Significance
Dengue virus is transmitted by mosquitoes and threatens more than half of the world population. In the absence of an effective vaccine and therapeutics and with a declining efficacy of insecticide, there is an urgent need to develop novel interventions. Here, we deployed an approach that combines metabolomics with molecular biology in mosquitoes. We revealed that dengue virus reconfigures the endomembrane phospholipids to form the structures required for its replication by activating the phospholipid remodeling cycle. We showed that a blood component that initiates de novo phospholipid production decreases mosquito infection. Overall, we revealed how and why dengue virus reconfigures phospholipids in mosquitoes. This understanding highlights intervention targets.
Keywords: mosquito, dengue, metabolomics, replication, phospholipid
Abstract
Dengue virus (DENV) subdues cell membranes for its cellular cycle by reconfiguring phospholipids in humans and mosquitoes. Here, we determined how and why DENV reconfigures phospholipids in the mosquito vector. By inhibiting and activating the de novo phospholipid biosynthesis, we demonstrated the antiviral impact of de novo–produced phospholipids. In line with the virus hijacking lipids for its benefit, metabolomics analyses indicated that DENV actively inhibited the de novo phospholipid pathway and instead triggered phospholipid remodeling. We demonstrated the early induction of remodeling during infection by using isotope tracing in mosquito cells. We then confirmed in mosquitoes the antiviral impact of de novo phospholipids by supplementing infectious blood meals with a de novo phospholipid precursor. Eventually, we determined that phospholipid reconfiguration was required for viral genome replication but not for the other steps of the virus cellular cycle. Overall, we now propose that DENV reconfigures phospholipids through the remodeling cycle to modify the endomembrane and facilitate formation of the replication complex. Furthermore, our study identified de novo phospholipid precursor as a blood determinant of DENV human-to-mosquito transmission.
Dengue virus (DENV) threatens more than half of the world population due to the global and expanding distribution of its mosquito vector, Aedes aegypti (1, 2). As an enveloped virus with a single-stranded positive-sense RNA genome [(+)gRNA], DENV relies on the host lipid membrane for its cellular cycle. Entry through endocytosis is mediated by attachment of envelope lipids to plasma membrane receptors (3). Upon internalization, the DENV envelope fuses with the plasma membrane, releasing the viral genome into the cytosol (4). The single open reading frame (ORF) is translated into a transmembrane polyprotein by ribosomes associated with the endoplasmic reticulum (ER) membrane (5). After cleavage by host and viral proteases, nonstructural viral proteins induce ER invaginations to form vesicle packets (VPs), encompassing clusters of double-membrane vesicles (Ve), which house the genome replication complex (6). The negative-sense RNA genome [(−)gRNA] is produced from the entering (+)gRNA and serves as a template for (+)gRNA production. VP and Ve structures enable efficient replication and protect the virus from host intracellular antiviral defense systems. On the ER, the nascent (+)gRNA is encapsidated and assembled into a lipid bilayer virion by host and virus structural proteins (7). Viral particles bud in the ER lumen and undergo maturation through the Golgi and trans-Golgi membrane networks (8), before release into extracellular space by fusion between endosome and plasma membranes.
Plasma and endosomal membranes are mainly composed of phospholipids (PL) (9). PLs contain one hydrophilic head group, a glycerol backbone, and two hydrophobic fatty acyl chains. Because of their amphiphilic nature, bilayer associations of PLs form cellular barriers. Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) represent more than 50% of cellular PL content, while in dipterans PE appears to be the predominant membrane constituent (10). PE and PC de novo biosynthesis occurs in the ER through parallel branches of the highly conserved Kennedy pathway: the cytidine 5′-diphosphate (CDP)–ethanolamine and CDP–choline pathways (9, 11, 12). In these pathways, one ethanolamine (Etn) or choline (Cho) is integrated into one diacylglycerol (DAG) to form one PE or PC, respectively (13). PE or PC then forms phosphatidylserine (PS) by a head group exchange reaction, while the reverse reaction of PS into PE also occurs (14, 15). PC, PE, and PS are called aminophospholipids (AminoPLs) based on their amine head group. Remodeling of PLs occurs through the Lands cycle (12, 16) by deacylation of one acyl chain to form lysophospholipid (LysoPL) and subsequent reacylation with a different acyl to generate a new PL species. While de novo PLs mostly contain saturated and monounsaturated acyls, remodeling is thought to increase unsaturation. Together, de novo synthesis and remodeling of PLs ensure maintenance and rearrangement of membrane composition, which determines membrane properties and structures (17). The polar head group and parallel acyl chains of PC result in a cylindrical geometry that forms planar bilayer membranes. The small head group of PE confers an inverted conical geometry, inducing negative curvature (18). Conversely, LysoPC and LysoPE with one acyl chain promote positive curvature and membrane defects that increase permeability (18), which regulates the integration of solutes to and from the cytosol. Composition of PL acyl chains also determines membrane behavior. High concentration of unsaturated PL leads to low lipid packing, increasing membrane fluidity (19, 20).
Cellular membranes undergo drastic modifications during DENV infection (6, 21), as illustrated by the large PL reconfiguration in both humans and mosquitoes (22–24). Understanding how DENV alters PL composition and the function of these alterations will provide insight into the virus biology that can be harnessed to design anti-DENV interventions. Recently, we described a major AminoPL reconfiguration throughout the DENV cycle in Ae. aegypti cells and mosquitoes (23). This reconfiguration is partially mediated by DENV-induced down-regulation of acylglycerolphosphate acyltransferase 1 (AGPAT1), which leads to a proviral environment. As AGPAT1 synthesizes the DAG precursor phosphatidic acid (PA), our preliminary study suggests that DENV-mediated inhibition of the de novo PL pathway favors infection. Here, we aim to decipher how DENV reconfigures AminoPL in mosquitoes—disentangling the role of de novo biosynthesis and remodeling—and the function of AminoPL reconfiguration in the virus cellular cycle. We used enzyme depletion to inhibit the de novo pathway and precursor supplementation to activate it. We confirmed these effects with metabolomic analysis and showed that de novo AminoPL synthesis is detrimental to DENV in vitro. Accordingly, we observed that DENV actively inhibits the de novo pathway and instead induces remodeling. Using pioneering lipid isotopic labeling methods in mosquito cells, we confirmed that the DENV AminoPL reconfiguration is initiated through remodeling. Importantly, we observed that activation of de novo PL biosynthesis in mosquitoes by supplementing infectious blood with de novo precursor reduced infection, thereby identifying a blood metabolite that reduces mosquito infection. Finally, we demonstrated that activation of the de novo pathway does not alter virus attachment, internalization, translation, or particle production but hampers replication, diminishing production of infectious particles.
Results
Inhibition of the De Novo Phospholipid Pathway Favors DENV Multiplication.
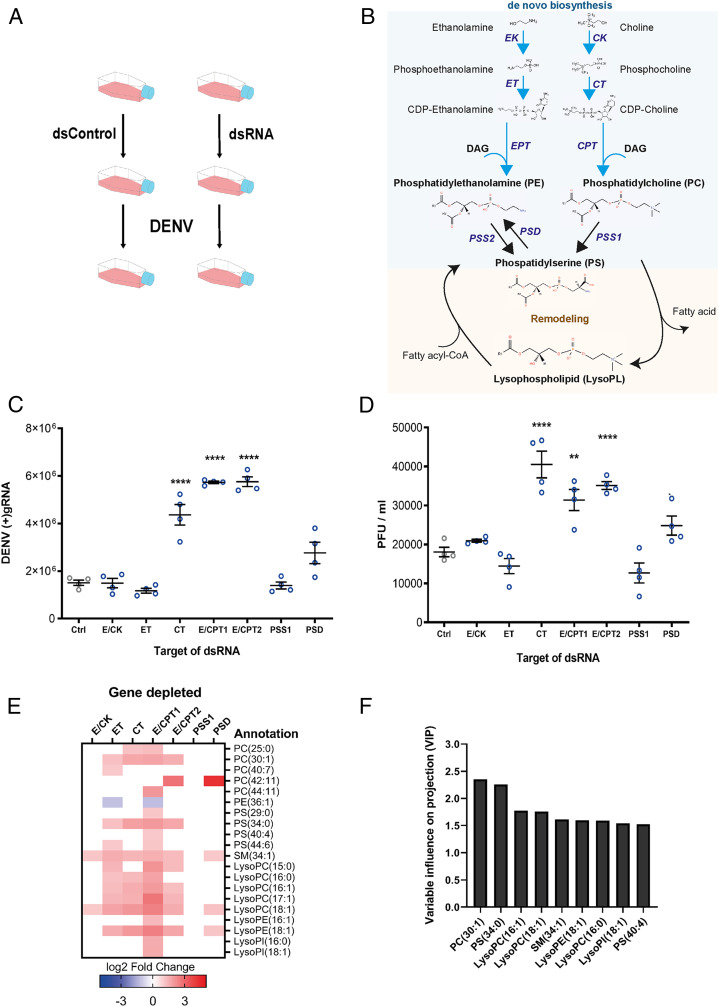
To determine how de novo AminoPL biosynthesis influences DENV infection, we individually silenced seven enzymes that catalyze different steps of the two branches (CDP–Etn and CDP–Cho) via RNA interference (RNAi) silencing in the Ae. aegypti cells, Aag2, before infecting with DENV at a multiplicity of infection (MOI) of 1 (Fig. 1 A and B and SI Appendix, Fig. S1). In insects, certain enzymes recognize the substrates from both branches, catalyzing the same step in the parallel biosyntheses (11). We depleted the kinase (E/CK) responsible for both Etn and Cho transformation into phosphoethanolamine and phosphocholine, respectively. We also depleted the two transferases (ET and CT) that synthesize CDP–Etn and CDP–Cho from the above products. We depleted the two transferases (E/CPT1 and E/CPT2) that then synthesize both PE and PC by introgression of one DAG. Both of these enzymes may recognize CDP–Etn and CDP–Cho based on their similarity to human CDP–amino alcohol phosphotransferase CEPT1, which catalyzes CDP–Etn and CDP–Cho (25, 26) (SI Appendix, Fig. S2 and Table S1). We also depleted the synthetase (PSS1) responsible for PC transformation into PS and the decarboxylase (PSD) that catalyzes PS transformation into PE. Cell survival was not affected by the enzyme depletion (SI Appendix, Fig. S3). At 48 h postinfection (hpi), we measured cellular infection by quantifying (+)gRNA in cells and production of infectious particles by determining plaque-forming units (PFUs) in supernatant. Both infection parameters showed an identical pattern (Fig. 1 C and D). Although the first step (E/CK) of the de novo pathway and the second step (ET) of the CDP–Etn branch did not alter virus multiplication, synthesis inhibition of CDP–Cho (CT) and PC and PE (E/CPT1 and E/CPT2) increased (+)gRNA and PFUs per milliliter. Of note, E/CPT1 and E/CPT2 enzymes catalyze the rate-limiting step in the pathway (11). Alteration of PS synthesis (PSS1) did not significantly influence virus production, whereas PSD depletion, resulting in a decrease of alternative PE synthesis, moderately increased (+)gRNA.
Fig. 1.
Impact of de novo PL pathway disruptions on DENV and PL reconfiguration. Aag2 cells were transfected with dsRNA targeting different enzymes of the de novo pathway or control dsRNA (Ctrl). Twenty-four hours later, cells were infected with DENV at a MOI of 1. Samples were collected at 48 hpi. (A) Experimental design. (B) Scheme of the PL de novo and remodeling pathways. Ethanolamine and choline are phosphorylated by E/CK and then integrate a cytidine diphosphate group by CTP–ethanolamine or CTP–phosphocholine cytidyltransferases (ET or CT). The CDP–ethanolamine and CDP–choline formed incorporate a DAG by DAG:CDP–ethanolamine ethanolaminephosphotransferase (EPT) or DAG:CDP–choline cholinephosphotransferase (CPT) to produce PE and PC, respectively. In mosquitoes, EPT and CPT catalyze both PE and PC and were named E/CPT. PS is produced by a head exchange reaction from PC or PE by PSS. PE is reversely produced by PSD. PL remodeling starts with deacylation by PLA2 to produce LysoPLs. LysoPLs are then reacylated by lysophospholipid acyltransferase via incorporation of another fatty acid to form a new PL species. (C and D) Impact of gene depletions on DENV (+)gRNA copies in cells (C) and on PFUs in supernatants (D). Lines show mean ± SEM from four biological repeats. **P < 0.01; ****P < 0.0001 as determined by Dunnett’s multiple comparison test. (E) DENV-induced phospholipidome reconfiguration upon enzyme depletion. Fold changes of annotated and significantly regulated metabolites (|log2 fold change| > 1 and P < 0.05) for each depleted enzyme compared to dsRNA control from three biological repeats. (F) VIP for each metabolite. Bars show VIP score > 1.5 as determined by PLS analysis with DENV production (PFU/mL) as the response variable and metabolite concentrations as predictors.
To characterize the proviral metabolic environment induced by inhibition of de novo AminoPL synthesis, we quantified changes in PLs upon DENV infection in enzyme-depleted mosquito cells compared to silencing control. Overall, proviral conditions (CT, E/CPT1, E/CPT2, and PSD depletions) were associated with an increase in PC, PS, sphingomyelin (SM), LysoPC, LysoPE, and lysophosphatidylinositol (LysoPI) compounds and a decrease in one PE, as illustrated by metabolite fold changes (Fig. 1E) and ion intensity (SI Appendix, Fig. S4 and Dataset S1). However, the neutral conditions—E/CK and ET depletions—were also associated with AminoPL increases, and several metabolites were similarly regulated as in proviral conditions. To correlate unique metabolite profiles with DENV titer, we performed a partial least squares regression (PLS) (27, 28) including the proviral and neutral conditions. Higher concentrations of PC (30:1) and PS (34:0) were the best predictors (highest variable importance in projection [VIP] scores) of higher DENV titer (Fig. 1F). Three LysoPCs, one LysoPE, one LysoPI, SM (34:1), and PS (40:4) then correlated best with DENV production. Together, those results indicate that disruption of the CDP–Etn and CDP–Cho branches and PS-derived PE synthesis favor DENV production (likely by allowing the virus to reconfigure AminoPLs for its benefit). This finding agrees with our previous observation that synthesis inhibition of a de novo pathway precursor favors infection in cells and mosquitoes (23). Moreover, up-regulation of LysoPLs suggests that DENV increased PL remodeling when de novo production of PLs is inhibited.
Activation of the De Novo Phospholipid Pathway Reduces DENV Multiplication.
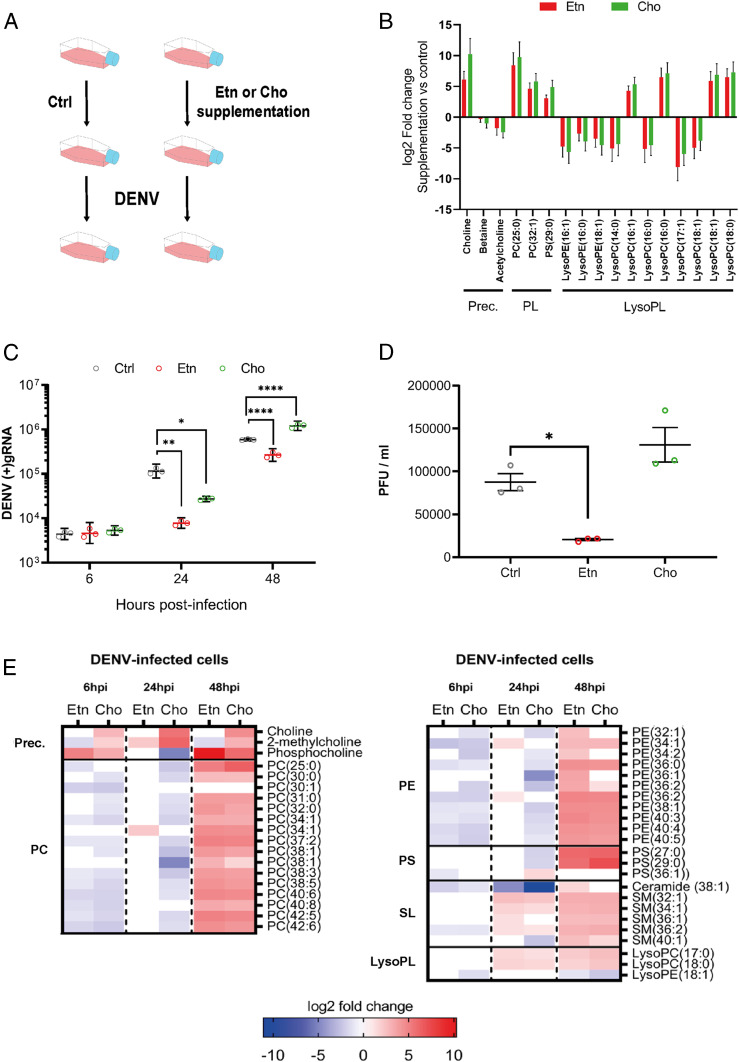
To test whether de novo PL biosynthesis restricts DENV multiplication, we activated one of the branches of the de novo pathway by supplementing mosquito cells with Etn or Cho precursor (Fig. 1B) for 24 h before and throughout infection (Fig. 2A and SI Appendix, Fig. S5) (29). Metabolomics analysis of supplemented cells before infection confirmed Cho uptake, while Etn was too small and nonpolar to be detected (Fig. 2B and Dataset S2). However, we observed an increase in Cho upon Etn supplementation. This can result from PC recycling for PL homeostasis (30) and is, therefore, indicative of Etn uptake. Overall, Cho and Etn supplementation similarly regulated AminoPLs before infection (Fig. 2B), suggesting interconnections and compensation mechanisms between the two branches of the de novo pathway (30). Two PCs, one PS, and four LysoPC species were increased, whereas four other LysoPCs and three LysoPEs were decreased. These results show that Etn and Cho uptakes activate AminoPL synthesis, most likely through the de novo pathway, while, as previously reported (31), de novo activation also alters PL remodeling, as indicated by changes in LysoPLs.
Fig. 2.
Impact of de novo PL production on DENV and phospholipid reconfiguration. Aag2 cells were supplemented with 2 mM of either Etn or Cho for 24 h before and throughout infection with DENV at a MOI of 1. (A) Experimental design. (B) Impact of the supplementations on the phospholipidome before infection. Fold changes of only significantly regulated metabolites (|log2 fold change| > 1 and P < 0.05) with a lipid or precursor (Prec.) annotation compared to control media are shown. Lines indicate means ± SEM from three biological repeats. (C) Impact of the supplementations on DENV (+)gRNA at 6, 24, and 48 hpi. Lines show geometric means ± 95% CI from three biological repeats. *P < 0.05; **P < 0.01; ****P < 0.0001, as determined by Dunnett’s multiple comparison test. (D) Impact of the supplementations on infectious particle production as determined by PFUs. Lines show mean ± SEM from three biological repeats. *P < 0.05, as determined by Dunnett’s multiple comparison test. (E) Impact of the supplementations on the phospholipidome at 6, 24, and 48 hpi. Fold changes of significantly regulated metabolites (|log2 fold change| > 1 and P < 0.05) compared to media control within the same time point for three biological replicates are shown. SL, sphingolipid.
We then quantified the effect of Etn and Cho supplementations on cellular (+)gRNA at 6, 24, and 48 hpi and on infectious particles (PFU/mL) at 48 hpi with a MOI of 1. Both supplementations had no impact at 6 hpi and reduced (+)gRNA at 24 hpi (Fig. 2C). However, at 48 hpi, whereas Etn maintained a lower (+)gRNA level, Cho negative impact was reverted. Accordingly, infectious particles at 48 hpi solely decreased upon Etn supplementation (Fig. 2D). We reasoned that the negative impact of Cho supplementation was compensated by DENV-induced reconfiguration. To increase DENV reconfiguring pressure, we infected with a higher MOI of 5. In accordance with DENV countering the negative impact of de novo PLs, at 24 hpi, Cho no longer reduced (+)gRNA, and the negative impact of Etn was reduced (SI Appendix, Fig. S6).
To understand how DENV countered de novo PL production, we profiled the phospholipidome of Etn- and Cho-supplemented cells at 6, 24, and 48 hpi with a MOI of 1. At 6 hpi, we noted an increase in the de novo pathway precursors (i.e., Cho, 2-methylcholine, and phosphocholine) and a decrease in numerous PCs and PEs and one PS with either Etn or Cho (Fig. 2E, SI Appendix, Fig. S7, and Dataset S3), suggesting inhibition of de novo PL production. At 24 hpi with either Etn or Cho, we observed an increase in two LysoPCs and several SMs, which derive from LysoPC, indicating PL deacylation. Upon Etn, increases in PC(34:1) and two PEs could have resulted from either de novo or remodeling, whereas upon Cho, sustained reduction of multiple PCs and PEs indicates inhibition of PL production. At 48 hpi, all regulated PCs, PSs, SMs, and LysoPCs and nine PEs were up-regulated in both supplementations, suggesting active PL production. However, three other PEs increased only with Etn and may account for the antiviral impact observed for Etn but not Cho supplementation at 48 hpi (Fig. 2 C and D). Altogether, these results demonstrate that de novo PLs are antiviral and that DENV counters their impact by reconfiguring AminoPLs through early de novo inhibition, followed by remodeling activation.
DENV Induces Early Phospholipid Remodeling.
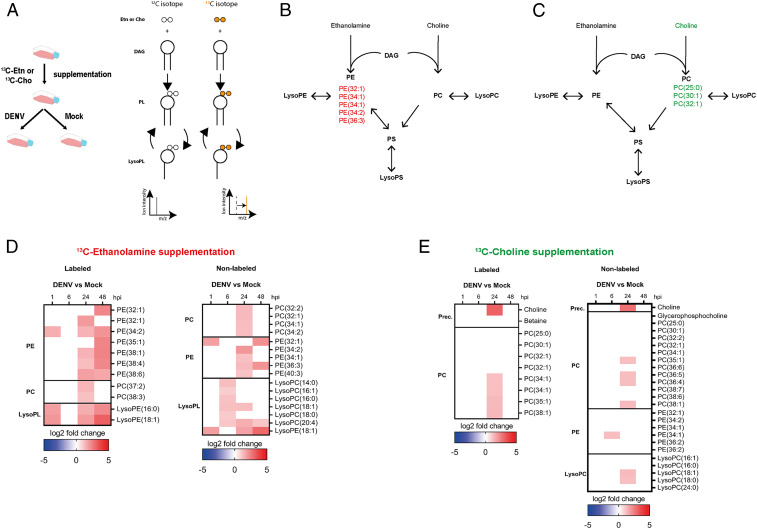
To determine how DENV regulates PL biosynthesis, we steadily supplemented mosquito cells with 100% 13C Etn or 99.9% 13C Cho and followed changes in isotope labeled and nonlabeled PEs and PCs at 1, 6, 24, and 48 hpi with a MOI of 1 compared to mock (Fig. 3A and SI Appendix, Fig. S8). The labels were integrated in de novo–produced PLs, which could then be further modified into labeled LysoPLs (Fig. 3A). Before infection, we supplemented cells with isotopes for 24 h to label only PEs upon Etn or PCs upon Cho supplementation (Fig. 3 B and C and SI Appendix, Table S2). In the same conditions, we also detected nonlabeled PLs and LysoPLs (Dataset S4), synthesized with unlabeled Etn and Cho that were available before the supplementations. Upon infection, we were able to differentiate de novo synthesis from remodeling. De novo synthesis was identified by 1) uptake of labeled precursor (only Cho could be detected), needed to fuel de novo production, and 2) increase in labeled PL without detecting labeled LysoPL, indicating the absence of remodeling. Remodeling was identified by 1) detection of labeled LysoPL, which can only result from deacylation of preinfection labeled PL, and 2) an increase in nonlabeled PL, which can only be synthesized through remodeling of PL produced before supplementation. Moreover, it is assumed that the de novo pathway produces saturated and monounsaturated PLs, whereas remodeling generates mono- and polyunsaturated PLs (32).
Fig. 3.
Impact of DENV on PL biosynthesis. Aag2 cells were supplemented with 2 mM of either 13C Etn or Cho for 24 h before and throughout infection with DENV at a MOI of 1 or mock. Metabolomics analysis was conducted at 1, 6, 24, and 48 hpi. (A) Experimental design and scheme of isotope tracing in PL biosynthesis. (B and C) Isotope-labeled PL before infection upon Etn (B) or Cho (C) supplementation. (D and E) Impact of DENV on labeled and nonlabeled PL at 1, 6, 24, and 48 hpi upon Etn (D) and Cho (E) supplementation. Fold changes of annotated and significantly regulated PLs (|log2 fold change| > 1 and P < 0.05) compared to mock.
Upon Etn supplementation at 1 hpi, labeled LysoPEs and PE (34:2) and nonlabeled LysoPE and PE(32:1) increased (Fig. 3D, SI Appendix, Fig. S9, and Dataset S5), all indicating remodeling induction. At 24 and 48 hpi, an increase in labeled LysoPEs and polyunsaturated PEs and nonlabeled PEs confirmed remodeling. Higher concentrations of labeled monounsaturated PEs might have resulted from either de novo synthesis or remodeling. Further supporting remodeling, nonlabeled LysoPCs increased at 6 hpi and were followed by an increase in nonlabeled unsaturated PCs at 24 hpi. Unexpectedly, there was an increase in labeled polyunsaturated PCs at 24 hpi that could not be produced from labeled Etn and may stem from PE methylation, which was observed in yeast and mammals (33).
Upon Cho supplementation, we did not observe changes in PC and LysoPC at 1 and 6 hpi (Fig. 3E, SI Appendix, Fig. S9, and Dataset S5). At 24 hpi, uptake of labeled and nonlabeled Cho and an increase in labeled PC without detection of labeled LysoPC indicated de novo pathway activation. However, an increase in nonlabeled LysoPC and polyunsaturated PC at the same time also suggests remodeling. At 6 hpi, we observed an increase in nonlabeled PE (34:1), which coincides with remodeling in the Etn supplementation assay. Altogether, isotopic tracing shows that DENV induces remodeling early during infection (within the first 24 h), followed by de novo synthesis.
High Blood Concentration of a De Novo Phospholipid Precursor Reduces DENV Infection in Mosquitoes.
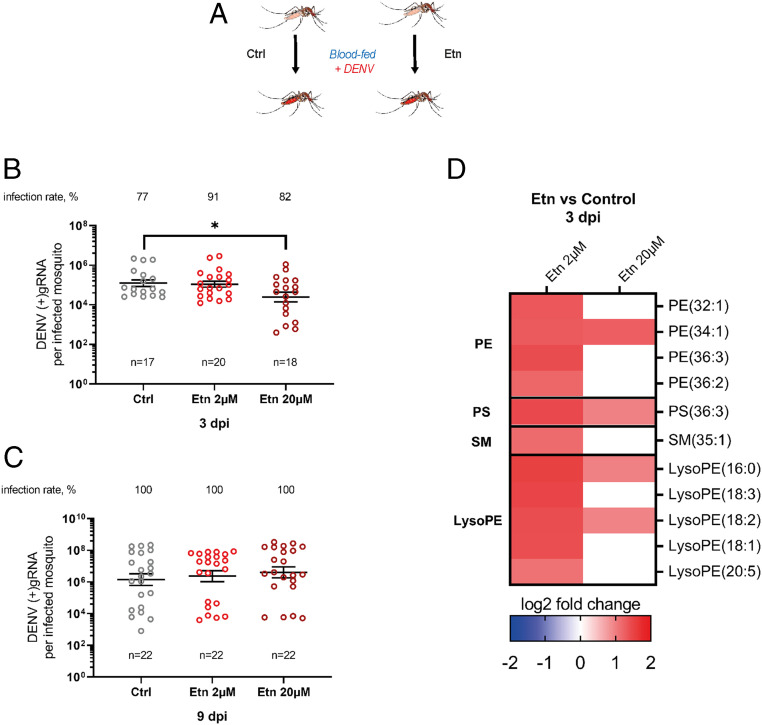
To test whether Etn also reduces DENV infection in vivo, we fed Ae. aegypti mosquitoes an infectious blood meal supplemented with either the Etn level found in healthy patients (2 µM) (34) or 10 times more (20 µM) and quantified (+)gRNA at 3 and 9 d postinfection (dpi) (Fig. 4A). Nonsupplemented blood was used as control, and Etn blood supplementation did not influence feeding rate (SI Appendix, Fig. S10). Interestingly, while 2 µM supplementation had no effect, supplementation with 20 µM of Etn decreased DENV infection at 3 dpi (Fig. 4B). At 9 dpi there were no differences in DENV infection levels (Fig. 4C), likely due to the ability of DENV to overcome an antiviral metabolic environment in mosquitoes.
Fig. 4.
Impact of ethanolamine blood supplementation on DENV infection in orally infected mosquitoes. Mosquitoes were fed a DENV infectious blood meal supplemented with 0 (Ctrl), 2, or 20 µM of Etn. (A) Experimental design. (B and C) DENV (+)gRNA copies and infection rate at 3 (B) and 9 (C) dpi in whole mosquitoes. Lines show geometric means ± 95% CI. n, number of mosquitoes. *P < 0.05, as determined by Dunnett’s multiple comparison test. (D) Phospholipidome of mosquitoes at 3 dpi. Fold changes of significantly regulated metabolites (|log2 fold change| > 1 and P < 0.05) compared to control from four biological replicates.
To understand how DENV reconfigures the metabolome for its benefit, we quantified changes in the mosquito phospholipidome at 3 dpi upon blood supplementation with 2 or 20 µM of Etn compared to no supplementation. First, although other PL classes were detected (Dataset S6), we observed an exclusive up-regulation of PEs, LysoPEs, PSs, and SM, the last two being synthesized from PE (Fig. 4D and SI Appendix, Fig. S11). This indicates that upon Etn supplementation DENV selectively regulated PE, most likely to compensate for the de novo PE production induced by the precursor supplementation. Second, with the higher Etn level, DENV up-regulated a smaller number of LysoPEs (two with 20 µM vs. five with 2 µM) and unsaturated PEs (one with 20 µM vs. four with 2 µM) than with the lower Etn concentration. As only the higher Etn concentration inhibited DENV (Fig. 4B), the metabolomics results indicate that DENV was unable to sufficiently reconfigure the lipidome with the higher concentration. This may be due to the previously reported antagonistic effect between de novo and remodeling pathways (31). Altogether, our in vivo data confirmed the antiviral effect of de novo PLs and suggested the DENV induction of remodeling.
De Novo Phospholipid Production Disrupts Replication.
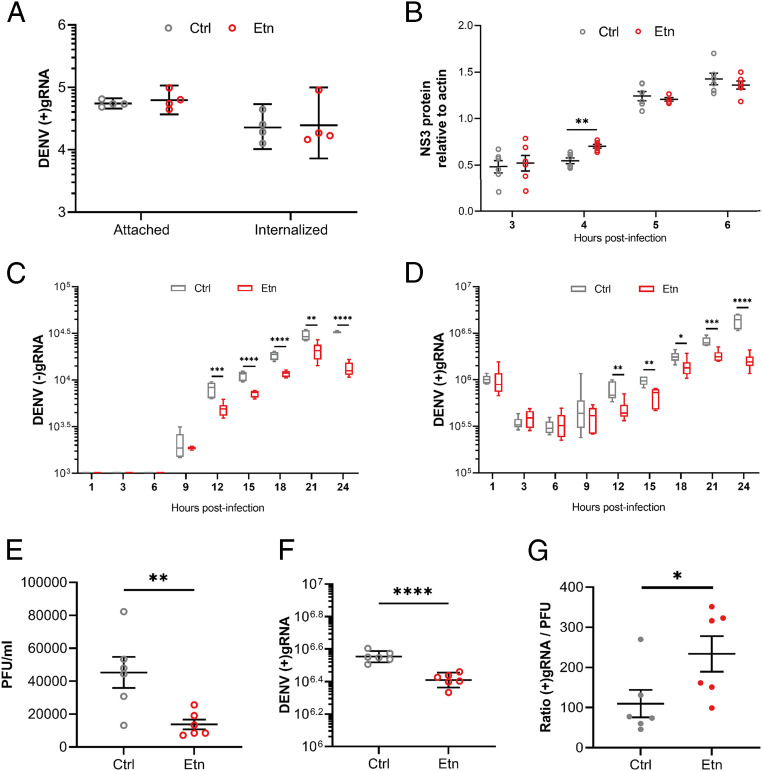
To determine what step(s) of the DENV cellular cycle requires PL reconfiguration, we activated de novo PLs by supplementing mosquito cells with Etn for 24 h before and throughout infection and quantified virus attachment, internalization, translation, replication, and particle infectivity compared to nonsupplemented conditions. At the onset of infection, attachment and internalization as quantified by numbers of attached and internalized (+)gRNA were not affected (Fig. 5A). Translation as measured by virus nonstructural 3 (NS3) protein quantity from 3 to 6 hpi was not altered either, except for an increase at 4 hpi, which does not match the repeatedly observed antiviral effect (Fig. 5B and SI Appendix, Fig. S12). However, replication estimated as the kinetics of antigenome [(−)gRNA] and (+)gRNA production from 1 to 21 hpi was inhibited by Etn (Fig. 5 C and D). (−)gRNA was detected from 9 hpi and was sustainably reduced by Etn from 12 to 24 hpi. In line with the appearance of its (−)gRNA template at 9 hpi, (+)gRNA amplification started at 12 hpi and was inhibited until 24 hpi. Eventually, infectious particles as quantified by PFUs per milliliter in supernatant were reduced at 32 hpi (Fig. 5E), similar to what we observed at 48 hpi (Fig. 2D). As replication deficiency is expected to produce deficient genomes, we calculated the ratio of genome/infectious particle by quantifying (+)gRNA copies in supernatant using qPCR amplification of a fragment of the envelope gene. In accordance with the higher number of defective particles, we observed a higher (+)gRNA:PFU ratio upon Etn supplementation (Fig. 5 F and G). To ensure that modification in the virion outer layers was not responsible for the observed defective particles, we quantified attachment and internalization for viruses produced in Etn-supplemented and nonsupplemented mosquito cells. Using the same volume for Etn- and control-produced virions, we did not observe an impact on attached and internalized genomes (SI Appendix, Fig. S13), indicating that Etn supplementation does not alter internalization capacity, thus supporting the production of deficient genomes, which reduces the infectious particles. Altogether, our results show that hindering the DENV-induced reconfiguration by de novo PL production disrupts replication.
Fig. 5.
Impact of de novo PL pathway on DENV cellular cycle. Aag2 cells were supplemented with Etn or control media (Ctrl) for 24 h before and throughout infection with DENV at a MOI of 1. (A) Numbers of attached and internalized (+)gRNA. (B) NS3 protein level at 3, 4, 5, and 6 hpi. Actin was used for normalization. (C and D) Numbers of cellular (−)gRNA (C) and (+)gRNA (D) per well at 1, 3, 6, 9, 12, 15, 18, 21, and 24 hpi. (E) PFU per milliliter in supernatant at 32 hpi. (F) Number of (+)gRNA per milliliter in supernatant at 32 hpi. (G) Ratio of (+)gRNA/PFU in supernatant at 32 hpi. (A, C, D, and F) Lines show geometric means ± 95% CI. Four (A) and six (B) biological repeats were collected from independent wells. (B, E, and G) Lines show mean ± SEM (C and D) Tukey box plots. (C, D, and E–G) Six biological replicates were collected from two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, as determined by Dunnett’s multiple comparisons test (A–D), an unpaired t test (E and F), or the Mann–Whitney U test (G).
Discussion
Several studies, including ours (23), have described a PL reconfiguration upon DENV infection in mosquitoes (22, 35) and mammals (36–38). Here, we determine how and why DENV reconfigures PLs in mosquitoes. There are two major ways PLs are generated: de novo synthesis and remodeling, each producing PLs with different biochemical properties, which could influence DENV infection. Our metabolomics results and isotope tracing assay clearly demonstrate that DENV induces remodeling in mosquito cells and mosquitoes. There exists an antagonistic interaction between de novo synthesis and remodeling of PLs, where activation or inhibition of one inversely regulates the other (31, 39). Our observations that inhibition of de novo synthesis was proviral and activation was antiviral indirectly support that remodeling benefits DENV. Our study demonstrates that DENV actively reconfigures the phospholipidome by inducing PL remodeling to increase its multiplication in mosquitoes.
Remodeled PLs have a higher number of unsaturations than those produced by de novo biosynthesis and thus should increase membrane fluidity (19, 20), which facilitates invagination required for VP and Ve formation (6). Accordingly, we determined that efficient replication but not translation and assembly depends on DENV-induced PL reconfiguration through remodeling. In human cells, activation of phospholipase A2 (PLA2), which produces LysoPLs, was previously shown to be important in Ve formation for West Nile virus, another mosquito-borne flavivirus (40). As LysoPCs colocalized with Ve and PLA2 depletion deformed Ve, the authors suggested that the conical shape of LysoPLs had a direct role in membrane curvature. Our discovery of the role of remodeling in replication indicates another model whereby DENV activates PL deacylation at the replication site for remodeling. This model is supported by the previously reported recruitment of fatty acyl synthase (FAS) by DENV NS3 at the replication site (41). Fatty acyls locally produced by FAS could be used locally for reacylation of LysoPLs to complete PL remodeling.
A better understanding of DENV metabolic requirements in mosquitoes will lead to identification of targets for transmission-blocking strategies. As mosquitoes cannot synthesize and must ingest Etn, we identified blood Etn level as a determinant of transmission from human to mosquito.
Materials and Methods
Aedes aegypti Mosquitoes and Cell Line.
The Aedes aegypti colony was established in 2010 from Singapore and was reared at 28 °C and 60% relative humidity with 12 h:12 h light:dark cycle. Eggs hatched in Milli-Q water were fed with a mix of fish food (TetraMin fish flakes), yeast, and liver powder (MP Biomedicals). Adults were held in rearing cages (Bioquip) supplemented with water and 10% sucrose solution.
Aedes aegypti Aag2 cells (42) were cultured in RPMI 1640 medium (Gibco) with 10% heat-inactivated fetal bovine serum (FBS) (HyClone) and 1% penicillin–streptomycin (Gibco). For media supplementation, either choline chloride (Sigma-Aldrich), ethanolamine (Sigma-Aldrich), choline–13C2 chloride (Sigma-Aldrich), or ethanolamine–13C2 (Sigma-Aldrich) was added to Aag2 growth medium 24 h before infection. Cells were maintained in vented culture flasks in a humidified incubator with 5% CO2 at 28 °C. BHK-21 (baby hamster kidney) (ATCC CCL-10) cells were cultured in the same media and maintained at 37 °C with 5% CO2.
Dengue Virus.
Dengue virus serotype 2 strain ST (DENV) was collected from the Singapore General Hospital in 1997 (43). DENV was propagated alternatively in Vero (ATCC CCL-81) and C6/36 (ATCC CRL-1660) cells. Virus titer was determined by plaque assay using BHK-21 cells (23).
Double-Stranded RNA-Mediated RNAi.
Templates for dsRNA against AAEL009765 (EK/CK, ethanolamine/choline kinase), AAEL011564 (CT, CTP:phosphocholine cytidyltransferase), AAEL014395 (E/CPT1, DAG:CDP–choline ethanolamine/cholinephosphotransferase 1), AAEL011841 (E/CPT2, DAG:CDP–choline ethanolamine/cholinephosphotransferase 2), AAEL005651 (ET, CTP:phosphoethanolamine cytidyltransferase), AAEL010223 (PSD, PS decarboxylase), and AAEL008393 (PSS, PS synthase) were PCR amplified from Ae. aegypti complementary DNA (cDNA) with primers (SI Appendix, Table S3) flanked with T7 promoter. dsRNA was synthetized with a megaScript T7 transcription kit (Thermo Fisher Scientific), purified with an E.Z.N.A. Total RNA Kit I (Omega Bio-tek) in diethyl pyrocarbonate (DEPC)-treated water, and annealed by slowly cooling down. Control dsRNA targeting LacZ was produced similarly from a plasmid (44); 2 × 105 Aag2 cells were transfected with 1 µg of dsRNA by using a TransIT-mRNA Transfection Kit (Mirusbio).
Protein Sequence Alignment.
Amino acid sequence homology was determined by MEGA X software (version 10.0.5) using maximum likelihood and bootstrapping. FASTA sequences were retrieved from https://www.ncbi.nlm.nih.gov/.
Quantification of Gene Expression.
Total RNA from Aag2 cells was extracted using an E.Z.N.A. Total RNA Kit I, treated with a RapidOut DNA Removal Kit (Thermo Fisher Scientific) and reverse transcribed with iScript cDNA Synthesis Kit (Bio-Rad). Gene expression was quantified using iTaq Universal SYBR Green Supermix (Bio-Rad) and primers detailed in SI Appendix, Table S4. The ΔΔCT method was used to calculate relative changes in gene expression. Actin expression was used for normalization. Quantification was conducted in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The thermal profile was 95 °C for 1 min and 40 cycles of 95 °C for 10 s and 60 °C for 15 s. Three biological replicates were conducted.
Quantification of DENV Genomic RNA [(+)gRNA].
Single mosquitoes were homogenized in 350 μL of TRK lysis buffer (Omega Bio-tek) with 1.0 mm silica beads (BioSpec) using Mini-Beadbeater-96 (BioSpec). Total RNA from mosquito or cells was extracted using an E.Z.N.A. Total RNA Kit I (Omega Bio-tek) and eluted in 30 µL of DEPC-treated water. Total RNA from supernatant was extracted using a QIAamp Viral RNA Mini Kit (Qiagen) and eluted in 40 µL of AVE buffer. gRNA was quantified with one-step qRT-PCR using an iTaq universal probe kit (Bio-Rad) and primers and probes targeting the DENV envelope (45). The 12.5 µL reaction mix contained 1 µM of forward and reverse primers, 0.125 μM of probe, and 4 μL of RNA extract. Quantification was conducted on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). The thermal profile was 50 °C for 10 min, 95 °C for 1 min, and 40 cycles of 95 °C for 10 s and 60 °C for 15 s An absolute standard curve was generated by amplifying the qPCR target using a forward primer tagged with a T7 promoter (forward: 5′-CAGGATAAGAGGTTCGTCTG-3′ and reverse: 5′-TTGACTCTTGTTTATCCGCT-3′), resulting in a 453 base-pair fragment. The fragment was reverse transcribed using a MegaScript T7 transcription kit (Ambion) and purified using an E.Z.N.A. Total RNA Kit I. The total amount of RNA was quantified using a NanoDrop (Thermo Fisher Scientific) and used to estimate copy number. Ten times serial dilutions were made and used to generate the absolute standard equation. In each subsequent qRT-PCR plate, five standards were added to adjust for threshold variation between plates.
Titration.
Titration was conducted by plaque assay with BHK-21 cells as described previously (46). Briefly, 80 to 90% confluent cells were inoculated with serial 10-fold dilutions of samples for 1 h. Cells were then incubated with 1% carboxylmethyl cellulose (Merck) media for 5 d, fixed with 4% formaldehyde (Merck) in PBS and stained with 1% crystal violet (Sigma-Aldrich) solution to count PFUs.
Metabolite Extraction.
Cells were washed with 0.9% NaCl (Sigma-Aldrich), collected in 500 µL of ice-cold methanol (liquid chromatography-mass spectrometry grade, Thermo Fisher Scientific) and water in the ratio of 80:20 (vol/vol) by scraping and sonicated in an ultrasonic bath (J.R. Selecta) for 15 min at 4 °C. Homogenates were centrifuged at 10,000 rpm for 1 min at 4 °C, and 400 µL of supernatant were collected. Pellets were extracted a second time by adding 500 µL of methanol:water (80:20) solution, then sonicated and centrifuged before collecting 400 µL of supernatant. Combined supernatants were vacuum-dried using Speed-Vac (Thermo Fisher Scientific) and stored at −20 °C. Three biological replicates were conducted per condition.
Mosquitoes were collected by group of 10 in 500 μL of ice-cold methanol:water (80:20), homogenized with 1.0 mm silica beads using Mini-Beadbeater-96, and sonicated in an ultrasonic bath (J.R. Selecta) for 15 min at 4 °C. Metabolites were then collected using the same method as above. Four biological replicates were conducted per condition.
Liquid Chromatography-High Resolution Mass Spectrometry (LC-HRMS) Metabolic Profiling.
Dry extracts were normalized to 2 mg/mL in an 80:20 methanol:water solution. Metabolites were profiled using a Q Exactive Plus quadrupole-mass spectrometer coupled to a U-HPLC Ultimate 3000 RSLC system (Thermo Fisher Scientific) set at 30,000 resolution with a Cortecs Hilic column (150 × 2.1 mm internal dimensions, 1.6 µm, Waters). Mobile phase A was 20 mM ammonium acetate buffered at pH 9, and B was acetonitrile. The flow rate was 200 μL/min, and the gradient ran from 90% B for 0.5 min to 40% B over 18 min, then was held at 40% B for an additional 3 min, returned in 0.5 min to initial condition (90% B), and finally held for 5 min before subsequent analysis. The m/z range was 100 to 1,500, and ISpray voltage was at 3.5 kV in the positive mode and 3.0 kV in the negative mode for the phospholipid profile upon PL pathway enzyme depletion. Each full MS scan was followed by data-dependent acquisition of MS/MS spectra for the six most intense ions using a stepped normalized collision energy of 20, 40, and 60 eV.
Data Analysis and Visualization.
Raw data were converted to abf files (Reifycs). Peak detection and alignment were performed using MS-DIAL (version 4.12) (47). Peak annotation was done using MS-finder (version 3.26) (48) with the Human Metabolome Database (HMDB), Chemical Entities of Biological Interest (ChEBI), Lipid MAPS structure (LMSD), and LipidBlast databases, allowing a level 2.2 of metabolite identification (49, 50). Data were normalized by total ion chromatogram, and features lower than twofold average blank were removed. Data were normalized by autoscaling before selecting regulated metabolites with more than twofold intensity change and P < 0.05, as indicated by an unpaired t test with false-discovery rate (FDR) adjustment using MetaboAnalyst (version 4.0) (51). Principal component analysis (PCA) was performed for quality control with MetaboAnalyst (version 4.0).
Isotope tracking analysis was performed using MS-DIAL. Peak detection and alignment were achieved first on the nonlabeled sample data to build a library of compounds with known mass (M), retention time (RT), and annotation (SI Appendix, Fig. S7). Peak detection and alignment were then performed on 13C-labeled sample data and compared with nonlabeled sample data. Peaks exclusively detected in 13C-labeled sample data with a similar RT and M+2 compared to a compound from nonlabeled samples were considered isotopically labeled.
Oral Infection of Mosquitoes.
Two- to four-day-old adult female mosquitoes were starved for 24 h before oral feeding on a blood meal containing a 40% volume of washed erythrocytes from serum-pathogen free (SPF) pig’s blood (PWG Genetics), 5% of 100 mM ATP (Thermo Fisher Scientific), 5% of human serum (Sigma-Aldrich), and 50% of DENV-2 in RPMI media. For supplementation, either choline chloride or ethanolamine was added in place of RPMI. The virus titer in the blood meal was 1 × 107 PFU/mL and was validated by plaque assay. Blood was maintained at 37 °C using a Hemotek membrane feeder system (Discovery Workshops) with sausage casing for 1.5 h. Engorged mosquitoes were visually selected and maintained at 28 °C with water and 10% sucrose solution.
Attachment and Internalization Assays.
The 2 × 105 Aag2 cells were chilled at 4 °C for 15 min and inoculated with DENV at a MOI of 1 in serum-free media for 30 min at 4 °C. Cells were washed with prechilled 2% FBS media. To quantify attached viruses, cells were lysed in 350 µL of TRK lysis buffer (E.Z.N.A. RNA Kit I), and total RNA was extracted to quantify gRNA. To quantify internalization, cells were incubated with 350 µL of 2% FBS medium at 28 °C, 5% CO2, for 1 h. The medium was then replaced with 200 µL of 2 mg/mL pronase (Sigma-Aldrich) in serum-free medium for 5 min on ice to remove virus particles on the cell surface. Cells were washed twice with 10% FBS media and lysed in 350 µL of TRK lysis buffer to extract RNA and quantify gRNA from internalized viruses.
Translation Assay.
The 106 Aag2 cells were inoculated with DENV at a MOI of 1 in serum-free media for 1 h. After infection, cells were washed with PBS before adding new fresh prewarmed complete media with respective supplementation. At 3, 4, 5, and 6 h postinfection, cells were washed twice with PBS, scrapped in 70 µL of radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher Scientific), and sonicated for 10 min in an ultrasonic bath (J.R. Selecta) at 4 °C. Protein concentration was measured by BCA assay (ThermoFisher), and 20 µg were fractioned under denaturing conditions in 10% polyacrylamide gel (Bio-Rad). Antibodies used were anti-DENV-2 NS3 (CTX124252, Genetex) and anti-beta-actin (SC-47778, Santa-Cruz Biotechnology).
Replication Assay.
The 2.5 × 105 Aag2 cells were inoculated with DENV at a MOI of 1 or 5 in serum-free RPMI for 1 h at 28 °C. After infection, cells were washed with PBS before adding new fresh prewarmed complete media with respective supplementation. At 1, 3, 6, 9, 12, 15, 18, 21, and 24 h postinfection, cells were washed twice with PBS and lysed in 350 µL of TRK lysis buffer to extract RNA and quantify +strand [(+)gRNA] and –strand [(−)gRNA] gRNA.
Quantification of DENV Antigenomic RNA [(−)gRNA].
Total RNA from cells was extracted using an E.Z.N.A. Total RNA Kit I. (−)gRNA was quantified using the same method as quantification of (+)gRNA but with the following primers: forward, 5′-GTCCAATACCGTGACAGTGCTA-3′, and reverse, 5′-GGTAGACGT-CGTTGTGGTAGAG-3. An absolute standard curve was generated by amplifying the qPCR target using the qPCR primers but with the forward primer tagged with a T7 promoter. The fragment was reverse transcribed using a MegaScript T7 transcription kit and purified using an E.Z.N.A. Total RNA Kit I. The total amount of RNA was quantified using a NanoDrop and used to estimate copy number. Ten times serial dilutions were made and used to generate the absolute standard equation. In each subsequent qRT-PCR plate, four standards were added to adjust for threshold variation between plates.
Statistical Analysis.
Differences in gRNA copies were tested on log-transformed values using an unpaired t test or Dunnett’s multiple comparisons test. Tests were performed with GraphPad PRISM software (version 8.02). For multivariate analysis, data were imported into SIMCA-P (version 15.0, Sartorius Stedim Biotech, Umetrics). To check for data consistency and outliers, data were analyzed with PCA. The data were then Pareto scaled for the partial least squares (PLS) regression analysis with DENV activity in PFU as the Y input. VIP scores were used to rank variables according to their correlation with DENV activity.
Supplementary Material
Acknowledgments
We are grateful to Prof. Mariano Garcia-Blanco for constant support and J.P. and Garcia-Blanco’s team for comments on a previous version. We thank Prof. Eng Eong Ooi for comments and for providing the virus strain. We also thank Prof. Jean-Paul Kovalic for comments on the manuscript. We thank the mass spectrometry Metatoul-AgromiX platform, Université de Toulouse, National Center for Scientific Research, Université Paul-Sabatier, Toulouse, France. Funding: This work was supported by a grant from National Medical Research Council, Singapore, (NMRC/ZRRF/0007/2017) awarded to J.P., a grant from the Ministry of Education, Singapore, (MOE2015-T3-1-003) partially awarded to J.P., a grant from the French embassy in Singapore for Singapore-France travel (2018 PHC Merlion project 6.01.18), and by the Duke-NUS Signature Research Programme funded by the Agency for Science, Technology and Research (A-STAR), Singapore, and the Ministry of Health, Singapore. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015095117/-/DCSupplemental.
Data Availability.
Metabolomics data have been deposited in Zenodo (http://doi.org/10.5281/zenodo.3923483).
References
- 1.Bhatt S.et al., The global distribution and burden of dengue. Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer M. U.et al., The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci. Data 2, 150035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perera-Lecoin M., Meertens L., Carnec X., Amara A., Flavivirus entry receptors: An update. Viruses 6, 69–88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smit J. M., Moesker B., Rodenhuis-Zybert I., Wilschut J., Flavivirus cell entry and membrane fusion. Viruses 3, 160–171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrows N. J.et al., Biochemistry and molecular biology of flaviviruses. Chem. Rev. 118, 4448–4482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junjhon J.et al., Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 88, 4687–4697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie X.et al., Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe 26, 606–622.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Yu I.-M.et al., Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008). [DOI] [PubMed] [Google Scholar]
- 9.McMaster C. R., From yeast to humans – Roles of the Kennedy pathway for phosphatidylcholine synthesis. FEBS Lett. 592, 1256–1272 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Fast P. G., A comparative study of the phospholipids and fatty acids of some insects. Lipids 1, 209–215 (1966). [DOI] [PubMed] [Google Scholar]
- 11.Gibellini F., Smith T. K., The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Moessinger C.et al., Two different pathways of phosphatidylcholine synthesis, the Kennedy pathway and the Lands cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 15, 43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vance J. E., Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Tamura Y.et al., Phosphatidylethanolamine biosynthesis in mitochondria: Phosphatidylserine (PS) trafficking is independent of a PS decarboxylase and intermembrane space proteins UPS1P and UPS2P. J. Biol. Chem. 287, 43961–43971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stone S. J., Vance J. E., Cloning and expression of murine liver phosphatidylserine synthase (PSS)-2: Differential regulation of phospholipid metabolism by PSS1 and PSS2. Biochem. J. 342, 57–64 (1999). [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B., Tontonoz P., Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 81, 165–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon H. T., Boucrot E., Membrane curvature at a glance. J. Cell Sci. 128, 1065–1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidsen J., Mouritsen O. G., Jørgensen K., Synergistic permeability enhancing effect of lysophospholipids and fatty acids on lipid membranes. Biochim. Biophys. Acta Biomembr. 1564, 256–262 (2002). [DOI] [PubMed] [Google Scholar]
- 19.van Meer G., Vaz W. L. C., Membrane curvature sorts lipids. Stabilized lipid rafts in membrane transport. EMBO Rep. 6, 418–419 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsch S.et al., Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chotiwan N.et al., Dynamic remodeling of lipids coincides with dengue virus replication in the midgut of Aedes aegypti mosquitoes. PLoS Pathog. 14, e1006853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vial T.et al., Dengue virus reduces AGPAT1 expression to alter phospholipids and enhance infection in Aedes aegypti. PLoS Pathog. 15, e1008199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L.et al., Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl. Trop. Dis. 7, e2373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henneberry A. L., Wistow G., McMaster C. R., Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J. Biol. Chem. 275, 29808–29815 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Henneberry A. L., McMaster C. R., Cloning and expression of a human choline/ethanolaminephosphotransferase: Synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem. J. 339, 291–298 (1999). [PMC free article] [PubMed] [Google Scholar]
- 27.Marti G.et al., Comparison of the phytochemical composition of Serenoa repens extracts by a multiplexed metabolomic approach. Molecules 24, 2208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z., Zhang Q., Dai S., Gao X., Discovering temporal patterns in longitudinal nontargeted metabolomics data via group and nuclear norm regularized multivariate regression. Metabolites 10, 33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakovic M., Fullerton M. D., Michel V., Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: The role of CTP:phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem. Cell Biol. 85, 283–300 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Li Z., Vance D. E., Phosphatidylcholine and choline homeostasis. J. Lipid Res. 49, 1187–1194 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Wang L.et al., Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 24, 4652–4669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita A.et al., Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 53, 18–81 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bobenchik A. M., Augagneur Y., Hao B., Hoch J. C., Ben Mamoun C., Phosphoethanolamine methyltransferases in phosphocholine biosynthesis: Functions and potential for antiparasite therapy. FEMS Microbiol. Rev. 35, 609–619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel D., Witt S. N., Ethanolamine and phosphatidylethanolamine: Partners in health and disease. Oxid. Med. Cell. Longev. 2017, 4829180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perera R.et al., Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 8, e1002584 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voge N. V.et al., Metabolomics-based discovery of small molecule biomarkers in serum associated with dengue virus infections and disease outcomes. PLoS Negl. Trop. Dis. 10, e0004449 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khedr A., Hegazy M. A., Kammoun A. K., Shehata M. A., Phospholipidomic identification of potential serum biomarkers in dengue fever, hepatitis B and hepatitis C using liquid chromatography-electrospray ionization-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1009–1010, 44–54 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Cui L.et al., Serum metabolomics reveals serotonin as a predictor of severe dengue in the early phase of dengue fever. PLoS Negl. Trop. Dis. 10, e0004607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butler P. L., Mallampalli R. K., Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J. Biol. Chem. 285, 6246–6258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebscher S.et al., Phospholipase A2 activity during the replication cycle of the flavivirus West Nile virus. PLoS Pathog. 14, e1007029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heaton N. S.et al., Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 17345–17350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barletta A. B. F., Silva M. C. L. N., Sorgine M. H. F., Validation of Aedes aegypti Aag-2 cells as a model for insect immune studies. Parasit. Vectors 5, 148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber M. J.et al., Genomic epidemiology of a dengue virus epidemic in urban Singapore. J. Virol. 83, 4163–4173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraiture M.et al., Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 5, 273–284 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Johnson B. W., Russell B. J., Lanciotti R. S., Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 43, 4977–4983 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manokaran G.et al., Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsugawa H.et al., MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsugawa H.et al., Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 88, 7946–7958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sumner L. W.et al., Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsugawa H.et al., A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Chong J.et al., MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabolomics data have been deposited in Zenodo (http://doi.org/10.5281/zenodo.3923483).