon Y, Sung P and Greene EC (2017) Human RAD52 interactions with Replication Protein A and the RAD51 presynaptic complex. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Patrick SM and Turchi JJ (2002) Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA-DNA interactions via enhanced complex stability and inhibition of strand separation activity. J Biol Chem, 277, 16096–16101. [DOI] [PubMed] [Google Scholar]
- 191.Collins KL and Kelly TJ (1991) Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Molecular and cellular biology, 11, 2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Taylor MRG and Yeeles JTP (2018) The Initial Response of a Eukaryotic Replisome to DNA Damage. Molecular cell, 70, 1067–1080.e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P and Krejci L (2008) Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. The Journal of biological chemistry, 283, 12166–12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Miller SD, Moses K, Jayaraman L and Prives C (1997) Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single-stranded DNA. Molecular and cellular biology, 17, 2194–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Romanova LY, Willers H, Blagosklonny MV and Powell SN (2004) The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene, 23, 9025–9033. [DOI] [PubMed] [Google Scholar]
- 196.Huttner D and Ulrich HD (2008) Cooperation of replication protein A with the ubiquitin ligase Rad18 in DNA damage bypass. Cell Cycle, 7, 3629–3633. [DOI] [PubMed] [Google Scholar]
- 197.Hedglin M, Aitha M, Pedley A and Benkovic SJ (2019) Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen. The Journal of biological chemistry, 294, 5157–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Patrone JD, Kennedy JP, Frank AO, Feldkamp MD, Vangamudi B, Pelz NF, Rossanese OW, Waterson AG, Chazin WJ and Fesik SW (2013) Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A. ACS Med Chem Lett, 4, 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Din S, Brill SJ, Fairman MP and Stillman B (1990) Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev, 4, 968–977. [DOI] [PubMed] [Google Scholar]
- 200.Oakley GG, Patrick SM, Yao J, Carty MP, Turchi JJ and Dixon K (2003) RPA phosphorylation in mitosis alters DNA binding and protein-protein interactions. Biochemistry, 42, 3255–3264. [DOI] [PubMed] [Google Scholar]
- 201.Liu VF and Weaver DT (1993) The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol, 13, 7222–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liaw H, Lee D and Myung K (2011) DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PloS one, 6, e21424–e21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carty MP, Zernik-Kobak M, McGrath S and Dixon K (1994) UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J, 13, 2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vassin VM, Wold MS and Borowiec JA (2004) Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol, 24, 1930–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Soniat MM, Myler LR, Kuo HC, Paull TT and Finkelstein IJ (2019) RPA Phosphorylation Inhibits DNA Resection. Mol Cell, 75, 145–153.e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sanford EJ, Faça VM, Vega SC, Comstock WJ and Smolka MB (2020) Phosphoproteomics Reveals a Distinct Mode of Mec1/ATR Signaling in Response to DNA End Hyper-Resection. bioRxiv, 2020.2004.2017.028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Brush GS, Morrow DM, Hieter P and Kelly TJ (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci U S A, 93, 15075–15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Brush GS and Kelly TJ (2000) Phosphorylation of the replication protein A large subunit in the Saccharomyces cerevisiae checkpoint response. Nucleic Acids Res, 28, 3725–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316, 1160–1166. [DOI] [PubMed] [Google Scholar]
- 210.Bartrand AJ, Iyasu D and Brush GS (2004) DNA stimulates Mec1-mediated phosphorylation of replication protein A. J Biol Chem, 279, 26762–26767. [DOI] [PubMed] [Google Scholar]
- 211.Dou H, Huang C, Singh M, Carpenter PB and Yeh ETH (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Molecular cell, 39, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Psakhye I and Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell, 151, 807–820. [DOI] [PubMed] [Google Scholar]
- 213.Zhao M, Geng R, Guo X, Yuan R, Zhou X, Zhong Y, Huo Y, Zhou M, Shen Q, Li Y et al. (2017) PCAF/GCN5-Mediated Acetylation of RPA1 Promotes Nucleotide Excision Repair. Cell Rep, 20, 1997–2009. [DOI] [PubMed] [Google Scholar]
- 214.He H, Wang J and Liu T (2017) UV-Induced RPA1 Acetylation Promotes Nucleotide Excision Repair. Cell Rep, 20, 2010–2025. [DOI] [PubMed] [Google Scholar]
- 215.van de Meent JW, Bronson JE, Wiggins CH and Gonzalez RL Jr. (2014) Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments. Biophys J, 106, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]