Abstract
We examined the prevalence and transmission of the fosA3 gene among Citrobacter freundii isolates from flowers and the retail environments. We identified 11 fosfomycin-resistant C. freundii strains (>256 μg/mL) from 270 samples that included petals (n = 7), leaves (n = 2), dust (n = 1) and water (n = 1). These 11 isolates were multidrug-resistant and most were simultaneously resistant to fosfomycin, cefotaxime, ciprofloxacin and amikacin. Consistently, all 11 isolates also possessed blaCTX–M–14, blaCMY–65/122, aac(6’)-Ib-cr, qnrS1, qnrB13/6/38 and rmtB. These fosA3-positive isolates were assigned to two distinct PFGE patterns and one (n = 9) predominated indicating clonal expansion of fosA3-positive isolates across flower markets and shops. Correspondingly, fosA3 was co-transferred with blaCTX–M–14 via two plasmid types by conjugation possessing sizes of 110 kb (n = 9) and 260 kb (n = 2). Two representatives were fully sequenced and p12-1 and pS39-1 possessed one and two unclassified replicons, respectively. These plasmids shared a distinctive and conserved backbone in common with fosA3-carrying C. freundii and other Enterobacteriaceae from human and food animals. However, the fosA3-blaCTX–M–14-containing multidrug resistance regions on these untypable plasmids were highly heterogeneous. To the best of our knowledge, this is the first report of fosA3 and blaCTX–M–14 that were present in bacterial contaminants from flower shops and markets. These findings underscore a public health threat posed by untypable and transferable p12-1-like and pS39-1-like plasmids bearing fosA3-blaCTX–M–14 that could circulate among Enterobacteriaceae species and in particular C. freundi in environmental isolates.
Keywords: flower, fosfomycin-resistance, Citrobacter freundii, fosA3, blaCTX–M–14
Introduction
There are two major problems in the treatment of bacterial infections: the spread of multidrug-resistant (MDR) or extensively drug-resistant (XDR) pathogens and lack of development of new antibiotics active against these bacteria (Falagas et al., 2016). This situation has renewed interest in older antibiotics such as fosfomycin as alternatives or “last resort” therapies (Falagas et al., 2016; Sastry and Doi, 2016). Fosfomycin is viewed as a suitable empirical drug that has retained activity against resistant strains (Popovic et al., 2010). The World Health Organization has reclassified fosfomycin as a “critically important antimicrobial” based on its broad-spectrum bactericidal reactivity and good pharmacological properties (World Health Organization [WHO], 2011).
Clinical cases of fosfomycin resistance have increased in the last decade especially due to inactivation of the drug via plasmid-mediated fosfomycin-modification (fos) genes in Enterobacteriaceae (Diez-Aguilar et al., 2013). Fosfomycin covalently binds to a cysteine thiol in the active site of MurA and interferes with peptidoglycan synthesis at an earlier step than the action of β-lactams or glycopeptides. FosA is a glutathione S-transferase that covalently modifies fosfomycin for inactivation. There are currently more than 10 fos types, and fosA, its subtypes, and fosC2 are primarily found in the Enterobacteriaceae (Yang et al., 2017). In China, plasmid-encoded fosA3 in Escherichia coli isolates from food and pets have been reported with high detection rates (Hou et al., 2012; Yang et al., 2014; Yao et al., 2016; Wang et al., 2017) although fosfomycin has not been approved for veterinary use. Interestingly, fosA3 is often found co-localized with blaCTX–M on epidemic plasmids and this most likely promotes the transfer and dissemination of fosA3 in humans and animals (Yang et al., 2014; Feng et al., 2015). Co-spread of fosA3 with other important antibiotic resistance genes (ARG) is concerning due to the potential to rapidly develop into MDR Enterobacteriaceae strains.
Additionally, E. coli strains carrying fosA3 and blaCTX–M–14 have been isolated from vegetables in Netherlands (Freitag et al., 2018), even more mcr-1 gene in E. coli was identified in fresh vegetables in Guangzhou, China (Luo et al., 2017). Literature also have claimed that plants could be contaminated by manure and wastewater from animal farming, contributing to the widely spread of AMR (Zeng et al., 2019; Sun et al., 2020). As a kind of plant, flowers are closely related to human life and possible vectors for AMR genes transformation. However, few reports have focused on the significance of flowers as a pathway for AMR spread.
Therefore, we investigated drug-resistant bacteria/drug-resistant genes from flowers and retail environment including water and dust in florists in Guangzhou, China. There are relatively few reports of plasmid-borne fosA3 in Citrobacter. freundii, a bacterium associated with opportunistic nosocomial infections of the respiratory and urinary tracts and blood (Feng et al., 2015; Li M. et al., 2018). Herein, we present the first report of the emergence of fosA3 in C. freundii isolates from flowers and the retail environments. We further investigated the molecular epidemiology of fosA3-carrying C. freundii isolates and characterized the fosA3-bearing plasmids.
Materials and Methods
Bacterial Strains and Detection of fos Genes Materials and Methods
A total of 270 samples were randomly collected from 3 flower markets and 6 flower shops in Guangzhou, China during March 2017. The samples included lily petals (n = 90), lily leaves (n = 90), dust (n = 45), and water (n = 45). Flowers and leaves were separately collected in sterile sealed bags. Water for watering flowers was collected in 50 mL centrifuge tubes. Dust samples were wiped with sterile cotton swabs in the surface dust from tables or floors (each 10-cm × 10-cm area) in retail shops, and then were rinsed in 2 mL sterile physiological saline solution. One petal and one leaf of each lily flowers were picked and washed with 10 mL of sterile saline solution. Then 100 μL of dust-resuspension, flower washed fluid and water samples were incubated in 4 mL drug-free LB broth for 12–16 h at 37°C and plated on MacConkey agar plates containing 256 μg/mL fosfomycin plus 25 μg/mL glucose-6-phosphate. After 18 h incubation at 37°C, 1–2 red colonies of different morphologies from each plate were selected. The bacterial species identification was performed using the MALDI-TOF MS (Shimadzu-Biotech, Japan) and 16S rRNA gene sequencing (16srRNA-F: AGAGTTTGATCATGGCTC; 16srRNA-R: GGTTACCTTGTTACGACTT). Fosfomycin-resistant C. freundii isolates (≥256 mg/L) were screened for the presence of the plasmid-mediated fosfomycin resistance genes fosA1, fosA2, fosA3, fosA4, fosA5, fosA6, fosA7, and fosC2 by PCR amplification and sequencing using primers as previously described (Huang et al., 2020).
Antimicrobial Susceptibility Testing and Resistance Genes Detecting
All fos-positive isolates were screened for the minimum inhibitory concentration (MIC) using the Mueller Hinton (MH) agar dilution method (MH agar was purchased from Huankai Co., Ltd., Guangzhou, China) and the results were interpreted according to CLSI (Clinical and Laboratory Standards Institute [CLSI], 2018) and veterinary CLSI (Clinical and Laboratory Standards Institute [CLSI], 2019) guidelines. The following antimicrobials were tested: fosfomycin (FOS), cefotaxime (CTX), amoxicillin-clavulanate (AMC), meropenem (MEM), tetracycline (TET), chlortetracycline (CTET), doxycycline (DOX), ciprofloxacin (CIP), gentamicin (GEN), amikacin (AMK), sulfamethoxazole/trimethoprim (SXT), florfenicol (FFC), chloramphenicol (CHL), and rifampicin (RIF) (above drugs were purchased from Sigma Chemical Co., St Louis, MO, United States). MICs of tigecycline and colistin were determined by MH broth microdilution (MH broth was purchased from Huankai Co., Ltd., Guangzhou, China) and the resistance breakpoint was interpreted according to EUCAST criteria (>2 μg/mL) and FDA criteria (≥8 μg/mL). E. coli ATCC 25922 was used as a quality control strain.
All fos-positive isolates were further screened for the presence of the extended spectrum β-lactamase (ESBL) gene blaCTX–M–9G/1G, the plasmid-mediated AmpC β-lactamase gene blaCMY, the plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB, qnrS, aac-(6′)-Ib-cr, qepA, oqxA and oqxB), floR as well as 16S-RMTase genes (armA/rmtB) using PCR amplification as previously described (Phuc Nguyen et al., 2009; Fang et al., 2016; Liu et al., 2016). Notably, blaCTX–M–9G-positive isolates were further tested by PCR amplification using previous primers of ESBL-encoding genes (ISEcp1-F: CTATCCGTACAAGGGAGTGT; IS903-R: TTTCCACTCGCCTTCACC) and protocols to confirm the subtypes of ESBL-encoding genes and other genotypes. All PCR products were sent for sequencing and the DNA sequences were blasted with GenBank database1.
Molecular Typing
Chromosomal DNA digested with XbaI restriction enzyme was used for PFGE (Gautom, 1997) to analyze the genetic relatedness of all isolates containing fos. PFGE patterns were analyzed with the Dice coefficient and the unweighted pair group method with average linkages (UPGMA) clustering method using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium). PFGE types were defined with >90% similarity between clusters.
Transfer of fos Genes, Gene Location and Plasmid Replicon Typing
To determine the transferability of fosA3 genes, isolates positive for fosA3 were selected for conjugation experiments using the broth-mating method and streptomycin-resistant E. coli strain C600 (MIC > 2000 μg/mL) as the recipient. The donor and recipient strains were inoculated into 4 mL LB broth (Huankai Co., Ltd., Guangzhou, China) and shaken at 37°C for 4 h, then the donors and recipients were mixed in a 1:4 (100 and 400 μL) ratio in a 2 mL EP tubes and incubated at 37°C for 20 h. Transconjugants were selected on MacConkey agar plates supplemented with 2000 μg/mL streptomycin and 256 μg/mL fosfomycin (Yang et al., 2014). Antimicrobial susceptibility testing of the transconjugants and co-transfer of other resistance genes were determined as mentioned above. Incompatibility (Inc) groups were determined using PCR-based replicon typing (PBRT) (Carattoli et al., 2005).
The bacterial cell of the transconjugants were lysed with the ESP buffer (0.5 M EDTA, pH 9.0; 1% sodium lauroyl sarcosinate; 1 mg of proteinase K per mL) and then the bacterial DNA was embedded in the gel block. The S1-PFGE protocol is detailed in previous reports (Barton et al., 1995). The fosA3 gene genomic locations were identified by linearization of plasmids from transconjugants using S1 nuclease followed by PFGE (Yang et al., 2014). Southern blotting was carried out from S1-PFGE gels using a digoxigenin-labelled probe specific for fosA3.
WGS Sequencing and Characterization of fosA3-Bearing Plasmids
Based on the results of plasmid analysis and PFGE typing, the total genomic DNA was extracted from two C. freundii strains S39 and H12-3-2 using a TIANamp Bacteria DNA Kit (Tiangen) and DNA libraries were constructed with 250-bp paired-end whole-genome sequencing using the Illumina HiSeq system (Illumina, San Diego, CA, United States) (Ma et al., 2020). The obtained paired-end Illumina reads were assembled de novo using SPAdes v3.6.2 (default parameters except –careful and –k 21,33,55,77,99,127) (Bankevich et al., 2012). In addition, to obtain long reads sequence, selected strains were further sequenced using Oxford Nanopore MinION flowcell R9.4 (Li R. et al., 2018). De novo hybrid assembly was performed using a combined Illumina HiSeq and Nanopore sequencing approach (Nextomics). Genome assembly was performed with Unicycler version 0.4.1 (Wick et al., 2017) using a combination of short and long reads, followed by error correction with Pilon version 1.12 (Walker et al., 2014). Gene prediction and annotation were performed using RAST (Overbeek et al., 2014)2 and BLAST3 and rechecked manually. Alignments with highly homologous sequences (with >90% coverage and >90% nucleotide identity) from the NCBI database and generation of plasmid maps were performed with BRIG (Alikhan et al., 2011) and Easyfig (Sullivan et al., 2011).
Nucleotide Sequence Accession Numbers
The representative fosA3-bearing genome sequences S39 and H12-1-2 were submitted to NCBI with the accession numbers CP045555 and CP045837, respectively, and fosA3-bearing plasmids sequences pS39-1 and pH12-1 with the accession numbers CP045556 and CP045838, respectively.
Results
Antimicrobial Susceptibility and Prevalence of fosA3 Genes
We isolated 17 fosfomycin-resistant Enterobacteriaceae from flower retail environments including 11 C. freundii and 6 E. coli from our group of 270 samples. The 11 C. freundii isolates were from 2 flowers markets and 3 flowers shops and samples included petals (n = 7), leaves (n = 2), dust (n = 1), and water (n = 1). All 11 isolates possessed the fosA3 gene and no other fos gene was detected by PCR. These 11 were all were concurrently resistant to fosfomycin, cefotaxim, amoxicillin-clavulanate, tetracycline, chlortetracycline, doxycycline, ciprofloxacin, gentamicin, sulfamethoxazole/trimethoprim, florfenicol, chloramphenicol, and rifampin (Table 1). In addition, most of these isolates were also resistant to amikacin with MICs > 256 μg/mL (n = 9). However, all fosA3-positive isolates were susceptible to meropenem and tigecycline.
TABLE 1.
Background information and characteristics of fosA3-positive Citrobacter freundii.
| Strains1 | Locations | Source | PFGE Typing | Resistance genes2 | Resistance profiles3 | Replicon type4 | Plasmids size (kb) |
| H12-3-2 | Flower market 1 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY–122, floR, aac(6’)-Ib-cr, rmtB, qnrB13, qnrS1 | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| H41-7-2 | Flower market 2 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| HR5 | Flower market1 | Dust | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| N21-1 | Flower shop 1 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| N52-2 | Flower shop 2 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| N89-1-1 | Flower shop 3 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| NW2-2 | Flower shop 1 | Water | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| S66-1-1 | Flower shop 3 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| S40-1-2 | Flower shop 2 | Lily petals | I | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, rmtB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, AMK, SXT, FFC, CHL, RFP | UT | ∼110 |
| S39 | Flower shop 2 | Lily leaf | II | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, qnrB38, qnrB6, qnrS1 | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, SXT, FFC, CHL, RFP | UT | ∼261 |
| S45-1 | Flower shop 3 | Lily leaf | II | fosA3, blaCTX–M–14, blaCMY, floR, aac(6’)-Ib-cr, qnrB, qnrB, qnrS | FOS, CTX, AMC, TET, CETE, DOX, CIP, GEN, SXT, FFC, CHL, RFP | UT | ∼261 |
1WGS sequence are underlined. 2Resistance phenotypes transferred to the recipient by conjugation are underlined. FOS, fosfomycin; CTX, cefotaxime; AMC, amoxicillin-clavulanate; MEM, meropenem; TIG, tigecycline; TET, tetracycline; CTET, chlortetracycline; DOX, doxycycline; CIP, ciprofloxacin; GEN, gentamicin; AMK, amikacin; SXT, sulfamethoxazole/trimethoprim; FFC, florfenicol; CHL, chloramphenicol; RIF, rifampicin. 3Transfer of fosA3 and co-transfer of this gene with other genotypes and phenotypes are underlined. 4UT, Untypeable.
We further screened these 11 fosA3-positive C. freundii isolates for the presence of other important ARGs and identified numerous gene combinations. These included blaCTX–M–14-blaCMY-rmtB-aac(6’)-Ib-cr-qnrB-qnrS-floR (n = 9) and blaCTX–M–14-blaCMY-aac(6’)-Ib-cr-qnrB-qnrB-qnrS-floR (n = 2) (Table 1).
Pulsed-Field Gel Electrophoresis (PFGE) Typing
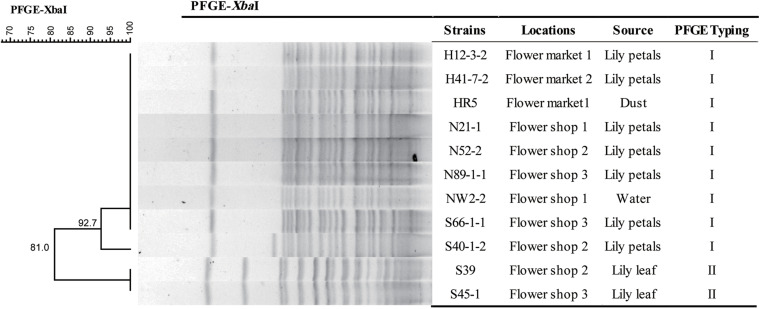
We successfully performed PFGE typing for all 11 fosA3-positive C. freundii isolates. We found two different PFGE patterns that retained >90% similarity and were designated types I and II. Type I predominated and contained nine C. freundii isolates recovered from Lily petals, dust and water from two markets and three flower shops (Figure 1).
FIGURE 1.
PFGE dendrogram showing the genetic relatedness of 13 Citrobacter freundii isolates including 2 fosA3-positive isolates.
Location of fosA3 Gene and Transferability of Plasmids Carrying fosA3
All 11 fosA3-positive isolates were able to mobilize and transfer fosA3 to recipient strain E. coli C600. These transconjugants were highly resistant to FOS (MIC > 512 μg/mL) with high-level resistance to CTX (MICs ≥ 64 μg/mL). In addition, two transconjugants also showed high-level resistance to TET, CETE, GEN, SXT, CHL, and RFP (MICs ≥ 64 μg/mL) and their MICs to DOX and CIP were increased 16- and 32-fold, respectively, when compared with the recipient strain (Table 1 and Supplementary Table S1). Correspondingly, blaCTX–M–14 (n = 11), aac(6’)-Ib-cr (2) and qnrB (n = 2) were co-transferred with fosA3 in 11 donors (Table 1). Gene location and plasmid replicon typing demonstrated that the transferable fosA3 gene coexisted with blaCTX–M–14 (n = 9) and blaCTX–M–14-aac(6’)-Ib-cr-qnrS (n = 2) that were present on the untypable plasmids with sizes of 110 kb (n = 9) and 260 kb (n = 2), respectively (Table 1 and Supplementary Figure S1).
fosA3-Carrying Plasmid Analysis
Based on the results of PFGE typing and plasmid analysis, we selected two representative C. freundii strains S39 and H12-3-2 for complete sequencing. Strain S39 contained one chromosome (4,806,164 bp) and 4 plasmids including one fosA3-carrying plasmid pS39-1. Strain H12-3-2 contained one chromosome (4,948,530 bp) and harbored 2 plasmids including a fosA3-carrying plasmid pH12-1. BWA (Li and Durbin, 2009) and Samtools (Li et al., 2009) software were used to obtain the sequence depth of each base, and the average sequencing depths of each nucleotide were calculated. The chromosome sequence of strain H12-1-2 was covered 96 × on average, and the chromosome sequence of strain S39 was covered 159 × on average. The plasmid sequence of pH12-1-1 was covered 106 × on average, and the plasmid sequence of pS39-1 was covered 90 × on average. In addition to the presence of fosA3, blaCTX–M–14, blaCMY–65, aac(6’)-Ib-cr, qnrB6, qnrS1 and qnrB38, strain S39 also harbored blaTEM–1B, catA2, floR, aac(3)-IIa, aadA16, aph(3”)-Ib, aph(6)-Id, sul1, sul2, dfrA27, tet(A), tet(D), and arr-3. In addition to fosA3, blaCTX–M–14, blaCMY–122, aac(6’)-Ib-cr, qnrS1, qnrB13, and rmtB, strain H12-3-2 also harbored blaOXA–1, blaTEM–1B, catB3, floR, aadA2b, aph(3”)-Ib, aph(6)-Id, sul1, sul2, dfrA12, and arr-3. These ARGs were all located on plasmids except for the chromosomal qnrB38 and blaCMY–65 in strain S39 and chromosomal qnrB13 and blaCMY–122 in strain H12-3-2. Furthermore, ARG carriage was consistent with the resistance phenotypes for these strains (Table 1).
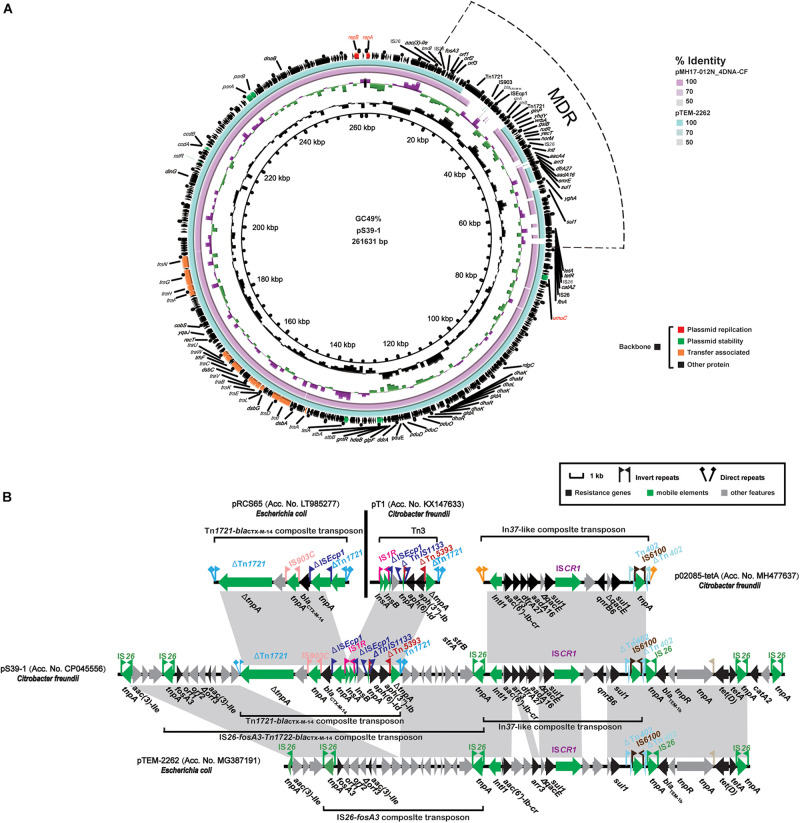
Plasmid pS39-1 was 261, 631 bp in length and included 332 coding sequences (CDS), an overall GC content of 49% and was untypable. The backbone sequence of pS39-1 was almost identical to the fosA3-bearing plasmids pMH17-012N_4 (Acc. No. AP018570) and pTEM-2262 (Acc. No. MG387191) (Figure 2A). These three plasmids contained 203 kb conserved backbone regions and the primary differences contained between repA (plasmid replication) and umuC (plasmid stability) that included large multidrug-resistance region (MRR). The conserved backbone regions encoding functions for plasmid replication (repA and repB), stability (parAB, stbAB, ccdAB, ralR, telA, umuC, and higA) and transfer (15 tra/trh genes). Of note, repA (837 bp) and repB (1119 bp) located on plasmid pS39-1 belonged to unclassified replicons and were nearly identical to the corresponding repA and repB genes of plasmids pMH17-012N_4 and pTEM-2262 with 0 and 1 SNP differences for repA and repB, respectively.
FIGURE 2.
Characteristics of the complete nucleotide sequences of the fosA3 plasmid pS39-1 identified in this study. (A) A comparison of pS39-1 with two untypable fosA3 plasmids pTEM-2262 (Acc. No. MG387191) and pMH17-012N_4DNA-CF (Acc. No. AP018570). (B) Comparison of the fosA3-containing MRR with the corresponding regions from pRCS65 (Acc. No. LT985277), pT1 (Acc. No. KX147633), p02085-tetA (Acc. No. MH477637), and pTEM-2262 (Acc. No. MG387191). Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on functional classification. Shaded regions denote shared DNA homology (>95% nucleotide identity).
In addition to the plasmid backbone region, plasmid pS39-1 possessed a 53.4 kb MDR containing 15 ARGs (fosA3, blaCTX–M–14, blaTEM–1B, catA2, aac(3)-IIa, aadA16, aac(6’)-Ib-cr, aph(3”)-Ib, aph(6)-Id, sul1, qnrB6, dfrA27, tet(A), tet(D), and arr-3) interspersed with different complete or truncated insertion sequences and transposons (IS26, ΔTn1721, IS903C, ΔISEcp1, IS1R, ISCR1, ΔTn402, and IS6100) (Figure 2A). This 53.4 kb MDR was highly similar to that in fosA3-bearing plasmid pTEM-2262. The primary difference between the two were full and partial deletions of the Tn1721-blaCTX–M–14-composite transposon (13,196 bp) and the In37-like composite transposon (12,959 bp), respectively. Of note, the Tn1721-blaCTX–M–14-composite transposon on pS39-1 was composed of two parts that were almost identical to the Tn1721-blaCTX–M–14-composite transposon (10,459 bp) on pRCS65 (Acc. No. LT985277) and the Tn3 transposon (4569 bp) on pT1 (Acc. No. KX147633), respectively. Furthermore, the In37-like composite transposon of plasmid pS39-1 was highly similar to that in plasmid p02085-tetA (Acc. No. MH477637) (Figure 2B).
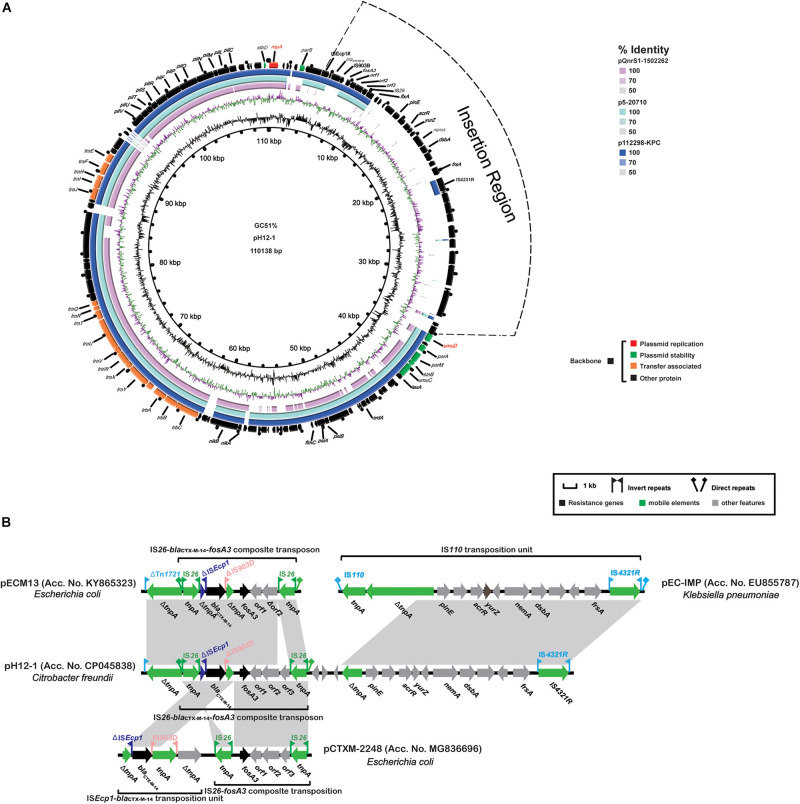
Plasmid pH12-1 was 110, 138 bp in length and contained 117 CDSs and an overall GC content of 51%. The plasmid was untypable although its replication gene repA(867 bp) was similar to orthologs of 261 bp on the IncFII plasmids pAMA1167-NDM-5 (Acc. No. CP024805) and pRSB107 (Acc. No. AJ851089) with a 69.7% coverage and 73.6% identity. Sequence comparisons demonstrated that the backbone region of plasmid p12-1 was highly similar to corresponding regions on plasmids pQnrS1-1502262 (Acc. No. CP031572) and p5-20710 (Acc. No. CP030079) and p112298-KPC (Acc. No. KP987215) except that pH12-1 possessed a 32.7 kb insertion between the parB (plasmid partition) and umuD (DNA polymerase V subunit) genes (Figure 3A). The conserved backbone regions in plasmid pH12-1 (91.4 kb) contained repA, parABM, umuCD, stbD, staB, and 16 tra/trh transfer gene modules. The 32.7 kb insertion region was primarily composed of hypothetical proteins except for the structure ΔtnpA-pinE-acrR-yurZ-dsbA-frsA-IS4321R (11.3 kb) and the resistance region containing blaCTX–M–14 and fosA3 (7.1 kb). The former was highly similar to that in pEC-IMP (Acc. No. EU855787) and the latter was highly similar to fosA3-blaCTX–M–14-carrying plasmids pECM13 (Acc. No. KY865323) and pCTXM-2248 (Acc. No. MG836696) except for partial gene deletions (Figure 3B).
FIGURE 3.
Characteristics of the complete nucleotide sequences of fosA3-carrying pH12-1 identified in this study. (A) A comparison with two fosA3-carrying untypeable plasmids p112298-KPC (Acc. No. KP987215), pQnrS1-1502262 (Acc. No. CP031572), and p5-20710 (Acc. No. CP030079). (B) Comparison of the fosA3-containing MRR in pH12-1 with the corresponding regions of pECM13 (Acc. No. KY865323), pEC-IMP (Acc. No. EU855787), and pCTXM-2248 (Acc. No. MG836696). See Figure 2 for annotation features.
Discussion
Fosfomycin plays a critical role against MDR and XDR Gram-negative pathogens and in particular, it commonly plays a synergistic role paired with β-lactams and aminoglycosides in treating urinary tract infections (Sastry and Doi, 2016). However, the occurrence and dissemination of plasmid-borne fosfomycin resistance genes, especially fosA3, has cast a shadow over the use of fosfomycin in clinics. In this study, we identified fosA3-carrying C. freundi isolates with a detection rate of 4.1% among 270 samples from flowers and the retail environments in Guangzhou, China. The prevalence of fosA3 among C. freundii strains from flowers was relatively low when compared with E. coli isolated from food animals (8.8%, during 2009–2011; 10.5%, during 2015–2016) in China (Yang et al., 2014; Wang et al., 2017). However, our fosA3 detection rate was consistent with the recent report that fosA3 sporadically occurs in C. freundii and other Enterobacteriaceae including Salmonella spp., Proteus mirabilis, and Enterobacter fergusonii from humans, food animals, pets, retail meat as well as wild birds (Lin and Chen, 2015; Villa et al., 2015; Wong et al., 2016; Yao et al., 2016; Fang et al., 2019). Perhaps of great concern is the emergence of the fosA3 gene in different Enterobacteriaceae species from these diverse origins.
Plants including vegetables and flowers can be contaminated with ARGs via wastewater irrigation or manure application. Interestingly, an MDR C. freundii strain (WCHCF65) in sewage from a Chinese hospital carried multiple clinically significant ARGs including blaCTX–M–12, blaCTX–M–14, blaSHV–12, blaNDM–1 and blaKPC–2 as well as fosA3 (Wu et al., 2016). Additionally, mcr-1 was present with fosA3 and blaCTX–M–14 in Raoultella ornithinolytica and E. coli isolates from retail vegetables in Guangzhou, China (Luo et al., 2017). MDR E. coli strains co-harboring fosA3 and blaCTX–M–14 also occurred in fresh vegetables in Netherlands (Freitag et al., 2018). These observations indicate that plants are potential ARG reservoirs including fosA3.
In the present study, we identified the presence of fosA3 on flowers and the retail environments. These fosA3-positive C. freundi isolates exhibited resistance to most of the tested antibiotics including cefotaxime, ciprofloxacin, and amikacin in addition to fosfomycin. Consistently we found that fosA3 coexisted with the ESBL blaCTX–M–14, the pAmpCs blaCMY–65 and blaCMY–122, the PMQR genes aac(6’)-Ib-cr, qnrS1, qnrB13/qnrB6/qnrB38 as well as rmtB. Additionally, WGS analysis demonstrated that blaCMY–65/blaCMY–122 and qnrB13/qnrB38 were located in the chromosome. This was consistent with the origin of plasmid-mediated qnrB and blaCMY–2 -like genes from the chromosome of Citrobacter spp. (Verdet et al., 2009; Jacoby et al., 2011; Liao et al., 2015). Importantly, flowers contaminated with MDR bacteria will most likely come into direct contact with humans complicating the treatment and management of disease.
In this study, the fosA3-carrying C. freundii isolates were genetically related as judged by their PFGE profiles. In particular, we found an epidemic PFGE type that was composed of isolates from diverse origins across flower markets and shops. This indicated possible clonal dissemination of MDR fosA3-positive C. freundii isolates from flower markets and shops in a local region.
The spread of bacterial plasmids is an increasing global problem contributing to widespread ARG dissemination (San Millan, 2018). Several plasmid types are associated with the spread of fosA3 and in particular, the epidemic IncF33:A-:B- and ST3-IncHI2 plasmids in Enterobacteriaceae from pets and food animals (Hou et al., 2012; Yang et al., 2014; Fang et al., 2019). However, we found that all our 11 fosA3-carrying plasmids including p12-1 and pS39-1 in C. freundi could not be assigned to any known Enterobacteriaceae incompatibility group. The single replicon gene repA possessed in plasmid p12-1 was highly similar to that in plasmid p112298-KPC where the repA was assigned to the IncFII RepA superfamily (Feng et al., 2015). Plasmid p12-1 showed a similar backbone region to the untypable fosA3-bearing plasmids in E. hormaechei and C. freundii isolates from humans in China and the United States (Wang et al., 2018; Figure 3).
Interestingly, a similar scenario was also observed for the other untypable fosA3-bearing plasmid pS39-1. Linear genomic comparisons revealed that a conserved backbone, including the two unclassified replicons repA and repB, were identified between pS39-1 and another two untypable fosA3-bearing plasmids pTEM-2262 and pMH17-012N_4 in C. freundii isolates of pig and human origin from China (Li M. et al., 2018; Zhang et al., 2019; Figure 2). Noticeably, pTEM-2262 also shared a conserved backbone with another four untypable non-fosA3-bearing plasmids from different species including C. freundii, Kosakonia radicincitans, and Citrobacter werkmanii with origins including the environment, vegetables and humans (Zhou et al., 2017; Zurfluh et al., 2017; Becker et al., 2018; Barry et al., 2019). These indicated that the untypable and conjugal p12-1-like and pS39-1-like plasmids could act as vectors for fosA3 transmission between different Enterobacteriaceae from different ecological niches.
In contrast to the conserved backbones, the fosA3-blaCTX–M–14-containing MDR of these untypable plasmids from the GenBank were highly heterogeneous. This was primarily due to acquisition or deletion of resistance determinants mediated by mobile genetic elements and recombination. Plasmid pS39-1 possessed a large fosA3-containing MRR composed of 15 ARGs and diverse insertion sequences and transposons. Furthermore, the large fosA3-containing MRRs in plasmid pS39-1 partially resembled analogous regions from different plasmids indicating the MRR likely originated from the recombination of genetic contents from different plasmids as previously described (Hammerum et al., 2016; Li M. et al., 2018; Jing et al., 2019; Maherault et al., 2019). Unlike pS39-1, an additional insertion region mainly composed of hypothetical proteins was also integrated into the variable region of plasmid p12-1 in addition to the fosA3-blaCTX–M–14-containing resistance region. Interestingly, heterogeneous fosA3-containing multidrug resistance regions have been also identified on the epidemic ST3-IncHI2 and F33:A-:B- plasmids with a conserved backbone in Salmonella (Yang et al., 2014). These data indicated that diverse and flexible transmission of fosA3 was associated with heterogeneous MRRs and conserved backbones of a specific group of plasmids including ST3-IncHI2 and F33:A-:B- as well as untypable replicons in Enterobacteriaceae.
In conclusion, this study revealed the presence of fosA3 that co-existed with blaCTX–M–14 in MDR C. freundii isolates from flowers and the retail environments. Clonal expansion and horizontal transmission of untypable plasmids were involved in the spread of fosA3 and blaCTX–M–14 among these C. freundi isolates. To the best of our knowledge, this is the first report of the identification of fosA3 in bacteria isolated from flower shops and markets. The fosA3- and blaCTX–M–14-bearing untypable and transferable p12-1-like and pS39-1-like plasmids could circulate among diverse Enterobacteriaceae species from diverse origins, including plants and humans. Future studies are necessary to monitor the prevalence and transmission of these plasmids in Enterobacteriaceae species especially C. freundi, to better understand the potential threat to public health.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
Y-HL, JS, and X-PL designed the study. KC, L-XF, DW, BH, Q-WG, J-QL, and Z-XZ performed the experiments and collected the data. KC, L-XF, DW, BH, Q-WG, J-QL, Z-XZ, X-LL, YY, and X-RW analyzed and interpreted the data. KC wrote the draft of the manuscript. L-XF, Y-HL, JS, X-PL, and X-LL edited and revised the manuscript. Y-HL and JS coordinated the whole project. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This research was jointly supported by the National Key Research and Development Program of China (2016YFD0501300), the Program for Innovative Research Team in the University of Ministry of Education of China (IRT_17R39), and the Foundation for Innovation and Strengthening School Project of Guangdong, China (2016KCXTD010).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.586504/full#supplementary-material
References
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry K. E., Wailan A. M., Sheppard A. E., Crook D., Vegesana K., Stoesser N., et al. (2019). Don’t overlook the little guy: an evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid 103 1–8. 10.1016/j.plasmid.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Barton B. M., Harding G. P., Zuccarelli A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226 235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- Becker M., Patz S., Becker Y., Berger B., Drungowski M., Bunk B., et al. (2018). Comparative genomics reveal a flagellar system, a type VI secretion system and plant growth-promoting gene clusters unique to the endophytic bacterium Kosakonia radicincitans. Front. Microbiol. 9:1997. 10.3389/fmicb.2018.01997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2018). Performance Standards for Antimicrobial Susceptibility Testing, 28th Edn Wayne, PA: Clinical and Laboratory Standards Institute; ClSI Suppl. M100. [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2019). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial Isolated from Animals, 5th Edn Wayne, PA: Clinical and Laboratory Standards Institute; VET01Ed5E. [Google Scholar]
- Diez-Aguilar M., Morosini M. I., Del Campo R., Garcia-Castillo M., Zamora J., Canton R. (2013). In vitro activity of fosfomycin against a collection of clinical Pseudomonas aeruginosa isolates from 16 Spanish hospitals: establishing the validity of standard broth microdilution as susceptibility testing method. Antimicrob. Agents Chemother. 57 5701–5703. 10.1128/aac.00589-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Vouloumanou E. K., Samonis G., Vardakas K. Z. (2016). Fosfomycin. Clin. Microbiol. Rev. 29 321–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Li X., Li L., Li S., Liao X., Sun J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L. X., Deng G. H., Jiang Q., Cen D. J., Yang R. S., Feng Y. Y., et al. (2019). Clonal expansion and horizontal transmission of epidemic F2:A1:B1 plasmids involved in co-spread of rmtB with qepA and blaCTX–M–27 in extensively drug-resistant Salmonella enterica serovar indiana isolates. J. Antimicrob. Chemother. 74 334–341. 10.1093/jac/dky441 [DOI] [PubMed] [Google Scholar]
- Feng J., Qiu Y., Yin Z., Chen W., Yang H., Yang W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J. Antimicrob. Chemother. 70 2987–2991. 10.1093/jac/dkv232 [DOI] [PubMed] [Google Scholar]
- Freitag C., Michael G. B., Li J., Kadlec K., Wang Y., Hassel M., et al. (2018). Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet. Microbiol. 219 63–69. 10.1016/j.vetmic.2018.03.028 [DOI] [PubMed] [Google Scholar]
- Gautom R. K. (1997). Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35 2977–2980. 10.1128/jcm.35.11.2977-2980.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum A. M., Hansen F., Nielsen H. L., Jakobsen L., Stegger M., Andersen P. S., et al. (2016). Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM–1 to Escherichia coli. Klebsiella pneumoniae and Klebsiella oxytoca. J. Antimicrob. Chemother. 71 3117–3124. [DOI] [PubMed] [Google Scholar]
- Hou J., Huang X., Deng Y., He L., Yang T., Zeng Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 56 2135–2138. 10.1128/aac.05104-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Lin Q., Zhou Q., Lv L., Wan M., Gao X., et al. (2020). Identification of fosA10, a novel plasmid-mediated fosfomycin resistance gene of Klebsiella pneumoniae origin, in Escherichia coli. Infect. Drug Resist. 13 1273–1279. 10.2147/idr.s251360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Griffin C. M., Hooper D. C. (2011). Citrobacter spp. as a source of qnrB Alleles. Antimicrob. Agents Chemother. 55 4979–4984. 10.1128/aac.05187-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Jiang X., Yin Z., Hu L., Zhang Y., Yang W., et al. (2019). Genomic diversification of IncR plasmids from China. J. Glob. Antimicrob. Resist. 19 358–364. 10.1016/j.jgar.2019.06.007 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li F., Mi Z., Zhao Y., Zhang X., Jiang Z., et al. (2018). Comparative genomics analysis of pTEM-2262, an MDR plasmid from Citrobacter freundii, harboring two unclassified replicons. Future Microbiol. 13 1657–1668. 10.2217/fmb-2018-0243 [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Wong M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Fang L., Li L., Sun J., Li X., Chen M., et al. (2015). Characterization of chromosomal qnrB and ampC alleles in Citrobacter freundii isolates from different origins. Infect. Genet. Evol. 35 214–220. 10.1016/j.meegid.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Lin D., Chen S. (2015). First detection of conjugative plasmid-borne fosfomycin resistance gene fosA3 in Salmonella isolates of food origin. Antimicrob. Agents Chemother. 59 1381–1383. 10.1128/aac.04750-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16 161–168. 10.1016/s1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Luo J., Yao X., Lv L., Doi Y., Huang X., Huang S., et al. (2017). Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 61:e1139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Feng Y., Liu L., Yao Z., Zong Z. (2020). A Cluster of colistin- and carbapenem-resistant Klebsiella pneumoniae carrying blaNDM–1 and mcr-8.2. J. Infect. Dis. 221 S237–S242. [DOI] [PubMed] [Google Scholar]
- Maherault A. C., Kemble H., Magnan M., Gachet B., Roche D., Le Nagard H., et al. (2019). Advantage of the F2:A1:B- IncF pandemic plasmid over IncC plasmids in in vitro acquisition and evolution of blaCTX–M gene-bearing plasmids in Escherichia coli. Antimicrob. Agents Chemother. 63 e1130–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42 D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuc Nguyen M. C., Woerther P. L., Bouvet M., Andremont A., Leclercq R., Canu A. (2009). Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 15 1648–1650. 10.3201/eid1510.090696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Steinort D., Pillai S., Joukhadar C. (2010). Fosfomycin: an old, new friend? Eur. J. Clin. Microbiol. Infect. Dis. 29 127–142. 10.1007/s10096-009-0833-2 [DOI] [PubMed] [Google Scholar]
- San Millan A. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26 978–985. 10.1016/j.tim.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Sastry S., Doi Y. (2016). Fosfomycin: resurgence of an old companion. J. Infect. Chemother. 22 273–280. 10.1016/j.jiac.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27 1009–1010. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liao X. P., D’souza A. W., Boolchandani M., Li S. H., Cheng K., et al. (2020). Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 11:1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdet C., Gautier V., Chachaty E., Ronco E., Hidri N., Decre D., et al. (2009). Genetic context of plasmid-carried blaCMY–2-like genes in Enterobacteriaceae. Antimicrob. Agents Chemother. 53 4002–4006. 10.1128/aac.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Guerra B., Schmoger S., Fischer J., Helmuth R., Zong Z., et al. (2015). IncA/C Plasmid Carrying bla(NDM–1), bla(CMY–16), and fosA3 in a Salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany. Antimicrob. Agents Chemother. 59 6597–6600. 10.1128/aac.00944-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yao X., Luo J., Lv L., Zeng Z., Liu J. H. (2018). Emergence of Escherichia coli co-producing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J. Antimicrob. Chemother. 73 252–254. 10.1093/jac/dkx335 [DOI] [PubMed] [Google Scholar]
- Wang X. M., Dong Z., Schwarz S., Zhu Y., Hua X., Zhang Y., et al. (2017). Plasmids of diverse inc groups disseminate the fosfomycin resistance gene fosa3 among Escherichia coli isolates from pigs, chickens, and dairy cows in Northeast China. Antimicrob. Agents Chemother. 61 e859–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2011). Critically Important Antimicrobials for Human Medicine, 3rd Revision 2011. WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Geneva: WHO. [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H., Chan E. W., Xie L., Li R., Chen S. (2016). IncHI2 plasmids are the key vectors responsible for oqxab transmission among Salmonella Species. Antimicrob. Agents Chemother. 60 6911–6915. 10.1128/aac.01555-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Espedido B., Feng Y., Zong Z. (2016). Citrobacter freundii carrying blaKPC–2 and blaNDM–1: characterization by whole genome sequencing. Sci. Rep. 6:30670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. Y., Lu P. L., Tseng S. P. (2017). Update on fosfomycin-modified genes in Enterobacteriaceae. J. Microbiol. Immunol. Infect. 52 9–21. 10.1016/j.jmii.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Yang X., Liu W., Liu Y., Wang J., Lv L., Chen X., et al. (2014). F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX–M–55/–14/–65 in Escherichia coli from chickens in China. Front. Microbiol. 5:688. 10.3389/fmicb.2014.00688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Wu D., Lei L., Shen Z., Wang Y., Liao K. (2016). The detection of fosfomycin resistance genes in Enterobacteriaceae from pets and their owners. Vet. Microbiol. 193 67–71. 10.1016/j.vetmic.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Zeng J., Pan Y., Yang J., Hou M., Zeng Z., Xiong W. (2019). Metagenomic insights into the distribution of antibiotic resistome between the gut-associated environments and the pristine environments. Environ. Int. 126 346–354. 10.1016/j.envint.2019.02.052 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lei C.-W., Wang H.-N. (2019). Identification of a novel conjugative plasmid carrying the multiresistance gene cfr in Proteus vulgaris isolated from swine origin in China. Plasmid 105:102440. 10.1016/j.plasmid.2019.102440 [DOI] [PubMed] [Google Scholar]
- Zhou G., Peng H., Wang Y. S., Huang X. M., Xie X. B., Shi Q. S. (2017). Complete genome sequence of Citrobacter werkmanii strain BF-6 isolated from industrial putrefaction. BMC Genomics 18:765. 10.1186/s12864-017-4157-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh K., Stephan R., Klumpp J., Nuesch-Inderbinen M., Hummerjohann J., Bagutti C., et al. (2017). Complete genome sequence of Citrobacter freundii 705SK3, an OXA-48-encoding wastewater isolate. Genome Announc. 5:e842-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.