Abstract
Background
The gut microbiome impacts human health through various mechanisms and is involved in the development of a range of non-communicable diseases. Diet is a well-known factor influencing microbe-host interaction in health and disease. However, very few findings are based on large-scale analysis using population-based studies. Our aim was to investigate the cross-sectional relationship between habitual dietary intake and gut microbiota structure in the Cooperative Health Research in the Region of Augsburg (KORA) FF4 study.
Results
Fecal microbiota was analyzed using 16S rRNA gene amplicon sequencing. Latent Dirichlet allocation (LDA) was applied to samples from 1992 participants to identify 20 microbial subgroups within the study population. Each participant’s gut microbiota was subsequently described by a unique composition of these 20 subgroups. Associations between habitual dietary intake, assessed via repeated 24-h food lists and a Food Frequency Questionnaire, and the 20 subgroups, as well as between prevalence of metabolic diseases/risk factors and the subgroups, were assessed with multivariate-adjusted Dirichlet regression models. After adjustment for multiple testing, eight of 20 microbial subgroups were significantly associated with habitual diet, while nine of 20 microbial subgroups were associated with the prevalence of one or more metabolic diseases/risk factors. Subgroups 5 (Faecalibacterium, Lachnospiracea incertae sedis, Gemmiger, Roseburia) and 14 (Coprococcus, Bacteroides, Faecalibacterium, Ruminococcus) were particularly strongly associated with diet. For example, participants with a high probability for subgroup 5 were characterized by a higher Alternate Healthy Eating Index and Mediterranean Diet Score and a higher intake of food items such as fruits, vegetables, legumes, and whole grains, while participants with prevalent type 2 diabetes mellitus were characterized by a lower probability for subgroup 5.
Conclusions
The associations between habitual diet, metabolic diseases, and microbial subgroups identified in this analysis not only expand upon current knowledge of diet-microbiota-disease relationships, but also indicate the possibility of certain microbial groups to be modulated by dietary intervention, with the potential of impacting human health. Additionally, LDA appears to be a powerful tool for interpreting latent structures of the human gut microbiota. However, the subgroups and associations observed in this analysis need to be replicated in further studies.
Video abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-020-00969-9.
Keywords: enable-Cluster, 16S rRNA gene sequencing, Nutrition, Dietary intake, Diabetes, Serum lipids, Obesity, Hypertension
Background
The last two decades of research have extensively studied the role of the human gut microbiome in host health [1]. The gut microbiome can be considered as a metabolically active organ that produces a multitude of metabolites that either positively or negatively affect human physiology [2]. Additionally, the composition and diversity of the gut microbiota have been associated with a wide range of diseases, ranging from gastrointestinal disorders such as Clostridium difficile infection to conditions such as type 2 diabetes mellitus (T2DM), cardiovascular disease, depression, and rheumatoid arthritis [3–7].
In addition to the many ways the gut microbiome influences its host, it has become clear that a variety of different factors influence the microbiome itself, including genetics, geographic environment, medication (especially antibiotics), nutrition, age, lifestyle, and mode of delivery at birth [8–13]. The number of modifiable factors associated with the gut microbiome, combined with its association with many disease states, presents the tempting possibility of influencing the development or progression of disease by modifying the gut microbiome. Yet this is complicated by wide interindividual variability [14, 15]. Furthermore, compositional changes do not necessarily translate into functional alterations, as different bacteria may perform similar functions [14]. As a result, it has so far proven difficult to characterize exactly what “healthy” microbiomes are [15].
Nutrition in particular is accepted as a major modifiable factor of the gut microbiome, yet it has also been independently associated with many of the same diseases that the gut microbiome is proposed to modulate [16–20]. In turn, the microbiota can also influence the absorption and metabolism of nutrients and other food components [15, 21]. This makes the task of unraveling the true nature of these associations challenging. While intense interest has produced a wealth of information on modifying the microbiome through nutrition, much of this still needs to be confirmed in large-scale, population-based cohorts.
We previously characterized the compositional and functional profiles of the prospective cohort Cooperative Health Research in the Region of Augsburg (KORA) and identified bacterial signatures of the development of T2DM [22]. Our aim in the present analysis was to evaluate the cross-sectional relationship between gut microbiota structure and habitual dietary intake in this large, population-based cohort. To this end, we applied an unsupervised machine learning method to identify latent structures (microbial subgroups) within the data and associations between habitual diet and/or metabolic diseases and these subgroups. In two subanalyses, we used the bacterial risk signatures of T2DM we previously established in this population and enterotype-like clusters to demonstrate the ability of LDA to identify latent features of the microbiota that would inherently be missed by other methods.
Methods
This analysis is reported according to the “Strengthening the Reporting of Observational Studies in Epidemiology - Nutritional Epidemiology (STROBE-nut)” recommendations [23].
Study population
The data used in this analysis originated from the KORA FF4 study (2013/2014), which is the second in a series of follow-up surveys of the original KORA S4 study, conducted from 1999 to 2001. KORA S4 is a population-based study that included 4261 participants recruited from the southern German city of Augsburg and its two surrounding counties. The FF4 survey included 2279 individuals from the original S4 study, ranging from 38 to 88 years old. Details concerning the design of the KORA studies have been published previously [24].
Collection and processing of biosamples
Participants in KORA FF4 collected a single stool sample according to paper-based instructions at home. They were also given instructions over the phone and were mailed a sterile stool collection kit. One spoonful taken from two different areas of the stool specimen was to be deposited into a tube containing 5 ml of DNA stabilizer (Stratec DNA Stool Stabilizer, No. 1038111100). The stool sample was to be collected on the morning of the study center visit and kept in the refrigerator until the appointment. If this was not possible, the stool sample could be mailed in afterward. Additionally, a short questionnaire was to be filled out regarding details of the sample collection (consistency, problems, time, storage conditions, etc.). After transport to the study center, samples were frozen at − 80 °C until further processing. Any participants who took antibiotics in the previous 2 months were excluded. Further details of the collection procedure and handling until analysis were described previously [25]. Samples were submitted from 2076 participants.
16S rRNA gene amplicon sequencing
Preparation and analysis of the gut microbiota samples were done as described in Reitmeier et al. [22]. Briefly, the metagenomic DNA was isolated from 600 μl of stool in DNA stabilizer solution using a modified version of the protocol by Godon et al. [26]. A FastPrep-24 instrument fitted with a cooling adapter and 0.1-mm glass beads was used to lyse microbial cells. NucleoSpin gDNA columns (Machery-Nagel, No. 740230.250) were used to purify the DNA. A robotic liquid handler was used for all pipetting steps after DNA extraction until sequencing to maximize reproducibility.
Polymerase chain reaction (PCR) runs were conducted in duplicates. The extracted DNA was diluted in PCR-grade water and 24 ng was used as a template for amplification (25 cycles) using a two-step process to minimize bias [27]. The V3/V4 regions of 16S rRNA genes were amplified using 341F-ovh and 785r-ovh primers [28]. During cleaning, PCR products were pooled using magnetic beads (Beckman Coulter). Fluorimetry was used to determine PCR fragment concentration, which was adjusted to 2 nM. An Illumina HiSeq was used to sequence multiplexed samples in paired-end mode (2 × 250 bp) using the Rapid v2 chemistry. Samples with a read count < 4700 (high-quality, chimera-checked) were re-sequenced on an Illumina MiSeq using v3. To identify potential inconsistencies between the platforms, identical samples were sequenced on both the HiSeq and MiSeq and were compared. No differences could be identified between the taxonomic compositions of the runs. Two negative controls (a PCR control without template DNA and a DNA extraction control containing 600 μl stool stabilizer but no sample) and one positive control (mock community; ZymoBIOMICS, No. D6300) were included in every batch of 45 samples (processed on a single 96-well plate) to control for artifacts.
Analysis of amplicon sequences
The 16S rRNA amplicon reads were preprocessed using the UPARSE-based IMNGS platform [29, 30]. Chimeras were removed using UCHIME [31]. Five nucleotides on both the 5′ and 3′end were trimmed for each of the R1 and R2 reads, respectively. The quality trim score was 5 and expected number of errors across assembled reads was 1. Sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity using UPARSE v8.1.1861_i86 [30]. OTUs occurring at a relative abundance < 0.25% across all samples were removed to prevent the analysis of spurious OTUs [32]. Taxonomies were assigned with adequate confidence (> 80%) to a maximum of genus level using the RDP classifier version 2.11 and confirmed using the SILVA database version 132 [33]. The OTU table containing 2091 OTUs was then normalized by total count per column to account for differences in library size. Taxonomic classification at the species levels for relevant OTUs was assigned using EzBioCloud (version 20200513) where possible [34].
Assessment of dietary intake
Habitual dietary intake was assessed in KORA FF4 participants using a two-step method combining information from up to three repeated 24-h food lists (24HFL) and one Food Frequency Questionnaire (FFQ) [35, 36]. The calculation of dietary intake in KORA FF4 is based on the estimation of consumption probability and consumption amount. Details have been published previously, but briefly, consumption probability is determined for each food item for each individual based on the 24HFLs and FFQ, while usual portion size for each item is estimated based on data from the Bavarian Food Consumption Survey II (BVS II) [37]. Consumption probability multiplied by consumption amount then results in the usual intake of each food item on any given day. Food items were then categorized into 16 food groups and 21 subgroups based on the European Prospective Investigation into Cancer and Nutrition (EPIC)-Soft classification scheme [38]. In addition to the standard food groups and subgroups that are specified by the EPIC-Soft criteria, the variable “whole grains” was created from the food items “whole grain bread,” “whole grain toast,” and “muesli.” The variable “refined grains” was constructed by subtracting intake of whole grains from the food group “grains and grain products.” For the purposes of this analysis, the food group “dairy” excludes cheese and yogurt, as they were investigated individually. Habitual nutrient intake was calculated based on usual food intake using the National Nutrient Database (Bundeslebensmittelschlüssel; BLS 3.02). Information about dietary supplement use was collected with the 24HFL, but was not included in the calculation of habitual dietary intake. Habitual dietary intake data was available for 1602 participants; however, both microbiota and diet information were available for only 1442 participants. Therefore, all nutrition-related analyses were limited to this sample size. The Alternate Healthy Eating Index 2010 (AHEI, modified to exclude trans fats) and Mediterranean Diet Score 2003 (MDS) were calculated for this subsample of 1442 individuals as performed by Wawro et al. [39–41].
Assessment of diseases/risk factors and covariates
On the day of the study center visit, a face-to-face interview was conducted, which gathered information on age (years), sex (male, female), education level (< 13 and ≥ 13 years, including vocational training), leisure time physical activity (> 1 h/week in summer and winter), smoking habits (current, ex-, never), medical diagnoses, and medication use, among other variables. Trained examiners took anthropometric measurements in standardized fashion. Blood pressure was measured and a fasting blood sample was drawn for the assessment of serum LDL-c, HDL-c, total cholesterol, and triglycerides. A self-reported diagnosis of diabetes or the use of antidiabetic medication was confirmed with the participant’s treating physician. An oral glucose tolerance test was carried out in all participants who did not have an existing diagnosis of diabetes. Participants were categorized based on their glucose tolerance status according to the 2003 American Diabetes Association diagnostic criteria as either (1) normal, (2) prediabetes, (3) undiagnosed diabetes mellitus, (4) known T2DM, or (5) other/unknown [42]. Participants’ hypertension status was classified according to existing diagnosis, if any, their blood pressure reading (according to the 1999 International Society of Hypertension-World Health Organization diagnostic criteria (≥ 140/90 mmHg) [43]), and use of antihypertensive medication into five groups: (1) normal (participant is normotensive); (2) known hypertension, controlled (participant is aware of hypertension, takes antihypertensive medication, and blood pressure is < 140/90 mmHg); (3) known hypertension, uncontrolled (participant is aware of hypertension and takes medication, but blood pressure is ≥ 140/90 mmHg); (4) known hypertension, not treated (participant is aware of hypertension, does not take medication, and blood pressure is ≥ 140/90 mmHg); and (5) undetected hypertension (participant is unaware of hypertension, does not take medication, and blood pressure is ≥ 140/90 mmHg). A detailed description of the assessment of the variables used in this analysis has already been presented in previous papers [25, 44].
Statistical analysis
Latent Dirichlet allocation
For the purposes of our analysis, it was necessary to perform either clustering or dimensionality reduction in order to reduce the number of OTUs from 2091 to a more practical number. The concept of “enterotypes,” typically a three-cluster solution, is probably the most commonly employed clustering strategy in regard to microbiome data [45]. However, the concept of only two to three clusters that can adequately describe any gut microbiota sample is very limited in its application and ability to describe interindividual differences, especially when it comes to microbiota structure in relation to health and disease. Therefore, we elected to implement latent Dirichlet allocation (LDA), a Bayesian probabilistic generative model proposed by Blei et al. in 2003, which is used to uncover latent structures present in unlabeled data [46]. This popular unsupervised machine learning method has been implemented most commonly in the field of natural language processing, where it can identify latent topics (e.g., “sports,” “politics,” “science”) present in a collection of documents. However, LDA has also been applied to a variety of biological data types, including population genetics data, protein sequence data, magnetic resonance imaging data, and microbiome data, where it can learn latent microbial subgroups [47–53].
Assuming there is a total number of O observations and K subgroups, the generative process modelled by LDA (described in relation to our analysis) assumes that the gut microbiota structure of each observation can be represented by a multinomial distribution, parametrized by θi, over latent subgroups, where θi is drawn from a latent Dirichlet distribution parametrized by α:
In turn, each subgroup is characterized by a multinomial distribution, parametrized by ϕk, over the OTUs, where ϕk is drawn from another latent Dirichlet distribution parametrized by β:
Given a total number of observations O and a total number of reads Ni for an observation i, the generation of all reads, where each read qi,j with the position, i and j, where i ∈ {1, …, O} and j ∈ {1, …, Ni}, is modelled consecutively by sampling the subgroup ki,j ~ Multinomial(θi) and a corresponding read qi,j ~ Multinomial(ϕki,j). In both cases, the Multinomial distributions refer to Multinomial distributions with one trial.
Given a fitted model, the variables θi and ϕk are of particular interest. For each observation, θi is a vector of probabilities over all subgroups, the sum of which is 1. A high probability of a subgroup means that it contributes to a large part of the microbiota structure of that observation. Likewise, for each subgroup, ϕk is a vector of probabilities over all OTUs, the sum of which is 1. In this case, a high probability of an OTU means that OTU contributes to a large part of that microbial subgroup.
When compared to traditional clustering or classification methods, θi can also be seen as fractional membership, meaning each observation’s microbiota structure can be described by a unique composition of several different microbial subgroups. For example, one observation may have a 45% probability for one subgroup, a 20% probability for a second subgroup, and a 5% probability for 7 more subgroups; another may have a 10% probability for 10 different subgroups. Likewise, each subgroup has a different probability of containing each of the 1713 OTUs. While fractional membership can also be achieved by fuzzy clustering methods, LDA differs from fuzzy clustering as well in that it learns patterns of co-occurrences of OTUs (ϕk) rather than clustering observations based on distance measures. This means that each subgroup represents a group of microbes that tend to appear together, due to similar environmental requirements, functions, or because they are modulated by a shared external factor.
Calculation of microbial subgroups
Microbiota data were available for 2033 participants. All participants who reported taking systemic antibiotics in the previous 2 months were excluded (n = 41), leaving 1992 participants available for analysis.
After quality controls and reads processing, 2091 OTUs were kept for analysis. Before performing LDA, the OTU table was filtered so that only OTUs occurring at a relative abundance > 0.1% and 1% prevalence were included in order to reduce sparsity in the data set, resulting in remaining 1713 OTUs. A relatively low cutoff was chosen due to the nature of the method. LDA is designed to process data sets with many words (OTUs). Additionally, because part of our goal was to identify associations between the gut microbiota and diseases, removing OTUs present only across e.g. less than 10% of samples could potentially result in the loss of OTUs present only in a certain disease state.
The LDA model was fitted using Gibbs’ sampling with the R package MetaTopics version 1.0 [54]. As this package requires the input matrix to contain count data in the form of integers, and our OTU table consists of normalized counts that add up to 1 for each participant, we multiplied the matrix by a factor of 1000 and rounded to the nearest whole number. The number of subgroups was selected using 5-fold cross-validation via the selectk() function (MetaTopics). LDA models for subgroup numbers between 5 and 190 were fitted and compared based on perplexity and loglikelihood values. Both parameters continued to improve with increasing subgroup number without a clear optimum, but the first jump in model performance was seen between 20 and 25 subgroups. As a relatively small subgroup number was necessary for this analysis to allow for interpretability, this subgroup number range was chosen for further analysis. In a sensitivity analysis performed with all 2091 OTUs, a small jump was seen between 15 and 20 subgroups. Five models with k = 20 were then fitted and compared.
Diet-subgroup and disease-subgroup associations
Associations between habitual diet and microbial subgroups were assessed using Dirichlet regression models (R package DirichletReg version 0.7.0), which are able to evaluate associations between predictor variables and multiple compositional outcome variables [55]. This was necessary in our case, as each subgroup corresponds to one variable, and together the 20 subgroup variables are compositional (i.e., values across the subgroups add up to 1 for each observation). One model was fitted for each of 29 selected food items or nutrients and two diet quality scores with all 20 subgroups as the response variables. These models were limited to the 1442 participants for whom both nutrition and microbiota data are available. Each model was adjusted for age, sex, energy intake, education, smoking, and physical activity. All 1442 participants had complete covariate information. Estimates were given per standard deviation for each dietary factor.
The associations between selected metabolic diseases or risk factors (body mass index (BMI), waist circumference, HDL-c, LDL-c, total cholesterol, triglycerides, diabetes, hypertension) and microbial subgroups were also evaluated using Dirichlet regression models. One model was fitted per disease or risk factor (8 models). All disease models were adjusted for age, sex, education, smoking, and physical activity. The serum lipid models were additionally adjusted for use of lipid-lowering medications. For each model, any participants with missing covariate information were excluded from the analysis; for the diabetes and serum lipid models, participants who were not fasted before the blood draw were also excluded (lipids, n = 20; BMI/waist circumference, n = 2; diabetes, n = 16; hypertension, n = 3). This resulted in a sample size of n = 1976 for the diabetes model, n = 1972 for the lipid models, n = 1990 for the BMI and waist circumference models, and n = 1989 for the hypertension model. Estimates were given per standard deviation for each continuous variable (BMI, waist circumference, HDL-c, LDL-c, total cholesterol, triglycerides). For the diabetes and hypertension models, the reference categories were normal glucose tolerance and normal blood pressure, respectively. P values for all associations were adjusted using the Bonferroni correction (α = 0.05 / 39 = 0.00128).
Subanalysis of arrhythmic OTUs
In a previous analysis, we identified time of defecation as one of the main factors responsible for interindividual differences in microbiota composition in the KORA FF4 cohort [22]. A heat map of the normalized relative abundances of 422 OTUs clearly showed daytime-dependent fluctuations in peak relative abundance. Strikingly, a subset of 87 of these OTUs lost their daytime-dependent fluctuations in relative abundance and became arrhythmic in T2DM and/or obesity. We identified a diabetes risk signature of 13 of these OTUs that were linked to disrupted circadian rhythmicity in microbial profiles. An additional 51 OTUs were identified as losing their rhythmicity in obesity specifically. A classification model including the diabetes-specific arrhythmic OTUs was able to predict type 2 diabetes in participants 5 years after the initial sampling. This indicates that the loss of rhythmicity may play a role in the development of these disease states and contributes significantly to the classification and prediction of type 2 diabetes mellitus.
Because LDA is able to identify hidden underlying patterns in a data set, and it was previously determined that time of defecation was one of the main factors responsible for interindividual differences in microbiota composition, we would expect LDA to pick up this effect and identify subgroups of microbes which are strongly influenced by circadian rhythm. Therefore, a subanalysis was done to determine the proportion of each subgroup comprised of OTUs identified as losing their circadian rhythmicity in either obesity, T2DM, or both. For each subgroup, the probabilities for each group of arrhythmic OTUs were summed up, both for all 87 arrhythmic OTUs and for the OTUs specific to obesity and T2DM (51 and 14 OTUs, respectively).
Subanalysis of enterotypes
To further explore the appropriateness of LDA in comparison to more traditional methods for identifying diet- and disease-microbiota relationships, three clusters, similar to enterotypes originally identified by Arumugam et al., were identified within the data set [45]. Further details of the clustering method and characteristics of the clusters utilized in the present subanalysis are reported in [22]. Briefly, the three clusters identified, C1 (n = 666), C2 (n = 1076), and C3 (n = 250), were dominated by the genus Bacteroides, Ruminococcus, and Prevotella, respectively. In the analyses restricted to participants with dietary data (n = 1442), clusters 1–3 contained 473, 798, and 171 participants, respectively. The relationships between diet, metabolic diseases, and enterotypes were evaluated using multinomial logistic regression models, with C2 (Ruminococcus) set as the reference level. The exposure and adjustment variables were identical to those in the Dirichlet regression models and again continuous variables were divided by standard deviation. Reference categories for the diabetes and hypertension variables were “normal glucose tolerance” and “normal blood pressure,” respectively. P values for all associations were adjusted using the Bonferroni correction (α = 0.05 / 39 = 0.00128).
Descriptive statistics and figures
Mean and standard deviation were calculated for continuous variables, while percentage and frequency were calculated for categorical variables for the descriptive tables, for the total population and stratified by sex. The violin plot, histogram, and bar plots were generated using ggplot2 version 3.3.1 in R. Hierarchical clustering of the subgroups was performed on log-transformed data using the agnes() function (cluster package version 2.1.0) and Ward’s method. The full matrix containing probabilities for each OTU was used for clustering to include potential differences in species, which would not be taken into account if probabilities were collapsed to the genus level. The dendrogram and cluster visualization were produced with the factoextra package version 1.0.7. A feature-expression heat map displaying the beta coefficients and P values from the Dirichlet regression models was created using the corrplot package version 0.84 in R and combined in Inkscape, as suggested by Haarman et al. [56]. All statistical analyses were conducted in RStudio Version 1.1.423 and R for Windows Version 3.5.1.
Results
Study population
The descriptive characteristics of the study population are shown in Table 1, for the total population and stratified by sex. On average, men (n = 969) were 61 years old and women (n = 1023) were 60 years old. Men had a higher mean waist circumference and BMI (103 cm and 28.3 kg/m2, respectively) than women (91 cm and 27.4 kg/m2, respectively). Women had a lower level of education on average (69.8% of women with < 13 years vs. 60.2% of men), but a higher percentage of women were physically active during leisure time (59.2% of women vs. 55.4% of men) and had never smoked (53.3% of women vs. 38.8% of men). In men, there was a 12.3% prevalence of T2DM compared to 7.9% in women (10.0% total). While this is higher than the prevalence T2DM in the general population, which was estimated to be between 6.9 and 7.1% in 2009 and 2010, respectively, this is to be expected as the average age of participants in our study population was 60.37 years, and the prevalence of T2DM rises sharply with increasing age [57].
Table 1.
Characteristics of the study population by sex
| Total | Men | Women | ||||
|---|---|---|---|---|---|---|
| n = 1992 | n = 969 | n = 1023 | ||||
| Continuous variables | Mean | SD | Mean | SD | Mean | SD |
| Age (years) | 60.37 | 12.24 | 60.83 | 12.53 | 59.94 | 11.95 |
| Waist circumference (cm) | 96.95 | 14.26 | 102.91 | 12.28 | 91.31 | 13.71 |
| BMI (kg/m2) | 27.85 | 5.01 | 28.30 | 4.52 | 27.43 | 5.40 |
| HDL-c (mmol/l) | 1.70 | 0.49 | 1.50 | 0.40 | 1.89 | 0.49 |
| LDL-c (mmol/l) | 3.48 | 0.92 | 3.45 | 0.90 | 3.51 | 0.94 |
| Total cholesterol (mmol/l) | 5.59 | 1.02 | 5.43 | 1.00 | 5.75 | 1.01 |
| Triglycerides (mmol/l) | 1.39 | 0.83 | 1.56 | 0.98 | 1.24 | 0.63 |
| Categorical variables | % | n | % | n | % | n |
| Education | ||||||
| < 13 years | 65.1 | 1297 | 60.2 | 583 | 69.8 | 714 |
| ≥ 13 years | 34.8 | 693 | 39.7 | 385 | 30.1 | 308 |
| NA | 0.1 | 2 | 0.1 | 1 | 0.1 | 1 |
| Physical activity | ||||||
| Active | 57.4 | 1143 | 55.4 | 537 | 59.2 | 606 |
| Inactive | 42.6 | 849 | 44.6 | 432 | 40.8 | 417 |
| Smoker | ||||||
| Current | 15.2 | 302 | 15.7 | 152 | 14.7 | 150 |
| Ex- | 38.6 | 769 | 45.5 | 441 | 32.1 | 328 |
| Never | 46.2 | 921 | 38.8 | 376 | 53.3 | 545 |
| Diabetes | ||||||
| Normal glucose tolerance | 47.8 | 952 | 37.4 | 362 | 57.7 | 590 |
| Prediabetes | 34.2 | 681 | 42.3 | 410 | 26.5 | 271 |
| UDM | 4.2 | 84 | 5.2 | 50 | 3.3 | 34 |
| Prevalent T2DM | 10.0 | 200 | 12.3 | 119 | 7.9 | 81 |
| Unknown/other | 3.8 | 75 | 2.9 | 28 | 4.6 | 47 |
| Hypertension | ||||||
| Normal blood pressure | 60.3 | 1202 | 55.3 | 536 | 65.1 | 666 |
| Known HTN, controlled | 26.3 | 523 | 26.6 | 258 | 35.9 | 265 |
| Known HTN, uncontrolled | 6.8 | 136 | 8.4 | 81 | 5.4 | 55 |
| Known HTN, not treated | 4.0 | 80 | 5.7 | 55 | 2.4 | 25 |
| Undiagnosed HTN | 2.5 | 50 | 3.9 | 38 | 1.2 | 12 |
| NA | 0.1 | 1 | 0.1 | 1 | 0.0 | 0 |
| Lipid-lowering medication (yes) | 16.5 | 328 | 19.4 | 188 | 13.7 | 140 |
SD, standard deviation; BMI, body mass index; HDL-c, high-density lipoprotein cholesterol; LDL-c, low-density lipoprotein; UDM, undiagnosed diabetes mellitus; T2DM, type 2 diabetes mellitus; HTN, hypertension
Hypertension was also more prevalent among men than women, particularly in the uncontrolled and untreated categories (8.4% vs. 5.4% and 5.7% vs. 2.4%, respectively).
A description of habitual dietary intake in the study population is presented in Table 2, for the total population and stratified by sex. On average, women consumed more fruits (161 g/day vs. 153 g/day), vegetables (192 g/day vs. 157 g/day), dairy products (128 g/day vs. 100 g/day), and yogurt (53.6 g/day vs. 40.0 g/day), whereas men had a notably higher energy intake (2127 kcal/day vs. 1619 kcal/day) and consumption of refined grains (170 g/day vs. 121 g/day), red and processed meat (56.0 g/day and 65.6 g/day respectively in men vs. 35.4 g/day and 34.5 g/day respectively in women), sugar and sweets (39.7 g/day vs. 15.8 g/day), sugar-sweetened beverages (SSB, 77.4 g/day vs. 30.6 g/day), beer (274 g/day vs. 21.9 g/day), and total alcohol (15.9 g/day vs. 4.5 g/day). Women had a higher AHEI score than men (45.6 vs. 40.7), but scored slightly lower on the MDS (4.3 vs. 4.6).
Table 2.
Habitual dietary intake of the study population by sex
| Total | Men | Women | ||||
|---|---|---|---|---|---|---|
| n = 1442 | n = 699 | n = 743 | ||||
| Mean | SD | Mean | SD | Mean | SD | |
| Food items | ||||||
| Potatoes (g/day) | 60.91 | 22.45 | 66.57 | 23.17 | 55.59 | 20.38 |
| Vegetables (g/day) | 174.66 | 57.78 | 157.03 | 47.36 | 191.24 | 61.68 |
| Legumes (g/day) | 6.13 | 4.43 | 5.41 | 3.89 | 6.82 | 4.79 |
| Fruit (g/day) | 156.86 | 80.16 | 152.71 | 80.75 | 160.77 | 79.45 |
| Nuts and seeds (g/day) | 7.54 | 8.09 | 8.16 | 8.66 | 6.95 | 7.47 |
| Dairy products (g/day) | 114.48 | 86.27 | 100.05 | 82.70 | 128.06 | 87.40 |
| Yogurt (g/day) | 47.00 | 45.56 | 39.97 | 43.26 | 53.62 | 46.69 |
| Cheese (g/day) | 35.00 | 18.47 | 36.63 | 19.07 | 34.40 | 17.89 |
| Refined grains (g/day) | 144.52 | 41.80 | 169.60 | 37.85 | 120.94 | 29.94 |
| Whole grains (g/day) | 23.50 | 20.63 | 24.05 | 22.90 | 22.97 | 18.23 |
| Fresh red meat (g/day) | 45.41 | 15.92 | 56.02 | 15.03 | 35.42 | 8.57 |
| Processed (red) meat (g/day) | 49.60 | 28.21 | 65.61 | 29.83 | 34.54 | 15.46 |
| Fish and shellfish (g/day) | 21.03 | 13.82 | 23.63 | 16.13 | 18.59 | 10.68 |
| Eggs (g/day) | 17.23 | 11.56 | 18.38 | 12.77 | 16.15 | 10.19 |
| Animal fats (g/day) | 15.92 | 6.75 | 18.73 | 7.32 | 13.28 | 4.87 |
| Plant oils (g/day) | 9.09 | 6.71 | 10.58 | 8.23 | 7.69 | 4.42 |
| Sugar and sweets (g/day) | 37.63 | 15.27 | 39.74 | 35.64 | 15.77 | 14.51 |
| Cakes (g/day) | 53.85 | 19.83 | 58.57 | 21.52 | 49.41 | 16.94 |
| Coffee (g/day) | 401.48 | 133.81 | 408.95 | 141.87 | 394.46 | 125.45 |
| SSB (g/day) | 53.28 | 132.34 | 77.40 | 168.77 | 30.59 | 78.45 |
| Wine (g/day) | 38.07 | 51.59 | 43.24 | 58.15 | 33.21 | 44.03 |
| Beer (g/day) | 144.30 | 208.56 | 274.39 | 233.58 | 21.92 | 47.24 |
| Nutrients | ||||||
| Energy intake (kcal/day) | 1865.35 | 405.36 | 2127 | 350 | 1619 | 281 |
| Total fat (g/day) | 78.08 | 17.06 | 88.41 | 15.52 | 68.37 | 12.00 |
| Total carbohydrates (g/day) | 199.66 | 49.43 | 223.27 | 48.31 | 177.45 | 39.11 |
| Total protein (g/day) | 69.89 | 14.85 | 78.31 | 13.73 | 61.97 | 11.00 |
| Total fiber (g/day) | 17.85 | 5.05 | 18.43 | 5.14 | 17.31 | 4.90 |
| Soluble fiber (g/day) | 5.79 | 1.60 | 6.11 | 1.64 | 5.49 | 1.51 |
| Insoluble fiber (g/day) | 11.96 | 3.44 | 12.29 | 3.48 | 11.65 | 3.37 |
| Alcohol (g/day) | 10.03 | 10.07 | 15.89 | 10.96 | 4.51 | 4.59 |
| Dietary patterns | ||||||
| Alternate Healthy Eating Index | 43.23 | 9.27 | 40.71 | 8.75 | 45.61 | 9.11 |
| Mediterranean Diet Score | 4.43 | 1.68 | 4.59 | 1.45 | 4.28 | 1.86 |
SD, standard deviation; SSB, sugar-sweetened beverages
Composition of the subgroups
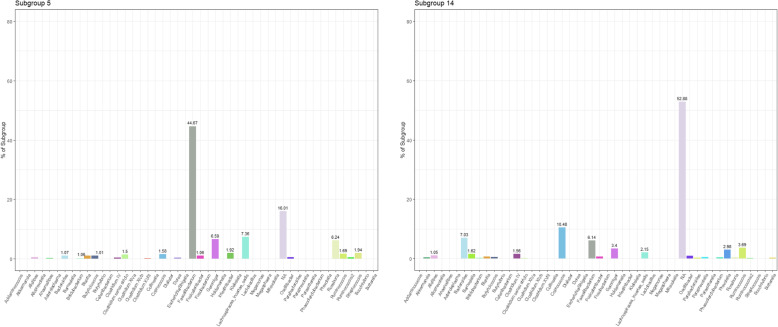
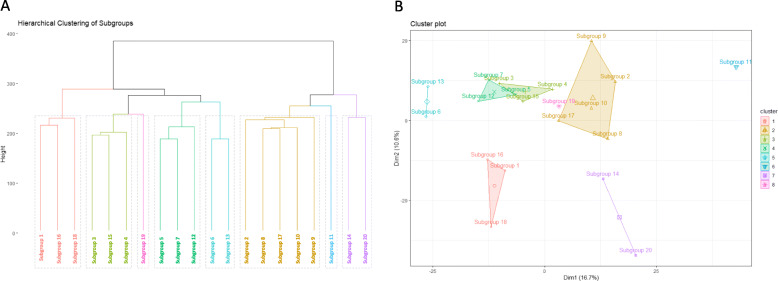
Figure 1 displays the top five genera for each of the 20 subgroups identified in the study population. The OTU probabilities were collapsed to the genus level here to allow for better interpretation of subgroup composition. Subgroup numbering is random and serves only as an identifier. While some subgroups (e.g., subgroups 1, 10, 11, 12, 14) were comprised of several different genera, others (e.g., subgroups 3, 4, 9, 13, 15, 19) were composed of more than 50% from a single genus. Figure 2 displays a visual comparison of the composition of subgroups 5 and 14, the two subgroups most strongly associated with both diet and disease, by genus. Only the 47 genera representing at least 1% of any subgroup plus are displayed, plus an NA category that encompassed all OTUs for which the genus was unknown. Some of the main genera in these two subgroups overlap (Faecalibacterium, Bacteroides, Coprococcus, Roseburia), though they are present in very different proportions in each subgroup. Additionally, some genera are prevalent in one subgroup but not the other (e.g., Intestinibacter and Streptococcus in subgroup 5, Barnesiella and Prevotella in subgroup 14). Notably, subgroup 14 was comprised 52.88% of OTUs that could not be identified to the genus level. Additional file 1 details the full composition of each subgroup by genus (97 genera in total were identified plus one overarching NA category). Figure 3 displays the results of hierarchical clustering of the subgroups. The clustering was performed on a matrix containing the probabilities of each of the 1713 unlabeled OTUs for each of the twenty subgroups. Eight clusters are each highlighted with a unique color to aid in comparison of subgroup similarity. Interestingly, although subgroups 3, 4, 6, 13, and 15 were all dominated by Bacteroides, they are split into two different clusters, which may be a result of different Bacteroides species which tend to appear in subgroups from one cluster vs. the other. Furthermore, although subgroups 13 and 15 appear very similar in regard to the composition of their top five genera, they also belong to different clusters. Additionally, though subgroups 5 and 14 appeared to have vaguely similar taxonomic compositions in Fig. 2, they were also assigned to different clusters, potentially also due to OTUs that differ at the species level. This highlights the importance of microbiota data that can reliably be identified beyond the genus level, but also the strength of LDA in identifying OTUs from a single genus that may represent different species and may, as a result, have differing functions or characteristics.
Fig. 1.
The top five genera (ranked by probability) per subgroup. For each subgroup, the five genera with the highest probability per subgroup are displayed. All OTUs for which the genus was unknown were grouped into an NA category; the percentage corresponding to NA indicates the percentage of the subgroup comprised of OTUs which could not be identified to the genus level
Fig. 2.
All 47 (of 97 total) genera that contributed to > 1% of any subgroup are shown, plus one NA category which encompasses all OTUs for which the genus was unknown
Fig. 3.
Hierarchical clustering of subgroups. a Dendrogram of subgroup clusters for comparison of subgroup similarity. Eight clusters are displayed, each represented by a different color and outlined by a gray rectangle. b Visualization of subgroup clusters. Cluster colors correspond to cluster colors from a
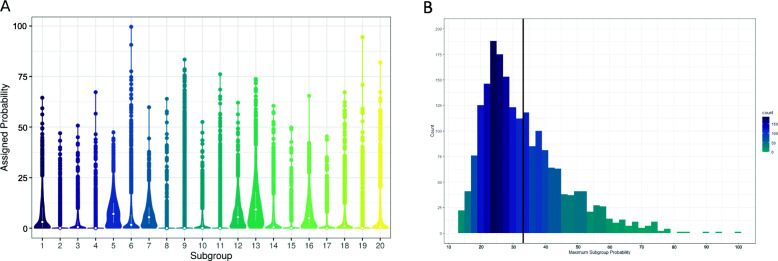
Figure 4a describes the distribution of subgroups among the study population. Some subgroups were quite prevalent, with a median probability across the study population of between 3.1 and 9.2% (subgroups 1, 5, 7, 12, 13, and 16), while others were rarer, with a median probability of < 0.1% (subgroups 2, 4, 8, 9, 10, 11, 15, 17, 19). Figure 4b shows the maximum subgroup probability across participants. Participants had an average maximum subgroup probability of 33.2%, although the highest maximum probability of any individual for any subgroup was 99.6%, and the lowest maximum probability of any individual was 13.1% (Fig. 4b). On average, each participant had 10 ± 2.2 subgroups with a probability of over 1%, 4.5 ± 1.1 subgroups with a probability of over 10%, and 1.5 ± 1.1 subgroups with a probability greater than 25%.
Fig. 4.
Distribution of subgroups across the study population. a Violin plot showing the probability per subgroup among the study population. Each violin displays the distribution of that subgroup across all participants. The median and interquartile range for each subgroup is represented by a white dot and bar. b Histogram showing the maximum probability for any subgroup per participant. The mean maximum probability was 33.24% ± 1.27% (indicated by vertical line). Each bin represents the participant count for that probability range. The darker the bin color, the higher the count
Habitual diet and subgroups
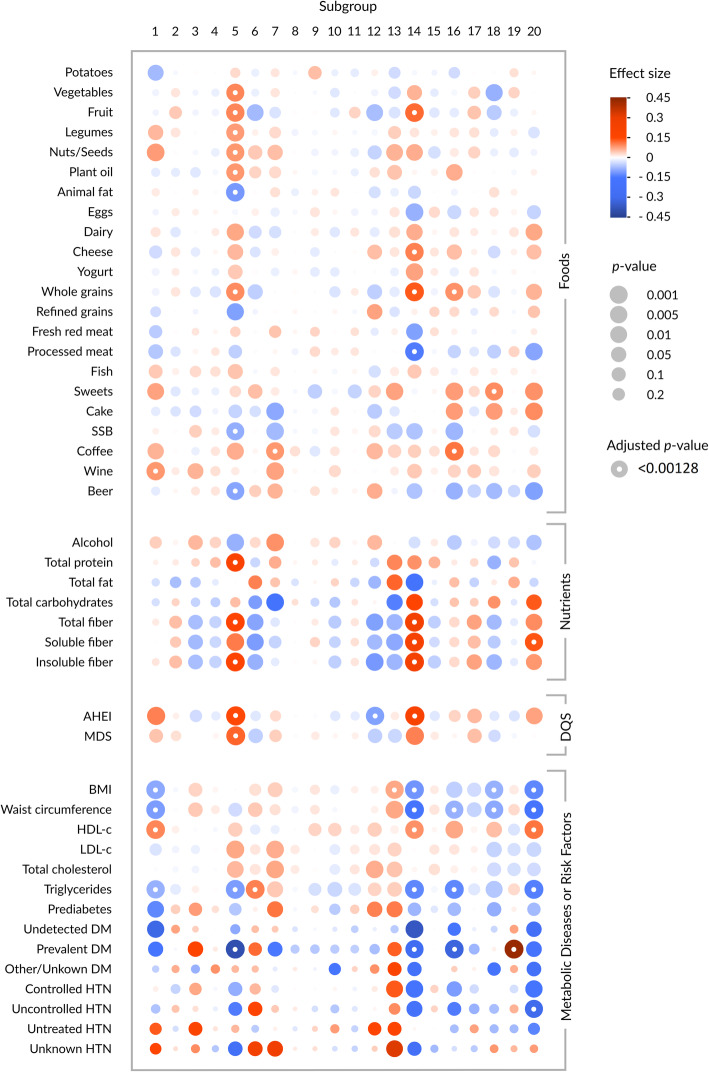
The associations between habitual diet (22 food items, seven nutrients, and two diet quality scores) and the 20 subgroups are displayed in Fig. 5. Subgroups 2, 4, 8, 9, 10, 11, 15, and 19 were not associated with any dietary factors. After adjustment for multiple testing, associations between diet and subgroups 1, 5, 7, 12, 14, 16, 18, and 20 remained significant. Subgroups 5 and 14 were most strongly and consistently associated with several nutrition factors. Participants with a high probability for subgroup 5 were characterized by a high intake of vegetables, fruits, legumes, nuts and seeds, plant oils, whole grains, total protein, total fiber, and insoluble fiber, and a low consumption of animal fat, SSB, and beer. A higher probability for subgroup 5 was also associated with a higher MDS and AHEI score. Those with a high probability for subgroup 14 were characterized by a higher AHEI score, as well as high consumption of fruit, cheese, whole grains, and all types of fiber, and a low consumption of processed meat. Individuals with a higher probability for subgroup 16 were marked by greater consumption of whole grains and coffee, while a higher probability for subgroup 20 was significantly associated with higher soluble fiber intake. Uniquely, subgroup 12 was significantly inversely associated with the AHEI, but not with any individual food item or the MDS after adjustment with the Bonferroni correction. Interestingly, many of the associations that lost significance after adjustment for multiple testing were with dietary factors that tend to be associated with adverse health effects, whereas many of the diet-subgroup associations that were the strongest were with dietary factors considered to be health-promoting. The full results of the Dirichlet regressions between habitual diet and the subgroups are shown in Additional file 2.
Fig. 5.
Feature expression heat map displaying results of the Dirichlet regression models. SSB, sugar-sweetened beverages; AHEI, Alternate Healthy Eating Index; MDS, Mediterranean Diet Score; DM, diabetes mellitus; HTN, hypertension; DQS, diet quality score. Foods, nutrients, or DQS and subgroups were analyzed in one set of models, while metabolic diseases or risk factors and subgroups were analyzed in another. Each association between a dietary factor or metabolic disease/risk factor and a subgroup (as identified in the Dirichlet regression models) is represented by a circle, where the size of the circle indicates the significance of the association (size inversely proportionate to P value), and intensity of the color (red, positive; blue, inverse) indicates the effect size. A white dot in the center of a circle indicates that the association remained significant after Bonferroni correction (P < 0.00128)
Metabolic diseases or risk factors and subgroups
The associations between selected metabolic diseases or risk factors and the 20 subgroups are also presented in Fig. 5 (full results in Additional file 3). Subgroups 1, 3, 5, 6, 7, 12, 13, 14, 16, 18, 19, and 20 were initially associated with one or more metabolic diseases or risk factors. After adjustment for multiple testing, only significant associations with subgroups 1, 5, 6, 13, 14, 16, 19, and 20 remained. The subgroups that showed the strongest/most numerous associations with diet (subgroup 5 (Faecalibacterium, Lachnospiracea incertae sedis, Gemmiger, Roseburia), subgroup 14 (Coprococcus, Bacteroides, Faecalibacterium, Ruminococcus), and, to a lesser extent, subgroup 16 (Gemmiger, Ruminococcus, Bacteroides, Dialister)) also showed strong associations with metabolic diseases or risk factors. Participants with lower serum triglyceride levels and a low prevalence of T2DM showed a higher probability for any of these three subgroups. Individuals with a lower BMI and/or waist circumference had a higher probability for subgroups 14 or 16, while higher serum HDL-c levels were associated with a higher percentage of subgroup 14.
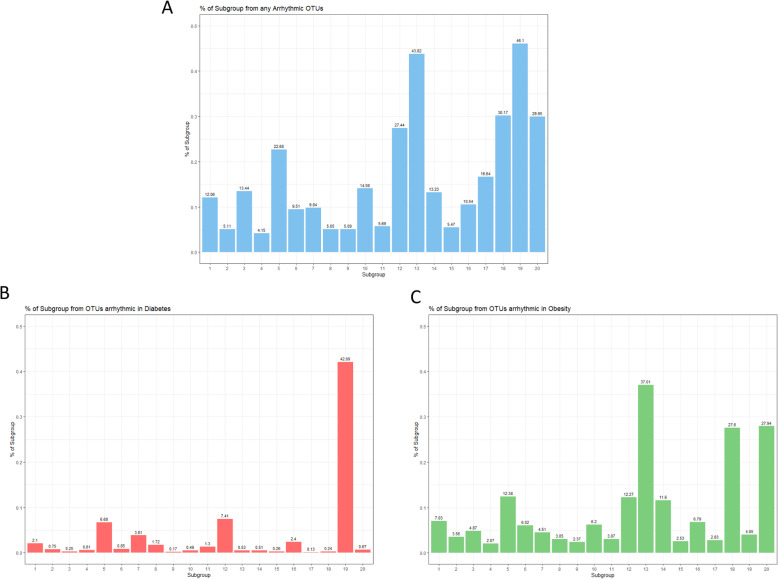
As a continuation of our previous analysis, in which we identified 87 OTUs that normally show daytime-dependent fluctuations in peak relative abundance but lose their rhythmicity in T2DM and/or obesity, we calculated the distribution of these arrhythmic OTUs among the subgroups (Fig. 6) [22]. Subgroups 19 (Escherichia/Shigella, Klebsiella, Streptococcus, Lactobacillus) and 13 (Bacteroides, Dialister, Parabacteroides, Alistipes) had a much higher percentage of arrhythmic OTUs than the rest (46% and 44%, respectively), followed by subgroups 18, 20, 12, and 5 (30%, 30%, 27%, and 23% arrhythmic OTUs, respectively). The remaining subgroups ranged between 5 and 17% arrhythmic OTUs. Regarding the 14 OTUs that were found to be arrhythmic in T2DM specifically, subgroup 19 had a notably higher proportion, with 42% of the subgroup being composed of diabetes-specific arrhythmic OTUs (all other subgroups contained only between 0 and 7%). Regarding the 51 OTUs found to be obesity-specific, subgroups 13, 18, and 20 had notably high percentages (37%, 28%, and 28%, respectively; the remaining subgroups ranged from 2 to 12%).
Fig. 6.
Percentage of each subgroup from arrhythmic OTUs. a Percentage of each subgroup from any arrhythmic OTUs. b Percentage of each subgroup from diabetes-specific arrhythmic OTUs. c Percentage of each subgroup from obesity-specific arrhythmic OTUs
Additionally, we conducted a subanalysis of the associations between habitual diet, metabolic diseases/risk factors, and three enterotype-like clusters. After correction for multiple testing, no dietary factors were significantly associated with enterotype (Additional file 4). BMI and waist circumference were positively associated with the Bacteroides cluster (OR (CI): 1.34 (1.21–1.49); 1.43 (1.27–1.61) respectively), as well as serum triglycerides (1.29 (1.16–1.43)) and undetected and prevalent diabetes (2.45 (1.47–4.07), 1.85 (1.28–2.67), respectively). Waist circumference was positively associated with the Prevotella enterotype (OR (CI): 1.31 (1.11–1.54)).
Discussion
We used LDA to identify 20 latent microbial subgroups in human gut microbiota samples from 1992 participants of the KORA FF4 study. We chose this method because of its wide use in other fields and its unique applicability to microbiota data [47]. Unlike clustering, LDA is a generative machine learning model, which is able to detect latent, or hidden, groups within data. Rather than using distance between observations as a measure, it identifies patterns of co-occurrence. As a result, each subgroup represents microbes that typically appear together in an environment (sample), likely due to similar environmental requirements (e.g., similar nutrient/substrate needs), similar roles or functions within an environment, or because the taxa within a subgroup are modulated by a shared, latent factor (e.g., medication, circadian rhythm, or other potentially unknown factors). The factors driving the co-occurrence of taxa within a subgroup may or may not be clear from a priori knowledge. As a result, LDA offers a unique advantage over traditional methods of clustering or drawing associations with individual taxa, as it can identify patterns within a data set that are driven by latent factors that researchers may or may not already be aware of and which can inherently not be detected by other methods of analysis. Additionally, traditional clustering methods are strongly influenced by dominant taxa, and are likely to miss more obscure features, which may be just as relevant. A further advantage of this strategy is that it is not limited to identifying individual taxa which may be beneficial or harmful, but rather a group of taxa that may act synergistically, have similar beneficial roles, produce similar metabolites, etc. Finally, LDA allows for partial membership, meaning each sample may contain several microbial subgroups, which is more appropriate to the biological situation and allows for a more complex description of the microbial environment than hard clustering methods. These unique advantages of LDA could prove useful for informing future studies by identifying subgroups of microbes within a population that are relevant for human health and disease, and which may be modulated by hidden factors that warrant further investigation. Several recent studies have demonstrated the advantages of LDA for metagenomic data. One excellent example of this is a recent analysis by Hosoda et al., in which LDA was utilized to identify four microbial “assemblages” (subgroups) within a metagenomic data set [50]. Three of these assemblages correlated strongly with the classic three-cluster enterotypes, but the fourth assemblage, which was present within each of the three enterotypes, was not as dominant within the data as the three more distinct assemblages and has therefore missed by classic analysis methods in the past.
Another paper, by authors Sankaran and Holmes, demonstrated the applicability of LDA to microbiome data using both a case study of the effect of two antibiotic courses on microbiota composition and a simulated microbiome data set [47]. The authors demonstrated that LDA was capable of appropriately modeling the simulation data set and was also able to identify four microbial “topics” (subgroups) in the data set, each of which responded uniquely to the antibiotic courses over time, despite a lack of temporal information.
In the present analysis, using multivariate Dirichlet regression models, one set that examined diet and subgroups, and another set of models that examined diseases/risk factors and subgroups, we identified a number of significant associations between habitual diet or metabolic diseases/risk factors and microbial subgroups. Subgroups 5 and 14 were most strongly and consistently associated with dietary factors and at the same time with metabolic diseases or risk factors. Many of these associations are consistent with current knowledge on diet-metabolic disease associations that were newly attributed to specific microbial subgroups in our analysis. This gives sound support to the hypothesis that diet-disease associations are at least partially modulated by the subject’s microbiota structure, as reflected by these two main subgroups—though causality remains to be confirmed.
The dietary associations we identified with subgroup 5 (Faecalibacterium, Lachnospiracea incertae sedis, Gemmiger, Roseburia) are consistent with that of a diet protective against T2DM. Interestingly, one study found that adhering to a Mediterranean diet increased occurrence of Faecalibacterium prausnitzii in feces and improved insulin sensitivity [58, 59]. This is consistent with our findings that closer adherence to a Mediterranean diet was associated with higher probability of subgroup 5 (44.67% Faecalibacterium; inversely associated with prevalent T2DM). Furthermore, several additional studies have shown an increase in F. prausnitzii, often as a result of modulation with a high-fiber diet, to be associated with improvement in T2DM parameters [19, 60, 61]. Subgroup 5 has the highest percentage of Faecalibacterium, but it is among the top five genera in other subgroups as well (e.g., subgroups 7, 10, 11). However, most of these subgroups showed no association with diet or disease markers, indicating the value of looking at a group of co-occurring bacteria rather than individual taxa.
Subgroup 14 (Coprococcus, Bacteroides, Faecalibacterium, Ruminococcus) was significantly positively associated with fruit, cheese, whole grains, and total, soluble and insoluble fiber intake after adjustment with the Bonferroni correction, and inversely with processed meat. These associations with subgroup 14 are logical, as Coprococcus species are fiber-fermenting butyrate producers, and their presence has generally been associated with positive health states [62–66].
Soluble fiber was the only dietary factor to remain associated with subgroup 20 after adjustment with the Bonferroni correction, although subgroup 20 was significantly associated with BMI, waist circumference, and several other metabolic parameters. Subgroup 20 was also one of the subgroups with a high percentage of arrhythmic OTUs; 28% of the subgroup was comprised of OTUs identified as behaving arrhythmically in obesity (Fig. 6). These findings support the associations we identified between subgroup 20 and BMI and waist circumference. Unfortunately, subgroup 20 is comprised of over 75% of OTUs that could not be reliably identified to the genus level, although its strong associations with disease and high percentage of OTUs that are arrhythmic in obesity suggest this subgroup could be of great interest for further research.
Subgroup 13 (Bacteroides, Dialister, Parabacteroides, Alistipes) comprised 74.76% of the genus Bacteroides and was significantly positively associated with BMI. After adjustment for multiple testing, subgroup 13 was no longer significantly associated with any food item or nutrient, which suggests that another factor may be its main driver. Interestingly, 37% of subgroup 13 is composed of OTUs that were identified in our previous analysis as losing their rhythmicity in obesity (Fig. 6) [22]. Because subgroup 13 is so strongly associated with BMI, it warrants further investigation. The high percentage of obesity-specific arrhythmic OTUs in this subgroup indicates that it may be worth further investigation regarding the importance of arrhythmic OTUs and obesity in general.
Subgroup 18 (Akkermansia, Alistipes, Ruminococcus, Bacteroides) was significantly inversely associated with BMI and waist circumference. Numerous studies have previously reported inverse associations between Akkermansia (27.13% of subgroup 18) and obesity, which is consistent with our results [67, 68]. Conversely, the genus Alistipes (19.52% of subgroup 18) has been associated with both positive and negative health states, potentially depending on the host environment. Our results suggest a potentially protective effect against obesity when co-occurring with other taxa in subgroup 18 in this study population. Additionally, 30% of subgroup 18 was comprised of OTUs previously identified as becoming arrhythmic in obesity (Fig. 6), again highlighting this subgroup as potentially highly relevant to obesity.
Subgroup 19 (Escherichia/Shigella, Klebsiella, Streptococcus, Lactobacillus) was not associated with any dietary factor. However, there was a significant positive association between prevalent T2DM and subgroup 19. It is possible that this association is driven by the intake of the drug metformin by participants with prevalent T2DM. Indeed, one previous study found an association between the severity of gastrointestinal side effects and the relative abundance of Escherichia-Shigella in participants receiving metformin after just 24 h [69], and several other studies have found an increase in Escherichia or E. coli in participants taking metformin [13, 70]. Additionally, 46% of subgroup 19 was comprised of arrhythmic OTUs, 91% of which were OTUs identified in our previous analysis as part of a diabetes risk signature [22]. This is a striking difference to the other subgroups, of which only 0–7% were composed of these diabetes-specific arrhythmic OTUs. The strong association of subgroup 19 with T2DM and its large percentage of diabetes-specific arrhythmic OTUs demonstrates how effectively LDA recognized a subgroup of OTUs previously demonstrated as specific to T2DM and which lose their rhythmicity in T2DM specifically. Other prominent OTUs in this subgroup should be investigated for their potential importance in T2DM as well.
Several additional subgroups were initially associated with one or more nutrition items/factors, but were no longer significant after adjustment for multiple testing (subgroups 3, 6, 13, 17). Additionally, many of the strongest diet-subgroup associations were with “healthy” dietary factors, such as whole grains and fruit, rather than those considered to have negative health effects, such as sweets, alcohol, and SSB. This indicates that the consumption of the food groups that remained statistically significant after correction for multiple testing may have a stronger impact on microbial subgroups than the ones that did not remain significant. In other words, an increase in consumption of healthy dietary constituents seems to be a more effective measure to modify gut microbiota composition (whereas the change in composition resulting from an increase in unhealthy foods is a more passive consequence.) Thus, reproduction of these findings in an independent study is urgently needed.
Several subgroups were not associated with any dietary factors (subgroups 2, 4, 8, 9, 10, 11, 15, 19) and, with the exception of subgroup 19, were also not associated with any of the covariates selected for adjustment. These independent subgroups are consistent with the idea that we still do not fully understand the factors responsible for shaping gut microbiome composition. Further analyses should seek to identify what latent factors may be driving these subgroups.
It is highly useful to be able to classify an individual’s highly complex microbiota structure into a manageable number of subgroups that can convey information about health and disease risk. The analysis of diet/metabolic disease and enterotype-like clusters demonstrated that, although habitual diet has been associated with enterotypes in the past, this three-cluster solution is not appropriate for identifying associations between habitual diet and microbiota structure in this study population [71]. While a few significant associations were seen between BMI or waist circumference, triglycerides, and T2DM and the clusters, no significant associations between diet and any enterotype cluster were identified after adjustment for multiple testing. The limitations of this clustering strategy have been discussed previously, and our results also indicate the need for a more sensitive and detailed approach to microbiota analysis [72, 73]. One major advantage that LDA offers over traditional clustering strategies is partial or fractional membership (similar to fuzzy clustering). A major criticism of enterotypes is the assignment of an individual into one cluster, ignoring often gradient-like differences in microbiota structure. Methods allowing for partial membership, such as LDA, enable an individual’s microbiota composition to be described by a combination of several microbial groups in varying proportions, which is more likely to reflect the actual complexity of an individual’s microbiome appropriately. Our results suggest that LDA, specifically, offers an appropriate alternative for the identification of latent structures within the microbiome that are of relevance to human health and disease. This method offers a practical way to characterize an individual’s microbiota and potentially to decipher more information about an individual’s health state and potential disease risk. However, the most appropriate use of this method may be in exploratory analyses, with the purpose of identifying subgroups of bacteria within a population that are driven by latent factors, and which should be investigated with further study.
Our analysis has several additional strengths, including high-quality nutrition data that characterizes habitual dietary intake, use of a large, originally population-based cohort with the opportunity for future longitudinal analyses, and the implementation of a popular, sophisticated unsupervised machine learning algorithm. Limitations include that only one stool sample was available per individual. Despite the use of a refined nutrition assessment method, measurement error is likely to persist to some degree. Due to the cross-sectional nature of this analysis, the causality of the associations cannot be determined. Generalizability of these results cannot be assumed without reproducing this analysis in other study populations and/or follow-ups of the present study population.
Conclusions
We described 20 microbial subgroups using an unsupervised machine learning model, several of which were associated with both habitual dietary intake and metabolic disease or relevant risk factors. The diet-microbiota and disease-microbiota associations identified in this analysis add insight to the complex relationship between diet, the human microbiome, and disease. Further analyses implementing LDA in other populations and in longitudinal studies are necessary to investigate the reproducibility of the present findings.
Supplementary Information
Acknowledgements
We thank the Core Facility “Statistical Consulting,” Institute for Computational Biology at the Helmholtz Zentrum München, for their helpful assistance with the statistical analysis and critical review of the manuscript. We also thank Caroline Ziegler and Angela Sachsenhauser from the ZIEL - Institute for Food & Health for their excellent technical assistance with amplicon sequencing.
Abbreviations
- 24HFL
24-h food list
- AHEI
Alternate Healthy Eating Index
- BMI
Body mass index
- DNA
Deoxyribonucleic acid
- EPIC
European Prospective Investigation into Cancer and Nutrition
- FFQ
Food Frequency Questionnaire
- HDL-c
High-density lipoprotein cholesterol
- KORA
Cooperative Health Research in the Region of Augsburg
- LDA
Latent Dirichlet allocation
- LDL-C
Low-density lipoprotein cholesterol
- MDS
Mediterranean Diet Score
- OTU
Operational taxonomic unit
- PCR
Polymerase chain reaction
- rRNA
Ribosomal ribonucleic acid
- T2DM
Type 2 diabetes mellitus
Authors’ contributions
J.L., T.A.B., and N.W. formulated the research question and designed the analysis. S. Reitmeier, T.C., J.S., H.G., W.R., A.P., D.H., and C.M. conducted research and/or provided essential materials. T.A.B. performed statistical analysis, interpreted results, and drafted the manuscript. N.W., J.B., S. Reitmeier, G.P., and S. Rohrmann contributed to the statistical analysis and interpretation of results. J.L., N.W., C.M., S. Reitmeier, G.P., S. Rohrmann, D.H., and T.C. critically reviewed the manuscript. J.L., D.H., and T.C. secured funding and J.L. supervised the entire project. All authors were given the opportunity to read and contribute to the text and approved the final manuscript.
Funding
The KORA study was initiated and financed by the Helmholtz Zentrum München - German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. Stool sample collection in KORA FF4 was supported by iMED, a research alliance within the Helmholtz Association, Germany. The preparation of this paper was supported by the enable-Cluster and is catalogued by the enable Steering Committee as enable 061 (http://enable-cluster.de). This work was funded by a grant of the German Federal Ministry for Education and Research (BMBF) (FK 01EA1409E). The funding agencies had no role in the design, analysis, or writing of this article. Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
The dataset analyzed during the current study is not publicly available due to restrictions imposed by the Ethics Committee of the Bavarian Medical Association to protect the privacy of the study participants. However, a request for use of the data can be made via a project agreement through the KORA.PASST platform (https://www.helmholtz-muenchen.de/en/kora/for-scientists/cooperation-with-kora/index.html). The code used in this analysis can be found in Additional file 5.
Declarations
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki. All procedures involving human subjects were approved by the ethics committee of the Bavarian Chamber of Physicians in Munich. All participants gave their written, informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Taylor A. Breuninger, Email: taylor.breuninger@helmholtz-muenchen.de
Nina Wawro, Email: nina.wawro@helmholtz-muenchen.de.
Jakob Breuninger, Email: jakob.breuninger@gmail.com.
Sandra Reitmeier, Email: sandra.reitmeier@tum.de.
Thomas Clavel, Email: tclavel@ukaachen.de.
Julia Six-Merker, Email: julia.six-merker@helmholtz-muenchen.de.
Giulia Pestoni, Email: giulia.pestoni@uzh.ch.
Sabine Rohrmann, Email: sabine.rohrmann@uzh.ch.
Wolfgang Rathmann, Email: wolfgang.rathmann@ddz.de.
Annette Peters, Email: peters@helmholtz-muenchen.de.
Harald Grallert, Email: harald.grallert@helmholtz-muenchen.de.
Christa Meisinger, Email: c.meisinger@unika-t.de.
Dirk Haller, Email: dirk.haller@tum.de.
Jakob Linseisen, Email: j.linseisen@unika-t.de.
References
- 1.Cresci GA, Bawden E. Gut microbiome: what we do and don’t know. Nutr Clin Pract. 2015;30:734–746. doi: 10.1177/0884533615609899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vadder FD, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 2016;24:151–157. doi: 10.1016/j.cmet.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 4.Jie Z, Xia H, Zhong S-L, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;8:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 6.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet. 2017;18:690–699. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 9.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 10.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson MA, Verdi S, Maxan M-E, Shin CM, Zierer J, Bowyer RCE, Martin T, Williams FMK, Menni C, Bell JT, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9:2655. [DOI] [PMC free article] [PubMed]
- 12.Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. 2017;66:515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Gentile CL, Weir TL. The gut microbiota at the intersection of diet and human health. Science. 2018;362:776–780. doi: 10.1126/science.aau5812. [DOI] [PubMed] [Google Scholar]
- 16.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeevi D, Zmora TKN, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, Suez J, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Filippis FD, Pasolli E, Tett A, Tarallo S, Naccarati A, Angelis MD, Neviani E, Cocolin L, Gobbetti M, Segata N, et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25:444–453. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Medina-Vera I, Sanchez-Tapia M, Noriega-López L, Granados-Portillo O, Guevara-Cruz M, Flores-López A, Avila-Nava A, Fernández ML, Tovar AR, Torres N. A dietary intervention with functional foods reduces metabolic endotoxaemia and attenuates biochemical abnormalities by modifying faecal microbiota in people with type 2 diabetes. Diabetes Metab. 2019;45:122–131. doi: 10.1016/j.diabet.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, Kim AD, Shmagel AK, Syed AN, Students PMC, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25:789–802. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reitmeier S, Kießling S, Clavel T, List M, Almeida EL, Ghosh TS, Neuhaus K, Grallert H, Linseisen J, Skurk T, et al. Arrhythmic gut microbiome signatures predict risk of type-2 diabetes. Cell Host Microbe. 2020;28:258–72.e6. doi: 10.1016/j.chom.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Lachat C, Hawwash D, Ocké MC, Berg C, Forsum E, Hörnell A, Larsson C, Sonestedt E, Wirfält E, Åkesson A, et al. Strengthening the Reporting of Observational Studies in Epidemiology—Nutritional Epidemiology (STROBE-nut): an extension of the STROBE statement. PLoS Med. 2016;13:e1002036. doi: 10.1371/journal.pmed.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holle R, Happich M, Löwel H, Wichmann HE, for the MONICA/KORA Study Group KORA—a research platform for population based health research. Gesundheitswesen. 2005;67:19–25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 25.Breuninger T, Wawro N, Meisinger C, Artati A, Adamski J, Peters A, Grallert H, Linseisen J. Associations between fecal bile acids, neutral sterols, and serum lipids in the KORA FF4 study. Atherosclerosis. 2019;288:1–8. doi: 10.1016/j.atherosclerosis.2019.06.911. [DOI] [PubMed] [Google Scholar]
- 26.Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Applied and Environmental Microbiology. 1997;63:2802–2813. doi: 10.1128/AEM.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry D, Mahfoudh KB, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, Clavel T. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reitmeier S, Hitch TC, Fikas N, Hausmann B, Ramer-Tait AE, Neuhaus K, Berry D, Haller D, Lagkouvardos I, Clavel T. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. 2020. Preprint at https://www.researchsquare.com/article/rs-11835/v1. [DOI] [PMC free article] [PubMed]
- 33.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017:1613–7. [DOI] [PMC free article] [PubMed]
- 35.Freese J, Feller S, Harttig U, Kleiser C, Linseisen J, Fischer B, Leitzmann M, Six-Merker J, Michels K, Nimptsch K, et al. Development and evaluation of a short 24-h food list as part of a blended dietary assessment strategy in large-scale cohort studies. Eur J Clin Nutr. 2014;68:324–329. doi: 10.1038/ejcn.2013.274. [DOI] [PubMed] [Google Scholar]
- 36.Bohlscheid-Thomas S, Hoting I, Boeing H, Wahrendorf J. Reproducibility and relative validity of energy and macronutrient intake of a food frequency questionnaire developed for the German part of the EPIC project. European Prospective Investigation into Cancer and Nutrition. Int J Epidemiol. 1997;26:S71–S81. doi: 10.1093/ije/26.suppl_1.S71. [DOI] [PubMed] [Google Scholar]
- 37.Mitry P, Wawro N, Six-Merker J, Zoller D, Jourdan C, Meisinger C, Thierry S, Nöthlings U, Knüppel S, Boeing H, et al. Usual dietary intake estimation based on a combination of repeated 24-h food lists and a food frequency questionnaire in the KORA FF4 cross-sectional study. Front Nutr. 2019;6:145. [DOI] [PMC free article] [PubMed]
- 38.Slimani N, Deharveng G, Charrondière RU, van Kappel AL, Ocké MC, Welch A, Lagiou A, van Liere M, Agudo A, Pala V, et al. Structure of the standardized computerized 24-h diet recall interview used as reference method in the 22 centers participating in the EPIC project. European Prospective Investigation into Cancer and Nutrition. Comput Methods Programs Biomed. 1999;58:251–266. doi: 10.1016/S0169-2607(98)00088-1. [DOI] [PubMed] [Google Scholar]
- 39.Chiuve SE, Fung TT, Rimm EB, Hu FB, McCullough ML, Wang M, Stampfer MJ, Willett WC. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr. 2012;142:1009–1018. doi: 10.3945/jn.111.157222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trichopoulou A, Costacou T, Bamia C, Trichopoulos D. Adherence to a Mediterranean diet and survival in a Greek population. New Engl J Med. 2003;348:2599–2608. doi: 10.1056/NEJMoa025039. [DOI] [PubMed] [Google Scholar]
- 41.Wawro N, Pestoni G, Riedl A, Breuninger TA, Peters A, Rathmann W, Koenig W, Huth C, Meisinger C, Rohrmann S, et al. Association of dietary patterns and type-2 diabetes mellitus in metabolically homogeneous subgroups in the KORA FF4 study. Nutrients. 2020;12:1684. doi: 10.3390/nu12061684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.American Diabetes Association. 2 Classification and diagnosis of diabetes. Diabetes Care. 2015;38:S8–S16. doi: 10.2337/dc15-S005. [DOI] [PubMed] [Google Scholar]
- 43.Guidelines Subcommittee 1999 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension. J Hypertens. 1999;17:151–183. [PubMed] [Google Scholar]
- 44.Kowall B, Rathmann W, Stang A, Bongaerts B, Kuss O, Herder C, Roden M, Quante A, Holle R, Huth C, et al. Perceived risk of diabetes seriously underestimates actual diabetes risk: the KORA FF4 study. PLoS One. 2017;12:e0171152. doi: 10.1371/journal.pone.0171152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. [Google Scholar]
- 47.Sankaran K, Holmes SP. Latent variable modeling for the microbiome. Biostatistics. 2019;20:599–614. doi: 10.1093/biostatistics/kxy018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, He T, Hu X, Zhou Y, An Y, Wu X. Estimating functional groups in human gut microbiome with probabilistic topic models. IEEE Trans Nanobioscience. 2012;11:203–215. doi: 10.1109/TNB.2012.2212204. [DOI] [PubMed] [Google Scholar]
- 49.Liu L, Tang L, Dong W, Yao S, Zhou W. An overview of topic modeling and its current applications in bioinformatics. Springerplus. 2016;5:1608. doi: 10.1186/s40064-016-3252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosoda S, Nishijima S, Fukunaga T, Hattori M, Hamada M. Revealing microbial assemblage structure in the human gut microbiome using latent Dirichlet allocation. Microbiome. 2020;8:95. [DOI] [PMC free article] [PubMed]
- 51.Sui X, Rajapakse JC. The Alzheimer’s Disease Neuroimaging Initiative. Profiling heterogeneity of Alzheimer’s disease using white-matter impairment factors. Neuroimage Clin. 2018;20:1222–1232. doi: 10.1016/j.nicl.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan X-Y, Zhang Y-N, Shen H-B. Large-scale prediction of human protein-protein interactions from amino acid sequence based on latent topic features. J Proteome Res. 2010;9:4992–5001. doi: 10.1021/pr100618t. [DOI] [PubMed] [Google Scholar]
- 53.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan J, Chuai G, Qi T, Shao F, Zhou C, Zhu C, Yang J, Yu Y, Shi C, Kang N, et al. MetaTopics: an integration tool to analyze microbial community profile by topic model. BMC Genomics. 2017;18:962. doi: 10.1186/s12864-016-3257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maier MJ. DirichletReg: Dirichlet regression for compositional data in R. Vienna: WU Vienna University of Economics and Business; 2014. [Google Scholar]
- 56.Haarman BCMB, Lek RFR-VD, Nolen WA, Mendes R, Drexhage HA, Burger H. Feature-expression heat maps—a new visual method to explore complex associations between two variable sets. J Biomed Inform. 2015;53:156–161. doi: 10.1016/j.jbi.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Tamayo T, Brinks R, Hoyer A, Kuß O, Rathmann W. The prevalence and incidence of diabetes in Germany: an analysis of statutory health insurance data on 65 million individuals from the years 2009 and 2010: an analysis of statutory health insurance data on 65 million individuals from the years 2009 and 2010. Dtsch Arztebl Int. 2016;113:177–182. doi: 10.3238/arztebl.2016.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwingshackl L, Hoffmann G, Lampousi A-M, Knüppel S, Iqbal K, Schwedhelm C, Bechthold A, Schlesinger S, Boeing H. Food groups and risk of type 2 diabetes mellitus: a systematic review and meta-analysis of prospective studies. Eur J Epidemiol. 2017;32:363–375. doi: 10.1007/s10654-017-0246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meslier V, Laiola M, Roager HM, Filippis FD, Roume H, Quinquis B, Giacco R, Mennella I, Ferracane R, Pons N, et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut Microbiota. 2020; ONLINE AHEAD OF PRINT. [DOI] [PMC free article] [PubMed]
- 60.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359:1151–6. [DOI] [PubMed]
- 61.Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, Guo Y, Zhang C, Zhou Q, Xue Z, et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. Int Soc Microb Ecol. 2015;9:552–562. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malinen E, Krogius-Kurikka L, Lyra A, Nikkilä J, Jääskeläinen A, Rinttilä T, Vilpponen-Salmela T, Wright AJV, Palva A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrov VA, Saltykova IV, Zhukova IA, Alifirova VM, Zhukova NG, Dorofeeva YB, Tyakht AV, Kovarsky BA, Alekseev DG, Kostryukova ES, et al. Analysis of gut microbiota in patients with Parkinson’s disease. Bulletin of Experimental Biology and Medicine. 2017;162:734–737. doi: 10.1007/s10517-017-3700-7. [DOI] [PubMed] [Google Scholar]
- 64.Peters BA, Wilson M, Moran U, Pavlick A, Izsak A, Wechter T, Weber JS, Osman I, Ahn J. Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Med. 2019;11:61. doi: 10.1186/s13073-019-0672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nylund L, Nermes M, Isolauri E, Salminen S. Vos WMd, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 66.Alessi AM, Gray V, Farquharson FM, Flores-López A, Shaw S, Stead D, Wegmann U, Shearman C, Gasson M, Collie-Duguid ESR, et al. β-Glucan is a major growth substrate for human gut bacteria related to Coprococcus eutactus. Environ Microbiol. 2020; ONLINE AHEAD OF PRINT. [DOI] [PubMed]
- 67.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yassour M, Lim MY, Yun HS, Tickle TL, Sung J, Song Y-M, Lee K, Franzosa EA, Morgan XC, Gevers D, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8:17. doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elbere I, Kalnina I, Silamikelis I, Konrade I, Zaharenko L, Sekace K, Radovica-Spalvina I, Fridmanis D, Gudra D, Pirags V, et al. Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS One. 2018;13:e0204317. doi: 10.1371/journal.pone.0204317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, Ståhlman M, Olsson LM, Serino M, Planas-Fèlix M, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 71.Wu G, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh S, Bewtra M, Knights D, Walters W, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. Rethinking “enterotypes”. Cell Host Microbe. 2014;16:433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chenga M, Ning K. Stereotypes about enterotype: the old and new ideas. Genomics Proteomics Bioinformatics. 2019;17:4–12. doi: 10.1016/j.gpb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset analyzed during the current study is not publicly available due to restrictions imposed by the Ethics Committee of the Bavarian Medical Association to protect the privacy of the study participants. However, a request for use of the data can be made via a project agreement through the KORA.PASST platform (https://www.helmholtz-muenchen.de/en/kora/for-scientists/cooperation-with-kora/index.html). The code used in this analysis can be found in Additional file 5.