Abstract
Histone deacetylases (HDACs) are conserved enzymes that regulate many cellular processes by catalyzing the removal of acetyl groups from lysine residues on histones and non-histone proteins. As appropriate for proteins that occupy such an essential biological role, HDAC activities and functions are in turn highly regulated. Overwhelming evidence suggests that the dysregulation of HDACs plays a major role in many human diseases. The regulation of HDACs is achieved by multiple different mechanisms, including posttranslational modifications. One of the most common posttranslational modifications on HDACs is reversible phosphorylation. Many HDAC phosphorylations are context-dependent, occurring in specific tissues or as a consequence of certain stimuli. Additionally, whereas phosphorylation can regulate some HDACs in a non-specific manner, many HDAC phosphorylations result in specific consequences. Although some of these modifications support normal HDAC function, aberrations can contribute to disease development. Here we review and critically evaluate how reversible phosphorylation activates or deactivates HDACs and, thereby, regulates their many functions under various cellular and physiological contexts.
Keywords: Histone deacetylase, Phosphorylation, Epigenetics, Cell signaling
Introduction
Histone deacetylases (HDACs) are a family of enzymes that have the ability to catalyze posttranslational modifications (PTMs) of their target substrates, of which the best known is lysine deacetylation. The human HDAC family is made up of 18 enzymes, which were identified and classified based on sequence homology to yeast deacetylases. The Classical HDAC family includes the Class I, II, and IV HDACs, whose deacetylase activity is dependent on Zn2+. The Class I HDACs (HDACs 1, 2, 3, and 8) are homologous to the yeast Rpd3 protein, whereas the Class II HDACs (HDACs 4, 5, 6, 7, 9, and 10) are similar to the yeast Hda1 enzyme. The sole Class IV HDAC is HDAC11, which is similar in sequence to both Classes I and II. The Sirtuin (SIRT) family, also referred to as the Class III HDACs, is similar to the yeast Sir2 protein and is dependent on NAD+ for its deacetylase activity [1].
PTMs occur as a response to changing conditions within the cellular microenvironment and can serve as a mechanism of intracellular communication to elicit biological functions mediated by the target proteins [2–4]. Although HDACs catalyze the addition and removal of PTMs from their target substrates [1, 5–15], they themselves can undergo the addition or removal of a variety of modifications on specific amino acid residues. These modifications include chemical groups or proteins, such as an acetyl group or ubiquitin, many of which have been characterized in regard to their effects on target protein characteristics [2]. Many posttranslational modifications of HDACs have been identified through high-throughput mass spectrometry approaches using different cell lines, tissue types, and drug treatments. Some of these sites have been validated and explored in a functional context.
Perhaps one of the best-studied modifications of HDACs is phosphorylation. In response to extracellular stimuli, signal transduction pathways are activated, which can lead to protein phosphorylation most commonly on target serine (Ser, S), threonine (Thr, T), and tyrosine (Tyr, Y) residues. In the context of phosphorylation, eukaryotic kinases catalyze the addition of phosphate from ATP to these target residues, proteins containing domains (e.g., SH2) that recognize the phosphorylated site bind to the modified protein to mediate signaling, and phosphatases reverse this modification [2, 3, 16–18]. Extensive studies have gone into understanding the mechanisms and functions of protein phosphorylation, and it is well established that due to the additional (− 2) negative charge introduced through the phosphate, phosphorylation can induce the protein to undergo a conformational change, which can alter the interactions and activity of the target protein to elicit a biological response to the stimulus [2–4, 19–21]. The response of cells to the cascade of signaling events and changes in protein functions contributes to the regulation of cellular functions. This is particularly critical for the maintenance of healthy cells, where dysregulation of signaling pathways and consequent changes in the phosphorylation state of the target protein can give rise to the development of disease states.
The mechanisms associated with HDAC phosphorylation have been studied in a variety of cellular contexts and disease states. This has led to the identification of class-wide characteristics such as the relationship between Class IIa HDAC phosphorylation and subcellular localization. However, given the complexity of phosphorylation as a regulatory mechanism and the unique roles of each HDAC, even for those of the same class, there is still much to be discovered and clarified regarding the mechanisms and cellular context of HDAC phosphorylation. Here we discuss the biochemical and physiological effects of phosphorylation of HDACs to address the similarities and differences among the known regulatory mechanisms and to highlight important areas for future research and therapeutic relevance. A summary of the effects of phosphorylation on HDAC attributes is provided in Table 1.
Table 1.
Effect of phosphorylation on histone deacetylase (HDAC) enzyme attributes
| Class | Protein | Affected protein attribute | References |
|---|---|---|---|
| Class I | HDAC1 | Deacetylase activity, protein interactions | [24, 26, 120] |
| HDAC2 | Deacetylase activity, transcriptional repression activity, protein interactions, acetylation, ubiquitination, protein stability | [26–29, 68, 110, 120, 130, 152] | |
| HDAC3 | Deacetylase activity, protein interactions | [31–34, 112] | |
| HDAC8 | Enzymatic activity, protein structure | [35–37] | |
| Class IIa | HDAC4 | Protein stability, protein interactions, subcellular localization | [52, 53, 55, 56, 61, 62, 64, 70, 78, 97, 98, 113, 153] |
| HDAC5 | Deacetylase activity, protein interactions, subcellular localization, protein structure | [54–59, 60, 62, 63, 78, 98, 100, 104, 107, 113, 114, 127, 128, 153–158] | |
| HDAC7 | Protein interactions, subcellular localization, phosphorylation (hierarchical) | [55, 56, 92, 94, 101, 102, 113, 154, 156, 159] | |
| HDAC9/MITR | Protein interactions, subcellular localization | [55, 62, 78, 99, 156] | |
| Class IIb | HDAC6 | Deacetylase activity | [38, 40, 41, 129] |
| Sirtuin | SIRT1 | Deacetylase activity, protein interactions, protein stability, protein structure, subcellular localization | [44–48, 65, 69, 75, 76, 79, 80, 84, 89, 118, 121, 122, 126, 160] |
| SIRT2 | Deacetylase activity, protein stability, protein interactions, subcellular localization | [50, 51, 72, 95, 96, 161] | |
| SIRT3 | Deacetylase activity | [87] | |
| SIRT6 | Mono-ADP ribosylation activity, protein interactions, ubiquitination, protein stability | [49, 73, 162] | |
| SIRT7 | Subcellular localization, protein stability | [74] |
Modulation of HDAC enzymatic and functional activities by phosphorylation
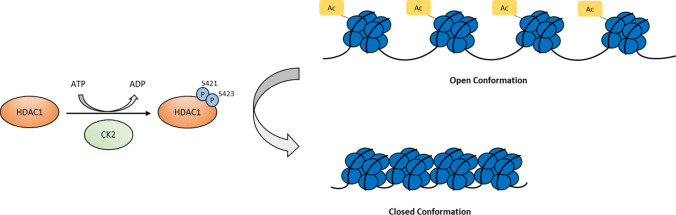
Phosphorylation can regulate the activity of HDACs in a variety of settings to generate responses that are specific to the enzyme and cellular environment. In early studies, serine phosphorylation of Class I HDACs was identified as a mode of regulating their enzymatic activity, although the mechanism and effects of phosphorylation are different for each protein. Class I HDACs are members of different co-repressor complexes, which augment their deacetylase activity and through which they could regulate transcription [1]. HDAC1 and HDAC2 are part of the NuRD (nucleosome remodeling and deacetylating), Sin3, CoREST (co-repressor for element-1-silencing transcription factor), and MiDAC (mitotic deacetylase) complexes, whereas HDAC3 is part of the SMRT (silencing mediator of retinoid and thyroid hormone receptors)/N-CoR (nuclear receptor co-repressor) complex [1, 22–24]. HDAC1 interactions with other members of the Sin3, CoREST, and NuRD co-repressor complexes (MTA2, RbAp48, mSin3a, and CoREST), and consequently its deacetylase activity, were found to be dependent on its phosphorylation at S421 and S423 by casein kinase 2 (CK2) [24]. Interestingly, discrepancies in the identified effect of phosphorylation on HDAC1 have been associated with the substrate and conditions tested. Pflum et al. found that S421 and S423 phosphorylation is necessary for HDAC1 activity towards isolated hyperacetylated core histones, where mutation of these sites to alanine to mimic the loss of phosphorylation diminished its activity (Fig. 1) [24]. In contrast, alkaline phosphatase treatment did not alter HDAC1 deacetylase activity towards a synthetic acetylated histone H4 N-terminal peptide [25]. However, Galasinski et al. found soon after that the activity of sensitive deacetylases—identified as HDAC1 and HDAC2—following okadaic acid phosphatase inhibitor treatment increased towards their acetylated histone H4 N-terminal peptide, but not isolated hyperacetylated histones [26].
Fig. 1.
Increased deacetylase activity of HDAC1 upon phosphorylation. Phosphorylation of HDAC1 at S421 and S423 by CK2 has been demonstrated to be necessary for its activity in deacetylating histones. Histone deacetylation leads to tighter binding of the DNA and histone proteins in a “closed conformation,” such that gene transcription is repressed [24]. P phosphate, Ac acetyl group
There is a similar discrepancy in the findings of the effect of HDAC2 phosphorylation on its activity. A study by Tsai et al. led to the finding that HDAC2 phosphorylation at S394 by CK2 is necessary for its deacetylase activity towards core histones and its interactions with mSin3 and Mi2, members of co-repressor complexes. However, they found that S394 phosphorylation does not impact its transcriptional repression activity [27]. In another study, Adenuga et al. corroborated that HDAC2 phosphorylation by CK2 is important for its interactions with other proteins, including HDAC1 and p53 [28]. In contrast, however, they found that HDAC2 phosphorylation stimulated by oxidative stress due to cigarette smoke extract treatment was associated with its reduced deacetylase activity and higher global histone H4 acetylation, as measured by a fluorometric assay, but higher transrepression activity using a reporter with Gal4-binding sites [28, 29]. The difference in the findings of these studies could depend on the sites that are modified within HDAC2, whereas Tsai et al. demonstrated that the phosphorylation of S394 affected HDAC2 activity [27], Adenuga et al. found that mutation of this site under the condition of oxidative stress did not notably affect HDAC2 phosphorylation levels [28]. Instead, they found that the double mutation of S422 and S424 to alanine (A) in conjunction with the S394A mutation or C-terminal truncation had a notable reduction in the overall phosphorylation level of HDAC2, suggesting that these are the key residues under this condition [28]. Of note, it is interesting that the phosphorylation of HDAC1 and HDAC2, which share 85% sequence similarity [30], at corresponding sites (S421 and S423 in HDAC1 and S422 and S424 in HDAC2) leads to opposite effects on their enzymatic activity. It may be that these proteins are specifically regulated, rather than through a common mechanism. Alternatively, since these studies were carried out in different contexts, it is possible that the environmental condition may have influenced the outcome and that the effect could vary by stimulus or cell type.
Unlike HDAC1 and HDAC2, there has been greater consensus regarding the effect of phosphorylation on the activity of the other Class I HDACs, HDAC3, and HDAC8. HDAC3 phosphorylation is important for its deacetylase activity towards core histones [31]. Specifically, phosphorylation of the HDAC3-H1.3 complex by CK2 during mitosis has been associated with histone H3K9 deacetylation, and phosphorylation of HDAC3 by leucine-rich repeat kinase 2 (LRRK2) in neurons was associated with the deacetylation of histone H4K5 and histone H4K12 [32, 33]. Similarly, tyrosine phosphorylation of HDAC3 by proto-oncogene tyrosine-protein kinase Src (c-Src) in the SKBR3 human epidermal growth factor receptor 2 (HER2)-positive breast cancer cell line increases its activity, as measured by a fluorometric assay kit specific for HDAC3 [34]. Unlike the other Class I HDACs, HDAC8 is phosphorylated by cyclic AMP (cAMP)-dependent protein kinase A (PKA), which is associated with its reduced deacetylase activity towards histones H3 and H4 [35, 36]. It has been suggested that this effect may be due to a structural change induced by the phosphorylation of HDAC8 at S39 [37].
In contrast to the effect of phosphorylation on Class I HDAC histone deacetylase activity, HDAC6 phosphorylation impacts its tubulin deacetylase activity, which further affects cellular functions dependent on intracellular trafficking. Phosphorylation of HDAC6 by epidermal growth factor receptor (EGFR) at Y570 was found to reduce its activity, such that changes in the trafficking of endosomes led to the lysosomal degradation of EGFR [38, 39]. Downstream of EGFR-mediated signaling, extracellular signal-regulated kinase (ERK) phosphorylates HDAC6 at S1035, which increases its alpha-tubulin deacetylase activity and thereby promotes cell migration [40]. Phosphorylation of HDAC6 by glycogen synthase kinase-3 beta (GSK3β) specifically in hippocampal neurons also promotes its tubulin deacetylase activity, but the effect is a reduction in the movement of mitochondria. According to what is known about the relationship between improper mitochondrial transport and neurodegenerative diseases, this pathway is suggested as a possible therapeutic target [41–43].
Phosphorylation also modulates the activity of sirtuins in modifying a variety of substrates. In particular, phosphorylation of SIRT1 influences its deacetylase activity to regulate DNA damage responses in different contexts and mechanisms. It has been noted that SIRT1 S152 phosphorylation is reduced in aging mice. Mechanistically, SIRT1 phosphorylation at this site is necessary for both its enzymatic activity and the expression of estrogen receptor beta (ERβ), which are involved in reducing DNA damage in the endothelium [44]. A study by Kang et al. found that SIRT1 is phosphorylated by CK2 at four serine residues following ionizing radiation-induced DNA damage, which increases its deacetylation rate and substrate-binding affinity. SIRT1 modification in this context increases the deacetylation of p53 and thereby protects cells against apoptosis [45]. The increased deacetylation of p53 following SIRT1 phosphorylation by the dual-specificity tyrosine-phosphorylated and regulated kinases (DYRK) DYRK1A and DYRK3 has been attributed to a modification-induced conformational change in SIRT1 that increases the turnover—defined as the deacetylation and release—of its substrates rather than changing its catalytic ability [46]. In contrast, modification of SIRT1 by homeodomain-interacting protein kinase 2 (HIPK2) following DNA damage and by AMP-activated protein kinase (AMPK) in hepatocellular carcinoma cells reduces its deacetylase activity towards p53, leading to apoptosis [47, 48].
Similar to SIRT1, the role of SIRT6 in DNA damage response can also be affected by its phosphorylation state. A study by Van Meter et al. showed that SIRT6 is phosphorylated at S10 by c-Jun N-terminal kinase (JNK) following the induction of oxidative stress by paraquat treatment. This modification increases the mono-ADP ribosylation activity of SIRT6 towards poly [ADP-ribose] polymerase 1 (PARP1) as a substrate and consequently promotes genome stability through DNA double-strand break repair [49]. Unlike SIRT1 and SIRT6, studies of SIRT2 have shown that its phosphorylation affects its deacetylase activity towards substrates that are involved in functions other than DNA damage-related processes. SIRT2 phosphorylation by Cyclin-dependent kinase (Cdk) proteins inhibits its enzymatic activity to promote functions such as mitotic entry and neuronal mobility [50, 51].
Influence of phosphorylation state on HDAC subcellular localization
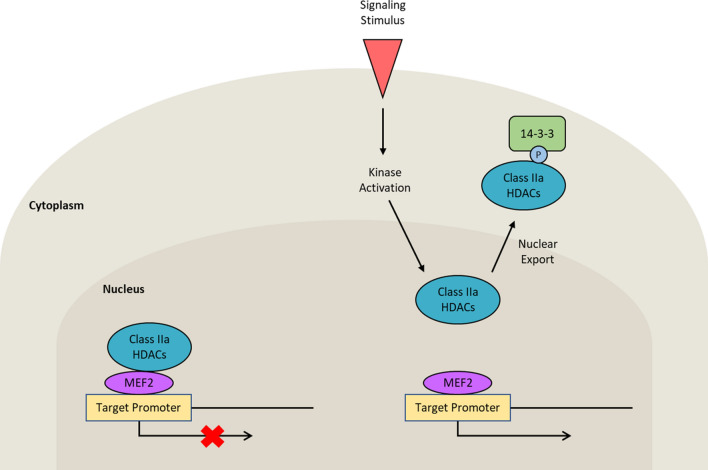
The regulation of Class IIa HDACs by phosphorylation and consequent change in subcellular localization has been studied extensively. In response to a variety of cellular signaling processes, Class IIa HDACs can be phosphorylated at sites that regulate either their nuclear export or import, which further influences their ability to regulate gene transcription. The change in subcellular localization can depend on the interaction between Class IIa HDACs and 14-3-3 proteins, chaperones that recognize specific phosphoserine- or phosphothreonine-containing motifs [52, 53]. McKinsey et al. identified a nuclear export sequence in HDAC5 that is conserved in HDAC4 and HDAC7 and is activated by Ca2+/calmodulin-dependent protein kinase (CaMK)-mediated phosphorylation in the 14-3-3 binding site of these HDACs [54]. Upon their interaction with 14-3-3 proteins, Class IIa HDACs undergo nuclear export, which is associated with reduced transcriptional repression due to the loss of their interaction with myocyte enhancer factor-2 (MEF2) transcription factors (Fig. 2).
Fig. 2.
14-3-3-mediated nuclear export of phosphorylated Class IIa HDACs. One of the general mechanisms by which Class IIa HDACs are regulated is phosphorylation-dependent change in subcellular localization. Though the phosphorylation of Class IIa HDACs can promote their nuclear import or export under specific stimuli and cellular contexts, many studies have presented the finding that the phosphorylation of Class IIa HDACs promotes their binding to 14-3-3 proteins and nuclear export under a wide variety of stimuli and contexts. As a consequence of their localization in the cytoplasm, Class IIa HDACs are unable to repress MEF2-mediated gene transcription [52–54]. P phosphate
Phosphorylation and consequent nuclear export of Class IIa HDACs occurs through both signaling-dependent and signaling-independent mechanisms to regulate gene expression in a cell type-specific manner. Several studies have demonstrated that some mechanisms can be extended to all Class IIa HDACs. Through the activation of the tumor suppressor liver kinase B1 (LKB1), the salt-inducible kinases (SIK) SIK2 and SIK3 phosphorylate the Class IIa HDACs in their 14-3-3 binding sites. Due to their subsequent nuclear export, these HDACs are unable to repress MEF2-dependent transcription as it pertains to myogenesis [55]. In their study of HDAC7 phosphorylation, Dequiedt et al. identified that Class IIa HDACs, by extension, are constitutively phosphorylated by the hPar-1/MARK (microtubule affinity-regulating kinase) members EMK and C-TAK1, which promote their binding to 14-3-3 proteins and nuclear export [56]. Other studies have shown that the mechanism of regulation is only applicable to a specific HDAC. For example, a study by McKinsey et al. led to the finding that 14-3-3 binding is constitutive for HDAC4 in yeast and mammalian cells. In contrast, the interaction between HDAC5 and 14-3-3 is dependent on CaMK signaling, as was simulated by the overexpression of constitutively active CaMKI in Cos cells and 10T1/2 fibroblasts [57]. CaMKII activation under the condition of genotoxic stress, when cellular reactive oxygen species levels are increased, also leads to HDAC5 phosphorylation and nuclear export, which has been associated with p53-dependent apoptosis due to reduced p53 deacetylation [58].
On the other hand, signaling-related mechanisms of phosphorylation can also promote the nuclear localization of Class IIa HDACs. For example, HDAC5 phosphorylation within its nuclear localization sequence is needed for its nuclear import, where the loss of its phosphorylation (e.g., S279A mutation) resulted in greater cytoplasmic localization [59]. In cardiomyocytes, phosphorylation of HDAC5 by PKA, activated by cAMP signaling, prevents the binding of HDAC5 with 14-3-3 proteins. As a result, HDAC5 is not exported from the nucleus and can perform in its role to transcriptionally repress cardiac fetal gene expression, genes that are related to cardiomyocyte hypertrophy [60]. Similarly, cAMP signaling and PKA activation in Schwann cells results in HDAC4 phosphorylation, which induces its nuclear localization. Through these series of events, HDAC4 is able to contribute to myelin sheath development by repressing c-Jun expression. While this study was focused on HDAC4, this model of cAMP-mediated nuclear shuttling in Schwann cells is extended to all Class IIa HDACs [61].
Cellular signaling can also lead to the dephosphorylation and nuclear import of Class IIa HDACs. Interestingly, whereas cAMP signaling has been associated with the inhibition of HDAC nuclear export through their phosphorylation as described above, it has also been found to promote nuclear import through other mechanisms. In myoblasts, cAMP signaling leads to nuclear import through the dephosphorylation of HDAC4, HDAC5, and HDAC9 in their conserved SP motif, referring to the serine phosphorylation site and the subsequent proline residue that respond to cAMP activation and change in localization. Notably, this mechanism is not extended to HDAC7 because it does not contain the SP motif in its sequence, demonstrating how the Class IIa HDACs can be differentially regulated [62]. Specific to HDAC5, it was identified that β1-adrenergic receptor activation and PKA activation in adult rat cardiomyocytes lead to its dephosphorylation by serine/threonine-protein phosphatase 2A (PP2A), promoting its nuclear accumulation and repression of MEF2-dependent gene transcription [63]. This is interesting because, as noted above, PKA can also directly phosphorylate HDAC5 to prevent its nuclear export in cardiomyocytes [60]. In another study, PP2A activity was also found to dephosphorylate HDAC4 to promote its nuclear import [64]. Thus, these studies demonstrate the complexity of HDAC regulation by phosphorylation and highlight the intricacies of the signaling pathways, cellular context, and protein-specific attributes that may contribute to the consequence of this modification.
Although the regulation of subcellular localization by phosphorylation has largely focused on Class IIa HDACs, there has been some investigation into the regulation of SIRT1 localization by phosphorylation. One study found that SIRT1 phosphorylation by JNK1 under conditions of oxidative stress increased its nuclear localization and affected its activity in a substrate-specific manner. Whereas SIRT1 phosphorylation promoted its enzymatic activity towards histone H3, the acetylation of p53, a SIRT1 substrate, was unaffected [65]. It is possible that the change in SIRT1 localization may regulate genes that protect cells from apoptosis due to oxidative stress [65–67].
Role of phosphorylation in regulating HDAC stability
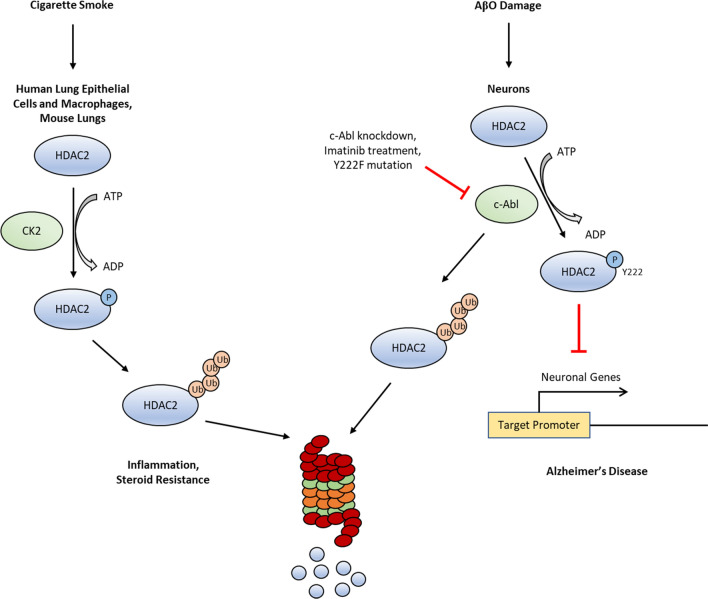
In several contexts, it has been demonstrated that HDAC phosphorylation can promote their stability through diverse and protein-specific mechanisms (Fig. 3). In neurons, phosphorylation of HDAC2 at Y222 by tyrosine-protein kinase ABL1 (c-Abl) is important for maintaining HDAC2 protein expression. The loss of its phosphorylation by site mutation or c-Abl inhibition resulted in HDAC2 poly-ubiquitination and proteasomal degradation, thereby reducing the transcriptional repression of its target synaptic and neuronal genes, such as Synaptotagmin and GluR1 [68]. Similarly, SIRT1 phosphorylation by Ca2+/calmodulin-dependent protein kinase kinase β (CaMKKβ) stimulated by pulsatile shear stress in vascular endothelial cells was associated with an increase in its stability and activity, consequently promoting an anti-oxidative stress response and anti-inflammatory effects that are protective against atherosclerosis [69] In a study focusing on osteoblastic cells, Shimizu et al. identified that PKA acting downstream of parathyroid hormone signaling serves not only to phosphorylate HDAC4 at S740 to promote its nuclear export, but also to stimulate PP2A-mediated dephosphorylation of HDAC4 at S355. Dephosphorylation of HDAC4 at this site then leads to its partial lysosomal degradation [53].
Fig. 3.
Phosphorylation of HDAC2 influences its stability. Phosphorylation of HDAC2 can have different effects on its stability depending on the cellular context. HDAC2 phosphorylation by CK2 in lung epithelial cells, macrophages, and mouse lungs following exposure to cigarette smoke leads to its ubiquitination and proteasomal degradation. Loss of HDAC2 through this mechanism is proposed to play a role in inflammation and steroid resistance associated with asthma and chronic obstructive pulmonary disease (COPD) [29]. In neurons, loss of HDAC2 phosphorylation at Y222 leads to the proteasomal degradation of HDAC2, releasing its repression of the transcription of neuronal genes that otherwise occurs in Alzheimer’s disease [68]. P phosphate, Ub ubiquitin
Phosphorylation can also directly destabilize these proteins under other conditions. HDAC4, in particular, contains a PEST1 sequence, a region that is enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) residues, that can link its interaction with E3 ubiquitin ligases [70, 71]. As demonstrated by the S298D mutation to mimic constitutive phosphorylation in this region, HDAC4 undergoes poly-ubiquitination and degradation [70]. A study by Adenuga et al. found that exposing human bronchial epithelial and primary small airway epithelial cells, macrophages, and mouse lungs to cigarette smoke extract led to HDAC2 phosphorylation through a CK2-mediated mechanism, and further to its ubiquitination and subsequent degradation. This finding provides a rationale and strategy to reverse the loss of HDAC2 in lung inflammation-related diseases [29].
The destabilization of several sirtuins by their phosphorylation has particularly been described in the context of cellular stress and death. Tyrosine phosphorylation of SIRT2 by c-Src reduces its protein stability and expression, which is associated with increased p53 expression and apoptosis [72]. SIRT6 phosphorylation by RAC-alpha serine/threonine-protein kinase (AKT1) leads to its ubiquitination by the E3 ubiquitin-protein ligase MDM2 and degradation. This reduces the ability of SIRT6 to promote cell cycle arrest and apoptosis, thus supporting the proliferation of breast cancer cells [73]. Another study found that upon energy starvation, SIRT7 undergoes phosphorylation by AMPK, leading to a change in its localization from the nucleolus to the nucleoplasm and its ubiquitin-independent degradation through the REGγ proteasome. This mechanism reduces the energy-consuming rDNA transcription mediated by SIRT7 to allow cell survival under the imposed stress conditions [74].
For SIRT1, differential effects of kinase activities on its stability demonstrate the importance of balance among the regulatory mechanisms. SIRT1 phosphorylation at S46 by JNK1 results in its proteasome-mediated degradation in murine macrophage cells during the lipopolysaccharide-activated macrophages inflammatory response, as well as in obese mice [75, 76]. However, in this study, Gao et al. found that both JNK1 inactivation and its persistent activation lead to SIRT1 degradation, where SIRT1 S46 phosphorylation first increases its enzymatic activity for a period of time, after which it eventually undergoes ubiquitination and proteasomal degradation [76]. Conversely, JNK2 activity has been found to promote SIRT1 stability and was correlated with SIRT1 phosphorylation at S27 in vitro [76, 77].
Regulation of HDAC attributes by phosphorylation during mitosis
Several studies have found that phosphorylation dynamics can regulate HDAC interactions, functions, and localization during mitosis. One study demonstrated that HDAC2, but not HDAC1, is hyperphosphorylated during mitosis following spindle checkpoint activation, where its hyperphosphorylation is associated with increased deacetylase activity [26]. Additionally, HDAC3 phosphorylation at Ser424 and interaction with linker histone subtype H1.3 in complex were found to be higher under mitotic conditions. The associated increase in HDAC3 activity towards histone H3K9 under these circumstances has been suggested to influence a more compact mitotic chromatin conformation and polar microtubule dynamics [32]. A study of HDAC5 phosphorylation during mitosis showed that modification in its nuclear localization sequence by Aurora kinase B (AurB) resulted in the sequestration of HDAC5 in the spindle midzone, such that it was unable to interact with members of the N-CoR co-repressor complex and to repress gene transcription. Though HDAC4 and HDAC9 can also be phosphorylated by this mitotic kinase, HDAC7 does not contain the AurB consensus sequence [78].
SIRT1 and SIRT2 are also subject to regulation by phosphorylation during mitosis. SIRT1 phosphorylation during mitosis suppresses centriole duplication. Specifically, to this end, its phosphorylation by the mitotic centrosomal kinase Aurora kinase A (AURKA) promotes its interaction with, and subsequent deacetylation and ubiquitin-dependent degradation of, the centrosome protein polo-like kinase 2 (Plk2). Thus, hypophosphorylation of SIRT1 during late G1 allows Plk2 accumulation and centriole duplication to be initiated [79]. SIRT1 phosphorylation by Cyclin B/Cdk1 is also important for cells to enter the G2 phase in the cell cycle and thus regulates cell proliferation [80]. SIRT2 phosphorylation, however, can either promote or inhibit its function in regulating cell proliferation depending on the kinase and phosphorylation site. One study proposed that the binding of group IVA cytosolic phospholipase A2 (cPLA2α) to SIRT2 results in Cdk2-mediated phosphorylation of SIRT2 at S331. This modification promotes mitotic entry by reducing SIRT2 activity, as detected towards acetylated histone H4 peptide and acetylated histone H4K16, and its recruitment to centrosomes and mitotic spindles [50]. In contrast, another study found that Cdk1-mediated phosphorylation of SIRT2 at S368 during mitosis does not affect its deacetylase activity, but instead supports its anti-proliferative function by delaying cell cycle progression in glioma cells [81]. It would be interesting to determine if this is a cell type-specific mechanism or if it can be applied to other contexts.
Physiological and disease relevance of HDAC phosphorylation
Cellular signaling and phosphorylation events are involved in the maintenance of normal physiological processes, as well as the development of disease states. HDACs, as well, play a role in regulating a wide range of cellular processes and have also been associated with diseases, such as cancer and metabolic disorders. Importantly, many HDAC phosphorylation sites have been identified in the context of certain diseases, tissue types, and signaling mechanisms. Some examples of HDAC phosphorylation in health and diseases are listed in Table 2. Phosphorylation of SIRT1 and its effect on the tumor suppressor p53 under various contexts are illustrated in Fig. 4. These examples provide insight into possible avenues by which to specifically target HDAC activity and function for therapeutic applications.
Table 2.
Histone deacetylase (HDAC) phosphorylation in physiological processes and disease
| Protein | Process/disease | References |
|---|---|---|
| HDAC1 | Osteoblast differentiation, Diet-induced obesity, Hepatosteatosis | [103, 120] |
| HDAC2 | Cardiac hypertrophy, Diet-induced obesity, Hepatosteatosis, Neurodegenerative diseases, Alzheimer’s disease, COPD-related inflammation | [28, 29, 68, 110, 120, 130] |
| HDAC3 | HER2-positive breast cancer, Parkinson’s disease, Glycolysis | [33, 34, 112] |
| HDAC4 | Glioblastoma, Skeletal myogenesis, Muscle cell differentiation, Cardiac hypertrophy, Gluconeogenesis, Myelin sheath development | [61, 85, 86, 97, 104, 105, 113] |
| HDAC5 | Glioblastoma, Immune signaling, Skeletal myogenesis, Muscle cell differentiation, Angiogenesis, Cardiac hypertrophy, Gluconeogenesis, Insulin resistance (Type II diabetes, obesity), Neuronal differentiation, Synaptic plasticity, Depression, Cocaine-reward behavior | [57, 60, 85, 86, 98, 99, 104, 107, 108, 113, 114, 127, 128, 158] |
| HDAC6 | Neurodegenerative diseases, Mitochondrial transport | [41, 129] |
| HDAC7 | Glioblastoma, Immune signaling, Muscle cell differentiation, Angiogenesis, Gluconeogenesis | [85, 86, 101, 102, 113] |
| HDAC9/MITR | Glioblastoma, Muscle cell differentiation | [85, 86, 99] |
| SIRT1 | Liver cancer, Osteosarcoma, Diet-induced obesity, NAFLD, Lipid homeostasis, Hepatosteatosis, Atherosclerosis, DNA damage response | [44, 48, 69, 84, 89, 118, 121, 122, 126] |
| SIRT2 | Microglial inflammation, Listeria monocytogenes infection | [95, 96] |
| SIRT3 | Colon carcinoma, Triple-negative breast cancer, Glioblastoma | [87] |
| SIRT6 | HER2-positive breast cancer | [73] |
| SIRT7 | Cardiac fibrosis | [111] |
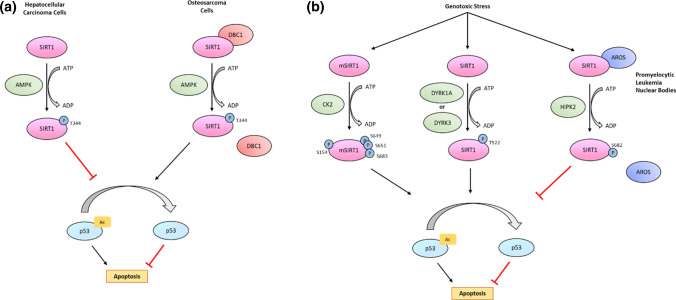
Fig. 4.
Relationship between SIRT1 phosphorylation and p53-mediated apoptosis. a Phosphorylation of SIRT1 at T344 by AMPK can have opposing effects on p53 acetylation, depending on the cellular context. SIRT1 phosphorylation in hepatocellular carcinoma cells inhibits its activity in deacetylating p53 and promotes apoptosis [48]. In contrast, its phosphorylation in osteosarcoma cells disrupts its interaction with its inhibitor, DBC1, allowing SIRT1 to deacetylate p53 and inhibit apoptosis [84]. b Genotoxic stress leads to the phosphorylation of SIRT1 at different sites by several kinases, including CK2, DYRK1A, DYRK3, and HIPK2, which influences the ability of SIRT1 to deacetylate p53. CK2- and DYRK-mediated phosphorylation of SIRT1 support its ability to deacetylate p53 and inhibit apoptosis [45, 46]. In contrast, SIRT1 phosphorylation by HIPK2 disrupts its interaction with its activator, AROS, which inhibits the deacetylation of p53 and leads to apoptosis [47]. P phosphate, Ac acetyl group
Cancer
HDACs have been studied extensively in relation to cancer. Several HDAC inhibitors have been approved by the United States Food and Drug Administration (FDA) and other drugs are under clinical investigation for the treatment of cancer [82, 83]. According to the complexity of the tumor microenvironment and the activated signaling pathway(s), it is critical to consider whether the role of HDAC phosphorylation supports oncogenic or tumor suppressor functions. For example, SIRT1 phosphorylation can have opposite effects depending on the tumor type. In U2OS cells, AMPK-mediated phosphorylation of SIRT1 at T344 causes its dissociation from Deleted in Breast Cancer 1 (DBC1), its inhibitor, to result in p53 deacetylation [84]. However, another group found that SIRT1 phosphorylation at the same site by AMPK in liver cancer cells led to its inactivation, although the role of DBC1 was not evaluated in this study [48, 84]. The activity of different HDACs can also be involved in regulating the same tumor type. In HER2-positive breast cancer cells, HDAC3 phosphorylation by c-Src increased its activity to promote cell proliferation, suggesting that this mechanism may potentially serve as a target in treatments for this breast cancer subtype [34]. SIRT6 promotes cell growth in HER2-positive breast cancer through another mechanism, by which it contributes to trastuzumab resistance. Specifically, SIRT6 phosphorylation by AKT1, interaction with the E3 ubiquitin ligase MDM2, and subsequent degradation reduces its inhibition of cell cycle arrest and apoptosis [73].
Phosphorylation of Class IIa HDACs has been associated with resistance to glioblastoma (GBM) therapies. It was initially found that mTOR Complex 2 (mTORC2) activation in EGFRvIII mutant GBM leads to the phosphorylation and subsequent inactivation of Class IIa HDACs, followed by higher acetylation of the forkhead box proteins FoxO1 and FoxO3, as well as Myc proto-oncogene protein (c-Myc) upregulation. The phosphorylation of Class IIa HDACs therefore has therapeutic relevance because activated mTORC2 signaling, acetylated FoxO proteins, and higher c-Myc levels are all associated with metabolic reprogramming and poor prognosis in glioblastoma patients [85]. Further study indicated that the auto-activation of mTORC2 and inactivating phosphorylation of Class IIa HDACs in EGFRvIII mutant GBM occur when nutrients, including glucose and acetate, are present at higher levels. Rictor acetylation was increased as a consequence of HDAC inactivation, resulting in an inability to inhibit mTORC2. In particular, Rictor acetylation in this process is connected with resistance to therapies that target EGFR, phosphoinositide 3-kinase (PI3K), and AKT [86].
Other drug resistance mechanisms related with phosphorylation status can be extended across tumor types. One study identified that radiation induces the co-localization of Cyclin B1-Cdk1 and SIRT3 at the mitochondria. Cdk1-mediated phosphorylation of SIRT3 at T150 and S159 promotes the deacetylation of mitochondrial proteins, as was tested in HCT116 colon carcinoma, MDA-MB-231 triple-negative breast cancer, and U87 glioblastoma cell lines. SIRT3 phosphorylation is important for mitochondrial functions, such as ATP production and maintenance of the mitochondrial membrane potential, and tumor adaptive resistance to radiation therapy [87]. Another mechanism of resistance is through interleukin 6 (IL-6)-mediated protection of cancer cells from chemotherapy drug-induced apoptosis [88, 89] This is detrimental, considering that the downstream Janus kinase-signal transducer and activator of transcription 3 (JAK-STAT3) pathway is activated in many tumor types, including lung adenocarcinoma and breast cancers [89, 90]. However, SIRT1 phosphorylation by JAK1 downstream of IL-6 stimulation serves as a negative feedback mechanism that targets STAT3 transcriptional activity and IL-6 activity to overcome the resistance to chemotherapy. In this way, SIRT1 phosphorylation can oppose IL-6-induced cancer cell survival [89]. Thus, a better understanding of the mechanisms that regulate specific HDACs in different tumor types can aid in the development of new therapeutic strategies and improve the design of current approaches to overcome drug resistance.
Infection and immune response
Several cases of HDAC phosphorylation in relation to the immune system have been described. One study found that B cell antigen receptor (BCR) activation in B lymphocytes leads to protein kinase D (PKD) enzymes-mediated phosphorylation of HDAC5 and HDAC7, followed by their nuclear export and reduced impact on transcriptional repression, as was demonstrated by nuclear receptor Nur77 reporter activation [91]. In thymocytes, T cell receptor (TCR) activation leads to HDAC7 phosphorylation by PKD1 through a calcium-independent mechanism. As a consequence, HDAC7 undergoes nuclear export, leading to the transcriptional activation of Nur77. This series of events causes thymocytes to undergo apoptosis [92–94]. In cytotoxic T lymphocytes (CTLs), however, HDAC7 has also been found to be constitutively phosphorylated and bound to 14-3-3 proteins through a mechanism that does not require TCR stimulation. Under these conditions, HDAC7 is localized in the cytoplasm and is unable to repress the transcription of cytokines and their receptors, as well as adhesion molecules. A demonstrated consequence of the loss of HDAC7 phosphorylation in CTLs was the repression of cytokine receptor CD25 expression and the consequent loss of effector cytokine interferon-gamma (IFNγ) production. Importantly, it was indicated that although HDAC7 phosphorylation is not a response to TCR activation, it still plays a role in mediating the effector response, as IFNγ was not produced upon TCR activation in CTLs expressing the HDAC7 phosphorylation-inactive mutant [94]. In totality, these findings demonstrate how a deacetylase enzyme, namely HDAC7, can be regulated by different mechanisms among various cell types to produce specific effects.
SIRT2 phosphorylation also plays several roles in infection and immune response. One study found that the loss of SIRT2 phosphorylation at S331, as was mimicked by mutation of the site to alanine, resulted in an increase in its deacetylase activity towards NFκB and consequent reduction in pro-inflammatory gene transcription in microglia. Accordingly, SIRT2 phosphorylation has been suggested as a target for microglial neurotoxicity and associated inflammation [95]. A role for the phosphorylation status of SIRT2 was additionally identified in the context of Listeria monocytogenes infection. Specifically, SIRT2 dephosphorylation at S25 following infection promotes its association with chromatin, deacetylation of H3K18, and transcriptional repression to support infection-related outcomes [96].
Differentiation and development
Class IIa HDACs are involved in differentiation and development processes due to their regulation of MEF2-mediated transcription in a variety of contexts. Phosphorylation, in particular, has been important for their function in these processes. As described above, phosphorylation of Class IIa HDACs can regulate their subcellular localization, which thereby influences their interaction with MEF transcription factors to regulate gene expression. A study by Liu et al. suggested crosstalk between the beta-adrenergic signaling pathway and muscle activity-dependent pathway in skeletal muscle fibers. In this study, HDAC4 phosphorylation at S265 and S266 by PKA following beta-adrenergic activation led to its nuclear influx and suppression of MEF2-mediated transcription of skeletal muscle genes, whereas its phosphorylation at S246, S267, and S232 by CaMKII following muscle fiber activity led to its nuclear efflux to have the opposite effect on transcription [97]. HDAC4 and HDAC5 phosphorylation downstream of CaMK signaling leads to their nuclear export, allowing MEF2 to interact with myoblast determination protein 1 (MyoD) and activate gene transcription for the muscle differentiation program for skeletal myogenesis [57, 98]. In myoblasts, phosphorylation of the nuclear localization sequence of Class IIa HDACs, as demonstrated in HDAC5 and the HDAC9 splice variant MITR, by Mirk in myoblasts reduces their nuclear accumulation and inhibition of MEF2C transcription factor, which allows myogenin transcription to promote muscle cell differentiation. Importantly, muscle cell differentiation is relevant to the therapeutic regeneration of damaged skeletal or cardiac muscle [99].
Angiogenesis is a process that is involved in vascular development. In endothelial cells, signaling downstream of vascular endothelial growth factor (VEGF) stimulation can lead to Class IIa HDAC phosphorylation, consequently influencing MEF2-mediated transcription of genes involved in angiogenesis. In this context, HDAC5 phosphorylation at S259 and S498 by PKD resulted in its nuclear export and activated transcription of MEF2-target genes related to endothelial cell migration and tube formation, processes that are representative of angiogenesis [100]. Similarly, downstream of VEGF stimulation, HDAC7 phosphorylation at S178, S344, and S479, was mediated by PKD1. HDAC7 cytoplasmic localization under this circumstance was associated with the transcriptional activation of the matrix metalloproteinases MMP10 and MT1-MMP, which support endothelial cell migration and tube formation [101]. Another study additionally identified that HDAC7 phosphorylation reduces its repression of angiogenesis-related genes, including Nur77 and RCAN2, accordingly promoting proliferation and migration [102].
Unlike the cases described above, HDAC1 phosphorylation has been suggested to play a role in osteoblast differentiation. Gemini-Piperni et al. performed a study to better understand the mechanisms involved in osteoblast adhesion, as is relevant to the clinical application of hydroxyapatite (HA) scaffolds for bone tissue regeneration. HDAC1 phosphorylation at S421 by cyclin-dependent kinase 5 (CDK5) was found to be associated with osteoblast differentiation. Further, HDAC1 phosphorylation levels were higher in cells that were adhering to HA surfaces, as opposed to the tissue culture plate, a difference that was attributed to osteoblast differentiation [103].
Cardiac hypertrophy
Class IIa HDAC regulation of MEF2-mediated transcription extends to the development of cardiac hypertrophy. Hypertrophic stimuli and their downstream signaling processes have been found to regulate the phosphorylation of HDAC4 and HDAC5 to influence their role in the fetal gene response, which is the reprogramming of gene expression in cardiomyocytes to allow the consequent hypertrophic phenotype [104]. Phosphorylation of HDAC4, but not other Class IIa HDACs, by the Ca2+/calmodulin-dependent protein kinase II form CaMKIIδB and of HDAC5 by PKD in cardiomyocytes reduces their repression of MEF2-mediated fetal cardiac gene expression, thereby contributing to the development of cardiac hypertrophy [104–106]. However, the different pathways leading to HDAC5 phosphorylation in cardiomyocytes must be balanced to appropriately regulate hypertrophy-related gene expression. Specifically, PKA activation downstream of acute beta-adrenergic signaling is associated with HDAC5 phosphorylation at S279 and its nuclear accumulation, in opposition to its nuclear export and transcriptional activation due to its phosphorylation at S259 and S498 by CaMKII and PKD downstream of G-protein coupled receptor (GPCR) signaling [107]. Notably, HDAC5 was also identified to be phosphorylated at S259 by protein kinase Cδ (PKCδ), a kinase that plays a role in myocardial hypertrophy and heart failure [108, 109].
Hypertrophic stimuli-mediated signaling can also induce the phosphorylation of other HDACs to promote the development of cardiac hypertrophy. Through immunohistochemistry and immunoblot methods, HDAC2 phosphorylation at S394 was detected to be higher in the heart tissue of patients with hypertrophic cardiomyopathy relative to normal heart tissue. HDAC2 phosphorylation by CK2 occurring downstream of hypertrophic stimuli leads to its increased enzymatic activity towards genes that promote disease development [110]. In cardiofibroblasts, angiotensin-II stimulation leads to an increase in the phosphorylation of SIRT7, which is suggested to influence changes in gene transcription through ERK and Mothers against decapentaplegic homolog 2 (Smad2) phosphorylation and activation. The changes in gene expression promote the differentiation of cardiac fibroblasts to myofibroblasts, thereby supporting the development of cardiac fibrosis [111].
Metabolic processes
HDAC phosphorylation has also been studied in the context of metabolic processes and diseases. For example, HDAC3 S424 is phosphorylated through the PI3K/AKT/mTOR pathway downstream of insulin stimulation, which promotes the deacetylation and activation of phosphoglycerate kinase 1 (PGK1) to produce ATP and 3-phosphoglycerate (3-PG) during glycolysis. HDAC3 modification is therefore suggested as a relevant target for diseases involving PGK1 dysfunction, such as hemolytic anemia [112]. A study by Mihaylova et al. showed that treatment of hepatocytes with the fasting hormone glucagon leads to the dephosphorylation of HDAC4, HDAC5, and HDAC7, which promotes their nuclear localization, deacetylation of FOXO, and subsequent activation of the FOXO family of transcription factors at the promoters of genes involved in gluconeogenesis. Given that the suppression of Class IIa HDACs was shown to improve hyperglycemia in mouse models of type 2 diabetes, it is suggested that the phosphorylation, and subsequent nuclear export, of Class IIa HDACs may be a mechanism by which to target metabolic syndrome [113].
Many studies have specifically focused on the role of HDAC phosphorylation in obesity and fatty liver. It was found that HDAC5 phosphorylation by AMPK in skeletal muscle is associated with its nuclear export, thereby reducing the transcriptional repression of glucose transporter type 4 (GLUT4) [114]. Given that higher GLUT4 expression is involved in overcoming insulin resistance related to obesity and type 2 diabetes, a therapeutic strategy in this context would be to promote HDAC5 phosphorylation in skeletal muscle [114–117]. SIRT1 phosphorylation has also been shown to have a protective effect against diet-induced obesity, as its consequent increased activity leads to the transcriptional activation of genes that promote higher thermogenic function and fatty acid oxidation. Therefore, another proposed therapeutic strategy is to promote SIRT1 phosphorylation at S434 downstream of beta-adrenergic signaling and the cAMP pathway in skeletal muscle and brown adipose tissue. [118, 119]. Additionally, HDAC1 and HDAC2 phosphorylation is also protective against diet-induced obesity, though through a different mechanism. Since dephosphorylation of HDAC1 S393 and HDAC2 S394 by mitogen-activated protein kinase phosphatase 3 (MKP-3) reduces their ability to repress lipogenic gene expression in the liver, regulation of MKP-3 activity presents a therapeutic target to oppose the development of hepatosteatosis [120].
Interestingly, SIRT1 can have differing effects on hepatosteatosis and fatty liver, depending on the context under which it is phosphorylated. CK2 expression and SIRT1 S164 phosphorylation levels were found to be higher in liver samples of non-alcoholic fatty liver disease (NAFLD) patients and were indicative of severity [121]. Under glucose starvation conditions, AMPK-mediated phosphorylation of SIRT1 at T530 reduces its degradation by the REGγ proteasome and allows its deacetylation of the autophagy proteins Atg5 and Atg7 to promote autophagy [122]. This balance between SIRT1 proteasomal degradation and autophagy is involved in regulating lipid homeostasis, further influencing attributes of metabolic disorders, where the consequence of SIRT1 phosphorylation is protective against steatosis and fatty liver [122–125]. Furthermore, Lu et al. generated mouse models that expressed knock-in T522A loss of phosphorylation and T522E (threonine to glutamic acid) phosphomimic mutations to show that the effect of SIRT1 T522 phosphorylation is tissue-dependent and must therefore be tightly regulated. Whereas T522 phosphorylation is necessary for systemic energy homeostasis, its dephosphorylation is needed for adipogenesis. Although constitutive phosphorylation of SIRT1 was related with an improvement in high-fat diet-induced dyslipidemia, loss of phosphorylation was associated with the development of hepatic steatosis under high-fat diet conditions because the expression of genes that promote hepatic fatty acid oxidation was lower [126]. These cases demonstrate how HDAC phosphorylation occurs in a variety of contexts to influence different outcomes. In the design of therapeutic approaches for metabolic disorders, it will be necessary to consider the intricacies of each pathway involved and the specific phosphorylation site that must be targeted without producing a harmful effect through another pathway.
Neurological disorders
Phosphorylation of HDACs also influences their roles in neurological development and disorders. HDAC4 phosphorylation downstream of cAMP signaling and PKA activation in Schwann cells leads to its nuclear localization and consequent repression of c-Jun to promote myelin sheath development [61]. Another study identified HDAC5 phosphorylation by SIK1 as a consequence of brain-derived neurotrophic factor (BDNF) signaling in cortical neurons. HDAC5 subsequently undergoes nuclear export, leading to an increase in MEF2 transcriptional activity. It is suggested that the transcriptional changes in this pathway are relevant to processes regulating neuronal differentiation and synaptic plasticity [127]. In hippocampal neurons, stimulation by ketamine, which is associated with antidepressant effects, leads to the activation of the CaMKII and PKD-dependent pathway and phosphorylation of HDAC5 at S259 and S498. MEF2 transcriptional activity is increased due to the nuclear export of HDAC5, presenting the possibility that HDAC5 phosphorylation induces anti-depressive effects. The phosphorylation of HDAC5 may therefore be relevant to the mechanism of action of ketamine [128].
In the context of neurodegenerative diseases, it was identified in the SH-SY5Y neuroblastoma cell line that HDAC6 phosphorylation by CK2 resulted in its increased cytoplasmic deacetylase activity, preventing protein aggregate accumulation and associated cellular stress that would otherwise lead to cytotoxicity [129]. In contrast, it has been proposed that HDAC6 may be phosphorylated by GSK3β, and that its increased tubulin deacetylase activity in hippocampal neurons could lead to issues in mitochondrial transport. Thus, Chen et al. proposed that this mechanism may serve as a therapeutic target for neurodegenerative disorders in which proper mitochondrial transport is affected [41]. HDAC2 phosphorylation has also been suggested as a therapeutic target for neurodegenerative diseases. HDAC2 phosphorylation at S394 was reduced under oxidative stress conditions, as demonstrated by cerebral ischemia in mice and hydrogen peroxide (H2O2) treatment of HT-22 hippocampal neurons. As a consequence of its loss of phosphorylation, HDAC2 was unable to interact with the transcription factor FOXO3a, leading to an increase in the expression of the cyclin-dependent kinase inhibitor p21, which has a protective effect against apoptosis under stress conditions [130]. Additionally, HDAC2 phosphorylation at Y222 by c-Abl is induced by Aβ oligomer damage in neurons, which promotes HDAC2-mediated repression of neuronal gene expression in Alzheimer’s disease [68]. HDAC3 phosphorylation contributes to cellular stress in the context of Parkinson’s disease. 6-hydroxydopamine (6-OHDA) treatment of neurons to model Parkinson’s disease conditions resulted in HDAC3 phosphorylation at S424 by LRRK2, which promoted HDAC3 activity. Consequences of HDAC3 modification included inhibition of MEF2D transcriptional activity, deacetylation of histone H4K12, and cytotoxicity [33]. Thus, HDACs produce different effects related to neurodevelopment and neurodegeneration, which can be targeted through their phosphorylation.
Looking to the future
As has been demonstrated and described above, phosphorylation can influence many different aspects of HDAC enzymes, including enzymatic activities, protein–protein interactions, protein localization, and protein stability, to affect physiological functions. Here, we have separated the effects of phosphorylation on protein regulation into different categories, such as activity and subcellular localization, but in many cases, these effects are not mutually exclusive and can influence one another. Accordingly, it is important to consider that phosphorylation can coordinate with other PTMs through crosstalk, such as ubiquitination, acetylation, and phosphorylation at other sites within the same protein, to regulate its characteristics and functions [28–30, 70]. Another manner of the temporal regulation of a protein is through its modification by different PTMs at the same residue under separate contexts [2]. These factors add layers of complexity to the identified mechanisms.
Although a few mechanisms of the effects of HDAC phosphorylation have been well-characterized, there are still many unknowns regarding intricacies in the cellular and molecular contexts. It is clear that while some sites within HDACs may be constitutively phosphorylated, many phosphorylation events occur as a response to a cellular stimulus. It would be valuable to determine whether identified mechanisms could be extended to other cell lines and conditions that actively undergo the same signaling processes. Though this has been explored to some extent, many studies focus on one protein within a given cellular context, which in itself could provide useful information regarding specificity. Additionally, numerous discoveries of HDAC phosphorylation sites have been made in certain cell lines under specific treatment conditions to mimic normal physiological processes or disease states. Elucidating the underlying conditions leading to HDAC phosphorylation, the downstream mechanism, and further, the biological outcome, is therefore particularly critical for the development of effective and specific targeted therapies that could target this modification.
A variety of tools and databases have helped to organize high-throughput PTM data in a comprehensive manner that is easily accessible to the public. Databases such as PhosphoSitePlus, Phospho.ELM, and Phosida provide an extensive range of information about predicted or validated sites, including their evolutionary conservation and the conditions under which they were detected [131–136]. Other databases, such as NetPhos 3.1, GPS (Group-based Prediction System), and Human Protein Reference Database (HPRD) PhosphoMotif Finder, offer predictions for phosphorylation sites and possible kinases or phosphatases based on sequence motifs [137–142]. These resources present a starting point for studies of protein phosphorylation to understand what is known and yet unknown. We anticipate that these resources will help uncover many more instances of HDAC phosphorylation, as well as point to their biological consequences.
There are many HDAC phosphorylation sites that have yet to be identified or validated. Though a protein may be phosphorylated at a certain site, this modification may elude detection if it occurs at low abundance or only transiently under specific conditions [4, 30]. Of note, the phosphorylation of HDAC10, HDAC11, SIRT4, and SIRT5 has not been extensively characterized, although several putative modified residues have been detected in mass spectrometry analyses (Table 3). Interestingly, HDAC10 Ser368 phosphorylation has been identified in several high-throughput analyses of cancer tissue and cell lines, including breast cancer and leukemia [131, 143–146]. HDAC11 threonine phosphorylation has been identified under the condition of ventricular tachycardia, and SIRT5 phosphorylation was predicted in acute myelogenous leukemia and lung cancer [131, 147]. Characterization of these predicted phosphorylation sites would provide some insight into the mechanisms by which these proteins are regulated, as well as their possible physiological relevance.
Table 3.
Putative phosphorylation sites on histone deacetylases (HDACs) that require further characterization
| Protein | Site | Cellular context | References |
|---|---|---|---|
| HDAC10 | S368 | Breast cancer, T-cell leukemia cells, Mitotic cells | [131, 143–146] |
| S373 | Mitotic cells | [144] | |
| S393 | Liver tissue | [163] | |
| S540 | T-cell leukemia cells | [131] | |
| HDAC11 | T5, T76 | Heart tissue, Ventricular tachycardia | [131] |
| Y199 | Mantle cell lymphoma cells | [164] | |
| SIRT4 | S255, S261, S262 | HeLa cells | [165] |
| SIRT5 | Y76 | Bone marrow tissue, Acute myelogenous leukemia | [131] |
| T87 | Lung cancer cells | [166] | |
| S160 | Lung cancer cells | [147] |
An additional point to be noted is that many characterized HDAC phosphorylation mechanisms involve the modification of serine and threonine residues, but fewer cases of tyrosine phosphorylation have been described. This difference can be attributed to the cellular or disease conditions under which the phosphorylation sites were detected and the transient nature of tyrosine phosphorylation [148]. Importantly, tyrosine phosphorylation has been taken into consideration and incorporated into the design of targeted therapeutic approaches due to its roles in growth factor-mediated signaling and regulation of cellular processes leading to disease development and progression [2, 3, 148]. The clinical relevance of tyrosine phosphorylation is supported by the many tyrosine kinase inhibitors that have been approved as treatment options, such as for cancer, in which aberrant tyrosine phosphorylation has been associated with cellular transformation and proliferation [148–151]. Therefore, although tyrosine phosphorylation may be more difficult to detect, it is a promising, yet largely unexplored, avenue of research regarding HDAC regulation and relationship with diseases.
Additional challenges in the study of protein phosphorylation arise as a result of the extensive variation in the effect of phosphorylation on the HDACs due to the complexity added by a multitude of factors, including the location of the modified site within the protein, such as the catalytic domain or nuclear localization sequence, and crosstalk between other PTMs. However, discrepancies in the conclusions made from experiments evaluating the effect of phosphorylation of the same site(s) within the same protein can also arise from differences between the methodologies used. As discussed above, HDAC1 phosphorylation at S421 and S423 was found to have different effects depending on whether core histones or the histone H4 peptide was used as the substrate to assess changes in deacetylase activity [24–26].
Further, it has been suggested that the results can vary depending on the approach used to induce or simulate a change in the phosphorylation state. Ideally, a study will employ 1) a genetic approach to generate mutants that mimic constitutive phosphorylation or loss of phosphorylation at the site and 2) a chemical approach through treating cells with activators or inhibitors of kinases or phosphatases. One such case that demonstrates the differences in results associated with genetic and chemical approaches is an evaluation of the phosphorylation state of HDAC1. In their study, Pflum et al. suggested that S421 and S423 may be constitutively phosphorylated because they are buried sites within the structure of HDAC1 and may thus be inaccessible to phosphatases [24]. In contrast, though, Galasinski et al. identified in their study that HDAC1 is basally phosphorylated, but that it can still be hyperphosphorylated upon treatment with the phosphatase inhibitor okadaic acid [26]. Also in contrast to the study by Pflum et al., Galasinski et al. found that hyperphosphorylation of HDAC1 through phosphatase inhibition in fact interfered with the ability of HDAC1 to interact with HDAC2, mSin3A, and YY1 [24, 26]. A possible explanation for this discrepancy may be in the methods employed in these studies, whereas Pflum et al. tested the effect of the loss of site-specific phosphorylation through mutations of S421 and S423 to alanine, Galasinski et al. evaluated the general phosphorylation of HDAC1 through treatment with the phosphatase inhibitor okadaic acid [24, 26].
Concluding remarks
HDACs regulate a variety of cellular processes, in addition to gene transcription, in different tissue types, thereby influencing normal physiological functions and disease development. Therefore, it is critical to define the mechanisms that regulate these enzymes and to identify the functional consequences associated with their dysregulation, particularly in the larger context of disease states. As has been discussed here, the modification of HDACs by phosphorylation can affect its protein characteristics and functions in different ways. Many class-wide, as well as protein-specific, mechanisms of phosphorylation have been described, but ample remain to be identified and validated. Further characterization of the intricacies regarding the regulation of HDACs by phosphorylation will be clinically relevant, as a better understanding of the underlying mechanisms will support the design of therapeutic strategies to selectively inhibit or activate any HDAC in a given physiological state.
Acknowledgements
We thank members of the Seto Lab for support and discussions. Additionally, we thank our past and present colleagues for discussions and insights on HDACs. We apologize to those whose work was not cited in this paper due to space constraints. Funding for this work was awarded to ES by the National Institutes of Health (R01CA187040, R01CA169210).
Author contributions
Both SB and ES conceived the idea for the article. The majority of the literature search and data analysis were performed by SB. SB wrote the first draft of the manuscript and ES critically revised the work.
Compliance with ethical standards
Conflict of interest
All authors declare that there are no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deribe Y, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 3.Spickett CM, Pitt AR, Morrice N, Kolch W. Proteomic analysis of phosphorylation, oxidation and nitrosylation in signal transduction. Biochim Biophys Acta. 2006;1764:1823–1841. doi: 10.1016/j.bbapap.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 5.Cao J, Sun L, Aramsangtienchai P, et al. HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. Proc Natl Acad Sci USA. 2019;116:5487–5492. doi: 10.1073/pnas.1815365116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roessler C, Nowak T, Pannek M, et al. Chemical probing of the human sirtuin 5 active site reveals its substrate acyl specificity and peptide-based inhibitors. Angew Chem Int Ed Engl. 2014;53:10728–10732. doi: 10.1002/anie.201402679. [DOI] [PubMed] [Google Scholar]
- 7.Tan M, Peng C, Anderson KA, et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Front Pharmacol. 2012;3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carafa V, Rotili D, Forgione M, et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenet. 2016;8:61. doi: 10.1186/s13148-016-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai H, Sinclair DA, Ellis JL, Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 12.Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman J, Dittenhafer-Reed K, Denu J. Sirtuin catalysis and regulation. J Biol Chem. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathias R, Greco TM, Oberstein A, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reichert N, Choukrallah M-A, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter T, Cooper J. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 17.Jin J, Pawson T. Modular evolution of phosphorylation-based signalling systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:2540–2555. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Laarse SAM, Leney AC, Heck AJR. Crosstalk between phosphorylation and O-GlcNAcylation: friend or foe. FEBS J. 2018;285:3152–3167. doi: 10.1111/febs.14491. [DOI] [PubMed] [Google Scholar]
- 19.Narayanan A, Jacobson M. Computational studies of protein regulation by post-translational phosphorylation. Curr Opin Struct Biol. 2009;19:156–163. doi: 10.1016/j.sbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Johnson L, Lewis R. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 21.Ryšlavá H, Doubnerová V, Kavan D, Vaněk O. Effect of posttranslational modifications on enzyme function and assembly. J Proteomics. 2013;92:80–109. doi: 10.1016/j.jprot.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 22.Kelly RDW, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 23.Milazzo G, Mercatelli D, Di Muzio G, et al. Histone deacetylases (HDACs): evolution, specificity, role in transcriptional complexes, and pharmacological actionability. Genes (Basel) 2020;11:556. doi: 10.3390/genes11050556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pflum MKH, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 25.Cai R, Kwon P, Yan-Neale Y, et al. Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem Biophys Res Commun. 2001;283:445–453. doi: 10.1006/bbrc.2001.4786. [DOI] [PubMed] [Google Scholar]
- 26.Galasinski SC, Resing KA, Goodrich JA, Ahn NG. Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J Biol Chem. 2002;277:19618–19626. doi: 10.1074/jbc.M201174200. [DOI] [PubMed] [Google Scholar]
- 27.Tsai S-C, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 28.Adenuga D, Rahman I. Protein kinase CK2-mediated phosphorylation of HDAC2 regulates co-repressor formation, deacetylase activity and acetylation of HDAC2 by cigarette smoke and aldehydes. Arch Biochem Biophys. 2010;498:62–73. doi: 10.1016/j.abb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am J Respir Cell Mol Biol. 2009;40:464–473. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segré CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol. 2011;2011:690848. doi: 10.1155/2011/690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Ozawa Y, Lee H, et al. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005;19:827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patil H, Wilks C, Gonzaelez RW, et al. Mitotic activation of a novel histone deacetylase 3-linker histone H1.3 protein complex by protein kinase CK2. J Biol Chem. 2016;291:3158–3172. doi: 10.1074/jbc.M115.643874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han KA, Shin WH, Jung S, et al. Leucine-rich repeat kinase 2 exacerbates neuronal cytotoxicity through phosphorylation of histone deacetylase 3 and histone deacetylation. Hum Mol Genet. 2017;26:1–18. doi: 10.1093/hmg/ddw363. [DOI] [PubMed] [Google Scholar]
- 34.Seo J, Guk G, Park S-H, et al. Tyrosine phosphorylation of HDAC3 by Src kinase mediates proliferation of HER2-positive breast cancer cells. J Cell Physiol. 2019;234:6428–6436. doi: 10.1002/jcp.27378. [DOI] [PubMed] [Google Scholar]
- 35.Lee H, Rezai-zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karolczak-Bayatti M, Sweeney M, Cheng J, et al. Acetylation of heat shock protein 20 (Hsp20) regulates human myometrial activity. J Biol Chem. 2011;286:34346–34355. doi: 10.1074/jbc.M111.278549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Somoza JR, Skene RJ, Katz BA, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 38.Deribe YL, Wild P, Chandrashaker A, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal. 2009;22:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- 39.Le Roy C, Wrana J. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 40.Williams KA, Zhang M, Xiang S, et al. Extracellular signal-regulated kinase (erk) phosphorylates histone deacetylase 6 (HDAC6) at serine 1035 to stimulate cell migration. J Biol Chem. 2013;288:33156–33170. doi: 10.1074/jbc.M113.472506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Owens G, Makarenkova H, Edelman D. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS ONE. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds I, Malaiyandi L, Coash M, Rintoul G. Mitochondrial trafficking in neurons: a key variable in neurodegeneration? J Bioenerg Biomembr. 2004;36:283–286. doi: 10.1023/B:JOBB.0000041754.78313.c2. [DOI] [PubMed] [Google Scholar]
- 43.Trimmer P, Borland M. Differentiated Alzheimer’s disease transmitochondrial cybrid cell lines exhibit reduced organelle movement. Antioxid Redox Signal. 2005;7:1101–1109. doi: 10.1089/ars.2005.7.1101. [DOI] [PubMed] [Google Scholar]
- 44.Kong D, Zhan Y, Liu Z, et al. SIRT1-mediated ERβ suppression in the endothelium contributes to vascular aging. Aging Cell. 2016;15:1092–1102. doi: 10.1111/acel.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang H, Jung J-W, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conrad E, Polonio-Vallon T, Meister M, et al. HIPK2 restricts SIRT1 activity upon severe DNA damage by a phosphorylation-controlled mechanism. Cell Death Differ. 2016;23:110–122. doi: 10.1038/cdd.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CW, Wong LL, Tse EY, et al. AMPK promotes p53 acetylation via phosphorylation and inactivation of SIRT1 in liver cancer cells. Cancer Res. 2012;72:4394–4404. doi: 10.1158/0008-5472.CAN-12-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Meter M, Simon M, Tombline G, et al. JNK phosphorylates SIRT6 to stimulate DNA double- strand break repair in response to oxidative stress by recruiting PARP1 to DNA breaks. Cell Rep. 2016;16:2641–2650. doi: 10.1016/j.celrep.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Movahedi Naini S, Sheridan AM, Force T, Shah JV, Bonventre JV. Group IVA cytosolic phospholipase A2 regulates the G2-to-M transition by modulating the activity of tumor suppressor SIRT2. Mol Cell Biol. 2015;35:3768–3784. doi: 10.1128/MCB.00184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pandithage R, Lilischkis R, Harting K, et al. The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J Cell Biol. 2008;180:915–929. doi: 10.1083/jcb.200707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang AH, Kruhlak MJ, Wu J, et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu E, Nakatani T, He Z, Partridge NC. Parathyroid hormone regulates histone deacetylase (HDAC) 4 through protein kinase A-mediated phosphorylation and dephosphorylation in osteoblastic cells. J Biol Chem. 2014;289:21340–21350. doi: 10.1074/jbc.M114.550699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walkinshaw DR, Weist R, Kim G-W, et al. The tumor suppressor kinase LKB1 activates the downstream kinases SIK2 and SIK3 to stimulate nuclear export of class IIa histone deacetylases. J Biol Chem. 2013;288:9345–9362. doi: 10.1074/jbc.M113.456996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dequiedt F, Martin M, Von Blume J, et al. New role for hPar-1 Kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol Cell Biol. 2006;26:7086–7102. doi: 10.1128/MCB.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen N, Kumari R, Singh MI, Das S. HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress. Mol Cell. 2013;52:406–420. doi: 10.1016/j.molcel.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteom. 2011;10(M110):004317. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha CH, Kim JY, Zhao J, et al. PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc Natl Acad Sci USA. 2010;107:15467–15472. doi: 10.1073/pnas.1000462107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomis-Coloma C, Velasco-Aviles S, Gomez-Sanchez JA, et al. Class IIa histone deacetylases link cAMP signaling to the myelin transcriptional program of Schwann cells. J Cell Biol. 2018;217:1249–1268. doi: 10.1083/jcb.201611150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walkinshaw DR, Weist R, Xiao L, et al. Dephosphorylation at a conserved SP motif governs cAMP sensitivity and nuclear localization of class IIa histone deacetylases. J Biol Chem. 2013;288:5591–5605. doi: 10.1074/jbc.M112.445668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weeks KL, Ranieri A, Karaś A, et al. β-adrenergic stimulation induces histone deacetylase 5 (HDAC5) nuclear accumulation in cardiomyocytes by B55α-PP2A-mediated dephosphorylation. J Am Heart Assoc. 2017;6:e004861. doi: 10.1161/JAHA.116.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paroni G, Cernotta N, Dello Russo C, et al. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:665–667. doi: 10.1091/mbc.e07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nasrin N, Kaushik VK, Fortier E, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berthiaume M, Boufaied N, Moisan A, Gaudreau L. High levels of oxidative stress globally inhibit gene transcription and histone acetylation. DNA Cell Biol. 2006;25:124–134. doi: 10.1089/dna.2006.25.124. [DOI] [PubMed] [Google Scholar]
- 67.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD + -dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 68.Gonzalez-Zuñiga M, Contreras PS, Estrada LD, et al. c-Abl stabilizes HDAC2 levels by tyrosine phosphorylation repressing neuronal gene expression in Alzheimer’s disease. Mol Cell. 2014;56:163–173. doi: 10.1016/j.molcel.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 69.Wen L, Chen Z, Zhang F, et al. Ca2 +/calmodulin-dependent protein kinase kinase β phosphorylation of Sirtuin 1 in endothelium is atheroprotective. Proc Natl Acad Sci USA. 2013;110:E2420–E2427. doi: 10.1073/pnas.1309354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cernotta N, Clocchiatti A, Florean C, Brancolini C. Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility. Mol Biol Cell. 2011;22:278–289. doi: 10.1091/mbc.E10-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. doi: 10.1016/S0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- 72.Choi YH, Kim H, Lee SH, Jin Y-H, Lee KY. Src regulates the activity of SIRT2. Biochem Biophys Res Commun. 2014;450:1120–1125. doi: 10.1016/j.bbrc.2014.06.117. [DOI] [PubMed] [Google Scholar]
- 73.Thirumurthi U, Shen J, Xia W, et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci Signal. 2014;7:71. doi: 10.1126/scisignal.2005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun L, Fan G, Shan P, et al. Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome. Nat Commun. 2016;7:12497. doi: 10.1038/ncomms12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hwang JS, Ham SA, Yoo T, et al. Upregulation of MKP-7 in response to rosiglitazone treatment ameliorates lipopolysaccharide-induced destabilization of SIRT1 by inactivating JNK. Pharmacol Res. 2016;114:47–55. doi: 10.1016/j.phrs.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 76.Gao Z, Zhang J, Kheterpal I, et al. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J Biol Chem. 2011;286:22227–22234. doi: 10.1074/jbc.M111.228874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 78.Guise AJ, Greco TM, Zhang IY, Yu F, Cristea IM. Aurora B-dependent regulation of class IIa histone deacetylases by mitotic nuclear localization signal phosphorylation. Mol Cell Proteom. 2012;11:1220–1229. doi: 10.1074/mcp.M112.021030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling H, Peng L, Wang J, Rahhal R, Seto E. Histone deacetylase SIRT1 targets Plk2 to regulate centriole duplication. Cell Rep. 2018;25:2851–2865. doi: 10.1016/j.celrep.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sasaki T, Maier B, Koclega KD, et al. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J Biol Chem. 2007;282:19546–19555. doi: 10.1074/jbc.M702990200. [DOI] [PubMed] [Google Scholar]
- 82.Eckschlager T, Plch J, Stiborova M, Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int J Mol Sci. 2017 doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 84.Lau AW, Liu P, Inuzuka H, Gao D. SIRT1 phosphorylation by AMP-activated protein kinase regulates p53 acetylation. Am J Cancer Res. 2014;4:245–255. [PMC free article] [PubMed] [Google Scholar]
- 85.Masui K, Tanaka K, Akhavan D, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Masui K, Tanaka K, Ikegami S, et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci USA. 2015;112:9406–9411. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu R, Fan M, Candas D, et al. CDK1-mediated SIRT3 activation enhances mitochondrial function and tumor radioresistance. Mol Cancer Ther. 2015;14:2090–2102. doi: 10.1158/1535-7163.MCT-15-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Conze D, Weiss L, Regen PS, et al. Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res. 2001;61:8851–8858. [PubMed] [Google Scholar]
- 89.Wang W, Li F, Xu Y, et al. JAK1-mediated Sirt1 phosphorylation functions as a negative feedback of the JAK1-STAT3 pathway. J Biol Chem. 2018;293:11067–11075. doi: 10.1074/jbc.RA117.001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao SP, Mark KG, Leslie K, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matthews SA, Liu P, Spitaler M, et al. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol Cell Biol. 2006;26:1569–1577. doi: 10.1128/MCB.26.4.1569-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parra M, Kasler H, McKinsey TA, Olson EN, Verdin E. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J Biol Chem. 2005;280:13762–13770. doi: 10.1074/jbc.M413396200. [DOI] [PubMed] [Google Scholar]
- 93.Dequiedt F, Van Lint J, Lecomte E, et al. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J Exp Med. 2005;201:793–804. doi: 10.1084/jem.20042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navarro MN, Goebel J, Feijoo-Carnero C, Morrice N, Cantrell DA. Phosphoproteomic analysis reveals an intrinsic pathway for the regulation of histone deacetylase 7 that controls the function of cytotoxic T lymphocytes. Nat Immunol. 2011;12:352–361. doi: 10.1038/ni.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pais TF, Szegő ÉM, Marques O, et al. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J. 2013;32:2603–2616. doi: 10.1038/emboj.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pereira JM, Chevalier C, Chaze T, et al. Infection reveals a modification of SIRT2 critical for chromatin association. Cell Rep. 2018;23:1124–1137. doi: 10.1016/j.celrep.2018.03.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu Y, Schneider MF. Opposing HDAC4 nuclear fluxes due to phosphorylation by β-adrenergic activated protein kinase A or by activity or Epac activated CaMKII in skeletal muscle fibres. J Physiol. 2013;591:3605–3623. doi: 10.1113/jphysiol.2013.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deng X, Ewton DZ, Mercer SE, Friedman E. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J Biol Chem. 2005;280:4894–4905. doi: 10.1074/jbc.M411894200. [DOI] [PubMed] [Google Scholar]
- 100.Ha CH, Wang W, Jhun BS, et al. Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J Biol Chem. 2008;283:14590–14599. doi: 10.1074/jbc.M800264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ha CH, Jhun BS, Kao HY, Jin ZG. VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating matrix metalloproteinase expression and angiogenesis. Arterioscler Thromb Vasc Biol. 2008;28:1782–1788. doi: 10.1161/ATVBAHA.108.172528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang S, Li X, Parra M, et al. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc Natl Acad Sci USA. 2008;105:7738–7743. doi: 10.1073/pnas.0802857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gemini-Piperni S, Milani R, Bertazzo S, et al. Kinome profiling of osteoblasts on hydroxyapatite opens new avenues on biomaterial cell signaling. Biotechnol Bioeng. 2014;111:1900–1905. doi: 10.1002/bit.25246. [DOI] [PubMed] [Google Scholar]
- 104.Carnegie GK, Soughayer J, Smith FD, et al. AKAP-Lbc mobilizes a cardiac hypertrophy signaling pathway. Mol Cell. 2008;32:169–179. doi: 10.1016/j.molcel.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Little GH, Bai Y, Williams T, Poizat C. Nuclear calcium/calmodulin-dependent protein kinase iidelta preferentially transmits signals to histone deacetylase 4 in cardiac cells. J Biol Chem. 2007;282:7219–7231. doi: 10.1074/jbc.M604281200. [DOI] [PubMed] [Google Scholar]
- 106.Huynh QK, McKinsey TA. Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism. Arch Biochem Biophys. 2006;450:141–148. doi: 10.1016/j.abb.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 107.Chang CW, Lee L, Yu D, et al. Acute β-adrenergic activation triggers nuclear import of histone deacetylase 5 and delays G(q)-induced transcriptional activation. J Biol Chem. 2013;288:192–204. doi: 10.1074/jbc.M112.382358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huynh QK. Evidence for the phosphorylation of serine259 of histone deacetylase 5 by protein kinase Cδ. Arch Biochem Biophys. 2011;506:173–180. doi: 10.1016/j.abb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 109.Chen L, Hahn H, Wu G, et al. Opposing cardioprotective actions and parallel hypertrophic effects of delta PKC and epsilon PKC. Proc Natl Acad Sci USA. 2001;98:11114–11119. doi: 10.1073/pnas.191369098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Eom GH, Cho YK, Ko JH, et al. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011;123:2392–2403. doi: 10.1161/CIRCULATIONAHA.110.003665. [DOI] [PubMed] [Google Scholar]
- 111.Wang H, Liu S, Liu S, et al. Enhanced expression and phosphorylation of Sirt7 activates smad2 and ERK signaling and promotes the cardiac fibrosis differentiation upon angiotensin-II stimulation. PLoS ONE. 2017;12:e0178530. doi: 10.1371/journal.pone.0178530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang S, Jiang B, Zhang T, et al. Insulin and mTOR pathway regulate HDAC3-mediated deacetylation and activation of PGK1. PLoS Biol. 2015;13:e1002243. doi: 10.1371/journal.pbio.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McGee SL, van Denderen BJ, Howlett KF, et al. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 115.Leturque A, Loizeau M, Vaulont S, Salminen M, Girard J. Improvement of insulin action in diabetic transgenic mice selectively overexpressing GLUT4 in skeletal muscle. Diabetes. 1996;45:23–27. doi: 10.2337/diab.45.1.23. [DOI] [PubMed] [Google Scholar]
- 116.Ren JM, Marshall BA, Mueckler MM, et al. Overexpression of Glut4 protein in muscle increases basal and insulin-stimulated whole body glucose disposal in conscious mice. J Clin Invest. 1995;95:429–432. doi: 10.1172/JCI117673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsao TS, Burcelin R, Katz EB, Huang L, Charron MJ. Enhanced insulin action due to targeted GLUT4 overexpression exclusively in muscle. Diabetes. 1996;45:28–36. doi: 10.2337/diab.45.1.28. [DOI] [PubMed] [Google Scholar]
- 118.Gerhart-Hines Z, Dominy JE, Jr, Blättler SM, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 120.Feng B, Jiao P, Helou Y, et al. Mitogen-activated protein kinase phosphatase 3 (MKP-3)-deficient mice are resistant to diet-induced obesity. Diabetes. 2014;63:2924–2934. doi: 10.2337/db14-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Choi SE, Kwon S, Seok S, et al. Obesity-linked phosphorylation of SIRT1 by casein kinase 2 inhibits its nuclear localization and promotes fatty liver. Mol Cell Biol. 2017;37:e00006–e00017. doi: 10.1128/MCB.00006-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dong S, Jia C, Zhang S, et al. The REGγ proteasome regulates hepatic lipid metabolism through inhibition of autophagy. Cell Metab. 2013;18:380–391. doi: 10.1016/j.cmet.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Czaja MJ. Autophagy in health and disease. 2. Regulation of lipid metabolism and storage by autophagy: pathophysiological implications. Am J Physiol Cell Physiol. 2010;298:C973–C978. doi: 10.1152/ajpcell.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y, Goldman S, Baerga R, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lu J, Xu Q, Ji M, et al. The phosphorylation status of T522 modulates tissue-specific functions of SIRT1 in energy metabolism in mice. EMBO Rep. 2017;18:841–857. doi: 10.15252/embr.201643803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Finsterwald C, Carrard A, Martin JL. Role of salt-inducible kinase 1 in the activation of MEF2-dependent transcription by BDNF. PLoS ONE. 2013;8:e54545. doi: 10.1371/journal.pone.0054545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi M, Lee SH, Wang SE, et al. Ketamine produces antidepressant-like effects through phosphorylation-dependent nuclear export of histone deacetylase 5 (HDAC5) in rats. Proc Natl Acad Sci USA. 2015;112:15755–15760. doi: 10.1073/pnas.1513913112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Watabe M, Nakaki T. Protein kinase CK2 regulates the formation and clearance of aggresomes in response to stress. J Cell Sci. 2011;124:1519–1532. doi: 10.1242/jcs.081778. [DOI] [PubMed] [Google Scholar]
- 130.Peng S, Zhao S, Yan F, et al. HDAC2 selectively regulates FOXO3a-mediated gene transcription during oxidative stress-induced neuronal cell death. J Neurosci. 2015;35:1250–1259. doi: 10.1523/JNEUROSCI.2444-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hornbeck PV, Zhang B, Murray B, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Diella F, Cameron S, Gemünd C, et al. Phospho.ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinform. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites–update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dinkel H, Chica C, Via A, et al. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gnad F, Ren S, Cox J, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 138.Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 139.Xue Y, Zhou F, Zhu M, et al. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–W187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xue Y, Ren J, Gao X, et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xue Y, Liu Z, Cao J, et al. GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng Des Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
- 142.Amanchy R, Periaswamy B, Mathivanan S, et al. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 143.Gu TL, Nardone J, Wang Y, et al. Survey of activated FLT3 signaling in leukemia. PLoS ONE. 2011;6:19169. doi: 10.1371/journal.pone.0019169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kettenbach AN, Schweppe DK, Faherty BK, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mertins P, Yang F, Liu T, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteom. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu G, Xiao CL, Lu CH, et al. Phosphoproteome profile of human lung cancer cell line A549. Mol BioSyst. 2011;7:472–479. doi: 10.1039/c0mb00055h. [DOI] [PubMed] [Google Scholar]
- 148.Hunter T. Tyrosine phosphorylation: thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hunter T. Treatment for chronic myelogenous leukemia: the long road to imatinib. J Clin Invest. 2007;117:2036–2043. doi: 10.1172/JCI31691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Arora A, Scholar EM. Role of tyrosine kinase inhibitors in cancer therapy. J Pharmacol Exp Ther. 2005;315:971–979. doi: 10.1124/jpet.105.084145. [DOI] [PubMed] [Google Scholar]
- 151.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci. 2015;36:422–439. doi: 10.1016/j.tips.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 152.Meng F, Han M, Zheng B, et al. All-trans retinoic acid increases KLF4 acetylation by inducing HDAC2 phosphorylation and its dissociation from KLF4 in vascular smooth muscle cells. Biochem Biophys Res Commun. 2009;387:13–18. doi: 10.1016/j.bbrc.2009.05.112. [DOI] [PubMed] [Google Scholar]
- 153.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14–3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chang J-K, Ni Y, Han L, et al. Protein kinase D1 (PKD1) phosphorylation on Ser203 by type I p21-activated kinase (PAK) regulates PKD1 localization. J Biol Chem. 2017;292:9523–9539. doi: 10.1074/jbc.M116.771394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guise AJ, Mathias RA, Rowland EA, Yu F, Cristea IM. Probing phosphorylation-dependent protein interactions within functional domains of histone deacetylase 5 (HDAC5) Proteomics. 2014;14:2156–2166. doi: 10.1002/pmic.201400092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Harrison BC, Huynh K, Lundgaard GL, et al. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 2010;584:1103–1110. doi: 10.1016/j.febslet.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 157.Lee C-W, Yang F-C, Chang H-Y, et al. Interaction between salt-inducible kinase 2 and protein phosphatase 2A regulates the activity of calcium/calmodulin-dependent protein kinase I and protein phosphatase methylesterase-1. J Biol Chem. 2014;289:21108–21119. doi: 10.1074/jbc.M113.540229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Taniguchi M, Carreira MB, Smith LN, Zirlin BC, Neve RL, Cowan CW. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron. 2012;73:108–120. doi: 10.1016/j.neuron.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.von Blume J, Knippschild U, Dequiedt F, et al. Phosphorylation at Ser244 by CK1 determines nuclear localization and substrate targeting of PKD2. EMBO J. 2007;26:4619–4633. doi: 10.1038/sj.emboj.7601891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shan P, Fan G, Sun L, et al. SIRT1 functions as a negative regulator of eukaryotic Poly(A)RNA transport. Curr Biol. 2017;27:2271–2284.e5. doi: 10.1016/j.cub.2017.06.040. [DOI] [PubMed] [Google Scholar]
- 161.Ramakrishnan G, Davaakhuu G, Kaplun L, et al. Sirt2 deacetylase Is a novel AKT binding partner critical for AKT activation by insulin. J Biol Chem. 2014;289:6054–6066. doi: 10.1074/jbc.M113.537266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miteva YV, Cristea IM. A proteomic perspective of sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol Cell Proteomics. 2014;13:168–183. doi: 10.1074/mcp.M113.032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bian Y, Song C, Cheng K, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 164.Pighi C, Gu T-L, Dalai I, et al. Phospho-proteomic analysis of mantle cell lymphoma cells suggests a pro-survival role of B-cell receptor signaling. Cell Oncol (Dordr) 2011;34:141–153. doi: 10.1007/s13402-011-0019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu L-R, Zhu Z, Chan KC, et al. Improved titanium dioxide enrichment of phosphopeptides from HeLa cells and high confident phosphopeptide identification by cross-validation of MS/MS and MS/MS/MS spectra. J Proteome Res. 2007;6:4150–4162. doi: 10.1021/pr070152u. [DOI] [PubMed] [Google Scholar]
- 166.Tsai C-F, Wang Y-T, Yen H-Y, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]