Abstract
Species of Diaporthe have often been reported as plant pathogens, endophytes or saprobes, commonly isolated from a wide range of plant hosts. Sixteen strains isolated from species of ten host genera in Yunnan Province, China, represented three new species of Diaporthe, D. chrysalidocarpi, D. machili and D. pometiae as well as five known species D. arecae, D. hongkongensis, D. middletonii, D. osmanthi and D. pandanicola. Morphological comparisons with known species and DNA-based phylogenies based on the analysis of a multigene (ITS, TUB, TEF, CAL and HIS) dataset support the establishment of the new species. This study reveals that a high species diversity of Diaporthe with wide host ranges occur in tropical rainforest in Yunnan Province, China.
Keywords: Diaporthaceae , Diaporthales , phylogeny, taxonomy, three taxa new to science
Introduction
The genus Diaporthe (DiaporthaceaeDiaporthales) with asexual morphs previously known as Phomopsis spp. is based on the type species Diaporthe eresNitschke (1870) from Ulmus sp. in Germany. Rossman et al. (2015) proposed to use the name Diaporthe over Phomopsis in the context of the one fungus – one name initiative, because it was described first, is encountered commonly in literature and includes the majority of known species. The sexual morph of Diaporthe is characterised by immersed ascomata and an erumpent pseudostroma with elongated perithecial necks; asci are unitunicate, clavate to cylindrical; and ascospores are fusoid, ellipsoid to cylindrical, hyaline, biseriate to uniseriate in the ascus, sometimes with appendages (Udayanga et al. 2011; Senanayake et al. 2017, 2018). The asexual morph is characterised by ostiolate pycnidia with cylindrical phialides often producing three types of hyaline, aseptate conidia called α-conidia, β-conidia and γ-conidia (Udayanga et al. 2011; Gomes et al. 2013). The α-conidia and β-conidia are produced frequently, but the γ-conidia are rarely observed (Gomes et al. 2013; Guarnaccia and Crous 2017; Guo et al. 2020).
Currently, more than 1100 epithets of Diaporthe are listed in Index Fungorum (http://www.indexfungorum.org/; accessed 1 Nov. 2020), but only one-fifth of these taxa have been well-studied with ex-type cultures and supplementary DNA barcodes (Guo et al. 2020; Yang et al. 2020; Zapata et al. 2020). Species of Diaporthe are widely distributed and have a broad range of hosts including economically significant agricultural crops and ornamental plants such as species of Camellia, Castanea, Citrus, Glycine, Helianthus, Juglans, Persea, Pyrus, Vaccinium, Vitis and many more (van Rensburg et al. 2006; Santos and Phillips 2009; Crous et al. 2011a, b, 2016; Santos et al. 2011; Thompson et al. 2011; Grasso et al. 2012; Huang et al. 2013; Lombard et al. 2014; Gao et al. 2015, 2016, 2017; Udayanga et al. 2012, 2015; Guarnaccia et al. 2016; Dissanayake et al. 2017; Guarnaccia and Crous 2017; Fan et al. 2018; Senanayake et al. 2018; Guo et al. 2020). Diaporthe species have been reported as destructive plant pathogens, harmless endophytes or saprobes (Murali et al. 2006; Udayanga et al. 2012; Gomes et al. 2013; Ménard et al. 2014; Guarnaccia et al. 2016; Torres et al. 2016; Senanayake et al. 2018). However, the biology and lifestyle of some of these fungi remain unclear (Vilka and Volkova 2015).
In the past, methods of species identification of Diaporthe had previously been based only on host as well as morphological characters such as the size and shape of ascomata and conidiomata. Nowadays, molecular phylogenetic studies demonstrate that determining species boundaries only by morphological characters is not possible due to lack of host specificity and their variability under changing environmental conditions (Gomes et al. 2013). Phylogenetic analysis using a five-locus dataset (ITS-TUB-TEF-CAL-HIS) has been determined to be the optimal combination to identify species of Diaporthe species, as revealed by Santos et al. (2017). Many Diaporthe species are described based on a polyphasic approach together with morphological characterisation (Rehner and Uecker 1994; Udayanga et al. 2011; Gao et al. 2017; Guarnaccia and Crous 2017; Yang et al. 2018a, 2020; Crous et al. 2020; Dayarathne et al. 2020; Guo et al. 2020; Hyde et al. 2020; Li et al. 2020; Zapata et al. 2020).
The aim of this study was to explore the diversity of Diaporthe species from symptomatic leaves of plants in Yunnan Province. We present three novel species and five known species of Diaporthe, collected from species belonging to ten host genera, based on morphological characters and phylogenetic analysis.
Materials and methods
Isolation and morphological studies
Leaves of samples were collected in Yunnan Province, China. Isolations from surface sterilized leaf tissues were conducted following the protocol of Gao et al. (2014). Tissue fragments (5 × 5 mm) were taken from the margin of leaf lesions and surface-sterilized by immersing them in 75% ethanol solution for 1 min, 5% sodium hypochlorite solution for 30 s, and then rinsing in sterile distilled water for 1 min. The pieces were dried with sterilized paper towels and placed on potato dextrose agar (PDA) (Cai et al. 2009). PDA plates (90 mm) were incubated in an incubator at 25 °C for 2–4 days, and hyphae were picked out of the periphery of the colonies and inoculated onto new PDA plates.
Following 2–3 weeks of incubation, photographs of colonies were taken at 7 days and 15 days using a Powershot G7X mark II digital camera. Colour notations was done using the colour charts of Rayner (1970). Micromorphological characters were observed using an Olympus SZX10 stereomicroscope and Olympus BX53 microscope, both fitted with Olympus DP80 high definition colour digital cameras to document fungal structures. All fungal strains were stored in 10% sterilized glycerin at 4 °C for further studies. Voucher and type specimens were deposited in the Herbarium of Plant Pathology, Shandong Agricultural University (HSAUP). Living cultures were deposited in the Shandong Agricultural University Culture Collection (SAUCC). Taxonomic information of the new taxa was submitted to MycoBank (http://www.mycobank.org).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelium on PDA, using a modified cetyltrimethylammonium bromide (CTAB) protocol as described in Guo et al. (2000). The internal transcribed spacer regions with intervening 5.8S nrRNA gene (ITS), part of the beta-tubulin gene region (TUB), partial translation elongation factor 1-alpha (TEF), histone H3 (HIS) and calmodulin (CAL) genes were amplified and sequenced by using primers pairs ITS4/ITS5 (White et al. 1990), Bt2a/Bt2b (Glass and Donaldson 1995), EF1-728F/EF1-986R (Carbone and Kohn 1999), CAL-228F/CAL-737R (Carbone and Kohn 1999) and CYLH3F/H3-1b (Glass and Donaldson 1995; Crous et al. 2004), respectively.
PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification reactions were performed in a 25 μL reaction volume, which contained 12.5 μL Green Taq Mix (Vazyme, Nanjing, China), 1 μL of each forward and reverse primer (10 μM) (Biosune, Shanghai, China), and 1 μL template genomic DNA in amplifier, and were adjusted with distilled deionized water to a total volume of 25 μL.
PCR parameters were as follows: 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at a suitable temperature for 30 s, extension at 72 °C for 1 min and a final elongation step at 72 °C for 10 min. Annealing temperature for each gene were 55 °C for ITS, 60 °C for TUB, 52 °C for TEF, 54 °C for CAL and 57 °C for HIS. The PCR products were visualised on 1% agarose electrophoresis gel. Sequencing was done bi-directionally, conducted by the Biosune Company Limited (Shanghai, China). Consensus sequences were obtained using MEGA 7.0 (Kumar et al. 2016). All sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
Species and Genbank accession numbers of DNA sequences used in this study. New sequences in bold.
| Species | Voucher | Host/Substrare | GeneBank accession number | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| ITS | TUB | TEF | CAL | HIS | ||||
| Diaporthe acuta | PSCG 046 | Pyrus pyrifolia | MK626958 | MK691224 | MK654803 | MK691124 | MK726162 | Guo et al. 2020 |
| PSCG 047* | Pyrus pyrifolia | MK626957 | MK691225 | MK654802 | MK691125 | MK726161 | Guo et al. 2020 | |
| D. acutispora | LC6160 | Camellia sasanqua | KX986763 | KX999194 | KX999154 | KX999273 | KX999234 | Gao et al. 2017 |
| LC6161 | Coffea sp. | KX986764 | KX999195 | KX999155 | KX999274 | KX999235 | Gao et al. 2017 | |
| D. amaranthophila | MAFF 246900 | Amaranthus tricolor | LC459575 | LC459579 | LC459577 | LC459583 | LC459581 | Rossman et al. 2015 |
| MAFF 246901 | Amaranthus tricolor | LC459576 | LC459580 | LC459578 | LC459584 | LC459582 | Rossman et al. 2015 | |
| D. angelicae | CBS 111592* | Heracleum sphondylium | KC343027 | KC343995 | KC343753 | KC343269 | KC343511 | Gomes et al. 2013 |
| D. anhuiensis | CNUCC 201901* | Cunninghamia lanceolata | MN219718 | MN227008 | MN224668 | MN224549 | MN224556 | Zhou and Hou 2019 |
| CNUCC 201902 | Cunninghamia lanceolata | MN219727 | MN227009 | MN224669 | MN224550 | MN224557 | Zhou and Hou 2019 | |
| D. arctii | DP0482 | Arctium sp. | KJ590736 | KJ610891 | KJ590776 | KJ612133 | KJ659218 | Udayanga et al. 2015 |
| D. arecae | CBS 161.64* | Areca catechu | KC343032 | KC344000 | KC343758 | KC343274 | KC343516 | Gomes et al. 2013 |
| CBS 535.75 | Citrus sp. | KC343033 | KC344001 | KC343759 | KC343275 | KC343517 | Gomes et al. 2013 | |
| SAUCC194.18 | Persea americana | MT822546 | MT855743 | MT855860 | MT855631 | MT855515 | This study | |
| D. arengae | CBS 114979* | Arenga engleri | KC343034 | KC344002 | KC343760 | KC343276 | KC343518 | Gomes et al. 2013 |
| D. aseana | MFLUCC 12-0299a* | On dead leaves | KT459414 | KT459432 | KT459448 | KT459464 | – | Dissanayake et al. 2017 |
| D. beilharziae | BRIP 54792* | Indigofera australis | JX862529 | KF170921 | JX862535 | – | – | Tan et al. 2013 |
| D. biconispora | ZJUD 60 | Citrus sinensis | KJ490595 | KJ490416 | KJ490474 | – | KJ490537 | Huang et al. 2017 |
| ZJUD 61 | Fortunella margarita | KJ490596 | KJ490417 | KJ490475 | – | KJ490538 | Huang et al. 2017 | |
| ZJUD 62 | Citrus grandis | KJ490597 | KJ490418 | KJ490476 | – | KJ490539 | Huang et al. 2017 | |
| D. brasiliensis | CBS 133183* | Aspidosperma tomentosus | KC343042 | KC344010 | KC343768 | KC343284 | KC343526 | Gomes et al. 2013 |
| D. caatingaensis | URM 7486* | Tacinga inamoena | KY085926 | KY115600 | KY115603 | KY115597 | KY115605 | Crous et al. 2017 |
| D. camporesii | JZB320143 | Urtica dioidca | MN535309 | MN561316 | MN984254 | – | – | Hyde et al. 2020 |
| D. caricae-papayae | NIBM-ABIJP | Carica papaya | MN335224 | – | – | – | – | Rossman et al. 2015 |
| D. caryae | CFCC 52563 | Carya illinoensis | MH121498 | MH121580 | MH121540 | MH121422 | MH121458 | Yang et al. 2018 |
| CFCC 52564 | Carya illinoensis | MH121499 | MH121581 | MH121541 | MH121423 | MH121459 | Yang et al. 2018 | |
| D. cercidis | CFCC 52565 | Cercis chinensis | MH121500 | MH121582 | MH121542 | MH121424 | MH121460 | Yang et al. 2018 |
| D. chrysalidocarpi | SAUCC194.33 | Chrysalidocarpus lutescens | MT822561 | MT855758 | MT855874 | MT855645 | MT855530 | This study |
| SAUCC194.35* | Chrysalidocarpus lutescens | MT822563 | MT855760 | MT855876 | MT855646 | MT855532 | This study | |
| D. cichorii | MFLUCC 17-1023* | Cichorium intybus | KY964220 | KY964104 | KY964176 | KY964133 | – | Dissanayake et al. 2017 |
| D. compacta | LC3083* | Camellia sinensis | KP267854 | KP293434 | KP267928 | – | KP293508 | Gao et al. 2016 |
| D. cucurbitae | CBS 136.25 | Cucumis sativus | KC343031 | KC343999 | KC343757 | KC343273 | KC343515 | Udayanga et al. 2014 |
| D. cuppatea | CBS 117499 | Aspalathus linearis | KC343057 | KC344025 | KC343783 | KC343299 | KC343541 | Udayanga et al. 2012 |
| D. decedens | CBS 109772 | Corylus avellana | KC343059 | KC344027 | KC343785 | KC343301 | KC343543 | Gomes et al. 2013 |
| D. eugeniae | CBS 444.82 | Eugenia aromatica | KC343098 | KC344066 | KC343824 | KC343340 | KC343582 | Gomes et al. 2013 |
| D. fraxini-angustifoliae | BRIP 54781* | Fraxinus angustifolius | JX862528 | KF170920 | JX862534 | – | – | Tan et al. 2013 |
| D. fulvicolor | PSCG 051* | Pyrus pyrifolia | MK626859 | MK691236 | MK654806 | MK691132 | MK726163 | Guo et al. 2020 |
| PSCG 057 | Pyrus pyrifolia | MK626858 | MK691233 | MK654810 | MK691131 | MK726164 | Guo et al. 2020 | |
| D. ganjae | CBS 180.91* | Cannabis sativa | KC343112 | KC344080 | KC343838 | KC343354 | KC343596 | Gomes et al. 2013 |
| D. guangxiensis | JZBH 320094* | Vitis vinifera | MK335772 | MK500168 | MK523566 | MK736727 | – | Manawasinghe et al. 2019 |
| D. gulyae | MF-Ha 17-042* | Helianthus annuus | MK024252 | MK033488 | MK039420 | – | – | Thompson et al. 2011 |
| D. hongkongensis | CBS 115448* | Dichroa febrifuga | KC343119 | KC344087 | KC343845 | KC343361 | KC343603 | Gomes et al. 2013 |
| CGMCC 3.17102 | Lithocarpus glaber | KF576275 | KF576299 | KF576250 | KF576227 | – | Gao et al. 2015 | |
| LC 3478 | Camellia sinensis | KP267904 | KP293484 | KP267978 | – | KP293553 | Gao et al. 2017 | |
| SAUCC194.81 | Millettia reticulata | MT822609 | MT855806 | MT855921 | MT855688 | MT855577 | This study | |
| SAUCC194.87 | Camellia sinensis | MT822615 | MT855812 | MT855927 | MT855694 | MT855583 | This study | |
| D. huangshanensis | CNUCC 201903 | Camellia oleifera | MN219729 | MN227010 | MN224670 | – | MN224558 | Zhou and Hou 2019 |
| CNUCC 201904 | Camellia oleifera | MN219730 | MN227011 | MN224671 | – | MN224559 | Zhou and Hou 2019 | |
| D. infecunda | CBS 133812* | Schinus terebinthifolius | KC343126 | KC344094 | KC343852 | KC343368 | KC343610 | Gomes et al. 2013 |
| D. krabiensis | MFLUCC 17-2481* | Bruguiera sp. | MN047101 | MN431495 | MN433215 | – | – | Dayarathne et al. 2020 |
| D. litchiicola | BRIP 54900* | Litchi chinensis | JX862533 | KF170925 | JX862539 | – | – | Tan et al. 2013 |
| D. limonicola | CPC 28200* | Citrus limon | MF418422 | MF418582 | MF418501 | MF418256 | MF418342 | Guarnaccia and Crous 2017 |
| D. lusitanicae | CBS 123212* | Foeniculum vulgare | KC343136 | KC344104 | KC343862 | KC343378 | KC343620 | Phillips and Santos 2009 |
| D. machili | SAUCC194.69 | Pometia pinnata | MT822597 | MT855794 | MT855909 | MT855677 | MT855565 | This study |
| SAUCC194.111* | Machilus pingii | MT822639 | MT855836 | MT855951 | MT855718 | MT855606 | This study | |
| D. malorum | CAA752* | Malus domestica | KY435643 | KY435671 | KY435630 | KY435661 | KY435651 | Santos et al. 2017 |
| CAA740 | Malus domestica | KY435642 | KY435670 | KY435629 | KY435660 | KY435650 | Santos et al. 2017 | |
| D. manihotia | CBS 505.76 | Manihot utilissima | KC343138 | KC344106 | KC343864 | KC343380 | KC343622 | Gomes et al. 2013 |
| D. mayteni | CBS 133185* | Maytenus ilicicolia | KC343139 | KC344107 | KC343865 | KC343381 | KC343623 | Gomes et al. 2013 |
| D. melitensis | CPC 27873* | Citrus limon | MF418424 | MF418584 | MF418503 | MF418258 | MF418344 | Guarnaccia and Crous 2017 |
| D. middletonii | BRIP 54884e* | Rapistrum rugostrum | KJ197286 | KJ197266 | KJ197248 | – | – | Thompson et al. 2015 |
| SAUCC194.27 | Litchi chinensis | MT822555 | MT855752 | MT855868 | MT855639 | MT855524 | This study | |
| SAUCC194.45 | Lithocarpus glaber | MT822573 | MT855770 | MT855886 | MT855654 | MT855542 | This study | |
| SAUCC194.46 | Lithocarpus glaber | MT822574 | MT855771 | MT855887 | MT855655 | MT855543 | This study | |
| SAUCC194.48 | Lithocarpus craibianus | MT822576 | MT855773 | MT855889 | MT855657 | MT855545 | This study | |
| D. millettiae | GUCC9167* | Millettia reticulata | MK398674 | MK502089 | MK480609 | MK502086 | – | Long et al. 2019 |
| D. multigutullata | ZJUD 98* | Citrus grandis | KJ490633 | KJ490454 | KJ490512 | – | KJ490575 | Huang et al. 2015 |
| D. musigena | CBS 129519* | Musa sp. | KC343143 | KC344111 | KC343869 | KC343385 | KC343627 | Crous et al. 2011 |
| D. myracrodruonis | URM7972 | Myracrodruon urundeuva | MK205289 | MK205291 | MK213408 | MK205290 | – | Silva et al. 2019 |
| D. neoarctii | CBS 109490* | Ambrosia trifida | KC343145 | KC344113 | KC343871 | KC343387 | KC343629 | Gomes et al. 2013 |
| D. novem | CBS 127270* | Glycine max | KC343156 | KC344124 | KC343882 | KC343398 | KC343640 | Santos et al. 2011 |
| D. osmanthi | GUCC9165* | Osmanthus fragrans | MK398675 | MK502091 | MK480610 | MK502087 | – | Long et al. 2019 |
| SAUCC194.21 | Litchi chinensis | MT822549 | MT855746 | MT855862 | MT855634 | MT855518 | This study | |
| D. oxe | CBS 133186* | Maytenus ilicifolia | KC343164 | KC344132 | KC343890 | KC343406 | KC343648 | Gomes et al. 2013 |
| CBS 133187 | Maytenus ilicifolia | KC343165 | KC344133 | KC343891 | KC343407 | KC343649 | Gomes et al. 2013 | |
| D. pandanicola | MFLUCC 17-0607 | Pandanus sp. | MG646974 | MG646930 | – | – | – | Tibpromma et al. 2018 |
| SAUCC194.82 | Millettia reticulata | MT822610 | MT855807 | MT855922 | MT855689 | MT855578 | This study | |
| D. paranensis | CBS 133184* | Maytenus ilicifolia | KC343171 | KC344139 | KC343897 | KC343413 | KC343655 | Gomes et al. 2013 |
| D. pascoei | BRIP 54847* | Persea americana | JX862532 | KF170924 | JX862538 | – | – | Tan et al. 2013 |
| D. perseae | CBS 151.73 | Persea gratissima | KC343173 | KC344141 | KC343899 | KC343415 | KC343657 | Gomes et al. 2013 |
| D. pescicola | MFLU 16-0105* | Prunus persica | KU557555 | KU557579 | KU557623 | KU557603 | – | Dissanayake et al. 2017 |
| D. podocarpi-macrophylli | LC6155* | Podocarpus macrophyllus | KX986774 | KX999207 | KX999167 | KX999278 | KX999246 | Gao et al. 2017 |
| LC6200 | Podocarpus macrophyllus | KX986769 | KX999201 | KX999161 | KX999276 | KX999240 | Gao et al. 2017 | |
| D. pometiae | SAUCC194.19 | Persea americana | MT822547 | MT855744 | MT855861 | MT855632 | MT855516 | This study |
| SAUCC194.72* | Pometia pinnata | MT822600 | MT855797 | MT855912 | MT855679 | MT855568 | This study | |
| SAUCC194.73 | Heliconia metallica | MT822601 | MT855798 | MT855913 | MT855680 | MT855569 | This study | |
| D. pseudomangiferae | CBS 101339* | Mangifera indica | KC343181 | KC344149 | KC343907 | KC343423 | KC343665 | Gomes et al. 2013 |
| D. pseudophoenicicola | CBS 462.69* | Phoenix dactylifera | KC343184 | KC344152 | KC343910 | KC343426 | KC343668 | Gomes et al. 2013 |
| D. pterocarpicola | MFLUCC 10-0580a* | Pterocarpus indicus | JQ619887 | JX275441 | JX275403 | JX197433 | – | Udayanga et al. 2012 |
| MFLUCC 10-0580b | Pterocarpus indicus | JQ619888 | JX275442 | JX275404 | JX197434 | – | Udayanga et al. 2012 | |
| D. pyracanthae | CAA487* | Pyracantha coccinea | KY435636 | KY435667 | KY435626 | KY435657 | KY435647 | Santos et al. 2017 |
| D. racemosae | CPC 26646* | Euclea racemosa | MG600223 | MG600227 | MG600225 | MG600219 | MG600221 | Marin-Felix et al. 2018 |
| D. raonikayaporum | CBS 133182* | Spondias mombin | KC343188 | KC344156 | KC343914 | KC343430 | KC343672 | Gomes et al. 2013 |
| D. rossmaniae | CAA 762* | Vaccinium corymbosum | MK792290 | MK837914 | MK828063 | MK883822 | MK871432 | Hilario et al. 2020 |
| D. sackstonii | BRIP 54669b* | Helianthus annuus | KJ197287 | KJ197267 | KJ197249 | – | – | Thompson et al. 2015 |
| D. salinicola | MFLU 18-0553* | Xylocarpus sp. | MN047098 | – | MN077073 | – | – | Dayarathne et al. 2020 |
| MFLU 17-2592 | Xylocarpus sp. | MN047099 | – | MN077074 | – | – | Dayarathne et al. 2020 | |
| D. schini | CBS 133181* | Schinus terebinthifolius | KC343191 | KC344159 | KC343917 | KC343433 | KC343675 | Gomes et al. 2013 |
| D. schoeni | MFLU 15-2609 | Schoenus nigricans | KY964229 | KY964112 | KY964185 | KY964141 | – | Dissanayake et al. 2017 |
| D. sennae | CFCC 51636* | Senna bicapsularis | KY203724 | KY228891 | KY228885 | KY228875 | – | Yang et al. 2017 |
| D. serafiniae | BRIP 55665a* | Helianthus annuus | KJ197274 | KJ197254 | KJ197236 | – | – | Thompson et al. 2015 |
| D. spinosa | PSCG 383* | Pyrus pyrifolia | MK626849 | MK691234 | MK654811 | MK691129 | MK726156 | Guo et al. 2020 |
| D. stewartii | CBS 193.36* | Cosmos bipinnatus | FJ889448 | JX275421 | GQ250324 | JX197415 | – | Santos et al. 2010; Udayanga et al. 2012 |
| D. subordinaria | CBS 101711 | Plantago lanceolata | KC343213 | KC344181 | KC343939 | KC343455 | KC343697 | Gomes et al. 2013 |
| CBS 464.90 | Plantago lanceolata | KC343214 | KC344182 | KC343940 | KC343456 | KC343698 | Gomes et al. 2013 | |
| D. taoicola | PSGG485 | Prunus persica | MK626869 | MK691227 | MK654812 | MK691120 | MK726173 | Dissanayake et al. 2017 |
| D. tarchonanthi | CPC 37479 | Tarchonanthus littoralis | MT223794 | – | – | – | – | Crous et al. 2020 |
| D. tectonigena | MFLUCC 12-0767* | Tectona grandis | KU712429 | KU743976 | KU749371 | KU749358 | – | Doilom et al. 2016 |
| D. terebinthifolii | CBS 133180* | Schinus terebinthifolius | KC343216 | KC344184 | KC343942 | KC343458 | KC343700 | Gomes et al. 2013 |
| D. undulate | LC6624* | Unknown host | KX986798 | KX999230 | KX999190 | – | KX999269 | Gao et al. 2017 |
| LC8110 | Unknown host | KY491545 | KY491565 | KY491555 | – | – | Gao et al. 2017 | |
| D. vawdreyi | BRIP 57887a* | Psidium guajava | KR936126 | KR936128 | KR936129 | – | – | Crous et al. 2015 |
| D. viniferae | JZBH 320071 | Vitis vinifera | MK341550 | MK500112 | MK500107 | MK500119 | – | Manawasinghe et al. 2019 |
| JZBH 320072 | Vitis vinifera | MK341551 | MK500113 | MK500108 | MK500120 | – | Manawasinghe et al. 2019 | |
| D. xishuangbanica | LC6707* | Camellia sinensis | KX986783 | KX999216 | KX999175 | – | KX999255 | Gao et al. 2017 |
| Diaporthella corylina | CBS 121124 | Corylus sp. | KC343004 | KC343972 | KC343730 | KC343246 | KC343488 | Gomes et al. 2013 |
Isolates marked with “*” are ex-type or ex-epitype strains.
Phylogenetic analyses
Novel sequences generated from the sixteen strains in this study, and all reference sequences of Diaporthe species downloaded from GenBank, were used for phylogenetic analyses. Alignments of the individual locus were determined using MAFFT v. 7.110 by default settings (Katoh et al. 2017) and manually corrected where necessary. To establish the identity of the isolates at species level, phylogenetic analyses were conducted first individually for each locus and then as combined analyses of five loci (ITS, TUB, TEF, CAL and HIS regions). Phylogenetic analyses were based on maximum likelihood (ML) and Bayesian inference (BI) for the multi-locus analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Nylander 2004) and incorporated into the analyses. ML and BI were run on the CIPRES Science Gateway portal (https://www.phylo.org/) (Miller et al. 2012) using RaxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014) and MrBayes on XSEDE (3.2.7a) (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012), respectively. For ML analyses the default parameters were used and BI was carried out using the rapid bootstrapping algorithm with the automatic halt option. Bayesian analyses included five parallel runs of 5,000,000 generations, with the stop rule option and a sampling frequency of 500 generations. The burn-in fraction was set to 0.25 and posterior probabilities (PP) were determined from the remaining trees. The resulting trees were plotted using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree) and edited with Adobe Illustrator CS5.1. New sequences generated in this study were deposited at GenBank (https://www.ncbi.nlm.nih.gov; Table 1) and the alignments and trees were deposited in TreeBASE: S27479 (http://treebase.org/treebase-web/home.html).
Results
Phylogenetic analyses
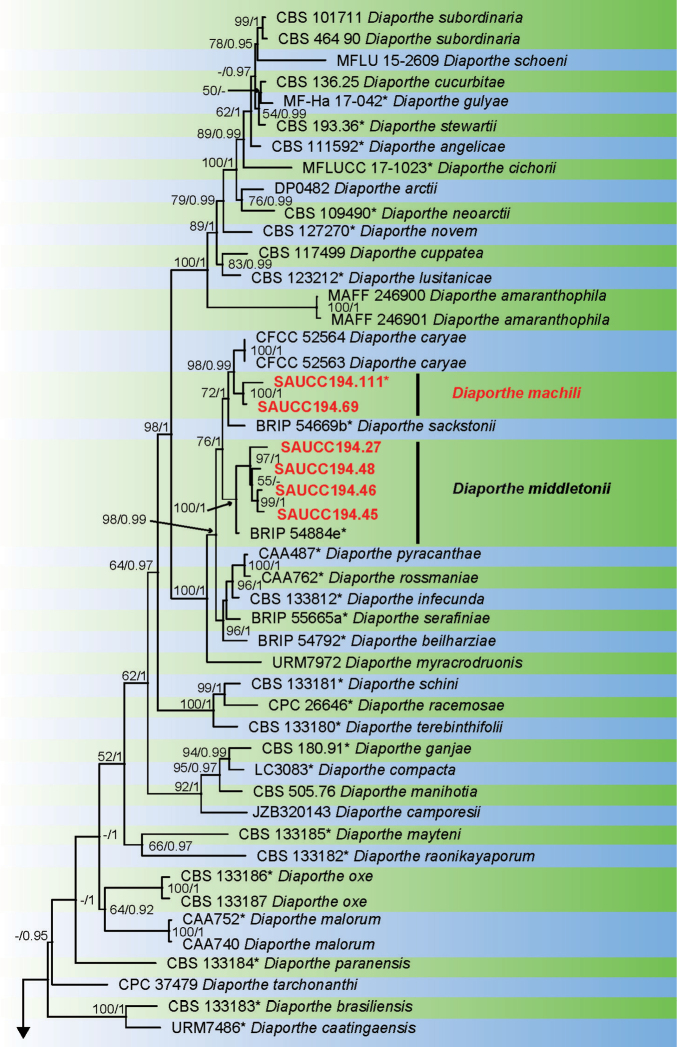
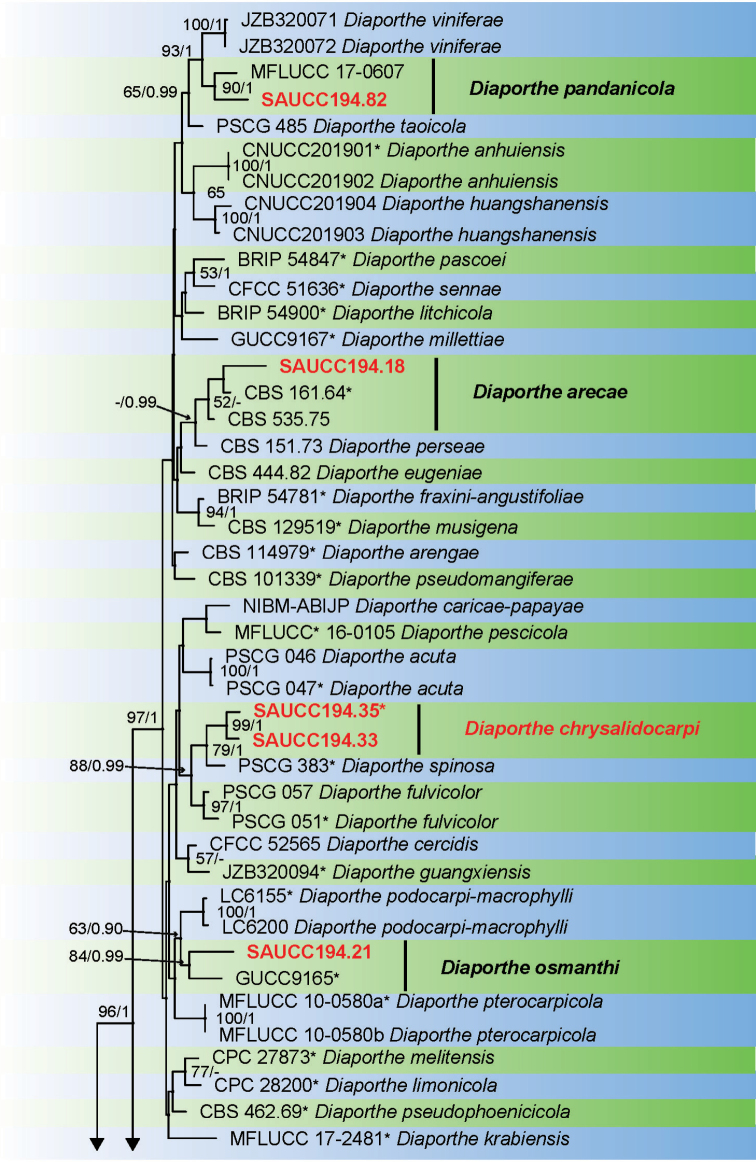
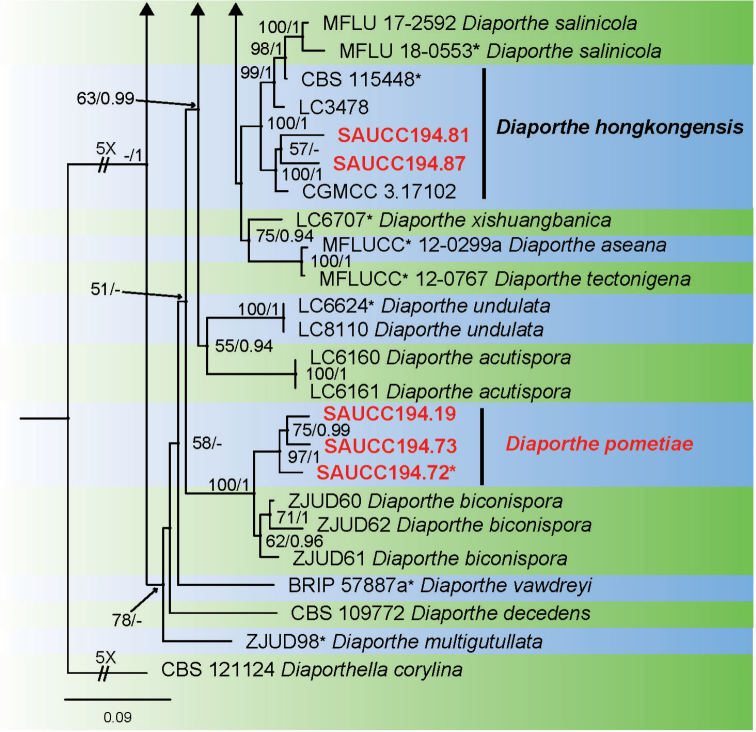
Sixteen strains of Diaporthe isolated from plant hosts from Yunnan, China, were grown in culture and used for analyses of molecular sequence data. Diaporthe spp. were analysed by using multilocus data (ITS, TUB, TEF, CAL and HIS) from 115 isolates of Diaporthe spp. and Diaporthella corylina (CBS 121124) as the outgroup taxon. A total of 3005 characters including gaps were obtained in the phylogenetic analysis, viz. ITS: 1–656, TUB: 657–1329, TEF: 1330–1860, CAL: 1861–2444, HIS: 2445–3005. Of these characters, 1349 were constant, 453 were variable and parsimony-uninformative, and 1203 were parsimony-informative. For the BI and ML analyses, the substitution model GTR+I+G for ITS, TUB, TEF and HIS, HKY+I+G for and CAL were selected and incorporated into the analyses. The ML tree topology confirmed the tree topologies obtained from the BI analyses, and therefore, only the ML tree is presented (Fig. 1).
Figure 1.
Phylogram of Diaporthe spp. based on combined sequence data of ITS, TUB, TEF, CAL and HIS genes. The ML and BI bootstrap support values above 50% and 0.90 BYPP are shown at the first and second position, respectively. Strains marked with “*” are ex-type or ex-epitype. Codes referring to strains from the current study are written in red. Some branches were shortened to fit them to the page as indicated by two diagonal lines with the number of times a branch was shortened indicated.
Figure 1.
Continued.
Figure 1.
Continued.
ML bootstrap support values (≥ 50%) and Bayesian posterior probability (≥ 0.90) are shown as first and second position above nodes, respectively. Based on the five-locus phylogeny and morphology, nine isolates were assigned to five species, including Diaporthe arecae (1), D. hongkongensis (2), D. middletonii (4), D. osmanthi (1) and D. pandanicola (1), whereas seven isolates formed distinct well supported clades, which refer to novel species named D. chrysalidocarpi (2), D. machili (2) and D. pometiae (3), respectively.
Taxonomy
Diaporthe arecae
(H.C. Srivast., Zakia & Govindar.) R.R. Gomes, Glienke & Crous, Persoonia 31: 16. (2013)
49466F7B-A01E-51FB-A01C-15E29B1B35D9
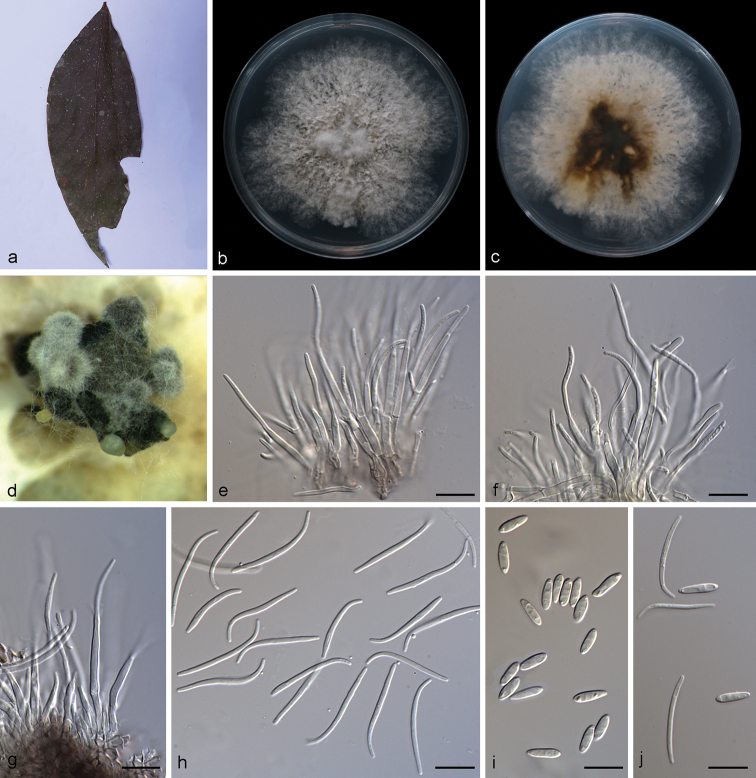
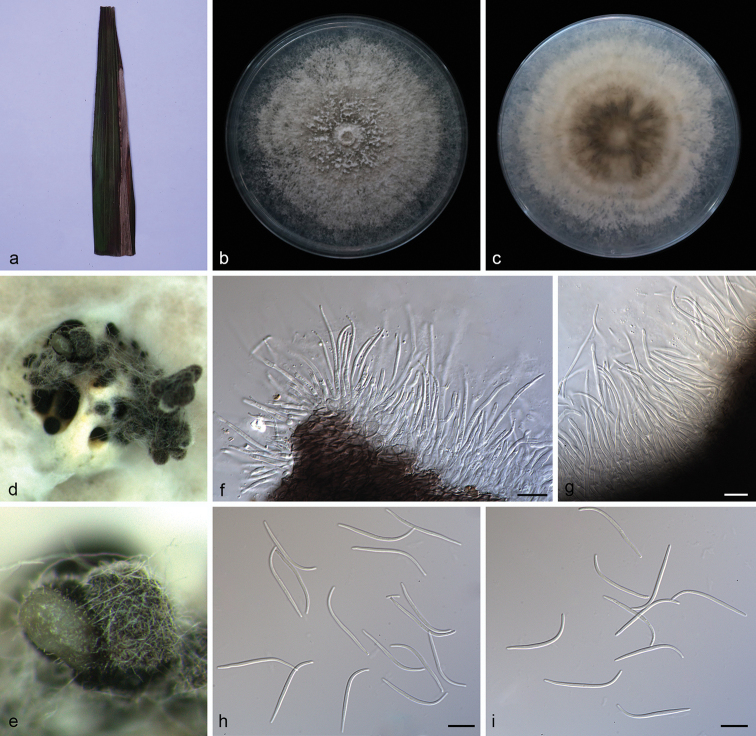
Figure 2.
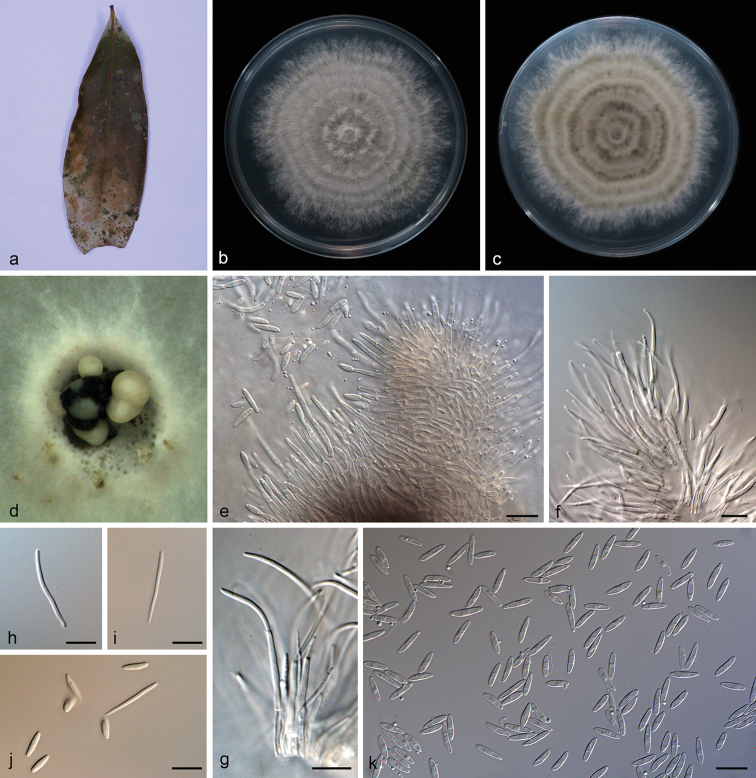
Diaporthe arecae (SAUCC194.18) a infected leaf of Persea americanab, c surface and reverse of a colony after 15 days on PDAd conidiomata e–g conidiophores and conidiogenous cells h beta conidia i alpha conidia j alpha conidia and beta conidia. Scale bars: 10 μm (e–j).
Subramanella arecae H.C. Srivast., Zakia & Govindar., in Srivastava, Banu and Govindarajan (1962). Basionym.
Description.
Asexual morph: Conidiomata pycnidial, several pycnidia grouped together, globose, black, erumpent, exuding creamy to yellowish conidial droplets from ostioles. Conidiophores hyaline, septate, branched, cylindrical, straight to sinuous, 25.0–32.0 × 1.4–2.5 μm. Conidiogenous cells 10.5–20.7 × 1.4–2.0 μm, phialidic, cylindrical, swollen at base, tapering towards apex, slightly curved. Alpha conidia hyaline, smooth, aseptate, ellipsoidal, guttulate, apex subobtuse, base subtruncate, 7.5–10.0 × 1.8–3.0 µm (mean = 8.2 × 2.4 μm, n = 20). Beta conidia hyaline, aseptate, filiform, slightly curved, tapering towards base, 18.5–26.5 × 1.0–1.8 µm (mean = 24.3 × 1.4 μm, n = 20). Gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 11.2–13.3 mm diam/day. Aerial mycelium white, cottony, feathery, abundant in center, sparse in margin, white on surface, reverse yellowish to tan.
Specimen examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Persea americana (Lauraceae). 19 April 2019, S.T. Huang, HSAUP194.18, living culture SAUCC194.18.
Notes.
Diaporthe arecae (CBS 161.64) was originally described as Subramanella arecae on fruit of Areca catechu in India (Srivastava et al. 1962) and placed in Diaporthe by Gomes et al. (2013). The Diaporthe isolate from fruits of Citrus sp. (CBS 535.75) in Suriname was also placed in D. arecae by Gomes et al. (2013). In the present study, strain (SAUCC194.18) from symptomatic leaves of Persea americana was congruent with D. arecae based on morphology and DNA sequences data (Fig. 1). We therefore consider the isolated strain as D. arecae.
Diaporthe chrysalidocarpi
S.T. Huang, J.W. Xia, W.X. Sun, & X.G. Zhang sp. nov.
F02A5541-B8BA-55D2-92D9-190AD164B410
837812
Figure 3.
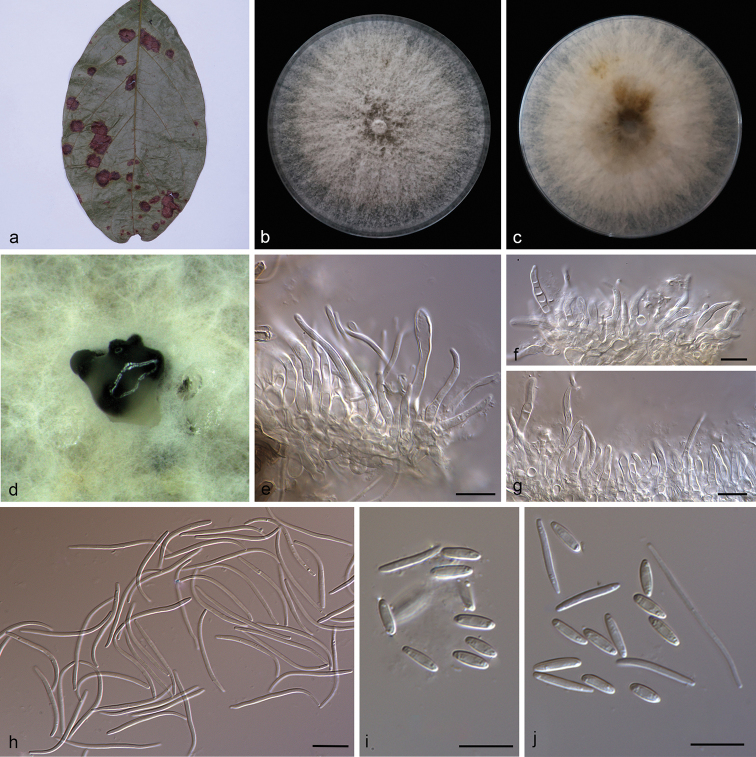
Diaporthe chrysalidocarpi (SAUCC194.35) a diseased leaf of Chrysalidocarpus lutescensb, c surface and reverse of a colony after 15 days on PDAd, e conidiomata f, g conidiophores and conidiogenous cells h, i beta conidia. Scale bars: 10 μm (f–i).
Etymology.
Named after the host genus on which it was collected, Chrysalidocarpus lutescens.
Diagnosis.
Diaporthe chrysalidocarpi can be distinguished from the phylogenetically most closely related species D. spinosa by longer beta conidia (28.0–32.5 × 1.2–1.6 vs. 18.5–30.5 × 1.0–1.5 μm), and from other species D. fulvicolor by the types of conidia (D. chrysalidocarpi produces only beta conidia, while D. fulvicolor produces only alpha conidia) and several loci (25/491 in the ITS region, 18/471 TUB, 4/298 TEF, 28/458 CAL and 13/441 HIS).
Type.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Chrysalidocarpus lutescens (Palmae). 19 April 2019, S.T. Huang, HSAUP194.35 holotype, ex-type living culture SAUCC194.35.
Description.
Asexual morph: Leaf spots irregular, pale brown in center, brown to tan at margin. Conidiomata pycnidial, scattered or aggregated, black, erumpent, raising above surface of culture medium, subglobose, exuding white or yellowish creamy conidial droplets from central ostioles after 30 days in light at 25 °C; pycnidial wall consists of black to dark brown, thin-walled cells. Conidiophores 27.5–35.0 × 1.4–2.0 μm, hyaline, slightly branched, swelling at base, subcylindrical, septate, smooth, straight or curved. Conidiogenous cells 10.5–23.0 × 1.4–1.8 μm, phialidic, cylindrical, terminal, straight to sinuous, tapering towards apex. Beta conidia 28.0–32.5 × 1.2–1.6 μm (mean = 30.3 × 1.3 μm, n = 20), filiform, hyaline, straight or slightly curved, aseptate, base subtruncate, tapering towards the base. Alpha conidia and gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 13.3–15.2 mm diam/day, initially white, becoming greyish, reverse pale brown, with concentric rings of dense, sparse hyphae, irregular margin, fluffy aerial mycelium at center, pycnidia forming after 15 days.
Additional specimen examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Chrysalidocarpus lutescens (Palmae). 19 April 2019, S.T. Huang, HSAUP194.33 paratype; living culture SAUCC194.33.
Notes.
Phylogenetic analysis of a combined five gene showed that D. chrysalidocarpi formed an independent clade (Fig. 1) and is phylogenetically distinct from D. spinosa and D. fulvicolor. This species can be distinguished from D. spinosa by 61 different nucleotides in the concatenated alignment (13/492 in the ITS region, 17/471 TUB, 4/298 TEF, 17/458 CAL and 10/441 HIS), and D. fulvicolor by 88 nucleotides (25/491 in the ITS region, 18/471 TUB, 4/298 TEF, 28/458 CAL and 13/441 HIS). Morphologically, D. chrysalidocarpi differs from D. spinosa in having longer beta conidia (28.0–32.5 × 1.2–1.6 vs. 18.5–30.5 × 1.0–1.5 μm) (Guo et al. 2020). Furthermore, Diaporthe chrysalidocarpi produces only beta conidia, while D. spinosa produces alpha conidia and beta conidia and D. fulvicolor produces only alpha conidia (Guo et al. 2020). Therefore, we establish this fungus as a novel species.
Diaporthe hongkongensis
R.R. Gomes, Glienke, Crous, Persoonia 31: 23. (2013)
D074B02D-1506-5551-AB26-43FB98A81B31
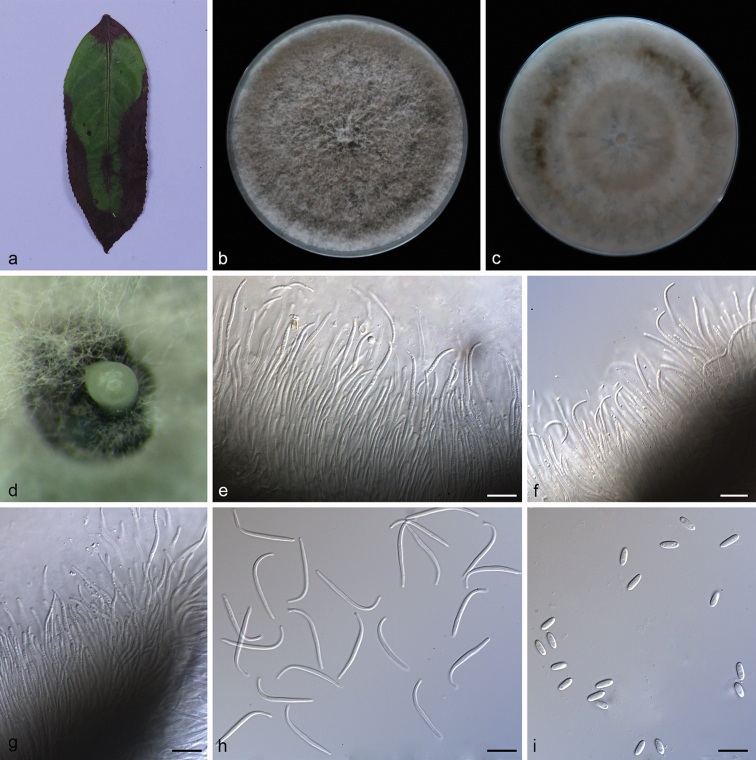
Figure 4.
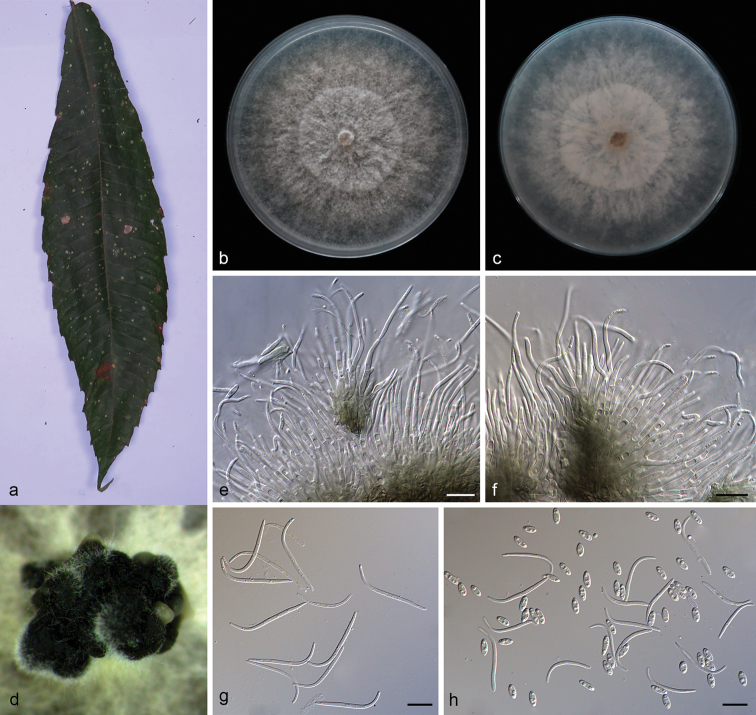
Diaporthe hongkongensis (SAUCC194.87) a diseased leaf of Camellia sinensisb, c surface and reverse of colony after 15 days on PDAd conidiomata e–g conidiophores and conidiogenous cells h beta conidia i alpha conidia. Scale bars: 10 μm (e–i).
Description.
Asexual morph: Conidiomata pycnidial, subglobose or globose, solitary, black, erumpent, coated with white hyphae, thick-walled, exuding creamy conidial droplets from central ostioles. Conidiophores hyaline, smooth, septate, unbranched, densely aggregated, cylindrical or clavate, straight to sinuous, swollen at base, tapering towards apex, 32.0–42.0 × 2.0–2.9 μm. Conidiogenous cells 20.0–24.2 × 1.3–2.3 μm, phialidic, cylindrical, terminal, slightly tapering towards apex. Alpha conidia, hyaline, smooth, aseptate, ellipsoidal or oval, 0–2 guttulate, apex subobtuse, base subtruncate, 5.5–7.0 × 2.0–2.5 µm (mean = 6.2 × 2.2 μm, n = 20). Beta conidia hyaline, aseptate, filiform, hamate, tapering towards both ends, mostly J-shaped, 21.5–27.0 × 1.4–1.8 µm (mean = 25.6 × 1.3 μm, n = 20). Gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 19.0–21.5 mm diam/day, cottony, radial with abundant aerial mycelium, sparse at margin, with an obvious pale brown concentric ring of dense hyphae, white to grayish on surface with age, white to pale brown on the reverse side.
Specimens examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, 19 April 2019, S.T. Huang. On diseased leaves of Millettia reticulata (Fabaceae) HSAUP194.81, living culture SAUCC194.81; on diseased leaves of Camellia sinensis (Theaceae) HSAUP194.87, living culture SAUCC194.87.
Notes.
In the present study, two strains (SAUCC194.81 and SAUCC194.87) from symptomatic leaves of Millettia reticulata and Camellia sinensis were similar to Diaporthe hongkongensis (CGMCC 3.17102) (Gomes et al. 2013) and D. salinicola (MFLU 18-0553) (Dayarathne et al. 2020) based on DNA sequences data (Fig. 1). Morphologically, our strains were similar to Diaporthe hongkongensis, which was originally described with an asexual morph on fruits of Dichroa febrifuga in China, but the asexual morph of D. salinicola was undetermined. We therefore identify our strains as D. hongkongensis.
Diaporthe machili
S.T. Huang, J.W. Xia, W.X. Sun, & X.G. Zhang sp. nov.
06BE2C60-1305-5EA2-87B4-B6E27EA3E874
837814
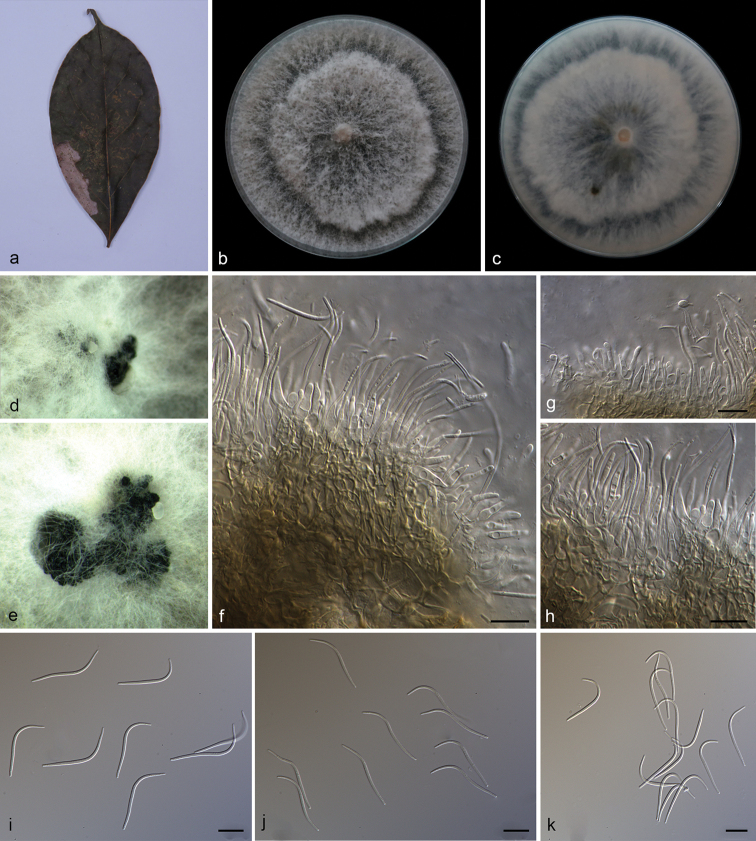
Figure 5.
Diaporthe machili (SAUCC194.111) a infected leaf of Machilus pingiib, c surface and reverse of colony after 15 days on PDAd, e conidiomata f–h conidiophores and conidiogenous cells i–k beta conidia. Scale bars: 10 μm (f–k).
Etymology.
Named after the host genus on which it was collected, Machilus pingii.
Diagnosis.
Diaporthe machili differs from D. caryae and D. sackstonii in the types of conidia (D. machili only produces beta conidia, while D. caryae produces alpha conidia and beta conidia, and D. sackstonii only produces alpha conidia), and from D. caryae in longer beta conidia (29.0–39.0 × 1.3–1.5 vs. 15.5–34.0 × 1.1–1.4 μm).
Type.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Machilus pingii (Lauraceae). 19 April 2019, S.T. Huang, HSAUP194.111 holotype, ex-holotype living culture SAUCC194.111.
Description.
Asexual morph: Conidiomata pycnidial, aggregated, black, erumpent, subglobose to globose, exuding creamy conidial droplets from central ostioles after 30 days in light at 25 °C. Conidiophores 7.0–11.4 × 1.8–2.8 μm, hyaline, unbranched, densely aggregated, mostly ampulliform, cylindrical, guttulate, septate, straight or slightly curved, swelling at base, tapering towards apex. Beta conidia 29.0–39.0 × 1.3–1.5 μm (mean = 32.5 × 1.4 μm, n = 20), filiform, hyaline, aseptate, mostly curved, J-shaped, swelling in middle, tapering towards both ends. Alpha and gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 16.3–17.5 mm diam/day, aerial mycelium abundant, white on surface, reverse white to pale yellow, with an obvious concentric zonation, pycnidia forming after 15 days.
Additional specimen examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Pometia pinnata (Sapindaceae). 19 April 2019, S.T. Huang, HSAUP194. 69 paratype; living culture SAUCC194. 69.
Notes.
In the phylogenetic tree, Diaporthe machili forms an independent clade and is phylogenetically distinct from D. caryae and D. sackstonii (Fig. 1). Diaporthe machili can be distinguished from D. caryae in ITS, TUB, TEF, CAL and HIS loci by 67 nucleotide differences in concatenated alignment (5/459 in ITS, 10/416 in TUB, 15/334 in TEF, 7/454 in CAL and 30/455 in HIS), and from D. sackstonii in ITS, TUB and TEF loci by 58 nucleotide differences (12/559 in ITS, 23/486 in TUB and 23/348 in TEF). Moreover, Diaporthe machili differs from D. caryae in having longer beta conidia (29.0–39.0 × 1.3–1.5 vs. 15.5–34.0 × 1.1–1.4 μm). Diaporthe machili only produces beta conidia, while D. caryae produces alpha conidia and beta conidia, and D. sackstonii only produces alpha conidia (Thompson et al. 2015; Yang et al. 2018b).
Diaporthe middletonii
R.G. Shivas, L. Morin, S.M. Thomps. & Y.P. Tan, Persoonia 35: 45. (2015)
F77900E1-889B-5291-AEC8-995A46C92870
Figure 6.
Diaporthe middletonii (SAUCC194.46) a infected leaf of Lithocarpus glaberb, c surface and reverse of colony after 15 days on PDAd, e conidiomata f–i conidiophores and conidiogenous cells j beta conidia k, l alpha conidia and beta conidia. Scale bars: 10 μm (f–l).
Description.
Asexual morph: Leaf spots discoid to irregular. Conidiomata pycnidial, scattered or aggregated in groups of 3–5 pycnidia, globose, black, erumpent, coated with white to greyish hyphae, thick-walled, exuding creamy translucent conidial droplets from central ostioles. Conidiophores hyaline, smooth, septate, unbranched, densely aggregated, cylindrical, straight to sinuous, tapering towards apex, 10.0–14.0 × 1.3–2.3 μm. Conidiogenous cells 5.0–9.5 × 1.3–1.7 μm, phialidic, cylindrical, terminal, slightly tapering towards apex. Alpha conidia hyaline, smooth, aseptate, biguttulate, ellipsoidal, oval, apex subobtuse, base subtruncate, 5.5–7.0 × 2.5–3.2 µm (mean = 6.3 × 2.8 μm, n = 20). Beta conidia hyaline, aseptate, filiform, mostly curved by 90–180°, tapering towards both ends, 26.0–36.5 × 1.0–1.6 µm (mean = 21.5 × 1.2 μm, n = 20). Gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 22.5–24.0 mm diam/day, fluffy with abundant aerial mycelium, margin fimbriate, white on surface, white to pale yellow on reverse.
Specimens examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, 19 April 2019, S.T. Huang. On diseased leaves of Litchi chinensis (Sapindaceae), HSAUP194.27, living culture SAUCC194.27; on diseased leaves of Lithocarpus glaber (Fagaceae), HSAUP194.45, living culture SAUCC194.45; on diseased leaves of Lithocarpus glaber (Fagaceae), 19 April 2019, S.T. Huang, HSAUP194.46, living culture SAUCC194.46; on diseased leaves of Lithocarpus craibianus (Fagaceae), HSAUP194.48, living culture SAUCC194.48.
Notes.
Diaporthe middletonii was originally described from the stem of Rapistrum rugosum (BRIP 54884e) (Brassicaceae) and Chrysanthemoides monilifera subsp. rotundata (BRIP 57329) (Asteraceae) in Australia (Thompson et al. 2015). In the present study, four strains (SAUCC194.27, SAUCC194.45, SAUCC194.46 and SAUCC194.48) are closely related to D. middletonii in the combined phylogenetic tree (Fig. 1). The differences between nucleotides in the concatenated alignment (17/565 in ITS, 9/494 in TUB and 10/340 in TEF) were minor. Morphologically, our strains were similar to D. middletonii by slightly shorter and wider alpha conidia (5.0–7.0 × 2.5–3.2 vs. 6.0–7.5 × 2.0–2.5 μm), and longer beta conidia (26.0–36.5 × 1.0–1.6 vs. 20.0–35.0 × 1.0–1.5 μm) (Thompson et al. 2015). We therefore identify our strains as Diaporthe middletonii.
Diaporthe osmanthi
H. Long, K.D. Hyde, & Yong Wang bis, MycoKeys 57: 120. (2019)
B8C2511F-5EDA-5C24-A327-D4A794BFA96E
Figure 7.
Diaporthe osmanthi (SAUCC194.21) a infected leaf of Litchi chinensisb, c surface and reverse of colony after 15 days on PDAd conidiomata e–g conidiophores and conidiogenous cells h, i beta conidia j, k alpha conidia. Scale bars: 10 μm (e–k).
Description.
Conidiomata pycnidial, globose, 5–10 pycnidia grouped together, dark brown to black, exuding creamy to yellowish conidial droplets from central ostioles. Conidiophores hyaline, smooth, densely aggregated, branched, cylindric-clavate, 20.5–32.0 × 1.8–2.4 μm. Conidiogenous cells phialidic, hyaline, terminal, cylindrical, straight, 14.0–20.5 × 1.5–2.0 μm, tapered towards apex. Alpha conidia hyaline, aseptate, fusiform, tapering towards both ends, guttulate, 7.3–9.3 × 1.8–2.3 μm (mean = 8.5 × 2.0 μm, n = 20). Beta conidia hyaline, aseptate, filiform, curved, 22.0–28.5 × 1.0–2.0 μm (mean = 27.2 × 1.3 μm, n = 20). Gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 12.0–13.5 mm diam/day, cottony with abundant aerial mycelium, sparse at margin. With several concentric rings of dense hyphae, white on surface, white to pale brown on reverse.
Specimen examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, 19 April 2019, S.T. Huang. On diseased leaves of Litchi chinensis (Sapindaceae) HSAUP194.21, living culture SAUCC194.21.
Notes.
Diaporthe osmanthi was originally described from the leaves of Osmanthus fragrans (Oleaceae) in Guangxi province, China (Long et al. 2019). In the present study, phylogenetic analyses (Fig. 1) indicated that the strain SAUCC194.21 is closely related to Diaporthe osmanthi and D. podocarpi-macrophylli (Gao et al. 2017). Morphological comparison indicated that this strain was most similar to D. osmanthi by the size of alpha conidia and beta conidia. We therefore identify this strain as belonging to D. osmanthi.
Diaporthe pandanicola
Tibpromma & K.D. Hyde, MycoKeys 33: 44 (2018)
6D78E7E2-75D9-50C1-A129-BC29A53C0A52
Figure 8.
Diaporthe pandanicola (SAUCC194.82) a infected leaf of Millettia reticulatab, c surface and reverse of colony after 15 days on PDAd conidiomata e–g conidiophores and conidiogenous cells h beta conidia i alpha conidia and gamma conidia j alpha conidia, beta conidia and gamma conidia. Scale bars: 10 μm (e–j).
Description.
Asexual morph: Conidiomata pycnidial, 3–5 pycnidia grouped together, superficial to embedded on PDA, erumpent, thin-walled, dark brown to black, globose or subglobose, exuding white creamy conidial mass from ostioles. Conidiophores hyaline, aseptate, cylindrical, smooth, straight to sinuous, unbranched, aggregated, 17.0–26.5 × 2.0–3.0 µm. Conidiogenous cells phialidic, cylindrical, terminal, 10.0–20.0 × 1.5–1.8 µm. Alpha conidia hyaline, smooth, aseptate, ellipsoidal, eguttulate, apex subobtuse, base subtruncate, 6.5–9.0 × 1.8–2.5 µm (mean = 7.5 × 2.0 μm, n = 20). Beta conidia hyaline, aseptate, filiform, curved, tapering towards apex, base truncate, 26.0–32.8 × 1.0–1.6 µm (mean = 29.0 × 1.3 μm, n = 20). Gamma conidia infrequent, aseptate, smooth, straight, hyaline, 12.5–14.5 × 1.3–1.8 µm (mean = 13.5 × 1.6 μm, n = 6). Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 12.8–15.0 mm diam/day, flat, cottony in centre, with aerial mycelium sparse toward margin, white on surface, white to pale yellow on reverse.
Specimen examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Millettia reticulata (Fabaceae). 19 April 2019, S.T. Huang, HSAUP194.82, living culture SAUCC194.82.
Notes.
Diaporthe pandanicola was originally described by Tibpromma et al. (2018) on healthy leaves of Pandanus sp. (Pandanaceae) as an endophytic fungus. Our strain (SAUCC194.82) is closely related to Diaporthe pandanicola based on phylogenetic analyses (Fig. 1). The differences of nucleotides in the concatenated alignment (19/533 in the ITS region and 11/351 in the TUB region) are less than 3%. Morphologically, our strain produces alpha conidia, beta conidia and gamma conidia, while Diaporthe pandanicola did not sporulate. We therefore identify our strains as Diaporthe pandanicola.
Diaporthe pometiae
S.T. Huang, J.W. Xia, W.X. Sun, & X.G. Zhang sp. nov.
D8C614FF-3589-5562-82FA-F44963136C2F
837815
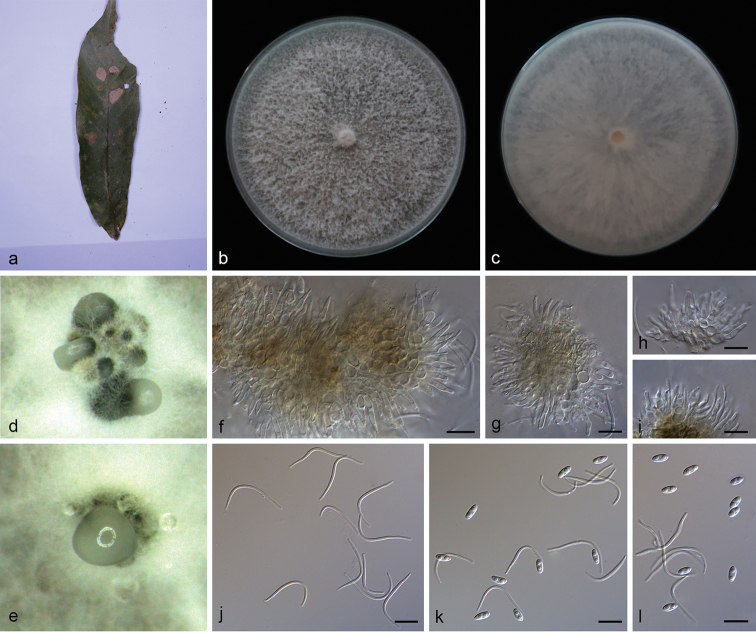
Figure 9.
Diaporthe pometiae (SAUCC194.72) a infected leaf of Pometia pinnatab, c surface and reverse of colony after 15 days on PDAd conidiomata e, f conidiophores and conidiogenous cells g beta conidia h alpha conidia and beta conidia. Scale bars: 10 μm (e–h).
Etymology.
Named after the host genus on which it was collected, Pometia pinnata.
Diagnosis.
Diaporthe pometiae is similar to D. biconispora but differs in having smaller alpha conidia (5.7–8.3 × 2.2–3.0 vs. 6.0–10.5 × 2–3.5 μm) and types of conidia (D. pometiae produces beta conidia unlike D. biconispora).
Type.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, on diseased leaves of Pometia pinnata (Sapindaceae). 19 April 2019, S.T. Huang, HSAUP194.72 holotype, ex-type living culture SAUCC194.72.
Description.
Asexual morph: Leaf spots subcircular, fawn to dark brown. Conidiomata pycnidial, subglobose to globose, aggregated in groups, black, coated with white hyphae, thick-walled, exuding creamy droplets from ostioles. Conidiophores hyaline, smooth, slightly septate, branched, densely aggregated, cylindric-clavate, straight to slightly sinuous, 22.5–32.5 × 1.0–2.0 μm. Conidiogenous cells 15.0–22.5 × 1.0–1.5 μm, phialidic, cylindrical, multi-guttulate, terminal, tapering towards apex. Alpha conidia abundant in culture, 2–4 guttulate, hyaline, smooth, aseptate, ellipsoidal to oblong ellipsoidal, with both ends obtuse, 5.7–8.3 × 2.2–3.0 µm (mean = 6.7 × 3.1 μm, n = 20). Beta conidia, hyaline, aseptate, filiform, multi-guttulate, slightly curved, tapering towards to apex, 27.8–34.5 × 1.0–1.7 µm (mean = 21.7 × 1.4 μm, n = 20). Gamma conidia not observed. Sexual morph not observed.
Culture characteristics.
Cultures incubated on PDA at 25 °C in darkness, growth rate 11.5–13.0 mm diam/day, cottony with abundant aerial mycelium, with a concentric zonation, white on surface, white to grayish on reverse.
Additional specimens examined.
China, Yunnan Province: Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, 19 April 2019, S.T. Huang. On diseased leaves of Persea americana (Lauraceae), HSAUP194.19 paratype, ex-paratype culture SAUCC194.19; on diseased leaves of Heliconia metallica (Musaceae), HSAUP194.73 paratype, ex-paratype culture SAUCC194.73.
Notes.
Diaporthe pometiae is introduced based on the multi-locus phylogenetic analysis, with three isolates clustering separately in a well-supported clade (ML/BI = 100/1). Diaporthe pometiae is most closely related to D. biconispora, but distinguished based on ITS, TUB, TEF and HIS loci by 74 nucleotide differences in the concatenated alignment, in which 2/492 are distinct in the ITS region, 8/353 in the TUB region, 49/370 in the TEF region and 15/471 in the HIS region. Morphologically, Diaporthe pometiae differs from D. biconispora in its smaller alpha conidia (5.7–8.3 × 2.2–3.0 vs. 6.0–10.5 × 2–3.5 μm). Furthermore, Diaporthe pometiae produces beta conidia unlike D. biconispora (Huang et al. 2015).
Discussion
The Yunnan Province in southeastern China has a unique geography where three climatic regions meet: the eastern Asia monsoon region, the Tibetan plateau region, and the tropical monsoon region of southern Asia and Indo-China. The environment is conducive to growth of unusual microbial species. Species diversity in Yunnan Province is high compared to other parts of China.
Previously, species identification of Diaporthe relied on the assumption of host-specificity, leading to the proliferation of names. The morphological characters of Diaporthe could be changeable, as most taxa in culture do not produce all spore states of the asexual (alpha, beta and gamma conidia) or the sexual morph (Gomes et al. 2013). Based on a polyphasic approach and morphology, more than one species of Diaporthe can colonize a single host, while one species can be associated with several hosts (Gomes et al. 2013; Gao et al. 2017; Guarnaccia and Crous 2017; Guarnaccia et al. 2018; Guo et al. 2020). These studies revealed a high diversity of Diaporthe species from different hosts. Our study supports this phenomenon. For example, Diaporthe arecae (SAUCC194.18) and D. pometiae (SAUCC194.19) were collected from Persea americana; In addition, isolates of D. middletonii were obtained from three hosts (Litchi chinensis, Lithocarpus craibianus, L. glaber). As for host specificity, in our study, four species of Diaporthe, D. machili (SAUCC194.69), D. middletonii (SAUCC194.27), D. osmanthi (SAUCC194.21), and D. pometiae (SAUCC194.72) were isolated from Litchi chinensis and Pometia pinnata belong to the Sapindaceae, and D. litchiicola also was reported from Litchi chinensis in Queensland (Tan et al. 2013); however, D. machili (SAUCC194.111) also was isolated from Machilus pingii (Lauraceae), D. middletonii (SAUCC194.45) from Lithocarpus glaber (Fagaceae), D. osmanthi (GUCC 9165) from leaves of Osmanthus fragrans (Oleaceae) (Long et al. 2019), and D. pometiae (SAUCC194.19 and SAUCC194.73) from Persea americana (Lauraceae) and Heliconia metallica (Musaceae). These results provide evidence that many species are able to colonise diverse hosts and several different species could co-occur on the same host. It seems obvious that specificity does not occur at the family level.
For the current study, sixteen strains isolated from ten host genera represented three new species and five known species, based on morphological characters and phylogenetic analyses of the five combined loci (ITS, TUB, TEF, CAL and HIS). The descriptions and molecular data for species of Diaporthe represent an important resource for plant pathologists, plant quarantine officials and taxonomists.
Supplementary Material
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (no. 31900014, 31770016, and 31750001) and the China Postdoctoral Science Foundation (no. 2018M632699).
Citation
Huang S, Xia J, Zhang X, Sun W (2021) Morphological and phylogenetic analyses reveal three new species of Diaporthe from Yunnan, China. MycoKeys 78: 49–77. https://doi.org/10.3897/mycokeys.78.60878
References
- Cai L, Hyde KD, Taylor PWJ, Weir B, Waller J, Abang MM, Zhang ZJ, Yang YL, Phoulivong S, Liu ZY, Prihastuti H, Shivas RG, McKenzie EHC, Johnston PR. (2009) A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hywel-Jones NL. (2004) Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Studies in Mycology 50: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, Edwards J, Seifert KA, Alfenas AC, Alfenas RF, Burgess TI, Carnegie AJ, Hardy GEStJ. (2011a) Fungal planet description sheets: 69–91. Persoonia 26(1): 108–156. 10.3767/003158511X581723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Swart L, Denman S, Taylor JE, Bezuidenhout CM, Palm ME, Marincowitz S, Groenewald JZ. (2011b) Fungal pathogens of Proteaceae. Persoonia 27(1): 20–45. 10.3767/003158511X606239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Roux JJL, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M. (2016) Fungal planet description sheets: 400–468. Persoonia 36(1): 316–458. 10.3767/003158516X692185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Schumacher RK, Akulov A, Bulgakov TS, Carnegie AJ, Jurjević Ž, Decock C, Denman S, Lombard L. (2020) New and interesting fungi. 3. Fungal Systematics and Evolution 6: 157–231. 10.3114/fuse.2020.06.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayarathne MC, Jones EBG, Maharachchikumbura SSN, Devadatha B, Sarma VV. (2020) Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 11(1): 1–188. 10.5943/mycosphere/11/1/1 [DOI] [Google Scholar]
- Dissanayake AJ, Phillips AJL, Hyde KD, Yan JY, Li XH. (2017) The current status of species in Diaporthe. Mycosphere 8: 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- Fan XL, Bezerra JDP, Tian CM, Crous PW. (2018) Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Persoonia 40: 119–134. 10.3767/persoonia.2018.40.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YH, Sun W, Su YY, Cai L. (2014) Three new species of Phomopsis in Gutianshan Nature Reserve in China. Mycological Progress 13(1): 111–121. 10.1007/s11557-013-0898-2 [DOI] [Google Scholar]
- Gao YH, Su YY, Sun W, Cai L. (2015) Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biology 119(5): 295–309. 10.1016/j.funbio.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Gao YH, Liu F, Cai L. (2016) Unravelling Diaporthe species associated with Camellia. Systematics and Biodiversity 14(1): 102–117. 10.1080/14772000.2015.1101027 [DOI] [Google Scholar]
- Gao YH, Liu F, Duan W, Crous PW, Cai L. (2017) Diaporthe is paraphyletic. IMA fungus 8: 153–187. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/AEM.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, Lombard L, Groenewald JZ, Crous PW. (2013) Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia: Molecular Phylogeny and Evolution of Fungi 31(1): 1–41. ttps:// 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed]
- Grasso FM, Marini M, Vitale A, Firrao G, Granata G. (2012) Canker and dieback on Platanus x acerifolia caused by Diaporthe scabra. Forest Pathology 42(6): 510–513. 10.1111/j.1439-0329.2012.00785.x [DOI] [Google Scholar]
- Guarnaccia V, Vitale A, Cirvilleri G, Aiello D, Susca A, Epifani F, Perrone G, Polizzi G. (2016) Characterisation and pathogenicity of fungal species associated with branch cankers and stem-end rot of avocado in Italy. European Journal of Plant Pathology 146(4): 963–976. 10.1007/s10658-016-0973-z [DOI] [Google Scholar]
- Guarnaccia V, Crous PW. (2017) Emerging citrus diseases in Europe caused by Diaporthe spp. IMA Fungus 8: 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V, Groenewald JZ, Woodhall J, Armengol J, Cinelli T, Eichmeier A, Ezra D, Fontaine F, Gramaje D, Gutierrez-Aguirregabiria A. (2018) Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in europe. Persoonia 40(6): 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Guo YS, Crous PW, Bai Q, Fu M, Yang MM, Wang XH, Du YM, Hong N, Xu WX, Wang GP. (2020) High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45: 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Hou X, Dewdney MM, Fu Y, Chen GQ, Hyde KD, Li HY. (2013) Diaporthe species occurring on citrus in China. Fungal Diversity 61(1): 237–250. 10.1007/s13225-013-0245-6 [DOI] [Google Scholar]
- Huang F, Udayanga D, Wang XH, Hou X, Mei XF, Fu YS, Hyde KD, Li HY. (2015) Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biology 119(5): 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: bayesian inference of phylogeny. Bioinformatics 17(17): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Dong Y, Phookamsak R, Jeewon R, Bhat DJ, Gareth Jones EB, Liu NG, Abeywickrama PD, Mapook A, Wei D. (2020) Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100(1): 1–273. 10.1007/s13225-020-00439-5 [DOI] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics: 1–7. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WJ, McKenzie EHC, Liu JK, Bhat DJ, Dai DQ, Camporesi E, Tian Q, Maharachchikumbura SSN, Luo ZL, Shang QJ. (2020) Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Diversity 100(1): 279–801. 10.1007/s13225-020-00440-y [DOI] [Google Scholar]
- Lombard L, van Leeuwen GCM, Guarnaccia V, Polizzi G, van Rijswick PCJ, Rosendahl KCHM, Gabler J, Crous PW. (2014) Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathologia Mediterranea 53(2): 287–299. 10.14601/PHYTOPATHOL_MEDITERR-14034 [DOI] [Google Scholar]
- Long H, Zhang Q, Hao YY, Shao XQ, Wei XX, Hyde KD, Wang Y, Zhao DG. (2019) Diaporthe species in south-western China. MycoKeys 57: 113–127. 10.3897/mycokeys.57.35448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard L, Brandeis PE, Simoneau P, Poupard P, Sérandat I, Detoc J, Robbes L, Bastide F, Laurent E, Gombert J, Morel E. (2014) First report of umbel browning and stem necrosis caused by Diaporthe angelicae on carrot in France. Plant Disease 98(3): 421–422. 10.1094/PDIS-06-13-0673-PDN [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery, USA, 8 pp. 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Murali TS, Suryanarayanan TS, Geeta R. (2006) Endophytic Phomopsis species: host range and implications for diversity estimates. Canadian Journal of Microbiology 52(7): 673–680. 10.1139/w06-020 [DOI] [PubMed] [Google Scholar]
- Nitschke T. (1870) Pyrenomycetes Germanici (2nd ed.). Eduard Trewendt, Breslau, 161–320.
- Nylander JAA. (2004) MrModeltest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Rayner RW. (1970) A mycological colour chart. CMI and British Mycological Society, Kew.
- Rehner SA, Uecker FA. (1994) Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Botany 72(11): 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D. (2015) Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 6(1): 145–154. 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JM, Phillips AJL. (2009) Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Diversity 34: 111–125. [Google Scholar]
- Santos JM, Vrandečić K, Ćosić J, Duvnjak T, Phillips AJL. (2011) Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 27(1): 9–19. 10.3767/003158511X603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L, Alves A, Alves R. (2017) Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5: e3120. 10.7287/peerj.preprints.2822v1 [DOI] [PMC free article] [PubMed]
- Senanayake IC, Crous PW, Groenewald JZ, Maharachchikumbura SSN, Jeewon R, Phillips AJL, Bhat DJ, Perera RH, Li QR, Li WJ. (2017) Families of Diaporthales based on morphological and phylogenetic evidence. Studies in Mycology 86: 217–296. 10.1016/j.simyco.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Jeewon R, Chomnunti P, Wanasinghe DN, Norphanphoun C, Karunarathna A, Pem D, Perera RH, Camporesi E, McKenzie EHC. (2018) Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93(1): 241–443. 10.1007/s13225-018-0410-z [DOI] [Google Scholar]
- Srivastava HC, Banu Z, Govindarajan VS. (1962) Fruit rot of arecanut caused by a new fungus. Mycologia 54(1): 5–11. 10.1080/00275514.1962.12024974 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Edwards J, Grice KRE, Shivas RG. (2013) Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Diversity 61(1): 251–260. 10.1007/s13225-013-0242-9 [DOI] [Google Scholar]
- Thompson SM, Tan YP, Young AJ, Neate SM, Aitken EAB, Shivas RG. (2011) Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27(1): 80–89. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Tan YP, Shivas RG, Neate SM, Morin L, Bissett A, Aitken EAB. (2015) Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia 35(1): 39–49. 10.3767/003158515X687506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibpromma S, Hyde KD, Bhat JD, Mortimer PE, Xu JC, Promputtha I, Doilom M, Yang JB, Tang AMC, Karunarathna SC. (2018) Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 33(33): 25–67. 10.3897/mycokeys.33.23670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C, Camps R, Aguirre R, Besoain XA. (2016) First report of Diaporthe rudis in Chile causing stem-end rot on hass avocado fruit imported from California, USA. Plant Disease 100(9): 1951–1951. 10.1094/PDIS-12-15-1495-PDN [DOI] [Google Scholar]
- Udayanga D, Liu X, McKenzie EH, Chukeatirote E, Bahkali AH, Hyde KD. (2011) The genus Phomopsis: biology, applications, species concepts and names of common phytopathogens. Fungal Diversity 50(1): 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Udayanga D, Liu XZ, Crous PW, McKenzie EHC, Chukeatirote E, Hyde KD. (2012) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Diversity 56(1): 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD. (2015) The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biology 119(5): 383–407. 10.1016/j.funbio.2014.10.009 [DOI] [PubMed] [Google Scholar]
- van Rensburg JCJ, Lamprecht SC, Groenewald JZ, Castlebury LA, Crous PW. (2006) Characterization of Phomopsis spp. associated with die-back of rooibos (Aspalathus linearis) in South Africa. Studies in Mycology 55: 65–74. 10.3114/sim.55.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilka L, Volkova J. (2015) Morphological diversity of Phomopsis vaccinii isolates from cranberry (Vaccinium macrocarpon Ait.) in Latvia. Proceedings of the Latvia University of Agriculture 33: 8–18. 10.1515/plua-2015-0002 [DOI] [Google Scholar]
- White T, Bruns T, Lee S, Taylor FJRM, White TJ, Lee SH, Taylor L, Shawe-Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A guide to methods and applications 18: 315–322. Academic Press, San Diego. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Yang Q, Du Z, Tian CM. (2018a) Phylogeny and morphology reveal two new species of Diaporthe from Traditional Chinese Medicine in Northeast China. Phytotaxa 336(2): 159–170. 10.11646/phytotaxa.336.2.3 [DOI] [Google Scholar]
- Yang Q, Fan XL, Guarnaccia V, Tian CM. (2018b) High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys 39(39): 97–149. 10.3897/mycokeys.39.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jiang N, Tian CM. (2020) Three new Diaporthe species from Shaanxi Province, China. MycoKeys 67: 1–18. 10.3897/mycokeys.67.49483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata M, Palma MA, Aninat MJ, Piontelli E. (2020) Polyphasic studies of new species of Diaporthe from native forest in Chile, with descriptions of Diaporthe araucanorum sp. nov., Diaporthe foikelawen sp. nov. and Diaporthe patagonica sp. nov. International Journal of Systematic and Evolutionary Microbiology 70(5): 3379–3390. 10.1099/ijsem.0.004183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.