Abstract
Gut microbiomes, such as the microbial community that colonizes the rumen, have vast catabolic potential and play a vital role in host health and nutrition. By expanding our understanding of metabolic pathways in these ecosystems, we will garner foundational information for manipulating microbiome structure and function to influence host physiology. Currently, our knowledge of metabolic pathways relies heavily on inferences derived from metagenomics or culturing bacteria in vitro. However, novel approaches targeting specific cell physiologies can illuminate the functional potential encoded within microbial (meta)genomes to provide accurate assessments of metabolic abilities. Using fluorescently labeled polysaccharides, we visualized carbohydrate metabolism performed by single bacterial cells in a complex rumen sample, enabling a rapid assessment of their metabolic phenotype. Specifically, we identified bovine-adapted strains of Bacteroides thetaiotaomicron that metabolized yeast mannan in the rumen microbiome ex vivo and discerned the mechanistic differences between two distinct carbohydrate foraging behaviors, referred to as “medium grower” and “high grower.” Using comparative whole-genome sequencing, RNA-seq, and carbohydrate-active enzyme fingerprinting, we could elucidate the strain-level variability in carbohydrate utilization systems of the two foraging behaviors to help predict individual strategies of nutrient acquisition. Here, we present a multi-faceted study using complimentary next-generation physiology and “omics” approaches to characterize microbial adaptation to a prebiotic in the rumen ecosystem.
Video abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-020-00975-x.
Keywords: Microbiome, Carbohydrate, Rumen, Glycoside hydrolase, Fluorescent polysaccharides, Yeast mannan, Bacteroides
Background
Ruminants have evolved foregut digestive systems specialized in the bioconversion of recalcitrant, complex carbohydrates into energy. These catabolic processes rely on a core bacterial community composed predominantly of the genera Prevotella, Butyrivibrio, Fibrobacter, and Ruminococcus, families Lachnospiraceae and Ruminococcaceae, and orders Bacteroidales and Clostridiales [1, 2]. The rumen microbiome is estimated to contain 69,000 carbohydrate-active enzyme (CAZyme) genes [3] that encode extensive catalytic activities. Despite this vast genetic repertoire and catalytic potential, the microbial conversion of plant fiber to host-accessible metabolites in the rumen is suboptimal and could be improved [4, 5]. For example, supplementation with direct fed microorganisms, such as Bacteroides spp., or prebiotic carbohydrates that modify the rumen microbiome to enhance feed conversion may help address the emerging challenges associated with sustainable production of food animals [6].
In diverse animal symbioses and environmental ecosystems, Bacteroides spp. and other members of Bacteroidetes are thought to play central roles in glycan digestion because they encode highly specialized carbohydrate metabolic systems called polysaccharide utilization loci (PULs) [7, 8]. The first described PUL was the starch utilization system of Bacteroides thetaiotaomicron (B. theta: BtVPI-5482) [9], and since its description, PULs that metabolize glycans with unique chemistries have been found in diverse ecosystems [8, 10–12]. PULs are distinguished by the presence of a TonB-dependent transporter coupled to a surface glycan-binding protein, known as the SusC/D-like complex, and other associated proteins that modify or bind the target glycan. These gene products function together in an orchestrated cascade to transport oligosaccharides into the periplasm where monosaccharides are released from polymeric substrates and used for primary metabolism. PULs can operate through a “distributive” mechanism, which releases products [13] to the microbial community or a “selfish” mechanism [14], which limits product loss by confining substrate depolymerization within the cell [8, 15]. Recently, PUL-prediction [16] and whole-PUL characterization [14, 17] have become common approaches for the discovery of new CAZyme families and catalytic activities at the species [18] and strain levels [19, 20]. The most common enzymes encoded within PULs are glycoside hydrolases (GHs), which cleave glycosidic bonds by acid-base catalysis [21]. GHs are divided into sequence-related families that display conserved folds, mechanisms, and catalytic residues. However, these features are not necessarily representative of function as many different GH families are polyspecific [22].
In addition to microorganisms that improve the efficiency of digestion, prebiotic glycans, such as yeast α-mannan (YM) and its derivative oligosaccharides (i.e., α-mannanoligosaccharides), are known to provide beneficial physiological outcomes to animals, such as cattle and pigs [23–25]. Prebiotics can enhance feed digestion and cattle health by becoming selective nutrients for symbiotic gut bacteria, such as Bacteroides spp. The digestion of YM requires a collection of CAZymes targeting distinct linkages using different modes of activity, including α-mannanases and α-mannosidases [15]. CAZymes that possess these activities are commonly found in family GH38, GH76, GH92, GH99, and GH125 [14, 26, 27], and correspondingly, these enzymes are present in PULs that target YM (i.e., MAN-PULs).
YM-specific CAZymes and PULs are widely distributed in Bacteroidetes; however, individual species differ in their abilities to consume α-mannans depending on the structural complexity of the substrate [14]. For example, Bacteroides xylanisolvens NLAE-zl isolated from pigs reared on a diet infused with distillers’ grains could only metabolize debranched YM [14]. The pathway responsible for YM catabolism in these strains (i.e., MAN-PUL1) was encoded on a transposable element, suggesting that aspects of YM metabolism can be exchanged between strains [14]. This finding is consistent with reports of specialized metabolic abilities being transferred to intestinal Bacteroides spp. from species that occupy ecologically distinct habitats [19, 20, 28], facilitating their persistence within highly competitive ecosystems and adaption to spatially and culturally diversified diets.
Although major advances have been made in understanding the diversity of metabolic potential in symbiotic bacteria and the mechanisms of prebiotic utilization, establishing stable engineered microbiomes in complex ecosystems, such as the rumen, will require more detailed knowledge of the competitive and complementary processes that drive metabolic phenotypes at the strain level. To achieve this, “next-generation physiology”-based [29] approaches that identify metabolic potentials of individual bacteria, thereby providing critical insights of cellular functions and assigning cellular phenotypes, must be developed. One such approach is fluorescently labeled polysaccharides (FLA-PS). FLA-PS were initially developed to demonstrate selfish uptake of marine polysaccharides in marine Bacteroidetes [30] and have also been applied to the gut bacterium BtVPI-5482 to confirm that YM metabolism also occurs through a selfish mechanism [31]. Fluorescent glucose analogs have been recently used to study glucose uptake by rumen bacteria [32]; however, use of fluorescent polysaccharides in the rumen has been limited until now.
Here, for the first time, we apply FLA-PS as a next-generation physiology approach to directly visualize YM metabolism by single cells in a complex rumen community and subsequently classify populations of cells using fluorescence in situ hybridization (FISH). We combine this analysis with a multi-tiered study of the evolution and function of YM metabolism in bovine-adapted B. theta strains (BtBov), which adopt one of two dichotomous growth phenotypes, referred to as “High Grower” (HG) or “Medium Grower” (MG), based on the optical density of cultures after 24 h. Despite displaying distinct growth profiles, the genetic, transcriptomic, or biochemical factors that contributed to the differential growth phenotypes of these strains remained to be defined. Using genomics, transcriptomics, and CAZyme fingerprinting, multiple MAN-PUL architectures were identified in this study that are consistent with reports for human-associated BtVPI-5482 [14] and key differences in the YM utilization systems between MGs and HGs were revealed. To define the mechanisms that contribute to these growth phenotypes, we present a new quantitative application of FLA-PS, which we believe has far-reaching implications for elucidating differences in substrate utilization of individual cells within complex microbial communities.
Results
Ex vivo visualization of YM-metabolizing taxa within the rumen community
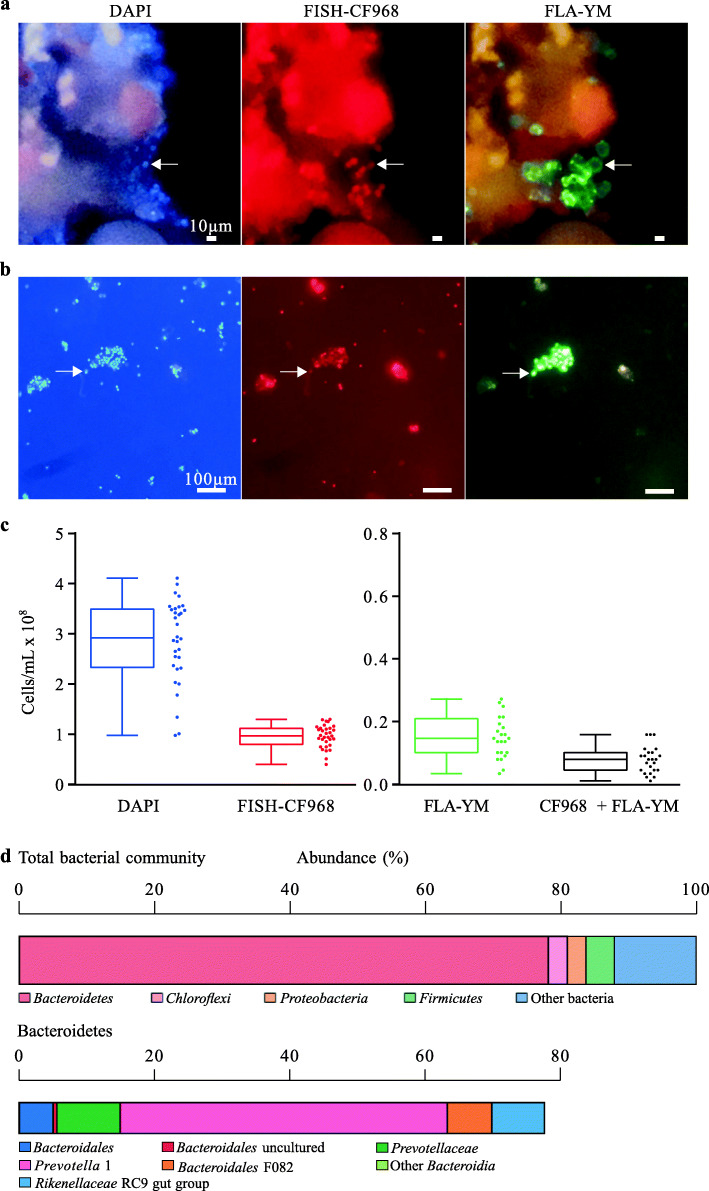
To assess the capability of rumen microbiota to metabolize YM, extracted rumen samples were incubated with FLA-YM and visualized on feed particles and in solution (Fig. 1a, b). The total cell density in 100 μm pre-filtered rumen fluid, as determined by enumerating DAPI-stained cells, was 2.98 × 108 ± 6.02 × 107 cells ml−1 (Fig. 1c). In these complex communities, on average 6.1% ± 0.5% of cells showed uptake of FLA-YM (0% after 15 min, 6% after 3 h, 7% after 1 day, 6% after 3 days). Fluorescence in situ hybridization (FISH) using the CF968 probe [33] specific for the phylum Bacteroidetes showed that 2.9 ± 0.5% of the cells showing FLA-YM uptake were members of the Bacteroidetes. In total, Bacteroidetes made up 34.8% ± 6.8% of the rumen bacterial community and only a fraction of these (~ 3%) showed uptake of FLA-YM (Fig. 1c). The microbial community composition of these rumen samples was determined by 16S rRNA metagenomics sequencing. The community was dominated by Bacteroidetes, specifically the genus Prevotella 1, which demonstrated that YM metabolism has penetrated distantly related members of the phylum (Fig. 1d).
Fig. 1.
YM utilization by BtBov isolates. Images of a 100 μm on food particles and b 10 μm filtered rumen extract incubated with FLA-YM and stained with DAPI and Bacteroides-FISH probe (FISH-CF968). Cells were visualized by epifluorescence microscopy. c Counts of cells from FLA-YM incubated rumen extract stained with DAPI, FISH-CF968, FLA-YM, and both FISH-CF968 and FLA-YM. Mean ± standard deviation shown. d 16S rRNA metagenomics sequencing data of extracted rumen communities showing most prevalent phyla (left) and the distribution of Bacteroidetes spp. (right). N = 4
Isolation and growth profiling of YM-utilizing BtBov strains
Targeted isolation approaches were performed to selectively isolate bovine-adapted-bacteria that utilize Saccharomyces cerevisiae YM from enriched rumen and fecal communities. Single colonies were observed within 24 h, with new colonies forming up to 96 h. In total, 50 bacterial isolates were collected and each mannan-degrading (MD) isolate was assigned a reference number (e.g., isolate #8 = MD8). The majority of these isolates were identified by 16S rRNA gene sequencing as strains of B. theta using the NCBI BLASTN database [34], and referred to as BtBov (Fig. 2a, Supplementary Table 1).
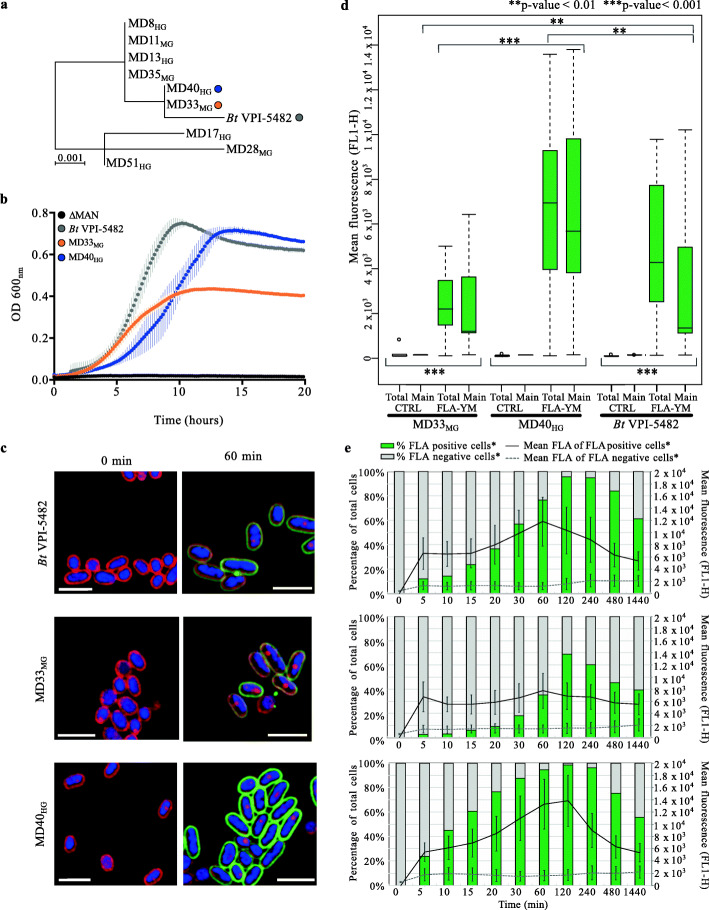
Fig. 2.
Characterization of BtBov isolates. a 16S rRNA gene sequence comparison of BtVPI-5482 and BtBov strains. Scale represents number of nucleotide changes across horizontal axis. b Growth profiles of BtVPI-5482, BtΔMAN-PUL1/2/3, MD33MG, and MD40HG grown on 0.5% YM-MM. c SR-SIM of B. theta strains 0-min and 60-min post-incubation with FLA-YM. Cells co-stained with Nile Red and DAPI. d Mean fluorescence of bacterial cells cultured in FLA-YM or YM-MM (control). **p value < 0.01; ***p value < 0.001; ns p value > 0.05. e Bars represent percentage of total cells showing an uptake of FLA-YM in cultures sampled over time. Green = increased FLA-YM uptake; grey = no uptake. Mean cell fluorescence represented by line graph; solid line = mean of cells with FLA-YM signal; dotted line = mean of cells showing no FLA-YM uptake
YM metabolism was confirmed for each MD strain by growth in liquid cultures using S. cerevisiae YM as the sole carbon source (Fig. 2b, Supplementary Fig. 1). Interestingly, based on their growth on S. cerevisiae YM, the BtBov isolates and BtVPI-5482 control strain were divided into two populations (Supplementary Table 2): “Medium Growers” (MGs; plateaued growth at OD600 ~ 0.4 after 24 h) and “High Growers” (HGs; plateaued growth at OD600 ~ 0.7 after 24 h). Notably, this growth phenotype is substrate specific and does not extend to other substrates, such as mannose in which all strains have a similar growth curve (data not shown), and YM from Schizosaccharomyces pombe (S. pombe; Supplementary Fig. 1b). In addition, the 16S rRNA gene topology of the BtBov isolates did not reveal a discernable relationship with the growth phenotype (Fig. 2a).
Visualization and quantification of differential FLA-YM uptake by BtBov isolates
To determine if the rate of glycan uptake varied between the two growth types, representative strains, one from each growth population and BtVPI-5482 as a control, were incubated with FLA-YM. BtVPI-5482, MD33MG, and MD40HG cells became fluorescent, whereas cells incubated with unlabeled YM did not (Fig. 2c). Phenotypic differences in FLA-YM uptake over time were determined by splitting cell populations by flow cytometric gating into FLA-positive and FLA-negative cells (Supplementary Fig. 2a). The total fluorescence intensity (rate of uptake) was significantly different between the representative strains (MD40HG vs. MD33MG t(10) = 4.4, p value = 0.001; MD40HG vs. BtVPI-5482 t(10) = 3.9, p value = 0.003; BtVPI-5482 vs. MD33MG t(10) = 4.0, p value = 0.002) (Fig. 2d). The change in mean fluorescence of the three strains showed a similar temporal pattern, increasing from 0 to 120 min, peaking at 120 min, and declining from 120 to 1440 min (Fig. 2e). Although all three strains showed uptake of FLA-YM after 60 min, MD33MG had the lowest fluorescence intensity at each time point. MD40HG displayed the highest fluorescence intensity, 2.7-fold higher than MD33MG. BtVPI-5482 displayed a fluorescence intensity between the two bovine strains for each time point, with a peak value 1.9-fold higher than MD33MG. Additionally, MD40HG cells showed a more rapid uptake (22% at 5 min), BtVPI-5482 cells showed an intermediate rate of uptake (9% at 5 min), and MD33MG had the slowest uptake rate (2% at 5 min) (Fig. 2e, Supplementary Fig. 2b).
To test if the phenotypic differences were inherited between generations, we measured the differences in FLA-YM uptake with and without prior exposure to YM. The cultures continued to display the same phenotypic uptake patterns (MD40HG and BtVPI-5482 higher uptake, MD33MG low uptake, Fig. 3a–c). However, previous exposure to YM resulted in a heightened cellular response as indicated by more rapid rates of FLA-YM uptake relative to cultures previously grown on mannose-MM (Fig. 3b–d). All cultures grown on mannose-MM reached a lower mean fluorescence and the temporal change in mean fluorescence was slower, with quantifiable uptake occurring only after 4 to 8 h.
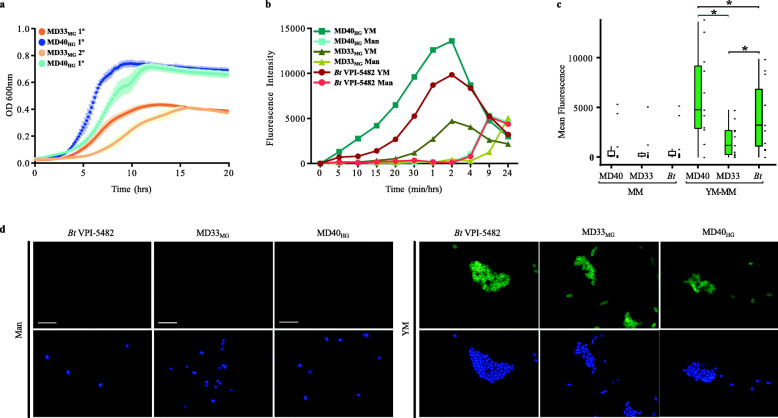
Fig. 3.
Reproducible YM foraging behaviors in BtBov isolates. a Growth profiles of MD33MG and MD40HG inoculated from cultures cultivated overnight on mannose-MM (1°) or YM-MM (2°). b Fluorescence intensity of individual cells of BtBov strains incubated in FLA-YM. Cells used to inoculate FLA-YM cultures were previously grown overnight in either YM-MM or Mannose-MM (Man) (N = 1). c Mean fluorescence of cells cultured in FLA-YM with significance (p < 0.05) between the cultures previously grown on YM (right) indicated by asterisk (N = 10,000). d SR-SIM images of BtVPI-5482, MD33MG, and MD40HG cultured in Man or YM and incubated with FLA-YM
Characterization of genotypes by PUL delineation
Whole-genome sequencing and de novo assembly were used to identify genes involved in YM metabolism. SPAdes [35] assembly output and average nucleotide identity based on BLAST+ (ANIb) [36] are shown in Supplementary Table 2. The ANIb results supported the 16S rRNA gene sequence data, confirming that each isolate was a strain of B. theta. Furthermore, comparative genomics revealed that these strains have acquired unique CAZome repositories (Fig. 4a) and PUL updates (Supplementary Fig. 3); features that may assist with their colonization of the bovine gut and represents opportunities for developing bovine-adapted probiotics (Supplementary Discussion).
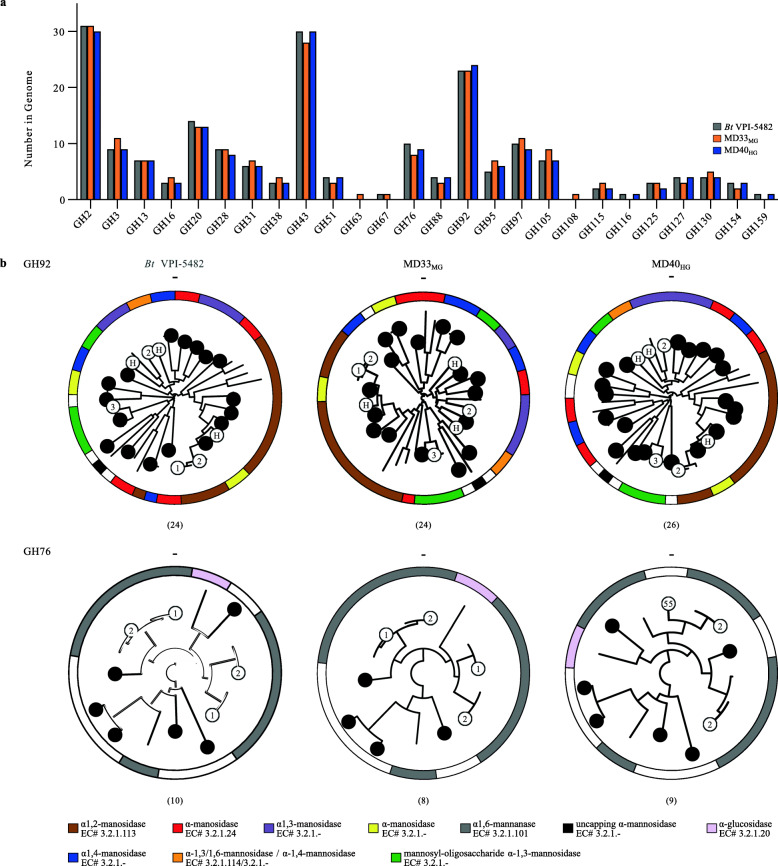
Fig. 4.
CAZyme fingerprinting of YM metabolism by BtBov isolates. a GH enzyme families encoded within the genomes of MD40HG and MD33MG that differ in total number of sequences. BtVPI-5482 sequences are provided as a reference for each GH family. b Phylogenetic trees of characterized GH92s and GH76s, and BtBov sequences generated with SACCHARIS [37]. Circles represent BtBov sequences and white circles highlight sequences from PULs with mannan activity: 1, 2, or 3 = MAN-PULs 1, 2, or 3, respectively; H = HMNG-PUL; 55 = BtMD40 PUL55. Activities assigned to characterized enzymes within each clade for GH92 and GH76 are depicted using the provided legend. Outer ring represents characterized specificities. Numbers in parenthesis indicate the total number of enzymes within each strain
Reconstruction of the three YM-specific PULs and alignment with BtVPI-5482 MAN-PULs determined that there was a high level of synteny among all strains in these pathways (Supplementary Fig. 4a). MAN-PUL2 and MAN-PUL3 were absolutely conserved, whereas MAN-PUL1, a PUL tailored for the consumption of mannan from S. pombe [14] (Supplementary Fig. 1b), was only present in BtVPI-5482, MD33MG, and MD35MG. The presence of MAN-PUL1 in two MGs indicated this pathway was not responsible for the HG phenotype. The HMNG-PUL, which is specific for digestion of high mannose N-glycans and not activated by YM in BtVPI-5482 [14], was also conserved in each of the BtBov genomes.
CAZome fingerprinting
To determine if there was amino acid sequence divergence within MAN-PULs, and potentially the function of homologous enzymes, polyspecific CAZyme families GH92 and GH76 were analyzed by SACCHARIS [37]. Enzyme sequences from GH92 and GH76 were embedded into phylogenetic trees comprised of all characterized enzyme sequences from each family (Supplementary Fig. 4b, c). Notably, every sequence within MAN-PUL1, MAN-PUL2, and MAN-PUL3 displayed the highest level of amino acid sequence conservation with its syntenic homolog. This suggested that each PUL is under strong selective pressure to function as an intact catabolic system. To determine if CAZyme sequences were conserved in other potential α-mannan-degrading PULs, a genome-wide approach (i.e., CAZome fingerprinting) was used [37]. Each isolate encoded between twenty-four and twenty-six GH92s and eight or nine GH76s (Fig. 4b, Supplementary Fig. 4b,c). Only MD17HG and MD51HG displayed identical conservation for GH76, whereas every GH92 tree was unique. Topological differences were observed for other α-mannan active enzyme families (e.g., GH38, GH99, and GH125), suggesting that despite the high level of functional conservation within the MAN-PULs, metabolic specialization in α-mannan consumption between these strains may be encoded within orphan PULs [38]. Therefore, the contributions of two exogenous GH76s to the foraging behavior of HGs and MGs were investigated. BtGH76-MD40 is a surface-exposed GH76 inserted into PUL55 of HGs (Fig. 4b) and is active on intact S. cerevisiae and S. pombe YM [39]. BT_3782 is a periplasmic endo-α-mannanase that generates small oligosaccharide products [14]. Addition of recombinant BtGH76-MD and BT_3782 to pure cultures of MD33MG did not augment the MG growth phenotype (Supplementary Fig. 5), suggesting that acquisition of BtGH76-MD40 or augmented endo-mannanase activity were not responsible for the HG phenotype.
Differences in YM import between phenotypes
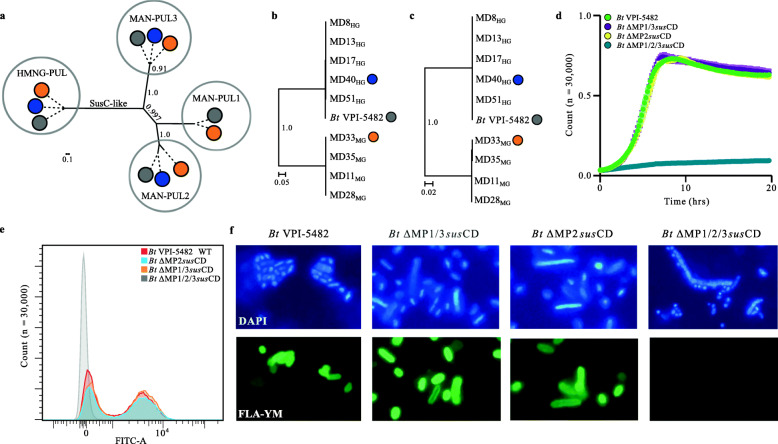
The differential transport kinetics of FLA-YM (Fig. 2c–e) and absence of genetic differences in PUL structure between phenotypes (Supplementary Fig. 4a) suggested that glycan transport processes may be responsible for the MG and HG growth phenotypes. Alignment of the SusC-like amino acid sequences from MAN-PULs 1, 2, and 3 and HMNG-PUL from BtVPI-5482, MD33MG, and MD40HG revealed that proteins cluster into functional clades (Fig. 5a). Furthermore, when the SusC-like and SusD-like transport proteins from each MD strain were aligned with BT_3788 (Fig. 5b) and BT_3789 (Fig. 5c) of BtVPI-5482, respectively, the proteins partitioned exclusively into clades associated with either the MG or HG phenotype. This result is in contrast with the 16S rRNA (Fig. 2a) and whole-genome (Supplementary Table 2) alignments, which showed no correlation with growth profiles. Interestingly, this pattern does not exist for MAN-PUL1 or MAN-PUL3 as the SusC-like proteins in these pathways are highly conserved (Fig. 5a), suggesting that syntenic conservation may not always reflect sequence-function relationships.
Fig. 5.
Divergence of MAN-PUL SusC/D-like proteins. a Phylogenetic tree of all the SusC-like MAN-PUL protein sequences from BtVPI-5482 (grey), MD33MG (orange), and MD40HG (blue). Phylogenetic trees of b SusC-like and c SusD-like amino acid sequences encoded in MAN-PUL2 of each BtBov strain. Bootstrap values above 70% are indicated at branch points. Scale represents number of amino acid substitutions per site. d Growth of BtVPI-5482 wild-type and MAN-PUL susC/D-like mutants on 0.5% YM-MM. e Enumeration (N = 30,000) and f epifluorescence microscopy images of the uptake of FLA-YM by BtVPI-5482 wild-type and mutant strains after 2 h incubation. Cells counterstained with DAPI (blue)
To study how transporters affect YM uptake, different combinations of susC/D-like genes were excised from the BtVPI-5482 MAN-PULs. Three mutant strains were produced: a MAN-PUL2 susC-like and susD-like gene knock-out strain (ΔMP2susCD), a MAN-PUL1 and 3 susC-like and susD-like deletion mutant (ΔMP1/3susCD), and a strain with all three sets of susC-like and susD-like genes deleted (ΔMP1/2/3susCD). Because MAN-PUL1 is absent in every HG except BtVPI-5482, we can conclude it has no effect on YM transport efficiency and that the ΔMP1/3susCD mutant essentially operates as a BtBov MAN-PUL3 susCD knock-out strain. The mutants, along with BtVPI-5482, were grown on YM-MM to assess how the loss of transport complexes impacted growth on YM (Fig. 5d). Surprisingly, the mutants retained an identical growth profile to the wild-type, with the exception of the triple knock-out mutant (ΔMP1/2/3susCD), which displayed no growth. Furthermore, when the mutants were incubated with FLA-YM, to study the impact on uptake rates, they displayed identical rates to the wild-type, with only the triple deletion mutant having a complete loss of FLA-YM import (Fig. 5e, f). These results demonstrated that the SusC-like/SusD-like proteins from MAN-PUL2 and MAN-PUL3 in BtVPI-5482 are functionally redundant. Although the absence of genetic tools prevented the investigation of the interplay between the MD33MG transporters, the sequence divergence existing between MAN-PUL2 SusC/D/E-like proteins from MD33MG and MD40HG (Supplementary Table 4) suggested that the dichotomous MG and HG growth phenotypes may result from differential transport through these complexes.
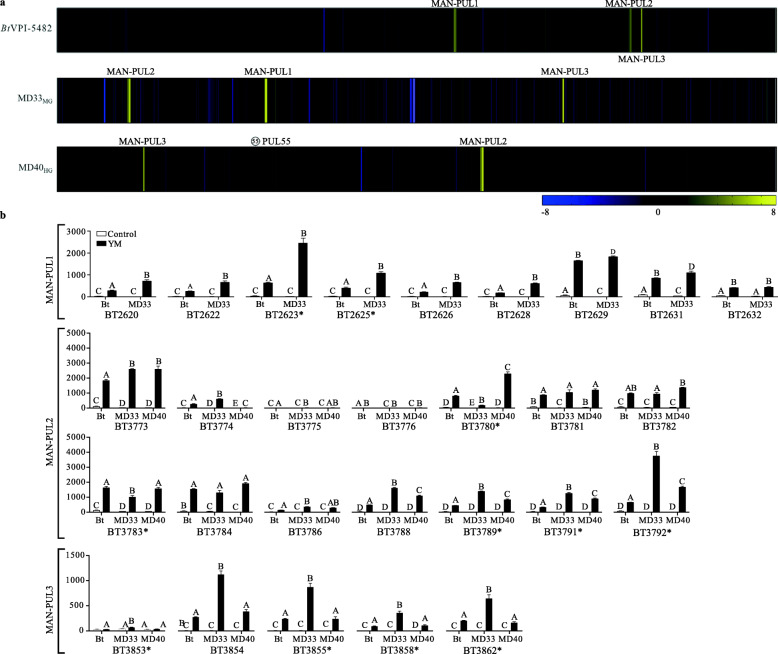
Comparative analysis of gene expression between BtBov growth phenotypes
RNA-seq was performed on BtVPI-5482, MD33MG, and MD40HG cultured on either mannose or YM to explore differential patterns in expression of the enzymes and transporters in the MAN-PULs and identify any distally expressed genes. MAN-PUL2 and MAN-PUL3 pathways were activated in all three bacteria, and MAN-PUL1 was activated in BtVPI-5482 and MD33MG (Fig. 6a, Supplementary Fig. 6) consistent with previous reports for BtVPI-5482 [18] (see Supplementary Discussion). To confirm that gene expression was representative of protein production, a C-Myc tag was fused to the C-terminal of the MAN-PUL2 SusD-like protein (BT_3789) in the chromosome of BtVPI-5482. Extracellular display of BT_3789 was demonstrated using antibodies directed at C-Myc when this bacterium was cultured on YM but not mannose (Supplementary Fig. 7).
Fig. 6.
RNA-seq analysis of BtVPI-5482, MD33MG, and MD40HG cultured in YM. a Log2 expression ratios of each transcript of BtVPI-5482, MD33MG, and MD40HG. Length of heap map is representative of genome size (BtVPI-5482 = 6.26 Mb, MD33MG = 6.28 Mb, MD40HG = 6.16 Mb). Location of MAN-PULs and MD40-PUL55 (white star) shown. Yellow indicates an increase in transcript expression compared to the control, while blue is decreased expression. b TPM values of susC-like genes from MAN-PUL1/2/3 showing if values within each gene are significantly different between the strains; different letters represent statistically significant values (e.g., A vs B) for each separate gene histogram; asterisk indicates SPII predicted proteins. Log2 fold change compares gene expression of YM cultures normalized to mannose, N = 3, p ≤ 0.01; except BT_3853 and HMNG-PUL, gene expression is not significantly (p > 0.05) different between the two treatments. Significantly different (p < 0.05) TPM expression of the susC-like genes between BtVPI-5482 and BtBov strains; statistical comparison of other MAN-PUL genes between strains were not calculated
The TPM values for every homologous gene transcript from MD33MG, MD40HG, and BtVPI-5482 were analyzed. Surprisingly, the sus-like genes (BT_3788 and BT_3789) and the surface enzyme transcripts (BT_3792, BT_2623, and BT_3858) of MD33MG consistently displayed significantly higher expression levels than the HG strains (Fig. 6b, Supplementary Fig. 6b). These values ranged between 6.2-log2 and 7.7-log2, suggesting that the expression level of gene products involved in outer membrane processing and intracellular transport is negatively correlated with growth proficiency on YM. The only example of an enzyme that is expressed at a significantly higher level in the MD40HG strain was BT_3780 (12.5-fold higher than MD33MG), which encodes a GH130 that is active on β-1,2-mannosides [40].
Differences in YM hydrolysis and import
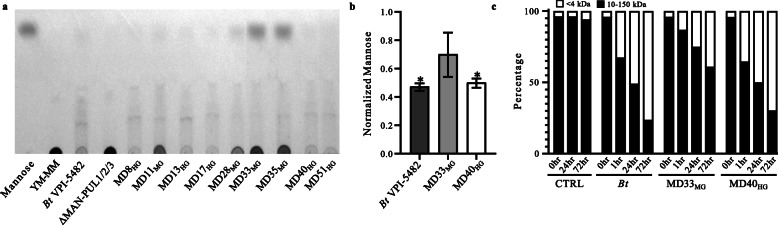
The enzymatic processing of YM (amount of YM products, extent of YM utilization, and total free mannose present in the post-growth supernatants) by each culture was analyzed using a combination of methods. Thin layer chromatography (TLC) (Fig. 7a) revealed that there was no detectable free mannooligosaccharides or mannose in the supernatant of the YM-MM negative control. Consistent with this observation, the BtMAN-PUL1/2/3 deletion mutant (∆MAN-PUL1/2/3) did not grow on YM and was unable to release products into the medium. BtVPI-5482 and each of the HGs generated a similar product profile, with a noticeable loss of YM signal and faint detection of oligosaccharides and mannose. In contrast, the post-growth media of MGs contained more mannose and had residual YM (Fig. 7a). Gas chromatography-mass spectrometry determined that the quantity of total mannosides (YM and oligosaccharides) in the supernatant was 1.48 and 1.40-fold higher for MD33MG (1.36 ± 0.29) than BtVPI-5482 (0.92 ± 0.04) and MD40HG (0.97 ± 0.05), respectively (Fig. 7b). Furthermore, post-growth BtVPI-5482 and MD40HG cultures, but not MD33MG, showed (p < 0.05) lower total mannose concentration in the media relative to the YM-MM negative control (Fig. 7b). This suggests that, consistent with their higher growth densities (Fig. 2b) and thin layer chromatography, BtVPI-5482 and MD40HG consume more YM.
Fig. 7.
Differential transport of YM by BtBov strains. a TLC analysis of post-growth supernatants (50 h) of MD isolates grown on 0.5% YM-MM. B. theta = BtVPI-5482, ΔMAN = BtΔMAN-PUL1/2/3, Man = mannose standard. b Quantification of total mannose (free and polymerized) in supernatants of BtVPI-5482, MD33MG, and MD40HG grown in 0.5% YM-MM for 21 h. Normalized to a no-cell control, N = 2, asterisk indicates significant difference (p < 0.05) relative to control. c Percent distribution of FLA-YM hydrolysis products in the supernatants of BtVPI-5482, MD33MG, and MD40HG grown in 0.2% FLA-YM over time. N = 3
To determine if surface α-mannanases generate different product profiles, cultures of BtVPI-5482, MD33MG, and MD40HG were incubated with FLA-YM and the products were analyzed by fluorescence-coupled high-performance liquid chromatography. In BtVPI-5482 and MD40HG, there was preferential hydrolysis of large FLA-YM products (> 10 kDa, ~ 55-mer), which was accompanied by a relative accumulation of lower molecular weight (< 4 kDa; ~ 22-mer) hydrolysis products (Fig. 7c, Supplementary Fig. 8).
Discussion
The gut microbiome plays an integral role in digestion and nutrient acquisition. Improved understanding of the functional potential encoded within members of the microbiota is still required to define metabolic abilities and microbial-prebiotic interactions. Next-generation physiology approaches represent promising strategies to rapidly assign cellular phenotypes and can consolidate genomic predictions [29]. By combining phenotypic and sequencing approaches, we have conducted a high-resolution study of differential YM utilization by isolated bovine-associated bacterial strains. In liquid culture, the isolates displayed one of two growth patterns: MG or HG; trends that were independent of taxonomic relationships (Fig. 2a, b, Supplementary Table 2). This showed that closely related Bacteroides spp. have evolved different foraging strategies for the same substrate. FLA-PS were successfully used to visualize (Fig. 2c) and quantify the accumulation (Fig. 2d) and uptake rate (Fig. 2e) of YM products in bacterial cells, confirming that HGs use a selfish mode of metabolism on this substrate, as previously reported for BtVPI-5482 [14, 31]. In contrast, the MG strains consumed less YM and released mannose into the medium (Fig. 7a, b), suggesting that MGs are less adept at YM catabolism and display some properties consistent with distributive metabolism (Figs. 2b and 7c).
Comparative genomics revealed genotypes with high synteny across genomes and MAN-PUL pathways, with few exceptions. Perhaps the most interesting genetic anomaly is the sequence variability of the MAN-PUL2 SusC/D-like proteins, which elegantly branch into two clades coinciding with the HG and MG growth phenotype (Fig. 5b, c), as well as differential rates and total levels of FLA-PS uptake (Fig. 2c–e, Supplementary Fig. 2b). Previously, it was shown that the amino acid homology of a SusD-like protein involved in utilization of two different fructans was low between two strains of B. theta, despite their taxonomic similarity [41], highlighting that syntenic genes within PULs can evolve independently. Here we report that SusC/D-like amino acid sequences from the major PUL involved in metabolism of YM correlate with differential utilization of a common substrate (Fig. 5b, c). MD40HG likely has a more efficient transport process (Fig. 7), as suggested by the following results: there is no perceived difference in the structures of surface enzymes encoded within the MAN-PULs (Supplementary Fig. 4), the outer surface endo-α-mannanases are expressed at lower levels in HGs (Fig. 6b), the addition of exogenous endo-GH76s to MG growth cultures did not augment MG growth (Supplementary Fig. 5), and the higher growth and faster disappearance of large YM products in MD40HG cultures (Fig. 7). Whether this is the direct result of higher transporter efficiency in MD40HG or indirect result from impoverished transport leading to product inhibition of surface enzymes in MD33MG is unclear. Intriguingly, deletion of MAN-PUL1/3 susC/D or the MAN-PUL2 susC/D did not impede growth of BtVPI-5482 on YM or uptake of YM (Fig. 5d–f), suggesting SusC/D-like pairs in MAN-PUL2 and 3 are functionally redundant in HGs. Based upon sequence identity (Supplementary Table 4), MGs possess one compromised SusC/D/E complex (MAN-PUL2) and one high-performing SusC/D complex (MAN-PUL3), which are regulated differently between the strains. In MD33MG, the MAN-PUL3 susC-like gene (bt3854 homolog) is expressed at a level similar to the MAN-PUL2 susC-like gene (bt3788 homolog), and at a level 2.9-fold higher than its homologous gene in MD40HG (Fig. 6b). Higher expression of outer surface proteins in MD33MG is a consistent pattern (Fig. 6b). Despite the higher expression levels of the MAN-PUL3 SusC/D-like complex in MD33MG, and the ability of the MAN-PUL3 SusC/D-like complex to compensate for deletion of the MAN-PUL2 transporter in BtVPI-5482 (Fig. 5d, e), the MAN-PUL3 SusC/D-like complex in MD33MG is unable to rescue the MG growth phenotype of the representative strain. Thus, the SusC/D-like complexes in MAN-PUL2 and MAN-PUL3 appear to compete for substrates and the inefficiencies of transport ascribed to the MAN-PUL2 complex are related to its ability to transport, but not recruit, YM substrates. Further biochemical and structural studies of the MAN-PUL2 SusC/D-like proteins are warranted to tease apart these results.
The “Nutrient Niche Hypothesis” [42] suggests that metabolic abilities are determined by the creation and filling of ecological nutrient niches. In theory, these relationships could be in response to the introduction of a new dietary glycan (i.e., prebiotic), resulting in the selection for or adaptation of a bacterium with the metabolic capacity to consume it. In this study, the MG and HG phenotypes represent a variation on this theme, as two closely related populations (> 98% identity) adapted to the colonization of a common host (Supplementary Fig. 3) display different (Fig. 2b, Supplementary Fig. 1), yet reproducible (Fig. 3a) and inducible (Fig. 3b–d) foraging behaviors on the same substrate. These findings raise several unsolved questions related to the existence and persistence of MGs, and potentially other glycan foragers that are less adept at substrate utilization, in the rumen. If HGs have a superior capacity for YM metabolism, why are MGs not eliminated by competitive exclusion? And if MGs have restricted abilities to digest YM and/or transport YM products (Fig. 7), why are these PULs not selected against and excised from the genome? The existence of multiple metabolic phenotypes suggests that ecological selection factors may be responsible. Firstly, Bacteroides spp. are generalists with the capacity to utilize a wide variety of substrates available in the diet of their hosts and glycan responses are prioritized in Bacteroides spp. [43, 44]. MGs may possess a different substrate hierarchy than HGs and, correspondingly, display more prowess for consuming chemically distinct glycans. Alternative substrate priorities would reduce the competitive burden on MGs when provided with complex diets. In this regard, the acquisition of new CAZymes or PULs that endow a microorganism with an ability to consume new substrates has been hypothesized to occur by horizontal gene transfer and is linked to spatial and dietary habits [20, 28]. YM from S. cerevisiae (Supplementary Fig. 1a) and S. pombe (Supplementary Fig. 1b) were the substrates used in this study and showcased that HGs are not consistently superior when it comes to glycan utilization. Further investigation into the ability of MGs and HGs to utilize other substrates is warranted to identify additional variability in substrate utilization and preference. Secondly, feeding strategies, such as distributive metabolism, may foster beneficial syntrophic relationships at multiple levels within a community [45, 46]. The generation of public goods [13] by MGs provides nutrients to species that are incapable of digesting YM. This event would increase the richness of the community and, potentially, result in the generation of additional secondary metabolites that benefit the lifestyle of MGs. Furthermore, it has been shown that both the concentrations and complexity of available substrate cause differential selection of distributive or selfish foraging strategies [47–49].
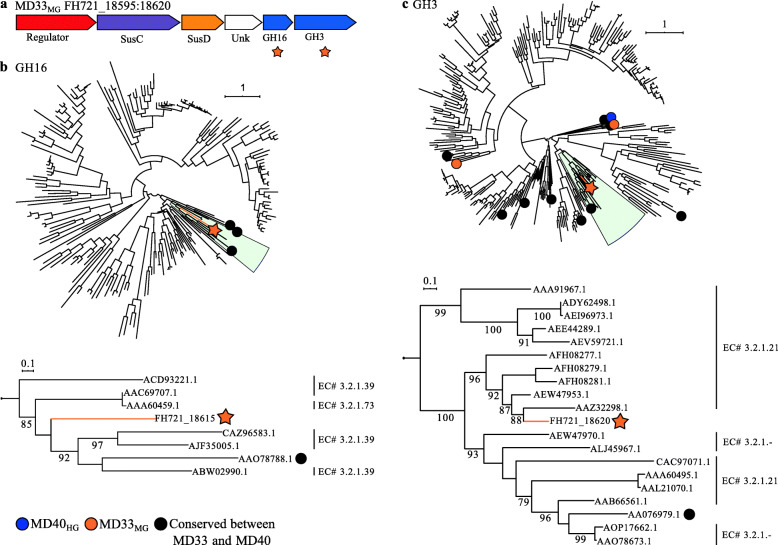
Comparison of the MD40HG and MD33MG CAZomes confirmed that there are many GH families, encoding different enzyme specificities that vary in number (Fig. 4a). Closer inspection of GH3 and GH16, two polyspecific GH families active on β-linkages, revealed CAZyme updates within a PUL in MD33MG (Fig. 8). This suggests that the acquisition of a putative β-glucan metabolic pathway, and potentially others, may provide a colonization advantage for MD33MG despite its weakened potential to metabolize YM. Recently, β-glucan utilization pathways were shown to have independently evolving genes that result in the expansion of protein specificity and glycan targets [50]. Thus, clustered mutations or differential acquisition of genes in PULs could unlock previously inaccessible nutrient niches. Conversely, however, there is the risk of impeding nutrient acquisition, as exhibited by the restricted β-glucan utilization of polyspecific proteins in Bacteroides fluxus [50] and, potentially, the inefficiencies of YM uptake governed by transporter specificity or efficiency. Further investigation of total CAZome function and transporter selectivity and efficiency encoded within genomes at the strain level will reveal how microorganisms living in partnership or competition within complex ecosystems tune their metabolic responses to complex dietary landscapes. Coupling “omics” methods and functional methods, such as FLA-PS, will help usher in a new frontier for the assignment of metabolic traits to bacterial populations within microecological food webs
Fig. 8.
CAZome analysis of MD33MG and MD40HG. Predicted PUL (FH721_18595-18620) found in the genome of MD33MG encoding GH16 and GH3 enzymes, which are not found in the MD40HG genome. Phylogenetic trees of GH16 (top) and GH3 (bottom) families in MD33MG (orange) and MD40HG (blue) genomes. (Left) Expanded clades with the unique GH16 and GH3 CAZymes are shown with known EC numbers
Materials and methods
Direct visualization of YM metabolism in rumen communities and cell identification by FISH
Rumen samples were collected from two cannulated cows fed a diet rich in barley grain. The rumen samples were filtered through cheesecloth under CO2 gas. Subsamples were taken, flash frozen, and stored at − 80 °C until genomic extractions could be completed. The rest of the sample was transferred into an anaerobic chamber (atmosphere: 85% N2, 10% CO2, 5% H2, at 37 °C) and filtered through a 100-μm pore size nylon net filter (Millipore, USA). The filtered samples from each cow were then aliquoted into three tubes. One tube was immediately fixed with 1% formaldehyde (FA) for 1 h at room temperature as the 0 h control. The other tubes were incubated with 20 μL FLA-YM for a final concentration of 3.1 nM and fixed with FA after 1 day and 3 days. Immediately after fixation, all samples were filtered through a 47 mm (0.2-μm pore size) polycarbonate filter (Millipore), using a 0.45-μm cellulose acetate support filter (Millipore) and a gentle vacuum of < 200 mbar. After drying, the filters were stored at − 20 °C.
Total cell counts were determined by staining with 4′,6-diamidino-2-phenylindole (DAPI) and visualizing on a Leica DMRX epifluorescence microscope (Leica, Germany). The number of FLA-YM stained cells was determined by enumerating cells which had a positive DAPI and FLA-YM signal (excitation at 405-nm and 488-nm wavelengths, respectively). For FISH, oligonucleotide probe CF968 targeting Bacteroidetes (5′-GGTAAGGTTCCTCGCGTA-3′) [33] was used, which was covalently labeled with four ATTO594 fluorochromes by Biomers (Konstanz, Germany). FISH was performed with slight alterations to the protocol of Manz et al. [51]. The hybridization buffer contained 900 mM NaCl, 20 mM TRIS-HCl (pH 7.5), 0.02% sodium dodecyl sulfate, 10% dextran sulfate (w/v), and 1% (w/v) blocking reagent (Boehringer, Germany) with a formamide concentration of 55%. Hybridizations were carried out at 35 °C in a humidity chamber overnight, with a subsequent 15-min wash in a buffer containing 10 mM NaCl, 20 mM TRIS-HCl (pH 7.5), 5 nM EDTA (pH 8), and 0.01% sodium dodecyl sulfate at 37 °C. After FISH, the abundance of Bacteroidetes as well as Bacteroidetes showing FLA-YM uptake was enumerated using a Leica DMRX epifluorescence microscope.
DNA from the frozen rumen samples was extracted using the Qiagen DNeasy PowerSoil Kit, and samples were sent to McGill GenomeQuebec for Illumina MiSeq PE250 16S rRNA metagenomics sequencing. The 16S rRNA sequences were merged and quality trimmed using the BBTools [52] software and subsequently classified using the standard settings of the SILVAngs pipeline using the SSU rRNA seed of the SILVA database release 132 [53]. All analysis and plotting of the microbial diversity data were done using RStudio version 3.6.3 using the Vegan package [54, 55]
Isolation of bovine-adapted mannan degraders
Bovine rumen and fecal samples were collected for in vitro batch culture experiments. Ruminal and fecal inoculants from cattle were enriched anaerobically (atmosphere: 85% N2, 10% CO2, 5% H2) at 37 °C with one of the following substrates: Bio-Mos® (1% w/v), corn distillers’ grains (1% w/v), or YM (1% w/v). Bacteria were isolated from the enriched batch cultures by streaking onto nutrient-restricted media supplemented with 0.5% YM to select for YM-degraders (supplementary methods). In total, 50 YM-degrading bacterial isolates were characterized for their propensity to metabolize YM. Nine of these isolates were selected for detailed analysis in this study.
Growth profiling of bovine isolates
Bovine isolates, wild-type BtVPI-5482, and a mutant Bt strain lacking MAN-PULs 1, 2, and 3 (ΔMAN-PUL1/2/3) [14] were cultured anaerobically (atmosphere: 85% N2, 10% CO2, 5% H2) at 37 °C overnight in tryptone-yeast-glucose (TYG) medium (supplementary methods). All incubations were performed in an anaerobic chamber at 37 °C. The overnight cultures (OD600 1.0–1.4) were diluted to an OD600 of 0.05 in 2X Bacteroides minimal medium (MM), pH 7.2 (supplementary methods). Wells of a 96-well microtiter plates (Falcon) were filled with 100 μL of sterilized 1% (w/v) YM (Sigma, St. Louis, USA; M7504) or mannose along with 100 μL inoculant (n = 4). Negative control wells consisted of 100 μL 2X MM combined with 100 μL 1% (w/v) of YM or mannose and were used to normalize growth curves. One hundred microliters of bacterial suspension was inoculated to get starting OD600 ~ 0.025. Plates were sealed with polyurethane Breathe-Easy gas-permeable membranes (Sigma; Z390059). Absorbance (600 nm) of each well was measured with a Biotek Eon microplate reader and recorded on Biotek Gen5 software every 10 min for 50 h. Mean (± standard deviation) of each condition (n = 4) was visualized using GraphPad Prism 6. Two replicates of each strain were also cultured on YM extracted from the cell wall of S. pombe (supplementary methods).
Post-growth cultures were harvested and centrifuged. Supernatants were taken and 6 μL was ran on a silica sheet in 2:1:1 (butanol to d2H2O to acetic acid) running buffer. The plate was dried at ambient temperature and stained with orcinol (diluted to 1% in a solution of 70:3 ethanol to sulfuric acid). Once the plate was dry, it was activated in an oven at 120 °C and imaged using a gel doc XR image system (Bio-Rad).
Genome sequencing, assembly, and annotation of BtBov strains
The 16S rRNA of 50 bovine bacterial isolates was sequenced to determine taxonomic classification using the universal primers 27F and 1492R (supplementary methods). Based on growth profiles (OD600 > 0.4) and 16S rRNA sequences, nine isolates were chosen for whole-genome sequencing using Illumina MiSeq PE150 bp. Genomes were assembled using SPAdes de novo assembly [35]. The K-mer value in SPAdes was chosen from (21, 33, 55, and 77 defaults for 150 bp reads). Quality reporting of the assemblies was done using Quast [56]. SPAdes assembly N50s, largest contigs, and number of contigs are shown in Supplementary Table 2. Genomes were uploaded to the NCBI submission portal and annotated using the NCBI Prokaryotic Annotation Pipeline. Isolate contigs were blasted against the reference genome BtVPI-5482 for MAN-PUL1/2/3 and the HMNG-PUL using NCBI BLAST (2.7.1) [34]. SPAdes contig assemblies were aligned with the JSpeciesWS reference BtVPI-5482 genomes to calculate average nucleotide identity based on BLAST+ (ANIb) [36].
Production of BtVPI-5482 MAN-PUL mutants
Flanking regions (~ 750 bp) of the susC/D-like genes from each MAN-PUL were PCR amplified, stitched together, and ligated into pExchange-tdk (pEx-tdk). The plasmids were transformed into E. coli strain S17-1λpir, which were donor cells used to conjugate the plasmids into the BtVPI-5482 ΔPUL75Δtdk recipient strain to delete the sus-like gene pairs [57]. Mutants with the MAN-PUL1 Sus genes deleted (ΔMP1susCD) were then conjugated with E. coli cells containing a plasmid with the flanking regions for the MAN-PUL3 Sus genes to create a dual mutant (ΔMP1/3susCD). The dual mutant was then conjugated with E. coli cells that contained a plasmid with the MAN-PUL2 Sus flanks to produce a triple mutant (ΔMP1/2/3susCD). Plasmids and mutants were sequenced at each step of this process.
The three knock-out strains, along with BtVPI-5482 wild-type, were grown on 0.5% YM-MM as described above. In addition, the strains were incubated with FLA-YM and sampled at 0 h, 1 h, 1 day, and 3 days. These samples were fixed and stored at 4 °C until analyzed by flow cytometry and epifluorescence microscopy (see below).
PUL delineation and comparative CAZomics
Isolate contigs were processed through EMBOSS GetORF [58] to determine open reading frames; these data were run through the dbCAN [59] HMMscan to identify CAZyme sequences. CAZyme sequences were then analyzed by SACCHARIS [37] (Sequence Analysis and Clustering of CarboHydrate Active enzymes for Rapid Informed prediction of Specificity). User CAZyme sequences were trimmed to their catalytic domain with dbCAN [59], aligned with MUSCLE [60], and fitted to a phylogenetic tree using ProtTest3 [61] to find the appropriate amino acid replacement model. RAxML [62] or FastTree [63] was used to generate the final tree. GHs from families 38, 76, 92, 99, and 125 identified in the genomes of the MD isolates were analyzed by SACCHARIS. Phylogenetic trees were developed using FastTree, and Newick file outputs were viewed and plotted using ITOL (doi.org/10.1093/nar/gkz239).
RNA-seq: assembly, quantitation, and comparative analysis
RNA from BtVPI-5482, MD33MG, and MD40HG grown in 0.5% mannose or YM (see supplementary methods) was extracted and purified using a GeneJET RNA Purification kit (Thermo Scientific) within 1 week of storage at − 80 °C. RNA was sent to Génome Québec for Illumina HiSeq 4000 PE100bp sequencing. Using Geneious v11.1.2 [64], each set of reads was mapped to their previously assembled genomic sequence or, in the case of BtVPI-5482, to the genomic sequence from the NCBI database (NC_004663). Expression levels were calculated as transcript expression (transcript per kb per million; TPM) for each growth treatment. Ambiguously mapped reads were counted as partial matches. The Geneious DESeq2 [65] plugin was used to compare the expression levels between the two treatments, producing log2 expression ratios and p values.
Generalized linear mixed models in SAS PROC GLIMMIX (SAS 9.4, SAS Institute, Cary, NC, USA) were used to estimate statistically significant (p < 0.05) differences of TPM means (least squares-means) for the MAN-PUL1/2/3 susC-like genes of each bacterial strain. Based on the Bayesian information criterion (BIC) of the generalized linear mixed models, the response was modeled using the log-normal distribution. The expression of gene transcripts was the dependent variable in models with two independent fixed factors: bacterial strain (i.e., BtVPI-5482, MD33MG, or MD40HG) and media treatment (i.e., YM or mannose). Mixed models of variance heterogeneity were selected based on the BIC. For the studied transcripts, the variance of expression was heterogeneous for the experimental treatments, bacteria, or their interaction. The statistical significance of the interaction between the TPM values of MAN-PUL genes for each bacterial strain and the media treatment was determined using an F test. Bonferroni’s method was used for multiple comparisons (Supplementary Table 3).
Production of SusD-like protein C-myc fusion B. theta strain
The C-myc epitope (EQKLISEEDL) was fused to the C-terminal domain of the MAN-PUL2 SusD-like protein (BT_3789) with a linker sequence (STSTST) between the SusD-like nucleotide sequence and the C-myc sequence BtVPI-5482 ∆tdk ∆pul75 (control), and BtVPI-5482 ∆tdk ∆pul75 SusD-like C-myc fusion mutant was inoculated in TYG and cultured as described above. The cells were centrifuged and resuspended in 1 mL 2X MM. One hundred microliters of the resuspension was inoculated into 0.5% YM-MM and incubated for 4 h at 37 °C. The cells were then centrifuged and washed three times in phosphate-buffered saline (PBS) pH 7.4 (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4), before resuspension in 2 mL 2X MM. Two hundred twenty-five microliters of the resuspended cells was added to 1.5 mL 0.2% FLA-YM or YM-MM and incubated for 3 h at 37 °C. One hundred microliters of each culture was collected and fixed in 1% formaldehyde for 1 h at room temperature. The samples were then incubated with 1:2500 rabbit IgG anti-C-myc polyclonal antibody (ThermoFisher #PA1-981) for 1 h at room temperature. The samples were then washed four times in PBS and resuspended in 1:2500 goat anti-rabbit DyLight 650 nm secondary antibody (ThermoFisher #84546) for 1 h at room temperature. The samples were then washed and stored in PBS until further analysis.
Sequence comparison and modeling of SusC/D/E-like proteins
MUSCLE was used to align MAN-PUL2 and 3 SusC-like, SusD-like (SGBPA), and SusE-like (SGBPB) amino acid sequences of each isolate and calculate percent identity (Supplementary Table 4). The 16S rRNA gene and MAN-PUL2 SusC-like and SusD-like amino acid phylogenetic trees were generated using the maximum likelihood method and Tamura-Nei model [66]. Evolutionary analyses were performed by MEGA X [67]. Trees with the highest log likelihood are shown in Fig. 4b and c.
Generation of FLA-YM conjugates
A previously defined protocol [30, 68] was used to generate fluorescently labeled YM (FLA-YM), with slight variations (supplementary methods).
Visualization of FLA-YM uptake by strains of BtBov
Wild-type BtVPI-5482, BtΔMAN-PUL1/2/3, and rumen isolates MD33MG and MD40HG were inoculated in TYG and grown as described above. Cells were harvested at OD600 ~ 1.0 and centrifuged (4700×g) for 5 min, the supernatant was removed, and pellets resuspended in 2 mL 2X MM for the first two washes. After the third centrifugation, pellets were resuspended in 2 mL MM with 0.5% YM (BtVPI-5482, MD33MG, and MD40HG) or 0.5% glucose + YM (BtΔMAN-PUL1/2/3) as the sole carbon source (not conjugated to FLA). After ~ 18-h incubation, cultures were centrifuged and washed three times in PBS, with the final resuspension in 2 mL 2X MM. Three hundred microliters of the resuspended pellet was aliquoted into 0.2% unlabeled YM or FLA-YM. Twenty microliters of the 2X MM resuspension was used as the 0-h time point, as the cells were not exposed to FLA-YM. Forty microliters aliquots of each condition were taken at time points: 5 min, 1 h, and 24 h. The cells were centrifuged (10 min; 2300×g), and the pellet was fixed in 1% formaldehyde (FA; Sigma; F8775) in PBS, at 4 °C for 18–24 h. The fixed cells were centrifuged (10 min; 2300×g) and washed in 1X PBS. The samples were centrifuged and stored at 4 °C in the dark until visualized by SR-SIM (supplementary methods).
Quantification of the rate of FLA-YM uptake by BtBov isolates
BtVPI-5482, MD33MG, and MD40HG were grown in TYG and prepared as described above. After 24 h of incubation, cultures were placed into 2 mL 0.5% YM. Cells were harvested in exponential phase (OD600 0.6–1.0), centrifuged (10 min; 2300×g), and resuspended in 2 mL 2X MM. Three hundred microliters of this suspension was added to 1 mL 2 X MM. Then 20 μL of each culture was aliquoted into 1 mL 1% FA and used as the T0 time point. Into the remaining 280 μL, 0.2% FLA-YM and 150 ng/mL fluoresceinamine (FLA) or YM was added and subsamples of 40 μL were taken at 5, 10, 15, 20, 30, and 60 min and 2, 4, 8, and 24 h. The subsamples were centrifuged (10 min, 2300×g), and the cell pellets were fixed in 1% FA in 1X PBS, at 4 °C for 18–24 h. The fixed cells were centrifuged (10 min; 2300×g) and resuspended in 1 ml 1X PBS and stored at 4 °C in the dark.
Cell fluorescence due to FLA or FLA-YM uptake was quantified in all samples using an Accuri C6 flow cytometer (BD Accuri Cytometers). The 8-peak and 6-peak validation bead suspensions (Spherotech, IL, USA) were used as internal references. All samples were measured under laser excitation at 488 nm from a blue-green laser, and the green fluorescence was collected in the FL1 channel (530 ± 30 nm). Using the medium as a background, an electric threshold of 17,000 FSC-H was set to reduce the background noise. All measurements were done at a slow flow rate and a total of 10,000 (FLA-YM) or 5000 (YM and FLA) events per sample were acquired. Bacteria were detected from the signature plot of SSC-H vs green fluorescence (FL1-H). The FCM output was analyzed using FlowJo v10-4-2 (Tree Star, USA). The FCM files were imported into FlowJo, and both the total population (all events) and main population (automated gating through event density) were determined. For each population (total and main), sample statistics (counts, mean fluorescence, and the standard deviation) were determined from the raw FL1-H data. The results were exported and analyzed using Welch’s t tests in R studio using the packages Vegan and Rioja [55, 69] to determine statistical difference between the control (YM and FLA) and FLA-YM incubation within each strain and between the FLA-YM incubation of each strain.
Quantification of mannose in minimal medium using GC-MS
Cell culture medium after incubation in 1% YM-MM was collected after 24 h and centrifuged (4700×g for 15 mins), and the supernatant was passed through a syringe filter (0.2 μm cellulose acetate membrane, VWR). The filtrate was kept frozen for 48 h at − 20 °C and then thawed and centrifuged (3000×g, 30 min) at room temperature. Concentration of mannose in the resulting supernatant was tested based on our previous report [70], with some modifications to cope with the relatively large amount of starting carbohydrate material and the presence of minimum medium. One milliliter of the supernatant was evaporated to dryness under a gentle flow of nitrogen. The residue was suspended and magnetically stirred in 3.5 mL of 6 M TFA at 100 °C for 6 h with headspace filled with nitrogen, followed by addition of internal standard myo-inositol (0.4 mg dissolved in 0.5 mL of water) and evaporation to dryness. Monosaccharides were reduced by magnetic stirring overnight in 10 mg of NaBD4 (99% D, Alfa Aesar) dissolved in 2 mL of 1 M ammonium oxide solution, followed by quenching excess reductant with acetic acid and evaporating to dryness. Boric acid was removed by evaporation to dryness five times in 3 mL of 10% (v/v) acetic acid in methanol followed by five times in 3 mL of absolute methanol. The residue was suspended in 4 mL of acetic anhydride, followed by magnetic stirring at 100 °C for 2 h with headspace filled with nitrogen, cooling to room temperature, and evaporation to dryness. The derivatives were purified by partitioning with water and dichloromethane, recovered by collecting and evaporating to dryness the organic phase after three changes of water, and re-dissolved and diluted in ethyl acetate for analysis on an Agilent 7890A-5977B GC-MS system (Agilent Technologies, Inc., CA, USA). Sample solution (1 μL) was splitless-injected to the system, and optimal analyte separation was achieved on a medium polarity SP2380 column (30 m × 0.25 mm × 0.20 μm, Sigma-Aldrich) with a constant helium flow of 0.8 mL/min and with oven temperature programmed to start at 55 °C (hold 1 min) followed by increasing at 30 °C/min to 120 °C then at 12 °C/min to 255 °C (hold 20 min). Two separate experiments were conducted for each sample. Mannose concentration was calculated based on calibration curve established from a series of mannose standard solution containing internal standard.
Measurement of YM hydrolysis
Samples from BtVPI-5482, MD33MG, and MD40HG cultures in 0.2% FLA-YM (as above) and a no-cell negative control were filtered through 0.2-μm cellulose acetate membrane syringe filter (VWR). The filtrates were flash frozen and stored at − 80 °C until analysis. Samples were analyzed as described in Arnosti 2003 [68]; in brief, samples were injected onto two columns of Sephadex G50 and G75 gel linked in series, with the column effluent passing through a Hitachi fluorescence detector set to excitation and emission wavelengths of 490 and 530 nm, respectively. The columns were standardized using FITC dextran standards (150 kDa, 70 kDa, 40kDA, 10 kDa, 4 kDa, FITC-glucose, and free fluorophore; Sigma), so the fraction of polysaccharide eluting in each molecular weight class at each time point could be calculated.
Supplementary Information
Additional file 1. Supplementary figures
Additional file 2. Supplementary materials
Acknowledgements
We wish to thank Sherif Ghobrial and Chad Lloyd for the assistance with the FLA-YM hydrolysis measurements, and Elizabeth Lowe, Newcastle University, for the kind gift of the MAN-PUL1/2/3 KO mutant.
Authors’ contributions
LK assisted with the rumen extractions and performed the rumen incubations with FLA-YM, selective bacterial growth profiling and TLC analysis, RNA sequencing, production of FLA-YM, FGC-bacterial incubations, construction of the SusD-C-myc mutant and evaluation of production, and extraction of S. pombe YM; prepared figures; and wrote manuscript. GR conducted the epifluorescence microscopy and SR-SIM analysis, FISH analysis, cell enumeration of rumen samples, flow cytometry sorting and analysis, and statistical analysis and assisted with figure and manuscript writing and preparation. JPT performed comparative genome and protein sequence analysis and assisted with figure preparation. DRJ assisted with comparative genome and protein analysis. JHH helped conceive of the study, interpreted the data, and assisted in the preparation of the manuscript. ADS performed isolations of rumen bacteria and 16S rRNA sequencing. TDS conducted the statistical analysis of RNA-seq data. CAr assisted with the generation of FLA-PS, analysis FLA-PS products, and preparation of the manuscript. LJ performed the rumen collections and assisted with the bacterial isolations. TWA assisted with the rumen extractions and preparation of the manuscript. CAm assisted with the RNA-seq analysis and figure generation. DT assisted with the CAZyme fingerprinting and maintenance of the SACCHARIS pipeline. RA helped conceive of the study and assisted with the SR-SIM microscopy and preparation of the manuscript. TAM assisted with the animal study, rumen extractions, and preparation of the manuscript. DPY assisted with the data analysis and preparation of the manuscript. DWA helped conceive of the study, secured the funding, designed the study, performed the data analysis, and assisted with figure and manuscript preparation. The authors read and approved the final manuscript.
Funding
This work was supported by funding from the Beef and Cattle Research Council awarded to DWA (Grant: FDE.13.15 & FDE.14.17). JHH, GR, and RA were supported by the Max Planck Society. RA and JHH acknowledge the support by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the research unit FOR2406 “Proteogenomics of Marine Polysaccharide Utilization (POMPU)” by grants AM 73/9-1 and HE 7217/1-1. GR has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No. 840804. CAr was supported by NSF OCE-1736772, DE-SC0013887, and DE-SC0019012.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the NCBI repository in the whole-genome sequencing BioProject No: PRJNA546576 (BioSample Accession Numbers: MD8HG, SAMN11961934; MD11MG, SAMN11961935; MD13HG, SAMN11961936; MD17HG, SAMN11961937; MD28MG, SAMN11961938; MD33MG, SAMN11961939; MD35MG, SAMN11961940; MD40HG, SAMN11961941; MD51HG, SAMN11961942), and RNA sequencing BioProject No: PRJNA658335 (BioSample Accession Numbers: BtVPI-5482, SAMN15866569; MD33MG, SAMN15866570; MD40HG SAMN15866571). The read mapping of RNA-seq data was done using the assembled genomic sequences from the BioProject or to the genomic sequence of BtVPI-5482 found in the NCBI database (NC_004663).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Leeann Klassen and Greta Reintjes contributed equally to this work.
References
- 1.Weimer PJ. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol. 2015;6:296. doi: 10.3389/fmicb.2015.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson G, Cox F, Ganesh S, Jonker A, Young W. Global Rumen Census C, Janssen PH: Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5:14567. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart RD, Auffret MD, Warr A, Wiser AH, Press MO, Langford KW, Liachko I, Snelling TJ, Dewhurst RJ, Walker AW, et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun. 2018;9:870. doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huws SA, Creevey CJ, Oyama LB, Mizrahi I, Denman SE, Popova M, Muñoz-Tamayo R, Forano E, Waters SM, Hess M, et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: past, present, and future. Front Microbiol. 2018;9:2161. doi: 10.3389/fmicb.2018.02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lima J, Auffret MD, Stewart RD, Dewhurst RJ, Duthie C-A, Snelling TJ, Walker AW, Freeman TC, Watson M, Roehe R. Identification of rumen microbial genes involved in pathways linked to appetite, growth, and feed conversion efficiency in cattle. Front Genet. 2019;10:701. doi: 10.3389/fgene.2019.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uyeno Y, Shigemori S, Shimosato T. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 2015;30:126–132. doi: 10.1264/jsme2.ME14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foley MH, Cockburn DW, Koropatkin NM. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci. 2016;73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grondin JM, Tamura K, Dejean G, Abbott DW, Brumer H. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199:e00860. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson KL, Salyers AA. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/JB.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruger K, Chafee M, Ben Francis T, Glavina Del Rio T, Becher D, Schweder T, Amann RI, Teeling H. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019;13:2800–2816. doi: 10.1038/s41396-019-0476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 13.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbott DW, Martens EC, Gilbert HJ, Cuskin F, Lowe EC. Coevolution of yeast mannan digestion: Convergence of the civilized human diet, distal gut microbiome, and host immunity. Gut Microbes. 2015;6:334–339. doi: 10.1080/19490976.2015.1091913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, Henrissat B. PULDB: the expanded database of polysaccharide utilization Loci. Nucleic Acids Res. 2018;46:D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hehemann JH, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci U S A. 2012;109:19786–19791. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pluvinage B, Grondin JM, Amundsen C, Klassen L, Moote PE, Xiao Y, Thomas D, Pudlo NA, Anele A, Martens EC, et al. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat Commun. 2018;9:1043. doi: 10.1038/s41467-018-03366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 22.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finck DN, Ribeiro FRB, Burdick NC, Parr SL, Carroll JA, Young TR, Bernhard BC, Corley JR, Estefan AG, Rathmann RJ, Johnson BJ. Yeast supplementation alters the performance and health status of receiving cattle. Prof Anim Sci. 2014;30:333–341. doi: 10.15232/S1080-7446(15)30125-X. [DOI] [Google Scholar]
- 24.Miguel JC, Rodriguez-Zas SL, Pettigrew JE. Efficacy of a mannan oligosaccharide (Bio-Mos®) for improving nursery pig performance. J Swine Health Prod. 2004;12:296–307. [Google Scholar]
- 25.Wohlt JE, Corcione TT, Zajac PK. Effect of yeast on feed intake and performance of cows fed diets based on corn silage during early lactation. J Dairy Sci. 1998;81:1345–1352. doi: 10.3168/jds.S0022-0302(98)75697-8. [DOI] [PubMed] [Google Scholar]
- 26.Gregg KJ, Zandberg WF, Hehemann JH, Whitworth GE, Deng L, Vocadlo DJ, Boraston AB. Analysis of a new family of widely distributed metal-independent alpha-mannosidases provides unique insight into the processing of N-linked glycans. J Biol Chem. 2011;286:15586–15596. doi: 10.1074/jbc.M111.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Suits MD, Thompson AJ, Chavan S, Dinev Z, Dumon C, Smith N, Moremen KW, Xiang Y, Siriwardena A, et al. Mechanistic insights into a Ca2+-dependent family of alpha-mannosidases in a human gut symbiont. Nat Chem Biol. 2010;6:125–132. doi: 10.1038/nchembio.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 29.Hatzenpichler R, Krukenberg V, Spietz RL, Jay ZJ. Next-generation physiology approaches to study microbiome function at single cell level. Nat Rev Microbiol. 2020;18:241–256. doi: 10.1038/s41579-020-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reintjes G, Arnosti C, Fuchs BM, Amann R. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 2017;11:1640–1650. doi: 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hehemann JH, Reintjes G, Klassen L, Smith AD, Ndeh D, Arnosti C, Amann R, Abbott DW. Single cell fluorescence imaging of glycan uptake by intestinal bacteria. ISME J. 2019;13:1883–1889. doi: 10.1038/s41396-019-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao J, McCourt C, Sultana H, Nelson C, Driver J, Hackmann TJ. Use of a fluorescent analog of glucose (2-NBDG) to identify uncultured rumen bacteria that take up glucose. Appl Environ Microbiol. 2019;85:e03018. doi: 10.1128/AEM.03018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acinas SG, Ferrera I, Sarmento H, Díez-Vives C, Forn I, Ruiz-González C, Cornejo-Castillo FM, Salazar G, Gasol JM. Validation of a new catalysed reporter deposition–fluorescence in situ hybridization probe for the accurate quantification of marine Bacteroidetes populations. Environ Microbiol. 2015;17:3557–3569. doi: 10.1111/1462-2920.12517. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics (Oxford, England) 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DR, Thomas D, Alger N, Ghavidel A, Inglis GD, Abbott DW. SACCHARIS: an automated pipeline to streamline discovery of carbohydrate active enzyme activities within polyspecific families and de novo sequence datasets. Biotechnol Biofuels. 2018;11:27. doi: 10.1186/s13068-018-1027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndeh D, Rogowski A, Cartmell A, Luis AS, Basle A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DR, Xing X, Tingley JP, Klassen L, King ML, Alexander TW, Abbott DW. Analysis of active site architecture and reaction product linkage chemistry reveals a conserved cleavage substrate within diverse yeast mannans for an endo-alpha-mannanase. J Mol Biol. 2020;432:1083-97. [DOI] [PubMed]
- 40.Cuskin F, Basle A, Ladeveze S, Day AM, Gilbert HJ, Davies GJ, Potocki-Veronese G, Lowe EC. The GH130 family of mannoside phosphorylases contains glycoside hydrolases that target beta-1,2-mannosidic linkages in Candida Mannan. J Biol Chem. 2015;290:25023–25033. doi: 10.1074/jbc.M115.681460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joglekar P, Sonnenburg ED, Higginbottom SK, Earle KA, Morland C, Shapiro-Ward S, Bolam DN, Sonnenburg JL. Genetic variation of the SusC/SusD homologs from a polysaccharide utilization locus underlies divergent fructan specificities and functional adaptation in Bacteroides thetaiotaomicron strains. mSphere. 2018;3:e00185-18. [DOI] [PMC free article] [PubMed]
- 42.Freter R, Brickner H, Botney M, Cleven D, Aranki A. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun. 1983;39:676–685. doi: 10.1128/IAI.39.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol. 2013;88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR. Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. mBio. 2017;8:e01068-17. [DOI] [PMC free article] [PubMed]
- 45.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 47.Reintjes G, Fuchs BM, Scharfe M, Wiltshire KH, Amann R, Arnosti C. Short-term changes in polysaccharide utilization mechanisms of marine bacterioplankton during a spring phytoplankton bloom. Environ Microbiol. 2020;22:1884–1900. doi: 10.1111/1462-2920.14971. [DOI] [PubMed] [Google Scholar]
- 48.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Baslé A, Morland C, Day AM, Zheng H, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarmento H, Morana C, Gasol JM. Bacterioplankton niche partitioning in the use of phytoplankton-derived dissolved organic carbon: quantity is more important than quality. ISME J. 2016;10:2582–2592. doi: 10.1038/ismej.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Déjean G, Tamura K, Cabrera A, Jain N, Pudlo NA, Pereira G, Viborg AH, Van Petegem F, Martens EC, Brumer H. Synergy between cell surface glycosidases and glycan-binding proteins dictates the utilization of specific beta(1,3)-glucans by human gut Bacteroides. mBio. 2020;11:e00095–e00020. doi: 10.1128/mBio.00095-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. doi: 10.1016/S0723-2020(11)80121-9. [DOI] [Google Scholar]
- 52.Bushnell B, Rood J, Singer E. BBMerge - Accurate paired shotgun read merging via overlap. PLoS One. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Team R . RStudio: integrated development for R. Boston: RStudio. PBC; 2020. [Google Scholar]
- 55.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M. The vegan package. Comm Ecol Package. 2007;10:719. [Google Scholar]
- 56.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones DR, Smith MB, McLean R, Grondin JM, Amundsen CR, Inglis GD, Selinger B, Abbott DW. Engineering dual-glycan responsive expression systems for tunable production of heterologous proteins in Bacteroides thetaiotaomicron. Sci Rep. 2019;9:17400. doi: 10.1038/s41598-019-53726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 59.Yin Y, Mao X, Yang J, Chen X, Mao F. Xu Y: dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnosti C. Fluorescent derivatization of polysaccharides and carbohydrate-containing biopolymers for measurement of enzyme activities in complex. J Chromatogr B. 2003;793:181–191. doi: 10.1016/S1570-0232(03)00375-1. [DOI] [PubMed] [Google Scholar]
- 69.Juggins S. rioja: analysis of quaternary science data. 2017. [Google Scholar]
- 70.MacMillan JL, Vicaretti SD, Noyovitz B, Xing X, Low KE, Inglis GD, Zaytsoff SJM, Boraston AB, Smith SP, Uwiera RRE, et al. Structural analysis of broiler chicken small intestinal mucin O-glycan modification by Clostridium perfringens. Poult Sci. 2019;98:5074–5088. doi: 10.3382/ps/pez297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary figures
Additional file 2. Supplementary materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI repository in the whole-genome sequencing BioProject No: PRJNA546576 (BioSample Accession Numbers: MD8HG, SAMN11961934; MD11MG, SAMN11961935; MD13HG, SAMN11961936; MD17HG, SAMN11961937; MD28MG, SAMN11961938; MD33MG, SAMN11961939; MD35MG, SAMN11961940; MD40HG, SAMN11961941; MD51HG, SAMN11961942), and RNA sequencing BioProject No: PRJNA658335 (BioSample Accession Numbers: BtVPI-5482, SAMN15866569; MD33MG, SAMN15866570; MD40HG SAMN15866571). The read mapping of RNA-seq data was done using the assembled genomic sequences from the BioProject or to the genomic sequence of BtVPI-5482 found in the NCBI database (NC_004663).