Significance
The inheritance of long (GAA)n repeats in the frataxin gene causes the debilitating neurodegenerative disease Friedreich’s ataxia. Subsequent expansions of these repeats throughout a patient’s lifetime in the affected tissues, like the nervous system, may contribute to disease onset. We developed an experimental model to characterize the mechanisms of repeat instability in nondividing cells to better understand how mutations can occur as cells age chronologically. We show that repeats can expand in nondividing cells. Notably, however, large deletions are the major type of repeat-mediated genome instability in nondividing cells, implicating the loss of important genetic material with aging in the progression of Friedreich’s ataxia.
Keywords: repeat expansion diseases, Friedreich’s ataxia, (GAA)n repeats, H-DNA, somatic instability
Abstract
Nearly 50 hereditary diseases result from the inheritance of abnormally long repetitive DNA microsatellites. While it was originally believed that the size of inherited repeats is the key factor in disease development, it has become clear that somatic instability of these repeats throughout an individual’s lifetime strongly contributes to disease onset and progression. Importantly, somatic instability is commonly observed in terminally differentiated, postmitotic cells, such as neurons. To unravel the mechanisms of repeat instability in nondividing cells, we created an experimental system to analyze the mutability of Friedreich’s ataxia (GAA)n repeats during chronological aging of quiescent Saccharomyces cerevisiae. Unexpectedly, we found that the predominant repeat-mediated mutation in nondividing cells is large-scale deletions encompassing parts, or the entirety, of the repeat and adjacent regions. These deletions are caused by breakage at the repeat mediated by mismatch repair (MMR) complexes MutSβ and MutLα and DNA endonuclease Rad1, followed by end-resection by Exo1 and repair of the resulting double-strand breaks (DSBs) via nonhomologous end joining. We also observed repeat-mediated gene conversions as a result of DSB repair via ectopic homologous recombination during chronological aging. Repeat expansions accrue during chronological aging as well—particularly in the absence of MMR-induced DSBs. These expansions depend on the processivity of DNA polymerase δ while being counteracted by Exo1 and MutSβ, implicating nick repair. Altogether, these findings show that the mechanisms and types of (GAA)n repeat instability differ dramatically between dividing and nondividing cells, suggesting that distinct repeat-mediated mutations in terminally differentiated somatic cells might influence Friedreich’s ataxia pathogenesis.
Nearly 50 human diseases have been linked to the inheritance of abnormally long DNA microsatellites—repetitive stretches of the genome where the repeating sequence motif is less than nine base pairs (bp) long (1, 2). In one such disease, Friedreich’s ataxia (FRDA), individuals inherit abnormally long (GAA)n trinucleotide repeats in the first intron of the human FXN gene (3). FRDA follows an autosomal recessive inheritance pattern with 97% of patients possessing two expanded (GAA)n alleles and the other 3% inheriting one expanded allele and a second, mutated copy of FXN. Disease-associated, expanded alleles contain 600 to 900 repeats on average (4). Patients with the disease display FXN messenger RNA deficiency as a result of localized heterochromatic silencing, which leads to shortage of the mitochondrial protein frataxin (3, 5).
Genetic anticipation is a characteristic hereditary pattern for many repeat expansion diseases: With each generation the inherited repeat increases in length, resulting in an earlier manifestation and increased severity of disease. Despite the correlation between repeat length and age of onset, the question remains as to the prevailing mechanism of disease onset and progression (6). One proposal is that the timing of disease manifestation is determined by the sequential accumulation of toxic repeat-associated molecules, typically RNA or protein, over the course of an individual’s lifetime. For example, in the case of Huntington’s disease (HD), an in-frame (CAG)n repeat causes the buildup of disruptive polyglutamine protein aggregates in the brain with age (7, 8). In FRDA, where the causative repetitive element is noncoding, it has been hypothesized that frataxin deficiency drives disease progression in a similar fashion via accumulation of toxic iron due to the dysregulation of iron homeostasis (9). An emerging alternative to the toxicity hypothesis is that the primary driver of symptom onset and progression could be age-related, somatic expansions of the inherited disease-size microsatellite. This model has been best supported in studies of HD, where 1) genetic modifiers of disease in humans tend to be mutations in DNA repair genes that influence somatic repeat expansion rate (10–12), 2) mouse models show that altering somatic expansion rates influences disease pathogenesis (13, 14), and 3) patients with interrupted repeats but the same polyglutamine length (i.e., CAA trinucleotides interspersed within a CAG microsatellite) present with significantly later age of onset (10, 12).
In addition to genetic anticipation, repeat expansion diseases tend to display prominent tissue specificity, with the majority manifesting as neurodegenerative disease. In the case of FRDA, despite the fact that frataxin is a ubiquitously expressed (5, 15) and essential protein (16) disease-related pathophysiology is largely limited to hypertrophic cardiomyopathy, degeneration of the central and peripheral nervous system and, in some cases, diabetes mellitus (17). In HD, where the mutant HTT gene is expressed throughout the brain (18), it is primarily the striatal white matter and caudate nucleus that degenerate over time (19). It has been posited, therefore, that somatic repeat expansions determine tissue specificity. Supporting this idea, (GAA)n repeats display greater levels of length instability in FRDA-affected tissues, such as the heart and spinal cord, in mice and humans (20–25). Similarly, postmortem HD brains show that the largest somatic (CAG)n expansions occur in the striatum (26, 27).
The molecular mechanisms governing the expansion of (GAA)n repeats have primarily been examined in dividing cells. In growing yeast, their instability can result from faulty DNA replication (28, 29), postreplicative repair (30), and homologous recombination (31, 32). In human cell cultures, (GAA)n expansions correlate directly with passage number and are dependent on DNA mismatch repair (MMR) and transcription (33–36). Thus, in dividing cells (GAA)n expansions are attributed to inaccurate DNA synthesis during replication or repair, which is in line with the findings that (GAA)n repeats stall DNA polymerization in vitro (37) and replication fork progression in vivo (30, 38–40). However, given the lack of active replication in human tissues most affected in repeat expansion diseases, it is difficult to extend these observations to the process of pathogenic somatic expansions. We have previously found that in stationary yeast cultures fragility mediated by (GAA)n repeats was increased in a time- and transcription-dependent manner (41). The only study of (GAA)n repeat expansions in nondividing neuronal tissues, which was done in a transgenic mouse model of FRDA, demonstrated that the Pms2 protein—the primary endonuclease of the MMR pathway—specifically suppresses large-scale expansions (42). This study also found that the upstream mismatch recognition complex MutSα plays a counteracting role by promoting somatic expansion.
We sought to characterize the nature and mechanism of replication-independent mutation at (GAA)n repeats by examining their mutability during chronological aging of homogeneous, nondividing quiescent cultures of Saccharomyces cerevisiae using our previously established genetic reporter for detecting repeat-associated mutagenesis (28, 30). We found that the spectrum of (GAA)n-associated mutations changes dramatically in nondividing cells as compared to dividing cells. In the course of their chronological aging, we observe a 20-fold increase in the incidence of repeat-mediated deletions with a median length of 500 bp. These deletions result from imprecise nonhomologous end-joining (NHEJ) repair of double-strand breaks (DSBs) at the repetitive and/or nearby sequences. Formation of these DSBs requires the MutSβ mismatch recognition complex, the MutLα MMR endonuclease and, possibly, the repair endonuclease Rad1. In addition to deletions, we observed gene conversion events between our cassette and a mutant ura3-52 allele present in our strain. Like deletions, gene conversion events depend on functional MutSβ, MutLα, and Rad1 protein complexes. In contrast to deletions, however, they occur via homologous recombination guided by Rad52 and Rad51 proteins. Finally, we observed an accumulation of repeat expansion events during chronological aging, which were particularly prominent in the absence of MutSβ and the 5′-3′ exonuclease Exo1. While replication-independent, these expansions depend on the processivity of DNA polymerase δ, which points to the role of nick or gap repair in the process. These findings establish that the mechanisms and types of mutagenesis at (GAA)n repeats differ between dividing and nondividing cell populations—suggesting that the cell-cycle status and DNA repair environment of somatic cells might influence FRDA pathogenesis.
Results
An Assay for Assessing (GAA)100-Associated Mutations in Quiescent Yeast.
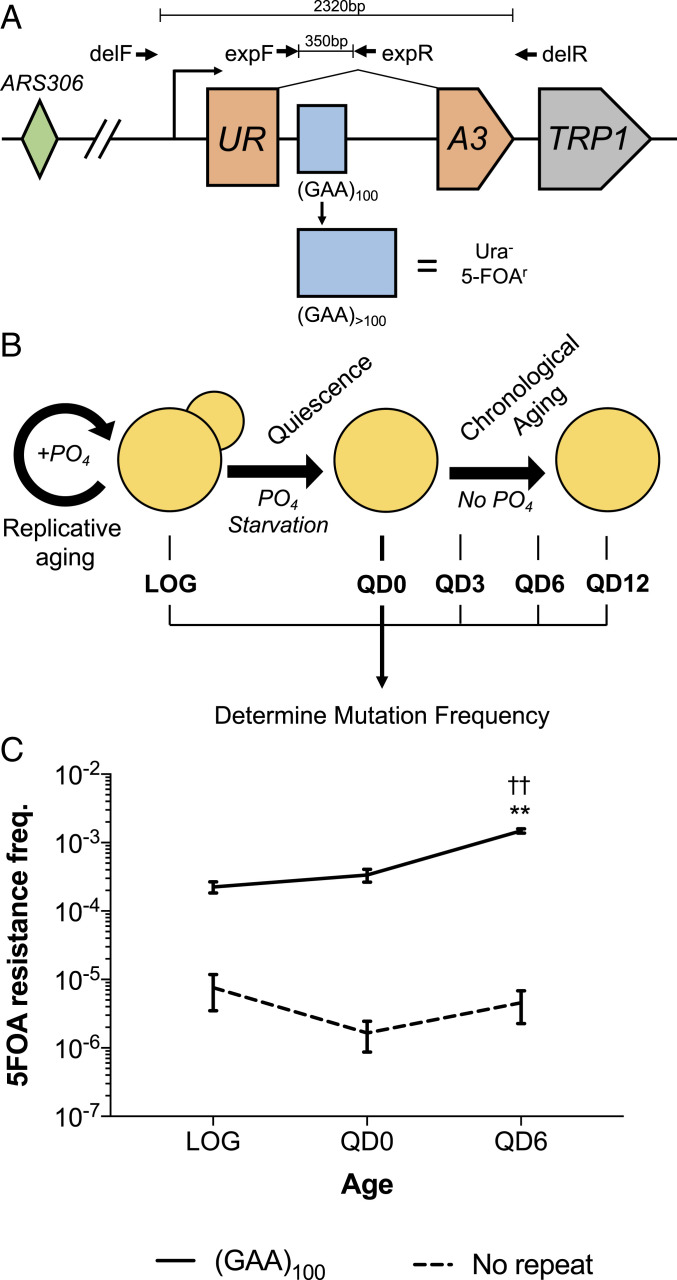
The primary objective of this study was to determine the spectrum and mechanisms of repeat-mediated mutations in nondividing cells. We used strains of S. cerevisiae bearing our previously established cassette on chromosome III, which allows for the selection of cells that have undergone various (GAA)n-associated mutation events (Fig. 1A). This cassette consists of a forward selection yeast URA3 gene that has been artificially split by an intron containing 100 (GAA)n repeats. When the repeats expand, it pushes the intron length beyond the yeast splicing threshold of ∼1 kb. This disrupts splicing of URA3 transcripts and renders cells resistant to the drug 5-fluoroorotic acid (5-FOA). Notably, any mutation that results in URA3 loss of function within our cassette makes cells 5-FOA–resistant. This allows for selection of any deleterious mutations, not just expansion events, by plating on media containing 5-FOA.
Fig. 1.
An assay for assessing (GAA)100-associated mutations in quiescent yeast. (A) A total of 100 (GAA)n repeats were cloned into an artificially split URA3 gene such that expansion events abrogate splicing and result in resistance to 5-FOA. ARS306: autonomously replicating sequence on Chr III. TRP1: auxotrophic marker for selection of strains bearing the construct. delF/R: primers to detect large-scale deletion events. expF/R: primers to detect expansion events. (B) Assay to determine mutation frequency in nondividing quiescent cells. Cells are grown in phosphate-limited synthetic media for 40 h, at which point mutation frequency is assessed by serial dilution and plating on 5-FOA–containing and complete media (LOG). After 64 h, when cells are fully starved of phosphate, they are run through a Percoll gradient to isolate Q cells that are resuspended in media devoid of phosphate (QD0). Aliquots of Q cells are then assessed for mutation frequency at the various time points indicated using notation QDX, where X represents the chronological day starting from QD0. (C) The 5-FOA resistance frequency in strains bearing the cassette from A or an identical cassette with no (GAA)n repeats and an intron of similar length. Error bars represent 95% confidence intervals. P < 0.01, two-way ANOVA with Tukey honestly significant difference for (GAA)100 compared to **LOG or ††QD0.
We detected mutations using this construct in a nondividing cell population by taking advantage of the fact that yeast enter quiescence when they are starved of an essential nutrient. Quiescent (Q) cells are nonproliferative cells that are arrested in “G0” and will only divide upon exposure to the original growth-limiting nutrient. They have longer chronological lifespans than nonquiescent cells and can be aged chronologically without dividing via maintenance in media that lacks the nutrient they were initially starved of (43, 44). We married our ability to select for (GAA)100-mediated mutations and isolate Q cells using Percoll density gradients by taking strains bearing our URA3 cassette and culturing them in media where phosphate was the growth-limiting nutrient. We assessed mutation frequencies by plating an aliquot of each culture on media containing 5-FOA (to select for cells that contained repeat-mediated mutations) and complete media (yeast extract peptone dextrose, YPD) (to determine total cell count) at various time points during logarithmic growth and quiescence (Fig. 1B). A major advantage of this approach is that it allows us to track two distinct modes of cellular aging across individual cultures: replicative aging (T0 to QD0) and chronological aging (QD0 to QD12).
Because much of the previous work with quiescent yeast was conducted in prototrophic strains (without nutrient requirements), we wanted to confirm that the Q cells we isolated from Percoll density gradients using our experimental auxotrophic strains were quiescent. We found that Q cell populations exhibited greater than 85% viability after 6 d of chronological aging for nearly all genotypes examined in our assay (SI Appendix, Fig. S1A). This level of viability is typical of quiescent yeast (45). Using a custom soft lithography-generated microfluidics chamber, we did not observe a single cell division in the 734 Q cells we tracked in media lacking phosphate over the course of 24 h (SI Appendix, Fig. S1B and Movies S1 and S2). We also found that over 85% of the Q cells we isolated using density gradients were unbudded daughter cells lacking bud scars, which is in line with previous studies (SI Appendix, Fig. S1C) (45).
(GAA)100 Repeats Increase the Frequency of 5-FOA–Resistant Mutations in Quiescent Cells.
We first assessed whether (GAA)n repeats enhance mutagenesis in quiescent cell populations by conducting our assay in strains bearing our cassette with (GAA)100 repeats (Fig. 1A) and a control cassette with no repeats but an intron of identical length. As expected, we found that the frequency of 5-FOA–resistant mutations was over an order of magnitude greater in the (GAA)100 strain than the no-repeat control at all time points (Fig. 1C). The increase in 5-FOA resistance frequency during the chronological aging portion of the assay for the no-repeat control (2.7-fold) was significantly less than the (GAA)100 strain (4.4-fold) (Fig. 1C).
Large Deletions Are the Predominant Mutation Type Observed at (GAA)100 Repeats during Chronological Aging.
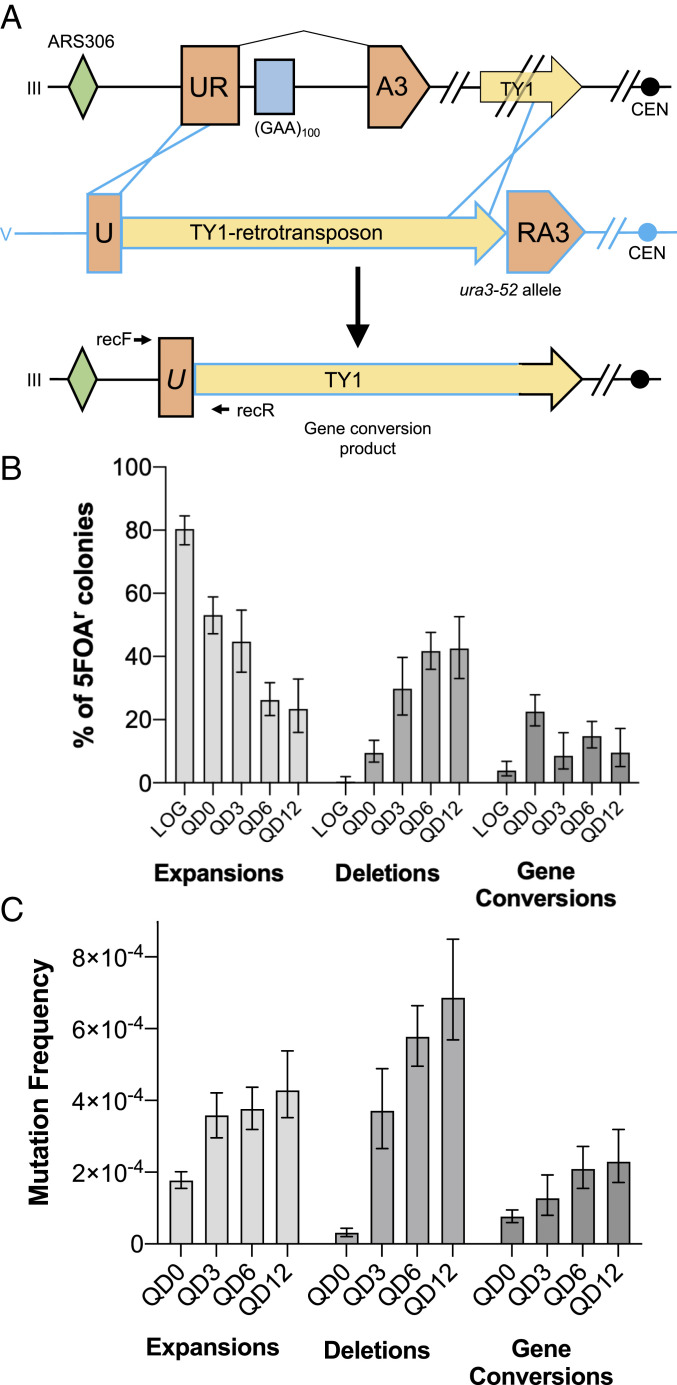
As previously mentioned, any mutation that inactivates URA3 in our construct will render cells resistant to 5-FOA. We characterized the various events that made cells 5-FOA–resistant in our assay by conducting a series of PCRs aimed at detecting three different mutation types: repeat expansions, large deletions, and gene conversion events. Expansions and deletions were detected as length changes in the PCR amplicons of the repetitive sequence (Fig. 1A, expF/expR) or the entire cassette (Fig. 1A, delF/delR), respectively. Gene conversion events were detected by amplification of a repair junction that occurs when our UR-GAA100-A3 construct recombines with the endogenous ura3-52 allele present on chromosome V in our strain. The ectopic ura3-52 donor sequence contains the wild-type (WT) URA3 sequence disrupted by a retroviral TY-1 element and, thus, bears significant homology to our cassette. Any recombination event in which the region on chromosome III upstream of our construct is joined specifically with the TY-1 element within ura3-52 on chromosome V results in a PCR product using primers recF (unique to chromosome III) and recR (unique to chromosome V) (Fig. 2A). Our previous whole-genome characterization using long-read sequencing of 22 5-FOA–resistant strains with repeat-associated gross chromosomal rearrangements (46) identified 16 as noncross-over gene conversions. They involved URA3 homologies on the centromere-distal end of the repair event and homologies between the ura3-52 TY element and one of two TY elements located roughly 8 kb downstream of our cassette on the centromere-proximal end (as illustrated in Fig. 2A) (46).
Fig. 2.
The predominant (GAA)n-mediated mutation in quiescence is large-scale deletions, but all mutation types increase throughout quiescence. (A) Schematic of previous long-read sequencing assessment of gene conversion events (46). Recombination occurs via homology between our artificial URA3-containing construct on Chr III (black) and the ura3-52 allele on Chr V (blue). Yellow arrows represent paralogous Ty-1 retrotransposable elements. RecF/R: primers to detect gene conversion events. (B) Percentage of 5-FOA–resistant colonies demonstrating each of the mutation types interrogated during logarithmic growth and quiescence. Error bars represent 95% confidence intervals calculated via the Wilson method. (C) Bars represent median mutation frequency across time points during chronological aging. Error bars represent 95% credible intervals from our model for estimating mutation frequency distributions via Bayesian logistic regression.
The type of mutation that made cells 5-FOA–resistant in our assay shifted dramatically during chronological aging. In logarithmically dividing cells, 80% of 5-FOA–resistant colonies harbored expansion events, with only a single deletion observed out of the 285 colonies analyzed (Fig. 2B). When cells entered quiescence, the proportion of 5-FOA resistance due to repeat expansion dropped to 53%. This corresponded with an increase in large deletions to 9% and gene conversions to 22% (Fig. 2B). After 6 and 12 d of chronological aging, the percentage of expansions fell to 26% and 23%, respectively, whereas the percentage of deletions was 42% at both time points (Fig. 2B). Altogether, these three mutation types account for roughly 85% of 5-FOA–resistant clones. The remaining events could not be classified using our PCR approach. This shift in mutational spectrum during chronological aging was reflected in the change in overall frequency for each mutation type. There was a substantial increase in median deletion (18-fold) frequency during the chronological aging portion of the assay between QD0 and QD6 (Fig. 2C). Over this same time period there was a more modest, but significant, increase in median gene conversion (2.8-fold) and expansion frequency (1.8-fold) (Fig. 2C). When the assay was carried out to QD12, we did not see a statistically significant increase in expansion, deletion or gene conversion frequency when compared with QD6 (Fig. 2C). This dramatic accumulation of deletions during chronological aging is in-line with our earlier observations of increased (GAA)n repeat fragility in stationary yeast cultures with time (41).
Repeat-Mediated DSBs Require Functional MutSβ and the Endonuclease Activity of MutLα.
Numerous previous studies have implicated MMR proteins as key instigators of mutagenesis at disease-associated microsatellites. Specifically, the expansion of (GAA)n and (CAG)n repeats is dependent on MMR in mice and human cell lines (reviewed in ref. 47). In yeast, we have shown that chromosomal fragility at (GAA)n repeats can occur in both G1 (48) and S phase of the cell cycle (49), and it is dependent upon the MutSβ complex (49)—a heterodimer of Msh2 and Msh3 that recognizes long (>2 nucleotides [nt]) insertion deletion loops in DNA (47, 50). In contrast, the MutSα complex, a heterodimer of Msh2 and Msh6 that preferentially binds to DNA base mismatches and short (<2 nt) insertion deletion loops (50), had less of an effect on fragility (49).
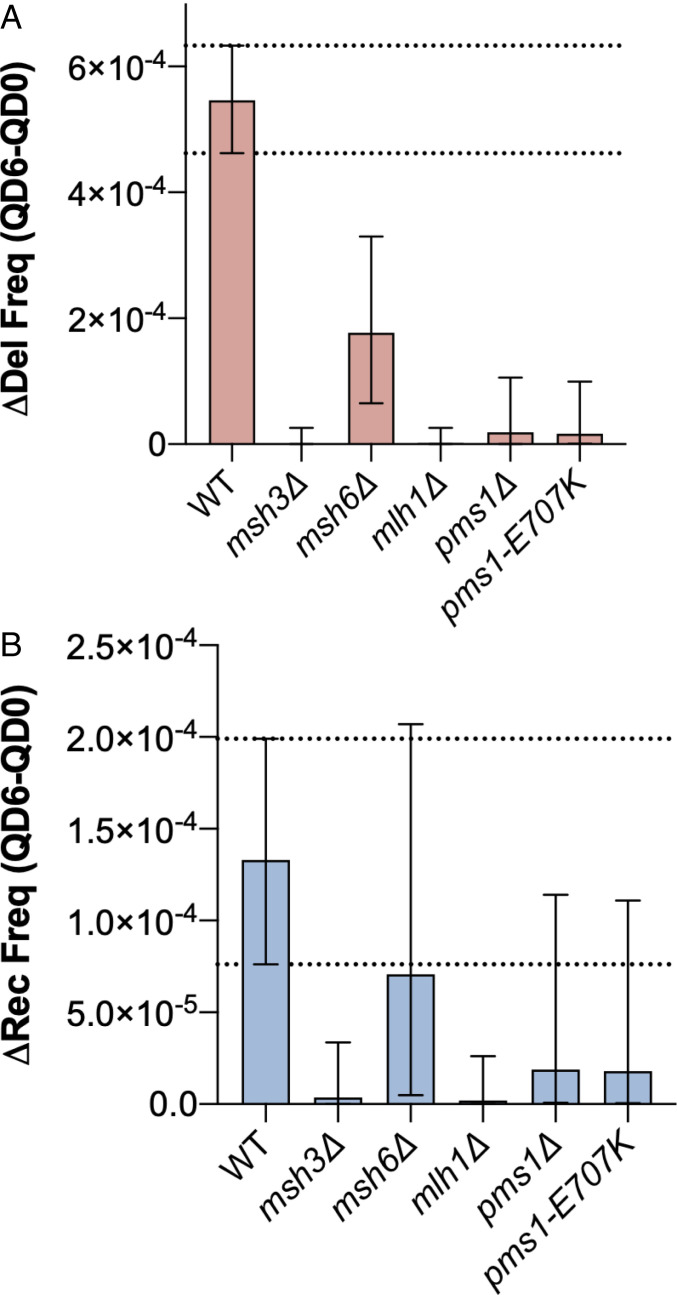
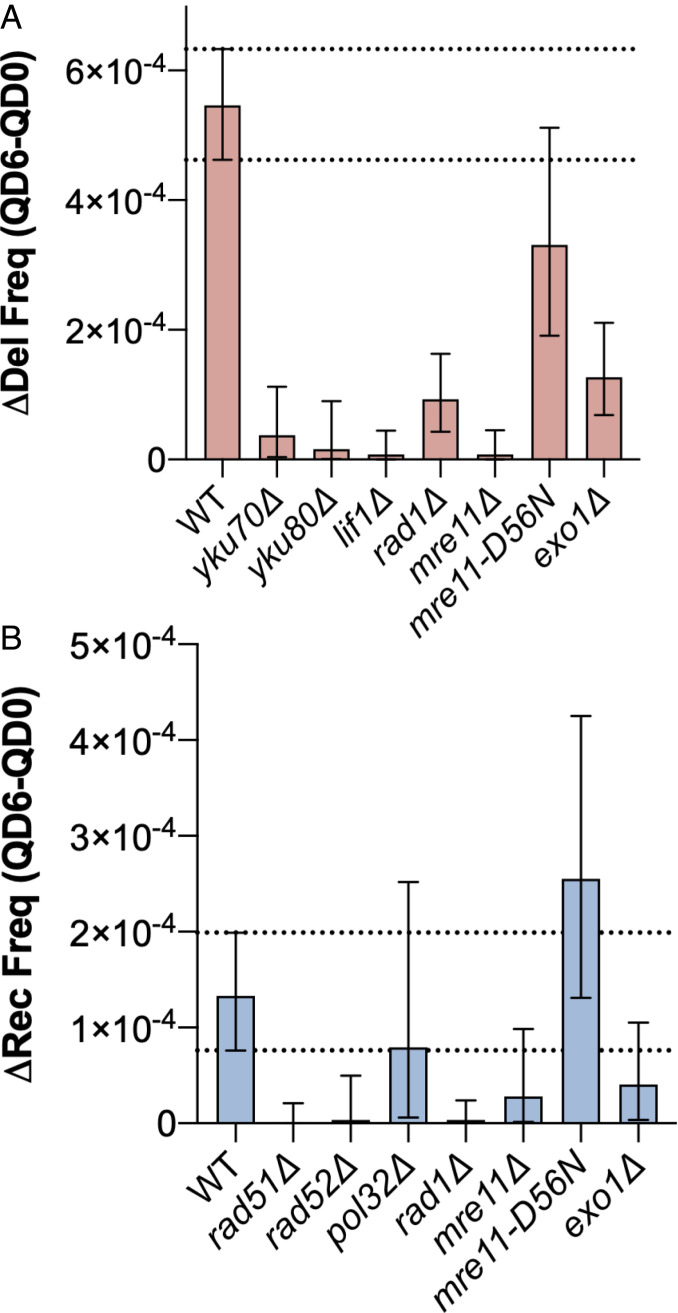
We hypothesized that the inciting event for the ectopic gene conversion and large deletion events we observed in our assay would be recognition of the (GAA)100 repeat by MutSβ. To assess the impact of gene knockout on repeat-associated mutation during chronological aging, we estimated the change in deletion frequency (Δdelfreq) and gene conversion frequency (Δrecfreq) between QD0 and QD6 using the Bayesian analysis described in Materials and Methods and SI Appendix. We found that knockout of MSH3 and to a lesser degree MSH6 reduced the change in deletion frequency between QD0 and QD6 (Δdelfreq) compared to WT (Fig. 3A). For gene conversions, only MSH3 knockout led to a substantial decrease in the change in recombination frequency between QD0 and QD6 (Fig. 3B). We also confirmed that Msh3 protein is present in quiescent cells via Western blot since gene expression is altered during quiescence (SI Appendix, Fig. S4C). Taken together, this suggests that DSB formation at (GAA)n repeats during chronological aging requires recognition by the MutSβ complex and, to a much lesser extent, MutSα.
Fig. 3.
Repeat-mediated DSBs require MutSβ and the endonuclease activity of MutLα. MMR is required for large-scale deletions (A) and gene conversions (B) during chronological aging. Bars represent the median change in deletion frequency (Δdel freq) (red) or gene conversion frequency (Δrec freq) (blue) between QD0 and QD6. Error bars represent 95% credible intervals of the calculated distributions. Biological significance was defined as no overlap between the WT and mutant 95% credible intervals. Dashed lines indicate WT 95% credible interval.
During postreplication MMR, the activity of MutLα endonuclease, composed of Mlh1 and Pms1 subunits, is latent and activated by a nick in the nascent strand. MutLα has also been shown to nick mismatch-free supercoiled DNA, suggesting that its nuclease activity can be activated by single-strandedness or non-B DNA structures present as a result of supercoiling (51). (GAA)n repeats are capable of forming a non-B DNA structure, H-DNA, when a portion of the repeat folds back to form a stable triplex with the neighboring repetitive duplex, leaving a segment of unpaired single-stranded DNA. MutLα was previously implicated in chromosomal fragility at (GAA)n repeats, and the importance of its endonuclease activity was confirmed by using an endonuclease-deficient pms1-E707K mutant. We hypothesized that MutLα can incise the single-stranded portion of H-DNA formed by supercoiled (GAA)n repeats, leading to a DSB. Knockout of members of the MutLα complex, MLH1 and PMS1, led to a dramatic reduction in Δdelfreq and Δrecfreq (Fig. 3 A and B). Furthermore, endonuclease-deficient pms1-E707K mutants had a similar reduction in Δdelfreq and Δrecfreq (Fig. 3 A and B), confirming the necessity of MutLα’s endonuclease activity in DSB formation.
Deletions during Chronological Aging Require Exo1.
Exo1 is a 5′-3′ exonuclease and flap endonuclease that functions in numerous DNA repair pathways including MMR, DSB repair, recombination, Okazaki fragment maturation, and telomere maintenance (52). In vitro MMR reconstitution experiments have revealed an Exo1-dependent pathway where multiple nicks in the mismatch-containing strand by MutLα allow for Exo1 loading and subsequent 5′-to-3′ resection of the mismatch-containing strand for over 800 bp away from the incised mismatch (53). Given this role of Exo1 in DNA resection during MMR, we predicted that it may be necessary for the MutLα-mediated large-scale deletion events we observed. In agreement with this hypothesis, knockout of EXO1 strongly reduced the accumulation of deletion events between QD0 and QD6 (see Fig. 5A). Knockout of EXO1 also decreased the accumulation of gene conversion events during chronological aging, albeit the difference was smaller and not statistically significant (see Fig. 5B).
Large Deletions Show Unique Sequence Patterns.
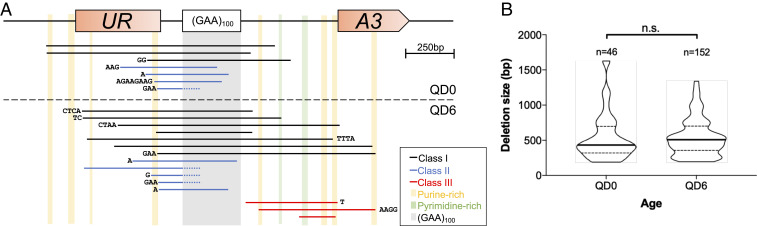
We sequenced PCR amplicons from 22 different deletion events observed at the QD0 or QD6 time points in WT cells and found that 16 (72%) of them contained short microhomologies (1 to 8 nt) that aligned to either side of the deletion (Fig. 4A). Most (15/16) of these microhomologies were not long enough to implicate the single-strand annealing or microhomology-mediated end-joining (MMEJ) (54–56) pathways of DSB repair. Also, there was no difference in the length of deletion events between QD0 and QD6 (Fig. 4B). We identified three distinct classes of deletion events based on sequencing. Ten out of 22 events (45%) were classified as Class I. These events included the entire repetitive sequence and some portion of the adjacent regions (Fig. 4A). Nine out of 22 (41%) deletions were Class II events. In these deletions, only a fraction of the repetitive sequence is lost as well as a portion of the 5′ upstream flanking sequence. All but one of these events contained some iteration of GAA microhomology at the deletion junction. The 5′ end of 5 Class II deletions was within 10 bp of a 30-bp-long sequence containing 87% purine content in the coding strand (Fig. 4A). In a different experimental system, similar purine rich sequences have been shown to interact with long (GAA)n repeats at a distance via intramolecular triplex loop formation (40). We also identified three deletions (14%) that were completely outside of the repetitive sequence (Class III). The 3′ end of all three events fell within a purine-rich sequence (Fig. 4A).
Fig. 4.
Three classes of repeat-mediated deletions whose size stays consistent throughout quiescence. (A) Scale representation of deletions from QD0 (above dashed line) and QD6 (below dashed line) ascertained via Sanger sequencing of delF/R PCR amplicons. Deleted genetic material is represented as solid black (Class I), blue (Class II), and red (Class III) lines aligned to the schematic of our UR-GAA100-A3 cassette (described in Fig. 1A). Sequences associated with each line represent microhomologies that can align to either side of the deleted sequence. Vertical bars represent (GAA)100 repeats (gray) or regions ≥15 bp with >80% purine (yellow) or pyrimidine (green) content in the coding strand. (B) Violin plot of deletion lengths measured in bp at QD0 and QD6 as determined by agarose gel electrophoresis of delF/R PCR amplicons. Bold lines indicate median. Dashed lines delineate quartiles. n.s. indicates P > 0.05, Mann–Whitney U test.
Large Deletions Require NHEJ.
It is well-established that the primary mechanism of DSB repair in G1 of the cell cycle is NHEJ (reviewed in ref. 57). Indeed, we found an increase in the expression of Yku70 protein in quiescent as compared to logarithmically dividing cells via Western blot (SI Appendix, Fig. S4C). In yeast, NHEJ is known to utilize very short microhomologies for healing breaks (58). Therefore, we suspected that inaccurate NHEJ was the repair pathway causing large deletions at (GAA)n repeats in Q cells. To examine this, we knocked out the highly conserved NHEJ genes YKU70, YKU80, and LIF1 and measured the frequency of mutation at (GAA)100 repeats. Deletion frequency was reduced dramatically in all three strains that lacked NHEJ (Fig. 5A), confirming NHEJ as the main mechanism of DSB repair leading to deletion formation during quiescence and essentially ruling out MMEJ. This conclusion is in line with a recent study of Top1-mediated deletion formation in quiescent yeast strains (59).
Fig. 5.
Genetic controls of deletion and recombination formation during quiescence. NHEJ and Exo1 are required for large-scale deletions (A) and HR is required gene conversions (B) during chronological aging. Bars represent the median change in deletion frequency (Δdel freq) (red) or gene conversion frequency (Δrec freq) (blue) between QD0 and QD6. Error bars represent 95% credible intervals of the estimated distributions. Biological significance was defined as no overlap between the WT and mutant 95% credible intervals. Dashed lines indicate WT 95% credible interval.
Gene Conversion Events Require Rad51 and Rad52 but Not Pol32.
There are two pathways of HR that might result in a noncross-over gene conversion event like those we have observed between our cassette and the endogenous ura3-52 allele. The first pathway, break-induced replication (BIR), typically resolves one-ended DSBs that result from collapsed replication forks (60). In BIR, strand invasion is followed by conservative migrating D-loop synthesis all the way to the end of the donor chromosome. One-ended BIR can lead to chromosomal translocations (61) and has been previously implicated in the expansion of (GAA)n and (CAG)n repeats in dividing yeast (32, 62). In some instances of BIR, template switching can lead to noncross-over gene conversions like those we observed in our assay (63). The second possible mechanism of gene conversion includes two classical forms of HR: synthesis-dependent strand annealing (SDSA) and double Holliday junction (dHJ) repair (64). Both pathways utilize homologous regions flanking either side of a DSB to direct repair. In dHJ, synthesis proceeds simultaneously from both ends of the break with synapsis resulting in the formation of a dHJ. In SDSA, typically only one end invades the donor and then anneals to homology on the opposite side of the break after synthesis and dissociation. In our case, the flanking homologies for these two classical HR pathways are the 5′ portion of the URA3 gene and the Ty elements downstream (Fig. 2A).
To address the pathway of HR responsible for gene conversion in our assay, we knocked out the recombination genes RAD51 and RAD52, which are necessary for all three of the aforementioned forms of HR. We also knocked out the POL32 gene, which encodes a small subunit of DNA polymerase δ that is absolutely required for BIR but is dispensable for SDSA or DSBR. We found that deletion of RAD51 and RAD52, but not POL32, suppressed Δrecfreq and led to reduction in gene conversion frequency to below the limit of detection of our assay at QD0 and QD6 (Fig. 5B and SI Appendix, Fig. S3B). Given that HR is typically employed during recombination between sister chromatids in S and G2 in mitotic cells, we confirmed the possibility of ectopic gene conversion in Q cells by detecting the presence of the Rad52 protein via Western blot (SI Appendix, Fig. S4C). Altogether, these data identify SDSA or dHJ as the likely pathway of gene conversion events at (GAA)100 repeats in dividing and quiescent cells.
NHEJ and HR Repair Pathways Operate Independently of Each Other.
Given the divergence of DSB repair events that occur at (GAA)100 repeats in our assay (HR vs. NHEJ), we were curious as to how eliminating one repair pathway influenced the frequency of the competing mutation type. Deletion of the NHEJ genes YKU70, YKU80, and LIF1 had little effect on Δrecfreq (SI Appendix, Fig. S4). Similarly, rad51Δ and rad52Δ had little effect on Δdelfreq (SI Appendix, Fig. S4 A and B). Thus, it appears that repair pathway choice at (GAA)100-mediated DSBs is not influenced by the presence or absence of the competing repair pathway. Instead, these events seem to proceed independently of one another—suggesting that there is no molecular switch governing pathway choice.
Mre11 Subunit of the MRX Complex, but Not Its Nuclease Activity, Is Required for Deletion and Gene Conversion Events during Quiescence.
Extensive end resection of DSBs formed at (GAA)100 repeats must occur to account for the large size of repeat-mediated deletions we observed during quiescence. Similarly, for HR to progress, extensive 5′ end resection must take place to allow for formation of Rad51 nucleoprotein filaments. In budding yeast, this end resection is initiated by MRX, a heterotrimer of Mre11, Xrs2, and Rad50. Short incision of the 5′ strand at DSBs by the Mre11 endonuclease is particularly important for resecting DSB ends bound by proteins or blocked by DNA hairpins (65). MRX also recruits important end resection machinery in a nuclease-independent fashion to so-called clean DSBs (66). We sought to examine the role of Mre11 at (GAA)100-mediated DSBs in quiescent cells by assessing mutation frequencies in mre11Δ strains and mre11-D56N mutants. mre11-D56N mutants lack both endonuclease and exonuclease activity, while leaving MRX complex formation intact (67, 68).
We found a significant reduction in Δdelfreq in mre11Δ strains, consistent with the essential role of Mre11 in NHEJ in S. cerevisiae (Fig. 5A) (69). In contrast, the mre11-D56N mutant showed no significant change in Δdelfreq compared to WT, which is consistent with previous studies demonstrating that Mre11 nuclease activity is not required for NHEJ (Fig. 5A) (70).
In terms of gene conversion, mre11Δ displayed the same frequency of recombination as the WT at QD0 (SI Appendix, Fig. S3B). However, at QD6, the frequency of gene conversion did not increase for mre11Δ (SI Appendix, Fig. S3B). This led to a significant reduction in Δrecfreq for mre11Δ (Fig. 5B), which contrasted from our findings in rad51Δ and rad52Δ strains where we were unable to detect gene conversion events at all at both time points (SI Appendix, Fig. S3B). These data suggest that Mre11 is specifically required for HR triggered by a repeat-induced DSB during chronological aging in Q cells but not in actively dividing cells. When we assessed recombination frequency in quiescent cells in the mre11-D56N mutant we found that there was no significant difference compared to WT, indicating that the role of Mre11 in quiescence was independent of its nuclease activity. It was, however, dependent on the Exo1 (Fig. 5B). This is consistent with previous work demonstrating no defect in HR in mre11-D56N mutants when DSBs contained “clean ends” generated by site-specific HO endonuclease within Ty elements (66). Taken together, it appears that repeat-associated DSBs generated during quiescence require MRX-mediated recruitment of HR factors for repair whereas those generated during logarithmic growth do not.
Deletions and Gene Conversions Require Rad1.
Rad1 is a flap endonuclease that recognizes branched DNA structures. It removes nonhomologous flaps during the alternative end-joining pathway MMEJ and HR in S. cerevisiae (71). It also plays an essential role in nucleotide excision repair. It is not required, however, for Ku-mediated end joining (54). Rad1 was recently shown to be necessary for chromosomal fragility at an H-DNA forming sequence in yeast (72). Since (GAA)n repeats can form H-DNA, we hypothesized that Rad1 may be involved in DSB formation at a repeat that subsequently leads to gene conversions and deletions. Knockout of RAD1 led to a substantial reduction in Δdelfreq and Δrecfreq in our assay (Fig. 5). These reductions were on par with those observed in MMR mutants. Given that Rad1 is not required for NHEJ, the reduction in deletion events in rad1Δ strains indicate it may contribute to DSB formation as opposed to break repair. In other words, Rad1 and the MMR machinery seem to overlap in generating DSBs at (GAA)n repeats.
Replication-independent (GAA)100 Expansion Is Suppressed by Exo1 and MutSβ/MutLα Complex.
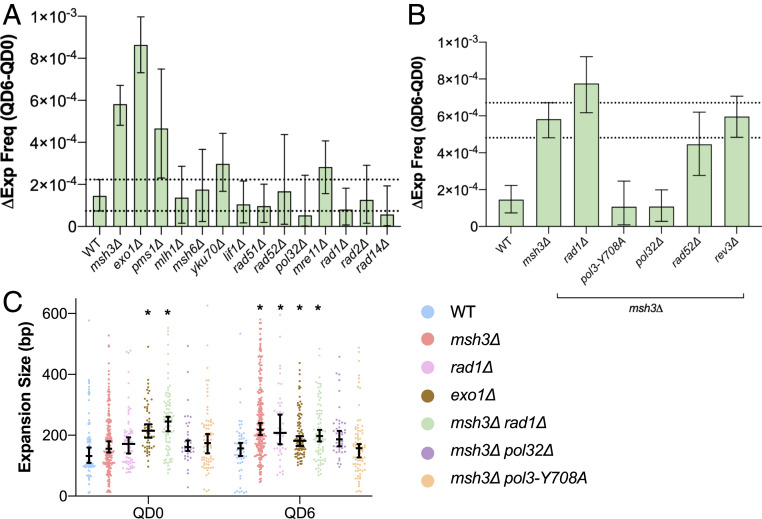
While deletions were reduced during quiescence in the exo1Δ strain, repeat expansions increased significantly in the absence of Exo1 throughout chronological aging (Fig. 6A), indicating that Exo1 suppresses expansions. Similarly, we found that the change in expansion frequency between QD0 and QD6 (Δexpfreq) was substantially elevated when compared to WT in msh3Δ strains despite the dramatic reduction in large deletions and gene conversion events in those strains (Fig. 6A). There was no difference between repeat expansion rate per cell division between WT and msh3Δ strains during logarithmic growth, proving that Msh3 specifically suppresses expansions during quiescence (SI Appendix, Fig. S5A). We also found that expansion events in msh3Δ strains were significantly longer at QD6 compared to WT (Fig. 6C). The effect of msh3Δ on Δexpfreq was recapitulated in pms1Δ strains (Fig. 6A), indicating the expansion suppression effect of MutSβ is somewhat dependent on incision by MutLα. Surprisingly, however, mlh1Δ caused no change in Δexpfreq.
Fig. 6.
Repeat expansions during quiescence are suppressed by MMR and require DNA synthesis by Polδ. (A) Msh3, Exo1, and Pms1 suppress expansions during chronological aging. (B) pol32Δ and pol3-Y708A rescue the expansion phenotype in msh3Δ. Bars represent median change in expansion frequency (ΔExp freq) between QD0 and QD6. Biological significance was defined as no overlap between the double mutant and msh3Δ 95% credible intervals. Dashed lines indicate msh3Δ 95% credible interval. (C) Expansion length as determined by agarose gel electrophoresis at QD0 and QD6. Black bars and whiskers represent medians and 95% confidence intervals. *P < 0.05, two-way ANOVA with Dunnett’s correction for multiple comparisons.
Replication-Independent Repeat Expansions Require Processive DNA Polymerase δ.
Given the requirement for DNA synthesis to generate new (GAA)n repeats in the absence of replication, we set out to identify the DNA polymerase responsible for these events. It has been previously shown that repair synthesis during G1 in yeast is dependent on Pol32, which can act as a subunit of both the DNA polymerase δ (73) and ζ (74) holoenzymes that perform lagging strand and translesion synthesis, respectively. An msh3 pol32Δ double mutant showed a dramatic reduction in Δexpfreq compared to msh3Δ, confirming that Pol32-dependent repair synthesis is responsible for generating long expanded repeats in msh3Δ cells during quiescence. Since Pol32 is also involved in DNA synthesis during some forms of HR (discussed above), we tested whether Rad52 was necessary for expansions during quiescence. An msh3Δ rad52Δ double mutant displayed the same pattern of expansion frequency as the msh3Δ mutant alone during quiescence, discounting this possibility (Fig. 6B).
We next set out to identify whether the Pol32 binding partners Pol3 or Rev3, the catalytic subunits of DNA polymerase δ and ζ, respectively, were responsible for replication-independent repeat synthesis. An msh3Δ rev3Δ double mutant showed Δexpfreq similar to msh3Δ, demonstrating no role for DNA polymerase ζ in expansions (Fig. 6B). To test the role of DNA polymerase δ, we generated a pol3-Y708A mutant that drastically decreases its processivity (75). Repeat expansion frequency in an msh3Δ pol3-Y708A double mutant showed a significant elevation in repeat expansion frequency at QD0 (SI Appendix, Fig. S5C), consistent with what we observed previously during replicative aging (28). However, there was a decrease in Δexpfreq of msh3Δ pol3-Y708A compared to msh3Δ during chronological aging, arguing that Polδ processivity is important for expansions specifically in quiescence. In addition, both the pol3-Y708A and pol32Δ mutations reversed the longer expansion length phenotype observed at QD6 in msh3Δ strains—confirming that DNA polymerase δ drives replication-independent expansions and its processivity is necessary for generating long expansions (Fig. 6C).
Replication-Independent (GAA)n Expansions Occur in Multiple Steps.
The (GAA)n expansion length required to inactivate the URA3 gene in our genetic reporter is roughly 30 bp. This makes the scale of expansion that the (GAA)100 tract must undergo in order to confer 5-FOA resistance longer than the expansion events that are typically evaluated at expandable microsatellite sequences, which tend to be 3 to 12 bp in length (6). We have previously demonstrated that these “large-scale” expansions occur in a single step—often due to faulty replication. We wondered if the same was true for (GAA)n expansion events during chronological aging. We hypothesized that the median size of (GAA)n expansions would be consistent across quiescent time points if expansions occurred in a single step. If, however, expansions were the result of multiple smaller events, we expected to see an increase in expansion length as cells aged chronologically. The length of expansion events in msh3Δ increased consistently from QD0 to QD12, with a statistically significant increase between multiple time points with nonoverlapping confidence intervals (SI Appendix, Fig. S5D). This points to the fact that in the absence of MutSβ (GAA)n expansion events accumulate in steps as cells age chronologically.
Discussion
We have previously identified several pathways that contribute to (GAA)n length instability in dividing cells (28, 30, 31, 32, 41). In this study, we found that the mechanisms of repeat instability in nondividing cells are distinct from those in actively dividing cells. In particular, DSBs at the (GAA)n repeat and its flanking sequences appear to be the principal driver of repeat-mediated instability in quiescent yeast. Preferential repair of these DSBs via NHEJ causes large deletions that are prevalent in nondividing cells. In addition, ectopic gene conversion via HR of the same DSBs contributes to repeat-mediated genome instability, albeit to a lesser extent. Given the paucity of deletions and gene conversions in logarithmically dividing cell populations (Fig. 2B), we conclude that these events are specific to nondividing cells. We have also identified a replication-independent mechanism of (GAA)n expansion in quiescent cells that is counteracted by Msh3 and Exo1. This mechanism requires DNA polymerase δ for repair synthesis and results in progressively longer expansions during chronological aging. Taken together, our data point to a model of (GAA)n repeat instability in quiescent cells whereby multiple DNA repair pathways act to influence the specific outcomes. This closely reflects data from transgenic mouse models for HD, myotonic dystrophy, and FRDA that have shown that the absence of repair proteins shapes the modes of somatic repeat instability in a tissue specific fashion (42, 76–78).
Our finding that DSB formation at (GAA)n repeats can lead to large-scale deletions of neighboring genetic material suggests that somatic mutations beyond simple changes in repeat length might influence FRDA onset and progression. Since all studies that have examined somatic mutagenesis in FRDA patients and mice have relied on PCR to detect repeat length changes, large-scale deletions involving repeat-adjacent genetic material could not have been detected. Considering that, at least in yeast, these deletions are most prevalent in nondividing cells, their occurrence in postmitotic neurons might explain tissue specificity in FRDA. Indeed, there is evidence to suggest that MMR is active in neural tissues and that adult neurons accumulate large deletions via NHEJ (79, 80). Furthermore, patient neural tissue samples show elevated levels of somatic (GAA)n contractions when compared to other areas of the body (20), which indicates that deletions may be the primary somatic mutation in those regions. It is also worth noting that FRDA is somewhat unique among the repeat expansion diseases in that it is autosomal-recessive, which means deletions are more likely to contribute to disease onset. This contrasts with autosomal-dominant disorders where somatic deletions would potentially mitigate disease processes that are the result of repeat-associated gain of function (e.g., HD). Supporting this reasoning, FRDA patients who are compound FXN heterozygotes possessing repeat expansion in one allele and exonic deletions in another allele have earlier disease onset compared to patients with expanded repeats in both FXN alleles (81, 82). As more FRDA patients’ DNA is sequenced using long-read technologies, it will be interesting to see if deletions or rearrangements occur at or around the repeats in affected tissues. In fact, long-read sequencing of patient lymphoblasts with another repeat expansion disease, familial adult myoclonic epilepsy, found repeat-associated microrearrangements in cells with the largest somatic expansions that are similar to the mutations we observe during chronological aging (83).
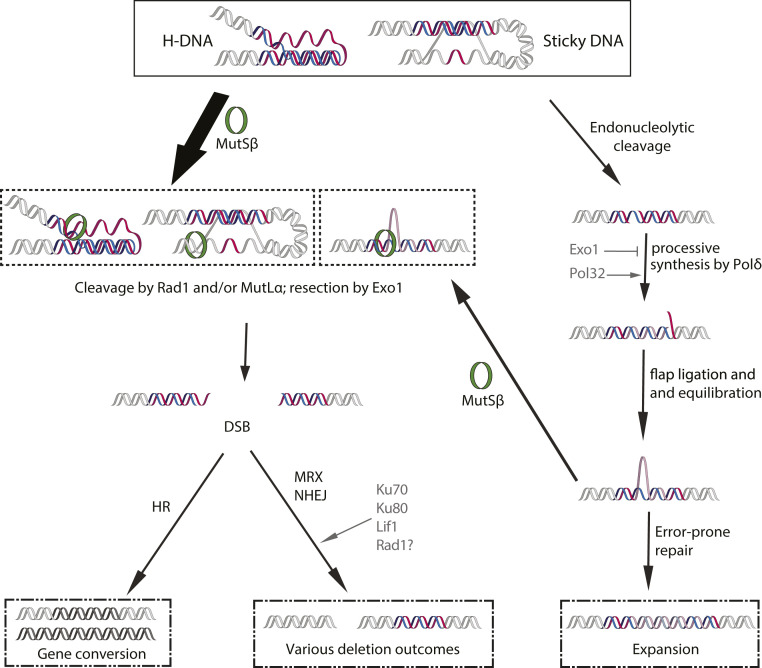
Mechanistically, our data indicate that the instability of (GAA)n repeats during quiescence is dependent on the type of DNA damage that occurs at the repeats and how that damage is repaired. In the case of gene conversions and deletions, the formation of a DSB is required. These DSBs are dependent on Msh3, Mlh1, and Pms1 as demonstrated by their requirement for deletions and gene conversion events in quiescent cells (Fig. 3). Rad1 appears to contribute to DSB formation as well given similar findings in rad1Δ strains (Fig. 5). Taken together, we conclude that H-DNA formation at (GAA)n repeats is a substrate for recognition by MutSβ and subsequent cleavage by the MutLα complex. Similarly, Rad1 appears capable of cleaving H-DNA, resulting in either DSB formation or the generation of a nick that would trigger the latent endonuclease activity of MutLα at the repeats. This agrees with our previous data showing cleavage of H-DNA formed by long (GAA)n repeats by MutLα in dividing yeast (49). It is also in line with studies showing that Rad1 knockout significantly reduces the frequency of chromosomal fragility at a triplex-forming mirror repeat in yeast (72) and that the human Rad1 ortholog XPF has been shown to incise single-stranded DNA (ssDNA) adjacent to Hoogsteen triads (72).
We also found that Exo1 plays a role in resecting DNA during deletion events based on the dramatic reduction in deletions during quiescence observed in exo1Δ strains. One plausible scenario of resection in Class I and Class II deletions is that nicks in both strands of an H-DNA–forming (GAA)100 repeat are introduced at a distance by MutLα and/or Rad1, which activates convergent exonucleolytic degradation by Exo1 to produce a DSB. These DSBs are then repaired by the NHEJ machinery, which is recruited to them by the MRX complex (Fig. 7). As for Class III events, the binding of MutSβ to regions flanking (as opposed to within) abnormally long (GAA)n repeats is not unprecedented—it has been previously shown downstream of the repetitive sequence in human induced pluripotent stems cells and mouse tissues—and could serve as the inciting event (84, 85).
Fig. 7.
Model for (GAA)n-repeat mediated instability during chronological aging. Transcription- or quiescence-induced superhelical stress drives the formation of three-stranded H-DNA by the (GAA)n repeat or sticky DNA between the repeats and a distant purine-rich sequence. These DNA structures can be recognized by MutSβ (Msh2, Msh3) and cleaved by MutLα (Mlh1, Pms1) or Rad1 to produce nick(s) either within the repeat, in the distant purine-rich sequence, or both. Exo1 5′-3′ resection leads to a DSB. In most cases, NHEJ of DSB ends yields distinct classes of deletions. Alternatively, homologous recombination yields a gene conversion event. In the absence of machineries producing multiple nicks, the resection of which yield a DSB (Msh3, Rad1, and Exo1), repair of a single-stranded nick can initiate an expansion event. In this case, additional repeats are generated by the processive synthesis of DNA polymerase δ (Pol3) in a Pol32-dependent manner followed by the ligation and equilibration of the repetitive flap. Error-prone repair can then expand the opposite strand to complete the repeat expansion. Multiple iterations of this repeat expansion process throughout chronological aging can lead to progressively longer repeats. Note that this expansion process can be reversed if MMR machinery converts an equilibrated flap(s) into a DSB, generating deletions or gene conversions.
Notably, we identified areas of high purine content within 15 bp of at least one repair junction in 20 out of 22 deletions sequenced (Fig. 4A). These data may point to an alternative deletion scenario: DSB formation triggered by the formation of sticky DNA between the (GAA)100 repeat and nearby purine-rich regions. Sticky DNA was originally identified in bacterial plasmids as a complex triplex formed by a pair of long, distant (GAA)n repeats (86, 87). In this structure, a purine-rich DNA strand from one repeat forms a stable triplex with a distant (GAA)n duplex. Based on the chemistry and topology of this structure it is also called distant H-DNA or tethered loop (88). Of a particular interest to us was an observation that tethered loops were shown to form between a long (GAA)n repeat and short nearby purine-rich motif(s) during episomal replication in mammalian cells (40). We speculate that an increase in the frequency and distance of intrachromosomal DNA interactions, which has been observed in quiescent cells (89), might promote similar sticky DNA formation at our cassette. This would explain the order of magnitude increase in the frequency of deletion events we see at (GAA)100 between QD0 and QD6 (Fig. 2C). The formation of sticky DNA causes significant unwinding of the duplex neighboring the region that donates a strand to the triplex (40), generating ssDNA–double-stranded DNA junctions that are preferred substrates for MutSβ (90). In particular, Class II deletion junctions tended to contain some iteration of GAA microhomology; the upstream junction of most of these deletions (five out of nine) clustered within or around a 30-bp-long sequence that contained 87% purines in the coding strand. If sticky DNA formed between a purine-rich hotspot and the (GAA)100 repeat is cleaved by MutLα, two DSBs at the base of an intramolecular triplex loop can join together, generating deletions that look similar to Class II events. Future biochemical experiments will distinguish between these different scenarios and the sequence of events leading to deletion formation.
If/when (GAA)n repeats are not converted into DSBs during quiescence, repeat expansions become their preferred mode of instability. This is clear from our data for msh3Δ mutant strain, which shows a greater increase in (GAA)n expansion frequency and length during chronological aging than the WT strain (Fig. 6 A and B). This increase was recapitulated in pms1Δ but did not carry over into mlh1Δ strains. Our data are consistent with findings in a FRDA mouse model, where deletion of the MMR endonuclease PMS2 (ortholog of yeast PMS1) promotes both intergenerational and somatic (GAA)n expansion but not contraction (42, 85). They contrast, however, with data showing that MSH2 and/or MSH3 are required for expansions that accumulate with passage number in human cell lines (35, 36, 84), as well as experiments showing MSH2 and MSH6 are necessary for (GAA)n somatic length instability in mouse cerebellum (42). The exact reasons for these differences in the effects of MMR enzymes on repeat instability between yeast and mammalian experimental systems are not yet clear. It should be noted that PMS2 knockout mice display a shift in somatic expansion size toward longer events—a pattern that was recapitulated in msh3Δ quiescent yeast specifically after chronological aging in our assay (Fig. 6C) (42). These events are also similar in length to those observed in the pancreas and heart of FRDA patients (>450 bp), which have some of the lowest levels of MSH3 RNA expression of any tissue according to data from The Human Protein Atlas (15) and show a predominance of (GAA)n expansions over contractions (20).
We suggest a unique mode of (GAA)n expansion that is suppressed by MutSβ and is specific to nondividing cells. Since Exo1 nuclease strongly suppresses repeat expansions as well, we conclude that these events are initiated by single-stranded breaks within the repetitive sequence that are left unresected. However, we could not yet identify a single specific nuclease responsible for these breaks (Fig. 6A and SI Appendix, Fig. S5B). Several recent studies implied that human MutLγ (MLH1–MLH3) heterodimer is an endonuclease that promotes expansions of several repeats, including FRDA (GAA)n repeats (91–95). In our system, however, Mlh3 knockout did not affect repeat expansions in quiescent cells (SI Appendix, Fig. S5B). Given the shear length of single-stranded segments in H-DNA, it is quite feasible that multiple nuclear endonucleases or endogenous damage sources could be involved.
Whatever the origin of the nick, our data show that the expansion process requires processive DNA polymerase δ, since Δexpfreq and expansion length decreases in msh3Δ pol3-Y708A and msh3Δ pol32Δ mutants. Pol32 has been implicated in G1-specific nucleotide excision repair and interstrand cross-link repair in yeast (96, 97), implying that it may be a crucial DNA polymerase subunit for repair synthesis during the G1 phase. Furthermore, it is required for displacement synthesis by Pol δ (98). We propose, therefore, that Pol δ repair synthesis would displace a repetitive flap, subsequent ligation of which would expand the repeat in the repaired strand (Fig. 7). A nick opposite to an equilibrated flap would convert it into a template for error-prone repair (99), resulting in repeat expansions in both DNA strands (Fig. 7). Repeated cycles of this process could result in long expansion events that accumulate during chronological aging. This process would be counteracted by the MMR machinery, which can recognize and cleave an equilibrated flap, ultimately leading to deletion formation or repeat contraction (Fig. 7).
In summary, we have analyzed the spectrum of (GAA)n-associated mutations that arise in homogeneous cultures of quiescent yeast that have been aged chronologically. We identified large scale deletions that result from imprecise NHEJ of nuclease-mediated DSBs as the predominant mutation type in nondividing cells. To a lesser extent, these DSBs are repaired by HR leading to ectopic gene conversions. In cells lacking Msh3, unusually long, replication-independent expansions occur that depend on the processivity of DNA polymerase δ. Taken as a whole, these data demonstrate that the type of instability mediated by expanded (GAA)n repeats depends greatly on the specific repair systems active in an individual tissue and at a given stage of the cell cycle. This puts forward a framework for somatic mutagenesis at (GAA)n repeats in patients with FRDA and stipulates that large-scale deletions at (GAA)n repeats in postmitotic cells may contribute to tissue specificity, disease onset and progression.
Materials and Methods
Yeast Strains.
All yeast strains used in this study were derived from the parent strain CH1585 (MATa, leu2-Δ1, trp1-Δ63, ura3-52) (30) and are listed in SI Appendix, Table S1. Knockouts were made using direct gene replacement and confirmed via PCR using pairs of internal and external primers against the targeted gene (SI Appendix, Table S2). Point mutations (mre11-D56N, pol3-Y708A and pms1-E707K) were made using CRISPR-Cas9 and primers listed in SI Appendix, Table S2 as previously described (100). The construction of our genetic cassette for measuring (GAA)n-associated mutagenesis was described in depth previously (28).
Quiescent Cell Mutation Frequency Assay.
Six 1.2-mL phosphate-limited cultures (media recipes in SI Appendix, Table S3, based off of ref. 101) were inoculated with less than 10 cells per well in a 24-well plate. The cell count of inoculates was determined by both hemocytometer and colony-forming units (CFUs) on complete media (YPD). Cultures were grown at 30 °C with rotation for 40 h, at which point aliquots were taken from each culture and serially diluted at 10× intervals for the “LOG” time point. One hundred microliters of 10× dilute samples were plated on synthetic complete media supplemented with 0.95% 5-FOA (Toronto Research Chemicals Inc.) to detect cells that contained repeat-associated mutations, while 50 µL of 10,000× dilute samples were plated on YPD to determine total cell count of the sample. After an additional 24 h (64 total), cultures were pelleted in Eppendorf tubes, washed once with water, and resuspended in 75 µL of sterile suspension buffer (50 mM Tris⋅HCl, pH 7.9). The suspension was mildly sonicated for 90 s and then applied to the top of a preformed 1.3 mL Percoll (Cytiva) gradient in a 1.5-mL microcentrifuge tube. Gradients consisted of 9:1 Percoll:1.5 M NaCl spun for 15 min at 19,250 rcf in a fixed-angle microcentrifuge (45). Cells were run through the gradient for 1 h at 400 × g on a swinging bucket centrifuge at 4 °C. Cells from the bottom 150 µL of the gradient were kept, washed twice in water, and resuspended in 1 mL of synthetic complete media with no phosphate in a 1.5-mL microcentrifuge tube (SI Appendix, Table S3). Aliquots were taken again after resuspension in the no-phosphate media and plated as described to determine mutation frequencies for QD0 (dilutions were identical to the LOG time point). Cells were then aged chronologically at 30 °C with nutation for up to 12 d and sampled as indicated to obtain mutation frequencies. For all time points and genotypes, colonies were counted on 5-FOA and YPD after 3 d of growth at 30 °C.
Mutation Type and Frequency Determination.
For each genotype at each time point, genomic DNA from at least 48 5-FOA–resistant colonies was isolated and analyzed. The number of colonies examined per culture correlated with the number of colonies on that 5-FOA plate as compared to others from the same time point and genotype (i.e., more colonies were sampled from plates with a higher colony count). In some cases, single cultures had clearly accumulated point mutations during the replicative aging portion of the assay and were excluded from subsequent mutation analysis. Expansions were defined as an addition of 10 or more repeats (30 bp) as determined via 1.5% agarose gel electrophoresis of expF/R PCR amplicons (SI Appendix, Table S2), as this was the shortest expansion size that could be clearly resolved via gel electrophoresis. Expansion length was determined using Image Lab 6.0 (Bio-Rad). Those samples that showed no product or no change in repeat length in the expansion PCR were subsequently evaluated for deletion events using primers delF and delR (Fig. 1A). Deletions were defined as length reductions of >200 bp on 1% agarose gel (>9% of the 2,319-bp PCR product). Note that the closest essential genes upstream and downstream are roughly 10 kb and 60 kb away from our cassette, respectively, limiting viable deletions to this 70-kb window. Samples that showed no product in the deletion PCR were assessed for recombination events using primers recF and recR (Fig. 2A). The presence of a PCR product in this case was evidence of recombination, since the forward and reverse primers are specific to chromosomes III and V, respectively. Microhomologies at deletion junctions were identified via Sanger sequencing of the deletion PCR product.
For the LOG time point, mutation rates were calculated as previously described using our FluCalc website (32). For chronological aging time points QD0 and QD6, we sampled six independent cultures for each genotype and used Bayesian logistic regression to infer a joint probability distribution for the theoretical values of the following parameters for the WT and mutant strains: 1) the frequency of 5-FOA resistance and 2) the frequency of each specific mutational event (expansion, recombination or deletion) among resistant cells. The model treats the number of 5-FOA–resistant cells and the number of colonies sampled from 5-FOA plates with the given mutation type as independent binomial data. Markov chain Monte Carlo simulation was used to generate a sample of 4,000 values for each of the four parameters from a distribution consistent with our experimental mutation frequencies. These simulated values were then used to generate a sample of mutation frequencies at each time point, by multiplying 1 and 2, and to estimate a change in mutation frequency during chronological aging (Δmutationfreq) by subtracting the mutation frequencies at QD0 from those at QD6. The end result of 4,000 estimates of Δmutationfreq represents a data-driven distribution that allows probabilistic statements about the theoretical value of Δmutationfreq. (102).
The first advantage of the Bayesian approach is that it allowed us to estimate the mutation frequencies even in cases where no mutations of a given type were observed, which we found in many mutant genotypes. A second benefit of the model is that it allowed us to incorporate the constraint that the mutation frequency and resistance frequency must increase over time, even if on a small scale. This constraint assumes that the viability of mutants and nonmutants is equal during chronological aging, which we confirmed by assessing viability in Ura+ and Ura− strains (SI Appendix, Fig. S1D). Incorporating such an assumption improves inference of frequencies at a given timepoint by making use of information from neighboring time points. Details of the statistical model and how it was fit are provided in SI Appendix.
Assessments of Quiescence.
Microfluidic channels were fabricated based on soft-lithography techniques (103). In short, a casting mold was prepared by spin coating SU-8 2075 (MicroChem Corp.) onto a silicon wafer. The chambers were patterned through photolithography using a mylar photomask (Advance Reproductions Corp.). Next, a silicon elastomer, polydimethylsiloxane (PDMS; Dow Corning), was prepared using a 10:1 ratio (wt/wt) of polymer to cross-linker. PDMS channels were cured in an oven at 65 °C for 1 h, and subsequently the PDMS channels were plasma-bonded to a standard glass microscope slide (25 mm × 75 mm × 1 mm; cleaned with ethanol) (104). The PDMS channels consisted of two circular 2-mm-diameter × 3-mm-length openings leading to a central chamber that is 3.5 mm in diameter × 100 µm in depth (total volume ∼20 μL). Four PDMS channels were cast per microscope slide. Chambers were first sterilized with 70% ethanol then rinsed with sterile deionized H2O. QD1 yeast samples were pelleted, resuspended in desired media, and injected into the central chamber. Capillaries from both ends of the channel were then removed and sealed with wax to prevent evaporation. Cells were recorded for 24 h at room temperature on a Nikon Ti-E microscope with a Zyla sCMOS camera (Andor Technology) utilizing phase-contrast microscopy with a 10×, 0.3 numerical aperture objective. Videos were captured using NIS Elements Advanced Research 4.3 and were processed using ImageJ. Three separate cultures were analyzed. Cells were considered “divided” if they underwent a single division in the 24-h time period.
For bud scar analysis, QD0 cells were pelleted and resuspended in a 10% calcofluor white (Sigma) solution for 5 min. They were rinsed once in water and whole cells were imaged as Z-stacks with 0.25 µM slices on a Zeiss Axio Imager.M1 microscope using DAPI filter set 49. Z-stacks were processed using extended depth of focus on default settings with Zeiss Zen software. Bud scar scoring was conducted using the cell counter tool in ImageJ. Viability across chronological aging was assessed via CFUs on YPD at each time point divided by YPD CFU at QD0 for that culture.
Protein Extraction and Western Blotting.
Protein extracts from dividing or quiescent yeast cultures were prepared and quantified as described in ref. 105 and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blotting with an anti-myc antibody (13-2500; Thermo Fisher).
Supplementary Material
Acknowledgments
We are indebted to Kirill Lobachev for prompting us to study repeat instability in nondividing cells and to Steve Fuchs for introducing us to the quiescent yeast model. We thank Sue Jinks-Robertson, Catherine Freudenreich, Mitch McVey, Wey Yang, Farid Kadyrov, and members of the S.M.M. and Freudenreich laboratories for invaluable discussions. We are grateful to Jeff Guasto for allowing us to use his laboratory materials and equipment to assess yeast quiescence in microfluidics chambers and Amin Dehkharghani for providing us instruction and protocols for these analyses. We recognize Tara Zhou for her work on this project. This study was supported by NIH Grant R35GM130322 and by a generous contribution from the White family to S.M.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013080118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
References
- 1.Mirkin S. M., Expandable DNA repeats and human disease. Nature 447, 932–940 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Zhao X. N., Usdin K., The repeat expansion diseases: The dark side of DNA repair. DNA Repair (Amst.) 32, 96–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campuzano V., et al. , Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Reetz K., et al. , Biological and clinical characteristics of the European Friedreich’s Ataxia consortium for translational studies (EFACTS) cohort: A cross-sectional analysis of baseline data. Lancet Neurol. 14, 174–182 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Campuzano V., et al. , Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6, 1771–1780 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Khristich A. N., Mirkin S. M., On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J. Biol. Chem. 295, 4134–4170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanker E. E., Ast A., Schindler F., Trepte P., Schnoegl S., The pathobiology of perturbed mutant huntingtin protein-protein interactions in Huntington’s disease. J. Neurochem. 151, 507–519 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Arrasate M., Finkbeiner S., Protein aggregates in Huntington’s disease. Exp. Neurol. 238, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorens J. V., Soriano S., Calap-Quintana P., Gonzalez-Cabo P., Moltó M. D., The role of iron in Friedreich’s Ataxia: Insights from studies in human tissues and cellular and animal models. Front. Neurosci. 13, 75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciosi M.et al.; TRACK-HD team; Enroll-HD team , A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. EBioMedicine 48, 568–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmans P. A., Massey T. H., Jones L., Genetic modifiers of Mendelian disease: Huntington’s disease and the trinucleotide repeat disorders. Hum. Mol. Genet. 26, R83–R90 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium. Electronic address: gusella@helix.mgh.harvard.edu; Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium, CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell 178, 887–900.e14 (2019). [DOI] [PMC free article] [PubMed]
- 13.Wheeler V. C., et al. , Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet. 12, 273–281 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Budworth H., et al. , Suppression of somatic expansion delays the onset of pathophysiology in a mouse model of Huntington’s disease. PLoS Genet. 11, e1005267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Cossée M., et al. , Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet. 9, 1219–1226 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Bidichandani S. I., Delatycki M. B., “Friedreich ataxia” in GeneReviews, Adam M. P., Ed. et al. (University of Washington, Seattle, WA, 1993). [Google Scholar]
- 18.Sapp E., et al. , Huntingtin localization in brains of normal and Huntington’s disease patients. Ann. Neurol. 42, 604–612 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Tabrizi S. J.et al.; TRACK-HD Investigators , Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: The 12-month longitudinal analysis. Lancet Neurol. 10, 31–42 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Long A., et al. , Somatic instability of the expanded GAA repeats in Friedreich’s ataxia. PLoS One 12, e0189990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Biase I., et al. , Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics 90, 1–5 (2007). [DOI] [PubMed] [Google Scholar]
- 22.De Biase I., et al. , Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Ann. Neurol. 61, 55–60 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Al-Mahdawi S., et al. , GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics 84, 301–310 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Anjomani Virmouni S., et al. , A novel GAA-repeat-expansion-based mouse model of Friedreich’s ataxia. Dis. Model. Mech. 8, 225–235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark R. M., et al. , The GAA triplet-repeat is unstable in the context of the human FXN locus and displays age-dependent expansions in cerebellum and DRG in a transgenic mouse model. Hum. Genet. 120, 633–640 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Shelbourne P. F.et al.; US-Venezuela Collaborative Research Group , Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 16, 1133–1142 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Kennedy L., et al. , Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 12, 3359–3367 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Shah K. A., et al. , Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2, 1088–1095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutakawa S., et al. , Phosphate steering by flap endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nat. Commun. 8, 15855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shishkin A. A., et al. , Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol. Cell 35, 82–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGinty R. J., et al. , A defective mRNA cleavage and polyadenylation complex facilitates expansions of transcribed (GAA)n repeats associated with Friedreich’s ataxia. Cell Rep. 20, 2490–2500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neil A. J., Liang M. U., Khristich A. N., Shah K. A., Mirkin S. M., RNA-DNA hybrids promote the expansion of Friedreich’s ataxia (GAA)n repeats via break-induced replication. Nucleic Acids Res. 46, 3487–3497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ditch S., Sammarco M. C., Banerjee A., Grabczyk E., Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet. 5, e1000704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polak U., Li Y., Butler J. S., Napierala M., Alleviating GAA repeat induced transcriptional silencing of the Friedreich’s ataxia gene during somatic cell reprogramming. Stem Cells Dev. 25, 1788–1800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du J., et al. , Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J. Biol. Chem. 287, 29861–29872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halabi A., Ditch S., Wang J., Grabczyk E., DNA mismatch repair complex MutSβ promotes GAA·TTC repeat expansion in human cells. J. Biol. Chem. 287, 29958–29967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gacy A. M., et al. , GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell 1, 583–593 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Krasilnikova M. M., Mirkin S. M., Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 24, 2286–2295 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerhardt J., et al. , Stalled DNA replication forks at the endogenous GAA repeats drive repeat expansion in Friedreich’s ataxia cells. Cell Rep. 16, 1218–1227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Follonier C., Oehler J., Herrador R., Lopes M., Friedreich’s ataxia-associated GAA repeats induce replication-fork reversal and unusual molecular junctions. Nat. Struct. Mol. Biol. 20, 486–494 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., et al. , Genome-wide screen identifies pathways that govern GAA/TTC repeat fragility and expansions in dividing and nondividing yeast cells. Mol. Cell 48, 254–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourn R. L., et al. , Pms2 suppresses large expansions of the (GAA·TTC)n sequence in neuronal tissues. PLoS One 7, e47085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miles S., Breeden L., A common strategy for initiating the transition from proliferation to quiescence. Curr. Genet. 63, 179–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Virgilio C., The essence of yeast quiescence. FEMS Microbiol. Rev. 36, 306–339 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Allen C., et al. , Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174, 89–100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGinty R. J., et al. , Nanopore sequencing of complex genomic rearrangements in yeast reveals mechanisms of repeat-mediated double-strand break repair. Genome Res. 27, 2072–2082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt M. H. M., Pearson C. E., Disease-associated repeat instability and mismatch repair. DNA Repair (Amst.) 38, 117–126 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Tang W., et al. , Friedreich’s ataxia (GAA)n•(TTC)n repeats strongly stimulate mitotic crossovers in Saccharomyces cerevisae. PLoS Genet. 7, e1001270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H. M., et al. , Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 27, 2896–2906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Acharya S., et al. , hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadyrov F. A., Dzantiev L., Constantin N., Modrich P., Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126, 297–308 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Goellner E. M., Putnam C. D., Kolodner R. D., Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst.) 32, 24–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsieh P., Zhang Y., The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 114, 3552–3554 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma J. L., Kim E. M., Haber J. E., Lee S. E., Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X., Gabriel A., Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163, 843–856 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K., Lee S. E., Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 176, 2003–2014 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathiasen D. P., Lisby M., Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 172–184 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Daley J. M., Wilson T. E., Rejoining of DNA double-strand breaks as a function of overhang length. Mol. Cell. Biol. 25, 896–906 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho J. E., Jinks-Robertson S., Deletions associated with stabilization of the Top1 cleavage complex in yeast are products of the nonhomologous end-joining pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 22683–22691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakofsky C. J., Malkova A., Break induced replication in eukaryotes: Mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol. 52, 395–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Symington L. S., Rothstein R., Lisby M., Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J. C., Harris S. T., Dinter T., Shah K. A., Mirkin S. M., The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat. Struct. Mol. Biol. 24, 55–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith C. E., Llorente B., Symington L. S., Template switching during break-induced replication. Nature 447, 102–105 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Yin Y., Dominska M., Yim E., Petes T. D., High-resolution mapping of heteroduplex DNA formed during UV-induced and spontaneous mitotic recombination events in yeast. eLife 6, e28069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh J., Symington L. S., Role of the Mre11 complex in preserving genome integrity. Genes (Basel) 9, E589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Llorente B., Symington L. S., The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 24, 9682–9694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreau S., Ferguson J. R., Symington L. S., The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19, 556–566 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krogh B. O., Llorente B., Lam A., Symington L. S., Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171, 1561–1570 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore J. K., Haber J. E., Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2164–2173 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langerak P., Mejia-Ramirez E., Limbo O., Russell P., Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartz E. K., Heyer W. D., Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120, 109–127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J., et al. , Distinct mechanisms of nuclease-directed DNA-structure-induced genetic instability in cancer genomes. Cell Rep. 22, 1200–1210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gerik K. J., Li X., Pautz A., Burgers P. M., Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273, 19747–19755 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Makarova A. V., Stodola J. L., Burgers P. M., A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 40, 11618–11626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pavlov Y. I., Shcherbakova P. V., Kunkel T. A., In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics 159, 47–64 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mason A. G., et al. , Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Hum. Mol. Genet. 23, 1606–1618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kovtun I. V., et al. , OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447, 447–452 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van den Broek W. J., et al. , Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 11, 191–198 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Hazen J. L., et al. , The complete genome sequences, unique mutational spectra, and developmental potency of adult neurons revealed by cloning. Neuron 89, 1223–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brooks P. J., Marietta C., Goldman D., DNA mismatch repair and DNA methylation in adult brain neurons. J. Neurosci. 16, 939–945 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galea C. A., et al. , Compound heterozygous FXN mutations and clinical outcome in friedreich ataxia. Ann. Neurol. 79, 485–495 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Anheim M., et al. , Exonic deletions of FXN and early-onset Friedreich ataxia. Arch. Neurol. 69, 912–916 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Florian R. T.et al.; FAME consortium , Unstable TTTTA/TTTCA expansions in MARCH6 are associated with familial adult myoclonic Epilepsy type 3. Nat. Commun. 10, 4919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ku S., et al. , Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA⋅TTC triplet repeat instability. Cell Stem Cell 7, 631–637 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ezzatizadeh V., et al. , The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol. Dis. 46, 165–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakamoto N., et al. , Sticky DNA: Self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol. Cell 3, 465–475 (1999). [DOI] [PubMed] [Google Scholar]
- 87.Vetcher A. A., et al. , Sticky DNA, a long GAA.GAA.TTC triplex that is formed intramolecularly, in the sequence of intron 1 of the frataxin gene. J. Biol. Chem. 277, 39217–39227 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Mirkin S. M., “Structure and biology of H DNA” in Triple Helix Forming Oligonucleotides, Malvy C., Harel-Bellan A., Pritchard L. L., Eds. (Springer US, Boston, MA, 1999), pp. 193–222. [Google Scholar]
- 89.Rutledge M. T., Russo M., Belton J. M., Dekker J., Broach J. R., The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 43, 8299–8313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gupta S., Gellert M., Yang W., Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 19, 72–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halabi A., Fuselier K. T. B., Grabczyk E., GAA•TTC repeat expansion in human cells is mediated by mismatch repair complex MutLγ and depends upon the endonuclease domain in MLH3 isoform one. Nucleic Acids Res. 46, 4022–4032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kadyrova L. Y., Gujar V., Burdett V., Modrich P. L., Kadyrov F. A., Human MutLγ, the MLH1-MLH3 heterodimer, is an endonuclease that promotes DNA expansion. Proc. Natl. Acad. Sci. U.S.A. 117, 3535–3542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hayward B. E., Steinbach P. J., Usdin K., A point mutation in the nuclease domain of MLH3 eliminates repeat expansions in a mouse stem cell model of the Fragile X-related disorders. Nucleic Acids Res. 48, 7856–7863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Su X. A., Freudenreich C. H., Cytosine deamination and base excision repair cause R-loop-induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 114, E8392–E8401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flores-Rozas H., Kolodner R. D., The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 95, 12404–12409 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kozmin S. G., Jinks-Robertson S., The mechanism of nucleotide excision repair-mediated UV-induced mutagenesis in nonproliferating cells. Genetics 193, 803–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sarkar S., Davies A. A., Ulrich H. D., McHugh P. J., DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 25, 1285–1294 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M., Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 283, 34129–34140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Panigrahi G. B., Lau R., Montgomery S. E., Leonard M. R., Pearson C. E., Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 12, 654–662 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Khristich A. N., Armenia J. F., Matera R. M., Kolchinski A. A., Mirkin S. M., Large-scale contractions of Friedreich’s ataxia GAA repeats in yeast occur during DNA replication due to their triplex-forming ability. Proc. Natl. Acad. Sci. U.S.A. 117, 1628–1637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saldanha A. J., Brauer M. J., Botstein D., Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 15, 4089–4104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gelman A. C., et al. , Bayesian Data Analysis (CRC Press, Boca Raton, ed. 3, 2014). [Google Scholar]
- 103.Xia Y., Whitesides G. M., Soft lithography. Angew. Chem. Int. Ed. Engl. 37, 550–575 (1998). [DOI] [PubMed] [Google Scholar]
- 104.Haubert K., Drier T., Beebe D., PDMS bonding by means of a portable, low-cost corona system. Lab Chip 6, 1548–1549 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Webb K. J., Xu T., Park S. K., J. R. Yates, 3rd, Modified MuDPIT separation identified 4488 proteins in a system-wide analysis of quiescence in yeast. J. Proteome Res. 12, 2177–2184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.