Abstract
Objectives:
Wild type RAS (RASWT) suppresses the function of oncogenic RAS mutants (RASMUT) in laboratory models. Loss of RASWT, which we termed loss of heterozygosity (LOH) for any RAS (LAR) or LAKR in the context of KRAS (LOH at KRAS), is found in patients with RASMUT cancers. However, the incidence and prognostic significance of LAR has not been studied in modern patient cohorts. LAR or LAKR in RASMUT cancers is attractive as a potential biomarker for targeted therapy.
Materials and methods:
We evaluated for associations between LAKR and cancer mortality in patients with RASMUT lung adenocarcinoma (LUAD). We also evaluated for associations between LAKR and the metabolic state of cancer cell lines, given that KRAS has been shown to regulate fatty acid synthesis. In line with this, we investigated fatty acid synthase (FASN) inhibitors as potential therapies for RASMUT LAKR, including combination strategies involving clinical KRASG12C and FASN inhibitors.
Results:
24% of patients with KRASMUT LUAD showed LAKR. KRASMUT LAKR cases had a median survival of 16 vs. 30 months in KRASMUT non-LAKR (p = 0.017) and LAKR was independently associated with death in this cohort (p = 0.011). We also found that KRASMUT LUAD cell lines with LAKR contained elevated levels of FASN and fatty acids relative to non-LAKR cell lines. KRASG12C LUAD cells with LAKR showed higher sensitivity to treatment with FASN inhibitors than those without. FASN inhibitors such as TVB-3664 showed synergistic effects with the KRASG12C inhibitor MRTX849 in LUAD cells with LAKR, including an in vivo trial using a xenograft model.
Conclusions:
LAKR in KRASMUT cancers may represent an independent negative prognostic factor for patients with KRASMUT LUAD. It also predicts for response to treatment with FASN inhibitors. Prospective testing of combination therapies including KRASG12C and FASN inhibitors in patients with KRASG12C LAKR is warranted.
Keywords: Lung adenocarcinoma, RAS, KRAS, loss of heterozygosity
1. Introduction
RAS mutations are a common genetic feature in human tumors and are enriched in aggressive cancers such as lung adenocarcinoma, cholangiocarcinoma, colorectal cancer, melanoma, and pancreatic ductal adenocarcinoma [1]. However, changes in RAS zygosity, especially loss of RASWT, which we refer to hereafter as loss of heterozygosity (LOH) at any RAS (LAR), or LAKR (LOH at KRAS) in the context of KRAS, also occur in malignancy [2–4]. Presumably this is because in the setting of mutant RAS (RASMUT), RASWT behaves as a tumor suppressor and loss of RASWT enhances tumor fitness. The incidence of LAR in cancer patients has not been characterized in modern cohorts and the prognostic value of LAR for patient care is unknown. LAR is also assocaiated with changes in cellular responses to perturbagens [5] but LAR has not yet shown efficacy as a predictive biomarker for patient selection of targeted therapies.
The RAS subfamily of small GTPases consists of 4 isoforms with a high degree of sequence identity: HRAS, NRAS, and two splice variants of KRAS, KRAS4A and KRAS4B. RAS mutations are generally thought to operate by increasing the fraction of GTP-bound RAS, thereby enhancing signaling pathways mediated by protein-protein interactions between RAS and its effectors [6, 7]. Nevertheless, these isoforms are mutated at different rates and preferentially at different locations according to cancer type [1]. This, together with accumulating data showing functional differences of specific RAS mutants [8–10] suggests that individual RAS isoforms and specific mutations should be considered separately for mechanistic studies and therapeutic targeting strategies.
RAS copy number also varies non-randomly between cancer types and RAS isoforms [11–14]. Copy number changes in RAS, such as LAR, can impact RAS-mediated signaling and modulate sensitivity to certain drugs, such as MEK inhibitors [11, 15–17]. In the presence of RASMUT, RASWT often behaves similarly to tumor suppressors, such as RB and BRCA1, where loss of RASWT leads to tumor progression [18]. Accordingly, mouse models demonstrated that LOH at KRAS (LAKR) increases the fitness of KRASMUT LUAD [5]. Similarly, LAKR in KRASMUT acute myeloid leukemia (AML) and colon adenocarcinoma cells enhances tumor aggressiveness [17]. Together these observations suggest that in the context of RASMUT cancer, LAR may be a clinically useful biomarker for prognostication and for patient selection of targeted therapies if appropriate therapies can be discovered.
In this study, we explored the relationship between LAKR and patient survival in KRASMUT LUAD using data from public databases [19]. These data suggest that LAKR is an independent prognostic factor that may help guide clinical management of LUAD. We also searched for targetable biological effects associated with LAKR and found an association with lipid metabolism, specifically fatty acid synthase (FASN). This association is important because FASN inhibitors are currently in clinical trials for KRASMUT LUAD, given that FASN is elevated in some RASMUT cancers [20]. We evaluated this potential therapeutic vulnerability for KRASMUT LUAD with LAKR and believe it represents an opportunity to overcome known therapeutic resistance to clinical KRASG12C inhibittors such as MRTX849.
2. Materials and method
2.1. Patients and clinical information
Patients with lung adenocarcinoma (LUAD), colon adenocarcinoma (COAD), pancreatic adenocarcinoma (PAAD) and skin cutaneous melanoma (SKCM) and their clinical information were obtained from the TCGA database. Samples lacking survival information, RAS status or LOH status were excluded from the study.
2.2. RAS and LOH status analysis
To determine RAS mutation status, “PASS” filter mutations in KRAS, NRAS, and HRAS were selected from the MC3 MAF file (v0.2.8) [21]. Intronic mutations, mutations in 3’ or 5’ UTRs or UTR flanking regions, and silent mutations were then further removed. LOH status of patient tumor tissues from TCGA was determined using the ABSOLUTE algorithm [22]. For each RAS gene and for each TCGA sample, the fraction of base pairs in each RAS gene composed of segments labeled with LOH by ABSOLUTE was computed. Cases where over 0.5 of the gene was labeled as LOH were considered to have undergone LOH.
2.3. Statistical methods
Descriptive statistics were used to characterize the patients at study entry. Chi-squared test was used to compare distributions of clinical characteristics across RASMUT patients with or without LAR. Fisher’s exact test was used when the sample size was smaller than 5. Two-sample t-test was used to compare the percentage of LAKR in wild type and mutated KRAS background. A p value < 0.05 was considered statistically significant. Cox Proportional Hazards Regression analysis was used to derive hazard ratios (HRs) for cancer-specific mortality in association with LAKR, with adjustment for age at diagnosis, gender, pathology stage, and smoking history. Kaplan-Meier survival curves were generated using “survival” and “survminer” R packages. Log Rank Test was used for the statistical analysis. All tests were two-tailed with a significance level of p < 0.05. All analyses were performed using R (version 3.6.0, R Foundation for Statistical Computing).
2.4. Cell lines and cell culture
Non-small lung cancer lines (NSCLC) H1150, H650, H2122, H2030, H23, HCC44, SW1573, H1373, H460, Calu-6, Calu-1, H358, H1792, H1573, and H441 were from the Hamon Center Cell Repository. Cells were cultured in RPMI-1640 supplemented with 10% FBS. Cell line identity was confirmed by DNA fingerprinting (PowerPlex 1.2 Kit, Promega) and mycoplasma-free status was verified by PCR (e-Myco Kit, Boca Scientific). The KRAS mutation status (from Cancer Cell Line Encylopedia, CCLE, data) and LAKR status of these cell lines are summarized in Table S1. LUAD cell lines with KRAS mutations that were homozygous for the mutations were scored as LAKR “positive”, while lines with KRASG12C and KRAS wild type sequences (heterozygous for KRAS allele) were scored as LAKR “negative”.
2.5. Chemical compounds
FASN inhibitors,TVB-3116 and TVB-3664 were gifts from Sagimet Biosciences (San Mateo, CA ). Cerulenin was purchased from Sigma-Aldrich (Burlington, MA) and Fasnall was purchased from Focus Biomolecules (Plymouth Meeting, PA). KRAS inhibitors, MRTX849 was purchased from DC Chemicals (Shanghai, China) and MRTX1257 were purchased from MedChemExpress (Monmouth Junction, NJ).
2.6. Cell proliferation assay
CellTiter-Glo was ordered from Promega (Madison, WI). Briefly, 50,000 cells were seeded per well in a 100 μl medium in 96-well format and were treated by a compound or transfected with a DNA construct. Assays were developed by addition of 100 μl of CellTiter-Glo reagent. Luminescence was measured using BioTek NEO plate reader.
2.7. Western Blotting
Whole cell lysates were separated by SDS-PAGE gel and protein was transferred onto Immobilon-P membrane. The membrane was blocked by 5% milk in PBS with 0.1% Tween-20. The blot was probed with the FASN antibody from Cell Signaling (Danvers, MA). Protein was detected by LumiGLO reagent and peroxide from Cell Signaling. Secondary antibody was from Cell Signaling.
2.8. Xenografts
8.5-week-old female nude mice (Jackson Laboratories Stock No: 007850 (J:NU), homozygous for Foxn1nu) were implanted at a single subcutaneous site with 5 × 106 H2122 NSCLC cells (0.1 ml). When tumors reached approximately 120 mm3 (day 4 post tumor cell implantation), therapy was initiated with 5 mg/kg TVB3664 only, 10 mg/kg MRTX849 only, 5 mg/kg TVB3664 + 10 mg/kg MRTX849, or vehicle only (30% fresh PEG400 + 70% 20 mm pH 4.5 citrate buffer) 0.2 ml/mouse (8 mice/group). Compounds were administered daily as clear solutions orally. Mice were weighed three times per week and tumors measured with calipers three times per week. Tumor volume was calculated as (L × W2 × pi)/6. When tumor volume reached >1250 mm3 or if mice appeared moribund, they were euthanized. Weight loss did not exceed 20% in any animal and overall was comparable between groups. All mice not previously euthanized, were sacrificed on day 23. For statistical analysis, TVB-3664 and MRTX849 arms were compared with the vehicle arm. The combination arm was compared with each single treatment arm. Statistical analyses were performed at day 19 (n = 8 mice/group). ANOVA was used to compare among multiple groups and each pair was analyzed by student t-test.
3. Results
3.1. LAKR is common in major cancer types
We evaluated four major cancer types, LUAD, COAD, PAAD, and SKCM, where RAS mutations are common and clinical data is available for associated TCGA specimens. Of the 501 evaluable patients with LUAD, 146 (29.1%) of them had mutations in KRAS (Table 1), consistent with other results [23]. Thirty-five (24%) of those had LAKR. As a control we also evaluated LAKR in the KRASWT background and found a similar rate of LAKR (26%; Table 1). In the 171 evaluable patients with PAAD and the 399 evaluable patients with COAD, the rates of KRASMUT LAKR were lower overall, at 14% for both. KRASMUT PAAD showed statistically similar rates for LAKR in KRASWT vs. KRASMUT backgrounds (5.6% vs. 13.7%, p = 0.12). However, in COAD, LAKR was lower in KRASWT (6.8% vs. 14.2%, p = 0.01; Table 1). Of the 469 evaluable patients with SKCM, 129 (27.5%) had mutations in NRAS. Twenty-eight (21.7%) of them had LANR, not statistically different from that observed in the NRASWT background (18.8%, p = 0.48; Table 1). Based on this analysis we focused on LUAD because both KRAS mutations and LAKR were more common in LUAD than other three cancer types.
Table 1.
Survey of LAR in four major cancer types.
| Cancer Type | LOH Status | KRASMUT (%) | KRASWT (%) | P |
|---|---|---|---|---|
| LUAD | LOH | 35 (24) | 91 (26) | 0.71 |
| no LOH | 111 (76) | 264 (74) | ||
| Total | 146 (100) | 355 (100) | ||
| COAD | LOH | 19 (14) | 18 (7) | 0.016 |
| no LOH | 115 (86) | 247 (93) | ||
| Total | 134 (100) | 265 (100) | ||
| PAAD | LOH | 16 (14) | 3 (6) | 0.12 |
| no LOH | 101 (86) | 51 (94) | ||
| Total | 117 (100) | 54 (100) | ||
| NRASMUT (%) | NRASWT (%) | |||
| SKCM | LOH | 28 (22) | 64 (19) | 0.48 |
| no LOH | 101 (78) | 276 (81) | ||
| Total | 129 (100) | 340 (100) | ||
3.2. LAKR is associated with increased mortality in patients with KRASMUT LUAD
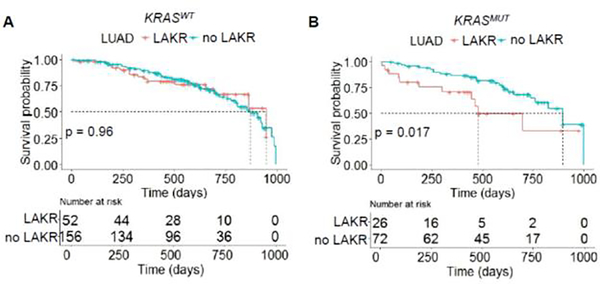
We performed Kaplan-Meier analysis to determine if LAKR is associated with mortality in LUAD cases contained in the TCGA. When comparing KRASMUT cancers with or without LAKR, the two populations were statistically balanced for age, gender, stage, and smoking history, although we noted that stage III-IV cases appeared numerically higher in the LAKR arm (Table 2). Kaplan-Meier analysis showed that LAKR was not associated with overall survival in the KRASWT background, (p = 0.96; Fig. 1A), but was associated with shorter overall survival in the KRASMUT background with a median survival of 16 vs. 30 months, (p = 0.017; Fig. 1B). The difference in survival suggests that LAKR-dependent effects on survival are mediated by an interaction between KRASMUT and KRASWT. To determine if LAKR is an independent prognostic factor, we performed univariate and multivariate analyses using LAKR, age, gender, stage, and smoking history. Of the factors analyzed, both LAKR and stage were associated with an increased risk of death on univariate and multivariate analysis (Table 3). However, in the KRASWT background, only tumor stage was associated with overall survival (Fig. S1), confirming that the association between survival and LAKR is dependent on KRASMUT. Overall, these results support that LAKR is associated with mortality in patients with KRASMUT LUAD.
Table 2.
Distribution of KRASMUT LUAD patients and treatment characteristics stratified by LAKR.
| Baseline characteristics | LAKR (%) | no LAKR (%) | p |
|---|---|---|---|
| Total patients Age at diagnosis | 26 (27) | 72 (73) | |
| Age at diagnosis | 0.75 | ||
| <65 | 15 (58) | 37 (51) | |
| ≥ 65 | 11 (42) | 35 (49) | |
| Gender | 0.74 | ||
| female | 15 (58) | 41 (57) | |
| male | 11 (42) | 31 (43) | |
| Stage | 0.21 | ||
| I | 11 (42) | 34 (47) | |
| II | 5 (19) | 23 (32) | |
| III–IV | 9 (35) | 14 (19) | |
| unknown | 1 (4) | 1 (1) | |
| Smoking history | 0.70 | ||
| <30 pack years | 8 (31) | 23 (32) | |
| ≥ 30 pack years | 14 (54) | 33 (46) | |
| unknown | 4 (15) | 16 (22) | |
Figure 1.
LAKR is associated with cancer mortality in KRASMUT LUAD. Kaplan-Meier analysis of patients with (A) KRASWT or (B) KRASMUT LUAD stratified by LAKR (LOH) status.
Table 3.
HRs of mortality for KRASMUT LUAD patients and clinical characteristics from univariate and multivariate Cox regression analysis.
| Covariate | No. of patients | No. of events | Univariate analysis HR (95% CI) | p | Multivariate analysis HR (95% CI) | p |
|---|---|---|---|---|---|---|
| LAKR: | 0.021 | 0.011 | ||||
| no | 72 | 24 | 1 (reference) | 1 (reference) | ||
| yes | 26 | 11 | 2.40 (1.1 – 4.90) | 2.86 (1.28 – 6.40) | ||
| Age at diagnosis | 0.61 | 0.77 | ||||
| <65 | 46 | 17 | 1 (reference) | 1 (reference) | ||
| ≥ 65 | 52 | 18 | 0.84 (0.43 – 1.60) | 1.07 (0.50 – 2.30) | ||
| Gender | 0.14 | 0.24 | ||||
| female | 56 | 17 | 1 (reference) | 1 (reference) | ||
| male | 42 | 18 | 1.70 (0.85 – 3.30) | 1.49 (0.70 – 3.20) | ||
| Stage | ||||||
| I | 45 | 6 | 1 (reference) | 1 (reference) | ||
| II | 28 | 16 | 3.50 (1.40 –8.50) | 0.007 | 5.00 (1.86 – 13.50) | 0.001 |
| III–IV | 23 | 12 | 5.10 (2.00 – 13.20) | < 0.001 | 6.25 (2.20 – 17.80) | < 0.001 |
| Smoking history | ||||||
| <30 pack years | 31 | 10 | 1 (reference) | 1 (reference) | ||
| ≥ 30 pack years | 47 | 16 | 1.10 (0.51 – 2.50) | 0.77 | 0.75 (0.32 – 1.80) | 0.51 |
| unknown | 20 | 20 | 1.10 (0.43 – 2.70) | 0.88 | 1.12 (0.43 – 2.90) | 0.81 |
3.3. LAKR is associated with upregulated lipogenesis in KRASMUT LUAD cell lines.
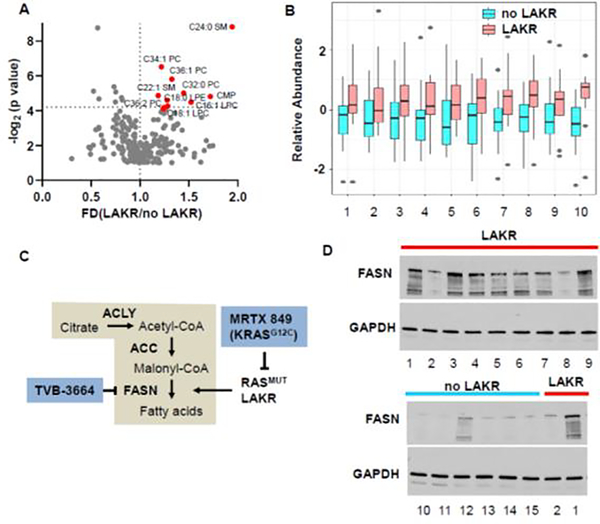
Previous studies showed that KRAS activation leads to lipogenesis through induction of fatty acid synthase (FASN) or acyl-coenzyme A synthetase long-chain family member 3 (ACSL3), and that KRASMUT cancers are sensitive to inhibitors of these enzymes [20, 24]. We speculated that LAKR might contribute to this phenomenon in KRASMUT LUAD if the suppressive effects of KRASWT occur upstream of mechanisms that lead to upregulation of FASN by KRAS. We analyzed cellular metabolites from KRASMUT LUAD lines and observed that KRASMUT LUAD cell lines with LAKR had higher levels of long chain fatty acid and phospholipids compared to those without LAKR (Figure 2A and 2B). This is consistent with established models showing that that KRAS activation can stimulate expression of FASN, which catalyzes the condensation of acetyl-CoA and malonyl-CoA, an early step in the synthesis of long-chain fatty acids (Figure 2C). To confirm that LAKR upregulates FASN, we evaluated FASN expression in KRASMUT LUAD cell lines with or without LAKR. FASN was almost uniformly upregulated in KRASMUT LAKR cancer cell lines compared to LAKR-negative cell lines (Figure 2D).
Figure 2.
LAKR correlates with increased lipogenesis in KRASMUT LUAD cell lines. (A) Metabolites in 31 KRASMUT LUAD cell lines were analyzed using the data from CCLE database. Mean fold change of metabolites in KRASMUT LUAD cell lines with LAKR (n = 15) vs. no LAKR (n = 16). Red circles represent significantly upregulated metabolites in LAKR positive cell lines. (B) Alternative view of metabolites shown in A. 1: aconitate; 2: CMP; 3: alpha-hydroxybutyrate; 4: C18:1 LPC; 5: C18: LPE; 6: C32:0 PC; 7: C34:1 PC; 8: C36:1 PC; 9: C16:1 SM; 10: C24:0 SM. (C) Schematic model of impact of KRASMUT LAKR on fatty acid (FA) synthesis. DAG, diacylglycerol; FA, fatty acid; LPA, lysophosphatidic acid; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine. (D) Expression of FASN is upregulated in KRASMUT LUAD cell lines with LAKR. H1155 and H650 were used as controls to compare FASN on two separate blots (lower blot). 1: H1155; 2: H650; 3: H2122; 4: H2030; 5: H23; 6: HCC44; 7: SW1573; 8: H1373; 9: H460;10: Calu-6; 11: Calu-1; 12: H358; 13: H1792; 14: H1573; 15: H441
3.4. KRASMUT LUAD cells with LAKR are sensitive to FASN inhibition.
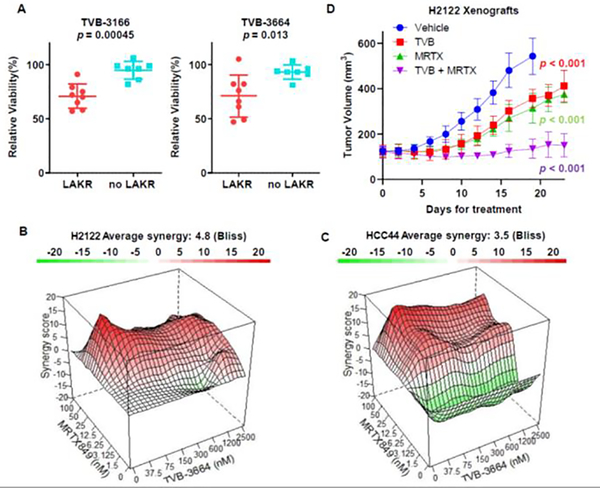
Inhibition of FASN in KRASG12D driven mouse models of lung cancer decreases tumor formation, leading to the hypothesis that FASN inhibitors may have therapeutic implications for RASMUT lung cancer [25]. In light of our findings that KRASMUT LAKR was associated with increased FASN expression, we considered that KRASMUT LAKR might show vulnerability to FASN inhibitors. We treated eight LAKR KRASMUT LUAD cell lines and seven non-LAKR KRASMUT lines with multiple FASN inhibitors, including TVB-3116, TVB-3664, fasnall and cerulenin. On average, FASN inhibitors showed better anti-proliferative activity in KRASMUT lung cancer cell lines with LAKR than lines without LAKR (Figures 3A, S2A and S2B).
Figure 3.
KRASG12C LAKR LUAD cell lines show increased sensitivity to FASN inhibition and show synergy with direct inhibition of KRASG12C. (A) KRASG12C LUAD cell lines were treated by FASN inhibitors, TVB-3116 or TVB-3664 for 7 days. Then cell viability at the dose of 0.075 μM was measured by CellTiter-Glo. (B,C) The KRASG12C inhibitor MRTX849 and FASN inhibitor TVB-3664 are synergistic in LUAD cells lines with KRASG12C LAKR. Synergy between MRTX849 and TVB-3664 was assessed at 5 days. Cell viability was determined by CellTiter-Glo. Bliss scores were computed by R synergyfinder package. (D) H2122 xenograft tumor models show synergy between MRTX849 and TVB-3664 in vivo. Therapy started when tumors reached approximately 120 mm3, therapy was initiated with 5 mg/kg TVB-3664, 10 mg/kg MRTX849 only, 5 mg/kg TVB-3664 + 10 mg/kg MRTX849, or vehicle (8 mice/group). Both inhibitors were delivered orally and were given daily. The results were shown as mean ± stdev.
KRASG12C inhibitors have shown activity in clinical trials [26–28]. However, development of resistance is an area of immediate concern, given that acquired resistance is common for targeted therapies in general and because cases of resistance to KRASG12C inhibitors have already been observed [29–32]. Although a prior report did not show clear associations between LAKR and sensitivity to KRASG12C inhibitors, SW1573, HCC44 and H2122, all KRASG12C-bearing tumors with LAKR, show some degree of resistance to KRASG12C inhibitors [33, 34]. We hypothesized that combining FASN and KRASG12C inhibitors might be synergistic in KRASG12C tumors with LAKR and provide an avenue to deal with therapeutic resistance. Combination treatment with MRTX849, a KRASG12C inhibitor in human trials, and TVB-3664, a close analogue of a FASN inhibitor in human trials, showed synergy as measured by the Bliss metric in H2122, HCC44, and SW1573 cell lines (Figure 3B and Figure S3A). Of note, we did not observe synergy for H358, a KRASG12C LUAD line without LAKR (Figure S3B) and H460, a KRASQ61H LUAD line (Figure S3C).
To evaluate if these effects could translate to in vivo cancer models, we performed a trial using an H2122 mouse xenograft model. Of note, this line was categorized as partially sensitive when mice were dosed at 100 mg/kg of MRTX849 [34]. We dosed mice with 5 mg/kg of TVB-3664 and 10 mg/kg of MRTX849 for approximately 3 weeks. All treatment regimens were well tolerated by the mice which showed stable weights throughout (Figure S4). Combination treatment was substantially better than the single agent arms and was able to halt tumor growth (Figure 3D). Of note, this is similar to effects previously achieved by single arm treatment of 100 mg/kg (10-fold higher) of MRTX849 [34]. These results suggest that concurrent inhibition of FASN and KRASG12C may be an effective strategy for overcoming resistance to KRASG12C inhibitors in patients with KRASG12C LAKR LUAD.
4. Discussion
Here we identify a high-risk lung cancer patient population, KRASMUT LUAD with LAKR. We also identify a potential strategy to intervene in this population, based on a new connection between LAKR and upregulation of lipid biogenesis at the level of FASN. Along these lines, we found that FASN dependence results in sensitivity to FASN inhibitors in KRASMUT LAKR. We further showed that inhibition of FASN was synergistic with direct KRASG12C inhibition in KRASG12C inhibitor-resistant cancer cells with RASG12C LAKR. These findings could quickly translate into a new targeted therapy paradigm for a subset of high-risk lung cancers but may have implications for other RAS-driven diseases as well.
Evaluating for LAKR in KRASMUT LUAD may improve the ability to prognosticate for LUAD, given the strong association between LAKR and mortality in our study, with an approximate doubling in median survival from 16 to 30 months for non-LAKR vs. LAKR KRASMUT NSCLC. If effective therapies targeted to KRASMUT LAKR can be discovered, it is conceivable this could alter clinical approaches to KRASMUT LUAD, especially given evolving attitudes and practices around other actionable molecular markers in LUAD, such as EGFR and ALK. Indeed, recent analysis showed that the presence of EGFR mutations or ALK rearrangements is associated with a quadrupling of median survival relative to no alterations (48 months vs. 12 months respectively) [35]. This has reshaped management of these patients with local therapies such as stereotactic radiation playing a more prominent role in the management of metastatic disease [36].
Based on our results, use of FASN inhibitors represents one possible therapeutic strategy for patients with RASMUT LAR. Using FASN inhibitors as an anti-cancer strategy is not a new idea. Cerulenin, an antifungal that irreversibly inhibits FASN, was identified in the 1970’s [37], and has been widely studied in multiple cancer models. However, cerulenin is too toxic for use in humans [38]. A number of other FASN inhibitors have subsequently been developed, such as GSK2194069, benzimidazole, 4-hydroxyquinoline, piperazine diamide, fasnall, and TVB-2640 [39]. Early phase trials of TVB-2640 have shown safety, but generally modest responses in patients with advanced KRASMUT NSCLC, breast and ovarian cancers [40]. Trials designed to evaluate the efficacy of TVB-2640 in specific cohorts of colon cancer, breast cancer, high grade astrocytoma and lung cancer are ongoing [41–43]. These trials primarily consider metabolic biomarkers, although NCT03808558 uses any KRAS mutation as an enrollment criterion. However, LAKR is not considered. Based on our results, adding LAKR as an evaluation criterion may enable identification of a high-risk subgroup that is more likely to benefit from TVB-2640 treatment. Additionally, our results showed synergy of FASN with MRTX1257 in KRASG12C inhibitor-resistant lines, suggesting a potential treatment strategy for patients with KRASG12C tumors who are resistant to KRASG12C therapies. Additional study on the potential value of LAKR as a predictive biomarker in these populations is warranted.
Aside from FASN, other therapeutic approaches may also be effective for RASMUT LAR. LAR is associated with an increase in sensitivity to MEK inhibition in lung cancer [5] and myeloid leukemia (AML) [17]. Nevertheless, these associations have not been validated in large sample sets, and it is unknown if other variables, such as other aspects of genetic background or cancer type, are important. This may be a significant issue in diseases such as NSCLC where genetic heterogeneity is high [44]. It also may be possible to discover new therapeutic innovations related to the physical mechanisms that underlie the tumor suppressive effects of RASWT. Current data suggests that suppression by RASWT is rooted in RAS oligomerization [5]. Nevertheless, RAS complexes have been difficult to isolate and characterize leading to the assumption that these complexes are likely dynamic and may take multiple forms, each with a different biological function. [15, 16, 45]. Related to that concept, it is possible that different RAS mutations impact the state of RAS assemblies and related LAR-dependent phenomenon in different ways. If this is true, RAS mutation-specific strategies may be required. Regardless, multiple studies now suggest methods by which RAS complexes might be perturbed [46, 47]. It is possible some of these could have therapeutic utility in the setting of KRASMUT LAKR.
In addition to finding LAKR in a sizable proportion of KRASMUT LUAD, we also found a similar proportion of LAR within the context of NRASMUT SKCM (melanoma). Whether LAR functions similarly in this setting will require additional study given that RAS isoforms and specific RAS mutations show biochemical and biological differences[48, 49]. Nevertheless, it is worth noting the potential to extend the utility of LAR-directed approaches beyond lung cancer. It is also worth noting that RAS genes are often amplified in cancers[17]. This phenomenon has the potential to show mechanistic similarities to LAKR, but further study is also required.
Because of its retrospective nature, the clinical associations observed in this study are subject to bias and should be verified by prospective study. Indeed, it is unclear what selection criteria may apply to cases within the clinical data set we evaluated. This study also suffers from small numbers, although this did not prevent us from finding statistical differences related to LAKR. Despite the above caveats, these results add evidence to the idea that KRASWT plays a suppressive role in human KRASMUT LUAD and that LAKR is a potentially useful predictive and prognostic biomarker [5, 12]. Moreover, these results could have immediate relevance in conjunction with KRASG12C inhibitors. Prospective studies to validate these findings are warranted.
Supplementary Material
Highlights.
Loss of wild type KRASWT (LAKR) in KRASMUT lung adenocarcinoma is common.
LAKR is associated with cancer mortality in KRASMUT lung adenocarcinoma.
FASN is upregulated in KRASMUT lung adenocarcinoma with LAKR, but this confers sensitivity to FASN inhibitors.
Combination treatment with FASN and KRASG12C inhibitors is synergistic and overcomes resistance to KRASG12C inhibitors in vivo.
Acknowledgments
Funding statement: This work was supported by CPRIT RP170373, ACS RSG-18-039-01-DMC and R01 CA244341 grants to K.D.W. and SPORE P50 CA070907, and CPRIT RP160652 to J.D.M. This work was also supported by NCI P30 CA 142543.
Abbreviations
- AML
acute myeloid leukemia
- COAD
colon adenocarcinoma
- FASN
fatty acid synthase
- LAKR
LOH at KRAS
- LAR
LOH at any RAS
- LOH
loss of heterozygosity
- LUAD
lung adenocarcinoma
- NSCLC
Non-small lung cancer lines
- PAAD
pancreatic adenocarcinoma
- SKCM
skin cutaneous melanoma
- TCGA
The Cancer Genome Atlas
Footnotes
Conflict of interest statement: K.D.W has received consulting fees from Sanofi Oncology and Vibliome Therapeutics. J.D.M. receives royalties from the NIH and UT Southwestern for distribution of cell lines. Other authors report no commercial conflicts of interest connected to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFEREENCES
- [1].Prior IA, Lewis PD, Mattos C, A comprehensive survey of Ras mutations in cancer, Cancer Res 72(10) (2012) 2457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhou B, Der CJ, Cox AD, The role of wild type RAS isoforms in cancer, Seminars in cell & developmental biology, Elsevier, 2016, pp. 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Network CGAR, Integrated genomic analyses of ovarian carcinoma, Nature 474(7353) (2011) 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, Konukiewitz B, Ollinger R, Zwiebel M, Strong A, Yen HY, Banerjee R, Louzada S, Fu B, Seidler B, Gotzfried J, Schuck K, Hassan Z, Arbeiter A, Schonhuber N, Klein S, Veltkamp C, Friedrich M, Rad L, Barenboim M, Ziegenhain C, Hess J, Dovey OM, Eser S, Parekh S, Constantino-Casas F, de la Rosa J, Sierra MI, Fraga M, Mayerle J, Kloppel G, Cadinanos J, Liu P, Vassiliou G, Weichert W, Steiger K, Enard W, Schmid RM, Yang F, Unger K, Schneider G, Varela I, Bradley A, Saur D, Rad R, Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes, Nature 554(7690) (2018) 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, Capelletti M, Caffarra C, Li S, Lv Q, Gondi S, Hunter JC, Lu J, Chiarle R, Santamaria D, Westover KD, Janne PA, KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS, Cell 172(4) (2018) 857–868 e15. [DOI] [PubMed] [Google Scholar]
- [6].Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A, The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants, Science 277(5324) (1997) 333–8. [DOI] [PubMed] [Google Scholar]
- [7].Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, Chen JH, Firdaus SJ, Babbar A, Ren P, Liu Y, Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State, Cancer Discov 6(3) (2016) 316–29. [DOI] [PubMed] [Google Scholar]
- [8].Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD, Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations, Mol Cancer Res 13(9) (2015) 1325–35. [DOI] [PubMed] [Google Scholar]
- [9].Poulin EJ, Bera AK, Lu J, Lin YJ, Strasser SD, Paulo JA, Huang TQ, Morales C, Yan W, Cook J, Nowak JA, Brubaker DK, Joughin BA, Johnson CW, DeStefanis RA, Ghazi PC, Gondi S, Wales TE, Iacob RE, Bogdanova L, Gierut JJ, Li Y, Engen JR, Perez-Mancera PA, Braun BS, Gygi SP, Lauffenburger DA, Westover KD, Haigis KM, Tissue-Specific Oncogenic Activity of KRAS(A146T), Cancer Discov 9(6) (2019) 738–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Parker JA, Mattos C, The K-Ras, N-Ras, and H-Ras Isoforms: Unique Conformational Preferences and Implications for Targeting Oncogenic Mutants, Cold Spring Harb Perspect Med 8(8) (2018) a031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, Edwards PA, Smith PD, Cook SJ, Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells, Sci Signal 4(166) (2011) ra17. [DOI] [PubMed] [Google Scholar]
- [12].Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, Anderson MW, Sills RC, Hong HL, Devereux TR, Jacks T, Guan KL, You M, Wildtype Kras2 can inhibit lung carcinogenesis in mice, Nat Genet 29(1) (2001) 25–33. [DOI] [PubMed] [Google Scholar]
- [13].Guerrero I, Villasante A, Corces V, Pellicer A, Loss of the normal N-ras allele in a mouse thymic lymphoma induced by a chemical carcinogen, Proc Natl Acad Sci U S A 82(23) (1985) 7810–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bremner R, Balmain A, Genetic changes in skin tumor progression: correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7, Cell 61(3) (1990) 407–17. [DOI] [PubMed] [Google Scholar]
- [15].Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S, Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway, Proc Natl Acad Sci U S A 112(26) (2015) 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen M, Peters A, Huang T, Nan X, Ras Dimer Formation as a New Signaling Mechanism and Potential Cancer Therapeutic Target, Mini Rev Med Chem 16(5) (2016) 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, Firestone AJ, Young A, Lacap JA, Crocker L, Asthana S, Davis EM, Xu J, Akagi K, Le Beau MM, Li Q, Haley B, Stokoe D, Sampath D, Taylor BS, Evangelista M, Shannon K, KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer, Cell 168(5) (2017) 817–829 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Merajver SD, Frank TS, Xu J, Pham TM, Calzone KA, Bennett-Baker P, Chamberlain J, Boyd J, Garber JE, Collins FS, et al. , Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer, Clin Cancer Res 1(5) (1995) 539–44. [PubMed] [Google Scholar]
- [19].Tomczak K, Czerwinska P, Wiznerowicz M, The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge, Contemp Oncol (Pozn) 19(1A) (2015) A68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gouw AM, Eberlin LS, Margulis K, Sullivan DK, Toal GG, Tong L, Zare RN, Felsher DW, Oncogene KRAS activates fatty acid synthase, resulting in specific ERK and lipid signatures associated with lung adenocarcinoma, Proc Natl Acad Sci U S A 114(17) (2017) 4300–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, Sofia HJ, Hutter C, Getz G, Wheeler D, Ding L, Group MCW, N Cancer Genome Atlas Research, Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines, Cell Syst 6(3) (2018) 271–281 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ, N. Cancer Genome Atlas Research, Cherniack AD, Beroukhim R, Meyerson M, Genomic and Functional Approaches to Understanding Cancer Aneuploidy, Cancer Cell 33(4) (2018) 676–689 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].El Osta B, Behera M, Kim S, Berry LD, Sica G, Pillai RN, Owonikoko TK, Kris MG, Johnson BE, Kwiatkowski DJ, Sholl LM, Aisner DL, Bunn PA, Khuri FR, Ramalingam SS, Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience, J Thorac Oncol 14(5) (2019) 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Padanad MS, Konstantinidou G, Venkateswaran N, Melegari M, Rindhe S, Mitsche M, Yang C, Batten K, Huffman KE, Liu J, Tang X, Rodriguez-Canales J, Kalhor N, Shay JW, Minna JD, McDonald J, Wistuba II, DeBerardinis RJ, Scaglioni PP, Fatty Acid Oxidation Mediated by Acyl-CoA Synthetase Long Chain 3 Is Required for Mutant KRAS Lung Tumorigenesis, Cell Rep 16(6) (2016) 1614–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Singh A, Ruiz C, Bhalla K, Haley JA, Li QK, Acquaah-Mensah G, Montal E, Sudini KR, Skoulidis F, Wistuba II, Papadimitrakopoulou V, Heymach JV, Boros LG, Gabrielson E, Carretero J, Wong KK, Haley JD, Biswal S, Girnun GD, De novo lipogenesis represents a therapeutic target in mutant Kras non-small cell lung cancer, FASEB J (2018) fj201800204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, Lanman BA, Werner J, Rapaport AS, San Miguel T, Ortiz R, Osgood T, Sun JR, Zhu X, McCarter JD, Volak LP, Houk BE, Fakih MG, O’Neil BH, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Hong DS, Ouyang W, Henary H, Arvedson T, Cee VJ, Lipford JR, The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity, Nature 575(7781) (2019) 217–223. [DOI] [PubMed] [Google Scholar]
- [27].Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA, The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients, Cancer discovery 10(1) (2020) 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fakih M, O’Neil B, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Rasmussen E, Morrow PKH, Ngang J, Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors, American Society of Clinical Oncology, 2019. [Google Scholar]
- [29].I. Mirati Therapeutics, Phase 1/2 Study of MRTX849 in Patients With Cancer Having a KRAS G12C Mutation, 2020. https://ClinicalTrials.gov/show/NCT03785249.
- [30].Janssen R, L.L.C. Development, First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation. https://ClinicalTrials.gov/show/NCT04006301.
- [31].Amgen A Phase 1/2, Study Evaluating the Safety, Tolerability, PK, and Efficacy of AMG 510 in Subjects With Solid Tumors With a Specific KRAS Mutation (CodeBreak 100). https://ClinicalTrials.gov/show/NCT03600883.
- [32].Amgen AMG 510 Activity in Subjects With Advanced Solid Tumors With KRAS p.G12C Mutation (CodeBreak 101). https://ClinicalTrials.gov/show/NCT04185883.
- [33].Hallin J, Aranda R, Baer BR, Briere DM, Burkard MR, Calinisan A, Chiang H, Engstrom LD, Fell JB, Fischer JP, Hargis L, Marx MA, Olson P, Sudhakar N, Christensen JG, Insight Towards Therapeutic Susceptibility of KRAS Mutant Cancers from MRTX1257: A Prototype Selective Inhibitor of KRAS G12C, 2018. https://www.mirati.com/wp-content/uploads/2018/12/AACR_RAS_Poster_-Insight.pdf. (Accessed March 12, 2020 2020).
- [34].Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA, Burkard MR, Fell JB, Fischer JP, Vigers GP, Xue Y, Gatto S, Fernandez-Banet J, Pavlicek A, Velastagui K, Chao RC, Barton J, Pierobon M, Baldelli E, Patricoin EF 3rd, Cassidy DP, Marx MA, Rybkin II, Johnson ML, Ou SI, Lito P, Papadopoulos KP, Janne PA, Olson P, Christensen JG, The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients, Cancer Discov 10(1) (2020) 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sperduto PW, Yang TJ, Beal K, Pan H, Brown PD, Bangdiwala A, Shanley R, Yeh N, Gaspar LE, Braunstein S, Sneed P, Boyle J, Kirkpatrick JP, Mak KS, Shih HA, Engelman A, Roberge D, Arvold ND, Alexander B, Awad MM, Contessa J, Chiang V, Hardie J, Ma D, Lou E, Sperduto W, Mehta MP, Estimating Survival in Patients With Lung Cancer and Brain Metastases: An Update of the Graded Prognostic Assessment for Lung Cancer Using Molecular Markers (Lung-molGPA), Jama Oncol 3(6) (2017) 827–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].N.C.C. Network, Non-Small Cell Lung Cancer (Version 5.2019), 2019. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. (Accessed July 24,2019.
- [37].Nomura S, Horiuchi T, Omura S, Hata T, The action mechanism of cerulenin. I. Effect of cerulenin on sterol and fatty acid biosynthesis in yeast, J Biochem 71(5) (1972) 783–96. [DOI] [PubMed] [Google Scholar]
- [38].Pizer ES, Chrest FJ, DiGiuseppe JA, Han WF, Pharmacological inhibitors of mammalian fatty acid synthase suppress DNA replication and induce apoptosis in tumor cell lines, Cancer Res 58(20) (1998) 4611–5. [PubMed] [Google Scholar]
- [39].Angeles TS, Hudkins RL, Recent advances in targeting the fatty acid biosynthetic pathway using fatty acid synthase inhibitors, Expert Opinion on Drug Discovery 11(12) (2016) 1187–1199. [DOI] [PubMed] [Google Scholar]
- [40].Dean EJ, Falchook GS, Patel MR, Brenner AJ, Infante JR, Arkenau H-T, Borazanci EH, Lopez JS, Pant S, Schmid P, Preliminary activity in the first in human study of the first-in-class fatty acid synthase (FASN) inhibitor, TVB-2640, American Society of Clinical Oncology, 2016. [Google Scholar]
- [41].Mark E, University of K, TVB 2640 for Resectable Colon Cancer Other Resectable Cancers; a Window Trial, 2020. [Google Scholar]
- [42].Mayo C, National Cancer I, FASN Inhibitor TVB-2640, Paclitaxel, and Trastuzumab in Treating Patients With HER2 Positive Advanced Breast Cancer.
- [43].C. University of Texas Southwestern Medical, Phase 2 Study of TVB-2640 in KRAS Non-Small Cell Lung Carcinomas, 2020.
- [44].C.G.A.R. Network, Comprehensive molecular profiling of lung adenocarcinoma, Nature 511(7511) (2014) 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Inouye K, Mizutani S, Koide H, Kaziro Y, Formation of the Ras dimer is essential for Raf-1 activation, J Biol Chem 275(6) (2000) 3737–40. [DOI] [PubMed] [Google Scholar]
- [46].Tran TH, Alexander P, Dharmaiah S, Agamasu C, Nissley DV, McCormick F, Esposito D, Simanshu DK, Stephen AG, Balius TE, The small molecule BI-2852 induces a nonfunctional dimer of KRAS, Proc Natl Acad Sci U S A 117(7) (2020) 3363–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, Cobbert J, Lavoie H, Smith M, Rajakulendran T, Dowdell E, Okur MN, Dementieva I, Sicheri F, Therrien M, Hancock JF, Ikura M, Koide S, O’Bryan JP, Inhibition of RAS function through targeting an allosteric regulatory site, Nat Chem Biol 13(1) (2017) 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Haigis KM, KRAS alleles: the devil is in the detail, Trends in cancer 3(10) (2017) 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Montalvo SK, Li L, Westover KD, Rationale for RAS mutation-tailored therapies, Future Oncology 13(3) (2017) 263–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.