Abstract
Purpose
Aurora kinase B (AURKB) plays a pivotal role in the regulation of mitosis and is gaining prominence as a therapeutic target in cancers; however, the role of AURKB in retinoblastoma (RB) has not been studied. The purpose of this study was to determine if AURKB plays a role in RB, how its expression is regulated, and whether it could be specifically targeted.
Methods
The protein expression of AURKB was determined using immunohistochemistry in human RB patient specimens and immunoblotting in cell lines. Pharmacological inhibition and shRNA-mediated knockdown were used to understand the role of AURKB in cell viability, apoptosis, and cell cycle distribution. Cell viability in response to AURKB inhibition was also assessed in enucleated RB specimens. Immunoblotting was employed to determine the protein levels of phospho-histone H3, p53, p21, and MYCN. Chromatin immunoprecipitation–qPCR was performed to verify the binding of MYCN on the promoter region of AURKB.
Results
The expression of AURKB was found to be markedly elevated in human RB tissues, and the overexpression significantly correlated with optic nerve and anterior chamber invasion. Targeting AURKB with small-molecule inhibitors and shRNAs resulted in reduced cell survival and increased apoptosis and cell cycle arrest at the G2/M phase. More importantly, primary RB specimens showed decreased cell viability in response to pharmacological AURKB inhibition. Additional studies have demonstrated that the MYCN oncogene regulates the expression of AURKB in RB.
Conclusions
AURKB is overexpressed in RB, and targeting it could serve as a novel therapeutic strategy to restrict tumor cell growth.
Keywords: aurora kinase B, retinoblastoma, optic nerve invasion, AURKB inhibition, therapeutic target, MYCN
Retinoblastoma (RB) is a malignant intraocular tumor in children that accounts for 4% of all childhood cancers.1 Although treatment outcomes have improved in developed countries, RB is still a major health concern in low- and middle-income nations, from which more than 80% of the world's cases are reported.2 For a significant number of children with RB (35%), enucleation is still the preferred treatment option.3 Extraocular spread and death are common in African countries,4 and histological risk factors including optic nerve and choroidal invasion are frequently found in Asian RB patients.5 Chemotherapy with drugs such as vincristine, etoposide, carboplatin, melphalan, and topotecan is usually administered for treatment of RB; however, not all children respond to therapy, and those who do respond may suffer long-term side effects.6,7 Further, some children develop secondary malignancies such as acute myeloid leukemia and osteosarcoma.8,9
RB initiation and progression require biallelic inactivation of the RB1 gene and subsequent events such as alterations in pathways involving MYCN, E2F3, DEK, and MDM4.10 Recently, a different subset of RB with no apparent RB1 mutations has been found to harbor amplification of the MYCN oncogene.11 RB1 plays a major role in the regulation of cell cycle progression. Other mitotic regulators such as Polo-like kinase 1 (PLK1) have been shown to be overexpressed in different tumors12 and associated with histological risk factors, such as choroidal invasion and poor differentiation in RB.13 PLK1 inhibition further enriches hyperthermia sensitivity, known to be important for focal therapy in RB.14 Recently, a few key proteins involved in cell cycle regulation such as aurora kinases have gained significance as plausible therapeutic targets in cancer.15,16
Aurora kinases are serine/threonine kinases that control mitosis (aurora kinases A and B) and meiosis (aurora kinase C).16,17 Aurora kinase B (AURKB) is part of the chromosomal passenger complex, and it facilitates chromosome condensation and segregation and promotes cytokinesis.18 Overexpression of AURKB causes aneuploidy, resulting in genomic instability and increasing the likelihood of tumor development (reviewed in Reference 16). Moreover, targeting aurora kinases has been shown to be beneficial in preclinical tumor models (reviewed in References 15 and 17). Currently, the role of AURKB in RB has not yet been determined. In this study, we tested the hypothesis that AURKB is overexpressed and linked to RB pathogenesis. First, the aberrant expression of AURKB in RB was evaluated using immunological approaches. Three inhibitors of AURKB (GSK1070916, ZM447439, and barasertib) were then selected, and their effect on RB cell survival, apoptosis, and cell cycle distribution was assessed. Finally, the regulation of AURKB expression by the MYCN oncogene was studied. Amplification and/or expression of the MYCN oncogene is considered to be important in the pathogenesis of retinoblastoma after RB1 inactivation.19,20
Materials and Methods
Ethics Approval
This study was approved by the institutional ethics committee of LV Prasad Eye Institute, Bhubaneswar, India, and adhered to the tenets of the Declaration of Helsinki.
Cell Culture
Commercially available human RB cell lines, Y79 and WERI-RB1, were procured from the American Type Culture Collection (Manassas, VA, USA) and maintained in RPMI-1640 (PAN-Biotech, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) and 1% (v/v) antibiotics (penicillin–streptomycin–amphotericin B mixture; PAN-Biotech). The patient-derived cells (LRB1 and LRB2) described previously21 and RB primary tumor cells isolated from enucleated eyes were maintained in a similar manner as the RB cell lines. Human retinal pigment epithelium cell line ARPE-19 (American Type Culture Collection) was grown in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12; PAN-Biotech) containing 10% FBS. Human embryonic kidney (HEK) 293T (American Type Culture Collection), MCF-7, and HCT-116 cells were cultured in DMEM (PAN-Biotech) supplemented with 10% FBS. MCF-7 and HCT-116 cells were a kind gift from Dr. Srinivas Patnaik's laboratory, KIIT School of Biotechnology, Bhubaneswar. The cell lines were utilized within 3 months of revival from frozen vials, and the presence of Mycoplasma was tested as described previously.22
Immunoblotting
Immunoblotting was performed as described previously.23 The primary antibodies used in the study were rabbit anti-AURKB monoclonal antibody (1:40000 dilution; Abcam, Cambridge, MA, USA), mouse anti-p53 monoclonal antibody (1:500 dilution; Santa Cruz Biotechnology, Dallas, TX, USA), mouse anti-β-actin monoclonal antibody (1:5000 dilution; Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-phospho-histone-H3 (Ser10) monoclonal antibody (1:1000 dilution; Cell Signaling Technology, Beverly, MA, USA), rabbit anti-p21 Waf1/Cip1 (p21) monoclonal antibody (1:1000 dilution; Cell Signaling Technology), rabbit anti-histone H3 polyclonal antibody (1:1000 dilution; Cell Signaling Technology), and rabbit anti-MYCN monoclonal antibody (1:1000 dilution; Cell Signaling Technology).
mRNA Expression Analysis of AURKB
Total RNA was isolated from different cell lines using TRIzol Reagent (Thermo Fisher Scientific). Equal quantities of RNA were reverse-transcribed using a cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA) and subjected to quantitative polymerase chain reaction (qPCR) analysis in the presence of specific primers—AURKB F, TCCTCTTCAAGTCCCAGATAGA; AURKB R, TAGATCCTCCTCCGGTCATAAA; and SYBR green master mix (Bio-Rad Laboratories). Beta 2 microglobulin (B2M) served as control (B2M F, TATCCAGCGTACTCCAAAGA; B2M R, GACAAGTCTGAATGCTCCAC).
Immunohistochemistry
A total of 51 enucleated RB tissue specimens were evaluated for the detection of AURKB protein expression. Formalin-fixed, paraffin-embedded tissue sections were placed on glass slides coated with (3-aminopropyl)triethoxysilane (Sigma-Aldrich), and immunohistochemistry (IHC) was performed using the EnVision FLEX Mini Kit (Agilent, Santa Clara, CA, USA) according to the manufacturer's protocol. The rabbit anti-AURKB monoclonal antibody was used at 1:250 dilution. Images were captured with an APERIO CS2 slide scanning microscope (Leica Microsystems, Wetzlar, Germany). Also, images were taken with a 100× objective on an Olympus BX53 microscope (Olympus, Tokyo, Japan) for scoring and data analysis.
Scoring of IHC Slides
AURKB expression in the tissue specimens was given an immunoreactivity score based on the intensity and extent of expression. This method of scoring has been described previously.24 The intensity of expression (i) was quantified using the following scores: 0 = negative, 1 = weak, 2 = medium, 3 = high. The extent of expression was evaluated as percentage of positively stained area in comparison to the entire tumor area, and the following scores were assigned: 0 for < 1%, 1 for 2% to 10%, 2 for 11% to 50%, 3 for 51% to 75% and 4 for >75% area showing expression. The final IHC score was determined as follows: Σi = 0 to 3 (i × score based on extent of expression). The results of the above expression would show a minimum IHC score of 0 to a maximum of 12. An IHC score of 10 to 12 was considered strong expression; 7 to 9, moderate expression; and 3 to 6, weak expression. An IHC score of <3 was taken as no expression.
shRNA-Mediated Knockdown
The knockdown of AURKB was carried out in Y79 and LRB1 cells using a lentiviral approach. The shRNA constructs used in the study were KD-1 (TRCN0000000777) and KD-2 (TRCN0000010547) (Dharmacon, Lafayette, CO, USA). Additionally, the shRNA constructs KD-3 (TRCN0000020695) and KD-4 (TRCN0000020698) (Dharmacon) were used for MYCN knockdown. Lentiviruses containing constructs targeting AURKB or scrambled controls were generated by transfecting HEK293T cells using Lipofectamine 3000 (Thermo Fisher Scientific). Viral supernatant was used for transducing Y79 or LRB1 cells in the presence of polybrene (4 µg/mL; Sigma-Aldrich). The transduced cells were selected in medium containing puromycin (3 µg/mL, Sigma-Aldrich), and the knockdown efficiency was confirmed by immunoblotting.
Pharmacological Inhibition of AURKB and Measurement of Cell Viability
AURKB was inhibited in RB cells by treating them with commercially available AURKB inhibitors: barasertib (AZD1152-HQPA), GSK1070916, and ZM447439 (Apex Bio, Houston, TX, USA). Cell viability was performed using the trypan blue dye (Sigma-Aldrich) exclusion method. The decrease in cell viability in response to inhibitor treatment or shRNA-mediated knockdown was compared with untreated or scrambled controls.
Apoptosis Assay
The percent apoptosis in inhibitor-treated and AURKB-knockdown RB cells was analyzed using a BD FACSCanto II Flow Cytometer (BD Biosciences, San Jose, CA, USA) using annexin V and propidium iodide (PI) staining as per the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Cell Cycle Analysis
The changes in cell cycle distribution upon AURKB inhibition were examined using PI staining. Briefly, 1 million cells were washed with Dulbecco's phosphate buffered saline (DPBS; PAN-Biotech) and fixed in 70% cold ethanol. After 1 hour of fixation and required washes, samples were treated with 50 µL of RNase (QIAGEN, Hilden, Germany) from a 100-µg/mL stock and incubated at 37°C for 30 minutes. Finally, 200 µL of PI was added to the samples before analysis using a CytoFLEX S Flow Cytometer (Beckman Coulter, Brea, CA, USA).
In Silico Analysis of AURKB Gene Promoter
The promoter sequence for the AURKB gene (2 kb upstream of transcription start site) was retrieved using Ensembl (release 100)25 and analyzed for the presence of MYCN binding motifs as reported previously.26–28
Chromatin Immunoprecipitation–qPCR
Chromatin immunoprecipitation (ChIP)–qPCR was performed to study whether MYCN binds on the promoter region of AURKB. Briefly, Y79 cells (40 × 106) were crosslinked with formaldehyde and neutralized with glycine. The cell pellet was washed with cold PBS and resuspended in nuclei extraction buffer (5-mM PIPES, pH 8.0; 85-mM KCl; and 0.5% NP-40) for 8 minutes on ice. The nuclei pellet was resuspended in radioimmunoprecipitation assay buffer and sonicated at 4°C for 15 minutes to generate chromatin fragments in the range of 100 to 600 bp. Chromatin (150 µg) was immunoprecipitated with ChIP-grade MYCN antibody (1:50 dilution; Cell Signaling Technology). The crosslinks of eluted protein/DNA complexes were reversed using NaCl and treated with proteinase K and RNase followed by purification using PCR purification kit (QIAGEN). The ChIP–qPCR primers are listed in Supplementary Table S1. Percent enrichment was calculated relative to negative control primers designed to amplify the region away from the AURKB promoter sequence.
Statistical Analysis
The baseline and disease characteristics were assessed using descriptive statistics. The expression of AURKB and clinicopathological associations were determined using the χ2 test and Student's t-test. The pharmacological, shRNA-knockdown, and ChIP–qPCR experiments were analyzed by Student's t-test.
Results
AURKB Is Significantly Overexpressed in Human RB Tumor Specimens and Cell Lines
The protein expression of AURKB was determined by immunoblotting and was found to be elevated in human RB cell lines compared to control retina (Fig. 1a). The levels of expression were comparable among the different RB cell lines. We have used commercially available Y79 cell line and patient-derived LRB1 cells in further experiments. To compare the expression of AURKB in RB with other tumor cell lines, we measured mRNA expression in MCF-7 (breast cancer) and HCT-116 (colon cancer) cells. Additionally, the expression of AURKB was determined in ARPE-19 cells. Of all the cell lines tested, RB cells had comparably higher expression of AURKB (Supplementary Fig. S1).
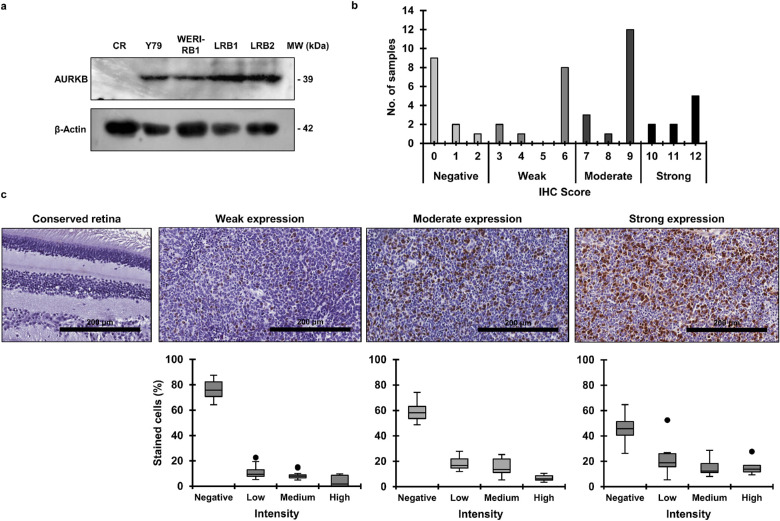
Figure 1.
AURKB is overexpressed in RB. (a) AURKB protein expression in RB cells compared to control retina (CR) as determined by immunoblotting. (b) The IHC score of AURKB expression among human RB patient specimens (n = 48). (c) Representative images of conserved retina showing no expression of AURKB and tumor specimens exhibiting weak, moderate, or strong expression (top panel). The bottom panel shows the distribution of percent stained cells in tumor specimens showing weak (n = 11), moderate (n = 16), or strong (n = 9) AURKB expression.
Next, we evaluated the expression of AURKB in human RB tissue specimens (n = 51) by immunohistochemistry. Of the 51 specimens, three samples were excluded from the study as there was extensive necrosis and calcification, with only focal viability. Out of the remaining 48 specimens, 16 were from patients who had received neo-adjuvant chemotherapy. The baseline characteristics of the patients are described in the Table. Immunohistochemical staining revealed positive AURKB staining in 36 samples (75%), among which there were 21 males and 15 females. The IHC slides were scored on the basis of intensity and extent of positive staining. Of the 36 specimens, weak, moderate, and strong expression was seen in 11 (30.56%), 16 (44.44%), and 9 (25%) specimens, respectively (Fig. 1b). Representative images of stained tissue specimens (top panel) and their respective distribution of stained cells (bottom panel) are shown in Figure 1c. Additionally, AURKB expression in the tumor area was compared to the adjacent conserved healthy retina, and it was observed that expression levels were negligible in the conserved retina (Fig. 1c). Further, the clinicopathological features were correlated with the presence of AURKB expression (Table). The statistical analysis showed a significant correlation between optic nerve invasion and positive AURKB expression (P < 0.05). In addition, the expression of AURKB was associated with anterior chamber invasion (P < 0.05). There was no statistical significance for other clinicopathological features (Table).
Table.
Correlation of Clinicopathological Features with Positive AURKB Expression in RB Patient Specimens
| N | Positive AURKB Expression | Negative AURKB Expression | Statistical Test | P | Significance | |
|---|---|---|---|---|---|---|
| Baseline characteristics | ||||||
| Age at diagnosis (mo), n | Student's t-test | 0.22 | NS | |||
| Median | 36 | 36 | 24 | |||
| Mean | 32.94 ± 22.39 | 35.25 ± 24.9 | 26 ± 10.02 | |||
| Range | 2–108 | 2–108 | 12–36 | |||
| Gender, n | χ2 test | 0.61 | NS | |||
| Male | 27 | 21 | 6 | |||
| Female | 21 | 15 | 6 | |||
| Laterality, n | χ2 test | 0.81 | NS | |||
| Unilateral | 41 | 31 | 10 | |||
| Bilateral | 7 | 5 | 2 | |||
| Association of histopathological features with AURKB expression | ||||||
| Choroid invasion, n | χ2 test | 0.50 | NS | |||
| Present | 24 | 19 | 5 | |||
| Absent | 24 | 17 | 7 | |||
| Optic nerve invasion, n | χ2 test | 0.01 | SS | |||
| Present | 34 | 29 | 5 | |||
| Absent | 14 | 7 | 7 | |||
| Anterior chamber invasion, n | χ2 test | 0.04 | SS | |||
| Present | 10 | 10 | 0 | |||
| Absent | 38 | 26 | 12 | |||
| Scleral invasion, n | χ2 test | 0.83 | NS | |||
| Present | 9 | 7 | 2 | |||
| Absent | 39 | 29 | 10 |
P < 0.05 represents statistical significance. SS, statistical significance; NS, no significance.
Pharmacological Inhibition of AURKB Leads to Decreased RB Cell Viability
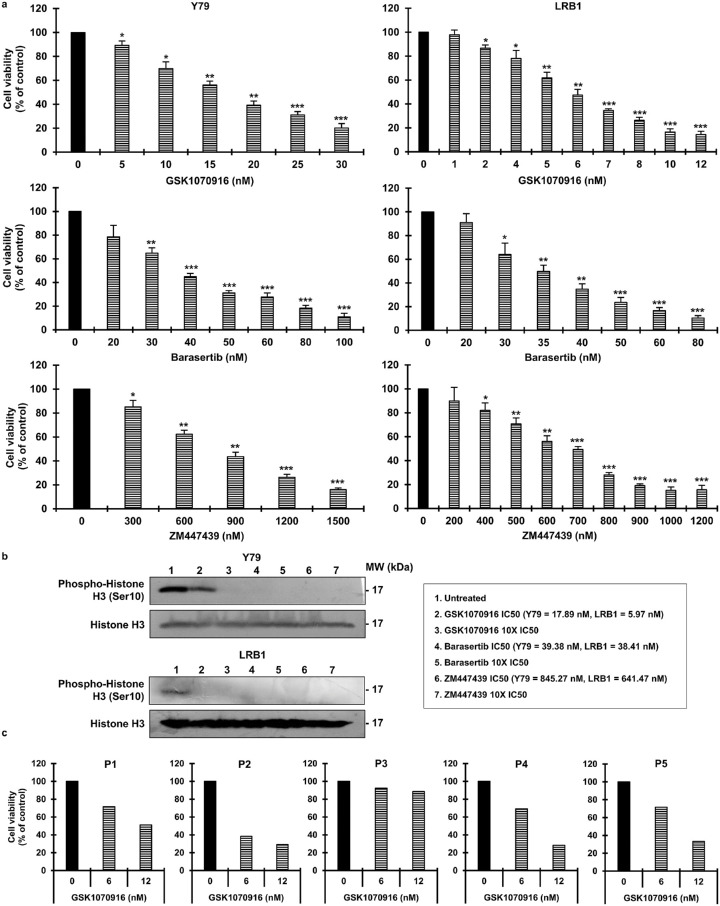
AURKB inhibition as a potential treatment strategy was investigated in RB cells (Y79, LRB1, and LRB2). The inhibitors used in the study were GSK1070916, barasertib, and ZM447439. Cell viability was assessed after 72 hours. It was seen that AURKB inhibition reduced cell viability in Y79 and LRB1 cells (Fig. 2a) and in LRB2 cells (Supplementary Fig. S2). For example, Y79 cells showed a 60.77% ± 3.44% reduction in cell viability relative to control at a 20-nM concentration of GSK1070916. In contrast, LRB1 cells were more sensitive to GSK1070916 and showed a 65.40% ± 1.44% reduction in cell viability at 7-nM. The IC50 values of all the inhibitors for each cell line were calculated (Supplementary Table S2). Overall, a consistent reduction in cell viability was detected with an increase in concentration of AURKB inhibitors. The specificity of AURKB inhibition was evaluated by measuring the phospho-histone H3 (Ser10) levels, and a profound decrease was observed in inhibitor-treated cells compared to control cells (Fig. 2b). The changes in cell viability in response to GSK1070916 were further tested on primary human RB patient specimens and were observed to be significantly reduced in treated cells (Fig. 2c). The effect of AURKB inhibition using GSK1070916 was also tested on a control human retinal pigment epithelial cell line (ARPE-19) and was found to be negligible at the concentrations that were effective against the human RB patient specimens, demonstrating a therapeutic window for targeting AURKB in RB (Supplementary Fig. S3).
Figure 2.
Reduction in cell viability upon pharmacological inhibition of AURKB in RB cells. (a) Cell viability of RB cells after 72 hours of treatment with AURKB inhibitors. The error bars represent standard deviation (*P < 0.05, **P < 0.01, ***P < 0.001 by Student's t-test). (b) Immunoblots showing phosphorylation of histone H3 (Ser 10) in AURKB-inhibited RB cells. (c) Cell viability of patient-derived primary tumor cells after 72 hours of treatment with GSK1070916.
Targeting AURKB in RB Promotes Apoptosis and Cell Accumulation at the G2/M Phase
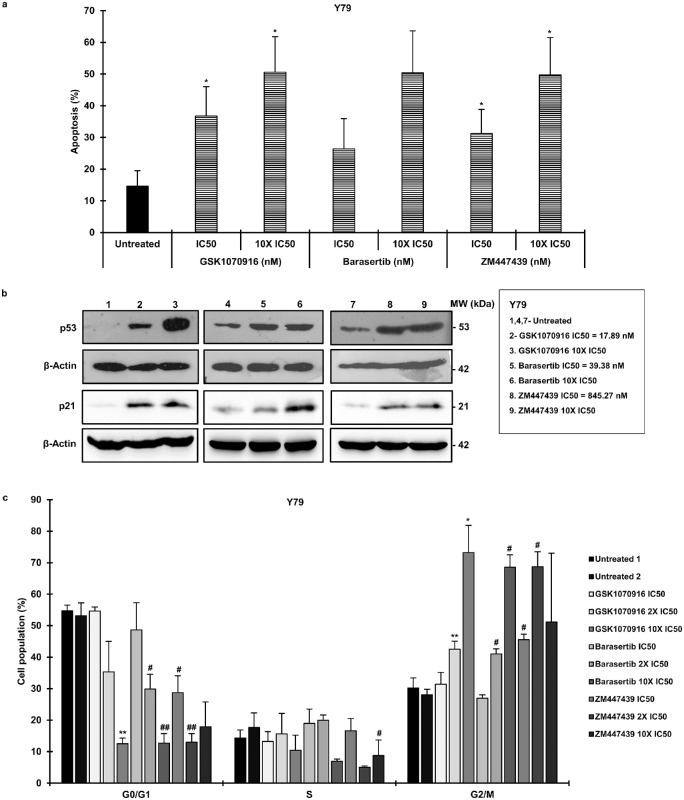
The extent of apoptosis upon AURKB inhibition was analyzed by using a flow cytometer. The inhibitor concentrations used in the study were IC50 and 10 times the IC50 as determined earlier (Supplementary Table S2). It was noticed that apoptosis increased in contrast to control in inhibitor-treated Y79 cells (Fig. 3a) and LRB1 cells (Supplementary Fig. S4a) in a dose-dependent manner. Additionally, the levels of p53 and p21 Waf1/Cip121 (p21) proteins were evaluated in inhibitor-treated RB cells by employing the same concentrations of inhibitors that were used in the apoptosis experiments. The protein expression of p53 and p21 was observed to be increased in both Y79 cells (Fig. 3b) and LRB1 cells (Supplementary Fig. S4b). Next, we evaluated the role of AURKB in RB cell cycle progression. The cells treated with AURKB inhibitors were stained with PI and analyzed by a flow cytometer. The drug concentrations used in the experiment were IC50, 2× IC50, and 10× IC50 (Supplementary Table S2). It was determined that AURKB inhibition resulted in a G2/M phase arrest in the cell cycle for Y79 cells (Fig. 3c) and LRB1 cells (Supplementary Fig. S4c).
Figure 3.
AURKB inhibition increases apoptosis and induces cell accumulation at the G2/M phase of cell cycle. (a) Total apoptosis in Y79 cells upon treatment with different AURKB inhibitors. (b) The p53 and p21 protein expression levels in inhibitor-treated Y79 cells. (c) Cell cycle distribution of Y79 cells treated with AURKB inhibitors. The cells treated with GSK1070916 were compared with Untreated 1 (*), and the remaining samples were compared with Untreated 2 (#). The error bars represent the standard error of the mean (*/#P < 0.05, **/##P < 0.01 by Student's t-test).
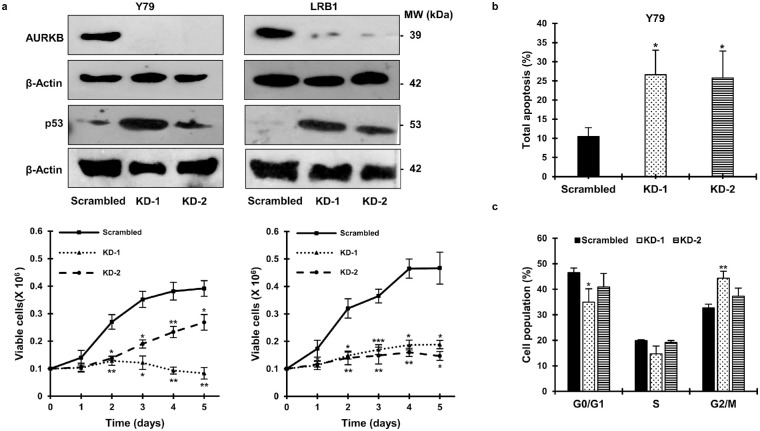
shRNA-Mediated Knockdown of AURKB Results in Decreased Cell Viability
To substantiate the role of AURKB in RB, shRNA-mediated lentiviral knockdown was performed in Y79 and LRB1 cells. The cells transduced with scrambled shRNA was used as control. The efficiency of AURKB-knockdown was confirmed by immunoblotting, and the levels of p53 were found to be elevated in AURKB-knockdown cells (Fig. 4a, top panel). Next, changes in cell viability were assessed in knockdown and scrambled control cells each day for 5 days. A marked reduction in cell viability was observed in knockdown cells compared to scrambled controls in Y79 and LRB1 cells (Fig. 4a, bottom panel). The results were consistent for both of the shRNA constructs used in the study. Further, there was a higher percentage of apoptosis in the cells with AURKB-knockdown in comparison with the cells that were transduced with a scrambled shRNA construct (Fig. 4b). In Y79 cells, 26.6% ± 6.4% and 25.76% ± 7.05% apoptosis was detected with KD-1 and KD-2 constructs, respectively, in comparison to 10.46% ± 2.31% apoptosis in the scrambled control cells (Fig. 4b). Similar to inhibition experiments, a G2/M phase cell cycle arrest was noticed in Y79 cells transduced with shRNA construct KD-1 (Fig. 4c).
Figure 4.
AURKB-knockdown in RB results in inhibition of cell growth, elevated apoptosis, and cell accumulation at the G2/M phase. (a) Confirmation of AURKB-knockdown and levels of p53 in RB cells with AURKB-knockdown (top panel). Cell viability in knockdown cells compared to scrambled control cells (bottom panel). Y79 cells with AURKB-knockdown were analyzed for (b) total apoptosis and (c) cell cycle distribution. The error bars denote SD (*P < 0.05, **P < 0.01, ***P < 0.001 determined by Student's t-test).
Regulation of AURKB Expression by the MYCN Oncogene
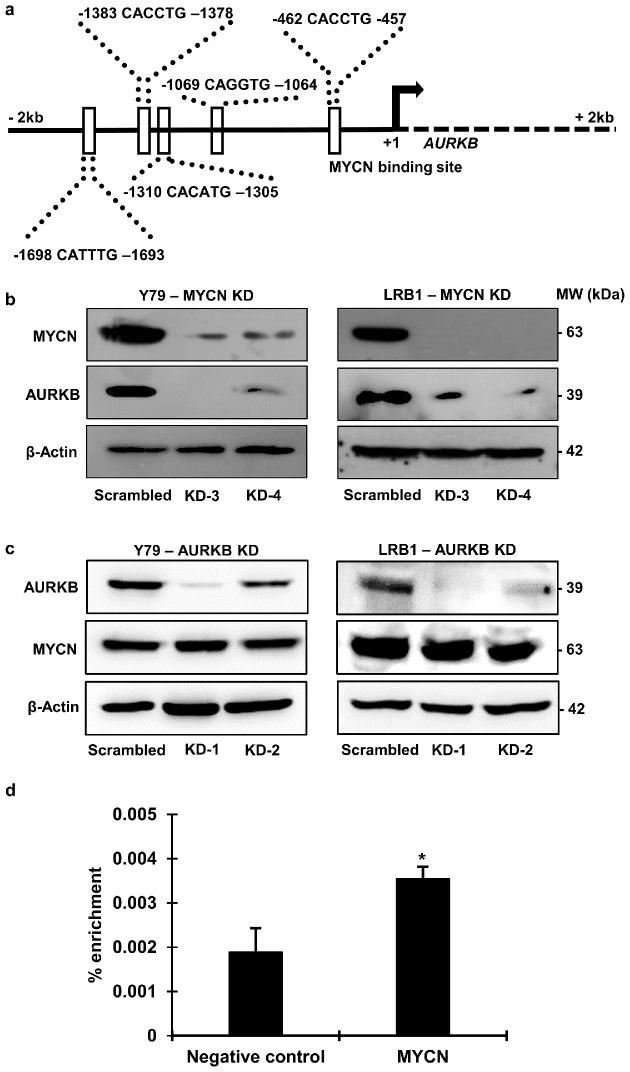
The MYCN oncogene is considered to be important in retinoblastoma development and progression after RB1 inactivation. MYCN is also known to play a role in the transcriptional regulation of various proteins.10,19,20 To examine the role of MYCN in the regulation of AURKB expression, we first analyzed the promoter sequence of AURKB for the presence of MYCN binding motifs. The AURKB promoter sequence was downloaded using Ensembl (release 100)25 and scanned for the presence of MYCN binding motifs based on a previously published study26 and the Eukaryotic Promoter Database.27,28 The analysis identified five MYCN binding motifs upstream of the transcription start site (Fig. 5a), suggesting a possible role for MYCN in the regulation of AURKB expression.
Figure 5.
The expression of AURKB is regulated by MYCN in RB cells. (a) Presence of MYCN binding motifs on AURKB gene promoter. The genomic contig NC_000017.11 and sequences from 8210574 to 8212573 were used. The +1 denotes the transcription start site. (b) AURKB protein levels in RB cells with MYCN-knockdown, and (c) MYCN protein levels in RB cells with AURKB-kncockdown. (d) ChIP–qPCR showing enrichment of the MYCN binding motif on the AURKB promoter.
To decipher a likely crosstalk between MYCN and AURKB, we studied the expression of AURKB in RB cells with MYCN-knockdown, as well as the expression of MYCN in AURKB-knockdown cells. We detected diminished AURKB protein levels in both Y79 and LRB1 cells compared to scrambled controls in MYCN-knockdown cells (Fig. 5b). However, we did not observe any significant change in the protein expression of MYCN in RB cells with AURKB-knockdown (Fig. 5c).
To further understand if MYCN binds to the promoter region of AURKB, we performed ChIP experiments in Y79 cells using ChIP-grade MYCN antibody, followed by qPCR using specific primers that would amplify the regions containing MYCN binding motifs. The results showed that the MYCN binding motif in the promoter region at –1064 from the transcription start site was enriched compared to negative control (Fig. 5d).
Discussion
Our study demonstrates that AURKB is markedly overexpressed in human RB patient specimens and further reveals that morphologically healthy-looking conserved retina adjoining the tumor is devoid of AURKB expression. Since it is challenging to target loss-of-function mutations such as RB1 inactivation, identification of druggable proteins involved in tumorigenesis can provide alternative therapeutic targets. The presence of elevated AURKB levels in human RB specimens compared to conserved retina suggests that AURKB inhibition could be a potential treatment strategy. The overexpression of AURKB has been implicated in a variety of malignancies (reviewed in Reference 15). For example, in non-small cell lung cancer, AURKB was found to be elevated in contrast to epithelial cells.29,30 Likewise, induced AURKB protein expression has been established in various other cancers, including acute lymphoblastic leukemia and acute myeloid leukemia.31 The role of AURKB in tumors was initially supported by its overexpression-mediated tumorigenesis in murine models.32 At the molecular level, AURKB modulates the repression of p21Cip1 via p5333 and, along with histone deacetylases, mediates cell proliferation via the AKT/mTOR signaling pathway.34 Additionally, the altered expression of AURKB has been implicated in the genomic instability of cancer cells.29
Optic nerve, choroidal, scleral, and anterior chamber invasion are important histological risk factors for disease prognosis in RB.35 In this study, we determined that overexpression of AURKB correlated with optic nerve and anterior chamber invasion. The association of AURKB with optic nerve and anterior chamber involvement indicates its possible role in RB invasion and metastasis. Similarly, AURKB overexpression was shown to be linked with tumor invasion, lymph node metastasis, and poor prognosis in non-small cell lung cancer.36
The significance of AURKB upregulation for RB cell survival has been demonstrated by the knockdown and pharmacological inhibition experiments in this study. This is especially relevant as AURKB inhibitors are in different phases of clinical development15,17,37 and can be translated for human application. Testing the inhibition of drug targets in primary RB tissues is crucial, as currently there are no suitable animal models that can replicate human RB.38 Because the tissue size is very small in RB, we could only employ one of the inhibitors. We used the inhibitor GSK1070916 to inhibit AURKB in RB patient specimens, as it was effective at a lower concentration compared to other inhibitors. The inhibition of AURKB decreased cell viability significantly in RB patient specimens but had little to no effect on normal retinal pigment epithelial cells at the same concentrations. Further, the patient sample that had the highest AURKB expression responded comparatively better to AURKB inhibition than the other specimens; however, whether the response to AURKB inhibition depends on the level of expression requires further investigation. Overall, our data suggest that we can specifically target AURKB to restrict RB cell growth. Consistent with impaired cell growth, this study demonstrates that AURKB inhibition led to increased apoptosis along with elevated levels of p53 and G2/M phase cell cycle arrest. The role of AURKB in decreasing p53 activity has been studied previously.39,40 Recent studies involving small cell lung cancer with RB1 mutations demonstrated a profound dependence on AURKB for their survival. The AURKB inhibition in these RB1 mutated tumors significantly affected their cell growth, possibly through a synthetic lethal relationship between RB1 mutations and AURKB inhibition.41 Moreover, AURKB was implicated in therapy-related drug resistance and treatment relapse in B-cell acute lymphoblastic leukemia.42 AURKB inhibition was further proposed as an intervention strategy in drug-resistant tumor cells, particularly against non-small cell lung cancer cells with drug-resistant mutations in the epidermal growth factor receptor.43
The molecular activators that regulate AURKB expression are not completely understood. We have identified consensus binding motifs for MYCN on AURKB promoter, which indicates that MYCN might regulate AURKB expression. The amplification and/or expression of MYCN has been considered to be an essential event in RB pathogenesis after RB1 mutations.19,20,44 Based on these observations, we designed a study to investigate the impact of MYCN-knockdown on AURKB expression. There was a consistent reduction in the expression of AURKB in response to MYCN-knockdown in RB cells. Further, ChIP–qPCR showed that MYCN binds on the promoter region of AURKB, and the extent of the binding is comparable to binding at other known gene promoter sequences.45,46 The regulation of AURKB expression by MYCN is particularly interesting, as it relates overexpression of AURKB to RB pathogenesis. A few other pathways have also been found to regulate AURKB expression in tumor cells. For example, MDM2, through AURKB expression, was shown to mediate cell cycle in prostate cancer cells.47 Likewise, in neuroblastoma, AURKB was identified as a direct transcriptional target of MYCN in MYCN-amplified neuroblastoma.48 The regulation of AURKB by MYCN would further hint at the possibility of therapeutic targeting of AURKB in MYCN-amplified RB tumors in addition to retinoblastoma with RB1 mutations.
Overall, our study indicates that the expression of AURKB is elevated in RB and positive AURKB expression is significantly correlated with optic nerve and anterior chamber invasion. More importantly, our study has demonstrated that AURKB could be specifically targeted in RB. Additionally, our results demonstrate a possible regulation of AURKB expression by MYCN.
Supplementary Material
Acknowledgments
The authors thank Bikash Ranjan Sahu, PhD (School of Biotechnology, KIIT University, Bhubaneswar, India), and Kalandi Charan Muduli (LV Prasad Eye Institute, Bhubaneswar, India) for their help with the flow cytometry and immunohistochemistry.
Supported by the Innovative Young Biotechnologist Award (to M.M.R.) from the Department of Biotechnology (DBT), Ministry of Science & Technology, Government of India (BT/09/IYBA/2015/10) and the Continuation of Centre of Excellence grant (BT/PR32404/MED/30/2136/2019; DBT). Additional support was received from Hyderabad Eye Research Foundation, LV Prasad Eye Institute.
Disclosure: N.A. Borah, None; S. Sradhanjali, None; M.R. Barik, None; A. Jha, None; D. Tripathy, None; S. Kaliki, None; S. Rath, None; S.K. Raghav, None; S. Patnaik, None; R. Mittal, None; M.M. Reddy, None
References
- 1. Shields CL, Shields JA. Diagnosis and management of retinoblastoma. Cancer Control. 2004; 11(5): 317–327. [DOI] [PubMed] [Google Scholar]
- 2. Fabian ID, Abdallah E, Abdullahi SU, et al.. Global retinoblastoma presentation and analysis by national income level. JAMA Oncol. 2020; 6(5): 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaliki S, Patel A, Iram S, Ramappa G, Mohamed A, Palkonda VAR. Retinoblastoma in India: clinical presentation and outcome in 1,457 patients (2,074 eyes). Retina. 2019; 39(2): 379–391. [DOI] [PubMed] [Google Scholar]
- 4. Waddell KM, Kagame K, Ndamira A, et al.. Clinical features and survival among children with retinoblastoma in Uganda. Br J Ophthalmol. 2015; 99(3): 387–390. [DOI] [PubMed] [Google Scholar]
- 5. Kaliki S, Shields CL, Eagle RC, Iram S, Shields JA. Comparison between Asian Indians and Americans from two major referral centers. Retina. 2018; 38(10): 2023–2029. [DOI] [PubMed] [Google Scholar]
- 6. Lambert MP, Shields C, Meadows AT. A retrospective review of hearing in children with retinoblastoma treated with carboplatin-based chemotherapy. Pediatr Blood Cancer. 2008; 50(2): 223–226. [DOI] [PubMed] [Google Scholar]
- 7. Leahey A. A cautionary tale: dosing chemotherapy in infants with retinoblastoma. J Clin Oncol. 2012; 30(10): 1023–1024. [DOI] [PubMed] [Google Scholar]
- 8. Gombos DS, Hungerford J, Abramson DH, et al.. Secondary acute myelogenous leukemia in patients with retinoblastoma. Is chemotherapy a factor? Ophthalmology. 2007; 114(7): 1378–1383. [DOI] [PubMed] [Google Scholar]
- 9. MacCarthy A, Bayne AM, Brownbill PA, et al.. Second and subsequent tumours among 1927 retinoblastoma patients diagnosed in Britain 1951-2004. Br J Cancer. 2013; 108(12): 2455–2463.23674091 [Google Scholar]
- 10. Dimaras H, Khetan V, Halliday W, et al.. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet. 2008; 17(10): 1363–1372. [DOI] [PubMed] [Google Scholar]
- 11. Rushlow DE, Mol BM, Kennett JY, et al.. Characterisation of retinoblastomas without RB1 mutations: genomic, gene expression, and clinical studies. Lancet Oncol. 2013; 14(4): 327–334. [DOI] [PubMed] [Google Scholar]
- 12. Gutteridge REA, Ndiaye MA, Liu X, Ahmad N. Plk1 inhibitors in cancer therapy: from laboratory to clinics. Mol Cancer Ther. 2016; 15(7): 1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh L, Pushker N, Sen S, Singh MK, Chauhan FA, Kashyap S. Prognostic significance of polo-like kinases in retinoblastoma: correlation with patient outcome, clinical and histopathological parameters. Clin Exp Ophthalmol. 2015; 43(6): 550–557. [DOI] [PubMed] [Google Scholar]
- 14. Yunoki T, Tabuchi Y, Hayashi A, Kondo T. Inhibition of Polo-like kinase 1 promotes hyperthermia sensitivity via inactivation of heat shock transcription factor 1 in human retinoblastoma cells. Invest Ophthalmol Vis Sci. 2013; 54(13): 8353–8363. [DOI] [PubMed] [Google Scholar]
- 15. Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017; 8(14): 23937–23954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Willems E, Dedobbeleer M, Digregorio M, Lombard A, Lumapat PN, Rogister B. The functional diversity of Aurora kinases: a comprehensive review. Cell Div. 2018; 13: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldenson B, Crispino JD.. The aurora kinases in cell cycle and leukemia. Oncogene. 2015; 34(5): 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001; 155(7): 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu XL, Fang Y, Lee TC, et al.. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009; 137(6): 1018–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu XL, Singh HP, Wang L, et al.. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 2014; 514(7522): 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sradhanjali S, Tripathy D, Rath S, Mittal R, Reddy MM. Overexpression of pyruvate dehydrogenase kinase 1 in retinoblastoma: a potential therapeutic opportunity for targeting vitreous seeds and hypoxic regions. PLoS One. 2017; 12(5): e0177744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jean A, Tardy F, Allatif O, Grosjean I, Blanquier B, Gerlier D. Assessing mycoplasma contamination of cell cultures by qPCR using a set of universal primer pairs targeting a 1.5 kb fragment of16S rRNA genes. PLoS One. 2017; 12(2): e0172358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhanot H, Reddy MM, Nonami A, et al.. Pathological glycogenesis through glycogen synthase 1 and suppression of excessive AMP kinase activity in myeloid leukemia cells. Leukemia. 2015; 29(7): 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han CP, Lee MY, Tzeng SL, et al.. Nuclear Receptor Interaction Protein (NRIP) expression assay using human tissue microarray and immunohistochemistry technology confirming nuclear localization. J Exp Clin Cancer Res. 2008; 27(1): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yates AD, Achuthan P, Akanni W, et al.. Ensembl 2020. Nucleic Acids Res. 2020; 48(D1): D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shohet JM, Ghosh R, Coarfa C, et al.. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011; 71(11): 3841–3851. [DOI] [PubMed] [Google Scholar]
- 27. Dreos R, Ambrosini G, Groux R, et al.. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 2017; 45(D1): D51–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dreos R, Ambrosini G, Périer RC, et al.. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015; 43(database issue): D92–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith SL, Bowers NL, Betticher DC, et al.. Overexpression of aurora B kinase (AURKB) in primary non-small cell lung carcinoma is frequent, generally driven from one allele, and correlates with the level of genetic instability. Br J Cancer. 2005; 93(6): 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Al-Khafaji ASK, Davies MPA, Risk JM, et al.. Aurora B expression modulates paclitaxel response in non-small cell lung cancer. Br J Cancer. 2017; 116(5): 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hartsink-Segers SA, Zwaan CM, Exalto C, et al.. Aurora kinases in childhood acute leukemia: the promise of aurora B as therapeutic target. Leukemia. 2013; 27(3): 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen HG, Makitalo M, Yang D, Chinnappan D, St. Hilaire C, Ravid K. Deregulated Aurora-B induced tetraploidy promotes tumorigenesis. FASEB J. 2009; 23(8): 2741–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. González-Loyola A, Fernández-Miranda G, Trakala M, et al.. Aurora B overexpression causes aneuploidy and p21 Cip1 repression during tumor development. Mol Cell Biol. 2015; 35(20): 3566–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang C, Chen J, Cao W, Sun L, Sun H, Liu Y. Aurora-B and HDAC synergistically regulate survival and proliferation of lymphoma cell via AKT, mTOR and Notch pathways. Eur J Pharmacol. 2016; 779: 1–7. [DOI] [PubMed] [Google Scholar]
- 35. Messmer EP, Heinrich T, Höpping W, de Sutter E, Havers W, Sauerwein W. Risk factors for metastases in patients with retinoblastoma. Ophthalmology. 1991; 98(2): 136–141. [DOI] [PubMed] [Google Scholar]
- 36. Takeshita M, Koga T, Takayama K, et al.. Aurora-B overexpression is correlated with aneuploidy and poor prognosis in non-small cell lung cancer. Lung Cancer. 2013; 80(1): 85–90. [DOI] [PubMed] [Google Scholar]
- 37. Falchook GS, Bastida CC, Kurzrock R. Aurora kinase inhibitors in oncology clinical trials: current state of the progress. Semin Oncol. 2015; 42(6): 832–848. [DOI] [PubMed] [Google Scholar]
- 38. Singh HP, Wang S, Stachelek K, et al.. Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proc Natl Acad Sci USA. 2018; 115(40): E9391–E9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu L, Ma CA, Zhao Y, Jain A. Aurora B interacts with NIR-p53, leading to p53 phosphorylation in its DNA-binding domain and subsequent functional suppression. J Biol Chem. 2011; 286(3): 2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gully CP, Velazquez-Torres G, Shin JH, et al.. Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci USA. 2012; 109(24): 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oser MG, Fonseca R, Chakraborty AA, et al.. Cells lacking the RB1 tumor suppressor gene are hyperdependent on aurora B kinase for survival. Cancer Discov. 2019; 9(2): 230–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poulard C, Kim HN, Fang M, et al.. Relapse-associated AURKB blunts the glucocorticoid sensitivity of B cell acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2019; 116(8): 3052–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bertran-Alamillo J, Cattan V, Schoumacher M, et al.. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun. 2019; 10(1): 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Villanueva MT. Tumorigenesis: establishing the origin of retinoblastoma. Nat Rev Cancer. 2014; 14(11): 706–707. [DOI] [PubMed] [Google Scholar]
- 45. Murphy DM, Buckley PG, Bryan K, et al.. Global MYCN transcription factor binding analysis in neuroblastoma reveals association with distinct E-box motifs and regions of DNA hypermethylation. PLoS One. 2009; 4(12): e8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cotterman R, Jin VX, Krig SR, et al.. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res . 2008; 68(23): 9654–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanagasabai T, Venkatesan T, Natarajan U, et al.. Regulation of cell cycle by MDM2 in prostate cancer cells through Aurora Kinase-B and p21WAF1(/CIP1) mediated pathways. Cell Signal. 2020; 66: 109435. [DOI] [PubMed] [Google Scholar]
- 48. Bogen D, Wei JS, Azorsa DO, et al.. Aurora B kinase is a potent and selective target in MYCN-driven neuroblastoma. Oncotarget. 2015; 6(34): 35247–35262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.