Abstract
Background
Recent epidemiological studies have suggested that sexual dimorphism influences treatment response and prognostic outcome in glioblastoma (GBM). To this end, we sought to (i) identify distinct sex-specific radiomic phenotypes—from tumor subcompartments (peritumoral edema, enhancing tumor, and necrotic core) using pretreatment MRI scans—that are prognostic of overall survival (OS) in GBMs, and (ii) investigate radiogenomic associations of the MRI-based phenotypes with corresponding transcriptomic data, to identify the signaling pathways that drive sex-specific tumor biology and treatment response in GBM.
Methods
In a retrospective setting, 313 GBM patients (male = 196, female = 117) were curated from multiple institutions for radiomic analysis, where 130 were used for training and independently validated on a cohort of 183 patients. For the radiogenomic analysis, 147 GBM patients (male = 94, female = 53) were used, with 125 patients in training and 22 cases for independent validation.
Results
Cox regression models of radiomic features from gadolinium T1-weighted MRI allowed for developing more precise prognostic models, when trained separately on male and female cohorts. Our radiogenomic analysis revealed higher expression of Laws energy features that capture spots and ripple-like patterns (representative of increased heterogeneity) from the enhancing tumor region, as well as aggressive biological processes of cell adhesion and angiogenesis to be more enriched in the “high-risk” group of poor OS in the male population. In contrast, higher expressions of Laws energy features (which detect levels and edges) from the necrotic core with significant involvement of immune related signaling pathways was observed in the “low-risk” group of the female population.
Conclusions
Sexually dimorphic radiogenomic models could help risk-stratify GBM patients for personalized treatment decisions.
Keywords: glioblastoma, machine learning, radiogenomics, sexual dimorphism
Key Points.
We developed “sex-controlled” prognostic MRI-based radiomic models for GBM patients.
Sex-specific” prognostic radiogenomic models provide cross-scale understanding of tumor biology differences.
Importance of the Study.
Recent epidemiological studies suggest that sexual dimorphism influences the prognostic outcome of GBM, and may have implications in designing targeted therapies. Using a large multi-institutional cohort, in this work, we present the first attempt at developed “sex-controlled” prognostic radiomic models using routinely acquired MRI scans that account for treatment variability across sexes, and thus may improve clinical stratification and treatment management of GBM patients. Further, by leveraging radiogenomic associations of radiomic models with transcriptomic data, we demonstrate that these “sex-specific” prognostic models may be used to provide a detailed cross-scale molecular understanding of transcriptomic differences across males and females, with clinical implications in building comprehensive and patient-centric treatment plans in GBM patients.
Patient sex is recognized as an important biological determinant that affects the risk, drug response, and prognosis across multiple cancer types.1,2 For instance, in glioblastoma (GBM) patients, the mortality rate differs between males and females, with females exhibiting longer overall survival (OS) and a survival advantage over males.3 These sex differences in GBM are known to be driven by hormonal, metabolic, and immune variances that result in poor prognosis.4,5 Further investigation of GBM at a molecular level has elucidated that the sexually dimorphic expression of genes also plays a significant role and influences treatment response and prognosis.1 In particular, the molecular subtypes of GBM, proneural, neural, and mesenchymal, have been found to be more prominent in males than females (2:1).6 Male-specific GBM astrocytes are known to undergo greater tumorigenesis and higher proliferation rates compared with female GBM astrocytes, and this has been linked to potentially unfavorable response to the conventional treatment in male GBM patients.6
In spite of the differences in treatment outcomes and responses, the standard of care for GBM (also known as the Stupp protocol) is consistent across both sexes, comprising maximal safe tumor resection, radiation therapy, and concomitant chemotherapy. Further, some of the current preclinical GBM studies, which attempt to elucidate the mechanism of glioma oncogenesis toward developing drug therapies, also do not account for the underlying sexual dimorphism.7,8
Recent advances in computational processing power and high-throughput algorithmic development have led to the development of novel quantitative image analysis methods, such as radiomics, which leverages routine pretreatment imaging (ie, MRI scans) to interrogate the tumor and the seemingly normal parenchyma around the tumor boundaries to predict patient prognosis as well as to evaluate treatment response in oncology.9 Similarly, radiogenomic models allow for studying significant associations between radiomic signatures and biological pathways for improved understanding of tumor biology for disease characterization. However, to our knowledge, none of the existing radiogenomic-based approaches have attempted to account for sex differences in creating prognostic models of OS in GBM tumors.10,11 There is hence an opportunity to create “sex-controlled” prognostic radiomic models using routinely acquired MRI scans that account for treatment variability across sexes and improve clinical stratification and treatment management of GBM patients. Further, by leveraging radiogenomic associations of radiomic models with transcriptomic data, these “sex-specific” prognostic models could be used to provide a detailed cross-scale molecular understanding of differences across males and females, with clinical implications in building comprehensive and patient-centric treatment plans in GBM patients.
In this work, we first aim to create sexually dimorphic radiomic risk score (RRS) models that are prognostic of OS in GBM patients using radiomic features from different GBM tumor subcompartments (necrosis, enhancing tumor, edema) on pretreatment MRI (gadolinium [Gd] T1-weighted [T1w], T2w, and T2w‒fluid attenuated inversion recovery [FLAIR]). Our first objective is based on the hypothesis that if there exist differences in overall prognosis across males and females, creating sex-specific radiomic models will allow for improved prediction of OS in GBM tumors. Secondly, we seek to obtain statistically significant associations of “sex-controlled” prognostic radiomic descriptors, with distinct signaling pathways that drive GBM tumor biology, treatment response, and prognosis. Establishing such associations may allow for providing a biological underpinning of the radiomic features that drive the prognosis of OS in GBM patients, and may ultimately impact clinical treatment decisions in GBM patients.
Materials and Methods
Dataset Description
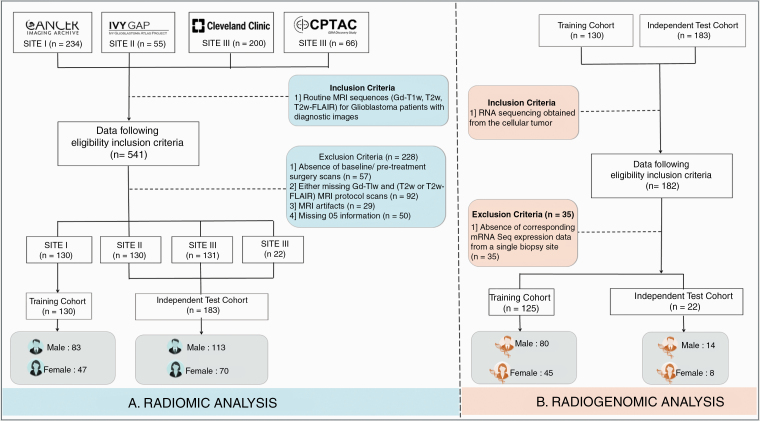
Pretreatment, multiparametric (Gd-T1w, T2w, and T2w-FLAIR) MRI protocols were retrospectively collected from 4 cohorts. Of these datasets, 3 were curated from publicly available sources—The Cancer Imaging Archive (TCIA),12 Ivy Glioblastoma Atlas Project (Ivy GAP),13 and the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) GBM Discovery Cohort.14 And lastly, the fourth dataset was curated from a Health Insurance Portability and Accountability Act (HIPAA) compliant and institution review board (IRB) approved participating institution—Cleveland Clinic Foundation (CCF), where the need for an informed consent from all patients was waived. Between December 1, 2011 and May 1, 2018, radiology image archives of CCF were consecutively searched to identify 200 histopathology-confirmed GBM patients.
These 541 collected GBM cases from these 3 sites were further triaged using inclusion criteria that involved the availability of (i) routine MRI sequences (Gd-T1w, T2w and/or T2w-FLAIR) for treatment-naive patients with diagnostic image quality, (ii) OS information for all individuals, and (iii) gene expression data. A patient enrollment flowchart is shown in Figure 1. Further details regarding data curation, patient characteristics, and the MRI acquisition for CCF site can be found in Supplementary Sections 1 and 2.
Fig. 1.
Patient enrollment and distribution. Flowchart of patient enrollment, inclusion and exclusion criteria for the radiomic and radiogenomic analysis in our study.
Thus, a total of 313 preoperative Gd-T1w MRI (1.5T/3T scans, multicenter) of GBM (196 males and 117 females) from TCIA (n = 130), Ivy GAP (n = 30), CPTAC (n = 22), and participating institution (n = 131), along with corresponding transcriptomic data (when available) were considered. Multisite data distribution is reported in Supplementary Table 1A. Complete information (when available) regarding age, OS, extent of resection, molecular status (MGMT, IDH) of individual patients across these 4 cohorts has been provided in Supplementary Sheet 1. The complete radiomic workflow can be found in Figure 2.
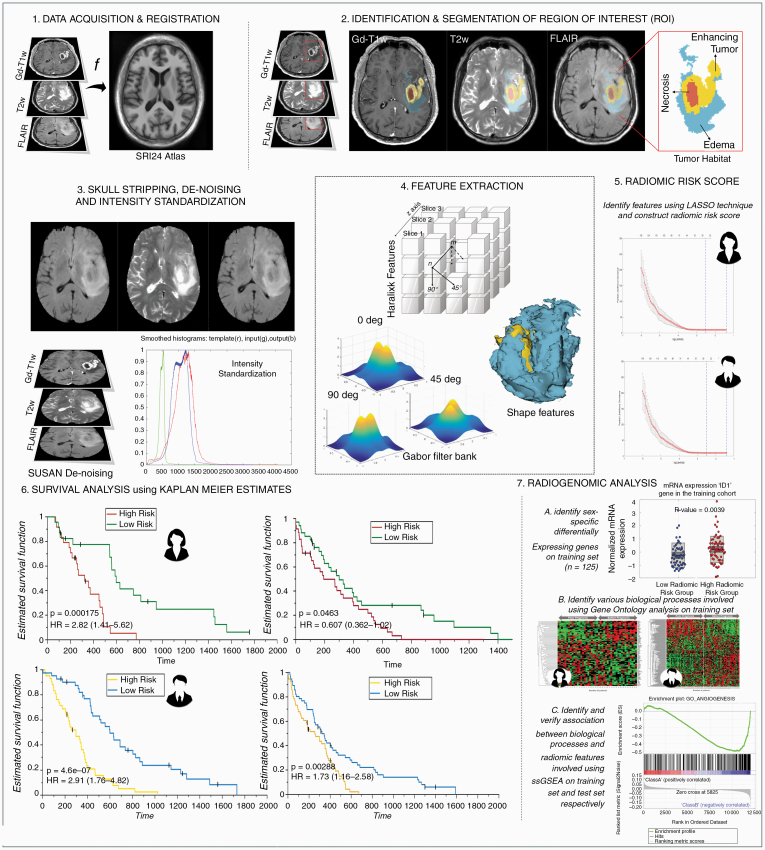
Fig. 2.
Experimental design. 1. MRI protocol (Gd-T1w, T2w, and/or T2w-FLAIR) image acquisition and registration to SRI24 atlas. 2. Region of interest (ROI) was identified and annotated. 3. Images were skull stripped, de-noised, and intensity-standardized. 4. 3D radiomic and shape features were extracted from every tumor sub-compartment. 5. Sex- specific radiomic risk scores were constructed using LASSO models. 6. Survival analysis using KM curves was investigated in sex-specific cohorts. 7. Radiogenomic analysis in sex-specific cohorts consisted of 3 main steps—A. Identification of sex-specific differentially expressing genes. B. Identification of biological processes implicated using Gene Ontology and implementation of single-sample Gene Set Enrichment Analysis (ssGSEA).
Further details about the preprocessing, tumor segmentation, and features extraction can be found in Supplementary Sections 3 and 4, respectively.
Development of Sexually Dimorphic Radiomic Risk Scores
For our training cohort, we developed 3 RRS from every MRI protocol (Gd-T1w, T2w, T2w-FLAIR): (i) male-specific RRS (m-RRS) using the male cohort, (ii) female-specific RRS (f-RRS) using the female cohort, and (iii) an “all-comers” RRS (all-RRS) consisting of both male and female cohorts. m-RRS and f-RRS were validated separately on the male and female test cohort respectively. For comparison purposes, all-RRS was similarly validated individually across the male and female test cohort.
Obtained were 2850 radiomic features from each of the 3 MRI protocols for every study, and due to the high dimensionality of our extracted features, feature selection was performed to avoid overfitting using univariate Cox regression method (P-value < 0.05). Next, using the training cohort, least absolute shrinkage and selection operator (LASSO) Cox regression models were developed, to create two groups of “low-risk” and “high-risk” based on patient prognosis of OS risk. Features selected by the LASSO models were then summed in a linear combination after multiplying with their respective coefficients to construct sex-specific RRS:
where α represents either male (m), female (f), or all-comers RRS, n is the number of features selected by LASSO for a given MRI protocol, β is the weighted coefficient of the selected feature, and τ is the selected radiomic feature.
Finally, survival outcome of these 2 risk groups, within the training and test cohorts, was evaluated using Kaplan–Meier (KM) estimates and log-rank test (P < 0.05 was considered significant). The 3 RRS models for: (i) male (n = 83) cohort, (ii) female cohort (n = 47) and (iii) all-comers cohort (n = 130) were individually created and validated on 3 independent test cohorts (m = 113, f = 70, all-comers = 183) using multivariate Cox regression. Further details regarding the survival analysis can be found in Supplementary Section 5.
Radiogenomic Analysis
To investigate the possible underlying biological processes of the m-RRS and f-RRS, within the training and CPTAC test cohort, transformed and normalized gene expression data was curated for 147 patients (Supplementary Table 1A, male = 94 patients, female = 53 patients). Further information regarding cross-platform normalization of RNA expression data can be found in Supplementary Section 6. For each sex within the training cohort, the most differentially expressing genes (DEGs) of the RRS were selected using Wilcoxon rank sum test, and the P-values were adjusted using Benjamini‒Hochberg false discovery rate (FDR) estimate.
These DEGs were then used to identify distinct Gene Ontology (GO) based biological processes (FDR P-value < 0.05).15,16 GO highlights the most overrepresented genes and finds the systematic linkages between those genes and biological processes. To further gain insight into the GO based biological processes and their association with the individual radiomic features from different subcompartments that were used to create the Gd-T1w RRS, we performed single-sample gene-set enrichment analysis (ssGSEA) on the training and CPTAC test cohort. For a predefined set of genes, ssGSEA captures the significantly enriched or depleted biological processes and calculates an enrichment score for every patient in the cohort. These predefined set of genes for the GO based biological processes were acquired from the Molecular Signatures Database platform. Thus, ssGSEA score was calculated for 147 GBM patients, and then a Spearman’s rank correlation coefficient (ρ) matrix was constructed to identify the contribution of the individual Gd-T1w radiomic features (which were used to create the RRS, 8 features for Male-RRS and 6 features for female-RRS) within the training cohort and confirm their association with the GO biological processes on the CPTAC test cohort.
Results
Sexually dimorphic radiomic risk scores are prognostic of OS on an independent test set compared with an “all-comers” survival model.
Male-Specific Gd-T1w Radiomic Risk Score
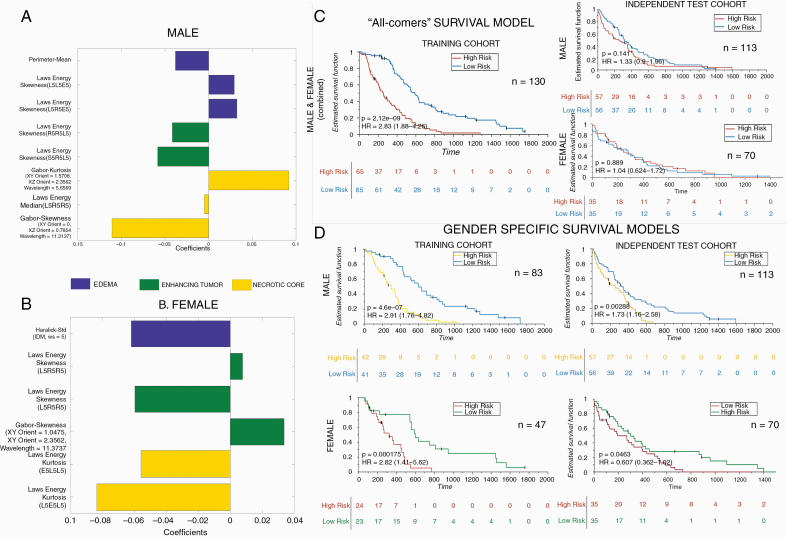
Obtained were 105 prognostic radiomic features from 2850 Gd-T1w radiomic features after feature pruning from univariable Cox regression. For the Gd-T1w protocol, LASSO Cox regression model selected 8 radiomic features. Details of the features selected and their coefficients have been listed in Figure 3A. Complete male Gd-T1w radiomic risk score (m-RRS) formulation can be found in Supplementary Section 7. Of the 8 Gd-T1w radiomic features from m-RSS, 3 were from the peritumoral edema region, 2 from the enhancing region, and the remaining 3 were from the necrotic core of GBM. The low-risk and high-risk groups of males were stratified based on the median m-RRS (cutoff = 0.0322). The differential expression of these 8 Gd-T1w radiomic features between the low-risk and high-risk groups of m-RRS has been illustrated in Supplementary Section 8 using violin plots (Supplementary Figure 1). It was observed that Laws energy features (capturing spots and ripples-like patterns) from the enhancing tumor and peritumoral edema region was elevated in the “high-risk” group of poor OS in the male population when compared with the low-risk group (p enhancing-tumor = 0.02 and p edema = 8.39 × 10−8 respectively). The m-RRS for every male patient in training and testing cohort is provided in Supplementary Sheet 1.
Fig. 3.
Sex-specific radiomic models and survival analysis. Forest plot of beta coefficients of the 8 and 6 radiomic features selected in the radiomic risk score for male (A) and female (B) respectively from Gd-T1w MRI. (C) Kaplan–Meier curves for male and female combined “all-comers” patients stratified into low-risk and high-risk groups according to the radiomic risk score in the training cohort and independent validation set respectively. (D) Kaplan–Meier curves for male and female patients stratified into low-risk and high-risk groups according to the radiomic risk score (male cutoff = 0.0322, female cutoff = 0.0221) in the sex-specific training cohort and independent validation set respectively. X-axis represents overall survival days, and Y-axis represents the estimated survival function.
In Cox analysis, m-RRS built on the training set resulted in statistically significant KM curves (n = 83, log-rank test, P < 0.00001, hazard ratio [HR] = 2.91; Figure 3D, top row). m-RRS was also found to be a prognostic indicator of OS in the independent test cohort (n = 113, log-rank test, P-value = 0.0028, HR = 1.73; Figure 3D, top row). Interestingly, the m-RRS obtained from the training set was not found to be prognostic in a female-specific validation cohort (n = 70, log-rank test, P = 0.263, HR = 1.39). Further details can be found in Supplementary Section 10 (Supplementary Figure 3, top row).
Details regarding the T2w and FLAIR radiomic based LASSO models can be found in Supplementary Section 8.
Female-Specific Gd-T1w Radiomic Risk Score
When creating a radiomic risk score for the female-specific training cohort across different MRI protocols (Gd-T1w, T2w, T2w-FLAIR), 287 prognostic radiomic features were obtained after feature pruning from univariable Cox regression. For the Gd-T1w protocol, LASSO model selected 6 radiomic features. Details of the features selected and their coefficients have been listed in Figure 3B. The complete female Gd-T1w radiomic risk score (f-RRS) formulation can be found in Supplementary Section 7. Of the 6 Gd-T1w radiomic features, 1 was from the peritumoral edema region, 3 were selected from the enhancing region, and 2 were from the necrotic core of GBM. The low-risk and high-risk groups of females were stratified based on the median f-RRS (cutoff = 0.0221). Differential expression of these 6 Gd-T1w radiomic features has been illustrated in Supplementary Section 8 using violin plots (Supplementary Figure 2). An increased expression of Laws energy features (that detect levels and edges) from the necrotic core and peritumoral edema was found in the relatively better OS group of “low-risk” female population when compared with the high-risk cohort (p necrosis = 0.01 and p edema = 0.0003 respectively). The f-RRS for every female patient within the training and testing cohort has been provided in Supplementary Sheet 1.
In Cox analysis, the Gd-T1w f-RRS resulted in statistically significant KM curves (n = 47, log-rank test, P < 0.0005, HR = 2.82; Figure 3D, bottom row). f-RRS was also found to be a prognostic indicator of OS in the independent test cohort (n = 70, log-rank test, P-value = 0.046, HR = 0.06; Figure 3D, bottom row). Interestingly, f-RRS obtained from the training set was not found to be prognostic in a male-specific validation cohort (n = 113, log-rank test, P-value = 0.75, HR = 1.06). These KM curves can be found in Supplementary Section 10 (Supplementary Figure 3, bottom row).
Additional details regarding the T2w and FLAIR radiomic based LASSO models can be found in Supplementary Section 8. Further, details regarding the development and validation of all-comers Gd-T1w radiomic risk score (all-RRS) can be found in Supplementary Section 11. The added clinical utility of sexually dimorphic RRS has also been demonstrated in Supplementary Section 12.
In Table 1 the C-index, HR, and P-value for age, molecular features (MGMT status, IDH status), extent of resection, and combined radiomic features for predicting OS both for the male and female cohort have also been listed.
Table 1.
Cox regression analysis of overall survival in sex-specific cohorts. Hazard ratios, concordance using clinical and radiomic risk score (RRS) from GBM tumor subcompartments on Gd-T1w MRI was evaluated and statistical significance (via P-value) has been reported. Cox regression analysis was implemented in a univariable setting for sex and molecular features (MGMT and IDH status), and EOR. Multivariable Cox regression model was implemented for the RRS as well as the combined features. Abbreviations: 95% CI—95% Confidence Interval, C-Index—Concordance Index, MGMT—O-6-Methylguanine-DNA Methyltransferase, IDH—Isocitrate dehydrogenase, EOR—Extent of resection
| A. Cox Regression Analysis in MALE cohort | ||||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Training Cohort | Independent Test Cohort | ||||||
| Hazard Ratio (95% CI) | C- Index (95% CI) | P-value | Hazard Ratio (95% CI) | C-Index (95% CI) | P-value | |||
| Univariable Analysis | Age | 1.67 (1.5–2.67) | 0.61 (0.53–0.69) | 0.02 * | 2.15 (1.42–3.25) | 0.63 (0.56–0.70) | <0.0001* | |
| Molecular features | MGMT Status | 0.92 (0.69–1.23) | 0.51 (0.45–0.58) | 0.58 | 1.25 (0.90–1.73) | 0.51 (0.45–0.57) | 0.17 | |
| IDH Status | 0.90 (0.66–1.24) | 0.52 (0.46–0.57) | 0.53 | 1.19 (0.79–1.78) | 0.52 (0.48–0.56) | 0.40 | ||
| Extent of Resection (EOR) | 1.16 (0.85–1.58) | 0.51 (0.48–0.54) | 0.34 | 1.38 (1.16–1.64) | 0.65 (0.59–0.71) | <0.0001* | ||
| Radiomic Risk Score (m-RRS) | 2.91 (1.76–4.82) | 0.71 (0.63–0.78) | <0.0001* | 1.73 (1.16–2.58) | 0.73 (0.57–0.89) | 0.0028 * | ||
| Multivariable Analysis | Age + Molecular features + EOR + Radiomic Risk Score | - | 0.73 (0.65–0.81) | <0.0001* | - | 0.88 (0.73–0.99) | <0.0001* | |
| B. Cox Regression Analysis in FEMALE cohort | ||||||||
| Feature | Training Cohort | Independent Test Cohort | ||||||
| Hazard Ratio (95% CI) | C- Index (95% CI) | P-value | Hazard Ratio (95% CI) | C-Index (95% CI) | P-value | |||
| Univariable Analysis | Age | 1.03 (1.01–1.06) | 0.626 (0.52–0.74) | 0.0155 * | 1.02 (1.00–1.04) | 0.60 (0.51–0.69) | 0.12 | |
| Molecular features | MGMT Status | 1.05 (0.75–1.9) | 0.52 (0.42–0.62) | 0.42 | 1.64 (1.11–2.42) | 0.57 (0.49–0.65) | 0.012* | |
| IDH Status | 0.82 (0.50–1.32) | 0.52 (0.45–0.56) | 0.40 | 0.90 (0.55–1.48) | 0.51 (0.44–0.58) | 0.70 | ||
| Extent of Resection (EOR) | 1.14 (0.70–1.84) | 0.50 (0.46–0.54) | 0.60 | 1.64 (1.28–2.09) | 0.67 (0.59–0.76) | <0.0001* | ||
| Radiomic Risk Score (f-RRS) | 2.82 (1.41–5.62) | 0.73 (0.62–0.83) | <0.0005* | 0.61 (0.36–1.02) | 0.67 (0.58–0.76) | 0.046* | ||
| Multivariable Analysis | Age + Molecular features + EOR + Radiomic Risk Score | - | 0.73 (0.62–0.84) | <0.0001* | - | 0.69 (0.61–0.79) | <0.01* | |
*Signifies statistically significance (P < 0.05).
Gene Ontology Identifies Distinct Biological Processes Associated with Sexually Dimorphic Radiomic Risk Scores
Biological processes associated with m-RRS
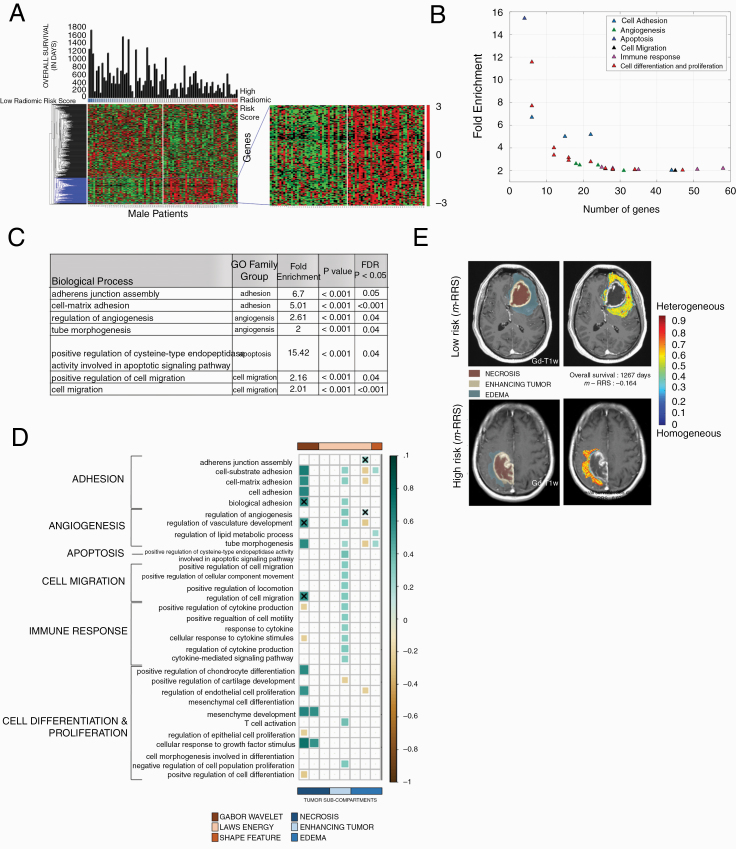
Differentially expressed were 495 genes (DEGs, P < 0.05, FDR = 5%) between the “high-risk” and “low-risk” groups of m-RRS. Figure 4A shows the supervised hierarchical clustering of these DEGs. Complete list of the 495 DEGs can be found in Supplementary Sheet 2.
Fig. 4.
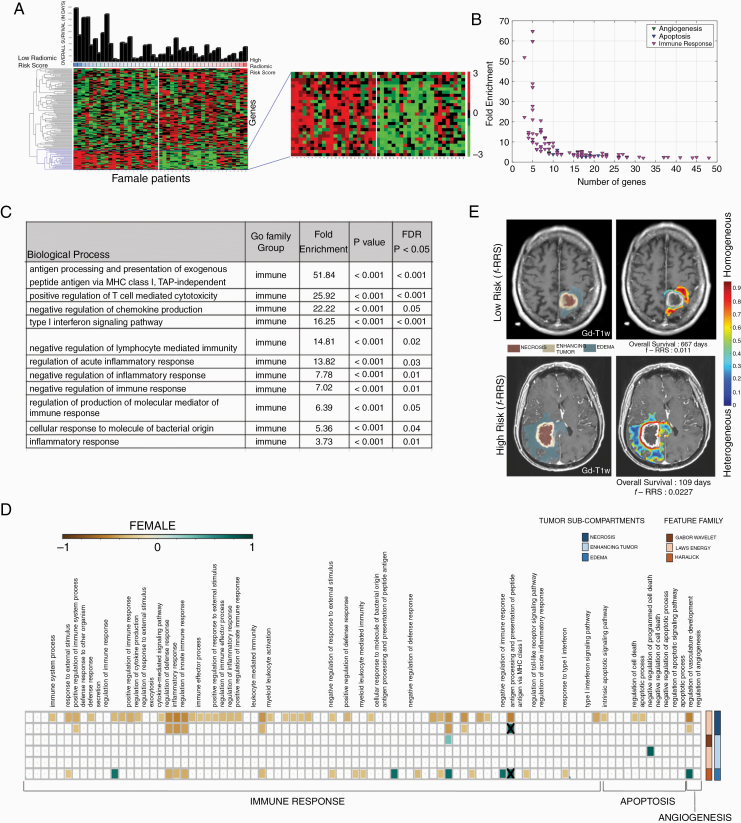
Radiogenomic analysis of male GBM patients. (A) Hierarchical clustering of the differentially expressing genes (DEG, n = 495, P < 0.05, false discovery rate = 5%) for sex-specific radiomic risk score within the training cohort. (B) 2D scatter plot to illustrates the number of genes involved in each biological process. (C) This table lists key GO biological processes in male GBM patients. Their respective enrichment score, P-values, and FDR corrected P-values are shown. (D) Spearman’s rank correlation coefficient (ρ) matrix of the single-sample gene set enrichment analysis (ssGSEA) scores and radiomic features extracted from the GBM tumor sub-compartments, where statistically significant associations (P < 0.05) were identified on the training cohort (as shown with colored boxed) and verified on the CPTAC test cohort (as shown with “X” symbol). (E) Radiomic heatmap of Laws energy (L5L5E5) in the peritumoral edema region of high-risk and low-risk male patients has been illustrated.
Radiogenomic analysis of the training cohort using for GO analysis revealed that the m-RRS was associated with 41 biological processes with enrichment fold greater than 2, as illustrated in Figure 4B. Implicated were 31 biological processes in cell adhesion, angiogenesis, cell proliferation, differentiation, and apoptosis. Figure 4C shows a few selected GO biological processes, but the complete list of all the biological processes is provided in Supplementary Sheet 3. A directed acyclic graph investigating the interrelationships between these 31 biological processes is provided in Supplementary Figure 5.
Biological processes associated with f-RRS
We identified 130 DEGs (P < 0.05, FDR = 5%) that were differentially expressed between the “high-risk” and “low-risk” groups of the f-RRS. Figure 5A shows the supervised hierarchical clustering of these DEGs. Complete list of these 130 DEGs can be found in Supplementary Sheet 2.
Fig. 5.
Radiogenomic analysis of female GBM patients. (A) Hierarchical clustering of the differentially expressing genes (DEG, n = 130, P < 0.05, false discovery rate = 5%) for sex-specific radiomic risk score within the training cohort. (B) 2-D scatter plot to illustrates the number of genes involved in each biological process. (C) This table lists key GO biological processes in female GBM patients. Their respective enrichment score, P-values, and FDR corrected P-values are shown. (D) Spearman’s rank correlation coefficient (ρ) matrix of the single-sample Gene set enrichment analysis (ssGSEA) scores and radiomic features extracted from the GBM tumor subcompartments, where statistically significant associations (P < 0.05) were identified on the training cohort (as shown with colored boxed) and verified on the CPTAC test cohort (as shown with “X” symbol). (E) Radiomic heatmap of Haralick feature (IDM) in the peritumoral edema region of high-risk and low-risk female patients has been illustrated.
Radiogenomic analysis of the DEGs obtained from the training cohort using GO revealed the association of f-RRS with 142 biological processes with enrichment fold greater than 2, as illustrated in Figure 5B. Figure 5C shows a few selected GO biological processes, but the complete list of all the biological processes is provided in Supplementary Sheet 4. It was observed that a total of 87 biological processes were implicated in immune response, angiogenesis, and apoptosis. A directed acyclic graph investigating the interrelationships between these 87 biological processes is provided in Supplementary Figure 6.
Single-Sample Gene Set Enrichment Analysis Provides Insights into Relationships of Sexually Dimorphic Biological Processes with Tumor Subcompartment-Based Radiomic Features
ssGSEA of biological pathways implicated in male GBM patients
Predefined gene set annotations for 31 GO based biological processes (associated with cell adhesion, angiogenesis, cell proliferation, differentiation and apoptosis) were available from Molecular Signatures Database (MSigDB), and thus were used to calculate the ssGSEA scores for the training and CPTAC test cohort. Supplementary Sheet 5 has the complete list of predefined set of genes that were used to describe these 31 GO based biological processes. The associations of these biological processes with individual 8 radiomic features, obtained from different tumor subcompartments and used to create the m-RRS, were investigated using ssGSEA. As observed in the correlation matrix of Figure 4D, statistically significant associations (P < 0.05) were identified on the training cohort (as shown with colored boxed) and verified on the CPTAC test cohort (as shown with “X” symbol). Positive correlations were found between Laws energy feature (ie, L5R5E5) of the peritumoral edematous region of the GBM and biological processes of cell adhesion and angiogenesis. Additionally, Gabor wavelet texture features from the necrotic core were found to be positively correlated with cell adhesion, angiogenesis, and cell migration.
ssGSEA of biological pathways implicated in female GBM patients
Similar to the male cohort analysis, predefined gene set annotations for 87 GO based biological processes (associated with immune response, angiogenesis, and apoptosis) were used to calculate ssGSEA scores for every female patient in the GBM training cohort. Supplementary Sheet 6 provides the complete list of predefined set of genes that were used to describe these biological processes. As may be observed in correlation matrix of Figure 5D, statistically significant (P < 0.05) and positive correlations were seen between Laws energy feature (L5E5L5) of the necrotic core with biological processes of immune response. Additionally, Haralick feature (inverse difference moment) from the peritumoral edema region was found to be positively correlated with immune response.
Sexually Dimorphic Drug-Gene Interactions: A Preliminary Analysis
The DEGs across the two groups of sex-specific “low-risk” and “high-risk” based on prognosis of OS were also used to evaluate the drug-gene interactions toward obtaining an understanding of their clinical implications in personalizing drug-therapy decisions in GBM patients. This preliminary analysis and the implications of these associated drugs for GBM treatment are briefly discussed in Supplementary Section 13.
Discussion
It is known that females tend to have better outcomes than males for GBM tumors.3 Existing clinical trials and preclinical studies may have been limited in their clinical translation on account of their “all-comers” patient-enrollment approach, instead of incorporating sex as a variable in patient selection for drug trials. Hence, developing “sex-specific” prognostic models, could allow for improved patient stratification in clinical trials as well as designing patient-centric treatment plans in GBM patients.
In this work, we presented the first approach at developing and independently evaluating sexually dimorphic RRS models that are prognostic of OS in primary GBM, on pretreatment Gd-T1w MRI. Further, to establish the molecular basis of these radiomic based imaging features and identify distinct signaling pathways that drive the underlying sexually dimorphic nature of tumor biology, treatment response and prognosis in GBM population, we implemented GO and ssGSEA on the corresponding transcriptomic data. Within the male cohort, a prognostic m-RRS, consisting of 8 radiomic features (5 Laws energy, 2 Gabor wavelets, and 1 shape based) from the 3 tumor subcompartments on Gd-T1w MRI, was found to be statistically significantly different across the “low risk” and “high risk” groups, both on training (P < 0.00001, n = 83) as well as the independent test set (P = 0.0028, n = 113). Laws energy features that capture ripples, and spots like pattern in an image have been previously shown to be prognostic of OS in multiple cancer types including GBMs.17,18 Along similar lines, we found Laws energy features from the enhancing tumor and peritumoral edema have an elevated expression in the “high-risk” of poor OS in GBM compared with the low-risk male patients. These textural features might potentially be capturing microvascular hyperplasia and proliferation that leads to local heterogeneity within the tumor. The extent of peritumoral edema has also been shown to be prognostic of OS in GBM (HR = 2.27, P-value = 0.015).19 We observed that the perimeter of edema was prognostic of OS in male GBM patients. More recently, Tixier et al have reported that GBM patients with large negative skewness of Gabor wavelets had a significantly longer median OS of 22.7 months (P = 0.004).11 In consensus, we found that skewness of a Gabor feature (wavelength, λ = 11.31) from the necrotic core was prognostic of OS in the male-specific GBM cohort. At this scale, Gabor wavelets might possibly be capturing the homogeneous appearance of hypercellular foci and dense regions of pseudopalisading cells around the necrotic core.20
Next, using the corresponding gene expression data available for the male-specific training set (n = 83), we identified a total of 495 DEGs across the “low risk” and “high risk” groups obtained from the m-RRS. Prognostic significance of some of these genes has been demonstrated within various cancers. For example, hypoxia-inducible factor 1 subunit α (HIF1A) is a known to produce vascular endothelial growth factor (VEGF), which in turn triggers angiogenesis.21 We found that HIF1A (with an adjusted P-value of 0.05) had a higher expression in the high-risk m-RRS group. When establishing radiogenomic associations of male-specific radiomic MRI features using GO analysis, consistently across the training and CPTAC test cohort, we found that cell adhesion and angiogenesis signaling pathways played an important role in tumor biology and prognosis of male GBM patients. These pathways are known to be associated with increased treatment resistance and poor prognosis. For example, increased angiogenesis is known to deteriorate patient outcome by amplifying tumor vasculature and, thus failing to efficiently deliver chemotherapeutic agents.22 Using ssGSEA we interestingly found that, higher expression of Laws energy (L5R5E5) from the peritumoral edema was positively correlated with biological pathways implicated in increased angiogenesis (ρ avg[TCGA+CPTAC GBM] = 0.28) and cell adhesion (ρ avg[TCGA+CPTAC GBM] = 0.21). In addition, higher expression of Gabor wavelet from the necrotic core were positively correlated with cell adhesion (ρ avg[TCGA+CPTAC GBM] = 0.58) and cell migration (ρ avg[TCGA+CPTAC GBM] = 0.59).
In a similar manner, f-RRS, developed on Gd-T1W MRI, was found to be statistically significantly different across the “low risk” and “high risk” groups, both on training (P < 0.0005, n = 47) as well as the hold-out test set (P = 0.046, n = 70). Colen et al have previously demonstrated that female GBM patients, unlike male patients, with high volumes of necrosis had significantly shorter survival (6.5 vs 14.5 months, P = 0.01).23 We found that Laws energy features (E5L5L5, L5E5L5) from the necrotic core are prognostic of OS in females, with low-risk patients exhibiting an increased expression compared with the high-risk cohort. Further, it is known that the heterogeneous regions on MR imaging manifest as local variation in image intensity, and results in lower Haralick IDM (inverse difference moment) values, whereas high expression of IDM suggests a more homogeneous appearance on MRI.19 We found that the peritumoral edema in low-risk female GBM patients exhibited higher IDM radiomic patterns, suggesting that the low- risk f-RRS cohort has a homogeneous tumor appearance compared with the high-risk group. This finding is consistent with previous findings,19,24,25 suggesting that heterogeneous radiomic textural changes of edema are potentially capturing the proliferative patterns of infiltrating tumor cells in the high-risk cohort of patients, who have poor OS.
Using the GO analysis, we found that multiple pathways of immune response were associated with the tumor biology and prognosis of female GBM patients. Immunological sex differences are known to predominantly originate from the human X chromosome, that contains genes associated with multiple immune functions.26 Multiple in vitro studies have demonstrated that the female immune system is more responsive to a variety of antigens, mitogens and other immunologic assays compared with males.27 To this end, the reduced incidence of certain tumors in females can potentially be attributed to their protective humoral and cell-mediated immune responses.28
On a cellular level, GBM tumor microenvironment is known to be characterized by production of cytokines, activation of regulatory T cells, presence of tumor-associated macrophages and tissue hypoxia.29 Interleukin-6 (IL-6), a pro-inflammatory cytokine, plays an important role in suppressing this immune microenvironment, leading to increased cell proliferation.30 In our radiogenomic analysis, we found that IL-6 was one of the DEGs that was statistically significantly different across the f-RRS groups (adjusted P-value = 0.01). Interestingly, inhibition of IL-6 signaling in preclinical GBM models has been previously shown to be associated with reduced tumor growth, and increased OS.8 In an attempt to further understand this dimorphism of immune responses in females, analysis using ssGSEA revealed that the laws energy (L5E5L5) based imaging feature from the necrotic core was positively correlated with a biological processes of immune response (ρ avg[TCGA+CPTAC GBM] = 0.76). On an imaging scale, these results suggest that “high-risk” f-RRS female patients, exhibiting lower expression of Laws energy from the necrotic core, are associated with low or impaired activity of the immune microenvironment, which potentially leads to shorter OS.
Beyond prognosis, differences in sex are also known to affect the degree of therapeutic response and drug induced toxicity in GBMs.5 For instance, Yang et al have demonstrated that the Stupp protocol is more effective for females compared with males with GBM.4 Other groups have reported that females are prone to higher chemotherapy based toxicity.4,5 Thus, sex-specific models could allow for creating more personalized treatment plans in GBM patients to target specific signaling pathways, instead of a “one-size-fits-all” Stupp protocol.
This retrospective study did have some limitations. The radiogenomic analysis was only possible on 2 of the 4 cohorts (due to unavailability of the corresponding gene expression data in the Cleveland Clinic cohort, and use of different gene expression pre-processing pipeline of Ivy GAP cohort). Thus, additional work is required to develop a radiogenomic approach that has been validated on a large co-localized, multiple site MRI and transcriptomic cohorts. We envision that obtaining co-localized datasets across imaging and omics data in future could allow for development of precise sex-specific radiogenomic prognostic and predictive markers that are biologically motivated and validated. Additionally, while the study consisted of 313 GBM studies, the stratification based on sex reduced the experimental design to 196 male GBM patients and 117 female GBM patients. This sex-stratification also resulted in only 53 female cases that were available for the radiogenomic analysis using GO and ssGSEA. Further, while our radiogenomic correlations were controlled by correcting the accepted criteria for significance (by using the FDR),31 it may be important to establish the casual inference of the identified radiogenomic relationships using preclinical or large multi-institutional prospective analysis. Our future work will focus on expanding the sex-controlled radiogenomic analysis on large prospectively collected MRI studies with corresponding co-localized gene expression data, toward optimizing treatment decisions in GBM tumors.
Supplementary Material
Acknowledgments
The CPTAC GBM Discovery Cohort data used in this publication was generated by the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The authors would like to thank the staff of Case Western Reserve University and Cleveland Clinic for setting up a Material transfer agreement (MTA) that allowed the transfer of medical imaging data (MRI scans) as per IRB regulations and HIPAA compliance.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Data Availability
Apart from the details below, all the other data supporting the findings of this study are available within the article and/or its associated supplementary information files. Please reach out to the corresponding author for further information, if required.
Images
-
The TCGA-GBM cases of 130 patients can be downloaded from here:
The CPTAC-GBM cases were downloaded from here: https://wiki.cancerimagingarchive.net/display/Public/CPTAC-GBM
The Ivy Gap cases of 30 patients can be downloaded from here: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=22515597
Since the MRI scans from Cleveland Clinic (CCF) are protected through institutional compliance, the clinical repository of 130 patient scans from CCF can only be shared per specific institutional review board (IRB) requirements. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines; Interested groups can contact the corresponding author for facilitating request regarding access to CCF MRI scans.
Preprocessing of the MR scans was performed using the CapTK software, which is available for download here—https://www.med.upenn.edu/cbica/captk/
Genomics Data
TCGA GBM expression data (Level3—Affymetrix HT HG U133A) and CPTAC GBM Discovery cohort read counts were downloaded using TCGABiolinks package on R. (https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html)
These predefined set of genes for the GO based biological processes can be acquired from the Molecular Signatures Database platform, using the following link: http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=BP
Code
The feature extraction pipeline MATLAB-based code has been publicly made available at https://github.com/ccipd/BrIC_Lab
Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, R01 CA220581-01A1, 1U01 CA239055-01, 1P20 CA233216-01, National Institute for Biomedical Imaging and Bioengineering 1R43EB028736-01, National Center for Research Resources under award number 1 C06 RR12463-01, VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the DOD Lung Cancer Investigator-Initiated Translational Research Award (W81XWH-18-1-0440), the DOD Peer Reviewed Cancer Research Program (W81XWH-16-1-0329), National Institute of Diabetes and Digestive and Kidney Diseases (1K25 DK115904-01A1), the Ohio Third Frontier Technology Validation Fund, the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University Department of Defense Peer Reviewed Cancer Research Program (PRCRP) Career Development Award, Dana Foundation David Mahoney Neuroimaging Program The V Translational Cancer Research Foundation.
Conflict of interest statement.
N. Beig is an employee of Tempus Labs, Inc.
V. B. Hill is an employee/paid consultant for Google.
V. Varadan is involved in sponsored research with Curis Inc. and Philips Healthcare. In addition, he currently serves or has served as a scientific advisory board member for Curis Inc.
A. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc. In addition, he currently serves or has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol-Meyers Squibb, Boehringer-Ingelheim and Merck. Currently he serves on the advisory board of Aiforia Inc. He also has sponsored research agreements with Philips and Inspirata Inc. His technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
M. Ahluwalia is an employee/paid consultant for AbbVie, AstraZeneca, BMS, Monteris, Bayer, Karyopharm, Forma Therapeutics, Varian Medical Systems, Flatiron, Tocagen, CBT Pharmaceuticals, and VBI Vaccines, reports receiving commercial research grants from Abbvie, AstraZeneca, Bayer, Bristol-Myers Squibb, Merck, Incyte, Novocure, Novartis, Pharmacyclics, Inspire, and holds ownership interest (including patents) in Mimivax and Doctible.
No potential conflicts of interest were disclosed by the other authors.
Authorship statement.
Each author has substantially made contributions to this work in the following way:
• Conception or design of the work: N.B., K.B., P.P., V.V., A.M., M.S.A., P.T.
• Acquisition of data: N.B., S.S., K.B., P.P., G.S., J.C., A.S., A.B., K.H., V.S., V.B.H., M.S.A.
• Analysis or interpretation of data: N.B., S.S., K.B., P.P., V.V., A.M., M.A.S., P.T. • All the authors have contributed to the writing and editing of the manuscript.
References
- 1. Yuan Y, Liu L, Chen H, et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell. 2016;29(5):711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li CH, Prokopec SD, Sun RX, Yousif F, Schmitz N, Boutros PC; PCAWG Tumour Subtypes and Clinical Translation; PCAWG Consortium . Sex differences in oncogenic mutational processes. Nat Commun. 2020;11(1):4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ostrom QT, Rubin JB, Lathia JD, Berens ME, Barnholtz-Sloan JS. Females have the survival advantage in glioblastoma. Neuro Oncol. 2018;20(4):576–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang D, Hanna DL, Usher J, et al. Impact of sex on the survival of patients with hepatocellular carcinoma: a surveillance, epidemiology, and end results analysis: impact of sex on survival in HCC. Cancer. 2014;120(23):3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim HI, Lim H, Moon A. Sex differences in cancer: epidemiology, genetics and therapy. Biomol Ther (Seoul). 2018;26(4):335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun T, Warrington NM, Luo J, et al. Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J Clin Invest. 2014;124(9):4123–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kratzsch T, Kuhn SA, Joedicke A, et al. Treatment with 5-azacitidine delay growth of glioblastoma xenografts: a potential new treatment approach for glioblastomas. J Cancer Res Clin Oncol. 2018;144(5):809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lamano JB, Lamano JB, Li YD, et al. Glioblastoma-derived IL6 induces immunosuppressive peripheral myeloid cell PD-L1 and promotes tumor growth. Clin Cancer Res. 2019;25(12):3643–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, Mohan S. Emerging applications of artificial intelligence in neuro-oncology. Radiology. 2019;290(3):607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park YW, Han K, Ahn SS, et al. Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. AJNR Am J Neuroradiol. 2018; 39(1):37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tixier F, Um H, Bermudez D, et al. Preoperative MRI-radiomics features improve prediction of survival in glioblastoma patients over MGMT methylation status alone. Oncotarget. 2019;10(6):660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shah N, Feng X, Lankerovich M, Puchalski R, Keogh B. Ivy GAP–The Cancer Imaging Archive (TCIA) public access–cancer imaging archive Wiki. Cancer Imaging Archive. 2016; doi: 10.7937/K9/TCIA.2016.XLwaN6nL. [DOI] [Google Scholar]
- 14. National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). Radiology data from the Clinical Proteomic Tumor Analysis Consortium Glioblastoma Multiforme [CPTAC-GBM] collection [Data set]. Cancer Imaging Archive. 2018; doi: 10.7937/k9/tcia.2018.3rje41q1. [DOI] [Google Scholar]
- 15. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. The Gene Ontology Consortium. Expansion of the Gene Ontology knowledge base and resources. Nucleic Acids Res. 2017;45(D1):D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braman N, Prasanna P, Whitney J, et al. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-positive Breast cancer. JAMA Netw Open. 2019;2(4):e192561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shiradkar R, Ghose S, Jambor I, et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: Preliminary findings. J Magn Reson Imaging. 2018;48(6):1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol. 2017; doi: 10.1007/s00330-016-4637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rong Y, Durden DL, Van Meir EG, Brat DJ. ‘Pseudopalisading’ necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J Neuropathol Exp Neurol. 2006;65(6):529–539. [DOI] [PubMed] [Google Scholar]
- 21. Beppu T, Kamada K, Yoshida Y, Arai H, Ogasawara K, Ogawa A. Change of oxygen pressure in glioblastoma tissue under various conditions. J Neurooncol. 2002;58(1):47–52. [DOI] [PubMed] [Google Scholar]
- 22. Rahman R, Smith S, Rahman C, Grundy R. Antiangiogenic therapy and mechanisms of tumor resistance in malignant glioma. J Oncol. 2010;2010. doi: 10.1155/2010/251231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215–227. [DOI] [PubMed] [Google Scholar]
- 24. Wu CX, Lin GS, Lin ZX, Zhang JD, Liu SY, Zhou CF. Peritumoral edema shown by MRI predicts poor clinical outcome in glioblastoma. World J Surg Oncol. 2015;13:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beig N, Bera K, Prasanna P, et al. Radiogenomic-based survival risk stratification of tumor habitat on Gd-T1w MRI Is associated with biological processes in glioblastoma. Clin Cancer Res. 2020; doi: 10.1158/1078-0432.CCR-19-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pérez-Carro R, Cauli O, López-Larrubia P. Multiparametric magnetic resonance in the assessment of the gender differences in a high-grade glioma rat model. EJNMMI Res. 2014;4. doi: 10.1186/s13550-014-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhatia A, Sekhon HK, Kaur G. Sex hormones and immune dimorphism. Sci World J. 2014;2014:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker PR, Calzascia T, Dietrich PY. All in the head: obstacles for immune rejection of brain tumours. Immunology. 2002;107(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schiffer D, Annovazzi L, Casalone C, Corona C, Mellai M. Glioblastoma: microenvironment and niche concept. Cancers. 2018;11(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Areeb Z, Stylli SS, Ware TM, et al. Inhibition of glioblastoma cell proliferation, migration and invasion by the proteasome antagonist carfilzomib. Med Oncol. 2016;33(5):53. [DOI] [PubMed] [Google Scholar]
- 31. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Apart from the details below, all the other data supporting the findings of this study are available within the article and/or its associated supplementary information files. Please reach out to the corresponding author for further information, if required.
Images
-
The TCGA-GBM cases of 130 patients can be downloaded from here:
The CPTAC-GBM cases were downloaded from here: https://wiki.cancerimagingarchive.net/display/Public/CPTAC-GBM
The Ivy Gap cases of 30 patients can be downloaded from here: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=22515597
Since the MRI scans from Cleveland Clinic (CCF) are protected through institutional compliance, the clinical repository of 130 patient scans from CCF can only be shared per specific institutional review board (IRB) requirements. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines; Interested groups can contact the corresponding author for facilitating request regarding access to CCF MRI scans.
Preprocessing of the MR scans was performed using the CapTK software, which is available for download here—https://www.med.upenn.edu/cbica/captk/
Genomics Data
TCGA GBM expression data (Level3—Affymetrix HT HG U133A) and CPTAC GBM Discovery cohort read counts were downloaded using TCGABiolinks package on R. (https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html)
These predefined set of genes for the GO based biological processes can be acquired from the Molecular Signatures Database platform, using the following link: http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=BP
Code
The feature extraction pipeline MATLAB-based code has been publicly made available at https://github.com/ccipd/BrIC_Lab