Abstract
Cyanobacteria are photosynthetic prokaryotes that inhabit diverse aquatic and terrestrial environments. However, the evolutionary mechanisms involved in the cyanobacterial habitat adaptation remain poorly understood. Here, based on phylogenetic and comparative genomic analyses of 650 cyanobacterial genomes, we investigated the genetic basis of cyanobacterial habitat adaptation (marine, freshwater, and terrestrial). We show: (1) the expansion of gene families is a common strategy whereby terrestrial cyanobacteria cope with fluctuating environments, whereas the genomes of many marine strains have undergone contraction to adapt to nutrient-poor conditions. (2) Hundreds of genes are strongly associated with specific habitats. Genes that are differentially abundant in genomes of marine, freshwater, and terrestrial cyanobacteria were found to be involved in light sensing and absorption, chemotaxis, nutrient transporters, responses to osmotic stress, etc., indicating the importance of these genes in the survival and adaptation of organisms in specific habitats. (3) A substantial fraction of genes that facilitate the adaptation of Cyanobacteria to specific habitats are contributed by horizontal gene transfer, and such genetic exchanges are more frequent in terrestrial cyanobacteria. Collectively, our results further our understandings of the adaptations of Cyanobacteria to different environments, highlighting the importance of ecological constraints imposed by the environment in shaping the evolution of Cyanobacteria.
Subject terms: Phylogenetics, Microbial ecology
Introduction
Cyanobacteria are pioneer organisms and the most important primary producers on our planet. The evolution of oxygenic photosynthesis in Cyanobacteria changed the Earth’s early environment, paving the way for the evolution of complex life [1–4]. In turn, their autotrophic lifestyle enabled Cyanobacteria to thrive in various habitats, ranging from terrestrial ecosystems to aquatic ecosystems, fresh waters to brackish waters, and hot springs to cold Arctic environments [5–8]. Previous ecological and genomic studies have provided insight into the genomic adaptation of Cyanobacteria to the local marine environment [5, 9, 10]. However, these studies majorly focus on marine picocyanobacteria groups, the genomic adaptation of Cyanobacteria to a wide range of environments remains poorly understood due to the lack of well-balanced genome sampling.
Here, we report 163 newly generated genomes in Cyanobacteria, which greatly expanded genomic representation from undersampled freshwater and terrestrial habitats. By combining data from publicly available cyanobacterial genomes with newly generated genomes, we built a reference phylogeny of 650 cyanobacterial genomes using 834 cyanobacterial-specific benchmarking universal single-copy orthologs (BUSCO). The resulting phylogenetic tree provided high-resolution relationships among cyanobacterial subclades. On the basis of the well-resolved cyanobacterial phylogeny, comparative genomic analysis was performed among phylogenetic related strains isolated from the three most prevalent habitat types: marine, freshwater, and terrestrial. In doing so, we found that genomic size variations among cyanobacterial strains were associated with habitat adaptation. Meanwhile, hundreds of genes were characterized to be correlated with the adaptation to various environments, including light wavelengths, trophic states, salinity, availability of water, etc. In addition, we further investigated the mechanisms underlying cyanobacterial habitat adaptation. Our results suggest that horizontal gene transfer (HGT), as a source of genes that confer cyanobacteria selective advantages for living in their habitats, has significant contributions to habitat adaptation. Altogether, our study provides insights into cyanobacterial genome evolution and adaptation to diverse ecosystems.
Materials and methods
Genome sequencing assembly
A total of 163 cyanobacterial strains were sequenced for this study. All axenic strains were provided by the Freshwater Algae Culture Collection of the Institute of Hydrobiology. Genomic DNA was prepared by the CTAB method followed by purification with E.Z.N.A. Bacterial DNA Kit. Paired-end libraries were constructed using the NEBNext Ultra DNA Library Prep Kit following the manufacturer’s instructions, and whole-genome sequencing (paired-end, 100 bp) was carried out on the Illumina HiSeq 2000 platform. After the removal of adapters and low-quality reads, sequencing reads were assembled using the SPAdes v3.1 genome assembler with k-mer lengths of 55, 77, and 100 [11]. Once assembled, the quality and accuracy of the genome assembly were evaluated using CheckM [12] and QUAST [13].
Genome datasets
The sequences of 727 Cyanobacteria/Melainabacteria genomes were downloaded from NCBI on January 2018. Together with the 163 genomes sequenced in this study, we performed quality control for all genomes according to the following criteria to reduce data redundancy and biased genome representation of Cyanobacteria.
First, we calculated average nucleotide identity (ANI) and alignment fraction values for each pair of genomes using the ANI calculator with default settings (http://enve-omics.ce.gatech.edu/ani/). Genomes from the same strain with an ANI greater than 99.9% and alignment fraction exceeding 95% were marked as redundant genomes, and they were then dereplicated by filtering out one of the genomes at random. The remaining genomes formed a representative Cyanobacteria genome dataset (hereafter referred to as the Cyano dataset; 650 strains). To obtain a more reliable genome dataset, we assessed the quality of each genome using CheckM, which determines the estimated completeness of a genome and detects possible contamination based on lineage-specific sets of single-copy genes. We further compiled a high-quality dataset comprising 519 Oxyphotobacteria strains and 7 Melainabacteria strains (hereafter referred to as the Cyano HQ dataset) in which genomes were only included if they were nearly complete (compeleteness ≥90%) with low contamination (less than 5% contamination).
Collection of metadata
A variety of habitats were included in the analysis. Habitats of the 650 cyanobacterial strains (Cyano dataset) were derived from their isolation source. The isolation source for each strain was determined manually by searching IMG metadata, NCBI Biosample, PCC, ATCC, and the scientific literature. On the basis of their isolation sources, we categorized the genomes into five major habitats: marine, freshwater, terrestrial, thermal spring, and host-associated; genomes whose source of isolation was not among these major habitats were labeled as belonging to other habitat; and genomes without isolation information were labeled as belonging to unknown habitat.
Genome annotation
We performed genome annotation with a rapid prokaryotic sequence annotation algorithm implemented in Prokka v1.12 [14]. For functional annotation, predicted ORFs were searched using DIAMOND against the Clusters of Orthologous Groups (COG) database and Kyoto Encyclopedia of Genes and Genomes database (KEGG) based on sequence similarities (identity ≥ 60%, coverage ≥ 50%). Pfam domains were identified by profiling predicted ORFs against the Pfam-A database (E-value ≤ 10–5) using hidden Markov models (HMM) implemented in HMMER 3.1b1 (http://hmmer.janelia.org/) [15].
Phylogenomic analyses
To obtain a more comprehensive evolutionary landscape of Cyanobacteria, the Cyano dataset was chosen to infer phylogenetic relationships. We generated two data matrices from the genomes of 604 Oxyphotobacteria strains and 46 Melainabacteria strains.
AMPHORA data matrix: 31 universal single-copy genes were identified in the cyanobacterial genomes using the AMPHORA2 pipeline [16]. The intermediate alignments were trimmed to remove poorly aligned positions using trimal [17] with default settings.
BUSCO data matrix: We used a HMM-based search to retrieve 834 cyanobacterial-specific BUSCOs (BUSCO cyanobacteria_odb9) [18] from each genome. Each BUSCO was aligned using MAFFT [19] with the options –localpair, –maxiterate = 1000, and the aligned data were trimmed with trimal with default settings.
For each data matrix, masked alignments were concatenated into a supermatrix, and phylogenetic analysis was conducted at the CIPRES Science Gateway v.3.3. web interface [20] using the maximum likelihood methods with the IQ-TREE [21] program under the LG + GAMMA model. Ultrafast bootstrap support values were calculated from 1000 replicates. Trees were rooted with the Melainabacteria group, which was recently recognized as the closest relative of Cyanobacteria [22, 23]. All trees were visualized using iTOL (http://itol.embl.de/) [24].
Comparison of genome sizes and analysis of gene category enrichment
As low-quality genomes might introduce biases in the analysis, we limited the current and subsequent analyses focused on the Cyano HQ dataset. We calculated the approximate genome size per strain based on the following formula: approximate genome size equals actual assembly genome size/estimated coverage/(1 + estimated contamination). Then, the estimated genome sizes for strains isolated from marine, freshwater, and terrestrial habitats were compared using t-tests and PhyloGLM tests through the R package phylolm [25]. The annotated KO (KEGG Orthology) numbers of each high-quality genome were mapped to gene categories as defined by KEGG. t-tests and PhyloGLM tests were used to infer the enrichment and depletion of 22 prokaryotic-related gene categories across genomes of marine, freshwater, and terrestrial cyanobacteria. P values were corrected for multiple testing with the Benjamini–Hochberg correction [26].
Identification of genes involved in light adaptation
We chose cpcAB encoding the phycocyanin (PC) pigment, cpeAB encoding the phycoerythrin (PE) pigment, pcb encoding accessory chlorophyll-binding proteins, and genes involved in far-red light photoacclimation (FaRLiP) as defined by Gan and Bryant [27] for analysis. The sets of protein sequences for each gene were downloaded from the NCBI database. To generate HMM profiles, the reference sequences for each gene family were aligned with MAFFT, followed by alignment trimming using trimal. We used hmmbuild to build HMM profiles based on the trimmed alignments, and hmmsearch was employed to search all high-quality genomes with the profiles (cutoff E-value = 1e−5). For each gene family, we set a score cutoff, and hits above the preset cutoff were further examined by manual inspection to confirm homology. We hypothesized that if a certain gene plays an important role in light adaptation to a specific habitat, that gene should be a core gene in that specific habitat but not in other habitats, or the copy number of that gene should significantly outnumber those in other habitats. Thus, we applied two statistical analyses to infer the enrichment and depletion of aforementioned gene families in genomes of marine, freshwater, and terrestrial cyanobacteria: the hypergeometric test and PhyloGLM. Both methods were performed through the R packages phytools [28] and phylolm [25] with two versions: one based on presence/absence data and the other based on gene/domain copy numbers. For these two methods, an FDR-corrected P value threshold of 0.05 was used in our analysis.
Genome-wide surveys of habitat-enriched genes/domains
Among the five habitat categories, marine, freshwater, and terrestrial habitats are the most prevalent habitats where cyanobacterial strains are found, and the Cyano HQ dataset comprised 99, 184, and 127 strains from these habitats, respectively. Pairwise comparisons were conducted for genes/domains derived from the genomes from these three habitats to determine habitat-associated genes/domains. We followed the method described in Levy et al. [29]. Briefly, we clustered genes/domains according to their functional annotation, and statistical analyses were subsequently performed to examine whether the clusters of genes/domains were significantly enriched or depleted in specific habitats. The hypergeometric test and PhyloGLM test were performed with two versions: one based on presence/absence data and the other based on gene/domain copy numbers. Hypergeometric testing identifies the overall enrichment of gene/domain copies from strains from specific habitat without taking phylogenetic relationships into account. PhyloGLM takes both situations into account to reduce false-positive enrichments resulting from strains’ shared ancestry. An FDR-corrected P value threshold of 0.05 was used in our analysis. We defined the gene or domain associated with habitat adaptation if at least one statistical test showed enrichment of the gene in the specific habitat (FDR adjusted P value < 0.05). To further distinguish universal adaptation to certain habitats from local adaptation to specific ecological niches, we considered genes/domains to be universally habitat-enriched if they were detected in more than 75% of strains belonging to that habitat. Here, we describe an example of the comparison of KO clusters (K00556; tRNA (guanosine-2′-O-)-methyltransferase) among marine genomes, freshwater genomes, and terrestrial genomes using the hypergeometric test based on gene copy number. We first retrieved all genes annotated as K00556 in cyanobacterial genomes. Next, we calculated the copy numbers of K00556 in marine genomes, freshwater genomes, and terrestrial genomes, respectively, by tracing genes back to their encoding genomes. If hypergeometric test showed that the copy number of K00556 in marine genomes was significantly larger than the copy numbers of freshwater genomes and the copy numbers of terrestrial genomes, the KO cluster was referred to as a marine-enriched cluster. Further, if K00556 were detected in more than 75% of marine strains, the KO cluster was considered as a universal marine-enriched cluster.
Identification of HGT candidates
We employed a modified BLAST-based HGT detection approach to identify genes acquired from noncyanobacterial strains. The fundamental principle of the approach is similar to existing HGTector software [30]. The accelerated BLAST-compatible software DIAMOND was used to speed up the process of aligning the query sequences against the reference database [31]. We constructed two custom databases: one derived from the NCBI nonredundant protein database (last accessed January 20, 2018), in which taxon IDs were mapped to protein accession numbers based on the protein accession to taxid file (ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/accession2taxid/prot.accession2taxid.gz), and the other consisting of all predicted protein sequences from 650 cyanobacterial genomes. For the protein sequences from each genome, two BLASTp searches were carried out against the constructed databases with the same settings: –evalue 1e−10, –max-target-seqs 5000. We next merged the results of these two BLASTp searches and sorted the BLAST hits according to the E-value. BLAST hits were filtered if multiple hits originated from one strain, and only the best hit was retained to overcome the putative taxon-sampling bias of the database. The top 500 hits exhibiting different taxon IDs were retained for further HGT detection analysis. Taxonomic classification was assigned for each hit with dump files downloaded from the NCBI Taxonomy database, and each hit was subsequently categorized into three different lineages (self-group: Oxyphotobacteria, close group: Melainabacteria, distal group: other taxonomy). We next calculated the percentage of strains from the distal group (distalg_pct). Protein sequences were reported as HGT candidates if they satisfied the following criteria: (1) the hit count cutoff was 50 to avoid sequences that might derive from assembly or annotation errors; and (2) the threshold of distalg_pct was 80%.
We additionally employed birth–death models to detect HGT events using the program count [32]. Since the gene family table was required, we built gene families with the following procedures: (1) all versus all BLASTp searches of the protein sequences of genomes were conducted with an E-value threshold of 1e−5 and a coverage threshold of 70%; and (2) homologs were clustered using the Markov clustering algorithm [33] with an inflation parameter of 1.6. Each homolog was considered a gene family. Then, the family history was inferred based on a Wagner parsimony approach using Count with the default settings.
Results and discussion
Genomic data yield a well-resolved phylogeny for Cyanobacteria
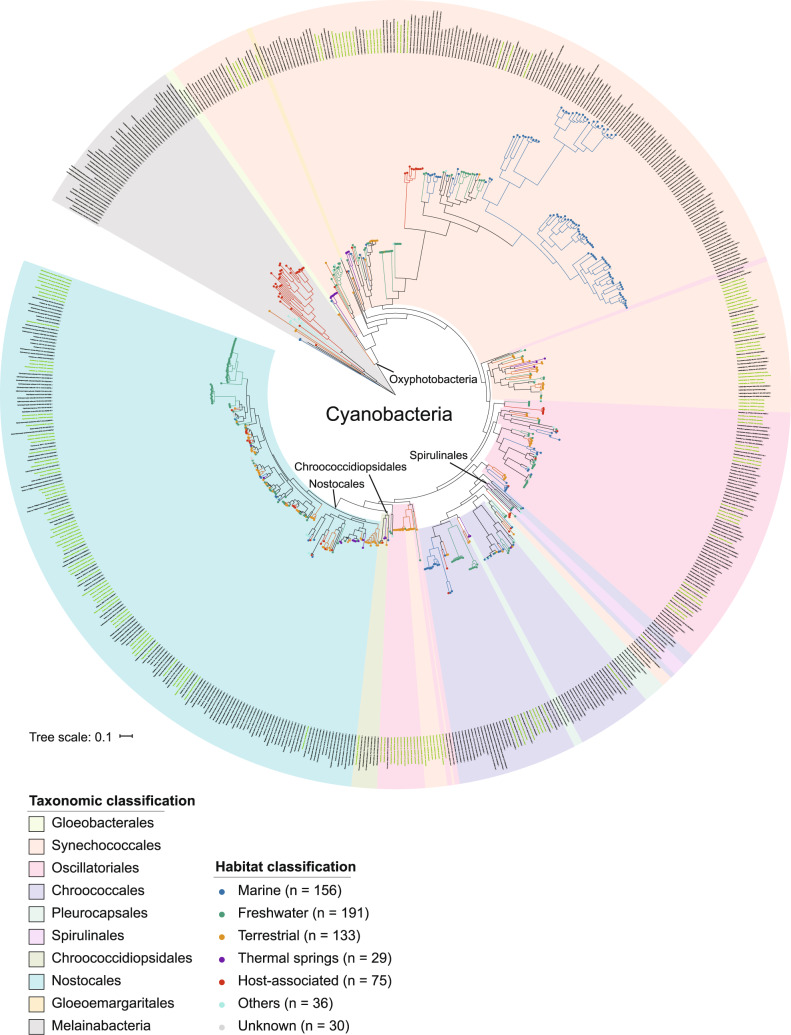
To infer the cyanobacterial phylogeny, two datasets compiled from a total of 650 taxa were analyzed (163 taxa reported in this study; Supplementary Table S1): (1) a multigene dataset including 31 universal core genes that has been adopted by many researchers and (2) a genomic dataset including 834 cyanobacterial-specific BUSCOs. Across the phylogenetic tree, the higher-order groups were generally in line with the described taxonomic scheme that proposed eight orders: Gloeobacterales, Synechococcales, Spirulinales, Chroococcales, Pleurocapsales, Oscillatoriales, Chroococcidiopsidales, and Nostocales [34, 35]. In addition, we assigned the newly characterized Gloeomargarita lithophora Alchichica-D10 to the Gloeomargaritales order, according to its special phenotypic characteristics and important phylogenetic position, reported as the closest relative to the original plastid endosymbiont (Fig. 1, Supplementary Fig. S1) [36–38].
Fig. 1. Phylogenomics of Cyanobacteria phyla.
The maximum likelihood phylogenetic tree was estimated on the basis of 834 cyanobacterial-specific benchmarking universal single-copy orthologs from 650 genomes implementing IQ-TREE under the LG + Γ model. The names of 163 genomes reported in this study are highlighted in green. Both colored circles at the tips of branches and colors of branches reflect the ecological habitats of the strains. The known oxygenic Cyanobacteria group was labeled as Oxyphotobacteria. Monophyletic taxonomic groups were also labeled for clarity. Bootstrap support values for internodes can be found in the trees deposited in iTOL.
Compared to previous phylogenomic studies, our analyses incorporated a greater number of phylogenetically diverse cyanobacterial strains [35, 36]. Especially the genomes sequenced in this study expanded genomic representation from undersampled freshwater and terrestrial environments (freshwater genomes: 73 out of 163; terrestrial genomes: 76 out of 163). The resulted phylogenies inferred from the multigene dataset (Amphora) and the genomic dataset (BUSCO) are largely congruent. Nevertheless, the resolution of phylogeny is lower for the multigene dataset than the genomic dataset. There are 73.0% of internodes (474 out of 649 internodes) in the multigene tree received high support values (ultrafast bootstrap value ≥ 95%), while 92.3% of internodes (599 out of 649 internodes) in the genomic tree received high support values (ultrafast bootstrap value ≥ 95%; Supplementary Fig. S2a). Topological discordances between two trees mainly arise when internodes of the multigene tree are weakly supported, whereas internodes of the genomic tree are highly supported (Supplementary Fig. S2b). Our genomic tree corroborated previously recognized basal relationships of Oxyphotobacteria, in which Gloeobacterales diverged first, followed by the early branching of Synechococcales, with Gloeomargaritales branching in a successive pattern [39, 40]. One major reorganization of our genomic tree stems from the placement of Crinalium epipsammum PCC 9333 and Chamaesiphon minutus PCC 6605 (subclade B3, Supplementary Fig. S2). Previous studies recovered a sister group relationship between subclade B3 and subclade B2 formed by Spirulinales. Pleurocapsales, and Chroococcales, with low statistical support [39]. Our results alternatively supported a sister group relationship between a clade formed by the subclade B3 plus 17 newly sequenced terrestrial genomes and a clade formed by Chroococcidiopsidales + Nostocales + Oscillatoriales, with strong statistical support (Supplementary Fig. S2). Overall, our study integrated a more comprehensive sampling of taxa and genes, and the improved resolution of the evolutionary history of Cyanobacteria demonstrated that the genome-scale dataset significantly improved the robustness of inference.
Habitat adaptation has shaped the genomic properties of Cyanobacteria
We retrieved habitat information for each strain and mapped the habitat traits onto the genomic tree (Fig. 1). Despite broad lifestyle diversity, we observed that strains that shared the same habitat frequently formed branching clusters, indicating that lifestyles are evolutionarily conserved rather than randomly distributed among cyanobacterial lineages. To further validate the conservative pattern of lifestyles, the habitat data were tested for phylogenetic signals by comparing the performance of models with and without phylogenetic signals [41]. We found a strong phylogenetic signal in the habitat data, suggesting that closely related strains tend to retain similar lifestyles over time (lambda = 0.975, P value < 0.01; one-sample t-test; lambda value ranges from 0 to 1, lambda equals one means maximum phylogenetic signal; Supplementary Table S2). The highly phylogenetically conservative mode of lifestyles implies that the genetic changes among cyanobacterial genomes might reflect the adaptation to different environment niches [42].
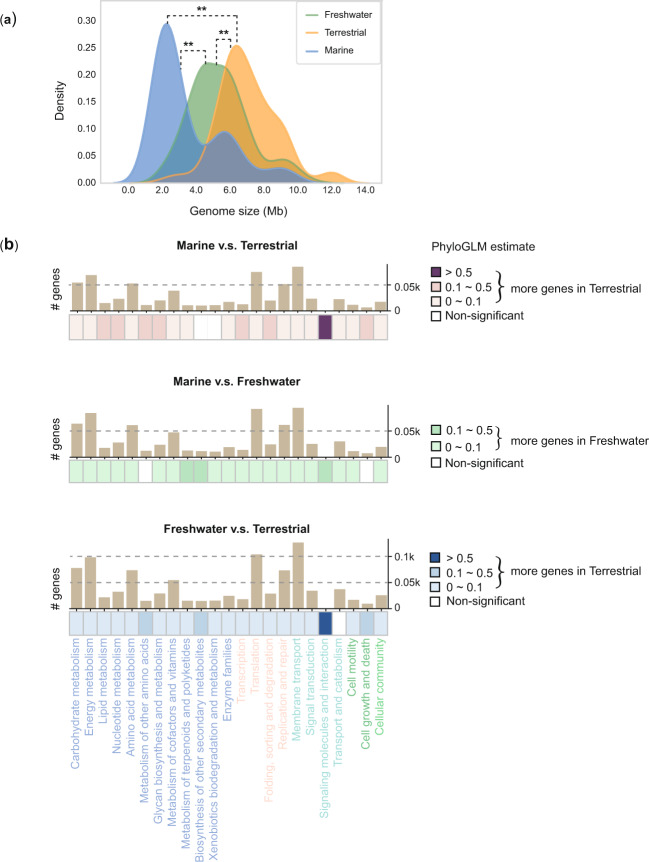
To assess the genomic variations among strains derived from different habitats, we performed comparative genomic analyses on strains isolated from the three most prevalent habitat types: marine, freshwater, and terrestrial. Low-quality genomes and genomes of the Melainabacteria group were excluded, resulting in 519 high-quality genomes. Strains derived from other habitats were not incorporated into further analyses due to their sparsely represented genomic data. Among the remaining strains, 99 live in marine habitats, 184 in freshwater habitats. and 127 in terrestrial habitats. We found that the genome sizes of terrestrial cyanobacteria were on average larger than the genomes of marine and freshwater cyanobacteria, while marine cyanobacteria generally exhibited smaller genome sizes than the other cyanobacteria (P value < 0.05, t-test). When the influence of evolutionary history was taken into account, the same pattern was observed (P value < 0.05, PhyloGLM; Fig. 2a). It has previously been hypothesized that prokaryotes with streamlined genome are likely to occupy low-complexity ecosystems, while prokaryotes with large genome are likely to occupy relatively turbulent ecosystems [43–46]. As the complexity of marine, freshwater, and terrestrial ecosystems is gradually increased considering the abiotic characteristics of corresponding habitats, such as winds are more turbulent than ocean currents, the temperature fluctuates more widely in the land than in the ocean, and the availability of water is more stable in the ocean than in the land, and the environmental variability of freshwater habitat falls in between [47], the differential pattern of genome sizes observed in marine, freshwater, and terrestrial cyanobacteria in our study further supports a hypothesis.
Fig. 2. A genome-wide comparison of marine, freshwater, and terrestrial strains.
a Density plot of genome sizes of marine, freshwater, and terrestrial strains. Double asterisks indicate a significant difference between two habitats inferred from both the t-test and PhyloGLM test (P value < 0.05). b Pairwise functional comparison among marine, freshwater, and terrestrial genomes using 22 gene categories based on KEGG annotation. Gene categories colored light blue, light pink, turquoise and spring green denote metabolism, genetic information processing, environmental information processing, and cellular processes, respectively. Bar charts represent the total number of counted genes for each gene category in statistical tests. The enrichment or depletion of each gene category based on PhyloGLM results is illustrated by heat maps. Blank cells indicate that there is no significant difference between genomes corresponding to the two habitats. Colored cells indicate significantly more genes in genomes from one habitat compared to another (P value < 0.05). It should be noted that some high estimated values (dark color cells) for categories including few genes were more likely to be subject to overestimation.
We next investigated whether certain functional categories were preferentially affected under the course of environmental adaptation. Enrichment analyses focusing on 22 functional groupings of KEGG annotations were carried out using statistical tests with and without phylogenetic signals (PhyloGLM and t-test). The enrichment analyses showed that the majority of functional categories were enriched in terrestrial cyanobacteria compared with strains from marine and freshwater environments (Fig. 2b). Categories such as regulatory, transport and motility have greatly been expanded in terrestrial strains (FDR adjusted P value < 0.05; Supplementary Fig. S3), consistent with the extensive interactions of these strains with the environment pressure factors [48]. There are more functional categories showing significant differences in marine–terrestrial genomic comparison than those seen in freshwater–terrestrial genomic comparison, which can be explained by more similar environmental conditions between terrestrial and freshwater habitats (Supplementary Fig. S3). Conversely, the free-living marine cyanobacteria have undergone reduction of gene families for the majority of functional categories in comparison with the functional categories of freshwater and terrestrial genomes (Fig. 2b), reinforcing the genome streamlining scenario for the evolution of marine cyanobacteria which inhabit less variable environments [43, 45, 49]. On the other hand, the majority of functional categories were enriched in freshwater genomes when comparing freshwater cyanobacteria to marine cyanobacteria, while the functional categories were underrepresented in freshwater genomes when comparing freshwater cyanobacteria to terrestrial cyanobacteria, suggesting that the intermediate degree of environmental fluctuation in freshwater habitat shaped the genomic content of freshwater cyanobacteria.
Genes associated with light sensing and absorption were habitat-enriched
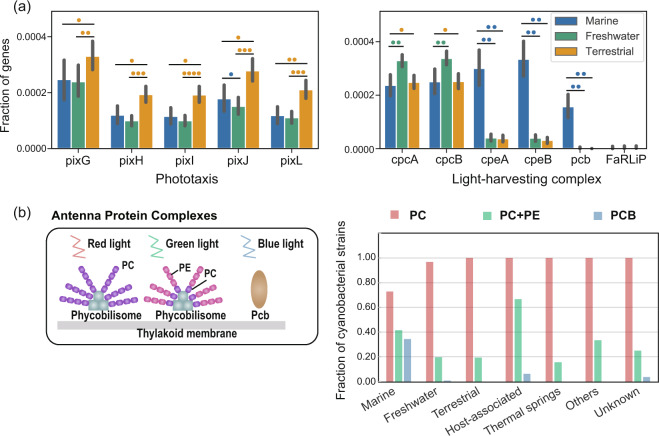
Cyanobacteria convert light energy into chemical energy through photosynthetic complexes. Thus, it is critical for Cyanobacteria to sense and respond to different light environments. Phototaxis has been characterized as one of the strategies that enables organisms to locate optimal light conditions and avoid UV irradiation [50]. Phototaxis studies on model organisms have shown that pixJ, which encodes a protein containing a light-sensing domain, might be a key gene mediating cyanobacterial phototaxis [51–53]. To investigate the potential capacity for phototaxis, we screened the pix gene cluster (pixJILHG) in cyanobacterial genomes. We observed that the pix gene cluster occurred in the majority of terrestrial genomes but was absent in major groups of freshwater and marine cyanobacteria (Supplementary Fig. S4). In addition, gene enrichment analyses showed that the pix gene cluster was significantly enriched in terrestrial cyanobacteria compared with marine and freshwater cyanobacteria (Fig. 3a). Our results suggest that the presence of pix gene cluster in terrestrial cyanobacteria enables them to move toward optimal light conditions and avoid UV irradiation, so that they can better cope with changing light conditions in the land.
Fig. 3. Distribution of genes associated with light sensing and absorption across marine, freshwater, and terrestrial genomes.
a The bars show the average gene frequencies of genes relevant to light adaptation in marine, freshwater, and terrestrial genomes, the error bars indicate 95% confidence interval (CI). Two statistical analyses were applied to infer the enrichment and depletion of light related genes in genomes of marine, freshwater, and terrestrial cyanobacteria: the hypergeometric test and PhyloGLM with two versions: one based on presence/absence data and the other based on gene/domain copy numbers. The number of colored circles denotes the number of statistical tests showing a significant difference (P value < 0.05). b The left panel presents the illustration of different forms of the antenna complexes. Phycobilisomes composed of the phycocyanin pigment (PC) encoded by the cpcA and cpcB genes are effective in absorbing red light, phycobilisomes composed of PC and the phycoerythrin pigment (PE) encoded by the cpeA and cpeB genes are better at absorbing green light, and antennas composed of prochlorophyte chlorophyll-binding protein (pcb genes) exhibit enhanced absorption of blue light. The right panel shows the proportion of genomes that have potential capacity to form different antenna complexes (phycobilisomes composed of PC, phycobilisomes composed of PC + PE and chlorophyll-binding antennas) in corresponding habitats.
An alternative well-known light adaptation strategy referred to as complementary chromatic adaptation is based on altering the pigmentation of phycobilisomes [54]. The PC pigment encoded by cpcAB genes mainly absorbs red light, and the PE pigment encoded by cpeAB mainly accumulates under green light [55]. Thus, cyanobacterium that produces PC could efficiently harvest red light, while cyanobacterium that produces both PC and PE could efficiently absorb green light. In addition, some strains of Prochlorococcus are known to produce a unique antennal pigment (divinyl chlorophyll) encoded by pcb genes, which can efficiently absorb blue light [56]. Based on the enrichment analysis of genes coding for antenna proteins, we observed that cpcAB genes were depleted in marine cyanobacteria. In contrast, cpeAB genes were enriched in marine cyanobacteria, and pcb genes encoding chlorophyll-binding proteins were exclusive to marine cyanobacteria (Fig. 3a, Supplementary Fig. S4). Additional assessment of light-harvest complex synthesis potential showed that the majority of freshwater and terrestrial cyanobacteria possess only PC pigment, and less than 20% of freshwater and terrestrial strains have the potential to form phycobilisomes composed of PC and PE pigments. In contrast, a substantial fraction of marine cyanobacteria showed the potential to form phycobilisomes composed of PC and PE pigments (marine: 41%). Meanwhile, 31% of marine cyanobacteria with the ability to synthesize chlorophyll-binding proteins as antenna complex. Given the fact that lights that could penetrate into the ocean are predominantly green and blue lights [51], our results suggest that marine cyanobacteria have adjusted their pigment components to adapt to the light conditions of the ocean.
Notably, it has recently been demonstrated that some cyanobacterial strains can use far-red light for growth through “FaRLiP” [27, 57]. Given that far-red wavelengths are enriched in soil or underplant-canopy environments, it might be expected that FaRLiP function exists in numerous terrestrial strains [58]. However, our analyses incorporated 127 high-quality terrestrial genomes, and our results showed that the distribution of the highly conserved FaRLiP cluster, comprising paralogs of genes encoding photosynthetic complexes, was unrelated to the terrestrial habitat: the FaRLiP cluster was found in 24 genomes of cyanobacteria, only four of them were isolated from terrestrial habitat, the rest of them originated from various habitats (marine: three, freshwater: five, thermal springs: eight, host-associated: two, others: one, unknown habitat: one; Fig. 3a, Supplementary Fig. S4, Supplementary Table S3). This observation indicated that the presence of FaRLiP might reflect the local adaptation of strains as previously suggested by Kühl et al. [59] that some terrestrial strains used the FaRLiP function to cope with shaded environment, rather than a universal strategy adopted by terrestrial cyanobacteria under the course of environmental adaptation. Taken together, these results indicate that genes related to light sensing and absorption are ecologically important genes that confer selective advantages for Cyanobacteria that inhabit distinct ecological niches.
Hundreds of genes/domains were involved in habitat adaptation
Besides the variation in light conditions, other environmental changes (e.g., trophic states, salinity, metal concentration, availability of water, and temperature) can also influence the survival and adaptation of organisms in specific habitats. To identify those genes associated with habitat adaptation, we conducted two statistical tests, the phylogeny-dependent PhlyloGLM test and the phylogeny-independent hypergeometric test, to infer the enrichment and depletion of genes/domains in genomes of marine, freshwater, and terrestrial cyanobacteria. In general, the genes/domains identified by the PhyloGLM test broadly overlapped with those identified by the hypergeometric test (Supplementary Figs S5–7). We identified up to 325 COGs (among which 181 genes were universally enriched), 231 (71) KOs, and 1139 (720) PFAMs that were significantly marine environment correlated; 178 (89) COGs, 93 (37) KOs, and 444 (232) PFAMs that were significantly freshwater environment correlated; and 748 (366) COGs, 544 (366) KOs, and 1066 (444) PFAMs that were significantly terrestrial environment correlated (Supplementary Tables S4–21).
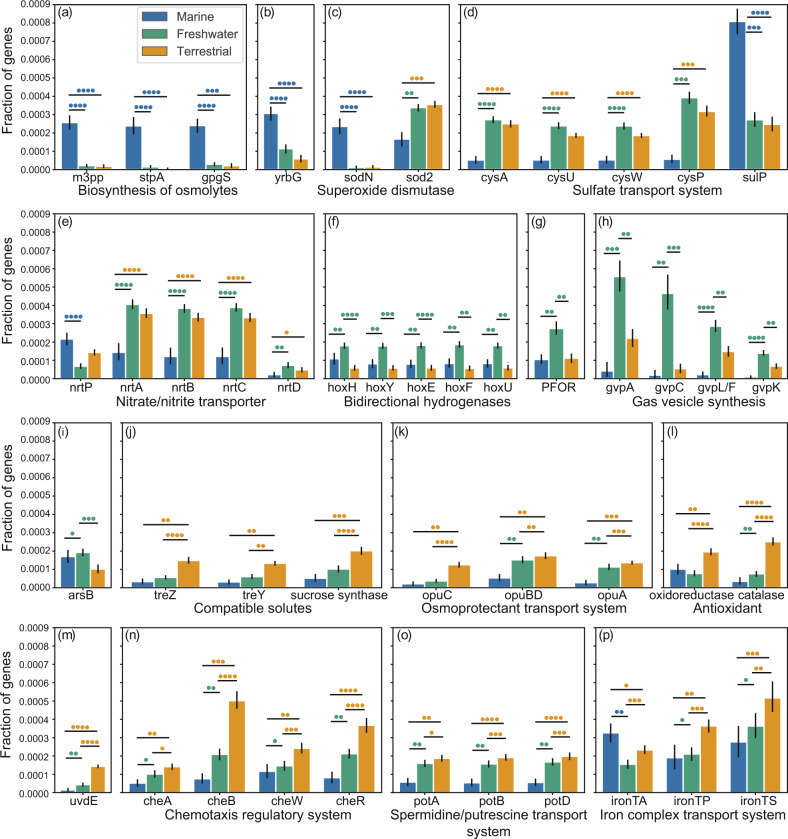
Previous studies have primarily focused on the molecular mechanisms by which marine picocyanobacteria survive in saline and oligotrophic waters [44]. In a further attempt to identify key biological mechanisms involved in habitat adaptation in diverse members of marine cyanobacteria, we performed gene enrichment analysis across genomes of marine, freshwater, and terrestrial strains to find out marine-enriched genes. Osmolytes are compatible solutes that regulate osmotic stress of organisms in high-salinity environments [60]. We observed that three marine-enriched genes involved in osmolyte biosynthesis were missing in many nonmarine genomes but present in the majority of marine genomes (related to the biosynthesis of mannosylglycerate, glucosylglycerol, and mannosylglucosylglycerate, see Table 1, Fig. 4a). In addition to the biosynthesis of osmolytes, we observed an putative ion transporter gene that might mediate sodium efflux for salt tolerance (yrbG) was widely distributed in marine cyanobacteria [61] (Table 1, Fig. 4b). We also noted that the sodN gene encoding nickel-containing superoxide dismutase was prevalent in marine cyanobacteria, whereas nonmarine strains harbored iron-containing superoxide dismutase (sod2). Compared with freshwater and terrestrial, the iron concentration of marine environments is relatively low. Thus, utilizing the sodN gene rather than the sod2 may have conferred a selective advantage in marine strains (Fig. 4c, Supplementary Tables S4 and S10). In addition, we found significant differences between marine and nonmarine strains in nutrient acquisition, such as sulfate/thiosulfate and nitrate/nitrite uptake. Marine cyanobacteria preferentially utilize monomer permease (sulP) [62] and transporter (nrtP) [63], rather than the ABC transporters used in nonmarine strains (cysAPUW and nrtABCD; Fig. 4d, e, Supplementary Tables S4 and S13).
Table 1.
Representative gene families associated with ecological adaptation in genomes of marine, freshwater, and terrestrial cyanobacteria.
| KO | EC number | Gene name | Marine (99) | Freshwater (184) | Terrestrial (127) | Pathway |
|---|---|---|---|---|---|---|
| Adaptation for salinity (marine enriched) | ||||||
| K07026 | 3.1.3.70 | Mannosyl-3-phosphoglycerate phosphatase | 68 | 20 | 11 | Mannosylglycerate biosynthesis |
| K05978 | 3.1.3.69 | stpA; glucosylglycerol 3-phosphatase | 59 | 11 | 2 | Glucosylglycerol biosynthesis |
| K13693 | 2.4.1.266 | gpgS; glucosyl-3-phosphoglycerate synthase | 64 | 25 | 11 | Mannosylglucosylglycerate biosynthesis |
| K07301 | – | yrbG; Ca2+/Na+ antiporter | 82 | 67 | 32 | Putative exclusion of Na+ |
| Tolerating anaerobic conditions (freshwater-enriched) | ||||||
| K00436 | 1.12.1.2 | hoxH; NAD-reducing hydrogenase large subunit | 38 | 140 | 43 | Hydrogen metabolism |
| K18007 | 1.12.1.2 | hoxY; NAD-reducing hydrogenase small subunit | 34 | 142 | 43 | Hydrogen metabolism |
| K05586 | 7.1.1.2 | hoxE; bidirectional [NiFe] hydrogenase diaphorase subunit | 34 | 141 | 41 | Hydrogen metabolism |
| K05587 | 7.1.1.2 | hoxF; bidirectional [NiFe] hydrogenase diaphorase subunit | 34 | 141 | 41 | Hydrogen metabolism |
| K05588 | 7.1.1.2 | hoxU; bidirectional [NiFe] hydrogenase diaphorase subunit | 36 | 143 | 44 | Hydrogen metabolism |
| Resistance to oxidative and osmotic stress (terrestrial enriched) | ||||||
| K11209 | 1.8.4.- | yfcG; GSH-dependent disulfide-bond oxidoreductase | 36 | 69 | 114 | Peroxidase activity |
| K07217 | – | Mn-containing catalase | 14 | 65 | 114 | Catalase activity |
| K01236 | 3.2.1.141 | treZ, glgZ; maltooligosyltrehalose trehalohydrolase | 17 | 59 | 101 | Trehalose biosynthesis |
| K06044 | 5.4.99.15 | treY, glgY; (1->4)-alpha-D-glucan 1-alpha-D-glucosylmutase | 17 | 63 | 102 | Trehalose biosynthesis |
| K00695 | 2.4.1.13 | Sucrose synthase | 20 | 73 | 102 | Sucrose biosynthesis |
Fig. 4. A representative set of genes enriched in specific habitats.
Bar plots show the differences in the estimated gene frequencies for the genomes of marine, freshwater, and terrestrial cyanobacteria, with the average gene frequency and 95% confidence interval (CI). We used PhyloGLM and hypergeometric tests to evaluate the differences in the copy numbers or presence of genes among the genomes from each habitat (see “Materials and methods”). Colored circles represent corresponding genomes that exhibited significantly more genes compared to genomes from another habitat. The number of colored circles denotes the number of statistical tests showing a significant difference (P value < 0.05). a–c Marine-enriched genes related to habitat adaptation. d–e Marine strains in which different genes perform similar functions compared to freshwater and terrestrial strains. f–i Genes enriched in the genomes of freshwater strains. j–p Terrestrial-enriched genes associated with habitat adaptation. m3pp mannosyl-3-phosphoglycerate phosphatase, oxidoreductase GSH-dependent disulfide-bond oxidoreductase, catalase Mn-containing catalase, ironTA iron complex transport system ATP-binding protein, ironTP iron complex transport system permease protein, ironTS iron complex transport system substrate-binding protein.
Although the genome sizes of freshwater Cyanobacteria were on average larger than the genomes of marine Cyanobacteria, the number of genes/domains that were specifically enriched in freshwater strains was much smaller than that in marine strains. We found that genes involved in the synthesis of bidirectional NiFe hydrogenase were significantly correlated with freshwater habitats (hoxHYEFU; Table 1, Fig. 4f, Supplementary Table S14). Bidirectional NiFe hydrogenase is a key enzyme in hydrogen metabolism that regulates reducing equivalent pools (NADH) to balance the oxidation/reduction state, especially when cells are under microaerobic or anaerobic conditions [64, 65]. Thus, we assume that freshwater cyanobacteria tend to be equipped with bidirectional NiFe hydrogenase to cope with microaerobic or anaerobic conditions such as those in bloom-forming periods. Moreover, the pyruvate ferredoxin oxidoreductase gene, which encodes an oxygen-sensitive fermentative enzyme was found enriched in freshwater cyanobacteria (Fig. 4g). This result also provides an evidence that the genomes of freshwater cyanobacteria are more prone to be shaped by microaerobic or anaerobic conditions. In addition to metabolic functional enrichment, the enriched genes/domains of freshwater lineages were indicated to be ecologically relevant, including genes associated with gas vesicle synthesis (gvpLFK; Fig. 4h, Supplementary Table S11), which can confer the capacity to float toward optimal light and oxygen conditions [66], and a gene encoding arsenite transporter (arsB; Fig. 4i, Supplementary Table S14), which can confer the resistance to the prevalent environmental toxin arsenic in aquatic systems [67].
Terrestrial habitats experience frequent environmental changes in terms of temperature, the availability of water and nutrients and ultraviolet irradiation [68]. The fluctuations in terrestrial ecosystems might impose strong selective pressures on microbial cells. In line with the scenario that terrestrial cyanobacteria expanded their gene repertoire to cope with fluctuating environments, the genomes of terrestrial cyanobacteria were found to harbor extensive specific enriched genes/domains (Supplementary Tables S15, S18, and S21). Among these genes/domains, we found that the treZY cluster and sucrose synthase gene were significantly enriched in terrestrial strains. These genes are related to the biosynthesis of trehalose and sucrose that are related to desiccation-protective in organisms (Table 1, Fig. 4j) [69]. Meanwhile, another desiccation-resistance-related gene cluster—opuACBD gene cluster, which encodes a putative ABC-type osmoprotectant uptake system, was also found to be enriched in terrestrial cyanobacteria (Fig. 4k) [70]. Collectively, these genes may have been utilized by terrestrial cyanobacteria to get through the dry periods in the land. On the other hand, two enzymatic antioxidant-encoding genes [71] (involved in the detoxification of reactive oxygen; Table 1, Fig. 4l) as well as a UV DNA damage repair endonuclease [72] (Fig. 4m, Supplementary Tables S18 and S21) were significantly correlated with terrestrial habitats, suggesting their important roles in adaptation to sun-exposed environments. In addition, increases in gene sets corresponding to the chemotaxis regulator system as well as putative transporters were found in terrestrial genomes in the majority of analyses. Examples of enriched transporters included the spermidine/putrescine transport system and putative iron complex transport system (Fig. 4n–p). This finding suggests that terrestrial cyanobacteria have adapted to survive in the terrestrial environment through the sensing and uptake of diverse nutrients. Accordingly, we found that the presence of multiple genes facilitating increased metabolic capacities was significantly correlated with terrestrial habitats (Supplementary Tables S15, S18, and S21), indicating that strains that can utilize abundant resources in the environment present a selective advantage in terrestrial ecosystems [46]. A detailed illustration of the distribution of the discussed enriched genes/domains is provided in Supplementary Figs. S8–10.
The role of HGT in habitat adaptation
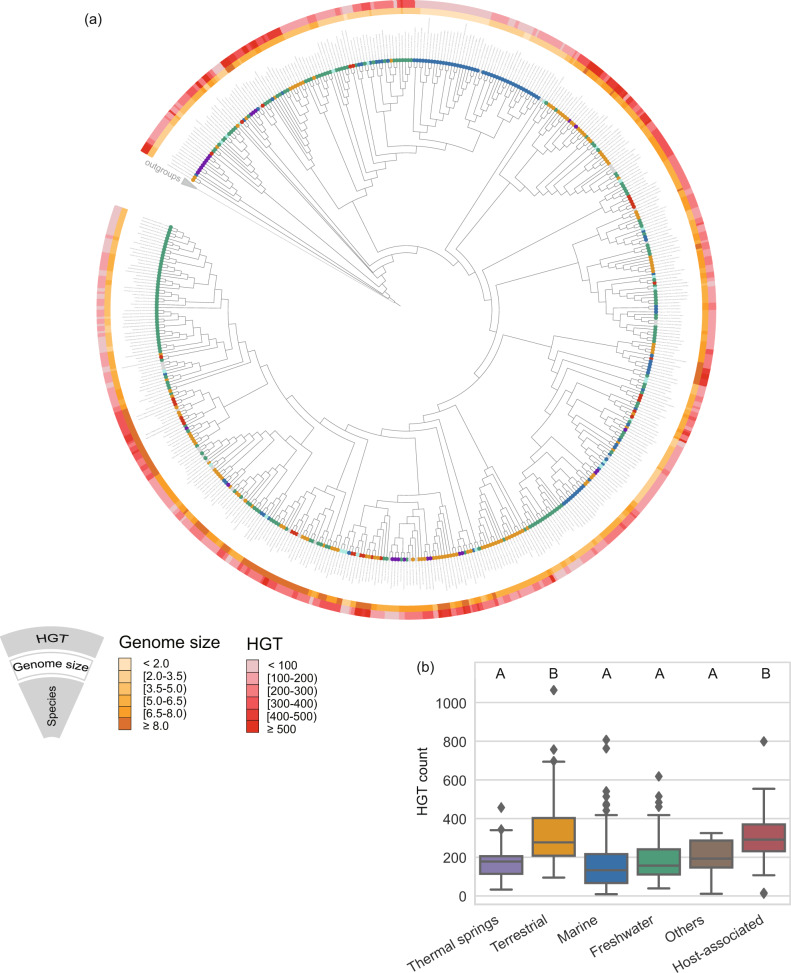
HGT is known to have a crucial impact on the evolution of bacterial genomes [73–75]. To assess whether HGT has also played an important role in explaining the genetic patterns observed in cyanobacterial habitat adaptation, we inferred HGT events in cyanobacterial genomes by using a BLAST-based HGT detection approach and COUNT software [30, 32] (see “Materials and methods”). The BLAST-based approach detected 117,938 gene transfer events in 519 high-quality cyanobacterial genomes, and 96,980 gene transfer events were found using COUNT. Although the predicted numbers of HGT events between these two methods were different, their results were positively correlated (Spearman’s ρ = 0.40, P = 5.01 × 10−21; Supplementary Fig. S11). Hence, we focused on the results of the BLAST-based approach, as it included genes that could not be assigned to any gene families. Predicted HGT events showed significant variation among the tested cyanobacterial genomes, ranging from 9 events to 1064 events (Fig. 5a, Supplementary Table S22). We compared the distributions of HGT count across cyanobacterial genomes from five defined habitats. Our results showed that host-associated genomes exhibit highest HGT frequency. This is in line with previous studies that frequent HGT event occurred in host-associated genomes as a sign of host adaptation [76]. Notably, terrestrial genomes have comparable HGT frequency to host-associated genomes, and exhibit higher HGT frequency than marine and freshwater genomes (Fig. 5b). Since genome size has a significant effect on the HGT rate [77] and a strong positive correlation between genome size and HGT frequency was observed in our analysis (Spearman’s ρ = 0.65, P = 6.47 × 10−63; Supplementary Fig. S12a), we further compared the rate of predicted HGT to genome size across different habitats. Similar trends were observed that terrestrial genomes show a comparable rate of HGT to host-associated genomes, and have a higher rate of HGT than marine and freshwater genomes (Supplementary Fig. S12b). Hence, we hypothesize that frequent HGT events occurred in terrestrial genomes also indicate a sign of habitat adaptation, and the genome expansions of terrestrial cyanobacteria might be attributed to comparatively large number of genes acquired through HGT, which help terrestrial cyanobacteria succeed in fluctuating environments.
Fig. 5. Predicted HGT events across the Cyanobacteria tree of life.
a The phylogenetic tree is a subtree of the genomic tree in Fig. 1 from which low-quality genomes were pruned. Colored circles at the tips of branches indicate the habitat from which the strains were isolated. The inner layer denotes the estimated genome size of each strain. The outer layer displays the predicted number of horizontal transfer events per genome using a BLAST-based approach. b Distribution of predicted HGT of high-quality genomes corresponding to marine (n = 99), freshwater (n = 184), terrestrial (n = 125), host-associated (n = 33), thermal springs (n = 26), and others (n = 24) environments. Boxes with different letters on top show statistically significant differences at a P value < 0.01 according to the t-test.
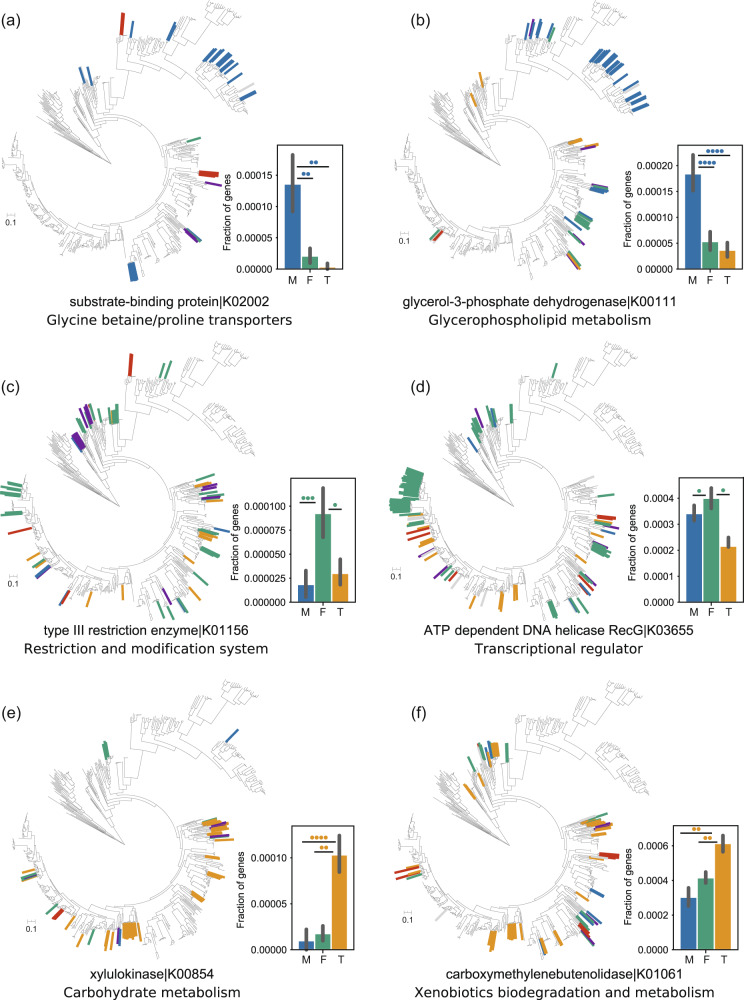
To assess whether HGT preferentially affected a certain function, we examined the functional annotations of putative transferred genes. Inspection of KEGG function revealed that among the putative transferred genes, the functions of many genes remained unknown (34%). Among the remaining 77,818 annotated transferred genes, more than half of the genes (51.2%) were categorized as being related to metabolic functions, 14.1% were related to environmental information processing, 13.2% were related to cellular processes, and 12.8% were related to genetic information processing (Supplementary Table S23). These results were consistent with the “complexity hypothesis” that fundamental genes are less likely than peripheral and operational genes to be horizontally transferred [78, 79]. Of note, it was observed that gene families enriched in specific habitat can be attributed to the frequent HGT from donors inhabiting the same habitat (Fig. 6, Supplementary Table S24). For example, the acquired gene encoding glycine betaine transporter showed tendency to have marine-specific activity, such a preference may reflect the necessity of protecting cells from osmotic stress in marine environments (Fig. 6a); a large number of acquired genes that encode type III restriction enzyme were detected in freshwater cyanobacteria, while the number is reduced in non-freshwater cyanobacteria (Fig. 6c), and many terrestrial cyanobacteria seems to have acquired xylB gene (xylulokinase) for xylose metabolism via HGT, in accordance with the need of efficient utilization of xylose in terrestrial ecosystem (Fig. 6e). The analysis of gene trees of the habitat-enriched genes further shows that the acquisition of adaptive genes occurred multiple times and from multiple donors (Supplementary Fig. S13). Taken together, our results indicate that the successful Cyanobacteria adaptation to specific habitat is associated with the acquisition and maintenance of foreign genes, which might confer fitness advantages to organisms in specific environments.
Fig. 6. Horizontal gene transfer events with habitat-specific activity facilitated the enrichment of gene families from specific ecosystem.
Gene families that have undergone a HGT have sparsely phylogenetic distributions but are concentrated in a certain type of habitats. The habitat-enriched gene families can be explained by gene acquisition via lateral transfer. Each panel is divided into two parts: the phylogenetic distribution of transferred gene families, leaf names shown in color are strains found that have acquired gene through HGT, and the color code using the same scheme as in Fig. 5b (left); the barplot shows the differences in the estimated gene frequencies for the genomes of marine (M), freshwater (F), and terrestrial (T) Cyanobacteria, bars colored blue, ocean green, and gamboge denote marine, freshwater, and terrestrial genomes, respectively. Colored circles represent corresponding genomes that exhibited significantly more genes compared to genomes from another habitat. The number of colored circles denotes the number of statistical tests showing a significant difference (P value < 0.05) (right). a, b HGT events with marine-specific activity. c, d HGT events with freshwater-specific activity. e, f HGT events with terrestrial-specific activity.
Supplementary information
Acknowledgements
This project was supported by the National Natural Science Foundation of China (NSFC) grants 31900002 and 41830318. We thank C. He and J.L. Liang for discussions on the results and comments on the manuscript.
Data availability
Cyanobacterial strains reported in this study could be found and ordered in Freshwater Algae Culture Collection at the Institute of Hydrobiology, China (http://algae.ihb.ac.cn/english/Cultrues.aspx). The genomes reported in this study are publicly available from the NCBI Bioproject database under the accession number PRJNA598298. The high-resolution phylogenetic trees of Cyanobacteria based on the BUSCO dataset and multigene dataset were deposited on iTOL (https://itol.embl.de/tree/1836397239155351578467673, https://itol.embl.de/tree/1836397145361531569311402). Additional analytic results are available through figshare (https://figshare.com/s/6b64fa50e1fa720e3587).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bo-Ping Han, Email: tbphan@jnu.edu.cn.
Li-Rong Song, Email: lrsong@ihb.ac.cn.
Wen-Sheng Shu, Email: shuwensheng@m.scnu.edu.cn.
Supplementary information
The online version of this article (10.1038/s41396-020-00775-z) contains supplementary material, which is available to authorized users.
References
- 1.Tomitani A, Knoll AH, Cavanaugh CM, Ohno T. The evolutionary diversification of Cyanobacteria: molecular-phylogenetic and paleontological perspectives. Proc Natl Acad Sci USA. 2006;103:5442–7. doi: 10.1073/pnas.0600999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schirrmeister BE, Gugger M, Donoghue PCJ. Cyanobacteria and the Great Oxidation Event: evidence from genes and fossils. Palaeontology. 2015;58:769–85. doi: 10.1111/pala.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer WW, Hemp J, Johnson JE. Evolution of oxygenic photosynthesis. Annu Rev Earth Planet Sci. 2016;44:647–83. [Google Scholar]
- 4.Soo RM, Hemp J, Parks DH, Fischer WW, Hugenholtz P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science. 2017;355:1436–40. doi: 10.1126/science.aal3794. [DOI] [PubMed] [Google Scholar]
- 5.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat Rev Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 6.Sánchez-Baracaldo P. Origin of marine planktonic Cyanobacteria. Sci Rep. 2015;5:14–17. doi: 10.1038/srep17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang JL, Chen M, Hou S, Li T, Yang YW, Li Q, et al. Genomic and transcriptomic insights into the survival of the subaerial cyanobacterium Nostoc flagelliforme in arid and exposed habitats. Environ Microbiol. 2019;21:845–63. doi: 10.1111/1462-2920.14521. [DOI] [PubMed] [Google Scholar]
- 8.Chrismas NAM, Anesio AM, Śanchez-Baracaldo P. The future of genomics in polar and alpine Cyanobacteria. FEMS Microbiol Ecol. 2018;94:fiy032. doi: 10.1093/femsec/fiy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–20. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 10.Larsson J, Celepli N, Ininbergs K, Dupont CL, Yooseph S, Bergman B, et al. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 2014;8:1892–903. doi: 10.1038/ismej.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–5. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 15.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Scott AJ. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics. 2012;28:1033–4. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 17.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35:543–8. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans (LA): IEEE; 2010. pp 1–8.
- 21.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013;2:e01102. doi: 10.7554/eLife.01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matheus Carnevali PB, Schulz F, Castelle CJ, Kantor RS, Shih PM, Sharon I, et al. Hydrogen-based metabolism as an ancestral trait in lineages sibling to the Cyanobacteria. Nat Commun. 2019;10:1–16. doi: 10.1038/s41467-018-08246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tung HoLS, Ané C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 2014;63:397–408. doi: 10.1093/sysbio/syu005. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 27.Gan F, Bryant DA. Adaptive and acclimative responses of Cyanobacteria to far-red light. Environ Microbiol. 2015;17:3450–65. doi: 10.1111/1462-2920.12992. [DOI] [PubMed] [Google Scholar]
- 28.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–23. [Google Scholar]
- 29.Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, et al. Genomic features of bacterial adaptation to plants. Nat Genet. 2018;50:138–50. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Q, Kosoy M, Dittmar K. HGTector: an automated method facilitating genome-wide discovery of putative horizontal gene transfers. BMC Genom. 2014;15:717. doi: 10.1186/1471-2164-15-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 32.Csurös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–2. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 33.Enright AJ. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komárek J. A polyphasic approach for the taxonomy of Cyanobacteria: principles and applications. Eur J Phycol. 2016;51:346–53. [Google Scholar]
- 35.Komárek J, Kaštovský J, Mareš J, Johansen JR. Taxonomic classification of cyanoprokaryotes (Cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 2014;86:295–335. [Google Scholar]
- 36.Ponce-Toledo RI, Deschamps P, López-García P, Zivanovic Y, Benzerara K, Moreira D. An early-branching freshwater Cyanobacterium at the origin of plastids. Curr Biol. 2017;27:386–91. doi: 10.1016/j.cub.2016.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries J, Archibald JM. Endosymbiosis: did plastids evolve from a freshwater Cyanobacterium? Curr Biol. 2017;27:R103–5. doi: 10.1016/j.cub.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Dagan T, Roettger M, Stucken K, Landan G, Koch R, Major P, et al. Genomes of stigonematalean Cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 2013;5:31–44. doi: 10.1093/gbe/evs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110:1053–8. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sánchez-Baracaldo P, Raven JA, Pisani D, Knoll AH. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc Natl Acad Sci USA. 2017;114:E7737–45. doi: 10.1073/pnas.1620089114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst Biol. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- 42.Monk JM, Charusanti P, Aziz RK, Lerman JA, Premyodhin N, Orth JD, et al. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci USA. 2013;110:20338–43. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripp HJ, Bench SR, Turk KA, Foster RA, Desany BA, Niazi F, et al. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–4. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- 44.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine Picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–99. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poulton NJ, Acinas SG, Lauro FM, Cavicchioli R, Swan BK, Hanson NW, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110:11463–8. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bentkowski P, Van Oosterhout C, Ashby B, Mock T. The effect of extrinsic mortality on genome size evolution in prokaryotes. ISME J. 2017;11:1011–8. doi: 10.1038/ismej.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steele JH, Brink KH, Scott BE. Comparison of marine and terrestrial ecosystems: suggestions of an evolutionary perspective influenced by environmental variation. ICES J Mar Sci. 2019;76:50–9. [Google Scholar]
- 48.Philippot L, Andersson SGE, Battin TJ, Prosser JI, Schimel JP, Whitman WB, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–9. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 49.Luo H, Csűros M, Hughes AL, Moran MA. Evolution of divergent life history strategies in marine Alphaproteobacteria. MBio. 2013;4:1–8. doi: 10.1128/mBio.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitton BA (editor). Ecology of Cyanobacteria II. Dordrecht, Netherlands: Springer; 2012.
- 51.Yoshihara S, Katayama M, Geng X, Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–37. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 52.Bhaya D, Takahashi A, Grossman AR. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc Natl Acad Sci USA. 2001;98:7540–5. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Lam V, Adomako M, Simkovsky R, Jakob A, Rockwell NC, et al. Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2018;115:E12378–87. doi: 10.1073/pnas.1812871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kehoe DM, Gutu A. Responding to color: the regulation of complementary chromatic adaptation. Annu Rev Plant Biol. 2006;57:127–50. doi: 10.1146/annurev.arplant.57.032905.105215. [DOI] [PubMed] [Google Scholar]
- 55.Sánchez-Baracaldo P, Bianchini G, Di Cesare A, Callieri C, Chrismas NAM. Insights Into the evolution of Picocyanobacteria and Phycoerythrin Genes (mpeBA and cpeBA) Front Microbiol. 2019;10:45. doi: 10.3389/fmicb.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ting CS, Rocap G, King J, Chisholm SW. Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 2002;10:134–42. doi: 10.1016/s0966-842x(02)02319-3. [DOI] [PubMed] [Google Scholar]
- 57.Gan F, Zhang S, Rockwell NC, Martin SS, Lagarias JC, Bryant DA. Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science. 2014;345:1312–7. doi: 10.1126/science.1256963. [DOI] [PubMed] [Google Scholar]
- 58.Thiel V, Tank M, Bryant DA. Diversity of chlorophototrophic bacteria revealed in the Omics Era. Annu Rev Plant Biol. 2018;69:21–49. doi: 10.1146/annurev-arplant-042817-040500. [DOI] [PubMed] [Google Scholar]
- 59.Kühl M, Trampe E, Mosshammer M, Johnson M, Larkum AWD, Frigaard N-U, et al. Substantial near-infrared radiation-driven photosynthesis of chlorophyll f-containing Cyanobacteria in a natural habitat. Elife. 2020;9:e50871. doi: 10.7554/eLife.50871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4:1–13. doi: 10.1186/1746-1448-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sääf A, Baars L, von Heijne G. The internal repeats in the Na+/Ca 2+ exchanger-related Escherichia coli protein YrbG have opposite membrane topologies. J Biol Chem. 2001;276:18905–7. doi: 10.1074/jbc.M101716200. [DOI] [PubMed] [Google Scholar]
- 62.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine Cyanobacteria. Proc Natl Acad Sci USA. 2004;101:18228–33. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakamoto T, Inoue-Sakamoto K, Bryant DA. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1999;181:7363–72. doi: 10.1128/jb.181.23.7363-7372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carrieri D, Wawrousek K, Eckert C, Yu J, Maness PC. The role of the bidirectional hydrogenase in Cyanobacteria. Bioresour Technol. 2011;102:8368–77. doi: 10.1016/j.biortech.2011.03.103. [DOI] [PubMed] [Google Scholar]
- 65.Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wunschiers R, Lindblad P. Hydrogenases and hydrogen metabolism of Cyanobacteria. Microbiol Mol Biol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huisman J, Codd GA, Paerl HW, Ibelings BW, Verspagen JMH, Visser PM. Cyanobacterial blooms. Nat Rev Microbiol. 2018;16:471–83. doi: 10.1038/s41579-018-0040-1. [DOI] [PubMed] [Google Scholar]
- 67.Ben Fekih I, Zhang C, Li YP, Zhao Y, Alwathnani HA, Saquib Q, et al. Distribution of arsenic resistance genes in prokaryotes. Front Microbiol. 2018;9:2473. doi: 10.3389/fmicb.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fürst-Jansen JMR, de Vries S, de Vries J. Evo-physio: on stress responses and the earliest land plants. J Exp Bot. 2020;71:3254–69. doi: 10.1093/jxb/eraa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murik O, Oren N, Shotland Y, Raanan H, Treves H, Kedem I, et al. What distinguishes Cyanobacteria able to revive after desiccation from those that cannot: the genome aspect. Environ Microbiol. 2017;19:535–50. doi: 10.1111/1462-2920.13486. [DOI] [PubMed] [Google Scholar]
- 70.Gul N, Poolman B. Functional reconstitution and osmoregulatory properties of the ProU ABC transporter from Escherichia coli. Mol Membr Biol. 2013;30:138–48. doi: 10.3109/09687688.2012.754060. [DOI] [PubMed] [Google Scholar]
- 71.Pathak J, Ahmed H, Singh PR, Singh SP, Häder D-P, Sinha RP. Mechanisms of photoprotection in Cyanobacteria. In: Mishra AK, Tiwari DN, Rai AN. editors. Cyanobacteria. Cambridge: Academic Press; 2019. pp. 145–171.
- 72.Meulenbroek EM, Peron Cane C, Jala I, Iwai S, Moolenaar GF, Goosen N, et al. UV damage endonuclease employs a novel dual-dinucleotide flipping mechanism to recognize different DNA lesions. Nucleic Acids Res. 2013;41:1363–71. doi: 10.1093/nar/gks1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richardson EJ, Bacigalupe R, Harrison EM, Weinert LA, Lycett S, Vrieling M, et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat Ecol Evol. 2018;2:1468–78. doi: 10.1038/s41559-018-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–76. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 75.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 76.Sheppard SK, Guttman DS, Fitzgerald JR. Population genomics of bacterial host adaptation. Nat Rev Genet. 2018;19:1–17. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 77.Oliveira PH, Touchon M, Rocha EPC. Regulation of genetic flux between bacteria by restriction-modification systems. Proc Natl Acad Sci USA. 2016;113:5658–63. doi: 10.1073/pnas.1603257113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–6. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–5. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cyanobacterial strains reported in this study could be found and ordered in Freshwater Algae Culture Collection at the Institute of Hydrobiology, China (http://algae.ihb.ac.cn/english/Cultrues.aspx). The genomes reported in this study are publicly available from the NCBI Bioproject database under the accession number PRJNA598298. The high-resolution phylogenetic trees of Cyanobacteria based on the BUSCO dataset and multigene dataset were deposited on iTOL (https://itol.embl.de/tree/1836397239155351578467673, https://itol.embl.de/tree/1836397145361531569311402). Additional analytic results are available through figshare (https://figshare.com/s/6b64fa50e1fa720e3587).