Abstract
The self-renewal capacity of multipotent haematopoietic stem cells (HSCs) supports blood system homeostasis throughout life and underlies the curative capacity of clinical haematopoietic stem cell transplantation therapies. However, despite extensive characterization of the HSC state in the adult bone marrow and embryonic fetal liver, the mechanism of HSC self-renewal has remained elusive. This Review presents our current understanding of HSC self-renewal in vivo and ex vivo, and discusses important advances in ex vivo HSC expansion that are providing new biological insights and offering new therapeutic opportunities.
Introduction
The haematopoietic system is essential for human health and is composed of numerous specialized cell types, including erythrocytes, megakaryocytes and platelets, adaptive immune cells (B lymphocytes and T lymphocytes) and innate immune cells (such as neutrophils and monocytes). Although some haematopoietic cell types are long-lived (such as memory T cells), others are short-lived (including neutrophils and platelets). These various blood lineages must be continuously regenerated to sustain homeostasis of the blood system, a process known as haematopoiesis1 (Box 1). Adult humans are estimated to require ~1011–1012 new blood cells each day2, although haematological stresses such as bleeding or infections can dramatically alter these requirements. Throughout life, haematopoiesis is maintained by haematopoietic stem cells (HSCs)3–5.
Box 1 |. Models of haematopoietic lineage commitment.
Haematopoietic stem cells (HSCs) generate all mature blood cell types via differentiation through a spectrum of haematopoietic progenitor cells (HPCs), during which self-renewal capacity and multipotency are progressively lost5,154. Based on haematopoietic system reconstitution kinetics in vivo and lineage potential in vitro, a hierarchy has been proposed with HSCs at the apex and the various progenitors downstream, with progenitors become progressively more lineage-committed before forming mature blood cell types. Self-renewal has been thought to be lost prior to multipotency, during differentiation into multipotent progenitors (MPPs). However, clonal transplantation assays have demonstrated that lineage commitment can occur within the HSC compartment, prior to loss of self-renewal13,34. From MPPs, multipotency is lost on differentiation into lymphoid-primed multipotent progenitors (LMPPs) or common myeloid progenitors (CMPs). Further lineage specification then occurs: LMPPs differentiate into common lymphoid progenitors (CLPs) or granulocyte–macrophage progenitors (GMPs), which further differentiate into B cells or T cells and neutrophils or monocytes, respectively. CMPs can also differentiate into either GMPs or megakaryocyte–erythrocyte progenitors (MEPs), which ultimately differentiate into erythrocytes or megakaryocyte progenitors (MkPs) prior to generating platelets. Although such simple graphical models of haematopoiesis imply that HSC differentiation occurs through discrete stepwise transitions, the evidence (reviewed elsewhere37,154,155) shows that haematopoiesis occurs as a continuous differentiation process. There remain considerable debate over the functional heterogeneity and relationships of the HSC and HPC populations described above.
The functional definition of an HSC is a cell with the potential for both self-renewal (that is, the generation of daughter HSCs by cell division) and multipotent differentiation (meaning the generation of any mature adult haematopoietic cell type)4,5. These two functional properties enable HSCs to persist throughout an individual’s lifespan. They also underpin the capacity of HSCs to reconstitute the entire haematopoietic system following transplantation into an irradiated (i.e. haematopoiesis-ablated) recipient, after which they continue to sustain haematopoiesis in the long term. This capacity provides the scientific basis for HSC transplantation (HSCT), which is widely used to reconstitute a healthy blood system within a patient6,7. Although pioneered over 60 years ago, HSCT still represents the only curative therapy for a number of haematological malignancies (such as leukaemia and lymphoma) as well as non-malignant blood disorders (such as immunodeficiency diseases, autoimmune conditions, hereditary blood disorders and anaemias)6,7. The clinical relevance of HSCs in the treatment of these human diseases and the need for large quantities of donor HSCs have been important driving forces behind the study of HSC self-renewal.
Despite substantial advances in our understanding of ex vivo HSC self-renewal, long-term maintenance and ex vivo expansion of HSCs remain challenging. This Review aims to provide an up-to-date synthesis of the field by discussing the experimental techniques developed to assay HSC self-renewal, the biological mechanisms known to regulate HSC self-renewal in vivo and ex vivo, and the therapeutic implications of this knowledge. We focus on self-renewal of mouse HSCs, as the predominant model system in HSC biology, but include discussion of human HSC self-renewal where relevant.
Identification of self-renewing HSCs
Prospective identification and isolation of functional HSCs is difficult, but progressive improvements over the past few decades now enable mouse and human HSCs to be purified at high (~50%) frequencies. During embryonic development, HSCs have been isolated from a number of anatomical regions (Box 2). Functional HSCs can also be isolated from a number of adult tissues8; however, most studies to date have focused on the purification and characterization of HSCs within the adult mouse bone marrow microenvironment. In human, bone marrow HSCs are a focus of research, although umbilical cord blood-derived HSCs are also commonly studied.
Box 2 |. HSC niches during development and adulthood.
Several waves of haematopoiesis occur during embryonic development156. However, only the final definitive wave of haematopoiesis generates functional haematopoietic stem cells (HSCs). Two earlier waves, known as primitive haematopoiesis and transient-definitive haematopoiesis, respectively, generate mature blood cells and certain embryonic haematopoietic progenitor cells but not adult HSCs. In mice, definitive HSCs have been shown to arise at embryonic day (E)10.5 in the aorta–gonad–mesonephros (AGM) region of the dorsal aorta156–158 and placenta66,159. The yolk sac is thought to also produce definitive HSCs156,160. These embryonic HSCs then migrate to and colonize the fetal liver, which seems to be the major site of HSC self-renewal in the developing embryo. Around the time of birth, HSCs move from the fetal liver and seed the bone marrow, which becomes the major site of haematopoiesis by around 3–4 weeks after birth156. The human umbilical cord also contains HSCs, which is routinely collected for research, ex vivo expansion, and HSCT6. HSCs have been found in a number of adult tissues8,69,70, but have been predominantly studied in the bone marrow, spleen, and liver. Two excellent reviews provide a comprehensive overview of developmental haematopoiesis in mice and humans152,156.
HSCs are typically isolated using multicolour fluorescence-activated cell sorting (FACS), which since the mid-1980s has been used to enrich bone marrow aspirates for HSCs on the basis of cell surface expression of specific proteins, detected using antibody labelling and/or fluorescent reporter molecules9,10. Several mouse HSC markers have been identified, but most FACS purification strategies use the CD150+CD48–CD34–/loCD117+Sca1+Lineage marker– (Lin–) bone marrow population to isolate adult mouse HSCs with long-term self-renewal capacity11–13. Additional positive expression markers in the mouse include Epcr14 (CD201), Fgd515, Hoxb516, Evi117, α-catulin18, and Krt719. By contrast, human HSCs are usually defined as CD49f+CD90+CD45RA–CD34+CD38–Lin– cells20. However, no immunophenotype can identify functional HSCs with 100% purity in either mice or humans20,21. Single-cell analyses of mouse and human HSCs (in vivo and ex vivo) have highlighted remarkable heterogeneity within these phenotypic populations13,22,23. Furthermore, the cell surface markers used for isolation of HSCs are often no longer accurate following ex vivo culture of HSCs. For example, CD49f and CD38 are not reliable markers for human HSCs that have been subjected to ex vivo culture; EPCR and ITGAM3 seem to be more reliable markers than CD49f and CD38 in this setting24,25.
At the cellular level, HSC self-renewal occurs during cell division. Conceptually, HSCs can undergo three types of cell division events: symmetric self-renewal generates two HSCs, symmetric differentiation generates two haematopoietic progenitor cells (HPCs), whereas asymmetric self-renewal generates one HSC and one HPC. Owing to the lack of accurate prospective markers for functional HSCs, self-renewal cell divisions have been difficult to track directly in real time. Instead, self-renewal of multipotent HSCs is most often determined retrospectively and indirectly from functional assays, as discussed in the next section.
Colony forming unit (CFU) assays.
Haematopoietic colony forming potential within liquid or semi-solid media have been widely used to study HSCs and HPCs in vitro3. Although quicker to perform than the in vivo assays described below, in vitro CFU assays usually only determine cell proliferation and myeloid, erythroid and megakaryocytic differentiation capacity, rather than bona fide HSC activity. However, these assays can be useful to identify self-renewing cells when performed in a serial re-plating approach.
In vivo spleen colony forming unit assays have also been developed, which measure the ability of donor cells to generate macroscopic haematopoietic colonies within the spleen of an irradiated mouse. This early CFU-based methods pioneered many important concepts in HSC biology26,27, although they probably did not accurately detect or measure multipotent HSCs.
HSC transplantation assay.
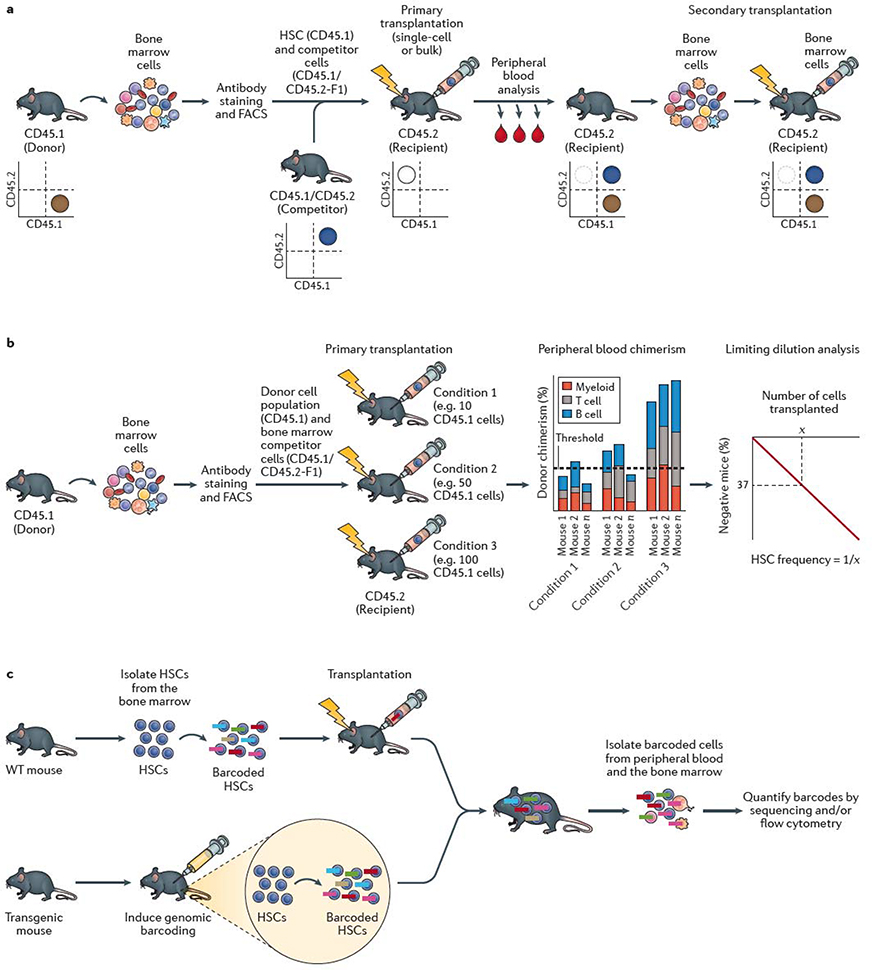
The now-standard mouse transplantation assay involves injection of the donor cells of interest into irradiated recipient mice, which lack endogenous haematopoiesis (Figure 1a). Only functionally multipotent HSCs are able to reconstitute the entire haematopoietic system within irradiated recipients and support long-term recipient survival3–5. Transplantation assays are often combined with limiting dilution analyses (Figure 1b), which enable the frequency of functional HSCs within the original cell population to be estimated28.
Figure 1 |. HSC self-renewal assays.
a | The standard mouse haematopoietic stem cell (HSC) transplantation assay uses the congenic CD45.1/CD45.2 system. Bone marrow cells are collected from a donor mouse bearing the CD45.1 allele from which HSCs are isolated using fluorescence-actiivated cell sorting (FACS). HSCs are mixed with competitor (also termed helper) bone marrow cells from CD45.1 × CD45.2 F1 mice and transplanted into a primary recipient mouse bearing the CD45.2 allele. Donor cell reconstitution kinetics can be determined within the peripheral blood and bone marrow using monoclonal antibodies and flow cytometric analysis. Flow cytometric quantification of donor chimerism within the peripheral blood is usually performed regularly over 16–24 weeks, and bone marrow analyzed at the study endpoint. As annotated in the representative flow cytometry plots, donor hematopoietic cells are CD45.1+, competitor hematopoietic cells are CD45.1+CD45.2+, while recipient hematopoietic cells are CD45.2+ (although irradation leads to loss of endogenous CD45.2+ haematopoietic cells in the transplant recipient). Bone marrow cells from the primary recipient mouse are transplanted into a secondary recipient mouse bearing the CD45.2 allele to confirm HSC self-renewal activity. Depending on the experimental design, CD45.1 recipient mice and CD45.2 donor mice may also be used. b | Limiting dilution analysis (LDA) can be used to estimate the frequency of HSCs within a cell population by transplantion of varying doses of cells into multiple mice. Presence or absence of an HSC(s) within the donor population is determined by assessing long-term peripheral blood reconstitution and LDA performed based on the number of positive versus negative mice as a function of the donor cell dose. Various reconstitution thresholds have been used in the field, but it is typically set at 1% donor peripheral blood chimerism. Based on Poisson statistics, the number of cells (X) that result in 37% negative mice is equal to the 1/HSC frequency. c | HSC self-renewal can be tracked using barcoding (or other genetic labelling) technologies. Barcoding can be performed ex vivo, by isolating HSCs from the bone marrow and labeling them in vitro with genetic barcodes (which can be applied using various methods, including transduction with a lentiviral library) and transplanted into recipient mice. Barcoding can also be performed directly in vivo, usually via inducing activity of a recombinase-based or transposon-based ‘shuffling’ of genetic sequences to generate unique genomic sequences in each HSC. The output of individual HSCs can be determined by quantifying the barcode abundance within peripheral blood and/or bone marrow cell populations. In vivo lineage tracing technologies can be used to track HSC self-renewal in the native bone marrow without the requirement for transplantation.
Genetic markers are commonly used to track the donor cells (Table 1) within recipients by enabling discrimination of donor-derived cells from any residual recipient cells as well as from competitor (also termed helper) bone marrow cells. Competitor bone marrow cells are usually transplanted along with purified HSCs to secure survival of the recipient by supplying transiently reconstituting HPCs, due to the relatively slow reconstitution kinetics of HSCs (Figure 1a). The first genetic method to confirm multilineage HSC reconstitution kinetics was retrovirus-mediated gene transfer29, which relied on unique integration sites identified by Southern blotting to detect reconstitution patterns and kinetics. However, the utility of retrovirus-mediated gene transfer was limited by low resolution of clone detection and non-random vector integration. Today, the most prevalent genetic HSC detection method uses congenic C57BL/6 mouse strains that carry different alleles of Ptprc, the gene encoding the pan-leukocyte surface marker CD459,30, known as CD45.1 and CD45.2. Expression of these two alleles can be distinguished by binding of monoclonal antibodies to the respective protein isoforms. In combination with flow cytometry, this approach affords simple yet robust quantification of donor chimerism within peripheral blood and/or bone marrow leukocyte populations. One minor limitation of this system is that CD45.2-bearing HSCs show slightly stronger reconstitution potential than do CD45.1-bearing HSCs31. This disparity seems to be due to other genetic differences between the two congenic mouse strains, which differ by a large 40×106 bp genomic region encoding ~300 genes31. However, another congenic CD45.1 C57BL/6 mouse strain termed CD45.1(STEM) has now been developed32, in which Ptprc differs by only a single point mutation within the native gene, overcoming this issue.
Table 1 |.
Genetic tools to track HSC activity
| Tool | Description | Evidence | Refs. |
|---|---|---|---|
| Retrovirus-mediated gene transfer | Donor haematopoietic cells marked by retrovirus-mediated gene transfer are tracked following transplantation into recipient mice | The first definitive evidence of HSC multipotency | 29 |
| CD45 alleles | HSCs from a donor mouse are transplanted into recipient mice each carrying a different allele of the pan-leukocyte marker CD45 (either CD45.1 or CD45.2). These isoforms are detected using monoclonal antibodies. Clonal analysis is possible after single cell transplantation | The most widely used system for tracking HSC activity following transplantation, in bulk and clonal assays | 9,30 |
| Fluorescent protein transgenesa | Transgenic donor mice are engineered to constitutively express fluorescent protein(s) within haematopoietic cells, thereby enabling assessment of platelet, erythrocyte and leukocyte chimerism. Clonal analysis is possible after single cell transplantation | Assessment of platelet and erythrocyte lineages led to the identification of lineage biases and lineage-restricted cell types | 13,34,35 |
| Retroviral or lentiviral barcoding | HSCs are transduced ex vivo with a barcode derived from a retroviral or lentiviral library. Barcodes are detected by sequencing at multiple time points to track the HSCs over time | These methods enabled reconstitution dynamics of numerous HSCs to be tracked within a single recipient | 39–41 |
| In vivo lineage tracing | HSCs are barcoded and genetically tagged in vivo, e.g. using inducible CreER recombination. The Polylox system enables single HSCs to be individually tagged by one of several hundred thousand barcodes | These experiments found that adult HSCs play a minor role in day-to-day haematopoiesis at steady-state | 43,51 |
| Confetti and rainbow labelling | In vivo Cre recombination ‘shuffles’ a cassette of fluorescent protein genes, such that different clones express different fluorescent proteins. Although the number of distinguishable fluorescent proteins limits the number of clones that can be traced, the expression of fluorescent proteins enables prospective identification of clones | These methods have facilitated molecular studies of in vivo intraclonal HSC heterogeneity, including at the genetic and epigenetic levels | 48,49 |
| Transposon-based lineage tracing | HSB transposase labels HSCs clonally in situ | These methods identified HPCs rather than HSCs as major drivers of day-to-day hematopoiesis at steady-state | 42 |
| Dye-dilution assays | Transient expression of a histone H2B tagged with GFP (H2B-GFP) labels dormant HSCs, enabling assessment of their cell division kinetics throughout life | Functional HSCs retained the H2B-GFP label in vivo for >22 months, confirming their dormant state | 50 |
| Whole genome sequencing | Rare somatic mutations within an individual’s haematopoietic system can predict HSC activity longitudinally in native haematopoiesis | Modelling of these datasets allowed population dynamics of the human HSC compartment to be estimated | 63 |
Such as Kusabira orange or green fluorescent protein (GFP). HPC, haematopoietic progenitor cell; HSB, hyperactive Sleeping Beauty; HSC, haematopoietic stem cell.
Although the CD45 congenic system can quantify chimerism within myeloid, T cell and B cell lineages, donor contribution to essential CD45– blood components such as platelets and erythrocytes cannot be tracked. Platelet and erythrocyte lineage chimerism can currently only be tracked using donor cells that express fluorescent proteins such as GFP or Kusabira orange transgenes13,33–35. However, fluorescent and CD45 markers can only distinguish donor cells from recipient cells, rather than enabling multiple individual donor cells to be distinguished from each other.
Multipotency is most commonly defined experimentally as myeloid (that is, neutrophil and/or monocyte), T cell, and B cell differentiation potential, whereas self-renewal capacity is demonstrated by the reconstitution kinetics. Stable multilineage reconstitution, which is usually assessed at >4 months post transplantation, is often the criterion used to define functional HSCs. By contrast, transferred HPCs can only partially and/or transiently reconstitute haematopoiesis. Serial transplantation, in which donor cells from the primary recipient are collected and transplanted into a secondary irradiated recipient mouse, is the current gold standard for defining HSCs with long-term retention of potent self-renewing activity (LT-HSCs). Only through self-renewal can the original donor HSCs reconstitute the primary recipient, and its progeny equivalently reconstitute the secondary recipient. So-called short-term HSCs (ST-HSCs) can only reconstitute primary recipients for ~4 months (and fail to reconstitute secondary recipients) and therefore lack potent self-renewal potential. A third class of HSCs has also been observed, intermediate-term HSCs (IT-HSCs), which are able to reconstitute multilineage haematopoiesis in primary recipients for up to 6–8 months36, but only transiently reconstitute secondary recipients13.
Clonal analysis.
Definitive evidence of multipotent HSC self-renewal requires the use of clonal transplantation analysis. Most transplantation assays transfer populations of cells, but only single-cell transplantation can definitively demonstrate HSC activity at the clonal level. In 1996, our group was able to demonstrate through such single-cell transplantation-based methods in mice that multipotent HSCs within a population of CD34–/loCD117+Sca1+ Lin– bone marrow cells did indeed have self-renewal capacity30.
More recently, single-cell transplantation analyses based on tracking of fluorescent reporter transgenes through cell differentiation events in mice have additionally identified self-renewing but lineage-restricted stem cells (notably platelet-restricted stem cells)13,33–35,37. The most stringent assay for self-renewal division events is the paired-daughter-cell transplantation assay, in which a single HSC is allowed to divide once before each daughter cell is transplanted into a different recipient, and the reconstitution kinetics are then analysed to evaluate the function of the transplanted cell13,38. To date, this assay has been performed only after ex vivo cell divisions because single-cell division events are difficult to track in vivo.
Lineage tracing.
Several groups have developed retroviral and lentiviral barcoding technologies that enable the activity of multiple individual HSCs to be tracked within a single recipient following transplantation39–41 (Figure 1c; Table 1). As transplantation assays cannot determine the self-renewal capacity of HSCs in the native bone marrow, in vivo lineage-tracing methods have also been developed that enable the study of native haematopoiesis42,43. Consistent with the observation that HSCs are mostly quiescent during homeostasis44–47, these lineage-tracing methods have suggested that HSCs have only a small role in day-to-day haematopoiesis during homeostasis42,43. Confetti and rainbow technologies have also been used for HSC lineage tracing48,49. These techniques enable individual clones to be prospectively distinguished based on the recombination and expression of genes encoding different fluorescent proteins. However, the limited number of available fluorescent markers means that the number of clones that can be simultaneously tracked directly by these methods is currently low (~4) compared to those that can be tracked by DNA barcoding technologies.
Dye-dilution assays, which are based on transient expression of GFP-tagged histone H2B, have been used as an elegant method of labelling non-dividing cells in vivo47,50. The results of these studies suggest that some HSCs do not enter the cell cycle during the host’s entire adulthood50. Interestingly, these studies also suggested that HSCs display limited self-renewal potential during homeostasis, and that their multipotency is lost after approximately four cell division events50. Technological advances in lineage tracing51,52 and live cell imaging53,54 methodologies might soon provide improved approaches capable of tracking HSCs through cell fate decisions both within multiple cells and across multiple generations in vivo and ex vivo.
Human HSC transplantation assays.
Although the long-term clinical success of HSCT provides definitive evidence of the self-renewal potential of HSCs, studying the function of human HSCs experimentally is challenging, largely because of the lack of assays that truly measure HSC function. The most widely-used human HSC assay to date is the xenograft transplantation assay, in which human cells are transplanted into sublethally irradiated immunodeficient NOD/PrkdcScid mice55 or subsequent adaptations of this mouse strain56. As in the mouse HSC transplantation assay, multilineage potential and long-term self-renewal capacity can be demonstrated by serial transplantation in these models. Xenograft transplantation has also been used in combination with clonal transplantation to confirm multipotency and self-renewal of human HSCs20.
Unfortunately, as the lifetime of a mouse is much shorter than that of a human, and human erythrocytes and platelets are poorly generated in this xenograft setting, these assays lack the elegance of mouse–mouse transplantation assays. However, new transgenic mouse models have helped to overcome some of these limitations. For example, human erythrocyte and platelet production is improved in immunodeficient mice carrying mutations in the important haematopoietic receptor, Kit (CD117)57–59. Additionally, human HSC reconstitution is improved in mice that express human haematopoietic cytokines60.
Lineage tracing in humans.
Understanding how HSCs function in vivo in humans also remains an important but challenging goal. Analysis of unique viral integration sites, which are equivalent to unique barcodes, in bone marrow and peripheral blood samples from patients who have received HSCT gene therapies has enabled clonal kinetics of the reconstituting cells to be traced over time61,62. Whole-genome sequencing has also been applied to study native haematopoiesis (that is, in the absence of transplantation): somatic mutations within human haematopoietic cells were used to identify clonal relationships that enabled the reconstruction of in vivo HSC dynamics63. These exciting new studies are broadening the study of human HSCs in vivo.
HSC self-renewal in vivo
The methods described above have been used to study the kinetics of HSC self-renewal at various stages of life. HSCs in the fetal liver seem to be highly proliferative; transplantation analyses estimated that HSCs expand by >100-fold within a short period (around 5 days) during embryonic development64–66. By contrast, during adulthood, the generally quiescent state of HSCs limits self-renewal activity44–47 and the number of functional HSCs remains largely stable33,49, although HSC function is also thought to decline with age67.
Considerable heterogeneity in HSC self-renewal capacity has been observed at the clonal level13,33,34. Quantitative analyses of functional HSCs after single-cell transplantation have revealed that ~1,000-fold expansion can be achieved in primary recipients, although self-renewal activity is generally reduced in secondary recipients68. Additionally, serial transplantation analyses have found that LT-HSCs initially have substantial expansion and reconstitution capacity, but that this capacity is lost after 5–6 successive transplantations33. These data indicate a limited HSC self-renewal capacity in vivo (at least within the damaged bone marrow microenvironment of irradiated recipients).
The mechanisms underlying the self-renewal capacity of HSCs have been a major focus of research over the years. As several comprehensive reviews describing our knowledge of the HSC microenvironment (niche) have been published in the past few years8,69,70, here we provide an overview of the molecular regulators of adult bone marrow HSC self-renewal in vivo (Figure 2). Many of the same mechanisms have been identified in fetal HSC self-renewal, although several interesting differences exist, as noted below.
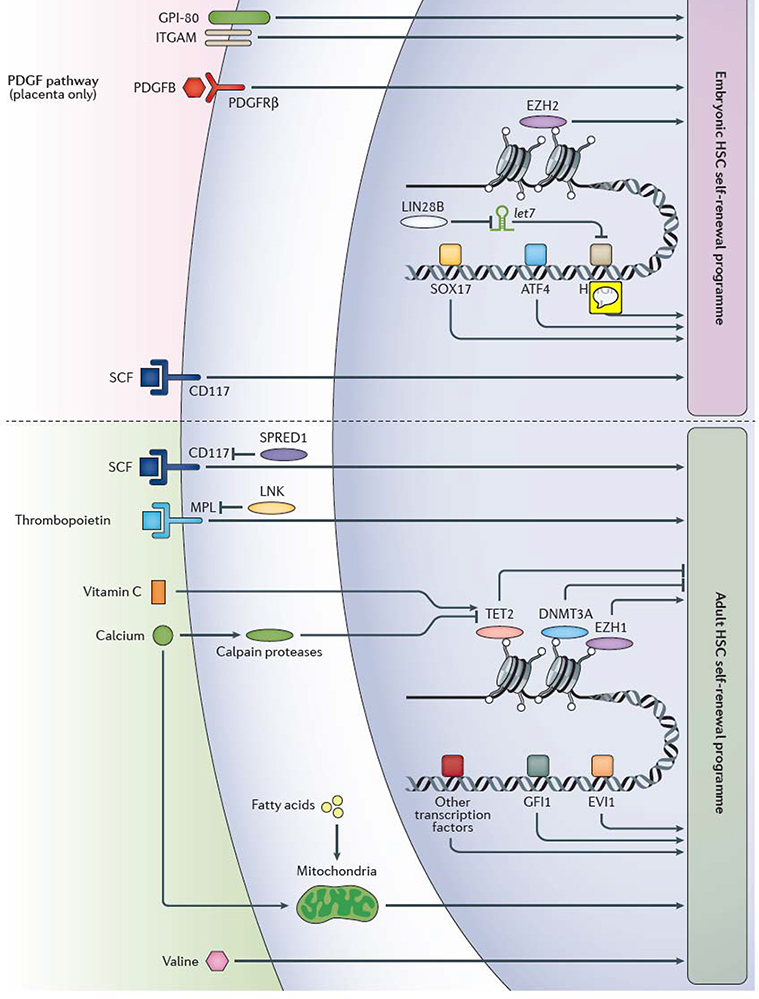
Figure 2 |. Extrinsic and intrinsic factors that regulate self-renewal in embryonic and adult haematopoiesis.
Various extrinsic and intrinsic regulators of embryonic and adult haematopoietic stem cell (HSC) self-renewal have been identified in mice and humans, however, our knowledge of the mechanism of HSC self-renewal remain incomplete. Factors that regulate HSC self-renewal in adult mice have been primarily studied in bone marrow HSCs, whereas factors that regulate HSC self-renewal in the mouse embryo have been mainly studied in fetal liver HSCs, except the platelet-derived growth factor (PDGF) pathway, which has been shown to regulate HSC differentiation only in the placenta. Our knowledge of the exact relationships between the various self-renewal factors is often lacking, although protein lin28 homolog B (Lin28b), the microRNA let-7 and high mobility group protein HMGIC (Hmga2) are known to participate in the same embryonic HSC self-renewal pathway. Multiple transcription factors including Sox17 and Atf4, as well as epigenetic regulators such as the polycomb repressive complex 2 component histone-lysine N-methyltransferase Ezh2, influence gene expression to promote HSC self-renewal in the mouse embryo. HSC self-renewal in adulthood is also regulated within the nucleus, including by the epigenetic regulatory molecules methylcytosine dioxygenase Tet2, DNA (cytosine-5)-methyltransferase 3A (Dnmt3a), the polycomb repressive complex 1 component Ezh1, and transcription factors including Gfi1 and Evi1, amongst others. CD117, Scf receptor Kit; Evi1, Ectropic virus integration site protein 1 homolog, histone-lysine N-methyltransferase MECOM; GPI-80, vanin 2; ITGAM, integrin αM; Lnk, SH2B adapter protein 3; Mpl, myeloproliferative leukemia protein, also known as thrombopoietin receptor; Pdgfb, platelet-derived growth factor subunit B; Pdgfrβ, platelet-derived growth factor receptor β; Scf, stem cell factor (also known as Kit ligand); Spred1, sprouty-related, EVH1-domain-containing protein 1; transcription factors, TFs.
Extrinsic regulation of HSC self-renewal.
Genetic perturbation of signalling pathways has revealed that a number of extrinsic factors regulate the self-renewal potential of HSCs within the mouse bone marrow microenvironment, including stem cell factor (Scf) and thrombopoietin71. In mouse, genetic deletion of Scf or its receptor Kit (CD117) results in loss of HSC self-renewal capacity72. In particular, Scf derived from bone marrow stromal cells seems to be essential for maintenance of HSCs in vivo73. Similarly, genetic deletion of either thrombopoietin or its receptor myeloproliferative leukemia protein (Mpl) in mouse results in reduced HSC self-renewal potential74. Interestingly, the mouse liver is now known to be the main source of the thrombopoietin required for HSC self-renewal in vivo75. Further evidence for the roles of Scf and thrombopoietin signalling in HSC self-renewal comes from mouse genetic perturbation studies of two negative regulators of these pathways, lymphocyte-specific adaptor protein (Lnk; also known as SH2B adapter protein 3)68 and sprouty-related EVH1 domain-containing protein 1 (Spred1)76. Lnk is a negative regulator of Mpl signalling in HSCs and its deletion causes substantial increases in self-renewal68,77. Single Lnk-deficient mouse HSCs can expand by ~3,000-fold following transplantation, which provides strong evidence for the in vivo self-renewal potential of HSCs68. Spred1 is a negative regulator of Kit signaling, and its genetic deletion similarly results in increased HSC self-renewal76.
Several other signalling pathways have been implicated in the extrinsic regulation of HSC self-renewal in vivo, particularly inflammatory signalling78–81. Diet and metabolic activity also have an important role in HSC self-renewal in vivo82. For example, fasting has also been implicated in the regulation of HSC self-renewal, with cycles of acute fasting (cycles of 48-h fasting, once per week) enhancing HSC self-renewal in mice, at least in part through reducing insulin-like growth factor (IGF) signalling pathways83. However, the consequences of chronic dietary restriction in mice seem to be complex, and different effects on HSC activity have been reported in different studies84,85. Additionally, other dietary perturbations can influence HSC self-renewal. For example, limiting dietary intake of the branched-chain amino acid valine inhibits HSC self-renewal86,87. The hypoxic nature of the bone marrow environment is also thought to be important for HSC maintenance88.
Although many self-renewal signalling pathways seem to be conserved between fetal and adult HSCs, some notable differences exist. For example, mouse fetal liver HSCs have an increased sensitivity to Scf compared with adult HSCs89. Glycosylphosphatidylinositol-anchored protein GPI80 (also known as vascular non-inflammatory molecule 2), a myeloid cell adhesion factor, along with integrin αM (ITGAM) are also thought to regulate fetal HSC expansion, at least in humans90. Self-renewal mechanisms have not been extensively studied in embryonic tissues other than the liver, although HSC differentiation in the placenta is thought to be prevented by trophoblast-derived platelet-derived growth factor subunit B (PDGFB) signalling, at least in part through inhibition of erythroid differentiation91.
Intrinsic regulation of HSC self-renewal.
Numerous cell-intrinsic factors regulate HSC self-renewal potential in vivo. These factors can be broadly categorized as relating to signalling, transcriptional, epigenetic and metabolic programs. Perturbation of intracellular signalling pathways downstream of the extrinsic self-renewal factors mentioned above regulate HSC self-renewal, presumably by modulating how HSCs sense and respond to these factors68,76,77.
Gene-deletion studies in mice have identified a number of epigenetic regulatory molecules that limit HSC self-renewal, in particular methylcytosine dioxygenase 2 (Tet2)92, DNA (cytosine-5)-methyltransferase 3A (Dnmt3a)93,94 and members of the polycomb repressive complexes95,96. Genetic mutation of TET2 and DNMT3A are also associated with clonal dominance within human hematopoiesis, suggesting they play similar roles in human HSC self-renewal97. More recently, the chromatin reader AF9 has also been identified as an important human HSC self-renewal factor98. Metabolic activity is thought to be tightly linked to HSC self-renewal, including fatty acid oxidation pathways99, mitochondrial activity100,101, mitophagy100 and autophagy102. These pathways and functions have been thoroughly described elsewhere82,103. Moreover, metabolism influences epigenetic mechanisms; for example, vitamin C (ascorbic acid) limits mouse HSC self-renewal via regulation of Tet2 activity104,105.
Gene-deletion studies have also implicated a number of transcription factors in the regulation of HSC self-renewal, including the zinc finger protein Gfi1 (also known as growth factor independent protein 1) and ecotropic virus integration site 1 protein homolog (Evi1, also known as histone-lysine N-methyltransferase Mecom), as documented elsewhere106. Furthermore, several transcription factors have been identified specifically as fetal HSC self-renewal regulators. For example, mouse Sox17 is only required for fetal HSC self-renewal and its expression is lost in adult HSCs107,108. Loss of cyclic AMP-dependent transcription factor Atf4 inhibits mouse fetal HSC self-renewal, although this effect seems to be partially mediated by the function of Atf4 in fetal stromal cells and endothelial cells109. Epigenetic regulation via histone-lysine N-methyltransferase Ezh2 (a subunit of polycomb repressive complex 2) contributes to mouse fetal HSC expansion110. However, one of the most notable mouse fetal HSC-specific pathways involves the RNA-binding protein lin28 homolog B (Lin28b), let7 microRNA and high mobility group protein AT-hook 2 (also termed HMGI-C), which is encoded by Hmga2. Lin28b is upregulated in fetal HSCs and promotes self-renewal activity by binding to let7 microRNA and thereby preventing its inhibition of Hmga2 expression111.
HSC self-renewal ex vivo
Given the clinical importance of HSCs in HSCT-based treatment of several haematological (and non-haematological) diseases, numerous studies have attempted to develop conditions that stimulate HSC self-renewal ex vivo, largely by extrapolation from knowledge of the above-described in vivo HSC self-renewal factors. However, despite extensive investigation, HSCs have long proved difficult to both maintain and expand ex vivo112. In this section, we discuss current strategies to expand HSCs ex vivo (Figure 3).
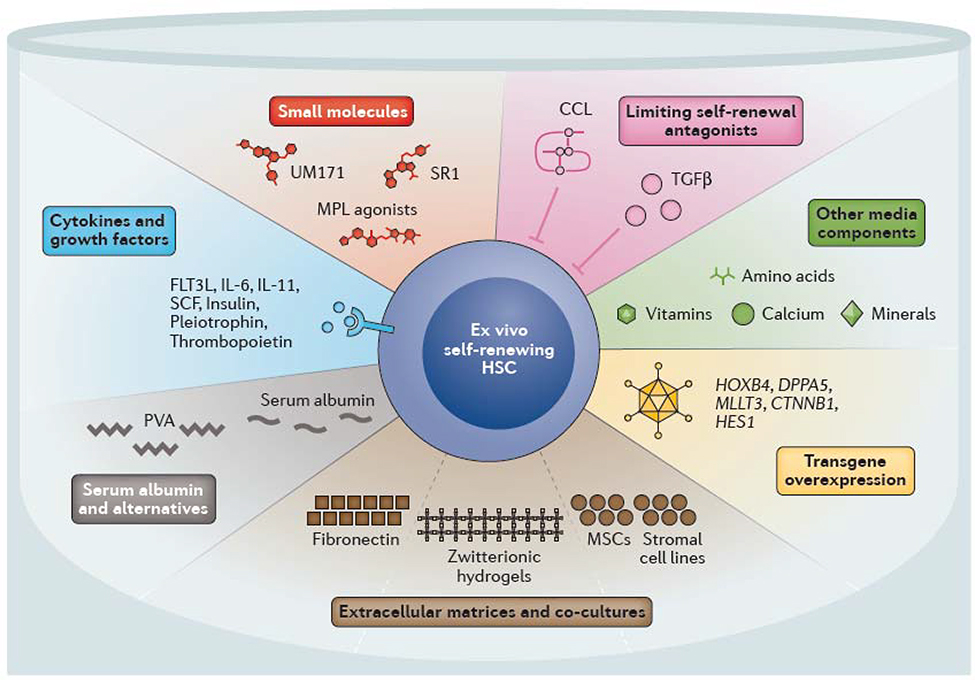
Figure 3 |. Components of ex vivo HSC culture systems.
Multiple strategies have been developed to support ex vivo haematopoietic stem cell (HSC) self-renewal. Many of the cytokines and growth factors that have been found to regulate in vivo self-renewal have been used to promote ex vivo self-renewal, including thrombopoietin and stem cell factor (SCF, also known as Kit ligand). Multiple small molecules and agonists have been identified that mimic or promote self-renewal in combination with these cytokines and growth factors. Specifically, StemRegenin 1 (SR1) and UM171 promote expansion of human cord blood HSCs. Other strategies include preventing the accumulation of endogenous self-renewal inhibitors, including transforming growth factor β (TGFβ) and C-C motif chemokine ligands (CCLs) by batch fed culture methods or complete media changes. Changes in other media components also influence ex vivo HSC stability (for example, low calcium levels promote HSC maintenance). Serum albumin has long been used in HSC culture and expansion systems, although it can can be replaced with polyvinyl alcohol (PVA). Extracellular matrices consisting of fibronectin or hydrogels and/or co-culture with mesenchymal stem cells (MSCs) or stromal cells can also help to promote HSC self-renewal. Genetic perturbations, including overexpression of transcription factors such as MTTL3 (AF9), homeobox protein Hoxb4 and developmental pluripotency-associated protein 5A (Dppa5) as well as upregulation of signaling pathways (Notch and Wnt) also promote ex vivo HSC self-renewal. CD117, Scf receptor Kit; FLT3L, FMS-like tyrosine kinase 3 ligand; IL, interleukin; MPL, myeloproliferative leukemia protein, also known as thrombopoietin receptor.
Cytokines and growth factors.
The most commonly used HSC self-renewal agonists are cytokines and growth factors, which are provided in liquid media. For mouse HSCs, Scf has frequently been used in combination with thrombopoietin113–115 or interleukin (IL)-11116. These molecules are used in standard human HSC culture systems, although usually in combination with FMS-related tyrosine kinase 3 ligand (FLT3L), IL-6 and/or other factors such as pleiotrophin117. In liquid growth media, the cytokines are usually added in combination with fetal bovine serum or human serum, or bovine serum albumin in combination with insulin112,118. Such culture conditions can generally maintain HSCs ex vivo for 1–2 weeks but provide minimal HSC expansion.
Extracellular matrices and co-cultured cells.
Several stromal cell lines have been developed to support HSCs ex vivo in co-cultures112. For example, endothelial cells, by providing Notch signalling, offer a supportive ex vivo microenvironment for HSCs119. Co-cultures of bone marrow mesenchymal stem (or stromal) cells (MSCs) have also been developed, including HSC-supportive ‘revitalized’ MSCs that overexpress the genes encoding five transcription factors: Klf7, Ostf1, Xbp1, Irf3 and Irf7120. This MSC–HSC co-culture system improves HSC maintenance ex vivo, apparently by reducing the accumulation of reactive oxygen species (ROS).
The use of 3D culture matrices formed from zwitterionic hydrogels has recently shown impressive results, suggesting that local cell density and perhaps surface texture might have a role in HSC self-renewal121. Interestingly, the beneficial effect of these hydrogels on HSC expansion was attributed to metabolic rewiring that resulted in inhibition of oxygen-related metabolism and a reduction in ROS production. Extracellular matrices formed of other materials, such as fibronectin, have also been used to improve ex vivo HSC expansion23, probably through integrin-signalling-mediated upregulation of the thrombopoietin pathway122.
Transgene overexpression.
Genetic perturbation has traditionally provided the best ex vivo HSC expansion results, for example, transgenic overexpression of Hoxb4 (encoding homeobox protein B4)123 or DPPA5 (encoding developmental pluripotency-associated 5)124. Genetic activation of Notch signalling125 and Wnt signalling126 also promote HSC self-renewal ex vivo. However, these methods have now been surpassed by use of small molecules and serum albumin-free medias, as discussed below.
Small molecules.
Small-molecule agonists of HSC self-renewal have provided some of the most impressive expansions of non-modified human HSCs to date. The two most frequently utilized small molecules are StemReginin-1 (SR1) and UM171127,128. Mechanistically, SR1 is an antagonist of the aryl hydrocarbon receptor, whereas UM171 is thought to promote homeostatic inflammatory-detoxification of ROS129. In combination with cytokines (thrombopoietin, SCF, FLT3L and IL-6), SR1 and UM171 are estimated to induce ~30-fold expansion of human HSCs ex vivo over 2 weeks127,128. Small-molecule mimics of thrombopoietin, also known as MPL agonists, also promote human HSC self-renewal ex vivo130,131. As these small molecules target human proteins and pathways, these agonists have been studied almost exclusively in human HSC cultures to date.
Minimizing self-renewal antagonists.
Preventing the accumulation of endogenously secreted inhibitors of self-renewal is critical in ex vivo HSC expansion. Several factors secreted into HSC culture media that negatively regulate self-renewal, including transforming growth factor β (TGFβ) and chemokine ligands, have important roles in inter-cell signalling23,132. Batch-fed culture systems have been developed to limit the buildup of these self-renewal antagonists132.
Serum albumin-free media.
A common confounding factor in HSC growth media has been the pervasive use of serum or serum albumin. Serum albumin has been thought to have an essential role in buffering culture media, stabilizing hydrophobic molecules, and providing various HSC-supportive bioactive molecules133. However, these biological materials introduce numerous unknown factors into the culture. Batch-testing of different serum lots led to the identification of sera that preferentially stimulate HSC self-renewal ex vivo118. However, it is largely unclear which self-renewal agonists are present in these samples. Serum albumin can be entirely replaced with the synthetic polymer polyvinyl alcohol (PVA) in HSC culture media23. In combination with optimized concentrations of Scf, thrombopoietin, insulin, and complete media changes (which reduce the accumulation of endogenous self-renewal inhibitors, as discussed above)23,134, the use of PVA-based media has afforded long-term and large-scale expansion of mouse HSCs ex vivo. Limiting dilution analyses, performed before and after ex vivo expansion of a purified population of mouse CD150+CD34–/loCD117+Sca1+ Lin– HSCs, indicate that a 236–899-fold expansion could be achieved by 4 weeks of culture under these conditions23. Split-clone transplantation assays further confirmed bona fide ex vivo mouse HSC self-renewal23. Mechanistically, PVA is thought to replace the cytokine-stabilizing activity of serum albumin135, but how PVA affects the metabolic activity of cultured HSCs is currently unknown. For example, serum albumin is a lipid carrier and also thought to act as an antioxidant in cell culture media, but whether PVA replicates these functions remains unclear. PVA in combination with SCF, thrompoietin, and insulin have also been successfully applied to the ex vivo maintenance of human HSCs23, although further work is needed to optimize human HSC culture conditions to promote equivalent levels of HSC self-renewal as seen in mouse HSC cultures.
Other media components.
Other culture media constitutents, such as amino acids86,87, vitamins105 and minerals136, are also thought to influence HSC self-renewal ex vivo. For example, HSCs are preferentially maintained in low-calcium growth media136, perhaps because calcium regulates HSCs through mitochondrial metabolism136,137. However, low-calcium conditions also stabilize Tet2, via calcium-based activation of Tet2-targeting calpain proteases136, observations that suggest low-calcium conditions might limit HSC self-renewal. Furthermore, hypoxia improves ex vivo maintenance of HSCs, both during initial isolation and subsequent culture88,138, probably through reducing oxidative stress. Therefore, many more aspects of culture media than just growth supplements need to be considered when optimizing the conditions for ex vivo HSC expansion.
Harnessing HSC self-renewal in therapy
HSCT can involve either allogeneic or autologous transplantation. Allogeneic HSCT is most widely used, and involves transplantation of donor HSCs collected from healthy individuals with an at least partial human leukocyte antigen (HLA) match with the recipient patient6,7. Autologous HSCT involves the collection and reintroduction of the patient’s own HSCs, and is used in patients receiving anticancer therapies involving high-dose radiation and/or high-dose chemotherapy, which would otherwise cause bone marrow failure. Gene therapies are also used in combination with autologous HSCT to provide safe and effective curative treatment for a number of congenital haematological diseases6,139. Current clinical gene therapy approaches rely on ex vivo retroviral or lentiviral vector-mediated genetic modification, and are currently only approved for a limited number of congenital diseases6,7,139. As discussed below, the availability and efficacy of both types of HSCT could be considerably improved by efficient ex vivo HSC expansion. Additionally, improved understanding of HSC self-renewal mechanisms would have applications in the development of novel therapies.
Allogeneic HSCT.
Allogeneic HSCT can be only performed if a suitable immune-matched healthy donor is available from whom sufficient HSC numbers can be collected. The lack of suitable donors is a major barrier to the widespread use of HSCT. If donor and patient are not sufficiently immune-matched, allogeneic HSCT can result in severe graft versus host disease (GvHD), in which an immune response is mounted by donor immune cells contaminating the transplanted cells. These immune cells recognize the host tissues as non-self and can cause considerable morbidity and even mortality140. However, it is worth noting that in certain contexts such as HSCT treatment of leukaemias, mild GvHD is desirable because targeting of any residual leukaemia cells by engrafted cells aids long-term remission140. Ex vivo expansion of donor HSCs could improve donor availability by requiring fewer HSCs to be harvested. The ex vivo culture of allogeneic donor cells before transplantation under conditions that are highly selective for HSC expansion might also help to deplete the contaminating immune cells that are responsible for GvHD.
Ex vivo HSC expansion could increase the availability of immune-matched HSCs. For example, it could help to facilitate the use of cord blood-derived HSCs for HSCT141, which represent a highly accessible source of HSCs. Allogeneic transplantation of cord blood-derived HSCs also causes less GvHD than transplantation of adult HSCs, due to the reduced allogeneic response of the contaminating immune cells that are co-transplanted with the donor HSCs141,142. Unfortunately, single units of cord blood often contain too few HSCs to be used for HSCT in adults; sufficient numbers of HSCs must be transplanted to prevent graft failure, a life-threatening complication of HSCT caused by too few donor HSCs engrafting and/or functioning following transplantation. As large HSC doses improve rates of donor engraftment and long-term reconstitution6, the ability to deliver more HSCs than were originally collected from the donor could improve therapeutic outcomes across many diseases treated by allogeneic HSCT.
The results of clinical trials in which UM171 was used to expand single units of cord blood HSCs for 7 days before transplantation highlight the safety and feasibility of ex vivo HSC expansion143. An average of 35-fold increases in total cell numbers were achieved in these 7-day cultures, and the transplanted cells supported haematopoietic reconstitution in all patients included in the study (as assessed at up to 18 months post-transplantation). Other clinical trials have used SR1 or MSC co-culture to expand HSCs ex vivo and their results also suggest that ex vivo HSC expansion could be a safe and effective method of improving haematopoietic recovery144,145, although these two trials both used two units of cord blood per patient, one fresh and one expanded ex vivo. Overall, these results are very promising for the clinical use of ex vivo HSC expansion. However, long-term follow up of these patients will be important to confirm that LT-HSCs were not depleted during ex vivo culture.
Autologous HSCT gene therapies.
Ex vivo gene editing of HSCs intended for use in autologous HSCT must currently be performed quickly (within days) to prevent substantial loss of HSCs139. Without ex vivo expansion, at best, the same number of HSCs that were collected from the patient can be returned. This situation not only limits the use of autologous HSCT gene therapies in diseases where few healthy HSCs can be collected, but also increases the risk of graft failure. Ex vivo HSC expansion could therefore greatly improve the availability of autologous HSCT gene therapies for patients in whom the collection of sufficient numbers of HSCs is currently a limiting factor. Ex vivo expansion also offers opportunities for enrichment of gene-corrected HSCs before transplantation, and might also improve safety, for example, by enabling additional quality control tests to be performed prior to transplantation. Exciting advances in HSC gene editing techniques using the CRISPR–Cas9 system also compound the need for clinical ex vivo HSC expansion protocols146,147. Lack of supportive ex vivo HSC culture conditions is likely to become a limiting factor in the success and availability of these potentially curative next-generation therapies.
Reduced need for bone marrow conditioning.
Recipient bone marrow conditioning, which is traditionally achieved through irradiation and/or chemotherapy, is normally required to destroy native HSCs and thereby enable the engraftment of donor HSCs6,7. HSCs will engraft in non-conditioned recipients, albeit only at very low frequencies148,149. However, we have demonstrated that mouse HSCs subjected to ex vivo expansion could robustly engraft in non-conditioned recipients23. The ability to transplant high numbers of HSCs might therefore reduce the need for bone marrow conditioning in specific disease contexts such as Fanconi anaemia. Patients with this disease lack an efficient DNA damage repair pathway, with the consequence that traditional bone marrow conditioning is highly toxic and thus contraindicated for these individuals150. Ex vivo HSC expansion, by increasing the number of HSCs available for transplantation, could help to reduce the requirement for bone marrow conditioning and might also enable the intensity of conditioning to be reduced, thereby limiting the considerable off-target toxicity of current conditioning methods.
Other therapies.
Just as stable expansion of pluripotent stem cells (PSCs) resulted in a number of exciting new therapeutic opportunities151, stable ex vivo expansion of HSCs is also likely to open the door to novel therapeutic paradigms. Culture conditions that stabilize the HSC state could also enable the generation of HSCs from unmodified PSCs152, which could overcome the lack of donor HSCs and offer opportunities for autologous HSCT using patient-derived PSCs without the risk of GvHD. Additionally, knowledge of HSC self-renewal mechanisms might find applications in other therapeutic paradigms, such as targeting the critical pathways responsible for self-renewal of malignant HSCs in leukaemias.
Conclusions
Ex vivo HSC expansion has been a holy grail in haematology for over 50 years, a fact that has stimulated much scientific research into HSC self-renewal. Successful generation of ex vivo culture conditions that are supportive of HSC self-renewal opens up new paradigms in both HSC biology and clinical haematology153. Progress in these fields has often been held up by a paucity of collected HSCs, but new HSC expansion methods now enable new biochemical and molecular assays to be applied to the study of HSCs and to the mechanisms of HSC self-renewal. Additionally, improved understanding of the conditions needed to achieve stable ex vivo self-renewal might aid in deciphering the complexities of the bone marrow HSC niche in vivo. Together, these new findings will help to establish novel technologies and new approaches for the treatment of haematological diseases. As haematopoiesis has long been an important paradigm in stem cell research, new knowledge of the mechanisms of HSC self-renewal and lineage commitment is also expected to contribute to the broader stem cell field and its applications within regenerative medicine.
Box 3 |. Remaining open questions in the HSC self-renewal field.
What are the endogenous signals for human haematopoietic stem cell (HSC) self-renewal in the bone marrow (and other HSC niche sites)?
Can human HSCs undergo sustained self-renewal ex vivo?
What is the molecular and cellular composition of in vivo HSC niches?
How many different HSC niches are there?
What are the relative contributions of intrinsic and extrinsic factors in the functional heterogeneity of the HSC compartment?
How is life-long HSC self-renewal implemented at the molecular level in vivo?
How is the HSC pool coordinated throughout life at the population level?
How do HSCs function during homeostasis vs. in an inflammatory environment?
Can we improve preclinical assays for ex vivo human HSC self-renewal?
Acknowledgements
A.C.W. is supported by the Leukemia & Lymphoma Society (grant 3385-19). K.J.I. is supported by the National Science Foundation (grant 2018261442). H.N. is supported by the California Institute for Regenerative Medicine (grants LA1_C12-06917; DISC1-10555); the National Instututes for Health (grants R01DK116944; R01HL147124; R21AG061487); and the Virginia and D.K. Ludwig Fund for Cancer Research (D.K. Ludwig Fund).
Footnotes
Competing interests
H.N. is a co-founder and shareholder in Megakaryon and Century Therapeutics.
References
- 1.Orkin SH & Zon LI Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644, doi: 10.1016/J.Cell.2008.01.025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon MY, Lewis JL & Marley SB Of mice and men...and elephants. Blood 100, 4679–4680, doi: 10.1182/blood-2002-08-2517 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Eaves CJ Hematopoietic stem cells: concepts, definitions, and the new reality. Blood 125, 2605–2613, doi: 10.1182/blood-2014-12-570200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissman IL & Shizuru JA The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood 112, 3543–3553, doi: 10.1182/blood-2008-08-078220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seita J & Weissman IL Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med 2, 640–653, doi: 10.1002/wsbm.86 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copelan EA Hematopoietic stem-cell transplantation. N Engl J Med 354, 1813–1826, doi: 10.1056/NEJMra052638 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Chabannon C et al. Hematopoietic stem cell transplantation in its 60s: A platform for cellular therapies. Sci Transl Med 10, doi: 10.1126/scitranslmed.aap9630 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Pinho S & Frenette PS Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol 20, 303–320, doi: 10.1038/s41580-019-0103-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spangrude GJ, Heimfeld S & Weissman IL Purification and characterization of mouse hematopoietic stem cells. Science 241, 58–62, doi: 10.1126/science.2898810 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Visser JW, Bauman JG, Mulder AH, Eliason JF & de Leeuw AM Isolation of murine pluripotent hemopoietic stem cells. J Exp Med 159, 1576–1590, doi: 10.1084/jem.159.6.1576 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C & Morrison SJ SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121, doi: 10.1016/j.cell.2005.05.026 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Morita Y, Ema H & Nakauchi H Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med 207, 1173–1182, doi: 10.1084/jem.20091318 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto R et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154, 1112–1126, doi: 10.1016/j.cell.2013.08.007 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Balazs AB, Fabian AJ, Esmon CT & Mulligan RC Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 107, 2317–2321, doi: 10.1182/blood-2005-06-2249 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazit R et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med 211, 1315–1331, doi: 10.1084/jem.20130428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227, doi: 10.1038/nature16943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka K et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med 208, 2403–2416, doi: 10.1084/jem.20110447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acar M et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130, doi: 10.1038/nature15250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tajima Y et al. Continuous cell supply from Krt7-expressing hematopoietic stem cells during native hematopoiesis revealed by targeted in vivo gene transfer method. Sci Rep 7, 40684, doi: 10.1038/srep40684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notta F et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221, doi: 10.1126/science.1201219 (2011).This paper describes the use of CD49f expression to purify human HSCs are high frequencies.
- 21.Wilson NK et al. Combined Single-Cell Functional and Gene Expression Analysis Resolves Heterogeneity within Stem Cell Populations. Cell Stem Cell 16, 712–724, doi: 10.1016/j.stem.2015.04.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notta F et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116, doi: 10.1126/science.aab2116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson AC et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature 571, 117–121, doi: 10.1038/s41586-019-1244-x (2019).Using polyvinyl alcohol, this paper describes a long-term ex vivo expansion culture system for mouse HSCs.
- 24.Fares I et al. EPCR expression marks UM171-expanded CD34. Blood 129, 3344–3351, doi: 10.1182/blood-2016-11-750729 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Tomellini E et al. Integrin-α3 Is a Functional Marker of Ex Vivo Expanded Human Long-Term Hematopoietic Stem Cells. Cell Rep 28, 1063–1073.e1065, doi: 10.1016/j.celrep.2019.06.084 (2019). [DOI] [PubMed] [Google Scholar]
- 26.TILL JE & McCULLOCH EA A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res 14, 213–222 (1961). [PubMed] [Google Scholar]
- 27.BECKER AJ, McCULLOCH EA & TILL JE Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454, doi: 10.1038/197452a0 (1963). [DOI] [PubMed] [Google Scholar]
- 28.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC & Eaves CJ Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci U S A 87, 8736–8740, doi: 10.1073/pnas.87.22.8736 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemischka IR, Raulet DH & Mulligan RC Developmental potential and dynamic behavior of hematopoietic stem cells. Cell 45, 917–927, doi: 10.1016/0092-8674(86)90566-0 (1986). [DOI] [PubMed] [Google Scholar]
- 30.Osawa M, Hanada K, Hamada H & Nakauchi H Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996).This paper describes the use of single cell transplantation assays to identify self-renewing multipotent HSCs within the mouse bone marrow.
- 31.Waterstrat A, Liang Y, Swiderski CF, Shelton BJ & Van Zant G Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood 115, 408–417, doi: 10.1182/blood-2008-03-143370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercier FE, Sykes DB & Scadden DT Single Targeted Exon Mutation Creates a True Congenic Mouse for Competitive Hematopoietic Stem Cell Transplantation: The C57BL/6-CD45.1(STEM) Mouse. Stem Cell Reports 6, 985–992, doi: 10.1016/j.stemcr.2016.04.010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto R et al. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell 22, 600–607.e604, doi: 10.1016/j.stem.2018.03.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrelha J et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106–111, doi: 10.1038/nature25455 (2018).This paper provided definitive evidence for platelet-restricted self-renewal stem cells.
- 35.Haas S et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell 17, 422–434, doi: 10.1016/j.stem.2015.07.007 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Benveniste P et al. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 6, 48–58, doi: 10.1016/j.stem.2009.11.014 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto R, Wilkinson AC & Nakauchi H Changing concepts in hematopoietic stem cells. Science 362, 895–896, doi: 10.1126/science.aat7873 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Ema H, Takano H, Sudo K & Nakauchi H In vitro self-renewal division of hematopoietic stem cells. J Exp Med 192, 1281–1288, doi: 10.1084/jem.192.9.1281 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerrits A et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood 115, 2610–2618, doi: 10.1182/blood-2009-06-229757 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Lu R, Neff NF, Quake SR & Weissman IL Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol 29, 928–933, doi: 10.1038/nbt.1977 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naik SH et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496, 229–232, doi: 10.1038/nature12013 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327, doi: 10.1038/nature13824 (2014).This paper described the use of Sleeping Beauty transposon-based barcoding of hematopoietic stem and progenitors in vivo.
- 43.Busch K et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546, doi: 10.1038/nature14242 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Bradford GB, Williams B, Rossi R & Bertoncello I Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp Hematol 25, 445–453 (1997). [PubMed] [Google Scholar]
- 45.Cheshier SH, Morrison SJ, Liao X & Weissman IL In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci U S A 96, 3120–3125 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudo K, Ema H, Morita Y & Nakauchi H Age-associated characteristics of murine hematopoietic stem cells. J Exp Med 192, 1273–1280 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson A et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129, doi: 10.1016/j.cell.2008.10.048 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Yu VWC et al. Epigenetic Memory Underlies Cell-Autonomous Heterogeneous Behavior of Hematopoietic Stem Cells. Cell 168, 944–945, doi: 10.1016/j.cell.2017.02.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganuza M et al. The global clonal complexity of the murine blood system declines throughout life and after serial transplantation. Blood 133, 1927–1942, doi: 10.1182/blood-2018-09-873059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernitz JM, Kim HS, MacArthur B, Sieburg H & Moore K Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell 167, 1296–1309.e1210, doi: 10.1016/j.cell.2016.10.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460, doi: 10.1038/nature23653 (2017).This paper described the development and validation of an in vivo Polylox barcode system that allows for numerous HSCs to be barcoded and tracked during native hematopoiesis.
- 52.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111, doi: 10.1038/nature20777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loeffler D et al. Asymmetric lysosome inheritance predicts activation of haematopoietic stem cells. Nature 573, 426–429, doi: 10.1038/s41586-019-1531-6 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Christodoulou C et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature 578, 278–283, doi: 10.1038/s41586-020-1971-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamel-Reid S & Dick JE Engraftment of immune-deficient mice with human hematopoietic stem cells. Science 242, 1706–1709, doi: 10.1126/science.2904703 (1988). [DOI] [PubMed] [Google Scholar]
- 56.Goyama S, Wunderlich M & Mulloy JC Xenograft models for normal and malignant stem cells. Blood 125, 2630–2640, doi: 10.1182/blood-2014-11-570218 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Yurino A et al. Enhanced Reconstitution of Human Erythropoiesis and Thrombopoiesis in an Immunodeficient Mouse Model with Kit(Wv) Mutations. Stem Cell Reports 7, 425–438, doi: 10.1016/j.stemcr.2016.07.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahmig S et al. Improved Human Erythropoiesis and Platelet Formation in Humanized NSGW41 Mice. Stem Cell Reports 7, 591–601, doi: 10.1016/j.stemcr.2016.08.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller PH et al. Analysis of parameters that affect human hematopoietic cell outputs in mutant c-kit-immunodeficient mice. Exp Hematol 48, 41–49, doi: 10.1016/j.exphem.2016.12.012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takagi S et al. Membrane-bound human SCF/KL promotes in vivo human hematopoietic engraftment and myeloid differentiation. Blood 119, 2768–2777, doi: 10.1182/blood-2011-05-353201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biasco L et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell 19, 107–119, doi: 10.1016/j.stem.2016.04.016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scala S et al. Dynamics of genetically engineered hematopoietic stem and progenitor cells after autologous transplantation in humans. Nat Med 24, 1683–1690, doi: 10.1038/s41591-018-0195-3 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478, doi: 10.1038/s41586-018-0497-0 (2018).This paper used whole genome sequencing to identify somatic mutations that could be used to infer lineage relationships with human hematopoiesis.
- 64.Morrison SJ, Hemmati HD, Wandycz AM & Weissman IL The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A 92, 10302–10306, doi: 10.1073/pnas.92.22.10302 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ema H & Nakauchi H Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 95, 2284–2288 (2000). [PubMed] [Google Scholar]
- 66.Gekas C, Dieterlen-Lièvre F, Orkin SH & Mikkola HK The placenta is a niche for hematopoietic stem cells. Dev Cell 8, 365–375, doi: 10.1016/j.devcel.2004.12.016 (2005). [DOI] [PubMed] [Google Scholar]
- 67.de Haan G & Lazare SS Aging of hematopoietic stem cells. Blood 131, 479–487, doi: 10.1182/blood-2017-06-746412 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Ema H et al. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell 8, 907–914, doi: 10.1016/j.devcel.2005.03.019 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Morrison SJ & Scadden DT The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334, doi: 10.1038/nature12984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crane GM, Jeffery E & Morrison SJ Adult haematopoietic stem cell niches. Nat Rev Immunol 17, 573–590, doi: 10.1038/nri.2017.53 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Zhang CC & Lodish HF Cytokines regulating hematopoietic stem cell function. Curr Opin Hematol 15, 307–311, doi: 10.1097/MOH.0b013e3283007db5 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edling CE & Hallberg B c-Kit--a hematopoietic cell essential receptor tyrosine kinase. Int J Biochem Cell Biol 39, 1995–1998, doi: 10.1016/j.biocel.2006.12.005 (2007). [DOI] [PubMed] [Google Scholar]
- 73.Ding L, Saunders TL, Enikolopov G & Morrison SJ Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462, doi: 10.1038/nature10783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Graaf CA & Metcalf D Thrombopoietin and hematopoietic stem cells. Cell Cycle 10, 1582–1589, doi: 10.4161/cc.10.10.15619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Decker M, Leslie J, Liu Q & Ding L Hepatic thrombopoietin is required for bone marrow hematopoietic stem cell maintenance. Science 360, 106–110, doi: 10.1126/science.aap8861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tadokoro Y et al. Spred1 Safeguards Hematopoietic Homeostasis against Diet-Induced Systemic Stress. Cell Stem Cell 22, 713–725.e718, doi: 10.1016/j.stem.2018.04.002 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Seita J et al. Lnk negatively regulates self-renewal of hematopoietic stem cells by modifying thrombopoietin-mediated signal transduction. Proc Natl Acad Sci U S A 104, 2349–2354, doi: 10.1073/pnas.0606238104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takizawa H et al. Pathogen-Induced TLR4-TRIF Innate Immune Signaling in Hematopoietic Stem Cells Promotes Proliferation but Reduces Competitive Fitness. Cell Stem Cell 21, 225–240.e225, doi: 10.1016/j.stem.2017.06.013 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Pietras EM et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol 18, 607–618, doi: 10.1038/ncb3346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takizawa H & Manz MG Impact of inflammation on early hematopoiesis and the microenvironment. Int J Hematol 106, 27–33, doi: 10.1007/s12185-017-2266-5 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Pietras EM Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood 130, 1693–1698, doi: 10.1182/blood-2017-06-780882 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilkinson AC & Yamazaki S The hematopoietic stem cell diet. Int J Hematol, doi: 10.1007/s12185-018-2451-1 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Cheng CW et al. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell 14, 810–823, doi: 10.1016/j.stem.2014.04.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lazare S et al. Lifelong dietary intervention does not affect hematopoietic stem cell function. Exp Hematol 53, 26–30, doi: 10.1016/j.exphem.2017.06.002 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Tang D et al. Dietary restriction improves repopulation but impairs lymphoid differentiation capacity of hematopoietic stem cells in early aging. J Exp Med 213, 535–553, doi: 10.1084/jem.20151100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taya Y et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 354, 1152–1155, doi: 10.1126/science.aag3145 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Wilkinson AC, Morita M, Nakauchi H & Yamazaki S Branched-chain amino acid depletion conditions bone marrow for hematopoietic stem cell transplantation avoiding amino acid imbalance-associated toxicity. Exp Hematol, doi: 10.1016/j.exphem.2018.04.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mantel CR et al. Enhancing Hematopoietic Stem Cell Transplantation Efficacy by Mitigating Oxygen Shock. Cell 161, 1553–1565, doi: 10.1016/j.cell.2015.04.054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowie MB, Kent DG, Copley MR & Eaves CJ Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood 109, 5043–5048, doi: 10.1182/blood-2006-08-037770 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Prashad SL et al. GPI-80 defines self-renewal ability in hematopoietic stem cells during human development. Cell Stem Cell 16, 80–87, doi: 10.1016/j.stem.2014.10.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chhabra A et al. Trophoblasts regulate the placental hematopoietic niche through PDGF-B signaling. Dev Cell 22, 651–659, doi: 10.1016/j.devcel.2011.12.022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moran-Crusio K et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20, 11–24, doi: 10.1016/j.ccr.2011.06.001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Challen GA et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44, 23–31, doi: 10.1038/ng.1009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jeong M et al. Loss of Dnmt3a Immortalizes Hematopoietic Stem Cells In Vivo. Cell Rep 23, 1–10, doi: 10.1016/j.celrep.2018.03.025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iwama A et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21, 843–851, doi: 10.1016/j.immuni.2004.11.004 (2004). [DOI] [PubMed] [Google Scholar]
- 96.Park IK et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305, doi: 10.1038/nature01587 (2003). [DOI] [PubMed] [Google Scholar]
- 97.Luis TC, Wilkinson AC, Beerman I, Jaiswal S & Shlush LI Biological implications of clonal hematopoiesis. Exp Hematol 77, 1–5, doi: 10.1016/j.exphem.2019.08.004 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Calvanese V et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 576, 281–286, doi: 10.1038/s41586-019-1790-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ito K et al. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med 18, 1350–1358, doi: 10.1038/nm.2882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ito K et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 354, 1156–1160, doi: 10.1126/science.aaf5530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ansó E et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol 19, 614–625, doi: 10.1038/ncb3529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ho TT et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210, doi: 10.1038/nature21388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ito K Hematopoietic stem cell fate through metabolic control. Exp Hematol 64, 1–11, doi: 10.1016/j.exphem.2018.05.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481, doi: 10.1038/nature23876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cimmino L et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 170, 1079–1095.e1020, doi: 10.1016/j.cell.2017.07.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilkinson AC & Gottgens B Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol 786, 187–212, doi: 10.1007/978-94-007-6621-1_11 (2013). [DOI] [PubMed] [Google Scholar]
- 107.Kim I, Saunders TL & Morrison SJ Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130, 470–483, doi: 10.1016/j.cell.2007.06.011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He S, Kim I, Lim MS & Morrison SJ Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes Dev 25, 1613–1627, doi: 10.1101/gad.2052911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao Y et al. ATF4 plays a pivotal role in the development of functional hematopoietic stem cells in mouse fetal liver. Blood 126, 2383–2391, doi: 10.1182/blood-2015-03-633354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mochizuki-Kashio M et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 118, 6553–6561, doi: 10.1182/blood-2011-03-340554 (2011). [DOI] [PubMed] [Google Scholar]
- 111.Copley MR et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol 15, 916–925, doi: 10.1038/ncb2783 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Kumar S & Geiger H HSC Niche Biology and HSC Expansion Ex Vivo. Trends Mol Med 23, 799–819, doi: 10.1016/j.molmed.2017.07.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ku H, Yonemura Y, Kaushansky K & Ogawa M Thrombopoietin, the ligand for the Mpl receptor, synergizes with steel factor and other early acting cytokines in supporting proliferation of primitive hematopoietic progenitors of mice. Blood 87, 4544–4551 (1996). [PubMed] [Google Scholar]
- 114.Sitnicka E et al. The effect of thrombopoietin on the proliferation and differentiation of murine hematopoietic stem cells. Blood 87, 4998–5005 (1996). [PubMed] [Google Scholar]
- 115.Ramsfjell V et al. Thrombopoietin, but not erythropoietin, directly stimulates multilineage growth of primitive murine bone marrow progenitor cells in synergy with early acting cytokines: distinct interactions with the ligands for c-kit and FLT3. Blood 88, 4481–4492 (1996). [PubMed] [Google Scholar]
- 116.Miller CL & Eaves CJ Expansion in vitro of adult murine hematopoietic stem cells with transplantable lympho-myeloid reconstituting ability. Proc Natl Acad Sci U S A 94, 13648–13653, doi: 10.1073/pnas.94.25.13648 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Himburg HA et al. Pleiotrophin regulates the expansion and regeneration of hematopoietic stem cells. Nat Med 16, 475–482, doi: 10.1038/nm.2119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ieyasu A et al. An All-Recombinant Protein-Based Culture System Specifically Identifies Hematopoietic Stem Cell Maintenance Factors. Stem Cell Reports 8, 500–508, doi: 10.1016/j.stemcr.2017.01.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Butler JM et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 6, 251–264, doi: 10.1016/j.stem.2010.02.001 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakahara F et al. Engineering a haematopoietic stem cell niche by revitalizing mesenchymal stromal cells. Nat Cell Biol 21, 560–567, doi: 10.1038/s41556-019-0308-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bai T et al. Expansion of primitive human hematopoietic stem cells by culture in a zwitterionic hydrogel. Nat Med, doi: 10.1038/s41591-019-0601-5 (2019).This paper described the novel use of zwitterionic hydrogel 3D-based cultures for expanding human HSCs ex vivo.
- 122.Umemoto T et al. Integrin-αvβ3 regulates thrombopoietin-mediated maintenance of hematopoietic stem cells. Blood 119, 83–94, doi: 10.1182/blood-2011-02-335430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Antonchuk J, Sauvageau G & Humphries RK HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109, 39–45, doi: 10.1016/s0092-8674(02)00697-9 (2002). [DOI] [PubMed] [Google Scholar]
- 124.Miharada K, Sigurdsson V & Karlsson S Dppa5 improves hematopoietic stem cell activity by reducing endoplasmic reticulum stress. Cell Rep 7, 1381–1392, doi: 10.1016/j.celrep.2014.04.056 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Kunisato A et al. HES-1 preserves purified hematopoietic stem cells ex vivo and accumulates side population cells in vivo. Blood 101, 1777–1783, doi: 10.1182/blood-2002-07-2051 (2003). [DOI] [PubMed] [Google Scholar]
- 126.Reya T et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414, doi: 10.1038/nature01593 (2003). [DOI] [PubMed] [Google Scholar]
- 127.Boitano AE et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348, doi: 10.1126/science.1191536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fares I et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345, 1509–1512, doi: 10.1126/science.1256337 (2014).This paper describes the identification of the small molecule UM171, a potent human HSC self-renewal agonist.
- 129.Chagraoui J et al. UM171 induces a homeostatic inflammatory-detoxification response supporting human HSC self-renewal. PLoS One 14, e0224900, doi: 10.1371/journal.pone.0224900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sun H, Tsai Y, Nowak I, Liesveld J & Chen Y Eltrombopag, a thrombopoietin receptor agonist, enhances human umbilical cord blood hematopoietic stem/primitive progenitor cell expansion and promotes multi-lineage hematopoiesis. Stem Cell Res 9, 77–86, doi: 10.1016/j.scr.2012.05.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nishino T et al. Ex vivo expansion of human hematopoietic stem cells by a small-molecule agonist of c-MPL. Exp Hematol 37, 1364–1377.e1364, doi: 10.1016/j.exphem.2009.09.001 (2009). [DOI] [PubMed] [Google Scholar]
- 132.Csaszar E et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 10, 218–229, doi: 10.1016/j.stem.2012.01.003 (2012). [DOI] [PubMed] [Google Scholar]
- 133.Francis GL Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology 62, 1–16, doi: 10.1007/s10616-010-9263-3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wilkinson AC, Ishida R, Nakauchi H & Yamazaki S Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat Protoc, doi: 10.1038/s41596-019-0263-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nishimura T et al. Use of polyvinyl alcohol for chimeric antigen receptor T-cell expansion. Exp Hematol 80, 16–20, doi: 10.1016/j.exphem.2019.11.007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luchsinger LL et al. Harnessing Hematopoietic Stem Cell Low Intracellular Calcium Improves Their Maintenance In Vitro. Cell Stem Cell, doi: 10.1016/j.stem.2019.05.002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Umemoto T, Hashimoto M, Matsumura T, Nakamura-Ishizu A & Suda T Ca. J Exp Med 215, 2097–2113, doi: 10.1084/jem.20180421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kobayashi H et al. Environmental Optimization Enables Maintenance of Quiescent Hematopoietic Stem Cells Ex Vivo. Cell Rep 28, 145–158.e149, doi: 10.1016/j.celrep.2019.06.008 (2019). [DOI] [PubMed] [Google Scholar]
- 139.Morgan RA, Gray D, Lomova A & Kohn DB Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell 21, 574–590, doi: 10.1016/j.stem.2017.10.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Negrin RS Graft-versus-host disease versus graft-versus-leukemia. Hematology Am Soc Hematol Educ Program 2015, 225–230, doi: 10.1182/asheducation-2015.1.225 (2015). [DOI] [PubMed] [Google Scholar]
- 141.Broxmeyer HE Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfus Apher Sci 54, 364–372, doi: 10.1016/j.transci.2016.05.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim YJ & Broxmeyer HE Immune regulatory cells in umbilical cord blood and their potential roles in transplantation tolerance. Crit Rev Oncol Hematol 79, 112–126, doi: 10.1016/j.critrevonc.2010.07.009 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cohen S et al. Hematopoietic stem cell transplantation using single UM171-expanded cord blood: a single-arm, phase 1–2 safety and feasibility study. Lancet Haematol 7, e134–e145, doi: 10.1016/S2352-3026(19)30202-9 (2020).This recent phase 1–2 clinical trial publication demonstrates the safety and feasibility of ex vivo UM171-expanded cord blood HSCs.
- 144.Wagner JE et al. Phase I/II Trial of StemRegenin-1 Expanded Umbilical Cord Blood Hematopoietic Stem Cells Supports Testing as a Stand-Alone Graft. Cell Stem Cell 18, 144–155, doi: 10.1016/j.stem.2015.10.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Lima M et al. Cord-blood engraftment with ex vivo mesenchymal-cell coculture. N Engl J Med 367, 2305–2315, doi: 10.1056/NEJMoa1207285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dever DP & Porteus MH The changing landscape of gene editing in hematopoietic stem cells: a step towards Cas9 clinical translation. Curr Opin Hematol 24, 481–488, doi: 10.1097/MOH.0000000000000385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gundry MC et al. Technical considerations for the use of CRISPR/Cas9 in hematology research. Exp Hematol 54, 4–11, doi: 10.1016/j.exphem.2017.07.006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bhattacharya D, Rossi DJ, Bryder D & Weissman IL Purified hematopoietic stem cell engraftment of rare niches corrects severe lymphoid deficiencies without host conditioning. J Exp Med 203, 73–85, doi: 10.1084/jem.20051714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shimoto M, Sugiyama T & Nagasawa T Numerous niches for hematopoietic stem cells remain empty during homeostasis. Blood 129, 2124–2131, doi: 10.1182/blood-2016-09-740563 (2017). [DOI] [PubMed] [Google Scholar]
- 150.Kitao H & Takata M Fanconi anemia: a disorder defective in the DNA damage response. Int J Hematol 93, 417–424, doi: 10.1007/s12185-011-0777-z (2011). [DOI] [PubMed] [Google Scholar]
- 151.Evans M Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol 12, 680–686, doi: 10.1038/nrm3190 (2011). [DOI] [PubMed] [Google Scholar]
- 152.Ivanovs A et al. Human haematopoietic stem cell development: from the embryo to the dish. Development 144, 2323–2337, doi: 10.1242/dev.134866 (2017). [DOI] [PubMed] [Google Scholar]
- 153.Wilkinson AC Hope for hematological diseases. Science 367, 1206, doi: 10.1126/science.aba6108 (2020). [DOI] [PubMed] [Google Scholar]
- 154.Laurenti E & Göttgens B From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426, doi: 10.1038/nature25022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haas S, Trumpp A & Milsom MD Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell 22, 627–638, doi: 10.1016/j.stem.2018.04.003 (2018). [DOI] [PubMed] [Google Scholar]
- 156.Dzierzak E & Speck NA Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 9, 129–136, doi: 10.1038/ni1560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Bruijn MF, Speck NA, Peeters MC & Dzierzak E Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 19, 2465–2474, doi: 10.1093/emboj/19.11.2465 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ivanovs A et al. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J Exp Med 208, 2417–2427, doi: 10.1084/jem.20111688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ottersbach K & Dzierzak E The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell 8, 377–387, doi: 10.1016/j.devcel.2005.02.001 (2005). [DOI] [PubMed] [Google Scholar]
- 160.Kumaravelu P et al. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development 129, 4891–4899 (2002). [DOI] [PubMed] [Google Scholar]