Abstract
Maternal obesity may lead to epigenetic alterations in the offspring and might thereby contribute to disease later in life. We investigated whether a lifestyle intervention in pregnant women with obesity is associated with epigenetic variation in cord blood and body composition in the offspring. Genome-wide DNA methylation was analyzed in cord blood from 208 offspring from the Treatment of Obese Pregnant women (TOP)-study, which includes pregnant women with obesity randomized to lifestyle interventions comprised of physical activity with or without dietary advice versus control subjects (standard of care). DNA methylation was altered at 379 sites, annotated to 370 genes, in cord blood from offspring of mothers following a lifestyle intervention versus control subjects (false discovery rate [FDR] <5%) when using the Houseman reference-free method to correct for cell composition, and three of these sites were significant based on Bonferroni correction. These 370 genes are overrepresented in gene ontology terms, including response to fatty acids and adipose tissue development. Offspring of mothers included in a lifestyle intervention were born with more lean mass compared with control subjects. Methylation at 17 sites, annotated to, for example, DISC1, GBX2, HERC2, and HUWE1, partially mediates the effect of the lifestyle intervention on lean mass in the offspring (FDR <5%). Moreover, 22 methylation sites were associated with offspring BMI z scores during the first 3 years of life (P < 0.05). Overall, lifestyle interventions in pregnant women with obesity are associated with epigenetic changes in offspring, potentially influencing the offspring’s lean mass and early growth.
Introduction
Obesity and type 2 diabetes are on the rise worldwide, as is the prevalence of obesity in pregnant women (1). Obesity during pregnancy increases the risk of adverse health outcomes in the offspring, including macrosomia and childhood obesity (2), which might be explained by a metabolically adverse intrauterine environment. The prevalence of childhood obesity, which is associated with an increased risk of adulthood obesity (3), metabolic syndrome (4), and early death (5), more than doubled between 1980 and 2015 (6). Greater increase in weight and height during infancy is associated with greater lean mass and lower risk of the metabolic syndrome in adulthood (7,8). Hence, greater lean mass during infancy might protect against future metabolic disease.
As gestational weight gain (GWG) affects the health of the mother and offspring, the Institute of Medicine recommends women with prepregnancy BMI >30 kg/m2 to limit their GWG to 5–9 kg (1). We have reported that GWG can be reduced by lifestyle interventions (9) and was positively associated with fat mass in infants born to mothers with obesity (10,11) and with carbohydrate intake in late pregnancy (12). Subsequently, lifestyle interventions might improve the cardiometabolic profile of pregnant mothers with obesity and their offspring.
Epigenetic alterations, such as DNA methylation, may occur following intrauterine perturbations caused by obesity and excess GWG (13,14). DNA methylation regulates gene expression, X-chromosome inactivation, imprinting, and cell differentiation (13). Intrauterine epigenetic alterations may therefore affect future health outcomes in the offspring (15). For example, early maternal exposures such as gestational diabetes, pregestational obesity, and famine were linked to dysregulated gene function in early life by altered DNA methylation (16–19), and dietary interventions during pregnancy impact the offspring epigenome (20). However, to our knowledge, it remains unknown whether interventions in pregnant women with obesity affect the epigenetic pattern in cord blood and whether this is associated with body composition and growth in their offspring.
Our aim was to investigate whether a lifestyle intervention including physical activity with and without advice on a low-energy Mediterranean-style diet in pregnant women with obesity from the Treatment of Obese Pregnant women (TOP)-study (9) is associated with DNA methylation alterations in offspring cord blood. We then tested whether specific epigenetic marks in cord blood are associated with body composition in the offspring at birth and growth during the first 3 years of life.
Research Design and Methods
Design and Clinical Data of the TOP-Study
The TOP-study was approved by the Ethics Committee for the Capital Region of Denmark (January 2009, H-D-2008–119; Hillerød, Denmark) and registered at ClinicalTrials.gov (NCT01345149). Before enrollment, written informed consent was obtained from all participants.
The TOP-study is a randomized controlled trial of 425 pregnant women with obesity including two lifestyle intervention groups—physical activity assessed with pedometer and dietary advice (PA + D) and physical activity assessed with pedometer (PA)—and a control arm receiving standard of care (C) (Fig. 1). The primary end point was to assess the impact of these lifestyle interventions on GWG (9). All participants, including control subjects, had a consultation with a dietitian who recommended a low-energy and low-fat Mediterranean-style diet of 1,200–1,675 kcal, based on Danish national recommendations, and they were encouraged to limit GWG to ≤5 kg. Participants in PA + D had regular contact/visits (every 2 weeks) with an experienced dietitian giving dietary advice and measuring weight. Participants in both PA + D and PA were encouraged to obtain 11,000 steps daily. We based our physical activity recommendation on our previous study of step counts among pregnant women and added 50% to increase physical activity (21). If this was not achievable, they were asked to set their own goal.
Figure 1.
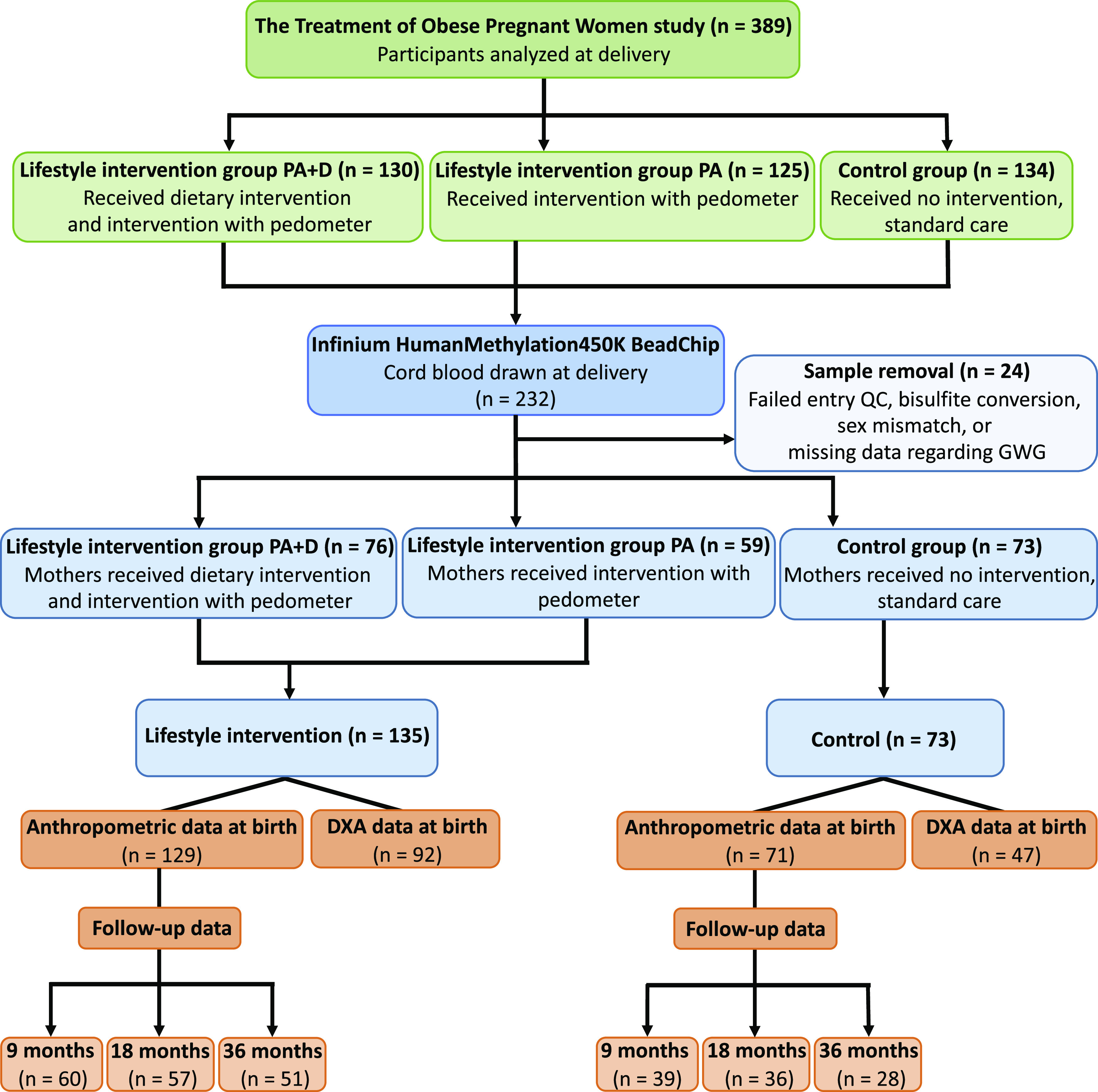
Flow diagram. QC, quality control.
Due to participants having miscarriages, withdrawing from the study, and moving from the region, 389 women completed the study (Fig. 1). Maternal age, prepregnancy BMI, maternal educational level, and previous childbirths (parity) were recorded at enrollment (weeks 11–14). Smoking during pregnancy was acquired through medical records, GWG was determined by subtracting self-reported prepregnancy weight with weight measured during weeks 36 and 37, and energy intake was attained from self-administered validated Food Frequency Questionnaires at weeks 11–14 and 36–38. Detailed information on enrollment, conduction of the trial, and clinical measurements is reported elsewhere (9,12).
DXA scans were performed within 48 h of birth to assess offspring body composition. Offspring measurements of interest for this study were: lean mass at birth and growth, in which BMI z scores at birth and at 9, 18, and 36 months of age were used (Fig. 1). Data collection and body composition assessment in the offspring are further described (10,22).
Cord blood was collected from the umbilical vein of the clamped umbilical cord at birth. Samples were immediately frozen (−80°C) and stored in a biobank at Copenhagen University Hospital Hvidovre. Whole-cord blood samples were available for 232 participants (Fig. 1).
DNA Methylation Analyses
Cord blood DNA was extracted using the QIAamp 96 DNA Blood Kit. DNA concentration and purity were determined using NanoDrop (NanoDrop Technologies, Inc.). Eight samples had DNA concentrations that were too low. Bisulfite conversion was performed using the EZ-96 DNA methylation kit (Zymo Research Corporation, Irvine, CA), according to the manufacturer’s instructions. Six samples failed bisulfite conversion. DNA methylation was measured using Illumina Infinium HumanMethylation450 BeadChips (Illumina, San Diego, CA), covering 485,577 sites (23). Illumina iScan was used to image the Infinium HumanMethylation450K BeadChips. Three samples were removed due to missing data regarding GWG (a covariate in the methylation model). Preprocessing was performed using R (24) (version 3.5.1), lumi (25), and methylumi (26) packages from Bioconductor. β-Values were calculated as β = intensity of the Methylated allele (M)/(intensity of the Unmethylated allele [U] + intensity of the Methylated allele [M] + 100). A total of 1,739 probes with mean detection P value >0.01, 64 rs probes, 3,089 ch-probes targeting non-CpG sites, 80 Y-chromosome probes, 26,772 cross-reactive probes, and 436 polymorphic probes with a minor allele frequency >0.1 were filtered out (27). Methylation data were obtained for 453,397 probes. For further analyses, M-values were used, calculated following the formula M = log2(β/[1−β]) (28). Background correction and quantile normalization were performed. Beta-Mixture Quantile normalization method (29) was applied. ComBat was applied to correct for batch effects (30,31). Principal component analysis (PCA) before and after applying ComBat ensures that between-array batch effects were removed. Seven participants were further excluded, as these samples clustered to the wrong sex in the PCA. Overall, DNA methylation data were available for 208 participants (Fig. 1 and Table 1). Additional gene annotation was performed using hg38, GENCODE version 22.
Table 1.
Parental and offspring baseline characteristics according to the lifestyle intervention and control groups for subjects with available cord blood of the TOP-study
| Lifestyle intervention | Control | P value | |
|---|---|---|---|
| Maternal characteristics | n = 135 | n = 73 | |
| Maternal age at enrollment (years)* | 30.90 (4.30) | 31.40 (4.74) | 0.440 |
| Prepregnancy BMI (kg/m2)* | 34.19 (4.00) | 34.36 (3.98) | 0.763 |
| Maternal educational level, n (%)† | 0.970 | ||
| Grammar school 10 years | 15 (11.1) | 6 (8.2) | |
| Secondary school 12 years | 16 (11.9) | 9 (12.3) | |
| Vocational training school | 13 (9.6) | 6 (8.2) | |
| Further education 1–2 years | 26 (19.3) | 12 (16.4) | |
| Tertiary education 3–4 years (Bachelor level) | 46 (34.1) | 29 (39.7) | |
| Advanced education (postgraduate) | 18 (13.3) | 10 (13.7) | |
| NA | 1 (0.7) | 1 (1.4) | |
| Smoking during pregnancy (yes/no), n (%)† | 10 (7.4) | 3 (4.1) | 0.524 |
| Parity (single/multi), n (%)† | 75 (55.6) | 39 (53.4) | 0.882 |
| Energy intake at enrollment, weeks 11–14 (kJ)‡§ | 8,019 (2,784) | 7,540 (3,246) | 0.829 |
| Paternal characteristics | n = 115 | n = 65 | |
| BMI (kg/m2) at enrollment, weeks 11–14* | 27.39 (4.51) | 27.01 (4.52) | 0.585 |
| Offspring characteristics | n = 135 | n = 73 | |
| Sex, n (%)† | 0.862 | ||
| Male | 69 (51.1) | 39 (53.4) | |
| Female | 66 (48.9) | 34 (46.6) | |
| GA (weeks)* | 40.17 (1.23) | 40.01 (1.31) | 0.393 |
| Weight (g), birth* | 3,724 (482) | 3,677 (513) | 0.515 |
| Weight (kg), 9 months*‖ | 9.61 (1.03) | 9.38 (1.15) | 0.299 |
| Weight (kg), 18 months*¶ | 11.86 (11.83) | 11.26 (10.27) | 0.014 |
| Weight (kg), 36 months*# | 15.30 (18.64) | 14.71 (12.97) | 0.141 |
| Length (cm), birth*** | 52.50 (2.17) | 52.48 (2.24) | 0.958 |
| Length (cm), 9 months*‖ | 73.14 (2.32) | 72.99 (1.97) | 0.740 |
| Length (cm), 18 months*†† | 82.75 (2.87) | 82.55 (2.48) | 0.724 |
| Height (cm), 36 months*‡‡ | 96.42 (4.21) | 95.95 (3.07) | 0.599 |
| Breastfeeding, exclusively (weeks)*§§ | 10.98 (9.41) | 8.38 (10.07) | 0.163 |
| Breastfeeding, partially (weeks)*§§ | 16.30 (11.05) | 14.88 (10.71) | 0.501 |
NA, not available.
Mean (SD), two-sided Student t test.
Frequencies, χ2 test.
Median (interquartile range), two-sided Mann-Whitney U test.
Lifestyle intervention, n = 133; control, n = 68.
Lifestyle intervention, n = 60; control, n = 39.
Lifestyle intervention, n = 58; control, n = 36.
Lifestyle intervention, n = 51; control, n = 29.
Lifestyle intervention, n = 129; control, n = 71.
Lifestyle intervention, n = 57; control, n = 36.
Lifestyle intervention, n = 51; control, n = 28.
Lifestyle intervention, n = 77; control, n = 42.
Statistical Analyses
We performed a power calculation using R package pwrEWAS (https://bioconductor.org/packages/devel/bioc/html/pwrEWAS.html). To find 10% difference in methylation between two groups (ratio 1:2) with 75% power, we needed 230 participants. Based on this power calculation, the fact that GWG, the primary end point of the TOP-study, did not differ between the two lifestyle intervention groups and the modest number of samples with cord blood in each lifestyle intervention group, we decided to combine the two lifestyle intervention groups. We subsequently investigated the impact of PA + D together with PA (lifestyle intervention group, n = 135) versus C (control group, n = 73) on all investigated parameters (Fig. 1).
To test if cord blood DNA methylation is associated with lifestyle intervention assignment, a linear regression model adjusted for maternal age (years), prepregnancy BMI (kilograms per meter squared), GWG (kilograms), gestational age (GA; weeks), and offspring sex was run. Adjustment for cell composition was done using the reference-free method from Houseman et al. (32). We also used the reference-based method to adjust for cell composition (33). We additionally performed linear regression including the same variables as above and calculated principal components (PCs) of the residuals from this model. The top five PCs were then used as covariates to correct for possible inflation, technical variation, and cell type composition.
Linear regression was used to assess whether cord blood DNA methylation (at sites significantly different between lifestyle intervention and control groups) is associated with lean mass at birth. Variables with a P < 0.25 in univariate analyses were incorporated in the final regression models (34), which included the following covariates: maternal smoking during pregnancy, GA, and offspring sex as well as factors with putative biological impact on DNA methylation (GWG and prepregnancy BMI). In this linear regression model, lean mass was the dependent variable and DNA methylation of respective site was the independent variable.
To assess if methylation in cord blood (at sites significantly different between the lifestyle intervention and control groups) is associated with growth in the offspring (at birth and at 9, 18, and 36 months of age), linear mixed models (LMMs) for repeated measurements were performed with random intercepts and different fixed slopes of BMI z scores for lifestyle intervention and control groups. BMI z scores, which are weight relative to height and adjusted for age and sex of the child, were calculated according to the World Health Organization using the anthro package in R (https://CRAN.R-project.org/package=anthro). Variables with P < 0.25 in univariate analyses were incorporated in final models (34). Models were adjusted for maternal education level, maternal smoking during pregnancy, GA, and parity, as well as factors with a likely biological impact on methylation (GWG, prepregnancy BMI, breastfeeding exclusively and partially [weeks], and offspring age at measurement). Association signals for specific methylation sites were considered significant (P < 0.05) if the direction of effect in the LMMs was consistent with at least three of the four time points in regular linear regression models.
Spearman correlations between CRP levels in pregnant mothers and cord blood DNA methylation of significant sites were performed. Grubbs test was used to detect outliers.
Statistical analyses were performed using the software R (24) (version 3.6.1) and RStudio (https://www.rstudio.com). Data are presented as mean ± SD, unless stated otherwise. Normalized methylation β-values were used for Spearman correlations and the analyses related to offspring body composition. Unless stated otherwise, models were corrected for multiple testing using false discovery rate (FDR) analysis (Benjamini-Hochberg) in which FDR <5% (q < 0.05) was considered significant. Bonferroni multiple-comparison post hoc correction was also used when testing if cord blood DNA methylation is associated with the lifestyle intervention, adjusting the linear model for maternal age, prepregnancy BMI, GWG, GA, and offspring sex.
Gene Ontology Analysis
To analyze possible biological functions of differential DNA methylation found in cord blood, we performed gene ontology (GO) mapping using Generic GO Term Mapper (35) and GO analysis using the gometh function in the missMethyl package (36). For GO mapping, we used Process Ontology in Homo sapiens, GOA slim, the list of the annotated genes, and P < 0.01. For GO analysis, we entered a list of the significantly associated methylation sites and removed redundant GO terms using REViGO (37). We allowed for 50% similarity between different GO terms, using Homo sapiens database and SimRel as the semantic similarity measure.
Causal Mediation Analysis
We performed a nonparametric causal mediation analysis, using the mediation R package and default settings (38), to investigate whether DNA methylation of any of the identified 25 sites in cord blood found to be associated with lean mass are part of a pathway through which the lifestyle intervention exerts its effects on lean mass. The effect is estimated for each association between treatment and outcome in participants with different methylation levels. DNA methylation of each respective site was designated as the mediator and lean mass as outcome. The models were adjusted for GWG, maternal BMI, GA, and offspring sex.
DNA Methylation in Muscle and Adipose Tissue
Sites showing differential cord blood methylation from the lifestyle intervention versus control group were also investigated in blood, muscle, and adipose tissue from participants in the Monozygotic Twin cohort. Infinium DNA methylation data from blood, adipose tissue, and muscle of the Monozygotic Twin cohort have been published (39,40). We used Spearman correlations to test whether methylation in blood correlates with methylation in muscle or adipose tissue for sites showing differential DNA methylation in cord blood from the TOP-study.
Data and Resource Availability
DNA methylation data from cord blood of the TOP-study (accession number LUDC2020.08.14) are deposited in the Lund University Diabetes Centre repository (https://www.ludc.lu.se/resources/repository) and are available upon request.
Results
Impact of a Lifestyle Intervention During Pregnancy on DNA Methylation in Cord Blood
To assess if a lifestyle intervention in pregnant mothers with obesity had an effect on the methylome in cord blood, we analyzed DNA methylation in participants of the TOP-study. Baseline characteristics of pregnant women with obesity included in the lifestyle intervention and control groups of the TOP-study, paternal BMI, as well as for their offspring at birth, are shown in Table 1. At enrollment, there was no difference in energy intake between the lifestyle intervention and control group (Table 1). Mothers in the lifestyle intervention group had a trend toward lower energy intake versus control subjects at weeks 36–38 supporting good adherence to the dietary intervention (Supplementary Fig. 1). During week 17 of pregnancy, daily step counts were 8,623 ± 2,615 for participants in the lifestyle intervention. As wearing the pedometer was part of the lifestyle intervention, no step counts were available for control subjects. Offspring were similar regarding weight, length, and GA at birth and there were no detectable differences between the groups regarding breastfeeding (Table 1) or BMI z scores at birth or 9 or 36 months of age (Table 2).
Table 2.
Estimated differences from linear regression models in offspring lean mass and BMI z scores and their associated 95% CIs when comparing lifestyle intervention (n = 92) vs. control (n = 47) groups for subjects with available cord blood of the TOP-study
| Phenotype | Estimated difference (95% CI) | P value |
|---|---|---|
| Lean mass (g), birth | 126.55 (−4.52; 257.62)* | 0.058* |
| Lean mass (%), birth | 1.36 (−0.05; 2.77)* | 0.059* |
| Abd. lean mass (g), birth | 59.09 (10.53; 107.65)* | 0.017* |
| Abd. lean mass (%), birth | 0.88 (0.24; 1.53)* | 0.008* |
| Fat mass (g), birth | 51.26 (−19.93; 122.44)* | 0.157* |
| Fat mass (%), birth | 1.35 (−0.06; 2.76)* | 0.061* |
| Abd. fat mass (g), birth | 6.88 (−3.51; 17.26)* | 0.192* |
| Abd. fat mass (%), birth | 0.49 (−0.58; 1.57)* | 0.365* |
| BMI z score, birth1 | 0.15 (−0.14; 0.43)† | 0.352† |
| BMI z score, 9 months2 | 0.31 (−0.14; 0.76)‡ | 0.315‡ |
| BMI z score, 18 months3 | 0.54 (0.14; 0.93)§ | 0.006§ |
| BMI z score, 36 months4 | 0.30 (−0.13; 0.74)§ | 0.169§ |
Lifestyle intervention, n = 129; control, n = 71.
Lifestyle intervention, n = 60; control, n = 39.
Lifestyle intervention, n = 57;control, n = 36.
Lifestyle intervention, n = 51; control, n = 28.
Adjusted for maternal education level, maternal smoking during pregnancy (yes/no), GWG (in kilograms), prepregnancy BMI, parity (single/multi), GA (in weeks), and offspring sex.
Adjusted for maternal education level, maternal smoking during pregnancy (yes/no), GWG (in kilograms), prepregnancy BMI, parity (single/multi), and GA (in weeks).
Adjusted for maternal education level, maternal smoking during pregnancy (yes/no), GWG (in kilograms), prepregnancy BMI, parity (single/multi), GA (in weeks), breastfeeding partially and exclusively, and BMI z score at birth.
Adjusted for maternal education level, maternal smoking during pregnancy (yes/no), GWG (in kilograms), prepregnancy BMI, parity (single/multi), breastfeeding partially and exclusively, and BMI z score at birth.
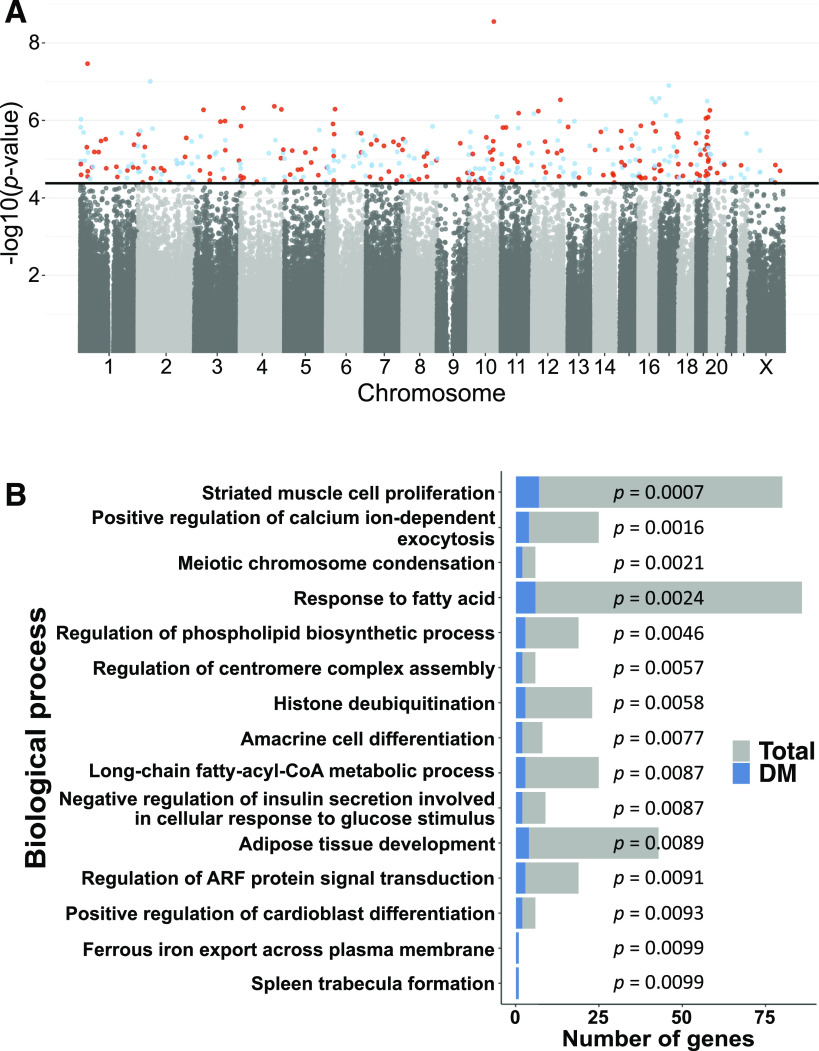
We next examined if cord blood DNA methylation at individual sites differed between lifestyle intervention and control groups. DNA methylation at 379 sites (q < 0.05), as seen in Fig. 2A representing the distribution of methylation sites across the genome, annotated to 370 unique genes, was different between the lifestyle intervention and control groups when adjustment for cell composition was done using the Houseman reference-free method (32) and Benjamini-Hochberg FDR analysis was used to correct for multiple testing (Supplementary Table 1). Three of these sites were significant based on Bonferroni correction. None of these 379 significant sites had been associated with DNA methylation signatures related to cell composition in cord blood, and methylation of 376 of the 379 sites was nominally associated with the lifestyle intervention when adjustment for cell composition was done using the reference-based method with P = 5.12 × 10−7–3 × 10−2 (33) (Supplementary Table 1). Moreover, methylation of 377 out of 379 sites was nominally associated with the lifestyle intervention after adjusting for the first top five PCs of the residuals, P = 3.08 × 10−9–4.7 × 10−2 (Supplementary Table 1). All of these 377 sites had FDR <5% when we performed post hoc Benjamini-Hochberg FDR analysis on 379 sites. The lifestyle intervention was associated with methylation of these sites also when adjusting for fewer covariates and when adjusting for smoking (P < 0.05) (Supplementary Table 1), suggesting that these covariates did not substantially influence the association. Moreover, since GWG has been associated with DNA methylation in cord blood (14), we tested if GWG was associated with methylation of the 379 sites. However, no methylation sites were associated with GWG (q < 0.05). We also performed a model in which we adjusted for maternal age, maternal BMI, smoking, and GA as well as offspring sex, and then methylation of 377 out of 379 sites was associated with the lifestyle intervention, with P = 1.3 × 10−8–2.4 × 10−4 (Supplementary Table 1).
Figure 2.
A: A Manhattan plot, representing the distribution of methylation sites across the genome, for the association between lifestyle intervention and offspring cord blood DNA methylation, after adjustment for covariates and cell composition adjustment. The black line shows the FDR threshold for multiple testing. Methylation sites that surpassed the FDR threshold (P < 4.17 × 10−5) are highlighted in color (red is hypermethylated and blue is hypomethylated sites in the lifestyle intervention group vs. the control group). B: Pathways from GO analysis after removal of redundant GO terms using REViGO (P < 0.05). The gray bars indicate the total number of genes in the pathway; the blue bars indicate the number of differently methylated (DM) genes in lifestyle intervention vs. control subjects. Data from A are also presented in Supplementary Table 1, and data from B are also presented in Supplementary Table 3.
To understand the biological role of the 370 genes, we used GO Term Mapper and found that ∼60% of the genes with differently methylated sites are involved in metabolic processes (Supplementary Table 2). Moreover, performing GO and REViGO analyses, we found 15 biological processes (P < 0.01). These include response to fatty acids, adipose tissue development, and negative regulation of insulin secretion involved in cellular response to glucose stimulus (35,37) (Fig. 2B and Supplementary Table 3).
Using the methylation quantitative trait loci (mQTL) database (https://mqtldb.org), we found that cord blood methylation of 110 out of our 379 sites has been associated with single nucleotide polymorphisms (SNPs), so-called mQTLs (Supplementary Table 4). These include cg21753618, which is among the three sites significant based on Bonferroni correction. Several of these sites also appear as mQTLs in peripheral blood in children and their mothers during childhood, adolescence, pregnancy, and middle age (Supplementary Tables 5 and 6). Among these mQTLs, 18 SNPs were associated with disease traits in the genome-wide association studies (GWAS) catalog (Supplementary Table 4). Moreover, we found that methylation at 56 of the 110 mQTLs are associated with type 2 diabetes, obesity, maternal stress, and sperm viability in previous epigenome-wide association studies (https://bigd.big.ac.cn/ewas/datahub/index) (Supplementary Table 7).
We then tested if SNPs that map to any of the 370 genes included in Supplementary Table 1 have been associated with birth weight, childhood obesity, obesity, adiposity, or type 2 diabetes in published GWAS (41). Sixteen genes annotated to 15 sites (Supplementary Table 8) were linked to SNPs associated with these traits in GWAS: 3 SNPs were associated with adiposity (MAP2K5, MEIS1, and IPO9) (42,43) (downloaded 19 December 2019), 4 with obesity (MAP2K5, PCDH9, SCNN1A, and TCF4) (EFO_0001073, downloaded 19 December 2019), 4 genes (ACSL1, HMGA2, RPSAP52, and SLC9B2) have SNPs associated with type 2 diabetes (EFO_0001360, downloaded 19 December 2019), and 7 genes (TENM4, HMGA2, MAP3K10, RB1, KLHL29, LRIG1, and PMFBP1) have SNPs associated with birth weight (EFO_0004344, downloaded 23 June 2020) (41). None of the discovered genes have SNPs associated with childhood obesity (41,44) (downloaded 22 June 2020). None of these 15 sites were among the 3 significant sites based on Bonferroni correction.
We further examined if our 379 sites were overrepresented within other epigenetic marks such as histone modifications representing active (H3K4me1 and H3K27ac) or inactive (H3K27me3) chromatin. We intersected the position of 379 sites with chromatin immunoprecipitation sequencing data of histone modifications in blood mononuclear cells from the Roadmap Epigenomics Consortium (45). A permutation distribution test using 10,000 permutations showed more significant methylation sites overlapping with H3K4me1 (P = 0.030) but not H3K27me3 (P = 0.356) and H3K27ac (P = 0.535), compared with what would have been expected by chance if all sites on the array were analyzed, indicating an enrichment of enhancer elements in sites differentially methylated in the lifestyle intervention.
We have previously shown that CRP levels were lower in pregnant mothers in lifestyle intervention versus control subjects (46). Therefore, we tested whether CRP levels in pregnant mothers correlated with cord blood methylation of our 379 sites. CRP levels correlated with methylation of two sites: cg17389519, which is annotated to PTF1A encoding a transcription factor involved in pancreas and neural tissue development (47), and cg27394563, annotated to SART3 (Supplementary Fig. 2A and B).
Impact of a Lifestyle Intervention During Pregnancy on Offspring Lean Mass
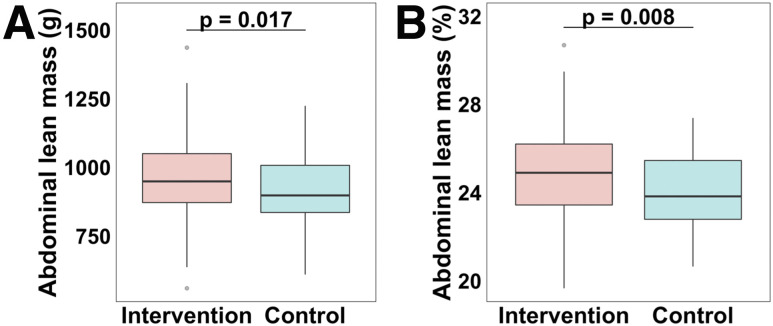
We proceeded to study the body composition of the offspring (Tables 1 and 2). Offspring to mothers included in the lifestyle intervention group were born with 59 g (95% CI 11; 108) (P = 0.017) and 0.88 percentage points (95% CI 0.24; 1.53) (P = 0.008) more abdominal lean mass versus control subjects (Fig. 3 and Table 2). We also observed a trend that offspring of mothers included in the lifestyle intervention group were born with 127 g (95% CI −5; 258) (P = 0.058) and 1.36 percentage points (95% CI −0.05; 2.77) (P = 0.059) more lean mass versus control subjects (Table 2). At birth, the offspring were similar in size (Tables 1 and 2), indicating that it is the body composition that differs between the groups. We found that offspring of both groups were similar in size at 9 and 36 months of age; however, at 18 months, children from the lifestyle intervention group were larger in size (Tables 1 and 2).
Figure 3.
Boxplots are showing abdominal lean mass (g) (A) and abdominal lean mass (%) (B) in the lifestyle intervention and control groups at birth in median (interquartile range). The P values are based on linear regression models adjusted for maternal education level, maternal smoking during pregnancy (yes/no), GWG (kg), prepregnancy BMI (kg/m2), parity (single/multi), GA (weeks), and offspring sex. Data are also presented in Table 2.
Associations Between DNA Methylation in Cord Blood and Offspring Lean Mass and Growth
Offspring in the lifestyle intervention group had more lean mass at birth and differential cord blood methylation at 379 sites versus control subjects (Table 2 and Supplementary Table 1); thus, we further tested whether there were associations between methylation of these 379 sites and lean mass (percentage) at birth in the offspring. Cord blood methylation of 25 sites was associated with lean mass (q < 0.05) (Supplementary Table 9). For the majority of these sites (80%), cord blood methylation levels were higher in the lifestyle intervention group and positively associated with greater lean mass.
We proceeded to assess whether cord blood methylation of the 379 sites was associated with growth over time in the offspring using LMMs and BMI z scores at birth and 9, 18, and 36 months of age. We found that methylation of 22 sites was associated with BMI z scores (P < 0.05). Next, we performed linear regression models to test if methylation at the 379 sites was associated with BMI z scores at each time point. The direction of effects for the LMMs was consistent with that for the linear regression models at all time points but for two sites; for these two sites, the direction altered at one time point (Supplementary Table 10). Included within the genes annotated to the 22 sites are ACSL1, which is involved in fatty acid β-oxidation and harboring a SNP associated with type 2 diabetes, and TCF4, encoding a transcription factor involved in Wnt signaling and harboring a SNP associated with obesity (EFO_0001073, downloaded 19 December 2019) in GWAS (41) (Supplementary Table 8).
Causal Mediation Analysis
We next used a causal mediation analysis (38) to investigate whether DNA methylation of any of the 25 sites in cord blood found to be associated with lean mass are part of a pathway through which the lifestyle intervention exerts its effects on offspring lean mass. The mediation analysis breaks down the total effect of treatment (lifestyle intervention) on outcome (lean mass) into two parts: first, the indirect effect acting via the mediator of interest (DNA methylation) and second, the direct effect acting directly or via a mediator other than what is under study. The analyses showed that 1) the lifestyle intervention has an overall effect of β = 1.35 (95% CI −0.092; 2.741) on lean mass, 2) that effect may operate via an indirect path (indirect effect), possibly through methylation, with a significant average causal mediator effect for 17 methylation sites (q < 0.05) (Table 3), and 3) consequently, the total effect of the lifestyle intervention on lean mass, 32.0–61.8%, is suggested to act via these 17 methylation sites. According to these results, we may call methylation of these sites partial mediators.
Table 3.
Causal mediation analysis on the significant associations between the lifestyle intervention and lean mass–related methylation (CpG) sites as mediators and lean mass (%) as outcome (ACME q value <0.05)
| CpG site | Gene | ACME estimate of mediator CpG (95% CI) | ACME q value | ADE estimate (95% CI) | Total effect (95% CI) | Proportion mediated by CpG (95% CI) |
|---|---|---|---|---|---|---|
| cg07405330 | MOBP | 0.84 (0.35; 1.39) | <0.001 | 0.52 (−0.84; 1.89) | 1.35 (−0.09; 2.74) | 0.62 (−0.73; 3.58) |
| cg06480224 | KIAA2012 | 0.78 (0.19; 1.51) | 0.013 | 0.57 (−0.97; 1.98) | 1.35 (−0.09; 2.74) | 0.58 (−1.11; 4.27) |
| cg11612786 | AC079135.1;GBX2 | 0.64 (0.13; 1.38) | 0.013 | 0.71 (−0.66; 2.03) | 1.35 (−0.09; 2.74) | 0.47 (−0.58; 2.78) |
| cg20982052 | 0.53 (0.14; 1.11) | 0.013 | 0.82 (−0.68; 2.18) | 1.35 (−0.09; 2.74) | 0.39 (−0.62; 2.63) | |
| cg00154557 | DSE | 0.50 (0.08; 1.08) | 0.021 | 0.86 (−0.64; 2.26) | 1.35 (−0.09; 2.74) | 0.37 (−0.58; 2.42) |
| cg13002044 | TMEM178B | 0.70 (0.14; 1.42) | 0.021 | 0.65 (−0.73; 1.98) | 1.35 (−0.09; 2.74) | 0.52 (−0.85; 2.87) |
| cg18088415 | LSM2 | 0.70 (0.19; 1.41) | 0.021 | 0.65 (−0.77; 2.07) | 1.35 (−0.09; 2.74) | 0.52 (−0.73; 3.42) |
| cg11594420 | TEX101 | 0.53 (0.10; 1.18) | 0.025 | 0.82 (−0.59; 2.15) | 1.35 (−0.09; 2.74) | 0.39 (−0.45; 2.84) |
| cg04678315 | 0.48 (0.08; 1.02) | 0.028 | 0.87 (−0.56; 2.20) | 1.35 (−0.09; 2.74) | 0.36 (−0.64; 2.05) | |
| cg08144675 | 0.70 (0.10; 1.42) | 0.032 | 0.66 (−0.85; 2.20) | 1.35 (−0.09; 2.74) | 0.52 (−0.98; 3.35) | |
| cg22454673 | HERC2 | 0.63 (0.16; 1.29) | 0.032 | 0.73 (−0.71; 2.18) | 1.35 (−0.09; 2.74) | 0.46 (−0.77; 3.10) |
| cg04058675 | HUWE1 | 0.45 (0.08; 1.03) | 0.033 | 0.90 (−0.46; 2.22) | 1.35 (−0.09; 2.74) | 0.34 (−0.48; 2.09) |
| cg03190725 | RP3–468B3.2 | 0.54 (0.10; 1.10) | 0.035 | 0.81 (−0.67; 2.19) | 1.35 (−0.09; 2.74) | 0.40 (−0.65; 2.58) |
| cg06799721 | TARS | 0.52 (0.05; 1.27) | 0.036 | 0.84 (−0.54; 2.07) | 1.35 (−0.09; 2.74) | 0.38 (−0.30; 2.00) |
| cg15157974 | DISC1 | 0.43 (0.04; 0.96) | 0.040 | 0.92 (−0.50; 2.21) | 1.35 (−0.09; 2.74) | 0.32 (−0.24; 2.10) |
| cg26142132 | AAT | 0.45 (0.06; 0.96) | 0.041 | 0.90 (−0.57; 2.40) | 1.35 (−0.09; 2.74) | 0.34 (−0.44; 2.26) |
| cg00354884 | ABR | 0.53 (0.03; 1.18) | 0.047 | 0.83 (−0.62; 2.17) | 1.35 (−0.09; 2.74) | 0.39 (−0.45; 2.32) |
Models adjusted for GWG (in kilograms), maternal BMI, GA (in weeks), and offspring sex. Based on 139 participants, lifestyle intervention, n = 92, and control, n = 47.
ACME, average causal mediator effect; ADE, average direct effect.
Cross-Tissue Methylation of Sites Associated With Lean Mass or Growth
We finally examined whether DNA methylation in blood of the 46 unique sites (one methylation site, cg11594420, overlap) associated with lean mass or growth in the offspring mirrors the methylation pattern in two other tissues of importance for obesity and type 2 diabetes: skeletal muscle and adipose tissue. We used available methylation array data from blood, muscle, and adipose tissue taken from the same individuals (39,40) (Supplementary Table 11). Among these sites, the methylation pattern in blood correlated positively with methylation of four sites in adipose tissue and two sites in muscle (P < 0.05) (Supplementary Table 12). Five correlations were nominal and one significant after FDR. These findings suggest that methylation of a few sites may have a biological role in tissues of relevance for obesity and type 2 diabetes.
Discussion
This is, to our knowledge, the first genome-wide epigenetic analysis in cord blood of pregnant women with obesity randomized to a lifestyle intervention including physical activity, with or without a hypocaloric Mediterranean-style diet, versus control subjects receiving standard of care. There are four key findings: first, DNA methylation at individual sites in cord blood differed between lifestyle intervention and control subjects. These sites were annotated to genes overrepresented in relevant GO terms (e.g., response to fatty acids and adipose tissue development). Second, we found that genes linked to SNPs associated with birth weight, obesity, adiposity, and type 2 diabetes by GWAS have been also annotated to sites that have altered DNA methylation in our study. Additionally, SNPs previously associated with DNA methylation in cord blood of our identified sites were linked to disease traits in the GWAS catalog. Third, offspring to mothers included in the lifestyle intervention were born with more lean mass. Finally, methylation at 17 sites partially mediates the effect of the lifestyle intervention on lean mass in the offspring. Together, these data provide evidence that the presented lifestyle intervention altered the epigenome of genes linked to metabolism and metabolic disease in offspring cord blood from pregnant mothers with obesity.
Previous studies have shown that tissues from people with obesity have different methylation profiles versus lean people (13,16,48); however, DNA methylation can be changed by lifestyle (40,49). Obesity during pregnancy increases the risk of metabolic disease in offspring (2), and obesity in pregnant mothers is associated with epigenetic alterations in cord blood (17). We demonstrate that exercise and healthy diets during pregnancy can change cord blood DNA methylation and that these epigenetic changes took place on genes involved in metabolic processes. It is possible that a healthier lifestyle during pregnancy and the consequential epigenetic changes help enhance the offspring’s health later in life. The epigenetic mechanisms linked to exercise and healthy diets in our study seem to be different compared with those previously associated with maternal BMI (17) and gestational diabetes (16,19).
We demonstrate that offspring to mothers in the lifestyle intervention have higher abdominal and a trend toward higher total lean mass versus control subjects. This result indicates a positive effect of the lifestyle intervention on the offspring as they were born with more metabolically active tissue, which might protect against future metabolic diseases. This is supported by studies showing that the body composition of newborns being born small for GA (SGA) differs more in terms of less lean mass than differences in fat mass, compared with appropriate-for-gestational-age newborns (50), and SGA increases the risk of metabolic disease later in life (51). Thereby, suggesting that negative effects of being born SGA could be due to decreased lean mass. Increased muscle mass and higher metabolic activity on the other hand may have beneficial effects on insulin sensitivity and protect from obesity and type 2 diabetes (7). We found associations between cord blood epigenetics and lean mass in the offspring at birth. For the majority of these sites, methylation levels were higher in cord blood of the lifestyle intervention group and positively associated with greater lean mass, and methylation at several sites seems to partially mediate the lifestyle effect on lean mass. Interestingly, the lifestyle intervention group had decreased methylation of SETD3, which encodes a methyltransferase. Hypomethylation of SETD3 correlates with increased expression and, in turn, increased muscle mass (52), which might in part explain the greater lean mass we see in the offspring of the lifestyle intervention group.
Finally, we found that blood-based methylation of sites associated with lean mass or growth in the offspring mirrors methylation patterns in muscle and adipose tissue, tissues of importance for metabolic disease (39,40). These include sites annotated to TCF4 and SYT9 encoding transcription factor 4 and synaptotagmin 9, respectively, which have been implicated in diabetes-related traits (53,54). Although these results are based on nominally significant P values, they give a possible indication that methylation of some identified sites may also have a biological role in tissues of importance for obesity and type 2 diabetes.
Strengths and Limitations
A strength of this study is the randomized design, the high rate of completers in the TOP-study, and the relatively homogenous study population in terms of prepregnancy BMI and ethnicity, which should reduce the risk of bias. A pedometer intervention is an inexpensive method for increasing daily physical activity and can easily be implemented into daily life. Other physical activity interventions often include attendance to classes, which can be difficult to implement. It is also a strength that we used several different methods to adjust the methylation data for cell composition and technical variation, which showed that DNA methylation of a large proportion of the identified sites was associated with the lifestyle intervention independent of the method used, although the strength of the associations was reduced for the reference-based compared with the reference-free method. It is important to adjust cord blood DNA methylation data for cell type composition. For discovery of our significant DNA methylation results, we used the reference-free method developed by Houseman et al. (32) for deconvolving heterogeneous cell mixtures. We then used the reference-based (33) and PCA-based methods to validate our results. These different methods (e.g., reference-based vs. reference-free) have their pros and cons. A reference-based method might provide robust estimations. However, it is usually based on few samples with limited clinical conditions. For umbilical cord blood, there is an available reference of 26 samples (33), which might not be able cover the variance in our data set, as it is eight times larger. In the publication by Houseman et al. (32), they show via a simulation study and several real data analyses that their method can perform as well as or better than methods that make explicit use of reference data sets. They also discuss that this reference-free method may adjust for detailed cell type differences that may be unavailable in existing reference data sets. Additionally, the algorithm estimates the number of cell types, meaning it should also consider the nucleated red blood cells that cord blood contains.
Furthermore, in this study, we used two methods to correct for multiple testing, Benjamini-Hochberg and Bonferroni. However, Bonferroni is known to be too conservative for epigenome-wide association studies since DNA methylation values at nearby probes are known to be correlated and many sites on the array are nonvariable (55,56). The alternative approach, Benjamini-Hochberg adjustment, is potentially a more powerful method identifying the associated sites with the phenotype of interest, but it may also generate some false-positive results. The PCA-based method that we used may, however, reduce false-positive results.
A limitation is that the two lifestyle intervention groups were merged. This was, however, necessary to have sufficient statistical power due to the modest number of samples with cord blood in each group. However, we have shown that lifestyle interventions in the TOP-study were effective and could reduce GWG (9), suggesting that both interventions successfully achieved the main primary end point, thus reducing potential bias when merging them. It should also be noted that the maternal energy intake is self-reported and may infer a type of reporting bias. Nevertheless, the maternal energy intake at late gestation was reduced in the lifestyle intervention group versus control subjects, indicating a successful intervention. Previous dietary analyses of participants in the TOP-study showed that participants having dietary and physical activity intervention changed their dietary composition in a healthier direction (12). Participants in the group only performing physical activity demonstrated a trend toward dietary changes in the same direction (12). This supports the approach of merging the intervention groups.
As in other intervention studies of pregnant women with obesity, it might be a challenge that the intervention intensity was too low, and a high proportion of the participants were noncompliant to the recommended diet and physical activity intervention. Few women achieved the target of the physical activity intervention, possibly indicating that this target was set high for this group and should be revised for future studies.
Of note, the methylation array covers ∼2% of sites in the human genome, and it is therefore possible that methylation of additional sites may be associated with the intervention. Future studies are needed to fully understand the biology behind the associations presented in this article and their possible health effects.
In summary, this study demonstrates that a lifestyle intervention in pregnant women with obesity is associated with the cord blood epigenome in offspring. We also provide evidence that epigenetic markers in cord blood associate with lean mass and growth in offspring. These results underline that the intrauterine environment in humans might have the ability to program the epigenome, which in turn may affect metabolism and growth later in life.
Article Information
Acknowledgments. The authors thank Marlena Maziarz, Department of Clinical Sciences, Lund University/CRC, Skåne University Hospital SUS, Malmö, Sweden, for guidance and expertise in biostatistics and Silja Schrader, Epigenetics and Diabetes Unit, Department of Clinical Sciences, Lund University Diabetes Centre, Lund University, Scania University Hospital, Malmö, Sweden, for help with power calculations. The authors also thank the Danish Diabetes Academy via the Novo Nordisk Foundation for support throughout this project and the National Bioinformatics Infrastructure Sweden, SciLifeLab (Uppsala, Sweden), for the bioinformatics long-term support with Nikolay Oskolkov.
Funding. Sygekassernes Helsefond, Hartmann Fonden, Hvidovre Hospital, and The Danish Council for Strategic Research supported the TOP-study. The work performed by A.C.E. and P.W.F. was supported by grants from the European Foundation for the Study of Diabetes, Vetenskapsrådet, Hjärt-Lungfonden, the H2020 European Research Council (CoG-2015_681742_NASCENT), and Novo Nordisk Fonden. The work performed by J.J., S.G.-C., A.P., and C.L. was supported by grants from the Novo Nordisk Fonden, Vetenskapsrådet, and Region Skåne (ALF), an H2020 European Research Council co-grant (PAINTBOX, number 725840), H2020 Marie Skłodowska-Curie Actions grant agreement 706081 (EpiHope), Hjärt-Lungfonden, EXODIAB, Stiftelsen för Strategisk Forskning (IRC15-0067), and Diabetesförbundet. All researchers from Lund University Diabetes Centre were supported by a research center grant from the Swedish Strategic Science Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.J. analyzed data, performed the statistical analyses, and drafted and revised the manuscript. K.M.R., A.V., P.W.F., and C.L. designed and planned the current study and participated in drafting the manuscript. K.M.R. and K.N. designed and planned the TOP-study. K.M.R. and E.M.C. conducted the TOP-study and collected data. S.G.-C. participated in analyzing data and drafting the manuscript. A.P. performed DNA methylation analysis. A.C.E. contributed to designing the study and performed statistical analyses. M.V.L. interpreted data and gave statistical support. M.V.L., K.F.M., and E.M.C. collected data on the offspring at follow-up after delivery. L.H. contributed to planning the epigenetic part of the study. E.M.C. analyzed the DXA scans. P.W.F. and C.L. supervised the analyses. All authors reviewed and provided critical comments on the manuscript. J.J. and K.M.R. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
J.J. and K.M.R. are shared first authors.
P.W.F. and C.L. are shared last authors.
This article contains supplementary material online at https://doi.org/10.2337/figshare.13501587.
References
- 1.Rasmussen KM, Yaktine AL (Eds). Weight Gain During Pregnancy: Reexamining the Guidelines. Washington (DC), National Academies Press, 2009 [PubMed] [Google Scholar]
- 2.Gu S, An X, Fang L, et al. Risk factors and long-term health consequences of macrosomia: a prospective study in Jiangsu Province, China. J Biomed Res 2012;26:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds M, Llewellyn A, Owen CG, Woolacott N. Predicting adult obesity from childhood obesity: a systematic review and meta-analysis. Obes Rev 2016;17:95–107 [DOI] [PubMed] [Google Scholar]
- 4.Ervin RB. Prevalence of metabolic syndrome among adults 20 years of age and over, by sex, age, race and ethnicity, and body mass index: United States, 2003-2006. Natl Health Stat Rep 2009;13:1–7 [PubMed] [Google Scholar]
- 5.Franks PW, Hanson RL, Knowler WC, Sievers ML, Bennett PH, Looker HC. Childhood obesity, other cardiovascular risk factors, and premature death. N Engl J Med 2010;362:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Global Burden of Disease Collaborative Network . Global Burden of Disease Study 2015 (GBD 2015) Obesity and Overweight Prevalence 1980-2015, 2017. Institute for Health Metrics and Evaluation. Accessed 11 March 2020. Available from http://ghdx.healthdata.org/record/ihme-data/gbd-2015-obesity-and-overweight-prevalence-1980-2015
- 7.Kim G, Lee SE, Jun JE, et al. Increase in relative skeletal muscle mass over time and its inverse association with metabolic syndrome development: a 7-year retrospective cohort study. Cardiovasc Diabetol 2018;17:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bann D, Wills A, Cooper R, et al.; NSHD Scientific and Data Collection Team . Birth weight and growth from infancy to late adolescence in relation to fat and lean mass in early old age: findings from the MRC National Survey of Health and Development. Int J Obes 2014;38:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renault KM, Nørgaard K, Nilas L, et al. The Treatment of Obese Pregnant Women (TOP) study: a randomized controlled trial of the effect of physical activity intervention assessed by pedometer with or without dietary intervention in obese pregnant women. Am J Obstet Gynecol 2014;210:134.e1–134.e9 [DOI] [PubMed] [Google Scholar]
- 10.Carlsen EM, Renault KM, Nørgaard K, et al. Newborn regional body composition is influenced by maternal obesity, gestational weight gain and the birthweight standard score. Acta Paediatr 2014;103:939–945 [DOI] [PubMed] [Google Scholar]
- 11.Renault KM, Carlsen EM, Nørgaard K, et al. Intake of carbohydrates during pregnancy in obese women is associated with fat mass in the newborn offspring. Am J Clin Nutr 2015;102:1475–1481 [DOI] [PubMed] [Google Scholar]
- 12.Renault KM, Carlsen EM, Nørgaard K, et al. Intake of sweets, snacks and soft drinks predicts weight gain in obese pregnant women: detailed analysis of the results of a randomised controlled trial. PLoS One 2015;10:e0133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab 2019;29:1028–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morales E, Groom A, Lawlor DA, Relton CL. DNA methylation signatures in cord blood associated with maternal gestational weight gain: results from the ALSPAC cohort. BMC Res Notes 2014;7:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jørgensen SW, Brøns C, Bluck L, et al. Metabolic response to 36 hours of fasting in young men born small vs appropriate for gestational age. Diabetologia 2015;58:178–187 [DOI] [PubMed] [Google Scholar]
- 16.Hjort L, Martino D, Grunnet LG, et al. Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight 2018;3:e122572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp GC, Salas LA, Monnereau C, et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum Mol Genet 2017;26:4067–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 2008;105:17046–17049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe CG, Cox B, Fore R, et al. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the pregnancy and childhood epigenetics consortium. Diabetes Care 2020;43:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geraghty AA, Sexton-Oates A, O’Brien EC, et al. A low glycaemic index diet in pregnancy induces DNA methylation variation in blood of newborns: results from the ROLO randomised controlled trial. Nutrients 2018;10:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renault K, Nørgaard K, Andreasen KR, Secher NJ, Nilas L. Physical activity during pregnancy in obese and normal-weight women as assessed by pedometer. Acta Obstet Gynecol Scand 2010;89:956–961 [DOI] [PubMed] [Google Scholar]
- 22.Ejlerskov KT, Christensen LB, Ritz C, Jensen SM, Mølgaard C, Michaelsen KF. The impact of early growth patterns and infant feeding on body composition at 3 years of age. Br J Nutr 2015;114:316–327 [DOI] [PubMed] [Google Scholar]
- 23.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288–295 [DOI] [PubMed] [Google Scholar]
- 24.R Core Team . A language and environment for statistical computing version 3.6.1. Vienna, Austria, R Foundation for Statistical Computing, 2019 [Google Scholar]
- 25.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548 [DOI] [PubMed] [Google Scholar]
- 26.Davis S, Du P, Bilke S, Triche T Jr., Bootwalla M. methylumi: Handle Illumina methylation data. R package version 2.28.0, Bioconductor, 2018
- 27.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data 2016;9:22–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 2013;29:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127 [DOI] [PubMed] [Google Scholar]
- 31.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics 2014;30:1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gervin K, Salas LA, Bakulski KM, et al. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenetics 2019;11:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bursac Z, Gauss CH, Williams DK, Hosmer DW. Purposeful selection of variables in logistic regression. Source Code Biol Med 2008;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyle EI, Weng S, Gollub J, et al. GO:TermFinder: open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004;20:3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 2016;32:286–288 [DOI] [PubMed] [Google Scholar]
- 37.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. mediation: R package for causal mediation analysis. J Stat Softw 2014;59:38 [Google Scholar]
- 39.Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 2014;63:2962–2976 [DOI] [PubMed] [Google Scholar]
- 40.Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 2012;61:3322–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005–D1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graff M, Scott RA, Justice AE, et al.; CHARGE Consortium; EPIC-InterAct Consortium; PAGE Consortium . Genome-wide physical activity interactions in adiposity: a meta-analysis of 200,452 adults. PLoS Genet 2017;13:e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson T, Rask-Andersen M, Pan G, et al. Contribution of genetics to visceral adiposity and its relation to cardiovascular and metabolic disease. Nat Med 2019;25:1390–1395 [DOI] [PubMed] [Google Scholar]
- 44.Bradfield JP, Vogelezang S, Felix JF, et al.; Early Growth Genetics Consortium . A trans-ancestral meta-analysis of genome-wide association studies reveals loci associated with childhood obesity. Hum Mol Genet 2019;28:3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kundaje A, Meuleman W, Ernst J, et al.; Roadmap Epigenomics Consortium . Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renault KM, Carlsen EM, Hædersdal S, et al. Impact of lifestyle intervention for obese women during pregnancy on maternal metabolic and inflammatory markers. Int J Obes 2017;41:598–605 [DOI] [PubMed] [Google Scholar]
- 47.Jin K, Xiang M. Transcription factor Ptf1a in development, diseases and reprogramming. Cell Mol Life Sci 2019;76:921–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davegårdh C, Broholm C, Perfilyev A, et al. Abnormal epigenetic changes during differentiation of human skeletal muscle stem cells from obese subjects. BMC Med 2017;15:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perfilyev A, Dahlman I, Gillberg L, et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr 2017;105:991–1000 [DOI] [PubMed] [Google Scholar]
- 50.Hediger ML, Overpeck MD, Kuczmarski RJ, McGlynn A, Maurer KR, Davis WW. Muscularity and fatness of infants and young children born small- or large-for-gestational-age. Pediatrics 1998;102:E60. [DOI] [PubMed] [Google Scholar]
- 51.Hattersley AT, Tooke JE. The fetal insulin hypothesis: an alternative explanation of the association of low birthweight with diabetes and vascular disease. Lancet 1999;353:1789–1792 [DOI] [PubMed] [Google Scholar]
- 52.Seaborne RA, Strauss J, Cocks M, et al. Human skeletal muscle possesses an epigenetic memory of hypertrophy. Sci Rep 2018;8:1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei H, Qu H, Wang H, et al. 1,25-Dihydroxyvitamin-D3 prevents the development of diabetic cardiomyopathy in type 1 diabetic rats by enhancing autophagy via inhibiting the β-catenin/TCF4/GSK-3β/mTOR pathway. J Steroid Biochem Mol Biol 2017;168:71–90 [DOI] [PubMed] [Google Scholar]
- 54.Shim YJ, Kim JE, Hwang SK, et al. Identification of candidate gene variants in Korean MODY families by whole-exome sequencing. Horm Res Paediatr 2015;83:242–251 [DOI] [PubMed] [Google Scholar]
- 55.Mansell G, Gorrie-Stone TJ, Bao Y, et al. Guidance for DNA methylation studies: statistical insights from the Illumina EPIC array. BMC Genomics 2019;20:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saffari A, Silver MJ, Zavattari P, et al. Estimation of a significance threshold for epigenome-wide association studies. Genet Epidemiol 2018;42:20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]