Abstract
MicroRNAs (miRNAs) have recently come under scrutiny for their role in various age-related diseases. Similarly, cellular senescence has been linked to disease and aging. MicroRNAs and senescence likely play an intertwined role in driving these pathologic states. In this review, we present the connection between these two drivers of age-related disease concerning mesenchymal stem cells (MSCs). First, we summarize key miRNAs that are differentially expressed in MSCs and other musculoskeletal lineage cells during senescence and aging. Additionally, we also reviewed miRNAs that are regulated via traditional senescence-associated secretory phenotype (SASP) cytokines in MSC. Lastly, we summarize miRNAs that have been found to target components of the cell cycle arrest pathways inherently activated in senescence. This review attempts to highlight potential miRNA targets for regenerative medicine applications in age-related musculoskeletal disease.
Keywords: MicroRNAs, senescence, Aging, Mesenchymal Stem Cells
Introduction
Cellular senescence was first reported by Hayflick and Moorhead in 1961 when they observed serial passaging of human fibroblasts results in irreversible cell cycle arrest (1). Since then, senescence has been well characterized as a stress response that prevents the proliferation of dysfunctional cells by inducing an irreversible cell cycle arrest (2). It has been well established that cellular senescence occurs in response to DNA damage, oncogene mutations, oxidative stress, damage-associated molecular pattern molecules from diseased tissue, and various other cellular insults (3, 4).
Senescent cells exhibit structural changes that include enlarged and flattened morphology, altered plasma membrane composition, accumulation of lysosomes and mitochondria, and nuclear chromatin changes (3). Although senescent cells are impaired of their normal function, they remain metabolically active and have increased resistance to apoptosis (5). These cells often exhibit persistent activation of the DNA damage response with concomitant increases in cyclin-dependent kinase (CDK) inhibitors (6, 7). CDKs regulate various proteins necessary for progression through the cell cycle and prevent advancement when inhibited. The CDK inhibitors p16INK4a (encoded by CDKN2A) and p21 (encoded by CDKN1A) have been highly implicated in senescence, along with their respective tumor suppressor pathway components retinoblastoma (Rb) and p53 (8). Specifically, p16 inhibits the G1 to S phase transition via inhibition of CDK4 and CDK6, thus preventing the phosphorylation of pRb (9). p21 acts similarly via inhibition of CDK2 among other CDKs (10). Senescent cells are also well characterized by their altered secretome consisting of proinflammatory cytokines, chemokines, and extracellular matrix proteins, collectively known as the senescence-associated secretory phenotype (SASP) (11, 12). When secreted into the extracellular milieu the SASP creates a toxic microenvironment that affects neighboring cells in a paracrine manner, likely inducing additional senescent cell accumulation (13).
Despite the growing implications of cellular senescence, identification of senescent cells remains challenging as the phenotypic expression is variable, and no marker is particularly specific to senescence (14). Of the tumor suppressor markers, p16 remains one of the most highly used cellular senescence markers (15). Furthermore, increased lysosomal content and activity of senescent cells have been leveraged for identification of senescent cells via the lysosomal enzyme senescence-associated β-galactosidase (SA β-Gal) (16). Detection of SA β-Gal has become one of the most widely accepted markers of senescence. Among other notable markers of senescence includes the downregulation of lamin B1, a structural component of the nuclear lamina (17). Impaired production of lamin A has also garnered attention for a potential role in senescence (18). Given the array of features mentioned above, there appears to be a growing need to develop biomarker panels specific for each senescence subtype (19).
Senescent cells’ ability to resist growth in the presence of growth stimuli and oncogenic insults highlights the widely accepted role of senescence as a tumor suppressor – a speculated theory for the evolutionary development of senescence (20–22). Senescent cells have also been well recognized for their beneficial role in various other physiologic contexts. Developmentally programmed senescence occurs during mammalian embryonic development, and its impairment leads to detectable developmental abnormalities (23, 24). In a similar tissue remodeling context, senescence has also been found to direct wound healing and tissue repair (25–27). Despite the established benefits of senescent cells, deleterious effects can occur when prolonged senescence persists and becomes chronic. For instance, senescent cells are recognized for their role in tissue dysfunction with aging and age-related diseases as well as their promotion of tumorigenesis (28).
Mounting evidence indicates these deleterious effects are normally prevented by the removal of senescent cells via the immune system, termed senescence surveillance (29–31). Natural killer cells target senescent cells via perforin exocytosis, bypassing the upregulated anti-apoptotic pathway, while macrophages and T cells known to clear senescent cells through less understood mechanisms (32, 33). The SASP has been implicated in the recruitment and activation of immune cells, highlighting one of its beneficial roles (32). However, chronic secretion of SASP drives tissue dysfunction and age-related pathologic conditions (28, 34). Interestingly, senescent cell numbers have been found to correlate with organism age across various species, including rodents, primates, and humans (35–37). Multiple proposed mechanisms for this observed phenomenon exist, likely acting together in concert. In advanced age, the immune system’s impaired function is thought to decrease the clearance of senescent cells (28). Ovadya et al. (2018) demonstrated immune function impairment in mice (perforin gene knockout) resulted in a higher burden of senescence and age-related disorders while lowering survival (31). Notably, senolytic therapy, a class of drugs that target the senescent cell anti-apoptotic pathway (SCAP) for removing senescent cells, reversed the age-related phenotype in these mice. Furthermore, chronic senescence is thought to result from stress-induced macromolecular damage that accumulates over an organism’s lifespan (28, 38). Oxidative stress levels are known to increase in aging and could very well contribute to increased senescence. Both scenarios lead to chronic elevation of the SASP, which induces paracrine senescence and further increases the overall burden of senescence.
Without exception, the musculoskeletal system has also been found to accumulate senescent cells with normal chronological aging. In aged bone osteoblast progenitors, osteoblasts, and osteocytes express increased senescent markers with osteocytes also express significantly higher SASP markers (39). In aged muscle, satellite cells shift from a quiescent resting state to a presenecent state that results in deep senescence under proliferative pressure (40). Many reports suggest that the accumulation of senescent cells in old age is detrimental and drives physiological aging while reducing a healthy lifespan (41–43). One study found that individuals with greater senescent cell numbers possess a higher risk of experiencing frailty (44). Consistent with this finding, suppression of the SASP has been shown to alleviate frailty in aged mice (45). Furthermore, transplantation of senescent cells into young and old mice causes persistent physical dysfunction and reduced survival (46).
Senolytic therapy in senescent transplanted mice and naturally aged mice reduces physical dysfunction and mortality while increasing lifespan. Senolytic therapy has also been shown to alleviate age-related bone loss in mice, resulting in anti-resorptive and anabolic effects on bone (47). These findings suggest a role for senescence in the pathophysiology of osteoporosis. Senescence is also implicated in the pathophysiology of osteoarthritis (48). Increased levels of senescent chondrocytes have been found in damaged cartilage of osteoarthritis patients (49). Furthermore, transplantation of senescent cells can induce an osteoarthritis-like state in mice (50). Additionally, senescent cells accumulate in the articular cartilage and synovium of mice after articular joint injury, and selective removal of these cells via senolytics attenuates the development of post-traumatic osteoarthritis (51). Senescence has also been implicated in sarcopenia’s pathophysiology, with clearance of senescent cells resulting in delayed onset of lordokyphosis (a measure of sarcopenia) in progeria mice (41, 42). In addition to their role in musculoskeletal disease, senescent cells are implicated in many other diseases ranging from atherosclerosis to idiopathic pulmonary fibrosis and diabetic nephropathy (52–54). Building upon the aforementioned results in animal models, clinical trials are underway to evaluate senolytics for their safety and efficacy in humans. The first-in-human trial of senolytics was performed on a small cohort of patients with idiopathic pulmonary fibrosis. It resulted in a significant and clinically meaningful improvement in physical function (55). Subsequent studies have demonstrated that senloytic treatment effectively reduces senescent cell burden and plasma levels of SASP factors in multiple disease states (56, 57). Ongoing trials aim to evaluate efficacy in the musculoskeletal application, such as the treatment of osteoarthritis (NCT04210986).
In the field of regenerative tissue engineering, mesenchymal stem cells (MSCs) have become a cornerstone of therapeutic potential. MSCs have vast implications in age-related diseases. Declines in MSC pools have been linked with premature aging disorders such as Werner progeria syndrome (58). Furthermore, the transplantation of young MSCs has been shown to increase the lifespan of progeria mice (59). More specifically, MSC dysfunction and attrition have been linked to age-related musculoskeletal pathology. Thus, MSCs play a vital role in aging and associated disease. While MSCs have been proposed for many therapies, limitations to their efficacy do exist. Many of these limitations, such as poor proliferation, migration, and differentiation properties, are potentially related to cellular senescence (60–62). Before autologous injection, MSCs often require expansion in vitro, which provides an opportunity for replicative senescence (63). Furthermore, MSCs taken from older individuals are likely to have a higher burden of senescence (64, 65). The ability to remove these detrimental cells before host grafting could significantly improve therapeutic potential.
The altered protein expression of senescent cells is well studied and characterized. Recent studies demonstrated the involvement of microRNAs (miRNAs) in MSCs senescences. MiRNAs are a form of epigenetic regulation that alters gene expression without changing genetic code. MiRNAs are short non-coding nucleotide sequences that selectively bind to mRNA sequences at the 3’-untranslated regions (3’-UTR). Upon binding, the miRNA inhibits translation or promotes the target mRNA’s degradation, thus negatively regulating the gene expression at the post-transcriptional level (66). MiRNAs have been studied for their role within cellular senescence (67, 68). However, their role in MSCs senescence and aging has not been fully characterized. With this review, we hope to highlight potential miRNAs modulating senescence in MSCs.
MiRNAs Differentially Expressed in Senescent MSCs
miRNA has been found to play critical roles in many age-related diseases, including cardiovascular and neurological diseases previously reviewed by Dimmeler and Nicotera (69). Studies have gone on to implicate various miRNA in musculoskeletal disease as well (70). As mentioned previously, there is a growing body of evidence showing senescent cells drive a wide spectrum of diseases and pathological states. Musculoskeletal disease and aging are without exception. Differentially expressed miRNA in cellular senescence has been well reported, with most results predominantly focusing on senescent fibroblasts (67, 68). In this review, we will turn our attention to MSCs. Numerous differentially expressed miRNA have been identified in senescent MSCs, and resultantly these likely contribute to disease and aging of the musculoskeletal system.
miRNA previously identified as cell cycle regulators have been found to correlate with senescence. For example, the miR-17 family is recognized as cell cycle regulators via direct targeting of p21 (71). miR-93 and miR-20a, which reside in the miR-17 family, have been found to decrease in senescent MSCs (72) and target p21 (73–76). The decline in these miRNAs levels is critical in the concomitant up-regulation of p21, and the overexpression of these miRNAs significently attenuate senescence (73–76). Indirect regulation of cell cycle arrest proteins p21 and p53 via miRNA has been reported in early senescence. Okada et al. (2016) reported elevated levels of miR-195 in old aged donor MSCs and senescent MSCs (77). This miRNA directly targets the 3’UTR of SIRT1 and TERT (77, 78). SIRT1 is a regulator of p53 deacetylation and known for its ability to inhibit senescence in various cell types (79). TERT encodes for telomerase, which prevents telomere shortening, a widely observed phenomenon in cellular senescence (80, 81). miR-195 inhibition in old MSCs induces telomere lengthening, increases SIRT1 expression, decreases p53 expression, and decreases SA B-galactosidase activity (77). miR-486-5p has also been found to increase in old MSCs derived from adipose tissue (82). Forced expression of this miRNA inhibits osteogenic and adipogenic differentiation and induces senescence of MSCs. Furthermore, this mRNA is known to target SIRT1, and miR-486-5p inhibitor results in the prevention of SIRT1 suppression. MicroRNA-204 is upregulated in stress-induced senescent chondrocytes and osteoarthritic cartilage (83). This miRNA has been shown to directly target SIRT1 in mouse embryonic stem cells (84). Ectopic expression of this miRNA is sufficient to trigger osteoarthritis development in mice, while knockdown ameliorates surgically induced osteoarthritis in mice and suppresses SASP development (83). Saunders et al. (2010) reported that miR-204 upregulated in senescent HUVECs (85).
Similarly, BMI1 is a known regulator of p16INK4a and inhibitor of senescence in various cell types (86). The miR-495 target BMI1 and induce the senescence of MSCs measured by increased SA B-galactosidase activity and elevated p16INK4a, p53, and p21 gene expressions (87). The conditioned media collected from miR-495 overexpressed MSCs has been found to inhibit the migration of multiple cell types, including MSCs. This is consistent with the known paracrine effect of the SASP to induce senescence in otherwise healthy neighboring cells.
Other miRNAs linked to senescence in MSCs are associated with lamin A. MiR-141-3p levels are elevated in senescent MSCs. This miRNA target ZMPSTE24, an enzyme that produces mature lamin by cleavage of prelamin A, leading to prelamin A accumulation within the nuclear envelope (88). Interestingly, the accumulation of prelamin A has been shown to drive senescence in MSCs. The other miRNAs that indirectly regulate lamin A are miR-543 and miR-590-3p. miR-543 and miR-590-3p target AIMP3/p18 to prevent the induction of cellular senescence in MSCs (89). These miRNAs decrease under senescence inducing conditions with a concomitant rise in AIMP3/p18. Supporting these findings, a group of researcher reported the overexpression of AIMP3/p18 cause progeria in mouse models and sufficient to induce senescence via downregulation of mature lamin A (89, 90).
Additional miRNAs are known to inhibit MSC proliferation, migration, and differentiation have also been implicated in senescence, such as miR-335 (91). This miRNA increases in aged MSCs as well as senescent MSCs induced by various insults such as γ-irradiation (92, 93). Forced miR-335 expression increases MSC senescence measured by SA B-galactosidase activity, p16INK4a levels, and SASP development with concomitant decreases in SOD2. It is also elevated in EVs of adult MSCs (94). When taken together, this suggests miR-335 may be a SASP component. This miRNA was shown to act through inhibition of AP-1 activity, a transcription factor known to promote bone formation (95). Several groups have also reported significant declines in miR-335* and miR-335-5p in aged MSCs (92, 96). As seen in Table 1, miR-335 is upregulated, while miR-335* is down-regulated across multiple studies.
Table 1.
Differentially expressed (up and downregulated) miRNA in senescent versus old MSCs/osteoblasts.
| Differentially Expressed - Up Regulated | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wagner et al (180) | Kundratos et al (181) | Yoo et al (118) | Meng et al (182) | Chen et al (183) | Li et al (184) | Okada et al (77) | Hackl et al (120) | Pandey et al (92) | |
| Senescent | Senescent | Senescent | Senescent | Senescent | Senescent | Old | Old | Old | |
| MSCs | MSCs | MSCs | MSCs | Osteoblasts | Osteoblasts | MSCs | MSCs | MSCs | |
| miR-132-3p | O | O | |||||||
| miR-136-5p | O | O | |||||||
| miR-140 | O** | O | |||||||
| miR-224 | O** | O | |||||||
| miR-27a-3p | O | O | |||||||
| miR-27b | O** | O | |||||||
| miR-30d | O | O** | |||||||
| miR-335 | O** | O | O | ||||||
| miR-34a | O** | O | |||||||
| miR-369-5p | O | O | |||||||
| Differentially Expressed - Down Regulated | |||||||||
| Wagner et al (180) | Kundratos et al (181) | Yoo et al (118) | Meng et al (182) | Chen et al (183) | Li et al (184) | Okada et al (77) | Hackl et al (120) | Pandey et al (92) | |
| Senescent | Senescent | Senescent | Senescent | Senescent | Senescent | Old | Old | Old | |
| MSCs | MSCs | MSCs | MSCs | Osteoblasts | Osteoblasts | MSCs | MSCs | MSCs | |
| let-7a | O** | O | |||||||
| miR-130b-5p | O | O | |||||||
| miR-15b-3p | O | O | O*** | ||||||
| miR-16 | O** | O | |||||||
| miR-17 | O | O | |||||||
| miR-188-3p | O | O | |||||||
| miR-199b-5p | O | O | |||||||
| miR-19b | O** | O | O | ||||||
| miR-20a-5p | O | O | O*** | ||||||
| miR-218-5p | O | O | |||||||
| miR-222-5p | O | O | |||||||
| miR-224-5p | O | O | |||||||
| miR-23b | O** | O | |||||||
| miR-29b | O | O | |||||||
| miR-335* | O | O | |||||||
| miR-337-3p | O | O | |||||||
| miR-424 | O | O | O | ||||||
| miR-483-3p | O | O | |||||||
| miR-7-5p | O | O | |||||||
| miR-935 | O | O | |||||||
Denotes the less predominant arm of the miRNA hairpin.
Denotes 3p or 5p arm was specified in the original literature but not reflected in this table.
Denotes miRNA arm not specified in the original literature.
Dimri et al. (2013) reported that miR-141 induces senescence in fibroblasts via targeting BMI1 with a concomitant rise in p16INK4a (97). Other known miR-141 gene targets in BMSCs includes BMI1, SDF-1, SVCT2, and DLX5 (98). These factors are known to regulate BMSC differentiation, migration, and stem cell proliferation (99–102). Previously, our group reported that miR-141 targets SDF-1 and vitamin C transporter (SVCT2) and inhibits osteogenic differentiation of MSCs. Furthermore, we showed that this miRNA expression increases with age in bone and MSCs of mice and humans. This suggests that miRNA-141 plays an important role in MSC aging (98, 102, 103).
MiR-1292 has been found to accelerate senescence and reduce osteogenesis in adipose-derived MSCs (104). The knockdown of this miRNA showed the opposite effect, decreasing senescence and enhancing osteogenesis. Similarly, in clinical bone samples, miR-1292 was positively correlated with senescence and negatively correlated with osteogenic markers ALP, OXN, and RUNX2. This miRNA mediates its effect through the Wnt/B-catenin pathway by targeting frizzled gene family member Frizzled-4 (FZD4). Another miRNA of interest is miR-188, which is elevated in BMSCs derived from aged mice and humans (105). This miRNA target MAP3K3 in lineage-negative bone marrow cells, whereas its overexpression enhances senescence (106). Moreover, knockout and inhibition of this miRNA demonstrated less age-related bone loss and bone marrow fat accumulation. Conversely, forced expression of miR-188 led to increased age-related bone loss and bone marrow fat accumulation. Histone deacetylase 9 (HDAC9) and RPTOR-independent companion of MTOR complex 2 (RICTOR) were identified as the direct targets of miR-188. This miRNA appears to be a key regulator in the age-related switch in BMSC differentiation from osteogenesis to adipogenesis with additional implications in senescence. MicroRNA-188 target various cyclins and CDKs (downstream of p21 and p16INK4a genes), but this has not yet been observed in MSCs (107).
Several studies utilize miRNA arrays to determine panels of differentially expressed miRNA in senescent MSCs. In Table 1, we present differentially expressed miRNA found in both senescent and aged MSCs/osteoblasts across multiple studies. Several miRNAs are found to be differentially expressed similarly in senescent versus aged MSCs and osteoblasts. These findings are consistent with the widely reported phenomenon that senescent cells accumulate in old age and suggest that aged MSCs exist in a presenescent state.
MiRNAs in the SASP of Senescent MSCs
It has been well recognized that senescent cells secrete a distinct profile of proteins, which has collectively been termed the senescence-associated secretory phenotype. The SASP factors serve to mediate an inflammatory response and contribute to various age-related pathological conditions (7, 11). Furthermore, the SASP is thought to possess the capability of inducing senescence in neighboring cells (13). If true, this would serve a protective effect within tumor microenvironments, as neoplastic cells would be turned off. However, it is known that the SASP induce many pro-neoplastic effects as well, such as angiogenesis and epithelial-mesenchymal transition (108, 109). There has been an indication that other molecules are also secreted in conjunction with the SASP, including miRNA within extracellular vesicles (EVs) (96, 110). Recently studies have shown that these secreted miRNA can stimulate senescence-like features in neighboring cells (111).
Our group recently identified that miR-183-5p increases with age in EVs isolated from bone marrow (112). We also reported that old bone marrow-derived EVs are taken up by young BMSCs and inhibit osteogenic differentiation. Furthermore, transfections of miR-183-5p mimic reduce cell proliferation, heme-oxygenase-1 (Hmox1) protein level, and increase senescence in BMSCS (60). Previously, another group also reported that the miR-183 cluster has anti-apoptotic roles in various malignancies, which is consistent with its suggested role in age-induced senescence (113). Moreover, we recently have shown that muscle-derived miR-34a increases with age in circulating extracellular vesicles and induces senescence in bone marrow stem cells (114). In vitro, myoblast and myotube derived EVs treated with hydrogen peroxide have elevated miR-34a-5p levels. Furthermore, these EVs home to and induce cellular senescence in BMSCs, likely through the miR-34a-5p targeting SIRT1. miR-34a targeting of SIRT1 has been previously well established across a variety of cell types. The overexpression of miR-34a in MSCs is known to induce apoptosis and senescence (5, 115). Further linking senescence to musculoskeletal disease, miR-34a has also been shown to contribute to osteoarthritis progression (116). This miRNA is upregulated across multiple age-related pathological conditions (see Table 1).
Terlecki-Zaniewicz and group (2018) reported that miR-17-3p and miR-199b-5p decrease in EVs of senescent fibroblasts (111). miR-199b-5p is also down-regulated in senescent MSCs and those from aged donors, as indicated in Table 1 (117, 118). miR-17-3p is down-regulated in senescent MSCs as well as skin fibroblasts aging models (119, 120). The miR-23a-5p expression upregulated in senescent fibroblasts and EVs secreted by these cells. In a separate study, miR-23a was reported to prevent TNFα induced osteoblast apoptosis via targeting Fas (121). Interestingly, miR-23a is also shown to regulate the osteogenic differentiation of BMSCs (122). Forced expression of this miRNA in MSCs promotes osteogenic differentiation by targeting TMEM64, whereas inhibition promotes adipogenic differentiation. However, another group found miR-23a cluster knockdown to decrease cellular differentiation of preosteoblasts (123). Furthermore, the miR-23a cluster has also target SATB2 directly and regulate TGF-B signaling in osteoblasts to promote terminal osteocyte differentiation (124). Further supporting its role in terminal differentiation, miR-23a levels are shown to increase throughout osteoblast differentiation (125). miR-23a is elevated in bone and serum of osteoporotic patients with fractures, possibly representing a rise in differentiating osteoblasts during fracture healing (126).
miR-31 is another circulating miRNA found to be differentially expressed with age. This miRNA is elevated in the plasma of elderly and osteoporotic patients (127). miR-31 is a master regulator of osteogenesis via targeting RUNX2, Osterix, and SATB2 (128, 129). It was recently found to be upregulated in replicative and stress-induced senescent endothelial cells of a variety of tissue origins (127). This miRNA also upregulated in senescent endothelial cells microvesicles. These microvesicles were found to inhibit osteogenic differentiation of MSCs by targeting Frizzled-3 (FZD3). Furthermore, miR-31 is known to induce senescence in cancer cells (130). In hepatocellular cancer, this miRNA directly targets CDK2 (131). This suggested that miR-31 is released as part of the SASP and potentially induce senescence in MSCs.
miRNAs in MSCs Regulated by Known SASP Factors
To date, the SASP is predominantly recognized for its secretory phenotype of several key proteins, previously reviewed by Coppe et al. (11). Most SASP factors are inflammatory cytokines, with prominent factors including IL1α, IL1β, IL6, and IL8. IL1β is known to induce p16INK4a in vitro, which is consistent with the ability of the SASP to induce paracrine senescence (132). The SASP is also comprised of shed cell surface molecules, including sTNFR1 and sTNFRII, known receptors of inflammatory cytokine TNF-α (11). TNF-α is known to elevate with aging; therefore the SASP could make cells more vulnerable to an age-associated increase in TNF-α (133). Some studies even included TNF-α as part of the SASP. We present several miRNAs that could be linked with senescence induction via key SASP proteins.
TNF-α, a SASP associated factor, suppresses miR-21 in MSCs. Furthermore, miR-21 has been found to decrease in senescent MSCs and EVs of adult MSCs (94, 119). This miRNA is also known to be down-regulated in BMSCs of postmenopausal osteoporosis patients and ovariectomized mice (134). Recombinant expression yields increased osteoblastogenesis in vitro and increased bone formation in vivo. These results suggest the SASP could help induce senescence in neighboring cells via intracellular regulation of miR-21. Interestingly, in breast cancer cells, this miRNA has been shown to target E2F2, a downstream effector of p21 and p16INK4a (135).
TNFa also regulates other miRNAs. miR-146a-5p is elevated in aged MSCs, EVs secreted from aged MSCs, senescent MSCs, and microvesicles of senescent MSCs (77, 94, 96). This miRNA is known to modulate NF-KB and the SASP of senescent cells (136). Anti-TNFα treatment decreases miR-146-5p in senescent cells with concomitant SASP reduction but does not reduce senescence levels measured by classic markers such as SA B-galactosidase and p16INK4a (137). MicroRNA-146a is also known to upregulate in osteoarthritis cartilage and rheumatoid arthritis synovial fluid. Furthermore, its expression is upregulated in human articular cartilage and rheumatoid arthritis synovial fibroblasts via IL1β, a known SASP component (138, 139).
IL1β is also known to regulated the expression of miR-24. This miRNA is noted to decrease with replicative senescence (140). Furthermore, miR-24 is known to target p16INK4a in various cell types, and ectopic expression reduces p16INK4a protein levels. It has also been reported that miR-24 repression correlates with p16INK4a upregulation in IL1β induced senescent OA chondrocytes (132). Furthermore, this miRNA is shown to target p16INK4a in several cell lines (140, 141). Taken together, this suggests IL1β could induce senescence by down-regulating miR-24, which in turn relieves pressure on p16INK4a in mesenchymal lineage cells.
miRNAs Known to Target Senescence Pathways
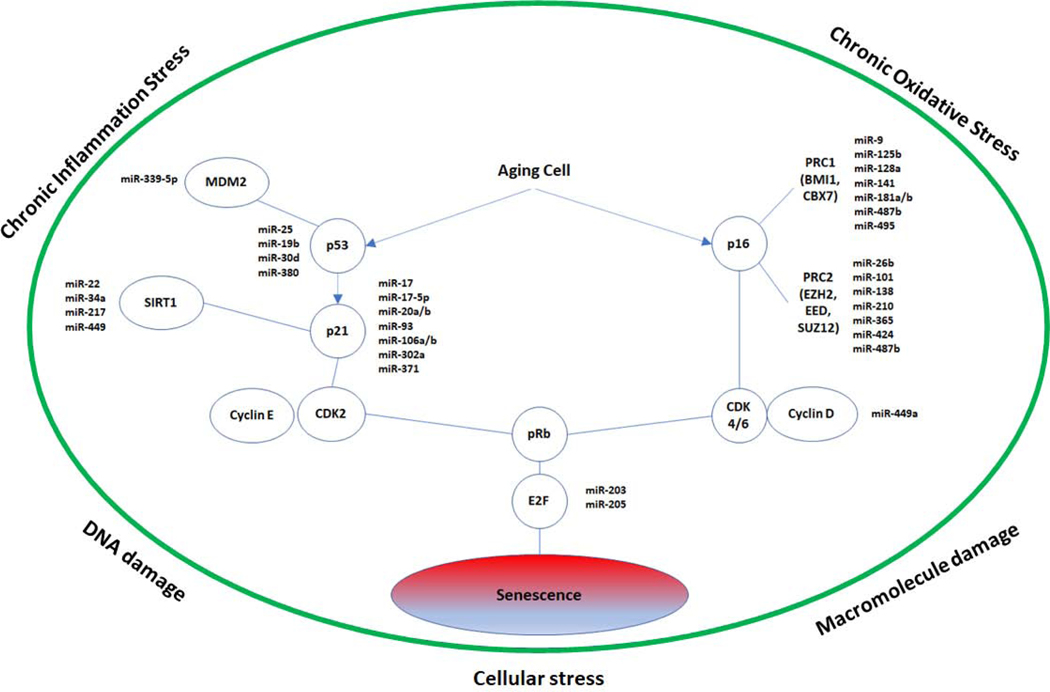
As elicited throughout this review, two well-established cell cycle arrest pathways are predominantly activated in cellular senescence: the p53 and p16INK4a pathways. In the p53 pathway, DNA insult leads to DNA damage response where ATM/ATR kinases phosphorylate p53, thus transforming it into its activated state (142). Activation and positive feedback also occur via the stress-induced p53 stabilizer ARF, known as p14 in humans and p19 in mice (143, 144). Once phosphorylated, p53 dissociates from MDM2, a ubiquitination protein, and translocates to the nucleus where p21 gene expression is induced. Subsequently, p21 inhibits CDK2 activity and prevents it from phosphorylating pRb (145). Hypophosphorylated pRb remains bound to the E2F transcription factors, and cell cycle arrest is maintained at the G1/S transition. The alternative pathway is mediated through an increase in p16INK4a. P16INK4A inhibits the activity of CDK4/6, which prevents these kinases from phosphorylating pRb. Resultantly, pRb remains associated with E2F transcription factors, thus preventing the cell from passing the G1/S transition (146). The rise in p53-activation, p21, p16INK4a, and pRb-activation are widely accepted hallmark signs of cellular senescence, and their inhibition shows promising results in relieving senescence.
Many miRNAs known to target these senescent associated cell cycle regulators in various cancer and other cell lines (See Table 2). It should be noted that several of these miRNA acts in feedback loops with intermediate regulators such as MDM2 and SIRT1. (147). One good example of this is miR-34a, which is upregulated by p53 and downregulates SIRT1, a p53 inhibitor, thus creating a positive feedback loop. Similar examples exist for MDM2, where various miRNAs such as miR-143 and miR-145 are upregulated by p53 and downregulate MDM2 (148). Conversely, other miRNA inhibitors of MDM2 such as miR-25 and miR-32 are repressed by p53 (149). Yet still, other miRNA inhibitors of MDM2, such as miR-17-3p, do not appear to have any effect on p53 levels (150). The need for further elucidation of these pathways is clear.
Table 2.
miRNA known to directly target nodes of the p53 and p16INK4a cell cycle arrest pathways confirmed via luciferase assay or similar methodology
| Target Gene | miRNA (and References) | |
|---|---|---|
| p53 | miR-15a (185), miR-16 (185), miR-19b (162), miR-25 (160), miR-30d (160), miR-33 (186), miR-92 (187), miR-98 (188), miR-125a (189), miR-125b (190), miR-150 (191), miR-138 (192), miR-141 (187), miR200a (193), miR-214 (194), miR-374 (195), miR-380–5p (161), miR453 (188), miR-504 (196), miR-518c* (197), miR-638 (197), miR-1285 (198), miR-3151 (199) | |
| MDM2 | miR-17–3p (150), miR-18b (200), miR-25 (149), miR-32 (149), miR- 143 (148), miR-145 (148), miR-192 (201), miR-193a (202), miR-194 (201), miR-215 (201), miR-221 (203), miR-339–5p (166), miR-509–5p (204), miR-605 (205), miR-660 (206), miR-661 (207) |
|
| SIRT1 | miR-9 (208), miR-22 (173), miR-34a (115, 171), miR-100 (209), miR132–3p (210), miR-135a (84), miR-137 (209), miR-138 (175), miR-143 (211), miR-145 (211), miR-181a (175), miR-181b (175), miR-181c (208), miR-195 (78), miR-199a (212), miR-199b (84), miR-200a (213), miR-204 (84), miR-217 (174), miR-449a (172, 214), miR-486–5p (82) |
|
| p21 | miR-17 (73), miR-17–5p (74), miR-18a (74), miR-20a (73, 76), miR20b (74), miR-93 (74, 75), miR-106a (74, 215), miR-106b (74, 75, 163), miR-302a (163), miR-372 (74), miR-663 (165) |
|
| p16INK4a | miR-24 (140, 141), miR-31 (216), miR-877–3p (167) | |
| BMI1 | miR-128a (217), miR-141 (97, 98), miR-194 (218), miR-218 (219), miR-487b (220), miR-495 (87) | |
| CBX7 | miR-9 (221), miR-125b (222), miR-181a (222, 223), miR-181b (222) | |
| EZH2 | miR-26a (224, 225), miR-26b (223), miR-101 (226, 227), miR-137 (228), miR-138 (229), miR-210 (223), miR-365 (230) | |
| EED | miR-26b (223), miR-101 (227), miR-210 (223), miR-424 (223) | |
| SUZ12 | miR-210 (223), miR-424 (223), miR-487b (220), miR-489 (231), miR767–5p (232) | |
| Cyclin D | miR-15b (233, 234), miR-34b (235), miR-34c (235), miR-188 (107), miR-195 (236), miR-449a (168, 235), miR-490–3p (237), miR-503 (238) | |
| Cyclin E | miR-15b (233, 234), miR-188 (107) | |
| CDK2 | miR-31 (131), miR-188 (107) | |
| CDK4 | miR-188 (107) | |
| CDK6 | miR-200a (239), miR-449a (240), miR-449b (240) | |
| pRb | miR-17 (176), miR-17–5p (241), miR-20a (176), miR-106b (176) | |
| E2F1 | miR-17–5p (242), miR-20a (242, 243), miR-205 (169), miR-331–3p (244), miR-493 (245) | |
| E2F2 | miR-20a (242, 243), miR-21 (135), miR-34b (235), miR-34c (235), miR-218 (246), miR-449a (235) | |
| E2F3 | miR-20a (242, 243), miR-34b (235), miR-34c (235), miR-203 (170), miR-203a (247), miR-210 (248), miR-214 (249), miR-449a (235), miR-503 (238), miR-577 (250) |
|
| E2F5 | miR-154 (251) |
Up to this point, only a select few miRNAs from Table 2 have been demonstrated to modulate senescence. These are presented in Figure 1. The remaining miRNAs in Table 2 can be correlated with senescence as they are shown to inhibit key proteins in senescent pathways. For example, E2F1 inhibition leads to senescence in cancer cells (151, 152). Inhibition of CDK4/6 known to induce senescence in multiple cancer cell lines, with similar results shown using FDA approved CDK4/6 inhibitor palbociclib (153, 154). Inhibition of CDK2 is sufficient to induce early senescence in vitro as well as in an in vivo tumor model (155). Depletion of Cyclin D1 disrupts the oxidative balance and induces senescence in cancer cells, but interestingly this occurs through a pRb-independent manner (156).
Figure 1.
p53 and p16INK4a senescence pathways with miRNA mediators previously shown to induce or rescue senescence via direct targeting of pathway nodes confirmed via luciferase assay
It should be noted that Cyclin D1 and Cyclin E expression elevated in senescence, but this could represent a compensatory mechanism (157). E2F1 is known to block the promoter of Cyclin D1, whereas E2F4 blocks the transcription of Cyclin E, possibly explaining the observed rise in these cyclins during senescence as pRb levels increase (158, 159). We highlight specific miRNAs shown to be directly associated with cellular senescence as follows.
MiRNAs targeting the p53 pathway
The miR-25 and miR-30d have been shown to reduce senescence in HCT116 cells by directly targeting the 3’UTR of p53 (160). The miR-380 prevent RAS-induced senescence in vivo by targeting p53 (161). Similarly, miR-19b also targets p53 and reduces senescence in HeLa and MCF7 cells (162). Another critical cell cycle regulatory protein p21 is inhibited by multiple miRNAs (such as the miR-106b family,miR-130b, miR-302a, miR-302b, miR-302c, miR-302d, miR-512-3p, and miR-515-3p) which have been shown to rescue HMECs from RasG12V-induced senescence (163). Some of these miRNAs are validated targets of p21 (Table 2). Furthermore, inhibition of p21 by miR-302a/b/c/d is conserved in human embryonic stem cells (164). Hong et al. (2010) reported that the miR-17-92 gene cluster inhibits oncogenic ras-induced senescence. They reveal that miR-17 and miR-20a are resistant to ras-induced senescence by directly targeting p21 (73). Yi et al. (2012) reported that miR-663 directly targeted p21 to promote the cellular G1/S transition in CNE1 and 5–8F cells (165). It have been previously demonstrated that p53 function is inhibited by Mouse double minute 2 homolog (MDM2) protein (166). Jansson et al. (2015) reported that miR-339-5p targets MDM2 and thus promotes p53 function. The overexpression of miRNA-339-5p leads to cell proliferation arrest and senescence by positively impacting p53 function, whereas inhibition of miR-339-5p reverses it (166).
miRNA targeting the p16INK4a pathway
p16INK4A plays a central role in cell cycle regulation, specifically inhibiting the G1 phase’s progression to s phase of the cell cycle. MicroRNAs targeting this important cell event affect normal cellular physiology. Philipot et al. (2014) reported that miR-24 is a negative regulator of p16INK4a. They reported that IL-1b induces the expression of p16INK4a and represses miR-24 level, in osteoarthritic cartilage and during terminal chondrogenesis (132). Similar findings have been reported in human diploid fibroblasts and cervical carcinoma cells that miR-24 suppresses p16 expression (140). Furthermore, in replicative senescence, a decreased level of miR-24 is associated with increased p16 expression. In contrast to the traditional inhibitory role if miRNAs, miR-877-3p has been found to bind the promotor site of p16INK4a to upregulate its mRNA and protein levels in bladder cancer cells lines T24 and UM-UC-3 (167). Several other miRNAs indirectly enhance p16 levels by targeting polycomb group complexes PRC1 and PRC2 and have been shown to induce senescence in various cell lines, as depicted in Figure 1. Cyclin D1 is targeted by miR-449a and induces senescence in DU-145 prostate cancer cells (168). MiR-205 targets E2F1 and induce senescence in melanoma cells, with a concomitant rise in p16INK4a likely representing a secondary role consistent with E2F1 inhibition (169). Furthermore, miR-203 targets E2F3 and induce senescence in melanoma cells (170). The inhibition of these pathways needs to be further explored in detail. For instance, studies have found that multiple pRb family members, including p107 and p130, act in redundancy (144). Thus, targeting a single pRb family member via miRNA might not inhibit senescence.
miRNA targeting SIRT1 dependent senescence
As mentioned above, SIRT1 plays an important role in senescence biology. SIRT1 is a validated target of miRNA-34a, a miRNA that has been reported to cause senescence in endothelial cells (171). The miR-449 is also known to cause senescence in gastric cell lines SNU638 and MKN74 by targeting SIRT1 (172). Similarly, miR-22 targets SIRT1 and induce senescence in human fibroblasts, senescence phenotypes in breast epithelial, and cancer cells (173). Menghini et al. (2009) reported that miR-217 inhibits expression of SIRT1 by targeting it’s 3’UTR and causes premature senescence-like phenotype in young human endothelial cells (174). Cervo et al. (2012) identified SIRT1 as a direct target of multiple miRNAs (miR-138, miR-181a, and miR-181b), which are upregulated with senescence in human epidermal keratinocytes (HEKn) (175). These miRNAs mentioned above play an important role in SIRT1 regulation and cellular senescence.
miRNA Candidates for Further Research
Many of the miRNAs we present in this study appear to target multiple nodes in the pathways associated with senescence (presented in Table 2). miRNA that targets a greater number of nodes could play a more prominent or complicated role in senescence. miR-499a and miR-20a both target five nodes whereas miR-188 and miR-210 target four nodes. miR-200a, miR-34b, miR-34c miR-17-5p, and miR-138 target three nodes. miR-9, miR-26b, miR-101, miR-125b, miR-137, miR-181a, miR-181b, miR-424, miR-487b, miR-106b, miR-141, miR-143, miR-145, miR-15b, miR-17, miR-194, miR-195, miR-214, miR-218, miR-25, miR-31, and miR-503 all target two nodes. In addition to targeting multiple nodes in the senescence pathways, the miR-17 and miR-20a were differentially expressed in multiple studies (Table 1). Similarly, miR-424, miR-132-3p, miR-16, miR-19b, miR-30d, and miR-34a are also differentially expressed across multiple studies (Table 1) and have a specific target (Table 2). Lastly, miR-15b-3p, miR-188-3p, miR-199b-5p, and miR-218-5p were differentially expressed across studies (Table 1) while also having an unspecific arm (3p or 5p) bind cell cycle regulators (as seen in Table 2). While several of these miRNAs have already known to modulate senescence, others have not and would be good candidates for further investigation.
For instance, one study found miR-17, miR-20a, and miR-106b target pRb in concert and promote cell progression via an increase in multiple E2F transcription factors, despite positive binding and likely inhibiting E2F1 mRNA (176). This is consistent with other findings that show miR-17-5p, miR-20a, and miR-106b target pRb (Table 2). However, these miRNAs also target p21 and E2Fs (Table 2). Furthermore, miR-17 and miR-20a have been reported to overcome Ras-induced senescence (73). miR-17 is also down-regulated across multiple studies in Table 1. Additionally, miR-17-3p targets MDM2 (Table 2) and is downregulated in senescent MSCs, and EVs secreted from senescent fibroblasts. The need for further clarification for the role of miR-17 in senescence is warranted. In general, we highlight the miR-17 family as good candidates for further research given their numerous targets in the cell cycle arrest pathway, especially miR-20a, differential expression in Table 1, and their demonstrated ability to rescue senescence. Modulation of these miRNA may yield novel findings in pre-clinical models of senescence-related musculoskeletal pathology.
The Current State of miRNA Therapeutics
While the understanding of miRNAs and their role in human disease has grown exponentially since their discovery in the mid-90s, the clinical application of miRNA still possesses significant development opportunities. Presently miRNAs are most used commercially for diagnostic purposes. There are numerous miRNA diagnostic panels in clinical practice for various cancers, neurodegenerative disorders (multiple sclerosis, Alzheimer’s, Parkinson), cardiovascular disease, and inflammatory bowel disease (177). OsteomiR is a commercially available panel of 19 blood circulating miRNA for determining fracture risk in the setting of osteoporosis, serving as an alternative to DXA scanning (178). Given the role of senescence in osteoporosis, osteoarthritis, and other musculoskeletal diseases, the detection of senescent cells could serve to identify these pathologic conditions. As established above, the role of miRNA as components of the SASP is becoming more evident and could provide a link to diagnostic applications. Studies have already pointed to the potential role of microvesicles and their contained miRNA as biomarkers to identify senescent MSCs (96). Future research is needed to determine if distinct panels of biomarkers exist for senescent cells of varying tissues and origins.
miRNA-based therapeutics are beginning to enter the market for clinical application. There are currently several therapeutics in Phase 1 and Phase 2 clinical trials, as reviewed by Bonneau et al. (177). These therapeutics fall into one of two categories: miRNA mimics or antagomiRs. Mimics are used to functionally replenish miRNA that decreases in a given pathologic condition while antagomiRs perform the inverse by suppressing overexpressed miRNA (179). The use of miRNA therapeutics to treat senescence-related musculoskeletal pathology is a logical next step. Currently, a small molecule inhibitor of the p53/MDM2 protein interaction (UBX0101) is undergoing clinical trials to treat osteoarthritis via localized injection (NCT03513016). Mimics and antagomiRs of various miRNA implicated above in osteoarthritis (miR-24, mir-34a, miR-146a, miR-204) should be evaluated for similar applications. For example, building off their pre-clinical findings, Kang et al. (2019) proposes the use of miR-204 antagomiRs to attenuate the SASP and prevent the transition of osteoarthritis to a rapidly progressing pathological phase (83).
Conclusion
Selectively removing senescent cells along with SASP attenuation is becoming a prominent goal in aging research. Given the vast quantity of studies implicating the critical role of miRNAs and senescence in tissue pathology, we postulate they are acting cooperatively to drive age-related musculoskeletal disease and dysfunction. Herein we focused on the characterization of miRNA predominantly in senescent MSCs, as these progenitors are fundamentally required for musculoskeletal tissue maintenance. Of note, throughout this characterization, we summarized emerging evidence that supports the novel concept that miRNA could be differentially secreted as part of the SASP. Building upon this information, therapeutic interventions leveraging miRNA to remove detrimental senescent cells and their inflammatory secretome show great potential for improving the way we age.
Highlights.
MicroRNAs and senescence play an intertwined role in driving age-related complications.
MicroRNAs are differentially expressed in MSCs during senescence and aging.
MicroRNAs in MSCs regulated by known SASP Factors.
MicroRNAs target number of Senescence pathways
MicroRNAs-based therapeutics have potential to prevent or reduce premature senescence.
Acknowledgments
Funding: This publication is based upon work supported in part by the National Institutes of Health AG036675 (National Institute on Aging-AG036675 S.F, W.D.H, M.H, C.S,). The above-mentioned funding did not lead to any conflict of interests regarding the publication of this manuscript.
Footnotes
Conflict of interest: The authors also declare that there is no other conflict of interest regarding the publication of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8(9):729–40. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018;28(6):436–53. [DOI] [PubMed] [Google Scholar]
- 4.Kirkland JL, Tchkonia T. Cellular Senescence: A Translational Perspective. EBioMedicine. 2017;21:21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childs BG, Baker DJ, Kirkland JL, Campisi J, van Deursen JM. Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 2014;15(11):1139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–15. [DOI] [PubMed] [Google Scholar]
- 9.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–7. [DOI] [PubMed] [Google Scholar]
- 10.Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, et al. inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6(4):387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15(8):978–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15(7):397–408. [DOI] [PubMed] [Google Scholar]
- 15.Liu JY, Souroullas GP, Diekman BO, Krishnamurthy J, Hall BM, Sorrentino JA, et al. Cells exhibiting strong p16 (INK4a) promoter activation in vivo display features of senescence. Proc Natl Acad Sci U S A. 2019;116(7):2603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5(2):187–95. [DOI] [PubMed] [Google Scholar]
- 17.Freund A, Laberge RM, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell. 2012;23(11):2066–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ao Y, Zhang J, Liu Z, Qian M, Li Y, Wu Z, et al. Lamin A buffers CK2 kinase activity to modulate aging in a progeria mouse model. Sci Adv. 2019;5(3):eaav5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, et al. Cellular Senescence: Defining a Path Forward. Cell. 2019;179(4):813–27. [DOI] [PubMed] [Google Scholar]
- 20.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436(7051):660–5. [DOI] [PubMed] [Google Scholar]
- 21.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):642. [DOI] [PubMed] [Google Scholar]
- 22.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10(1):51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155(5):1104–18. [DOI] [PubMed] [Google Scholar]
- 24.Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, et al. senescence is adevelopmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155(5):1119–30. [DOI] [PubMed] [Google Scholar]
- 25.Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jun JI, Lau LF. Cellular senescence controls fibrosis in wound healing. Aging (Albany NY). 2010;2(9):627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, et al. senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134(4):657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21(12):1424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479(7374):547–51. [DOI] [PubMed] [Google Scholar]
- 30.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445(7128):656–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovadya Y, Landsberger T, Leins H, Vadai E, Gal H, Biran A, et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat Commun. 2018;9(1):5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prata L, Ovsyannikova IG, Tchkonia T, Kirkland JL. Senescent cell clearance by the immune system: Emerging therapeutic opportunities. Semin Immunol. 2018;40:101275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagiv A, Biran A, Yon M, Simon J, Lowe SW, Krizhanovsky V. Granule exocytosis mediates immune surveillance of senescent cells. Oncogene. 2013;32(15):1971–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basisty N, Kale A, Jeon OH, Kuehnemann C, Payne T, Rao C, et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18(1):e3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311(5765):1257. [DOI] [PubMed] [Google Scholar]
- 37.Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–6. [DOI] [PubMed] [Google Scholar]
- 38.Liguori I, Russo G, Curcio F, Bulli G, Aran L, Della-Morte D, et al. Oxidative stress, aging, and diseases. Clin Interv Aging. 2018;13:757–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farr JN, Fraser DG, Wang H, Jaehn K, Ogrodnik MB, Weivoda MM, et al. Identification of Senescent Cells in the Bone Microenvironment. J Bone Miner Res. 2016;31(11):1920–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sousa-Victor P, Gutarra S, Garcia-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506(7488):316–21. [DOI] [PubMed] [Google Scholar]
- 41.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker DJ, Perez-Terzic C, Jin F, Pitel KS, Niederlander NJ, Jeganathan K, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10(7):825–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng TP, Camous X, Nyunt MSZ, Vasudev A, Tan CTY, Feng L, et al. Markers of T-cell senescence and physical frailty: insights from Singapore Longitudinal Ageing Studies. NPJ Aging Mech Dis. 2015;1:15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu M, Tchkonia T, Ding H, Ogrodnik M, Lubbers ER, Pirtskhalava T, et al. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc Natl Acad Sci U S A. 2015;112(46):E6301–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, et al. Senolytics improve physical function and increase lifespan in old age. Nat Med. 2018;24(8):1246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farr JN, Xu M, Weivoda MM, Monroe DG, Fraser DG, Onken JL, et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat Med. 2017;23(9):1072–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCulloch K, Litherland GJ, Rai TS. Cellular senescence in osteoarthritis pathology. Aging Cell. 2017;16(2):210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price JS, Waters JG, Darrah C, Pennington C, Edwards DR, Donell ST, et al. The role of chondrocyte senescence in osteoarthritis. Aging Cell. 2002;1(1):57–65. [DOI] [PubMed] [Google Scholar]
- 50.Xu M, Bradley EW, Weivoda MM, Hwang SM, Pirtskhalava T, Decklever T, et al. Transplanted Senescent Cells Induce an Osteoarthritis-Like Condition in Mice. J Gerontol A Biol Sci Med Sci. 2017;72(6):780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verzola D, Gandolfo MT, Gaetani G, Ferraris A, Mangerini R, Ferrario F, et al. Accelerated senescence in the kidneys of patients with type 2 diabetic nephropathy. Am J Physiol Renal Physiol. 2008;295(5):F1563–73. [DOI] [PubMed] [Google Scholar]
- 53.Schafer MJ, White TA, Iijima K, Haak AJ, Ligresti G, Atkinson EJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. 2017;8:14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I. Endothelial cell senescence in human atherosclerosis: role of telomere in endothelial dysfunction. Circulation. 2002;105(13):1541–4. [DOI] [PubMed] [Google Scholar]
- 55.Justice JN, Nambiar AM, Tchkonia T, LeBrasseur NK, Pascual R, Hashmi SK, et al. Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study. EBioMedicine. 2019;40:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hickson LJ, Langhi Prata LGP, Bobart SA, Evans TK, Giorgadze N, Hashmi SK, et al. Senolytics decrease senescent cells in humans: Preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine. 2019;47:446–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martyanov V, Whitfield ML, Varga J. Senescence Signature in Skin Biopsies From Systemic Sclerosis Patients Treated With Senolytic Therapy: Potential Predictor of Clinical Response? Arthritis Rheumatol. 2019;71(10):1766–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348(6239):1160–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavasani M, Robinson AR, Lu A, Song M, Feduska JM, Ahani B, et al. Muscle-derived stem/progenitor cell dysfunction limits healthspan and lifespan in a murine progeria model. Nat Commun. 2012;3:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng H, Qiu L, Ma J, Zhang H, Cheng M, Li W, et al. Replicative senescence of human bone marrow and umbilical cord derived mesenchymal stem cells and their differentiation to adipocytes and osteoblasts. Mol Biol Rep. 2011;38(8):5161–8. [DOI] [PubMed] [Google Scholar]
- 61.Sepulveda JC, Tome M, Fernandez ME, Delgado M, Campisi J, Bernad A, et al. Cell senescence abrogates the therapeutic potential of human mesenchymal stem cells in the lethal endotoxemia model. Stem Cells. 2014;32(7):1865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7(3):335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Turinetto V, Vitale E, Giachino C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int J Mol Sci. 2016;17(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choudhery MS, Khan M, Mahmood R, Mehmood A, Khan SN, Riazuddin S. Bone marrow derived mesenchymal stem cells from aged mice have reduced wound healing, angiogenesis, proliferation and anti-apoptosis capabilities. Cell Biol Int. 2012;36(8):747–53. [DOI] [PubMed] [Google Scholar]
- 65.Block TJ, Marinkovic M, Tran ON, Gonzalez AO, Marshall A, Dean DD, et al. Restoring the quantity and quality of elderly human mesenchymal stem cells for autologous cell-based therapies. Stem Cell Res Ther. 2017;8(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. [DOI] [PubMed] [Google Scholar]
- 67.Suh N MicroRNA controls of cellular senescence. BMB Rep. 2018;51(10):493–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdelmohsen K, Gorospe M. Noncoding RNA control of cellular senescence. Wiley Interdiscip Rev RNA. 2015;6(6):615–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dimmeler S, Nicotera P. MicroRNAs in age-related diseases. EMBO Mol Med. 2013;5(2):180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore BT, Xiao P. MiRNAs in bone diseases. Microrna. 2013;2(1):20–31. [DOI] [PubMed] [Google Scholar]
- 71.Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C, et al. suppression of p21 by c-Myc through members of miR-17 family at the post-transcriptional level. Int J Oncol. 2010;37(5):1315–21. [PubMed] [Google Scholar]
- 72.Guo J, Zhao Y, Fei C, Zhao S, Zheng Q, Su J, et al. Dicer1 downregulation by multiple myeloma cells promotes the senescence and tumor-supporting capacity and decreases the differentiation potential of mesenchymal stem cells. Cell Death Dis. 2018;9(5):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hong L, Lai M, Chen M, Xie C, Liao R, Kang YJ, et al. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70(21):8547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28(7):2167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13(3):272–86. [DOI] [PubMed] [Google Scholar]
- 76.Sokolova V, Fiorino A, Zoni E, Crippa E, Reid JF, Gariboldi M, et al. The Effects of miR-20a on p21: Two Mechanisms Blocking Growth Arrest in TGF-beta-Responsive Colon Carcinoma. J Cell Physiol. 2015;230(12):3105–14. [DOI] [PubMed] [Google Scholar]
- 77.Okada M, Kim HW, Matsu-ura K, Wang YG, Xu M, Ashraf M. Abrogation of Age-Induced MicroRNA-195 Rejuvenates the Senescent Mesenchymal Stem Cells by Reactivating telomerase. Stem Cells. 2016;34(1):148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mortuza R, Feng B, Chakrabarti S. miR-195 regulates SIRT1-mediated changes in diabetic retinopathy. Diabetologia. 2014;57(5):1037–46. [DOI] [PubMed] [Google Scholar]
- 79.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim Biophys Acta. 2010;1804(8):1684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernadotte A, Mikhelson VM, Spivak IM. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging (Albany NY). 2016;8(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, Ritchie A, et al. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ Res. 2006;99(2):156–64. [DOI] [PubMed] [Google Scholar]
- 82.Kim YJ, Hwang SH, Lee SY, Shin KK, Cho HH, Bae YC, et al. miR-486–5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev. 2012;21(10):1749–60. [DOI] [PubMed] [Google Scholar]
- 83.Kang D, Shin J, Cho Y, Kim HS, Gu YR, Kim H, et al. Stress-activated miR-204 governs senescent phenotypes of chondrocytes to promote osteoarthritis development. Sci Transl Med. 2019;11(486). [DOI] [PubMed] [Google Scholar]
- 84.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY). 2010;2(7):415–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olivieri F, Lazzarini R, Recchioni R, Marcheselli F, Rippo MR, Di Nuzzo S, et al. MiR-146a as marker of senescence-associated pro-inflammatory status in cells involved in vascular remodelling. Age (Dordr). 2013;35(4):1157–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113(2):175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X, Song Y, Liu D, Zhao J, Xu J, Ren J, et al. MiR-495 Promotes Senescence of Mesenchymal Stem Cells by Targeting Bmi-1. Cell Physiol Biochem. 2017;42(2):780–96. [DOI] [PubMed] [Google Scholar]
- 88.Yu KR, Lee S, Jung JW, Hong IS, Kim HS, Seo Y, et al. MicroRNA-141–3p plays a role in human mesenchymal stem cell aging by directly targeting ZMPSTE24. J Cell Sci. 2013;126(Pt 23):5422–31. [DOI] [PubMed] [Google Scholar]
- 89.Lee S, Yu KR, Ryu YS, Oh YS, Hong IS, Kim HS, et al. miR-543 and miR-590–3p regulate human mesenchymal stem cell aging via direct targeting of AIMP3/p18. Age (Dordr). 2014;36(6):9724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oh YS, Kim DG, Kim G, Choi EC, Kennedy BK, Suh Y, et al. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell. 2010;9(5):810–22. [DOI] [PubMed] [Google Scholar]
- 91.Tome M, Lopez-Romero P, Albo C, Sepulveda JC, Fernandez-Gutierrez B, Dopazo A, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18(6):985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pandey AC, Semon JA, Kaushal D, O’Sullivan RP, Glowacki J, Gimble JM, et al. MicroRNA profiling reveals age-dependent differential expression of nuclear factor kappaB and mitogen-activated protein kinase in adipose and bone marrow-derived human mesenchymal stem cells. Stem Cell Res Ther. 2011;2(6):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tome M, Sepulveda JC, Delgado M, Andrades JA, Campisi J, Gonzalez MA, et al. miR-335 correlates with senescence/aging in human mesenchymal stem cells and inhibits their therapeutic actions through inhibition of AP-1 activity. Stem Cells. 2014;32(8):2229–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fafian-Labora J, Lesende-Rodriguez I, Fernandez-Pernas P, Sangiao-Alvarellos S, Monserrat L, Arntz OJ, et al. Effect of age on pro-inflammatory miRNAs contained in mesenchymal stem cell-derived extracellular vesicles. Sci Rep. 2017;7:43923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lei Q, Liu T, Gao F, Xie H, Sun L, Zhao A, et al. Microvesicles as Potential Biomarkers for the Identification of Senescence in Human Mesenchymal Stem Cells. Theranostics. 2017;7(10):2673–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dimri M, Carroll JD, Cho JH, Dimri GP. microRNA-141 regulates BMI1 expression and induces senescence in human diploid fibroblasts. Cell Cycle. 2013;12(22):3537–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fariyike B, Singleton Q, Hunter M, Hill WD, Isales CM, Hamrick MW, et al. Role of MicroRNA-141 in the Aging Musculoskeletal System: A Current Overview. Mech Ageing Dev. 2019;178:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lopez-Arribillaga E, Rodilla V, Pellegrinet L, Guiu J, Iglesias M, Roman AC, et al. Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development. 2015;142(1):41–50. [DOI] [PubMed] [Google Scholar]
- 100.Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol. 2008;18(14):1094–9. [DOI] [PubMed] [Google Scholar]
- 101.Sangani R, Pandya CD, Bhattacharyya MH, Periyasamy-Thandavan S, Chutkan N, Markand S, et al. Knockdown of SVCT2 impairs in-vitro cell attachment, migration and wound healing in bone marrow stromal cells. Stem Cell Res. 2014;12(2):354–63. [DOI] [PubMed] [Google Scholar]
- 102.Sudharsan Periyasamy-Thandavan P, John Burke M, Bharati Mendhe M, Galina Kondrikova M, Ravindra Kolhe M, Monte Hunter M, et al. MicroRNA-141–3p Negatively Modulates SDF-1 Expression in Age-Dependent Pathophysiology of Human and Murine Bone Marrow Stromal Cells. J Gerontol A Biol Sci Med Sci. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sangani R, Periyasamy-Thandavan S, Kolhe R, Bhattacharyya MH, Chutkan N, Hunter M, et al. MicroRNAs-141 and 200a regulate the SVCT2 transporter in bone marrow stromal cells. Mol Cell Endocrinol. 2015;410:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fan J, An X, Yang Y, Xu H, Fan L, Deng L, et al. MiR-1292 Targets FZD4 to Regulate Senescence and Osteogenic Differentiation of Stem Cells in TE/SJ/Mesenchymal Tissue System via the Wnt/beta-catenin Pathway. Aging Dis. 2018;9(6):1103–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng Y, Liu H, Kong Y. miR-188 promotes senescence of lineage-negative bone marrow cells by targeting MAP3K3 expression. FEBS Lett. 2017;591(15):2290–8. [DOI] [PubMed] [Google Scholar]
- 107.Wu J, Lv Q, He J, Zhang H, Mei X, Cui K, et al. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coppe JP, Kauser K, Campisi J, Beausejour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 2006;281(40):29568–74. [DOI] [PubMed] [Google Scholar]
- 109.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98(21):12072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weilner S, Schraml E, Redl H, Grillari-Voglauer R, Grillari J. Secretion of microvesicular miRNAs in cellular and organismal aging. Exp Gerontol. 2013;48(7):626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Terlecki-Zaniewicz L, Lammermann I, Latreille J, Bobbili MR, Pils V, Schosserer M, et al. Small extracellular vesicles and their miRNA cargo are anti-apoptotic members of the senescence-associated secretory phenotype. Aging (Albany NY). 2018;10(5):1103–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davis C, Dukes A, Drewry M, Helwa I, Johnson MH, Isales CM, et al. MicroRNA-183–5p Increases with Age in Bone-Derived Extracellular Vesicles, Suppresses Bone Marrow Stromal (Stem) Cell Proliferation, and Induces Stem Cell Senescence. Tissue Eng Part A. 2017;23(21–22):1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dambal S, Shah M, Mihelich B, Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 2015;43(15):7173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fulzele S, Mendhe B, Khayrullin A, Johnson M, Kaiser H, Liu Y, et al. Muscle-derived miR-34a increases with age in circulating extracellular vesicles and induces senescence of bone marrow stem cells. Aging (Albany NY). 2019;11(6):1791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang F, Cui J, Liu X, Lv B, Liu X, Xie Z, et al. Roles of microRNA-34a targeting SIRT1 in mesenchymal stem cells. Stem Cell Res Ther. 2015;6:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abouheif MM, Nakasa T, Shibuya H, Niimoto T, Kongcharoensombat W, Ochi M. Silencing microRNA-34a inhibits chondrocyte apoptosis in a rat osteoarthritis model in vitro. Rheumatology (Oxford). 2010;49(11):2054–60. [DOI] [PubMed] [Google Scholar]
- 117.Peffers MJ, Collins J, Fang Y, Goljanek-Whysall K, Rushton M, Loughlin J, et al. Age-related changes in mesenchymal stem cells identified using a multi-omics approach. Eur Cell Mater. 2016;31:136–59. [DOI] [PubMed] [Google Scholar]
- 118.Yoo JK, Kim CH, Jung HY, Lee DR, Kim JK. Discovery and characterization of miRNA during cellular senescence in bone marrow-derived human mesenchymal stem cells. Exp Gerontol. 2014;58:139–45. [DOI] [PubMed] [Google Scholar]
- 119.Savickiene J, Baronaite S, Zentelyte A, Treigyte G, Navakauskiene R. Senescence-Associated Molecular and Epigenetic Alterations in Mesenchymal Stem Cell Cultures from Amniotic Fluid of Normal and Fetus-Affected Pregnancy. Stem Cells Int. 2016;2016:2019498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9(2):291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong J, Cui X, Jiang Z, Sun J. MicroRNA-23a modulates tumor necrosis factor-alpha-induced osteoblasts apoptosis by directly targeting Fas. J Cell Biochem. 2013;114(12):2738–45. [DOI] [PubMed] [Google Scholar]
- 122.Guo Q, Chen Y, Guo L, Jiang T, Lin Z. miR-23a/b regulates the balance between osteoblast and adipocyte differentiation in bone marrow mesenchymal stem cells. Bone Res. 2016;4:16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Godfrey TC, Wildman BJ, Beloti MM, Kemper AG, Ferraz EP, Roy B, et al. The microRNA-23a cluster regulates the developmental HoxA cluster function during osteoblast differentiation. J Biol Chem. 2018;293(45):17646–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, et al. A network connecting Runx2, SATB2, and the miR-23a~27a~24–2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107(46):19879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108(24):9863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, et al. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res. 2014;29(8):1718–28. [DOI] [PubMed] [Google Scholar]
- 127.Weilner S, Schraml E, Wieser M, Messner P, Schneider K, Wassermann K, et al. Secreted microvesicular miR-31 inhibits osteogenic differentiation of mesenchymal stem cells. Aging Cell. 2016;15(4):744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Deng Y, Wu S, Zhou H, Bi X, Wang Y, Hu Y, et al. Effects of a miR-31, Runx2, and Satb2 regulatory loop on the osteogenic differentiation of bone mesenchymal stem cells. Stem Cells Dev. 2013;22(16):2278–86. [DOI] [PubMed] [Google Scholar]
- 129.Baglio SR, Devescovi V, Granchi D, Baldini N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene. 2013;527(1):321–31. [DOI] [PubMed] [Google Scholar]
- 130.Cho JH, Dimri M, Dimri GP. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J Biol Chem. 2015;290(16):10555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q, Park SJ, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6(10):8089–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Philipot D, Guerit D, Platano D, Chuchana P, Olivotto E, Espinoza F, et al. p16INK4a and its regulator miR-24 link senescence and chondrocyte terminal differentiation-associated matrix remodeling in osteoarthritis. Arthritis Res Ther. 2014;16(1):R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Paolisso G, Rizzo MR, Mazziotti G, Tagliamonte MR, Gambardella A, Rotondi M, et al. Advancing age and insulin resistance: role of plasma tumor necrosis factor-alpha. Am J Physiol. 1998;275(2):E294–9. [DOI] [PubMed] [Google Scholar]
- 134.Yang N, Wang G, Hu C, Shi Y, Liao L, Shi S, et al. Tumor necrosis factor alpha suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res. 2013;28(3):559–73. [DOI] [PubMed] [Google Scholar]
- 135.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37(14):4850–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Olivieri F, Albertini MC, Orciani M, Ceka A, Cricca M, Procopio AD, et al. DNA damage response (DDR) and senescence: shuttled inflamma-miRNAs on the stage of inflamm-aging. Oncotarget. 2015;6(34):35509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prattichizzo F, Giuliani A, Recchioni R, Bonafe M, Marcheselli F, De Carolis S, et al. Anti-TNF-alpha treatment modulates SASP and SASP-related microRNAs in endothelial cells and in circulating angiogenic cells. Oncotarget. 2016;7(11):11945–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58(5):1284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60(4):1035–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R Jr., Srikantan S, et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3(3):e1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giglio S, Cirombella R, Amodeo R, Portaro L, Lavra L, Vecchione A. MicroRNA miR-24 promotes cell proliferation by targeting the CDKs inhibitors p27Kip1 and p16INK4a. J Cell Physiol. 2013;228(10):2015–23. [DOI] [PubMed] [Google Scholar]
- 142.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell. 2004;14(4):501–13. [DOI] [PubMed] [Google Scholar]
- 143.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24(17):2899–908. [DOI] [PubMed] [Google Scholar]
- 144.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37(5):961–76. [DOI] [PubMed] [Google Scholar]
- 145.Beausejour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22(16):4212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dai CY, Enders GH. p16 INK4a can initiate an autonomous senescence program. Oncogene. 2000;19(13):1613–22. [DOI] [PubMed] [Google Scholar]
- 147.Liu J, Zhang C, Zhao Y, Feng Z. MicroRNA Control of p53. J Cell Biochem. 2017;118(1):7–14. [DOI] [PubMed] [Google Scholar]
- 148.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32(1):61–9. [DOI] [PubMed] [Google Scholar]
- 149.Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S, Lee TJ, et al. MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci U S A. 2012;109(14):5316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li H, Yang BB. Stress response of glioblastoma cells mediated by miR-17–5p targeting PTEN and the passenger strand miR-17–3p targeting MDM2. Oncotarget. 2012;3(12):1653–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rouaud F, Hamouda-Tekaya N, Cerezo M, Abbe P, Zangari J, Hofman V, et al. E2F1 inhibition mediates cell death of metastatic melanoma. Cell Death Dis. 2018;9(5):527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Park C, Lee I, Kang WK. E2F-1 is a critical modulator of cellular senescence in human cancer. Int J Mol Med. 2006;17(5):715–20. [PubMed] [Google Scholar]
- 153.Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun. 2017;8:15916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Knudsen ES, Witkiewicz AK. The Strange Case of CDK4/6 Inhibitors: Mechanisms, Resistance, and Combination Strategies. Trends Cancer. 2017;3(1):39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zalzali H, Nasr B, Harajly M, Basma H, Ghamloush F, Ghayad S, et al. CDK2 transcriptional repression is an essential effector in p53-dependent cellular senescence-implications for therapeutic intervention. Mol Cancer Res. 2015;13(1):29–40. [DOI] [PubMed] [Google Scholar]
- 156.Laphanuwat P, Likasitwatanakul P, Sittithumcharee G, Thaphaengphan A, Chomanee N, Suppramote O, et al. Cyclin D1 depletion interferes with oxidative balance and promotes cancer cell senescence. J Cell Sci. 2018;131(12). [DOI] [PubMed] [Google Scholar]
- 157.Lucibello FC, Sewing A, Brusselbach S, Burger C, Muller R. Deregulation of cyclins D1 and E and suppression of cdk2 and cdk4 in senescent human fibroblasts. J Cell Sci. 1993;105 ( Pt 1):123–33. [DOI] [PubMed] [Google Scholar]
- 158.Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, et al. inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18(6):3212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Le Cam L, Polanowska J, Fabbrizio E, Olivier M, Philips A, Ng Eaton E, et al. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 1999;18(7):1878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, et al. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene. 2011;30(7):843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, et al. miR-380–5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16(10):1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fan Y, Yin S, Hao Y, Yang J, Zhang H, Sun C, et al. miR-19b promotes tumor growth and metastasis via targeting TP53. RNA. 2014;20(6):765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1). Oncogene. 2010;29(15):2262–71. [DOI] [PubMed] [Google Scholar]
- 164.Dolezalova D, Mraz M, Barta T, Plevova K, Vinarsky V, Holubcova Z, et al. MicroRNAs regulate p21(Waf1/Cip1) protein expression and the DNA damage response in human embryonic stem cells. Stem Cells. 2012;30(7):1362–72. [DOI] [PubMed] [Google Scholar]
- 165.Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31(41):4421–33. [DOI] [PubMed] [Google Scholar]
- 166.Jansson MD, Damas ND, Lees M, Jacobsen A, Lund AH. miR-339–5p regulates the p53 tumor-suppressor pathway by targeting MDM2. Oncogene. 2015;34(15):1908–18. [DOI] [PubMed] [Google Scholar]
- 167.Li S, Zhu Y, Liang Z, Wang X, Meng S, Xu X, et al. Up-regulation of p16 by miR-877–3p inhibits proliferation of bladder cancer. Oncotarget. 2016;7(32):51773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Noonan EJ, Place RF, Basak S, Pookot D, Li LC. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1(5):349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286(19):16606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Noguchi S, Mori T, Otsuka Y, Yamada N, Yasui Y, Iwasaki J, et al. Anti-oncogenic microRNA-203 induces senescence by targeting E2F3 protein in human melanoma cells. J Biol Chem. 2012;287(15):11769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398(4):735–40. [DOI] [PubMed] [Google Scholar]
- 172.Bou Kheir T, Futoma-Kazmierczak E, Jacobsen A, Krogh A, Bardram L, Hother C, et al. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol Cancer. 2011;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193(2):409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120(15):1524–32. [DOI] [PubMed] [Google Scholar]
- 175.Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109(4):1133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Trompeter HI, Abbad H, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, et al. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 2011;6(1):e16138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bonneau E, Neveu B, Kostantin E, Tsongalis GJ, De Guire V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114–27. [PMC free article] [PubMed] [Google Scholar]
- 178.Walter E, Dellago H, Grillari J, Dimai HP, Hackl M. Cost-utility analysis of fracture risk assessment using microRNAs compared with standard tools and no monitoring in the Austrian female population. Bone. 2018;108:44–54. [DOI] [PubMed] [Google Scholar]
- 179.Metias SM, Lianidou E, Yousef GM. MicroRNAs in clinical oncology: at the crossroads between promises and problems. J Clin Pathol. 2009;62(9):771–6. [DOI] [PubMed] [Google Scholar]
- 180.Wagner W, Horn P, Castoldi M, Diehlmann A, Bork S, Saffrich R, et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3(5):e2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kundrotas G, Gasperskaja E, Slapsyte G, Gudleviciene Z, Krasko J, Stumbryte A, et al. Identity, proliferation capacity, genomic stability and novel senescence markers of mesenchymal stem cells isolated from low volume of human bone marrow. Oncotarget. 2016;7(10):10788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Meng X, Xue M, Xu P, Hu F, Sun B, Xiao Z. MicroRNA profiling analysis revealed different cellular senescence mechanisms in human mesenchymal stem cells derived from different origin. Genomics. 2017;109(3–4):147–57. [DOI] [PubMed] [Google Scholar]
- 183.Chen YJ, Chang WA, Huang MS, Chen CH, Wang KY, Hsu YL, et al. identification of novel genes in aging osteoblasts using next-generation sequencing and bioinformatics. Oncotarget. 2017;8(69):113598–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li XH, Ha CT, Fu D, Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS One. 2012;7(11):e48700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Herrera-Merchan A, Cerrato C, Luengo G, Dominguez O, Piris MA, Serrano M, et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9(16):3277–85. [DOI] [PubMed] [Google Scholar]
- 187.Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell. 2010;7(6):671–81. [DOI] [PubMed] [Google Scholar]
- 188.Zhang S, Zhang C, Li Y, Wang P, Yue Z, Xie S. miR-98 regulates cisplatin-induced A549 cell death by inhibiting TP53 pathway. Biomed Pharmacother. 2011;65(6):436–42. [DOI] [PubMed] [Google Scholar]
- 189.Chen J, Ouyang H, An X, Liu S. miR-125a is upregulated in cancer stem-like cells derived from TW01 and is responsible for maintaining stemness by inhibiting p53. Oncol Lett. 2019;17(1):87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587(15):2346–51. [DOI] [PubMed] [Google Scholar]
- 192.Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, et al. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells. 2012;30(8):1645–54. [DOI] [PubMed] [Google Scholar]
- 193.Becker LE, Lu Z, Chen W, Xiong W, Kong M, Li Y. A systematic screen reveals MicroRNA clusters that significantly regulate four major signaling pathways. PLoS One. 2012;7(11):e48474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Xu CX, Xu M, Tan L, Yang H, Permuth-Wey J, Kruk PA, et al. MicroRNA miR-214 regulates ovarian cancer cell stemness by targeting p53/Nanog. J Biol Chem. 2012;287(42):34970–8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 195.Liu Y, Xing R, Zhang X, Dong W, Zhang J, Yan Z, et al. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair (Amst). 2013;12(9):741–50. [DOI] [PubMed] [Google Scholar]
- 196.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, et al. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38(5):689–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tay Y, Tan SM, Karreth FA, Lieberman J, Pandolfi PP. Characterization of dual PTEN and p53-targeting microRNAs identifies microRNA-638/Dnm2 as a two-hit oncogenic locus. Cell Rep. 2014;8(3):714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3’ untranslated region. Biochem Biophys Res Commun. 2010;396(2):435–9. [DOI] [PubMed] [Google Scholar]
- 199.Lankenau MA, Patel R, Liyanarachchi S, Maharry SE, Hoag KW, Duggan M, et al. MicroRNA-3151 inactivates TP53 in BRAF-mutated human malignancies. Proc Natl Acad Sci U S A. 2015;112(49):E6744–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, et al. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst. 2013;105(6):433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, et al. downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010;18(4):367–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 202.Li Y, Gao L, Luo X, Wang L, Gao X, Wang W, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509. [DOI] [PubMed] [Google Scholar]
- 203.Kim D, Song J, Jin EJ. MicroRNA-221 regulates chondrogenic differentiation through promoting proteosomal degradation of slug by targeting Mdm2. J Biol Chem. 2010;285(35):26900–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 204.Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F, et al. Mir-509–5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis. 2014;5:e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xiao J, Lin H, Luo X, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30(3):524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fortunato O, Boeri M, Moro M, Verri C, Mensah M, Conte D, et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis. 2014;5:e1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hoffman Y, Bublik DR, Pilpel Y, Oren M. miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell Death Differ. 2014;21(2):302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Schonrock N, Humphreys DT, Preiss T, Gotz J. Target gene repression mediated by miRNAs miR-181c and miR-9 both of which are down-regulated by amyloid-beta. J Mol Neurosci. 2012;46(2):324–35. [DOI] [PubMed] [Google Scholar]
- 209.Tarantino C, Paolella G, Cozzuto L, Minopoli G, Pastore L, Parisi S, et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24(9):3255–63. [DOI] [PubMed] [Google Scholar]
- 210.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23(11):1876–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10(8):1470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104(7):879–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q. miR-200a regulates SIRT1 expression and epithelial to mesenchymal transition (EMT)-like transformation in mammary epithelial cells. J Biol Chem. 2011;286(29):25992–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17(3):452–8. [DOI] [PubMed] [Google Scholar]
- 215.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130(11–12):731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Malhas A, Saunders NJ, Vaux DJ. The nuclear envelope can control gene expression and cell cycle progression via miRNA regulation. Cell Cycle. 2010;9(3):531–9. [DOI] [PubMed] [Google Scholar]
- 217.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5(6):e10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J, et al. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer. 2011;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63(4):886–95. [DOI] [PubMed] [Google Scholar]
- 220.Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S, et al. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest. 2013;123(3):1241–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.O’Loghlen A, Brookes S, Martin N, Rapisarda V, Peters G, Gil J. CBX7 and miR-9 are part of an autoregulatory loop controlling p16(INK) (4a). Aging Cell. 2015;14(6):1113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.O’Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 2012;10(1):33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Overhoff MG, Garbe JC, Koh J, Stampfer MR, Beach DH, Bishop CL. Cellular senescence mediated by p16INK4A-coupled miRNA pathways. Nucleic Acids Res. 2014;42(3):1606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Li G, Liu H, Zhang X, Liu X, Zhang G, Liu Q. The protective effects of microRNA-26a in steroid-induced osteonecrosis of the femoral head by repressing EZH2. Cell Cycle. 2020;19(5):551–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112(10):4202–12. [DOI] [PubMed] [Google Scholar]
- 226.Chen L, Jia J, Zang Y, Li J, Wan B. MicroRNA-101 regulates autophagy, proliferation and apoptosis via targeting EZH2 in laryngeal squamous cell carcinoma. Neoplasma. 2019;66(4):507–15. [DOI] [PubMed] [Google Scholar]
- 227.Xiaoping L, Zhibin Y, Wenjuan L, Zeyou W, Gang X, Zhaohui L, et al. CPEB1, a histone-modified hypomethylated gene, is regulated by miR-101 and involved in cell senescence in glioma. Cell Death Dis. 2013;4:e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Cui S, Sun Y, Liu Y, Liu C, Wang J, Hao G, et al. MicroRNA137 has a suppressive role in liver cancer via targeting EZH2. Mol Med Rep. 2017;16(6):9494–502. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 229.Liang J, Zhang Y, Jiang G, Liu Z, Xiang W, Chen X, et al. MiR-138 induces renal carcinoma cell senescence by targeting EZH2 and is downregulated in human clear cell renal cell carcinoma. Oncol Res. 2013;21(2):83–91. [DOI] [PubMed] [Google Scholar]
- 230.Wang C, Su K, Zhang Y, Zhang W, Chu D, Zhao Q, et al. MicroRNA-365 targets multiple oncogenes to inhibit proliferation, invasion, and self-renewal of aggressive endometrial cancer cells. Cancer Manag Res. 2018;10:5171–85. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 231.Xie Z, Cai L, Li R, Zheng J, Wu H, Yang X, et al. Down-regulation of miR-489 contributes into NSCLC cell invasion through targeting SUZ12. Tumour Biol. 2015;36(8):6497–505. [DOI] [PubMed] [Google Scholar]
- 232.Zhang J, Xu S, Xu J, Li Y, Zhang J, Zhang J, et al. miR7675p inhibits glioma proliferation and metastasis by targeting SUZ12. Oncol Rep. 2019;42(1):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Sun G, Shi L, Yan S, Wan Z, Jiang N, Fu L, et al. MiR-15b targets cyclin D1 to regulate proliferation and apoptosis in glioma cells. Biomed Res Int. 2014;2014:687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, et al. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380(2):205–10. [DOI] [PubMed] [Google Scholar]
- 235.Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem. 2012;287(26):21686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wang N, Wei H, Yin D, Lu Y, Zhang Y, Zhang Q, et al. MicroRNA-195 inhibits proliferation of cervical cancer cells by targeting cyclin D1a. Tumour Biol. 2016;37(4):4711–20. [DOI] [PubMed] [Google Scholar]
- 237.Gu H, Yang T, Fu S, Chen X, Guo L, Ni Y. MicroRNA-490–3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochem Biophys Res Commun. 2014;444(1):104–8. [DOI] [PubMed] [Google Scholar]
- 238.Xiao F, Zhang W, Chen L, Chen F, Xie H, Xing C, et al. MicroRNA-503 inhibits the G1/S transition by downregulating cyclin D3 and E2F3 in hepatocellular carcinoma. J Transl Med. 2013;11:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Xiao F, Zhang W, Zhou L, Xie H, Xing C, Ding S, et al. microRNA-200a is an independent prognostic factor of hepatocellular carcinoma and induces cell cycle arrest by targeting CDK6. Oncol Rep. 2013;30(5):2203–10. [DOI] [PubMed] [Google Scholar]
- 240.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23(20):2388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310(2):442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. [DOI] [PubMed] [Google Scholar]
- 243.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282(4):2135–43. [DOI] [PubMed] [Google Scholar]
- 244.Guo X, Guo L, Ji J, Zhang J, Zhang J, Chen X, et al. miRNA-331–3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem Biophys Res Commun. 2010;398(1):1–6. [DOI] [PubMed] [Google Scholar]
- 245.Gu Y, Cheng Y, Song Y, Zhang Z, Deng M, Wang C, et al. MicroRNA-493 suppresses tumor growth, invasion and metastasis of lung cancer by regulating E2F1. PLoS One. 2014;9(8):e102602. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 246.Dong Y, Zou J, Su S, Huang H, Deng Y, Wang B, et al. MicroRNA-218 and microRNA-520a inhibit cell proliferation by downregulating E2F2 in hepatocellular carcinoma. Mol Med Rep. 2015;12(1):1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang H, Wang L, Tang X, Bai W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol Lett. 2017;14(6):7687–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7(2):255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yang Y, Chang S, Zhao Z, Hou NI, He K, Wang X, et al. MicroRNA-214 suppresses the proliferation of human hepatocellular carcinoma cells by targeting E2F3. Oncol Lett. 2015;10(6):3779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yu Z, Zhang W, Deng F. MicroRNA-577 inhibits gastric cancer growth by targeting E2F transcription factor 3. Oncol Lett. 2015;10(3):1447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8(6):2620–30. [PMC free article] [PubMed] [Google Scholar]