Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) causes serious infections in both community and hospital settings, with high mortality rates. Treatment of MRSA infections is challenging because of the rapidly evolving resistance mechanisms combined with the protective biofilms of S. aureus. Together, these characteristic resistance mechanisms continue to render conventional treatment modalities ineffective. The use of nanoformulations with unique modes of transport across bacterial membranes could be a useful strategy for disease-specific delivery. In this review, we summarize treatment approaches for MRSA, including novel techniques in nanoparticulate designing for better therapeutic outcomes; and facilitate an understanding that nanoparticulate delivery systems could be a robust approach in the successful treatment of MRSA.
Introduction
Staphylococcus aureus is a Gram-positive bacterium responsible for many complex infections, including skin and soft tissue, pneumonia, bone and joint infections, osteomyelitis, and infective endocarditis [1]. A high mortality rate resulting from S. aureus was observed when antibiotic treatments were not available [2]. Penicillin came into use as a treatment for these infections, but resistance developed to this antibiotic within 2 years. Soon, ~80% of S. aureus were penicillin resistant [3]. Following this development, methicillin was introduced, with stability against degradation by the penicillinase enzyme. However, methicillin resistance began as soon as it was used to treat the infections [4] and became a major clinically relevant concern from 1960 onwards [5]. MRSA infection is acquired predominately in two settings: secondary to exposure in a healthcare facility, such as hospitals, nursing homes, or dialysis centers [termed health care-associated MRSA (HA-MRSA)]; or exposure acquired in otherwise healthy individuals in the community [termed community-associated MRSA (CA-MRSA)]. In 2017, the Center for Disease Control (CDC) recorded 323 700 cases and 10 600 deaths as ‘incident hospitalized positive clinical cultures, including hospital- & community-onset MRSA infections’ [6]. Chamber and DeLeo illustrated the trends of this resistance, which was first identified during the 1940s and is ongoing [3] (Fig. 1). The most recent wave of vancomycin resistance was first identified during the 2000s and remains prevalent. Although treatment of MRSA remains challenging, the much needed progress in MRSA infection management has slowed significantly [7].
FIGURE 1.
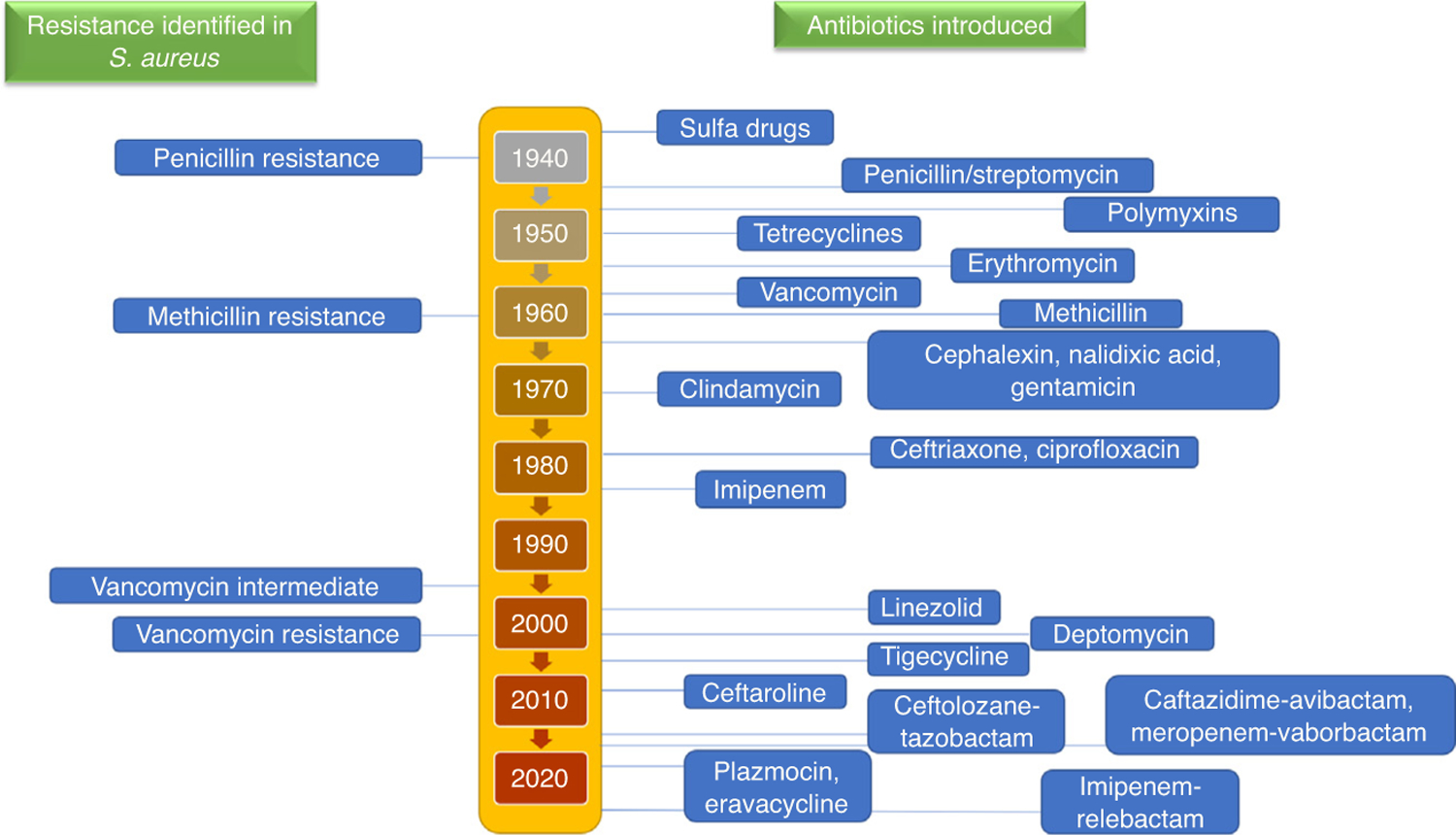
Timeline of identification of resistance in Staphylococcus aureus. Resistance to treatment in methicillin-resistant S. aureus (MRSA) occurs in waves with the introduction of novel antibiotics.
Challenges in the treatment of MRSA
Resistance mechanisms
Resistance to antimicrobial agents is a major reason for the failure of MRSA treatment. There are various mechanisms that contribute to antimicrobial resistance. These can be inherent to MRSA or acquired with the time and length of treatment. Each antibiotic combats MRSA by different mechanisms of action characteristic of the antibiotic class. Similarly, the subsequent resistance mechanisms against each of these drug classes in MRSA can also manifest by different mechanisms (Fig. 2). Table 1 lists selected types of resistance mechanism for various antimicrobial agents for MRSA [8,9].
FIGURE 2.
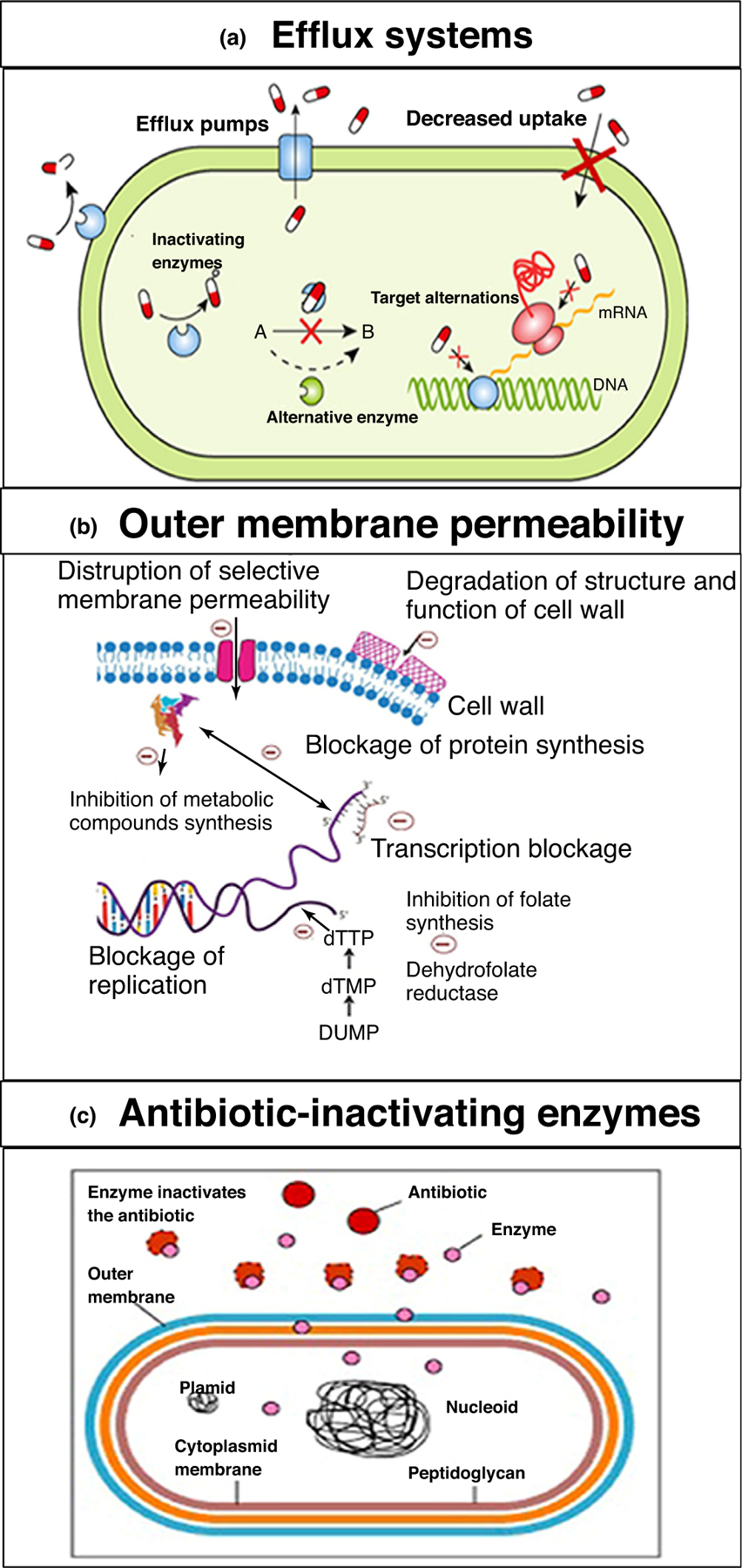
Examples of important endogenous resistance mechanisms in methicillin-resistant Staphylococcus aureus (MRSA). Reproduced with permission, from [9]. (a) Active efflux mechanisms; (b) decreases in the permeability of the outer membrane of MRSA to drugs; (c) and enzymatic degradation of drugs by MRSA. All three mechanisms are responsible for the development of antibiotic drug resistance in MRSA.
TABLE 1.
Examples of antimicrobial resistance mechanisms in MRSA
| Resistance mechanism | Antimicrobial agents |
|---|---|
| Limiting drug uptake | Glycopeptides |
| Modification of drug target | β-lactams; glycopeptides; lipopeptides; aminoglycosides; tetracyclines; macrolides; lincosamides; oxazolidinones; streptogramins; fluoroquinolones; metabolic pathway inhibitors |
| Inactivation of drug | β-lactams; chloramphenicol |
| Active drug efflux | Tetracyclines |
Antibiotic resistance because of biofilms
Biofilms are defined as ‘an organized population of bacteria encapsulated in self-produced extracellular polymeric dense matrix that adheres to biotic and abiotic surfaces’ [10–12], are responsible for protecting the bacteria from the surrounding environment, and also aid antibiotic resistance. Hence, biofilm formation is a mechanism of survival for the colony. These mechanisms arise from environmental factors, such as opsonization, phagocytosis, and antimicrobial agents [13]. Biofilms become a chronic and recurrent challenge in healthcare-associated infections, especially in infections originating from medical devices, such as catheters, heart valves, pacemakers, contact lens, and shunts [14–17]. Such infections are often chronic and recurrent. One of the most prevalent biofilms develops from variants of S. aureus, including MRSA. The lifecycle of a biofilm in infections is detailed in Fig. 3 [12]. Bacterial cultures in intact biofilms are up to a thousand-fold more resistant to antibiotics than those cultures growing as planktonic (free floating) forms. Biofilms contribute to the development of antibiotic resistance primarily by two mechanisms [18,19]: (i) by limiting the penetration of drugs through the highly protective biofilm and the associated matrix to reach the bacterium; and (ii) physiological changes in the bacteria in the biofilms, such as a drastic slowdown of the growing phase leading to a state close to dormancy. It is postulated that differences in metabolic activity of the bacterial cells between the different biofilm layers and drug resistance in biofilm-embedded cells contribute to the differences in the extent of resistance to antibiotics. Furthermore, the dense nature of the biofilms and the matrix create a condition of hypoxia in the deeper biofilm layers, which is also considered a reason for the poor performance of antibiotics. Consequently, bacteria can develop a resistant phenotype, altering drug targets for antibiotic activity [20]. Hence, eradication of bacterial colonies in biofilms requires concentrations of antibiotics above their minimum inhibitory concentrations (MIC, which is defined as the lowest concentration in µg/mL of an antibiotic that inhibits the growth of a given strain of bacteria) [21]. Higher doses of antibiotics are a cause of concern owing to the probability of multidrug resistance and dose-associated toxicities. For instance, vancomycin requires a tenfold increase in dose for MRSA biofilms compared with free planktonic states [22], increasing the risk of toxicity. Daptomycin has been found to be effective to some extent in the removal of biofilms [23,24] in various methicillin-susceptible and methicillin-resistant variants of S. aureus [25,26]. Although the results of such studies are encouraging, other researches have reported an inferior activity of vancomycin and daptomycin against biofilms [27–30]; thus, there is a need to develop new antibiotics or combinations thereof.
FIGURE 3.
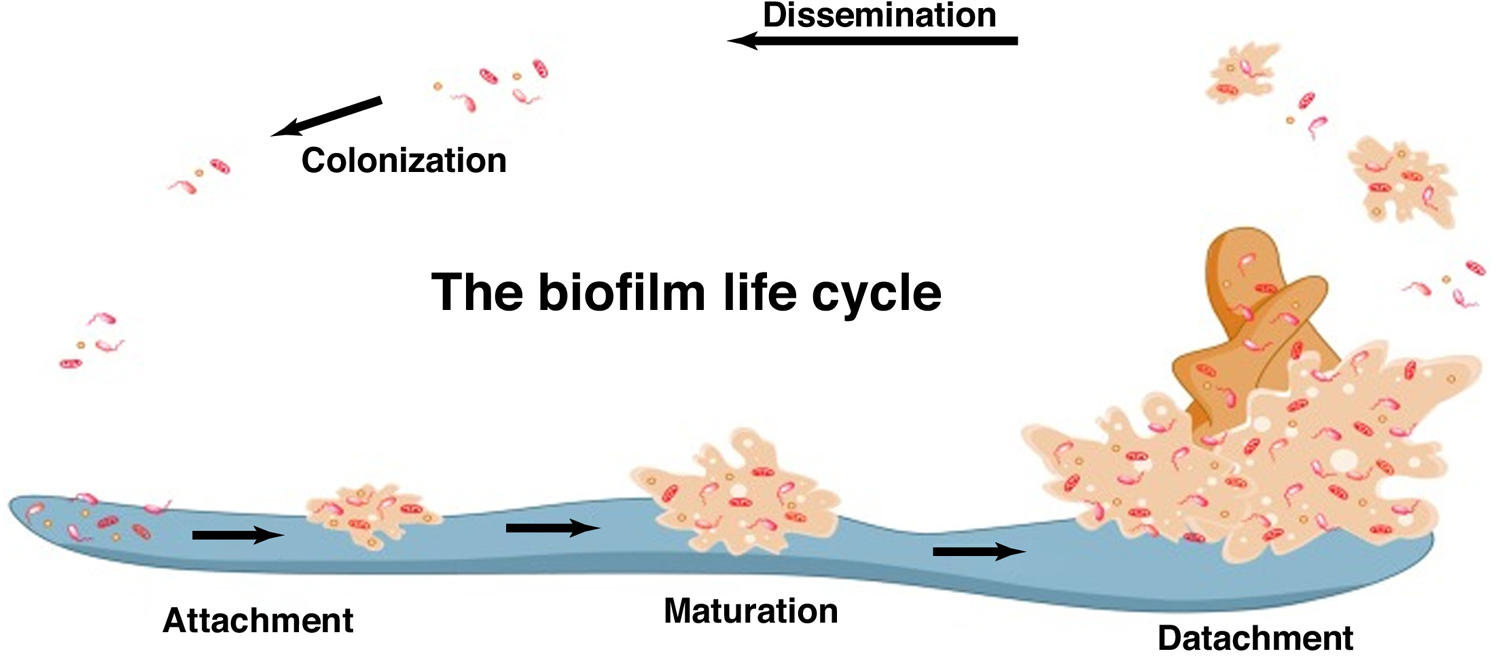
The life cycle of a biofilm Reproduced with permission, from [12].
Drawbacks of current conventional therapies
There are various treatment options available clinically for MRSA. Although vancomycin remains the frontline treatment option, it has several limitations, such as high dosage, longer treatment duration, renal toxicity, and low oral absorption, with intravenous injection, although inconvenient, the only available treatment method [31,32]. In addition to vancomycin, there are other drugs available on the market that could circumvent the drug resistance seen in MRSA therapy. However, when administered intravenously, each of these therapies is more likely to result in adverse effects (Table 2). Furthermore, MRSA antibiotic treatments also show nonspecificity in that they kill useful human bacteria, which results in dysbiosis (microbial imbalance) [33]. The other major challenge is attributed to the abuse of broad-spectrum antibiotics, which causes drug resistance over time [34]. Unfortunately, some bacteria, such as MRSA, develop cross resistance to second-line therapies [35,36]. Bacteria develop drug resistance via various mechanisms, such as multidrug efflux pumps, DNA alteration, cellular membrane modification, and activation of specific enzymes that are able to degrade drugs intracellularly. Thus, traditional antibiotics fail to treat resistant bacteria because they cannot achieve a suitable therapeutic level intracellularly [37,38].
TABLE 2.
Drugs currently available for MRSA treatment and their potency, advantages, disadvantages and average treatment costsa
| Drug | Susceptible MIC (µg/mL) | Dosing regimen | Disadvantages | Refs |
|---|---|---|---|---|
| Vancomycin | ≤2 | 25–30 mg/kg IV load, then 15 mg/kg IV every 8–12 h | Higher doses cause nephrotoxicity, Red Man syndrome; long-term use might develop resistance | [43–45] |
| Daptomycin | ≤1 | 6–10 mg/kg IV every 24 h | Not active for pneumonia | [43–45] |
| Ceftaroline | ≤1 | 600 mg IV every 8–12 h | Similar to other ?-lactams, its MIC varies; can lead to autoimmune hemolytic anemia | [43–45] |
| Dalbavancin | ≤0.12 | 1500 mg IV as single dose | ALT elevations; limited evidence of efficacy against VRSA | [43–45] |
| Oritavancin | ≤0.12 | 1200 mg IV as single dose | Artificially prolongs coagulation tests (INR, PT, aPTT) for ~48 h after administration: use is contraindicated with heparin IV | [43–45] |
| Telavancin | ≤0.12 | 10 mg/kg IV every 24 h | Nephrotoxicity (boxed warning); QTc prolongation; interferes with coagulation tests (INR, PT, aPTT, ACT) for ~18 h after administration | [18–21,23–27, 29–31,33,35,36, 39–46] |
| Tedizolid | ≤0.5 | 200 mg IV/PO daily | Bacteriostatic | [43–45] |
| Linezolid | ≤4 | 600 mg IV/PO every 12 h | Peripheral and optic neuropathy; reversible myelosuppression; serotonin syndrome because of MAO-mediated drug interactions | [43–45] |
| Tigecycline | ≤0.25 | 100 mg IV load, then 50 mg IV every 12 h | Bacteriostatic; high protein binding; controversial use in bacteremia because of low serum concentrations with standard dosing; pancreatitis; hepatotoxicity. | [43–45] |
| Delafloxacin | ≤0.25 | 300 mg every 12 h | Serious adverse reactions, including tendinitis, tendon rupture, peripheral neuropathy, central nervous system effects, and exacerbation of Myasthenia gravis | [23–26] |
Abbreviations: ACT, activated clotting time; ALT, alanine aminotransferase; aPTT, activated partial thromboplastin time; INR, international normalized ratio; IV, intravenous; MAO, monoamine oxidase inhibitors; PO, per oral; PT, prothrombin time; VRSA, vancomycin-resistant Staphylococcus aureus.
Although many novel antibiotics are being tested as treatment options to circumvent the ever-evolving resistance mechanisms, the efficacy and application of such agents is still under review in clinical trials. US Food and Drug Administration (FDA) initiatives have encouraged the search for infectious disease products [39], including effective combination therapies. Combination therapies used so far involve glycopeptides or lipopeptides (i.e., vancomycin and daptomycin). The combination of vancomycin with β-lactams has shown promising results. The synergistic mechanism is still being investigated but is thought to be a ‘see-saw’ effect on the susceptibility of the bacterium to the two drug classes and an overall induction of the host defense mechanism [40]. The ‘See-saw’ effect is the phenomenon where MRSA demonstrates an inverse relationship of resistance to antibiotics of the glycopeptide/lipopeptide and β-lactam classes [41]. This particular combination was effective in clinical trials, with a lower manifestation of adverse effects of either drug [42]. Various other combinations (e.g., aminoglycosides) with vancomycin were explored but discontinued because of severe nephrotoxicity [43,44]. The other most explored combination is daptomycin with β-lactams. This combination enhanced the effects of daptomycin in the treatment of MRSA by improving cell wall binding of daptomycin and resulted in the eventual decrease in the development of resistance [45–47]. Several other combinations have also been explored, such as that of rifampin with vancomycin or daptomycin and other antibiotics [48–50]. Studies with rifampin showed mixed results requiring further evaluation of its efficacy in combination with first-line therapies [51,52]. By contrast, such combinations of antibiotics for MRSA also have more severe adverse effects, as shown by a randomized clinical trial that reported that the combination of vancomycin with β-lactams had a higher incidence of nephrotoxicity [53,54]. Thus, although such combinations are being explored, there is still an immense need for more research for better management modalities.
Several clinical studies for novel antibiotics or repurposed drugs are in the pipeline for antibacterial drugs for MRSA [55] (Table 3). However, the number of novel drugs reaching clinical trials for bacterial infections is limited [56–58]. This might be primarily because of the rapidly developing drug-resistant strains of the bacteria, an overall lack of the right approach in this area of research [59–61] because of the weak market for such antibiotics, the very low rate of return, high investments/costs incurred, few novel lead molecules proceeding into advanced research, and naive regulatory systems for such drugs [62,63].
TABLE 3.
Examples of recent clinical trials for antibiotics against MRSA
| Drug | NCT number | Phase | Status |
|---|---|---|---|
| CG400549 | NCT01593761 | Phase II | Completed |
| CG400549 | NCT01593761 | Phase II | Completed |
| Delafloxacin | NCT01811732, NCT01984684 | Phase III | Completed |
| Iclaprim | NCT02600611, NCT02607618 | Phase III | Completed |
| Lefamulin | NCT01119105, NCT02559310, NCT02813694 | Phase II/III | Completed |
| LTX-109 | NCT01158235 | Phase I/II | Completed |
| MRX-1 | NCT02269319 | Phase II | Completed |
| Nemonoxacin | NCT01944774, NCT00685698 | Phase II | Completed |
| Retapamulin | NCT03304873 | Phase III | Completed |
| XF-73 | NCT03915470 | Phase II | Recruiting |
Nanoparticles for the delivery of anti-MRSA agents
Given that current treatment strategies for MRSA are either rendered ineffective or have shown severe adverse effects, there is a need to find alternative strategies besides novel drug discovery and repurposing of current drugs. This alternative strategy could also contribute to improving the bioavailability and safety of current treatments. Research endeavors have been made in this direction to enhance the bioactivity and bioavailability of different pharmaceutical agents by using nanoparticle (NP)-based delivery strategies. This approach exploits the NP size, shape, charge, or protective internal environment as a way to efficiently deliver drug molecules into cells or tissues through a sustained release action, with the end goal that the drug can exert its therapeutic impact more successfully. The scope of using NPs for accomplishing this is broad, because the agent can be delivered through entrapment or encapsulation, adsorption, or chemical bonding inside the NP structure. NPs can be used to improve the serum stability of the drug, delay the circulation time, and release drugs at specific sites of action in a sustained and controlled manner [64] compared with available platforms such as tablets or capsule-based delivery. NPs can also accumulate in the infected tissue either passively by enhanced permeation and retention (EPR) effect and/or actively by targeting specific receptors, minimizing off-target drug release and subsequent adverse effects [65,66]. In addition, NPs can enter cells via endocytosis, a mechanism that can be utilized to treat intracellular microbial infections [67]. Several studies have shown that different types of NP overcome the bacterial resistance of several antibiotics by escaping from drug degradation enzymes, such as β-lactamase [68,69], inhibiting efflux pumps [70,71], or penetrating thick cell walls [71,72]. A few NP-based drug delivery systems that have been designed for MRSA treatment are now either candidates for preclinical and clinical trials or have recently been approved for clinical use [73,74].
Nanoparticles for MRSA imaging
Nonspecific MRSA drugs and the unavailability of methods for tracking the progress of infection have been a challenge in MRSA treatment. Developing a robust imaging technique to detect the infection site was one strategy to aid the choice of efficient treatment modalities. NP-aided MRSA imaging techniques provide superior theranostic models by incorporating anti-MRSA agents along with imaging probes into the nanocarrier system. Lim et al. developed a peptidoglycan-binding protein-modified magnetic NP system that is able to bind S. aureus in the blood during sepsis because of its affinity to the peptidoglycan layer of the bacterium. The NPs are surface modified with a peptidoglycan-binding protein tagged with a fluorophore to specifically target the bacterial strain and, once they are bound by the magnetic properties of the NP, the bacterium can be imaged (Fig. 4) [75]. Zhao et al. developed a silica-based NP system in which a modified vancomycin-cypate complex is entrapped; this is then released and activated on contact with bacteria and emits near-infrared fluorescence (NIRF) upon laser irradiation. This novel strategy proved to have a longer treatment tracking system to locate the specific infection sites in vivo (Fig. 5) [76]. Hence, it is evident that NPs not only render the current therapies more efficient, but can also be multifaceted in their overall utility.
FIGURE 4.
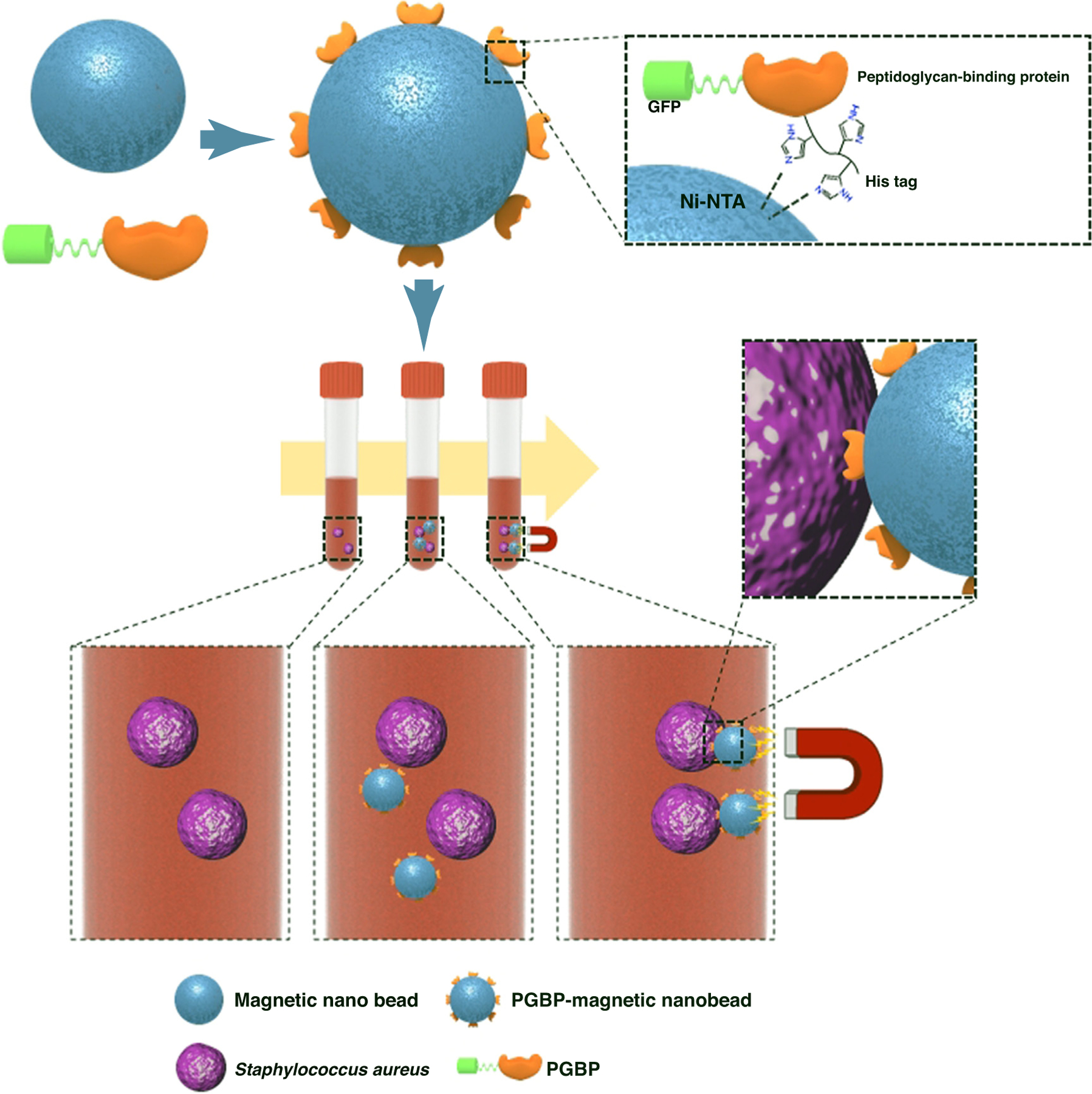
Peptidoglycan binding protein (PGBP)-modified magnetic nanobeads for efficient capturing of gram-positive MRSA. Reproduced with permission, from [75].
FIGURE 5.
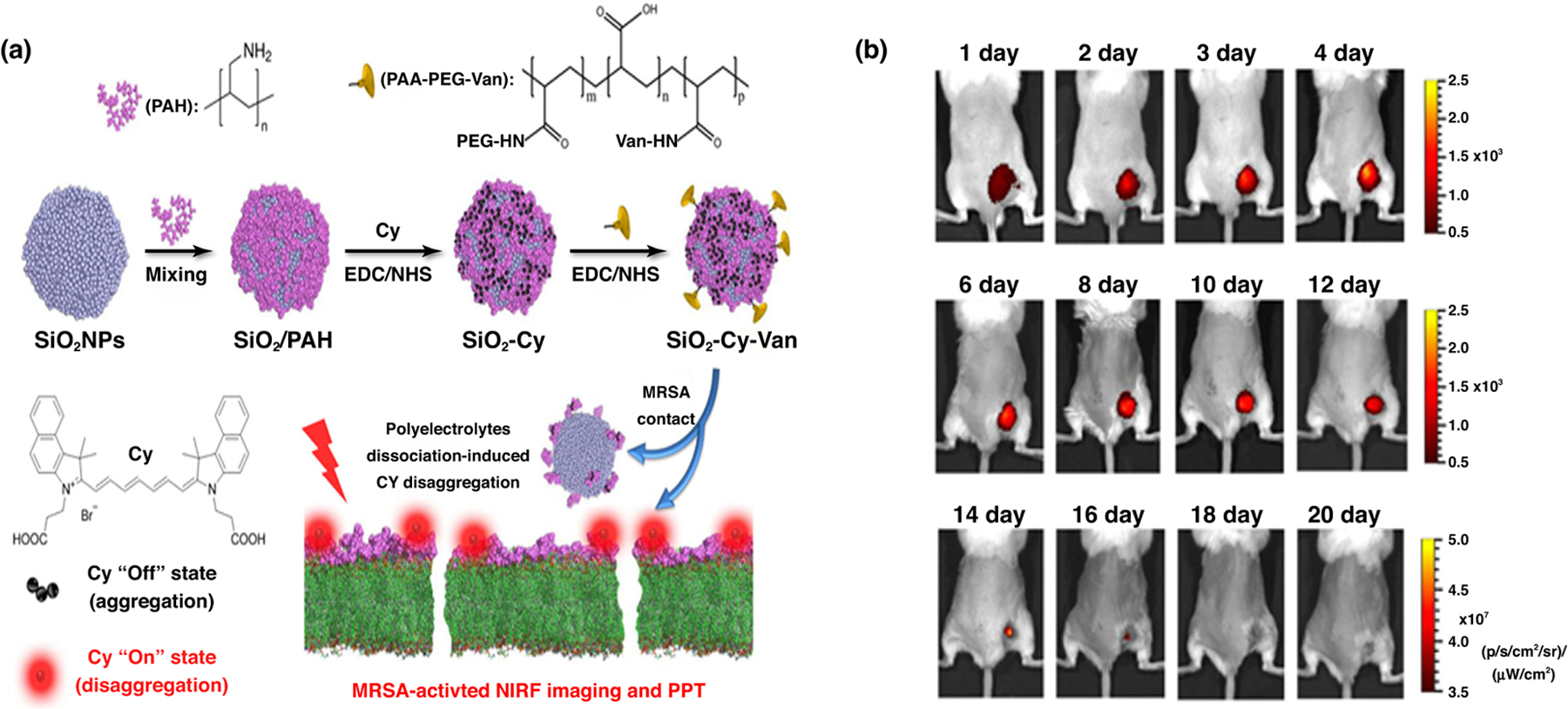
Example for application of nanoparticles as theranostic agents. (a) Preparation and mechanism of vancomycin-cypate silica nanoprobes (Si-Cy-Van). Si-Cy-Van in an aggregated state shows no near-infrared (NIRF) signal; when in contact with bacteria, the nanoparticle disintegrates and releases polyelectrolyte Van complexes onto the bacteria surface; this disaggregated form emits NIRF signals. (b) Images of methicillin-resistant Staphylococcus aureus (MRSA)-infected rats demonstrating the ability of Si-Cy-Van nanoparticles to remain at the infected site for longer time periods. Reproduced with permission, from [76].
Types of nanoparticles used to overcome resistance
Lipid nanoparticles
Lipid NPs are the most widely explored nanoformulations for delivering drugs for MRSA treatment. Many alterations have been made to the composition, preparation method, surface charge, and structural arrangement of lipid NPs to obtain an optimized formulation to deliver both hydrophilic and hydrophobic drugs.
The first report of an anti-MRSA drug in a NP form came in 1994. Multilamellar liposomes, either with vancomycin or teicoplanin, were found to have enhanced in vitro action against MRSA infection inside human macrophages [77]. Another study showed that treatment of MRSA-infected macrophages with surface-PEGylated liposomes did not show any activity [78]. This might be the consequence of the ‘stealth’ effect created by PEGylation of the NPs impeding the recognition of the liposomes by macrophages [78]. More recently, an alternate liposomal vancomycin was seen as progressively viable in vitro against MRSA and, in a murine MRSA disease model, contrasted with a comparative portion of the nonliposomal vancomycin control. In addition, the liposomal delivery of vancomycin reduced its accumulation in kidney, thus avoiding the nephrotoxicity associated with vancomycin [79].
Solid lipid NPs (SLNPs) emerged as a novel technique for the delivery of poorly water-soluble drugs. SLNPs also showed higher encapsulation efficiency for water-soluble antibiotics. The SLNP-based formulation can hold vancomycin for up to 54 h, leading to an ~22-fold increase in MRSA clearance in murine models [80].
Polymer nanoparticles
Although polymer NPs, both natural and synthetic, are a more recent avenue being explored as delivery systems, they are gaining importance rapidly. The most researched and characterized polymer is poly(lactic-co-glycolic acid) (PLGA) because of its non-toxicity, tunable properties, such as erosion times, and biodegradability, which contribute to flexibility in its design and use; it is also approved by the FDA [9]. However, PLGA has a disadvantage in terms of its sensitivity to changes in formulation conditions, such as solvents and drug-loading methods. Rifampicin is regularly used to treat Mycobacterium diseases, including TB, and is a promising drug when used in combination with drugs such as fusidic acid for MRSA [81–83]. Esmaeili et al. developed PLGA-rifampicin NPs that had a normal size of 250 nm but showed low loading capacity. in vitro antibacterial data against a clinical strain of MRSA suggested that MIC was reduced by a factor of 4 (0.002 µg/mL) in contrast to free drug (0.008 µg/mL), showing that the NPs improved the delivery of the compound to bacterial cells [84]. In a comparable report, it was shown that rifampicin loaded in PLGA microparticles of 20–60 µm size range both maintained anti-MRSA activity and reduced the cytotoxicity of rifampicin [85].
Hydroxyapatite-based hollow NPs have additionally been used to deliver vancomycin against MRSA associated with chronic osteomyelitis. Vancomycin-loaded hydroxyapatite pellets showed increased discharge levels of vancomycin over a sustained period of time and provided higher stability. Furthermore, they also showed bone regeneration in rabbit models of chronic osteomyelitis [86].
Poly(ɛ-caprolactone) (PCL)-based vancomycin microparticles (normal particle size measurement >1000 nm) reduced the required dose of vancomycin to treat MRSA-associated osteomyelitis [87,88] and infective discitis. Furthermore, PCL NPs reduced systemic effects by improving the localization of the drug at the site of infection [89]. Another study reported that daptomycin-loaded PCLs microparticles had targeted activity against MRSA biofilms and also inhibited the recovery of the biofilm [90]. Fang et al. reported the use of PCL to formulate a chitosan-based sustained-release delivery system that could maintain a plasma concentration of vancomycin suitable for cytotoxicity of MRSA for a period of 6 weeks, highlighting the usefulness of this polymer for improving MRSA therapy [91].
Han et al. developed nitric oxide (NO) NPs for delivery in bacterial cells [92]. NO is a naturally available lipophilic molecule with a short half-life and its high reactivity produces free radicals. It is responsible for the modulation of intracellular cytokines involved in the wound-healing process by modulating zinc metalloproteins involved in replication and cellular respiration. The study involved silane hydrogels loaded with NO in a dry matrix. The application of NO-based NPs reduced the bacterial burden on the open skin wounds caused by MRSA. The NPs fused with the lipid bilayer of the microbial cells and released the antibiotics directly into the cells.
Durairaj et al. developed a dendrimer-based delivery system for the fluoroquinolone analog, gatifloxacin, which is endorsed for treatment of bacterial conjunctivitis [93]. The dendrimer, ~350 nm in size, increased the water solubility of the antibiotic up to fourfold, enabling it to quickly enter human corneal epithelial cells (<5 min). In addition, the dendrimer–gatifloxacin complex killed MRSA more quickly compared with free gatifloxacin, achieved higher drug concentrations at the target site after in vivo delivery, and maintained good drug levels for 24 h.
Metallic nanoparticles
Metallic NPs disrupt the bacterial cell membrane disruption and/or result in the formation of reactive oxygen species (ROS). Different types of metal ion have been used to investigate their efficacy against MRSA, of which the most explored are silver NPs (AgNPs). The efficacy of these NPs can be further changed by altering the particle size or pH; ultrasound, NIR, or UV stimulation, adding potassium iodide or sodium bromide, and peptide or antibody conjugation. Some metal NPs were used to load anti-infective agents to achieve an additional killing effect. AgNPs have application as antibacterial agents and are utilized in various fields to impede bacterial growth and reduce infection in burn treatments. AgNPs showed effective anti-infective activity against a range of bacteria, including MRSA [94]. Recently, AgNPs were combined with the protein apoferritin to form a stable Ag(I) complex, which was then measured for in vitro antibiotic activity against MRSA [95]. AgNPs release ions that interact with thiol groups on cysteine to disrupt the cell wall and DNA replication. AgNPs in combination with different metallic forms known to form nanocomposites have been widely used for antibacterial purposes. Provaznik et al. reported the use of iron and silver nanocomposites to actively suppress the growth of MRSA under in vitro conditions [96]. Guzman et al. reported that the adsorption of ampicillin on AgNPs significantly inhibited MRSA compared with AgNPs alone [97].
In addition to silver NPs, copper NPs showed effective killing of MRSA under in vitro conditions. The enhanced inhibitory activity of copper and copper oxide NPs results from the copper ions, which can change the local pH and conductivity at the cellular level, damaging the cell membrane and affecting the respiratory enzymes [98].
Gold NPs (AuNPs) have major advantages over other NPs, in that they can be easily modified chemically with an amine or thiol coating that enhances their biological use. Lambert reported that attaching amino saccharides to AuNPs enabled the latter to penetrate the bacterial peptidoglycan layer, inhibiting cell wall synthesis and resulting in bacterial death [99]. Yang et al. conjugated AuNPs to several types of amino saccharide, and found that these AuNPs inhibited the proliferation of multidrug resistance (MDR) bacterial strains. Interestingly, neither AuNPs nor amino saccharides alone showed any antibacterial activity, suggesting that conjugating AuNPs to multivalent amino saccharides is essential to achieve the antibacterial activity [100].
Similarly, the usage of zinc oxide (ZnO) NPs to reduce the bacterial burden in MRSA-associated skin infection in murine models has proven effective [101,102], with one study reporting antibacterial activity of ZnO NPs in MRSA at a concentration of 1875 mg/mL [103]. Similarly, another study reported the bactericidal activity of ZnO NPs with additional insights into the mechanisms of these NPs, which inhibit multiple metabolic pathways, such as amino acid synthesis, in S. aureus [104]. These observations were strengthened by other reported observations, such as activity against bacterial biofilms [105,106] and decreased resistance to drugs [107].
Titanium dioxide (TiO2) NPs have also been explored as metallic NPs and have been successfully applied in MRSA therapy. Their application with different combinations of antibiotics, such as cephalosporins, glycopeptides, and azalides, shown anti-MRSA activity in a disk diffusion assay. Under UV photoactivation, TiO2 NPs form free radicals that lead to their enhanced killing of MRSA [108].
Chitosan
Chitosan is a polysaccharide that has the ability to disrupt the bacterial cell wall in a similar way to metal ions (i.e., bacterial cell wall disruption) and has proven to be an effective product to use against MRSA infection. In addition NPs made of chitosan showed synergistic effects with the antibacterial encapsulated within them. This feature is affected by numerous factors, including pH, type of microorganism, metal cation presence or absence, pKa, molecular weight, and degree of deacetylation (DD) [109].
Cell-penetrating peptides
Cell-penetrating peptides (CPPs) and antimicrobial peptides (AMPs) are an attractive and intriguing area for the development of treatments for MRSA. AMPs, such as buforin and magainin 2, are efficient CPPs in human cells. CPP penetration showed potent antimicrobial activity against both Gram-positive and Gram-negative bacteria without any toxicity towards mammalian cells. The development of NPs that comprise amphiphilic peptides with a TAT fragment (YGRKKRRQRRR) showed an MIC value of 11.4 mM against MRSA as well as the ability to cross the blood–brain barrier to suppress MRSA growth in the infected brain. CyLoP-1, another CPP (CRWRWKCCKK), also showed efficient antimicrobial activity and aids the internalization of the drug [110].
Treatment methods using NPs in MRSA therapy
Photodynamic therapy
The use of nontoxic dyes or photosensitizers (PS) in conjunction with harmless visible light, known as photodynamic therapy (PDT), has become popular [111] and can be used successfully to eradicate the growth of tumors. A similar strategy was used to suppress the growth of bacterial cells. Perni described the implementation of light-activated NPs to enhance the delivery of agents to the infection site [112].
Liposomes have been widely explored for PDT using hematoporphyrin to enhance activity and improve the permeability of cell membranes [113]. Another feature of NPs is the use of targeted ligands for specific receptors of the bacteria, which deliver higher doses of existing drugs to the bacterial infection site. For example, AuNPs surface modified with vancomycin under irradiation with NIR light lead to a mass reduction in the bacterial growth. Similarly, iron oxide NPs modified with vancomycin and porphyrin-platinum led to thermal degradation of MRSA [112].
Transcription factor decoys
Transcription factor decoys (TFDs) are oligonucleotide copies of DNA-binding sites for transcription factors. TFDs represent a potential strategy for suppressing microbial growth without triggering typical resistance mechanisms. TFDs aid the inhibition of gene expression by sequestering transcription factors, ultimately leading to inhibition of protein expression. Chitosan-based TFDs and vancomycin nanocomplexes were developed by Hibbitts et al. and showed enhanced anti-MRSA effects [114].
siRNA therapy
Recent research involves the use of NPs for the delivery of small interfering RNA (siRNA) and CRISPR technology for enhancing MRSA inhibition. These systems include the delivery of siRNA and miRNA to downregulate the expression of essential virulence proteins [115]. Dicer-substrate siRNAs (DsiRNA, formed by the activity of Dicer endonuclease on long double-stranded RNA) were used to develop C16-DsiRNA by conjugating them with palmitic acid; the resulting molecules exhibited a potent gene silencing effect that inhibited MRSA growth via site-specific cleavage and gene silencing. C16-DsiRNA also improved the effects of RNA interference (RNAi) technology in MRSAs cell with high efficiency [116]. Despite being a promising therapy model, siRNA has a short distribution half-life and is prone to lysosomal degradation; thus, NPs are used to overcome these limitations. Liu et al. described the liposomal delivery of antisense siRNA for the mecA gene can effectively restore MRSA susceptibility to oxacillin under both in vitro and in vivo conditions [117].
Given the constant evolution of bacterial strains, new resistance mechanisms can develop for any new antibacterial drugs. Immunotherapy has proven to be a novel way to counteract the resistance pathways of bacteria. Reprogramming macrophages to induce phagocytosis and downregulate proinflammatory macrophages is a recent application of siRNA treatment in infectious diseases [118]. Figure 6 details an application of siRNA NP treatment by modulation of macrophages in staphylococcal pneumonia [119]. Kim et al. developed a hybrid NP system comprising a fusogenic liposome shell on the surface conjugated to a targeting peptide, encapsulated within porous calcium silicate NPs, which contain siRNA. This treatment model was able to kill and clear bacteria in vivo from murine lungs within 7 days of treatment [119].
FIGURE 6.
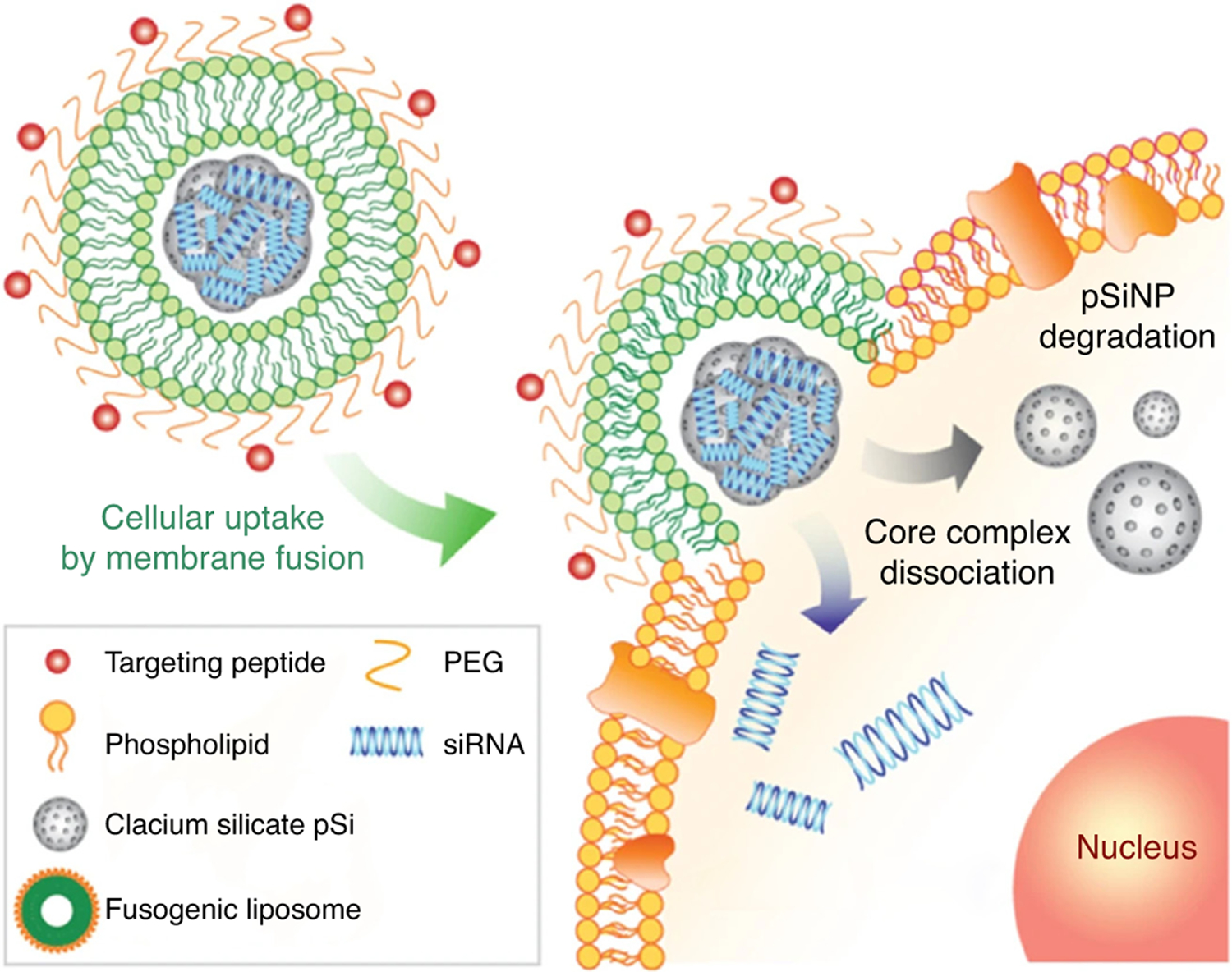
Mechanism of fusogenic hybrid nanoparticles delivering siRNA into macrophages. Reproduced with permission, from[119].
Although there is extensive research ongoing to develop nano-delivery systems, we are still in need of an optimized targeted delivery system to disrupt the biofilm. The ultimate goal for any nanoformulation is to overcome drug resistance, achieve maximum biological activity, and to reduce toxicity. In this regard, there is an immense need to create a delivery system using nanotechnology that is not affected by progressive mutations in the bacteria.
Challenges for nanodrug delivery for MRSA treatment
The success of any nanoparticulate treatment method depends on the ability of the formulation to translate to the clinical setting. However, there are many obstacles in the clinical translation of therapies to the clinic. The first is standardization of the in vitro analyses that are applied at every step of the development of a formulation. These include a variety of experimental methods, from the evaluation of MIC of an antibiotic to toxicity assays, biofilm imaging assays, and so on [120]. Although these experiments are standardized and well established, they falter when they are applied in a preclinical setting. In addition, it is a challenge to harmonize them with other aspects of clinical trials. Once the preclinical criteria are established, the next challenge is to choose the right strain of MRSA to test those criteria. It is evident that there are multiple strains of MRSA in different population groups. Hence, for clinical trials, it is important to choose the right strain of bacterium for the right population [121,122]. The next challenge is the choice of the animal model, which should be in line with the appropriate clinical MRSA scenario. Each source of infection has to be evaluated and all the related aspects of the infections should be included in these preclinical and clinical models [123].
Another obstacle in translating nanodrug delivery technologies into a clinical setting are the limitations arising during the scale-up from the laboratory scale to industrial production. The successful scale-up of a nanomedicine product from bench to market has many component, such as the structure of the material, changes associated with the size and shape of NPs, in vivo biodegradability of nanocarriers, and large-scale set-up for balancing the multicomponent system. One has to be careful about selecting materials, solvents, and the NP production process, as well as being aware of the associated costs and finished product acceptability by the market [124]. Sometimes, the desired features of NPs are lost during laboratory scale-up. Hence, it is also necessary to select the right method of production of NPs to save time during the scale-up process [125].
Prospects and applications of nanotechnology
A characteristic feature of nanotechnology is its ability to make existing products more efficient by introducing new functionality. Nanodrugs are now available in the clinic to treat various cancers, fungal infections, iron deficiency, and macular degeneration, have also been used as image contrast agents, vaccines, and anesthesia, and have successfully demonstrated greater efficacy compared with conventional models of therapy [126]. Increasing research, development, and translation of novel antimicrobial therapies to counter drug resistance is a priority across the pharmaceutical industry and its partners [127]. Pharmaceutical and other research organizations are encouraging the investigation of repurposed drugs for MRSA therapy because this saves both time and money and provides promising advantages [128]. Thus, nanodrug delivery systems have great potential in the translation of MRSA treatments towards better therapeutic outcomes and are a significant approach for overcoming bacterial MDR.
Concluding remarks
MRSA treatment faces several challenges with limited therapeutic options. Although some drugs have been introduced to replace or work in synergy with the most widely used drugs, such as vancomycin, all have seen developing resistance by MRSA and, hence, increasing MIC values and dosage required. Moreover, these drugs are also associated with other adverse effects, such as renal toxicity and hepatotoxicity, in addition to their ineffectiveness against bacterial biofilms, which remain one of the most virulent resistance mechanisms. These problems could be resolved either by faster discovery of new antibiotics to keep up with evolving resistance mechanisms or delivery of the drugs using a method that can circumvent the resistance mechanisms of the bacteria. Nanodrug delivery systems provide an effective and efficient way to deliver drugs to treat the infections by overcoming the drug resistance in the bacteria. It could also be a potential method to reduce the inherent adverse effects of the drug. There are various types of NP used to encapsulate the existing drugs, which have demonstrated promising results in vitro and in vivo. Despite their potential, there are several reasons for the failure of NP strategies in clinical settings. Difficulty in translating successful in vitro results into in vivo settings and in scaling up production of NPs to industrial batch-sizes are some hurdles that need to be addressed. However, research is progressing to develop improved delivery platforms and to optimize nanoformulations to make them a viable long-term strategy to overcome antibiotic resistance.
Acknowledgments
K.V. would like to acknowledge the Department of Pharmaceutical Sciences, Wayne State University for a Frank O Taylor scholarship. A.K.I. acknowledges the Michigan Translational Research and Commercialization (MTRAC) award (#380121), EACPHS FRAP award (#477482) and Wayne State University start-up award (#176575) for funding the MRSA projects in his lab.
Biographies

Kushal Vanamala is pursuing a MSc in the Iyer lab in the Department of Pharmaceutical Sciences at Wayne State University (WSU). His current research focuses on developing targeted nanoparticle delivery systems for infectious diseases and cancers.

Marc Scheetz is a professor at Midwestern University in the Chicago College of Pharmacy and holds a joint appointment in the Department of Pharmacology, College of Graduate Studies. Dr Scheetz was awarded a Doctorate of Pharmacy from Butler University, earned a MSc in Clinical Investigation at Northwestern University, and completed his pharmacy practice residency and an infectious diseases fellowship at Northwestern Memorial Hospital. Dr Scheetz is also the Director of the Pharmacometric Center of Excellence at Midwestern University. He currently practices clinically as an infectious diseases pharmacist at Northwestern Memorial Hospital in Chicago, IL and serves as the Director for the Post-Doctoral Fellowship Program in Infectious Diseases Pharmacotherapy.

Michael J. Rybak is a professor of pharmacy in the Department of Pharmacy Practice, adjunct professor of pharmaceutical sciences in, and Director of the Anti-Infective Research Laboratory, Eugene Applebaum College of Pharmacy & Health Sciences, WSU. He is also adjunct Professor of Medicine, Division of Infectious Diseases, School of Medicine, WSU and adjunct Clinical Professor of Pharmacy, College of Pharmacy, University of Michigan. His research focus is antimicrobial pharmacokinetics and pharmacodynamics and the assessment of infectious diseases outcomes, including their relationship to bacterial resistance. His most recent work focuses on the use of combination therapy, including the use of bacteriophages plus antibiotics to prevent resistance.

David Andes is the William Craig Professor in the Departments of Medicine and Medical Microbiology, Head of the Division of Infectious Diseases, and Director of the Wisconsin Antimicrobial Drug Discovery and Development Center. His research strives to identify strategies to combat antimicrobial drug resistance. His study tactics span from the bench to the clinic, including delineating the optimal dosing strategies for the treatment of drug-resistant infections, identifying new resistance mechanisms, discovering new antimicrobial drugs and targets, and clinical study of resistance epidemiology.

Arun K. Iyer is an associate professor and the Director of the U-BiND Systems Laboratory at the Department of Pharmaceutical Sciences, WSU. Dr. Iyer received his PhD from Sojo University, Japan, under Hiroshi Maeda. In 2012, Dr. Iyer received the prestigious CRS T. Nagai Research Achievement Award. He has authored >100 publications in peer-reviewed international journals and books and has wide expertise in biomaterials and nanomedicine for treating diseases such as infection, dementia, and cancer.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tong SYC et al. (2015) Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev 28, 603–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skinner D and Keefer C (1941) Significance of bacteremia caused by Staphylococcus aureus. Arch. Intern. Med 68, 851–875 [Google Scholar]
- 3.Chambers HF and DeLeo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev Microbiol 7, 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumfitt W and Hamilton-Miller J (1989) Methicillin-resistant Staphylococcus aureus. N. Engl. J. Med 320, 1188–1196 [DOI] [PubMed] [Google Scholar]
- 5.Deurenberg RH and Stobberingh EE (2008) The evolution of Staphylococcus aureus. Infect. Genet Evol 8, 747–763 [DOI] [PubMed] [Google Scholar]
- 6.CDC (2019) Antibiotic Resistance Threats in the United States CDC [Google Scholar]
- 7.Diekema DJ et al. (2019) Twenty-year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY Antimicrobial Surveillance Program. Open Forum Infect Dis 6, S47–S53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reygaert WC (2018) An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol 4, 482–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirenkumar M and Steven S (2011) Poly Lactic-co-glycolic acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers 3, 1377–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies D (2003) Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov 2, 114–122 [DOI] [PubMed] [Google Scholar]
- 11.Piechota M et al. (2018) Biofilm formation by methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains from hospitalized patients in Poland. Biomed. Res. Int 2018, 4657396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craft KM et al. (2019) Methicillin-resistant: Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. MedChemComm 10, 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aslam S (2008) Effect of antibacterials on biofilms. Am. J. Infect. Control 36, S175.e9–S175.e11 [DOI] [PubMed] [Google Scholar]
- 14.Hanke ML et al. (2013) Targeting macrophage activation for the prevention and treatment of Staphylococcus aureus biofilm infections. J. Immunol 190, 2159–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Günther F et al. (2017) MRSA decolonization failure-are biofilms the missing link? Antimicrob. Resist. Infect. Control 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemming HC et al. (2016) Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol 14, 563–575 [DOI] [PubMed] [Google Scholar]
- 17.El-Azizi M et al. (2005) In vitro activity of vancomycin, quinupristin/dalfopristin, and linezolid against intact and disrupted biofilms of staphylococci. Ann. Clin. Microbiol. Antimicrob 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall-Stoodley L et al. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 19.Miyaue S et al. (2018) Bacterial memory of persisters: Bacterial persister cells can retain their phenotype for days or weeks after withdrawal from colony-biofilm culture. Front Microbiol 9, 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart PS and Costerton JW (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 [DOI] [PubMed] [Google Scholar]
- 21.Idexx (2019) Microbiology Guide to Interpreting Minimum Inhibitory Concentration Idexx [Google Scholar]
- 22.Salem AH et al. (2011) Pharmacodynamic assessment of vancomycin-rifampicin combination against methicillin resistant Staphylococcus aureus biofilm: a parametric response surface analysis. J. Pharm. Pharmacol 63, 73–79 [DOI] [PubMed] [Google Scholar]
- 23.Ojea-Jiménez I et al. (2012) Facile preparation of cationic gold nanoparticle-bioconjugates for cell penetration and nuclear targeting. ACS Nano 6, 7692–7702 [DOI] [PubMed] [Google Scholar]
- 24.Stewart PS et al. (2009) Daptomycin rapidly penetrates a Staphylococcus epidermidis biofilm. Antimicrob. Agents Chemother 53, 3505–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roveta S et al. (2008) Activity of daptomycin on biofilms produced on a plastic support by Staphylococcus spp. Int. J. Antimicrob. Agents 31, 321–328 [DOI] [PubMed] [Google Scholar]
- 26.Telles JP et al. (2019) Daptomycin to bone and joint infections and prosthesis joint infections: a systematic review. Braz. J. Infect. Dis 23, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudjemaa R et al. (2016) New insight into daptomycin bioavailability and localization in Staphylococcus aureus biofilms by dynamic fluorescence imaging. Antimicrob. Agents Chemother 60, 4983–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer J et al. (2013) A combined pharmacodynamic quantitative and qualitative model reveals the potent activity of daptomycin and delafloxacin against Staphylococcus aureus biofilms. Antimicrob. Agents Chemother 57, 2726–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barber KE et al. (2014) A novel approach utilizing biofilm time-kill curves to assess the bactericidal activity of ceftaroline combinations against biofilm-producing methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother 58, 2989–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaPlante KL and Woodmansee S (2009) Activities of daptomycin and vancomycin alone and in combination with rifampin and gentamicin against biofilm-forming methicillin-resistant Staphylococcus aureus isolates in an experimental model of endocarditis. Antimicrob. Agents Chemother 53, 3880–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybak MJ (2006) The pharmacokinetic and pharmacodynamic properties of vancomycin activity. Clin. Infect. Dis 42, 35–39 [DOI] [PubMed] [Google Scholar]
- 32.Micek ST (2007) Alternatives to vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin. Infect. Dis 45, S184–S190 [DOI] [PubMed] [Google Scholar]
- 33.Rashid M et al. (2012) Effect of new antimicrobial agents on the ecological balance of human microflora. Anaerobe 18, 249–253 [DOI] [PubMed] [Google Scholar]
- 34.Bell M (2014) Antibiotic misuse: a global crisis. JAMA 174, 1920–1921 [DOI] [PubMed] [Google Scholar]
- 35.Singh R et al. (2014) The role of nanotechnology in combating multi-drug resistant bacteria. J. Nanosci. Nanotechnol 14, 4745–4756 [DOI] [PubMed] [Google Scholar]
- 36.WHO (2000) Antimicrobial Resistance WHO [Google Scholar]
- 37.Alekshun M and Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050 [DOI] [PubMed] [Google Scholar]
- 38.Briones E et al. (2008) Delivery systems to increase the selectivity of antibiotics in phagocytic cells. J. Control Release 125, 210–227 [DOI] [PubMed] [Google Scholar]
- 39.Tillotson J and Tillotson GS (2015) The regulatory pathway for antifungal drugs: a US perspective. Clin. Infect. Dis 61, S678–S683 [DOI] [PubMed] [Google Scholar]
- 40.Choo EJ and Chambers HF (2016) Treatment of methicillin-resistant Staphylococcus aureus bacteremia. Infect. Chemother 48, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vignaroli C et al. (2011) Striking ‘seesaw effect’ between daptomycin nonsusceptibility and beta-lactam susceptibility in Staphylococcus haemolyticus. Antimicrob. Agents Chemother 55, 2496–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dilworth TJ et al. (2014) β-Lactams enhance vancomycin activity against methicillin-resistant Staphylococcus aureus bacteremia compared to vancomycin alone. Antimicrob. Agents Chemother 58, 102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malacarne P et al. (2006) Nephrotoxicity due to combination antibiotic therapy with vancomycin and aminoglycosides in septic critically III patients. Chemotherapy 52, 178–184 [DOI] [PubMed] [Google Scholar]
- 44.Timpe EM (2005) Nephrotoxicity with combination vancomycin-aminoglycoside therapy. J Pediatr. Pharmacol. Ther 10, 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers HF et al. (2016) Daptomycin-nonsusceptible methicillin-resistant Staphylococcus aureus endocarditis. Can. J. Infect. Dis. Med. Microbiol 60, 3976–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jahanbakhsh S et al. (2020) Impact of daptomycin dose exposure alone or in combination with β-lactams or rifampin against vancomycin-resistant enterococci in an in vitro biofilm model. Antimicrob. Agents Chemother 64, e02074–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siala W et al. (2016) Synergistic activity between an antimicrobial polyacrylamide and daptomycin versus Staphylococcus aureus biofilm. Pathog. Dis 74, ftw042. [DOI] [PubMed] [Google Scholar]
- 48.Jørgensen NP et al. (2016) Rifampicin-containing combinations are superior to combinations of vancomycin, linezolid and daptomycin against Staphylococcus aureus biofilm infection in vivo and in vitro. Pathog. Dis 74, ftw019. [DOI] [PubMed] [Google Scholar]
- 49.Hall Snyder AD et al. (2015) Evaluation of high-dose daptomycin versus vancomycin alone or combined with clarithromycin or rifampin against Staphylococcus aureus and S. epidermidis in a novel in vitro PK/PD model of bacterial biofilm. Infect. Dis. Ther 4, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Broussou DC et al. (2018) Differential activity of the combination of vancomycin and amikacin on planktonic vs. biofilm-growing Staphylococcus aureus bacteria in a Hollow Fiber infection model. Front Microbiol 9, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang HJ et al. (2013) In vitro efficacies and resistance profiles of rifampin-based combination regimens for biofilm-embedded methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57, 5717–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiter KC et al. (2012) Rifampicin fails to eradicate mature biofilm formed by methicillin-resistant Staphylococcus aureus. Rev. Soc. Bras. Med. Trop 45, 471–474 [DOI] [PubMed] [Google Scholar]
- 53.Tong SYC et al. (2020) Effect of vancomycin or daptomycin with vs without an antistaphylococcal β-lactam on mortality, bacteremia, relapse, or treatment failure in patients with MRSA bacteremia: a randomized clinical trial. JAMA 323, 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis J et al. (2016) Combination of vancomycin and β-lactam therapy for methicillin-resistant Staphylococcus aureus bacteremia: a pilot multicenter randomized controlled trial. Clin. Infect. Dis 62, 173–180 [DOI] [PubMed] [Google Scholar]
- 55.Theuretzbacher U et al. (2020) The global preclinical antibacterial pipeline. Nat. Rev. Microbiol 18, 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.WHO (2017) Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline, Including Tuberculosis WHO [Google Scholar]
- 57.CDC (2017) Drug Resistance Threats CDC [Google Scholar]
- 58.Tacconelli E et al. (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis 18, 318–327 [DOI] [PubMed] [Google Scholar]
- 59.WHO (2020) Lack of New Antibiotics Threatens Global Efforts to Contain Drug-Resistant Infections WHO [Google Scholar]
- 60.Boucher HW et al. (2013) 10×’20 progress - development of new drugs active against Gram-negative bacilli: an update from the infectious diseases society of America. Clin. Infect. Dis 56, 1685–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradford PA (2018) Antibiotics — from there to where? How the antibiotic miracle is threatened by resistance and a broken market and what we can do about it. Pathog Immun 3, 19–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payne DJ et al. (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov 6, 29–40 [DOI] [PubMed] [Google Scholar]
- 63.Schlaes D (2010) Antibiotics: The Perfect Storm Springer [Google Scholar]
- 64.Kingsley JD et al. (2006) Nanotechnology: A focus on nanoparticles as a drug delivery system. J. Neuroimmune. Pharmacol 1, 340–350 [DOI] [PubMed] [Google Scholar]
- 65.Edagwa BJ et al. (2014) Long-acting antituberculous therapeutic nanoparticles target macrophage endosomes. FASEB J 28, 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie S et al. (2014) Biodegradable nanoparticles for intracellular delivery of antimicrobial agents. J. Control Release 187, 101–117 [DOI] [PubMed] [Google Scholar]
- 67.Destache CJ et al. (2010) Antiretroviral release from poly(DL-lactide-co-glycolide) nanoparticles in mice. J. Antimicrob. Chemother 65, 2183–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown AN et al. (2012) Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl. Environ. Microbiol 78, 2768–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y and Jiang X (2013) Multiple strategies to activate gold nanoparticles as antibiotics. Nanoscale 5, 8340–8350 [DOI] [PubMed] [Google Scholar]
- 70.Shaik N et al. (2008) Interactions of pluronic block copolymers on P-gp efflux activity: experience with HIV-1 protease inhibitors. J. Pharm. Sci 97, 5421–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pelgrift R and Friedman A (2013) Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv Rev 65, 1803–1815 [DOI] [PubMed] [Google Scholar]
- 72.Chakraborty S et al. (2012) In vitro antimicrobial activity of nanoconjugated vancomycin against drug resistant Staphylococcus aureus. Int. J. Pharm 436, 659–676 [DOI] [PubMed] [Google Scholar]
- 73.Smith NM et al. (2016) An unexpected transient breakdown of the blood brain barrier triggers passage of large intravenously administered nanoparticles. Sci. Rep 6, 22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang L et al. (2018) Immune response effects of diverse vaccine antigen attachment ways based on the self-made nanoemulsion adjuvant in systemic MRSA infection. RSC Adv 8, 10425–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim J et al. (2019) Peptidoglycan binding protein (PGBP)-modified magnetic nanobeads for efficient magnetic capturing of Staphylococcus aureus associated with sepsis in blood. Sci. Rep 9, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Z et al. (2017) Bacteria-activated theranostic nanoprobes against methicillin-resistant Staphylococcus aureus infection. ACS Nano 11, 4428–4438 [DOI] [PubMed] [Google Scholar]
- 77.Onyeji CO et al. (1994) Enhanced killing of methicillin-resistant Staphylococcus aureus in human macrophages by liposome-entrapped vancomycin and teicoplanin. Infection 22, 338–342 [DOI] [PubMed] [Google Scholar]
- 78.Pumerantz A et al. (2011) Preparation of liposomal vancomycin and intracellular killing of meticillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 37, 140–144 [DOI] [PubMed] [Google Scholar]
- 79.Sande L et al. (2012) Liposomal encapsulation of vancomycin improves killing of methicillin-resistant Staphylococcus aureus in a murine infection model. J. Antimicrob. Chemother 67, 2191–2194 [DOI] [PubMed] [Google Scholar]
- 80.Huh AJ and Kwon YJ (2011) ‘Nanoantibiotics’: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control Release 156, 128–145 [DOI] [PubMed] [Google Scholar]
- 81.Garske LA et al. (2004) Rifampicin and sodium fusidate reduces the frequency of methicillin-resistant Staphylococcus aureus (MRSA) isolation in adults with cystic fibrosis and chronic MRSA infection. J. Hosp. Infect 56, 208–214 [DOI] [PubMed] [Google Scholar]
- 82.Kim B-N et al. (2014) Oral antibiotic treatment of staphylococcal bone and joint infections in adults. J. Antimicrob. Chemother 69, 309–322 [DOI] [PubMed] [Google Scholar]
- 83.Wu WS et al. (2013) Efficacy of combination oral antimicrobial agents against biofilm-embedded methicillin-resistant Staphylococcus aureus. J. Microbiol. Immunol. Infect 46, 89–95 [DOI] [PubMed] [Google Scholar]
- 84.Esmaeili F et al. (2007) Preparation and antibacterial activity evaluation of rifampicin-loaded poly lactide-co-glycolide nanoparticles. Nanomedicine 3, 161–167 [DOI] [PubMed] [Google Scholar]
- 85.Martins D et al. (2011) Evaluation of the antibacterial activity of poly-(D,L-lactide-co-glycolide) nanoparticles containing violacein. J. Nanoparticle Res 13, 355–363 [Google Scholar]
- 86.Jiang J-L et al. (2012) Vancomycin-loaded nano-hydroxyapatite pellets to treat MRSA-induced chronic osteomyelitis with bone defect in rabbits. Inflamm. Res 61, 207–215 [DOI] [PubMed] [Google Scholar]
- 87.Pardridge WM (2003) Blood-brain barrier drug targeting: the future of brain drug development. Mol. Interv 3, 90–105 [DOI] [PubMed] [Google Scholar]
- 88.Cevher E et al. (2006) Characterization of biodegradable chitosan microspheres containing vancomycin and treatment of experimental osteomyelitis caused by methicillin-resistant Staphylococcus aureus with prepared microspheres. Int. J. Pharm 317, 127–135 [DOI] [PubMed] [Google Scholar]
- 89.le Ray AM et al. (2005) In vitro and In vivo bactericidal activities of vancomycin dispersed in porous biodegradable poly(ɛ-caprolactone) microparticles. Antimicrob. Agents Chemother 49, 3025–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferreira I et al. (2015) Activity of daptomycin- and vancomycin-loaded poly-epsilon-caprolactone microparticles against mature staphylococcal biofilms. Int. J. Nanomed 10, 4351–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang T et al. (2012) Poly (ɛ-caprolactone) coating delays vancomycin delivery from porous chitosan/β-tricalcium phosphate composites. J. Biomed. Mater. Res. B 100, 1803–1811 [DOI] [PubMed] [Google Scholar]
- 92.Han G et al. (2009) Nitric oxide releasing nanoparticles are therapeutic for Staphylococcus aureus abscesses in a murine model of infection. PLoS One 4, e7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Durairaj C et al. (2010) Nanosized dendritic polyguanidilyated translocators for enhanced solubility, permeability, and delivery of gatifloxacin. Invest. Ophthalmol. Vis. Sci 51, 5804–5816 [DOI] [PubMed] [Google Scholar]
- 94.Morones JR et al. (2005) The bactericidal effect of silver nanoparticles. Nanotechnology 16, 2346. [DOI] [PubMed] [Google Scholar]
- 95.Nabikhan A et al. (2010) Synthesis of antimicrobial silver nanoparticles by callus and leaf extracts from saltmarsh plant, Sesuvium portulacastrum. L. Colloids Surf. B Biointerfaces 79, 488–493 [DOI] [PubMed] [Google Scholar]
- 96.Provaznik I et al. (2012) Electrochemical behaviour of apoferritin encapsulating of silver (i) ions and its application for treatment of Staphylococcus aureus. Int. J. Electrochem. Sci 7, 6378–6395 [Google Scholar]
- 97.Guzman M et al. (2012) Synthesis and antibacterial activity of silver nanoparticles against gram-positive and gram-negative bacteria. Nanomedicine 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 98.Strauch BM et al. (2017) Comparison between micro-and nanosized copper oxide and water soluble copper chloride: interrelationship between intracellular copper concentrations, oxidative stress and DNA damage response in human lung cells. Part Fibre Toxicol 14, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lambert PA (2002) Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol 92, 46S–54S [PubMed] [Google Scholar]
- 100.Yang X et al. (2018) Aminosaccharide-gold nanoparticle assemblies as narrow-spectrum antibiotics against methicillin-resistant Staphylococcus aureus. Nano Res 11, 6237–6243 [Google Scholar]
- 101.Rauf MA et al. (2017) Biomimetically synthesized ZnO nanoparticles attain potent antibacterial activity against less susceptible S. aureus skin infection in experimental animals. RSC Adv 7, 36361–36373 [Google Scholar]
- 102.Ansari MA et al. (2012) Characterization of clinical strains of MSSA, MRSA and MRSE isolated from skin and soft tissue infections and the antibacterial activity of ZnO nanoparticles. World J. Microbiol Biotechnol 28, 1605–1613 [DOI] [PubMed] [Google Scholar]
- 103.Umamageswari SSM et al. (2018) Evaluation of antibacterial activity of zinc oxide nanoparticles against biofilm producing methicillin resistant Staphylococcus aureus (MRSA). Res. J. Pharm. Technol 11, 1884–1888 [Google Scholar]
- 104.Kadiyala U et al. (2018) Unexpected insights into antibacterial activity of zinc oxide nanoparticles against methicillin resistant: Staphylococcus aureus (MRSA). Nanoscale 10, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jesline A et al. (2015) Antimicrobial activity of zinc and titanium dioxide nanoparticles against biofilm-producing methicillin-resistant Staphylococcus aureus. Appl. Nanosci 5, 157–162 [Google Scholar]
- 106.Ashajyothi C et al. (2016) Antibiofilm activity of biogenic copper and zinc oxide nanoparticles-antimicrobials collegiate against multiple drug resistant bacteria: a nanoscale approach. J. Nanostruct. Chem 6, 329–341 [Google Scholar]
- 107.Iram S et al. (2015) Potentiating efficacy of antibiotic conjugates with zinc oxide nanoparticles against clinical isolates of Staphylococcus aureus. Dig. J. Nanomater. Biostruct 10, 901–914 [Google Scholar]
- 108.Roy AS et al. (2010) Effect of nano-titanium dioxide with different antibiotics against methicillin-resistant Staphylococcus aureus. J. Biomater. Nanobiotechnol 01, 37–41 [Google Scholar]
- 109.Asli A et al. (2017) Antibiofilm and antibacterial effects of specific chitosan molecules on Staphylococcus aureus isolates associated with bovine mastitis. PLoS One 12, e0176988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zou L et al. (2013) Cell-penetrating peptide-mediated therapeutic molecule delivery into the central nervous system. Curr. Neuropharmacol 11, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castano AP et al. (2004) Mechanisms in photodynamic therapy: Part one - Photosensitizers, photochemistry and cellular localization. Photodiagnosis Photodyn. Ther 1, 279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Perni S et al. (2011) Antimicrobial properties of light-activated polyurethane containing indocyanine green. J. Biomater. Appl 25, 387–400 [DOI] [PubMed] [Google Scholar]
- 113.Pornpattananangkul D et al. (2011) Bacterial toxin-triggered drug release from gold nanoparticle-stabilized liposomes for the treatment of bacterial infection. J. Am. Chem. Soc 133, 4132–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hibbitts A et al. (2019) Co-delivery of free vancomycin and transcription factor decoy-nanostructured lipid carriers can enhance inhibition of methicillin resistant Staphylococcus aureus (MRSA). PLoS One 14, e0220684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Marraffini LA and Sontheimer EJ (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet 11, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kubo T et al. (2012) Amino-modified and lipid-conjugated dicer-substrate siRNA enhances RNAi efficacy. Bioconjug. Chem 23, 164–173 [DOI] [PubMed] [Google Scholar]
- 117.Liu C et al. (2017) Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 266, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burke B et al. (2002) Macrophages in gene therapy: cellular delivery vehicles and in vivo targets. J. Leukoc. Biol 72, 417–428 [PubMed] [Google Scholar]
- 119.Kim B et al. (2018) Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus. Nat. Commun 9, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hibbitts A and O’Leary C (2018) Emerging nanomedicine therapies to counter the rise of Methicillin-resistant Staphylococcus aureus. Materials 11, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Calfee DP (2017) Trends in community versus health care-acquired methicillin-resistant Staphylococcus aureus infections. Curr. Infect. Dis. Rep 19, 48. [DOI] [PubMed] [Google Scholar]
- 122.Hudson LO et al. (2013) Diversity of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from inpatients of 30 hospitals in Orange County, California. PLoS One 8, e62117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marra A (2014) Animal models in drug development for MRSA. Methods Mol. Biol 1085, 333–345 [DOI] [PubMed] [Google Scholar]
- 124.Buzea C et al. (2007) Nanomaterials and nanoparticles: sources and toxicity. Biointerphases 2, MR17–MR71 [DOI] [PubMed] [Google Scholar]
- 125.Paliwal R et al. (2014) Nanomedicine scale-up technologies: feasibilities and challenges. Ageing Int 15, 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anselmo AC and Mitragotri S (2016) Nanoparticles in the clinic. Bioeng. Transl. Med 1, 10–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simpkin VL et al. (2017) Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J. Antibiot 70, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thangamani S et al. (2015) Drug repurposing for the treatment of staphylococcal infections. Curr. Pharm. Des 21, 2089–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]


