Abstract
Retinoblastoma protein (RB) encoded by Rb1 is a prominent inducer of cell cycle arrest (CCA). The hormone progesterone (P4) promotes CCA in the uterine epithelium and previous studies indicated that P4 activates RB by reducing the phosphorylated, inactive form of RB. Here, we show that embryo implantation is impaired in uterine‐specific Rb1 knockout mice. We observe persistent cell proliferation of the Rb1‐deficient uterine epithelium until embryo attachment, loss of epithelial necroptosis, and trophoblast phagocytosis, which correlates with subsequent embryo invasion failure, indicating that Rb1‐induced CCA and necroptosis of uterine epithelium are involved in embryo invasion. Pre‐implantation P4 supplementation is sufficient to restore these defects and embryo invasion. In Rb1‐deficient uterine epithelial cells, TNFα‐primed necroptosis is impaired, which is rescued by the treatment with a CCA inducer thymidine or P4 through the upregulation of TNF receptor type 2. TNFα is expressed in the luminal epithelium and the embryo at the embryo attachment site. These results provide evidence that uterine Rb1‐induced CCA is involved in TNFα‐primed epithelial necroptosis at the implantation site for successful embryo invasion.
Keywords: embryo implantation, progesterone, TNFα signaling, trophoblast phagocytosis, uterine luminal epithelium
Subject Categories: Autophagy & Cell Death, Cell Cycle, Development & Differentiation
Uterine Rb1‐induced cell cycle arrest is involved in TNFα‐primed epithelial necroptosis at the implantation site, and these processes are critical for embryo invasion. These findings may help to better understand the molecular factors underlying human recurrent implantation failure.

Introduction
Infertility is estimated to influence up to 30% of couples worldwide (Inhorn & Patrizio, 2015). The number of infertile patients undergoing in vitro fertilization and embryo transfer (IVF‐ET) is increasing, and some of these patients repeatedly fail to conceive even after ET using good quality embryos. This pathological condition is called recurrent implantation failure, which becomes a major problem in IVF‐ET treatment. Embryo implantation, a series of processes from embryo apposition and attachment to the uterus to embryo invasion into the uterus, is doubtlessly regulated by a precise and intricate molecular interaction between the embryo and uterus (Dey et al, 2004; Cha et al, 2012; Egashira & Hirota, 2013; Fukui et al, 2019; Hirota, 2019). In mice, embryo apposition is followed by intimate adherence of the blastocyst trophectoderm to the luminal epithelium, marking the first discernible sign of implantation from the midnight of day 4 to day 5 morning (day 1 = vaginal plug). This interaction starts when embryonic development is synchronized with uterine preparation to become receptive for implantation. The physiological contact between the blastocyst and the luminal epithelium affects the neighboring stroma, where vascular permeability increases under the control of several factors such as COX2 and VEGF (Matsumoto et al, 2002). Blastocyst attachment coincides with increased stromal vascular permeability at the site of the blastocyst called as attachment reaction and can be demarcated by distinct blue bands along the uterus after intravenous injection of Chicago blue dye solution (Das et al, 1994). With the implantation process in progress, stromal cells surrounding the implanting embryo undergo decidualization. Decidual growth reaches its peak on day 8 of pregnancy. The luminal epithelium surrounding the implantation sites disappears on day 5 evening, and embryo invasion into the stroma starts from day 5 night to day 6 morning (Matsumoto et al, 2018). Our previous observation has indicated that direct contact between embryo and uterine stroma is involved in embryonic cell survival and invasion (Matsumoto et al, 2018).
Ovarian steroid hormone progesterone (P4) governs the entire process of pregnancy, including embryo implantation. By the stage of embryo apposition, P4 guides the uterus to the receptive state (Dey et al, 2004). In mice, pre‐ovulatory ovarian estrogen secretion induces proliferation of the luminal and glandular epithelium during days 1–2 of pregnancy. On day 3, the newly formed corpora luteum produces P4. By day 4, increasing P4 causes stromal cell proliferation as well as epithelial cell differentiation detected by epithelial cell cycle arrest (CCA), which we call proliferation–differentiation switching (PDS) in the receptive uterus (Haraguchi et al, 2014; Hirota, 2019). PDS regulated by P4 is associated with the expression of several genes critical for embryo implantation on day 4, followed by attachment of blastocyst to the luminal epithelium from day 4 midnight to day 5 morning. In addition, P4 acts via the nuclear P4 receptor (PGR) for transcriptional activation of genes involved in uterine receptivity including PDS and embryo implantation (Lydon et al, 1995; Tranguch et al, 2007; Haraguchi et al, 2014). Thus, P4‐PGR signaling is implicated in the preparation of uterine receptivity and embryo implantation, and uterine epithelial CCA with stromal proliferation is a hallmark of uterine receptivity to the embryo (Dey et al, 2004; Cha et al, 2012; Haraguchi et al, 2014). However, it remains uncertain how P4‐induced epithelial CCA is involved in the process of embryo implantation. The aim of this study is to analyze the underlying mechanisms of embryo implantation by focusing on CCA regulators in the uterus.
A known tumor suppressor retinoblastoma protein (RB) is encoded by Rb1 gene, and its classical function is to induce CCA by binding and inactivating the transcription factors E2F, repressing the transcription of genes which are required for the S phase (Giacinti & Giordano, 2006; Dick & Rubin, 2013). Therefore, Rb1 deletion promotes cell cycle progression, which results in loss of cell differentiation. RB acts as a cell cycle suppressor, but it also has the ability to prevent excess action. Unphosphorylated RB (uRB), the active form of RB, has the activity of CCA until a cell is ready to divide. Once cell division is ready, uRB becomes phosphorylated RB (pRB), the inactive form of RB, which releases its binding to E2F factors and loses the function as a cell cycle suppressor (Giacinti & Giordano, 2006; Dick & Rubin, 2013). The previous studies have shown that estrogen increases pRB and promotes cell proliferation in the uterine epithelium, which is suppressed by P4, indicating that uterine RB controls ovarian hormone‐dependent cell proliferation status (Tong & Pollard, 1999; Chen et al, 2005). In the present study using female mice with uterine Rb1 deficiency (Rb1d/d mice), we aimed to investigate the fundamental machinery in which uterine RB and P4 are involved in embryo implantation through epithelial CCA.
Results
Mice with uterine deletion of Rb1 show subfertility
We first examined the expression of RB in the pre‐implantation mouse uterus and found that RB is expressed in the uteri of wild‐type (WT) mice from day 1 to day 6 of pregnancy (Fig 1A). In contrast, phosphorylated RB (pRB), the inactive form of RB, is expressed in uterine epithelium on days 2 and 3 but not on day 4 (Fig 1B), suggesting that the suppression of RB activity is lost on day 4. These findings indicate that uterine RB becomes functionally active on day 4 when uterine PDS takes place. To explore the role of uterine Rb1 in pregnancy, we generated female mice with deletion of Rb1 in the uterus (Rb1d/d mice) by crossing Rb1‐loxP mice with Pgr‐Cre mice. We next confirmed that RB expression is efficiently deleted in Rb1d/d uteri (Fig 1C). In contrast to the uterus, RB expression is normal in the granulosa and lutein cells of Rb1d/d ovary (Fig 1D). Rb1d/d mice and their littermate controls (Rb1f/f mice) were mated with WT fertile male mice. We found that the number of pups delivered by Rb1d/d dams is reduced (Fig 1E), suggesting that uterine Rb1 is critical for successful pregnancy. Based on these results, we next focused on the role of uterine Rb1 in embryo implantation.
Figure 1. Mice with uterine deletion of Rb1 show subfertility.
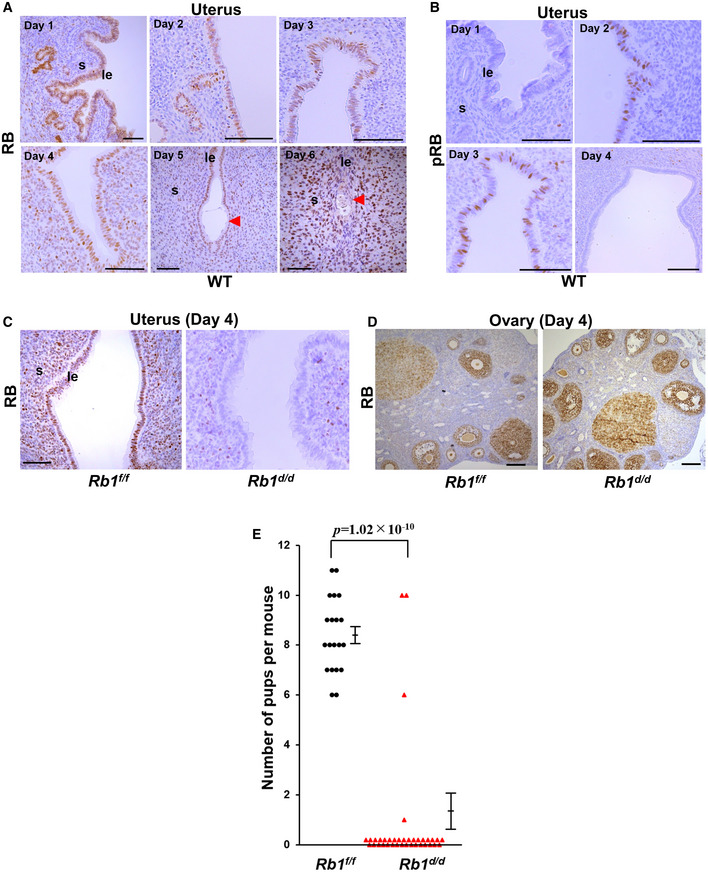
- Rb1 protein (RB) was expressed in uterine luminal epithelium and stroma during early pregnancy in mice. Scale bar = 100 μm; arrowhead, embryo.
- Phosphorylated RB (pRB) protein, the inactive form of RB, was expressed in the luminal epithelium on days 2 and 3 of pregnancy. Scale bar = 100 μm.
- RB was efficiently deleted in the uterus of mice with uterine deletion of Rb1 (Rb1d/d mice) on day 4 of pregnancy. Scale bar = 100 μm.
- RB expression was normal in the ovary of Rb1d/d mice on day 4 of pregnancy. Scale bar = 100 μm.
- The number of new‐born pups was decreased in Rb1d/d dams (n = 37 different dams) compared with their littermate controls (Rb1f/f mice, n = 20 different dams) (mean ± SEM, Student’s t‐test).
Data information: le, luminal epithelium; s, stroma.
Rb1d/d mice show normal ovulation and pre‐implantation embryo development, but cell cycle arrest of luminal epithelium and embryo implantation are impaired
We examined ovulation and fertilization in Rb1d/d mice by flushing eggs and/or embryos on day 2 of pregnancy and found that both processes are normal (Fig 2A and B). We also confirmed normal development of pre‐implantation embryos and their timely transport from the oviduct into uterine lumens of Rb1d/d mice by recording the number of blastocysts retrieved by flushing on day 4 of pregnancy (Fig 2C), suggesting that pre‐implantation ovarian and oviductal functions are normal in Rb1d/d mice. PDS, luminal epithelial CCA, and stromal proliferation, in the uterus on day 4 morning, are hallmarks of uterine receptivity in embryo implantation (Haraguchi et al, 2014; Hirota, 2019). Immunostaining of Ki67, a marker of cell cycle progression, revealed that Rb1d/d mice have increased numbers of proliferating cells in the luminal epithelium on day 4 compared with Rb1f/f mice (Fig 2D and E), suggesting impaired epithelial CCA and uterine receptivity in Rb1d/d mice. We found that the number of distinct implantation sites in Rb1d/d mice is comparable to Rb1f/f mice on day 5 morning (Fig 2F and G). By hematoxylin and eosin (H&E) staining, we confirm that embryo attachment occurs normally in Rb1d/d mice (Fig EV1A), and the expression of COX2, a marker of attachment reaction, is also normal (Fig EV1B). LIF, a key inducer of embryo attachment, activates and phosphorylates STAT3. The expression of Lif mRNA and phosphorylated STAT3 (pSTAT3) protein was also normal in Rb1d/d mice (Fig EV1C and D), suggesting that embryo attachment is normal in Rb1d/d mice. To determine whether the implantation process after embryo attachment is normal in Rb1d/d mice, we counted the number of implantation sites in Rb1d/d and Rb1f/f mice on day 8 of pregnancy. The number of implantation sites remained comparable between the two groups on day 8, but increased resorption sites with internal bleeding were observed only in implantation sites of Rb1d/d mice, not in those of Rb1f/f mice on day 8 (Fig 2H and I). By H&E staining, we confirmed that embryos are completely degraded in Rb1d/d mice on day 8 of pregnancy (Fig 2J), suggesting that embryo implantation fails after normal embryo attachment in Rb1d/d mice. Concomitantly with embryo resorption, the weight of implantation sites decreased in Rb1d/d mice (Fig 2K). The mRNA expression of decidualization markers Cox2 and Bmp2 was comparable between Rb1d/d and Rb1f/f mice on day 8 (Fig 2L and M), suggesting that degradation of embryos in Rb1d/d mice is not due to decidualization failure.
Figure 2. Rb1d/d mice show impaired pre‐implantation epithelial cell cycle arrest and implantation failure.
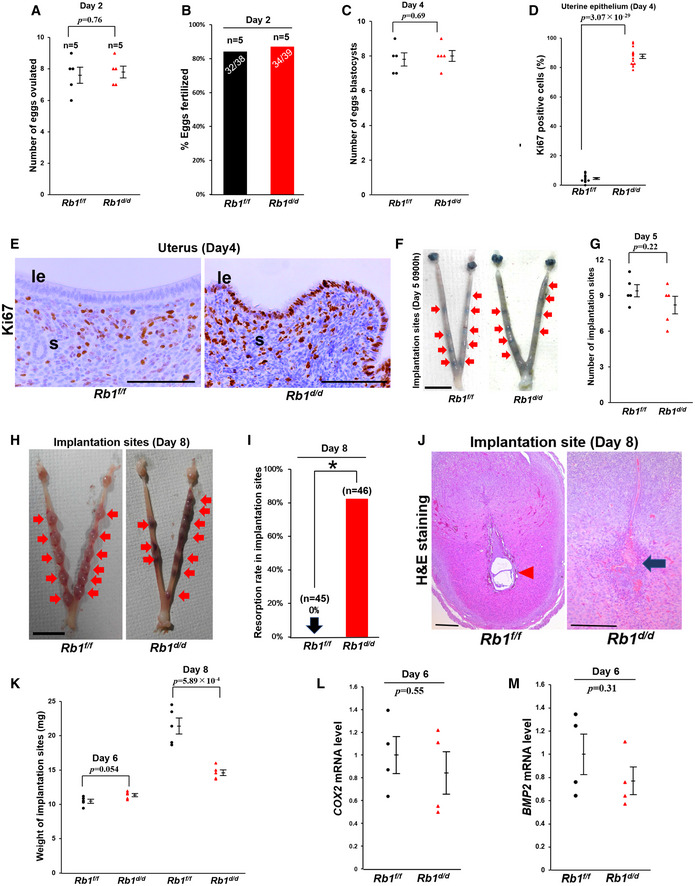
-
A–COvulation, fertilization, and development of pre‐implantation embryos were normal in Rb1d/d mice (A and C, mean ± SEM, Student’s t‐test; n = 5 mice for each group; B, Fisher's exact test; n = 5 mice for each group).
-
D, ENumber of Ki67‐positive cells on day 4 in uterine epithelium of Rb1d/d mice was higher than that of Rb1f/f mice (mean ± SEM, Student’s t‐test; n = 5 mice for each group). Scale bar = 100 μm; le, luminal epithelium; s, stroma.
-
F, GEmbryo attachment occurred normally in Rb1d/d mice at 09:00 h on day 5 (mean ± SEM, Student’s t‐test; n = 5 mice for each group). Arrow, implantation site; scale bar = 1 cm.
-
H, IRb1d/d mice showed higher rate of embryo resorption on day 8 (*P < 0.05; Fisher's exact test; n = 45 implantation sites for Rb1f/f mice and n = 46 implantation sites for Rb1d/d mice). Arrow, implantation site; scale bar = 1 cm.
-
JH&E staining showed embryo resorption in Rb1d/d mice on day 8 of pregnancy. Scale bar = 200 μm; arrowhead, embryo; arrow, broken embryo with blood cell infiltration.
-
KWeight of implantation site was comparable between Rb1f/f and Rb1d/d mice on day 6, but reduced on day 8 in Rb1d/d mice compared with Rb1f/f mice (mean ± SEM, Student’s t‐test; n = 6 mice for each group).
-
L, MExpression of Cox2 and Bmp2, markers of decidualization, was comparable in between Rb1d/d and Rb1f/f mice at 09:00 h on day 6 in implantation sites (mean ± SEM, Student’s t‐test; n = 4 mice for each group).
Data information: le, luminal epithelium; s, stroma; tr, trophoblast; n, nucleus.
Figure EV1. Embryo attachment occurs normally in Rb1d/d mice.
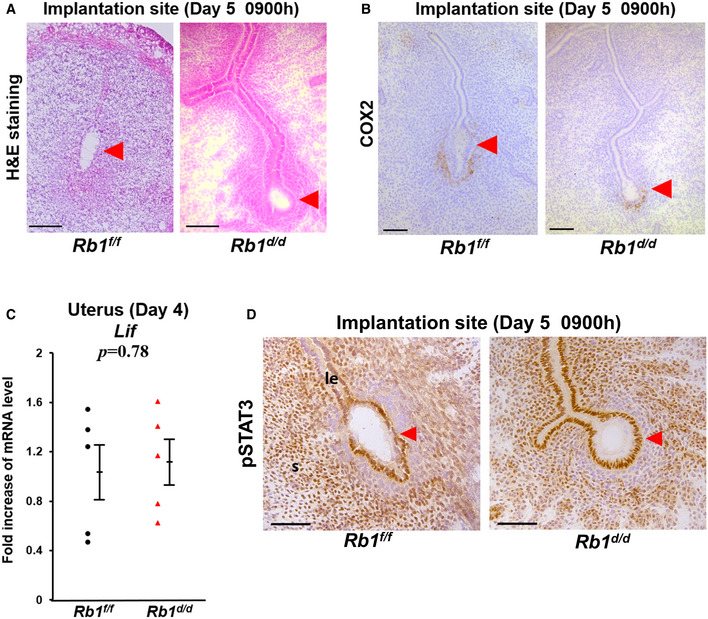
- H&E staining of implantation sites in Rb1f/f and Rb1d/d mice at 09:00 h on day 5 of pregnancy. Scale bar = 100 μm; arrowhead, embryo.
- Immunostaining of COX2, a marker of embryo attachment reaction in the implantation sites of Rb1f/f and Rb1d/d mice at 09:00 h on day 5 of pregnancy. Scale bar = 100 μm; arrowhead, embryo.
- Uterine expression of Lif mRNA on day 4 of pregnancy was comparable between Rb1f/f and Rb1d/d mice (mean ± SEM, Student’s t‐test; n = 5 mice for each group).
- Immunostaining of pSTAT3, a marker of activated LIF signaling in the implantation sites of Rb1f/f and Rb1d/d mice at 09:00 h on day 5 of pregnancy. Scale bar = 100 μm; le, luminal epithelium; s, stroma; arrowhead, embryo.
Data information: le, luminal epithelium; s, stroma.
Intact alignment of luminal epithelium surrounding the embryo and impaired trophoblast phagocytosis are associated with embryo invasion failure in Rb1d/d mice
We next assessed embryo invasion by investigating the elimination of luminal epithelium alignment and trophoblast invasion at the implantation site, which is a critical process for embryo invasion (Matsumoto et al, 2018). Cytokeratin 8 (CK8), a marker of epithelial cell lineage, was used to identify the uterine epithelium and trophoblast. Rb1f/f mice demonstrated that luminal epithelial alignment is collapsed and eliminated around the implantation site at 20:00 h on day 5, and the area of trophoblast invasion is expanded at 09:00 h on day 6 (Fig 3A). In contrast, Rb1d/d mice show that alignment of the luminal epithelium is persistent without trophoblast invasion at implantation sites at 20:00 h on day 5, and embryo invasion is compromised and limited at 09:00 h on day 6 (Fig 3A). Transmission electron microscopic analyses (TEM) of embryo–uterine interface at 20:00 h on day 5 demonstrated direct contact between trophoblast and stroma at the area of eliminated luminal epithelium in Rb1f/f mice but not in Rb1d/d mice (Fig 3B), suggesting the role of uterine Rb1 in trophoblast invasion by eliminating luminal epithelium alignment. These findings indicate that intact alignment of luminal epithelium is associated with embryo invasion failure in Rb1d/d mice. Surprisingly, TEM revealed that the cytoplasm of the luminal epithelium with lipid droplets is trimmed and engulfed by trophoblast cells, and its nucleus is not apparently condensed in control mice (Figs 3C and D), suggesting luminal epithelium elimination at implantation sites is due to trophoblast phagocytosis of the luminal epithelium. Dying cells exude phosphatidylserine (PS) in their outer membranes to enable phagocytic cell detection. To evaluate the presentation of PS in the outer cellular membrane of the luminal epithelium at the implantation site, we performed immunofluorescence of MFG‐E8 as a marker of PS in the outer membrane. Indeed, we found that MFG‐E8 is not present in the outer membrane of Rb1d/d mice but in that of Rb1f/f mice immediately before embryo invasion (Fig 3E). Cytoplasmic lipid droplets, which are present specifically in programmed cell death such as apoptosis and necroptosis (Boren & Brindle, 2012; Yamanaka et al, 2014; Zhu et al, 2016), were not observed in the luminal epithelium of Rb1f/f mice on day 4, but were apparent on day 5 evening (Fig 3D). In contrast, this phenomenon was not detected in the epithelium of Rb1d/d mice on day 5 evening (Fig 3B), indicating that cytoplasmic lipid droplets are characteristic findings of programmed cell death of the luminal epithelium during normal embryo invasion.
Figure 3. Rb1d/d mice show persistent alignment of luminal epithelium surrounding the embryo, impaired trophoblast phagocytosis, and defected embryo invasion.
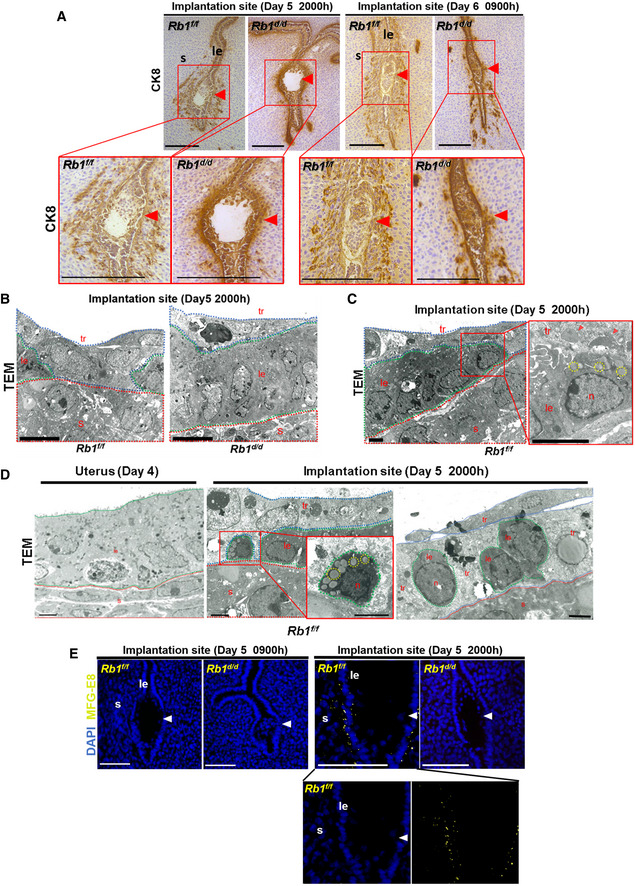
- Immunostaining of cytokeratin 8 (CK8), a marker of epithelial cell lineage, showed that epithelial alignment is collapsed and the trophoblast invades the stroma at implantation sites in Rb1f/f mice, while epithelial alignment is persistent and trophoblast invasion into the stroma is compromised in Rb1d/d mice. Scale bar = 200 μm; arrowhead, an embryo.
- Transmission electron microscopic analyses (TEM) showed that the uterine epithelium surrounding the embryo disappears and the trophoblast contacts directly with the stroma at 20:00 h on day 5 in Rb1f/f mice, while the uterine epithelium is persistent and the embryo does not attach to the stroma in Rb1d/d mice. Scale bar = 10 µm; red dotted line, stroma; green dotted line, luminal epithelium; blue dotted line, trophoblast.
- TEM showed that fragmented luminal epithelial cells with cytoplasmic lipid droplets are engulfed by the trophoblast in the control mice (Rb1f/f mice). Scale bar = 2 µm. Arrowhead, cytoplasmic fragments engulfed by trophoblast; yellow dotted line circle, cytoplasmic lipid droplets in the luminal epithelium; red dotted line, stroma; green dotted line, luminal epithelium; blue dotted line, trophoblast.
- TEM showed that the microvilli, markers of uterine receptivity, were observed at the surface of intact luminal epithelium on day 4 of pregnancy and epithelial cells with cytoplasmic lipid droplets were observed at 20:00 h on day 5 of pregnancy in the control mice (Rb1f/f mice). Scale bar = 2 µm; red dotted line, stroma; green dotted line, luminal epithelium; blue dotted line, trophoblast.
- Immunofluorescence of MFG‐E8, a marker of programmed cell death, showed that MFG‐E8 is not expressed at the implantation sites of Rb1f/f mice at 09:00 h on day 5 and Rb1d/d mice at 09:00 h and 20:00 h on day 5, but is expressed at the implantation sites of Rb1f/f mice at 20:00 h on day 5. Scale bar = 100 μm; blue signal, nuclei stained by DAPI; yellow signal, MFG‐E8; arrowhead, embryo.
Data information: le, luminal epithelium; s, stroma; tr, trophoblast; n, nucleus.
Pre‐implantation P4 supplementation is sufficient to normalize epithelial CCA and embryo invasion in Rb1d/d mice
To confirm whether ovarian functions of Rb1d/d mice other than ovulation and fertilization are normal, we first examined the expression of a key steroidogenic enzyme Hsd3b1 mRNA in the ovary and found that the levels of Hsd3b1 mRNA are comparable between Rb1d/d and Rb1f/f ovaries on day 4 of pregnancy (Fig EV2A). We next examined the circulating levels of estradiol‐17β (E2) and P4 during embryo implantation, and found that serum levels of E2 and P4 are comparable between Rb1d/d and Rb1f/f mice on days 4 and 6 (Fig 4A and B), suggesting that ovarian functions are normal in Rb1d/d mice in early pregnancy and the reproductive phenotypes of Rb1d/d mice are due to uterine Rb1 deletion. Since E2 and P4 act via nuclear receptors estrogen receptor α (ERα) and PGR, respectively, and we next investigated uterine ERα and PGR by immunostaining. The expression pattern of ERα and PGR in Rb1d/d uteri looked comparable to Rb1f/f ones (Fig EV2B and C). The expression of Forkhead Box O1 (FOXO1), which interacts with P4‐PGR signaling and regulates embryo implantation (Vasquez et al, 2018), was also comparable between these mouse uteri (Fig EV2D). In addition, uterine mRNA levels of E2‐ and P4‐ responsive genes, all of which were reported previously (Tranguch et al, 2007; Franco et al, 2012; Wetendorf et al, 2017), were comparable between Rb1d/d and Rb1f/f uteri on day 4 of pregnancy (Fig EV3A and B), suggesting uterine P4‐PGR signaling is normal. Since we have shown that epithelial CCA in the receptive uterus is induced by P4, we tried to examine whether exogenous supplementation of P4 in Rb1d/d mice recovers abnormal epithelial cell cycle status on day 4 morning. As expected, the injection of P4 (2 mg/day/mouse) on days 2 and 3 normalized epithelial cell cycle status in Rb1d/d mice on day 4 (Fig 4C–E). The administration of exogenous P4 on days 2 and 3 lead to the circulating P4 levels on day 4 of pregnancy as highly as those in Rb1f/f and Rb1d/d mice without P4 treatment on day 6 (Fig 4B). Daily P4 administration from day 2 to day 7 throughout the implantation period in Rb1d/d mice recovered resorption on day 8, suggesting P4 normalizes embryo invasion in Rb1d/d mice (Fig 4F and G; Group 1). Next, we examined whether the timing of P4 supplementation is critical for recovering embryo invasion in Rb1d/d mice. In Rb1d/d mice on day 8, the resorption rate in mice with post‐implantation P4 treatment on days 5–7 was as high as Rb1d/d mice without P4 treatment (Fig 4F and G; Group 3). In contrast, pre‐implantation P4 treatment on days 2 and 3 completely rescued the implantation sites of Rb1d/d mice (Fig 4F and G; Group 2). More importantly, pre‐implantation P4 treatment on days 2 and 3 recovered subfertility of Rb1d/d mice completely (Fig 4H). This pre‐implantation P4 treatment did not increase blood P4 levels on day 6 (Fig 4B), suggesting that the pre‐implantation P4 administration influences the uterus until embryo attachment. These findings indicate that pre‐implantation P4 supplementation is sufficient not only to normalize epithelial CCA but to recover embryo invasion and full‐term pregnancy in Rb1d/d mice.
Figure EV2. Normal expressions of ovarian Hsd3b1 mRNA and uterine estrogen receptor α (ERα), progesterone receptor (PGR), and FOXO1 proteins in Rb1d/d mice.
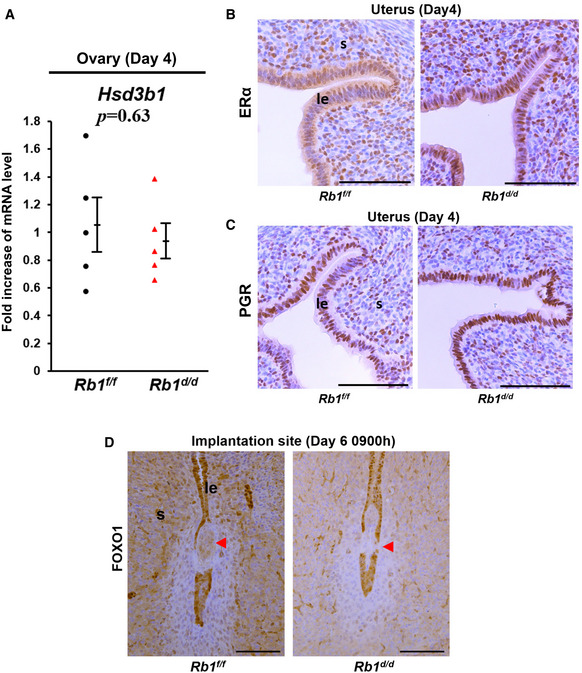
-
AQuantitative PCR demonstrated that ovarian expression of ovarian steroidogenic enzyme Hsd3b1 mRNA was comparable between Rb1f/f and Rb1d/d mice on day 4 of pregnancy (mean ± SEM, Student’s t‐test; n = 5 mice for each group).
-
B, CImmunohistochemistry demonstrated that uterine expression of ERα and PGR was comparable between Rb1f/f and Rb1d/d mice on day 4 of pregnancy. Scale bar = 100 μm.
-
DImmunohistochemistry demonstrated that uterine expression of FOXO1 was comparable between Rb1f/f and Rb1d/d mice on day 6 of pregnancy. Scale bar = 100 μm; arrowhead, embryo.
Data information: le, luminal epithelium; s, stroma.
Figure 4. Pre‐implantation progesterone (P4) supplementation is sufficient to normalize epithelial CCA and embryo invasion in Rb1d/d mice.
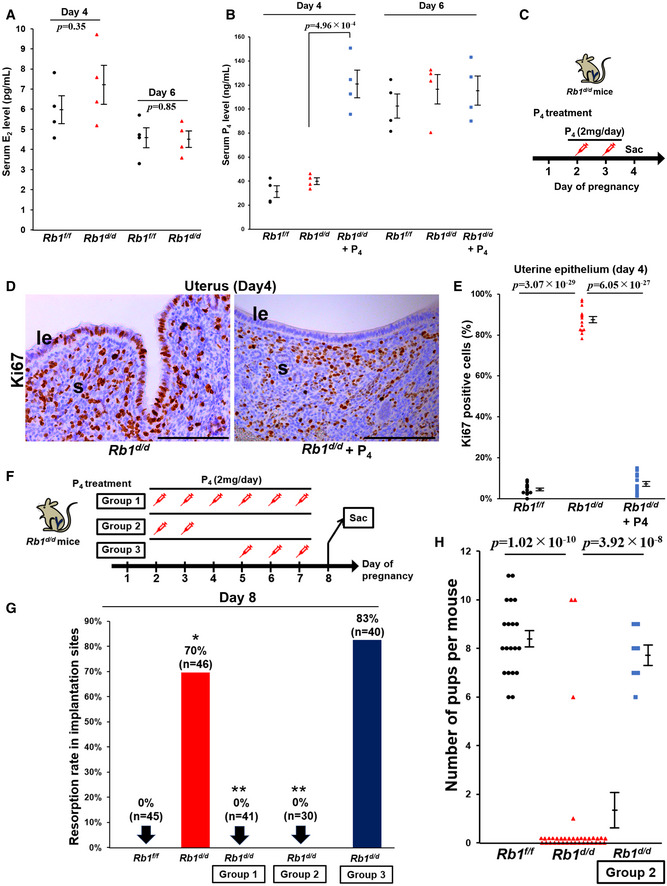
- Serum estradiol‐17β (E2) levels on days 4 and 6 of pregnancy were comparable between Rb1f/f and Rb1d/d mice (mean ± SEM, Student’s t‐test; n = 4 mice for each group).
- Serum P4 levels on days 4 and 6 of pregnancy were comparable between Rb1f/f and Rb1d/d mice. In Rb1d/d mice, pre‐implantation P4 treatment on days 2 and 3 increased serum P4 levels on day 4 but did not affect those on day 6 (mean ± SEM, Student’s t‐test; n = 4 mice for each group).
- Protocol of pre‐implantation P4 supplementation for the recovery of epithelial CCA in Rb1d/d mice. Rb1d/d mice underwent subcutaneous injection of P4 (2 mg/day/mouse) on days 2 and 3, and sacrificed on day 4.
- Persistent Ki67 expression in the epithelium of Rb1d/d mice on day 4 was recovered by pre‐implantation P4 treatment on days 2 and 3. Scale bar = 100 μm; le, luminal epithelium; s, stroma.
- Ratio of Ki67‐positive epithelial cells in Rb1d/d mice was completely suppressed by pre‐implantation P4 supplementation on days 2 and 3. The same data of Rb1f/f and Rb1d/d mice without P4 treatment in Fig 2D were used (mean ± SEM, Student’s t‐test; n = 5 different mice for each group). Three different high‐powered fields per mouse were analyzed, and each of the Ki67‐positive ratios was demonstrated.
- Protocol of P4 treatment for the recovery of embryo invasion and subfertility in Rb1d/d mice. P4 (2 mg/day/mouse) was injected to Rb1d/d mice on days 2–7 in Group 1, on days 2 and 3 in Group 2, and on days 5–7 in Group 3.
- Resorption rate was reduced on day 8 in Groups 1 (n = 41 different implantation sites) and 2 (n = 30 different implantation sites), but not in Group 3 (n = 40 different implantation sites). The same data of Rb1f/f and Rb1d/d mice without P4 treatment in Fig 2I were used. H&E staining was performed using all sections that show the implantation sites, and resorption rate was evaluated. *P < 0.05 vs. Rb1f/f mice; **P < 0.05 vs. Rb1d/d mice; Fisher's exact test.
- Number of pups delivered by Rb1d/d dams was normalized by pre‐implantation P4 treatment on days 2 and 3 (Group 2) (mean ± SEM, Student’s t‐test). The same data of Rb1f/f and Rb1d/d dams without P4 treatment in Fig 1E were used. The numbers of pups in 20 Rb1f/f and 37 Rb1d/d dams without P4 treatment and 7 Rb1d/d dams with P4 treatment were evaluated.
Figure EV3. Comparable uterine expressions of E2‐responsive and P4‐responsive genes in Rb1f/f and Rb1d/d mice on day 4 of pregnancy.
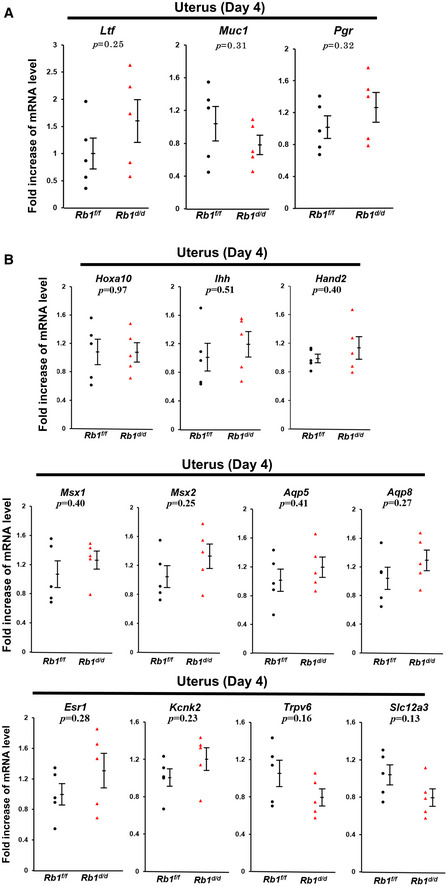
-
A, BQuantitative PCR demonstrated that uterine expression of E2‐responsive (A) and P4‐responsive genes (B) was comparable between Rb1f/f and Rb1d/d mice on day 4 of pregnancy (mean ± SEM, Student’s t‐test; n = 5 mice for each group).
TNFα‐related necroptosis of the luminal epithelium at the implantation site is impaired in Rb1d/d mice
Since our recent study has revealed that apoptosis is not a key mechanism for the initial step of the luminal epithelium disappearance around the embryo (Matsumoto et al, 2018), we hypothesized that the major machinery of luminal epithelium elimination is necroptosis, another type of programmed cell death. To clarify our hypothesis, we performed immunofluorescence of pRIP3, a marker of necroptosis, and found that pRIP3 is expressed in the membrane of Rb1f/f mice but not in that of Rb1d/d mice (Fig 5A). We also found that TNFα stimulated phosphorylation of MLKL, a critical signaling pathway of necroptosis, in primary mouse uterine epithelial cells (Fig 5B). Furthermore, uterine expression levels of TNFα was increased after embryo attachment in control mice (Rb1f/f mice) (Fig 5C), and both embryo and uterus at the implantation site produced TNFα in Rb1d/d mice as intensely as in Rb1f/f mice (Fig 5D), suggesting that TNFα‐induced necroptosis is compromised by the mechanism which is downstream of TNFα signaling in Rb1d/d mice.
Figure 5. Necroptosis of luminal epithelium at the implantation sites is observed in Rb1d/d mice.
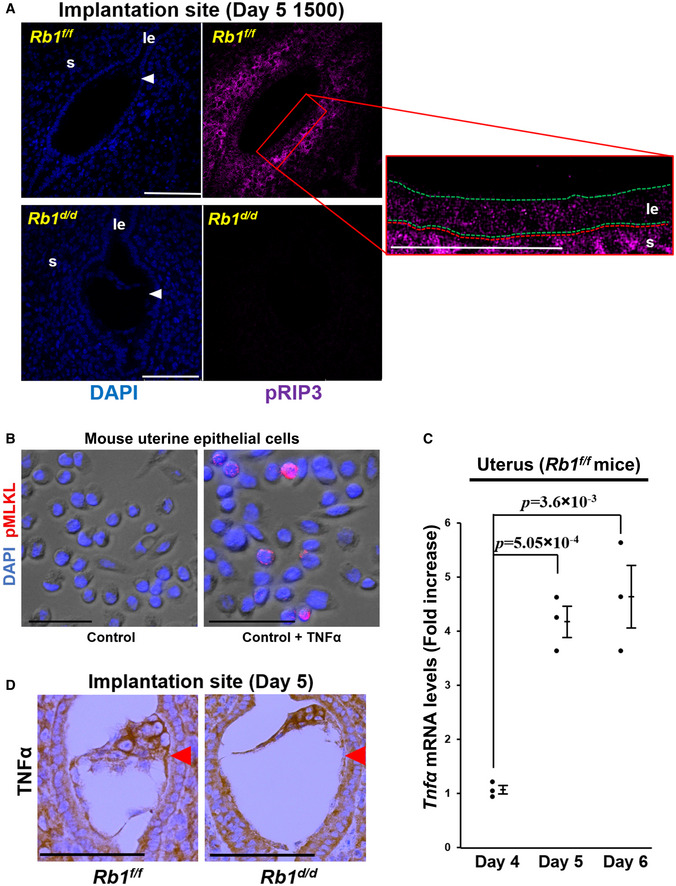
- pRIP3, a central mediator of necroptosis, was not expressed at the implantation sites of Rb1d/d mice but was expressed at those of Rb1f/f mice at 15:00 h on day 5. Scale bar = 100 μm; blue signal, nuclei stained by DAPI; purple signal, pRIP3; le, luminal epithelium; s, stroma; arrowhead, embryo; green dotted line, luminal epithelium; red dotted line, stroma.
- TNFα stimulated the expression of pMLKL, a critical mediator of necroptosis, in the primary mouse uterine epithelial cells. Scale bar = 50 μm; blue signal, nuclei stained by DAPI; red signal, pMLKL.
- Uterine expression of TNFα was elevated in the control mice (Rb1f/f mice) on days 5 and 6 compared with on day 4 (mean ± SEM, Student’s t‐test; n = 3 mice for each group). As for the uterine samples on days 5 and 6, those with implantation sites were examined.
- The implanting embryo and the uterus produced TNFα at the implantation site of both Rb1f/f and Rb1d/d mice. Immunostaining of TNFα at the implantation sites of Rb1f/f and Rb1d/d mice were performed. Scale bar = 100 μm; arrowhead, embryo.
P4‐induced CCA of uterine epithelial cells contributes to susceptibility to TNFα and TNFα‐primed necroptosis through TNF receptor type 2
PS presentation in the outer membrane is associated with necroptosis (Guo et al, 2009; Ehrhardt et al, 2013; Zargarian et al, 2017). We evaluated the effects of TNFα on necroptosis of primary mouse uterine epithelial cells obtained from Rb1f/f and Rb1d/d mice. The expression of annexin V, a marker of PS presentation in the outer membrane, was not stimulated by TNFα in Rb1‐deficient uterine epithelial cells but in the control cells (Fig 6A and B), suggesting that TNFα‐primed necroptosis in the uterine epithelium is controlled by Rb1. We investigated whether P4, which promotes CCA in uterine epithelial cells during embryo implantation (Haraguchi et al, 2014), rescues TNFα‐primed necroptosis in Rb1‐deficient uterine epithelial cells. Supplementation of P4 restored the responsiveness to TNFα and induced necroptosis in Rb1‐deficient uterine epithelial cells (Fig 6A and B), suggesting that TNFα‐primed epithelial necroptosis is controlled by CCA. Thymidine stimulates CCA in G1/S phase in many cell types (Harper, 2005; Chen & Deng, 2018), which was confirmed in primary mouse uterine epithelial cells (Fig EV4A). We next used thymidine to evaluate the role of CCA on TNFα‐primed necroptosis in the uterine epithelial cells. We found that TNFα administration to Rb1‐deleted cells does not increase the expression of annexin V, but the concomitant treatment of thymidine restores TNFα‐primed expression of annexin V (Fig 6C and D), indicating that TNFα‐primed epithelial necroptosis is regulated by CCA. Furthermore, we found that the expression of TNF receptor type 2 (TNFR2) is downregulated in Rb1‐deleted epithelial cells, and the treatment of CCA inducers thymidine or P4 upregulates its expression (Fig 6E). We confirmed that CCA promotes TNFα‐primed necroptosis in a human endometrial epithelial cell line HEC151 (Fig EV4, EV5), suggesting CCA‐induced necroptosis is a common phenomenon in the cells whose growth is ovarian hormone‐dependent. These findings indicate that Rb1‐induced CCA of uterine epithelial cells contributes to TNFα‐primed necroptosis by the upregulation of TNFR2. We believe that this is a novel physiological machinery of luminal epithelium elimination during embryo invasion.
Figure 6. CCA of uterine epithelial cells contributes to susceptibility to TNFα and TNFα‐primed necroptosis through TNF receptor type 2.
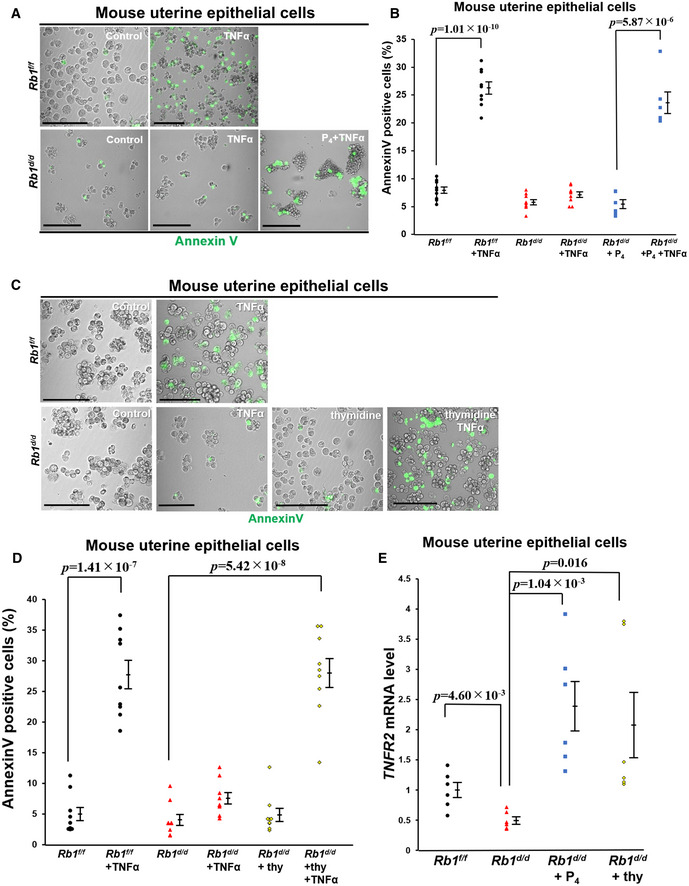
-
A, BIn vitro analyses using primary mouse uterine epithelial cells demonstrated that TNFα induces the expression of annexin V, a marker of PS presentation at the outer membrane of Rb1f/f epithelial cells but does not in Rb1d/d ones, and P4 restores TNFα‐primed annexin V expression in Rb1d/d uterine epithelial cells (mean ± SEM, Student’s t‐test). Using three lines of primary mouse epithelial cells obtained from both Rb1f/f and Rb1d/d mice, the in vivo experiments were performed three times. Each of the annexin V‐positive ratios was demonstrated. Scale bar = 100 μm; green signal, annexin V.
-
C, DAnnexin V assay using primary mouse uterine epithelial cells showed that TNFα administration increases the expression of annexin V in growth‐arrested mouse epithelial cells with thymidine treatment, but does not in the control cells (mean ± SEM, Student’s t‐test). Using three lines of primary mouse epithelial cells obtained from both Rb1f/f and Rb1d/d mice, the in vivo experiments were performed three times. Each of the annexin V‐positive ratios was demonstrated. Scale bar = 100 μm; green signal, annexin V.
-
EThe expression of TNF receptor type 2 (TNFR2) was upregulated in growth‐arrested epithelial cells with thymidine and P4 supplementation (mean ± SEM, Student’s t‐test). Using three lines of primary mouse epithelial cells obtained from both Rb1f/f and Rb1d/d groups, qPCR was performed in duplicate.
Figure EV4. Thymidine‐induced cell cycle arrest in primary mouse epithelial cells and HEC151 endometrial epithelial cell line.

-
AThymidine‐induced cell cycle arrest in primary mouse epithelial cells. Mouse uterine epithelial cells were stained with propidium iodine, and cell fractions were quantified by flow cytometry.
-
B, CAnnexin V assay using a human endometrial epithelial cell line HEC151 showed that TNFα administration increases the expression of annexin V in growth‐arrested HEC151 cells with thymidine treatment, but does not in the control cells (mean ± SEM, Student’s t‐test). The assay was performed in duplicate for each group, and the examination was performed three times.
-
DThe expression of TNF receptor type 2 (TNFR2) was upregulated in growth‐arrested HEC151 cells with thymidine treatment (mean ± SEM, Student’s t‐test). Quantitative PCR was performed in quadruplicate for each group.
Figure EV5. RNA‐seq analyses using uterine epithelium of Rb1f/f and Rb1d/d mice on day 4 of pregnancy.
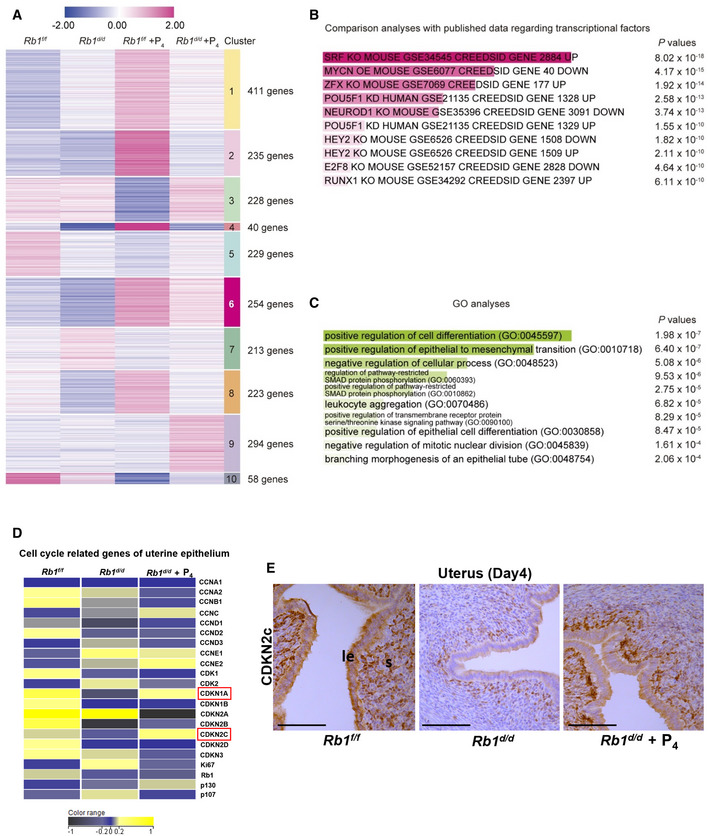
- Day 4 uteri were collected from Rb1f/f, Rb1d/d, P4‐primed Rb1f/f, and P4‐primed Rb1d/d mice. As for P4‐primed Rb1f/f and Rb1d/d mice, pre‐implantation P4 treatment (2 mg/mouse/day) was performed on days 2 and 3 of pregnancy. RNA‐seq was performed using the uterine luminal epithelium dissected out by laser capture microdissection. The heatmaps depicted log2 fold enrichment of differentially expressed genes (DEGs) among the uterine epithelium. Clusters were defined by k‐means clustering. Each DEG has a log2 fold change > |1| in at least one group comparing to others. Numbers shown in right indicate different gene clusters and numbers of DEGs in each cluster.
- Gene regulatory systems in the gene cluster 6 in which the transcripts are poorly expressed in Rb1d/d mice without P4 supplementation compared with other groups were predicted in Enrichr by comparing our data with publicly available datasets as for transcriptional factors. DEGs in the cluster 6 were highly correlated with cell cycle‐related TFs such as serum response factor (SRF) and E2F8.
- GO terms analyses by Enrichr for the cluster 6 demonstrated that the genes related to epithelial differentiation and inhibition of cell proliferation are downregulated in the luminal epithelium of Rb1d/d mice without P4 treatment.
- The heatmap of cell cycle‐related genes in the luminal epithelium of Rb1f/f and Rb1d/d mice with and without pre‐implantation P4 treatment was demonstrated. Cyclin‐dependent kinase inhibitors CDKN2A (p21) and CDKN2C (p18) were upregulated in Rb1f/f and Rb1d/d mice with P4 treatment compared with Rb1d/d mice without P4 treatment. Yellow, blue, and black indicate high, intermediate, and low gene expression, respectively.
- Immunohistochemistry of CDKN2C (p18) protein in the uterus of Rb1f/f and Rb1d/d mice with and without P4 treatment. Same as transcriptome analysis, CDKN2C staining was intense in the uteri of Rb1f/f and Rb1d/d mice with P4 treatment compared with Rb1d/d mice without P4 treatment. Scale bar = 100 μm; le, luminal epithelium; s, stroma.
Pre‐implantation P4 rescues necroptosis of the luminal epithelium at the implantation site in Rb1d/d mice
We examined whether P4 supplementation recovers epithelial necroptosis in Rb1d/d mice. Pre‐implantation administration of P4 recovered the fragmented cytoplasm with lipid droplets in the luminal epithelium and trophoblast engulfment (Fig 7A) and normalized the expression of MFG‐E8 and pRIP3 in the luminal epithelium of Rb1d/d mice (Fig 7B and C). These findings suggest that P4 rescues epithelial CCA and necroptosis, trophoblast engulfment, and subsequent embryo invasion in Rb1d/d mice.
Figure 7. Pre‐implantation P4 administration recovers epithelial necroptosis and trophoblast phagocytosis at the implantation site in Rb1d/d mice.
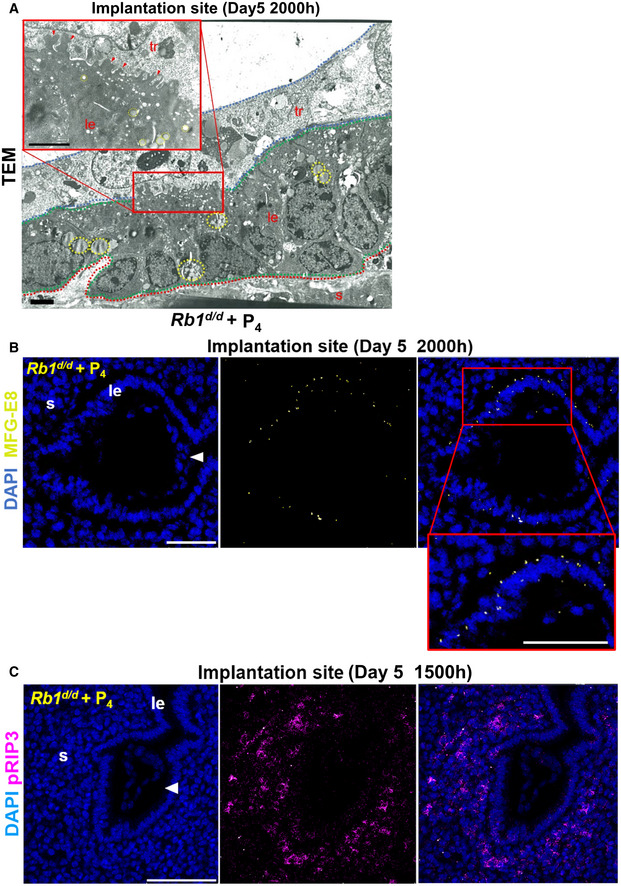
- In Rb1d/d mice with pre‐implantation P4 treatment, the fragmented uterine epithelial cells with the cytoplasmic lipid droplets were engulfed by the trophoblast cells. Scale bar = 2 µm. Arrowhead, cytoplasmic fragments engulfed by trophoblast; dotted line circle, lipid droplets in the cytoplasm; tr, trophoblast; le, luminal epithelium; s, stroma; red dotted line, stroma; green dotted line, luminal epithelium; blue dotted line, trophoblast.
- Pre‐implantation P4 treatment rescued the expression of MFG‐E8 on day 5 of pregnancy in Rb1d/d uteri. Scale bar = 100 μm. Blue signal, nuclei stained by DAPI; yellow signal, MFG‐E8; le, luminal epithelium; s, stroma; arrowhead, embryo.
- Pre‐implantation P4 treatment recovered the expression of pRIP3, a mediator of necroptosis, on day 5 of pregnancy in Rb1d/d uteri. Scale bar = 100 μm; blue signal, nuclei stained by DAPI; purple signal, pRIP3; le, luminal epithelium; s, stroma; arrowhead, embryo.
Rb1 deletion in the luminal epithelium skews cell cycle‐related genes
To examine the influence of Rb1 and P4 on transcriptome in the luminal epithelium, we performed RNA‐seq using day 4 luminal epithelium dissected out from Rb1f/f mice and Rb1d/d mice with and without P4 supplementation by laser capture microdissection (LCM). We found 2,185 differentially expressed genes (DEGs) which showed log2‐fold changes ≥ 1 in at least one group compared with others (Fig EV5A). Among 10 unique clusters defined by K‐means clustering, we focused on the gene cluster 6 in which the transcripts are poorly expressed especially in Rb1d/d mice without P4 supplementation compared with other groups. To examine the feature of the cluster 6, we examined the profile of the genes which belong to this cluster using publicly available database of RNA‐seq and ChIP‐seq regarding transcriptional factors (TFs). We found that DEGs in this cluster are poorly correlated with PGR but highly correlated with cell cycle‐related TFs such as serum response factor (SRF) and E2F8 (Fig EV5B). In addition, gene ontology (GO) analyses also revealed that the cluster 6 is related to epithelial differentiation and inhibition of cell proliferation (Fig EV5C). These results indicate that P4 administration to Rb1d/d mice rescues embryo invasion failure by specifically influencing the cell proliferation pathway of the luminal epithelium. Among cell cycle‐associated genes, mRNAs of cyclin‐dependent kinase inhibitors Cdkn1a and Cdkn2c were downregulated by Rb1 deletion and upregulated by P4 treatment, suggesting that CCA in the luminal epithelium is regulated by Cdkn1a and Cdkn2c (Fig EV5D). Consistent with RNA‐seq results, CDKN2c protein was expressed less in Rb1d/d mice compared with Rb1f/f mice, and exogenous P4 increased CDKN2c expression in Rb1d/d mice (Fig EV5E). Cyclin‐dependent kinase inhibitors Cdkn1a and Cdkn2c might work together in epithelial necroptosis.
Stromal Rb1 controls epithelial proliferation and embryo invasion in cooperation with epithelial Rb1
We found that RB is localized in the epithelium and stroma on day 4. These findings indicate the roles of Rb1 not only in the epithelium but in the stroma during the peri‐implantation period. To clarify the role of epithelial Rb1 in embryo implantation, we generated epithelial Rb1‐deleted mice (Rb1 eKO) by crossing Rb1‐loxP mice with Ltf‐Cre mice. Cre‐negative homozygous littermate female mice for the conditional alleles were used as controls (Rb1 eControl). Although RB was efficiently deleted in the uterine epithelium of Rb1 eKO mice (Fig 8A), Rb1 eKO female mice showed normal fertility (Fig 8B). Notably, epithelial CCA on day 4 was normal in Rb1 eKO mice (Fig 8C), suggesting that stromal Rb1 controls epithelial cell proliferation and embryo invasion in cooperation with epithelial Rb1 during embryo implantation.
Figure 8. Stromal Rb1 controls epithelial proliferation and embryo invasion in cooperation with epithelial Rb1 .
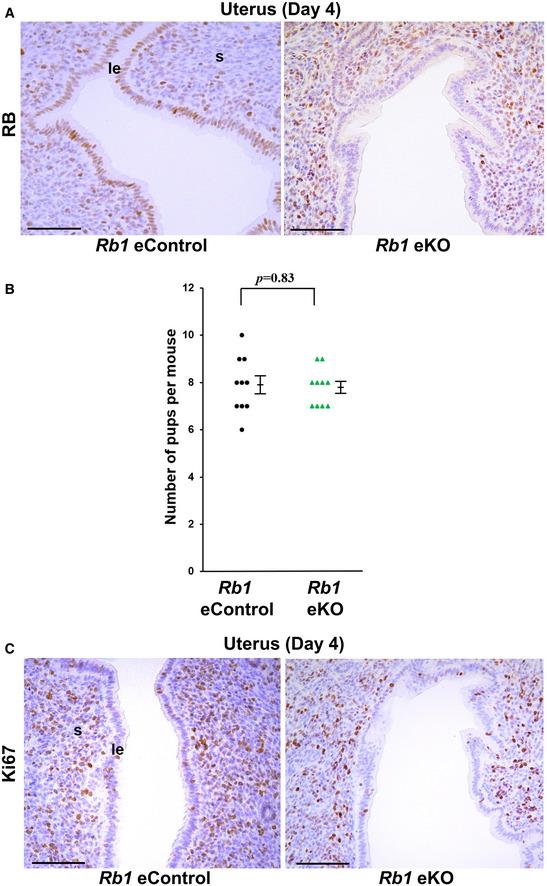
- RB was efficiently deleted in the uterine epithelium of uterine epithelium‐specific conditional knockout mice (Rb1 eKO mice). Scale bar = 100 μm. Rb1 eKO mice, Rb1loxP/loxPLtfCre /+ mice; Rb1 eControl mice (littermate controls of Rb1 eKO mice), Rb1loxP/loxP mice; le, luminal epithelium; s, stroma.
- Number of new‐born pups was comparable between Rb1 eKO and eControl dams (mean ± SEM, Student’s t‐test; n = 10 different dams for each group).
- The expression patterns of Ki67 on day 4 in uterine epithelium were comparable between Rb1 eKO and Rb1 eControl mice. Scale bar = 100 μm; le, luminal epithelium; s, stroma.
Appropriate CCA of the luminal epithelium is associated with embryo implantation outcome in patients undergoing infertility treatment
We investigated cell cycle status at the time of implantation in human endometria obtained from patients undergoing IVF‐ET treatment to evaluate the correlation between the extent of epithelial CCA and the clinical outcome of embryo transfer following endometrial biopsy. The ratio of Ki67‐positive staining was low in the group of patients with successful clinical pregnancy compared with that of patients without pregnancy (Fig 9A and B), suggesting that CCA in the uterine luminal epithelium is involved in human embryo implantation.
Figure 9. CCA of uterine luminal epithelial cells is a hallmark of embryo implantation in mice and humans.
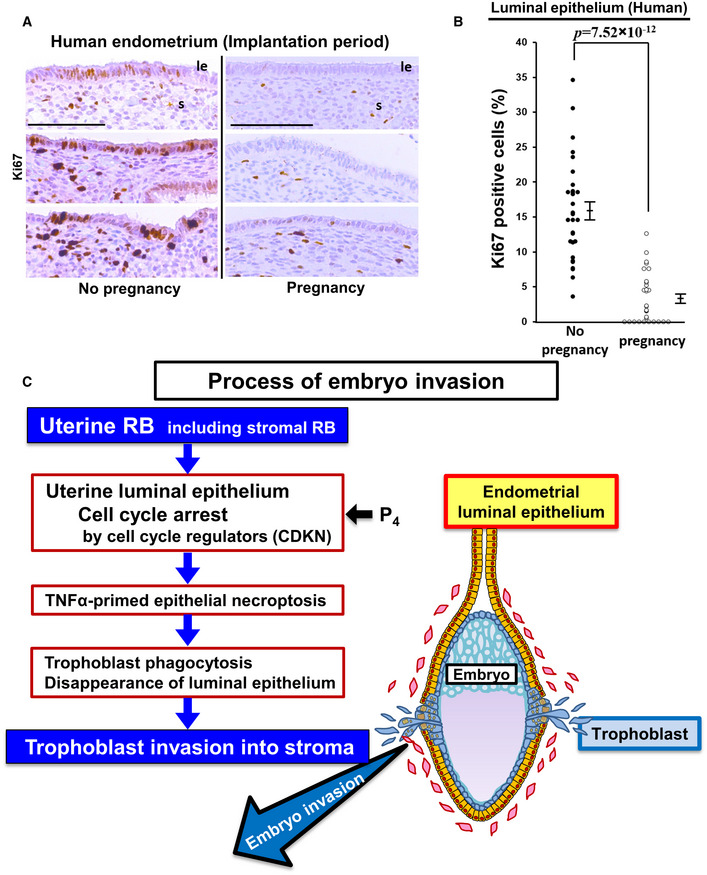
- Representative pictures of Ki67 staining in the human endometria obtained from women at the time of embryo implantation. Samples were divided into two groups according to the outcome of clinical pregnancy following endometrial biopsy. Scale bar = 100 μm; le, luminal epithelium; s, stroma.
- Ratio of Ki67‐positive cells in the human uterine luminal epithelium during the implantation period was lower in the group with pregnancy than the group without pregnancy (mean ± SEM, Student’s t‐test; n = 10 different individuals for each group). Three different high‐powered fields per sample were analyzed and each of the Ki67‐positive ratios was demonstrated.
- Potential processes that contribute to embryo invasion through uterine RB‐induced CCA and necroptosis of luminal epithelial cells.
Discussion
In the present study, we found that uterine deficiency of Rb1 results in persistent cell cycle progression of the luminal epithelium until embryo attachment, failure in the disappearance of luminal epithelial cells during embryo invasion, defects in the presentation of PS on the outer membrane of luminal epithelial cells, and loss of luminal epithelium necroptosis with trophoblast phagocytosis, suggesting that CCA of the uterine luminal epithelium stimulates necroptosis during embryo invasion. Importantly, we also found that thymidine‐ and P4‐induced CCA recovers TNFα‐primed necroptosis through the induction of TNFR2 in Rb1‐deficient epithelial cells. Pre‐implantation P4 administration normalizes the cell cycle status of the luminal epithelium at the time of embryo attachment, induces epithelial necroptosis with trophoblast phagocytosis during embryo invasion, and finally rescues embryo invasion in uterine Rb1 null mice. These findings indicate that proper CCA and necroptosis in uterine luminal epithelium are precisely regulated by Rb1 and P4 during implantation, which results in successful embryo invasion (Fig 9C). In addition, epithelial CCA at the time of attachment is associated with successful implantation in humans.
Necroptosis, a relatively new concept of non‐apoptotic programmed cell death, is augmented by TNFα which activates the RIP1‐RIP3‐MLKL axis by phosphorylation (Guo et al, 2009), and phosphorylated MLKL (pMLKL) drills the cellular membrane like necrosis (Zhu et al, 2016). The initial disappearance of the luminal epithelium barrier surrounding the blastocyst occurs at the lateral sides of the implantation chamber. Our previous study has revealed that apoptosis of the luminal epithelium is not a key mechanism for the initial step of the luminal epithelium disappearance around the embryo (Matsumoto et al, 2018). A recent study demonstrated that entosis, a non‐apoptotic form of cell death with cell‐in‐cell formation (Overholtzer et al, 2007), is involved in this process in which a live trophoblast cell engulfs and kills an entire live luminal epithelial cell to remove the luminal epithelium barrier (Li et al, 2015). The present study revealed that at the lateral sides of the implantation chamber, PS is exposed at the outer membranes of luminal epithelium, pRIP3 is expressed in the cytoplasm of luminal epithelium, and dying epithelial cells are engulfed by the neighboring trophoblast cells. It has been reported that PS exposure is observed in not only apoptotic but non‐apoptotic cell death including necroptosis and entosis, and pRIP3 is a specific marker of necroptosis, and phagocytes engulf live cells in the process of entosis and dying cells in the processes of apoptosis and necroptosis (Segawa et al, 2018; Shlomovitz et al, 2019; Tang et al, 2019). Our findings indicate that necroptosis is involved in luminal epithelium elimination at the lateral sides of the implantation chamber during the initial disappearance of luminal epithelium, which is a similar physiological phenomenon to synaptic pruning (Chechik et al, 1999). Given that epithelial entosis occurs at the lateral sides of the implantation chamber where epithelial necroptosis takes place, at least it is certain that non‐apoptotic cell death mechanisms including necroptosis and entosis play major roles in the initial step of luminal epithelium barrier removal during embryo invasion. In the following step, apoptosis occurs in the luminal epithelium at the anti‐mesometrial side (Joswig et al, 2003; Li et al, 2015). Apoptotic and non‐apoptotic cell death mechanisms regulate the luminal epithelium disappearance in the spaciotemporal manners.
Uterine stromal–epithelial communication contributes to successful embryo implantation. The basic helix–loop–helix transcription factor Hand2 in the uterine stroma suppresses the production of fibroblast growth factors that act as paracrine inducers of estrogen‐dependent epithelial proliferation in the peri‐implantation period (Li et al, 2011), suggesting that stromal Hand2 is in charge of P4‐dependent communication from stroma to epithelium. Previous studies reported by us and others suggested that uterine stroma regulates embryo invasion through detachment of the luminal epithelium (Schlafke & Enders, 1975; Matsumoto et al, 2018). Our recent study demonstrated that stromal HIF2α, a major transcriptional factor inducible by low oxygen tension, contributes to trophoblast invasion by inducing the expression of MT2‐MMP, lysyl oxidase, VEGF, and adrenomedullin, provoking the luminal epithelium detachment from the stroma, and activating the embryonic PI3K‐AKT pathway (Matsumoto et al, 2018). The present study showed that stromal Rb1 works in cooperation with epithelial Rb1 to regulate epithelial CCA and embryo invasion. These findings reinforce the critical stromal functions for embryo invasion. GO analysis demonstrated that the gene cluster in which the transcripts were downregulated in the luminal epithelium of Rb1d/d mice without P4 treatment compared with other groups was not related to inflammation but to epithelial differentiation and inhibition of cell proliferation (Fig EV5C), suggesting the pathological processes other than inflammation in the luminal epithelium may cause embryo invasion failure in Rb1d/d mice.
Dying cells exude PS at their outer membranes to enable detection by phagocytic cells, which is called an “eat‐me signal” (Hanayama et al, 2002). MFG‐E8 secreted by phagocytic cells binds PS at the outer membrane and is therefore used for a marker of eat‐me signals (Hanayama et al, 2002). In addition, eat‐me signals are activated in not only apoptosis but necroptosis which TNFα promotes (Guo et al, 2009; Ehrhardt et al, 2013; Zargarian et al, 2017). This study revealed that the implanting embryo and uterus at the implantation site produce TNFα which promotes the presentation of PS at the outer membrane in the luminal epithelium, and MFG‐E8 is present at the outer membrane of the luminal epithelium. This indicates that TNFα produced at the implantation site stimulates necroptosis of the luminal epithelial cells, and MFG‐E8 facilitates the trophoblast seeking out dying luminal epithelial cells and finally engulfing the cytoplasmic fragments of luminal epithelial cells at the embryo–uterine interface. Therefore, we believe that TNFα and MFG‐E8 are involved in epithelial necroptosis, and these molecular and cellular interactions between the luminal epithelium and trophoblast are critical for embryo invasion.
Ki67 is not expressed in the nucleus during the G0 stage when the cell cycle is arrested (Schwarting et al, 1986). We found that Ki67‐positive cells of the luminal epithelium decrease in the process of normal implantation on day 4, and the proliferating epithelial cells remain persistent in Rb1 null mice with failed epithelial necroptosis and embryo invasion. Cells with CCA are susceptible to TNFα, which promotes cell death (Ehrhardt et al, 2013; Zargarian et al, 2017). The current study revealed that TNFα is abundantly produced at the implantation site and CCA inducers thymidine and P4 rescue TNFα‐primed necroptosis in Rb1‐deficient growth‐arrested uterine epithelial cells through the upregulation of TNFR2. These findings indicate that the luminal epithelium with Rb1‐ and P4‐induced CCA is susceptible to implantation site‐derived TNFα, which induces epithelial necroptosis and embryo invasion.
Influence of P4 during pregnancy remains elusive, and it is unclear how P4 supports successful embryo implantation. P4 inhibits estrogen‐induced cell cycle progression of endometrial epithelial cells, which is a fundamental role of P4 to prevent endometrial cancer as well as an infertility treatment for the luteal phase of hormonal support. Our study revealed novel mechanisms in which RB and P4 regulate endometrial epithelial CCA and necroptosis precisely and lead to successful embryo invasion. This study may improve understanding of both molecular and cellular aspects of embryo implantation in humans.
Materials and Methods
Mice
WT mice (C57BL/6J, SLC, Hamamatsu, Japan), Pgr‐Cre mice (Soyal et al, 2005), Ltf‐Cre mice (Daikoku et al, 2014), and Rb1‐floxed mice (Marino et al, 2000) were used in this study. Ltf and Pgr are expressed in the luminal epithelium and all the layers of uterus, respectively. In Ltf‐Cre female mice, Cre recombinase is specifically expressed in the uterine epithelium (Daikoku et al, 2014). In the previous study, the floxed gene was specifically deleted in the uterine epithelium of the pregnant mouse model using Ltf‐Cre female mice (Matsumoto et al, 2018). In Pgr‐Cre female mice, Cre recombinase is expressed not only in the uterus but in the ovary (Soyal et al, 2005). Although Pgr‐Cre mouse line can be used for deletion of the floxed gene in the entire uterus (Matsumoto et al, 2018), the events regulated by ovarian functions such as ovulation and fertilization need to be evaluated in the pregnant mouse models using Pgr‐Cre females (Haraguchi et al, 2019). Mice with deletion of Rb1 in the whole uterus (Rb1d/d mice) were generated by crossing Pgr‐Cre with Rb1‐floxed mice. Mice with deletion of Rb1 in the uterine epithelium (Rb1 eKO mice) were generated by crossing Rb1‐floxed mice with Ltf‐Cre mice. Cre‐negative homozygous littermates for the conditional alleles were used as controls. All mice used in this investigation were housed in the University of Tokyo Animal Care Facility according to the institutional guidelines for the use of laboratory animals. The experimental procedures were approved by the institutional animal experiment committee.
Analysis of pregnancy events
For the evaluation of pregnancy phenotypes, female mice of each line (2‐to 5‐month‐old) were mated with fertile males, and day 1 of pregnancy was defined as the day when we recognized a vaginal plug. Ovulation, fertilization, pre‐implantation embryo development, and implantation were assessed as described (Hirota et al, 2010; Matsumoto et al, 2018). Daily subcutaneous injections of P4 (2 mg/mouse/day) to Rb1d/d mice were performed from day 2 of pregnancy or from day 5 of pregnancy. Daily P4 injection at the dose of 2 mg/day/mouse was used for the induction of embryo implantation in delayed implantation mouse model (Yoshinaga & Adams, 1966; Huet & Dey, 1987; Ma et al, 2003). In our previous study, we confirmed that daily administration of P4 at the dose of 2 mg/day/mouse from day 2 is not harmful for embryo implantation in WT mice (Matsuo et al, 2020).
Human tissues
Endometrial tissues were obtained from infertile patients planning to undergo IVF‐ET treatment. As the limitation of the experiments using human subjects, it is practically impossible to perform human endometrial biopsy in the implantation period of the embryo transfer cycle. Since the endometrial genes related to uterine receptivity are expressed in a reproducible fashion among the cycles (Diaz‐Gimeno et al, 2013), we consider that the uterine receptivity‐associated uterine cell proliferation status in the cycle prior to the embryo transfer cycle corresponds to that in embryo transfer cycle. Thus, we substituted the endometria in the cycle prior to embryo transfer instead. Endometrial biopsy was performed at the 7th day of P4 treatment in an artificial cycle which is considered as the implantation period in humans. To minimize the differences of hormonal conditions among the individual patients, the same protocol of artificial hormonal treatment in the cycle of endometrial biopsy was used for all the patients. The patients underwent embryo transfer in the following artificial cycles after endometrial biopsy. We investigated the relationship between the peri‐implantation uterine cell proliferation status and the pregnancy outcome. The samples were divided into two groups according to the outcome of clinical pregnancy following endometrial biopsy. Endometrial samples were evaluated by immunostaining of Ki67. Experimental procedures were approved by the institutional review board of the University of Tokyo (IRB number: 10991), and signed informed consent for the use of tissues was obtained from each patient. The experiments were performed according to the principles in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
H&E staining, immunostaining, and fluorescence assay
H&E staining, immunohistochemistry, and fluorescence assay using frozen sections (12 μm), or formalin‐fixed paraffin‐embedded sections (6 μm), were performed as described previously (Matsumoto et al, 2018). Antibodies to Ki67 (SP6, Thermo Fisher Scientific, Cheshire, UK 1:400), and pSTAT3 (EP2147Y, Abcam, Cambridge, UK 1:300), COX2 (AA570‐598, Cayman, Ann Arbor, MI, USA 1:200), CK8 (AB531826, DSHB, Iowa City, IA, USA 1:100), RB (ab181616, Abcam, 1:200), pRB (8516, Cell Signaling Technology, Beverly, MA, USA, 1:200), TNFα (BS1857, Bioworld Technology, Nanjing, China 1:500), ERα (ab80922, Abcam, 1:400), PGR (ab63605, Abcam, 1:200), and FOXO1 (2880S, Cell Signaling Technology, 1:200) were used.
Measurement of serum P4 and E2 levels
Blood samples from mice were collected on the indicated days of pregnancy. Serum P4 and E2 levels were measured as described previously (Matsumoto et al, 2018). Serum P4 levels were measured by Progesterone EIA kit (582601, Cayman), and E2 levels were measured by Estradiol ELISA kit (ES180S‐100, CalBiotech, Spring Valley, CA, USA).
Transmission Electron Microscopy (TEM)
TEM was performed as described previously (Matsumoto et al, 2018).
Culture of primary mouse uterine epithelial cells
Mouse uterine epithelial cells are obtained from Rb1f/f and Rb1d/d mice on day 4 of pregnancy. Briefly, uterine tissues were incubated with pancreatin (Sigma‐Aldrich, St. Louis, MO, USA) and dispase (Fuji Film Wako Pure Chemicals, Osaka, Japan) in HBSS at 4°C for 30 min, at room temperature for 30 min and then, at 37°C for 4 min. The tissue debris was removed by centrifugation, and the supernatant was passed through 70 μm cell strainer, and the epithelial cells were collected on the mesh of the cell strainer. The cells were seeded in collagen I‐coated plate and cultured in MEM with 10% charcoal‐stripped FBS for 2 days. Cultured epithelial cells were pre‐incubated in the media with or without 10−7 M of P4 for 1 h and then incubated in those with or without 60 ng/ml of mouse recombinant TNFα (Sigma‐Aldrich) for 4 h at 37°C. To induce CCA in the primary mouse uterine epithelial cells, the method of double thymidine block was used (Chen & Deng, 2018). Briefly, the cells were seeded in plastic plate and cultured in MEM with 5% charcoal‐stripped FBS. As the first round of thymidine block, the cultured cells were incubated in the media with or without 2 mM thymidine (Sigma‐Aldrich) for 18 h and incubated in the fresh media for 9 h. As the second round of thymidine block, the cells were incubated again in the media with or without 2 mM thymidine (Sigma‐Aldrich) for 18 h. Then, the cells were incubated in those with or without 60 ng/ml of mouse recombinant TNFα (Sigma‐Aldrich) for at 37°C.
Culture of human epithelial cell line
HEC151, an endometrial epithelial cell line, was used. To induce CCA in HEC151 cells, the protocol described in the methods of culture of primary mouse uterine epithelial cells was employed.
Fluorescence assay using the cultured cells
The fluorescence assay using the cultured uterine epithelial cells was performed as described previously (Hirota et al, 2003). The antibodies to pMLKL (37333, Cell Signaling Technology, 1:400), MFG‐E8 (D161‐3, MBL, Nagoya, Japan 1:500), and pRIP3 (ab195117, Abcam, 1:200) were used.
Cell cycle analysis and annexin V assay
As for cell cycle analysis, the cells were stained with propidium iodine (Abcam), and cell fractions were quantified by flow cytometry (FACS Calibur, BD Bioscience, Bedford, MA, USA). Presentation of PS at the outer membrane of the cells was investigated using Annexin V assay kit (MBL) with flow cytometry (FACS Calibur). To evaluate annexin V‐positive cells, we counted the numbers of cells on 3 randomly selected sections manually by microscopic images at × 40 magnification.
Laser capture microdissection (LCM)
LCM was performed as described previously (Haraguchi et al, 2014; Matsumoto et al, 2018). The uterine epithelium of each section was microdissected with an LMD7000 system (Leica Microsystems).
RNA‐seq
RNA extracted from LCM samples was processed by SMART‐seq v4 Ultra Low Input RNA Kit (Takara Bio USA, Mountain View, CA, USA) and was subjected to RNA‐seq using BGI RNA‐Seq service (BGI, Hong Kong, China; www.bgi.com) according to the standard protocol. The reads per kilobase of exon per million mapped sequence reads (RPKMs) were counted by FeatureCounts function in Subread (version 2.0.0) (Liao et al, 2013). Genes with log2 fold change (FC) > |1| were defined as differentially expressed genes (DEGs). Heatmaps of DEGs were created by Morpheus (https://software.broadinstitute.org/morpheus/). K‐means clustering and visualizations of data were performed by trinityrnaseq (version 2.0.6) (Haas et al, 2013). Genes in each cluster were then proceeded to comparison analysis and gene ontology (GO) analysis in Enrichr (https://amp.pharm.mssm.edu/Enrichr/).
RT–qPCR
RT–qPCR was performed as described previously. Total RNA extraction from frozen whole tissues was performed as described previously (Matsumoto et al, 2018). A housekeeping gene (Actb) was used as an internal standard for normalizing the relative mRNA expression. Sequences of qPCR primers are listed in Table EV1.
Statistical analysis
Statistical analyses were performed using Fisher's exact test and a two‐tailed Student t‐test.
Author contributions
Study design: SAk and YH; Experiments and data collection: SAk, YH, YF, TK, MG, THirat, THirao, MM, HH, and MS‐K; Data analysis: SAk, YH, YF, and SAi; Result discussion and interpretation: RS‐H, NT, TF, and YO; Manuscript writing: SAk, YH, SAi, and RS‐H; Manuscript reviewing: RS‐H; Study supervision: YH.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank NCI mouse repository for providing Rb1‐floxed mice, Dr. Francesco J. DeMayo (National Institute of Environmental Health Sciences, Research Triangle Park, NC, USA) and Dr. John P. Lydon (Baylor College of Medicine, Houston, TX, USA) for providing Pgr‐Cre mice, Dr. Sudhansu K. Dey (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA) for providing Ltf‐Cre mice, Dr. Satoru Fukuda (The University of Tokyo, Japan) for technical assistance of TEM, Ms. Katie A. Gerhardt for efficient editing of the manuscript, and Ms. Atsumi Miura for technical assistance. This work was supported by JSPS KAKENHI (grant numbers 19H03144, 18K19601, 19H03796, 18K19600, 18H02943, 19K16022, 19K18631, 19K18630, 20K08894) and AMED‐Force (20gm4010010h0001), AMED‐Wise (20gk0210021h0002), Takeda Science Foundation, and the fund of joint research with NIPRO corporation.
EMBO reports (2021) 22: e50927.
Data availability
The RNA‐seq data from this study have been deposited to the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/Traces/study1/?acc=PRJNA658848&o=acc_s%3Aa) and assigned the identifiers (SRA: SRP278560, BioProject: PRJNA658848).
References
- Boren J, Brindle KM (2012) Apoptosis‐induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ 19: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Sun X, Dey SK (2012) Mechanisms of implantation: strategies for successful pregnancy. Nat Med 18: 1754–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, Meilijson I, Ruppin E (1999) Neuronal regulation: a mechanism for synaptic pruning during brain maturation. Neural Comput 11: 2061–2080 [DOI] [PubMed] [Google Scholar]
- Chen B, Pan H, Zhu L, Deng Y, Pollard JW (2005) Progesterone inhibits the estrogen‐induced phosphoinositide 3‐kinase–>AKT–>GSK‐3beta–>cyclin D1–>pRB pathway to block uterine epithelial cell proliferation. Mol Endocrinol 19: 1978–1990 [DOI] [PubMed] [Google Scholar]
- Chen G, Deng X (2018) Cell synchronization by double thymidine block. Bio Protoc 8: e2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Ogawa Y, Terakawa J, Ogawa A, DeFalco T, Dey SK (2014) Lactoferrin‐iCre: a new mouse line to study uterine epithelial gene function. Endocrinology 155: 2718–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Wang XN, Paria BC, Damm D, Abraham JA, Klagsbrun M, Andrews GK, Dey SK (1994) Heparin‐binding EGF‐like growth factor gene is induced in the mouse uterus temporally by the blastocyst solely at the site of its apposition: a possible ligand for interaction with blastocyst EGF‐receptor in implantation. Development 120: 1071–1083 [DOI] [PubMed] [Google Scholar]
- Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H (2004) Molecular cues to implantation. Endocr Rev 25: 341–373 [DOI] [PubMed] [Google Scholar]
- Diaz‐Gimeno P, Ruiz‐Alonso M, Blesa D, Bosch N, Martinez‐Conejero JA, Alama P, Garrido N, Pellicer A, Simon C (2013) The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril 99: 508–517 [DOI] [PubMed] [Google Scholar]
- Dick FA, Rubin SM (2013) Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 14: 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egashira M, Hirota Y (2013) Uterine receptivity and embryo–uterine interactions in embryo implantation: lessons from mice. Reprod Med Biol 12: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt H, Wachter F, Grunert M, Jeremias I (2013) Cell cycle‐arrested tumor cells exhibit increased sensitivity towards TRAIL‐induced apoptosis. Cell Death Dis 4: e661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Rubel CA, Large MJ, Wetendorf M, Fernandez‐Valdivia R, Jeong JW, Spencer TE, Behringer RR, Lydon JP, Demayo FJ (2012) Epithelial progesterone receptor exhibits pleiotropic roles in uterine development and function. FASEB J 26: 1218–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Hirota Y, Matsuo M, Gebril M, Akaeda S, Hiraoka T, Osuga Y (2019) Uterine receptivity, embryo attachment, and embryo invasion: multistep processes in embryo implantation. Reprod Med Biol 18: 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C, Giordano A (2006) RB and cell cycle progression. Oncogene 25: 5220–5227 [DOI] [PubMed] [Google Scholar]
- Guo Y, Cordes KR, Farese RV Jr, Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122: 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al (2013) De novo transcript sequence reconstruction from RNA‐seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S (2002) Identification of a factor that links apoptotic cells to phagocytes. Nature 417: 182–187 [DOI] [PubMed] [Google Scholar]
- Haraguchi H, Saito‐Fujita T, Hirota Y, Egashira M, Matsumoto L, Matsuo M, Hiraoka T, Koga K, Yamauchi N, Fukayama M et al (2014) MicroRNA‐200a locally attenuates progesterone signaling in the cervix, preventing embryo implantation. Mol Endocrinol 28: 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi H, Hirota Y, Saito‐Fujita T, Tanaka T, Shimizu‐Hirota R, Harada M, Akaeda S, Hiraoka T, Matsuo M, Matsumoto L et al (2019) Mdm2‐p53‐SF1 pathway in ovarian granulosa cells directs ovulation and fertilization by conditioning oocyte quality. FASEB J 33: 2610–2620 [DOI] [PubMed] [Google Scholar]
- Harper JV (2005) Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol Biol 296: 157–166 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Osuga Y, Yoshino O, Koga K, Yano T, Hirata T, Nose E, Ayabe T, Namba A, Tsutsumi O et al (2003) Possible roles of thrombin‐induced activation of protease‐activated receptor 1 in human luteinized granulosa cells. J Clin Endocrinol Metab 88: 3952–3957 [DOI] [PubMed] [Google Scholar]
- Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK (2010) Uterine‐specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest 120: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y (2019) Progesterone governs endometrial proliferation‐differentiation switching and blastocyst implantation. Endocr J 66: 199–206 [DOI] [PubMed] [Google Scholar]
- Huet YM, Dey SK (1987) Role of early and late oestrogenic effects on implantation in the mouse. J Reprod Fertil 81: 453–458 [DOI] [PubMed] [Google Scholar]
- Inhorn MC, Patrizio P (2015) Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update 21: 411–426 [DOI] [PubMed] [Google Scholar]
- Joswig A, Gabriel HD, Kibschull M, Winterhager E (2003) Apoptosis in uterine epithelium and decidua in response to implantation: evidence for two different pathways. Reprod Biol Endocrinol 1: 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Kannan A, DeMayo FJ, Lydon JP, Cooke PS, Yamagishi H, Srivastava D, Bagchi MK, Bagchi IC (2011) The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 331: 912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun X, Dey SK (2015) Entosis allows timely elimination of the luminal epithelial barrier for embryo implantation. Cell Rep 11: 358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed‐and‐vote. Nucleic Acids Res 41: e108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA Jr, Shyamala G, Conneely OM, O'Malley BW (1995) Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev 9: 2266–2278 [DOI] [PubMed] [Google Scholar]
- Ma WG, Song H, Das SK, Paria BC, Dey SK (2003) Estrogen is a critical determinant that specifies the duration of the window of uterine receptivity for implantation. Proc Natl Acad Sci USA 100: 2963–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A (2000) Induction of medulloblastomas in p53‐null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 14: 994–1004 [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Ma WG, Daikoku T, Zhao X, Paria BC, Das SK, Trzaskos JM, Dey SK (2002) Cyclooxygenase‐2 differentially directs uterine angiogenesis during implantation in mice. J Biol Chem 277: 29260–29267 [DOI] [PubMed] [Google Scholar]
- Matsumoto L, Hirota Y, Saito‐Fujita T, Takeda N, Tanaka T, Hiraoka T, Akaeda S, Fujita H, Shimizu‐Hirota R, Igaue S et al (2018) HIF2α in the uterine stroma permits embryo invasion and luminal epithelium detachment. J Clin Invest 128: 3186–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M, Hirota Y, Fukui Y, Fujita H, Saito‐Fujita T, Kaku T, Gebril M, Hirata T, Akaeda S, Hiraoka T et al (2020) Levonorgestrel inhibits embryo attachment by eliminating uterine induction of leukemia inhibitory factor. Endocrinology 161: bqz005 [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS (2007) A nonapoptotic cell death process, entosis, that occurs by cell‐in‐cell invasion. Cell 131: 966–979 [DOI] [PubMed] [Google Scholar]
- Schlafke S, Enders AC (1975) Cellular basis of interaction between trophoblast and uterus at implantation. Biol Reprod 12: 41–65 [DOI] [PubMed] [Google Scholar]
- Schwarting R, Gerdes J, Niehus J, Jaeschke L, Stein H (1986) Determination of the growth fraction in cell suspensions by flow cytometry using the monoclonal antibody Ki‐67. J Immunol Methods 90: 65–70 [DOI] [PubMed] [Google Scholar]
- Segawa K, Yanagihashi Y, Yamada K, Suzuki C, Uchiyama Y, Nagata S (2018) Phospholipid flippases enable precursor B cells to flee engulfment by macrophages. Proc Natl Acad Sci USA 115: 12212–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomovitz I, Speir M, Gerlic M (2019) Flipping the dogma – phosphatidylserine in non‐apoptotic cell death. Cell Commun Signal 17: 139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP (2005) Cre‐mediated recombination in cell lineages that express the progesterone receptor. Genesis 41: 58–66 [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G (2019) The molecular machinery of regulated cell death. Cell Res 29: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Pollard JW (1999) Progesterone inhibits estrogen‐induced cyclin D1 and cdk4 nuclear translocation, cyclin E‐ and cyclin A‐cdk2 kinase activation, and cell proliferation in uterine epithelial cells in mice. Mol Cell Biol 19: 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranguch S, Wang H, Daikoku T, Xie H, Smith DF, Dey SK (2007) FKBP52 deficiency‐conferred uterine progesterone resistance is genetic background and pregnancy stage specific. J Clin Invest 117: 1824–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez YM, Wang X, Wetendorf M, Franco HL, Mo Q, Wang T, Lanz RB, Young SL, Lessey BA, Spencer TE et al (2018) FOXO1 regulates uterine epithelial integrity and progesterone receptor expression critical for embryo implantation. PLoS Genet 14: e1007787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetendorf M, Wu S‐P, Wang X, Creighton CJ, Wang T, Lanz RB, Blok L, Tsai SY, Tsai M‐J, Lydon JP et al (2017) Decreased epithelial progesterone receptor A at the window of receptivity is required for preparation of the endometrium for embryo attachment†. Biol Reprod 96: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Urano Y, Takabe W, Saito Y, Noguchi N (2014) Induction of apoptosis and necroptosis by 24(S)‐hydroxycholesterol is dependent on activity of acyl‐CoA:cholesterol acyltransferase 1. Cell Death Dis 5: e990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Adams CE (1966) Delayed implantation in the spayed, progesterone treated adult mouse. J Reprod Fertil 12: 593–595 [DOI] [PubMed] [Google Scholar]
- Zargarian S, Shlomovitz I, Erlich Z, Hourizadeh A, Ofir‐Birin Y, Croker BA, Regev‐Rudzki N, Edry‐Botzer L, Gerlic M (2017) Phosphatidylserine externalization, "necroptotic bodies" release, and phagocytosis during necroptosis. PLoS Biol 15: e2002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Cui H, Xia Y, Gan H (2016) RIPK3‐mediated necroptosis and apoptosis contributes to renal tubular cell progressive loss and chronic kidney disease progression in rats. PLoS One 11: e0156729 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File
Data Availability Statement
The RNA‐seq data from this study have been deposited to the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/Traces/study1/?acc=PRJNA658848&o=acc_s%3Aa) and assigned the identifiers (SRA: SRP278560, BioProject: PRJNA658848).


