Abstract
N-Glycosylation is one of the most important post-translational protein modifications in eukaryotic cells. Although more than 200 N-glycogenes contributing to N-glycan biosynthesis have been identified and characterized, the information on insect N-glycosylation is still limited. Here, focusing on insect N-glycosylation, we characterized Bombyx mori N-acetylgalactosaminyltransferase (BmGalNAcT) participating in complex N-glycan biosynthesis in mammals. BmGalNAcT localized at the Golgi and was ubiquitously expressed in every organ and in the developmental stage of the middle silk gland of fifth instar larvae. Analysis of recombinant BmGalNAcT expressed in Sf9 cells showed that BmGalNAcT transferred GalNAc to non-reducing terminals of GlcNAcβ1,2-R with β1,4-linkage. In addition, BmGalNAcT mediated transfer of galactose and N-acetylglucosamine residues but not transfer of either glucose or glucuronic acid from the UDP-sugar donor substrate to the N-glycan. Despite this tri-functional sugar transfer activity, however, most of the endogenous glycoproteins of insect cells were present without GalNAc, Gal, or GlcNAc residues at the non-reducing terminal of β1,2-GlcNAc residue(s). Moreover, overexpression of BmGalNAcT in insect cells had no effect on N-acetylgalactosaminylation, galactosylation, or N-acetylglucosaminylation of the major N-glycan during biosynthesis. These results suggested that B. mori has a novel multifunctional glycosyltransferase, but the N-glycosylation is highly and strictly regulated by the endogenous N-glycosylation machineries.
Subject terms: Glycobiology, Post-translational modifications
Introduction
The final step of gene expression involves the functional protein biosynthesis. In higher eukaryotic cells, most proteins are hardly functional in polypeptide chain and further require post-translational modifications to express their diversity of physiological and biological activities. Among the post-translational modifications, N-glycosylation, the addition of sugar residues to asparagine residues in the context of N-X-S/T (where X is any amino acid except proline) on the peptide backbone, plays important roles in protein folding, assembly, and control of the metabolic rates of proteins by protecting proteins from proteolysis or antigenic recognition1–4. N-Glycans are synthesized by the actions of highly conserved glycosyltransferases (GT) and glycosylhydrolases in the endoplasmic reticulum (ER) and Golgi. The structure of N-glycan biosynthesized in the ER is universal, but GTs functioning in medial- to trans-Golgi have species specificities, resulting in the structural diversity of species-specific N-glycans.
The details of N-glycosylation in insects, especially within Golgi, have been controversial. The distinctive N-glycan structure referred to as insect type is 3 mannoses (Man3) with α1,3- and α1,6-fucose residues (M3FF). Thus, insects have active α1,3-fucosyltransferase (FUCT) and α1,6-FUCT, which are particularly distributed in plants and mammals, respectively5–8. However, except in honeybees, M3FF is not general and is hardly detected in total N-glycan9, even though Man2s and/or Man3s with α1,3-fucose (Fuc) or α1,6-Fuc are abundant, indicating that α1,3-FUCT and α1,6-FUCT compete for an identical acceptor substrate10,11. Another curious feature of insect GT is α2,6-sialyltransferase (α2,6-ST). Drosophila and silkworm possess the α2,6-ST gene, but the reaction product of endogenous sialylated N-glycans has not been detected except for Drosophila embryos, suggesting that insect α2,6-STs are almost non-functional enzymes in vivo12,13. Remarkably, even though insect α2,6-STs are inactive in vivo, insect α2,6-STs show an in vitro substrate preference different from that of mammals; insect α2,6-STs prefer β-linked N-acetylgalactosamine (GalNAc) residue(s) to galactose (Gal) residue at the non-reducing terminus, and efficiently transfer N-acetylneuraminic acid (NeuAc) to arylglycosides substrates rather than N-glycans13–15. These results suggested that insects might have specific GT(s), i.e., insect β1,4-N-acetylgalactosaminyltransferase(s) (GalNAcT(s)) that are more suitable for sialylation of N-glycans than β1,4- and β1,3-galactosyltransferase (GALT). Actually, endogenous β1,4-N-acetylgalactosaminylated N-glycans have been detected in insects, such as Drosophila, mosquito, honeybees, and lepidopteran larvae16–21 and some insect GalNAcTs were identified22–24, suggesting that insects have a potential for synthesizing LacdiNAc structures on N-glycans.
N-Acetylgalactosaminyltransferases are divided into several families. One of the major GalNAcTs contributing to N-glycan biosynthesis are categorized as a Glycosyltransferase Family 7 (GT7) proteins in the Carbohydrate-Active enZYmes database and are mainly distributed in invertebrates, unlike β1,4-GALTs, which are distributed in vertebrates. The other well-known GalNAcTs are UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases (αppGalNAcTs), which are categorized as GT27 proteins responsible for the biosynthesis of mucin-type O-glycans25. GalNAcTs and αppGalNAcTs have different substrate specificities and catalyze the GalNAc transfer to glycan to produce a LacdiNAc structure or a polypeptide acceptor from an activated sugar, UDP-GalNAc, respectively. The protein structure and sequences of catalytic domains lead us to propose that GalNAcTs and αppGalNAcTs were emerged from a common ancestral gene26. Interestingly, a single amino acid residue on GalNAcTs, Ile/Leu, is a key determinant affecting the donor sugar specificity; GalNAcT converts to β1,4-GALT27. In fact, GalNAcTs possess weak Gal transferase activity24,27, supporting the notion that GalNAcTs could play the same role as β1,4-GALTs. Thus, the evolutional substitution of Ile/Leu on GalNAcTs resulting in β1,4-GALT emergence, is indispensable for changing the glycosylation of glycoproteins and glycolipids in vertebrates. In other words, similar to β1,4-GALTs in vertebrates28–30, GalNAcTs might play critical role(s) in physiological and/or biological functions in invertebrates. Indeed, a mutation in Drosophila GalNAcT resulted in different behavioral phenotypes in adult flies, and the neural and muscle phenotypes in larvae23,31 and Caenorhabditis elegans GalNAcT mutant, ngat-1, showed temperature sensitivity and defects in cell migration32. These facts demonstrate that GalNAcTs play important functions in vivo and might explain their presence. However, information on the function of GalNAcTs and the LacdiNAc structure on N-glycans in vivo is still limited. Therefore, additional information on GalNAcTs and their in vivo function is needed. Information on the genome sequence of the silkworm, Bombyx mori, has been made available33. B. mori is an invertebrate, and thus it is likely to possess GalNAcT rather than β1,4-GALT23,24,26. The above facts should help us to identify a B. mori GalNAcT homologue and to elucidate the physiological and developmental roles of LacdiNAc structure in insects. Moreover, the recent finding that α2,6-ST is present in B. mori also supports the idea that B. mori possesses an active GalNAcT like that in Drosophila possessing both GalNAcT and α2,6-ST. Recently, B. mori GalNAcT was identified and shown to be enzymatically active34. However, its donor substrate preference, its subcellular localization and contribution to in vivo N-glycan biosynthesis were not extensively addressed.
Here, focusing on the insect GalNAcT, we characterized B. mori GalNAcT (BmGalNAcT). BmGalNAcT encoded a protein of 420 amino acids and was localized in the Golgi as N-glycoproteins. BmGalNAcT transferred GalNAc residues to the various acceptor substrates that contained terminal N-acetylglucosamine (GlcNAc) residue(s) with β1,4-linkage. BmGalNAcT transferred Gal residues as expected, but it also transferred GlcNAc residues. Although BmGalNAcT was successfully expressed in insect cells, the enhancement of LacdiNAc formation on major N-glycans in vivo was not observed despite in vitro activity; this suggests some additional requirement for GalNAc transfer during N-glycan processing.
Results
GALT family topology of the putative B. mori N-acetylgalactosaminyltransferase
In insects, β1,4-GALT orthologs GalNAcT from Drosophila, Trichoplusia ni, Spodoptera frugiperda, and Mamestra brassicae have been identified and characterized22–24. Based on the amino acid sequences of human β1,4-GALT, a database search using the KAIKOBase (http://sgp.dna.affrc.go.jp/KAIKObase/) led to the identification of a putative B. mori GalNAcT on the genome. The sequence was encoded on chromosome 3 and assigned as BGIBMGA007485 in KAIKOBase. The B. mori GalNAcT (BmGalNAcT) is a protein of 420 amino acids with a calculated molecular mass of 48.5 kDa. BmGalNAcT possessed 10 potential N-glycosylation sites. BmGalNAcT had a hydrophobic transmembrane sequence at the N-terminus, suggesting that BmGalNAcT localizes at the ER or Golgi as a type II transmembrane protein. The amino acid alignments with Drosophila melanogaster GalNAcTs (DmGalNAcTA and DmGalNAcTB) and Trichoplusia ni GalNAcT (TnGalNAcT) are shown (Supplementary Fig. S1). Though the N-termini of the sequences were dissimilar to each other, the C-termini of the sequences had higher similarities. This agreed with the idea that the catalytic domain of glycosyltransferases categorized into the same family should be conserved. In fact, BmGalNAcT also had the highly conserved sequences including the catalytic domain, binding sites of the donor and acceptor substrates and metal ions, which were observed at the C-terminal regions of GT 7-categolized GALT family proteins and responsible for showing the GALT activity27,35,36. Interestingly, BmGalNAcT had a putative long stem region, Leu37 to Gly164, which agreed well with the characteristics of β1,4-GALT37. In addition, most of the putative N-glycosylation sites, 8 of 10, were positioned on the stem region, indicating that the N-glycan on BmGalNAcT had little effect on its activity.
Ramakrishnan et al. showed that Ile289 of DmGalNAcT was an important factor for GalNAcT transfer activity26,27. The substitution of Ile289 to Tyr resulted in showing major β1,4-galactose transfer activity, and therefore the site is a key determinant and a hallmark of GALT family protein for sugar transfer. The amino acid of the corresponding site, Ile289 of DmGalNAcT, was Ile in BmGalNAcT (Ile310). In addition, the phylogenetic analysis using mammalian GALTs and invertebrate GalNAcTs suggested that BmGalNAcT transfers N-acetylgalactosamine rather than galactose to N-glycan (Supplementary Fig. S2).
Expression and localization of BmGalNAcT
In order to confirm the specificities of BmGalNAcT expression in B. mori, the expression levels were examined by quantitative RT-PCR using various kinds of cDNAs isolated from different organs and developmental stages of the middle silk gland (MSG) in 5th instar larvae (Supplementary Fig. S3 and the source data for the figure is provided in Supplementary Data 1). The expression level was different to a varying degree and showed organ-dependent expression, but BmGalNAcT was constitutively expressed among all organs and in the developmental stage of MSG. These results suggested that if the BmGalNAcT was active, the reaction product, the LacdiNAc structure, was detected in N-glycoprotein(s), O-glycoprotein(s) or glycolipid(s).
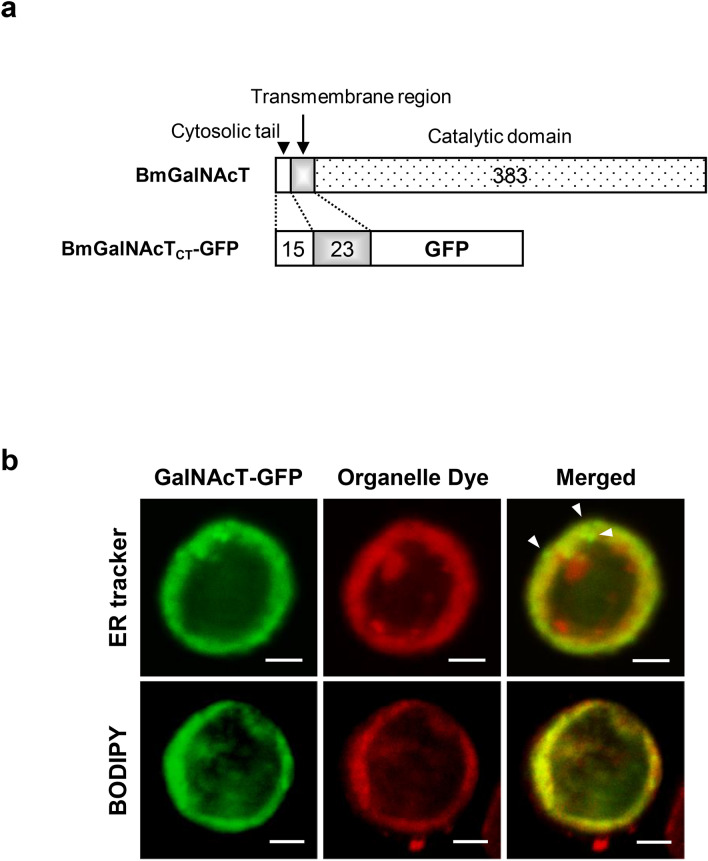
Next, the subcellular localization of BmGalNAcT was examined using a chimeric protein consisting of the putative cytosolic and transmembrane (CT) regions of BmGalNAcT and GFP (BmGalNAcTCT-GFP) (Fig. 1a). BmGalNAcTCT-GFP was transiently expressed in Sf9 cells and the cells were co-stained with the ER or Golgi marker. BmGalNAcTCT-GFP showed tight co-localization with the Golgi marker (Fig. 1b). On the other hand, some of the GFP signal overlapped a little with the ER marker, some of BmGalNAcTCT-GFP did not co-localize with the ER marker and most of the signal was observed around the ER. These results were similar to the localization pattern of B. mori α2,6-sialyltransferase13. Collectively, they showed that the putative CT regions were sufficient for Golgi localization and that BmGalNAcT was a Golgi-localized membrane protein.
Figure 1.
Subcellular localization of BmGalNAcT. (a) Schematic representation of the fusion protein of BmGalNAcTCT-GFP. The putative cytosolic and transmembrane regions were fused to the N-terminus of GFP. Black dotted, gray, and white boxes indicate the putative cytosolic region, transmembrane region, and catalytic region including the stem region of BmGalNAcT. The numbers indicate the length of each region. (b) Dual-color-imaging of BmGalNAcTCT-GFP-expressing Sf9 cells stained with organelle dyes. (Top) Imaging of BmGalNAcTCT-GFP-expressing cells stained with ER marker. BmGalNAcTCT-GFPs without overlapping ER marker are indicated by white triangles (bottom) Imaging of BmGalNAcTCT-GFP expressing cells stained with BODIPY TR Ceramide as Golgi marker. Bars: 10 μm.
β1,4-N-acetylgalactosaminyltransferase activity of BmGalNAcT
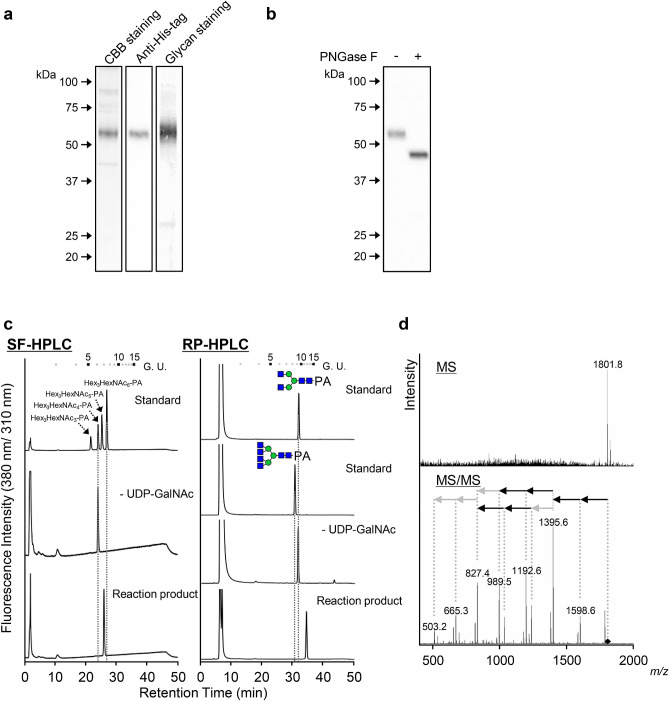
The open reading frame of BmGalNAcT was isolated using 5th instar cDNA. The truncated form of BmGalNAcT without the CT region was prepared from the full-length cDNA, followed by insertion into a baculovirus expression vector with the N-terminal gp67 sequence and C-terminal His-tag sequence to produce BmGalNAcT as a soluble secreted protein in Sf9 cells. The His-tagged BmGalNAcT was purified from the medium using Co2+ affinity chromatography. CBB staining, His-tagged protein detection, and N-glycoprotein staining of the purified protein revealed that BmGalNAcT was expressed as an N-glycoprotein (Fig. 2a and Supplementary Data 2). Actually, the BmGalNAcT that was de-N-glycosylated by PNGase F was approximately 45 kDa (Fig. 2b), which agreed well with the calculated molecular mass of BmGalNAcT without the CT region.
Figure 2.
Analysis of the reaction products of BmGalNAcT. (a) CBB staining, His-tag staining, N-glycoprotein staining, and (b) de-glycosylation analysis of purified BmGalNAcT. Black and white triangles indicate the purified N-glycosylated form and de-glycosylated BmGalNAcTs, respectively. (c) SF- and RP-HPLC analysis of the reaction products. BmGalNAcT reaction was carried out using UDP-GalNAc and GN2M3 as a donor and an acceptor substrate, respectively. The elution position of the product was compared with authentic PA-sugar chains. Numbers at the top represent the elution positions of glucose units on the basis of the elution times of PA-isomalto-oligosaccharides with degrees of polymerization from 3 to 15. Green circles and blue boxes indicate Man and GlcNAc, respectively. (d) MS and MS/MS analysis of the reaction product. The MS signal represents (M + H)+ ions of the reaction product. The value of m/z 503 detected in the MS/MS spectra agrees with the calculated mass of HexNAc2-PA. The mass of the precursor ion, m/z 1801.8, was considered to correspond to HexNAc4Hex3HexNAc2-PA. The black and gray arrows represent N-acetylhexosamine and hexose, respectively. A black diamond indicates the precursor ion of MS/MS fragmentation.
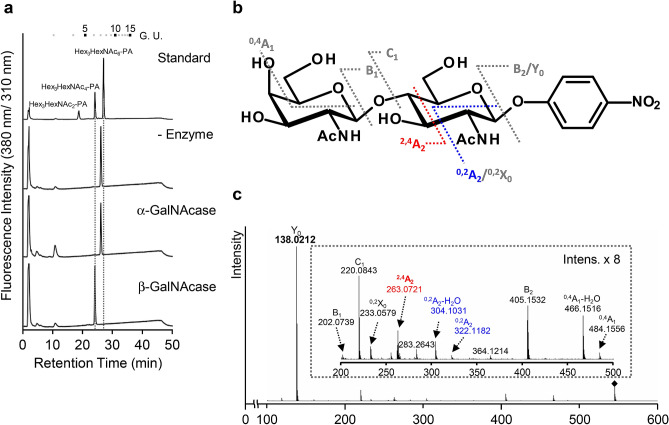
The catalytic activity of the purified BmGalNAcT was examined using UDP-GalNAc and the PA-labeled sugar chain GlcNAc2Man3GlcNAc2 (GN2M3-PA) as donor and acceptor substrate, respectively, and the reaction product was analyzed by size-fractionation (SF)-high performance liquid chromatography (HPLC) and reverse phase (RP)-HPLC. The reaction product of BmGalNAcT had a peak that was shifted from the acceptor substrate, which did not correspond to an authentic sugar chain, Hex3HexNAc6-PA, in either HPLC analysis (Fig. 2c). However, the molecular mass of the reaction product at m/z 1801.8 agreed with the calculated mass of Hex3HexNAc6-PA (Fig. 2d). In addition, the product ions caused by fragmentation in the MS/MS analysis revealed that the reaction product consisted of three Hex and six HexNAc residues (Fig. 2d), demonstrating that BmGalNAcT transferred two GalNAc residues from a donor substrate to the non-reducing terminal of glycan. α- or β-linkage-specific N-acetylgalactosaminidase digestion revealed that the GalNAc residues were transferred with β-linkage (Fig. 3a). To ascertain the linkage type by MS/MS analysis in negative mode, the acceptor substrate was changed to p-nitrophenyl-GlcNAc (GlcNAcβ-pNP). This analysis enabled us to identify the linkage type from the signal pattern of product ions13. The molecular mass of the reaction product was m/z 544.18, which corresponded to the calculated mass of HexNAc2-pNP (Fig. 3c). In addition, fragmentation signals from the precursor ion revealed specific signals, i.e., 1,4-linkage m/z 263.07 of 2,4A2 , m/z 322.11 of 0,2A2 and m/z 304.1 of 0,2A2-H2O38,39 and showed that the fragmentation pattern corresponded to authentic HexNAc1,4-HexNAc-pNP rather than HexNAc1,3-HexNAc-pNP (Fig. 3b,c). Thus, HexNAc was linked with 1,4-linkage at the non-reducing terminal of GlcNAcβ-pNP. These results provided the evidence that BmGalNAcT was the β1,4-N-acetylgalactosaminyltransferase contributing to N-glycan biosynthesis.
Figure 3.
Linkage analysis of the reaction product. (a) Linkage-specific N-acetylgalactosaminidase digestion of the reaction product. The reaction product purified by SF-HPLC were digested independently by two N-acetylgalactosaminidases specific for α- and β- linkage. The digested products were separated by SF-HPLC. Numbers at the top represent the elution positions of glucose units. (b) Structural representation and fragmentation scheme of GalNAcβ1,4-GlcNAc-pNP. Diagnostic ions are represented in gray characters. (c) Negative ion MS/MS fragmentation spectra of N-acetylgalactosaminylated GlcNAc-pNP. The corresponding m/z values of the CID-derived fragment ions and their nomenclatures are assigned in the spectra. The small window shows the enlarged fragment ions in the mass range m/z 200–500. m/z values shown in black and gray indicate 1,4-linked specific and GalNAcβ-GlcNAc-pNP signals, respectively.
Acceptor preference of BmGalNAcT
To determine the characteristics of BmGalNAcT, GNM3A (Manα1,6-(GlcNAcβ1,2-Manα1,3)Manβ1,4-GlcNAcβ1,4-GlcNAc-PA; Table 1, Supplementary Fig. S4) was used as a standard acceptor substrate because there was only one terminal GlcNAc residue that was a possible acceptor site, and this assignment facilitated an evaluation of BmGalNAcT activities. The enzymatic properties of BmGalNAcT were as follows: the specific activity was 15.4 nmol/h/mg toward GNM3A, and Km and Vmax were 7.8 ± 1.7 µM and 282.8 ± 49.2 nmol/h/mg for GNM3A and 0.77 ± 0.14 mM and 19.4 ± 1.2 nmol/h/mg for UDP-GalNAc (Supplementary Fig. S5). The activities and stabilities under various conditions were analyzed and are summarized in Table 2. Optimal temperature and pH were 20–35 °C and pH 5.5–8.0, respectively. BmGalNAcT showed decreased activities at temperatures higher than 50 °C and pH 7.0 and required Mn2+ or Co2+ as a cofactor (Supplementary Fig. S6). A similar Mn2+ requirement for activity was observed for GT family 7 proteins, especially β1,4-GALT, whereas a Co2+ requirement was a unique feature of BmGalNAcT.
Table 1.
Acceptor substrate specificities of the recombinant BmGalNAcT.
| Substrate, structure | Number of terminal GlcNAc | Number of terminal GlcNAc | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||||
| Specific activity (nmol/h/mg) | Relative activity (%) | Specific activity (nmol/h/mg) | Relative activity (%) | Specific activity (nmol/h/mg) | Relative activity (%) | ||||
| pNP-sugars | |||||||||
| Monosaccharide | Glcβ-pNP | 12.3 ± 2.12 | 0.33 | ||||||
| Galα-pNP | – | – | |||||||
| Galβ-pNP | – | – | |||||||
| GlcNAcβ-pNP | 3.70 ± 0.05 × 103 | 100 | |||||||
| GalNAcβ-pNP | – | – | |||||||
| Disaccharide | Type I | Galβ1,3-GlcNAcβ-pNP | – | – | |||||
| Type II (LacNAc) | Galβ1,4-GlcNAcβ-pNP | – | – | ||||||
| Type III | Galβ1,3-GalNAcβ-pNP | – | – | ||||||
| GalNAcβ1,3-GlcNAcβ-pNP | – | – | |||||||
| (LacdiNAc) | GalNAcβ1,4-GlcNAcβ-pNP | – | – | ||||||
| N-Glycans | |||||||||
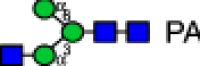
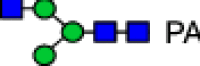
| Terminal GlcNAc type |
|
GNM3A | 1 | 15.4 ± 0.50 | 100 | ||||
|
GNM3B | 1 | 7.92 ± 0.33 | 51.4 | |||||
|
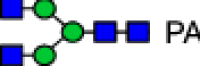
GN2M3 | 2 | 16.7 ± 0.17 | 108.2 | 2.1 ± 0.03 | 13.6 | |||

|
GN3M3 | 3 | 21.9 ± 0.20 | 142.2 | 10.1 ± 0.40 | 65.9 | |||
|
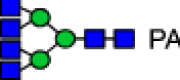
GN4M3 | 4 | 23.3 ± 0.47 | 151.4 | 8.3 ± 0.57 | 53.8 | 1.0 ± 0.10 | 6.8 | |
|
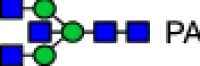
GN2M3 + bisect GlcNAc | 3 | – | – | |||||

|
GN3M3 + bisect GlcNAc | 4 | 8.27 ± 0.20 | 53.7 | |||||
| Core Fucose type |
|
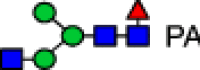
GNM3FA | 1 | 10.2 ± 0.07 | 66.2 | ||||
|
GNM3FB | 1 | 9.70 ± 0.27 | 63.0 | |||||
|
GN2M3F | 2 | 11.3 ± 0.07 | 73.5 | 1.4 ± 0.2 | 8.8 | |||
|
GN2M3F + bisect GlcNAc | 3 | – | – | |||||
| Terminal Gal type |
|
GalGN2M3A | 1 | 3.53 ± 0.20 | 22.9 | ||||
|
|
GalGN2M3B | 1 | 8.29 ± 2.13 | 53.8 | |||||
|
|
Gal2GN2M3 | 0 | – | – | |||||
|
|
GalGN2M3FA | 1 | – | – | |||||
|
|
GalGN2M3FB | 1 | 8.02 ± 1.47 | 52.1 | |||||
| Mannose type |
|
M3 | 0 | – | – | ||||
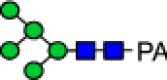
|
M5 | 0 | – | – | |||||
Relative rates were calculated on the basis of the activity toward GNM3A (100%).
Table 2.
Enzyme properties of recombinant BmGalNAcT expressed in Sf9 cells.
| Optimal condition | |
|---|---|
| Optimum pH | 5.5–8.0 |
| pH stability | 4.0–7.0 |
| Optimum temperature | 20–35 °C |
| Temperature stability | 0–50 °C |
| Metal-ion dependence : relative activity in the presence of | |
| No addition | 2.3 ± 1.4% |
| 10 mM MgCl2 | 3.9 ± 1.4% |
| 10 mM MnCl2 | 100% |
| 10 mM CaCl2 | 12.4 ± 0.7% |
| 10 mM CoCl2 | 91 ± 0.1% |
| 10 mM ZnCl2 | 1.2 ± 0.2% |
The relative activities in metal-ion dependency were calculated on the basis of the presence of Mn2+ (100%).
The acceptor substrate specificities were examined using various kinds of synthetic pNP substrates or PA-labeled sugar chain candidates (Table 1, Supplementary Fig. S4). BmGalNAcT transferred GalNAc to Glcβ-pNP and GlcNAcβ-pNP, but the transfer efficiency was much higher efficiency toward GlcNAcβ-pNP than toward Glcβ-pNP. In case of N-glycan substrate, BmGalNAcT preferred GlcNAc residue(s) at the non-reducing terminal of the core Manα1,3-Man-R residue, but all GlcNAc residues were utilized as target residues for GalNAc transfer(s). In comparison with GNM3A and GalGNM3B or GNM3B and GalGNM3A, the relative activities were approximately half due to the presence of the β1,4-linked Gal residue. It is noteworthy that the transfer activity was decreased by the α1,6-fucose (Fuc) residue and was strictly inhibited by the bisected GcNAc residue even though the α1,6-Fuc residue, in particular, was not proximal to the non-reducing terminal of GlcNAc residue(s). Three-dimensional N-glycan modeling revealed that α1,6-Fuc was apart from the β1,2-linked GlcNAc residue(s) and that the bisected GlcNAc residue was clearly localized in close proximity to the target GlcNAc residues and faced the same side of target β1,2-linked GlcNAc residues (Supplementary Fig. S7). These findings suggested that α1,6-Fuc, when close to the active site, interfered with the access of the acceptor N-glycan to BmGalNAcT, whereas the bisected GlcNAc residue prevented the access of donor substrates to BmGalNAcT. On the other hand, GN3M3 with bisected GlcNAc was utilized as an acceptor. Moreover, the specific activity toward GN3M3 was higher than that toward GN2M3. These results suggested that the GalNAc was preferentially transferred to the β1,4-linked GlcNAc residue at the non-reducing terminal of the core Manα1,3-Man residue. This substrate preference was also observed for GN4M3. This result was supported by the two results. First, GN2M3 with bisected GlcNAc was not available as an acceptor substrate, but GN3M3 with bisected GlcNAc did function as an acceptor substrate by the additional β1,4-linked GlcNAc on the core Manα1,3-Man residue of GN2M3. Second, only the β1,4-linked GlcNAc residue was located outside of the face of GN2M3 with bisected GlcNAc (Supplementary Fig. S7b,c). These results indicated that the β1,4-linked GlcNAc residue was permitted to access the donor substrate in an active site and was recognized as a target residue of GalNAc transfer. Therefore, GalNAc was transferred selectively to the β1,4-linked GlcNAc residue.
Gal and GlcNAc transfer activity of BmGalNAcT
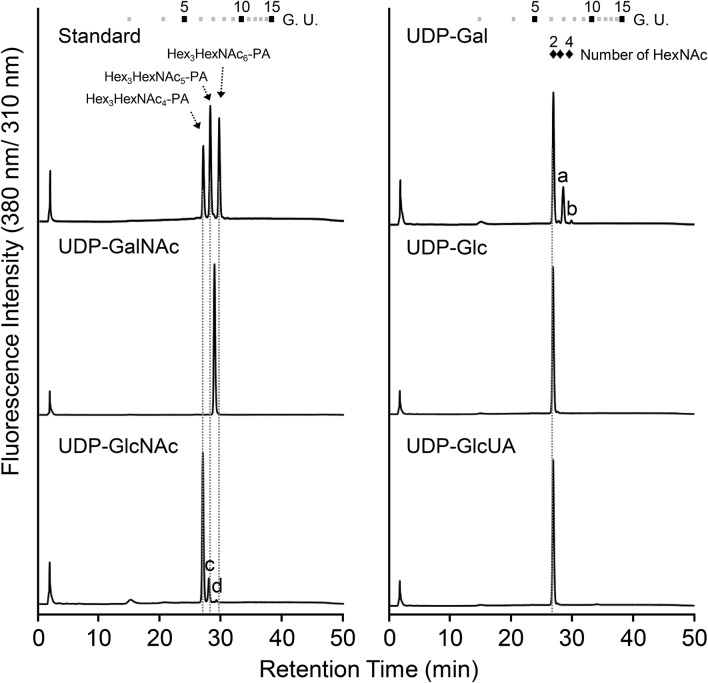
A previous report revealed that TnGalNAcT transferred Gal and GlcNAc to a synthetic acceptor substrate but had little or no activity toward N-glycan24. To ascertain whether this property of TnGalNAcT applied to BmGalNAcT, BmGalNAcT reaction was performed in the presence of GN2M3 or GlcNAcβ-pNP as acceptor substrates and a series of UDP-sugar nucleotides, i.e., UDP-Gal, UDP-GlcNAc, UDP-glucose (Glc), or UDP- glucuronic acid (GlcUA) as donor substrates. Surprisingly, BmGalNAcT mediated sugar transfer from UDP-Gal or UDP-GlcNAc and synthesized chain-length elongated N-glycan or GlcNAcβ-pNP, thereby resulting in antennal extension of N-glycans and formation of chitobiosyl-pNP (Fig. 4, Supplementary Fig. S8), however, the specific activities of BmGalNAcT using UDP-Gal and UDP-GlcNAc as a donor substrate were quite different from that of UDP-GalNAc. Specific activities using UDP-GalNAc, UDP-Gal, and UDP-GlcNAc as donor substrates were 3.70 ± 0.05 × 103 nmol/h/mg, 158 ± 3.2 nmol/h/mg, 65.8 ± 1.89 nmol/h/mg, respectively. Focusing on the reaction product using UDP-Gal as a donor substrate, MS and MS/MS analysis and SF-HPLC analysis of the reaction product using GN2M3 as an acceptor substrate exhibited that the molecular mass and the retention of the predominant product (peak a in Fig. 4) corresponded to Galβ1,4-GN2M3 (Supplementary Fig. S9a,b). Linkage-specific galactosidase digestion also revealed that Gal residues were transferred with β1,4-linkage (Supplementary Fig. S9c). In addition, the N-acetylhexosaminidase (HEXO)-digested peak a corresponded to authentic PA-sugar chains, GalGNM3A and GalGNM3B, at approximately the same ratio (Supplementary Fig. S9d), indicating that BmGalNAcT had the potential to transfer a Gal residue to both non-reducing termini of bi-antenna GlcNAc on sugar chain. Indeed, BmGalNAcT synthesized Gal2GN2M3 (peak b in Fig. 4, Supplementary Fig. S10).
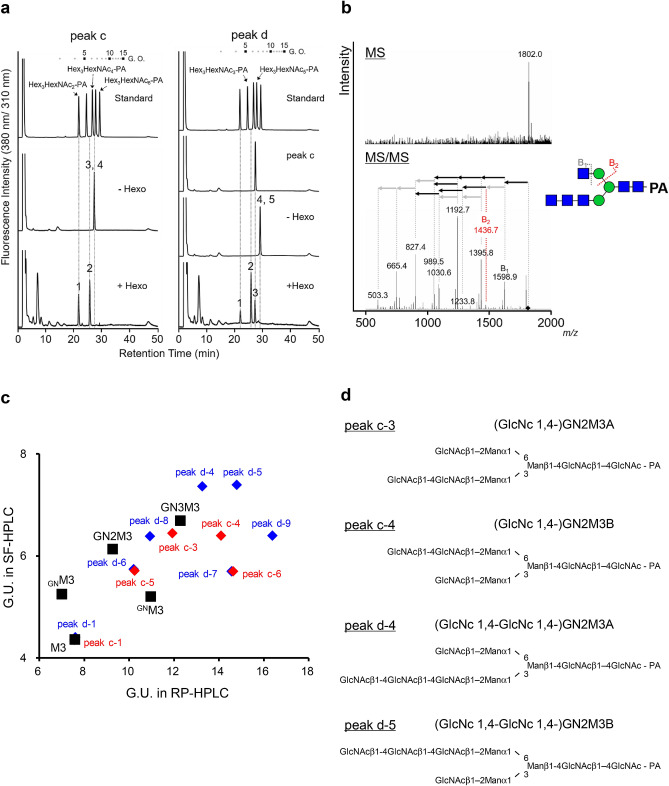
Figure 4.
RP-HPLC analysis of the products of BmGalNAcT reaction using various donor substrates. Reaction products were separated by SF-HPLC. Peaks a and b in UDP-Gal and peaks c and d in UDP-GlcNAc were collected and used for further analysis. Boxes and diamonds with numbers at the top represent the elution positions of glucose units and N-acetylglucosaminylated PA-sugar chains, respectively.
In the GlcNAc transfer, the reaction product showed that BmGalNAcT also mediated GlcNAc transfer to GN2M3 and GlcNAcβ-pNP (peak c and d in Fig. 4, Supplemental Fig. S8). However, the retention time of the products and the HEXO-digested products in SF-HPLC were different from those of authentic bi-, tri-, and tetra-antennary GlcNAc carrying PA-sugar chains (Fig. 5a), suggesting that BmGalNAcT transferred GlcNAc residue(s) not to Man residue(s), but possibly to terminal GlcNAc residue(s). MS, MS/MS analysis, and RP-HPLC analysis of the predominant product peak c demonstrated that the reaction product carried GlcNAc residue(s) at two different positions on GN2M3 (Supplementary Fig. S11a,b). The reaction products did not correspond to either bisected GN2M3 or GN3M3 as shown in Supplementary Fig. S11b. This suggested that BmGalNAcT transferred two GlcNAc residues to the non-reducing terminal of GlcNAcβ1,2-Man-R on N-glycan. An unknown linkage of the GlcNAc residue was HEXO-sensitive, but the structure of the reaction product did not correspond to GN2M3 in SF-HPLC (Fig. 5a). The HEXO-digested reaction product also showed two peaks in RP-HPLC, which had different hydrophobicities from GN2M3 (Supplementary Fig. S11c), indicating that the GlcNAc residue linked at two different positions and that HEXO preferentially digested the β1,2-GlcNAc residue on GN2M3. These results led to speculation that the putative HEXO-digested structures were (GlcNAcβ1,4-)GNM3A and (GlcNAcβ1,4-)GNM3B (Fig. 5d). Thus, the GlcNAc-transferred structures of peak c were (GlcNAcβ1,4-)GN2M3A and (GlcNAcβ1,4-)GN2M3B. The ratio of (GlcNAcβ1,4-)GN2M3A and (GlcNAcβ1,4-)GN2M3B synthesized by BmGalNAcT was approximately the same: a molar ratio of 54.8%:45.2%. Importantly, BmGalNAcT transferred two GlcNAc residues to different positions on GN2M3 (peak d in Fig. 4, Supplementary Fig. S11d). This was quite different from the results for the reaction of BmGalNAcT using UDP-GalNAc as a donor substrate, which yielded GalNAc2GN2M3 as the only reaction product (Fig. 2c). To determine these structures, additional RP-HPLC analysis and MS/MS analysis were performed (Fig. 5b and Supplementary Fig. S11d,e). The peak d had two peaks in RP-HPLC analysis, indicating that two GlcNAc residues were transferred to a GN2M3 position different form that for tetra-antenna GN4M3. Furthermore, the second product ion from the precursor, B2 m/z 1436.7, on MS/MS analysis demonstrated that core mannose residues were masked with only one GlcNAc residue and that at least one terminal GlcNAc residue on GN2M3 was modified with serially concatenated GlcNAc residues. Interestingly, 2D-mapping of PA-sugar chains based on retention times in SF-HPLC and RP-HPLC demonstrated that the HEXO product of peak d did not correspond to any PA-sugar chains of the BmGalNAcT reaction product or the HEXO products (Fig. 5c). Thus, two GlcNAc residues were not transferred to either β1,2-linked GlcNAc residue but rather were utilized to synthesize (GlcNAcβ1,4-GlcNAcβ1,4-)GN2M3A or (GlcNAcβ1,4-GlcNAcβ1,4-)GN2M3B (Fig. 5d). Unfortunately, the amount was too small, the further structural analysis was unable to be carried out, however, it is predicted that the structures contained a novel linkage type of GlcNAc. These results demonstrated that although the specific activities of Gal and GlcNAc transfer were not determined due to the quite low activities, BmGalNAcT clearly functioned as β1,4-GALT and β1,4-N-acetylglucosaminyltransferase.
Figure 5.
Structural determination of N-acetylglucosaminylated sugar chains. (a) SF-HPLC analysis of N-acetylhexosaminidase-digested peaks c and d. (b) MS and MS/MS analysis of peak d-4. The putative structure of peak d-4, fragmentation scheme, and diagnostic ions are represented. (c) Two-dimensional mapping of peak c, hexosaminidase-digested peak c, peak d, hexosaminidase-digested peak d, and authentic PA-sugar chains. Elution positions were calculated from the glucose unit in RP-HPLC and SF-HPLC. The red diamond, blue diamond, and black box indicate peak c, peak d in Figure, their N-acetylhexosaminidase (HEXO)-digested products, and the authentic PA-sugar chain, respectively. (d) Deduced structures of N-acetylglucosaminylated GN2M3.
N-Glycan analysis of BmGalNAcT-expressing insect cells
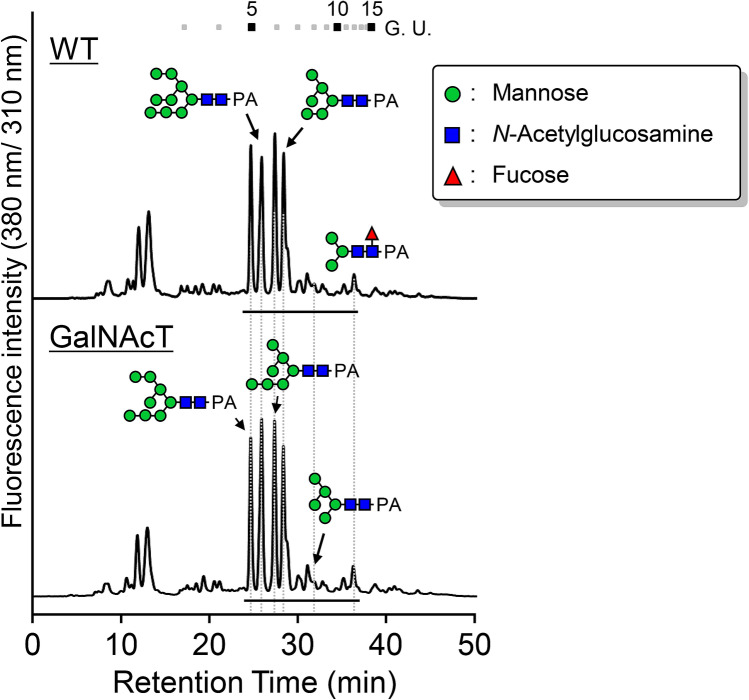
Previous studies had identified β1,4-N-acetylgalactosaminylated N-glycans in insect cells15–21. Interestingly, expression of T. ni GalNAcT in Sf9 cells resulted in the production of terminal β1,4-N-acetylgalactosaminylated N-glycans in vivo24. We therefore reasoned that, if BmGalNAcT has a functional in vivo activity in Sf9 cells, the structure of N-glycan would be terminally modified with GalNAc residue(s). To investigate this hypothesis, full length BmGalNAcT was introduced into Sf9 cells. BmGalNAcT was successfully expressed in Sf9 cells as an N-glycoprotein and the cells possessed the same level of GalNAcT activity as seen in secreted forms of BmGalNAcT (Supplementary Fig. S12 and Supplementary Data 3). To determine the major N-glycan structures in greater detail, the neutral N-glycan structures attached at total soluble N-glycoproteins were determined by hydrazinolysis of N-glycoprotein, followed by PA-labeling, RP-HPLC, and LC–MS/MS analysis (Fig. 6, and Supplementary Table S1). There was no significant N-glycan structural difference between mock cells and BmGalNAcT-expressing cells: the predominant structures were of high mannose type, whereas complex-type structures were hardly detected. Moreover, BmGalNAcT-expressing cells did not synthesize detectable levels of endogenous terminal β1,4-N-acetylgalactosaminylated N-glycoprotein. These results indicated that BmGalNAcT has β1,4-N-acetylgalactosamine activity in vitro but the overexpression of BmGalNAcT itself did not result in a N-glycosylation change in insect cells.
Figure 6.
N-Glycan analysis of WT and BmGalNAcT-expressing Sf9 cells. Total soluble sugar chains prepared from glycoproteins and labeled with PA were analyzed by RP-HPLC. The major structures are shown in chromatographs. Underlining indicates the PA-sugar chain fraction applied to LC–MS/MS analysis.
Discussion
In general, complex-type N-glycan exists in a state in which sialic acid is transferred to galactose. Sialylated N-glycan in mammals is synthesized by sequential reactions mediated by galactosyltransferase and sialyltransferase. Neither gene presents alone, such that the presence of either gene indicates the possibility of the presence of the other gene. Previously, the presence and the activity of B. mori ST were identified, suggesting that B. mori has the transferase classified as the GT7 family protein and transfers the Gal and/or GalNAc residue to N-glycan. In addition, Dojima et al. showed that B. mori larvae had β1,4-galactosylated N-glycan, even though B. mori β1,4-GALT has not been identified40. Thus, the enzyme contributing to β1,4-galactosylation in B. mori larvae was speculated to be GalNAcT, instead of β1,4-GALT, from the evolution of the enzyme and the presence of another insect GalNAcT. Besides, to date, the LacdiNAc structure has also been found not only on human glycoproteins41, but in insects i.e., in Drosophila, mosquito, honeybees, and lepidopteran15–21 and in glycolipids42. Recently, B. mori GalNAcT was identified34, and therefore, BmGalNAcT might synthesize uncharacterized and unique N-glycan structure(s). These pieces of evidence encouraged us to further characterize B. mori GalNAcT as also the enzyme was already identified.
Here, focusing on the potential for synthesizing potential complex N-glycan(s) in silkworms, we identified and characterized B. mori Golgi-localized N-acetylgalactosaminyltransferase. The donor substrate determinant of GT7 in BmGalNAcT was Ile310, which resulted in the transfer of GalNAc preferentially to the sugar residue on the sugar chain by β1,4-linkage rather than Gal. To reveal the presence of all BmGalNAcT homologues, we reinvestigated B. mori genome. Putative β1,4-N-acetylgalactosaminyltransferase (KWMTBOMO10034) encoding bre-4 has higher homology to BmGalNAcT than any other candidates. B. mori bre-4 has some of the highly conserved sequences in GT family 7 proteins, such as the site binding both the donor and acceptor substrates, and the ion binding site. However, the other sequences except for the conserved sequences did not corresponded to other insect GalNAcTs. In addition, XP_004926306, assigned as β1,4-galactosyltransferase 1-like on the database, is another candidate for B. mori GalNAcT, but it also lacked most of the conserved sequences of GalNAcT. These results indicated that B. mori possesses a single GalNAcT gene with a potential role in N-glycosylation.
BmGalNAcT activity was suppressed by the presence of α1,6-Fuc and/or β1,4-Gal residue(s) and strictly inhibited by a bisected GlcNAc residue. Even though weaker efficiency than GalNAc transfer, BmGalNAcT could also produce the β1,4-galactosylated sugar chain, which is distinct from the properties of TnGalNAcT, which shows no ability to transfer Gal to N-glycan24. In addition to GalNAc and Gal, BmGalNAcT transferred GlcNAc to the sugar chain and pNP-sugar, but not Glc and glucuronic acid GlcUA. This broad substrate specificity was not observed in the previous report34. Since this GlcNAc transfer to N-glycan has not been reported, it could be a unique feature of BmGalNAcT. These results suggest that the donor substrate is required; BmGalNAcT recognizes both the axial OH group at the C4-position and/or acetoamide group at the C2-position on the sugar residue of the donor substrate for transferring the sugar residue to the acceptor substrate.
TnGalNAcT had in vivo activity and produced N-acetylgalactosaminylated and β1,4-galactosylated N-glycoprotein, suggesting that insect cells had the potential to biosynthesize N-acetylgalactosaminylated and β1,4-galactosylated N-glycan even though TnGalNAcT had no in vitro activity toward N-glycan24. In fact, at least one donor substrate, UDP-Gal, in insect cells is abundant43. Therefore, overexpression of GT7 proteins in insect cells results in successful biosynthesis of β1,4-galactosylated N-glycan without the addition of UDP-Gal44–46. However, our detailed total N-glycan analysis was unable to detect either N-acetylgalactosaminylated or β1,4-galactosylated N-glycan in major neutral glycan fraction. This may have been because the weak β1,4-GALT activity of BmGalNAcT was insufficient to produce β1,4-galactosylated N-glycan in comparison with GalNAcT activity. In addition, both mock and BmGalNAcT-expressing Sf9 cells had high-mannose type structures as a predominant N-glycan. It is possible that high-mannose N-glycans were immature in N-glycosylation. Indeed, though mock cells had high-mannose type N-glycan only, stable expression of a mammalian glycosyltransferase, N-acetylglucosaminyltransferase II (GNTII), in Sf9 cells resulted in the production of complex-type N-glycan47. Therefore, even for purposes of elucidating the effects of N-acetylgalactosaminylation of N-glycan on physiological and biological functions in insects, introduction of both BmGalNAcT and GNTII would be necessary for the efficient production of N-acetylgalactosaminylated N-glycan. Interestingly, previous studies represented that insect has N-acetylgalactosaminylated N-glycans16–21. In this study, we investigated the major N-glycan structures and could not detect N-acetylgalactosaminylated N-glycans. However, BmGalNAcT-expressing Sf9 cells might have anionic N-glycans with glucuronic acid, sulphation, phosphorylcholine, and/or phosphoethanolamine as observed in other insects18–21 and N-acetylgalactosaminylated O-glycans and glycolipid because of the possibility that some GT7 proteins contribute to O-glycans and glycosphingolipid biosynthesis48.
It is worth noting that BmGalNAcT transferred GlcNAc residues in a different manner from GalNAc residues; BmGalNAcT synthesized structures of (GlcNAcβ1,4-GlcNAcβ1,4-)GN2M3 (Fig. 5d) instead of bi-antenna (GlcNAcβ1,4-GlcNAc)2M3. This suggests that BmGalNAcT prefers β1,4-GlcNAc residue to the β1,2-GlcNAc residue at the non-reducing terminus as an acceptor in the case of GlcNAc transfer. However, we observed that the reaction product transferred only two GlcNAc residues (Fig. 4). This was considered to be due to the weak GlcNAc transfer activity. On the other hand, GalNAcβ1,4-GlcNAc was detectable, whereas GalNAcβ1,4-GalNAc was not detected. These results provided evidence that BmGalNAcT recognizes the GlcNAc residue as an acceptor sugar residue. This idea was further supported by the result that BmGalNAcT transfers a GalNAc residue to the β1,4-linked GlcNAc residue on GN3M3 (Table 1). However, such recognition would not apply to bisected GlcNAc due to N-glycan conformation (Supplementary Fig. S7). Here, one question arises. Can BmGalNAcT could the repeating unit(s) of GalNAcβ1,4-GlcNAc, (GalNAcβ1,4-GlcNAc)n, on GN2M3? It is assumed that various conditions must be met to biosynthesize the repeating unit, but introduction of Caenorhabditis elegans GalNAcT into mammalian CHO cells resulted in the successful production of α1,3-fucosylated (GalNAcβ1,4-GlcNAc)nM349. Thus, BmGalNAcT also possesses the potential to synthesize the repeating unit(s) of (GalNAcβ1,4-GlcNAc)n on N-glycan. To achieve this and analyze the in vivo function of not only N-glycan N-acetylgalactosaminylation but also poly GalNAcβ1,4-GlcNAc on N-glycan, the creation of enzymes with higher activity of GlcNAc transfer, i.e., protein engineering, and establishment of a supplemental pathway for a donor substrate of UDP-GalNAc are required. There are no reports focusing on the production of UDP-GalNAc in B. mori, but as a first effort in this direction, the presence of UDP-GalNAc and the genes involved in biosynthesis of UDP-GalNAc should first be clarified.
Complete loss of UDP-galactose 4′ epimerase of Drosophila is embryotic lethal in fruit flies50. On the other hand, GalNAcT deficiency in insects leads to the display of differentiation and developmental phenotypes, but not lethality31,51, suggesting that UDP-GalNAc is essential for biosynthesis of glycolipids and O-glycans rather than N-glycans. What is the in vivo function of BmGalNAcT? To investigate this question, there is need of an effective knockout strategy. Genome editing technologies, such as the ZFN, TALEN and CRISPR/Cas9 platforms, have been established for silkworm, and have led to a remarkable acceleration of B. mori research52–55. However, these technologies have not been applied to knockout of the glycosyltransferases responsible for N-glycan biosynthesis and maturation, including BmGalNAcT and BmST. Furthermore, overexpression of these proteins in insects has not been performed. Such overexpressions or knockouts of glycogenes of unknown function might lead to the uncovering of a positive and/or negative effect on B. mori growth, morphological features and physiological changes. Our recent study demonstrated that only MSG shows drastic change though the stage of fifth larvae, but accumulates a high level of N-acetylglucosaminylated N-glycan at the stage of fifth larvae, which accounted for more than 50% of total N-glycan in some cases, whereas other organs, such as the gut, posterior silk gland, fat body, and body fluid, accumulate high-mannose type structures as predominant structures and have a small amount of N-acetylglucosaminylated N-glycan (data not shown). Thus, in the same manner as the nervous system, MSG in B. mori fifth larvae seems to be the organ rendered most susceptible by overexpression and knockout of BmGalNAcT and BmST. The UAS/GAL4 system, which was widely used for organ-specific gene expression, has also been considered to be useful for both genetic approaches to specifically control gene expression in MSG. There is still room for elucidation of N-glycosylation in B. mori, and the N-glycosylation function(s) related to insect differentiation and growth should be further elucidated.
Methods
Materials
UDP-GalNAc, UDP-Gal, UDP-GlcNAc, UDP-Gal, and UDP-GlcUA were from YAMASA (Chiba, Japan). p-Nitrophenyl (pNP)-GlcNAc was purchased from Tokyo Chemical Industry (Tokyo, Japan). 2-Pyridylaminated (PA)-sugar chains were purchased from TaKaRa Bio (Shiga, Japan) and Masuda Chemical Industry (Kagawa, Japan). Streptococcus pneumoniae β-N-Acetylgalactosaminidase was purchased from Sigma (St. Louis, MO). ER-Tracker Red and BODIPY TR Ceramide Complex to BSA was purchased from Molecular Probes (Eugene, OR).
Insect cells
Spodoptera frugiperda Sf9 cells were maintained at 25 °C in Sf-900 III SFM (Gibco, Eggenstein, Germany) containing 10% fetal calf serum (PAA Laboratories GmbH, Pashing, Austria).
Identification, isolation and cloning of BmGalNAcT
BmGalNAcT was identified using KAIKOBase (http://sgp.dna.affrc.go.jp/KAIKO/jp/index.ht ml) and human β1,4-Galactosyltransferase I (NP_001488), Drosophila (AAD34746 and AAF56843), and Trichoplusia ni (AAT11926) GalNAcTs as queries.
Bombyx mori fifth instar larvae total RNA was isolated using an RNeasy Plant Mini Kit (QIAGEN, Chatsworth, CA), followed by reverse transcription using a PrimeScript RT reagent Kit (TaKaRa). The full-length BmGalNAcT cDNA was amplified using KOD plus polymerase (ToYoBo, Osaka, Japan), cDNA, and the following primer set: BmGalNAcT-Fw, 5′-GCTTGGTCATCGCATGCG-3′; BmGalNAcT-Re, 5′-ATACCTCGCCAAGCTGCTGT-3′. The PCR product was subcloned into pGEM T-Easy vector (Promega, Madison, WI), followed by sequencing using an ABI PRISM Big Dye Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA).
RNA extraction and quantification of BmGalNAcT expression
Total RNA was isolated from the middle silkgland, posterior silkgland, fat body, body liquid, middle gut, Malpighian tubule, and testis in 5th instar larvae, and cDNAs were synthesized as described above. The expressions of BmGalNAcT were estimated by amplification using the following primer sets: forward, 5′-AGAGATCGCCAACAGCACTTG-3′; reverse, 5′-TCCAGTCTCTGGCTCTCGAT-3′. As a control, Bmrp49 was amplified using following primer set: forward, 5′-CAGGCGGTTCAAGGGTCAATAC-3′; reverse, 5′-TGCTGGGCTCTTTCCACGA-3′.
Subcellular localization analysis
A chimeric construct of BmGalNAcTCT-GFP was generated by PCR using full-length GmGalNAcT and GFP as templates. The fusion construct was ligated into pFastBac vector (Invitrogen, Carlsbad, CA). Baculovirus for the production of BmGalNAcTCT-GFP was prepared using the Bac-to-Bac expression system and FuGENE Transfection Reagent (Promega). Sf9 cells were transfected using P2 baculovirus and grown at 25 °C for 3 days. The BmGalNAcTCT-GFP cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, followed by washing with PBS three times, and staining using organelle dye ER-Tracker Red or BODIPY TR Ceramide in PBS for 15 min. The cells were washed with PBS three times and then the fluorescence signals were observed under a Leica DMI4000B equipped with TCS-SPE (Leica Microsystems, Heidelberg, Germany), and LAS AF software (Leica Microsystems). Fluorescence was excited with the 488-nm and the 532-nm lines of solid lasers. Image processing for GFP and organelle dye coloration was performed using Adobe Photoshop CS4.
Heterologous expression and purification of BmGalNAcT
Full-length BmGalNAcT with or without a putative cytoplasmic and transmembrane region was amplified using KOD plus polymerase, full-length cDNA as a template and the following primer set (forward: 5′-ATGGATCCATGGGAGCGGCGCG-3′ for full-length BmGalNAcT expression and ATTGAATTCGACGCCTCGCCGCTC for expression of the truncated form of BmGalNAcT; reverse: 5′-ATAAGCTTCTAGTGATGGTGATGGTGATGGCTGCGCTCATCGATG-3′). The truncated form of BmGalNAcT with a GP67 signal at the 5′-upstream region was introduced into the pFastBac vector (Invitrogen) to produce BmGalNAcT as a soluble and secreted protein.
Baculovirus for the production of BmGalNAcT was prepared as described above. After amplification of the P2 baculovirus, BmGalNAcT was expressed in Sf9 cells as previously reported13. In brief, a total of 1.0 × 108 Sf9 cells in 100 ml medium were transfected with P2 baculovirus and cultivated at 25 °C, 120 rpm, for 4 days. The medium was collected and centrifuged at 4 °C, 1000g for 5 min. To precipitate proteins, ammonium sulfate precipitation was performed. The precipitant was dissolved in 50 mM Tris–HCl buffer, pH 7.5, 500 mM NaCl, and 5 mM imidazole (buffer A), followed by dialysis against buffer A overnight. The solution was subject to a TALON Resin column (TaKaRa) equilibrated with buffer A. After washing with a 50-times-column volume of buffer A, the recombinant enzyme was eluted with buffer A containing 200 mM imidazole. The eluate was dialyzed against 40 mM Tris–HCl pH 7.5, 300 mM NaCl (buffer B) for 3 h, followed by measurement of the protein concentration and an addition of glycerol and buffer B to prepare 0.15 mg/ml of BmGalNAcT at a final concentration of 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 50% glycerol. The enzyme solution was stored at – 20 °C.
N-Glycosylation analysis of BmGalNAcT
The purified BmGalNAcT was separated by 5–20% SDS-PAGE and detected by CBB staining, glycan detection using G.P.Sensor (J-OIL MILLS, Tokyo, Japan), or western blotting. In western blotting analysis, anti-HRP antibody and anti-rabbit antibody conjugated to horseradish peroxidase (GE Healthcare, Tokyo, Japan) were used as a primary antibody and a secondary antibody, respectively, and specific signals were visualized using Luminate Forte western HRP Substrate (MILLIPORE, Billerica, MA) and a ChemiDoc MP system (Bio-Rad, Hercules, CA).
The purified BmGalNAcT were digested with Peptide:N-glycosidase F (PNGase F, TaKaRa) or following the manufacturer's protocols, followed by separation by 5–20% SDS-PAGE and visualization by western blotting.
BmGalNAcT assay
Basic BmGalNAcT assays were performed in 20 µl of total reaction volume containing 10 mM cocodylic acid buffer, pH 7.5, 10 mM MnCl2, 5 mM UDP-GalNAc, 10 pmol 2-aminoprydine (PA)-labeled GNM3A, GN2M3 or 5 mM GlcNAcβ-pNP and 0.30 µg of purified BmGalNAcT at 25 °C for 2 h. The reaction was terminated by incubation at 100 °C for 5 min. The samples were centrifuged at 4 °C, 20,000g for 5 min. The supernatant was subject to HPLC analysis.
HPLC analysis
The reaction products of the PA-sugar chains and GlcNAcβ-pNP were detected by an SF-HPLC and/or RP-HPLC using a HITACHI LaChrom HPLC System equipped with a fluorescence or a UV detector, respectively. The PA-sugar chains of reaction products were separated using the mobile phase of acetonitrile/acetic acid (solvent A: 98/2, v/v) and water/acetic acid/triethylamine (solvent B: 92/5/3, v/v/v). The PA-sugar chain was separated using a Shodex Asahipak NH2P-50 2D column (2.0 mm ID × 150 mm; SHOWA DENKO Co., Ltd.) by linearly increasing the solvent B concentration from 20 to 55% over 35 min at a flow rate of 0.2 ml/min. The eluted fractions were monitored by measuring the fluorescence intensity using excitation and emission wavelengths of 310 and 380 nm, respectively. The reaction products of pNP derivatives were separated using 0.02% trifluoroacetic acid (TFA) (solvent C) and methanol/0.02% TFA (solvent D: 10/90, v/v) using a Mightysil RP-18 GP column (4.6 mm × 250 mm, Kanto Chemical Co., Tokyo, Japan) with an HITACHI LaChrom HPLC System by linearly increasing the solvent D concentration from 0 to 15% over 7 min. The eluted fractions were monitored by measuring the UV intensity at a wavelength of 300 nm. The reaction product of GlcNAcβ-pNP for MS/MS analysis was purified using the mobile phase of 50 mM CH3COONH4/acetonitrile (87/13, v/v) by Cosmosil 5C18-AR-II column (6.0 × 250 mm; Nacalai Tesque, Kyoto, Japan) at an isocratic flow rate of 1.2 ml/min.
The structural determination was performed using RP-HPLC. The mobile phase was composed of 0.02% TFA (solvent E) and acetonitrile/0.02% TFA (solvent F) (20/80, v/v). RP-HPLC was performed using a Cosmosil 5C18-AR-II column (4.6 × 250 mm; Nacalai Tesque, Kyoto, Japan) with an HITACHI LaChrom HPLC System by linearly increasing the solvent F concentration from 0 to 20% over 35 min at a flow rate of 0.7 ml/min. The eluted fractions were monitored by measuring the fluorescence intensity using excitation and emission wavelengths of 310 and 380 nm, respectively.
Linkage analysis by mass spectrometry and exoglycosidase digestion
In the RP-HPLC analysis, the reaction product, GalNAc-GlcNAcβ-pNP, was collected and lyophilized. The resultant product was dissolved in 50% acetonitrile prior to the mass spectrometry (MS) analysis. The molecular mass of the reaction product was determined by direct infusion into a micrOTOF-QII (Bruker Daltonics, Bremen, Germany)13. The MS data were analyzed using Data Analysis 4.0 software (Bruker Daltonics).
The lyophilized reaction product samples of PA-sugar chain and pNP were dissolved in dH2O and digested with either β-N-Acetylglucosaminidase from Streptococcus pneumoniae (Sigma), α-N-Acetylgalactosaminidase (NEB, Beverly, MA), or β-N-Acetylgalactosaminidase56,57 following the manufacturer’s protocols. The digested samples were analyzed by SF- or RP-HPLC.
Determination of kinetic parameters
Kinetic parameters of BmGalNAcT were determined under standard reaction condition with varying the concentrations of UDP-GalNAc and GNM3A. The velocities under the various concentrations of substrates were measured by the time-dependent changes of the reaction products, then reaction products were analyzed as described above. The velocities versus the corresponding UDP-GalNAc or GNM3A concentrations were plotted and determined using a nonlinear regression analysis program of SigmaPlot software (Systat Software Inc., San Jose, CA).
Preparation and structural analysis of neutral N-glycan
The Sf9 cells expressing BmGalNAcT were defatted with acetone and completely dried out. The sugar chains were released from crude glycoproteins by hydrazinolysis at 100 °C for 10 h. After N-acetylation of the hydrazinolysate with saturated sodium bicarbonate and acetic anhydride, the N-acetylated hydrazinolysate was desalted with Dowex 50 × 2 (Muromachi Kagaku Kogyo Kaisha), and fractionated on a TSK gel Toyopearl HW-40 (Tosoh) column (2.5 × 30 cm) in 0.1 N ammonia. The released sugar chains were 2-aminopyridine (PA)-labeled, as described previously, followed by fractionation on a TSK gel Toyopearl HW-40 column (2.5 × 30 cm) in 0.1 N ammonia. The sugar chains were detected by RP-HPLC as described above.
The molecular masses of the PA-sugar chains and the number of their sugar moieties were estimated by LC–MS/MS using an Agilent Technologies 1200 series instrument (Agilent Technologies, Santa Clara, CA) equipped with HCT plus software (Bruker Daltonics) as previously reported13.
N-Glycan modeling
N-Glycan structures of GN2M3F, GN3M3, and GN3M3 with bisect GlcNAc were from a 3D structure libraries on a GLYCAM server (http://glycam.org/). Figures were prepared using PyMOL Molecular Graphics System, Version 1.7.1.1. (http://www.pymol.org/).
Supplementary Information
Acknowledgements
We thank National Institute of Agrobiological Sciences for providing us B. mori larvae and technical advices. This study was supported by a grant from the Agri-Genome Project of the Ministry of Agriculture, Forestry and Fisheries of Japan and by a Scientific Technique Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry.
Author contributions
H.K. and K.F. designed the research with assistance from T.O. and R.Misaki. H.K. and R.Miyauchi. performed N-glycan analyses. H.K., R.Miyauchi. and A.K. prepared a recombinant and purified protein. H.K. performed all the other experiments. H.K and K.F. performed all the statistics analysis. H.K., K.F. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84771-z.
References
- 1.Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Intracellular function of N-Linked Glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Hebert DN, Lamriben L, Powers ET, Kelly JW. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat. Chem. Biol. 2014;10:902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson IB. Glycosylation of proteins in plants and invertebrates. Curr. Opin. Struct. Biol. 2002;12:569–577. doi: 10.1016/S0959-440X(02)00367-6. [DOI] [PubMed] [Google Scholar]
- 6.Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Curr. Drug Targets. 2007;8:1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strasser R. Plant protein glycosylation. Glycobiology. 2016;26:926–939. doi: 10.1093/glycob/cww023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tjondro HC, Loke I, Chatterjee S, Thaysen-Andersen M. Human protein paucimannosylation: Cues from the eukaryotic kingdoms. Biol. Rev. Camb. Philos. Soc. 2019 doi: 10.1111/brv.12548. [DOI] [PubMed] [Google Scholar]
- 9.Kubelka V, et al. Primary structures of the N-linked carbohydrate chains from honeybee venom phospholipase A2. Eur. J. Biochem. 1993;213:1193–1204. doi: 10.1111/j.1432-1033.1993.tb17870.x. [DOI] [PubMed] [Google Scholar]
- 10.Staudacher E, März L. Strict order of (Fuc to Asn-linked GlcNAc) fucosyltransferases forming core-difucosylated structures. Glycoconj. J. 1998;15:355–360. doi: 10.1023/A:1006969701231. [DOI] [PubMed] [Google Scholar]
- 11.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IB. Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology. 2007;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 12.Aoki K, et al. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J. Biol. Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 13.Kajiura H, Hamaguchi Y, Mizushima H, Misaki R, Fujiyama K. Sialylation potentials of the silkworm, Bombyx mori; B. mori possesses an active α2,6-sialyltransferase. Glycobiology. 2015;25:1441–1453. doi: 10.1093/glycob/cwv060. [DOI] [PubMed] [Google Scholar]
- 14.Krzewinski-Recchi MA, et al. Identification and functional expression of a second human β-galactoside α2,6-sialyltransferase, ST6Gal II. Eur. J. Biochem. 2003;270:950–961. doi: 10.1046/j.1432-1033.2003.03458.x. [DOI] [PubMed] [Google Scholar]
- 15.Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. J. Biol. Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 16.Kubelka V, Altmann F, März L. The asparagine-linked carbohydrate of honeybee venom hyaluronidase. Glycoconj. J. 1995;12:77–83. doi: 10.1007/BF00731872. [DOI] [PubMed] [Google Scholar]
- 17.Kimura M, et al. Occurrence of GalNAcβ1-4GlcNAc unit in N-glycan of royal jelly glycoprotein. Biosci. Biotechnol. Biochem. 2002;66:1985–1989. doi: 10.1271/bbb.66.1985. [DOI] [PubMed] [Google Scholar]
- 18.Aoki K, Tiemeyer M. The glycomics of glycan glucuronylation in Drosophila melanogaster. Methods Enzymol. 2010;480:297–321. doi: 10.1016/S0076-6879(10)80014-X. [DOI] [PubMed] [Google Scholar]
- 19.Stanton R, et al. The underestimated N-glycomes of lepidopteran species. Biochim. Biophys. Acta Gen. Subj. 2017;1861:699–714. doi: 10.1016/j.bbagen.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurz S, et al. Targeted release and fractionation reveal glucuronylated and sulphated N- and O-glycans in larvae of dipteran insects. J. Proteomics. 2015;126:72–88. doi: 10.1016/j.jprot.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hykollari A, Malzl D, Stanton D, Eckmair B, Paschinger K. Tissue-specific glycosylation in the honeybee: Analysis of the N-glycomes of Apis mellifera larvae and venom. Biochim. Biophys. Acta Gen. Subj. 2019;1863:129409. doi: 10.1016/j.bbagen.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Die I, van Tetering A, Bakker H, van den Eijnden DH, Joziasse DH. Glycosylation in lepidopteran insect cells: Identification of a β1→4-N-acetylgalactosaminyltransferase involved in the synthesis of complex-type oligosaccharide chains. Glycobiology. 1996;6:157–164. doi: 10.1093/glycob/6.2.157. [DOI] [PubMed] [Google Scholar]
- 23.Haines N, Irvine KD. Functional roles for β1,4-N-acetlygalactosaminyltransferase-A in Drosophila larval neurons and muscles. Genetics. 2007;175:671–679. doi: 10.1534/genetics.106.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vadaie N, Jarvis DL. Molecular cloning and functional characterization of a Lepidopteran insect β4-N-acetylgalactosaminyltransferase with broad substrate specificity, a functional role in glycoprotein biosynthesis, and a potential functional role in glycolipid biosynthesis. J. Biol. Chem. 2004;279:33501–33518. doi: 10.1074/jbc.M404925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases: completion of the family tree. Glycobiology. 2012;22:768–777. doi: 10.1093/glycob/cwr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramakrishnan B, Qasba PK. Structure-based evolutionary relationship of glycosyltransferases: A case study of vertebrate β1,4-galactosyltransferase, invertebrate β1,4-N-acetylgalactosaminyltransferase and α-polypeptidyl-N-acetylgalactosaminyltransferase. Curr. Opin. Struct. Biol. 2010;20:536–542. doi: 10.1016/j.sbi.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan B, Qasba PK. Role of a single amino acid in the evolution of glycans of invertebrates and vertebrates. J. Mol. Biol. 2007;365:570–576. doi: 10.1016/j.jmb.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asano M, et al. Growth retardation and early death of β-1,4-galactosyltransferase knockout mice with augmented proliferation and abnormal differentiation of epithelial cells. EMBO J. 1997;16:1850–1857. doi: 10.1093/emboj/16.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Q, Hasty P, Shur BD. Targeted mutation in β1,4-galactosyltransferase leads to pituitary insufficiency and neonatal lethality. Dev. Biol. 1997;181:257–267. doi: 10.1006/dbio.1996.8444. [DOI] [PubMed] [Google Scholar]
- 30.Hansske B, et al. Deficiency of UDP-galactose:N-acetylglucosamine β-1,4-galactosyltransferase I causes the congenital disorder of glycosylation type IId. J. Clin. Investig. 2002;109:725–733. doi: 10.1172/JCI0214010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haines N, Irvine KD. Functional analysis of Drosophila β1,4-N-acetlygalactosaminyltransferases. Glycobiology. 2005;15:335–346. doi: 10.1093/glycob/cwi017. [DOI] [PubMed] [Google Scholar]
- 32.Veyhl J, et al. The directed migration of gonadal distal tip cells in Caenorhabditis elegans requires NGAT-1, a β1,4-N-acetylgalactosaminyltransferase enzyme. PLoS ONE. 2017;12(8):e0183049. doi: 10.1371/journal.pone.0183049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimomura M, et al. KAIKObase: An integrated silkworm genome database and data mining tool. BMC Genom. 2009;10:486. doi: 10.1186/1471-2164-10-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki T, et al. Biochemical characterization and mutational analysis of silkworm Bombyx mori β-1,4-N-acetylgalactosaminyltransferase and insight into the substrate specificity of β-1,4-galactosyltransferase family enzymes. Insect Biochem. Mol. Biol. 2019;115:103254. doi: 10.1016/j.ibmb.2019.103254. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishnan B, Boeggeman E, Ramasamy V, Qasba PK. Structure and catalytic cycle of β-1,4-galactosyltransferase. Curr. Opin. Struct. Biol. 2004;14:593–600. doi: 10.1016/j.sbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Togayachi A, Sato T, Narimatsu H. Comprehensive enzymatic characterization of glycosyltransferases with a β3GT or β4GT motif. Methods Enzymol. 2006;416:91–102. doi: 10.1016/S0076-6879(06)16006-1. [DOI] [PubMed] [Google Scholar]
- 37.Qasba PK, Ramakrishnan B, Boeggeman E. Structure and function of β-1,4-galactosyltransferase. Curr. Drug Targets. 2008;9:292–309. doi: 10.2174/138945008783954943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey DJ. Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 2005;16:647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Wuhrer M. Glycomics using mass spectrometry. Glycoconj. J. 2013;30:11–22. doi: 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dojima T, et al. Comparison of the N-linked glycosylation of human β1,3-N-acetylglucosaminyltransferase 2 expressed in insect cells and silkworm larvae. J. Biotechnol. 2009;143:27–33. doi: 10.1016/j.jbiotec.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Dell A, et al. Structural analysis of the oligosaccharides derived from glycodelin, a human glycoprotein with potent immunosuppressive and contraceptive activities. J. Biol. Chem. 1995;270:24116–24126. doi: 10.1074/jbc.270.41.24116. [DOI] [PubMed] [Google Scholar]
- 42.Seppo A, Moreland M, Schweingruber H, Tiemeyer M. Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur. J. Biochem. 2000;267:3549–3558. doi: 10.1046/j.1432-1327.2000.01383.x. [DOI] [PubMed] [Google Scholar]
- 43.Palcic MM, Hindsgaul O. Flexibility in the donor substrate specificity of β1,4-galactosyltransferase: Application in the synthesis of complex carbohydrates. Glycobiology. 1991;1:205–209. doi: 10.1093/glycob/1.2.205. [DOI] [PubMed] [Google Scholar]
- 44.Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: Sugar nucleotide contents in cultured insect cells and mammalian cells. Anal. Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 45.Chang KH, et al. Expression of recombinant cyclooxygenase 1 in Drosophila melanogaster S2 cells transformed with human β1,4-galactosyltransferase and Galβ1,4-GlcNAc α2,6-sialyltransferase. Biotechnol. Lett. 2007;29:1803–1809. doi: 10.1007/s10529-007-9489-0. [DOI] [PubMed] [Google Scholar]
- 46.Kim YK, et al. Expression of β-1,4-galactosyltransferase and suppression of β-N-acetylglucosaminidase to aid synthesis of complex N-glycans in insect Drosophila S2 cells. J. Biotechnol. 2011;153:145–152. doi: 10.1016/j.jbiotec.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Geisler C, Mabashi-Asazuma H, Kuo CW, Khoo KH, Jarvis DL. Engineering β1,4-galactosyltransferase I to reduce secretion and enhance N-glycan elongation in insect cells. J. Biotechnol. 2015;193:52–65. doi: 10.1016/j.jbiotec.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomono T, Kojima H, Fukuchi S, Tohsato Y, Ito M. Investigation of glycan evolution based on a comprehensive analysis of glycosyltransferases using phylogenetic profiling. Biophys. Physicobiol. 2015;12:57–68. doi: 10.2142/biophysico.12.0_57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawar ZS, Haslam SM, Morris HR, Dell A, Cummings RD. Novel poly-GalNAcβ1-4GlcNAc (LacdiNAc) and fucosylated poly-LacdiNAc N-glycans from mammalian cells expressing β1,4-N-acetylgalactosaminyltransferase and α1,3-fucosyltransferase. J. Biol. Chem. 2005;280:12810–12819. doi: 10.1074/jbc.M414273200. [DOI] [PubMed] [Google Scholar]
- 50.Sanders RD, Sefton JM, Moberg KH, Fridovich-Keil JL. UDP-galactose 4' epimerase (GALE) is essential for development of Drosophila melanogaster. Dis. Model Mech. 2010;3:628–638. doi: 10.1242/dmm.005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haines N, Stewart BA. Functional roles for β1,4-N-acetlygalactosaminyltransferase-A in Drosophila larval neurons and muscles. Genetics. 2007;175:671–679. doi: 10.1534/genetics.106.065565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takasu Y, et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem. Mol. Biol. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Sajwan S, et al. Efficient disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem. Mol. Biol. 2013;43:17–23. doi: 10.1016/j.ibmb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Tsubota T, Sezutsu H. Genome editing of silkworms. Methods Mol. Biol. 2017;1630:205–218. doi: 10.1007/978-1-4939-7128-2_17. [DOI] [PubMed] [Google Scholar]
- 55.Ma SY, Smagghe G, Xia QY. Genome editing in Bombyx mori: New opportunities for silkworm functional genomics and the sericulture industry. Insect Sci. 2019;26:964–972. doi: 10.1111/1744-7917.12609. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka A, Ozaki S. Purification and characterization of β-N-acetylgalactosaminidase from Bacillus sp. AT173-1. J. Biochem. 1997;122:330–336. doi: 10.1093/oxfordjournals.jbchem.a021757. [DOI] [PubMed] [Google Scholar]
- 57.Kimura Y, et al. Tumor antigen occurs in N-glycan of royal jelly glycoproteins: Honeybee cells synthesize T-antigen unit in N-glycan moiety. Biosci. Biotechnol. Biochem. 2006;70:2583–2587. doi: 10.1271/bbb.60331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.