Abstract
The recent development of neuroimaging technology and network theory allows us to visualize and characterize the whole-brain functional connectivity in vivo. The importance of conventional structural image atlas widely used in population-based neuroimaging studies has been well verified. Similarly, a “common” brain connectivity map (also called connectome atlas) across individuals can open a new pathway to interpreting disorder-related brain cognition and behaviors. However, the main obstacle of applying the classic image atlas construction approaches to the connectome data is that a regular data structure (such as a grid) in such methods breaks down the intrinsic geometry of the network connectivity derived from the irregular data domain (in the setting of a graph). To tackle this hurdle, we first embed the brain network into a set of graph signals in the Euclidean space via the diffusion mapping technique. Furthermore, we cast the problem of connectome atlas construction into a novel learning-based graph inference model. It can be constructed by iterating the following processes: (1) align all individual brain networks to a common space spanned by the graph spectrum bases of the latent common network, and (2) learn graph Laplacian of the common network that is in consensus with all aligned brain networks. We have evaluated our novel method for connectome atlas construction in comparison with non-learning-based counterparts. Based on experiments using network connectivity data from populations with neurodegenerative and neuropediatric disorders, our approach has demonstrated statistically meaningful improvement over existing methods.
Keywords: Brain network, atlas construction, graph learning
1. Introduction
The human brain is known to contain more than 100 trillion connections over 100 billion neurons [1, 2], making it one of the greatest mysteries in science and the biggest challenges in medicine. Due to this complexity, the underlying causes of many neurological and psychiatric disorders, such as Alzheimer’s disease, Parkinson’s disease, autism, epilepsy, schizophrenia, and depression, are mostly unknown. Recent advances in neuroimaging technology now allow us to visualize a large-scale in vivo map of structural and functional connections in the whole brain at the individual level. The ensemble of macroscopic brain connections can then be described as a complex network - the connectome [3–6].
In the last two decades, many atlas construction algorithms have been proposed for various neuroimaging studies using structural images (e.g., MRI) [7–11]. In general, the construction process of structural atlases consists of two key steps [11]: (1) register all individual images into a common space using deformable image registration (warping) methods [12–16]; and (2) average intensity values across the warped images at each voxel to produce the atlas representing the common anatomical structures for the entire population [9, 17]. The structural image atlas plays a critical role in neuroimaging studies since it provides a common reference space to compare and discover the alteration caused by neurological disorders in the brain [7, 11, 18, 19]. In the same regard, the connectome atlas, a population-wide average of functional brain networks, is of high necessity to reveal and characterize the network differences across clinical cohorts yet has not been explored.
Compared to the structural atlas construction, there exist several different technical challenges of building connectome atlases as follows. First, the data structure of brain networks is irregular. A graph can be a solution since it has been considered as a powerful tool to represent both structured and unstructured data. As a regular data example, a typical image plane (shown as a lattice in Fig. 1(a)) can be easily represented with the graph structure, where each node corresponds to a pixel, and each pixel value is related to the values of its four adjacent spatial neighbors. Note that the spatial neighborhood pattern is regular in the lattice while keeping the same weights for all edges. On the contrary, mounting evidence shows that the human brain network (Fig. 1(b)) holds an irregular “small world” topology [3]. In particular, it is characterized by dense local clustering of connections between neighboring nodes yet a short path length between any (distant) pair of nodes due to the existence of relatively few long-range links [20]. Such irregularity of connection makes network analysis much more complicated than the fixed neighborhood pattern in a 3D regular grid. Second, a graph algebra to fuse the connectome information across brain networks has not been generalized yet. The structural atlas is essentially the voxel-wise average across the aligned individual images defined in the Euclidean space. However, such arithmetic operation is not appropriate to average the connectivity strength at each link since simple link-wise averaging does not fully reflect the geometry of the entire network. For example, let us assume that there are two networks (Networks #1 and #2 in Fig. 1(c)) obtained from a network population. If we look at them in Euclidean space (the square area inside dashed blue line in Fig. 1(c)), it is difficult to ensure the existence of a connection between nodes A and B in the network population. It is caused by the link-wise arithmetic average typically used in Euclidean space since there should be votes both for the absence (Network #1) and presence (Network #2) of connectivity between the two nodes. In contrast, the global network geometry (i.e., Diffusion space) indicates that nodes A and B are strongly connected even for Network # 1 via several pathways along which form a set of short and high probability jumps (red arrows in Fig. 1(c)) on the graph. In this regard, it is more reasonable to represent such population-wise connectivity in the connectome atlas to the extent of global network geometry.
Fig. 1.
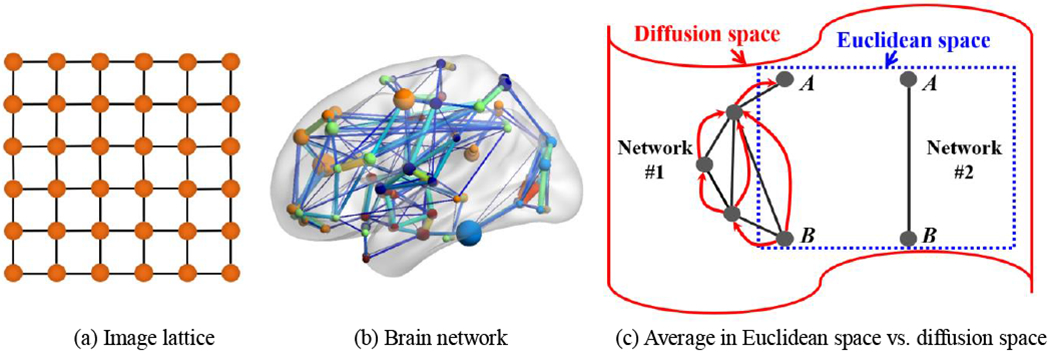
The major challenge for constructing connectome atlas is that brain network (b) has an irregular data structure (often encoded in a graph), which is much more complicated than regular data representations such as an image lattice (a). To address this challenge, we introduce diffusion distance (c) to characterize the network connectivity, which also allows us to embed the brain network to the Euclidian space.
1.1. Related Works
Currently, most of the work on brain connectome atlas focuses on the construction of nodes in the brain network [21–23], instead of discovering the most representative node-to-node connectivities in the population. For example, a connectivity-based parcellation framework is proposed to better characterize functional connectivity by using fine-grained network nodes, thus resulting in a node-wise connectome atlas consisting of 210 cortical and 36 subcortical regions [21], Wig et al. [23] applied snowball sampling on the resting-state functional connectivity data to identify the centers of cortical areas, the subdivision of subcortical nuclei, and the cerebellum in a population. However, region-to-region brain connectivity in the population remains unexplored.
Among very few connectome atlas works that address the embedding of the most representative characteristics of brain connectivities in the population, the most popular approach is to average the individual connectivity values at each link separately by treating the brain network as a data matrix [24]. Albeit simple, such averaging-based solution might break down the network topology since the brain network is a structured data representation with complicated data geometry. It is worth noting that the similarity network fusion technique was used in [25] to propagate the connectivity information by random walk method from one subject to another until all networks become similar [26], Thus, the connectome atlas is the average over the diffused matrices, instead of the original network matrices. However, matrix diffusion suffers from the issue of network degeneration where each node has an equal probability of connecting with all other nodes after applying the excessive amount of matrix diffusions.
1.2. Our Contributions
To address the above challenges, we propose a first-time learning-based graph inference framework to unravel the connectome atlas in the population. To do so, we first introduce diffusion distance to measure the network connectivity, a time-dependent connectivity measurement, reflecting both local and global network geometry at different scales. Next, we employ a diffusion mapping technique [27] to embed the brain network into the Euclidean space, wherein each node of the network is represented as a vector in the Euclidean space, and the Euclidean distance between two vectors corresponds to the diffusion distance between two nodes they represent. After that, we propose a learning-based approach to discover the shared network that is prevalent in the population. Specifically, we regard the diffusion map vectors of each brain network as the graph signals [28] that reside on the latent common network. Thus, the graph-based inference is optimized to learn a graph shift operator such that the topology of the common network has the largest consensus with all aligned graph signals. Since the diffusion distance can integrate local connectivity to the global geometry of the whole network, our learning-based framework offers a novel solution for constructing multi-scale connectome atlas that can “think globally, fit locally” [28, 29],
We evaluate the accuracy and robustness of our learning-based connectome atlas construction method on both functional and structural network data in terms of various group comparisons including Autism Spectrum Disorders (ASD), Fronto-Temporal Dementia (FTD), and Alzheimer’s Disease (AD). Compared to the non-learning based counterparts (i.e., a network averaging method [24] and network diffusion method [25]), our method shows more reasonable atlas construction results.
2. Backgrounds
2.1. Definition of Graph and Graph Laplacian
Suppose each brain network consists of N nodes V ={vi|i = 1,…, N}. Affinity matrix W =[wij]i,j=1,…,N is often used to encode brain network in the form of a matrix, where each element wij measures the strength of connectivity between nodes vi and vj. In the structural network, wij is related to the number of fibers traveling between two regions [4], On the other hand, in the functional network, wij measures the synchronization of functional activities in terms of statistical correlation between BOLD (blood oxygen level dependent) signals [30, 31]. For convenience, it is reasonable to assume that each adjacent matrix is symmetric (i.e., wij = wji), non-negative ( wij ≥ 0), and no self-connection (wij =0). To that end, each brain network can be represented by a graph structure The corresponding Laplacian matrix L is also a N × N matrix, i.e., L = D – W where D is the degree matrix that contains the degrees of the vertices along the diagonal with the ith diagonal element equals .
Since the graph Laplacian L is symmetric, it has a complete set of orthonormal eigenvectors, where we use denote the left eigenvector matrix of the Laplacian matrix L Each χc is a column vector corresponding the cth eigenvalue of L. Each χc is associated to the eigenvalue ωc satisfying Lχc = ωcχc. Since we consider connected graphs here, we assume the eigenvalues of graph Laplacian matrix L are ordered as 0 = ω1 < ω2 ≤ ω3 …≤ ωN. We denote the entire graph spectrum by Ω(L) = {ω1, ω2,…ωN}.
2.2. A Graph Fourier Transform and Notion of Frequency
For any signal f ∈ ℝN, the classic Fourier transform is the expansion of a function f in terms of complex exponentials, which are the eigenfunctions of the one-dimensional Laplacian operator −Δ(e−2πiξt)=(2πξ)2e2πiξt. In analogy, the graph Fourier transform of signal f on the vertices of can be defined as the expansion of f in terms of the eigenvectors of the graph Laplacian L [28]: , where is the column vector in the right eigenvector matrix of L. The inverse graph Fourier graph transform is given by .
The eigenvalues {(2πξ)2} in the classic Fourier signal analysis carries a specific meaning of frequency. Specifically, the complex exponential eigenfunction associated with smaller ξ exhibits slower oscillating patterns, while higher ξ is often associated with complex exponential eigenfunction with much more rapid oscillations. In the setting of a graph, each element in χ1 (associated with ω1 = 0) is constant and equals to at each graph vertex. As pointed in [28], the eigenvector χc with smaller eigenvalue ωc (lower graph frequency) varies slowly across graph in term of that χc(i) and χc(j) are more likely having similar values if vertices Vi and Vj are connected in the graph. On the other hand, the eigenvectors associated with larger eigenvalues (higher graph frequency) oscillated more rapidly and more likely to have distinct values on the vertices connected in the graph even with high connectivity degree.
2.3. Diffusion Distance and Diffusion Mapping
Diffusion distance.
Here, we go one step further to normalize each row of adjacency matrix by P = D−1W, resulting in a Markov matrix P. Each element pij of P can be interpreted as transition probability of a single step taken from node i to j in the network. Since P is a symmetric matrix, it has a complete set of orthonormal eigenvectors and associated eigenvalues by P = ΦΛΦT, where Φ = [φc]c=1,…,N is the eigenvector matrix that contains eigenvectors as columns, and Λ = diag(λ1, λ2,… λN) is the diagonal eigenvalue matrix with λ1≥λ2≥⋯≥ λN. It is worth noting that each φc represents an oscillation mode of network. In general, φc corresponding to the smaller eigenvalues shows more high-frequency changes in the network [27]. By taking the power of the Markov matrix, we can increase the number of steps taken. For instance, each element in Pt sums up all paths of length t from node i to node j. As we increase the value of t, we can characterize the connectivity of the brain network in the local to the global manner, as shown in the top row of Fig. 2(c).
Fig. 2.
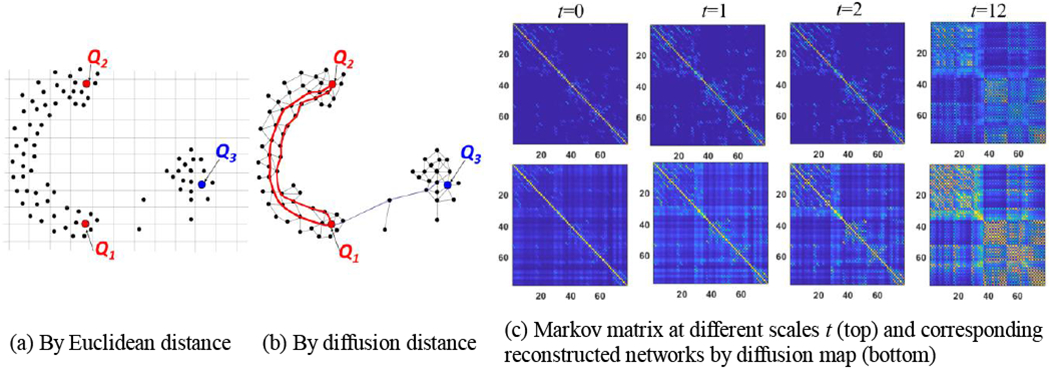
Measuring node-to-node distance using Euclidean distance (a) and diffusion distance (b). (c) The power of Markov matrix (top) and the corresponding reconstructed networks (bottom) using the diffusion distance via diffusion map technique.
Given the Markov matrix P, the diffusion distance dt between two nodes is a time-dependent metric that measures the connectivity strength at the specific scale t [27]:
| (1) |
Unlike Euclidean distance, the diffusion distance becomes short if there are many high probabilistic paths of length t between nodes vi and vj. As illustrated in Fig. 2(a), the Euclidean distance between points Q1 and Q2 is roughly the same as the distance between Q1 and Q3. However, as the diffusion process runs forward, the diffusion distance between Q1 and Q3 is much longer since the pathway between Q1 and Q3 has to go through the bottleneck between two clusters (Fig. 2(b)). Since the diffusion distance encodes the topology of the whole network, it is more robust to noise and spurious connections than the classic Euclidean distance.
Diffusion mapping.
Given , the diffusion map is formed by a set of orthogonal column vectors, where each is a time-dependent basis and form the spectrum bases of the underlying network. Hereafter, we term as diffusion coordinate at scale t. Meanwhile, each row vector in ψt can be regarded as the diffusion embedding vector of the corresponding network node where each element is the projection coefficient of each spectrum basis. Thus, the diffusion distance dt(vi, vj) in Eq. (1) can be approximated by the ℓ2-norm distance in the Euclidean space as:
| (2) |
Note that nodes that are strongly connected in the network should have similar diffusion embedding vectors. Thus, the diffusion map allows us to embed the network data to the Euclidian space. In the bottom row in Fig. 2(c), we display the reconstructed networks using the approximated diffusion distance at scales t = 0, t = 1, t = 2, and t = 12. Compared to the corresponding power of Markov matrix in the top row, diffusion mapping offers an efficient way to characterize network connectivity with the algebraic operation defined in Euclidian space.
2.4. Smooth Graph Signals
Graph signal processing [28] is an emerging research area for analyzing the structured data, where the signal values are defined on the vertex of a weighted graph. Taking the brain network as an example, we assume to have a mean cortical thickness for each node in the network. It is common to arrange the centralized cortical thickness degrees into a data array by following the order of brain parcellation, as shown in Fig. 3(a), where red and green denote for positive (higher than the mean) and negative (lower than the mean) values respectively. Note that the data array alone is unable to characterize the relationship of cortical thickness between two regions. In the setting of a graph, however, it is natural to regard the set of whole-brain cortical thickness as a graph signal since the interplay of cortical thickness between two brain regions can be interpreted in the context of the associated network connectivity.
Fig. 3.
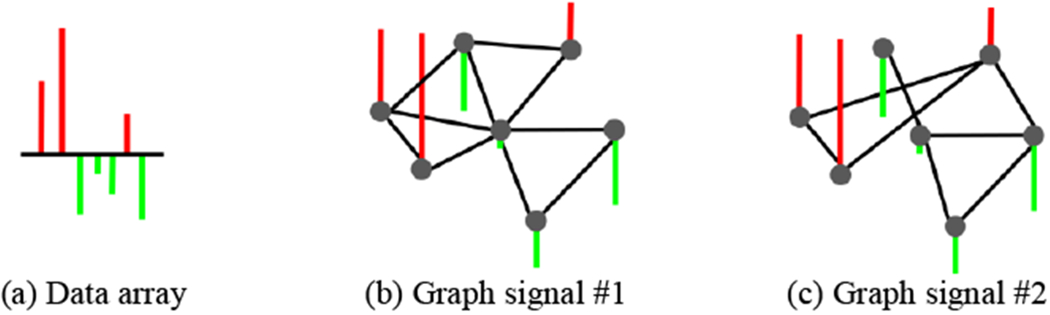
A data array (a) can potentially reside on different graphs and form different graph signals (b)-(c). However, only one leads to the smooth graph signal where the strongly connected nodes have similar values. Hence, graph signal in (b) is more reasonable than graph signal in (c) after inspecting the values for each pair of connected nodes, where the red and green denote for positive and negative degrees.
Without a doubt, we can map the same data array to multiple graphs and potentially lead to different graph signals like the examples of two graph signals shown in Fig. 3(b)–(c). Although these two graph signals are both valid, the graph signal shown in Fig. 3(b) makes more sense since the signal is smooth in terms that two connected nodes have similar scalar values along with the connection defined by the underlying network. Given the network , the smoothness of graph signal f can be measured in terms of a quadratic form of the graph Laplacian:
| (3) |
where f(i) and f(j) are the signal values associated to node vi and vj, respectively. The intuition behind Eq. (2) is that the graph signal f is considered to be smooth if the strongly connected nodes (with a large connectivity strength) generally have similar values. The smaller the quadric term in Eq. (3), the smoother the graph signal f on the network . As we will explain in the next section, we lavage this smooth graph model to find the latent common network from a set of individual brain networks.
2.5. Topological Distance Between Brain Networks
Since brain network is usually encoded in an adjacency matrix, many existing brain network distances are defined based on matrix norm that uses the sum of element-wise difference. For example, the L∞ distance between two network χ1 and χ2 is defined as , where and measure the correlation distance dij = 1 – ρij where ρij is the correlation between two regions. Such distance measurements have limited capability to quantify the topological differences such as connected components and modules in brain networks [32], Gromov-Hausdorff (GH) distance is proposed in [33, 34] to measure the topological difference two brain networks. Specifically, the shape of network is first defined using the graph filtration technique that iteratively build a nested subgraphs of the original network. As shown in the box of Fig. 4, we start the distance threshold ε from the smallest correlation distance and gradually increase ε until ε reaches the largest value of correlation distance. We connect two nodes xi and xj if their distance is less than current ε (i.e., dij < ε). If two nodes are already connected directly or indirectly via other intermediate nodes by smaller ε, we do not connect them. For example, we do not connect x2 and x5 in Fig. 4 at ε = 0.32 since we are already connected through other nodes at ε = 0.3. As ε increases, we can obtain a sequence of nested graphs which is called graph fdtration in algebraic topology [35]. After that, we construct a single linkage matrix S where each element quantifies the single linkage distance between nodes xi and xj in the network. Note, the single linkage distance sij is the minimum ε that makes nodes xi and xj are connected on the dengrogram (shown in the bottom of Fig. 4). For example, the single linkage distance between x1 and x6 in Fig. 4 is 0.3, although there is a direct link between x1 and x6 (the distance is 0.4 on the edge). GH distance between two networks X1 and X2 is then defined through the corresponding single linkage matrix L1 and L2 as . It has been demonstrated in [33] that GH distance, characterizing the network topology, has superior performance over other graph theory based network similarity measures.
Fig. 4.
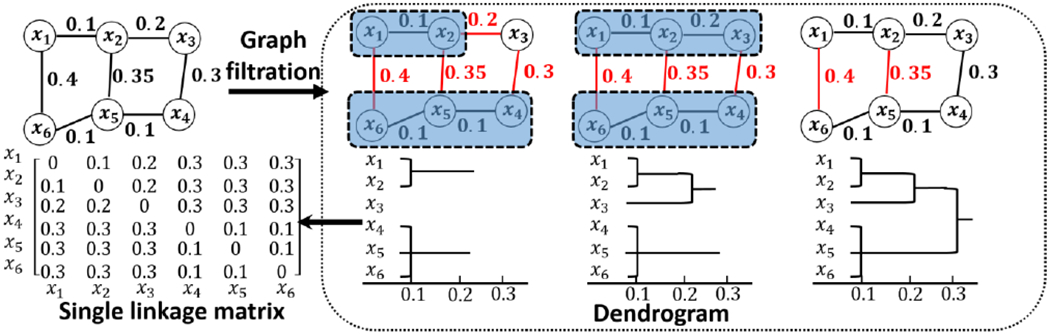
The toy example shows the iterative procedure of graph filtration and calculating the single linkage matrix based on the dendrogram.
3. Methods
Suppose we have a set of brain networks from M individual subjects. Each brain network (m = 1,…, M). The connectome atlas represents the center of all individual brain networks in the population, where the population-wise whole-brain connectivity is encoded in the corresponding adjacency matrix . In the following, we present a novel learning-based approach to find the common network by learning its graph Laplacian.
3.1. Overcome Irregular Data Geometry: from Network to Graph Signals
First, we address the challenge of irregular data structure by embedding each individual brain network into its own native diffusion space. Specifically, we obtain a set of diffusion mappings {Ψt,m} by applying eigen decomposition on the adjacency matrix Wm and constructing the diffusion mapping at each scale t. As shown in Fig. 2I, a larger scale t renders more global connectivity characteristics in the entire network. In order to maintain the details of connectivity profiles in the common network, we propose to learn the common network at a relatively local scale (i.e., t = 1) based on the set of diffusion coordinates . After that, it is straightforward to construct a multi-scale connectome atlas based on the learned adjacency matrix . For simplicity, we omit the superscript t to the variables related to scale t in the following text. Note, there are alternative approaches that can be used here to transform a graph (network) into a collection of signals such as multi-dimensional scaling [36] used in [37]. The reason we use the diffusion mapping technique here is mainly because the diffusion distance is a function of scale t, which allows exploring the multi-scale connectome atlas in a local to global manner.
Fig. 5(a)–(c) demonstrate the procedure for embedding each network data to graph signals. For each individual network , its diffusion coordinates consist of N column eigenvectors, where each element in is associated with the corresponding node in the network. Instead of considering each as a column vector of diffusion coordinate, we further regard each as the graph signal residing on the latent common network (shown in Fig. 5(c)), where the element-to-element relationship in each is interpreted in the context of (shown in Fig. 5(d)). In addition, the graph signal , which is structured data representation, allows us to quantify the representativeness of common network by inspecting the consistency between the diffusion coordinates {Ψm} that derive from individual networks and the common network that supports these diffusion coordinates.
Fig. 5.
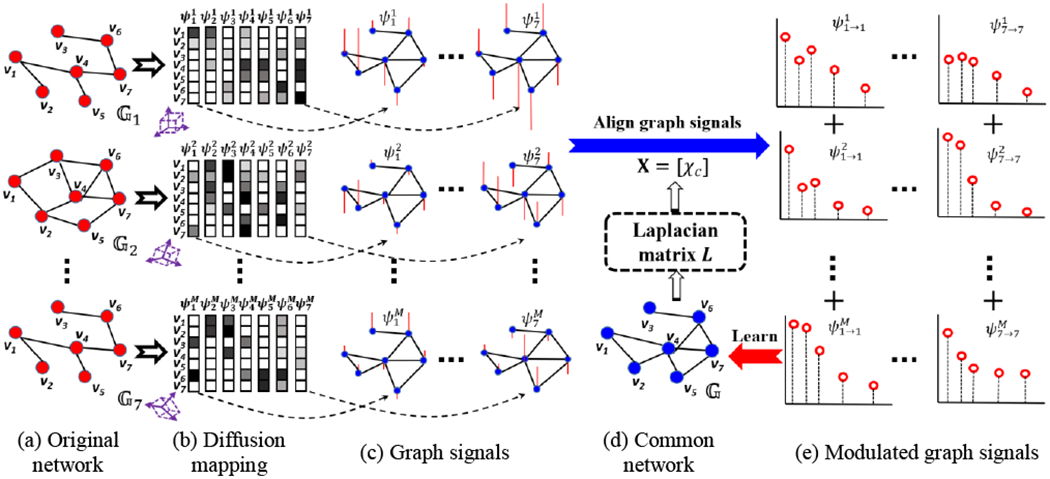
The overview of our learning-based graph inference model for connectome atlas. In this example, we assume each network consists of seven nodes, i.e., N = 7.
3.2. Learn Graph Laplacian of Common Network: from Graph Signals Back to Network
Since the graph signals are defined in Euclidean space, the naïve approach to construct a common network consists of two steps: (1) calculate the mean graph signal yc at each oscillation mode by arithmetic averaging, i.e., , and (2) reconstruct the adjacency matrix where each element (the inverse of diffusion distance in Eq. (2)) for i ≠ j. wii = 0 since we do not allow for self-connection.
However, such averaging-based approach has two limitations. (1) Since the graph signals associated with cth largest eigenvalue might be associated with different eigenvalues across M individual networks, direct averaging over in each oscillation mode might introduce the spurious connectivity information that is not related to the connectome atlas. In analogy to the scenario that one cubic box undergoes various rotations and translations in the 3D Cartesian coordinates (the purple boxes in Fig. 5(b)), the success to unravel the accurate atlas shape (i.e., cubic box) is dispensable to the removal of the external variations (i.e., translations and rotations) which is not related to the intrinsic nature of the underlying shape. To that end, it is of necessity to align each graph signal in the common space, such that the graph signals at the same oscillation mode are associated with the same eigenvalue. (2) Simply averaging individual diffusion maps ignores the interplay between data and the latent common network. As shown in Fig. 3, a data array consisting of unordered scalar values can potentially reside on different graphs and then lead to entirely different graph signals. In this regard, the common network should be consistent to all graph signals at all network frequencies. To tackle this issue, we propose a learning-based approach to learn the Laplacian matrix of common network from a set of graph signals which mainly consists of two alternative steps (shown in Fig. 5(c)–(e)).
Step 1: Modulate the frequency of graph signals to the common graph spectrum space.
We use L denote the Laplacian matrix of latent common network . is the left eigenvector matrix of the Laplacian matrix L, where χc is the column vector corresponding the cth eigenvalue ωc of L Note, we calculate the diffusion mapping by applying eigen decomposition to the adjacency matrix, where larger eigenvalues reflect the low-frequency network oscillation. Here, we apply eigen decomposition to the Laplacian matrix. Hence, χc with smaller eigenvalue ωc characterizes the low-frequency network oscillation.
Recall that each graph signal is associated with the oscillation mode. In each cth mode, and may not have the same eigenvalue (frequency) to each other. Therefore, we first need to align each graph signals at the cth oscillation mode by modulating the frequency to the corresponding eigen basis χc of latent common network by (see Figure 10 in [28]). Suppose the graph signal is associated with eigenvalue , the above modulation operator represents a translation in the graph Fourier domain, i.e., (see Section 2.2 for the graph Fourier transform). Thus, all modulated graph signals are aligned to the same oscillation mode where the common network holds at cth mode, as shown in Fig. 5(e).
Fig. 10.
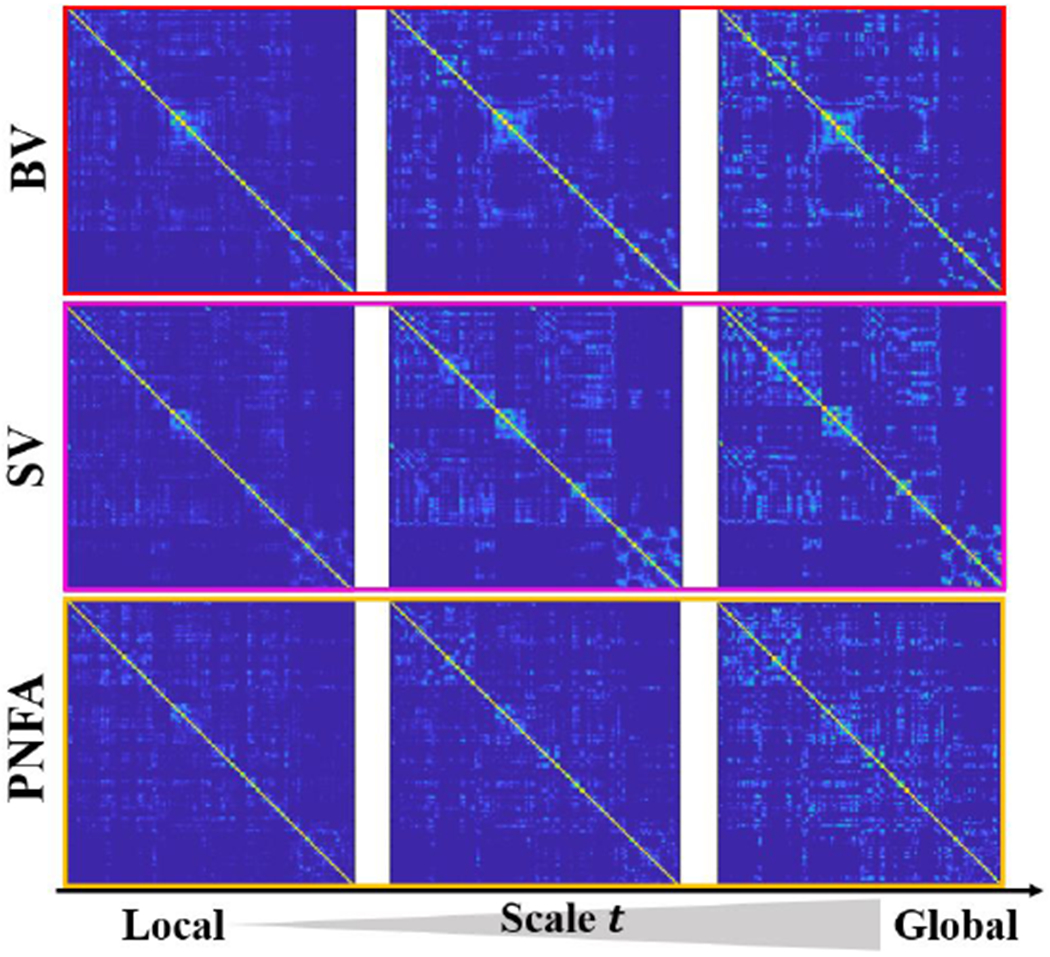
The multi-scale connectome atlas for variants of FTD (top: behavior variant FTD, middle: semantic variant FTD, bottom: progressive nonfluent aphasia).
Step 2: Learn Laplacian matrix of common network.
First, we seek for the representative graph signal yc, which is close to all aligned graph signals at the corresponding cth oscillation mode. Second, we assign the relevance weight to each modulated graph signal where the classic nonlocal constraint is used to penalize the outlier exponentially. Third, we examine the consensus between the common network geometry and the current representative graph signal yc. For this, we minimize the value of graph smoothness in the sense that the ith and jth elements in yc should have similar values, if nodes vi and vj are strongly connected in the common network . The graph smoothness under the condition of can be measured in terms of a quadratic form of the graph Laplacian (Eq. 3). Third, we apply the Frobenius norm and the trace norm constraint (i.e., tr(L) = s) to the matrix for avoiding trivial solutions. Note, s > 0 is a scaling factor, and we set s to the number of nodes in the network, i.e., s = N. By combining these terms, the overall energy function for learning the Laplacian matrix of common network is:
| (4) |
where α, β and ρ are three scalars controlling the strength of the constraints on graph smoothness, the shape of Laplacian matrix L, and the influence of non-local constraint.
3.3. Optimization
Although the energy function in Eq. 4 is non-convex, we opt to optimize the representative graph signals {yc}, weights and the common graph Laplacian L in an alternative manner.
Optimization of representative graph signal.
By discarding unrelated terms w.r.t. yc in Eq. (4), the optimization of each yc is a classic Tikhonov regularization problem:
| (5) |
We have the closed form solution for yc:
| (6) |
The intuition behind Eq. (6) can be interpreted as filtering the weighted average of individual modulated graph signals via the latent graph spectrum encoded in L. In graph signal processing [38], filtering a graph signal μc by a filter h is defined as the operation:
| (7) |
where is the graph Fourier representation of μc with respect to common network oscillation mode is a set of low-pass filters, where the spectrum of each filter is characterized by . In this regard, our learning-based method offers a new window to adaptively smooth the non-local mean of graph signal via the learned smoothing kernel h which is defined in the graph spectrum domain. Since χk with smaller ωk reflects the low-frequency network change, h(L) highlights the contributions of major low-frequency components in the population and attenuates those of high-frequency components, which usually minor patterns of individual network.
Optimization of weighting vector.
Given yc, we can optimize the weighting vector for each oscillation mode by
| (8) |
Graph Laplacian learning.
By fixing {yc} and , we can optimize L by minimizing
| (9) |
where r = α/β. Since all elements in the adjacency matrix are non-negative, it is relatively easier to find the valid , instead of L. In [39], it has been proven that the minimization of equals to minimizing where 1 is the all-ones vector. Thus, we energy function of optimizing becomes:
| (10) |
where is a NN matrix with each element zij = ‖yi – yj‖2. ‖W‖1,1 is the elementwise L1-norm of , i.e., . Note, for γ = 0 we have a very sparse solution that only assigns weight s to the connection corresponding to the smallest pairwise distance in Z and zeros everywhere else. On the other hand, we intend to penalize large degrees in the node degree vector as setting γ to big values, and in the limit γ →∞, we obtain a dense adjacency matrix with a constant degree.
We use primal-dual techniques [40] that scale to solve in Eq. 10. In order to make the optimization easier, we use the vector forms and η = vec(Z) to represent matrix and Z. Following the principle in [40], we convert Eq. 10 into the following general objective function as:
| (11) |
where h1 and h2 are the functions that we can efficiently compute proximal operators, and h3 is differentiable with a gradient that has Lipschitz constant ζ ∈ (0, ∞). K is a linear operator that satisfies (recall θ is the vectorized form of ). As shown in Eq. 12, we first group the non-negative constraint and the first term in Eq. 10 into h1(θ). Note, 𝕀{·} is the indicator function that becomes zero when the condition in the brackets is satisfied and infinite otherwise. Second, we incorporate the constraint of in Eq. 10 into h2 by letting K = 21T so that the dual variable is . Lastly, we group the second and the third term in Eq. 10 into f since they are dilferentiable.
| (12) |
Using these functions, we provide the solution for θ by primal-dual algorithm [40] in the Appendix.
Summary of connectome atlas construction by graph Laplacian learning.
The input of our learning-based connectome atlas construction method is M individual brain networks with corresponding adjacency matrices {Wm}. The output is the common network encoded in adjacency matrix which describes the most representative network structure in the population. The learning procedure is summarized below:
Input: M adjacency matrices {Wm|m=1,…,M}, three parameters α, β. and ρ in graph learning, and a pre-defined total iteration number Δ.
-
0.
Set δ = 1;
-
1.
For each Wm, calculate the diffusion coordinates Ψm in the local scale t = 1;
-
2.
Initialize our graph Laplacian learning algorithm with the arithmetic mean network ;
-
3.
Given , calculate the spectrum basis function of common network X by applying singular value decomposition (SVD) on the Laplacian matrix L;
-
4.
Modulate each native diffusion coordinates Ψm into the graph signal in the common space;
-
5.
Update the weighting vector πc for each oscillation mode by Eq. 8;
-
6.
Update each representative graph signal yc by Eq. 6;
-
7.
Update the adjacency matrix of the common network by solving θ the objective function in Eq. 12 (solution in the Appendix);
-
8.
δ = δ + 1 if δ < Δ, go to step 3; otherwise, stop.
Output: The common network at a local scale. It is straight forward to construct multi-scale connectome atlas by increasing the scale using the diffusion mapping technique (detailed in Section 2.3).
4. Experiments
Description of dataset.
There are four datasets used in the following experiments in total. (1) ASD dataset. We use the resting-state fMRI (rs-fMRI) data of 45 ASD patients and 47 healthy controls (HC) from the NYU Langone Medical Center of Autism Brain Imaging Data Exchange (ABIDE) database (http://preprocessed-connectomes-proiect.org/abide/). (2) FTD dataset. We processed the rs-fMRI data of 90 FTD subjects and 101 age-matched HC subjects from the recent NIFD database, which is managed by the frontotemporal lobar degeneration neuroimaging initiative (https://cind.ucsf.edu/research/grants/frontotemporal-lobar-degeneration-neuroimaging-initiative-0). Furthermore, there are three major clinic subtypes in the FTD cohort: behavioral variant of FTD (BV), semantic variant of FTD (SV), and progressive nonfluent aphasia (PNFA). For the above functional neuroimaging data, we follow the partitions of AAF template [41] and construct the function network for each subject with 90 nodes 1. The region-to-region connection is measured by Pearson’s correlation coefficient between the mean time courses. (3) ADNI dataset. In total, 51 subjects (31 HC and 20 AD) are selected from the ADNI database (http://adni.loniusc.edu/). For each scan, we parcellate the cortical surface into 148 regions [42] using FreeSurfer on T1-weighted MRI scans. Then we apply the probabilistic fiber tractography on DWI (difiusion weighted imaging) and T1-weighted image using FSF software library to obtain a 148×148 connectivity matrices of the structural networks, where each element reflects the number of fibers traveling between two brain regions. (4) HCP dataset. Resting stage fMRI data from 90 healthy young adults from Human Connectome Project (HCP) are used in the replicability test in Section 4.2. MRI scanning was done using a customized 3T Siemens Connectome Skyra with a standard 32-channel Siemens receive head coil and a body transmission coil. Resting state fMRI data were collected using a gradient-echo echo-planner imaging (EPI) with 2.0mm isotropic resolution (FOV=208×180mm, matrix=104×90, 72 slices, TR=720ms, TE=33.1ms, FA=52°, multi-band factor=8, 1200 frames, ~15min/run). Runs with left-right and right-left phase encoding were done to correct for EPI distortions. Here, we consider the left-right and right-left runs as test/retest data. For each fMRI data, we first partition whole brain into 268 regions using the spatially aligned T1 image. Then, the function connectivity network is constructed based on the mean BOLD signals of each parcellated region. The demographic data of these three datasets is summarized in Table 1.
Table 1.
The demographic information of the neuroimaging data from ABIDE, NIFD, and ADNI databases.
| Data | Subj. # | Gender | Age | Modality | Diagnosis label |
|---|---|---|---|---|---|
| ABIDE | 92 | F:18/M:74 | 11.0±2.2 | rs-fMRI | HC(45), ASD(47) |
| NIFD | 191 | F:91/M:100 | 64.1±7.8 | rs-fMRI | HC(101), BV(31), SV(23),PNFA(36) |
| ADNI | 51 | F:20/M:31 | 73.3±6.8 | DWI | HC(31), AD(20) |
| HCP | 180 | F:46/M:44 | 31.2±7.8 | rs-fMRI | Normal |
Experiment setup.
As shown in Eq. 4, α, β, and ρ are three critical parameters required in the optimization. Due to the lack of ground truth, we determine the optimal set of parameters based on the simulated network data in Section 4.1. Then, we evaluate the replicability of connectome atlas on the ASD dataset in Section 4.2. The quality of connectome atlas is inspected visually and quantitatively in Section 4.3 for ASD, FTD, and AD population separately. Finally, we demonstrate the augmented statistical power of network analyses using our learning-based connectome atlas construction method. For comparison, the counterpart methods include (1) matrix averaging on original networks (called “matrix averaging”), and (2) graph-based averaging with matrix diffusion [25] (called “graph diffusion”). Note that none of the counterpart methods is learning-based.
4.1. Validation on Simulated Network Data
In this experiment, we first generate a set of networks from a predefine node distribution where the node locations and affiliations (clusters membership) are fixed. However, we randomly mislabel the node affiliations in each network instance. Since the mislabeling process is performed in a random manner, the average of these individual networks should respect the original node distribution, which becomes the criterion of measuring the accuracy of connectome atlas construction methods. As shown in Fig. 6, a set of simulated networks were generated from the predefined node distribution (Fig. 6(a)), with three separable clusters (displayed in red, blue, and purple). At each iteration, we selectively swap several data points (randomly, near the cluster boundaries) to another cluster. For the data points with correct cluster labels, we construct the connection based on the distance between two points. However, we specifically leave some points not sampled, resulting in the isolated nodes in the network. For the data points with the incorrect cluster labels, we only allow very few connections to the points with the same label. We show three typical simulated networks in Fig. 6(b).
Fig. 6.
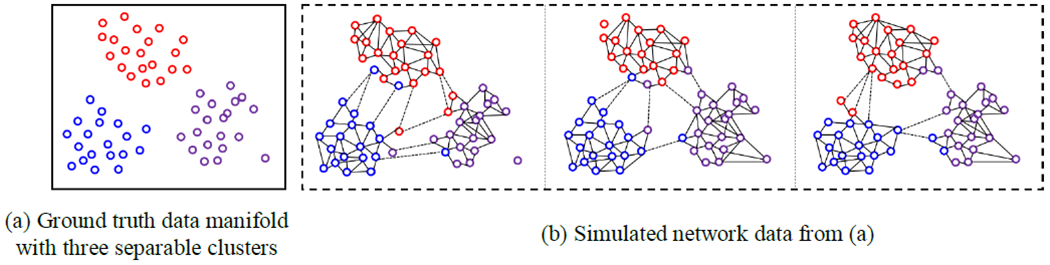
Simulated network data (b), which is generated based on the ground truth data manifold with three separable clusters (a).
We simulate networks with different proportions of node swapping percentage, each with 50 simulations. Given the swapping percentages, we apply our learning-based connectome atlas construction method to the simulated networks with different sets of parameters. Specifically, we apply a grid search strategy for α, β, and ρ separately, where the search range is set to [0.005,2.0] for each parameter, and the search step is set to 0.005. For each setting, we apply the classic k-mean clustering method to determine three clusters based on the common network. The criterion of parameter selection is the dice ratio between the ground truth and the clustering result based on common network . In this way, we fix using α = 0.5, β = 1.0, and ρ = 0.5 in the following experiments.
Furthermore, we apply matrix averaging (no parameter needed) and graph diffusion (using the recommended parameters in [25]) methods on the same simulated network with respect to different swap percentages. We show the dice ratio between the ground truth (Fig. 6(a)) and the clustering result based on the common networks in Table 2. Since the clustering result closely depends on the node-to-node connectivity encoded in the common network, it is reasonable to consider that our graph learning-based connectome atlas construction method achieves higher accuracy than the counterpart non-leaming-based methods.
Table 2.
The overlap ratio between the ground truth and the clustering result based on common network estimated using matrix averaging, graph diffusion, and our graph learning approach.
| Swap percentage | 2.5% | 5.0% | 7.5% | 10.0% | 12.5% | 15.0% |
|---|---|---|---|---|---|---|
| Matrix averaging | 0.91 | 0.87 | 0.85 | 0.81 | 0.79 | 0.76 |
| Graph diffusion | 0.94 | 0.92 | 0.89 | 0.86 | 0.84 | 0.81 |
| Our method | 0.95 | 0.93 | 0.90 | 0.89 | 0.87 | 0.83 |
4.2. Evaluation of Functional Connectome Atlas
4.2.1. Evaluate the Replicability of Functional Connectome Atlas
In this experiment, we use the ASD dataset to evaluate the replicability of connectome atlas. For each connectome atlas construction method, we apply the following resample procedure to generate simulated test/retest cases for 1,000 times: (1) randomly sample 70 brain networks from the entire 92 networks; (2) continue to randomly sample ten brain networks from the remaining networks; (3) randomly divide the ten networks into two equal groups (each have five brain networks), (4) assemble the test and re-test dataset by combining the networks selected in step (1) and step (3), respectively; and (5) run the underlying atlas construction method on two datasets independently. Since two paired cohorts only have 6.7% differences in terms of subjects, we can evaluate the replicability of the connectome atlas method by examining whether the resulting connectome atlases show significant difference via paired two-sample t-test at each link (p < 0.01). Fig. 7 visualizes the connectivities that present a statistically significant difference between two largely overlapped datasets by three different connectome atlas construction methods, where the thickness and color of an edge indicate the magnitude of difference after paired t-test. To display the locations of connectivities showing the statistical difference in the whole brain, we map the indices (yellow dots) of connectives showing significant difference under the resampling test. Since our learning-based method leverages global information such as network geometry (encoded in the Laplacian matrix L) during atlas construction, our method shows better replicability than the other two methods.
Fig. 7.
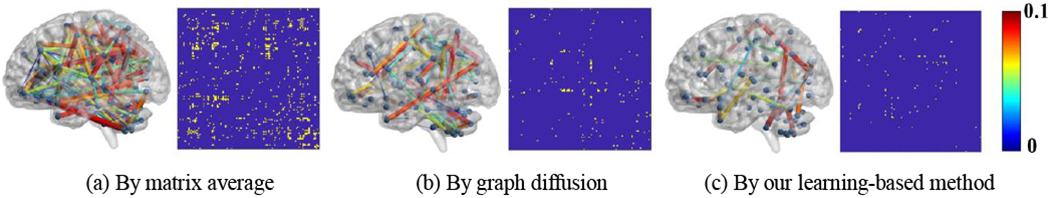
The replicability test by matrix averaging (a), graph diffusion (b), and our graph learning-based method (c). The thickness of each link indicates the magnitude of the difference between two paired network data cohorts. The links showing the significant difference after resampling test are also indicated in yellow in the matrix.
Furthermore, we evaluate the replicability on the test/retest resting-stage fMRI data from HCP dataset. Specifically, we first apply matrix average, graph diffusion, and our learning-based method to 90 left-right runs (considered as testing data) and 90 right-left runs (considered as retesting data) separately. Since the left-right and right-left run scans are performed to same subject one after another, the two connectome atlases are expected to be very similar in test/retest data. Fig. 8 show the connectome atlases of left-right run (top) and right-left run (middle) by matrix average (a), graph diffusion (b), and our method (c), respectively. Next, we repeat this test/retest experiment on a subset (randomly pick 60 subjects each time) for 1,000 times. Then we perform two-sample t-test at each link to examine whether the connectivity degree exhibits significant difference between the connectome atlases constructed using left-right and right-left fMRI data. The red dots in the third row of Fig. 8 indicate the links showing significant difference (p < 0.01) in 1,000 test/retest experiments. Furthermore, we calculate the ICC (intra-class correlation) coefficient to measure the reliability of common connectome atlases, where higher ICC indicates better reliability [43], A one-way ANOVA is applied to measure to the measures of the two common connectome atlases across resampling tests, to calculate the between-test mean square of total connectivities and within-test mean square error of total connectivities. The ICC coefficients are 0.53 by matrix average, 0.56 by graph diffusion, and 0.62 by our learning-based method.
Fig. 8.
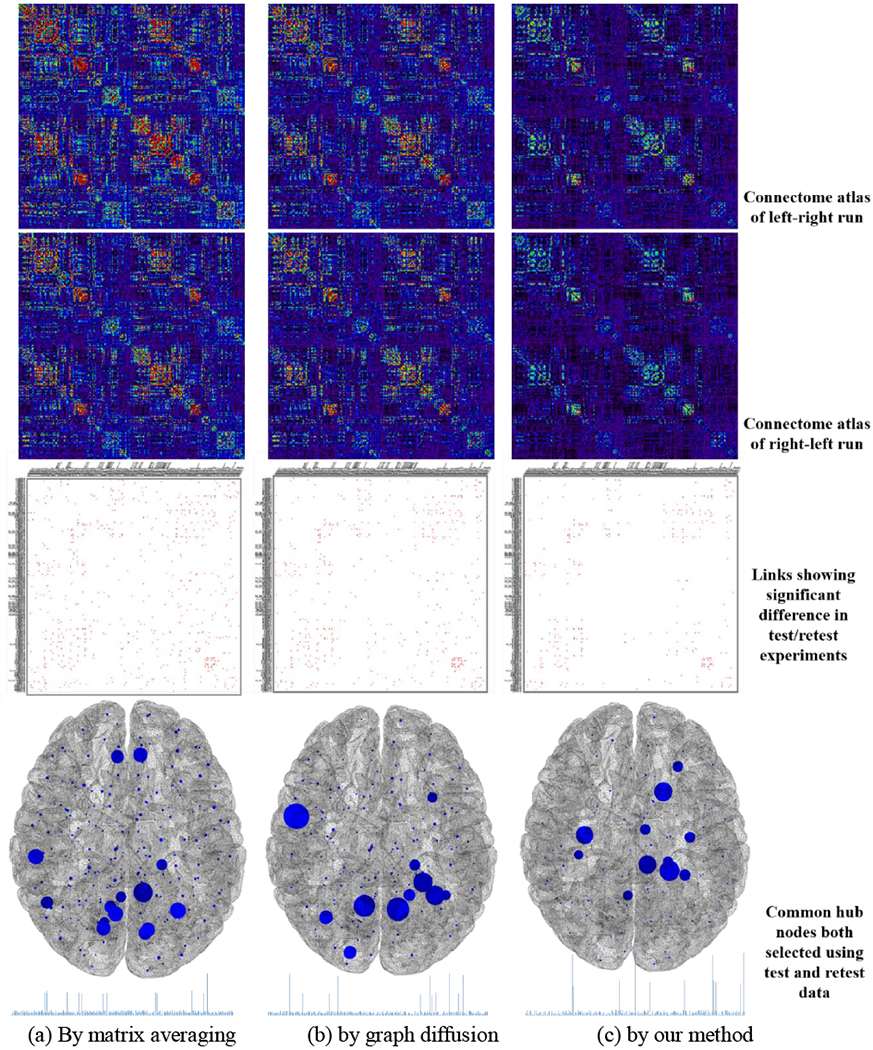
The replicability test on HCP dataset by matrix averaging (a), graph diffusion (b), and our graph learning-based method (c). The first and second row show the connectome atlas constructed using all left-right run and right-left run rs-fMRI data, respectively. The third row show the links (red dots) showing significant differences in 1,000 test/retest experiments. The last row shows the common hub nodes that are selected in each test/retest experiment, where larger node size reflect higher frequency of being both selected.
In each test/retest experiment, we select 8 hub nodes from the connectome atlas of left-right run and the connectome of right-left run separately and then examine the unanimous of hub identification results in test/retest experiment. The frequency of common hub nodes being selected in both testing and retesting data is reflected by the node size in the fourth row of Fig. 8. Based on the frequency histogram of common hubs (in the bottom of Fig. 8), we can calculate the entropy degree, which are 5.25 by matrix average, 5.03 by graph diffusion, and 4.72 by our learning-based method. Note, smaller entropy value indicates better replicability of hub identification results which are obtained from the more consistent connectome atlases in test/retest experiments.
4.2.2. Evaluate the Quality of Functional Connectome Atlas
Visual inspection.
We apply matrix averaging, graph diffusion, and our learning-based connectome atlas construction method to overall 92 functional networks from the ABIDE database and 191 functional networks from the NIFD database. The connectome atlases by matrix averaging, diffusive shrinking graphs, and our learning-based method are shown in Fig. 9(a)–(c), respectively. Through visual inspection, the connectome atlases built by our learning-based method show a more distinct modular structure than the counterpart methods. Based on the age information shown in Table 1, the connectome atlas built upon ABIDE and NIFD dataset should reflect the distinct representative connectivity profdes of young and aging populations, where the age-related alterations have been reported in many neuroimaging studies [44, 45], However, fewer changes between the young brains and aging brains have been observed in the connectome atlases constructed by matrix averaging and graph diffusion methods.
Fig. 9.
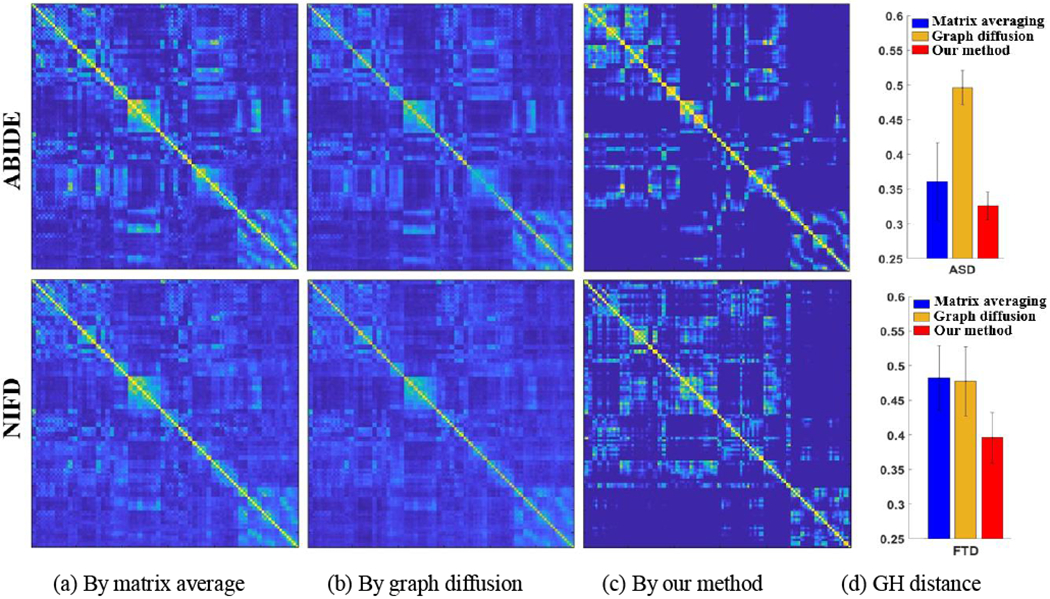
The connectome atlas built on ABIDE and NIFD datasets using matrix averaging (a), graph diffusion (b), and our graph learning method (c). The quantitative results of GH distance between the connectome atlas and each individual brain network are shown in (d).
Quantitative evaluation.
Furthermore, we calculate the network distance between each network to the connectome atlas using the Gromov-Hausdorff (GH) distance [46], which shows superiority over other network distance metrics. As shown in Fig. 9(d), the overall network distance with respect to the connectome atlas by our learning-based method is much smaller than the other two counterpart methods, indicating that our connectome atlas is more reasonable in terms of the closeness to the population center.
Multi-scale connectome atlas.
The diffusion distance used in our learning-based method allows us to characterize the connectome atlas in a multi-scale manner. In Fig. 10, we display the multi-scale connectome atlases for 31 behavior variant FTD, 23 semantic variant FTD, and 36 progressive nonfluent aphasia subjects, respectively, where the scale t ranges from 0.5 to 10. It is evident that high-level information of network organization such as modules has been captured at the global scale. Since FTD syndromes comprise a heterogeneous group of neurodegeneration conditions, characterized by atrophy in the frontal and temporal lobes, different variants of FTD exhibit more and more distinct morphological patterns of network organization as the scale t increases.
4.3. Evaluation of Structural Connectome Atlas
4.3.1. Evaluate the Replicability of Structural Connectome Atlas
For each subject selected from the ADNI database, we obtain the structural network based on not only the baseline DTI scan (shown in Table 1) but also the follow-up DTI (within 6–10 months range). Since the time interval between two scans is less than one year, and there is no change of diagnostic label, it is reasonable to assume that there should be no statistical significance between the structural connectome atlases constructed at baseline and follow-up time point. We apply random sampling strategy to withdraw 40 pair of networks from in total 51 networks pairs. At each time, we deploy matrix averaging, graph diffusion, and our learning-based method to construct the baseline connectome atlas and follow-up connectome atlas, separately. After repeating this procedure for the sufficient number of times, we run the significant test on the network difference between baseline and follow-up, at each link, to examine whether we can observe statistically significant difference across the random sampling tests. In the bottom of Fig. 11, we display the links showing a significant difference (less replicable) by matrix averaging (a), graph diSusion (b), and our method (c), where the larger thickness of the link indicates the higher variation of connectivity strength across the resampling tests. It is observed that the connectome atlas constructed by our learning-based method shows better replicability in representing the connectivity profiles for the population.
Fig. 11.
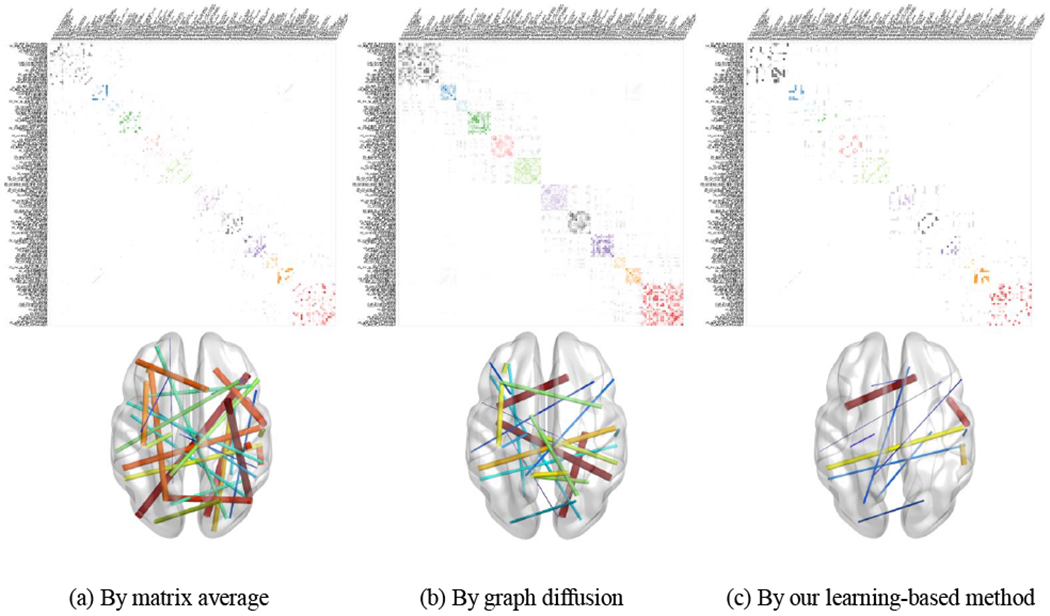
Top: The structural connectome atlas constructed by matrix averaging (a), graph diffusion (b), and our learning-based method (c). Bottom: The replicability results after resampling tests where the link designates the statistically significant difference, and the link thickness reflects the variation of connectome strength across resampling tests.
4.3.2. Evaluate the Classification Accuracy using Structural Connectome Atlas
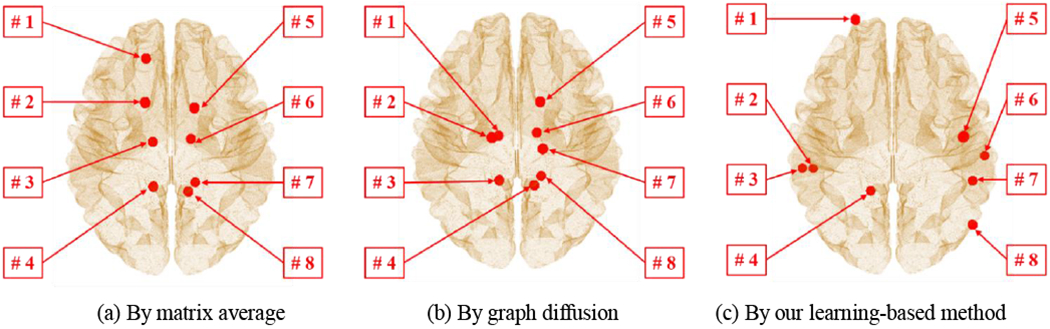
The connectome atlases constructed by matrix averaging, graph diSusion and our method are shown in the top of Fig. 11. To further demonstrate the application of connectome atlas, we first identify a set of hub nodes from the connectome atlas using a module-based methods that identify hub nodes based on the a priori definitions of network modularity [47, 48], Follow the distribution of connectivity strength as suggested in [48], we set the number of hub nodes to eight. The identified hub nodes from three connectome atlases by different construction approaches are displayed in Fig. 12 and the anatomical information is shown in Table 3.
Fig. 12.
The identified hub nodes based on the structural connectome atlas constructed by matrix averaging (a), graph diSusion (b), and our learning-based method (c).
Table 3.
The anatomical information of identified hub nodes from three connectome atlases. The ‘*’ indicates statistical significance between NC and AD cohort at the corresponding hub node.
| Node | Hubs by matrix averaging | Hubs by graph diffusion | Hubs by our method |
|---|---|---|---|
| #1 | G_and_S_cingul_ant (L) | S_pericallosal (L) * | G_and_S_transv_frontopol (L) |
| #2 | G_and_S_cingul_mid_ant (L) | G_and_S_cingul_mid_post (L) | Stemporalinf (L) * |
| #3 | S_pericallosal (L) * | G_cingul_post_dorsal (L) * | Gtemporalmiddle (L) |
| #4 | G_cingul_post_dorsal (L) * | G_cingul_post_dorsal (R) | G_cingul_post_ventral (L) * |
| #5 | G_and_S_cingul_mid_ant (R) | G_and_S_cingul_mid_ant (R) | G_presentral (R) * |
| #6 | S_pericallosal (R) * | S_pericallosal (R) * | G temporal middle (R) * |
| #1 | G_cingul_post_ventral (R) | G_and_S_cingul_mid_post (R) | Stemporalsup (R) * |
| #8 | G_cingul_post_ventral (R) * | G_cingul_post_ventral (R) * | G_pariet_inf_angular (R) * |
Although the connectome atlases shown in Fig. 11 are similar, the locations of the identified hub nodes vary across atlas construction methods. Since the identified hub nodes from the connectome atlas provides a common setting of hubs across individual networks, we extract nodal features at the identified hub nodes such as local clustering coefficient [49] for each individual network. Then we apply two-sample t-test at each hub nodes and examine the statistical network difference between NC and AD cohorts, where the significant differences are indicated by ‘*’ in Table 3. The connectome atlas by our method has identified six hub nodes showing distinct difference of network profile while the counterpart atlases by matrix averaging and graph diffusion method only found four and three hub nodes with significant network difference between NC and AD cohorts. Since the hub identification result is highly dispensable to the network data, our connectome atlas construction method exhibits enhanced statistical power over the other two methods.
5. Discussions and Conclusions
Discussions.
As shown in Fig. 2 and Fig. 10, our connectome atlas construction offers the way to construct multi-scale atlas by adjusting the scales in diflusion mappings. It is worth noting that our proposed method constructs the connectome atlas at a given scale t separately. Although it is always favorable to jointly model the dependency of connectome atlas across scales and simultaneously construct the multi-scale connectome atlas, it is computationally prohibitive to do so in our current optimization framework as illustrated in Section 3.3. However, there might be several possible solutions to explore the new regime of multi-scale connectome atlas. (1) Unify multi-scale connectome atlases by fusing the connectivity information from local to global manner. First, we generate a set of connectome atlas with different setting of scale t. Then, we can apply the matrix fusion method in [50] to combine into single network which includes both local and global connectivity information. (2) Use supervised/unsupervised feature selection method to find the multi-scale connectome feature representation. Since most of the network-based studies is interested in finding network feature representations which can separate two or multiple cohorts, we can extend the current approach into the scenario of multi-scale by using feature selection technique such as sparse feature selection [51, 52] to determine the best feature representations from multi-scale connectome atlases.
Conclusions.
The construction of connectome atlas is much more challenging than image atlas, due to the irregular graph structure. To address this problem, we proposed a learning-based approach to discover the common network in the setting of graphs. Our proposed method offers a new window to investigate the population-wise whole-brain connectivity profiles in the human brain. Promising results have been obtained in various brain network studies such as ASD, FTD, and AD, which indicates the applicability of our proposed learning-based connectome atlas construction methods in neuroimaging applications.
Acknowledgments
This work was supported in part by NIH R21AG059065 and K01AG049089.
Appendix
To solve Eq. 12, we need to determine the proximity operator for h1 and h2, and the derivative of h3 as follows:
Besides, we introduce a step-size variable τ ∈ (0,1 + ζ + ‖K‖2) to control the convergence, where .The primal dual algorithm for Eq. 12 is summarized below:
Input: η, γ, S, θ0, ϑ0, τ, and tolerance ϵ.
-
0.
for i = 1,…, imax do;
-
1.
oi = θi − τ(2γ(2θi + KT Kθi) + 2ϑi)
-
2.
-
3.
pi = max(0, θi − 2τη)
-
4.
-
5.
qi = pi − τ(2γ(2pi + KTKpi)+ 2pi)
-
6.
-
7.
θi+1 = θi − oi + qi
-
8.
-
9.
if ‖θi+1− θi‖/‖θi‖< ϵ and θϑi+1− ϑi‖/‖ϑi‖< ϵ then
-
10.
Break
-
11.
end if
-
12.
end for
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
It is worth noting that the parcellation of the AAL template is defined based on anatomical structure. Although there are still quite a few fMRI studies using the AAL template, the interest is shifting towards using functional atlases which follow the insight of brain functions. However, we demonstrate the common functional brain networks using both structural and functional parcellations in order to show the proposed computational method can work with different atlases.
Information Sharing Statement
The source code, experimental data, and produced data will be publicly distributed through NITRC (Neuroimaging Informatics Tools and Resources Clearinghouse, https://www.nitrc.org/).
References
- 1.Herculano-Houzel S, The Human Brain in Numbers: A Linearly Scaled-up Primate Brain. Frontiers in Human Neuroscience, 2009. 3(31). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowakowski RS, Stable neuron numbers from cradle to grave. Proceedings of the National Academy of Sciences of the United States of America, 2006. 103(33): p. 12219–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spoms O, Networks of the brain. 2011: MIT Press. [Google Scholar]
- 4.Spoms O, Structure and function of complex brain networks. Dialogues Clin Neurosci., 2013. 15(3): p. 247–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heuvel M.P.v.d. and Spoms O, Rich-club organization of the human connectome. Journal of Neuroscience, 2011. 31(44): p. 15775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan C, et al. , Standardizing the intrinsic brain: towards robust measurement of inter-individual variation in 1000functional connectomes. Neuroimage, 2013. 80(10): p. 246–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blezek DJ and Miller JV, Atlas stratification. Medical Image Analysis, 2007. 11(5): p. 443–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serag A, et al. , Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. Neuroimage, 2012. 59(3): p. 2255–2265. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher PT, Venkatasubramanian S, and Joshi S, The geometric median on Riemannian manifolds with application to robust atlas estimation. Neuroimage, 2009. 45(1, Supplement 1): p. S143–S152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y, Ho J, and Vemuri BC, Image Atlas Construction via Intrinsic Averaging on the Manifold of Images, in IEEE Conference on Computer Vision and Pattern Recognition. 2010: San Francisco, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joshi S, et al. , Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage, 2004. 23(Supplement 1): p. S151–S160. [DOI] [PubMed] [Google Scholar]
- 12.Avants BB, et al. , Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis, 2008. 12(1): p. 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vercauteren T, et al. , Diffeomorphic demons: efficient non-parametric image registration. Neuroimage, 2009. 45(1, Supplement 1): p. S61–S72. [DOI] [PubMed] [Google Scholar]
- 14.Wu G, et al. , S-HAMMER: Hierarchical Attribute-Guided, Symmetric Diffeomorphic Registration for MR Brain Images. Human Brain Mapping, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, Qi F, and Shen D, Learning-based deformable registration of MR brain images. Medical Imaging, IEEE Transactions on, 2006. 25(9): p. 1145–1157. [DOI] [PubMed] [Google Scholar]
- 16.Zitova B and Flusser J, Image registration methods: a survey. Image and Vision Computing, 2003. 21(11): p. 977–1000. [Google Scholar]
- 17.Shi F, et al. , Neonatal Atlas Construction Using Sparse Representation. Human Brain Mapping, 2014. 35(9): p. 4663–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shattuck DW, et al. , Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage, 2008. 39(3): p. 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi F, et al. , Infant Brain Atlases from Neonates to 1- and 2-year-olds. PLoS ONE, 2011. 6(4): p. el8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassett DS and Bullmore E, Small-world brain networks. Neuroscientist, 2006. 12(6): p. 512–23. [DOI] [PubMed] [Google Scholar]
- 21.Fan L, et al. , The Human Brainnetome Atlas: A New Brain Atlas Based on Connectional Architecture. Cerebral Cortex, 2016. 26(8): p. 3508–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. , Groupwise whole-brain parcellation from resting-state fMRl data for network node identification. Neuroimage, 2013. 82(11): p. 403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wig GS, et al. , Parcellating an Individual Subject’s Cortical and Subcortical Brain Structures Using Snowball Sampling of Resting-State Correlations. Cerebral Cortex, 2013. 24(8): p. 2036–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arsigny V, et al. , Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Magnetic Resonance in Medicine, 2006. 56(2): p. 411–21. [DOI] [PubMed] [Google Scholar]
- 25.Rekik I, et al. , Estimation of Brain Network Atlases using Diffusive-Shrinking Graphs: Application to Developing Brains, in International Conference on Information Processing in Medical Imaging. 2017. p. 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, et al. , Similarity network fusion for aggregating data types on a genomic scale. Nature Methods, 2014. 11: p. 333–7. [DOI] [PubMed] [Google Scholar]
- 27.Coifman RR and Lafon S, Diffusion Maps. Applied and Computational Harmonic Analysis, 2006. 21(1): p. 5–30. [Google Scholar]
- 28.Shuman DI, et al. , The emerging field of signal processing on graphs: Extending high-dimensional data analysis to networks and other irregular domains. IEEE Signal Processing Magazine, 2013. 30(3): p. 83–98. [Google Scholar]
- 29.Talmon R, et al. , Diffusion Maps for Signal Processing: A Deeper Look at Manifold-Learning Techniques Based on Kernels and Graphs. IEEE Signal Processing Magazine, 2013. 30(4): p. 75–86. [Google Scholar]
- 30.Hlinka J, et al. , Functional Connectivity in Resting-State fMRl: Is Llinear Correlation Sufficient. Neuroimage, 2011. 54(3): p. 2212–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchison RM, et al. , Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage, 2013. 80(10): p. 360–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung MK, et al. , Topological Distances Between Brain Networks, in International Workshop on Connectomics in Neuroimaging. 2017, LNCS, Springer: Quebec City, Canada. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H, et al. , Computing the shape of brain network using graph filtration and Gromov-Haudorff metric, in MICCAI 2011. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, et al. , Persistent brain network homology from the perspective of dendrogram. IEEE Trans, on Medical Imaging, 2012. 31(12): p. 2267–77. [DOI] [PubMed] [Google Scholar]
- 35.Edelsbrunner H and Harer J, Persistent Homology — a Survey. Contemporary Mathematics, 2008. 453: p. 257–82. [Google Scholar]
- 36.Cox TF and Cox MAA, Multidimensional Scaling. 2001: Chapman and Hall. [Google Scholar]
- 37.Shimada Y, Ikeguchi T, and Shigehara T, From Networks to Time Series. Physical Review Letters, 2012. 109(15): p. 158701. [DOI] [PubMed] [Google Scholar]
- 38.Shuman D, et al. , The Emergence Field of Signal Processing on Graphs. IEEE Signal Processing Magazine, 2013. 30(3): p. 83–90. [Google Scholar]
- 39.Kalofolias V, How to learn a graph from smooth signals, in The 19th International Conference on Artificial Intelligence and Statistics (AISTATS 2016). 2016: Cadiz, Spain. [Google Scholar]
- 40.Komodakis N and Pesquet J-C, Playing with Duality: An overview of recent primal?dual approaches for solving large-scale optimization problems. IEEE Signal Processing Magazine, 2015. 32(6): p. 31–54. [Google Scholar]
- 41.Tzourio-Mazoyer N, et al. , Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNIMRI single-subject brain. Neuroimage, 2002. 15(1): p. 273–89. [DOI] [PubMed] [Google Scholar]
- 42.Destrieux C, et al. , Automatic parcellation of human cortical gvri and sulci using standard anatomical nomenclature. Neuroimage, 2010. 53(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo CC, et al. , One-vear test-retest reliability of intrinsic connectivity network fMRL in older adults. Neuroimage, 2012. 61(4): p. 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu K, et al. , Structural Brain Network Changes across the Adult Lifespan. Frontiers in Aging Neuroscience, 2017. 9(275). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geerligs L, et al. , A Brain-Wide Studv of Age-Related Changes in Functional Connectivitv Cerebral Cortex, 2015. 25: p. 1987–99. [DOI] [PubMed] [Google Scholar]
- 46.Lee H, et al. , Persistent brain network homology from the perspective of dendrogram. IEEE Transactions on Medical Imaging, 2012. 31(12): p. 1387–1402. [DOI] [PubMed] [Google Scholar]
- 47.Guimera R and Nunes Amaral LA, Functional cartography of complex metabolic networks. Nature, 2005. 433(7028): p. 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van den Heuvel MR and Spoms O, Network hubs in the human brain. Trends in cognitive sciences, 2013. 17(12): p. 683–696. [DOI] [PubMed] [Google Scholar]
- 49.Rubinov M and Spoms O, Complex network measures of brain connectivity: uses and interpretations. Neuroimage, 2010. 52(3): p. 1059–69. [DOI] [PubMed] [Google Scholar]
- 50.Wang B, et al. , Similarity network fusion for aggregating data types on a genomic scale. Nature Methods, 2014. 11(1): p. 333–7. [DOI] [PubMed] [Google Scholar]
- 51.Huang H, et al. , A New Sparse Simplex Model for Brain Anatomical and Genetic Network Analysis, in MICCAI 2013: Nagoya, Japan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zu C, et al. , Identifying Disease-related Connectome Biomarkers by Sparse Hvpergraph Learning. Brain Imaging and Behavior, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]


