Abstract
Background
ZNF385B, a zinc finger protein, has been known as a potential biomarker in some neurological and hematological studies recently. Although numerous studies have demonstrated the potential function of zinc finger proteins in tumor progression, the effects of ZNF385B in breast cancer (BC) are less studied.
Methods
The Oncomine database and “ESurv” tool were used to explore the differential expression of ZNF385B in pan-cancer. Furthermore, data of patients with BC were downloaded from The Cancer Genome Atlas (TCGA). The receiver operating characteristic (ROC) curve of ZNF385B expression was established to explore the diagnostic value of ZNF385B and to obtain the cut-off value of high or low ZNF385B expression in BC. The chi-square test as well as Fisher exact test was used for identification of the relationships between clinical features and ZNF385B expression. Furthermore, the effects of ZNF385B on BC patients' survival were evaluated by the Kaplan-Meier and Cox regression. Data from the Gene Expression Omnibus (GEO) database were employed to validate the results of TCGA. Protein expression of ZNF385B in BC patient specimens was detected by immunohistochemistry (IHC) staining.
Results
ZNF385B expression was downregulated in most types of cancer including BC. Low ZNF385B expression was related with survival status, overall survival (OS), and recurrence of BC. ZNF385B had modest diagnostic value, which is indicated by the area under the ROC curve (AUC = 0.671). Patients with lower ZNF385B expression had shorter OS and RFS (relapse-free survival). It had been demonstrated that low ZNF385B expression represented independent prognostic value for OS and RFS by multivariate survival analysis. The similar results were verified by datasets from the GEO database as well. The protein expression of ZNF385B was decreased in patients' samples compared with adjacent tissues by IHC.
Conclusions
Low ZNF385B expression was an independent predictor for worse prognosis of BC patients.
1. Introduction
Breast cancer (BC) is the most common female carcinoma [1] and has become a concerned health issue globally in recent years. In fact, BC has been reported as the first cause of cancer death among women across the world now [2]. With the groundbreaking medical advances during the past decades, people have realized that BC is a complicated disease with various stages and subtypes [3]. Approaches to dealing with BC are moving in a more effective and accurate direction. However, detection and prognosis of BC are still challenging. Thus, identifying biomarkers with clinical significance for the diagnosis and prognosis of BC is strongly demanded.
Malignant proliferation is always shadowed by the essential alterations in genome changes. ZNF385B, also called ZNF533 [4], belongs to a member of the zinc finger gene (ZNF) family. ZNF385B played a vital role in gene expression, working as encoding transcription factors [5]. Henriette found that trimethylation of lysine 9 (H3me3K9), which is associated with silenced chromatin, was highly enriched in ZNFs. ZNFs, as promoters of transcription factor, were also combined with the corepressor KAP1. Briefly, Krüppel-associated box domain zinc finger (KRAB-ZNF) family members were involved in a self-regulatory loop and led to H3K9 trimethylation and transcriptional inhibition which indicated that this mRNA may be a transcription suppressor [6]. In recent studies, it has already been reported in several systemic disease, including Burkitt' s lymphoma, cardiac arrest, nonsyndromic orofacial clefts, and obesity [4, 7–9]. However, the potential effects of ZNF385B expression in BC have not been defined. In the present research, we found differential expression of ZNF385B in multiple types of cancer including BC compared to normal tissues using the Oncomine database [10] and significant prognostic value of ZNF385B in BC based on a web-based tool “ESurv” [11]. In addition, a research based on the prognostic value of 8-gene for early BC found that ZNF385B may show some value in the prognostic scoring of early BC, but it remains to be further validated [12]. However, the diagnostic and prognostic value of ZNF385B for BC and the associations between ZNF385B and clinicopathological parameters in BC patients have not been reported. We discussed ZNF385B mRNA expression between BC patients and healthy human beings by analyzing TCGA (The Cancer Genome Atlas) and GEO (Gene Expression Omnibus) databases, and its expression was verified by immunohistochemistry (IHC) staining using patients' samples. To identify the diagnostic value of ZNF385B, patients were split into high or low group based on ZNF385B expression. Besides, we also explored the connections between ZNF385B expression and clinicopathologic factors as well as survival times including overall survival (OS) and relapse-free survival (RFS) of BC patients. Our results showed that ZNF385B might serve as a potential diagnostic and prognostic biomarker of BC.
2. Methods
2.1. Pan-Cancer Analysis of ZNF385B Based on Oncomine Database and “ESurv” Tool
The Oncomine database (http://www.oncomine.com) was adopted to evaluate the difference of ZNF385B mRNA expression levels between tumor and normal tissues in various tumor types. The analysis conditions were set as follows: p < 0.0001, fold change > 2, gene rank (top 10%), and data type (all). The web-based tool “ESurv” (https://www.giantonline.org) was used to perform the survival analysis of ZNF385B in pan-cancer systemically as well.
2.2. Data Mining and Analysis in TCGA and GEO Databases
The level 3 expression data of BC was obtained from TCGA database (https://cancergenome.nih.gov/), and the data underwent log2 (x + 1) transformation for RSEM normalized counts. Clinical data of BC patients were also obtained from TCGA database. The processing and analysis of all data were performed with R software [13]. To validate the results in TCGA database, the GSE21422 [14] and GSE20711 [15] datasets were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). Correlations between ZNF385B expression and pathological variables such as ER, HER-2, grade, and subtype were explored. The effects of ZNF385B on OS and RFS were assessed as well.
2.3. Immunochemistry (IHC) Staining
A total of 10 paired tumor and adjacent tissues of patients with BC were collected. All patients received surgery at the China-Japan Union Hospital of Jilin University and were histologically diagnosed by 2 independent pathologists based on WHO criteria. IHC staining was performed to identify the expression level of ZNF385B in BC patients' tissues. Briefly, 3 μm thick sections were deparaffinized, rehydrated, and submerged into EDTA for antigen retrieval. All sections were next dealt with hydrogen, heated, incubated in bovine serum albumin, and then followed by incubation with ZNF358B antibody (Abcam, Cambridge, MA, USA) at 4°C overnight. Normal goat serum served as negative control. The sections were washed and subsequently cultivated with secondary antibody and incubated with horseradish peroxidase-streptavidin complex (Invitrogen, CA, USA). Each section was immersed in 3-amino-9-ethyl carbazole followed by counterstaining with Mayer's hematoxylin, dehydrating, and mounting. Nuclear staining was considered positive.
2.4. Statistical Analysis
The expression of ZNF385B in patients was evaluated through box plots. The value of ZNF385B in diagnosis was assessed with AUC (area under the curve) by establishing the ROC (receiver operating characteristic) curve through the pROC package. According to the threshold value confirmed by the ROC curve, patients were classified into low or high ZNF385B expression group. The chi-square test as well as Fisher exact test was used for analysis of the associations between clinicopathologic features and ZNF385B expression in BC. Using the survival package in R, OS and RFS were compared via Kaplan-Meier analysis between the high and low ZNF385B expression groups, and the p value was calculated via log-rank test. Potential prognostic factors were screened via univariate Cox analysis followed by further survival evaluation using multivariate Cox analysis. p value < 0.05 signified statistical significance.
3. Results
3.1. Pan-Cancer Analysis of ZNF385B mRNA Expression
Firstly, the Oncomine database was used to assess the differential expression of ZNF385B mRNA levels in various cancers, and low ZNF385B expression was found in multiple types of cancer, such as BC, brain malignant tumor, lung cancer, kidney cancer, and liver cancer (Supplementary Figure 1A). Then, single gene analysis was performed in pan-cancer about ZNF385B using the web-based tool “ESurv” [11], and it was found that aberrant expression of ZNF385B was related to poor prognosis of BC, renal cancer, liver cancer, and brain cancer (Supplementary Figures 1B–1E).
3.2. Characteristics of the Study Population
Clinical data of 1104 BC patients and 114 normal controls were obtained from TCGA database. All data are shown in Table 1, including age, gender, menopause status, histological type, T classification, N classification, M classification, TNM stage, molecular subtype, ER, PR, HER-2, margin status, vital status, radiation therapy, neoadjuvant treatment, targeted molecular therapy, sample type, OS, RFS and ZNF385B expression of BC. And all the patients were assigned to different groups based on these characters.
Table 1.
Characteristics of the study population.
| Characteristics | Numbers of cases (%) |
|---|---|
| Age | |
| NA | 2 (0.18) |
| <60 | 589 (53.35) |
| ≥60 | 513 (46.47) |
| Gender | |
| NA | 2 (0.18) |
| Female | 1090 (98.73) |
| Male | 12 (1.09) |
| Histological type | |
| NA | 3 (0.27) |
| Infiltrating ductal carcinoma | 790 (71.56) |
| Infiltrating lobular carcinoma | 204 (18.48) |
| Other | 107 (9.69) |
| Molecular subtype | |
| NA | 255 (23.1) |
| Basal | 142 (12.86) |
| HER-2 | 67 (6.07) |
| Luminal A | 422 (38.22) |
| Luminal B | 194 (17.57) |
| Normal | 24 (2.17) |
| ER | |
| NA | 50 (4.53) |
| Indeterminate | 2 (0.18) |
| Negative | 239 (21.65) |
| Positive | 813 (73.64) |
| PR | |
| NA | 51 (4.62) |
| Indeterminate | 4 (0.36) |
| Negative | 345 (31.25) |
| Positive | 704 (63.77) |
| HER-2 | |
| NA | 183 (16.58) |
| Equivocal | 180 (16.3) |
| Indeterminate | 12 (1.09) |
| Negative | 565 (51.18) |
| Positive | 164 (14.86) |
| Menopause status | |
| NA | 93 (8.42) |
| Indeterminate | 34 (3.08) |
| Peri | 40 (3.62) |
| Post | 706 (63.95) |
| Pre | 231 (20.92) |
| T classification | |
| NA | 2 (0.18) |
| T1 | 281 (25.45) |
| T2 | 640 (57.97) |
| T3 | 138 (12.5) |
| T4 | 40 (3.62) |
| TX | 3 (0.27) |
| N classification | |
| NA | 2 (0.18) |
| N0 | 516 (46.74) |
| N1 | 367 (33.24) |
| N2 | 120 (10.87) |
| N3 | 79 (7.16) |
| NX | 20 (1.81) |
| M classification | |
| NA | 2 (0.18) |
| M0 | 917 (83.06) |
| M1 | 22 (1.99) |
| MX | 163 (14.76) |
| TNM stage | |
| NA | 10 (0.91) |
| I | 182 (16.49) |
| II | 626 (56.7) |
| III | 252 (22.83) |
| IV | 20 (1.81) |
| X | 14 (1.27) |
| Margin status | |
| NA | 72 (6.52) |
| Close | 31 (2.81) |
| Negative | 922 (83.51) |
| Positive | 79 (7.16) |
| Vital status | |
| NA | 2 (0.18) |
| Deceased | 155 (14.04) |
| Living | 947 (85.78) |
| Radiation therapy | |
| NA | 102 (9.24) |
| No | 445 (40.31) |
| Yes | 557 (50.45) |
| Neoadjuvant treatment | |
| NA | 3 (0.27) |
| No | 1088 (98.55) |
| Yes | 13 (1.18) |
| Targeted molecular therapy | |
| NA | 525 (47.55) |
| No | 46 (4.17) |
| Yes | 533 (48.28) |
| Sample type | |
| Metastatic | 7 (0.63) |
| Primary tumor | 1097 (99.37) |
| OS | |
| NA | 17 (1.54) |
| 0 | 933 (84.51) |
| 1 | 154 (13.95) |
| RFS | |
| NA | 192 (17.39) |
| 0 | 816 (73.91) |
| 1 | 96 (8.70) |
| ZNF385B | |
| High | 804 (72.83) |
| Low | 300 (27.17) |
ER: estrogen receptor; PR: progesterone receptor; HER-2: human epidermal growth factor-2; T: tumor; M: metastasis; N: node; OS: overall survival; RFS: relapse-free survival; NA: not available.
3.3. ZNF385B Expression in BC
ZNF385B expression status between BC and normal tissues is shown in Figure 1, which demonstrated downregulation of ZNF385B expression in BC (p < 0.0001). Furthermore, differences in ZNF385B expression were also observed in molecular subtype (p < 0.0001), ER (p < 0.0001), PR (p < 0.0001), TNM stage (p = 0.023), HER-2 (p = 0.0056), and vital status (p = 0.0043). To better determine the role of ZNF385B expression in predicting prognosis in BC patients, the ROC curves were performed to determine the optimal cut-off values for high and low ZNF385B expression. The cut-off value for high and low ZNF385B expression was 3.439 for TCGA data and 4.403 for GEO dataset (Supplementary Figure 2).
Figure 1.
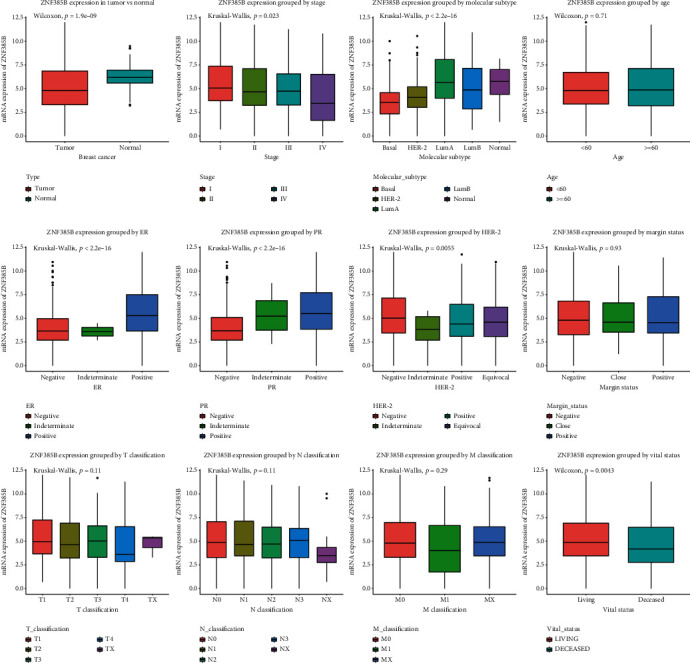
ZNF385B expression in breast tumor. The expression of ZNF385B is significantly lower in tumor than normal tissue. Low ZNF385B expression closely related with TNM stage, molecular subtype, ER, PR, HER-2 and vital status.
3.4. Diagnostic Value of ZNF385B Expression in BC
The ROC curve was plotted to evaluate the diagnostic value of ZNF385B based on the expression data from healthy individuals and BC patients (Figure 2(a)). It showed modest diagnostic value concluded from AUC with 0.671. We can observe that ZNF385B expression in different stages of BC also showed certain diagnostic value via subgroup analysis with AUC value of 0.618 for stage I, 0.669 for stage II, 0.698 for stage III, and 0.739 for stage IV (Figures 2(b)–2(e)).
Figure 2.
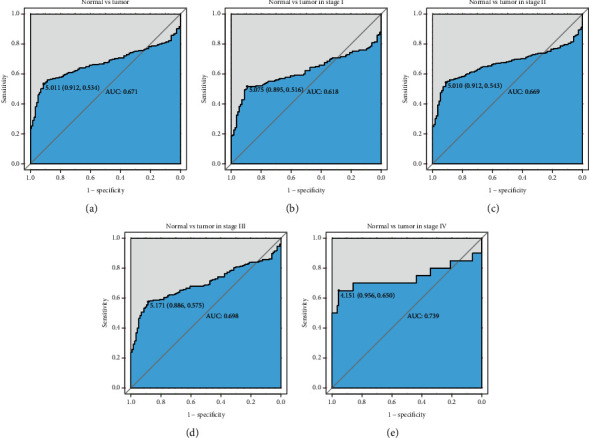
The diagnostic value of ZNF385B expression. (a) ROC curve for ZNF385B expression in breast cancer and normal tissue. (b–e) Different stages of breast cancer also showed certain diagnostic value.
3.5. Connection between Clinicopathologic Features and ZNF385B Expression in BC
The association between clinicopathologic parameters and ZNF385B expression of BC cases from TCGA database is shown in Table 2. The following features had significant correlation with ZNF385B expression including histological type, molecular subtype, ER, PR, vital status, OS, RFS (all p < 0.001), and TNM stage (p = 0.039).
Table 2.
Correlation between clinicopathological characteristics and ZNF385B expression in breast cancer patients' samples.
| Parameters | Variable | N | ZNF385B expression | χ 2 | p value | |
|---|---|---|---|---|---|---|
| High (N%) | Low (N%) | |||||
| Age | <60 | 589 | 436 (54.3) | 153 (51.17) | 0.856 | 0.355 |
| ≥60 | 513 | 367 (45.7) | 146 (48.83) | |||
|
| ||||||
| Gender | Female | 1090 | 795 (99) | 295 (98.66) | 0.025 | 0.873 |
| Male | 12 | 8 (1) | 4 (1.34) | |||
|
| ||||||
| Histological type | Infiltrating ductal carcinoma | 790 | 551 (68.7) | 239 (79.93) | 22.841 | <0.001 |
| Infiltrating lobular carcinoma | 204 | 176 (21.95) | 28 (9.36) | |||
| Other | 107 | 75 (9.35) | 32 (10.7) | |||
|
| ||||||
| Molecular subtype | Basal | 142 | 76 (12.1) | 66 (29.86) | 61.651 | <0.001 |
| HER-2 | 67 | 44 (7.01) | 23 (10.41) | |||
| LumA | 422 | 355 (56.53) | 67 (30.32) | |||
| LumB | 194 | 132 (21.02) | 62 (28.05) | |||
| Normal | 24 | 21 (3.34) | 3 (1.36) | |||
|
| ||||||
| ER | Indeterminate | 2 | 1 (0.13) | 1 (0.35) | 43.781 | <0.001b |
| Negative | 239 | 134 (17.4) | 105 (36.97) | |||
| Positive | 813 | 635 (82.47) | 178 (62.68) | |||
|
| ||||||
| PR | Indeterminate | 4 | 3 (0.39) | 1 (0.35) | 71.900 | <0.001b |
| Negative | 345 | 194 (25.19) | 151 (53.36) | |||
| Positive | 704 | 573 (74.42) | 131 (46.29) | |||
|
| ||||||
| HER-2 | Equivocal | 180 | 127 (18.84) | 53 (21.46) | 3.960 | 0.266 |
| Indeterminate | 12 | 7 (1.04) | 5 (2.02) | |||
| Negative | 565 | 425 (63.06) | 140 (56.68) | |||
| Positive | 164 | 115 (17.06) | 49 (19.84) | |||
|
| ||||||
| Menopause status | Inde | 34 | 21 (2.86) | 13 (4.71) | 3.811 | 0.283 |
| Peri | 40 | 32 (4.35) | 8 (2.9) | |||
| Post | 706 | 509 (69.25) | 197 (71.38) | |||
| Pre | 231 | 173 (23.54) | 58 (21.01) | |||
|
| ||||||
| T classification | T1 | 281 | 220 (27.4) | 61 (20.4) | 8.685 | 0.069 |
| T2 | 640 | 455 (56.66) | 185 (61.87) | |||
| T3 | 138 | 102 (12.7) | 36 (12.04) | |||
| T4 | 40 | 24 (2.99) | 16 (5.35) | |||
| TX | 3 | 2 (0.25) | 1 (0.33) | |||
|
| ||||||
| N classification | N0 | 516 | 374 (46.58) | 142 (47.49) | 6.606 | 0.158 |
| N1 | 367 | 276 (34.37) | 91 (30.43) | |||
| N2 | 120 | 85 (10.59) | 35 (11.71) | |||
| N3 | 79 | 58 (7.22) | 21 (7.02) | |||
| NX | 20 | 10 (1.25) | 10 (3.34) | |||
|
| ||||||
| M classification | M0 | 917 | 669 (83.31) | 248 (82.94) | 4.062 | 0.131 |
| M1 | 22 | 12 (1.49) | 10 (3.34) | |||
| MX | 163 | 122 (15.19) | 41 (13.71) | |||
|
| ||||||
| TNM stage | I | 182 | 145 (18.19) | 37 (12.46) | 10.100 | 0.039 |
| II | 626 | 448 (56.21) | 178 (59.93) | |||
| III | 252 | 184 (23.09) | 68 (22.9) | |||
| IV | 20 | 10 (1.25) | 10 (3.37) | |||
| X | 14 | 10 (1.25) | 4 (1.35) | |||
|
| ||||||
| Margin status | Close | 31 | 24 (3.2) | 7 (2.47) | 0.596 | 0.742 |
| Negative | 922 | 666 (88.92) | 256 (90.46) | |||
| Positive | 79 | 59 (7.88) | 20 (7.07) | |||
|
| ||||||
| Vital status | Deceased | 155 | 91 (11.33) | 64 (21.4) | 18.287 | <0.001 |
| Living | 947 | 712 (88.67) | 235 (78.6) | |||
|
| ||||||
| Radiation therapy | No | 445 | 319 (43.58) | 126 (46.67) | 0.762 | 0.383 |
| Yes | 557 | 413 (56.42) | 144 (53.33) | |||
|
| ||||||
| Neoadjuvant treatment | No | 1088 | 795 (99) | 293 (98.32) | 0.380 | 0.538 |
| Yes | 13 | 8 (1) | 5 (1.68) | |||
|
| ||||||
| Targeted molecular therapy | No | 46 | 34 (7.96) | 12 (7.89) | 0.001 | 0.979 |
| Yes | 533 | 393 (92.04) | 140 (92.11) | |||
|
| ||||||
| Sample type | Metastatic | 7 | 6 (0.75) | 1 (0.33) | 0.118 | 0.732 |
| Primary tumor | 1097 | 798 (99.25) | 299 (99.67) | |||
|
| ||||||
| OS | 0 | 933 | 702 (88.52) | 231 (78.57) | 17.473 | <0.001 |
| 1 | 154 | 91 (11.48) | 63 (21.43) | |||
|
| ||||||
| RFS | 0 | 816 | 617 (91.54) | 199 (83.61) | 11.743 | <0.001 |
| 1 | 96 | 57 (8.46) | 39 (16.39) | |||
ER: estrogen receptor; PR: progesterone receptor; HER-2: human epidermal growth factor-2; T: tumor; M: metastasis; N: node; OS: overall survival; RFS: relapse-free survival. bFisher's exact test.
3.6. Low ZNF385B Expression Showed Independent Prognostic Value of BC Patients for OS
The impact of ZNF385B expression on OS was evaluated through the Kaplan-Meier curve which indicated that low ZNF385B expression related with worse OS (p < 0.0001) (Figure 3). Furthermore, low ZNF385B expression significantly affected the OS in infiltrating ductal carcinoma (p = 0.00031), ER-negative BC (p = 0.04), ER-positive BC (p = 0.0025), PR-negative BC (p = 0.00015), HER-2-negative BC (p = 0.0038), and HER-2-positive BC (p = 0.0013) through subgroup analysis. Univariate and multivariate analyses were carried out to evaluate the prognostic value of clinicopathologic parameters. As shown in Table 3, the univariate analysis showed that age, menopause status, T classification, N classification, M classification, TNM stage, margin status, and ZNF385B expression were all meaningful prognostic factors for OS. Furthermore, multivariate analysis indicated that ZNF385B expression was an independent predictor for OS of BC patients (HR = 3.04, 95% CI: 1.894-4.877, p < 0.001). Besides, age, N classification, M classification, and margin status also showed independent prognostic value for OS.
Figure 3.
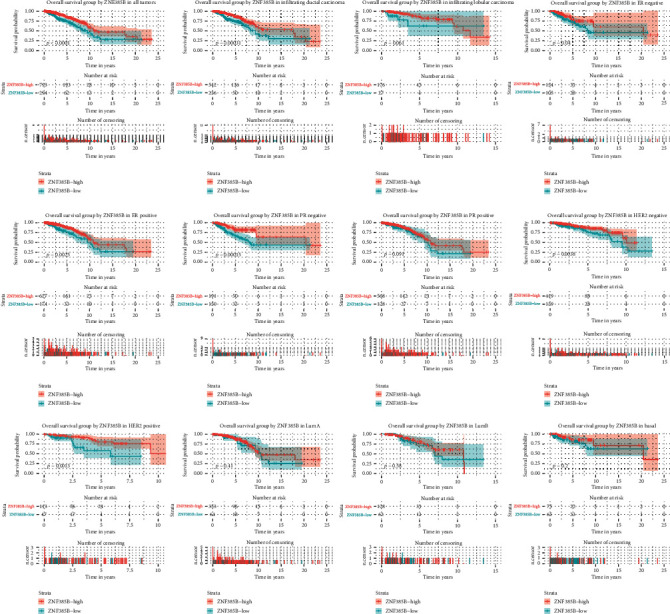
Kaplan-Meier curves of OS in breast cancer. Kaplan-Meier curves for OS in all tumors, different histological types (infiltrating ductal carcinoma and infiltrating lobular carcinoma), ER, PR, HER-2, and molecular subtype (LumA, LumB, and basal).
Table 3.
Univariate and multivariate Cox regression analyses of various prognostic parameters of overall survival in patients with breast cancer.
| Parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Age (years) (≥60 vs.<60) | 1.912 (1.390-2.631) | <0.001 | 2.249 (1.219-4.148) | 0.009 |
| Menopause status | ||||
| Peri vs. pre | 0.369 (0.049-2.761) | 0.332 | 0.822 (0.108-6.284) | 0.850 |
| Post vs. pre | 2.105 (1.281-3.459) | 0.003 | 1.228 (0.585-2.581) | 0.587 |
| T classification | ||||
| T2 vs. T1 | 1.331 (0.887-1.998) | 0.167 | 1.198 (0.632-2.271) | 0.580 |
| T3 vs. T1 | 1.578 (0.936-2.658) | 0.087 | 1.484 (0.605-3.643) | 0.389 |
| T4 vs. T1 | 3.967 (2.138-7.362) | <0.001 | 1.721 (0.534-5.547) | 0.363 |
| N classification | ||||
| N1 vs. N0 | 1.874 (1.273-2.758) | 0.001 | 1.231 (0.638-2.372) | 0.535 |
| N2 vs. N0 | 2.719 (1.627-4.545) | <0.001 | 3.195 (0.974-10.478) | 0.055 |
| N3 vs. N0 | 4.081 (2.256-7.381) | <0.001 | 4.463 (1.401-14.222) | 0.011 |
| M classification (M1 vs. M0) | 4.784 (2.858-8.007) | <0.001 | 3.504 (1.393-8.815) | 0.008 |
| TNM stage (III-IV vs. I-II) | 2.605 (1.869-3.630) | <0.001 | 1.187 (0.415-3.391) | 0.749 |
| Margin status | ||||
| Close vs. negative | 1.876 (0.818-4.303) | 0.137 | 2.740 (1.028-7.306) | 0.044 |
| Positive vs. negative | 1.951 (1.186-3.212) | 0.009 | 0.647 (0.296-1.417) | 0.277 |
| ZNF385B (low vs. high) | 2.021 (1.465-2.788) | <0.001 | 3.040 (1.894-4.877) | <0.001 |
HR: hazard ratio; CI: confidence interval; M: metastasis; N: node; T: tumor.
3.7. Low ZNF385B Expression Showed Independent Prognostic Value of BC Patients for RFS
The Kaplan-Meier curves indicated that low ZNF385B expression had relation to poorer RFS (p < 0.0001) (Figure 4). In addition, low ZNF385B expression significantly affected the RFS in infiltrating ductal carcinoma (p < 0.0001), ER-negative BC (p = 0.016), ER-positive BC (p = 0.025), PR-negative BC (p = 0.00044), HER-2-negative BC (p = 0.00069), HER-2-positive BC (p = 0.048), and basal-like BC (p = 0.039) through subgroup analysis. Univariate and multivariate analyses were used to confirm the prognostic value of clinicopathologic parameters. As shown in Table 4, the univariate analysis showed that ER, PR, T classification, N classification, M classification, TNM stage, margin status, and ZNF385B expression were all meaningful prognostic factors for RFS. Furthermore, multivariate analysis manifested that ZNF385B was an independent predictor for RFS of BC patients (HR = 2.609, 95% CI: 1.531-4.449, p < 0.001). Besides, N classification and margin status also showed independent prognostic value for RFS.
Figure 4.
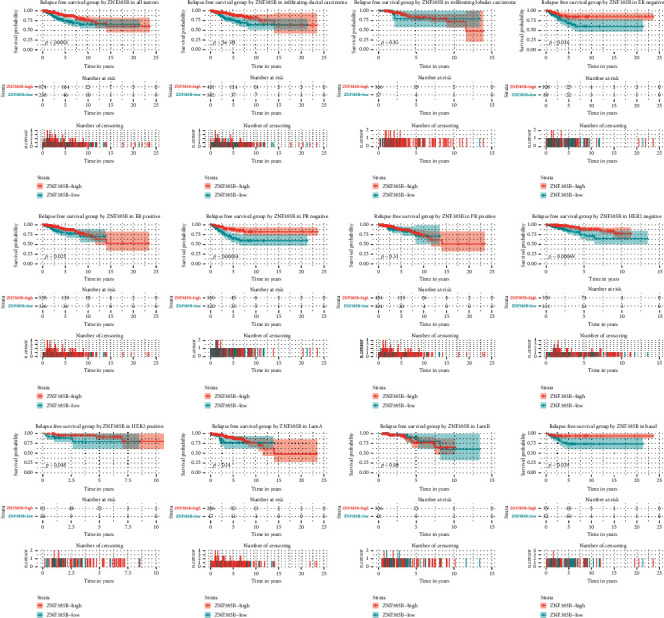
Kaplan-Meier curves of RFS in breast cancer. Kaplan-Meier curves for RFS in all tumors, different histological types (infiltrating ductal carcinoma and infiltrating lobular carcinoma), ER, PR, HER-2, and molecular subtype (LumA, LumB, and basal).
Table 4.
Univariate and multivariate Cox regression analyses of various prognostic parameters of relapse-free survival in patients with breast cancer.
| Parameters | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| ER (negative vs. positive) | 1.643 (1.062-2.542) | 0.026 | 1.340 (0.611-2.937) | 0.465 |
| PR (negative vs. positive) | 1.641 (1.088-2.477) | 0.018 | 1.402 (0.657-2.994) | 0.383 |
| T classification | ||||
| T2 vs. T1 | 1.635 (0.944-2.833) | 0.079 | 1.235 (0.631-2.417) | 0.537 |
| T3 vs. T1 | 2.524 (1.321-4.824) | 0.005 | 1.267 (0.487-3.299) | 0.628 |
| T4 vs. T1 | 6.72 (2.885-15.656) | <0.001 | 2.013 (0.553-7.321) | 0.288 |
| N classification | ||||
| N1 vs. N0 | 2.255 (1.357-3.748) | 0.002 | 1.560 (0.822-2.958) | 0.173 |
| N2 vs. N0 | 2.642 (1.326-5.264) | 0.006 | 2.741 (0.749-10.035) | 0.128 |
| N3 vs. N0 | 6.54 (3.415-12.524) | <0.001 | 7.106 (1.998-25.278) | 0.002 |
| M classification (M1 vs. M0) | 4.024 (1.742-9.292) | 0.001 | 1.294 (0.442-3.784) | 0.638 |
| TNM stage (III-IV vs. I-II) | 3.190 (2.106-4.831) | <0.001 | 1.147 (0.375-3.508) | 0.809 |
| Margin status | ||||
| Close vs. negative | 2.228 (0.966-5.141) | 0.060 | 3.229 (1.319-7.905) | 0.010 |
| Positive vs. negative | 2.453 (1.441-4.177) | 0.001 | 1.738 (0.836-3.611) | 0.139 |
| ZNF385B (low vs. high) | 2.227 (1.480-3.349) | <0.001 | 2.609 (1.531-4.449) | <0.001 |
CI: confidence interval; ER: estrogen receptor; HR: hazard ratio; M: metastasis; N: node; PR: progesterone receptor; T: tumor.
3.8. ZNF385B Expression and Pathological Characteristics of BC Patients in GEO Databases
We obtained GEO datasets to further investigate the role of ZNF385B in BC (Figure 5). The results were broadly consistent with our findings above. The validation via the microarray GSE21422 demonstrated that ZNF385B expression levels in tumor tissues, which included invasive ductal carcinoma (ICD) and ductal carcinoma in situ (DCIS), were lower than those in healthy samples (p = 0.03; Figure 5(a)). Differential levels of ZNF385B expression were presented in the following variables (validated by microarray GSE20711): ER status (p = 0.00015), HER-2 status (p = 0.047), grade (p = 0.00026), and subtype (p < 0.0001) (Figures 5(b)–5(e)). Besides, ZNF385B expression showed differences in survival analysis as well. Patients with lower ZNF385B expression level had shorter OS (p = 0.044; Figure 5(f)).
Figure 5.
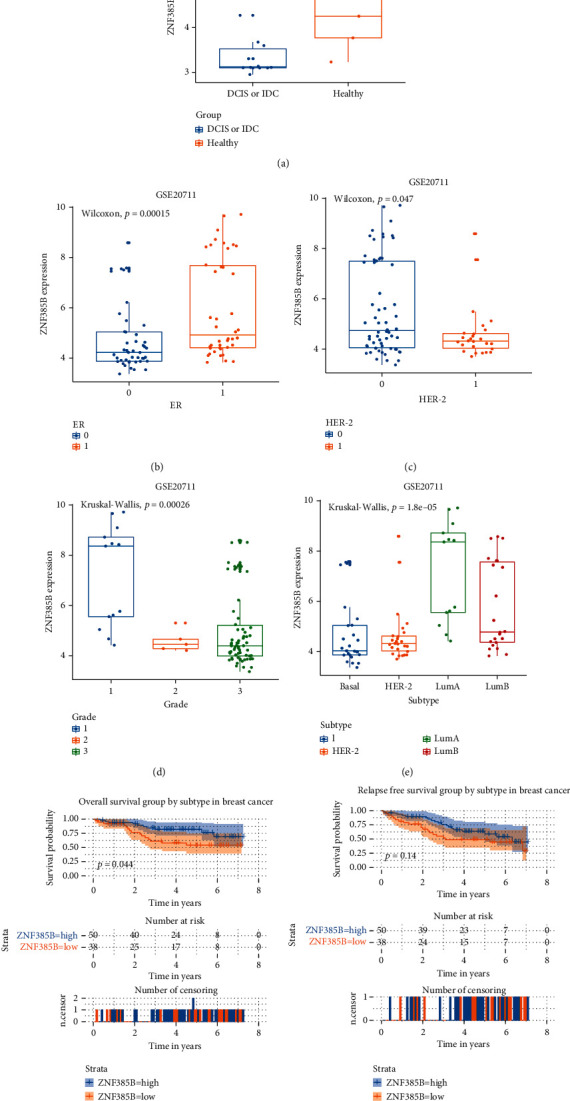
Association between mRNA expression levels of ZNF385B and pathological parameters of breast cancer. The parameters included ER, HER-2, grade, and subtype. Low ZNF385B expression related with short OS. The validation data from GEO database were analyzed. p < 0.05 was considered as statistically significant.
3.9. Low ZNF385B Expression in BC Patient Tissue Samples
IHC staining was performed to verify the expression of ZNF385B in BC patients' tissue samples. ZNF385B presented high expression in paratumor tissues when detected by IHC staining (Figures 6(a) and 6(b)), while low expression in BC tissues (Figures 6(c) and 6(d)). All paratumor breast tissues presented positive nuclear staining (n = 10), and most of the tumor tissues presented negative nuclear staining (n = 8) and others with weak positive staining (n = 2).
Figure 6.
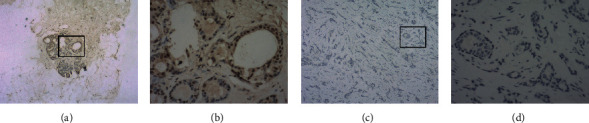
Immunohistochemistry staining for ZNF385B. The expression of ZNF385B in breast cancer cell was decreased. (a, b) IHC staining of paratumor breast tissues ((a) ×40, (b) ×400); (c, d) IHC staining of breast cancer tissues ((c) ×40, (d) ×400).
4. Discussion
Our present research was the first to find the difference of ZNF385B expression between BC patients and healthy individuals. To our knowledge, our research was the first to link ZNF385B to BC. It was found that aberrant expression of ZNF385B was associated with BC significantly based on a web-based tool “ESurv” [11] and Oncomine database [10]. We then further validated the relationship between ZNF385B expression and diagnosis as well as prognosis of BC using data from TCGA and GEO databases. We found that low ZNF385B expression related with poor survival status and recurrence of patients with BC. Besides, ZNF385B expression represented modest diagnostic value in BC using ROC analysis. The Kaplan-Meier curves for OS and RFS revealed that lower ZNF385B expression had relation to worse prognosis in BC patients. We obtained the similar results verified by datasets from the GEO database as well. Besides, the low expression of ZNF385B was verified in BC patients' samples using IHC. It has been showed that ZNF385B could work as an independent biomarker for BC prognosis through univariate and multivariate Cox analysis.
In human genome, zinc finger proteins have been known as one of the largest transcription factor family [16]. The functions were diverse, such as RNA packaging, DNA recognition, transcriptional activation, protein folding assembly, regulation of apoptosis, and lipid binding [17]. Zinc finger proteins showed diversity in multiple biological processes as a result of a complex combination and function of zinc finger motifs, which included development, differentiation, metabolism, and autophagy. In recent years, more and more reports have revealed the potential function of zinc finger proteins in tumor progression; however, the mechanisms were different, even in different types of cancer and the same cancer under different types of stress.
ZNF385B, whose encoded protein sequences with 4 matrin-type zinc fingers are highly conserved, is located in the AUTS5 region (2q31.2-q31.3) [18]. ZNF385B was considered to be a potential transcriptional repressor, while its target genes have not been identified [6]. It has been reported that ZNF385B had three isoforms. Isoform- (IF-) 1, containing four ZF domains, was the longest transcript variant. And IF-2/3 was shorter which contained three ZF domains. ZNF385B IF-1 could influence p53 and mediate apoptosis through upregulating PERP and FAS/CD95 and then activated caspase-8 and caspase-3. We supposed that transcriptional inhibition decreased neoplasia and metastasis of tumor cells in some way. Thus, tumor growth and metastasis could be promoted when the transcriptional repression of ZNF385B reduced and mRNA levels raised and resulted in shorter OS.
ZNF385B was expressed in adult brain tissue widely; besides, it related with the development of the lip and palate as well [19]. In the field of oncology, Elgaaen BV et al. first reported that in serous ovarian carcinomas, ZNF385B was lowly expressed and correlated with survival [5]. In addition, ZNF385B is expressed only in Burkitt lymphoma cells, but hardly in diffuse large B-cell lymphoma [4]. In the current genome-wide copy number variation analysis, containing 2319 individuals with BRCA1 pathogenic variants, it was reported that ZNF385B might relate with BC via qPCR and/or nanosorting analysis [20]. However, the position of ZNF385B in diagnosis and prognosis of BC has not been defined. Our present study showed that ZNF385B was expressed at low level in BC. ROC analysis has provided evidence for ZNF385B as a potential diagnostic biomarker of BC. We found a feasible relationship between ZNF385B and survival in BC by connecting ZNF385B expression with survival status as well. Subgroup analyses showed that low ZNF385B expression significantly affected the OS and RFS in many BC subtypes such as infiltrating ductal carcinoma, ER-negative BC, ER-positive BC, PR-negative BC, HER-2-negative BC, and HER-2-positive BC, and its low expression showed worse prognosis in those patients. As we all know, molecular subtype has become the standard for guiding treatment options of BC patients [21], and different therapies such as chemotherapy and endocrine therapy have been developed to better treat patients with BC based on it [22]. Proper molecular typing and accurate prognosis are essential for cancer treatment [23]. Therefore, it is of great significance to research new prognosis-related genes for prognosis assessment and treatment guidance for BC and its different subtypes. In our study, low expression of ZNF385B has shown important prognostic value in different subtypes, which might be used to evaluate the risk and prognosis of subgroups and guide beneficial individual treatment options for patients with high risk of death or worse prognosis [24].Currently, surgery has been regarded as an important measure for malignant tumor. However, the possibility of recurrence especially in basal-like BC could adversely influence the outcome of patients. In this report, the connections between ZNF385B expression and recurrence in different subtypes of BC were explored. Low expression of ZNF385B affected RFS in patients who suffered basal-like BC, while it less affected luminal A or luminal B subtype, which indicated the specific prognostic value of ZNF38B.
Overall, our study provided evidence that ZNF385B showed important value in diagnosis and prognosis of patients with BC. However, further experimental confirmation is required to verify the conclusion.
5. Conclusion
Our research indicated that ZNF385B was downregulated in BC. Low ZNF385B expression might be related to clinical progression and work as a potential biomarker in diagnosis and prognosis of BC patients.
Acknowledgments
This study was supported by the 11th Young Fund of the First Hospital of Jilin University (JDYY11202008).
Abbreviations
- AUC:
Area under the curve
- BC:
Breast cancer
- GEO:
Gene Expression Omnibus
- HR:
Hazard ratio
- IHC:
Immunohistochemistry
- OS:
Overall survival
- RFS:
Relapse-free survival
- ROC:
Receiver operating characteristic
- TCGA:
The Cancer Genome Atlas.
Contributor Information
Yan Jiao, Email: lagelangri1@126.com.
Miao He, Email: hemiao@jlu.edu.cn.
Data Availability
The data used to support the findings of this study are included within the article.
Ethical Approval
All tissues enrolled in our study were agreed by patients with informed consents and approved by the China-Japan Union Hospital ethics committee of Jilin University.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
Yan Jiao and Miao He have contributed equally to the work and should be considered co-corresponding authors.
Supplementary Materials
Supplementary Figure 1: pan-cancer analysis of ZNF385B mRNA expression based on Oncomine database and “ESurv” tool.
Supplementary Figure 2: the ROC curves determining the optimal cut-off values of high and low ZNF385B expression in TCGA and GEO databases.
References
- 1.Yan Y., Xu H., Wang J., et al. Inhibition of breast cancer cells by targeting E2F-1 gene and expressing IL15 oncolytic adenovirus. Bioscience Reports. 2019;39(7) doi: 10.1042/bsr20190384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seely J. M., Alhassan T. Screening for breast cancer in 2018—what should we be doing today? Current Oncology. 2018;25(Supplement 1):S115–S124. doi: 10.3747/co.25.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaby V., Tawk R., Sanogo V., Xiao H., Montero A. J. A review of systematic reviews of the cost-effectiveness of hormone therapy, chemotherapy, and targeted therapy for breast cancer. Breast Cancer Research and Treatment. 2015;151(1):27–40. doi: 10.1007/s10549-015-3383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iijima K., Yamada H., Miharu M., et al. ZNF385B is characteristically expressed in germinal center B cells and involved in B-cell apoptosis. European Journal of Immunology. 2012;42(12):3405–3415. doi: 10.1002/eji.201242530. [DOI] [PubMed] [Google Scholar]
- 5.Elgaaen B. V., Olstad O. K., Sandvik L., et al. ZNF385B and VEGFA are strongly differentially expressed in serous ovarian carcinomas and correlate with survival. PLoS One. 2012;7(9, article e46317) doi: 10.1371/journal.pone.0046317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Geen H., Squazzo S. L., Iyengar S., et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genetics. 2007;3(6):e89–0926. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aouizerat B. E., Vittinghoff E., Musone S. L., et al. GWAS for discovery and replication of genetic loci associated with sudden cardiac arrest in patients with coronary artery disease. BMC Cardiovascular Disorders. 2011;11(1) doi: 10.1186/1471-2261-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J., Lee T., Kim T. H., Lee K. T., Kim H. An integrated approach of comparative genomics and heritability analysis of pig and human on obesity trait: evidence for candidate genes on human chromosome 2. BMC Genomics. 2012;13(1):p. 711. doi: 10.1186/1471-2164-13-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J., Zheng Q., Huang Y. Q., et al. Significant evidence of association between polymorphisms in ZNF533, environmental factors, and nonsyndromic orofacial clefts in the western Han Chinese population. DNA and Cell Biology. 2011;30(1):47–54. doi: 10.1089/dna.2010.1082. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes D. R., Kalyana-Sundaram S., Mahavisno V., et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pak K., Oh S. O., Goh T. S., et al. A user-friendly, web-based integrative tool (ESurv) for survival analysis: development and validation study. Journal of Medical Internet Research. 2020;22(5, article e16084) doi: 10.2196/16084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Navarro I., Gámez-Pozo A., Pinto Á., et al. An 8-gene qRT-PCR-based gene expression score that has prognostic value in early breast cancer. BMC Cancer. 2010;10(1):p. 336. doi: 10.1186/1471-2407-10-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y., Li Y., Lu Z., Liu Y. High trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Digestive Diseases and Sciences. 2019;64(1):137–143. doi: 10.1007/s10620-018-5315-x. [DOI] [PubMed] [Google Scholar]
- 14.Kretschmer C., Sterner-Kock A., Siedentopf F., Schoenegg W., Schlag P. M., Kemmner W. Identification of early molecular markers for breast cancer. Molecular Cancer. 2011;10(1):p. 15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedeurwaerder S., Desmedt C., Calonne E., et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Molecular Medicine. 2011;3(12):726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jen J., Wang Y. C. Zinc finger proteins in cancer progression. Journal of Biomedical Science. 2016;23(1):p. 53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laity J. H., Lee B. M., Wright P. E. Zinc finger proteins: new insights into structural and functional diversity. Current Opinion in Structural Biology. 2001;11(1):39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 18.Monfort S., Rosello M., Orellana C., et al. Detection of known and novel genomic rearrangements by array based comparative genomic hybridisation: deletion of ZNF533 and duplication of CHARGE syndrome genes. Journal of Medical Genetics. 2008;45(7):432–437. doi: 10.1136/jmg.2008.057596. [DOI] [PubMed] [Google Scholar]
- 19.Liang S., Wang X. L., Zou M. Y., et al. Family-based association study of ZNF533, DOCK4 and IMMP2L gene polymorphisms linked to autism in a northeastern Chinese Han population. Journal of Zhejiang University: Science B. 2014;15(3):264–271. doi: 10.1631/jzus.B1300133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker L. C., BCFR L. M., Pearson J. F., et al. Evaluation of copy-number variants as modifiers of breast and ovarian cancer risk for _BRCA1_ pathogenic variant carriers. European Journal of Human Genetics. 2017;25(4):432–438. doi: 10.1038/ejhg.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen G., Zhang J. S., Zhang Y. J., Zhu Y. J., Huang X. B., Guan X. X. Predictive value of molecular subtyping for locoregional recurrence in early-stage breast cancer with N1 without postmastectomy radiotherapy. Journal of Breast Cancer. 2016;19(2):176–184. doi: 10.4048/jbc.2016.19.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao X., Huang X., Ye F., et al. The MIR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Scientific Reports. 2016;6 doi: 10.1038/srep21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Z. J., Zhou X. H., Zhang H. Y. DNA methylation module network-based prognosis and molecular typing of cancer. Genes. 2019;10(8):p. 571. doi: 10.3390/genes10080571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel H., Dunican C., Cunnington A. J. Predictors of outcome in childhood Plasmodium falciparum malaria. Virulence. 2020;11(1):199–221. doi: 10.1080/21505594.2020.1726570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: pan-cancer analysis of ZNF385B mRNA expression based on Oncomine database and “ESurv” tool.
Supplementary Figure 2: the ROC curves determining the optimal cut-off values of high and low ZNF385B expression in TCGA and GEO databases.
Data Availability Statement
The data used to support the findings of this study are included within the article.


