Abstract
Signal transducer and activator of transcription 3 (STAT3) is a transcriptional regulator of a number of biological processes including cell differentiation, proliferation, survival, and angiogenesis, while cyclin-dependent kinases (CDKs) are a critical regulator of cell cycle progression. These proteins appear to play central roles in angiogenesis and cell survival and are widely implicated in tumor progression. In this study, we used the well-characterized US National Cancer Institute 60 (NCI60) human tumor cell lines to screen the in vitro anti-cancer activities of our novel small molecule derivatives (NSC765690 and NSC765599) of salicylanilide. Furthermore, we used the DTP-COMPARE algorithm and in silico drug target prediction to identify the potential molecular targets, and finally, we used molecular docking to assess the interaction between the compounds and prominent potential targets. We found that NSC765690 and NSC765599 exhibited an anti-proliferative effect against the 60 panels of NCI human cancer cell lines, and dose-dependent cytotoxic preference for NSCLC, melanoma, renal, and breast cancer cell lines. Protein–ligand interactions studies revealed that NSC765690 and NSC765599 were favored ligands for STAT3/CDK2/4/6. Moreover, cyclization of the salicylanilide core scaffold of NSC765690 mediated its higher anti-cancer activities and had greater potential to interact with STAT3/CDK2/4/6 than did NSC765599 with an open-ring structure. NSC765690 and NSC765599 met the required safety and criteria of a good drug candidate, and are thus worthy of further in-vitro and in-vivo investigations in tumor-bearing mice to assess their full therapeutic efficacy.
Keywords: protein-ligand interaction, molecular docking simulation, target identification, small-molecule derivatives of salicylanilide, drug discovery, drug development
1. Introduction
Despite advances in biomedical research, cancer remains a public health concern and is currently ranked the second leading cause of global mortality [1,2]. The etiology of cancer is often multifactorial, involving an interplay between genetic and epigenetic factors which amount to dysregulation of molecular networks, proteins, RNA, and DNA in favor of cell growth and proliferation [3,4]. The survival, growth, and metastasis of tumor cells depend on cellular differentiation, proliferation, angiogenic, and apoptotic mechanisms [5], which are controlled by a range of protein kinases and signal transduction pathways [6,7]. Cyclin-dependent kinases (CDKs) are a family of serine/threonine protein kinases with catalytic and regulatory subunits [8]. Out of the nine CDKs so far identified, four (CDK1, CDK2, CDK4, and CDK6) play important regulatory roles in the cell cycle; CDK4/D-cyclin, CDK2/E-cyclin, and CDK6/D-cyclin regulate the G1 to S phase of the cell cycle transition, while the complex of CDK1 or CDK2 with cyclin A regulates the S to G2 transition [9]. Under physiological conditions, the catalytic and regulatory (cyclin) subunits of CDKs remain dissociated. However, periodic complexation of a cyclin with its catalytic unit leads to its activation and phosphorylation of a variety of downstream target proteins required for cell cycle progression [10,11]. CDKs regulate cell cycle transitions via phosphorylation and subsequent inactivation of the retinoblastoma (Rb) protein, a tumor suppressor that prevents cell cycle transition [12]. Thus, the inactivation of this Rb protein allows the free flow and progression of cells into the cell cycle, leading to multi-cell cycles, cell proliferation, and eventual development into cancer cells [12].
A number of key oncogenic abnormalities, including amplification of cyclin D; inactivation of CDKN2A (p16); and deletions/mutations upstream of cyclin D, such as activating mutations of phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA)/the B-raf proto-oncogene, serine/threonine kinase (BRAF), and phosphatase and tensin homolog (PTEN) deletion [13,14], were identified as contributing factors to hyperactive CDKs and consequently deregulated the cell cycle. Therefore, since dysfunctional cell cycle regulation via oncogenic aberrations of CDKs is a hallmark of all human cancers [15], pharmacological targeting of CDKs will undoubtedly affect cancer proliferation and survival [16,17]. Hence, CDK inhibitors have been developed and evaluated; however, disappointingly, while first-generation inhibitors of CDK were non-selective and besieged with toxicity [18,19], second-generation CDK4/6 inhibitors, although showing promising outcomes, are plagued with acquired resistance, which develops in almost all cases [20] due to the activation of other oncogenic pathways, including c-Myc, signal transducer and activator of transcription 3 (STAT3), and phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling pathways [21,22].
STAT3 is a cytoplasmic transcription factor involved in a number of biological processes including cell differentiation, proliferation, survival, and angiogenesis [23]. Overexpression of STAT3 is associated with poor clinical prognoses of cancer patients [24]. It is therefore not surprising that the STAT3 signaling axis has long been explored in cancer therapy owing to its roles in tumor formation, metastasis, and therapeutic failure [25,26]. Preclinical studies revealed that aberrant STAT3 expression mediates immunosuppression of tumor cells [27,28], while inhibition of STAT3 re-sensitizes therapy-resistance breast cancer cells to palbociclib treatment [29]. Summing up the above clinical and preclinical evidence, it is convincing that identifying and validating novel CDK inhibitors capable of simultaneously targeting STAT3 signaling may open up new windows for long-lasting and multilayered tumor control [30].
The translational value of knowledge of cancer biology into developing effective prognostic/diagnostic and therapeutic strategies for clinical practice remains disappointing [31]. However, increasing knowledge of the molecular basis of tumorigenesis, applications of multi-omics approaches, and molecular simulations based on a structural analysis of receptor–ligand interactions have jointly contributed to the identification of more-reliable predictive markers and the discovery of novel, less-toxic, and target-specific anticancer agents [31,32,33,34]. At present, substantial attention is being focused on small molecules for targeted therapy in cancer treatments [35].
NSC765599 and NSC765690 are small molecule derivatives of salicylanilides (PubChem CID: 60202556; NDMC101), which were previously synthesized and evaluated for biological activities in our Lab [36]. We had previously conducted a series of chemical modifications of the lead molecule, NDMC101, to yield the open ring (NSC765599) [37] and close ring (NSC765690) [38] derivatives. Herein we demonstrated that both NSC765690 and NSC765599 exerted antitumor activities in vitro against panels of NCI60 human tumor cell lines. We further identified and validated CDK2/4/6 and STAT3 as druggable candidates for the compounds, through in silico and molecular simulation of ligand–receptor interaction studies. Hence, our data provide evidence that expressions of CDK2/4/6 and STAT3 can be directly regulated by NSC765690 and NSC765599 with consequent antitumor implications in multiple cancer types.
2. Materials and Methods
2.1. In Vitro Anticancer Screening against 60 Full NCI Cell Panels of Human Tumor Cell Lines
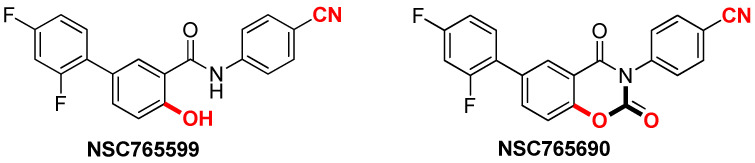
NSC765690 and NSC765599 (Figure 1) were submitted to the National Cancer Institute (NCI) for the screening of its panel of NCI60 cancer cell lines. The preliminary single-dose screening of the two compounds was conducted against 60 full NCI cell line panels comprising melanomas, leukemia, central nervous system (CNS) cancers, NSCLC, renal cancer, breast cancer, ovarian cancer, and prostate cancer in agreement with the protocol of the NCI. Following single-dose testing at 10 μM, the two compounds were selected for five-dose screening against the same panels of cancer cell lines. As described previously [39,40], protocols for NCI60 cell five-dose screening involved seeding of about 5000~40,000 cells/well (depending on the doubling time of individual cell lines) in 96-well plates, followed by treatment with NSC765690 or NSC765599 at concentrations of 0.01, 0.1, 1.0, 10, and 100 μM and incubation at 37 °C in 5% humidified CO2 for 48 h. Cells were fixed with a sulforhodamine B (SRB) solution [41] followed by a series of washing and staining to determine their viability. Growth inhibition was calculated relative to cells without drug treatment and time-zero control. Results of the five-dose assay are represented in terms of a dose-dependent curve, tumor growth inhibition (TGI), LC50 (concentration needed to kill 50% of cells by cytotoxic activity) for each cell line tested, and GI50 (concentration needed to inhibit 50% of cancer cell growth) [42]. Results were presented as cell growth relative to the untreated cell control and to the time zero number of cells. Growth inhibitions were indicated by values between 0 and 100, while lethality (cytotoxic effect) was indicated by values less than 0.
Figure 1.
Chemical structures of NSC765599 (N-(4-cyanophenyl)-2′,4′-difluoro-4-hydroxy-[1,10-biphenyl]-3-carboxamide) and NSC765690 (4-(6-(2,4-Difluorophenyl)-2,4-dioxo-2H-benzo[e][1,3]-oxazin-3(4H)-yl) benzonitrile).
2.2. Identifying the Molecular Targets and Therapeutic Classes of NSC765599 and NSC765690
NSC765599 and NSC765690 were screened for potential drug target using SwissTarget prediction algorithm, which predicts potential drug target based on the principle of similarity [43]. In addition, we also employ the computer-aided Prediction of Biological Activity Spectra (PASS) web resources to predict the potential drug targets [44]. The activity patterns (fingerprints) of both NSC765599 and NSC765690 were correlated to NCI synthetic compounds, standard agents and molecular targets using the DTP-COMPARE algorithms [45]. The NSC numerical IDs were used as “seed” while GI50, TGI and LC50 were set as the endpoints.
2.3. In Silico Molecular Docking Analyses
The three-dimensional (3D) structure of palbociclib (CID: 5330286) and SH-4-54 (CID: 72188643) were retrieved in SDF file format from the PubChem database, while the 3D structures of NSC765690 and NSC765599 were drawn out in sybyl mol2 format using the Avogadro molecular builder and visualization tool vers. 1.XX [46] and were subsequently transformed into the protein data bank (PDB) format using the PyMOL Molecular Graphics System, vers. 1.2r3pre (Schrödinger, LLC). The PDB file of the 3D structure of the receptors and crystal structures of apo CDK2 (PDB; 4EK3), CDK4/cyclin D3 (PDB; 3G33), CDK6/cyclin (PDB; 1JOW), and STAT3 (PDB; 4ZIA), were retrieved from the Protein Data Bank. The PDB file formats of the ligands (NSC765690, NSC765599, and palbociclib) and the receptors (STAT3; CDK2, 4, and 6) were subsequently converted into the Auto Dock Pdbqt format using AutoDock Vina (vers. 0.8, The Scripps Research Institute, La Jolla, CA, USA) [47]. Pre-docking preparation of the receptors followed the removal of water molecules, while hydrogen atoms and Kolmman charges were added accordingly. Molecular docking studies were performed using Autodock VINA software and by following the protocols described in our previous study [3]. The docking results based on hydrogen bonds and electrostatic and hydrophobic interactions of the best pose of the ligand–receptor complexes were expressed as binding energy values (kcal/mol). PyMOL software was used to visualize H-bond interactions, binding affinities, interacting amino acid residues, binding atoms on the ligands and receptors, and 3D graphical representations of ligand-receptor complexes, while 2D graphical illustrations of ligand-binding interactions were further visualized using Discovery studio visualizer vers. 19.1.0.18287 (BIOVIA, San Diego, CA, USA) [48].
2.4. Pharmacokinetics, Drug-Likeness, Toxicity and Medicinal Chemical Analyses
The drug-likeness, medicinal chemistry, and pharmacokinetics, including the adsorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of NSC765690 and NSC765599 were analyzed using SwissADME software developed by the Swiss Institute of Bioinformatics [49]. The drug-likeness properties were analyzed in terms of Ghose (Amgen), Egan (Pharmacia), and Veber (GSK), and more importantly, the Lipinski (Pfizer) rule-of-five, as well as cLogP, molecular mass, hydrogen acceptor, hydrogen donor, and molar refractive index [50] for drug-likeness and drug discovery. The Abbot Bioavailability Score was calculated based on the probability of the compound to have at least 0.1 (10%) oral bioavailability in rats or measurable Caco-2 permeability [51], while gastrointestinal absorption and brain penetration properties were analyzed using the Brain Or IntestinaL EstimateD permeation (BOILED-Egg) model [52]. The acute toxicity (LD50) in rats and environmental toxicity were predicted using GUSAR software [53].
2.5. Data Analysis
Spearman’s rank correlation was used to assess the correlations of NSC765599 and NSC765690 fingerprints with NCI synthetic compounds, standard agents, and molecular targets. The COMPARE correlation threshold was set to ≥45 common cell lines, ≥0.1 correlation coefficient, and ≥0.05 standard deviation. The growth inhibition by NSC765690 and NSC765599 in single-dose assay was obtained by subtracting the positive value on the plot from 100, i.e., a value of 40 would mean 60% growth inhibition.
3. Results
3.1. NSC765690 and NSC765599 Exhibited Anti-Proliferative Effects on NCI60 Human Cancer Cell Lines
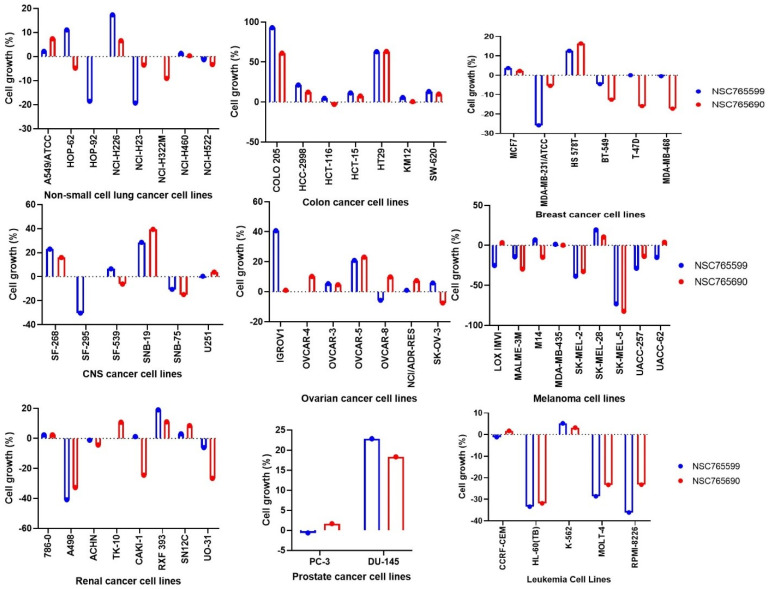
Both NSC765690 and NSC765599 exhibited anti-proliferative effect against all the 60 panels of NCI human cancer cell lines (Figure 2). Furthermore, single-dose treatment with NSC765690 and NSC765599 also demonstrated cytotoxic effects against some cell lines. As indicated by the percentage growth altered by treatment, melanoma (SK-MEL-5, SK-MEL-2, and MALME-3M), renal (A498, UO-31, and CAKI-1), leukemia (HL-60, molt-4, and RPMI-8226), and breast (MDA-MB-468, T-47D, and HS 578T) cancer cell lines were the most responsive to NSC765690 treatment. For NSC765599, melanoma, renal cancer, leukemia, and the breast cancer cell line were also sensitive to cytotoxic effects of NSC765599 at 10 μM. However, panels of prostate, ovarian, and colon cancer cell lines were less responsive to NSC765690 and NSC765599 treatment (Figure 2). These primary single dose screening results clearly indicated the anti-proliferative activities of NSC765690 and NSC765599 against different kinds of human cancer cell lines, and thus are worthy of further evaluation for dose-dependent activities.
Figure 2.
Anti-proliferative effect of NSC765690 and NSC765599 against panels of 60 human cancer cell lines. Each cell line was treated with a single dose of (10 μM) of each compound. The zero points denote the mean percentage of cell growth. The percentage growth inhibition of each cell line relative to the mean is represented by values under 100, whereas those values below 0 indicate cell death.
3.2. NSC765690 and NSC765599 Exhibited Dose-Dependent Cytotoxic Effects against NCI 60 Human Cancer Cell Lines
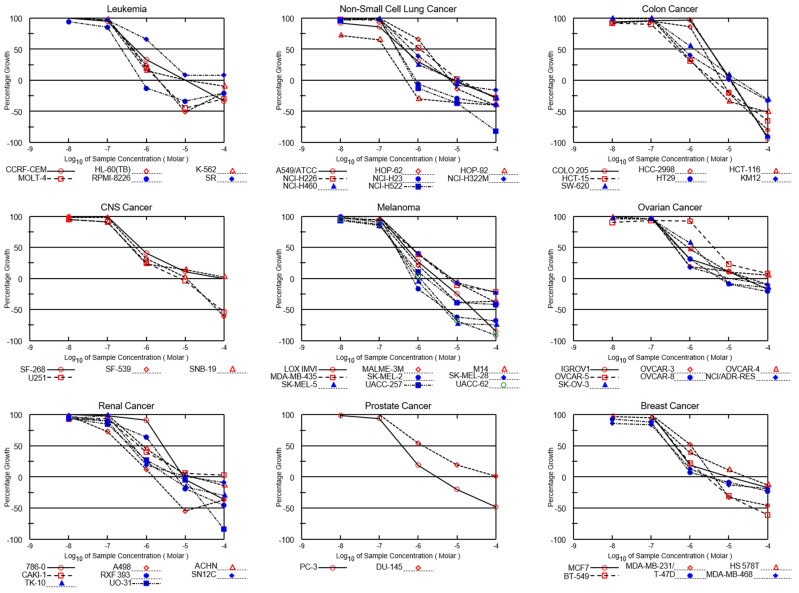
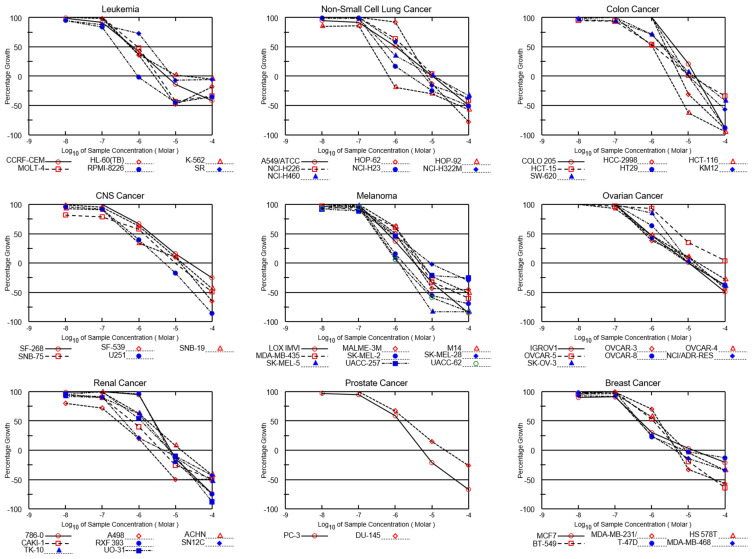
Both NSC765690 and NSC765599 exhibited dose-dependent cytotoxic activities against melanoma (SK-MEL-5, SK-MEL-2, and MALME-3M), NSCLC (HOP-92, A549/ATCC, NCI-H23, NCI-H522, and NCI-H522), CNS (SF-268, SNB-19, and U251), renal (A498, UO-31, and CAKI-1), leukemia (HL-60, molt-4, and RPMI-8226), and breast (MDA-MB-468, T-47D, and HS 578T) cancer cell lines (Figure 3 and Figure 4). The GI50 values ranged from 0.14~2.79 μM in NSC76569-treated cell lines and 0.219~5.55 μM in NSC765599-treated cell lines. Consistent with the activities demonstrated in single-dose treatments, the NSCLC, melanoma, renal, and breast cancer cell lines were most sensitive to NSC765690 treatment, while leukemia, CNS, ovarian, and colon cancer cell lines were the least sensitive to NSC765690 (Table 1). Analysis of LC50 values also indicated that the melanoma cell lines were the most sensitive to drug treatments, with NSC765690 being the most active (Table 1).
Figure 3.
Dose–cytotoxic response curves of NSC765690 against panels of 60 NCI human cancer cell lines. The growth percentage value of +100 on the Y-axis represents the growth of untreated cells, the 0 value represents no net growth, while −100 represents the complete death of cells.
Figure 4.
Dose–cytotoxic response curves of NSC765599 against panels of 60 NCI human cancer cell lines. The growth percentage value of +100 on the Y-axis represents the growth of untreated cells, the 0 value represents no net growth, while −100 represents the complete death of cells.
Table 1.
Anti-proliferative and cytotoxic activities of NSC765690 and NSC765599 against panels of NCI60 human cancer cell lines.
| Cancer Type | Panel/Cell Line | GI50 (μM) | TGI (μM) | LC50 (μM) | |||
|---|---|---|---|---|---|---|---|
| NSC765690 | NSC765599 | NSC765690 | NSC765599 | NSC765690 | NSC765599 | ||
| Leukemia | CCRF-CEM | 0.53 | 0.703 | 9.73 | 5.6 | >100 | >100 |
| HL-60(TB) | 0.51 | 0.586 | 2.09 | 2.68 | >100 | >100 | |
| K-562 | 0.37 | 0.628 | 10.2 | 19.1 | >100 | >100 | |
| MOLT-4 | 0.46 | 0.928 | 2.05 | 3.39 | >100 | >100 | |
| RPMI-8226 | 0.23 | 0.248 | 0.74 | 0.951 | >100 | >100 | |
| SR | 1.91 | 1.95 | >100 | 8.27 | >100 | >100 | |
| NSCLC | A549/ATCC | 0.44 | 1.05 | 7.96 | 10.6 | >100 | 99.4 |
| HOP-62 | 1.61 | 2.51 | 6.78 | 7.48 | >100 | 37 | |
| HOP-92 | 0.14 | 0.219 | 0.48 | 0.655 | >100 | 53.7 | |
| NCI-H226 | 1.07 | 1.73 | 11.2 | 12 | >100 | >100 | |
| NCI-H23 | 0.29 | 0.395 | 0.88 | 2.54 | >100 | 94.2 | |
| NCI-H322M | 0.68 | 1.3 | 6.7 | 6.13 | >100 | >100 | |
| NCI-H460 | 0.45 | 0.569 | 8.78 | 10.7 | >100 | >100 | |
| NCI-H522 | 0.28 | 0.77 | 19.4 | ||||
| Colon Cancer | COLO 205 | 3.23 | 4.48 | 11.2 | 15.6 | 35.8 | 44.4 |
| HCC-2998 | 2.16 | 2.42 | 6.31 | 5.83 | 30.6 | 21.2 | |
| HCT-116 | 0.55 | 1.05 | 3.1 | 2.84 | 93.1 | 7.7 | |
| HCT-15 | 0.48 | 1.22 | 4.05 | 11.4 | 45.9 | >100 | |
| HT29 | 3.63 | 3.43 | 10.6 | 9.93 | 36.5 | 36.6 | |
| KM12 | 0.7 | 1.99 | 10.5 | 10.1 | >100 | 75.5 | |
| SW-620 | 1.28 | 2.1 | 16.5 | 13.7 | >100 | >100 | |
| CNS Cancer | SF-268 | 0.71 | 2.17 | >100 | 24.6 | >100 | >100 |
| SF-539 | 0.53 | 1.78 | 10.8 | 13.7 | 67.8 | 63.8 | |
| SNB-19 | 0.41 | 0.529 | >100 | 15.2 | >100 | >100 | |
| SNB-75 | 1.39 | 9.75 | >100 | ||||
| U251 | 0.42 | 0.633 | 7.23 | 4.97 | 82.9 | 29.7 | |
| Melanoma | LOX IMVI | 0.5 | 0.64 | 3.28 | 3.36 | 26.3 | 21 |
| MALME-3M | 0.4 | 1.02 | 2.26 | 3.48 | >100 | >100 | |
| M14 | 0.68 | 1.38 | 7.16 | 5.37 | >100 | 93.7 | |
| MDA-MB-435 | 0.63 | 1.31 | 6.04 | 4.5 | >100 | 42.7 | |
| SK-MEL-2 | 0.28 | 0.443 | 0.73 | 1.69 | 5.51 | 8.39 | |
| SK-MEL-28 | 0.68 | 0.865 | 7.05 | 9.28 | >100 | >100 | |
| SK-MEL-5 | 0.26 | 0.345 | 0.88 | 1.26 | 4.61 | 4.39 | |
| UACC-257 | 0.29 | 0.815 | 1.59 | 4.84 | >100 | >100 | |
| UACC-62 | 0.28 | 0.319 | 1.14 | 1.22 | 5.59 | 7.21 | |
| Ovarian Cancer | IGROV1 | 0.55 | 0.909 | 24.3 | 10.7 | >100 | >100 |
| OVCAR-3 | 0.44 | 0.674 | 33 | 15.3 | >100 | >100 | |
| OVCAR-4 | 0.86 | 0.841 | >100 | 18.5 | >100 | >100 | |
| OVCAR-5 | 4.04 | 5.55 | >100 | >100 | >100 | >100 | |
| OVCAR-8 | 0.51 | 1.64 | 6.01 | 10.1 | >100 | >100 | |
| NCI/ADR-RES | 0.42 | 0.741 | 10.4 | 9.96 | >100 | >100 | |
| SK-OV-3 | 1.29 | 2.68 | 7.3 | 11.8 | >100 | >100 | |
| Renal cancer | 786-0 | 2.72 | 2.58 | 9.07 | 7.45 | >100 | 39.7 |
| A498 | 0.24 | 0.263 | 1.52 | 1.93 | >100 | >100 | |
| ACHN | 0.82 | 1.73 | 15.5 | 14.4 | >100 | >100 | |
| CAKI-1 | 0.65 | 0.637 | >100 | 4.07 | >100 | >100 | |
| RXF 393 | 1.47 | 2.48 | 5.93 | 6.72 | >100 | 35.2 | |
| SN12C | 0.37 | 0.384 | 12.7 | 4.73 | >100 | >100 | |
| TK-10 | 0.45 | 1.46 | 4.01 | 6.87 | >100 | 82.7 | |
| UO-31 | 0.41 | 1.14 | 7.18 | 6.95 | 37.4 | 32.4 | |
| Prostate Cancer | PC-3 | 0.38 | 1.3 | 3.04 | 5.43 | >100 | 42.8 |
| DU-145 | 1.28 | 2.16 | >100 | 23.4 | >100 | >100 | |
| Breast Cancer | MCF7 | 0.42 | 0.471 | 9.11 | 13.4 | >100 | >100 |
| MDA-MB231/ATCC | 1.06 | 4.76 | 4.08 | 50.5 | >100 | 0 | |
| HS 578T | 0.65 | 1.31 | 28.7 | 8.65 | >100 | >100 | |
| BT-549 | 0.45 | 1.11 | 2.59 | 5.5 | 43.6 | 48.8 | |
| T-47D | 0.29 | 0.449 | 2.74 | 7.49 | >100 | >100 | |
| MDA-MB-468 | 0.3 | 0.425 | 3.17 | 4.42 | >100 | >100 | |
The GI50 = 50% growth inhibition, TGI = total growth inhibition, LC50 = 50% loss of cells.
3.3. NSC765599 and NSC765690 Shared Similar NCI Anti-Cancer Fingerprints and Molecular Targets of Cell Cycle Transition Proteins
DTP-COMPARE analysis results showed that the 10 NCI synthetic compounds that are most correlated with NSC765690 and NSC765599 anti-cancer fingerprints are small molecules (MW: 168.19~506.5 g/mol), with p values in the range of 0.44~0.62. In addition, NSC765690 (p-value range of 0.33~0.74) and NSC765599 (p-value range of 0.15~0.36) shared anticancer mechanism with a number of standardized drugs in the NCI database (Table 2). The molecular target fingerprints generated also showed that both compounds shared a similar positive correlation (p = 0.1~0.3) with the expression of multiple genes, most of which are proteins involved in cell cycle progressions (Table 3).
Table 2.
NCI synthetic compounds and standard anticancer agent sharing similar anti-cancer fingerprints and mechanistic correlation with NSC765599 and NSC765690.
| NCI-Synthetic Compounds | NCI-Standard Agents | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Drugs | Rank | P | CCLC | Target Descriptor | MW (g/mol) | P | CCLC | Target Descriptor | Mechanism of Action |
| NSC765690 Fingerprints | 1 | 0.61 | 42 | Antineoplastic-643812 | 419.3 | 0.74 | 55 | Actinomycin D | Transcription inhibitor |
| 2 | 0.58 | 49 | Combretastatin A-4 | 316.3 | 0.61 | 58 | Mitramycin | Transcription inhibitor | |
| 3 | 0.55 | 49 | N-(3-chloro-2-methylphenyl)-2-hydroxy-3-nitrobenzamide | 306.7 | 0.6 | 59 | Thioguanine | Inhibit cell cycle transition | |
| 4 | 0.51 | 46 | 2-Methyl-4-(phenylimino)naphth(2,3-d)oxazol-9-one | 288.3 | 0.58 | 58 | Cisplatin | Inhibit DNA replication | |
| 5 | 0.51 | 46 | Resibufogenin, Methacrylate De | 452.6 | 0.58 | 49 | Morpholino-ADR | Tubulin inhibitor | |
| 6 | 0.5 | 43 | 3-Nitro-2′,4′-Salicyloxylidide | 286.2 | 0.56 | 59 | 5-Azacytidine | Tubulin inhibior | |
| 7 | 0.48 | 49 | 5,7-Dichloro-3-hydroxy-3-[2-(4-nitrophenyl)-2-oxoethyl]-1,3-dihydro-2H-indol-2-one | 381.2 | 0.53 | 58 | Topotecan | DNA damage inducer | |
| 8 | 0.46 | 58 | 4-ipomeanol | 168.1 | 0.48 | 59 | Doxorubicin (Adriamycin) | DNA damage inducer | |
| 9 | 0.46 | 48 | 3,3′-Diethyl-9-methylthiacarbocyanine iodide | 506.5 | 0.39 | 59 | 5-Fluorouracil | Inhibitor of DNA replication. | |
| 10 | 0.44 | 44 | 2,2-Dibutyl-3-(p-tolylsulfonyl)-1,3,2-thiazastannolidine | 462.3 | 0.33 | 59 | Abemaciclib | Inhibit cell cycle transition | |
| NSC765599 Fingerprints | 1 | 0.62 | 47 | N-(3-chloro-2-methylphenyl)-2-hydroxy-3-nitrobenzamide | 306.7 | 0.36 | 57 | Trametinib | MEK Inhibitor |
| 2 | 0.57 | 44 | Uvaretin | 378.4 | 0.3 | 57 | Erlotinib HCL | Growth factor receptor inhibitor | |
| 3 | 0.54 | 44 | Eunicin | 334.4 | 0.29 | 55 | Vandetanib | Growth factor receptor inhibitor | |
| 4 | 0.54 | 44 | Resibufogenin derivative | 452.6 | 0.28 | 56 | Topotecan | DNA damage inducer | |
| 5 | 0.54 | 45 | 1H,3H-Thiazolo(3,4-a)benzimidazole, 1-(2-chloro-6-fluorophenyl)- | 304.8 | 0.27 | 55 | Ixabepilone | microtubule inhibitor | |
| 6 | 0.53 | 44 | Combretastatin A-4 | 316.3 | 0.23 | 57 | Abemaciclib | Inhibitor of cell cycle transition | |
| 7 | 0.52 | 44 | Nagilactone C | 362.4 | 0.25 | 56 | Idelalisib | phosphoinositide 3-kinase | |
| 8 | 0.51 | 56 | dichloroallyl lawsone | 283.1 | 0.23 | 57 | Pazopanib Hydrochloride | Growth factor receptor inhibitor | |
| 9 | 0.51 | 56 | Merbarone | 263.2 | 0.21 | 57 | Doxorubicin (Adriamycin) | DNA damage inducer | |
| 10 | 0.48 | 55 | 5-Bromo-1-[[4-methylidene-5-oxo-2-(4-phenylphenyl)oxolan-2-yl]methyl]pyrimidine-2,4-dione | 453.3 | 0.15 | 57 | Palbociclib | Inhibitor of cell cycle transition | |
P: Pearson’s correlation coefficient. CCLC: Common cell lines count. MW: molecular weight.
Table 3.
Molecular targets correlated to NSC765599 and NSC765690 activity.
| P | CCLC | P | CCLC | Target ID | Gene Card Code | Target Description |
|---|---|---|---|---|---|---|
| NSC765599 | NSC765690 | |||||
| 0.30 | 53 | 0.29 | 55 | CG2399 | CCNB1 | Cyclin B1 |
| 0.30 | 56 | 0.32 | 58 | CG2465 | CCND1 | Cyclin D1 |
| 0.29 | 51 | 0.31 | 52 | CG2440 | RARB | Retinoic Acid Receptor Beta |
| 0.28 | 54 | 0.13 | 55 | CG2585 | CDH1 | Cadherin-1 |
| 0.28 | 48 | 0.17 | 50 | CG2558 | FGFR1 | Fibroblast growth factor receptor 1 |
| 0.28 | 55 | 0.28 | 57 | CG2369 | RAF1 | Raf-1 Proto-Oncogene |
| 0.23 | 50 | - | - | CG2405 | E2F4 | E2F Transcription Factor 4 |
| 0.22 | 52 | - | - | CG2555 | CDKN2A | cyclin-dependent kinase inhibitor 2A |
| 0.21 | 56 | 0.15 | 58 | CG2357 | MYCN | N-myc proto-oncogene protein |
| 0.22 | 50 | CG2499 | CDC25A | Activator of cyclin dependent kinase 2/4 | ||
| 0.21 | 56 | 0.11 | 58 | CG2531 | CDK4 | Cyclin dependent kinase 4 |
| 0.21 | 56 | - | - | CG2448 | TCL1A | T-cell leukemia/lymphoma protein 1A |
| 0.20 | 49 | - | - | CG2269 | BTK | Bruton Tyrosine Kinase |
| 0.15 | 56 | 0.16 | 58 | CG2311 | PIK3CB | Phosphatidylinositol-4,5-bisphosphate 3-kinase |
| 0.14 | 55 | - | - | CG2466 | CCND2 | Cyclin D2 |
| 0.12 | 56 | 0.1 | 58 | CG2327 | CDK6 | Cyclin dependent kinase 6 |
| - | - | 0.13 | 57 | CG2467 | CDC25B | Activator of cyclin dependent kinase CDC2 |
| - | - | 0.18 | 58 | CG2468 | CDKN1A | Cyclin Dependent Kinase Inhibitor 1A |
P: Pearson’s correlation coefficient. CCLC: Common cell lines count. Pearson’s correlation coefficient ranges from high −1 (negative correlation) to +1 (high positive correlation). The higher the positive value, the more positive correlation between gene expression and NSC765599 and NSC765690 activities.
3.4. CDK2/4/6 and STAT3 Are Potential Druggable Candidates for NSC765690 and NSC765599
Using NSC765690 and NSC765599 as query molecules on the SwissTargetPrediction algorithm, a computer-based drug target prediction tool that identifies the most probable macromolecular targets of a small molecule, on the basis of similarity with a known actives compound in the library [54], we identified a number of targetable proteins, most of which are CDKs and associated proteins. Among these predicted targets, we found that kinases, cytochrome P450, enzymes, and electrochemical transporters were the most occurring targeted classes (Table 4, Supplementary Figure S1). Specifically, STAT3, four members of CDKs (CDK1/2/4/9), and three cyclins were among the top-ranked targeted proteins, while NSC765599 was predicted to target CDK2/4/5/6. Other top-ranked proteins targetable by NSC765599 are shown in Table 4. Coherent with the SwissTarget prediction, the PASS analysis of NSC765690 and NSC765699 also predicted (all pa > pi) inhibitions of CDKs and STAT3 amongst other activities (Table 5).
Table 4.
SwissTarget prediction of potential protein targets for NSC765690 and NSC765599.
| Gene Name | Common Name | Uniprot ID | ChEMBL ID | Target Class |
|---|---|---|---|---|
| NSC765690 Targets | ||||
| CDK9/cyclin T1 | CDK9, CCNT1 | P50750 O60563 | CHEMBL2111389 | Other cytosolic protein |
| Cyclin-dependent kinase 1 | CDK1 | P06493 | CHEMBL308 | Kinase |
| Cyclin-dependent kinase 1/cyclin B | CCNB3 CDK1 CCNB1 CCNB2 | Q8WWL7 P06493 P14635 O95067 | CHEMBL2094127 | Other cytosolic protein |
| Cyclin-dependent kinase 2 | CDK2 | P24941 | CHEMBL301 | Kinase |
| Cyclin-dependent kinase 4/cyclin D1 | CCND1 CDK4 | P24385 P11802 | CHEMBL1907601 | Kinase |
| Epidermal growth factor receptor erbB1 | EGFR | P00533 | CHEMBL203 | Kinase |
| Signal transducer and activator of transcription 3 | STAT3 | P40763 | CHEMBL4026 | Transcription factor |
| Fibroblast growth factor receptor 1 | FGFR1 | P11362 | CHEMBL3650 | Kinase |
| Insulin receptor | INSR | P06213 | CHEMBL1981 | Kinase |
| MAP kinase ERK2 | MAPK1 | P28482 | CHEMBL4040 | Kinase |
| PI3-kinase p110-gamma subunit | PIK3CG | P48736 | CHEMBL3267 | Enzyme |
| Platelet-derived growth factor receptor | PDGFRA PDGFRB | P16234 P09619 | CHEMBL2095189 | Kinase |
| Rho-associated protein kinase 1 | ROCK1 | Q13464 | CHEMBL3231 | Kinase |
| Serine/threonine-protein kinase 11/16/Chk1/MST2 | STK11/3/16/CHEK1 | Q15831 | CHEMBL5606 | Kinase |
| Tyrosine-protein kinase ABL/ITK/JAK1/JAK2 | ABL1 | P00519 | CHEMBL1862 | Kinase |
| NSC765599 Targets | ||||
| Cyclin-dependent kinase 5/CDK5 activator 1 | CDK5R1 CDK5 | Q15078 Q00535 | CHEMBL1907600 | Kinase |
| Epidermal growth factor receptor erbB1 | EGFR | P00533 | CHEMBL203 | Kinase |
| Cyclin-dependent kinase 2 | CDK2 | P24941 | CHEMBL301 | Kinase |
| Cyclin-dependent kinase 4/cyclin D1 | CCND1, CDK4 | P24385, P11802 | CHEMBL1907601 | Kinase |
| Hepatocyte growth factor receptor | MET | P08581 | CHEMBL3717 | Kinase |
| Inhibitor of NF-kappa-B kinase (IKK) | CHUK | O15111 | CHEMBL3476 | Kinase |
| Insulin-like growth factor I receptor | IGF1R | P08069 | CHEMBL1957 | Kinase |
| MAP kinase p38 alpha | MAPK14 | Q16539 | CHEMBL260 | Kinase |
| CDK6/cyclin D1 | CCND1, CDK6 | P24385, Q00534 | CHEMBL2111455 | Kinase |
| PI3-kinase p110-alpha/p85-alpha | PIK3CA, PIK3R1 | P42336, P27986 | CHEMBL2111367 | Enzyme |
| Receptor protein-tyrosine kinase erbB-2 | ERBB2 | P04626 | CHEMBL1824 | Kinase |
| Receptor protein-tyrosine kinase erbB-4 | ERBB4 | Q15303 | CHEMBL3009 | Kinase |
| Rho-associated protein kinase 1 | ROCK1/2 | Q13464 | CHEMBL3231 | Kinase |
| Signal transducer and activator of transcription 3 | STAT3 | P40763 | CHEMBL4026 | Transcription factor |
| Serine/threonine-protein kinase RIPK2 | RIPK2 | O43353 | CHEMBL5014 | Kinase |
| Tankyrase−1/2 | TNKS, TNKS2 | O95271, Q9H2K2 | CHEMBL6164, 6154 | Enzyme |
| Tyrosine-protein kinase JAK1 | JAK1 | P23458 | CHEMBL2835 | Kinase |
| Tyrosine-protein kinase SRC | SRC | P12931 | CHEMBL267 | Kinase |
| Vascular endothelial growth factor receptor 1/2 | FLT1, KDR | P17948, P35968 | CHEMBL1868, 279 | Kinase |
Targets were predicted using Swiss Target Prediction, which operates on the principle of ‘similarity’.
Table 5.
Prediction of Biological Activity Spectra (PASS) of NSC765690 and NSC765699 Targets.
| NSC765690 PASS Predicted Targets | NSC765699 PASS Predicted Targets | ||||
|---|---|---|---|---|---|
| Pa | Pi | Activity | Pa | Pi | Activity |
| 0.446 | 0.034 | CDK6/cyclin D1 inhibitor | 0.505 | 0.012 | Transcription factor inhibitor |
| 0.430 | 0.037 | Transcription factor STAT inhibitor | 0.430 | 0.037 | Transcription factor STAT inhibitor |
| 0.266 | 0.085 | Transcription factor STAT3 inhibitor | 0.391 | 0.020 | CDK6 inhibitor |
| 0.167 | 0.038 | CDK9/cyclin T1 inhibitor | 0.255 | 0.072 | Transcription factor STAT3 inhibitor |
| 0.151 | 0.046 | CDK2/cyclin A inhibitor | 0.162 | 0.029 | CDK2/cyclin A inhibitor |
| 0.114 | 0.021 | Transcription factor STAT6 inhibitor | 0.114 | 0.021 | Transcription factor STAT6 inhibitor |
| 0.101 | 0.094 | CDK1/cyclin B inhibitor | 0.020 | 0.011 | CDK4/cyclin D3 inhibitor |
| 0.019 | 0.012 | CDK4/cyclin D3 inhibitor | 0.046 | 0.007 | CDK5 inhibitor |
Key: Pa > Pi, Pa: probability to be active, Pi: probability to be inactive.
3.5. Molecular Docking Reavealed Favoured Ligandability of NSC765690 and NSC765599 for CDK2/4/6 and STAT3
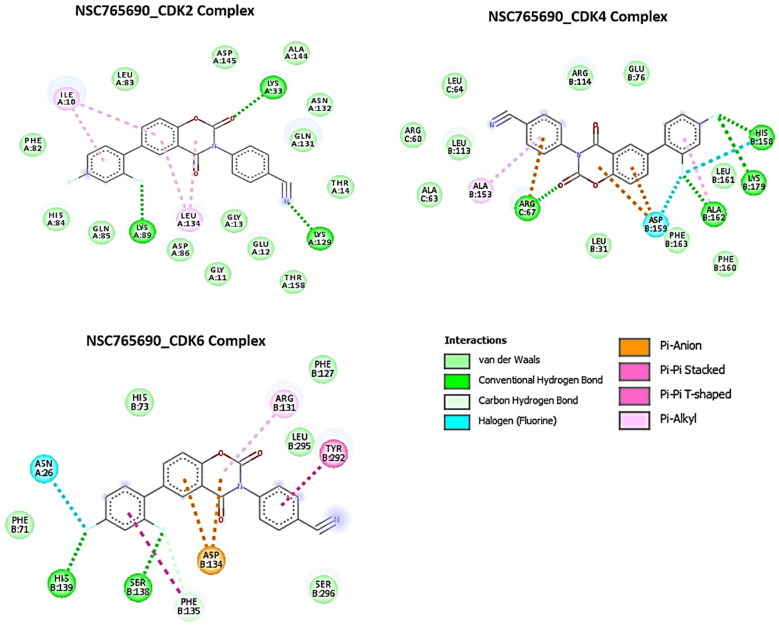
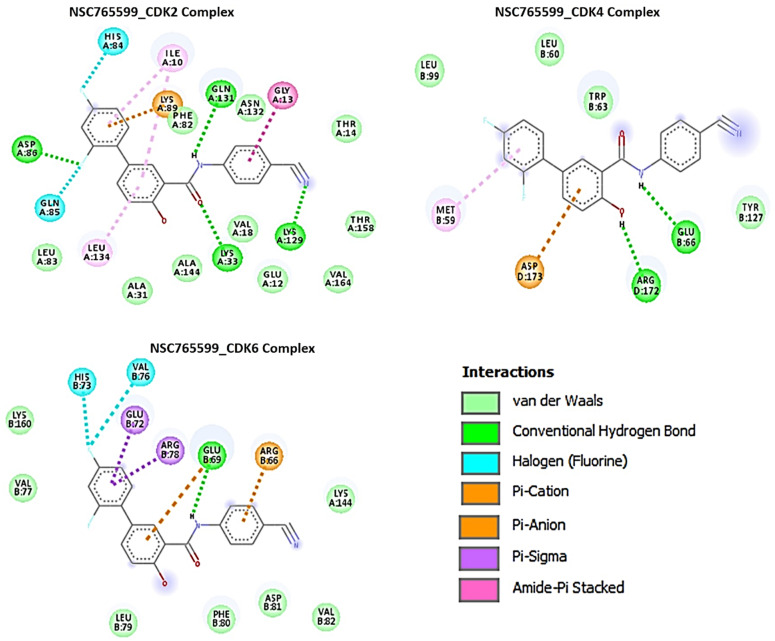
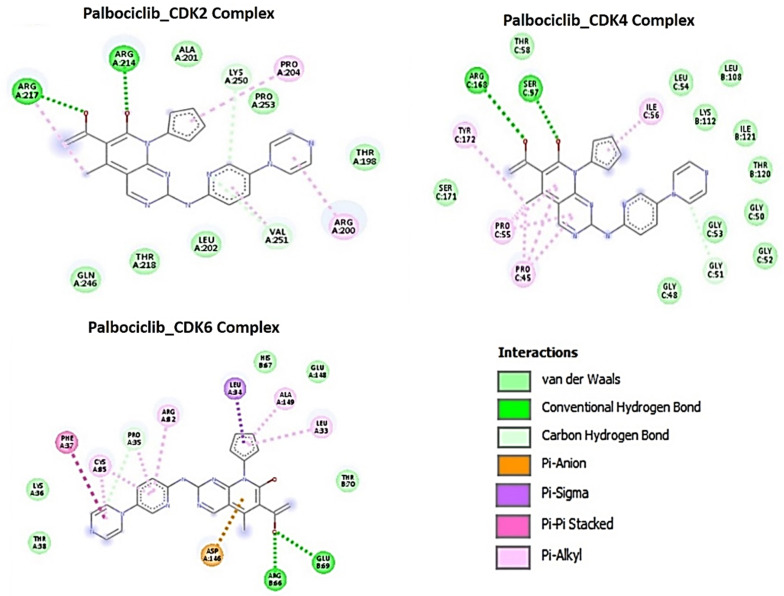
Using molecular docking studies, we found that both NSC765690 and NSC765599 exhibited strong interactions with the crystal structure of apo CDK2 (PDB; 4EK3), CDK4/cyclin D3 (PDB; 3G33), CDK6/cyclin (PDB; 1JOW), and STAT3 (PDB; 4ZIA). The observed interactions and binding affinities indicated that CDK2 is the most favored receptor for both compounds (Figure 5 and Figure 6, Table 6, Supplementary Figures S2–S4), while NSC765690 exhibited stronger interactions with CDK2/4/6 than does NSC765599. Furthermore, we compared the docking profiles of the two compounds with a standard CDK inhibitor, palbociclib (Figure 7), and found that NSC765690 demonstrated stronger binding affinities with CDK2/4/6 compared to NSC765599 and palbociclib. Docking of all ligands (NSC765690, NSC765599, and palbociclib) exhibited short binding distances with the receptors CDK2 (2.14~3.36 Å), CDK4 (1.87~3.23 Å), and CDK6 (2.31~3.17 Å) (Table 6).
Figure 5.
Docking profiles of NSC765690 with CDK2, CDK4, and CDK6. Two dimensional (2D) representations of ligand–receptor interactions occurring between NSC765690 and the target receptors (CDK2/4/6). The deep-green color indicates the strongest ligand–receptor interaction (due to conventional hydrogen bonds) of the best docking pose.
Figure 6.
Docking profiles of NSC765599 with CDK2, CDK4, and CDK6. Two dimensional (2D) representations of ligand–receptor interactions occurring between NSC765599 and the target receptors (CDK2/4/6). The deep-green color indicates the strongest ligand–receptor interaction (due to conventional hydrogen bonds) of the best docking pose.
Table 6.
Comparative docking profile of NSC765690, NSC765599 and standard STAT3/CDK2/4/6 inhibitors.
| Cyclin Dependent Kinase 2 | ||||||
| Docking Parameters | NSC765599_CDK2 Complex | NSC765690_CDK2 Complex | Palbociclib_CDK2 Complex | |||
| ΔG = (Kcal/mol) | −11 | −11.6 | −9.1 | |||
| Type of Interactions | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) |
| Conventional H-bond | 4 | LYS129 (2.67), LYS33 (2.73), GLN131 (2.72) ASP86 (3.36), | 3 | LYS129 (2.75), LYS89 (2.15) LYS33 (2.14) | 2 | ARG214(2.16), ARG217 (2.65) |
| C-H bond | 2 | VAL251(3.70) LYS250 (3.70) | ||||
| Halogen bond | 2 | HIS84 (3.69),GLN85 (3.36) | ||||
| Pi-cation | 1 | LYS89 (3.11) | ||||
| Pi-anion | ||||||
| Pi-alkyl | 2 | LEU134, ILE10 | 2 | PRO204, ARG200 | ||
| Pi-pi stack | ||||||
| Amide-pi stack | 1 | GLY13 (3.34) | ||||
| Van der waal forces | 10 | LEU83, PHE82, ALA144, VAL18, GLU12, VAL164, THR158, ALA31, THR14, ASN132, | 14 | PHE82, LEU83, ASP145, ALA144, ASN132, GLN131, THR14, THR158, GLU12, GLY13, GLY11, ASP86, GLN85, HIS84 | 5 | GLN246, THR218, LEU202, THR198, LYS250 |
| Cyclin Dependent Kinase 4 | ||||||
| NSC765599_CDK4 Complex | NSC765690_CDK4 Complex | Palbociclib_CDK4 Complex | ||||
| ΔG = (Kcal/mol) | −7.5 | −8.5 | −8.3 | |||
| Type of Interactions | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) |
| Conventional H-bond | 2 | ARG172 (2.07) GLU66 (2.06) | 4 | ARG67(1.84), ALA162 (2.62) LYS179 (2.90) HIS158(2.87) | 4 | ARG168(3.23), SER57(2.49) |
| Halogen bond | 2 | ASP159 (2.90), HIS158(3.62) | ||||
| C-H bond | 1 | GLY51 (3.46) | ||||
| Pi-cation | 1 | ARG67(3.89) | ||||
| Pi-anion | 1 | ASP173 (4.48) | 1 | ASP159 (3.78) | ||
| Pi-alkyl | 1 | MET59 | 1 | ALA153, ALA162 | 4 | TYR172, PRO55, PRO45, ILE56 |
| Van der waal forces | 3 | LEU60, LEU99, TRP63 | 10 | LEU64, ARG60, LEU113, ALA63, LEU31, PHE163, PHE160, LEU161, GLU76, ARG114 | 11 | SER171, GLY48, GLY53, GLY52, GLY50, THR120, ILE121, LYS112, LEU108, LEU54, THR58 |
| Cyclin Dependent Kinase 6 | ||||||
| NSC765599_CDK6 Complex | NSC765690_CDK6 Complex | Palbociclib_CDK6 Complex | ||||
| ΔG = (Kcal/mol) | −9.0 | −9.6 | −8.1 | |||
| Type of Interactions | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) |
| Conventional H-bond | 1 | GLU69 (2.22) | 2 | HIS139 (2.29) SER138 (2.31) | 2 | ARG66 (2.50) GLU69 (3.17) |
| C-H bond | 1 | PHE135(3.69) | 1 | PRO35(3.71) | ||
| Halogen bond | 2 | HIS73 (2.88) VAL76 (2.99) | 1 | ASN26 (3.32) | ||
| Pi-cation | 1 | ARG66 (3.40) | ||||
| Pi-anion | 1 | GLU69 (4.90) | 1 | ASP134(3.99) | 1 | ASP146(3.59) |
| Pi-sigma | 1 | LEU34 | ||||
| Pi-alkyl | 1 | ARG131 | 4 | ALA149, LEU33, ARG82, CYS85 | ||
| Pi-pi stack | 1 | GLU72, ARG78 | 1 | TYR292 | 1 | PHE37292 |
| Pi-pi T-shape | 1 | PHE135 | ||||
| Van der waal forces | 7 | Lys160, Val77, Leu79, Phe80, Asp81, Val82, Lys144 | 5 | Phe71, Ser296, Leu295, Phe127, His73 | 5 | Lys36, Thr38, Thr70, Glu148, His67 |
| Signal Transducer and Activator of Transcription 3 | ||||||
| NSC765599_STAT3 Complex | NSC765690_STAT3 Complex | SH-4-54_STAT3 Complex | ||||
| ΔG = (Kcal/mol) | −8.0 | −8.3 | −7.3 | |||
| Type of Interaction | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) | n-Bond | Interacting AA (Distance (Ă)) |
| Conventional H-bond | 2 | ARG107(2.73) TRP110 (3.79) | 1 | SER113 (2.57) | ||
| C-H bond | 1 | ARG107 (3.23) | 2 | TRP110 (3.69) ALA106 (3.47) | ||
| Halogen | 1 | GLN3 (3.52) | 1 | ALA106 (3.67) | 3 | ARG107(3.65), ARG103 (3.05), ALA44 (3.26) |
| Pi-pi stacked | 1 | TRP110 | ||||
| Pi-sigma | 1 | ALA44 | ||||
| Pi-sulfur | 1 | TRP110 (4.49) | ||||
| Pi-alkyl | 3 | LEU109, ALA106, ALA44 | 3 | LEU109, ARG103, ALA44 | 1 | ALA106 |
| Pi-pi T-shaped | 1 | TRP110 | ||||
| Van der waal forces | 5 | GLU39, GLN41, ASP42, TYR45, TRP43 | 4 | TRP43, GLU39, GLN3, SER113 | 4 | GLU39, GLN3, GLN117, ALA47 |
Interacting AA: Interacting Amino acids.
Figure 7.
Docking profiles of palbociclib with CDK2, CDK4, and CDK6. Two dimensional (2D) representations of ligand–receptor interactions occurring between palbociclib and the target receptors (CDK2/4/6). The deep-green color indicates the strongest ligand–receptor interaction (due to conventional hydrogen bonds) of the best docking pose.
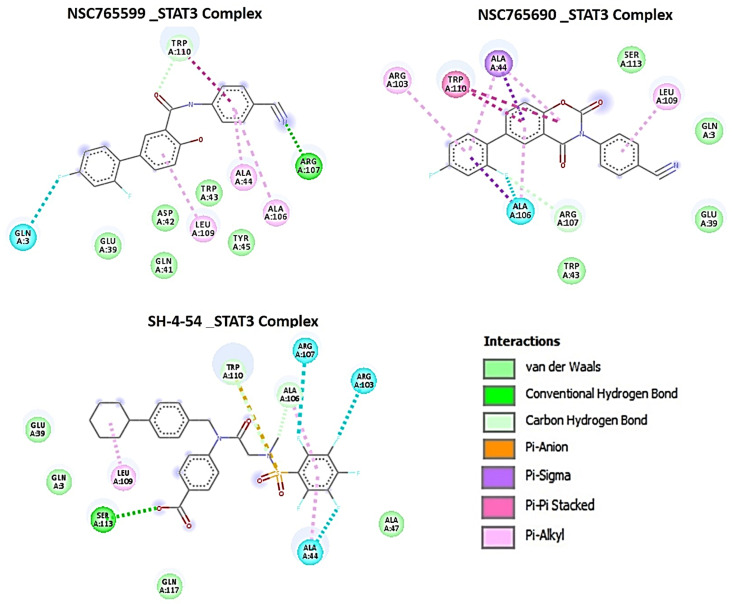
Our careful analysis of the interactions between NSC765690 and the receptors revealed a higher number of conventional H-bonding, pi interactions, and Van der Waal forces created on the ligand backbone with a higher number of amino acids residues than does NSC765599 and palbociclib, in the CDK2/4/6 binding cavities (Table 6). NSC765690-STAT3 and NSC765599-STAT3 complexes were stabilized by similar interactions. However, the overall binding affinity of STAT3 with a known inhibitor, SH-4-54, was less negative than the values observed for NSC765690-STAT3 and NSC765599-STAT3 complexes (Figure 8, Supplementary Figure S5).
Figure 8.
Comparative docking profiles of STAT3 with NSC765599, NSC765690, and SH-4-54 (a known STAT3 inhibitor). A 2D representation of ligand–receptor interactions of STAT3 with NSC765599, NSC765690, and SH-4-54 in the receptor binding pocket.
3.6. NSC765599 and NSC765690 Met the Required Criteria of Drug-Likeness and Safety
NSC765690 met the required criteria of a good drug candidate in terms of lipophilicity, polarity, flexibility, solubility, saturation, and molecular weight, while NSC765599 slightly violated the required range of solubility (Log S(ESOL) = 0~6) and lipophilicity (XlogP3 = −0.7~5) having slight outlying values of −5.71 and 5.47 respectively. Both compounds demonstrated good synthetic accessibilities, highly probable GIA absorption, and bioavailability, but poor BBB permeation (Supplementary Figures S6 and S7, Table 7). Predicted ecotoxicity and acute toxicity for different administration routes of NSC765690 and NSC765599 produce class 4 and 5 levels of acute toxicity (LD50) according to OECD classification (Table 7). Collectively, NSC765599 and NSC765690 met the criteria for a drug-likeness candidate and are relatively known to be toxic.
Table 7.
Drug likeness, ADME and safety/toxicity profile of NSC765690 and NSC765599.
| Properties | NSC765690 | NSC765599 | Reference Value |
|---|---|---|---|
| Formula | C21H10F2N2O3 | C20H12F2N2O2 | - |
| Molecular weight | 376.31 g/mol | 350.32 g/mol | 150–500 g/mol |
| Num. rotatable bonds | 2 | 4 | 0–9 |
| Num. H-bond acceptors | 6 | 5 | 0–10 |
| Num. H-bond donors | 0 | 2 | 0–5 |
| Molar Refractivity | 98.15 | 92.75 | |
| TPSA | 76.00 Ų | 73.12 Ų | 20–130 Ų |
| Fraction Csp3 | 0.00 | 0.00 | 0.25~<1 |
| Log Po/w (XLOGP3) | 4.31 | 5.47 | −0.7~5 |
| Consensus Log Po/w | 4.11 | 4.29 | |
| Log S (ESOL) | 0.34 | −5.71 | 0–6 |
| Lipinski, Ghose, Veber and Egan’s rule | Yes; 0 violation | Yes; 0 violation | |
| Bioavailability Score | 0.56 | 0.55 | >0.1 (10%) |
| Synthetic accessibility | 3.33 | 2.37 | 1 (very easy) to 10 (very difficult). |
| Acute toxicity | |||
| LD50 for Intraperitoneal (mg/kg) | 446.100 (OECD:4) | 593.200 (OECD:5) | |
| LD50 for Intravenous (mg/kg) | 127.200 (OECD:4) | 251.200 (OECD:4) | |
| LD50 for Oral (mg/kg) | 494.000 (OECD:4) | 836.900 (OECD:4) | |
| LD50 for Subcutaneous (mg/kg) | 398.300 (OECD:4) | 740.900 (OECD:4) | |
| Environmental toxicity | |||
| Bioaccumulation factor Log10 (BCF) | 1.204 | 1.210 | |
| Daphnia magna LC50Log10 (mol/L) | 7.294 | 6.713 | |
| Fathead Minnow LC50Log10 (mmol/L) | −3.088 | −3.099 | |
| Tetrahymena pyriformis IGC50Log10 (mol/L) | 2.016 | 2.033 | |
4. Discussion
Despite advances in treatment modalities, cancer survival ratios are still disappointing; thus, developing a novel chemotherapeutic strategy may improve the prognosis of the cancer patient [55]. At present, substantial attention is being focused on small molecules for targeted therapy in cancer treatments [35]. In the present study, we demonstrated the anti-proliferative and dose-dependent cytotoxic effect of NSC765690 and NSC765599 against panels of NCI human cancer cell lines, with NSC765690 demonstrating higher activities than NSC765599. However, there was the least amount of cell lethality in panels of leukemia, CNS, ovarian, and colon cancer cell lines, suggesting that NSC765690 and NSC765599 are not generally toxic to growing cell lines, but display some degree of cytotoxic preference for NSCLC, melanoma, renal, and breast cancer cell line.
The NCI60 cell lines have been well-characterized for genetic and protein expression patterns, and, a computational tool, the DTP-COMPARE algorithm, has been developed to identify the molecular targets and mechanism(s) of action for the unknown compound from known drugs as well as the known molecular targets in the NCI databases [56]. Interestingly, our analysis of correlation patterns of NSC765690 anticancer fingerprint with mechanistically known NCI-Standard Agents suggests that NSC765690 might be targeting DNA replication and cell cycle. NSC765599, on the other hand, shares similar (p = 0.15~0.36) antitumor mechanistic fingerprints with known inhibitors of growth factor receptor and cell cycle transition, and DNA damage inducer. However, the modest correlation with NCI-standard agents suggests that NSC765599 may have a unique mechanism of action not common to standard agent database. The strong antitumor pattern correlation (p = 0.44~0.6) between NSC765690 and NSC765599 fingerprint with NCI synthetic compounds suggests a mechanism of action similar to that of the seed compounds. Unfortunately, the top-ranked correlated compounds antineoplastic-643812 (p = 0.61) and N-(3-chloro-2-methylphenyl)-2-hydroxy-3-nitrobenzamide (p = 0.62) are yet to be mechanistically investigated, while combretastatin A-4 with p-value 0.58 (NSC765690) and p-value 0.53 (NSC765599) have been reported for the mechanism of action, indicating cell cycle arrest and inhibition of tubulin [57], hence providing hypothesis about the possible mechanism of actions of NSC765690 and NSC765599. The high number of cell cycle proteins and cyclin and cyclin-dependent kinases identified as molecular target fingerprints of both NSC765690 and NSC765599 strongly suggest that cyclin and cyclin-dependent kinase pathways are the core molecular targets of NSC765690 and NSC765599 predicted by COMPARE. However, the correlations (p = 0.1~0.3) were low, indicating the possibility of a new mechanism not captured by COMPARE.
The COMPARE prediction, together with in-silico SwissTarget and PASS prediction collectively identified STAT3/CDK2/4/6 as the most probable target for NSC765690 and NSC765599, and thus were further studied for ligand–receptor interactions. Interestingly, we found that NSC765690 and NSC765599 docked well into the receptor cavities of STAT3, CDK2, CDK4, and CDK6, with CDK2 being the most favored receptor while CDK4 appeared to be the least favored. Comparatively, the higher binding affinities and closer proximity of NSC765690 to the receptors suggested that NSC765690 is a better ligand for CDK2, CDK4, and CDK6 than is NSC765599.
NSC765690 and NSC765599 are both derivatives of NDMC101. A close look at the structure-related activities of NSC765690 and NSC765599 revealed that cyclization of salicylanilide core scaffold (Figure 1) was the structural feature associated with enhanced anticancer activity and better docking profiles of NSC765690 with STAT3/CDK2/4/6 compared to NSC765599 with an open-ring structure (Figure 7 and Figure 8). Interestingly, when the same receptors (CDK2/4/6) were docked with palbociclib, a clinical CDK inhibitor, higher binding energies (lower binding affinities) were recorded for palbociclib compared to NSC765690, thus making the latter the more-favored ligand for CDK2/4/6.
The unique stability of NSC765690 in the binding sites of CDK2, CDK4, and CDK6 could be attributed to the larger number of H-bond and pi interactions including pi–cation, pi–anion, pi–alkyl, pi–pi stack, and pi–pi T-shaped interactions between NSC765690 and the receptors. The large numbers of pi interactions, which mostly involve charge transfer, help in intercalating the NSC765690 in the receptors’ binding cavities. The higher affinity of NSC765690 was also associated with the presence of a larger number of Van der Waal forces created on its backbone with respective amino acids Phe82, Leu83, Asp145, Ala144, Asn132, Gln131, Thr14, Thr158, Glu12, Gly13, Gly11, Asp86, Gln85 and His84 of CDK2; Leu64, Arg60, Leu113, Ala63, Leu31, Phe163, Phe160, Leu161, Glu76 and Arg114 of CDK4; and Phe71, Ser296, Leu295, Phe127, and His73 of CDK6, which undoubtedly created a strong cohesive environment, thereby stabilizing the complex formed [58]. These higher numbers of interactions undoubtedly contributed to the higher affinity that NSC765690 has for CDK2, CDK4, and CDK6 than do NSC765599 and palbociclib. Palbociclib has been actively applied in multiple preclinical models and was approved for targeting CDK4/6 as anticancer therapy for breast cancer; however, acquired resistance occurs in almost all cases [20]. Although docking STAT3 with a known inhibitor, SH-4-54, revealed a higher number of conventional H-bond and halogen bond interactions with STAT3, the pi interactions were fewer, and consequently, the overall binding affinity was less negative. Therefore, higher pi interactions also contributed to the higher binding affinity that NSC765690 has for STAT3 than does SH-4-54. Our molecular simulations of ligand–receptor interactions validated the computational prediction of STAT3/CDK2/4/6 as druggable candidates for NSC765690 and NSC765599. The present study, therefore, provides useful molecular docking-based evidence of alternative small molecules that could target CDK4/6 as well as CDK2 and STAT3. They thus offer a wider targeted therapy with less probability of suffering drug resistance
The concept of drug-likeness gives useful guidelines for identifying potential drug candidates during the early stage of drug discovery and development [59]. Estimation of rodent acute toxicity (LD50), an adverse effect that follows a single dose exposure to a substance, is an important task in drug design and risk assessment of chemicals [60,61,62]. The LD50 values predicted for oral, intravenous, intraperitoneal, and subcutaneous administration indicated that both NSC765690 and NSC765599 were non-toxic and will be well-tolerated by the experimental animals. In addition, NSC765690 and NSC765599 met the criteria of a good drug candidate (Table 7), suggesting that the compounds were drug-like molecules and thus possessed the potential to be considered oral drug candidates. Collectively, findings from this study have opened the door for new research direction for NSC765690 and NSC765599 as a novel and potent anti-cancer agent. We found these compounds worthy of further investigation, and both in vitro and in vivo studies in tumor-bearing mice are currently ongoing in our lab to assess the full therapeutic efficacies of these compounds.
5. Conclusions
In conclusion, this study demonstrated the anti-proliferative effect of NSC765690 and NSC765599 against the 60 panels of NCI human cancer cell lines, and dose-dependent cytotoxic preference for NSCLC, melanoma, renal, and breast cancer cell lines. STAT3/CDK2/4/6 signatures appear to be potential druggable candidates for the small molecules and interacted with high binding affinity, indicating the potential of NSC765690 and NSC765599 to dysregulate expressions of STAT3/CDK2/4/6 signature and, consequently, compromised cell survival. Further in-vitro and in-vivo confirmation studies are ongoing in our lab.
Acknowledgments
The NCI Developmental Therapeutics Program (DTP) for the 60-cancer-cell-line screening of selected compounds described in this paper, funded by the National Cancer Institute, National Institutes of Health (NIH-NCI).
Supplementary Materials
The following are available online at https://www.mdpi.com/2227-9059/9/1/92/s1. Figure S1: Pie chart showing the repartition of protein classes of potential druggable candidates for NSC765690 and NSC765599. Figures S2–S5: Docking profiles of NSC765690, NSC765599, palbociclib with CDK2/4/6 and STAT3. Figure S6: BOILED-Egg model of brain or intestinal estimated permeation of NSC765599 and NSC765690. Figure S7: Bioavailability Radar showing suitable physicochemical spaces of the oral bioavailability of NSC765690 and NSC765599.
Author Contributions
B.L. wrote the manuscript; Y.-L.L., N.M., H.K. helped with data collection and analyses; H.-S.H. and M.R.S. synthesized and provided NSC765690 and NSC765599; H.-S.H. and A.T.H.W. designed and oversaw the study. All authors have read and agreed to the published version of the manuscript.
Funding
Hsu-Shan Huang is funded by the Ministry of Science and Technology MOST 109-2113-M-038-003. Alexander TH Wu and Yen-Lin Liu were funded by Taipei Medical University Hospital, Taipei, Taiwan (Grant No. 104TMU-TMUH-11).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The datasets generated and/or analyzed in this study are available on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim I., He Y.-Y. Targeting the AMP-Activated Protein Kinase for Cancer Prevention and Therapy. Front. Oncol. 2013;3:15. doi: 10.3389/fonc.2013.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.-C., Wu A.T.H., Chen J.-H., Huang W.-Y., Lawal B., Mokgautsi N., Huang H.-S., Ho C.-L. HNC0014, a Multi-Targeted Small-Molecule, Inhibits Head and Neck Squamous Cell Carcinoma by Suppressing c-Met/STAT3/CD44/PD-L1 Oncoimmune Signature and Eliciting Antitumor Immune Responses. Cancers. 2020;12:3759. doi: 10.3390/cancers12123759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanwal R., Gupta S. Epigenetic modifications in cancer. Clin. Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., Zheng P., Zhu W. Research Progress of Small Molecule VEGFR/c-Met Inhibitors as Anticancer Agents (2016–Present) Molecules. 2020;25:2666. doi: 10.3390/molecules25112666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhuo L.-S., Xu H.-C., Wang M.-S., Zhao X.-E., Ming Z.-H., Zhu X.-L., Huang W., Yang G.-F. 2, 7-naphthyridinone-based MET kinase inhibitors: A promising novel scaffold for antitumor drug development. Eur. J. Med. Chem. 2019;178:705–714. doi: 10.1016/j.ejmech.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Robinson D.R., Wu Y.-M., Lin S.-F. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen K., Van Bockstaele D.R., Berneman Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires M.S., Feltell R.E., Wallis N.G., Lewis E.J., Smith D.-M., Cross D.M., Lyons J.F., Thompson N.T. Biological characterization of AT7519, a small-molecule inhibitor of cyclin-dependent kinases, in human tumor cell lines. Mol. Cancer Ther. 2009;8:324–332. doi: 10.1158/1535-7163.MCT-08-0890. [DOI] [PubMed] [Google Scholar]
- 10.Brown V.D., Phillips R.A., Gallie B.L. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol. Cell. Biol. 1999;19:3246–3256. doi: 10.1128/MCB.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J., Dynlacht B., Imai T., Hori T.-A., Harlow E. Expression of NPAT, a novel substrate of cyclin E–CDK2, promotes S-phase entry. Genes. Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel B., Tripathi N., Bhardwaj N., Jain S.K. Small Molecule CDK Inhibitors for the Therapeutic Management of Cancer. Curr. Top. Med. Chem. 2020;20:1535–1563. doi: 10.2174/1568026620666200516152756. [DOI] [PubMed] [Google Scholar]
- 13.Kim S., Loo A., Chopra R., Caponigro G., Huang A., Vora S., Parasuraman S., Howard S., Keen N., Sellers W., et al. Abstract PR02: LEE011: An orally bioavailable, selective small molecule inhibitor of CDK4/6– Reactivating Rb in cancer. Mol. Cancer Ther. 2013;12:PR02. doi: 10.1158/1535-7163.targ-13-pr02. [DOI] [Google Scholar]
- 14.Okada Y., Kato S., Sakamoto Y., Oishi T., Ishioka C. Synthetic lethal interaction of CDK inhibition and autophagy inhibition in human solid cancer cell lines. Oncol. Rep. 2017;38:31–42. doi: 10.3892/or.2017.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vijayaraghavan S., Moulder S., Keyomarsi K., Layman R.M. Inhibiting CDK in Cancer Therapy: Current Evidence and Future Directions. Target. Oncol. 2018;13:21–38. doi: 10.1007/s11523-017-0541-2. [DOI] [PubMed] [Google Scholar]
- 16.Kodym E., Kodym R., Reis A.E., Habib A.A., Story M.D., Saha D. The small-molecule CDK inhibitor, SNS-032, enhances cellular radiosensitivity in quiescent and hypoxic non-small cell lung cancer cells. Lung Cancer. 2009;66:37–47. doi: 10.1016/j.lungcan.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Asghar U., Witkiewicz A.K., Turner N.C., Knudsen E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswamy B., Phelps M.A., Baiocchi R., Bekaii-Saab T., Ni W., Lai J.-P., Wolfson A., Lustberg M.E., Wei L., Wilkins D. A dose-finding, pharmacokinetic and pharmacodynamic study of a novel schedule of flavopiridol in patients with advanced solid tumors. Investig. New Drugs. 2012;30:629–638. doi: 10.1007/s10637-010-9563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan A.R., Yang X., Berman A., Zhai S., Sparreboom A., Parr A.L., Chow C., Brahim J.S., Steinberg S.M., Figg W.D. Phase I trial of the cyclin-dependent kinase inhibitor flavopiridol in combination with docetaxel in patients with metastatic breast cancer. Clin. Cancer Res. 2004;10:5038–5047. doi: 10.1158/1078-0432.CCR-04-0025. [DOI] [PubMed] [Google Scholar]
- 20.Pandey K., Park N., Park K.-S., Hur J., Cho Y.B., Kang M., An H.-J., Kim S., Hwang S., Moon Y.W. Combined CDK2 and CDK4/6 Inhibition Overcomes Palbociclib Resistance in Breast Cancer by Enhancing Senescence. Cancers. 2020;12:3566. doi: 10.3390/cancers12123566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien N.A., McDermott M.S.J., Conklin D., Luo T., Ayala R., Salgar S., Chau K., Di Tomaso E., Babbar N., Su F., et al. Targeting activated PI3K/mTOR signaling overcomes acquired resistance to CDK4/6-based therapies in preclinical models of hormone receptor-positive breast cancer. Breast Cancer Res. 2020;22:89. doi: 10.1186/s13058-020-01320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey K., An H.-J., Kim S.K., Lee S.A., Kim S., Lim S.M., Kim G.M., Sohn J., Moon Y.W. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int. J. Cancer. 2019;145:1179–1188. doi: 10.1002/ijc.32020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishibashi K., Koguchi T., Matsuoka K., Onagi A., Tanji R., Takinami-Honda R., Hoshi S., Onoda M., Kurimura Y., Hata J. Interleukin-6 induces drug resistance in renal cell carcinoma. Fukushima J. Med. Sci. 2018;64:103–110. doi: 10.5387/fms.2018-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priego N., Zhu L., Monteiro C., Mulders M., Wasilewski D., Bindeman W., Doglio L., Martínez L., Martínez-Saez E., y Cajal S.R. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018;24:1024–1035. doi: 10.1038/s41591-018-0044-4. [DOI] [PubMed] [Google Scholar]
- 27.Kortylewski M., Kujawski M., Wang T., Wei S., Zhang S., Pilon-Thomas S., Niu G., Kay H., Mulé J., Kerr W.G. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 28.Villarino A.V., Kanno Y., O’Shea J.J. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat. Immunol. 2017;18:374. doi: 10.1038/ni.3691. [DOI] [PubMed] [Google Scholar]
- 29.Kettner N.M., Vijayaraghavan S., Durak M.G., Bui T., Kohansal M., Ha M.J., Liu B., Rao X., Wang J., Yi M., et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019;25:3996–4013. doi: 10.1158/1078-0432.CCR-18-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou S., Tong Q., Liu B., Huang W., Tian Y., Fu X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phosrithong N., Ungwitayatorn J. Molecular docking study on anticancer activity of plant-derived natural products. Med. Chem. Res. 2010;19:817–835. doi: 10.1007/s00044-009-9233-5. [DOI] [Google Scholar]
- 32.Ekins S., Mestres J., Testa B. In silico pharmacology for drug discovery: Methods for virtual ligand screening and profiling. Br. J. Pharm. 2007;152:9–20. doi: 10.1038/sj.bjp.0707305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortega S.S., Cara L.C.L., Salvador M.K. In silico pharmacology for a multidisciplinary drug discovery process. Drug Met. Pers. Ther. 2012;27:199–207. doi: 10.1515/dmdi-2012-0021. [DOI] [PubMed] [Google Scholar]
- 34.Kening L., Yuxin D., Lu L., Dong-Qing W. Bioinformatics Approaches for Anti-cancer Drug Discovery. Curr. Drug Target. 2020;21:3–17. doi: 10.2174/1389450120666190923162203. [DOI] [PubMed] [Google Scholar]
- 35.Coussens N.P., Braisted J.C., Peryea T., Sittampalam G.S., Simeonov A., Hall M.D. Small-Molecule Screens: A Gateway to Cancer Therapeutic Agents with Case Studies of Food and Drug Administration—Approved Drugs. Pharm. Rev. 2017;69:479–496. doi: 10.1124/pr.117.013755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng C.-P., Huang H.-S., Hsu Y.-C., Sheu M.-J., Chang D.-M. A Benzamide-linked Small Molecule NDMC101 Inhibits NFATc1 and NF-κB Activity: A Potential Osteoclastogenesis Inhibitor for Experimental Arthritis. J. Clin. Immunol. 2012;32:762–777. doi: 10.1007/s10875-012-9660-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee C.-C., Liu F.-L., Chen C.-L., Chen T.-C., Chang D.-M., Huang H.-S. Discovery of 5-(2′,4′-difluorophenyl)-salicylanilides as new inhibitors of receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Eur. J. Med. Chem. 2015;98:115–126. doi: 10.1016/j.ejmech.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 38.Lee C.-C., Liu F.-L., Chen C.-L., Chen T.-C., Liu F.-C., Ahmed Ali A.A., Chang D.-M., Huang H.-S. Novel inhibitors of RANKL-induced osteoclastogenesis: Design, synthesis, and biological evaluation of 6-(2,4-difluorophenyl)-3-phenyl-2H-benzo[e][1,3]oxazine-2,4(3H)-diones. Bioorg. Med. Chem. 2015;23:4522–4532. doi: 10.1016/j.bmc.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 40.Holbeck S.L., Collins J.M., Doroshow J.H. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 2010;9:1451–1460. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vichai V., Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 42.Boyd M.R., Paull K.D. Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev. Res. 1995;34:91–109. doi: 10.1002/ddr.430340203. [DOI] [Google Scholar]
- 43.Gfeller D., Michielin O., Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29:3073–3079. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
- 44.Poroikov V.V., Filimonov D.A., Gloriozova T.A., Lagunin A.A., Druzhilovskiy D.S., Rudik A.V., Stolbov L.A., Dmitriev A.V., Tarasova O.A., Ivanov S.M., et al. Computer-aided prediction of biological activity spectra for organic compounds: The possibilities and limitations. Russ. Chem. Bull. 2019;68:2143–2154. doi: 10.1007/s11172-019-2683-0. [DOI] [Google Scholar]
- 45.Paull K., Shoemaker R., Hodes L., Monks A., Scudiero D., Rubinstein L., Plowman J., Boyd M. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. JNCI J. Natl. Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 46.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Chem. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visualizer D.S. BIOVIA, Dassault Systèmes, BIOVIA Workbook, Release 2020. Dassault Systèmes; San Diego, CA, USA: 2020. BIOVIA Pipeline Pilot, Release 2020. [Google Scholar]
- 49.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipinski C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Martin Y.C. A bioavailability score. J. Med. Chem. 2005;48:3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 52.Daina A., Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagunin A.A., Zakharov A.V., Filimonov D.A., Poroikov V.V. A new approach to QSAR modelling of acute toxicity. SAR QSAR Environ. Res. 2007;18:285–298. doi: 10.1080/10629360701304253. [DOI] [PubMed] [Google Scholar]
- 54.Daina A., Michielin O., Zoete V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rathnagiriswaran S., Wan Y.-W., Abraham J., Castranova V., Qian Y., Guo N.L. A population-based gene signature is predictive of breast cancer survival and chemoresponse. Int. J. Oncol. 2010;36:607–616. doi: 10.3892/ijo_00000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bates S.E., Fojo A.T., Weinstein J.N., Myers T.G., Alvarez M., Pauli K.D., Chabner B.A. Molecular targets in the National Cancer Institute drug screen. J. Cancer Res. Clin. Oncol. 1995;121:495–500. doi: 10.1007/BF01197759. [DOI] [PubMed] [Google Scholar]
- 57.Hamze A., Rasolofonjatovo E., Provot O., Mousset C., Veau D., Rodrigo J., Bignon J., Liu J.-M., Wdzieczak-Bakala J., Thoret S. B-ring-modified isocombretastatin A-4 analogues endowed with interesting anticancer activities. ChemMedChem. 2011;6:179–191. doi: 10.1002/cmdc.201100325. [DOI] [PubMed] [Google Scholar]
- 58.Arthur D.E., Uzairu A. Molecular docking studies on the interaction of NCI anticancer analogues with human Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. J. King Saud Univ. Sci. 2019;31:1151–1166. doi: 10.1016/j.jksus.2019.01.011. [DOI] [Google Scholar]
- 59.Keller T.H., Pichota A., Yin Z. A practical view of ‘druggability’. Curr. Opin. Chem. Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Lawal B., Shittu O.K., Oibiokpa F.I., Mohammed H., Umar S.I., Haruna G.M. Antimicrobial evaluation, acute and sub-acute toxicity studies of Allium sativum. J. Acute Dis. 2016;5:296–301. doi: 10.1016/j.joad.2016.05.002. [DOI] [Google Scholar]
- 61.Shittu O.K., Lawal B., Alozieuwa B.U., Haruna G.M., Abubakar A.N., Berinyuy E.B. Alteration in biochemical indices following chronic administration of methanolic extract of Nigeria bee propolis in Wistar rats. Asian Pac. J. Trop. Dis. 2015;5:654–657. doi: 10.1016/S2222-1808(15)60907-0. [DOI] [Google Scholar]
- 62.Lagunin A., Zakharov A., Filimonov D., Poroikov V. QSAR Modelling of Rat Acute Toxicity on the Basis of PASS Prediction. Mol. Inf. 2011;30:241–250. doi: 10.1002/minf.201000151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in this study are available on reasonable request.