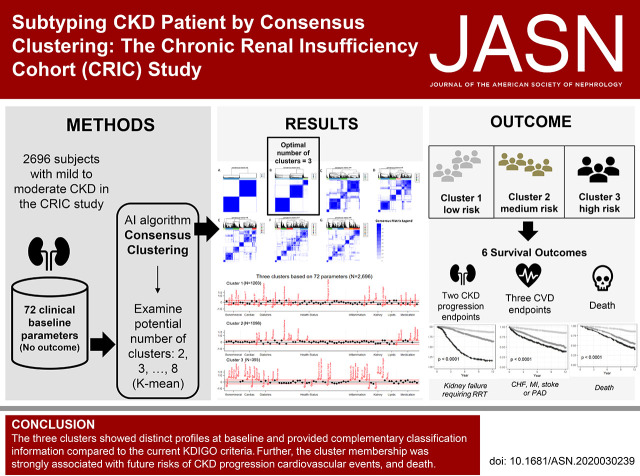
Significance Statement
Staging CKD by eGFR and urine albumin-creatinine ratio does not fully capture underlying patient heterogeneity. Applying machine learning consensus clustering to multidimensional patient data, including demographics, biomarkers from blood and urine, health status and behaviors, and medication use, enables subtyping of patients with CKD into three distinct subgroups defined by 72 baseline characteristics. These subgroups are strongly associated with future risks of kidney disease, cardiovascular events, and death, independent of established CKD risk factors. Identification of clinically meaningful subgroups among patients with CKD provides an important step toward patient classification and precision medicine in nephrology.
Keywords: CKD subgroups, clustering analysis, patient heterogeneity, survival
Visual Abstract
Abstract
Background
CKD is a heterogeneous condition with multiple underlying causes, risk factors, and outcomes. Subtyping CKD with multidimensional patient data holds the key to precision medicine. Consensus clustering may reveal CKD subgroups with different risk profiles of adverse outcomes.
Methods
We used unsupervised consensus clustering on 72 baseline characteristics among 2696 participants in the prospective Chronic Renal Insufficiency Cohort (CRIC) study to identify novel CKD subgroups that best represent the data pattern. Calculation of the standardized difference of each parameter used the cutoff of ±0.3 to show subgroup features. CKD subgroup associations were examined with the clinical end points of kidney failure, the composite outcome of cardiovascular diseases, and death.
Results
The algorithm revealed three unique CKD subgroups that best represented patients’ baseline characteristics. Patients with relatively favorable levels of bone density and cardiac and kidney function markers, with lower prevalence of diabetes and obesity, and who used fewer medications formed cluster 1 (n=1203). Patients with higher prevalence of diabetes and obesity and who used more medications formed cluster 2 (n=1098). Patients with less favorable levels of bone mineral density, poor cardiac and kidney function markers, and inflammation delineated cluster 3 (n=395). These three subgroups, when linked with future clinical end points, were associated with different risks of CKD progression, cardiovascular disease, and death. Furthermore, patient heterogeneity among predefined subgroups with similar baseline kidney function emerged.
Conclusions
Consensus clustering synthesized the patterns of baseline clinical and laboratory measures and revealed distinct CKD subgroups, which were associated with markedly different risks of important clinical outcomes. Further examination of patient subgroups and associated biomarkers may provide next steps toward precision medicine.
CKD is a major public health problem that affects >15% of adults or about 37 million people in the United States.1 According to the 2012 Kidney Disease Improving Global Outcome (KDIGO) guideline, CKD is staged by patients’ eGFR and the urine albumin excretion-urine creatinine ratio (UACR).2 Although useful for screening and monitoring, classification by eGFR and UACR alone does not capture the cause(s) or differences in pathophysiology of CKD in individual patients.3,4 CKD is a highly heterogeneous condition that is closely related to systemic diseases and conditions, like diabetes,5 hypertension,6–8 autoimmune diseases,9 genetic disposition, or congenital abnormalities,10 as well as a number of other etiologies, including inflammation,11 toxic environmental,12 and drug exposures.13,14 The underlying causes of CKD are not always known in clinical practice.
There are clear differences across individuals with CKD that can be captured by a comprehensive examination of phenotypic data, such as laboratory results, medical history, medications, and social factors. More sophisticated phenotyping may reveal different and more reliable CKD clusters and underlying disease pathologies, which could help better understand different mechanisms for disease pathways and progression. With detailed data from a well-characterized cohort of individuals with CKD, we hypothesized that distinct CKD subpopulations can be identified using multidimensional phenotypic data, and that these subpopulations have different risks of future CKD progression, development of kidney failure requiring replacement therapy, cardiovascular diseases, and death. To do this, we performed consensus clustering analysis, an unsupervised class discovery and validation tool that has been used in genetic and cancer studies,15,16 on the baseline characteristics of subjects in the Chronic Renal Insufficiency Cohort (CRIC) study.
The primary goal of this study is to examine phenotype heterogeneity in patients with CKD utilizing multidimensional data of patients’ demographics, biomarkers, and clinical characteristics collected at study entry. The secondary goal is to study the relationship between the revealed cluster membership and future clinical events. We anticipated that identifying clinically distinct CKD subgroups could be an important step to understanding the heterogeneity of CKD and its causes and to promoting precision medicine for CKD management.
Methods
Study Design and Population
The CRIC study is a prospective ongoing cohort study of adults with stages 2–4 CKD. During the initial phase of the study, 3939 participants were recruited between June 2003 and September 2008 from clinical centers in seven United States cities (Ann Arbor and Detroit, MI; Baltimore, MD; Chicago, IL; Cleveland, OH; New Orleans, LA; Philadelphia, PA; and Oakland, CA).17–19 Main inclusion criteria were ages 21–74 years with age-based eGFR range of 20–70 ml/min per 1.73 m2 at enrollment. Patients were excluded if they had previous dialysis (for >1 month), New York Heart Association class 3 or 4 heart failure, polycystic kidney disease, or other primary kidney diseases requiring active immunosuppression. Study participants were followed through annual clinic visits during which investigators collected the updated health information and biospecimens of urine and blood. Additional details on study design, participants’ eligibility criteria, and characteristics can be found in previous publications.17,18 Written informed consent was gathered from study participants, and the study protocol was approved by institutional review boards at each of the clinical centers. For this study, we used August 31st, 2015 as the administrative censoring date for the survival outcomes.
Data Collection and Preparation
Measured Baseline Characteristics
We selected 72 clinically available and novel factors from over 822 variables that were measured at CRIC study baseline. Variables were selected on the basis of literature review for those that are most clinically relevant to CKD.20–24 We excluded variables with over 10% missing data or small variability (e.g., binary variable with <5%). The 72 variables are specified in Table 1, including variables of sociodemographic characteristics (n=6), health status and behavior (n=15), biomarkers of inflammation (n=10), lipid metabolism (n=4), cardiac function (n=7), kidney function (n=8), bone and mineral metabolism (n=7), diabetes (n=6), and medications (n=9). Sociodemographic characteristics (age, sex, education level, marital status, and income levels), health history (any cardiovascular disease at baseline, family history of kidney disease, any cancer diagnosis or treatment 5 years prior to baseline, acidosis, and history of chronic obstructive pulmonary disease), health behavior (smoking status, alcohol use, physical activity, and attempt to lose weight), and medication use were on the basis of self-reported information. Anthropometric measures of height, weight, and waist circumference and measurements of BP were obtained following study standardized protocols. Serum and urinary biomarkers were also measured using standard assays.17,18 We combined race/ethnicity categories (including non-Hispanic White, non-Hispanic Black, Hispanic, and others) with participants’ APOL1 gene risk alleles and divided Blacks into groups with high or low APOL1 risk.10
Table 1.
Variables input to the consensus clustering algorithm by categories
| Category | Markers |
|---|---|
| Social demographic factors (n=6) | Age, sex, race, Apol1 risk alleles, education level, marital status, and income levels |
| Bone and mineral markers (n=7) | Serum calcium, FGF-23, serum phosphate, serum chloride, alkaline phosphatase, serum total PTH, aldosterone, urinary sodium, and urinary potassium |
| Cardiac markers (n=7) | Troponin I, high-sensitivity troponin T, high-sensitivity CRP, NTproBNP, bicarbonate, systolic BP, and diastolic BP |
| Diabetes markers (n=6) | HbA1C, C-peptide, CBC hemoglobin, glucose, fetuin-A, and mean cell hemoglobin concentration |
| Inflammation markers (n=10) | IL-10, IL-1RA, IL-6, IL-1β, TNF- α, TGF-β, CXCL12, fibrinogen, white blood cell, and serum albumin |
| Lipids markers (n=4) | Total cholesterol, triglycerides, HDL, and LDL |
| Kidney markers (n=8) | eGFR, UACR, UPCR, serum urea nitrogen, uric acid, NGAL |
| Health status and behaviors (n=15) | Diabetes, hypertension, smoking status, alcohol use, try to lose weight, waist circumference, physical activities, weight, BMI, family history of kidney disease, diagnosed or treated for any cancer 5 yr before enrollment, cardiovascular disease history, history of congestive heart failure, history of COPD, and acidosis |
| Medications (n=9) | NSAID, diuretics, ACE inhibitor, β-blockers, calcium blockers, statins, steroids, antidiabetes medications, and antiplatelet |
FGF-23, fibroblast growth factor 23; PTH, parathyroid hormone; CRP, C-reactive protein; NTproBNP, N-terminal prohormone of brain natriuretic peptide; HbA1C, hemoglobin A1C; CBC, complete blood count; CXCL12, C-X-C motif chemokine ligand 12; UPCR, urine protein-creatinine ratio; NGAL, neutrophil gelatinase–associated lipocalin; BMI, body mass index; COPD, chronic obstructive pulmonary disease; NSAID, nonsteroidal anti-inflammatory drug; ACE, angiotensin-converting enzyme.
Clinical End Points
After identifying the CKD subgroups with patients’ characteristics measured at baseline, we prospectively examined six clinical end points: (1) the composite outcome of CKD progression (including incidence kidney failure requiring replacement therapy or 50% decrease of eGFR from baseline); (2) incident kidney failure requiring replacement therapy; (3) incident congestive heart failure (CHF); (4) the composite outcome myocardial infarction (MI), stroke, and peripheral artery disease (PAD; composite cardiovascular disease 1); (5) the composite outcome of CHF, MI, stroke, and PAD (composite cardiovascular disease 2); and (6) death. Kidney failure requiring replacement therapy was defined as receiving dialysis or kidney transplantation, which was ascertained primarily through self-report and was supplemented by information abstracted from the United States Renal Data System. eGFRs used for defining the composite outcome of CKD progression were calculated using the CRIC study equation25 on the basis of serum creatinine and cystatin C levels. When estimating the time point of 50% eGFR decrease, we assumed a linear decline between two successive eGFR measurements.26 Participants were followed until withdrawal, loss to follow-up, death, or the administrative cutoff date of August 31st, 2015.
Statistical Analyses
We excluded individuals with any missing data in the 72 selected input variables. We compared those with missing data with the overall CRIC population using standardized mean difference. For continuous variables with normal distribution, we calculated mean and SD; for continuous variables with skewed distributions (skewness more than two), we calculated median and interquartile range; and for categorical variables, we presented count and percentage. We performed consensus clustering analysis15 on the whole study population, as well as among subgroups defined by eGFR and UACR. The clustering algorithm is to maximize the potential number of clusters while maintaining high cluster consensus. The optimal number of clusters was determined by examining the consensus matrix heat map, the within-cluster consensus scores, and the proportion of ambiguously clustered pairs (Supplemental Material, Supplemental Figure 1). We compared the cluster membership with the subgroups defined by eGFR (<45 and ≥ 45 ml/min per 1.73 m2) and UACR (≤ 30, 30–300, and > 300 mg/g) according to KDIGO 2012 guideline through a three-way crosstabulation and a scatterplot of individuals’ eGFR and UACR colored by the cluster membership. We compared the distribution of the 72 input variables across clusters using the ANOVA test and the Chi2 test. To examine the cluster profile, we calculated and graphically displayed the standardized mean differences of the 72 input variables between each cluster and the overall study population. We examined the robustness of cluster membership due to missing data. We generated three imputed datasets of full CRIC participants (n=3921) through multiple imputation by chain equations and repeated the consensus clustering analysis. We additionally performed consensus clustering analysis excluding socioeconomic status (SES) variables of race, income, education, and marital status.
We examined the associations between cluster membership (derived from baseline information only) and six clinical end points stated above using Kaplan–Meier curves and Cox proportional hazard models. In the Cox regression, we considered the cluster membership as “exposure” and adjusted for baseline age, sex, race, APOL1 risk genotype, education level, diabetes status, smoking status, alcohol use, physical activity level, body mass index, eGFR, urinary protein-creatinine ratio, systolic BP, family history of kidney disease, and history of prior cardiovascular disease event. We checked the proportional hazard assumption for each end point. For outcomes that violated the assumption, we divided the follow-up time into 0–2, 2–5, and >5 years and additionally reported time-specific hazard ratios (HRs). We compared the discrimination performance for the following four models: (1) model that included only cluster membership, (2) model that included only eGFR and UACR, (3) model that included only the traditional risk factors, and (4) model that included traditional risk factors plus cluster membership. The time-dependent receiver operating characteristics curve was calculated for each outcome and compared at 10 years after study entry for the different models.27
In the subgroup analyses, we examined CKD heterogeneity within predefined subgroups with similar baseline kidney function. We performed consensus clustering among the subgroup of individuals with impaired baseline function (eGFR<45 ml/min per 1.73 m2), individuals with preserved kidney function (eGFR≥ 45 ml/min per 1.73 m2), and individuals with mild baseline albuminuria (UACR≤30 mg/g). After determining the optimal number of clusters in each of the three additional analyses, we applied the previously described survival analysis to correlate the discovered CKD subgroups with the clinical end points of CKD progression, cardiovascular disease, and death.
All analyses were performed using R, version 3.5.2 (RStudio, Inc., Boston, MA; http://www.rstudio.com/), with the packages of ConsensusClusterPlus (version 1.46.0) and survival (version 2.44–1.1).
Results
We included 2696 individuals who had no missing data over the 72 baseline factors. The mean age of our study population was 58.4 years, 44% of the subjects were women, 28.8% of the subjects were Black with low-risk APOL1 genotype, and 6.8% were Black with high-risk APOL1 genotype. The overall mean eGFR was 45.4 ml/min per 1.73 m2 (SD 15.9 ml/min/ per 1.73 m2), and the overall median UACR was 44.7 mg/g (interquartile range, 7.8–382.3 mg/g) (Table 2). The standardized differences for all baseline characteristics between our study population and the overall CRIC population were smaller than 0.1 (Supplemental Table 1).
Table 2.
Baseline characteristics of overall study population (n=2696) across three clusters revealed by the 72 baseline parameters
| Baseline Characteristics | Overall | Cluster 1 | Cluster 2 | Cluster 3 | P Value |
|---|---|---|---|---|---|
| N | 2696 | 1203 | 1098 | 395 | |
| Age, yr | 58 (11) | 55 (12) | 62 (8) | 57 (11) | <0.001 |
| Women (%) | 1186 (44.0) | 527 (43.8) | 506 (46.1) | 153 (38.7) | 0.04 |
| Race and Apol1 risks (%) | <0.001 | ||||
| White | 1107 (41.1) | 624 (51.9) | 422 (38.4) | 61 (15.4) | |
| Black, low Apol1 risk | 776 (28.8) | 274 (22.8) | 366 (33.3) | 136 (34.4) | |
| Black, high Apol1 risk | 184 (6.8) | 78 (6.5) | 77 (7.0) | 29 (7.3) | |
| Hispanic or others | 629 (23.3) | 227 (18.9) | 233 (21.2) | 169 (42.8) | |
| Education level (%) | <0.001 | ||||
| Less than high school | 517 (19.2) | 105 (8.7) | 259 (23.6) | 153 (38.7) | |
| High school graduate | 515 (19.1) | 175 (14.6) | 257 (23.4) | 83 (21.0) | |
| Some college | 770 (28.6) | 347 (28.8) | 319 (29.1) | 104 (26.3) | |
| College graduate or higher | 894 (33.2) | 576 (47.9) | 263 (24.0) | 55 (13.9) | |
| Household income (%) | <0.001 | ||||
| $20,000 or under | 788 (29.2) | 188 (15.6) | 385 (35.1) | 215 (54.4) | |
| $20,001–$50,000 | 678 (25.1) | 291 (24.2) | 304 (27.7) | 83 (21.0) | |
| $50,000–$100,000 | 537 (19.9) | 330 (27.4) | 163 (14.8) | 44 (11.1) | |
| >$100,000 | 285 (10.6) | 205 (17.0) | 69 (6.3) | 11 (2.8) | |
| Do not wish to answer | 408 (15.1) | 189 (15.7) | 177 (16.1) | 42 (10.6) | |
| Marital status (%) | <0.001 | ||||
| Currently married | 1558 (57.8) | 753 (62.6) | 588 (53.6) | 217 (54.9) | |
| Never married | 346 (12.8) | 159 (13.2) | 124 (11.3) | 63 (16.0) | |
| Formerly married | 792 (29.4) | 291 (24.2) | 386 (35.2) | 115 (29.1) | |
| Diabetes (%) | 1249 (46.3) | 146 (12.1) | 855 (77.9) | 248 (62.8) | <0.001 |
| Hypertension (%) | 2316 (85.9) | 898 (74.7) | 1044 (95.1) | 374 (94.7) | <0.001 |
| Smoke now (%) | 335 (12.4) | 130 (10.8) | 113 (10.3) | 92 (23.3) | <0.001 |
| Alcohol (%) | 1720 (63.8) | 920 (76.5) | 573 (52.2) | 227 (57.5) | <0.001 |
| Try to lose weight (%) | 1906 (70.7) | 757 (62.9) | 903 (82.2) | 246 (62.3) | <0.001 |
| Waist circumference, cm | 105.6 (16.2) | 98.7 (13.7) | 114.0 (15.0) | 103.2 (16.0) | <0.001 |
| Any activities with MET score ≥6 (%) | 653 (24.2) | 439 (36.5) | 149 (13.6) | 65 (16.5) | <0.001 |
| Weight, kg | 90.9 (21.1) | 84.1 (18.2) | 99.7 (20.8) | 87.1 (21.5) | <0.001 |
| BMI, kg/m2 | 31.8 (6.9) | 29.0 (5.4) | 35.2 (6.9) | 30.9 (6.9) | <0.001 |
| eGFR, ml/min per 1.73 m2 | 45 (16) | 54 (16) | 42 (12) | 31 (10) | <0.001 |
| UPCR,a mg/mg | 0.14 [0.06–0.65] | 0.08 [0.04–0.25] | 0.15 [0.06–0.48] | 2.57 [0.77–4.67] | <0.001 |
| UACR,a μg/mg | 44.7 [7.8–382.3] | 15.0 [5.0–116.6] | 47.2 [11.4–278.7] | 1524.5 [416.1–3178.6] | <0.001 |
| Serum urea nitrogen,a mg/dl | 26.0 [20.0–35.0] | 21.0 [17.0–27.0] | 29.0 [23.0–39.0] | 37.0 [28.0–46.0] | <0.001 |
| Uric acid, mg/dl | 7.38 (1.85) | 6.79 (1.76) | 7.86 (1.82) | 7.84 (1.71) | <0.001 |
| NGAL,a ng/ml | 13.86 [6.40–32.33] | 10.42 [5.10–20.84] | 13.63 [6.42–26.16] | 49.45 [24.25–109.60] | <0.001 |
| Urinary sodium, mmol/24 h | 161.8 (76.4) | 158.4 (78.4) | 168.6 (74.8) | 153.2 (73.1) | <0.001 |
| Urinary potassium, mmol/24 h | 55.7 (26.5) | 57.0 (26.0) | 57.0 (27.6) | 48.4 (24.1) | <0.001 |
| Calcium, mg/dl | 9.20 (0.46) | 9.28 (0.42) | 9.22 (0.46) | 8.91 (0.49) | <0.001 |
| FGF-23, RU/ml | 193.4 (186.4) | 128.9 (98.9) | 215.7 (185.8) | 327.8 (281.9) | <0.001 |
| Phosphate, mg/dl | 3.67 (0.61) | 3.47 (0.53) | 3.74 (0.59) | 4.09 (0.66) | <0.001 |
| Chloride, mmol/L | 104.8 (3.7) | 104.2 (3.5) | 104.5 (3.6) | 107.3 (3.8) | <0.001 |
| Alkaline phosphatase,a U/L | 83.0 [69.0–102.0] | 76.0 [65.0–91.0] | 87.0 [73.0–106.0] | 101.0 [84.0–121.5] | <0.001 |
| Total parathyroid hormone,a pg/ml | 52.0 [34.0–83.9] | 41.0 [29.6–60.6] | 58.1 [38.7–89.0] | 101.5 [61.0–165.6] | <0.001 |
| Aldosterone,a pg/ml | 100.9 [71.8–151.1] | 98.8 [70.2–152.3] | 101.83 [72.0–146.4] | 106.2 [77.7–157] | 0.10 |
| Hemoglobin A1C,a % | 6.1 [5.6–7.1] | 5.7 [5.3–6.10 | 6.8 [6.0–7.9] | 6.4 [5.7–7.8] | <0.001 |
| C-peptide,a ng/ml | 2.95 [1.90–4.15] | 2.60 [1.80–3.55] | 3.40 [2.19–4.80] | 3.05 [1.80–4.45] | <0.001 |
| CBC hemoglobin, g/dl | 12.67 (1.73) | 13.45 (1.55) | 12.22 (1.54) | 11.53 (1.65) | <0.001 |
| Glucose,a mg/dl | 97.0 [86.0–121.0] | 90.0 [84.0–99.0] | 113.0 [94.0–145.0] | 100.0 [86.0–138.0] | <0.001 |
| Fetuin-A,a mg/ml | 0.53 (0.11) | 0.55 (0.11) | 0.53 (0.11) | 0.50 (0.10) | <0.001 |
| Mean cell hemoglobin concentration, g/dl | 33.56 (1.07) | 33.83 (0.98) | 33.30 (1.09) | 33.45 (1.05) | <0.001 |
| Troponin I,a ng/ml | 0.00 [0.00–0.01] | 0.00 [0.00–0.00] | 0.00 [0.00–0.01] | 0.01 [0.00–0.02] | <0.001 |
| High-sensitivity troponin T,a pg/ml | 11.46 [5.65–21.92] | 6.86 [1.50–11.92] | 15.55 [9.01–25.75] | 28.29 [14.67–49.22] | <0.001 |
| High-sensitivity CRP,a mg/L | 2.52 [1.01–6.20] | 1.72 [0.85–4.02] | 3.25 [1.32–7.59] | 3.73 [1.13–9.15] | <0.001 |
| N-terminal prohormone of brain natriuretic peptide,a pg/ml | 142.5 [61.6–374.0] | 84.5 [36.9–178.8] | 183.2 [80.7–440.6] | 463.8 [207.1–1377.0] | <0.001 |
| Bicarbonate, mmol/L | 24.51 (3.10) | 25.13 (2.86) | 24.58 (2.97) | 22.43 (3.30) | <0.001 |
| Systolic BP, mm Hg | 127.1 (20.6) | 120.8 (17.7) | 128.5 (20.0) | 142.6 (21.5) | <0.001 |
| Diastolic BP, mm Hg | 70.9 (12.2) | 72.8 (11.4) | 67.1 (11.7) | 75.6 (13.0) | <0.001 |
| IL-10,a pg/ml | 0.00 [0.00–0.00] | 0.00 [0.00–0.00] | 0.00 [0.00–0.00] | 0.00 [0.00–0.00] | <0.001 |
| IL-1RA,a pg/ml | 704.5 [386.1–1501.2] | 561.5 [322.5–1239.2] | 775.1 [444.1–1576.8] | 1103.9 [486.4–2125.0] | <0.001 |
| IL-6,a pg/ml | 1.82 [1.12–2.91] | 1.25 [0.83–1.99] | 2.25 [1.53–3.27] | 2.74 [1.83–4.61] | <0.001 |
| IL-1β,a pg/ml | 0.18 [0.06–1.23] | 0.06 [0.06–0.86] | 0.15 [0.06–1.07] | 1.08 [0.06–2.95] | <0.001 |
| TNF-α,a pg/ml | 2.20 [1.50–3.20] | 1.70 [1.20–2.50] | 2.30 [1.70–3.20] | 3.40 [2.50–4.75] | <0.001 |
| TGF-β,a ng/ml | 10.67 [6.24–17.60] | 10.29 [5.87–17.48] | 10.52 [6.29–16.57] | 12.01 [7.70–19.85] | <0.001 |
| CXCL12, pg/ml | 2449 (532) | 2294 (484) | 2510 (519) | 2750 (544) | <0.001 |
| Fibrinogen, g/L | 4.11 (1.08) | 3.63 (0.86) | 4.32 (1.00) | 4.98 (1.15) | <0.001 |
| White blood cell, 1000/μl | 6.48 (1.88) | 6.02 (1.66) | 6.74 (1.94) | 7.13 (1.99) | <0.001 |
| Total cholesterol, mg/dl | 180.4 (40.3) | 189.7 (38.4) | 168.4 (37.1) | 185.5 (45.9) | <0.001 |
| Triglycerides,a mg/dl | 128.0 [89.0–183.0] | 115.0 [81.0–163.0] | 138.0 [98.0–195.0] | 138.0 [99.0–205.0] | <0.001 |
| HDL, mg/dl | 46.8 (13.7) | 50.9 (14.6) | 43.3 (11.3) | 44.2 (13.5) | <0.001 |
| LDL, mg/dl | 101.3 (32.6) | 110.3 (31.7) | 90.4 (29.5) | 104.0 (34.9) | <0.001 |
| Serum albumin, g/dl | 4.0 (0.4) | 4.1 (0.4) | 4.0 (0.4) | 3.6 (0.4) | <0.001 |
| Family history of kidney disease (%) | 412 (15.3) | 154 (12.8) | 180 (16.4) | 78 (19.7) | 0.002 |
| Diagnosed or treated for any cancer (%) | 147 (5.5) | 69 (5.7) | 64 (5.8) | 14 (3.5) | 0.19 |
| Cardiovascular disease (%) | 878 (32.6) | 168 (14.0) | 543 (49.5) | 167 (42.3) | <0.001 |
| Congestive heart failure (%) | 242 (9.0) | 23 (1.9) | 158 (14.4) | 61 (15.4) | <0.001 |
| History of chronic obstructive pulmonary disease (%) | 80 (3.0) | 23 (1.9) | 51 (4.6) | 6 (1.5) | <0.001 |
| Acidosis (%) | 2251 (83.5) | 1091 (90.7) | 938 (85.4) | 222 (56.2) | <0.001 |
| NSAID (%) | 1422 (52.7) | 515 (42.8) | 746 (67.9) | 161 (40.8) | <0.001 |
| Diuretics (%) | 1556 (57.7) | 456 (37.9) | 832 (75.8) | 268 (67.8) | <0.001 |
| ACE inhibitor (%) | 1327 (49.2) | 509 (42.3) | 637 (58.0) | 181 (45.8) | <0.001 |
| β-blockers (%) | 1312 (48.7) | 388 (32.3) | 697 (63.5) | 227 (57.5) | <0.001 |
| Calcium blockers (%) | 1081 (40.1) | 344 (28.6) | 525 (47.8) | 212 (53.7) | <0.001 |
| Statins (%) | 1510 (56.0) | 461 (38.3) | 832 (75.8) | 217 (54.9) | <0.001 |
| Steroids (%) | 261 (9.7) | 129 (10.7) | 98 (8.9) | 34 (8.6) | 0.26 |
| Antidiabetes (%) | 783 (29.0) | 69 (5.7) | 587 (53.5) | 127 (32.2) | <0.001 |
| Antiplatelet (%) | 1271 (47.1) | 401 (33.3) | 708 (64.5) | 162 (41.0) | <0.001 |
MET, metabolic equivalent; BMI, body mass index; UPCR, urine protein-creatinine ratio; NGAL, neutrophil gelatinase–associated lipocalin; FGF-23, fibroblast growth factor 23; CBC, complete blood count; CRP, C-reactive protein; CXCL12, C-X-C motif chemokine ligand 12; NSAID, nonsteroidal anti-inflammatory drug; ACE, angiotensin-converting enzyme.
Continuous variables that were summarized medium [interquartile range].
CKD Clusters
Using the 72 baseline variables, the consensus clustering algorithm identified three clusters that best represent the data pattern of our CKD population. The consensus matrix heat maps for each cluster size, visualizing the pairwise consensus of all participants, are shown in Figure 1. There were 1203, 1098, and 395 individuals in clusters 1, 2, and 3, respectively. The mean consensus score was 0.96 for cluster 1, 0.90 for cluster 2, and 0.92 for cluster 3, with a larger value indicating better stability of cluster membership (Figure 2A). The changes of proportion of ambiguously clustered pair values under each cluster size are shown in Figure 2B. The “elbow point” for increase value of proportion of ambiguously clustered pairs was observed at a cluster size of three using the relaxed criterion. The distribution of the 72 baseline variables was significantly different between the three clusters, except for aldosterone level, history of diagnosed or treated cancer, and use of steroids (Table 2). Additional sensitivity analyses performed without including the four SES variables (Supplemental Figure 2) and among the three imputed datasets (Supplemental Figures 3–5, Supplemental Table 2) showed almost identical results as the main analysis.
Figure 1.
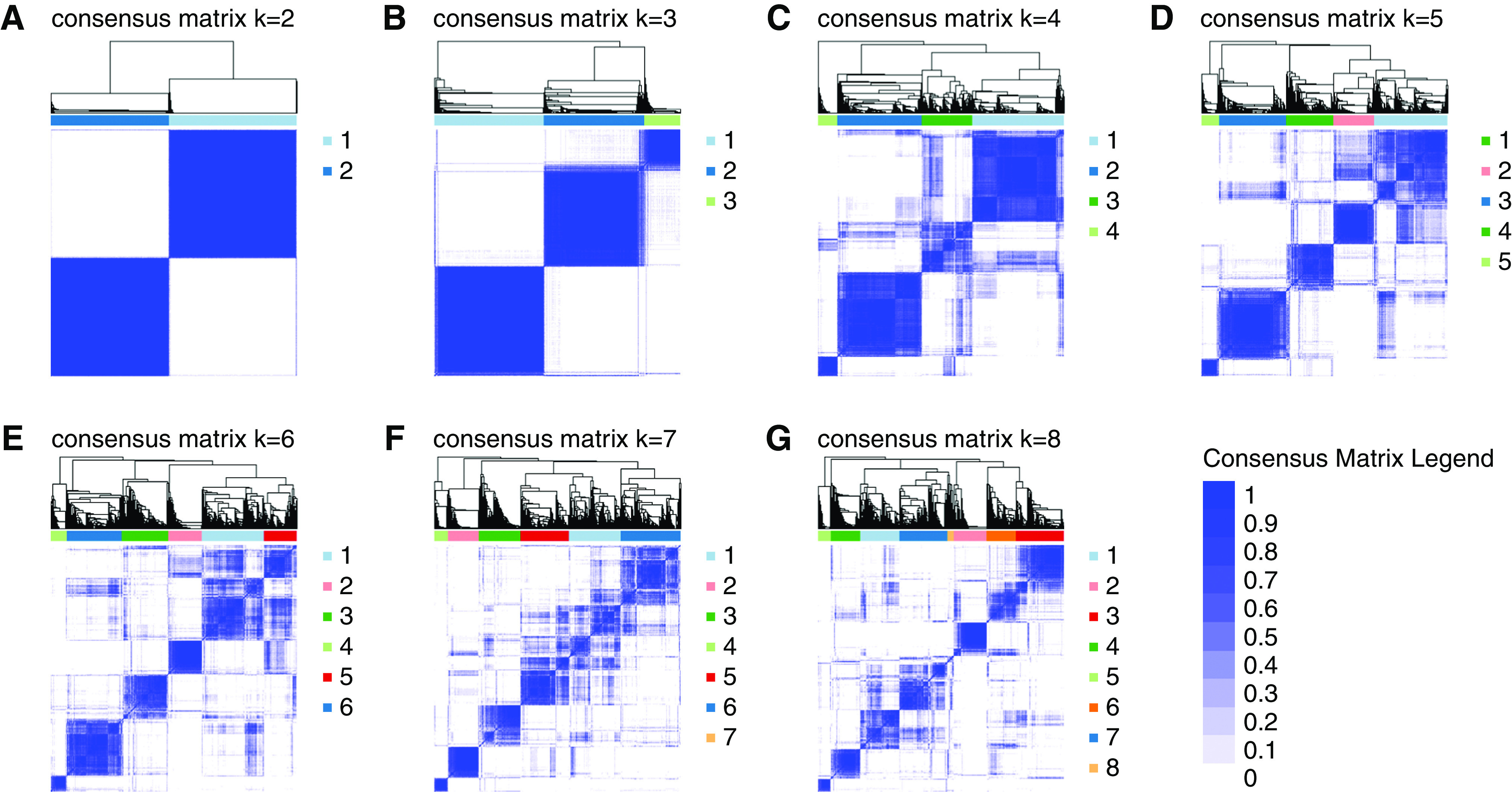
Consensus matrix heatmaps using all baseline predictors. The consensus matrix heat maps of K=2 to K=8 using all 72 baseline parameters (n=2696) The darkest blue color represents perfect consensus where two individuals always group together, the white color represents perfect consensus where two individuals always group separately, and the blue color scales in between represent ambiguous consensus where two individuals are grouped together in some runs but separately in others. (A) K=2. (B) K=3. (C) K=4. (D) K=5. (E) K=6. (F) K=7. (G) K=8.
Figure 2.
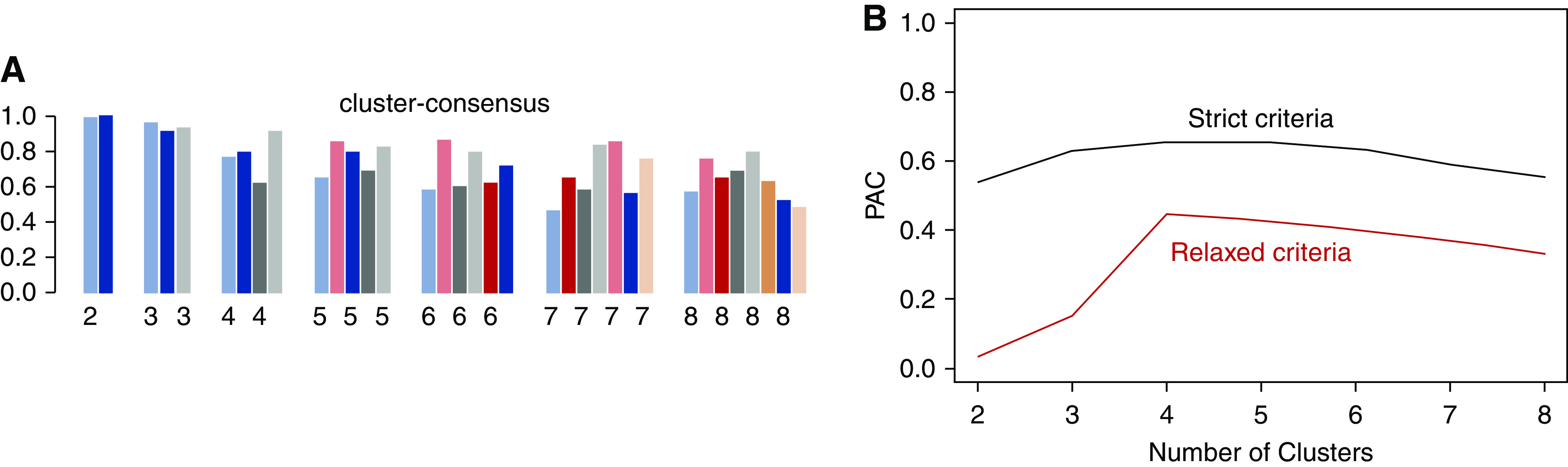
Cluster consensus score and proportion of ambiguously clustered (PAC) pair for consensus clustering using 72 predictors. The bar plot in (A) represents the mean consensus score for different numbers of clusters (K ranges from two to eight) on the basis of 100 repeated resamplings of 80% of the 2696 CRIC participants. The black line in (B) shows the PAC values using the strict criteria with the predetermined boundary of (0, 1) as the definition for ambiguously clustered pairs, and the red line in (B) represents the PAC values using the relaxed criteria with the predetermined boundary of (0.1, 0.9) as the definition for ambiguously clustered pairs.
Standardized difference plots visualized the key characteristics of each cluster. Variables with absolute standardized difference >0.3 are highlighted as the key features for each cluster (Figure 3). Cluster 1 (n=1203) was made up of generally well-educated individuals with relatively high baseline kidney function (mean eGFR =53.6 ml/min per 1.73 m2, SD 15.8) and lower levels of nonfavorable risk factors of bone and mineral markers and cardiac markers. Cluster 2 (n=1098) included individuals more likely to be older, be obese/overweight, have diabetes, and use more medications. Cluster 3 (n=395) had individuals with the worst health status, including higher levels of inflammation, cardiac, and bone and mineral markers; lower levels of serum albumin, serum bicarbonate, and serum calcium; and a lower level of kidney function (mean eGFR =31.0 ml/min per 1.73 m2, SD 10.0), along with lower SES.
Figure 3.
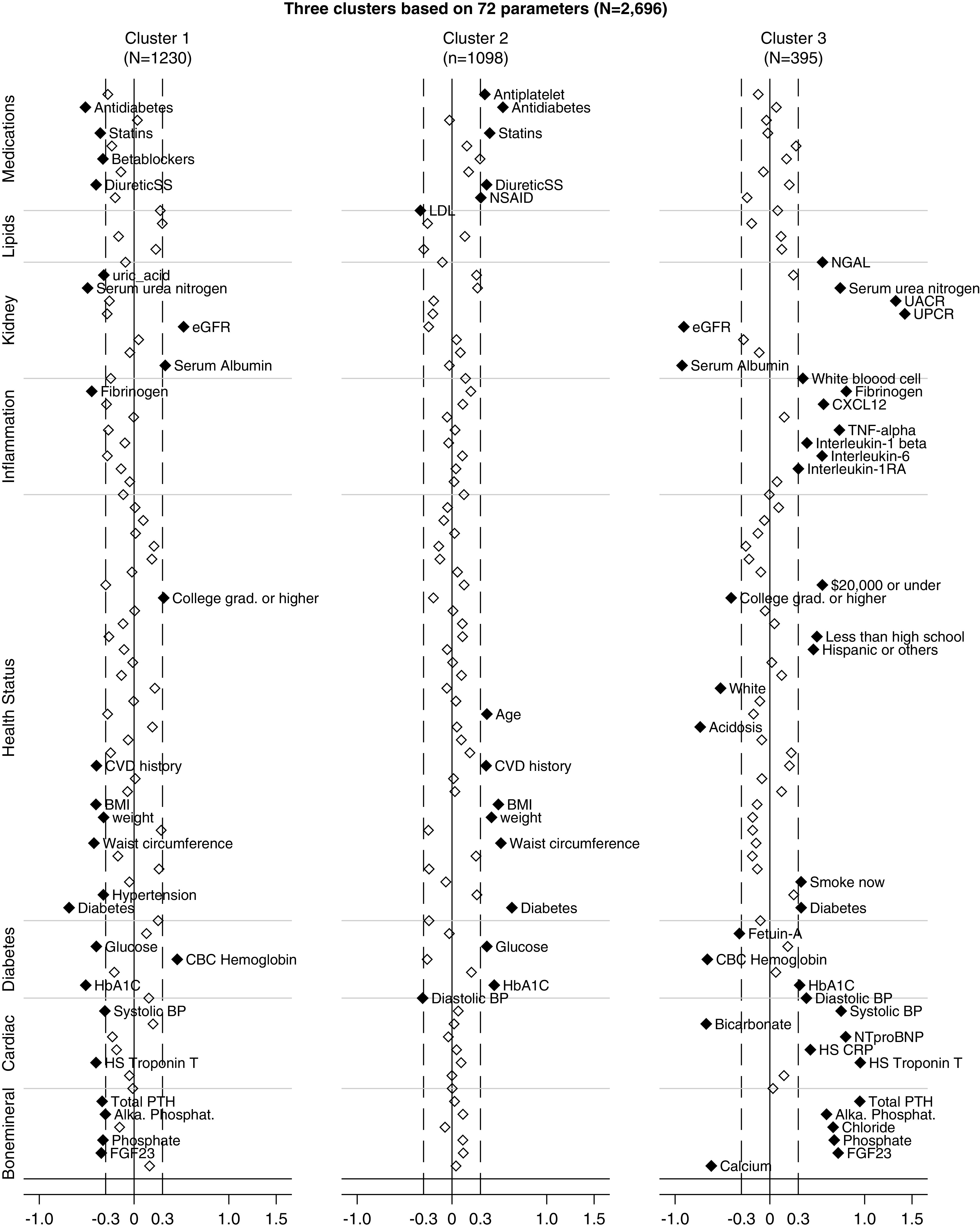
The Manhattan plot of the standardized differences across three CKD subgroups for each of the 72 baseline parameters. The y axis is the standardized differences value, and the x axis shows the eight categories of the baseline parameters. The dashed vertical lines represent the standardized differences cutoffs of >0.3 or <−0.3. The gray horizontal lines lay out the category to which each marker belongs, including bone and mineral markers, cardiac markers, diabetes markers, factors of health status, inflammation markers, kidney markers, lipids markers, and use of medications. Cluster 1: well-educated individuals with relatively high baseline kidney function and lower levels of nonfavorable risk factors of bone and mineral markers and cardiac markers. Cluster 2: individuals who are older in age, are obese/overweight, have diabetes, and use more medications. Cluster 3: individuals with higher levels of inflammation, cardiac, and bone and mineral markers; lower levels of serum albumin, serum bicarbonate, and serum calcium; and a lower level of kidney function, along with lower SES. Alka. Phosphat., alkaline phosphatase; BMI, body mass index; CBC, complete blood count; CRP, high-sensitivity C-reactive protein; CVD, cardiovascular disease; CXCL12, C-X-C motif chemokine ligand 12; FGF-23, fibroblast growth factor 23; HbA1C, hemoglobin A1C; HS, high-sensitivity; NGAL, neutrophil gelatinase–associated lipocalin; NSAID, nonsteroidal anti-inflammatory drug; NTproBNP, N-terminal prohormone of brain natriuretic peptide; PTH, parathyroid hormone; UPCR, urine protein-creatinine ratio.
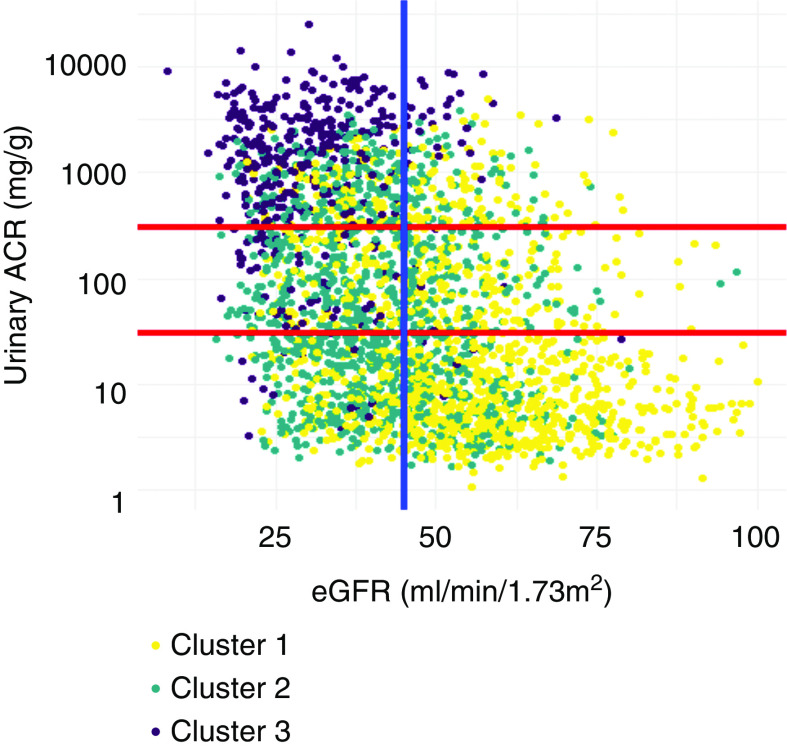
Figure 4 displays the crosstabulation of the CKD clusters and the subgroups on the basis of the KDIGO 2012 guideline, defined by eGFR (<45 and ≥ 45 ml/min per 1.73 m2) and UACR (UACR≤ 30 mg/g, 30<UACR≤300 mg/g, and UACR> 300 mg/g). Among 578 patients with very high risks defined by the 2012 KDIGO guideline (G3b, G4, and G5: eGFR<45 ml/min per 1.73 m2 and A3: UACR>300 mg/g), 16.8%, 33.7%, and 49.5% individuals were classified into cluster 1, cluster 2, and cluster 3, respectively. Among 781 patients with relatively low risks (KDIGO G3a: eGFR=45–59 ml/min per 1.73 m2 and A1: UACR<30 mg/g), 74% and 26% were classified into cluster 1 and cluster 2, respectively (Supplemental Table 3).
Figure 4.
The scatterplot of eGFR and UACR, colored by CKD cluster membership. Each dot represents one individual and is colored by the CKD cluster membership. The two horizontal lines represent the conventional UACR cutoffs of 30 and 300 mg/g; the vertical line represents the conventional eGFR cutoff of 45 ml/min per 1.73 m2. Cluster 1: well-educated individuals with relatively high baseline kidney function and lower levels of nonfavorable risk factors of bone and mineral markers and cardiac markers. Cluster 2: individuals who are older in age, are obese/overweight, have diabetes, and use more medications. Cluster 3: individuals with higher levels of inflammation, cardiac, and bone and mineral markers; lower levels of serum albumin, serum bicarbonate, and serum calcium; and a lower level of kidney function, along with lower SES. ACR, albumin-creatinine ratio.
Survival Analyses
Over a median of 9 years of follow-up from study entry, 900 individuals developed the composite outcome of CKD progression (event rate: 53.9 per 1000 person-year follow-up). Additionally, 689 individuals experienced kidney failure requiring replacement therapy (RRT; event rate: 33.0); 469 individuals had CHF (event rate: 22.6); 460 developed the composite outcome of MI, stroke, and PAD (event rate: 22.1); 720 individuals developed the composite outcome of MI, stroke, PAD, and CHF (event rate: 36.7); and 724 individuals died (event rate: 30.4).
Kaplan–Meier survival plots showed that the cluster membership was significantly associated with the risks of CKD progression, kidney failure requiring replacement therapy, cardiovascular disease outcomes, and death. Clusters 1, 2, and 3 were associated with low, medium, and high risks for the above clinical outcomes, respectively (Figure 5). In the fully adjusted models, belonging to cluster 2 was independently associated with increased risks for CKD progression (HR, 1.32; 95% CI, 1.05 to 1.67); CHF (HR, 2.40; 95% CI, 1.67 to 3.46); MI, stroke, or PAD (HR, 1.46; 95% CI, 1.06 to 2.02); CHF, MI, stroke, or PAD (HR, 1.59; 95% CI, 1.22 to 2.07); and death (HR, 1.77; 95% CI, 1.37 to 2.29). The HRs associated with cluster 3 were 1.63 (95% CI, 1.27 to 2.09) for CKD progression, 1.42 (95% CI, 1.07 to 1.89) for kidney failure requiring RRT, 3.11 (95% CI, 2.05 to 4.74) for CHF, 1.85 (95% CI, 1.25 to 2.74) for MI, stroke, or PAD, 2.15 (95% CI, 1.57 to 2.96) for CHF, MI, stroke, or PAD, and 2.39 (95% CI, 1.76 to 3.25) for death, respectively (Table 3). The proportional hazard assumption over the full follow-up time was violated for the outcomes of CKD progression; kidney failure requiring RRT; CHF; and the composite cardiovascular disease outcome of CHF, MI, stroke, and PAD. Supplemental Table 4 reports the HRs stratified by follow-up time. The association between the cluster membership and outcomes was stronger early on during the study follow-up.
Figure 5.
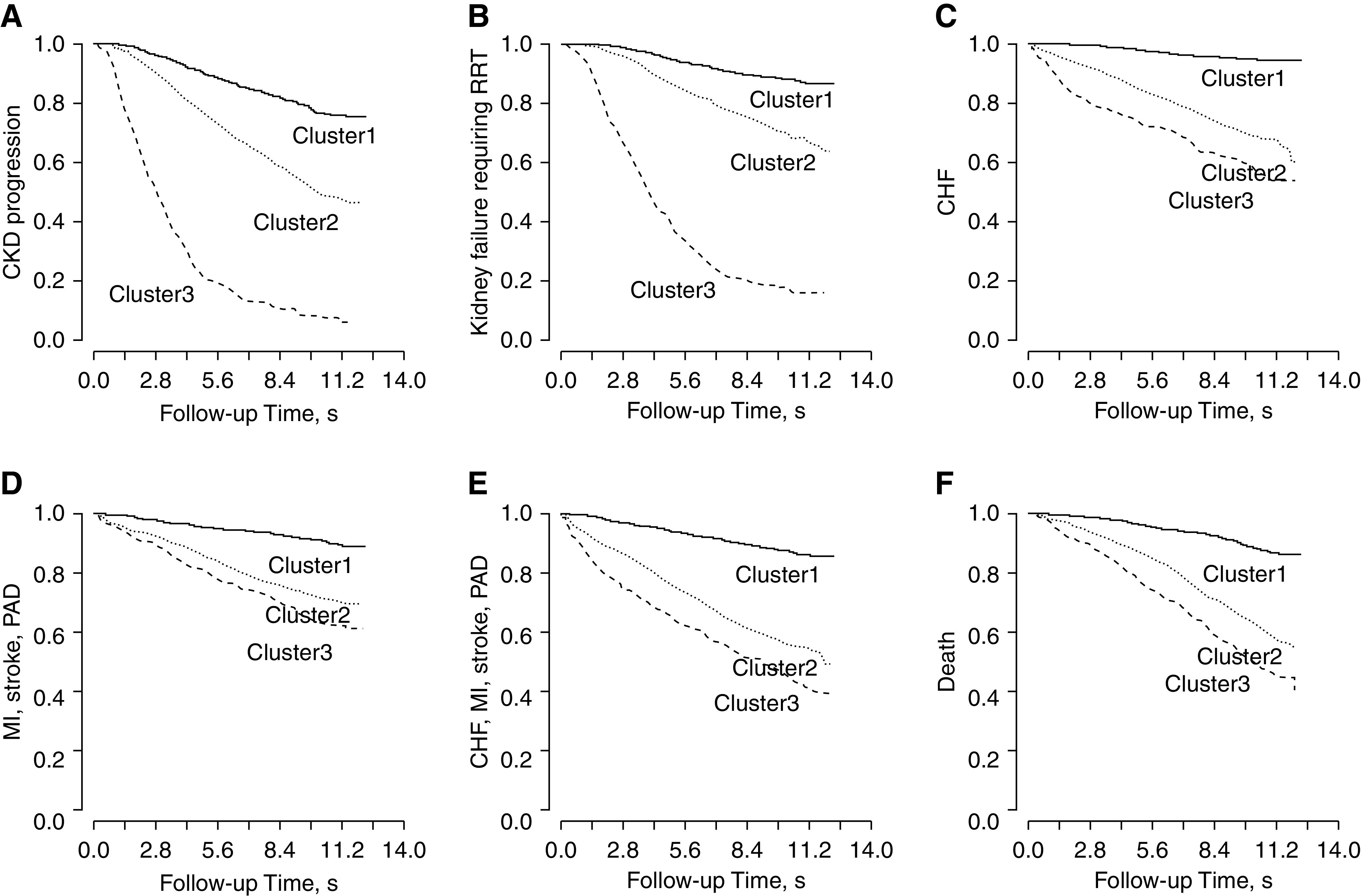
Kaplan–Meier survival plots of the three clusters defined by 72 baseline variables and six clinical outcomes. (A) CKD progression; (B) ESKD; (C) CHF; (D) composite cardiovascular disease outcome of MI, stroke, or PAD; (E) composite cardiovascular disease outcome of CHF, MI, stroke, or PAD; and (F) death. The log rank P values for all comparisons were <0.001. Cluster 1: well-educated individuals with relatively high baseline kidney function and lower levels of nonfavorable risk factors of bone and mineral markers and cardiac markers. Cluster 2: individuals who are older in age, are obese/overweight, have diabetes, and use more medications. Cluster 3: individuals with higher levels of inflammation, cardiac, and bone and mineral markers; lower levels of serum albumin, serum bicarbonate, and serum calcium; and a lower level of kidney function, along with lower SES.
Table 3.
The risks of CKD progression, cardiovascular disease, and death that were associated with cluster membership using 72 baseline parameters
| Outcome | No. of Events | Average Follow-Up (Yr) | Event Rate per 1000 PY | Unadjusted | Adjusted | ||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P Value | HR (95% CI) | P Value | ||||
| CKD progression | |||||||
| Overall | 900 | 6.2 | 53.9 | — | — | ||
| Cluster 1 | 217 | 7.6 | 23.8 | Reference | Reference | ||
| Cluster 2 | 385 | 5.8 | 60.0 | 2.62 (2.21 to 3.09) | <0.001 | 1.32 (1.05 to 1.67) | 0.02 |
| Cluster 3 | 298 | 3 | 254.3 | 13.16 (10.98 to 15.79) | <0.001 | 1.63 (1.27 to 2.09) | <0.001 |
| Kidney failure requiring RRT | |||||||
| Overall | 689 | 7.7 | 33.0 | — | — | ||
| Cluster 1 | 137 | 9.2 | 12.4 | Reference | Reference | ||
| Cluster 2 | 278 | 7.5 | 33.8 | 2.78 (2.26 to 3.41) | <0.001 | 1.26 (0.95 to 1.67) | 0.10 |
| Cluster 3 | 274 | 4.1 | 170.9 | 16.15 (13.08 to 19.94) | <0.001 | 1.42 (1.07 to 1.89) | 0.02 |
| CHF | |||||||
| Overall | 469 | 7.7 | 22.6 | — | — | ||
| Cluster 1 | 56 | 8.9 | 5.2 | Reference | Reference | ||
| Cluster 2 | 281 | 7.1 | 36.0 | 6.92 (5.19 to 9.22) | <0.001 | 2.40 (1.67 to 3.46) | <0.001 |
| Cluster 3 | 132 | 5.7 | 58.4 | 11.22 (8.20 to 15.36) | <0.001 | 3.11 (2.05 to 4.74) | <0.001 |
| MI, stroke, or PAD | |||||||
| Overall | 460 | 7.7 | 22.1 | — | — | ||
| Cluster 1 | 100 | 8.7 | 9.5 | Reference | Reference | ||
| Cluster 2 | 253 | 7.1 | 32.4 | 3.42 (2.71 to 4.31) | <0.001 | 1.46 (1.06 to 2.02) | 0.02 |
| Cluster 3 | 107 | 6.2 | 43.6 | 4.61 (3.50 to 6.06) | <0.001 | 1.85 (1.25 to 2.74) | 0.002 |
| CHF, MI, stroke, or PAD | |||||||
| Overall | 720 | 7.3 | 36.7 | — | — | ||
| Cluster 1 | 137 | 8.6 | 13.2 | Reference | Reference | ||
| Cluster 2 | 408 | 6.6 | 56.7 | 4.27 (3.52 to 5.18) | <0.001 | 1.59 (1.22 to 2.07) | <0.001 |
| Cluster 3 | 175 | 5.3 | 84.3 | 6.32 (5.04 to 7.91) | <0.001 | 2.15 (1.57 to 2.96) | <0.001 |
| Death | |||||||
| Overall | 724 | 8.8 | 30.4 | — | — | ||
| Cluster 1 | 139 | 9.7 | 11.9 | Reference | Reference | ||
| Cluster 2 | 398 | 8.4 | 43.4 | 3.84 (3.16 to 4.66) | <0.001 | 1.77 (1.37 to 2.29) | <0.001 |
| Cluster 3 | 187 | 7.5 | 63.4 | 5.80 (4.66 to 7.23) | <0.001 | 2.39 (1.76 to 3.25) | <0.001 |
Model adjusts for age, sex, Black race, education level, diabetes, smoking status, alcohol use, physical activities, body mass index levels, eGFR, log-transformed UACR, systolic BP, family history of kidney disease, and history of cardiovascular disease. Cox model proportional hazard assumption test for variable of cluster membership. PY, person year.
The time-dependent receiver operating characteristics evaluated at 10 years was used to quantify the improvement in model discrimination performance for predicting different clinical outcomes with and without the cluster membership (Supplemental Figure 6). Compared with the model that included only eGFR and UACR (model 1), the model with only CKD cluster membership (model 0) had worse discrimination performance in predicting outcomes of CKD progression and incident kidney failure requiring RRT and better performance in predicting cardiovascular disease outcomes and death. The multivariable models that included the cluster membership (model 3) showed minimal improvement on top of the model that included 14 traditional risk factors (model 2) for cardiovascular outcomes and death.
Subgroup CKD Clustering
In three predefined subgroups, we repeated the unsupervised consensus clustering with 72 baseline variables, followed by survival analyses. Among the 1411 individuals with impaired baseline kidney function (eGFR<45 ml/min per 1.73 m2), we identified three clusters (Supplemental Figure 7) with relatively high cluster consensus scores and low proportion of ambiguously clustered values (Supplemental Figure 8, A and C). We observed similar cluster patterns as in the main analyses. Clusters 1, 2, and 3 were associated with low, medium, and high risks of developing the six clinical end points, respectively (Supplemental Figures 9 and 10, Supplemental Tables 5 and 6).
Among the 1285 individuals with eGFR≥45 ml/min/1.73 m2, two clusters (Supplemental Figure 11) were identified (Supplemental Figure 8, B and D). Compared with cluster 1 (n=608), individuals in cluster 2 (n=677) had higher levels of high-sensitivity troponin T, diabetes, hypertension, obesity markers, fibrinogen, and serum urea nitrogen; used more medications; and had lower complete blood count hemoglobin and eGFR at baseline (Supplemental Figure 12). Individuals in cluster 2 also had higher risks of all six clinical end points (Supplemental Figure 13, Supplemental Tables 7 and 8).
Finally, among the 1207 individuals with baseline UACR<30 mg/g, we identified two clusters (Supplemental Figure 14) with relatively high cluster consensus score values and low proportion of ambiguously clustered values (Supplemental Figure 15). Compared with cluster 1 (n=678), individuals in cluster 2 (n=529) had higher levels of fibroblast growth factor 23, fibrinogen, serum urea nitrogen, uric acid, diabetes, and obesity markers; had lower levels of complete blood count hemoglobin and eGFR; and used more medications (Supplemental Figure 16). Similar to the previous survival analysis results, individuals in cluster 2 had higher risks of developing all six clinical end points (Supplemental Figure 17, Supplemental Tables 9 and 10).
Discussion
In this study, we applied the unsupervised consensus clustering algorithm and identified three clusters on the basis of 72 selected baseline variables. The three clusters represented clinically distinct subgroups that summarized the multidimensional data pattern at baseline and differed significantly with regard to their risks of kidney disease progression, cardiovascular events, and death. Further stratification by eGFR and UACR confirmed that patient heterogeneity exists not only in the overall patient population but also, among individuals with relatively high kidney function or low proteinuria levels. Being able to characterize this heterogeneity early is an important step toward individualizing follow-up strategies for these patients.
Although patients in cluster 1 were primarily in early stages of CKD, patients in cluster 2 and cluster 3 had worse kidney function, were more often diabetic with worse glycemic control, and were more likely to have hypertension and severe mineral and bone disorders. Recent studies suggest that inflammatory mechanisms and the upregulation of corresponding pathways in response to kidney injury are involved not only in the development but also in the progression of CKD and its comorbidities, including cardiovascular disease.28,29 The identified clusters may thus represent different states of inflammation, which could, in part, explain the differences in risks of developing adverse clinical events. Sensitivity analyses of consensus clustering performed without including SES variables revealed the same CKD clusters as the main analyses (Supplemental Table 2). This indicates that the cluster membership found in this study is mainly driven by patients’ clinical/subclinical manifestations, and the data pattern that underlies the multidimensional medical factors could sufficiently capture patients’ SES variability.
We showed a strong independent association between the cluster membership and future adverse events after controlling for established CKD risk factors, such as eGFR, UACR, BP, diabetes status, etc. The adjusted HRs comparing cluster 3 versus cluster 1 on cardiovascular disease outcomes and death (HRs range from 1.85 [95% CI, 1.25 to 2.74] to 3.11 [95% CI, 2.05 to 4.74]) were greater than many established risk factors, such as diabetes (HRs range from 1.23 [95% CI, 0.98 to 1.56] to 1.38 [95% CI, 1.09 to 1.75]) and male sex (HR range from 1.15 [95% CI, 0.94 to 1.41] to 1.61 [95% CI, 1.37 to 1.90]), in the same model. The cluster membership provided a simple metric of summarizing patient heterogeneity and comorbidity profiles encoded in the 72 baseline variables. In addition, it implicitly handled the measurement error possibly associated with each individual variable and thus, can potentially be used as a more reliable measure for evaluating risks of clinical events. The cluster membership also provided additional information to the 2012 KDIGO CKD classification guideline.30 Given our main clustering results of three CKD subgroups, we were able to classify patients with very high risks defined by the 2012 KDIGO guideline (G3b, G4, and G5: eGFR<45 ml/min per 1.73 m2 and A3: UACR>300 mg/g) further into three clusters—17% in cluster 1 with low risk and 34% in cluster 2 with medium risk. Among patients with moderately increased risks defined by the 2012 KDIGO guideline (G3a: eGFR between 45 and 59 ml/min per 1.73 m2 and A1: UACR<30 mg/g), 26% were classified in cluster 2 with substantial increased risks of both kidney and cardiovascular outcomes (Figure 4, Supplemental Table 3).
From a risk prediction perspective, the additional discrimination value of CKD subgroups added to a model with known risk factors, such as eGFR, UACR, demographics, diabetes, etc., was limited, possibly due to redundancy of information. However, the model with cluster membership alone had better discrimination values compared with the model with only eGFR and UACR for differentiating the risks for cardiovascular disease outcomes (Supplemental Figure 6). This finding highlights the potential of applying consensus clustering for detecting subgroups of patients with CKD who are at higher risk of developing adverse cardiovascular disease events.
To our knowledge, this is the first study to investigate the heterogeneity of patients with CKD from patterns of laboratory-enriched clinical attributes using unsupervised clustering methods. In other disease settings, these phenotyping tools have been used to better characterize heterogenous conditions, such as heart failure,31,32 type 2 diabetes,33 and different forms of cancer.16,34–36 Scherzer et al.32 found that unsupervised cluster analysis on the basis of selected serum biomarkers helped to differentiate cardiopulmonary abnormalities in HIV-infected patients. In addition, the assigned cluster membership was then used to predict mortality in these patients. A study by Ahlqvist et al.33 identified five subgroups of patients with type 2 diabetes using a data-driven cluster analysis approach, which provided subgroups of patients with significantly different patient characteristics that also differed with respect to their risk of developing diabetic complications. In kidney disease, cluster analysis has also been used to identify differing pathogenetic patterns of membranoproliferative GN and permitted differentiating patients by underlying pathobiologic mechanisms, clinical features, and disease survival.37 A study in patients with CKD analyzed data from the 500 Cities Project and defined distinct geographic regions across the United States clustered by unhealthy behaviors, prevention measures, and CKD-related outcomes, such as hypertension and diabetes.38
Our findings provide clear evidence that substantial heterogeneity exists under the umbrella term of “CKD,” which encompasses a number of distinct pathologies and etiologies. Consensus clustering using discrete data elements is one approach to uncover that heterogeneity. As further ‘omics interrogation technologies are developed and applied to the clinic, approaches like ours could translate large amounts of data to clinically relevant groupings of patients with distinct clinical trajectories. Further stratification of patients with similar and relatively “preserved” or “impaired” kidney function is of particular interest for guiding clinical interventions toward precision medicine. Whether this approach can lead to more focused interventions and improved outcomes—the promise of precision medicine—will require further clinical studies.
The study has several limitations. First, cluster analysis belongs to the category of exploratory data analysis. It is a data-driven approach so that the cluster membership depends on the input of the data (e.g., the 72 baseline variables in our analysis). The algorithm finds clusters of individuals that share the same or similar characteristics in terms of the input variables. As a result, if the input variables were to be changed to other patient characteristics, we would expect to find different CKD subgroups. Second, the study used a subset of the CRIC participants due to missing variable information. Because the study population is comparable with the CRIC population (i.e., the standardized mean differences of all covariates were smaller than 0.1), the internal validity of the clustering result is not a concern. Additional sensitivity analyses performed using the three imputed datasets further confirmed the robustness of the identified cluster membership. However, it is not clear how the results are generalizable to other CKD populations. Subsequent studies are required to examine its external validity. This will require that the external studies collect the same information as was done in the CRIC study. Third, although cluster analysis is useful to explore large datasets and heterogeneity among individuals, it summarizes patients’ heterogeneity, which is a continuous measure by nature, into discrete categories and may result in loss of information in exchange for better clinical interpretability.32 To ensure the stability of discovered cluster membership, we chose the consensus clustering algorithm, a more robust clustering approach than the traditional hierarchical or K-mean clustering approach.
Using a data-driven approach, we identified three clusters of patients with CKD on the basis of 72 variables, including various biomarkers and clinical measures. The three clusters showed distinct profiles at baseline and provided complementary classification information compared with the current KDIGO criterion. Further, the cluster membership was strongly associated with future risks of kidney disease, cardiovascular events, and death. Phenotyping of patients with CKD using unsupervised machine learning algorithms provides a powerful tool for identifying patient heterogeneity. Future studies are required for external validation of the identified clusters and to investigate CKD subgroups by incorporating novel biomarker data (e.g., genomics, metabolomics, proteomics, etc.) as an important next step toward identifying subgroup-specific therapeutic targets for better patient care and precision medicine in nephrology.
Disclosures
A.H. Anderson reports fees from Kyowa Hakko Kirin, outside the submitted work. H.I. Feldman reports consultancy agreements with DLA Piper, LLP, InMed, Inc., Kyowa Hakko Kirin Co, Ltd. (ongoing), and the National Kidney Foundation; research funding from Regeneron; honoraria from Rogosin Institute (invited speaker); and scientific advisor or membership as a member of the Steering Committee for the CRIC Study and Editor-in-Chief of the National Kidney Foundation (member of advisory board). C.-y. Hsu reports personal fees from EcoR1 Capital Fund, Health Advances, Ice Miller LLP, Reata, Satellite Healthcare, and UpToDate and grants from Satellite Healthcare, outside the submitted work. T. Isakova reports personal fees from Akebia Therapeutics, Kyowa Kirin Co, and LifeSci Capital, outside the submitted work. P.S. Rao reports honoraria from AstraZeneca and scientific advisor or membership with the AstraZeneca Nephrology Fellowship Advisory Board and the Renal Research Institute. G. Saab serves as a hemodialysis unit medical director for Fresenius Medical Care, outside the submitted work. T. Shafi reports consultancy agreements with Siemens; research funding from Baxter (clinical trial); honoraria from National Institute of Health, Siemens; being a scientific advisor or member with CJASN and American Journal of Kidney Diseases. S. S. Waikar reports personal fees from Barron and Budd (versus Fresenius), Bunch and James, Cerus, CVS, GE Health Care, GSK, Harvard Clinical Research Institute (also known as Baim), JNJ, Kantum Pharma, Mallinckrodt, Mass Medical International, Pfizer, Public Health Advocacy Institute, Roth Capital Partners, Strataca, Takeda, Venbio, and Wolters Kluewer and grants and personal fees from Allena Pharmaceuticals, outside the submitted work. F.P. Wilson reports consultancy agreements with Translational Catalyst, LLC; ownership interest as owner of Efference, LLC; scientific advisor or membership as an Editorial Board member of American Journal of Kidney Disease and an Editorial Board member of CJASN; and other interests/relationships as a member of the Board of Directors of Gaylord Health Care and a medical commentator for Medscape. Z. Zheng reports consultancy agreement and personal fees from Akebia Therapeutics outside the submitted work. All remaining authors have nothing to disclose.
Funding
Funding for the CRIC study was obtained under a cooperative agreement with National Institute of Diabetes and Digestive and Kidney Diseases grants U01DK060990, U01DK060984, U01DK061022, U01DK061021, U01DK061028, U01DK060980, U01DK060963, U01DK060902, and U24DK060990. In addition, this work was supported in part by Perelman School of Medicine at the University of Pennsylvania Clinical and Translational Science Award National Institutes of Health (NIH)/National Center for Advancing Translational Sciences grant UL1TR000003; Johns Hopkins University National Center for Advancing Translational Sciences grant UL1TR-000424; University of Maryland General Clinical Research Center National Center for Advancing Translational Sciences grant M01 RR-16500; the Clinical and Translational Science Collaborative of Cleveland; National Center for Advancing Translational Sciences component of the NIH and NIH roadmap for Medical Research grant UL1TR000439; Michigan Institute for Clinical and Health Research National Center for Advancing Translational Sciences grant UL1TR000433; University of Illinois at Chicago Clinical and Translational Science Award National Center for Research Resources grant UL1RR029879; Tulane Center of Biomedical Research Excellence for Clinical and Translational Research in Cardiometabolic Diseases grant P20 GM109036; Kaiser Permanente NIH/National Center for Research Resources University of California San Francisco Clinical and Translational Science Institute grant UL1 RR-024131; and Department of Internal Medicine, University of New Mexico School of Medicine Albuquerque, New Mexico National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK119199. I. M. Schmidt is supported by the American Philosophical Society Daland Fellowship in Clinical Investigation.
Supplementary Material
Acknowledgments
We thank the CRIC participants for time and commitment to the study.
S.S. Waikar, W. Yang, and Z. Zheng designed the study; W. Yang and Z. Zheng analyzed the data; Z. Zheng made the figures; C.-y. Hsu, I.M. Schmidt, S.S. Waikar, W. Yang, and Z. Zheng drafted and revised the paper; and A.H. Anderson, J. Chen, H.I. Feldman, J.C. Fink, C.-y. Hsu, T. Isakova, R. Kallem, J.R. Landis, P.S. Rao, A.C. Ricardo, H. Rincon-Choles, G. Saab, I.M. Schmidt, T. Shafi, S.S. Waikar, F.P. Wilson, D. Xie, W. Yang, and Z. Zheng reviewed and approved the final version of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Contributor Information
Collaborators: CRIC Study Investigators, Lawrence J. Appel, Alan S. Go, Jiang He, James P. Lash, Mahboob Rahman, and Raymond R. Townsend
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020030239/-/DCSupplemental.
Supplemental Material. Consensus clustering algorithm.
Supplemental Table 1. Baseline characteristics of individuals in the study population (N=2696) and the overall CRIC population (N=3921).
Supplemental Table 2. Cluster features and standardized difference in the main and sensitivity analyses.
Supplemental Table 3. The distribution of CKD subgroups revealed by 72 baseline parameters within the CKD classes defined by KDIGO guideline.
Supplemental Table 4. The risks of CKD progression, cardiovascular disease, and death across overall or stratified periods of follow-up time that were associated with cluster membership using 72 baseline parameters.
Supplemental Table 5. Baseline characteristics of individuals with eGFR<45 ml/min per 1.73 m2 (N=1411) across three subgroups revealed by 72 baseline parameters.
Supplemental Table 6. The adjusted risks of CKD progression, cardiovascular disease, and death associated with CKD subgroups among patients with eGFR<45 ml/min per 1.73 m2 (N=1411).
Supplemental Table 7. Baseline characteristics of individuals with eGFR≥45 ml/min per 1.73 m2 (N=1285) across two subgroups revealed by 72 baseline parameters.
Supplemental Table 8. The adjusted risks of CKD progression, cardiovascular disease, and death associated with CKD subgroups among patients with eGFR≥45 ml/min per 1.73 m2 (N=1285).
Supplemental Table 9. Baseline characteristics of individuals with UACR<30mg/g (N=1207) across two subgroups revealed by 72 baseline parameters.
Supplemental Table 10. The adjusted risks of CKD progression, cardiovascular disease, and death associated with CKD subgroups among patients with UACR<30 mg/g (N=1207).
Supplemental Figure 1. The consensus clustering algorithm flow chart.
Supplemental Figure 2. The Manhattan plot of the standardized differences across three subgroups on the basis of 68 non-SES baseline parameters in the main analysis population (N=2696).
Supplemental Figure 3. The Manhattan plot of the standardized differences across three subgroups on the basis of 72 baseline parameters in imputed dataset 1 (N=3921).
Supplemental Figure 4. The Manhattan plot of the standardized differences across three subgroups on the basis of 72 baseline parameters in imputed dataset 2 (N=3921).
Supplemental Figure 5. The Manhattan plot of the standardized differences across three subgroups on the basis of 72 baseline parameters in imputed dataset 3 (N=3921).
Supplemental Figure 6. Time-dependent ROC curves of Cox regression for six clinical end points at year 10: (A) CKD progression; (B) kidney failure requiring RRT; (C) CHF; (D) MI, stroke, and PAD; (E) CHF, MI, stroke, and PAD; and (F) death.
Supplemental Figure 7. The consensus matrix heat maps of K=2 to K=8 among individuals with eGFR<45 ml/min per 1.73 m2 (N=1411) using the 72 baseline parameters.
Supplemental Figure 8. Cluster consensus score and proportion of ambiguously clustered pair for consensus clustering using all parameters (A and C) among individuals with baseline eGFR <45 ml/min per 1.73 m2 (N=1411) and (B and D) among individuals with baseline eGFR ≥45 ml/min per 1.73 m2 (N=1285).
Supplemental Figure 9. The Manhattan plot of the standardized differences across three subgroups among individuals with eGFR<45 ml/min per 1.73 m2 (N=1411) for each of the 72 baseline parameters.
Supplemental Figure 10. Kaplan–Meier survival plots of the three subgroups defined by 72 baseline parameters among individuals with baseline eGFR <45 ml/min per 1.73 m2 (N=1411) and the outcomes of (A) CKD progression; (B) kidney failure requiring RRT; (C) composite outcome of MI, stroke, and PAD; (D) composite outcome of CHF, MI, stroke, and PAD; (E) CHF; and (F) death.
Supplemental Figure 11. The consensus matrix heat maps of K=2 to K=8 among individuals with eGFR≥45 ml/min per 1.73 m2 (N=1285) using the 72 baseline parameters
Supplemental Figure 12. The Manhattan plot of the standardized differences across two subgroups among individuals with eGFR≥45 ml/min per 1.73 m2 (N=1285) for each of the 72 baseline parameters.
Supplemental Figure 13. Kaplan–Meier survival plots of the two subgroups defined by 72 baseline parameters among individuals with baseline eGFR ≥45 ml/min per 1.73 m2 (N=1285) and the outcomes of (A) CKD progression; (B) kidney failure requiring RRT; (C) composite outcome of MI, stroke, and PAD; (D) composite outcome of CHF, MI, stroke, and PAD; (E) CHF; and (F) death.
Supplemental Figure 14. The consensus matrix heat maps of K=2 to K=8 among individuals with UACR<30 mg/g (N=1207) using the 72 baseline parameters.
Supplemental Figure 15. Cluster consensus score and proportion of ambiguously clustered pair for consensus clustering using all parameters among individuals with baseline UACR<30 mg/g (N=1207).
Supplemental Figure 16. The Manhattan plot of the standardized differences across two subgroups among individuals with UACR<30 mg/g (N=1207) for each of the 72 baseline parameters.
Supplemental Figure 17. Kaplan–Meier survival plots of the two subgroups defined by 72 baseline parameters among individuals with baseline UACR <30 mg/g (N=1207) and the outcomes of (A) CKD progression; (B) kidney failure requiring RRT; (C) composite outcome of MI, stroke, and PAD; (D) composite outcome of CHF, MI, stroke, and PAD; (E) CHF; and (F) death.
Supplemental Figure 18. An illustration of portion of ambiguously clustered relaxed criteria versus strict criteria.
References
- 1.Centers for Disease Control and Prevention: Chronic Kidney Disease in the United States, 2019, Atlanta, GA, US Department of Health and Human Services, Centers for Disease Control and Prevention, 2019 [Google Scholar]
- 2.Levin A, Stevens PE: Summary of KDIGO 2012 CKD Guideline: Behind the scenes, need for guidance, and a framework for moving forward. Kidney Int 85: 49–61, 2014 [DOI] [PubMed] [Google Scholar]
- 3.Levey AS, Becker C, Inker LA: Glomerular filtration rate and albuminuria for detection and staging of acute and chronic kidney disease in adults: A systematic review. JAMA 313: 837–846, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levey AS, Inker LA: GFR as the “gold standard”: Estimated, measured, and true. Am J Kidney Dis 67: 9–12, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Tervaert TWC, Mooyaart AL, Amann K, Cohen AH, Cook HT, Drachenberg CB, et al.; Renal Pathology Society: Pathologic classification of diabetic nephropathy. J Am Soc Nephrol 21: 556–563, 2010 [DOI] [PubMed] [Google Scholar]
- 6.Nelson RG, Bennett PH, Beck GJ, Tan M, Knowler WC, Mitch WE, et al.; Diabetic Renal Disease Study Group: Development and progression of renal disease in Pima Indians with non-insulin-dependent diabetes mellitus. N Engl J Med 335: 1636–1642, 1996 [DOI] [PubMed] [Google Scholar]
- 7.Bidani AK, Griffin KA: Pathophysiology of hypertensive renal damage: Implications for therapy. Hypertension 44: 595–601, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Cheung AK, Rahman M, Reboussin DM, Craven TE, Greene T, Kimmel PL, et al.; SPRINT Research Group: Effects of intensive BP control in CKD. J Am Soc Nephrol 28: 2812–2823, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurts C, Panzer U, Anders H-J, Rees AJ: The immune system and kidney disease: Basic concepts and clinical implications. Nat Rev Immunol 13: 738–753, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, et al.; AASK Study Investigators; CRIC Study Investigators: APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avesani C, Carrero J, Axelsson J, Qureshi A, Lindholm B, Stenvinkel P: Inflammation and wasting in chronic kidney disease: Partners in crime. Kidney Int 70: S8–S13, 2006 [Google Scholar]
- 12.Soderland P, Lovekar S, Weiner DE, Brooks DR, Kaufman JS: Chronic kidney disease associated with environmental toxins and exposures. Adv Chronic Kidney Dis 17: 254–264, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Wright JT Jr, Bakris G, Greene T, Agodoa LY, Appel LJ, Charleston J, et al.; African American Study of Kidney Disease and Hypertension Study Group: Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: Results from the AASK trial [published correction appears in JAMA 295: 2726, 2006]. JAMA 288: 2421–2431, 2002 [DOI] [PubMed] [Google Scholar]
- 14.Xie Y, Bowe B, Li T, Xian H, Balasubramanian S, Al-Aly Z: Proton pump inhibitors and risk of incident CKD and progression to ESRD. J Am Soc Nephrol 27: 3153–3163, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monti S, Tamayo P, Mesirov J, Golub T: Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52: 91–118, 2003 [Google Scholar]
- 16.Soria D, Garibaldi JM, Ambrogi F, Green AR, Powe D, Rakha E, et al.: A methodology to identify consensus classes from clustering algorithms applied to immunohistochemical data from breast cancer patients. Comput Biol Med 40: 318–330, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Feldman HI, Appel LJ, Chertow GM, Cifelli D, Cizman B, Daugirdas J, et al.; Chronic Renal Insufficiency Cohort (CRIC) Study Investigators: The chronic renal insufficiency cohort (CRIC) study: Design and methods. J Am Soc Nephrol 14[Suppl 2]: S148–S153, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Lash JP, Go AS, Appel LJ, He J, Ojo A, Rahman M, et al.; Chronic Renal Insufficiency Cohort (CRIC) Study Group: Chronic Renal Insufficiency Cohort (CRIC) Study: Baseline characteristics and associations with kidney function [published correction appears in Clin J Am Soc Nephrol 6: 2548–2553, 2011]. Clin J Am Soc Nephrol 4: 1302–1311, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer MJ, Go AS, Lora CM, Ackerson L, Cohan J, Kusek JW, et al.; CRIC and H-CRIC Study Groups: CKD in Hispanics: Baseline characteristics from the CRIC (Chronic Renal Insufficiency Cohort) and Hispanic-CRIC studies. Am J Kidney Dis 58: 214–227, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isakova T, Wahl P, Vargas G, Gutiérrez OM, Scialla J, Xie H, et al.: Fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease [published correction appears in Kidney Int 82: 498, 2012]. Kidney Int 79: 1370–1378, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munoz Mendoza J, Isakova T, Cai X, Bayes LY, Faul C, Scialla JJ, et al.; CRIC Study Investigators: Inflammation and elevated levels of fibroblast growth factor 23 are independent risk factors for death in chronic kidney disease. Kidney Int 91: 711–719, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal N, Hyre Anderson A, Yang W, Christenson RH, deFilippi CR, Deo R, et al.: High-sensitivity troponin T and N-terminal pro-B-type natriuretic peptide (NT-proBNP) and risk of incident heart failure in patients with CKD: The Chronic Renal Insufficiency Cohort (CRIC) Study. J Am Soc Nephrol 26: 946–956, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fotheringham J, Weatherley N, Kawar B, Fogarty DG, Ellam T: The body composition and excretory burden of lean, obese, and severely obese individuals has implications for the assessment of chronic kidney disease. Kidney Int 86: 1221–1228, 2014 [DOI] [PubMed] [Google Scholar]
- 24.Liu KD, Yang W, Go AS, Anderson AH, Feldman HI, Fischer MJ, et al.; CRIC Study Investigators: Urine neutrophil gelatinase-associated lipocalin and risk of cardiovascular disease and death in CKD: Results from the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis 65: 267–274, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson AH, Yang W, Hsu CY, Joffe MM, Leonard MB, Xie D, et al.; CRIC Study Investigators: Estimating GFR among participants in the Chronic Renal Insufficiency Cohort (CRIC) study. Am J Kidney Dis 60: 250–261, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Xie D, Pan Q, Feldman HI, Guo W: Joint modeling of repeated measures and competing failure events in a study of chronic kidney disease. Stat Biosci 9: 504–524, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heagerty PJ, Lumley T, Pepe MS: Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 56: 337–344, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Mihai S, Codrici E, Popescu ID, Enciu A-M, Albulescu L, Necula LG, et al.: Inflammation-related mechanisms in chronic kidney disease prediction, progression, and outcome. J Immunol Res 2018: 2180373, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carracedo J, Alique M, Vida C, Bodega G, Ceprián N, Morales E, et al.: Mechanisms of cardiovascular disorders in patients with chronic kidney disease: A process related to accelerated senescence. Front Cell Dev Biol 8: 185, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group: KDIGO 2012. clinical practice guideline for the evaluation and management of chronic kidney disease. Available at: https://kdigo.org/guidelines/ckd-evaluation-and-management/. Accessed December 5, 2020
- 31.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, et al.: Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 131: 269–279, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherzer R, Shah SJ, Secemsky E, Butler J, Grunfeld C, Shlipak MG, et al.: Association of biomarker clusters with cardiac phenotypes and mortality in patients with HIV infection. Circ Heart Fail 11: e004312, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahlqvist E, Storm P, Käräjämäki A, Martinell M, Dorkhan M, Carlsson A, et al.: Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6: 361–369, 2018 [DOI] [PubMed] [Google Scholar]
- 34.Sakr L, Small D, Kasymjanova G, Suissa S, Ernst P: Phenotypic heterogeneity of potentially curable non-small-cell lung cancer: Cohort study with cluster analysis. J Thorac Oncol 10: 754–761, 2015 [DOI] [PubMed] [Google Scholar]
- 35.Shukla N, Hagenbuchner M, Win KT, Yang J: Breast cancer data analysis for survivability studies and prediction. Comput Methods Programs Biomed 155: 199–208, 2018 [DOI] [PubMed] [Google Scholar]
- 36.Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC, et al.: Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 9: 1777, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iatropoulos P, Daina E, Curreri M, Piras R, Valoti E, Mele C, et al.; Registry of Membranoproliferative Glomerulonephritis/C3 Glomerulopathy; Nastasi: Cluster analysis identifies distinct pathogenetic patterns in C3 glomerulopathies/immune complex-mediated membranoproliferative GN. J Am Soc Nephrol 29: 283–294, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu SH, Li Y, Liu B: Exploratory cluster analysis to identify patterns of chronic kidney disease in the 500 cities project. Prev Chronic Dis 15: E60, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.