Abstract
Many diseases exhibit population-specific causal effect sizes with trans-ethnic genetic correlations significantly less than 1, limiting trans-ethnic polygenic risk prediction. We develop a new method, S-LDXR, for stratifying squared trans-ethnic genetic correlation across genomic annotations, and apply S-LDXR to genome-wide summary statistics for 31 diseases and complex traits in East Asians (average N = 90K) and Europeans (average N = 267K) with an average trans-ethnic genetic correlation of 0.85. We determine that squared trans-ethnic genetic correlation is 0.82× (s.e. 0.01) depleted in the top quintile of background selection statistic, implying more population-specific causal effect sizes. Accordingly, causal effect sizes are more population-specific in functionally important regions, including conserved and regulatory regions. In regions surrounding specifically expressed genes, causal effect sizes are most population-specific for skin and immune genes, and least population-specific for brain genes. Our results could potentially be explained by stronger gene-environment interaction at loci impacted by selection, particularly positive selection.
Subject terms: Evolutionary biology, Genome-wide association studies, Heritable quantitative trait, Medical genetics
Trans-ethnic genetic correlation is significantly less than 1 for many diseases. Here, the authors stratify this correlation by genomic annotations, finding that loci whose causal disease effect sizes differ between ethnicities are likely impacted by selection, particularly positive selection.
Introduction
Trans-ethnic genetic correlations are significantly less than 1 for many diseases and complex traits1–6, implying that population-specific causal disease effect sizes contribute to the incomplete portability of genome-wide association study (GWAS) findings and polygenic risk scores (PRSs) to non-European populations6–12. However, current methods for estimating genome-wide trans-ethnic genetic correlations assume the same trans-ethnic genetic correlation for all categories of SNPs2,5,13, providing little insight into why causal disease effect sizes are population-specific. Understanding the biological processes contributing to population-specific causal disease effect sizes can help inform polygenic risk prediction in non-European populations and alleviate health disparities6,14,15.
Here, we introduce a new method, S-LDXR, for estimating enrichment of stratified squared trans-ethnic genetic correlation across functional categories of SNPs using GWAS summary statistics and population-matched linkage disequilibrium (LD) reference panels (e.g. the 1000 Genomes Project (1000G)16); we stratify the squared trans-ethnic genetic correlation across functional categories to robustly handle noisy heritability estimates. S-LDXR analyzes GWAS summary statistics of HapMap317 SNPs with minor allele frequency (MAF) greater than 5% in both East Asian (EAS) and European (EUR) populations (regression SNPs) to draw inferences about causal effects of all SNPs with MAF greater than 5% in both populations (heritability SNPs). We confirm that S-LDXR yields robust estimates in extensive simulations. We apply S-LDXR to 31 diseases and complex traits with GWAS summary statistics available in both East Asian (EAS) and European (EUR) populations, leveraging recent large studies in East Asian populations from the CONVERGE consortium and Biobank Japan18–20; we analyze a broad set of genomic annotations from the baseline-LD model21–23, as well as tissue-specific annotations based on specifically expressed gene (SEG) sets24. Most results are meta-analyzed across the 31 traits to maximize power (analogous to refs. 21–23), as we expect to see similar patterns of enrichment/depletion across traits (even though the underlying biological processes differ across traits). We also investigate trait-specific enrichments/depletions for the tissue-specific annotations (analogous to ref. 24).
Results
Overview of methods
Our method (S-LDXR) for estimating stratified trans-ethnic genetic correlation is conceptually related to stratified LD score regression21,22 (S-LDSC), a method for partitioning heritability from GWAS summary statistics while accounting for LD. S-LDXR determines that a category of SNPs is enriched for trans-ethnic genetic covariance if SNPs with high LD to that category have a higher product of Z-scores than SNPs with low LD to that category. Unlike S-LDSC, S-LDXR models per-allele effect sizes (accounting for differences in MAF between populations).
In detail, the product of Z-scores of SNP j in two populations, Z1jZ2j, has the expectation
| 1 |
where Np is the sample size for population p; ℓ×(j, C) = ∑kr1jkr2jkσ1jσ2jaC(k) is the trans-ethnic LD score of SNP j with respect to annotation C, whose value for SNP k, aC(k), can be either binary or continuous; rpjk is the LD (Pearson correlation) between SNP j and k in population p; σpj is the standard deviation of SNP j genotypes in population p; and θC represents the per-SNP contribution to trans-ethnic genetic covariance of the per-allele causal disease effect size of annotation C. Here, rpjk and σpj can be estimated from population-matched reference panels (e.g. 1000 Genomes Project16). We estimate θC for each annotation C using weighted least square regression. Subsequently, we estimate the trans-ethnic genetic covariance of each binary annotation C (ρg(C)) by summing trans-ethnic genetic covariance of each SNP in annotation C as , using coefficients () for all binary and continuous-valued annotations included in the analysis; the heritabilities in each population ( and ) are estimated analogously. We then estimate the stratified squared trans-ethnic genetic correlation, defined as
| 2 |
We define the enrichment/depletion of squared trans-ethnic genetic correlation as , where is the genome-wide squared trans-ethnic genetic correlation; λ2(C) can be meta-analyzed across traits with different . S-LDXR analyzes GWAS summary statistics of common HapMap317 SNPs (regression SNPs) to estimate λ2(C) for (causal effects of) all common SNPs (heritability SNPs). Further details (quantities estimated, analytical bias correction, shrinkage estimator to reduce standard errors, estimation of standard errors, significance testing, and factors impacting power) of the S-LDXR method are provided in the Methods section and the Supplementary Notes; we have publicly released open-source software implementing the method (see “Code availability”).
We apply S-LDXR to 62 annotations (defined in both EAS and EUR populations) from our baseline-LD-X model (“Methods”, Supplementary Data 1, Supplementary Figs. 1, 2), primarily derived from the baseline-LD model21–23 (v1.1), and 53 SEG annotations (Supplementary Data 2). We have publicly released all baseline-LD-X model annotations and LD scores for EAS and EUR populations (see “Data availability”).
Simulations
We evaluated the accuracy of S-LDXR in simulations using genotypes that we simulated using HAPGEN225 from phased haplotypes of 481 EAS and 489 EUR individuals from the 1000 Genomes Project16, preserving population-specific MAF and LD patterns (18,418 simulated EAS-like and 36,836 simulated EUR-like samples, after removing genetically related samples, the ratio of sample sizes similar to empirical data; ~2.5 million SNPs on chromosomes 1–3) (“Methods”); we did not have access to individual-level EAS data at sufficient sample size to perform simulations with real genotypes. For each population, we randomly selected a subset of 500 simulated samples to serve as the reference panel for estimating LD scores. We performed both null simulations (heritable trait with functional enrichment but no enrichment/depletion of squared trans-ethnic genetic correlation; λ2(C) = 1) and causal simulations (λ2(C) ≠ 1). In our main simulations, we randomly selected 10% of the SNPs as causal SNPs in both populations, set genome-wide heritability to 0.5 in each population, and adjusted genome-wide genetic covariance to attain a genome-wide rg of 0.60 (unless otherwise indicated). In the null simulations, we used heritability enrichments from analyses of real traits in EAS samples to specify per-SNP causal effect size variances and covariances. In the causal simulations, we directly specified per-SNP causal effect size variances and covariances to attain λ2(C) ≠ 1 values from analyses of real traits, as these were difficult to attain using the heritability and trans-ethnic genetic covariance enrichments from analyses of real traits.
First, we assessed the accuracy of S-LDXR in estimating genome-wide trans-ethnic genetic correlation (rg); we note that S-LDXR does not use the shrinkage estimator for genome-wide estimates. Across a wide range of simulated rg values (0.0 to 1.0), S-LDXR yielded approximately unbiased estimates and well-calibrated jackknife standard errors (Supplementary Table 1, Supplementary Fig. 3).
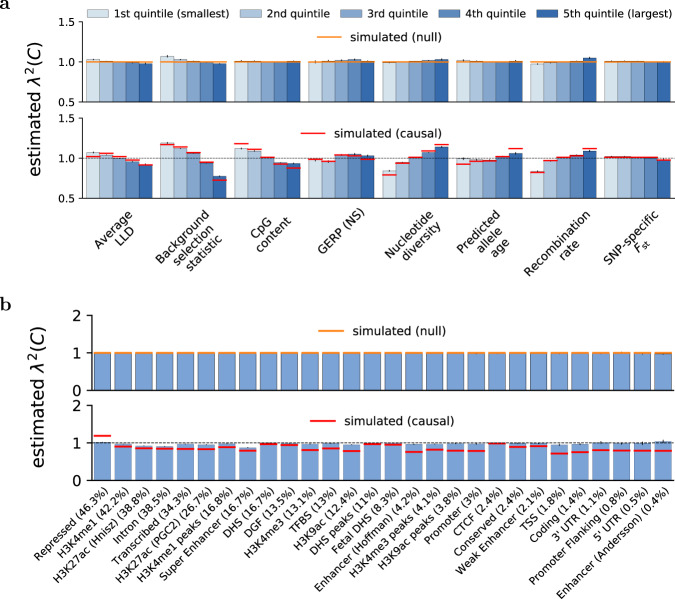
Second, we assessed the accuracy of S-LDXR in estimating λ2(C) in quintiles of the 8 continuous-valued annotations of the baseline-LD-X model. We performed both null simulations (λ2(C) = 1) and causal simulations (λ2(C) ≠ 1). Results are reported in Fig. 1a and Supplementary Data 3–8. In both null and causal simulations, S-LDXR yielded approximately unbiased estimates of λ2(C) for most annotations, validating our analytical bias correction. As a secondary analysis, we tried varying the S-LDXR shrinkage parameter, α, which has a default value of 0.5. In null simulations, results remained approximately unbiased; in causal simulations, reducing α led to less precise (but less biased) estimates of λ2(C), whereas increasing α biased results towards the null (λ2(C) = 1), demonstrating a bias-variance tradeoff in the choice of α (Supplementary Fig. 4, Supplementary Data 4, 7). Results were similar at other values of the proportion of causal SNPs (1% and 100%; Supplementary Data 3, 5, 6, 8). We also confirmed that S-LDXR produced well-calibrated jackknife standard errors (Supplementary Data 3–8).
Fig. 1. Accuracy of S-LDXR in null and causal simulations.
We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) in both null and causal simulations, for (a) quintiles of 8 continuous-valued annotations and (b) 28 main binary annotations (sorted by proportion of SNPs, displayed in parentheses). Results are averaged across 1000 simulations. Error bars denote ± 1.96 × standard error of the mean. Numerical results are reported in Supplementary Data 4 and 7.
Third, we assessed the accuracy of S-LDXR in estimating λ2(C) for the 28 main binary annotations of the baseline-LD-X model (inherited from the baseline model of ref. 21). We discarded λ2(C) estimates with the highest standard errors (top 5%), as estimates with large standard errors (which are particularly common for annotations of small size) are uninformative for evaluating unbiasedness of the estimator (in analyses of real traits, trait-specific estimates with large standard errors are retained, but contribute very little to meta-analysis results, and would be interpreted as inconclusive when assessing trait-specific results). Results are reported in Fig. 1b and Supplementary Data 4, 7. In null simulations, S-LDXR yielded unbiased estimates of λ2(C), further validating our analytical bias correction. In causal simulations, estimates were biased towards the null (λ2(C) = 1)—particularly for annotations of small size (proportion of SNPs < 1%)—due to our shrinkage estimator; increasing the shrinkage parameter above its default value of 0.5 further biased the estimates towards the null (λ2(C) = 1) in causal simulations (Supplementary Data 6–8). To ensure robust estimates, we focus on the 20 main binary annotations of large size (>1% of SNPs) in analyses of real traits (see below); although results for these annotations may still be biased towards the null, we emphasize that S-LDXR is unbiased in null data. Results were similar at other values of the proportion of causal SNPs (1% and 100%; Supplementary Data 3, 5, 6, 8). We also confirmed that S-LDXR produced well-calibrated jackknife standard errors (Supplementary Data 3–8) and conservative p-values (Supplementary Fig. 7, Supplementary Data 3–8).
Fourth, we performed additional null simulations in which causal variants differed across the two populations (Methods). S-LDXR yielded robust estimates of λ2(C), well-calibrated standard errors and conservative p-values in these simulations (Supplementary Fig. 11, Supplementary Data 11).
Fifth, we performed additional null simulations with annotation-dependent MAF-dependent genetic architectures26–28, defined as architectures in which the level of MAF-dependence is annotation-dependent, to ensure that estimate of λ2(C) remains unbiased. We disproportionately sampled low-frequency causal variants from the top quintile of background selection statistic, and set the variance of per-allele effect sizes of a causal SNP to be inversely proportional to its maximum MAF across both populations (Methods). Results are reported in Supplementary Figs. 8–10, and Supplementary Data 9–10. S-LDXR yielded nearly unbiased estimates of λ2(C) for the 28 binary functional annotations (Supplementary Fig. 8) and nearly unbiased estimates of λ2(C) for most quintiles of continuously valued annotations (Supplementary Fig. 9); estimates were slightly biased estimates in the top and bottom quintile of the average level of LD annotation and the recombination-rate annotation, likely due to less accurate reference LD scores at SNPs with extreme levels of LD. We repeated these simulations with five MAF bin annotations added to the baseline-LD-X model and obtained similar results (Supplementary Figs. 8a, 9b), supporting our decision not to include MAF bin annotations into the baseline-LD-X model.
Sixth, we performed additional null simulations, in which we increased or decreased the reference panel size from 500 to 250 or 1000, to assess the impact of reference panel size on the accuracy of S-LDXR (Methods). We simulated GWAS summary statistics based on the baseline-LD-X model as well as the model with annotation-dependent MAF-dependent genetic architectures. We determined that the small systematic biases in null simulations of continuous-valued annotations were on the same order of magnitude as for 500 reference samples (Supplementary Figs. 12, 13 and Supplementary Data 12 for 250 reference samples; Supplementary Figs. 14, 15 and Supplementary Data 13 for 1000 reference samples). We also performed simulations in which we reduced the simulated GWAS sample size by half, from NEAS = 18K, NEUR = 37K to NEAS = 9K, NEUR = 18K (while fixing the reference panel size at 500). We again determined that the small systematic biases were generally on the same order of magnitude as for NEAS = 18K, NEUR = 37K (although estimates were less stable and sometimes subject to larger biases, likely because our analytical bias correction starts to break down when the GWAS has low power) (Supplementary Figs. 16, 17 and Supplementary Data 14). Although it was not computationally feasible to perform simulations at larger GWAS sample sizes, these analyses do not provide a reason to believe that the small systematic biases that we observed in some of our null simulations of continuously valued annotations would substantially increase at larger GWAS sample sizes.
In summary, S-LDXR produced approximately unbiased estimates of enrichment/depletion of squared trans-ethnic genetic correlation in null simulations, and conservative estimates in causal simulations of both quintiles of continuous-valued annotations and binary annotations.
Analysis of baseline-LD-X model annotations across 31 diseases and complex traits
We applied S-LDXR to 31 diseases and complex traits with summary statistics in East Asians (average N = 90K) and Europeans (average N = 267K) available from Biobank Japan, UK Biobank, and other sources (Supplementary Table 2 and Methods). First, we estimated the trans-ethnic genetic correlation (rg) (as well as population-specific heritabilies) for each trait. Results are reported in Supplementary Fig. 18 and Supplementary Table 2. The average rg across 31 traits was 0.85 (s.e. 0.01) (average = 0.72 (s.e. 0.02)). 28 traits had rg < 1, and 11 traits had rg significantly less than 1 after correcting for 31 traits tested (P < 0.05/31); the lowest rg was 0.34 (s.e. 0.07) for Major Depressive Disorder (MDD), although this may be confounded by different diagnostic criteria in the two populations29. Several other complex traits, including Age at Menopause (rg = 0.57 (s.e. 0.09)) and LDL (rg = 0.66 (s.e. 0.11)) also had low trans-ethnic rg, likely due to pervasive gene-environment interaction across the genome. These estimates were consistent with estimates obtained using Popcorn2 (Supplementary Fig. 19) and those reported in previous studies2,5,6. We note that our estimates of trans-ethnic genetic correlation for 31 complex traits are higher than those reported for gene expression traits2 (average estimate of 0.32, increasing to 0.77 when restricting the analysis to gene expression traits with (cis) heritability greater than 0.2 in both populations), which are expected to have different genetic architectures.
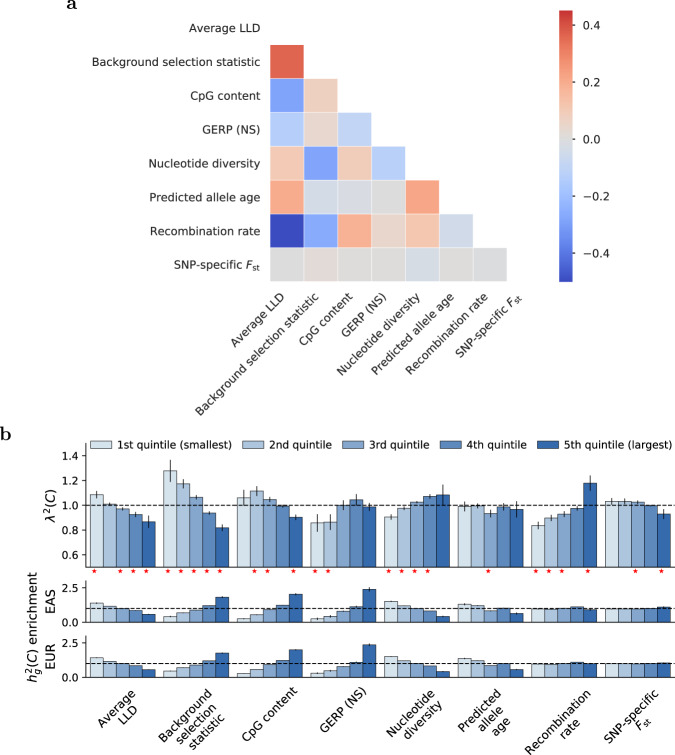
Second, we estimated the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) in quintiles of the 8 continuous-valued annotations of the baseline-LD-X model, meta-analyzing results across traits; these annotations are moderately correlated (Fig. 2a and Supplementary Data 1). We used the default shrinkage parameter (α = 0.5) in all analyses. Results are reported in Fig. 2b and Supplementary Data 15. We consistently observed a depletion of (λ2(C) < 1, implying more population-specific causal effect sizes) in functionally important regions. For example, we estimated λ2(C) = 0.82 (s.e. 0.01) for SNPs in the top quintile of background selection statistic (defined as 1 − McVicker B statistic/100030; see ref. 22); λ2(C) estimates were less than 1 for 29/31 traits, including 2 traits (Height and EGFR) with two-tailed p < 0.05/31. The background selection statistic quantifies the genetic distance of a site to its nearest exon; regions with high background selection statistic have higher per-SNP heritability, consistent with the action of selection, and are enriched for functionally important regions22. We observed the same pattern for CpG content and SNP-specific Fst (which are positively correlated with background selection statistic; Fig. 2a) and the opposite pattern for nucleotide diversity (which is negatively correlated with background selection statistic). We also estimated λ2(C) = 0.87 (s.e. 0.03) for SNPs in the top quintile of average LLD (which is positively correlated with background selection statistic), although these SNPs have lower per-SNP heritability due to a competing positive correlation with predicted allele age22. We caution that average LLD was the annotation most susceptible to bias in our simulations; see “Simulations”. Likewise, we estimated λ2(C) = 0.84 (s.e. 0.02) for SNPs in the bottom quintile of recombination rate (which is negatively correlated with background selection statistic), although these SNPs have average per-SNP heritability due to a competing negative correlation with average LLD22. However, λ2(C) < 1 estimates for the bottom quintile of GERP (NS) (which is positively correlated with both background selection statistic and recombination rate) and the middle quintile of predicted allele age are more difficult to interpret. For all annotations analyzed, heritability enrichments did not differ significantly between EAS and EUR, consistent with previous studies20,31. Results were similar at a more stringent shrinkage parameter value (α = 1.0; Supplementary Fig. 20), and for a meta-analysis across a subset of 20 approximately independent traits (Methods; Supplementary Fig. 21).
Fig. 2. S-LDXR results for quintiles of 8 continuous-valued annotations across 31 diseases and complex traits.
a We report correlations between each continuous-valued annotation; diagonal entries are not shown. Numerical results are reported in Supplementary Data 1. b We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)), as well as population-specific estimates of heritability enrichment, for quintiles of each continuous-valued annotation. Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error of the meta-analyzed λ2(C). P-values are obtained from the standard normal distribution. Red stars (⋆) denote two-tailed p < 0.05/40. Numerical results are reported in Supplementary Data 15.
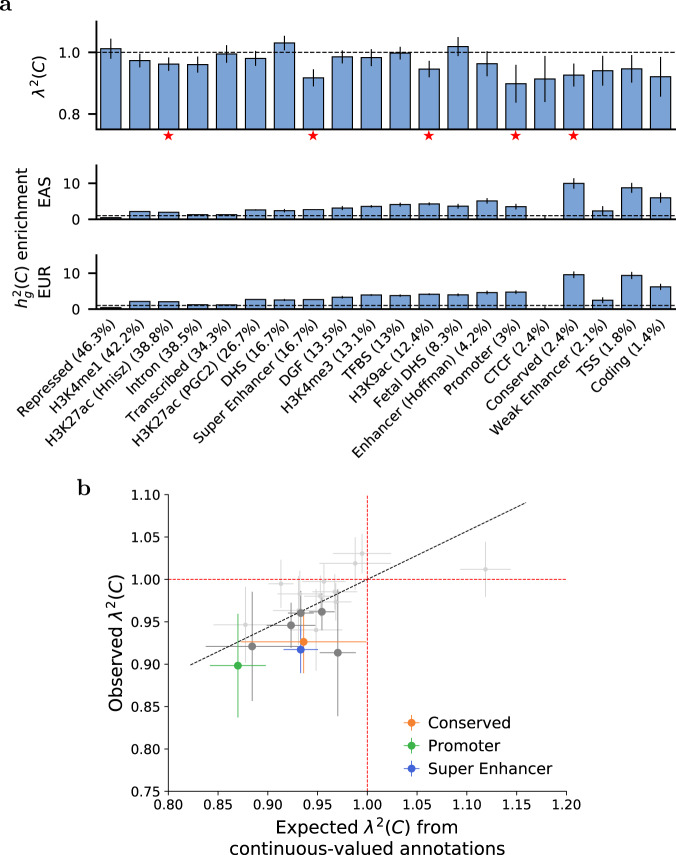
Finally, we estimated λ2(C) for the 28 main binary annotations of the baseline-LD-X model (Supplementary Data 1), meta-analyzing results across traits (as we did not observe significant trait-specific enrichment/depletion of squared trans-ethnic genetic correlation for these annotations due to limited power). Results are reported in Fig. 3a and Supplementary Data 16. Our primary focus is on the 20 annotations of large size (>1% of SNPs), for which our simulations yielded robust estimates; results for remaining annotations are reported in Supplementary Data 16. We consistently observed a depletion of λ2(C) (implying more population-specific causal effect sizes) within these annotations: 17 annotations had λ2(C) < 1, and 5 annotations had λ2(C) significantly less than 1 after correcting for 20 annotations tested (P < 0.05/20). These annotations included Conserved (λ2(C) = 0.93 (s.e. 0.02)), Promoter (λ2(C) = 0.85 (s.e. 0.04)) and Super Enhancer (λ2(C) = 0.93 (s.e. 0.02)), each of which was significantly enriched for per-SNP heritability, consistent with ref. 21. For all annotations analyzed, heritability enrichments did not differ significantly between EAS and EUR (Fig. 3a), consistent with previous studies20,31. Results were similar at a more stringent shrinkage parameter value (α = 1.0; Supplementary Fig. 20), and for a meta-analysis across a subset of 20 approximately independent traits (Methods; Supplementary Fig. 22). As a secondary analysis, we also estimated λ2(C) across 10 MAF bin annotations; we did not observe variation in λ2(C) estimates across MAF bins (Supplementary Table 3), further supporting our decision to not include MAF bin annotations in the baseline-LD-X model.
Fig. 3. S-LDXR results for 20 binary functional annotations across 31 diseases and complex traits.
a We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)), as well as population-specific estimates of heritability enrichment, for each binary annotation (sorted by proportion of SNPs, displayed in parentheses). Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error. Red stars (⋆) denote two-tailed p < 0.05/20. Numerical results are reported in Supplementary Data 16. b We report observed λ2(C) vs. expected λ2(C) based on 8 continuous-valued annotations, for each binary annotation. Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error of the meta-analyzed λ2(C). P-values are obtained from the standard normal distribution. Annotations for which λ2(C) is significantly different from 1 (p < 0.05/20) are denoted in color (see legend) or dark gray. The dashed black line (slope = 0.57) denotes a regression of observed λ(C) − 1 vs. expected λ(C) − 1 with intercept constrained to 0. Numerical results are reported in Supplementary Data 17.
Since the functional annotations are moderately correlated with the 8 continuous-valued annotations (Supplementary Data 1c, Supplementary Fig. 1), we investigated whether the depletions of squared trans-ethnic genetic correlation (λ2(C) < 1) within the 20 binary annotations could be explained by the 8 continuous-valued annotations. For each binary annotation, we estimated its expected λ2(C) based on values of the 8 continuous-valued annotations for SNPs in the binary annotation (Methods), meta-analyzed this quantity across traits, and compared observed vs. expected λ2(C) (Fig. 3b and Supplementary Data 17). We observed strong concordance, with a slope of 0.57 (correlation of 0.61) across the 20 binary annotations. This implies that the depletions of (λ2(C) < 1) within binary annotations are largely explained by corresponding values of continuous-valued annotations.
In summary, our results show that causal disease effect sizes are more population-specific in functionally important regions impacted by selection. Further interpretation of these findings, including the role of positive and/or negative selection, is provided in the “Discussion” section.
Analysis of SEG annotations
We analyzed 53 SEG annotations, defined in ref. 24 as ±100kb regions surrounding the top 10% of genes specifically expressed in each of 53 GTEx32 tissues (Supplementary Data 2), by applying S-LDXR with the baseline-LD-X model to the 31 diseases and complex traits (Supplementary Table 2). We note that although SEG annotations were previously used to prioritize disease-relevant tissues based on disease-specific heritability enrichments20,24, enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) is standardized with respect to heritability (i.e. increase in heritability in the denominator would lead to increase in trans-ethnic genetic covariance in the numerator (Eq. (2))), hence not expected to produce exceedingly disease-specific signals. Thus, we first assess meta-analyzed λ2(C) estimates across the 31 diseases and complex traits (trait-specific estimates are assessed below).
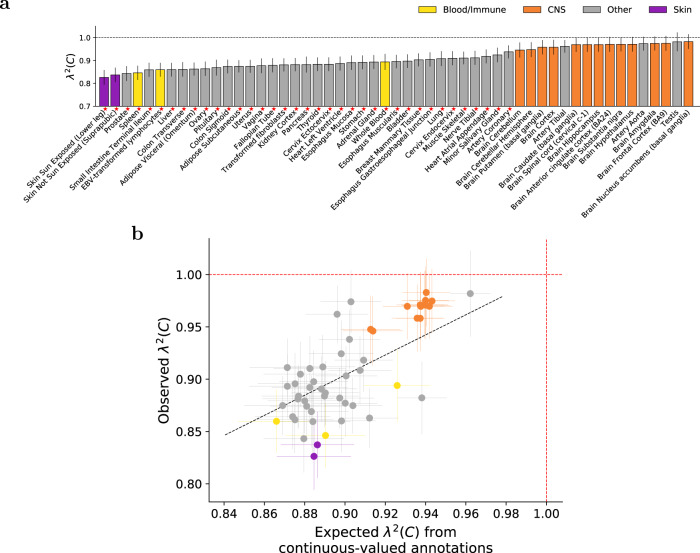
Results are reported in Fig. 4a and Supplementary Data 18. λ2(C) estimates were less than 1 for all 53 tissues and significantly less than 1 (p < 0.05/53) for 37 tissues, with statistically significant heterogeneity across tissues (p < 10−20; Methods). The strongest depletions of squared trans-ethnic genetic correlation were observed in skin tissues (e.g. λ2(C) = 0.83 (s.e. 0.02) for Skin Sun Exposed (Lower Leg)), Prostate and Ovary (λ2(C) = 0.84 (s.e. 0.02) for Prostate, λ2(C) = 0.86 (s.e. 0.02) for Ovary) and immune-related tissues (e.g. λ2(C) = 0.85 (s.e. 0.02) for Spleen), and the weakest depletions were observed in Testis (λ2(C) = 0.98 (s.e. 0.02); no significant depletion) and brain tissues (e.g. λ2(C) = 0.98 (s.e. 0.02) for Brain Nucleus Accumbens (Basal Ganglia); no significant depletion). Results were similar at less stringent and more stringent shrinkage parameter values (α = 0.0 and α = 1.0; Supplementary Figs. 23, 24 and Supplementary Data 18). A comparison of 14 blood-related traits and 16 other traits yielded highly consistent λ2(C) estimates (R = 0.82; Supplementary Fig. 25, Supplementary Data 19), confirming that these findings were not exceedingly disease-specific.
Fig. 4. S-LDXR results for 53 specifically expressed gene (SEG) annotations across 31 diseases and complex traits.
a We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) for each SEG annotation (sorted by λ2(C)). Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error. P-values are obtained from the standard normal distribution. Red stars (⋆) denote two-tailed p < 0.05/53. Numerical results are reported in Supplementary Data 21. b We report observed λ2(C) vs. expected λ2(C) based on 8 continuous-valued annotations, for each SEG annotation. Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error of the meta-analyzed λ2(C). Annotations are color-coded as in (a). The dashed black line (slope=0.96) denotes a regression of observed λ(C) − 1 vs. expected λ(C) − 1 with intercept constrained to 0. Numerical results and population-specific heritability enrichment estimates are reported in Supplementary Data 20.
These λ2(C) results were consistent with the higher background selection statistic30 in Skin Sun Exposed (Lower Leg) (R = 0.17), Prostate (R = 0.16), and Spleen (R = 0.14) as compared to Testis (R = 0.02) and Brain Nucleus Accumbens (Basal Ganglia) (R = 0.08) (Supplementary Fig. 26, Supplementary Data 2), and similarly for CpG content (Supplementary Fig. 27, Supplementary Data 2). Although these results could in principle be confounded by gene size33, the low correlation between gene size and background selection statistic (R = 0.06) or CpG content (R = −0.20) (in ±100kb regions) implies limited confounding. We note the well-documented action of recent positive selection on genes impacting skin pigmentation34–38, the immune system34–37,39, and Ovary40; we are not currently aware of any evidence of positive selection impacting Prostate. We further note the well-documented action of negative selection on fecundity- and brain-related traits26,28,41, but it is possible that recent positive selection may more closely track differences in causal disease effect sizes across human populations, which have split relatively recently42 (see “Discussion”).
More generally, since SEG annotations are moderately correlated with the 8 continuous-valued annotations (Supplementary Fig. 28, Supplementary Data 2), we investigated whether these λ2(C) results could be explained by the 8 continuous-valued annotations (analogous to Fig. 3b). Results are reported in Fig. 4b and Supplementary Data 20. We observed strong concordance, with a slope of 0.96 (correlation of 0.76) across the 53 SEG annotations. This implies that the depletions of λ2(C) within SEG annotations are explained by corresponding values of continuous-valued annotations.
The strong depletion of squared trans-ethnic genetic correlation in tissues impacted by positive selection (as opposed to negative selection) suggests a possible connection between positive selection and population-specific causal effect sizes. To further assess this, we estimated the enrichment/depletion of squared trans-ethnic genetic correlation in SNPs with high integrated haplotype score (iHS)43,44, which quantifies the action of positive selection (“Methods”). We observed a significant depletion (λ2(C) = 0.88 (s.e. 0.03)), further implicating positive selection (however, it is difficult to assess whether the iHS annotation contains unique information about λ2(C) conditional on other annotations; see “Discussion”). In addition, we observed a high genome-wide trans-ethnic genetic correlation for schizophrenia (rg = 0.95 (s.e. 0.04) vs. average of 0.85 (s.e. 0.01) across traits), a psychiatric disorder hypothesized to be strongly impacted by negative selection45,46, suggesting that negative selection may play a limited role in population-specific causal effect sizes. As noted above, these estimates pertain to parameters that were defined based on common variants (see “Overview of methods”); we note that although negative selection has the strongest impact on low-frequency variants26, common variants are also impacted by negative selection and can inform inferences about negative selection22. The role of positive selection (as opposed to negative selection) in population-specific causal effect sizes is discussed further in the Discussion section.
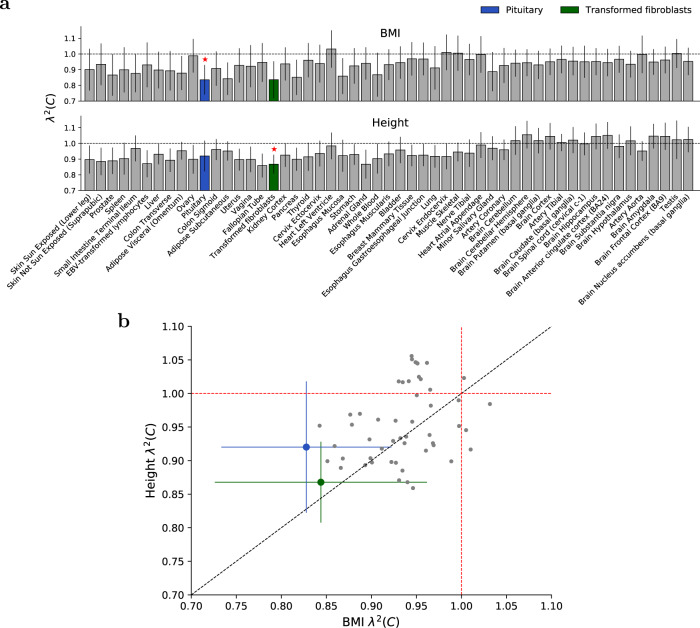
We investigated the enrichment/depletion of λ2(C) in the 53 SEG annotations for each individual trait (Supplementary Data 21). We identified six significantly depleted (vs. 0 significantly enriched) trait-tissue pairs at per-trait p < 0.05/53. The limited number of statistically significant results was expected, due to the reduced power of trait-specific analyses; however, λ2(C) estimates were generally consistent across traits. Results for BMI and height, two widely studied anthropometric traits, are reported in Fig. 5. For BMI, we observed significant depletion of squared trans-ethnic genetic correlation (λ2(C) = 0.84 (s.e. 0.05)) in Pituitary. Previous studies have highlighted the role of Pituitary in obesity47–49; our results suggest that this tissue-specific mechanism is population-specific. For height, we observed significant depletion of squared trans-ethnic genetic correlation for Transformed fibroblasts (λ2(C) = 0.87 (s.e. 0.03)), a connective tissue linked to human developmental disorders50; again, our results suggest that this tissue-specific mechanism is population-specific. Although Pituitary was significantly depleted for BMI but not height, and Transformed fibroblasts were significantly depleted for height but not BMI, we caution that for both tissues our λ2(C) estimates did not differ significantly between BMI and height.
Fig. 5. S-LDXR results for 53 specifically expressed gene (SEG) annotations for BMI and height.
a We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) for each SEG annotation for BMI and height. SEG annotations are ordered as in Fig. 4. Error bars denote ± 1.96 × standard error. P-values are obtained from the standard normal distribution. Red stars (⋆) denote two-tailed p < 0.05/53 for each respective trait (p = 8.2 × 10−4 for BMI and p = 3.3 × 10−4 for Height). b We report λ2(C) estimates for height vs. BMI for each SEG annotation. These estimates were moderately correlated (R = 0.35). Annotations are color-coded as in (a). For Pituitary, height λ2(C) is left-shifted by 0.008 and BMI λ2(C) is right-shifted by 0.008 for easier visualization of standard errors. Error bars denote ± 1.96 × standard error. The dashed black line denotes the y vs x line. Numerical results for all 31 diseases and complex traits are reported in Supplementary Data 21.
In summary, our results show that causal disease effect sizes are more population-specific in regions surrounding SEGs.
Discussion
We developed a new method (S-LDXR) for stratifying squared trans-ethnic genetic correlation across functional categories of SNPs that yields approximately unbiased estimates in extensive simulations. By applying S-LDXR to East Asian and European summary statistics across 31 diseases and complex traits, we determined that SNPs with high background selection statistic30 have substantially depleted squared trans-ethnic genetic correlation (vs. the genome-wide average), implying that causal effect sizes are more population-specific. Accordingly, squared trans-ethnic genetic correlations were substantially depleted for SNPs in many functional categories and enriched in less functionally important regions (although the power of S-LDXR to detect enrichment of squared trans-ethnic genetic correlation is limited due to depletion of heritability in less functionally important regions). In analyses of SEG annotations, we observed substantial depletion of squared trans-ethnic genetic correlation for SNPs near skin and immune-related genes, which are strongly impacted by recent positive selection, but not for SNPs near brain genes. We also observed trait-specific depletions of squared-trans-ethnic genetic correlation for SEG annotations, which indicate population-specific disease mechanisms.
Reductions in trans-ethnic genetic correlation have several possible underlying explanations, including gene-environment (G × E) interaction, gene–gene (G × G) interaction, and dominance variation (but not differences in heritability across populations, which would not affect trans-ethnic genetic correlation and were not observed in our study). Given the increasing evidence of the role of G × E interaction in complex trait architectures51, and evidence that G × G interaction and dominance variation explain limited heritability52–54, we hypothesize that depletion of squared trans-ethnic genetic correlation in the top quintile of background selection statistic and in functionally important regions may be primarily attributable to stronger G × E interaction in these regions. Interestingly, a recent study on plasticity in Arabidopsis observed a similar phenomenon: lines with more extreme phenotypes exhibited stronger G × E interaction55. Although depletion of squared trans-ethnic genetic correlation is often observed in regions with higher per-SNP heritability, which may often be subject to stronger G × E, depletion may also occur in regions with lower per-SNP heritability that are subject to stronger G × E; we hypothesize that this is the case for SNPs in the top quintile of average LLD and the bottom quintile of GERP (NS) (Fig. 2).
Distinguishing between stronger G × E interaction in regions impacted by selection and stronger G × E interaction in functionally important regions as possible explanations for our findings is a challenge, because functionally important regions are more strongly impacted by selection. To this end, we constructed an annotation that is similar to the background selection statistic but does not make use of recombination rate, instead relying solely on a SNP’s physical distance to the nearest exon (“Methods”). Applying S-LDXR to the 31 diseases and complex traits using a joint model incorporating baseline-LD-X model annotations and the nearest exon annotation, the background selection statistic remained highly conditionally informative for trans-ethnic genetic correlation, whereas the nearest exon annotation was not conditionally informative (Supplementary Table 4). This result implicates stronger G × E interaction in regions with a reduced effective population size that are impacted by the selection, and not just proximity to functional regions, in explaining depletions of squared trans-ethnic genetic correlation; however, we emphasize that selection acts on allele frequencies rather than causal effect sizes, and could help explain our findings only in conjunction with other explanations such as G × E interaction. Our results on SEGs implicate stronger G × E interaction near skin, immune, and ovary genes and weaker G × E interaction near brain genes, potentially implicating positive selection (as opposed to negative selection). This conclusion is further supported by the significant depletion of squared trans-ethnic genetic correlation in the integrated haplotype score (iHS) annotation that specifically reflects positive selection, high genome-wide trans-ethnic genetic correlation for schizophrenia (Supplementary Table 2), and lack of variation in squared trans-ethnic genetic correlation across genes in different deciles of probability of loss-of-function intolerance56 (Methods, Supplementary Figs. 29, 30, Supplementary Table 5). We conclude that depletions of squared trans-ethnic genetic correlation could potentially be explained by stronger G × E interaction at loci impacted by positive selection. We caution that other explanations are also possible; in particular, evolutionary modeling using an extension of the Eyre–Walker model57 to two populations suggests that our results for the background selection statistic could also be consistent with negative selection (Supplementary Notes, Supplementary Figs. 31, 32, Supplementary Table 6). Additional information, such as genomic annotations that better distinguish different types of selection or data from additional diverse populations, may help elucidate the relationship between selection and population-specific causal effect sizes.
Our study has several implications. First, PRSs in non-European populations that make use of European training data6,9,11 may be improved by reweighting SNPs based on the expected enrichment/depletion of squared trans-ethnic genetic correlation, helping to alleviate health disparities6,14,15. For example, when applying LD-pruning + p-value thresholding methods58,59, both the strength of association and trans-ethnic genetic correlation should be accounted for when prioritizing SNPs for trans-ethnic PRS, as our results suggest that trans-ethnic genetic correlation is likely depleted near functional SNPs with significant p-values (due to stronger G × E). In particular, when multiple SNPs have a similar level of significance, the SNPs enriched for trans-ethnic genetic correlation should be prioritized. Analogously, when applying more recent methods that estimate posterior mean causal effect sizes60–66 (including functionally informed methods62,66), these estimates should subsequently be weighted according to the expected enrichment/depletion for squared trans-ethnic genetic correlation based on their functional annotations. Second, modeling population-specific genetic architectures may improve trans-ethnic fine-mapping. Our results suggest that causal effect sizes and/or causal variants are likely to differ across different populations, contrary to standard assumptions31,67. Thus, incorporating information about trans-ethnic genetic correlations in trans-ethnic fine-mapping may lead to more accurate identification of both population-specific and shared causal variants68. Third, modeling population-specific genetic architectures may also increase power in trans-ethnic meta-analysis69, e.g. by adapting MTAG70 to two populations (instead of two traits), leveraging trans-ethnic (instead of cross-trait) genetic correlation between pairs of populations to improve the estimation of SNP effect sizes in both populations. Fourth, it may be of interest to stratify G × E interaction effects51 across genomic annotations. Fifth, modeling and incorporating environmental variables, where available, may provide additional insights into population-specific causal effect sizes. In our simulations, we did not explicitly simulate G × E. However, G × E would induce population-specific causal effect sizes, which we did explicitly simulate. Sixth, the S-LDXR method could potentially be extended to stratify squared cross-trait genetic correlations71 across genomic annotations72.
We note several limitations of this study that pertain to the S-LDXR method. First, S-LDXR is designed for populations of homogeneous continental ancestry (e.g. East Asians and Europeans) and is not currently suitable for analysis of admixed populations73 (e.g. African Americans or admixed Africans from UK Biobank74), analogous to LDSC and its published extensions21,71,75. However, a recently proposed extension of LDSC to admixed populations76 could be incorporated into S-LDXR, enabling its application to the growing set of large studies in admixed populations10. Second, S-LDXR estimates of enrichment of stratified squared trans-ethnic genetic correlation (λ2(C)) are slightly downward biased in null simulations of the top quintile of the background selection statistic and average LLD annotations, especially in simulations involving annotation-dependent MAF-dependent genetic architectures. However, these biases are small compared to the depletions of λ2(C) observed in the analysis of real traits. We further note that our estimates are unbiased in null simulations of binary annotations, implying that our results on real traits for binary annotations are robust. Third, since S-LDXR applies shrinkage to reduce standard error in estimating stratified squared trans-ethnic genetic correlation and its enrichment, estimates are conservative—true depletions of squared trans-ethnic genetic correlation in functionally important regions may be stronger than the estimated depletions. However, we emphasize that S-LDXR is approximately unbiased in null data. Fourth, the optimal value of the shrinkage parameter α may be specific to the pair of populations analyzed. In our simulations, we determined that α = 0.5 provides a satisfactory bias-variance tradeoff across a wide range of values of polygenicity and power. Thus, α = 0.5 may also be satisfactory for other pairs of populations. However, we recommend that one should ideally perform simulations on the pair of populations being analyzed to selection the optimal value of α. Fifth, it is difficult to assess whether a focal annotation contains unique information about λ2(C) conditional on other annotations, as squared trans-ethnic genetic correlation is a non-linear quantity defined by the quotient of squared trans-ethnic genetic covariance and the product of heritabilities in each population.
We also note several limitations of this study that pertain to our analysis of real traits. First, we focused on comparisons of East Asians and Europeans, due to the limited availability of very large GWAS in other populations. For other pairs of continental populations, if differences in the environment are similar, then we would expect similar genome-wide trans-ethnic genetic correlation and similar enrichment/depletion of squared trans-ethnic genetic correlation, based on our hypothesis that imperfect trans-ethnic genetic correlation is primarily attributable to G × E. We also note that different sets of SNPs, with different MAF and LD patterns, would be analyzed for different pairs of populations. However, we expect that these differences would not contribute to differences in trans-ethnic genetic correlation, if G × E is the fundamental factor impacting trans-ethnic genetic correlation. Second, the SEG annotations analyzed in this study are defined predominantly (but not exclusively) based on gene expression measurements of Europeans24. We hypothesize that results based on SEG annotations defined in East Asian populations would likely be similar, as heritability enrichment of functional annotations (predominantly defined in Europeans) are consistent across continental populations20,31, despite the fact that gene expression patterns and genetic architectures of gene expression differ across diverse populations12,77,78. Thus, SEG annotations derived from gene expression data from diverse populations may provide additional insights into population-specific causal effect sizes. Third, we restricted our analyses to SNPs that were relatively common (MAF > 5%) in both populations (estimating parameters that were defined based on common SNPs), due to the lack of a large LD reference panel for East Asians. Extending our analyses to lower-frequency SNPs may provide further insights into the role of negative selection in shaping population-specific genetic architectures, as negative selection has the strongest impact on variants with low frequency26,27. Fourth, we did not consider population-specific variants in our analyses, due to the difficulty in defining trans-ethnic genetic correlation for population-specific variants2,5, a more fundamental challenge than analyzing low-frequency SNPs; a recent study79 has reported that population-specific variants substantially limit trans-ethnic genetic risk prediction accuracy. Fifth, estimates of genome-wide trans-ethnic genetic correlation may be confounded by different trait definitions or diagnostic criteria in the two populations, particularly for major depressive disorder. However, this would not impact estimates of enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)), which is defined relative to genome-wide values. Sixth, we have not pinpointed the exact underlying phenomena (e.g. environmental heterogeneity coupled with gene-environment interaction) that lead to population-specific causal disease effect sizes at functionally important regions. Despite these limitations, our study provides an improved understanding of the underlying biology that contribute to population-specific causal effect sizes, and highlights the need for increasing diversity in genetic studies.
Methods
Definition of stratified squared trans-ethnic genetic correlation
We model a complex phenotype in two populations using linear models, Y1 = X1β1 + ϵ1 and Y2 = X2β2 + ϵ2, where Y1 and Y2 are vectors of phenotype measurements of population 1 and population 2 with sample size N1 and N2, respectively; X1 and X2 are mean-centered but not normalized genotype matrices at M SNPs in the two populations; β1 and β2 are per-allele causal effect sizes of the M SNPs; and ϵ1 and ϵ2 are environmental effects in the two populations. We assume that in each population, genotypes, causal effect sizes, and environmental effects are independent of each other. We assume that the per-allele effect size of SNP j in the two populations has variance and covariance,
| 3 |
where aC(j) is the value of SNP j for annotation C, which can be binary or continuous-valued; τ1C and τ2C are the net contribution of annotation C to the variance of β1j and β2j, respectively; and θC is the net contribution of annotation C to the covariance of β1j and β2j.
We define stratified trans-ethnic genetic correlation of a binary annotation C (e.g. functional annotations21 or quintiles of continuous-valued annotations22) as
| 4 |
where is the trans-ethnic genetic covariance of annotation C; and is the "allelic-scale heritability" (sum of per-SNP variance of per-allele causal effect sizes; different from heritability on the standardized scale) of annotation C in population p. Here, includes all binary and continuous-valued annotations included in the analysis. Since estimates of can be noisy (possibly negative), we estimate squared stratified trans-ethnic genetic correlation,
| 5 |
to avoid bias or undefined values in the square root. In this work, we only estimate for SNPs with minor allele frequency (MAF) greater than 5% in both populations. To assess whether causal effect sizes are more or less correlated for SNPs in annotation C compared with the genome-wide average, , we define the enrichment/depletion of stratified squared trans-ethnic genetic correlation as
| 6 |
We meta-analyze λ2(C) instead of across diseases and complex traits. For continuous-valued annotations, defining and λ2(C) is challenging, as squared correlation is a non-linear term involving a quotient of squared covariance and a product of variances; we elected to instead estimate λ2(C) for quintiles of continuous-valued annotations (analogous to ref. 22). We note that the average value of λ2(C) across quintiles of continuous-valued annotations is not necessarily equal to 1, as squared trans-ethnic genetic correlation is a non-linear quantity.
We provide a more detailed definition of the estimands in the Supplementary Notes.
S-LDXR method
S-LDXR is conceptually related to stratified LD score regression21,22 (S-LDSC), a method for stratifying heritability from GWAS summary statistics, to two populations. The S-LDSC method determines that a category of SNPs is enriched for heritability if SNPs with high LD to that category have higher expected χ2 statistic than SNPs with low LD to that category. Analogously, the S-LDXR method determines that a category of SNPs is enriched for trans-ethnic genetic covariance if SNPs with high LD to that category have higher expected product of Z-scores than SNPs with low LD to that category.
S-LDXR relies on the regression equation
| 7 |
to estimate θC, where Zpj is the Z-score of SNP j in population p; ℓ×(j, C) = ∑kr1jkr2jkσ1jσ2jaC(k) is the trans-ethnic LD score of SNP j with respect to annotation C, whose value for SNP k, aC(k), can be either binary or continuous; rpjk is the LD between SNP j and k in population p; and σpj is the standard deviation of SNP j in population p. We obtain unbiased estimates of ℓ×(j, C) using genotype data of 481 East Asian and 489 European samples in the 1000 Genomes Project16. To account for heteroscedasticity and increase statistical efficiency, we use weighted least square regression to estimate θC. We use regression equations analogous to those described in ref. 21 to estimate τ1C and τ2C. We include only well-imputed (imputation INFO > 0.9) and common (MAF > 5% in both populations) SNPs that are present in HapMap 317 (irrespective of GWAS significance level) in the regressions (regression SNPs), analogous to our previous work21,71,75. We use all SNPs present in either population in 1000 Genomes16 to estimate trans-ethnic LD scores ℓ×(j, C) (reference SNPs; analogous to S-LDSC21), so that the resulting coefficients θC also pertain to these SNPs. However, we estimate and λ2(C) (see below; defined as a function of causal effect sizes) for all SNPs with MAF > 5% in both populations (heritability SNPs), accounting for tagging effects (analogous to S-LDSC21).
Let , , and be the estimates of τ1C, τ2C, and θC, respectively. For each binary annotation C, we estimate the stratified heritability of annotation C in each population, and , and trans-ethnic genetic covariance, ρg(C), as
| 8 |
respectively, restricting to causal effects of SNPs with MAF > 5% in both populations (heritability SNPs), using coefficients (, , and ) of both binary and continuous-valued annotations. We estimate genome-wide trans-ethnic genetic correlation as , where represents the set of all SNPs with MAF > 5% in both populations. We then estimate as
| 9 |
where . The correction to in Eq. (9) is necessary for obtaining an unbiased estimate of , as computing quotients of two random variables introduces bias (Supplementary Notes). (We do not constrain the estimate of to its plausible range of [−1, 1], as this would introduce bias.) Subsequently, we estimate enrichment of stratified squared trans-ethnic genetic correlation as
| 10 |
where , the ratio between estimated stratified () and genome-wide () squared trans-ethnic genetic correlation. We use block jackknife over 200 non-overlapping and equally sized blocks to obtain the standard error of all estimates. The standard error of λ2(C) primarily depends on the total allelic-scale heritability of SNPs in the annotation (sum of per-SNP variances of causal per-allele effect sizes), which appears as the denominator () in the estimation of a stratified squared trans-ethnic genetic correlation (); if this denominator is small, estimation of becomes noisy. The standard error of λ2(C) indirectly depends on the size of the annotation, because larger annotations tend to have larger total heritability. However, estimates of λ2(C) for a large annotation may have a large standard error if the annotation is depleted for heritability.
To assess the informativeness of each annotation in explaining disease heritability and trans-ethnic genetic covariance, we define standardized annotation effect size on heritability and trans-ethnic genetic covariance for each annotation C analogous to ref. 22,
| 11 |
where , , and represent proportionate change in per-SNP heritability in population 1 and 2 and trans-ethnic genetic covariance, respectively, per standard deviation increase in annotation C; τ1C, τ2C, and θC are the corresponding unstandardized effect sizes, defined in Eq. (3); and σC is the standard deviation of annotation C.
We provide a more detailed description of the method, including derivations of the regression equation and unbiased estimators of the LD scores, in the Supplementary Notes.
S-LDXR shrinkage estimator
Estimates of can be imprecise with large standard errors if the denominator, , is close to zero and noisily estimated. This is especially the case for annotations of small size (<1% SNPs). We introduce a shrinkage estimator to reduce the standard error in estimating .
Briefly, we shrink the estimated per-SNP heritability and trans-ethnic genetic covariance of annotation C towards the genome-wide averages, which are usually estimated with smaller standard errors, prior to estimating . In detail, let MC be the number of SNPs in annotation C, we shrink , , and towards , , and , respectively, where , , are the genome-wide estimates, and M the total number of SNPs. We obtain the shrinkage as follows. Let , , and be the shrinkage obtained separately for , and , respectively, where α ∈ [0, 1] is the shrinkage parameter adjusting magnitude of shrinkage. We then choose the most stringent shrinkage, , as the final shared shrinkage for both heritability and trans-ethnic genetic covariance.
We shrink heritability and trans-ethnic genetic covariance of annotation C using γ as, , , and , where , , and are the shrunk counterparts of , , and , respectively. We shrink by substituting , , and with , , , respectively, in Eq. (9), to obtain its shrunk counterpart, . Finally, we shrink , by plugging in in Eq. (10) to obtain its shrunk counterpart, . We recommend α = 0.5 as the default shrinkage parameter value, as this value provides robust estimates of λ2(C) in simulations. We note that S-LDXR does not use the shrinkage estimator when estimating genome-wide rg and .
Significance testing
To assess whether an annotation C is enriched or depleted of squared trans-ethnic genetic correlation for a trait, we test the null hypothesis . Since is not normally distributed80, we instead test the equivalent null hypothesis , where is the genome-wide squared trans-ethnic genetic correlation. We obtain test statistic as , and obtain p-value under t-distribution with B − 1 degrees of freedom, where B is the number of jackknife blocks. Since the statistic does not involve division by , we do not apply any shrinkage to .
Baseline-LD-X model
We include a total of 54 binary functional annotations in the baseline-LD-X model. These include 53 annotations introduced in ref. 21, which consists of 28 main annotations including conserved annotations (e.g. Coding, Conserved) and epigenomic annotations (e.g. H3K27ac, DHS, Enhancer) derived from ENCODE81 and Roadmap82, 24 500-base-pair-extended main annotations, and 1 annotation containing all SNPs. We note that although chromatin accessibility can be population-specific, the fraction of such regions is small83. Following ref. 22, we created an additional annotation for all genomic positions with number of rejected substitutions84 greater than 4. Further information for all functional annotations included in the baseline-LD-X model is provided in Supplementary Data 1a.
We also include a total of 8 continuous-valued annotations in the baseline-LD-X model. First, we include 5 continuous-valued annotations introduced in ref. 22 (see “Data availability”), without modification: background selection statistic30, CpG content (within a ± 50 kb window), GERP (number of substitutation) score84, nucleotide diversity (within a ± 10 kb window), and Oxford map recombination rate (within a ± 10 kb window)85. Second, we include 2 minor allele frequency (MAF) adjusted annotations introduced in ref. 22, with modification: level of LD (LLD) and predicted allele age. We created analogous annotations applicable to both East Asian and European populations. To create an analogous LLD annotation, we estimated LD scores for each population using LDSC75, took the average across populations, and then quantile-normalized the average LD scores using 10 average MAF bins. We call this annotation “average level of LD”. To create analogous predicted allele age annotation, we quantile-normalized allele age estimated by ARGweaver86 across 54 multi-ethnic genomes using 10 average MAF bins. Finally, we include 1 continuous-valued annotation based on FST estimated by PLINK287, which implements the Weir & Cockerham estimator of FST88. Further information for all continuous-valued annotations included in the baseline-LD-X model is provided in Supplementary Data 1b.
Simulations
We used simulated East Asian (EAS) and European (EUR) genotype data to assess the performance of our method, as we did not have access to real EAS genotype data at a sufficient sample size to perform simulations with real genotypes. We simulated genotype data for 100,000 East-Asian-like and 100,000 European-like individuals using HAPGEN225 (see “Code availability”), which preserves population-specific MAF and LD patterns, starting from phased haplotypes of 481 East Asians and 489 Europeans individuals available in the 1000 Genomes Project16 (see “Data availability”), restricting to ~2.5 million SNPs on chromosome 1 – 3 with minor allele count greater than 5 in either population. Since the direct output of HAPGEN2 includes substantial relatedness2, we used PLINK287 (see “Code availability”) to remove simulated individuals with genetic relatedness greater than 0.05, resulting in 35,378 EAS-like and 36,836 EUR-like individuals. From the filtered set of individuals, we randomly selected 500 individuals in each simulated population to serve as reference panels. We used 18,418 EAS-like and 36,836 EUR-like individuals to simulate GWAS summary statistics, capturing the imbalance in sample size between EAS and EUR GWAS in the analysis of real traits. In our secondary simulations, we also decreased or increased the reference panel size or decreased the GWAS sample size, to evaluate the robustness of our method with respect to reference panel size and GWAS sample size.
We performed both null simulations, where enrichment of squared trans-ethnic genetic correlation, λ2(C), is 1 across all functional annotations, and causal simulations, where λ2(C) varies across annotations, under various degrees of polygenicity (1%, 10%, and 100% causal SNPs). In the null simulations, we set τ1C, τ2C, θC to be the meta-analyzed τC in real-data analyses of EAS GWASs, and followed Eq. (3) to obtain a variance, Var[β1j] and Var[β2j], and covariance, Cov[β1j, β2j], of per-SNP causal effect sizes β1j, β2j, setting all negative per-SNP variance and covariance to 0. In the causal simulations, we directly specified per-SNP causal effect size variances and covariances using self-devised τ1C, τ2C, and θC coefficients, to attain λ2(C) ≠ 1, as these were difficult to attain using the coefficients from analyses of real traits.
We randomly selected a subset of SNPs to be causal for both populations, and set Var[β1j], Var[β2j], and Cov[β1j, β2j] to be 0 for all remaining non-causal SNPs. We scaled the trans-ethnic genetic covariance to attain a desired genome-wide rg. Next, we drew causal effect sizes of each causal SNP j in the two populations from the bi-variate Gaussian distribution,
| 12 |
and scaled the drawn effect sizes to match the desired total heritability and trans-ethnic genetic covariance. We also performed null simulations in which imperfect genome-wide trans-ethnic genetic correlation is due to population-specific causal variants. In these simulations, we randomly selected 10% of the SNPs to be causal in each population, with 80% of causal variants in each population shared with the other population, and sampled perfectly correlated causal effect sizes for shared causal variants using Eq. (12). We simulated the genetic component of the phenotype in population p as Xpβp, where Xp is column-centered genotype matrix, and drew environmental effects, ϵp, from the Gaussian distribution, , such that the total phenotypic variance in each population is 1. Finally, we simulated GWAS summary association statistics for population p, Zp, as , where σpj is the standard deviation of SNP j in population p. We have publicly released Python code for simulating GWAS summary statistics for 2 populations (see “Code availability”). Fifth, we performed additional null simulations with annotation-dependent MAF-dependent genetic architectures26–28, defined as architectures in which the level of MAF-dependence is annotation-dependent.
We also performed null simulations with annotation-dependent MAF-dependent genetic architectures26–28, defined as architectures in which the level of MAF-dependence is annotation-dependent, to assess the impact on estimates of enrichment of stratified squared trans-ethnic genetic correlation, (λ2(C)). In these simulations, we set the variance of causal effect size of each SNP j in both populations to be proportional to , where pj,max is the maximum MAF of SNP j in the two populations. (We elected to use maximum MAF because a SNP that is rare in one population but common in the other is less likely to be impacted by negative selection.) We set α to − 0.38, as previously estimated for 25 UK Biobank diseases and complex traits in ref. 28. We sampled causal effect sizes using Eq. (12), with Var[β1j], Var[β2j], and Cov[β1j, β2j] scaled to attain a desired genome-wide heritability and trans-ethnic genetic correlation. We randomly selected 10% of SNPs to be causal in both populations. Additionally, in the top quintile of background selection statistic, we selected 1.8 × more low-frequency causal variants (pj,max < 0.05) than common variants (pj,max ≥ 0.05), capturing the action of negative selection across low-frequency and common variants27.
Summary statistics for 31 diseases and complex traits
We analyzed GWAS summary statistics of 31 diseases and complex traits, primarily from UK Biobank74, Biobank Japan20, and CONVERGE18. All summary statistics were based on genotyping arrays with imputation to an appropriate LD reference panel (e.g. Haplotype Reference Consortium89 and UK10K90 for UK Biobank74, the 1000 Genomes Project16 for Biobank Japan20), except those of the MDD GWAS in the East Asian population, which was based on low-coverage whole genome sequencing data18. These include: atrial fibrillation (AF)91,92, age at menarche(AMN)93,94, age at menopause (AMP)93,94, basophil count(BASO)20,95, body mass index (BMI)20,96, blood sugar(BS)20,96, diastolic blood pressure (DBP)20,96, eosinophil count(EO)20,96, estimated glomerular filtration rate (EGFR)20,97, hemoglobin A1c(HBA1C)20,96, height (HEIGHT)96,98, high density lipoprotein (HDL)20,96, hemoglobin (HGB)20,95, hematocrit (HTC)20,95, low density lipoprotein (LDL)20,96, lymphocyte count(LYMPH)20,96, mean corpuscular hemoglobin (MCH)20,96, mean corpuscular hemoglobin concentration (MCHC)20,95, mean corpuscular volume (MCV)20,95, major depressive disorder (MDD)18,99, monocyte count (MONO)20,96, neutrophil count(NEUT)20,95, platelet count (PLT)20,96, rheumatoid arthritis(RA)100, red blood cell count (RBC)20,96, systolic blood pressure (SBP)20,96, schizophrenia (SCZ)101, type 2 diabetes (T2D)102,103, total cholesterol (TC)20,96, triglyceride (TG)20,96, and white blood cell count (WBC)20,96. Further information for the GWAS summary statistics analyzed is provided in Supplementary Table 2. In our main analyses, we performed random-effect meta-analysis to aggregate results across all 31 diseases and complex traits. To test if the meta-analyzed is significantly different from 1, we computed a test statistic as , where is the standard error of meta-analyzed , and obtained a p-value under the normal distribution. We also defined a set of 20 approximately independent diseases and complex traits with cross-trait (estimated using cross-trait LDSC71) less than 0.25 in both populations: AF, AMN, AMP, BASO, BMI, EGFR, EO, HBA1C, HEIGHT, HTC, LYMPH, MCHC, MCV, MDD, NEUT, PLT, RA, SBP, TC, TG.
Expected enrichment of stratified squared trans-ethnic genetic correlation from 8 continuous-valued annotations
To obtain expected enrichment of squared trans-ethnic genetic correlation of a binary annotation C, λ2(C), from 8 continuous-valued annotations, we first fit the S-LDXR model using these 8 annotations together with the base annotation for all SNPs, yielding coefficients, , , and , for a total of 9 annotations. We then use Eq. (3) to obtain per-SNP variance and covariance of causal effect sizes, β1j and β1j, substituting τ1C, τ2C, θC with , , and , respectively. We apply shrinkage with default parameter setting (α = 0.5), and use Eqs. (9) and (10) to obtain expected stratified squared trans-ethnic genetic correlation, , and subsequently λ2(C).
Analysis of SEG annotations
We obtained 53 SEG annotations, defined in ref. 24 as ±100k-base-pair regions surrounding genes specifically expressed in each of 53 GTEx32 tissues. A list of the SEG annotations is provided in Supplementary Data 2. Correlations between SEG annotations and the 8 continuous-valued annotations are reported in Supplementary Fig. 28 and Supplementary Data 2. Most SEG annotations are moderately correlated with the background selection statistic and CpG content annotations.
To test whether there is heterogeneity in enrichment of squared trans-ethnic genetic correlation, λ2(C), across the 53 SEG annotations, we first computed the average λ2(C) across the 53 annotations, , using fixed-effect meta-analysis. We then computed the test statistic , where Ci is the ith SEG annotation, and the estimated λ2(C). We computed a p-value for this test statistic based on a χ2 distribution with 53 degrees of freedom.
Analysis of distance to nearest exon annotation
We created a continuous-valued annotation, named “distance to nearest exon annotation”, based on a SNP’s physical distance (number of base pairs) to its nearest exon, using 233,254 exons defined on the UCSC genome browser104 (see “Data availability”). This annotation is moderately correlated with the background selection statistic annotation22 (R = −0.21), defined as (1 - McVicker B statistic/1000), where the McVicker B statistic quantifies a site’s genetic distance to its nearest exon30. We have publicly released this annotation (see “Data availability”).
To assess the informativeness of functionally important regions versus regions impacted by selection in explaining the depletions of squared trans-ethnic genetic correlation, we applied S-LDXR on the distance to nearest exon annotation together with the baseline-LD-X model annotations. We used both enrichment of squared trans-ethnic genetic correlation (λ2(C)) and standardized annotation effect size (, , and ) to assess informativeness.
Analysis of probability of loss-of-function intolerance decile gene annotations
We created 10 annotations based on genes in deciles of the probability of being loss-of-function intolerant (pLI) (see “Data availability”), defined as the probability of assigning a gene into haplosufficient regions, where protein-truncating variants are depleted56. Genes with high pLI (e.g. >0.9) have highly constrained functionality, and therefore mutations in these genes are subject to negative selection. We included SNPs within a 100kb-base-pair window around each gene, following ref. 24. A correlation heat map between pLI decile gene annotations and the 8 continuous-valued annotations is provided in Supplementary Fig. 29. All pLI decile gene annotations are moderately correlated with the background selection statistic and CpG content annotations.
Analysis of the integrated haplotype score annotation
We created a binary annotation (proportion of SNPs: 6.3%) that includes all SNPs whose maximum absolute value of the integrated haplotype score (iHS)43,44 (see “Data availability”) across all 1000 Genomes Project EAS and EUR sub-populations are greater than 2.0, the recommended threshold to detect positive selection in ref. 43. This annotation is positively correlated with the top quintile of the background selection statistic annotation (R = 0.077). We note that although the iHS is a recombination-rate-adjusted quantity to detect the action of recent positive selection, it may also capture actions of negative selection43,44.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors are grateful to L. O’Connor, H. Finucane, D. Kassler, S. Mallick, N. Patterson, B. Neale, R. Walters, A. Martin, B. Brown, F. Hormozdiari, M. Hujoel, K. Burch, and B. Pasaniuc for helpful discussions. This research was conducted using the UK Biobank Resource under Application 16549 and was funded by NIH grants R01 HG006399, U01 HG009379, R37 MH107649, R01 MH101244, and R01 CA222147.
Author contributions
H.S. and A.L.P. designed the experiments. H.S. performed the experiments. H.S., S.G., M.K., E.M.K., A.P.S., and A.L.P. analyzed the data. H.S. and A.L.P. wrote the paper with assistance from all authors.
Data availability
All baseline-LD-X model annotations and other annotations used in this work are available at https://data.broadinstitute.org/alkesgroup/S-LDXR/. We used exon definitions from the UCSC Genome Browser (https://genome.ucsc.edu/). We used gene pLI scores from the Exome Aggregation Consortium (ExAC) (https://exac.broadinstitute.org/). The integrated haplotype scores (iHS) are available at http://coruscant.itmat.upenn.edu/data/JohnsonEA_iHSscores.tar.gz. The 1000 Genomes Project Phase 3 data are available at https://www.internationalgenome.org/. The baseline-LD model annotations are available at https://alkesgroup.broadinstitute.org/LDSCORE/.
Code availability
Python code implementing S-LDXR is available at https://github.com/huwenboshi/s-ldxr. Python code for simulating GWAS summary statistics under the baseline-LD-X model is available at https://github.com/huwenboshi/s-ldxr-sim. Python code implementing the two-population Eyre-Walker model is available at https://github.com/huwenboshi/two-population-Eyre-Walker-model. Python code for creating the distance to nearest exon annotation is available at https://github.com/huwenboshi/distance-to-nearest-exon. We used HAPGEN2 (https://mathgen.stats.ox.ac.uk/genetics_software/hapgen/hapgen2.html) to simulated genotype data. We used PLINK2 (https://www.cog-genomics.org/plink/2.0/) to remove related individuals in the simulated genotype data.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Jian Zeng, and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huwenbo Shi, Email: hshi@hsph.harvard.edu.
Alkes L. Price, Email: aprice@hsph.harvard.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-21286-1.
References
- 1.de Candia TR, et al. Additive genetic variation in schizophrenia risk is shared by populations of african and european descent. Am. J. Hum. Genet. 2013;93:463–470. doi: 10.1016/j.ajhg.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown BC, et al. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 2016;99:76–88. doi: 10.1016/j.ajhg.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancuso N, et al. The contribution of rare variation to prostate cancer heritability. Nat. Genet. 2016;48:30. doi: 10.1038/ng.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikeda M, et al. Genome-wide association study detected novel susceptibility genes for schizophrenia and shared trans-populations/diseases genetic effect. Schizophr. Bull. 2018;45:824–834. doi: 10.1093/schbul/sby140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galinsky KJ, et al. Estimating cross-population genetic correlations of causal effect sizes. Genet. Epidemiol. 2019;43:180–188. doi: 10.1002/gepi.22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson CS, et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the page study. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin AR, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Márquez-Luna C, Loh P-R, Consortium SATDS, Consortium STD, Price AL. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 2017;41:811–823. doi: 10.1002/gepi.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature570, 514–518 (2019). [DOI] [PMC free article] [PubMed]
- 11.Duncan L, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keys, K. L. et al. On the cross-population portability of gene expression prediction models. bioRxiv10.1101/552042 (2019).
- 13.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurdasani, D., Barroso, I., Zeggini, E. & Sandhu, M. S. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet.20, 520–535 (2019). [DOI] [PubMed]
- 16.Consortium GP, et al. A global reference for human genetic variation. Nature. 2015;526:68. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium IH, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai N, et al. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagai A, et al. Overview of the biobank japan project: study design and profile. J. Epidemiol. 2017;27:S2–S8. doi: 10.1016/j.je.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 21.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazal S, et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazal S, Marquez-Luna C, Finucane HK, Price AL. Reconciling s-LDSC and LDAK functional enrichment estimates. Nat. Genet. 2019;51:1202–1204. doi: 10.1038/s41588-019-0464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, Marchini J, Donnelly P. Hapgen2: simulation of multiple disease SNPs. Bioinformatics. 2011;27:2304–2305. doi: 10.1093/bioinformatics/btr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng J, et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018;50:746. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- 27.Gazal S, et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 2018;50:1600. doi: 10.1038/s41588-018-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoech AP, et al. Quantification of frequency-dependent genetic architectures in 25 UK biobank traits reveals action of negative selection. Nat. Commun. 2019;10:790. doi: 10.1038/s41467-019-08424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai, N., Kendler, K. & Flint, J. Minimal phenotyping yields GWAS hits of low specificity for major depression. BioRxiv10.1101/440735 (2018).
- 30.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kichaev G, Pasaniuc B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 2015;97:260–271. doi: 10.1016/j.ajhg.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium G, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raychaudhuri S, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007;8:857. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laland KN, Odling-Smee J, Myles S. How culture shaped the human genome: bringing genetics and the human sciences together. Nat. Rev. Genet. 2010;11:137. doi: 10.1038/nrg2734. [DOI] [PubMed] [Google Scholar]
- 38.Wilde S, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc. Natl Acad. Sci. 2014;111:4832–4837. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Boehmer H. Positive selection of lymphocytes. Cell. 1994;76:219–228. doi: 10.1016/0092-8674(94)90330-1. [DOI] [PubMed] [Google Scholar]
- 40.Li J, et al. Natural selection has differentiated the progesterone receptor among human populations. Am. J. Human Genet. 2018;103:45–57. doi: 10.1016/j.ajhg.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connor, L. J. et al. Extreme polygenicity of complex traits is explained by negative selection. Am. J. Hum. Genet.105, 456–476 (2019). [DOI] [PMC free article] [PubMed]
- 42.Veeramah KR, Hammer MF. The impact of whole-genome sequencing on the reconstruction of human population history. Nat. Rev. Genet. 2014;15:149. doi: 10.1038/nrg3625. [DOI] [PubMed] [Google Scholar]
- 43.Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol.4, e72 (2006). [DOI] [PMC free article] [PubMed]
- 44.Johnson KE, Voight BF. Patterns of shared signatures of recent positive selection across human populations. Nat. Ecol. Evol. 2018;2:713–720. doi: 10.1038/s41559-018-0478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dongen J, Boomsma DI. The evolutionary paradox and the missing heritability of schizophrenia. Am. J. Med. Genet. B: Neuropsychiatric Genet. 2013;162:122–136. doi: 10.1002/ajmg.b.32135. [DOI] [PubMed] [Google Scholar]
- 46.Pardiñas AF, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vicennati V, Pasquali R. Abnormalities of the hypothalamic-pituitary-adrenal axis in nondepressed women with abdominal obesity and relations with insulin resistance: evidence for a central and a peripheral alteration. J. Clin. Endocrinol. Metab. 2000;85:4093–4098. doi: 10.1210/jcem.85.11.6946. [DOI] [PubMed] [Google Scholar]
- 48.Vgontzas A, et al. Hypothalamic-pituitary-adrenal axis activity in obese men with and without sleep apnea: effects of continuous positive airway pressure therapy. J. Clin. Endocrinol. Metab. 2007;92:4199–4207. doi: 10.1210/jc.2007-0774. [DOI] [PubMed] [Google Scholar]
- 49.Bose M, Oliván B, Laferrère B. Stress and obesity: the role of the hypothalamic–pituitary–adrenal axis in metabolic disease. Curr. Opin. Endocrinol. Diabetes Obes. 2009;16:340. doi: 10.1097/MED.0b013e32832fa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itoh N, Ornitz DM. Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J. Biochem. 2011;149:121–130. doi: 10.1093/jb/mvq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson MR, et al. Genotype–covariate interaction effects and the heritability of adult body mass index. Nat. Genet. 2017;49:1174. doi: 10.1038/ng.3912. [DOI] [PubMed] [Google Scholar]
- 52.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mäki-Tanila A, Hill WG. Influence of gene interaction on complex trait variation with multilocus models. Genetics. 2014;198:355–367. doi: 10.1534/genetics.114.165282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am. J. Hum. Genet. 2015;96:377–385. doi: 10.1016/j.ajhg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jong M, et al. Natural variation in arabidopsis shoot branching plasticity in response to nitrate supply affects fitness. PLoS Genet. 2019;15:e1008366. doi: 10.1371/journal.pgen.1008366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eyre-Walker, A. Genetic architecture of a complex trait and its implications for fitness and genome-wide association studies. Proc. Natl Acad. Sci. USA107, 1752–1756 (2010). [DOI] [PMC free article] [PubMed]
- 58.Consortium IS, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012;44:483. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loh P-R, et al. Efficient bayesian mixed-model analysis increases association power in large cohorts. Nature genetics. 2015;47:284. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilhjálmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Y, et al. Leveraging functional annotations in genetic risk prediction for human complex diseases. PLoS Comput. Biol. 2017;13:e1005589. doi: 10.1371/journal.pcbi.1005589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung W, et al. Efficient cross-trait penalized regression increases prediction accuracy in large cohorts using secondary phenotypes. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lloyd-Jones LR, et al. Improved polygenic prediction by bayesian multiple regression on summary statistics. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-12653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Márquez-Luna, C. et al. LDpred-funct: incorporating functional priors improves polygenic prediction accuracy in UK biobank and 23andme data sets. bioRxiv10.1101/375337 (2020). [DOI] [PMC free article] [PubMed]
- 67.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 2018;19:491–504. doi: 10.1038/s41576-018-0016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi, H. et al. Localizing components of shared transethnic genetic architecture of complex traits from gwas summary data. Am. J. Hum. Genet. 106, 805–817 (2020). [DOI] [PMC free article] [PubMed]
- 69.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turley P, et al. Multi-trait analysis of genome-wide association summary statistics using mtag. Nat. Genet. 2018;50:229. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Q, et al. A powerful approach to estimating annotation-stratified genetic covariance via gwas summary statistics. Am. J. Hum. Genet. 2017;101:939–964. doi: 10.1016/j.ajhg.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat. Rev. Genet. 2011;12:523. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bulik-Sullivan BK, et al. Ld score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo, Y. et al. Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. bioRxiv10.1101/503144 (2019). [DOI] [PMC free article] [PubMed]
- 77.Martin AR, et al. Transcriptome sequencing from diverse human populations reveals differentiated regulatory architecture. PLoS Genet. 2014;10:e1004549. doi: 10.1371/journal.pgen.1004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mogil LS, et al. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14:e1007586. doi: 10.1371/journal.pgen.1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Durvasula, A. & Lohmueller, K. E. Negative selection on complex traits limits genetic risk prediction accuracy between populations. bioRxiv10.1101/721936 (2019). [DOI] [PMC free article] [PubMed]
- 80.Curtiss J. On the distribution of the quotient of two chance variables. Annal. Math. Statistics. 1941;12:409–421. doi: 10.1214/aoms/1177731679. [DOI] [Google Scholar]
- 81.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using gerp++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 86.Rasmussen MD, Hubisz MJ, Gronau I, Siepel A. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 2014;10:e1004342. doi: 10.1371/journal.pgen.1004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang CC, et al. Second-generation plink: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weir BS, Cockerham CC. Estimating f-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 89.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.consortium U, et al. The uk10k project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Low S-K, et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat. Genet. 2017;49:953. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- 92.Nielsen JB, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nature Genet. 2018;50:1234. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horikoshi M, et al. Elucidating the genetic architecture of reproductive ageing in the japanese population. Nat. Commun. 2018;9:1977. doi: 10.1038/s41467-018-04398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Day FR, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and brca1-mediated DNA repair. Nat. Genet. 2015;47:1294. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pattaro C, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Akiyama M, et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-12276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lam M, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nature Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suzuki K, et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat. Genet. 2019;51:379. doi: 10.1038/s41588-018-0332-4. [DOI] [PubMed] [Google Scholar]
- 103.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr. Protocols Bioinformatics. 2012;40:1–4. doi: 10.1002/0471250953.bi0104s40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All baseline-LD-X model annotations and other annotations used in this work are available at https://data.broadinstitute.org/alkesgroup/S-LDXR/. We used exon definitions from the UCSC Genome Browser (https://genome.ucsc.edu/). We used gene pLI scores from the Exome Aggregation Consortium (ExAC) (https://exac.broadinstitute.org/). The integrated haplotype scores (iHS) are available at http://coruscant.itmat.upenn.edu/data/JohnsonEA_iHSscores.tar.gz. The 1000 Genomes Project Phase 3 data are available at https://www.internationalgenome.org/. The baseline-LD model annotations are available at https://alkesgroup.broadinstitute.org/LDSCORE/.
Python code implementing S-LDXR is available at https://github.com/huwenboshi/s-ldxr. Python code for simulating GWAS summary statistics under the baseline-LD-X model is available at https://github.com/huwenboshi/s-ldxr-sim. Python code implementing the two-population Eyre-Walker model is available at https://github.com/huwenboshi/two-population-Eyre-Walker-model. Python code for creating the distance to nearest exon annotation is available at https://github.com/huwenboshi/distance-to-nearest-exon. We used HAPGEN2 (https://mathgen.stats.ox.ac.uk/genetics_software/hapgen/hapgen2.html) to simulated genotype data. We used PLINK2 (https://www.cog-genomics.org/plink/2.0/) to remove related individuals in the simulated genotype data.