Abstract
Despite the advent of immunotherapy, metastatic melanoma represents an aggressive tumor type with a poor survival outcome. The successful application of immunotherapy requires in-depth understanding of the biological basis and immunosuppressive mechanisms within the tumor microenvironment. In this study, we conducted spatially explicit analyses of the stromal-immune interface across 400 melanoma hematoxylin and eosin (H&E) specimens from The Cancer Genome Atlas. A computational pathology pipeline (CRImage) was used to classify cells in the H&E specimen into stromal, immune, or cancer cells. The estimated proportions of these cell types were validated by independent measures of tumor purity, pathologists’ estimate of lymphocyte density, imputed immune cell subtypes, and pathway analyses. Spatial interactions between these cell types were computed using a graph-based algorithm (topological tumor graphs, TTG). This approach identified two stromal features, namely stromal clustering and stromal barrier, which represented the melanoma stromal microenvironment. Tumors with increased stromal clustering and barrier were associated with reduced intratumoral lymphocyte distribution and poor overall survival independent of existing prognostic factors. To explore the genomic basis of these TTG-derived stromal phenotypes, we used a deep learning approach integrating genomic (copy number) and transcriptomic data, thereby inferring a compressed representation of copy number-driven alterations in gene expression. This integrative analysis revealed that tumors with high stromal clustering and barrier had reduced expression of pathways involved in naïve CD4 signaling, MAPK, and PI3K signaling. Taken together, our findings support the immunosuppressive role of stromal cells and T-cell exclusion within the vicinity of melanoma cells.
Introduction
Cutaneous melanoma remains one of the most aggressive cancer types with global increase in incidence and mortality, despite the advent of immunotherapy (1, 2). Late-stage diagnosis is associated with poor survival related to metastatic spread of disease to distant anatomical sites (3–5). Despite durable responses in patients treated with systemic anti-CTLA-4 and anti-PD-1 immunotherapy in the past decade, the fact remains that a significant proportion of patients do not respond to treatment (6). To this effect, diverse intrinsic mechanisms of tumor immune evasion have been described, including loss of tumor antigenicity (7), upregulation of coinhibitory immune checkpoint molecules, and recruitment of regulatory T cells. On the other hand, immunosuppression is modulated by the tumor microenvironment, which is composed of cell types including fibroblasts, endothelial cells, immune cells, and tissue-resident interstitial cells. Fibroblasts can act as immunosuppressive agents through paracrine signaling, modulating the cytotoxic activity of natural killer cells (8), and inhibition of the antitumoral T-cell response (9). For example, remodeling of the extracellular matrix (ECM) by fibroblasts has been shown to result in increased ECM rigidity because of thickening and linearization of collagen fibers (10, 11). This modified ECM could restrict access of immune cells to cancer cells, serving as a physical barrier (12, 13). Understanding the mechanisms by which this stroma network is generated has the potential to aid the development of new therapeutics. However, to date there is no established method to provide systems characterization of the stroma-mediated immunosuppression.
We propose a new method to study the spatial structure of the tumor microenvironment, referred to as topological tumor graphs (TTG), which establishes spatial interactions among cell types as phenotypes of tumor–host interactions. We focus our analyses on specific cell types broadly defined by nuclear morphology, namely tumor cells (large, round nuclei), lymphocytes (smaller, darker nuclei), and stromal cells (spindle-shaped cells likely to be fibroblasts, with possible inclusion of endothelial cells).
On the basis of TTGs inferred directly from pathologic slide images, algorithms that are typically used for social network analysis can be directly applied to the investigation of architectural complexity of the tumor microenvironment. Building on the idea that the TTGs transform seemingly chaotic cell distribution data in pathologic slides into highly structured microenvironmental phenotypes, integration with high-dimensional genomic data may inform cancer adaptive strategies that enable it to overcome external constraints. In this work, we (i) developed a new graph-based method to characterize two distinct spatial features of the stromal microenvironment, stromal cell clustering and stromal barrier between cancer cells and lymphocytes; (ii) developed a new unsupervised deep learning approach for simultaneous dimension reduction and integration of copy-number alterations (CNA) and gene expression data, which further enables genotype–phenotype integration; and (iii) investigated the clinical implications and biological processes underpinning stromal recruitment and their effects on immunosuppression.
Materials and Methods
Patients and samples
This study is based on 400 patients from The Cancer Genome Atlas (TCGA) SKCM cohort with no previous systemic therapy (except that adjuvant interferon-α ≥90 days prior was permitted; ref. 14), wherein all specimens were obtained with written informed consent from the relevant institutional review board participated in TCGA. Hematoxylin and eosin (H&E)-stained whole-tumor sections (obtained from the public TCGA Data Portal) from diagnostic samples were used owing to better preserved morphology compared with frozen samples. Clinical endpoint was overall survival (OS), which was censored at the date of death or the date of last contact (for living patients). Samples were randomly split into equal-sized discovery and validation cohorts.
A second cohort that included 12 patients with metastatic melanoma who underwent serial biopsies was analyzed using the same methods (Supplementary Table S1). Patients were treated with antibodies targeting CTLA4, PD-1, or a combination of both. Treatment was administered until disease progression or unacceptable toxicity. All specimens were obtained with consent from the patient in accordance to ethical approval 111/2010 granted by the Institutional Review Board of the University of Navarra.
H&E image processing
An previously described computational pathology pipeline CRImage was used for image processing (15). In brief, watershed segmentation and Otsu thresholding for hematoxylin positive nuclei was performed. For every cell nucleus morphological features such as shape, intensity and texture features were calculated. Cell classification was based on a support vector (15, 16) using 97 morphological and textural features. Each cell was classified into artefact, lymphocyte, cancer, or stromal cell. The following measures were used to validate the automated cellular classifications for melanoma:
LScore
Pathologist-provided LScore was defined on the basis of lymphocyte distribution and density within the tumors (14). LScore was calculated as the sum of two scores: lymphocyte distribution (0–3, 0 = no lymphocytes within the tissue, 1 = lymphocytes present involving <25% of the tissue cross sectional area, 2 = lymphocytes present in 25–50% of the tissue, 3 = lymphocytes present in >50% of tissue), and lymphocyte density (0–3; 0 = absent, 1 = mild, 2 = moderate, 3 = severe).
Consensus measure of purity estimation
Molecular measure of tumor purity value based on ref. 17.
Imputed lymphocyte scores
From TIMER (18).
Immune phenotype clusters
Immune phenotype clusters were derived from previously described (19) six immune phenotype groups as follows:
Immune phenotype 1 and 2 (“low cytotoxicity”): Cluster 1
Immune phenotype 3 and 4 (“intermediate cytotoxicity”): Cluster 2
Immune phenotype 5 and 6 (“high cytotoxicity”): Cluster 3
Differential expression analyses
TCGA Biolinks (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html) was used for differential gene expression, using the function TCGAanalyze_EAcomplete for enrichment analysis.
TTGs
On the basis of cell spatial mapping provided by automated image analysis, each cell was treated as a node and edges between cells were drawn if they were in spatial proximity (<35 μm). This threshold was determined by calculating the distance of cancer cells to their closest stromal neighbor in regions of the tumor–stroma interface. The mean value of this distribution was used. Cancer supernodes were created by deleting all edges of cancer cells to noncancer cells. Afterwards the connected components in the remaining network were calculated. Connected components with more than 50 cells have been summarized in cancer supernodes and the network was connected again. For every network we kept only the largest connected component.
Network centrality measures
The node degree measures the number of neighbors (edges) of a node. The node degree for the whole network was calculated, regardless of the node’s cell type. Cell type–specific node degrees were created by separating the nodes by their cell type (the neighbors were counted regardless of the cell type).
The clustering coefficient measures the degree in which the neighborhood of a node is connected. For a particular node, clustering , where a triplet consists of three nodes with edges connecting all of them.
Stromal clustering was defined as the average clustering coefficient of stromal cells within a tumor, whereas only the stromal neighbors of the stromal cell were considered in the calculation. To avoid biased results at the border of tumor clusters, we only calculated the clustering coefficient for stromal cells that did not directly border a cancer supernode.
Stromal barrier was calculated by counting the number of stromal cells that a lymphocyte has to cross to reach a cancer cluster. Lymphocytes that directly bordered a cancer cluster were excluded. The overall stromal barrier of a sample was calculated as the average of the individual stromal barriers of the lymphocytes in the sample.
Betweenness centrality measures the network flow or traffic in a communication network or graph based on shortest paths. The betweenness centrality for a node in the network is the number of the shortest paths between every pair of nodes that pass through this node. For each type of cells, betweenness was computed and averaged to represent the betweenness centrality of this cell type within the cell network.
Survival analysis
Univariate Cox proportional hazards model was used to test the differential OS between patients whose tumors had low or high stromal clustering and barrier. Patient stratification into low (n = 97) or high (n = 97) stromal barrier was defined using the upper/lower quartile of stromal barrier, because this produced the best group separation. In the case of stromal clustering, since OS in the upper/lower quartile did not vary significantly, an optimal cutoff was identified. This cutoff (stromal clustering ≤0.590253) was identified in the discovery subset and found to be significant in the validation subset, stratifying the tumors into low (n = 156) or high (n = 230) stromal clustering. Multivariate Cox regression was performed using the same patient stratification, after adjusting for clinical parameters. The above-described stratification of low/high stromal barrier and clustering were used to create the combination groups. Kaplan–Meier curves were used for visual representation.
Genomic and transcriptomic data
RNA sequencing (RNA-seq)–derived transcriptomic data were downloaded from TCGA biolinks (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html; ref. 20). Thresholded copy number data were downloaded from the Broad Institute TCGA Genome Data Analysis Centre (Broad Institute TCGA Genome Data Analysis Centre (2016): SNP6 Copy number analysis (GIS-TIC2). Copy number data and the gene expression data were scaled between zero and one using min–max scaling.
For differential gene expression analysis, we used RNA-seq data that were aligned against the human reference genome hg19. The data were normalized using within-lane and between-between lane normalization and filtered using quantile filtering with a threshold of 0.25 (20).
CNx
Integration of transcriptomic and copy number data was done using CNx, which was inspired by the stacked denoising autoencoders (SDA) previously applied to gene expression data (21, 22). Transcriptomic data were binarized using Gaussian mixture clustering and used in the loss function of the unsupervised deep learning network. In contrast to an SDA, the input for CNx was copy number data and the loss function was the cross-entropy between copy number data and binarized gene expression data. Genes that contributed to the nodes were determined by multiplying the weight matrices (22) of the individual layers and selecting the genes that have a weight that is more than two SDs from the mean.
The bottleneck layer of CNx was visualized as a network, by binarizing the node activities (23). We identified each node’s highest and lowest activity values and defined 10 equally spaced activation thresholds between these values. We evaluated the agreement in the binarized activation values between two nodes at each threshold. Two nodes were connected if they had an agreement larger than 55% of the binarized activation values for one of the thresholds.
Software and data
Image processing was performed using the Bioconductor packages CRImage (15) and EBImage (24). Topological Tumor Graphs were created using the Python package NetworkX (25). The encoder–decoder network was implemented using Python and Tensorflow (4). Genomic analysis was performed in R using the packages TCGAbiolinks (20).
Data availability
Pathologic images and clinicopathologic information of the TCGA samples are available in a public repository from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). All other data supporting the findings of this study are available as part of the reproducible Sweave package.
Code availability
CRImage is available from Bioconductor. Code used to generate topological tumor graphs and R code for performing relevant analyses are provided (https://github.com/Henrik86/Topological-tumour-graphs) for reproducibility.
Results
Mapping cell spatial distribution in whole-tumor histology melanoma sections
To spatially map the locations and types of cells, 400 full face, H&E-stained sections from formalin-fixed paraffin-embedded (FFPE) diagnostic blocks of patients with melanoma from TCGA were subjected to fully automated image analysis (Fig. 1A; Supplementary Fig. S1A, Methods). Samples consisted of primary tumors (n = 91), regional lymph node metastases (n = 191), metastases from regional skin (n = 61), and from distant sites (n = 51), with six samples being unclassified. Our quantitative morphologic classifier identified the following cell types from whole-section images: cancer cells (an average of 662,276 cells with SD ±489,852), 210,658 lymphocytes (±213,832), and 123,081 stromal cells that included fibroblasts and endothelial cells (±121,035). Individual cellular percentages were computed as a proportion of all cell types (stromal%, cancer%, and lymphocyte% henceforth).
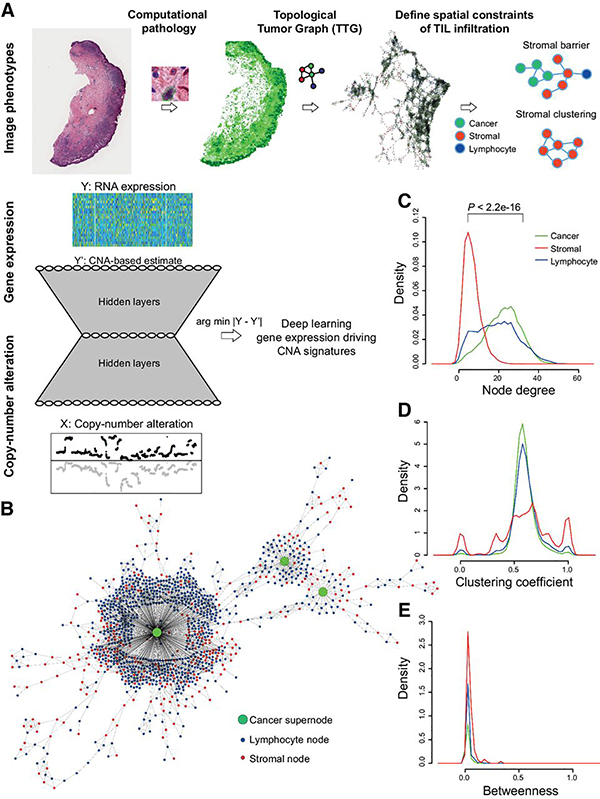
Figure 1.
Computational pipeline schema. A, Schema of automated computational pathology pipeline to analyze H&E slides classified at single-cell resolution and converted to TTGs. TTG-derived tumor phenotypes were integrated with omic data using a convolutional neural network with an hourglass-shaped architecture. This network was designed to learn a sparse representation of CNAs that drive gene expression changes. B, Illustration of a part of a TTG from a whole-tumor section, with cancer cluster summarized into a supernode. C–E, Cell-type-specific distribution of node degree, clustering coefficient, and betweenness. P value from t test.
The cellular classifications derived from image analyses were validated using single-cell annotations on the H&E images, pathologist scores, imputed immune subtypes, tumor purity measure, and pathway analysis. Using a set of 3,230 single-cell annotations, the balanced accuracy of our classifier was found to be 84.9% (81.9% recall, 90.9% precision). Lymphocyte% was significantly higher in tumors with higher pathologist-measured LScore (P < 0.001; Supplementary Fig. S1B) whereas cancer% was significantly higher in tumors with increased tumor purity (as measured by consensus measure of purity estimation score, P < 0.001; Supplementary Fig. S1C). Lymphocyte% was significantly higher in tumors with high imputed lymphocyte score (Supplementary Fig. S1D, P < 0.001) and in imputed immune subtypes, which were described as “high-immune” subtype (“Cluster 3,” P < 0.014; Supplementary Fig. S1E; ref. 24). Differential gene expression analysis revealed that samples with high lymphocyte% ratio were enriched for immune-related pathways, including B-cell and T-cell receptor signaling (Supplementary Fig. S2, Methods). Collectively, these orthogonal analyses supported the validity of automated H&E image analysis when applied to the TCGA melanoma dataset.
Constructing TTGs for structural mapping of the melanoma microenvironment
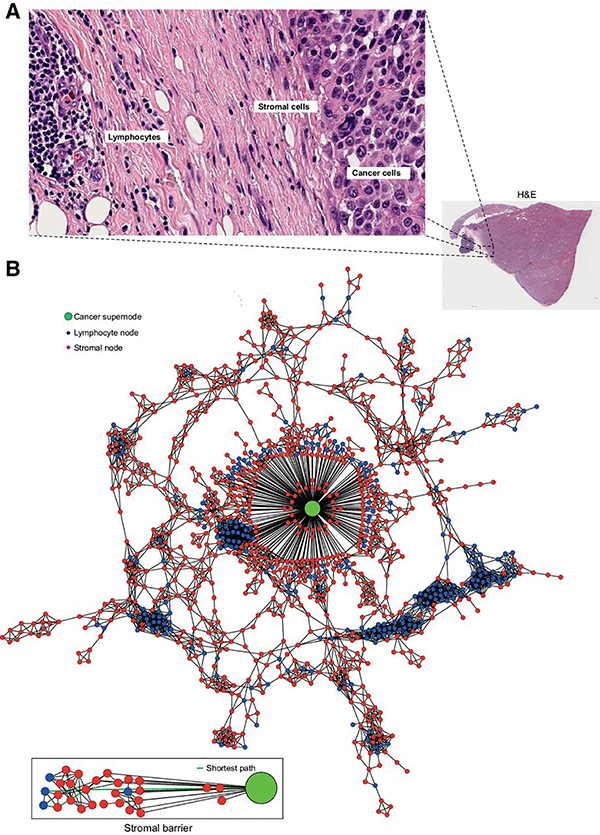
We quantitatively dissected the stromal architecture using graph-based algorithms. This enabled transformation of the three cell types (lymphocytes, stromal, and cancer cells) and their spatial relationships into a new representation, namely TTGs (see Materials and Methods). In the TTGs, each cell (irrespective of the type) was represented as a node, whereas the spatial proximity between cells were represented as edges (Fig. 1B). Because cancer cells were frequently found to be clustered, those with low edge length (“connected” cancer cells) were merged into “cancer supernodes.” The TTG produced the following network centrality measures, which gauged the spatial relationship between cell types: (i) node degrees that quantified the number of edges connected to a node; (ii) clustering coefficient that measured the degree of clustering among cells; and (iii) betweenness centrality that quantified the amount of control over communications among cells in a network. The mean node degree of stromal cells was significantly lower than that of cancer cells (P < 2.2e−16), indicating increased spatial interactions of cancer cells with neighboring cells than the stromal cells. Lymphocytes had a wider spread of node degree, indicating a mixture of connection patterns (Fig. 1C). Importantly, the clustering coefficient of stromal cells had higher variability compared with cancer cells and lymphocytes, which share similar distribution of clustering coefficient (Fig. 1D). In contrast, there was no significant difference in betweenness centrality among the cell types, indicating little difference in cellular position relative to the network center (Fig. 1E). Thus, we further investigated the clustering coefficient of stromal cells (“stromal clustering” henceforth), from among the aforementioned network features. In addition, in an effort to gauge lymphocytes’ accessibility to cancer cells (a frequent histopathologic observation in melanomas), we measured the “stromal barrier,” which is a measure of the barrier presented by stromal cells to cancer infiltrating lymphocytes (Fig. 2A and B; see Materials and Methods).
Figure 2.
Conceptual illustration of stromal barrier for lymphocytic infiltration. A, Example of the stromal barrier in a region of an H&E slide, where a fibrous stroma can be seen between a lymphoid aggregate and a tumor islet. B, The corresponding TTG, in which cancer cell nodes have been summarized to a cancer supernode, whereas each of the lymphocyte/stromal node represent a single lymphocyte/stromal cell. The number of stromal nodes on the shortest path from a lymphocyte to the nearest cancer supernode was calculated and later averaged to derive the quantitative measure of stromal barrier.
In comparing stromal features with clinicopathologic features in the TCGA melanoma cohort (Table 1), stromal clustering and barrier did not vary significantly with AJCC stage or NRAS/BRAF mutation subtype, but stromal clustering was significantly inversely associated with Breslow thickness and ulceration status. In comparing with tumor type, stromal clustering was significantly lower in primary melanomas compared with metastases, with no significant difference in stromal barrier between primary and metastatic melanomas. Notably, stromal clustering and barrier were positively associated with stromal% and inversely associated with lymphocyte%, respectively. The inverse correlation of lymphocyte% with stromal features persisted in a stratified analysis of tumor type (primary tumors, regional lymph node metastases, and distant regional cutaneous metastases), suggesting that stromal features are inversely correlated with lymphocytes, irrespective of the tumor type.
Table 1:
Association of stromal features with clinicopathologic measures in the TCGA melanoma cohort.
| Association of stromal phenotype with | Stromal clustering | Stromal barrier |
|---|---|---|
| AJCC stage | p=0.78 | p=0.98 |
| NRAS/BRAF status | p=0.64 | p=0.88 |
| Breslow thickness | R=−0.167, p=0.003 | R= 0.01, p=0.80 |
| Ulceration | p=0.02 | p=0.23 |
| Tumour type (primary vs metastases) | p=0.0037 | p=0.237 |
| Stromal% | R=0.33, p=1.7e-11 | R=0.58, 2.2e-16 |
| Lymphocyte% | ||
| Whole data set | R=−0.31, p=2.2e-10 | R=−0.18, p=0.00018 |
| Primary tumours only | R=−0.32, p=0.0012 | R=−0.31, p=0.0016 |
| Distant regional cutaneous metastases | R=−0.51, p=4.6e-09 | R=−0.25, p=0.0058 |
| Regional lymph node metastases | R=−0.34, p=1.2e-06 | R=−0.10, p=0.15 |
Quantitative measures of tumor microenvironmental architecture are independent predictors of patient survival
Using univariate survival analyses, high stromal clustering (Fig. 3A) and barrier (Fig. 3B) were both associated with poor 10-year OS (Table 2) whereas stromal% was not significantly associated with survival. Multivariable survival analyses revealed that the poor prognosis associated with high stromal clustering and stromal barrier was independent of ulceration status and Breslow depth (Table 2). Notably, the prognostic significance of both stromal clustering and barrier was also independent of stromal% and lymphocyte% (Table 2). Even among tumors with high lymphocyte%, high stromal clustering and stromal barrier were associated with poor 10-year OS (Supplementary Fig. S3A). In tumor site-specific analyses, high stromal clustering was associated with poor prognosis within regional lymph nodes and distant metastasis (Supplementary Fig. S3B). High stromal barrier was associated with poor prognosis within regional lymph nodes but not in distant metastases (Supplementary Fig. S3C).
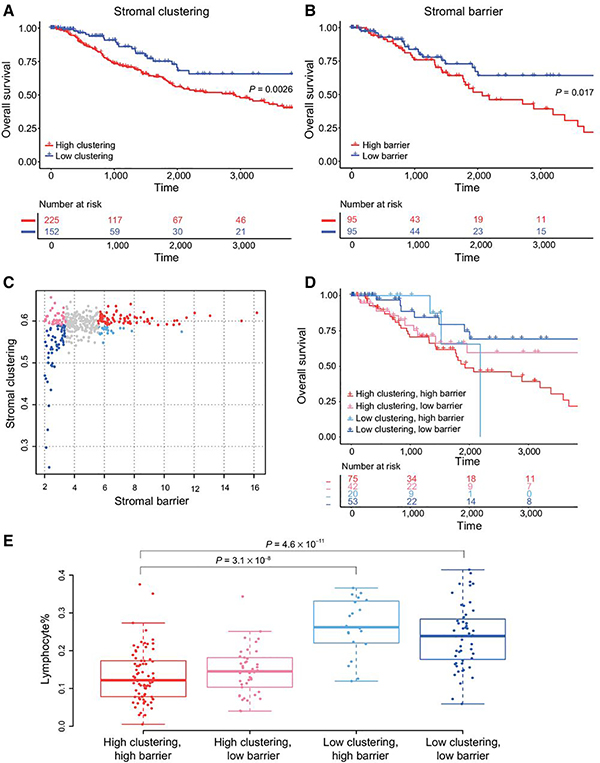
Figure 3.
Stromal barrier and clustering are associated with patient survival. Effect of stromal barrier (A) and stromal clustering (B) on 10-year OS. P values are from univariate Cox proportional hazards model. C, Scatter plot of stromal barrier versus clustering, with colors corresponding to the four combination groups. Comparison of 10-year OS (D) and lymphocyte% (E) between four combination groups. P values in E are from Tukey multiple comparisons of means test.
Table 2:
Association of stromal features with patient survival using univariate and multivariate analyses.
| P-value | HR | HR 95% CI | |
|---|---|---|---|
| Stromal clustering | |||
| Univariate (lowa vs. high) | 0.002 | 1.97 | 1.26–3.07 |
| Adjusted for Breslow depth and ulceration | 0.003 | 2.50 | 1.35–4.62 |
| Adjusted for Lymphocyte% | 0.006 | 1.99 | 1.21–3.27 |
| Adjusted for Stromal % | 0.005 | 1.93 | 1.22–3.06 |
| Stromal barrier | |||
| Univariate (low vs. high) | 0.017 | 1.92 | 1.12–3.29 |
| Adjusted for Breslow depth and ulceration | 0.017 | 1.78 | 1.10–2.86 |
| Adjusted for Lymphocyte% | 0.03 | 1.82 | 1.05–3.166 |
| Adjusted for Stromal% | 0.04 | 1.87 | 1.02–3.43 |
| Mixed model: Stromal barrier + Stromal clustering high clustering/high barrieravs. | |||
| high clustering/low barrier | 0.62 | 0.158 | 0.31–1.20 |
| low clustering/high barrier | 0.53 | 0.308 | 0.16–1.77 |
| low clustering/low barrier | 0.35 | 0.007 | 0.16–0.76 |
Note: Low and high stromal clustering are denoted by optimal cutoff (stromal clustering≤0.590253); lowand high stromal barrier are denoted by the first and fourth quartiles.
Baseline.
We then interrogated the combination of stromal clustering and barrier by defining four combination groups: low-clustering/low-barrier, low-clustering/high-barrier, high-clustering/high-barrier, and high-clustering/low-barrier; Fig. 3C). Tumors with high-clustering/high-barrier had significantly worse 10-year OS compared with low-clustering/low-barrier tumors (Fig. 3D; Table 2). Lymphocyte% was significantly lower in tumors with high-clustering/high-barrier compared with those with low-clustering/low-barrier (Fig. 3E).
Taken together, histology-based measures of stromal architecture were significant predictors of melanoma prognosis, independent of the common clinicopathologic predictors of OS as well as the content of stroma and lymphocytes.
Immunosuppressive potential of stromal barrier and clustering
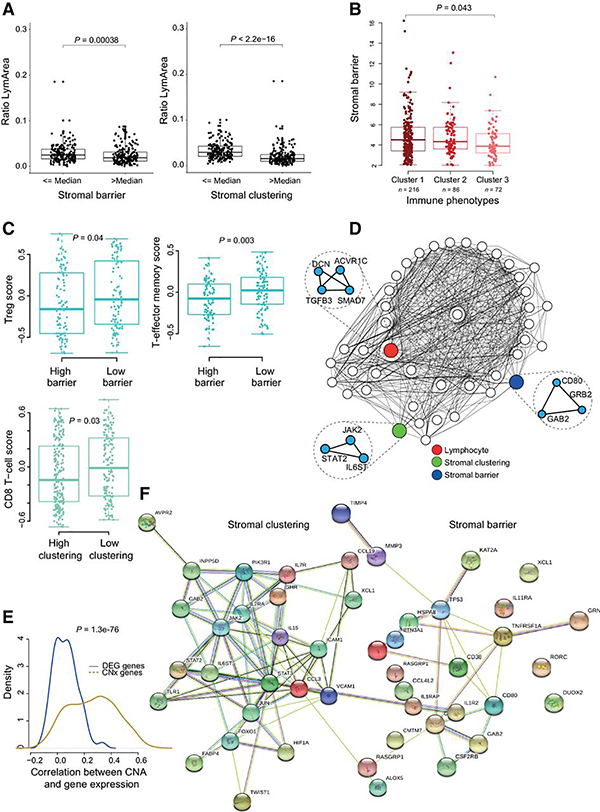
Given that high stromal clustering and barrier were independently predictive of poor prognosis, we tested the hypothesis that stromal barrier and clustering could promote immunosuppression by impeding cancer–lymphocyte interactions. To this effect, we observed a significant negative correlation of lymphocyte-to-tumor area ratio with stromal barrier and clustering (Fig. 4A). Concordantly, stromal barrier was significantly higher in tumors belonging to the “low-cytotoxicity” immune phenotype (“Cluster 1”) compared with “high-cytotoxicity” immune subtypes (“Cluster 3”; Fig. 4B), which also had the highest lymphocyte%. However, stromal clustering and barrier did not vary significantly across the previously reported melanoma molecular subtypes (Supplementary Fig. S4A). To delineate the immune contexture associated with stromal features, we compared 16 previously reported (19) immune cell signatures between tumors with low or high stromal features. Although T-regulatory cells (T-reg) and T-effector memory cells were significantly higher in tumors with low stromal barrier, CD8 T cells scores were significantly higher in tumors with low stromal clustering (Fig. 4C). Taken together, this suggested that increased stromal cell barrier and spatial clustering are associated with reduced distribution of lymphocytes over the entire tumor tissue.
Figure 4.
Immunosuppressive potential of stromal barrier and clustering. A, Boxplot showing the relationship between the ratio of lymphocyte to tissue area and stromal barrier. The same for stromal clustering (right). B, Difference in stromal barrier between clusters derived from previously described immune phenotypes (19). Cluster 1, “Low-cytotoxicity”; Cluster 2, “intermediate-cytotoxicity”; Cluster 3, “High-cytotoxicity.” C, Difference in immune cell signatures between low/high stromal features. D, Visualization of the bottleneck layer of CNx. Nodes in the bottleneck/compressed layer were connected if they shared similar activity values (Materials and Methods) and visualized as a network. The nodes with highest correlation with different image phenotypic features are colored. Genes with highest activation for these nodes are shown as nested network. E, Correlation between CNA and gene expression of genes identified by CNx and by differential gene expression analysis. F, Cytokine genes from each gene list illustrated as protein–protein interaction networks (string interaction score >0.4).
To further explore the evolution of the stromal features over time, we used a small melanoma cohort with serial biopsies (Supplementary Table S1). There was a significant decrease in stromal clustering but not in stromal barrier (Supplementary Fig. S5A). Since stromal clustering was significantly higher in metastasis tissues compared with primary tumors (Supplementary Fig. S5B) in the TCGA melanoma cohort, it is unlikely that this decrease can be attributed to the difference in the type of samples. Therefore, in this very limited, sequential cohort of melanoma patients, our preliminary analysis on melanoma evolution over time suggest that stromal clustering decreases with disease progression, possibly indicating a higher diversity in immune evasion strategies in progression compared with the initial stage of the disease.
Taken together, these data support the immunosuppressive potential and prognostic value of stromal organization, identified by the proposed stromal graph-based measures, to suppress anti-tumoral activity of lymphocytes.
Deciphering the molecular basis of stromal features from omics-based deep learning approaches
To identify biological processes underpinning histologically-defined stromal recruitment, matching copy-number and transcriptomic data were integrated and analyzed using an unsupervised deep-learning framework (named CNx). This enabled the identification of gene expression changes driven by CNA in cancer cells, within the context of the microenvironmental measures described thus far. CNx consisted of an input layer (15,667 nodes), an encoding layer (7,000 nodes), a bottleneck layer (200 nodes), a decoding layer (7,000 nodes), and an output layer (15,667 nodes). To integrate the histology-based stromal architecture measures into this model, we identified the nodes in the bottleneck layer that had the highest Spearman correlation between their activation and stromal clustering or barrier. These nodes and their relationship with other nodes were visualized as a network, along with the most weighted genes as nested nodes (Fig. 4D). Because this compressed layer captures copy-number-driven gene expression (both cis and trans), the list of genes contributing to the activation of nodes associated with stromal feature therefore constitute a catalogue of putative genes implicated in cancer–stromal interactions. To this effect, CNx identified 578 genes (Supplementary Table S2) and 633 genes (Supplementary Table S3) associated with stromal barrier and clustering, respectively.
The genes identified by CNx had significantly higher correlation between copy number and gene expression data, compared with the genes identified by differential expression analysis as being correlated with stromal clustering and barrier (Fig. 4E).
Stroma-mediated downregulation of lymphocyte cytotoxic program
The genes downregulated in tumors with high stromal barrier (268 genes at FC < 0, red-labeled; Supplementary Table S2) and high stromal clustering (324 genes at FC < 0, red-labeled; Supplementary Table S3) were enriched for pathways such as TCR signaling in naïve CD4 T cells, Wnt, IL, PI3K-Akt, and MAPK signaling (Supplementary Tables S4 and S5). Conversely, genes upregulated in tumors with high stromal barrier (310 genes at FC > 0, green-labeled; Supplementary Table S2) and clustering (309 genes at FC > 0, green-labeled; Supplementary Table S3) were enriched for mitochondrial translation, nonalcoholic fatty liver disease, and processing of Capped intron-containing pre-mRNA (Supplementary Tables S6 and S7).
We compared these CNx-derived findings with a lasso regression-based approach (detailed in Supplementary Materials and Methods), which identified copy number of 396 cytobands associated with gene expression events, analogous to the bottleneck nodes identified by CNx. These “nodal cytobands” pertained to 692 and 723 constituent genes correlated significantly with stromal barrier and clustering, respectively. In comparing the pathways enriched for these genes with those identified by CNx, we report that CNx identified genes enriched for immune-related pathways such as IL12-, IL2-, CXCR4-, and TCR-mediated signaling, to be downregulated in tumors with high stromal clustering and barrier, as opposed to the lasso regression-based approach (Supplementary Tables S8 and S9).
Since chemokines and cytokines direct cell trafficking and mediate cell recruitment in the tumor microenvironment (26), we investigated them and related simulator/regulator in the CNx gene lists (Fig. 4F). There were only three genes shared between the cytokine/chemokine networks for stromal clustering and barrier: GAB2, RASGRP1, XCL1. Exclusive to stromal clustering were genes involved in JAK/STAT signaling (JAK2, STAT2, STAT3), chemokines and interleukin receptors (CCL3, IL2RA, IL7R, IL15), adhesion proteins (VCAM1, ICAM1), key hypoxia and differentiation markers (HIF1A, TWIST1, FOXO1, JUN). Among genes exclusive to the stromal barrier network were CD80, CD38 and TNFRSF1A. The apparent lack of overlap between these two gene networks suggests functional mutual exclusivity, which was also consistent with the independent prognostic value of these two features.
Thus, CNx enabled the curation of a catalog of genomic alterations in cancer cells significantly over-represented in specific stromal phenotypes, thereby linking genes with microenvironmental context and providing a catalog of putative genomic features of stromal recruitment.
Discussion
In this study, we present TTGs: a new approach to study the spatial architecture of the tumor microenvironment in primary and metastatic melanoma. Although cell–cell network analysis has previously been used for tasks such as aiding nucleus identification (27), it has not been explicitly used to study tumor–host interactions. To construct TTGs, the melanoma tumor microenvironment learned from whole-tumor pathologic section images were transformed into a network of cells using computational pathology. An understanding of the spatial interplay of heterogeneous cell types in the melanoma tumor microenvironment was derived from network centrality measures such as node degree (the number of neighbors), clustering coefficient (a measure for the connectedness of a node), and betweenness (the number of shortest path in the network that cross this node). The node distribution of a melanoma TTG was similar to random graphs (Supplementary Fig. 1F), suggesting that most cells have similar numbers of neighbors and there are not many hubs in the network. However, there were strong differences in node degree distribution between the cell types, particularly stromal cells having smaller node degrees than epithelial cells and more widespread clustering coefficients. The variable distribution patterns for different cell types suggested that spatial interaction patterns among cancer, lymphocyte, and stromal cells are dependent on cell identity.
The roles of stromal cells, in particular cancer-associated fibroblasts (CAF), in regulating T-cell activity and function are well known. As an important source of TGFβ, CAFs could promote cell death of effector CD8+ T cells by inhibiting expression of the prosurvival protein Bcl-2 (28), or alter cytotoxic CD8+ T-cell function by inhibiting the expression of perforin, granzymes A and B, Fas ligand, and IFNγ (29, 30). Alternatively, CAFs could restrict CD4+ and CD8+ T cells motility in dense matrix areas. Aligned fibers around tumor epithelial cell regions could restrict T cells from entering tumor islets (31). These studies and others, highlight the importance of stromal spatial positioning in the study of tumor immune response.
To this effect, we defined spatially explicit measurements based on TTGs to define two phenotypes of stromal recruitment, spatial clustering of stromal cells within the tumor and stromal barrier, which quantified the average number of stromal cells that a lymphocyte has to cross along a shortest path to infiltrate a cancer cluster. High degree of both stroma clustering and barrier significantly and independently correlated with decreased lymphocytic infiltration and poor survival in melanoma. These support the immunosuppressive potential of stromal cells in melanoma and their clinical relevance.
To identify intrinsic factors in cancer cells that drive stromal recruitment and enhance cell fitness, we investigated genomic alterations and gene expression in the context of stromal architecture. We applied a new deep learning approach, CNx, to identify CNAs that most influence the transcriptome and therefore best predict gene expression data even when reduced in dimension. There is a surge of interest in applying deep learning to cancer omics (32, 33). Approaches that assess the association between gene expression and copy number changes, while being informative, do not always account for indirect effects of copy number change such as alterations in transcriptional factors. To this effect, we have adopted an unsupervised deep learning-based approach to directly integrate genome-wide copy number and tumor transcriptomics data. Furthermore, this study describes an integration of stromal phenotypes derived from whole-tumor section digital pathology, with omics data, in contrast to studies that focus on characterizing microenvironmental phenotypes using transcriptional immune/stromal signatures (19, 34, 35, 36). Therefore, we propose that CNx could be a novel solution to infer independent, compressed representation of copy-number-driven features from high-dimensional and highly intercorrelated genomic data in search of molecular features underpinning specific morphologic phenotype.
Limitations of this study include the limit of access to more extensive, independent clinical cohorts, and the availability of data on response to immunotherapy, and the lack of specific marker staining for immune cell subsets. Nevertheless, we have been able to identify novel features of the genomic imprint of tumors that may potentially enable specific stromal features. New phenotypes of stroma-mediated immunosuppression identified using spatial histology analysis of stroma–cancer interface could help illuminate potential molecular machineries that allow tumors to engage with this interface. Furthermore, the concept of TTGs could easily be adapted for the analysis of other cell–cell interactions in the tumor microenvironment, for example tumor–lymphocyte interactions or lymphocyte–lymphocyte clusters.
Supplementary Material
Significance:
Computational histology-based stromal phenotypes within the tumor microenvironment are significantly associated with prognosis and immune exclusion in melanoma.
Acknowledgments
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. Y. Yuan acknowledges funding from Cancer Research UK Career Establishment Award (C45982/A21808), Breast Cancer Now (2015NovPR638), Children’s Cancer and Leukemia Group (CCLGA201906), NIH U54 CA217376 and R01 CA185138, CDMRP Breast Cancer Research Program Award BC132057, European Commission ITN (H2020-MSCA-ITN-2019), Wellcome Trust (105104/Z/14/Z), and The Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre. S. Muralidhar is funded by NIH U54 CA217376, R01 CA185138. C.E. de Andrea was funded by the Instituto de Salud Carlos III (AC14/00034) and cofinanced by Fondos FEDER (Spain). A. Rullan and E. Sahai are supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001144), the UK Medical Research Council (FC001144), and the Wellcome Trust (FC001144). A. Rullan was additionally supported by the Spanish Society for Medical Oncology (Beca Fundación SEOM).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
Y. Yuan is a consultant at Merck & Co. and has received speakers bureau honoraria from Roche. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Karlsson AK, Saleh SN. Checkpoint inhibitors for malignant melanoma: a systematic review and meta-analysis. Clin Cosmet Investig Dermatol 2017;10: 325–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali Z, Yousaf N, Larkin J. Melanoma epidemiology, biology and prognosis. EJC Suppl 2013;11:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database Syst Rev 2018;2:CD011123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. 2016. arXiv:1603.04467. [Google Scholar]
- 5.Rodríguez-Cerdeira C, Carnero Gregorio M, López-Barcenas A, Sánchez-Blanco E, Sánchez-Blanco B, Fabbrocini G, et al. Advances in immunotherapy for melanoma: a comprehensive review. Mediators Inflamm 2017;2017:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ziani L, Ben Safta-Saadoun T, Gourbeix J, Cavalcanti A, Robert C, Favre G, et al. Melanoma-associated fibroblasts decrease tumor cell susceptibility to NK cell-mediated killing through matrix-metalloproteinases secretion. Oncotarget 2017; 8:19780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 2012;196:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sangaletti S, Chiodoni C, Tripodo C, Colombo MP. The good and bad of targeting cancer-associated extracellular matrix. Curr Opin Pharmacol 2017;35: 75–82. [DOI] [PubMed] [Google Scholar]
- 12.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74–80. [DOI] [PubMed] [Google Scholar]
- 13.Sorokin L The impact of the extracellular matrix on inflammation. Nat Rev Immunol 2010;10:712–23. [DOI] [PubMed] [Google Scholar]
- 14.Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012;4:157ra143. [DOI] [PubMed] [Google Scholar]
- 16.Heindl A, Khan AM, Rodrigues DN, Eason K, Sadanandam A, Orbegoso C, et al. Microenvironmental niche divergence shapes BRCA1-dysregulated ovarian cancer morphological plasticity. Nat Commun 2018;9:3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Severson E, Pignon J-C, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamborero D, Rubio-Perez C, Muiños F, Sabarinathan R, Piulats JM, Muntasell A, et al. Biology of human tumors a pan-cancer landscape of interactions between solid tumors and infiltrating immune cell populations. Clin Cancer Res 2018;24:3717–28. [DOI] [PubMed] [Google Scholar]
- 20.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 2018;24: 1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danaee P, Ghaeini R, Hendrix DA. A deep learning approach for cancer detection and relevant gene identification. Pac Symp Biocomput 2017;22: 219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J, Ung M, Cheng C, Greene CS. Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders. Pac Symp Biocomput 2015;20:132–43. [PMC free article] [PubMed] [Google Scholar]
- 24.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage–an R package for image processing with applications to cellular phenotypes. Bioinformatics 2010; 26:979–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagberg A, Schult D, Schult P. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python Science Conference (SciPy); Pasadena, CA 2008. [Google Scholar]
- 26.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17: 559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Janowczyk A, Zhou Y, Thawani R, Fu P, Schalper K, et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep 2017;7:13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjabi S, Mosaheb MM, Flavell RA. Opposing Effects of TGF-β and IL-15 cytokines control the number of short-lived effector CD8+ T Cells. Immunity 2009;31:131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas DA, Massagué J.TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005;8:369–80. [DOI] [PubMed] [Google Scholar]
- 30.Ahmadzadeh M, Rosenberg SA.TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol 2005;174:5215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean M-C, Validire P, Trautmann A, et al. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J Clin Invest 2012;122:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning–based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 2018;24: 1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018;115: E2970–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhaak RGW, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 2013;123:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakravarthy A, Furness A, Joshi K, Ghorani E, Ford K, Ward MJ, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun 2018;9:3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarthy A, Furness A, Joshi K, Ghorani E, Ford K, Ward MJ, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun 2018;9:3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pathologic images and clinicopathologic information of the TCGA samples are available in a public repository from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). All other data supporting the findings of this study are available as part of the reproducible Sweave package.