Abstract
Background: Thyroxine (T4) is generally considered to be a prohormone that requires conversion to triiodothyronine (T3) to exert biological activity. Although evidence suggests that T4 has intrinsic activity, it is questionable if this activity has any physiological relevance.
Methods: To answer this question, triple knockout (KO) mice (Triples) that cannot express the types 1 (D1) and 2 (D2) deiodinase and the Pax8 genes were generated. Thus, they lack a thyroid and cannot convert T4 to T3. Triples were injected on alternate days with either vehicle or physiological doses of T4, T3, or T3+T4 from postnatal days 2–14. They were euthanized at P15, and RNA-seq was employed to profile gene expression in the liver. In another experiment, Pax8KO mice were injected with T3, T4, or T4+T3, and growth rate and survival to P84 were determined.
Results: The growth retardation of Triples was not improved by either T3 or T4 alone but was significantly improved by T4+T3. In the liver, T4 significantly regulated the expression of genes that were also regulated by T3, but the proportion of genes that were negatively regulated was higher in mice treated with T4 than in mice treated with T3. Treatment with T4+T3 identified genes that were regulated synergistically by T3 and T4, and genes that were regulated only by T4+T3. Analysis of these genes revealed enrichment in mechanisms related to cell proliferation and cholesterol physiology, suggesting a unique contribution of T4 to these biological functions. Pax8KO mice all survived to P84 when injected with T4 or T4+T3. However, survival rate with T3 was only 50% and 10% at 3.5 and 12 weeks of life, respectively.
Conclusions: T4 has intrinsic activity in vivo and is critical for survival and growth. At a physiological level, T4 per se can upregulate or downregulate many T3 target genes in the neonatal liver. While most of these genes are also regulated by T3, subsets respond exclusively to T4 or demonstrate enhanced or normalized expression only in the presence of both hormones. These studies demonstrate for the first time a complex dependency on both T4 and T3 for normal mammalian growth and development.
Keywords: thyroid hormone, thyroxine, survival, growth, transcription, cholesterol
Introduction
Thyroxine (T4) is the major thyroid hormone (TH) in the thyroid gland and the circulation. However, it is widely accepted on the basis of abundant evidence that triiodothyronine (T3) is responsible for most, if not all, of the physiological effects of TH in extrathyroidal tissues, and consequently, T4 functions as the prohormone. Indeed, in 1972, on the basis of their calculations regarding the extent of T4 to T3 conversion in the rat, Schwartz et al. suggested the possibility that T4 has no metabolic activity other than that arising through the formation of T3 (1).
However, several in vitro studies provide compelling evidence that T4 does have intrinsic TH activity. Thus, Samuels et al. found that a growth hormone response was readily induced in cultured GH1 cells when a supraphysiological concentration of T4 was present in the medium. The effect was unlikely to be due to T3 generated from T4 during incubation as the TH content associated with the thyroid hormone receptors (TRs) after incubation comprised 90% T4 and 10% T3, and this degree of TR occupancy by T3 was insufficient to elicit the observed response (2). An effect of T4 per se has also been demonstrated in tadpole red blood cells (RBCs) in vitro. Tadpole RBCs have nuclei and their TR number is markedly upregulated by TH both in vivo (3) and in vitro (4). Furthermore, these cells do not express any 5′deiodinase (5′D) activity (5). TR number was increased twofold when these RBCs were cultured in the presence of T4 for two days (6). An effect of T4 has also been demonstrated in a transient transfection assay system. CAT reporter constructs containing the nucleotide sequence for the TH response element (TRE) of either the malic enzyme gene (ME-TRE-TK-CAT) or the TSHβ gene (TSH-TRE-TK-CAT) were transfected with or without TRα into NIH3T3 cells. Addition of T4 to the medium in the presence of TRα resulted in a more than fourfold increase in the basal level of ME-TRE-TK-CAT expression and a 35% reduction in the level of TSH-TRE-TK-CAT expression. This effect was not due to conversion of the T4 to T3 since it persisted in the presence of iopanoic acid, which inhibited any 5′D activity as assessed by the absence of T3 in extracts of the transfected cells (7). More recently, Gil-Ibanez et al., in a study of gene expression in primary cerebrocortical and neuroblastoma cells studies, also obtained compelling evidence that T4 has intrinsic genomic activity (8).
Recent research also shows that T3 and T4 exert rapid nongenomic biological actions that do not require the classical type I mechanism (9) of binding to the DNA-bound receptor in the nucleus to produce biological effects (10–14). T3 and T4 can also bind to integrin αvβ3 in the cell membrane and activate this pathway, which influences angiogenesis, the proliferation of cancer cells (15,16) and brain cell progenitors (17), and the cytoskeleton of astrocytes and neuronal migration (18,19).
Considered together, these findings strongly support the concept that T4 has intrinsic hormonal activity. However, the actions of T4 in in vivo physiological systems have been difficult to dissociate from those of T3. This is due to the inability of pharmacological approaches (e.g., use of 6-n-propylthiouracyl) to selectively and completely inhibit T4 to T3 conversion and thyroidal T3 secretion independent of T4. Thus, the question of whether T4 has sufficient intrinsic activity for it to have physiological relevance in vivo has not been resolved.
To answer this question, we have used a genetic approach by creating a mouse model that can neither synthesize TH nor produce T3 from administered T4. This has been achieved by cross-breeding our types 1 and 2 double deiodinase knockout (D1/D2KO) mouse, which is completely devoid of 5′D activity in all tissues (20), with the Pax8KO mouse, which manifests thyroid agenesis and thus cannot produce TH (21), to yield the triple knockout D1/D2/Pax8KO mouse (Triple). Like the Pax8KO mouse, the Triple dies before weaning unless given T4 (21). It is notable that this is the first animal model whereby circulating and tissue T4 and T3 levels can be manipulated in vivo independent of each other.
Using this triple KO mouse model, we have demonstrated that most survive to P15 when given either T4 or T3, but growth is significantly improved only when the mice are given a combination of both hormones. Gene expression profiling has shown that T4 per se modulates the expression of many T3-responsive genes in the neonatal liver. However, some genes were responsive only to T4, and others exhibit full modulation of expression only when both hormones were present. In addition, we have shown that Pax8KO mice survive well when treated with T4, but only rarely survive to adulthood when treated with T3 alone.
Materials and Methods
Animals
D1/D2KO mice from our colony were crossed with mice that were heterozygous for the Pax8 gene (Pax8Het mice). These mice were kindly provided by Dr. Anthony Hollenberg (Boston, MA) with the permission of Dr. Ahmed Mansouri (Göttingen, Germany) who generated the genotype in 1998 (21). The first generation of offspring were all heterozygous for both the D1 and D2 genes, and those that were also Pax8+/− mice were determined by genotyping tail-tip DNA by polymerase chain reaction (PCR) at postpartum day 15 (P15). By breeding these triple heterozygous mice, D1/D2KO/Pax8Het mice were eventually obtained. Breeding pairs of these latter mice were maintained to produce triple D1/D2/Pax8KO pups (Triples). All mice were housed in the barrier section of the Geisel School of Medicine's animal facility, under conditions of controlled lighting, 12-hour light, 12-hour dark cycle, and temperature (22°C ± 1°C). Animal protocols were approved by the Institutional Animal Care and Use Committee (Protocol No. 2100).
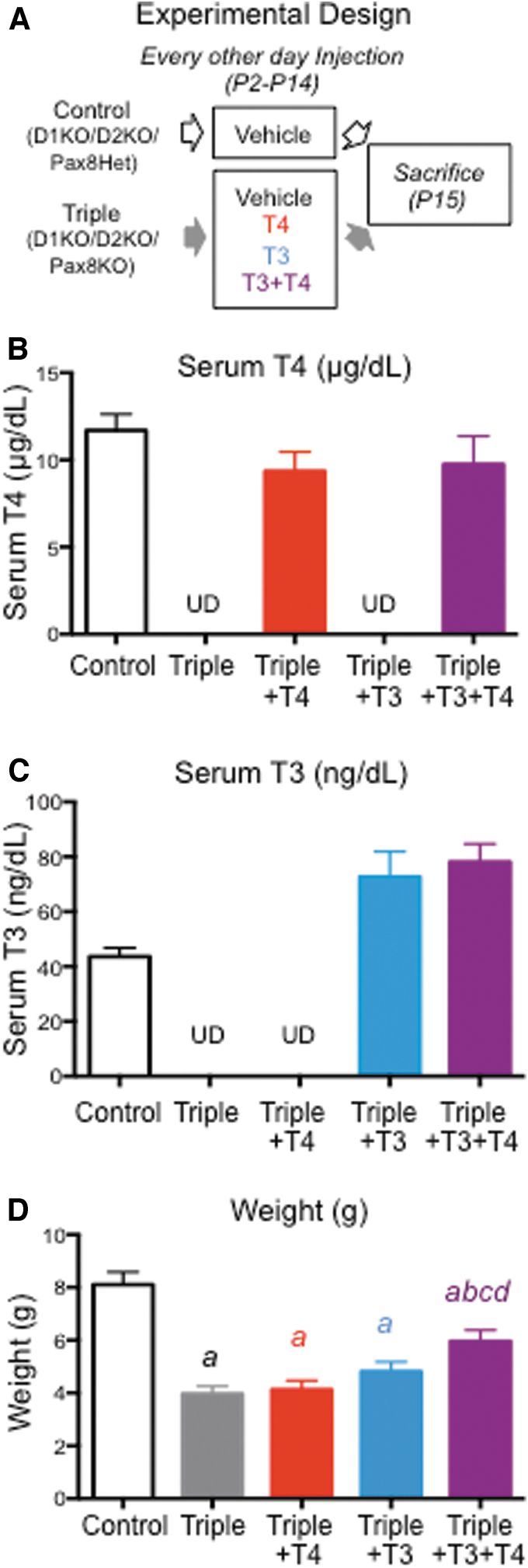
The experimental design of the main study is illustrated in Figure 1A. Likely Triples were selected at postnatal day 2 (P2) on the basis of their low body weight for their age. These potential Triple pups received one of four treatments: T4, T3, T4 plus T3 (T4+T3), or phosphate-buffered saline (PBS). D1/D2KO/Pax8Het pups given PBS served as controls. The T4 employed was the ultrapure T4 (T3 < 0.1%) from Henning (Germany). T3 was obtained from the Sigma Chemical Co. (St. Louis, MO). The hormones or PBS were injected subcutaneously every other day starting at P2, with the final injection being given at P14, 24 hours before sacrifice at P15. Each dose of T4 was 0.06 μg/g body weight and that of T3 was 0.02 μg/g body weight, and the hormones were administered in PBS in a volume of 0.01 mL/g body weight. These treatments had been shown in a pilot experiment to yield serum T4 and T3 levels that were, respectively, approximately normal and elevated in Triple mice at P15, 24 hours after injection. At P15, the mice were weighed, euthanized with CO2, decapitated, and trunk blood collected. The serum was stored frozen for subsequent determination of serum T4 and T3 levels. The livers were harvested, snap frozen, and stored at −80°C for subsequent isolation of RNA. Genotypes were confirmed by PCR using DNA prepared from tail-tip tissue.
FIG. 1.
Body weight and serum TH levels in experimental animals. (A) Experimental design and treatments of mice in the main experiment. (B) Serum T4 level. (C) Serum T3 level. (D) Body weight. The first four experimental groups were assigned a letter (a, b, c, d) in sequential order. Letter(s) above bars corresponding to one experimental group indicate statistical significance (p < 0.05) between that group and the group that the letter represents, as based on ANOVA and Fisher's LSD test. ANOVA, analysis of variance; LSD, least significance difference; T3, triiodothyronine; T4, thyroxine; TH, thyroid hormone; UD, undetectable.
A second study was carried out using Pax8KO mice in which the two 5′D genes had not been disrupted. These mice were obtained from breeding pairs of Pax8Het mice. Likely Pax8KO pups were selected on the basis of retarded growth and treated as described above except that treatment was not started until P4 and was continued until the genotype was confirmed and the mice were weaned. After weaning, the hormones were administered in the drinking water at a concentration of 250 μg/L for T4 and 100 μg/L for T3. Mice drink on average 4 mL of water per day, and thus, it is estimated that they received ∼1.0 μg T4 and/or 0.4 μg T3 per day. We have found that these treatments yield serum T4 and T3 levels close to physiological for these mice. The mice were weighed weekly until they were 12 weeks old when they were euthanized with CO2.
Tissue preparation
Total RNA was isolated from the liver using a commercial RNA isolation reagent (TRIzol solution; Invitrogen, Carlsbad, CA), according to the manufacturer's instructions. Then, aliquots of RNA from each sample were adsorbed onto QIAamp columns included in the Qiagen (Valencia, CA) RNeasy Mini Kit and subjected to DNase treatment with the Qiagen RNase-Free DNase Set. RNA integrity was assessed on a Fragment Analyzer instrument (Advanced Analytical Technologies, Inc.) using the standard sensitivity RNA kit.
T4 and T3 determinations
Total T4 and T3 concentrations in serum were determined using the Coat-A-Count RIA total T4 and total T3 kits (Diagnostic Systems Laboratories, Inc., Webster, TX). The T4 assay was performed according to the manufacturer's instructions. For the T3 assay, the instructions were modified to correct for the small nonspecific effect of mouse serum in this assay (22). The minimal detectable concentrations of T4 and T3 in the assays were, respectively, 0.25 μg and 10 ng/100 mL serum. Values obtained in the T3 assay were corrected for the cross-reactivity of the T4, which was ∼0.38%.
RNA sequencing procedure
This procedure was carried out by the University of Vermont's Advanced Genome Technologies Core. Each RNA sample was prepared from a separate liver obtained from male and female mice at P15. The number of RNA samples per group was: five Controls, five Triples, eight Triples+T4; six Triples+T3, and seven Triples+T4+T3. The facility rechecked the RNA for concentration and quality using the Bioanalyzer 2100 and Qubit spectrofluorometer. Library synthesis for RNA-Seq was performed with 1 μg of high-quality RNA (RNA integrity number 7.5 or higher) using the Illumina TruSeq mRNA Stranded library synthesis as described by the manufacturer. Sequencing was performed in an Illumina HiSeq 1500 using single-end 100 base pair sequencing. RNA sequencing raw data from the experiment have been deposited on Gene Expression Omnibus (GEO, accession No. GSE154156).
RNA sequencing data analysis
The RNA-Seq data analysis was carried out by the Bioinformatics Shared Resource of the University of Vermont College of Medicine, following the method described by Trapnell et al. (23). Briefly, Illumina HiSeq reads were trimmed and clipped for quality control in Trimmomatic v0.27. (24). Read quality was checked for each sample using FastQC v0.10.1. High-quality reads were then imported into TopHat v2.0.8 for alignment initially to the transcriptome (hg19, GRCch37.72) and then to the genome (hg19, GRCch37). BAM files were used as input to the SummarizeOverlaps function of the GenomicAlignments package (25) in Bioconductor to create a read counts matrix, where each cell (i,j) in the matrix specifies how many reads of gene i appear in sample j. The resulting matrix was then used as input to the RNA-Seq pipeline of DESeq2 (26).
Identification of differential expression
Sample-based differential expression
Multivariate principal component analysis was performed on the normalized data set. The analysis used the covariance matrix.
Gene-based differential expression
DESeq2 removed genes with very low chance of being differentially expressed (e.g., genes with 0 read counts across samples) as well as outliers. DESeq2 applied a generalized linear model where the negative binomial distribution is used for the counts. It also included other features that allow for “shrinking” the log2-fold change estimates of low count genes. The Wald test of significance was used to find differentially expressed genes (DEGs), and a “step-up,” adjusted p-value was used for the purpose of controlling the false discovery rate (27). Heat maps were made on the scaled gene counts using the Complex Heatmap package (28). Summaries of DEGs and analyses are included in the Supplemental Data.
Functional ontology and pathway analyses of DEGs Disease and functional ontology analyses were performed on selected groups of genes using the INGENUITY software (Qiagen) and the Database for Annotation, Visualization and Integrated Discovery (DAVID).
Real-time quantitative PCR
Validation by quantitative PCR (qPCR) was carried out on the original RNA samples plus additional samples from additional mice. The number of RNA samples per group was: 12 Controls, 10 Triples, 11 Triples+T4, 11 Triples+T3, and 11 Triples+T4+T3. Total RNA (1 μg) was reverse transcribed for 1 hour at 37°C with 1 μL of MMLV reverse transcriptase (Life Technologies, Waltham, MA). The mix was heated at 75°C for 15 minutes to inactivate the reverse transcriptase and diluted appropriately with water. Aliquots of the mixes from a given experiment were pooled before dilution to establish the first point of an internal standard. Three consecutive 1 to 4 dilutions of this standard were made to generate three additional standard points. Ten microliters of each of the diluted samples were mixed with 12.5 μL of SYBR Select Master Mix (Life Technologies) and 2.5 μL of the appropriate gene-specific primer mix (3.33 pmol/μL for each primer). The mixture was subjected to PCR cycling using a MyiQ Single Color Real-Time PCR Detection System from BioRad (Hercules, CA). PCRs were performed in triplicate. Either Gapdh or Rn18 was used as the control gene. Expression levels were read from the standard curve and are reported in arbitrary units, after correction for the expression of an appropriate housekeeping gene, whose expression did not change among experimental groups. The sequences of the primers utilized are listed in Supplementary Table S1. Data were analyzed statistically by analysis of variance and Fisher's least significance difference (Protected t-tests). p-Value <0.05 was considered statistically significant.
Results
Animals
The number of Triple pups per litter did not reach the anticipated 25% Mendelian proportion, indicating that partial fetal and/or perinatal death occurred. Furthermore, potential Triples were easy to identify by P1 or P2 by their small size and general poor appearance compared with their littermates. These observations indicate that Triple neonates were already hypothyroid at P2. If no TH was given, most (∼90%) of them died before P15. In contrast, most of those that were given TH survived to P15, regardless of whether the hormone administered was T4 or T3. This observation is consistent with the concept that T4 per se has activity in vivo when present at a close to physiological level.
Neonatal growth and serum TH levels
Values for serum T4 and T3 levels and body weight at P15, 24 hours after the final injection, are shown in Figure 1B–D. The mean serum T4 level in the Triple+T4 and Triple+T4+T3 mice was close to that observed in the control (D1/D2KO/Pax8Het) mice (Fig. 1B). In a pilot study using a separate set of T4-treated pups, T4 levels at 6, 12, and 36 hours after injection were 15.3 ± 1.23, 13.6 ± 1.82, and 8.12 μg/100 mL, respectively. Thus, although the serum level was inevitably higher closer to the time of injection, these finding suggest that for most of the 48-hour period between injections the serum T4 level was not excessively elevated. No T3 was detected in the serum of mice in either the Triple or the Triple+T4 group (Fig. 1C). In the two groups treated with T3, the serum T3 level was somewhat elevated compared with that of the controls (Fig. 1C). No T4 was detected in the serum of mice in either the Triple or the Triple+T3 group (Fig. 1B).
Somatic growth was severely compromised in the Triple mice, and by P15, they had achieved only about 60% of the weight of the control mice (Fig. 1D). It was also noted that hair growth was minimal. Treatment of Triple mice with either T4 or T3 did not restore somatic or hair growth. Indeed, the average weight of these treated mice was not significantly different from that of the untreated Triple mice. Growth was significantly improved when the Triple mice were given T4+T3 (Fig. 1D). Furthermore, their hair growth appeared comparable to that of D1/D2KO/Pax8Het littermates.
Analysis of RNA-seq data from P15 liver
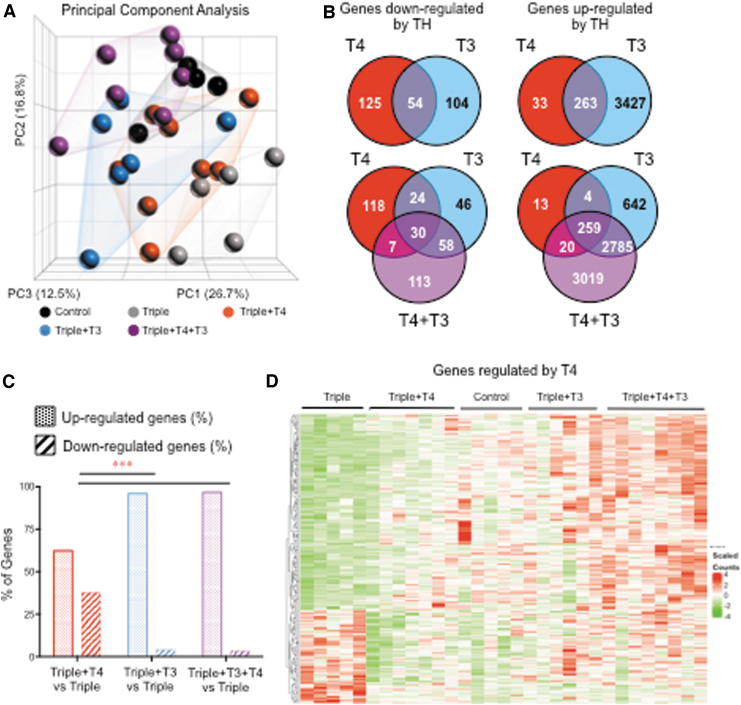
The RNA-seq analysis included 43,629 genes or transcripts. Principal component analysis of the data revealed three major parameters, with variations of 27%, 17%, and 12.5% (Fig. 2A). Experimental samples largely clustered with samples of the same experimental group, but there is some overlapping with other groups. Importantly, experimental groups were clearly distributed along the two principal components of variation (represented in the X and Y axes), suggesting that most of the source of variation depends on the experimental group to which the sample belongs. The data were analyzed for genes modulated by T4 and/or T3 (DEGs) at different statistical stringencies by comparing (i) Triples+T4 versus Triples; Triples+T3 versus Triples; Triples+T4+T3 versus Triples. The resulting gene subsets were then sorted for genes that were positively or negatively modulated by TH. At a nonadjusted p < 0.01, 179 genes were negatively regulated in the Triples+T4 group, while 296 were positive regulated in the same group (Fig. 2B). In the Triples+T3 group, 158 genes were negatively regulated, while 3690 genes were positively regulated. In the Triples+T3+T4 group, 208 genes were negatively regulated and 6083 genes were positively regulated (Fig. 2B). In both the Triples+T3 and the Triples+T3+T4 groups, the vast majority of DEGs were positively regulated. However, there was a significantly increased proportion of negatively regulated genes in the Triples+T4 group (Fig. 2C). This suggests that T4 targets a higher proportion of negatively regulated genes than T3. A heat map representing the genes significantly regulated by T4 is shown in Figure 2D. In that heat map, it can be appreciated that genes downregulated by T4 are not significantly regulated by T3.
FIG. 2.
RNA sequencing results in the liver from mice at P15. (A) Principal component analyses of data from individual samples. (B) Venn diagram of upregulated and downregulated genes in the different TH-treated Triples vs. Triples treated with vehicle (p < 0.01). (C) Percentage distribution of upregulated and downregulated genes in the different TH-treated groups. (D) Heat map of genes significantly regulated by T4 based on a p < 0.01. ***p < 0.001 as determined by chi-square test.
In the Triples+T3+T4 group at the same statistical stringency, there were 113 negatively regulated genes and 3019 positively regulated genes that were not significantly changed in the group treated with T3 or T4 alone (Fig. 2B). Thus, a large subset of both the TH negatively and positively regulated genes is uniquely responsive only to the combination of T4+T3. This suggests a synergistic contribution between T4 and T3 to overall gene expression that goes well beyond the effects of each of the individual hormones.
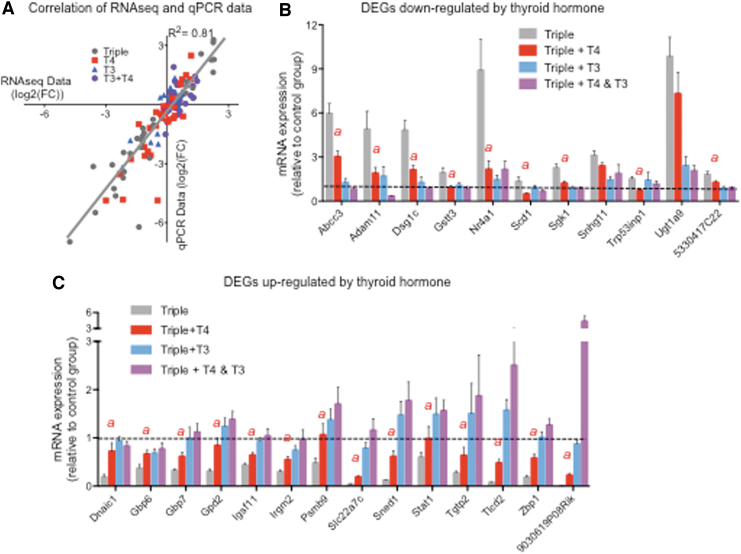
An extended number of samples were used to validate RNA-seq results using qPCR. The results obtained using the latter techniques exhibited a strong correlation with those from the RNA-seq experiment (R2 = 0.81) (Fig. 3A). We confirmed the significant effects of T4 in the negative and positive regulation of 11 and 14 genes, respectively (Fig. 3B, C). Although significantly regulated by T4, these genes were also regulated by T3. For most of these genes, the effect of T3 was stronger than that of T4, while for some others (e.g., Adam11, Sgk1, Dnaic1, Gbp6), the effect was the same (Fig. 3B, C). These data suggest that T4 is largely regulating these genes through the same mechanism as T3.
FIG. 3.
Validation by qPCR of genes regulated by T4 alone. (A) Correlation of RNA-seq and qPCR data for all genes validated. (B) Genes downregulated by T4. (C). Genes upregulated by T4. The statistical significances between the Triple+T4 group vs. the Triple group are the only ones indicated (with an “a” above) as determined by ANOVA and Fisher's LSD test. Dotted lines indicated control values. qPCR, quantitative polymerase chain reaction.
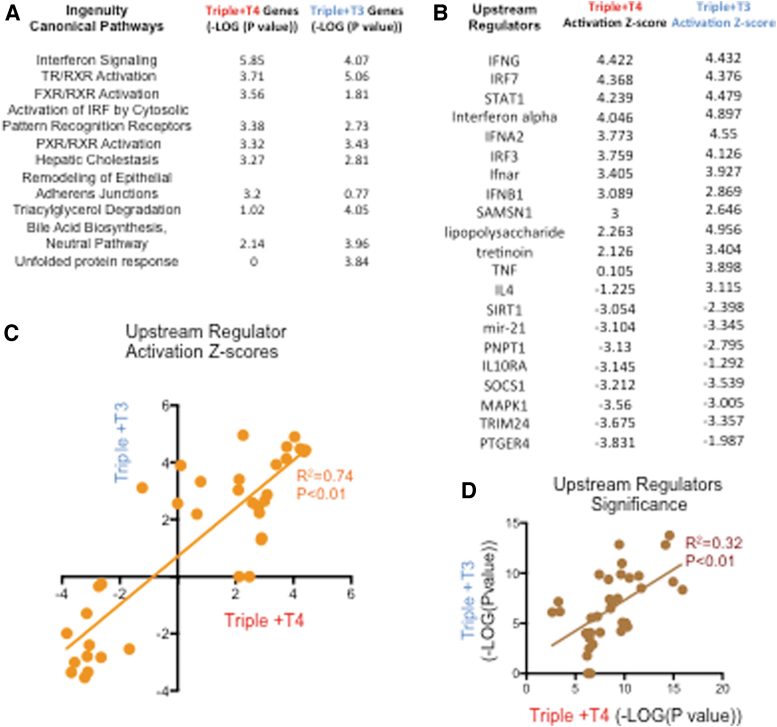
Ingenuity Pathway Analysis (IPA) was used to compare the effects of T4 and T3 on biological functions and molecular pathways. Included were 262 genes regulated by T4 (p < 0.01) and 535 genes regulated by T3 (adjusted p < 0.05). The canonical pathways affected by each hormone with highest statistical significance are shown in Figure 4A for both of them. Consistent with the experiment, TR/RXR activation is shown as significantly affected by both T3 and T4. Activation is also observed in pathways related to immune response and inflammation (interferon signaling) and lipid physiology (FXR/RXR activation, hepatic cholestasis, bile acid biosynthesis, etc.). Pathways related to “unfolded protein response” and “triacylglycerol degradation” appeared to be significantly affected in T3-treated mice but not in T4-treated mice (Fig. 4A). In contrast, “remodeling of epithelial adherens junctions,” a pathway involved in promoting epithelial to mesenchymal transition, was significantly affected by T4, but not by T3.
FIG. 4.
Compared results of IPA of genes regulated by T4 and those regulated by T3. (A) Most significantly enriched canonical pathways. (B) Activation z-scores of upstream regulators whose pathways are most significantly affected in T4- and T3-treated triples. (C, D) Correlation of upstream regulator activation z-scores (C) and p-values (D) between experimental groups treated with T3 or T4. IPA, Ingenuity Pathway Analysis.
IPA also identified upstream regulators whose pathways are most significantly affected by T3 and T4 (Fig. 4B). In this case, both T3 and T4 treatments resulted in the activation of pathways related to immune cells (IFNG, IFNA2, lipopolysaccharide, and others), and retinoic acid (Tretinoin). Pathways related to cell proliferation such as those driven by MAPK1 and mir-21 were downregulated (Fig. 4B). A strong correlation was observed between T4 and T3 in the molecular pathways that they affect and the extent to which they are affected, although exceptions were noted (Fig. 4C, D). These analyses also support the idea that T4 primarily affects the same genes and pathways that are affected by T3. However, it should be emphasized that T4 downregulated or upregulated 158 unique gene products that were not responsive to T3 administered alone (Fig. 2B).
Specific effects of treatment with both hormones
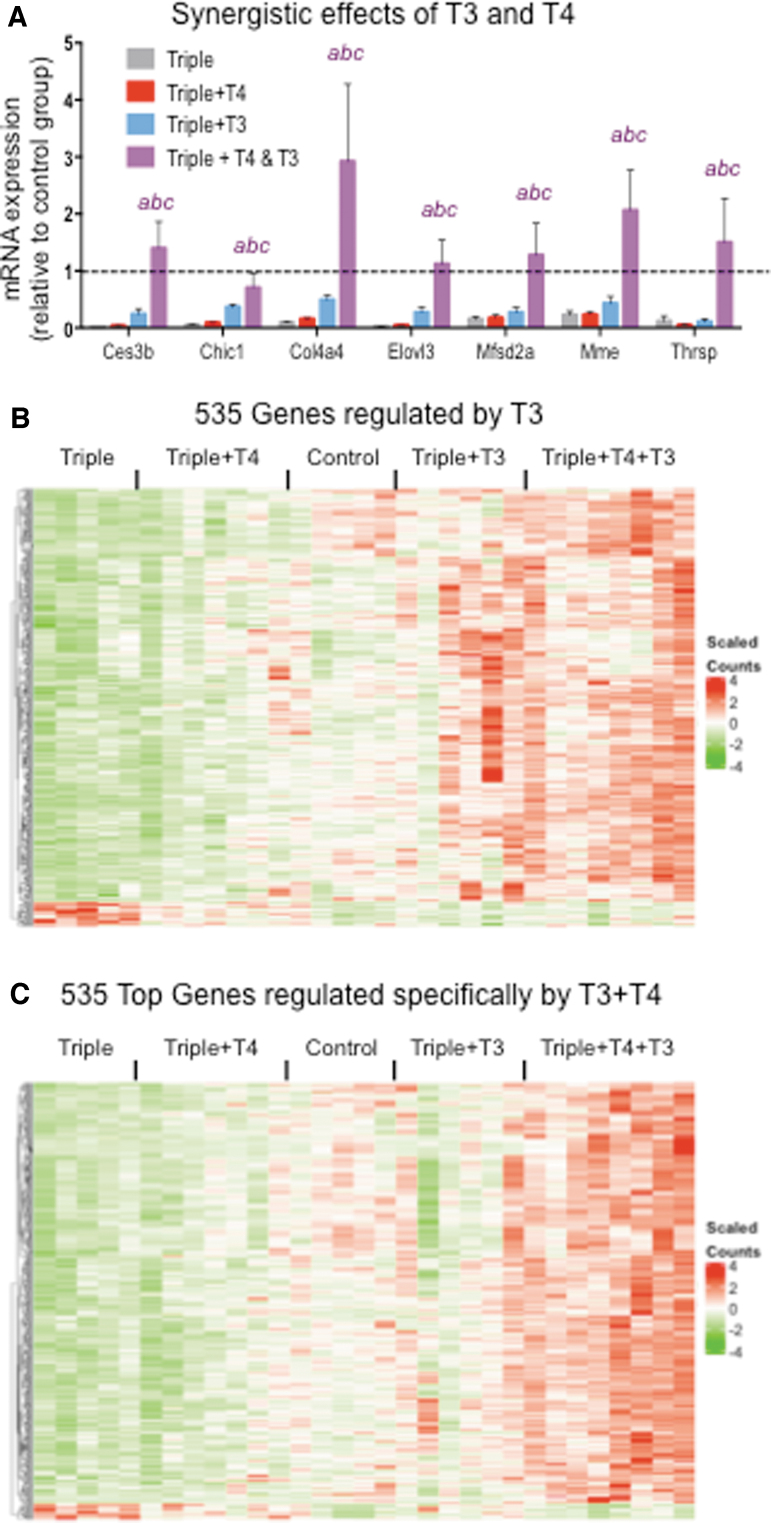
Treatment with T4+T3 identified genes that were synergistically regulated by both hormones (Fig. 5A). Although some of these genes were regulated significantly by T3 alone (Ces3b, Elovl3), only the combined treatment with both hormones normalized or enhanced their expression levels. Known targets of TH, Thrsp and Mme, were only regulated by the combination but not by either hormone alone (Fig. 5A).
FIG. 5.
Comparison of genes regulated by T3 and specifically by T3+T4. (A) qPCR validation of some genes showing synergistic regulation by T3 and T4. (B) Heat map of the 535 genes significantly regulated by T3. (C) Heat map of 535 genes most significantly and specifically regulated by T3+T4. “abc” indicate p < 0.05 triples+T3+T4 vs. triples, triples+T4, and triples+T3, respectively, as determined by ANOVA and Fisher's LSD test. Dotted lines indicated control values (D1/D2KOPax8Het).
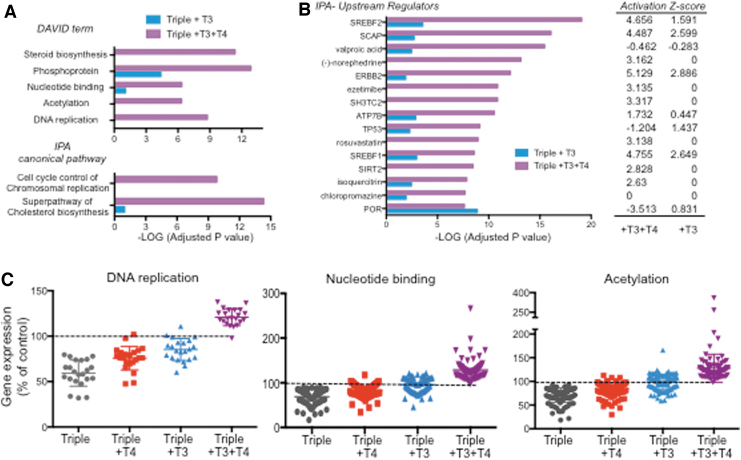
While at an adjusted p < 0.05, only 535 genes were regulated by T3 alone, 1485 genes were significantly regulated by T3+T4 treatment but not by T3 alone (Fig. 5B). This suggests that T4 has additional contributions per se to the liver gene regulation that depends on the presence of T3. To gain biological insight into those contributions, we performed biological function and pathway analyses on genes specifically regulated by T4+T3 (adjusted p < 0.05). For best comparison to those regulated by T3, we selected the 535 genes specifically regulated by T4+T3 at the highest significance. The results were then compared with those obtained from the analyses of the 535 genes significantly regulated by T3. Both DAVID and IPA analyses were consistent and identified biological functions that were affected by T4+T3 but not by T3 alone. These included pathways related to “steroid biosynthesis,” “superpathway of cholesterol biosynthesis,” nucleotide binding,“ acetylation,” “DNA replication,” and “cell cycle control of chromosomal replication” (Fig. 6A). Upstream regulators identified by IPA (Fig. 6B) also show differences between T3 and T3+T4 treatments in the statistical significance and activation scores in pathways related to cholesterol physiology (SREBF2, SREBF1, rosuvastatin, ezetimibe, SCAP) and cell proliferation (TP53, ERBB2).
FIG. 6.
Synergistic and specific effects of the T3+T4 treatment. (A) DAVID terms and IPA canonical pathways exhibiting most differential significance between 535 genes regulated by T3 (adjusted p < 0.05) and 535 genes specifically regulated by T4+T3 showing top statistical significance. (B) IPA upstream regulators showing highest differences in statistical significance and z-activation between the same set of genes. (C) RNA-sequence data as a percentage of control values of genes identified by DAVID as related to DNA replication, nucleotide binding, and acetylation. DAVID, Database for Annotation, Visualization and Integrated Discovery.
The expression profile of genes related to DNA replication, nucleotide binding, and acetylation was normalized to control values and is represented in Figure 6C for all experimental groups. T3 treatment normalizes the expression of some of these genes, but the presence of physiological levels of T4 significantly enhanced the effect, fully normalizing the expression of all genes.
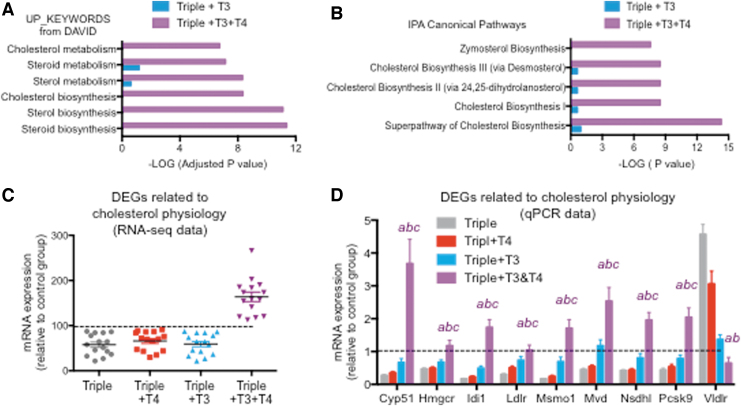
Given the well-established relationship between thyroid status and serum cholesterol levels, it was interesting to note the specific and strong effects of the T4+T3 treatment on genes involved in cholesterol physiology. Both keywords identified by DAVID and canonical pathways identified by IPA show a marked difference between T3 and T3+T4 treatments in the statistical significance of cholesterol-related terms (Fig. 7A, B). Representation of the expression profile of 15 of these genes in each mouse group, as obtained from RNA-seq and normalized to control values, indicates that neither T4 nor T3 alone had any significant effect on those genes, whose expression reaches normal values and above only with the T4+T3 treatment (Fig. 7C). These results were confirmed by qPCR in the extended sample set (Fig. 7D).
FIG. 7.
Specific effects of the T3+T4 treatment on cholesterol physiology genes. (A) Statistical significance of DAVID terms related to cholesterol in 535 genes affected by T3 compared with 535 top genes affected by T3+T4 treatment. (B) Statistical significance of IPA canonical pathways related to cholesterol in 535 genes affected by T3 compared with 535 top genes affected by T3+T4. (C) RNA-sequence data as a percentage of control values of genes identified by IPA as related to the superpathway of cholesterol biosynthesis. (D) qPCR validation of most genes shown in (C). “abc” indicate p < 0.05 triples+T3+T4 vs. triples, triples+T4, and triples+T3, respectively, as determined by ANOVA and Fisher's LSD test. Dotted lines indicated control values (D1/D2KOPax8Het). Significance for triple+T3 and triple 4 groups is not indicated. Data for Vldlr are not shown in (C).
Finally, it is important to note that the levels of mRNA expression of the TRs Thra and Thrb were comparable in all experimental groups, suggesting that the results observed are not due to differences in receptor number.
Survival of Pax8KO mice depends on T4
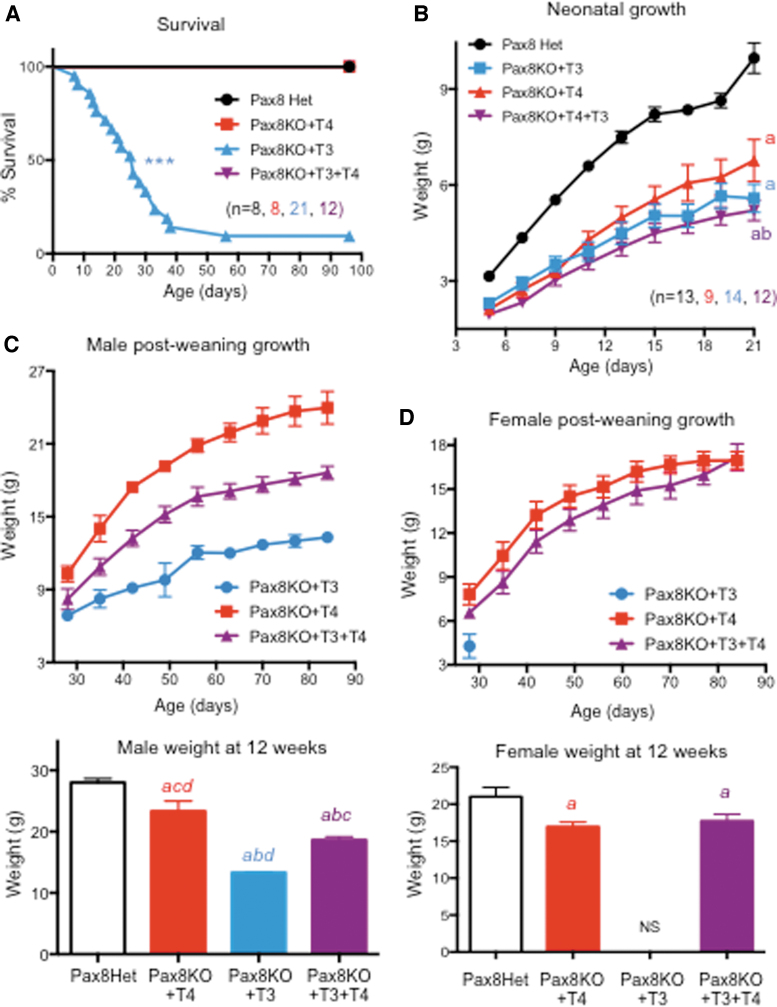
Despite lacking a thyroid gland, Pax8KO mice can survive into adulthood with the administration of T4 (21), but it has not been determined if T3 alone is sufficient for survival. Based on the results described above, we hypothesized that T3 replacement alone would be insufficient to support growth and survival in these athyreotic animals. To test this hypothesis, starting at P3, Pax8KO mice were treated with T3, T4, or T4+T3 (Pax8Het mice were injected with vehicle and used as controls) and analyzed for their growth and survival. We observed that 100% of the animals treated with T4 or with T4+T3 survived into adult age, as did control animals (Fig. 8A). However, only 2 of 21 mice treated with T3 alone (10%) survived 8 weeks and into adulthood. At 24 days of age, we observed a 50% survival of mice treated with T3 (Fig. 8A), demonstrating that T3 alone is not sufficient to support life in neonatal mice, but that T4 is also needed.
FIG. 8.
Survival and growth of Pax8KO mice. (A) Survival curves of Pax8KO mice treated with T4, T3, or T4+T3 compared with Pax8Het controls. (B) Neonatal growth. (C, D) Postweaning growth in TH-treated Pax8KO males (C) and females (D). “abcd” indicate p < 0.01 vs. Pax8Het controls, Pax8KO+T4, and Pax8KO+T3+T4, respectively, as determined by ANOVA and Fisher's LSD test. ***Indicates p < 0.001 vs. all other groups as determined by a log-rank Mantel–Cox test. (Note that only two Pax8KO+T3 mice, both males, survived to 12 weeks of age). NS, none survived.
Neonatal growth of all Pax8KO mice was retarded, independent of treatment, when compared with those of controls (Fig. 8B). Neonatal growth was slightly better in mice treated with T4 alone. After weaning, we observed that differences in growth among experimental groups were sexually dimorphic. Pax8KO males treated with T4 showed markedly better growth than those treated with T4+T3, while the growth of males treated with T3 alone was the weakest (Fig. 8C). In contrast, the postweaning growth of Pax8KO females treated with T4 was only slightly better than those treated with T4+T3 (Fig. 8D). Despite the poor survival rate of mice treated with T3 alone, male mice tended to survive longer than females after weaning (Fig. 8C, D).
Discussion
Whether T4 exerts any biological effects that are independent of its conversion to T3 has been debated by thyroidologists for several decades. Although available data, obtained both in vitro and in vivo, support the view that T4 has intrinsic activity by classical (7,8) or nonclassical mechanisms (15,16), it has not been demonstrated whether this activity occurs in a physiological setting with physiological consequences and in a manner that is completely independent of T4 to T3 conversion. To address this question, we have created and used a unique model: a triple D1/D2/Pax8KO mouse that can neither synthesize T3 nor generate T3 from T4 in any tissue. We have used highly purified hormones, a large number of animals and samples, and extensive validating determinations to produce robust results.
In the Triple model, the only TH available is that provided by exogenous administration, as no T4 to T3 conversion occurs in vivo when D1 and D2 are absent (22). Thus, T3 and T4 were only detected in mice treated with T3 and T4, respectively. The dose regimens for both hormones were aimed at achieving close to physiological serum TH levels. Since the THs were administered every other day, achieving constant serum TH levels was not possible. Nevertheless, as stated above, although the serum T4 level was inevitably higher immediately following an injection, compared with the level in control mice, at 6 and 12 hours, it was only elevated by ∼30% and 16%, respectively, and was slightly reduced at 24 hours when the tissues were harvested. It is also important to note that the serum T3 level was still increased about 70% when the tissues were harvested at 24 hours.
Consistent with observations in Pax8KO mice (21), Triple mice were markedly growth retarded and the rate of growth was not significantly altered by treatment with either of the two hormones. Only treatment with T3+T4 significantly improved neonatal growth, suggesting that T4 is playing a biological role that is distinct from that of T3. The combined treatment did not fully normalize growth in Triple mice possibly because part of the growth retardation had occurred already during late fetal and early neonatal life, before any hormonal treatment started.
Gene expression profiling of the liver at P15 identified genes that were regulated by T4, a larger number that was regulated by T3 and a much larger number of genes regulated by the combination of both hormones. Results from 41 genes were confirmed by qPCR in an extended number of samples, validating the results obtained from RNA sequencing. Most of the genes regulated by T4 were also regulated by T3, suggesting that in severe hypothyroidism a physiological dose of T4 has sufficient intrinsic activity to regulate some T3 target genes, most likely through the classical type I mechanism involving the binding to the TR in the nucleus. This is further supported by the fact that functional ontology and pathway analyses reveal that many, but not all, of the pathways influenced by T4 were also affected by T3 with comparable levels of activation and statistical significance.
Although T4 alone regulated the least number of genes, the proportion of genes that were downregulated was much higher than in groups treated with T3 or with the T4+T3 combination. This also supports the hypothesis that T4 is also having some biological effects that are distinct from those initiated by T3 and possibly involve different mechanisms.
Remarkably, in mice treated with T3+T4, we identified a number of DEGs that was much higher than the number of genes regulated by either of the hormones alone. Within this subset of apparently obligate co-regulated genes, there were 1485 genes that were demonstrated to be specifically regulated by the T4+T3 combination but not by either of the hormones alone. This observation suggests a synergistic or additive effect of T3 and T4 in the regulation of liver gene expression. For example, Thrsp, a well-established TH target gene, was synergistically regulated by T3+T4, but not regulated by either hormone alone. It is possible that the additive or synergistic effects achieved by the combination of hormones simply reflect the increased dose of generic TH and that a higher dose of T3 alone would have yielded similar results. This possibility may apply to a subset of genes. However, we used a dose of T3 that already resulted in an elevated level of T3 and yet many genes did not respond unless T4 is present, suggesting a unique contribution from T4 to the regulatory effects.
Synergistic effects of physiological doses of T3+T4 in the regulation of gene expression suggest that the two hormones are acting via different mechanisms and that they may depend on each other for full biological effects. Analysis of genes regulated by T3 and the same number of genes with top significance and exclusively regulated by T4+T3 identified a number of pathways that are specifically altered when both hormones are present. One of the principal biological functions regulated by the T4+T3 is cholesterol physiology (29,30). Although T3 alone was close to normalizing some cholesterol-related genes, this set of genes was fully regulated only when both hormones were present. This observation provides novel insight into the role of each of the hormones in regulating this important biological process and may have important implications for the regulation of serum cholesterol in the clinical context of thyroid disease.
Another biological function exclusively affected by T4+T3 was DNA replication. This is interesting in the context of recent reports associating T4 and tumor progression in cancer models (31–34). In a model of lung cancer, the effects of T4 on promoting tumor growth are mediated by signaling through cell membrane integrin αvβ3 (34). This is consistent with our present findings indicating that T4 exerts intrinsic effects on the expression of genes involved in DNA replication and involved in epithelial-to-mesenchymal transition.
The present study focuses on the liver in the context of neonatal development. Whether and to what extent the observations can be extrapolated to other systems and ages remains unclear. Considering our current knowledge of TH action, it is likely that T4-specific effects will be dependent on tissue type and developmental stage. In this regard, preliminary observations in brain tissue support this idea. Presumably, the significance and biological effects of T4 per se will be highly dependent on mechanisms of action that are unique to T4 and distinct from those of T3. In addition, the critical role of T4 in ensuring survival may not be dependent only on its effects on the liver but also on its effects in other tissues, effects that need to be investigated.
Consistent with the current literature, our data substantiate the view that T4 plays a role in cell proliferation, possibly as a result of signaling through αvβ3 integrin. If this was the major pathway utilized by T4 in vivo, one would anticipate that most T4 target genes will partially overlap with those of αvβ3 integrin signaling and will be largely independent of TRs. Furthermore, the synergistic effects of T4 and T3 on gene expression in the liver may also be the consequence of T4 affecting cell proliferation and the cellular composition of the liver. It may also result from increased T3 sensitivity secondary to changes in the molecular determinants of T3 signaling. A role of T4 in cell proliferation, together with our observation on the poor survival of mice lacking T4, suggests that the unique role of T4 is particularly important during the developmental period.
In our experimental model, the type 3 deiodinase (D3) is functional. Although D3 expression is changed (elevated in the liver and probably reduced in other tissues), the generation of triiodothyronine (reverse T3, rT3) from T4 and of 3,3′-diiodothyronine (3.3′-T2) from T3 is possible. This raises the question of whether any of these metabolites mediate some of the effects observed. Although physiological effects of 3,5-diiodothyronine in vivo have been described (35–39), no comparable observations have been reported for 3,3′-T2. However, rT3 can also bind to αvβ3 integrin (40,41) and cause biological effects on brain cell cytoskeleton (42), proliferation of cancer cells (41), and calcium mobilization in Sertoli cells (43). This allows for the possibility that in our model rT3 partially contributes to the effects observed in experimental mice treated with T4.
The critical importance of T4 activity per se is also strongly illustrated by the results with Pax8KO mice. Since Pax8KO mice have intact deiodinases, Pax8KO mice survived into adult age when administered T4 alone just as well as with T4+T3. However, 90% of Pax8KO mice treated with T3 alone did not. In fact, these mice exhibited a 50% lethality shortly after weaning, underscoring the importance of T4 to support survival during the developmental period.
Much has been written regarding the pros and cons of administering various TH regimens in the treatment of hypothyroidism (44,45). Our findings herein, indicating that both T4 and T3 have unique and complementary effects on gene expression, demonstrate a delicate interplay between these two hormones in maintaining homeostasis. The clinical implications of this may be significant. For example, our findings strongly suggest that T3 alone is inappropriate as a physiological replacement in patients with hypothyroidism, at least during the developmental period, as studied here. Conversely, the administration of T4 alone fails to exactly mimic the serum TH profile in humans with primary hypothyroidism (45,46) potentially failing to exactly mimic the physiological conditions of euthyroidism. The clinical implications of such subtle changes in TH action remain uncertain.
Further complicating this matter is the sexual dimorphism observed in the patterns of growth in response to TH replacement reported herein. Thus, postweaning growth of Pax8KO males was much better when treated with T4 alone than when treated with the combination of hormones, while the growth of Pax8KO females was similar in T4- and T3+T4-treated groups. Whether such sex-base responses occur in other of the many developmental processes and physiological systems affected by TH require further investigation.
In summary, using an optimal in vivo experimental model, we have obtained the first evidence that T4 has intrinsic and unique hormonal activity when circulating at a physiological level. T4 is critical for neonatal growth and survival and plays a role on gene expression probably via both classical (type I) and nonclassical (possibly type 4) mechanisms (9). Additional research is needed to better understand the mechanisms of T4 action in vivo and the biological processes and physiological systems that are affected as a result.
Supplementary Material
Acknowledgment
We are grateful to Ramiro Barrantes for assistance with heat map generation and GEO submission. This work used the Maine Medical Center Research Institute Molecular Phenotyping Core Facility, which is partially supported by grants P30GM106391 and U54GM115516 from the National Institute of General Medical Sciences.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by grants HD072526 and DK095908 from the U.S. National Institutes of Health.
Supplementary Material
References
- 1. Schwartz HL, Surks MI, Oppenheimer JH. 1971. Quantitation of extrathyroidal conversion of l-thyroxine to 3,5,3′-triiodo-l-thyronine in the rat. J Clin Invest 50:1124–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Samuels HH, Stanley F, Casanova J. 1979. Relationship of receptor affinity to the modulation of thyroid hormone nuclear receptor levels and growth hormone synthesis by l-triiodothyronine and iodothyronine analogues in cultured GH1 cells. J Clin Invest 63:1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schneider MJ, Galton VA. 1991. Regulation of c-erbAa mRNA species in tadpole erythrocytes by thyroid hormone. Mol Endocrinol 5:201–208 [DOI] [PubMed] [Google Scholar]
- 4. Thomas RT, Drake J, Frieden E. 1992. Thyroid hormone receptor induction by triiodothyronine in tadpole erythrocytes in vivo and in vitro and the effect of cycloheximide and actinomycin-D. Gen Comp Endocrinol 86:42–51 [DOI] [PubMed] [Google Scholar]
- 5. Galton VA, Hiebert A. 1988. The ontogeny of iodothyronine 5′-monodeiodinase activity in Rana catesbeiana tadpoles. Endocrinology 122:640–645 [DOI] [PubMed] [Google Scholar]
- 6. Schneider MJ, Galton VA. 1995. Effect of glucocorticoids on thyroid hormone action in cultured red blood cells from Rana catesbeiana tadpoles. Endocrinology 136:1435–1440 [DOI] [PubMed] [Google Scholar]
- 7. Bogazzi F, Bartalena L, Brogioni S, Burelli A, Grasso L, Dell'Unto E, Manetti L, Martino E. 1997. l-Thyroxine directly affects expression of thyroid hormone-sensitive genes: regulatory effect of RXRbeta. Mol Cell Endocrinol 134:23–31 [DOI] [PubMed] [Google Scholar]
- 8. Gil-Ibanez P, Belinchon MM, Morte B, Obregon MJ, Bernal J. 2017. Is the intrinsic genomic activity of thyroxine relevant in vivo? Effects on gene expression in primary cerebrocortical and neuroblastoma cells. Thyroid 27:1092–1098 [DOI] [PubMed] [Google Scholar]
- 9. Flamant F, Cheng SY, Hollenberg AN, Moeller LC, Samarut J, Wondisford FE, Yen PM, Refetoff S. 2017. Thyroid hormone signaling pathways. Time for a more precise nomenclature. Endocrinology 158:2052–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis PJ, Ashur-Fabian O, Incerpi S, Mousa SA. 2019. Editorial: non genomic actions of thyroid hormones in cancer. Front Endocrinol 10:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cayrol F, Sterle HA, Diaz Flaque MC, Barreiro Arcos ML, Cremaschi GA. 2019. Non-genomic actions of thyroid hormones regulate the growth and angiogenesis of T cell lymphomas. Front Endocrinol 10:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gnoni GV, Rochira A, Leone A, Damiano F, Marsigliante S, Siculella L. 2012. 3,5,3′Triiodo-l-thyronine induces SREBP-1 expression by non-genomic actions in human HEP G2 cells. J Cell Physiol 227:2388–2397 [DOI] [PubMed] [Google Scholar]
- 13. Iordanidou A, Hadzopoulou-Cladaras M, Lazou A. 2010. Non-genomic effects of thyroid hormone in adult cardiac myocytes: relevance to gene expression and cell growth. Mol Cell Biochem 340:291–300 [DOI] [PubMed] [Google Scholar]
- 14. Guigon CJ, Cheng SY. 2009. Novel non-genomic signaling of thyroid hormone receptors in thyroid carcinogenesis. Mol Cell Endocrinol 308:63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis PJ, Leonard JL, Lin HY, Leinung M, Mousa SA. 2018. Molecular basis of nongenomic actions of thyroid hormone. Vitam Horm 106:67–96 [DOI] [PubMed] [Google Scholar]
- 16. Hercbergs A 2019. Clinical implications and impact of discovery of the thyroid hormone receptor on integrin alphavbeta3-a review. Front Endocrinol 10:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stenzel D, Wilsch-Brauninger M, Wong FK, Heuer H, Huttner WB. 2014. Integrin alphavbeta3 and thyroid hormones promote expansion of progenitors in embryonic neocortex. Development 141:795–806 [DOI] [PubMed] [Google Scholar]
- 18. Leonard JL 2008. Non-genomic actions of thyroid hormone in brain development. Steroids 73:1008–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farwell AP, Dubord-Tomasetti SA, Pietrzykowski AZ, Stachelek SJ, Leonard JL. 2005. Regulation of cerebellar neuronal migration and neurite outgrowth by thyroxine and 3,3′,5′-triiodothyronine. Brain Res Dev Brain Res 154:121–135 [DOI] [PubMed] [Google Scholar]
- 20. Galton VA, Schneider MJ, Clark AS, St Germain DL. 2009. Life without thyroxine to triiodothyronine conversion: studies in mice devoid of the 5′-deiodinases. Endocrinology 150:2957–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mansouri A, Chowdhury K, Gruss P. 1998. Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet 19:87–90 [DOI] [PubMed] [Google Scholar]
- 22. Galton VA, Wood ET, St Germain EA, Withrow CA, Aldrich G, St Germain GM, Clark AS, St Germain DL. 2007. Thyroid hormone homeostasis and action in the type 2 deiodinase-deficient rodent brain during development. Endocrinology 148:3080–3088 [DOI] [PubMed] [Google Scholar]
- 23. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300 [Google Scholar]
- 28. Gu Z, Eils R, Schlesner M. 2016. HilbertCurve: an R/Bioconductor package for high-resolution visualization of genomic data. Bioinformatics 32:2372–2374 [DOI] [PubMed] [Google Scholar]
- 29. Astapova I, Ramadoss P, Costa-e-Sousa RH, Ye F, Holtz KA, Li Y, Niepel MW, Cohen DE, Hollenberg AN. 2014. Hepatic nuclear corepressor 1 regulates cholesterol absorption through a TRbeta1-governed pathway. J Clin Invest 124:1976–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN. 2008. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci U S A 105:19544–19549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chin YT, Wei PL, Ho Y, Nana AW, Changou CA, Chen YR, Yang YS, Hsieh MT, Hercbergs A, Davis PJ, Shih YJ, Lin HY. 2018. Thyroxine inhibits resveratrol-caused apoptosis by PD-L1 in ovarian cancer cells. Endocr Relat Cancer 25:533–545 [DOI] [PubMed] [Google Scholar]
- 32. Chan YX, Knuiman MW, Divitini ML, Brown SJ, Walsh J, Yeap BB. 2017. Lower TSH and higher free thyroxine predict incidence of prostate but not breast, colorectal or lung cancer. Eur J Endocrinol 177:297–308 [DOI] [PubMed] [Google Scholar]
- 33. Puhr HC, Wolf P, Berghoff AS, Schoppmann SF, Preusser M, Ilhan-Mutlu A. 2020. Elevated free thyroxine levels are associated with poorer overall survival in patients with gastroesophageal cancer: a retrospective single center analysis. Horm Cancer 11:42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Latteyer S, Christoph S, Theurer S, Hones GS, Schmid KW, Fuhrer D, Moeller LC. 2019. Thyroxine promotes lung cancer growth in an orthotopic mouse model. Endocr Relat Cancer 26:565–574 [DOI] [PubMed] [Google Scholar]
- 35. Lanni A, Moreno M, Lombardi A, Goglia F. 1996. Calorgenic effect of diiodothyronines in the rat. J Physiol 494:831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ball SG, Sokolov J, Chin WW. 1997. 3,5-Diiodo-l-thyronine (T2) has selective thyromimetic effects in vivo and in vitro. J Mol Endocrinology 19:137–147 [DOI] [PubMed] [Google Scholar]
- 37. Padron AS, Neto RAL, Pantaleao TU, de Souza Dos Santos MC, Araujo RL, de Andrade BM, da Silva Leandro M, de Castro JPSW, Ferreira ACF, de Carvalho DP. 2014. Administration of 3,5-diiodothyronine (3,5-T2) causes central hypothyroidism and stimulates thyroid-sensitive tissues. J Endocrinol 221:415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jonas W, Lietzow J, Wohlgemuth F, Hoefig CS, Wiedmer P, Schwitzer U, Köhrle J, Schürmann A. 2015. 3,5-Diiodo-l-thyronine (3,5-T2) exerts thyromimetic effects on hypothalamus-pituitary-thyroid axis, body composition, and energy metabolism in male diet-induced obese mice. Endocrinology 156:389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Lange P, Cioffi F, Senese R, Moreno M, Lombardi A, Silvestri E, De Matteis R, Lionetti L, Mollica MP, Goglia F, Lanni A. 2011. Nonthyrotoxic prevention of diet-induced insulin resistance by 3,5-diiodo-l-thyronine in rats. Diabetes 60:2730–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Domingues JT, Cattani D, Cesconetto PA, Nascimento de Almeida BA, Pierozan P, Dos Santos K, Razzera G, Mena Barreto Silva FR, Pessoa-Pureur R, Zamoner A. 2018. Reverse T(3) interacts with αvβ3 integrin receptor and restores enzyme activities in the hippocampus of hypothyroid developing rats: insight on signaling mechanisms. Mol Cell Endocrinol 470:281–294 [DOI] [PubMed] [Google Scholar]
- 41. Lin HY, Tang HY, Leinung M, Mousa SA, Hercbergs A, Davis PJ. 2019. Action of reverse T3 on cancer cells. Endocr Res 44:148–152 [DOI] [PubMed] [Google Scholar]
- 42. Farwell AP, Dubord-Tomasetti SA, Pietrzykowski AZ, Leonard JL. 2006. Dynamic nongenomic actions of thyroid hormone in the developing rat brain. Endocrinology 147:2567–2574 [DOI] [PubMed] [Google Scholar]
- 43. Zanatta AP, Zanatta L, Gonçalves R, Zamoner A, Silva FR. 2013. Rapid responses to reverse T3 hormone in immature rat Sertoli cells: calcium uptake and exocytosis mediated by integrin. PLoS One 8:e77176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jonklaas J, Bianco AC, Bauer AJ, Burman KD, Cappola AR, Celi FS, Cooper DS, Kim BW, Peeters RP, Rosenthal MS, Sawka AM. 2014. Guidelines for the treatment of hypothyroidism: prepared by the American Thyroid Association Task Force on Thyroid Hormone Replacement. Thyroid 24:1670–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peterson SJ, McAninch EA, Bianco AC. 2016. Is a normal TSH synonymous with “euthyroidism” in levothyroxine monotherapy? J Clin Endocrinol Metab 101:4964–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stock JM, Surks MI, Oppenheimer JH. 1974. Replacement dosage of l-thyroxine in hypothyroidism. A re-evaluation. N Engl J Med 290:529–533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.