Abstract
Primary cell culture in vitro suffers from cellular senescence. We hypothesized that expansion on decellularized extracellular matrix (dECM) deposited by simian virus 40 large T antigen (SV40LT) transduced autologous infrapatellar fat pad stem cells (IPFSCs) could rejuvenate high-passage IPFSCs in both proliferation and chondrogenic differentiation. In the study, we found that SV40LT transduced IPFSCs exhibited increased proliferation and adipogenic potential but decreased chondrogenic potential. Expansion on dECMs deposited by passage 5 IPFSCs yielded IPFSCs with dramatically increased proliferation and chondrogenic differentiation capacity; however, this enhanced capacity diminished if IPFSCs were grown on dECM deposited by passage 15 IPFSCs. Interestingly, expansion on dECM deposited by SV40LT transduced IPFSCs yielded IPFSCs with enhanced proliferation and chondrogenic capacity but decreased adipogenic potential, particularly for the dECM group derived from SV40LT transduced passage 15 cells. Our immunofluorescence staining and proteomics data identify matrix components such as basement membrane proteins top candidates for matrix mediated IPFSC rejuvenation. Both cell proliferation and differentiation were endorsed by transcripts measured by RNASeq during the process. This study provides a promising model for in-depth investigation of the matrix protein influence on surrounding stem cell differentiation.
Keywords: Infrapatellar fat pad-derived stem cells, Simian virus 40, Proliferation, Chondrogenesis, Decellularized extracellular matrix
1. Introduction
As cartilage does not readily self-heal, articular cartilage has difficulty recovering from trauma or degenerative disease. Given that autologous chondrocyte implantation, the most promising cell therapy for articular cartilage defects, has limited cell sources for clinical application [1], mesenchymal stromal/stem cells (MSCs), especially tissue-specific stem cells deposited by synovial tissue (SDSCs), have received much attention as possible cartilage repair therapies [2–4]. Considering the difficulty in harvesting synovium without contamination by surrounding connective tissue, the infrapatellar fat pad (IPFP), an easily accessible adjacent tissue, might serve as an alternative, large quantity autologous tissue-specific stem cell source for cartilage regeneration and repair [5,6]. However, increasing evidence shows that IPFSCs also suffer from replicative senescence after long-term ex vivo expansion [7].
To acquire a sufficient number of cells for cartilage engineering and regeneration, modification strategies, including modification of internal genomics or the external matrix microenvironment, have been proposed for cell-based therapy [8]. Simian virus 40 (SV40), a well-known oncogene, has been commonly utilized for cell immortalization. SV40 is restricted to two proteins, the large T (LT) antigen and small t antigen (ST). The former mainly influences the SV40-extended lifespan due to its ability to bind to pRb and p53 to inactivate these two tumor suppressors, causing cells to move from G1 phase into S phase thus promoting DNA replication [9]. However, malignant transformation is one of the potential risks caused by genetic manipulation [10].
Decellularized extracellular matrix (dECM), an integral part of the external matrix microenvironment, can be prepared from cell or tissue sources but they play a different role in cell functionality [11]. Briefly, cell-derived dECM has a distinct role in rejuvenation of adult stem cells, mainly promoting adult stem cells’ proliferation and differentiation capacity [12–14], which is different from tissue-derived dECM that largely guides tissue-specific behaviors [15]. In this scenario, cells isolated from patients themselves for dECM preparation provide another benefit in avoiding potential immune issues [16], despite the fact that most patients are either elderly or older adults and the isolated stem cells are prone to premature senescence.
Few reports have investigated the influence of SV40LT transduction alone or a combined dECM approach on stem cells’ chondrogenic potential. SV40LT transduction in “old” IPFSCs might be a novel cell source to provide “young” autologous cells for dECM preparation, in which a patient’s IPFSCs could be rejuvenated. In this study, we investigated whether SV40LT transduction could promote IPFSCs’ proliferation and chondrogenic potential and whether high-passage IPFSCs could be rejuvenated after expansion on dECM deposited by SV40LT transduced IPFSCs from either early or late passage IPFSCs. Given the close relationship between chondrogenesis and adipogenesis [17], we also aimed to explore whether the influence of SV40LT transduction on chondrogenesis also applied to adipogenesis. The main purpose of this study was to identify key matrix components guiding adult stem cells’ differentiation preference by using both dECM and immortalization approaches, which might facilitate engineering of smart matrix materials for cartilage engineering and regeneration in the near future.
2. Materials and methods
2.1. IPFSC culture and SV40LT transduction
The study was approved by the Institutional Review Board. Adult human infrapatellar fat pads were collected from six young patients with acute meniscus or anterior cruciate ligament tears (four male and two female, 22 years old on average). Minced infrapatellar fat pads were digested with 0.1% trypsin (Roche, Indianapolis, IN) for 30 min followed by 0.1% collagenase P (Roche) for 2 h at 37°C before filtration. After centrifugation, IPFSCs were cultured in alpha minimum essential medium (αMEM) with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL fungizone (Invitrogen, Carlsbad, CA) at 37°C in a humidified 21% O2 and 5% CO2 incubator.
Passage 1 IPFSCs (CTR group) were transduced with either SV40LT (pRSC-EF1-SV40LT-E2A-Puro-wpre) or green fluorescence protein (GFP) (pRSC‐EF1‐Puro‐GFP‐wpre) lentiviral vectors (SV40 and GFP group, respectively) in the presence of 4 μg/mL protamine sulfate (MilliporeSigma, St. Louis, MO). After 24 h, the medium was changed to αMEM with 10% FBS and 2 μg/mL puromycin (MilliporeSigma) for a four-day selection. Passage 5 and passage 15 IPFSCs from the CTR, GFP and SV40 groups were evaluated for transduction efficiency using reverse transcription (RT) polymerase chain reaction (PCR). Briefly, total RNA was extracted from expanded cells using an RNase-free TRIzol® (Invitrogen). After RT with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems Inc., Foster, CA), an initial denaturation at 95°C for 1 min was followed by 34 cycles of denaturation at 94°C for 30 sec; annealing was done at 55°C for 30 sec followed by extension at 72°C for 1 min with a final extension at 72°C for 5 min. An SV40LT specific primer (forward: 5’-CCT GAC TTT GGA GGC TTC TG-3’; reverse: 5’–GGA AAG TCC TTG GGG TCT TC-3’) was used for detection. The amplification PCR products were resolved on 2% agarose/TAE (Tris-acetate-EDTA) gel (Nacalai tesque, Inc., Japan), electrophoresed at 100 mV and visualized by ethidium bromide staining.
Passage 5 IPFSCs were also used for karyotype analysis according to standard cytogenetic techniques [18]. In brief, cells were arrested in the metaphase with 0.05 μg/mL colchicine reagents (MilliporeSigma), swollen in a hypotonic 0.075 M KCl solution and fixed in freshly prepared Carnoy’s fixative (3:1 absolute methanol/glacial acetic acid) before washing. One drop of the solution was placed onto pre-cooled cleaned slides, which were then air-dried. The metaphase mitotic cells were observed after staining with Giemsa solution.
2.2. dECM preparation and immunofluorescence staining
dECM was prepared following a protocol described previously [19]. Briefly, tissue culture plastic (TCP) was pre-coated with 0.2% gelatin (MilliporeSigma) at 37°C for 1 h, followed by 1% glutaraldehyde (MilliporeSigma) and 1 M ethanolamine (MilliporeSigma) for 30 min. Passage 5 and passage 15 IPFSCs from the CTR and SV40 groups were seeded on pre-coated TCP until 90% confluence was reached, and then 250 μM of L-ascorbic acid phosphate (Wako Chemicals, Richmond, VA) were added to the culture medium [20]. Seven days later, 0.5% Triton X-100 (MilliporeSigma) containing 20 mM ammonium hydroxide (Sargent-Welch, Skokie, IL) was added at 37°C for 5 min. After cell removal, dECM was stored at 4°C in phosphate buffered solution (PBS) containing 100 U/mL penicillin, 100 μg/mL streptomycin and 0.25 μg/mL fungizone until use.
dECM was fixed in 4% paraformaldehyde, blocked with 1% bovine serum albumin (BSA) and incubated with primary antibodies against human fibronectin (catalog number HFN 7.1; Developmental Studies Hybridoma Bank, Iowa City, IA), type IV collagen (catalog number M3F7; Developmental Studies Hybridoma Bank) and laminin (catalog number PA5–16287, Invitrogen) at 4°C overnight. After rinsing with PBS, dECM was incubated with a secondary antibody [Donkey anti-Mouse or anti-Rabbit IgG (H+L) Alexa Fluor 488, Invitrogen] at room temperature for 1 h. A Zeiss Axiovert 40 CFL Inverted Microscope (Zeiss, Oberkochen, Germany) was used to observe fluorescence intensity.
2.3. IPFSC culture and evaluation of proliferation, surface markers and stemness genes
Two experiments were designed as follows: (1) TCP culture regimen (Experiment 1): passage 5, 10 and 15 IPFSCs were expanded on TCPs, including three cell groups (“CTR”, “GFP” and “SV40”) and (2) dECM culture regimen (Experiment 2): passage 15 IPFSCs were grown on dECMs deposited by passage 5 versus passage 15 cells and SV40LT non-transduced (C) versus transduced (S) cells with TCP culture substrate as a control including four dECM groups (“C5E”, “C15E”, “S5E” and “S15E”) and five expanded cell groups (“C5C”, “C15C”, “S5C”, “S15C” and “CC”).
Population doubling (PD) level was calculated for cell growth (5 T175 flasks each group) in Experiment 1 using a formula as follows: n = 3.32 (log UCY – log I) + X, where n = the final PD level number (PDN) at the end of a given subculture, UCY= the cell yield at that point, I = the cell number used as inoculum to begin that subculture and X = the doubling level of the inoculum used to initiate the subculture being quantified. The amount of cell growth in Experiment 2 was calculated using Countess® (Invitrogen).
Expanded cells from Experiment 1 and Experiment 2 were also evaluated for cell proliferation using Click-iT 5-ethynyl-2’-deoxyuridine (EdU) cell Proliferation Assay kit (Invitrogen) and cell cycle. After expanded cells reached 50% confluence, EdU was added to the culture medium with a final concentration of 10 μM and the cells were incubated at 37°C for 18 h before being fixed with 4% paraformaldehyde. Collected cells (2 × 105 each group) were incubated with Click-iT reaction cocktail at room temperature for 30 min. Fluorescence was analyzed by a FACS Calibur (BD Biosciences, San Jose, CA) using the FCS Express software package (De Novo Software, Los Angeles, CA). For cell cycle analysis, collected cells were fixed with 70% ethanol and stained with propidium iodide (MilliporeSigma). DNA contents were measured by a FACS Calibur (BD Biosciences) using the FCS Express software package (De Novo Software).
Cell surface markers were evaluated using the following primary antibodies: CD73-APC (catalog number 17–0739-42; eBioScience, Fisher Scientific, Waltham, MA), CD90-APC-Vio770 (catalog number 130–114-863; Miltenyi Biotec, San Diego, CA), CD105-PerCP-Vio700 (catalog number 130–112-170; Miltenyi Biotec), CD146-PE (catalog number 12–1469-42; eBioScience) and the stage-specific embryonic antigen 4-PE (SSEA4-PE; catalog number 330406; BioLegend, Dedham, MA). Expanded cells (2 × 105 each group) were incubated in cold PBS containing 0.1% ChromPure Human IgG whole molecule (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 min. Then cells were incubated with primary antibodies at 4°C for 30 min. Fluorescence was analyzed by a FACS Calibur (BD Biosciences) using the FCS Express software package (De Novo Software).
For TaqMan® real-time quantitative PCR (qPCR), about 1 μg of RNA from expanded cells was used for RT. Stemness genes [NANOG (assay ID: Hs02387400_g1), SOX2 (SRY-box 2; assay ID: Hs01053049_s1), KLF4 (Kruppel-like factor 4; assay ID: Hs00358836_m1), BMI1 (B lymphoma Mo-MLV insertion region 1 homolog; assay ID: Hs00180411_m1), MYC (MYC proto-oncogene; assay ID: Hs00153408_m1), NOV (nephroblastoma overexpressed; assay ID: Hs00159631_m1), POU5F1 (POU class 5 homeobox 1; assay ID: Hs04260367_gH) and NES (nestin; assay ID: Hs04187831_g1) ] and senescent genes [CDKN1A (cyclin dependent kinase inhibitor 1A; assay ID: Hs00355782_m1), CDKN2A (cyclin dependent kinase inhibitor 2A; assay ID: Hs00923894_m1) and TP53 (tumor protein p53; assay ID: Hs01034249_m1)] were customized by Applied Biosystems as part of the Custom TaqMan® Gene Expression Assays. GAPDH (glyceraldehyde-3-phosphate dehydrogenase; assay ID: Hs02758991_g1) was carried out as the endogenous control gene. qPCR was performed using Applied Biosystems™ 7500 Fast Real-Time PCR System (Applied Biosystems). Relative transcript levels were calculated as χ =2−ΔΔCt, in which ΔΔCt=ΔE −ΔC, ΔE=Ctexp−CtGAPDH, and ΔC=Ctct1−CtGAPDH.
2.4. Chondrogenic induction and evaluation
For chondrogenic differentiation, aliquots of 0.3 × 106 expanded cells were centrifuged at 500 g for 7 min in a 15-ml polypropylene tube to form a pellet. Pellets were cultured in a serum-free high-glucose Dulbecco’s Modified Eagle’s Medium (DMEM) with 100 U/mL penicillin, 100 μg/mL streptomycin, 40 μg/mL proline (MilliporeSigma), 100 nM dexamethasone (MilliporeSigma), 0.1 mM ascorbic acid-2-phosphate, ITS™ Premix (BD Biosciences) and 10 ng/mL transforming growth factor beta 3 (TGFβ3; PeproTech, Rocky Hill, NJ). Chondrogenic differentiation was evaluated using histology, immunohistochemical staining and qPCR for the expression of chondrogenic marker genes.
Representative pellets (n=3) were fixed in 4% paraformaldehyde at 4°C overnight, followed by dehydrating in a gradient ethanol series, clearing with xylene and embedding in paraffin blocks. Five-μm thick sections were stained with Alcian blue (MilliporeSigma) for sulfated glycosaminoglycan (GAG). For immunohistochemical staining (IHC), sections were incubated with primary antibodies against type II collagen (catalog number II-II6B3; Developmental Studies Hybridoma Bank) followed by the secondary antibody of biotinylated horse anti-mouse IgG (Vector, Burlingame, CA). Immunoactivity was detected using Vectastain ABC reagent (Vector).
About 2 μg of RNA isolated from each pellet (n=3) were used for RT followed by a qPCR procedure to evaluate chondrogenic marker related genes [SOX9 (SRY (sex determining region Y)-box 9; assay ID: Hs00165814_m1), ACAN (aggrecan; assay ID: Hs00153936_m1), COL2A1 (type II collagen; assay ID: Hs00156568_m1) and PRG4 (proteoglycan 4; assay ID: Hs00981633_m1)] and hypertrophic marker genes [COL10A1 (type X collagen; assay ID: Hs00166657_m1)]. GAPDH was carried out as the endogenous control gene.
2.5. Adipogenic differentiation and evaluation
Expanded cells around 95% confluence in Experiment 1 and Experiment 2 were incubated for 21 days in adipogenic medium (αMEM supplemented with 10% FBS, 1 μM dexamethasone, 0.5 mM isobutyl-1-methyxanthine, 200 μM indomethacin and 10 μM insulin). For intracellular lipid-filled droplet staining, samples (n=3) were fixed in 4% paraformaldehyde and stained with a 0.6% (w/v) Oil Red O (ORO) solution (60% isopropanol, 40% water) for 10 min. Adipogenic marker genes were quantified using qPCR. About 2 μg of RNA isolated from each sample (n=3) were used for RT followed by a qPCR procedure to evaluate adipogenic marker genes [LPL (lipoprotein lipase; assay ID: Hs00173425_m1), PPARG (peroxisome proliferator-activated receptor gamma; assay ID: Hs01115513_m1) and CEBPA (CCAAT enhancer binding protein alpha; assay ID: Hs00269972_s1)]. GAPDH was carried out as the endogenous control gene.
Proteins were extracted from cells using lysis buffer (Cell Signaling Technology, Inc., Danvers, MA) with protease inhibitors. Total proteins were quantified using Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Forty μg of protein from each sample were separated using NuPAGE™ Bis-Tris Mini Gels (Invitrogen) in the XCell SureLock™ Mini-Cell (Life Technologies, Carlsbad, CA) at 120 V at room temperature for 2.5 h. Bands were transferred onto a nitrocellulose membrane using an XCell II™ Blot module (Life Technologies) at 70 V at 4°C overnight. The membrane was incubated with primary monoclonal antibodies targeting LPL (Santa Cruz Biotechnology, Dallas, TX) and GAPDH (Thermo Fisher Scientific) in 5% BSA in TBST buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.05% Tween-20) at 4°C overnight, followed by the secondary antibody of horseradish peroxidase-conjugated goat anti-mouse (Invitrogen) for 1 h. ECL™ Prime Western Blotting Detection Reagents (Amersham Biosciences, Waltham, MA) were used for exposure.
2.6. Proteomics analysis
dECMs (C5E, S5E, C15E and S15E), expanded cells (CC, C5C, S5C, C15C and S15C) and pellet samples (CP, C5P, S5P, C15P and S15P) from Experiment 2 were chemically and enzymatically digested prior to liquid chromatography tandem mass spectrometry (LC-MS/MS) as previously described [21] with the following exceptions. Digested samples were desalted and concentrated on a Thermo Scientific Pierce C18 tip and analyzed on a Thermo nanoEasy LC II coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated on an in-house made 20 cm C18 analytical column (100 μm ID) packed with 2.7 μm Phenomenex Cortecs C18 resin. Gradient conditions and MS acquisition parameters were described previously [22]. Raw files were directly loaded into the Proteome Discoverer 2.2 and searched against the human uniprotKB database (release date 2018.08) utilizing the Mascot search engine. Mass tolerances were set to +/− 10 ppm for MS parent ions and +/− 25 ppm for MS/MS fragment ions. Trypsin specificity was used allowing for 1 missed cleavage. Cys carbamidomethylation was set as a fixed modification and variable modifications included Met oxidation, Pro hydroxylation, protein N-terminal acetylation and peptide N-terminal pyroglutamic acid formation. Search results were exported into a spreadsheet for further processing with figures generated utilizing GraphPad. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [23] partner repository with the dataset identifier PXD021069 and 10.6019/PXD021069.
2.7. RNASeq analysis
Total RNAs were isolated from cells using Trizol (Invitrogen) followed by additional purification using an RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. RNASeq library was prepared by the Genomics Core of West Virginia University using KAPA mRNA HyperPrep Kit (KAPA Biosystems, Wilmington, MA) and sequenced by the Genomics Core of Marshall University using HiSeq 2500. We followed the procedures described previously [6] for RNASeq data analysis: subread [24] for reads alignment (hg38), Rsubread [25] for calculation of number of reads for protein coding genes from RefSeq and Excel for calculation of Reads Per Kilobase of transcript per Million mapped reads (RPKM) of the exon model, which measures gene expression from RNASeq. Gene set enrichment analysis (GSEA) was performed against gene sets annotated in MSigDB with GSEA [26] using genes sorted based on fold change of expression from two conditions of interest; genes not expressed in any of the two compared conditions were excluded. Gene expression visualization with heat map was done by MeV (http://mev.tm4.org/). RNASeq data sets were deposited to GEO with accession number GSE148632.
2.8. Statistical analysis
Mann-Whitney U test was used for pairwise comparison. All statistical analyses were performed with SPSS 20.0 statistical software (SPSS Inc., Chicago, IL). P values less than 0.05 were considered statistically significant.
3. Results
3.1. Evaluation of Cas9-SV40LT transduction and effect on IPFSC proliferation
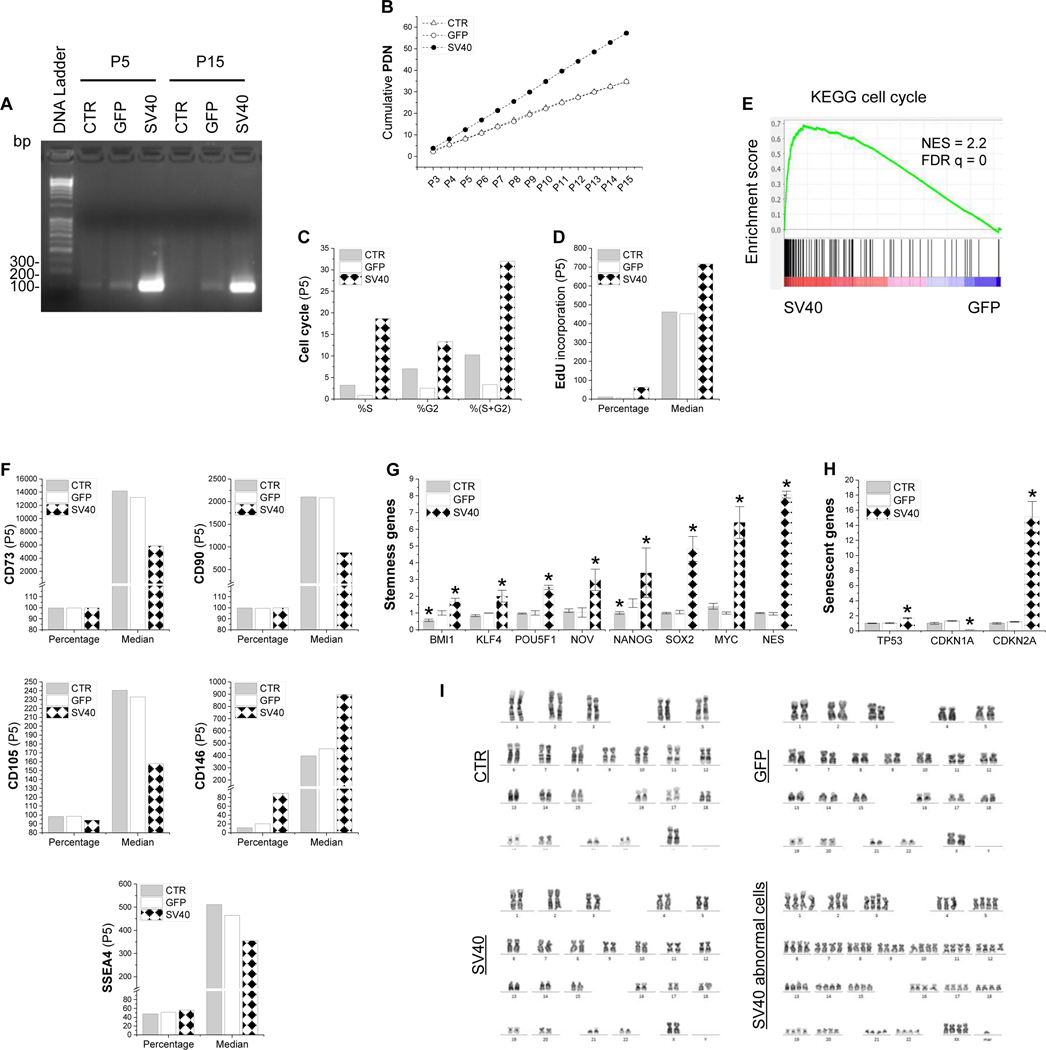
To make an immortalized cell line of human IPFSCs, SV40LT lentiviral vector was used for transduction with GFP as a control. RT-PCR data confirmed successful transduction of Cas9-SV40LT in IPFSCs, evidenced by strong expression of SV40LT in cells from the SV40 group at both passages 5 and 15, followed by weak expression in the GFP group and negligible expression in the CTR group (Fig. 1A). To determine whether SV40LT transduction affects the proliferation ability of IPFSCs, PDN results showed, compared to similar growth rate of cells in the CTR and GFP groups, SV40LT transduction yielded more rapidly growing cells for up to passage 15 (Fig. 1B). Consistently, cell cycle data revealed a ten-fold, 9-fold and 2-fold increase in “S+G2” of passage 5 (Fig. 1C), passage 10 (Supplementary Fig. 1A) and passage 15 cells (Supplementary Fig. 1D), respectively, from the SV40 group compared to the corresponding GFP group, which was further confirmed by relative EdU incorporation data in both percentage and median fluorescence intensity (median) of passage 5 (Fig. 1D), passage 10 (Supplementary Fig. 1B) and passage 15 cells (Supplementary Fig. 1E). Additional GSEA using RNASeq data further confirmed that cell cycle genes were generally upregulated in the SV40 group as compared to the GFP control (Fig. 1E).
Figure 1.
SV40LT transduction promoted human IPFSCs in cell proliferation and stemness. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Successful transduction of SV40LT in IPFSCs (A), evidenced by RT-PCR data in both passage 5 and 15 cells. Evaluation of proliferation used cumulative population doubling number (PDN) in IPFSCs from passage 3 to 15 (B) along with flow cytometry for cell cycle analysis (C) and relative EdU incorporation (D). GSEA for expressed genes ranked by the fold change of expression (SV40/GFP) at passage 5 against the MSigDB gene set KEGG cell cycle (NES: normalized enrichment score) (E). Flow cytometry was employed to evaluate MSC surface markers including CD73, CD90, CD105, CD146 and SSEA4 (F). TaqMan® real-time quantitative PCR was used to evaluate stemness genes (BMI1, KLF4, POU5F1, NOV, NANOG, SOX2, MYC and NES) (G) and senescence genes (TP53, CDKN1A and CDKN2A) (H). Data are exhibited as bar charts. * means a statistically significant difference compared to the corresponding GFP control (p<0.05). Evaluation of chromosome abnormality of SV40LT transduced IPFSCs using karyotype analysis (I).
To further characterize molecular phenotype changes following SV40LT transduction, flow cytometry was used to quantify both percentage and median of typical MSC surface markers in expanded cells. Following SV40LT transduction, we found that, for all passage 5 (Fig. 1F), passage 10 (Supplementary Fig. 1C) and passage 15 IPFSCs (Supplementary Fig. 1F), the expression of CD73, CD90 and CD105 dropped in median and only CD105 slightly decreased in percentage; interestingly, compared to an increase of CD146 expression in both percentage and median (Fig. 1F; Supplementary Fig. 1C/F), SSEA4 expression decreased in median in passage 5 IPFSCs (Fig. 1F) and in percentage and median in passage 10 IPFSCs but increased in median and decreased in percentage in passage 15 IPFSCs (Supplementary Fig. 1C/F).
Cell morphology observation demonstrated a spindle-like appearance of cells from the CTR and GFP groups while those from the SV40 group were smaller. qPCR data showed that all tested stemness genes including BMI1, KLF4, POU5F1, NOV, NANOG, SOX2, MYC and NES were upregulated in IPFSCs following SV40LT transduction (Fig. 1G). For senescence associated genes, TP53 (along with TP53 protein expression assessed by western blot in Supplementary Fig. 2) and CDKN2A were upregulated while CDKN1A was downregulated (Fig. 1H). Given that SV40 is an oncogenic DNA virus, karyotype analysis was performed to investigate chromosome abnormality after SV40LT transduction. Results showed that, compared to diploid cells from the CTR and GFP groups, both diploid and abnormal karyotypes (about 5%) were observed in IPFSCs from the SV40 group (Fig. 1I).
3.2. Effect of SV40LT transduction on IPFSCs’ chondrogenic and adipogenic capacities
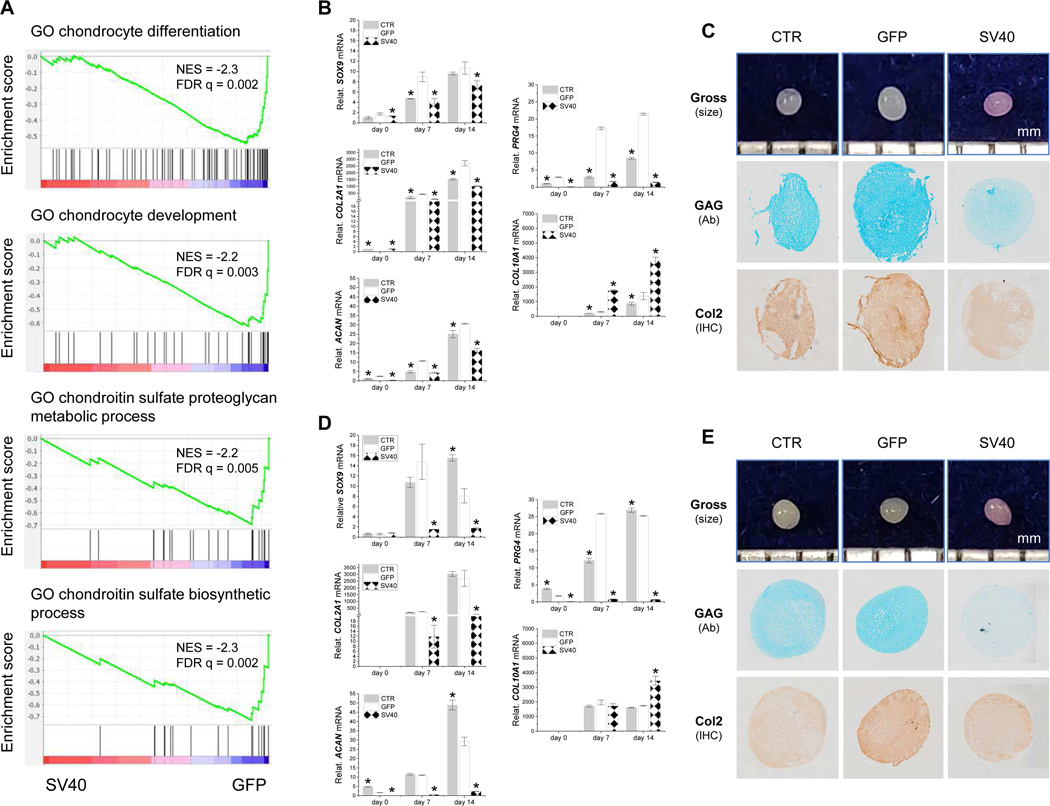
GSEA with RNASeq data demonstrated that genes related to chondrocyte development and differentiation were downregulated in the SV40 group as compared to the GFP control (Fig. 2A). This finding was confirmed by an in vitro induction experiment, in which IPFSCs from the CTR, GFP and SV40 groups were chondrogenically induced for up to 14 days in a pellet culture system followed by analyses using qPCR for chondrogenic marker gene expression, and Alcian blue staining (Ab) for sulfated GAGs and immunohistochemical staining (IHC) for type II collagen. Consistent with GSEA, qPCR data showed that all tested chondrogenic genes (SOX9, COL2A1, ACAN and PRG4) revealed a similar trend - a dramatic decrease with time after SV40LT transduction. Interestingly, COL10A1, a hypertrophic marker gene, greatly increased in IPFSCs of passage 5 (Fig. 2B), passage 10 (Supplementary Fig. 3) and passage 15 cells (Fig. 2D). Staining data further confirmed that pellets from P5 (Fig. 2C) and P15 cells (Fig. 2E) of the SV40 group exhibited less staining intensity of sulfated GAG and type II collagen than those from the GFP group.
Figure 2.
Chondrogenic evaluation of SV40LT transduced IPFSCs. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). SV40LT transduction on IPFSCs downregulated genes related to chondrocyte development and differentiation, evidenced by GSEA for expressed genes ranked by the fold change of expression (SV40/GFP) at passage 5 against MSigDB Gene Ontology (GO) gene sets annotated for chondrocyte differentiation, chondrocyte development, chondroitin sulfate proteoglycan metabolic process and chondroitin sulfate biosynthetic process (A). Fourteen-day chondrogenically induced pellets from passage 5 IPFSCs (B/C) and passage 15 IPFSCs (D/E) were evaluated using TaqMan® real-time quantitative PCR (B/D) for chondrogenic marker genes (SOX9, COL2A1, ACAN and PRG4) and hypertrophic marker gene (COL10A1) (GAPDH served as an internal control), photographs for the size of pellet (mm), Alcian blue staining (Ab) for sulfated GAGs and immunohistochemical staining (IHC) for type II collagen (C/E). Data are exhibited as bar charts. * indicates a statistically significant difference compared to the corresponding GFP control (p<0.05).
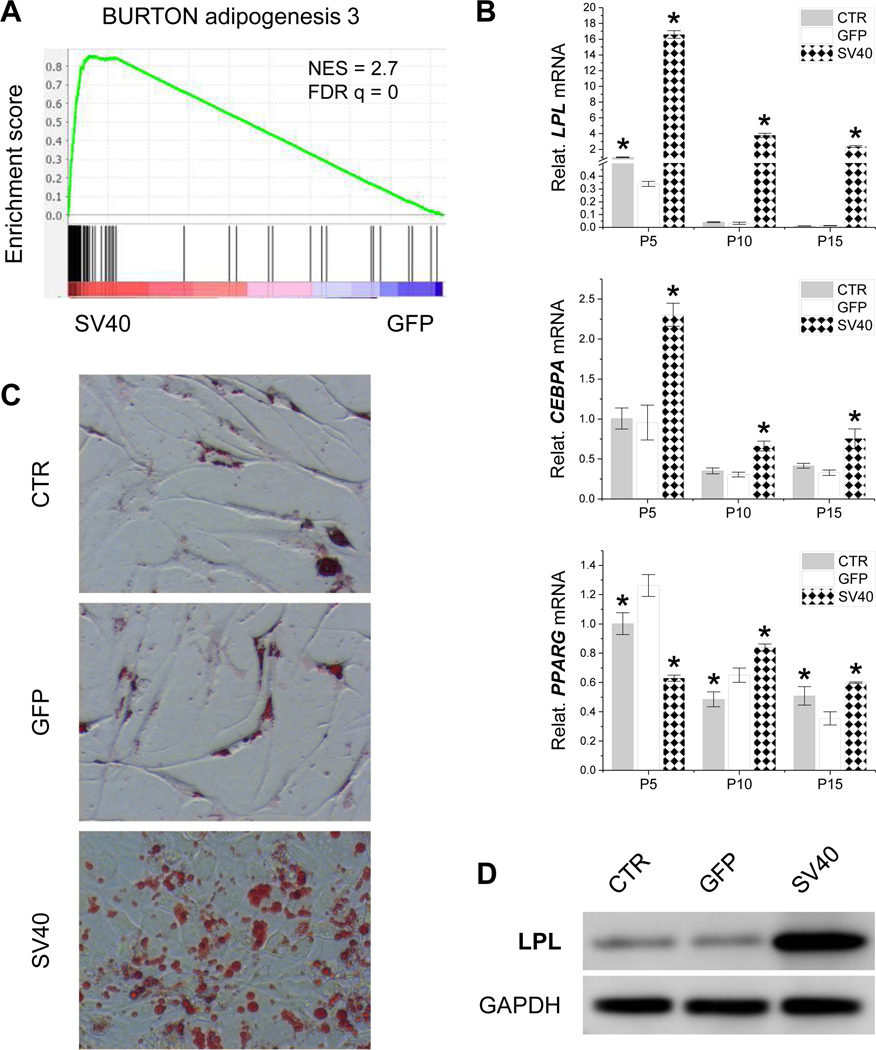
GSEA with RNASeq data also demonstrated a substantial increase in expression for genes related to adipogenesis by SV40LT transduction (Fig. 3A). This result was confirmed by an in vitro induction study, in which IPFSCs from the CTR, GFP and SV40 groups were adipogenically induced for 21 days. The data revealed a robust increase in IPFSCs’ adipogenic differentiation following SV40LT transduction, evidenced by higher expression of the adipogenic marker genes LPL, CEBPA and PPARG compared to those from passages 5, 10 and 15 in the CTR and GFP groups (Fig. 3B). We also found that adipogenic capacity decreased in a passage-dependent manner (Fig. 3B). The expression change at the mRNA level was confirmed by that at the protein level, in which the strongest intensity of Oil Red O staining indicating the most lipid droplets (Fig. 3C) and highest expression of LPL by western blot analysis were found in IPFSCs after SV40LT transduction (Fig. 3D).
Figure 3.
Adipogenic evaluation of SV40LT transduced IPFSCs. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). GSEA for expressed genes ranked by the fold change of expression (SV40/GFP) at passage 5 against the MSigDB gene set “BURTON adipogenesis 3”, which includes genes strongly upregulated during differentiation from fibroblasts (3T3) into adipocytes (A). Twenty-one day adipogenically induced IPFSCs from passages 5, 10 and 15 were evaluated using TaqMan® real-time quantitative PCR for adipogenic marker genes (LPL, CEBPA and PPARG) (B). GAPDH served as an internal control gene. Data are exhibited as bar charts. * indicates a statistically significant difference compared to the corresponding GFP control (p<0.05). Adipogenically induced passage 5 IPFSCs were also evaluated using Oil Red O staining for lipid droplets (C) and western blot for LPL expression at the protein level (D).
3.3. Evaluation of dECM deposited by SV40LT transduced cells on IPFSCs’ proliferation
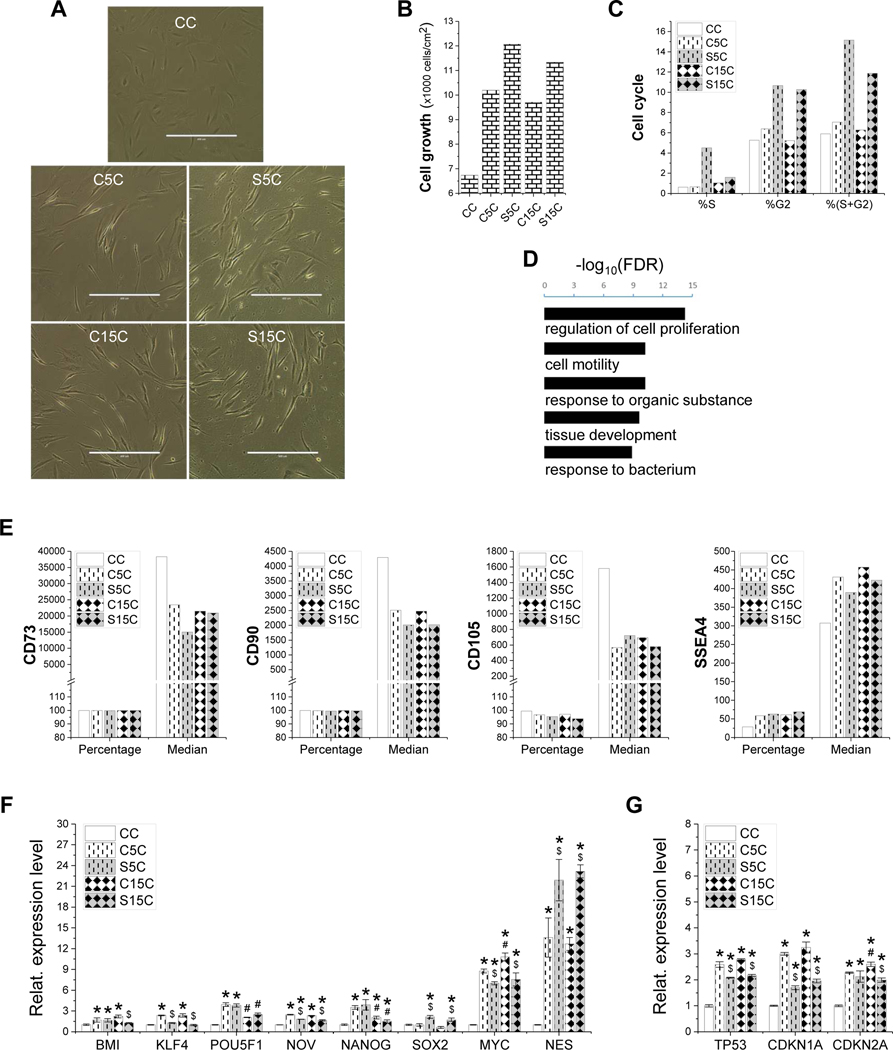
To determine the influence on proliferation capacity following expansion on dECM deposited by SV40LT transduced cells, IPFSCs were assessed using cell morphology, cell growth rate, cell cycle, MSC surface markers and stemness and senescence associated genes. Compared to those grown on TCP, dECM expanded IPFSCs exhibited a glistening cell surface (Fig. 4A). dECM expansion increased cell number, particularly for that deposited by SV40LT transduced cells (Fig. 4B), which is in line with cell cycle data (Fig. 4C). Consistently, genes upregulated in the dECM expanded IPFSCs as compared to those on TCP were enriched in gene ontologies related to biological processes including cell proliferation and cell motility (Fig. 4D). Intriguingly, flow cytometry results exhibited a decrease of CD73, CD90 and CD105 expression and an increase of SSEA4 expression in IPFSCs following expansion on dECMs (Fig. 4E). qPCR data showed that dECM expanded cells exhibited generally higher levels of stemness gene expression; interestingly, IPFSCs had higher levels of SOX2 and NES and lower expression of KLF4 and MYC following expansion on dECM deposited by SV40LT transduced cells compared to non-SV40LT transduced cells (Fig. 4F). In addition, dECM expanded cells had higher levels of senescence associated gene expression including TP53, CDKN1A and CDKN2A compared to those grown on TCP while expansion on dECM deposited by SV40LT transduced cells decreased IPFSCs’ senescence associated gene expression (Fig. 4G).
Figure 4.
Evaluation of high-passage IPFSCs’ proliferation after growth on dECMs and TCP. Cell morphology of passage 15 IPFSCs (A) was photographed on TCP (CC) and dECMs deposited by passage 5 IPFSCs (C5C), passage 15 IPFSCs (C15C), SV40LT transduced passage 5 IPFSCs (S5C) and SV40LT transduced passage 15 IPFSCs (S15C). Scale bar: 400 μm. Cell proliferation was evaluated using cell counting for cell growth (B), flow cytometry for cell cycle (C) and RNASeq analysis (D), in which Gene Ontology enrichment analysis on biological processes was done for genes upregulated in the dECM expanded IPFSCs (C5C, C15C, S5C and S15C) as compared to those cultured on TCP (CC) (t-test p-value<0.01 and fold change>2) (D). Surface markers were evaluated using flow cytometry including CD73, CD90, CD105 and SSEA4 (E). TaqMan® real-time quantitative PCR was used to evaluate stemness genes (BMI1, KLF4, POU5F1, NOV, NANOG, SOX2, MYC and NES) (F) and senescence genes (TP53, CDKN1A and CDKN2A) (G). GAPDH served as an internal control. Data are exhibited as bar charts. Statistically significant differences are indicated using * when compared to CC, using $ when compared to corresponding C5C or C15C and using # when compared to corresponding C5C or S5C (p<0.05).
3.4. Effect of dECM deposited by SV40LT transduced cells on IPFSCs’ chondrogenic capacity
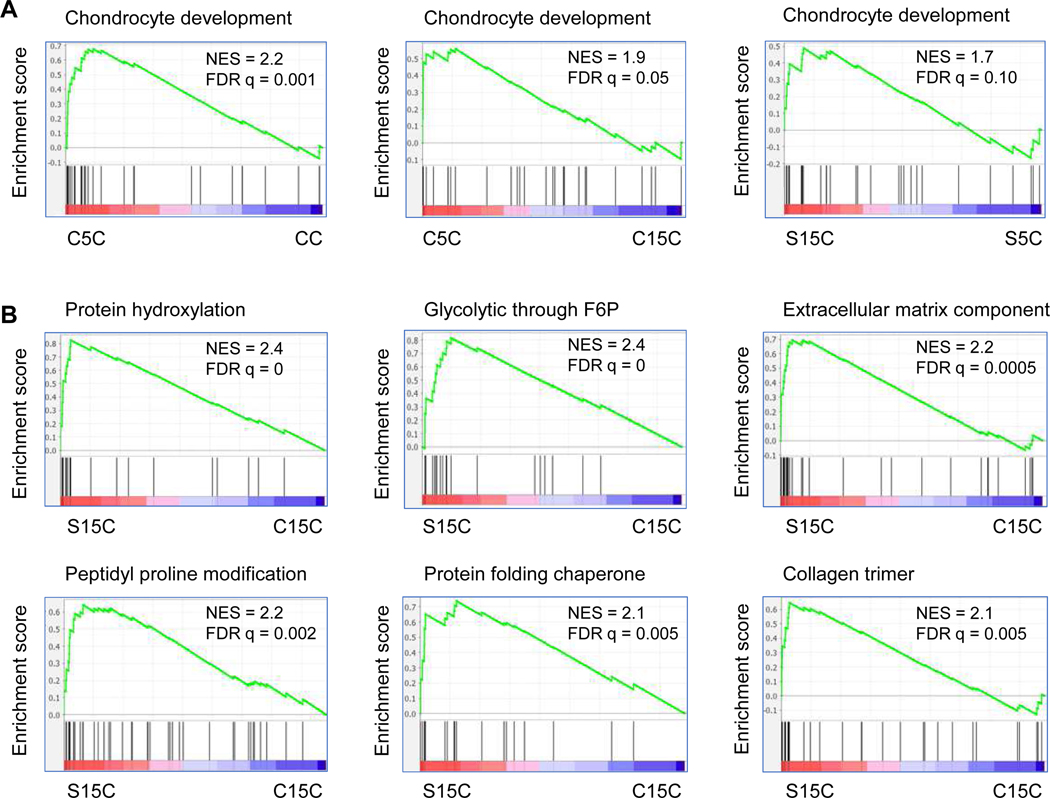
Changes of expression on gene sets related to chondrogenesis were examined across IPFSCs expanded on TCP or on dECMs (Fig. 5). The analysis revealed that the Gene Ontology set annotated for chondrocyte development was upregulated in the C5C group as compared to the CC group or to the C15C group and in the S15C group as compared to S5C (Fig. 5A). Remarkably, the S15C group also exhibited a higher level of expression than the C15C group on Gene Ontologies related to chondrogenesis including protein hydroxylation, glycolytic through fructose 6 phosphate (F6P), extracellular matrix component, peptidyl proline modification, protein folding chaperone and collagen trimer (Fig. 5B).
Figure 5.
RNASeq validation on the expression changes of genes related to chondrogenesis for IPFSCs cultured on dECM deposited by SV40LT transduced cells. GSEA for expressed genes ranked by the fold change of expression (C5C/CC for the leftmost panel, C5C/C15C for the middle panel and S15C/S5C for the rightmost panel) against the MSigDB gene set annotated for Gene Ontology chondrocyte development (A). GSEA for expressed genes ranked by the fold change of expression (S15C/C15C) against the MSigDB gene sets related to chondrogenesis including protein hydroxylation, glycolytic through fructose 6 phosphate (F6P), extracellular matrix component, peptidyl proline modification, protein folding chaperone and collagen trimer (B).
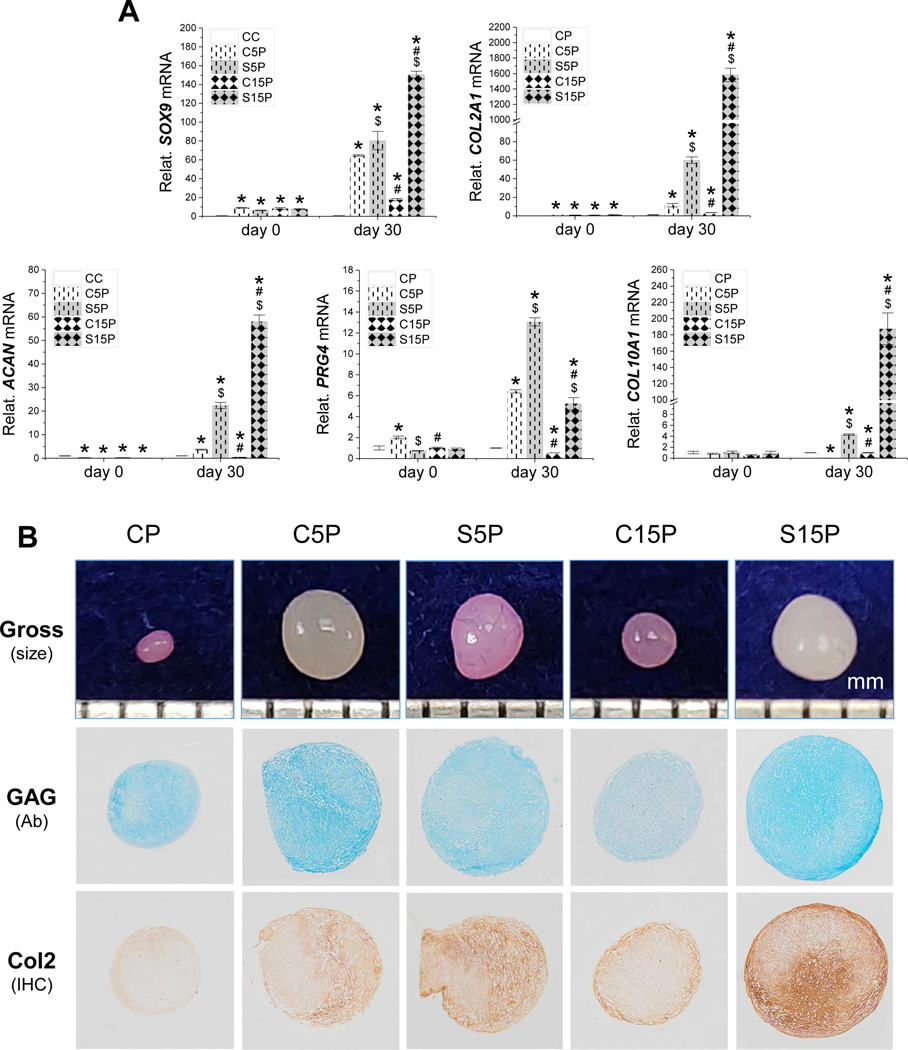
To confirm the observations, expanded cells were incubated in a pellet culture under chondrogenic induction for 30 days; pellets were assessed using qPCR for chondrogenic marker gene expression, Alcian blue staining for sulfated GAG and immunohistochemical staining for type II collagen. Despite an upregulation of SOX9 and COL2A1 and a downregulation of ACAN in day 0 pellets of IPFSCs following dECM expansion, expansion on dECMs deposited by low-passage cells (C5C) or SV40LT transduced cells (S5C and S15C) yielded pellets (termed C5P, S5P and S15P, respectively) with higher expression of SOX9, COL2A1, ACAN and PRG4 compared to those from the TCP groups (termed CP) after 30-day chondrogenic induction (Fig. 6A). With dECM pretreatment, the pellets from the S15C group exhibited the highest level of chondrogenic marker gene expression followed by those from the S5C group with the C15C group being the least (Fig. 6A).
Figure 6.
Chondrogenic evaluation of high-passage IPFSCs after growth on dECMs and TCP. Following expansion on TCP (CC) and dECMs deposited by passage 5 IPFSCs (C5C), passage 15 IPFSCs (C15C), SV40LT transduced passage 5 IPFSCs (S5C) and SV40LT transduced passage 15 IPFSCs (S15C), passage 15 IPFSCs were grown in a pellet culture system (termed CP, C5P, C15P, S5P and S15P, respectively) supplemented with chondrogenic medium. Thirty-day pellets (along with 0-day pellets) were assessed using TaqMan® real-time quantitative PCR for chondrogenic marker genes (SOX9, COL2A1, ACAN, PRG4) and hypertrophic marker gene (COL10A1) (A). GAPDH served as an internal control. Data are exhibited as bar charts. Statistically significant differences are indicated using * when compared to CC, using $ when compared to corresponding C5C or C15C and using # when compared to corresponding C5C or S5C (p<0.05). Histologically, 30-day pellets were evaluated using photographs for the size (mm), Alcian blue staining (Ab) for sulfated GAGs and immunohistochemical staining (IHC) for type II collagen (Col2) (B).
In line with the above mRNA data, dECM expanded cells yielded pellets with a larger size and more intensive staining of sulfated GAG and type II collagen compared to those grown on TCP (Fig. 6B). In chondrogenic induction, dECM deposited by high-passage cells (C15P) diminished the rejuvenation effect on expanded IPFSCs compared to that deposited by low-passage cells (C5P); dECM deposited by SV40LT transduced cells (S5P and S15P) further promoted the rejuvenation effect on expanded IPFSCs compared to the corresponding group deposited by non-SV40LT transduced cells (C5P and C15P); intriguingly, expansion on dECM deposited by high-passage SV40LT transduced cells (S15P) contributed to IPFSCs with the largest pellet size and most intensive staining for sulfated GAG and type II collagen (Fig. 6B).
3.5. Effect of dECM deposited by SV40LT transduced cells on IPFSCs’ adipogenic capacity
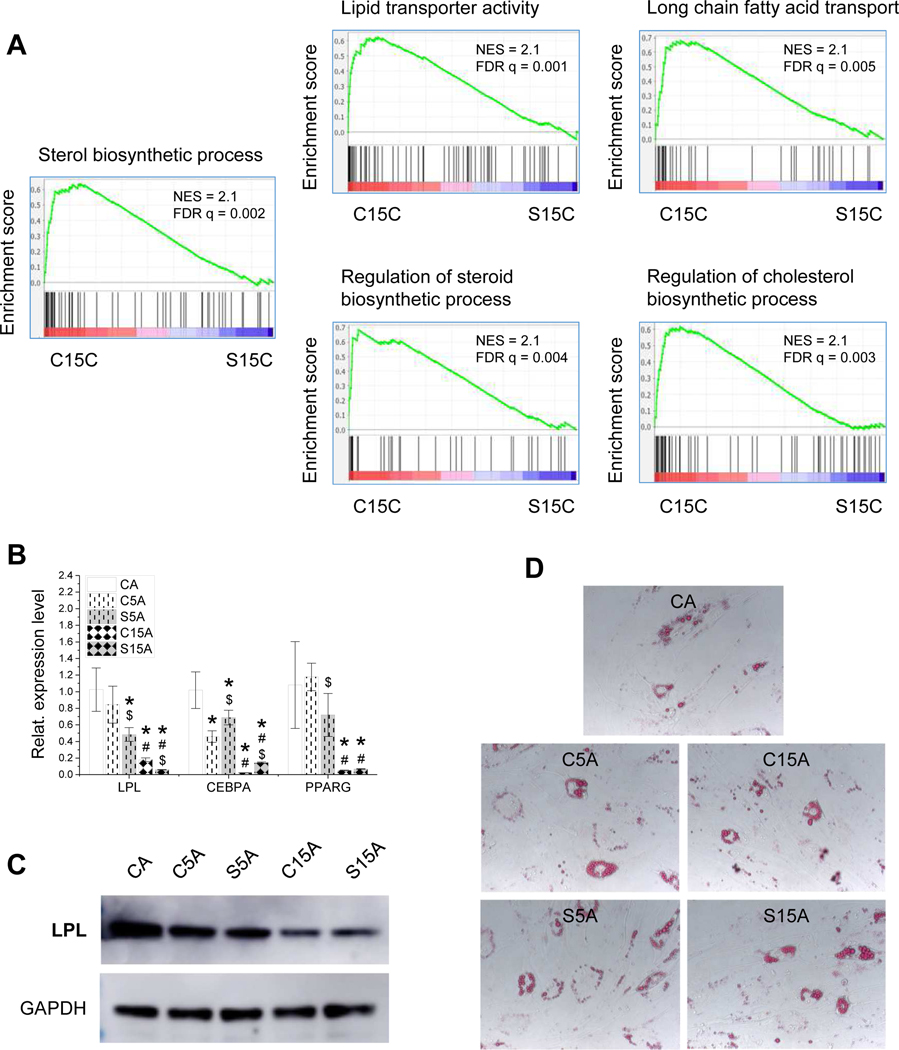
GSEA with RNASeq data suggested that IPFSCs grown on dECM deposited by high-passage SV40LT transduced IPFSCs exhibited reduced expression level on gene sets related to adipogenesis compared with those on dECM from high-passage IPFSCs (Fig. 7A). This finding was confirmed by an in vitro induction study, in which expanded cells were induced in adipogenic medium for 21 days followed by assessment using qPCR for adipogenic marker gene expression and western blot for LPL expression at a protein level and Oil Red O staining for lipid droplets. qPCR data suggested that dECM expansion decreased IPFSCs’ adipogenic marker gene expression including LPL, CEBPA and PPARG, particularly for the dECMs deposited by high-passage cells (C15A) or by SV40LT transduced cells (S5A and S15A) (Fig. 7B). Western blot data showed that, compared to growth on TCP, expansion on dECMs yielded IPFSCs with less LPL expression after adipogenic induction, particularly for dECM deposited by high-passage cells (Fig. 7C), despite there being not much difference in the staining of lipid droplets among groups (Fig. 7D).
Figure 7.
Adipogenic evaluation of high-passage IPFSCs after growth on dECMs and TCP. GSEA for expressed genes ranked by the fold change of expression (C15C/S15C) against MSigDB gene sets related to adipogenesis (A). Following expansion on TCP (CC) and dECMs deposited by passage 5 IPFSCs (C5C), passage 15 IPFSCs (C15C), SV40LT transduced passage 5 IPFSCs (S5C) and SV40LT transduced passage 15 IPFSCs (S15C), passage 15 IPFSCs were incubated in adipogenic induction medium for 21 days. To confirm adipogenic capacity of IPFSCs, TaqMan® real-time quantitative PCR was used to assess expression of adipogenic marker genes (LPL, CEBPA and PPARG) (B), western blot was used to evaluate LPL expression at a protein level (C), and Oil Red O (ORO) was used to stain lipid droplets (D). Gene expression data are exhibited as bar charts. Statistically significant differences are indicated using * when compared to CC, using $ when compared to corresponding C5C or C15C and using # when compared to corresponding C5C or S5C (p<0.05).
3.6. Potential mechanisms underlying matrix and SV40 mediated stem cell fate determination
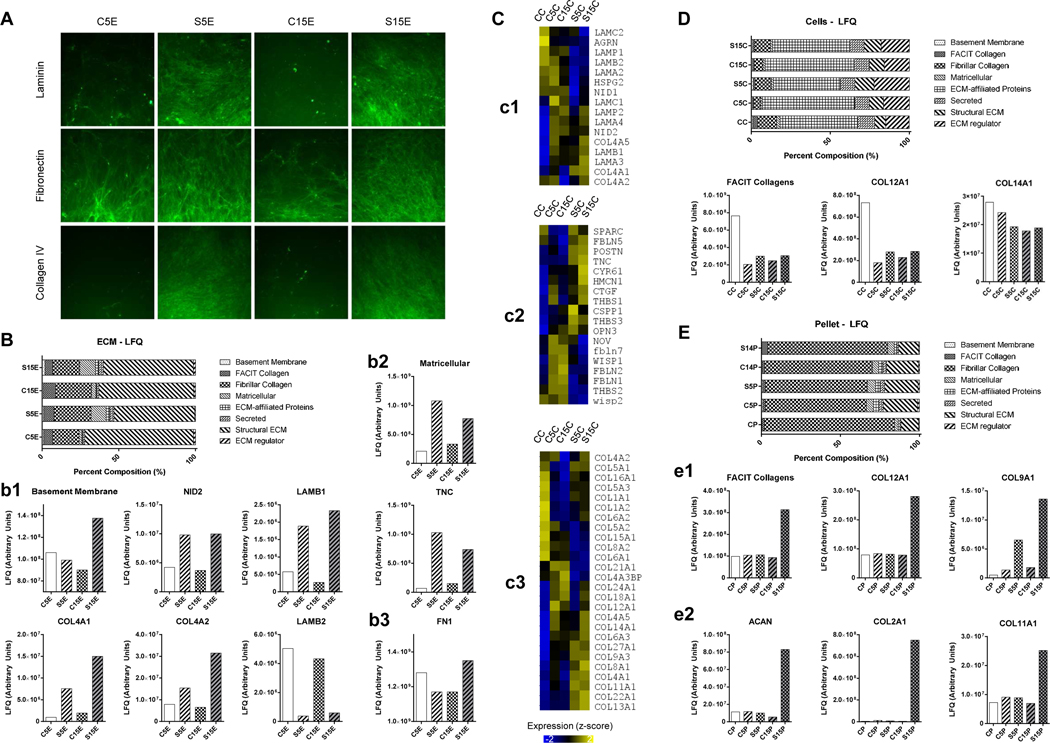
Untargeted proteomic analysis identified 3,060 proteins from 23,156 unique peptides in the dECMs, expanded cells and pellet samples, cumulatively. With a focus on the ECM, 188 matrisome-associated proteins were identified including ECM-affiliated proteins, secreted factors and ECM regulators; label-free quantitative (LFQ) values were used for relative comparisons between the samples. Immunofluorescence staining of typical matrix proteins indicated that both laminin and type IV collagen expression significantly increased while fibronectin expression moderately increased in the dECMs deposited by SV40LT IPFSCs, particularly for those deposited by high-passage cells (S15E) (Fig. 8A). The staining result was mirrored by proteomics data, in which basement membrane proteins (Fig. 8B/b1) markedly increased in dECM deposited by SV40LT transduced cells, notably a two- to ten-fold increase in COL4A1, COL4A2, nidogen 2 (NID2) and laminin beta1 (LAMB1), relative to their corresponding control. Interestingly, LAMB2 trends were opposite to both the gross laminin signal by immunostaining (Fig. 8A) and LAMB1 signaling (Fig. 8B/b1), possibly indicating a shift in laminin heterotrimer composition with immortalization. Additionally, the multi-modular matricellular protein TNC increased at least four-fold in dECM deposited by SV40LT cells relative to the corresponding controls (Fig. 8B/b2). LFQ values for FN1 (Fig. 8B/b3) confirmed an immunofluorescence signal with a modest increase in dECM deposited by SV40LT cells (Fig. 8A).
Figure 8.
Matrix component identification using immunostaining, proteomics and RNASeq analyses. Immunofluorescence staining of matrix proteins included laminin, fibronectin and type IV collagen in dECMs deposited by passage 5 IPFSCs (C5E), passage 15 IPFSCs (C15E), SV40LT transduced passage 5 IPFSCs (S5E) and SV40LT transduced passage 15 IPFSCs (S15E) (A). Proteomics analysis of dECMs (B) showed advantageous expression of basement membrane proteins (b1) and matricellular proteins (b2) in the dECM deposited by SV40 transduced IPFSCs, particularly for high-passage cells and fibronectin (b3). Heat map visualization of RNASeq gene expression level in passage 15 IPFSCs after growth on TCP (CC), C5E, C15E, S5E, S15E (C) for representative genes encoding basement membrane proteins (c1), matricellular proteins (c2) and collagens (c3). Expression values normalized into z-score across groups. Proteomics analysis of expanded cells (D) showed advantageous expression of FACIT collagens in passage 15 IPFSCs after growth on TCP (CC) compared to those grown on dECMs. Proteomics analysis of chondrogenic differentiated pellets (E) showed advantageous expression of FACIT collagens (COL9A1 and COL12A1) (e1) along with chondrogenic markers (ACAN, COL2A1 and COL11A1) (e2) in those from expansion on dECM deposited by SV40 transduced passage 15 IPFSCs.
Interestingly, the above dECM proteomics data (Fig. 8B) were also reflected in high-passage IPFSCs after growth on corresponding dECM in basement membrane protein genes (Fig. 8C/c1), matricellular protein genes (Fig. 8C/c2), and type IV collagen genes (Fig. 8C/c3) shown by expression level with RNASeq data. Fibril Associated Collagens with Interrupted Triple helices (FACIT) signal was the highest in the expanded cell control group (CC) (Fig. 8D); however, after chondrogenic induction, IPFSCs expanded on dECM deposited by high-passage SV40LT transduced cells yielded pellets (S15P) with the highest expression of FACIT collagen (Fig. 8E/e1) as well as chondrogenic markers ACAN (more than four-fold increase) and COL2A1 (more than two orders of magnitude) (Fig. 8E/e2), indicating that the FACIT collagen signal might correlate with chondrogenic differentiation.
4. Discussion
Donor age and long-term in vitro culture are both responsible for MSC senescence and are a challenge for stem cell-based tissue engineering and regeneration [7,27]. Previous reports indicate that SV40 transduction could immortalize primary cells but there were conflicting results in differentiation capacities [8]. In this study, we found that SV40LT transduction yielded human IPFSCs with significantly improved proliferation and adipogenic differentiation ability; however, chondrogenic capacity was dramatically decreased. Our data also demonstrated that SV40LT transduced IPFSCs had 5% abnormal chromosomes, making it unrealistic to use SV40LT transduced cells for clinical treatment. Increasing evidence indicates that dECM is a potential culture substrate to expand MSCs for rejuvenation and tissue regeneration [13,14]. dECM from young donors had a better rejuvenation effect than that from old donors [21]; however, most patients are elderly and allogenic/xenogenic sources of dECM from young donors might raise immune concerns [16]. In this study, we hypothesized that dECM deposited by SV40LT transduced autologous IPFSCs might be a better option to expand human adult IPFSCs. As expected, we found that dECM expansion significantly improved high-passage IPFSC proliferation, which was further promoted if dECM was deposited by SV40LT transduced cells. Surprisingly, expanded human IPFSCs on dECM yielded a reversed differentiation trend, that is, a decreased adipogenic but increased chondrogenic capacity, particularly for those deposited by SV40LT transduced high-passage IPFSCs.
One of our findings is that SV40LT transduction resulted in an upregulation of p53 at both the RNA and protein levels, together with a significantly increased expression of CDKN2A (encodes p16 and p14ARF). As a key player in regulation of cell cycle, p14ARF was reported to be activated in response to an oncogene, leading to p53 stabilization and accumulation [28]. By contrast, CDKN1A (encodes p21) showed a completely opposite trend in SV40LT transduced IPFSCs as the expression was significantly decreased compared with control groups. p21, a cyclin-dependent kinase inhibitor, can cause efficient G1 arrest and blockage of DNA replication from either the G1 or G2 phase [29]. p21 is also a major target of p53 in cell cycle arrest [30]. In this scenario, transduced SV40LT stimulation could upregulate the expression of CDKN2A, resulting in TP53 accumulation and consequent decrease of CDKN1A expression, leading to remarkably enhanced proliferation potential, which was also supported by increased cell percentages in the G2 and S phases as well as upregulated expression of stemness genes.
Immortalization using genetic modification is a useful way to combat cell senescence; however, there is the possibility of carcinogenicity. In this study, we found abnormal karyotypes in human IPFSCs after SV40LT transduction, which raised concerns for the safety of using the transduced cells themselves for clinical application. Instead, we found that expansion on dECM, especially that from SV40LT transduced cells, yielded IPFSCs with better proliferation abilities than TCP expansion, as evidenced by greater levels of cell growth, cell cycle and SOX2 and NES expression than the corresponding dECM groups, indicating that SV40LT transduction rendered dECM an appropriate microenvironment in favor of cell expansion. The beneficial effect of dECM on stem cell proliferation has been validated in human SDSCs [19], human bone marrow-derived MSCs (BMSCs) [31], human urine-derived stem cells [32] and porcine SDSCs [33] and IPFSCs [34,35]. Our proteomics data showed that more basement membrane proteins such as nidogen 2, laminins and type V collagen were expressed in dECMs deposited by SV40LT transduced IPFSCs, particularly for those from SV40LT transduced high-passage IPFSCs, supporting that basement membrane proteins may be actively involved in stem cell proliferation and stemness maintenance [36–38]. Interestingly, a study showed that dECM deposited by BMSCs and adipose-derived stromal cells enhanced the proliferation capacity of MSCs instead of cancer cell lines (HeLa, MCF7 and MDA-MB-231) [39], which further guarantees the safety of using these expansion substrates for stem cell rejuvenation.
Interestingly, SV40LT immortalization was found to dramatically decrease chondrogenic potential but increase hypertrophy in human IPFSCs. Despite the fact that it is the first report to demonstrate the compromised chondrogenic capacity of MSCs due to SV40LT transduction, our finding is consistent with previous reports focusing on the effect of SV40LT on redifferentiation of articular chondrocytes, in which SV40LT transduced equine articular chondrocytes exhibited decreased expression of Sox9, aggrecan, and type II collagen but increased type X collagen at both the mRNA and protein levels [40] and SV40LT immortalized rabbit articular chondrocytes lost their ability to re-express chondrocyte phenotypes during chondrogenic induction [41]. The inhibition of redifferentiation capacity might be associated with the modulation of protein kinase in SV40LT transduced chondrocytes [41–43]. Intriguingly, we also found that SV40LT immortalization significantly increased adipogenic potential in human IPFSCs. This finding is inconsistent with previous reports in which primary adipocyte precursor cells isolated from inguinal subcutaneous white adipose tissue (WAT) of SV40 transgenic mice exhibited a decreased adipogenic capacity compared with wildtype mice in response to external induction [44] and wildtype SV40LT effectively blocked adipogenic differentiation of preadipocytes (3T3-F442A cells) [45]. Evidence showed that preadiocytes from brown adipose tissue (BAT) reacted differently than those from WAT. For example, SV40 transduced BAT preadipocytes could differentiate into adipocytes [46]; however, SV40 transduced WAT preadipocytes would lose their adipogenic capacity [44,45], probably due to the inactivation of p300/CBP cAMP-response element-binding-protein in SV40LT transduced cells [47]. As a unique WAT, IPFP not only has inflammatory and immunology properties [48] but also has mechanical function and is relatively unresponsive to nutritional cues [49]. Our finding provides further evidence to support that the IPFP is a special kind of WAT given the enhanced capacity toward adipogenesis of SV40LT transduced preadipocytes from IPFP.
For the dECM experiment, we found that expansion on dECM deposited by low-passage IPFSCs promoted expanded cells’ chondrogenic potential while expansion on high-passage IPFSCs deposited dECM yielded IPFSCs with dramatically decreased chondrogenic differentiation, though both dECM expansion increased cell proliferation compared to expansion on TCP. This finding is consistent with a previous report in which adult SDSCs yielded a significant increase in proliferation and chondrogenic capacity after expansion on dECM deposited by fetal SDSCs versus adult SDSCs [21]. Interestingly, expansion on dECM deposited by SV40LT transduced cells yielded IPFSCs with significantly higher chondrogenic capacity, particularly for dECM deposited by high-passage IPFSCs. This finding is possibly related to a potential involvement of post-translation modification in the regulation of chondrogenesis as evidenced by upregulated expression of gene ontologies related to protein hydroxylation and protein folding chaperone in high-passage IPFSCs cultured on dECM deposited by SV40LT transduced cells compared to that by non-transduced cells. This internal gene alteration is possibly induced by external microenvironment changes as our proteomic result demonstrated more deposition of basement membrane proteins such as laminin and matricellular proteins such as tenascin and other proteins such as transforming growth factor-beta-induced protein ig-h3 (TGFBI) than the corresponding dECM groups. Increasing evidence showed that basement membrane proteins such as laminins are involved in chondrogenesis [50], indicating the importance of basement membrane proteins in stem cells’ chondrogenic differentiation. Tenascins are ECM glycoproteins, which can regulate cell adhesion either alone or by interacting with fibronectin [51]. Evidence showed that tenascin expression was significantly decreased in mature cartilage and almost completely disappeared in adult articular cartilage, indicative of the close association of tenascin with articular cartilage development [52]. TGFBI, a versatile matrix molecule induced by TGFβ, was reported to promote prechondrogenic cell adhesion and growth as well as to inhibit endochondral ossification [53]. In this scenario, the enrichment of these proteins creates a chondrogenic lineage-specific “niche” for the expanded IPFSCs, which might be responsible for the superior capacity of high-passage IPFSCs after expansion on SV40LT transduced high-passage cells in differentiating toward a chondrogenic lineage.
Despite the fact that SV40LT transduced IPFSCs exhibited an enhanced capacity for adipogenic differentiation, high-passage IPFSCs grown on dECMs achieved an opposite result, that is, a compromised adipogenic capacity. This conclusion is in accordance with our previous findings that expansion on dECM deposited by either IPFSCs or SDSCs suppressed the adipogenic differentiation of IPFSCs [34] and expansion on dECM deposited by BMSCs yielded human BMSCs with decreased adipogenic capacity [31]. This decreased adipogenic differentiation might be associated with the ‘spontaneous’ differentiation inhibiting effect of ECM as ECM was shown to interact with cell surface receptors and regulate growth factor activity, suppressing the absorbance of growth factors from serum [54,55].
5. Conclusion
For the first time, this study indicated that SV40LT transduction in human IPFSCs not only promoted proliferation but also increased adipogenic potential and decreased chondrogenic potential. Furthermore, we found that dECM deposited by SV40LT transduced IPFSCs reversed the differentiation preference of expanded IPFSCs by promoting chondrogenic potential but decreasing adipogenic capacity. Despite the unelucidated mystery underlying the rejuvenation effect of dECM deposited by high-passage SV40LT transduced cells, this study provided a promising model for in-depth investigation of ECM proteins in determining how these matrix proteins affect surrounding stem cells’ differentiation preference.
Supplementary Material
SV40LT transduction promoted human IPFSCs of passage 10 (A-C) and passage 15 (D-F) in cell proliferation. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Evaluation of proliferation used flow cytometry for cell cycle analysis (A/D) and relative EdU incorporation (B/E) as well as surface markers including CD73, CD90, CD105, CD146 and SSEA4 (C/F).
SV40LT transduction upregulated TP53 expression at a protein level in human IPFSCs. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Western blot was used to evaluate TP53 expression with GAPDH as an internal control.
Chondrogenic evaluation of SV40LT transduced IPFSCs of passage 10. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Fourteen-day chondrogenically induced pellets from passage 10 IPFSCs were evaluated using TaqMan® real-time quantitative PCR for chondrogenic marker genes (SOX9, COL2A1, ACAN and PRG4) and hypertrophic marker gene (COL10A1) (GAPDH served as an internal control). Data are exhibited as bar charts. * indicates a statistically significant difference compared to the corresponding GFP control (p<0.05).
Acknowledgements
We thank Suzanne Danley and Amanda Stewart for editing the manuscript. This project was supported by Research Grants from the National Institutes of Health (1R01AR067747) and the Musculoskeletal Transplant Foundation (MTF). We also would like to acknowledge the WVU Flow Cytometry & Single Cell Core Facility and the Bioinformatics Core as well as the grants that support the facility, WV-INBRE grant P20 GM103434, TME CoBRE grant P20GM131322 and the WV CTS grant 5U54 GM104942–04.
Footnotes
Conflict of Interest
None.
Credit Author Statement
Yiming Wang: Methodology, Validation, Formal analysis, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing. Gangqing Hu: Methodology, Software, Validation, Formal analysis, Writing – Review & Editing. Ryan C. Hill, Monika Dzieciatknowska, and Kirk C. Hansen: Software, Validation, Formal analysis, Writing – Review & Editing. Xiao-Bing Zhang and Zuoqin Yan: Methodology, Validation, Resources, Writing – Review & Editing. Ming Pei: Conceptualization, Methodology, Validation, Formal analysis, Writing – Review & Editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karnes J, Zhang Y, Pei M, Cell Therapy for the Creation of Cartilage and Related Clinical Trials, in Templeton NS (Ed), Gene and Cell Therapy: Therapeutic Mechanisms and Strategies, 4th Edition, Taylor & Francis/CRC Press, 2014, pp. 1123–1135. [Google Scholar]
- 2.Barry F, Murphy M, Mesenchymal stem cells in joint disease and repair, Nat. Rev. Rheumatol. 9 (2013) 584–594. [DOI] [PubMed] [Google Scholar]
- 3.Jones BA, Pei M, Synovium-derived stem cells: a tissue-specific stem cell for cartilage engineering and regeneration, Tissue Eng. Part B Rev. 18 (2012) 301–311. [DOI] [PubMed] [Google Scholar]
- 4.Pizzute T, Lynch K, Pei M, Impact of tissue-specific stem cells on lineage-specific differentiation: a focus on the musculoskeletal system, Stem Cell Rev. 11 (2015) 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Chen S, Pei M, Comparative advantages of infrapatellar fat pad: an emerging stem cell source for regenerative medicine, Rheumatology (Oxford) 57 (2018) 2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang TL, Hill RC, Dzieciatkowska M, Zhu L, Infante AM, Hu GQ, Hansen KC, Pei M, Site-dependent lineage preference of adipose stem cells, Front. Cell Dev. Biol. 8 (2020) 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Pei M, Cell senescence: a challenge in cartilage engineering and regeneration, Tissue Eng. Part B Rev. 18 (2012) 270–287. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Chen S, Yan Z, Pei M, A prospect of cell immortalization combined with matrix microenvironmental optimization strategy for tissue engineering and regeneration, Cell Biosci. 9 (2019) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shay JW, Pereira-Smith OM, Wright WE, A role for both RB and p53 in the regulation of human cellular senescence, Exp. Cell Res. 196 (1991) 33–39. [DOI] [PubMed] [Google Scholar]
- 10.Li N, Yang R, Zhang W, Dorfman H, Rao P, Gorlick R, Genetically transforming human mesenchymal stem cells to sarcomas: changes in cellular phenotype and multilineage differentiation potential, Cancer 115 (2009) 4795–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Yan L, Chen S, Pei M, Functionality of decellularized matrix in cartilage regeneration: a comparison of tissue versus cell sources, Acta Biomater. 74 (2018) 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Narayanan K, Zhang Y, Hill RC, He F, Hansen KC, Pei M, Role of lineage-specific matrix in stem cell chondrogenesis, Biomaterials 231 (2020) 119681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei M, Li J, Shoukry M, Zhang Y, A review of decellularized stem cell matrix: a novel cell expansion system for cartilage tissue engineering, Eur. Cell Mater. 22 (2011) 333–343; discussion 343. [DOI] [PubMed] [Google Scholar]
- 14.Pei M, Environmental preconditioning rejuvenates adult stem cells’ proliferation and chondrogenic potential, Biomaterials 117 (2017) 10–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han W, Singh NK, Kim JJ, Kim H, Kim BS, Park JY, Jang J, Cho DW, Directed differential behaviors of multipotent adult stem cells from decellularized tissue/organ extracellular matrix bioinks, Biomaterials 224 (2019) 119496. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Pizzute T, Li J, He F, Pei M, sb203580 preconditioning recharges matrix-expanded human adult stem cells for chondrogenesis in an inflammatory environment - A feasible approach for autologous stem cell based osteoarthritic cartilage repair, Biomaterials 64 (2015) 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S, Chen S, Jiang Q, Pei M, Determinants of stem cell lineage differentiation toward chondrogenesis versus adipogenesis, Cell Mol. Life Sci. 76 (2019) 1653–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohtani K, Suzumura A, Sawada M, Marunouchi T, Nakashima I, Takahashi A, Establishment of mouse oligodendrocyte/type-2 astrocyte lineage cell line by transfection with origin-defective simian virus 40 DNA, Cell Struct. Funct. 17 (1992) 325–333. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Pei M, A protocol to prepare decellularized stem cell matrix for rejuvenation of cell expansion and cartilage regeneration, Methods Mol. Biol. 1577 (2018) 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pizzute T, Zhang Y, He F, Pei M, Ascorbate-dependent impact on cell-derived matrix in modulation of stiffness and rejuvenation of infrapatellar fat derived stem cells toward chondrogenesis, Biomed. Mater. 11 (2016) 045009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Hansen KC, Zhang Y, Dong C, Dinu CZ, Dzieciatkowska M, Pei M, Rejuvenation of chondrogenic potential in a young stem cell microenvironment, Biomaterials 35 (2014) 642–653. [DOI] [PubMed] [Google Scholar]
- 22.Reisz JA, Nemkov T, Dzieciatkowska M, Culp-Hill R, Stefanoni D, Hill RC, Yoshida T, Dunham A, Kanias T, Dumont LJ, Busch M, Eisenmesser EZ, Zimring JC, Hansen KC, D’Alessandro A, Methylation of protein aspartates and deamidated asparagines as a function of blood bank storage and oxidative stress in human red blood cells, Transfusion 58 (2018) 2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data, Nucleic Acids Res. 47 (2019) D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W, The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote, Nucleic Acids Res. 41 (2013) e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y, Smyth GK, Shi W, The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads, Nucleic Acids Res. 47 (2019) e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl. Acad. Sci. USA 102 (2005) 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch K, Pei M, Age associated communication between cells and matrix: a potential impact on stem cell-based tissue regeneration strategies, Organogenesis 10 (2014) 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Xiong Y, Yarbrough WG, ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways, Cell 92 (1998) 725–734. [DOI] [PubMed] [Google Scholar]
- 29.Niculescu AB 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI, Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication, Mol. Cell Biol. 18 (1998) 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B, Requirement for p53 and p21 to sustain G2 arrest after DNA damage, Science 282 (1998) 1497–1501. [DOI] [PubMed] [Google Scholar]
- 31.Pei M, He F, Kish VL , Expansion on extracellular matrix deposited by human bone marrow stromal cells facilitates stem cell proliferation and tissue-specific lineage potential, Tissue Eng. Part A 17 (2011) 3067–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei M, Li J, Zhang Y, Liu G, Wei L, Zhang Y, Expansion on a matrix deposited by nonchondrogenic urine stem cells strengthens the chondrogenic capacity of repeated-passage bone marrow stromal cells, Cell Tissue Res. 356 (2014) 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Chen X, Pei M, Reconstruction of an in vitro tissue-specific microenvironment to rejuvenate synovium-derived stem cells for cartilage tissue engineering, Tissue Eng. Part A 15 (2009) 3809–3821. [DOI] [PubMed] [Google Scholar]
- 34.He F, Pei M, Extracellular matrix enhances differentiation of adipose stem cells from infrapatellar fat pad toward chondrogenesis, J. Tissue Eng. Regen. Med. 7 (2013) 73–84. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Fu Y, Yan Z, Zhang XB, Pei M, Impact of fibronectin knockout on proliferation and differentiation of human infrapatellar fat pad-derived stem cells, Front. Bioeng. Biotechnol. 7 (2019) 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs E, Finding one’s niche in the skin, Cell Stem Cell 4 (2009) 499–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs E, Tumbar T, Guasch G, Socializing with the neighbors: stem cells and their niche, Cell 116 (2004) 769–778. [DOI] [PubMed] [Google Scholar]
- 38.Schlötzer-Schrehardt U, Dietrich T, Saito K, Sorokin L, Sasaki T, Paulsson M, Kruse FE, Characterization of extracellular matrix components in the limbal epithelial stem cell compartment, Exp. Eye Res. 85 (2007) 845–860. [DOI] [PubMed] [Google Scholar]
- 39.Marinkovic M, Block TJ, Rakian R, Li Q, Wang E, Reilly MA, Dean DD, Chen XD, One size does not fit all: developing a cell-specific niche for in vitro study of cell behavior, Matrix Biol. 52–54 (2016) 426–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gurusinghe S, Hilbert B, Trope G, Wang L, Bandara N, Strappe P, Generation of immortalized equine chondrocytes with inducible Sox9 expression allows control of hypertrophic differentiation, J. Cell Biochem. 118 (2017) 1201–1215. [DOI] [PubMed] [Google Scholar]
- 41.Thenet S, Benya PD, Demignot S, Feunteun J, Adolphe M, SV40-immortalization of rabbit articular chondrocytes: alteration of differentiated functions, J. Cell Physiol. 150 (1992) 158–167. [DOI] [PubMed] [Google Scholar]
- 42.Benya PD, Padilla SR, Staurosporine, an inhibitor of protein kinase C, enhances reexpression of the differentiated chondrocyte collagen phenotype, Trans. Orthop. Res. Soc. 15 (1990) 183. [Google Scholar]
- 43.Scheidtmann KH, Haber A, Simian virus 40 large T antigen induces or activates a protein kinase which phosphorylates the transformation-associated protein p53, J. Virol. 64 (1990) 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Church C, Brown M, Rodeheffer MS, Conditional immortalization of primary adipocyte precursor cells, Adipocyte 4 (2015) 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherington V, Brown M, Paucha E, St Louis J, Spiegelman BM, Roberts TM, Separation of simian virus 40 large-T-antigen-transforming and origin-binding functions from the ability to block differentiation, Mol. Cell Biol. 8 (1988) 1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zilberfarb V, Piétri-Rouxel F, Jockers R, Krief S, Delouis C, Issad T, Strosberg AD, Human immortalized brown adipocytes express functional beta3-adrenoceptor coupled to lipolysis, J. Cell Sci. 110 (1997) 801–807. [DOI] [PubMed] [Google Scholar]
- 47.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM, Binding and modulation of p53 by p300/CBP coactivators, Nature 387 (1997) 823–827. [DOI] [PubMed] [Google Scholar]
- 48.Hui W, Litherland GJ, Elias MS, Kitson GI, Cawston TE, Rowan AD, Young DA, Leptin produced by joint white adipose tissue induces cartilage degradation via upregulation and activation of matrix metalloproteinases, Ann. Rheum. Dis. 71 (2012) 455–462. [DOI] [PubMed] [Google Scholar]
- 49.Gimble JM, Guilak F, Differentiation potential of adipose derived adult stem (ADAS) cells, In: Schatten GP (Ed), Current Topics in Developmental Biology, Elsevier Academic Press, London WC1X 8RR, UK, 2003, pp 140. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y, Wang T, Toh WS, Pei M, The role of laminins in cartilaginous tissues: from development to regeneration, Eur. Cell Mater. 34 (2017) 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumra H, Reinhardt DP, Fibronectin-targeted drug delivery in cancer, Adv. Drug Deliv. Rev. 97 (2016) 101–110. [DOI] [PubMed] [Google Scholar]
- 52.Hasegawa M, Yoshida T, Sudo A, Role of tenascin-C in articular cartilage, Mod. Rheumatol. 28 (2018) 215–220. [DOI] [PubMed] [Google Scholar]
- 53.Ohno S, Doi T, Tsutsumi S, Okada Y, Yoneno K, Kato Y, Tanne K, RGD-CAP ((beta)ig-h3) is expressed in precartilage condensation and in prehypertrophic chondrocytes during cartilage development, Biochim. Biophys. Acta 1572 (2002) 114–122. [DOI] [PubMed] [Google Scholar]
- 54.Chen XD, Dusevich V, Feng JQ, Manolagas SC, Jilka RL, Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts, J. Bone Miner. Res. 22 (2007) 1943–1956. [DOI] [PubMed] [Google Scholar]
- 55.Dallas SL, Rosser JL, Mundy GR, Bonewald LF, Proteolysis of latent transforming growth factor-beta (TGF-beta)-binding protein-1 by osteoclasts. A cellular mechanism for release of TGF-beta from bone matrix, J. Biol. Chem. 277 (2002) 21352–21360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SV40LT transduction promoted human IPFSCs of passage 10 (A-C) and passage 15 (D-F) in cell proliferation. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Evaluation of proliferation used flow cytometry for cell cycle analysis (A/D) and relative EdU incorporation (B/E) as well as surface markers including CD73, CD90, CD105, CD146 and SSEA4 (C/F).
SV40LT transduction upregulated TP53 expression at a protein level in human IPFSCs. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Western blot was used to evaluate TP53 expression with GAPDH as an internal control.
Chondrogenic evaluation of SV40LT transduced IPFSCs of passage 10. IPFSCs transduced with lentivirus carrying GFP served as a viral transduction control (GFP) and those without transduction served as a blank control (CTR). Fourteen-day chondrogenically induced pellets from passage 10 IPFSCs were evaluated using TaqMan® real-time quantitative PCR for chondrogenic marker genes (SOX9, COL2A1, ACAN and PRG4) and hypertrophic marker gene (COL10A1) (GAPDH served as an internal control). Data are exhibited as bar charts. * indicates a statistically significant difference compared to the corresponding GFP control (p<0.05).