Supplemental Digital Content is available in the text.
Keywords: arrhythmogenic right ventricular dysplasia, cardiomyopathies, deep learning, mutation
Abstract
Background:
ECG interpretation requires expertise and is mostly based on physician recognition of specific patterns, which may be challenging in rare cardiac diseases. Deep neural networks (DNNs) can discover complex features in ECGs and may facilitate the detection of novel features which possibly play a pathophysiological role in relatively unknown diseases. Using a cohort of PLN (phospholamban) p.Arg14del mutation carriers, we aimed to investigate whether a novel DNN-based approach can identify established ECG features, but moreover, we aimed to expand our knowledge on novel ECG features in these patients.
Methods:
A DNN was developed on 12-lead median beat ECGs of 69 patients and 1380 matched controls and independently evaluated on 17 patients and 340 controls. Differentiating features were visualized using Guided Gradient Class Activation Mapping++. Novel ECG features were tested for their diagnostic value by adding them to a logistic regression model including established ECG features.
Results:
The DNN showed excellent discriminatory performance with a c-statistic of 0.95 (95% CI, 0.91–0.99) and sensitivity and specificity of 0.82 and 0.93, respectively. Visualizations revealed established ECG features (low QRS voltages and T-wave inversions), specified these features (eg, R- and T-wave attenuation in V2/V3) and identified novel PLN-specific ECG features (eg, increased PR-duration). The logistic regression baseline model improved significantly when augmented with the identified features (P<0.001).
Conclusions:
A DNN-based feature detection approach was able to discover and visualize disease-specific ECG features in PLN mutation carriers and revealed yet unidentified features. This novel approach may help advance diagnostic capabilities in daily practice.
What Is Known?
Deep neural networks can be used to interpret raw electrocardiograms with high accuracy in, for example, hypertrophic cardiomyopathy patients.
Deep neural networks are able to discover complex features in ECG signals but are considered black box algorithms that lack interpretability.
What the Study Adds?
A deep learning-based feature detection approach is able to discover and visualize disease-specific ECG features.
In PLN (phospholamban) p.Argdel14 mutation carriers, visualizations revealed established ECG features (low QRS voltages and T-wave inversions), specified these features (eg, R- and T-wave attenuation in V2/V3) and identified novel PLN-specific ECG features (eg, increased PR-duration).
Interpretation of the ECG requires expertise and is mainly based on physician recognition of patterns that are known to belong to a particular disorder. However, for rare and relatively unknown cardiac diseases, this may be challenging since ECG features are often unknown and require expert knowledge to recognize. By automating the discovery and expanding the knowledge on disease-specific ECG features, interpretation of ECGs by physicians could be improved. Such a support tool could be of particular importance when expert knowledge is not readily available or in research settings to automate the detection of disease-specific ECG features.
Recently, ECGs have been analyzed using deep neural networks (DNNs), which are computer algorithms that are based on the structure and functioning of the human brain.1 Their layers can be trained to discover complex patterns in ECGs, without requiring hand-crafted feature extraction. Several studies have applied DNNs for automated predictions from ECGs, and one recent study showed that it is feasible to diagnose hypertrophic cardiomyopathy on the ECG.2–4 However, the methods used in these studies all require very large data sets, which are often not available for rare diseases. Furthermore, these previous studies all focus on prediction, but specific ECG patterns used by DNNs are rarely visualized.3,5–8 Visualization of such features takes advantage of the feature discovery embedded in DNNs and will help clinicians to interpret ECGs more accurately and possibly facilitate discovery of novel features.
Cardiomyopathy-related genetic mutations are rare but are often associated with typical ECG features. An example is the deletion of 3 base pairs (c.40_42delAGA) in the phospholamban (PLN) gene, leading to the deletion of Arginine 14 in the PLN protein (p.Arg14del).9–11 Prevalence of the PLN p.Arg14del mutation is estimated to be 0.07% in the northern regions of the Netherlands and is present in 12% of Dutch patients developing a phenotype of arrhythmogenic right ventricular cardiomyopathy and in 15% of patients developing dilated cardiomyopathy.11–13 With regard to ECG characteristics, in these mutation carriers, typical features that have previously been described are attenuated QRS-amplitudes and inverted T-waves in the right and left precordial leads.12,14,15
Beside using DNNs merely for prediction or diagnosis, we hypothesize that DNNs can also be used for feature visualization itself. This will potentially enable discovery of novel ECG features that belong to a particular disease. In this study, we used a cohort of PLN mutation carriers to investigate whether a novel DNN-based approach can (1) identify the already well-established ECG features in these mutation carriers and (2) possibly expand our knowledge on ECG features in these mutation carriers.
Methods
Data Availability
The data used in this study are not publicly available due to privacy restrictions. The code for training the DNN and for generating the visualizations and tables in this article is available upon request from the corresponding author.
Data Source and Study Participants
The data set consisted of 12-lead ECGs from patients between 18 and 85 years old acquired in the University Medical Center Utrecht from January 2000 to August 2019. All extracted data were deidentified in accordance with the EU General Data Protection Regulation, and written informed consent was, therefore, not required by the ethical committee. All ECGs were interpreted by a physician as part of the clinical workflow, and these free-text annotations were structured using a text-mining algorithm described before.3 We excluded all ECGs of insufficient quality and all ECGs with supraventricular and ventricular arrhythmias (excluding premature atrial and ventricular complexes), paced rhythms, undefined rhythms, and signs of acute ischemia.
All index patients in the data set who carry the genetic PLN p.Arg14del mutation and their relatives that tested positive were identified. ECGs acquired after the implantation of a left ventricular assist device or heart transplantation were excluded. Only the first acquired ECG of each mutation carrier was used for development of the model.
The control group was derived from the remaining data set and consisted of 365 173 ECGs of 147 098 unique patients. Per mutation carrier, 20 controls were matched using propensity score matching on age and sex. This number was chosen to have sufficient samples to train the DNN without having a too severe class imbalance. Only one ECG per control subject, sampled without replacement, was used to make sure every subject was only used once. The matched groups were randomly split in an 80:20 manner to training and test sets.
Data Acquisition
For all ECGs, the median beats were exported from the MUSE ECG system (MUSE version 8, GE Healthcare, Chicago, IL). The median beat data are constructed by aligning all QRS-complexes of the same shape (eg, excluding premature ventricular complexes) and generating a representative QRS-complex by taking the median voltage.16 Acquisition and feature extraction of the included ECGs is described in more detail in the Data Supplement.
Baseline Logistic Regression Model
To demonstrate the capability of DNN in identifying novel relevant features, we first developed a baseline logistic regression model, only based on the established ECG features of PLN mutation carriers. The matching variables, age and sex, and the established PLN-specific ECG features (low QRS voltage and right [V2–V3] and left [V4–V6] precordial T-wave inversion) were included as predictors in the model.17 The model was trained on the training data set and evaluated on the test set.
DNN Development
We constructed a deep convolutional neural network with exponentially dilated causal convolutions. The proposed architecture, inspired by the method described by van den Oord et al18 and Franceschi et al,19 compromises several 1-dimensional dilated causal convolution blocks. Eight-fold cross validation on the training data set was used for optimization of the hyperparameters of the network. The simplest network with the highest geometric mean of area under the receiver operating curve and F2 score averaged over all folds was chosen and trained on the complete training data set. The performance of this network was estimated on the test subset. Network training was performed using the PyTorch package (version 1.3).20 A detailed description of the architecture of the DNN can be found in the Expanded Methods in the Data Supplement, and an overview of the network architecture is shown in Figure I in the Data Supplement.
Feature Visualization
To identify the parts of the ECG that are important for the DNNs prediction, we applied Guided Gradient Class Activation Mapping++ (Guided Grad-CAM++), a technique for explanations in convolutional neural networks, to 1-dimensional data.5,6 Guided Grad-CAM++ combines the fine-grained and lead-specific visualizations of guided backpropagation with the class-discriminative and global Grad-CAM technique. The median beat visualization methodology is described in more detail in the Data Supplement.
Validation of Newly Identified Features in an Updated Model
Based on inspection of the visualization output, we identified distinctive features with an arbitrary prevalence above 25%. The detected important features were translated to quantitative features (eg, R-wave amplitude) and added to the baseline logistic regression model, starting with the most prevalent. If multiple similar features were found in leads belonging to the same region, the most prevalent feature in that region was used. Leads I, aVL and V4-V6 were grouped as lateral leads and II, III and aVF as inferior leads. To evaluate the added value of the newly identified ECG features, we determined if the nested baseline logistic regression model fit improved using the likelihood ratio test and Akaike information criterion.
Subgroup Analyses
In subgroup analyses, we analyzed whether predictive performance and detected features differed between subsets of patients. Due to the small sample size, these exploratory subgroup analyses were performed on the combined training and test data sets. We investigated the performance in presymptomatic PLN p.Arg14del mutation carriers. Presymptomatic was defined as follows: no cardiac symptoms as per judgment of the treating physician, no history of (non)sustained ventricular arrhythmia, premature ventricular complex burden of <500 beats per 24 hours and left ventricular ejection fraction ≥45%.
Statistical Analysis
The baseline characteristics were expressed as mean ± SD or median with interquartile range, where appropriate. Categorical variable differences were tested using the χ2 test or Fisher exact test and continuous variables using the Student t test or Mann-Whitney U test. Multiple testing correction was performed for the baseline characteristics using Bonferroni method. The overall discriminatory performance of the DNN, baseline and updated models were assessed in the test set with the concordance-statistic (c-statistic) or area under the receiver operating characteristic curve, sensitivities, specificities, positive, and negative predictive values. The models were compared at a prespecified specificity of 94%. The 95% CI around the performance measures and odds ratios were obtained using 2000 bootstrap samples. All statistical analyses were performed using R version 3.5 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Study Population
A total of 93 PLN p.Arg14del mutation carriers were identified, of which 86 were eligible for this study. Four patients were excluded as all their ECGs were acquired after left ventricular assist device or heart transplantation and 3 patients as all their ECGs were nonsinus rhythm. The control group consisted of 135 353 patients after exclusions, of which 1720 patients were matched. The flowchart is shown in Figure 1 and the baseline characteristics in Table 1.
Figure 1.
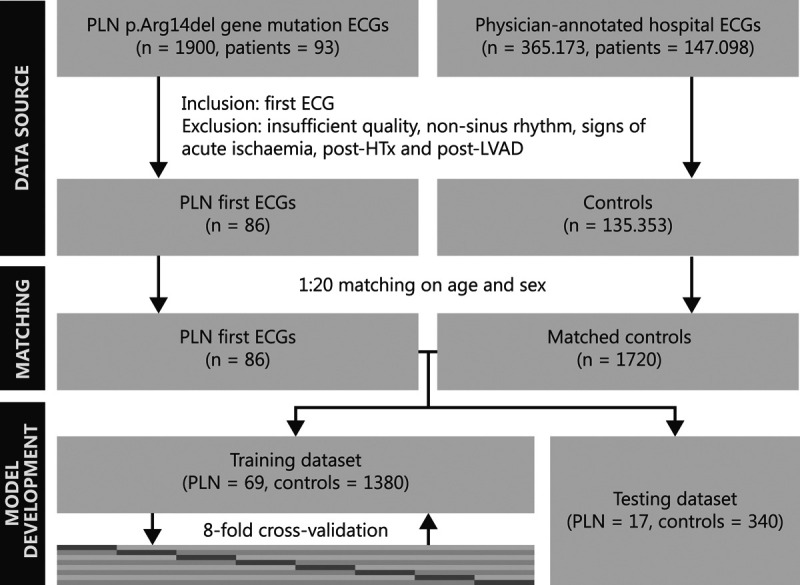
Flowchart of the patient selection and model development process. HTx indicates heart transplantation; LVAD, left ventricular assist device; and PLN, phospholamban.
Table 1.
Baseline Demographics and ECG Characteristics of All Patients and Patients in the Training and Test Splits, Stratified by PLN Mutation Carriers and Their Matched Controls

Baseline Logistic Regression Performance
The discriminative performance (by c-statistic) of the baseline logistic regression model was 0.84 (95% CI, 0.73–0.92) in the test set. The most important predictor of the PLN mutation was the presence of low QRS voltage, followed by left precordial inverted T-waves. No significant effect of age, sex, or right precordial negative T-waves was found.
DNN Performance
The cross-validated mean c-statistic, sensitivity, specificity, and F2 score obtained in the training data set were 0.86±0.07, 0.73±0.13, 0.91±0.04, and 0.56±0.05, respectively. The c-statistic of the DNN, trained on the complete training data set, was 0.95 (95% CI, 0.91–0.99) in the independent test set. The mean ECG beats for the complete data set with a superimposed Guided Grad-CAM visualization can be found in Figure 2. Figure 3 shows a representative example of a mutation carrier and a control subject that shows similar preestablished features (low QRS voltage and inverted T-waves) but is correctly identified by the DNN.
Figure 2.
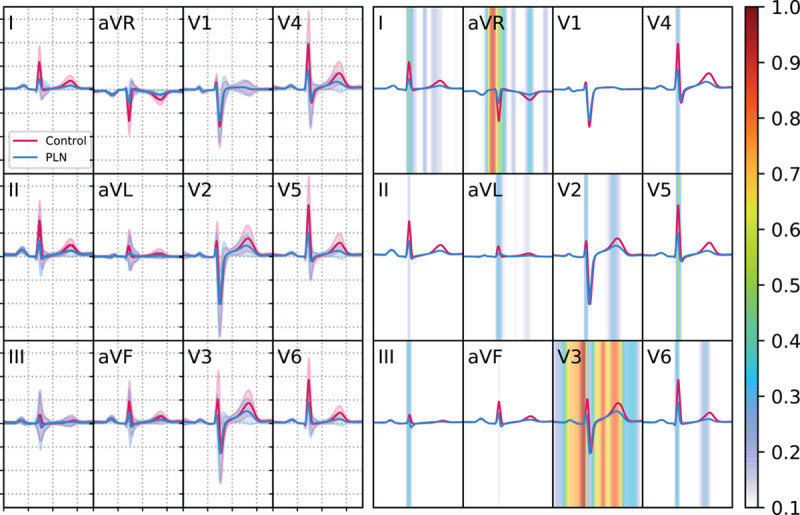
Output of the Guided Grad-CAM visualization algorithm for all PLN (phospholamban) mutation carriers and their controls. Left: Mean of temporally normalized median 12-lead ECGs of both the PLN mutation carriers (blue) and control patients (red) with their respective standard deviations. Right: The same median ECG beat with the Guided Gradient Class Class Activation Mapping output of the deep neural network (DNN) superimposed to indicate the importance of a specific temporal segment for the classification of the DNN. The colormap represents the proportion of patients where that region was important (ie, had a Guided Gradient Class Class Activation Mapping value above the threshold).
Figure 3.

Representative examples of an ECG of a PLN (phospholamban) mutation carrier (top) and a control subject (bottom) with their respective deep neural network (DNN) probability score for having the PLN mutation. Note that the control subject ECG also exhibits the established PLN features (low QRS voltages and the presence of inverted T-waves in the left precordial leads) but is classified correctly as a control subject. The features as detected by the DNN (decreased R- and T-wave voltage in V3) can be used to distinguish the PLN mutation carriers and control subject.
Feature Detection
Based on the Guided Grad-CAM maps (Figure 2), we identified the following 6 most prevalent combined ECG segments: (1) R-waves in V2/V3 (58%–99%), (2) PR interval (98%), (3) T-waves in V2/V3 (36%–89%), (4) R-waves in I/aVL/V4-V6 (34%–59%), (5) R-waves in II/III/aVF (22%–46%) and (6) T-waves in I/aVL/V6 (22%–36%). Figure 4 shows correlation between the Grad-CAM maps and the human interpretation, on an individual level.
Figure 4.
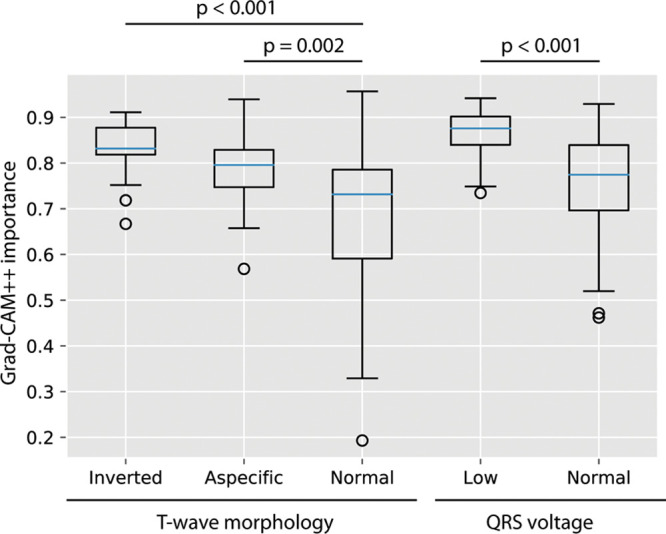
Relationship of the mean gradient class activation mapping ++ (Grad-CAM++) importance value of the T-wave area with the human interpretation of the T-wave and of the QRS-complex area with the human classification of low QRS voltage in PLN (phospholamban) patients. In the temporally aligned Grad-CAM++ curves, the mean is taken for the area of the QRS-complex and the T-wave. A boxplot of the importance values (between 0 and 1) of that region for the network for predicting PLN are shown in relationship with the human interpretation of the corresponding segments.
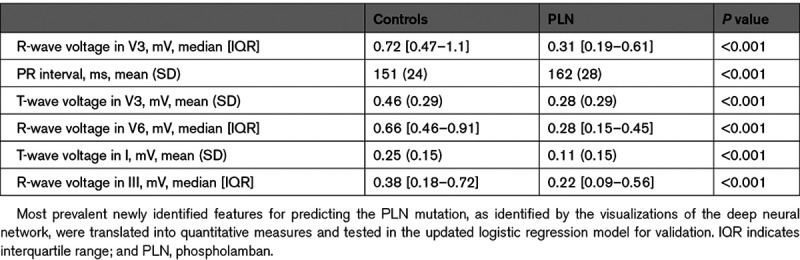
After inspection of the median beat and its SD at these locations, the following most prevalent features per region were extracted from the ECG and added to the baseline logistic regression model: (1) maximum R-wave amplitude in V3, (2) PR interval, (3) T-wave peak voltage in V3, (4) maximum R-wave amplitude in V6, (5) maximum R-wave amplitude in III, and (6) T-wave peak voltage in I.
The updated logistic regression models c-statistic was 0.91 (95% CI, 0.83–0.97). The significantly associated baseline variables low QRS voltage and inverted left precordial T-waves remained significant in the updated model. The newly identified features were maximum R-wave amplitude in V3 and V6, the T-wave amplitude in I and V3 and the PR interval. The updated model had a better fit than the baseline model with an Akaike information criterion of 388, compared with 461 for the baseline model (likelihood ratio test P<0.001). The performance measures of all 3 models are shown in Table 2. The odds ratios of the variables in the baseline and updated models are appreciated in Table 3. The summary measures for the quantitative translations of the newly identified features, that are added to the baseline logistic regression model, are shown in Table 4.
Table 2.
Discriminatory Performance of the Baseline and Updated Logistic Regressions Models and the DNN in the Independent Test Set

Table 3.
Odds Ratios and 95% CI for the Variables in the Baseline and Updated Logistic Regression Models for Prediction of PLN Mutation Carrier Status in the Training Data Set
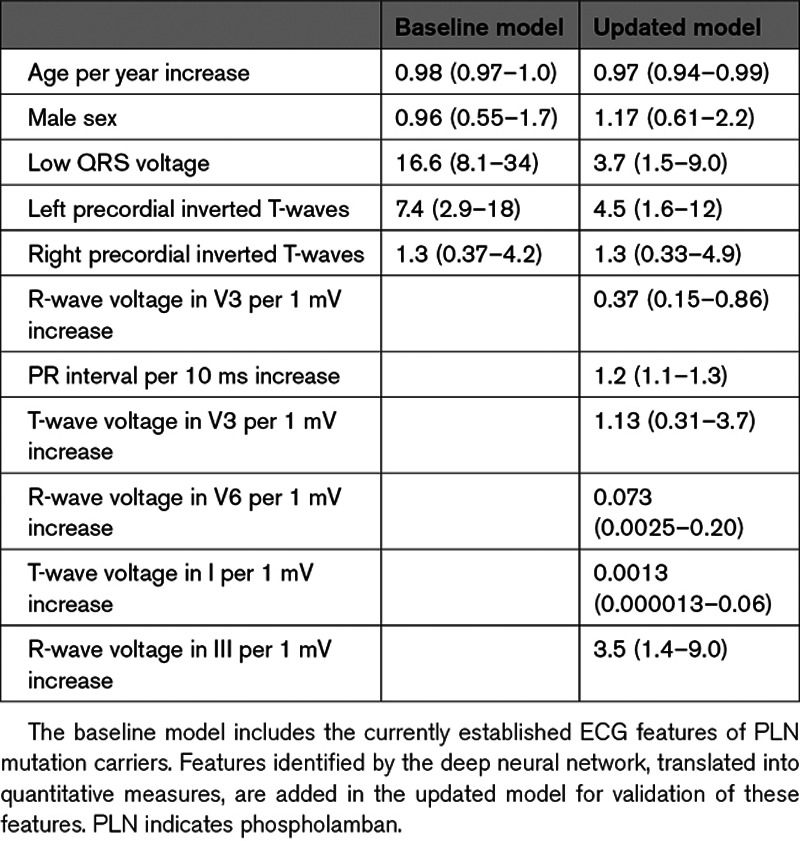
Table 4.
Summary Measures of the Quantitative Translations of the Newly Identified ECG Features of PLN Mutation Carriers
Subgroup Analyses
Performance was higher for symptomatic than presymptomatic patients, with C statistics of 0.97 (95% CI, 0.95–0.98) and 0.95 (95% CI, 0.91–0.98), respectively. Sensitivity was 86% for symptomatic patients (n=75) and 64% for presymptomatic patients (n=11), at a similar specificity of 94%. Guided Grad-CAM maps showed a difference in features between symptomatic and asymptomatic patients, where the prolonged PR interval, attenuated R- and T-wave in V3 and attenuated T-wave in V6 were more important in presymptomatic patients while the overall attenuated R-waves were more prominent in symptomatic patients. The Guided Grad-CAM maps for presymptomatic and symptomatic patients are shown in Figures II and III in the Data Supplement.
Discussion
In this study, we demonstrate a novel DNN-based end-to-end approach that allows for detection and visualization of disease-specific ECG features. To the best of our knowledge, this is the first time DNNs have successfully been applied as an ECG feature detector, in contrast to previously developed prediction algorithms. Using a unique combination of median ECG beats and visualizations, the algorithm was able to automatically reveal established ECG features in PLN p.Arg14del mutation carriers (low QRS voltages and T-wave inversions), specify these features (R- and T-wave attenuation in V2 and V3), and find novel features (increased PR-duration). Applying this promising concept in more cardiac diseases (especially rare or unknown ones) can potentially support physicians while reviewing ECGs, thereby improving ECG interpretation in daily clinical practice.
Previous Literature
Several studies showed that DNNs can be used to make predictions from ECGs with a high performance.2–4,7,8 An example is the recent study by Ko et al,4 who developed a DNN to detect hypertrophic cardiomyopathy, resulting in an area under the curve of 0.96. From a clinical point of view, this network is very attractive because this would allow the clinician to easily and automatically distinguish hypertrophic cardiomyo in a screening setting. However, clinical implementation of such a network is still challenging for several reasons. First, such networks are often seen as black boxes, and, second, the validity of these high-dimensional networks in external data sets is still unproven.
Similarly, we developed a DNN that recognizes ECGs of a specific patient population (ie, PLN mutation carriers) with high diagnostic performance.4 A different architecture was chosen, as it has an increased diagnostic performance in PLN mutation carriers and allows for more detailed visualizations. Unique to our study is the use of hard outcome data and the focus on feature detection, which may directly support clinicians with ECG interpretation in daily practice. Moreover, we show that these features can be used in a relatively simple logistic regression model, which might be easier generalizable.
Disease-Specific ECG Features in PLN Mutation Carriers
This novel approach was validated in PLN mutation carriers because typical ECG characteristics in these subjects have been described extensively before.10,12,14,15 PLN mutation carriers are at risk of developing an often biventricular phenotype of arrhythmogenic right ventricular cardiomyopathy and dilated cardiomyopathy and are typically characterized by subepicardial fibrofatty replacement.21 This leads to an ECG with low QRS voltages, which can be seen both in the limb leads and in precordial leads.10,14 In addition, negative T-waves were previously described in both the right precordial leads and in the left precordial leads.12,15
Using this novel approach, we could correctly identify all of these previously described ECG features (Figure 2) and show that the network also uses the preestablished features for diagnosis (Figure 4). In addition, we could specify the leads in which these features are typically present. With the visualization tool, we found attenuated R-waves to be particularly present in the lateral leads I, aVL and V6, and in the right precordial leads V2 and V3. While the low voltages in these mutation carriers are often measured as QRS peak-to-peak amplitude, we observed that these low voltages were only based on R-wave attenuation, while the S-wave seemed unaltered. Furthermore, we found attenuated/inverted T-waves to be typically present in leads V2, V3 and V6 (as described previously) but also in leads I and aVL. Besides the ECG characteristics that were already identified before in PLN mutation carriers, we also found an ECG feature, the PR interval, that was not described before in these subjects. This was confirmed in the updated logistic regression model. Interestingly, a recent meta-analysis of genome-wide association studies also showed an association between a locus in the PLN gene and PR interval, which already suggested that PLN plays a role in atrioventricular conduction.22
In an exploratory analysis, the DNN performed well in both presymptomatic and symptomatic mutation carriers. Our approach also suggested that particular features were more important in presymptomatic mutation carriers (PR interval and R- and T-wave attenuation in V2 and V3), as compared to symptomatic carriers. This might indicate that our approach can be used in subgroups who are in different stages of a disease, to gain knowledge on the sequence in which ECG abnormalities naturally occur. In particular for PLN mutation carriers, it is important to gain knowledge on the first electrical changes because this may improve early screening and risk stratification of presymptomatic mutation-carrying family members.
Employed Methodology
The use of DNN for the analysis of data generally requires large amounts of balanced data but the group of PLN mutation carriers studied in this investigation contained only 86 patients. The focus on features detection instead of prediction in this article, however, allowed the use of such small data sets, as we were able to reduce the highly dimensional DNN to a few important features. Moreover, to allow training on this extremely imbalanced data set, while also correcting for age and sex differences between mutation carriers and controls, we applied propensity score matching.
In the present study, we used ECG median beats as input for the DNN model, which allowed the network to focus on morphology rather than rhythm. The use of median beats prohibits detection of rhythm specific ECG features (eg, premature contractions or heart rate variability) and can also not be used for detection of beat-to-beat ECG variations. To our best knowledge, this is the first study in which median beats are used for deep learning.
Limitations
First, although the proposed approach is feasible in small data sets, care should be taken while interpreting results derived from small cohort studies as findings may not hold up when evaluated on other cohorts. Especially, the number of patients in the test set is a major limitation. To show clinical applicability of the ECG features and algorithm as described in this study, external validation studies should be performed. Second, for the PLN mutation carriers, the clinical phenotype may be variable among mutation carriers. Therefore, it should be noted that this approach helps to visualize the most common ECG features on a group level, but important ECG features that are present in small subgroups may be missed. Subgroup analyses in more homogeneous subgroups (eg, presymptomatic relatives) can be used to reveal important features in these specific subgroups. Third, the ECGs of the control group were extracted from a large database in which additional patient-specific characteristics are not available. Therefore, no comparisons or matching between both groups were possible to correct for other influencing factors. However, the case-control matching ratio of 1:20 used in this study presumably equalized the groups, and the detected features align with literature on other PLN mutation carriers. Fourth, the conduction intervals and P-, QRS-, and T-wave boundaries are based on the automated GE algorithm, which might cause inaccuracies. Boundary measurements on median beats have proven to be very accurate, however.23 Fifth, the proposed approach is not possible for ECGs with arrhythmias or acute ischemia, as these (temporary) conditions have a large influence on the morphology of the median beat. In this study, the algorithm is not intended to be used in these situations and only 3 patients were excluded for this reason. Finally, the visualization technique used in this article, Guided Grad-CAM++, is one of the most frequently used techniques for fine-grained heatmaps but has limitations of its own.6 For example, guided backpropagation might be independent on the choice of the model or data generating process.24 Therefore, we validated the detected features in a logistic regression model and showed that Grad-CAM++ values agree with the preestablished PLN ECG features. Feature visualization in DNNs is a new and developing field and future research should focus on improving visualization techniques for DNNs and applying them in ECGs.
Future Perspectives
Future studies should be conducted applying this novel approach to other less well-characterized diseases, such as new genetic mutations, to discover novel ECG characteristics. The visualizations provide the end-user with feedback on the importance and location of detected ECG features. Moreover, future studies should consider elucidating the pathophysiological mechanisms of newly identified ECG features by using other experimental methods such as (non)invasive electrophysiological mapping. The influence of the discovered ECG features on disease penetrance in asymptomatic carriers or progression of disease in symptomatic carriers should be examined with longitudinal ECG or outcome data. Finally, combining our approach and a DNN trained on other cohorts with a focus on screening, such as family members of mutation carriers or large healthy population cohorts, might be of interest in clinical practice. Detection and visualization of possible carrier status in the ECG even before the genetic diagnosis is done could determine which family members or healthy individuals require genetic testing or follow-up.
Conclusions
This study demonstrated a novel DNN-based end-to-end approach that allows for detection and visualization of disease-specific ECG features. In a cohort of PLN p.Arg14del mutation carriers, the algorithm showed excellent diagnostic performance and revealed already established ECG features. Moreover, we were able to specify these features and to detect novel features. This novel way to use DNNs may help advance diagnostic capabilities in daily practice, especially in rare and new cardiac diseases.
Acknowledgments
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research. Folkert Asselbergs is supported by UCL Hospitals NIHR Biomedical Research Centre.
Sources of Funding
This study was financed by the Netherlands Organisation for Health Research and Development (ZonMw) with grant number 104021004, the Dutch Heart Foundation and the PLN Genetic Heart Disease Foundation. Part of this work is funded by Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation (grant number CVON2015-12 eDETECT).
Disclosures
None.
Supplementary Material
Footnotes
Nonstandard Abbreviations and Acronyms
- DNN
- deep neural network
- Guided Grad-CAM
- Guided Gradient Class Activation Mapping
- PLN
- Phospholamban
Drs van de Leur and Taha contributed equally to this work.
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCEP.120.009056.
For Sources of Funding and Disclosures, see page 146.
Contributor Information
Rutger R. van de Leur, Email: r.r.vandeleur@umcutrecht.nl.
Karim Taha, Email: k.taha-2@umcutrecht.nl.
Max N. Bos, Email: maxn.bos@gmail.com.
Jeroen F. van der Heijden, Email: J.F.vanderHeijden-2@umcutrecht.nl.
Deepak Gupta, Email: guptadeepak2806@gmail.com.
Maarten J. Cramer, Email: M.J.M.Cramer@umcutrecht.nl.
Rutger J. Hassink, Email: r.j.hassink@umcutrecht.nl.
Pim van der Harst, Email: p.van.der.harst@umcg.nl.
Pieter A. Doevendans, Email: P.Doevendans@umcutrecht.nl.
Folkert W. Asselbergs, Email: f.w.asselbergs@umcutrecht.nl.
References
- 1.Goodfellow I, Bengio Y, Courville A. Deep Learning. 2016. The MIT Press [Google Scholar]
- 2.Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–69. doi: 10.1038/s41591-018-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van de Leur RR, Blom LJ, Gavves E, Hof IE, Van der Heijden JF, Clappers NC, Doevendans PA, Hassink RJ, Van Es R. Automatic triage of 12-lead electrocardiograms using deep convolutional neural networks. J Am Heart Assoc. 2020;9:e015138 doi: 10.1161/JAHA.119.015138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko WY, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, Demuth SJ, Ackerman MJ, Gersh BJ, Arruda-Olson AM, et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol. 2020;75:722–733. doi: 10.1016/j.jacc.2019.12.030 [DOI] [PubMed] [Google Scholar]
- 5.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. Proc IEEE Int Conf Comput Vis. 2017618–626. doi: 10.1109/ICCV.2017.74 [Google Scholar]
- 6.Chattopadhay A, Sarkar A, Howlader P, Balasubramanian VN.Grad-CAM++: generalized gradient-based visual explanations for deep convolutional networks.. In: Proc - 2018 IEEE Winter Conf Appl Comput Vision. 2018;839–847. 2018 IEEE Winter Conference on Applications of Computer Vision (WACV); March 12–15; Lake Tahoe, NV. [Google Scholar]
- 7.Raghunath S, Ulloa Cerna AE, Jing L, vanMaanen DP, Stough J, Hartzel DN, Leader JB, Kirchner HL, Stumpe MC, Hafez A, et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat Med. 2020;26:886–891. doi: 10.1038/s41591-020-0870-z [DOI] [PubMed] [Google Scholar]
- 8.Kwon JM, Lee SY, Jeon KH, Lee Y, Kim KH, Park J, Oh BH, Lee MM. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J Am Heart Assoc. 2020;9:e014717 doi: 10.1161/JAHA.119.014717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Zwaag PA, van Rijsingen IA, de Ruiter R, Nannenberg EA, Groeneweg JA, Post JG, Hauer RN, van Gelder IC, van den Berg MP, van der Harst P, et al. Recurrent and founder mutations in the Netherlands-Phospholamban p.Arg14del mutation causes arrhythmogenic cardiomyopathy. Neth Heart J. 2013;21:286–293. doi: 10.1007/s12471-013-0401-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haghighi K, Kolokathis F, Gramolini AO, Waggoner JR, Pater L, Lynch RA, Fan GC, Tsiapras D, Parekh RR, Dorn GW, 2nd, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hof IE, van der Heijden JF, Kranias EG, Sanoudou D, de Boer RA, van Tintelen JP, van der Zwaag PA, Doevendans PA. Prevalence and cardiac phenotype of patients with a phospholamban mutation. Netherlands Hear J. 2019;27:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, Cox MG, van Lochem LT, de Boer RA, Hofstra RM, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14:1199–1207. doi: 10.1093/eurjhf/hfs119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milano A, Blom MT, Lodder EM, van Hoeijen DA, Barc J, Koopmann TT, Bardai A, Beekman L, Lichtner P, van den Berg MP, et al. Sudden cardiac arrest and rare genetic variants in the community. Circ Cardiovasc Genet. 2016;9:147–153. doi: 10.1161/CIRCGENETICS.115.001263 [DOI] [PubMed] [Google Scholar]
- 14.Posch MG, Perrot A, Geier C, Boldt LH, Schmidt G, Lehmkuhl HB, Hetzer R, Dietz R, Gutberlet M, Haverkamp W, et al. Genetic deletion of arginine 14 in phospholamban causes dilated cardiomyopathy with attenuated electrocardiographic R amplitudes. Heart Rhythm. 2009;6:480–486. doi: 10.1016/j.hrthm.2009.01.016 [DOI] [PubMed] [Google Scholar]
- 15.Groeneweg JA, van der Zwaag PA, Olde Nordkamp LR, Bikker H, Jongbloed JD, Jongbloed R, Wiesfeld AC, Cox MG, van der Heijden JF, Atsma DE, et al. Arrhythmogenic right ventricular dysplasia/cardiomyopathy according to revised 2010 task force criteria with inclusion of non-desmosomal phospholamban mutation carriers. Am J Cardiol. 2013;112:1197–1206. doi: 10.1016/j.amjcard.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 16.GE Healthcare. Marquette 12SL ECG Analysis Program Physician’s Guide. 2012. Chicago, United States: Accessed April 30, 2020. https://www.gehealthcare.com/products/diagnostic-cardiology/marquette-12sl [Google Scholar]
- 17.Pearce N. Analysis of matched case-control studies. BMJ. 2016;352:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oord A van den, Dieleman S, Zen H, Simonyan K, Vinyals O, Graves A, Kalchbrenner N, Senior A, Kavukcuoglu K. WaveNet: a generative model for raw audio. Neural Comput. 2016;21:793–830. [Google Scholar]
- 19.Franceschi J-Y, Dieuleveut A, Jaggi M. Unsupervised scalable representation learning for multivariate time series. In: Advances in Neural Information Processing Systems. 2019;32:4650–4661. 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, Canada, December 8–14, 2019 [Google Scholar]
- 20.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. PyTorch: an imperative style, high-performance deep learning library. In: Advances in Neural Information Processing Systems. 2019;32:8026–8037. 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, Canada, December 8–14, 2019 [Google Scholar]
- 21.Gho JMIH, Van Es R, Stathonikos N, Harakalova M, Te Rijdt WP, Suurmeijer AJH, Van Der Heijden JF, De Jonge N, Chamuleau SAJ, et al. High resolution systematic digital histological quantification of cardiac fibrosis and adipose tissue in phospholamban p.Arg14del mutation associated cardiomyopathy. PLoS One. 2014;9:e94820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Setten J, Verweij N, Mbarek H, Niemeijer MN, Trompet S, Arking DE, Brody JA, Gandin I, Grarup N, Hall LM, et al. Genome-wide association meta-analysis of 30,000 samples identifies seven novel loci for quantitative ECG traits. Eur J Hum Genet. 2019;27:952–962. doi: 10.1038/s41431-018-0295-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willems JL, Zywietz C, Arnaud P, van Bemmel JH, Degani R, Macfarlane PW. Influence of noise on wave boundary recognition by ECG measurement programs. Recommendations for preprocessing. Comput Biomed Res. 1987;20:543–562. doi: 10.1016/0010-4809(87)90025-5 [DOI] [PubMed] [Google Scholar]
- 24.Adebayo J, Gilmer J, Muelly M, Goodfellow I, Hardt M, Kim B. Sanity checks for saliency maps. In: Advances in Neural Information Processing Systems. 20189505–9515. 32nd Conference on Neural Information Processing Systems (NeurIPS 2018), Montréal, Canada, December 2–8, 2018 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are not publicly available due to privacy restrictions. The code for training the DNN and for generating the visualizations and tables in this article is available upon request from the corresponding author.


