Abstract
RNA modifications have recently emerged as a widespread and complex facet of gene expression regulation. Counting more than 170 distinct chemical modifications with far-reaching implications for RNA fate, they are collectively referred to as the epitranscriptome. These modifications can occur in all RNA species, including messenger RNAs (mRNAs) and noncoding RNAs (ncRNAs). In mRNAs the deposition, removal, and recognition of chemical marks by writers, erasers and readers influence their structure, localization, stability, and translation. In turn, this modulates key molecular and cellular processes such as RNA metabolism, cell cycle, apoptosis, and others. Unsurprisingly, given their relevance for cellular and organismal functions, alterations of epitranscriptomic marks have been observed in a broad range of human diseases, including cancer, neurological and metabolic disorders. Here, we will review the major types of mRNA modifications and editing processes in conjunction with the enzymes involved in their metabolism and describe their impact on human diseases. We present the current knowledge in an updated catalog. We will also discuss the emerging evidence on the crosstalk of epitranscriptomic marks and what this interplay could imply for the dynamics of mRNA modifications. Understanding how this complex regulatory layer can affect the course of human pathologies will ultimately lead to its exploitation toward novel epitranscriptomic therapeutic strategies.
Keywords: RNA modifications, epitranscriptomics, mRNA, posttranscriptional regulation of gene expression, human disease, cancer
INTRODUCTION
RNA molecules can undergo more than 170 different chemical modifications (Boccaletto et al. 2018). These marks can decorate many types of RNA species, both coding and noncoding RNA (ncRNA), including messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA) and others. This ever-expanding set of RNA modifications, collectively referred to as the epitranscriptome, has recently emerged as a widespread facet of cotranscriptional and posttranscriptional gene expression regulation (Laurencikiene et al. 2006; Saletore et al. 2012; Nachtergaele and He 2017; Roundtree et al. 2017; Martinez and Gilbert 2018; Zhao et al. 2018). These regulatory layers are key determinants of protein levels and cellular phenotypes (Halbeisen et al. 2008; Vogel et al. 2010; Schwanhäusser et al. 2011; Corbett 2018).
A broad set of RNA-binding proteins (RBPs) determines the mRNA epitranscriptome: Modifications are induced by writers, and several can be reverted by erasers. Eventually, some modifications need readers to be decoded. (Kadumuri and Janga 2018; Nachtergaele and He 2018; Delaunay and Frye 2019; Quinones-Valdez et al. 2019). Through the action of these RBPs, the epitranscriptome controls processes ranging from alternative splicing and polyadenylation to RNA stability, localization, and translation (Gerstberger et al. 2014; Bartel 2018). These regulators form complex networks of interaction leading to a dynamic control of gene expression with deep implications for cellular physiology and pathology (Wurth and Gebauer 2015; Dassi 2017; Quattrone and Dassi 2019; Zanzoni et al. 2019). Given their relevance in multiple cellular functions, alterations of RNA modifications and their modifying enzymes have been observed in a broad range of human diseases, including cancer, neurological disorders and several others (Meier et al. 2016; Jonkhout et al. 2017; Angelova et al. 2018; Jain et al. 2018; Christofi and Zaravinos 2019; Huang et al. 2020b).
In this review, we will describe mRNA modifications and their increasingly appreciated role as drivers of human pathologies. We will give particular focus on the most abundant ones, namely RNA editing (A-to-I and C-to-U), N6-methyladenosine (m6A), and pseudouridine (Ψ), for which we provide flashcards (Figs. 1–4) summarizing their most important features and disease associations, and a comprehensive list of disease-related modified sites (Supplemental Table S1). Furthermore, we will provide an overview of detection methods and discuss emerging evidence on the interplay of different modifications, proposing potential avenues to improve our understanding of these pervasive RNA regulators.
FIGURE 1.
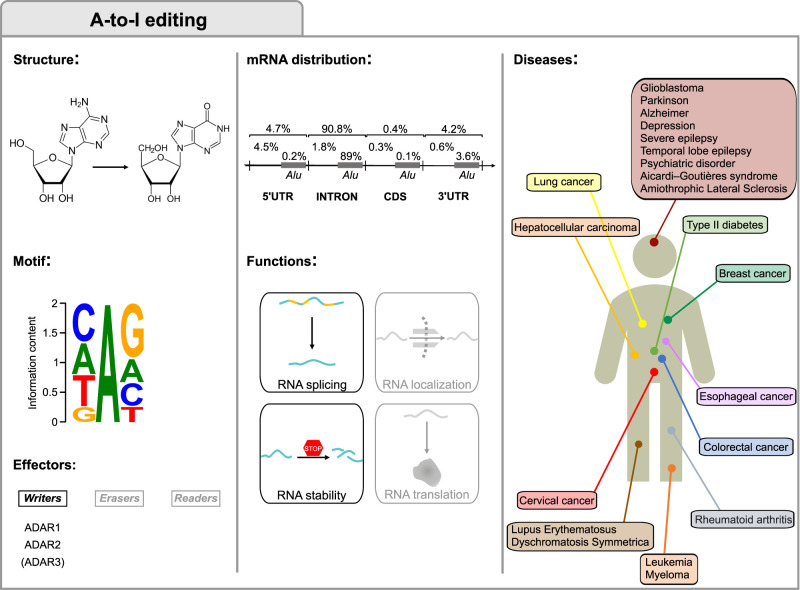
A-to-I editing. The first column displays the structures of adenosine and inosine involved in the deamination, the consensus motif and the A-to-I editing main effectors. The motif was obtained by data in Cohen-Fultheim and Levanon (2021) and plotted with WebLogo (Crooks et al. 2004). The central column shows the percentage of editing at nonrepetitive regions and Alu repeats and the functions in mRNA fate. The third column displays A-to-I editing-associated disorders and the organs to which they are associated.
FIGURE 2.
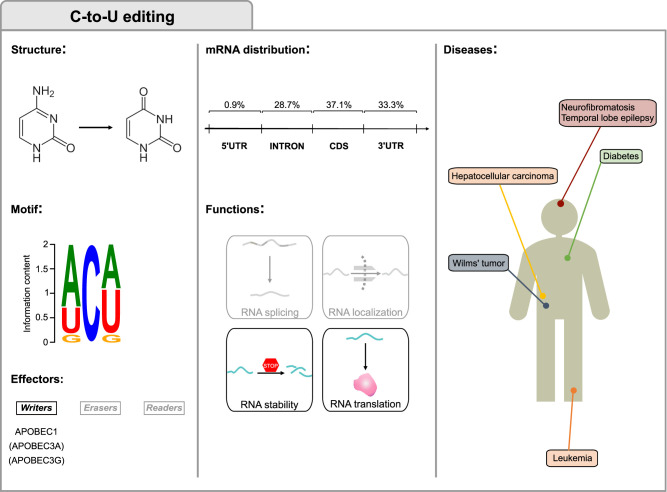
C-to-U editing. The first column displays the structures of cytidine and uridine involved in the deamination, the consensus motif and the C-to-U editing main effectors. The motif was obtained by data in Rosenberg et al. (2011) and plotted with WebLogo (Crooks et al. 2004). The central column shows the percentage of editing in the mRNA regions and the functions in mRNA fate. The third column displays C-to-U editing-associated disorders and the organs to which they are associated. Considering that little is known on the significance of RNA editing by APOBEC3A and APOBEC3G, all features in the figure relate to APOBEC1, and APOBEC3A/APOBEC3G are only mentioned in parentheses.
FIGURE 3.
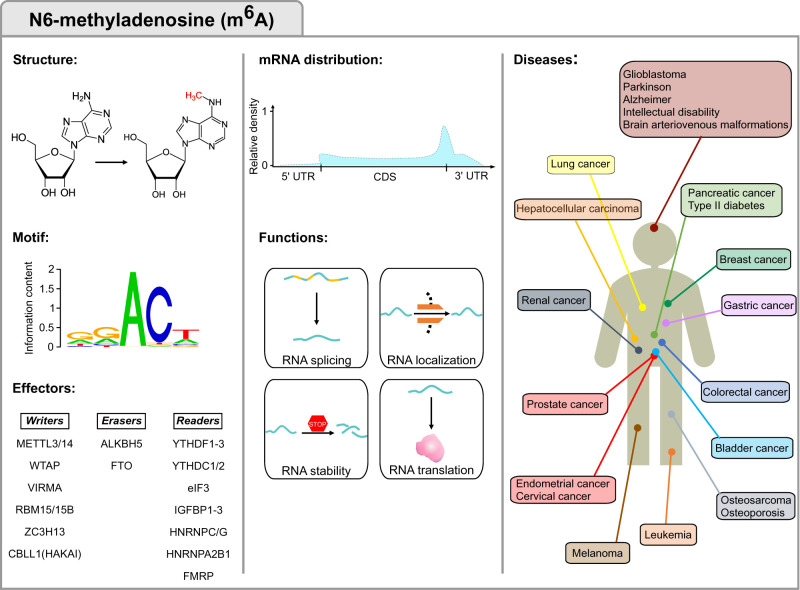
N6-methyladenosine (m6A) modification. The first column displays the m6A structure, consensus motif and m6A machinery factors. The motif was obtained by data in Linder et al. (2015) and plotted with WebLogo (Crooks et al. 2004). The central column highlights the m6A distribution and functions in mRNA fate, while the third column displays m6A-associated disorders and the organs to which they are associated.
FIGURE 4.
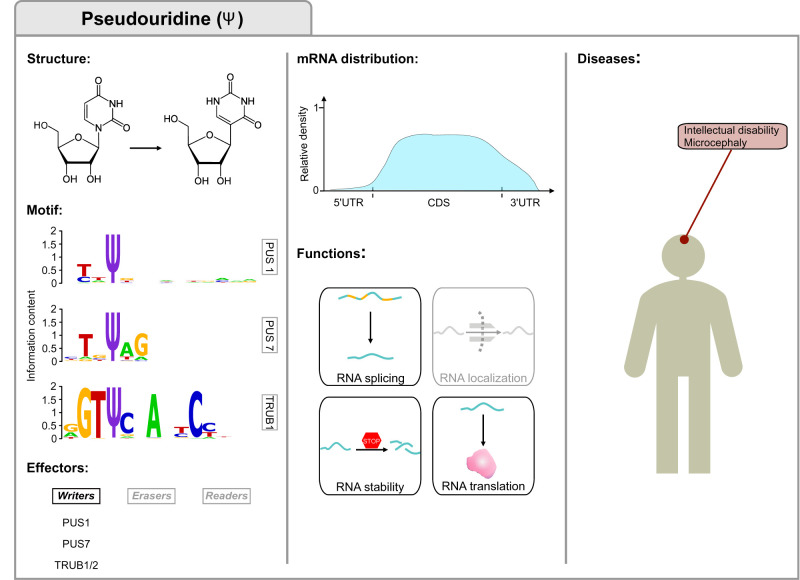
Pseudouridine (Ψ) modification. The first column displays the Ψ structure, consensus motifs and lists the main writers of Ψ in mRNAs. The motif was obtained by data in Carlile et al. (2019) and plotted with WebLogo (Crooks et al. 2004). The central column highlights Ψ distribution (Carlile et al. 2019) and functions in mRNA fate. The third column displays the disorders associated with dysregulated pseudouridylation.
EPITRANSCRIPTOMIC MARKS
RNA editing by deamination
RNA editing, mediated by several enzymes belonging to a zinc-binding superfamily of deaminases, targets most types of cellular RNAs. A-to-I is the most common form of editing in human cells and is performed by the adenosine deaminase acting on RNA (ADAR) enzymes (Bass 2002; Mannion et al. 2015; Eisenberg and Levanon 2018). Alongside A-to-I editing, C-to-U editing is performed by the Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) family of enzymes. Both ADAR and APOBEC gene families likely originate from the adenosine deaminase acting on tRNA (ADAT) genes (Gerber and Keller 2001; Conticello et al. 2007), whose encoded proteins edit the wobble position of many tRNAs (Torres et al. 2014).
A-to-I editing
In humans, A-to-I editing (Fig. 1) is mediated by ADAR1 and ADAR2, while the catalytically inactive ADAR3 can modulate the process. These enzymes act as homodimers and deaminate adenosines within double-stranded regions of RNA (Gallo et al. 2003; Thuy-Boun et al. 2020). Binding to the target region is mediated by double-stranded RNA (dsRNA) binding domains. Since inosines that result from editing are read as guanosines by the translational machinery, editing can recode the mRNA and lead to the translation of proteins different from those specified by the genome, thus increasing the complexity of the transcriptome.
The first edited sites were discovered on the transcripts of the glutamate receptor 2 (GRIA2) and the serotonin 5-hydroxytryptamine (2C) (5-HT2c) receptors (Sommer et al. 1991; Higuchi et al. 1993; Burns et al. 1997). GRIA2 editing is essential for brain development as it allows formation of heteromeric complexes modulating neuronal function. Historically, the main role of A-to-I editing was considered to be recoding, mainly due to the importance of ADAR2-mediated editing in brain development (Brusa et al. 1995; Higuchi et al. 2000). However, it soon became evident that many edited sites lie outside the coding regions (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004; Li et al. 2009; Bazak et al. 2014; Picardi et al. 2016, 2017; Eisenberg and Levanon 2018). Most A-to-I RNA editing sites occur on noncoding sequences such as 5′ and 3′ untranslated regions (UTRs) (Chen and Carmichael 2012), introns, and microRNAs (miRNAs) (Luciano et al. 2004; Blow et al. 2006; Yang et al. 2006). In humans, most of these sites lie in Alu sequences, ancient retrotransposons whose repeated sequences facilitate formation of double-stranded structures (Athanasiadis et al. 2004; Kim et al. 2004; Levanon et al. 2004). ADAR-mediated editing of noncoding regions can modulate the RNA fate and function. For example, changes in their primary sequence can affect how they are targeted by miRNAs (Roberts et al. 2018) or alter transcript splicing (Rueter et al. 1999). More importantly, insertion of I:U mismatches in place of A:U pairs can alter the structure of the RNA itself, affecting transcript interactions and stability (Wang et al. 2013). Indeed, ADAR1 deficiency leads to accumulation of cytoplasmic dsRNA that, being interpreted as a sign of viral infection, leads to the activation of the cellular response to dsRNA through RIG-I and MDA5 (Mannion et al. 2014; Liddicoat et al. 2015; Pestal et al. 2015). ADAR1 homozygous deficiency in mice induces embryonic lethality (Wang et al. 2000).
ADAR1 also plays a role in the physiological interferon-mediated cellular response, as widespread editing prevents translational shutdown and cell death (Hartner et al. 2009; Chung et al. 2018). Missense mutations in ADAR1 cause Aicardi–Goutières Syndrome, a childhood autoimmune encephalitis characterized by increased interferon (Rice et al. 2012; Gallo et al. 2017). Mutations in ADAR1 are also associated with dyschromatosis symmetrica hereditaria (DSH), a rare autosomal genetic disorder of the skin, but the pathogenetic mechanisms are not yet clear (Miyamura et al. 2003; Kono and Akiyama 2019).
Deficiencies of A-to-I RNA editing mediated by ADAR2 have instead been associated with diseases of the central nervous system (Costa Cruz and Kawahara 2021). Increased levels of GRIA2 editing have been found in epileptic patients (Vollmar et al. 2004). In amyotrophic lateral sclerosis (ALS), alterations in editing levels of GRIA2 and other transcripts may contribute to the disease (Kawahara et al. 2004; Kwak et al. 2008; Donnelly et al. 2014). Similarly, decreases in editing levels of the 5-HT2C serotonin receptor affect serotonin production and are involved in several psychiatric disorders (Sodhi et al. 2001; Grohmann et al. 2010; O'Neil and Emeson 2012; Weissmann et al. 2016), and it has also been found in the prefrontal cortex of suicide victims (Gurevich et al. 2002a,b). Reduced editing was also observed in Alzheimer's patients (Khermesh et al. 2016; Franzén et al. 2018). Moreover, probably due to their involvement in interferon response, ADAR enzymes may play a role in autoimmune diseases, such as lupus erythematosus (Laxminarayana et al. 2002, 2007; Orlowski et al. 2008; Vlachogiannis et al. 2020).
Alterations in A-to-I editing have also been associated with cancer (Kung et al. 2018). On one hand, hypo-editing in Alu repeats has been observed in several tumor types (Paz et al. 2007). Low levels of GRIA2 editing were observed in human gliomas (Maas et al. 2001) and overall editing levels have been used to stratify glioblastoma patients (Tomaselli et al. 2015; Silvestris et al. 2019). On the other hand, increased levels of ADAR1 have been observed in esophageal, lung carcinomas (Qin et al. 2014; Anadón et al. 2016) in lymphoproliferative diseases (Beghini et al. 2000; Jiang et al. 2013; Lazzari et al. 2017) and in hepatocellular carcinoma (Chen et al. 2013), sometimes associated with poor prognosis. Editing of AZIN1 is correlated to hepatocellular carcinoma and it is involved in cell proliferation and invasion by maintaining polyamine homeostasis (Chen et al. 2013; Qin et al. 2014; Shigeyasu et al. 2018) and high levels of A-to-I editing of the Ras homolog family member Q increase tumor invasion in colorectal cancer (Han et al. 2014). Intriguingly, editing targets of ADAR2 with opposite effects have been identified in esophageal squamous cell carcinoma (Chen et al. 2017; Fu et al. 2017). Composite effects have also been observed as up-regulation of ADAR1 and down-regulation of ADAR2 promote hepatocellular carcinoma (Chan et al. 2014).
Finally, viruses can hijack the A-to-I editing machinery to trigger a proviral phenotype through editing of viral transcripts (Phuphuakrat et al. 2008; Doria et al. 2009) or cellular transcripts that modulate the cellular response (Gélinas et al. 2011; Pfaller et al. 2011; Samuel 2012).
C-to-U editing
C-to-U RNA editing (Fig. 2) was the first form of editing described in humans when a discrepancy between transcript and genetic sequence of the Apolipoprotein B (APOB) mRNA was identified in the small intestine (Chen et al. 1987). Recoding of the APOB transcript leads to the translation of a truncated form -ApoB48-, that allows synthesis of chylomicrons (Lo and Coschigano 2020).
Soon after, a deaminase -APOBEC1- was identified as the enzymatic core of the editing complex (Navaratnam et al. 1993; Teng et al. 1993; Blanc and Davidson 2010). APOBEC1 is a member of the AID/APOBEC family of deaminases that target cytosines in the context of single-stranded nucleic acids (Salter et al. 2016).
Contrary to ADARs, whose targeting presents only a slight sequence preference (guanines 5′ to the edited sites are not favored—e.g., Cohen-Fultheim and Levanon 2021), APOBEC1 targets nucleic acids within a stronger sequence context (adenine/uracil 5′ to the edited site, A/U rich regions—e.g., Rosenberg et al. 2011; Blanc et al. 2014). While APOBEC1 can target RNA autonomously, the editing specificity and efficiency are largely determined by its cofactors APOBEC1 complementation factor (A1CF) (Lellek et al. 2000; Mehta et al. 2000) and RNA-binding-motif-protein-47 (RBM47) (Fossat et al. 2014; Fossat and Tam 2014). Until advent of high-throughput sequencing, only a few APOBEC1-edited transcripts had been identified beyond APOB (Skuse et al. 1996; Yamanaka et al. 1997; Meier et al. 2005). The availability of APOBEC1 deficient murine models allowed the identification of several other editing targets of APOBEC1 in mice (Rosenberg et al. 2011; Blanc et al. 2014; Harjanto et al. 2016; Cole et al. 2017; Rayon-Estrada et al. 2017). Most of these editing sites lie in the 3′UTRs and at this time it is still difficult to envision a global role for APOBEC1 editing beyond its effect on selected specific transcripts. For example, APOBEC1 deficiency in mice promotes a proinflammatory environment in the brain, likely mediated by lack of editing in microglia, which is correlated to progressive central nervous system pathophysiology (Cole et al. 2017). A fascinating hypothesis posits that APOBEC1-mediated RNA editing might increase variability among cellular subpopulations (Harjanto et al. 2016). More important, APOBEC1-mediated RNA editing could also act as a restriction factor against viruses and mobile elements (Bishop et al. 2004; Petit et al. 2009; Ikeda et al. 2011; Di Giorgio et al. 2020). Independently from its editing activity, APOBEC1 binding to RNA could regulate transcript stability (Anant and Davidson 2000; Prohaska et al. 2014).
A potential role in disease for APOBEC1 has been envisioned since its discovery: Overexpression of APOBEC1 in the liver of several animal models induces cancer (Yamanaka et al. 1995) and its deficiency in cancer-prone mice reduces the onset of neoplastic lesions (Blanc et al. 2007). Indeed, recoding of the tumor-suppressor NF1 (Skuse et al. 1996; Mukhopadhyay et al. 2002) leads to its inhibition, and editing of NAT1 (Yamanaka et al. 1997) could deregulate p21 (Kung et al. 2018). Noteworthy, APOBEC1 can also target DNA and therefore, its oncogenic potential could also be derived from its mutagenic activity (Saraconi et al. 2014).
APOBEC1 might also be involved in the progression of temporal lobe epilepsy through editing of glycine receptors (Meier et al. 2005; Kankowski et al. 2018).
Considering how central APOB editing is for cholesterol transport in the blood—Apobec1 deficient mice display hypercholesterolemia—mutations/polymorphisms in Apobec1 might increase the risk of cardiovascular diseases. Yet, no inactivating mutations have been identified in humans so far.
Beyond APOBEC1, other AID/APOBECs target RNA (Sharma et al. 2015, 2016; Asaoka et al. 2019; Jalili et al. 2020). Among them, APOBEC3A is induced by hypoxia and interferon in monocytes and macrophages (Sharma et al. 2015). While its biological significance is not yet clear, increased levels of C-to-U RNA editing in tumors have been associated with improved survival, likely due to a better immune response (Asaoka et al. 2019).
N6-methyladenosine
m6A, or methylation at the N6 position in adenosine (Fig. 3), is the most abundant internal modification in mRNAs and long noncoding RNAs (lncRNAs) in eukaryotes, regulating transcriptional and posttranscriptional processes that control gene expression. m6A was first discovered in mRNAs in 1974, and shortly after, the RRACH motif was identified as a highly conserved m6A consensus sequence in mammals (Desrosiers et al. 1974; Perry and Kelley 1974; Lavi and Shatkin 1975; Wei et al. 1975; Schibler et al. 1977; Wei and Moss 1977; Csepany et al. 1990; Harper et al. 1990; Linder et al. 2015). Furthermore, it has been shown that m6A can be added cotranscriptionally (Ke et al. 2017), and it has been predominantly located at long internal exons, near stop codons and along 3′UTRs (Dominissini et al. 2012; Meyer et al. 2012; Linder et al. 2015).
In mammalian cells, the core m6A methyltransferase complex, hereafter referred to as “writers,” consists of the heterodimer Methyltransferase-Like 3 (METTL3) and 14 (METTL14) (Liu et al. 2014), effecting the enzymatic activity and serving as an RNA-binding scaffold, respectively (Śledź and Jinek 2016; Wang et al. 2016b). Other writer components, reviewed elsewhere (Lence et al. 2019), are important for the deposition of m6A at specific transcripts. Fat mass and obesity-associated protein (FTO) and Alkb homolog 5 (ALKBH5) are the demethylases or “erasers” of m6A (Jia et al. 2011; Zheng et al. 2013), suggesting that this chemical modification can be formed and removed in a reversible manner. The m6A-mark is recognized by a group of proteins categorized as “readers” which bind and decode transcripts harboring the m6A modification into distinct RNA fates (Zaccara et al. 2019).
m6A plays a central role in several biological and pathological processes (Fig. 3; Aguilo and Walsh 2017; Malla et al. 2019). Hence, the dysregulated expression of writers, erasers, and readers, leading to aberrant m6A patterns, plays a role in metabolic disease, neurodegeneration, and tumorigenesis, among others. For instance, m6A is necessary for the function of the pancreatic β-cell, as depletion of m6A impairs insulin secretion by decreasing AKT phosphorylation and PDX1 protein levels (Jesus et al. 2019). Noticeably, in type 2 diabetes patients, decreased METTL3/14 expression in β cells has been observed (Jesus et al. 2019).
Furthermore, its diverse implications in neurobiological processes have been highlighted in many studies (Widagdo and Anggono 2018; Du et al. 2019; Li et al. 2019; Rockwell and Hongay 2019; Chokkalla et al. 2020). Thus, perturbations of the m6A machinery have been observed in numerous neuropathological states, including Alzheimer's disease, depression, and gliomas. Increased m6A and METTL3 levels promote the development of Alzheimer's disease (Han et al. 2020; Huang et al. 2020a). Moreover, decreased FTO expression has been correlated with increased risk of Alzheimer's disease in different ethnic populations (Reitz et al. 2012). Polymorphisms in m6A erasers have also been linked with increased disease risk for major depressive disorder, and attention deficit/hyperactivity disorder (Choudhry et al. 2013; Du et al. 2015; Huang et al. 2020c). However, conflicting evidence has been observed in Parkinson's disease. While a study has shown association of polymorphism in ALKBH5 with Parkinson's disease (Qiu et al. 2020), another one, conducted on the Han Chinese population was unable to identify a significant correlation between this disease and gene variation of m6A players (Qin et al. 2020). Whether these antithetical results are based on ethnic differences in gene variants between the study populations or are given by gene specific m6A modifications remains to be clarified.
The m6A modification is associated with cancer. m6A writers, erasers, and readers act either as oncogenes or tumor suppressors in several types of cancer, although the mechanisms behind are still poorly understood (Lan et al. 2019). For instance, findings for the m6A machinery in breast cancer can seem controversial. Hence, high and low levels of m6A modification have been reported to be both oncogenic and tumor-suppressive. Recent studies have shown that METTL3 is highly expressed in breast cancer tissue compared to normal tissue and that silencing METTL3 could lead to a decrease in proliferation, increased apoptosis, and thereby inhibit tumor growth in vivo and in vitro. Mechanistically, METTL3 promoted the expression of the oncoprotein hepatitis B virus X-interacting protein (HBXIP) which in turn, facilitated METTL3 expression by inhibiting miRNA let-7g which targets METTL3 for subsequent degradation (Cai et al. 2018). In addition, METTL3-mediated deposition of m6A at the BCL-2 transcript increased its translation. BCL-2 is one of the most important anti-apoptotic genes which facilitates the survival of tumor cells enhancing the breast cancer phenotype (Wang et al. 2020a). However, another study revealed that the expression of METTL3, together with METTL14 and WTAP, was significantly decreased in breast cancer (Wu et al. 2019). Therefore, according to this study, low m6A levels would promote breast tumorigenesis. Indeed, depletion of m6A at large internal exons results in prematurely polyadenylated transcripts, leading to nonfunctional tumor suppressor genes (Ni et al. 2018). In addition, the eraser FTO is up-regulated in breast cancer where it down-regulates the pro-apoptotic factor BNIP3 to mediate breast cancer proliferation, progression, and metastasis (Niu et al. 2019). Hypoxic environments and dysregulation of hypoxia-inducible factors (HIFs) lead to an adaptive response playing a central role in tumor progression and therapy resistance (Masson and Ratcliffe 2014). The expression of the eraser ALKBH5 was induced by hypoxia/HIF-dependent mechanisms, leading to decreased m6A levels that promoted the specification of breast cancer stem cells (CSC) (Zhang et al. 2016a). In addition, hypoxia also induced the expression of the oncogenic transcription factor ZNF217, promoting the breast CSC phenotype (Zhang et al. 2016b). The mouse orthologue ZFP217 has been shown to recruit the methyltransferase METTL3 into an inactive complex in embryonic stem cells (Aguilo et al. 2015), and hence, would cooperate with ALKBH5 in negatively regulating m6A levels and promoting breast tumorigenesis.
In summary, both high and deficient m6A levels might influence global expression programs that lead to malignant phenotypes, and the crosstalk among m6A readers, erasers and writers critically regulates the expression of key transcripts to maintain cellular homeostasis (Panneerdoss et al. 2018).
Pseudouridine
Ψ (Fig. 4) was the first RNA modification identified in the early 1950s (Cohn and Volkin 1951; Davis and Allen 1957). It was originally described in tRNAs and has been detected in rRNAs, small nuclear RNAs (snRNAs), other ncRNAs, and mRNAs, representing the most abundant of all known RNA marks (Davis and Allen 1957; Reddy et al. 1972; Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014; Li et al. 2015; Adachi et al. 2019a,b). It consists of a posttranscriptional isomerization of uridine, resulting in the addition of an extra carbon–carbon bond between the base and the sugar, and a hydrogen bond donor (Cohn 1960; Charette and Gray 2000).
Ψ is an irreversible modification which can occur through two different mechanisms. The first is RNA-dependent, mediated by the H/ACA box small nucleolar ribonucleoproteins (snoRNPs) complex comprised of four conserved proteins, namely NHP2, GAR1, NOP10, and dyskerin (Yu et al. 2005; Hamma and Ferré-D'Amaré 2010; Ge and Yu 2013; Yu and Meier 2014; Adachi et al. 2019b). In contrast, the second is a highly conserved RNA-independent mechanism and involves different types of pseudouridine synthases (PUS enzymes) (Koonin 1996; Kaya and Ofengand 2003). These enzymes possess a conserved catalytic domain which enables them to recognize uridine substrates and convert them to Ψ (Hamma and Ferré-D'Amaré 2006; Rintala-Dempsey and Kothe 2017). Some members of the PUS enzyme family were identified as catalyzers of this dynamic, stress-induced modification on mRNA, specifically, PUS1, PUS7, and the mammalian homologs of yeast Pus4—TRUB1, TRUB2 (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014; Li et al. 2015; Safra et al. 2017a; Carlile et al. 2019). For some of these enzymes, namely PUS7 and TRUB1, conserved consensus sequence motifs were detected in both yeast and human cells through an in vitro pseudouridylation assay (Carlile et al. 2019). Instead, only a weak, three-nucleotide sequence motif (HRU) was identified for PUS1, for which indeed a shared structure motif was detected (Carlile et al. 2019).
To date, only a few biological functions of Ψ on mRNAs have been identified (Adachi et al. 2019b; Borchardt et al. 2020). Previous studies have shown that pseudouridylation contributes to mRNA stabilization and the enhancement of translational capability in some mRNAs in vitro (Karikó et al. 2008; Anderson et al. 2010; Schwartz et al. 2014; Adachi et al. 2019b). In addition, artificial changes of U to Ψ in premature stop codons resulted in stop codon read-through both in vitro and in vivo, and in suppression of nonsense-mediated mRNA decay (Karijolich and Yu 2011; Adachi and Yu 2020). However, Ψ-containing mRNAs have also been shown to impede translation elongation and alter tRNA selection by the ribosome (Eyler et al. 2019). Thus, further work is needed to fully understand the role of pseudouridine in determining endogenous mRNAs fate.
The importance of Ψ in human pathology was highlighted by numerous studies associating its dysregulation in ncRNAs with diseases such as X-linked dyskeratosis congenita, cancer, diabetes, viral infections, heart defects, and inherited and mitochondrial disorders (Montanaro et al. 2006; Alter et al. 2009; Sieron et al. 2009; Liu et al. 2012; Fernandez-Garcia et al. 2013; Shaheen et al. 2016; Wang et al. 2016a; Zhao et al. 2016; Penzo et al. 2017; de Brouwer et al. 2018; Darvish et al. 2019; Shaheen et al. 2019; Watanabe et al. 2019; Nagasawa et al. 2020). Whether alteration of Ψ sites on mRNAs is also involved in these or other pathologies remains to be elucidated. Recent studies indicate, however, a potential connection. For instance, mutations in PUS7 that segregate with intellectual disability and microcephaly lead to the abolishment of pseudouridylation not only in tRNAs but also in mRNAs (de Brouwer et al. 2018; Shaheen et al. 2019). Collectively, these results highlight the necessity for a deeper understanding of how mRNA pseudouridylation is related to human pathologies.
Other mRNA modifications
N6, 2′-O-di-methyladenosine
Adjacent to the N7-methylguanosine (m7G) cap, the second nucleotide in many mRNAs can be methylated at the 2′-hydroxyl group; if the transcription start nucleoside is 2′-O-methyladenosine (Am), its N6 position can be further methylated to form N6, 2′-O-dimethyladenosine (m6Am) (Keith et al. 1978).
This modification stabilizes the mRNA by preventing DCP2-mediated decapping and microRNA-mediated mRNA degradation (Mauer et al. 2017). Unlike m6A, the biological function of m6Am and its role in cellular homeostasis are still poorly understood.
m6Am is a reversible modification catalyzed by the writer PCIF1/CAPAM (Akichika et al. 2019). PCIF1/CAPAM knockout cells are viable, but sensitive to oxidative stress (Akichika et al. 2019), a common adaptive advantage found in many types of cancer. Indeed, a genetic screen identified PCIF1/CAPAM as a putative tumor growth suppressor in bladder cancer (Hensel et al. 2015). m6Am is also erased by the demethylase FTO (Mauer et al. 2017). Hence, whether a given phenotype resulting from the loss of FTO is due to defects in m6A or m6Am metabolism is ambiguous and controversial: Whereas FTO has higher demethylase activity toward m6Am, the number of m6A sites in mRNA is at least 10-fold higher than the number of m6Am sites. It has been proposed that FTO localization within the cellular compartments can vary between cell types and pathological states, being FTO-mediated demethylation of m6A and m6Am prominent in the nucleus and in the cytoplasm, respectively (Wei et al. 2018a). In agreement with this observation, cytoplasmic FTO inhibits the CSC phenotype in colorectal cancer through its m6Am demethylase activity. Hence, low FTO expression in patient-derived cell lines leads to increased m6Am mRNA levels, resulting in enhanced tumorigenesis and chemoresistance (Relier et al. 2020).
N1-methyladenosine
Methylation at the N1 position in adenosine (m1A) confers a positive charge that can influence the local structure of the RNA or its interaction with RBPs. It can be found in several RNA species, including tRNA, rRNA and mRNA (Dominissini et al. 2016; Li et al. 2016b, 2017b; Safra et al. 2017b). In the particular case of this mark, the methyl group is added by distinct isoforms of the TRMT family of proteins, namely TRMT6/61A and TRMT10C, depending on the cytoplasmic or mitochondrial localization of the target mRNA (Safra et al. 2017b). The methyl group blocks the normal Watson–Crick base-pairing, resulting in erroneous incorporation and translation blocking. Early transcriptome-wide m1A-mapping studies suggested that thousands of transcripts could be decorated with the m1A mark (Dominissini et al. 2016; Li et al. 2016b). In addition, it was proposed that this modification correlated with higher translation efficiency when located in the 5′UTR of mRNA (Dominissini et al. 2016; Li et al. 2016b). However, a later m1A base-resolution mapping study revealed that m1A is not widespread on mRNAs and identified only ten and five cytosolic and mitochondrial m1A-modified transcripts, respectively (Safra et al. 2017b). The enzyme NADH dehydrogenase-5 (ND5) was among the m1A-marked mitochondrial mRNAs identified. ND5 contains a single-nucleotide polymorphism that prevents the formation of m1A in the ND5 mRNA. This mutation is linked to Leber's hereditary optic neuropathy, a hereditary disease leading to acute loss of central vision (Safra et al. 2017b).
The removal of m1A from mRNA is catalyzed by ALKBH3 (Li et al. 2016b). ALKBH3 is highly expressed in human tumors including prostate (Koike et al. 2012), non-small-cell lung (Tasaki et al. 2011), pancreatic (Yamato et al. 2012), and renal cell carcinoma (Hotta et al. 2015), and elevated ALKBH3 expression is associated with poor prognosis. However, whether high expression of ALKBH3 leads to aberrant m1A in cancer patients remains elusive. Notably, ALKBH3 also targets other substrates than m1A-marked RNA which also include abasic sites and methylated nucleosides of DNA (Westbye et al. 2008; Müller et al. 2010). The readers of the m1A modification include YTHDF1-3 and YTHDC1, although the downstream effect on the RNA fate and, therefore, disease outcome, remains to be elucidated (Dai et al. 2018b).
5-methylcytosine
The methylation of carbon 5 in cytosine (m5C) was initially discovered in rRNAs and tRNAs. More recently, high-throughput techniques have revealed its presence in mRNAs, although its prevalence is limited (Yang et al. 2017).
In multicellular organisms, m5C is catalyzed by at least seven conserved RNA m5C methyltransferases of the NOL1/NOP2/SUN domain (NSUN) family of proteins (NSUN1–7) and DNMT2, being all of them specific for distinct RNA species. Although NSUN2 was originally described as a tRNA methyltransferase (Frye and Watt 2006; Goll et al. 2006; Tuorto et al. 2012), it can also methylate other ncRNA species (Khoddami and Cairns 2013) and mRNA (Yang et al. 2017). Several studies have shown that m5C sites are not randomly distributed—they are most abundant in proximity to the translation start codon, 3′UTRs, and near Argonaute-binding regions (Squires et al. 2012; Amort et al. 2017; Legrand et al. 2017; Yang et al. 2017). NSUN2-mediated m5C deposition influences mRNA translation (Tang et al. 2015; Bohnsack et al. 2019) nuclear-cytoplasmic shuttling (Yang et al. 2017), and mRNA stabilization (Chen et al. 2019). For instance, m5C deposition on the HDGF oncogene mRNA promotes its stabilization, therefore driving urothelial carcinoma of the bladder (Chen et al. 2019). NSUN2 is a direct target of the oncogene Myc, and it is required for Myc-induced proliferation (Frye and Watt 2006). Consistently, NSUN2 is highly expressed in a range of tumors such as breast cancer, lymph-node metastases, and colorectal cancer (Frye et al. 2010; Okamoto et al. 2012; Yi et al. 2017). In gastric cancer, NSUN2 can suppress p57Kip2 and therefore promote tumor growth (Mei et al. 2020). In head and neck carcinoma, high NSUN2 expression adversely affects other tumor suppressors such as TP53, p16, and p27, increasing the risk of mortality (Lu et al. 2018). Depletion of NSUN2 results in decreased growth of human squamous-cell-carcinoma xenografts, suggesting that NSUN2 could be potentially targeted for cancer therapy (Frye and Watt 2006). Importantly, whether the oncogenic phenotypes resulting from NSUN2 overexpression are specifically due to aberrant m5C deposition at mRNA, tRNA or both RNA species, needs to be further investigated.
2′-O-methylation
2′-O-methylation (2′-O-Me) consists in the transfer of a methyl group at the 2′-hydroxyl of the ribose of all RNA species, predominantly of rRNA and tRNA (reviewed elsewhere in Ayadi et al. 2019; Dimitrova et al. 2019; Höfler and Carlomagno 2020).
Its deposition on mRNA was thought to occur only at the first 2 ribonucleotides (N1 and N2) within the 5′ cap structure (Langberg and Moss 1981; Inesta-Vaquera et al. 2018) being deposited solely by cap methyltransferase 1 (CMTR1) and 2 (CMTR2), respectively (Bélanger et al. 2010; Werner et al. 2011; Smietanski et al. 2014). Although, thousands of potential 2′-O-Me sites on coding regions of human mRNAs were identified by a detection technique known as Nm-seq (Dai et al. 2017), the same authors published a later corrigendum stating that this method was suitable to identify 2′-O-Me sites in rRNA but had led to false positives sites in mRNA due to mispriming contamination. According to the authors, a refined Nm-seq version was able to detect a similar distribution pattern as the original version, results confirmed by other techniques (Dai et al. 2018a). Nevertheless, further work to validate these newly identified sites is required.
In addition, the mechanism of 2′-O-Me deposition on coding transcripts is still to be revealed, but it might be guided by box C/D snoRNAs (SNORDs) as in the case of rRNA or directly mediated by single methyltransferases known to methylate other RNA species, such as FTSJ3 (Ge et al. 2010; Bartoli et al. 2018; Elliott et al. 2019).
The most studied function of 2′-O-Me on mRNA is recognition of self RNA by the immune system during viral infections (Daffis et al. 2010; Züst et al. 2011; Devarkar et al. 2016; Leung and Amarasinghe 2016; Encinar and Menendez 2020; Krafcikova et al. 2020; Morales et al. 2020). In addition, 2′-O-Me affects the stabilization of mRNA and translation, including the codon reading (Choi et al. 2018; Elliott et al. 2019). Moreover, 2′-O-Me at N1 prevents transcripts’ degradation by blocking decapping and exoribonuclease activities of DXO which degrades defectively capped pre-mRNAs (Jiao et al. 2013; Picard-Jean et al. 2018).
To date, no direct association has yet been established between 2′-O-Me on mRNA and human pathologies other than infections with viral agents, such as HIV or coronaviruses (Szretter et al. 2012; Ringeard et al. 2019; Krafcikova et al. 2020). However, some mediators of this mark, as for instance CMTR1, are involved in diverse pathologies, among which are asthma and cancer (Dahlin et al. 2015; Degryse et al. 2018; Du et al. 2018). In cancer, CMTR1 is overexpressed in T-cell acute lymphoblastic leukemia with JAK3 mutations and undergoes gene rearrangements with ALK, producing a fusion protein promoting non-small-cell lung cancer development (Degryse et al. 2018; Du et al. 2018). The role of CMTR1 in these cancers is still to be determined, but its overexpression may cause increased stability or translation of specific oncogene transcripts, leading to tumor development. Furthermore, CMTR2 was shown to be mutated in patients with lung adenocarcinomas (Campbell et al. 2016). Despite these observations, it has yet to be answered whether the disease phenotype derives from altered mRNA 2′-O-Me patterns induced by aberrant expression of such enzymes. Further work will be necessary to elucidate the mechanism of 2′-O-Me deposition and understand whether specific factors can act on all RNA species.
DETECTION METHODS
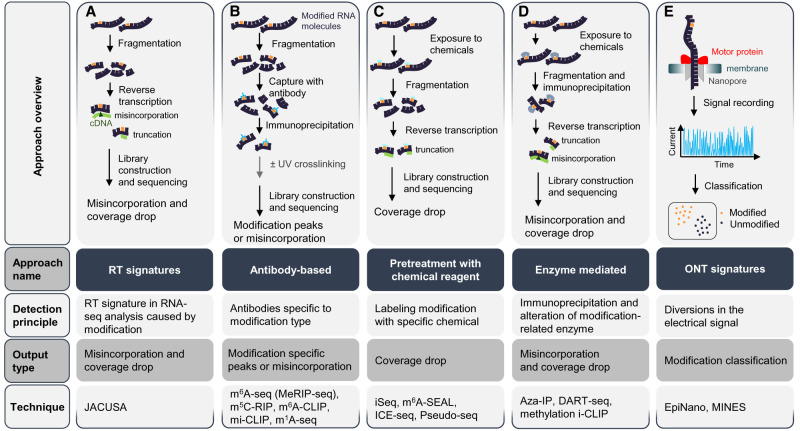
The efforts to detect, map, and quantify epitranscriptomic marks revealed many systematic properties of these marks, such as abundance, evolutionary conservation, reversibility, and biological function. Detection of A-to-I and C-to-U editing exploits a straightforward principle: Since the reverse transcriptase (RT) signatures can process both inosines (reading them as guanines) and uracils, so a discrepancy between RNA and DNA sequences can be detected by RT-PCR or high-throughput sequencing (Athanasiadis et al. 2004; Levanon et al. 2004; Rosenberg et al. 2011; Blanc et al. 2014; Oakes et al. 2017; Piechotta et al. 2017; Malik et al. 2021; Srinivasan et al. 2021). Moreover, bioinformatic approaches have been developed for improving quantitation (Fig. 5A; Piechotta et al. 2017; Cohen-Fultheim and Levanon 2021; Lerner et al. 2021; Lo Giudice et al. 2021), chemical modifications have been used to increase specificity (Fig. 5C; Cattenoz et al. 2013; Okada et al. 2019; Sakurai et al. 2021), and other approaches allow quantitation of specific editing in live cells (Garncarz et al. 2013; Chieca et al. 2021). Transcriptome-wide m6A-mapping methods, mostly represented by antibody-based techniques (Fig. 5B) such as methyl RNA immunoprecipitation followed by sequencing (MeRIP-seq or m6A-seq) and m6A individual-nucleotide-resolution crosslinking and immunoprecipitation (mi-CLIP or m6A-CLIP) (Dominissini et al. 2012; Meyer et al. 2012; Linder et al. 2015), have revealed a unique topology for this mark. However, these techniques can detect both m6A and m6Am through the same antibody recognizing 6-methyladenine and hence, it is difficult to distinguish between the two marks within the mRNA 5′UTR (Fig. 5B; Linder et al. 2015; Hawley and Jaffrey 2019; McIntyre et al. 2020). To overcome potential biases of antibody-based techniques, pretreatment with chemical reagents (m6A-SEAL, Fig. 5C), or enzyme-mediated techniques (DART-seq, Fig. 5D; Liu et al. 2013; Meyer 2019; Vandivier et al. 2019; Wang et al. 2020b) have been developed, although they have not been widely used yet. Recently, nanopore-based sequencing developed by Oxford Nanopore Technologies (ONT), which allows the direct sequencing of native RNA, has also been used to investigate m6A (EpiNano and MINES, Fig. 5E). ONT signatures will advance our knowledge of m6A biology as this technology allows the novo identification of this and other marks at single-nucleotide resolution without RNA immunoprecipitation or pretreatment (Liu et al. 2019; Lorenz et al. 2020).
FIGURE 5.
Approaches for the detection of RNA modifications. The figure presents the experimental approaches to the detection of RNA modifications, one per column and left to right: (A) Detection of nonrandom mismatch signatures (RT-signature), (B) capture with antibody, (C) pretreatment with chemical reagents, (D) capture with modification-related enzymes, and (E) detection of modification-specific signals using the Oxford Nanopore Technologies (ONT) platform (ONT-signature). Each column includes the approach schema, with rows below it indicating the approach name, its detection principle, output type, and techniques implementing that approach.
Ψ sites are detected by single-nucleotide resolution transcriptome-wide techniques based on combining high-throughput sequencing with chemical reagents pretreatment such as N-Cyclohexyl-N′-(2-morpholinoethyl) carbodiimide methyl-p-toluenesulfonate (CMC), a covalent adduct that blocks the RT activity (Fig. 5C; Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014; Li et al. 2015). Among these methods are pseudo-seq, Ψ-seq, PSI-seq, and CeU-seq (Li et al. 2016a; Penzo et al. 2017; Adachi et al. 2019a). Non-CMC based methods, such as RNA bisulfite sequencing (RBS-seq), have also been developed (Khoddami et al. 2019).
For the detection of the other mRNA modifications, transcriptome-wide mapping either by coupling an antibody-based approach to Dimroth rearrangement or by using an m1A-induced RT mismatch signature (namely m1A-seq, Fig. 5B), are used to detect the m1A mark (Dominissini et al. 2016; Li et al. 2016b). m5C can be determined by bisulfite sequencing (Fig. 5C) in which unmodified cytosines are converted to uracils after bisulfite treatment, whereas m5C sites are protected from deamination allowing the detection by high-throughput sequencing (Wei et al. 2018b). Additionally, m5C can be also detected by antibody-based techniques (Fig. 5B) such as m5C-seq and m5C-RIP or enzyme-mediated approaches (Fig. 5D) such as Aza-IP and methylation i-CLIP (Edelheit et al. 2013; Hussain et al. 2013; Khoddami and Cairns 2013). In conclusion, the detection of 2′-O-Me is currently performed by methods based on pretreatment with chemical reagents (Fig. 5C) such as RibOxi-seq and Nm-seq (Dai et al. 2017; Zhu et al. 2017; Motorin and Marchand 2018).
PERSPECTIVES
RNA modifications have risen as major factors in posttranscriptional regulation of gene expression. However, the breadth of their action and its implications for cell physiology and pathology are still far from being sufficiently understood.
Mechanism
The first aspect in need of further attention is the composition of the machinery behind these modifications. While writers and erasers were, in general, more thoroughly characterized, our knowledge of reader proteins is limited, and we are likely missing those proteins with secondary or moonlighting roles as modification readers. This knowledge will be instrumental in fully appreciating how these marks influence RNA fate. Secondly, while there is evidence supporting the possibility of a dynamic life cycle for several modifications, a neglected aspect is the magnitude of this dynamicity. Are RNA modifications continuously written and erased, or subject to less frequent cycles of deposition and degradation? Understanding this aspect could help elucidate their role in cellular processes requiring a fast response and in those with a slower unfolding.
Interplay
Up to now, most of the epitranscriptomics literature focused on individual RNA marks, their physiological functions, and consequences of their dysregulation. Recently, new data suggested that different epitranscriptomic marks could coexist on the same transcript, and that a potentially widespread cooperative and competitive interplay could control the RNA fate, likely through RBPs (Dassi 2017). While the extent of this crosstalk is still unclear, several research groups described potential, mostly correlative, occurrences of such mechanisms (Li et al. 2017a; Dai et al. 2018b, 2020; Sokołowski et al. 2018; Wei et al. 2018a; Xiang et al. 2018; Huang et al. 2019; Seo and Kleiner 2020). For instance, both m6A and m5C methylation sites were found in a specific region of the p21 mRNA. Furthermore, it has been shown that the m6A modification can facilitate m5C methylation and vice versa. This cooperation can synergistically enhance p21 translation in a model of oxidative stress-induced cellular senescence (Li et al. 2017a). This study also suggests that since NSUN2 (m5C) and METTL3/14 (m6A) methylate many coding and ncRNA species (Hussain et al. 2013; Liu et al. 2014; Kadumuri and Janga 2018), their interplay may act beyond p21 (Li et al. 2017b) and affect several other transcripts. However, the prevalence of m5C in mRNAs is limited (Yang et al. 2017), and considerably lower than that of m6A. Thus, it is unclear whether this interplay can happen at a broader scale beyond p21 mRNA, and further work will be necessary to understand its amplitude. Similarly, mRNAs encoding the four Yamanaka factors, exogenously modified with both Ψ and m5C, showed an increased efficiency in cellular reprogramming to a pluripotent state with respect to unmodified mRNAs (Warren et al. 2010). In both studies, the proximity of the modifications has been identified as the basis of their interplay. However, the molecular mechanisms behind this cooperation are still to be described. Also, a negative correlation between A-to-I editing and m6A methylation was observed (Xiang et al. 2018). In contrast to previous studies, this mutually exclusive interaction has been investigated and attributed to RNA structural features preventing ADAR1 binding, rather than direct competition (Xiang et al. 2018). Globally, while these studies highlight the co-occurrence of multiple marks on the same RNA molecules, the existence of direct cooperative and competitive mechanisms between those still needs to be demonstrated.
On the other end, RBPs controlling the life cycle of RNA modifications appear to interact with marks other than their canonical one. For instance, the YTHDF2 m6A reader may “integrate” epitranscriptomics marks by also reading m1A and m5C (Dai et al. 2018b, 2020; Lao and Barron 2019; Seo and Kleiner 2020). A conserved residue of YTHDF2 (Trp432) is required for the recognition of all three modifications, albeit the affinities for m1A and m5C are lower than those for m6A. This lower affinity, coupled to the scarcity of both marks in mRNAs, leaves the actual occurrence and phenotypic impact of this “integration” as a question still to be answered. If confirmed, also other YTH domain-family proteins could behave similarly (Dai et al. 2018b, 2020; Seo and Kleiner 2020), as might be expected given their observed redundancy (Zaccara and Jaffrey 2020). One may thus wonder how these mechanisms could induce or affect pathological states (Meier et al. 2016; Kadumuri and Janga 2018; Christofi and Zaravinos 2019; Huang et al. 2020b). Few studies have explored the relation between cancer and the crosstalk of different enzymes controlling the same modification (Panneerdoss et al. 2018), and how multiple modifications can concurrently control disease states has yet to be established. In Supplemental Table S1, we collect alterations of RNA modifications and editing in several diseases, obtained from the literature on this topic. As shown there, several tumor types are associated with the altered deposition of multiple modifications. This catalog could allow the identification of disorders associated with multiple RNA modifications and thus possibly affected by their interplay.
Overall, it appears that RNA marks may “talk” through direct cooperation or competition, and through the integration of multiple modifications by their reader proteins. Nevertheless, further work will be necessary to demonstrate the actual occurrence and elucidate the amplitude of this interplay, the complexity of the induced regulatory networks and its importance in shaping cell physiology. Is this behavior a form of “epitranscriptomic signaling,” allowing to coordinate the outcome of different pathways? And do changes in the RNA secondary structure interact to alter the transcript life cycle or are reader enzymes required for this crosstalk to modulate cell phenotypes? The answers could bring a new layer of complexity to epitranscriptomics, leading the field into uncharted avenues of even greater possibilities.
Disease
Since RNA modifications represent such a basic layer in the biology of the cell, loss of these modifications is often fatal (Brusa et al. 1995; Wang et al. 2000; Geula et al. 2015) and might have profound effects on cell viability. This is probably the reason why only inactivating mutations in few genes associated with the epitranscriptome have so far been causatively linked to genetic diseases (Miyamura et al. 2003; Bykhovskaya et al. 2004; Rice et al. 2012). Yet, association of RNA modifications with the onset and progression of several human diseases is increasingly being uncovered. Particularly in cancer, several marks appear to play a key role in shaping the prognosis. However, the molecular mechanisms are still not fully understood. Are these modifications disease drivers or mere passengers? It is clear that in some cases alterations in the mediators of these RNA marks have direct effects on cellular tumor-promoting features (Frye et al. 2010; Qin et al. 2014). As such, alterations in these pathways could be selected in the cancer evolutionary process. On the other hand, a direct involvement in the onset of tumors has not been conclusively shown yet. Could these modifications be targeted to alter the course of the pathology? In Supplemental Table S1 we have highlighted the presence of opposite roles that can be played by RNA marks-regulating factors at an intra- and intertumor level. Can the affected pathways explain these disease-specific behaviors? Furthermore, tumors with potential alterations in multiple RNA marks have been identified. Can alterations to multiple enzymes controlling the life cycle of the different modifications be regarded as “double hits” leading to oncogenesis?
Answering these questions will greatly expand our knowledge on the role of epitranscriptomics in disease onset and progression, ultimately enabling us to design novel, highly specific therapeutic strategies against still incurable diseases.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely apologize to authors whose work could not be included due to space limitations. This article is based upon work from COST Action EPITRAN—CA16120, supported by COST (European Cooperation in Science and Technology, www.cost.eu). P.G. and F.A. are supported by grants from the Knut and Alice Wallenberg Foundation, Umeå University, Västerbotten County Council, Swedish Research Council, and Cancerfonden.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077271.120.
Freely available online through the RNA Open Access option.
REFERENCES
- Adachi H, Yu YT. 2020. Pseudouridine-mediated stop codon readthrough in S. cerevisiae is sequence context–independent. RNA 26: 1247–1256. 10.1261/rna.076042.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H, DeZoysa MD, Yu Y-T. 2019a. Detection and quantification of pseudouridine in RNA. Methods Mol Biol 1870: 219–235. 10.1007/978-1-4939-8808-2_17 [DOI] [PubMed] [Google Scholar]
- Adachi H, De Zoysa MD, Yu Y-T. 2019b. Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim Biophys Acta Gene Regul Mech 1862: 230–239. 10.1016/j.bbagrm.2018.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilo F, Walsh MJ. 2017. The N6-methyladenosine RNA modification in pluripotency and reprogramming. Curr Opin Genet Dev 46: 77–82. 10.1016/j.gde.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee D-F, Chen C-H, Rengasamy M, Andino B, et al. 2015. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17: 689–704. 10.1016/j.stem.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. 2019. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363: eaav0080. 10.1126/science.aav0080 [DOI] [PubMed] [Google Scholar]
- Alter BP, Giri N, Savage SA, Rosenberg PS. 2009. Cancer in dyskeratosis congenita. Blood 113: 6549–6557. 10.1182/blood-2008-12-192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T, Rieder D, Wille A, Khokhlova-Cubberley D, Riml C, Trixl L, Jia X-Y, Micura R, Lusser A. 2017. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol 18: 1. 10.1186/s13059-016-1139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anadón C, Guil S, Simó-Riudalbas L, Moutinho C, Setien F, Martínez-Cardús A, Moran S, Villanueva A, Calaf M, Vidal A, et al. 2016. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene 35: 4407–4413. 10.1038/onc.2015.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anant S, Davidson NO. 2000. An AU-rich sequence element (UUUN[A/U]U) downstream of the edited C in apolipoprotein B mRNA is a high-affinity binding site for Apobec-1: binding of Apobec-1 to this motif in the 3′ untranslated region of c-myc increases mRNA stability. Mol Cell Biol 20: 1982–1992. 10.1128/MCB.20.6.1982-1992.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Karikó K. 2010. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 38: 5884–5892. 10.1093/nar/gkq347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova MT, Dimitrova DG, Dinges N, Lence T, Worpenberg L, Carré C, Roignant J-Y. 2018. The emerging field of epitranscriptomics in neurodevelopmental and neuronal disorders. Front Bioeng Biotechnol 6: 46. 10.3389/fbioe.2018.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka M, Ishikawa T, Takabe K, Patnaik SK. 2019. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int J Mol Sci 20: 5621. 10.3390/ijms20225621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. 10.1371/journal.pbio.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. 2019. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochim Biophys Acta 1862: 253–269. 10.1016/j.bbagrm.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2018. Metazoan microRNAs. Cell 173: 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli KM, Schaening C, Carlile TM, Gilbert WV. 2018. Conserved methyltransferase Spb1 targets mRNAs for regulated modification with 2′-O-methyl ribose. bioRxiv 10.1101/271916 [DOI] [Google Scholar]
- Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846. 10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. 2000. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 9: 2297–2304. 10.1093/oxfordjournals.hmg.a018921 [DOI] [PubMed] [Google Scholar]
- Bélanger F, Stepinski J, Darzynkiewicz E, Pelletier J. 2010. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem 285: 33037–33044. 10.1074/jbc.M110.155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Malim MH. 2004. APOBEC-mediated editing of viral RNA. Science 305: 645. 10.1126/science.1100658 [DOI] [PubMed] [Google Scholar]
- Blanc V, Davidson NO. 2010. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med 2: 594–602. 10.1002/wsbm.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Henderson JO, Newberry RD, Xie Y, Cho S-J, Newberry EP, Kennedy S, Rubin DC, Wang HL, Luo J, et al. 2007. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apcmin mice. Cancer Res 67: 8565–8573. 10.1158/0008-5472.CAN-07-1593 [DOI] [PubMed] [Google Scholar]
- Blanc V, Park E, Schaefer S, Miller M, Lin Y, Kennedy S, Billing AM, Ben Hamidane H, Graumann J, Mortazavi A, et al. 2014. Genome-wide identification and functional analysis of Apobec-1-mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol 15: R79. 10.1186/gb-2014-15-6-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. 2006. RNA editing of human microRNAs. Genome Biol 7: R27. 10.1186/gb-2006-7-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack KE, Höbartner C, Bohnsack MT. 2019. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel) 10: 102. 10.3390/genes10020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt EK, Martinez NM, Gilbert WV. 2020. Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet 54: 309–336. 10.1146/annurev-genet-112618-043830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. 1995. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270: 1677–1680. 10.1126/science.270.5242.1677 [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. 10.1038/387303a0 [DOI] [PubMed] [Google Scholar]
- Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. 2004. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 74: 1303–1308. 10.1086/421530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, Liu Y, Zhang X, Zhang W, Ye L. 2018. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett 415: 11–19. 10.1016/j.canlet.2017.11.018 [DOI] [PubMed] [Google Scholar]
- Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. 2016. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48: 607–616. 10.1038/ng.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Martinez NM, Schaening C, Su A, Bell TA, Zinshteyn B, Gilbert WV. 2019. mRNA structure determines modification by pseudouridine synthase 1. Nat Chem Biol 15: 966–974. 10.1038/s41589-019-0353-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattenoz PB, Taft RJ, Westhof E, Mattick JS. 2013. Transcriptome-wide identification of A>I RNA editing sites by inosine specific cleavage. RNA 19: 257–270. 10.1261/rna.036202.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan THM, Lin CH, Qi L, Fei J, Li Y, Yong KJ, Liu M, Song Y, Chow RKK, Ng VHE, et al. 2014. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 63: 832–843. 10.1136/gutjnl-2012-304037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351. 10.1080/152165400410182 [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. 2012. Nuclear editing of mRNA 3′-UTRs. Curr Top Microbiol Immunol 353: 111–121. 10.1007/82_2011_149 [DOI] [PubMed] [Google Scholar]
- Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M. 1987. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 238: 363–366. 10.1126/science.3659919 [DOI] [PubMed] [Google Scholar]
- Chen L, Li Y, Lin CH, Chan THM, Chow RKK, Song Y, Liu M, Yuan Y-F, Fu L, Kong KL, et al. 2013. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 19: 209–216. 10.1038/nm.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-B, Liao X-Y, Zhang J-B, Wang F, Qin H-D, Zhang L, Shugart YY, Zeng Y-X, Jia W-H. 2017. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int J Oncol 50: 622–630. 10.3892/ijo.2016.3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li A, Sun B-F, Yang Y, Han Y-N, Yuan X, Chen R-X, Wei W-S, Liu Y, Gao C-C, et al. 2019. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol 21: 978–990. 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- Chieca M, Torrini S, Conticello SG. 2021. Live-cell quantification of APOBEC1-mediated RNA editing: a comparison of RNA editing assays. Methods Mol Biol 2181: 69–81. 10.1007/978-1-0716-0787-9_5 [DOI] [PubMed] [Google Scholar]
- Choi J, Indrisiunaite G, DeMirci H, Ieong K-W, Wang J, Petrov A, Prabhakar A, Rechavi G, Dominissini D, He C, et al. 2018. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat Struct Mol Biol 25: 208–216. 10.1038/s41594-018-0030-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalla AK, Mehta SL, Vemuganti R. 2020. Epitranscriptomic regulation by m6A RNA methylation in brain development and diseases. J Cereb Blood Flow Metab 23: 271678X20960033. 10.1177/0271678X20960033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry Z, Sengupta SM, Grizenko N, Thakur GA, Fortier M-E, Schmitz N, Joober R. 2013. Association between obesity-related gene FTO and ADHD. Obesity 21: E738–E744. 10.1002/oby.20444 [DOI] [PubMed] [Google Scholar]
- Christofi T, Zaravinos A. 2019. RNA editing in the forefront of epitranscriptomics and human health. J Transl Med 17: 319. 10.1186/s12967-019-2071-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann H-H, Rosenberg BR, et al. 2018. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172: 811–824.e14. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fultheim R, Levanon EY. 2021. Detection of A-to-I hyper-edited RNA sequences. Methods Mol Biol 2181: 213–227. 10.1007/978-1-0716-0787-9_13 [DOI] [PubMed] [Google Scholar]
- Cohn WE. 1960. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem 235: 1488–1498. [PubMed] [Google Scholar]
- Cohn WE, Volkin E. 1951. Nucleoside-5′-phosphates from ribonucleic acid. Nature 167: 483–484. 10.1038/167483a0 [DOI] [Google Scholar]
- Cole DC, Chung Y, Gagnidze K, Hajdarovic KH, Rayon-Estrada V, Harjanto D, Bigio B, Gal-Toth J, Milner TA, McEwen BS, et al. 2017. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc Natl Acad Sci 114: 13272–13277. 10.1073/pnas.1710493114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Langlois M-A, Yang Z, Neuberger MS. 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94: 37–73. 10.1016/S0065-2776(06)94002-4 [DOI] [PubMed] [Google Scholar]
- Corbett AH. 2018. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol 52: 96–104. 10.1016/j.ceb.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Cruz PH, Kawahara Y. 2021. RNA editing in neurological and neurodegenerative disorders. Methods Mol Biol 2181: 309–330. 10.1007/978-1-0716-0787-9_18 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ Jr, Beemon K. 1990. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem 265: 20117–20122. [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Zust R, Dong H, et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468: 452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE, Linneman JG, Shaffer CM, Weeke P, Xu H, et al. 2015. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis 3: 350–359. 10.1002/iid3.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C. 2017. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods 14: 695–698. 10.1038/nmeth.4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C. 2018a. Correction: Corrigendum: Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods 15: 226–227. 10.1038/nmeth0318-226c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Wang T, Gonzalez G, Wang Y. 2018b. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem 90: 6380–6384. 10.1021/acs.analchem.8b01703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Gonzalez G, Li L, Li J, You C, Miao W, Hu J, Fu L, Zhao Y, Li R, et al. 2020. YTHDF2 binds to 5-methylcytosine in RNA and modulates the maturation of ribosomal RNA. Anal Chem 92: 1346–1354. 10.1021/acs.analchem.9b04505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvish H, Azcona LJ, Alehabib E, Jamali F, Tafakhori A, Ranji-Burachaloo S, Jen JC, Paisán-Ruiz C. 2019. A novel PUS7 mutation causes intellectual disability with autistic and aggressive behaviors. Neurol Genet 5: e356. 10.1212/NXG.0000000000000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassi E. 2017. Handshakes and fights: the regulatory interplay of RNA-binding proteins. Front Mol Biosci 4: 67. 10.3389/fmolb.2017.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FF, Allen FW. 1957. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem 227: 907–915. [PubMed] [Google Scholar]
- de Brouwer APM, Abou Jamra R, Körtel N, Soyris C, Polla DL, Safra M, Zisso A, Powell CA, Rebelo-Guiomar P, Dinges N, et al. 2018. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am J Hum Genet 103: 1045–1052. 10.1016/j.ajhg.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degryse S, de Bock CE, Demeyer S, Govaerts I, Bornschein S, Verbeke D, Jacobs K, Binos S, Skerrett-Byrne DA, Murray HC, et al. 2018. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 32: 788–800. 10.1038/leu.2017.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay S, Frye M. 2019. RNA modifications regulating cell fate in cancer. Nat Cell Biol 21: 552–559. 10.1038/s41556-019-0319-0 [DOI] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci 71: 3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, Patel SS, Marcotrigiano J. 2016. Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci 113: 596–601. 10.1073/pnas.1515152113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio S, Martignano F, Torcia MG, Mattiuz G, Conticello SG. 2020. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 6: eabb5813. 10.1126/sciadv.abb5813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DG, Teysset L, Carré C. 2019. RNA 2′-O-methylation (Nm) modification in human diseases. Genes (Basel) 10: 117. 10.3390/genes10020117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. 2016. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446. 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Grima JC, Sattler R. 2014. Aberrant RNA homeostasis in amyotrophic lateral sclerosis: potential for new therapeutic targets? Neurodegener Dis Manag 4: 417–437. 10.2217/nmt.14.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria M, Neri F, Gallo A, Farace MG, Michienzi A. 2009. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res 37: 5848–5858. 10.1093/nar/gkp604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Rao S, Wu L, Ye N, Liu Z, Hu H, Xiu J, Shen Y, Xu Q. 2015. An association study of the m6A genes with major depressive disorder in Chinese Han population. J Affect Disord 183: 279–286. 10.1016/j.jad.2015.05.025 [DOI] [PubMed] [Google Scholar]
- Du X, Shao Y, Qin H-F, Tai Y-H, Gao H-J. 2018. ALK-rearrangement in non-small-cell lung cancer (NSCLC). Thorac Cancer 9: 423–430. 10.1111/1759-7714.12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Zhang L, Lee T, Sun T. 2019. m6A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol 56: 1596–1606. 10.1007/s12035-018-1138-1 [DOI] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. 2013. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9: e1003602. 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2018. A-to-I RNA editing—immune protector and transcriptome diversifier. Nat Rev Genet 19: 473–490. 10.1038/s41576-018-0006-1 [DOI] [PubMed] [Google Scholar]
- Elliott BA, Ho H-T, Ranganathan SV, Vangaveti S, Ilkayeva O, Abou Assi H, Choi AK, Agris PF, Holley CL. 2019. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat Commun 10: 3401. 10.1038/s41467-019-11375-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar JA, Menendez JA. 2020. Potential drugs targeting early innate immune evasion of SARS-Coronavirus 2 via 2′-O-methylation of viral RNA. Viruses 12: 525. 10.3390/v12050525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler DE, Franco MK, Batool Z, Wu MZ, Dubuke ML, Dobosz-Bartoszek M, Jones JD, Polikanov YS, Roy B, Koutmou KS. 2019. Pseudouridinylation of mRNA coding sequences alters translation. Proc Natl Acad Sci 116: 23068–23074. 10.1073/pnas.1821754116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia I, Marcos T, Muñoz-Barrutia A, Serrano D, Pio R, Montuenga LM, Ortiz-de-Solorzano C. 2013. Multiscale in situ analysis of the role of dyskerin in lung cancer cells. Integr Biol 5: 402–413. 10.1039/c2ib20219k [DOI] [PubMed] [Google Scholar]
- Fossat N, Tam PPL. 2014. Re-editing the paradigm of Cytidine (C) to Uridine (U) RNA editing. RNA Biol 11: 1233–1237. 10.1080/15476286.2014.996054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Tourle K, Radziewic T, Barratt K, Liebhold D, Studdert JB, Power M, Jones V, Loebel DAF, Tam PPL. 2014. C to U RNA editing mediated by APOBEC 1 requires RNA-binding protein RBM 47. EMBO Rep 15: 903–910. 10.15252/embr.201438450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén O, Ermel R, Sukhavasi K, Jain R, Jain A, Betsholtz C, Giannarelli C, Kovacic JC, Ruusalepp A, Skogsberg J, et al. 2018. Global analysis of A-to-I RNA editing reveals association with common disease variants. PeerJ 6: e4466. 10.7717/peerj.4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Watt FM. 2006. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol 16: 971–981. 10.1016/j.cub.2006.04.027 [DOI] [PubMed] [Google Scholar]
- Frye M, Dragoni I, Chin S-F, Spiteri I, Kurowski A, Provenzano E, Green A, Ellis IO, Grimmer D, Teschendorff A, et al. 2010. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett 289: 71–80. 10.1016/j.canlet.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Fu L, Qin Y-R, Ming X-Y, Zuo X-B, Diao Y-W, Zhang L-Y, Ai J, Liu B-L, Huang T-X, Cao T-T, et al. 2017. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci 114: E4631–E4640. 10.1073/pnas.1703178114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Keegan LP, Ring GM, O'Connell MA. 2003. An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J 22: 3421–3430. 10.1093/emboj/cdg327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Vukic D, Michalík D, O'Connell MA, Keegan LP. 2017. ADAR RNA editing in human disease; more to it than meets the I. Hum Genet 136: 1265–1278. 10.1007/s00439-017-1837-0 [DOI] [PubMed] [Google Scholar]
- Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. 2013. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol 10: 192–204. 10.4161/rna.23208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Yu YT. 2013. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci 38: 210–218. 10.1016/j.tibs.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Liu H, Yu YT. 2010. Regulation of pre-mRNA splicing in Xenopus oocytes by targeted 2′-O-methylation. RNA 16: 1078–1085. 10.1261/rna.2060210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gélinas J-F, Clerzius G, Shaw E, Gatignol A. 2011. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J Virol 85: 8460–8466. 10.1128/JVI.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Keller W. 2001. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 26: 376–384. 10.1016/S0968-0004(01)01827-8 [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398. 10.1126/science.1120976 [DOI] [PubMed] [Google Scholar]
- Grohmann M, Hammer P, Walther M, Paulmann N, Büttner A, Eisenmenger W, Baghai TC, Schüle C, Rupprecht R, Bader M, et al. 2010. Alternative splicing and extensive RNA editing of human TPH2 transcripts. PLoS ONE 5: e8956. 10.1371/journal.pone.0008956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C. 2002a. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci 22: 10529–10532. 10.1523/JNEUROSCI.22-24-10529.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. 2002b. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34: 349–356. 10.1016/S0896-6273(02)00660-8 [DOI] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP. 2008. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65: 798–813. 10.1007/s00018-007-7447-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2006. Pseudouridine synthases. Chem Biol 13: 1125–1135. 10.1016/j.chembiol.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2010. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem 285: 805–809. 10.1074/jbc.R109.076893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S-W, Kim H-P, Shin J-Y, Jeong E-G, Lee W-C, Kim KY, Park SY, Lee D-W, Won J-K, Jeong S-Y, et al. 2014. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med 211: 613–621. 10.1084/jem.20132209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Liu Z, Xu Y, Liu X, Wang D, Li F, Wang Y, Bi J. 2020. Abnormality of m6A mRNA methylation is involved in Alzheimer's disease. Front Neurosci 14: 98. 10.3389/fnins.2020.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjanto D, Papamarkou T, Oates CJ, Rayon-Estrada V, Papavasiliou FN, Papavasiliou A. 2016. RNA editing generates cellular subsets with diverse sequence within populations. Nat Commun 7: 12145. 10.1038/ncomms12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL. 1990. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res 18: 5735–5741. 10.1093/nar/18.19.5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10: 109–115. 10.1038/ni.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley BR, Jaffrey SR. 2019. Transcriptome-wide mapping of m6A and m6Am at single-nucleotide resolution using miCLIP. Curr Protoc Mol Biol 126: e88. 10.1002/cpmb.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel J, Duex JE, Owens C, Dancik GM, Edwards MG, Frierson HF, Theodorescu D. 2015. Patient mutation directed shRNA screen uncovers novel bladder tumor growth suppressors. Mol Cancer Res 13: 1306–1315. 10.1158/1541-7786.MCR-15-0130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. 1993. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75: 1361–1370. 10.1016/0092-8674(93)90622-W [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. 10.1038/35017558 [DOI] [PubMed] [Google Scholar]
- Höfler S, Carlomagno T. 2020. Structural and functional roles of 2′-O-ribose methylations and their enzymatic machinery across multiple classes of RNAs. Curr Opin Struct Biol 65: 42–50. 10.1016/j.sbi.2020.05.008 [DOI] [PubMed] [Google Scholar]
- Hotta K, Sho M, Fujimoto K, Shimada K, Yamato I, Anai S, Harada H, Tsujikawa K, Konishi N, Shinohara N, et al. 2015. Clinical significance and therapeutic potential of prostate cancer antigen-1/ALKBH3 in human renal cell carcinoma. Oncol Rep 34: 648–654. 10.3892/or.2015.4017 [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, et al. 2019. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567: 414–419. 10.1038/s41586-019-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Camats-Perna J, Medeiros R, Anggono V, Widagdo J. 2020a. Altered expression of the m6A methyltransferase METTL3 in Alzheimer's disease. eNeuro 7: ENEURO.0125-20.2020. 10.1523/ENEURO.0125-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Deng X, Chen J. 2020b. RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annu Rev Cancer Biol 4: 221–240. 10.1146/annurev-cancerbio-030419-033357 [DOI] [Google Scholar]
- Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B, Yang L, Wang Y, Zhang H, Zhang H, et al. 2020c. N6-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry 88: 392–404. 10.1016/j.biopsych.2020.02.018 [DOI] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. 2013. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261. 10.1016/j.celrep.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T, Abd El Galil KH, Tokunaga K, Maeda K, Sata T, Sakaguchi N, Heidmann T, Koito A. 2011. Intrinsic restriction activity by apolipoprotein B mRNA editing enzyme APOBEC1 against the mobility of autonomous retrotransposons. Nucleic Acids Res 39: 5538–5554. 10.1093/nar/gkr124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inesta-Vaquera F, Chaugule VK, Galloway A, Chandler L, Rojas-Fernandez A, Weidlich S, Peggie M, Cowling VH. 2018. DHX15 regulates CMTR1-dependent gene expression and cell proliferation. Life Sci Alliance 1: e201800092. 10.26508/lsa.201800092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Mann TD, Stulić M, Rao SP, Kirsch A, Pullirsch D, Strobl X, Rath C, Reissig L, Moreth K, et al. 2018. RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J 37: e94813. 10.15252/embj.201694813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili P, Bowen D, Langenbucher A, Park S, Aguirre K, Corcoran RB, Fleischman AG, Lawrence MS, Zou L, Buisson R. 2020. Quantification of ongoing APOBEC3A activity in tumor cells by monitoring RNA editing at hotspots. Nat Commun 11: 2971. 10.1038/s41467-020-16802-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesus DFD, De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, Gupta MK, He C, Kulkarni RN. 2019. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab 1: 765–774. 10.1038/s42255-019-0089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Crews LA, Barrett CL, Chun H-J, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, et al. 2013. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci 110: 1041–1046. 10.1073/pnas.1213021110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. 2013. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell 50: 104–115. 10.1016/j.molcel.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. 2017. The RNA modification landscape in human disease. RNA 23: 1754–1769. 10.1261/rna.063503.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadumuri RV, Janga SC. 2018. Epitranscriptomic code and its alterations in human disease. Trends Mol Med 24: 886–903. 10.1016/j.molmed.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankowski S, Förstera B, Winkelmann A, Knauff P, Wanker EE, You XA, Semtner M, Hetsch F, Meier JC. 2018. A novel RNA editing sensor tool and a specific agonist determine neuronal protein expression of RNA-edited glycine receptors and identify a genomic APOBEC1 dimorphism as a new genetic risk factor of epilepsy. Front Mol Neurosci 10: 439. 10.3389/fnmol.2017.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu Y-T. 2011. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474: 395–398. 10.1038/nature10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. 2008. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16: 1833–1840. 10.1038/mt.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. 2004. Glutamate receptors: RNA editing and death of motor neurons. Nature 427: 801. 10.1038/427801a [DOI] [PubMed] [Google Scholar]
- Kaya Y, Ofengand J. 2003. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 9: 711–721. 10.1261/rna.5230603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Jr DJ, Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JM, Ensinger MJ, Moss B. 1978. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J Biol Chem 253: 5033–5039. [PubMed] [Google Scholar]
- Khermesh K, D'Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, Picardi E, Eisenberg E. 2016. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. RNA 22: 290–302. 10.1261/rna.054627.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR. 2013. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31: 458–464. 10.1038/nbt.2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, Cairns BR. 2019. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci 116: 6784–6789. 10.1073/pnas.1817334116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DDY, Kim TTY, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725. 10.1101/gr.2855504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike K, Ueda Y, Hase H, Kitae K, Fusamae Y, Masai S, Inagaki T, Saigo Y, Hirasawa S, Nakajima K, et al. 2012. Anti-tumor effect of AlkB homolog 3 knockdown in hormone-independent prostate cancer cells. Curr Cancer Drug Targets 12: 847–856. 10.2174/156800912802429283 [DOI] [PubMed] [Google Scholar]
- Kono M, Akiyama M. 2019. Dyschromatosis symmetrica hereditaria and reticulate acropigmentation of Kitamura: an update. J Dermatol Sci 93: 75–81. 10.1016/j.jdermsci.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Koonin EV. 1996. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res 24: 2411–2415. 10.1093/nar/24.12.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafcikova P, Silhan J, Nencka R, Boura E. 2020. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun 11: 3717. 10.1038/s41467-020-17495-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C-P, Maggi LB Jr, Weber JD. 2018. The role of RNA editing in cancer development and metabolic disorders. Front Endocrinol 9: 762. 10.3389/fendo.2018.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S, Nishimoto Y, Yamashita T. 2008. Newly identified ADAR-mediated A-to-I editing positions as a tool for ALS research. RNA Biol 5: 193–197. 10.4161/rna.6925 [DOI] [PubMed] [Google Scholar]
- Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. 2019. The critical role of RNA m6A methylation in cancer. Cancer Res 79: 1285–1292. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- Langberg SR, Moss B. 1981. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methyltransferases from HeLa cells. J Biol Chem 256: 10054–10060. [PubMed] [Google Scholar]
- Lao N, Barron N. 2019. Cross-talk between m6A and m1A regulators, YTHDF2 and ALKBH3 fine-tunes mRNA expression. bioRxiv 10.1101/589747 [DOI] [Google Scholar]
- Laurencikiene J, Källman AM, Fong N, Bentley DL, Öhman M. 2006. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep 7: 303–307. 10.1038/sj.embor.7400621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi S, Shatkin AJ. 1975. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci 72: 2012–2016. 10.1073/pnas.72.6.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxminarayana D, Khan IU, Kammer GM. 2002. Transcript mutations of the α regulatory subunit of protein kinase A and up-regulation of the RNA-editing gene transcript in lupus T lymphocytes. Lancet 360: 842–849. 10.1016/S0140-6736(02)09966-X [DOI] [PubMed] [Google Scholar]
- Laxminarayana D, O'Rourke KS, Maas S, Olorenshaw I. 2007. Altered editing in RNA editing adenosine deaminase ADAR2 gene transcripts of systemic lupus erythematosus T lymphocytes. Immunology 121: 359–369. 10.1111/j.1365-2567.2007.02582.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzari E, Mondala PK, Santos ND, Miller AC, Pineda G, Jiang Q, Leu H, Ali SA, Ganesan A-P, Wu CN, et al. 2017. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat Commun 8: 1922. 10.1038/s41467-017-01890-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F. 2017. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res 27: 1589–1596. 10.1101/gr.210666.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. 2000. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem 275: 19848–19856. 10.1074/jbc.M001786200 [DOI] [PubMed] [Google Scholar]
- Lence T, Paolantoni C, Worpenberg L, Roignant J-Y. 2019. Mechanistic insights into m6A RNA enzymes. Biochim Biophys Acta Gene Regul Mech 1862: 222–229. 10.1016/j.bbagrm.2018.10.014 [DOI] [PubMed] [Google Scholar]
- Lerner T, Kluesner M, Tasakis RN, Moriarity BS, Papavasiliou FN, Pecori R. 2021. C-to-U RNA editing: from computational detection to experimental validation. Methods Mol Biol 2181: 51–67. 10.1007/978-1-0716-0787-9_4 [DOI] [PubMed] [Google Scholar]
- Leung DW, Amarasinghe GK. 2016. When your cap matters: structural insights into self vs non-self recognition of 5′ RNA by immunomodulatory host proteins. Curr Opin Struct Biol 36: 133–141. 10.1016/j.sbi.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. 10.1038/nbt996 [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. 10.1126/science.1170995 [DOI] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. 2015. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597. 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- Li X, Ma S, Yi C. 2016a. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol 33: 108–116. 10.1016/j.cbpa.2016.06.014 [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. 2016b. Transcriptome-wide mapping reveals reversible and dynamic N 1-methyladenosine methylome. Nat Chem Biol 12: 311–316. 10.1038/nchembio.2040 [DOI] [PubMed] [Google Scholar]
- Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M, Wang W. 2017a. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem 118: 2587–2598. 10.1002/jcb.25957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen X-W, et al. 2017b. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005.e9. 10.1016/j.molcel.2017.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang X, Qi Z, Sang Y, Liu Y, Xu B, Liu W, Xu Z, Deng Y. 2019. The role of mRNA m6A methylation in the nervous system. Cell Biosci 9: 66. 10.1186/s13578-019-0330-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349: 1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zhang J, Huang C, Liu H. 2012. Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS ONE 7: e43147. 10.1371/journal.pone.0043147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. 2013. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19: 1848–1856. 10.1261/rna.041178.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM. 2019. Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10: 4079. 10.1038/s41467-019-11713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CC, Coschigano KT. 2020. ApoB48 as an efficient regulator of intestinal lipid transport. Front Physiol 11: 796. 10.3389/fphys.2020.00796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice C, Pesole G, Picardi E. 2021. High-throughput sequencing to detect DNA-RNA changes. Methods Mol Biol 2181: 193–212. 10.1007/978-1-0716-0787-9_12 [DOI] [PubMed] [Google Scholar]
- Lorenz DA, Sathe S, Einstein JM, Yeo GW. 2020. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26: 19–28. 10.1261/rna.072785.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO. 2014. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9: e110799. 10.1371/journal.pone.0110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. 2018. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Invest 36: 246–253. 10.1080/07357907.2018.1466896 [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. 2004. RNA editing of a miRNA precursor. RNA 10: 1174–1177. 10.1261/rna.7350304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A. 2001. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci 98: 14687–14692. 10.1073/pnas.251531398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik TN, Cartailler J-P, Emeson RB. 2021. Quantitative analysis of adenosine-to-inosine RNA editing. Methods Mol Biol 2181: 97–111. 10.1007/978-1-0716-0787-9_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla S, Melguizo-Sanchis D, Aguilo F. 2019. Steering pluripotency and differentiation with N6-methyladenosine RNA modification. Biochim Biophys Acta Gene Regul Mech 1862: 394–402. 10.1016/j.bbagrm.2018.10.013 [DOI] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N, Arieti F, Gallo A, Keegan LP, O'Connell MA. 2015. New insights into the biological role of mammalian ADARs; the RNA editing proteins. Biomolecules 5: 2338–2362. 10.3390/biom5042338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Gilbert WV. 2018. Pre-mRNA modifications and their role in nuclear processing. Quant Biol 6: 210–227. 10.1007/s40484-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Ratcliffe PJ. 2014. Hypoxia signaling pathways in cancer metabolism: the importance of co-selecting interconnected physiological pathways. Cancer Metab 2: 3. 10.1186/2049-3002-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur J-J, Chen Q, et al. 2017. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541: 371–375. 10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre ABR, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, Mason CE. 2020. Limits in the detection of m6A changes using MeRIP/m6A-seq. Sci Rep 10: 6590. 10.1038/s41598-020-63355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Kinter MT, Sherman NE, Driscoll DM. 2000. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol 20: 1846–1854. 10.1128/MCB.20.5.1846-1854.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Shen C, Miao R, Wang J-Z, Cao M-D, Zhang Y-S, Shi L-H, Zhao G-H, Wang M-H, Wu L-S, et al. 2020. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57Kip2 by an m5C-dependent manner. Cell Death Dis 11: 270. 10.1038/s41419-020-2487-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JC, Henneberger C, Melnick I, Racca C, Harvey RJ, Heinemann U, Schmieden V, Grantyn R. 2005. RNA editing produces glycine receptor alpha3(P185L), resulting in high agonist potency. Nat Neurosci 8: 736–744. 10.1038/nn1467 [DOI] [PubMed] [Google Scholar]
- Meier JC, Kankowski S, Krestel H, Hetsch F. 2016. RNA editing—systemic relevance and clue to disease mechanisms? Front Mol Neurosci 9: 47338. 10.3389/fnmol.2016.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD. 2019. DART-seq: an antibody-free method for global m6A detection. Nat Methods 16: 1275–1280. 10.1038/s41592-019-0570-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y. 2003. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet 73: 693–699. 10.1086/378209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, Taffurelli M, Calienni M, Teruya-Feldstein J, Trerè D, et al. 2006. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol 210: 10–18. 10.1002/path.2023 [DOI] [PubMed] [Google Scholar]
- Morales P, Curtis NL, Zárate SG, Bastida A, Bolanos-Garcia VM. 2020. Interfering with mRNA methylation by the 2′O-methyltransferase (NSP16) from SARS-CoV-2 to tackle the COVID-19 disease. Catalysts 10: 1023. 10.3390/catal10091023 [DOI] [Google Scholar]
- Motorin Y, Marchand V. 2018. Detection and analysis of RNA ribose 2′-O-methylations: challenges and solutions. Genes (Basel) 9: 642. 10.3390/genes9120642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Anant S, Lee RM, Kennedy S, Viskochil D, Davidson NO. 2002. C→U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA–editing enzyme. Am J Hum Genet 70: 38–50. 10.1086/337952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller TA, Meek K, Hausinger RP. 2010. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 9: 58–65. 10.1016/j.dnarep.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, He C. 2017. The emerging biology of RNA post-transcriptional modifications. RNA Biol 14: 156–163. 10.1080/15476286.2016.1267096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S, He C. 2018. Chemical modifications in the life of an mRNA transcript. Annu Rev Genet 52: 349–372. 10.1146/annurev-genet-120417-031522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa CK, Kibiryeva N, Marshall J, O'Brien JE Jr, Bittel DC. 2020. scaRNA1 levels alter pseudouridylation in spliceosomal RNA U2 affecting alternative mRNA splicing and embryonic development. Pediatr Cardiol 41: 341–349. 10.1007/s00246-019-02263-4 [DOI] [PubMed] [Google Scholar]
- Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng BB, Davidson NO, Scott J. 1993. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem 268: 20709–20712. [PubMed] [Google Scholar]
- Ni TK, Elman JS, Jin DX, Gupta PB, Kuperwasser C. 2018. Premature polyadenylation of MAGI3 is associated with diminished N6-methyladenosine in its large internal exon. Sci Rep 8: 1415. 10.1038/s41598-018-19916-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong X-F, Wei B, et al. 2019. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer 18: 46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes E, Vadlamani P, Hundley HA. 2017. Methods for the detection of adenosine-to-inosine editing events in cellular RNA. Methods Mol Biol 1648: 103–127. 10.1007/978-1-4939-7204-3_9 [DOI] [PubMed] [Google Scholar]
- Okada S, Ueda H, Noda Y, Suzuki T. 2019. Transcriptome-wide identification of A-to-I RNA editing sites using ICE-seq. Methods 156: 66–78. 10.1016/j.ymeth.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, Ogawa I, Takata T, Shimamoto F, Tatsuka M. 2012. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol 31: 660–671. 10.1089/dna.2011.1446 [DOI] [PubMed] [Google Scholar]
- O'Neil RT, Emeson RB. 2012. Quantitative analysis of 5HT2C receptor RNA editing patterns in psychiatric disorders. Neurobiol Dis 45: 8–13. 10.1016/j.nbd.2011.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RJ, O'Rourke KS, Olorenshaw I, Hawkins GA, Maas S, Laxminarayana D. 2008. Altered editing in cyclic nucleotide phosphodiesterase 8A1 gene transcripts of systemic lupus erythematosus T lymphocytes. Immunology 125: 408–419. 10.1111/j.1365-2567.2008.02850.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, Abdelfattah N, Onyeagucha BC, Cui X, Lai Z, et al. 2018. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci Adv 4: eaar8263. 10.1126/sciadv.aar8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. 2007. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17: 1586–1595. 10.1101/gr.6493107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo M, Guerrieri AN, Zacchini F, Treré D, Montanaro L. 2017. RNA pseudouridylation in physiology and medicine: for better and for worse. Genes (Basel) 8: 301. 10.3390/genes8110301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE. 1974. Existence of methylated messenger RNA in mouse L cells. Cell 1: 37–42. 10.1016/0092-8674(74)90153-6 [DOI] [Google Scholar]
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. 2015. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43: 933–944. 10.1016/j.immuni.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit V, Guétard D, Renard M, Keriel A, Sitbon M, Wain-Hobson S, Vartanian J-P. 2009. Murine APOBEC1 is a powerful mutator of retroviral and cellular RNA in vitro and in vivo. J Mol Biol 385: 65–78. 10.1016/j.jmb.2008.10.043 [DOI] [PubMed] [Google Scholar]
- Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23: 573–582. 10.1016/j.coi.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphuakrat A, Kraiwong R, Boonarkart C, Lauhakirti D, Lee T-H, Auewarakul P. 2008. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol 82: 10864–10872. 10.1128/JVI.00238-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, Manzari C, Mastropasqua F, Aiello I, D'Erchia AM, Pesole G. 2016. Corrigendum: profiling RNA editing in human tissues: towards the inosinome Atlas. Sci Rep 6: 20755. 10.1038/srep20755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardi E, D'Erchia AM, Lo Giudice C, Pesole G. 2017. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res 45: D750–D757. 10.1093/nar/gkw767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Jean F, Brand C, Tremblay-Létourneau M, Allaire A, Beaudoin MC, Boudreault S, Duval C, Rainville-Sirois J, Robert F, Pelletier J, et al. 2018. 2′-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS ONE 13: e0193804. 10.1371/journal.pone.0193804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechotta M, Wyler E, Ohler U, Landthaler M, Dieterich C. 2017. JACUSA: site-specific identification of RNA editing events from replicate sequencing data. BMC Bioinformatics 18: 7. 10.1186/s12859-016-1432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaska KM, Bennett RP, Salter JD, Smith HC. 2014. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip Rev RNA 5: 493–508. 10.1002/wrna.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y-R, Qiao J-J, Chan THM, Zhu Y-H, Li F-F, Liu H, Fei J, Li Y, Guan X-Y, Chen L. 2014. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res 74: 840–851. 10.1158/0008-5472.CAN-13-2545 [DOI] [PubMed] [Google Scholar]
- Qin L, Min S, Shu L, Pan H, Zhong J, Guo J, Sun Q, Yan X, Chen C, Tang B, et al. 2020. Genetic analysis of N6-methyladenosine modification genes in Parkinson's disease. Neurobiol Aging 93: 143.e9–143.e13. 10.1016/j.neurobiolaging.2020.03.018 [DOI] [PubMed] [Google Scholar]
- Qiu X, He H, Huang Y, Wang J, Xiao Y. 2020. Genome-wide identification of m6A-associated single-nucleotide polymorphisms in Parkinson's disease. Neurosci Lett 737: 135315. 10.1016/j.neulet.2020.135315 [DOI] [PubMed] [Google Scholar]
- Quattrone A, Dassi E. 2019. The architecture of the human RNA-binding protein regulatory network. iScience 21: 706–719. 10.1016/j.isci.2019.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones-Valdez G, Tran SS, Jun H-I, Bahn JH, Yang E-W, Zhan L, Brümmer A, Wei X, Van Nostrand EL, Pratt GA, et al. 2019. Regulation of RNA editing by RNA-binding proteins in human cells. Commun Biol 2: 19. 10.1038/s42003-018-0271-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayon-Estrada V, Harjanto D, Hamilton CE, Berchiche YA, Gantman EC, Sakmar TP, Bulloch K, Gagnidze K, Harroch S, McEwen BS, et al. 2017. Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function. Proc Natl Acad Sci 114: 13296–13301. 10.1073/pnas.1714227114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Ro-Choi TS, Henning D, Shibata H, Choi YC, Busch H. 1972. Modified nucleosides of nuclear and nucleolar low molecular weight ribonucleic acid. J Biol Chem 247: 7245–7250. [PubMed] [Google Scholar]
- Reitz C, Tosto G, Mayeux R, Luchsinger JA, NIA-LOAD/NCRAD Family Study Group; Alzheimer's Disease Neuroimaging Initiative. 2012. Genetic variants in the fat and obesity associated (FTO) gene and risk of Alzheimer's disease. PLoS ONE 7: e50354. 10.1371/journal.pone.0050354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relier S, Ripoll J, Guillorit H, Amalric A, Boissière F, Vialaret J, Attina A, Debart F, Choquet A, Macari F, et al. 2020. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. bioRxiv 10.1101/2020.01.09.899724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. 2012. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genet 44: 1243–1248. 10.1038/ng.2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. 2019. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565: 500–504. 10.1038/s41586-018-0841-4 [DOI] [PubMed] [Google Scholar]
- Rintala-Dempsey AC, Kothe U. 2017. Eukaryotic stand-alone pseudouridine synthases–RNA modifying enzymes and emerging regulators of gene expression? RNA Biol 14: 1185–1196. 10.1080/15476286.2016.1276150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JT, Patterson DG, King VM, Amin SV, Polska CJ, Houserova D, Crucello A, Barnhill EC, Miller MM, Sherman TD, et al. 2018. ADAR mediated RNA editing modulates microRNA targeting in human breast cancer. Processes (Basel) 6: 42. 10.3390/pr6050042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell AL, Hongay CF. 2019. The m6A dynamics of profilin in neurogenesis. Front Genet 10: 987. 10.3389/fgene.2019.00987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. 2011. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat Struct Mol Biol 18: 230–236. 10.1038/nsmb.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter SM, Dawson TR, Emeson RB. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80. 10.1038/19992 [DOI] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. 2017a. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27: 393–406. 10.1101/gr.207613.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. 2017b. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255. 10.1038/nature24456 [DOI] [PubMed] [Google Scholar]
- Sakurai M, Okada S, Ueda H, Yang Y. 2021. Discovering A-to-I RNA editing through chemical methodology “ICE-seq.” Methods Mol Biol 2181: 113–148. 10.1007/978-1-0716-0787-9_8 [DOI] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S, Mason CE. 2012. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biol 13: 175. 10.1186/gb-2012-13-10-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter JD, Bennett RP, Smith HC. 2016. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 41: 578–594. 10.1016/j.tibs.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. 2012. ADARs: viruses and innate immunity. Curr Top Microbiol Immunol 353: 163–195. 10.1007/82_2011_148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraconi G, Severi F, Sala C, Mattiuz G, Conticello SG. 2014. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol 15: 417. 10.1186/s13059-014-0417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. 1977. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol 115: 695–714. 10.1016/0022-2836(77)90110-3 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo KW, Kleiner RE. 2020. YTHDF2 recognition of N1-methyladenosine (m1A)-modified RNA is associated with transcript destabilization. ACS Chem Biol 15: 132–139. 10.1021/acschembio.9b00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, Alkuraya FS. 2016. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 135: 707–713. 10.1007/s00439-016-1665-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Tasak M, Maddirevula S, Abdel-Salam GMH, Sayed ISM, Alazami AM, Al-Sheddi T, Alobeid E, Phizicky EM, Alkuraya FS. 2019. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum Genet 138: 231–239. 10.1007/s00439-019-01980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, Baysal BE. 2015. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 6: 6881. 10.1038/ncomms7881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Patnaik SK, Taggart RT, Baysal BE. 2016. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci Rep 6: 39100. 10.1038/srep39100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeyasu K, Okugawa Y, Toden S, Miyoshi J, Toiyama Y, Nagasaka T, Takahashi N, Kusunoki M, Takayama T, Yamada Y, et al. 2018. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 3: e99976. 10.1172/jci.insight.99976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieron P, Hader C, Hatina J, Engers R, Wlazlinski A, Müller M, Schulz WA. 2009. DKC1 overexpression associated with prostate cancer progression. Br J Cancer 101: 1410–1416. 10.1038/sj.bjc.6605299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestris DA, Picardi E, Cesarini V, Fosso B, Mangraviti N, Massimi L, Martini M, Pesole G, Locatelli F, Gallo A. 2019. Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma. Genome Biol 20: 33. 10.1186/s13059-019-1647-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. 1996. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res 24: 478–485. 10.1093/nar/24.3.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śledź P, Jinek M. 2016. Structural insights into the molecular mechanism of the m6A writer complex. Elife 5: e18434. 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smietanski M, Werner M, Purta E, Kaminska KH, Stepinski J, Darzynkiewicz E, Nowotny M, Bujnicki JM. 2014. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat Commun 5: 3004. 10.1038/ncomms4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi MS, Burnet PWJ, Makoff AJ, Kerwin RW, Harrison PJ. 2001. RNA editing of the 5-HT 2C receptor is reduced in schizophrenia. Mol Psychiatry 6: 373–379. 10.1038/sj.mp.4000920 [DOI] [PubMed] [Google Scholar]
- Sokołowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. 2018. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta Gene Regul Mech 1861: 409–418. 10.1016/j.bbagrm.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. 10.1016/0092-8674(91)90568-J [DOI] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. 2012. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033. 10.1093/nar/gks144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan H, Kok EPL, Tan MH. 2021. RNA editing in human and mouse tissues. Methods Mol Biol 2181: 163–176. 10.1007/978-1-0716-0787-9_10 [DOI] [PubMed] [Google Scholar]
- Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M, Virgin HW, Klein RS, Sen GC, Diamond MS. 2012. 2′-O methylation of the viral mRNA cap by West Nile virus evades Ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog 8: e1002698. 10.1371/journal.ppat.1002698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Fan X, Xing J, Liu Z, Jiang B, Dou Y, Gorospe M, Wang W. 2015. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging 7: 1143–1158. 10.18632/aging.100860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. 2011. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. Br J Cancer 104: 700–706. 10.1038/sj.bjc.6606012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260: 1816–1819. 10.1126/science.8511591 [DOI] [PubMed] [Google Scholar]
- Thuy-Boun AS, Thomas JM, Grajo HL, Palumbo CM, Park S, Nguyen LT, Fisher AJ, Beal PA. 2020. Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition. Nucleic Acids Res 48: gkaa532. 10.1093/nar/gkaa532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S, Galeano F, Alon S, Raho S, Galardi S, Polito VA, Presutti C, Vincenti S, Eisenberg E, Locatelli F, et al. 2015. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol 16: 5. 10.1186/s13059-014-0575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Piñeyro D, Filonava L, Stracker TH, Batlle E, de Pouplana LR. 2014. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett 588: 4279–4286. 10.1016/j.febslet.2014.09.025 [DOI] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. 2012. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Vandivier LE, Anderson ZD, Gregory BD. 2019. HAMR: high-throughput annotation of modified ribonucleotides. Methods Mol Biol 1870: 51–67. 10.1007/978-1-4939-8808-2_4 [DOI] [PubMed] [Google Scholar]
- Vlachogiannis NI, Gatsiou A, Silvestris DA, Stamatelopoulos K, Tektonidou MG, Gallo A, Sfikakis PP, Stellos K. 2020. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J Autoimmun 106: 102329. 10.1016/j.jaut.2019.102329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, de Sousa Abreu R, Ko D, Le S-Y, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. 10.1038/msb.2010.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmar W, Gloger J, Berger E, Kortenbruck G, Köhling R, Speckmann E-J, Musshoff U. 2004. RNA editing (R/G site) and flip-flop splicing of the AMPA receptor subunit GluR2 in nervous tissue of epilepsy patients. Neurobiol Dis 15: 371–379. 10.1016/j.nbd.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768. 10.1126/science.290.5497.1765 [DOI] [PubMed] [Google Scholar]
- Wang IX, So E, Devlin JL, Zhao Y, Wu M, Cheung VG. 2013. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep 5: 849–860. 10.1016/j.celrep.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu H, Zheng J, Chen B, Zhou M, Fan W, Wang H, Liang X, Zhou X, Eriani G, et al. 2016a. A deafness- and diabetes-associated tRNA mutation causes deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J Biol Chem 291: 21029–21041. 10.1074/jbc.M116.739482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. 2016b. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 534: 575–578. 10.1038/nature18298 [DOI] [PubMed] [Google Scholar]
- Wang H, Xu B, Shi J. 2020a. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 722: 144076. 10.1016/j.gene.2019.144076 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao Y, Dong S, Yu Q, Jia G. 2020b. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat Chem Biol 16: 896–903. 10.1038/s41589-020-0525-x [DOI] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7: 618–630. 10.1016/j.stem.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Yamamoto G, Fujiyoshi K, Akagi Y, Kakuta M, Nishimura Y, Akagi K. 2019. Development of metachronous rectal cancers in a young man with dyskeratosis congenita: a case report. J Med Case Rep 13: 117. 10.1186/s13256-019-2044-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Moss B. 1977. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16: 1672–1676. 10.1021/bi00627a023 [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. 1975. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4: 379–386. 10.1016/0092-8674(75)90158-0 [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. 2018a. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71: 973–985.e5. 10.1016/j.molcel.2018.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Panneerdoss S, Timilsina S, Zhu J, Mohammad TA, Lu Z-L, de Magalhães JP, Chen Y, Rong R, Huang Y, et al. 2018b. Topological characterization of human and mouse m5C epitranscriptome revealed by bisulfite sequencing. Int J Genomics Proteomics 2018: 1351964. 10.1155/2018/1351964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann D, van der Laan S, Underwood MD, Salvetat N, Cavarec L, Vincent L, Molina F, Mann JJ, Arango V, Pujol JF. 2016. Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Transl Psychiatry 6: e878. 10.1038/tp.2016.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, Zamudio JR, Sturm NR, Jaworski J, Bujnicki JM. 2011. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res 39: 4756–4768. 10.1093/nar/gkr038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk N-B, et al. 2008. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem 283: 25046–25056. 10.1074/jbc.M803776200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Anggono V. 2018. The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J Neurochem 147: 137–152. 10.1111/jnc.14481 [DOI] [PubMed] [Google Scholar]
- Wu L, Wu D, Ning J, Liu W, Zhang D. 2019. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer 19: 326. 10.1186/s12885-019-5538-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth L, Gebauer F. 2015. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta 1849: 881–886. 10.1016/j.bbagrm.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Xiang J-F, Yang Q, Liu C-X, Wu M, Chen L-L, Yang L. 2018. N6-methyladenosines modulate A-to-I RNA editing. Mol Cell 69: 126–135.e6. 10.1016/j.molcel.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. 1995. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci 92: 8483–8487. 10.1073/pnas.92.18.8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. 1997. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev 11: 321–333. 10.1101/gad.11.3.321 [DOI] [PubMed] [Google Scholar]
- Yamato I, Sho M, Shimada K, Hotta K, Ueda Y, Yasuda S, Shigi N, Konishi N, Tsujikawa K, Nakajima Y. 2012. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res 72: 4829–4839. 10.1158/0008-5472.CAN-12-0328 [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21. 10.1038/nsmb1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, Li A, Wang X, Bhattarai DP, Xiao W, et al. 2017. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 27: 606–625. 10.1038/cr.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Gao R, Chen Y, Yang Z, Han P, Zhang H, Dou Y, Liu W, Wang W, Du G, et al. 2017. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget 8: 20751–20765. 10.18632/oncotarget.10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Meier UT. 2014. RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA Biol 11: 1483–1494. 10.4161/15476286.2014.972855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Terns RM, Terns MP. 2005. Mechanisms and functions of RNA-guided RNA modification. In Fine-tuning of RNA functions by modification and editing (ed. Grosjean H), pp. 223–262. Springer, Berlin/ Heidelberg. [Google Scholar]
- Zaccara S, Jaffrey SR. 2020. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181: 1582–1595.e18. 10.1016/j.cell.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. 2019. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20: 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zanzoni A, Spinelli L, Ribeiro DM, Tartaglia GG, Brun C. 2019. Post-transcriptional regulatory patterns revealed by protein-RNA interactions. Sci Rep 9: 4302. 10.1038/s41598-019-40939-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Samanta D, Lu H, Bullen JW. 2016a. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci 113: E2047–E2056. 10.1073/pnas.1602883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, Semenza GL. 2016b. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 7: 64527–64542. 10.18632/oncotarget.11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. 2016. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep 17: 1441–1451. 10.15252/embr.201642682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C. 2018. Publisher correction: post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 19: 808. 10.1038/s41580-018-0075-1 [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pirnie SP, Carmichael GG. 2017. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 23: 1303–1314. 10.1261/rna.061549.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137–143. 10.1038/ni.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.